- 1Key Laboratory for Microbial Resources of the Ministry of Education, School of Life Sciences, School of Medicine, School of Chemical Science and Technology, Yunnan Institute of Microbiology, Yunnan University, Kunming, China
- 2School of Agriculture and Life Sciences, Kunming University, Kunming, China
A novel actinobacterium, strain YIM 96748T, was isolated from a saline soil sample collected from the south bank of Aiding Lake in Xinjiang Province, Northwest China. Phylogenetic analysis based on 16S rRNA gene sequences revealed that strain YIM 96748T is closely related to Amycolatopsis cihanbeyliensis BNT52T (98.9%) and Amycolatopsis jiangsuensis KLBMP 1262T (97.2%). The DNA–DNA relatedness between strain YIM 96748T and its closest type strain A. cihanbeyliensis BNT52T was 59.6%. The average nucleotide identity between strain YIM 96748T and its neighbor strain was 88.97%. Based on the genotypic and phenotypic characteristics, it is concluded that strain YIM 96748T represents a novel species of the genus Amycolatopsis, whose name was proposed as Amycolatopsis aidingensis sp. nov. The type strain is YIM 96748T. To investigate the biosynthetic potential of producing secondary metabolites, the complete genome of YIM 96748T was sequenced and analyzed. The complete genome sequence of YIM 96748T consists of a 7,657,695-bp circular chromosome, comprising 7,162 predicted genes with a DNA G + C content of 70.21 mol%. Fifty-one putative biosynthetic gene clusters of secondary metabolites were found, including the antibacterial/antitumor agent TLN-05220, the antibacterial agent nocardicin A, the antifungal agent nystatin A1, and the osmolyte ectoine. The investigation of the secondary metabolites of A. aidingensis YIM96748T led to the discovery of two new phenylpropyl acetate enantiomers, amycoletates A (1) and B (2), and five known compounds: 4-hydroxy phenethyl acetate (3), 2-p-acetoxyphenylethanol (4), (S)-ethyl indole-3-lactate (5), (R)-ethyl indole-3-lactate (6), and p-hydroxybenzoic acid (7). One of the gene clusters 14, 36, and 43, which contain a single module of polyketide synthase, might be responsible for the biosynthesis of compounds 1 and 2 from compound 7 as a precursor. Further studies, including the one strain many compounds approach (OSMAC) and genetic modification, are needed to explore novel compounds from this talented halophilic Amycolatopsis strain.
Introduction
Strains of the genus Amycolatopsis are valuable sources, which could produce many active secondary metabolites. Glycopeptide antibiotic vancomycin from Amycolatopsis orientalis has been used to treat bacterial infections, including methicillin-resistant Staphylococcus aureus (MRSA), for decades (Foldes et al., 1983). The ansamycin antibiotic rifamycin from Amycolatopsis mediterranei S699 is one of the first-line therapies used for the treatment of pathogenic Gram-positive cocci and mycobacteria infections for more than half a century (Sepkowitz et al., 1995). Besides, many powerful antibiotics, such as barrymycin, balhimycin, chloroeremomycin, telavancin, oritavancin, and dethymicin, have been discovered from this genus. The earliest discovered species of Amycolatopsis is Streptomyces orientalis (Brigham and Pittenger, 1956). It was renamed as Amycolatopsis orientalis in 1986 (Lechevalier et al., 1986). Eighty-eight species have been validly published with the correct name in this genus until now. Members of the genus Amycolatopsis are aerobic or facultative aerobic, Gram-positive, catalase-positive, non-acid-fast, non-motile actinomycetes that contain meso-diaminopimelic acid in the wall peptidoglycan (Lechevalier and Lechevalier, 1970). Fatty acids are rich in iso- and anteiso-branched components and show a lack of mycolic acids (Takahashi, 2001). The predominant menaquinone type is MK-9(H4), and the G + C content of the genomic DNA ranges from 66 to 75 mol%.
Halophilic actinomycete strains are regarded as one of the valuable resources for the discovery of useful metabolites, such as antibiotics, compatible solutes, and potential industrial useful polymers. During our research on the resources of actinomycetes in the high-salt environment in Xinjiang Province, a novel actinobacterial strain, YIM 96748T, with siderophore activity was isolated from the south bank of Aiding Lake, which is situated in the southern part of the Turpan Basin in Xinjiang Province, Northwest China. In this study, YIM 96748T was recognized as a novel species of the genus Amycolatopsis through a polyphasic approach, and its name was proposed as Amycolatopsis aidingensis sp. nov. Seven metabolites, including two new ones, were obtained from fermentation extracts. The complete genome of this talented halophilic actinomycete strain was sequenced and analyzed. Putative secondary metabolite biosynthetic gene clusters were investigated to guide the discovery of novel natural products.
Materials and Methods
Bacterial Isolation
Strain YIM 96748T was isolated on cellulose–casein multi-salt (CCMS) medium (Tang et al., 2008) supplemented with 5% NaCl (w/v), which had been inoculated with a soil suspension and incubated at 37°C for 4 weeks from soil samples isolated from a saline soil sample collected from the south bank of Aiding Lake, which is situated in the southern part of the Turpan Basin in Xinjiang Province, Northwest China. The isolate was purified on and maintained on yeast extract–malt extract agar (ISP 2) and preserved as a suspension of mycelial fragments in glycerol (20%, v/v) at −80°C.
16S Ribosomal RNA Gene Sequence, Phylogenetic Analysis, and G + C Content
DNA isolation, 16S ribosomal RNA (rRNA) gene amplification, and sequencing were done as described by Feng et al. (2020). Identification of the phylogenetic neighbors and the calculation of pairwise 16S rRNA gene sequence identities were achieved using the EzTaxon-e database1 (Yoon et al., 2017). Phylogenetic analyses were carried out using three treeing algorithms—the neighbor-joining (Saitou and Nei, 1987), maximum likelihood (Felsenstein, 2005), and maximum parsimony (Fitch, 1971) methods—with MEGA version 7.0 (Kumar et al., 2016) and bootstrap values based on 1,000 replications (Felsenstein, 1985). The DNA G + C mole percent value was obtained from the genomic sequence. DNA–DNA hybridization (DDH) was determined using the in silico method with the Genome-to-Genome Distance Calculator server version 2.1 (Meier-Kolthoff et al., 2012). The average nucleotide identity (ANI) was calculated with OrthoANI (Richter et al., 2016). The average amino acid identity (AAI) values were calculated from the protein sequences using an online AAI calculator.2 Protein sequences were predicted from the genome sequences using GeneMarkS (Besemer et al., 2001). The strain Amycolatopsis cihanbeyliensis BNT52T was used as the reference strain in the ANI value calculation and digital DDH (Stamatakis, 2014).
Physiological, Morphological, and Biochemical Tests
The cultural and morphological characteristics of the isolate were determined after growth for 7 and 14 days at 37°C on the following media: ISP 2, ISP 3, ISP 4, and ISP 5, Czapek’s agar, potato dextrose agar (PDA), and nutrient agar (NA) media with 5% (w/v) NaCl concentration. Cell morphology was examined using a light microscope (DM2000; Leica, Wetzlar, Germany) and a scanning electron microscope (XL30 ESEM-TMP; Philips-FEI, Eindhoven, Netherlands), with cells grown on ISP 2 medium for 14 days at 37°C. Growth was tested at different temperatures (4, 10, 15, 20, 25, 28, 30, 35, 37, 40, 45, 50, and 55°C) and at pH 4.0–12.0 (at intervals of 0.5 pH unit) using the buffer system described by Xu et al. (2005) and for NaCl tolerance (0–30%, w/v) using the ISP 2 medium after 14 days of incubation at 37°C. Carbon source tests for growth were carried out on ISP 9 containing 5% (w/v) NaCl, as described by Shirling and Gottlieb (1966). Nitrogen source utilization tests were carried out as described by Gordon et al. (1974). Catalase activity was determined by bubble production in 3% (v/v) H2O2, and oxidase activity was determined using 1% (w/v) oxidation of tetramethyl-p-phenylenediamine. Gelatinase activities, starch hydrolysis, nitrate reduction, urease, milk peptonization and coagulation, and H2S and melanin production were assessed as described by Smibert and Krieg (1994). The other enzyme activities were determined using the API ZYM and API 20NE systems (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions.
Biochemical Characteristics
Biomass for quantitative fatty acid analysis was obtained from cultures grown in tryptic soy broth (TSB) for 7 days at 37°C and 200 rpm. Biomass for other chemotaxonomic studies was obtained after cultivation at 37°C for 7 days in shaken cultures with ISP 2 containing 5% (w/v) NaCl. The whole-cell sugar pattern and peptidoglycan amino acids were identified by high-performance liquid chromatography (HPLC) according to the methods used by Tang et al. (2009). The isomer of diaminopimelic acid in whole-cell hydrolyzates was determined using thin-layer chromatography (TLC) as described by Lechevalier and Lechevalier (1970) and Hasegawa et al. (1983). Polar lipids were extracted, examined by two-dimensional TLC, and identified using previously described procedures (Minnikin et al., 1984). Menaquinones were extracted and purified as described by Collins et al. (1977) and analyzed using HPLC (Groth et al., 1996). Cellular fatty acid analysis was performed using the microbial identification system (Sherlock version 6.1; MIDI database: TSBA6).
