
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 25 November 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.738784
This article is part of the Research TopicAntimicrobial Resistance in Foodborne Pathogens: ecology, epidemiology, and mechanismsView all 12 articles
 Norhan K. Abd El-Aziz1†
Norhan K. Abd El-Aziz1† Yasmine H. Tartor1*†
Yasmine H. Tartor1*† Rasha M. A. Gharieb2
Rasha M. A. Gharieb2 Ahmed M. Erfan3
Ahmed M. Erfan3 Eman Khalifa4
Eman Khalifa4 Mahmoud A. Said5
Mahmoud A. Said5 Ahmed M. Ammar1
Ahmed M. Ammar1 Mohamed Samir2
Mohamed Samir2The emergence of extensive drug-resistant (XDR) Salmonella in livestock animals especially in poultry represents a serious public health and therapeutic challenge. Despite the wealth of information available on Salmonella resistance to various antimicrobials, there have been limited data on the genetic determinants of XDR Salmonella exhibiting co-resistance to ciprofloxacin (CIP) and tigecycline (TIG). This study aimed to determine the prevalence and serotype diversity of XDR Salmonella in poultry flocks and contact workers and to elucidate the genetic determinants involved in the co-resistance to CIP and TIG. Herein, 115 Salmonella enterica isolates of 35 serotypes were identified from sampled poultry (100/1210, 8.26%) and humans (15/375, 4.00%), with the most frequent serotype being Salmonella Typhimurium (26.96%). Twenty-nine (25.22%) Salmonella enterica isolates exhibited XDR patterns; 25 out of them (86.21%) showed CIP/TIG co-resistance. Exposure of CIP- and TIG-resistant isolates to the carbonyl cyanide 3-chlorophenylhydrazone (CCCP) efflux pump inhibitor resulted in an obvious reduction in their minimum inhibitory concentrations (MICs) values and restored the susceptibility to CIP and TIG in 17.24% (5/29) and 92% (23/25) of the isolates, respectively. Molecular analysis revealed that 89.66% of the isolates contained two to six plasmid-mediated quinolone resistance genes with the predominance of qepA gene (89.66%). Mutations in the gyrA gene were detected at codon S83 (34.62%) or D87 (30.77%) or both (34.62%) in 89.66% of XDR Salmonella. The tet(A) and tet(X4) genes were detected in 100% and 3.45% of the XDR isolates, respectively. Twelve TIG-resistant XDR Salmonella had point mutations at codons 120, 121, and 181 in the tet(A) interdomain loop region. All CIP and TIG co-resistant XDR Salmonella overexpressed ramA gene; 17 (68%) out of them harbored 4-bp deletion in the ramR binding region (T-288/A-285). However, four CIP/TIG co-resistant isolates overexpressed the oqxB gene. In conclusion, the emergence of XDR S. enterica exhibiting CIP/TIG co-resistance in poultry and humans with no previous exposure to TIG warrants an urgent need to reduce the unnecessary antimicrobial use in poultry farms in Egypt.
Salmonella enterica subspecies enterica serovars are the leading cause of food-borne zoonoses worldwide, accounting for 93.8 million cases of gastroenteritis and 155000 deaths annually (Majowicz et al., 2010). S. enterica is common in chickens, turkeys, quails, pheasants, and other game birds and has been isolated at high percentages from commercially reared poultry. Hence, multiple S. enterica serovars originating from poultry have been considered the potential source of human salmonellosis through the consumption of contaminated bird meat (Hoelzer et al., 2011; Thomas et al., 2017; Anbazhagan et al., 2019). Furthermore, farms represent a direct hazard to public health (Hoelzer et al., 2011), while indirect infections occur through the contaminated food products from carrier birds harboring Salmonella in the vicinity of food production units (Thomas et al., 2017). The spread of multidrug-resistant (MDR, i.e., resistant to at least one agent in three or more antimicrobial categories) Salmonella usually stems from the unjustified use of antimicrobials particularly in the poultry industry. Thus, the potential risk to public health posed by transmission of MDR non-typhoidal Salmonella (NTS) from poultry (Anbazhagan et al., 2019) warrants the need for an integrative “One Health” approach for NTS surveillance among human, poultry, and animal populations. More alarming than the MDR is the recent emergence of the extensive drug-resistant [XDR, i.e., resistant to one or more agents in all antimicrobial categories except two or fewer from the worksheet for defining and categorizing the isolates according to Magiorakos et al. (2012)] Salmonella such as the previously reported Salmonella Indiana (Wang et al., 2017) and Salmonella Typhi (Klemm et al., 2018; Saeed et al., 2019). There have been scarce data on the prevalence of such threatening XDR isolates in humans and poultry. It is worthy to note that the available studies on the mechanisms of antimicrobial resistance in S. enterica and in particular the XDR isolates have focused on the resistance to a single antibiotic (Chen et al., 2017; Zhang et al., 2017) without deciphering the molecular rationale behind the emergence of cross-antibiotic resistance, including to unrelated drugs.
Fluoroquinolone (e.g., ciprofloxacin, CIP) resistance in Salmonella species is mainly attributed to chromosomal mutations or the existence of plasmid-mediated quinolone resistance (PMQR) determinants or both (Robicsek et al., 2006). The chromosome-mediated quinolone resistance is commonly arbitrated by two main mechanisms: (1) spontaneous point mutations in the quinolone-resistant determining regions (QRDRs) of topoisomerase subunits, mainly gyrA (Hopkins et al., 2005; Aldred et al., 2014; Wasyl et al., 2014) and (2) overexpression of the major efflux pump (AcrAB-TolC) in Salmonella efflux systems, which usually leads to reduced cellular drug accumulation (Zhang et al., 2017).
The expression of AcrAB efflux pump is known to be regulated by global regulators (Usui et al., 2013), including the ramR gene. RamR mutations could increase the expression of ramA and acrAB genes resulting in the appearance of efflux-mediated MDR phenotype (Abouzeed et al., 2008), including CIP resistance. Because the resistance of Salmonella to CIP is often associated with cross-resistance to other antimicrobials (e.g., tetracyclines, beta-lactams, and chloramphenicol) (Hopkins et al., 2005), seeking novel antimicrobials that could inactivate XDR Salmonella is becoming a top priority. In this regard, glycylcycline (e.g., tigecycline, TIG) has come out as a promising, novel, and last-resort broad-spectrum agent for managing XDR Salmonella infections. However, decreased susceptibility of S. enterica to TIG has been observed (Chen et al., 2017). It was hypothesized that TIG resistance may be attributed to three main tetracycline resistance mechanisms (Chopra and Roberts, 2001): (i) mutations within tetracycline-specific efflux proteins, mainly tet(A), (ii) mutations in ribosomal protection proteins, tet(M), and (iii) enzymatic inactivation, tet(X). Additional resistance mechanisms as AcrAB-TolC and OqxAB efflux pumps overexpression may be also involved in such resistance (Tuckman et al., 2000; Hentschke et al., 2010; Chen et al., 2017).
The available information on the resistance mechanisms of Salmonella to CIP and TIG suggests a complex mechanistic view that needs to be delineated.
This study was designed to determine the prevalence, serotype diversity, and antimicrobial resistance phenotypes of S. enterica isolated from poultry and contact workers. Moreover, we combined molecular approaches with data analyses to investigate the genetic determinants involved in the co-resistance of XDR Salmonella isolates to CIP and TIG. Further, we precisely pinpointed the mutational alterations in ramRA regulatory gene that lie behind the overexpression of AcrAB-TolC efflux pump, which could confer CIP and TIG co-resistance.
This study was conducted during the period from January 2019 to April 2020. A total of 1210 clinically diseased and recently dead 1-day to 4-week-old poultry including broiler chickens (n = 650), ducks (n = 150), pigeons (n = 160), quails (n = 130), and turkeys (n = 120) from 25 poultry flocks located in five governorates in Egypt were sampled aseptically. Diseased birds were diarrheic, depressed, and weak with ruffled feathers. Flight inability was observed on pigeons and quails. Gross lesions including enteritis, enlarged liver with necrotic foci, perihepatitis, pericarditis, and unabsorbed yolk sac were observed in recently dead birds. The samples included fresh fecal dropping and cloacal swabs from diarrheic birds as well as liver, spleen, cecum, gall bladder, and yolk sac from recently dead birds. Fluoroquinolones, sulfonamides, penicillins, aminoglycosides, and tetracyclines were administered to the entire flocks for treatment. For prophylaxis, ESB3 (sulfaclozine 30%), lincomycin, oxytetracycline, and tylan premix (tylosin) were frequently used in poultry farms. Moreover, 375 human stool samples were collected from diarrheic poultry workers who were employed in the same flocks from which the poultry samples were obtained. The samples were collected in sterile separate containers and transported immediately in an icebox to the Microbiology Laboratory, Faculty of Veterinary Medicine, Zagazig University, for bacteriological analysis.
The study was approved by Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) (approval number ZU-IACUC/2/F/12/2019). Written informed consent was obtained from the owners for the participation of their animals in this study. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable data included in this article.
Isolation of S. enterica was carried out according to ISO (ISO, 2017). Briefly, 1 g of each sample was suspended in 9 ml of buffered peptone water (BPW, Oxoid, United Kingdom) and incubated at 37°C for 18 ± 2 h. An aliquot of 0.1 ml of the pre-enrichment culture was inoculated into 10 ml of Rappaport Vassiliadis soy broth (RV; Oxoid, United Kingdom) then incubated at 42°C for 24 h. Selective plating was done on xylose lysine deoxycholate agar (XLD, Oxoid, United Kingdom) and salmonella shigella agar (SS; Oxoid, United Kingdom) followed by incubation at 37°C for 24 h. For biotyping, fresh colonies from each pure culture were examined for oxidase, methyl red, Voges–Proskauer, and citrate utilization tests as well as their characteristic reactions on triple sugar iron and lysine decarboxylase agar (Oxoid, United Kingdom) media. Serotyping of Salmonella isolates was conducted using commercially available antisera (Denka Seiken Co., Ltd., United Kingdom) according to the antigenic profile (Kauffmann, 1957). Polymerase chain reaction (PCR) of the invA gene was performed to confirm Salmonella identification (Oliveira et al., 2003). The reference strain S. enterica serovar Typhimurium ATCC® 14028TM was used for quality control.
The antimicrobial susceptibilities of all Salmonella isolates were evaluated against 24 commercially available antimicrobial agents (Oxoid, Hampshire, England, United Kingdom) using the disk diffusion method (Bauer et al., 1966). The tested antimicrobials were ampicillin (10 μg), amoxycillin-clavulanic acid (20/10 μg), ampicillin-sulbactam (20/10 μg), cefazolin (30 μg), cefuroxime (30 μg), ceftriaxone (30 μg), cefepime (30 μg), cefoxitin (30 μg), ertapenem (10 μg), imipenem (10 μg), meropenem (10 μg), doripenem (10 μg), gentamicin (10 μg), tobramycin (10 μg), amikacin (30 μg), nalidixic acid (30 μg), CIP (5 μg), tetracycline (30 μg), TIG (15 μg), fosfomycin (50 μg), chloramphenicol (30 μg), sulfamethoxazole-trimethoprim (23.75/1.25 μg), aztreonam (30 μg), and colistin (10 μg). The inhibition zone diameters were measured and interpreted according to Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (European Committee Antimicrobial Susceptibility Testing (EUCAST), 2018; Clinical Laboratory Standards Institute (CLSI), 2019). The multiple antibiotic resistance (MAR) indices were calculated as described elsewhere (Tambekar et al., 2006). The MDR and XDR Salmonella isolates were categorized according to Magiorakos et al. (2012). The analyzed antimicrobials were synchronized with veterinary guidelines (Plumb, 2015). Justification of the selected antimicrobial agents was for monitoring the XDR Salmonella isolates (Magiorakos et al., 2012) and with the end goal of public health concern, e.g., TIG, fluoroquinolones, carbapenems, aminoglycosides, and cephalosporins.
