
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 October 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.731425
 Haijiang Chen1,2,3†
Haijiang Chen1,2,3† Lijuan Mao4†
Lijuan Mao4† Nan Zhao2†
Nan Zhao2† Chenyang Xia2
Chenyang Xia2 Jian Liu3
Jian Liu3 Christian P. Kubicek5*
Christian P. Kubicek5* Wenneng Wu1
Wenneng Wu1 Su Xu1
Su Xu1 Chulong Zhang2*
Chulong Zhang2*Trichodermin, a trichothecene first isolated in Trichoderma species, is a sesquiterpenoid antibiotic that exhibits significant inhibitory activity to the growth of many pathogenic fungi such as Candida albicans, Rhizoctonia solani, and Botrytis cinerea by inhibiting the peptidyl transferase involved in eukaryotic protein synthesis. Trichodermin has also been shown to selectively induce cell apoptosis in several cancer cell lines and thus can act as a potential lead compound for developing anticancer therapeutics. The biosynthetic pathway of trichodermin in Trichoderma has been identified, and most of the involved genes have been functionally characterized. An exception is TRI3, which encodes a putative acetyltransferase. Here, we report the identification of a gene cluster that contains seven genes expectedly involved in trichodermin biosynthesis (TRI3, TRI4, TRI6, TRI10, TRI11, TRI12, and TRI14) in the trichodermin-producing endophytic fungus Trichoderma taxi. As in Trichoderma brevicompactum, TRI5 is not included in the cluster. Functional analysis provides evidence that TRI3 acetylates trichodermol, the immediate precursor, to trichodermin. Disruption of TRI3 gene eliminated the inhibition to R. solani by T. taxi culture filtrates and significantly reduced the production of trichodermin but not of trichodermol. Both the inhibitory activity and the trichodermin production were restored when native TRI3 gene was reintroduced into the disruption mutant. Furthermore, a His-tag-purified TRI3 protein, expressed in Escherichia coli, was able to convert trichodermol to trichodermin in the presence of acetyl-CoA. The disruption of TRI3 also resulted in lowered expression of both the upstream biosynthesis TRI genes and the regulator genes. Our data demonstrate that T. taxi TRI3 encodes an acetyltransferase that catalyzes the esterification of the C-4 oxygen atom on trichodermol and thus plays an essential role in trichodermin biosynthesis in this fungus.
Trichothecenes are a group of naturally occurring sesquiterpenoids capable of inhibiting the function of eukaryotic ribosomes, thereby causing hemorrhagic lesions, dermatitis, and immunological problems to people and animals (Pace et al., 1998; McLaughlin et al., 2009; McCormick et al., 2011). All trichothecenes contain a core tricyclic 12,13-epoxytrichothec-9-ene (EPT) structure with an epoxide substituent and could be divided into four different types according to the pattern of esterification and oxygenation (Ueno, 1984; Ueno and Hsieh, 1985; Cole et al., 2003). Trichodermin belongs to the type that lacks the oxygen substitution at C-8 while containing a hydroxyl residue at C-4 and is renowned as a very potent inhibitor of protein synthesis in eukaryotic cells (Carrasco et al., 1973; Cundliffe and Davies, 1977; Kubicek and Druzhinina, 2015). It primarily interferes in the chain-termination process on the ribosome by blocking the interaction of the peptidyl transferase with the peptide release factor (Wei et al., 1974). Trichodermin was first isolated from a Trichoderma species as an antifungal antibiotic that is active against a variety of pathogenic fungi including Candida albicans (Godtfredsen and Vangedal, 1965). This sesquiterpenoid metabolite is known to be produced by only two Trichoderma species (Trichoderma brevicompactum and Trichoderma arundinaceum; Kubicek and Druzhinina, 2015; Nielsen et al., 2005) and exhibits significant inhibition to several human pathogenic Candida spp. and a nosocomial strain of the filamentous fungus Aspergillus fumigatus (Tijerino et al., 2011). The ability of these Trichoderma species to produce trichodermin, to secrete extracellular hydrolytic enzymes, and to directly interact with plants has made them effective biocontrol agents against plant pathogens like Rhizoctonia solani, Botrytis cinerea, and some soil pathogens (e.g., Fusarium spp.; Howell, 2003; Kulikov et al., 2006; Malmierca et al., 2012). A silkworm infection assay demonstrated that trichodermin yielded therapeutic effects on silkworms infected by C. albicans (Uchida et al., 2016). Recently, trichodermin has drawn attentions from cancer biologists because it can induce cell apoptosis through mitochondrial dysfunction and endoplasmic reticulum stress in human chondrosarcoma cells and by mitotic arrest and DNA damage in human p53-mutated pancreatic cancer cells (Su et al., 2013; Chien et al., 2016). These new findings suggest that trichodermin is a potential therapeutic agent worthy of further development to a clinical trial candidate for treating cancer. To this end, a thorough understanding of the biosynthetic pathway leading to trichodermin is essential.
The study of trichothecene biosynthesis has been pioneered in Fusarium spp. In Fusarium, most of TRI genes are located in the so-called TRI5 cluster, which consists of TRI3, TRI4, TRI5, TRI6, TRI8, TRI9, TRI10, TRI11, TRI12 and TRI13 (Brown et al., 2004). Three other genes occur at separate distant positions: TRI101 and TRI1–TRI16 cluster, which are physically close (Kimura et al., 2007). Thereby, TRI6 and TRI10 genes encode transcriptional regulators (Tag et al., 2001; Kimura et al., 2007; Nasmith et al., 2011), and TRI12 encodes a transport protein for export of trichothecenes (Alexander et al., 1999), while the remaining genes in TRI5 cluster encode enzymes involved in trichothecene biosynthesis (Alexander et al., 2004; Kimura et al., 2007).
Trichothecene biosynthesis in Trichoderma shares several features with that in Fusarium yet exhibits some distinct differences in gene clustering, gene arrangement, and gene functions (Malmierca et al., 2013). For example, orientation of part of TRI genes is opposite to that in Fusarium (Cardoza et al., 2011), and TRI5 gene—which encodes a trichodiene synthase that in T. arundinaceum coverts farnesyl pyrophosphate (FPP) into trichodiene, consistent with the function of FsTRI5 in trichothecenes biosynthesis (Hohn and Middlesworth, 1986)—is not present in the TRI cluster (Cardoza et al., 2011). In addition, some enzymes encoded by the TRI cluster show several catalytic differences: the cytochrome P450 monooxygenase TaTRI4 exhibits a different pattern of oxidation: in T. arundinaceum, TaTRI4 introduces oxygen into trichodiene at positions C-2, C-11, and C-13 (Cardoza et al., 2011), whereas the Fusarium orthologue TRI4 further targets position C-3 (McCormick et al., 2006; Tokai et al., 2007); TaTRI11 of T. arundinaceum functions as a hydroxylase, which catalyzes the hydroxylation of EPT at the C-4 position in contrast to that of the C-15 position in Fusarium sporotrichioides (Alexander et al., 1998). TRI3 of Fusarium is responsible for acetylation of hydroxyl group at C-15 in T-2 toxin biosynthesis (McCormick et al., 1996; Cardoza et al., 2011), while the role of TRI3 in Trichoderma has not been characterized until recently. TRI3 had been proposed to take part in the acetylation of C-4 hydroxyl on trichodermol in Trichoderma (Cardoza et al., 2011) and was evidenced to be responsible for trichodermin biosynthesis in an endophytic T. brevicompactum by mutational analysis (Shentu et al., 2018). Yet evidence showing direct participation of TRI3 in trichodermol biotransformation is still lacking, and study of trichodermin biosynthetic pathway in other Trichoderma species rather than T. arundinaceum and T. brevicompactum has not been reported.
We have previously identified a novel trichodermin-producing endophytic fungus from Chinese yew Taxus mairei, Trichoderma taxi ZJUF0986 (Zhang et al., 2007). It was then shown to inhibit the growth of several fungal plant pathogens, including (but not limited to) B. cinerea and R. solani (Zhang et al., 2007; Chen et al., 2008; Shentu et al., 2013). The objective of this study was thus to identify the core genes responsible for the synthesis of trichodermin in the species and—more importantly—to investigate the role of TRI3 in producing trichodermin in Trichoderma. The results reveal that TRI clusters in T. taxi are significantly similar to those in T. arundinaceum and T. brevicompactum and that TtTRI3 encodes an acetyltransferase that performs the step by acetylating trichodermol at C-4 to trichodermin.
Endophytic T. taxi ZJUF0986 was isolated from T. mairei (Zhang et al., 2007) and is available in the China Center for Type Culture Collection under CGMCC No. 1672. R. solani (ACCC 3614) was used as a test pathogen as described previously (Wang et al., 2012). T. taxi and its derivatives and R. solani were maintained on potato dextrose agar (PDA) plates at 25°C in the dark. Escherichia coli DH5α was used for regular cloning and plasmid maintenance. E. coli M15 (Qiagen, Hilden, Germany) was adopted as protein expression host.
