
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 November 2021
Sec. Infectious Agents and Disease
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.748147
This article is part of the Research Topic Molecular Adaptations of Vibrionaceae to Changing Environments, Volume II View all 5 articles
The high-pathogenicity island irp-HPI is widespread among Vibrionaceae encoding the piscibactin siderophore system. The expression of piscibactin genes in the fish pathogen Vibrio anguillarum is favored by low temperatures. However, information about the regulatory mechanism behind irp-HPI gene expression is scarce. In this work, in-frame deletion mutants of V. anguillarum defective in the putative regulators AraC1 and AraC2, encoded by irp-HPI, and in the global regulators H-NS and ToxRS, were constructed and their effect on irp-HPI gene expression was analyzed at 15 and 25°C. The results proved that only AraC1 (renamed as PbtA) is required for the expression of piscibactin biosynthesis and transport genes. PbtA inactivation led to an inability to grow under iron restriction, a loss of the outer membrane piscibactin transporter FrpA, and a significant decrease in virulence for fish. Inactivation of the global repressor H-NS, which is involved in silencing of horizontally acquired genes, also resulted in a lower transcriptional activity of the frpA promoter. Deletion of toxR-S, however, did not have a relevant effect on the expression of the irp-HPI genes. Therefore, while irp-HPI would not be part of the ToxR regulon, H-NS must exert an indirect effect on piscibactin gene expression. Thus, the temperature-dependent expression of the piscibactin-encoding pathogenicity island described in V. anguillarum is the result of the combined effect of the AraC-like transcriptional activator PbtA, harbored in the island, and other not yet defined regulator(s) encoded by the genome. Furthermore, different expression patterns were detected within different irp-HPI evolutionary lineages, which supports a long-term evolution of the irp-HPI genomic island within Vibrionaceae. The mechanism that modulates piscibactin gene expression could also be involved in global regulation of virulence factors in response to temperature changes.
Bacteria of the genus Vibrio are among the most predominant infectious agents threatening marine wildlife and aquaculture (Toranzo et al., 2005). Vibrio anguillarum is the etiological agent of classical vibriosis in fish, a typical hemorrhagic septicemia that causes high mortalities and economic losses in aquaculture worldwide (Toranzo et al., 2017). Increments in sea water temperature are associated with the proliferation of Vibrio species (Maeda et al., 2003), and hence, the subsequent occurrence of fish disease outbreaks (Le Roux et al., 2015). However, V. anguillarum is also able to cause vibriosis at cold temperatures (5–18°C) (Austin and Austin, 2007; Bellos et al., 2015; Ma et al., 2017). Numerous factors including motility, chemotaxis, LPS, extracellular products with hemolytic and proteolytic activities, and several iron-uptake systems have a role in V. anguillarum virulence (Rodkhum et al., 2006; Li and Ma, 2017; Toranzo et al., 2017). V. anguillarum adjusts the expression of some of these virulence factors by unknown mechanisms, responding to environmental signals such as iron levels and temperatures (Denkin and Nelson, 1999; Crisafi et al., 2014; Lages et al., 2019).
Bacteria possess tools to silence the expression of horizontally acquired genes. H-NS, an important global repressor of transcription in Gram-negative bacteria, functions in the process of xenogenetic silencing and also in the regulation of temperature-dependent gene expression (Stoebel et al., 2008; Prajapat and Saini, 2012; Mou et al., 2013). Another conserved global regulator is ToxR, which functions as a sensor and signal transducer controlling the expression of multiple virulence genes (ToxR regulon) in response to environmental cues (Skorupski and Taylor, 1997). Conversely, horizontally acquired DNA usually contains genes encoding transcriptional regulators that promote their own expression (Stoebel et al., 2008).
Production of siderophores is a key virulence factor for most bacterial pathogens including V. anguillarum (Miethke and Marahiel, 2007; Balado et al., 2018; Kramer et al., 2020). Highly virulent strains of V. anguillarum can simultaneously synthesize two siderophores: vanchrobactin and piscibactin (Balado et al., 2018). Vanchrobactin is considered the ancestral siderophore system of V. anguillarum since it is encoded by a chromosomal gene cluster (vab genes) that is widespread in all V. anguillarum isolates either environmental or pathogenic (Balado et al., 2006). The synthesis and transport of piscibactin is encoded by a high-pathogenicity island (irp-HPI element) (Osorio et al., 2015; Balado et al., 2018). In addition, irp-HPI encodes two conserved AraC-like regulators not studied so far (Figure 1), which are among the most upregulated genes of V. anguillarum when temperature decreases (Lages et al., 2019). This HPI was firstly identified within a plasmid in Photobacterium damselae subsp. piscicida, for which it is also a key virulence factor (Osorio et al., 2006, 2015). However, recent works showed that it is widespread among many species of the Vibrionaceae family, including relevant animal pathogens of the Splendidus and Harveyi clades and human pathogens like V. cholerae (Thode et al., 2018).
Siderophore production in V. anguillarum is balanced in a temperature-dependent manner since piscibactin genes have a dual requirement for iron starvation and low temperatures (<20°C) to be significantly expressed (Balado et al., 2018; Lages et al., 2019). Thus, the acquisition of irp-HPI seems to play a role in the adaptation of V. anguillarum to changing environments enhancing niche flexibility and enabling it to infect cold- and warm-water adapted fish (Balado et al., 2018; Lages et al., 2019).
In this work, we analyzed the effect of the two AraC-like regulators, araC1 and araC2, contained in irp-HPI, and the global regulators H-NS and ToxR-S on the temperature-dependent expression of piscibactin genes in V. anguillarum. The results showed that AraC1 is the main transcriptional regulator that modulates the expression of the siderophore piscibactin system. In addition, although the global regulators, ToxR-S and H-NS, do not have major effects on irp-HPI expression, they indirectly intervene in the regulatory circuit of this genomic island through the modulation of the frpA promoter activity. Thus, the temperature-dependent expression pattern of irp-HPI genes results from the combined effect of regulatory factor(s) encoded outside the genomic island and the AraC1 transcriptional regulator within the genomic island.
The bacterial strains and plasmids used in this work are listed in Table 1. V. anguillarum strains were grown at 25°C or 15°C in tryptic soy agar (TSA) or broth (TSB) (Cultimed) supplemented with 1% NaCl. Escherichia coli strains were grown in Luria-Bertani broth or agar (Cultimed) at 37°C. When required, antibiotics were added at the following final concentrations: kanamycin 50 μg ml–1, ampicillin sodium salt 60 μg ml–1 or 100 μg ml–1, and gentamycin 15 μg ml–1.
In-frame deletions of araC1, araC2, h-ns, and toxR-S were constructed by allelic exchange in V. anguillarum RV22 strain in a ΔvabF background (impaired to synthesize vanchrobactin) as previously described (Balado et al., 2006). The flanking regions of each gene were amplified by PCR and cloned into the vector pWKS30; the resulting constructions were ligated into the suicide vector pNidKan. The resulting plasmid was conjugated with RV22ΔvabF strain and selected based on ampicillin and kanamycin resistance. A second event of recombination was performed, and the mutants were selected based on sucrose (15%) resistance. A PCR was performed to confirm the allelic exchange event. This process led to the formation of V. anguillarum mutant strains RV22ΔvabFΔaraC1, RV22ΔvabFΔaraC2, RV22ΔvabFΔh-ns, and RV22ΔvabFΔtoxR-S. For araC1 mutant complementation, araC1 was amplified by PCR, cloned into the vector pSEVA651 in E. coli S17-1 λpir, and mobilized to the appropriate mutant strain by conjugation. To restore the original phenotype, the WT genes h-ns and toxR-S were cloned into the suicide vector pNidKan and the complementation was accomplished as indicated above for the construction of mutants. The oligonucleotides used are listed in Supplementary Table 1.
Growth ability assays were performed in CM9 medium supplemented with 10 μM FeCl3 to achieve iron excess or with 25 or 75 μM 2,2′-dipyridyl (TCI) to achieve iron deficiency. V. anguillarum strains RV22ΔvabF, RV22ΔvabFΔaraC1, RV22 ΔvabFΔaraC2, RV22ΔvabFΔh-ns, RV22ΔvabFΔtoxR-S, and complemented strains were grown overnight in TSB-1. Each culture was adjusted to an OD600 = 0.5 and a 1:50 dilution was inoculated in CM9 medium. The resulting cultures were incubated at 15°C with shaking at 120 rpm. Growth was recorded after 48 h.
Bacterial cultures grown in CM9 medium with 25 μM 2,2′-dipyridyl and at an OD600 ∼ 0.8 were used to measure siderophore production with the chrome azurol-S (CAS) liquid assay (Schwyn and Neilands, 1987). Briefly, supernatants were obtained by pelleting bacterial cells using centrifugation and equal volumes of these supernatants were incubated with the CAS reagent at room temperature for 15 min. The quantification was performed by measuring A630 in a spectrophotometer (Hitachi).
The regions immediately upstream of Photobacterium damselae subsp. piscicida (DI21 strain) araC1 and frpA genes were amplified by PCR and fused to a promoterless lacZ gene in the low copy number plasmid pHRP309. This process leads to the construction of plasmid pLP28 and plasmid pLP9 carrying constructs araC1pdp::lacZ (ParaC1pdp) and frpApdp::lacZ (PfrpApdp), respectively. Plasmids pMB277 and pMB276 carrying the lacZ fusions of V. anguillarum araC1ang::lacZ and frpAang::lacZ (ParaC1ang and PfrpAang promoters) were previously constructed (Balado et al., 2018). The constructs were mobilized from E. coli S17-1 λpir to V. anguillarum (RV22ΔvabF and its derivative mutants) and P. damselae subsp. piscicida (DI21 strain) by conjugation. The presence of the promoters was confirmed by PCR. The resulting V. anguillarum strains and P. damselae subsp. piscicida carrying the promoter fusions were grown under weak iron restriction using CM9 medium supplemented with 25 μM 2,2′-dipyridyl. When the bacterial cultures reached an OD600 = 0.3, the β-galactosidase activities were measured by the method of Miller (1992). The results shown are means of three independent experiments.
