
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 29 November 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.741164
The biofloc system has recently attracted great attention as a cost-effective, sustainable, and environmentally friendly technology and expected to contribute toward human food security (Zero Hunger SDG 2). It is also expected that this endeavor can be adopted widely because of its characteristics of zero water exchange and reduced artificial feeding features. In the biofloc system, the flocs which are generally formed by aggregation of heterotrophic microorganisms, serve as natural bioremediation candidates. These microbes effectively maintain water quality by utilizing the nutrient wastes, mostly originated from digested, unconsumed, and metabolic processes of feed. Additionally, the flocs are important sources of nutrients, mainly a protein source, and when these are consumed by aquaculture animals they improve the growth performance, immunity, and disease tolerance of host against pathogenic microbial infection. Here in this review, we focus on recent advances that could provide a mechanistic insight on how the microbial community developed in the biofloc system helps in the bioremediation process and enhances the overall health of the host. We have also tried to address the possible role of these microbial communities against growth and virulence of pathogenic microbes.
Aquaculture, the farming of aquatic animals and plants, continues to dominate the food producing sector in the world (FAO, 2019). The global aquaculture production increased to 4.9% as compared to 2016 and reached to 111.9 million tons in 2017. The aquaculture share in total global aquatic animal production including both capture and aquaculture has risen sharply from 25.7% in 2000 to 46.4% in 2017, with an average annual growth rate of 4.8% during 2011–2017 (Kumar, 2020; Roy, 2020; Tacon, 2020). The 10 top aquaculture producing countries including China, India, Indonesia, Vietnam, Bangladesh, Egypt, Norway, Chile, Myanmar, and Thailand, contribute over 88% by quantity of global aquaculture production in 2017. Interestingly, the aquaculture industry is playing a major role in economic development and together with capture fisheries it supports the livelihood of more than 10% of the world population (Kumar et al., 2021b). Moreover, as the global human population continues to expand at a high rate and is expected to reach over 9 billion by 2030, the aquaculture industry could be crucial for food and nutritional security with high-quality animal protein in both inland and coastal regions, and providing livelihood and income source generation to millions of people (Roy et al., 2019, 2020; Ngasotter et al., 2020). However, due to the global demand increase, the pressure for intensification and further expansion of culture systems has created many problems including scarcity of natural resources, increased environmental pollution and losses due to disease outbreak. Disease outbreaks caused by bacterial infections, considered as the primary cause of production loss in fish farming, have moved to the forefront in recent years and brought socio-economic and environmental unsustainability to the aquaculture industry (Costa-Pierce et al., 2010; Verdegem, 2013; Kumar V. et al., 2018, Kumar et al., 2020a). The economic losses in the aquaculture sector from disease outbreak has been calculated by FAO to be over US$9 billion per year, that is approximately 15% of world aquaculture fish and shellfish production, by value (Kumar et al., 2019a, b; Tran et al., 2020). Additionally, the fish feed, the prices vary from a few hundred dollars a ton to more than US $1000 a ton depending on the species being fed, is the major operational cost for most fish farms accounting for 50–70% of the variable cost (FAO, 2019). In addition, in developing countries like Indonesia, Vietnam, or Bangladesh, the commercial feed is simply beyond the reach of most marginal and landless farmers, limiting their ability to intensify aquaculture production. In this context, development of culture protocol/technology that can reduce the input cost and enhance the immunity and disease tolerance of cultured animals seems to be a preferable alternative for aquaculture systems.
In recent years, growing aquaculture species in the biofloc system is becoming more popular and has shown promising results in improving water quality, fish health, and production. This technology could become essential not only to cover the growing demand for dietary animal proteins but also for the water scarcity, environmental issues, and animal health and disease. The biofloc system’s basic principle is to recycle and transform waste and excessive nutrients, in particular inorganic nitrogen (NH3-N and NO2-N), generated from feces and uneaten feed into microbial biomass. These biomasses are generally high in protein content and utilized by cultured animals in situ or if harvested they could be processed and used as a nutrient source for feeds. This is achieved by steering the carbon and nitrogen ratio (C/N ratio) through modification of feed carbohydrate content or carbon source addition in water (Avnimelech, 1999; Kuhn et al., 2009; Crab et al., 2012; Ekasari et al., 2014; Fatimah et al., 2019; Hostins et al., 2019). For instance, Schneider et al. (2005) reported that if the C:N ratio is maintained between 10 and 15:1, the organic nitrogenous ammonium waste are converted into bacterial biomass (Schneider et al., 2005). In another study, Crab et al. (2007) noted that the use of biofloc system in intensive tilapia culture significantly improved the nitrogen recovery from 23 to 43% and non-toxic levels of ammonia/ammonium concentration could be maintained, without water exchange (Crab et al., 2007). There are also few reports that suggest that biofloc contains microbe-associated molecular pattern (MAMP) and microbially bioactive components such as carotenoids, vitamins, glutathione, antioxidants, and minerals, which nutritionally modulate the fish health and immune response and resulted in better growth performance and increased resistance against microbial pathogens (Xu and Pan, 2013; Hostins et al., 2019; Kumar et al., 2020b) (Figure 1). Since the biofloc system has several beneficial effects that contribute to the maintenance of optimum water quality in the culture system and improvement of feed utilization and nutrition of the cultured animals, the technology has a wide range of acceptability across several aquaculture producing countries. However, still refinements of the biofloc culture protocol in terms of growth and nutrition requirements of aquaculture species is required. In addition, more importantly, information on biofloc derived microbes and how these microbial origins affect the external aquatic pathogens and beneficially improve the aquatic environment and host survival is needed for better understanding and scientific application of the biofloc system. Hence, in this review at first an overview of the current knowledge on the effect of the biofloc system microbial community on aquatic environment and host is given. Later, the possible role of these microbes on the activity and virulence of pathogenic microorganisms with respect to aquaculture are summarized.
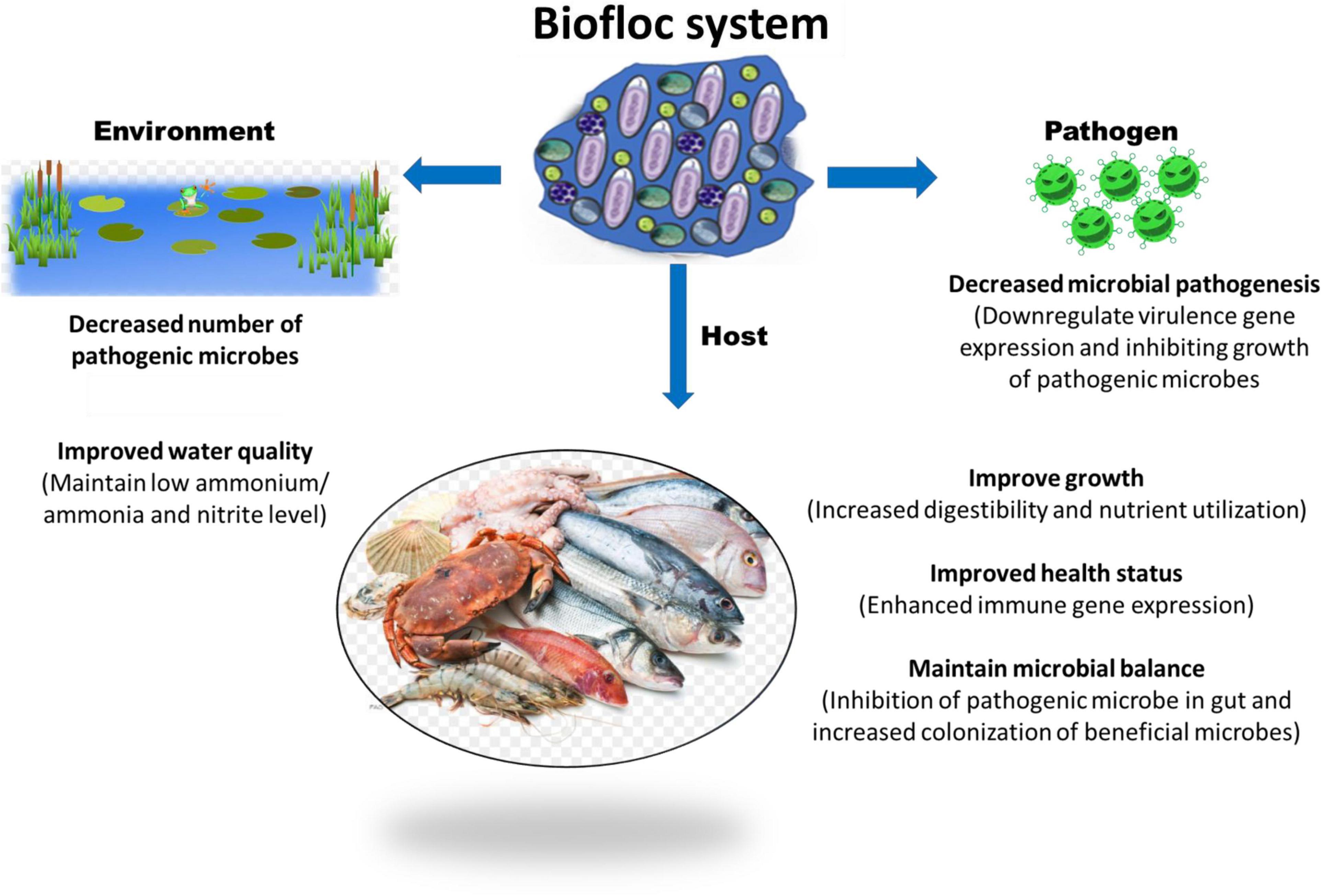
Figure 1. Potential role of biofloc system in host, pathogen, and environment in a culture facility.
The biofloc system is a relatively new aquaculture technology that allows high-density culture at a limited or zero water exchange facility. Interestingly, once the C/N ratio reaches between 10 and 15:1, by addition of exogenous carbon source, the available microbe in the system utilizes the accumulated nitrogenous wastes originated from unconsumed feed and animal excretion, including fecal materials and metabolic products and accelerates the growth of microbial communities that conglomerate together and produce flocs (Ju et al., 2008; de Jesús Becerra-Dorame et al., 2014). The potential of this system in increasing the resource utilization efficiency has raised the attention for both research and application during the past decade. As a consequence, more beneficial effects of the biofloc system have been discovered including the nutritional properties, exogenous digestive enzymes contribution, potential control of pathogens, and immunostimulatory effects (Betanzo-Torres et al., 2020; Vyas, 2020; El-Sayed, 2021). Few reports have also suggested that the biofloc formation is mediated by one of the microbial phenotypes, which are involved in the production of small membrane diffusible metabolites (Chong et al., 2012; Hawver et al., 2016). Therefore, it becomes very intriguing to investigate and characterize the biofloc microbiome stem from their ability to maintain water quality and confer immunostimulatory effect on the cultured animals.
To develop a biofloc, at first the tanks (might be circular or rectangular, however, circular tanks are preferred) were filled with water, and nitrogenous material (fish feed and urea fertilizer) and a carbon source (molasses, wheat flour, or starch, about 0.7% of feed) were added to the water (Table 1). In some cases, soil clay particles, after mixing thoroughly and filtering with a sieve, were also added to the tanks, it helps in biofloc formation and further mass continuity. Afterward, the primary inoculum of the microbial biomass and necessary elements, including proper aeration, were added to the system, it improves the microbial flocs formation in the new culture system (Zemor et al., 2019). As a standard, 20 g of clay, 10 mg of ammonium sulfate, and 200 mg of carbonaceous organic matter such as molasses can stimulate biofloc formation in 1 L of water. The presence of carbonated organic matter enables heterotrophic bacteria to become more active than other bacteria, and they remove nitrogen and carbon from water by an absorption process and produce microbial biomass/flocs. These biomasses were subsequently combined/fed by other organisms (viz., algae, detritus, ciliates, or yeast) and collectively form biofloc in the culture system (Khanjani et al., 2017). During the biofloc formation, algae were first developed and form foam, and eventually microbial biomass. The development of a brown state in culture tanks indicates the presence and activity of heterotrophic bacteria. Moreover, once the experimental animals were added to the biofloc system, the physicochemical parameters (temperature, oxygen, pH, alkalinity, total nitrogen, ammonium, nitrite, and nitrate) should be measured weekly. In the case of any deviation from standard water quality parameters, subsequently appropriate responses including water exchange, stopped artificial feeding, etc. should be adopted quickly (Avnimelech, 1999; Khanjani and Sharifinia, 2020). For instance, if the ammonia levels were high, then input carbon source might be increased while decreasing the feed content. Similarly, if the nitrite level is high, then increase the carbon input and check oxygenation and sludge collection. Moreover, if microbial biomass is low, add carbon source and if the volume of the biofloc is too high, then do a partial water exchange.
