
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 October 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.743831
This article is part of the Research Topic Diversity of Beetles and Associated Microorganisms View all 22 articles
The gut microflora of insects plays important roles throughout their lives. Different foods and geographic locations change gut bacterial communities. The invasive wood-borer Agrilus mali causes extensive mortality of wild apple, Malus sieversii, which is considered a progenitor of all cultivated apples, in Tianshan forests. Recent analysis showed that the gut microbiota of larvae collected from Tianshan forests showed rich bacterial diversity but the absence of fungal species. In this study, we explored the antagonistic ability of the gut bacteria to address this absence of fungi in the larval gut. The results demonstrated that the gut bacteria were able to selectively inhibit wild apple tree-associated fungi. Among them, Pseudomonas synxantha showed strong antagonistic ability, producing antifungal compounds. Using different analytical methods, such as column chromatography, mass spectrometry, HPLC, and NMR, an antifungal compound, phenazine-1-carboxylic acid (PCA), was identified. Activity of the compound was determined by the minimum inhibitory concentration method and electron microscopy. Moreover, our study showed that the gut bacteria could originate from noninfested apple microflora during infestation. Overall, the results showed that in newly invaded locations, A. mali larvae changed their gut microbiota and adopted new gut bacteria that prevented fungal colonization in the gut.
Insects have diverse bacterial associations including relationships ranging from parasitism to mutualism (Douglas, 2015). There are a number of studies on the gut microbiota of wood-boring beetles, due to its essential roles in food digestion, compensation for dietary deficiencies, compound detoxification, nutrient production, and more, in the literature (Egert et al., 2003; Douglas, 2015). In addition, insects have a very close relationship with their gut microbiome, and symbiotic interactions can result in host survival under extreme environmental conditions (Gupta and Nair, 2020). There is an unexplored question in invasion ecology, and a study highlighted by Pennisi (2017) raised a new hypothesis: Could the gut microbiome determine the invasion success of phytophagous insects? (Lefort et al., 2017). Recent reports support the crucial role of the gut microflora in insect growth and development and in environmental adaptation (Bahrndorff et al., 2016; Lefort et al., 2017). Gut-associated microorganisms such as bacteria, fungi, protozoa, and viruses can be transiently or permanently transmitted to insects, and this relationship can be either beneficial or harmful (Feldhaar, 2011; Hammer et al., 2017). For example, the gut microflora has a symbiotic relationship with hosts to obtain nutrients, aid digestion, and promote host immunity by protecting against pathogens (Engel and Moran, 2013; Douglas, 2015; Mereghetti et al., 2017). Therefore, studying the microbiome of the insect digestive tract is essential to improve beneficial insect digestive capabilities or pest management programs (Morales-Jiménez et al., 2012).
The interaction between fungi and bacteria in the invasive insect gut has not been thoroughly studied. Fungi and bacteria are frequent in most wood-feeding insect guts and likely play a role in the digestion of plant-cell polymers due to their symbiotic interaction with the host. Microbial composition and abundance in the insect gut vary; wood-feeding insects have a rich bacterial diversity, whereas sap-feeding insects have poor diversity (Engel and Moran, 2013) due to insect gut compartmentalization (Dillon and Dillon, 2004). The bacterial composition of the gut could either be specifically adapted to the gut, maternally transmitted, or acquired from the environment (Ruby et al., 2004). Additionally, the microbial diversity of the gut also depends on the geographic location, which has been shown for honeybees (Yun et al., 2014). The composition of the myco- and microflora of the larval gut are dependent on the developmental stage of the insect and the location, as was shown for the citrus pest Bactrocera minax (Yao et al., 2019). However, during larval growth, the gut bacteria may influence fungal diversity (Yun et al., 2014; Sam et al., 2017; Yao et al., 2019). Moreover, gut bacteria can compete with fungi during gut colonization (Sam et al., 2017).
Wild apple Malus sieversii (Ledeb.) Roem. is a species native to Tianshan forests and usually found in sub-mountain areas (Lin and Lin, 2000), and it is considered the primary progenitor of all cultivated apples (Richards et al., 2009). Because of its rich genetic diversity, it remains a globally significant genetic resource. Unfortunately, wild apple faces extensive abiotic and biotic stresses. Among them, the invasive insect, a wood-borer Agrilus mali (Coleoptera: Buprestidae) has heavily attacked trees since its first detection in the early 1990s and has extensively damaged the wild apple forests of Tianshan (Ji et al., 2004; Bozorov et al., 2019a); since then, 40% of the forest area has been damaged (Wang, 2013). Mainly, mortality of tree is caused by the larval stage of insect that feed cambium, phloem, and outer xylem parts resulting in serpentine galleries that prevent nutrient movement (Ji et al., 2004; Bozorov et al., 2019a). In situ or ex situ conservation of wild apple via propagation nurseries and the development of biotechnological tools to address this problem are necessary.
In our recent study, we showed the microbial diversity of the larval gut of invasive A. mali collected from a Tianshan wild apple forest using throughput sequencing and microbiological approaches to detect fungal species. However, the analysis demonstrated the absence of fungal species (Bozorov et al., 2019b). Recently, Zhang et al. (2018) demonstrated that both bacterial and fungal species colonized adult and larval guts of A. mali (collected from Ili Kazakh autonomous prefecture, Xinjiang Province, China) when fed different apple species in a laboratory condition. In this study, we explored the origin of A. mali larval gut bacteria and their antagonistic interaction with wild apple-associated fungi using microbiological, physiological, and analytical chemistry approaches in the Tianshan Mountain Forest. We hypothesized that the larvae-acquired gut bacteria compete with fungi for colonization in invaded locations. Environmentally acquired bacteria may contribute to shaping the gut microbiome of this important agricultural pest.
The wild apple twigs of randomly chosen trees were collected from the forest in Mohe Village (43°51N, 82°15W), Gongliu County, Ili-Kazakh District, Xinjiang-Uyghur Autonomous Province, China, which is located in the Ili Valley of the Tianshan Mountains. Since insect larvae are specialized in feeding in the phloem/cambium part and rarely in the outer xylem twig, each tree uninfested, larvae-infested, dead twigs, and as well as larval frass were randomly collected. For this purpose, if more than two fourth to fifth instar larvae were found in 50-sm twig (often 1–5 sm in diameter), then it was considered as infested. Moreover, it also considered the healthiness of the infested twig if the upper part of the infested twigs shows more than 50% of dryness. Often, appearance of cracks and coloration change in the bark indicate the presence of larva under it. For twigs that are dried completely but are still on the tree, we considered them as dead twigs. Larval frass was collected from the respective collected infested twig.
The gut bacterial species [Pseudomonas synxantha (#283), Ps. orientalis (#24), Erwinia billingiae (#32), E. persicina (#12), and four Pantoea species strains (#2, 43, 153, 287)] were used from the bacterial cryopreserved stock from the previous study (Bozorov et al., 2019b). The bacteria were grown in Luria–Bertani broth medium (Sigma).
Five replicates of uninfested, infested, and dead wild apple twigs, as well as larval frass were used to isolate fungi and bacteria. Larval frass was collected from serpentine galleries made by larvae after removal of the twig bark. Next, healthy, infested, and dead twigs with xylem, bark, and frass were ground with a home blender in sterile conditions. Ground tissues were placed on the respective PDA medium (potato starch 4 gm L–1, dextrose 20 gm L–1, and agar 15 gm L–1, pH 5.6) (Potato Dextrose Agar, Solarbio, P8931-250G) and nutrient agar (NA) (0.5% peptone, 0.3% beef extract, 1.5% agar, pH 6.8) (Difco, France) for fungi and bacteria isolation, and incubated at 25°C. Single colonies were isolated and re-cultivated to classify their morphological features.
Fungal isolates were cultivated on PDA medium for 7–14 days upon sufficient production of mycelia. Mini-preparation of fungal DNA method (Liu et al., 2000) with minor modification was used to extract the fungal DNA. Briefly, the cell walls of fungi mycelia were ground with mortar and pestle in the presence of liquid nitrogen. One milliliter of lysis buffer [400 mM Tris-HCl (pH 8.0), 60 mM EDTA (pH 8.0), 150 mM NaCl, 1% sodium dodecyl sulfate] was added into the fine powdered fungal mycelia. Next, the mix was transferred into a 2-ml Eppendorf tube, and thoroughly mixed, and left at room temperature for 10 min. Then, 0.3 ml of potassium acetate (pH 4.8; which is made of 60 ml of 5 M potassium acetate, 11.5 ml of glacial acetic acid, and 28.5 ml of distilled water) was added, vortexed briefly, and centrifuged at 10,000 × g for 1 min. The supernatant was transferred into a new 2-ml Eppendorf tube, and an equal volume of isopropyl alcohol was added and mixed by inversion. Tubes were centrifuged at 10,000 g for 2 min, and the supernatant was discarded. Pellet was washed with 0.3 ml of 70% ethanol and was spun at 10,000 × g for 1 min, then the supernatant was discarded. The DNA pellet was air dried and dissolved in 50 ml of 1 × Tris-EDTA buffer. Bacteria DNA extraction was carried out following our earlier study (Bozorov et al., 2019b).
For apple-associated fungi isolate identification, the internal transcribed spacer (ITS) region was amplified using primer pairs ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). Amplifications were performed in a total volume of 50 μl containing 10 μl of PrimeSTAR HS (Premix) (Takara, Japan) with an appropriate concentration of dNTPs (0.2 mM) and Taq polymerase (5 U), 1 μl (0.2 μM) of each primer, and 1 μl of diluted DNA. The PCR conditions included 5 min at 95°C for the initial step followed by 35 cycles at 94°C for 15 s (denaturation), 55°C for 30 s (annealing), and 72°C for 2 min (elongation), with a final extension at 72°C for 10 min. PCR products were visualized on a 1.0% agarose gel. PCR products were sequenced bidirectionally with the Sanger method in Beijing Genomics Institute (Shenzhen, China).
PCR amplification of 20-fold-diluted bacterial DNA was performed on a Veriti thermocycler (Applied Biosystems, United States) using forward 27F 5′-AGAGTTTGATCATGG CTCAG-3′ and reverse 1492R 5′-TACGGCTACCTTGTTA CGACTT-3′ primers (Broderick et al., 2004). PCR reaction and condition were performed following our earlier study (Bozorov et al., 2019b).
Sequences were assembled using SeqMan (DNASTAR Lasergene 7). Sequences of ITS and 16S RNA were compared with respective other orthologous fungal and bacterial sequences deposited in GenBank using the BLASTN algorithm. Representative OTUs and sequences from the Sanger method were aligned with CLUSTALW. Maximum likelihood (ML) phylogenetic tree was constructed based on the neighbor-joining algorithm following the Tajima–Nei model with 1,000 bootstrap replicates in MEGA7.
Each gut bacteria was cultured in 2-L Erlenmayer flasks with 5 L of LB liquid medium. After 5 days, the culture was centrifuged at 8,000 rpm for 10 min to obtain a cell-free supernatant. Supernatants were dehydrated under a fume hood. Initially, a small part of the dried content was divided into three parts and dissolved in either petroleum ether, dichlormethane, or methanol to determine the efficient extracting solvent for antifungal compound. Next, these extracts were vortexed and centrifuged at 10,000 rpm for 5 min. Supernatants were concentrated with a rotary evaporator, and contents were dissolved in 1 ml of the appropriate solution and was examined for its antifungal ability using agar diffusion assay against selected fungi.