Overnight cultures of RV22ΔvabF and RV22ΔvabFΔaraC1 were grown in TSB-1 and adjusted to OD600 = 0.5. A 1:50 dilution was inoculated in 10 ml CM9 medium supplemented with 25 μM 2,2′-dipyridyl. As the bacterial cultures reached an OD600 = 0.8, they were pelleted at 4,000 rpm, for 30 min at 4°C. The pellet was resuspended in 5 ml of 10 mM Tris-HCl and 0.3% NaCl, pH 8.0. Cellular disruption was accomplished by sonication on ice (five cycles of 30 s). Then, the samples were centrifuged at 4,000 rpm for 30 min at 4°C to eliminate the cellular debris. For the isolation of membrane proteins, 1% sarkosyl was added to the supernatant and incubated at room temperature for 30 min. The samples were centrifuged at 40,000 rpm for 30 min at 4°C and the pellet was resuspended in 20 μl of water. The samples were mixed 1:1 with SDS-PAGE loading buffer and loaded into a 12% polyacrylamide gel. After the separation by SDS-PAGE, the proteins were transferred onto a PVDF membrane as previously described. The membrane was blocked in blocking buffer (5% skim milk in TBST, Tris Buffered Saline with Tween 20) for 1 h at room temperature with shaking. The membrane was incubated overnight in the primary antibody solution against the target protein (1:10,000 dilution of the anti-FrpA antibody). After rinsing the membrane with TBST, it was incubated with the secondary antibody (1:10,000 anti-rabbit IgG HRP conjugate antibody) for 1 h at room temperature with shaking. The signal was detected using the ClarityTM Western ECL substrate (Bio-Rad). Rabbit polyclonal antibodies against the external loop 6 (short peptide PGGFSPAPRSSGDKNGYSP) of FrpA (anti-FrpA) were purchased from GenScript.
Experimental infections were performed using Senegalese sole (Solea senegalensis) fingerlings of approximately 15 g of weight. Fish were divided into four groups of 30 animals, one per tested strain, and were maintained into 50-L seawater tanks at 18°C with aeration and water recirculation. Colonies from a fresh 24-h plate were resuspended in saline solution (0.85% NaCl) to achieve an OD600 of 0.5. Fish were intraperitoneally injected with 100 μl of the bacterial suspension (2–3 × 103 CFU/fish). The number of bacterial cells injected was determined by plating serial dilutions on TSA-1. A control group was injected with saline solution. Mortalities were followed for 12 days after injection and dead events were daily registered. Statistical differences in survival curves were determined using the Kaplan–Meier method with Mantel-Cox log-rank test using SPSS (version 20; IBM SPSS Inc., Chicago, IL). p-values were significant when P was < 0.05. The protocols for animal experimentation follow the current legislation and have been approved by the Bioethics Committee of the University of Santiago de Compostela.
To analyze the diversity of piscibactin gene promoters, we performed BlastN searches in the nucleotide collection (nr/nt) and whole-genome shotgun (wgs) NCBI databases using as a query the nucleotide sequence of V. anguillarum RV22 irp-HPI between 250 bp upstream pbtA and the frpA stop codon (sequence ID. AEZB01000030, from position 30,437 to 35,095). Homologous sequences were clustered in promoter types by similarity; thus, each type included sequences sharing 100% of coverage and≥99.5% of nucleotide identity. Representative sequences of each type were downloaded from NCBI and aligned using MUSCLE (MEGA X suite). Phylogenetic trees were inferred using p-distances (Transitions + Transversions) and Neighbor-Joining method. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 3,377 positions in the final dataset. Sequence alignments, nucleotide pairwise p-distances, and evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
To analyze the role of the two AraC-like regulators encoded by irp-HPI in piscibactin production, in-frame deletion mutants for araC1 and araC2 genes (Figure 1) were constructed in a V. anguillarum RV22ΔvabF background, a strain that produces only piscibactin as siderophore (Balado et al., 2018). Then, the RV22ΔvabF parental strain and its derivative ΔaraC1 and ΔaraC2 mutants were challenged to grow under iron excess (CM9 supplemented with FeCl3 10 μM) and under iron-deprivation conditions by adding the iron chelator 2,2′-dipyridyl at 25 μM (weak iron-deprivation) or at 75 μM (strong iron-deprivation).
Under iron excess, or weak iron-deprivation conditions, no significant differences were observed between parental and mutant strains. However, under strong iron-deprivation conditions, the ΔaraC1 defective mutant was impaired in growth (Figure 2). The ΔaraC1 mutant showed a phenotype like that observed for the Δirp1 defective mutant that is unable to produce piscibactin (Balado et al., 2018). Under this condition, the parental strain and the ΔaraC2 defective mutant showed indistinguishable growth ability. Subsequent evaluation of siderophore content in the cell-free culture supernatants showed a strong reduction in the ΔaraC1 defective mutant (Figure 2). When the ΔaraC1 mutant was complemented with a functional version of araC1, the parental phenotype was restored. This result suggests that araC1 could encode a transcriptional regulator essential for siderophore piscibactin synthesis. Consequently, araC1 and araC2 were renamed as piscibactin transcriptional regulator pbtA (piscibactin regulator A) and pbtB, respectively.
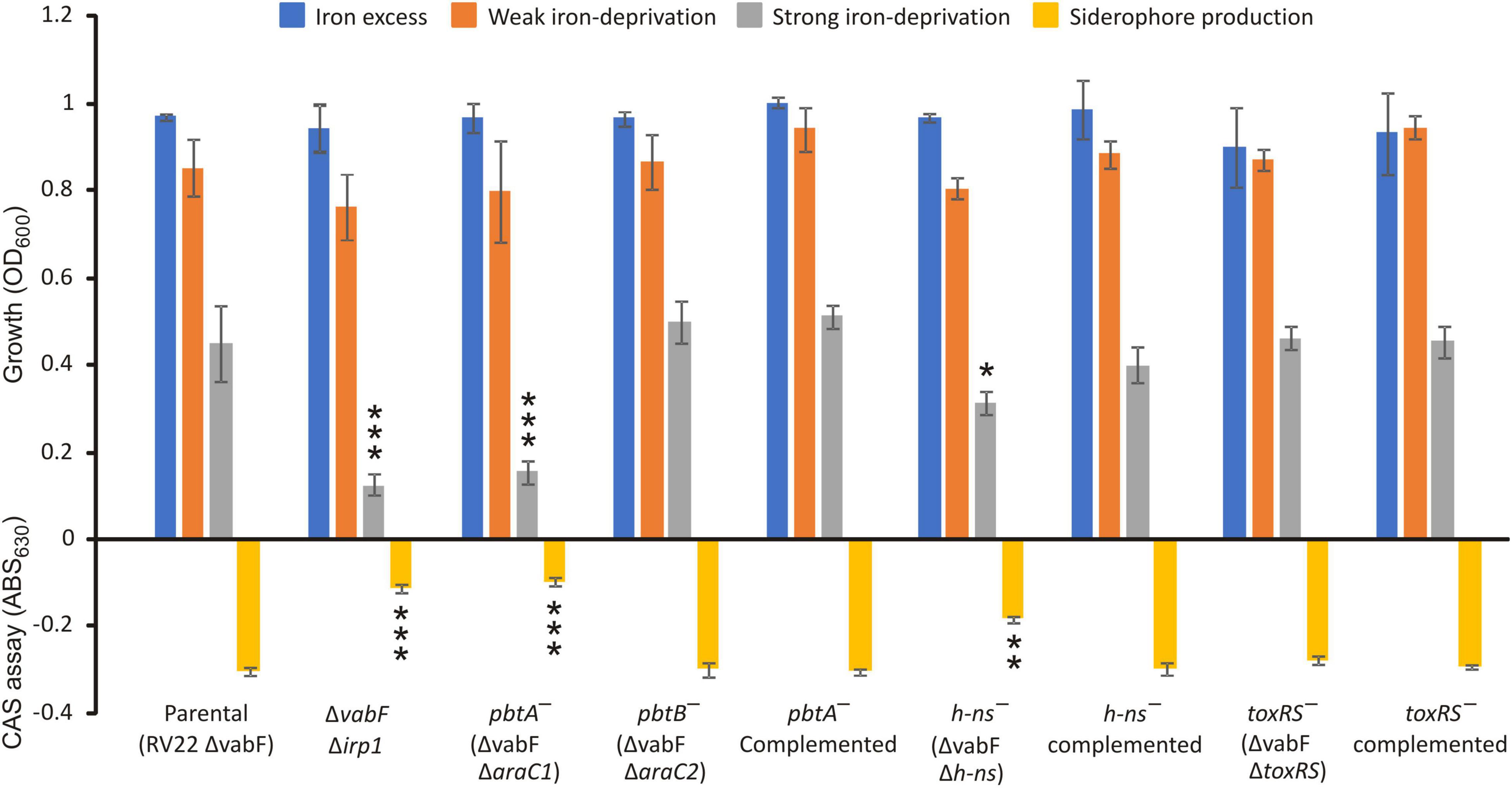
Figure 2. Growth ability under different iron availability conditions of V. anguillarum parental strain compared to its derivative araC1, araC2, h-ns, and toxR-S defective mutants and complemented strains. V. anguillarum strains were cultured in CM9 minimal medium supplemented with 10 μM FeCl3 (iron excess conditions), or the iron chelator 2,2′-dipyridyl at 25 μM (weak iron-deprivation), or at 75 μM 2,2′-dipyridyl (strong iron deprivation). Siderophore production was evaluated in cell-free supernatants after growth of each V. anguillarum strain in weak iron deprivation (CM9 with 25 μM 2,2′-dipyridyl) up to OD600 ca. 0.8. t-test was used to detect significant differences between each mutant and the parental strain. *p < 0.05; **p < 0.01; ***p < 0.001.
