
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 February 2022
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.738818
This article is part of the Research Topic Bacterial Secretion Systems, Volume II View all 17 articles
Yersinia adhesin A (YadA) is a key virulence factor of Yersinia enterocolitica and Yersinia pseudotuberculosis. YadA is a trimeric autotransporter adhesin, a class of adhesins that have been shown to enable many Gram-negative pathogens to adhere to/interact with the host extracellular matrix proteins such as collagen, vitronectin, and fibronectin. Here, we show for the first time that YadA of Yersinia enterocolitica serotype O:9 not only interacts with proteinaceous surface molecules but can also attach directly to glycan moieties. We show that YadA from Y. enterocolitica serotype O:9 does not interact with the vitronectin protein itself but exclusively with its N-linked glycans. We also show that YadA can target other glycan moieties as found in heparin, for example. So far, little is known about specific interactions between bacterial autotransporter adhesins and glycans. This could potentially lead to new antimicrobial treatment strategies, as well as diagnostic applications.
Yersinia adhesin A (YadA), a type Vc trimeric autotransporter adhesin of Yersinia spp. is crucial for virulence. YadA is encoded on a virulence plasmid, the pYV plasmid. Expression of the YadA gene is temperature controlled, and upon a temperature shift to 37°C, once the bacterium enters the host, the expression of YadA is initiated (Toivanen and Skurnik, 1992).
YadA is a surface-exposed adhesin that is anchored in the bacterial outer membrane via a trimeric β-barrel domain (Shahid et al., 2015). The passenger domain of YadA, a trimeric coiled-coil stalk, and an N-terminal β-roll head domain are channeled through the barrel in an unfolded state during the autotransport process (Chauhan et al., 2019). Upon autotransport, the passenger domain starts folding, building a rigid structure protruding toward the outside of the cell (Chauhan et al., 2019). Here, the head domain has been shown to be responsible for many of YadA’s adhesion properties (Leo et al., 2008; Mühlenkamp et al., 2015).
While YadA is typically classified as an adhesin that aids in pathogen–host interactions via interactions with the extracellular matrix (ECM) (Tamm et al., 1993; Westerlund and Korhonen, 1993; Leo et al., 2008; Keller et al., 2015), YadA has also been shown to be involved in immune evasion (Tamm et al., 1993; Westerlund and Korhonen, 1993; Grosskinsky et al., 2007; Leo et al., 2008; Schindler et al., 2012; Keller et al., 2015). YadA-knockout mutants are avirulent (Pepe et al., 1995; Schütz et al., 2010). This is only partially due to the adhesion properties of YadA as Yersinia spp. have additional adhesins which can replace its function (Mallick et al., 2012; Chauhan et al., 2016). During an infection with Yersinia enterocolitica, YadA is involved in surface adhesion and has been shown to interact with a variety of proteinaceous ECM molecules such as collagen, fibronectin, and vitronectin (Vn) (Tertti et al., 1992; Schulze-Koops et al., 1993; Leo et al., 2008; Mühlenkamp et al., 2017). The interaction with ECM varies in strength and depends on environmental shear forces (Müller et al., 2011). While the YadA head domain is conserved among Yersinia species, some Y. enterocolitica strains and Yersinia pseudotuberculosis exhibit an additional stretch of approximately 31 residues at the N-terminus of each monomer of the YadA head domain (Figure 1A). This stretch has been shown to be responsible for Vn binding (Mühlenkamp et al., 2017).
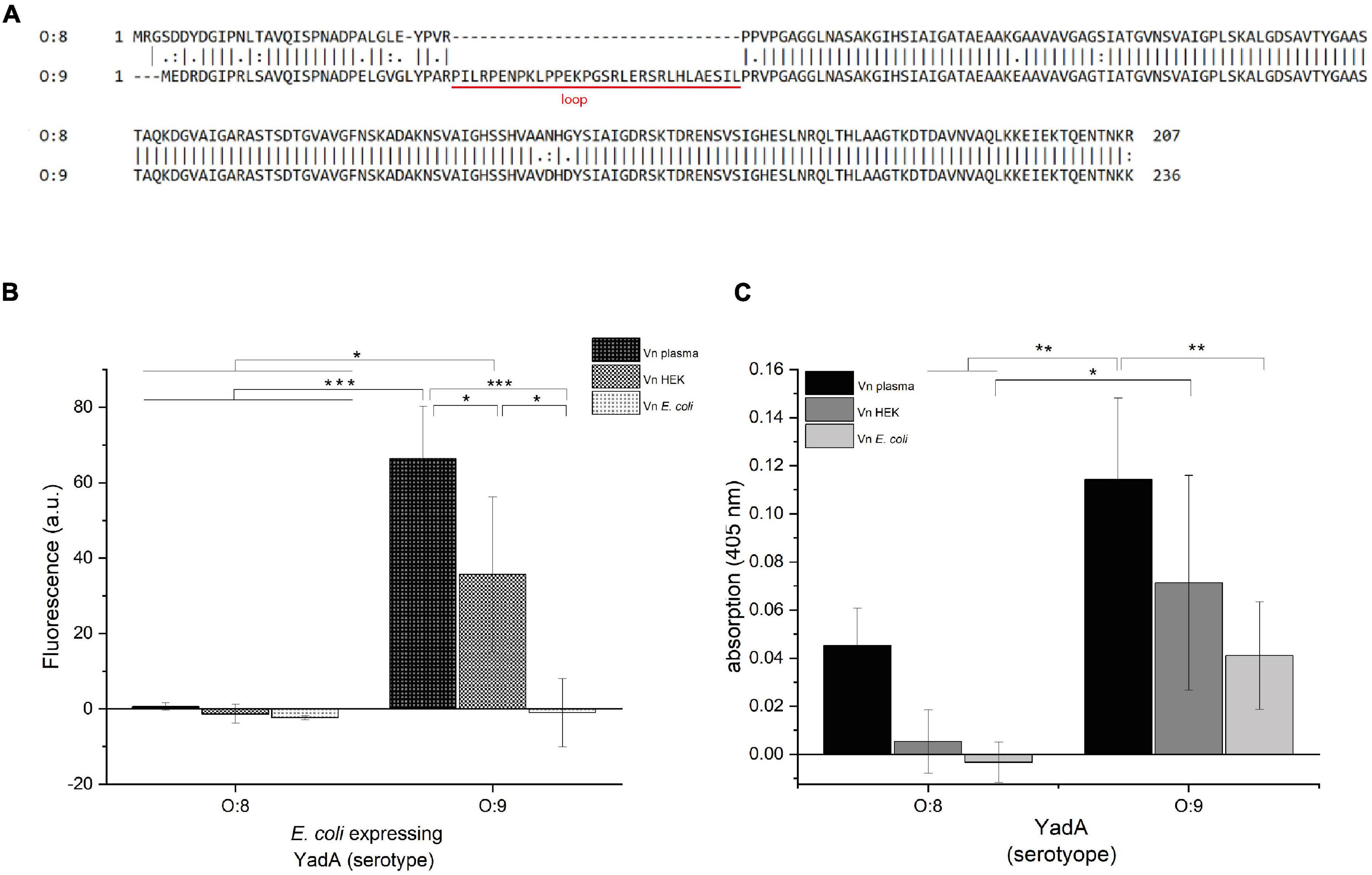
Figure 1. YadAO:9 binds to Vnplasma and VnHEK but shows reduced binding to VnEc. (A) Alignment of the N-terminal head domains of the YadA head domain from Y. enterocolitica serotypes O:8 and O:9. The alignments include the head domain of YadA, the neck region, and the first 20 residues of the coiled-coil stalk domain. YadAO:9 has an insertion of 31 residues toward the N-terminus of the head domain. (B) Whole-cell binding assay to Vn using E. coli expressing either YadAO:8 or YadAO:9. (C) Vn binding assay with purified YadAO:8 and YadAO:9 head domains. Significance levels are indicated with *p < 0.05, **p < 0.01, ***p < 0.001.