The genome of ZJUF0986 was obtained by sequencing (manuscript in preparation). Nucleotide and amino acid sequences of TRI genes from T. arundinaceum and T. brevicompactum were retrieved from National Center for Biotechnology Information (NCBI). Initially, TaTRI3 gene was used for BLASTn search against ZJUF0986 genome to identify the contig containing the TRI3 orthologue in ZJUF0986. The contig was then used as a query to search for other TRI genes by BLASTx.1 The organization of the encoded open reading frames (ORFs) was illustrated by pDRAW32.2 Alignment of the amino sequences of all the putative T. taxi TRI proteins with those from T. arundinaceum and T. brevicompactum was performed by MEGA5.0 (Tamura et al., 2011) and then reviewed by Genedoc (Nicholas et al., 1997). All TRI amino sequences were subjected to protein domain analysis by Pfam3 (Finn et al., 2014).
The gene deletion vector was constructed based on the double-joint PCR strategy (Yu et al., 2004). Briefly, approximate 1 kb upstream and 1 kb downstream sequences flanking TtTRI3 locus were amplified with the primers TRI3-upF/R and TRI3-downF/R, respectively. A 1.4-kb hygromycin B resistance (HPH) cassette was cloned from pCB1003 (Carrol et al., 1994) with primers HPH-F/R. The three fragments were mixed together in the second-round PCR, the product of which acted as template for the final amplification with nested primers TRI3-NestF/R. The double-jointed PCR product was purified and subcloned into the commercial TA-cloning vector pMD18-T (TaKaRa, Dalian, China). The resulting plasmid and the Agrobacterium binary vector for plant transformation, pCAMBIA1300 (GenBank: AF234296; Hajdukiewicz et al., 1994), were both cut with HindIII/XbaI and then joined together by T4 DNA ligase (TaKaRa), leading to the final TtTRI3 gene knockout vector, pCAMBIA-TRI3. To obtain the TtTRI3 null mutants, pCAMBIA-TRI3 was transformed into the wild strain using Agrobacterium tumefaciens-mediated transformation (ATMT) (Rho et al., 2001). The transformants with resistance to hygromycin B (40 μg/ml) were selected and screened by PCR with the primers ATMT-F/R and ATMT2-F/R to verify deletion by replacement. To complement the deletion strain, a fragment containing TtTRI3 gene locus with its native promoter and terminator region was amplified by primers TRI3comp-F/R and ligated into a modified pCAMBIA1300 vector in which the original hygromycin B resistance gene was replaced with a G418-resistant gene. The final complementation vector was introduced in the mutant by ATMT and randomly inserted into the genome.
Vegetative mycelia were harvested from 5-day-old culture in a 250-ml flask containing 100 ml of liquid potato dextrose broth (PDB) incubated at 25°C with shaking (200 rpm). Genomic DNA and total RNA were extracted by the cetyl trimethylammonium bromide (CTAB) method (Del Sal et al., 1989) and the TRIzol reagent (TaKaRa), respectively. qRT-PCR was performed using SYBR Premix ExTaq (TaKaRa) on a Mastercycler Ep realplex thermo cycler (Eppendorf, Hamburg, Germany). Actin gene was chosen as the normalizing gene, and the expression levels of TRI genes were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001). Each reaction was repeated three times independently.
The wild-type ZJUF0986 and the mutant ΔTtTRI3 were cultured in 150 ml of PDB medium in 500-ml Erlenmeyer flasks at 25°C in the dark. After 7 days’ incubation with shaking (200 rpm), the fermentation broth was filtered through three layers of lens paper. The filtered medium was extracted three times with equal volumes of ethyl acetate. The extract was dried by evaporation and dissolved in methanol and then analyzed by gas chromatography (GC)–MS as described by Cardoza et al. (2011).
The fermentation broth of T. taxi strains was harvested from 7-day-old cultures grown at conditions described above. R. solani was incubated on PDA containing 10 or 20% (v/v) harvested fermentation broth, using PDA containing 10 or 20% of non-inoculated fermentation broth as a reference set. Growth of R. solani on plates was monitored and recorded after 3 days’ incubation.
Reverse transcription and subsequent PCR was carried out with the PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa) and the primers TRI3cDNA-F/R. Purified PCR product was double-digested by BamHI and HindIII and ligated into pQE30 (Qiagen) (Figure 6A). The resulted plasmid pQE30-TRI3 was sequencing verified and introduced into E. coli M15 (Qiagen), leading to strain M15-TRI3. Culture of M15-TRI3 was initiated and allowed to grow to OD600 at 0.6 with aeration at 37°C; 1 mM of IPTG was then added, and the culture was incubated overnight at 25°C with shaking. Cells were harvested by centrifugation at 10,000 g for 5 min at 4°C, re-suspended in buffer (pH 7.5) containing 50 mM of Tris, 200 mM of NaCl, 1 mM of β-mercaptoethanol, and 5 mM of imidazole. The cell suspension was subjected to sonication for a total of 15 min on ice, with 40% duty cycle (2-s working and 3-s pause). The cell lysate was clarified at 20,000 g, and cell debris was discarded. The His-tagged TtTRI3 protein in the lysate was enriched using Ni-NTA beads (Invitrogen, Carlsbad, CA, United States) as recommended in the suppliers’ manual and concentrated and buffer-changed to 0.1 M of potassium phosphate buffer (pH 8.0) by centrifugation using Amicon® Ultra centrifugal filters (Millipore, Billerica, MA, United States). Protein concentration was determined by the NanoDropTM 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
The activity of the acetyltransferase assay was performed according to Garvey et al. (2009) with slight modifications. Briefly, reaction mixtures were prepared at room temperature by combining 150 μl of 1.5 mM acetyl-CoA (Sigma-Aldrich Corp., St. Louis, MO, United States) with 50 μl of 4 mM trichodermol and 2 μl of 5 mg/ml bovine serum albumin (BSA). All components were dissolved in 0.1 M of potassium phosphate buffer (pH 8.0). The reaction was initiated by the addition of 10 μl of 290 μg/ml enriched enzyme described above. After incubation at 25°C for 5 h, the reaction mixture was extracted with an equal volume of ethyl acetate, and the extract was subjected to GC-MS analysis to verify the formation of trichodermin.
All sequences of the primers used in the study are listed in Table 1. The nucleotide sequence of the contig containing TRI genes was directly submitted to GenBank under the accession number KY670722. The coding sequences of the individual TRI genes were also deposited with the following assigned accession numbers: KY860616 (TtTRI3), KY860617 (TtTRI4), KY860618 (TtTRI6), KY860619 (TtTRI10), KY860620 (TtTRI11), KY860621 (TtTRI12), and KY860622 (TtTRI14).
In order to understand the underlying biosynthetic pathway of trichodermin in T. taxi, we made use of the recently sequenced genome of T. taxi (manuscript in preparation). With the use of T. arundinaceum TRI3 gene as a query to search against the genome, BLASTn identified the TtTRI3-containing contig as a 42.5-kb-long assembly (Figure 1). Further BLASTp search found that the contig encodes several other TRI orthologues, namely, TtTRI4, TtTRI6, TtTRI10, TtTRI11, TtTRI12, and TtTRI14, and that they were located in immediate vicinity on the contig. Along with TtTRI3, they appeared in the same order and orientation as those in T. arundinaceum and T. brevicompactum (Figure 1). Notably, the size of the genomic region between TRI11 and TRI12 in T. taxi is more like in T. arundinaceum than in T. brevicompactum, where the size is much larger (Figure 1). Consistent with previous observation in Trichoderma (Cardoza et al., 2011), TRI5 is absent from the core TRI cluster in T. taxi. It is noteworthy that we are unable to recover the complete TtTRI5 gene sequence from the contigs since querying TaTRI5 against the assembled genome resulted in three separated short contigs (data not shown).
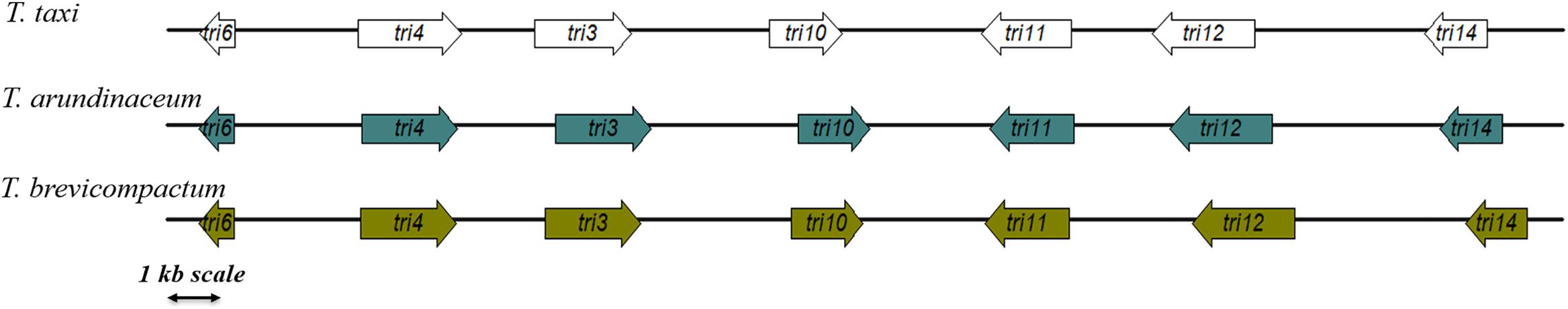
Figure 1. The TRI gene cluster in Trichoderma taxi ZJUF0986, Trichoderma brevicompactum, and Trichoderma arundinaceum. The orientations of arrows represented the direction of transcription, and the relative position of genes was proportionally scaled.