The piscibactin TonB-dependent outer membrane transporter frpA gene is located in the piscibactin operon upstream the genes encoding the biosynthetic functions (Balado et al., 2018; Figure 1). To determine whether inactivation of pbtA blocks the piscibactin siderophore system, the presence of FrpA in the V. anguillarum ΔvabF mutant (used as parental strain) and in its derivative pbtA defective mutant was determined by Western blot (Figure 3). While a unique protein band of ca. 70 kDa was detected (which is congruent with the 68-kDa molecular weight of FrpA) in the outer membrane sample (Figure 3A) of the V. anguillarum parental strain (RV22ΔvabF) and in the corresponding Western blot using anti-FrpA (Figure 3B), only a residual amount of FrpA was detected in the sample of the outer membrane proteins and Western blot of V. anguillarum pbtA defective mutant (Figure 3B). These findings greatly suggest that inactivation of the AraC-like transcriptional regulator PbtA disables iron uptake via the siderophore piscibactin.
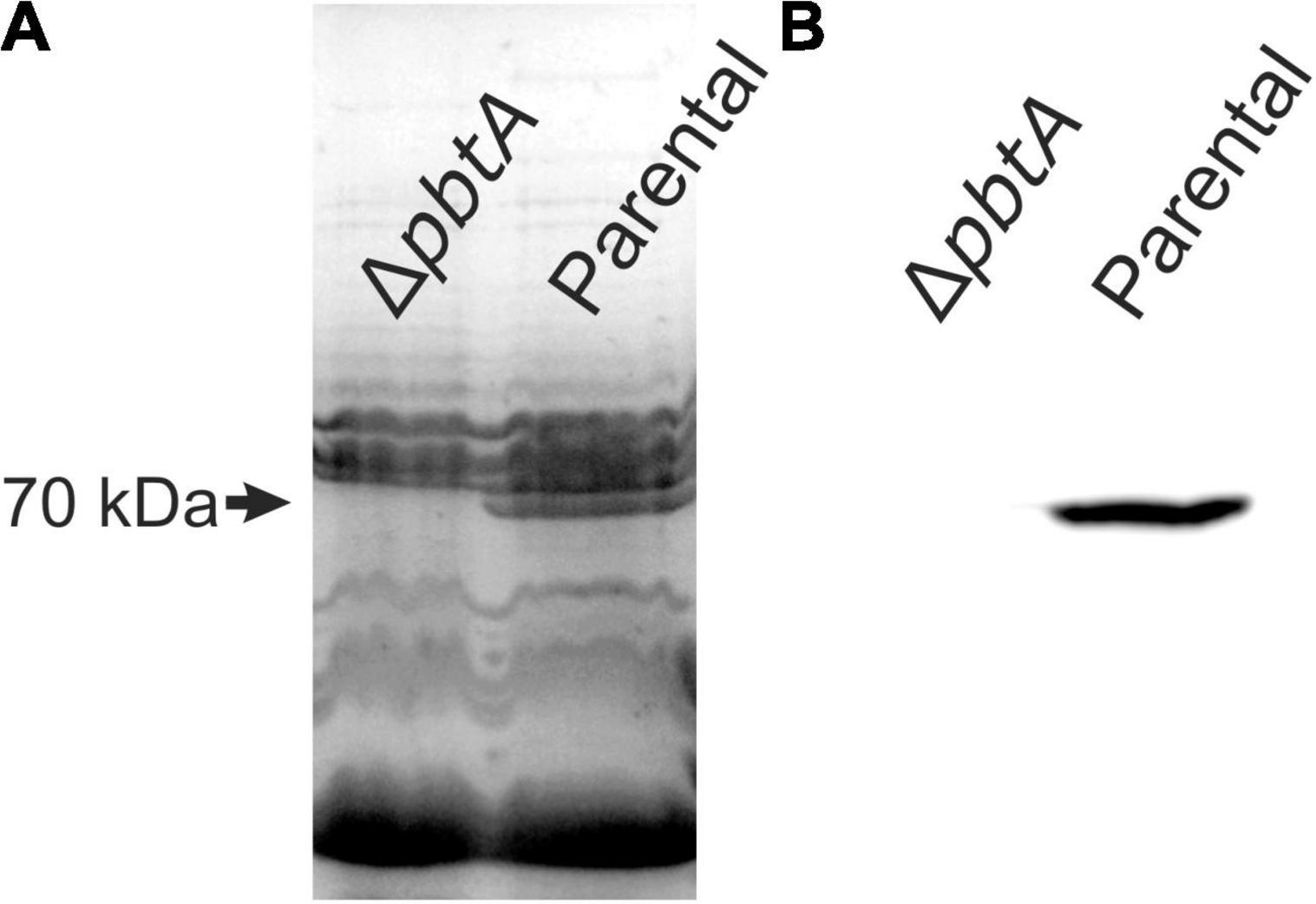
Figure 3. Outer membrane protein (OMP) band patterns (A) and detection of TonB-dependent piscibactin transporter FrpA by Western blot (B) in the V. anguillarum parental strain RV22ΔvabF and in the ΔpbtA (ΔaraC1) mutant cultured under low-iron availability.
To evaluate a putative role of the global regulators H-NS and ToxR-S in piscibactin production, single h-ns or toxR-S mutants were constructed. Then, growth ability and siderophore production were assayed in each mutant and compared to the parental strain RV22ΔvabF (Figure 2). While the phenotype of the ΔtoxR-S mutant (RV22 ΔvabFΔtoxR-S) was indistinguishable from that of the parental strain, the Δh-ns mutant showed a slight reduction of growth ability under strong iron-restricted conditions. The diminution of growth observed in the Δh-ns mutant correlates with a decrease in piscibactin production. Finally, when the Δh-ns mutant was complemented with a functional h-ns gene, the parental phenotype was restored. These results suggest that H-NS is required for a maximum piscibactin production. By contrast, ToxR-S did not exhibit a role in siderophore production.
To evaluate the role of PbtA and H-NS in V. anguillarum virulence, groups of 30 sole fingerlings were inoculated with a dose of 2–3 × 103 CFU per fish of either V. anguillarum parental strain or one of its derivatives ΔpbtA or Δh-ns mutant strains. A control group that was inoculated with saline solution did not show any signs of infection and no mortality was observed. The survival curves of each group of fish are shown in Figure 4. While the fish group challenged with the parental strain (RV22ΔvabF) showed a 60% mortality 7 days after infection, the V. anguillarum pbtA defective strain (ΔpbtA) showed a significant reduction of virulence since it caused approximately 20% mortality. Notably, survival curves of pbtA defective mutant and the ΔvabFΔirp1 double mutant, impaired to produce siderophores (neither piscibactin nor vanchrobactin) (Balado et al., 2018), were statistically indistinguishable. In addition, inactivation of h-ns also resulted in a significant reduction of virulence for fish.
