
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 September 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.735732
This article is part of the Research TopicInnovative Biocontrol Strategies to Manage Crop and Pest DiseasesView all 17 articles
 Xiaojuan Li1,2,3†
Xiaojuan Li1,2,3† Miaoyi Zhang1†
Miaoyi Zhang1† Dengfeng Qi1
Dengfeng Qi1 Dengbo Zhou1
Dengbo Zhou1 Chunlin Qi3
Chunlin Qi3 Chunyu Li4
Chunyu Li4 Siwen Liu4
Siwen Liu4 Dandan Xiang4
Dandan Xiang4 Lu Zhang2*
Lu Zhang2* Jianghui Xie1*
Jianghui Xie1* Wei Wang1*
Wei Wang1*Strawberry is a very popular fruit with a special taste, color, and nutritional value. Anthracnose caused by Colletotrichum fragariae severely limits fruit shelf life during post-harvest storage. Use of traditional chemical fungicides leads to serious environment pollution and threatens food safety. Biocontrol is considered as a promising strategy to manage the post-harvest fruit diseases. Here, strain QN1NO-4 isolated from noni (Morinda citrifolia L.) fruit exhibited a high antifungal activity against C. fragariae. Based on its physicochemical profiles and phylogenetic tree of the 16S rRNA sequence, strain QN1NO-4 belonged to the genus Bacillus. The average nucleotide identity (ANI) calculated by comparing two standard strain genomes was below 95–96%, suggesting that the strain might be a novel species of the genus Bacillus and named as Bacillus safensis sp. QN1NO-4. Its extract effectively reduced the incidence of strawberry anthracnose of harvested fruit. Fruit weight and TSS contents were also maintained significantly. The antifungal mechanism assays indicated that the extract of the test antagonist inhibited mycelial growth and spore germination of C. fragariae in vitro. Cells of strain QN1NO-4 demonstrated the cytoplasmic heterogeneity, disappeared organelles, and ruptured ultrastructure. Notably, the strain extract also had a broad-spectrum antifungal activity. Compared with the whole genome of strain QN1NO-4, several functional gene clusters involved in the biosynthesis of active secondary metabolites were observed. Fifteen compounds were identified by gas chromatography–mass spectrometry (GC-MS). Hence, the fruit endophyte B. safensis sp. QN1NO-4 is a potential bio-agent identified for the management of post-harvest disease of strawberry fruit.
Strawberry (Fragaria × ananassa Duch.) is one of the most popular fruits with more than 6.1 million tons of annual production in the world (Han et al., 2016; Zhimo et al., 2021). Strawberry fruit is also highly perishable due to mechanical injury and pathogen infection, which limits its shelf life during storage (Rico et al., 2019). Strawberry anthracnose is one of the most serious fungal diseases. It is caused by different Colletotrichum fungal species including Colletotrichum fragariae, C. acutatum, and C. gloeosporioides (Denoyes-Rothan et al., 2003; Han et al., 2016). Among the causal agents, C. fragariae is an important fungus which causes the disease named anthracnose crown rot (Miller-Butler et al., 2018). The fungi can infect the whole strawberry plant containing fruit, and it has become more destructive in the past decade (Chen et al., 2019; Chung et al., 2020). Currently, the control of strawberry anthracnose is done primarily through chemical fungicides to reduce post-harvest losses (Dukare et al., 2019). However, excessive use of fungicides causes environment pollution and pathogenic resistance (Li et al., 2021; Wang et al., 2021). In some developed countries, use of fungicides has become increasingly limited and even banned (Wisniewski et al., 2016; Wang et al., 2018). Therefore, it is necessary to develop a safer and eco-friendly strategy to manage post-harvest diseases of fruit.
Application of biocontrol agents was considered as an alternative and promising strategy to control plant pathogens (Feliziani et al., 2016; Ye et al., 2021). Biocontrol contributed to minimize the use of chemical pesticides and reduce environmental pollution. In the past few decades, some microorganisms such as yeasts, Pseudomonas fluorescens, Streptomyces spp., and Bacillus species were accepted as important biocontrol agents (Rong et al., 2020; Einloft et al., 2021). They had potential as an alternative to synthetic fungicides. The well-studied Pichia membranefaciens and Pichia guilliermondii were effective in controlling rhizopus rot of peaches and anthracnose of loquat fruit (Zhao et al., 2019; Zhang et al., 2020). Recent studies demonstrated that Pseudomonas synxantha had a biocontrol efficacy against Monilinia fructicola and Monilinia fructigena in stone fruit (Aiello et al., 2019). P. fluorescens can suppress post-harvest gray mold in apples (Zhang et al., 2011; Wallace et al., 2018).
Especially, the Bacillus genera were evaluated as outstanding biocontrol agents against fungal pathogens, such as Bacillus halotolerans against Botrytis cinerea in strawberry fruit (Wang et al., 2021; Ye et al., 2021), Bacillus sp. w176 against post-harvest green mold in citrus (Tian et al., 2020), Bacillus atrophaeus against anthracnose in soursop and avocado (Guardado-Valdivia et al., 2018), and Bacillus amyloliquefaciens against other phytopathogens of fruits (Calvo et al., 2017; Gotor-Vila et al., 2017; Wang et al., 2020; Ye et al., 2021). Several Bacillus spp. had been applied as biofertilizers or biopesticides in different crops (Li et al., 2019). They inhibited the pathogenic growth and improved the plant resistance by competition for space or nutrients with pathogens and production of bioactive substances and cell wall-degrading compounds (Ye et al., 2021). Although various Bacillus sp. strains are reported in biocontrol of post-harvest fruit, little work is conducted to identify a broad-spectrum antifungal Bacillus strain with an increased efficiency.
In our study, a strain marked with QN1NO-4 with a wide-spectrum antifungal ability is newly isolated from noni fruit. Based on the physicochemical characteristics as well as average nucleotide identity (ANI) assay, the strain is identified as a species of the genus Bacillus, called after Bacillus safensis sp. QN1NO-4. The isolated extracts successfully inhibit the infection of C. fragariae on strawberry fruit during post-harvest and could effectively keep the weight loss and TSS of fruit. The antifungal mechanism is investigated by assaying its effects on spore germination and morphological profile by scanning electron microscopy (SEM) as well as hyphal ultrastructure of C. fragariae by transmission electron microscopy (TEM). The complete genome of strain QN1NO-4 reveals a number of key gene clusters of active secondary metabolites. Antimicrobial compounds of strain QN1NO-4 extracts are further identified by gas chromatography–mass spectrometry (GC-MS). These results indicate that the fruit endophyte B. safensis sp. QN1NO-4 is a promising biocontrol agent for controlling post-harvest diseases of strawberry fruit.
Strawberry (Fragaria × ananassa Duch var. Zhang Ji) fruit with similar size (approximately 5-cm diameter), color, and shape was selected from a supermarket. No visible injury and pathogen infection were detected on the fruit surface. After being surface-sterilized with 75% (v/v) of ethanol for 2 min, these fruit samples were rinsed with sterile water for three times and air-dried for 2 h at 25°C.
Noni (Morinda citrifolia L.) fruit was collected from a planting farm in Chengmai city (19°58′35″N, 109°55′35″E) of Hainan province, China. Fruit samples were surface-sterilized with 75% (v/v) of ethanol for 5 min, disinfected using 2% of sodium hypochlorite for 20 min, and washed using sterile water for five times.
Ten grams of noni fruit was ground in a sterile mortar until paste. One milliliter of homogenate was mixed with 4 ml of Luria–Bertani (LB) medium into a 50-ml conical flask. The mixture was cultured with shaking at 180 rpm for 1 h at room temperature. The suspension was diluted from 10–1 to 10–3 fold with sterile distilled water. Two hundred microliters of dilution was spread on the LB solid medium and incubated at 28°C for 2 days. A single colony was obtained by repeatedly streaking on LB plates and was kept in 20% (v/v) glycerol at −80°C.
Fungal pathogen C. fragariae (ATCC 58718) was kindly provided by the Institute of Environment and Plant Protection, China Academy of Tropical Agricultural Sciences, Haikou, China. The antifungal ability of each isolated bacterium was tested against C. fragariae using our reported method (Li et al., 2021). Briefly, fungal pathogen was first cultured on a potato dextrose agar (PDA) plate for 7 days. A 5-mm-diameter pathogenic disk was prepared and placed in the middle of the plate. Each isolated bacterium was spotted on the four symmetrical points of the pathogen. The fungal disk alone was used as a control. Five Petri dishes were used for each replicate with a diameter of 90 mm. After culture at 28°C for 7 days, antifungal activity of the isolated bacterium was assessed by determining the radial mycelial growth of fungal pathogen. To further detect the antifungal activity of the isolated bacterium supernatant, the isolate was inoculated in 50 ml of LB liquid medium in a 250-ml conical flask. After being cultured at 150 rpm and 28°C for 3 days, the supernatant was filtered using the Whatman no. 1 qualitative filter paper and sterilized through a 0.22-μm sterile filter (Millipore, Bedford, MA, United States). Twenty microliters of supernatant was spotted at four symmetrical points of the tested pathogens, respectively. An equal amount of LB was used as a control. The antagonistic experiment was kept at 28°C for 7 days. The antifungal activity of the isolated bacterium supernatant was assessed by determining the radial mycelial growth of the fungal pathogen. All experiments were carried out in triplicate.
Morphological profiles of the selected isolate were observed after growth at 37°C for 3–4 days. Physiological and biochemical indexes were tested including resistance to pH, temperature and NaCl, enzymatic characteristics, and utilization of carbon and nitrogen sources (Wei et al., 2020). In addition, a disk diffusion method was applied to test the sensitivity of the selected isolate to different antibiotics (Kumar et al., 2014).
Extract was prepared according to the previous description with a minor modification (Qi et al., 2019). Briefly, the selected isolate was inoculated in a 5-l Erlenmeyer flask containing 1 l of LB liquid medium. After culture in a rotary shaker (180 rpm) for 3 days at 28°C, an equal volume of ethyl acetate was added to the supernatant. The mixture was subjected to ultrasound for 1 h and then injected into a separating funnel. The organic solvent in the collected extract was evaporated using a rotary vacuum evaporator (N-1300, EYELA, Ailang Instrument Co., Ltd., Shanghai, China). The dried extract was then dissolved in 100% of methanol, and 10 g l–1 of stock solution was prepared. To detect the antifungal activity assays of the extract, a 5-mm-diameter pathogenic disk was placed in the middle of the PDA plate containing 50 mg l–1 of extract. After culture at 28°C for 7 days, antifungal activity was assessed by determining the radial mycelial growth of fungal pathogen. All experiments were carried out in triplicate.
