
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 20 January 2022
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.738470
This article is part of the Research Topic Microbial Stress: From Model Organisms to Applications in Food, Microbiotechnology and Medicine View all 17 articles
The pathogenic microorganism Listeria monocytogenes is ubiquitous and responsible for listeriosis, a disease with a high mortality rate in susceptible people. It can persist in different habitats, including the farm environment, the food production environments, and in foods. This pathogen can grow under challenging conditions, such as low pH, low temperatures, and high salt concentrations. However, L. monocytogenes has a high degree of strain divergence regarding virulence potential, environmental adaption, and stress response. This review seeks to provide the reader with an up-to-date overview of clonal and serotype-specific differences among L. monocytogenes strains. Emphasis on the genes and genomic islands responsible for virulence and resistance to environmental stresses is given to explain the complex adaptation among L. monocytogenes strains. Moreover, we highlight the use of advanced diagnostic technologies, such as whole-genome sequencing, to fine-tune quantitative microbiological risk assessment for better control of listeriosis.
Listeria monocytogenes is a Gram-positive, facultative anaerobe, non-spore-forming, and psychro- and salt-tolerant organism. It is also a facultative intracellular pathogen, both for humans and animals. In susceptible people, including immunocompromised persons, infants, pregnant women, and older people, it can cause clinical manifestations with high mortality rates (Desai et al., 2019; Kayode et al., 2019; Schlech, 2019). Cases of human listeriosis often can be traced back to food products contaminated during production, on which the microorganism grows to high numbers. Special concern is in particular given to ready-to-eat (RTE) products, such as salads, deli meat, or smoked salmon, because these are consumed without a further heating step (Leong et al., 2015; EFSA Panel on Biological Hazards [Efsa Biohaz Panel]et al., 2018). Given the ubiquitous distribution of this microorganism, its transmission into food-processing facilities occurs either via raw materials or via equipment and employees. Once introduced in the facilities, several factors have been suggested to contribute to the ability of a strain to establish long-lasting colonization (Holch et al., 2013; Leong et al., 2014; Lakicevic and Nastasijevic, 2017; Stoller et al., 2019). Some authors hypothesized that a particular feature that makes the control of L. monocytogenes difficult in the processing environment is its capacity to survive or even to grow in different stressful conditions and to form biofilms (Gandhi and Chikindas, 2007; Ferreira et al., 2014; Gahan and Hill, 2014; Ariza-Miguel et al., 2015). The fast ability of L. monocytogenes to colonize food-processing facilities and the formation of persisters of some L. monocytogenes strains in various niches along the food chain have been described (Berrang et al., 2010; Leong et al., 2014; Bolocan et al., 2016). This suggests that persister cells provide reservoirs for contamination, ultimately increasing the likelihood of infecting humans. An alternative hypothesis is that there are no strains of L. monocytogenes with unique attributes but hard-to-reach areas (also known as harborage sites) in food industry environments and equipment where L. monocytogenes can reside (Carpentier and Cerf, 2011). Furthermore, tolerance to sanitizers and disinfectants such as benzalkonium chloride (BC) was observed in L. monocytogenes isolates from food-processing environments. This tolerance may be attributed to subinhibitory concentrations of a disinfectant, which are caused by insufficient cleaning and improper sanitation, thus probably contributing to biofilm formation and leading to Listeria persistence (Martínez-Suárez et al., 2016). The same authors have hypothesized that these subinhibitory concentrations cause the expression of stress response genes leading to a reduction in cell permeability to these compounds. These genetic traits include resistance genes such as the qacH gene of transposon Tn6188 and the resistance cassette bcrABC (Elhanafi et al., 2010; Müller et al., 2013, 2014; Zuber et al., 2019), described in Persistence mechanisms. Moreover, it is crucial to identify the interactions between stress response and virulence and to know how this microorganism survives, adapts to adverse conditions, and triggers genes involved in virulence or promoting persistence. This would help to explain the observed inverse correlation between strains with a higher prevalence of genes involved in BC tolerance, as well as other stress-related genes, amongst hypovirulent (i.e., low virulence) lineage II strains (Quereda et al., 2021). All this knowledge may contribute to the development of new intervention strategies for better control of the level of L. monocytogenes in the food chain.
L. monocytogenes evolves slowly but has been characterized by a significant level of diversity (Ragon et al., 2008; Orsi et al., 2011). It can be grouped into four major evolutionary lineages indicated by the roman numbers from I to IV, by 14 lineage-related serotypes and more than 170 clonal complexes (CCs),1 geographically and temporally widespread, as defined by multilocus sequence typing, and whole-genome phylogenetic analysis (Doumith et al., 2004; Orsi et al., 2011; Haase et al., 2014; Doijad et al., 2015; Chen et al., 2016; Moura et al., 2016; Bergholz et al., 2018). It belongs to the genus that currently includes 26 recognized species, of which notably 20 have been described since 2009 (Graves et al., 2010; Leclercq et al., 2010, 2019; Bertsch et al., 2013; Lang Halter et al., 2013; den Bakker et al., 2014; Weller et al., 2015; Doijad et al., 2018; Nuñez-Montero et al., 2018; Quereda et al., 2020; Carlin et al., 2021). Lineage I of L. monocytogenes encompasses serotypes 1/2b, 3b, 4b, 4d, 4e, and 7, lineage II includes serotypes 1/2a, 1/2c, 3a, 3c, and 4h, lineage III comprises serotypes 4a, atypical 4b, and 4c, whereas lineage IV encompasses serotypes 4a and 4c (Seeliger and Hohne, 1979; Ragon et al., 2008; Maury et al., 2016; Painset et al., 2019; Yin et al., 2019).
Previous studies have found that some hypervirulent clones such as CC1, CC2, CC4, and CC6 (all of lineage I and predominant in Western countries) were strongly associated with listeriosis, whereas hypovirulent clones, including CC8, CC9, CC101, CC121, and CC204 (lineage II), were strongly associated with food product contamination but less with human infections, in part due to loss-of-function mutations in virulence genes (Fagerlund et al., 2016; Maury et al., 2016, 2019). In accordance with that, all CC2 isolates carried a full-length inlA gene (see later), whereas CC9 and CC121 presented a premature stop codon mutation in this gene that correlated with reduced virulence (Gelbíčová et al., 2015; Guidi et al., 2021). Maury et al. (2019) demonstrated that differences in product associations among clones might be attributed to adaptation differences between clones in distinct ecological niches and/or different food product contamination routes during processing. They showed that CC1 was more representative for dairy products, whereas hypovirulent clones, mainly CC9 and CC121, were strongly associated with meat and fish products and produced more biofilm in the presence of low BC concentrations. Hypervirulent strains of L. monocytogenes sequence type (ST) 6 have been associated with outbreaks, including an outbreak linked to frozen vegetables in five countries in Europe during 2015–2018, an outbreak associated with contaminated meat pâté in Switzerland during 2016, listeriosis outbreak that occurred in South Africa during 2017–2018 with a 27% mortality rate, and the largest outbreak of listeriosis in Germany linked to blood sausages in 2018–2019 (Althaus et al., 2017; EFSA and ECDC, 2018; Halbedel et al., 2020; Thomas et al., 2020). More recently, an outbreak of listeriosis was caused by the persistence of L. monocytogenes serotype 4b ST6 (lineage I) in a cheese-processing facility in Switzerland (Nüesch-Inderbinen et al., 2021).
To cause listeriosis in humans and animals, important genes must be present in L. monocytogenes genome and expressed under the appropriate conditions. The inlAB locus and the pathogenicity islands LIPI-1, LIPI-3, and LIPI-4 encode such key virulence factors (Gelbíčová et al., 2015; Maury et al., 2016; Quereda et al., 2018). In particular, inlA and inlB code for internalin A (InlA) and internalin B (InlB) that bind the host cell receptors E-cadherin and Met, respectively. The transcriptional regulator positive regulator factor A (PrfA) controls the expression of both inlAB and LIPI-1 (Quereda et al., 2018).
LIPI-1, present in all L. monocytogenes, is found between the genes prs and orfX, is 9-kb long, and consists of six genes, i.e., prfA, plcA, hly, mpl, actA, and plcB (Dussurget, 2008). Notably, Listeria innocua, a non-pathogenic Listeria species, lacks both LIPI-1 and inlAB genes; however, LIPI-1 and inlA, functional both in vivo and in vitro, are present in rare, natural, atypical L. innocua species (Johnson et al., 2004; Volokhov et al., 2007; den Bakker et al., 2010; Moreno et al., 2012; Moura et al., 2019). This suggested that L. monocytogenes and L. innocua likely evolved from a common ancestor where the virulence loci LIPI-1 and inlAB were both present (Volokhov et al., 2007). L. innocua FSL J1-023 is one such aberrant strain, described as a rare, natural, non-pathogenic, hemolytic-positive, rhamnose and xylose fermentation-negative strain; its genome sequence is the reference one for linking horizontal gene transfer and recombination as drivers in the evolution of Listeria pathogenicity (Johnson et al., 2004; Lakicevic et al., 2014). Notably, all L. innocua genomes lack other internalins (i.e., inlCEFGHJKP), and this suggested that, unlike inlAB, the former genes were not present in the common ancestor and acquired at a later stage by L. monocytogenes (Moura et al., 2019).
As for LIPI-2, this pathogenicity island was discovered in Listeria ivanovii, a species pathogenic for feedstocks, mostly ovines and bovines, and humans, though rarely (Guillet et al., 2010). Originally, it was described as a LIPI specific to L. ivanovii. It encompasses the smcL gene coding for sphingomyelinase (involved in phagosome disruption) and 10 genes coding for proteins of the internalin family (Domínguez-Bernal et al., 2006). However, recent studies showed that L. monocytogenes isolates, belonging to a new sub-lineage of the major lineage II with hypervirulent features (SL626/CC33, serovar 4 h), contain a truncated LIPI-2, i.e., carrying only smcL and two internalins genes, namely i-inlF and i-inlE, likely acquired by transposon-mediated horizontal gene transfer from L. ivanovii (Yin et al., 2019; Feng et al., 2020).
LIPI-3 is an additional sub-lineage pathogenicity island encoding listeriolysin S (LLS), a bacteriocin. LLS (coded by llsA), a hemolytic toxin secreted by L. monocytogenes, is present only in a subset of isolates from lineage I epidemic strains that specifically secrete it in the gut (Quereda et al., 2017). Notably, Quereda et al. (2016) demonstrated that in an orally infected mouse model, L. monocytogenes lls mutants exhibited reduced bacterial load in the intestinal content at 6 h post-infection as compared with the wild-type strain, and the differences were also evident at 24 and 48 h post-infection and correlated with the reduced number of intracellular bacteria. Moreover, the same authors showed that the presence of L. monocytogenes-produced LLS in the intestine of the infected mice caused a significant decrease in the occurrence of different bacterial genera, such as Alloprevotella, Allobaculum, and Streptococcus. These results, for the first time, provided evidence that LLS plays an important role in the interaction with other species in the gut microbiota. In a study conducted by Matle et al. (2020), LIPI-3 was detected in isolates of which the majority were from lineage I, i.e., CC1, CC2, CC3, and CC228. Also, Roedel et al. (2019) found LIPI-3 in L. monocytogenes isolates belonging to CC1, CC3, CC4, CC6, and CC288. Tavares et al. (2020) described single-nucleotide polymorphism (SNP) of eight LIPI-3 genes (llsAGHXBYDP) of the four different STs (ST1, ST3, ST218, and ST288) compared with reference strain F2365 (lineage I). The authors revealed that LIPI-3 genes are well conserved in ST1 (serogroup IVb), whereas a number of SNPs were identified in ST3 (serogroup IIb), ST218 (serogroup IVb-v1), and ST288 (serogroup IIb). Within the LIPI-3 island, llsX is the only gene that is highly conserved among different LIPI-3-positive L. monocytogenes CC (Figure 1) and even in atypical hemolytic L. innocua (Figure 1). However, under acid stress conditions, only reference strain F2365 (4b) presented expression of llsX comparable with ST3 and ST288 (serogroup IIb, lineage I) strains, which points to acidic pH as an important environmental trigger (Clayton et al., 2014).
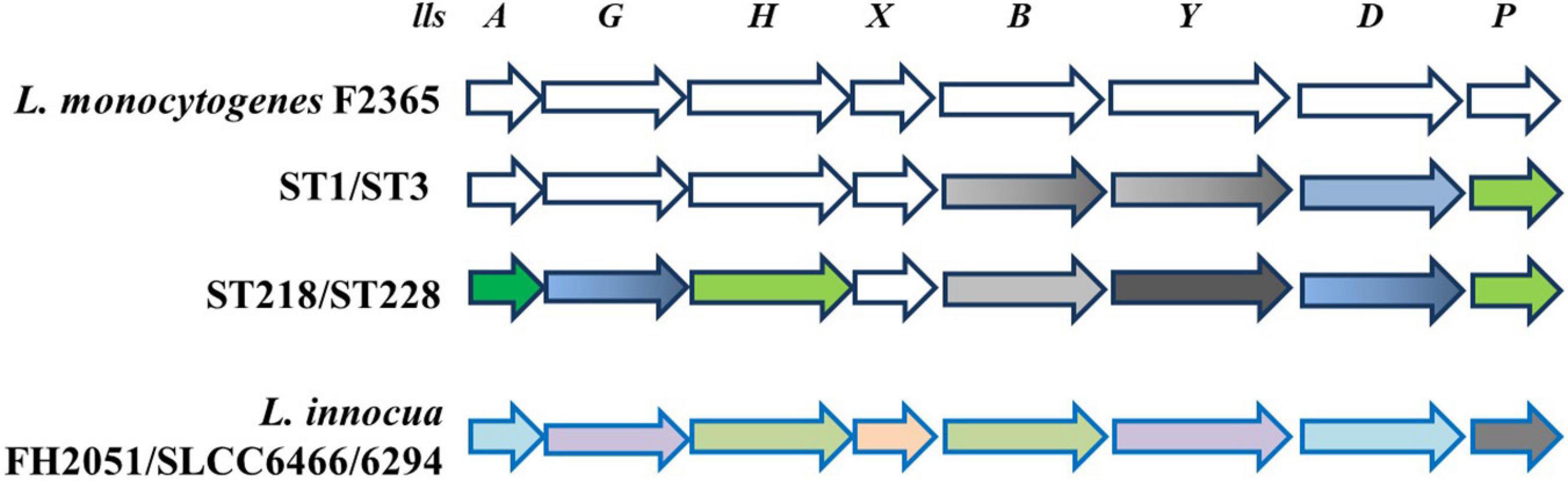
Figure 1. Schematic representation of LIPI-3 pathogenicity island. In L. monocytogenes reference strain F2365 and different STs, all LIPI-3 genes display SNPs, except llsX, which is strongly conserved. SNPs are shown as follows: 0—white; 1—gray; 2—green; 4—dark green; 10–17—blue; 18—black. When color is shaded, SNPs are present only in one of two ST indicated. Bottom line shows that LIPI-3 is also present in some strains of L. innocua, where sequence identity with L. monocytogenes reference strain F2365 ranges from 91% (llsP, gray) to 95% (llsY and llsG, violet), 97% (llsX, pink), 98% (llsA and llsD, light blue), and 99% (llsH and llsB, light green) (adapted from Clayton et al., 2014; Tavares et al., 2020).
