- 1Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece
- 2Institute of Technology of Agricultural Products, Hellenic Agricultural Organization DIMITRA, Athens, Greece
Lactobacillus is a diverse genus that includes species of industrial and biomedical interest. Lactiplantibacillus pentosus, formerly known as Lactobacillus pentosus, is a recently reclassified species, that contains strains isolated from diverse environmental niches, ranging from fermented products to mammalian gut microbiota. Importantly, several L. pentosus strains present health-promoting properties, such as immunomodulatory and antiproliferative activities, and are regarded as potential probiotic strains. In this study, we present the draft genome sequence of the potential probiotic strain L. pentosus L33, originally isolated from fermented sausages. Comprehensive bioinformatic analysis and whole-genome annotation were performed to highlight the genetic loci involved in host-microbe interactions and the probiotic phenotype. Consequently, we found that this strain codes for bile salt hydrolases, adhesins and moonlighting proteins, and for Class IIb bacteriocin peptides lacking the GxxxG and GxxxG-like motifs, crucial for their inhibitory activity. Its adhesion ability was also validated in vitro, on human cancer cells. Furthermore, L. pentosus L33 contains an exopolysaccharide (EPS) biosynthesis cluster, and it does not carry transferable antibiotic resistance genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and CAZymes analyses showed that L. pentosus L33 possesses biosynthetic pathways for seven amino acids, while it can degrade a wide array of carbohydrates. In parallel, Clusters of Orthologous Groups (COGs) and KEGG profiles of L. pentosus L33 are similar to those of 26 L. pentosus strains, as well as of two well documented L. plantarum probiotic strains. Conclusively, L. pentosus L33 exhibits good probiotic potential, although further studies are needed to elucidate the extent of its biological properties.
Introduction
Lactobacillus is a diverse genus that includes Gram-positive, facultatively anaerobic, non-spore-forming, hetero-, or homo-fermentative bacteria that inhabit a broad range of nutrient-rich environmental niches (Duar et al., 2017). The species of this genus have been recently reclassified to 25 genera, based on shared ecological and metabolic properties (Zheng et al., 2020). Lactobacillus strains can be found as autochthonous or allochthonous, mainly in the mammalian gastrointestinal tract, fresh fruit and vegetable microbiota, as well as in fermented foodstuffs (Inglin et al., 2018). In this context, several strains exhibit great biotechnological interest, due to their fermentation capacity and are being incorporated as starter cultures in a broad range of dairy and non-dairy products (Kok and Hutkins, 2018). Furthermore, specific strains are considered probiotic, meaning that they can confer health benefits to the host, when consumed in adequate quantities (FAO/WHO, 2002). Regarding the proposed positive impact of probiotics on host health, preclinical and clinical studies have shown that they can exhibit antimicrobial (Yu et al., 2015), immunomodulatory (Chondrou et al., 2020), antioxidant (Wu et al., 2019), antiproliferative (Tiptiri-Kourpeti et al., 2016), and even psychobiotic activity (Tian et al., 2020). Today, probiotics are commercially available in supplements or in functional products, comprising a rapidly growing global market, currently worth more than $50 billion, as market reports indicated.1
The commercialization of probiotic strains is strictly monitored. Indeed, several guidelines have been set in place for the characterization of novel probiotic strains by the World Health Organization (WHO), the Food and Agriculture Organization of the United Nations (FAO), and the European Food Safety Authority (EFSA) (FAO/WHO, 2002; EFSA, 2018). First, new isolates should be molecularly assigned to a specific taxonomic group. EFSA also requires full genome sequencing and annotation of strains that are intended for biotechnological applications (EFSA, 2018). Importantly, probiotics must be safe for consumption; they should not exhibit hemolytic or virulence activity. Consequently, they should be characterized by either the Food and Drug Administration (FDA) or EFSA with the “Generally Recognized as Safe” (GRAS) or of “Qualified Presumption of Safety” status, respectively (Rodrigo-Torres et al., 2019). Furthermore, probiotic microorganisms should be able to tolerate the gastrointestinal tract conditions; be resistant to low pH, gastric enzymes, and bile acids and, also, adhere to and, at least transiently, colonize the gastrointestinal mucosa (Hill et al., 2014). The proposed health effects of new isolates should be thoroughly explored in vitro and in vivo, to finally be validated in the clinical setting (FAO/WHO, 2002). Mechanistic studies on host-probiotic interactions have flourished recently, with the advent of multi-omics technologies, facilitating a better understanding of their properties and biological functions (Kiousi et al., 2021).
The introduction of genomics in the microbiology field has restructured the characterization of novel Lactobacillus strains as probiotics. As next-generation sequencing platforms are becoming increasingly accessible, the taxonomic and functional characterization of new isolates can be performed with greater accuracy. One of the species that has been reclassified recently is Lactiplantibacillus pentosus, formerly known as Lactobacillus pentosus (Zheng et al., 2020). The bacteria of this species are mainly associated with environmental samples, such as fruit and vegetable microbiota, however, several strains harbor genes for mammalian host adaptation (Abriouel et al., 2017). Genome mining in L. pentosus strains and comparative genomic analysis with the closely related L. plantarum species have revealed functional characteristics involved in the probiotic phenotype, such as the presence of genes involved in stress response (Ye et al., 2020), metabolic capacity (Abriouel et al., 2017), adhesion on the intestinal mucosa and bacteriocin production (Maldonado-Barragán et al., 2011).
L. pentosus L33 is a LAB (Lactic Acid Bacteria) strain, with desirable probiotic properties, as previously demonstrated in a series of established in vitro tests (Pavli et al., 2016). The aim of this study was to further investigate the probiotic potential of the strain by characterizing the genetic basis of the probiotic phenotype. Firstly, whole-genome sequencing was performed to reveal the genomic characteristics of the strain. Then, genome annotation and comparative genomic analysis with other L. pentosus, as well as, L. plantarum genome sequences were executed to detect strain-specific genes and pinpoint genes of interest. More specifically, the presence of gene clusters involved in the biosynthesis of bacteriocins, adhesion proteins and exopolysaccharides were investigated. Lastly, KEGG pathway and CAZymes analyses were performed to evaluate the metabolic capabilities of L. pentosus L33.
Materials and Methods
Bacterial Strain, Culture Conditions, and DNA Isolation
L. pentosus L33 was originally isolated from fermented sausages (Pavli et al., 2016), and was acquired by the Institute of Technology of Agricultural Products, Hellenic Agricultural Organization DIMITRA (Athens, Greece). It was maintained in de Man, Rogosa, and Sharpe (MRS) broth (Condalab, Madrid, Spain) at 37°C for 16–18 h under anaerobic conditions, prior to DNA extraction. Bacterial cells were collected by centrifugation at 8,000 g for 4 min. Total genomic DNA was extracted from the cell pellets using the NucleoSpin® Tissue kit (Macherey-Nagel, Düren, Germany), according to manufacturer’s instructions. DNA purity and quantity were confirmed spectrophotometrically at 260 nm using NanoDrop® ND-1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Whole-Genome Sequencing and Genome Annotation
The genomic DNA of L. pentosus L33 was sequenced using Illumina NovaSeq6000 (2 × 151 paired ends) platform. A total of 8,806,648 paired-end reads were obtained. The quality of the reads was estimated using FASTQC (version 0.11.9) (Andrews, 2010), while low-quality reads were removed via Trimmomatic (version 0.39) (Bolger et al., 2014). De novo assembly process was executed with SPAdes (version 3.15.1) (Bankevich et al., 2012), selecting the “–careful” option to reduce mismatches and SSPACE_Standard (version 3.0) (Boetzer et al., 2011) with the parameter to filter out contigs with length below 500 base pairs.
Genome annotation was carried out locally, using the Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016) algorithm with default parameters. EggNOG-mapper (version 2.0) tool from the online EggNOG database (version 5.0) (Huerta-Cepas et al., 2019) was used for functional classification of proteins into COGs. BlastKOALA (version 2.2) was utilized for Kyoto Encyclopedia of Genes and Genomes Orthology (KO) assignment and KEGG mapping of the predicted genes (Kanehisa et al., 2016). Carbohydrate-active enzymes (CAZymes) were searched against the CAZy database (Lombard et al., 2014). Clustered regularly interspaced palindromic repeats (CRISPR) inside the assembly were evaluated using CRISPRDetect (version 2.4) (Biswas et al., 2016). PHAge Search Tool Enhanced Release (PHASTER) (Arndt et al., 2016) was utilized for identification and annotation of putative prophage sequences inside the bacterial assembly. Visualization of the genome assembly was performed by Artemis tool (version 18.1.0) (Carver et al., 2012), while its metrics were calculated with the Quality Assessment Tool (QUAST) (version 5.2.0) (Gurevich et al., 2013).
Phylogenetic and Comparative Analysis
Average Nucleotide Identity (ANI) analysis was performed on the complete genome assembly, using a python module called Pyani (version 0.2.10) (Pritchard et al., 2016), to verify the taxonomic identity of L. pentosus L33. Pangenome analysis of the available L. pentosus strains (May 2021), was operated by Roary (version 3.13.0) (Page et al., 2015). The phylogenomic analysis, including 1,000 bootstrap replicates (Maximum Composite Likelihood model), was performed by MEGAX (version 10.1.8) (Kumar et al., 2018). The publicly available online EMBL tool called “Interactive Tree of Life” (iTol) (version 6.1.1) (Letunic and Bork, 2016) was used for phylogenetic tree construction. Strain-specific genes were determined via an in-house Python script.
Detection of Genetic Elements Associated With Probiotic Characteristics
BAGEL (version 4) (de Jong et al., 2006) was employed for detection and visualization of gene clusters that are involved in bacteriocin biosynthesis. The presence of antibiotic resistance genes was verified by Resistance Gene Identifier (RGI) (version 5.1.1) (Jia et al., 2017). BLAST (basic local alignment search tool) was used for the search of genes that are involved in EPS production, bile salt hydrolysis and cell adhesion.
Quantitative Adhesion Assay
The assay was performed as described before, with minor modifications (Plessas et al., 2020). Briefly, human colon adenocarcinoma HT-29 cells were seeded in 24-well plates at a density of 40 × 104 cells per well and incubated for 14 days to form a monolayer. The cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium enriched with GlutaMAXTM, 10% fetal bovine serum (FBS), 100 μg/mL streptomycin and 100 U/mL penicillin (Thermo Fisher Scientific, Waltham, MA, United States) and incubated at 37°C, 5% CO2 in a humidified atmosphere. 107 or 108 CFU/mL of viable L. pentosus L33 or L. rhamnosus GG cells were added to each well. After 4 h of co-incubation at 37°C, the cells were washed with PBS and lysed with 1% Triton X-100 (Sigma-Aldrich, Taufkirchen, Germany). The lysates were serially diluted in Ringer’s solution (Lab M, Lancashire, United Kingdom), plated on 2% MRS agar, and incubated at 37°C, until the formation of visible colonies. For the calculation of adhesion values the following formula was applied: % Adhesion = (VB/VA) × 100, where VA is the initial viable count of bacteria tested, and VB is the viable bacteria count attached on HT-29 cells. Colony forming units per milliliter (CFU/mL) was used as viable count measure that was determined using the formula: CFU/mL = (number of colonies × dilution factor)/volume of culture plate.