As expectable from the close phylogenetic relationship between T. taxi and T. brevicompactum/T. arundinaceum (Jaklitsch and Voglmayr, 2015), the proteins encoded by the TRI cluster display a high degree of identity, suggesting that they are structurally and functionally conserved (Table 2). Results from protein domain search identified TtTRI3 as a TRI3 family 15-O-acetyltransferase, TtTRI4 and TtTRI11 as cytochrome P450 domain-containing oxygenases, TtTRI12 as a fungal trichothecene efflux pump, TtTRI6 as a C2H2-type zinc finger domain-containing transcriptional regulator, and TtTRI10 likely to be a transcription factor. No match in protein domain database was found for TtTRI14. Though orthologues of TRI14 have been found in other Hypocreales, such as Fusarium spp. (Peplow et al., 2003), Stachybotrys spp., and Cordyceps spp., the function of the protein is currently unknown.
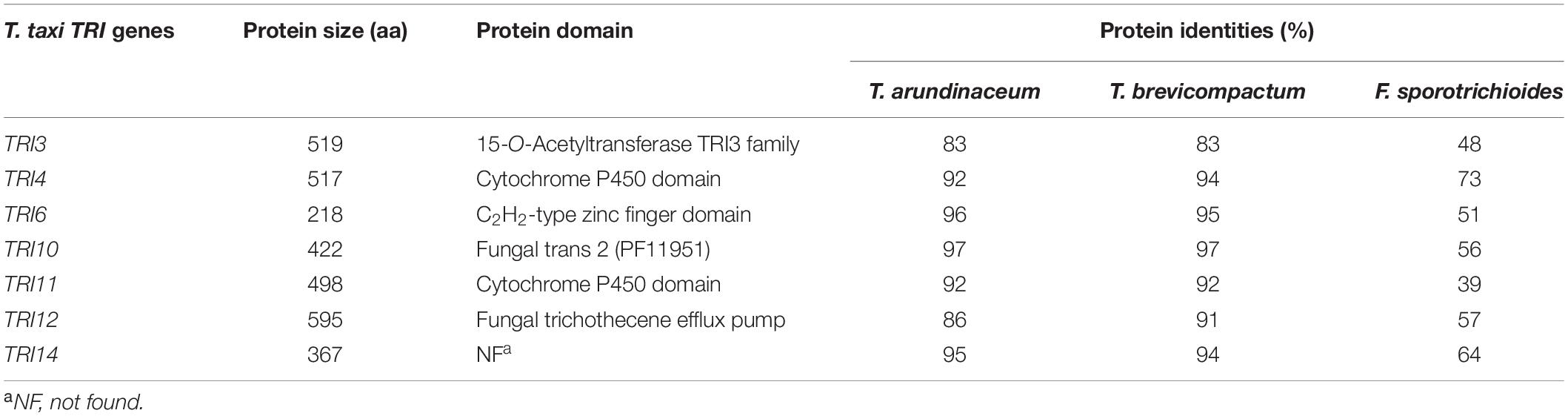
Table 2. Comparison of TRI genes from Trichoderma taxi with orthologues from Trichoderma arundinaceum, Trichoderma brevicompactum, and Fusarium sporotrichioides.
TRI3 had not been characterized in Trichoderma spp. when we initiated this study. It is highly conserved in the genus Trichoderma and other Sordariomycetes, but the distance (i.e., branch length) between Trichoderma TRI3 and other fungi is high, suggesting potential differences in function (Figure 2). In Fusarium, TRI3 catalyzes the acetylation of the hydroxy group at C-15 of the trichothecene skeleton (McCormick et al., 1996). In T. brevicompactum, TRI3 was proposed to be responsible for the acetylation of the hydroxy group at C-4 of the trichothecene skeleton (Cardoza et al., 2011), but this hypothesis has not been experimentally tested yet. We therefore generated a TtTRI3 null mutant by replacing the TRI3 ORF by a 1.4-kb HPH cassette. Successful gene replacement was confirmed by PCR screening (Figure 3).
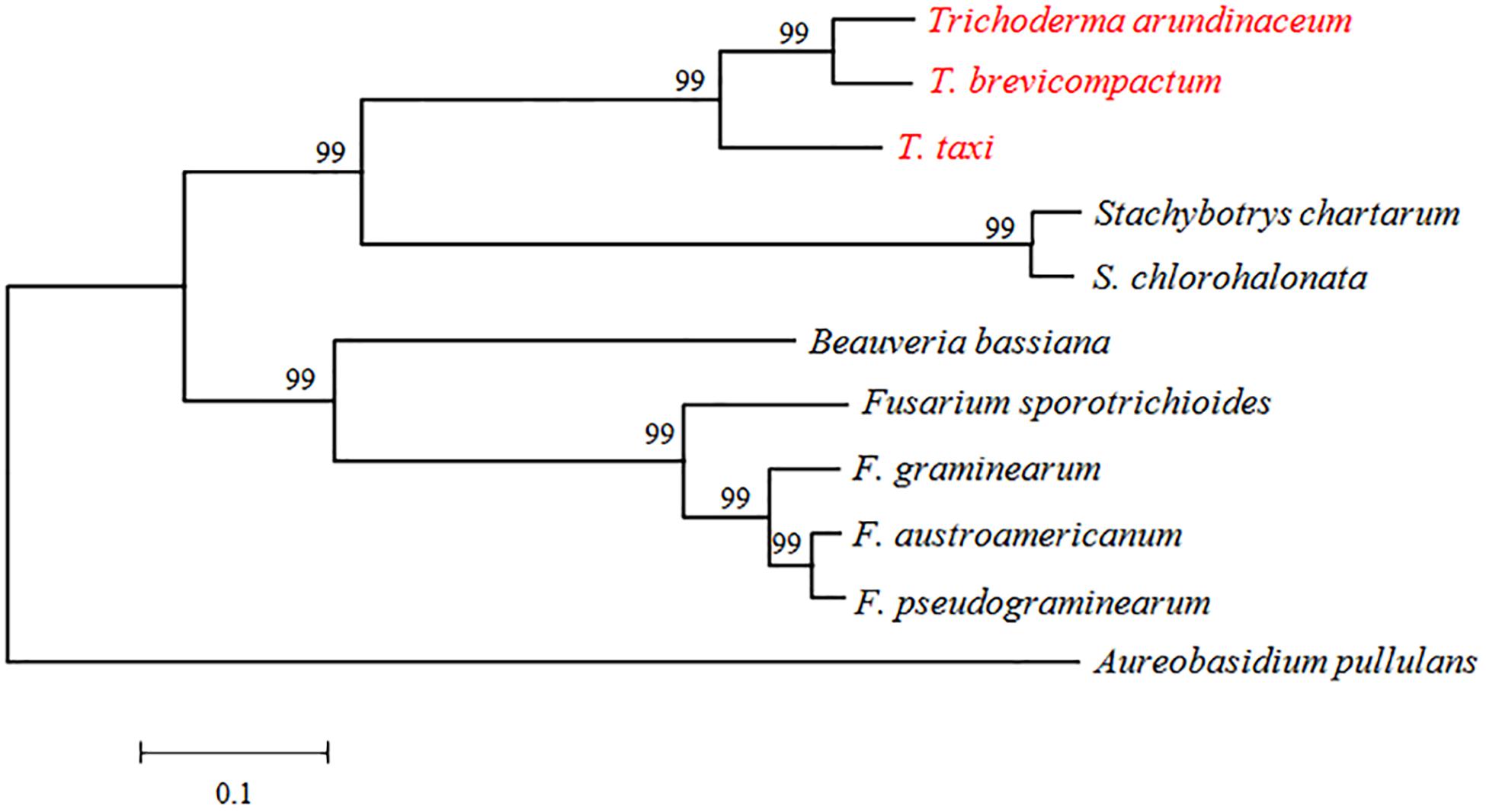
Figure 2. Phylogenetic tree based on alignment of TRI3 from Trichoderma, Stachybotrys, Beauveria, Fusarium, and Aureobasidium. The species were chosen as representatives of hits from a BLASTp search. Sequence alignment and tree building were performed by MEGA5.0 using the neighbor-joining method, and phylogeny was tested by 500 bootstrap replications. The numbers on branches were calculated bootstrap values. The accession numbers for TRI3 genes were as follows: TaTRI3 (CAY87361), TbTRI3 (CBY93780), Stachybotrys chartarum TRI3 (KEY64037), Stachybotrys chlorohalonata TRI3 (KFA68878), FgTRI3 (BAC22114), FpTRI3 (XP_009263555), FaTRI3 (AAM48923), FsTRI3 (AAK33072), BbTRI3 (XP_008602023), and ApTRI3 (KEQ86789).