LIPI-4 pathogenic island is a cluster of six genes encoding a putative cellobiose family phosphotransferase system and shown to confer hypervirulence by enhancing invasion of the CNS and placenta (Maury et al., 2016). It is only found in some isolates of lineage I, i.e., CC2, CC4, and CC87 (Maury et al., 2016; Chen et al., 2018, 2020; Hilliard et al., 2018; Painset et al., 2019; Roedel et al., 2019; Matle et al., 2020; Zhang et al., 2020). It is known that CC2 has a worldwide distribution, whereas CC4 and CC87 are the most prevalent clones in France and China (Maury et al., 2016; Zhang et al., 2020). The latter CC was also responsible for two outbreaks in Guipúzcoa (Northern Spain) in 2013 and 2014 (Pérez-Trallero et al., 2014; Wang et al., 2019). In addition to LIPI-4, all CC87 strains contained a novel type II restriction–modification system with unknown significance (Wang et al., 2019).
Anaerobiosis represents an important trigger for virulence determinants because, in the gastrointestinal tract, the oxygen level gradually decreases and favors facultative anaerobic microorganisms such as L. monocytogenes (Horn and Bhunia, 2018). Indeed, Müller-Herbst et al. (2014) detected 28 non-essential genes that were upregulated only anaerobically, of which a subset were virulence-related genes, e.g., inlB and Listeria adhesion protein that is essential for full virulence (Burkholder et al., 2009). A complementary screening of an insertion mutant library of L. monocytogenes demonstrated that F1F0-ATPase (see FoF1-ATPase, Glutamate decarboxylase, and Arginine and Agmatine deiminases) is essential for anaerobic proliferation of L. monocytogenes (Müller-Herbst et al., 2014). Anaerobiosis also induced an acid tolerance response (Sewell et al., 2015), providing L. monocytogenes robustness to survive the stomach acidity and transit to the intestine. According to that, some CCs are more tolerant to low pH and colonize better the intestinal lumen (also known as host-associated hypervirulent clones) than those of hypovirulent CCs (Hingston et al., 2017; Maury et al., 2019).
All this information highlight that hypervirulent clones (CC1, CC2, CC4, and CC6) greatly connect with clinical cases, are predominantly found in lineage I, colonize better in the intestine lumen, cause listeriosis in a healthy host, and are strongly associated with dairy products. On the contrary, hypovirulent clones (e.g., CC9 and CC121), greatly connected with food and food-related isolates, are predominantly found in lineage II, causing listeriosis in immunocompromised patients, and are strongly associated with meat and fish products. All lineages of L. monocytogenes possess highly conserved LIPI-1 and inlA/B locus, which fits with the fact that the majority of hypovirulent clones and other lineage II isolates present truncated InlA, leading to virulence attenuation. Furthermore, comparative genomics analysis between hypo- and hypervirulent clones have uncovered the specific virulence clusters such as LIPI-3 (present approximately in 50% of lineage I strains) and LIPI-4. Regardless of these facts, the regulatory authorities consider all strains of L. monocytogenes to be equally pathogenic (Vázquez-Boland et al., 2020; Quereda et al., 2021), whereas the food safety risk attributed to different subgroups differ (Chen et al., 2006).
The resistance to environmental stresses such as acidic environment, nisin, bile acids, and high osmolarity is conferred to L. monocytogenes by stress resistance determinants located on the Stress Survival Islet 1 (SSI-1), whereas stress resistance determinants to alkaline and oxidative stresses are located on the SSI-2 (Hein et al., 2011; Guidi et al., 2021).
Generally, uncovering the resistance mechanisms to stressful conditions in food matrices and the environment is regarded as important to contribute to the development of novel and efficient measures to prevent contamination through the whole food chain continuum and control the growth of L. monocytogenes during food storage (Bucur et al., 2018).
SSI-1, a five-gene stress survival islet [lmo0444, lmo0445, lmo0446 (pva), lmo0447 (gadD1), and lmo0448 (gadT1)], has an equal distribution in human clinical isolates and in strains isolated from food and food-processing environments (Ryan et al., 2010). The presence of SSI-1, as well as the ability to form biofilms, correlated with the persistence of L.monocytogenes strains (Keeney et al., 2018). Notably, the strongest biofilms were formed by strains from serotype 1/2b, such as CC3 and CC5, the majority of which contained SSI-1, whereas serotype 4b (such as CC2 and CC6), most of which do not contain SSI-1, formed the weakest biofilms (Keeney et al., 2018).
According to Harter et al. (2017), SSI-2, consisting of the genes lin0464 and lin0465, encoding a putative transcriptional regulator and an intracellular PfpI protease, respectively, is predominantly found in the hypovirulent strains of L. monocytogenes ST121. SSI-2 contributes to survival upon oxidative and alkaline stress conditions, thus potentially favoring L. monocytogenes adaptation and persistence in the food-processing environments. Notably, in addition to SSI-2, the genome of L. monocytogenes ST121 possesses plasmids and the transposon Tn6188, which were hypothesized to be responsible for supporting its survival in food-processing environments (Müller et al., 2014; Schmitz-Esser et al., 2015; Pasquali et al., 2018). SSI-2 occasionally is found in other L. monocytogenes strains, such as ST1033 (CC1, serotype 4b).
Interestingly, SSI-2-positive strains were detected in L. monocytogenes lineage I, lineage III, and also in L. innocua but with a slightly shorter islet harboring only 1,947 bp (Harter et al., 2017). Phylogenetic analysis indicated that L. innocua SSI-2 shares the highest similarity with those of L. monocytogenes strains ST13 and CC193 that belong to lineage II (i.e., all food isolates) (Harter et al., 2017).
Notably, SSI-1 contains gadD1 and gadT1, which are among the genes protecting from acid stress. Indeed, the ability to tolerate a low pH environment is an important feature of L. monocytogenes because it allows survival in acidic environments encountered in the gastrointestinal tract of the host, in the macrophage phagosome, and in natural and food-processing environments (Gahan and Hill, 2014; Lund et al., 2014; Arcari et al., 2020; Lund et al., 2020). L. monocytogenes harbors membrane-associated systems and intracellular systems to resist acidic environments and to control intracellular pH. Several mechanisms, schematically depicted in Figure 2, are known to maintain intracellular pH (pHi) to values compatible with L. monocytogenes vitality. These include the F0F1-ATPase, the glutamate decarboxylase (GAD) system, and the arginine and agmatine deiminases (ADI and AgDI, respectively) (Cotter et al., 2000; Feehily et al., 2014; Cheng et al., 2017). The F0F1-ATPase, a multi-subunit enzyme system, is involved in the acid tolerance response initiation upon mild acidic pH stress. The GAD system, on the other hand, can affect survival under mild acid stress but also under harsher acidic conditions (Karatzas et al., 2012). Notably, the GAD system is also activated under low oxygen availability, typically encountered by L. monocytogenes when exposed to food packaging atmosphere (Francis et al., 2007; Sewell et al., 2015). The system consists of three homologous glutamate decarboxylases, namely GadD1, GadD2, and GadD3, and of the cognate glutamate/GABA antiporters GadT1 and GadT2 (Figure 2). The decarboxylases and the antiporters are encoded by the relevant genes at three distinct genetic loci. Four genes are organized in the following operons: gadD1T1 and gadT2D2, whereas the gadD3 gene, the fifth gene, is an independent unit (Cotter et al., 2005). In particular, the gadD1T1 operon enhances growth under mildly acidic conditions (see also Stress survival islets 1 and 2), whereas gadT2D2 plays an important role in conferring survival under extremely acidic conditions (Cotter et al., 2001, 2005). The gadD3 gene, positively regulated by the stationary-phase sigma factor σB, in addition to being part of the GAD system, was shown to be involved in nisin resistance (Begley et al., 2010). According to Chen et al. (2012), gadD2 and gadD3 are present in all L. monocytogenes strains, whereas gadD1 is present in only 36.6% of the strains, including all those belonging to serovar 1/2c, and 68.5% of the strains of serovar 1/2a. Notably, only a small fraction of the strains of L. monocytogenes serovar 1/2b and lineage III strains (including J2-071 and HCC23) possess the gadD1 gene. Furthermore, the gadD1T1 operon is absent in most serotype 4 L. monocytogenes clinical strains (Cotter et al., 2005).
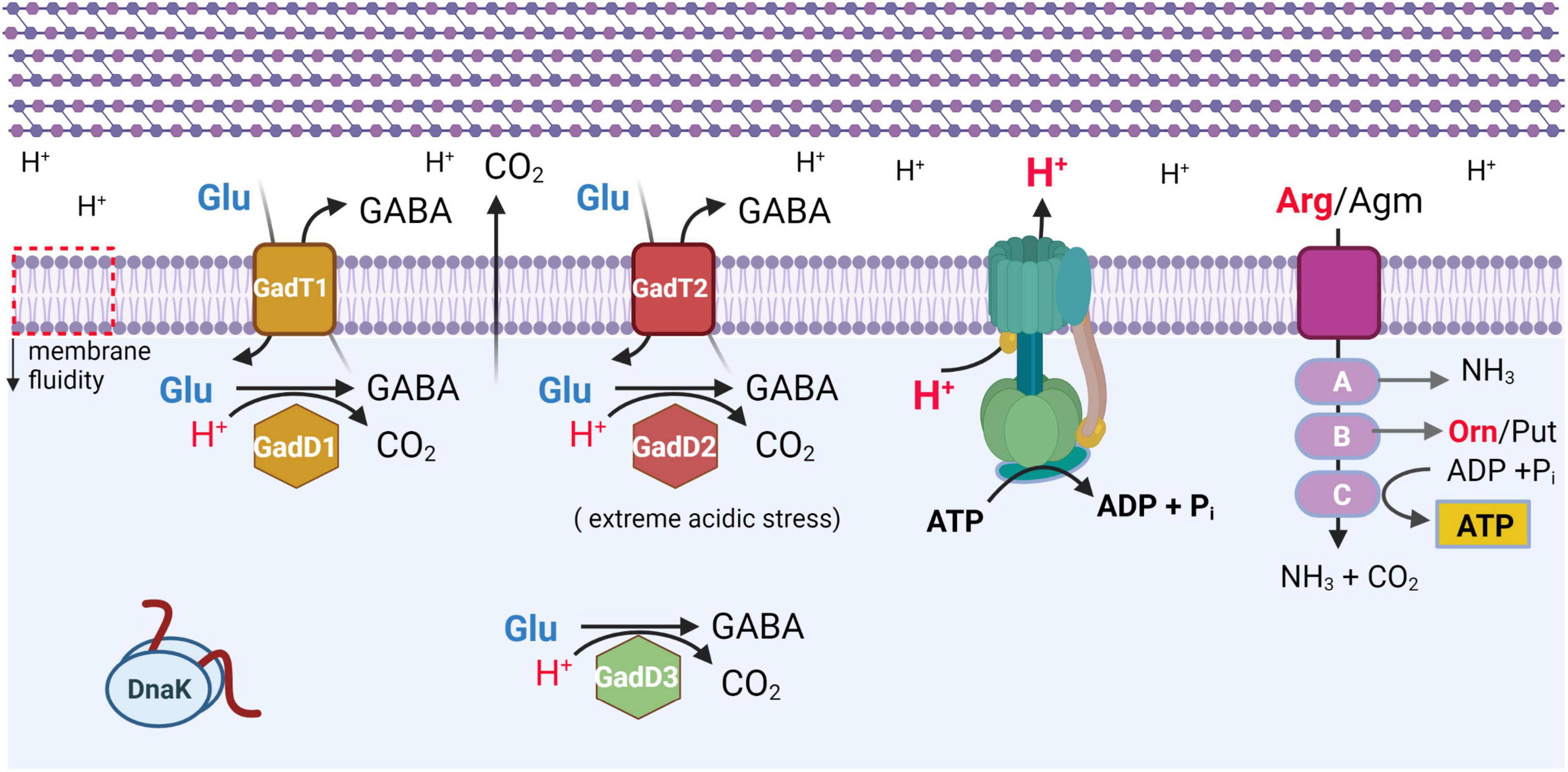
Figure 2. Schematic representation of most effective systems protecting L. monocytogenes from acidic stress. All systems are activated under mild acidic stress, with GadD2T2 system mostly effective under extreme acidic stress (adapted from Lund et al., 2020). F0F1-ATPase is shown in blue-green. ArgD/AguD transporter is shown in purple. A, ArcA (arginine), AguA1 (agmatine) deiminase; B, ArcB/AguB carbamoyl-transferase; C, ArcC/AguC carbamate kinase. GadT1 and GadT2 refer to glutamate (Glu)/GABA antiporters. GadD1, GadD2, and GadD3 are glutamate decarboxylase isoforms. Orn, ornithine. Put, putrescine. DnaK, chaperone. All membrane proteins are localized in lipid bilayer of plasma membrane, which also undergoes a decrease in membrane fluidity (downward pointing arrow, on leftmost side). Multilayered peptidoglycan of cell wall is schematically represented above plasma membrane. Created with BioRender.com.