To determine the minimum inhibitory concentrations (MIC) of CIP against XDR Salmonella isolates, VITEK® 2 (bioMérieux, Marcy L’Étoile, France) testing was performed using AST-GN91 cards (SKU Number: 414780) according to the manufacturer’s instructions. Interpretive correlation of the VITEK® 2 MIC results was applied using the Advanced Expert System (AESTM) rules. Moreover, the MICs of TIG and colistin were determined using broth microdilution method, and the interpretive criteria were those reported in the above mentioned CLSI and EUCAST documents. The MIC breakpoints for CIP (≥1 mg/L) and TIG (>2 mg/L) were considered accordingly.
The efflux pump activity of XDR Salmonella isolates was determined using the ethidium bromide (EtBr) cartwheel method as described previously (Martins et al., 2011). In brief, trypticase soy agar (TSA, Oxoid, United Kingdom) plates containing 0.0 to 2.5 mg/L of EtBr (Sigma-Aldrich, Germany) concentrations were prepared on the same day of the experiment and kept away from light. Each XDR Salmonella isolate (approximately 108 CFU/ml) was streaked on an EtBr plate in a cartwheel pattern. The plates were wrapped in aluminum foil and incubated at 37°C overnight. The minimum concentration of EtBr that produced fluorescence of bacterial colonies under AccurisTM E3000 UV Transilluminator (Accuris Instruments, United States) was recorded. A pan-susceptible Salmonella Tamale isolate generated during this study was used as a comparative control for fluorescence analysis. The capacity of each XDR Salmonella isolate to expel EtBr substrate was graded relative to the control isolate according to the following equation:
where MCEtBr (XDR) represents the minimum EtBr concentration that produces fluorescence of the XDR test isolate. Meanwhile, MCEtBr (REF) indicates the minimum EtBr concentration that produces fluorescence of the reference isolate.
The MICs of CIP and TIG were determined by broth microdilution method in the presence of the carbonyl cyanide 3-chlorophenylhydrazone (CCCP, 5 μg/ml; Sigma-Aldrich, United Kingdom) efflux pump inhibitor (EPI), which is known to dissipate the proton-motive force essential for the activity of resistance-nodulation division (RND) family efflux pumps. A significant efflux inhibition activity was considered when a ≥ 4-fold decrease in the MIC values was reported in the presence of EPI (Rushdy et al., 2013; Deng et al., 2014).
Plasmid DNA was extracted from XDR Salmonella isolates using QIAprep Spin Miniprep Kits (Qiagen, Germany) following the manufacturer’s recommendations. PMQR genes, including qnrA, qnrB, and qnrS were amplified through a multiplex PCR assay, while the qepA, aac(6′)-Ib-cr, oqxA, and oqxB genes were amplified by uniplex PCRs using previously published oligonucleotide primers (Supplementary Table 1) and cycling conditions (Robicsek et al., 2006; Cattoir et al., 2008; Kim et al., 2009; Lunn et al., 2010). Positive (S. enterica serovar Typhimurium ATCC® 14028TM) and negative (a reaction mixture without DNA template) controls were included in each run.
The plasmid-encoded tet(X1 to X5) genes conferring TIG resistance were amplified using multiplex PCR (Ji et al., 2020). The presence of tet(A), tet(B), and tet(M) genes were also examined (Van et al., 2008) using primer sequences depicted in Supplementary Table 1.
The QRDR of gyrA, tet(A), and ramRA genes were amplified using PCR followed by DNA sequencing using oligonucleotide primers listed in Supplementary Table 1. Genomic DNA was extracted from overnight cultures of the XDR Salmonella isolates using QIAamp DNA Mini kit (Qiagen, Germany) following the manufacturer’s instructions. The PCR amplicons were purified using the QIAquick PCR purification kit (Qiagen, Germany) and sequenced in an ABI 3130 automated DNA Sequencer (Applied Biosystems, United States) using the BigDyeR Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, United States) following the supplier protocol. Nucleotide sequences were compared with those previously deposited at GenBank using Basic Local Alignment Search Tool (BLAST1). Alignment of the nucleotide sequences was performed using the MEGA6 program (Tamura et al., 2013). The amino acid sequences were deduced using the ExPASy (Expert Protein Analysis System) Translate Tool2. The respective regions of nucleotide and polypeptide sequences were analyzed for mutational changes by comparison with the complete genome of S. enterica serovar Typhimurium LT2 (GenBank accession number NC_003197).
Quantitative PCR (qPCR) was used to determine the relative expression levels of ramA, acrB, and oqxB genes using previously published oligonucleotide primers (Supplementary Table 1). Total RNA was extracted from XDR Salmonella isolates using QIAamp RNeasy Mini kit (Qiagen, Germany) following the manufacturer’s instructions. The relative quantification was done in triplicates using QuantiTect SYBR Green real-time PCR Kit (Qiagen, Germany) in MX3005P real-time PCR thermal cycler (Agilent, La Jolla, CA, United States) following the manufacturer’s recommendations. Melting curve analysis was conducted to confirm the specificity of the tested assays. The 16S rRNA housekeeping gene was used as a normalizer (Fàbrega et al., 2016), and the fold change values were estimated using 2–ΔΔCT method (Livak and Schmittgen, 2001). A pan-susceptible Salmonella Tamale isolate was used as a comparative control.
Fisher’s exact test was used to determine if there were significant differences between the infection rates with Salmonella in the two age groups (i.e., 1–5 days and 1–4 weeks) in bird species. To visualize the overall distribution of the XDR Salmonella isolates based on their resistance patterns, a heatmap supported by hierarchical clustering (dendrogram) was generated (Kolde, 2019). To determine the significance of the association between a certain genetic marker and the resistance phenotype in Salmonella isolates, Fisher’s exact test and odds ratio (confidence intervals = 95%) were estimated on contingency tables considering the presence of genetic markers as independent variables and the resistance phenotypes as dependent variables. These analyses were done using GraphPad Prism version 8 for Windows, San Diego, CA, United States3. To reveal the significance of the genetic markers as classifiers of the isolates as resistant or susceptible to a certain drug, we applied a random forest classification model using an ensemble of 500 trees. The optimal number of random train predictor variables was determined using the tuneRF R function at an “out of bag error,” OOB (prediction error) = 0. The mean decrease in the Gini index was used as an indicator of variable importance (the higher the mean decrease in the Gini index, the more important the marker). This analysis was done using R package “Random Forest” (Breiman, 2001) in the R environment (v. 3.6.2). To calculate the correlation, the raw data were converted into binary outcomes, and the correlation significance was determined (p-value significance level = 0.05). These analyses were done using R packages corrplot and Hmisc (Harrell, 2020). Binary distances between isolates of the same host were measured using dist function in R software. Non-metric multidimensional scaling was used to visualize the clustering of isolates belonging to different hosts based on binary distance. This analysis was done using VEGAN package and the function metaMDS in R software4.
The nucleotide sequences of the genes under study were deposited into the GenBank under the following accession numbers: MT725561–MT725589 for gyrA, MT725590–MT725614 and MT740093–MT740096 for tet(A), and MT743008–MT743036 for ramRA genes.
As shown in Table 1, out of 1210 diarrheic and recently dead poultry sampled, 100 (8.26%) were positive for Salmonella species. The prevalence of Salmonella was 11.54% (75/650), 6.67% (10/150), 3.13% (5/160), 3.85% (5/130), and 4.17% (5/120) in chickens, ducks, pigeons, quails, and turkeys, respectively. Meanwhile, 15 S. enterica isolates were isolated from 375 contact worker stool samples (4.00%). The recovered Salmonella isolates were confirmed based on conventional phenotypic and molecular identification methods. Statistical analysis revealed that the difference in the infection rates of all birds with Salmonella species was significant (p-value = 0.04), whereas it was non-significant among each poultry species (p-value > 0.05). Typical Salmonella colonies were pink with or without black centers on XLD agar, while white colonies with black centers were characteristic on SS agar medium. Biochemical reactions for presumptive identification of Salmonella isolates indicated that all tested isolates were positive for methyl red, Simmons’ citrate, and oxidase tests and displayed characteristic reactions on triple sugar iron and lysine decarboxylase agar media, whereas the analyzed isolates were negative for indole and Voges–Proskauer tests. All Salmonella isolates were further confirmed by PCR detection of the invA gene (fragment size = 284 bp).
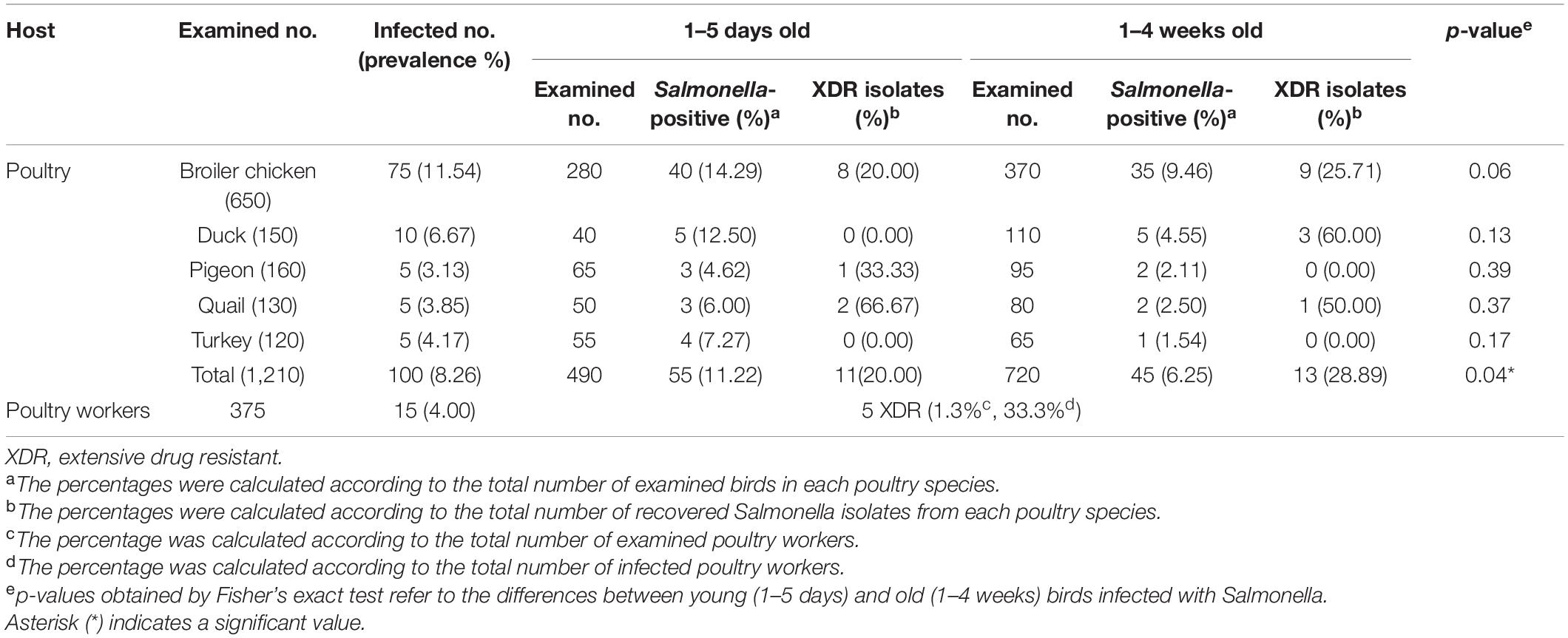
Table 1. Salmonella prevalence and XDR isolates recovered from clinically diseased, recently dead birds, and contact poultry workers.
