
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 December 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.748416
This article is part of the Research Topic Industrial Perspectives of Food Biopreservation View all 5 articles
Changes are currently being made to winemaking processes to reduce chemical inputs [particularly sulfur dioxide (SO2)] and adapt to consumer demand. In this study, yeast growth and fungal diversity were investigated in merlot during the prefermentary stages of a winemaking process without addition of SO2. Different factors were considered, in a two-year study: vintage, maturity level and bioprotection by the adding yeast as an alternative to SO2. The population of the target species was monitored by quantitative-PCR, and yeast and filamentous fungi diversity was determined by 18S rDNA metabarcoding. A gradual decrease of the α-diversity during the maceration process was highlighted. Maturity level played a significant role in yeast and fungal abundance, which was lower at advanced maturity, while vintage had a strong impact on Hanseniaspora spp. population level and abundance. The presence of SO2 altered the abundance of yeast and filamentous fungi, but not their nature. The absence of sulfiting led to an unexpected reduction in diversity compared to the presence of SO2, which might result from the occupation of the niche by certain dominant species, namely Hanseniaspora spp. Inoculation of the grape juice with non-Saccharomyces yeast resulted in a decrease in the abundance of filamentous fungi generally associated with a decline in grape must quality. Lower abundance and niche occupation by bioprotection agents were observed at the overripened stage, thus suggesting that doses applied should be reconsidered at advanced maturity. Our study confirmed the bioprotective role of Metschnikowia pulcherrima and Torulaspora delbrueckii in a context of vinification without sulfites.
Grape must contains a number of microorganisms: yeast, bacteria and filamentous fungi (Barata et al., 2012). Various parameters have been shown to impact grape must microbial diversity, such as the farming system (Comitini and Ciani, 2008; Martins et al., 2014; Grangeteau et al., 2017; Agarbati et al., 2019), vintage (Grangeteau et al., 2017), geographical area (Bokulich et al., 2014), cultivar (Fleet, 2008; Zhang et al., 2019), ripening stage (Martins et al., 2012, 2014), sanitary status of the grape berries (Barata et al., 2012), prefermentary operations (Epifanio et al., 1999; Grangeteau et al., 2017), or the use of SO2 (Bokulich et al., 2015). Filamentous fungi most often found on grape berries and in must are generally undesirable: Cladosporium spp. causes rot, which reduces yield and affects the quality of wines (Briceño et al., 2009; Latorre et al., 2011), Alternaria spp., Aspergillus spp. and Penicillium spp. produce mycotoxins (Serra et al., 2003; Bui-Klimke and Wu, 2015; Rantsiou et al., 2020). Additionally, the association of B. cinerea with other molds like Penicillium spp. and Rhizopus spp. can lead to result in production of wine defects (Barata et al., 2012). Non-Saccharomyces yeasts, such as Hanseniaspora, Starmerella, Pichia, Metschnikowia, Zygoascus, Issatchenkia or Rhodotorula genus, are considered dominant in the prefermentary stages (Fleet, 2008; Zott et al., 2010; Barata et al., 2012; Albertin et al., 2014). Some non-Saccharomyces species/strains are considered detrimental for wine quality, others represent resources to promote innovation in the oenological sector (Padilla et al., 2016; Berbegal et al., 2017; Tufariello et al., 2021).
Sulfur dioxide (SO2) has been used in enology since the 1800s (Pasteur, 1866; Ladrey, 1871) for its antimicrobial, antioxidant (Waterhouse et al., 2016) and antioxydasic (Ough and Crowell, 1987; Son et al., 2001) properties, throughout the winemaking process, from vatting to bottling. During the winemaking process, free SO2 has been reported to neutralize and eliminate spoilage microorganisms, delay growth of lactic acid bacteria, and limit growth of non-Saccharomyces yeast (Henick-Kling et al., 1998; Takahashi et al., 2014). A negative effect of SO2 has been demonstrated on maximum populations of Hanseniaspora, followed by a faster decline during fermentation (Andorrà et al., 2008; Albertin et al., 2014; Takahashi et al., 2014). Sulfur dioxide also facilitates implantation of the sulfite-tolerant Saccharomyces cerevisiae species (Constantì et al., 1998; Henick-Kling et al., 1998; Albertin et al., 2014), in particular due to the presence of sulfite pumps (Park and Bakalinsky, 2004; Zimmer et al., 2014; Marullo et al., 2020).
Nowadays, the use of preservatives is a source of controversy. Use of SO2 in producing wines is pointed to as a cause of intolerance (Warner et al., 2000; Timbo et al., 2004; Vally et al., 2009). In addition, consumer views of the agrifood products they ingest have changed, as they have become more aware of preservative compounds and thus wish to eat and drink more “healthily” (D’Amico et al., 2016; Apaolaza et al., 2017). Consumers are willing to pay more for so-called “sustainable” wines, which they consider more environmentally friendly, such as sulfite-free wines (Poveda et al., 2005; Forbes et al., 2009; Pérès et al., 2018; Galati et al., 2019). Furthermore, faced with global warming, as wines become richer with higher pH levels, the effectiveness of SO2 will be reduced, with significant consequences for fermentation and the management of microbial populations (De Orduna, 2010; Drappier et al., 2019).
Faced with these issues, winemakers have had to reduce the use of sulfur dioxide in their wine production, as wine is the main food source of sulfites in terms of doses. Different alternatives are available on the market, such as adjuvants like chitosan and dimethyl decarbonate, or physical methods such as filtration or heat treatments (Lisanti et al., 2019). Recently, bioprotection by adding yeasts has been proposed as a new alternative. This involves adding antagonist cultures (or their metabolic products) to inhibit pathogens and/or extend shelf life, while minimizing the impact on the sensory properties of the treated product (Lücke, 2000). Simonin et al. (2018) showed the effectiveness of Torulaspora delbrueckii in colonizing the medium in an aligoté must, a white Burgundy grape variety. In a preliminary study, the use of two species as bioprotection agents was tested in a context of Bordeaux region: Torulaspora delbrueckii and Metschnikowia pulcherrima in merlot must (Windholtz et al., 2021a). Their implantation was confirmed as limiting the abundance of filamentous fungi.
Furthermore, Johnson et al. (2020) showed the impact of different bioprotection products on growth of Hanseniaspora spp. during a cold prefermentary maceration.
Bioprotection by adding non-Saccharomyces yeasts would therefore seem to be a promising alternative to sulfur dioxide during the vinification process. The impact of the absence of sulfur dioxide and use of bioprotection yeasts on the grape juice microbial community should be evaluated in several contexts in terms of grape must microbial and chemical composition and of ripening stages, particularly advanced maturity in a context of global warming. Regardless of grape juice chemical composition, the maturity stage has been shown to impact the size and composition of the microbial communities on the grape-berry surface, with significantly higher population levels and diversity at the overripened stage (Martins et al., 2012, 2014). Through two complementary molecular biology approaches (Q-PCR and high throughput sequencing), this study aims to provide indications about the impact of the harvest date, absence of SO2 and bioprotection agents on yeast and filamentous fungi. The impact of the harvest date, absence of SO2 and use of a bioprotection alternative on yeast dynamics and diversity was studied in merlot during the prefermentary stages, in comparison with the traditional use of SO2, over two consecutive vintages. From the results of high throughput sequences for the filamentous fungi, intra- and inter-sample biodiversity indices were determined, thus allowing yeast and fungal community composition to be characterized.
In 2017 and 2018, merlot N. grapes (Vitis vinifera L.) were harvested manually from a wine estate applying an organic farming system in the “Entre-deux-Mers” area. They were picked at two ripening stages: technological maturity as defined by the winemaker (maturity “I”) and advanced maturity after which harvest was carried out one week later (maturity “II”). Precipitation during the ripening period (August, September, and October) was 115 and 111 mm in 2017 and 2018, respectively. Average minimal temperatures were 11.5–16.6°C in 2017 and 7–17.1°C in 2018, and average maximal temperatures were 21.2–28°C for 2017 and 14.5–29.5°C in 2018. The 2017 vintage was characterized as an early vintage due to rains at the beginning of September (72 mm) causing an early start to the harvest (Geny et al., 2018), while 2018 had ideal conditions for the harvest (precipitation in September: 3 mm) with higher sugar content (Geny et al., 2019).
The same plot was harvested in 2017 and 2018, collecting the grapes from every other row for the two maturity stages. Three winemaking processes were followed: 50 mg/L of bioprotection [Torulaspora delbrueckii and Metschnikowia pulcherrima species (Zymaflore® Egide – Laffort)] applied directly to the grapes following the manufacturer’s indications for rehydration and without addition of SO2 (“BP” modality), 50 mg/L of SO2 at vatting (“SO2” modality), and without any addition (“Without SO2” modality). Grapes were distributed evenly for each treatment and were then crushed and destemmed. Winemaking was conducted in tanks of 35L in triplicate (maturity I) and duplicate (maturity II) for each modality. Prefermentary maceration was carried out at 10°C before inoculation (200 mg/L) with commercial Active Dry Yeast (ADY) Saccharomyces cerevisiae after 48H. During prefermentary maceration, 10 mL of must was sampled in sterile conditions at three times (Vatting, 24H of maceration, 48H of maceration corresponding to “Stage” parameter) and transported immediately on ice to the laboratory for processing (Figure 1).