Results
Genome Features
Whole-genome sequencing and comprehensive bioinformatic analysis were employed for the investigation of the genomic features of L. pentosus L33 (Table 1), ultimately leading to the construction of its genome map (Figure 1). The complete genome of L. pentosus L33 has a length of 3,923,201 bp with a GC content of 46.01%. Among the 3,630 predicted genes, 3,429 were found to be protein-coding sequences (CDS). Furthermore, 127 pseudogenes, 58 tRNAs, 6 rRNAs, and 5 ncRNAs were identified. The 58 tRNA encoding sequences correspond to all 20 amino acids (Supplementary Table 1). In addition, 3 clustered regularly interspaced short palindromic repeats (CRISPR) arrays (Supplementary Table 2), as well as 4 intact prophage regions (Supplementary Table 3) were recognized.
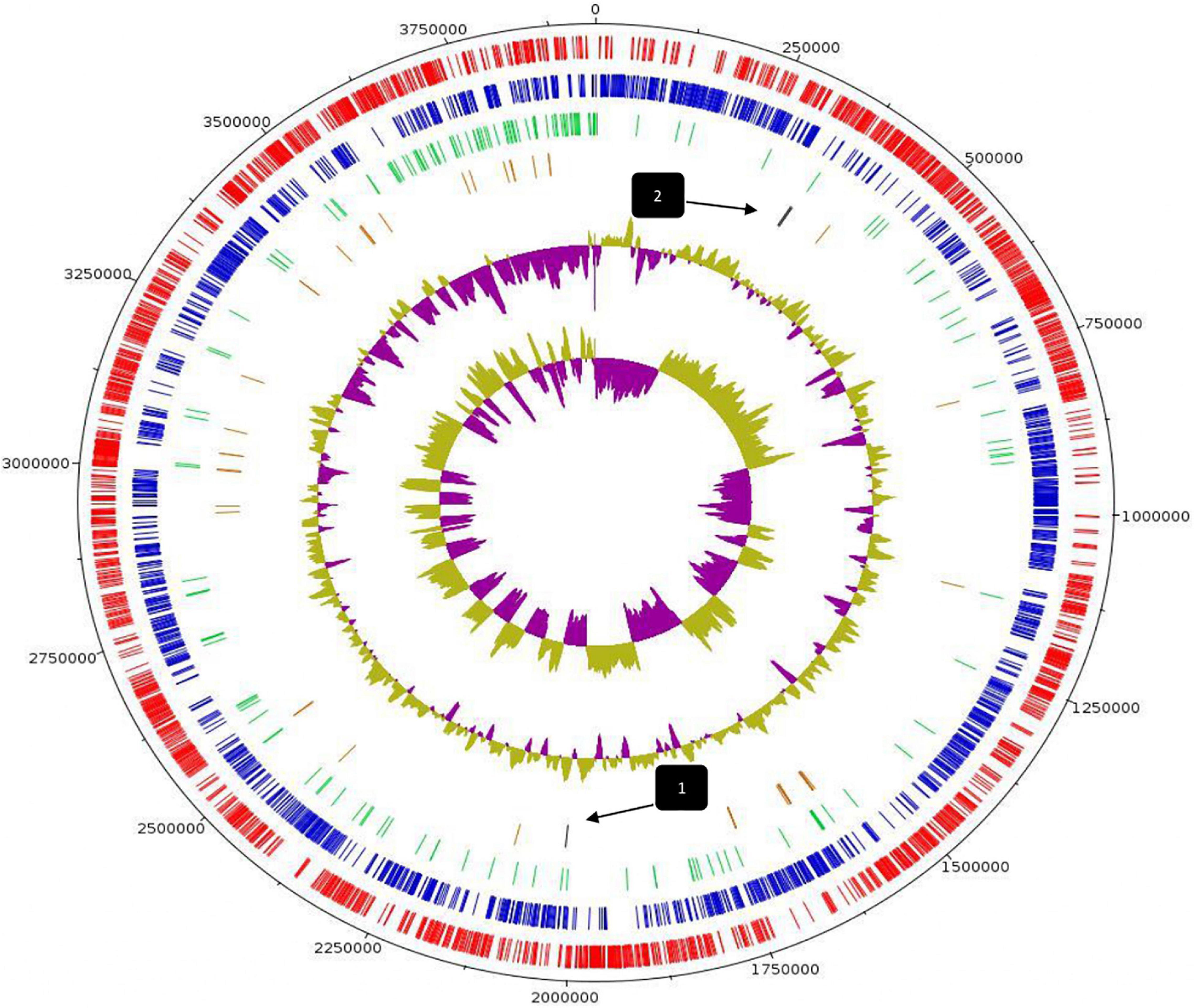
Figure 1. Circular genome map of L. pentosus L33. From outer circle to inner, information is displayed as follows: Genome Size (black), Forward strand CDS (red), Reverse strand CDS (blue), Pseudogenes (green), tRNA genes (brown), GC Content, GC Skew. Arrows indicate the position of CRISPR arrays.
Phylogenetic Analysis and Unique Genome Characteristics of Lactiplantibacillus pentosus L33
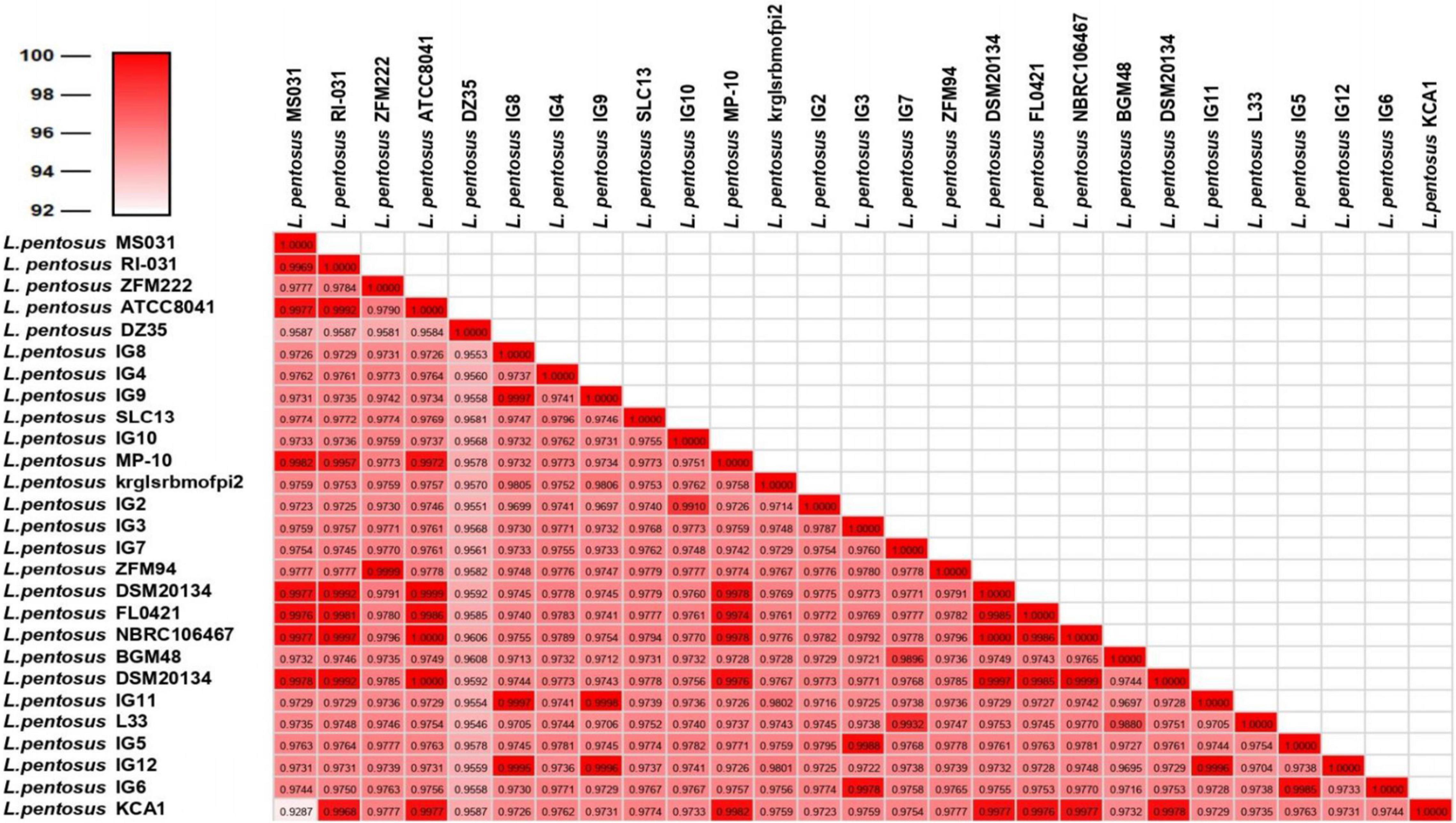
For the characterization of strain L33, sequencing of the V1-V3 region of 16S rRNA gene, followed by multiplex PCR targeting the recA gene was performed. Strain L33 was assigned to the species of Lactobacillus pentosus (Pavli et al., 2016), currently known as Lactiplantibacillus pentosus (Zheng et al., 2020). A neighbor-joining phylogenetic tree, including 1,000 bootstrap replications, based on orthologous gene clusters, was built to reveal the exact phylogenetic position of L. pentosus L33 within L. pentosus species (Figure 2). Moreover, we have used 26 L. pentosus strains, 2 well documented L. plantarum probiotic strains; L. plantarum WCFS1 (van den Nieuwboer et al., 2016) and L. plantarum 299v (Nordström et al., 2021), as well as Staphylococcus aureus NCTC8325 and Streptococcus pneumoniae NCTC11032, as controls (Supplementary Figure 1). The closest evolutionary relatives of L. pentosus L33 are L. pentosus IG7, which was isolated from the brine of natural Spanish-style green olive fermentation (Calero-Delgado et al., 2018), and L. pentosus BGM48, originated from laboratory scale Sicilian-style green olive fermentation (Golomb et al., 2013; Figure 2). Furthermore, when compared to L. pentosus L33, ANI analysis found that L. pentosus IG7 and L. pentosus BGM48 exhibit the greatest ANI scores, with 99.3 and 98.8%, respectively. The full ANI matrix, including all genomes, is presented in Figure 3. Additionally, L. pentosus L33, when comparing to 26 L. pentosus analyzed genomes, has 243 genes (6.60%) that were found to be strain-specific. The proteins encoded by unique genes were classified into COG functional categories (Figure 4). A total of 190 (78.18%) unique proteins were assigned to 18 COG functional categories. The majority (96 proteins) were categorized as “poorly characterized.”