Vn has been described as an incidental component of the ECM (Leavesley et al., 2013). The ECM is a matrix composed of a variety of proteins, such as collagen, fibronectin, and laminin, and also proteoglycans and glycosaminoglycans (GAGs) forming a hydrogel. This matrix surrounds cellular components of the cell surface and provides strength and elasticity (Frantz et al., 2010). Vn is an approximately 75 kDa glycoprotein involved in tissue repair. It is heavily glycosylated, exhibiting three N-linked glycans (N86, N169, and N242) (Hwang et al., 2014). Vn shows great flexibility, and its conformational state is greatly dependent on interaction partners such as heparin (Izumi et al., 1989; Stockmann et al., 1993). The ability of Vn to associate with GAGs like heparin and heparan sulfate, which are in turn part of the ECM, contributes to the function of Vn in tissue repair (Leavesley et al., 2013). The ECM and its components are an attractive binding target for Y. enterocolitica and Y. pseudotuberculosis as surface adhesion is crucial for subsequent tissue invasion (Pepe et al., 1995).
Here, we report that YadA from Y. enterocolitica strain E40, serotype O:9 (YadAO:9) interacts with Vn via its glycosylations. We furthermore show that YadAO:9 can directly interact with heparin. Up until now, an interaction with glycan moieties like the glycosylations of ECM proteins or GAGs has not been described for YadA.
Plasmids and constructs used in this study are listed in Table 1, and sequences can be found in the supplements (Table 1). Constructs made in this study were cloned using the Gibson assembly (Gibson et al., 2009).
Bacteria were cultivated in Lysogeny broth (LB, Miller formulation). For whole-cell assays with fluorescence detection, Escherichia coli Top10 glmS:sfGFP (AS75) was used and grown in the presence of arabinose (Saragliadis and Linke, 2019). For protein purification, genes encoding the proteins of interest were expressed in E. coli BL21 (DE3) Gold. Generally, bacteria were grown at 37°C to the desired OD600. During overexpression, the temperature was shifted to 23°C after induction.
pASK-IBA3_YadAO:8/O:9 was transformed into E. coli BL21 (DE3) Gold and grown on ampicillin plates. A single colony was inoculated into 20 ml of LB medium supplemented with 100 μg/ml ampicillin and grown at 37°C overnight (o/n). The following day, a 2 L subculture was prepared and grown in a home-built fermentation system (a system where air is bubbling through bottles of growth medium that stand in a temperature-controlled water bath) until an OD600 of 0.5–0.7 was reached. The temperature was shifted to 23°C, and expression was induced with 0.2 μg/ml anhydrotetracycline (AHTC). Protein expression was allowed for 16 h. The culture was harvested by centrifugation at 4,000 × g. Afterward, the pellet was resuspended in Tris-buffered saline (TBS) buffer (20 mM Tris pH 7.5, 300 mM NaCl, 20 mM imidazole) with 8 μg/ml lysozyme and a pinch of DNAse. The suspension was subjected to cell lysis using a French press after addition of a HALT protease inhibitor mix (1:500, Thermo Fisher Scientific; 1861278). The lysate was centrifuged for 1 h at 69,600 × g, and the supernatant was then filtered through a 0.2 μm filter and subjected to Ni-NTA affinity chromatography (Cytiva, 17531901). As YadA with C-terminal His6-tag elutes at high imidazole concentrations (160–500 mM), the protein was pure enough for binding experiments after Ni-affinity chromatography. The protein was subjected to dialysis against TBS buffer (20 mM Tris pH 7.5, 150 mM NaCl).
E. coli AS75 with pASK-Iba4C_YadAO:8 or pASK-Iba4C_YadAO:9 was grown o/n in LB medium supplemented with 0.02% (w/v) arabinose and 100 μg/ml ampicillin. The next day, the cultures were diluted 1:100 in 20 ml LB medium supplemented as before and grown at 37°C to an OD600 of 0.5. YadA expression was then induced by the addition of AHTC to a final concentration of 0.2 μg/ml and grown for another 3 h at 37°C. YadA expression was checked for by visual inspection for auto-aggregation (Trunk et al., 2018). In the meantime, clear flat-bottom 96-well plates were coated with 100 μl of a 10 μg/ml Vn solution, from either plasma (Gibco, PHE0011), recombinantly expressed in HEK cell cultures (Merck/Millipore, SRP3186), or E. coli (Thermo Fisher Scientific, A14700), by incubation for 1 h at room temperature (RT). The Vn solution was discarded from the plates, and the wells were washed three times with TBS (20 mM Tris pH 7.5, 150 mM NaCl). Afterward, the wells were blocked using 3% bovine serum albumin (BSA) in TBS. The bacteria were then harvested, washed twice with TBS, and resuspended in TBS with 0.1% BSA to achieve an OD600 of 0.2. One hundred microliters of the bacterial suspension was added per well and incubated for 1 h at RT. After that, the wells were washed three times using TBS. Lastly, the wells were filled with 100 μl TBS buffer, and fluorescence was measured using an excitation wavelength of 488 nm and recording the emission at 533 nm (BioTek Synergy H). For experiments with deglycosylated Vn, the experiment was performed the same way, but deglycosylated Vn (see section “Deglycosylation of Vitronectin”) was used for coating.
A clear 96-well plate was coated with Vn and blocked as described before (section “Vitronectin Binding Experiments With Whole Bacteria”). Then 100 μl of a 10 μg/ml YadA solution in 0.1% (w/v) BSA in TBS (20 mM Tris pH 7.5, 150 mM NaCl) was added to the wells and incubated for 1 h at RT. After the wells were washed twice with 0.1% BSA in TBS and once with TBS with Tween-20 (TBS-T), Ni-horseradish peroxidase (HRP) conjugates were used for the detection of bound, His6-tagged YadA. One hundred microliters of a Ni-HRP conjugate solution at a final concentration of 5 μg/ml (Thermo Fisher Scientific, 15165) was incubated for 1 h at RT in 3% BSA in TBS per well. This was discarded, and the wells were washed three times with TBS-T and once with TBS. Binding was detected using 150 μl of a 1 mg/ml ABTS solution in ABTS buffer (2.43 ml of 100 mM citric acid, 2.57 ml of 200 mM Na2HPO4, 5.0 ml H2O, 10 μl H2O2) (Thermo Fisher Scientific, 34026; VWR, ICNA0219502305). The color development was stopped by adding 100 μl of 1% (w/v) SDS after incubation at RT, and the absorption was measured at 405 nm in a BioTek Synergy H plate reader.
For deglycosylation of Vn, 20 μg of Vn from the respective sources (in water) was mixed with 2 μg of the glycopeptidase PNGase F (500 U) (Promega, V4831) and incubated at 37°C for 19 h. The non-deglycosylated control samples of Cn were incubated at 37°C for 19 h, omitting the PNGase F. For PNGase F control samples, 2 μl of PNGase F was added to water and incubated as described before. Successful deglycosylation was checked for on a SDS-PAGE gel with subsequent silver staining (Nesterenko et al., 1994).
Glass coverslips were coated with 50 μl Vn (10 μg/ml) at 4°C o/n. An o/n culture of E. coli AS75 harboring pASK-IBA4C_YadAO:8/O:9 was inoculated into LB supplemented with 20 μg/ml chloramphenicol and 0.02% w/v arabinose. The next day, the culture was diluted 1:100 in the same broth, and the culture was grown to OD600 of 0.5 followed by induction with 0.2 μg/ml AHTC and yadA expression for 3 h at 37°C. In the meantime, Vn-coated coverslips were incubated with TBS or 100 μM heparin-disaccharide I-S (Merck, H9267-1MG) for 1 h at RT where applicable. After that, all coverslips were blocked with 3% (w/v) BSA in TBS for 1 h at RT. One hundred microliters of 5 × 108 bacteria in suspension were centrifuged at 4,000 × g for 5 min and resuspended in either TBS (20 mM Tris pH 7.5, 150 mM NaCl) or 100 μM heparin-disaccharide in TBS and incubated for 1 h at RT. After that, the bacteria were centrifuged down again and washed three times in 100 μl TBS. Finally, the bacteria were resuspended in 1 ml 3% BSA in TBS. Three hundred microliters of the bacteria was added to the coverslips and incubated for 30 min at RT. The supernatant was discarded, and the coverslips were washed three times with TBS and fixed with 500 μl of 4% (w/v) paraformaldehyde in TBS for 20 min at RT. Finally, the coverslips were mounted in 5 μl ProLong Glass Antifade Mountant (Invitrogen, P36980) and dried o/n. Microscopy was performed using a fluorescent microscope (Zeiss Axioplan 2) and a 100× oil immersion objective. For quantification, images were converted into binary files, and the area of the particles was calculated using Fiji (Supplementary Figure 3). Mean areas were plotted including the standard error of the mean.