The stock solution (10 g l–1) was diluted into 5, 2.5, 1.25, 0.625, 0.313, 0.156, and 0.078 g l–1 using a double continuous dilution method (Wei et al., 2020). When 50 ml of the sterilized PDA liquid medium was precooled to 40–50°C, 1 ml of each extract diluent was added to generate different treatment concentrations (1.563, 3.125, 6.25, 12.5, 25, 50, 100, and 200 mg l–1, respectively). The inhibition ability of each extract concentration on the mycelial growth of C. fragariae was analyzed according to our previous method (Li X.J. et al., 2020). Antifungal activity was measured according to the growth diameter of C. fragariae at 28°C for 7 days. The morphological profile of pathogenic mycelia treated with 200 mg l–1 of extract was observed using an optical microscope (Nikon, E200MV, Japan). The half-maximal effective concentration (EC50) of the extract against C. fragariae was calculated in the light of a toxicity regression equation established by a least square method (Vanewijk and Hoekstra, 1993).
Different fold EC50 concentrations (2 × EC50, 4 × EC50, and 8 × EC50) were first prepared. Based on the extract quantity of the 1 × EC50 value, the proper volume of the stock solution (10 mg ml–1) was diluted into 8 × EC50 with sterile distilled water. Then, the double continuous dilution method was used to obtain 4 × EC50, 2 × EC50, and 1 × EC50, respectively. The antagonistic ability of the extract was tested for controlling post-harvest anthracnose decay of strawberry fruit according to our previous description (Li et al., 2021). A 2-mm-wide and 1-mm-deep wound was manufactured on the surface of the selected fruit samples using a sterilized needle. Ten microliters of different concentrations of extracts (1 × EC50, 2 × EC50, 4 × EC50, and 8 × EC50, respectively) was inoculated into fruit wound. An equal volume of sterile water was used as a control. After the treated fruits were air-dried, 10 μl of conidial suspension (1.0 × 106 CFU ml–1) was added to the wounded site. All fruit samples were kept in an artificial climate cabinet (Ever Scientific Instrument Ltd., Shanghai, China) at 28°C and 85% relative humidity under 12 h light/12 h darkness for 7 days. Twenty-four fruit samples of strawberry were selected for each treatment in three replicates. The disease incidence (DI) was calculated according to the following formula:
where D1 and D2 represent the number of decayed fruits and total fruits, respectively.
The weight loss and total soluble solids (TSS) of strawberry fruit were determined after treatment with C. fragariae and different concentration extracts of the isolated strain for 7 days (Li et al., 2021). Weight loss was expressed as a percentage of fruit weight before and after storage. TSS content was measured using the refractive index of fruit juice with a MASTER Refractometer (PAL-1, Atago, Japan).
The fungal spores were collected from the PDA medium by rubbing and washing the surface of each petri dish with a sterile L-shaped spreader. The mycelia were removed using a sterile muslin cloth. The spore concentration was adjusted to 1.0 × 106 CFU ml–1 with a hemocytometer (Neubauer, Superior Ltd., Marienfeld, Germany). An equal volume of pathogenic spore suspension (1.0 × 106 CFU ml–1) and each concentration extract (1 × EC50, 2 × EC50, 4 × EC50, or 8 × EC50, respectively) was mixed at 28°C and 85% relative humidity for 12 h. More than 200 spores of C. fragariae were used to detect germination using an optical microscope (Nikon, E200MV, Japan) (Pei et al., 2020). Based on the spore germination, 4 × EC50 of extract concentration was selected for the following experiment.
To detect the effect of extract on the spore morphology of C. fragariae, equal volumes of spores (1.0 × 106 CFU ml–1) and 4 × EC50 of extracts were completely mixed in a 10-ml sterile centrifugal tube. The mixture was incubated at 28°C for 6 h. After centrifugation at 10,000 rpm for 5 min at 4°C, spores were collected and fixed with 2.5% (v/v) of glutaraldehyde overnight. Then, these fixed spores were washed for three times with a phosphate-buffered saline (PBS, 0.1 mol l–1, pH 7.2) for 15 min. The sample was dehydrated using a gradient ethanol solution (30, 50, 70, 80, 90, 95, and 100%, 15 min for each time). The dried samples were coated with gold powder using the ion sputtering instrument (Qi et al., 2019). Platinum was used as the plating material. Samples coated with a film of gold–palladium alloy under vacuum were detected by a scanning electron microscope (SEM, Sigma 500/VP, Zeiss, Germany).
C. fragariae was cultured in the sterilized PDA liquid medium at 28°C and 200 rpm for 5 days. Cells of fungal pathogen were collected and fixed as described in Section “Morphological Profile of C. fragariae Spores Treated With Extract.” The pathogenic samples were embedded in Epon 812 resin. An 80-nm-thick section was manufactured by an ultramicrotome (EM UC6, Leica, Germany). The sections were stained with 3% (v/v) of lead citrate for 10 min and 3% (v/v) of saturated uranyl acetate for 30 min. The cell ultrastructure of C. fragariae was observed by transmission electron microscopy (TEM, HT7700, Hitachi, Japan).
To test the safety of the extract for human, a hemolysis experiment was performed (Li et al., 2021). The human red blood cells were collected by centrifugation at 1,000 g for 5 min and then were washed with 0.85% (w/v) of normal saline for three times. Two percent of red blood cells was prepared as in our previous description (Li et al., 2021). Four hundred fifty microliters of blood cell solution was mixed with 50 μl of extract (1 × EC50, 2 × EC50, 4 × EC50, or 8 × EC50) at 37°C for 1 h. The red blood cells treated with 0.85% (w/v) of normal saline and 0.1% (v/v) of Triton X-100 were used as negative and positive controls, respectively. The release of hemoglobin was monitored by absorbance at 540 nm. Hemolytic activity was expressed as the percentage of total hemoglobin released by extract treatment in comparison with that released by Triton X-100 treatment (Wang et al., 2018).
In order to test whether the selected isolate extract had a broad-spectrum antifungal activity, we selected 10 phytopathogenic fungi, including C. fragariae (ATCC 58718), Fusarium graminearum (ATCC MYA-4620), C. acutatum (ATCC 56815), F. oxysporum f. sp. cucumerinum (ATCC 204378), Curvularia lunata (ATCC 42011), Pyricularia oryzae (ATCC 52352), C. gloeosporioides Penz. (ATCC 58222), Alternaria sp. (ATCC 20492), C. capsici (ATCC 48574), and C. gloeosporioides (ATCC 16330). These phytopathogenic fungi were kept in our lab. The antifungal activity of the extract was detected against each phytopathogenic fungus according to our previous method (Wei et al., 2020).
The genomic DNA of the selected isolate was extracted using the BioTeke Bacterial Genomic DNA Rapid Extraction Kit (DP1301, Beijing Biotech Co., Ltd., China) and quantified using the TBS-380 fluorometer (Turner BioSystems Inc., Sunnyvale, CA, United States). Sequencing was performed on the platform of the Illumina high-throughput sequencing platform (HiSeq). The complete genome was assembled by Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. Bioinformatics analysis was carried out using the I-Sanger Cloud Platform 2.0. The ANI was analyzed using the online OrthoANI (Yoon et al., 2017). The G + C content of the strain genome was calculated based on the complete genome sequence. The protein-coding genes were predicted by the Glimmer v3.02 software (Delcher et al., 2007). The unigenes were annotated using the public databases of Gene Ontology (GO1), Clusters of Orthologous Groups of proteins (COG2), and Kyoto Encyclopedia of Genes and Genomes (KEGG3) (Zhang et al., 2020). Gene cluster analysis of secondary metabolite synthesis was carried out by the online antiSMASH v4.2.0 software (Huang et al., 2019).
The chemical compounds of the extract were identified by gas chromatography–mass spectrometry (GC-MS) (Ogundajo et al., 2017). The analysis was performed on gas chromatography (5973 Inert XL MSD, Agilent, United States) plus with DB-FFAP (30 m × 0.25 mm × 0.25 μm) and a triple quadrupole mass spectrometer. Helium was used as a carrier gas at 1 ml min–1. The temperature program was kept at 70°C for 2 min and increased to 100°C at 5°C min–1 for 5 min, 250°C at 10°C min–1 for 35 min, and 280°C at 4°C min–1 for 5 min. The mass spectrometer was operated in an optimized condition as in our previous description (Li et al., 2021); these data of mass spectra were determined by alignment with the National Institute of the Standards and Technology (NIST) spectral library version 2.0.
A one-way analysis of variance (ANOVA) was applied to analyze all data using the SPSS software version 22 (SPSS Inc., Chicago, IL, United States). Data from three independent experiments were expressed as the mean ± standard deviation. A significant difference was evaluated in the light of the LSD multiple-range test at the level of p < 0.05.
A total of 35 endophytic bacteria were isolated from noni fruit. Twenty-nine bacteria had an antifungal activity against C. fragariae in vitro in Supplementary Table 1. The fermentation broth of 10 endophytic bacteria exhibited antagonistic activity against C. fragariae (Supplementary Table 2). Especially, a strain labeled with QN1NO-4 had the strongest antagonistic activity. Compared with 85.50 mm ± 1.43 of growth diameter of C. fragariae in the control plate, the growth diameter was 25.50 mm ± 0.79 in the presence of strain QN1NO-4 (Figure 1A). The inhibition activity of mycelial growth was up to 70%. To further analyze antifungal components of strain QN1NO-4, extracts were isolated using 100% of methanol. The inhibition activity was approximately 51% in the PDA solid medium with 50 mg l–1 of the final extract concentration (Figure 1B). Hence, strain QN1NO-4 was selected for the following study.