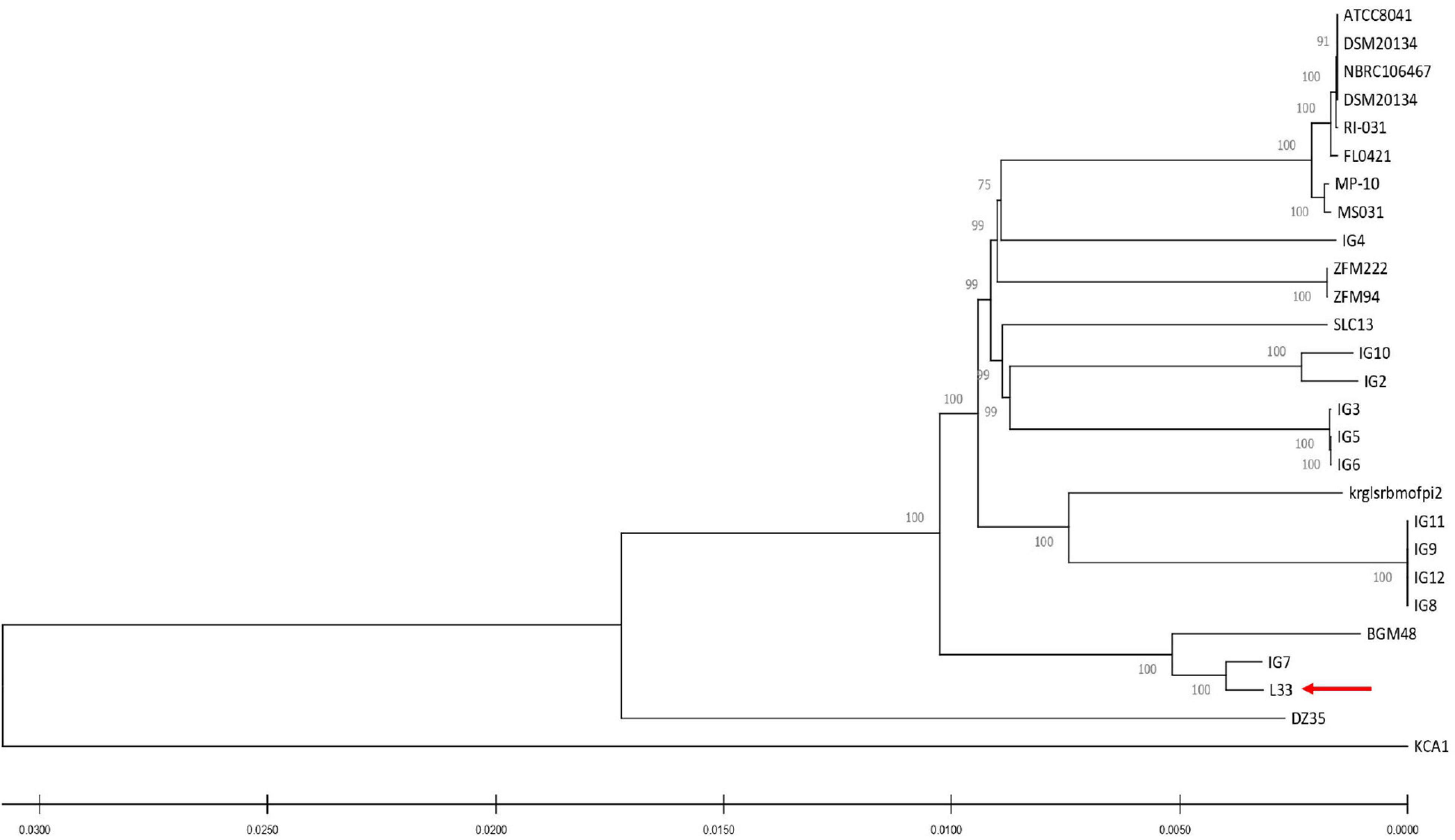
Figure 2. Neighbor-joining phylogenetic tree of L. pentosus L33 and 26 L. pentosus strains based on orthologous genes found by Roary (version 3.13.0). Values of 1,000 bootstrap replicates calculated by MEGAX (version 10.1.8) are depicted.
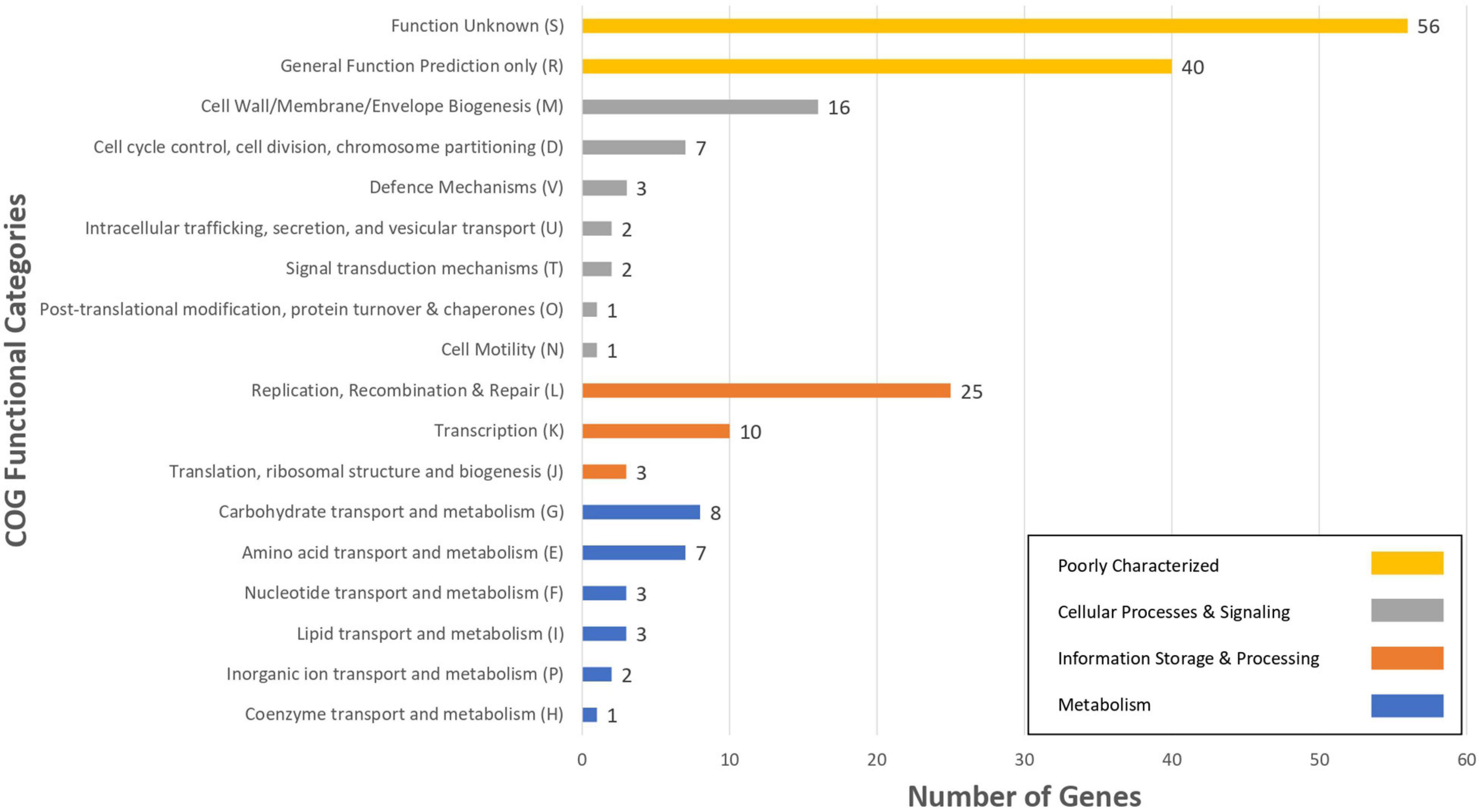
Figure 4. The number of L. pentosus L33 strain specific genes, compared to 26 L. pentosus strains, assigned to COGs functional categories. Different bar colors represent the further classification of all functional categories into four major classes—Poorly Characterized (Yellow bars), Cellular Processes and Signaling (Gray bars), Information Storage and Processing (Orange bars) and Metabolism (Blue bars).
Functional Classification
We sought to perform in silico functional classification of L. pentosus L33 and applied various interconnected approaches to achieve a well-rounded categorization of its genes/CDSs. The COG database is a valuable tool for describing the functional characteristics of newly sequenced genomes, as well as, comparing microbial communities (Galperin et al., 2019). Moreover, KEGG analysis is used to examine the diversity, as well as, the functionality of the proteins. Therefore, we performed a comprehensive analysis and comparison of the COG and KEGG profiles for L. pentosus L33, 26 L. pentosus strains, L. plantarum WCFS1 and L. plantarum 299v. The vast majority (94.66%) of the CDSs of L. pentosus L33, were allocated to 20 COG functional categories (Figure 5). The category “Function Unknown” was the most abundant (21.1%), followed by “General Function Prediction only” (12.3%), “Transcription” (9.0%), “Replication, Recombination, and Repair” (6.2%), “Carbohydrate transport and metabolism” (6.1%). Furthermore, comparison of the COG profile of L. pentosus L33 with the respective COG profiles of the 26 L. pentosus strains, L. plantarum WCFS1, and L. plantarum 299v, highlighted its similarity in respect to the percentage of the genes allocated in each of the COG functional categories (Figure 5 and Supplementary Table 4). The abovementioned similarity is irrelevant of the isolation source of the analyzed strains, since they have been derived from a variety of ecological niches such as meat samples, olive brines, milk products, fermented vegetables, and human intestine etc.
