
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 December 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.740983
This article is part of the Research Topic Women In Microbiology View all 24 articles
Although conventionally considered an animal pathogen, recent evidence increasingly suggests that fresh produce may act as significant transmission vehicles and alternative hosts to Salmonella. This study reports the differential survivability of two genetically similar Salmonella Thompson strains (ST 889B and ST 688C) on the adaxial surface of pre-harvest basil (Ocimum basilicum) leaves. Upon inoculation, two distinct phenomena, a dried water-print or a macroscopic lesion, were observed within 24 h. ST 889B survived better than ST 688C on healthy-looking leaves without lesions, possibly due to its higher biofilm-forming ability. Both strains survived better on the leaves with lesions than on the healthy-looking leaves (ST 688C: 4.39 ± 0.68 vs. 2.18 ± 0.29; ST 889B: 4.78 ± 0.12 vs. 2.83 ± 0.18 log CFU per sample at 6 days post-inoculation). ST 889B caused the formation of lesions at a higher frequency [70/117 leaves (59.8%)] than ST 688C [35/96 leaves (36.5%)]. Thus, we highlighted two distinct Salmonella survival strategies in the basil pathosystem and demonstrated gene expression polymorphism (variations in the expression of the same set of genes) as an indispensable strategy in the colonization of plants as hosts by the human pathogens.
Salmonella enterica, which causes non-typhoidal salmonellosis, is an important foodborne pathogen estimated to cause up to 11% of the 9.4 million foodborne illnesses in the United States annually. It is also one of the most virulent and fatal foodborne pathogens, causing up to 35 and 28% of all foodborne-related hospitalizations and deaths, respectively, in the United States (Scallan et al., 2011). Its impact is also felt in smaller nations like Singapore, where 68% of all reported food- and waterborne disease cases consisted of non-typhoidal salmonellosis (Zwe and Yuk, 2017). Fresh produce has been identified as an important vehicle for the transmission of Salmonella. Culinary herbs such as basil have drawn particular attention for their food safety since once fresh culinary herbs are contaminated, completely removing or killing pathogens is unlikely. Large-scale S. Senftenberg and S. Anatum outbreaks have been reported in recent history due to contaminated fresh basil consumption in multiple countries (Pezzoli et al., 2008; Pakalniskiene et al., 2009). Therefore, it is crucial to understand how Salmonella survives in the pre-harvest basil plants for its food safety implications.
In this study, we report the differential capabilities of two S. Thompson isolates in colonizing and surviving on the basil plants. ST 889B was isolated from basil in Belgium in 2013 (Delbeke et al., 2015), and ST 688C was a clonal subculture of the 1999 California coriander outbreak clinical-isolate RM1894 (Li and Uyttendaele, 2018). These two S. Thompson isolates were of near-identical genetic makeup and thus served as valuable study materials to examine the plant–pathogen interactions between Salmonella and fresh produce, offering insights that may be critical to improving the microbial safety of plant produce.
ST 688C is a clonal subculture of the S. Thompson strain RM1984 clinically isolated from a salmonellosis patient during the California coriander outbreak in 1999, kindly provided by Dr. Maria Brandl (U.S. Department of Agriculture, Agricultural Research Service, Albany, CA, United States). ST 889B was previously isolated from basil (Delbeke et al., 2015), kindly provided by Prof. Mieke Uyttendaele (Ghent University). The frozen cultures of bacteria were activated by transferring twice consecutively (37°C, 24 h) in sterile tryptic soy broth (TSB, Oxoid, Basingstoke, Hampshire, England). The stock cultures of bacteria were maintained on tryptic soy broth (TSA, Oxoid) plates stored at 4°C. A single colony from TSA was transferred into 10-ml TSB and incubated at 37°C for 24 h. A 1-ml portion of the bacterial suspension was centrifuged at 9,000 × g at 4°C, washed twice in phosphate-buffered saline (PBS; Vivantis, Inc., Oceanside, CA, United States), serially diluted in 0.1% peptone water (PW; Oxoid) to appropriate concentration to obtain the working cultures of bacteria for downstream experiments.
The DNA from the 24-h cultures of bacteria were extracted using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Vilnius, Lithuania) following the manufacturer’s instructions. Whole genome sequencing (WGS) was performed by NovogeneAIT Genomics Singapore Pte Ltd. using Illumina Hiseq4000, and 350 bp insert DNA library preparation.
Raw reads were assembled into contigs using Assembler 1.2 publicly available at the Center for Genomic Epidemiology by the Technical University of Denmark1 using default parameters. Assembled contigs were analyzed for resistance determinants, multilocus sequence type (MLST), and core genome multilocus sequence type (cgMLST) using ResFinder 4.0 (Zankari et al., 2012; ResFinder Database 2020-04-08 and PointFinder Database 2019-07-02), MLST 2.0 (Larsen et al., 2012), and cgMLSTFinder 1.1 (Alikhan et al., 2018), respectively, using default parameters. The average nucleotide identity between ST 688C and ST 889B was analyzed using the ANI Calculator (Yoon et al., 2017). The contigs were deposited to the GenBank, National Center for Biotechnology Information (NCBI)2 under accession numbers JAFFRV000000000 (ST 688C) and JAFDOS000000000 (ST 889B) of BioProject PRJNA697834.