In all, 35 Salmonella serotypes were identified among the 115 isolates originating from poultry and humans using a classical agglutination assay. Regardless of the isolate source, the most frequent serotype was Salmonella Typhimurium (26.96%) followed by Salmonella Enteritidis (11.30%), Salmonella Infantis (6.96%), Salmonella Kentucky (5.22%), and Salmonella Newport (3.48%); other Salmonella serotypes were reported by lower frequencies (Supplementary Table 2). The distribution of Salmonella serotypes varied within host species. Typhoidal Salmonella serotypes Typhi and Paratyphi C were detected among the human isolates only (3/15; 20% each), whereas all the subspecies enterica of poultry origin were categorized as NTS. It was noted that Salmonella Typhimurium was the predominant serotype in chickens (22/75; 29.33%) followed by Salmonella Enteritidis (12/75; 16.00%). However, certain Salmonella serovars were reported and characterized only for ducks (Salmonella Derby and Salmonella Larochelle), pigeons (Salmonella Alfort and Salmonella Wingrove), turkeys (Salmonella Vejle and Salmonella Apeyeme), or quails (Salmonella Shangani and Salmonella Jedburgh).
As depicted in Supplementary Table 3, the highest level of resistance was recorded for amoxycillin-clavulanic acid (99.13%) followed by cefazolin (94.78%), nalidixic acid (77.39%), cefoxitin (76.52%), tetracycline (74.78%), and cefepime (73.04%). More than 50% of the isolates exhibited resistance to each of tobramycin, cefuroxime, chloramphenicol, ceftriaxone, sulfamethoxazole-trimethoprim, and ampicillin. CIP and TIG resistance were observed in 51.30 and 24.35% of S. enterica isolates, respectively. However, the carbapenems including ertapenem, imipenem, meropenem, and doripenem displayed the maximum activity against the isolates (70.43, 79.13, 89.57, and 93.04%, respectively). Only one isolate of chicken origin (Salmonella Tamale; 0.87%), was pan-susceptible to the tested antimicrobials. According to the antimicrobial resistance profile, 73.9% (n = 85) of the isolates were MDR (MAR index = 0.13–0.71) and 25.22% (n = 29) were XDR (MAR index = 0.63–0.88), of which 24 originated from poultry and only 5 were of human origin (Table 1). Salmonella Typhimurium accounts for almost half of the XDR isolates (12/29; 41.38%), of which 8 (66.67%) were human isolates and 4 (33.33%) were of chicken origin.
As shown in Table 2 and Supplementary Table 4, all XDR Salmonella isolates (n = 29) were CIP-resistant (MIC = 2–128 μg/ml), and 25 (86.21%) were TIG-resistant (MIC = 4–32 μg/ml). It is worthy of note that four (13.79%) XDR isolates were uniquely resistant to CIP (MIC = 4–8 μg/ml), while 25 isolates (86.21%) showed CIP/TIG co-resistance (Figure 1).
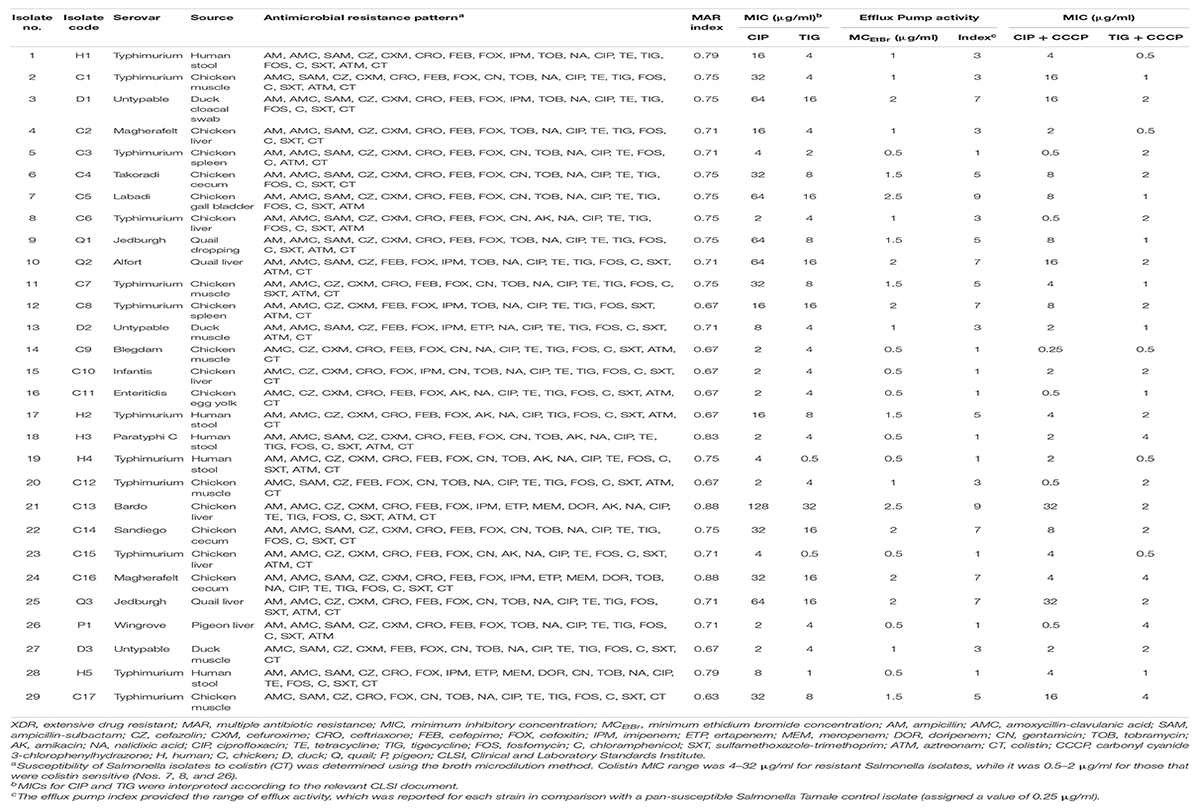
Table 2. Phenotypic characterization of XDR Salmonella isolates recovered from poultry and human origins.
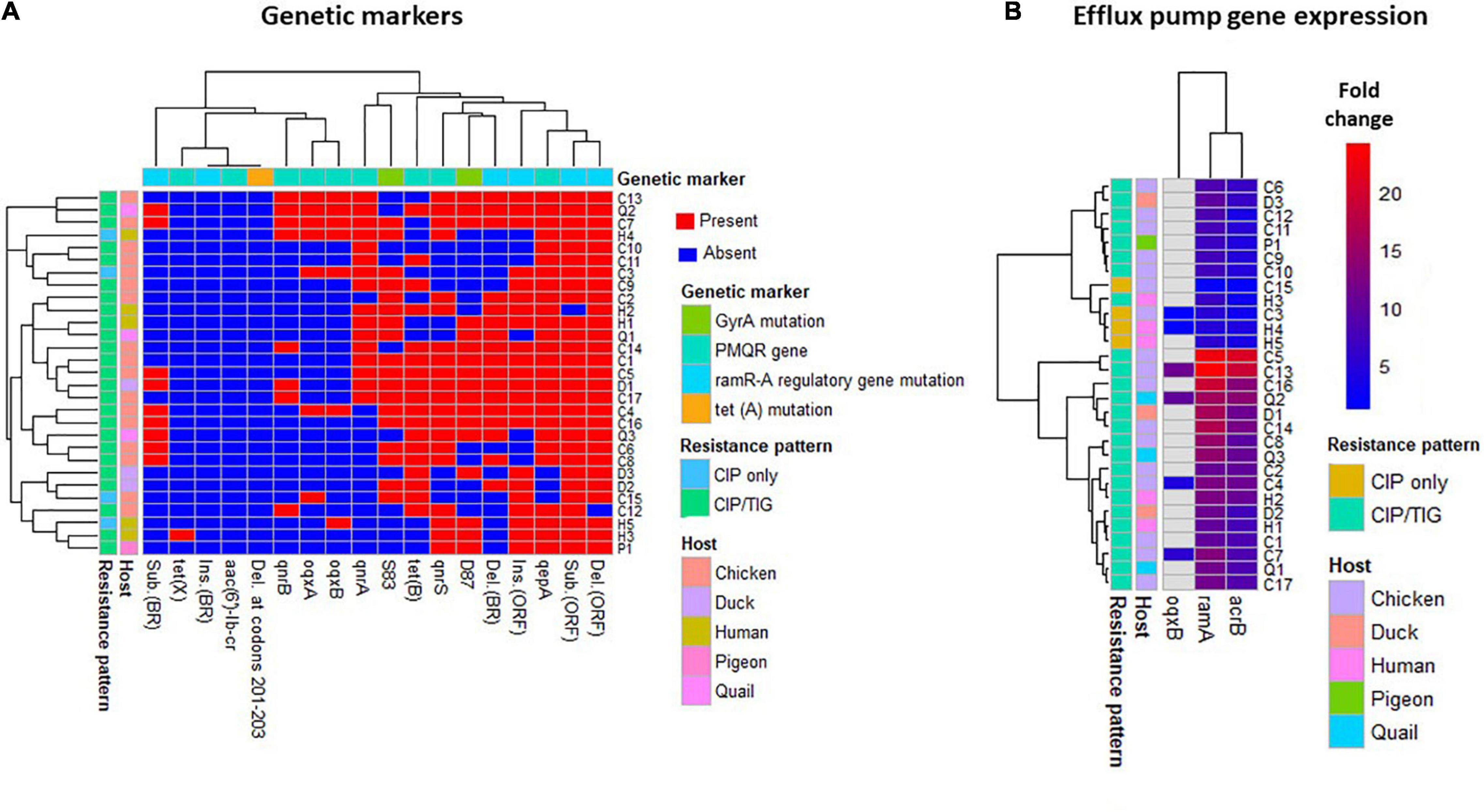
Figure 1. Heatmap supported by hierarchical clustering (dendrogram) depicting the overall distribution of genetic markers (A) and efflux pumps gene expression (B) in the 29 XDR Salmonella isolates. The isolates are color-annotated based on their hosts (C, chicken; D, duck; H, human; P, pigeon, and Q, quails) and resistance pattern [i.e., to CIP, ciprofloxacin (n = 4) and to both CIP and TIG (n = 25)]. The genetic markers are color-annotated based on their type. BR, binding region; ORF, open reading frame.
