
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 September 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.738058
This article is part of the Research Topic Plant Microbiome: Interactions, Mechanisms of Action, and Applications, Volume II View all 20 articles
Endophytes are well-acknowledged inoculants to promote plant growth, and extensive research has been done in different plants. However, there is a lacuna about the endophytes associated with tea clones and their benefit to promote plant growth. The present study focuses on isolating and characterizing the beneficial endophytic bacteria (EnB) prevalent in commercially important tea clones cultivated in North Eastern India as plant growth promoters. Diversity of culturable EnB microbiome, in vitro traits for plant growth promotion (PGP), and applicability of potent isolates as bioinoculant for in vivo PGP abilities have been assessed in the present study. A total of 106 EnB identified as members of phyla Proteobacteria, Firmicutes, and Actinobacteria were related to 22 different genera and six major clusters. Regarding PGP traits, the percentage of isolates positive for the production of indole acetic acid, phosphate solubilization, nitrogen fixation siderophore, ammonia, and 1-aminocyclopropane-1-carboxylic acid deaminase production were 86.8, 28.3, 78.3, 30.2, 95.3, and 87.7, respectively. In total, 34.0, 52.8, and 17.0% of EnB showed notable production of hydrolytic enzymes like cellulase, protease, and amylase, respectively. Additionally, based on the bonitur score, the top two isolates K96 identified as Stenotrophomonas sp. and M45 identified as Pseudomonas sp. were evaluated for biofilm formation, motility, and in vivo plant growth promoting activity. Results suggested strong biofilm formation and motility in K96 and M45 which may attribute to the colonization of the strains in the plants. Further in vivo plant growth promotion experiment suggested sturdy efficacy of the K96 and M45 as plant growth promoters in nursery condition in commercial tea clones Tocklai vegetative (TV) TV22 and TV26. Thus, this study emphasizes the opportunity of commercialization of the selected isolates for sustainable development of tea and other crops.
Plants interact with a broad range of diverse bacteria having a beneficial, neutral, or pathogenic impact on their hosts, in both natural and managed ecosystems. The majority of bacterial symbionts known as epiphytes colonize the rhizosphere (Compant et al., 2005; Lugtenberg and Kamilova, 2009). But a subset of bacteria from the rhizosphere are capable to enter and proliferate within plants as endophytes establishing a mutualistic association with the plants (Rosenblueth and Martínez-romero, 2006; Hallmann and Berg, 2007). Endophytes (bacteria or fungi) are an endosymbiotic group of microorganisms, ubiquitous in nature and well-known to dwell inside the plant endosphere without causing any apparent harmful effect on the host plant (Hallmann et al., 1997). Endophytic bacteria (EnB) have been isolated from almost all plants and plant parts such as roots, stems, leaves, barks, floral organs, and even seeds (Ryan et al., 2008; Compant et al., 2011; Dudeja et al., 2012). EnB studies in rice (Fujita et al., 2017), wheat (Emami et al., 2019), tomato (Khan et al., 2014), potato (Sessitsch et al., 2004), sugarcane (Mendes et al., 2007), and many other crops, medicinal plants, halophytes, and woody trees were mainly focussed, but endophytes from tea plants were rarely considered. To maintain stable symbiosis, endophytes ameliorate the host plant to tolerate abiotic and biotic stresses through phosphate solubilization (Liu et al., 2014; Oteino et al., 2015), nitrogen fixation (Hurek and Reinhold-Hurek, 2003; Zhu et al., 2012), indole-3-acetic acid (IAA) (Patten and Glick, 1996; Hassan, 2017), siderophore (Zhang et al., 2011), and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase (Zhang et al., 2011; Khan et al., 2014) synthesis and augment the resistance of plants to insects, pests, and pathogens by producing bioactive metabolites (Etminani and Harighi, 2018) and polymer hydrolyzing enzymes such as cellulase, amylase, chitinase, and protease (El-Deeb et al., 2012) and promote plant growth and development. Bacterial endophytes that have potentially beneficial effects on plant growth and development has been found in many genera including Arthrobacter, Azoarcus, Azospirillum, Azotobacter, Bacillus, Burkholderia, Curtobacterium, Erwinia, Gluconobacter, Klebsiella, Serratia, Pantoea, Herbaspirillum, Rahnella, Pseudomonas, and Xanthomonas (Zinniel et al., 2002; Berg et al., 2005; Compant et al., 2011).
Camellia sinensis (L) O. Kuntze (Tea), an economically important crop, is the world’s most-consumed non-alcoholic beverage which has been commercially produced by more than 30 countries worldwide (Hayat et al., 2015), with 1283.3 M. Kgs produced by India making it the world’s second-largest producer of tea (Sarani, 2020). In India, tea is extensively cultivated in Assam, Darjeeling, Nilgiri, and other North-Eastern (NE) states of India (Mondal et al., 2004). Assam plays a pivotal role in the national economy by producing 623.23 M. Kgs of tea, being the leading producer and exporter of commercial tea (Sarani, 2020). With the favorable climatic and topographic conditions of Meghalaya, suitable tea varieties brought from Assam and Darjeeling were transplanted at tea gardens located at Umsning, Meghalaya, India, in 1978 which were acclaimed by tea makers to be of very high quality (Directorate of Food Processing Meghalaya, n.d.). Tocklai Tea Research Institute (TTRI), Jorhat, Assam, India, has released 33 Tocklai Vegetative (TV) series tea clones, 153 region-specific garden series clones, and 16 biclonal seed stocks to the tea industry (Patel et al., 2019) and classified them based on their yield and quality into a standard clone (greater than average quality and yield), quality clone (high quality with average yield), and yield clone (average quality with high yield) (Das et al., 2012).
Owing to the production of piles of nutraceuticals in tea plants, consumption of tea drinks is related to numerous medicinal and nutritional benefits (Sharangi, 2009) such as antioxidant (Frei and Higdon, 2003), anti-inflammatory (Hsu et al., 2003), anti-carcinogenic (Kris-Etherton et al., 2002; Cooper et al., 2005), anti-allergic (Maeda-Yamamoto et al., 2007), and anti-microbial effect (Hamilton-Miller, 1995; Siddiqui et al., 2016). Although studies on tea phytochemicals and their potent effects have been well characterized, research on endophytic bacteria associated with tea clones is rather sparse.
Compared to the rhizospheric bacterial community (Dutta and Thakur, 2017) and endophytic fungal community (Win et al., 2018), EnB community of tea plants has received less attention. The interaction studies between EnB microflora associated with different tea clones cultivated especially in the NE region of India have not yet been investigated thoroughly. The culturable microbial diversity of EnB associated with tea clones has barely been explored. Also, the ability to utilize the EnB as plant growth promoters has not been well reported. Thus, the current study aims to investigate EnB community prevalent in selected commercially popular tea clones (TV1, TV22, TV9, Teenali 17, and TV25) using molecular, phylogenetic, and functional approaches. To evaluate the plant growth stimulation by plant growth-promoting bacteria (PGPB), phosphate solubilization, production of phytohormone, siderophore and ammonia, biological nitrogen fixation, and stress alleviation by production of ACC deaminase were carried out. To identify and explore the microbial diversity of the endophytic isolates molecular techniques such as 16S rRNA and phylogenetic analysis were used. Further, to determine the efficacy of the potent plant growth-promoting isolates, a nursery experiment was performed using two different yield clones, TV22 and TV26, which are extensively used for commercial cultivation in NE India. Thus, the results endowed the importance of EnB microflora as plant growth promoters that can be used to develop microbial formulations to reduce the use of agrochemicals and for sustainable development of tea plants including the soil.
Random sampling method was used to collect leaf and root samples from selected healthy tea plants of five clones, TV1, TV9, Teenali 17, TV22, and TV25 (Table 1), and all clones were selected based on their commercial importance. The samples were collected from two locations: Kopati Tea estate, Assam, India, and MEG tea estate, Meghalaya, India. The leaves and roots were collected aseptically in sterile ziplock bags as composite samples for each tea clone, transported to the laboratory in an ice box, and further processing for isolation was done within 1 month.
EnB was isolated by using surface sterilization and serial dilution method as described (Schulz et al., 1993; Sturz et al., 1998) with slight modifications. In brief, plant materials were washed for 3–5 min in running tap water to remove any debris and rinsed in 70% ethanol for 30 s followed by surface sterilization for 5 min using 1.5 and 2% v/v sodium hypochlorite solution for leaves and roots, respectively, followed by washing with sterile distilled water thrice for 5 min. The efficacy of surface sterilization was validated by spreading 100 μL aliquots of distilled water on nutrient agar (NA) plates taken from the last wash of the samples.
The surface-sterilized samples were excised and macerated in 0.9% sodium chloride (NaCl) with a sterile mortar pestle. EnB isolation was done by spread plate method, 100 μL of the aliquots were spread on selective media, namely Nutrient Agar, King’s B agar, Pikovskaya’s agar, Pseudomonas agar (HiMedia, India) followed by incubation at 28 ± 2°C for 48–72 h. Distinguished colonies based on phenotypic characters (size, elevation, color, form, and margin) were selected and pure culture of the isolates were made to determine the culturable bacterial population. For further studies and long-term preservation pure cultures were maintained at 4°C and −80°C as 20% (w/v) glycerol stocks, respectively.
Genomic DNA was extracted from isolates according to the protocol in the maker’s instructions as mentioned in QIAamp DNA mini kit (Qiagen, Hilden, Germany). The purity and concentration of the eluted DNA were quantified by NanoDrop spectrophotometer (Thermo Fisher Scientific, United States) and stored at 4°C until required for PCR.
In addition, 16S rRNA gene of the isolates was amplified using 1 μL (10 μmol L–1) forward (27F 5′-AGA GTT TGA TCC TGG CTC AG-3′) and reverse primers (1492R 5′- GGT TAC CTT GTT ACG ACT T-3′), 5 μL (10x) Taq buffer, 2.5 μL (2.5 mmol L–1) dNTPs, 0.5 μL (2.5 U) Taq polymerase in a total volume of 50 μL reaction mixture. PCR cycling conditions were set up in a Thermal cycler (Eppendorf, United States) as follows: 94°C for 5 min for initial denaturation of DNA, followed by 35 cycles of denaturation at 94°C for 30 s, annealing of primer at 55°C for 40 s, extension of DNA strand at 72°C for 1 min and final extension at 72°C for 10 min (Passari et al., 2015). The amplicons were then separated on a 1.2% agarose gel with ethidium bromide, visualized and documented in a BioRad Gel Doc XRS+ (Hercules, CA, United States). The PCR amplicons were then outsourced to 1st Base DNA Sequencing service, Malaysia for Sanger sequencing to determine the nucleotide sequence of DNA.
The 16S rRNA sequences were subjected to de-novo assembly using UGENE 37.0 and contigs generated were queried for homology using NCBI BLASTn and EZ taxon server 2.1 programs. The identified accessions were then submitted to GenBank and accession numbers were collected for each isolate. The multiple sequence alignment was carried out using MAFFT v7.45 (Katoh and Standley, 2013) and corrected by trimAl v1.4 using the gappyout algorithm (Capella-Gutiérrez et al., 2009) followed by a validation step using pmodel test tool of ETE3 toolkit (Huerta-Cepas et al., 2016). The evolutionary relationship between the accessions was analyzed by the maximum likelihood method using RAxML with 1,000 bootstrap steps (Stamatakis, 2014) and a phylogenetic tree was constructed using. Fig Tree 1.4.4 (Rambaut, 2018).
The EnB isolates were screened in vitro for identifying the traits required for nutrient acquisition and plant growth, like production of phytohormone, ammonia, and iron chelating agents, fixation of nitrogen from the atmosphere, production of stress alleviating enzymes, and hydrolytic enzymes. For all the PGP assays Bacillus pseudomycoides strain SN29 (GenBank accession no. KJ767523) (Dutta et al., 2015) was used as a positive control.
Gordon and Weber’s (1951) method was used to determine IAA production quantitatively and qualitatively with modifications. EnB isolates were inoculated in minimal salt (MS) medium amended with 2 mg mL–1 L-Tryptophan and incubated in an orbital shaker (170 rpm) at 28 ± 2°C for 48 h. To obtain bacterial cells free supernatant centrifugation at 10,000 rpm for 15 min was done. To the supernatant Salwoski reagent (0.5 M FeCl3 and 70% perchloric acid) was added in 1:2 ratio (v/v) and incubated in dark for 25 min at room temperature. After incubation optical density was measured at 530 nm using a multimode reader (Varioskan, Thermo Fisher Scientific). Commercial IAA (Sigma-Aldrich) was used as a standard to quantify IAA production. The experiment was done in triplicates.
Phosphate solubilization by EnB isolates was screened by spot inoculation on Pikovskaya’s agar medium (PKA) followed by incubation at 28 ± 2°C for 48 h. To quantify the potential of isolates to solubilize tricalcium phosphate, they were grown in NBRIP (National Botanical Research Institute’s Phosphate Growth) medium at 28°C for 48 h as described by Fiske and Subbarow (1925) and centrifuged at 10,000 rpm for 20 min to obtain supernatant. The amount of phosphate content in the supernatant was determined by adding ammonium molybdate reagent in equal volume and measured by colorimetry at 650 nm. A standard curve of KH2PO4 was used to determine the amount of phosphate solubilized which was expressed in mg L–1. The experiment was done in triplicates.