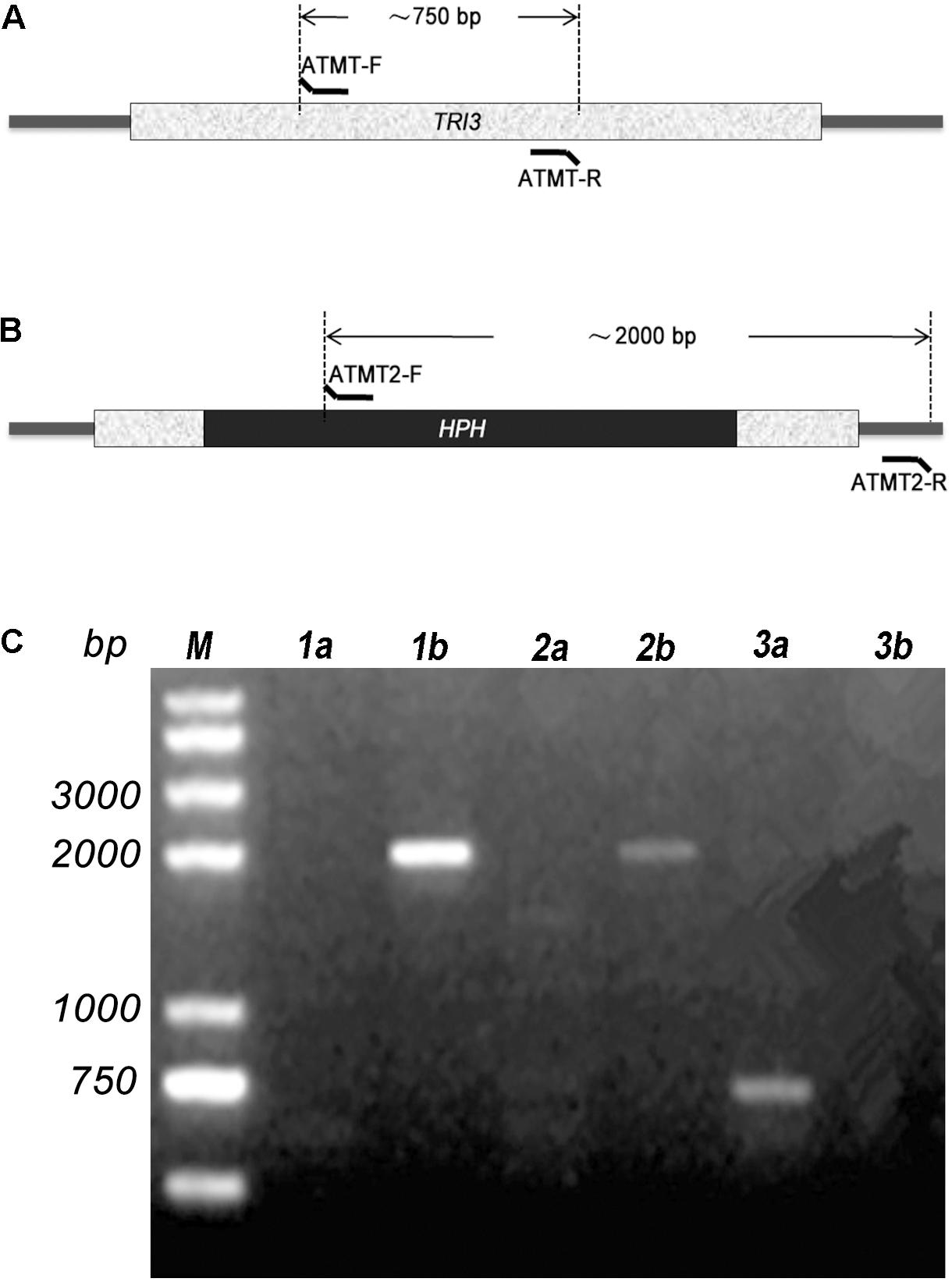
Figure 3. PCR confirmation of insertion of the HPH cassette in TRI3 gene. The relative positions of primers ATMT-F/R (A) and ATMT2-F/R (B) are shown. (C) Two transformants were confirmed by PCR. Primer pair ATMT-F/R was used for lane 1a, 2a, and 3a; while ATMT2-F/R was used for lane 1b, 2b, and 3b. 1a, 1b and 2a, 2b were designated for the two transformants. Wild-type strain was used in 3a and 3b.
There were no significant phenotypic changes observed in colonial morphology and growth rate between wild type and ΔTtTRI3 (data not shown). However, in contrast to the wild type, ΔTtTRI3 lost the ability to secrete molecules that inhibited growth of R. solani (Figure 4): when 10% of the filtered fermentation broth from the parent strain was added to a PDA plate, growth of R. solani on the plate was strongly inhibited, and complete inhibition was observed by addition of 20%. In contrast, even adding 20% of the fermentation broth of the knockout strain ΔTtTRI3 to the plate did not impair growth of R. solani. This suggests that TRI3 catalyzes an essential step in the generation of the compound that inhibits R. solani growth.
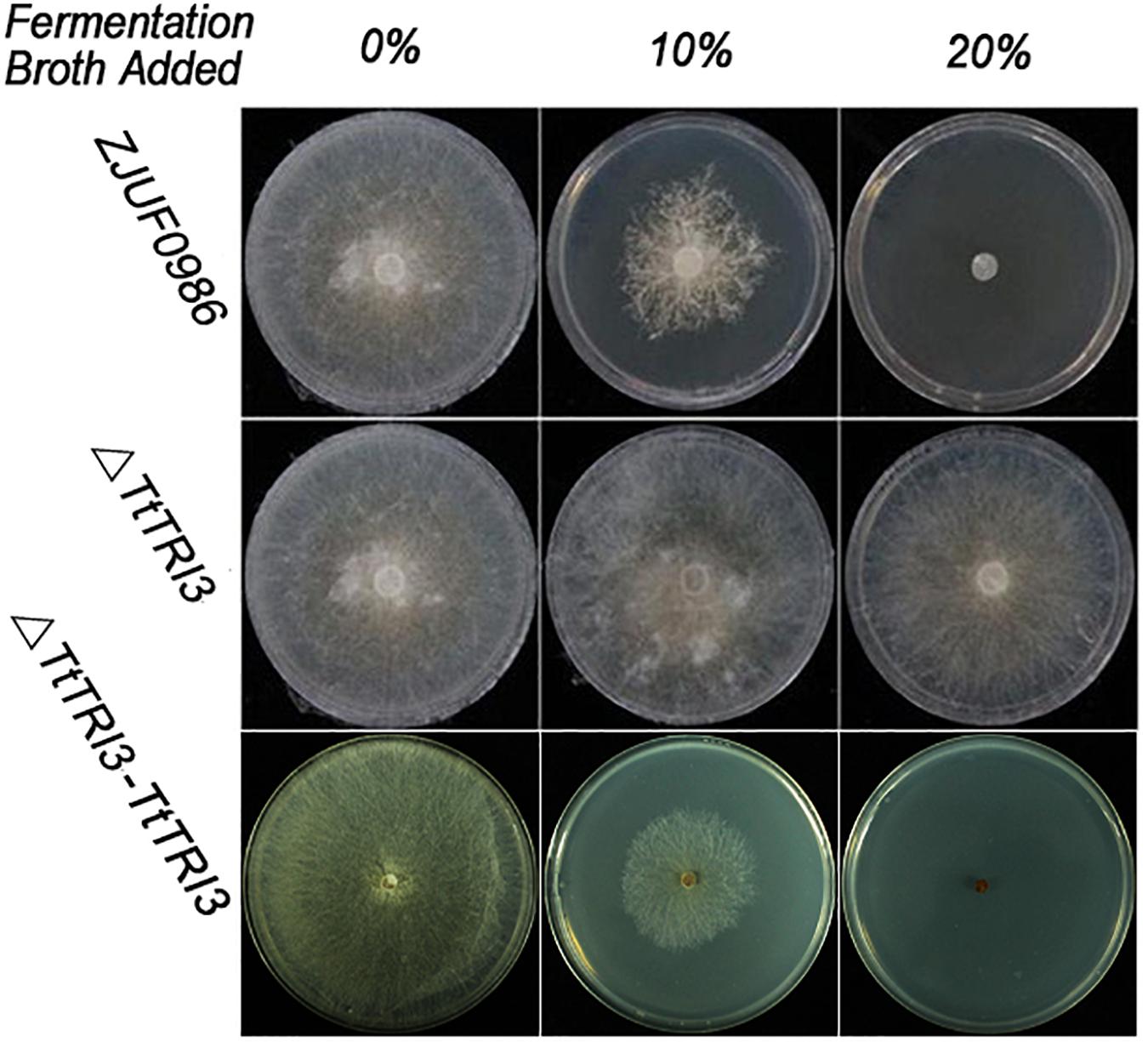
Figure 4. Disruption of TRI3 relieved growth inhibition to Rhizoctonia solani. From left to right: 0% (v/v), 10% (v/v), or 20% (v/v) of harvested fermentation broth from wild-type strain or TtTRI3 disruption strain, or complementation strain was added in potato dextrose agar (PDA). Photos were taken 3 days after incubation.