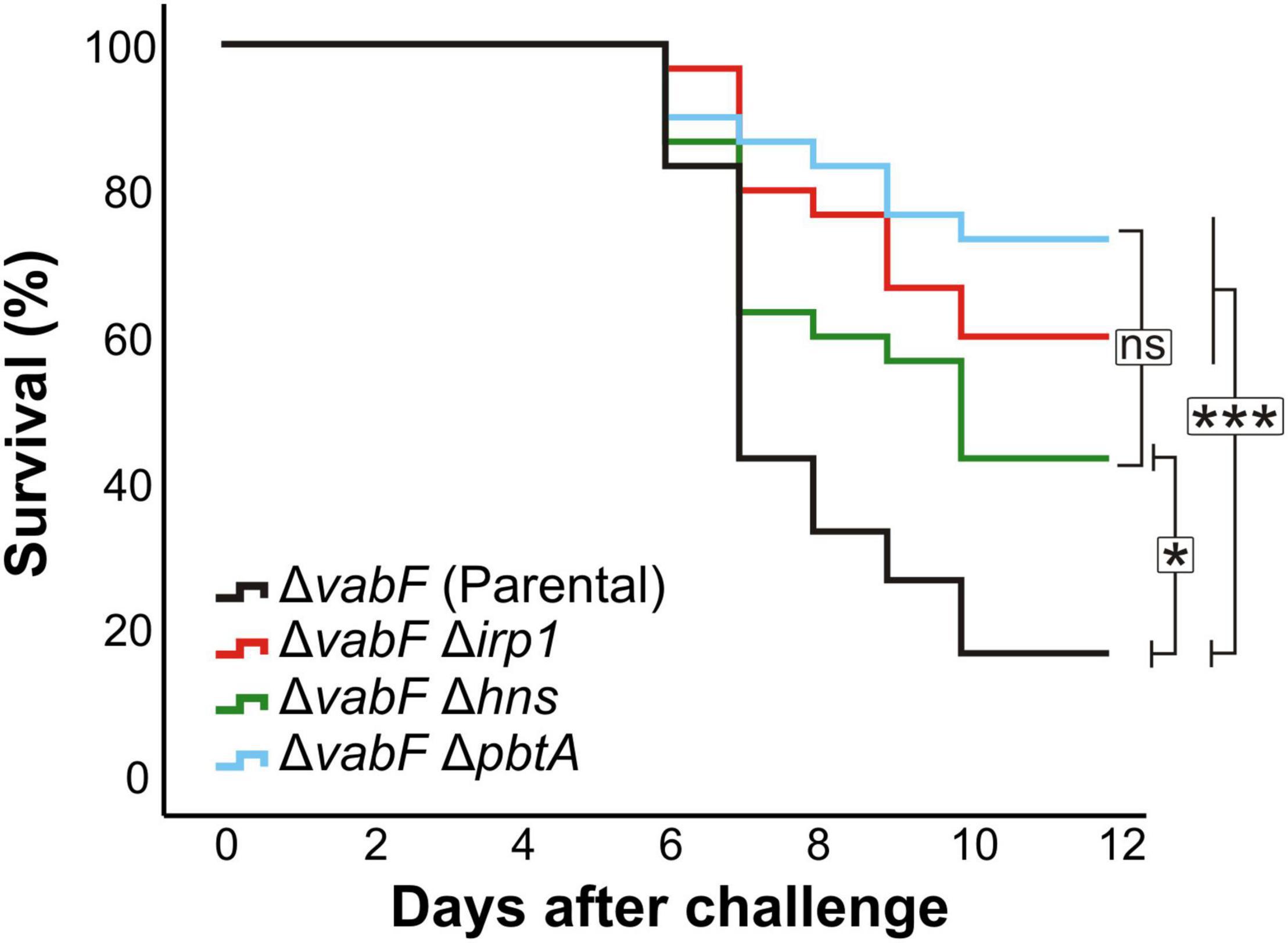
Figure 4. Survival abilities (Kaplan–Meier survival functions) of fish infected with the V. anguillarum pbtA– (RV22 ΔvabFΔpbtA) or h-ns– (RV22ΔvabFΔh-ns) strains compared to parental strain (RV22ΔvabF) and the non-siderophore producer strain RV22ΔvabFΔirp1. Asterisks denote statistical significance, * p < 0.05; *** p < 0.001, ns, no statistically significant differences.
Sequences upstream of pbtA (formerly araC1) and frpA (Figure 1) were previously characterized as the main promoter regions that control the expression of piscibactin synthesis and transport genes (Balado et al., 2018). Thus, to evaluate the role of the putative transcriptional regulator PbtA in the expression of piscibactin genes, the transcriptional activity of pbtA and frpA promoters (PpbtA and PfrpA, respectively) was measured in a pbtA– background (ΔpbtA mutant). At both temperatures tested, 15°C or 25°C, inactivation of pbtA results in the loss of PfrpA activity (Figure 5). Notably, the expression pattern of PpbtA still follows a temperature-dependent pattern in a pbtA– background, showing identical activity levels as the parental strain at the same temperatures. On the other hand, pbtB (formerly araC2) deletion did not alter the transcriptional levels of PpbtA or PfrpA. PpbtA and PfrpA promoter activity evaluation in the pbtA– complemented strain could not be done since the plasmid used to complement (pSEVA651) and the plasmid used to obtain the LacZ fusions (pHRP309) both confer gentamicin resistance. However, as was shown above, complementation of ΔpbtA mutant with the wild-type gene restored a phenotype identical to the parental strain.
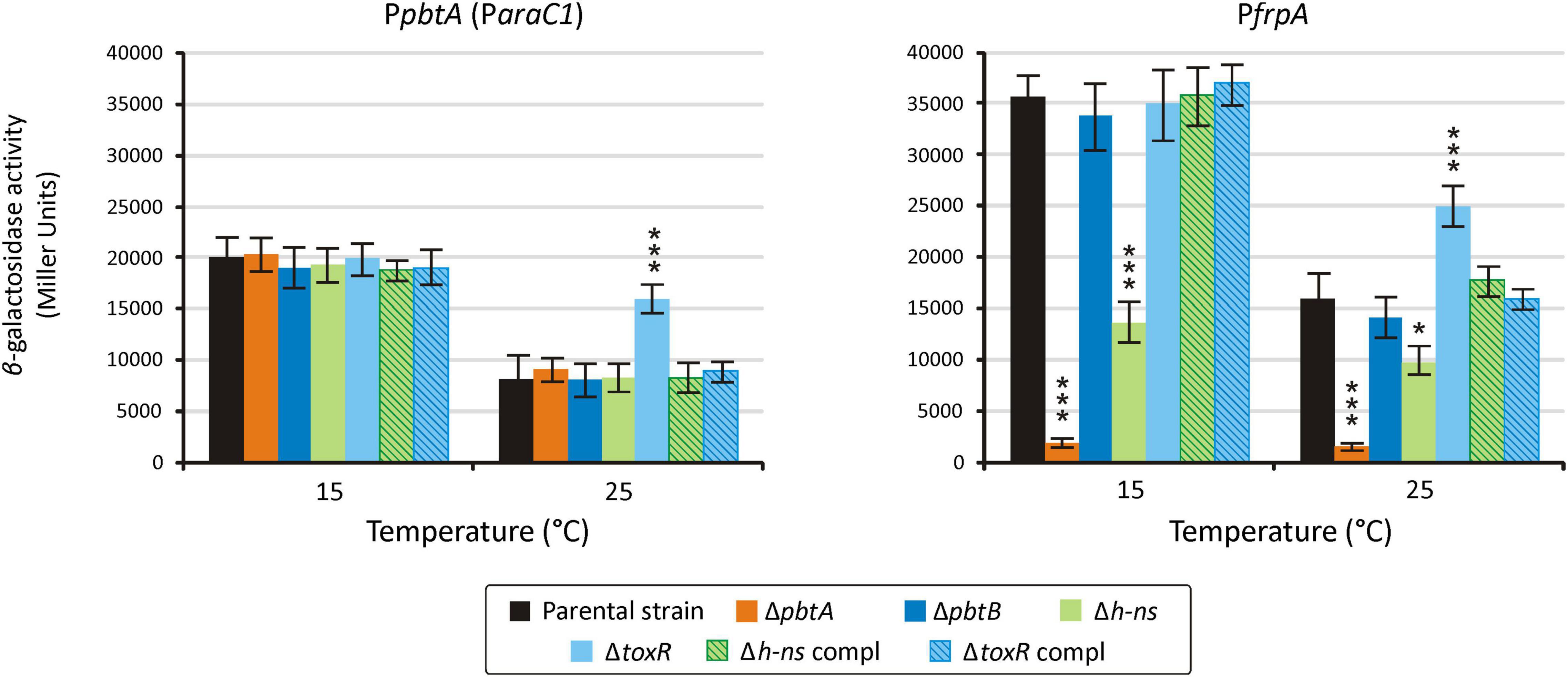
Figure 5. Transcriptional activity of the promoters PpbtA and PfrpA in either pbtA, pbtB, toxR-S, and h-ns mutant strains measured under weak iron deprivation (CM9 supplemented with 25 μM 2,2′-dipyridyl). A t-test was used to detect significant differences between each mutant and the parental strain. * p < 0.05; *** p < 0.001.
Transcriptional activity of PpbtA and PfrpA was also evaluated in the h-ns and toxR-S defective mutants. Unexpectedly, while the activity of the pbtA promoter in the h-ns mutant was indistinguishable from that of the parental strain at both temperatures tested, the activity of frpA promoter (PfrpA) in a Δh-ns background was reduced ca. 60% at 15°C and ca. 20% at 25°C (Figure 4). The evaluation of piscibactin promoters activity in the ΔtoxR-S mutant showed that the expression levels of PpbtA and PfrpA are increased two-fold and 30%, respectively, at 25°C, when compared to the parental strain (Figure 5). No significant changes in expression were observed at 15°C. Complemented strains, either of h-ns or toxR-S, showed expression patterns indistinguishable from those of the parental strain.
Several versions of the irp-HPI genomic island are found within Vibrionaceae showing identical gene structure and an overall nucleotide diversity (p-distance) of 0.3 substitutions per site (Figure 6). Not surprisingly, nucleotide diversity is much higher in intergenic regions than within protein-coding sequences. Alignment of representative irp-HPI genomic islands from different species showed that the sequences immediately upstream of pbtA ATG start codon (PpbtA region) (Supplementary Figure 1) and the pbtB-frpA intergenic region (PfrpA region) (Supplementary Figure 2) showed major differences between Vibrio species and thus different types of piscibactin promoters PpbtA and PfrpA could be defined according to their similarity. Notably, the distribution of each piscibactin promoter type (PpbtA and PfrpA sequences) does not match with irp-HPI phylogenetic lineages (Figure 7). The most variable sequences are found in the pbtB-frpA intergenic region (Supplementary Figures 3,4). Although all pbtB-frpA sequences share a conserved region of ca. 100 bp located immediately upstream of frpA start codon, the region downstream of pbtB stop codon shows higher differences since deletions and/or insertions events would have occurred (Supplementary Figure 2). Thus, the irp-HPI genomic islands of species like V. anguillarum, V. ordalli, and V. qinghaiensis contain a long pbtB-frpA intergenic region with a size of ca. 360 bp, which includes a low complexity sequence between positions 137 and 167 with six repeats of an AAAAT motif (Figure 7 and Supplementary Figure 2). By contrast, P. damselae, V. ostreicida, V. sonorensis, and V. cholerae harbor shorter intergenic sequences with nucleotide lengths between 100 and 140 bp due to the lack of the segment immediately downstream of pbtB stop codon (Figure 7 and Supplementary Figure 1). In addition, there are some intermediate versions such as those found in V. mimicus and V. neptunius (Supplementary Figure 2). The high variability found upstream of pbtA (PpbtA) and in the pbtB-frpA intergenic region (PfrpA) sequences could imply the existence of different expression patterns among those Vibrio spp. harboring the irp-HPI genomic island.