An o/n culture of E. coli AS75 harboring pASK-IBA4C_YadAO:8/O:9 was inoculated into LB supplemented with 100 μg/ml ampicillin and 0.02% (w/v) arabinose. The next day, the culture was diluted 1:100 in the same broth and grown to OD600 of 0.5 followed by induction with 0.2 μg/ml AHTC and yadA expression for 3 h at 37°C. The culture was diluted to an OD600 of 1.0, and 50 μl was centrifuged down at 4,000 × g for 5 min. The pellets were then resuspended in 50 μl TBS or TBS supplemented with 100 μM heparin-disaccharide (Merck, H9267-1MG). This was incubated at 37°C in a shaking incubator for 30 min. Five microliters of each solution was wet-mounted onto microscope slides, and the edges were sealed using a CoverGrip coverslip sealant (Biotium, 23005). Microscopy was performed using a fluorescence microscope (Zeiss Axioplan 2) and a ×100 oil immersion objective. For quantification, images were converted into binary files, and the particle sizes were calculated using Fiji (Supplementary Figure 4). The area of each individual particle was plotted in a column scatter plot.
Nitrocellulose membranes were cut and transferred into a six-well plate. Three 2 μl drops of a 700 μg/ml purified YadAO:8 or YadAO:9 solution were applied onto the membrane and air-dried. Then, the membrane was blocked with 5% BSA in TBS-T (20 mM Tris pH 7.5, 150 mM NaCl, 0.2% Tween-20) for 1 h at RT. Five hundred microliters of a 100 μM biotinylated heparin (Merck, B9806-10MG) solution in TBS-T was incubated on the membrane for 1 h at RT. The membrane was washed three times with TBS-T and afterward incubated with 500 μl of 1:10,000 diluted Strep-Tactin–HRP conjugate (IBA Lifesciences, 2-1502-001) in 5% BSA in TBS-T for 30 min at RT. After the membrane was washed three times with TBS-T and once with TBS (20 mM Tris pH 7.5, 150 mM NaCl), a 500 μl ECL reagent (Thermo Fisher Scientific, 320106) was added, and the membrane was immediately imaged using a Kodak Image Station 4000R.
An o/n culture of E. coli AS75 pASK-IBA4C_YadAO:8/O:9 was grown in the presence of 0.2% (w/v) arabinose and 100 μg/ml ampicillin. This culture was diluted 1:100 the next morning and grown to an OD600 of 0.5. YadA full-length expression was induced by addition of 0.2 μg/ml AHTC. Expression was allowed for 3 h at 37°C. Uninduced bacteria were used as a control. The bacteria were diluted to an OD600 of 0.2, spun down, and resuspended in phosphate-buffered saline (PBS). One hundred microliters of that bacterial solution was pipetted into 96-well plates and centrifuged at 4,000 × g. After that, 100 μl of a 10 μg/ml biotinylated heparin (Merck, B9806-10MG) solution in 3% (w/v) BSA in TBS was added and incubated at RT for 0.5 h. The plate was washed three times with TBS. The plate was centrifuged as before after every wash before discarding the washing buffer. Strep-Tactin–HRP conjugates (IBA Lifesciences, 2-1502-001) at 1:1,000 in 3% (w/v) BSA in TBS were added and incubated for 30 min at RT. The plate was washed as described before. The ABTS solution was prepared, and color development was stopped as described before. Wells that did not contain any bacteria were used as background controls. Absorbance at 405 nm was measured in a plate reader (BioTek Synergy H).
One hundred microliters of 10 μg/ml YadA in TBS was coated into a 96-well plate by incubation at RT for 1 h. The plate was washed three times with 200 μl TBS (20 mM Tris pH 7.5, 150 mM NaCl) and blocked using 200 μl of 3% BSA in TBS. Afterward, 100 μl of biotinylated heparin dilution (0–6.75 μg/ml) in TBS was added to the wells and incubated for 1 h at RT. The wells were washed three times with TBS as described above and blocked with 3% BSA in TBS for 1 h at RT. Strep-Tactin–HRP (IBA Lifesciences, 2-1502-001) at 1:1,000 was added in 3% BSA in TBS and incubated for 1 h at RT. The wells were washed again as described earlier, and an ABTS solution was used for detection as described before. After color development, the reaction was stopped by adding 100 μl of a 1% SDS solution. Absorbance at 405 nm was measured in a plate reader (BioTek Synergy H).
Gold screen-printed electrodes (BVT-AC1.W1.RS.Dw2) from BVT Technologies were employed for biosensor fabrication. The electrodes were pre-treated by washing with 97% v/v ethanol for 30 min, rinsed with deionized water, and dried with N2. Twenty-five microliters of 2.5 mM octopamine in 10 mM phosphate buffer pH 7.2 was spread across the working electrode and electro-polymerized for two cycles at a scan rate of 100 mV/s from +0.0 to +1.6 V. The electrodes were rinsed with 10 mM PBS and dried with Ar. The electrodes were functionalized by binding of biotinylated NeutrAvidin (Ahmed et al., 2013). After that, 10 μl of a 1 mg/ml biotinylated heparin was coated onto the surface for 1 h at RT. The surface was washed with 10 mM PBS and dried with Ar.
For binding measurements, 10 μl of E. coli AS75 expressing either YadAO:8 or YadAO:9 full length at OD600 of 2, 0.2, and 0.02 was applied to the working electrodes and incubated for 30 min at RT. Blanks were acquired by measuring 10 mM phosphate buffer, omitting the bacteria. Electrical impedance measurements were carried out in a three-cell system of a PalmSens4 potentiostat, galvanostat, and frequency response analyzer (PalmSens BV, Netherland), adding 10 mM [Fe(CN)6]3–/4– in 10 mM PBS pH 7.2 onto the electrodes. EIS measurements were recorded at 0 V over a frequency range of 5–0.1 Hz, with a modulation voltage of + 10 mV. Measurements corresponding to finite Warburg impedances were excluded from the Nyquist plots (Nguyen and Breitkopf, 2018). PSTrace (5.8) was used to record the EIS measurements. Metrohm Autolab Nova 2.1.4. was used to fit the Nyquist plots into Randles’ equivalent circuits. From the fitting, the charge-transfer resistance (Rct) was obtained. The biosensor was assessed before and after analyte addition. Changes in Rct (%) were obtained to analytically assess bacterial binding with Equation 1:
All experiments were replicated n ≥ 6. Layer-by-layer construction can be found in Supplementary Figure 2.
For binding data analysis, data are shown as means ± SD and were analyzed using a one-way ANOVA including Tukey’s test. For data plotting and statistical analysis, OriginPro and “R” were used. For microscopy analysis, mean particle areas ± SEM were plotted. As the particles sizes were not normally distributed, non-parametric testing including a Kruskal–Wallis test and subsequent Wilcox testing were applied to test for significance. Significance levels are indicated in the graphs with p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
We first aimed to describe the molecular details of the interaction between the YadA head domain of Y. enterocolitica serotype O:9 (YadAO:9) and Vn. YadAO:9 harbors an additional, N-terminal, 31-residue stretch (Figure 1A) that has been described to interact with Vn (Mühlenkamp et al., 2017). We started out replicating the experiments done by Mühlenkamp et al. (2017). For these enzyme-linked immunosorbent assay (ELISA)-like binding experiments, Vn from different sources was used. Vn purified from plasma (Vnplasma), Vn expressed in HEK cell culture (VnHEK), and Vn recombinantly expressed in E. coli (VnEc) were tested for their capacity to be bound by E. coli expressing either full-length YadAO:8 or YadAO:9 (Figure 1B). Additionally, Vn binding by purified YadA head domains was tested (Figure 1C). We reasoned that, if YadA indeed bound to a conserved sequence within Vn, the binding should happen irrespective of the origin of Vn and only with YadAO:9. Indeed, the whole-cell assays show that only YadAO:9-expressing bacteria bound to Vnplasma and VnHEK (Figure 1B). Binding between VnEc and YadAO:9 could not be observed. Bacteria expressing YadAO:8 did not bind to either Vn variant (Figure 1B). These findings were corroborated by assays using purified YadA head domains. While we observed some binding of purified YadAO:8 to Vnplasma, no binding to VnHEK and VnEc was observed (Figure 1C). The weak binding of purified YadAO:8 can be explained by Vnplasma being contaminated with other proteins (Supplementary Figure 1). YadAO:9 on the other hand showed clear binding to all Vn variants, with reduced binding to VnEc. Based on these findings, we sought to investigate the difference between YadAO:9 binding to Vnplasma/HEK and binding to VnEc.
