
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 September 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.739995
This article is part of the Research TopicBioactive Compounds with Potential Medicinal Properties Derived from Fungi: Recent and Future Developments in Microbial BiotechnologyView all 15 articles
 Alexander B. J. Grey1
Alexander B. J. Grey1 Melissa M. Cadelis1,2
Melissa M. Cadelis1,2 Yiwei Diao1
Yiwei Diao1 Duckchul Park3
Duckchul Park3 Thomas Lumley4
Thomas Lumley4 Bevan S. Weir3
Bevan S. Weir3 Brent R. Copp2†
Brent R. Copp2† Siouxsie Wiles1*†
Siouxsie Wiles1*†There is a real and urgent need for new antibiotics able to kill Mycobacteria, acid-fast bacilli capable of causing multiple deadly diseases. These include members of the Mycobacterium tuberculosis complex, which causes the lung disease tuberculosis (TB) as well as non-tuberculous Mycobacteria (NTM) a growing cause of lung, skin, soft tissue, and other infections. Here we describe a medium-throughput bioluminescence-based pipeline to screen fungi for activity against Mycobacteria using the NTM species Mycobacterium abscessus and Mycobacterium marinum. We used this pipeline to screen 36 diverse fungal isolates from the International Collection of Microorganisms from Plants (ICMP) grown on a wide variety of nutrient-rich and nutrient-poor media and discovered that almost all the tested isolates produced considerable anti-mycobacterial activity. Our data also provides strong statistical evidence for the impact of growth media on antibacterial activity. Chemical extraction and fractionation of a subset of the ICMP isolates revealed that much of the activity we observed may be due to the production of the known anti-mycobacterial compound linoleic acid. However, we have identified several ICMP isolates that retained their anti-mycobacterial activity in non-linoleic acid containing fractions. These include isolates of Lophodermium culmigenum, Pseudaegerita viridis, and Trametes coccinea, as well as an unknown species of Boeremia and an isolate of an unknown genus and species in the family Phanerochaetaceae. Investigations are ongoing to identify the sources of their anti-mycobacterial activity and to determine whether any may be due to the production of novel bioactive compounds.
There is a real and urgent need for new antibiotics able to kill Mycobacteria, acid-fast bacilli capable of causing multiple deadly diseases. Because of their slow growth and hydrophobic, lipid-rich outer membrane, treatment of mycobacterial infections can take months to years and require multiple antibiotics (Seaworth and Griffith, 2017; Pontali et al., 2019). The major mycobacterial human pathogens are members of the Mycobacterium tuberculosis complex, which causes the lung disease tuberculosis (TB) (Gagneux, 2018), described by the World Health Organization as a global epidemic. Also of concern are the non-tuberculous Mycobacteria (NTM), free living opportunistic pathogens that are ubiquitous in the environment and able to cause lung, skin, and soft tissue infections (Mirsaeidi et al., 2014; Gonzalez-Santiago and Drage, 2015; Koh and Schlossberg, 2017). Almost two hundred NTM species have been identified to date, which were recently found to divide into five clades based on phylogenetic characteristics (Gupta et al., 2018). Rates of NTM infections are increasing globally, including in hospital settings (Al-Mahruqi et al., 2009; Roux et al., 2009; Moore et al., 2010; Morimoto et al., 2014; Donohue and Wymer, 2016; Donohue, 2018; Ratnatunga et al., 2020). NTM are natural inhabitants of water and their inclusion in implanted devices such as catheters, prosthetics, and pacemakers, have resulted in cases of bacteremia and disseminated infection, while NTM outbreaks have been associated with invasive procedures such as cosmetic surgeries, intramuscular injections, and tattooing (Griffin et al., 2019; Jabbour et al., 2020). Recently, some cases of pulmonary infections with Mycobacterium chimaera were traced back to site of manufacture of heater-cooler units routinely used during open heart surgery (Williamson et al., 2017).
Aotearoa New Zealand is an archipelago which split from the Gondwanan supercontinent approximately 85 million years ago and has since gradually become more isolated from other land masses (Wallis and Trewick, 2009). This geographical separation has led to the evolution of iconic native flora, fauna, and fungi. The Crown Research Institute Manaaki Whenua is the custodian of the International Collection of Microorganisms from Plants (ICMP) (Johnston et al., 2017). The ICMP contains over 10,000 fungal cultures derived from plants and soil from Aotearoa New Zealand and the South Pacific. The collection has a great diversity of fungal species, host substrates, and collection localities, with the earliest cultures dating from the 1960s. While the collection contains some of the fungal genera traditionally used for antibiotic production it has not been rigorously tested for antimicrobial activity against mycobacterial species. In our view, this makes the ICMP an excellent and untapped resource for antibiotic discovery.
The search for new antibiotics with activity against Mycobacteria is complicated by their slow growth, with species like M. tuberculosis having a doubling time of approximately 24 h. Mycobacteria also tend to clump in liquid culture due to their hydrophobic cell envelope. These properties make the two most common methods of measuring antibacterial activity, the production of zones of inhibition when grown on agar, or degree of turbidity when grown in liquid culture, slow and unreliable. Tagging bacteria with the genes that encode for luciferase-based reporters allows light to be used as a rapid surrogate marker for bacterial viability (Andreu et al., 2012). We and others have shown that bioluminescence is an excellent non-destructive real-time reporter to assay for anti-mycobacterial activity in microtiter plate formats using a luminometer (Andreu et al., 2012; Dalton et al., 2016; Early et al., 2019; Chengalroyen et al., 2020; Jain et al., 2020) or in vivo using sensitive imaging equipment (Andreu et al., 2013).
Here we describe a medium-throughput bioluminescence-based pipeline to screen fungi for activity against Mycobacteria using the NTM species Mycobacterium abscessus and Mycobacterium marinum. Our results indicate that many of the ICMP fungal isolates are anti-mycobacterial and have identified isolates of Lophodermium culmigenum, Pseudaegerita viridis, and Trametes coccinea, as well as an unknown species of Boeremia and an isolate of an unknown genus and species in the family Phanerochaetaceae as suitable for further study.
In this study, we used M. abscessus BSG301 (Cadelis et al., 2021) and M. marinum BSG101 (Dalton et al., 2017) which are stable bioluminescent derivatives transformed with the integrating plasmid pMV306G13ABCDE (Andreu et al., 2010). We grew mycobacterial cultures with shaking (200 rpm) in Middlebrook 7H9 broth (Fort Richard, New Zealand) supplemented with 10% Middlebrook ADC enrichment media (Fort Richard, New Zealand), 0.4% glycerol (Sigma-Aldrich, New Zealand) and 0.05% tyloxapol (Sigma-Aldrich, New Zealand). We grew M. abscessus at 37°C and M. marinum at 28°C.
Fungal isolates (Table 1) were provided by Manaaki Whenua – Landcare Research, a New Zealand Crown Research Institute responsible for the curation of the International Collection of Microorganisms from Plants (ICMP). We stored fungal isolates individually in cryotubes at −80°C. We made freezer stocks by growing each fungus on 1.5% Potato Dextrose Agar (PDA) and excising small cubes of agar (5–6 mm in length) from the fungus’ growing edge. We placed these cubes within a cryovial containing 1 mL of 10% glycerol and rested them for 1 h after which we removed the remaining liquid glycerol and stored the tubes at -80°C.
We used a small portion of mycelium from growing fungi and extracted DNA using the REDExtract-N-AmpTM Plant PCR Kit (Sigma-Aldrich) according to the manufacturer’s protocol. We diluted DNA samples five-fold and amplified using the ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) primer set in a 10 μl reaction volume using the REDExtract-N-Amp Plant PCR Kit (Sigma-Aldrich) according to the manufacturer’s instructions. We used the following PCR conditions: initial denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s and extending at 72°C for 30 s. The final extension was performed at 72°C for 7 min. We checked the amplified DNA by gel electrophoresis before sequencing using an Applied BiosystemsTM 3500xL Genetic Analyzer using both ITS1F and ITS4 primers. We trimmed and combined the sequence data using Geneious (Geneious Biologics), removed any low-quality reads and used BLAST to check fungal identification. Optimized sequence data were aligned using MEGA7 (Kumar et al., 2016).
We grew fungal isolates on PDA (Fort Richard, New Zealand) prior to screening for antibacterial activity using a 24 well plate assay. Briefly, we added 0.5 mL aliquots of agar to triplicate wells of a black 24 well plate (4titude, Millennium Science, New Zealand) and allowed them to set. We obtained all media from Fort Richard (New Zealand). In addition to PDA, these comprised: Czapek Solution Agar (CSA), Czapek Yeast Extract Agar (CYA), Malt Extract Agar (MEA), Malt Yeast Extract Agar (MYA), Oatmeal Agar (OA), Rice Extract Agar (REA), and Tryptone Yeast Extract Agar (TYA). With the aid of a sterile scalpel blade, we sectioned fungal isolates grown on PDA into cubes ≤5 mm in diameter, and then transferred the cubes to the agar-filled wells of the 24-well plates ensuring that each cube was placed fungus-side down and touching the agar. We covered the inoculated 24-well screening plates, sealed them with parafilm, and incubated them at room temperature.