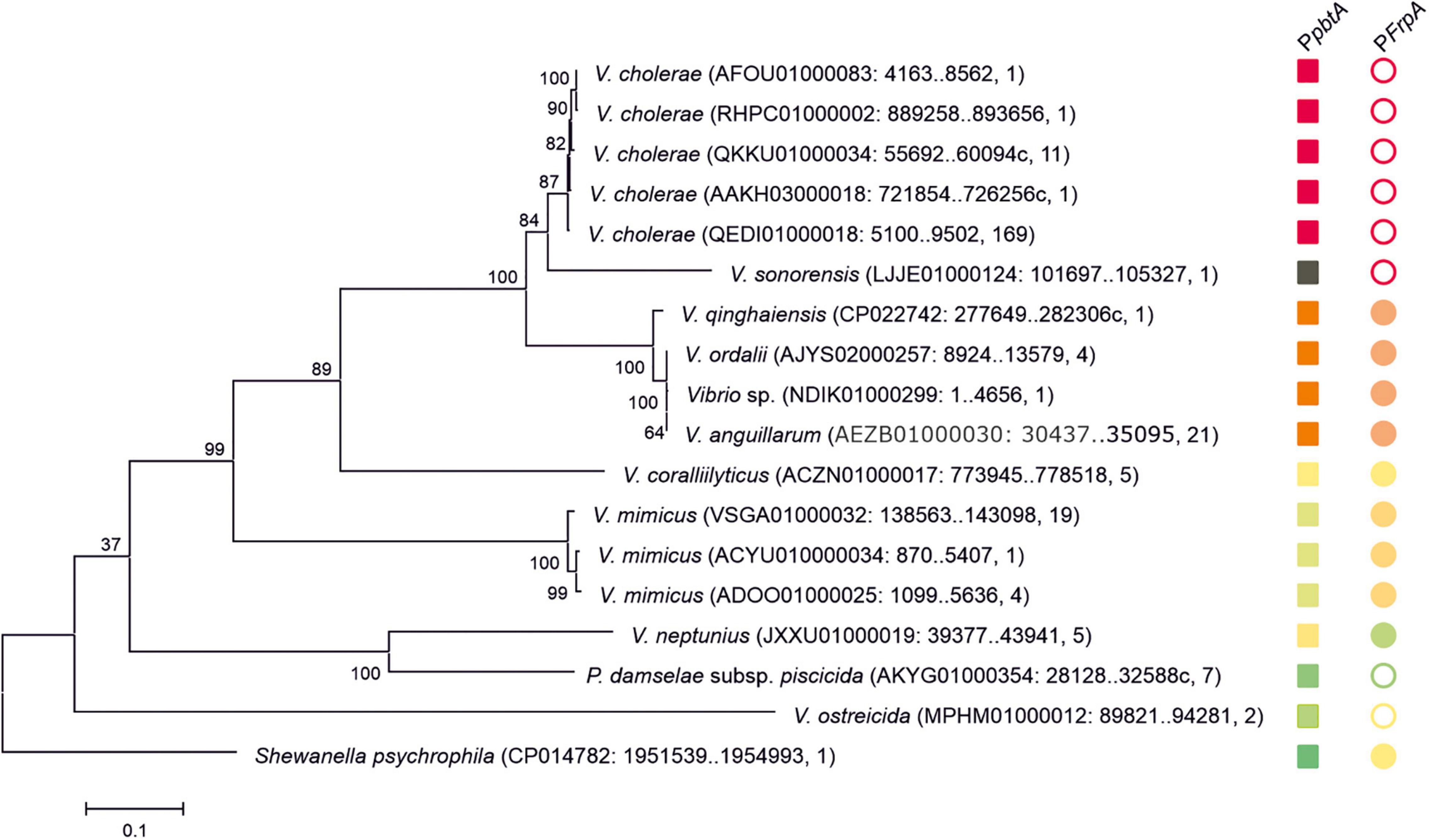
Figure 6. Phylogenetic relationships of the pbtA-frpA region of irp-HPI genomic island and distribution of PpbtA and PfrpA piscibactin promoter versions. Sequences are identified with species name. Sequence ID, sequence region, and the number of closely related sequences deposited in GenBank sharing coverage of 100% and identity nucleotide sequence≥99.5% are shown between parentheses. The tree is drawn to scale, with branch lengths representing the evolutionary distances (number of base substitutions per site). The different versions of the piscibactin promoters PpbtA and PfrpA are represented with squares and circles, respectively. Filled circles denote “long versions” of frpA promoter while empty circles denote “short versions.” Closely related promoter sequences, according to pairwise nucleotide p-distances (Supplementary Figures 3, 4) are represented with the same color. PfrpA promoter of V. sonorensis is represented with a black square since it does not align with the other sequences.
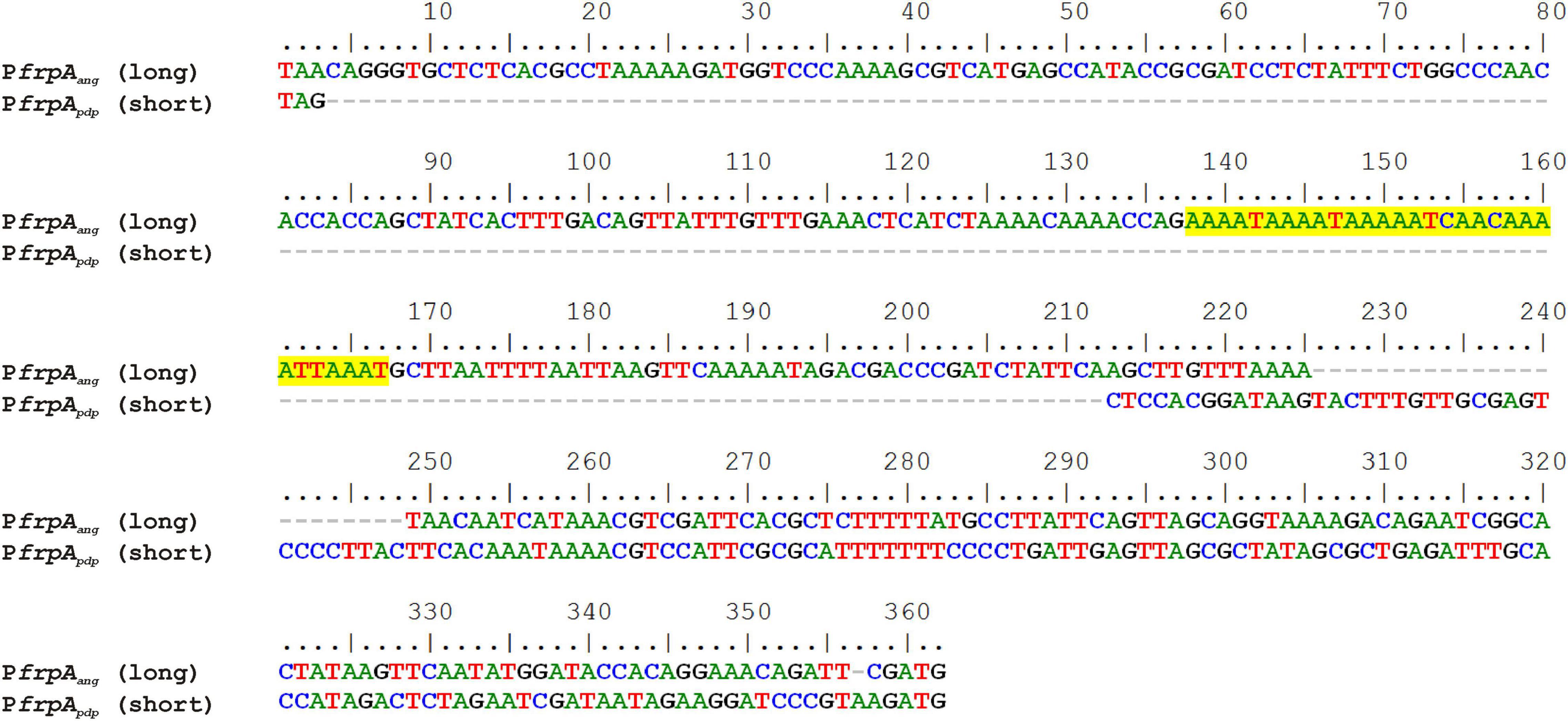
Figure 7. Alignment of a long and a short pbtB-frpA intergenic region (between pbtB stop codon and frpA ATG start codon) containing frpA promoter of V. anguillarum (PfrpAang) and P. damselae subsp. piscicida (PfrpApdp). Low complexity AAAAT repeat motifs are highlighted in yellow.