Single-nucleotide polymorphism tree for phylogenetic analysis was performed using the CSI Phylogeny 1.4 (Kaas et al., 2014) using default parameters. All known genome assemblies of 46 S. Thompson strains were downloaded from the NCBI. Three genomes were found to be outgroups from the others in an initial trial and were subsequently excluded. A group of 43 genomes, ST 688C, and ST 889B, were used to construct a phylogenetic tree based on SNPs with S. Thompson RM6836 as the reference. The resulting tree was visualized using the public available FigTree 1.4.4 software.3 Bootstrap values displayed were automatically calculated by the proprietary algorithm of the CSI Phylogeny 1.4 software.
A 100 g portion of Ocimum basilicum sweet basil leaves and stems from the freshly bought basil plants were immediately harvested and added to 200 ml of deionized (DI) water and homogenized in a food processor. The solid particles in the resultant suspension were removed by first filtering through a mesh strainer, followed by multiple rounds of centrifugation (9,000 × g, 4°C, 10 min) until no visible solid particles were present throughout the liquid. The basil juice was sterilized by passing through 0.22 μm filters.
A 100 μl portion of the working culture of ST 688C or ST 889B (ca. 109 CFU/ml) in 0.1% PW was inoculated into 30 ml of sterile basil juice in 50-ml falcon tubes. The samples were stored loosely capped at 25°C over 3 days. A 100 μl portion of the sample was drawn from each sample at 0 (immediately after inoculation), 1, 2, and 3 days post-inoculation (dpi), serially diluted in 0.1% PW and enumerated on xylose lysine deoxycholate (XLD; Oxoid) agar.
As described by Lee et al. (2011), crystal violet biofilm assay was employed with slight modifications to determine the biofilm-forming ability of ST 688C and ST 889B. Briefly, working cultures of bacterial cells were serially diluted in 0.1% PW while the final dilution was performed in either 1/10 TSB (TSB:water of 1:9) or sterile basil juice to a final concentration of 105 CFU/ml. For every combination of bacterial strain and medium, a 200 μl portion of the sample per well was added to six wells in a 96-well plate. After incubating at 25°C for 24 h, each well was washed three times with sterile DI water, stained with 200 μl of 0.1% (wt/vol) crystal violet (Sigma-Aldrich, St. Louis, MO, United States) solution for 20 min at room temperature, rewashed three times with DI water, and incubated with 200 μl of 95% ethanol at 4°C for 45 min to extract dye from the biofilm. Absorption at 595 nm was measured using a Multiskan FC microplate photometer (Thermo Fisher Scientific, Shanghai, China).
The procedures for preparing competent Salmonella cells and the construction of GFP-tagged Salmonella strains through the transformation with pGFPuv plasmid have been previously described (Ma et al., 2011). Briefly, a 250 μl portion of overnight cultures of ST 889B incubated at 37°C in at 37°C Luria-Bertani (LB, Invitrogen, CA, United States) broth was transferred into 25 ml of LB broth and incubated at 37°C for 3 h in the shaker incubator shaking at 200 rpm. The resultant bacterial suspension was then cooled in ice for 10 min, centrifuged (4,000 rpm, 4°C, 10 min), resuspended in 10 ml of 0.1 M CaCl2, centrifuged, and resuspended again in 2 ml of 0.1 M CaCl2 with 15% glycerol medium. The pGFPuv plasmid was extracted using the Qiagen plasmid miniprep kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The transformation was performed by the heat-shock method. Briefly, 5 μl of the plasmid was mixed into 50 μl of competent cells and kept on ice for 30 min before transferring to a 42°C incubator for 30 s and transferring back on ice for 5 min. A 950 μl portion of LB broth was added to the heat-shocked bacterial-plasmid mix and incubated at 37°C for 1 h while shaking at 250 rpm. The suspension was plated onto LB agar with 100 μg/ml ampicillin to recover the transformants. The GFP-tagged bacteria were grown in TSB with 100 μg/ml ampicillin before washing and appropriately diluted as previously described for inoculation. The observation of GFP-tagged bacteria was performed with fluorescence confocal microscopy Olympus Fluoview FV1000 using the emission band 505–525 nm and the autofluorescence from the leaves, using emission band 525–650 nm.
Pre-harvest O. basilicum sweet basil plants (ca. 2 months of age) were purchased from a local supermarket. A 300-W LED lamp (Melonfarm) was used in the 32″ × 32″ × 63″ grow tent (MelonFarm, China) to supply light with a photoperiod of 18 h and a dark period of 6 h. The temperature (26 ± 2°C) and relative humidity (58 ± 8%) in the grow tent were monitored using an EL-USB-2-LCD+ logger (Lascar Electronics Ltd., United Kingdom). Newly purchased plants were subjected to a 24-h acclimatization period to the new environment in the grow tent before inoculation with bacteria.
A 100 μl portion working culture of ST 688C or ST 889B (ca. 109 CFU/ml) in PBS was inoculated onto the adaxial side of an inverted basil leaf (to form a “cup-like” depression to assist inoculation) in a roughly 1 cm2 area and dried ambiently. Immediately after drying (0 dpi) and at other time points (1 and 6 dpi), leaves for each bacterial strain were harvested to be enumerated. The 1 cm2 portion from each leaf where the inoculum has dried was cut and mixed with 5 ml of PBS in a small vacuum-sealed stomacher pouch. The leaf’s surface was lightly massaged for 1 min, followed by a light crushing using a pestle to release any bacterial cells that may be internalized within the leaf structure. The resultant suspension was then serially diluted to the appropriate concentration in 0.1% PW and spread plated on XLD (Oxoid) agar plates and incubated at 37°C for 24 h.