Due to the observation that YadAO:9 binds Vnplasma and VnHEK but shows at least reduced binding to VnEc, we wanted to investigate whether YadAO:9 actually binds a stretch within Vn or whether it either recognized a folded binding site or the glycosylations of Vn. Vn is heavily glycosylated with at least three N-linked glycans at residues N86, N169, and N242 (Figure 2A). As eukaryotic proteins recombinantly expressed in E. coli are usually not glycosylated, we first tested the latter hypothesis. We used PNGase F, a glycopeptidase that selectively removes glycans directly at the N-linkage by cleaving the glycosidic bond between asparagine and the core GlcNAc. With the deglycosylated Vn, the binding assays were repeated to see whether binding could be abrogated by removal of the N-linked glycosylations. In Figure 2B, the fluorescence-based whole-cell assay using E. coli AS75 expressing either full-length YadAO:8 or full-length YadAO:9 is shown. No binding was observed with cells expressing YadAO:8, which fits the hypothesis, as the postulated Vn binding stretch is not present in YadA from Y. enterocolitica serotype O:8. In the case of binding of bacteria expressing full-length YadAO:9, a clear difference in binding to Vnplasma was observed between the glycosylated Vnplasma and deglycosylated Vnplasma (Figure 2B). For VnHEK, no change in binding of YadAO:9-expressing E. coli AS75 before and after glycosylation was observed. We can at this point not say as to why no change was observed for bacterial binding of VnHEK compared to deglycosylated VnHEK. VnEc was bound in neither the glycosylated nor the deglycosylated state. While this supported our hypothesis that the glycan residues of Vn might be involved in the YadAO:9–Vn interaction rather than the proteinaceous part of Vn, we also repeated the binding assay using purified YadA head domains from both serotypes of Y. enterocolitica (Figure 2C). While, as expected, YadAO:8 did not bind to Vn-coated plates, neither to the untreated nor to the deglycosylated version, YadAO:9 bound to both untreated Vnplasma and untreated VnHEK (Figure 2C). Untreated VnEc was not bound as already shown in Figure 1C. After deglycosylation with PNGase F, neither Vnplasma nor VnHEK was bound by YadAO:9 anymore, further supporting our hypothesis of YadAO:9 interacting with the N-linked glycans.
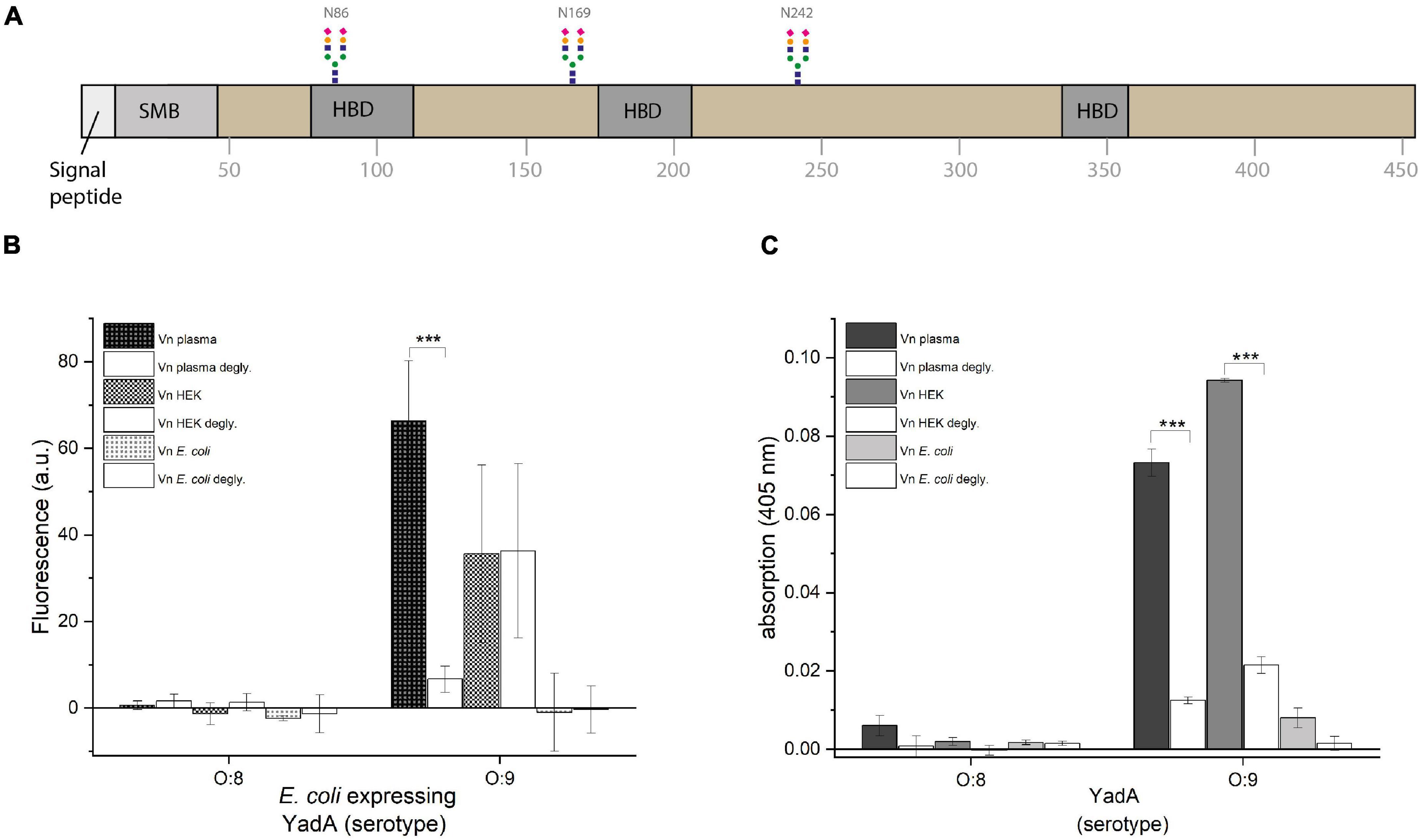
Figure 2. YadAO:9 binding to deglycosylated Vn is reduced. (A) Schematic representation of full-length Vn. Vn has three N-linked glycosylations in positions N86, N169, and N242. (B) Whole-cell binding assay to glycosylated and deglycosylated Vn. (C) Binding assay using purified YadA head domains to glycosylated and deglycosylated Vn. Significance levels are indicated with *p < 0.05, **p < 0.01, ***p < 0.001.
Heparin was described to abrogate the interaction between Vn and YadAO:9 (Mühlenkamp et al., 2017). We next wanted to investigate whether the potential YadAO:9 glycan interaction might be the cause for this observation. It was hypothesized before that heparin blocks the YadA binding site on Vn. As in the globular state, the heparin binding site in Vn is mostly hidden inside the core of the protein; this seemed unlikely to be the reason for YadAO:9 binding inhibition (Hayashi et al., 1985; Izumi et al., 1989; Zhuang et al., 1996; Leavesley et al., 2013). Coverslips were coated with untreated Vnplasma, VnHEK, or VnEc. YadAO:9 (full length)-expressing, fluorescent bacteria were checked for binding (Figure 3, left column). To check for the influence of heparin on this interaction, we also prepared samples where we either preincubated Vn with heparin (Figure 3, middle column) or preincubated YadAO:9-expressing bacteria with heparin (Figure 3, right column). In the fluorescence microscopy adhesion assay, we observed only minimal adhesion of bacteria to VnEc (Figure 3, bottom row). When coverslips had been coated with Vnplasma or VnHEK, adhesion was observed only in the absence of heparin. In cases where Vn was preincubated with heparin, bacteria expressing YadAO:9 adhered to Vn to a comparable level as in the untreated samples (Figure 3, left and middle columns). When YadAO:9-expressing bacteria were preincubated with heparin, reduced binding to untreated Vnplasma and VnHEK was observed (Figure 3, right column). Quantifications of the area of the particles reflect the tendencies seen in the experiment, where preincubation of the bacteria expressing YadAO:9 with heparin seems to reduce binding to Vn whereas preincubation of Vn with heparin did not change the adhesion of YadAO:9-expressing bacteria. This observation further strengthened our hypothesis that the YadA Vn-binding loop aids in adhesion of YadAO:9 to glycan moieties.