The rest of the dehydrated supernatant was extracted with dichlormethane. Dichlormethane phase was concentrated using a rotary evaporator (IKA RV8V, Germany). Crude extract was fractionated with silica gel or sephadex columns chromatography. The crude extract was mixed with an equal mass of silica gel (200–300 mesh) (Qingdao Marine Chemical Company, China), mixed, and loaded on top of the chromatography column (80 cm length and 5 cm diameter) containing 280 g of silica gel. The chromatography column was washed with a mobile phase (v/v) with different proportions of petroleum ether: methanol (100:0, 36:1, 18:1, 9:1, 4:1, 2:1, 1:1, and 0:100 v/v) and ethyl actetate: methanol (9:1, 4:1, 2:1, 1:1, and 0:1 v/v). About 7 ml of eluate was collected in glass vials, monitored with thin layer chromatography (TLC), and each fraction was examined for antifungal activity by agar diffusion. Active fractions were combined based on TLC and diffusion agar results and concentrated using the rotary evaporator. Next, combined fractions were loaded on to a Sephadex column (50 cm length and 1.5 cm diameter). The Sephadex column (Sephadex LH-20, Amersham Pharmacia Biotech, Sweden) was washed with a mobile phase of chloroform: methanol proportion (1:1 v/v). Fractions with 7 ml were collected to glass vials and monitored with TLC. Antifungal activity was examined by agar diffusion against fungi. Next, based on TLC and activity results, active fractions were combined and re-extracted again by silica gel column. Furthermore, the column was washed with a mobile phase with different proportions of hexane: ethyl acetate (100:0, 40:1, 30:1, 20:1, 15:1, 12:1, 10:1, and 0:100 v/v). The fractions were examined by TLC and for antifungal activity. Positive fractions were purified with a Sephadex column by washing with different proportions of dichlormethane:methanol mobile phase (100:0, 70:1, and 0:100).
To investigate the qualitative compositions of the antifungal compound, TLC was applied. TLC analysis was used to monitor the fractions from column chromatography, and spots on silica gel plates were visualized by spraying with a solution (1.5% of aluminum chloride in ethyl alcohol, ammonia vapor, and 5% sulfuric acid in ethyl alcohol pre-heated at 105°C). Next, purity of compound was measured by an UV spectrophotometer (UV-2550 Shimadzu, Japan).
HPLC analysis was performed using a Hitachi Chromaster HPLC system consisting of an 1,110 pump, DT-230 column oven, 1,430 diode array detector, and a YMC C18 column (250 × 4.6 mm, 5 μm). HPLC analysis was performed with the EZChrom Elite software. The mobile phases were water, acetonitrile, and methanol. The mass spectra were measured in a 2690-ZQ 4000 Water-Alliance LC-MS spectrometer (Applied Biosystems/MDS Sciex Concord, ON, Canada). 1H NMR, 13C NMR, and 2D NMR spectra were recorded on Varian MR-400 and VNMRS-600 NMR spectrometers with TMS as an internal standard.
To determine the antagonistic abilities of the gut bacteria, a mix of half of the ISP2 medium (yeast extract 4 g L–1, malt extract 10 g L–1, dextrose 4 g L–1, and agar 20 g L–1, pH7.2) and half of the PDA were used to pour onto a plastic petri dish (90 cm in diameter). Next, each gut bacteria was co-cultured with different fungi isolates. Co-cultivation experiments in single plate was repeated, and the antagonistic ability of bacteria was determined by evaluation of distance between the bacterial growth edge (from the fungal side) and the fungi growth edge (from the bacterial side). Inhibition of fungal growth was calculated by using the following equation (Alenezi et al., 2016):
where a is the distance from the center of the fungal colony to the fungi growth edge (from the bacterial side and bacterial growth edge), and b is the radius of control of the fungal colony.
To examine antifungal activity of the chromatography fractions, 3-mm diameter holes were punched out in PDA plates with a hole puncher. Fractions from each column chromatography were loaded into a well under sterile flow cabinet. A 5-mm piece of fungi mycelium grown on PDA was punched out and transferred onto the middle of the PDA plate. The plates were cultured for 7 days at 25°C, and fungi growth was recorded every day post cultivation.
To evaluate different enzyme activities of lignocellulolytic pathways that are involved in the degradation of plant cell wall compounds such as cellulose, lignin, glucans, cellobiose, and xylan, for cellulose degradation, steps were performed following the report by Vasanthakumar et al. (2008). For ligninolytic activity and lignin oxidation assay, reports from Machado et al. (2005) and Vasanthakumar et al. (2008) were followed. Activities of xylanase, cellobiase, and glucanase were determined by evaluation of the coloration of the respective substrates (Vargas-Asensio et al., 2014; Rojas-Jiménez and Hernández, 2015). For lipolytic and proteolytic activities, respective tween 20/80 and milk powder were used as substrates. Enzyme activities of the gut bacteria were evaluated by the appearance of clear halos (Mayerhof et al., 1973; Kumar et al., 2012).
The MIC for selected fungi were determined using a 10-fold serial dilution method. Petri dishes with PDA medium were prepared. Diluted pure compound with respective concentrations (20, 10, 5, 2.5, 1.25, 0.63, 0.31, 0.16, and 0.08 μg) was mixed with the PDA medium. The punched-out fungi mycelium with 0.7-mm diameter gel piece was transferred onto the middle of the agar plate, and mycelial growth was evaluated after 3, 6, 8, 10, and 12 days of post incubation at 25°C. Each treatment was performed in triplicate, and the diameter of the mycelial growth inhibition (MGI) was measured with a caliper, and calculated according to the following equation:
where, Dc (mm) is the mean of the colony diameter in the control, and Dt (mm) is the mean of the colony diameter in each treatment.
To examine the effect of the antifungal compound on the fungi hyphae structure, the wild apple-associated Dothiorella sarmentorum fungi was selected. The aerial mycelium of the fungi, a glass coverslip was placed on the surface of the PDA medium inoculated with a mycelial plug and cultured for 4, 6, or 12 days. Glass coverslips covered with mycelium that were directly coated with platinum for 120 s by an Iron Sputter Coater E1045 (Hitachi, Japan) and were viewed under a scanning electron microscope (Carl Zeiss Jena, SUPRA 55VP, Germany) as described earlier in our study (Liu et al., 2020).
All the experimental data consisted of the means of at least three independent replicates, and comparisons of data were performed using one-way ANOVA with Fisher’s PLSD post-hoc test. A p-value of <0.05 was considered statistically significant. All statistical analyses were performed using StatView software packages (SAS Institute Inc., Cary, NC, United States). All figures were generated in Adobe Illustrator CS3 Version 13.0.0.
To elucidate competition between wild apple-associated fungi and bacteria in terms of the larval gut of A. mali colonization, several molecular, microbiological, and analytical tools were applied. For this purpose, we developed an exploratory strategy to conduct stepwise identification of the host tree-associated microorganisms and screen the gut bacteria for antagonistic ability as well as determine the origin of the gut bacteria, as depicted in Figure 1.
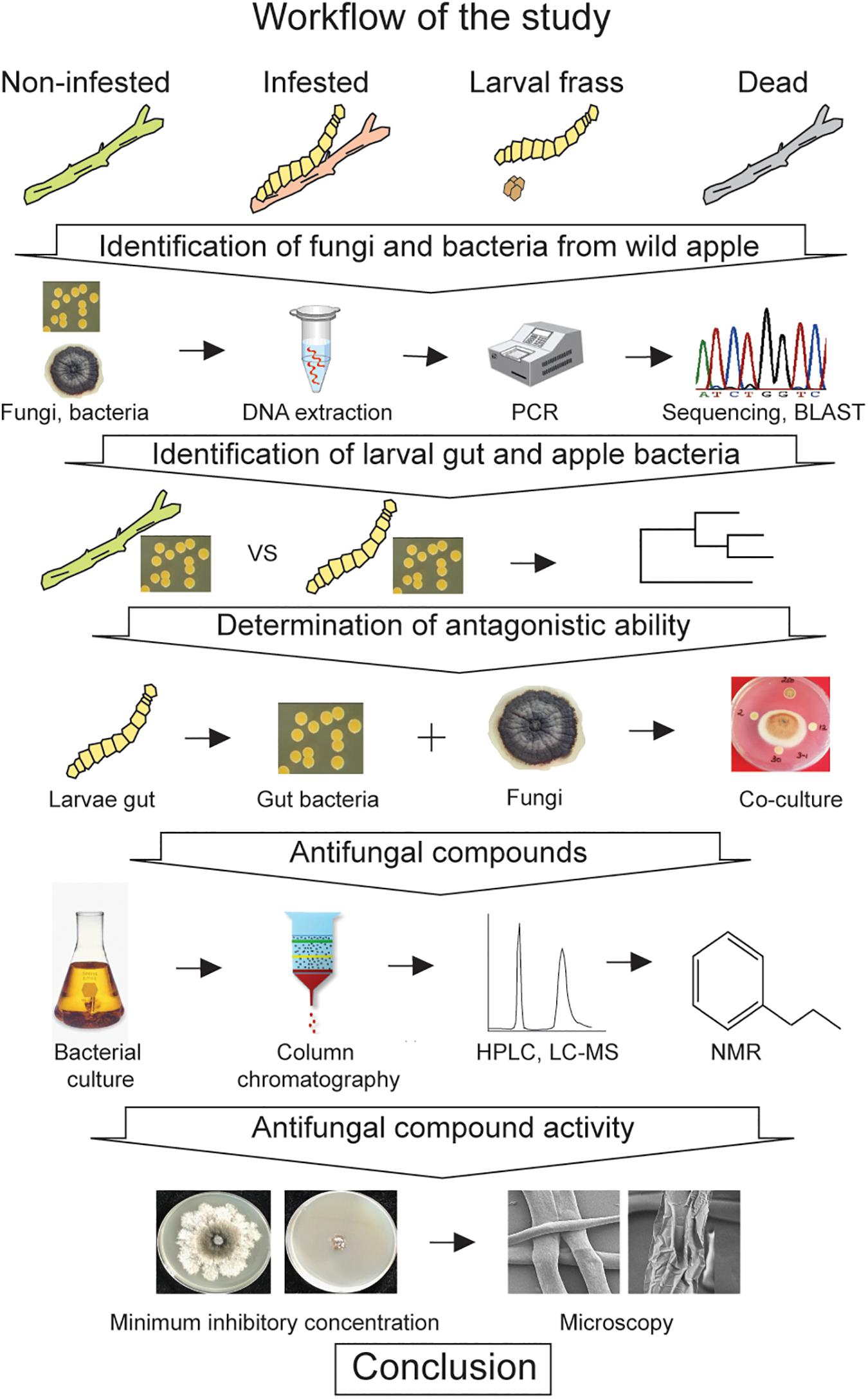
Figure 1. A workflow depicting the strategy used to investigate the gut microbiota against the wild apple-associated fungi.