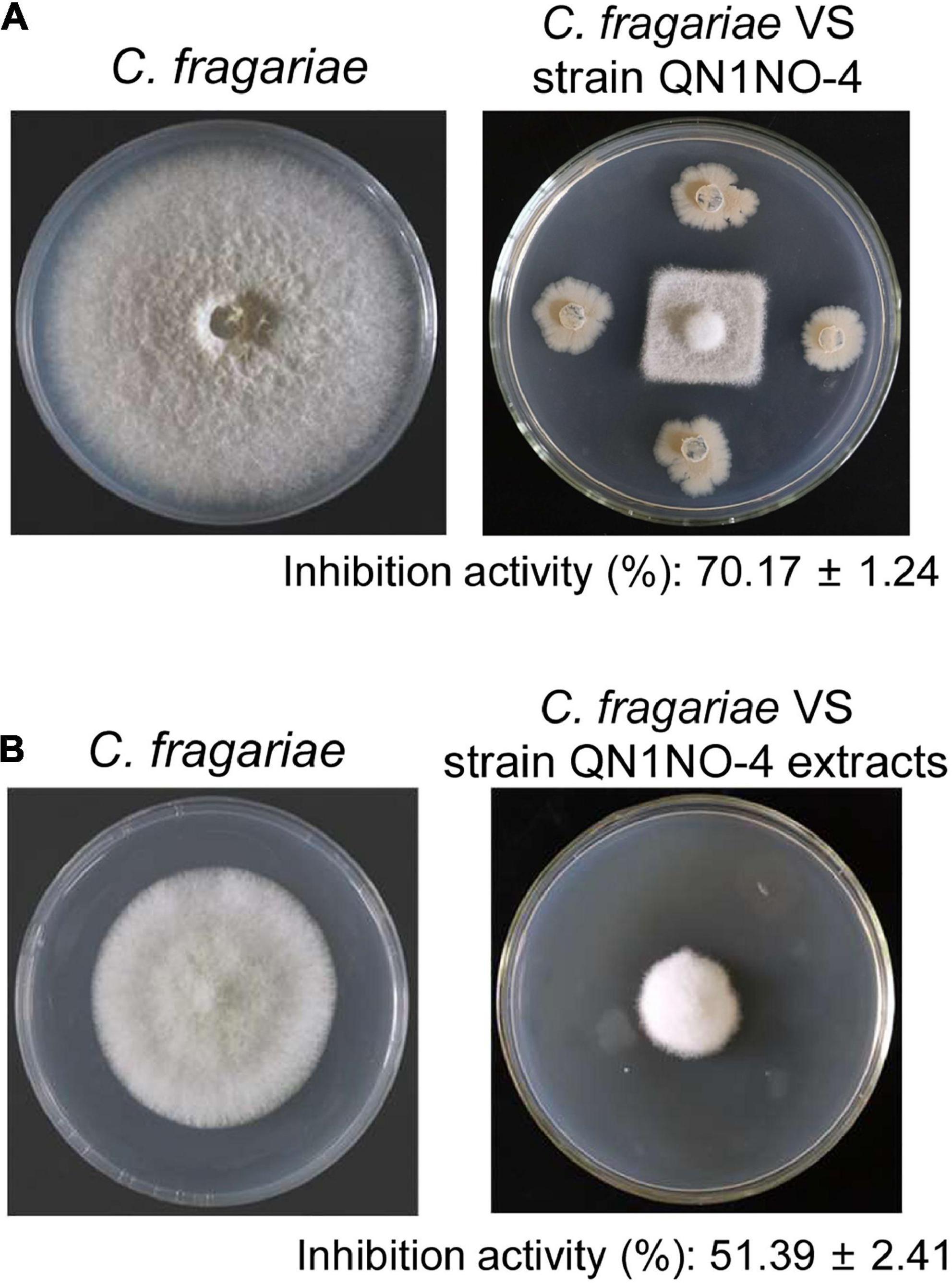
Figure 1. Antifungal activity of strain QN1NO-4 and its extract against C. fragariae of strawberry. (A) In vitro inhibition activity of strain QN1NO-4 on mycelial growth of C. fragariae. (B) Effects of extract isolated with 100% of methanol on mycelial growth of C. fragariae.
Based on the analysis of the morphological properties, strain QN1NO-4 was a round and primrose yellow colony on the LB plate. The colonial margin became much rougher and more irregular along with aging. The physiological and biochemical characteristics showed that strain QN1NO-4 could produce urease, catalase, and nitrate reductase but could not generate hydrogen sulfide. Positive results were detected under analysis of gelatin liquefaction, starch hydrolysis, or V-P test. The strain could grow on the medium with up to 13% of NaCl, temperature from 30 to 65°C, and pH from 5 to 10. It could utilize all carbon sources and nitrogen sources tested except for L-glutamate and L-tyrosine. In addition, strain QN1NO-4 was sensitive to 18 antibiotics tested except for penicillin and piperacillin (Table 1; Figure 2A and Supplementary Tables 3, 4).
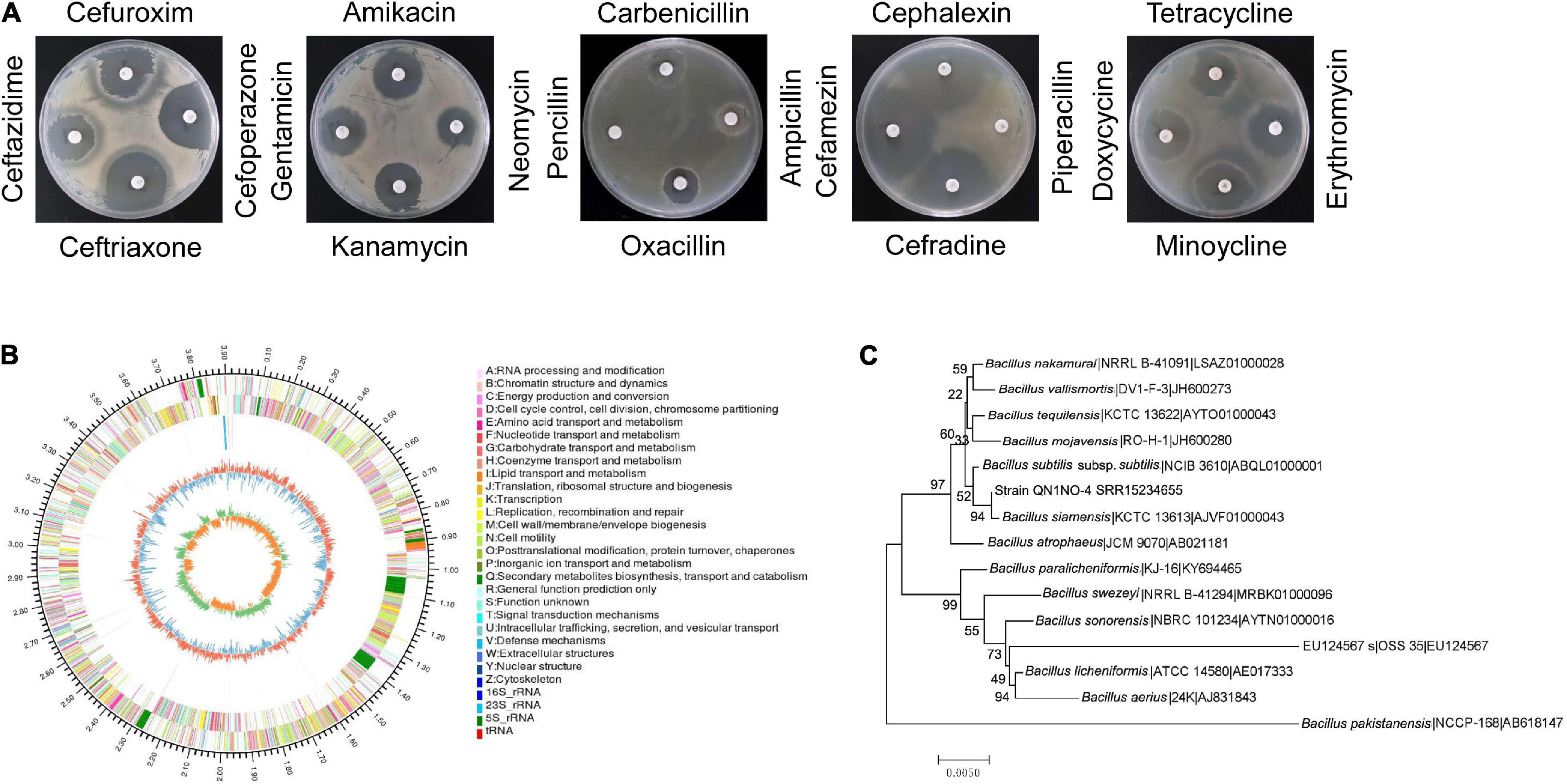
Figure 2. Identification of strain QN1NO-4. (A) Antibiotic sensitivity test of strain QN1NO-4. (B) Circular map of the strain QN1NO-4 genome. From outside to center, ring 1 represented the genome-size marker. Rings 2 and 3 showed coding sequences on forward and reverse strands, respectively. Ring 4 demonstrated rRNAs and tRNAs. Ring 5 represented the G + C content, followed by G + C skew in ring 6. (C) Phylogenetic tree of strain QN1NO-4 based on the 16S rRNA sequence. The tree was constructed using the neighbor-joining method in the MEGA software. The level of bootstrap support (1,000 repetitions) was indicated at all nodes.
The complete genome of strain QN1NO-4 was sequenced and submitted to the GenBank with accession number of SRR15234655. It contained 3,923,715 bp with 46% of G + C content. The genome consisted of three rRNA genes, 52 tRNA genes, and 3,917 protein-coding genes (Figure 2B and Table 2). Based on 16S rRNA gene sequences, a phylogenetic tree was constructed using the neighbor-joining method (Figure 2C). Strain QN1NO-4 showed a 100% of similarity with the standard strain B. siamensis (KCTC 13613). In addition, the ANI value was calculated to further identify the species of strain QN1NO-4. Genomic data of the highest homology standard strains (B. siamensis KCTC 13613 and B. subtilis NCIB 3610) were downloaded from the EzBioCloud public genome database4 and then were submitted to the ANI computing platform.5 The ANI values of strain QN1NO-4 were 94 and 77% in comparison with two standard strains B. siamensis KCTC 13613 and B. subtilis NCIB 3610, respectively (Supplementary Table 5). Both ANI values were below 95–96% of the threshold value for novel species definition (Richter and Rosselló-Móra, 2009). Thus, strain QN1NO-4 might be a novel species of the genus Bacillus, called after B. safensis sp. QN1NO-4.
The effects of strain QN1NO-4 extracts on mycelial growth of C. fragariae were assayed. The inhibitory efficiency showed a dose-dependent manner (Figure 3A). After the strain QN1NO-4 extract was co-incubated with C. fragariae at 28°C for 7 days, significant growth inhibition of the pathogen was observed in all the concentration groups (1.563, 3.125, 6.25, 12.5, 25, 50, 100, and 200 mg l–1). The inhibitory efficiency was 16, 23, 28, 41, 55, 69, and 83%, respectively (Figure 3B). Compared with the growth diameter of C. fragariae (68.00 mm ± 0.1) in the control plate, the half-maximal effective concentration (EC50) value of the strain QN1NO-4 extract was 33.81 ± 0.46 mg l–1 using the toxicity regression equation. The concentration was defined as 1 × EC50 for the following study.