The efflux activity of XDR Salmonella isolates was assessed by testing the ability of the bacteria to pump EtBr out of the cell using the cartwheel test. Fluorescence of Salmonella isolates which grew as a confluent mass along a radial line of TSA plates containing increasing concentrations of EtBr was reported. The minimum concentration of EtBr and the efflux activity index for each Salmonella isolate are illustrated in Table 2. The fluorescence of tested Salmonella was lower than that produced by the control (pan-susceptible Salmonella Tamale), which fluoresced at 0.25 μg/ml EtBr. Two chicken isolates (2/29; 6.90%) began to fluoresce at an EtBr concentration of 2.5 μg/ml, whereas 6 isolates from various poultry types fluoresced at 2 μg/ml EtBr. The remaining Salmonella isolates (n = 21) fluoresced at EtBr concentration range of 0.5–1.5 μg/ml. As shown in Table 2, the MIC values of CIP were reduced by one–threefold in the presence of CCCP in 25 out of 29 (86.21%) CIP-resistant isolates; among them, only 5 isolates (17.24%) showed reverse CIP resistance patterns (MIC = 0.5 μg/ml), while one–fourfold decrease in the TIG MICs was reported in 23 out of 25 (92%) TIG-resistant isolates. Based on these results, we continued the study by further molecular evaluation of the contribution of different mechanisms to the resistance to CIP and TIG, known to be substrates for the efflux pump system, in the 29 XDR and EtBr cartwheel positive Salmonella clinical isolates.
The existence of PMQR genes, qnrA, qnrB, qnrS, qepA, oqxA, oqxB, and aac(6′)-lb-cr, was examined in XDR Salmonella using conventional PCRs. Data analyses revealed that three XDR Salmonella isolates (10.34%) did not have any of the studied PMQR genes, while none of the isolates harbored all the PMQR genes. The majority of the isolates (89.66%) contained two to six PMQR genes. The qepA gene was the most frequent PMQR determinant (89.66%) followed by qnrS (62.07%), qnrA (55.17%), and qnrB (27.59%). Six isolates (20.69%) were tested positive for both oqxA and oqxB genes, whereas one isolate (3.45%) was tested positive for each of oqxA and oqxB genes. The aac(6′)-lb-cr was not detected in any of the tested Salmonella isolates (Table 3).
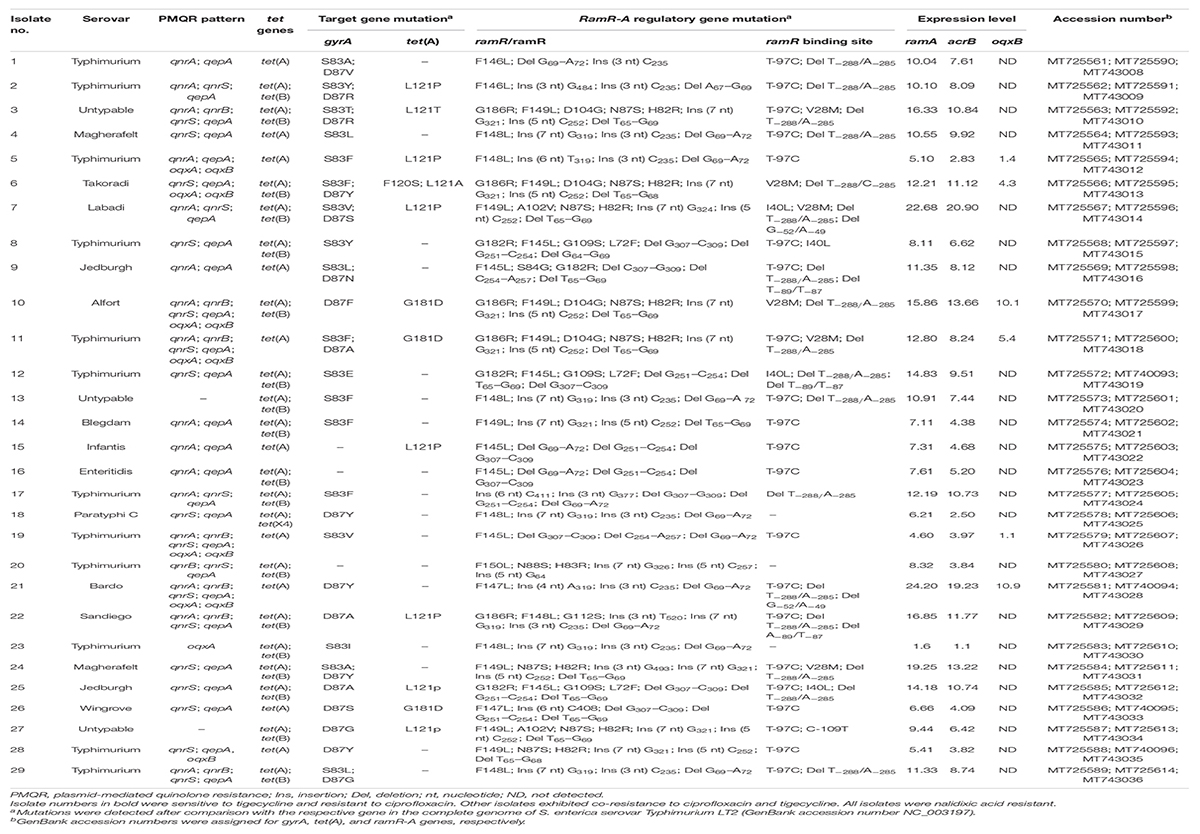
Table 3. Existence of plasmid-mediated quinolone resistance genes, tet genes, mutations in the target [gyrA, tet(A)] and regulatory (RamR-A) genes, and expression of efflux pump genes (ramA and acrB) in XDR Salmonella enterica serovars showing ciprofloxacin and/or tigecycline resistance.
DNA sequencing of the QRDRs of gyrA gene revealed specific stepwise point mutations in the topoisomerase target gene; those were associated with nalidixic acid and CIP resistance in 26 out of 29 (89.66%) XDR Salmonella isolates. As presented in Table 3, single mutations in gyrA gene either at S83 (9/26; 34.62%) or D87 (8/26; 30.77%) were detected in either low (MIC = 2 μg/ml; 5/29), moderate (MIC = 4–16; 8/29), or high (MIC = 32–128 μg/ml; 4/29) levels of CIP resistance. However, nine XDR isolates (CIP MIC = 16–64 μg/ml) were associated with double mutations at both positions. No mutation was reported in the QRDR of gyrA among three Salmonella isolates that showed nalidixic acid resistance and low-level CIP resistance (MIC = 2 μg/ml). The amino acid substitution at S83F was the most frequent mutation being detected in 23.08% of the isolates followed by D87Y (19.23%).
As depicted in Tables 2 and 3, Salmonella isolates (code Nos. 3, 11, and 29) contained double amino acid substitutions at S83 and D87 in QRDR of gyrA along with four to six PMQR determinants that had elevated MIC values for CIP (32–64 μg/ml). However, isolates of code Nos. 19 and 21 harbored single amino acid substitutions in QRDR of gyrA gene (S83V and D87Y, respectively) as well as six PMQR genes with CIP MICs of 4 and 128 μg/ml, respectively. Some isolates (code Nos. 15, 16, and 20) had up to three PMQR genes with no mutations in gyrA gene and showed a low level of CIP resistance (MIC = 2 μg/ml). On the other hand, other isolates of code Nos. 13, 23, and 27 had a single amino acid substitution, and no or just one PMQR gene showed moderate (MIC = 4–8 μg/ml) or low (MIC = 2 μg/ml) levels of CIP resistance. These results suggest that the acquisition of gyrA mutations as well as PMQR determinants were not the primary cause of the resistance to nalidixic acid and CIP in S. enterica serovars, and additional resistance mechanisms, such as enhanced efflux pump activity, may be involved.
The tet(A) and tet(B) genes were detected in 29 (100%) and 18 (62.07%) of the XDR Salmonella isolates, whereas tet(X4) gene was detected in only one isolate (3.45%) (code No. 18, Tables 2, 3), and tet(M) gene was not detected. To determine whether tet(A)-positive isolates had mutations in the tet(A) interdomain loop region, the amplicons (402 bp) from all isolates were sequenced. As shown in Tables 2, 3, comparisons of tet(A) gene sequences of Salmonella isolates with that of the wild type Salmonella Typhimurium reference strain (accession number NC_003197) revealed two-point mutations in the interdomain loop region of the efflux pump at codons 120 and 121 in eight TIG-resistant (MIC = 4–16 μg/ml) and one TIG-sensitive (MIC = 2 μg/ml) isolates. Moreover, a point mutation at codon 181 was observed in three isolates (MIC = 4–16 μg/ml); this important residue is located near the transmembrane domains of the efflux pump (codons 201–203) where mutations accumulated in these domains have a functional role in the efflux of TIG.
To investigate the impact of non-target mutations on the expression of the AcrAB-TolC efflux system and consequently on the level of CIP and TIG resistance, DNA sequencing of the ramRA regulatory locus of the 29 XDR Salmonella isolates was performed then compared with the wild-type ramRA gene of Salmonella Typhimurium reference strain (accession number NC_003197). As depicted in Table 3 and Supplementary Figure 1, screening for non-target mutations of the ramRA region revealed various amino acids alterations and frameshift mutations. In ramR gene open reading frame (ORF), the amino acid substitution F145–150 → L and the nucleotide deletion at G64-A72 were the predominant mutations, being detected in 28 (96.5% each) XDR isolates, followed by the insertion at the nucleotide T319–326 (62.07%). Meanwhile, deletion at T–288/A–285 was the highest detected mutation (17/29; 58.5%) in ramR binding region (BR). As shown in Supplementary Table 5, deletions in ORF and BR of ramR gene were found in 96% (24/25) and 68% (17/25) of the co-resistant XDR isolates, respectively, whereas amino acids substitutions in both regions were found in 96 and 36% of these isolates, respectively. It is interesting that the in-frame 4-bp deletion of T–288/A–285 dominated in ramR BR of the 17 (68%) co-resistant XDR Salmonella isolates displaying 2- to 32-fold and 2- to 4-fold increased resistance to CIP and TIG, respectively, than the non-mutated isolates.