Based on the workflow, we first isolated the fungi and bacteria from different states of twigs, such as noninfested, larvae-infested, and dead twigs, as well as larval frass, to obtain as many species as possible, since the A. mali larvae especially the Malus species feeds on twigs, especially in the younger twigs. A total of 204 monoconidial fungi and 320 bacterial isolates were obtained. Each fungus and bacterium were individually isolated. Individual DNA was extracted from each fungal and bacterial colony. For preliminary identification of fungal and bacterial isolates, the respective internal transcribed spacer (ITS) and 16S RNA regions were amplified. The traditional Sanger method was performed to sequence the regions. Next, sequences of host tree-associated bacterial isolates and sequences of the gut bacteria from our earlier study (Bozorov et al., 2019b) were compared to understand the origin of the gut bacteria. Then, the antifungal activity of the gut bacteria against the host tree-associated fungi was determined. Antifungal compounds were isolated from the strongest gut bacteria, which were hypothesized to prevent fungal growth in the gut. Furthermore, the activity of the antifungal compounds was analyzed.
Sequence analysis among the 204 culturable fungal and 320 bacterial isolates demonstrated 23 and 54 operational taxonomic units (OTUs), respectively (Figure 2). Fungal isolates were distributed into two phyla, seven classes, 13 orders, 15 families, and 19 genera, whereas bacterial isolates corresponded to four phyla, eight classes, 14 orders, 17 families, and 27 genera (Figure 2 and Tables 1, 2).
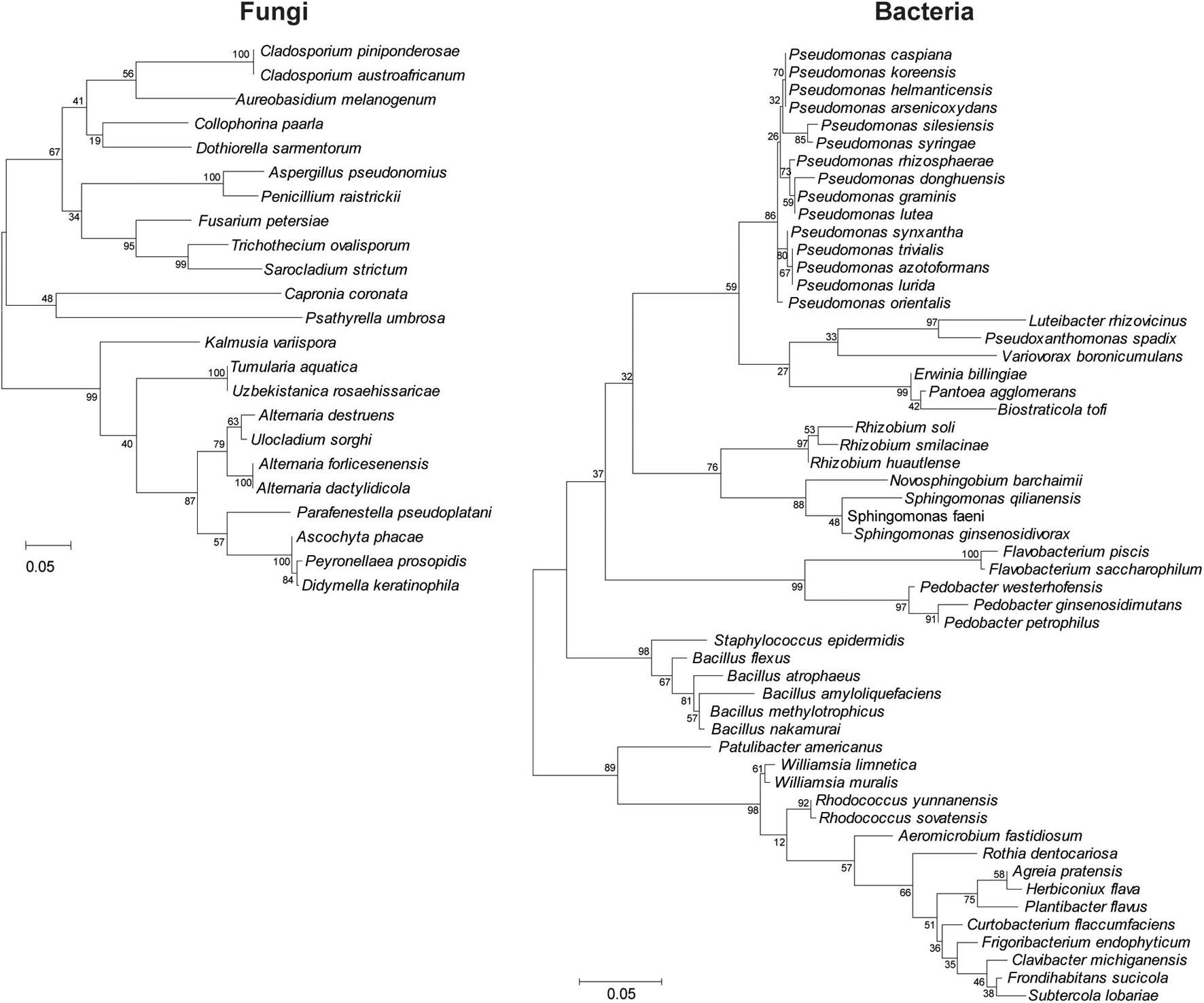
Figure 2. Clustering analysis of the wild apple-associated fungal and bacterial diversity. Maximum likelihood was inferred using the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the Tajima–Nei method and are in units of the number of base substitutions per site. Distance scale represents the number of differences between the sequences. Evolutionary analyses were conducted in MEGA 7.
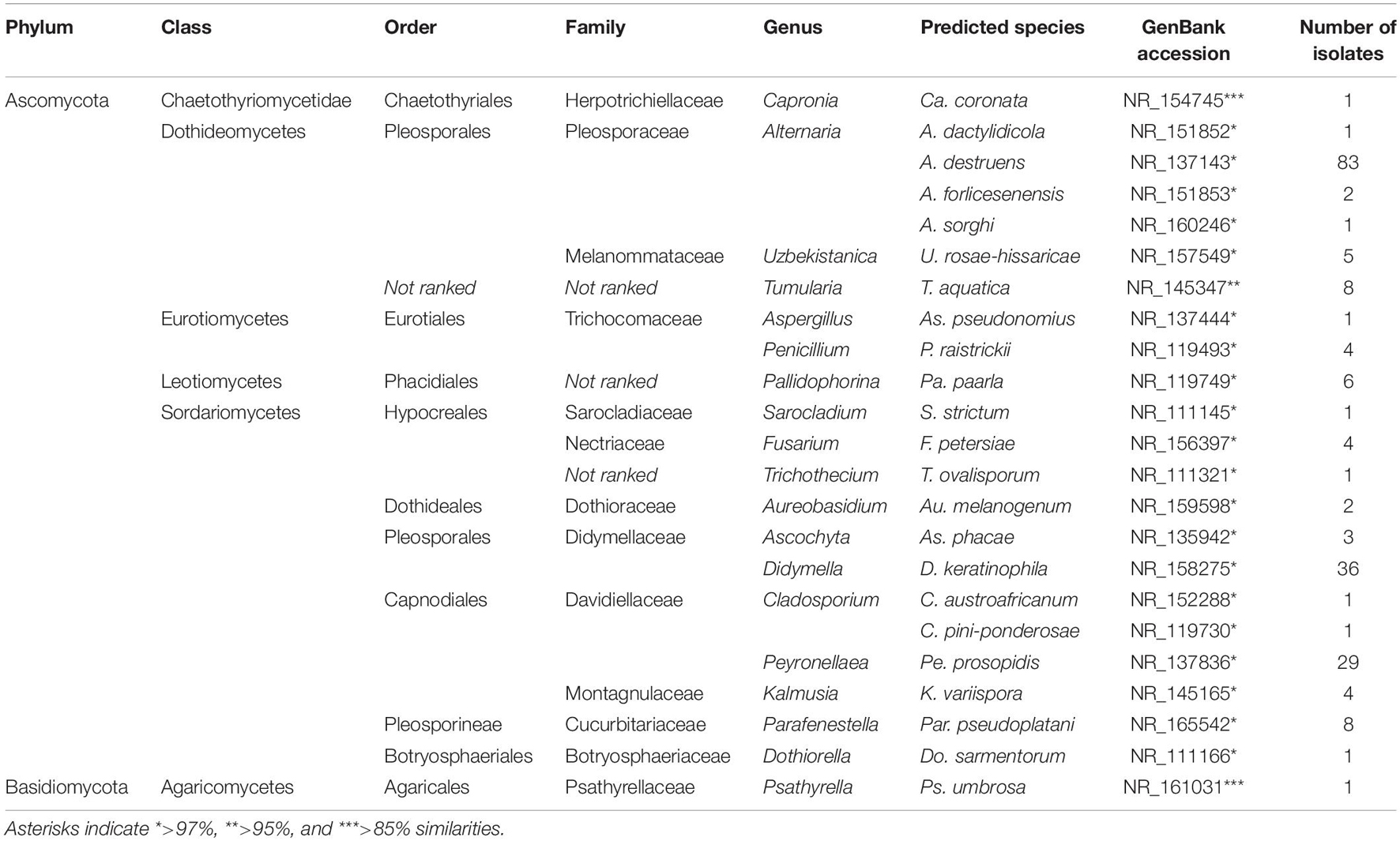
Table 1. Identified fungal isolates from wild apple stems by internal transcribed spacer (ITS) sequence analysis and their taxonomic status.
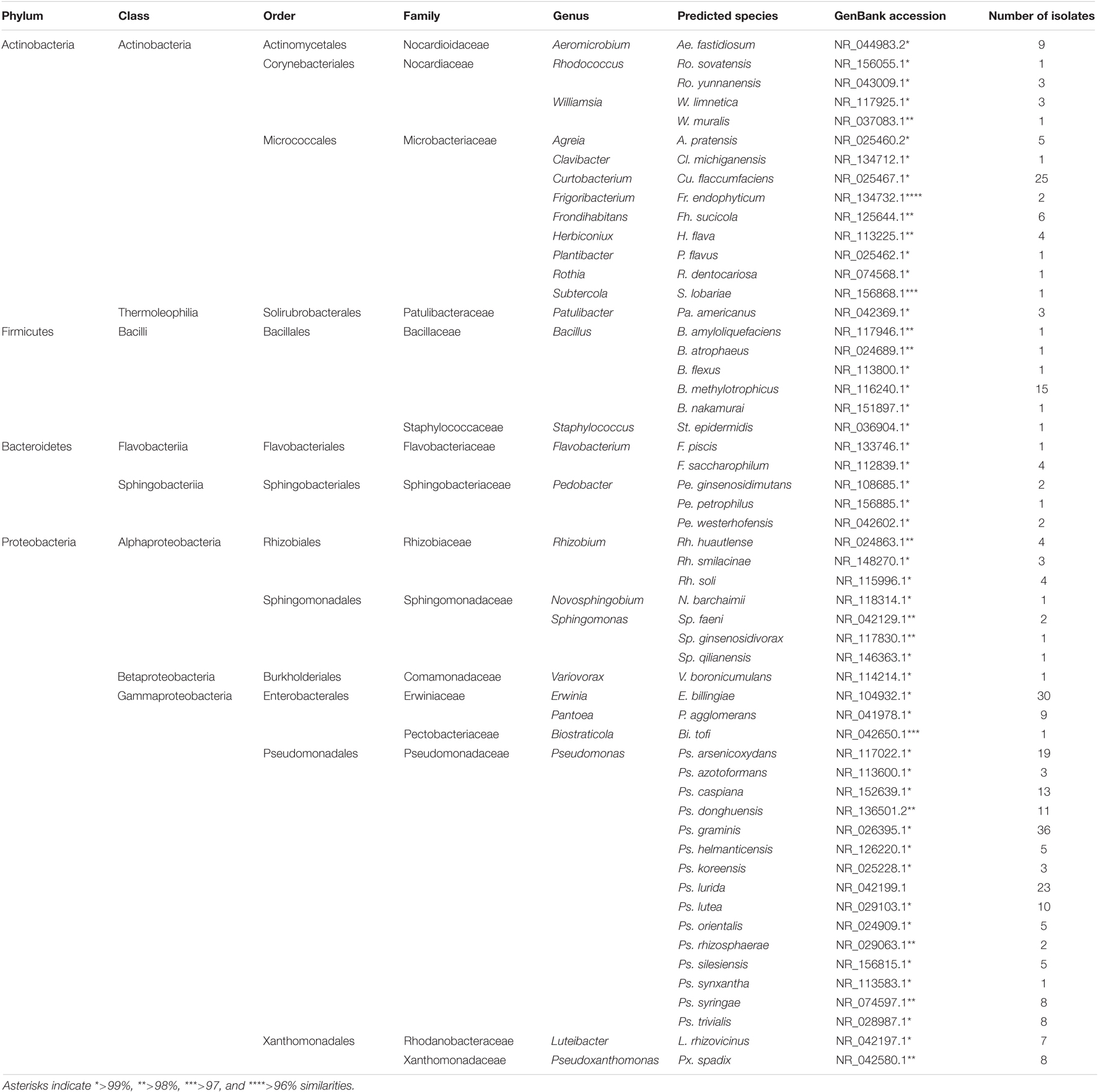
Table 2. Identified bacterial isolates from wild apple stems by 16S RNA sequence analysis and their taxonomic status.