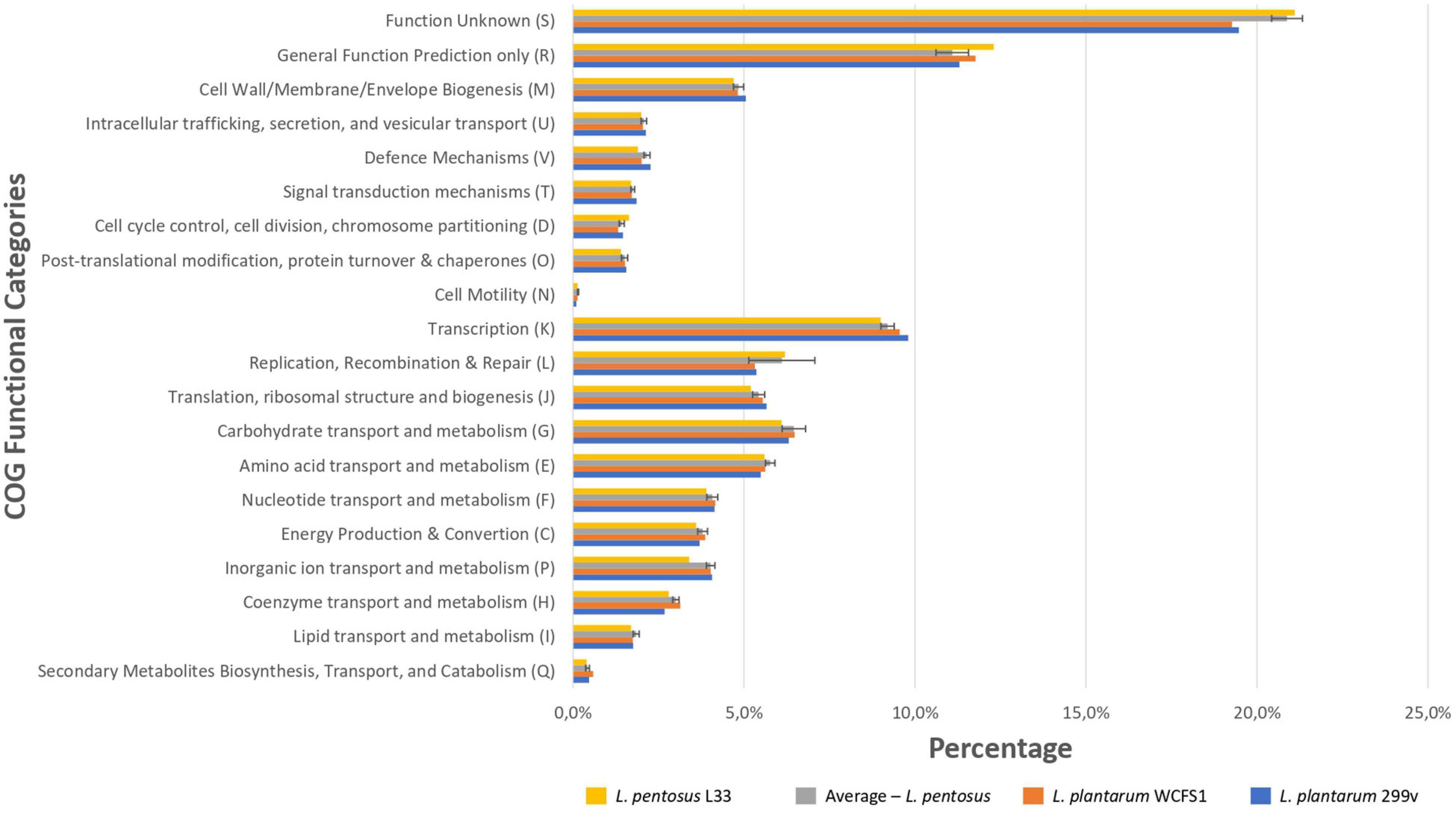
Figure 5. Comparison of the percentage of genes assigned to the COG functional categories of L. pentosus L33 (Yellow bars), L. plantarum WCFS1 (Orange bars), L. plantarum 299v (Blue bars) and of 26 L. pentosus strains (Gray bars). We calculated the percentage of genes for each COG functional category, for each one of the 26 L. pentosus strains, and depict their average values alongside the standard deviation (Gray bars).
To unveil the functional characterization of the CDSs of L. pentosus L33, we performed KEGG analysis. More precisely, approximately half of the L. pentosus L33 CDSs (52.10%) were assigned to 39 KEGG functional categories and 189 pathways. These pathways are mainly involved in the biosynthesis of secondary metabolites (ko: 01110; 180 genes), microbial metabolism in diverse environments (ko: 01120; 100 genes), and biosynthesis of amino acids (ko: 01230; 86 genes). Similarly to COG profiles, the number of genes assigned to each of the KEGG functional categories, is similar between L. pentosus L33, the other 26 L. pentosus strains, L. plantarum WCFS1, and L. plantarum 299v (Figure 6 and Supplementary Table 5). In addition, five virulence factors were identified in L. pentosus L33, including, a molecular chaperone (Hsp33), a translocase (YidC), two proteins of Mycobacterium tuberculosis with poorly defined function (Jag and YidD) (Yu et al., 2011), and a hemolysin iii family protein. However, the functionality of the detected hemolysin remains questionable, due to reports which indicate that L. pentosus L33 does not exhibit hemolytic activity in vitro (Pavli et al., 2016).
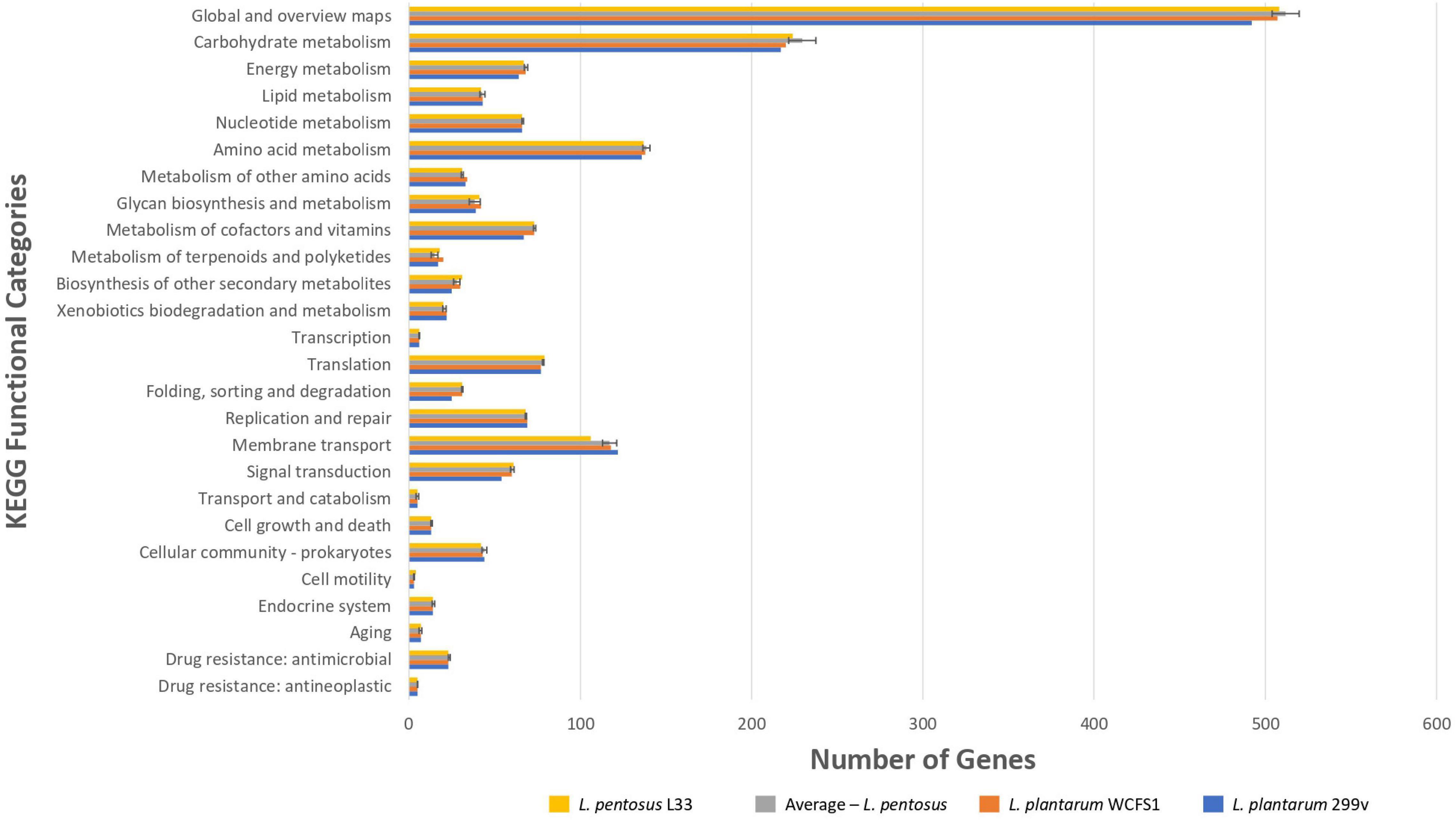
Figure 6. Comparison of the number of genes assigned to the most abundant KEGG functional categories of L. pentosus L33 (Yellow bars), L. plantarum WCFS1 (Orange bars), L. plantarum 299v (Blue bars), and of 26 L. pentosus strains (Gray bars). We calculated the number of genes for each KEGG functional category, for each one of the 26 L. pentosus strains, and depict their average values alongside the standard deviation (Gray bars).
Furthermore, employing CAZymes analysis, we classified the genes into respective CAZymes gene families (Supplementary Table 6). Thus, we showed that the L. pentosus L33 genome contains 92 genes, which were categorized into four CAZymes gene classes: 48 glycoside hydrolase (GH) genes, 34 glycosyltransferase (GT) genes, 7 carbohydrate-binding modules (CBMs), 3 carbohydrate esterase (CE) genes.
Identification of Genes Implicated in the Probiotic Potential of Lactiplantibacillus pentosus L33
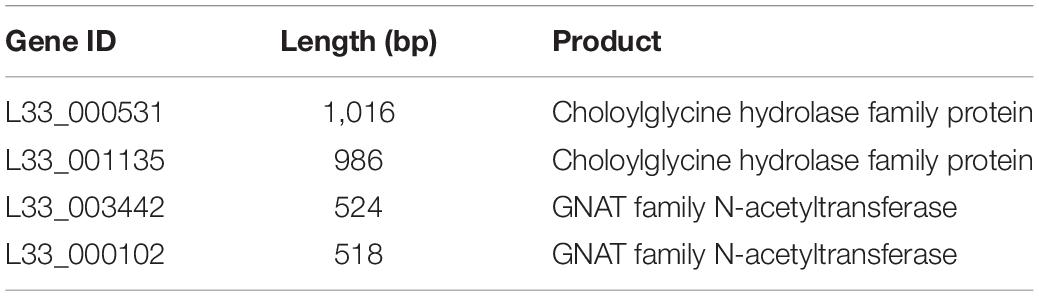
Finally, we performed comparative and comprehensive bioinformatical analysis to analyze in depth the L. pentosus L33 genome and locate genes and/or regions endowing a probiotic potential. Prokaryotic Genome Annotation Pipeline (PGAP) predicted that L. pentosus L33 contains 4 genes related to bile salt resistance; two bile salt hydrolases and two enzymes that are members of the GCN5-related N-acetyltransferases family (GNAT) (Table 2). Furthermore, RGI showed that the resistome of L. pentosus L33 does not contain transferable antibiotic resistance genes. Furthermore, a gene cluster consisting of 18 genes, involved in EPS biosynthesis, was identified during genome annotation. The aforementioned cluster has been, previously, described in the potential probiotic strain L. pentosus SLC13 and it is also present in the probiotic strain L. plantarum WCFS1 (Huang et al., 2018). Comparison between the EPS gene clusters indicated that the genes carried by L. pentosus L33 are homologous to those of strain SLC13, which is a potent exopolysaccharide producing strain (Figure 7; Huang et al., 2018).