We monitored fungal growth visually at regular intervals and recorded the time taken for them to either cover the entire well or to stop visibly growing. At twice this time, we removed a 6 mm plug of agar from each well using a biopsy punch. To screen for antibacterial activity, we resuspended M. abscessus BSG301 and M. marinum BSG101 in 0.8% Middlebrook 7H9 broth (Fort Richard, New Zealand) supplemented with 10% Middlebrook ADC enrichment media (Fort Richard, New Zealand) to a final concentration of 107 colony forming units (CFU)/mL for M. abscessus and 108 CFU/mL for M. marinum. With the aid of a pipette, we pipetted 50 μL of the bacterial-agar mixture into the cylindrical holes left after removal of the fungal-agar plugs and allowed the mixture to set. We measured bacterial luminescence at regular intervals using a Victor X-3 luminescence plate reader (PerkinElmer) with an integration time of 1 s. Between measurements, plates were covered, placed in a plastic box lined with damp paper towels, and incubated static at 37°C for M. abscessus and 28°C for M. marinum. We performed these assays three times. We have published a more detailed description of our methods on the protocol repository website protocols.io (Wiles and Grey, 2021a,b).
We grew fungal cultures either in liquid media or on solid media at room temperature and then freeze-dried them. We extracted the dry cultures with MeOH (Sigma-Aldrich, New Zealand) for 4 h followed by CH2Cl2 (Sigma-Aldrich, New Zealand) overnight. We concentrated the combined organic extracts under reduced pressure and subjected the crude extracts to C8 reversed-phase column chromatography eluting with a gradient of H2O/MeOH (Sigma-Aldrich, New Zealand) to afford five fractions (F1–F5). Full details are provided in Supplementary Material.
We grew mycobacterial cultures until they reached stationary phase (approximately 3–5 days for M. abscessus BSG301 and 7–10 days for M. marinum BSG101) and then diluted these in Mueller Hinton broth II (MHB) (Fort Richard) supplemented with 10% Middlebrook ADC enrichment media and 0.05% tyloxapol to give an optical density at 600 nm (OD600) of 0.001 which is the equivalent of ∼106 bacteria per mL. We dissolved the fungal fractions in DMSO (Sigma-Aldrich, New Zealand) and added these in duplicate to the wells of a black 96-well plate (Nunc, Thermo Scientific) at doubling dilutions with a maximum concentration of 50 mg/mL. Then we added 50 μL of diluted bacterial culture to each well of the fraction containing plates giving final extract concentrations of 0–1000 μg/mL and a cell density of ∼5 × 105 CFU/mL.
We used the antibiotic rifampicin (Sigma-Aldrich, New Zealand) as a positive control at 1000 μg/mL for M. abscessus and 10 μg/mL for M. marinum. Between measurements, plates were covered, placed in a plastic box lined with damp paper towels, and incubated with shaking at 100 rpm at 37°C for M. abscessus and 28°C for M. marinum. We measured bacterial luminescence at regular intervals using a Victor X-3 luminescence plate reader (PerkinElmer) with an integration time of 1 s. We have defined the MIC as causing a 1 log reduction in light production, as previously described (Dalton et al., 2016, 2017). We have published a more detailed description of our methods on the protocol repository website protocols.io (Wiles and Grey, 2021c,d).
We recorded NMR spectra using a Bruker Avance DRX-400 spectrometer or an Avance III-HD 500 spectrometer operating at 400 MHz or 500 MHz for 1H nuclei and 100 MHz or 125 MHz for 13C nuclei utilizing standard pulse sequences at 298 K. We recorded high resolution mass spectra on a Bruker micrOTOF QII (Bruker Daltonics, Bremen, Germany). We carried out analytical thin layer chromatography (TLC) on 0.2 mm thick plates of DC-plastikfolien Kieselgel 60 F254 (Merck). We carried out reversed-phase column chromatography on C8 support with a pore size of 40–63 μm (Merck). We carried out gel filtration chromatography on Sephadex LH-20 (Pharmacia). We carried out flash chromatography on Diol-bonded silica with a pore size of 40–63 micron (Merck). We used solvents that were of analytical grade or better and/or purified according to standard procedures.
We fitted a logistic mixed model for activity with a random effect for biologic replicates. We tested the main effects and second-order interactions of the variables, using the lme4 and car packages in R (Bates et al., 2015; Fox and Weisberg, 2019; R Core Team, 2020).
The 36 ICMP fungal isolates used in this study were collected between 1961 and 2016 and from locations across Aotearoa New Zealand, including the North, South, and Chatham Islands (Figure 1). Of the 36 isolates, nine were not able to be identified as a known species and one is from both an unknown genus and species (Table 2). These isolates likely represent novel taxa endemic to Aotearoa New Zealand. As with the broader collection, they cover a range of isolation dates, the earliest being isolated in 1973 (Cylindrobasidium sp. ICMP 16397) and the most recent in 2014 (Mortierella sp. ICMP 20597). They also cover a broad range of isolation locations within New Zealand, from Kerikeri in the North Island (Agaricales sp. ICMP 17554) to Christchurch in the South Island (Xylariaceae sp. ICMP 16006).
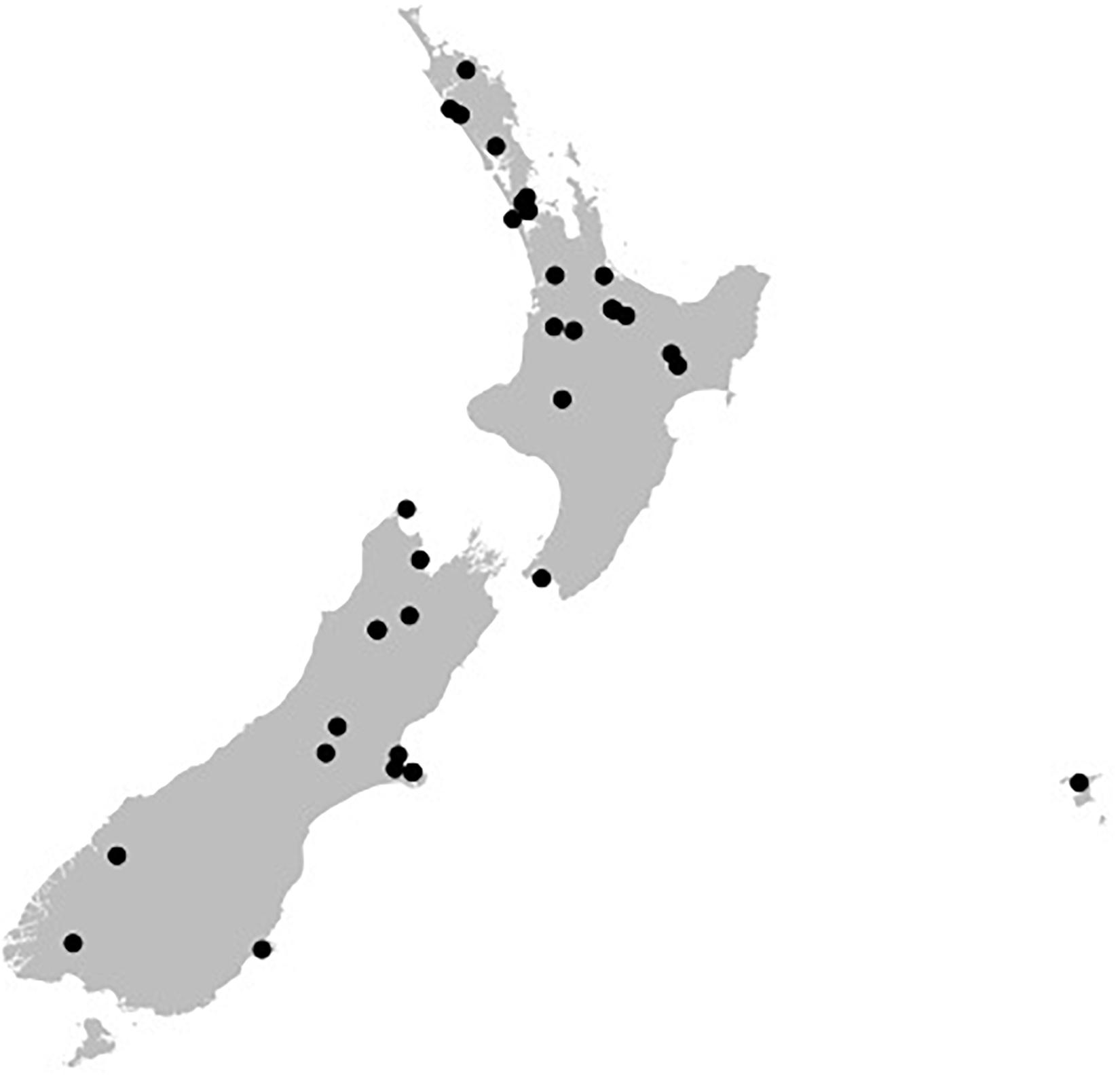
Figure 1. Geographical spread of the isolation locations for the fungal isolated used in this study within the archipelago of Aotearoa New Zealand. Black dots are individual ICMP isolates.
