
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 September 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.743981
A correction has been applied to this article in:
Corrigendum: Genetic and Phenotypic Characterization of the Novel Metallo-β-Lactamase NDM-29 From Escherichia coli
Objectives: The New Delhi metallo-β-lactamase (NDM) can hydrolyze almost all clinically available β-lactam antibiotics and has widely spread all over the world. NDM-29, a novel carbapenemase, was discovered in an Escherichia coli (19NC225) isolated from a patient with biliary tract infection in 2019 in China.
Methods: Conjugation, transformation, cloning test, fitness cost, PacBio Sequel, and Illumina sequencing were performed to analyze the genetic and phenotypic characterization of blaNDM–29.
Results: The susceptibility testing results showed 19NC225 was resistant to cephalosporins, carbapenems, combinations of β-lactam and β-lactamase inhibitors, and levofloxacin. Conjugation and transformation were performed to verify the transferability of NDM-29-encoding plasmid, and cloning test was conducted to prove the function of blaNDM–29 to increase carbapenem resistance. Furthermore, fitness cost test confirmed that the presence of NDM-29 exerts no survival pressure on bacteria. PacBio Sequel and Illumina sequencing were performed to analyze the genetic characterization of 19NC225, which contains two plasmids (pNC225-TEM1B and pNC225-NDM-29). pNC225-NDM-29, exhibiting 99.96% identity and 100% coverage with pNDM-BTR (an IncN1 plasmid from an E. coli in urine specimen from Beijing in 2013), showed responsibility for the multidrug-resistant (MDR) phenotype. Compared with blaNDM–1, blaNDM–29, located on pNC225-NDM-29, carries a G388A (D130N) mutation. The region harboring blaNDM–29 is located in an ISKpn19-based transposon, and two Tn6292 remnants are symmetrically located upstream and downstream of the transposon. The sequence results also indicated several important virulence genes.
Conclusion: The findings of the novel carbapenemase NDM-29 could pose a threat to the control of antimicrobial resistance and arouse attention about the mutation of bacteria.
- The first detailed report of the carbapenemase NDM-29 from E. coli and its genetic environment in China.
- A whole genome sequence analysis of the newly found plasmid pNC225-NDM-29 and the gene blaNDM–29.
- Complete functional verification experiments, including conjugation, transformation, cloning, and fitness cost.
One popular resistant mechanism of gram-negative bacteria is producing hydrolytic enzymes, especially β-lactamases, which can hydrolyze β-lactam ring to break the amide bond and inactivate the antibacterial activity of drugs (Dever and Dermody, 1991). New Delhi metallo-β-lactamase (NDM) is classified as Ambler molecular class B β-lactamase, which can hydrolyze almost all clinically available β-lactam antibiotics (Khan et al., 2017; Liu et al., 2017). NDM-positive strains have widely spread all over the world, including India, Europe, China, Brazil, Australia, etc. (Khan et al., 2017; van Duin and Doi, 2017). Since NDM was first reported in a Klebsiella pneumoniae (K. pneumoniae) strain from a Swedish patient in New Delhi in 2009 (Yong et al., 2009), 31 variants of NDM (NDM-1–31) have been detected (NCBI) until now. Among them, NDM-1, 4, 5, and 7 are most prevalent around the world (Khan et al., 2017). NDM variant was discovered in not only human but also food animals and environment (van Duin and Doi, 2017; Liu et al., 2018). The highest distribution of NDM-positive species is observed in K. pneumoniae and Escherichia coli (E. coli) (Gamal et al., 2020).
In a recent study, we discovered an NDM-29 carbapenemase-producing E. coli strain (19NC225), isolated from a patient’s bile in 2019. The gene sequence of blaNDM–29 from a Klebsiella pneumoniae strain has been submitted to the NCBI database (NCBI), by researchers from Saint Petersburg, Russia (Starkova et al., 2021). Our study is the first detailed report of NDM—29 from E. coli in China, including genetic and phenotypic characterization, and confirms the potential threat of this new NDM-type carbapenemase to be a cause of extensively drug-resistant organism spread.
The E. coli isolate 19NC225 was collected from the bile of a 65-year-old Chinese male patient with biliary tract infection in Nanchang, Jiangxi, China on March 2, 2019, and was identified by Vitek MS MALDI-TOF (BioMérieux) system in Peking Union Medical College Hospital.
E. coli DH5α, rifampin-resistant E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603 were used in construction of transconjugants, transformants, and functional cloning experiments. The ampicillin-resistant pHSG575 plasmid was used in functional cloning experiments.
For donor isolates, recipient isolates, and transconjugants/transformants, the minimum inhibitory concentrations (MICs) of ertapenem, cefoxitin, imipenem, ceftriaxone, piperacillin/tazobactam, ceftazidime, ceftazidime/avibactam, meropenem, ceftolozane/tazobactam, cefepime, amikacin, levofloxacin, aztreonam, and imipenem-relebactam were determined by broth microdilution method according to CLSI M100 documents (Institute Clinical and Laboratory Standards, 2015). The MICs were measured only when the quality controls were acceptable. E. coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used as quality controls.
The transferability of pNC225-NDM-29 among isolates was determined using 19NC225 as donor and E. coli DH5α, E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603 as the recipients. The procedure of conjugation was performed according to the protocol previously described (Yang, 2015). Into 40 ml antibiotic-free Luria–Bertani (LB) broth, 100 μl overnight culture of clinical isolate (19NC255) in LB broth containing meropenem (2 μg/ml) and 200 μl overnight culture of recipients DH5α, EC600, ATCC 13883, or ATCC 700603 in LB broth containing rifampin (100 μg/ml) were mixed and were incubated at 37°C for 18 h. Then, the conjugant mixtures were placed on selective LB agar containing 100 μg/ml rifampin and 2 μg/ml meropenem. PCR analysis and agarose gel electrophoresis were performed to confirm the effects of transconjugation.
Transformation was performed through electrophoration. Competent cells of recipients for electrophoration were prepared according to the procedure previously described (Wang R. et al., 2018; Wang Y. et al., 2018). Overnight cultures of recipients (E. coli DH5α, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603) in LB broth were diluted 1:100 into 100 ml LB broth and were incubated at 37°C. The culture was immediately cooled on ice for 20 min once the optical density at 600 nm (OD600) of the cell culture reached 0.5–0.7. Then, the culture was centrifuged at 4,500 rpm for 5 min. The supernatant was discarded, and 15 ml of sterile ice-cold 10% glycerol was pipetted into the cells. The mixture was resuspended gently. The centrifugation and pipetting steps were repeated thrice in all. Finally, the supernatant was discarded, and the cells were resuspended with 1 ml of ice-cold 10% glycerol. All cell-preparing procedures were performed on ice. Competent cells should be stored at –80°C for long-term use.