Quantitative analysis of siderophore production was determined by Chrome Azurol S (CAS) shuttle assay as described (Schwyn and Neilands, 1987; Alexander and Zuberer, 1991). The EnB isolates were grown in MS media at 28 ± 2°C for 48 h and after incubation culture was centrifuged at 10,000 rpm for 10 min and mixed with an equal volume of CAS reagent. The un-inoculated MS medium served as a control. The colorimetric measurements were performed by using a multimode reader at 630 nm. The percentage of siderophore produced (psu) by isolates was calculated by the following formula as described (Payne, 1994).
where Ar = absorbance of reference and As = absorbance of the sample at 630 nm
Ammonia production was determined as described (Yuen and Pollard, 1954; Singh et al., 2014). EnB isolates were inoculated into Peptone Water Broth medium and incubated at 28 ± 2°C, 150 rpm for 48 h. The bacteria cells were removed by centrifugation at 8,000 rpm for 5 min. To the supernatant, Nessler’s reagent was added in a 2:1 ratio (v/v). Yellow to brown color development indicated ammonia production. Ammonium sulfate (0.1–5.0 μmol mL–1) was used as a standard to quantify the amount of ammonia production, the absorbance of which was measured at 450 nm by a multimode reader and expressed as μmol mL–1. The experiment was done in triplicates.
Nitrogen-fixing abilities of the EnB isolates were determined qualitatively by culturing in Nitrogen-free (NF) medium containing (g L–1) mannitol 20 g, K2HPO4 0.2 g, NaCl 0.2 g, MgSO4.7H2O 0.2 g, K2SO4 0.1 g, CaCO3 5 g, and agar 20 g and Jensen’s agar medium by spot inoculation method as described (Kifle and Laing, 2016). The growth of the isolates was monitored after incubation at 28 ± 2°C for 48 h.
ACC deaminase activity of the EnB isolates was qualitatively assessed by spot inoculation method in Dworkin and Foster (DF) salt medium (Dworkin and Foster, 1958) amended with 3 mM ACC as sole nitrogen source. The inoculated plates were incubated for 48 h at 28 ± 2°C and monitored. The colonies growing on the plates were recognized as ACC deaminase producers (Glick et al., 2007).
For the production of carboxymethyl cellulase (CMCase), EnB isolates were spot inoculated on M9-CMC agar plates containing (g L–1), NaNO3 1 g, K2HPO4 1 g, KCl 1 g, MgSO4 0.5 g, yeast extract 5 g, agar 15 g, and CMC 2 g as substrate and incubated for 48 h at 28 ± 2°C. The incubated plates were flooded with 0.2% congo red solution for 15 min and de-stained with 0.1 M NaCl for 15 min. The formation of clear halo zones around colonies after 3–5 min confirmed cellulose hydrolysis (Meddeb-mouelhi et al., 2014).
The starch degrading ability of isolates to produce amylolytic enzymes was determined by spot inoculation on starch agar plates containing (g L–1) meat extract 3 g, peptic digest of animal tissue 5 g, starch soluble 2 g, and agar 15 g. Lugol solution (5 g iodine crystals and 10 g potassium iodide in 100 mL of distilled water) was flooded in plates for 2 min after incubation at 28 ± 2°C for 48 h. On starch hydrolysis, a clear zone around the colonies indicated amylase production (Iverson and Millis, 1974).
Protease production by EnB isolates was evaluated by spot inoculation on skim milk agar (SMA) medium (Sgroy et al., 2009). The inoculated plates were incubated at 28 ± 2°C for 48 h. Proteolytic activity was confirmed by the formation of clear halo zones around colonies.
In vitro biofilm formation was quantified using microtiter plate assay method described (O’Toole and Kolter, 1998) with minor modifications. Overnight grown cultures of EnB were re-suspended in nutrient broth (NB) medium and incubated in a rotary shaker (180 rpm) at 28°C to obtain the final OD600 = 0.2 (107 CFU mL–1) bacterial cells. Aliquots of 200 μL of EnB cultures were moved to 96 well microtiter plate and incubated at 28 ± 2°C for 48 h in static condition. NB broth without inoculation was taken as control. To remove the planktonic bacteria, the wells were emptied and washed thrice with 1X phosphate-buffered saline (PBS), pH 7.2 followed by staining with 0.1% crystal violet solution for 30 min. Excess bound stains on wells were removed and the wells were washed with 1X PBS and allowed to air dry. The bound crystal violet was then solubilized using 100% ethanol and the intensity of biofilm formation was measured at 570 nm using a multimode plate reader (Gupta et al., 2019).
Light microscopy and confocal microscopy were used to evaluate bacterial biofilm development. Freshly grown overnight bacterial cultures (20 μL) of K96 and M45 were inoculated in sterile glass coverslip (20 mm) and placed in a 60 mm culture plate and incubated at 28 ± 2°C for 24–48 h under static condition. Loosely adhered cells were removed from the surface by washing glass coverslip thrice with 1X PBS and stained with 0.1% crystal violet (CV) for 10 min and 0.1% acridine orange (AO) for 20 min followed by removal of excess stain by washing with 1X PBS and allowed to dry. To examine the extent of microbial attachment or biofilm formation on the glass surface the slides stained with CV were visualized under a compound microscope.
Confocal Laser Scanning Microscope (Clsm) imaging was performed using a Leica Tcs Sp8 confocal laser scanning microscope (Leica, Mannheim, Germany) equipped with a visible light laser, Leica Dmi 6000B inverted light microscope, and an Hc Pl Apo Cs2 63X/1.40 Oil immersion objective. Acridine Orange (Ao) was excited using a 488 nm argon laser line, while the fluorescent emission was detected from 502 to 526 nm. The pinhole aperture size was adjusted to 1.8 Airy units and detector gain was set to 630 for visualization of the biofilm. In the z-plane, 36 fluorescent optical images were collected to span the full depth of the biofilms. Leica Application Suite X software was used for visualization and ImageJ software was used for the analysis of the images.
Microbial Adhesion to Hydrocarbons (MATH) method was used to establish CSH by K96 and M45 EnB isolates as described (Rosenberg, 2006). The percentage of hydrophobicity was described as follows:
where A0 is the initial absorbance of bacterial culture at 400 nm and A1 is the final absorbance of the aqueous phase at 400 nm.
Bacterial motility assays (swimming and swarming) were performed by the protocol described (Gupta et al., 2019) with slight modification. Luria broth (LB) medium with 0.3% agar (w/v) and 0.5% (w/v) agar supplemented with 5 g L–1 D-glucose was used for swimming motility and swarming motility assay, respectively. To study the bacterial migration freshly grown bacterial culture (5 μL, OD600 = 0.2) of cell density (107 cells mL–1) was inoculated on each assay plate and incubated in an inverted position at 28 ± 2°C for 48 h and the swarm diameter for both swimming and swarming motilities was measured at 24 and 48 h of incubation and expressed in mm. The twitching motility assay was done by inoculation of freshly grown bacterial isolates in LB medium with 0.6% agar followed by incubation at 28 ± 2°C for 48 h (Henrichsen, 1983). The bacterial twitching motility was visualized under the microscope (Motic BA410).
For the plant growth promotion experiment in nursery conditions, 3-months-old tea vegetative clones TV22 (Indo-China) and TV26 (Ayapthar D × Ayapathar A) were collected from Bateli Tea estate, Assam. These TV clones are Cambod type yield clones (high yield average quality). The procured tea clones were propagated mainly through vegetative propagation using cuttings for commercial cultivation. The cuttings consist of a 3–4 cm long single leaf with a stem below the node about 2.5 cm and about 0.5 cm of stem above it. Before propagation, the cuttings were first treated with 0.1% solution of zinc sulfate. For callusing or rooting of cuttings they are transferred to nursery beds. After rooting (6–8 weeks), the cuttings were transferred to polythene sleeves where they were grown and maintained in nursery condition. For vegetative propagation of tea in the tea estate, the sleeves were prepared following standard protocol described in Field Management Notes, TTRI, Jorhat. The polythene sleeves prepared are 15 ± 17.7 cm layflat wide, 20 ± 25 cm long, and 150 gauge thick in size. The soil used in the sleeves contains well-drained sandy loam (15–20% clay less than 0.002 mm, 0–50% silt 0.002–0.2 mm, and 50–70% sand greater than 0.2 mm) of good tilth having ideal pH of 4.5.
For the PGP experiment in nursery condition, two potent isolates K96 (Stenotrophomonas sp., GenBank Accession number MW905624) and M45 (Pseudomonas sp., GenBank Accession number MZ008002) were selected based on in vitro PGP abilities. For mass production, the pure bacterial cultures were inoculated in NB (HiMedia, India) and grown for 24 h at 28 ± 2°C at 180 rpm. The cultures were centrifuged at 10,000 rpm for 15 min and the pellets were washed twice with PBS to obtain a final suspension of 108 CFU mL–1 (Xu et al., 2014). Then, 10 mL of the resulting cell suspension was used to treat the tea clones in the nursery condition by soil drenching method in 15 days interval for 6 months (Dutta and Thakur, 2017; Borah et al., 2019). The nursery experiment was divided into three treatments, namely T1:K96 inoculation, T2 :M45 inoculation, T3 :inoculated with consortia of K96 and M45, and control, without bacterial inoculum, with three replicates each. To retain the soil moisture the saplings were watered once daily until harvesting. Harvesting of the tea saplings was done after 6 months of the last inoculum treatment. Physiological parameters of plants (number of leaves, shoot and root length along with their fresh and dry weight, and chlorophyll content) were recorded and compared to control.
Mature tea leaves were obtained from the treated and untreated tea clones to quantitatively determine the chlorophyll content in the leaf tissues as described by Hiscox and Israelstam (1979). To facilitate the extraction of chlorophyll, 100 mg of fresh leaf tissue was cut into small pieces and immersed in 7 mL DMSO (HiMedia). After extraction at 65°C for 30 min in dark using a water bath, the supernatant was decanted and absorbance was determined at 645 and 663 nm against DMSO as blank using a multimode reader. Chlorophyll a and b were calculated by the following formula (Arnon, 1949):
where A = absorbance at specific wavelengths; V = final volume of chlorophyll extract; W = fresh weight of tissue extracted.
The Vennture software (Martin et al., 2012) was used to make a Venn diagram to represent all the isolates producing different PGP traits. The plant growth parameters were analyzed statistically by subjecting to two-way Anova (analysis of variance). Statistically significant data was determined at p ≤ 0.05. The interrelationships among the treatments and vegetative parameters were evaluated using principal component analysis (PCA) in Matlab R (2017a). In addition, to determine the absolute value difference between group means of control and treatment fold change analysis was done. The resulting values are generated in a log2 scale such that the up or down-regulation will be equidistant to the baseline.
Altogether 106 culturable EnB were isolated from leaves and roots of five different tea clones using different enriched media and identified based on their morphological characteristics. The population density of EnB isolates in the selected tea clones varied from 3.2 to 4.4 log10 colony forming units g–1 (CFU g–1) per fresh weight (Supplementary Figure 1). In addition, 59.43% EnB (n = 63) isolates were obtained from roots and 40.56% (n = 43) from leaves. The bacterial isolates were confirmed to be endophytes as no bacterial colonies were observed in the control plates. The relative abundance of each EnB isolated was determined at the genus level in both leaf and root samples of the five different tea clones. In leaf samples, as evident from Figure 1A, Alcaligenes sp. (42.58%) was most abundant in TV1 clone, Brevibacillus sp. (100%) in TV9, Bacillus sp. (28.83%) and Alcaligenes sp. (28.83%) in TV22, Brevundimonas sp. (67.14%) in TV25, and Alcaligenes sp. (32.05%) in Teenali17. In root samples, as shown in Figure 1B, Pseudomonas sp. (79.97%) was most abundant in TV1, Bacillus sp. (47.38%) in TV9, Microbacterium sp. (36.67%) in TV22, Stenotrophomonas sp. (62.52%) in TV25, and Bacillus sp. (49.98%) in Teenali 17.