We screened 36 ICMP fungal isolates for antibacterial activity against M. abscessus BSG301 and M. marinum BSG101. The isolates belong to three different fungal Phyla, and we grew them on eight different media giving us a total of 288 fungi-media combinations tested for each bacterium (Figures 2–4).
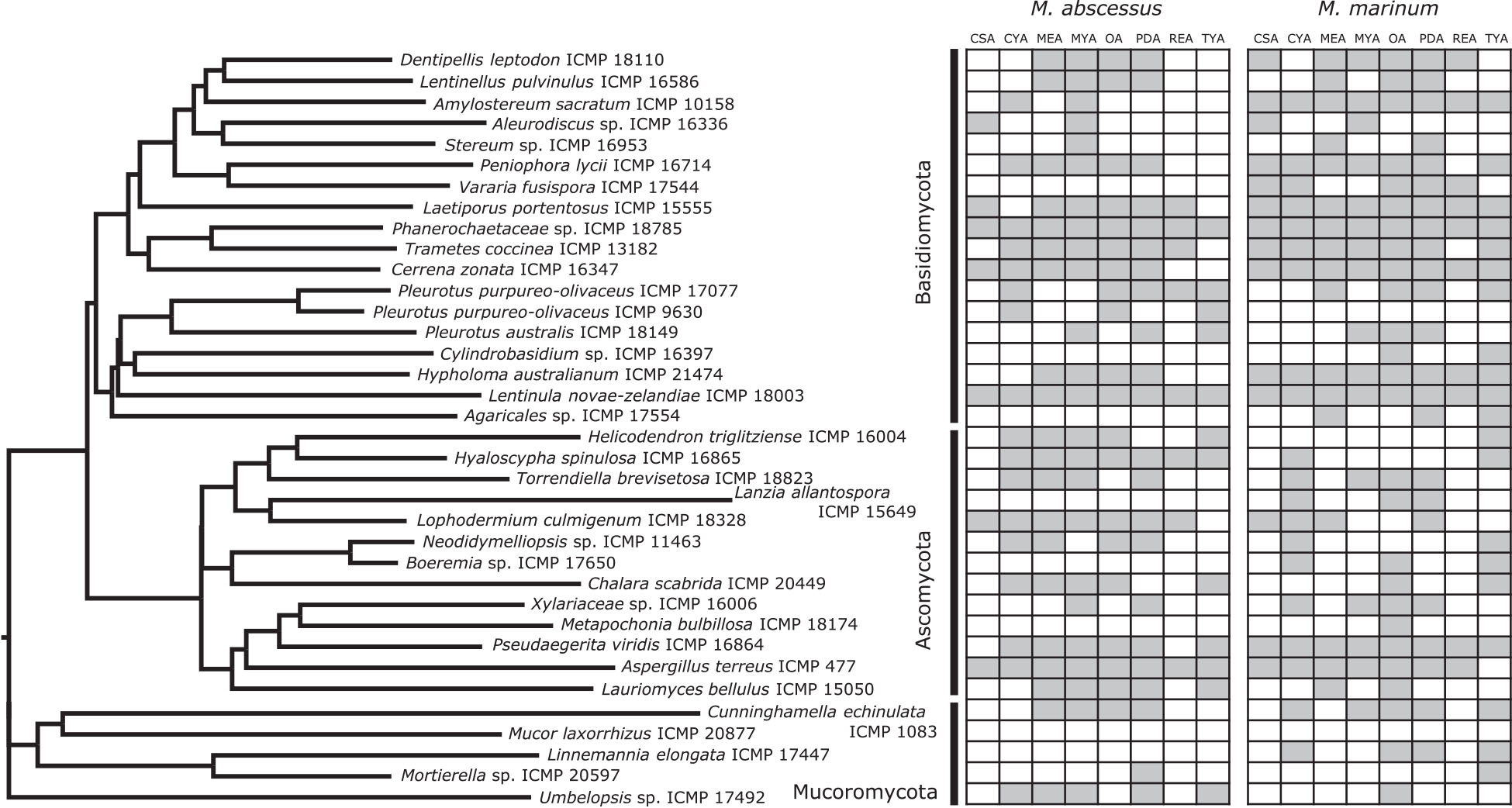
Figure 2. Phylogeny and activity of ICMP isolates grown in different media against M. abscessus BSG301 and M. marinum BSG101. Active media for each isolate are shown in gray. CSA, Czapek Solution Agar; CYA, Czapek Yeast extract Agar; MEA, Malt Extract Agar; MYA, Malt Yeast extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; REA, Rice Extract Agar; TYA, Tryptone Yeast extract Agar. ICMP isolates are described as active if they caused a minimum 1-log (90%) reduction in the bioluminescence of the mycobacterial strains. The phylogenetic tree was constructed by comparing ITS sequences to those in GenBank and the phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis version 7.0 software (MEGA7) using the neighbor-joining method and p-distance as the substitution model. Each phylogeny was tested using the bootstrap method with 500 replications.
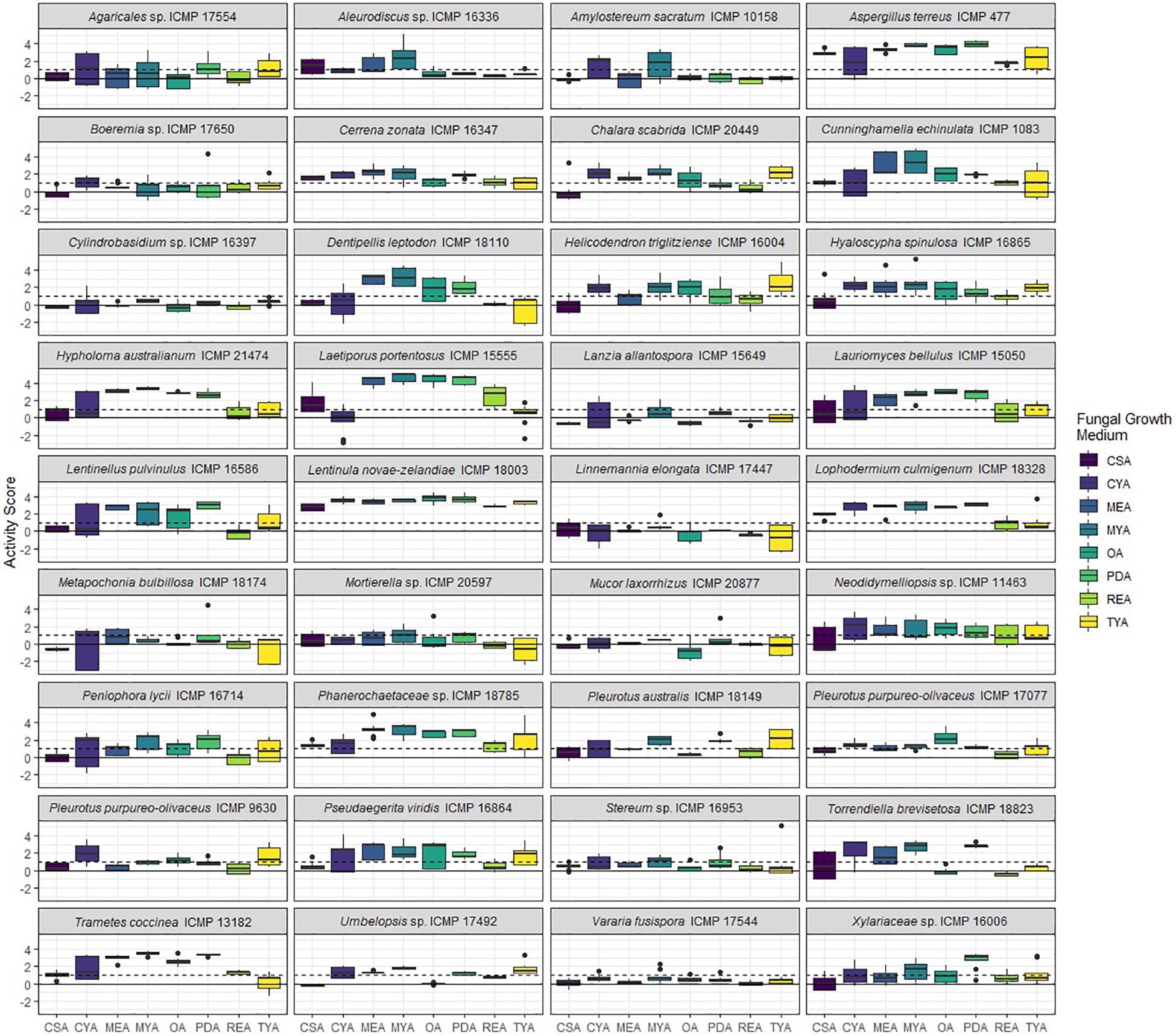
Figure 3. Antibacterial activity of ICMP fungal isolates against Mycobacterium abscessus BSG301 when grown on different media. Data is presented as box and whisker plots of activity scores. The solid line shown at 0 is the median control value while the dotted line at 1 is the activity threshold. Scores above 1 correspond to a >90% reduction in bacterial bioluminescence compared to the corresponding no-fungi control. Similarly, an activity score above 2 means corresponds to a >99% reduction. CSA, Czapek Solution Agar; CYA, Czapek Yeast Extract Agar; MEA, Malt Extract Agar; MYA, Malt Yeast Extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; REA, Rice Extract Agar; TYA, Tryptone Yeast Extract Agar. Boxes are upper and lower quartiles with median shown. The whiskers extend up to 1.5× the inter-quartile range and any dots beyond those bounds are outliers.