Genome Sequencing and Mining
A single colony of YIM 96748T was inoculated into 50 ml TSB liquid (BD Biosciences, Franklin Lakes, NJ, United States) for 40 h at 28°C with 250 rpm vigorous shaking. DNA isolation and purification in YIM 96748T were carried out according to standard procedures (Pospiech and Neumann, 1995). The quality and quantity of purified genomic DNA were analyzed using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and 0.8% agarose gel electrophoresis.
Genomic DNA was sheared to 8–10-kb length fragments randomly and a genomic DNA library constructed for PacBio sequencing. PacBio clean data were generated by the sequencing platform, and all reads were assembled and checked using HGAP software (Rhoads and Au, 2015). The protein coding sequences were predicted with Glimmer (v3.02) on NCBI (Delcher et al., 2007), and the gene functions were annotated using the NCBI Prokaryotic Genome Annotation Pipeline. The Clusters of Orthologous Groups of proteins (COG) and Gene Ontology (GO) programs (Tatusov et al., 2003; The Gene Ontology Consortium, 2015) were used to analyze the function of the annotated genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Kanehisa and Goto, 2000) was carried out to determine the key potential pathways in YIM 96748T.
The CRISPR finder platform (Grissa et al., 2007) was used to identify the CRISPR-Cas sequences on the chromosome. ResFinder (Zankari et al., 2012) and the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al., 2013) were used to predict resistance genes. The VFDB database (Chen et al., 2016) was used to predict the bacterial virulence factors, and PHAST (Zhou et al., 2011) was used to identify putative prophages on the chromosome. The biosynthetic gene clusters for putative secondary metabolites were identified using the antiSMASH 5.0 program (Blin et al., 2019) and verified by manual inspection.
Fermentation
Strain YIM96748T was activated in ISP 2 medium (4 g soytone, 10 g malt extract, 4 g glucose, 1,000 ml H2O, pH 7.1–7.2) at 37°C for 5 days. The activated strain was inoculated into 500-ml Erlenmeyer flasks containing 100 ml of the ISP 2 liquid medium and cultured for 7 days at 37°C and 200 rpm. Then, 5.0 ml of the seed cultures were transferred into 500-ml Erlenmeyer flasks containing 100 ml of fermentation medium [20 ml glycerol, 20 g dextrin, 10 g soytone, 2 g yeast extract, 2 g (NH4)2SO4, 2 g CaCO3, 1,000 ml H2O, pH 7.0] and cultured for 10 days at 37°C and 200 rpm (total, 10 L).
Isolation and Identification of Compounds
The broth was extracted with 10 L ethyl acetate (EtOAc) three times, and the solvent was removed under vacuum to obtain the EtOAc extract (5.12 g). The crude extract was separated into five fractions (Fr.1–Fr.5) with column chromatography (CC) on silica gel, eluting stepwise with a CHCl3–MeOH gradient [CHCl3, CHCl3–MeOH = 30:1 (v/v), CHCl3–MeOH = 10:1 (v/v), CHCl3–MeOH = 1:1 (v/v), MeOH]. Fr.2 was divided into three parts (Fr.2-1–Fr.2-3) with a silica gel column with petroleum ether–ethyl acetate (50:1–1:1, v/v), and then Fr.2-2 was separated by a Sephadex LH-20 column with MeOH to obtain compound 3 (8.9 mg). Fr.2-3 was fractionated by a silica gel column with dichloromethane–ethyl acetate (50:1–1:1, v/v) to afford compounds 5 (1.0 mg) and 6 (1.2 mg). Fr.3 was divided into three parts (Fr.3-1–Fr.3-3) by a silica gel column with dichloromethane–ethyl acetate (80:–1:1, v/v). Then, Fr.3-1 was separated using preparative HPLC to yield compound 7 (10.2 mg). Fr.3-2 was fractionated using a Sephadex LH-20 column with MeOH to obtain compounds 1/2 (0.9 mg) and 4 (3.4 mg).
NMR spectra, including 1D and 2D spectra, were acquired at room temperature on a Bruker Avance 400-MHz instrument (Bruker, Karlsruhe, Germany). High-resolution electrospray ionization mass spectroscopy (HRESIMS) data were obtained on an Agilent G3250AA (Agilent, Santa Clara, CA, United States). Reversed-phase HPLC was performed on an Agilent Eclipse XDB-C18 (5 μm) column. Silica gel (200–300 mesh; Qingdao Marine Chemical Group Co., Qingdao, China), Lichroprep RP-18 gel (40–63 mm; Merck, Darmstadt, Germany), and Sephadex LH-20 (GE Healthcare Bio-Science AB, Uppsala, Sweden) were used for CC. Analytical TLC was carried out using HSGF 254 plates (Qingdao Marine Chemical Group Co., Qingdao, China) and visualized by spraying with an anisaldehyde–H2SO4 reagent.
Results and Discussion
Molecular Phylogenetic Analysis
The sequence of the 16S rRNA gene of strain YIM 96748T (1,517 bp, accession no. MZ348539) was used for phylogenetic analysis. Phylogenetic analyses of the 16S rRNA gene sequences showed that strain YIM 96748T was a member of the genus Amycolatopsis. It is evident from the neighbor-joining tree (Figure 1) that strain YIM 96748T formed a cluster with A. cihanbeyliensis BNT52T, Amycolatopsis jiangsuensis KLBMP 1262T, and Amycolatopsis suaedae 8-3EHSuT within members of the genus Amycolatopsis. The isolate showed the highest 16S rRNA gene sequence similarity with A. cihanbeyliensis BNT52T (98.9%). The genome tree demonstrated that YIM 96748T is steadily clustered in a branch with strain A. cihanbeyliensis BNT52T (GCA_006715045.1) under the 100 bootstrap values (Supplementary Figure 1). However, the level of DNA–DNA relatedness between strains YIM 96748T and A. cihanbeyliensis BNT52T (GenBank accession no. GCA_006715045.1) was only 59.60%, a value well below the 70% threshold recommended for the delineation of bacterial species by Wayne (1988). The ANI value between these two strains was 88.97%. Two-way AAI analysis was used. Strain YIM 96748T and the closely related strain A. cihanbeyliensis BNT52T had an AAI value of 88.9%, lower than the 95–96% threshold for species demarcation, confirming that strain YIM 96748T represents a novel species within the genus Amycolatopsis. The G + C content of the genomic DNA of the type strain was 70.21 mol%.
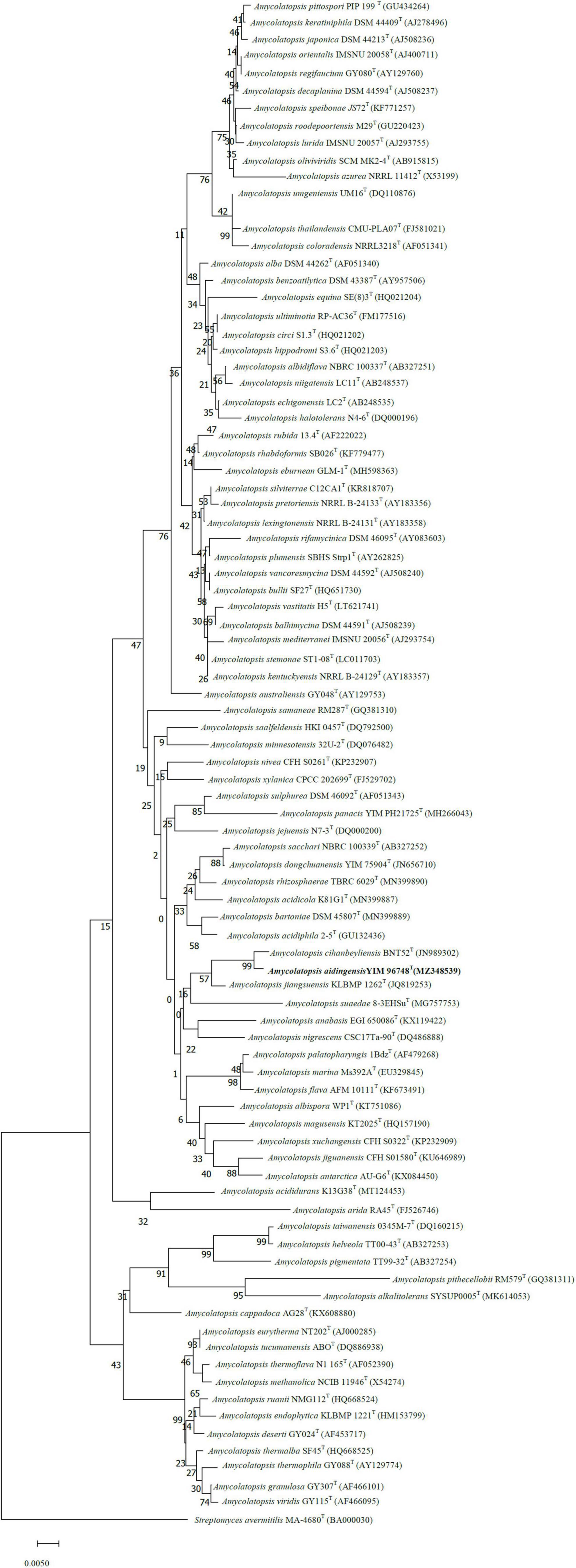
Figure 1. Neighbor-joining tree (MEGA 7.0) showing the relationship between strain YIM 96748T and related taxa. Streptomyces avermitilis MA-4680T was used as the outgroup.