RNA from freshly dried bacterial inoculum on basil leaves was extracted using the RNeasy Mini Kit (Qiagen) and was reverse transcribed using the GoScript™ Reverse Transcriptase kit (Promega Corp., Madison, WI, United States) following the manufacturer’s instructions to obtain the cDNA. The primers used in this study were designed using the Primer-BLAST tool4 and are described in Table 1. The single-copy chromosomal gene hcaT was used as the reference gene (Barak et al., 2007). The assay was performed in 10 μl reaction volumes with 5 μl of GoTaq™ qPCR Master Mix (Promega), 0.25 μl each of the forward and reverse primer at 10 μmol/l concentration, and 1 μl of cDNA template. Threshold cycle (CT) values were measured using the StepOnePlus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, United States). The thermal cycling conditions were as previously described (Yang et al., 2014). The fold change in gene expression was calculated as previously described (Pfaffl, 2001).
Each bacterial count data point was derived from three independent experiments with two biological replicates (n = 6). Biofilm-forming assay data consisted of two independent experiments with six wells each. Fold change in gene expression data was derived from two independent experiments, each consisting of RNA extracted from three biological samples which were polled together. Unpaired t-test, which can be accessed online at https://www.graphpad.com was used to compare bacterial count, biofilm-forming ability, and fold change in gene expression data set.
Whole Genome Sequencing analysis revealed that ST 688C and ST 889B had identical MLST (Larsen et al., 2012) and cgMLST (Alikhan et al., 2018) of 26 and 5529, respectively. They also carried the identical chromosomal mutations gyrA S83Y and parC T57S. The acquired resistance gene aac(6′)-Iaa was present with an ID match of 97.72%, and both genomes featured identical point mutations in ten bases. The exact sequence of the aac(6′)-Iaa gene in the form of ResFinder 4.0 (Zankari et al., 2012) output text file from both ST 688C and ST 889B is available as Supplementary Figure S1. The two genomes shared an average nucleotide identity of 99.98% and an identical GC content of 52.23%. A SNP tree was constructed by including all known S. Thompson genomes from the NCBI database to investigate their phylogeny (Figure 1). ST 688C was genetically similar to the RM1894 outbreak strain and ST 889B. The SNP differences between the genomes were as follows: ST 688C and RM1894 (6 SNPs), ST 688C and ST 889B (9 SNPs), and ST 889B and RM1894 (9 SNPs). These three isolates formed a distinct clade (Figure 1; colored) whose most genetically similar clade consists of S. Thompson strains isolated exclusively from the United States from 2009 to 2012 except for the S. Thompson ATCC BAA 1738. In short, ST 688C and ST 889B are genetically similar to each other and the original RM1894 outbreak strain.
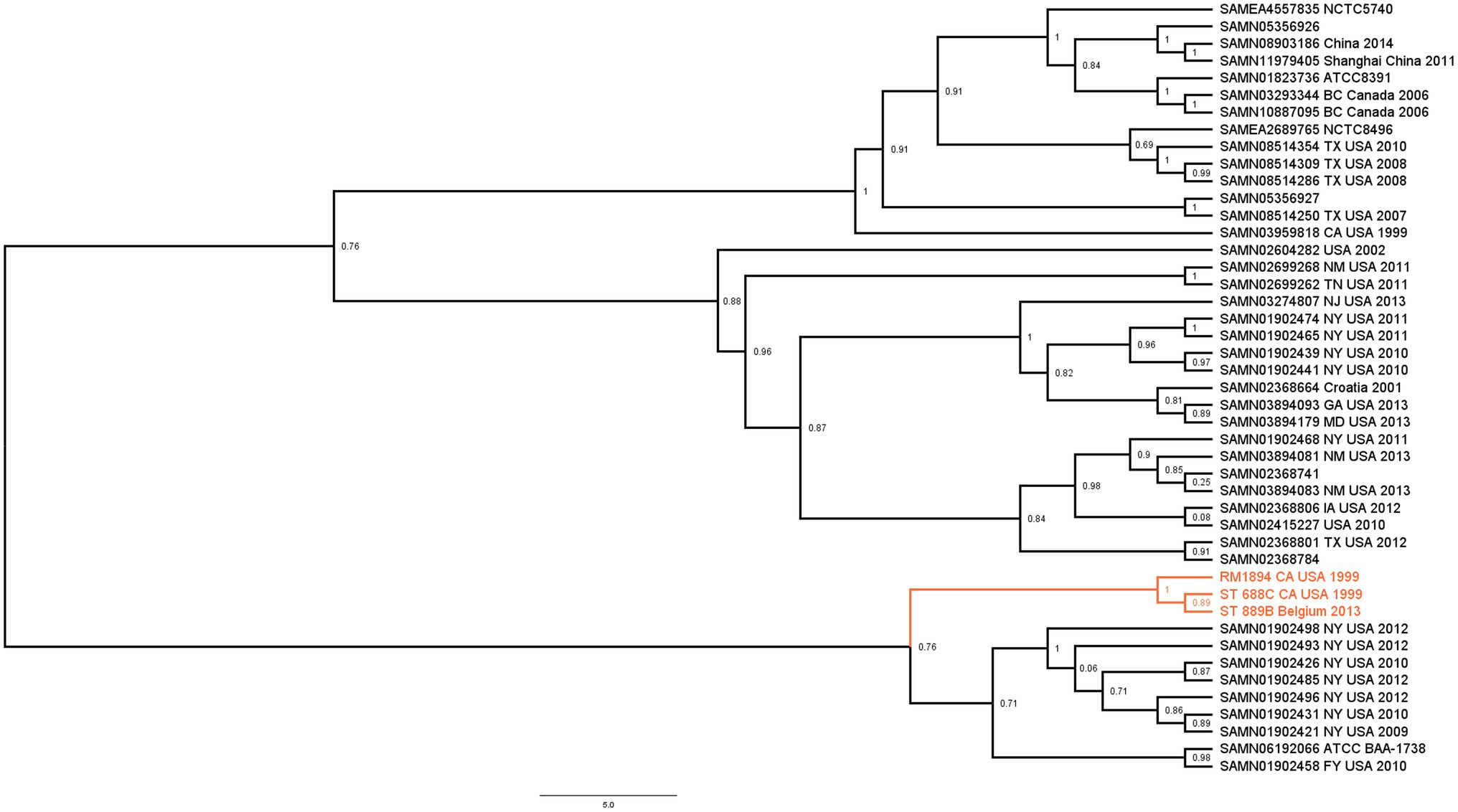
Figure 1. Maximum likelihood SNP tree illustrating the phylogenetic relationship between ST 688C, ST 889B, the 1999 California coriander outbreak clinical strain RM1984 (three isolates of interest highlighted in color) and all other publicly available closely related members of S. Thompson isolates. Genomes downloaded from NCBI were denoted with their Biosample accession numbers followed by the place and year of isolation, if available. Numbers denote bootstrap values.