Paudyal et al. (2020) have shown that casamino acids, peptone, and tryptone are major GAD system activators resulting in upregulation of the transcription of gadD2. Furthermore, Paudyal et al. (2018) demonstrated that maleic acid inhibits the GAD of L. monocytogenes significantly, enhancing its sensitivity to acidic conditions, and thus, together with the ability to remove biofilms, maleic acid has been proposed to make a good candidate for disinfection regimes.
Moreover, Boura et al. (2020) recently showed a secondary novel role of the GAD system in protection against oxidative stress. The authors hypothesized that under oxidative stress GABA, instead of being exported by the transporter (Figure 2), is transferred to the GABA shunt enzymes that provide NADPH to contrast oxidative stress and allow to bypass two missing steps in the TCA cycle. They also suggested that this knowledge could find application in food hurdle technology to eliminate L. monocytogenes.
In addition to the GAD system, also the ADI system can be activated in response to low pH. This latter system consists of three enzymatic activities, namely arginine deiminase, ornithine carbamoyl-transferase, and carbamate kinase, encoded by the arcA, arcB, and arcC genes, respectively (Cheng et al., 2017). Through these enzymatic activities, arginine is converted to ornithine, CO2, and ammonia, with concomitant production of adenosine triphosphate (Figure 2). It was previously reported that this gene cluster is present in lineage I and lineage II but absent from lineage III and non-pathogenic L. innocua and Listeria welshimeri (Ryan et al., 2009). However, Deng et al. (2010) showed that the ADI gene cluster is also highly conserved in lineage IIIB.
The AgDI pathway is a less known system of acid stress tolerance in L. monocytogenes. AgDI converts agmatine into putrescine, ammonia, and CO2 and produces adenosine triphosphate (Chen J. et al., 2011; Soares and Knuckley, 2016). Cheng et al. (2013) found that L. monocytogenes harbors two putative AgDIs (aguA1 and aguA2), but only AguA1 functionally participates in the AgDI pathway and mediates acid tolerance in L. monocytogenes.
Also, the thiamine uptake system, encoded by the thiT gene (formerly lmo1429), was shown to be required for full acid tolerance in L. monocytogenes (Madeo et al., 2012). According to the authors’ findings, a thiT mutant strain resulted in significantly higher acid sensitivity than the control strain. It was suggested that the acid sensitivity is due to the lack of thiamine that does not allow the reaction of acetolactate synthase to occur, and therefore, acetoin synthesis, which involves proton consumption, is impaired (Madeo et al., 2012).
When different L. monocytogenes strains are exposed to acidic environments, the acid tolerance displayed varies significantly, and this might contribute to the observed strains’ differences in robustness and pathogenicity (Conte et al., 2000). The virulent reference strains L. monocytogenes EGD-e (1/2a, lineage II), 850658 (4a, lineage III), and 10403S (1/2a, lineage II) were more resistant to acidic stress than the avirulent M7 (4a, lineage III), which showed a defect in maintaining pHi homeostasis (Cheng et al., 2015). Despite the observations mentioned, EGD-e cannot export GABA and relies exclusively on GadD3, whereas strain 10403S relies upon GadD2 (Feehily et al., 2014). This suggested that the GAD system in the commonly used reference strains originated from separate lines of evolution. Strain-specific patterns of acid resistance are also recognizable in other datasets (Ramalheira et al., 2010).
The sigma factor of RNA polymerase, responsive to general stress, namely σB, plays an important role in L. monocytogenes, both in the adaptation to different stress conditions and in virulence. As shown by many studies conducted using the lineage II reference strains (mainly the 10403S as mentioned earlier and EGD-e), σB regulates approximately 300 genes important for virulence and responses to stresses (Severino et al., 2007; Hain et al., 2008; Raengpradub et al., 2008). Also, the literature review revealed that the σB regulon played a significant role in the resistance of L. monocytogenes strains belonging to lineages I, II, and IIIB and a limited role in the resistance of the L. monocytogenes lineage IIIA strain to acid and oxidative stresses (Oliver et al., 2010; Liu et al., 2019). Similarly, σB plays a significant role in resistance to acid and salt stresses also in L. innocua (Raengpradub et al., 2008).
A systematic review of the σB regulon in L. monocytogenes identifies several regulon members that include genes involved, or putatively involved, in stress response: osmotic (18 genes), oxidative (14 genes), acid (12 genes), antibiotic (6 genes), bile (3 genes), and others (24 genes) (Liu et al., 2019). However, σB is not the only alternative sigma factor that has been shown to play a role in the stress tolerance of L. monocytogenes. Other alternative sigma factors, including σC (previously implicated in nisin response), σH, and σL (RpoN), also regulate transcription of genes important for virulence and response to various stress and growth conditions (Glaser et al., 2001; Chaturongakul et al., 2011). Some of the genes needed in the stress responses of L. monocytogenes are regulated by more than one sigma factor. Overlaps have been reported between σB and σH, σB and σL, and σC and σB regulons (Chaturongakul et al., 2011). Notably, the regulation of SSI-2 (see Stress survival islets 1 and 2) is independent of the σB, but σH or σL could be involved (Harter et al., 2017). Interestingly, of the 51 genes classified in the “virulence” group of the σB regulon, 23 were also classified in the “stress response” group and 4 in the “metabolism” group (Liu et al., 2019). The genes that belonged both to the “virulence” and “stress response” groups included those required for survival and multiplication under host-imposed stress conditions such as acidic pH and bile acids in the gastrointestinal tract of the host and those contrasting oxidative stress in the mammalian host phagosome. These observations reinforced the hypothesis that stress response and virulence are strongly associated. Among the 24 genes of the σB regulon that belong only to the “virulence” group, there is the gene coding for the PrfA, located in LIPI-1 (see L. monocytogenes diversity and heterogeneity of the virulence determinants), which can be transcribed from multiple promoters, regulated by σB, σA, and PrfA itself (De Las Heras et al., 2011).
Natural antimicrobials, such as nisin (which belongs to the class I bacteriocins), sakacins, pediocin PA-1, plantaricin BM-1, and leucocin A (which belong to the class II bacteriocins), can be used to control L. monocytogenes on RTE foods (Ferreira and Lund, 1996; Nilsson et al., 1997; Franklin et al., 2004; Trinetta et al., 2010; Woraprayote et al., 2013; Ortiz et al., 2014; Balay et al., 2017; Xie et al., 2018). However, nisin is the only bacteriocin approved as a preserving additive in food. It should be highlighted that RTE foods add up further stress to L. monocytogenes when these foods are exposed to cold stress, organic acid stress, and osmotic stress, and some of these environmental stresses may affect bacteriocin resistance, e.g., nisin resistance increases if L. monocytogenes is preexposed to acid stress (van Schaik et al., 1999; Bonnet and Montville, 2005).
VirR, the response regulator of the VirRS two-component system, with a role in defense against cell envelope stress (Mandin et al., 2005), is directly controlling nisin resistance. In this specific case, instead of the receptor histidine kinase VirS, an ABC-transporter encoded by virAB seems to be responsible for sensing the stressor (Grubaugh et al., 2018). VirR mediates nisin and other cell envelope stress resistance by regulating the dltABCD operon (Kang et al., 2015), which modifies lipoteichoic acids (Abachin et al., 2002). In addition to VirR, the two-component systems LiaRS and LisRK were shown to be involved in resistance to nisin (Cotter et al., 2002; Collins et al., 2012; Bergholz et al., 2013; Nikparvar et al., 2021). In particular, these two-component systems regulate the expression of anrAB, dltABCD, lmo2229, mprF, and telA (Abachin et al., 2002; Gravesen et al., 2004; Thedieck et al., 2006; Collins et al., 2010a,b, 2012). With the exception of the latter, all the listed genes have an assigned role in the biosynthesis/metabolism of components of the membrane and the cell wall. Using a laboratory cheese model, temperature and pH were shown to be among the environmental conditions that affected the sensitivity of L. monocytogenes to nisin, the efficacy of which was stronger when the cheese was stored at low temperatures and prepared at pH close to neutrality (i.e., pH 6 and 6.5), due to the activity of dltA and mprF (Henderson et al., 2020).
The resistance to nisin varies among the various lineages, with lineage II strains being more tolerant than lineage I strains. These differences extended to serotype and CC levels. For example, serotypes 1/2a and 1/2c were more tolerant than serotype 1/2b, whereas serotype 4b showed the least tolerance to nisin, as also supported by studies showing that serotype 1/2a and 4b strains were more tolerant and sensitive to nisin, respectively (Buncic et al., 2001; Katla et al., 2003; Szendy et al., 2019; Wambui et al., 2020). Comparing clonal types, Wambui et al. (2020) concluded that CC7 (lineage II, 1/2a) strains displayed the highest nisin resistance, whereas CC2 (lineage I, 4b) and CC3 (lineage I, 1/2b) strains showed the lowest nisin resistance levels, similar to previous observations (Malekmohammadi et al., 2017). In addition, among CCs belonging to lineage II serotype 1/2a, CC155 was the most nisin tolerant, whereas CC14, CC199, and CC403 (lineage II, serotype 1/2a) had increased nisin sensitivity. Differences in the ability to respond to stress among the L. monocytogenes genotypes are probably linked to differences in the expression of proteins associated with the membrane, such as the penicillin-binding proteins coded by the lmo0441, lmo0540, lmo1892 genes, and lmo2229 as mentioned earlier, as well as the gene coding for σB, all of which were reported to be more expressed in lineage II than lineage I (Gravesen et al., 2004; Begley et al., 2006; Severino et al., 2007).
In food factories, sanitizer tolerance or resistance and an enhanced ability to form biofilms have been suggested as typical strain characteristics contributing to persistence (Norwood and Gilmour, 1999; Aase et al., 2000; Lundén et al., 2000, 2003; Borucki et al., 2003; Heir et al., 2004; Pan et al., 2006; Lourenço et al., 2009). However, the exact mechanisms behind persistence are not fully understood. Difficulties in eradicating L. monocytogenes contamination in food-processing settings may be conferred by the BC tolerance that some strains harbor and which provides an advantage for survival under stress and in food-processing settings, allowing the bacteria to persist in the environment (Mullapudi et al., 2008). A recent publication by Guérin et al. (2021) reported the capacity of L. monocytogenes strains to adapt to biocides, in particular ammonium quaternary compounds (commonly known as quats or QACs), and proposed the possible link between this adaptation and the selection of resistance regarding the fluoroquinolone antibiotic ciprofloxacin. This link was investigated also by others (Martínez-Suárez et al., 2016; Kode et al., 2021). The current hypothesis is that dilution in the environment and biodegradation give rise to QAC concentration gradients, which means that microorganisms (including L. monocytogenes) become frequently exposed to sub-inhibitory concentrations of QACs (Martínez-Suárez et al., 2016). Indeed, Møretrø et al. (2017) measured and found residues of QACs after sanitation in meat- and salmon-processing plants in Norway and suggested that this may result in a growth advantage for L. monocytogenes harboring the QAC resistance genes (i.e., qacH and bcrABC; see later). Therefore, the low-level resistance to QACs in L. monocytogenes has been proposed to be a contributing factor to its environmental adaptation and persistence. This may explain why the minimal inhibitory concentration of QAC tolerant strains way below user concentrations of QAC may still be of practical relevance. More studies are needed to confirm this hypothesis. Another study conducted by Castro et al. (2021) demonstrated that mobile genetic elements (MGEs) support the persistence of L. monocytogenes on dairy farms and may be spread through the food industry. It is believed that MGEs are pivotal in increasing the antimicrobial resistance of L. monocytogenes strains. MGEs can be exchanged between Listeria and other species leading to the creation of novel resistance phenotypes (Matereke and Okoh, 2020; Castro et al., 2021).
Several BC tolerance determinants have been identified in L. monocytogenes, including BC efflux pumps qacH (Tn6188), bcrABC, and emrE, which are located on MGEs and mostly present in lineage II isolates, i.e., CC9, CC13, CC14, CC31, and CC121 (Dutta et al., 2013; Müller et al., 2013, 2014; Ebner et al., 2015; Kovacevic et al., 2015; Maury et al., 2016; Ortiz et al., 2016; Zuber et al., 2019). Additional BC tolerance genes have been identified, such as emrC, identified on a plasmid in some ST6 isolates, qacA and qacC, which are both located on plasmids, and multidrug resistance Listeria mdrL (lmo1409), negatively regulated by ladR (lmo1408) (Xu et al., 2014; Kremer et al., 2017; Jiang et al., 2019). Literature data on the presence of resistance genes are sometimes contradictory and not always related to all strains of a particular L. monocytogenes CC. For example, Hurley et al. (2019) and Chen et al. (2020) did not find emrC in ST6, as previously described by Kremer et al. (2017) in ST6.
Several very recent investigations showed that gene qacH can occasionally be present in some different CC primarily of serotype 1/2a (CC8, CC20, CC31, CC101, and CC121) but also in serotypes 4b (CC2) and 1/2c (CC9) (Ebner et al., 2015; Meier et al., 2017; Horlbog et al., 2018; Roedel et al., 2019; Stoller et al., 2019; Zuber et al., 2019; Wieczorek et al., 2020; Gelbicova et al., 2021; Guidi et al., 2021; Palaiodimou et al., 2021; Pérez-Baltar et al., 2021). In general, Cherifi et al. (2018) recommended that only one genetic determinant should not be taken into consideration when strain persistence is investigated.