The abundance and composition of the apple twig fungi and bacteria differed depending on the material categories (Figure 3). The abundance of fungal isolates was increased in infested twigs compared with non-infested and dead twigs, whereas bacterial diversity was richer in non-infested twigs than in either infested or dead twigs. Additionally, the number of fungi and bacterial isolates in larval frass also differed from the other material categories (Figure 3A). Fungi rather than bacteria mostly colonized dead twigs, whereas a higher abundance of bacterial isolates was found in the larval frass.
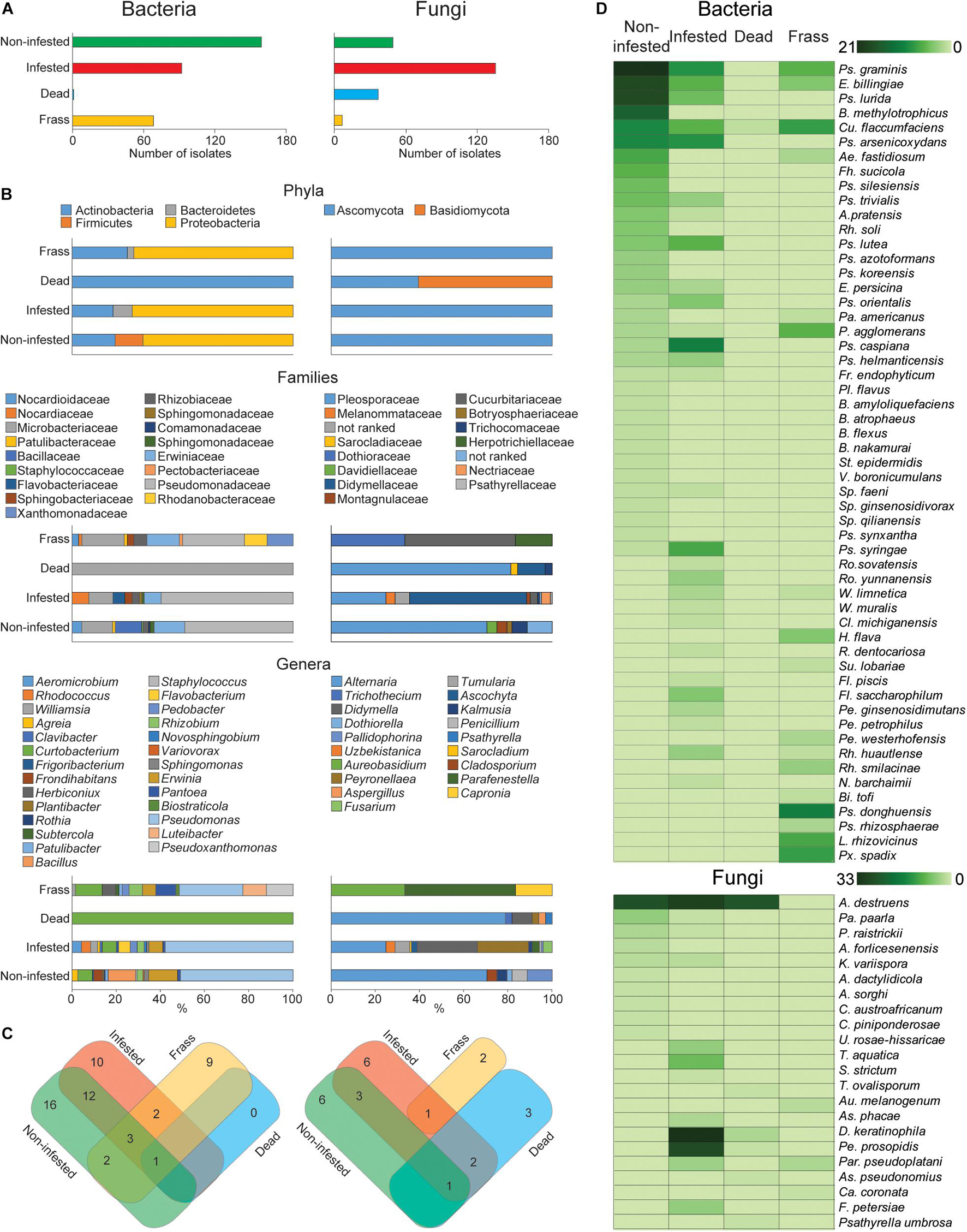
Figure 3. Fungal and bacterial diversity and abundance in different wild apple twigs and larval frass. (A) Number of isolates of fungi and bacteria of different twigs and larval frass. (B) Percentage distribution of isolates by their phylum, family, and genus. (C) Venn diagram summarizing the composition of fungal and bacterial isolates at the species level. (D) Heat map analysis displaying a comparison of fungal and bacterial abundance by different twigs and larval frass.
The composition of isolates demonstrated that the fungi and bacteria can colonize twigs commonly or specifically. The distribution of isolates by phylum, family, and genus revealed diverse colonization of different twigs and larval frass (Figure 3B). For example, an additional fungal phylum, Basidiomycota, was found in dead twigs, whereas bacteria were reduced to one phylum. This suggests that increased saprophytic fungi such as Alternaria and Psathyrella in dead twigs could antagonistically reduce bacterial species.
Larval attack made a disappearance of the bacterial phylum Firmicutes but emerged Bacteroidetes. Similarly, it was observed in larval frass (Figure 3B). The Bacillaceae, Staphylococcaceae, and Comamonadaceae families were specific to non-infested twigs, whereas, the Flavobacteriaceae, Nocardiaceae, and Sphingobacteriaceae families were specific to infested twigs, and the Pectobacteriaceae, Rhodanobacteraceae, and Xanthomonadaceae were specific to the larval frass. At the genus level, Frondihabitans, Plantibacter, Bacillus, Staphylococcus, Sphingomonas, and Variovorax were replaced by Rhodococcus, Clavibacter, Williamsia, Rothia, Flavobacterium, Pedobacter, and Novosphingobium in the infested twigs and Luteibacter, Pseudoxanthomonas, Biostraticola, Subtercola, and Herbiconiux in the larval frass. Dead twigs showed only one species, Curtobacterium flaccumfaciens, with the lowest abundance (Figure 3B).
The distribution of fungal species by their taxonomy differed from that of bacteria. The genera Cladosporium and Dothiorella specifically colonized noninfested twigs while Uzbekistanica, Tumularia, Sarocladium, Ascochyta, Didymella, Peyronellaea, and Fusarium were detected from infested twigs. Trichothecium and Psathyrella as well as Aureobasidium and Capronia were specific to dead twigs and larval frass, respectively. Distribution by species composition of fungi and bacteria revealed that 16 bacterial species were specific to noninfested twigs, 10 to infested twigs, and nine to larval frass, whereas six fungal species were specific to noninfested twigs, five to infested twigs, three to dead twigs, and two to larval frass. Taken together, during larval infestation, host tree-associated bacterial composition was reduced, whereas fungal species were increased.
In our previous study, we demonstrated the rich diversity of the Agrilus mali larval gut microbiota, and approximately 99% of the taxa were cultivable bacteria (Bozorov et al., 2019b). To determine the antagonistic ability of the larval gut bacteria toward apple-associated fungi, in vitro screening was performed using cocultivation (Figure 4A). For this experiment, 25 fungi belonging to different species isolated from apple twigs were randomly selected. The results indicated that all the gut bacteria demonstrated antagonistic ability in a selective manner (Figure 4B). The distribution of the share of total antifungal activity by each gut bacteria differed. Among them, Ps. synxantha was able to inhibit fungal isolates with a 41.8% share (24 fungal species), Ps. orientalis with an 11.0% share (12 fungal species), E. billingiae with a 10.4% share (16 fungal species), E. persicina with a 13.3% share (17 fungal species), and Pantoea sp. with 2.5% share (two fungi species). Three P. aqqlomerans strains, 2, 153, and 43, inhibited fungi with a 14.0% (16 fungal species), 3.5% (six fungal species), and 3.5% share (five fungal species), respectively (Figures 4C,D). The results suggest that Ps. synxantha was the strongest antagonist in the larval gut and was able to inhibit 90.9% of the selected host tree-associated fungal species. The difference in antifungal activity of bacteria could be due to the level of fungi ability to neutralize the effect of bacterial toxins and the level of bacterial compound.