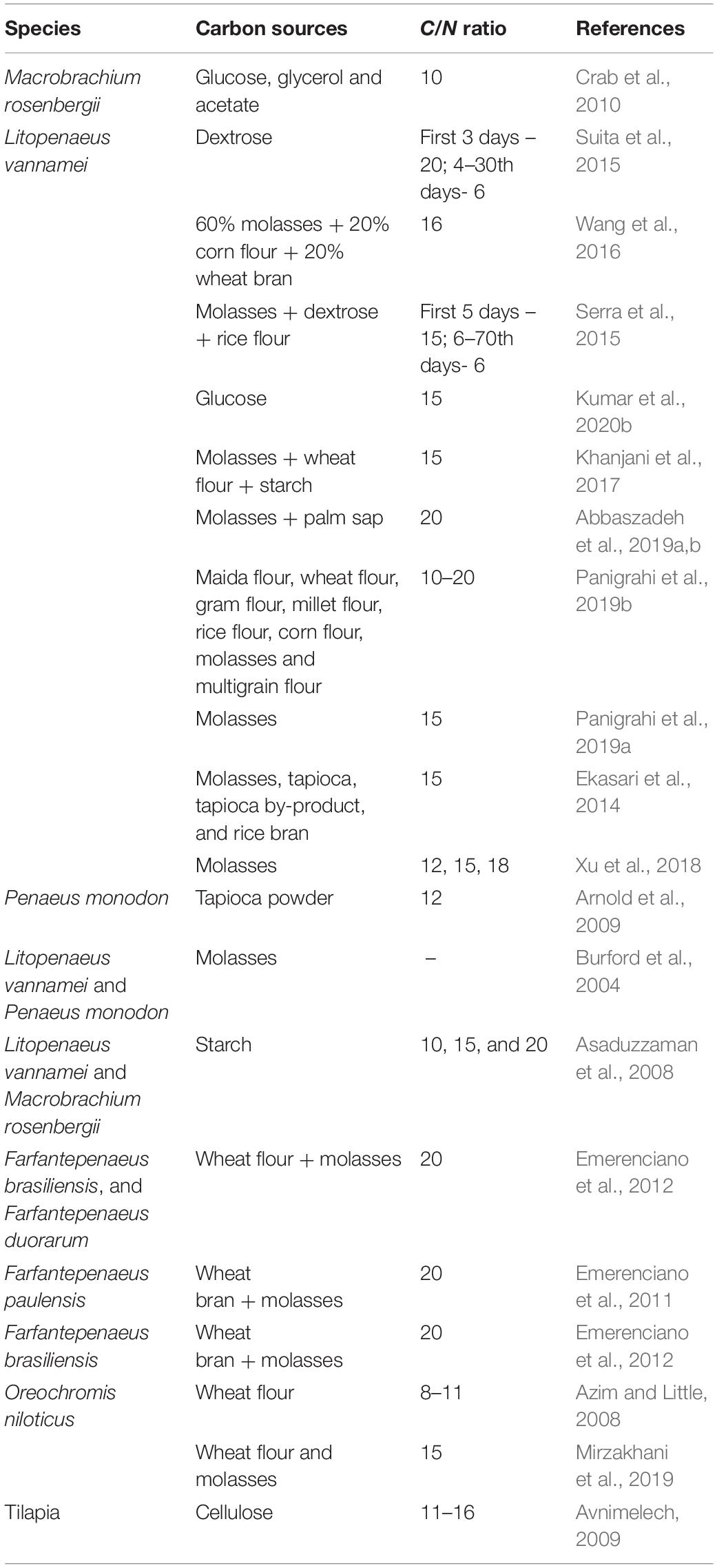
Table 1. The use of different carbon sources and C/N ratio for development of stable biofloc culture system.
From the studies mentioned in the above paragraphs, it is clear that the bacterial population, i.e., heterotrophic bacteria, are the predominant group of biofloc microbiota; however, fungi, algae (dinoflagellates and diatoms), flagellates, rotifers, ciliates, and detritus also constitute the microbial components (Ju et al., 2008; Kim et al., 2014; Kasan et al., 2017; Tepaamorndech et al., 2020). For instance, Cardona et al. (2016) analyzed the biofloc system water samples used for culture of Litopenaeus stylirostris using 16S rRNA amplicon sequencing. The results showed that bacteria taxa belonging to Proteobacteria, Bacteroidetes, and Cyanobacteria groups have the highest relative abundance (Cardona et al., 2016). Later, Tepaamorndech et al. (2020) performed 16S rRNA amplicon sequencing and shotgun metagenomic analysis to characterize the complex of bacterial communities in the biofloc system culturing Litopenaeus vannamei. The analysis revealed that 90% biofloc microbial population comprised of Vibrio sp., while Bacillus, Lactobacillus, Pseudoalteromonas, Clostridium, Shewanella, Acinetobacter, Photobacterium, Alteromonas, Marinifilum, and Pseudomonas were also identified. In another study, Wei et al. (2020) investigated the microbial communities in the biofloc ecosystem with high-throughput sequencing and quantified the 16S rRNA gene. Findings suggest that bacterial groups belonging to Flavobacteriaceae (e.g., Marivita, Ruegeria, and Maribacter) and Rhodobacteraceae were the key bacterial taxa in the biofloc system (Wei et al., 2020). Recently, Islam et al. (2021) evaluated the microbial community structure in biofloc culturing Macrobrachium rosenbergii. It was observed that biofloc systems maintained in 15–20:1 C:N ratio have a higher count of Lactobacillus spp. followed by Enterococcus spp. (Islam et al., 2021). In another study, Meenakshisundaram et al. (2021) characterize the microbial composition of grown-out biofloc used for culture of genetically improved farmed tilapia (GIFT). The metagenomic profile analyzed through Illumina Nextseq500 platform shotgun sequencing showed that microbial composition in biofloc includes 70.80% bacteria, 5.08% eukarya, 0.62% archaea, 0.16% virus and 23.35% are unclassified. Further classification reveals that abundant genus in biofloc microbiome are Proteobacteria and Caldilinea aerophila (Meenakshisundaram et al., 2021). Taking together, these studies highlight that dominant microbiota in biofloc system, i.e., Lactobacillus, Bacillus, and Vibrio along with other bacterial groups, e.g., Halomonas, Providencia, Nitratireductor, Pseudoalteromonas, etc. might be responsible for inducing beneficial effect on host, environment, and pathogenic microbes, respectively.
Moreover, it is important to realize that quorum sensing (QS), a bacterial intercommunication system that controls the expression of numerous genes, regulates the activities of a large group of bacterial cells (Fuqua et al., 1994; Bassler et al., 1997; Xu et al., 2006). A QS system uses small signal molecules called autoinducers (AIs) to control directly or indirectly bacterial bioluminescence, virulence factor expression, biofilm formation, motility, entry into stationary phase, sporulation, and mating (Miller and Bassler, 2001; Schauder and Bassler, 2001). Additionally, the QS mediated formation of extracellular polymeric substances (EPS) matrix leads to transformation of planktonic bacterial cells into sessile mode of growth and form microbial clusters or aggregates (Kumar et al., 2020a, 2021a). In clusters or biofilm mode of life, bacteria play a vital role in removing or converting harmful compounds and is considered as an excellent biosorbent material for the remediation of toxic substances. Additionally, these aggerates are excellent microbial protein sources and help to improve growth, immunity, and survival of consuming host animal against both biotic and abiotic stressors. Interestingly, one recent study has demonstrated that microbial quorum sensing plays important roles in biofloc characteristics and functionality from aquaculture perspective. The QS regulated the biofloc formation, protein contents, total ammonium nitrogen (TAN) removal capacity and growth of cultured African catfish, Clarias gariepinus (Fatimah et al., 2019). However, further characterization on dominant microbial species involved in QS-regulated microbial gene expressions will be helpful to find the possibility to modulate QS activity in the biofloc microcommunity. In addition, it will be also useful in identification of microbial phenotypes that are beneficial in aquaculture perspective, such as the excretion of various digestive enzymes that may contribute to the increased food digestibility of fish and production of essential nutrients that could improve the nutritional value of bioflocs.
The functions of biofloc are strongly related to the interaction of the microbial community in the spatial cohabitation involved in the acquisition of nutrients and the biochemical processes (Ekasari et al., 2014; Bossier and Ekasari, 2017). These communities play an essential role as natural bioremediation candidates in maintenance of water quality and conversion of nitrogenous waste materials. Additionally, they also play a significant part in development of nutrient rich flocs that serve as food sources and contribute in nutrition to support high density growth of aquaculture animals (Figures 1, 2) (Zhao et al., 2014; Zhang et al., 2016).
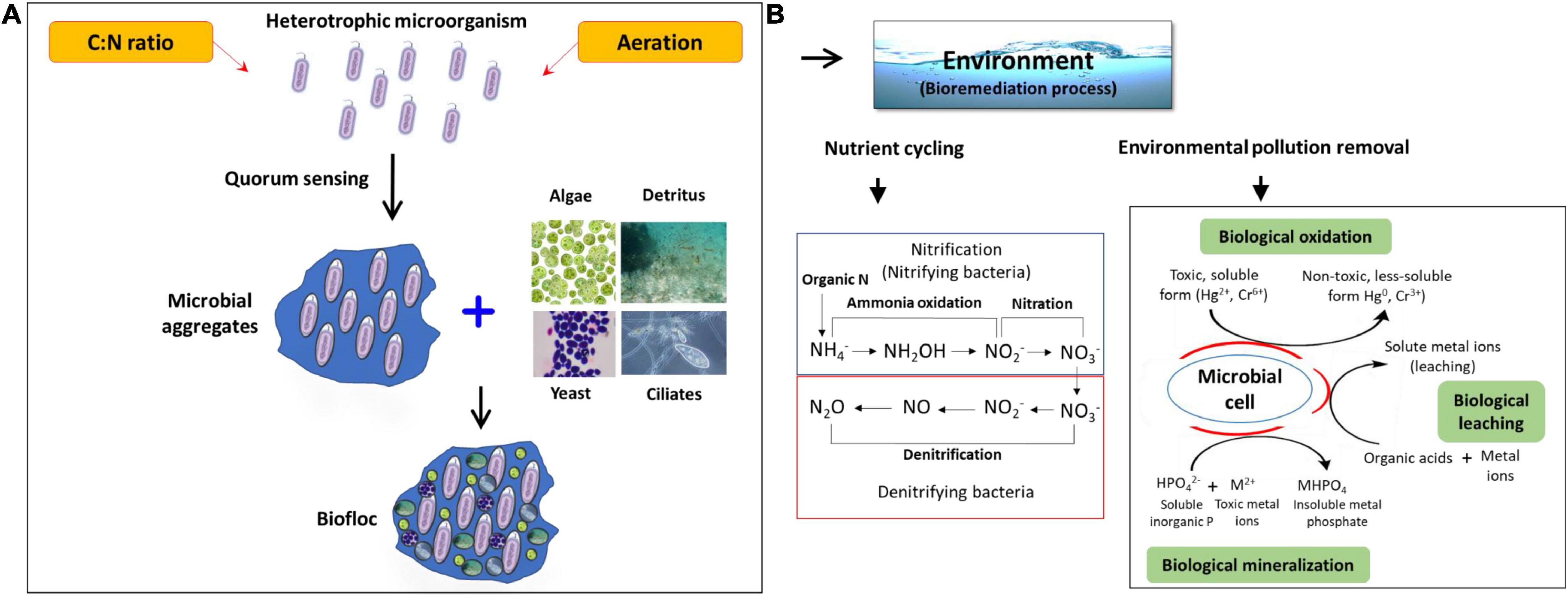
Figure 2. Schematic overview on the possible role of the biofloc microbiome. (A) Development of a biofloc system; (B) potential role of the biofloc system in the bioremediation process.
Bioremediation is a process of contaminated water or waste treatment into less toxic form using beneficial microorganisms (Das et al., 2019, 2020; Behera et al., 2020a, b). It is achieved by inducing the biological process that leads to reduction, removal, and conversion of the contaminated compounds (Table 2) (Divya et al., 2015; Jasmin et al., 2020). Moreover, the treatment method relies on content and toxicity of contaminants, hydrogeological conditions, ecology of microbial communities, and other temporal and spatial factors (Sarkar et al., 2021). The microbial bioremediation process is among the most preferred ways to remove contamination from a system, as it is cost effective and able to immobilize or destroy the contaminants efficiency (Gadd, 2000). Interestingly, these microbial communities utilize the contaminants as their energy source, e.g., phosphorus and nitrogen forms in contaminants are utilized by microbes as their nutrient source (Jones et al., 2018). It is noteworthy to mention that, it is not always necessary to utilize the existing natural microbial population for bioremediation process, the exogenous microbes or genetically engineered population can also be used for the process (Brim et al., 2003). Based on the carbon source utilization, microorganisms involved in the bioremediation process are mainly classified in two groups, autotrophs and heterotrophs. The autotrophic microbes are able to utilize the inorganic substances and synthesize their own food, which makes the autotrophs a good bioremediator (Musyoka, 2016). The commonly known nitrite-oxidizing and ammonia-oxidizing bacteria are classified under autotrophic microbes (Merchant and Helmann, 2012). The second group under bioremediation microbes are heterotrophic bacteria, that immobilize or destroy the non-living organic material and generate carbon sources to build their own cells. These microbes act as electron donors in catalyzing reactions, during the oxidation of harmful contaminants. Moreover, unlike autotrophs, the heterotrophic bacteria contribute comparatively less in the process of nitrification and denitrification, however, they breakdown the organic waste including feces, uneaten feed, and dead materials and transfer the nitrogenous ammonia into non-harmful products known as microbial aggregates or mass (Ebeling et al., 2006).
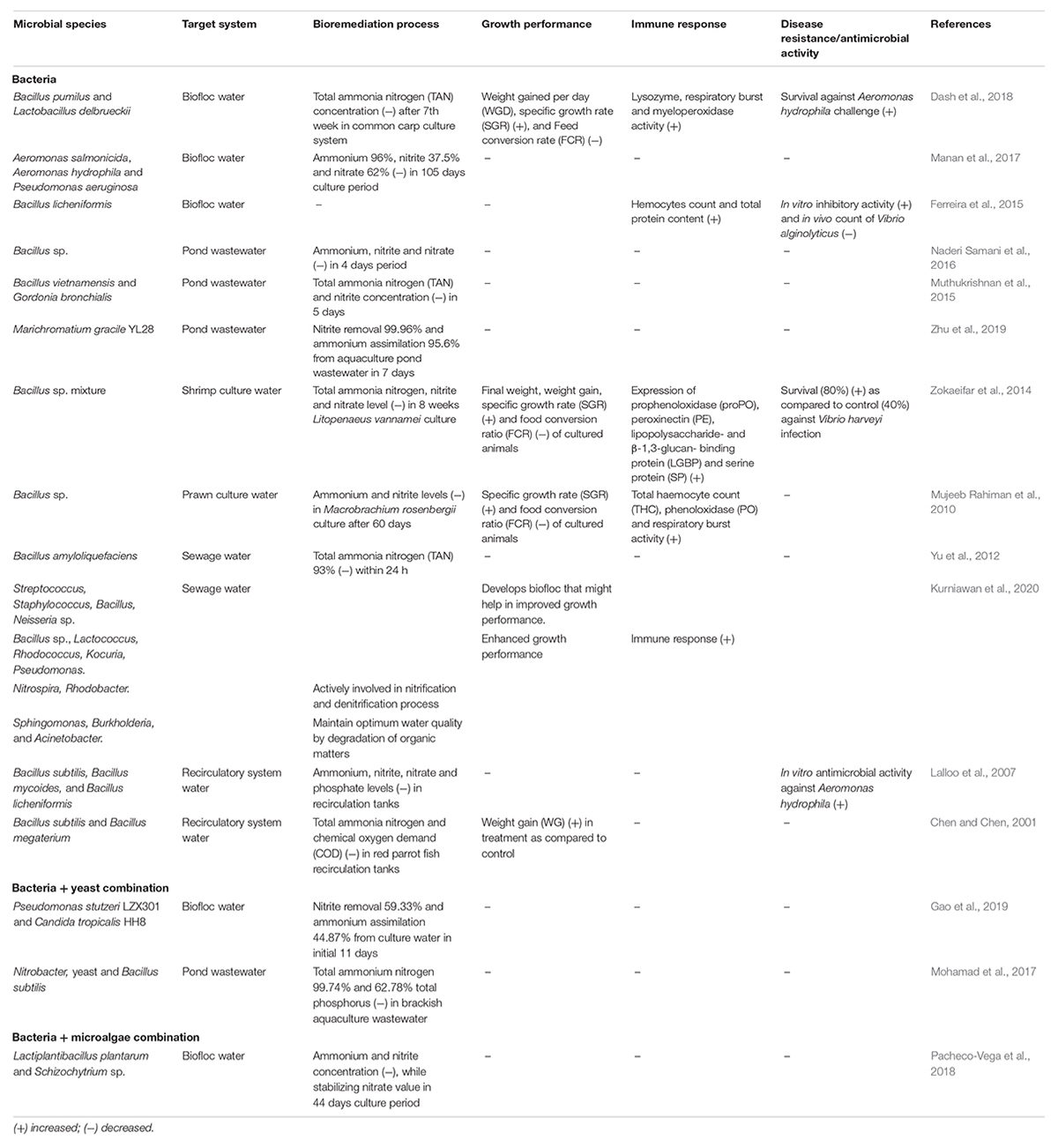
Table 2. The list of microorganisms reported to be involved in the natural bioremediation process, growth, immunity, and disease tolerance of aquaculture animals.