Physiological, Morphological, and Biochemical Tests
The cells of strain YIM 96748T were Gram-reaction-positive, aerobic, non-motile, catalase-positive actinomycetes, which formed a septal substrate mycelium that fragmented into rod-like elements. Irregular swelling appeared in the part of the aerial mycelium (Figure 2) when grown on ISP 2 media with 5% (w/v) NaCl for 14 days. Strain YIM 96748T grew well on ISP 2, ISP 3, ISP 4, and ISP 5 agar, Czapek’s agar, NA, and PDA media. The color of the aerial mycelium was white on ISP 2, ISP 3, ISP 4, and ISP 5 agar, Czapek’s agar, NA, and PDA media. The color of the substrate mycelium was purple on PDA, NA, and ISP 2 media. Diffusible pigments were not produced. The growth of strain YIM 96748T was observed at pH 5–12 (optimum, 8) and in the presence of 1–15% (w/v) NaCl. The temperature range for growth was 20–45°C, with optimum growth at 37°C. The strain was positive for nitrate reduction, gelatin, milk peptonization, and para-nitro-β-D-methyl galactose. It was negative for urease, indole, H2S, and hydrolysis of aesculin and starch. Tweens 20, 40, and 80 were hydrolyzed. In the API 20NE test, the strain was positive for urease, malic acid, and maltose. In the API ZYM test, it was positive for alkaline phosphatase, esterase C4, esterase lipase C8, lipase C14, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-glucosidase, and N-acetyl-β-glucosaminidase. In the API 20NE test, there was assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, maltose, gluconate, adipic acid, and citric acid. The physiological properties that distinguished strain YIM 96748T from closely related species of the genus Amycolatopsis are listed in Table 1.
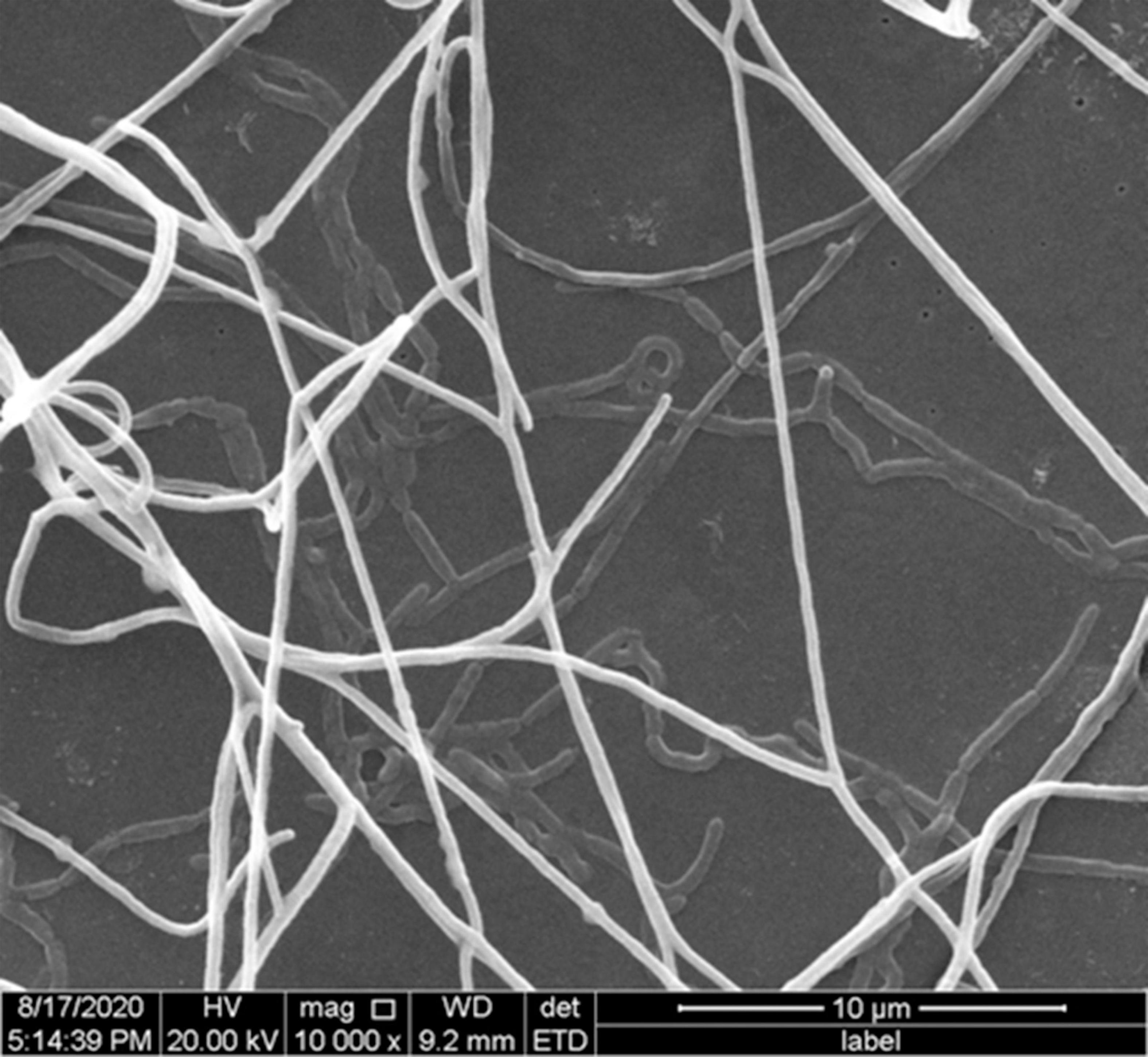
Figure 2. Scanning electron micrograph of the aerial mycelium and substrate mycelium of strain YIM 96748T on ISP 2 with 5% (w/v) NaCl concentration after incubation for14 days. Bar, 10 μm.