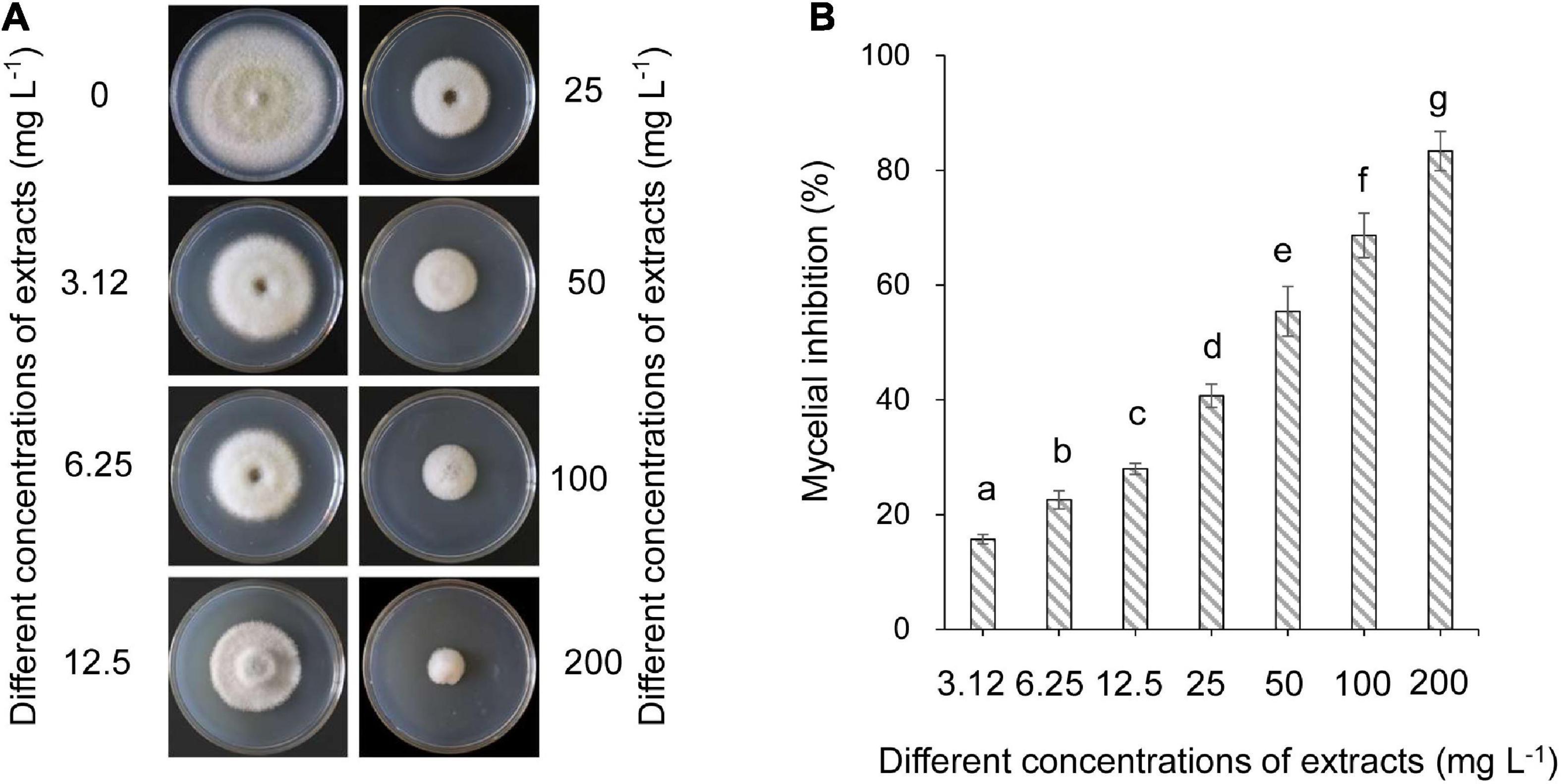
Figure 3. Determination of the half-maximal effective concentration value of extract against C. fragariae. (A) Growth inhibition of C. fragariae on the PDA medium after treatment with different dose extracts. (B) Quantitative analysis of C. fragariae growth diameters. Error bars indicated standard errors of the means of three repeated experiments. Different lowercase letters represented a significant difference (p < 0.05) of mycelial growth diameter at the same time point using the LSD multiple-range test.
To investigate the biocontrol efficiency of strain QN1NO-4 against C. fragariae in post-harvest strawberry, each concentration extract (1 × EC50, 2 × EC50, 4 × EC50, or 8 × EC50, respectively) was used to treat the selected fruit samples (Figure 4A). After artificially co-incubating with C. fragariae (1.0 × 106 CFU ml–1) for 7 days, the DIs and quality parameters of strawberry fruit were measured. Compared with the control group, all treatments alleviated significantly the infection of C. fragariae in post-harvest strawberry fruit at 7 days post-inoculation (dpi). The DIs were 67, 46, 29, and 13% in treatment groups of 1 × EC50, 2 × EC50, 4 × EC50, and 8 × EC50, respectively (Figure 4B). Therefore, strain QN1NO-4 extract can inhibit effectively the infection of C. fragariae in post-harvest strawberry fruit.
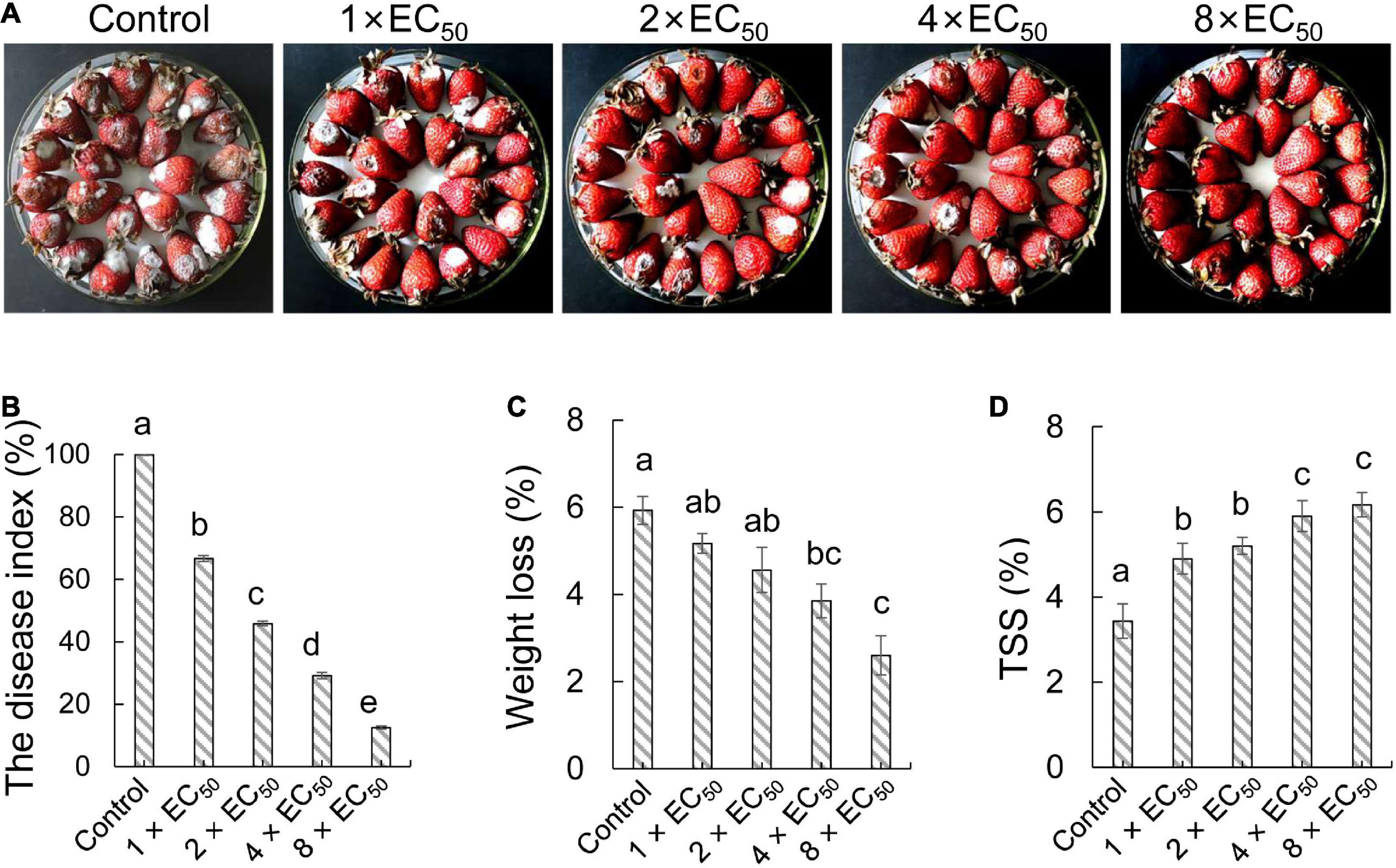
Figure 4. In vivo effects of strain QN1NO-4 extract on fruit decays and disease incidence caused by C. fragariae. (A) Disease inhibition of extract on strawberry fruit. Four extract concentrations (1 × EC50, 2 × EC50, 4 × EC50, and 8 × EC50) were selected. The treated fruit samples were stored at 28°C for 7 days. (B) Quantitative analysis of disease incidence of strawberry fruit treated with different dose extracts. (C) Effects of strain QN1NO-4 extract on weight loss of strawberry fruit. (D) Effects of strain QN1NO-4 extract on TSS contents of strawberry fruit. Error bars indicated standard errors of the means of three repeated experiments. Different letters indicated a significant difference (p < 0.05) among different dose extracts using the LSD multiple-range test.
Weight loss and TSS were measured to evaluate the effects of the strain QN1NO-4 extract on the fruit quality of strawberry. Weight loss showed a continuous decrease along with the increasing dose of extract (Figure 4C). Compared with the control group, weight loss was reduced by 4 and 3% after treatment with 4 × EC50 and 8 × EC50, respectively. No obvious difference was observed between 1 × EC50 and 2 × EC50 treatment groups. Moreover, strain QN1NO-4 extract also delayed the decrease in TSS content during storage. Compared with the control group at 7 dpi, high TSS contents were kept in the treated fruit samples (Figure 4D).