Electroporation was also conducted according to the procedure previously described (Wang R. et al., 2018; Wang Y. et al., 2018). Fifty microliters of electrocompetent cells, which have been thawed on ice, were mixed with no more than 5 μl plasmid. The mixture was transferred to a prechilled, sterile electroporation cuvette (0.2 cm) and placed in the electroporation apparatus (Bio-Rad Laboratories, Richmond, CA) (parameter setting: 2.5 kV, 200 Ω, and 25 μF). After being pulsed, the cells were recovered in 1 ml antibiotic-free LB broth and incubated at 37°C for 1.5 h before being selected by LB agar plates containing meropenem (2 mg/l). Then, the plates were incubated at 37°C overnight.
To confirm the drug-resistant phenotypes conferred by the suspected resistance gene, blaNDM–29 and blaNDM–1 were ligated into the α-complementation plasmid vector pHSG575 (Takeshita et al., 1987). The recombinant plasmid was introduced into E. coli DH5α, E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603 by electroporation as described before. The transformants were selected on LB agar containing 50 mg/l chloramphenicol, and PCR was conducted to confirm the presence of plasmid pHSG575-NDM-29. Subsequently, antimicrobial susceptibility testing was performed to determine the microbiology phenotype of the transformants. Primers used in the experiments are listed in Supplementary Table 1.
Growth curve was used to assess the fitness impact of plasmid carriage under non-competitive conditions as Wang R. et al., 2018 described previously. The recipients and transconjugants/transformants carrying pNC225/NDM-29 plasmid from 19NC225 isolate were cultured overnight in LB broth containing 2 mg/l meropenem at 37°C. Bacterial suspensions were diluted to a 0.5 McFarland standard and then diluted 1:100 in LB medium (approximately 106 CFU/ml) and grown at 37°C in triplicate for 48 h by EpochTM 2 Microplate Spectrophotometer from BioTek Instruments. The OD600 of each culture was measured every 30 min, and the plates were shaken for 15 s before measuring. The growth curves were estimated by GraphPad Prism version 8 (GraphPad Software, Inc., United States). Statistical significance was defined for an overall error at 0.05 level (95% confidence interval) using one-way analysis of variance (ANOVA) followed by Tukey tests.
Genomic DNA was extracted using UltraClean® Microbial DNA Isolation Kit (MOBIO Laboratories, Inc.). Whole-genome sequencing was implemented using the PacBio Sequel platform. A 20-kb SMRTbell library was prepared from sheared genomic DNA (≥5 g) with an additional bead clean-up step before primer annealing (Zhu et al., 2016).
To correct the polymer errors produced during PacBio sequencing, we re-sequenced this isolate using next-generation sequencing. Paired-end libraries were prepared from 5 μg of isolated genomic DNA using a TruSeq DNA sample prep kit (Illumina Inc., San Diego, California, United States) and sequenced with a read length of 2 × 150 nucleotides using an Illumina Genome Analyzer IIx. Raw Illumina sequencing reads were trimmed at a threshold of 0.01 (Phred score of 20). De novo assembly of the genome was performed using Unicycler (Wick et al., 2017) from short and long sequencing reads.
Genome sequences were annotated with the rapid prokaryotic genome annotation software Prokka (Seemann, 2014). For the assembled plasmids, blastn was first carried out to search the reference plasmids according to the similarity of sequence. The plasmids and its references were submitted to RAST, Glimmer, GeneMarkS, and prodigal for gene prediction, after finding replication origin (repR or repA). The plasmids with rough annotation from Prokka (Seemann, 2014) were added with detailed comments on its CDS and MGE using NCBI, ExPASy, ISFinder, and INTEGRALL. Then, the genome structure of plasmids was compared to analyze their evolutionary relationships. The genome structure comparisons among plasmids were performed to analyze the sequence homology. Plasmid circular structure maps were generated with BRIG software.
Virulence genes were downloaded from virulence factor database (VFDB). FASTA sequences of these genes were used to search for corresponding genes by using BLAST with a coverage of 50% and identity of 90%. Antimicrobial resistance genes were identified using ResFinder 4.0 with the whole genomic sequences.
Sequence types (STs) were determined using SRST2 (Inouye et al., 2014) based on the Illumina reads. Serotype was analyzed using SerotypeFinder 2.0 (Joensen et al., 2015) based on the assembled contigs. In order to obtain the phylogenetic features of 19NC225, the genome sequences of 99 E. coli strains containing blaNDM gene were downloaded from NCBI. The phylogenetic analysis of 19NC225 and 99 downloaded strains was based on the core-genome SNPs detected by MUMmer 3.23 using the assembled genome of 19NC225 (NC_000962) as the reference (Delcher et al., 2002). The MAFFT (Nakamura et al., 2018) was adopted to align the concatenated SNP sequences, and phylogenetic tree was generated by FastTree (Price et al., 2009).
The genome data have been deposited in NCBI with the GenBank accession number CP066844.
The clinical isolate 19NC225 was an E. coli strain isolated from bile of a 65-year-old Chinese male patient with biliary tract infection in Nanchang, Jiangxi, China collected on March 2, 2019. The patient was hospitalized with hepatolithiasis coupled with cholangitis and had a history of diabetes, hypoproteinemia, incursion treatment, operation, and antibiotic use. In the duration of hospitalization, the patient was treated with cefoperazone/sulbactam (3 g, q 12 h, February 19, 2019–February 27, 2019), ornidazole (0.75 g, q 12 h, March 4, 2019–March 5, 2019), and deoxycephalosporin (1 g, q 12 h, March 4, 2019–March 5, 2019). Antimicrobial chemotherapy did not take effect. The pathogen still existed in bile after therapy, and the patient was uncured when discharged from hospital (March 12, 2019).
The susceptibility testing results showed that the clinical isolate 19NC225 was resistant to almost all β-lactams examined, including cephalosporins (cefoxitin, ceftriaxone, ceftazidime, and cefepime), carbapenems (ertapenem, imipenem, and meropenem), combinations of β-lactams and β-lactamase inhibitors (piperacillin/tazobactam, ceftazidime/avibactam, ceftolozane/tazobactam, and imipenem-relebactam), and levofloxacin. The strain was only susceptible to aztreonam and amikacin (Table 1).
In the study, E. coli DH5α, E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603 were used for conjugation of plasmids in 19NC225. Due to the presence of two plasmids (pNC225-NDM-29 and pNC225-TEM1B) in donor 19NC225, we performed agarose gel electrophoresis and PCR to confirm that all the conjugants (E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603) only obtained NDM-29 gene without TEM-1 gene. It confirmed that the plasmid harboring blaNDM–29 in the clinical strain 19NC225 has the potential for horizontal transfer. Considering the low efficiency of the transconjugation of K. pneumoniae ATCC 13883 and K. pneumoniae ATCC 700603, the transformants of ATCC 13883 and ATCC 700603 were constructed through electroporation. The transformants of DH5α are also constructed by electroporation. As shown in Table 1, the antimicrobial susceptibility of transformants was similar with that of corresponding transconjugants. Compared to the plasmid-free counterparts, plasmid pNC225-NDM-29 reduced the susceptibility of transconjugants/transformants to almost all β-lactams examined. Except for ATCC 700603 that was resistant to cefoxitin, ceftriaxone, ceftazidime, piperacillin/tazobactam, and aztreonam naturally, the susceptibility results of all transformants were consistent with the donor (clinical isolate 19NC225): resistant to cephalosporins, carbapenems, combinations of penicillin and β-lactamase inhibitors, and levofloxacin.