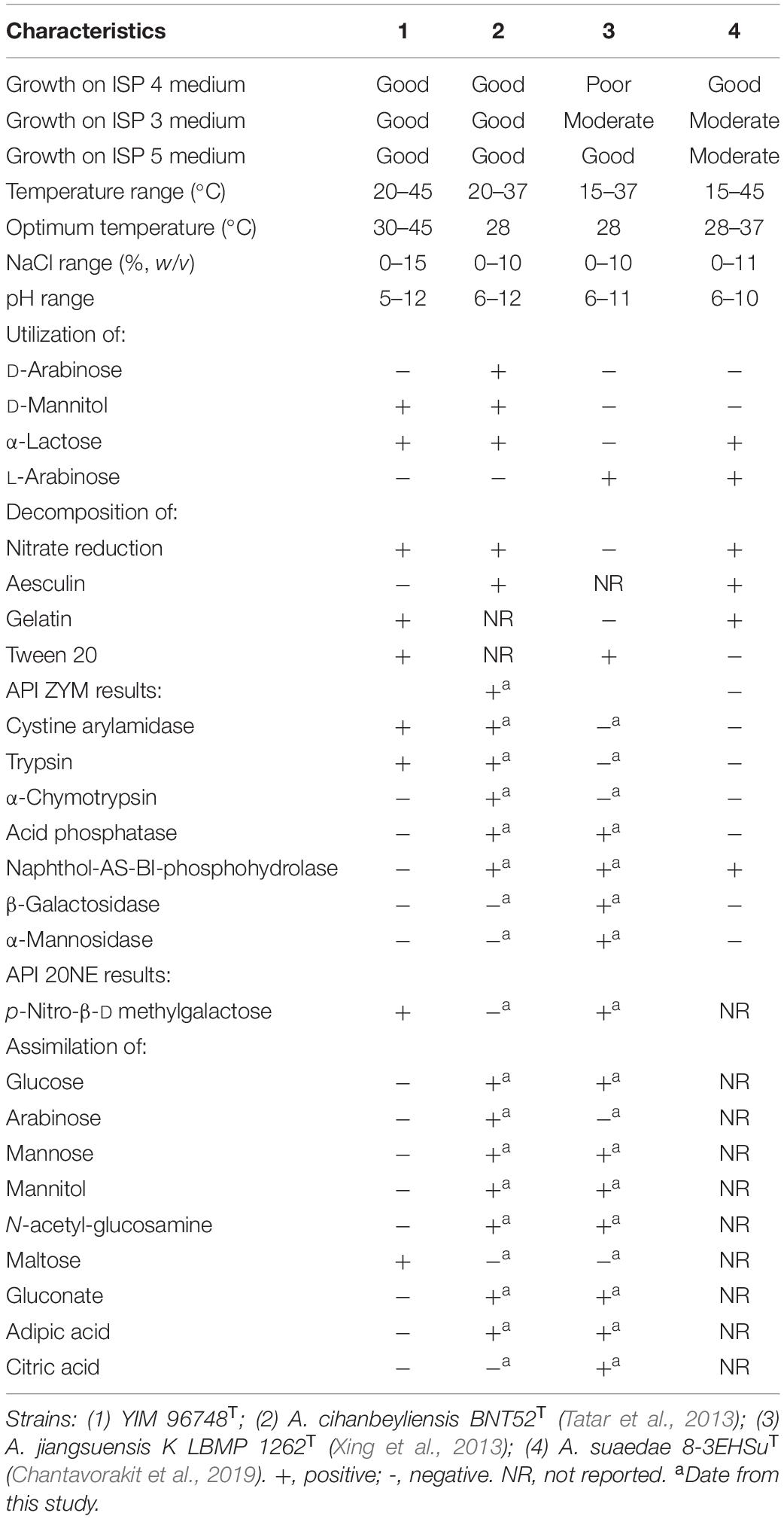
Table 1. Differentiation of the physiological characteristics of strain YIM 96748T, Amycolatopsis cihanbeyliensis BNT52T, Amycolatopsis jiangsuensis KLBMP 1262T, and Amycolatopsis suaedae 8-3EHSuT.
Biochemical Characteristics
The cell wall diamino acid in the peptidoglycan of strain YIM 96748T was determined to be meso-diaminopimelic acid, and the whole-cell sugars include manose (6.8%), ribose (5.3%), glucose (57.8%), and galactose (19.8%) as the major components. The polar lipids were found to include diphosphatidylglycerol, unidentified polar lipids, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipids, an unidentified amino lipid, and phosphatidylmonomethylethanolamine (Supplementary Figure 2). MK-9(H4) was detected as the major menaquinone. The major fatty acids were determined to be iso-C16:0 (54.8%) and C17:1 w6c (15.6%), with C16:0 (2.8%), C18:0 (2.7%), anteiso-C17:0 (2.6%), iso-C14:0 (2.3%), iso-C15:0 (2.8%), Summed Feature 3 (16:1 w6c/16:1 w7c) (1.8%), iso-C16:1 H (1.7%), C17:1 w8c (1.6%), C19:0 10-methyl (1.1%), and iso-C17:0 (1.1%) present as minor components. The major fatty acid is the same as that of A. cihanbeyliensis BNT52T, but its content is really different. The chemotaxonomic characteristics of strain YIM 96748T are therefore considered to be typical of members of the genus Amycolatopsis. Further detailed biochemical characteristics are listed in Table 2.
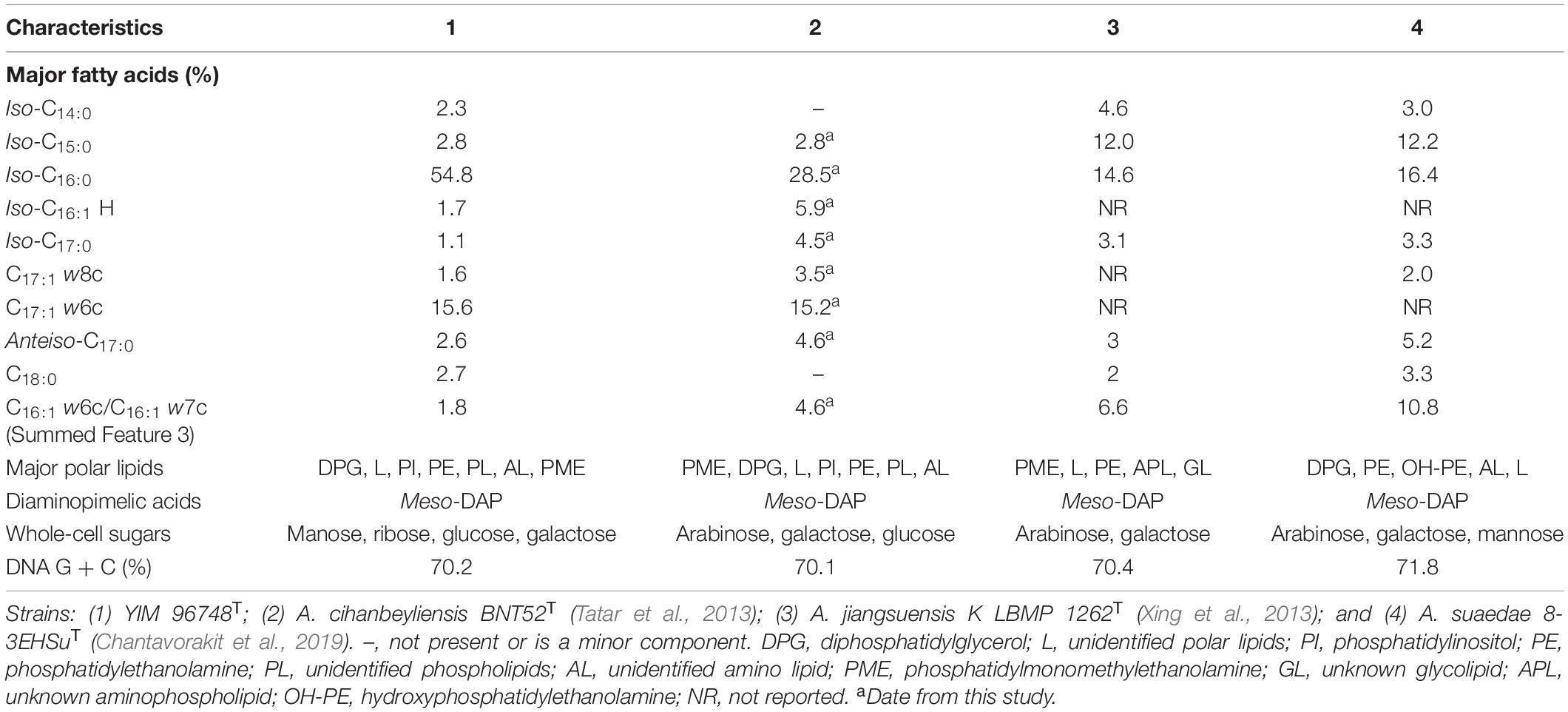
Table 2. Differentiation of the chemotaxonomic characteristics between strain YIM 96748T, Amycolatopsis cihanbeyliensis BNT52T, Amycolatopsis jiangsuensis KLBMP 1262T, and Amycolatopsis suaedae 8-3EHSuT.
Description of Amycolatopsis aidingensis sp. nov.
Amycolatopsis aidingensis (ai.ding.en’sis), N.L. fem. adj. aidingensis of or belonging to Aiding Lake, a salt lake in China (where the type strain was isolated). This strain is a Gram-positive, aerobic, non-motile actinobacterium that formed a septal substrate mycelium that fragmented into rod-like elements, with irregular swelling appearing in the part of the aerial mycelium. It grows well on ISP 2, ISP 3, ISP 4, and ISP 5 agar, Czapek’s agar, NA, and PDA media. The color of the aerial mycelium was white on ISP 2, ISP 3, ISP 4, and ISP 5 agar, Czapek’s agar, NA, and PDA media. The color of the substrate mycelium was purple on PDA, NA, and ISP 2 media. Growth occurs at pH 5–12 (optimum, pH 8) at 20–45°C (optimum, 37°C) and with 1–15% (w/v) NaCl tolerance (optimum, 5% NaCl). D-Trehalose, mannitol, xylitol, D-cellulose, D-mannitol, D-sorbitol, raffinose, D-xylose, maltose, α-lactose, fructose, sodium citrate, amber acid, and α-D-glucose were utilized as sole carbon and energy sources, but not D-galactose, melezitose, oligofructose, L-arabinose, α-methyl glucoside, D-salicylin, β-cyclodextrin, or maltitol. It utilizes xanthine, L-phenylalanine, glycine, L-threonine, L-lysine, L-tyrosine, glutamine, L-asparagine, alanine but not adenine, L-methionine, aspartic acid, arginine, or hypoxanthine as sole nitrogen sources.