To test whether deletion of TtTRI3 indeed affects the production of trichodermin, the fermentation broth from both the wild-type culture and the ΔTtTRI3 culture was subject to GC-MS analysis. This showed that the production of trichodermin was significantly decreased in the TtTRI3 strains (Figure 5A). Interestingly, the concentration of trichodermol, the immediate trichodermin precursor, did not accumulate to higher levels in the ΔTtTRI3 strain and remained at the same level as in the wild-type strain (Figure 5B), suggesting the operation of an effective feedback control in its biosynthesis. These data demonstrate that TRI3 plays an essential role in the conversion of trichodermol to trichodermin and that trichodermin confers T. taxi most (if not all) of its ability to inhibit growth of R. solani.
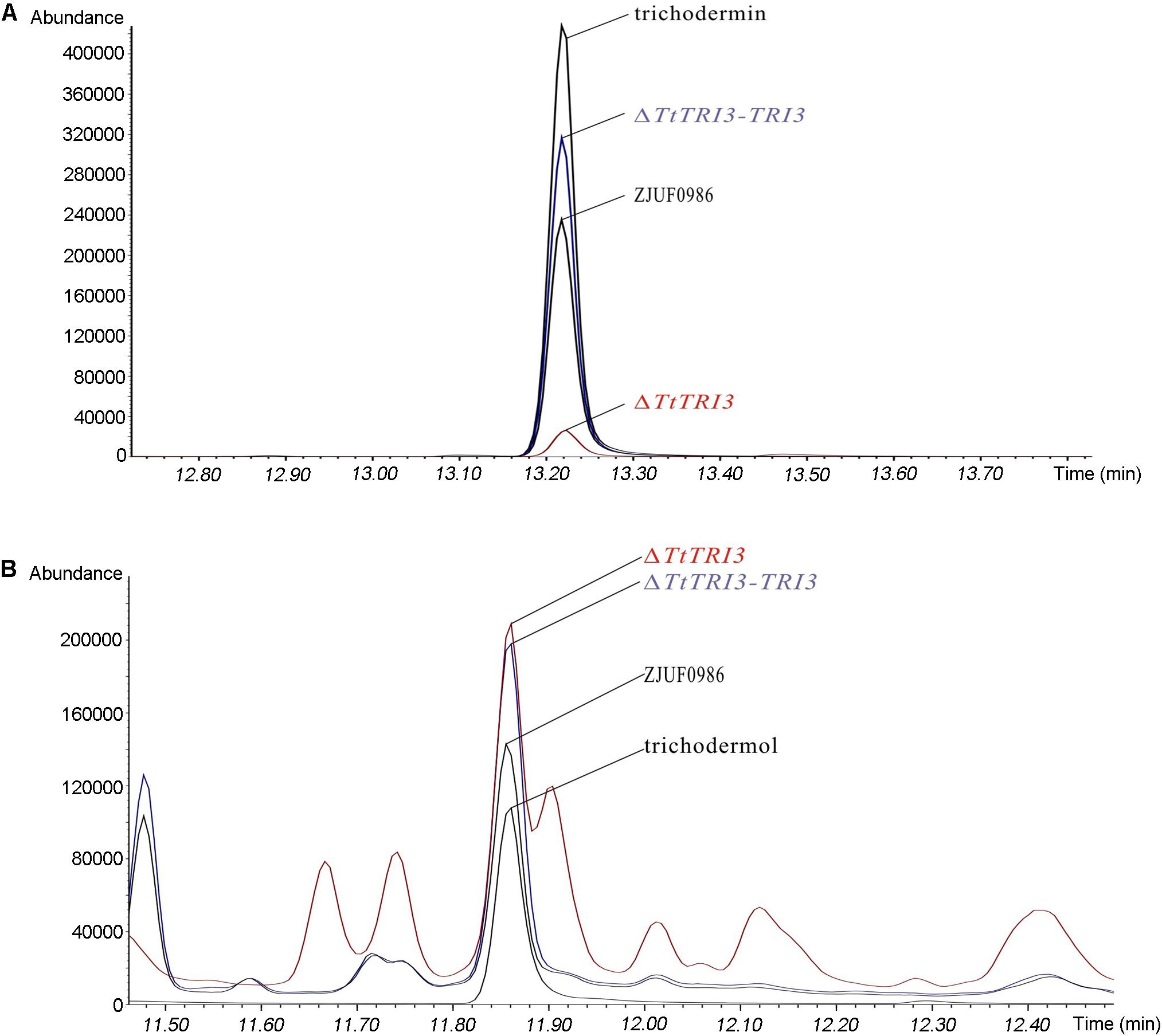
Figure 5. Trichodermin and trichodermol production in strain ZJUF0986, ΔTtTRI3, and ΔTtTRI3-TRI3. Amount of trichodermin (A) and trichodermol (B) in fermentation broth was detected by gas chromatography (GC)–MS. Peaks for trichodermin and trichodermol were determined by the corresponding reference chemicals. The concentration of reference chemicals in the prepared samples was 188 ppm (part per million) for trichodermin and 42 ppm for trichodermol.
To further confirm that TRI3 is responsible for converting trichodermol to trichodermin in T. taxi and that trichodermin is the main component conferring inhibitory effect to R. solani, wild-type TtTRI3 gene with its native promoter and terminator was introduced into the ΔTtTRI3 mutant, resulting in the complementation strain ΔTtTRI3–TRI3.
The TRI3 complementation restored the knockout strain’s ability to inhibit growth of R. solani to that of the wild-type level; i.e., 10% of added fermentation broth largely inhibited growth of R. solani and 20% resulted in complete inhibition (Figure 4). In addition, GC-MS analysis of the fermentation broth from ΔTtTRI3–TRI3 showed that trichodermin biosynthesis was recovered and that its abundance in the complementation strain was even higher than that in the wild-type strain (Figure 5A). These data confirmed that the results obtained with the TtTRI3 knockout strain are valid, and TRI3 plays an essential role in converting trichodermol into trichodermin in T. taxi.
To examine the potential conversion of trichodermol to trichodermin by TtTRI3 also in vitro, TtTRI3 was recombinantly produced with a His-tag in E. coli and purified to >70% homogeneity (estimated by ImageJ: https://cnij.imjoy.io/) by Ni-NTA resin batch treatment (Figure 6B). A biochemical assay combining trichodermol, TtTRI3, and acetyl-CoA was established to test if this enriched TtTRI3 was able to catalyze acetylation of trichodermol in vitro. GC-MS analysis of the ethyl acetate extract of the reaction mixture showed that a new peak appeared at the position of trichodermin, while the peak was missing when the cell lysate was prepared from E. coli cells where the empty vector was introduced instead (Figure 6C). Exclusion of acetyl-CoA or the recombinantly produced TtTRI3 protein from the mixture also prevented the formation of trichodermin (data not shown), thus proving that TRI3 and acetyl-CoA are required for the reaction to happen.