To determine the specific function of carbapenemase NDM-29, the genes blaNDM–29 and blaNDM–1 were cloned into pHSG575 (pHSG575/NDM-29 and pHSG575/NDM-1), and the corresponding transconjugants/transformants were detected to determine their antimicrobial susceptibility. As shown in Table 2, compared to the transformants with the plasmid pHSG575, transformants with plasmid pHSG575-NDM-29 showed higher resistance to antibiotics including cephalosporins, carbapenems, and combinations of penicillin and β-lactamase inhibitors. The transformants with plasmid pHSG575/NDM-29 showed no difference in susceptibility to the drugs detected with the transformants with plasmid pHSG575/NDM-1. It is worth noticing that transformants with cloned plasmid pHSG575/NDM-29 maintained the susceptibility to levofloxacin, while transformants with plasmid pNC225/NDM-29 showed an increased resistance to levofloxacin, which suggested that the drug resistance gene for levofloxacin is located on the plasmid pNC225-NDM-29, which was in accordance with the WGS analysis as the gene qnrS1 can imply quinolone resistance.
To future analyze the impact of the plasmid to the survival of strains, fitness cost assay was conducted, and growth curve is shown in Figure 1. The growth rates of transconjugants/transformants showed no difference compared to their donor strains. No significant difference in growth rates was found within pairs (p > 0.05). Therefore, the antibiotics resistance mediated by pNC225-NDM-29 did not increase the growth fitness cost of the resistant strains compared to its susceptible counterparts.
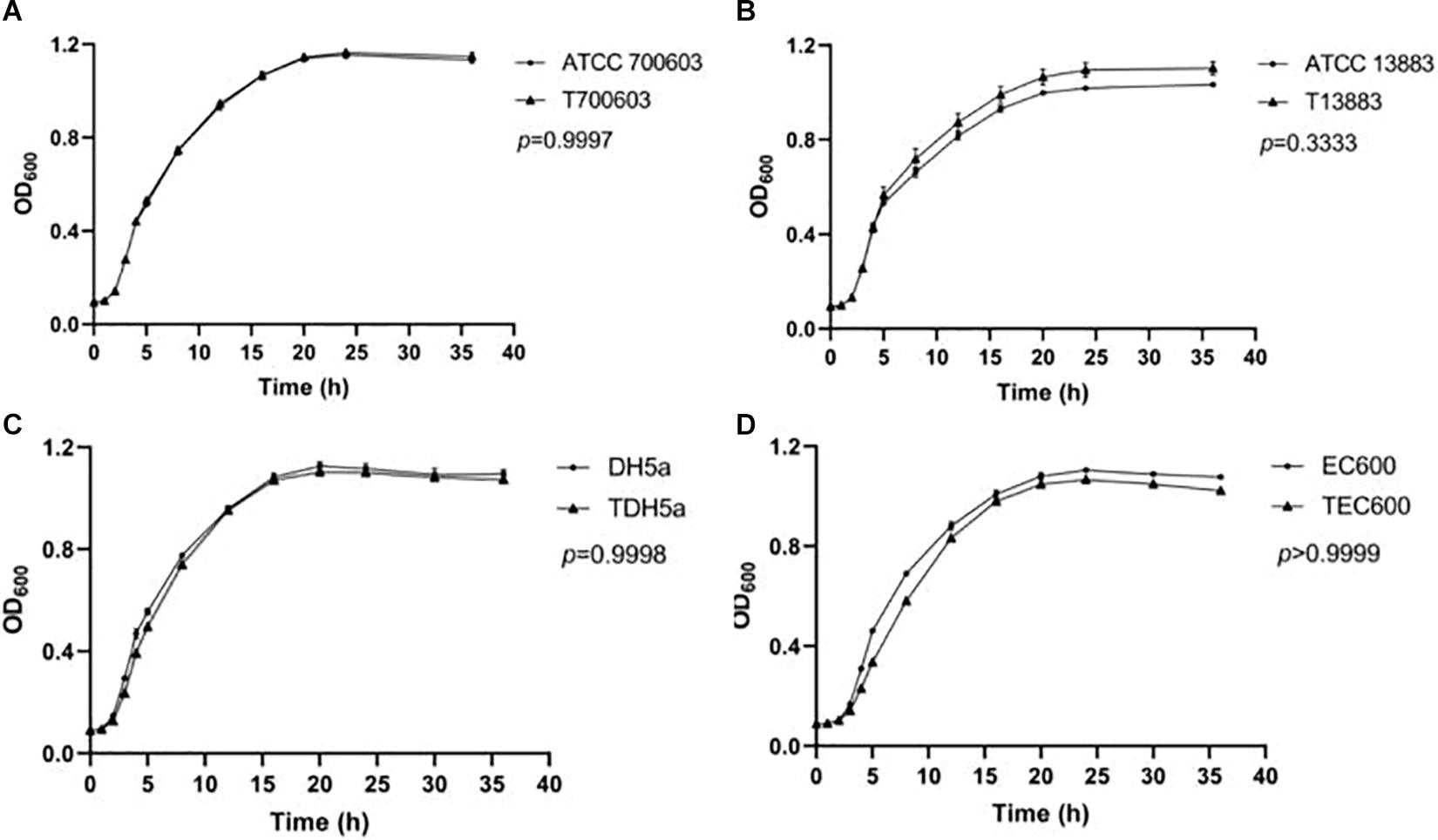
Figure 1. Growth curve of transformants containing pNC225-NDM-29. One-way analysis of variance (ANOVA) followed by Tukey tests was used to define the statistical significance. P > 0.05 indicates no fitness cost was observed within pairs. (A) Growth curve of ATCC 700603 and corresponding transformant containing pNC225-NDM-29. (B) Growth curve of ATCC 13883 and corresponding transformant containing pNC225-NDM-29. (C) Growth curve of DH5a and corresponding transformant containing pNC225-NDM-29. (D) Growth curve of EG600 and corresponding transformant containing pNC225-NDM-29. T700603, transformant of ATCC 700603; T13883, transformant of ATCC 13883; TDH5α, transformant of DH5α; TEC600, transformant of EC600.
To reveal the genetic basis of the multidrug-resistant (MDR) phenotype, we obtained the complete genome of the clinical isolate 19NC225 including a 4.8-Mb chromosome, a 128-kb plasmid (pNC225-TEM1B), and a 60-kb plasmid (pNC225-NDM-29) (Table 3 and Figure 2). Bioinformatics analysis provided the general chromosome genome information, including GC% content (50.63%), and predicted protein coding genes (4,576), average gene length (938 bp), and coding region (89.39%). The two plasmids possess a higher GC content (∼52%), lower ratio of coding regions, and shorter average gene length. The phylogenetic analysis (Figure 3) showed that 19NC225, belonging to ST1485 and O83:H42, is clustered together with E. coli strain MS6198 from Australia (Hancock et al., 2017), which contains a blaNDM–1-positive IncA/C plasmid.