The cell wall contained meso-diaminopimelic acid, and the predominant menaquinone was MK-9 (H4). The major phospholipids consisted of diphosphatidylglycerol, unidentified polar lipids, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipids, an unidentified amino lipid, and phosphatidylmonomethylethanolamine. The major fatty acids (>10.0%) were iso-C16:0 (54.8%) and C17:1 w6c (15.6%). The DNA G + C content of the type strain is 70.2 mol%. The type strain is YIM 96748T (= KCTC 49720T = CGMCC 4.7734T = CCTCC AA 2021025T), isolated from Turpan City, Xinjiang Province, Northwest China.
Genome Sequencing and Analysis
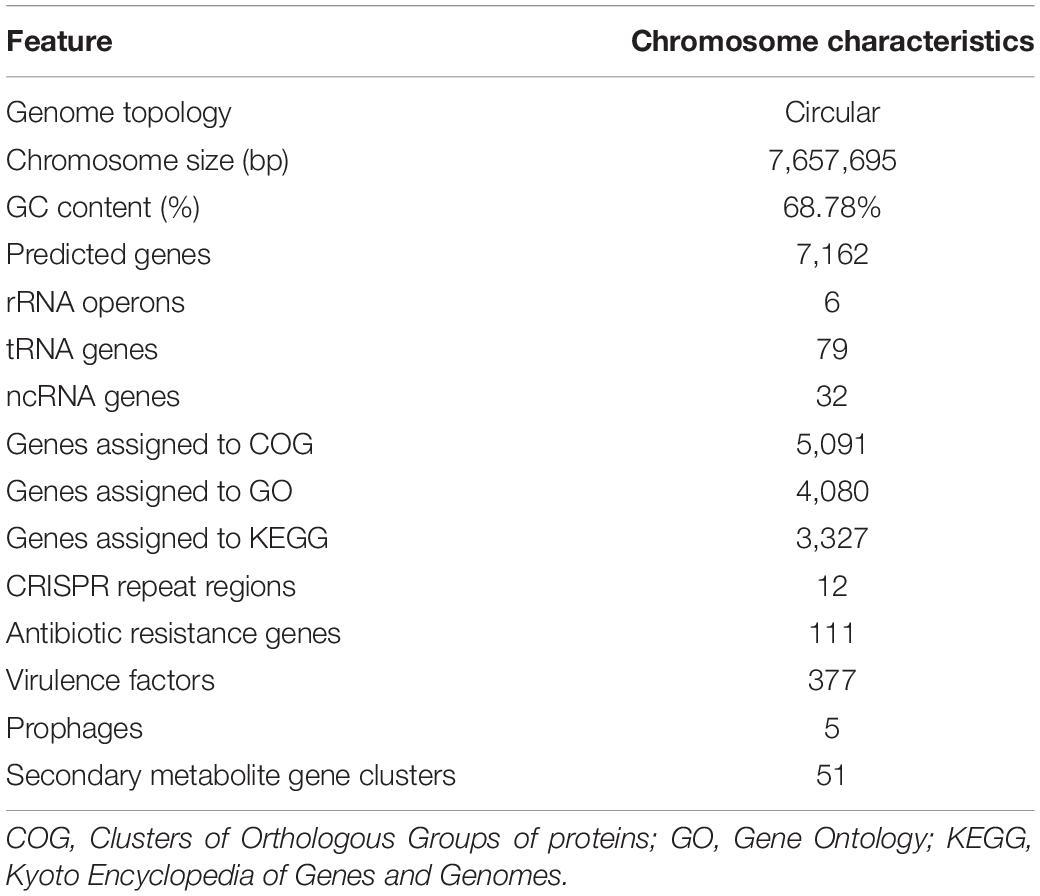
The complete genome of YIM 96748T was obtained using the PacBio RSII platform. Approximately 714 Mb PacBio clean data were generated. The average depth of genome coverage was 93-fold. The complete genome of YIM 96748T was composed of a circular chromosome of 7,657,695 bp with a GC content of 70.21 mol% (accession no. GCA_018885265.1). The chromosome contained 7,162 predicted genes, including 6 rRNA genes, 49 transfer RNA (tRNA) genes, and 32 small RNA (sRNA) genes (Figure 3 and Table 3). Among the identified genes, 5,091 and 4,080 were classified into functional categories based on COG and GO designation, respectively (Supplementary Figures 6, 7). Of the KEGG pathways, 3,327 genes were assigned (Supplementary Figure 8). The CRISPR-Cas systems are used by bacteria for defense against the invasion of foreign genetic elements such as viruses and alien plasmids (Barrangou et al., 2007). Twelve putative CRISPR repeat regions were identified on the chromosome of YIM 96748T. The length of CRISPR ranged from 89 to 518 bp, and the number of spacers ranged from 1 to 8 (Table 3). One hundred eleven putative antibiotic resistance genes and 377 putative virulence factors were found (Table 3). Five incomplete prophage remnants were detected on the chromosome, and the length of the prophages ranged from 8,264 to 28,169 bp (Table 3).
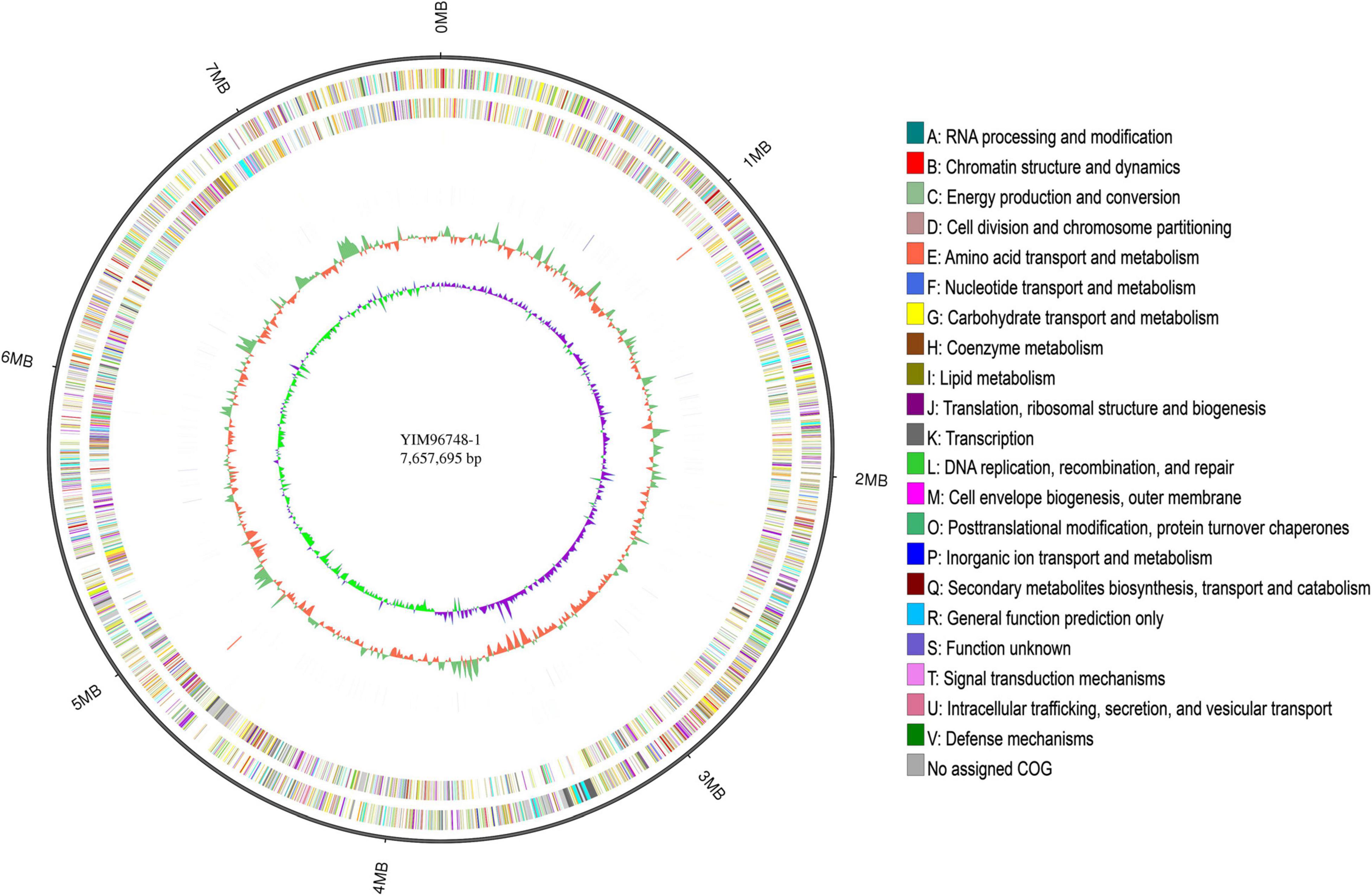
Figure 3. Circular genome map of YIM 96748T. Tracks (from inner to outer): 1, Genome size; 2, forward strand gene, colored according to Clusters of Orthologous Groups of proteins (COG) classification; 3, reverse strand gene, colored according to COG classification; 4, forward strand ncRNA; 5, reverse strand ncRNA; 6, repeat; 7, GC content; and 8, GC skew.