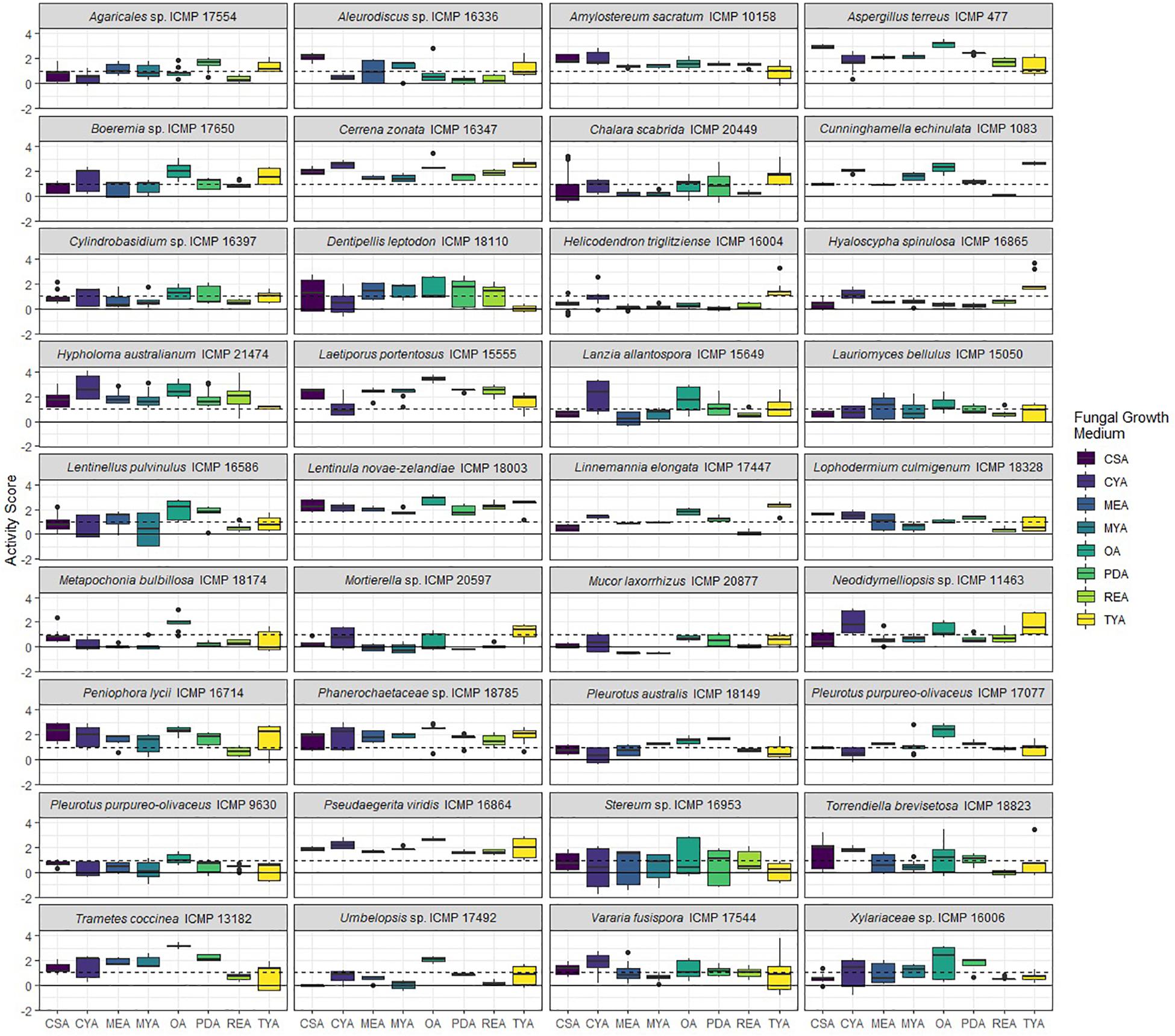
Figure 4. Antibacterial activity of ICMP fungal isolates against Mycobacterium marinum BSG101 when grown on different media. Data is presented as box and whisker plots of activity scores. The solid line shown at 0 is the median control value while the dotted line at 1 is the activity threshold. Scores above 1 correspond to a >90% reduction in bacterial bioluminescence compared to the corresponding no-fungi control. Similarly, an activity score above 2 means corresponds to a >99% reduction. CSA, Czapek Solution Agar; CYA, Czapek Yeast Extract Agar; MEA, Malt Extract Agar; MYA, Malt Yeast Extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; REA, Rice Extract Agar; TYA, Tryptone Yeast Extract Agar. Boxes are upper and lower quartiles with median shown. The whiskers extend up to 1.5× the inter-quartile range and any dots beyond those bounds are outliers.
We measured antibacterial activity as reductions in light output of our bioluminescent mycobacterial strains over a 72-h period. We calculated activity scores by first converting the luminescence measurement at each time-point into an area under the curve (AUC) value for each well. We then divided this number by the median AUC of a sterile control plate inoculated and incubated at the same time as the fungus-containing plates. The negative log of this value corresponds to the activity score. We define a fungus-media combination as active/antibacterial if the median activity score is above 1 which corresponds to a > 90% reduction in light compared to the control. Similarly, an activity score above 2 means corresponds to a > 99% reduction.
We observed no consistent difference in activity by mycobacterial strain, but there is strong statistical evidence of differences in activity between media and fungal Phyla, and that these vary by mycobacterial strain (Table 3).
We observed that 28/36 (77%) fungal isolates were active against M. abscessus when grown in at least one medium, with 130/288 (45%) fungi-medium combinations being anti-mycobacterial against this bacterium (Figures 2, 3). In contrast, 34/36 (94%) fungal isolates were active against M. marinum, with 146/288 (51%) fungi-medium combinations being anti-mycobacterial against this bacterium (Figures 2, 4). Of the two fungal isolates that were not active against M. marinum when grown in any of the media tested, only the Mucoromycota fungus Mucor laxorrhizus ICMP 20877 displayed no activity against M. abscessus. The second isolate, Pleurotus purpureo-olivaceus ICMP 9630 was active against M. abscessus when grown on CYA, MYA, and TYA. However, a second isolate of P. purpureo-olivaceus we tested, ICMP 17077, was active against both mycobacterial strains. We could discern no obvious pattern between fungal species or genus for those isolates that were only active against M. marinum, namely Agaricales sp. ICMP 17554, Boeremia sp. ICMP 17650, Cylindrobasidium sp. ICMP 16397, Lanzia allantospora ICMP 15649, Linnemannia elongata ICMP 17447, Metapochonia bulbillosa ICMP 18174, and Vararia fusispora ICMP 17544.
The fungal isolates we tested belong to three Phyla: the Basidiomycota (18 ICMP isolates), the Ascomycota (13 ICMP isolates), and the Mucoromycota (5 ICMP isolates) (Figure 2). We observed that the group with the most active fungi-medium combinations was the Basidiomycota (55%), followed by the Mucoromycota (52%), and then the Ascomycota (46%%) (Figure 2). More Basidiomycota-medium combinations were active against M. marinum [92/144 (64%)] (Figures 2, 4) than M. abscessus [67/144 (47%)] (Figures 2, 3). In contrast, more Ascomycota-medium combinations were active against M. abscessus [53/104 (51%)] (Figures 2, 3) than M. marinum [43/104 (41%)] (Figures 2, 4).
We observed that many of the ICMP fungi displayed differential activity depending on their growth medium with the majority being active on more than one medium. An isolate of the native New Zealand “shiitake” mushroom Lentinula novae-zelandiae (ICMP 18003) and an isolate of unknown genus and species in the family Phanerochaetaceae (ICMP 18785) were active against both M. abscessus and M. marinum when grown on an all 8 media (Figures 2–4). An isolate of Aspergillus terreus (ICMP 477) was also active against M. abscessus regardless of growth media (Figures 2, 4), while Amylostereum sacratum ICMP 10158, Cerrena zonata ICMP 16347, Hypholoma australianum ICMP 21474, Laetiporus portentosus ICMP 15555, and Pseudaegerita viridis ICMP 16864 were active against M. marinum when grown on all media (Figures 2, 4).