Samples of 10 mL of must were centrifuged at 9,000 rpm for 10 min, and then pellets were rinsed twice with 2 mL 0.05 M EDTA pH8 and conserved at −20°C until subsequent DNA extraction. A FastPrep-24 instrument (MP Biomedicals, Illkirch, France) was used for DNA extraction: 200 μl of glass beads (acid-washed, Ø 0.1 mm, Sigma, Lyon, France) and 1 ml of EDTA 0.05 M were added to the frozen pellet. The protocol described by Zott et al. (2010) was followed until complete extraction. DNA was conserved at −20°C.
The quantitative PCR (Q-PCR) method was used to monitor population dynamics of Torulaspora delbruecki and Hanseniaspora spp. with primers described in Zott et al. (2010), and Metschnikowia pulcherrima with primers described by García et al. (2017) (Supplementary Table 1). For quantification, standard curves were built for each yeast species in triplicate, using 10-fold serial dilutions of fresh culture in pasteurized red must. On must samples, quantitative PCRs to monitor Botrytis cinerea spores were obtained using Bc3R and Bc3F primers (Diguta et al., 2010).
FR1 and FF390 primers were used to target 18S rDNA (David et al., 2014). First, the 18S rDNA gene fragment was amplified with an adapter for Amplicon PCR Reverse Primer (5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACA + FR1) and for Amplicon PCR Forward (5′TCGTCGGCA GCGTCAGATGTGTATAAGAGACAGCG + FF390). This PCR was taken in the laboratory: reactions were cycled for 3 min at 95°C, then for 35 cycles of 98°C for 30 s, 52°C for 30 s and 72°C for 60 s, then followed by a final extension period of 8 min at 72°C. The mix PCR was composed of 2.5 μL dilute template (DNAs standardized to 5 ng/μL), 5 μL each Amplicon PCR Primer 1 μM, 12.5 μL 2X KAPA HiFi HotStart Ready Mix (Roche, Bâle, Suisse). After the first amplification, a second PCR was carried out by the Plateforme Genome-Transcriptome of Bordeaux added indices and Illumina sequencing adapters with the Nextera®XT Index Kit. For the Illumina paired-end library, normalized pool libraries were prepared and clusters generated, and 2*250 bp paired-end sequencing (MiSeq Kit NANO v2) was performed on an Illumina MiSeq instrument. A total of 90 samples (3 modalities at 3 stages in triplicate for maturity I and 3 modalities at 3 stages in duplicate for maturity II, during two vintages) were sequenced by Illumina Miseq.
Sequence cleaning was carried out on the FROGS (Find Rapidly OTUs with Galaxy Solution) pipeline (Escudié et al., 2017) with the pre-process steps (pared end assembled with 5′ primer and 3′ primer, with expected length (<300 and >400 bps) and without N), SWARM clustering (Mahé et al., 2015), removal of chimera with VSEARCH (Rognes et al., 2016), and removal of singletons and contaminants. The Silvia132 18S database (Quast et al., 2012) was used as the database for the taxonomic assignment of OTUs (Operational Taxonomic Units). Sequences with <97% identity and <95% of coverage were deleted by filtration on BLAST. An affiliation postprocess was used to resolve inclusive amplicon ambiguities and aggregate OTUs based on alignment metrics. Finally, OTUs corresponding to Vitis sp. were removed.
The Phyloseq package (McMurdie and Holmes, 2013) was used on the Rstudio software (RStudio Team, 2020) to calculate α-diversity [Shannon index, inv-Simpson index and the species richness estimator (chao1)]. The ANOVA test and T-test were used to determine the impact of different parameters on α-diversity after data was assumed to be normally distributed (Shapiro-Wilks normality test, p > 0.05) and variance homogeneity was verified (Leven test, p > 0.05). Then, the Tukey post hoc test (p < 0.05) was used to find significant differences between the modalities (represented by different letters). The Ggplot2 package (Wickham et al., 2016, 2) was used for graphic representations.
From normalized sequences based on the sample which had the least sequences, β-diversity was determined with Jaccard (qualitative approach) and Bray-Curtis distances (quantitative approach). A principal coordinate analysis (PCoA) was performed to visualize the matrix obtained according to these two distances. A permutational multivariate analysis of variance (Manova) was applied with the Adonis function from the vegan package to determine the significant effect of the parameters on β-diversity (Oksanen et al., 2013).
Merlot grapes at two grape ripening stages (technological and advanced maturity) were processed applying three different prefermentary treatments, Bioprotection without SO2 (“BP” modality), 50 mg/L of SO2 at vatting (“SO2” modality), and without any addition (“Without SO2” modality), over two consecutive vintages. Then, different combinations in triplicate for maturity I and in duplicate for maturity II (due to the low harvest quantity) were considered for further analysis. Analysis of the must and quantification of Botrytis cinerea spores are presented in the Supplementary Table 2. Sugar content and pH were higher for maturity II than maturity I for both vintages (Supplementary Table 2). No significant differences were found between the two vintages regarding the quantification of the spores of Botrytis cinerea; however, on average, the quantity of spores was higher at technological maturity than at advanced maturity in both 2017 and 2018 (ANOVA, p-value = 0.003754**). In addition, the quantity of spores was significantly higher in the presence of SO2 than in the control without SO2 for both vintages (ANOVA, p-value = 0.03703*). Chemical analyses of the wines after alcoholic fermentation and total SO2 after malolactic fermentation are given in Supplementary Table 3. As expected, ethanol and pH were higher for advanced maturity and total SO2 was quantified only for the SO2 treatment; volatile acidity (0.09–0.14 g/L of acetic acid) was low for all experiments.
Sampling was carried out at vatting, 24H and 48H of maceration at 10°C and population levels of Hanseniaspora spp. (A), Metschnikowia pulcherrima (B) and Torulaspora delbrueckii (C) were evaluated by Q-PCR (Supplementary Figure 1). Data are given in Figure 2 according to different parameters (stage, treatment, maturity and vintage). Population levels of Hanseniaspora spp. increased during prefermentary maceration (Figure 2A1). A strong vintage effect (Figure 2A4) was noted, with 25.8% of the variance explained (Supplementary Table 4). Population level was higher in 2018 (median: above 107 cells/mL) than 2017 (median: below 106 cells/mL). The treatment explained 8.6% of the total variance, with significantly lower population levels for the SO2 modality compared to the others. Use of bioprotection yeast did not limit growth of Hanseniaspora spp. (Figure 2A2) compared to SO2 addition. It is important to note a significant interaction for Vintage:Stage (10.5%) and Treatment:Stage (6.8%).
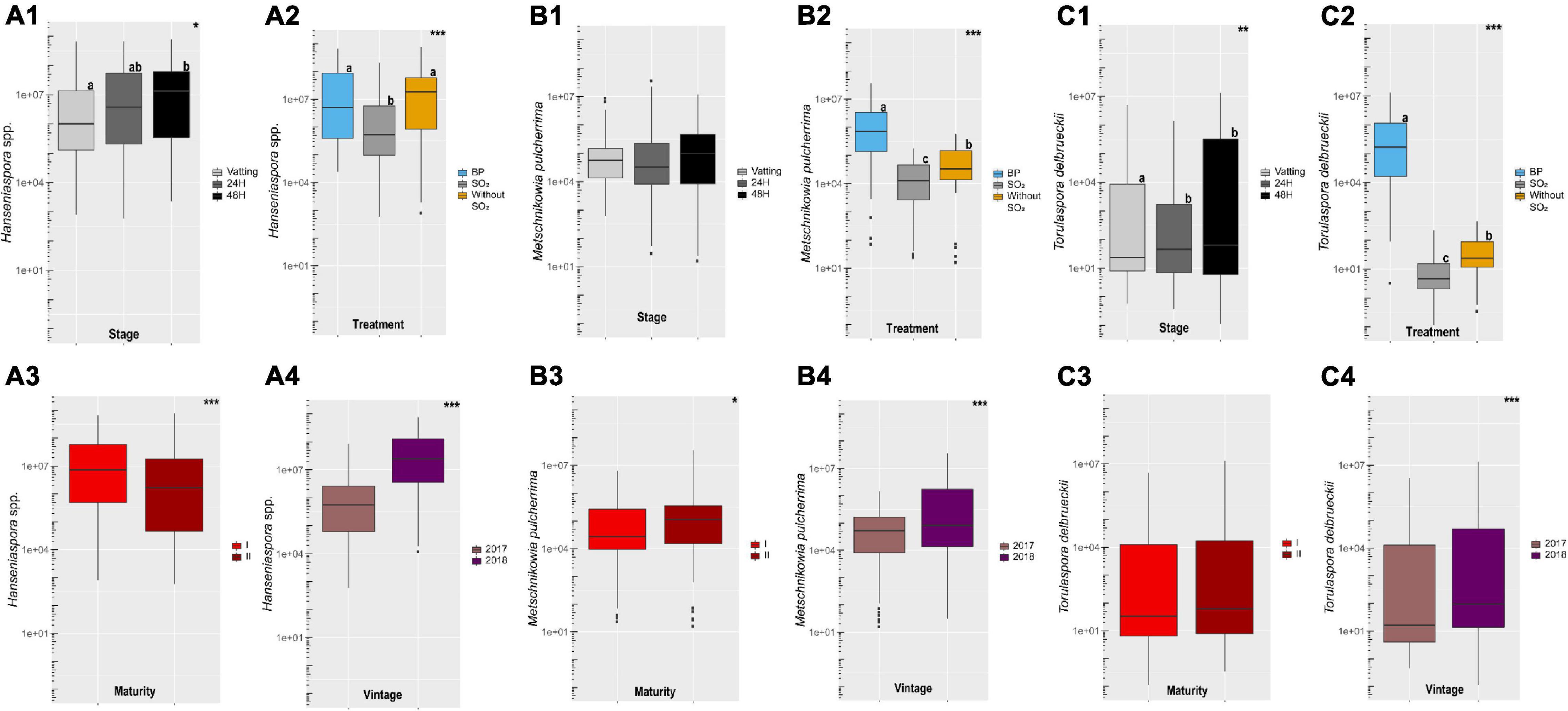
Figure 2. Boxplot of population levels obtained by Q-PCR (cells/mL) for Hanseniaspora spp. (A), Metschnikowia pulcherrima (B), Torulaspora delbrueckii (C) according to Stage (1) (Vatting, 24H and 48H of maceration), Treatment (2) (Bioprotection (BP), SO2 and Without SO2), Maturity (3) [technological (I) and advanced maturities (II)] and Vintage (4) (2017 and 2018) parameters. Significance is indicated as follow: * significant at 5%, ** significant at 1%, *** significant at 0.1% (ANOVA and T-test) and boxplot with different letters differ significantly (HSD-test).