Upon inoculation of ST 688C and ST 889B on the adaxial surface of pre-harvest basil leaves, it was observed that the bacterial inoculum might either dry up on the leaf surface without eliciting any visible response from the leaf, i.e., no macroscopic lesions (referred to as healthy-looking leaf samples hereafter) as shown in Figure 2A or it may result in the formation of macroscopic lesions as shown in Figure 3A. The lesions fully developed within 24-h post-inoculation and were restricted to the leaf’s portion in contact with the inoculum. Once fully formed, no further enlargement of the lesions was observed throughout the experiment (6 days).

Figure 2. (A) A representative healthy-looking basil leaf as seen unaided at 1 dpi, and under confocal microscopy at 1 and 6 dpi (from left to right) where red signals represent leaf cells while green signal represents GFP-tagged Salmonella cells; (B) bacterial counts of ST 688C and ST 889B on healthy-looking leaf tissue at 0, 1, and 6 dpi; and (C) in sterile basil juice at 0, 1, 2, and 3 dpi; (D) biofilm forming ability of ST 688C and ST 889B in 1/10 TSB and sterile basil juice; (E) fold change in gene expression of biofilm formation genes in ST 889B relative to ST 688C. Values linked by lines and denoted by * and ** represent significant difference at p < 0.05 and < 0.01, respectively, except in (E), where * denotes significant difference (p < 0.05) to a theoretical mean of 1, which is the relative gene expression level of ST 688C. Bacterial counts in (B,C) were the average of three experiments, while that of biofilm forming ability in (D) and fold change in gene expression in (E) were the average of two experiments. Error bars denote standard deviation.
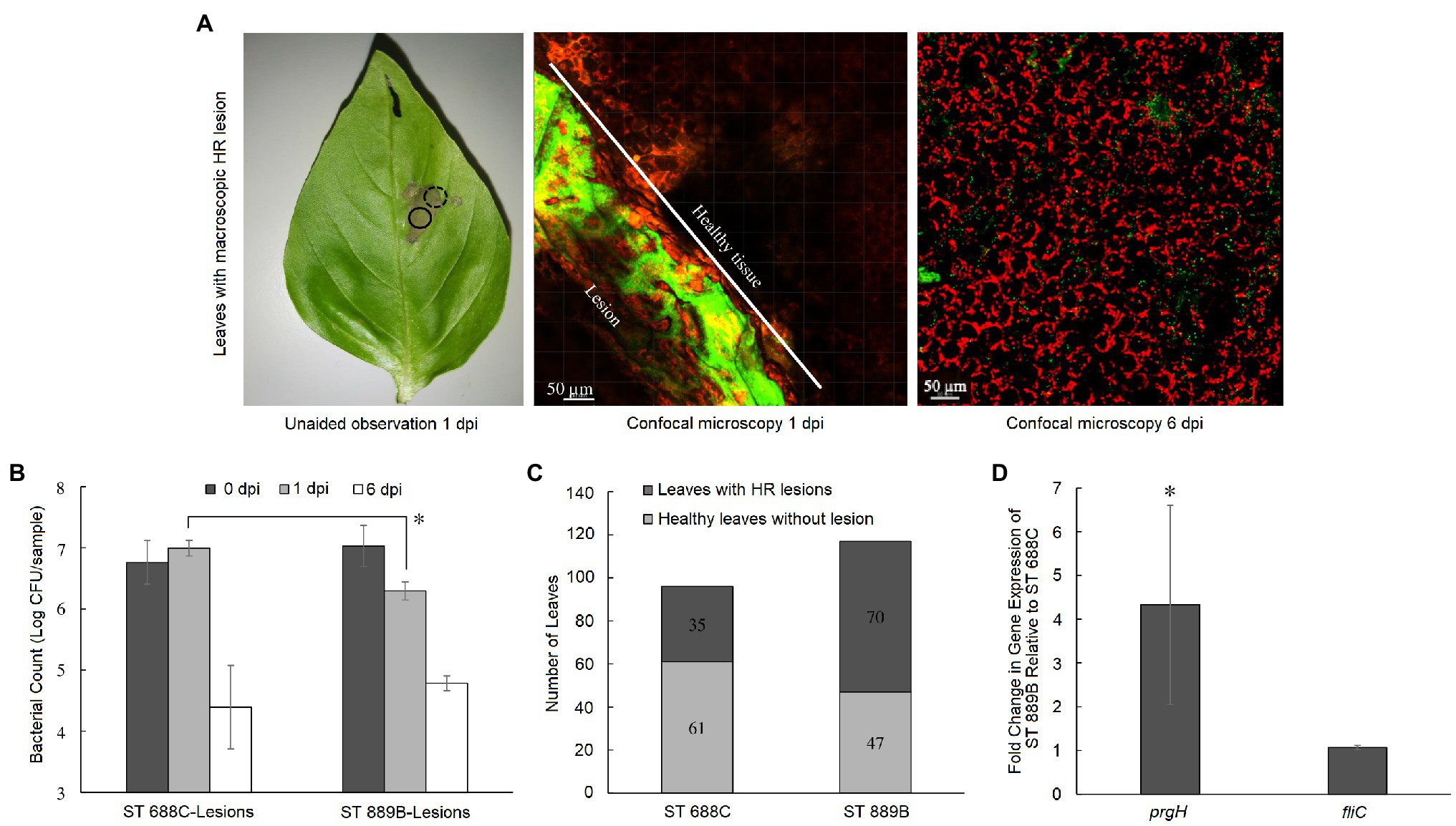
Figure 3. (A) A representative basil leaf with macroscopic HR lesion as seen unaided at 1 dpi, and under confocal microscopy at 1 (dotted circle showing the border between healthy tissue and the lesion separated by the white line) and 6 dpi (full circle showing the lesion; from left to right) where red signals represent leaf cells while green signal represents GFP-tagged Salmonella cells; (B) bacterial counts of ST 688C and ST 889B on leaf tissue with HR lesion at 0, 1, and 6 dpi; (C) number of resultant healthy-looking leaves without HR and unhealthy leaves with HR following inoculation with ST 688C or ST 889B; (D) fold change in gene expression of select HR-related genes in ST 889B relative to ST 688C. Values linked by lines and denoted by * represent significant difference at p < 0.05 except in (D), where * denotes significant difference (p < 0.05) to a theoretical mean of 1, which is the relative gene expression level of ST 688C. Bacterial counts were the average of three experiments in (B), while that of the fold change in gene expression in (D) was of two experiments. Data on the number of leaves with and without HR lesions upon inoculation with ST 688C and ST 889B (C) were derived across 10 and 12 different pre-harvest basil plants, respectively. Error bars denote standard deviation.