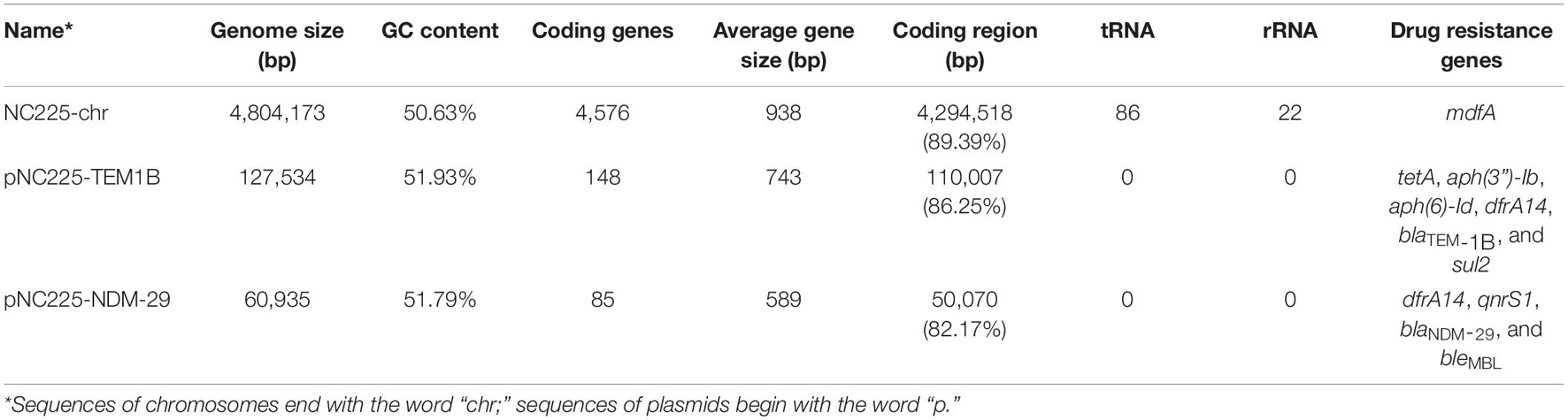
Table 3. The genomic characteristics and antimicrobial resistance genotype analysis of the sequences of chromosomes and plasmids of 19NC225.
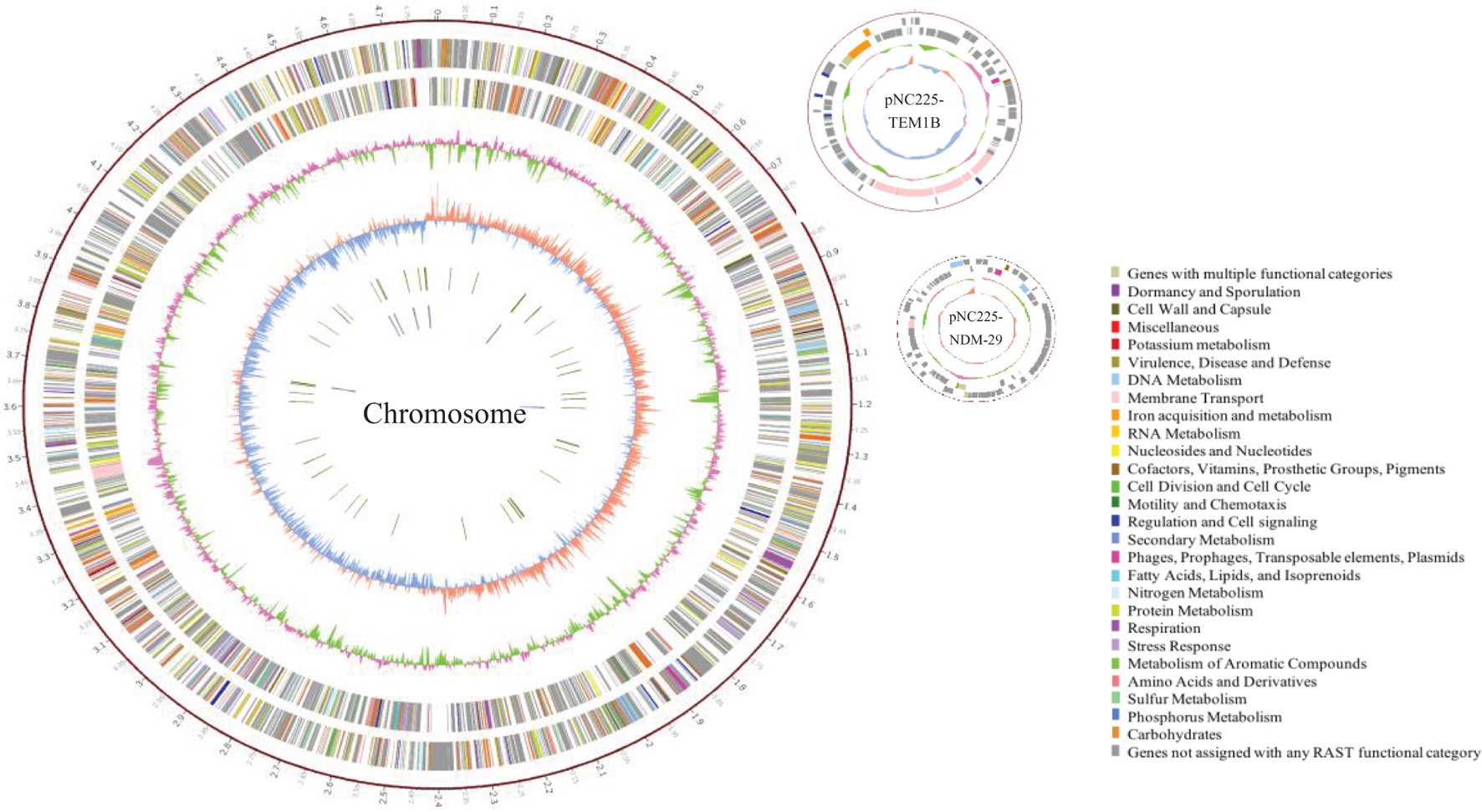
Figure 2. Circular representation of 19NC225 genome (chromosome and two plasmids). The complete genome of the clinical isolate 19NC225 includes a 4,804,173-bp chromosome, a 127,534-bp plasmid (pNC225-TEM1B), and a 60,935-bp plasmid (pNC225-NDM-29). Circles are shown as follows (outside to inside): (1) A physical map scaled in megabases (Mb) from base 1 (the start of the putative replication origin); (2) coding sequences transcribed in a clockwise direction; (3) coding sequences transcribed in a counter-clockwise direction; (4) G + C content; (5) GC skew; (6) tRNA genes; and (7) rRNA genes. Genes displayed in (2) and (3) are color-coded according to different functional categories shown on the right.