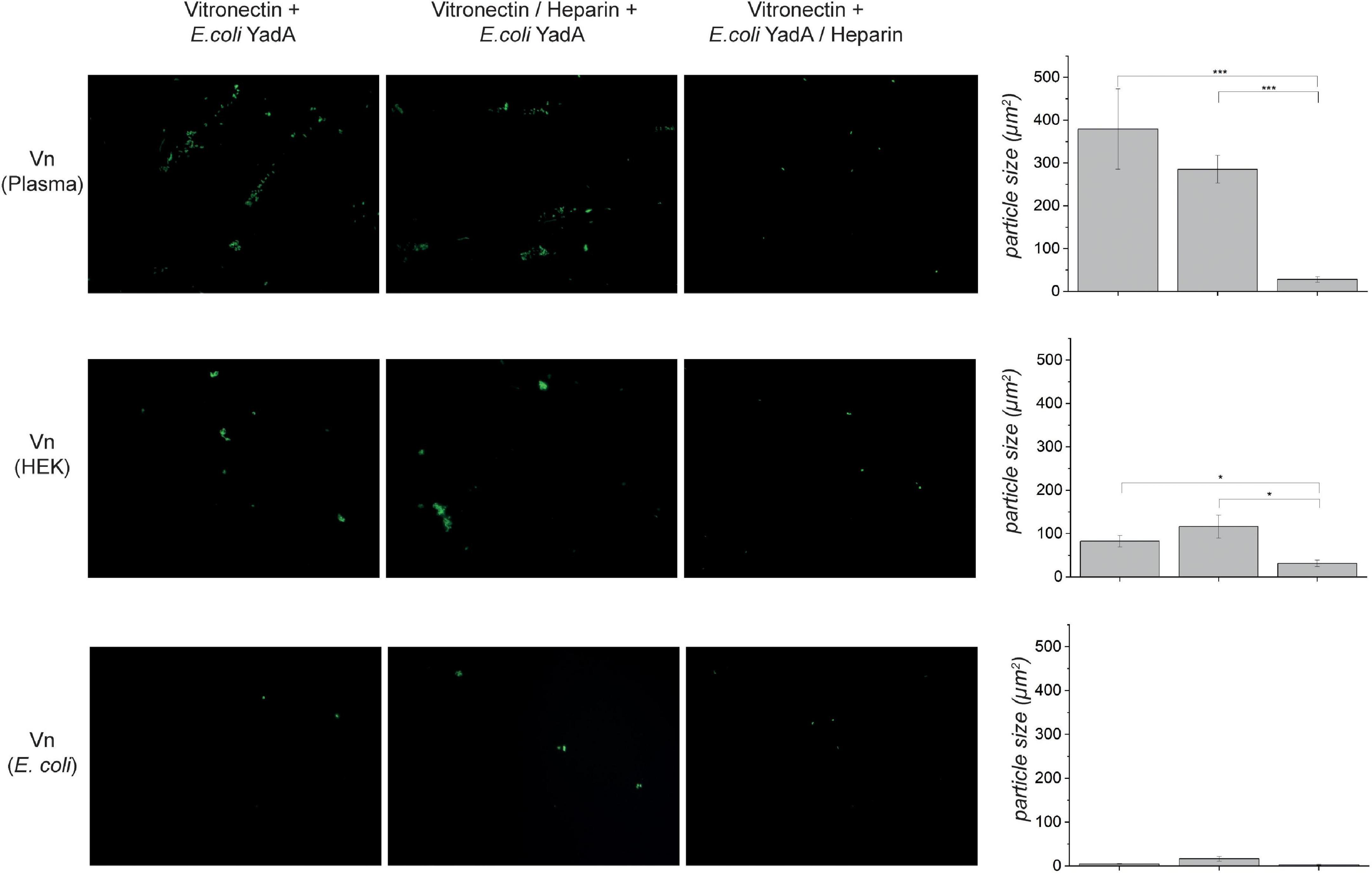
Figure 3. Binding of E. coli expressing YadAO:9 is reduced when the bacteria are preincubated with heparin-disaccharide. Vn from different sources was coated onto coverslips, and binding of fluorescence E. coli expressing YadAO:9 was measured (left column). Images in the middle column show micrographs of Vn-coated coverslips that were preincubated with 100 μM heparin-disaccharide. Afterward, binding of fluorescent E. coli expressing YadAO:9 was monitored. When Vn was coated and binding of fluorescent E. coli expressing YadAO:9 that were preincubated with heparin was (column 3) reduced, adhesion was measured for both Vnplasma and VnHEK. E. coli expressing YadAO:9 did only weakly bind to VnEc as already observed in previous experiments. In the last column, the mean area of particles (μm2) ± SEM was calculated and plotted as bar graphs. Significance levels are indicated with *p < 0.05, **p < 0.01, ***p < 0.001.
YadA, as an adhesin, is involved in autoaggregation, which has been described as an important mechanism for immune evasion during infection as well as for biofilm formation (Trunk et al., 2018). We have observed earlier that the interaction with other adhesin targets, such as ECM molecules, interferes with autoaggregation (manuscript in preparation). We thus wanted to investigate what effect heparin might have on autoaggregation mediated by YadAO:9. We expressed YadAO:8 or YadAO:9 full length in fluorescent E. coli AS75 and allowed for autoaggregation of these samples. Uninduced samples served as a control. Half of the samples were then preincubated with heparin-disaccharide. The uninduced samples did not show any autoaggregation behavior, either in the presence or in the absence of heparin (Figure 4, rows 1 and 3). The induced YadAO:8 samples autoaggregated to similar degrees both in the absence and in the presence of heparin (Figure 4, row 2). Fluorescent bacteria expressing YadAO:9 showed autoaggregation in the absence of heparin but reduced autoaggregation in the presence of heparin (Figure 4, lower row). This indicates that, indeed, heparin binding of YadAO:9 dissolves the autoaggregation tendencies caused by surface expression of YadAO:9. The dispersion of particles sizes (μm2) is shown in Figure 4B. The scatter plots reflect the difference in aggregate (particle) sizes. A reduction in area between aggregates of E. coli expressing YadAO:9 with and without addition of heparin-disaccharide can be seen shifting from large aggregates to smaller aggregates or fully disaggregated samples.