To compare the expression pattern of different versions of the irp-HPI genomic island found in different Vibrionaceae, the expression pattern of the irp-HPI present in Photobacterium damselae subsp. piscicida (irp-HPIpdp) was compared with the expression pattern of the V. anguillarum island (irp-HPIang). Particularly, irp-HPIpdp harbors the short version of frpA promoter (PfrpApdp) described above (Figures 6, 7). LacZ fusions of the sequences immediately upstream of pbtA and frpA from P. damselae subsp. piscicida (denoted as pbtApdp and frpApdp, respectively), homologous to their counterparts of V. anguillarum (Figures 1, 7), were obtained and the transcriptional activity of each promoter (PpbtApdp and PfrpApdp) was assayed at 15 and 25°C under low iron availability (Figure 8A). The expression pattern of each piscibactin promoter from P. damselae subsp. piscicida (Figure 8A) showed extensive differences with piscibactin promoters from V. anguillarum (Figure 8C). Thus, PpbtApdp promoter is expressed in P. damselae subsp. piscicida although it showed almost the same transcriptional activity at 15°C and at 25°C (Figure 8A), which suggests that piscibactin genes, in contrast with the behavior described in V. anguillarum (Balado et al., 2018), do not show a temperature-dependent expression in P. damselae subsp. piscicida. In addition, PpbtApdp showed a threefold lower activity in P. damselae subsp. piscicida (Figure 8A) than its counterpart PpbtAang in V. anguillarum (Figure 8C). Unexpectedly, while PfrpAang reached ca. 50,000 β-galactosidase units at 15°C, the activity displayed by PfrpApdp was almost undetectable (<750 U), suggesting that the short pbtA-frpA intergenic region found in irp-HPIpdp does not contain a transcriptional promoter.
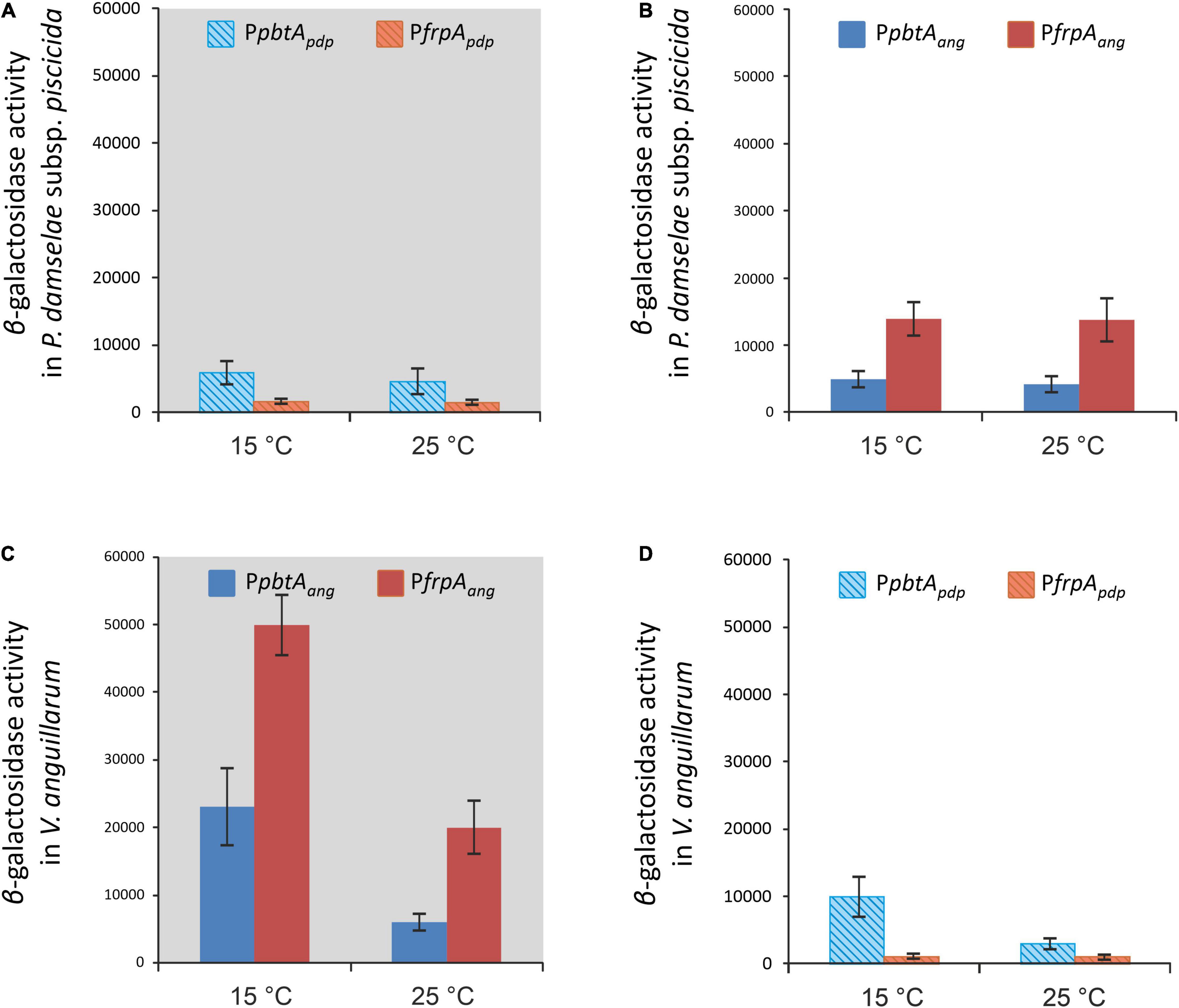
Figure 8. Native (A,C, gray background) and heterologous (B,D) transcriptional activity evaluation (β-galactosidase activity) of both versions of the piscibactin promoters PfrpA (PfrpApdp or PfrpAang) and PpbtA (PpbtApdp or PpbtAang) in P. damselae subsp. piscicida (A,B) or in V. anguillarum (C,D) under iron restriction at 15 or 25°C.
To ascertain whether the differences observed between the expression patterns of each version of irp-HPI promoters would be due to the differences in the promoter versions or to other factors encoded in the respective recipient genomes, a heterologous evaluation of each LacZ fusion was done (Figures 8B,D). The results showed that both versions of the irp-HPI promoters, either PpbtApdp and PfrpApdp or PpbtAang and PfrpAang, have a 2.5-fold less transcriptional activity in P. damselae subsp. piscicida (Figures 8A,B) than PpbtAang and PfrpAang in V. anguillarum (Figure 8C). The expression level achieved by PpbtAang in P. damselae subsp. piscicida was equivalent to that shown by the native evaluation of PpbtApdp. Notably, while the transcriptional activity of PpbtAang and PfrpAang measured within P. damselae subsp. piscicida did not show differences between temperatures (Figure 8B), the PpbtApdp promoter showed a three-fold higher transcriptional activity at 15°C than at 25°C when its activities were measured within V. anguillarum (Figure 8D). By contrast, PfrpApdp did not show any activity within V. anguillarum, which greatly suggests that the short version of pbtB-frpA intergenic sequence present in irp-HPIpdp does not contain a transcriptional promoter (Figure 8A).
Horizontally acquired DNA usually encodes transcriptional regulators that promote their own expression (Stoebel et al., 2008). To ascertain the regulatory mechanism that controls the expression of irp-HPI genes, we constructed and analyzed V. anguillarum mutants defective in the two irp-HPI encoded AraC-like regulators PbtA and PbtB, and in the global regulators H-NS and ToxR-S. Inactivation of pbtA (araC1) disables piscibactin biosynthesis and transport, greatly reducing growing capacities under low iron availability. It also resulted in a marked decrease on the degree of virulence, confirming the relevance of piscibactin synthesis in the pathogenesis of V. anguillarum (Balado et al., 2018). By contrast, neither phenotypic nor piscibactin gene expression changes were found by the inactivation of pbtB (araC2). Generally, the regulation of iron uptake systems in Gram-negative bacteria is mediated by the negative transcriptional regulator Fur (ferric uptake regulator) (Fillat, 2014). Conversely, some AraC-like transcriptional regulators have been identified as exerting positive regulation in the expression of the siderophore biosynthetic and transport genes such as AlcR in Bordetella pertussis that upregulates the expression of alcaligin siderophore biosynthesis and transport (Brickman et al., 2001), the ferrioxamine B transport regulator DesR in V. vulnificus (Tanabe et al., 2005), or PchR in Pseudomonas aeruginosa, which is required for maximal expression of the pyochelin biosynthetic and transport genes (Michel et al., 2005).
AraC-like regulators can act as autoinducers by inducing its own expression, and/or activating the expression of other promoters in trans (Gallegos et al., 1997; Galperin, 2006). The piscibactin system is transcribed within a large operon whose transcription could start from alternative promoters located upstream of pbtA (araC1) and frpA (Figure 1; Balado et al., 2018). The piscibactin siderophore system is structurally and functionally related to the yersiniabactin system (Osorio et al., 2006; Souto et al., 2012). In Yersinia pestis and E. coli, the AraC-like protein named YbtA, encoded in the yersiniabactin gene cluster, activates the expression of yersiniabactin genes, including its own promoter (Fetherston et al., 1996; Anisimov et al., 2005); however, the AraC-like PbtA described here does not enhance the expression of its own promoter (PpbtA). Although deletion of ybtA blocks yersiniabactin production, the ybtA mutant has a virulence phenotype intermediate between the wild type and a yersiniabactin biosynthesis null mutant (Smati et al., 2017). Our results clearly show that inactivation of PbtA in V. anguillarum results in an attenuation of the degree of virulence equivalent to that observed in a V. anguillarum strain unable to produce siderophores (RV22ΔvabFΔirp1). This finding, together with the absence of FrpA in the outer membrane of the V. anguillarum pbtA– strain and the extremely low levels of transcriptional activity found in PfrpA promoter, strongly suggests that (1) PbtA is a transcriptional activator required to express the transport and biosynthesis components of the piscibactin siderophore system, (2) effective expression of piscibactin functions only occurs from frpA promoter (PfrpA), and (3) the piscibactin regulator PbtA plays a key role in V. anguillarum virulence.