The bcrABC cassette was first described by Elhanafi et al. (2010) and isolated from L. monocytogenes strains associated with the 1998–1999 listeriosis outbreak in the United States caused by hotdog contamination. It consists of a TetR family transcriptional regulator (bcrA) and two small multidrug resistance genes (bcrB and bcrC). This cassette is located in the pLM80 plasmid, but a chromosomal location was also reported. The occurrence of the bcrABC cassette in non-pathogenic species of Listeria, such as L. innocua and L. welshimeri, suggests that these species may be the reservoirs of BC and other resistance determinants that are transferred to L. monocytogenes by conjugation (Katharios-Lanwermeyer et al., 2012). The bcrABC cassette was significantly associated with L. monocytogenes isolates belonging to CC321, CC155, CC204, and CC199 but can be present in other CCs such as ST14, CC288, ST9, ST121, CC5, L. welshimeri, and L. innocua (Meier et al., 2017; Møretrø et al., 2017; Pasquali et al., 2018; Chen et al., 2020; Naditz, 2020; Cooper et al., 2021; Gelbicova et al., 2021; Palaiodimou et al., 2021).
Another putative efflux pump gene responsible for increased tolerance to QACs is the emrE gene. This gene is located on a mobile genomic island LGI1 and was found in one clone (CC8) that includes strains implicated in the 2008 deli meat outbreak in Canada (Kovacevic et al., 2015). Meier et al. (2017) concluded that the emrE is associated with a serotype 1/2a (CC8) and seems to be limited to sublineage 8 strains. The expression of emrE was found to be upregulated in the presence of BC, demonstrating that emrE-harboring strains are likely to adapt in food-processing environments better (Kovacevic et al., 2015).
The link between BC resistance and cadmium (Cd) resistance in Listeria spp. strains has been reported in several studies (Mullapudi et al., 2008; Ratani et al., 2012; Korsak and Szuplewska, 2016). In particular, in L. monocytogenes strains, five Cd resistance determinants (cadAC efflux systems) were identified (Chmielowska et al., 2021). The cadA1 gene is located on the transposon Tn5422 and often plasmid-borne, and predominates (as operon cadA1C1) in CC3, CC8, and CC121 (Lebrun et al., 1994; Gelbicova et al., 2021). Also, the cadA2 gene is usually found on plasmids and is typically accompanied by the bcrABC cassette (Kuenne et al., 2010; Dutta et al., 2013). As for the genes cadA3, cadA4 (also involved in biofilm formation), and cadA5, their location is typically on chromosomes, as part of integrative conjugative elements and genomic islands (LGI2 and LGI2-1), respectively (Kuenne et al., 2013; Lee et al., 2017; Parsons et al., 2017, 2019). Notably, LGI2, in addition to carrying cadA4, also carries a cassette for resistance to arsenic, which encompasses the arsR1D2R2A2B1B2 operon and the upstream arsA1D1 (Kuenne et al., 2013). The arsenic resistance is primarily associated with L. monocytogenes strains belonging to 4b serotype, particularly CC1, CC2, and CC4 hypervirulent clonal clones (Lee et al., 2017). On the contrary, Cd resistance typically is found in L. monocytogenes strains belonging to serotypes 1/2a and 1/2b, from food and food-processing environments (Mullapudi et al., 2008; Ratani et al., 2012). In addition, a specific association with lineages was found, namely cadA1C1 cassette with lineage II and cadA2C2 cassette with lineage I. On the other hand, strains containing both cadA1 and cadA2 were more frequent in lineage I than in lineage II (Mullapudi et al., 2010).
As said at the beginning of this section, the enhanced ability to form a biofilm that is hard to remove mechanically and less sensitive to sanitizers was proposed as a mechanism for persistence, given that the biofilm provides a clear advantage for surviving in food-processing or retail environments. However, other studies have not found a clear link between the biofilm-forming ability of some isolates and their persistence, and differences in the experimental setup and in the strains used have been ascribed as the reasons for the observed different results (Djordjevic et al., 2002; Holch et al., 2013; Kadam et al., 2013; Lee et al., 2019). Although some authors reported a correlation between lineages and biofilm-forming ability, with lineage II strains presenting higher levels of biofilm production, other results did not support these findings (Norwood and Gilmour, 2001; Djordjevic et al., 2002; Borucki et al., 2003; Di Bonaventura et al., 2008; Takahashi et al., 2009; Combrouse et al., 2013; Bai et al., 2021). Maury et al. (2019) found that hypovirulent genotypes, CC121 and CC9, were more efficient in biofilm production than hypervirulent clones (such as lineage I clones: CC1, CC2, CC4, and CC6) under sub-lethal concentrations of BC, implying that lineage II hypovirulent clones were associated with persistence features. Also, Pérez-Baltar et al. (2021) found that CC121 strains are strong biofilm formers, and some harbored the transposon Tn6188, related to increased tolerance to QACs. Interestingly, the lmo0435 homolog biofilm-associated protein, BapL, putative peptidoglycan bound protein involved in biofilm formation, but not essential, is truncated in ST121 strains, which belong to CC121 (Jordan et al., 2008; Schmitz-Esser et al., 2015). Some authors suggested that CC8 strains possess a strong capacity for biofilm formation, which may support persistence within food production environments and subsequent contamination of foods (Verghese et al., 2011; Zuber et al., 2019). A study done in Canada by Upham et al. (2019) found that the formation of biofilms is associated with serotype 1/2a isolates in lineage II, as well as the presence of SSI-1. SSI-1, rare in clinical isolates, has been shown to be associated with a survival advantage in the environment, thus supporting the link between SSI-1 and persistence in L. monocytogenes (Hilliard et al., 2018). Furthermore, SSI-1 was strongly correlated with biofilm formation and a truncation (stop codon) in inlA (Franciosa et al., 2009; Keeney et al., 2018). More recent findings confirmed the influence of SSI-1 and a truncated inlA in increased biofilm levels in L. monocytogenes (Ciccio et al., 2019).
Regarding niche preference, reasons why the so-called persistent isolates are recurrently isolated in the same food-processing premises over long periods remain elusive. Persistent isolates could belong to specific STs particularly well adapted to the environmental conditions of the food manufacturing environment (Knudsen et al., 2017). It is, however, difficult to pinpoint adaptive traits directly correlated to persistence.
Notably, intra-genotype variation was observed in some CCs, suggesting that minor genetic variants within a genotype may impact biofilm phenotype (Lee et al., 2019; Zuber et al., 2019). Nevertheless, it should be pointed out that next to genetic determinants, biofilm formation is influenced by factors such as temperature, nutrient availability, and biofilm formation maturity (de Oliveira et al., 2010; Barbosa et al., 2013; Kadam et al., 2013). The limitation of most biofilm studies is that they have been done in monocultures, which may not be most relevant, as this bacterium is not alone in the food industry. Therefore, research on multispecies community might be more suitable to gain better insights into interactions among different species within the biofilms and the formation of the biofilm itself.
Whole-genome sequencing (WGS) of strains that have been isolated from different ecological niches has become more and more standard practice and demonstrates to be powerful in outbreak investigations at national and international levels (ECDC, 2019). These outbreak investigations rely on cross-sectorial cooperation between epidemiologists, microbiologists, and bioinformaticians to link clinical isolates to outbreak isolates. The availability of WGS data gives new opportunities to explain intraspecific variability and to find genetic biomarkers that predict microbial behavior. Multilocus sequence typing and whole-genome phylogenetic analyses demonstrated that pathogenic subtypes vary in their virulence and association with food (L. monocytogenes diversity and heterogeneity of the virulence determinants), and this intraspecific variability is relevant for risk assessments in general and, more specifically, also for quantitative microbiological risk assessments (QMRA). A QMRA is a structured and quantitative process for determining the risk associated with microbiological hazards in a food (CAC, 1999). The basic steps of a QMRA include hazard identification, exposure assessment, hazard characterization, and risk characterization, and this formalized approach has been adopted by regulators globally and is also used by industry. Various QMRA studies have been performed for different product/pathogen combinations (e.g., FDA/CFSAN and USDA/FSIS, 2003; Tirloni et al., 2018; EFSA, 2019) aiming to characterize and quantify the risk of a pathogen associated with a product (category). In these studies, the exposure assessment and hazard characterization steps of QMRAs are performed for a pathogenic species as a whole. The current advances in the field of omics technology give opportunities to make use of the greater understanding of intraspecific variability based on various recently published bioinformatics tools (Brul et al., 2012; Den Besten et al., 2018; Haddad et al., 2018; Rantsiou et al., 2018; Fritsch et al., 2019; Njage et al., 2020). Instead of considering all-hazard strains of a species as equally likely to cause disease or equally likely to survive the food chain, WGS data could give support to rank subtypes with respect to their virulence potential (Chen Y. et al., 2011; Collineau et al., 2019) or to groups subtypes with respect to their differences in robustness or fitness to reach the consumer stage (Den Besten et al., 2018). The QMRA input distributions can be tailored to each subgroup accordingly, making it possible to fine-tune the QMRA output. The studies of Chen Y. et al. (2011) and Fritsch et al. (2018) illustrate the potential to refine QMRA studies when considering pheno-genotype associations for specific properties of L. monocytogenes. The authors described the variability of L. monocytogenes’ growth characteristics more accurately using two different distributions for the minimum temperature of growth (Tmin). For risk characterization, three different groups of virulence were considered according to the CCs, making use of reported differences in clinical frequencies of different CCs (Maury et al., 2016, see also L. monocytogenes diversity and heterogeneity of the virulence determinants). The QMRA output showed that CCs that are contributing the most to consumer exposure were not those that contributed the most to listeriosis cases. Chen Y. et al. (2011) followed a similar approach for the hazard characterization step where they attributed different dose-response models for L. monocytogenes subtypes with genes encoding a full-length and a truncated InlA, respectively. These examples of fine-tuning a QMRA highlight the potential impact of implementing genomic data in QMRA. This field is still young and relies on high efforts to phenotypically characterize strain variability. Grouping of strains with shared characteristics is only possible when subgroups of strains have different phenotypes. This pushes the need to characterize various aspects of strains, such as fitness and stress robustness, because these details are needed to quantitatively describe intraspecific variability in the exposure assessment part of a QMRA. Also, it is important to note that routine collection of WGS data is more standardized across regulatory and public health agencies and more limited in surveillance by industry (Jagadeesan et al., 2019; Cohn et al., 2021), and this introduces a bias in isolate characterization, whereas representative WGS data from food and human isolates are critical to assess the likelihood of subtypes to cause disease. When these challenges are recognized and taken up, it will open avenues to make use of pheno-genotype associations in the next generation of QMRA and to incorporate subtype-specific assessment of public health significance in food control strategies and regulations.
The notorious foodborne pathogen L. monocytogenes is ubiquitous in nature and can be found in soil, in the farm environment, in the food production environment, and in food products (Figure 3) (Kallipolitis et al., 2020). As highlighted in this review, there is a high degree of strain divergence regarding virulence potential, environmental adaption, and stress response. In addition, persistent L. monocytogenes subtypes have the ability to survive and persist for months and even years in food-processing environments and to keep contaminating food products. It is, however, difficult to correlate adaptive traits directly to persistence. Hence, pheno-genotype association studies are promising approaches to increase our mechanistic understanding of how this pathogen survives along the food chain and infects the human host. Notably, most published studies do not assess the presence/absence of specific genes (or sets of genes) in all currently known L. monocytogenes lineages, CCs/sequence types. Hence, experimental and holistic approaches based on WGS and environmental studies may play a role in determining the distribution and diversity of Listeria species. The use of advanced diagnostic technologies such as WGS can open avenues to fine-tune risk assessments, which is of great importance in the prevention and control of both animal and human listeriosis (Den Besten et al., 2018; Rantsiou et al., 2018). Successful implementation and use of WGS needs, however, an appropriate and functioning infrastructure and resources (Grace, 2015), i.e., functional control and an already established surveillance system to collect isolates and metadata from clinical, food, and environmental samples (EFSA, 2008; FAO, 2016). This will support an unbiased assessment of the likelihood of subtypes to be present in food and to cause disease to come to risk-based interventions at the intraspecific level.