To further investigate the possible mechanism of the strain QN1NO-4 extract inhibiting fruit decay of post-harvest strawberry, the spore germination of C. fragariae was first analyzed after treatment with extract. The results showed that spore germination was inhibited, and the inhibitory efficiency was gradually increased with the increase of extract concentrations (Figure 5A). In the control group, 95% of spore can germinate, but only 36% and 18% of germination rates were detected in 1 × EC50 and 2 × EC50 treatment groups for 12 h, respectively. Especially, more than 4 × EC50 extract completely inhibited the spore germination of C. fragariae. Dense and short hypha at the colony edges were observed in comparison with the sparse and slender mycelia in the control plate (Figure 5B).
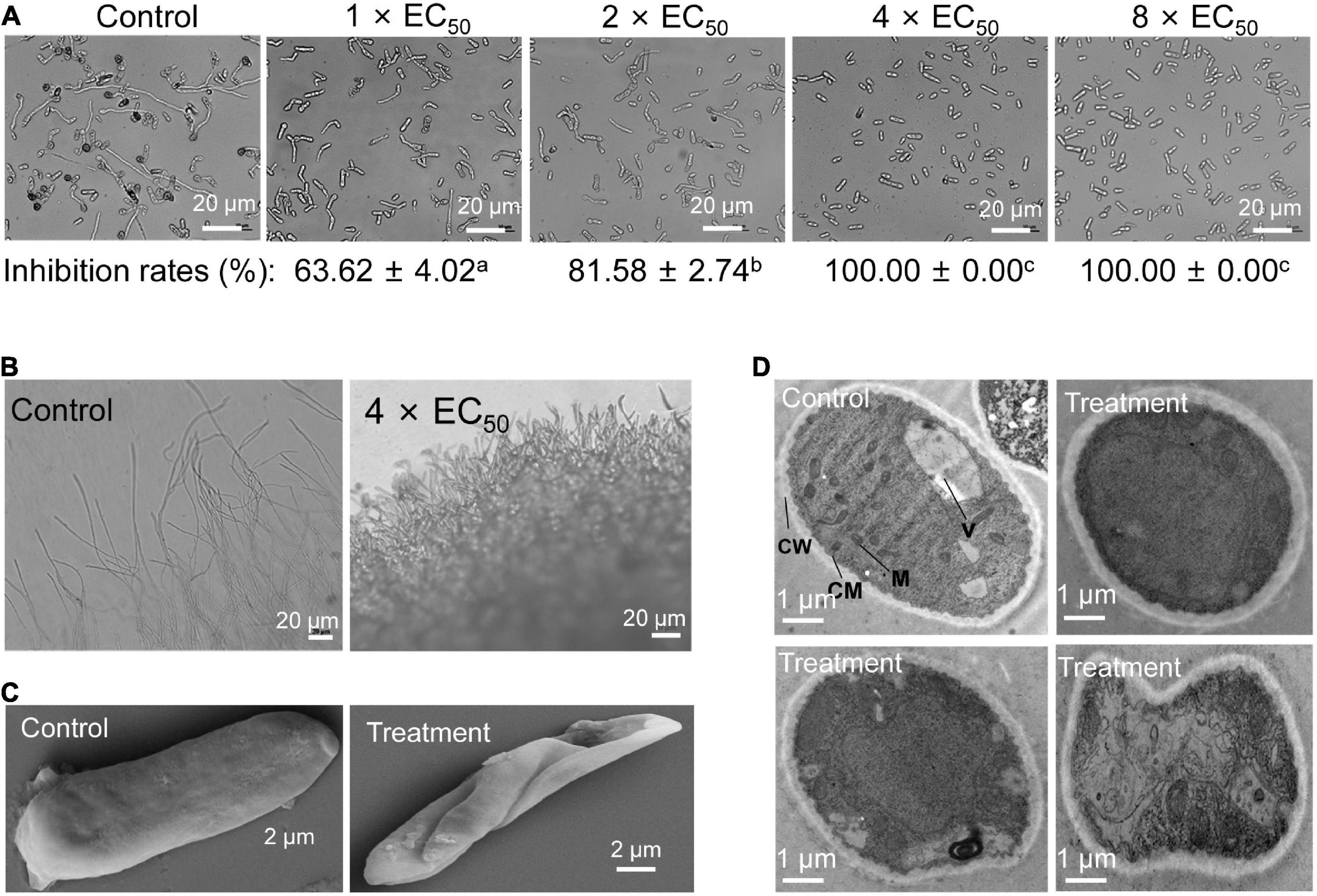
Figure 5. Inhibitory effects of strain QN1NO-4 extract on conidial germination, mycelial morphology, and ultrastructure of C. fragariae. (A) Conidial germination assay of C. fragariae after treatment with 1×, 2×, 4×, or 8 × EC50 extracts. Bar = 20 μm. CW: cell wall; CM: cell membrane; M: mitochondria; V: vacuole. (B) Growth characteristics of C. fragariae mycelia treated with 4 × EC50 of extract. Bar = 20 μm. (C) Representative pictures showing the mycelial morphology of C. fragariae after treatment with 4 × EC50 of extracts. Bar = 20 μm. (D) Representative pictures showing the ultrastructure of C. fragariae after treatment with 4 × EC50 of extract. Bar = 1 μm.
We further analyzed the morphological characteristics of C. fragariae spores before and after extract treatment. A wizened and distorted surface of spores was observed in the treatment group of 4 × EC50 extracts for 12 h. The regular and smooth morphology of hypha was displayed in the control group (Figure 5C). By contrast, vacuoles became bigger and disintegrated gradually after treatment with strain H4 extract. The organelle integrity was also broken, and cells showed a cytoplasmic heterogeneity (Figure 5D). In the control group, some complete organelles such as vacuoles and mitochondria could be clearly detected by TEM (Figure 5D).
To investigate the toxicity of strain QN1NO-4 extract on eukaryotic cells, the hemolytic activity of extract (1 × EC50, 2 × EC50, 4 × EC50, or 8 × EC50) on human red blood cells was assayed using the release of hemoglobin after treatment at 37°C for 1 h (Supplementary Figure 1). A percentage of 0.1% of Triton X-100 exhibiting 100% of hemolytic activity was selected as a positive control. No obvious hemolytic activity appeared in all treatment groups. Therefore, extract had non-specific cell lytic activity and toxicity to eukaryotic cells.
Based on the prediction of secondary metabolites by genomic sequencing, we further analyzed whether strain QN1NO-4 extract had a broad-spectrum antifungal activity against 10 phytopathogenic fungi selected in our study. After co-incubating for 7 days, the extract exhibited an excellent inhibitory activity for mycelial growth of the selected fungi, ranging from 35 to 56% (Figure 6). The best inhibition activity with 56% was found against Alternaria sp., suggesting that the extract had the strongest antifungal activity against the fungi, followed by C. lunata (54%) and F. graminearum (50%). The minimal inhibition activity of the extract was 35% against C. gloeosporioides, indicating that the fungus had the highest tolerable ability for extract treatment.
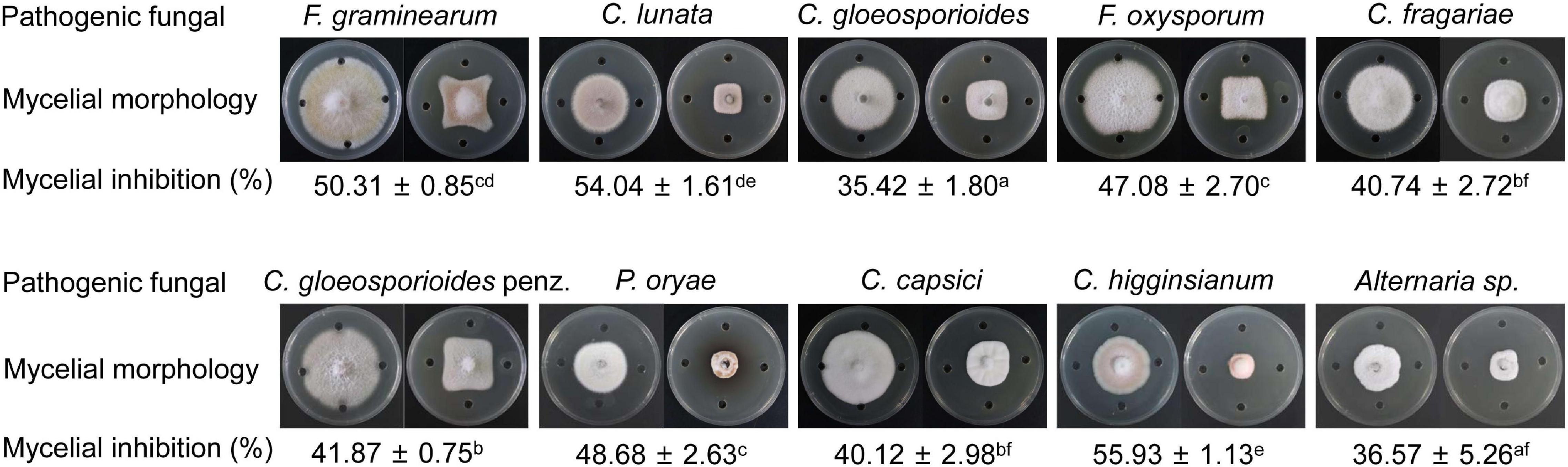
Figure 6. A broad-spectrum antifungal activity of strain QN1NO-4 extract against the selected phytopathogenic fungi.
Functional analysis showed that 2,554, 2,967, and 2,190 out of the identified 3,917 protein-coding genes were annotated into GO, COG, and KEGG categories, respectively (Supplementary Figure 2). For GO categories, the predicted genes were divided into three classes including biological process (27%), cellular components (27%), and molecular function (45%) (Supplementary Figure 2A). For COG categories, these genes were classified into 20 types of four categories. The largest category was metabolism (41%), followed by cellular processes and signaling (18%) as well as information storage and processing (16%). Twenty-five percent of the proportion was classified into the poorly characterized category (Supplementary Figure 2B). KEGG analysis showed that 2,190 of identified protein-coding genes were annotated into 41 pathways (Supplementary Figure 2C). Notably, some pathways including signal transduction, biosynthesis of other secondary metabolites, and metabolism of terpenoids and polyketides were pivotal for disease resistance.