When assessing for activity against M. abscessus, 2/36 fungal isolates were only active when grown on one of the eight media, an unknown species of Mortierella (ICMP 20597) when grown on PDA and an unknown species of Stereum (ICMP 16953) on MYA (Figures 2, 3). ICMP 20597 was also only active against M. marinum when grown on one of the eight media, though in this case it was TYA (Figures 2, 4). Three other fungal isolates were only active against M. marinum when grown on one of the eight media, Helicodendron triglitziense ICMP 16004 on TYA, and Metapochonia bulbillosa ICMP 18174 and an unknown species of Umbelopsis (ICMP 17492) on OA (Figures 2, 4).
We observed that PDA was the most active culture medium with 29/36 (80%) fungal isolates active against either bacterium (Figure 2). For M. marinum, the most active culture medium was OA, followed by PDA (24 and 23 active fungi, respectively) while for M. abscessus it was MYA followed by PDA (24 and 22 active fungi, respectively). We observed that isolates grown on REA and CSA were the least active, with only 14/36 isolates (39%) being active against either mycobacterium species when cultured on these media (Figure 2). MYA and MEA were the two media that favored M. abscessus activity, with 24 fungal isolates active when grown on MYA and 20 isolates active when grown on MEA, compared to 17 and 16 being active against M. marinum, respectively (Figure 2).
We prepared extracts from 41 fungus-medium combinations which were further separated into 5 fractions, designated F1–F5. Fraction F1 (100% water) is generally comprised of sugars while fraction F5 (100% methanol) contains predominantly fatty acids and sterols. Fractions F2, F3, and F4 typically contain the chemical compounds we are most interested in pursuing, with the potential to be bioactive.
We tested, at a single concentration of 1000 μg/mL, the crude extracts and fractions F1–F5 from all 41 fungus-medium combinations for activity against M. marinum BSG101 (Figures 5, 6) and for 38 of the combinations for activity against M. abscessus BSG301 (Figures 5, 7). As described previously, we measured antibacterial activity as reductions in light output of our bioluminescent mycobacterial strains over a 72-h period and calculated activity scores as the negative log of the ratio of the AUC values of the fungus-containing measurements and the control measurements. We define an extract/fraction as active/antibacterial if the median activity score is above 1, which corresponds to a >90% reduction in light compared to the control, as previously described. Similarly, an activity score above 2 means corresponds to a > 99% reduction.
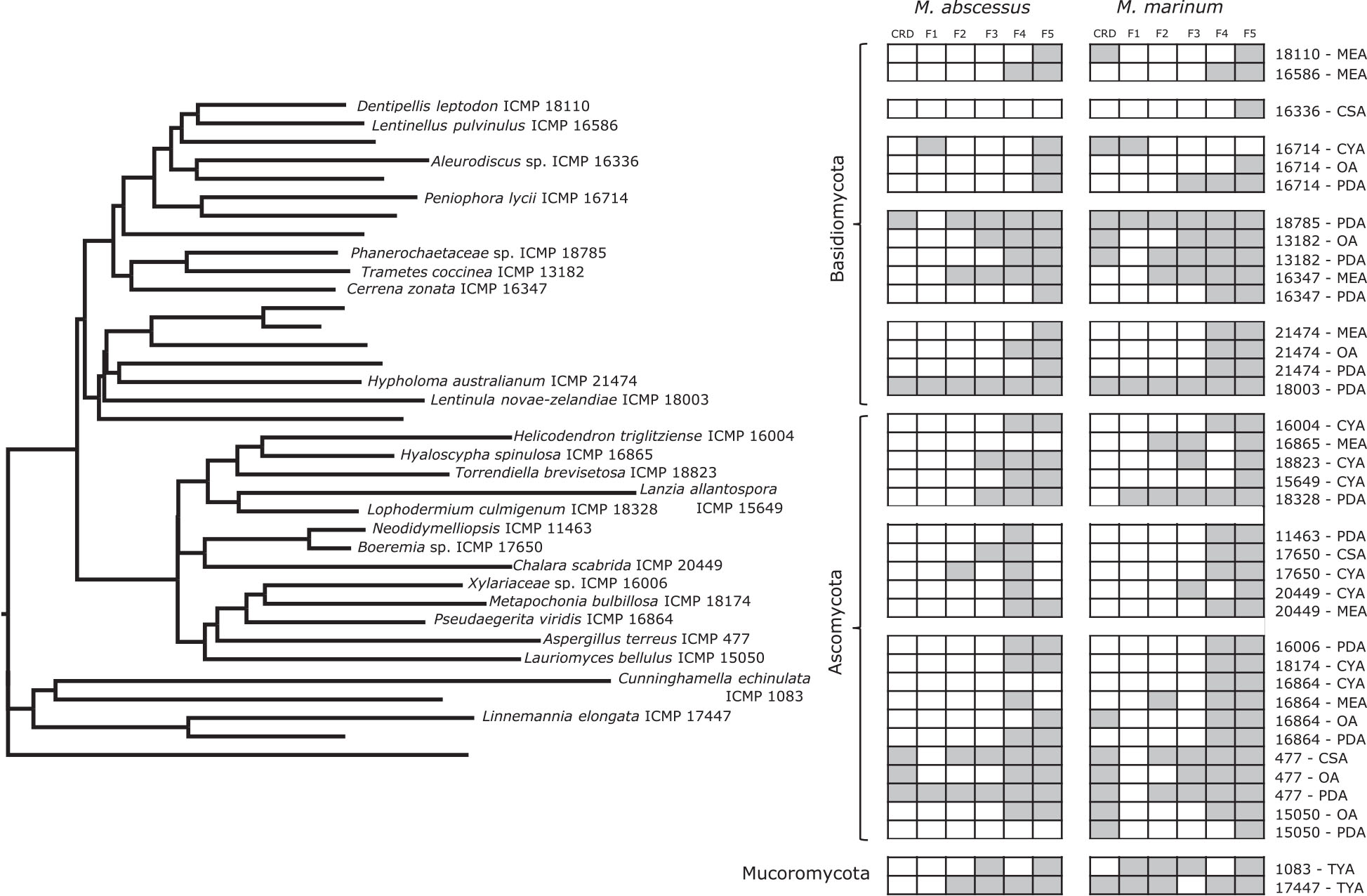
Figure 5. Phylogeny and activity of crude extracts and fractions from ICMP isolates grown in different media against M. abscessus BSG301 and M. marinum BSG101. Active extracts and fractions for each isolate are shown in gray. CSA, Czapek Solution Agar; CYA, Czapek Yeast extract Agar; MEA, Malt Extract Agar; MYA, Malt Yeast extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; REA, Rice Extract Agar; TYA, Tryptone Yeast extract Agar. ICMP isolates are described as active if they caused a minimum 1-log (90%) reduction in the bioluminescence of the mycobacterial strains. The phylogenetic tree was constructed by comparing ITS sequences to those in GenBank and the phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis version 7.0 software (MEGA7) using the neighbor-joining method and p-distance as the substitution model. Each phylogeny was tested using the bootstrap method with 500 replications.

Figure 6. Antibacterial activity of crude extracts and fractions 1–5 from ICMP fungal isolates against Mycobacterium marinum BSG101. Data is presented as box and whisker plots of activity scores. The solid line shown at 0 is the median control value while the dotted line at 1 is the activity threshold. Scores above 1 correspond to a >90% reduction in bacterial bioluminescence compared to the corresponding no-fungi control. Similarly, an activity score above 2 means corresponds to a >99% reduction. CSA, Czapek Solution Agar; CYA, Czapek Yeast Extract Agar; MEA, Malt Extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; TYA, Tryptone Yeast Extract Agar. Boxes are upper and lower quartiles with median shown. The whiskers extend up to 1.5× the inter-quartile range and any dots beyond those bounds are outliers.

Figure 7. Antibacterial activity of crude extracts and fractions 1–5 from ICMP fungal isolates against Mycobacterium abscessus BSG301. Data is presented as box and whisker plots of activity scores. The solid line shown at 0 is the median control value while the dotted line at 1 is the activity threshold. Scores above 1 correspond to a >90% reduction in bacterial bioluminescence compared to the corresponding no-fungi control. Similarly, an activity score above 2 means corresponds to a >99% reduction. CSA, Czapek Solution Agar; CYA, Czapek Yeast Extract Agar; MEA, Malt Extract Agar; OA, Oatmeal Agar; PDA, Potato Dextrose Agar; TYA, Tryptone Yeast Extract Agar. Boxes are upper and lower quartiles with median shown. The whiskers extend up to 1.5× the inter-quartile range and any dots beyond those bounds are outliers.
We observed that only 3/38 of the fungus-medium combinations we tested for activity against M. abscessus BSG301 did not retain any activity in either the crude extract or any of the 5 fractions. These belonged to Aleurodiscus sp. ICMP 16336 grown on CSA, Pseudaegerita viridis ICMP 16864 grown on CYA, and Lauriomyces bellulus ICMP 15050 grown on PDA (Figures 5, 7). Of the remaining 14 Basidiomycota-medium combinations tested, the most active fraction was F5 [14/14 (100%)], followed by F4 [7/14 (50%)]. Four Basidiomycota-medium combinations also displayed some activity from fractions F2 and/or F3. Of the 17 Ascomycota-medium combinations we observed to be active, the most active fraction was F4 [17/17 (100%)], followed by F5 [13/17 (76%)]. Six Ascomycota-medium combinations also displayed some activity from fractions F2 and/or F3. Both Mucoromycota-medium combinations tested had multiple active fractions.