The AraC-like regulators can be part of a larger regulon that can act in a cascade manner as a response to environmental signals. ToxR is a regulatory protein that is essential for virulence in a range of different pathogenic vibrios (Herrington et al., 1988; Peterson and Mekalanos, 1988; Lee et al., 2000; Whitaker et al., 2012). In Vibrio cholerae, the ToxR regulon is responsible for the transcriptional activation of the AraC-like regulator ToxT, which responds to environmental signals to directly activate many virulence-related genes (Higgins and DiRita, 1994; Skorupski and Taylor, 1997; Childers and Klose, 2007). However, some works suggest that in V. anguillarum, ToxR is not a major regulator of virulence factors (Okuda et al., 2001; Wang et al., 2002). Our results showed that deletion of the global regulator ToxR-S system in V. anguillarum did not cause appreciable changes in growth ability under iron restriction nor apparently had effects on siderophore production at cold temperatures. Although ToxR-S could have an indirect effect in piscibactin genes expression at warm temperatures, irp-HPI would not be part of the ToxR regulon.
Gram-negative bacteria possess tools to silence the expression of horizontally acquired genes such as the global repressor H-NS, which is implicated in xenogenetic silencing (Stoebel et al., 2008; Prajapat and Saini, 2012). The V. anguillarum H-NS mutant strain showed a slight reduction in growth potential under low-iron conditions, which is the result of a lower activity of the piscibactin promoter PfrpA at both cold and warm temperatures (15 and 25°C). Thus, our results suggest that H-NS must exert an indirect effect on the expression of piscibactin biosynthetic and transport genes. However, the transcriptional activity of PpbtA promoter does not change when H-NS is inactivated. Therefore, H-NS would not have any direct or indirect regulatory effect on the expression of the piscibactin transcriptional activator PbtA. Although H-NS can also mediate processes of temperature-dependent expression regulation by upregulating target genes when temperature increases (Mou et al., 2013), this type of regulatory effect was not observed in our data. Thus, although H-NS would not mediate in the temperature-dependent modulation of piscibactin genes, it is required to achieve the maximum activity of piscibactin promoter PfrpA (Figure 5). Consequently, and since our previous works demonstrated that piscibactin has a great impact in V. anguillarum virulence (Balado et al., 2018), inactivation of h-ns results in a significant reduction of V. anguillarum virulence for fish (Figure 4).
The role of evolutionary dynamics in bacterial disease is not well understood, but it is known that intensive aquaculture selects for increased virulence, which may trigger the emergence of novel diseases (Pulkkinen et al., 2010; Sundberg et al., 2016). In addition, the rapid spread of selectively favored virulence factors by horizontal gene transfer (HGT) facilitates the emergence of new bacterial diseases (Bruto et al., 2017; Le Roux and Blokesch, 2018). However, horizontally transferred genes are not always expressed in the recipient genome, because of possible incompatibilities in promoter sequences, different codon usages, and/or excessive energy cost (Ochman et al., 2000; Park and Zhang, 2012). They must be subjected to the precise regulatory control by the recipient’s genome that allows genes to be expressed under the control of specific signals (Stoebel et al., 2008). The piscibactin high pathogenicity island (irp-HPI) is widespread among Vibrionaceae including the human pathogen V. cholerae (Thode et al., 2018). Phylogenetic analysis clearly shows that several irp-HPI lineages exist and that extensive sequence differences are present in the intergenic regions where piscibactin promoters PpbtA and PfrpA are found. The piscibactin siderophore system was firstly identified in the marine pathogen P. damselae subsp. piscicida and it is also a major virulence factor in this bacterium (Osorio et al., 2006, 2015). However, the expression pattern of irp-HPI genes described in V. anguillarum is almost incompatible with the ecology of P. damselae subsp. piscicida, which mainly cause disease outbreaks when water temperature is above 20°C (Romalde, 2002; Toranzo et al., 2005; Bellos et al., 2015). Notably, P. damselae subsp. piscicida irp-HPI genomic island (irp-HPIpdp) lacks a promoter in frpA. Thereby, all piscibactin genes must be necessarily expressed from the pbtA promoter (PpbtA) in a large operon that includes a regulator, and biosynthetic and transport functions (Figure 1; Osorio et al., 2006). Interestingly, irp-HPIpdp did not follow a temperature-dependent expression pattern in P. damselae subsp. piscicida and the transcriptional activity is significantly lower than in V. anguillarum, but its expression is activated at low temperatures when it is inserted in V. anguillarum. Conversely, the transcriptional activity of V. anguillarum promoters PpbtA and PfrpA do not respond to temperature changes when they are inserted in P. damselae subsp. piscicida. Thus, irp-HPI temperature-dependent expression pattern would be modulated by yet unknown activator(s) present in the recipient genome, which would enhance transcription of PpbtA when temperature decreases, rather than by an inherent property of the irp-HPI genomic island.
As a conclusion, the temperature-dependent expression pattern of the piscibactin-encoding high-pathogenicity island irp-HPI described in V. anguillarum is the result of the combined effect of the AraC-like transcriptional activator PbtA encoded by this genomic island and regulatory factor(s) encoded by the recipient bacterial genome. Thus, the horizontally acquired piscibactin genes would have been subjected to global cell control, maximizing bacterial fitness advantages (Stoebel et al., 2008). Notably, the existence of different expression patterns within different irp-HPI evolutionary lineages supports the hypothesis of the long-term evolution of the irp-HPI genomic islands within the Vibrionaceae (Thode et al., 2018). The mechanism that modulates piscibactin gene expression through PbtA and/or the putative regulatory effect(s) exerted by PbtA outside the genomic island could result in global modulation of virulence factors in response to temperature changes. Thus, further studies focused on the PbtA regulation mechanism will be highly valuable to decipher the adaptation of V. anguillarum and other members of Vibrionaceae to environmental temperature changes.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Bioethics Committee of the University of Santiago de Compostela, Spain.
MAL and MB performed the lab experiments, analyzed the data, and wrote the first draft of the manuscript. MB and MLL corrected the draft and built the final version of the manuscript. All authors conceived and designed the study, contributed to manuscript revision, and read and approved the submitted version.
This work was supported by grants PID2019-103891RJ-100 (AEI) to MB and RTI2018-093634-B-C21 (AEI/FEDER, EU) to MLL, from the State Agency for Research (AEI) of Spain, and by grant GRC2018/018 from Xunta de Galicia, Spain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.748147/full#supplementary-material
Anisimov, R., Brem, D., Heesemann, J., and Rakin, A. (2005). Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int. J. Med. Microbiol. 295, 19–28. doi: 10.1016/j.ijmm.2004.11.007
Austin, B., and Austin, D. A. (2007). “Epizootiology: gram-negative bacteria,” in Bacterial fish Pathogens. Springer Praxis Books, (Dordrecht: Springer). doi: 10.1007/978-1-4020-6069-4_8
Balado, M., Lages, M. A., Fuentes-Monteverde, J. C., Martínez-Matamoros, D., Rodríguez, J., Jiménez, C., et al. (2018). The siderophore piscibactin is a relevant virulence factor for Vibrio anguillarum favored at low temperatures. Front. Microbiol. 9:1766. doi: 10.3389/fmicb.2018.01766
Balado, M., Osorio, C. R., and Lemos, M. L. (2006). A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 152, 3517–3528. doi: 10.1099/mic.0.29298-0
Bellos, G., Angelidis, P., and Miliou, H. (2015). Effect of temperature and seasonality principal epizootiological risk factor on vibriosis and photobacteriosis outbreaks for European sea bass in Greece (1998-2013). J. Aquac. Res. Dev. 6:338. doi: 10.4172/2155-9546.1000338
Brickman, T. J., Kang, H. Y., and Armstrong, S. K. (2001). Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183, 483–489. doi: 10.1128/JB.183.2.483-489.2001
Bruto, M., James, A., Petton, B., Labreuche, Y., Chenivesse, S., Alunno-Bruscia, M., et al. (2017). Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052. doi: 10.1038/ismej.2016.162
Childers, B. M., and Klose, K. E. (2007). Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol. 2, 335–344. doi: 10.2217/17460913.2.3.335
Crisafi, F., Denaro, R., Genovese, M., Yakimov, M., and Genovese, L. (2014). Application of relative real-time PCR to detect differential expression of virulence genes in Vibrio anguillarum under standard and stressed growth conditions. J. Fish Dis. 37, 629–640. doi: 10.1111/jfd.12158
Denkin, S. M., and Nelson, D. R. (1999). Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65, 3555–3560. doi: 10.1128/AEM.65.8.3555-3560.1999
Fetherston, J. D., Bearden, S. W., and Perry, R. D. (1996). YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22, 315–325. doi: 10.1046/j.1365-2958.1996.00118.x
Fillat, M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52. doi: 10.1016/j.abb.2014.01.029
Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997). AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410. doi: 10.1128/mmbr.61.4.393-410.1997
Galperin, M. Y. (2006). Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182. doi: 10.1128/JB.01887-05
Herrero, M., de Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K., and Levine, M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492. doi: 10.1084/jem.168.4.1487
Higgins, D. E., and DiRita, V. J. (1994). Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol. Microbiol. 14, 17–29. doi: 10.1111/J.1365-2958.1994.TB01263.X
Kramer, J., and Özkaya, Ö, and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lages, M. A., Balado, M., and Lemos, M. L. (2019). The expression of virulence factors in Vibrio anguillarum is dually regulated by iron levels and temperature. Front. Microbiol. 10:2335. doi: 10.3389/FMICB.2019.02335
Le Roux, F., and Blokesch, M. (2018). Eco-evolutionary dynamics linked to horizontal gene transfer in Vibrios. Annu. Rev. Microbiol. 72, 89–110. doi: 10.1146/annurev-micro-090817-062148
Le Roux, F., Wegner, K. M., Baker-Austin, C., Vezzulli, L., Osorio, C. R., Amaro, C., et al. (2015). The emergence of Vibrio pathogens in Europe: ecology, evolution, and pathogenesis (Paris, 11–12th March 2015). Front. Microbiol. 6:830. doi: 10.3389/fmicb.2015.00830
Lee, S. E., Shin, S. H., Kim, S. Y., Kim, Y. R., Shin, D. H., Chung, S. S., et al. (2000). Vibrio vulnificus has the transmembrane transcription activator toxRS stimulating the expression of the hemolysin gene vvhA. J. Bacteriol. 182, 3405–3415. doi: 10.1128/jb.182.12.3405-3415.2000
Li, Y., and Ma, Q. (2017). Iron acquisition strategies of Vibrio anguillarum. Front. Cell. Infect. Microbiol. 7:342. doi: 10.3389/fcimb.2017.00342
Ma, Y., Wang, Q., Gao, X., and Zhang, Y. (2017). Biosynthesis and uptake of glycine betaine as cold-stress response to low temperature in fish pathogen Vibrio anguillarum. J. Microbiol. 55, 44–55. doi: 10.1007/s12275-017-6370-2
Maeda, T., Matsuo, Y., Furushita, M., and Shiba, T. (2003). Seasonal dynamics in a coastal Vibrio community examinedby a rapid clustering method based on 16S rDNA. Fish. Sci. 69, 385–394. doi: 10.1046/j.1444-2906.2003.00633.x
Martínez-García, E., Goñi-Moreno, A., Bartley, B., McLaughlin, J., Sánchez-Sampedro, L., Del Pozo, H. P., et al. (2020). SEVA 3.0: an update of the standard European vector architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Res. 48, D1164–D1170. doi: 10.1093/nar/gkz1024
Michel, L., González, N., Jagdeep, S., Nguyen-Ngoc, T., and Reimmann, C. (2005). PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58, 495–509. doi: 10.1111/j.1365-2958.2005.04837.x
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/MMBR.00012-07
Miller, J. H. (1992). A short Course in Bacterial Genetics – A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press.