Genome Mining of Secondary Metabolites
A large number of halophilic actinomycetes were isolated in our previous work. Most strains of this group grow very slowly, and little product could be isolated under traditional fermentation conditions. To evaluate the ability of these strains to produce secondary metabolites, the genomes of more than 40 strains were sequenced. Most of them contained very few gene clusters. Among these strains, YIM 96748T contained significantly more gene clusters than did the other strains. Fifty-one putative biosynthetic gene clusters were found (Supplementary Figure 9), including 14 saccharides, 6 polyketides, 6 polyketide–non-ribosomal peptides, 6 non-ribosomal peptides, 3 terpenes, 3 fatty acids, 3 lantipeptides, 2 lasso peptides, 2 NAPAAs (non-alpha poly-amino acids), 2 RiPPs (ribosomally synthesized and post-translationally modified peptides), 1 guanidinotide, 1 sactipeptide, 1 siderophore, and 1 ectoine. Seven putative gene clusters showed high similarity (>70% of genes showed similarity) to ectoine, Ery, geosmin, citrulassin B, mirubactin, nocardicin A, and TLN-05220 gene clusters. The presence of one putative ectoine biosynthetic gene cluster consisted of high levels of saline environment where YIM 96748T was isolated. A high concentration of compatible solute ectoine, which serves as a protective osmolyte, has been found in many halophilic microorganisms (Pastor et al., 2010). Seven putative gene clusters showed moderate similarity (30–70% of genes showed similarity) to nystatin A1, salinichelins, ochronotic pigment, landepoxcin, fortimicin, WS9326, and diazaquinomycin gene clusters. The existence of these putative gene clusters indicated that YIM 96748T offers the opportunity to produce these important antibiotics or their analogs. Twenty-two putative gene clusters showed low similarity (<30% of genes showed similarity) to reported scleric acid, xiamycin, allylmalonyl-CoA, atratumycin, arginomycin, xiamycin, rifamorpholine, ML-449, staphylobactin, echinomycin, BE-43547, arginomycin, teicoplanin, chlorizidine A, chejuenolide, armeniaspirol, rubrolone, phosphonoglycans, rifamorpholine, A54145, and kijanimicin gene clusters. Fifteen putative gene clusters were not conserved relative to any known cluster. The existence of these cryptic secondary metabolite biosynthetic gene clusters implied that YIM 96748T could be a potential source for novel antibiotic discovery. Therefore, we fermented it in 15 different media and found two new compounds and five known ones.
Compounds Produced by Strain YIM 96748T
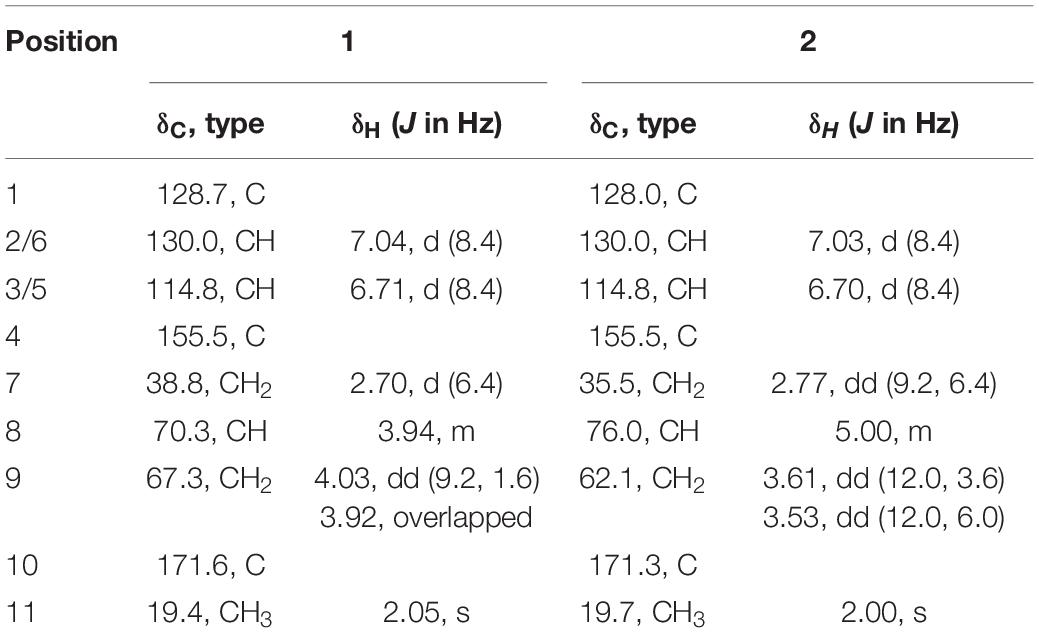
Compounds 1/2, a yellowish solid, were isolated as a mixture, which was confirmed by analytical HPLC (Supplementary Figure 3). However, we failed to purify compounds 1 and 2 due to the small amount. The same molecular formula for compounds 1 and 2 was assigned as C11H4O4 by interpretation of positive HRESIMS (m/z 233.0783 [M + Na]+, calculated for 233.0784) (Supplementary Figure 4), indicating 5 degrees of unsaturation. Analysis of the NMR spectra (Supplementary Figure 5) suggested that compounds 1 and 2 shared similar chemical structures. The NMR data were almost identical for compounds 1 and 2, except that the chemical shift values for the C-7, C-8, and C-9 in compound 1 (δH/δC = 2.70/38.8, 3.94/70.3, and 4.03, 3.92/67.3) were different from those in compound 2 (δH/δC = 2.77/35.5, 5.00/76.0, and 3.61, 3.53/62.1) (Table 4).
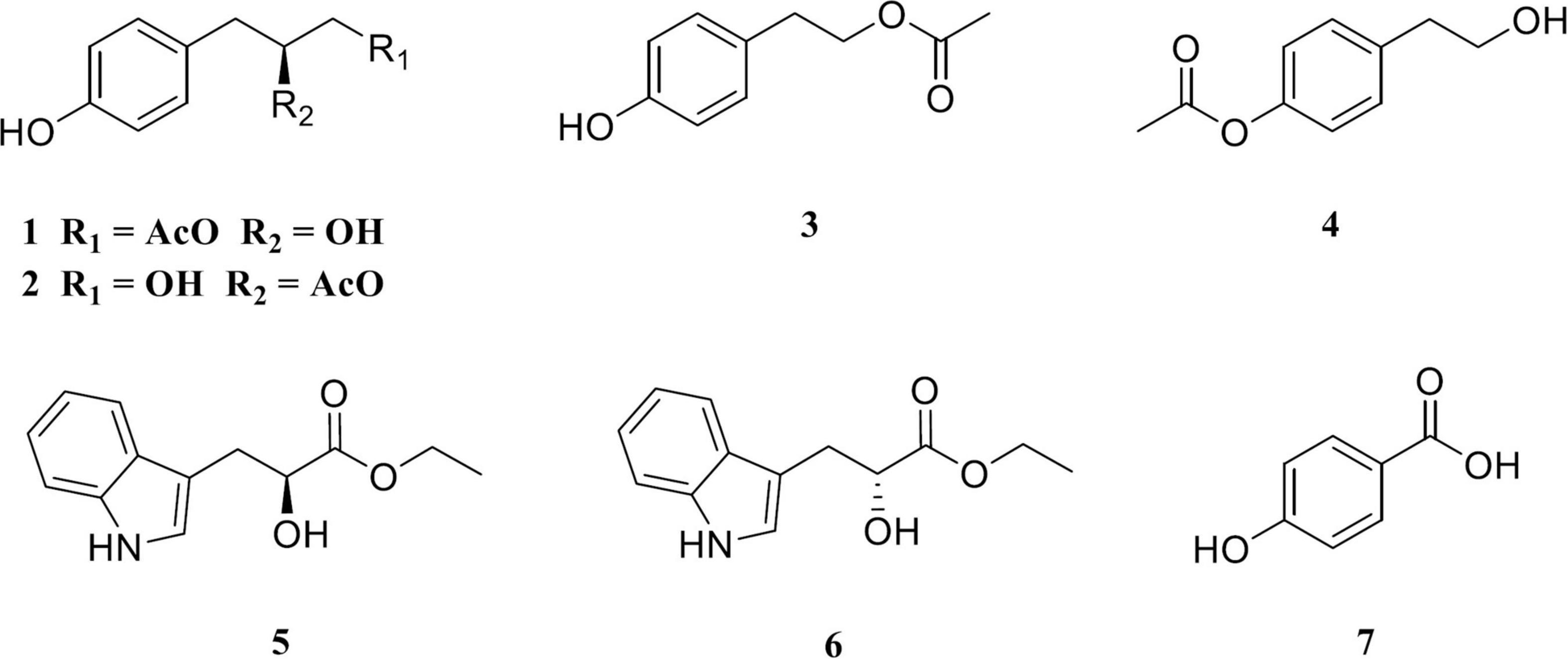
Then, the structure of compound 1 was ascertained with 2D NMR experiments (Supplementary Figure 5). The 1H–1H correlated spectroscopy (COSY) correlations of H-2/H-3 and H-5/H-6 and the heteronuclear multiple bond correlations (HMBCs) from H-2, H-3, H-5, and H-6 to C-1 and C-4 indicated the presence of a 1,4-disubstituted benzene in compound 1 (Figure 4). The 1H–1H COSY correlations of H-7/H-8/H-9 and the HMBCs from H-7 to C-1, C-2, and C-6, together with the chemical shift values of C-4, C-8, and C-9, revealed that the p-substituted phenyl group was hydroxyphenyl propane-diol. Furthermore, the HMBCs from H-9 and H-11 to C-10 elucidated that the acetyl linked to C-9. Meanwhile, the 2D NMR data of compound 2 were highly similar to those of compound 1, except for the HMBC from H-8 to C-10, which indicated that the acetyl linked to C-8 in compound 2. Consequently, the planar structures of compounds 1 and 2 are established in Figure 4. The relative configurations of compounds 1 and 2 were deduced from the rotating-frame Overhauser effect spectroscopy (ROESY) experiment (Figure 4). The key ROESY correlations of H-8/H-2 and H-8/H-6 in both compounds 1 and 2 proved the H-8 β-orientation. On the basis of the above, the relative configurations of compounds 1 and 2 were deduced to be 8S (Figure 5) and named as amycoletates A and B, respectively.