To elucidate the role of enhanced efflux activity in CIP and TIG resistance, the relative expression levels of the global transcriptional regulator ramA and the transporter gene acrB of the AcrAB-TolC efflux system were assessed via qPCR. As shown in Table 3, the relative mRNA levels of ramA gene were maximal (exceeded 10-fold; range = 10.0–24.2, median = 12.8) in 17 out of 25 CIP/TIG co-resistant Salmonella isolates (68%) when compared to the pan-susceptible Salmonella Tamale control isolate (assigned a value of 1). Of these 17 co-resistant Salmonella isolates, 9 showed high expression levels for the acrB gene (range = 10.73–20.9; median = 11.8). None of the isolates that were uniquely resistant to CIP (n = 4) exhibited this overexpression in both genes. To further confirm the role of OqxAB efflux pump, we examined the relative expression levels of the oqxB gene in the oqxAB-positive isolates. Six isolates overexpressed the oqxB (range = 1.1–10.9; median = 4.85), that was more pronounced in four XDR isolates. Taking together the increased MIC values of CIP (2- to 32-fold) and TIG (2- to 16-fold), MAR and efflux activity indices (Table 2), and the maximal ramA and acrB expression levels for the above mentioned 17 CIP/TIG co-resistant isolates (Table 3), it is well suggested that enhanced AcrAB-TolC efflux pump activity was the most likely mechanism underlying CIP/TIG co-resistance in XDR S. enterica serovars.
Figure 2 shows the ranked significance of each genetic marker as a contributor to the occurrence of certain resistance patterns. The random forest classification model (Figure 2A) showed that ramA gene expression and deletion in ramR BR were the top genetic markers differentiating the CIP/TIG co-resistant isolates (n = 25) from those that showed resistance to CIP only (n = 4). The odds ratio calculation and Fisher’s exact test applied on the 25 co-resistant XDR isolates (Figure 2B) showed that the expression of ramA and acrB genes and deletion in the ramR BR were importantly (odds ratio = infinite) and significantly (p < 0.05) associated with the appearance of this phenotype.
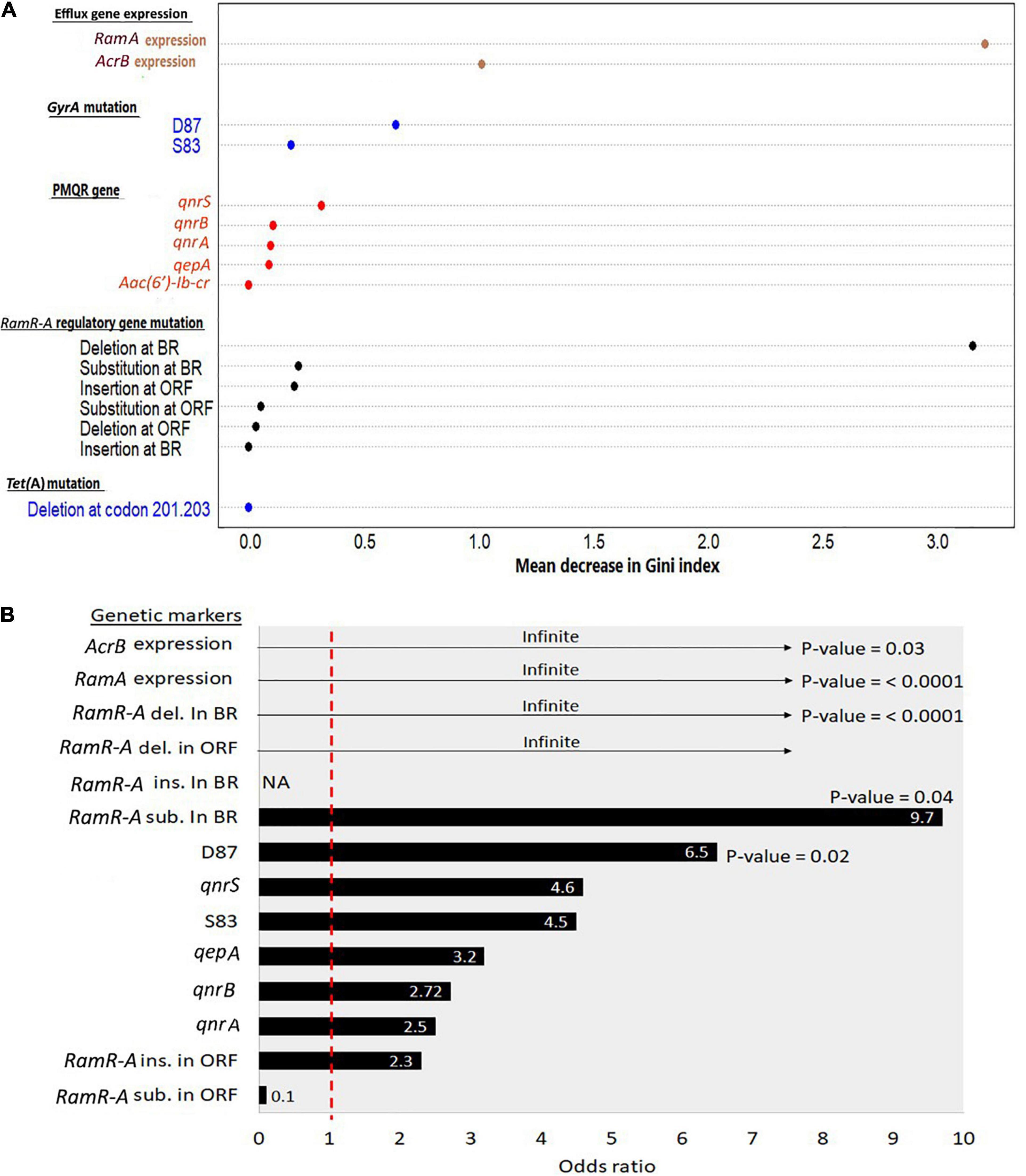
Figure 2. Ranked significance of genetic markers in the XDR Salmonella isolates that showed co-resistance to ciprofloxacin and tigecycline (n = 25). (A) Random forest classification analyses showing the importance of genetic markers in differentiating the co-resistant XDR Salmonella isolates from those that were resistant to CIP only (n = 4). The X-axis shows the mean decrease in Gini index for each marker. The higher the mean decrease in the Gini index, the more important the marker. (B) Significance of association of genetic markers and the co-resistance phenotype. The X-axis shows the odds ratio (at confidence intervals = 95%) of the respective genetic features that are shown on the Y-axis. Numbers within the bars refer to odds value. Numbers beneath the bars show the significance of association (p-value based on Fisher’s exact test at a cutoff level of 0.05) between the occurrence of certain genetic features and the appearance of the co-resistance to CIP and TIG in the 25 XDR isolates. The odds ratios that were shown as headed arrow indicate an infinite (∞) or maximum odds. The odds ratio of 1 (the vertical dashed red line) indicates no contribution of the respective genetic feature to the occurrence of the co-resistant phenotype. Del, deletion; Sub, substitution; Ins, insertion; BR, binding region; ORF, open reading frame.
The correlations among pairs of the genetic markers are shown in Figure 3. In the 25 co-resistant XDR Salmonella isolates, certain PMQR genes were significantly positively correlated as seen in qnrB-qnrA and qnrS-qepA (r = 0.5 each). In the four isolates that were uniquely resistant to CIP, the highest positive correlation was found between ramA expression and the deletion in its BR (r = 1; p < 0.05), followed by an intermediate positive significant correlation between some members of PMQR genes. The insertion and substitution in ramR gene ORF were highly negatively correlated (r = −0.5; p = 0.02).
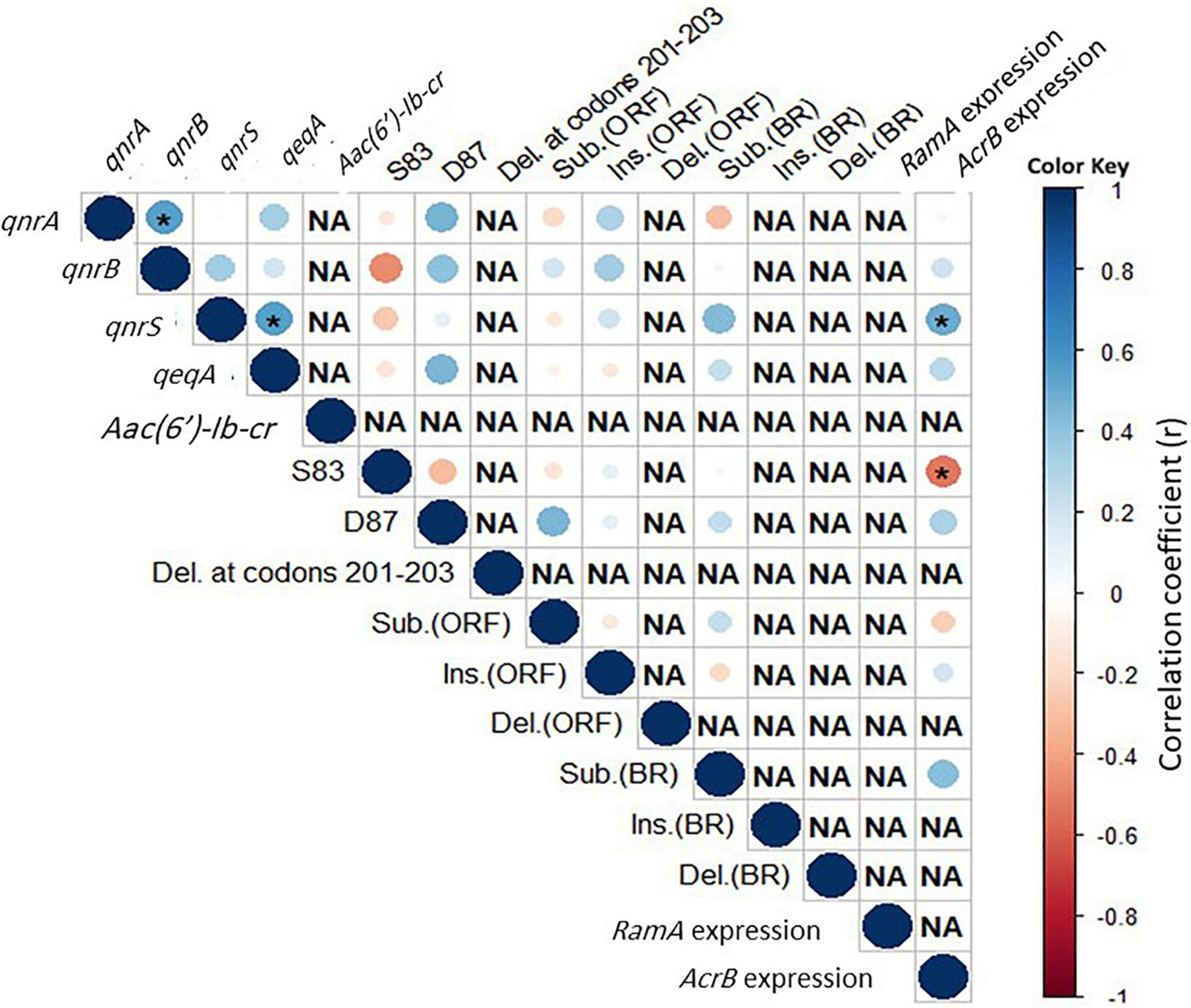
Figure 3. Correlation among genetic features in the co-resistant XDR Salmonella isolates (n = 25). Asterisks (*) indicate a significant correlation at a 0.05 p-value. The color scale refers to the correlation coefficient (blue and red colors indicate negative and positive correlations, respectively).