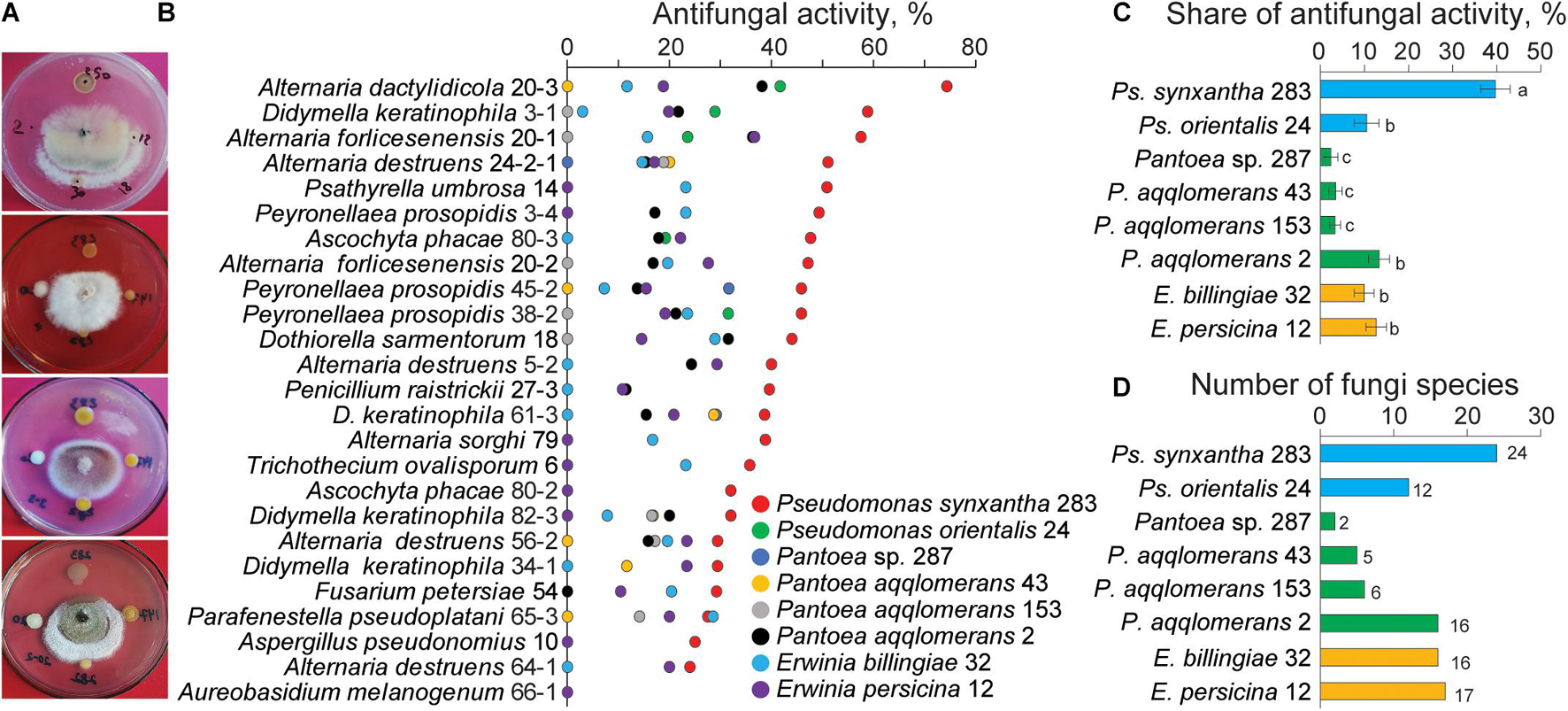
Figure 4. Antifungal activity of the gut bacteria against different fungal species isolated from wild apple twigs. (A) Examples of the gut bacterial inhibition of wild apple fungi. (B) Antifungal activity of the gut bacteria against selected fungi from the different types of apple twigs. (C) Share of antifungal activity of each gut bacteria. Different letters indicate significant differences determined by one-way ANOVA, followed by a Fisher PLSD post-hoc test (p < 0.05). (D) Number of fungal isolates inhibited by each gut bacterium.
The gut bacteria perform various enzymatic functions in the degradation of cell wall compounds, lipids, and proteins (Table 3). In a previous report, A. mali larvae gut collected from wild apple twigs represented 99.1% of cultivable Pantoea, Erwinia, and Pseudomonas species. Other 36 OTUs were non-culturable bacteria (Bozorov et al., 2019a). We showed that only Pantoea, Erwinia, and Pseudomonas species consisted of the examined ability of the gut bacteria to degrade plant cell wall components, but in this study, we showed the levels of bacterial lysocellulotic and other enzymatic activities. Pantoea species were able to highly degrade lignocellulosics such as cellulose, xylanase, glucanase, and cellobiose but not lignin, lipids, or proteins. Both Erwinia species showed weak cellulolytic activity, and only E. persicina demonstrated xylanase, glucanase, and lipase activities. Pseudomonas species were not able to degrade plant cell wall compounds but showed strong protease activity. Moreover, Ps. orientalis showed weak lignocellulolytic and strong lipolytic activities. Lignin oxidation was not observed by any gut bacteria.
To isolate antifungal metabolites from gut bacteria, dehydrated bacterial cultures were extracted using different solvents to determine the extraction efficiency. The results showed that methanolic extracts from Ps. synxantha, Ps. orientalis, P. aqqlomerans 2, E. billingiae, and E. persicina demonstrated the highest antifungal activities compared with other solvents (Table 4). The strongest antifungal activity was observed for Ps. synxantha and P. aqqlomerans 2 compared with other gut bacteria. Since Ps. synxantha was able to strongly inhibit almost all selected fungal species and could be a potential bacterium preventing fungal growth in the gut, further work along these lines will be continued.
An estimated 280-mg crude methanolic extract from Ps. synxantha was re-extracted with silica and Sephadex gel column chromatographies using different solvent systems. Each fraction was examined with an agar diffusion to determine its antifungal activity and monitored with TLC. For the final purification of the active fraction, Sephadex column chromatography was used with a dichloromethane mobile phase. The purity of the active fraction was analyzed with preparative HPLC showing a single symmetrical peak at 250 nm with a retention time of 9.1 min. The dried pure compound was greenish-yellow, needle-crystalline, and soluble in chloroform, DMSO, and methanol but insoluble in water.
Pure antifungal compounds were characterized by ESI-MS, 1H-NMR, and 13C-NMR. The molecular mass spectra demonstrated ion peaks at m/z 225.06544 [M + H]+ (base peak) and m/z 247.04738 [M + NA]+ (Figure 5). The 1H and 13C NMR spectral data of the compounds and their assignments are shown in Table 5. After NMR and mass spectrometry analyses, the compound of interest was determined to be phenazine-1-carboxylic acid.
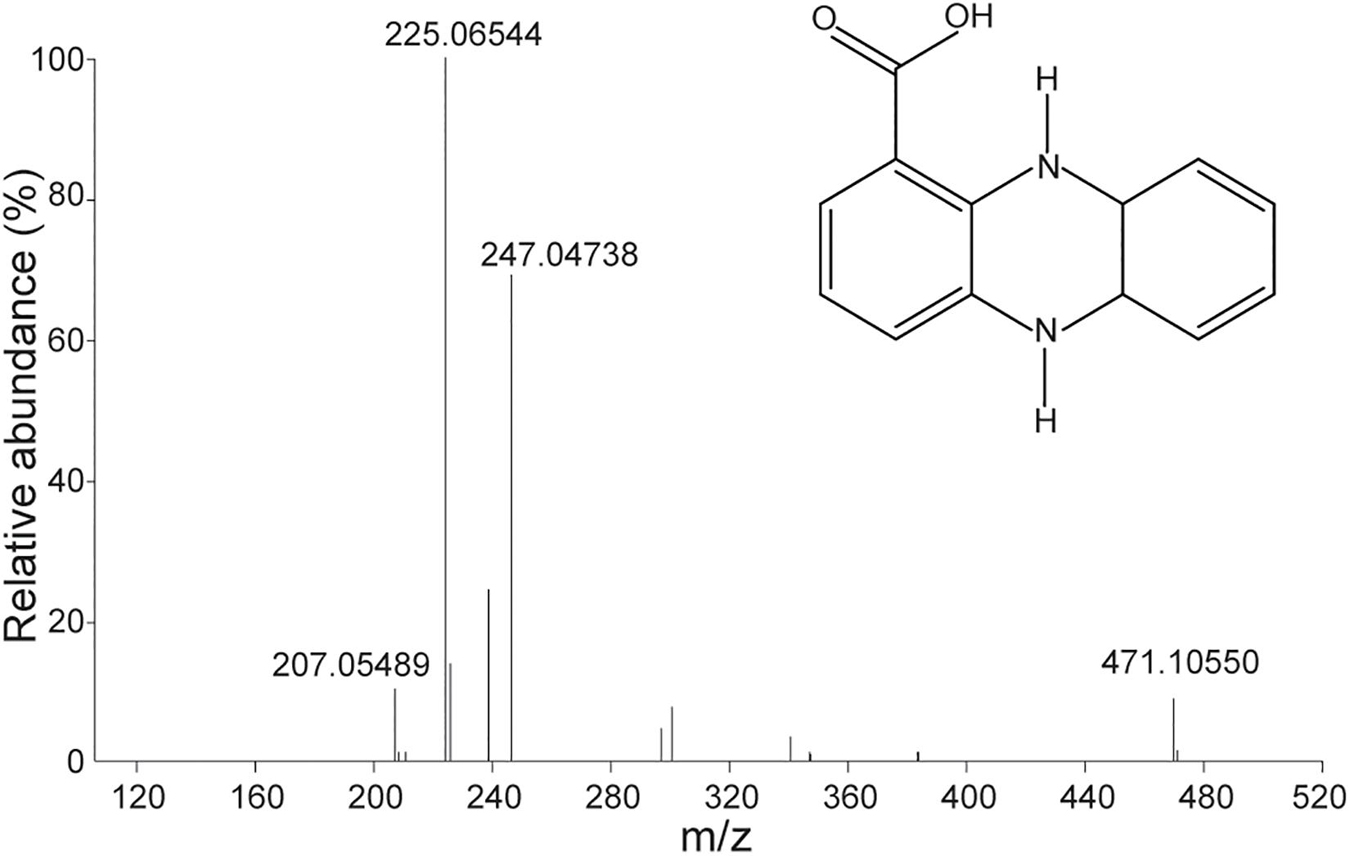
Figure 5. Mass spectra of antifungal compounds and the structure of phenazine-1-carboxylic acid (PCA).
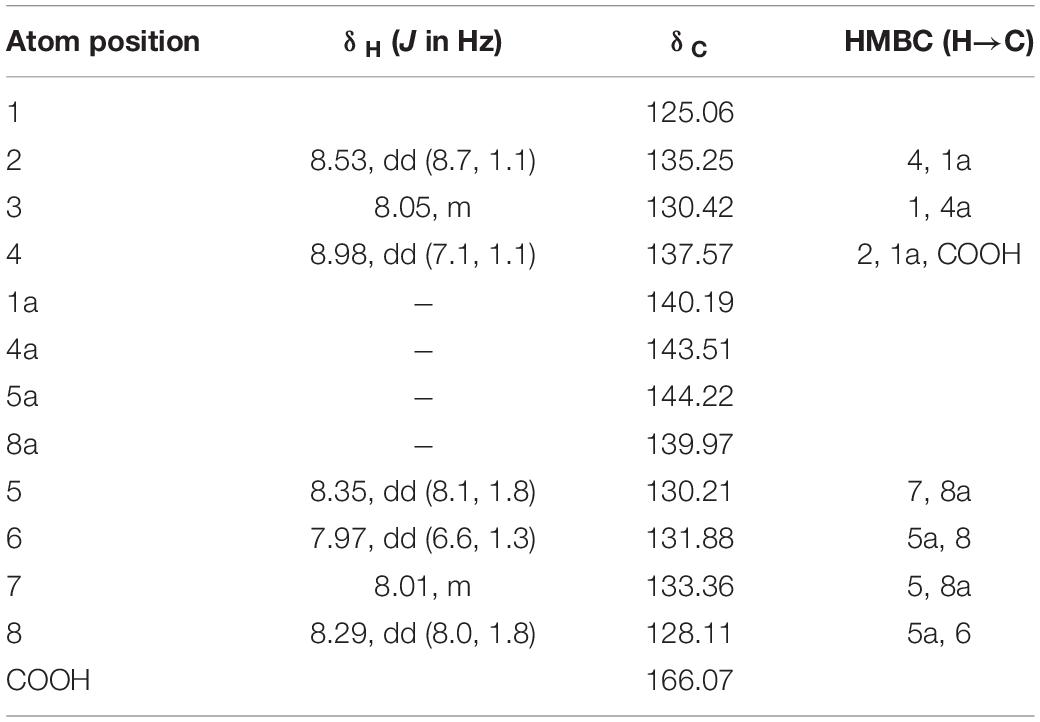
Table 5. 1H and 13C NMR chemical shifts of phenazine-1-carboxylic acid and HMBC data in CDCL3, δ, ppm at 400 MHz.