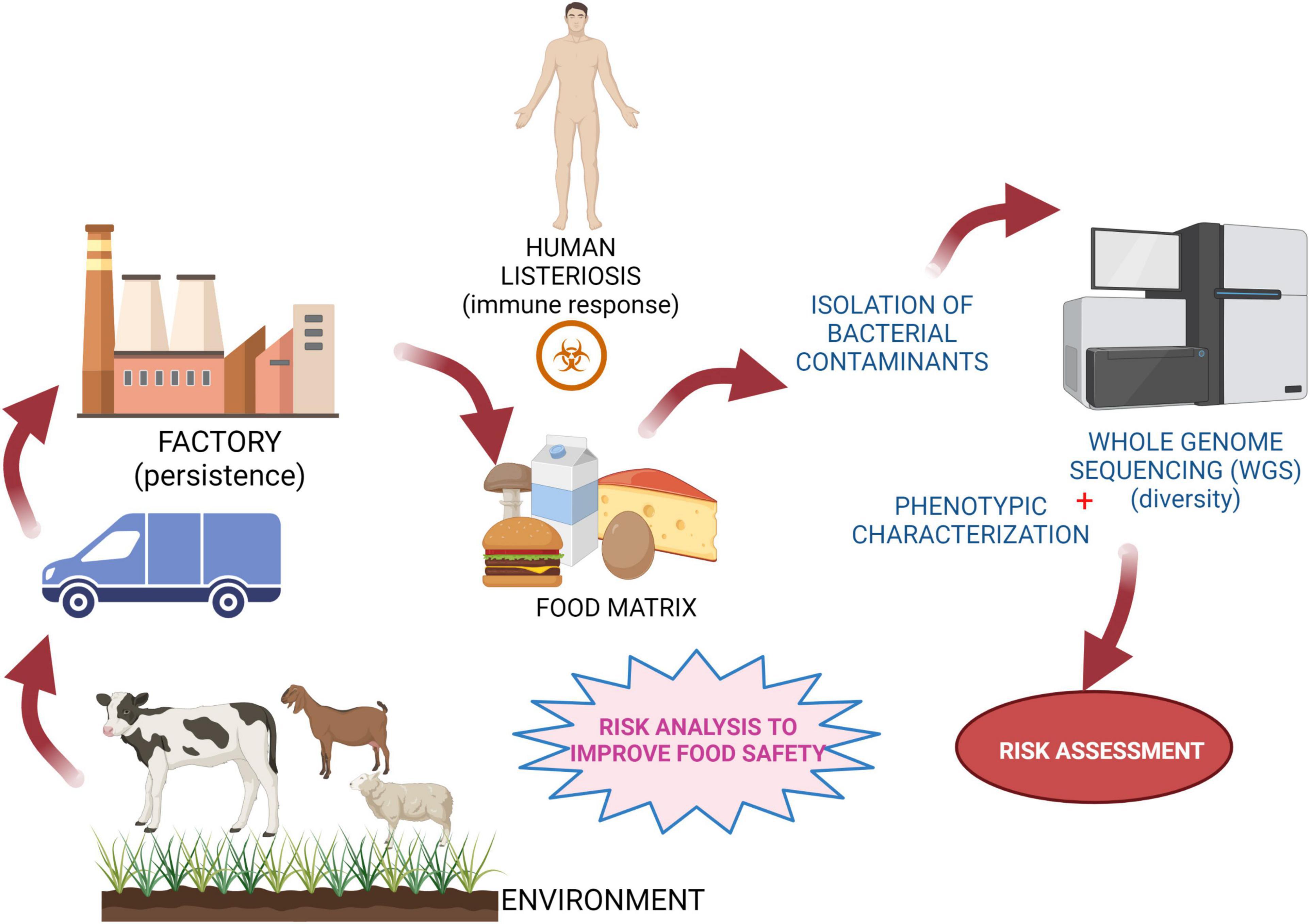
Figure 3. Schematic representation of risk control strategies. Experimental concept to assess risk and monitor diversity among L. monocytogenes strains for prevention/control of food chain contamination. Created with BioRender.com.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This work was funded through the COST Action “Understanding and exploiting the impacts of low pH on microorganisms” (EuroMicropH) CA18113. BL visited the DD laboratory as the recipient of a Short-Term Scientific Mission grant supported by the COST Action CA18113 EuroMicropH (https://euromicroph.eu/stsm/).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aase, B., Sundheim, G., Langsrud, S., and Rørvik, L. M. (2000). Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62, 57–63. doi: 10.1016/s0168-1605(00)00357-3
Abachin, E., Poyart, C., Pellegrini, E., Milohanic, E., Fiedler, F., Berche, P., et al. (2002). Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43, 1–14. doi: 10.1046/j.1365-2958.2002.02723.x
Althaus, D., Jermini, M., Giannini, P., Martinetti, G., Reinholz, D., Nüesch-Inderbinen, M., et al. (2017). Local outbreak of Listeria monocytogenes serotype 4b sequence type 6 due to contaminated meat pâté. Foodborne Pathog. Dis. 14, 219–222. doi: 10.1089/fpd.2016.2232
Arcari, T., Feger, M. L., Guerreiro, D. N., Wu, J., and O’Byrne, C. P. (2020). Comparative review of the responses of Listeria monocytogenes and Escherichia coli to low pH stress. Genes (Basel) 11:1330. doi: 10.3390/genes11111330
Ariza-Miguel, J., Fernandez-Natal, M. I., Soriano, F., Hernandez, M., Stessl, B., and Rodríguez Lazaro, D. (2015). Molecular epidemiology of invasive listeriosis due to Listeria monocytogenes in a Spanish hospital over a nine-year study period, 2006-2014. BioMed Res. Int. 2015:191409. doi: 10.1155/2015/191409
Bai, X., Liu, D., Xu, L., Tenguria, S., Drolia, R., Gallina, N. L. F., et al. (2021). Biofilm-isolated Listeria monocytogenes exhibits reduced systemic dissemination at the early (12–24 h) stage of infection in a mouse model. NPJ Biofilms Microbiomes 7:18. doi: 10.1038/s41522-021-00189-5
Balay, D. R., Dangeti, R. V., Kaur, K., and McMullen, L. M. (2017). Purification of leucocin A for use on wieners to inhibit Listeria monocytogenes in the presence of spoilage organisms. Int. J. Food Microbiol. 255, 25–31.
Barbosa, J., Borges, S., Camilo, R., Magalhães, R., Ferreira, V., Santos, I., et al. (2013). Biofilm formation among clinical and food isolates of Listeria monocytogenes. Int. J. Microbiol. 2013:524975. doi: 10.1155/2013/524975
Begley, M., Cotter, P. D., Hill, C., and Ross, R. P. (2010). Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes. Appl. Environ. Microbiol. 76, 6541–6546. doi: 10.1128/aem.00203-10
Begley, M., Hill, C., and Ross, R. P. (2006). Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl. Environ. Microbiol. 72, 2231–2234. doi: 10.1128/AEM.72.3.2231-2234.2006
Bergholz, T. M., Shah, M. K., Burall, L. S., Rakic-Martinez, M., and Datta, A. R. (2018). Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 102, 3475–3485. doi: 10.1007/s00253-018-8852-5
Bergholz, T. M., Tang, S., Wiedmann, M., and Boor, K. J. (2013). Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl. Environ. Microbiol. 79, 5682–5688. doi: 10.1128/AEM.01797-13
Berrang, M. E., Meinersmann, R. J., Frank, J. F., and Ladely, S. R. (2010). Colonization of a newly constructed commercial chicken further processing plant with Listeria monocytogenes. J. Food Prot. 73, 286–291. doi: 10.4315/0362-028X-73.2.286
Bertsch, D., Rau, J., Eugster, M. R., Haug, M. C., Lawson, P. A., Lacroix, C., et al. (2013). Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 63, 526–532.
Bolocan, A. S., Nicolau, A. I., Álvarez-Ordóñez, A., Borda, D., Oniciuc, E. A., Stessl, B., et al. (2016). Dynamics of Listeria monocytogenes colonisation in a newly-opened meat processing facility. Meat Sci. 113, 26–34. doi: 10.1016/j.meatsci.2015.10.016
Bonnet, M., and Montville, T. J. (2005). Acid-tolerant Listeria monocytogenes persist in a model food system fermented with nisin-producing bacteria. Lett. Appl. Microbiol. 40, 237–242. doi: 10.1111/j.1472-765X.2005.01661.x
Borucki, M. K., Peppin, J. D., White, D., Loge, F., and Call, D. R. (2003). Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69, 7336–7342. doi: 10.1128/AEM.69.12.7336-7342.2003
Boura, M., Brensone, D., and Karatzas, K. A. G. (2020). A novel role for the glutamate decarboxylase system in Listeria monocytogenes; protection against oxidative stress. Food Microbiol. 85:103284. doi: 10.1016/j.fm.2019.103284
Brul, S., Bassett, J., Cook, P., Kathariou, S., McClure, P., Jasti, P. R., et al. (2012). ‘Omics’ technologies in quantitative microbial risk assessment. Trends Food Sci. Technol. 27, 12–24.
Bucur, F. I., Grigore-Gurgu, L., Crauwels, P., Riedel, C. U., and Nicolau, A. I (2018). Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 9:2700. doi: 10.3389/fmicb.2018.02700
Buncic, S., Avery, S. M., Rocourt, J., and Dimitrijevic, M. (2001). Can food-related environmental factors induce different behaviour in two key serovars, 4b and 1/2a, of Listeria monocytogenes? Int. J. Food Microbiol. 65, 201–212. doi: 10.1016/s0168-1605(00)00524-9
Burkholder, K. M., Kim, K.-P., Mishra, K. K., Medina, S., Hahm, B.-K., Kim, H., et al. (2009). Expression of LAP, a SecA2-dependent secretory protein, is induced under anaerobic environment. Microbes Infect. 11, 859–867. doi: 10.1016/j.micinf.2009.05.006
CAC (1999). “Principles and guidelines for the conduct of a microbiological risk assessment. 1999,” in Proccedings of the Joint FAO/WHO Food Standards Programme, CAC/GL-30, Rome (1999) (Rome).
Carlin, C. R., Liao, J., Weller, D., Guo, X., Orsi, R., and Wiedmann, M. (2021). Listeria cossartiae sp. nov. Listeria immobilis sp. nov., Listeria portnoyi sp. nov. and Listeria rustica sp. nov., isolated from agricultural water and natural environments. Int. J. Syst. Evol. Microbiol. 71:004795. doi: 10.1099/ijsem.0.004795
Carpentier, B., and Cerf, O. (2011). Review–persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145, 1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005
Castro, H., Douillard, F. P., Korkeala, H., and Lindström, M. (2021). Mobile elements harboring heavy metal and bacitracin resistance genes are common among Listeria monocytogenes strains persisting on dairy farms. mSphere 6:00383-21. doi: 10.1128/mSphere.00383-21
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Hu, Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes. Appl. Environ. Microbiol. 7, 187–200. doi: 10.1128/AEM.00952-10
Chen, J., Cheng, C., Xia, Y., Zhao, H., Fang, C., Shan, Y., et al. (2011). Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology 157, 3150–3161. doi: 10.1099/mic.0.049619-0
Chen, J., Fang, C., Zheng, T., Zhu, N., Bei, Y., and Fang, W. (2012). Genomic presence of gadD1 glutamate decarboxylase correlates with the organization of ascB-dapE internalin cluster in Listeria monocytogenes. Foodborne Pathog. Dis. 9, 175–178. doi: 10.1089/fpd.2011.1022
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H., et al. (2018). Prevalence, potential virulence, and genetic diversity of Listeria monocytogenes isolates from edible mushrooms in chinese markets. Front Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, Y., Chen, Y., Pouillot, R., Dennis, S., Xian, Z., Luchansky, J. B., et al. (2020). Genetic Diversity and profiles of genes associated with virulence and stress resistance among isolates from the 2010-2013 interagency market basket survey. PLos One 15:e0231393. doi: 10.1371/journal.pone.0231393
Chen, Y., Gonzalez-Escalona, N., Hammack, T. S., Allard, M. W., Strain, E. A., and Brown, E. W. (2016). Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl. Environ. Microbiol. 282, 6258–6672. doi: 10.1128/AEM.01532-16
Chen, Y., Ross, W. H., Gray, M. J., Wiedmann, M., Whiting, R. C., and Scott, V. N. (2006). Attributing risk to Listeria monocytogenes subgroups: dose response in relation to genetic lineages. J. Food Prot. 69, 335–344. doi: 10.4315/0362-028x-69.2.335
Chen, Y., Ross, W. H., Whiting, R. C., Van Stelten, A., Nightingale, K. K., Wiedmann, M., et al. (2011). Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl. Environ. Microbiol. 77, 1171–1180. doi: 10.1128/AEM.01564-10
Cheng, C., Chen, J., Fang, C., Xia, Y., Shan, Y., Liu, Y., et al. (2013). Listeria monocytogenes aguA1, but Not aguA2, encodes a functional agmatine deiminase. J. Biol. Chem. 288, 26606–26615. doi: 10.1074/jbc.M113.477380
Cheng, C., Dong, Z., Han, X., Sun, J., Wang, H., Jiang, L., et al. (2017). Listeria monocytogenes 10403S arginine repressor ArgR finely tunes arginine metabolism regulation under acidic conditions. Front. Microbiol. 8:145. doi: 10.3389/fmicb.2017.00145
Cheng, C., Yang, Y., Dong, Z., Wang, X., Fang, C., Yang, M., et al. (2015). Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometry. Front. Microbiol. 6:15. doi: 10.3389/fmicb.2015.00015
Cherifi, T., Carrillo, C., Lambert, D., Miniaï, I., Quessy, S., Larivière-Gauthier, G., et al. (2018). Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 18:220. doi: 10.1186/s12866-018-1363-9
Chmielowska, C., Korsak, D., Szuplewska, M., Grzelecka, M., Maćkiw, E., Stasiak, M., et al. (2021). Benzalkonium chloride and heavy metal resistance profiles of Listeria monocytogenes strains isolated from fish, fish products and food-producing factories in Poland. Food Microbiol. 98:103756. doi: 10.1016/j.fm.2021.103756
Ciccio, P. D., Chiesa, F., Rubiola, S., and Civera, T. (2019). “Genetic determinants associated with biofilm formation of Listeria monocytogenes from food and food processing environment,” in Proceedings of the 33rd EFFoST International Conference Sustainable Food Systems-Performing by Connecting, (Rotterdam).
