Corrigendum: Molecular Epidemiology and Characteristics of CTX-M-55 Extended-Spectrum β-Lactamase-Producing Escherichia coli From Guangzhou, China
- 1KingMed School of Laboratory Medicine, Guangzhou Medical University, Guangzhou, China
- 2Department of Clinical Laboratory, Fifth Affiliated Hospital, Southern Medical University, Guangzhou, China
- 3State Key Laboratory of Respiratory Disease, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
In recent years, the CTX-M-55 extended-spectrum β-lactamase (ESBL)-positive rate has gradually increased in the clinic. To identify the molecular epidemiology and characteristics of blaCTX–M–55-positive isolates, a total of 374 non-repetitive ESBL-producing Escherichia coli strains were collected from patients in two hospitals in Guangzhou, and 89 blaCTX–M–55-positive isolates were selected by CTX-M-1-group PCR amplification and confirmed by DNA sequencing. Whole-genome sequencing was used to analyze the resistance phenotype, plasmid types, phylogenetic relationships and genetic environment of the blaCTX–M–55 gene. Conjugation experiments and PCR were performed to confirm whether the plasmid harboring blaCTX–M–55 gene could be transferred. The results showed that all blaCTX–M–55-positive isolates were resistant to ceftriaxone, and 88.76 and 76.40% were resistant to ceftazidime and cefepime, respectively. The resistance rates to levofloxacin and sulfamethoxazole were 66.29 and 59.55%, respectively. However, the sensitivity rate of piperacillin/tazobactam, amoxicillin/clavulanate, and amikacin exceeded 90%. All blaCTX–M–55-positive isolates were sensitive to carbapenems. Thirty-two STs were detected in the blaCTX–M–55-positive isolates, among which the detection rate of ST1193 was relatively high (19.10%, 17/89), and other ST types were scattered. It remains to be seen whether ST1193 carrying the blaCTX–M–55 gene can become a popular clone strain in this region in the future. The plasmid types carrying the blaCTX–M–55 gene included IncI1, IncFII, IncFIC, IncFIB, IncHI2, IncI2, and IncX/Y, among which the IncI1 and IncFII plasmids were the main plasmids, accounting for 37.80 and 28.09%, respectively. Among them, 11 strains of the IncI1 plasmid existed in ST1193 strains. The blaCTX–M–55 gene was found on chromosomes of 13 isolates, and seemed to be increasing annually. Up to five distinct types of genetic environments surrounding the blaCTX–M–55 gene were analyzed. The most common structure was type II “ISEcp1-blaCTX–M–55-ORF477.” In conclusion, whether ST1193, which carries blaCTX–M–55 gene, will be an epidemic clone of this region in the future remains to be concerned. The plasmids IncI1 and IncFII, and mobile elements such as ISEcp1 and IS26 may be the main factors leading to the spread and prevalence of CTX-M-55 genotypes.
Introduction
Extended-spectrum β-lactamases (ESBLs), including TEM, SHV, CTX-M, and OXA enzymes, are the main resistance mechanism of Enterobacteriaceae against β-lactam antibiotics. Among them, CTX-M type β-lactamase, which was first found in 1990 by Bauernfeind et al. (1990), could preferentially hydrolyze cefotaxime (CTX) compared with TEM- and SHV-type enzymes. The CTX-M type has been reported to be the main type of ESBL in Enterobacteriaceae spreading worldwide and is also widely distributed in zoonotic pathogens (Bernard et al., 1992; Rossolini et al., 2008). CTX-M-14 and CTX-M-15 have been reported to be the most common genotypes in China in recent years. However, the CTX-M-55 positive rate has gradually increased in China, especially in the area of South China (Zhang et al., 2014). In Zhang et al. (2014), Cao et al. (2011), and Zhao and Hu (2013), the detected positive rate of CTX-M-55 was even higher than that of its derivative type, CTX-M-15. CTX-M-55 is a variant of CTX-M-15 with only one amino acid substitution (Ala-80-Val). Both CTX-M-15 and CTX-M-55 belong to the CTX-M-1 group, but CTX-M-55 exhibits high hydrolytic activity to ceftazidime (Kiratisin et al., 2007). Since CTX-M-55 was first reported in Thailand in 2006, it has been identified in Escherichia coli, Klebsiella pneumoniae, Salmonella, and Morganella morganii (Kiratisin et al., 2007; Kim J. S. et al., 2017; Xia et al., 2017; Hu et al., 2018). Plasmids are known as an important reason for the rapid spread of blaCTX–M. Recently, CTX-M-55 has been reported to appear on the IncI1, IncF, IncP, and IncA/C plasmids (Kim J. S. et al., 2017; Xia et al., 2017; Hounmanou et al., 2021). Additionally, the transmission of blaCTX–M is also related to many mobile genetic elements. For example, ISEcp1, IS26, and IS903 are often detected around blaCTX–M (Lartigue et al., 2006; Poirel et al., 2008; Hu et al., 2018). In this study, 89 blaCTX–M–55-producing E. coli isolates isolated from patients in two hospitals in Guangzhou in recent years were selected, and the epidemiology and characteristics of these isolates were analyzed.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