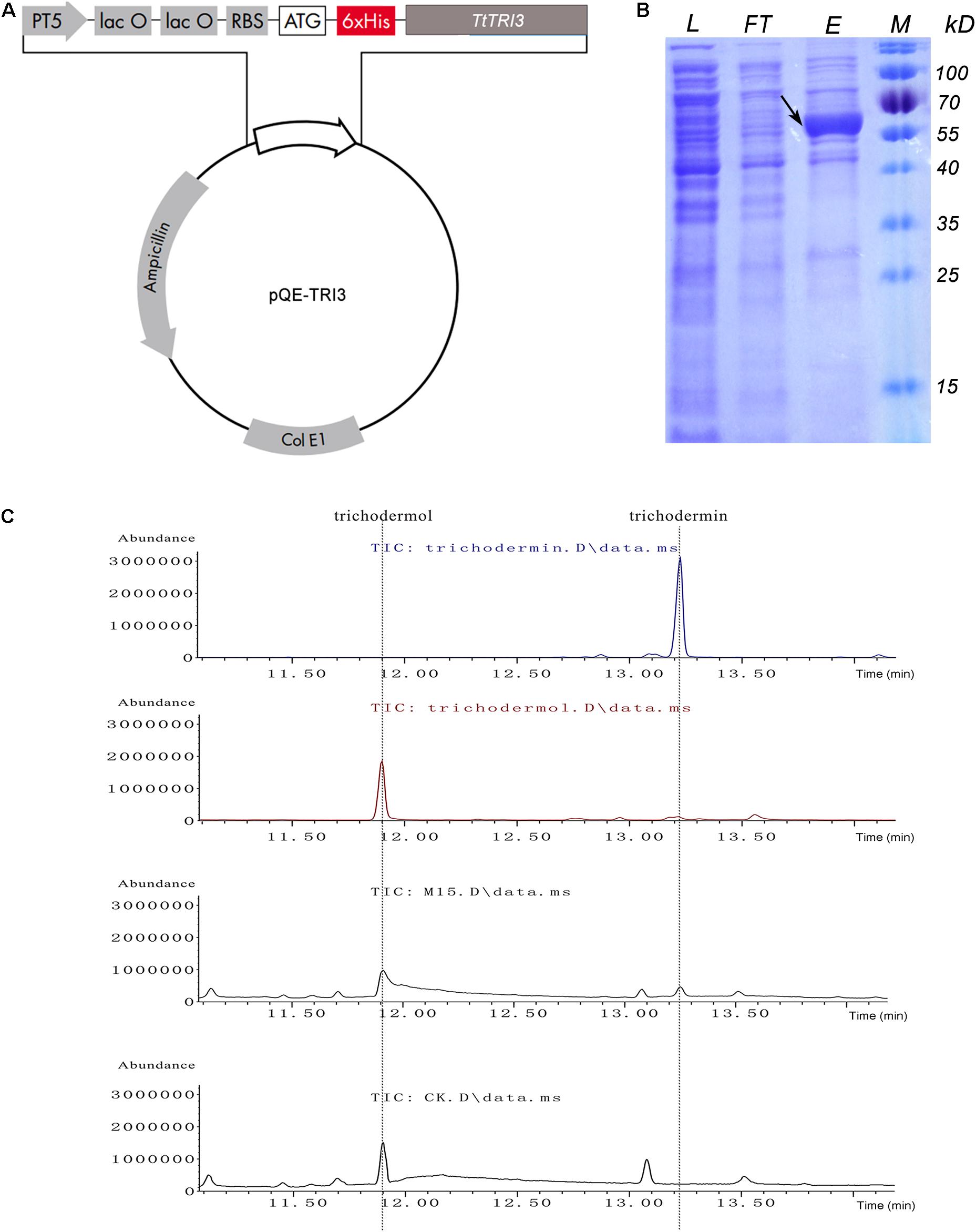
Figure 6. In vitro conversion of trichodermol to trichodermin catalyzed by bacterial expressed TtTRI3. (A) The structure of the constructed plasmid for expression of TtTRI3. The 6xHis tag was engineered to fuse immediately upstream of the TtTRI3 coding region. (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) shows the purification of Escherichia coli-expressed TtTRI3. The black arrow indicates the band of the target protein. The molecular weight (MW) of this band on the gel is consistent with the calculated theoretical MW, 60 kDa. L: cell lysate; FT: flowthrough from Ni-NTA column after cell lysate was loaded; E: eluate from TtTRI3-bound column; M: protein marker. (C) GC-MS analysis results showed that addition of TtTRI3 resulted in the appearance of trichodermin in reaction mixture. The upper two curves show the respective retention time of trichodermin and trichodermol. The two curves at the bottom represent curves obtained from ethyl acetate extract from trichodermol reaction mixture, with combination of either TtTRI3 (sample M15) or cell lysate prepared from Escherichia coli cell bearing empty vector pQE30 (sample CK).
We have noted above that the deletion of TtTRI3 did not result in significant over-accumulation of trichodermol, suggesting a tight feedback control. To test whether this would involve regulation of gene expression, we therefore quantified the expression level of TtTRI4, TtTRI6, TtTRI10, TtTRI11, and TtTRI12 in the parent strain ZJU0986 and its knockout mutant ΔTtTRI3. As illustrated in Figure 7, we indeed found that the expression of all structural genes was decreased in the mutant by more than 50%, whereas the expression of the regulator genes TtTRI10 was unaffected and that of TtTRI6 reduced by only 30%. These data suggest the operation of a feedback loop in the trichodermin biosynthesis pathway in T. taxi.
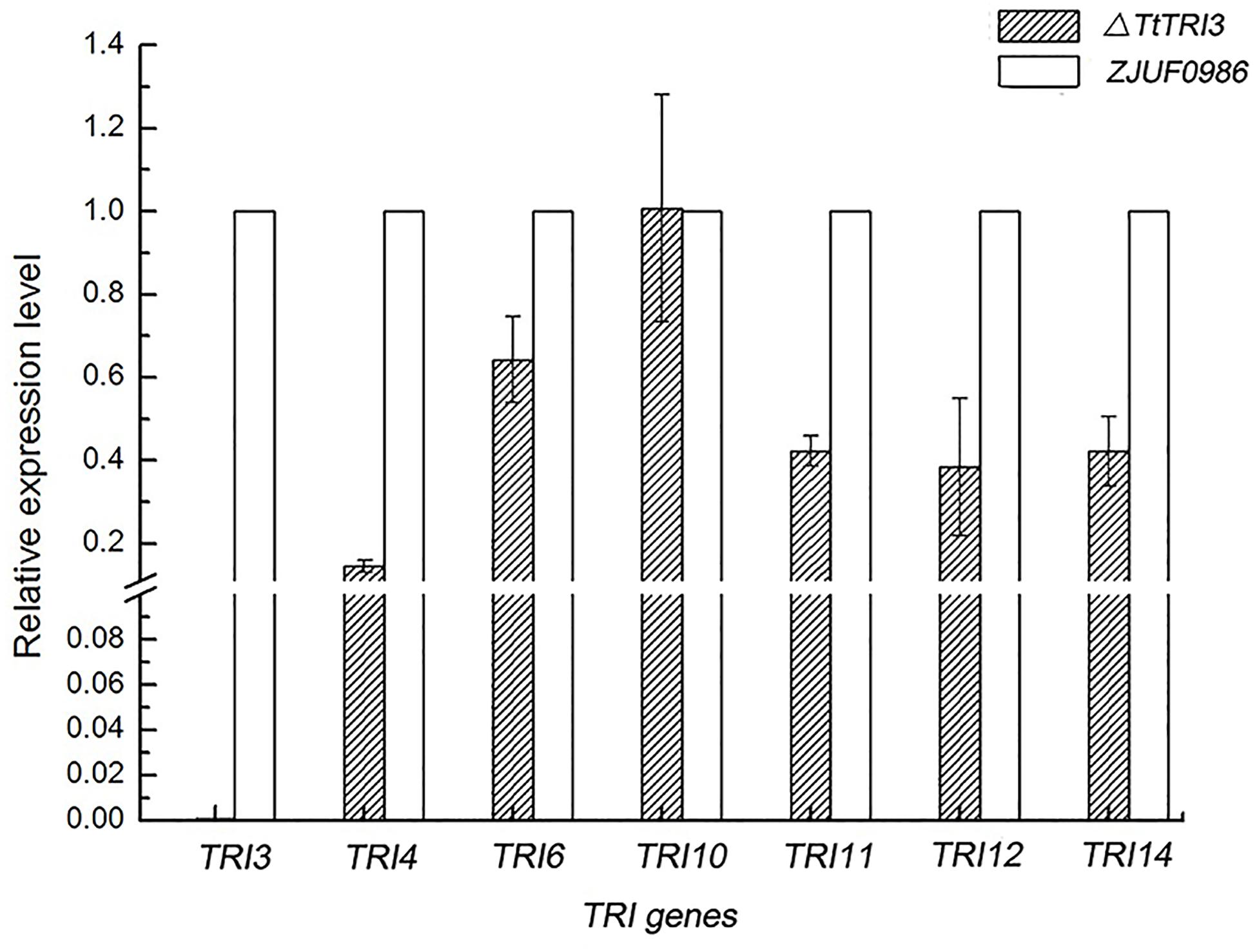
Figure 7. Comparison of TRI genes expression level between wild-type strain and ΔTtTRI3. Expression level for TRI genes was determined by qRT-PCR. Expression level of genes in ZJUF0986 was normalized as 1. Calculation was based on data obtained from three independent experiments.
Trichodermin can be produced by T. taxi, T. brevicompactum, and T. arundinaceum (Nielsen et al., 2005; Zhang et al., 2007). Since its discovery, this fungal secondary metabolite has been shown to have several important bioactivities such as growth inhibition to fungal pathogens, cytotoxicity, and recently anticancer activity (Godtfredsen and Vangedal, 1965; Wei et al., 1974; Su et al., 2013; Chien et al., 2016; Uchida et al., 2016). Although chemical synthetic routes for trichodermin had been developed not long after its discovery (Colvin et al., 1973), it was not until recently that the biosynthetic pathway was elucidated (Cardoza et al., 2011). Like other trichothecenes, biosynthesis of trichodermin starts from the cyclization of FPP to form trichodiene by the TRI5 trichodiene synthase. TRI4 then oxidizes trichodiene to isotrichodiol, which subsequently forms EPT by non-enzymatic dehydration. TRI11 then hydroxylates EPT to trichodermol (Figure 8), and TRI3 was proposed to catalyze the acetylation of trichodermol to trichodermin (Cardoza et al., 2011). Recently, TRI3 has been shown to be involved in trichodermin synthesis in an endophytic T. brevicompactum by mutational analysis (Shentu et al., 2018). In this study, for the first time, we present direct evidence that TtTRI3 is capable of performing the acetylation of trichodermol to trichodermin, using acetyl-CoA as acetyl donor. Our results also imply that biosynthetic pathway of trichodermin is conserved in Trichoderma species capable of producing it.