On the healthy-looking leaves, fluorescence confocal microscopy revealed that despite many GFP-tagged bacteria (emitting green fluorescence signal) at 1-day post-inoculation (dpi), very few remained by 6 dpi (Figure 2A). ST 889B was found to have survived significantly (p < 0.05) better at both 1 (6.68 ± 0.17 vs. 6.01 ± 0.51 log CFU/sample) and 6 dpi (2.83 ± 0.18 vs. 2.18 ± 0.29 log CFU/sample) than ST 688C (Figure 2B).
As basil is known to produce antimicrobial substances such as linalool (Bagamboula et al., 2004; Beier et al., 2014), we first investigated if the two S. Thompson isolates had differential resistance to the potential antimicrobial basil-derived substances in freshly prepared sterile basil juice. The survivability of the two S. Thompson strains in sterile basil juice was determined over 3 days, and no significant differences in bacterial counts between the two strains were observed (p > 0.05, Figure 2C). On average, the counts of both strains reduced from ~5.6 log CFU/ml initially to ~4.2 log CFU/ml by 3 dpi.
Biofilm formation can increase Salmonella’s resistance toward stresses such as desiccation (Gibson et al., 2006; White et al., 2006, 2010) and, consequently, its ability to persist on abiotic surfaces and non-host environments (Gibson et al., 2006; White et al., 2006, 2010; Vestby et al., 2009). There is accumulating evidence showing that Salmonella can adhere to and from biofilms on plant surfaces, leading to persistent survival and colonization (Yaron and Römling, 2014; Maruzani et al., 2019). Accordingly, we compared the two S. Thompson isolates’ biofilm-forming ability in both bacterial culture broth (1/10 tryptone soya broth, TSB; 1:9 TSB:DI water) and freshly prepared basil juice. Indeed, ST 889B showed significantly higher (p < 0.01) biofilm-forming ability than ST 688C in both 1/10 TSB and basil juice (Figure 2D). Accordingly, ST 889B also showed a higher mean fold change in gene expression level across the four biofilm formation genes tested in this study: bapA (1.95-fold), bcsA (2.49-fold), csgB (3.15-fold), and wcaD (1.86-fold) as compared to ST 688C after the inoculum have dried on pre-harvest basil leaves (Figure 2E).
These results suggest that in the scenario where Salmonella failed to elicit a visible reaction from the basil leaves upon inoculation, their survival strategy is more comparable to that of on an inert or abiotic surface, i.e., mainly characterized by biofilm formation to resist desiccation and starvation.