The population level of Metschnikowia pulcherrima was stable during cold maceration (Figure 2B1), unlike Torulaspora delbrueckii, which increased significantly (Figure 2C1). The indigenous population of Metschnikowia pulcherrima (median: above 104 cells/L, Figure 2B2) was high compared to Torulaspora delbrueckii (median: below 102 cells/L, Figure 2C2) and both species were impacted negatively by SO2 addition (Figure 2B2 and Figure 2C2). As expected, population levels of both non-Saccharomyces yeast in the bioprotection modality were significantly higher, due to the inoculation at 50 mg/L, except for Metschnikowia pulcherrima in 2017, maturity II (Supplementary Figure 1).
The growth of Torulaspora delbrueckii during prefermentary maceration and its low indigenous population level resulted in a higher percentage of variance in population levels explained by the treatment (77.4%) than for Metschnikowia pulcherrima (29.7%).
To evaluate yeast and fungal microbial diversity during prefermentary stages, the 18S rDNA gene was analyzed by high-throughput sequencing (HTS). 1,134,820 sequences were obtained, i.e., 12,609 sequences per sample. 1,042,017 sequences were retained after a pre-process step containing paired-end assembled, with 5′ and with 3′ primers, with expected length (between 300 and 400 bps) and without ambiguous characters (N) in their sequences. The SWARM clustering step generated 48,968 OTUs. After cleaning of chimera (−11,222 OTUs and −118,732 sequences), singletons (−36,190 OTUs, 36,785 sequences) and the OTU affiliations, 1,554 OTUs and 886,481 sequences were retained. A filtration on BLAST with the identity (>97%) and coverage (>95%) parameters was used and Vitis sp. OTUs were deleted from the dataset (−1374 OTUs and −51,377 sequences). Finally, an aggregation of OTUs based on alignment metrics was performed to obtain 180 OTUs and 835,104 sequences, i.e., 9,279 sequences per sample.
Ascomycota phylum, the mostly widely represented in the OTU dataset (99.46%), was made up of 4 major Classes: Saccharomycetes at 72.28% (represented by Hanseniaspora (58.9%), Saccharomyces (19.79%); Torulaspora (14.91%) and Metschnikowia (5.7%) genus), Dothideomycetes at 13.70% (represented by Aureobasidium (67%) and Cladosporium (29.3%) genus mostly), Eurotiomycetes at 8.66% [represented by Aspergillus genus (98.9%)] and Leotiomycetes at 4.84% represented by Botrytis genus (95.8%).
The relative abundance obtained for each experiment is presented in Figure 3 and Supplementary Figure 2. Overall, filamentous fungi abundance gradually decreased during the prefermentary stages. The diversity and abundance of OTUs differed from one vintage to another. Alternaria and Neophaerosphaeria were identified only in 2018, with an unexpected high abundance for maturity I after 48H. The relative abundance of Aureobasidium was higher in 2018 than in 2017, and Aspergillus was identified systematically in 2018 but not in 2017 (maturity II). The relative abundance of Botrytis cinerea spores showed a greater presence of Botrytis cinerea at technological maturity than at advanced maturity, according to the Q-PCR analysis and lower abundance at maturity I at vatting and 24H for bioprotection for both vintages. A spontaneous population of Saccharomyces colonized the grape juice at the end of maceration in 2017, but was absent in 2018. Hanseniaspora was the dominant OTU whatever the modality, stage and vintage considered. Both bioprotection species were present at the vatting stage for all the trials, with a higher abundance of Torulaspora delbrueckii than Metschnikowia pulcherrima during prefermentary maceration. The abundance of OTUs related to filamentous fungi (Aspergillus, Cladosporium and Botrytis) was lower when bioprotection yeasts was applied (Supplementary Figure 2), as was Saccharomyces abundance at the end of maceration in 2017. Between the two maturity levels, both vintages combined, the relative abundance of both non-Saccharomyces species were lower at maturity II and the abundance of Hanseniaspora was higher at advanced maturity.
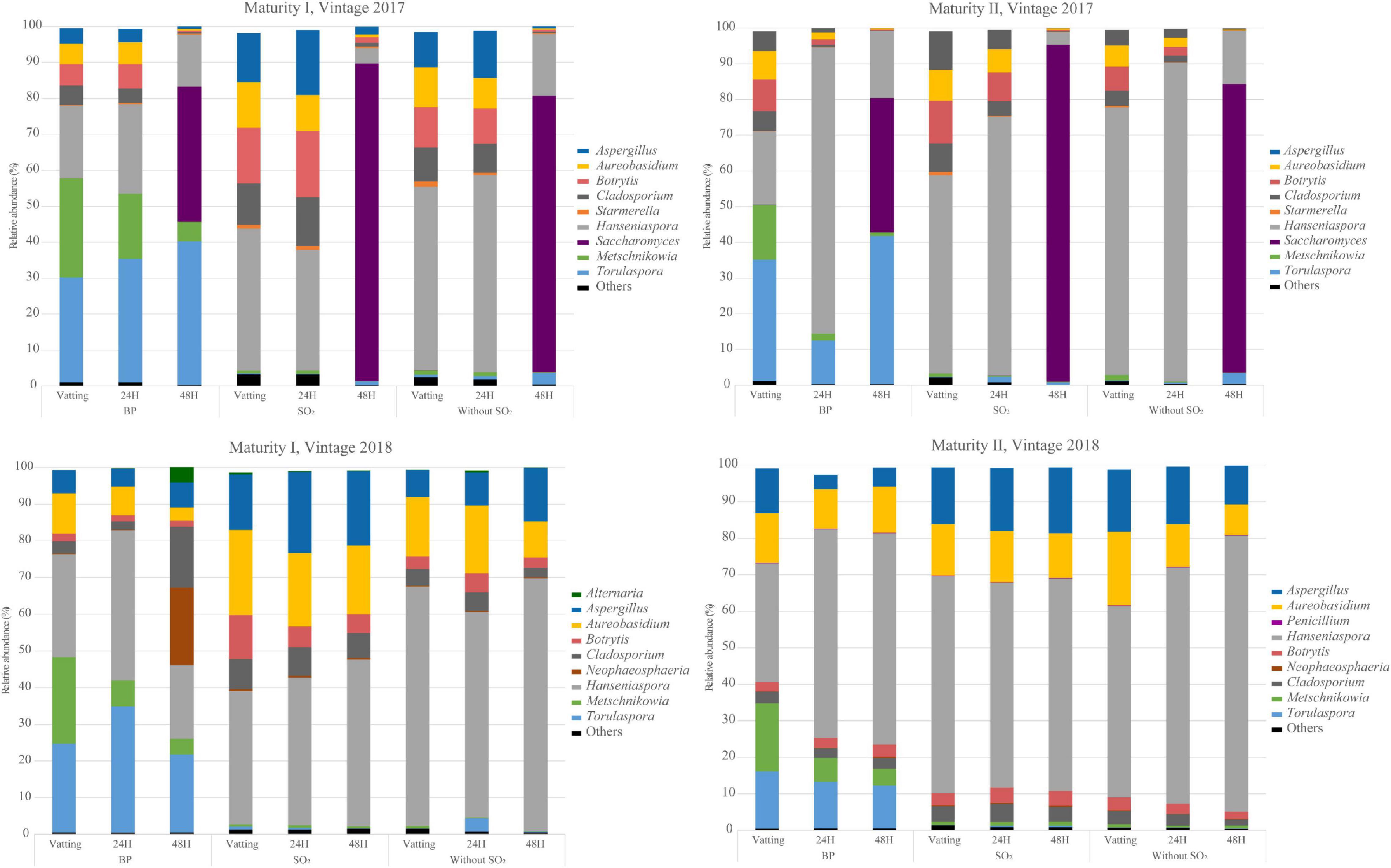
Figure 3. Relative abundances (%) of 9 major genus of Ascomycota phylum for all sample during prefermentary stages (Vatting, 24H and 48H of maceration) in two vintages (2017 and 2018) and two maturities [technological (I) and advanced maturities (II)]. Bioprotection (BP), SO2 and Without SO2 treatments in early stages of winemaking. Results of each sample is the mean of biological replicats (n = 3 technological maturity, n = 2 advanced maturity).
In order to characterize the interspecific biodiversity, different indices were calculated: Observed and Chao1 providing qualitative information only, then the Shannon and InvSimpson indices, which take into account both the abundance and nature of the OTUs (Figure 4). By combining all the data (Figure 4A), significant differences were obtained only for InvSimpson. Indeed, BP – Vatting had a significantly higher index, and the Without SO2 – 48H modality was significantly the lowest.