Interestingly, a biofloc system promotes the growth of both autotrophic and heterotrophic microbes (Manan et al., 2017; Pacheco-Vega et al., 2018). However, bioflocs contain a high number of heterotrophic beneficial microbial communities, including Bacillus, Acinetobacter, Sphingomonas, Pseudomonas, Rhodopseudomonas, Micrococcus, Nitrosomonas, Nitrospira, Nitrobacter, Cellulomonas, and yeast. These microorganisms act as potential bioremediation agents in biofloc culture systems, leading to improving water quality, growth performance, and health of cultured aquatic animals (Thomas et al., 1992; Monroy-Dosta et al., 2013; Das and Dash, 2014; Adel et al., 2017) (Table 2 and Figure 2). The build-up of particulate and dissolved organic matter is a common phenomenon observed in biofloc systems, however, high levels of heterotrophic microbes efficiently minimize the organic nitrogen and carbon levels in the system. These heterotrophic microbes, as potential bioremediators, produce diverse metabolic enzymes which assist in safe removal of contaminants either by converting to safer or less toxic substances or direct destruction (Dash and Das, 2012). For instance, Manan et al. (2017) carried out an experiment to determine the role of aggregating biofloc in the bioremediation process including degradation and decomposition of organic matter. The results showed that heterotrophic bacteria identified from Aeromonas (Aeromonas salmonicida and Aeromonas hydrophila) and Pseudomonas family (Pseudomonas aeruginosa) consumed the bottom organic matter of shrimp (L. vannamei) culture biofloc tanks. In addition, after converting these bottom wastes through chemical processes, they help in the production of high protein flocs that are utilized by the cultured shrimp (Manan et al., 2017). It is important to mention that Aeromonas and Pseudomonas sp. could be pathogenic to shrimp species (Ramalingam and Ramarani, 2007; Zhou et al., 2019); hence, it becomes important to validate the pathogenicity of these bacterial strains before concluding it as beneficial microbes. In another study, Hostins et al. (2019) designed an experiment to investigate the effect of autotrophic (with or without probiotics) and heterotrophic biofloc (with or without probiotics) cultured with L. vannamei against AHPND bacterial strain. The results showed that heterotrophic biofloc (with and without probiotics) and autotrophic biofloc (with probiotics) can decrease the impact of AHPND-causing Vibrio parahaemolyticus. However, in heterotrophic biofloc, there was significant improvement in water quality and L. vannamei showed the highest survival with and without probiotic supplementation, when challenged in the presence of their respective biofloc suspensions (Hostins et al., 2019). There were also few reports that suggest that association of bacteria (Pseudomonas stutzeri LZX301/Nitrobacter/Bacillus subtilis) with yeast (Candida tropicalis HH8) or microalgae (Schizochytrium sp.) is effective in maintaining optimum water quality by decreasing the total ammonium nitrogen, nitrite, and nitrate concentration (Mohamad et al., 2017; Pacheco-Vega et al., 2018; Gao et al., 2019). Recently, Kurniawan et al. (2020) investigated the diversity and abundance of biofloc forming bacteria in the river waters using 16S rDNA sequencing method. The analysis revealed that seven bacterial phyla including Proteobacteria, Cyanobacteria, Verrucomicrobia, Actinobacteria, Bacteriodetes, Chloroflex, and Planctomycetes and 14 bacterial genera Streptococcus, Staphylococcus, Bacillus, Neisseria sp., Bacillus sp., Lactococcus, Rhodococcus, Kocuria, Pseudomonas, Nitrospira, Rhodobacter, Sphingomonas, Burkholderia, and Acinetobacter have potential biofloc forming abilities (Kurniawan et al., 2020).
The beneficial effects of bioflocs on growth and immunity of farmed animals have been widely documented. Bioflocs can enhance innate/non-specific immune systems of cultured species through providing a wide range of immunostimulatory effects against microbial infections. The heterotrophic microbial cell walls could contain either lipopolysaccharides, glucans, or peptidoglycans. These microbe-associated molecular patterns (MAMPs) can activate the non-specific immune mechanisms, leading to significant enhanced immune response in farmed species (Aguilera-Rivera et al., 2019; Panigrahi et al., 2019a, 2020). For example, the heterotrophic biofloc reared L. vannamei showed enhanced immune response and increased level of total hemocyte count (THC) and prophenoloxidase (ProPO) activity as compared to control animals (Panigrahi et al., 2019b). Bioflocs are also capable of accumulating bacterial compound, namely, poly-β-hydroxybutyrate (PHB), which has been reported to improve growth performance, food digestibility, and development of resistance against bacterial infections in farmed aquatic animals (Kuhn et al., 2009; Khanjani and Sharifinia, 2020). These beneficial microbes can positively modulate the gut microbiota resulting in enhanced growth performance and immune response of the host (Balcázar et al., 2006; Pérez et al., 2010). In addition, the biofloc microbial species contain several nutritional factors and digestive enzymes, e.g., amylase and proteases, which could contribute in the natural digestive process and improve the food digestion and absorption, resulting in efficient utilization of feed and enhanced growth performance of the host (Wang, 2007; Liu et al., 2009). For instance, Bacillus sp. has been reported to contribute in host nutrition, especially by supplying vitamins and fatty acids and improve the growth and survival of aquaculture animals (e.g., Penaeus monodon postlarvae) in zero water exchange facility (Devaraja et al., 2013; NavinChandran et al., 2014; Kumar et al., 2016). In another study, Zokaeifar et al. (2012, 2014) demonstrated that Bacillus sp. significantly enhances the activity of digestive enzyme, growth performance, immunity, and resistance of shrimp toward bacterial infection. These results highlight that beneficial microbe in the system, namely, Bacillus sp., could significantly improve the amylase and protease activity and subsequently increase the final weight and weight gain of shrimp juveniles. Interestingly, in a few other studies, Bacillus sp. have been demonstrated to stimulate the immune response of L. vannamei juveniles, resulting in enhanced disease resistance and survival of shrimp against Vibrio harveyi challenge (Zokaeifar et al., 2012, 2014). Later, Nimrat et al. (2012) and Sadat Hoseini Madani et al. (2018) evaluated the role of commercial Bacillus sp. application on feed efficiency, growth performance, bacterial number, body composition, water quality, and immune response in L. vannamei. The results highlight that administration of Bacillus sp. to the experiential units significantly enhances the weight gain %, length gain %, specific growth rate %, average daily gain, and FCR of L. vannamei as compared to the control group. The beneficial bacteria also improves the water quality parameters, feed utilization, immune response, and survival of L. vannamei postlarvae (Nimrat et al., 2012; Sadat Hoseini Madani et al., 2018). In another study, Bachruddin et al. (2018) demonstrated that white leg shrimp (L. vannamei) culture water supplemented with beneficial bacterial species, i.e., Bacillus sp., significantly increase the feed utilization and improve the total length, weight gain, FCR, and survival of shrimp species (Bachruddin et al., 2018). Kongnum and Hongpattarakere (2012) and Chai et al. (2016) performed an experiment using indigenous Bacillus sp., isolated from wild and healthy shrimp intestine, and investigated its effect on shrimp health. The finding suggests that Bacillus sp. significantly improves the growth performance, immune system, and resistance of L. vannamei against microbial pathogens (Kongnum and Hongpattarakere, 2012; Chai et al., 2016).
The bacterial biomass developed as aggregates in the biofloc system can also serve as a nutrient for aquatic animals especially as a protein source and thus improve the growth and overall health of farmed aquatic animals (Table 2 and Figure 3) (Cohen et al., 2005; Azim and Little, 2008; Cardona et al., 2015; Lee et al., 2017). Additionally, these microbes were also demonstrated to produce transduction signaling molecules that have the ability to alert the immune system and protect the host from pathogenic microbial infection (Rendón and Balcazar, 2003; Cerezuela et al., 2013). Hence, administration of these health-benefitting microbes in feed or any sort of incorporation can boost the cellular and humoral component of the innate immune response in both fish and shellfish species (Kuhn et al., 2009; Ahmad et al., 2016; Anand et al., 2017; Kamilya et al., 2017; Kheti et al., 2017; Lee et al., 2017; Fauji et al., 2018; Kumar V.S. et al., 2018). Xu and Pan (2013) reported that shrimp cultured in the biofloc system had higher total hemocyte count and phagocytic activity. Additionally, significantly upregulated total antioxidant and superoxide dismutase capacity and reduced/oxidized glutathione ratio were observed in biofloc cultured shrimp (Xu and Pan, 2013). A study from Kim et al. (2014) suggested that biofloc water contains an abundant number of bacteria biomass and the bacterial cell wall consists of various components such as bacterial peptidoglycan lipopolysaccharide and β-1, 3-glucans, which are potential immunostimulatory agents, stimulating the non-specific immune activity of shrimp. In the experiment, it was demonstrated that the biofloc system significantly enhances shrimp survival, final body weights, and prophenoloxidase (proPO) cascade, which is one of the major innate immune responses in crustaceans, by upregulating prophenoloxidase activation enzyme, masquerade-like proteinase, prophenoloxidase1, prophenoloxidase2, serine proteinase1, and ras-related nuclear protein expression (Kim et al., 2014). Similar findings were also observed in Labeo rohita (Ahmad et al., 2016; Kamilya et al., 2017), Oreochromis niloticus (Mansour and Esteban, 2017; Mirzakhani et al., 2019; Hwihy et al., 2021; Shourbela et al., 2021), Cyprinus carpio (Bakhshi et al., 2018; Aalimahmoudi and Mohammadiazarm, 2019), and Apostichopus japonicus (Chen et al., 2018), where animals reared in biofloc water display higher growth performance including improved feed efficiency ratio (FER), specific growth rate (SGR), and feed conversion ratio (FCR). Additionally, the cultured animals have enhanced non-specific immune response, highlighted by significantly increased serum protein, serum albumin, total immunoglobulin, lysozyme, respiratory burst, and myeloperoxidase activity. Taken together, it can be concluded that availability of beneficial microbes in the system can positively improve the growth, immune response, and disease tolerance of cultured animals.
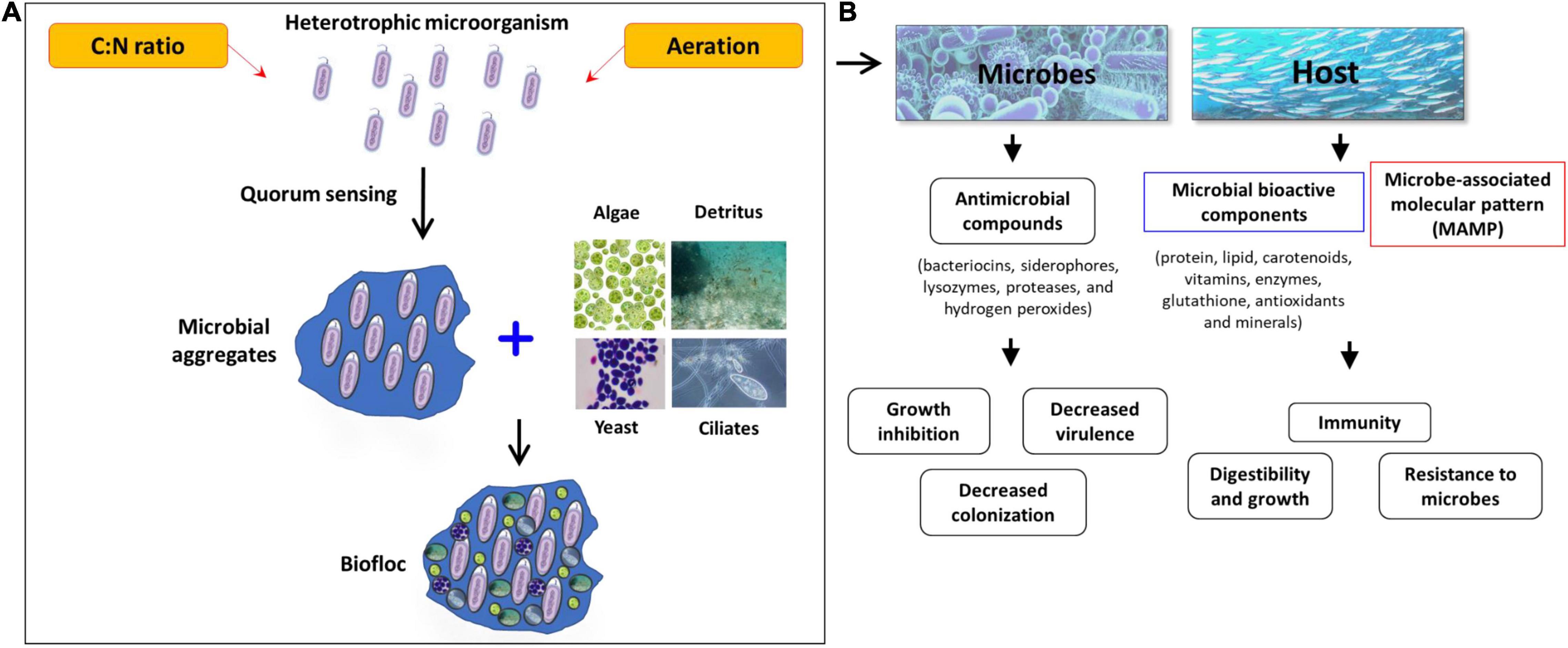
Figure 3. Schematic overview on the possible role of the biofloc microbiome. (A) Development of a biofloc system; (B) potential role of the biofloc system against pathogenic microbes and in a host.