We observed that all 41 of the fungus-medium combinations we tested for activity against M. marinum BSG101 retained some activity in either the crude extract or at least one of the 5 fractions (Figures 5, 6). Of the 18 Basidiomycota-medium combinations tested, the most active fraction was F5 [17/18 (94%)], followed by F4 [11/17 (65%)]. Seven Basidiomycota-medium combinations also displayed some activity from fractions F2 and/or F3. Like the Basidiomycota-medium combinations, of the 21 Ascomycota-medium combinations we observed to be active, the most active fraction was F5 [21/21 (100%)], followed by F4 [16/21 (76%)]. Eight Ascomycota-medium combinations also displayed some activity from fractions F2 and/or F3. Both Mucoromycota-medium combinations tested had multiple active fractions.
Given the anti-mycobacterial activity we observed from the F5 fractions of so many of the ICMP isolates, we analyzed this fraction in more detail from four phylogenetically diverse ICMP isolates: the Basidiomycota Aleurodiscus sp. ICMP 16336, the Ascomycota Hyaloscypha spinulosa ICMP 16865 and Lanzia allantospora ICMP 15649, and the Mucoromycota Cunninghamella echinulata ICMP 1083. NMR spectroscopic and Mass spectrometric analysis confirmed the presence of linoleic acid in fraction F5 of these fungi.
To prioritize the most active anti-mycobacterial fungus-medium combinations for further NMR spectroscopic and Mass spectrometric analysis, we tested those fractions F2, F3, and F4 that were active at 1000 mg/mL to obtain their minimum inhibitory concentration (MIC) (Table 4).
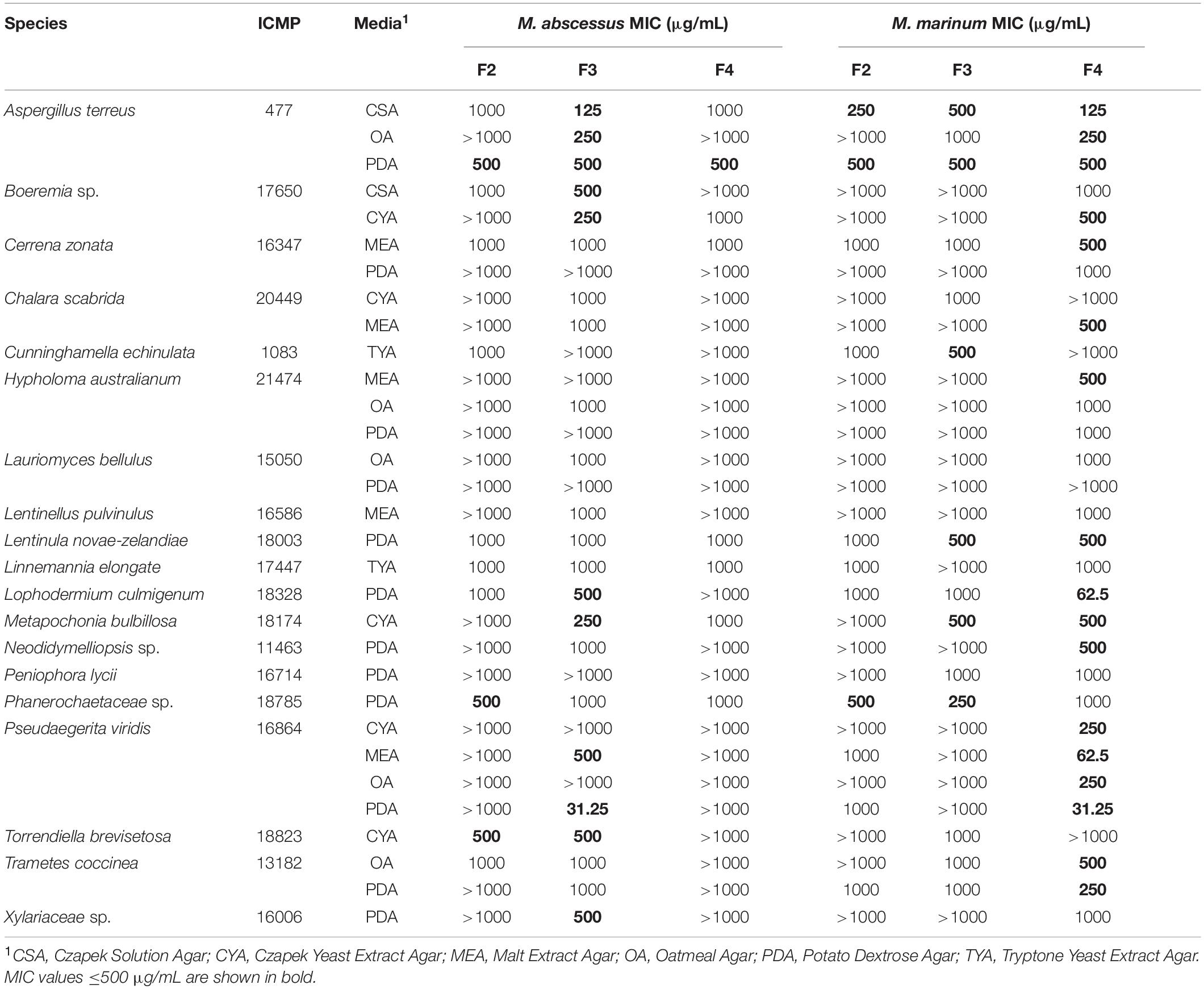
Table 4. Minimum inhibitory concentrations (MIC) of ICMP fungal fractions (F2, F3, and F4) against M. abscessus and M. marinum.
Of the 31 fungus-medium combinations tested against M. abscessus BSG301, 11/31 (35%) F3 fractions had an MIC ≤ 500 μg/mL. Of these, the most active fungus-medium combinations were Pseudaegerita viridis ICMP 16864 grown on PDA with an MIC of 31.25 μg/mL, and Aspergillus terreus ICMP 477 grown on CSA, with an MIC of 125 μg/mL (Table 4). In contrast, only three F4 fractions and one F2 fraction had an MIC ≤ 500 μg/mL.
In contrast to M. abscessus, of the 31 fungus-medium combinations tested against M. marinum BSG101, the most active fractions were F4 rather than F3, with 17/31 (55%) F4 fractions having an MIC ≤ 500 μg/mL. Of these, the most active fungus-medium combinations were Pseudaegerita viridis ICMP 16864 grown on PDA and MEA, with an MIC of 31.25 and 62.5 μg/mL, respectively, and Lophodermium culmigenum ICMP 18328, with an MIC of 62.5 μg/mL (Table 4). Three F2 fractions and six F3 fractions had an MIC ≤ 500 μg/mL.
In this study, we describe a medium-throughput bioluminescence-based pipeline to screen fungi for activity against Mycobacteria using bioluminescent derivates of M. abscessus and M. marinum as the testing strains. We included M. abscessus as it is a relatively fast-growing non-tuberculous mycobacterial species and is a cause of opportunistic infections in patients with cystic fibrosis or chronic pulmonary disease, and of skin and soft tissue infections, for which treatment options are limited (To et al., 2020; Victoria et al., 2021). We also included M. marinum as, despite being a pathogen of fish, amphibians, and reptiles, it shares conserved virulence determinants with M. tuberculosis (Bouz and Al Hasawi, 2018; Ramakrishnan, 2020) and is a Biosafety Level (BSL) 2 rather than an airborne BSL 3 organism.
We screened 36 ICMP fungal isolates using our assay and discovered that almost all produced considerable anti-mycobacterial activity. This is in contrast with our experience screening ICMP isolates for activity against other human pathogens such as Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, where we find just 5–20% of fungal isolates have some antibacterial activity (unpublished data). We also observed some differences between the two mycobacterial species, with more ICMP isolates being active against M. marinum than M. abscessus. This is not unsurprising given their different ecological niches and divergent genomes (Malhotra et al., 2017). Identification of the chemical compounds responsible may shed some further light on these differences.
Our screening pipeline involves growing fungi in 24 well plates on multiple growth media. It has previously been shown that different culture conditions can alter the expression of biosynthetic gene clusters and therefore the structural diversity and quantity of secondary metabolites produced by microorganisms, including fungi (Bills et al., 2008). For example, growing Fusarium tricinctum on Rice medium supplemented with fruit and vegetable juices led to the discovery of Fusarielin J (Hemphill et al., 2017) while growing Asteromyces cruciatus on Czapek-Dox medium with an altered nitrogen source led to the discovery of Lajollamide A (Gulder et al., 2012). We selected media that cover a broad range of carbon and nitrogen sources, as well as different pH and metal ions. We have also included media with less chemically defined elements, including from potatoes, rice, and oatmeal. Our data provides strong statistical evidence for the impact of growth media on antibacterial activity of the ICMP fungi we tested, with some fungal isolates only active when grown on one medium and others active when grown on several, or even all. The highest proportion of ICMP isolates were active when grown on the nutritionally rich Oatmeal Agar (OA), Potato Dextrose Agar (PDA), and (MYA) while the lowest proportion were active when grown on the more nutritionally poor Rice Extract Agar (REA) and Czapek Solution Agar (CSA).