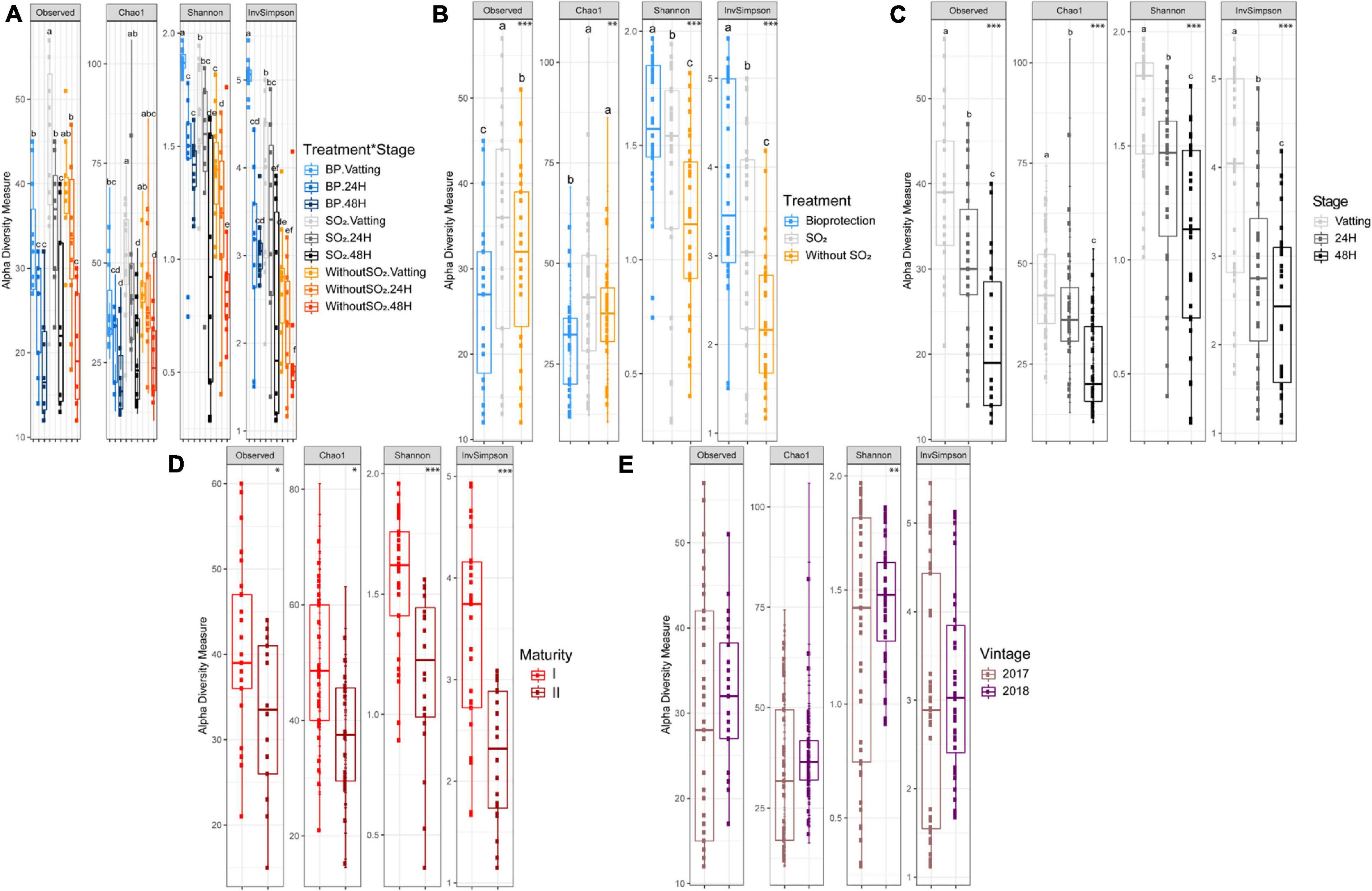
Figure 4. α – diversity by four indices (Observed, Chao1, Shannon, InvSimpson) according to Treatment∗Stage (A), Treatment (B) (Bioprotection (BP), SO2 and Without SO2), Stage (C) (Vatting, 24H and 48H of maceration), Maturity (D) [technological (I) and advanced maturities (II)] and Vintage (E) (2017 and 2018). Significance is indicated as follows: * significant at 5%, ** significant at 1%, *** significant at 0.1% (ANOVA and T-test), and boxplot with different letters differ significantly (HSD-test).
Considering the Treatment parameter (Figure 4B), significant differences could be observed for all indices: the number of OTUs was lower with bioprotection yeasts, but due to the significant presence of Torulaspora delbrueckii and Metschnikowia pulcherrima, its Shannon and InvSimpson index was significantly higher. Interestingly, the absence of sulfiting led to a decrease in both the Shannon and InvSimpson indices.
The Stage parameter affected all the indices that were calculated (Figure 4C): biodiversity of the yeast and fungal microbial community decreased gradually during the maceration process. Concerning maturity (Figure 4C), the Shannon and InvSimpson indices were significantly lower with advanced maturity (II), as were the observed α-diversity and Chao index, but to a lesser extent (p-value = 0.05). Finally, the Vintage parameter did not have a significant impact on α-diversity, except for the Shannon index, which was lower for the 2017 vintage (Figure 4E).
The proportion of variance explained by each parameter and their interactions is presented in Table 1. The Stage parameter explained the largest part of variance for the four indices, followed by Treatment. Maturity accounted for variance of the quantitative indices (Shannon: 11.9% and InvSimpson: 13.6%). The Vintage*Stage interaction was significant and had an impact on the qualitative biodiversity indices, and to a lesser extent, on the Shannon index. The proportion of variance not explained by the parameters considered here (Residuals) was relatively low, except for the Chao1 index for which it was 21%.

Table 1. Percentage of variance explained by Treatment (Without SO2, SO2 and Bioprotection), Stage, Vintage, Maturity factors and these interactions for different α biodiversity indices.
In order to study intra sample biodiversity (β-diversity), only the “SO2” and “Without SO2” treatments were taken into account, and 48H of maceration in the 2017 vintage was not taken into consideration as the ubiquity of Saccharomyces cerevisiae biased the data set for covariance analysis.
Figure 5 presents the PCoA according to different parameters, with their significance obtained by MANOVA test (999 permutations). As for α-diversity, the use of the Jaccard matrix is qualitative, in that it considers only the nature of the OTUs, whereas the Bray-Curtis matrix takes their abundance into account. Concerning the Treatment parameter, the Jaccard matrix explained 13% of the variance on axis 1 and 7.9% on axis 2, whereas the Bray-Curtis matrix explained 71.3% on axis 1 and 14.9% on axis 2. The use of SO2 did not modify the relative composition of the OTUs (Figure 5A1), but did change their abundance (Figure 5B1). Conversely, Stage parameter affected the nature of the OTUs (Figure 5A2) but not their abundance (Figure 5B2). The fungal community structure, in terms of abundance, differed strongly between Maturity I and Maturity II (Figure 5B3) and between 2017 and 2018 (Figure 5B4), and to a lesser extent in terms of relative composition (Figure 5B4).
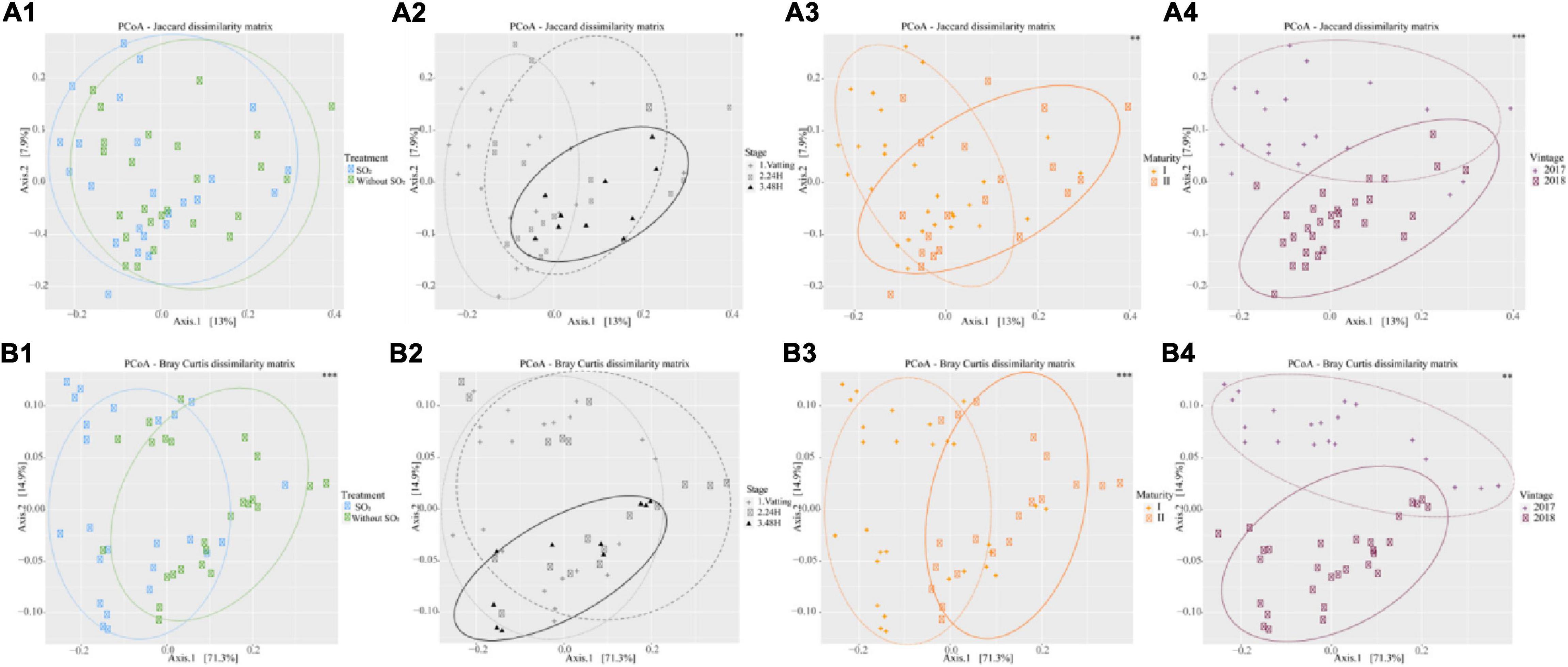
Figure 5. β – diversity by two matrix [Jaccard (A) and Bray Curtis (B)] in function of SO2 addition or not (1), Stage (Vatting, 24H and 48H of maceration, except 48 h maceration in 2017) (2), maturity (3) [technological (I) and advanced maturities (II)] and vintage (4) (2017 and 2018). Signif. codes for p-value (Manova): ∗∗∗0.001, ∗∗0.01.