The maintenance of intestinal immunity and metabolic homeostasis is regulated by interaction between host mucosa and the intestinal microbiota. The beneficial microbes were reported to establish a balanced and healthy microbiome within the gastrointestinal tract and minimize harmful bacteria (Kiron, 2012; Gupta et al., 2019). The microbial communities were reported to contain microbe-associated molecular patterns (MAMPs), for instance, bacterial cell wall components like peptidoglycan, lipopolysaccharide, and lipoteichoic acid, which are involved in the modulation of receptor signaling cascades and plays a crucial role in the activation of host immune response and protection from a host from infectious diseases (Cerenius and Soderhall, 2013; Wang and Wang, 2013; Song and Li, 2014; Im et al., 2016). Moreover, there is some evidence that highlights that improved growth performance and non-specific immunity results in development of resistance in cultured animals (Table 2). For example, studies in O. niloticus (Elayaraja et al., 2020), C. gariepinus (Dauda et al., 2017, 2018; Fauji et al., 2018), L. rohita (Kheti et al., 2017), C. carpio (Dash et al., 2018), L. vannamei (Liu et al., 2017), Fenneropenaeus indicus (Megahed et al., 2018), and M. rosenbergii (Miao et al., 2017) demonstrated that aquatic animals cultured in biofloc water have significantly enhanced growth and immune response that leads to increased protection against pathogenic microbial infection. Overall, it can be concluded that microbes, mainly heterotrophic bacteria, developed within the biofloc system contribute as bioremediation agents by ensuring optimum water quality and in the process help in the generation of new biomass which are further consumed by cultured animals resulting in improved growth, health and disease tolerance in cultured animals (Liu et al., 2017; Miao et al., 2017; Dash et al., 2018; Megahed et al., 2018; Elayaraja et al., 2020).
Several researchers have pointed out that biofloc grown aquatic animals have enhanced resistance to pathogenic microbial infections (Figure 3) (Azim and Little, 2008; Crab et al., 2012; Pérez-Rostro et al., 2014; Luis-villaseñor et al., 2016; Anand et al., 2017; Bossier and Ekasari, 2017; Lee et al., 2017; Kasan et al., 2018; Pacheco-Vega et al., 2018; Fatimah et al., 2019; Kumar et al., 2020b). One of the possible scenarios that might be involved behind this induced resistance is improved immunity and health of cultured animals in biofloc as also suggested in an earlier paragraph. It is noted that shrimp in the biofloc system consumes up to 29% flocculating particles of their daily feed intake (Burford et al., 2003), hence the consumed biofloc might nutritionally modulate the health status of shrimp resulting in increased protection of host against microbial diseases. However, another possible mechanism that could also possibly be involved in the protective effect of the biofloc system is intermicrobial interaction that leads to direct growth inhibition or modulation of pathogenic microbe virulence by biofloc derived microbial communities.
The microbes, including both beneficial and pathogenic for cultured species, are ubiquitously present in aquatic environments. The presence of one group in the ecosystem might interact (synergistically or antagonistically) and substantially affects the behavior and abundance of the second group of the microbial community (Matsui et al., 2000; Das et al., 2006, 2014). Bacterial interaction is a common phenomenon that occurs naturally in an aquatic environment. Interestingly, these microbial interactions play a major role in keeping the equilibrium between potentially pathogenic and competing beneficial microorganisms. Nonetheless, the microbial communities composition can be altered by environmental conditions and husbandry practices that stimulate the multiplication of selected bacterial species (Kumar et al., 2016). It is well established that aquatic animals gastrointestinal microbiota can be altered and modified by ingestion of other microorganisms; hence, the microbial manipulation constitutes a promising tool to eliminate or reduce the incidence of opportunistic microbial pathogens (Balcázar et al., 2006). The direct effect, for example, inhibiting the growth and proliferation of other microorganisms, could be the main mode of action of beneficial bacteria that can be observed in cultured systems (Kesarcodi-Watson et al., 2008; Giri et al., 2013; Jiang et al., 2013), and studies have demonstrated that autochthonous microorganisms have significantly high potential because these microbes can easily adapt to the same ecological niche and competitively exclude the pathogenic microbes from the system (Lalloo et al., 2010). Apart from direct inhibition, competition for binding site and nutrition inside the host by adhesion and colonization on the mucosal surfaces are other indirect possible protective mechanisms of beneficial bacteria against pathogenic microbes (Pickard et al., 2017). For instance, study on rainbow trout (Oncorhynchus mykiss) illustrated that lactobacilli administration significantly decrease the adhesion and colonization of pathogenic Carnobacterium piscicola, Yersinia ruckeri, and A. salmonicida in intestinal mucus of the host (Balcázar et al., 2007). The beneficial bacteria produce a variety of wide-spectrum chemical compounds including siderophores, bacteriocins, hydrogen peroxides, proteases, and lysozymes in the intestine of the host, thus constituting a barrier against the proliferation of opportunistic pathogenic microbes. Additionally, they produce organic acids that leads to alteration of the intestinal pH due and inhibition of microbial pathogens (Oppegård et al., 2007; Zai et al., 2009; Korkea-aho et al., 2011, 2012; Ström-Bestor and Wiklund, 2011).
Interestingly, there are several studies that have used pathogenic Listeria monocytogenes as a model organism, a Gram-positive pathogenic bacterium reported to cause severe problems in the food industry, to study the possible interaction mechanism on growth and virulence of another bacterial community (Zilelidou and Skandamis, 2018). The microbial species, e.g., Carnobacterium piscicola (produces bacteriocins) (Buchanan and Bagi, 1997; Nilsson et al., 1999, 2004, 2005; Yamazaki et al., 2003); lactic acid bacteria (LAB)/mainly Lactobacillus (pH reduction, lactic acid production) (Callon et al., 2011); Lactobacillus plantarum (produces bacteriocins, pH reduction) (Nielsen et al., 2010; Aguilar et al., 2011); Lactococcus lactis (produces bacteriocins, pH reduction) (Breidt and Fleming, 1998; Rodríguez et al., 2005; Coelho et al., 2014); Lactobacillus sakei (produces bacteriocins) (Vermeiren et al., 2006; Gao et al., 2015; Martinez et al., 2015; Quinto et al., 2016), and Lactococcus piscium (Saraoui et al., 2016) are reported to inhibit/reduce the growth of L. monocytogenes. There are also few studies that highlight that microbial community groups can antagonistically interact with L. monocytogenes and modulate the virulence of pathogens. The microbial species, e.g., L. lactis (inhibits biofilm formation) (García-Almendárez et al., 2008; Habimana et al., 2011); Lactobacillus paracasei, Listeria innocua (inhibits adherence and biofilm formation) (Bendali et al., 2014; Koo et al., 2014); Staphylococcus sciuri (production of siderophores and extracellular polysaccharide) (Leriche et al., 2000), and Lactobacillus acidophilus (produce antimicrobial compounds) (Woo and Ahn, 2013) were demonstrated to act antagonistically, while synergistic interaction was observed in Staphylococcus aureus and Flavobacterium spp. (higher biofilm formation) (Bremer et al., 2001; Rieu et al., 2008). Apart from studies on L. monocytogenes, there are also few reports, which indicates the antagonistic properties of bacterial species against pathogenic microbes. For instance, Lalloo et al. (2007) demonstrated that B. subtilis, Bacillus mycoides, and Bacillus licheniformis act antagonistically and significantly inhibit the growth and virulence of A. hydrophila.
Moreover, there is not much work done on the effect of biofloc derived microbial communities on the growth and virulence of pathogenic microorganisms. However, few studies like Ferreira et al. (2015) demonstrated that bioflocs are a rich source of beneficial microbial communities, more specifically microbes with probiotics activity. The study showed that Gram-positive bacteria B. licheniformis, isolated from biofloc water, have antagonistically inhibited the growth of pathogenic Vibrio alginolyticus. Additionally, the B. licheniformis modulates growth and immunity and reduces in vivo V. alginolyticus count in L. vannamei (Ferreira et al., 2015). Recently, Kumar et al. (2020a, b) demonstrated that acute hepatopancreatic necrosis disease (AHPND) causing V. parahaemolyticus shift phenotype from virulent state to non-virulent phenotype in a biofloc environment. The results showed that biofloc environment induces adaptive changes in pathogenic V. parahaemolyticus strain, observed by downregulation of growth, motility, and virulence related genes. Additionally, the bacterial strain loses the ability to kill the host and when the pathogenic V. parahaemolyticus strain added to cultured water it fails to induce significantly high mortality in shrimp species (Kumar et al., 2020a, b, 2021a). Although there was no direct correlation on the possible involvement of biofloc derived microbial communities and observed effect on pathogenic V. parahaemolyticus, the results indicate the pathogenic microbe becomes non-virulent in the biofloc system. Taken together, the studies highlight that biofloc associated microbial communities influence the growth and virulence of pathogenic microbes and are involved, at least partially, in providing protection to cultured animals; however, the mechanism by which this phenomenon occurs needs further investigation.
The adverse climatic condition, namely, droughts, scarcity, and expensive water for the development of aquaculture and the adverse effects of aquaculture effluents on the environment, pollutions, and the spread of infectious diseases have drawn the attention for farm biosecurity and development of alternate technology to reduce the amount of water exchange in the farms (Khanjani and Sharifinia, 2020). The biofloc technology (BFT) is based on the principle of flocculation or co-culture of heterotrophic bacteria and algae within the system (Crab et al., 2007; Ahmad et al., 2017). Interestingly, production in biofloc in the large-scale aquaculture can have environmental benefits in marine and coastal ecosystems. For instance, it can help in minimizing the potential negative effect of artificial commercial feed containing soybean or fish meal on aquaculture wastewater and environmental by serving as a nutrient source in both in situ and ex situ systems.
The successful demonstration of BFT for several aquaculture species has made this technology a promising endeavor for future fish production. In general, a commercial biofloc system varies in size, between 0.1 and 2 ha and the essential components including aspirators and paddle wheels are installed to aerate and mix the culture water to keep floc particles in suspension. Currently, commercial, large-scale, and small-scale BFT-based fish farms are expanding in a number of countries, especially in Asia (e.g., Indonesia, Malaysia, Thailand, South Korea, China, and India) and the Americas (e.g., United States and Brazil) (Emerenciano et al., 2013). For instance, in Indonesia about 20–25% of shrimp farms have employed the biofloc system, resulting in an average production of more than 20 mt ha–1 per cycle in 0.5-ha lined ponds (Crab et al., 2012). Interestingly the lining in ponds, partially or completely lined with high density polyethylene (HDPE) sheets, may affect the overall fish production in the biofloc system. For example, in Malaysia, lower production was achieved (12 mt ha–1 per crop) when only pond dikes were lined, while fully lined ponds produced 16.2–22.5 mt ha–1 (Bossier and Ekasari, 2017). However, despite the high potential of biofloc technology in aquaculture, the system has some major associated drawbacks. The most important economic problem is the excessive use of energy for continuously high aeration and water mixing. This system also requires regular monitoring, alarms, and emergency power supply. This means that the biofloc systems could increase the operating costs, due to the cost of the aeration system and the carbon source added to the system (Pérez-Rostro et al., 2014). Additionally, after some period the flocs particles tend to become old and increase in size, which might be not acceptable to the cultured fish. Hence, continuous monitoring of flocs volume and particle size is necessary to harness the maximum beneficial effect of the biofloc system. Therefore, it is necessary that economic analyses must be performed before adopting the BFT on commercial scales. In this regard, a species-specific study must be carried out to determine the effectiveness of BFT in aquaculture system.
Although the biofloc system has several attributes that have increased wide adoption for commercial intensive and super-intensive fish culture, data and information on the economy of these systems in Indian major cultured species are almost lacking, or not accessible to the public. Additionally, experience and information generated by private BFT enterprises are generally not open to the public. These companies tend to keep information and their own know how proprietary. Even when the data are available, proper dissemination channels are almost lacking. Hence, studies must be carried out in the future to demonstrate the application of this technology for commercial fish species of India, in order to persuade the farmers to set it up to justify the BFT technology rather than conventional culture methods.
Biofloc technology allows high-density culture and offers the possibility to maintain good water quality with no or minimal water exchange by recycling of nutrient, in particular, nitrogenous waste into microbial biomass that can be utilized in situ by the cultured animals. The addition of a carbon source, e.g., molasses or tapioca in C:N ratio of 12–15:1, promotes aggregation of microbial mass, i.e., biofloc, that helps in natural bioremediation process by converting toxic nutrient from the system and these flocs are subsequently consumed and utilized as a nutrient source for growth, immunity, and developing tolerance against disease by cultured animal. In fact, these microbial masses, which mainly consist of heterotrophic microorganisms, hold enormous potential to promote biofloc development and improve water quality, host immunity and resistance to microbial pathogens. However, still not much work or information is available on the microbial species or diversity responsible for developing flocs and maintaining optimum water quality and health of cultured animals. Hence, isolation of biofloc derived microbial community, mainly heterotrophic microbes, and further characterizing their possible interaction mechanism with environment, host, and pathogenic microbes will open new avenues and will be a promising aquaculture technology for future aquatic environments and pathogen management and possibly result in an overall increase in the aquaculture production with high-density and minimal or no water exchange culture.
Apart from serving as a potential tool to maintain good water quality and the health of the host, the biofloc system derived microbial community can also be useful to manage wastewater by serving as natural bioremediation agents. Wastewater from industry and daily household discharge, which mainly contains nitrogenous compounds, phosphorous, and other dissolved organic carbons, has created havoc in aquatic environments and even destroyed many ecosystems. Interestingly, there is a growing interest in using the microbial community as potential bioremediators to treat the culture water discharge and wastewater. This suggests that identification of a microbial species or consortia from the biofloc system that regulates natural bioremediation processes would be a promising strategy for wastewater treatment. In conclusion, we can say that maintaining a beneficial microbial diversity could be a promising approach to manage wastewater and aquaculture systems.
VK: conceptualization, writing original draft, and editing. SR: conceptualization and editing. HSS: editing. BB: conceptualization and supervision. BD: overall supervision.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to Director ICAR-Central Inland Fisheries Research Institute (ICAR-CIFRI) for financial support and other supporting staff for their technical support.