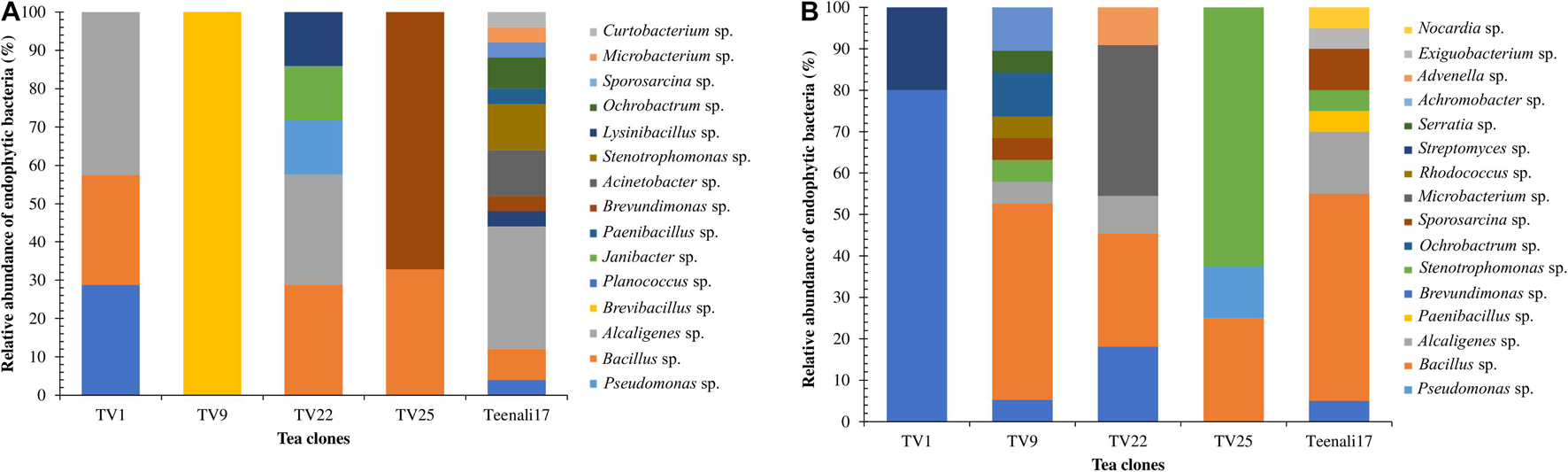
Figure 1. Relative abundance (in percentage) of endophytic bacterial genera procured from (A) leaf and (B) roots of five different tea clones (TV1, TV9, TV22, TV25, and Teenali 17).
The Shannon diversity indices of EnB were more in leaf (H = 2.3) as compared to the root (H = 2.14). Simpson diversity index was 0.125 in leaf and 0.177 in the root. Species richness (S) was found to be 63 in root and 43 in leaf samples. The species evenness value was found to be 0.610 in leaf and 0.516 in the root. The average population size is found to be 2.87 and 3.94 in leaf and root samples, respectively. The population of EnB isolates with their diversity indices is shown in Table 2.
The present study revealed a diverse population of bacterial isolates within 22 different genera based on 16S rRNA gene sequencing. The sequence results procured from BLAST validated the identity with 91–100% similarity. The 16s rRNA sequence GenBank accession numbers of individual isolates and sample details are listed in Supplementary Table 1. The phylogenetic analysis by maximum likelihood revealed 6 major clusters (Figure 2). Cluster II was the largest consisting of 42 isolates of different genera belonging to Bacillus sp., Sporosarcina sp., Paenibacillus sp., Lysinibacillus sp., Planococcus sp., Brevibacillus sp., and Exiguobacterium sp. Actinobacteria species viz. Microbacterium sp., Streptomyces sp., Curtobacterium sp., Janibacter sp., Rhodococcus sp., and Nocardia sp. were distributed separately in cluster I which consisted of 11 isolates. Pseudomonas sp., Acinetobacter sp., and Serratia sp. were found to be in cluster III containing 15 isolates. Cluster IV comprised of 21 isolates, where it was mostly dominated by Alcaligenes sp. followed by Achromobacter sp. and Advenella sp. All 10 isolates in cluster V belongs to Stenotrophomonas sp. and 7 isolates in cluster VI belong to the genera Ochrobactrum sp. and Brevundimonas sp. Cluster I, II, and the rest of the clusters are found to be divided into two distinct sub-nodes from the origin, following a separate hierarchy. Taxa within the same genera and similar tissue (viz. leaf and root) can be distinguished separately in the subclusters with minimum evolutionary distance. In most of the cases, a diverse phylogenetic relationship was observed between accessions originating from different tea clones.
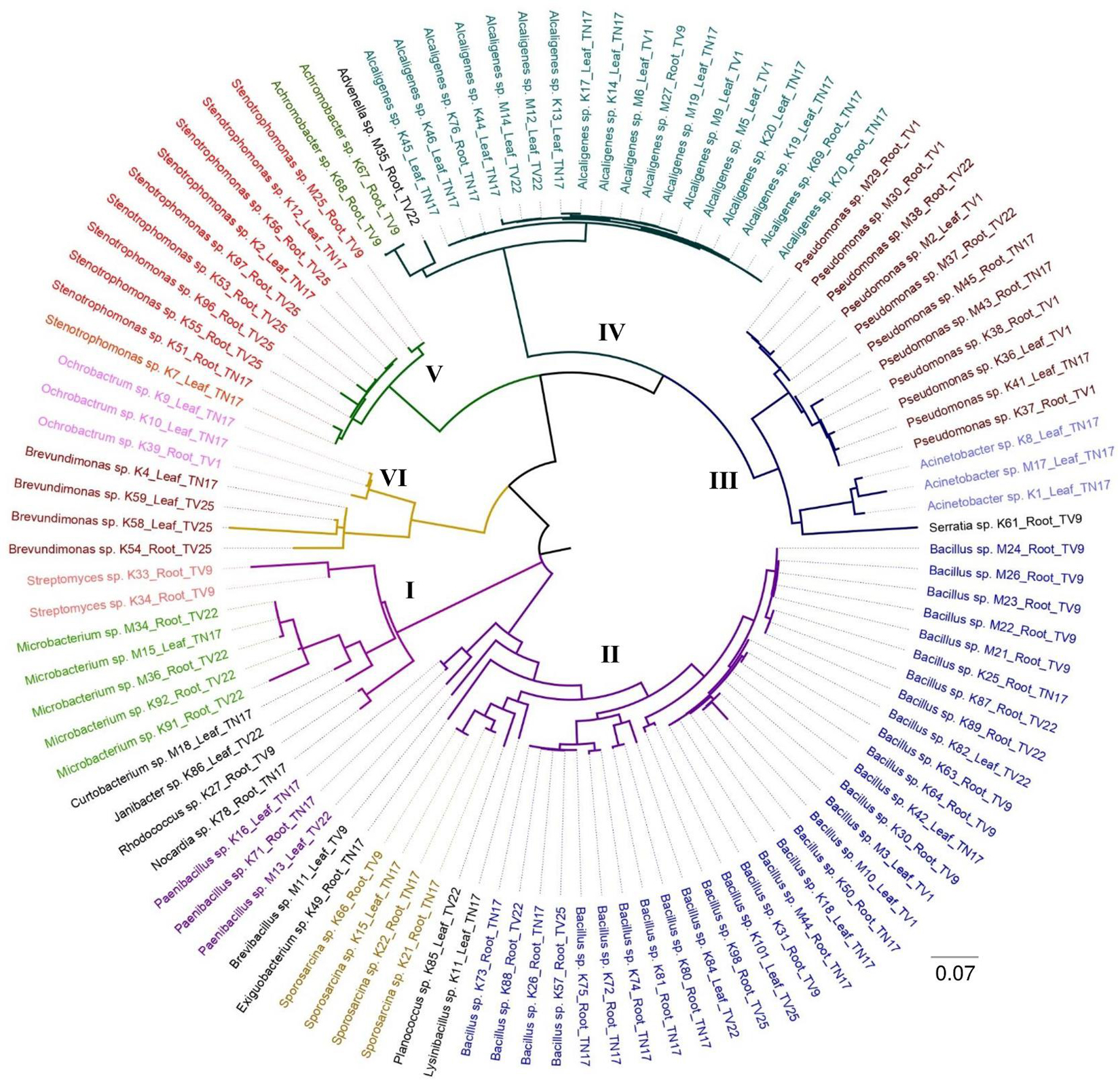
Figure 2. The maximum likelihood phylogenetic tree of endophytic bacterial isolates obtained from five different tea clones based on 16S rRNA gene sequences; the scale bar represents the distance from the origin to the most distant taxa. Clades that are distinctly clustered are represented by six different groups.
All the 106 isolates when screened for various PGP traits in vitro showed at least one trait as shown in Figure 3A and Supplementary Table 2. On the combined evaluation of all the six PGP traits, seven isolates (K63, K71, K96, M2, M29, M37, and M45) revealed the capacity to produce all the traits (Phosphate solubilization, IAA, Ammonia and Siderophore production, Nitrogen fixation and ACC deaminase activity) which is shown in the Venn diagram in Figure 3B.
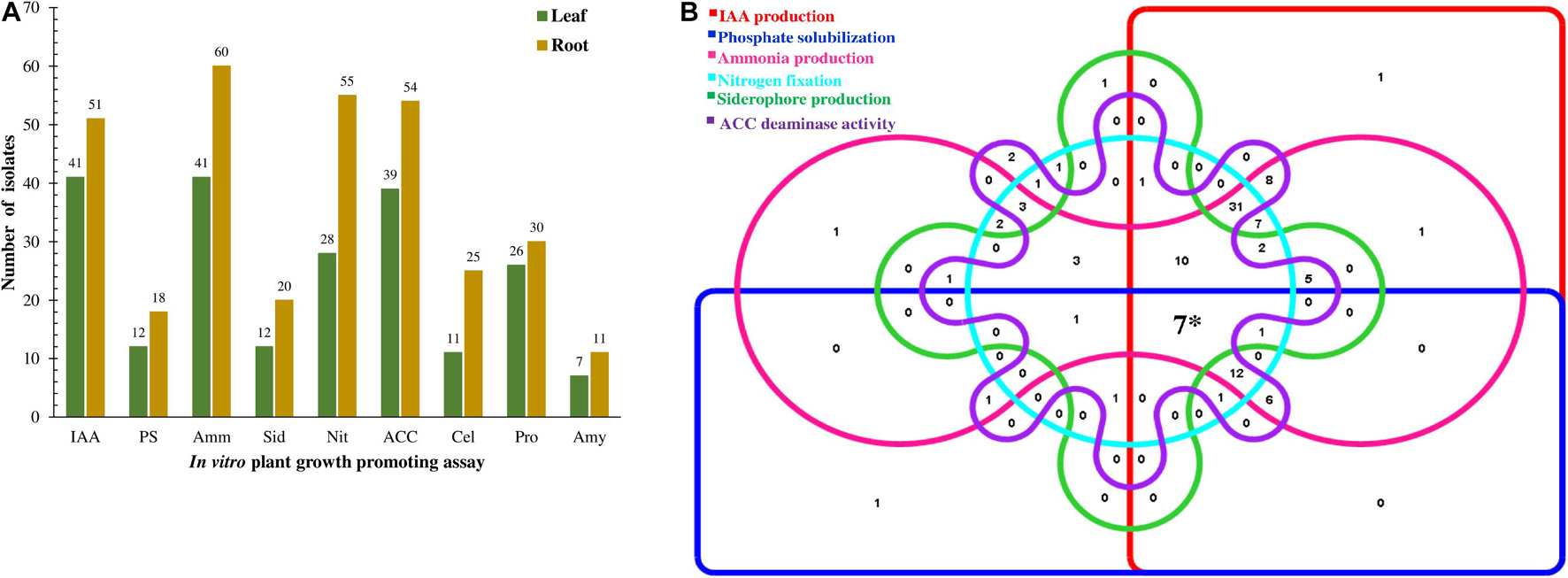
Figure 3. Representation of plant growth-promoting traits showed by endophytic bacteria isolated from the tea clones. (A) Quantitative distribution of bacterial isolates according to PGP traits and isolation source and (B) Venn diagram illustrating the number of isolates showing PGP traits. The 106 isolates were allocated to 6 different PGP traits. (*7 showed PGP traits for all the attributes).
The ability to biosynthesize IAA was observed in 92 isolates, 41 isolates from leaf, and 51 from the root. The majority of isolates to produce IAA belong to the genera Bacillus sp. (26 isolates) followed by Alcaligenes sp. (16 isolates), Pseudomonas sp. (11 isolates), and Stenotrophomonas sp. (10 isolates). Quantitatively IAA production in presence of L-Tryptophan ranges from 3.00 ± 1.19 to 129.84 ± 4.72 μg mL–1. Twelve (11.32%) produced more than 40 μg mL–1. Isolate Ochrobactrum sp. (K39) showed the highest production of IAA of 129.84 ± 4.72 μg mL–1 followed by Stenotrophomonas sp. (K96) 123.55 ± 2.50 μg mL–1.
In addition, 28.30% of all the isolates (n = 30), among which 12 from leaf and 18 from roots, were identified to solubilize inorganic phosphate based on the development of clear zones around colonies in Pikovskaya’s agar medium. Among the isolates Bacillus sp. (10 isolates) followed by Pseudomonas sp. (6 isolates) showed maximum phosphate solubilizing activity. The efficiency of phosphate solubilization varies from 2.79 ± 0.67 to 81.42 ± 5.73 mg L–1. The highest phosphate solubilizing ability was shown by Bacillus sp. (K98) which solubilized 81.42 ± 5.73 mg L–1 calcium phosphate available in the medium followed by Serratia sp. (K61) 73.24 ± 0.38 mg L–1 and Stenotrophomonas sp. (K96) 71.62 ± 2.2 mg L–1.