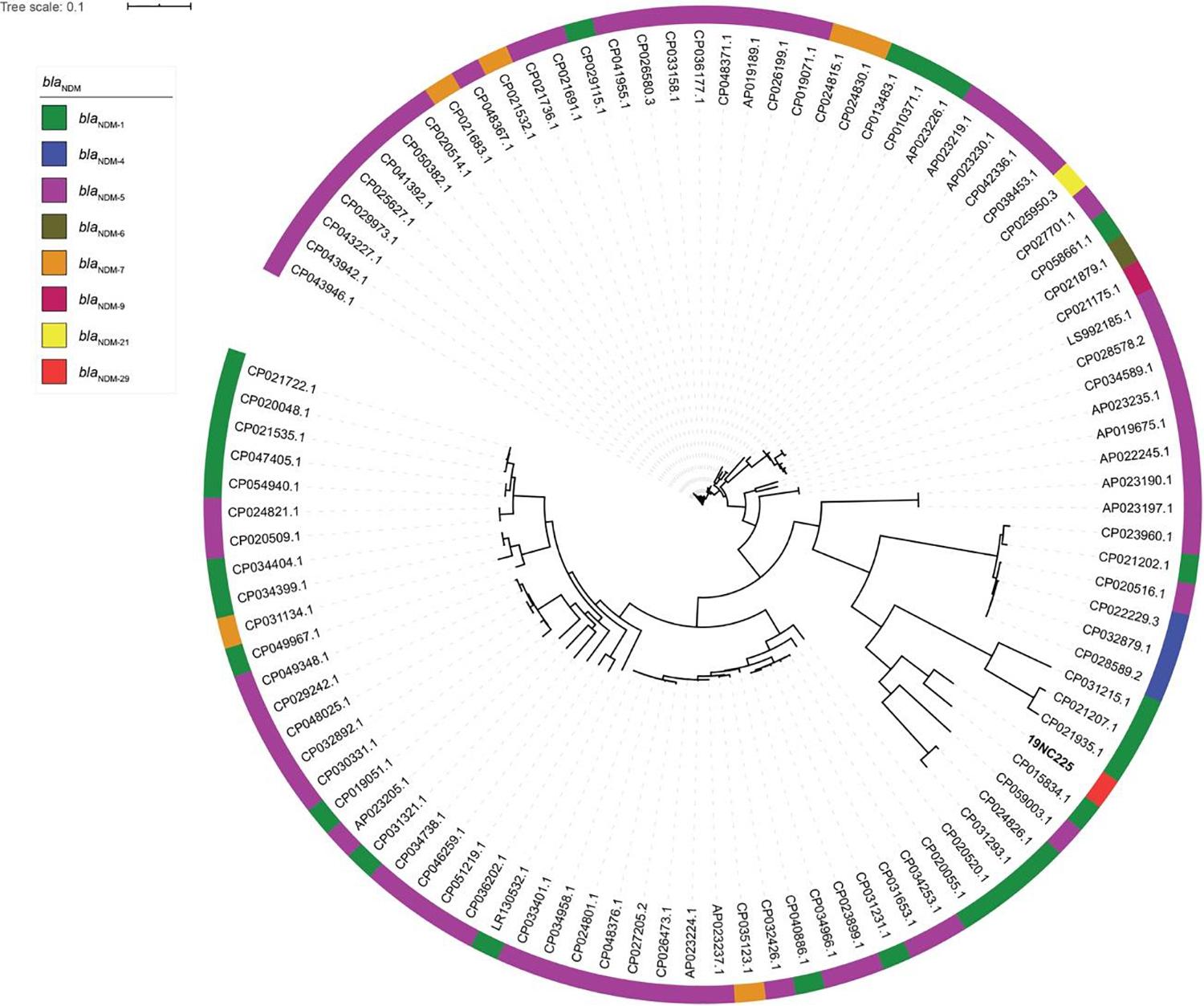
Figure 3. Phylogenic tree of E. coli strains with complete genomes containing blaNDM variants. The colored strips on the outside ring indicate the eight blaNDM variants.
Virulence genes (Supplementary Table 2) analysis showed that the clinical isolate 19NC225 contains 235 virulence genes, 12 of which are located on the 128-kb plasmid (pNC225-TEM1B), including aerobactin synthesis and transport protein (iutA, iucABCD, and ECVR50_3327), ferrous iron transport proteins (sitABCD), and aaiQR. No virulence genes were found on the 60-kb plasmid (pNC225-NDM-29).
Nine categories of antimicrobial-resistance genes were discovered in 19NC225 including one antimicrobial resistance gene on the chromosome: multidrug transporter mdfA, an efflux pump driven by the proton motive force that confers resistance to a broad spectrum of chemically unrelated drugs; six antimicrobial resistance genes on the 128-kb plasmid (pNC225-TEM1B): a tetracycline resistance gene (tetA), two aminoglycoside resistance genes [aph(3″)-Ib and aph(6)-Id], one sulfonamide resistance gene (sul2), one trimethoprim resistance gene (dfrA14), and one β-lactamase gene (blaTEM–1B); four antimicrobial resistance genes on the 60-kb plasmid (pNC225-NDM-29): one quinolone resistance gene (qnrS1), one trimethoprim resistance gene (dfrA14), one bleomycin resistance gene (bleMBL), and importantly one carbapenemase resistance gene blaNDM–29 (a very rare subtype of blaNDM, that will be discussed in detail in subsequent sections, Table 2). These antimicrobial-resistance genes, especially blaNDM–29, are responsible for the MDR phenotype, including most β-lactam drugs, which is in accordance with antimicrobial susceptibility testing.
Antimicrobial-resistance genes analysis showed a blaNDM gene located on the 60-kb plasmid (pNC225-NDM-29). Sequence alignment presented that this blaNDM gene shares 100% identity with blaNDM–29 [carrying a G388A (D130N) mutation compared to blaNDM–1] (Supplementary Figure 1), a very rare subtype of blaNDM found in K. pneumoniae. This is the first report of this subtype in E. coli in the world.
The predicted model of NDM-29 was generated by SWISS-MODEL according to homology modeling using NDM-1 as a template (PDB accession no. 4EY2) (Figure 4). There is a metal binding at the position 124, which is near the mutation site (D130N, Berman et al., 2000).
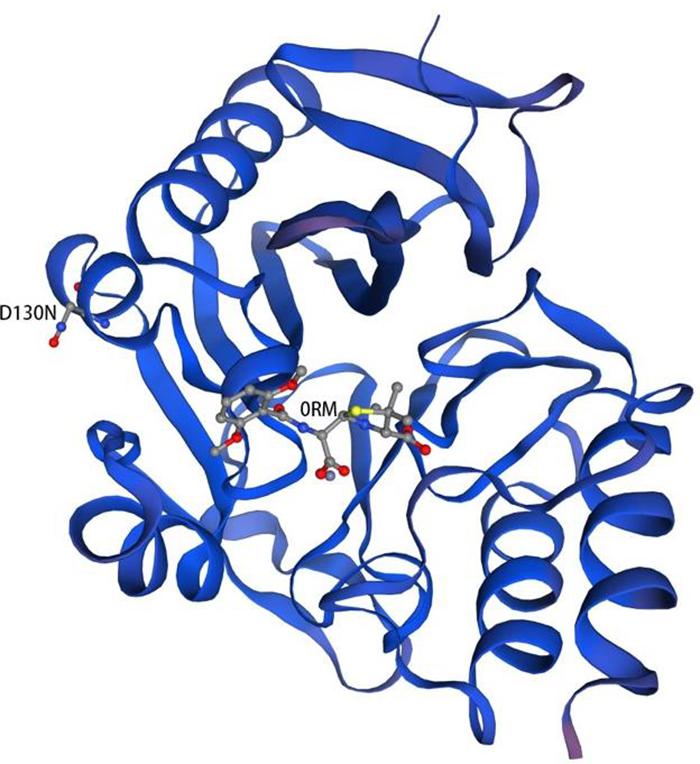
Figure 4. Homology model of NDM-29. Protein backbone of NDM-29 was shown with the helices and strands. 0RM ((2R,4S)-2-{(R)-carboxy[(2,6-dimethoxybenzoyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid) was bound to NDM-29 protein. The mutation (D130N) was labeled on the left helix (Guex et al., 2009; Bertoni et al., 2017; Bienert et al., 2017; Waterhouse et al., 2018; Studer et al., 2020).