Aalimahmoudi, M., and Mohammadiazarm, H. (2019). Dietary protein level and carbon/nitrogen ratio manipulation in bioflocs rearing of Cyprinus carpio juvenile: evaluation of growth performance, some blood biochemical and water parameters. Aquaculture 513:734408. doi: 10.1016/j.aquaculture.2019.734408
Abbaszadeh, A., Keyvanshokooh, S., Yavari, V., and Naderi, M. (2019a). Proteome modifications of Pacific white shrimp (Litopenaeus vannamei) muscle under biofloc system. Aquac. Nutr. 25, 358–366. doi: 10.1111/anu.12861
Abbaszadeh, A., Yavari, V., Hoseini, S. J., Nafisi, M., and Torfi Mozanzadeh, M. (2019b). Effects of different carbon sources and dietary protein levels in a biofloc system on growth performance, immune response against white spot syndrome virus infection and cathepsin L gene expression of Litopenaeus vannamei. Aquac. Res. 50, 1162–1176. doi: 10.1111/are.13991
Adel, M., El-Sayed, A.-F. M., Yeganeh, S., Dadar, M., and Giri, S. S. (2017). Effect of potential probiotic Lactococcus lactis Subsp. lactis on growth performance, intestinal microbiota, digestive enzyme activities, and disease resistance of Litopenaeus vannamei. Probiotics Antimicrob. Proteins 9, 150–156. doi: 10.1007/s12602-016-9235-9
Aguilar, C., Vanegas, C., and Klotz, B. (2011). Antagonistic effect of Lactobacillus strains against Escherichia coli and Listeria monocytogenes in milk. J. Dairy Res. 78, 136–143. doi: 10.1017/S0022029910000877
Aguilera-Rivera, D., Escalante-Herrera, K., Gaxiola, G., Prieto-Davó, A., Rodríguez-Fuentes, G., Guerra-Castro, E., et al. (2019). Immune response of the Pacific white shrimp, Litopenaeus vannamei, previously reared in biofloc and after an infection assay with Vibrio harveyi. J. World Aquac. Soc. 50, 119–136. doi: 10.1111/jwas.12543
Ahmad, H. I., Verma, A. K., Babitha Rani, A. M., Rathore, G., Saharan, N., and Gora, A. H. (2016). Growth, non-specific immunity and disease resistance of Labeo rohita against Aeromonas hydrophila in biofloc systems using different carbon sources. Aquaculture 457, 61–67. doi: 10.1016/j.aquaculture.2016.02.011
Ahmad, I., Babitha Rani, A. M., Verma, A. K., and Maqsood, M. (2017). Biofloc technology: an emerging avenue in aquatic animal healthcare and nutrition. Aquac. Int. 25, 1215–1226. doi: 10.1007/s10499-016-0108-8
Anand, P. S. S., Kumar, S., Kohli, M. P. S., Sundaray, J. K., Sinha, A., Pailan, G. H., et al. (2017). Dietary biofloc supplementation in black tiger shrimp, Penaeus monodon: effects on immunity, antioxidant and metabolic enzyme activities. Aquac. Res. 48, 4512–4523. doi: 10.1111/are.13276
Arnold, S. J., Coman, F. E., Jackson, C. J., and Groves, S. A. (2009). High-intensity, zero water-exchange production of juvenile tiger shrimp, Penaeus monodon: an evaluation of artificial substrates and stocking density. Aquaculture 293, 42–48. doi: 10.1016/j.aquaculture.2009.03.049
Asaduzzaman, M., Wahab, M. A., Verdegem, M. C. J., Huque, S., Salam, M. A., and Azim, M. E. (2008). C/N ratio control and substrate addition for periphyton development jointly enhance freshwater prawn Macrobrachium rosenbergii production in ponds. Aquaculture 280, 117–123. doi: 10.1016/j.aquaculture.2008.04.019
Avnimelech, Y. (1999). Carbon / nitrogen ratio as a control element in aquaculture systems. Aquaculture 176, 227–235.
Avnimelech, Y. (2009). Biofloc Technology: A Practical Guide Book. Baton Rouge, LA: World Aquaculture Society.
Azim, M. E., and Little, D. C. (2008). The biofloc technology (BFT) in indoor tanks: water quality, biofloc composition, and growth and welfare of Nile tilapia (Oreochromis niloticus). Aquaculture 283, 29–35. doi: 10.1016/j.aquaculture.2008.06.036
Bachruddin, M., Sholichah, M., Istiqomah, S., and Supriyanto, A. (2018). “Effect of probiotic culture water on growth, mortality, and feed conversion ratio of Vaname shrimp (Litopenaeus vannamei Boone),” in Proceedings of the 7th Asean-Fen International Fisheries Symposium, Batu. doi: 10.1088/1755-1315/137/1/012036
Bakhshi, F., Najdegerami, E. H., Manaffar, R., Tokmechi, A., Rahmani Farah, K., and Shalizar Jalali, A. (2018). Growth performance, haematology, antioxidant status, immune response and histology of common carp (Cyprinus carpio L.) fed biofloc grown on different carbon sources. Aquac. Res. 49, 393–403. doi: 10.1111/are.13469
Balcázar, J. L., de Blas, I., Ruiz-Zarzuela, I., Cunningham, D., Vendrell, D., and Múzquiz, J. L. (2006). The role of probiotics in aquaculture. Vet. Microbiol. 114, 173–186. doi: 10.1016/j.vetmic.2006.01.009
Balcázar, J. L., Vendrell, D., de Blas, I., Ruiz-Zarzuela, I., Gironés, O., and Múzquiz, J. L. (2007). In vitro competitive adhesion and production of antagonistic compounds by lactic acid bacteria against fish pathogens. Vet. Microbiol. 122, 373–380. doi: 10.1016/j.vetmic.2007.01.023
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Behera, B. K., Chakraborty, H. J., Patra, B., Rout, A. K., Dehury, B., Das, B. K., et al. (2020a). Metagenomic analysis reveals bacterial and fungal diversity and their bioremediation potential from sediments of river Ganga and Yamuna in India. Front. Microbiol. 11:556136. doi: 10.3389/fmicb.2020.556136
Behera, B. K., Patra, B., Chakraborty, H. J., Sahu, P., Rout, A. K., Sarkar, D. J., et al. (2020b). Metagenome analysis from the sediment of river Ganga and Yamuna: in search of beneficial microbiome. PLoS One 15:e0239594. doi: 10.1371/journal.pone.0239594
Bendali, F., Hébraud, M., Sadoun, D., Bendali, F., Hébraud, M., and Anti-bacterial, D. S. (2014). Anti-bacterial and anti-adherence activities of a probiotic strain of Lactobacillus paracasei subsp. paracasei against Listeria monocytogenes. Int. J. Appl. Microbiol. Biotechnol. Res. 2, 52–63.
Betanzo-Torres, E. A., Piñar-álvarez, M., de los, Á, Sandoval-Herazo, L. C., Molina-Navarro, A., Rodríguez-Montoro, I., et al. (2020). Factors that limit the adoption of biofloc technology in aquaculture production in mexico. Water (Switzerland) 12:2775. doi: 10.3390/w12102775
Bossier, P., and Ekasari, J. (2017). Biofloc technology application in aquaculture to support sustainable development goals. Microb. Biotechnol. 10, 1012–1016. doi: 10.1111/1751-7915.12836
Breidt, F., and Fleming, H. P. (1998). Modeling of the competitive growth of Listeria monocytogenes and Lactococcus lactis in vegetable broth. Appl. Environ. Microbiol. 64, 3159–3165. doi: 10.1128/aem.64.9.3159-3165.1998
Bremer, P. J., Monk, I. A. N., and Osborne, C. M. (2001). Survival of Listeria monocytogenes attached to stainless steel surfaces in the presence or absence of Flavobacterium spp. J. Food Prot. 64, 1369–1376. doi: 10.4315/0362-028X-64.9.1369
Brim, H., Venkateswaran, A., Kostandarithes, H. M., Fredrickson, J. K., and Daly, M. J. (2003). Engineering Deinococcus geothermalis for vioremediation of high-temperature radioactive waste environments. Appl. Environ. Microbiol. 69, 4575L–4582. doi: 10.1128/AEM.69.8.4575-4582.2003
Buchanan, R. L., and Bagi, L. K. (1997). Microbial competition: effect of culture conditions on the suppression of Listeria monocytogenes Scott A by Carnobacterium piscicola†. J. Food Prot. 60, 254–261. doi: 10.4315/0362-028X-60.3.254
Burford, M. A., Thompson, P. J., Bauman, H., and Pearson, D. C. (2003). Microbial communities affect water quality, shrimp performance at Belize Aquaculture. Glob. Aquac. Advocate 6, 64–65.
Burford, M. A., Thompson, P. J., McIntosh, R. P., Bauman, R. H., and Pearson, D. C. (2004). The contribution of flocculated material to shrimp (Litopenaeus vannamei) nutrition in a high-intensity, zero-exchange system. Aquaculture 232, 525–537. doi: 10.1016/S0044-8486(03)00541-6
Callon, C., Saubusse, M., Didienne, R., Buchin, S., and Montel, M. C. (2011). Simplification of a complex microbial antilisterial consortium to evaluate the contribution of its flora in uncooked pressed cheese. Int. J. Food Microbiol. 145, 379–389. doi: 10.1016/j.ijfoodmicro.2010.12.019
Cardona, E., Gueguen, Y., Magré, K., Lorgeoux, B., Piquemal, D., Pierrat, F., et al. (2016). Bacterial community characterization of water and intestine of the shrimp Litopenaeus stylirostris in a biofloc system. BMC Microbiol. 16:157. doi: 10.1186/s12866-016-0770-z
Cardona, E., Saulnier, D., Lorgeoux, B., Chim, L., and Gueguen, Y. (2015). Rearing effect of biofloc on antioxidant and antimicrobial transcriptional response in Litopenaeus stylirostris shrimp facing an experimental sub-lethal hydrogen peroxide stress. Fish Shellfish Immunol. 45, 933–939. doi: 10.1016/j.fsi.2015.05.041
Cerenius, L., and Soderhall, K. (2013). Variable immune molecules in invertebrates. J. Exp. Biol. 216, 4313–4319. doi: 10.1242/jeb.085191
Cerezuela, R., Fumanal, M., Tapia-Paniagua, S. T., Meseguer, J., Moriñigo, M. Á., and Esteban, M. A. (2013). Changes in intestinal morphology and microbiota caused by dietary administration of inulin and Bacillus subtilis in gilthead sea bream (Sparus aurata). Fish Shellfish Immunol. 34, 1063–1070. doi: 10.1016/j.fsi.2012.12.026
Chai, P. C., Song, X. L., Chen, G. F., Xu, H., and Huang, J. (2016). Dietary supplementation of probiotic Bacillus PC465 isolated from the gut of Fenneropenaeus chinensis improves the health status and resistance of Litopenaeus vannamei against white spot syndrome virus. Fish Shellfish Immunol. 54, 602–611. doi: 10.1016/j.fsi.2016.05.011
Chen, C.-C., and Chen, S.-N. (2001). Water quality management with Bacillus spp. in the high-density culture of red-parrot fish Cichlasoma citrinellum × C. synspilum. N. Am. J. Aquac. 63, 66–73. doi: 10.1577/1548-84542001063<0066:wqmwbs<2.0.co;2
Chen, J., Ren, Y., Wang, G., Xia, B., and Li, Y. (2018). Dietary supplementation of biofloc influences growth performance, physiological stress, antioxidant status and immune response of juvenile sea cucumber Apostichopus japonicus (Selenka). Fish Shellfish Immunol. 72, 143–152. doi: 10.1016/j.fsi.2017.10.061
Chong, G., Kimyon, O., Rice, S. A., Kjelleberg, S., and Manefield, M. (2012). The presence and role of bacterial quorum sensing in activated sludge. Microb. Biotechnol. 5, 621–633. doi: 10.1111/j.1751-7915.2012.00348.x
Coelho, M. C., Silva, C. C. G., Ribeiro, S. C., Dapkevicius, M. L. N. E., and Rosa, H. J. D. (2014). Control of Listeria monocytogenes in fresh cheese using protective lactic acid bacteria. Int. J. Food Microbiol. 191, 53–59. doi: 10.1016/j.ijfoodmicro.2014.08.029
Cohen, J. M., Samocha, T. M., Fox, J. M., Gandy, R. L., and Lawrence, A. L. (2005). Characterization of water quality factors during intensive raceway production of juvenile Litopenaeus vannamei using limited discharge and biosecure management tools. Aquac. Eng. 32, 425–442. doi: 10.1016/j.aquaeng.2004.09.005
Costa-Pierce, B. A., Bartley, D. M., Hasan, M., Yusoff, F., Kaushik, S. J., Rana, K., et al. (2010). “Responsible use of resources for sustainable aquaculture. Farming Waters People Food,” in Proceedings of the Global Conference on Aquaculture 2010, Phuket, 113–147.
Crab, R., Avnimelech, Y., and Defoirdt, T. (2007). Nitrogen removal techniques in aquaculture for a sustainable production. Aquaculture 270, 1–14. doi: 10.1016/j.aquaculture.2007.05.006
Crab, R., Chielens, B., Wille, M., Bossier, P., and Verstraete, W. (2010). The effect of different carbon sources on the nutritional value of bioflocs, a feed for Macrobrachium rosenbergii postlarvae. Aquac. Res. 41, 559–567. doi: 10.1111/j.1365-2109.2009.02353.x
Crab, R., Defoirdt, T., Bossier, P., and Verstraete, W. (2012). Biofloc technology in aquaculture: beneficial effects and future challenges. Aquaculture 356–357, 351–356. doi: 10.1016/j.aquaculture.2012.04.046
Das, B. K., Behera, B. K., Chakraborty, H. J., Paria, P., Gangopadhyay, A., Rout, A. K., et al. (2020). Metagenomic study focusing on antibiotic resistance genes from the sediments of River Yamuna. Gene 758:144951. doi: 10.1016/j.gene.2020.144951
Das, B. K., Nidhi, R. G. N., Roy, P., Muduli, A. K., Swain, P., Mishra, S. S., et al. (2014). Antagonistic activity of cellular components of Bacillus subtilis AN11 against bacterial pathogens. Int. J. Curr. Microbiol. App. Sci. 3, 795–809.