Some Trichoderma species such as T. brevicompactum were reported to produce both trichodermin and harzianum A (HA), depending on the strains and the cultivation conditions (Nielsen et al., 2005). HA differs from trichodermin in that it is esterified at C-4 with octa-2Z,4E,6E-trienedioic acid while trichodermin is esterified with acetic acid. In HA-producing strains, octa-2Z,4E,6E-trienedioic acid was also detected, suggesting that HA is synthesized in one single step from trichodermol and an octa-2Z,4E,6E-trienedioic group donor (Nielsen et al., 2005), which has been proved in T. arundinaceum by gene disruption (Proctor et al., 2018). Here, we showed that acetylation of trichodermol is also catalyzed by TRI3.
In Fusarium, TRI3 functions to acetylate the hydroxyl group at C-15 of the trichothecene skeleton (McCormick et al., 1996). Gene TRI3 in F. sporotrichioides (FsTRI3) shares 48% identity in translated amino acid sequence with TRI3 from Trichoderma (Table 2; Cardoza et al., 2011), which is rather low but consistent with a functional difference (shift of acetylation site from C-15 in Fusarium to C-4 in Trichoderma) between the two TRI3 enzymes. Intriguingly, a similar situation has been observed in the case of TRI11/TRI11, where amino acid sequence identity between the two genera is 39%, much lower than that of other TRI proteins, such as TRI4 (73%), TRI5 (57%), and TRI12 (55%) (Table 2; Cardoza et al., 2011), consistent with the fact that Fusarium TRI11 oxygenates trichothecene skeleton at C-15, while Trichoderma TRI11 does so at C-4 position and that other TRI proteins are functionally more conserved (Alexander et al., 1998; Cardoza et al., 2011). It is reasonable that the two genes are co-evolved since the substrate upon which TRI3 acts depends on TRI11.
In this study, the disruption of TtTRI3 gene severely reduced the expression of other trichodermin biosynthesis genes, while the regulatory gene TtTRI6 was less and TtTRI10 not at all affected. The results were in accordance with the observation that deletion or silencing of TaTRI4 in T. arundinaceum reduced expression level of TaTRI5 (Malmierca et al., 2012). Given the sampling time points in our study, our results are also in good compliance with those obtained by Shentu et al. (2018) where TRI genes were downregulated at mid–late growth phase. Surprisingly, the disruption of TaTRI5 in the HA biosynthetic pathway upregulated all other TaTRI genes except TaTRI4 (Malmierca et al., 2013). Our data suggest that the expression of TRI genes is subjected to metabolite feedback repression, a common phenomenon in biosynthetic pathways (Vasanthakumar et al., 2013; Pratelli and Pilot, 2014; Ness, 2015), likely by TRI10. In T. taxi, a decrease in the concentration of trichodermin may elicit this regulation, because the concentration of trichodermol was not significantly altered in the knockout mutant. However, other metabolites of the biosynthetic pathway, which have not been measured, may also be involved. In Fusarium, TRI6 and TRI10 function as regulatory genes to control expression of other genes in trichothecene biosynthesis (Proctor et al., 1995; Tag et al., 2001; Nasmith et al., 2011). FsTRI6 binds to the consensus sequence TNAGGCCT that is found in the promoter of most TRI genes (Hohn et al., 1999). The disruption of Fusarium TRI6 greatly reduced the expression of TRI5 and TRI4, and the mutant failed to accumulate T-2 toxin (Proctor et al., 1995). FsTri10 was also found to regulate all other TRI genes and several putative isoprenoid biosynthetic genes (Peplow et al., 2003). How TRI6 and TRI10 cooperate to regulate the expression of other TRI genes in Trichoderma, possibly in coordination with metabolite feedback, would be an interesting topic for the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
HC performed TRI3 expression, purification, and the in vitro reaction and prepared the original manuscript. LM carried out all the GC-MS experiments and analyzed the data. NZ sequenced the genome and constructed the mutant strains. CX performed the antifungal activity test. JL and CZ designed and supervised the research and revised the manuscript. WW assisted with chemical structures and manuscript revision. SX and CK perfected the language and revised the manuscript. All authors discussed, edited, and approved the final version.
This study was funded by the National High Technology Research and Development Program of China (863 Program) (No. 2011AA10A205-03) from the Ministry of Science and Technology of the People’s Republic of China, the Young Sci-Tech Talents Growth Program from the Department of Education of Guizhou Province (No. QJHKYZ[2018]297), the special Project for New Academic Seedlings and Innovative Exploration (GYU-KJT[2019]-21) and the Basic Research Program (No. QKHJC[2020]1Y089) from the Department of Sci-Tech of Guizhou Province, and the NNSFC Breeding Plan from Guiyang Science and Technology Bureau and Guiyang University (GYU-KY-[2021]).
HC and JL was employed by China Tobacco Guizhou Industrial Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Li Boyan from Guizhou Medical University and Zhang Xiaotao from China Tobacco Guizhou Industrial Co., Ltd. for carefully revising the original manuscript.
Alexander, N. J., Hohn, T. M., and McCormick, S. P. (1998). The TRI11 gene of Fusarium sporotrichiodes encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64, 221–225. doi: 10.1128/aem.64.1.221-225.1998
Alexander, N. J., McCormick, S. P., and Hohn, T. M. (1999). TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol. Gen. Genet. 261, 977–984. doi: 10.1007/s004380051046
Alexander, N. J., McCormick, S. P., Larson, T. M., and Jurgenson, J. E. (2004). Expression of TRI15 in Fusarium sporotrichioides. Curr. Genet. 45, 157–162. doi: 10.1007/s00294-003-0467-3
Brown, D. W., Dyer, R. B., McCormick, S. P., Kendra, D. F., and Plattner, R. D. (2004). Functional demarcation of the Fusarium. Fungal. Genet. Biol. 41, 454–462. doi: 10.1016/j.fgb.2003.12.002 core trichothecene gene cluster
Cardoza, R. E., Malmierca, M. G., Hermosa, M. R., Alexander, N. J., McCormick, S. P., Proctor, R. H., et al. (2011). Identification of loci and functional characterization of trichothecene biosynthetic genes in the filamentous fungus Trichoderma. Appl. Environ. Microbiol. 77, 4867–4877. doi: 10.1128/aem.00595-11
Carrasco, L., Barbacid, M., and Vazquez, D. (1973). The trichodermin group of antibiotics, inhibitors of peptide bond formation by eukaryotic ribosomes. Biochim. Biophys. Acta 312, 368–376. doi: 10.1016/0005-2787(73)90381-x
Carrol, A. M., Sweigard, J. A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal. Gen. Newslett. 41:22. doi: 10.4148/1941-4765.1367
Chen, S. Y., Zhang, C. L., Chen, Y. Z., and Lin, F. C. (2008). Trichodermin (4beta-acet-oxy-12,13-epoxy-trichothec-9-ene). Acta Crystallogr. Sect. E Struct. Rep. Online 64:702. doi: 10.1107/s1600536808006168
Chien, M. H., Lee, T. H., Lee, W. J., Yeh, Y. H., Li, T. K., Wang, P. C., et al. (2016). Trichodermin induces c-Jun N-terminal kinase-dependent apoptosis caused by mitotic arrest and DNA damage in human p53-mutated pancreatic cancer cells and xenografts. Cancer Lett. 388, 249–261. doi: 10.1016/j.canlet.2016.12.002
Cole, R. A., Jarvis, B. B., and Schweikert, M. A. (2003). Handbook of Secondary Metabolites. New York, NY: Academic Press, 199–560.
Colvin, E. W., Malchenko, S., Raphael, R. A., and Roberts, J. S. (1973). Total synthesis of (+)-trichodermin. J. Chem. Soc. Perkin 117, 1989–1997. doi: 10.1039/p19730001989
Cundliffe, E., and Davies, J. E. (1977). Inhibition of initiation, elongation, and termination of eukaryotic protein synthesis by trichothecene fungal toxins. Antimicrob. Agents Chemother 11, 491–499. doi: 10.1128/aac.11.3.491
Del Sal, G., Manfioletti, G., and Schneider, C. (1989). The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7, 514–520.
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Garvey, G. S., McCormick, S. P., Alexander, N. J., and Rayment, I. (2009). Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides. Protein Sci. 18, 747–761. doi: 10.1002/pro.80
Godtfredsen, W. O., and Vangedal, S. (1965). Trichodermin, a new sesquiterpene antibiotic. Acta Chem. Scand. 19, 1088–1102.
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/bf00014672
Hohn, T. M., Krishna, R., and Proctor, R. H. (1999). Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26, 224–235. doi: 10.1006/fgbi.1999.1122
Hohn, T. M., and Middlesworth, F. (1986). Purification and characterization of the sesquiterpene cyclase trichodiene synthetase from Fusarium sporotrichioides. Arch. Biochem. Biophys. 251, 756–761. doi: 10.1016/0003-9861(86)90386-3
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4–10. doi: 10.1094/pdis.2003.87.1.4
Jaklitsch, W. M., and Voglmayr, H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud. Mycol. 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Kimura, M., Tokai, T., Takahashi-Ando, N., Qhsato, S., and Fujimura, M. (2007). Molecular and genetic studies of Fusarium trichothecene biosynthesis pathways, gene, and evolution. Biosci. Biotechnol. Biochem 71, 2015–2123.