To determine the minimum inhibitory concentration, a filamentous fungus, Dothiorella sarmentorum #18, was chosen because of its faster growth on PDA. This species was also used during all purification steps to determine the antifungal activity of the fractions. Purified phenazine-1-carboxylic acid at an MIC of 12.5 mg ml–1 was able to clearly inhibit mycelial growth of D. sarmentorum. Serial dilutions of the compound showed that the effective doses at 50% and 80% were 1.25 and 5 mg ml–1, respectively (Figures 6A,B).
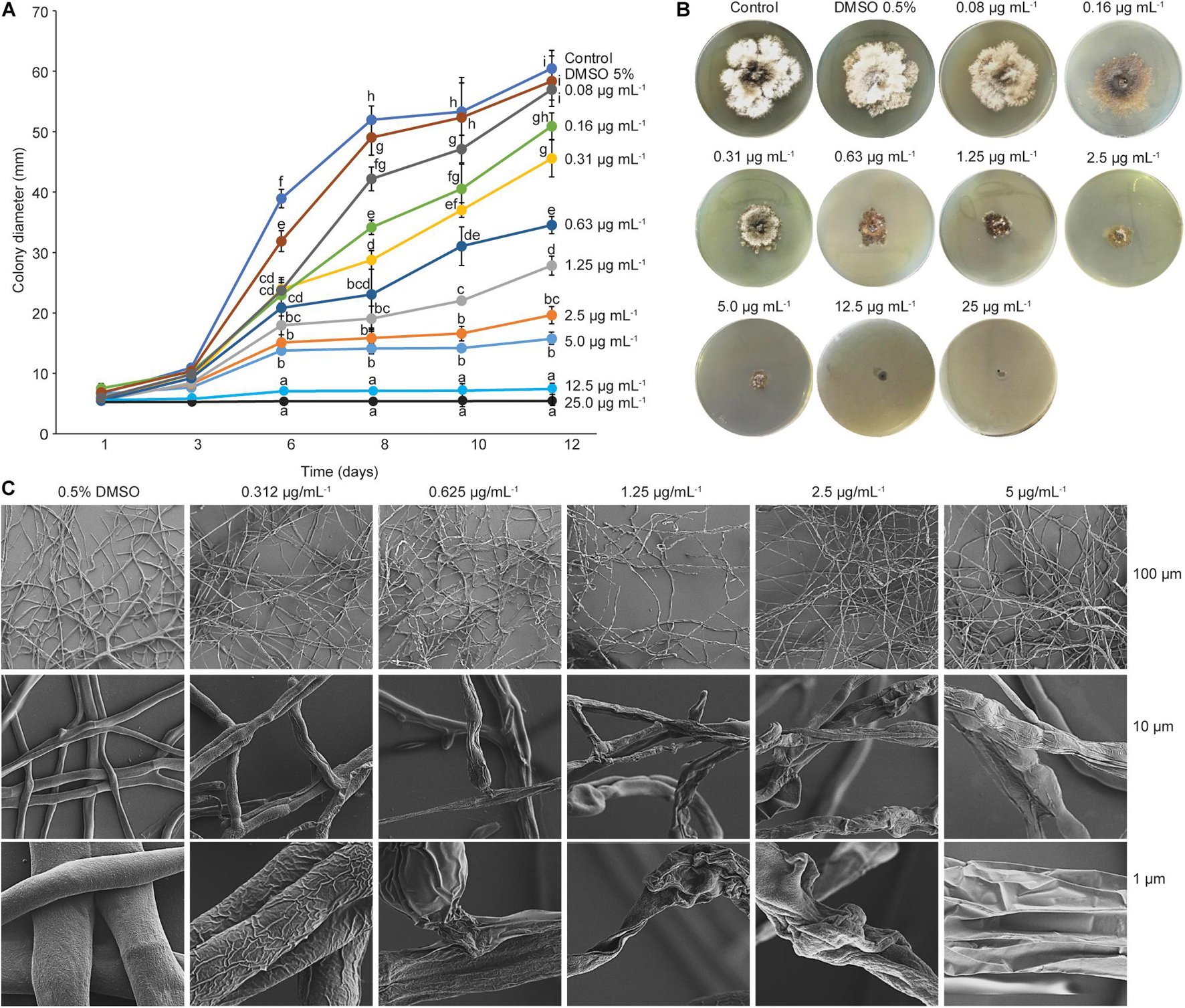
Figure 6. Minimum inhibitory concentration and scanning electron microscopy. (A) Minimum inhibitory concentration (MIC) of PCA. Graphic represents the effects of different PCA concentrations on fungal growth at different incubation time. Fungal growth decreased at a concentration of 0.08 mg ml–1 PCA, and complete inhibition of mycelial growth was observed at 25 and 12.5 mg ml–1. Different letters indicate significant differences determined by one-way ANOVA, followed by a Fisher PLSD post-hoc test (p < 0.05). (B) MIC assay after 12 days of incubation at 28°C. (C) Scanning electron microscopy (SEM) images of the effect of PCA against D. sarmentorum.
To further understand the effect of phenazine-carboxylic acid on fungal growth, mycelial growth was observed with SEM. Mycelia obtained from the edge of the D. sarmentorum colony growing in antifungal compound-free medium (control) produced hyphae with smooth surfaces (Figure 6C). With the addition of several concentrations of PCA ranging from 0.312 to 5 μg ml–1 into the medium, fungal hyphae lost smoothness depending on the PCA concentration. A high amount of PCA caused the failure to form a hyphal network thus inhibiting fungal growth.
After hatching, insect larvae can adopt bacteria either directly parentally or from the host during feeding. To understand the origin of gut bacteria, we compared 16S RNA sequences of the same species found in apple twigs and gut bacteria (Figure 7A). For this purpose, we analyzed bacteria from noninfested and infested twigs and larval frass. Since gut bacteria could be excreted, they could be found in larval frass. Sequence analysis and the neighbor-joining tree revealed that the gut bacterial gene sequences were identical to those of related apple bacteria (Figure 7A). Erwinia billingiae, E. perscina, Ps. orientalis, Ps. synxantha, and P. agglomerans specifically colonized non-infested twigs were still found in infested twigs but not in dead twigs. Erwinia billingiae and P. agglomerans were detected in the larval frass since these bacteria were highly abundant in the gut and thus could be excreted (Figure 7B).
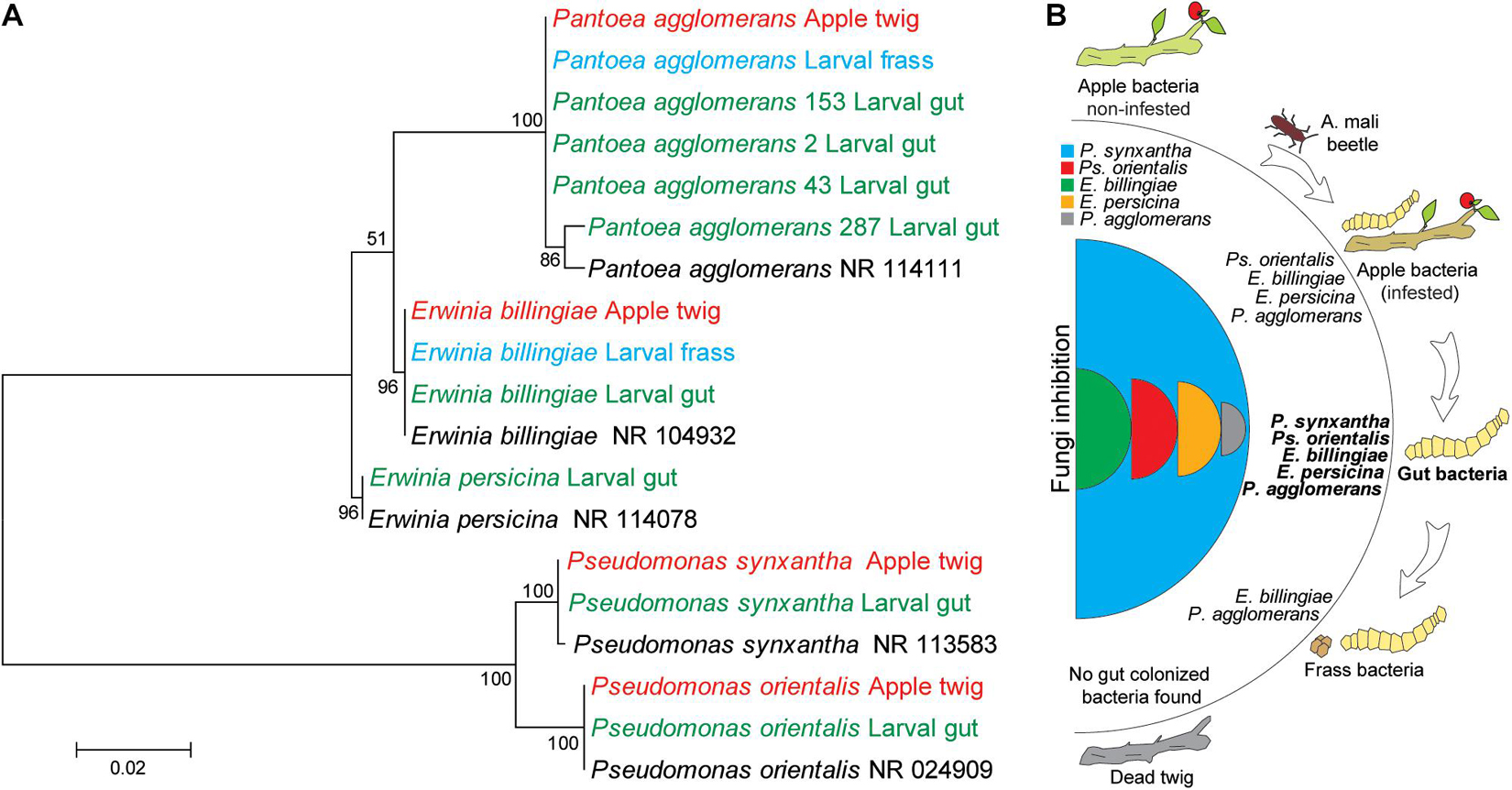
Figure 7. Identity analysis of the gut bacteria and possible route of bacterial transmission to larvae. (A) Clustering of bacterial species from the larval gut, apple twig-associated bacteria, and frass. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The evolutionary distances were computed using the Tajima–Nei method and are in units of the number of base substitutions per site. Distance scale represents the number of differences between the sequences. Evolutionary analyses were conducted in MEGA 7. Bacterial species in black, red, and green words relate to GenBank, apple twigs, and the gut bacteria, respectively. (B) Possible mechanism of transmission of tree bacteria to the larval gut.