The percentage of variance explained by each parameter for β-diversity (Jaccard and Bray-Curtis) is presented in Table 2. Regarding the Jaccard matrix, each parameter impacted diversity significantly but at low percentages, and 54.8% of the variance remain unexplained by the parameters. For the Bray-Curtis matrix, Maturity, Treatment and Vintage impacted diversity significantly (22.7, 22.0, and 11.5% respectively) and 19.8% of the variance of the Bray-Curtis matrix was not explained by our parameters. Furthermore, the interaction of Vintage*Maturity was significant.
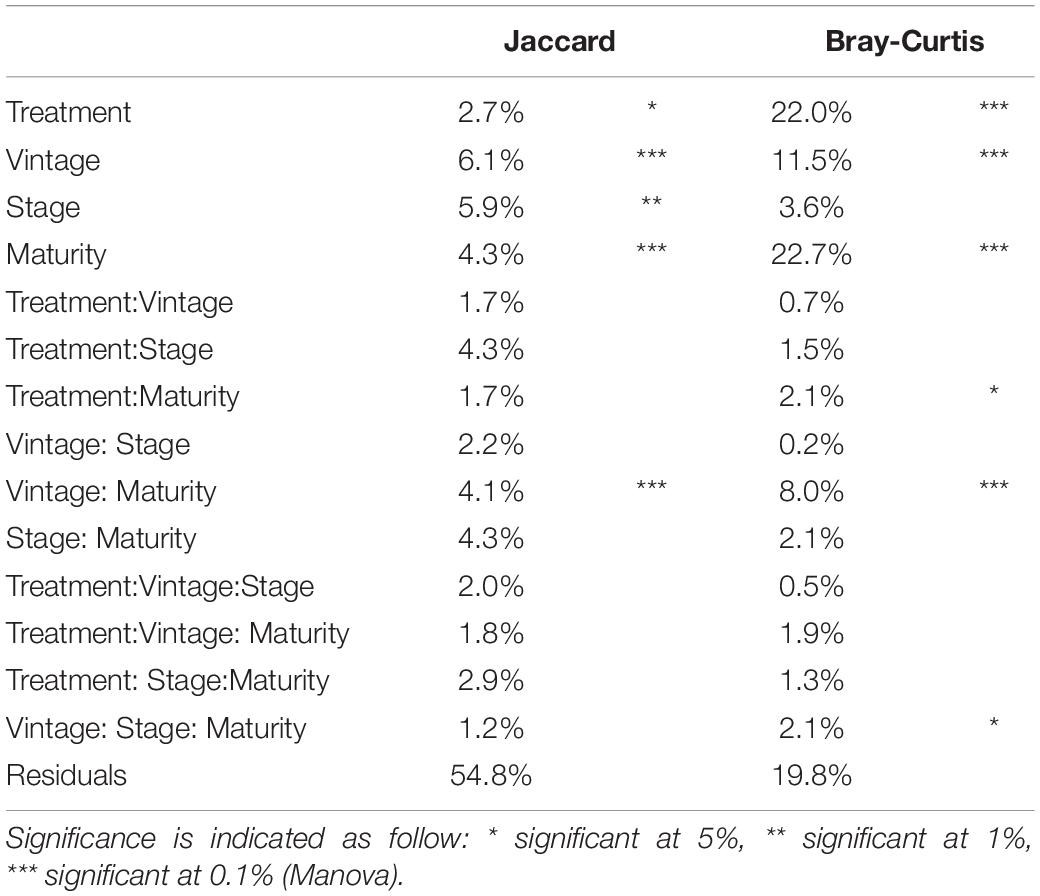
Table 2. Percentage of variance explained by Treatment (Without SO2 and SO2), Stage, Vintage, Maturity factors and the interactions for different β-biodiversity indices.
Nowadays, the use of sulfur dioxide is controversial due to consumers’ perceptions of its harmful effect and their demand for more “healthy food and beverages.” Moreover, with global warming, advanced maturity is a recurrent situation for harvests, and, as a consequence, fermentation management must take account of drastic changes in the matrix composition: high sugar content and pH, and elevated population levels of yeast and bacteria when grape berries become overripe (Martins et al., 2014). In this context, winemakers are being forced to redefine their winemaking processes, by reducing the use of SO2 and controlling the microbial community and alcoholic fermentation kinetics more effectively in order to preserve the sensorial quality of the wines. In the red winemaking process, the first addition of SO2 comes at the moment of vatting. Changes in the microbial yeast and fungal community during the first stages of the red winemaking process in the absence of SO2 may therefore have an impact on the alcoholic fermentation process and final wine quality.
In this work, the yeast and filamentous fungi microbial community was studied at different stages during prefermentary maceration (Vatting, 24H and 48H) using two complementary methodologies, Q-PCR to evaluate the population levels of target yeasts and metabarcoding to analyze microbial diversity. Different factors were considered: vinification practices (SO2, without SO2, Bioprotection), maturity level [technological (I), advanced (II)] and vintages (2017 and 2018).
The α-diversity was strongly impacted by the Stage, with a gradual decrease during the maceration process (Conacher et al., 2021). The yeast and filamentous fungi α-diversity have previously been shown to decrease during alcoholic fermentation, with the selection of dominant species (Nisiotou et al., 2007; David et al., 2014; Conacher et al., 2021). Our results showed that a decrease in α-diversity also occurred during prefermentary maceration; different factors, such as semi-anaerobiosis and low temperature (10°C), can explain the decrease in α-diversity, with the selection of yeast and filamentous fungi that are better suited to this environment.
It is well known that the microbial community is conditioned by the vintage, climate, and as a consequence, the sanitary status of the grape berries (Nisiotou et al., 2007; Bisson and Joseph, 2009; Bokulich et al., 2014; Steenwerth et al., 2021). The climatic conditions differed between the 2017 and 2018 vintages, with rainfall during September in 2017, while the weather was hot and dry during the harvest in 2018. This may explain the differences between the intra-sample diversity indices and their abundance. Hanseniaspora spp. dominated the fungal grape must community. Q-PCR and HTS analysis showed that Hanseniaspora spp. was strongly impacted by the vintage, with a higher population level and abundance in 2018 than in 2017. Previous studies showed a correlation between precipitation and humidity and population of Hanseniaspora spp. (Jara et al., 2016; Steenwerth et al., 2021). In the present work, precipitation during the harvest period was higher in 2017 than in 2018, and this climatic parameter therefore cannot explain our results. Another difference was highlighted between the two vintages: an indigenous population of S. cerevisiae colonized the grape juice at the end of the prefermentary maceration in 2017, whatever the treatment considered, but not in 2018. Since S. cerevisiae was not detected at vatting and after 24H of maceration, we can hypothesize that the origin of this population was the cellar, but not the grape berries.
The nature and abundance of the OTUs differed between the two maturity levels. The impact of the maturity level on the abundance of the yeast and filamentous fungi community was high, with 22.7% of variance explained (p-value 0.001, Bray-Curtis). In the vineyard, modifications in the nutrient composition of grape berry exudates, particularly sugar exudation with advanced maturity, are likely to explain the changes in grape berry microbial community. Following this process, the grape must composition also differs both in sugar content and in pH between the two ripening stages, which can also have an impact on the diversity of microorganisms, including bacteria. For instance, some yeasts are able to persist and occupy the niche preferentially to others, resulting in a reduction in α-diversity in the grape must (Martins et al., 2012, 2014). Sanitary status should also be considered; in our study, the higher number of spores of Botrytis cinerea at technological maturity may explain why the native population levels of Hanseniaspora spp. in the initial must were higher at technological maturity than at advanced maturity (Rabosto et al., 2006; Liu et al., 2010).