Das, B. K., Samal, S. K., Samantaray, B. R., Sethi, S., Pattnaik, P., and Mishra, B. K. (2006). Antagonistic activity of cellular components of Pseudomonas species against Aeromonas hydrophila. Aquaculture 253, 17–24. doi: 10.1016/j.aquaculture.2005.01.028
Das, P., Chatterjee, S., Behera, B. K., Dangar, T. K., Das, B. K., and Mohapatra, T. (2019). Isolation and characterization of marine bacteria from East Coast of India: functional screening for salt stress tolerance. Heliyon 5:e01869. doi: 10.1016/j.heliyon.2019.e01869
Das, S., and Dash, H. R. (2014). “1–Microbial bioremediation: a potential tool for restoration of contaminated areas,” in Microbial Biodegradation and Bioremediation, ed. S. Das (Oxford: Elsevier), 1–21. doi: 10.1016/B978-0-12-800021-2.00001-7
Dash, H. R., and Das, S. (2012). Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegradation 75, 207–213. doi: 10.1016/j.ibiod.2012.07.023
Dash, P., Tandel, R. S., Bhat, R. A. H., Mallik, S., Pandey, N. N., Singh, A. K., et al. (2018). The addition of probiotic bacteria to microbial floc: water quality, growth, non-specific immune response and disease resistance of Cyprinus carpio in mid-Himalayan altitude. Aquaculture 495, 961–969. doi: 10.1016/j.aquaculture.2018.06.056
Dauda, A. B., Romano, N., Ebrahimi, M., Karim, M., Natrah, I., Kamarudin, M. S., et al. (2017). Different carbon sources affects biofloc volume, water quality and the survival and physiology of African catfish Clarias gariepinus fingerlings reared in an intensive biofloc technology system. Fish. Sci. 83, 1037–1048. doi: 10.1007/s12562-017-1144-7
Dauda, A. B., Romano, N., Ebrahimi, M., Teh, J. C., Ajadi, A., Chong, C. M., et al. (2018). Influence of carbon/nitrogen ratios on biofloc production and biochemical composition and subsequent effects on the growth, physiological status and disease resistance of African catfish (Clarias gariepinus) cultured in glycerol-based biofloc systems. Aquaculture 483, 120–130. doi: 10.1016/j.aquaculture.2017.10.016
de Jesús Becerra-Dorame, M., Martinez-Cordova, L. R., Martínez-Porchas, M., Hernández-López, J., López–Elías, J. A., and Mendoza–Cano, F. (2014). Effect of using autotrophic and heterotrophic microbial-based-systems for the pre-grown of Litopenaeus vannamei, on the production performance and selected haemolymph parameters. Aquac. Res. 45, 944–948. doi: 10.1111/are.12033
Devaraja, T., Banerjee, S., Yusoff, F., Shariff, M., and Khatoon, H. (2013). A holistic approach for selection of Bacillus spp. as a bioremediator for shrimp postlarvae culture. Turkish J. Biol. 37, 92–100. doi: 10.3906/biy-1203-19
Divya, M., Aanand, S., Srinivasan, A., and Ahilan, B. (2015). Bioremediation–an eco-friendly tool for effluent treatment: a review. Int. J. Appl. Res. 1, 530–537.
Ebeling, J. M., Timmons, M. B., and Bisogni, J. J. (2006). Engineering analysis of the stoichiometry of photoautotrophic, autotrophic, and heterotrophic removal of ammonia–nitrogen in aquaculture systems. Aquaculture 257, 346–358. doi: 10.1016/j.aquaculture.2006.03.019
Ekasari, J., Hanif, M., Surawidjaja, E. H., Nuryati, S., De Schryver, P., and Bossier, P. (2014). Immune response and disease resistance of shrimp fed biofloc grown on different carbon sources. Fish Shellfish Immunol. 41, 332–339. doi: 10.1016/j.fsi.2014.09.004
Elayaraja, S., Mabrok, M., Algammal, A., Sabitha, E., Rajeswari, M. V., Zágoršek, K., et al. (2020). Potential influence of jaggery-based biofloc technology at different C:N ratios on water quality, growth performance, innate immunity, immune-related genes expression profiles, and disease resistance against Aeromonas hydrophila in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 107, 118–128. doi: 10.1016/j.fsi.2020.09.023
El-Sayed, A.-F. M. (2021). Use of biofloc technology in shrimp aquaculture: a comprehensive review, with emphasis on the last decade. Rev. Aquac. 13, 676–705. doi: 10.1111/raq.12494
Emerenciano, M., Ballester, E. L. C., Cavalli, R. O., and Wasielesky, W. (2011). Effect of biofloc technology (BFT) on the early postlarval stage of pink shrimp Farfantepenaeus paulensis: growth performance, floc composition and salinity stress tolerance. Aquac. Int. 19, 891–901. doi: 10.1007/s10499-010-9408-6
Emerenciano, M., Ballester, E. L. C., Cavalli, R. O., and Wasielesky, W. (2012). Biofloc technology application as a food source in a limited water exchange nursery system for pink shrimp Farfantepenaeus brasiliensis (Latreille, 1817). Aquac. Res. 43, 447–457. doi: 10.1111/j.1365-2109.2011.02848.x
Emerenciano, M., Gaxiola, G., and Cuzon, G. (2013). “Biofloc technology (BFT): a review for aquaculture application and animal food industry,” in Biomass Now–Cultivation and Utilization, ed. M. D. Matovic (London: InTech). doi: 10.5772/53902
FAO (2019). State of Fisheries and Aquaculture in the World. Rome: FAO. doi: 10.1109/BMEI.2010.5639447
Fatimah, N., Pande, G. S. J., Natrah, F. M. I., Meritha, W. W., Widanarni, Sucipto, A., et al. (2019). The role of microbial quorum sensing on the characteristics and functionality of bioflocs in aquaculture systems. Aquaculture 504, 420–426. doi: 10.1016/j.aquaculture.2019.02.022
Fauji, H., Budiardi, T., and Ekasari, J. (2018). Growth performance and robustness of African Catfish Clarias gariepinus (Burchell) in biofloc-based nursery production with different stocking densities. Aquac. Res. 49, 1339–1346. doi: 10.1111/are.13595
Ferreira, G. S., Bolívar, N. C., Pereira, S. A., Guertler, C., Vieira, F., do, N., et al. (2015). Microbial biofloc as source of probiotic bacteria for the culture of Litopenaeus vannamei. Aquaculture 448, 273–279. doi: 10.1016/j.aquaculture.2015.06.006
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density- responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gadd, G. M. (2000). Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr. Opin. Biotechnol. 11, 271–279. doi: 10.1016/S0958-1669(00)00095-1
Gao, F., Liao, S., Liu, S., Bai, H., Wang, A., and Ye, J. (2019). The combination use of Candida tropicalis HH8 and Pseudomonas stutzeri LZX301 on nitrogen removal, biofloc formation and microbial communities in aquaculture. Aquaculture 500, 50–56. doi: 10.1016/j.aquaculture.2018.09.041
Gao, Y., Li, D., and Liu, X. (2015). Effects of Lactobacillus sakei C2 and sakacin C2 individually or in combination on the growth of Listeria monocytogenes, chemical and odor changes of vacuum-packed sliced cooked ham. Food Control 47, 27–31. doi: 10.1016/j.foodcont.2014.06.031
García-Almendárez, B. E., Cann, I. K. O., Martin, S. E., Guerrero-Legarreta, I., and Regalado, C. (2008). Effect of Lactococcus lactis UQ2 and its bacteriocin on Listeria monocytogenes biofilms. Food Control 19, 670–680. doi: 10.1016/j.foodcont.2007.07.015
Giri, S. S., Sukumaran, V., and Oviya, M. (2013). Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish Immunol. 34, 660–666. doi: 10.1016/j.fsi.2012.12.008
Gupta, S., Lokesh, J., Abdelhafiz, Y., Siriyappagouder, P., Pierre, R., Sørensen, M., et al. (2019). Macroalga-derived alginate oligosaccharide alters intestinal bacteria of Atlantic Salmon. Front. Microbiol. 10:2037. doi: 10.3389/fmicb.2019.02037
Habimana, O., Guillier, L., Kulakauskas, S., and Briandet, R. (2011). Spatial competition with Lactococcus lactis in mixed-species continuous-flow biofilms inhibits Listeria monocytogenes growth. Biofouling 27, 1065–1072. doi: 10.1080/08927014.2011.626124
Hawver, L. A., Jung, S. A., and Ng, W. L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752. doi: 10.1093/femsre/fuw014
Hostins, B., Wasielesky, W., Decamp, O., Bossier, P., and De Schryver, P. (2019). Managing input C/N ratio to reduce the risk of acute hepatopancreatic necrosis disease (AHPND) outbreaks in biofloc systems–a laboratory study. Aquaculture 508, 60–65. doi: 10.1016/j.aquaculture.2019.04.055
Hwihy, H., Zeina, A., Husien, M. A., and El-Damhougy, K. (2021). Impact of Biofloc technology on growth performance and biochemical parameters of Oreochromis niloticus. Egypt. J. Aquat. Biol. Fish. 25, 761–774.
Im, D., Na, K., Vv, G., and Na, R. (2016). Encapsulation and nodulation in insects. Invertebrate Surviv. J. 13, 229–246.
Islam, M. A., Islam, S. S., Debnath, P., Bir, J., and Huq, K. A. (2021). Effect of different C/N ratios on volume and potential microbial composition of flocs in freshwater prawn Macrobrachium rosenbergii culture system. Int. J. Fish. Aquat. Stud. 9, 310–317. doi: 10.22271/fish.2021.v9.i1d.2417
Jasmin, M. Y., Syukri, F., Kamarudin, M. S., and Karim, M. (2020). Potential of bioremediation in treating aquaculture sludge: review article. Aquaculture 519:734905. doi: 10.1016/j.aquaculture.2019.734905
Jiang, H.-F., Liu, X.-L., Chang, Y.-Q., Liu, M.-T., and Wang, G.-X. (2013). Effects of dietary supplementation of probiotic Shewanella colwelliana WA64, Shewanella olleyana WA65 on the innate immunity and disease resistance of abalone, Haliotis discus hannai Ino. Fish Shellfish Immunol. 35, 86–91. doi: 10.1016/j.fsi.2013.04.009
Jones, D. L., Magthab, E. A., Gleeson, D. B., Hill, P. W., Sánchez-Rodríguez, A. R., Roberts, P., et al. (2018). Microbial competition for nitrogen and carbon is as intense in the subsoil as in the topsoil. Soil Biol. Biochem. 117, 72–82. doi: 10.1016/j.soilbio.2017.10.024
Ju, Z. Y., Forster, I., Conquest, L., Dominy, W., Kuo, W. C., and David Horgen, F. (2008). Determination of microbial community structures of shrimp floc cultures by biomarkers and analysis of floc amino acid profiles. Aquac. Res. 39, 118–133. doi: 10.1111/j.1365-2109.2007.01856.x
Kamilya, D., Debbarma, M., Pal, P., Kheti, B., Sarkar, S., and Singh, S. T. (2017). Biofloc technology application in indoor culture of Labeo rohita (Hamilton, 1822) fingerlings: the effects on inorganic nitrogen control, growth and immunity. Chemosphere 182, 8–14. doi: 10.1016/j.chemosphere.2017.05.021
Kasan, N. A., Dagang, A. N., and Abdullah, M. I. (2018). Application of biofloc technology (BFT) in shrimp aquaculture industry. IOP Conf. Ser. Earth Environ. Sci. 196:012043.
Kasan, N. A., Ghazali, N. A., Ikhwanuddin, M., and Ibrahim, Z. (2017). Isolation of potential bacteria as inoculum for biofloc formation in Pacific Whiteleg Shrimp, Litopenaeus vannamei culture ponds. Pak. J. Biol. Sci. PJBS 20, 306–313. doi: 10.3923/pjbs.2017.306.313
Kesarcodi-Watson, A., Kaspar, H., Lategan, M. J., and Gibson, L. (2008). Probiotics in aquaculture: the need, principles and mechanisms of action and screening processes. Aquaculture 274, 1–14. doi: 10.1016/j.aquaculture.2007.11.019
Khanjani, M. H., Sajjadi, M. M., Alizadeh, M., and Sourinejad, I. (2017). Nursery performance of Pacific white shrimp (Litopenaeus vannamei Boone, 1931) cultivated in a biofloc system: the effect of adding different carbon sources. Aquac. Res. 48, 1491–1501. doi: 10.1111/are.12985
Khanjani, M. H., and Sharifinia, M. (2020). Biofloc technology as a promising tool to improve aquaculture production. Rev. Aquac. 12, 1836–1850. doi: 10.1111/raq.12412
Kheti, B., Kamilya, D., Choudhury, J., Parhi, J., Debbarma, M., and Singh, S. T. (2017). Dietary microbial floc potentiates immune response, immune relevant gene expression and disease resistance in rohu, Labeo rohita (Hamilton, 1822) fingerlings. Aquaculture 468, 501–507. doi: 10.1016/j.aquaculture.2016.11.018
Kim, S., Pang, Z., Seo, H., Cho, Y., Samocha, T., and Jang, I. (2014). Effect of bioflocs on growth and immune activity of Pacific white shrimp, Litopenaeus vannamei postlarvae. Aquac. Res. 45, 362–371. doi: 10.1111/are.12319
Kiron, V. (2012). Fish immune system and its nutritional modulation for preventive health care. Anim. Feed Sci. Technol. 173, 111–133. doi: 10.1016/j.anifeedsci.2011.12.015
Kongnum, K., and Hongpattarakere, T. (2012). Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol. 32, 170–177. doi: 10.1016/j.fsi.2011.11.008
Koo, O. K., Ndahetuye, J. B., O’Bryan, C. A., Ricke, S. C., and Crandall, P. G. (2014). Influence of Listeria innocua on the attachment of Listeria monocytogenes to stainless steel and aluminum surfaces. Food Control 39, 135–138. doi: 10.1016/j.foodcont.2013.11.008
Korkea-aho, T. L., Heikkinen, J., Thompson, K. D., von Wright, A., and Austin, B. (2011). Pseudomonas sp. M174 inhibits the fish pathogen Flavobacterium psychrophilum. J. Appl. Microbiol. 111, 266–277. doi: 10.1111/j.1365-2672.2011.05044.x
Korkea-aho, T. L., Papadopoulou, A., Heikkinen, J., von Wright, A., Adams, A., Austin, B., et al. (2012). Pseudomonas M162 confers protection against rainbow trout fry syndrome by stimulating immunity. J. Appl. Microbiol. 113, 24–35. doi: 10.1111/j.1365-2672.2012.05325.x
Kuhn, D. D., Boardman, G. D., Lawrence, A. L., Marsh, L., and Flick, G. J. (2009). Microbial floc meal as a replacement ingredient for fish meal and soybean protein in shrimp feed. Aquaculture 296, 51–57. doi: 10.1016/j.aquaculture.2009.07.025
Kumar, V. (2020). Acute hepatopancreatic necrosis disease (AHPND) in shrimp: virulence, pathogenesis and mitigation strategies. Toxins 13:524.