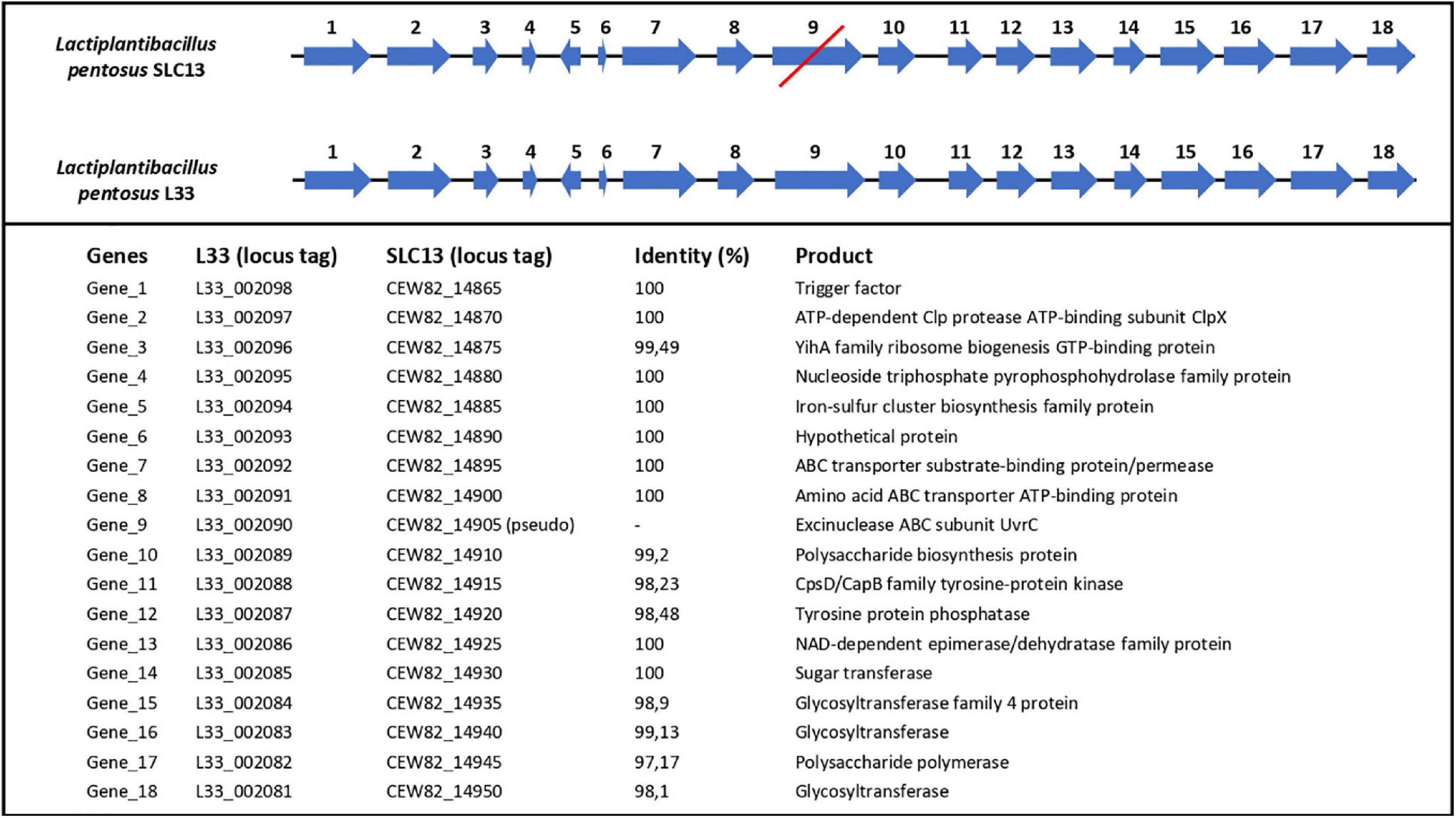
Figure 7. Comparison of length, position and direction of genes comprising the EPS biosynthesis cluster of L. pentosus L33 and L. pentosus SLC13. Protein identities between the two strains are also displayed. The red line indicates that gene number 9 in L. pentosus SLC13 is a pseudogene.
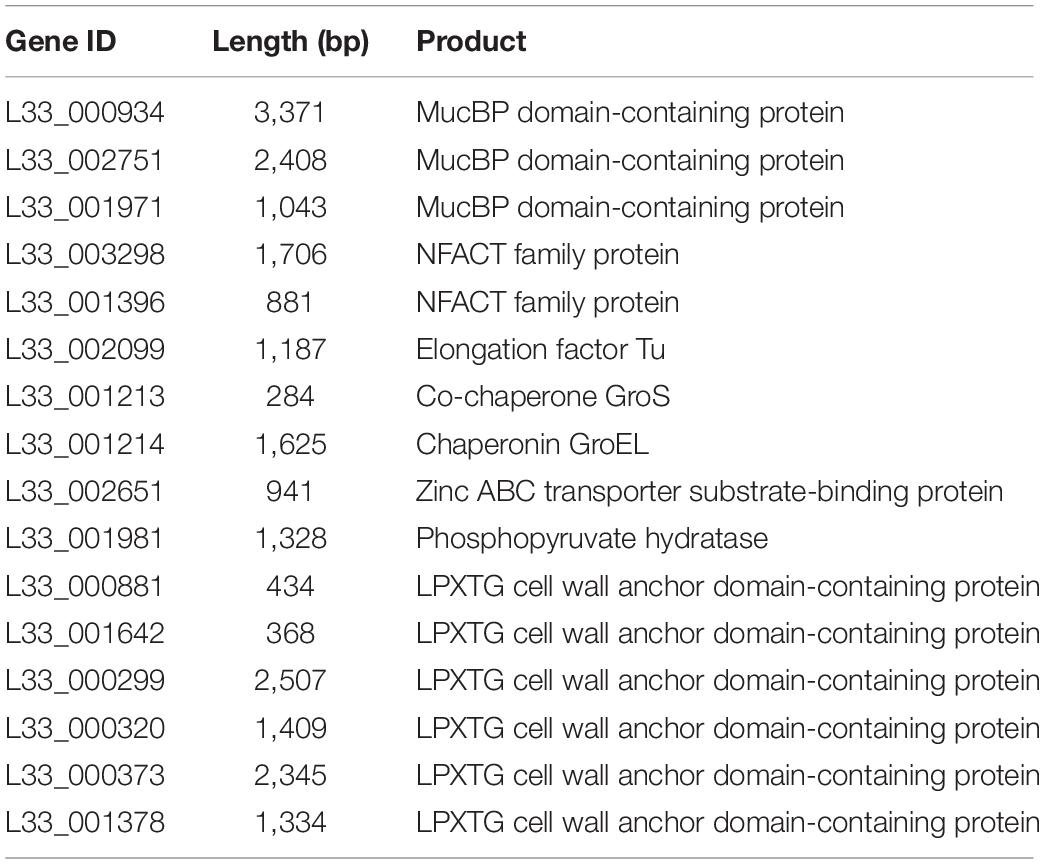
In addition, L. pentosus L33 contains 3 mucus-binding domain containing proteins and 2 proteins with fibronectin-binding domains along with NFACT domains (Table 3). Furthermore, 6 surface proteins carrying LPxTG cell wall anchored motifs were identified (Table 3). Moreover, moonlighting proteins with adhesin-like activity, elongation factor Tu, chaperonin GroEL, and co-chaperone GroES, are also present in the genome of L. pentosus L33 (Table 3). Notably, the adhesion capacity of L. pentosus L33 was validated in vitro utilizing HT-29 cells. Importantly, the strain exhibited similar adhesion capacity to that of the reference strain, L. rhamnosus GG (Figure 8).
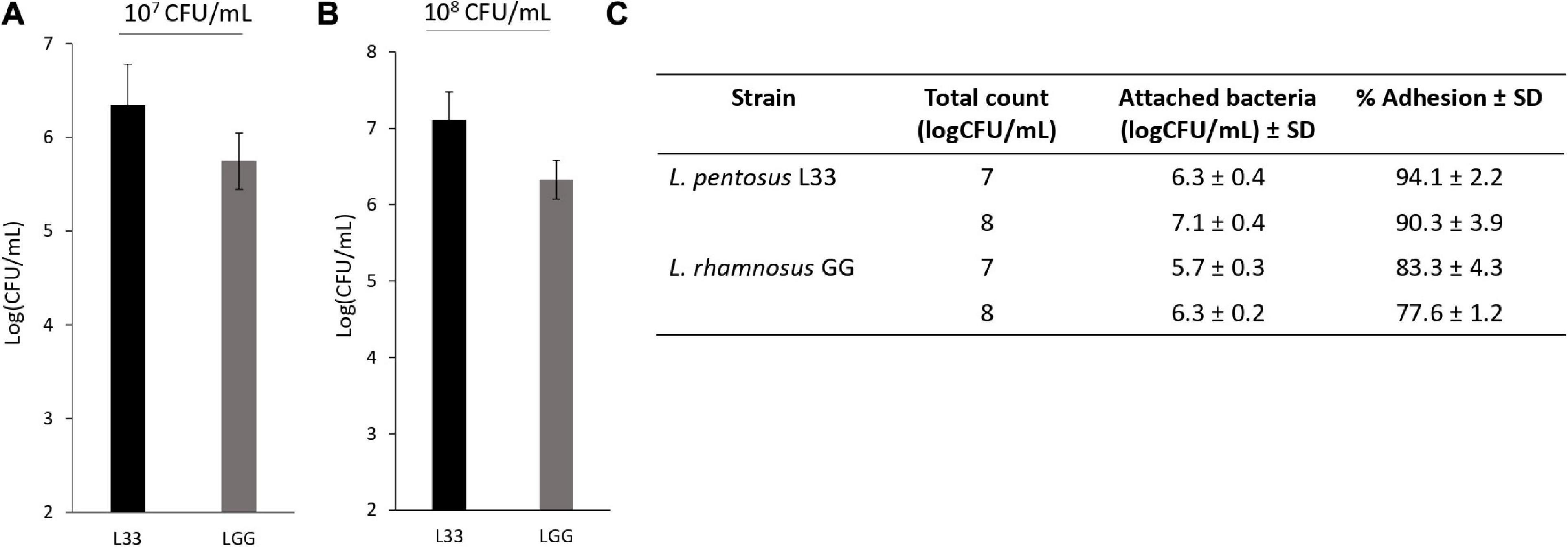
Figure 8. Assessment of the adhesion capacity of L. pentosus L33 by quantitative analysis. L. rhamnosus GG was used as a reference strain. (A) Adhesion capacity of viable cells at a concentration of 107 CFU/mL to HT-29 cells after 4-h co-incubation. (B) Adhesion capacity of viable cells at a concentration of 108 CFU/mL to HT-29 cells after 4-h co-incubation. (C) Adhesion (%) of attached bacteria to initial added total count. The data presented are the mean ± standard deviation of three independent experiments performed in duplicates.