The complete nucleotide sequence of the plasmid pNC225-NDM-29 carrying blaNDM–29 was 60,935 bp in length, constituting a circular DNA with an average G + C content of 51.79%. Eighty-five open reading frames were annotated (Table 2) because it contained an IncN-type repA (plasmid replication initiation) gene (D130N).
pNC225-NDM-29 was assigned to the IncN group. We further performed a full-plasmid BLAST comparative analysis. The result showed that pNC225-NDM-29 exhibited 99% identity with six blaNDM containing plasmids: four plasmids of K. pneumoniae (pSCH6109-NDM, pNDM1_LL34, pC2414-3-NDM, and CP040178.1_p3), one plasmid of Enterobacter cloacae (pNDM1-CBG), and one plasmid of E. coli (pNDM-BTR), with high coverages (99, 99, 99,100, 99, and 100%, respectively, Figure 5). These six plasmids are blaNDM–1-carrying ones. A linear genomic comparison was further conducted between pNC225-NDM-29 and pNDM-BTR (blaNDM–1-carrying plasmid) in E. coli (Figure 6A) since they share high identity and both belong to E. coli. The backbone region and accessory modules containing dfrA14 show high similarity to that of pNDM-BTR with 99.96% identity and 100% coverage, indicating the high-level conservation of this IncN backbone. However, there is an inversion of the accessory modules containing blaNDM–29 compared with the blaNDM–1 region of pNDM-BTR. As shown in Figure 6B, the region harboring blaNDM–29 is located in an ISKpn19-based transposon, with two ISKpn19 elements at terminal regions (in opposite directions). This region also contained a bleomycin resistance protein (bleMBL), trpF, dsbC, cutA, groS, and groL. Additionally, two Tn6292 remnants, carrying quinolone resistance gene qnrS1, are symmetrically located upstream and downstream of this ISKpn19-based transposon. Moreover, the whole region containing the above elements is located inside two opposite-directed IS26 elements. This provided evidence of insertion of this region into the backbone of the plasmid, based on IS26- or ISkpn19-mediated intermolecular replicative transposition.
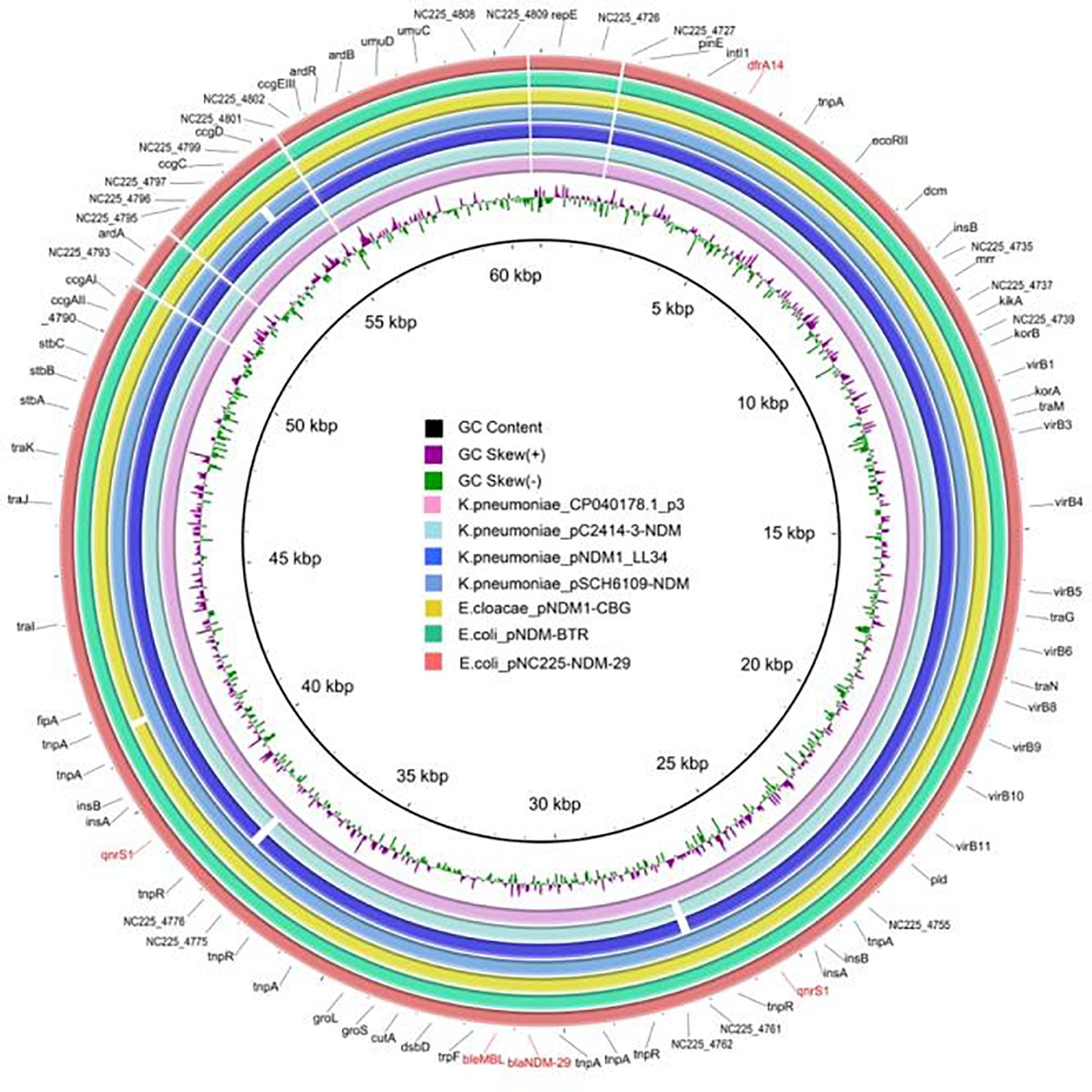
Figure 5. Genetic structure of pNC225-NDM-29 and six blaNDM–1-harboring plasmids (pSCH6109-NDM, pNDM1_LL34, pC2414-3-NDM, and CP040178.1_p3 from Klebsiella pneumoniae; pNDM1-CBG from Enterobacter cloacae; and pNDM-BTR from Escherichia coli). Alignments of resemble plasmids are shown as concentric rings. The outermost shows the main coding genes of pNC225-NDM-29. Antimicrobial-resistance genes are highlighted in red.