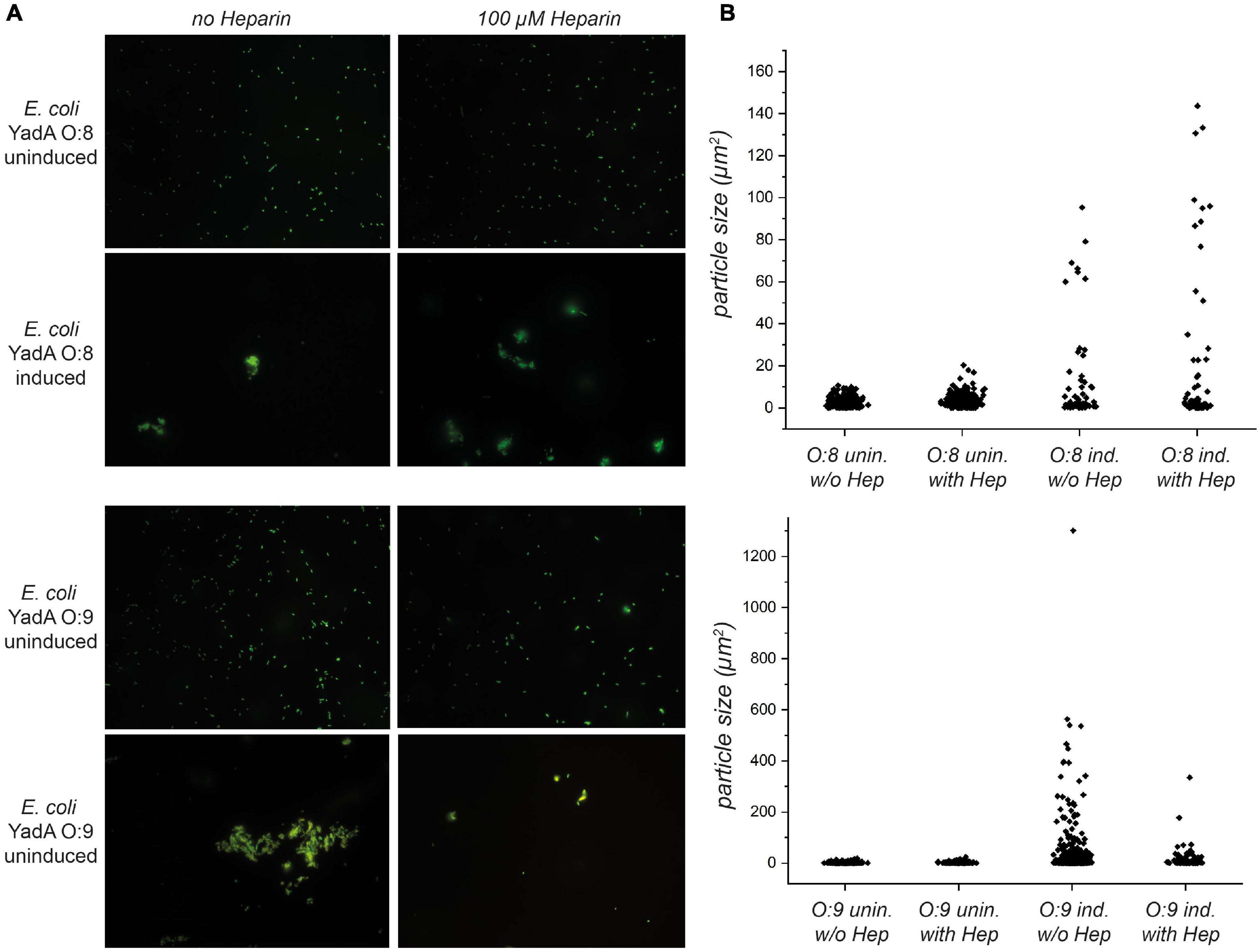
Figure 4. Autoaggregation of E. coli expressing YadAO:9 in the presence of 100 μM heparin-disaccharide is reduced. (A) In the upper row, uninduced bacteria expressing YadAO:8 in the presence and absence of heparin-disaccharide are shown. Uninduced bacteria (no YadAO:8 expressed) were used as a negative control for aggregation. After induction, E. coli expressing YadAO:8 show autoaggregation (row 2). In rows 3 and 4, the autoaggregation behavior of E. coli expressing YadAO:9 was tested in the presence and absence of 100 μM heparin-disaccharide. Uninduced E. coli expressing YadAO:9 are shown in row 3, and induced samples are shown in row 4. (B) Quantifications of the area of the particles are shown in the bar scatter plots for E. coli expressing YadAO:8 samples in the upper graph and E. coli expressing YadAO:9 samples in the lower graph.
To test for a direct interaction between YadAO:9 and heparin, we used E. coli AS75 cells expressing either YadAO:8 or YadAO:9. The bacteria were immobilized in a 96-well plate to capture biotinylated heparin. Bound biotinylated heparin was detected using Strep-Tactin–HRP. While no binding of biotinylated heparin to YadAO:8-expressing bacteria was observed, bacteria expressing YadAO:9 clearly showed heparin binding (Figure 5A). To support these results, we used electrochemical impedance measurements to measure bacterial binding to a heparin-coated surface. Biotinylated heparin was coated onto a biosensor using matrix-embedded NeutrAvidin (Ahmed et al., 2013). The change in impedance was then measured upon binding of E. coli AS75 expressing either YadAO:8 or YadAO:9 (Figure 5B). Please note that negative binding values are due to stronger adhesion of E. coli expressing YadAO:8/O:9 to uncoated electrodes that were used as a background and subtracted. While, for uninduced E. coli AS75 and E. coli expressing YadAO:8, no change in impedance was observed, we could clearly measure binding of E. coli expressing YadAO:9 by a significant change of impedance (Figure 5B). We then aimed to test for binding of heparin to purified YadA head domains. A dot blot using immobilized YadAO:8 and YadAO:9 head domains to detect binding of biotinylated heparin was performed. While no heparin binding was observed for either the buffer control or the YadAO:8 head domain, a signal could be observed for the binding of biotinylated heparin to the immobilized YadAO:9 head domain (Figure 5C). To quantify the binding, we immobilized the head domains of YadAO:8 and YadAO:9 in a 96-well plate and tested for binding at various concentrations. We observed that at 450 μg/ml of heparin, binding between YadAO:8 or YadAO:9 head domains and heparin is significantly different (Figures 5D,E). Repeating the assay with a dilution series of biotinylated heparin allowed us to investigate the concentration dependency of the binding. Using a fifth-party logistics fit, we estimate the (apparent) KD to be approximately 30 nM. Furthermore, this experiment allows for an estimation of the binding ratio between YadAO:9 and biotinylated heparin. The binding ratio is estimated to be 1:1 (YadAO:9 monomer to biotinylated heparin). We can at this point not claim an accurate KD or binding ratio as heparin varies in length but averages at 15 kDa (Shriver et al., 2012).

Figure 5. YadAO:9 directly binds to heparin. (A) Whole-cell binding assay of heparin to E. coli expressing YadAO:8 or YadAO:9 shows binding of YadAO:9-expressing bacterial binding to heparin. (B) Impedimetric biosensor experiment showing the adhesion of bacteria expressing YadAO:8 or YadAO:9 to surface-coated heparin. (C) Dot blot using immobilized YadAO:8 or YadAO:9 head domains shows direct binding to biotinylated heparin. (D) ELISA-like binding assay using immobilized YadA head domains to capture biotinylated heparin. (E) ELISA-like binding assay showing the interaction between immobilized, purified YadA head domains and biotinylated heparin in a concentration-dependent manner. Significance levels are indicated with *p < 0.05, **p < 0.01, ***p < 0.001.
With this work, we present evidence that a 31-residue loop insertion specifically found in YadAO:9 is responsible for the interaction between YadAO:9 and glycan moieties. All experiments presented in this work were done using YadA from Y. enterocolitica strains WA-314 (serotype O:8) or E40 (serotype O:9). While Vn binding results published previously indicate that all Y. enterocolitica strains of serotype O:9 harbor this N-terminal 31-residue loop (Mühlenkamp et al., 2017), we cannot be sure that the presence or absence of this loop correlates with the serotypes in all cases. To our knowledge, sequence variations of YadA have never been reported to contribute directly to serotyping.
This loop region aids in the interaction between YadAO:9 and the glycosylated host protein Vn as well as heparin, which, like Vn, is part of the ECM. We show that this interaction is not specific for one type of glycan residue but rather for a variety of glycans. This is supported by the observation that YadAO:9 interacts not only with the glycan residues of the glycoprotein Vn but also with the carbohydrate polymer heparin. Interactions with glycans are employed by many pathogens for adhesion and invasion, especially in viruses (Marks et al., 2001; Guan et al., 2017; Sorin et al., 2021). Also, bacterial virulence factors like UpaB and Pili have been shown to interact with the glycosylations of glycoproteins and GAGs (Rajas et al., 2017; Paxman et al., 2019; Sauer et al., 2019; Vizarraga et al., 2021). To our knowledge, this is the first time that a trimeric autotransporter adhesin is described to bind glycans.