By alignment of antiSMASH software and GenBank, 14 biosynthetic gene clusters containing 457 genes were predicted in the strain QN1NO-4 genome, including three NRPS gene clusters (54 genes), one PKS-like gene cluster (48 genes), one transAT-PKS gene cluster (48 genes), two transAT-PKS-like gene clusters (36 genes), two terpene gene clusters (48 genes), one LAP gene cluster (20 genes), one PKS III gene cluster (48 genes), one beta-lactone gene cluster (50 genes), and one MerR family transcriptional gene cluster (58 genes) (Table 3 and Supplementary Table 6). Among them, eight chemicals produced by gene clusters were involved in the biosynthesis of antimicrobial metabolites, such as bacillibactin, bacilysin, bacillaene, macrolactin, butirosin, difficidin, plantazolicin, and kalimantacin A (Supplementary Figure 3). Four gene clusters (cluster1, cluster2, cluster4, and cluster5) presented a 100% similarity with the known compounds. However, 2 out of 14 clusters exhibited a 13% similarity with the predicted compounds, suggesting that these clusters might be involved in the biosynthesis of novel metabolites.
GC-MS was employed to analyze the bioactive compounds of the strain QN1NO-4 extract. A total ion current chromatogram is shown in Supplementary Figure 4. Based on retention time, molecular mass, and molecular formula, 15 chemical compounds were identified by the alignment of their mass spectra against the NIST library (Supplementary Table 7). These compounds mainly contained hydrocarbons, acids, pyrrolizidine, esters, and phenol. The predicted chemical structures are shown in Figure 7, including 2-methyloctanoic acid (1), 2,3-butylene glycol diacetate (2), phenylacetaldehyde (3), 7-hexadecene (4), pantolactone (5), propanoic acid, 3-(methylthio) (6), 2(3H)-benzofuranone (7), 1-octadecene (8), 2,4-bis(1,1-dimethylethyl)-phenol (9), indolizine (10), ethyl palmitate (11), 2-tetradecanol (12), trichloroacetic acid myristyl ester (13), 17b-methyl-5a-androstan-3a,17b-diol (14), and butyl isobutyl phthalate (15). The peak areas of compounds represented the portions of different compounds in the strain QN1NO-4 extract. Among these compounds, the peak area of 17b-methyl-5a-androstan-3a,17b-diol was 42%, suggesting that it was a dominant compound in strain QN1NO-4 extracts.
Anthracnose caused by C. fragariae is a common and destructive disease of strawberry (Mehmood et al., 2021). Biocontrol has received an increasing attention in controlling post-harvest fruit diseases (Li X.J. et al., 2020). Several endophytes play a vital role in plant protection and plant growth promotion. Many promising endophytic bacteria have been reported as biocontrol candidates against plant pathogens (Morales-Cedeño et al., 2021). Especially, Bacillus strains draw more and more attention due to their ability to produce resistant endospores and bioactive metabolites (Zhou et al., 2021). In our present study, a strain labeled QN1NO-4 was isolated from noni fruit and exhibited a strong antifungal ability against C. fragariae (Figure 1). According to morphological characteristics as well as genome alignment, strain QN1NO-4 was identified as a novel species of the genus Bacillus (Table 1, Figure 2 and Supplementary Tables 3, 4). As potential biocontrol agents, Bacillus species had also been successfully used to control other post-harvest diseases of different fruit, such as Bacillus species against B. cinerea and green mold in strawberry fruit and citrus (Tian et al., 2020; Wang et al., 2021), B. amyloliquefaciens against Penicillium in oranges and apples (Calvo et al., 2017; Wang et al., 2020; Ye et al., 2021), and B. atrophaeus against anthracnose on soursop and avocado (Guardado-Valdivia et al., 2018).
To further elucidate the antifungal mechanism of the strain QN1NO-4 extract against C. fragariae, we examined the spore germination, morphology, and ultrastructure of pathogenic cells after extract treatment. Consistent with antifungal activity in vivo, more than 4 × EC50 of strain QN1NO-4 extract completely inhibited the spore germination of C. fragariae. In addition, the extract also caused the irregular morphology, cell wall thickening, extensive vacuolization, and intracellular organelle degradation of C. fragariae (Figure 5D). Inhibition of spore germination was essential for protecting post-harvest fruit from pathogenic infection (Miyara et al., 2010). A similar germination inhibition of spores was detected in P. digitatum treated with C. lusitaniae and P. fermentans, P. horianna treated with Bacillus species, and F. incarnatum treated with B. amyloliquefaciens (Dheepa et al., 2016; Perez et al., 2019; Li Y.G. et al., 2020). Moreover, the toxicity and broad antimicrobial activity of the strain QN1NO-4 extract were also determined (Supplementary Figure 1 and Figure 6), suggesting that it is a promising alternative biological fungicide to control post-harvest diseases.
Natural antimicrobials were an attractive source of controlling post-harvest diseases, mitigating the reliance on synthetic fungicides (Kim et al., 2020). We found that the strain QN1NO-4 extract effectively inhibited mycelial growth of C. fragariae in vitro, and the mycelial inhibition rate depended on a dose-dependent manner (Figure 3A). The previous study showed that a novel peptide produced by B. amyloliquefaciens W10 had a strong antifungal activity (Zhang et al., 2017). It suggested that high antagonistic components were different from diverse Bacillus species. Fox example, extracts from different Bacillus species significantly inhibited different mycelial growths of Fusarium solani, P. digitatum, and Fusarium incarnatum (Ahmad et al., 2012; Tian et al., 2020). Moreover, the extract effectively kept the fruit quality of post-harvest strawberry by inhibiting fruit weight loss and TSS decrease (Figures 4C,D). Similarly, weight loss in L. plantarum-treated litchis was lower than the control fruit (Martínez-Castellanos et al., 2011).
GC-MS was used to further identify antifungal metabolites of strain QN1NO-4. A total of 15 chemical compounds including hydrocarbons, acids, pyrrolizidine, esters, and phenol were identified (Supplementary Table 7 and Figure 7). Among them, 2(3H)-benzofuranone, 2,4-bis(1,1-dimethylethyl)-phenol, and butyl isobutyl phthalate exhibited high antifungal and antibacterial activities. 2(3H)-benzofuranone produced by the indigenous fungus remarkably inhibited the mycelial growth of M. oryzae (Fan et al., 2019). 2,4-bis(1,1-Dimethylethyl)-phenol effectively inhibited the spore germination of P. cinnamomi and Aspergillus and controlled tomato fungal diseases such as early blight and gray mold (Rangel-Sánchez et al., 2014; Gao et al., 2017). Butyl isobutyl phthalate exhibited an antibacterial activity with an MIC value of 128 g l–1 (Hussain et al., 2017). Interestingly, the compounds in the strain QN1NO-4 extract identified by GC-MS were different from those involved in the synthesis of the above gene clusters (Supplementary Tables 6, 7). It might be because the identification method is not enough to identify all metabolites of strain QN1NO-4 (Wei et al., 2020). Thus, further study was necessary to identify specific active metabolites of strain QN1NO-4, contributing to the broad-spectrum antifungal activity.
Considering the deficiency of the GC-MS method, whole-genome analysis was used to predict rare and novel secondary metabolites, which cannot be detected directly in the fermentation broth using the current technology. In our study, the genome of strain QN1NO-4 was sequenced, revealing numerous unknown and known gene clusters of secondary metabolites (Table 3 and Supplementary Figure 3). These gene clusters might be responsible for the production of microbial natural products. For example, the biosynthetic gene clusters encoding for polyketides (PKS) and non-ribosomal peptides (NRPS) were identified. They belonged to a structurally varied group of compounds and participated in the biosynthesis of antifungal compounds (Nimaichand et al., 2015; Prasad et al., 2015). In addition, some antimicrobials were also found such as bacillibactin, bacilysin, fengycin, bacillaene, macrolactin, butirosin, difficidin, plantazolicin, and kalimantacin A (Supplementary Table 6 and Supplementary Figure 3). Bacillibactin was a specific transport system enabling Bacillus cells to accumulate and take up limited iron ions from their natural environment (Chen et al., 2009). Bacilysin participated in the antagonistic activity of B. velezensis against Gram-negative foodborne pathogens (Nannan et al., 2021). As a potent inhibitor of filamentous fungi, fengycin owned a strong antifungal ability against V. dahliae and B. cinerea (Li Q.R. et al., 2020; Su et al., 2020). Bacillaene can inhibit prokaryotic protein synthesis (Li Q.R. et al., 2020). Macrolactin effectively inhibited the replication of mammalian Herpes simplex virus and HIV in lymphoblast cells (Gustafson et al., 1989). Butirosin was an aminoglycoside antibiotic produced by Bacillus circulans (Llewellyn et al., 2007). Difficidin exhibited a broad-spectrum antibacterial activity (Anthony et al., 2009). Plantazolicin belonged to a type of polyheterocyclic natural product with highly selective and potent activity against anthrax-causing bacteria (Hao et al., 2015). Thistlethwaite et al. (2017) found that kalimantacin showed a selective and high activity against staphylococci with an MIC value of 0.064 g l–1. Considering the novel species of strain QN1NO-4 in our study, some unknown function gene clusters might encode some new secondary metabolites. It will be further proved that strain QN1NO-4 has a broad-spectrum resistance to phytopathogen fungi.
In the present study, a newly isolated strain QN1NO-4 from noni fruit was identified as B. safensis sp. and exhibited a strong antifungal ability against C. fragariae. Its extract can effectively reduce the fungal disease index and preserve the post-harvest quality of strawberry fruit. Moreover, strain QN1NO-4 extract can significantly inhibit mycelial growth and spore germination of C. fragariae, causing morphological and ultrastructure changes of fungal pathogen. Fifteen chemical compounds were identified from strain QN1NO-4 extracts by GC-MS. In addition, the whole-genome analysis revealed that some key function gene clusters in the strain QN1NO-4 genome were involved in the biosynthesis of active secondary metabolites. These diverse gene clusters and metabolites might contribute to its broad-spectrum antifungal activity. Therefore, strain QN1NO-4 is an effective bio-agent for controlling strawberry post-harvest diseases caused by C. fragariae.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XL, LZ, JX, and WW developed the ideas and designed the experimental plans. LZ, JX, and WW supervised the research and provided the funding support. XL, MZ, DZ, DQ, CQ, CL, SL, and DX performed the experiments. MZ, DZ, and DQ provided the materials. XL, LZ, and WW analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2020YFD1000104), the National Natural Science Foundation of China (31770476, 32072504), the Natural Science Foundation of Hainan (2019RC293, 320CXTD441), and the China Agriculture Research System (CARS-31).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.735732/full#supplementary-material
Supplementary Figure 1 | Hemolytic activity of red blood cells treated with strain QN1NO-4 extract.