In this study, we observed the formation of distinctive macroscopic cell death lesions upon direct inoculation of S. Thompson on some basil leaves within 24 h (Figure 3A). In such samples, ST 688C survived slightly better than ST 889B at 1 dpi initially (6.99 ± 0.13 vs. 6.29 ± 0.15 log CFU/sample, p < 0.05) but this difference in survivability was no longer present at 6 dpi (p > 0.05, Figure 3B). More notably, for both Salmonella strains, the bacterial counts were drastically higher in the leaf samples with lesions than the healthy-looking leaf samples at 6 dpi (ST 688C: 4.39 ± 0.68 vs. 2.18 ± 0.29; ST 889B: 4.78 ± 0.12 vs. 2.83 ± 0.18 log CFU/sample, p < 0.01, Figures 2B, 3B). The frequency at which the lesions formed on pre-harvest basil leaves was significantly lower (p < 0.05) for the clinical-isolate ST 688C at 36.5% (35/96 tested leaves across 10 basil plants) as compared to that of the basil-isolate ST 889B at 59.8% (70/117 tested leaves across 12 basil plants; Figure 3C). In other words, the basil-isolate ST 889B could induce the formation of these cell death lesions in basil leaves significantly better than the clinical counterpart ST 688C. Our fluorescence confocal microscopy of the lesions confirmed that although Salmonella grew luxuriantly, they were confined within the lesions while the healthy tissue was devoid of Salmonella cells (Figure 3A).
Plants, lacking the highly specific acquired immune systems of higher animals, feature a generalized multilayer immune system designed to resist invasion from a large variety of potential pathogens. The first layer of the plant immune system is the recognition of pathogen-associated molecular patterns (PAMPs), after which the plant mounts an immune response called the PAMP-triggered immunity (PTI; Monaghan and Zipfel, 2012; Garcia and Hirt, 2014). In response, pathogens well-adapted to the host plant can use bacterial delivery systems such as the type III secretion system (T3SS) to suppress the host PTI response (Garcia and Hirt, 2014). Upon failure of PTI, plants typically resort to the second layer of their immune system called the effector-triggered immunity (ETI). This typically leads to the hypersensitive response (HR), characterized by localized cell death lesions, designed to restrict the spread and growth of pathogens (Morel and Dangl, 1997; Heath, 2000a,b; Heidrich et al., 2012).
We next dedicated our efforts to shedding light on the possible underlying basis of ST 889B’s ability to induce the formation of lesions in basil leaves more successfully. Since PTI precedes ETI, which leads to HR, a successful evasion or suppression of PTI must occur for the subsequent induction of HR. It has been reported that in Salmonella, flg22 epitome at the N-terminal region of bacterial flagellin is the most well-characterized PAMP (Gómez-Gómez and Boller, 2000; Zipfel et al., 2004) that leads to PTI in plants. Hence, we investigated if any downregulation of the flagellin gene fliC by ST 889B to mask its presence and evade PTI was at work. No significant difference was identified in the fold change in the expression level of the fliC gene between the two strains (Figure 3D) upon inoculation onto the basil leaves. Hence, the difference in fliC gene expression levels between the two strains was promptly eliminated as a possible basis for their differential ability to induce HR in basil leaves.
Subsequently, we explored the possible involvement of the T3SS in the formation of these cell death lesions. Although the role of T3SS and its effectors of a plant pathogen like Pseudomonas syringae is very well established (Jones and Dangl, 2006) that of the Salmonella T3SS in colonization, proliferation and eliciting the HR in plants is, unfortunately, less unambiguous. The spaS− and sipB− T3SS mutants showed increased colonization in the alfalfa roots and wheat seedlings (Iniguez et al., 2005) while the prgH− and ssaV− T3SS mutant counts decreased in Arabidopsis thaliana (Schikora et al., 2011). Chalupowicz et al. (2018), on the other hand, observed no significant difference in survivability of wild type S. Typhimurium ATCC 14028 to those of various T3SS mutants (invA−, ssaV−, sifA−, and sipB−) in lettuce leaves.
More pertinently, the wild type S. Typhimurium ATCC 14028 also failed to elicit HR in beetroot tissue and pepper leaves when directly inoculated. However, its T3SS effectors SipB and SifA successfully elicited HR when they were delivered instead by plant pathogens Erwinia amylovora and Xanthomonas euvesicatoria, suggesting that Salmonella T3SS effectors, if successfully translocated into plant cells, can cause HR, but the Salmonella T3SS machinery is incapable of doing so (Chalupowicz et al., 2018). In contrast, we observed the development of HR cell death lesions within 24 h by direct inoculation of S. Thompson onto the basil leaves. This may be due to the high initial inoculum (108 CFU per leaf sample) used in this study. The T3SS has been shown to be an important factor for virulence at high (~108 CFU per plant) but not low titers of Salmonella in tomato plants (De Moraes et al., 2017) and Arabidopsis (Schikora et al., 2011), although the underlying reason was unclear. High bacterial inoculum may have allowed Salmonella to secrete T3SS effectors at levels sufficient to suppress PTI and elicit HR from the basil leaf without translocating into the plant cells directly. Alternatively, the exact outcome of the interaction may also be contingent upon the specific plant species, cultivar., and the Salmonella serovar in question (Klerks et al., 2007; Barak et al., 2011). Further research involving multiple plant pathosystems with multiple Salmonella serovars need to be conducted to shed light on this matter.
Despite prevailing mixed opinions and results, particularly on the Salmonella T3SS and the plant HR as outlined above, generally, the role of the T3SS in triggering HR is well established in many plant pathosystems (Jones and Dangl, 2006). Hence, in this study, we compared the gene expression level of Salmonella T3SS gene prgH, which has been employed as the reporter indicative of the overall T3SS activity in Salmonella (Clark et al., 2009; Wang et al., 2017) in the two S. Thompson strains immediately upon inoculation to the basil leaves. We observed a significantly higher (p < 0.05) 4.33-fold change in the mean expression level of prgH in ST 889B than ST 688C (Figure 3D). First, these results allude to possible involvement of the T3SS in suppressing the PTI and subsequently eliciting HR, specifically in the S. Thompson and basil plant pathosystem. Second, a higher T3SS activity may be responsible for the enhanced ability of ST 889B over ST 688C to cause HR in basil leaves.