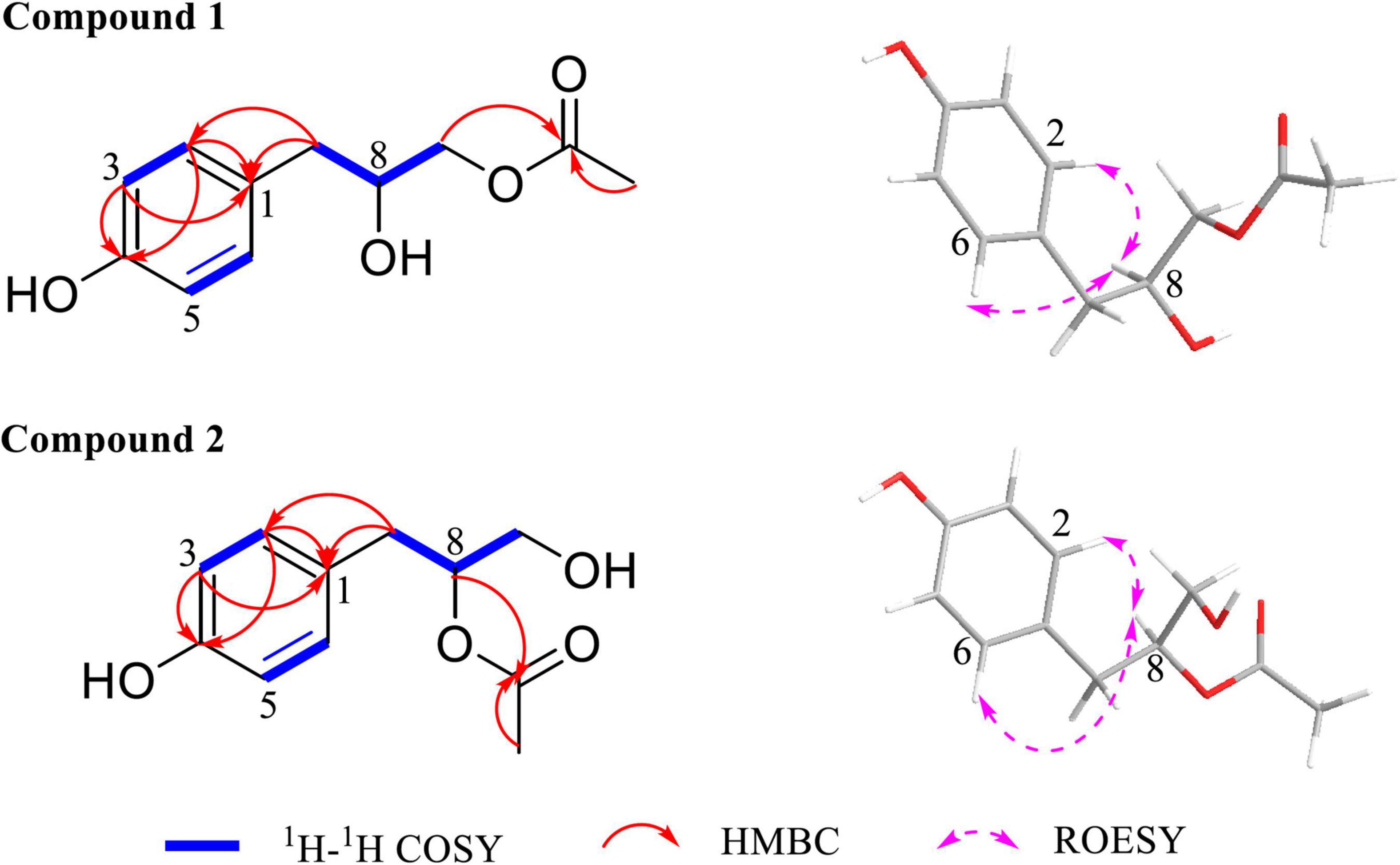
Figure 4. Key 1H–1H correlated spectroscopy (COSY), heteronuclear multiple bond correlation (HMBC), and rotating-frame Overhauser effect spectroscopy (ROESY) correlations of amycoletates A (1) and B (2).
The structures of the other five purified compounds were confirmed as 4-hydroxy phenethyl acetate (compound 3) (Wei et al., 2018), 2-p-acetoxyphenylethanol (compound 4) (Singh et al., 2014), (S)-ethyl indole-3-lactate (compound 5), (R)-ethyl indole-3-lactate (compound 6) (Marinos et al., 1992), and p-hydroxybenzoic acid (compound 7) (Tan et al., 2013) by comparing their NMR data with those of previously published ones. Compounds 1 and 2 were probably produced from compound 7 as a precursor through the one-module plyketide synthase (PKS) pathway and unknown enzymes. Each of gene cluster 14 (rifamorpholine, 12% similarity), gene cluster 36 (atratumycin, 18% similarity), and gene cluster 43 (ML-449, 12% similarity) contained a single module of PKS. Which gene cluster is responsible for the biosynthesis of amycoletates A and B is yet to be verified.
The putative siderophore biosynthetic gene cluster from YIM 96748T showed low similarity (12%) to the staphylobactin biosynthetic gene cluster from S. aureus NCTC 8325 (Dale et al., 2004). YIM 96748T showed siderophore activity in our previous studies. The presence of a novel siderophore cluster indicated that this strain is likely to produce new siderophores. The absence of a novel siderophore in the compounds we isolated indicated that this gene cluster might be silent under the tested fermentation conditions. Possibly due to the characteristics of halophilic actinomycetes, other gene clusters might be silenced under traditional fermentation conditions. Intensive studies on fermentation condition optimization, heterologous expression, and genetic modification need to be tried in the future.
Conclusion
The saline soil-derived halotolerant actinobacterial strain YIM 96748T is a novel species of the genus Amycolatopsis whose name was proposed as Amycolatopsis aidingensis sp. nov., and the type strain is YIM 96748T. It could produce two new compounds, amycoletates A and B, and five known compounds: 4-hydroxy phenethyl acetate, 2-p-acetoxyphenylethanol, (S)-ethyl indole-3-lactate, (R)-ethyl indole-3-lactate, and p-hydroxybenzoic acid. The complete genome was sequenced and analyzed. Fifty-one putative secondary metabolite gene clusters were found. Many of them were significantly different from known gene clusters, indicating that this strain could produce many novel compounds. One of the gene clusters 14, 36, and 43 might be responsible for the biosynthesis of the new compounds amycoletates A and B through the one-module PKS pathway and other unknown enzymes. A siderophore cluster is likely to produce novel siderophores because of YIM 96748T having shown siderophore activity. The one strain many compounds (OSMAC) approach and genetic modification including heterologous expression are needed to obtain more novel compounds from this talented antibiotic producer. In conclusion, strain YIM 96748T is a promising candidate for the discovery of novel secondary metabolites.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MY, HZ, and S-KT designed the study, carried out the data analysis, and wrote the manuscript. RL, MW, ZR, and YJ carried out the experiments and participated in the data analysis. All authors have read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China grants 31860017 to MY and 31760003 to S-KT, the Yunnan Natural Science Foundation 202001BB050069 and YNWR-QNBJ-2019-031 to MY, and the Major Science and Technology Projects of Yunnan Province (Digitalization, Development, and Application of Biotic Resource, 202002AA100007) to S-KT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank Jung Sook Lee and Kyuhee Kang for providing the reference strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.743116/full#supplementary-material
Footnotes
References
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712.