The majority of isolates (101) produced ammonia, accounting for 41 isolates from leaf and 60 isolates from roots. The majority of ammonia producers belong to the genera Bacillus sp. (29 isolates) followed by Alcaligenes sp. (17 isolates) and Pseudomonas sp. (11 isolates). The amount of ammonia produced falls in the range 0.61 ± 0.26–4.92 ± 0.19 μmol mL–1 with 39 isolates (36.79%) having the capacity to produce ammonia more than 3.0 μg mL–1. Sporosarcina sp. (M15) produced the highest amount of ammonia of 4.92 ± 0.19 μmol mL–1 in the medium.
Also, 78.30% of the total isolates were capable of growing on NF media and Jensen’s agar media, where 28 were from leaf and 55 from roots. Maximum nitrogen fixation was observed in Bacillus sp. (31 isolates) subsequently by Pseudomonas sp. (11 isolates), Stenotrophomonas sp. (10 isolates), and Alcaligenes sp. (10 isolates). NF media and Jensen’s agar media are defined medium without nitrogen used for detection of nitrogen-fixing bacteria. The growth of EnB on medium indicates that these bacteria can fix atmospheric nitrogen.
Siderophore production in liquid media was detected in 32 (30.18%) isolates among which 12 isolates were from leaf and 20 from roots, where maximum production was shown by genera Bacillus sp. (9 isolates). Quantitative estimation of siderophore production showed that isolate Brevundimonas sp. (K4) and Bacillus sp. (K87) produced the highest quantities of siderophore units (SU) of 83.22% followed by 81.2% SU by Pseudomonas sp. (M45) and 79.06% SU by Stenotrophomonas sp. (K96). A total of 17 isolates produced significantly higher quantities of siderophores above 40% siderophore units.
Out of 93 EnB isolates, 39 from leaf and 54 from roots utilized ACC as the sole nitrogen source and exhibit ACC deaminase activity. Genera Bacillus sp. (29 isolates) showed maximum ACC deaminase activity followed by Alcaligenes sp. (15 isolates) and Stenotrophomonas sp. (10 isolates). The isolates showing similar diameters as the control plate was considered to be weak or negative isolates and the isolates showing greater diameter than control plates were considered to be positive.
Qualitative analysis of 106 EnB isolates for hydrolysis of extracellular enzymes such as cellulase, protease, and amylase was performed. A total of 56 (52.83%) isolates hydrolyzed proteins by the formation of clear zones around colonies in SMA medium. For amylase screening, 20 (18.87%) isolates hydrolyzed starch by the production of amylase in starch agar medium by the formation of clear zones around bacterial isolates and cellulase enzyme production by 37 (34.90%) isolates were confirmed by the development of clear halo zones in the vicinity of colonies in M9-CMC medium (Figure 3A, Table 3, and Supplementary Table 2).
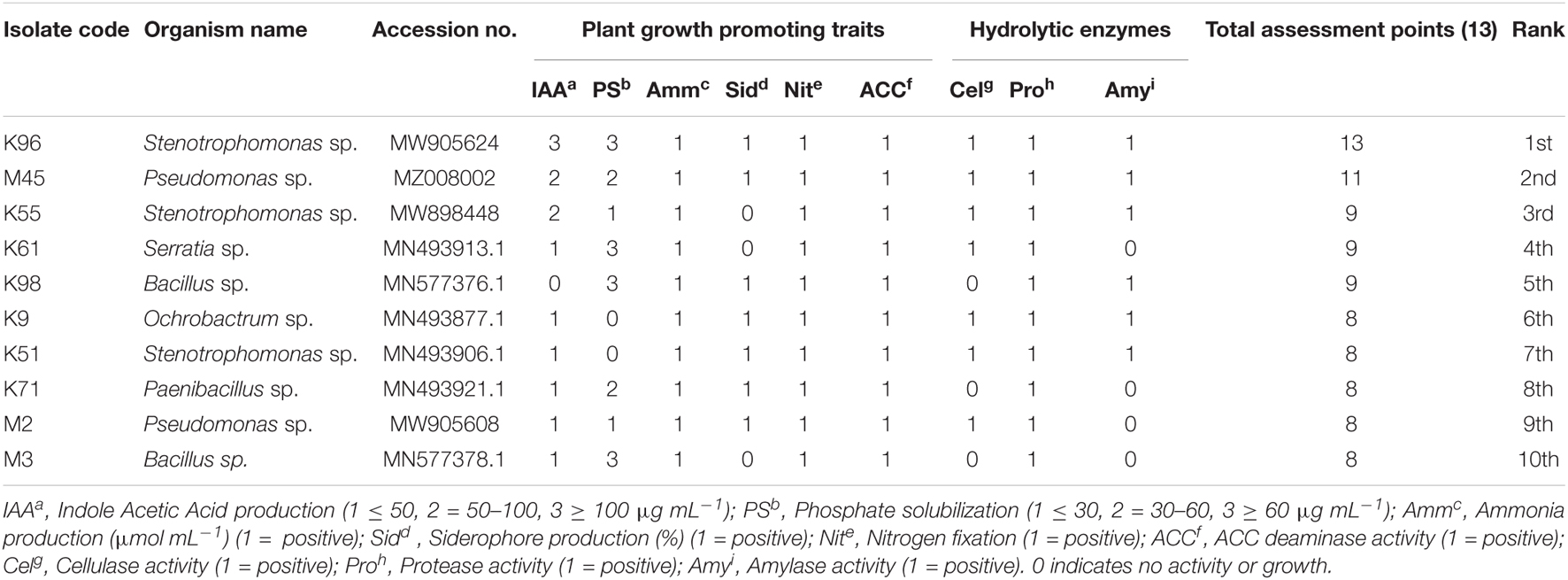
Table 3. Endophytic bacterial isolates with their PGP traits and general ranking for their ability to function as plant growth promoters based on bonitur scale.
A bonitur scale was generated similar to that described by El-Sayed et al. (2014) to select the best EnB isolates having high plant growth-promoting potential. In this scale PGP traits examined are scored. The maximum possible score in the traits examined here is 13 points: 3 points each for IAA production and phosphate solubilization; 1 point each for nitrogen fixation, ammonia, siderophore, and ACC deaminase production, and production of hydrolytic enzymes was given as the assessment values. The assessment revealed that isolate Stenotrophomonas sp. (K96) was the foremost isolate exhibiting the highest score of 13 points followed by Pseudomonas sp. (M45) exhibiting 11 points. Based on the bonitur scale the top 10 EnB isolates with the ranking for their ability to function as plant growth promoters are shown in Table 3 and the ranking of all the isolates are shown in Supplementary Table 2.
Bacterial isolate M45 showed strong biofilm formation and isolate K96 showed moderate biofilm formation at 48 h, i.e., during the stationary growth phase. The same was observed when biofilm was developed on a glass slide and observed under a compound microscope at 24 and 48 h after staining with crystal violet (Figures 4A–D). The attachment of bacterial cells increased significantly after 12 h (initial period of lag phase) as assessed using crystal violet (Figure 4E).
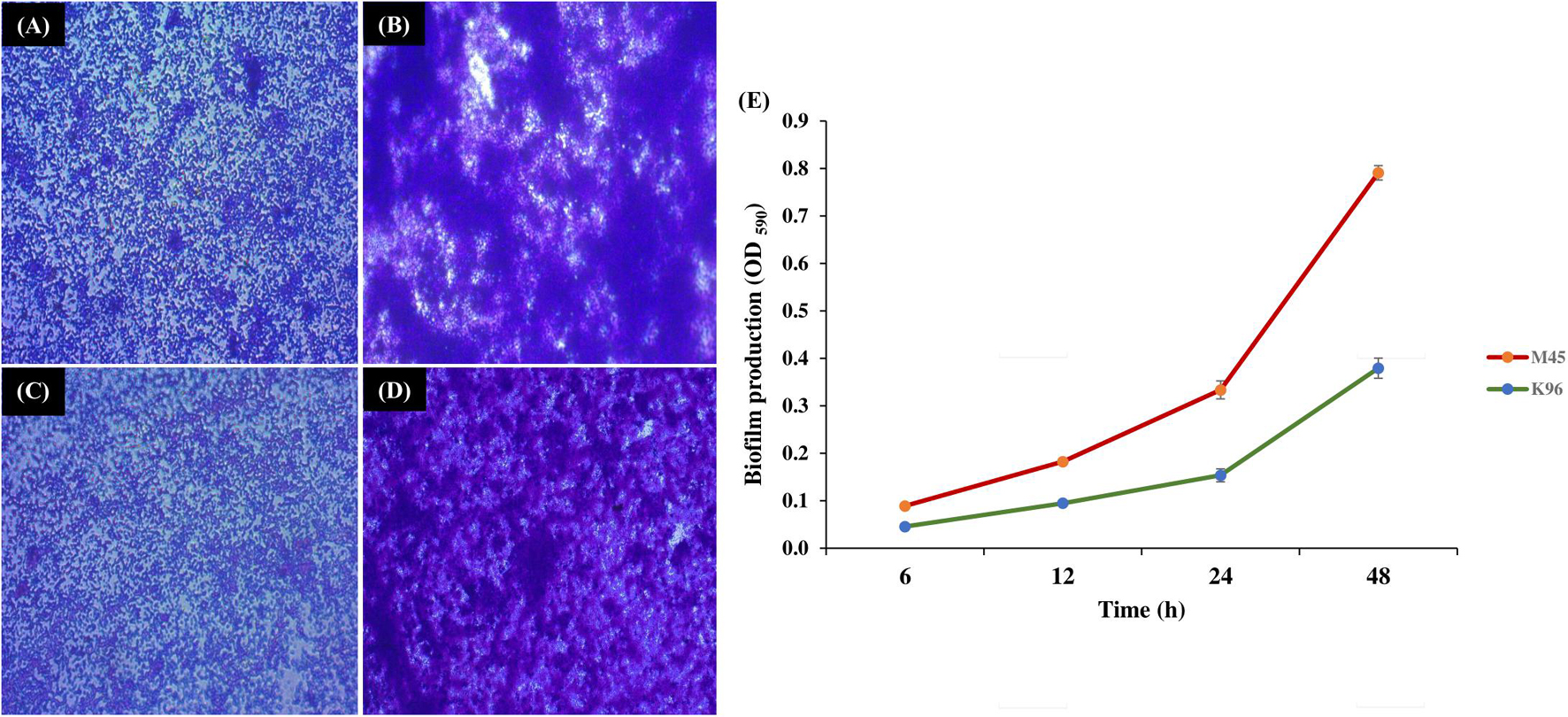
Figure 4. Biofilm formation by potent bacterial isolates. Light microscopy images of biofilm formation by (A,B) M45 (Pseudomonas sp.), and (C,D) K96 (Stenotrophomonas sp.) at 24 and 48 h, respectively. The biofilms were stained by crystal violet; (E) Quantification of biofilm production for 48 h by M45 and K96 after elution of crystal violet stain by ethanol.
CLSM microscopy observations revealed the thickness of the biofilms in the z-plane as 9 μm for K96 and 10 μm M45, respectively. The biofilm growth revealed that the cells of K96 and M45 attached to the glass surface and colonized the whole surface (Figure 5 and Supplementary Figure 4).