In nature, many microorganisms interact with each other to coexist, and their ecological relationships range from parasitism to mutualism (Prosser et al., 2007). Depending on the species, insects have symbiotic associations with diverse and complex resident and transient microorganisms (Hooper and Gordon, 2001; Egert et al., 2003; Dillon and Dillon, 2004; Douglas, 2015). In previous work, the larval gut of the invasive wood-borer A. mali demonstrated a diverse bacterial community, but among them, the most abundant Panthoea spp. (99%), some Erwinia species, and Pseudomonas species, were able to degrade plant cell wall compounds (Bozorov et al., 2019a). The larval gut did not show any fungal species using either high-throughput sequencing or culture-dependent methods (Bozorov et al., 2019b). In this study, we explored the interaction between the gut bacteria of invasive wood-borer insect larvae and wild apple-associated fungi to elucidate the absence of fungi in the larval gut.
Fungi and bacteria can be transiently or permanently associated with the host and transmitted to the gut from their parents or the host (Feldhaar, 2011; Hammer et al., 2017). In contrast to the results of Zhang et al. (2018), who showed that A. mali larvae can be colonized by fungi and bacteria, our earlier report demonstrated the absence of fungi in the larval gut of A. mali (Bozorov et al., 2019b). However, both fungi and bacteria can colonize the congeneric A. planipennis (Mogouong et al., 2020). Hypothetically, the initial A. mali invasion of the Tianshan forests could have involved both bacterial and fungal species but eventually, competition by local microorganisms within the gut could have orchestrated the gut community, resulting in the elimination of fungal species from the gut. This could result in recompositing the larval gut microbiota to adapt to new environments for better insect survival. The diversity of the insect gut microbiota can be determined by the environmental habitat, diet, developmental stage, and phylogeny of the host (Yun et al., 2014). The gut microbiota could play an important role in the larval overwintering process at low temperature (Wang et al., 2017; Ferguson et al., 2018), and the climate of the natural habitat of A. mali, the Far East, Eastern China, and the Korean Peninsula, is not colder than that in Tianshan (Yang et al., 2017; Yao et al., 2020). The most abundant Panthoea bacteria of the larval gut with strong lygnocellulosic ability might provide sugars as energy resources and as cryoprotectants. During overwintering, insects accumulate sugars and polyols (Khani et al., 2007; Kostal et al., 2007) that can act as cryoprotectants and enhance cold hardiness for winter survival (Kost’al et al., 2001). However, to deeper understand the invasive insect larvae adaptation, it requires conducting additional in vitro experiments.
Both fungal and bacterial compositions varied in different twigs. Bacterial abundance was high in noninfested twigs, but fungal abundance was higher in infested twigs. This indicates that plant immunity reduced because larvae feed on the phloem of trees by creating serpentine galleries, thus preventing nutrient movement. The transition from twig infestation to twig death, thus, creates the growth condition for saprophytic wood decaying fungi (WDF) (Lundell et al., 2014; Park et al., 2020). Lignin is known to be extensively degraded by WDF and other fungi (Lundell et al., 2014; Park et al., 2020). Eversince, A. mali larvae specialized in less lignified phloem tissue compared with lignified xylem and that the gut bacteria of larvae degrade cellulose but not lignin (Bozorov et al., 2019b). Moreover, the lignin composition and content vary between xylem and phloem (Lourenço et al., 2016). Wood-decaying saprophytic fungi are the majority of species belonging to Basidiomycota and Ascomycota (Lundell et al., 2014). Bacteria and WDF must interact with each other to coexist, although WDF are well known for being highly competitive (Boddy, 2000). This is consistent with our result that bacterial composition was highly reduced in dead twigs. Saprophytic fungi can be inhibited by symbiotic bacteria when co-cultured together on artificial medium (Figure 4B) but in nature, dead twigs cannot be a food source for bacteria. This indicates that tree-associated bacteria found in this study could be symbiotic bacteria growth of which rely on relationship with the living plant, but in dead twigs bacteria might not grow because of the interrupted symbiotic link. Likely, bacterial species from non-infested trees must be transmitted into the larval gut because the originated gut bacteria were reduced or not found in the infested twigs (Figure 7B). Additionally, fungal isolates could also be transmitted into the A. mali gut, as reported by Zhang et al. (2018), but our study showed that this might not have occurred in the Tianshan forests (Bozorov et al., 2019b). Probably, presence of gut antagonistic bacteria might prevent fungi colonization of the gut. However, additional in vitro experiments are required to clarify bacterial communities of insect host trees of natural habitat and invaded area.
The current study demonstrated that all gut bacteria were able to antagonize tree-associated fungi with different inhibition levels. However, Ps. synxantha inhibited almost all fungal species with the highest inhibition rate. Possibly, this bacterium might play as key competitor against fungi in the gut. Apparently, fungi also had a low composition and abundance in larval frass that might be caused by the presence of antifungal compounds produced by the gut bacteria. However, no Pseudomonas species were detected in the larval frass. In contrast to Pantoea aqqlomerans and E. billingiae that were detected in the larval frass, it could be that Pseudomonas species might be localized in one of the upper compartments of the gut, such as the foregut or midgut. Chen et al. (2018) demonstrated that a more diverse microbial community was found in the foregut than in the midgut and hindgut in silkworms (Bombyx mori). However, experimental validation of bacterial species localization by gut compartments is required.
Several studies have reported the antagonistic ability of Pseudomonas species against fungi by the production of various types of antifungal metabolites (Jayaswal et al., 1993). Phenazine carboxylic acid (PCA) is one of the antifungal compounds produced by Pseudomonas chlororaphis, Ps. Fluorescens, and Ps. protegens that is able to kill some lepidopteran larvae (Flury et al., 2017) but the current study showed that PCA produced by Ps. synxantha was not lethal to A. mali larvae. Likely, PCA might prevent fungal colonization in the gut, which likely might explain the absence of fungi in the larval gut. This contrasts with the results of Zhang et al. (2018), who showed that both bacteria and fungi were found in A. mali larvae, including one Pseudomonas sp., but identification in species level was not clear. However, this species could be Ps. orientalis identified in our study as possessing weak inhibition of some apple-associated fungi. It was reported that these bacteria produce various types of antifungal compounds, such as pyoverdine, safracin, and phenazine, which are effective against bacteria and fungi (Kron et al., 2020). Moreover, Ps. orientalis has been isolated from most coleopteran insects due to its encoded cellulolytic enzyme involved in terpene transformation of plant resin compounds (Rojas-Jiménez and Hernández, 2015); this is consistent with our result that Ps. orientalis plays a role in cell-wall degradation (Bozorov et al., 2019b).
Other gut bacteria, such as Panthoea and Erwinia, also showed average antagonistic abilities against some apple-associated fungi despite their strong lignocellulolytic activities (Bozorov et al., 2019b). The most abundant gut bacteria, P. aqqlomerans, also had antifungal activity but was not able to inhibit all endophytic and saprophytic fungi. However, its antagonistic ability is reported in the literature (Iturritxa et al., 2017; Thissera et al., 2020). It was reported that E. billingiae also has antifungal ability against pathogenic Heterobasidion annosum, Armillaria mellea, and Fusarium circinatum fungi infecting Pinus radiata trees (Mesanza et al., 2016; Iturritxa et al., 2017), and E. persicina has activity against A. alternata (Goryluk-Salmonowicz et al., 2016). It is likely that bacterial species within the genus could produce similar antifungal compounds, such as herbicolin, pulicatin, and pyrrolnitrin (Greiner and Winkelmann, 1991; Chernin et al., 1996; Thissera et al., 2020).
We analyzed wild apple-associated bacteria, and some of them were transmitted to the gut of A. mali larvae. Among them, Ps. synxantha produced the antifungal phenasine carboxylic acid compound that strongly inhibited apple-associated fungal species; thus, it might inhibit fungal colonization in the larval gut. Other gut bacterial species, Panthoea and some Erwinia species, participated in plant cell wall cellulose degradation. Taken together, in the invaded area, insect larvae adopt selectively the apple-associated fungi and bacteria to build its own gut microflora. The gut bacterial community might participate in intensive plant cellulosic degradation to provide an energy source for the larvae to better adapt and survive in new regions.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.
TB wrote the manuscript and prepared all the tables and figures. TB, ZT, and GK contributed to the compound extraction and purification. ZT contributed to the compound analysis and structural determination. TB and YG contributed to fungi analysis. TB, DZ, and HS contributed to the manuscript writing and corrections. TB and DZ supervised the experiment and manuscript writing. All authors reviewed the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the NSFC-Xinjiang Key Project (No. U1903206), the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0502030403), and a CAS PIFI fellowship (#2019VBC0007) that supported this research.
Alenezi, F. N., Rekik, I., Belka, M., Ibrahim, A. F., Luptakova, L., Jaspars, M., et al. (2016). Strain-level diversity of secondary metabolism in the biocontrol species Aneurinibacillus migulanus. Microbiol. Res. 182, 116–124. doi: 10.1016/j.micres.2015.10.007
Bahrndorff, S., Alemu, T., Alemneh, T., and Lund Nielsen, J. (2016). The microbiome of animals: implications for conservation biology. Int. J. Genomics 2016:5304028. doi: 10.1155/2016/5304028
Boddy, L. (2000). Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 31:18594. doi: 10.1111/j.1574-6941.2000.tb00683.x
Bozorov, T. A., Luo, Z. H., Li, X. S., and Zhang, D. Y. (2019a). Agrilus mali Matsumura (Coleoptera: Buprestidae), a new invasive pest of wild apple in western China: DNA barcoding and life cycle. Ecol. Evol. 9, 1160–1172. doi: 10.1002/ece3.4804
Bozorov, T. A., Rasulov, B. A., and Zhang, D. (2019b). Characterization of the gut microbiota of invasive Agrilus mali Matsumura (Coleoptera: Buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci. Rep. 9:4923. doi: 10.1038/s41598-019-41368-x
Broderick, N. A., Raffa, K. F., Goodman, R. M., and Handelsman, J. (2004). Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70, 293–300. doi: 10.1128/AEM.70.1.293-300.2004
Chen, B., Du, K., Sun, C., Vimalanathan, A., Liang, X., Li, Y., et al. (2018). Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 12, 2252–2262. doi: 10.1038/s41396-018-0174-1
Chernin, L., Brandis, A., Ismailov, Z., and Chet, I. (1996). Pyrrolnitrin production by an Enterobacter agglomerans strain with a broad spectrum of antagonistic activity towards fungal and bacterial phytopathogens. Curr. Microbiol. 32, 208–212. doi: 10.1007/s002849900037
Dillon, R. J., and Dillon, V. M. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Egert, M., Wagner, B., Lemke, T., Brune, A., and Friedrich, M. W. (2003). Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69, 6659–6668. doi: 10.1128/AEM.69.11.6659-6668.2003
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Feldhaar, H. (2011). Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543. doi: 10.1111/j.1365-2311.2011.01318.x
Ferguson, L. V., Dhakal, P., Lebenzon, J. E., Heinrichs, D. E., Bucking, C., and Sinclair, B. J. (2018). Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct. Ecol. 32, 2357–2368. doi: 10.1111/1365-2435.13153
Flury, P., Vesga, P., Péchy-Tarr, M., Aellen, N., Dennert, F., Hofer, N., et al. (2017). Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 8:100. doi: 10.3389/fmicb.2017.00100
Goryluk-Salmonowicz, A., Piorek, M., Rekosz-Burlaga, H., Studnicki, M., and Blaszczyk, M. (2016). Endophytic detection in selected European herbal plants. Pol. J. Microbiol. 65, 369–375. doi: 10.5604/17331331.1215617
Greiner, M., and Winkelmann, G. (1991). Fermentation and isolation of herbicolin A, a peptide antibiotic produced by Erwinia herbicola strain A 111. Appl. Microbiol. Biotechnol. 34, 565–569. doi: 10.1007/BF00167899
Gupta, A., and Nair, S. (2020). Dynamics of insect–microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11:1357. doi: 10.3389/fmicb.2020.01357
Hammer, T. J., Janzen, D. H., Hallwachs, W., Jaffe, S. P., and Fierer, N. (2017). Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 114, 9641–9646. doi: 10.1073/pnas.1707186114
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. doi: 10.1126/science.1058709
Iturritxa, E., Trask, T., Mesanza, N., Raposo, R., Elvira-Recuenco, M., and Patten, C. L. (2017). Biocontrol of Fusarium circinatum infection of young Pinus radiata trees. Forests 8:32. doi: 10.3390/f8020032
Jayaswal, R. K., Fernandez, M., Upadhyay, R. S., Visintin, L., Kurz, M., Webb, J., et al. (1993). Antagonism of Pseudomonas cepacia against phytopathogenic fungi. Curr. Microbiol. 26, 17–22. doi: 10.1007/BF01577237
Ji, Y., Ji, R., and Huang, R. X. (2004). Invasive species - Agrilus Mali matsumura and damage in Xinjiang. Xinjiang Agric. Sci. 41, 31–33.