The indigenous population level of Metschnikowia pulcherrima and Torulaspora delbrueckii was impacted negatively by SO2 addition at vatting. The first is one of the dominant species in the must, unlike the latter (Bokulich et al., 2014; David et al., 2014; Kecskeméti et al., 2016; Conacher et al., 2021). Non-Saccharomyces yeast is known to be sensitive to SO2, unlike Saccharomyces cerevisiae (Divol et al., 2012; Vicente et al., 2020). Surprisingly, the absence of sulfiting resulted in a decrease in yeast and fungal α-diversity during prefermentary maceration. Previous results have reported alteration of wine microbial diversity with sulfur dioxide treatment, but for white wine vinification, in which the antiseptic effect of SO2 is higher than in red wines (Bokulich et al., 2015; Morgan et al., 2019). In our study, the growth of species that are normally controlled by sulfur dioxide, in particular Hanseniaspora spp. (Albertin et al., 2014), could explain the decrease in α-diversity, by becoming the dominant species in the microbial community. Morgan et al. (2019) reported similar results on pinot gris, showing that the relative abundance of Hanseniaspora spp. was significantly higher in fermentations to which little or no SO2 had been added. The same result was observed by Grangeteau et al. (2017) in chardonnay, where the absence of SO2 led to a preponderance of indigenous yeasts such as Hanseniaspora spp., even preventing the occupation of the must by Saccharomyces cerevisiae and then leading to sluggish alcoholic fermentation. Hanseniaspora spp., particularly H. uvarum, can produce large amounts of ethyl acetate and acetic acid (Domizio et al., 2011; Jolly et al., 2014) and its presence at high levels is not desirable during prefermentary stages. However, used in mixed fermentation with S. cerevisiae, H. uvarum was shown to reduce volatile acidity and increase wine organoleptic quality (Tristezza et al., 2016; Capozzi et al., 2019). It is important to highlight that the population levels of the species were high (106–107 cells/mL) in our different experiments.
When comparing SO2 vs without SO2, there was little difference in β-diversity regarding the nature of the OTUs (Jaccard), but a significant impact of the treatment was highlighted when abundance was taken into consideration (22% of the variance explained, p-value 0.001). Our results, therefore, showed that SO2 altered the abundance of yeast and filamentous fungi, but not their nature.
Until now, few studies have reported the effect of bioprotection on the grape juice yeast and filamentous fungi community as an alternative to sulfites (Simonin et al., 2018, 2020; Windholtz et al., 2021a,b). In the present study, a mix of two species, Metschnikowia pulcherrima and Torulaspora delbrueckii, was used as an alternative to sulfites. The two species had different behavior after inoculation throughout the prefermentary stages: Torulaspora delbrueckii grew to a high population level (>106 cells/L), while the population of Metschnikowia pulcherrima remained stable. Low temperature during the maceration process (10°C), and a negative interaction with Torulaspora delbrueckii, through toxin killers for example (Villalba et al., 2016) or with the indigenous population of Metschnikowia pulcherrima, could explain the lower colonization of grape juice by Metschnikowia pulcherrima. The two species represented half of the relative abundance at technological maturity, but only 20–30% at advanced maturity, irrespective of the vintage considered. Lower abundance of bioprotection yeasts at advanced maturity could be explained by grape berries and grape juice microbial communities (yeast, filamentous fungi and bacteria) that differ from those at technological maturity. The ripening stage has been reported as impacting population level and diversity, with the highest abundance and diversity being found at the overripe stage (Martins et al., 2012, 2014). The greater occupation of the niche by some yeast species (hence the lower α-diversity at advanced maturity) through interaction phenomena could explain the lower abundance of bioprotective non-Saccharomyces at advanced maturity. As a consequence, it could be necessary to reconsider the doses applied at advanced maturity to manage bioprotection yeasts implantation and efficiency more effectively.
In a preliminary experiment on merlot, we reported on the occupation of the must by Torulaspora delbrueckii and Metschnikowia pulcherrima used as bioprotection, limiting the relative abundance of filamentous fungi, and in particular of Aureobasidium and Botrytis (Windholtz et al., 2021a). Our present data confirm that the use of these two species led to a decrease in the number of OTUs (Observed and Chao1), and especially in the numbers of OTUs of filamentous fungi that are systematically associated with a decline in grape must quality, such as Aspergillus spp. is likely to produce ochratoxin A in grape must, the main mycotoxin occurring in wine (Cabañes et al., 2002; Gil-Serna et al., 2018). On the other hand, biodiversity increased with the presence of bioprotection yeasts in the community, contrary to the use and the absence of SO2 (Shannon and InvSimpson). Bioprotection treatment has been shown to limit the colonization of Saccharomyces indigenous yeast at the end of the prefermentary stage when the population level was high (in 2017). This could facilitate the implantation of industrial Active Dry Yeast when added at the end of the cold maceration.
In previous studies, Torulaspora delbrueckii and Metschnikowia pulcherrima were recognized as species with antimicrobial potential (Oro et al., 2014; Ramírez et al., 2015; Villalba et al., 2016; Kántor et al., 2019). Johnson et al. (2020) showed a 44% reduction in acetic acid and 39% in population levels of Hanseniaspora spp. in cold maceration of grape juice with the use of these two species. In addition, they have shown that the impact of non-Saccharomyces yeast on the growth of Hanseniaspora spp. could depend on the strains present in the medium, with a high intra-specific genetic diversity being reported for this species (Albertin et al., 2016). In our experimental conditions, the use of bioprotection had no impact on Hanseniaspora spp. population levels but did have a negative effect on its abundance. One hypothesis to explain this result is the very high initial population level of indigenous Hanseniaspora spp. (>106 cells/ml) compared to our previous study in the 2018 vintage with merlot grapes from the Bordeaux area (104 cells/mL) (Windholtz et al., 2021a).
The impact of various parameters on the yeast and filamentous fungi microbial community during the prefermentary stages of the winemaking process without sulfites was highlighted by this study. Intra-sample diversity of the yeast and filamentous fungi community remained stable from one vintage to another, but its intrinsic composition changed. A decrease in diversity could be observed during the prefermentary stage, as the species present on the grape berry finally gave way to those better suited to the grape must environment. Finally, the absence of sulfiting led to an occupation of the population niche by some dominant species, but which were not particularly desirable (such as Hanseniaspora spp.), leading to an unexpected reduction in diversity compared to the presence of SO2. On the other hand, the OTUs were similar in both treatments, with and without sulfites, and only their abundance varied. Niche colonization by bioprotection yeasts was effective in all of the trials, although Torulaspora delbrueckii showed growth, unlike Metschnikowia pulcherrima, which remained stable. Moreover, their relative abundance was lower at advanced maturity. The use of bioprotective non-Saccharomyces yeast limited the abundance of filamentous fungi that are systematically associated with a decline in grape must quality. Study of the bacterial community should make it possible to complete these results. It would also be interesting to further include grapes from others varieties and with different sanitary status in the same experiment for additional vintages to expand our knowledge of the prefermentary microbial community in a context of vinification without sulfites.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author. The datasets generated for this study can be found in NCBI (ID PRJNA772556).
SW: conceptualization, methodology, investigation, data curation, formal analysis, visualization, and writing – original draft. EV: resources and investigation. LF: investigation. CT: conceptualization and supervision. IM-P: conceptualization, project administration, supervision, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This research was supported by Nouvelle Aquitaine region and Biolaffort.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank the Technology Transfert Unit Microflora for work on Botrytis cinerea spores, IFV of Blanquefort (France) for winemaking process and Château Chillac for merlot grapes. The authors are grateful to the INRAE MIGALE bioinformatics facility (MIGALE, INRAE, 2020. Migale bioinformatics Facility, doi: 10.15454/1.5572390655343293E12) for providing help and computing and storage resources.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.748416/full#supplementary-material
Agarbati, A., Canonico, L., Mancabelli, L., Milani, C., Ventura, M., Ciani, M., et al. (2019). The influence of fungicide treatments on mycobiota of grapes and its evolution during fermentation evaluated by metagenomic and culture-dependent methods. Microorganisms 7:114. doi: 10.3390/microorganisms7050114
Albertin, W., Miot-Sertier, C., Bely, M., Marullo, P., Coulon, J., Moine, V., et al. (2014). Oenological prefermentation practices strongly impact yeast population dynamics and alcoholic fermentation kinetics in Chardonnay grape must. Int. J. Food Microbiol. 178, 87–97. doi: 10.1016/j.ijfoodmicro.2014.03.009
Albertin, W., Setati, M. E., Miot-Sertier, C., Mostert, T. T., Colonna-Ceccaldi, B., Coulon, J., et al. (2016). Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front. Microbiol. 6:1569. doi: 10.3389/fmicb.2015.01569
Andorrà, I., Landi, S., Mas, A., Guillamón, J. M., and Esteve-Zarzoso, B. (2008). Effect of oenological practices on microbial populations using culture-independent techniques. Food Microbiol. 25, 849–856. doi: 10.1016/j.fm.2008.05.005
Apaolaza, V., Hartmann, P., Echebarria, C., and Barrutia, J. M. (2017). Organic label’s halo effect on sensory and hedonic experience of wine: a pilot study. J. Sens. Stud. 32:e12243. doi: 10.1111/joss.12243
Barata, A., Malfeito-Ferreira, M., and Loureiro, V. (2012). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153, 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Berbegal, C., Spano, G., Tristezza, M., Grieco, F., and Capozzi, V. (2017). Microbial resources and innovation in the wine production sector. South Afr. J. Enol. Vitic. 38, 156–166. doi: 10.21548/38-2-1333
Bisson, L. F., and Joseph, C. M. L. (2009). “Fungi of grapes. biology of microorganisms on grapes,” in Must and in Wine, eds H. Konig, G. Unden, and J. Frohlich (Berlin: Springer), 47–60.