Concerning the antimicrobial activity of the studied strain, L. pentosus L33 encodes for a class IIb bacteriocin, which is homologous to plantaricin NC8 αβ (Bengtsson et al., 2020). Class IIb bacteriocins consist of two peptides, which mediate their action by the interaction of their GxxxG and GxxxG-like (SxxxS and GxxxS) motifs with the membrane of the target pathogen (Maldonado et al., 2003; Bengtsson et al., 2020). The peptides coded by this strain lack a GxxxG-like motif and as a result, the functionality of the final product might be seriously affected (Supplementary Figure 2).
Discussion
In this study, we present the draft genome sequence of Lactiplantibacillus pentosus L33, a strain isolated from traditional meat products (Pavli et al., 2016). The genome of this strain consists of a circular chromosome; with no plasmid sequences detected. The complete genomic length (3,923,201 bp) and GC content (46.01%) of L. pentosus L33 were found to be similar to that of other L. pentosus strains, such as the closely related L. pentosus IG7 (3,802,404 bp, GC content: 45.79%, Accession: GCA_002993395.1) (Calero-Delgado et al., 2018), and the potential probiotic strain L. pentosus MP-10 (3,698,214 bp, GC content: 46.00%, Accession: GCA_900092635.1) (Abriouel et al., 2017). The genomic size and GC content of strains could be indicative of their lifestyle and preferred environmental niche. Strains of the Lactobacillus sensu lato that are free-living or nomadic usually possess a larger genome with an approximate length of 3–4 Mb, whereas host-adapted strains have a drastically smaller genome due to gene loss (Duar et al., 2017). The genetic traits that can be affected by this event are clusters for amino acid synthesis, and genes involved in metabolism regulation (Zheng et al., 2015). In this study, we found that the genes of L. pentosus L33 are involved in the complete biosynthesis of seven amino acids (Supplementary Table 7 and Supplementary Figures 3–8) and encode part of the required proteins necessary for the biosynthesis of the rest 13 amino acids. Interestingly, comprising of 100 genes, the “microbial metabolism in diverse environments” pathway (ko: 01120) was the second most common. The modules of this pathway include carbohydrate, methane, nitrogen, co-factor, and vitamin metabolism, among others. Concerning carbohydrate metabolism, CAZymes analysis showed that L. pentosus L33 does, indeed, code for enzymes involved in the synthesis and degradation of a broad array of simple and complex carbohydrates, such as glucose, galactose, mannose, trehalose, xylose, chitin, and cellulose. Additionally, it codes for galactose-, lactose-, starch-, and glycogen- binding modules that facilitate the catalytic activity of hydrolases (Boraston et al., 2004). These findings suggest that L. pentosus L33 may be able to inhabit a broad range of environmental niches.
Concerning the functional properties of this strain, L. pentosus L33 has presented desirable attributes in a previous in vitro study, where a total of 48 Lactobacillus strains were assessed for their susceptibility to common antibiotics, hemolytic activity, tolerance to gastrointestinal conditions, and antimicrobial properties (Pavli et al., 2016). It was found that L. pentosus L33 exhibited good tolerance to bile salts, that was not accompanied by bile hydrolase activity. Interestingly, in the present study, we detected two coding sequences for bile salt hydrolases (Table 2); however, their functionality is questioned based on the in vitro findings. Nevertheless, it is important to note that bile salt resistance is a complex phenotype that can be mediated by several mechanisms, such as bile-efflux systems, changes in EPS and S-layer protein production (Ruiz et al., 2013). This character should be explored in greater depth in future studies.
In this study, we found that this strain does not carry transferable antibiotic resistance genes. In fact, with the exception of vancomycin, L. pentosus L33 was not able to survive treatments with common antibiotics (Pavli et al., 2016). Vancomycin resistance in Lactobacillus strains is considered to be intrinsic (Guo et al., 2017); therefore, it is no surprise that L. pentosus L33 presents resistance to vancomycin. Consequently, the demonstrated resistance does not raise any safety concerns, as there is no implication of horizontal gene transfer (Shao et al., 2015). The mode of action of this antibiotic involves its interaction with peptidoglycan precursors, leading to the inhibition of cell wall synthesis. More specifically, vancomycin binds to the D-alanine/D-alanine terminus of the muramyl pentapeptide and inhibits the polymerization of the peptidoglycan precursor. In this context, we found that L. pentosus L33 possesses a gene (VanX) encoding a D-ala-D-ala dipeptidase which hydrolyzes D-alanine/D-alanine residues (Liu et al., 2009). Moreover, in several LAB species, the D-alanine residue located at the end of the pentapeptide is substituted by D-lactate or D-serine and thus blocking vancomycin binding (Delcour et al., 1999). KEGG analysis showed that L. pentosus L33 encodes a Ddl ligase, responsible for the D-alanine to D-lactate substitution in Lactobacilli (Tuyarum et al., 2021). Furthermore, there are reports that indicate that many LAB genera exhibit intrinsic resistance to other antibiotics, such as bacitracin, kanamycin, teicoplanin, and quinolones (Imperial and Ibana, 2016). Additionally, it should be noted, that transfer of the vancomycin resistance cluster from Enterobacteriaceae to commercial probiotic strains has been reported in vitro and in vivo, during transit in the murine gastrointestinal tract (Mater et al., 2008).
The cellular surface of Lactobacilli is decorated by a plethora of cell surface proteins that can interact with host receptors and give rise to a variety of probiotic effects (Teame et al., 2020). Indeed, probiotics can interact with the gastrointestinal mucosa of mammalian hosts utilizing pilli, mucin-, and fibronectin- binding proteins, as well as surface-layer (S-layer) proteins (Siciliano et al., 2019). These interactions are necessary for the transient attachment of ingested probiotics in the intestinal mucosa, while they can also facilitate important probiotic functions, including antimicrobial (Tytgat et al., 2016) and immunomodulatory activity (Monteagudo-Mera et al., 2019). In this study, we found that L. pentosus L33 carries mucus- and fibronectin- binding proteins (Table 3), however, it does not encode for spaCBA pilli, commonly found in other LAB strains, such as L. rhamnosus GG (Reunanen et al., 2012). The adhesins are covalently anchored to the peptidoglycan layer by a C-terminal Leu-Pro-any-Thr-Gly (LPxTG) motif, which is, also, used for their identification in silico (Siegel et al., 2017). Moreover, cytoplasmic proteins that participate in important housekeeping functions such as carbohydrate metabolism, translation regulation and protein folding, can be found, anchorless, in the cellular envelope acting as adhesins (Celebioglu et al., 2017). These multifunctional proteins, also known as moonlighting proteins, have been identified in animals, plants, yeast and bacteria. L. pentosus L33 encodes some of these proteins; elongation factor Tu (EF-Tu), chaperonin GroEL, and co-chaperonin GroES. Previous reports have shown that L. plantarum and L. pentosus strains utilize EF-Tu (Choudhary et al., 2019) and GroEL (Calasso et al., 2013) for the adhesion on the intestinal epithelium. The adhesion capacity of the strain was further validated in vitro. We showed that L. pentosus L33 can efficiently adhere to HT-29 cells, exhibiting similar behavior to L. rhamnosus GG, a reference strain, whose capacity to attach to and colonize the gastrointestinal mucosa has been previously described (Chondrou et al., 2018; Pagnini et al., 2018). Further studies are required to evaluate this finding and elucidate its contribution to probiotic efficacy.
Furthermore, we report that the L. pentosus L33 genome includes five virulence factors. Hemolysin iii family protein is very common among Lactiplantibacillus genomes, including the probiotic strains L. plantarum 299v and L. plantarum ST-III. The abovementioned strains have an established safety profile, and they are widely used as probiotics (Chokesajjawatee et al., 2020). Heat shock protein 33 (Hsp33) is a redox-regulated molecular chaperone that binds to unfolded proteins and prevents protein aggregation (Winter et al., 2005). YidC gene encodes a translocase that regulates respiratory metabolism in Mycobacterium tuberculosis (Thakur et al., 2016). YidD and Jag belong to the same gene cluster along with YidC, but their function remains unclear (Yu et al., 2011). Nevertheless, the impact of these factors in the safety profile of L. pentosus L33 has to be further examined.
The probiotic character has also been linked to EPS biosynthesis, as it is well established that EPS play a key role in the dynamic interaction of bacteria with their environment (Angelin and Kavitha, 2020). EPS can be found loosely attached to the cell surface or excreted in the growth medium of the producer strain, while the yield of production can fluctuate based on growth conditions. In addition, the produced exopolysaccharides can vary in terms of monosaccharide constitution, charge, linkage, and existence of repeated sidechains (Caggianiello et al., 2016). EPS can facilitate niche adaptation, as they promote auto-aggregation (Aslim et al., 2007), attachment to abiotic or biotic surfaces and biofilm formation (Castro-Bravo et al., 2018). Furthermore, there are several physiological functions attributed to EPS such as anti-inflammatory, antioxidant, antiviral, and antiproliferative activity (Nguyen et al., 2020). Lastly, the production of EPS in high concentrations can alter the organoleptic characteristics of fermented products (Ale et al., 2020). In this study, we found an EPS biosynthesis cluster, homologous to that of L. pentosus SLC13, a LAB strain known for its capacity to produce high yields of EPS (Huang et al., 2018). In this context, the EPS fraction of L. pentosus L33, is currently being studied for its antimicrobial and antibiofilm potential.