Regarding the isolation source, XDR Salmonella isolates belonging to human and poultry hosts were largely overlapped based on the resistance phenotypes and molecular characterization of the isolates (Figure 4). This is numerically evidenced by binary distance measured between pairs of isolates (Supplementary Table 6) belonging to different hosts or based on the average distances among hosts (0.3).
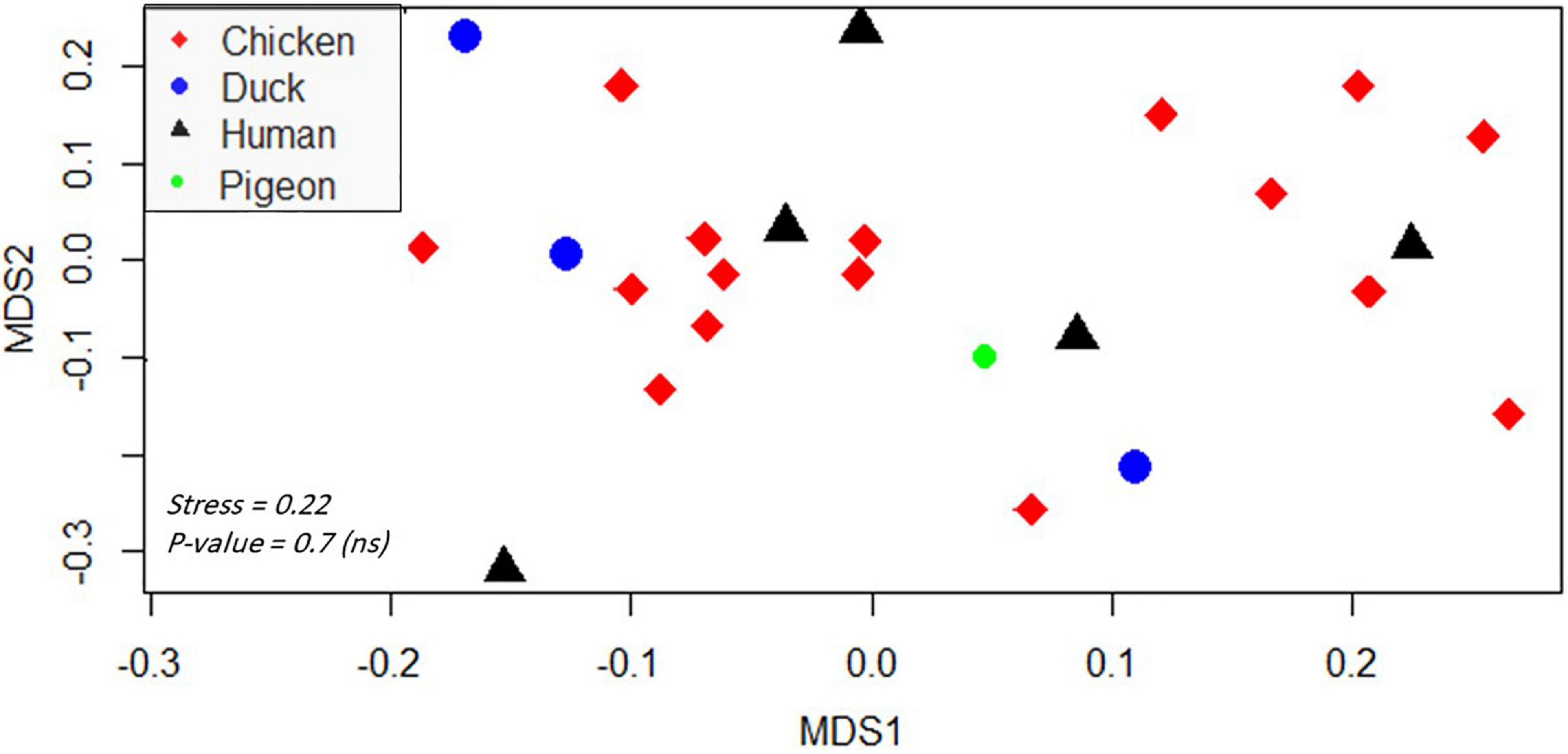
Figure 4. Non-metric multidimensional scaling showing the clustering and overlap of various isolates belonging to different hosts. Different hosts are color- and shape-coded. Stress refers to the performance of the multidimensional scaling. The p-value was calculated by PERMANOVA test and refers to the significant clustering of the groups. A p-value < 0.05 indicates that isolates were not significantly clustered based on their hosts (thus there is an overlap among hosts). The abbreviations of codes are described in Table 2 in detail.
Salmonellosis is one of the most frequent food-borne zoonoses globally, and the consumption of contaminated poultry meat is considered the main source of NTS infections in humans (Antunes et al., 2016). A tremendous increase in the antimicrobial resistance in S. enterica poses a significant global concern. While resistance to CIP in Salmonella isolates has been intensively studied, limited data are available on how resistance to CIP could confer resistance to other unrelated antimicrobials such as TIG, which is the last-resort anti-Salmonella drug. In the current study, we exploited molecular and data analysis approaches to directly investigate the molecular rationale behind the occurrence of co-resistance to CIP and TIG in XDR Salmonella isolates recovered from poultry and contact workers in Egypt.
The prevalence of Salmonella in chickens (11.54%) and pigeons (3.13%) in this study was lower than that reported in Northern China, where 20% of 1- to 4-week-old broiler chickens and 21.82% of pigeons were positive for Salmonella. Meanwhile, the prevalence among 1- to 4-week-old ducks (4.51%) (Wang et al., 2020) was nearly similar to that reported in our study (6.67%). The observed Salmonella prevalence among turkeys (4.17%) was lower than that previously reported in German fattening flocks (10.3%) (Käsbohrer et al., 2013). In contrast to the reported prevalence of Salmonella among quails (3.85%), a very high rate (75%) has been declared in Brazil (de Freitas Neto et al., 2013). These variations may be attributed to the geographical areas, climatic conditions, poultry species or breed, sample types, and disparity in sampling procedure and Salmonella isolation protocol. Serological identification of Salmonella isolates (n = 115) demonstrated that Salmonella Typhimurium (26.96%) and Salmonella Enteritidis (11.30%) were the most frequent serotypes in poultry and contact workers, which is consistent with previous reports from China (Wang et al., 2020), Egypt (Gharieb et al., 2015), and some European countries (Osimani et al., 2016).
Our data showed high resistance to amoxycillin-clavulanic acid (99.13%), cefazolin (94.78%), nalidixic acid (77.39%), cefoxitin (76.52%), tetracycline (74.78%), and cefepime (73.04%). Modest resistance rates were observed for CIP and TIG (51.30 and 24.35%, respectively). Plausible explanations of resistance are the indiscriminate use of antibiotics both in human and poultry husbandry, in addition to the easy accessibility to antibiotics in many countries worldwide, notably in Egypt. In contrast, lower resistance rates were observed for ertapenem (7.83%), imipenem (19.13%), meropenem (5.22%), and doripenem (2.61%) as there is no history of using the carbapenems for prevention or treatment in poultry farms in Egypt.
While MDR Salmonella represented the majority of our isolates (73.9%), we rather focused our analyses on the XDR isolates (n = 29; 25.22%) from poultry (n = 24), and contact workers (n = 5) because of the scarcity of data on this particular Salmonella phenotype in Egypt [despite its presence in other countries, e.g., Salmonella Typhi in Pakistan (Klemm et al., 2018; Saeed et al., 2019) and Salmonella Indiana in China (Wang et al., 2017)] and due to its seriousness in cases of outbreaks, as compared to MDR strains.
In the present study, we attempted to elucidate the co-resistance mechanism contributed to 25 CIP/TIG co-resistant Salmonella isolates. Our results revealed that the CCCP EPI was able to reverse the CIP and TIG resistance patterns for some examined Salmonella isolates. Therefore, we could not exclude the prospect that the AcrAB-TolC efflux pump might play a role in CIP/TIG resistance. In accordance with these findings, reduction in CIP and TIG MICs have been documented while using the CCCP EPI in previous studies (Deng et al., 2014; Zhong et al., 2014; Razavi et al., 2020). The twofold or greater decrease in the MIC levels for CIP- and TIG-resistant Salmonella isolates affirmed the involvement of active efflux components (Marimón et al., 2004). In a previous study, the CIP MICs decreased by twofold and those of nalidixic acid decreased by fivefold in the presence of EPIs, establishing the inclusion of an efflux pump in conversing quinolone resistance (Keddy et al., 2010). However, no complete reversion of the CIP resistance phenotype was detected with CCCP EPI in the work of Rushdy et al. (2013) implying the contribution of other mechanisms to this resistance, mainly, mutations in the target genes.
Bacterial resistance to fluoroquinolones is usually mediated by the acquisition of PMQR determinants (Robicsek et al., 2006) or mutations in bacterial DNA gyrase, particularly gyrA (Hopkins et al., 2005; Wasyl et al., 2014) or active efflux (Zhang et al., 2017). Herein, PMQR determinants were found in 27 XDR Salmonella isolates resistant to CIP (MICs ranging from 2 to 128 μg/ml), indicating the frequent occurrence of transferable quinolone resistance (Wasyl et al., 2014). This corresponds to the recent findings that all Enterobacteriaceae strains with high and intermediate resistance phenotypes to CIP (MICs = 1.5–512 μg/ml) harbored one or more PMQR genes (Kotb et al., 2019). This differs from the results of Szabó et al. (2018) who found that PMQR-positive Enterobacteriaceae strains were susceptible or intermediately resistant to CIP (MIC values between 0.06 and 1 mg/L). Notably, chromosomal copies of PMQR genes such as oqxAB, aac(6′)-Ib-cr, and qnrS genes have recently been discovered in CIP-resistant Salmonella strains (Chang et al., 2021). Thus, the presence of PMQR genes on the chromosome should not be excluded.
Previous results showed that fluoroquinolone resistance was attributed to amino acids substitutions at S83 or D87 codons within gyrA subunit, and these substitutions were not specific to certain Salmonella serovars (Robicsek et al., 2006; Chen et al., 2017). Our study revealed 16 different amino acids substitutions at both codons in 26 XDR S. enterica isolates representing 16 serovars, which were not associated with specific Salmonella serovar or CIP MIC value. Although quinolone resistance in Salmonella is firstly attributed to point mutations in the gyrA gene, other mutations of gyrB and parC genes may exist especially in higher-level resistance to fluoroquinolones. Oethinger et al. (2000) stated that the presence of gyrA mutations in the absence of active AcrAB efflux did not confer CIP resistance. This study’s shortcoming is not detecting gyrB, parC, and parE mutations; whether mutations within these genes also contribute to CIP resistance here is unknown. The distribution of gyrA mutations in the 25 co-resistant XDR isolates did not follow a specific pattern, and they were present in the other 4 XDR isolates showing resistance to CIP. This agrees with the results of random forest classification (i.e., having a low mean decrease in Gini index) and odds values.