Clayton, E. M., Daly, K. M., Guinane, C. M., Hill, C., Cotter, P. D., and Ross, P. R. (2014). Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol. 14:58. doi: 10.1186/1471-2180-14-58
Cohn, A. R., Cheng, R. A., Orsi, R. H., and Wiedmann, M. (2021). Moving past species classifications for risk-based approaches to food safety: Salmonella as a case study. Front. Sustain. Food Syst. 5:652132. doi: 10.3389/fsufs.2021.652132
Collineau, L., Boerlin, P., Carson, C. A., Chapman, B., Fazil, A., Hetman, B., et al. (2019). Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: a review of opportunities and challenges. Front. Microbiol. 10:1107. doi: 10.3389/fmicb.2019.01107
Collins, B., Curtis, N., Cotter, P. D., Hill, C., and Ross, R. P. (2010a). The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423. doi: 10.1128/AAC.00503-10
Collins, B., Guinane, C. M., Cotter, P. D., Hill, C., and Ross, P. R. (2012). Assessing the contributions of the lias histidine kinase to the innate resistance of Listeria monocytogenes to nisin, cephalosporins, and disinfectants. Appl. Environ. Microbiol. 78, 2923–2929. doi: 10.1128/AEM.07402-11
Collins, B., Joyce, S., Hill, C., Cotter, P. D., and Ross, R. P. (2010b). TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob. Agents Chemother. 54, 4658–4663. doi: 10.1128/AAC.00290-10
Combrouse, T., Sadovskaya, I., Faille, C., Kol, O., Guérardel, Y., and Midelet-Bourdin, G. (2013). Quantification of the extracellular matrix of the Listeria monocytogenes biofilms of different phylogenic lineages with optimization of culture conditions. J. Appl. Microbiol. 114, 1120–1131. doi: 10.1111/jam.12127
Conte, M. P., Petrone, G., Di Biase, A. M., Ammendolia, M. G., Superti, F., and Seganti, L. (2000). Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb. Pathog. 29, 137–144. doi: 10.1006/mpat.2000.0379
Cooper, A. L., Carrillo, C. D., Deschênes, M., and Blais, B. W. (2021). Genomic markers for quaternary ammonium compound resistance as a persistence indicator for Listeria monocytogenes contamination in food manufacturing environments. J. Food Prot. 84, 389–398. doi: 10.4315/JFP-20-328
Cotter, P. D., Gahan, C. G. M., and Hill, C. (2000). Analysis of the role of the Listeria monocytogenesF0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 60, 137–146. doi: 10.1016/S0168-1605(00)00305-6
Cotter, P. D., Gahan, C. G. M., and Hill, C. (2001). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. doi: 10.1046/j.1365-2958.2001.02398.x
Cotter, P. D., Guinane, C. M., and Hill, C. (2002). The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46, 2784–2790. doi: 10.1128/AAC.46.9.2784-2790.2002
Cotter, P. D., Ryan, S., Gahan, C. G. M., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/AEM.71.6.2832
De Las Heras, A., Cain, R. J., Bielecka, M. K., and Vazquez-Boland, J. A. (2011). Regulation of Listeria virulence: prfa master and commander. Curr. Opin. Microbiol. 14, 118–127. doi: 10.1016/j.mib.2011.01.005
de Oliveira, M. M., Brugnera, D. F., Alves, E., and Piccoli, R. H. (2010). Biofilm formation by Listeria monocytogenes on stainless steel surface and biotransfer potential. Braz. J. Microbiol. 41, 97–106. doi: 10.1590/S1517-838220100001000016
den Bakker, H. C., Cummings, C. A., Ferreira, V., Vatta, P., Orsi, R. H., Degoricija, L., et al. (2010). Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genom. 11:688. doi: 10.1186/1471-2164-11-688
den Bakker, H. C., Warchocki, S., Wright, E. M., Allred, A. F., Ahlstrom, C., Manuel, C. S., et al. (2014). Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 64, 1882–1889.
Den Besten, H. M. W., Amézquita, A., Bover-Cid, S., Dagnas, S., Ellouze, M., Guillou, S., et al. (2018). Next generationof microbiological risk assessment: potential of omics data for exposure assessment. Int. J. Food Microbiol. 287, 18–27.
Deng, X., Phillippy, A. M., Li, Z., Salzberg, S. L., and Zhang, W. (2010). Probing the pan-genome of Listeria monocytogenes: new insights into intraspecific niche expansion and genomic diversification. BMC Genom. 11:500. doi: 10.1186/1471-2164-11-500
Desai, A. N., Anyoha, A., Madoff, L. C., and Lassmann, B. (2019). Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 84, 48–53. doi: 10.1016/j.ijid.2019.04.021
Di Bonaventura, G., Piccolomini, R., Paludi, D., D’Orio, V., Vergara, A., Conter, M., et al. (2008). Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 104, 1552–1561. doi: 10.1111/j.1365-2672.2007.03688.x
Djordjevic, D., Wiedmann, M., and McLandsborough, L. A. (2002). Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68, 2950–2958. doi: 10.1128/aem.68.6.2950-2958.2002
Doijad, S., Weigel, M., Barbuddhe, S., Blom, J., Goesmann, A., Hain, T., et al. (2015). Phylogenomic grouping of Listeria monocytogenes. Can. J. Microbiol. 61, 637–646. doi: 10.1139/cjm-2015-0281
Doijad, S. P., Poharkar, K. V., Kale, S. B., Kerkar, S., Kalorey, D. R., Kurkure, N. V., et al. (2018). Listeria goaensis sp. nov. Int. J. Syst. Evol. Microbiol. 68, 3285–3291. doi: 10.1099/ijsem.0.002980
Domínguez-Bernal, G., Müller-Altrock, S., González-Zorn, B., Scortti, M., Herrmann, P., Monzó, H. J., et al. (2006). A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol. Microbiol. 59, 415–432. doi: 10.1111/j.1365-2958.2005.04955.x
Doumith, M., Cazalet, C., Simoes, N., Frangeul, L., Jacquet, C., Kunst, F., et al. (2004). New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72, 1072–1083. doi: 10.1128/IAI.72.2.1072-1083.2004
Dussurget, O. (2008). New insights into determinants of Listeria monocytogenes virulence. Int. Rev. Cell Mol. Biol. 270, 1–38. doi: 10.1016/S1937-6448(08)01401-9
Dutta, V., Elhanafi, D., and Kathariou, S. (2013). Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 79, 6067–6074. doi: 10.1128/AEM.01751-13
Ebner, R., Althaus, S. R., Brisse, D., Maury, S., and Tasara, M. T. (2015). Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 57, 321–326. doi: 10.1016/j.foodcont.2015.04.030
ECDC (2019). ECDC Strategic Framework for the Integration of Molecular and Genomic Typing into European Surveillance and Multi-Country Outbreak Investigations - 2019-2021. Stockholm:ECDC; 2019. Solna Municipality: ECDC.
EFSA (2008). Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents. Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin. Microbiol. Infect. 14, 522–533.
EFSA (2019). Quantitative risk assessment of Listeria monocytogenes in a traditional RTE product. EFSA J. 17:e170906. doi: 10.2903/j.efsa.2019.e170906
EFSA and ECDC (2018). Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections probably linked to frozen corn. EFSA Support Publ. 15:1402E.
EFSA Panel on Biological Hazards [Efsa Biohaz Panel]</snm> Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., et al. (2018). Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 16:5134. doi: 10.2903/j.efsa.2018.5134
Elhanafi, D., Dutta, V., and Kathariou, S. (2010). Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 76, 8231–8238. doi: 10.1128/AEM.02056-10
Fagerlund, A., Langsrud, S., Schirmer, B. C. T., Møretrø, T., and Heir, E. (2016). Genome analysis of Listeria monocytogenes sequence type 8 strains persisting in salmon and poultry processing environments and comparison with related strains. PLoS One. 11:e0151117. doi: 10.1371/journal.pone.0151117
FAO (2016). Applications of Whole Genome Sequencing (WGS) in Food Safety Management. Technical Background Paper, Draft Version 6: 11 April 2016. Rome: FAO.
FDA/CFSAN and USDA/FSIS (2003). Quantitative Assessment of Relative Risk to Public Health From Foodborne Listeria monocytogenes Among Selected Categories of Ready-to-Eat Foods. Silver Spring, MD: FDA.
Feehily, C., Finnerty, A., Casey, P. G., Hill, C., Gahan, C. G. M., O’Byrne, C. P., et al. (2014). Divergent evolution of the activity and regulation of the glutamate decarboxylase systems in Listeria monocytogenes EGD-e and 10403S: roles in virulence and acid tolerance. PLoS One 9:e112649. doi: 10.1371/journal.pone.0112649
Feng, Y., Yao, H., Chen, S., Sun, X., Yin, Y., and Jiao, X. (2020). Rapid detection of hypervirulent serovar 4h Listeria monocytogenes by multiplex PCR. Front. Microbiol. 11:1309. doi: 10.3389/fmicb.2020.01309
Ferreira, M. A., and Lund, B. M. (1996). The effect of nisin on Listeria monocytogenes in culture medium and long-life cottage cheese. Lett. Appl. Microbiol. 22, 433–438. doi: 10.1111/j.1472-765x.1996.tb01197.x
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Franciosa, G., Maugliani, A., Scalfaro, C., Floridi, F., and Aureli, P. (2009). Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int. J. Immunopathol. Pharmacol. 22, 183–193. doi: 10.1177/039463200902200121
Francis, G. A., Scollard, J., Meally, A., Bolton, D. J., Gahan, C. G. M., Cotter, P. D., et al. (2007). The glutamate decarboxylase acid resistance mechanism affects survival of Listeria monocytogenesLO28 in modified atmosphere-packaged foods. J. Appl. Microbiol. 103, 2316–2324. doi: 10.1111/j.1365-2672.2007.03466.x
Franklin, N. B., Cooksey, K. D., and Getty, K. J. (2004). Inhibition of Listeria monocytogenes on the surface of individually packaged hot dogs with a packaging film coating containing nisin. J. Food Prot. 67, 480–485. doi: 10.4315/0362-028x-67.3.480
Fritsch, L., Felten, A., Palma, F., Mariet, J.-F., Radomski, N., Mistou, M.-Y., et al. (2019). Insights from genome-wide approaches to identify variants associated to phenotypes at pan-genome scale: application to L. monocytogenes’ ability to grow in cold conditions. Int. J. Food Microbiol. 291, 181–188. doi: 10.1016/j.ijfoodmicro.2018.11.028
Fritsch, L., Guillier, L., and Augustin, J. C. (2018). Next generation quantitative microbiological risk assessment: refinement of the cold smoked salmon-related listeriosis risk model by integrating genomic data. Microb. Risk Anal. 10, 20–27.
Gahan, C. G., and Hill, C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 4:9. doi: 10.3389/fcimb.2014.00009
Gandhi, M., and Chikindas, M. L. (2007). Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113, 1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008
Gelbicova, T., Florianova, M., Hluchanova, L., Kalova, A., Korena, K., Strakova, N., et al. (2021). Comparative analysis of genetic determinants encoding cadmium, arsenic, and benzalkonium chloride resistance in Listeria monocytogenes of human, food, and environmental origin. Front. Microbiol. 11:599882. doi: 10.3389/fmicb.2020.599882
Gelbíčová, T., Kolácková, I., Pantucek, R., and Karpíšková, R. (2015). A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 38, 293–296.
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Grace, D. (2015). Food safety in low and middle income countries. Int. J. Environ. Res. Public Health 12, 10490–10507. doi: 10.3390/ijerph120910490
Graves, L. M., Helsel, L. O., Steigerwalt, A. G., Morey, R. E., Daneshvar, M. I., Roof, S. E., et al. (2010). Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60, 1280–1288.
Gravesen, A., Kallipolitis, B., Holmstrøm, K., Høiby, P. E., Ramnath, M., and Knøchel, S. (2004). pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl. Environ. Microbiol. 70, 1669–1679. doi: 10.1128/AEM.70.3.1669
Grubaugh, D., Regeimbal, J. M., Ghosh, P., Zhou, Y., Lauer, P., Dubensky, T. W., et al. (2018). The VirAB ABC transporter is required for VirR regulation of Listeria monocytogenes virulence and resistance to nisin. Infect. Immun. 86, e901–e917. doi: 10.1128/IAI.00901-17
Guérin, A., Bridier, A., Le Grandois, P., Sévellec, Y., Palma, F., Félix, B., et al. (2021). Exposure to quaternary ammonium compounds selects resistance to ciprofloxacin in Listeria monocytogenes. Pathogens 10:220. doi: 10.3390/pathogens10020220
Guidi, F., Orsini, M., Chiaverini, A., Centorame, P., Acciari, V. A., Salini, R., et al. (2021). Hypo - and hyper-virulent Listeria monocytogenes clones persisting in two different food processing plants of central Italy. Microorganisms 9:376.
Guillet, C., Join-Lambert, O., Le Monnier, A., Leclercq, A., Mechaï, F., Mamzer-Bruneel, M. F., et al. (2010). Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 16, 136–138. doi: 10.3201/eid1601.091155
Haase, J. K., Didelot, X., Lecuit, M., Korkeala, H., and Group, LmMS, et al. (2014). The ubiquitous nature of Listeria monocytogenes clones: a large-scale multilocus sequence typing study. Environ. Microbiol. 16, 405–416. doi: 10.1111/1462-2920.12342
Haddad, N., Johnson, N., Kathariou, S., Métris, A., Phister, T., Pielaat, A., et al. (2018). Next generation microbiological risk assessment-Potential of omics data for hazard characterisation. Int. J. Food Microbiol. 287, 28–39.
Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., et al. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 8:20. doi: 10.1186/1471-2180-8-20
Halbedel, S., Wilking, H., Holzer, A., Kleta, S., Fischer, M. A., Lüth, S., et al. (2020). Large nationwide outbreak of invasive listeriosis associated with blood sausage, Germany, 2018-2019. Emerg. Infect. Dis. 26, 1456–1464. doi: 10.3201/eid2607.200225
Harter, E., Wagner, E. M., Zaiser, A., Halecker, S., Wagner, M., and Rychli, K. (2017). Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 83:e00827-17. doi: 10.1128/AEM.00827-17
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stess, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in Sequence Type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi: 10.1128/AEM.02159-10
Heir, E., Lindstedt, B.-A., Røtterud, O.-J., Vardund, T., Kapperud, G., and Nesbakken, T. (2004). Molecular epidemiology and disinfectant susceptibility of Listeria monocytogenes from meat processing plants and human infections. Int. J. Food Microbiol. 96, 85–96. doi: 10.1016/j.ijfoodmicro.2004.03.014
Henderson, L. O., Erazo Flores, B. J., Skeens, J., Kent, D., Murphy, S. I., Wiedmann, M., et al. (2020). Nevertheless, she resisted - Role of the environment on Listeria monocytogenes sensitivity to nisin treatment in a laboratory cheese model. Front. Microbiol. 11:635. doi: 10.3389/fmicb.2020.00635
Hilliard, A., Leong, D., Callaghan, A. O., Culligan, E. P., Morgan, C. A., Delappe, N., et al. (2018). Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9:171. doi: 10.3390/genes9030171
Hingston, P., Chen, J., Dhillon, B. K., Laing, C., Bertelli, C., Gannon, V., et al. (2017). Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 8:369. doi: 10.3389/fmicb.2017.00369
Holch, A., Webb, K., Lukjancenko, O., Ussery, D., Rosenthal, B. M., and Gram, L. (2013). Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 79, 2944–2951. doi: 10.1128/AEM.03715-12
Horlbog, J. A., Kent, D., Stephan, R., and Guldimann, C. (2018). Surviving host- and food relevant stresses: phenotype of L. monocytogenes strains isolated from food and clinical sources. Sci. Rep. 8:12931. doi: 10.1038/s41598-018-30723-z
Horn, N., and Bhunia, A. K. (2018). Food-associated stress primes foodborne pathogens for the gastrointestinal phase of infection. Front. Microbiol. 9:1962. doi: 10.3389/fmicb.2018.01962
Hurley, D., Luque-Sastre, L., Parker, C. T., Huynh, S., Eshwar, A. K., Nguyen, S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4:e00252-19. doi: 10.1128/mSphere.00252-19
Jagadeesan, B., Gerner Smidt, P., Allard, M. W., Leuillet, S., Winkler, A., Xiao, Y., et al. (2019). The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol. 79, 96–115. doi: 10.1016/j.fm.2018.11.005
Jiang, X., Yu, T., Xu, Y., Wang, H., Korkeala, H., and Shi, L. (2019). MdrL, a major facilitator superfamily efflux pump of Listeria monocytogenes involved in tolerance to benzalkonium chloride. Appl. Microbiol. Biotechnol. 103, 1339–1350. doi: 10.1007/s00253-018-9551-y
Johnson, J., Jinneman, K., Stelma, G., Smith, B. G., Lye, D., Messer, J., et al. (2004). Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 70, 4256–4266. doi: 10.1128/AEM.70.7.4256-4266.2004
Jordan, S. J., Perni, S., Glenn, S., Fernandes, I., Barbosa, M., Sol, M., et al. (2008). Listeria monocytogenes biofilm-associated protein (BapL) may contribute to surface attachment of L. monocytogenesbut is absent from many field isolates. Appl. Environ. Microbiol. 74, 5451–5456. doi: 10.1128/AEM.02419-07
Kadam, S. R., Den Besten, H. M. W., van der Veen, S., Zwietering, M. H., Moezelaar, R., and Abee, T. (2013). Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 165, 259–264. doi: 10.1016/j.ijfoodmicro.2013.05.025
Kallipolitis, B., Gahan, C. G. M., and Piveteau, P. (2020). Factors contributing to Listeria monocytogenes transmission and impact on food safety. Curr. Opin. Food Sci. 36, 9–17.