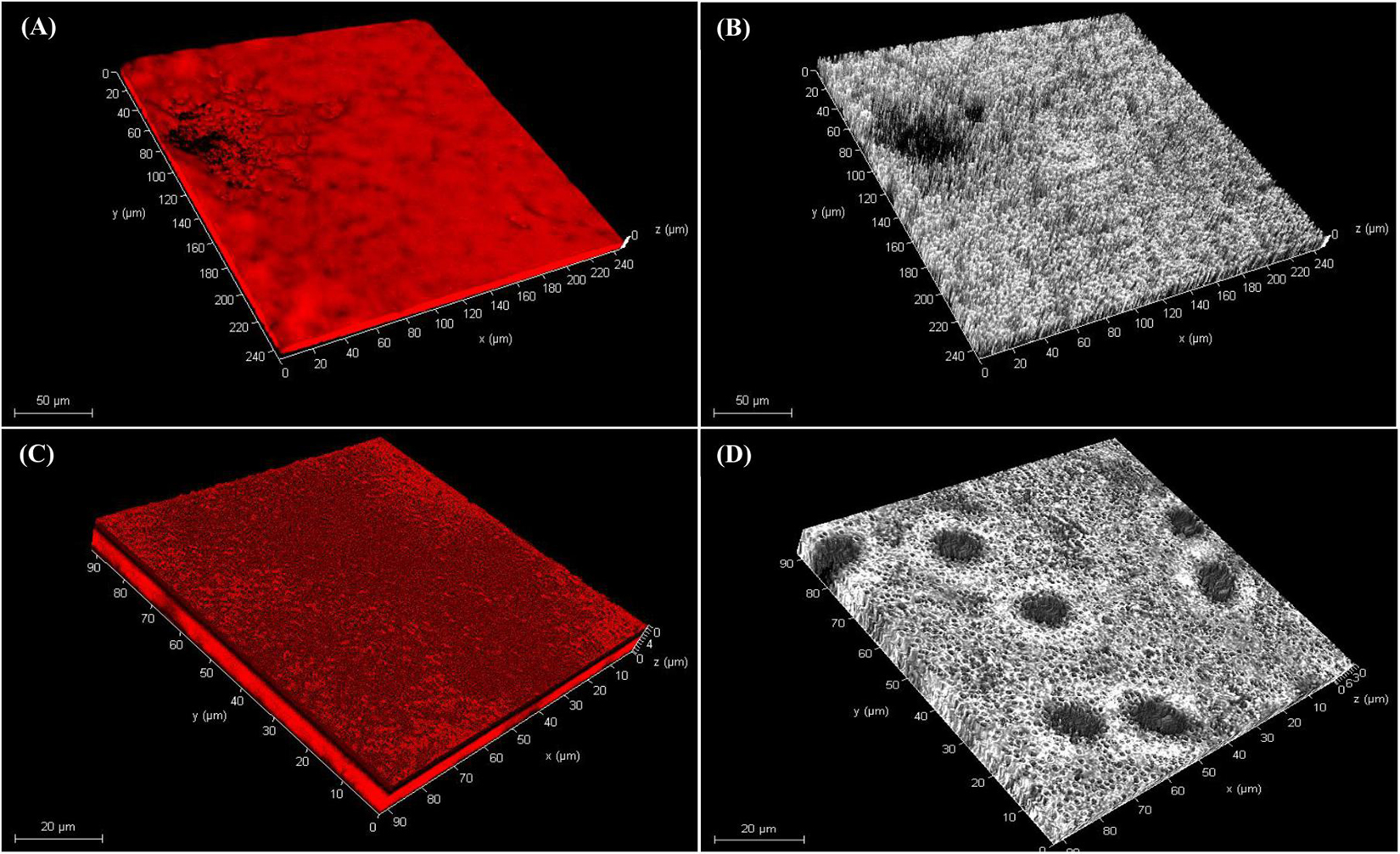
Figure 5. Confocal laser scanning microscopy of biofilm. Three-dimensional (3D) views of (A,B) M45 and (C,D) K96 biofilms grown for 48 h. Bacteria were stained with acridine orange and observed by CLSM.
The bacterial CSH plays a crucial role in bacterial aggregation which sequentially promotes bacterial dispersion and survival in soil and aid in microbial adhesion to plant surfaces during biofilm formation. In this study, 38.29% CSH was shown by K96 and 27.77% CSH by M45.
The bacterial motility was studied over time for bacterial isolates M45 and K96. Both the isolates were seen to exhibit twitching, swimming, and swarming motility. Isolates M45 and K96 isolates produced a flat, widely spread, irregularly shaped colony at 100X magnification in media-specific for twitching motility (Figures 6A,B). The isolates were seen to exhibit swarming motility with diameter 53 mm ± 1.2 and 45 mm ± 0.25 for K96 and M45, respectively, within a time period of 48 h (Figures 6C–E). Swimming assays revealed that M45 and K96 isolates are motile presenting concentric halos on 0.3% agar plates at 48 h (Figures 6F–H).
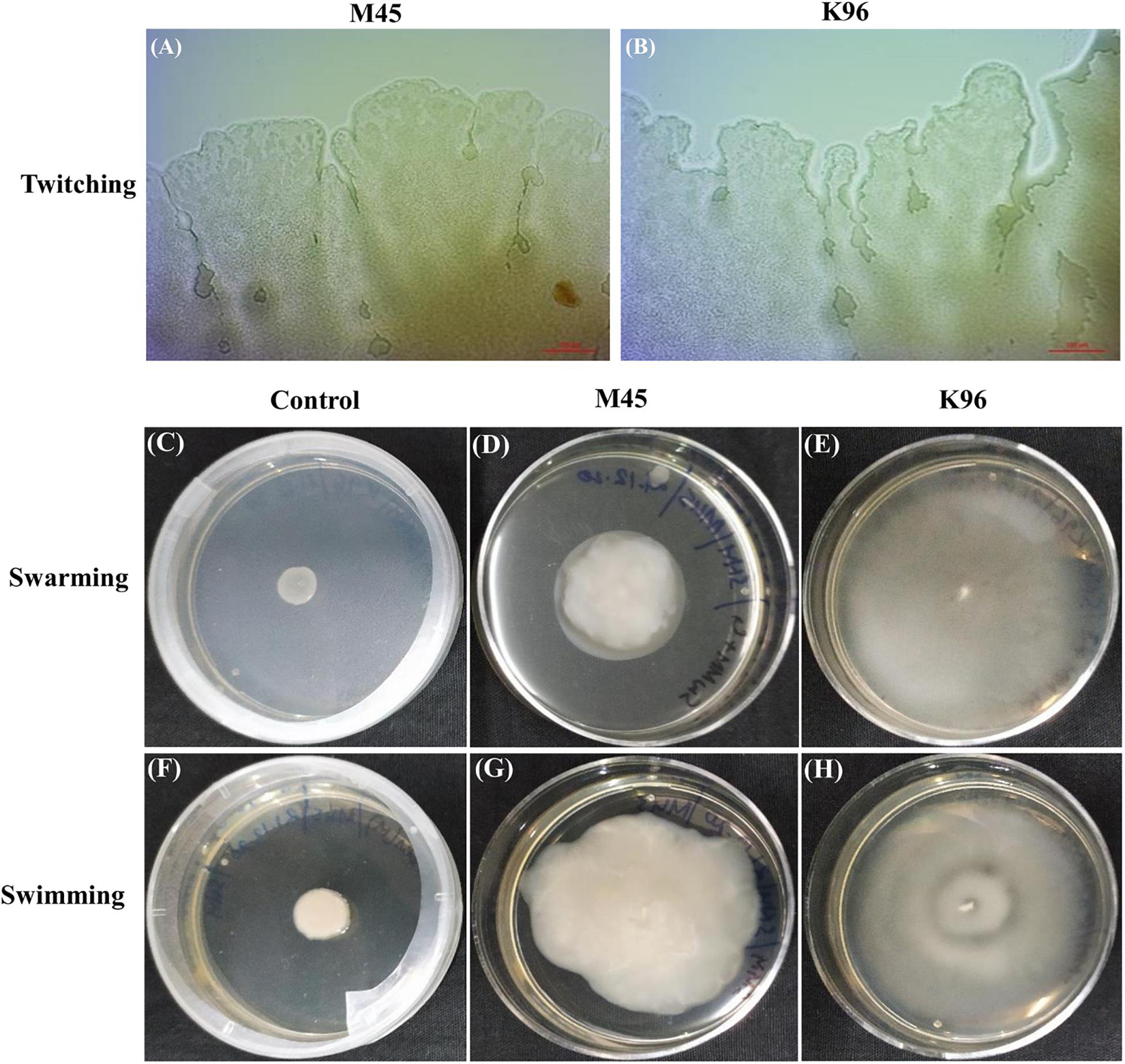
Figure 6. Motility assays. (A,B) Light microscopy of colony edges of M45 and K96 in twitching motility. Both M45 and K96 isolates produced a flat, widely spread, irregularly shaped colony at 100X magnification. (C–E) Swarming motility [NA plate with 5 g glucose/L and 0.5% (w/v) agar] and (F–H) Swimming motility [LB with 0.3% (w/v)] of Control, M45 and K96 incubated at 28°C for 48 h.
Based on bonitur score, isolates with the top two scores namely K96 and M45 were selected to observe their effective contribution to promote plant growth in the treated plant group compared to the control group in vivo experiments in nursery conditions (Supplementary Figure 2). All the treated plant groups showed a significant (p < 0.05) increase in different plant growth-promoting vegetative parameters in comparison to the control plant group (Figure 7) on inoculation with the potent EnB isolates. PCA resulted in two major principal components (PC) with a variance of 70.96% (PC1) and 15.21% (PC2) on reduction of the dimension of the various PGP parameters (Figure 8). The analysis revealed a major distinction between treatment and control groups throughout the PC1. Among the treatment groups viz. T1 and T3 the interrelationship among the vegetative parameters in clone TV26 and TV22 were reflected by PC2 and PC1, respectively, where a minor distinction was observed in the case of the T1 group for both the clones. Likewise, PC1 represented a higher degree of correlation of parameters dry weight root and shoot (DWR and DWS), fresh weight shoot and root (FWS and FWR), and chlorophyll b (Chlb) toward clone TV22 in T1 and T2 group. Likewise, parameters chlorophyll a (Chla), root length (RL), number of leaves (NL), and shoot length (SL) were correlated mostly toward clone TV26 in both the treatment groups.
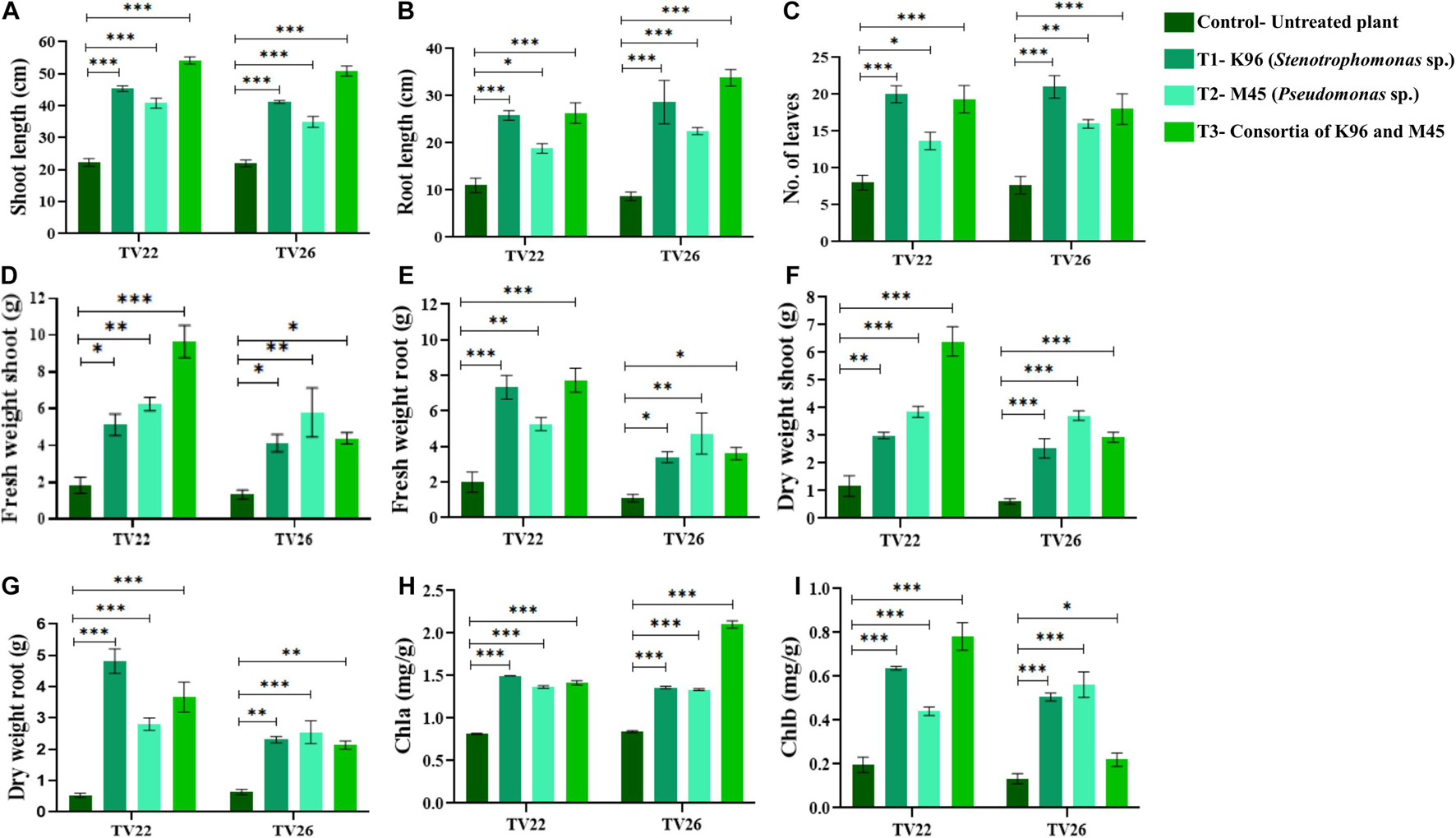
Figure 7. Evaluation of PGP experiment to show the effect of treatments of endophytic bacterial inoculum in two tea clones TV22 and TV26 for (A) shoot length; (B) root length; (C) number of leaves (D) fresh weight shoot; (E) fresh weight root; (F) dry weight shoot; (G) dry weight root; (H) chlorophyll a; and (I) chlorophyll b. Error bar represents the standard error of means of replicates (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
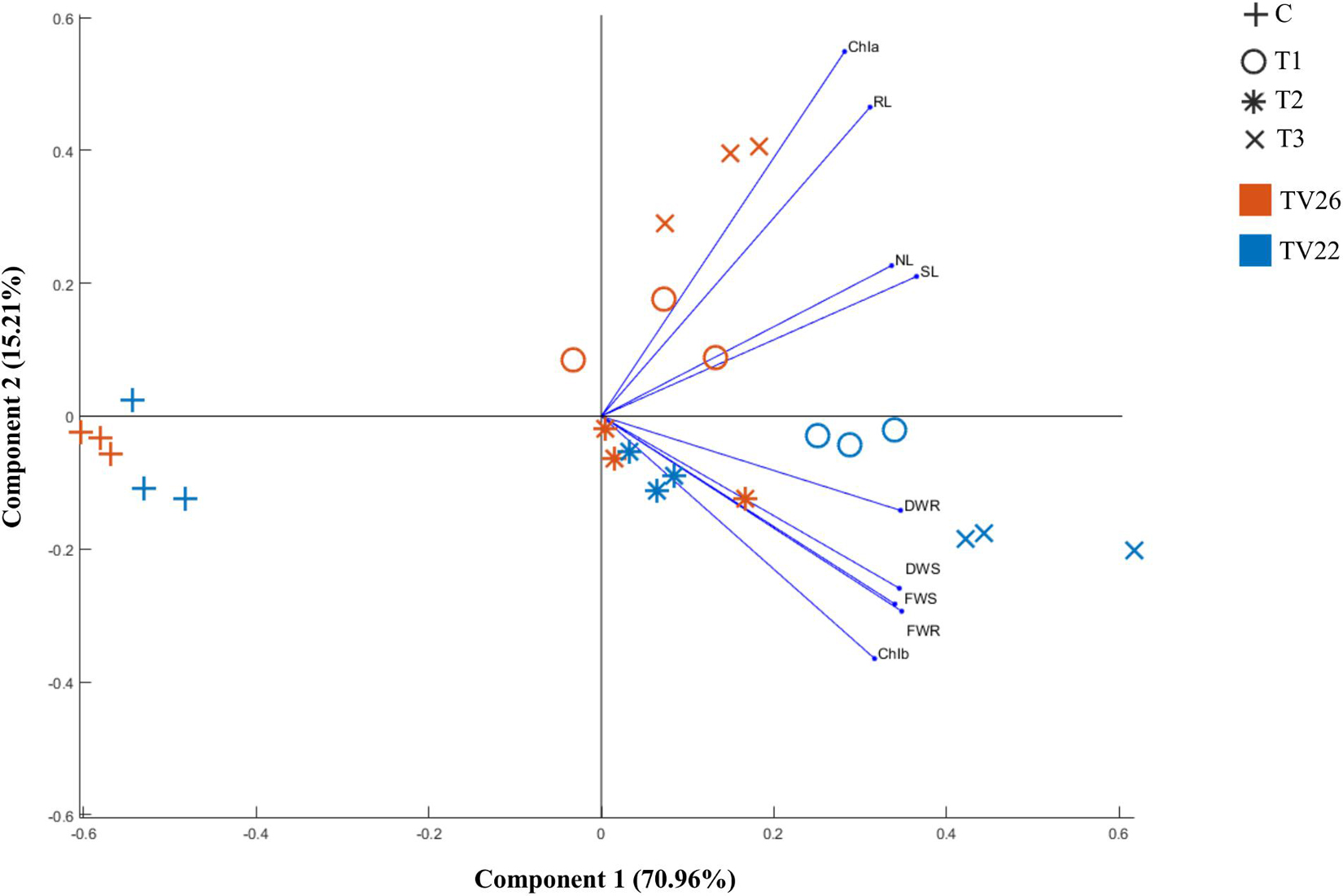
Figure 8. Principle component analysis biplot based on correlation matrices of the PGP dataset of growth parameters and treatment groups of tea clones TV22 and TV26. C- Control, Untreated plant; T1-Treatment 1, K96 (Stenotrophomonas sp.); T2- Treatment 2, M45 (Pseudomonas sp.); T3- Treatment 3, Consortia of K96 and M45.
The level of growth in vegetative parameters of treated plants compared to untreated plants was measured by fold change analysis. T3 showed considerable increase in the number of fold change compared to T1 and T2 in TV22 tea clone with an increase in shoot length by 4.99-fold, root length by 3.93-fold, fresh shoot weight by 2.96-fold, dry shoot weight by 2.38-fold, fresh root weight by 2.51 fold, and dry root weight by 1.64-fold. However, T1 showed a maximum increase in the number of leaves of TV22 clone by 3.59-fold. In the case of clone TV26, the degree of growth promotion was more significant when treated with T2 that showed a 2.16 fold increase for fresh shoot weight, 1.63-fold for dry weight shoot, 1.86-fold for fresh root weight, and 0.94 fold for dry root weight. On the other hand, T3 showed maximum shoot length and root length with an increase of 4.85 and 4.65, respectively, and T1 showed a maximum increase in the number of leaves by 3.73 fold (Supplementary Figures 3A–F).
Over the past few decades to prevent food shortage worldwide and escalate crop yield, extensive amounts of chemical fertilizers, pesticides, and herbicides have been used that resulted in environmental hazards and is one of the major areas of concern in tea and other agricultural production (Sharma and Singhvi, 2017; Xie et al., 2019). It has become important to seek a substitute to take over these chemicals. The beneficial bacteria that inhabit inside plants can improve plant growth under natural and challenging conditions (Glick, 2012; Santoyo et al., 2016). They are capable of improving plant nutrient uptake, amending soil health, and priming plant defense to develop quality clones of tea with higher yield potential and confer sustainability in agriculture (Jain and Kumar, 2017). Consequently, understanding the EnB community associated with tea to delineate their function in PGP has become important. To endeavor beneficial effects on plant growth, PGPB has to colonize the endosphere after colonization in the rhizosphere. Many hypotheses including motility, attachment, plant-polymer degradation, production of growth phytohormones, phosphate solubilization, siderophore production, and evasion of plant defenses have been described to be involved in growth promotion by PGPB (Glick, 2014; Liu et al., 2017).
Camellia sinensis known for its therapeutic aid in several diseases is being widely consumed worldwide. Tea plants harbor a broad range of beneficial EnB which can be used as bioinoculants in promoting safe and sustainable agriculture. With the possibility to isolate diverse culturable EnB, leaf, and roots of five different tea clones were selected as variation in the endophytic community depends on the plant. Most t of the EnB isolates was recovered from root tissues as compared to leaf tissues which are unanimous with several studies that indicated the dominance of EnB in root tissues (Ma et al., 2013; Kandel et al., 2017).