Bokulich, N. A., Swadener, M., Sakamoto, K., Mills, D. A., and Bisson, L. F. (2015). Sulfur dioxide treatment alters wine microbial diversity and fermentation progression in a dose-dependent fashion. Am. J. Enol. Vitic. 66, 73–79. doi: 10.5344/ajev.2014.14096
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Briceño, E. X., Latorre, B. A., and Schwarze, E. B. (2009). Effect of Cladosporium rot on the composition and aromatic compounds of red wine. Span. J. Agric. Res. 7, 119–128. doi: 10.5424/sjar/2009071-404
Bui-Klimke, T. R., and Wu, F. (2015). Ochratoxin a and human health risk: a review of the evidence. Crit. Rev. Food Sci. Nutr. 55, 1860–1869. doi: 10.1080/10408398.2012.724480
Cabañes, F. J., Accensi, F., Bragulat, M. R., Abarca, M. L., Castellá, G., Minguez, S., et al. (2002). What is the source of ochratoxin A in wine? Int. J. Food Microbiol. 79, 213–215. doi: 10.1016/s0168-1605(02)00087-9
Capozzi, V., Berbegal, C., Tufariello, M., Grieco, F., Spano, G., and Grieco, F. (2019). Impact of co-inoculation of Saccharomyces cerevisiae, Hanseniaspora uvarum and Oenococcus oeni autochthonous strains in controlled multi starter grape must fermentations. LWT 109, 241–249. doi: 10.1016/j.lwt.2019.04.045
Comitini, F., and Ciani, M. (2008). Influence of fungicide treatments on the occurrence of yeast flora associated with wine grapes. Ann. Microbiol. 58, 489–493. doi: 10.1007/bf03175547
Conacher, C. G., Luyt, N. A., Naidoo-Blassoples, R. K., Rossouw, D., Setati, M. E., and Bauer, F. F. (2021). The ecology of wine fermentation: a model for the study of complex microbial ecosystems. Appl. Microbiol. Biotechnol. 105, 3027–3043. doi: 10.1007/s00253-021-11270-6
Constantì, M., Reguant, C., Poblet, M., Zamora, F., Mas, A., and Guillamón, J. M. (1998). Molecular analysis of yeast population dynamics: effect of sulphur dioxide and inoculum on must fermentation. Int. J. Food Microbiol. 41, 169–175. doi: 10.1016/s0168-1605(98)00041-5
D’Amico, M., Di Vita, G., and Monaco, L. (2016). Exploring environmental consciousness and consumer preferences for organic wines without sulfites. J. Clean. Prod. 120, 64–71. doi: 10.1016/j.jclepro.2016.02.014
David, V., Terrat, S., Herzine, K., Claisse, O., Rousseaux, S., Tourdot-Maréchal, R., et al. (2014). High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 41, 811–821. doi: 10.1007/s10295-014-1427-2
De Orduna, R. M. (2010). Climate change associated effects on grape and wine quality and production. Food Res. Int. 43, 1844–1855. doi: 10.1016/j.foodres.2010.05.001
Diguta, C. F., Rousseaux, S., Weidmann, S., Bretin, N., Vincent, B., Guilloux-Benatier, M., et al. (2010). Development of a qPCR assay for specific quantification of Botrytis cinerea on grapes. FEMS Microbiol. Lett. 313, 81–87. doi: 10.1111/j.1574-6968.2010.02127.x
Divol, B., du Toit, M., and Duckitt, E. (2012). Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 95, 601–613. doi: 10.1007/s00253-012-4186-x
Domizio, P., Romani, C., Lencioni, L., Comitini, F., Gobbi, M., Mannazzu, I., et al. (2011). Outlining a future for non-Saccharomyces yeasts: selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 147, 170–180. doi: 10.1016/j.ijfoodmicro.2011.03.020
Drappier, J., Thibon, C., Rabot, A., and Geny-Denis, L. (2019). Relationship between wine composition and temperature: impact on Bordeaux wine typicity in the context of global warming. Crit. Rev. Food Sci. Nutr. 59, 14–30. doi: 10.1080/10408398.2017.1355776
Epifanio, S. I., Gutierrez, A. R., Santamaría, M. P., and López, R. (1999). The influence of enological practices on the selection of wild yeast strains in spontaneous fermentation. Am. J. Enol. Vitic. 50, 219–224.
Escudié, F., Auer, L., Bernard, M., Mariadassou, M., Cauquil, L., Vidal, K., et al. (2017). FROGS: find, rapidly, OTUs with galaxy solution. Bioinformatics 34, 1287–1294. doi: 10.1093/bioinformatics/btx791
Forbes, S. L., Cohen, D. A., Cullen, R., Wratten, S. D., and Fountain, J. (2009). Consumer attitudes regarding environmentally sustainable wine: an exploratory study of the New Zealand marketplace. J. Clean. Prod. 17, 1195–1199. doi: 10.1016/j.jclepro.2009.04.008
Galati, A., Schifani, G., Crescimanno, M., and Migliore, G. (2019). “Natural wine” consumers and interest in label information: an analysis of willingness to pay in a new Italian wine market segment. J. Clean. Prod. 227, 405–413. doi: 10.1016/j.jclepro.2019.04.219
García, M., Esteve-Zarzoso, B., Crespo, J., Cabellos, J. M., and Arroyo, T. (2017). Yeast monitoring of wine mixed or sequential fermentations made by native strains from DO “Vinos de Madrid” using real-time quantitative PCR. Front. Microbiol. 8:2520. doi: 10.3389/fmicb.2017.02520
Geny, L., Marchal, A., Lavigne, V., Guittard, E., and Danede, N. (2018). Le millésime 2017 à Bordeaux. Available online at: https://www.bordeauxraisins.fr/images/millesimes/Millesime-2017-ISVV.pdf (accessed October 27, 2020).
Geny, L., Marchal, A., Lavigne, V., Guittard, E., Danede, N., Barsacq, A., et al. (2019). Le millésime 2018 à Bordeaux. Available online at: https://www.bordeauxraisins.fr/images/millesimes/Millesime_2018_ISVV.pdf (accessed October 27, 2020).
Gil-Serna, J., Vázquez, C., González-Jaén, M. T., and Patiño, B. (2018). Wine contamination with ochratoxins: a review. Beverages 4:6. doi: 10.3390/beverages4010006
Grangeteau, C., Roullier-Gall, C., Rousseaux, S., Gougeon, R. D., Schmitt-Kopplin, P., Alexandre, H., et al. (2017). Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 10, 354–370. doi: 10.1111/1751-7915.12428
Henick-Kling, T., Edinger, W., Daniel, P., and Monk, P. (1998). Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J. Appl. Microbiol. 84, 865–876. doi: 10.1046/j.1365-2672.1998.00423.x
Jara, C., Laurie, V. F., Mas, A., and Romero, J. (2016). Microbial terroir in chilean valleys: diversity of non-conventional yeast. Front. Microbiol. 7:663. doi: 10.3389/fmicb.2016.00663
Johnson, J., Fu, M., Qian, M., Curtin, C., and Osborne, J. P. (2020). Influence of select non-saccharomyces yeast on Hanseniaspora uvarum growth during prefermentation cold maceration. Am. J. Enol. Vitic. 71, 278–287. doi: 10.5344/ajev.2020.20004
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Kántor, A., Hutková, J., Petrová, J., Hleba, L., and Kačániová, M. (2019). Antimicrobial activity of pulcherrimin pigment produced by Metschnikowia pulcherrima against various yeast species. J. Microbiol. Biotechnol. Food Sci. 2019, 282–285. doi: 10.15414/jmbfs.2015/16.5.3.282-285
Kecskeméti, E., Berkelmann-Löhnertz, B., and Reineke, A. (2016). Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS One 11:e0160852. doi: 10.1371/journal.pone.0160852
Latorre, B. A., Briceño, E. X., and Torres, R. (2011). Increase in Cladosporium spp. populations and rot of wine grapes associated with leaf removal. Crop Prot. 30, 52–56. doi: 10.1016/j.cropro.2010.08.022
Lisanti, M. T., Blaiotta, G., Nioi, C., and Moio, L. (2019). Alternative methods to SO2 for microbiological stabilization of wine. Compr. Rev. Food Sci. Food Saf. 18, 455–479. doi: 10.1111/1541-4337.12422
Liu, H. M., Guo, J. H., Cheng, Y. J., Luo, L., Liu, P., Wang, B. Q., et al. (2010). Control of gray mold of grape by Hanseniaspora uvarum and its effects on postharvest quality parameters. Ann. Microbiol. 60, 31–35. doi: 10.1007/s13213-010-0018-3
Lücke, F.-K. (2000). Utilization of microbes to process and preserve meat. Meat Sci. 56, 105–115. doi: 10.1016/S0309-1740(00)00029-2
Mahé, F., Rognes, T., Quince, C., de Vargas, C., and Dunthorn, M. (2015). Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3:e1420. doi: 10.7717/peerj.1420
Martins, G., Miot-Sertier, C., Lauga, B., Claisse, O., Lonvaud-Funel, A., Soulas, G., et al. (2012). Grape berry bacterial microbiota: impact of the ripening process and the farming system. Int. J. Food Microbiol. 158, 93–100. doi: 10.1016/j.ijfoodmicro.2012.06.013
Martins, G., Vallance, J., Mercier, A., Albertin, W., Stamatopoulos, P., Rey, P., et al. (2014). Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 177, 21–28. doi: 10.1016/j.ijfoodmicro.2014.02.002
Marullo, P., Claisse, O., Raymond Eder, M. L., Börlin, M., Feghali, N., Bernard, M., et al. (2020). SSU1 checkup, a rapid tool for detecting chromosomal rearrangements related to the SSU1 promoter in Saccharomyces cerevisiae: an ecological and technological study on wine yeast. Front. Microbiol. 11:1331. doi: 10.3389/fmicb.2020.01331
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Morgan, S. C., Tantikachornkiat, M., Scholl, C. M., Benson, N. L., Cliff, M. A., and Durall, D. M. (2019). The effect of sulfur dioxide addition at crush on the fungal and bacterial communities and the sensory attributes of Pinot gris wines. Int. J. Food Microbiol. 290, 1–14. doi: 10.1016/j.ijfoodmicro.2018.09.020
Nisiotou, A. A., Spiropoulos, A. E., and Nychas, G.-J. E. (2007). Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl. Environ. Microbiol. 73, 6705–6713. doi: 10.1128/AEM.01279-07
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., et al. (2013). Package ‘vegan.’ Community Ecology Package, Version Vol. 2, 1–295.