Kumar, V., Baruah, K., and Bossier, P. (2021a). Bamboo powder protects gnotobiotically-grown brine shrimp against AHPND-causing Vibrio parahaemolyticus strains by cessation of PirAB VP toxin secretion. Aquaculture 5539:736624. doi: 10.1016/j.aquaculture.2021.736624
Kumar, V., Roy, S., Behera, B. K., Bossier, P., and Das, B. K. (2021b). Acute hepatopancreatic necrosis disease (Ahpnd): virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins (Basel) 13, 1–28. doi: 10.3390/toxins13080524
Kumar, V., Baruah, K., Nguyen, D. V., Smagghe, G., Vossen, E., and Bossier, P. (2018). Phloroglucinol mediated Hsp70 production in crustaceans: protection against Vibrio parahaemolyticus in Artemia franciscana and Macrobrachium rosenbergii. Front. Immunol. 9:1091. doi: 10.3389/fimmu.2018.01091
Kumar, V. S., Pandey, P. K., Anand, T., Bhuvaneswari, G. R., Dhinakaran, A., and Kumar, S. (2018). Biofloc improves water, effluent quality and growth parameters of Penaeus vannamei in an intensive culture system. J. Environ. Manage. 215, 206–215. doi: 10.1016/j.jenvman.2018.03.015
Kumar, V., Bels, L., De, Couck, L., Baruah, K., Bossier, P., et al. (2019a). PirABVP toxin binds to epithelial cells of the digestive tract and produce pathognomonic AHPND lesions in germ-free brine shrimp. Toxins (Basel) 11:717. doi: 10.3390/toxins11120717
Kumar, V., Viet, D., Baruah, K., and Bossier, P. (2019b). Probing the mechanism of VP AHPND extracellular proteins toxicity purified from Vibrio parahaemolyticus AHPND strain in germ-free Artemia test system. Aquaculture 504, 414–419. doi: 10.1016/j.aquaculture.2019.02.029
Kumar, V., Roy, S., Baruah, K., Van Haver, D., Impens, F., and Bossier, P. (2020a). Environmental conditions steer phenotypic switching in acute hepatopancreatic necrosis disease-causing Vibrio parahaemolyticus, affecting PirAVP/PirBVP toxins production. Environ. Microbiol. 22, 4212–4230. doi: 10.1111/1462-2920.14903
Kumar, V., Wille, M., Lourenço, T. M., and Bossier, P. (2020b). Biofloc-based enhanced survival of Litopenaeus vannamei upon AHPND-causing Vibrio parahaemolyticus challenge is partially mediated by reduced expression of its virulence genes. Front. Microbiol. 11:1270. doi: 10.3389/fmicb.2020.01270
Kumar, V., Roy, S., Meena, D. K., and Sarkar, U. K. (2016). Application of probiotics in shrimp aquaculture: importance, mechanisms of action, and methods of administration. Rev. Fish. Sci. Aquac. 24, 342–368. doi: 10.1080/23308249.2016.1193841
Kurniawan, K., Wheeler, R., Dann, L. M., and Mitchell, J. G. (2020). “Investigating the effects of urban input on the abundance and diversity of potential bio-floc forming bacteria in the River Murray, South Australia,” in Proceedings of the IOP Conference Series. Earth and Environmental Science, Bristol, 521. doi: 10.1088/1755-1315/521/1/012015
Lalloo, R., Moonsamy, G., Ramchuran, S., Görgens, J., and Gardiner, N. (2010). Competitive exclusion as a mode of action of a novel Bacillus cereus aquaculture biological agent. Lett. Appl. Microbiol. 50, 563–570. doi: 10.1111/j.1472-765X.2010.02829.x
Lalloo, R., Ramchuran, S., Ramduth, D., Görgens, J., and Gardiner, N. (2007). Isolation and selection of Bacillus spp. as potential biological agents for enhancement of water quality in culture of ornamental fish. J. Appl. Microbiol. 103, 1471–1479. doi: 10.1111/j.1365-2672.2007.03360.x
Lee, C., Kim, S., Lim, S., and Lee, K. (2017). Supplemental effects of biofloc powder on growth performance, innate immunity, and disease resistance of Pacific white shrimp Litopenaeus vannamei. Fish. Aquat. Sci. 20, 1–7. doi: 10.1186/s41240-017-0059-7
Leriche, V., Sibille, P., and Carpentier, B. (2000). Use of an enzyme-linked lectinsorbent assay to monitor the shift in polysaccharide composition in bacterial biofilms. Appl. Environ. Microbiol. 66, 1851–1856.
Liu, C. H., Chiu, C. S., Ho, P. L., and Wang, S. W. (2009). Improvement in the growth performance of white shrimp, Litopenaeus vannamei, by a protease-producing probiotic, Bacillus subtilis E20, from natto. J. Appl. Microbiol. 107, 1031–1041. doi: 10.1111/j.1365-2672.2009.04284.x
Liu, G., Zhu, S., Liu, D., Guo, X., and Ye, Z. (2017). Effects of stocking density of the white shrimp Litopenaeus vannamei (Boone) on immunities, antioxidant status, and resistance against Vibrio harveyi in a biofloc system. Fish Shellfish Immunol. 67, 19–26. doi: 10.1016/j.fsi.2017.05.038
Luis-villaseñor, I. E., Voltolina, D., Audelo-naranjo, J. M., Pacheco-marges, M. R., Herrera-espericueta, V. E., and Romero-beltrán, E. (2016). Effects of biofloc promotion on water quality, growth, biomass yield and heterotrophic community in Litopenaeus Vannamei (Boone, 1931) experimental intensive culture. Ital. J. Anim. Sci. 14:3726. doi: 10.4081/ijas.2015.3726
Manan, H., Moh, J. H. Z., Kasan, N. A., Suratman, S., and Ikhwanuddin, M. (2017). Identification of biofloc microscopic composition as the natural bioremediation in zero water exchange of Pacific white shrimp, Penaeus vannamei, culture in closed hatchery system. Appl. Water Sci. 7, 2437–2446. doi: 10.1007/s13201-016-0421-4
Mansour, A. T., and Esteban, M. Á (2017). Effects of carbon sources and plant protein levels in a biofloc system on growth performance, and the immune and antioxidant status of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 64, 202–209. doi: 10.1016/j.fsi.2017.03.025
Martinez, R. C. R., Staliano, C. D., Vieira, A. D. S., Villarreal, M. L. M., Todorov, S. D., Saad, S. M. I., et al. (2015). Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol. 48, 143–152. doi: 10.1016/j.fm.2014.12.010
Matsui, K., Kono, S., Saeki, A., Ishii, N., Min, M. G., and Kawabata, Z. (2000). Direct and indirect interactions for coexistence in a species-defined microcosm. Hydrobiologia 435, 109–116. doi: 10.1023/A:1004016907260
Meenakshisundaram, M., Sugantham, F., Muthukumar, C., and Chandrasekar, M. S. (2021). Metagenomic characterization of biofloc in the grow-out culture of genetically improved farmed tilapia (GIFT). Aquac. Res. 52, 4249–4262. doi: 10.1111/are.15263
Megahed, M. E., Elmesiry, G., Ellithy, A., and Mohamed, K. (2018). Genetic, nutritional and pathological investigations on the effect of feeding low protein diet and biofloc on growth performance, survival and disease prevention of Indian white shrimp Fenneropenaeus indicus. Aquac. Int. 26, 589–615. doi: 10.1007/s10499-017-0231-1
Merchant, S. S., and Helmann, J. D. (2012). “Chapter 2 - Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation,” in Advances in Microbial Physiology Advances in Microbial Physiology, ed. R. K. Poole (Cambridge, MA: Academic Press), 91–210. doi: 10.1016/B978-0-12-398264-3.00002-4
Miao, S., Zhu, J., Zhao, C., Sun, L., Zhang, X., and Chen, G. (2017). Effects of C/N ratio control combined with probiotics on the immune response, disease resistance, intestinal microbiota and morphology of giant freshwater prawn (Macrobrachium rosenbergii). Aquaculture 476, 125–133. doi: 10.1016/j.aquaculture.2017.04.027
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199.
Mirzakhani, N., Ebrahimi, E., Jalali, S. A. H., and Ekasari, J. (2019). Growth performance, intestinal morphology and nonspecific immunity response of Nile tilapia (Oreochromis niloticus) fry cultured in biofloc systems with different carbon sources and input C:N ratios. Aquaculture 512:734235. doi: 10.1016/j.aquaculture.2019.734235
Mohamad, K. A., Mohd, S. Y., Sarah, R. S., Mohd, H. Z., and Rasyidah, A. (2017). Total nitrogen and total phosphorus removal from brackish aquaculture wastewater using effective microorganism. AIP Conf. Proc. 1885:20127. doi: 10.1063/1.5002321
Monroy-Dosta, M., del, C., de Lara, R. A., Castro-Mejía, J., Castro-Mejía, G., and Coelho-Emerenciano, M. G. (2013). Microbiology community composition and abundance associated to biofloc in tilapia aquaculture. Rev. Biol. Mar. Oceanogr. 48, 511–520. doi: 10.4067/s0718-19572013000300009
Mujeeb Rahiman, K. M., Jesmi, Y., Thomas, A. P., and Mohamed Hatha, A. A. (2010). Probiotic effect of Bacillus NL110 and Vibrio NE17 on the survival, growth performance and immune response of Macrobrachium rosenbergii (de Man). Aquac. Res. 41, e120–e134. doi: 10.1111/j.1365-2109.2009.02473.x
Musyoka, S. (2016). Concept of microbial bioremediation in aquaculture wastes; review. Int. J. Adv. Sci. Tech. Res. 5, 1–10.
Muthukrishnan, S., Sabaratnam, V., Tan, G. Y. A., and Chong, V. C. (2015). Identification of indigenous bacteria isolated from shrimp aquaculture wastewater with bioremediation application: total ammoniacal nitrogen (TAN) and nitrite removal. Sains Malays. 44, 1103–1110. doi: 10.17576/jsm-2015-4408-04
Naderi Samani, M., Jafaryan, H., Gholipour, H., Harsij, M., and Farhangi, M. (2016). Effect of different concentration of profitable Bacillus on bioremediation of common carp (Cyprinus carpio) pond discharge. Iran. J. Aquat. Anim. Heal. 2, 44–54. doi: 10.18869/acadpub.ijaah.2.2.44
NavinChandran, M., Iyapparaj, P., Moovendhan, S., Ramasubburayan, R., Prakash, S., Immanuel, G., et al. (2014). Influence of probiotic bacterium Bacillus cereus isolated from the gut of wild shrimp Penaeus monodon in turn as a potent growth promoter and immune enhancer in P. monodon. Fish Shellfish Immunol. 36, 38–45. doi: 10.1016/j.fsi.2013.10.004
Ngasotter, S., Panda, S. P., Mohanty, U., Akter, S., Mukherjee, S., Waikhom, D., et al. (2020). Current scenario of fisheries and aquaculture in india with special reference to Odisha: a review on its status, issues and prospects for sustainable development. Int. J. Bio Resour. Stress Manag. 11, 370–380. doi: 10.23910/1.2020.2126a
Nielsen, D. S., Cho, G.-S., Hanak, A., Huch, M., Franz, C. M. A. P., and Arneborg, N. (2010). The effect of bacteriocin-producing Lactobacillus plantarum strains on the intracellular pH of sessile and planktonic Listeria monocytogenes single cells. Int. J. Food Microbiol. 141, S53–S59. doi: 10.1016/j.ijfoodmicro.2010.03.040
Nilsson, L., Gram, L., and Huss, H. H. (1999). Growth control of Listeria monocytogenes on cold-smoked salmon using a competitive lactic acid bacteria flora. J. Food Prot. 62, 336–342. doi: 10.4315/0362-028X-62.4.336
Nilsson, L., Hansen, T. B., Garrido, P., Buchrieser, C., Glaser, P., Knøchel, S., et al. (2005). Growth inhibition of Listeria monocytogenes by a nonbacteriocinogenic Carnobacterium piscicola. J. Appl. Microbiol. 98, 172–183. doi: 10.1111/j.1365-2672.2004.02438.x
Nilsson, L., Ng, Y. Y., Christiansen, J. N., Jørgensen, B. L., Grótinum, D., and Gram, L. (2004). The contribution of bacteriocin to inhibition of Listeria monocytogenes by Carnobacterium piscicola strains in cold-smoked salmon systems. J. Appl. Microbiol. 96, 133–143. doi: 10.1046/j.1365-2672.2003.02129.x
Nimrat, S., Suksawat, S., Boonthai, T., and Vuthiphandchai, V. (2012). Potential Bacillus probiotics enhance bacterial numbers, water quality and growth during early development of white shrimp (Litopenaeus vannamei). Vet. Microbiol. 159, 443–450. doi: 10.1016/j.vetmic.2012.04.029
Oppegård, C., Rogne, P., Emanuelsen, L., Kristiansen, P. E., Fimland, G., and Nissen-Meyer, J. (2007). The two-peptide class II bacteriocins: structure, production, and mode of action. Microb. Physiol. 13, 210–219. doi: 10.1159/000104750
Pacheco-Vega, J. M., Cadena-Roa, M. A., Leyva-Flores, J. A., Zavala-Leal, O. I., Pérez-Bravo, E., and Ruiz-Velazco, J. M. J. (2018). Effect of isolated bacteria and microalgae on the biofloc characteristics in the Pacific white shrimp culture. Aquac. Rep. 11, 24–30. doi: 10.1016/j.aqrep.2018.05.003
Panigrahi, A., Esakkiraj, P., Jayashree, S., Saranya, C., Das, R. R., and Sundaram, M. (2019a). Colonization of enzymatic bacterial flora in biofloc grown shrimp Penaeus vannamei and evaluation of their beneficial effect. Aquac. Int. 27, 1835–1846. doi: 10.1007/s10499-019-00434-x
Panigrahi, A., Sundaram, M., Saranya, C., Swain, S., Dash, R. R., and Dayal, J. S. (2019b). Carbohydrate sources deferentially influence growth performances, microbial dynamics and immunomodulation in Pacific white shrimp (Litopenaeus vannamei) under biofloc system. Fish Shellfish Immunol. 86, 1207–1216. doi: 10.1016/j.fsi.2018.12.040
Panigrahi, A., Saranya, C., Ambiganandam, K., Sundaram, M., Sivakumar, M. R., and Kumaraguru vasagam, K. P. (2020). Evaluation of biofloc generation protocols to adopt high density nursery rearing of Penaeus vannamei for better growth performances, protective responses and immuno modulation in biofloc based technology. Aquaculture 522:735095. doi: 10.1016/j.aquaculture.2020.735095
Pérez, T., Balcázar, J. L., Ruiz-Zarzuela, I., Halaihel, N., Vendrell, D., De Blas, I., et al. (2010). Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 3, 355–360. doi: 10.1038/mi.2010.12
Pérez-Rostro, C. I., Pérez-Fuentes, J. A., and Hernández-Vergara, M. P. (2014). “Biofloc, a technical alternative for culturing malaysian prawn Macrobrachium rosenbergii,” in Sustainable Aquaculture Techniques, eds J. A. Pérez-Fuentes and M. P. Hernández-Vergara (London: IntechOpen). doi: 10.5772/57501
Pickard, J. M., Zeng, M. Y., Caruso, R., and Núñez, G. (2017). Gut microbiota: role in pathogen colonization, immune responses and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567.Gut
Quinto, E. J., Marín, J. M., and Schaffner, D. W. (2016). Effect of the competitive growth of Lactobacillus sakei MN on the growth kinetics of Listeria monocytogenes Scott A in model meat gravy. Food Control 63, 34–45. doi: 10.1016/j.foodcont.2015.11.025
Ramalingam, K., and Ramarani, S. (2007). Effect of Pseudomonas aeruginosa on the giant freshwater prawn, Macrobrachium rosenbergii–histopathological and electron microscopic study. J. Environ. Biol. 28, 627–635.