PCR amplification followed by 16S rRNA gene sequencing and phylogenetic analysis determined the molecular identity and genetic diversity of these isolates. Similarity search using NCBI GenBank and EZ Taxon identified all the isolates at genus level showing 91–100% identity with reference sequences. Also, 16S rRNA gene sequencing is regarded as a robust technique for the identification of bacteria at genus and species level which differentiates between closely related bacterial species (Johnson et al., 2019). EnB were grouped into three phyla which consisted of 22 different genera that were grouped into six clusters based on phylogenetic analysis. Proteobacteria (n = 53, 50%) was most dominant which is represented by Acinetobacter sp., Stenotrophomonas sp., Brevundimonas sp., Pseudomonas sp., Ochrobactrum sp., Alcaligenes sp., Serratia sp., Achromobacter sp., and Advenella sp. followed by Firmicutes (n = 42, 39.62%) including Lysinibacillus sp., Sporosarcina sp., Paenibacillus sp., Bacillus sp., Exiguobacterium sp., Planococcus sp., and Brevibacillus sp. and Actinobacteria (n = 11, 10.37%) which is represented by Rhodococcus sp., Janibacter sp., Curtobacterium sp., Streptomyces sp., Nocardia sp., and Microbacterium sp. which is similar to the findings as reported by Verma et al. (2015). Bacillus was found to be the dominant genus (n = 31, 29.24%) followed by Alcaligenes (n = 18, 17.0%), Pseudomonas (n = 11,10.37%), and Stenotrophomonas (n = 10, 9.43%). Bacillus sp. was reported previously as dominant genera as an endophyte in different crops. In Pisarska and Pietr, (2015) and Passari et al. (2016) Bacillus was reported as the dominant genus among the EnB isolated from maize and Clerodendrum colebrookianum Walp which agrees with our findings.
For the judicious selection of potential indigenous PGP strains, the isolates were evaluated for a range of PGP traits by direct (include phosphate solubilization, production of IAA, ACC deaminase, and nitrogen fixation) and indirect (include siderophore, ammonia, and extracellular enzyme production) methods. All the isolates showed multifarious PGP traits which may be beneficial for the growth of the tea plant.
Phosphorus is an essential nutrient for plant growth which is required for numerous key plant functions inclusive of photosynthesis, nutrient uptake, and energy transfer, and so on. But it is unavailable to plants because most of the phosphorus exists in the soil in an insoluble form as organic phosphate or as mineral salts emerging as a major limiting factor in agricultural systems (Miller et al., 2010; Oteino et al., 2015). Thus, to determine the ability of EnB isolates to solubilize tri-calcium phosphate present in PKA and NBRIP medium quantitative and qualitative analysis was performed. A significant decrease in pH of culture medium due to production of organic acids confirmed phosphate solubilization by 30 of the isolates. As reported (Oteino et al., 2015) the acidity and concentration of soluble phosphate in the medium are inversely proportional. The phosphate solubilization values in this study fall in the range 2.79 ± 0.67- 81.42 ± 5.73 mg L–1 which is in agreement with the typical findings in PSB range from 10 to 800 mg L–1 as reported (Rodríguez and Fraga, 1999; Passari et al., 2016).
Auxin IAA production by beneficial bacteria is very essential for plant growth-promoting microorganisms as it is one of the most important phytohormones regulating growth and several developmental processes in plants (Egamberdieva et al., 2017). In the present study, noticeably more IAA was produced by isolates in presence of precursor L-tryptophan than previously reported by Hassan (2017) ranging from 4.1 to 23.4 μg mL–1 in Teucrium polium L. Similarly, Woźniak et al. (2019) reported IAA production by bacteria isolated from wheat, maize, rye, broad bean, burdock, and horsetail in the range 0.13–22.51 μg mL–1. Since IAA production is governed by multiple pathways, the amount of IAA produced may vary among isolates (Moliszewska and Nabrdalik, 2020). Studies revealed that most of the IAA producing EnB belong to phyla Proteobacteria, Firmicutes, and Actinobacteria (Nabrdalik et al., 2018; Hazarika and Thakur, 2020) which is in agreement with this study.
Production of ammonia by EnB potentially serves as a source for fixation of atmospheric nitrogen which stimulates plant growth and contributes to plant defense. Almost all the isolates produced ammonia among which Bacillus sp. produced a higher amount of ammonia which is similar with the report (Brígido et al., 2019; Kang et al., 2020) that can escalate plant biomass by the accumulation of nitrogen.
Siderophore production by plant-associated bacteria assists in plant growth by scavenging iron from the environment because of the low iron bioavailability (Ahmed and Holmström, 2014). The EnB isolates under iron limiting conditions produced a high amount of siderophore in this study. Siderophore production by Brevundimonas sp. has rarely been reported earlier which has been confirmed in our study, although it has been isolated from various sources such as rhizospheric soil of sugarcane, potato, and maize (Montañez et al., 2012; Kumar and Gera, 2014; Naqqash et al., 2020). Previous studies reported Bacillus sp. (Ferreira et al., 2019), Pseudomonas sp., Stenotrophomonas sp., Acinetobacter sp., and Ochrobactrum sp. (Yu et al., 2018; Ferreira et al., 2019) as producers of siderophore which is similar with our study.
Drought is one of the most common stresses induced in tea plants (Lin, 2011). Thus, one of the vital roles of beneficial bacteria associated with tea plants is to alleviate stress and promote growth. One such mechanism exploited by EnB is lowering plant ACC levels, a precursor of ethylene by the production of ACC deaminase (Sessitsch and Nowak, 2008). In the present study, 87.73% of isolates produced ACC deaminase which was much higher than reported by Borah et al. (2019). Production of other extracellular enzymes like cellulase, protease, and amylase plays a significant role in the decay of organic matter and plant growth promotion. In our study, Bacillus sp. exhibited the highest number of exoenzyme activities among tested enzymes, which is in accordance with the results reported (El-Deeb et al., 2012). Among other isolates Pseudomonas sp., Stenotrophomonas sp., Ochrobactrum sp., and Alcaligenes sp. produced all the enzymes tested. Among isolates, Serratia sp. and Acinetobacter sp. had cellulase and protease activity; Streptomyces sp. and Brevundimonas sp. displayed cellulase and amylase activity. Bacterial strains Brevibacillus sp., Lysinibacillus sp., and Microbacterium sp. showed only cellulase activity; Achromobacter sp. showed only amylase activity; and Planococcus sp., Janibacter sp., and Paenibacillus sp. produced only protease. This result is in accordance with the study of El-Deeb et al. (2012); Liu et al. (2016), and Passari et al. (2016). Many plant beneficial bacteria produces extracellular enzymes through indirect mechanism for PGP (Gupta et al., 2015). The production of hydrolytic enzymes by EnB emerges as important for colonization of plant roots and movement of bacteria into the interior of plants rendering help in plant growth (Al-Mallah et al., 1987; Quadt-Hallmann et al., 1997; Choubane et al., 2016).