The observed differences in the gene expression levels despite remarkably similar genetic makeup, which ultimately led to the differences in phenotype and survivability on pre-harvest basil leaves, might in part be attributed to the inherently different natures of the two S. Thompson strains. It is known that pathogenic bacteria such as Salmonella spp. possesses two distinct “lifestyles,” one tuned for virulence and one for environmental persistence, with each its own set of gene expression profiles as comprehensively reviewed elsewhere (Desai and Kenney, 2019). ST688C, being isolated from a human host with clinical salmonellosis, might have its gene expression profile tuned to that of human host virulence, which may partially explain its lower biofilm formation ability. Conversely, ST889B, being isolated from basil, might have its expression profile tuned to that of environmental persistence. However, whether allowing the clinical ST688C strain to be grown and adapted in basil over an extended time will improve its survivability to match that of ST889B is currently unknown and is a subject of further study.
In this study, we demonstrated that S. Thompson was capable of causing cell death lesions congruent with HR elicited from the basil plant. Furthermore, given the remarkable genetic similarity between ST 889B and ST 688C, the observed differences in phenotypes such as biofilm-forming ability, virulence (lesion formation rate), and survivability on pre-harvest basil could be attributed to the variations in expression levels of the same set of genes rather than to the differences in their genetic makeup. We also highlighted the two distinct Salmonella survival strategies in the basil pathosystem (biofilm formation on the healthy-looking leaf surface and elicitation of cell death lesions with subsequent invasion). Gene expression polymorphism may be an important strategy in the adaptation by human pathogens to survive and persist in plants, as demonstrated in this S. Thompson-basil model.
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA697834.
YZ and DL designed the experiments and wrote the manuscript. YZ carried out the experiments with contributions from MT, XP, and CW. All authors have read and reviewed the manuscript.
This study was supported by an Advanced Manufacturing and Engineering (AME) Young Individual Research Grant (YIRG; A1984c0042) funded by A-STAR, Singapore.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.740983/full#supplementary-material
Supplementary Figure S1 | aac(6')-Iaa gene sequences of ST 688C (template) and ST 889B (query).
1. ^https://cge.cbs.dtu.dk/services/Assembler/
2. ^https://www.ncbi.nlm.nih.gov/
Alikhan, N.-F., Zhou, Z., Sergeant, M. J., and Achtman, M. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14:e1007261. doi: 10.1371/journal.pgen.1007261
Bagamboula, C., Uyttendaele, M., and Debevere, J. (2004). Inhibitory effect of thyme and basil essential oils, carvacrol, thymol, estragol, linalool and p-cymene towards Shigella sonnei and S. flexneri. Food Microbiol. 21, 33–42. doi: 10.1016/S0740-0020(03)00046-7
Barak, J. D., Jahn, C. E., Gibson, D. L., and Charkowski, A. O. (2007). The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant-Microbe Interact. 20, 1083–1091. doi: 10.1094/MPMI-20-9-1083
Barak, J. D., Kramer, L. C., and Hao, L.-Y. (2011). Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77, 498–504. doi: 10.1128/AEM.01661-10
Beier, R. C., Byrd Ii, J. A., Kubena, L. F., Hume, M. E., Mcreynolds, J. L., Anderson, R. C., et al. (2014). Evaluation of linalool, a natural antimicrobial and insecticidal essential oil from basil: effects on poultry. Poult. Sci. 93, 267–272. doi: 10.3382/ps.2013-03254
Chalupowicz, L., Nissan, G., Brandl, M. T., Mcclelland, M., Sessa, G., Popov, G., et al. (2018). Assessing the ability of Salmonella enterica to translocate type III effectors into plant cells. Mol. Plant-Microbe Interact. 31, 233–239. doi: 10.1094/MPMI-07-17-0166-R
Clark, L., Martinez-Argudo, I., Humphrey, T. J., and Jepson, M. A. (2009). GFP plasmid-induced defects in Salmonella invasion depend on plasmid architecture, not protein expression. Microbiology 155, 461–467. doi: 10.1099/mic.0.025700-0
De Moraes, M. H., Desai, P., Porwollik, S., Canals, R., Perez, D. R., Chu, W., et al. (2017). Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. Appl. Environ. Microbiol. 83:e03028-16. doi: 10.1128/AEM.03028-16
Delbeke, S., Ceuppens, S., Jacxsens, L., and Uyttendaele, M. (2015). Microbiological analysis of pre-packed sweet basil (Ocimum basilicum) and coriander (Coriandrum sativum) leaves for the presence of Salmonella spp. and Shiga toxin-producing E. coli. Int. J. Food Microbiol. 208, 11–18. doi: 10.1016/j.ijfoodmicro.2015.05.009
Desai, S. K., and Kenney, L. J. (2019). Switching lifestyles is an in vivo adaptive strategy of bacterial pathogens. Front. Cell. Infect. Microbiol. 9:421. doi: 10.3389/fcimb.2019.00421
Garcia, A. V., and Hirt, H. (2014). Salmonella enterica induces and subverts the plant immune system. Front. Microbiol. 5:141. doi: 10.3389/fmicb.2014.00141
Gibson, D., White, A., Snyder, S., Martin, S., Heiss, C., Azadi, P., et al. (2006). Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188, 7722–7730. doi: 10.1128/JB.00809-06
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Heath, M. C. (2000a). “Hypersensitive response-related death,” in Programmed Cell Death in Higher Plants. eds. E. Lam, H. Fukuda, and J. Greenberg (Springer: Netherlands), 77–90.