While it has been established in earlier work that YadAO:9 interacts directly with human Vn (Mühlenkamp et al., 2017), we show that recombinant Vn expressed in E. coli is not bound by YadAO:9. Eukaryotic proteins expressed in E. coli often lack glycosylations, as E. coli does not possess the glycosyltransferases and glycosidases present in eukaryotes (Sahdev et al., 2008; Khow and Suntrarachun, 2012). We further show in deglycosylation experiments that properly deglycosylated Vn was not bound by YadAO:9 any longer. This, and the fact that binding does not occur when using YadAO:8, further supports our model that the YadAO:9 loop is responsible for interactions with glycans. Furthermore, as Mühlenkamp et al. (2017) had described that heparin could inhibit the interaction between YadAO:9 and Vn, we set out to investigate whether heparin binding to Vn was actually the reason for this inhibition or whether a more direct interaction of heparin with YadAO:9 was the reason for this effect. While Vn indeed harbors a heparin binding site, this site is hidden in globular Vn (Izumi et al., 1989). We thus checked for binding of fluorescent E. coli expressing full-length YadAO:9 after preincubating either Vn with heparin-disaccharide or after preincubating fluorescent E. coli expressing YadAO:9 with heparin-disaccharide. Heparin preincubation of fluorescent E. coli expressing YadAO:9 inhibited Vn interaction, while preincubation of Vn with heparin-disaccharide did not. This is in agreement with literature stating that only 2% of the overall plasma Vn is present in a heparin-binding-competent state (Izumi et al., 1989), as well as with our model stating that the YadAO:9 31-aa loop might be responsible for glycan binding. Furthermore, heparin-disaccharide was able to dissolve the YadA-mediated autoaggregation of bacteria expressing YadAO:9, which again indicates that there might be a direct interaction between heparin and YadAO:9. Finally, we were able to directly show the interaction using YadAO:9-expressing E. coli as well as purified YadAO:9 head domains in dot blots and ELISA-like assays. When looking at the YadAO:9 sequence, one can see that the loop contains seven positively charged residues (Arg and Lys). We hypothesize that the interaction between YadAO:9 could be explained by charge interactions with these residues, as the terminal sugar of the glycosylation of Vn is in most cases negatively charged sialic acid (Hwang et al., 2014). In heparin, sulfate moieties render this oligosaccharide heavily negatively charged (Rabenstein, 2002). It is worth noting that many known heparin binding motifs exhibit multiple, evenly spaced basic residues (Capila and Linhardt, 2002). Overall, we thus suggest that electrostatic interactions between the positively charged YadAO:9 loop residues and negatively charged functional groups on glycans are key to the binding affinity between YadAO:9 and glycans.
In terms of biological relevance, we hypothesize the interaction with YadAO:9 to be an additional mechanism for binding to host cell surfaces. As many secreted eukaryotic proteins are glycosylated for protein stability in the extracellular space (Varki et al., 2009), glycosylated ECM proteins could make a prime adhesion target during infection. Furthermore, a major group of molecules found in the ECM are GAGs such as heparin and heparan sulfate (Frantz et al., 2010). In addition to glycan adhesion being beneficial for the pathogen, it can conceptually be used in diagnostic workflows, e.g., to enrich pathogens from biological samples and potentially to develop anti-infective drugs. As glycans play a crucial role in pathogen adhesion, they have been used previously as therapeutics. Examples are the use of D-mannose in the treatment of urinary tract infections or of glycan derivatives to treat influenza (Domenici et al., 2016; Rustmeier et al., 2019; Weiss et al., 2020).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
IM: data acquisition, data visualization, methodology, writing of the original draft, and project conceptualization. JL-B: data acquisition, methodology, data visualization, and draft writing and review. PM, MS, and SP: conceptualization, and draft review and editing. DL: project administration, project conceptualization, and draft writing and editing. All authors contributed to the article and approved the submitted version.
This work was funded by the Horizon 2020 Innovative Training Network “ViBrANT” (to DL, SP, and PM) (funding ID: 765042). Contributions by the University of Oslo are gratefully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank D. Hatlem and Ana Lucía Campaña (University of Oslo) for helpful discussions and T. Späth (Institute for Medical Microbiology, Tubingen) for technical assistance. We thank Kristian Prydz for helpful discussions concerning this manuscript. We furthermore thank the imaging platform (NorMIC), especially Frode Miltzow Skjeldal, for help with the image analysis and quantification.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.738818/full#supplementary-material
Ahmed, A., Rushworth, J. V., Wright, J. D., and Millner, P. A. (2013). Novel impedimetric immunosensor for detection of pathogenic bacteria Streptococcus pyogenes in human Saliva. Anal. Chem. 85, 12118–12125. doi: 10.1021/ac403253j
Capila, I., and Linhardt, R. J. (2002). Heparin-protein-wechselwirkungen. Angew. Chem. 114, 426–450. doi: 10.1002/1521-3757(20020201)114:3<426::aid-ange426>3.0.co;2-q
Chauhan, N., Hatlem, D., Orwick-Rydmark, M., Schneider, K., Floetenmeyer, M., van Rossum, B., et al. (2019). Insights into the autotransport process of a trimeric autotransporter, Yersinia Adhesin A (YadA). Mol. Microbiol. 111, 844–862. doi: 10.1111/mmi.14195
Chauhan, N., Wrobel, A., Skurnik, M., and Leo, J. C. (2016). Yersinia adhesins: an arsenal for infection. Proteomics Clin. Appl. 10, 949–963. doi: 10.1002/prca.201600012
Domenici, L., Monti, M., Bracchi, C., Giorgini, M., Colagiovanni, V., Muzii, L., et al. (2016). D-mannose: a promising support for acute urinary tract infections in women. A pilot study. Eur. Rev. Med. Pharmacol. Sci. 20, 2920–2925.
Frantz, C., Stewart, K. M., and Weaver, V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200. doi: 10.1242/jcs.023820
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Grosskinsky, U., Schütz, M., Fritz, M., Schmid, Y., Lamparter, M. C., Szczesny, P., et al. (2007). A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin a autotransport. J. Bacteriol. 189, 9011–9019. doi: 10.1128/JB.00985-07
Guan, J., Bywaters, S. M., Brendle, S. A., Ashley, R. E., Makhov, A. M., Conway, J. F., et al. (2017). Cryoelectron microscopy maps of human papillomavirus 16 reveal L2 densities and heparin binding site. Structure 25, 253–263. doi: 10.1016/j.str.2016.12.001
Hayashi, M., Akama, T., Kono, I., and Kashiwagi, H. (1985). Activation of vitronectin (serum spreading factor) binding of heparin by denaturing agents. J. Biochem. 98, 1135–1138. doi: 10.1093/oxfordjournals.jbchem.a135363
Hwang, H., Lee, J. Y., Lee, H. K., Park, G. W., Jeong, H. K., Moon, M. H., et al. (2014). In-depth analysis of site-specific n-glycosylation in vitronectin from human plasma by tandem mass spectrometry with immunoprecipitation. Anal. Bioanal. Chem. 406, 7999–8011. doi: 10.1007/s00216-014-8226-5
Izumi, M., Yamada, K. M., and Hayashi, M. (1989). Vitronectin exists in two structurally and functionally distinct forms in human plasma. Biochim. Biophys. Acta Gen. Subj. 990, 101–108. doi: 10.1016/S0304-4165(89)80019-4
Keller, B., Mühlenkamp, M., Deuschle, E., Siegfried, A., Mössner, S., Schade, J., et al. (2015). Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell. Microbiol. 17, 1179–1204. doi: 10.1111/cmi.12429
Khow, O., and Suntrarachun, S. (2012). Strategies for production of active eukaryotic proteins in bacterial expression system. Asian Pac. J. Trop. Biomed. 2, 159–162. doi: 10.1016/S2221-1691(11)60213-X