As described by Chen et al. (2013) the ability of bacteria to form a stable biofilm for attachment and persistence on the plant root has been proven to be highly advantageous for plant growth promotion. Colonization of bacteria is crucial for plant-PGPB interactions to aid in plant growth. Thus, for biofilm formation and competitive colonization in the plant-root system bacterial motility plays an important role (Harshey, 2003; Danhorn and Fuqua, 2007). Motility and biofilm formation exhibited by the tea-associated bacteria M45 and K96 denotes their potentiality as PGPB and may colonize and establish on plant root system with ease, aiding in plant growth promotion.
Several reports have shown endophytic bacteria can boost plant biomass in different plants rendering multifarious advantages to host plants (Dutta and Thakur, 2017; Finkel et al., 2017). The PGP experiment was performed in an established natural condition using non-sterile soil. The soil microflora associated with tea is diverse and complex which portrays a substantial challenge in tea cultivation. To assess the efficacy of the indigenous bacterial inoculum and to constitute a compatible interaction with the host the treated tea plants were compared with untreated tea plants in the same condition. Multifarious experiments on PGP traits and bonitur scale assessment ameliorated the selection of potent isolates which augmented all growth parameters tested in the nursery experiment. The most promising effect on the tea plants tested were displayed by consortia of the isolates than the individual inoculum. The multivariate PCA analysis showed accord with the treatment and vegetative growth parameters. The efficiency of the selected isolates was further confirmed as the treated plants deviated significantly from the untreated plants. This was validated by two-way ANOVA analysis. Therefore, the bacteria inoculums utilized in this nursery experiment were assumed to show synergistic PGP effect, colonization, and competence with the other native microbial population in the treated tea clones.
In vivo, the growth of the tea plants is not directly related to the growth-promoting traits such as IAA production, phosphate solubilization, ACC deaminase production, and so on. The specificity of the bacteria’s interaction with the plants influenced the growth-promotion. The efficiency of the PGP strains was possibly dependent on the genotype of the host plant or the bacterial genotype. Thus, the genetic factors of both partners appear to influence plant colonization and growth promotion by the EnB (Afzal et al., 2019).
Endophytic bacteria residing within the host plant are potential natural bioresources aiding in plant growth promotion. The exploration of endophytes indigenous to tea plants is limited as compared to endophytes from various agricultural systems. This current study helped in establishing the benefits of endophytes associated with tea clones endowed with plant growth-promoting traits, motility, and biofilm-forming ability and suggesting its applicability in tea gardens and other agricultural crops as biofertilizer or as an alternative for chemical fertilizers and pesticides, thereby promoting sustainability. However, further research is needed to establish the colonization of the endophytes within the host and study the correlation with the native soil microflora.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SH conceptualized, planned, designed, and conducted the laboratory and field experiments, acquired and analyzed the data, interpreted the results, and wrote and reviewed the original draft. KS contributed to data analysis. AB helped in an experimental setup. DT conceptualized and supervised the research work and reviewed and edited the original draft. All authors have read and approved the final manuscript.
This work was partially supported by DST, Government of India in-house R&D core budget funded project for the year 2021–2022.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Director, Institute of Advanced Study in Science and Technology (IASST), Guwahati, Assam, India, for providing facilities for this work; Kopati Tea estate, Assam, India, and Meg Tea estate, Umsning, Meghalaya, for approving and assisting in sample collection; Bioinformatics Facility (BIF), IASST for providing facilities and support for data analysis; and Central Instrumentation Facility (CIF), IASST for providing infrastructure facilities to carry out part of the work. We would like to thank Somarani Dash, IASST for great help in imaging using CLSM. We would like to thank the Department of Science and Technology (DST), Government of India for the fellowship to SH.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.738058/full#supplementary-material
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49.
Ahmed, E., and Holmström, S. J. M. (2014). Siderophores in environmental research : roles and applications. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Alexander, D. B., and Zuberer, D. A. (1991). Use of chrome Azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Al-Mallah, M. K., Davey, M. R., and Cocking, E. C. (1987). Enzymatic treatment of clover root hairs removes a barrier to rhizobium-host specificity. Nat. Biotechnol. 5, 1319–1322. doi: 10.1038/nbt1287-1319
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Berg, G., Krechel, A., Ditz, M., Sikora, R. A., Ulrich, A., and Hallmann, J. (2005). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microb. Ecol. 51, 215–229. doi: 10.1016/j.femsec.2004.08.006
Borah, A., Das, R., Mazumdar, R., and Thakur, D. (2019). Culturable endophytic bacteria of Camellia species endowed with plant growth promoting characteristics. J. Appl. Microbiol. 127, 825–844. doi: 10.1111/jam.14356
Brígido, C., Singh, S., Menéndez, E., Tavares, M. J., Glick, B. R., Félix, M. D. R., et al. (2019). Diversity and functionality of culturable endophytic bacterial communities in chickpea plants. Plants 8, 1–21. doi: 10.3390/plants8020042
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15, 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Choubane, S., Cheba, B. A., and Benourrad, A. (2016). Screening and phenotypic diversity of amylase producing rhizospheric bacteria from some North African plants. Proc. Technol. 22, 1197–1204. doi: 10.1016/j.protcy.2016.01.168
Compant, C., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197. doi: 10.1007/s00248-011-9883-y
Compant, S., Duffy, B., Nowak, J., Clement, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases : principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Cooper, R., Morré, D. J., and Morré, D. M. (2005). Medicinal benefits of green tea: part ii. review of anticancer properties. J. Altern. Complement. Med. 11, 639–652. doi: 10.1089/acm.2005.11.639
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Das, S. C., Das, S., and Hazarika, M. (2012). “Breeding of the tea plant (Camellia sinensis) in India,” in Global Tea Breeding, eds L. Chen, Z. Apostolides, and Z. M. Chen (Hangzhou: Zhejiang University Press), 69–124. doi: 10.1007/978-3-642-31878-8_3
Directorate of Food Processing Meghalaya (n.d.). Meg Tea. Available online at: dofpmeghalaya.org/meg-tea/ (accessed June 26, 2021).
Dudeja, S. S., Giri, R., Saini, R., Suneja-Madan, P., and Kothe, E. (2012). Interaction of endophytic microbes with legumes. J. Basic Microbiol. 52, 248–260. doi: 10.1002/jobm.201100063
Dutta, J., Handique, P. J., and Thakur, D. (2015). Assessment of culturable tea rhizobacteria isolated from tea estates of Assam, India for growth promotion in commercial tea cultivars. Front. Microbiol. 6:1252. doi: 10.3389/fmicb.2015.01252
Dutta, J., and Thakur, D. (2017). Evaluation of multifarious plant growth promoting traits, antagonistic potential and phylogenetic affiliation of rhizobacteria associated with commercial tea plants grown in Darjeeling, India. PLoS One 12:e0182302. doi: 10.1371/journal.pone.0182302
Dworkin, M., and Foster, J. W. (1958). Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 75, 592–603. doi: 10.1128/jb.75.5.592-603.1958
Egamberdieva, D., Stephan, J., Wirth, S. J., Alqarawi, A. A., Abd-Allah, E. F., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8:2104. doi: 10.3389/fmicb.2017.02104
El-Deeb, B., Bazaid, S., Gherbawy, Y., and Elhariry, H. (2012). Characterization of endophytic bacteria associated with rose plant (Rosa damascena trigintipeta) during flowering stage and their plant growth promoting traits. J. Plant Interact. 7, 248–253. doi: 10.1080/17429145.2011.637161
El-Sayed, W. S., Akhkha, A., El-Naggar, M. Y., and Elbadry, M. (2014). In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 5:651. doi: 10.3389/fmicb.2014.00651
Emami, S., Alikhani, H. A., Pourbabaei, A. A., Etesami, H., Sarmadian, F., and Motessharezadeh, B. (2019). Effect of rhizospheric and endophytic bacteria with multiple plant growth promoting traits on wheat growth. Environ. Sci. Pollut. Res. 26, 19804–19813. doi: 10.1007/s11356-019-05284-x
Etminani, F., and Harighi, B. (2018). Isolation and identification of endophytic bacteria with plant growth promoting activity and biocontrol potential from wild pistachio trees. Plant Pathol. J. 34, 208–217. doi: 10.5423/PPJ.OA.07.2017.0158
Ferreira, C. M. H., Vilas-Boas, Â., Sousa, C. A., Soares, H. M. V. M., and Soares, E. V. (2019). Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express 9:78. doi: 10.1186/s13568-019-0796-3
Finkel, O. M., Castrillo, G., Paredes, S. H., González, I. S., and Dangl, J. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Fiske, C. H., and Subbarow, Y. J. (1925). The colorimetric determination of phosphorus. Biol. Chem. 66, 375–400. doi: 10.1007/s13398-014-0173-7.2
Frei, B., and Higdon, J. V. (2003). Proceedings of the third international scientific symposium on tea and human health : role of flavonoids in the diet antioxidant activity of tea polyphenols in vivo : evidence from animal studies. J. Nutr. 133, 3275S–3284S.
Fujita, M., Kusajima, M., Okumura, Y., Nakajima, M., Minamisawa, K., and Hideo Nakashita, H. (2017). Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in tomato. Biosci. Biotechnol. Biochem. 81, 1657–1662. doi: 10.1080/09168451.2017.1329621
Glick, B. R. (2012). Plant growth-promoting bacteria : mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Glick, B. R., Todorovic, B., Czarny, J., Cheng, Z., Duan, J., and McConkey, B. (2007). Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 26, 227–242. doi: 10.1080/07352680701572966
Gordon, S. A., and Weber, R. P. (1951). Colorimetric estimation of indoleacetic Acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Gupta, G., Parihar, S. S., Ahirwar, N. K., Snehi, S. K., and Singh, V. (2015). Microbial and biochemical technology plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 7, 96–102. doi: 10.4172/1948-5948.1000188
Gupta, K., Singh, S. P., Manhar, A. K., Saikia, D., Namsa, N. D., Konwar, B. K., et al. (2019). Inhibition of Staphylococcus aureus and Pseudomonas aeruginosa biofilm and virulence by active fraction of Syzygium cumini (L.) skeels leaf extract: in-vitro and in silico studies. Indian J. Microbiol. 59, 13–21. doi: 10.1007/s12088-018-0770-9
Hallmann, J., and Berg, G. (2007). “Spectrum and population dynamics of bacterial root endophytes,” in Microbial Root Endophytes, eds B. J. E. Schulz, C. J. C. Boyle, and T. N. Sieber (Heidelberg: Springer), 15–31.
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., and Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Hamilton-Miller, J. M. (1995). Antimicrobial properties of tea. Antimicrob. Agents Ch. 39, 2375–2377. doi: 10.1128/aac.39.11.2375
Harshey, R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Hassan, S. E. (2017). Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res. 8, 687–695. doi: 10.1016/j.jare.2017.09.001
Hayat, K., Iqbal, H., Malik, U., Bilal, U., and Mushtaq, S. (2015). Tea and its consumption: benefits and risks. Crit. Rev. Food Sci. Nutr. 55, 939–954. doi: 10.1080/10408398.2012.678949
Hazarika, S. N., and Thakur, D. (2020). “Actinobacteria,” in Beneficial Microbes in Agro-Ecology: Bacteria and Fungi, eds N. Amaresan, M. Senthil Kumar, K. Annapurna, K. Kumar, and A. Sankaranarayanan (Cambridge, MA: Academic Press), 443–476.
Henrichsen, J. (1983). Twitching motility. Annu. Rev. Microbiol. 37, 81–93. doi: 10.1146/annurev.mi.37.100183.000501
Hiscox, J. D., and Israelstam, G. F. (1979). A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334. doi: 10.1139/b79-163
Hsu, S., Bollag, W. B., Lewis, J., Huang, Q., Singh, B., Sharawy, M., et al. (2003). Green tea polyphenols induce differentiation and proliferation in epidermal keratinocytes. J. Pharmacol. Exp. Ther. 306, 29–34. doi: 10.1124/jpet.103.049734
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Hurek, T., and Reinhold-Hurek, B. (2003). Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes. J. Biotechnol. 106, 169–178. doi: 10.1016/j.jbiotec.2003.07.010
Iverson, W. G., and Millis, N. F. (1974). a method for the detection of starch hydrolysis by bacteria. J. App. Bacteriol. 37, 443–446. doi: 10.1111/j.1365-2672.1974.tb00460.x
Jain, P., and Kumar, P. R. (2017). “Potential role of endophytes in sustainable agriculture-recent developments and future prospects,” in Endophytes: Biology and Biotechnology. Sustainable Development and Biodiversity, ed. D. K. Maheshwari (Cham: Springer), 15.