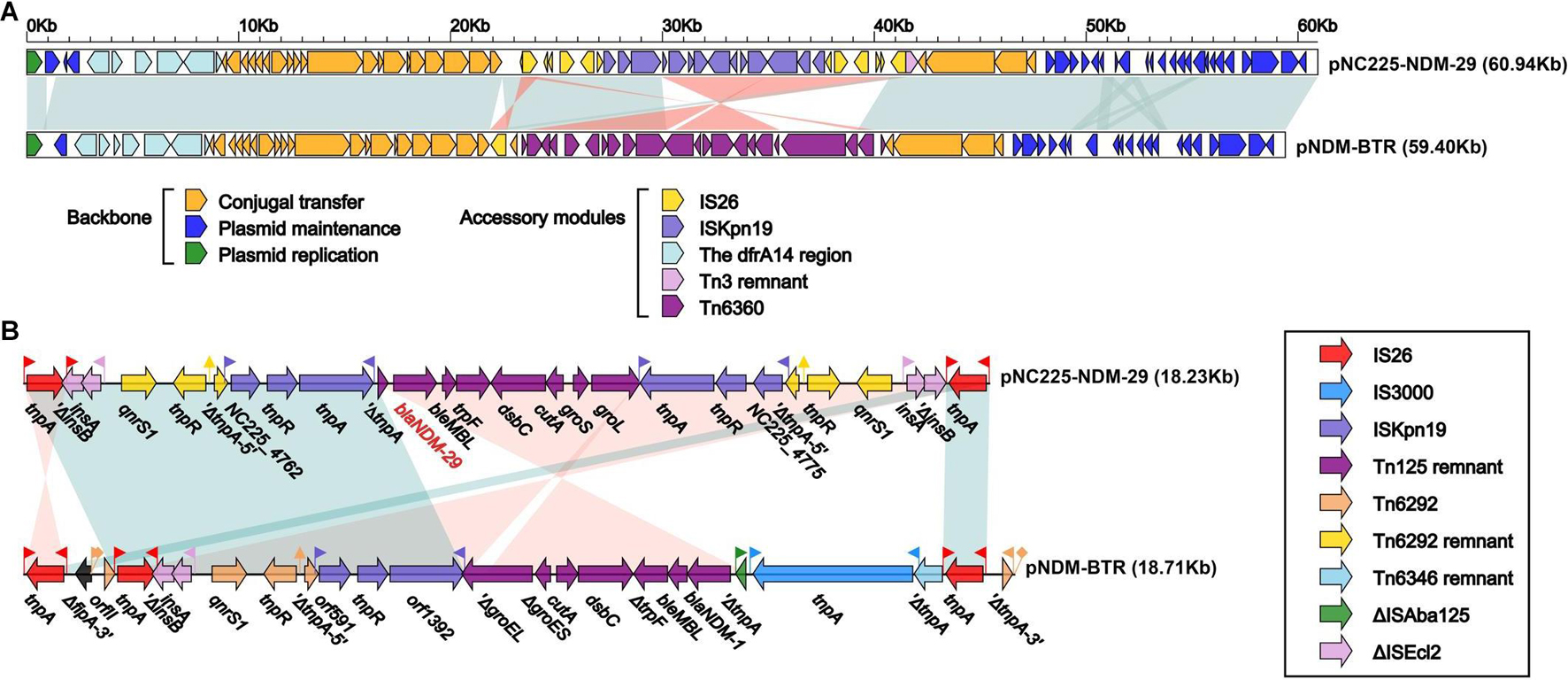
Figure 6. Structure of pNC225-NDM-29 and related transposon. (A) Linear comparison of plasmid pNC225-NDM-29 with the most similar plasmid pNDM-BTR. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. Shading denotes the regions with high homology (95% nucleotide identity). (B) The linear genomic comparison of the drug resistance region in plasmids between pNC225-NDM-29 and pNDM-BTR. The whole region containing main functional areas of pNC225-NDM-29 is located inside two opposite-directed IS26 elements, formatting an IS26-based transposon structured as IS16-ΔISEcl2-qnrS1-Tn6292 remnant-ISKpn19-blaNDM–29-bleMBL-trpF-dsbC-cutA-groS-groL-ISKpn19-Tn6292-remnant-qnrS1-ΔISEcl2-IS16.
Here we described a newly found carbapenemase, NDM-29, isolated from a clinical strain of E. coli. The success of conjugation from clinical isolation to E. coli EC600, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 700603 indicated the transferability of pNC225-NDM-29. The presence of the ISKpn19-based transposon where the region harboring blaNDM–29 is located suggested the risk of the spread of resistance caused by NDM-29. Meanwhile, no fitness cost was observed in the transconjugants/transformants containing plasmid pNC225-NDM-29, which demonstrated a limited burden from the transferable plasmid. However, the conjugation from donor to recipients (ATCC 13883, ATCC 700603, and DH5α) showed low efficiency, which may be due to high energy cost or multibarrier in recipients (Llosa et al., 2002). Although the growth curves did not show a significant impact of transferred plasmid on recipients, a limitation of growth curve that the competitive fitness of donor over recipients was not estimated and needs further study (Hanafi et al., 2016). Anyhow, the dissemination and adaptability of the NDM-29-harboring plasmid among clinical bacteria would be a threat to infectious control.
The pNC225-NDM-29 shows high identity with six NDM-1-carrying plasmids (≥ 99%). The highest identity (99.96%) appears between pNC225-NDM-29 and pNDM-BTR, an NDM-1-encoding plasmid from an E. coli strain BTR isolated from the urine specimen of an 89-year-old female patient with chronic obstructive pulmonary disease in Beijing in 2013 (Zhao et al., 2017). Considering the transposon near NDM-29, we tend to believe NDM-29 is likely a mutation of NDM-1 produced during propagation. This suggests some genomic functions such as high transmission capacity and pathogenicity, and influence on patient prognosis of blaNDM–29 may be similar to blaNDM–1. Whether the mutation will cause the change of carbapenem resistance needs further validation. The prevalence of NDM-29 existing in animals also needs more surveillance and research.
Currently, the mutation of D130G has been reported in NDM-14, and kinetic analysis indicated that NDM-14 has greater carbapenem resistance with a higher affinity for imipenem and meropenem (Zou et al., 2015). Based on our research, there is no difference between NDM-1 and NDM-29 in terms of susceptibility to carbapenems, which is in accordance with the findings of Starkova et al. (2021).
In conclusion, we report the identification of a novel class B enzyme with carbapenemase activity, NDM-29, in a clinical E. coli isolate. The novel blaNDM–29 is first detected in China and was obtained from a MDR E. coli strain isolated from bile of a patient with biliary tract infection. The strain, containing two plasmids (pNC225-TEM1B and pNC225-NDM-29), belongs to ST1485 and O83:H42, showed homology with E. coli MS6198 from Australian (Hancock et al., 2017), which harbors blaNDM–1. The plasmid, pNC225-NDM-29, which encods blaNDM–29, exhibited 99% identity with six blaNDM–1-carrying plasmids, especially a IncN1 plasmid pNDM-BTR from an E. coli in urine specimen (99.96% identity and 100% coverage), and showed responsibility for the MDR phenotype.