Heath, M. C. (2000b). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. doi: 10.1016/s1369-5266(00)00087-x
Heidrich, K., Blanvillain-Baufumé, S., and Parker, J. E. (2012). Molecular and spatial constraints on NB-LRR receptor signaling. Curr. Opin. Plant Biol. 15, 385–391. doi: 10.1016/j.pbi.2012.03.015
Iniguez, A. L., Dong, Y., Carter, H. D., Ahmer, B. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant-Microbe Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Klerks, M., Van Gent-Pelzer, M., Franz, E., Zijlstra, C., and Van Bruggen, A. (2007). Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl. Environ. Microbiol. 73, 4905–4914. doi: 10.1128/AEM.02522-06
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lee, J. H., Cho, M. H., and Lee, J. (2011). 3‐Indolylacetonitrile decreases Escherichia coli O157: H7 biofilm formation and Pseudomonas aeruginosa virulence. Environmental microbiology 13:62–73.
Li, D., and Uyttendaele, M. (2018). Potential of human norovirus surrogates and Salmonella enterica contamination of pre-harvest basil (Ocimum basilicum) via leaf surface and plant substrate. Front. Microbiol. 9:1728. doi: 10.3389/fmicb.2018.01728
Ma, L., Zhang, G., and Doyle, M. P. (2011). Green fluorescent protein labeling of Listeria, Salmonella, and Escherichia coli O157: H7 for safety-related studies. PLoS One 6:e18083. doi: 10.1371/journal.pone.0018083
Maruzani, R., Sutton, G., Nocerino, P., and Marvasi, M. (2019). Exopolymeric substances (EPS) from Salmonella enterica: polymers, proteins and their interactions with plants and abiotic surfaces. J. Microbiol. 57, 1–8. doi: 10.1007/s12275-019-8353-y
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Morel, J.-B., and Dangl, J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. doi: 10.1038/sj.cdd.4400309
Pakalniskiene, J., Falkenhorst, G., Lisby, M., Madsen, S., Olsen, K., Nielsen, E., et al. (2009). A foodborne outbreak of enterotoxigenic E. coli and Salmonella Anatum infection after a high-school dinner in Denmark, November 2006. Epidemiol. Infect. 137, 396–401. doi: 10.1017/S0950268808000484
Pezzoli, L., Elson, R., Little, C. L., Yip, H., Fisher, I., Yishai, R., et al. (2008). Packed with Salmonella—investigation of an international outbreak of Salmonella Senftenberg infection linked to contamination of prepacked basil in 2007. Foodborne Pathog. Dis. 5, 661–668. doi: 10.1089/fpd.2008.0103
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schikora, A., Virlogeux-Payant, I., Bueso, E., Garcia, A. V., Nilau, T., Charrier, A., et al. (2011). Conservation of Salmonella infection mechanisms in plants and animals. PLoS One 6:e24112. doi: 10.1371/journal.pone.0024112
Vestby, L. K., Møretrø, T., Langsrud, S., Heir, E., and Nesse, L. L. (2009). Biofilm forming abilities of Salmonella are correlated with persistence in fish meal-and feed factories. BMC Vet. Res. 5:20. doi: 10.1186/1746-6148-5-20
Wang, L., Cai, X., Wu, S., Bomjan, R., Nakayasu, E. S., Händler, K., et al. (2017). InvS coordinates expression of PrgH and FimZ and is required for invasion of epithelial cells by Salmonella enterica serovar Typhimurium. J. Bacteriol. 199:e00824-16. doi: 10.1128/JB.00824-16
White, A., Gibson, D., Kim, W., Kay, W., and Surette, M. (2006). Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188, 3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006
White, A. P., Weljie, A. M., Apel, D., Zhang, P., Shaykhutdinov, R., Vogel, H. J., et al. (2010). A global metabolic shift is linked to Salmonella multicellular development. PLoS One 5:e11814. doi: 10.1371/journal.pone.0011814
Yang, Y., Khoo, W. J., Zheng, Q., Chung, H.-J., and Yuk, H.-G. (2014). Growth temperature alters Salmonella Enteritidis heat/acid resistance, membrane lipid composition and stress/virulence related gene expression. Int. J. Food Microbiol. 172, 102–109. doi: 10.1016/j.ijfoodmicro.2013.12.006
Yaron, S., and Römling, U. (2014). Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 7, 496–516. doi: 10.1111/1751-7915.12186
Yoon, S.-H., Ha, S.-M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E. J., Jones, J. D., Felix, G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. doi: 10.1038/nature02485
Keywords: Salmonella Thompson, pre-harvest basil, hypersensitive response, expression polymorphism, type III secretion system
Citation: Zwe YH, Ten MMZ, Pang X, Wong CH and Li D (2021) Differential Survivability of Two Genetically Similar Salmonella Thompson Strains on Pre-harvest Sweet Basil (Ocimum basilicum) Leaves. Front. Microbiol. 12:740983. doi: 10.3389/fmicb.2021.740983
Received: 14 July 2021; Accepted: 12 November 2021;
Published: 07 December 2021.
Edited by:
Sara Hallin, Swedish University of Agricultural Sciences, SwedenReviewed by:
Sunil D. Saroj, Symbiosis International University, IndiaCopyright © 2021 Zwe, Ten, Pang, Wong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Li, ZGFuLmxpQG51cy5lZHUuc2c=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.