Supplementary Figure 2 | Genome annotation of strain QN1NO-4.
Supplementary Figure 3 | Genome-wide analysis of gene clusters related to the biosynthesis of secondary metabolites in the strain QN1NO-4 genome using the online antiSMASH v4.2.0 software.
Supplementary Figure 4 | A total ion current chromatogram of strain QN1NO-4 extract.
Supplementary Table 1 | Antifungal activities of 29 isolated bacteria on mycelial growth of C. fragariae.
Supplementary Table 2 | Antifungal activities of fermentation broth of 10 selected bacteria on mycelial growth of C. fragariae.
Supplementary Table 3 | Carbon source utilization and nitrogen source utilization of strain QN1NO-4.
Supplementary Table 4 | Sensitivity of strain QN1NO-4 to antibiotics.
Supplementary Table 5 | Results of average nucleotide identity (ANI).
Supplementary Table 6 | Prediction and functional annotation of secondary metabolites of strain QN1NO-4 by alignment with the online antiSMASH v4.2.0 software.
Supplementary Table 7 | Compounds identified of extracts from strain QN1NO-4 by GC-MS.
Ahmad, B., Khan, I., Bashir, S., and Azam, S. (2012). Chemical composition and antifungal, phytotoxic, brine shrimp cytotoxicity, insecticidal and antibacterial activities of the essential oils of Acacia modesta. J. Med. Plants Res. 6, 4653–4659. doi: 10.5897/JMPR12.016
Aiello, D., Restuccia, C., Stefani, E., Vitale, A., and Cirvilleri, G. (2019). Postharvest biocontrol ability of Pseudomonas synxantha against Monilinia fructicola and Monilinia fructigena on stone fruit. Postharvest Biol. Technol. 149, 83–89. doi: 10.1016/j.postharvbio.2018.11.020
Anthony, A. A., Marc, O., Badre, H., Yannick, L., Alain, B., Bernard, J., et al. (2009). Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 8:63. doi: 10.1186/1475-2859-8-63
Calvo, H., Marco, P., Blanco, D., Oria, R., and Venturini, M. (2017). Potential of a new strain of Bacillus amyloliquefaciens BUZ-14 as a biocontrol agent of postharvest fruit diseases. Food Microbiol. 63, 101–110. doi: 10.1016/j.fm.2016.11.004
Chen, X. H., Koumoutsi, A., Scholz, R., and Borriss, R. (2009). More than anticipated – production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 16, 14–24. doi: 10.1159/000142891
Chen, X. Y., Dai, D. J., Zhao, S. F., Shen, Y., Wang, H. D., and Zhang, C. Q. (2019). Genetic diversity of Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China. Plant Dis. 104, 1351–1357. doi: 10.1094/PDIS-09-19-2026-RE
Chung, P. C., Wu, H. Y., Wang, Y. W., Ariyawansa, H. A., Hu, H. P., Hung, T. H., et al. (2020). Diversity and pathogenicity of Colletotrichum species causing strawberry anthracnose in Taiwan and description of a new species, Colletotrichum miaoliense sp. nov. Sci. Rep. 10:14664. doi: 10.1038/s41598-020-70878-2
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Denoyes-Rothan, B., Guérin, G., Délye, C., Smith, B., Minz, D., Maymon, M., et al. (2003). Genetic diversity and pathogenic variability among isolates of Colletotrichum species from strawberry. Phytopathology 93, 219–228. doi: 10.1094/PHYTO.2003.93.2.219
Dheepa, R., Vinodkumar, S., Renukadevi, P., and Nakkeeran, S. (2016). Phenotypic and molecular characterization of chrysanthemum white rust pathogen Puccinia horiana (Henn) and the effect of liquid based formulation of Bacillus spp. for the management of chrysanthemum white rust under protected cultivation. Biol. Control 103, 172–186. doi: 10.1016/j.biocontrol.2016.09.006
Dukare, A. S., Paul, S., Nambi, V. E., Gupta, R. K., Singh, R., Sharma, K., et al. (2019). Exploitation of microbial antagonists for the control of postharvest diseases of fruit: a review. Crit. Rev. Food Sci. Nutr. 59, 1498–1513. doi: 10.1080/10408398.2017.1417235
Einloft, T. C., Oliveira, P., Radunz, L. L., and Dionello, R. G. (2021). Biocontrol capabilities of three Bacillus isolates towards aflatoxin B1 producer A. flavus in vitro and on maize grains. Food Control 125:107978. doi: 10.1016/j.foodcont.2021.107978
Fan, X. Y., Matsumoto, H., Wang, Y., Hu, Y., Liu, Y. F., Fang, H. D., et al. (2019). Microenvironmental interplay predominated by beneficial Aspergillus abates fungal pathogen incidence in paddy environment. Environ. Sci. Technol. 53, 13042–13052. doi: 10.1021/acs.est.9b04616
Feliziani, E., Lichter, A., Smilanick, J. L., and Ippolito, A. (2016). Disinfecting agents for controlling fruit and vegetable diseases after harvest. Postharvest Biol. Technol. 122, 53–69. doi: 10.1016/j.postharvbio.2016.04.016
Gao, Z., Zhang, B., Liu, H., Han, J., and Zhang, Y. (2017). Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control 105, 27–39. doi: 10.1016/j.biocontrol.2016.11.007
Gotor-Vila, A., Teixidó, N., Casals, C., Torres, R., De Cal, A., Guijarro, B., et al. (2017). Biological control of brown rot in stone fruit using Bacillus amyloliquefaciens CPA-8 under field conditions. Crop Prot. 102, 72–80. doi: 10.1016/j.cropro.2017.08.010
Guardado-Valdivia, L., Tovar-Pérez, E., Chacón-López, A., López-García, U., Gutiérrez-Martínez, P., Stoll, A., et al. (2018). Identification and characterization of a new Bacillus atrophaeus strain B5 as biocontrol agent of postharvest anthracnose disease in soursop (Annona muricata) and avocado (Persea americana). Microbiol. Res. 210, 26–32. doi: 10.1016/j.micres.2018.01.007
Gustafson, K., Roman, M., and Fenical, W. (1989). The macrolactins, a novel class of antiviral and cytotoxic macrolides from a deep-sea marine bacterium. J. Am. Chem. Soc. 111, 7519–7524. doi: 10.1021/ja00201a036
Han, Y. C., Zeng, X. G., Xiang, F. Y., Ren, L., Chen, F. Y., and Gu, Y. C. (2016). Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei, China. Plant Dis. 100, 996–1006. doi: 10.1094/PDIS-09-15-1016-RE
Hao, Y., Blair, P. M., Sharma, A., Mitchell, D. A., and Nair, S. K. (2015). Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin. ACS Chem. Biol. 10, 1209–1216. doi: 10.1021/cb501042a
Huang, X. L., Kong, F. D., Zhou, S. Q., Huang, D. Y., Zheng, J. P., and Zhu, W. M. (2019). Streptomyces tirandamycinicus sp. nov., a novel marine sponge-derived actinobacterium with antibacterial potential against Streptococcus agalactiae. Front. Microbiol. 10:482. doi: 10.3389/fmicb.2019.00482
Hussain, A., Rather, M. A., Dar, M. S., Aga, M. A., Ahmad, N., Manzoor, A., et al. (2017). Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 27, 2579–2582. doi: 10.1016/j.bmcl.2017.03.075
Kim, J. D., Kang, J. E., and Kim, B. S. (2020). Postharvest disease control efficacy of the polyene macrolide lucensomycin produced by Streptomyces plumbeus strain CA5 against gray mold on grapes. Postharvest Biol. Technol. 162, 111–115.