Here, plasmid-encoded tet(X4) gene was detected in only one out of the 29 (3.45%) XDR Salmonella isolates (MIC = 4 μg/ml). Consistent with our results, the mobile tet(X) gene was recently detected in diverse pathogens resulting in TIG resistance, which is considered a public health concern (Li et al., 2020). Tuckman et al. (2000) stated that tet(A) mutations could induce TIG resistance in S. enterica isolates. Considering mutations in tet(A) gene, sequence analysis indicated that the point mutations of codons 120, 121, and 181 in 12 tet(A)-carrying isolates differed from the frameshift mutations of codons 201, 202, and 203 that reduced the sensitivity to TIG (Hentschke et al., 2010). Therefore, the contribution of these mutations to TIG resistance needs further investigation.
According to Linkevicius et al. (2016), the chemical change in position C-9 of the tet(B) efflux transporter gene cannot expel TIG out of the cytoplasm. In this study, the MICs of TIG for tet(B) positive isolates (n = 18) ranged from 0.5 to 16 μg/ml. TIG resistance in such tet(B)-carrying isolates may be attributed to tet(A) mutations or upregulation of ramA, acrB, and oqxB efflux pump genes (Table 3). A previous study indicated that the MIC for tet(B) positive S. enterica isolates (n = 13/49) ranged from 0.064 to 0.5 mg/L (Akiyama et al., 2013). They concluded that the mutations in tet(A) gene promote a low-level resistance of S. enterica isolates to TIG (MIC ranged from 0.19 to 3 mg/L), and additional resistance mechanisms such as ramA and ramR mutations could increase the TIG MIC to reach the resistance breakpoint.
Since the ramRA region have been reported to play a major role in the regulation of ramR and acrB genes expression in Salmonella and, consequently, in the efflux-mediated decreased susceptibility to antimicrobials (Abouzeed et al., 2008; Fàbrega et al., 2016), we investigated how mutations of ramRA could influence the transcription level of ramR and acrB genes and how this is linked to the occurrence of CIP/TIG co-resistance in XDR Salmonella. While this mechanism has been shown to play roles in Salmonella resistance to CIP (Fàbrega et al., 2016) or TIG (Chen et al., 2017), limited data are available on its contribution to the concurrent Salmonella resistance to these two drugs. A reason behind this information shortage could be the fact that TIG is a newly launched drug that has not yet received much attention. Another reason could be the lack of studies on XDR Salmonella, where such resistance phenotype infrequently occurs. In our analyses, deletions in ramR BR and ramA overexpression existed in 17 XDR co-resistant isolates. In addition, the random forest classification model, odds ratio, and Fisher’s exact test lend further support that these two determinants were the top two mechanisms occurring in the co-resistant XDR isolates. These data are consistent with a previous report (Fàbrega et al., 2016), although it is an in vitro study and involves MDR isolates. This suggests the importance of these two determinants as characteristic features of those co-resistant isolates (Figures 2A,B) and might also hint at a link between both markers as shown previously (Fàbrega et al., 2016). Coupling our data with the knowledge that ramR binds, through its BR, directly to the promoter of the ramA gene and thus controls its expression (Baucheron et al., 2012) puts forward the assumption that deletions in the ramR BR could impair its binding efficiency, leading to overexpression of ramA gene, and that the constitutive or concurrent occurrence of these two events are important for the occurrence of co-resistance to CIP and TIG in XDR Salmonella. This also in turn could enhance the expression of acrB, albeit to a low extent, and, thus, the increased MICs, MAR, and efflux activity indices. It is worth noting that although certain mutations (e.g., nucleotide deletions and amino acid substitutions in ramR ORF) were significantly associated with the appearance of the co-resistant phenotype and present in 96% of the 25 co-resistant isolates, respectively, their importance as determinants for such co-resistance is questionable because they also were present in the other four XDR isolates (Figure 1 and Table 3). While deletions in ramRA region have been linked to acrAB overexpression and could confer CIP (Abouzeed et al., 2008; Kehrenberg et al., 2009; Fàbrega et al., 2016) and TIG (Hentschke et al., 2010) resistance in Salmonella, it does not seem to be the case for the co-resistant isolates as acrB overexpression existed in only 36% of the co-resistant isolates.
Our analyses also pinpointed specific novel mutations at ramR BR in the co-resistant XDR isolates. This included deletion at T–288/A–285, which was detected in 68% of the co-resistant isolates (odds ratio = infinite and p-value of Fisher’s exact test ≤ 0.0001), suggesting the importance of this particular mutation in inducing such phenotype. We could not compare our results to others due to the scarcity of data on isolates exhibiting co-resistance to CIP and TIG. Similar results have also highlighted the role of ramA overexpression in the efflux-mediated MDR phenotype in Salmonella species (Fàbrega et al., 2016) and the positive correlation with the increased MICs of the antibiotics and expression of AcrAB efflux pump (Abouzeed et al., 2008). It is worthy of note that the genetic alterations reported in our study differed from those reported in the aforementioned studies and confirmed the hypothesis that sequence alterations may occur at various positions in the ramRA gene (Kehrenberg et al., 2009). Therefore, these mutations may have switched on the transcription of the efflux pump genes constitutively and subsequently triggered the appearance of CIP/TIG co-resistance phenotype of XDR Salmonella. Wong et al. (2015) reported that oqxAB, an RND efflux pump, is one of several endogenous efflux systems found in Klebsiella pneumoniae and Enterobacter species, with a role functionally comparable to that of acrAB in other Enterobacteriaceae members. A recent research reported that the oqxAB-bearing plasmid might cause Salmonella Typhimurium to develop a TIG-resistant phenotype. This phenomenon was probably related to the overexpression of MDR efflux pumps AcrAB-TolC and OqxAB (Chen et al., 2017). In this study, overexpression of the oqxB gene in four CIP/TIG co-resistant isolates indicated that the OqxAB efflux pump might be also involved in CIP and TIG co-resistance in XDR Salmonella. This is consistent with the findings of Zhong et al. (2014) that AcrAB-TolC efflux pump plays an important role in TIG resistance in K. pneumoniae strains with MICs of 8 μg/ml, whereas both the AcrAB-TolC and OqxAB efflux pumps contributed to the TIG resistance in strains with an MIC of 16 μg/ml.
This study highlights the prevalence of S. enterica serovars in poultry flocks and their contact workers, particularly of the alarming XDR phenotype. The knowledge gained from this study is highly relevant in the field of antimicrobial resistance in Salmonella in many aspects. First, our data are deemed as an initial step in delineating the molecular rationale behind co-resistance of Salmonella to CIP/TIG, opening doors for the experimental validation of the proposed role of AcrAB-TolC and OqxAB efflux pumps in TIG and CIP resistance. Second, it makes it possible to understand the emergence of TIG resistance in the absence of respective selection pressure, which could be seen clinically in cases where TIG-resistant isolates emerge in Salmonella-infected humans or animals with no previous exposure to TIG (Hentschke et al., 2010; Chen et al., 2017).
This is the first report that concludes a variety of XDR Salmonella serotypes circulate in poultry flocks and their contact workers in Egypt, with Salmonella Typhimurium having the highest frequency. Enhanced efflux pump activity, in particular the overexpression of ramA, plays a fundamental role in acrAB overexpression and facilitates the efflux-mediated CIP/TIG co-resistance with no previous exposure to TIG. These genetic alterations suggest a potential public health concern possibly associated with the poultry-to-human transfer of resistant bacteria on farms using the antimicrobials for treatment or non-therapeutic use or both.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) (approval number ZU-IACUC/2/F/12/2019). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) (approval number ZU-IACUC/2/F/12/2019). Written informed consent was obtained from the owners for the participation of their animals in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
NA and YT contributed equally to the conception and design of the study and participated with RG in the application of classical microbiological techniques. AE carried out all PCR assays and sequencing approaches and participated in the analysis of the sequences. MS performed the bioinformatics, established the figures, and participated with MAS in statistical analyses of the data. RG, AE, MS, EK, MAS, and AA conceived the study and participated in the design. NA and YT carried out the sequence analysis and participated in the data analysis. NA, YT, and MS wrote the initial draft of the manuscript. All authors revised the manuscript critically for important intellectual content. All authors gave the final approval of the version to be published.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.738784/full#supplementary-material
Supplementary Figure 1 | Frequency of different mutations in the ramR-A regulatory gene as identified in the 2- investigated isolate’s group (those resistant to CIP or to both CIP and TIG).
Abouzeed, Y. M., Baucheron, S., and Cloeckaert, A. (2008). ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52, 2428–2434. doi: 10.1128/AAC.00084-08
Akiyama, T., Presedo, J., and Khan, A. A. (2013). The tetA gene decreases tigecycline sensitivity of Salmonella enterica isolates. Int. J. Antimicrob. Agents. 42, 133–140. doi: 10.1016/j.ijantimicag.2013.04.017
Aldred, K. J., Kerns, R. J., and Osheroff, N. (2014). Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574. doi: 10.1021/bi5000564
Anbazhagan, P. V., Thavitiki, P. R., Varra, M., Annamalai, L., Putturu, R., Lakkineni, V. R., et al. (2019). Evaluation of efflux pump activity of multidrug-resistant Salmonella Typhimurium isolated from poultry wet markets in India. Infect. Drug Resist. 12, 1081–1088. doi: 10.2147/IDR.S185081
Antunes, P., Mourão, J., Campos, J., and Peixe, L. (2016). Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 110–121. doi: 10.1016/j.cmi.2015.12.004
Baucheron, S., Coste, F., Canepa, S., Maurel, M. C., Giraud, E., Culard, F., et al. (2012). Binding of the RamR repressor to wild-type and mutated promoters of the ramA gene involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 56, 942–948. doi: 10.1128/AAC.05444-11
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496.
Botteldoorn, N., Van Coillie, E., Grijspeerdt, K., Werbrouck, H., Haesebrouck, F., Donné, E., et al. (2006). Real-time reverse transcription PCR for the quantification of the mntH expression of Salmonella enterica as a function of growth phase and phagosome-like conditions. J. Microbiol. Methods 66, 125–135. doi: 10.1016/j.mimet.2005.11.003
Cattoir, V., Poirel, L., and Nordmann, P. (2008). Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804. doi: 10.1128/AAC.00638-08
Chang, M.-X., Zhang, J.-F., Sun, Y.-H., Li, R.-S., Lin, X.-L., Yang, L., et al. (2021). Contribution of different mechanisms to ciprofloxacin resistance in Salmonella spp. Front. Microbiol. 12:663731. doi: 10.3389/fmicb.2021.663731
Chen, Y., Hu, D., Zhang, Q., Liao, X. P., Liu, Y. H., and Sun, J. (2017). Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell Infect. Microbiol. 7:1–8. doi: 10.3389/fcimb.2017.00037
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Clinical Laboratory Standards Institute (CLSI) (2019). Performance Standard for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100. Wayne, PA: CLSI.
de Freitas Neto, O. C., da Angela, H. L., Soares, N. M., Guastalli, E. A. L., de Almeida, A. M., and Berchieri Junior, A. (2013). Salmonella spp. in meat-type quails (coturnix coturnix coturnix) in the state of são paulo, brazil. Rev. Bras. Cienc. Avic. 15, 277–281. doi: 10.1590/S1516-635X2013000300016
Deng, M., Zhu, M. H., Li, J. J., Bi, S., Sheng, Z. K., Hu, F. S., et al. (2014). Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob. Agents Chemother. 58, 297–303. doi: 10.1128/AAC.01727-13
European Committee Antimicrobial Susceptibility Testing (EUCAST) (2018). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.1.