Kubicek, C. P., and Druzhinina, I. S. (2015). “Trichoderma mycoses and mycotoxins,” in Molecular Biology of Food and Water Borne Mycotoxigenic and Mycotic Fungi, eds R. Russell, M. Paterson, and N. Lima (Boca Raton, FL: CRC Press), 521–538.
Kulikov, S. N., Alimova, F. K., Zakharova, N. G., Nemtsev, S. V., and Varlamov, V. P. (2006). Biological preparations with different mechanism of action for protecting potato against fungal diseases. Prikl. Biokhim. Mikrobiol. 42, 86–92.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta DeltaC(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Malmierca, M. G., Cardoza, R. E., Alexander, N. J., McCormick, S. P., Collado, I. G., Hermosa, R., et al. (2013). Relevance of trichothecenes in fungal physiology: disruption of Tri5 in Trichoderma arundinaceum. Fungal Genet. Biol. 53, 22–33. doi: 10.1016/j.fgb.2013.02.001
Malmierca, M. G., Cardoza, R. E., Alexander, N. J., McCormick, S. P., Hermosa, R., Monte, E., et al. (2012). Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl. Environ. Microbiol. 78, 4856–4868. doi: 10.1128/aem.00385-12
McCormick, S. P., Alexander, N. J., and Proctor, R. H. (2006). Fusarium Tri4 encodes a multifunctional oxygenase required for trichothecene biosynthesis. Can. J. Microbiol. 52, 636–642. doi: 10.1139/w06-011
McCormick, S. P., Hohn, T. M., and Desjardins, A. E. (1996). Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62, 353–359. doi: 10.1128/aem.62.2.353-359.1996
McCormick, S. P., Stanley, A. M., Stover, N. A., and Alexander, N. J. (2011). Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814. doi: 10.3390/toxins3070802
McLaughlin, J. E., Bin-Umer, M. A., Tortora, A., Mendeze, N., McCormic, S., and Tumer, N. E. (2009). A genome-wide screen in Saccharomyces cerevisiae reveals a critical role for the mitochondria in the toxicity of a trichothecene mycotoxin. Proc. Natl. Acad. Sci. U.S.A. 106, 21883–21888. doi: 10.1073/pnas.0909777106
Nasmith, C. G., Walkowiak, S., Wang, L., Leung, W. W. Y., Gong, Y., Johnston, A., et al. (2011). Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum. PLoS Pathog. 7:e1002266. doi: 10.1371/journal.ppat.1002266
Ness, G. C. (2015). Physiological feedback regulation of cholesterol biosynthesis: role of translational control of hepatic HMG-CoA reductase and possible involvement of oxylanosterols. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1851, 667–673. doi: 10.1016/j.bbalip.2015.02.008
Nicholas, K. B., Nicholas, H. B. Jr., and Deerfield, D. W. I. I. (1997). GeneDoc: Analysis and Visualization of Genetic Variation. Embnews News 4:14. Available online at: http://www.nrbsc.org/gfx/genedoc/ebinet.htm. (accessed June 9, 2020).
Nielsen, K. F., Gräfenhan, T., Zafari, D., and Thrane, U. (2005). Trichothecene production by Trichoderma brevicompactum. J. Agric. Food Chem. 53, 8190–8196. doi: 10.1021/jf051279b
Pace, J. G., Watts, M. R., and Canterbury, W. J. (1998). T-2 mycotoxin inhibits mitochondrial protein synthesis. Toxicon 26, 77–85. doi: 10.1016/0041-0101(88)90139-0
Peplow, A. W., Tag, A. G., Garifullina, G. F., and Beremand, M. N. (2003). Identification of new genes positively regulated by Tri10 and a regulatory network for trichothecene mycotoxin production. Appl. Environ. Microbiol. 69, 2731–2736. doi: 10.1128/aem.69.5.2731-2736.2003
Pratelli, R., and Pilot, G. (2014). Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 65, 5535–5556. doi: 10.1093/jxb/eru320
Proctor, R. H., Hohn, T. M., McCormick, S. P., and Desjardins, A. E. (1995). Tri6 encodes an unusual zinc-finger protein involved in regulation of trichothecene biotynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61, 1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995
Proctor, R. H., Mccormick, S. P., Hye-Seon, K., Cardoza, R. E., Stanley, A. M., Laura, L., et al. (2018). Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 14:e1006946. doi: 10.1371/journal.ppat.1006946
Rho, H. S., Kang, S., and Lee, Y. H. (2001). Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus Magnaporthe grisea. Mol. Cells 12, 705–765.
Shentu, X., Yao, J., Yuan, X., He, L., Sun, F., Ochi, K., et al. (2018). Tri11, tri3, and tri4 genes are required for trichodermin biosynthesis of Trichoderma brevicompactum. AMB Express 8:58. doi: 10.1186/s13568-018-0585-4
Shentu, X. P., Liu, W. P., Zhan, X. H., Yu, X. P., and Zhang, C. X. (2013). The elicitation effect of pathogenic fungi on trichodermin production by Trichoderma brevicompactum. Sci. World. J. 2013, 607102. doi: 10.1155/2013/607102
Su, C. M., Wang, S. W., Lee, T. H., Tzeng, W. P., Hsiao, C. J., Liu, S. C., et al. (2013). Trichodermin induces cell apoptosis through mitochondrial dysfunction and endoplasmic reticulum stress in human chondrosarcoma cells. Toxicol. Appl. Pharmacol. 272, 335–344. doi: 10.1016/j.taap.2013.06.010
Tag, A. G., Garifullina, G. F., Peplow, A. W., Ake, C., Phillips, J. R., Hohn, T. M., et al. (2001). A novel regulatory gene, TRI10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67, 5294–5302. doi: 10.1128/aem.67.11.5294-5302.2001
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tijerino, A., Cardoza, R. E., Moraga, J., Malmierca, M. G., Vicente, F., Aleu, J., et al. (2011). Overexpression of the trichodiene synthase gene tri5 increases trichodermin production and antimicrobial activity in Trichoderma brevicompactum. Fungal Genet. Biol. 48, 285–296. doi: 10.1016/j.fgb.2010.11.012
Tokai, T., Koshino, H., Takahashi-Ando, N., Sato, M., Fujimura, M., and Kimura, M. (2007). Fusarium tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem. Biophys. Res. Commun. 353, 412–417. doi: 10.1016/j.bbrc.2006.12.033
Uchida, R., Namiguchi, S., Ishijima, H., and Tomoda, H. (2016). Therapeutic effects of three trichothecenes in the silkworm infection assay with Candida albicans. Drug Discov. Ther. 10, 44–48. doi: 10.5582/ddt.2016.01013
Ueno, Y. (1984). Toxicological features of T-2 toxin and related trichothecenes. Fundam. Appl. Toxicol. 4, S124–S132. doi: 10.1016/0272-0590(84)90144-1
Ueno, Y., and Hsieh, D. P. H. (1985). The toxicology of mycotoxins. CRC Crit. Rev. Toxicol. 14, 99–132.
Vasanthakumar, A., Kattusamy, K., and Prasad, R. (2013). Regulation of daunorubicin biosynthesis in Streptomyces peucetius–feed forward and feedback transcriptional control. J. Basic Microbiol. 53, 636–644. doi: 10.1002/jobm.201200302
Wang, L. W., Xu, B. G., Wang, J. Y., Su, Z. Z., Lin, F. C., Zhang, C. L., et al. (2012). Bioactive metabolites from Phoma species, an endophytic fungus from the Chinese medicinal plant Arisaema erubescens. Appl. Microbiol. Biotechnol. 93, 1231–1239. doi: 10.1007/s00253-011-3472-3
Wei, C. M., Hansen, B. S., Vaughan, M. H. Jr., and McLaughlin, C. S. (1974). Mechanism of action of the mycotoxin trichodermin, a 12,13-epoxytrichothecene. Proc. Natl. Acad. Sci. U.S.A. 71, 713–717.
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchiob, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Keywords: Trichoderma, TRI3, trichodermin biosynthesis, antifungal activity, trichothecene
Citation: Chen H, Mao L, Zhao N, Xia C, Liu J, Kubicek CP, Wu W, Xu S and Zhang C (2021) Verification of TRI3 Acetylation of Trichodermol to Trichodermin in the Plant Endophyte Trichoderma taxi. Front. Microbiol. 12:731425. doi: 10.3389/fmicb.2021.731425
Received: 27 June 2021; Accepted: 16 September 2021;
Published: 25 October 2021.
Edited by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaReviewed by:
Fengming Lin, Southeast University, ChinaCopyright © 2021 Chen, Mao, Zhao, Xia, Liu, Kubicek, Wu, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chulong Zhang, Y2x6aGFuZ0B6anUuZWR1LmNu; Christian P. Kubicek, cGV0ZXIua3ViaWNla0B0dXdpZW4uYWMuYXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.