Recently, the reports of new β-lactamase genes have increased, especially those with carbapenemase activity. Because of the travel of infected or colonized individuals between countries, NDM-1 became a global epidemic in fewer than 5 years since the first discovery (Bush and Bradford, 2020). With more and more NDM variants found in animals, we further need to attach great importance to the threat of antimicrobial resistance from food. Thus, the research of the original newly discovered variants is valuable for surveillance of resistance outbreak and the genetic evolution of bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP066844.
The Human Research Ethics Committee of Peking Union Medical College Hospital approved this study and waived the need for consent (Ethics Approval Number: S-K238).
YZ and XJ wrote the manuscript. PJ, XL, and QY revised the manuscript. YZ, XJ, PJ, and XL performed the experiments. QY conceived and designed the study. All authors contributed to the article and approved the submitted version.
This study was sponsored by funding from the National Natural Science Foundation of China (No. 82072318) and Beijing Key Clinical Specialty for Laboratory Medicine—Excellent Project (No. ZK201000). The funders were not involved in the study design, collection, analysis, interpretation of data, writing of this article, or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Jianhua Liu from the College of Veterinary Medicine, South China Agricultural University, for providing the plasmid, pHSG575, as a gift. We are also extremely grateful to Longhua Hu from the Clinical Laboratory of The Second Affiliated Hospital of Nanchang University for providing the clinical strain 19NC225.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.743981/full#supplementary-material
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. doi: 10.1093/nar/28.1.235
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., and Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7:10480. doi: 10.1038/s41598-017-09654-8
Bienert, S., Waterhouse, A., de Beer, T. A., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45, D313–D319. doi: 10.1093/nar/gkw1132
Bush, K., and Bradford, P. A. (2020). Epidemiology of beta-lactamase-producing pathogens. Clin. Microbiol. Rev. 33:e00047-19. doi: 10.1128/CMR.00047-19
Delcher, A. L., Phillippy, A., Carlton, J., and Salzberg, S. L. (2002). Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30, 2478–2483. doi: 10.1093/nar/30.11.2478
Dever, L. A., and Dermody, T. S. (1991). Mechanisms of bacterial resistance to antibiotics. Arch. Intern. Med. 151, 886–895. doi: 10.1001/archinte.1991.00400050040010
Gamal, D., Egea, P., Elías, C., Fernández-Martínez, M., Causse, M., Pérez-Nadales, E., et al. (2020). High-risk clones and novel sequence type ST4497 of Klebsiella pneumoniae clinical isolates producing different alleles of NDM-type and other carbapenemases from a single tertiary-care centre in Egypt. Int. J. Antimicrob. Agents 56:106164. doi: 10.1016/j.ijantimicag.2020.106164
Guex, N., Peitsch, M. C., and Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl. 1), S162–S173. doi: 10.1002/elps.200900140
Hanafi, A., Lee, W. C., Loke, M. F., Teh, X., Shaari, A., Dinarvand, M., et al. (2016). Molecular and proteomic analysis of levofloxacin and metronidazole resistant Helicobacter pylori. Front. Microbiol. 7:2015. doi: 10.3389/fmicb.2016.02015
Hancock, S. J., Phan, M. D., Peters, K. M., Forde, B. M., Chong, T. M., Yin, W. F., et al. (2017). Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents Chemother. 61:e01740–e01716. doi: 10.1128/AAC.01740-16
Inouye, M., Dashnow, H., Raven, L. A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90. doi: 10.1186/s13073-014-0090-6
Institute Clinical and Laboratory Standards (2015). Methods for Dilution Antimicrobial Susceptibility Test for Bacteria that Grow Aerobically Document, 10th Edn. Philadelphia, PA: Clinical and Laboratory Standards Institute.
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi Metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Liu, Z., Li, J., Wang, X., Liu, D., Ke, Y., Wang, Y., et al. (2018). Novel variant of New Delhi Metallo-beta-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 9:248. doi: 10.3389/fmicb.2018.00248
Liu, Z., Wang, Y., Walsh, T. R., Liu, D., Shen, Z., Zhang, R., et al. (2017). Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 61:e02233-16. doi: 10.1128/AAC.02233-16
Llosa, M., Gomis-Rüth, F. X., Coll, M., and de la Cruz Fd, F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8. doi: 10.1046/j.1365-2958.2002.03014.x
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Starkova, P., Lazareva, I., Avdeeva, A., Sulian, O., Likholetova, D., Ageevets, V., et al. (2021). Emergence of hybrid resistance and virulence plasmids harboring New Delhi metallo-beta-lactamase in Klebsiella pneumoniae in Russia. Antibiotics 10:691. doi: 10.3390/antibiotics10060691
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, R., Haas, J., and Schwede, T. (2020). QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. doi: 10.1093/bioinformatics/btz828
Takeshita, S., Sato, M., Toba, M., Masahashi, W., and Hashimoto-Gotoh, T. (1987). High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Genes 61, 63–74. doi: 10.1016/0378-1119(87)90365-9
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Wang, R., Liu, Y., Zhang, Q., Jin, L., Wang, Q., Zhang, Y., et al. (2018). The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents 51, 739–744. doi: 10.1016/j.ijantimicag.2018.01.023
Wang, Y., Wang, S., Chen, W., Song, L., Zhang, Y., Shen, Z., et al. (2018). CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 84:e01834-18. doi: 10.1128/AEM.01834-18
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yang, Q. (2015). The Epidemiology and Resistance Mechanisms of Carbapenem-Resistant Enterobacteriaceae in China. Ph. D. thesis. Beijing: Peking Union Medical College.
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhao, Y., Wang, L., Zhang, Z., Feng, J., Kang, H., Fang, L., et al. (2017). Structural genomics of pNDM-BTR harboring In191 and Tn6360, and other bla NDM-carrying IncN1 plasmids. Future Microbiol. 12, 1271–1281. doi: 10.2217/fmb-2017-0067
Zhu, L., Zhong, J., Jia, X., Liu, G., Kang, Y., Dong, M., et al. (2016). Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 44, 730–743. doi: 10.1093/nar/gkv1498
Keywords: carbapenemase, NDM, Escherichia coli, whole-genome sequence, antimicrobial resistance
Citation: Zhu Y, Jia X, Jia P, Li X and Yang Q (2021) Genetic and Phenotypic Characterization of the Novel Metallo-β-Lactamase NDM-29 From Escherichia coli. Front. Microbiol. 12:743981. doi: 10.3389/fmicb.2021.743981
Received: 19 July 2021; Accepted: 02 September 2021;
Published: 29 September 2021.
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomReviewed by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesCopyright © 2021 Zhu, Jia, Jia, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Yang, eWFuZ3Fpd2VuODFAdmlwLjE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.