Fàbrega, A., Ballesté-Delpierre, C., and Vila, J. (2016). Differential impact of ramRA mutations on both ramA transcription and decreased antimicrobial susceptibility in Salmonella Typhimurium. J. Antimicrob. Chemother. 71, 617–624. doi: 10.1093/jac/dkv410
Gharieb, R. M., Tartor, Y. H., and Khedr, M. H. E. (2015). Non-typhoidal Salmonella in poultry meat and diarrhoeic patients: Prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 7:34. doi: 10.1186/s13099-015-0081-1
Hentschke, M., Christner, M., Sobottka, I., Aepfelbacher, M., and Rohde, H. (2010). Combined ramR mutation and presence of a Tn1721-associated tet(A) variant in a clinical isolate of Salmonella enterica serovar hadar resistant to tigecycline. Antimicrob. Agents Chemother. 54, 1319–1322. doi: 10.1128/AAC.00993-09
Hoelzer, K., Switt, A. I. M., and Wiedmann, M. (2011). Animal contact as a source of human non-typhoidal salmonellosis. Vet. Res. 42:34. doi: 10.1186/1297-9716-42-34
Hopkins, K. L., Davies, R. H., and Threlfall, E. J. (2005). Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25, 358–373. doi: 10.1016/j.ijantimicag.2005.02.006
ISO (2017). Microbiology of the Food ChainHorizontal Method for the Detection, Enumeration and Serotyping of Salmonella - ISO 6579-1.
Ji, K., Xu, Y., Sun, J., Huang, M., Jia, X., Jiang, C., et al. (2020). Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 11, 49–56. doi: 10.1080/21505594.2019.1706913
Käsbohrer, A., Schroeter, A., Helmuth, R., and Tenhagen, B. A. (2013). Salmonella prevalence in turkey flocks before and after implementation of the control program in Germany. Agriculture 3, 342–361. doi: 10.3390/agriculture3030342
Kauffmann, F. (1957). The Kauffmann-White Schema; Diagnostic Salmonella Antigen Schema. Copenhagen: Munksgaard, 126.
Keddy, K. H., Smith, A. M., Sooka, A., Ismail, H., and Oliver, S. (2010). Fluoroquinolone resistant typhoid, south Africa. Emerg. Infect. Dis. 16, 879–880.
Kehrenberg, C., Cloeckaert, A., Klein, G., and Schwarz, S. (2009). Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J. Antimicrob. Chemother. 64, 1175–1180. doi: 10.1093/jac/dkp347
Khoshbakht, R., Derakhshandeh, A., Jelviz, L., and Azhdari, F. (2018). Tetracycline resistance genes in Salmonella enterica serovars with animal and human origin. Int. J. enteric. Pathog. 6, 60–64. doi: 10.15171/ijep.2018.17
Kim, H. B., Wang, M., Park, C. H., Kim, E. C., Jacoby, G. A., and Hooper, D. C. (2009). oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. doi: 10.1128/AAC.01574-08
Klemm, E. J., Shakoor, S., Page, A. J., Qamar, F. N., Judge, K., Saeed, D. K., et al. (2018). Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio 9, 1–10. doi: 10.1128/mBio.00105-18
Kotb, D. N., Mahdy, W. K., Mahmoud, M. S., and Khairy, R. M. M. (2019). Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis. 19:979. doi: 10.1186/s12879-019-4606-y
Li, Q., Sherwood, J. S., and Logue, C. M. (2007). Characterization of antimicrobial resistant Escherichia coli isolated from processed bison carcasses. J. Appl. Microbiol. 103, 2361–2369. doi: 10.1111/j.1365-2672.2007.03470.x
Li, R., Lu, X., Peng, K., Liu, Z., Li, Y., Liu, Y., et al. (2020). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems 5, e134–e120. doi: 10.1128/msystems.00134-20
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2016). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/AAC.02465-15
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lunn, A. D., Fàbrega, A., Sánchez-Céspedes, J., and Vila, J. (2010). Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int. Microbiol. 13, 15–20. doi: 10.2436/20.1501.01.107
Magiorakos, A., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268281. doi: 10.1111/j.14690691.2011.03570.x
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of non-typhoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Marimón, J. M., Gomáriz, M., Zigorraga, C., Cilla, G., and Pérez, T. E. (2004). Increasing prevalence of quinolone resistance in human nontyphoid Salmonella enterica isolates obtained in Spain from 1981 to 2003. Antimicrob. Agents Chemother. 48, 3789–3793.
Martins, M., Viveiros, M., Couto, I., Costa, S. S., Pacheco, T., Fanning, S., et al. (2011). Identification of efflux pump-mediated multidrug-resistant bacteria by the ethidium bromide-agar cartwheel method. In Vivo 25, 171–178.
Oethinger, M., Kern, W. V., Jellen-Ritter, A. S., McMurry, L. M., and Levy, S. B. (2000). Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44, 10–13. doi: 10.1128/AAC.44.1.10-13.2000
Oliveira, S. D., Rodenbusch, C. R., Cé, M. C., Rocha, S. L. S., and Canal, C. W. (2003). Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Lett. Appl. Microbiol. 36, 217–221. doi: 10.1046/j.1472-765x.2003.01294.x
Osimani, A., Aquilanti, L., and Clementi, F. (2016). Salmonellosis associated with mass catering: a survey of European union cases over a 15-year period. Epidemiol. Infect. 144, 3000–3012. doi: 10.1017/S0950268816001540
Razavi, S., Mirnejad, R., and Babapour, E. (2020). Involvement of AcrAB and OqxAB efflux pumps in antimicrobial resistance of clinical isolates of Klebsiella pneumonia. J. Appl. Biotechnol. Reports 7, 251–257. doi: 10.30491/jabr.2020.120179
Robicsek, A., Strahilevitz, J., Jacoby, G. A., Macielag, M., Abbanat, D., Chi, H. P., et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. doi: 10.1038/nm1347
Rushdy, A. A., Mabrouk, M. I., Abu-Sef, F. A., Kheiralla, Z. H., Mohamed Abdel-All, S., and Saleh, N. M. (2013). Contribution of different mechanisms to the resistance to fluoroquinolones in clinical isolates of Salmonella enterica. Braz. J. Infect. Dis. 17, 431–437. doi: 10.1016/j.bjid.2012.11.012
Saeed, N., Usman, M., and Khan, E. A. (2019). An overview of extensively drug-resistant Salmonella Typhi from a tertiary care hospital in Pakistan. Cureus 11:e5663. doi: 10.7759/cureus.5663
Szabó, O., Gulyás, D., Szabó, N., Kristóf, K., Kocsis, B., and Szabó, D. (2018). Plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from urine clinical samples. Acta Microbiol. Immunol. Hung. 65, 255–265. doi: 10.1556/030.65.2018.012
Tambekar, D., Dhanorkar, D., Gulhane, S., Khandelwal, V., and Dudhane, M. (2006). Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. Afr. J. Biotechnol. 5, 1562–1265. doi: 10.4314/AJB.V5I17.43162
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thomas, M., Fenske, G. J., Antony, L., Ghimire, S., Welsh, R., Ramachandran, A., et al. (2017). Whole genome sequencing-based detection of antimicrobial resistance and virulence in non-typhoidal Salmonella enterica isolated from wildlife. Gut Pathog. 9, 1–9. doi: 10.1186/s13099-017-0213-x
Tuckman, M., Petersen, P., and Projan, S. (2000). Mutations in the interdomain loop region of the tetA (A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines. Microb. Drug Resist. 6, 277–282. doi: 10.1089/mdr.2000.6.277
Usui, M., Nagai, H., Hiki, M., Tamura, Y., and Asai, T. (2013). Effect of antimicrobial exposure on AcrAB expression in Salmonella enterica subspecies enterica serovar Choleraesuis. Front. Microbiol. 4:1–6. doi: 10.3389/fmicb.2013.00053
Van, T. T., Chin, J., Chapman, T., Tran, L. T., and Coloe, P. J. (2008). Safety of raw meat and shellfish in vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 124, 217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029
Wang, J., Li, J., Liu, F., Cheng, Y., and Su, J. (2020). Characterization of Salmonella enterica isolates from diseased poultry in northern China between 2014 and 2018. Pathogens 9:95. doi: 10.3390/pathogens9020095
Wang, W., Peng, Z., Baloch, Z., Hu, Y., Xu, J., Zhang, W., et al. (2017). Genomic characterization of an extensively-drug resistance Salmonella enterica serotype Indiana strain harboring blaNDM–1 gene isolated from a chicken carcass in China. Microbiol. Res. 204, 48–54. doi: 10.1016/j.micres.2017.07.006
Wasyl, D., Hoszowski, A., and Zajac, M. (2014). Prevalence and characterization of quinolone resistance mechanisms in Salmonella spp. Vet. Microbiol. 171, 307–314. doi: 10.1016/j.vetmic.2014.01.040
Wong, M. H., Chan, E. W., and Chen, S. (2015). Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob. Agents Chemother. 59, 3290–3297. doi: 10.1128/AAC.00310-15
Yang, B., Xi, M., Cui, S., Zhang, X., Shen, J., Sheng, M., et al. (2012). Mutations in gyrase and topoisomerase genes associated with fluoroquinolone resistance in Salmonella serovars from retail meats. Food Res. Int. 45, 935–939. doi: 10.1016/j.foodres.2011.01.031
Zhang, W. H., Zhang, C. Z., Liu, Z. J., Gu, X. X., Li, W., Yang, L., et al. (2017). In vitro development of ciprofloxacin resistance of Salmonella enterica serovars Typhimurium, enteritidis, and Indiana isolates from food animals. Microb. Drug Resist. 23, 687–694. doi: 10.1089/mdr.2016.0119
Zheng, J., Cui, S., and Meng, J. (2009). Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J. Antimicrob. Chemother. 63, 95–102. doi: 10.1093/jac/dkn448
Keywords: XDR, Salmonella enterica, AcrAB efflux pump, ciprofloxacin, tigecycline
Citation: Abd El-Aziz NK, Tartor YH, Gharieb RMA, Erfan AM, Khalifa E, Said MA, Ammar AM and Samir M (2021) Extensive Drug-Resistant Salmonella enterica Isolated From Poultry and Humans: Prevalence and Molecular Determinants Behind the Co-resistance to Ciprofloxacin and Tigecycline. Front. Microbiol. 12:738784. doi: 10.3389/fmicb.2021.738784
Received: 21 July 2021; Accepted: 14 October 2021;
Published: 25 November 2021.
Edited by:
Jing Wang, Yangzhou University, ChinaReviewed by:
Liang-xing Fang, South China Agricultural University, ChinaCopyright © 2021 Abd El-Aziz, Tartor, Gharieb, Erfan, Khalifa, Said, Ammar and Samir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasmine H. Tartor, amFzbWVuMjFAeWFob28uY29t; eWFzbWluZWh0YXJ0b3JAenUuZWR1LmVn; orcid.org/0000-0003-1246-6548
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.