Kang, J., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2015). VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl. Environ. Microbiol. 81, 4553–4562. doi: 10.1128/AEM.00648-15
Karatzas, K. A. G., Suur, L., and O’Byrne, C. P. (2012). Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 78, 3571–3579. doi: 10.1128/AEM.00227-12
Katharios-Lanwermeyer, S., Rakic-Martinez, M., Elhanafi, D., Ratani, S., Tiedje, J. M., and Kathariou, S. (2012). Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 78, 7549–7556. doi: 10.1128/AEM.02245-12
Katla, T., Naterstad, K., Vancanneyt, M., Swings, J., and Axelsson, L. (2003). Differences in susceptibility of Listeria monocytogenes strains to sakacin P, sakacin A, pediocin PA-1, and nisin. Appl. Environ. Microbiol. 69, 4431–4437. doi: 10.1128/AEM.69.8.4431-4437.2003
Kayode, A. J., Igbinosa, E. O., and Okoh, A. I. (2019). Overview of listeriosis in the Southern African hemisphere-review. J. Food Saf. 40:e12732. doi: 10.1111/jfs.12732
Keeney, K., Trmcic, A., Zhu, Z., Delaquis, P., and Wang, S. (2018). Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 67, 530–536. doi: 10.1111/lam.13072
Knudsen, G. M., Nielsen, J. B., Marvig, R. L., Ng, Y., Worning, P., Westh, H., et al. (2017). Genome-wide-analyses of Listeria monocytogenes from food-processing plants reveal clonal diversity and date the emergence of persisting sequence types. Environ. Microbiol. Rep. 9, 428–440. doi: 10.1111/1758-2229.12552
Kode, D., Nannapaneni, R., Bansal, M., Chang, S., Cheng, W. H., Sharma, C. S., et al. (2021). Low-level tolerance to fluoroquinolone antibiotic ciprofloxacin in QAC-adapted subpopulations of Listeria monocytogenes. Microorganisms 9:1052. doi: 10.3390/microorganisms9051052
Korsak, D., and Szuplewska, M. (2016). Characterization of nonpathogenic Listeria species isolated from food and food processing environment. Int. J. Food Microbiol. 238, 274–280. doi: 10.1016/j.ijfoodmicro.2016.08.032
Kovacevic, J., Ziegler, J., Wałecka-Zacharska, E., Reimer, A., Kitts, D. D., and Gilmour, M. W. (2015). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953. doi: 10.1128/AEM.03741-15
Kremer, P. H., Lees, J. A., Koopmans, M. M., Ferwerda, B., Arends, A. W., Feller, M. M., et al. (2017). Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 23, 265.e1–265.e7. doi: 10.1016/j.cmi.2016.12.008
Kuenne, C., Billion, A., Mraheil, M. A., Strittmatter, A., Daniel, R., Goesmann, A., et al. (2013). Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 14:47. doi: 10.1186/1471-2164-14-47
Kuenne, C., Voget, S., Pischimarov, J., Oehm, S., Goesmann, A., Daniel, R., et al. (2010). Comparative analysis of plasmids in the genus Listeria. PLoS One 5:e12511. doi: 10.1371/journal.pone.0012511
Lakicevic, B., Buncic, O., Katic, V., Lepsanovic, Z., Petrovic, L., Jankovic, V., et al. (2014). Detection of Listeria spp. during production and ripening of Petrovská klobása. Acta Vet. 64, 367–377. doi: 10.2478/acve-2014-0035
Lakicevic, B., and Nastasijevic, I. (2017). Listeria monocytogenes in retail establishments: contamination routes and control strategies. Food Rev. Int. 33, 247–269. doi: 10.1080/87559129.2016.1175017
Lang Halter, E., Neuhaus, K., and Scherer, S. (2013). Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int. J. Syst. Evol. Microbiol. 63, 641–647.
Lebrun, M., Audurier, A., and Cossart, P. (1994). Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 176, 3049–3061.
Leclercq, A., Clermont, D., Bizet, C., Grimont, P. A., Le Fleche-Mateos, A., Roche, S. M., et al. (2010). Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60, 2210–2214. doi: 10.1099/ijs.0.017376-0
Leclercq, A., Moura, A., Vales, G., Tessaud-Rita, N., Aguilhon, C., and Lecuit, M. (2019). Listeria thailandensis sp. nov. Int. J. Syst. Evol. Microbiol. 69, 74–81. doi: 10.1099/ijsem.0.003097
Lee, B.-H., Cole, S., Badel-Berchoux, S., Guillier, L., Felix, B., Krezdorn, N., et al. (2019). Biofilm formation of Listeria monocytogenes strains under food processing environments and pan-genome-wide association study. Front. Microbiol. 10:2698. doi: 10.3389/fmicb.2019.02698
Lee, S., Ward, T. J., Jima, D. D., Parsons, C., and Kathariou, S. (2017). The arsenic resistanceassociated Listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 83: e01189-17. doi: 10.1128/AEM.01189-17
Leong, D., Alvarez-Ordóñez, A., and Jordan, K. (2014). Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front. Microbiol. 5:436. doi: 10.3389/fmicb.2014.00436
Leong, D., Alvarez-Ordóñez, A., Zaouali, S., and Jordan, K. (2015). Examination of Listeria monocytogenes in seafood processing facilities and smoked salmon in the Republic of Ireland. J. Food Prot. 78, 2184–2190. doi: 10.4315/0362-028X.JFP-15-233
Liu, Y., Orsi, R. H., Gaballa, A., Wiedmann, M., Boor, K. J., and Guariglia Oropeza, V. (2019). Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 14, 801–828. doi: 10.2217/fmb-2019-0072
Lourenço, A., Neves, E., and Brito, L. (2009). Susceptibility of Listeria monocytogenes from traditional cheese-dairies to in-use sanitizers. Food Control 20, 585–589. doi: 10.1016/j.foodcont.2008.08.009
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS 38, 1091–1125. doi: 10.1111/1574-6976.12076
Lund, P. A., De Biase, D., Liran, O., Scheler, O., Mira, N. P., Cetecioglu, Z., et al. (2020). Understanding how microorganisms respond to acid pH is central to their control and successful exploitation. Front. Microbiol. 11:556140. doi: 10.3389/fmicb.2020.556140
Lundén, J., Autio, T., Markkula, A., Hellström, S., and Korkeala, H. (2003). Adaptive and cross-adaptive responses of persistent and non-persistent Listeria monocytogenes strains to disinfectants. Int. J. Food Microbiol. 82, 265–272. doi: 10.1016/s0168-1605(02)00312-4
Lundén, J. M., Miettinen, M. K., Autio, T. J., and Korkeala, H. J. (2000). Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J. Food Prot. 63, 1204–1207. doi: 10.4315/0362-028X-63.9.1204
Madeo, M., O’Riordan, N., Fuchs, T. M., Utratna, M., Karatzas, K. A., and O’Byrne, C. P. (2012). Thiamine plays a critical role in the acid tolerance of Listeria monocytogenes. FEMS Microbiol. Lett. 326, 137–143. doi: 10.1111/j.1574-6968.2011.02442.x
Malekmohammadi, S., Kodjovi, K. K., Sherwood, J., and Bergholz, T. M. (2017). Genetic and environmental factors influence Listeria monocytogenes nisin resistance. J. Appl. Microbiol. 123, 262–270. doi: 10.1111/jam.13479
Mandin, P., Fsihi, H., Dussurget, O., Vergassola, M., Milohanic, E., Toledo-Arana, A., et al. (2005). VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57, 1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x
Martínez-Suárez, J. V., Ortiz, S., and López-Alonso, V. (2016). Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front. Microbiol. 7:638. doi: 10.3389/fmicb.2016.00638
Matereke, L. T., and Okoh, A. I. (2020). Listeria monocytogenes virulence, antimicrobial, resistance and environmental persistence: a review. Pathogens 9:528. doi: 10.3390/pathogens9070528
Matle, I., Mafuna, T., Madoroba, E., Mbatha, K. R., Magwedere, K., and Pierneef, R. (2020). Population structure of non-ST6 Listeria monocytogene isolated in the red meat and poultry value chain in South Africa. Microorganisms 8:1152. doi: 10.3390/microorganisms8081152
Maury, M. M., Bracq-Dieye, H., Huang, L., Vales, G., Lavina, M., Thouvenot, P., et al. (2019). Hypervirulent L. monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 10:2488. doi: 10.1038/s41467-019-10380-0
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 8, 308–313. doi: 10.1038/ng.3501
Meier, A. B., Guldimann, C., Markkula, A., Pöntinen, A., Korkeala, H., and Tasara, T. (2017). Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8:397. doi: 10.3389/fmicb.2017.00397
Moreno, L. Z., Paixão, R., Gobbi, D. D., Raimundo, D. C., Ferreira, T. P., Hofer, E., et al. (2012). Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Res. Microbiol. 163, 268–271. doi: 10.1016/j.resmic.2012.02.004
Møretrø, T., Schirmer, B. C. T., Heir, E., Fagerlund, A., Hjemli, P., and Langsrud, S. (2017). Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 241, 215–224. doi: 10.1016/j.ijfoodmicro.2016.10.025
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Moura, A., Disson, O., Lavina, M., Thouvenot, P., Huang, L., Leclercq, A., et al. (2019). Atypical hemolytic Listeria innocua isolates are virulent, albeit less than Listeria monocytogenes. Infect. Immun. 87:e00758-18. doi: 10.1128/IAI.00758-18
Mullapudi, S., Siletzky, R. M., and Kathariou, S. (2008). Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74, 1464–1468. doi: 10.1128/AEM.02426-07
Mullapudi, S., Siletzky, R. M., and Kathariou, S. (2010). Diverse cadmium resistance determinants in Listeria monocytogenes isolates from the turkey processing plant environment. Appl. Environ. Microbiol. 76, 627–630.
Müller, A., Rychli, K., Muhterem-Uyar, M., Zaiser, A., Stessl, B., Guinane, C. M., et al. (2013). Tn6188 – a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835. doi: 10.1371/journal.pone.0076835
Müller, A., Rychli, K., Zaiser, A., Wieser, C., Wagner, M., and Schmitz-Esser, S. (2014). The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 361, 166–173. doi: 10.1111/1574-6968.12626
Müller-Herbst, S., Wüstner, S., Mühlig, A., Eder, D., Fuchs, T. M., Held, C., et al. (2014). Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology 160, 752–765. doi: 10.1099/mic.0.075242-0
Naditz, A. (2020). A Comparative Analysis of Listeria monocytogenes Plasmids: Presence, Contribution to Stress and Conservation. Graduate Theses and Dissertations. Ames, IA: Iowa State University.