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Brigham, R. B., and Pittenger, R. C. (1956). Streptomyces orientalis, n. sp., the source of vancomycin. Antibiotics Chemotherapy 6:642.
Chantavorakit, T., Suksaard, P., Matsumoto, A., and Duangmal, K. (2019). Amycolatopsis suaedae sp. nov., an endophytic actinomycete isolated from Suaeda maritima roots. Int. J. Syst. Evol. Microbiol. 69, 2591–2596. doi: 10.1099/ijsem.0.003546
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Collins, M., Pirouz, T., Goodfellow, M., and Minnikin, D. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. General Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Dale, S. E., Doherty-Kirby, A., Lajoie, G., and Heinrichs, D. (2004). Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72, 29–37. doi: 10.1128/IAI.72.1.29-37.2004
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Felsenstein, J. (2005). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Feng, Y.-Z., Chunyu, W.-X., Liang, R., Hahnke, R., Schumann, P., Zhao, Y.-R., et al. (2020). Vallicoccus soli gen. nov., sp. nov., a novel actinobacterium isolated from soil, and description of Vallicoccaceae fam. nov., Motilibacterales ord. nov. Antonie van Leeuwenhoek 113, 2155–2165. doi: 10.1007/s10482-020-01484-5
Fitch, W. (1971). Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Biol. 20, 406–416.
Foldes, M., Munro, R., Sorrell, T. C., Shanker, S., and Toohey, M. (1983). In-vitro effects of vancomycin, rifampicin, and fusidic acid, alone and in combination, against methicillin-resistant Staphylococcus aureus. J. Antimicrob Chemother. 11, 21–26. doi: 10.1093/jac/11.1.21
Gordon, R., Barnett, D. A., Handerhan, J. E., and Pang, C. (1974). Nocardia coeliaca, Nocardia autotrophica, and the Nocardin Strain. Int. J. Syst. Evol. Microbiol. 24, 54–63.
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57.
Groth, I., Schumann, P., Weiss, N., Martin, K., and Rainey, F. (1996). Agrococcus jenensis gen. nov., sp. nov., a new genus of actinomycetes with diaminobutyric acid in the cell wall. Int. J. Systematic Bacteriol. 46, 234–239. doi: 10.1099/00207713-46-1-234
Hasegawa, T., Takizawa, M., and Tanida, S. (1983). A rapid analysis for chemical grouping of aerobic actinomycetes. J. General Appl. Microbiol. 29, 319–322.
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lechevalier, M., and Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443. doi: 10.1128/jb.94.4.875-883.1967
Lechevalier, M., Prauser, H., Labeda, D., and Ruan, J. (1986). Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int. J. Systematic Evol. Microbiol. 36, 29–37.
Marinos, V. A., Tate, M., and Williams, P. J. (1992). Glucosides of ethyl indole-3-lactate and uroterpenol in riesling wine. Phytochemistry 31, 2755–2759.
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357.
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H., and Göker, M. (2012). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Minnikin, D., O’donnell, A., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Pastor, J. M., Salvador, M., Argandona, M., Bernal, V., Reina-Bueno, M., Csonka, L. N., et al. (2010). Ectoines in cell stress protection: uses and biotechnological production. Biotechnol. Adv. 28, 782–801. doi: 10.1016/j.biotechadv.2010.06.005
Pospiech, A., and Neumann, B. (1995). A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11, 217–218. doi: 10.1016/s0168-9525(00)89052-6
Rhoads, A., and Au, K. F. (2015). PacBio sequencing and its applications. Genom. Proteom. Bioinform. 13, 278–289.
Richter, M., Rosselló-Móra, R., Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sepkowitz, K. A., Raffalli, J., Riley, L., Kiehn, T. E., and Armstrong, D. (1995). Tuberculosis in the AIDS era. Clin. Microbiol. Rev. 8, 180–199.
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16, 313–340.
Singh, D., Kumar, R., and Chaudhuri, P. (2014). A new phenolic compound from the flowers of Jasminum multiflorum. Chem. Nat. Compounds 50, 48–49.
Smibert, R., and Krieg, N. R. M. (1994). “Phenotypic characterization,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. Murray, W. A. Wood, N. R. Krieg (Washington, DC: American Society for Microbiology), 607–654.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Takahashi, Y. (2001). “Family pseudonocardiaceae,” in Identification Manual of Actinomycetes, ed. The Society for Actinomycetes Japan (Tokyo: The Business Centre for Academic Societies).
Tan, D., Yan, Q., and Kang, H. (2013). Chemical constituents from Blumea balsamifera. Chem. Nat. Comp. 48, 1072–1073. doi: 10.1007/s10600-013-0468-5
Tang, S.-K., Tian, X., Zhi, X.-Y., Cai, M., Wu, J.-Y., Yang, L., et al. (2008). Haloactinospora alba gen. nov., sp. nov., a halophilic filamentous actinomycete of the family Nocardiopsaceae. Int. J. System. Evol. Microbiol. 58(Pt 9), 2075–2080. doi: 10.1099/ijs.0.65531-0
Tang, S.-K., Wang, Y., Chen, Y., Lou, K., Cao, L.-L., Xu, L.-H., et al. (2009). Zhihengliuella alba sp. nov., and emended description of the genus Zhihengliuella. Int. J. System. Evol. Microbiol. 59(Pt 8), 2025–2031. doi: 10.1099/ijs.0.007344-0
Tatar, D., Sazak, A., Guven, K., Cetin, D., and Sahin N. (2013). Amycolatopsis cihanbeyliensis sp. nov., a halotolerant actinomycete isolated from a salt mine. Int. J. Syst. Evol. Microbiol. 63, 3739–3743. doi: 10.1099/ijs.0.050963-0
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
The Gene Ontology Consortium (2015). Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056. doi: 10.1093/nar/gku1179
Wayne, L. (1988). International committee on systematic bacteriology: announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 268, 433–434.
Wei, W. D., Hao, E. W., Qin, J. F., Xue, J. L., Luo, H., Deng, Y. T., et al. (2018). Chemical constituents from Ethyl acetate extract of scoparia dulcis linn. Vietnam J. Sci. Technol. 54:278.
Xing, K., Liu, W., Zhang, Y. J., Bian, G. K., Zhang, W. D., Tamura, T., et al. (2013). Amycolatopsis jiangsuensis sp. nov., a novel endophytic actinomycete isolated from a coastal plant in Jiangsu, China. Ant. Leeuw. 103:4339. doi: 10.1007/s10482-012-9823-y
Xu, P., Li, W.-J., Tang, S.-K., Zhang, Y.-Q., Chen, G., Chen, H.-H., et al. (2005). Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int. J. Systematic Evol. Microbiol. 55(Pt 3), 1149–1153. doi: 10.1099/ijs.0.63407-0
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob Chemother. 67, 2640–2644.
Keywords: halophilic actinomycete, amycoletate, genome sequence, secondary metabolites, Amycolatopsis aidingensis sp. nov.
Citation: Li R, Wang M, Ren Z, Ji Y, Yin M, Zhou H and Tang S-K (2021) Amycolatopsis aidingensis sp. nov., a Halotolerant Actinobacterium, Produces New Secondary Metabolites. Front. Microbiol. 12:743116. doi: 10.3389/fmicb.2021.743116
Received: 26 July 2021; Accepted: 28 October 2021;
Published: 06 December 2021.
Edited by:
Pankaj Kumar Arora, Babasaheb Bhimrao Ambedkar University, IndiaReviewed by:
Christopher Milton Mathew Franco, Flinders University, AustraliaRamprasad E. V. V., University of Hyderabad, India
Copyright © 2021 Li, Wang, Ren, Ji, Yin, Zhou and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Yin, eWlubWluQHludS5lZHUuY24=; Hao Zhou, aGFvemhvdUB5bnUuZWR1LmNu; Shu-Kun Tang, dGFuZ3NodWt1bkB5bnUuZWR1LmNu
†These authors have contributed equally to this work
 Rui Li1†
Rui Li1† Meng Wang
Meng Wang Zhen Ren
Zhen Ren Yang Ji
Yang Ji Min Yin
Min Yin Hao Zhou
Hao Zhou Shu-Kun Tang
Shu-Kun Tang