Kumar, V., Naik, B., Gusain, O., and Bisht, G. S. (2014). An actinomycete isolate from solitary wasp mud nest having strong antibacterial activity and kills the candida cells due to the shrinkage and the cytosolic loss. Front. Microbiol. 5:446. doi: 10.3389/fmicb.2014.00446
Li, Q. R., Liao, S. T., Wei, J. H., Xing, D. X., Xiao, Y., and Yang, Q. (2020). Isolation of Bacillus subtilis strain SEM-2 from silkworm excrement and characterization of its antagonistic effect against Fusarium spp. Can. J. Microbiol. 66, 401–412. doi: 10.1139/cjm-2019-0621
Li, X. J., Jing, T., Zhou, D. B., Zhang, M. Y., Qi, D. F., Zang, X. P., et al. (2021). Biocontrol efficacy and possible mechanism of Streptomyces sp. H4 against postharvest anthracnose caused by Colletotrichum fragariae on strawberry fruit. Postharvest Biol. Technol. 175:111401. doi: 10.1016/j.postharvbio.2020.111401
Li, X. J., Li, K., Zhou, D. B., Zhang, M. Y., Qi, D. F., Jing, T., et al. (2020). Biological control of banana wilt disease caused by Fusarium oxyspoum f. sp. cubense using Streptomyces sp. H4. Biol. Control 155:104524. doi: 10.1016/j.biocontrol.2020.104524
Li, Y., Héloir, M., Zhang, X., Geissler, M., Trouvelot, S., Jacquens, L., et al. (2019). Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 20, 1037–1050. doi: 10.1111/mpp.12809
Li, Y. G., Cai, Y. A., Liang, Y. B., Ji, P. S., and Xu, L. K. (2020). Assessment of antifungal activities of a biocontrol bacterium BA17 for managing postharvest gray mold of green bean caused by Botrytis cinerea. Postharvest Biol. Technol. 161:111086. doi: 10.1016/j.postharvbio.2019.111086
Llewellyn, N. M., Li, Y., and Spencer, J. B. (2007). Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem. Biol. 14, 379–386. doi: 10.1016/j.chembiol.2007.02.005
Martínez-Castellanos, G., Pelayo-Zaldivar, C., Perez-Flores, L. J., Lopez-Luna, A., Gimeno, M., Barzana, E., et al. (2011). Postharvest litchi (Litchi chinensis Sonn.) quality preservation by Lactobacillus plantarum. Postharvest Biol. Technol. 59, 172–178. doi: 10.1016/j.postharvbio.2010.09.005
Mehmood, N., Yuan, Y., Ali, M., Ali, M., Iftikhar, J., Cheng, C. Z., et al. (2021). Early transcriptional response of terpenoid metabolism to Colletotrichum gloeosporioides in a resistant wild strawberry Fragaria nilgerrensis. Phytochemistry 18:112590. doi: 10.1016/j.phytochem.2020.112590
Miller-Butler, M. A., Smith, B. J., Babiker, E. M., Kreiser, B. R., and Blythe, E. K. (2018). Comparison of whole plant and detached leaf screening techniques for identifying anthracnose resistance in strawberry plants. Plant Dis. 102, 2112–2119. doi: 10.1094/pdis-08-17-1138-re
Miyara, I., Shafran, H., Davidzon, M., Sherman, A., and Prusky, D. (2010). pH regulation of ammonia secretion by Colletotrichum gloeosporioides and its effect on appressorium formation and pathogenicity. Mol. Plant Microbe Interact. 23, 304–316. doi: 10.1094/mpmi-23-3-0304
Morales-Cedeño, L. R., del CarmenOrozco-Mosqueda, M., Loeza-Lara, P. D., Parra-Cota, F. I., Santos-Villalobos, S., and Santoyo, G. (2021). Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: fundamentals, methods of application and future perspectives. Microbiol. Res. 242:126612. doi: 10.1016/j.micres.2020.126612
Nannan, C., Vu, H. Q., Gillis, A., Caulier, S., Nguyen, T. T. T., and Mahillon, J. (2021). Bacilysin within the Bacillus subtilis group: gene prevalence versus antagonistic activity against Gram-negative foodborne pathogens. J. Biotechnol. 327, 28–35. doi: 10.1016/j.jbiotec.2020.12.017
Nimaichand, S., Devi, A. M., Tamreihao, K., Ningthoujam, D. S., and Li, W. J. (2015). Actinobacterial diversity in limestonedeposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front. Microbiol. 6:413. doi: 10.3389/fmicb.2015.00413
Ogundajo, A., Okeleye, B., and Ashafa, A. O. (2017). Chemical constituents, in vitro antimicrobial and cytotoxic potentials of the extracts from Macaranga barteri Mull-Arg. Asian Pac. J. Trop. Biomed. 7, 654–659. doi: 10.1016/j.apjtb.2017.06.014
Pei, S. P., Liu, R. L., Gao, H. Y., Chen, H. J., Wu, W. J., Fang, X. J., et al. (2020). Inhibitory effect and possible mechanism of carvacrol against Colletotrichum fructicola. Postharvest Biol. Technol. 163:111126. doi: 10.1016/j.postharvbio.2020.111126
Perez, M. F., Díaz, M. A., Pereyra, M. M., Córdoba, J. M., Isas, A. S., Sepúlveda, M., et al. (2019). Biocontrol features of Clavispora lusitaniae against Penicillium digitatum on lemons. Postharvest Biol. Technol. 155, 57–64. doi: 10.1016/j.postharvbio.2019.05.012
Prasad, P., Varshney, D., and Adholeya, A. (2015). Whole genome annotation and comparative genomic analyses of bio-control fungus Purpureocillium lilacinum. BMC Genomics 16:1004. doi: 10.1186/s12864-015-2229-2
Qi, D., Zou, L., Zhou, D., Chen, Y., Gao, Z., Feng, R., et al. (2019). Taxonomy and broad-spectrum antifungal activity of Streptomyces sp. SCA3-4 isolated from rhizosphere soil of Opuntia stricta. Front. Microbiol. 10:1390. doi: 10.3389/fmicb.2019.01390
Rangel-Sánchez, G., Castro-Mercado, E., and García-Pineda, E. (2014). Avocado roots treated with salicylic acid produce phenol-2,4-bis (1,1-dimethylethyl), a compound with antifungal activity. J. Plant Physiol. 171, 189–198. doi: 10.1016/j.jplph.2013.07.004
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rico, D., Barcenilla, B., Meabe, A., Gonz’alez, C., and Ana, B. (2019). Mechanical properties and quality parameters of Chitosan-edible algae (Palmaria palmata) on ready-to-eat strawberries. J. Sci. Food Agric. 99, 2651–2921. doi: 10.1002/jsfa.9504
Rong, S., Hong, X., Li, L., Chen, R., Gao, X., and Xu, Z. (2020). Antifungal activity of endophytic Bacillus safensis b21 and its potential application as a biopesticide to control rice blast - sciencedirect. Pestic. Biochem. Phys. 162, 69–77. doi: 10.1016/j.pestbp.2019.09.003
Su, Z. H., Chen, X. Y., Liu, X. M., Guo, Q. G., Li, S. Z., Lu, X. Y., et al. (2020). Genome mining and UHPLC-QTO-MS/MS to identify the potential antimicrobial compounds and determine the specificity of biosynthetic gene clusters in Bacillus subtilis NCD-2. BMC Genomics 21:767. doi: 10.1186/s12864-020-07160-2
Thistlethwaite, I. R. G., Bull, F. M., Cui, C. S., Walker, P. D., Gao, S. S., Wang, L. Y., et al. (2017). Elucidation of the relative and absolute stereochemistry of the kalimantacin/batumin antibiotics. Chem. Sci. 8, 6196–6201. doi: 10.1039/C7SC01670K
Tian, Z. H., Chen, C. W., Chen, K., Liu, P., Fan, Q. J., Zhao, J., et al. (2020). Biocontrol and the mechanisms of Bacillus sp. w176 against postharvest green mold in citrus. Postharvest Biol. Technol. 159:111022. doi: 10.1016/j.postharvbio.2019.111022
Vanewijk, P., and Hoekstra, J. (1993). Calculation of the EC50 and its confidence interval when subtoxic stimulus is present. Ecotoxicol. Environ. Saf. 25, 25–32. doi: 10.1006/eesa.1993.1003
Wallace, R. L., Hirkala, D. L., and Nelson, L. M. (2018). Mechanisms of action of three isolates of Pseudomonas fluorescens active against postharvest grey mold decay of apple during commercial storage. Biol. Control 117, 13–20.
Wang, D. D., Li, J. H., Zhu, G. L., Zhao, K., Jiang, W. W., Li, H. D., et al. (2020). Mechanism of the potential therapeutic candidate Bacillus subtilis BSXE-1601 against shrimp pathogenic Vibrios and Multifunctional metabolites biosynthetic capability of the strain as predicted by genome analysis. Front. Microbiol. 11:581802. doi: 10.3389/fmicb.2020.581802
Wang, F., Xiao, J., Zhang, Y. Z., Li, R. Y., Liu, L., and Deng, J. (2021). Biocontrol ability and action mechanism of Bacillus halotolerans against Botrytis cinerea causing grey mould in postharvest strawberry fruit. Postharvest Biol. Technol. 174:111456. doi: 10.1016/j.postharvbio.2020.111456
Wang, W., Deng, L., Yao, S., and Zeng, K. (2018). Control of green and blue mold and sour rot in citrus fruits by the cationic antimicrobial peptide PAF56. Postharvest Biol. Technol. 136, 132–138. doi: 10.1016/j.postharvbio.2017.10.015
Wei, Y., Zhao, Y., Zhou, D., Qi, D., Li, K., and Tang, W. (2020). A newly isolated Streptomyces sp.YYS-7 with a broad-spectrum antifungal activity improves the banana plant resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol 11:1712. doi: 10.3389/fmicb.2020.01712
Wisniewski, M., Droby, S., Norelli, J., Liu, J., Schena, L., Agraria, D., et al. (2016). Alternative management technologies for postharvest disease control: the journey from simplicity to complexity. Postharvest Biol. Technol. 122, 3–10. doi: 10.1016/j.postharvbio.2016.05.0120925-5214
Ye, W. Q., Sun, Y. F., Tang, Y. J., and Zhou, W. W. (2021). Biocontrol potential of a broad-spectrum antifungal strain Bacillus amyloliquefaciens B4 for postharvest loquat fruit storage. Postharvest Biol. Technol. 174:111439. doi: 10.1016/j.postharvbio.2020.111439
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., and Seo, H. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhang, D., Spadaro, D., Garibaldi, A., and Lodovica, M. (2011). Potential biocontrol activity of a strain of Pichia guilliermondii against grey mold of apples and its possible modes of action. Biol. Control 57, 193–201.
Zhang, Q. X., Zhang, Y., Shan, H. H., Tong, Y. H., Chen, X. J., and Liu, F. C. (2017). Isolation and identification of antifungal peptides from Bacillus amyloliquefaciens W10. Sci. Pollut. Res. 24, 25000–25009. doi: 10.1007/s11356-017-0179-8
Zhang, X. Y., Wu, F., Gu, N., Yan, X. L., Wang, K. L., Dhanasekaran, S., et al. (2020). Postharvest biological control of Rhizopus rot and the mechanisms involved in induced disease resistance of peaches by Pichia membranefaciens. Postharvest Biol. Technol. 163, 111–146. doi: 10.1016/j.postharvbio.2020.111146
Zhao, L., Zhou, Q., Yang, H., Zheng, H., Wen, X., and Sun, W. (2019). Inhibitory effect of Pichia guilliermondii Y35-1 against postharvest anthracnose infection in loquat fruit and its effect on quality preservation. Food Sci. 40, 170–177.
Zhimo, V. Y., Ajay, K., Antonio, B., Shoshana, S., Oleg, F., Mohamad, A. T., et al. (2021). Compositional shifts in the strawberry fruit microbiome in response to near-harvest application of Metschnikowia fructicola, a yeast biocontrol agent. Postharvest Biol. Technol. 175:111469. doi: 10.1016/j.postharvbio.2021.111469
Keywords: Bacillus safensis, Colletotrichum fragariae, biological control, antifungal properties, strawberry fruit, whole genome sequencing
Citation: Li X, Zhang M, Qi D, Zhou D, Qi C, Li C, Liu S, Xiang D, Zhang L, Xie J and Wang W (2021) Biocontrol Ability and Mechanism of a Broad-Spectrum Antifungal Strain Bacillus safensis sp. QN1NO-4 Against Strawberry Anthracnose Caused by Colletotrichum fragariae. Front. Microbiol. 12:735732. doi: 10.3389/fmicb.2021.735732
Received: 03 July 2021; Accepted: 18 August 2021;
Published: 17 September 2021.
Edited by:
Florence Fontaine, Université de Reims Champagne-Ardenne, FranceReviewed by:
Dewa Ngurah Suprapta, Udayana University, IndonesiaCopyright © 2021 Li, Zhang, Qi, Zhou, Qi, Li, Liu, Xiang, Zhang, Xie and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, d2FuZ3dlaXN5c0BhaGF1LmVkdS5jbg==; Jianghui Xie, eGllamlhbmdodWlAaXRiYi5vcmcuY24=; Lu Zhang, bHV6aGFuZ3Rlc3RAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.