Leavesley, D. I., Kashyap, A. S., Croll, T., Sivaramakrishnan, M., Shokoohmand, A., Hollier, B. G., et al. (2013). Vitronectin - master controller or micromanager? IUBMB Life 65, 807–818. doi: 10.1002/iub.1203
Leo, J. C., Elovaara, H., Brodsky, B., Skurnik, M., and Goldman, A. (2008). The Yersinia Adhesin YadA binds to a collagenous triple-helical conformation but without sequence specificity. Protein Eng. Design Sel. 21, 475–484. doi: 10.1093/protein/gzn025
Mallick, E. M., Brady, M. J., Luperchio, S. A., Vanguri, V. K., Magoun, L., Liu, H., et al. (2012). Allele- and Tir-independent functions of intimin in diverse animal infection models. Front. Microbiol. 3:11. doi: 10.3389/fmicb.2012.00011
Marks, R. M., Lu, H., Sundaresan, R., Toida, T., Suzuki, A., Imanari, T., et al. (2001). Probing the interaction of dengue virus envelope protein with heparin: assessment of glycosaminoglycan-derived inhibitors. J. Med. Chem. 44, 2178–2187. doi: 10.1021/jm000412i
Mühlenkamp, M., Oberhettinger, P., Leo, J. C., Linke, D., and Schütz, M. S. (2015). Yersinia Adhesin A (YadA) - beauty & beast. Int. J. Med. Microbiol. 305, 252–258. doi: 10.1016/j.ijmm.2014.12.008
Mühlenkamp, M. C., Hallström, T. I, Autenrieth, B., Bohn, E., Linke, D., Rinker, J., et al. (2017). Vitronectin binds to a specific stretch within the head region of Yersinia Adhesin a and thereby modulates yersinia enterocolitica host interaction. J. Innate Immun. 9, 33–51. doi: 10.1159/000449200
Müller, N. F., Kaiser, P. O., Linke, D., Schwarz, H., Riess, T., Schäfer, A., et al. (2011). Trimeric Autotransporter Adhesin-dependent adherence of Bartonella henselae, Bartonella quintana, and Yersinia enterocolitica to Matrix components and endothelial cells under static and dynamic flow conditions. Infect. Immun. 79, 2544–2553. doi: 10.1128/IAI.01309-10
Nesterenko, M. V., Tilley, M., and Upton, S. J. (1994). A simple modification of blum’s silver stain method allows for 30 minite detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28, 239–242. doi: 10.1016/0165-022x(94)90020-5
Nguyen, T. Q., and Breitkopf, C. (2018). Determination of diffusion coefficients using impedance spectroscopy data. J. Electrochem. Soc. 165, E826–E831. doi: 10.1149/2.1151814jes
Paxman, J. J., Lo, A. W., Sullivan, M. J., Panjikar, S., Kuiper, M., Whitten, A. E., et al. (2019). Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-09814-6
Pepe, J. C., Wachtel, M. R., Wagar, E., and Miller, V. L. (1995). Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect. Immun. 63, 4837–4848. doi: 10.1128/iai.63.12.4837-4848.1995
Rabenstein, D. L. (2002). Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 19, 312–331. doi: 10.1039/b100916h
Rajas, O., Quirós, L. M., Ortega, M., Vazquez-Espinosa, E., Merayo-Lloves, J., Vazquez, F., et al. (2017). Glycosaminoglycans are involved in bacterial adherence to lung cells. BMC Infect. Dis. 17:319. doi: 10.1186/s12879-017-2418-5
Rustmeier, N. H., Strebl, M., and Stehle, T. (2019). The symmetry of viral sialic acid binding sites-implications for antiviral strategies. Viruses 11, 1–15. doi: 10.3390/v11100947
Sahdev, S., Khattar, S. K., and Saini, K. S. (2008). Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol. Cell. Biochem. 307, 249–264. doi: 10.1007/s11010-007-9603-6
Saragliadis, A., and Linke, D. (2019). Assay development for the discovery of small-molecule inhibitors of YadA adhesion to collagen. Cell Surface 5:100025. doi: 10.1016/j.tcsw.2019.100025
Sauer, M. M., Jakob, R. P., Luber, T., Canonica, F., Navarra, G., Ernst, B., et al. (2019). Binding of the bacterial Adhesin FimH to its natural, multivalent high-mannose type glycan targets. J. Am. Chem. Soc. 141, 936–944. doi: 10.1021/jacs.8b10736
Schindler, M. K. H., Schutz, M. S., Muhlenkamp, M. C., Rooijakkers, S. H. M., Hallstrom, T., Zipfel, P. F., et al. (2012). Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J. Immunol. 189, 4900–4908. doi: 10.4049/jimmunol.1201383
Schulze-Koops, H., Burkhardt, H., Heesemann, J., Kirsch, T., Swoboda, B., Bull, C., et al. (1993). Outer membrane protein YadA of enteropathogenic yersiniae mediates specific binding to cellular but not plasma fibronectin. Infect. Immun. 61, 2513–2519. doi: 10.1098/rspb.2017.0729
Schütz, M., Weiss, E. M., Schindler, M., Hallström, T., Zipfel, P. F., Linke, D., et al. (2010). Trimer stability of YadA is critical for virulence of Yersinia enterocolitica. Infect. Immun. 78, 2677–2690. doi: 10.1128/IAI.01350-09
Shahid, S. A., Nagaraj, M., Chauhan, N., Franks, T. W., Bardiaux, B., Habeck, M., et al. (2015). Solid-state NMR study of the YadA Membrane-anchor domain in the bacterial outer membrane. Angew. Chem. Int. Edn. 54, 12602–12606. doi: 10.1002/anie.201505506
Shriver, Z., Capila, I., Venkataraman, G., and Sasisekharan, R. (2012). Heparin and heparan sulfate: analyzing structure amd microheterogeneity. Handb. Exp. Pharmacol. 207, 159–176. doi: 10.1007/978-3-642-23056-1_3
Sorin, M. N., Kuhn, J., Stasiak, A. C., and Stehle, T. (2021). Structural insight into non-enveloped virus binding to glycosaminoglycan receptors: a review. Viruses 13, 1–11. doi: 10.3390/v13050800
Stockmann, A., Hess, S., Declerck, P., Timpl, R., and Preissner, K. T. (1993). Multimeric vitronectin. Identification and characterization of conformation-dependent self-association of the adhesive protein. J. Biol. Chem. 268, 22874–22882. doi: 10.1016/s0021-9258(18)41608-0
Tamm, A., Tarkkanen, A. M., Korhonen, T. K., Kuusela, P., Toivanen, P., and Skurnik, M. (1993). Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA Protein of Yersinia enterocolitica. Mol. Microbiol. 10, 995–1011. doi: 10.1111/j.1365-2958.1993.tb00971.x
Tertti, R., Skurnik, M., Vartio, T., and Kuusela, P. (1992). Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60, 3021–3024. doi: 10.1128/iai.60.7.3021-3024.1992
Toivanen, P., and Skurnik, M. (1992). LcrF is the temperature-regulated activator of the YadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174, 2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992
Trunk, T., Khalil, H. S., and Leo, J. C. (2018). Bacterial autoaggregation. AIMS Microbiol. 4, 140–164. doi: 10.3934/microbiol.2018.1.140
Varki, A., Esko, J. D., and Colley, K. J. (2009). Essentials of Glycobiology, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Vizarraga, D., Torres-Puig, S., Aparicio, D., and Pich, O. Q. (2021). The sialoglycan binding adhesins of Mycoplasma genitalium and Mycoplasma pneumoniae. Trend Microbiol. 29, 477–481. doi: 10.1016/j.tim.2021.01.011
Weiss, G. L., Stanisich, J. J., Sauer, M. M., Lin, C.-W., Eras, J., Zyla, D. S., et al. (2020). Filaments in urinary tract infections. Science 369, 1005–1010. doi: 10.1126/science.aaz9866
Westerlund, B., and Korhonen, T. K. (1993). Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9, 687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x
Zhuang, P., Li, H., Williams, J. G., Wagner, N. V., Seiffert, D., and Peterson, C. B. (1996). Characterization of the denaturation and renaturation of human plasma vitronectin II. Investigation into the mechanism of formation of multimers. J. Biol. Chem. 271, 14333–14343. doi: 10.1074/jbc.271.24.14333
Keywords: trimeric autotransporter adhesin, bacterial adhesion, virulence, extracellular matrix (ECM), adhesion, glycan
Citation: Meuskens I, Leva-Bueno J, Millner P, Schütz M, Peyman SA and Linke D (2022) The Trimeric Autotransporter Adhesin YadA of Yersinia enterocolitica Serotype O:9 Binds Glycan Moieties. Front. Microbiol. 12:738818. doi: 10.3389/fmicb.2021.738818
Received: 09 July 2021; Accepted: 06 December 2021;
Published: 01 February 2022.
Edited by:
Eric Cascales, Aix-Marseille Université, FranceReviewed by:
Amit Kumar, Laboratory Corporation of America Holdings (LabCorp), United StatesCopyright © 2022 Meuskens, Leva-Bueno, Millner, Schütz, Peyman and Linke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dirk Linke, ZGlyay5saW5rZUBpYnYudWlvLm5v
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.