Mou, X., Spinard, E. J., Driscoll, M. V., Zhao, W., and Nelson, D. R. (2013). H-NS is a negative regulator of the two hemolysin/cytotoxin gene clusters in Vibrio anguillarum. Infect. Immun. 81, 3566–3576. doi: 10.1128/IAI.00506-13
Mouriño, S., Osorio, C. R., and Lemos, M. L. (2004). Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186, 6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004
Ochman, H., Lawrence, J. G., and Grolsman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. doi: 10.1038/35012500
Okuda, J., Nakai, T., Chang, P. S., Oh, T., Nishino, T., Koitabashi, T., et al. (2001). The toxR gene of Vibrio (Listonella) anguillarum controls expression of the major outer membrane proteins but not virulence in a natural host model. Infect. Immun. 69, 6091–6101. doi: 10.1128/IAI.69.10.6091-6101.2001
Osorio, C. R., Juiz-Rio, S., and Lemos, M. L. (2006). A siderophore biosynthesis gene cluster from the fish pathogen Photobacterium damselae subsp. piscicida is structurally and functionally related to the Yersinia high-pathogenicity island. Microbiology 152, 3327–3341. doi: 10.1099/mic.0.29190-0
Osorio, C. R., Rivas, A. J., Balado, M., Fuentes-Monteverde, J. C., Rodríguez, J., Jiménez, C., et al. (2015). A transmissible plasmid-borne pathogenicity island encodes piscibactin biosynthesis in the fish pathogen Photobacterium damselae subsp. piscicida. Appl. Environ. Microbiol. 81, 5867–5879. doi: 10.1128/AEM.01580-15
Parales, R. E., and Harwood, C. S. (1993). Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram-bacteria. Gene 133, 23–30. doi: 10.1016/0378-1119(93)90220-w
Park, C., and Zhang, J. (2012). High expression hampers horizontal gene transfer. Genome Biol. Evol. 4, 523–532. doi: 10.1093/gbe/evs030
Peterson, K. M., and Mekalanos, J. J. (1988). Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun 56, 2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988
Prajapat, M. K., and Saini, S. (2012). Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium. Comput. Biol. Med. 42, 1133–1140. doi: 10.1016/j.compbiomed.2012.09.005
Pulkkinen, K., Suomalainen, L.-R., Read, A. F., Ebert, D., Rintamäki, P., and Valtonen, E. T. (2010). Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc. R. Soc. B Biol. Sci. 277, 593–600. doi: 10.1098/rspb.2009.1659
Rodkhum, C., Hirono, I., Stork, M., Di Lorenzo, M., Crosa, J. H., and Aoki, T. (2006). Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. J. Fish Dis. 29, 157–166.
Romalde, J. L. (2002). Photobacterium damselae subsp. piscicida: an integrated view of a bacterial fish pathogen. Int. Microbiol. 5, 3–9. doi: 10.1007/s10123-002-0051-6
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Skorupski, K., and Taylor, R. K. (1997). Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25, 1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x
Smati, M., Magistro, G., Adiba, S., Wieser, A., Picard, B., Schubert, S., et al. (2017). Strain-specific impact of the high-pathogenicity island on virulence in extra-intestinal pathogenic Escherichia coli. Int. J. Med. Microbiol. 307, 44–56. doi: 10.1016/j.ijmm.2016.11.004
Souto, A., Montaos, M. A. M. A., Rivas, A. J. A. J., Balado, M., Osorio, C. R., Rodríguez, J., et al. (2012). Structure and biosynthetic assembly of piscibactin, a siderophore from Photobacterium damselae subsp. piscicida, predicted from genome analysis. Eur. J. Org. Chem. 2012, 5693–5700. doi: 10.1002/ejoc.201200818
Stoebel, D. M., Free, A., and Dorman, C. J. (2008). Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154, 2533–2545. doi: 10.1099/mic.0.2008/020693-0
Sundberg, L.-R., Ketola, T., Laanto, E., Kinnula, H., Bamford, J. K. H., Penttinen, R., et al. (2016). Intensive aquaculture selects for increased virulence and interference competition in bacteria. Proc. R. Soc. B Biol. Sci. 283:20153069. doi: 10.1098/rspb.2015.3069
Tanabe, T., Takata, N., Naka, A., Moon, Y. H., Nakao, H., Inoue, Y., et al. (2005). Identification of an AraC-like regulator gene required for induction of the 78-kDa ferrioxamine B receptor in Vibrio vulnificus. FEMS Microbiol. Lett. 249, 309–314. doi: 10.1016/j.femsle.2005.06.025
Thode, S. K., Rojek, E., Kozlowski, M., Ahmad, R., and Haugen, P. (2018). Distribution of siderophore gene systems on a Vibrionaceae phylogeny: database searches, phylogenetic analyses and evolutionary perspectives. PLoS One 13:e0191860. doi: 10.1371/journal.pone.0191860
Toranzo, A. E., Barreiro, S., Casal, J. F., Figueras, A., Magariños, B., and Barja, J. L. (1991). Pasteurellosis in cultured gilthead seabream (Sparus aurata): first report in Spain. Aquaculture 99, 1–15. doi: 10.1016/0044-8486(91)90284-E
Toranzo, A. E., Magariños, B., and Avendaño-Herrera, R. (2017). “Vibriosis: Vibrio anguillarum, V. ordalii and Aliivibrio salmonicida,” in Fish Viruses and Bacteria: Pathobiology and Protection, eds P. T. K. Woo and R. C. Cipriano (Wallingford: CABI), 314–333. doi: 10.1079/9781780647784.0314
Toranzo, A. E., Magariños, B., and Romalde, J. L. (2005). A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246, 37–61. doi: 10.1016/j.aquaculture.2005.01.002
Wang, R. F., and Kushner, S. R. (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199. doi: 10.1016/0378-1119(91)90366-j
Wang, S.-Y., Lauritz, J., Jass, J., and Milton, D. L. (2002). A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J. Bacteriol. 184, 1630–1639. doi: 10.1128/JB.184.6.1630-1639.2002
Keywords: fish pathogens, virulence factors, piscibactin, high pathogenicity island, aquaculture, Vibrio anguillarum, horizontal gene transfer
Citation: Lages MA, Lemos ML and Balado M (2021) The Temperature-Dependent Expression of the High-Pathogenicity Island Encoding Piscibactin in Vibrionaceae Results From the Combined Effect of the AraC-Like Transcriptional Activator PbtA and Regulatory Factors From the Recipient Genome. Front. Microbiol. 12:748147. doi: 10.3389/fmicb.2021.748147
Received: 27 July 2021; Accepted: 20 October 2021;
Published: 19 November 2021.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Michele Kiyoko Nishiguchi, University of California, Merced, United StatesCopyright © 2021 Lages, Lemos and Balado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel L. Lemos, bWFudWVsLmxlbW9zQHVzYy5lcw==; Miguel Balado, bWlndWVsLmJhbGFkb0B1c2MuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.