Rendón, L., and Balcazar, J. (2003). Inmunología de camarones: conceptos básicos y recientes avances. Revista AquaTIC 19, 27–33.
Rieu, A., Lemaître, J. P., Guzzo, J., and Piveteau, P. (2008). Interactions in dual species biofilms between Listeria monocytogenes EGD-e and several strains of Staphylococcus aureus. Int. J. Food Microbiol. 126, 76–82. doi: 10.1016/j.ijfoodmicro.2008.05.006
Rodríguez, E., Calzada, J., Arqués, J. L., Rodríguez, J. M., Nuñez, M., and Medina, M. (2005). Antimicrobial activity of pediocin-producing Lactococcus lactis on Listeria monocytogenes, Staphylococcus aureus and Escherichia coli O157:H7 in cheese. Int. Dairy J. 15, 51–57. doi: 10.1016/j.idairyj.2004.05.004
Roy, S. (2020). Modulating Innate Immune Memory in Brine Shrimp (Artemia franciscana) and in Giant Freshwater Prawn (Macrobrachium rosenbergii). Ph.D. thesis. Ghent: Ghent University.
Roy, S., Bossier, P., Norouzitallab, P., and Vanrompay, D. (2020). Trained immunity and perspectives for shrimp aquaculture. Rev. Aquac. 12, 2351–2370. doi: 10.1111/raq.12438
Roy, S., Kumar, V., Bossier, P., Norouzitallab, P., and Vanrompay, D. (2019). Phloroglucinol treatment induces transgenerational epigenetic inherited resistance against Vibrio infections and thermal stress in a brine shrimp (Artemia franciscana) model. Front. Immunol. 10:2745. doi: 10.3389/fimmu.2019.02745
Sadat Hoseini Madani, N., Adorian, T. J., Ghafari Farsani, H., and Hoseinifar, S. H. (2018). The effects of dietary probiotic Bacilli (Bacillus subtilis and Bacillus licheniformis) on growth performance, feed efficiency, body composition and immune parameters of whiteleg shrimp (Litopenaeus vannamei) postlarvae. Aquac. Res. 49, 1926–1933. doi: 10.1111/are.13648
Saraoui, T., Fall, P. A., Leroi, F., Antignac, J. P., Chéreau, S., and Pilet, M. F. (2016). Inhibition mechanism of Listeria monocytogenes by a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 53, 70–78. doi: 10.1016/j.fm.2015.01.002
Sarkar, D. J., Das Sarkar, S., Das, B. K., Sahoo, B. K., Das, A., Nag, S. K., et al. (2021). Occurrence, fate and removal of microplastics as heavy metal vector in natural wastewater treatment wetland system. Water Res. 192:116853. doi: 10.1016/j.watres.2021.116853
Schauder, S., and Bassler, B. L. (2001). The langauges of bacteria. Genes Dev. 15, 1468–1480. doi: 10.1101/gad.899601.attracting
Schneider, O., Sereti, V., Eding, E. H., and Verreth, J. A. J. (2005). Analysis of nutrient flows in integrated intensive aquaculture systems. Aquac. Eng. 32, 379–401. doi: 10.1016/j.aquaeng.2004.09.001
Serra, F. P., Gaona, C. A. P., Furtado, P. S., Poersch, L. H., and Wasielesky, W. (2015). Use of different carbon sources for the biofloc system adopted during the nursery and grow-out culture of Litopenaeus vannamei. Aquac. Int. 23, 1325–1339. doi: 10.1007/s10499-015-9887-6
Shourbela, R. M., Khatab, S. A., Hassan, M. M., Van Doan, H., and Dawood, M. A. O. (2021). The effect of stocking density and carbon sources on the oxidative status, and nonspecific immunity of nile tilapia (Oreochromis niloticus) reared under biofloc conditions. Animals 11:184.
Song, Y. L., and Li, C. Y. (2014). Shrimp immune system–special focus on penaeidin. J. Mar. Sci. Technol. 22, 1–8. doi: 10.6119/JMST-013-0813-1
Ström-Bestor, M., and Wiklund, T. (2011). Inhibitory activity of Pseudomonas sp. on Flavobacterium psychrophilum, in vitro. J. Fish Dis. 34, 255–264. doi: 10.1111/j.1365-2761.2010.01232.x
Suita, S. M., Ballester, E. L. C., Abreu, P. C., and Wasielesky, W. (2015). Dextrose as carbon source in the culture of Litopenaeus vannamei (Boone, 1931) in a zero exchange system. Lat. Am. J. Aquat. Res. 43, 526–533. doi: 10.3856/vol43-issue3-fulltext-13
Tacon, A. G. J. (2020). Trends in global aquaculture and aquafeed production: 2000–2017. Rev. Fish. Sci. Aquac. 28, 43–56. doi: 10.1080/23308249.2019.1649634
Tepaamorndech, S., Nookaew, I., Higdon, S. M., Santiyanont, P., Phromson, M., Chantarasakha, K., et al. (2020). Metagenomics in bioflocs and their effects on gut microbiome and immune responses in Pacific white shrimp. Fish Shellfish Immunol. 106, 733–741. doi: 10.1016/j.fsi.2020.08.042
Thomas, G. M., Ward, C. H., Raymond, R. L., Wilson, J. T., and Loehr, R. C. (1992). “Bioremediation,” in Encyclopedia of Microbiology, ed. J. Leperberg (London: Academic Press), 369–385.
Tran, P. T. N., Kumar, V., and Bossier, P. (2020). Do acute hepatopancreatic necrosis disease-causing PirABVP toxins aggravate vibriosis? Emerg. Microbes Infect. 9, 1919–1932. doi: 10.1080/22221751.2020.1811778
Verdegem, M. C. J. (2013). Nutrient discharge from aquaculture operations in function of system design and production environment. Rev. Aquac. 5, 158–171. doi: 10.1111/raq.12011
Vermeiren, L., Devlieghere, F., Vandekinderen, I., and Debevere, J. (2006). The interaction of the non-bacteriocinogenic Lactobacillus sakei 10A and lactocin S producing Lactobacillus sakei 148 towards Listeria monocytogenes on a model cooked ham. Food Microbiol. 23, 511–518. doi: 10.1016/j.fm.2005.10.005
Vyas, A. (2020). “Chapter 3–Biofloc systems in aquaculture: global status and trends,” in Trends of Microbial Biotechnology for Sustainable Agriculture and Biomedicine Systems: Perspectives for Human Health, eds A. A. Rastegari, A. N. Yadav, and N. Yadav (Amsterdam: Elsevier), 31–42. doi: 10.1016/B978-0-12-820528-0.00004-1
Wang, C., Pan, L., Zhang, K., Xu, W., Zhao, D., and Mei, L. (2016). Effects of different carbon sources addition on nutrition composition and extracellular enzymes activity of bioflocs, and digestive enzymes activity and growth performance of Litopenaeus vannamei in zero-exchange culture tanks. Aquac. Res. 47, 3307–3318. doi: 10.1111/are.12784
Wang, X. W., and Wang, J. X. (2013). Pattern recognition receptors acting in innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. 34, 981–989. doi: 10.1016/j.fsi.2012.08.008
Wang, Y. B. (2007). Effect of probiotics on growth performance and digestive enzyme activity of the shrimp Penaeus vannamei. Aquaculture 269, 259–264. doi: 10.1016/j.aquaculture.2007.05.035
Wei, G., Shan, D., Li, G., Li, X., Tian, R., He, J., et al. (2020). Prokaryotic communities vary with floc size in a biofloc-technology based aquaculture system. Aquaculture 529:735632. doi: 10.1016/j.aquaculture.2020.735632
Woo, J., and Ahn, J. (2013). Probiotic-mediated competition, exclusion and displacement in biofilm formation by food-borne pathogens. Lett. Appl. Microbiol. 56, 307–313. doi: 10.1111/lam.12051
Xu, L., Li, H., Vuong, C., Vadyvaloo, V., Wang, J., Yao, Y., et al. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496. doi: 10.1128/IAI.74.1.488-496.2006
Xu, W.-J., Morris, T. C., and Samocha, T. M. (2018). Effects of two commercial feeds for semi-intensive and hyper-intensive culture and four C/N ratios on water quality and performance of Litopenaeus vannamei juveniles at high density in biofloc-based, zero-exchange outdoor tanks. Aquaculture 490, 194–202. doi: 10.1016/j.aquaculture.2018.02.028
Xu, W. J., and Pan, L. Q. (2013). Enhancement of immune response and antioxidant status of Litopenaeus vannamei juvenile in biofloc-based culture tanks manipulating high C/N ratio of feed input. Aquaculture 412–413, 117–124. doi: 10.1016/j.aquaculture.2013.07.017
Yamazaki, K., Suzuki, M., Kawai, Y., Imque, N., Montvila, I., and Thomas, J. (2003). Inhibition of Listeria monocytogenes in cold-smoked salmon by Carnobacterium piscicola CS526 isolated from frozen surimi. J. Food Prot. 66, 1420–1425. doi: 10.4315/0362-028X-66.8.1420
Yu, C.-H., Wang, Y., Guo, T., Shen, W.-X., and Gu, M.-X. (2012). Isolation and identification of ammonia nitrogen degradation strains from industrial wastewater. Engineering 04, 790–793. doi: 10.4236/eng.2012.411101
Zai, A. S., Ahmad, S., and Rasool, S. A. (2009). Bacteriocin production by indigenous marine catfish associated Vibrio spp. Pak. J. Pharm. Sci. 22, 162–167.
Zemor, J. C., Wasielesky, W., Fóes, G. K., and Poersch, L. H. (2019). The use of clarifiers to remove and control the total suspended solids in large-scale ponds for production of Litopenaeus vannamei in a biofloc system. Aquac. Eng. 85, 74–79. doi: 10.1016/j.aquaeng.2019.03.001
Zhang, N., Luo, G., Tan, H., Liu, W., and Hou, Z. (2016). Growth, digestive enzyme activity and welfare of tilapia (Oreochromis niloticus) reared in a biofloc-based system with poly-β-hydroxybutyric as a carbon source. Aquaculture 464, 710–717. doi: 10.1016/j.aquaculture.2016.08.013
Zhao, Z., Xu, Q., Luo, L., Wang, C., Li, J., and Wang, L. (2014). Effect of feed C/N ratio promoted bioflocs on water quality and production performance of bottom and filter feeder carp in minimum-water exchanged pond polyculture system. Aquaculture 434, 442–448. doi: 10.1016/j.aquaculture.2014.09.006
Zhou, H., Gai, C., Ye, G., An, J., Liu, K., Xu, L., et al. (2019). Aeromonas hydrophila, an emerging causative agent of freshwater-farmed whiteleg shrimp Litopenaeus vannamei. Microorganisms 7:450. doi: 10.3390/microorganisms7100450
Zhu, B., Chen, S., Zhao, C., Zhong, W., Zeng, R., and Yang, S. (2019). Effects of Marichromatium gracile YL28 on the nitrogen management in the aquaculture pond water. Bioresour. Technol. 292:121917. doi: 10.1016/j.biortech.2019.121917
Zilelidou, E. A., and Skandamis, P. N. (2018). Growth, detection and virulence of Listeria monocytogenes in the presence of other microorganisms: microbial interactions from species to strain level. Int. J. Food Microbiol. 277, 10–25. doi: 10.1016/j.ijfoodmicro.2018.04.011
Zokaeifar, H., Babaei, N., Saad, C. R., Kamarudin, M. S., Sijam, K., and Balcazar, J. L. (2014). Administration of Bacillus subtilis strains in the rearing water enhances the water quality, growth performance, immune response, and resistance against Vibrio harveyi infection in juvenile white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 36, 68–74. doi: 10.1016/j.fsi.2013.10.007
Zokaeifar, H., Balcázar, J. L., Saad, C. R., Kamarudin, M. S., Sijam, K., Arshad, A., et al. (2012). Effects of Bacillus subtilis on the growth performance, digestive enzymes, immune gene expression and disease resistance of white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 33, 683–689. doi: 10.1016/j.fsi.2012.05.027
Keywords: bioremediation, pathogenic microbes, heterotrophic microbes, biofloc system, host immunity
Citation: Kumar V, Roy S, Behera BK, Swain HS and Das BK (2021) Biofloc Microbiome With Bioremediation and Health Benefits. Front. Microbiol. 12:741164. doi: 10.3389/fmicb.2021.741164
Received: 14 July 2021; Accepted: 25 October 2021;
Published: 29 November 2021.
Edited by:
Ashwani Kumar, Dr. Harisingh Gour Central University, IndiaReviewed by:
Joginder Singh, Lovely Professional University, IndiaCopyright © 2021 Kumar, Roy, Behera, Swain and Das. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bijay Kumar Behera, YmVoZXJhYmsxOEB5YWhvby5jby5pbg==; Basanta Kumar Das, YmFzYW50YWt1bWFyZEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.