Johnson, J. S., Spakowicz, D. J., Hong, B. Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10:5029. doi: 10.1038/s41467-019-13036-1
Kandel, S., Joubert, P., and Doty, S. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. doi: 10.3390/microorganisms5040077
Kang, S. M., Bilal, S., Shahzad, R., Kim, Y. N., Park, C. W., Lee, K. E., et al. (2020). Effect of ammonia and indole-3-acetic acid producing endophytic Klebsiella pneumoniae YNA12 as a bio-herbicide for weed inhibition: special reference with evening primroses. Plants 9:761. doi: 10.3390/plants9060761
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khan, A. L., Waqas, M., Kang, S. M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52, 689–695. doi: 10.1007/s12275-014-4002-7
Kifle, M. H., and Laing, M. D. (2016). Isolation and screening of bacteria for their diazotrophic potential and their influence on growth promotion of maize seedlings in greenhouses. Front. Plant Sci. 6:1225. doi: 10.3389/fpls.2015.01225
Kris-Etherton, P. M., Hecker, K. D., Bonanome, A., Coval, S. M., Binkoski, A. E., Hilpert, K. F., et al. (2002). Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 113, 71–88. doi: 10.1016/s0002-9343(01)00995-0
Kumar, V., and Gera, R. (2014). Isolation of a multi-trait plant growth promoting Brevundimonas sp. and its effect on the growth of Bt-Cotton. 3 Biotech. 4, 97–101. doi: 10.1007/s13205-013-0126-4
Lin, B. B. (2011). Resilience in agriculture through crop diversification: adaptive management for environmental change. BioScience 61, 183–193. doi: 10.1525/bio.2011.61.3.4
Liu, F. P., Liu, H. Q., Zhou, H. L., Dong, Z. G., Bai, X. H., Bai, P., et al. (2014). Isolation and characterization of phosphate-solubilizing bacteria from betel nut (Areca catechu) and their effects on plant growth and phosphorus mobilization in tropical soils. Biol. Fertil. Soils 50, 927–937. doi: 10.1007/s00374-014-0913-z
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values : diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552. doi: 10.3389/fmicb.2017.02552
Liu, Y. H., Guo, J. W., Salam, N., Li, L., Zhang, Y. G., Han, J., et al. (2016). Culturable endophytic bacteria associated with medicinal plant Ferula songorica: molecular phylogeny, distribution and screening for industrially important traits. 3 Biotech. 6:209. doi: 10.1007/s13205-016-0522-7
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Ma, B., Lv, X., Warren, A., and Gong, J. (2013). Shifts in diversity and community structure of endophytic bacteria and archaea across root, stem and leaf tissues in the common reed, Phragmites australis, along a salinity gradient in a marine tidal wetland of northern China. Anton. Leeuw Int J Gen. Mol. Microbiol. 104, 759–768. doi: 10.1007/s10482-013-9984-3
Maeda-Yamamoto, M., Ema, K., and Shibuichi, I. (2007). In vitro and in vivo anti-allergic effects of “benifuuki” green tea containing O-methylated catechin and ginger extract enhancement. Cytotechnology 55, 135–142. doi: 10.1007/s10616-007-9112-1
Martin, B., Chadwick, W., Yi, T., Park, S.-S., Lu, D., Ni, B., et al. (2012). VENNTURE–a Novel Venn Diagram investigational tool for multiple pharmacological dataset analysis. PLoS One 7:e36911. doi: 10.1371/annotation/27f1021c-b6f2-4b90-98bc-fcacd2679185
Meddeb-mouelhi, F., Kelly, J., and Beauregard, M. (2014). Enzyme and microbial technology a comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme Microb. Technol. 66, 16–19. doi: 10.1016/j.enzmictec.2014.07.004
Mendes, R., Pizzirani-kleiner, A. A., Araujo, W. L., and Raaijmakers, J. M. (2007). Diversity of cultivated endophytic bacteria from sugarcane : genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 73, 7259–7267. doi: 10.1128/AEM.01222-07
Miller, S. H., Browne, P., Prigent-Combaret, C., Combes-Meynet, E., Morrissey, J. P., and O’Gara, F. (2010). Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ. Microbiol. Rep. 2, 403–411. doi: 10.1111/j.1758-2229.2009.00105.x
Moliszewska, E. B., and Nabrdalik, M. (2020). “Application and biological impact of endophytic bacteria as IAA producers,” in Molecular Aspects of Plant Beneficial Microbes in Agriculture, Vol. 438, eds V. Sharma, R. Salwan, and L. Tawfeeq (Cambridge, MA: Academic Press), 77–87.
Mondal, T. K., Bhattacharya, A., Laxmikurmaran, M., and Ahuja, P. S. (2004). Recent advances of tea (Camellia sinensis) biotechnology. Plant Cell Tiss.Org. Cult. 76, 195–254. doi: 10.1023/B:TICU.0000009254.87882.71
Montañez, A., Blanco, A. R., Barlocco, C., Beracochea, M., and Sicardi, M. (2012). Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (Zea mays L.) and their inoculation effects in vitro. Appl. Soil Ecol. 58, 21–28. doi: 10.1016/j.apsoil.2012.02.009
Nabrdalik, M., Moliszewska, E., and Wierzba, S. (2018). Importance of endophytic strains pantoea agglomerans in the biological control of rhizoctonia solani. Ecol. Chem. Eng. S 25, 331–342. doi: 10.1515/eces-2018-0023
Naqqash, T., Imran, A., Hameed, S., Shahid, M., Majeed, A., Iqbal, J., et al. (2020). First report of diazotrophic Brevundimonas spp. as growth enhancer and root colonizer of potato. Sci. Rep. 10:12893. doi: 10.1038/s41598-020-69782-6
Oteino, N., Lally, R. D., Kiwanuka, S., Lloyd, A., Ryan, D., Germaine, K. J., et al. (2015). Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 6:745. doi: 10.3389/fmicb.2015.00745
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Passari, A. K., Mishra, V. K., Leo, V. V., Gupta, V. K., and Singh, B. P. (2016). Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 193, 57–73. doi: 10.1016/j.micres.2016.09.006
Passari, A. K., Mishra, V. K., Saikia, R., Gupta, V. K., and Singh, B. P. (2015). Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 6:273. doi: 10.3389/fmicb.2015.00273
Patel, P. K., Zhang, D., Borthakur, D., Hazarika, M., Boruah, P., Barooah, R., et al. (2019). Quality green tea (Camellia sinensis L.) clones marked through novel traits. Beverages 5:63. doi: 10.3390/beverages5040063
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Payne, S. M. (1994). Detection, isolation, and characterization of siderophores. Meth. Enzymol. 235, 329–344. doi: 10.1016/0076-6879(94)35151-1
Pisarska, K., and Pietr, S. (2015). Biodiversity of dominant cultivable endophytic bacteria inhabiting tissyes of six different cultivar of Maize (Zea mays L. ssp mays). Pol. J. Microbiol. 64, 163–170.
Quadt-Hallmann, A., Benhamou, N., and Kloepper, J. W. (1997). Bacterial endophytes in cotton: mechanisms of entering the plant. Can. J. Microbiol. 43, 577–582. doi: 10.1139/m97-081
Rambaut, A. (2018). FigTree v1.4.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed May 12, 2021).
Rodríguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rosenberg, M. (2006). Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol. Lett. 262, 129–134. doi: 10.1111/j.1574-6968.2006.00291.x
Rosenblueth, M., and Martínez-romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Santoyo, G., Moreno-hagelsieb, G., Orozco-mosqueda, C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sarani, B. T. M. (2020). State / Region Wise and Month Wise Tea Production Data for the Year 2020-21. Available online at: www.teaboard.gov.in/pdf/Production_20202_21_Apr_Dec_and_2020_Jan _Dec_pdf7013.pdf (accessed June 26, 2021).
Schulz, B., Wanke, U., Draeger, S., and Aust, H. J. (1993). Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol. Res. 97, 1447–1450. doi: 10.1016/S0953-7562(09)80215-3
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sessitsch, A., and Nowak, J. (2008). Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN : from the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 63, 84–93. doi: 10.1111/j.1574-6941.2007.00410.x
Sessitsch, A., Reiter, B., and Berg, G. (2004). Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can. J. Microbiol. 50, 239–249. doi: 10.1139/w03-118
Sgroy, V., Cassán, F., Masciarelli, O., Del Papa, M. F., Lagares, A., and Luna, V. (2009). Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 85, 371–381. doi: 10.1007/s00253-009-2116-3
Sharangi, A. B. (2009). Medicinal and therapeutic potentialities of tea (Camellia sinensis L.) - A review. Food Res. Int. 42, 529–535. doi: 10.1016/j.foodres.2009.01.007
Sharma, N., and Singhvi, R. (2017). Effects of chemical fertilizers and pesticides on human health and environment: a review. Int. J. Agr. Environ. Biotechnol. 10:675. doi: 10.5958/2230-732x.2017.00083.3
Siddiqui, M. W., Sharangi, A. B., Singh, J. P., Thakur, P. K., Ayala-Zavala, J. F., Singh, A., et al. (2016). Antimicrobial properties of teas and their extracts in vitro. Crit. Rev. Food Sci. Nutr. 56, 1428–1439. doi: 10.1080/10408398.2013.769932
Singh, P., Kumar, V., and Agrawal, S. (2014). Evaluation of phytase producing bacteria for their plant growth promoting activities. Int. J. Microbiol. 2014:426483. doi: 10.1155/2014/426483
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sturz, A. V., Christie, B. R., and Matheson, B. C. (1998). Associations of bacterial endophyte populations from red clover and potato crops with potential for beneficial allelopathy. Can. J. Microbiol. 44, 162–167. doi: 10.1139/w97-146
Verma, P., Yadav, A. N., Khannam, K. S., Panjiar, N., Kumar, S., Saxena, A. K., et al. (2015). Assessment of genetic diversity and plant growth promoting attributes of psychrotolerant bacteria allied with wheat (Triticum aestivum) from the northern hills zone of India. Ann. Microbiol. 65, 1885–1899. doi: 10.1007/s13213-014-1027-4
Win, P. M., Matsumura, E., and Fukuda, K. (2018). Diversity of tea endophytic fungi: cultivar- and tissue preferences. Appl. Ecol. Environ. Res. 16, 677–695. doi: 10.15666/aeer/1601_677695
Woźniak, M., Gała̧zka, A., Tyśkiewicz, R., and Jaroszuk-ściseł, J. (2019). Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their phenotypic/physiological profiles in the biolog gen iii microplateTM test. Int. J. Mol. Sci. 20:5283. doi: 10.3390/ijms20215283
Xie, S., Feng, H., Yang, F., Zhao, Z., Hu, X., Wei, C., et al. (2019). Does dual reduction in chemical fertilizer and pesticides improve nutrient loss and tea yield and quality? A pilot study in a green tea garden in Shaoxing, Zhejiang Province, China. Environ. Sci. Pollut. Res. 26, 2464–2476. doi: 10.1007/s11356-018-3732-1
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S., et al. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 30, 835–845. doi: 10.1007/s11274-013-1486-y
Yu, Z., Kang, X., Liu, H., Liu, Y., Li, Y., Yu, X., et al. (2018). Endophytes isolated from ginger rhizome exhibit growth promoting potential for Zea mays. Arch. Agron. Soil Sci. 64, 1302–1314. doi: 10.1080/03650340.2018.1430892
Yuen, S. H., and Pollard, A. G. (1954). Determination of nitrogen in agricultural materials by the nessler reagent. II.—micro-determinations in plant tissue and in soil extracts. J. Sci. Food Agr. 5, 364–369. doi: 10.1002/jsfa.2740050803
Zhang, Y. F., He, L. Y., Chen, Z. J., Wang, Q. Y., Qian, M., and Sheng, X. F. (2011). Characterization of ACC deaminase-producing endophytic bacteria isolated from copper-tolerant plants and their potential in promoting the growth and copper accumulation of Brassica napus. Chemosphere 83, 57–62. doi: 10.1016/j.chemosphere.2011.01.041
Zhu, B., Chen, M., Lin, L., Yang, L., Li, Y., and An, Q. (2012). Genome sequence of Enterobacter sp. strain SP1, an endophytic nitrogen-fixing bacterium isolated from sugarcane. J. Bacteriol. 194, 6963–6964. doi: 10.1128/JB.01933-12
Keywords: endophytic bacteria, Camellia sinensis, plant growth promotion, bio-inoculum, 16S rRNA gene, diversity, biofilm
Citation: Hazarika SN, Saikia K, Borah A and Thakur D (2021) Prospecting Endophytic Bacteria Endowed With Plant Growth Promoting Potential Isolated From Camellia sinensis. Front. Microbiol. 12:738058. doi: 10.3389/fmicb.2021.738058
Received: 08 July 2021; Accepted: 20 August 2021;
Published: 30 September 2021.
Edited by:
Gustavo Santoyo, Universidad Michoacana de San Nicolás de Hidalgo, MexicoReviewed by:
Pramod Kumar Sahu, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2021 Hazarika, Saikia, Borah and Thakur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Debajit Thakur, ZGViYWppdHRoYWt1ckB5YWhvby5jby51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.