Our analysis showed that L. pentosus L33 does not code for functional bacteriocins, due to the lack of motifs crucial for their inhibitory action. This finding agrees with previous in vitro studies, where no bacteriocin-like activity was detected (Pavli et al., 2016). However, probiotics can exert antimicrobial effects through various mechanisms. such as competition for nutrients, inhibition of pathogen adhesion (Walsham et al., 2016) and immune system stimulation (Tuo et al., 2018). Moreover, they can produce other inhibitory compounds, fatty acids, hydrogen peroxide, ethanol (Chen et al., 2019) and biosurfactants (Sharma and Saharan, 2016), or induce indirect antimicrobial effects by lowering of intestinal pH, due to production of high amounts of lactic and acetic acids. Thus, ongoing studies focus on the potential of this strain to interfere with proliferation and biofilm formation of clinically relevant strains, such as Staphylococcus aureus, Salmonella enteritidis, and Escherichia coli, by alternative mechanisms to bacteriocin production.
Conclusively, these findings in combination with previous in vitro work, support that L. pentosus L33 is a good probiotic candidate. This strain fulfills the main criteria for probiotic selection; tolerance to gastrointestinal tract conditions, susceptibility to common antibiotics and γ-hemolytic activity. In the present study, we introduced new traits that add to the characterization of L. pentosus L33 as a novel probiotic strain, the capacity to produce adhesins and exopolysaccharides. Whole-genome sequencing and comprehensive bioinformatic analysis facilitate targeted laboratory validation of traits of newly isolated strains, streamlining their characterization as probiotic. In this context, future studies will demonstrate the in situ performance of L. pentosus L33 strain as a starter/adjunct culture for the production of fermented dry meat products (as this strain was previously isolated from fermented sausages), to signify its effectiveness for application in sausage manufacturing. Additionally, future researches will explore the L. pentosus L33-host interactome, and especially gut colonization mechanisms. Overall, L. pentosus L33 exhibits a great interest as a potential probiotic strain and forthcoming studies will further unravel its characteristics in vitro, in vivo, and in situ.
Data Availability Statement
The datasets presented in this study have been submitted to DDBJ/ENA/GenBank under the accession number JAHKRU000000000. The version described in this manuscript is the JAHKRU010000000.
Author Contributions
AP, NC, PK, and AG designed the study. OS, KT, DK, and MT performed genome analysis and participated in the writing of the manuscript. AP, CT, NC, PK, and AG contributed to editing and critical reviewing of the manuscript. CT and NC took charge of the resources. All authors had read and approved the final manuscript.
Funding
This research has been financed by the project “A Model Smart Quality Assurance and Safety System for Fresh Poultry Products” (QAPP: T1EDK-04344) that is co-financed by the EU and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, RESEARCH–CREATE–INNOVATE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the support of the projects: “InTechThrace: Integrated Technologies in biomedical research: multilevel biomarker analysis in Thrace” (MIS Code 5047285), under the Operational Program “Competitiveness, Entrepreneurship, and Innovation” (EPAnEK), co-funded by the European Regional Development Fund (ERDF) and national resources (Partnership Agreement 2014-2020) and “ELIXIR-GR: Hellenic Research Infrastructure for the Management and Analysis of Data from the Biological Sciences” (MIS 5002780) under the Action Reinforcement of the Research and Innovation Infrastructure, funded by the Operational Program Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund) and of the M.Sc. course «Translational Research in Biomedicine», of the Department of Molecular Biology and Genetics, Democritus University of Thrace.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.746659/full#supplementary-material
Footnotes
- ^ https://www.grandviewresearch.com/industry-analysis/probiotics-market (Grand View Research, 2021).
References
Abriouel, H., Pérez Montoro, B., Casimiro-Soriguer, C. S., Pérez Pulido, A. J., Knapp, C. W., Caballero Gómez, N., et al. (2017). Insight into potential probiotic markers predicted in Lactobacillus pentosus MP-10 genome sequence. Front. Microbiol. 22:891. doi: 10.3389/fmicb.2017.00891
Ale, E. C., Rojas, M. F., Reinheimer, J. A., and Binetti, A. G. (2020). Lactobacillus fermentum: Could EPS production ability be responsible for functional properties? Food Microbiol. 90:103465. doi: 10.1016/j.fm.2020.103465
Andrews, S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed May 2, 2021).
Angelin, J., and Kavitha, M. (2020). Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 162, 853–865. doi: 10.1016/j.ijbiomac.2020.06.190
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Aslim, B., Onal, D., and Beyatli, Y. (2007). Factors influencing autoaggregation and aggregation of Lactobacillus delbrueckii subsp. bulgaricus isolated from handmade yogurt. J. Food Prot. 70, 223–227. doi: 10.4315/0362-028x-70.1.223
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bengtsson, T., Selegård, R., Musa, A., Hultenby, K., Utterström, J., Sivlér, P., et al. (2020). Plantaricin NC8 αβ exerts potent antimicrobial activity against Staphylococcus spp. and enhances the effects of antibiotics. Sci. Rep. 10:3580. doi: 10.1038/s41598-020-60570-w
Biswas, A., Staals, R. H., Morales, S. E., Fineran, P. C., and Brown, C. M. (2016). CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 17:356. doi: 10.1186/s12864-016-2627-0
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781. doi: 10.1042/BJ20040892
Caggianiello, G., Kleerebezem, M., and Spano, G. (2016). Exopolysaccharides produced by lactic acid bacteria: from health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 100, 3877–3886. doi: 10.1007/s00253-016-7471-2
Calasso, M., Di Cagno, R., De Angelis, M., Campanella, D., Minervini, F., and Gobbetti, M. (2013). Effects of the peptide pheromone plantaricin A and cocultivation with Lactobacillus sanfranciscensis DPPMA174 on the exoproteome and the adhesion capacity of Lactobacillus plantarum DC400. Appl. Environ. Microbiol. 79, 2657–2669. doi: 10.1128/AEM.03625-12
Calero-Delgado, B., Martín-Platero, A. M., Pérez-Pulido, A. J., Benítez-Cabello, A., Casimiro-Soriguer, C. S., Martínez-Bueno, M., et al. (2018). Draft genome sequences of six Lactobacillus pentosus strains isolated from brines of traditionally fermented spanish-style green table olives. Genome Announc. 6:e379–18. doi: 10.1128/genomeA.00379-18
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Castro-Bravo, N., Wells, J. M., Margolles, A., and Ruas-Madiedo, P. (2018). Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front. Microbiol. 9:2426. doi: 10.3389/fmicb.2018.02426
Celebioglu, H. U., Olesen, S. V., Prehn, K., Lahtinen, S. J., Brix, S., Hachem, M. A., et al. (2017). Data regarding the growth of Lactobacillus acidophilus NCFM on different carbohydrates and recombinant production of elongation factor G and pyruvate kinase. Data Brief 14, 118–122. doi: 10.1016/j.dib.2017.07.021
Chen, C. C., Lai, C. C., Huang, H. L., Huang, W. Y., Toh, H. S., Weng, T. C., et al. (2019). Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 10:789. doi: 10.3389/fmicb.2019.00789
Chokesajjawatee, N., Santiyanont, P., Chantarasakha, K., Kocharin, K., Thammarongtham, C., Lertampaiporn, S., et al. (2020). Safety assessment of a nham starter culture Lactobacillus plantarum BCC9546 via whole-genome analysis. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-66857-2
Chondrou, P., Karapetsas, A., Kiousi, D. E., Tsela, D., Tiptiri-Kourpeti, A., Anestopoulos, I., et al. (2018). Lactobacillus paracasei K5 displays adhesion, anti-proliferative activity and apoptotic effects in human colon cancer cells. Benef. Microb. 9, 975–983. doi: 10.3920/BM2017.0183
Chondrou, P., Karapetsas, A., Kiousi, D. E., Vasileiadis, S., Ypsilantis, P., Botaitis, S., et al. (2020). Assessment of the immunomodulatory properties of the probiotic strain Lactobacillus paracasei K5 in vitro and in vivo. Microorganisms 8:709. doi: 10.3390/microorganisms8050709
Choudhary, J., Dubey, R. C., Sengar, G., and Dheeman, S. (2019). Evaluation of probiotic potential and safety assessment of Lactobacillus pentosus MMP4 isolated from Mare’s lactation. Probiot. Antimicrob. Proteins 11, 403–412. doi: 10.1007/s12602-018-9431-x
de Jong, A., van Hijum, S. A., Bijlsma, J. J., Kok, J., and Kuipers, O. P. (2006). BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34, W273–W279. doi: 10.1093/nar/gkl237
Delcour, J., Ferain, T., Deghorain, M., Palumbo, E., and Hols, P. (1999). The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Anton. Leeuw. 76, 159–184.
Duar, R. M., Lin, X. B., Zheng, J., Martino, M. E., Grenier, T., Pérez-Muñoz, M. E., et al. (2017). Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 41, S27–S48. doi: 10.1093/femsre/fux030
EFSA (2018). Guidance on the characterisation of microorganisms used as feed additives or 30 as production organisms. EFSA J. 16:5206. doi: 10.2903/j.efsa.2018.5206
FAO/WHO (2002). Guidelines for the Evaluation of Probiotics in Food. Available online at: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed May 2, 2021).
Galperin, M. Y., Kristensen, D. M., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2019). Microbial genome analysis: the COG approach. Brief. Bioinform. 20, 1063–1070. doi: 10.1093/bib/bbx117
Golomb, B. L., Morales, V., Jung, A., Yau, B., Boundy-Mills, K. L., and Marco, M. L. (2013). Effects of pectinolytic yeast on the microbial composition and spoilage of olive fermentations. Food Microbiol. 33, 97–106.
Grand View Research (2021). Probiotics Market Size, Share and Trends Analysis Report by Product (Food and Beverages, Dietary Supplements), by Ingredient (Bacteria, Yeast), by End Use (Human, Animal), by Distribution Channel, and Segment Forecasts, 2021 – 2028. San Francisco, CA: Grand View Research.
Guo, H., Pan, L., Li, L., Lu, J., Kwok, L., Menghe, B., et al. (2017). Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 82, 724–730. doi: 10.1111/1750-3841.13645
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Huang, M. L., Huang, J. Y., Kao, C. Y., and Fang, T. J. (2018). Complete genome sequence of Lactobacillus pentosus SLC13, isolated from mustard pickles, a potential probiotic strain with antimicrobial activity against foodborne pathogenic microorganisms. Gut. Pathog. 10:1.