Khani, A., Moharamipour, S., and Barzegar, M. (2007). Cold tolerance and trehalose accumulation in overwintering larvae of the codling moth, Cydia pomonella (Lepidoptera: Tortricidae). Eur. J. Entomol. 104, 385–392. doi: 10.14411/eje.2007.057
Kost’al, V., Slachta, M., and Simek, P. (2001). Cryoprotective role of polyols independent of the increase in supercooling capacity in diapausing adults of Pyrrhocoris apterus (Heteroptera: Insecta). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130, 365–374. doi: 10.1016/S1096-4959(01)00441-9
Kostal, V., Zahradnickova, H., Simek, P., and Zeleny, J. (2007). Multiple component system of sugars and polyols in the overwintering spruce bark beetle, Ips typographus. J. Insect. Physiol. 53, 580–586. doi: 10.1016/j.jinsphys.2007.02.009
Kron, A. S., Zengerer, V., Bieri, M., Dreyfuss, V., Sostizzo, T., Schmid, M., et al. (2020). Pseudomonas orientalis F9 pyoverdine, safracin, and phenazine mutants remain effectiveantagonists against Erwinia amylovora in appleflowers. Appl. Environ. Microbiol. 86, e02620–19. doi: 10.1128/AEM.02620-19
Kumar, D., Kumar, L., Nagar, S., Raina, C., Parshad, R., and Gupta, V. K. (2012). Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch. Appl. Sci. Res. 4, 1763–1770.
Lefort, M. C., Boyer, S., and Glare, T. R. (2017). A response to Pennisi - “How do gut microbiomes help herbivores”, a hint into next-generation biocontrol solutions. Rethinking Ecol. 1, 9–13. doi: 10.3897/rethinkingecology.1.12932
Lin, P., and Lin, Y. (2000). “The survey of the distribution of Tianshan Mountain wild fruit forests and it distribution in Ili prefecture,” in Wild Fruit Forests Resources in Tianshan Mountains - Comprehensive Research on Wild Fruit Forests in Ili, Xinjiang, China, eds P. Lin and N. Cui (Beijing: China Forestry Publishing House), 46–62.
Liu, D., Coloe, S., Baird, R., and Pederson, J. (2000). Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38:471. doi: 10.1128/JCM.38.1.471-471.2000
Liu, X., Li, X., Bozorov, T. A., Ma, R., Ma, J., Zhang, Y., et al. (2020). Characterization and pathogenicity of six Cytospora strains causing stem canker of wild apple in the Tianshan Forest. China For. Pathol. 50:e12587. doi: 10.1111/efp.12587
Lourenço, A., Rencoret, J., Chemetova, C., Gominho, J., Gutiérrez, A., Del Río, J. C., et al. (2016). Lignin composition and structure differs between xylem, phloem and phellem in Quercus suber L. Front. Plant Sci. 7:1612. doi: 10.3389/fpls.2016.01612
Lundell, T. K., Mäkelä, M. R., De Vries, R. P., and Hildén, K. S. (2014). “Genomics, lifestyles and future prospects of wood-decay and litter-decomposing Basidiomycota,” in Advances in Botanical Research, ed. F. M. Martin (Cambridge MA: Academic Press), 329–370.
Machado, K. M. G., Matheus, D. R., and Bononi, V. L. R. (2005). Ligninolytic enzymes production and Remazol Brilliant Blue R decolorization by tropical Brazilian basidiomycetes fungi. Braz. J. Microbiol. 36, 246–252. doi: 10.1590/S1517-83822005000300008
Mayerhof, H., Marshall, R. T., White, C. H., and Lu, M. (1973). Characterization of a heat-stable protease of Pseudomonas fluorescens P26. Appl. Microbiol. 25, 44–48. doi: 10.1128/am.25.1.44-48.1973
Mereghetti, V., Chouaia, B., and Montagna, M. (2017). New insights into the microbiota of moth pests. Int. J. Mol. Sci. 18:2450. doi: 10.3390/ijms18112450
Mesanza, N., Iturritxa, E., and Patten, C. L. (2016). Native rhizobacteria as biocontrol agents of Heterobasidion annosum s.s. and Armillaria mellea infection of Pinus radiata. Biol. Control 101, 8–16. doi: 10.3390/f8070235
Mogouong, J., Constant, P., Lavallee, R., and Guertin, C. (2020). Gut microbiome of the emerald ash borer, Agrilus planipennis Fairmaire, and its relationship with insect population density. FEMS Microbiol. Ecol. 96:141. doi: 10.1093/femsec/fiaa141
Morales-Jiménez, J., Zúñiga, G., Ramírez-Saad, H. C., and Hernández-Rodríguez, C. (2012). Gut-associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microbial. Ecol. 64, 268–278. doi: 10.1007/s00248-011-9999-0
Park, J. H., Pavlov, I. N., Kim, M. J., Park, M. S., Oh, S. Y., Park, K. H., et al. (2020). Investigating wood decaying fungi diversity in Central Siberia, Russia using ITS sequence analysis and interaction with host trees. Sustainability 12:2535. doi: 10.3390/su12062535
Pennisi, E. (2017). How do gut microbes help herbivores? Counting the ways. Science 355, 236–236. doi: 10.1126/science.355.6322.236
Prosser, J. I., Bohannan, B. J. M., Curtis, T. P., Ellis, R. J., Firestone, M. K., Freckleton, R. P., et al. (2007). The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392. doi: 10.1038/nrmicro1643
Richards, C. M., Volk, G. M., Reilley, A. A., Henk, A. D., Lockwood, D. R., Reeves, P. A., et al. (2009). Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet. Genomes 5, 339–347. doi: 10.1007/s11295-008-0190-9
Rojas-Jiménez, K., and Hernández, M. (2015). Isolation of fungi and bacteria associated with the guts of tropical wood-feeding Coleoptera and determination of their lignocellulolytic activities. Int. J. Microbiol. 2015:285018. doi: 10.1155/2015/285018
Ruby, E., Henderson, B., and Mcfall-Ngai, M. (2004). Microbiology. We get by with a little help from our (little) friends. Science 303, 1305–1307. doi: 10.1126/science.1094662
Sam, Q. H., Chang, M. W., and Chai, L. Y. (2017). The fungal mycobiome and its interaction with gut bacteria in the host. Int. J. Mol. Sci. 18:330. doi: 10.3390/ijms1802.0330
Thissera, B., Alhadrami, H. A., Hassan, M. H. A., Hassan, H. M., Bawazeer, M., Yaseen, M., et al. (2020). Induction of cryptic antifungal pulicatin derivatives from Pantoea agglomerans by microbial co-culture. Biomolecules 10:268. doi: 10.3390/biom10020268
Vargas-Asensio, G., Pinto-Tomas, A., Rivera, B., Hernandez, M., Hernandez, C., Soto-Montero, S., et al. (2014). Uncovering the cultivable microbial diversity of costa rican beetles and its ability to break down plant cell wall components. PLoS One 9:e113303. doi: 10.1371/journal.pone.0113303
Vasanthakumar, A., Handelsman, J. O., Schloss, P. D., Bauer, L. S., and Raffa, K. F. (2008). Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ. Entomol. 37, 1344–1353.
Wang, J., Chen, H., and Tang, M. (2017). Community structure of gut bacteria of Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae) larvae during overwintering stage. Sci. Rep. 7:14242. doi: 10.1038/s41598-017-14724-y
Wang, Z. Y. (2013). Research on Biological Control of Agrilus mali Matsumura (Coleoptera: Buprestidae) in stands of Malus sieversii in Xinjiang. Doctor Doctoral Dissertation. Beijing: Chinese Academy of Forestry.
White, T. J., Bruns, T., Lee, S., and Taylor, J. H. (1990). “Amplification and directsequencing of fungal ribosomal RNA gene for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. S. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Yang, J., Fang, G., Chen, Y., and De Maeyer, P. (2017). Climate change in the Tianshan and northern Kunlun Mountains based on GCM simulation ensemble with Bayesian model averaging. J. Arid Land. 9, 622–634. doi: 10.1007/s40333-017-0100-9
Yao, T., Zhao, Q., Li, X.-Y., Shen, Z.-T., Ran, P.-Y., and Wu, W. (2020). Spatiotemporal variations of multi-scale drought in Shandong Province from 1961 to 2017. Water Supply 21, 525–541. doi: 10.2166/ws.2020.332
Yao, Z., Ma, Q., Cai, Z., Raza, M. F., Bai, S., Wang, Y., et al. (2019). Similar shift patterns in gut bacterial and fungal communities across the life stages of Bactrocera minax larvae from two field populations. Front Microbiol 10:2262. doi: 10.3389/fmicb.2019.02262
Yun, J.-H., Roh, S. W., Whon, T. W., Jung, M.-J., Kim, M.-S., Park, D.-S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Keywords: Agrilus mali, larval gut microbiota, Pseudomonas synxantha, invasive insect, wild apple, antifungal compound
Citation: Bozorov TA, Toshmatov ZO, Kahar G, Zhang D, Shao H and Gafforov Y (2021) Wild Apple-Associated Fungi and Bacteria Compete to Colonize the Larval Gut of an Invasive Wood-Borer Agrilus mali in Tianshan Forests. Front. Microbiol. 12:743831. doi: 10.3389/fmicb.2021.743831
Received: 19 July 2021; Accepted: 13 September 2021;
Published: 15 October 2021.
Edited by:
Takema Fukatsu, National Institute of Advanced Industrial Science and Technology (AIST), JapanReviewed by:
Kohei Kubota, The University of Tokyo, JapanCopyright © 2021 Bozorov, Toshmatov, Kahar, Zhang, Shao and Gafforov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tohir A. Bozorov, dG9oaXJib3pvcm92QHlhaG9vLmNvbQ==; Daoyuan Zhang, emhhbmdkeUBtcy54aWIuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.