The ICMP fungi we screened in this project included isolates of several species well known to produce antimicrobial compounds. For example, Aspergillus terreus produces terrein (Goutam et al., 2017), maunakeanolic acid A and B (Zaman et al., 2020), and helvolic acid (Zaman et al., 2020), amongst other compounds. Chemical extraction and fractionation of a subset of the ICMP isolates revealed that much of the activity we observed may be due to the production of the known anti-mycobacterial compound linoleic acid (Kanetsuna, 1985; Choi, 2016). However, we have identified several ICMP isolates that retained their anti-mycobacterial activity in non-linoleic acid containing fractions. These include isolates of Lophodermium culmigenum, Pseudaegerita viridis, and Trametes coccinea, as well as an unknown species of Boeremia and an isolate of an unknown genus and species in the family Phanerochaetaceae. Investigations are ongoing to identify the sources of their anti-mycobacterial activity and to determine whether any may be due to the production of novel bioactive compounds. Species of Lophodermium and Pseudaegerita have previously been found to produce several antifungal compounds (Hosoya et al., 2007; Sumarah et al., 2011; McMullin et al., 2015), while T. coccinea is predicted to have secondary metabolite pathways though genomic analysis (Zhang et al., 2020).
An interesting observation we have made, is of the abundant anti-mycobacterial activity of fungi we tested in the order Polyporales. The isolates Cerrena zonata ICMP 16347, Laetiporus portentosus ICMP 15555, Phanerochaetaceae sp. ICMP 18785, and T. coccinea ICMP 13182, were active against both mycobacterial species when grown in almost all media. These fungi are bracket-like fungi with pores on the under surface. To fulfill their ecological niche of digesting moist wood, these fungi first need to colonize the wood. To do this they need to compete with other microorganisms, including bacteria, and producing antimicrobial compounds would be beneficial on this process. An alternative hypothesis could be that their antibacterial activity is a by-product of these fungi producing the peroxidases and oxidases they need to digest wood (Sulej et al., 2019). Should the activity prove not to be the result of peroxidase/oxidase production, this would suggest that focusing future screening efforts on Polyporales fungi could prove fruitful for the discovery of new antibacterial compounds.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT107903, MZ325955, MZ325952, MW862777, MW862790, MW862786, MK432752, MZ325951, MZ325956, MZ325966, MK432688, MK432695, MZ325972, MZ325953, AY755334, EF029218, MW862787, MZ325965, MZ325962, MZ325968, MZ325967, MZ325970, MZ325971, MW862783, MZ325959, MZ325969, MH395972, MH395959, GQ411512, MZ325960, MZ325961, JN225946, MW862784, EU770239, MZ325963, and MZ325954 and https://figshare.com/, https://auckland.figshare.com/articles/dataset/Antibacterial_activity_of_fungal_isolates_from_the_International_Collection_of_Microorganisms_from_Plants_ICMP_against_Mycobacterium_abscessus_and_Mycobacterium_marinum_/14937894.
BW, BC, and SW contributed to conception and design of the study. AG, MC, YD, and DP performed the experiments. AG, MC, TL, BW, and SW were involved in data analysis. AG and SW wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by funds from Cure Kids (9102 3715810), NZ Carbon Farming (9102 3718092), Maurice Wilkins Centre for Molecular Biodiscovery (9159 3715235), and donations from the New Zealand public. BW, DP, and the ICMP culture collection were funded by the SSIF infrastructure investment fund of the New Zealand Ministry of Business, Innovation and Employment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.739995/full#supplementary-material
Al-Mahruqi, S. H., van Ingen, J., Al-Busaidy, S., Boeree, M. J., Al-Zadjali, S., Patel, A., et al. (2009). Clinical relevance of nontuberculous mycobacteria, Oman. Emerg. Infect. Dis. 15, 292–294. doi: 10.3201/eid1502.080977
Andreu, N., Fletcher, T., Krishnan, N., Wiles, S., and Robertson, B. D. (2012). Rapid measurement of antituberculosis drug activity in vitro and in macrophages using bioluminescence. J. Antimicrob. Chemother. 67, 404–414. doi: 10.1093/jac/dkr472
Andreu, N., Zelmer, A., Fletcher, T., Elkington, P. T., Ward, T. H., Ripoll, J., et al. (2010). Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777. doi: 10.1371/journal.pone.0010777
Andreu, N., Zelmer, A., Sampson, S. L., Ikeh, M., Bancroft, G. J., Schaible, U. E., et al. (2013). Rapid in vivo assessment of drug efficacy against Mycobacterium tuberculosis using an improved firefly luciferase. J. Antimicrob. Chemother. 68, 2118–2127. doi: 10.1093/jac/dkt155
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting Linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48.
Bills, G. F., Platas, G., Fillola, A., Jiménez, M. R., Collado, J., Vicente, F., et al. (2008). Enhancement of antibiotic and secondary metabolite detection from filamentous fungi by growth on nutritional arrays. J. Appl. Microbiol. 104, 1644–1658. doi: 10.1111/j.1365-2672.2008.03735.x
Bouz, G., and Al Hasawi, N. (2018). The zebrafish model of tuberculosis – no lungs needed. Crit. Rev. Microbiol. 44, 779–792. doi: 10.1080/1040841X.2018.1523132
Cadelis, M. M., Gordon, H., Grey, A., Geese, S., Mulholland, D. R., Weir, B. S., et al. (2021). Isolation of a novel polyketide from Neodidymelliopsis sp. Molecules 26:3235. doi: 10.3390/molecules26113235
Chengalroyen, M. D., Jordaan, A., Seldon, R., Ioerger, T., Franzblau, S. G., Nasr, M., et al. (2020). Biological profiling enables rapid mechanistic classification of phenotypic screening hits and identification of KatG activation-dependent pyridine carboxamide prodrugs with activity against Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 10:699. doi: 10.3389/fcimb.2020.582416
Choi, W. H. (2016). Evaluation of anti-tubercular activity of linolenic acid and conjugated-linoleic acid as effective inhibitors against Mycobacterium tuberculosis. Asian Pac. J. Trop. Med. 9, 125–129. doi: 10.1016/j.apjtm.2016.01.021
Dalton, J. P., Uy, B., Okuda, K. S., Hall, C. J., Denny, W. A., Crosier, P. S., et al. (2017). Screening of anti-mycobacterial compounds in a naturally infected zebrafish larvae model. J. Antimicrob. Chemother. 72, 421–427. doi: 10.1093/jac/dkw421
Dalton, J. P., Uy, B., Phummarin, N., Copp, B. R., Denny, W. A., Swift, S., et al. (2016). Effect of common and experimental anti-tuberculosis treatments on Mycobacterium tuberculosis growing as biofilms. PeerJ 4:e2717. doi: 10.7717/peerj.2717
Donohue, M. J. (2018). Increasing nontuberculous mycobacteria reporting rates and species diversity identified in clinical laboratory reports. BMC Infect. Dis. 18:163. doi: 10.1186/s12879-018-3043-7
Donohue, M. J., and Wymer, L. (2016). Increasing prevalence rate of nontuberculous mycobacteria infections in five states, 2008–2013. Ann. Am. Thorac. Soc. 13, 2143–2150. doi: 10.1513/AnnalsATS.201605-353OC
Early, J. V., Mullen, S., and Parish, T. (2019). A rapid, low pH, nutrient stress, assay to determine the bactericidal activity of compounds against non-replicating Mycobacterium tuberculosis. PLoS One 14:e0222970. doi: 10.1371/journal.pone.0222970
Fox, J., and Weisberg, S. (2019). An R Companion to Applied Regression, 3rd Edn. Thousand Oaks, CA: Sage.
Gagneux, S. (2018). Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213. doi: 10.1038/nrmicro.2018.8
Gonzalez-Santiago, T. M., and Drage, L. A. (2015). Nontuberculous mycobacteria: skin and soft tissue infections. Granulomat. Disord. Adult Skin 33, 563–577. doi: 10.1016/j.det.2015.03.017
Goutam, J., Sharma, G., Tiwari, V. K., Mishra, A., Kharwar, R. N., Ramaraj, V., et al. (2017). Isolation and characterization of “Terrein” an antimicrobial and antitumor compound from endophytic fungus Aspergillus terreus (JAS-2) associated from Achyranthus aspera Varanasi, India. Front. Microbiol. 8:1334. doi: 10.3389/fmicb.2017.01334
Griffin, I., Schmitz, A., Oliver, C., Pritchard, S., Zhang, G., Rico, E., et al. (2019). Outbreak of tattoo-associated nontuberculous mycobacterial skin infections. Clin. Infect. Dis. 69, 949–955. doi: 10.1093/cid/ciy979
Gulder, T. A. M., Hong, H., Correa, J., Egereva, E., Wiese, J., Imhoff, J. F., et al. (2012). Isolation, structure elucidation and total synthesis of lajollamide A from the marine fungus Asteromyces cruciatus. Mar. Drugs 10, 2912–2935. doi: 10.3390/md10122912
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the Genus Mycobacterium into an emended Genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Hemphill, C. F. P., Sureechatchaiyan, P., Kassack, M. U., Orfali, R. S., Lin, W., Daletos, G., et al. (2017). OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J. Antibiot. (Tokyo) 70, 726–732. doi: 10.1038/ja.2017.21
Hosoya, T., Ohsumi, J., Hamano, K., Ono, Y., and Miura, M. (2007). Method for Producing Cercosporamide. Worldwide Patent No WO2007018194A1.