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Imperial, I. C. V. J., and Ibana, J. A. (2016). Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 7:1983. doi: 10.3389/fmicb.2016.01983
Inglin, R. C., Meile, L., and Stevens, M. J. A. (2018). Clustering of Pan- and Core-genome of Lactobacillus provides novel evolutionary insights for differentiation. BMC Genom. 19:284. doi: 10.1186/s12864-018-4601-5
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kiousi, D. E., Rathosi, M., Tsifintaris, M., Chondrou, P., and Galanis, A. (2021). Pro-biomics: Omics technologies to unravel the role of probiotics in health and disease. Adv. Nutr. 12, 1802–1820. doi: 10.1093/advances/nmab014
Kok, C. R., and Hutkins, R. (2018). Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 76, 4–15. doi: 10.1093/nutrit/nuy056
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Liu, C., Zhang, Z. Y., Dong, K., Yuan, J. P., and Guo, X. K. (2009). Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 22, 401–412. doi: 10.1016/S0895-3988(10)60018-9
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Maldonado, A., Ruiz-Barba, J. L., and Jiménez-Díaz, R. (2003). Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69, 383–389. doi: 10.1128/AEM.69.1.383-389.2003
Maldonado-Barragán, A., Caballero-Guerrero, B., Lucena-Padrós, H., and Ruiz-Barba, J. L. (2011). Genome sequence of Lactobacillus pentosus IG1, a strain isolated from spanish-style green olive fermentations. J. Bacteriol. 193:5605. doi: 10.1128/JB.05736-11
Mater, D. D., Langella, P., Corthier, G., and Flores, M. J. (2008). A probiotic Lactobacillus strain can acquire vancomycin resistance during digestive transit in mice. J. Mol. Microbiol. Biotechnol. 14, 123–127. doi: 10.1159/000106091
Monteagudo-Mera, A., Rastall, R. A., Gibson, G. R., Charalampopoulos, D., and Chatzifragkou, A. (2019). Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 103, 6463–6472. doi: 10.1007/s00253-019-09978-7
Nguyen, P. T., Nguyen, T. T., Bui, D. C., Hong, P. T., Hoang, Q. K., and Nguyen, H. T. (2020). Exopolysaccharide production by lactic acid bacteria: the manipulation of environmental stresses for industrial applications. AIMS Microbiol. 6, 451–469. doi: 10.3934/microbiol.2020027
Nordström, E. A., Teixeira, C., Montelius, C., Jeppsson, B., and Larsson, N. (2021). Lactiplantibacillus plantarum 299v (LP299V®): three decades of research. Benef. Microbes 12, 441–465. doi: 10.3920/BM2020.0191
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pagnini, C., Corleto, V. D., Martorelli, M., Lanini, C., D’Ambra, G., Di Giulio, E., et al. (2018). Mucosal adhesion and anti-inflammatory effects of Lactobacillus rhamnosus GG in the human colonic mucosa: A proof-of-concept study. World J. Gastroenterol. 24, 4652–4662. doi: 10.3748/wjg.v24.i41.4652
Pavli, F. G., Argyri, A. A., Papadopoulou, O. S., Nychas, J. G. E., Chorianopoulos, N. G., and Tassou, C. C. (2016). Probiotic potential of lactic acid bacteria from traditional fermented dairy and meat products: assessment by in vitro tests and molecular characterization. J. Prob. Health 4:1000157. doi: 10.4172/2329-8901.1000157
Plessas, S., Kiousi, D. E., Rathosi, M., Alexopoulos, A., Kourkoutas, Y., Mantzourani, I., et al. (2020). Isolation of a Lactobacillus paracasei strain with probiotic attributes from kefir grains. Biomedicines 8:594.
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G., and Toth, I. K. (2016). Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/c5ay02550h
Reunanen, J., von Ossowski, I., Hendrickx, A. P., Palva, A., and de Vos, W. M. (2012). Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 78, 2337–2344. doi: 10.1128/AEM.07047-11
Rodrigo-Torres, L., Yépez, A., Aznar, R., and Arahal, D. R. (2019). Genomic insights into five strains of Lactobacillus plantarum with biotechnological potential isolated from chicha, a traditional maize-based fermented beverage from Northwestern Argentina. Front. Microbiol. 10:2232. doi: 10.3389/fmicb.2019.02232
Ruiz, L., Margolles, A., and Sánchez, B. (2013). Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4:396. doi: 10.3389/fmicb.2013.00396
Shao, Y., Zhang, W., Guo, H., Pan, L., Zhang, H., and Sun, T. (2015). Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control 50, 250–258. doi: 10.1016/j.foodcont.2014.09.003
Sharma, D., and Saharan, B. S. (2016). Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Rep. 11, 27–35. doi: 10.1016/j.btre.2016.05.001
Siciliano, R. A., Lippolis, R., and Mazzeo, M. F. (2019). Proteomics for the investigation of surface-exposed proteins in probiotics. Front. Nutr. 6:52. doi: 10.3389/fnut.2019.00052
Siegel, S. D., Reardon, M. E., and Ton-That, H. (2017). Anchoring of LPXTG-like proteins to the gram-positive cell wall envelope. Curr. Top. Microbiol. Immunol. 404, 159–175. doi: 10.1007/82_2016_8
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Teame, T., Wang, A., Xie, M., Zhang, Z., Yang, Y., Ding, Q., et al. (2020). Paraprobiotics and postbiotics of probiotic Lactobacilli, Their Positive Effects on the Host and Action Mechanisms: A Review. Front. Nutr. 7:570344. doi: 10.3389/fnut.2020.570344
Thakur, P., Gantasala, N. P., Choudhary, E., Singh, N., Abdin, M. Z., and Agarwal, N. (2016). The preprotein translocase YidC controls respiratory metabolism in Mycobacterium tuberculosis. Sci. Rep. 6:24998. doi: 10.1038/srep24998
Tian, P., O’Riordan, K. J., Lee, Y. K., Wang, G., Zhao, J., Zhang, H., et al. (2020). Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice. Neurobiol. Stress 12:100216. doi: 10.1016/j.ynstr.2020.100216
Tiptiri-Kourpeti, A., Spyridopoulou, K., Santarmaki, V., Aindelis, G., Tompoulidou, E., Lamprianidou, E. E., et al. (2016). Lactobacillus casei exerts anti-proliferative effects accompanied by apoptotic cell death and Up-Regulation of TRAIL in colon carcinoma cells. PLoS One 11:e0147960. doi: 10.1371/journal.pone.0147960
Tuo, Y., Song, X., Song, Y., Liu, W., Tang, Y., Gao, Y., et al. (2018). Screening probiotics from Lactobacillus strains according to their abilities to inhibit pathogen adhesion and induction of pro-inflammatory cytokine IL-8. J. Dairy Sci. 101, 4822–4829. doi: 10.3168/jds.2017-13654
Tuyarum, C., Songsang, A., and Lertworapreecha, M. (2021). In vitro evaluation of the probiotic potential of Lactobacillus isolated from native swine manure. Vet. World 14, 1133–1142. doi: 10.14202/vetworld.2021.1133-1142
Tytgat, H. L., Douillard, F. P., Reunanen, J., Rasinkangas, P., Hendrickx, A. P., Laine, P. K., et al. (2016). Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via Mucus-Binding Pili: evidence for a novel and heterospecific probiotic mechanism. Appl. Environ. Microbiol. 82, 5756–5762. doi: 10.1128/AEM.01243-16
van den Nieuwboer, M., van Hemert, S., Claassen, E., and de Vos, W. M. (2016). Lactobacillus plantarum WCFS1 and its host interaction: a dozen years after the genome. Microb. Biotechnol. 9, 452–465. doi: 10.1111/1751-7915.12368
Walsham, A. D., MacKenzie, D. A., Cook, V., Wemyss-Holden, S., Hews, C. L., Juge, N., et al. (2016). Lactobacillus reuteri inhibition of enteropathogenic Escherichia coli adherence to human intestinal epithelium. Front. Microbiol. 7:244. doi: 10.3389/fmicb.2016.00244
Winter, J., Linke, K., Jatzek, A., and Jakob, U. (2005). Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell. 17, 381–392. doi: 10.1016/j.molcel.2004.12.027
Wu, Y., Wang, B., Xu, H., Tang, L., Li, Y., Gong, L., et al. (2019). Probiotic Bacillus attenuates oxidative stress- induced intestinal injury via p38-mediated autophagy. Front. Microbiol. 10:2185. doi: 10.3389/fmicb.2019.02185
Ye, K., Li, P., and Gu, Q. (2020). Complete genome sequence analysis of a strain Lactobacillus pentosus ZFM94 and its probiotic characteristics. Genomics 112, 3142–3149. doi: 10.1016/j.ygeno.2020.05.015
Yu, H. J., Chen, Y. F., Yang, H. J., Yang, J., Xue, J. G., Li, C. K., et al. (2015). Screening for Lactobacillus plantarum with potential inhibitory activity against enteric pathogens. Ann. Microbiol. 65, 1257–1265. doi: 10.1007/s13213-014-0963-3
Yu, Z., Lavèn, M., Klepsch, M., de Gier, J. W., Bitter, W., van Ulsen, P., et al. (2011). Role for Escherichia coli YidD in membrane protein insertion. J. Bacteriol. 193, 5242–5251. doi: 10.1128/JB.05429-11
Zheng, J., Ruan, L., Sun, M., and Gänzle, M. A. (2015). Genomic view of Lactobacilli and Pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 81, 7233–7243. doi: 10.1128/AEM.02116-15
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: Lactiplantibacillus pentosus, whole-genome sequencing, probiotics, comparative genomics, phylogenetic analysis
Citation: Stergiou OS, Tegopoulos K, Kiousi DE, Tsifintaris M, Papageorgiou AC, Tassou CC, Chorianopoulos N, Kolovos P and Galanis A (2021) Whole-Genome Sequencing, Phylogenetic and Genomic Analysis of Lactiplantibacillus pentosus L33, a Potential Probiotic Strain Isolated From Fermented Sausages. Front. Microbiol. 12:746659. doi: 10.3389/fmicb.2021.746659
Received: 24 July 2021; Accepted: 08 October 2021;
Published: 26 October 2021.
Edited by:
Flavia Figueira Aburjaile, Federal University of Minas Gerais, BrazilReviewed by:
Cristian Botta, University of Turin, ItalyChristian U. Riedel, University of Ulm, Germany
Copyright © 2021 Stergiou, Tegopoulos, Kiousi, Tsifintaris, Papageorgiou, Tassou, Chorianopoulos, Kolovos and Galanis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petros Kolovos, cGtvbG92b3NAbWJnLmR1dGguZ3I=; Alex Galanis, YWdhbGFuaXNAbWJnLmR1dGguZ3I=
†These authors have contributed equally to this work and share first authorship
 Odysseas Sotirios Stergiou
Odysseas Sotirios Stergiou Konstantinos Tegopoulos
Konstantinos Tegopoulos Despoina Eugenia Kiousi
Despoina Eugenia Kiousi Margaritis Tsifintaris
Margaritis Tsifintaris Aristotelis C. Papageorgiou
Aristotelis C. Papageorgiou Chrysoula C. Tassou
Chrysoula C. Tassou Nikos Chorianopoulos
Nikos Chorianopoulos Petros Kolovos
Petros Kolovos Alex Galanis
Alex Galanis