Nikparvar, B., Andreevskaya, M., Duru, I. C., Bucur, F. I., Grigore-Gurgu, L., Borda, D., et al. (2021). Analysis of temporal gene regulation of Listeria monocytogenes revealed distinct regulatory response modes after exposure to high pressure processing. BMC Genom. 22:266. doi: 10.1186/s12864-021-07461-0
Nilsson, L., Huss, H. H., and Gram, L. (1997). Inhibition of Listeria monocytogenes on cold-smoked salmon by nisin and carbon dioxide atmosphere. Int. J. Food Microbiol. 38, 217–227. doi: 10.1016/s0168-1605(97)00111-6
Njage, P. M. K., Leekitcharoenphon, P., Hansen, L. T., Hendriksen, R. S., Faes, C., Aerts, M., et al. (2020). Quantitative microbial risk assessment based on whole genome sequencing data: case of Listeria monocytogenes. Microorganisms 8:1772. doi: 10.3390/microorganisms8111772
Norwood, D. E., and Gilmour, A. (1999). Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86, 576–582. doi: 10.1046/j.1365-2672.1999.00694.x
Norwood, D. E., and Gilmour, A. (2001). The differential adherence capabilities of two Listeria monocytogenes strains in monoculture and multispecies biofilms as a function of temperature. Lett. Appl. Microbiol. 33, 320–324. doi: 10.1046/j.1472-765x.2001.01004.x
Nüesch-Inderbinen, M., Bloemberg, G. V., Müller, A., Stevens, M., Cernela, N., Kollöffel, B., et al. (2021). Listeriosis caused by persistence of Listeria monocytogenes serotype 4b sequence type 6 in cheese production environment. Emerg. Infect. Dis. 27, 284–288. doi: 10.3201/eid2701.203266
Nuñez-Montero, K., Leclercq, A., Moura, A., Vales, G., Peraza, J., Pizarro-Cerda, J., et al. (2018). Listeria costaricensis sp. nov. Int. J. Syst. Evol. Microbiol. 68, 844–850. doi: 10.1099/ijsem.0.002596
Oliver, H. F., Orsi, R. H., Wiedmann, M., and Boor, K. J. (2010). Listeria monocytogenessigmaB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76, 4216–4232. doi: 10.1128/AEM.00031-10
Orsi, R. H., den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Ortiz, S., López, V., Garriga, M., and Martínez-Suárez. (2014). Antilisterial effect of two bioprotective cultures in a model system of Iberian chorizo fermentation. Int. J. Food Sci. Technol. 49, 753–758. doi: 10.1111/ijfs.12362
Ortiz, S., Lopez-Alonso, V., Rodriguez, P., and Martinez-Suarez, J. V. (2016). The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: evidence from comparative genome analysis. Appl. Environ. Microbiol. 82, 308–317. doi: 10.1128/AEM.02824-15
Painset, A., Björkman, J. T., Kiil, K., Guillier, L., Mariet, J. F., Félix, B., et al. (2019). LiSEQ - whole genome sequencing of a cross - sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 5:e000257. doi: 10.1099/mgen.0.000257
Palaiodimou, L., Fanning, S., and Fox, E. M. (2021). Genomic insights into persistence of Listeria species in the food processing environment. J. Appl. Microbiol. 131, 2082–2094. doi: 10.1111/jam.15089
Pan, Y., Breidt, F., and Kathariou, S. (2006). Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 72, 7711–7717. doi: 10.1128/AEM.01065-06
Parsons, C., Lee, S., Jayeola, V., and Kathariou, S. (2017). Novel cadmium resistance determinant in Listeria monocytogenes. Appl. Environ. Microbiol. 83:e02580-16. doi: 10.1128/AEM.02580-16
Parsons, C., Lee, S., and Kathariou, S. (2019). Heavy metal resistance determinants of the foodborne pathogen Listeria monocytogenes. Genes 10:11. doi: 10.3390/genes10010011
Pasquali, F., Palma, F., Guillier, L., Lucchi, A., De Cesare, A., and Manfreda, G. (2018). Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: persistence and ecophysiology. Front. Microbiol. 9:596. doi: 10.3389/fmicb.2018.00596
Paudyal, R., Barnes, C. P., and Karatzas, K. A. (2018). A novel approach in acidic disinfection through inhibition of acid resistance 2 mechanisms; maleic acid-mediated inhibition of glutamate decarboxylase activity 3 enhances acid sensitivity of Listeria monocytogenes. Food Microbiol. 69, 96–104. doi: 10.1016/j.fm.2017.07.013
Paudyal, R., O’Byrne, R. H., and Karatzas, K. A. (2020). Amino acids other than glutamate affect the expression of the GAD system in Listeria monocytogenes enhancing acid resistance. Food Microbiol. 90:103481. doi: 10.1016/j.fm.2020.103481
Pérez-Baltar, A., Pérez-Boto, D., Medina, M., and Montiel, R. (2021). Genomic diversity and characterization of Listeria monocytogenes from dry-cured ham processing plants. Food Microbiol. 99:103779. doi: 10.1016/j.fm.2021.103779
Pérez-Trallero, E., Zigorraga, C., Artieda, J., Alkorta, M., and Marimón, J. M. (2014). Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg. Infect. Dis 20, 2155–2157. doi: 10.3201/eid2012.140993
Quereda, J. J., Andersson, C., Cossart, P., Johansson, J., and Pizarro-Cerda, J. (2018). Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet. Res. 49:13. doi: 10.1186/s13567-017-0496-4
Quereda, J. J., Dussurget, O., Nahori, M. A., Ghozlane, A., Volant, S., Dillies, M. A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5706–5711. doi: 10.1073/pnas.1523899113
Quereda, J. J., Leclercq, A., Moura, A., Vales, G., Gomez-Martin, A., Garcia-Munoz, A., et al. (2020). Listeria valentina sp. nov., isolated from a water trough and the faeces of healthy sheep. Int. J. Syst. Evol. Microbiol. 70, 5868–5879. doi: 10.1099/ijsem.0.004494
Quereda, J. J., Meza-Torres, J., Cossart, P., and Pizarro-Cerdá, J. (2017). Listeriolysin S: a bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microb. 8, 384–391. doi: 10.1080/19490976.2017.1290759
Quereda, J. J., Morón-García, A., Palacios-Gorba, C., Dessaux, C., García-del Portillo, F., Graciela Pucciarelli, M., et al. (2021). Pathogenicity and virulence of Listeria monocytogenes: a trip from environmental to medical microbiology. Virulence 12, 2509–2545. doi: 10.1080/21505594.2021.1975526
Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171. doi: 10.1128/AEM.00951-07
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Ramalheira, R., Almeida, M., Azeredo, J., Brandão, T. R., Almeida, G., Silva, J., et al. (2010). Survival of clinical and food isolates of Listeria monocytogenes through simulated gastrointestinal tract conditions. Foodborne Pathog. Dis. 7, 121–128. doi: 10.1089/fpd.2009.0319
Rantsiou, K., Kathariou, S., Winkler, A., Skandamis, P., Saint-Cyr, M. J., Rouzeau-Szynalski, K., et al. (2018). Next generation microbiological risk assessment: opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int. J. Food Microbiol. 287, 3–9.
Ratani, S. S., Siletzky, R. M., Dutta, V., Yildirim, S., Osborne, J. A., Lin, W., et al. (2012). Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 78, 6938–6945. doi: 10.1128/AEM.01553-12
Roedel, A., Dieckmann, R., Brendebach, H., Hammerl, J. A., Kleta, S., Noll, M., et al. (2019). Biocide-tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 85:e01253-19. doi: 10.1128/AEM.01253-19
Ryan, S., Begley, M., Gahan, C. G., and Hill, C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. doi: 10.1111/j.1462-2920.2008.01782.x
Ryan, S., Begley, M., Hill, C., and Gahan, C. G. M. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi: 10.1111/j.1365-2672.2010.04726.x
Schlech, W. (2019). Epidemiology and clinical manifestations of Listeria monocytogenes infection. Microbiol. Spectr. 7, 1–12. doi: 10.1128/9781683670131.ch50
Schmitz-Esser, S., Müller, A., Stessl, B., and Wagner, M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6:380. doi: 10.3389/fmicb.2015.00380
Seeliger, H. P. R., and Hohne, K. (1979). Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13, 31–49.
Severino, P., Dussurget, O., Vêncio, R. Z., Dumas, E., Garrido, P., Padilla, G., et al. (2007). Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 73, 6078–6088. doi: 10.1128/AEM.02730-06
Sewell, D., Allen, S. C. H., and Phillips, C. A. (2015). Oxygen limitation induces acid tolerance and impacts simulated gastro-intestinal transit in Listeria monocytogenes J0161. Gut Pathog. 7, 1–5. doi: 10.1186/s13099-015-0058-0
Soares, C. A., and Knuckley, B. (2016). Mechanistic studies of the agmatine deiminase from Listeria monocytogenes. Biochem. J. 473, 1553–1561. doi: 10.1042/BCJ20160221
Stoller, A., Stevens, M., Stephan, R., and Guldimann, C. (2019). Characteristics of Listeria monocytogenes strains persisting in a meat processing facility over a 4-year period. Pathogens 8:32. doi: 10.3390/pathogens8010032
Szendy, M., Kalkhof, S., Bittrich, S., Kaiser, F., Leberecht, C., Labudde, D., et al. (2019). Structural change in GadD2 of Listeria monocytogenes field isolates supports nisin resistance. Int. J. Food Microbiol. 305:108240. doi: 10.1016/j.ijfoodmicro.2019.108240
Takahashi, H., Miya, S., Igarashi, K., Suda, T., Kuramoto, S., and Kimura, B. (2009). Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J. Food Prot. 72, 1476–1480. doi: 10.4315/0362-028X-72.7.1476
Tavares, R. M., Silva, D. A. L. D., Camargo, A. C., Yamatogi, R. S., and Nero, L. A. (2020). Interference of the acid stress on the expression of llsX by Listeria monocytogenes pathogenic island 3 (LIPI-3) variants. Food Res. Int. 132:109063. doi: 10.1016/j.foodres.2020.109063
Thedieck, K., Hain, T., Mohamed, W., Tindall, B. J., Nimtz, M., Chakraborty, T., et al. (2006). The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62, 1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x
Thomas, J., Govender, N., McCarthy, K. M., Erasmus, L. K., Doyle, T. J., Allam, M., et al. (2020). Outbreak of listeriosis in South Africa associated with processed meat. N. Engl. J. Med. 382, 632–643. doi: 10.1056/NEJMoa1907462
Tirloni, E., Stella, S., de Knegt, L. V., Gandolfi, G., Bernardi, C., and Nauta, M. J. (2018). A quantitative microbial risk assessment model for Listeria monocytogenes in RTE sandwiches. Microb. Risk Anal. 9, 11–21.
Trinetta, V., Floros, J. D., and Cutter, C. N. (2010). Sakacin a-containing pullulan film: an active packaging system to control epidemic clones of Listeria monocytogenes in ready-to-eat foods. J. Food Saf. 30, 366–381. doi: 10.1111/j.1745-4565.2010.00213.x
Upham, J., Chen, S., Boutilier, E., Hodges, L., Eisebraun, M., Croxen, M. A., et al. (2019). Potential ad hoc markers of persistence and virulence in Canadian Listeria monocytogenes food and clinical isolates. J. Food Prot. 82, 1909–1921. doi: 10.4315/0362-028X.JFP-19-028
van Schaik, W., Gahan, C. G., and Hill, C. (1999). Acid-adapted Listeria monocytogenes displays enhanced tolerance against the lantibiotics nisin and lacticin 3147. J. Food Prot. 62, 536–539. doi: 10.4315/0362-028x-62.5.536
Vázquez-Boland, J. A., Wagner, M., and Scortti, M. (2020). Why are some Listeria monocytogenes genotypes more likely to cause invasive (brain, placental) infection? mBio 11:e03126-20. doi: 10.1128/mBio.03126-20
Verghese, B., Lok, M., Wen, J., Alessandria, V., Chen, Y., Kathariou, S., et al. (2011). comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 77, 3279–3292. doi: 10.1128/AEM.00546-11
Volokhov, D. V., Duperrier, S., Neverov, A. A., George, J., Buchrieser, C., and Hitchins, A. D. (2007). The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 73, 1928–1939. doi: 10.1128/AEM.01796-06
Wambui, J., Eshwar, A. K., Aalto-Araneda, M., Pöntinen, A., Stevens, M. J. A., Njage, P. M. K., et al. (2020). The analysis of field strains isolated from food, animal and clinical sources uncovers natural mutations in Listeria monocytogenes nisin resistance genes. Front. Microbiol. 6:549531. doi: 10.3389/fmicb.2020.549531
Wang, Y., Luo, L., Li, O., Wang, H., Wang, Y., Sun, H., et al. (2019). Genomic dissection of the most prevalent Listeria monocytogenes clone, sequence type ST87, in China. BMC Genom. 20:1014. doi: 10.1186/s12864-019-6399-1
Weller, D., Andrus, A., Wiedmann, M., and den Bakker, H. C. (2015). Listeria booriae sp. nov. and Listeria newyorkensis sp. nov., from food processing environments in the USA. Int. J. Syst. Evol. Microbiol. 65, 286–292. doi: 10.1099/ijs.0.070839-0
Wieczorek, K., Bomba, A., and Osek, J. (2020). Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Int. J. Mol. Sci. 21:9419. doi: 10.3390/ijms21249419
Woraprayote, W., Kingcha, Y., Amonphanpokin, P., Kruenate, J., Zendo, T., Sonomoto, K., et al. (2013). Anti-listeria activity of poly(lactic acid)/sawdust particle biocomposite film impregnated with pediocin PA-1/AcH and its use in raw sliced pork. Int. J. Food Microbiol. 167, 229–235. doi: 10.1016/j.ijfoodmicro.2013.09.009
Xie, Y., Zhang, M., Gao, X., Shao, Y., Liu, H., Jin, J., et al. (2018). Development and antimicrobial application of plantaricin BM-1 incorporating a PVDC film on fresh pork meat during cold storage. J. Appl. Microbiol. 125, 1108–1116. doi: 10.1111/jam.13912
Xu, D., Li, Y., Zahid, M. S., Yamasaki, S., Shi, L., Li, J. R., et al. (2014). Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 190, 24–30. doi: 10.1016/j.ijfoodmicro.2014.08.017
Yin, Y., Yao, H., Doijad, S., Kong, S., Shen, Y., Cai, X., et al. (2019). A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat. Commun. 10:4283. doi: 10.1038/s41467-019-12072-1
Zhang, H., Chen, W., Wang, J., Xu, B., Liu, H., Dong, Q., et al. (2020). 10-year molecular surveillance of Listeria monocytogenes using whole-genome sequencing in Shanghai. China, 2009-2019. Front. Microbiol. 11:551020. doi: 10.3389/fmicb.2020.551020
Keywords: L. monocytogenes, stress genes, genomic islands, diversity, lineages, clonal complexes, low pH, persistence
Citation: Lakicevic BZ, Den Besten HMW and De Biase D (2022) Landscape of Stress Response and Virulence Genes Among Listeria monocytogenes Strains. Front. Microbiol. 12:738470. doi: 10.3389/fmicb.2021.738470
Received: 08 July 2021; Accepted: 30 November 2021;
Published: 20 January 2022.
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Beatrix Stessl, University of Veterinary Medicine Vienna, AustriaCopyright © 2022 Lakicevic, Den Besten and De Biase. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brankica Z. Lakicevic, YnJhbmtpY2EubGFraWNldmljQGlubWVzLnJz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.