Jabbour, S. F., Malek, A. E., Kechichian, E. G., Tomb, R. R., and Nasr, M. W. (2020). Nontuberculous mycobacterial infections after cosmetic procedures: a systematic review and management algorithm. Dermatol. Surg. 46, 116–121.
Jain, P., Garing, S., Verma, D., Saranathan, R., Clute-Reinig, N., Gadwa, J., et al. (2020). Nanoluciferase reporter mycobacteriophage for sensitive and rapid detection of Mycobacterium tuberculosis drug susceptibility. J. Bacteriol. 202:e00411-20. doi: 10.1128/JB.00411-20
Johnston, P. R., Weir, B. S., and Cooper, J. A. (2017). Open data on fungi and bacterial plant pathogens in New Zealand. Mycology 8, 59–66. doi: 10.1080/21501203.2016.1278409
Kanetsuna, F. (1985). Bactericidal effect of fatty acids on mycobacteria, with particular reference to the suggested mechanism of intracellular killing. Microbiol. Immunol. 29, 127–141. doi: 10.1111/j.1348-0421.1985.tb00811.x
Koh, W.-J., and Schlossberg, D. (2017). Nontuberculous mycobacteria-overview. Microbiol. Spectr. 5:5.1.11. doi: 10.1128/microbiolspec.TNMI7-0024-2016
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Malhotra, S., Vedithi, S. C., and Blundell, T. L. (2017). Decoding the similarities and differences among mycobacterial species. PLoS Negl. Trop. Dis. 11:e0005883. doi: 10.1371/journal.pntd.0005883
McMullin, D. R., Green, B. D., and Miller, J. D. (2015). Antifungal sesquiterpenoids and macrolides from an endophytic Lophodermium species of Pinus strobus. Phytochem. Lett. 14, 148–152. doi: 10.1016/j.phytol.2015.10.006
Mirsaeidi, M., Farshidpour, M., Allen, M. B., Ebrahimi, G., and Falkinham, J. O. (2014). Highlight on advances in nontuberculous mycobacterial disease in North America. BioMed Res. Int. 2014:919474. doi: 10.1155/2014/919474
Moore, J. E., Kruijshaar, M. E., Ormerod, L. P., Drobniewski, F., and Abubakar, I. (2010). Increasing reports of non-tuberculous mycobacteria in England, Wales and Northern Ireland, 1995-2006. BMC Public Health 10:612. doi: 10.1186/1471-2458-10-612
Morimoto, K., Iwai, K., Uchimura, K., Okumura, M., Yoshiyama, T., Yoshimori, K., et al. (2014). A steady increase in nontuberculous mycobacteriosis mortality and estimated prevalence in Japan. Ann. Am. Thorac. Soc. 11, 1–8. doi: 10.1513/AnnalsATS.201303-067OC
Pontali, E., Raviglione, M. C., and Migliori, G. B. (2019). Regimens to treat multidrug-resistant tuberculosis: past, present and future perspectives. Eur. Respir. Rev. 28:190035. doi: 10.1183/16000617.0035-2019
R Core Team. (2020). R: A Language And Environment for Statistical Computing. Available online at: https://www.R-project.org/ (accessed July 12, 2021)
Ramakrishnan, L. (2020). Mycobacterium tuberculosis pathogenicity viewed through the lens of molecular Koch’s postulates. Curr. Opin. Microbiol. 54, 103–110. doi: 10.1016/j.mib.2020.01.011
Ratnatunga, C. N., Lutzky, V. P., Kupz, A., Doolan, D. L., Reid, D. W., Field, M., et al. (2020). The rise of non-tuberculosis mycobacterial lung disease. Front. Immunol. 11:303. doi: 10.3389/fimmu.2020.00303
Roux, A.-L., Catherinot, E., Ripoll, F., Soismier, N., Macheras, E., Ravilly, S., et al. (2009). Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J. Clin. Microbiol. 47, 4124–4128. doi: 10.1128/JCM.01257-09
Seaworth, B. J., and Griffith, D. E. (2017). Therapy of multidrug-resistant and extensively drug-resistant Tuberculosis. Microbiol. Spectr. 5, doi: 10.1128/microbiolspec.TNMI7-0042-2017
Sulej, J., Osińska-Jaroszuk, M., Jaszek, M., Gra̧z, M., Kutkowska, J., Pawlik, A., et al. (2019). Antimicrobial and antioxidative potential of free and immobilised cellobiose dehydrogenase isolated from wood degrading fungi. Fungal Biol. 123, 875–886. doi: 10.1016/j.funbio.2019.09.007
Sumarah, M. W., Kesting, J. R., Sørensen, D., and Miller, J. D. (2011). Antifungal metabolites from fungal endophytes of Pinus strobus. Phytochemistry 72, 1833–1837. doi: 10.1016/j.phytochem.2011.05.003
To, K., Cao, R., Yegiazaryan, A., Owens, J., and Venketaraman, V. (2020). General overview of nontuberculous mycobacteria opportunistic pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 9:2541. doi: 10.3390/jcm9082541
Victoria, L., Gupta, A., Gómez, J. L., and Robledo, J. (2021). Mycobacterium abscessus complex: a review of recent developments in an emerging pathogen. Front. Cell. Infect. Microbiol. 11:338. doi: 10.3389/fcimb.2021.659997
Wallis, G. P., and Trewick, S. A. (2009). New Zealand phylogeography: evolution on a small continent. Mol. Ecol. 18, 3548–3580. doi: 10.1111/j.1365-294X.2009.04294.x
Wiles, S., and Grey, A. (2021a). Bioluminescence-Based 24 Well Plate Assay for Screening Fungi for Activity Against Mycobacterium abscessus. Protocols.io. Available online at: https://dx.doi.org/10.17504/protocols.io.bvqtn5wn (accessed July 12, 2021).
Wiles, S., and Grey, A. (2021b). Bioluminescence-Based 24 Well Plate Assay For Screening Fungi for Activity Against Mycobacterium marinum. Protocols.io. Available online at: https://dx.doi.org/10.17504/protocols.io.bvnbn5an (accessed July 12, 2021).
Wiles, S., and Grey, A. (2021c). Bioluminescence-Based Minimum Inhibitory Concentration (MIC) Testing of Fungal Extracts Against Mycobacterium abscessus. Protocols.io. Available online at: https://dx.doi.org/10.17504/protocols.io.bvm9n496 (accessed July 12, 2021).
Wiles, S., and Grey, A. (2021d). Bioluminescence-based Minimum Inhibitory Concentration (MIC) Testing of Fungal Extracts Against Mycobacterium marinum. Protocols.io. Available online at: https://dx.doi.org/10.17504/protocols.io.bvnan5ae (accessed July 12, 2021).
Williamson, D., Howden, B., and Stinear, T. (2017). Mycobacterium chimaera spread from heating and cooling units in heart surgery. N. Engl. J. Med. 376, 600–602. doi: 10.1056/NEJMc1612023
Zaman, K. A. U., Hu, Z., Wu, X., Hou, S., Saito, J., Kondratyuk, T. P., et al. (2020). NF-κB inhibitory and antibacterial helvolic and fumagillin derivatives from Aspergillus terreus. J. Nat. Prod. 83, 730–737. doi: 10.1021/acs.jnatprod.9b01190
Keywords: mycobacteria, Mycobacterium marinum, Mycobacterium abscessus, bioluminescence, luciferase, minimum inhibitory concentration, screening, antibacterial
Citation: Grey ABJ, Cadelis MM, Diao Y, Park D, Lumley T, Weir BS, Copp BR and Wiles S (2021) Screening of Fungi for Antimycobacterial Activity Using a Medium-Throughput Bioluminescence-Based Assay. Front. Microbiol. 12:739995. doi: 10.3389/fmicb.2021.739995
Received: 12 July 2021; Accepted: 10 August 2021;
Published: 06 September 2021.
Edited by:
Paola Angelini, University of Perugia, ItalyReviewed by:
Joseph Oliver Falkinham, Virginia Tech, United StatesCopyright © 2021 Grey, Cadelis, Diao, Park, Lumley, Weir, Copp and Wiles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siouxsie Wiles, cy53aWxlc0BhdWNrbGFuZC5hYy5ueg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.