Oro, L., Ciani, M., and Comitini, F. (2014). Antimicrobial activity of M Etschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 116, 1209–1217. doi: 10.1111/jam.12446
Ough, C. S., and Crowell, E. A. (1987). Use of sulfur dioxide in winemaking. J. Food Sci. 52, 386–388. doi: 10.1111/j.1365-2621.1987.tb06620.x
Padilla, B., Gil, J. V., and Manzanares, P. (2016). Past and future of Non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol 7:411. doi: 10.3389/fmicb.2016.00411
Park, H., and Bakalinsky, A. T. (2004). Evidence for Sulfite Proton Symport in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 14, 967–971.
Pasteur, L. (1866). Études sur le vin, ses maladies, causes qui les provoquent, procédés nouveaux pour le conserver et pour le vieillir [Studies on wine, its diseases and their causes, and new processes to conserve and age wine]. Imprimerie Impériale, V. L’Imprimerie impériale. Paris: Impr. Impeériale, 264.
Pérès, S., Raineau, Y., Tempere, S., Pons, A., and Giraud-Heraud, E. (2018). Vins bio, vins sans sulfites ajoutés, vins nature: quelles demandes réelles des consommateurs? Revue Droit et Patrimoine 281, 26–29.
Poveda, A. M., Pérez, M. R., Mollá-Bauza, M. M. B., and Martínez, L. M.-C. (2005). Determination of the surplus that consumers are willing to pay for an organic wine. Span. J. Agric. Res. 3, 43–51. doi: 10.5424/sjar/2005031-123
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rabosto, X., Carrau, M., Paz, A., Boido, E., Dellacassa, E., and Carrau, F. M. (2006). Grapes and vineyard soils as sources of microorganisms for biological control of Botrytis cinerea. Am. J. Enol. Vitic. 57, 332–338.
Ramírez, M., Velázquez, R., Maqueda, M., López-Piñeiro, A., and Ribas, J. C. (2015). A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 6:983. doi: 10.3389/fmicb.2015.00983
Rantsiou, K., Giacosa, S., Pugliese, M., Englezos, V., Ferrocino, I., Río Segade, S., et al. (2020). Impact of chemical and alternative fungicides applied to grapevine cv Nebbiolo on microbial ecology and chemical-physical grape characteristics at harvest. Front. Plant Sci. 11:700. doi: 10.3389/fpls.2020.00700
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
RStudio Team (2020). RStudio: Integrated Development Environment for R. Boston, MA: RStudio, PBC. Available online at: http://www.rstudio.com/ (accessed July 24, 2020).
Serra, R., Abrunhosa, L., Kozakiewicz, Z., and Venâncio, A. (2003). Black Aspergillus species as ochratoxin a producers in Portuguese wine grapes. Int. J. Food Microbiol. 88, 63–68. doi: 10.1016/s0168-1605(03)00085-0
Simonin, S., Alexandre, H., Nikolantonaki, M., Coelho, C., and Tourdot-Maréchal, R. (2018). Inoculation of Torulaspora delbrueckii as a bio-protection agent in winemaking. Food Res. Int. 107, 451–461. doi: 10.1016/j.foodres.2018.02.034
Simonin, S., Roullier-Gall, C., Ballester, J., Schmitt-Kopplin, P., Quintanilla-Casas, B., Vichi, S., et al. (2020). Bio-Protection as an alternative to sulphites: impact on chemical and microbial characteristics of red wines. Front. Microbiol. 11:1308. doi: 10.3389/fmicb.2020.01308
Son, S. M., Moon, K. D., and Lee, C. Y. (2001). Inhibitory effects of various antibrowning agents on apple slices. Food Chem. 73, 23–30. doi: 10.1016/s0308-8146(00)00274-0
Steenwerth, K. L., Morelan, I., Stahel, R., Figueroa-Balderas, R., Cantu, D., Lee, J., et al. (2021). Fungal and bacterial communities of ‘Pinot noir’must: effects of vintage, growing region, climate, and basic must chemistry. PeerJ 9:e10836. doi: 10.7717/peerj.10836
Takahashi, M., Ohta, T., Masaki, K., Mizuno, A., and Goto-Yamamoto, N. (2014). Evaluation of microbial diversity in sulfite-added and sulfite-free wine by culture-dependent and -independent methods. J. Biosci. Bioeng. 117, 569–575. doi: 10.1016/j.jbiosc.2013.10.012
Timbo, B., Koehler, K. M., Wolyniak, C., and Klontz, K. C. (2004). Sulfites-a food and drug administration review of recalls and reported adverse events. J. Food Prot. 67, 1806–1811. doi: 10.4315/0362-028X-67.8.1806
Tristezza, M., Tufariello, M., Capozzi, V., Spano, G., Mita, G., and Grieco, F. (2016). The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front. Microbiol. 7:670. doi: 10.3389/fmicb.2016.00670
Tufariello, M., Fragasso, M., Pico, J., Panighel, A., Castellarin, S. D., Flamini, R., et al. (2021). Influence of Non-Saccharomyces on wine chemistry: a focus on aroma-related compounds. Molecules 26:644. doi: 10.3390/molecules26030644
Vally, H., Misso, N. L. A., and Madan, V. (2009). Clinical effects of sulphite additives. Clin. Exp. Allergy 39, 1643–1651. doi: 10.1111/j.1365-2222.2009.03362.x
Vicente, J., Ruiz, J., Belda, I., Benito-Vázquez, I., Marquina, D., Calderón, F., et al. (2020). The Genus Metschnikowia in enology. Microorganisms 8:1038. doi: 10.3390/microorganisms8071038
Villalba, M. L., Susana Sáez, J., del Monaco, S., Lopes, C. A., and Sangorrín, M. P. (2016). TdKT, a new killer toxin produced by Torulaspora delbrueckii effective against wine spoilage yeasts. Int. J. Food Microbiol. 217, 94–100. doi: 10.1016/j.ijfoodmicro.2015.10.006
Warner, C. R., Diachenko, G. W., and Bailey, C. J. (2000). Sulfites: an important food safety issue. Food Test. Anal. 6, 8–10.
Waterhouse, A. L., Sacks, G. L., and Jeffery, D. W. (2016). Understanding Wine Chemistry. Hoboken, NJ: Wiley Online Library.
Wickham, H., Chang, W., and Wickham, M. H. (2016). Package ‘ggplot2.’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version 2.
Windholtz, S., Dutilh, L., Lucas, M., Maupeu, J., Vallet-Courbin, A., Farris, L., et al. (2021a). Population dynamics and yeast diversity in early winemaking stages without sulfites revealed by three complementary approaches. Appl. Sci. 11:2494. doi: 10.3390/app11062494
Windholtz, S., Redon, P., Lacampagne, S., Farris, L., Lytra, G., Cameleyre, M., et al. (2021b). Non-Saccharomyces yeasts as bioprotection in the composition of red wine and in the reduction of sulfur dioxide. LWT 149:111781. doi: 10.1016/j.lwt.2021.111781
Zhang, J., Wang, E. T., Singh, R. P., Guo, C., Shang, Y., Chen, J., et al. (2019). Grape berry surface bacterial microbiome: impact from the varieties and clones in the same vineyard from central China. J. Appl. Microbiol. 126, 204–214. doi: 10.1111/jam.14124
Zimmer, A., Durand, C., Loira, N., Durrens, P., Sherman, D. J., and Marullo, P. (2014). QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One 9:e86298. doi: 10.1371/journal.pone.0086298
Keywords: vinification without SO2, bioprotection, biodiversity, prefermentary stages, maturity level
Citation: Windholtz S, Vinsonneau E, Farris L, Thibon C and Masneuf-Pomarède I (2021) Yeast and Filamentous Fungi Microbial Communities in Organic Red Grape Juice: Effect of Vintage, Maturity Stage, SO2, and Bioprotection. Front. Microbiol. 12:748416. doi: 10.3389/fmicb.2021.748416
Received: 27 July 2021; Accepted: 01 December 2021;
Published: 24 December 2021.
Edited by:
Kate Howell, The University of Melbourne, AustraliaReviewed by:
Agustin Aranda, Institute for Integrative Systems Biology, University of Valencia, SpainCopyright © 2021 Windholtz, Vinsonneau, Farris, Thibon and Masneuf-Pomarède. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara Windholtz, c2FyYS53aW5kaG9sdHpAdS1ib3JkZWF1eC5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.