A total of 374 non-repetitive ESBLs-producing E. coli strains isolated from patients in two hospitals in Guangzhou were identified by the advanced expert system (AES) of the VITEK-2 COMPACT Automatic Microbial Identification System (bioMérieux, Marcy-l’Étoile, France). Minimal inhibitory concentrations (MICs) including the ESBLs-resistant phenotype were determined using the VITEK-2 Automated Susceptibility System (Spanu et al., 2006). The MICs of the isolates against amoxicillin/clavulanic acid (AMC), piperacillin/tazobactam (TZP), amikacin (AMK), compound sulfamethoxazole (SXT), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), ertapenem (ETP), imipenem (IPM), and levofloxacin (LVX) were determined by agar dilution method, and the results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI/NCCLS M100-S30) [Clinical and Laboratory Standards Institute [CLSI], 2021]. All isolates were collected in two tertiary hospitals from 2012 to 2017 and 2020, while the isolates from 2018 to 2019 were not collected, so they were not included in this study. E. coli ATCC 25922 was used as a quality control strain. E. coli C600 was used as the recipient strain in the conjugation experiments.
Detection of the CTX-M-1-Group β-Lactamase Gene by PCR Assays
Primers for detecting all CTX-M-1-group type genes were designed are shown in Supplementary Table 1. Bacterial genomic DNA was extracted for PCR amplification and the positive PCR products were subsequently sequenced and to confirm their phenotype. Only blaCTX–M–55-positive isolates were selected for subsequent testing.
Conjugation Experiments
The transferability of the blaCTX–M–55 gene was determined by conjugation experiments with rifampin-resistant E. coli C600 as the recipient strain. Transconjugants were selected on Luria–Bertani agar plates containing rifampin (100 μg/mL) and CRO (4 μg/mL). PCR using CTX-M-1-group primers and sequencing were used to confirm whether the transconjugants carrying the blaCTX–M–55 gene were successfully acquired. Antimicrobial susceptibility testing and plasmid replicon typing (primers are in Supplementary Table 1) were conducted on transconjugants. The presence of resistant genes from transconjugants was also investigated by PCR (primers showed in Supplementary Table 1).
Whole-Genome Sequencing and Analysis
The genomic DNA of blaCTX–M–55-positive E. coli was extracted by a bacterial genomic DNA extraction kit (Tiangen, Beijing, China) and sequenced by next-generation sequencing (NGS) on an Illumine platform (Nuohezhiyuan, Tianjin, China). The quality of the raw readings was controlled by the interactive program FastQC (Wingett and Andrews, 2018), and the genomes were assembled using SPAdes 3.13.1 (Bankevich et al., 2012) and annotated using Prokka 1.14.5 (Seemann, 2014) on the Lunix system. Multilocus sequence typing analysis of the E. coli isolates was executed using MLST 2.18.0 (Larsen et al., 2012). The core genome multilocus sequence typing (cgMLST) of 17 ST1193 isolates was performed using Ridom SeqSphere+ 4.1.9 (Junemann et al., 2013). The resistance genes and plasmids type were determined based on the CGE server (Thomsen et al., 2016), and the plasmid circle map illustrates with BRIG (Alikhan et al., 2011). In all second generation genome annotation files, contigs harboring the blaCTX–M–55 gene were analyzed, and the blaCTX–M–55 gene locations were roughly determined combined with BLAST.1 Representative isolates with unclear blaCTX–M–55 gene locations or with different blaCTX–M–55-harboring plasmid types were selected and sequenced for long read sequencing on the Nanopore platform (Nuohezhiyuan, Tianjin, China). Finally, the location and genetic environment of the blaCTX–M–55 gene in blaCTX–M–55-positive E. coli were analyzed based on second- and third-generation genomic data. The genetic environment of the blaCTX–M–55 genes was drawn by Easyfig (Sullivan et al., 2011).
Results
blaCTX–M–55-Positive Escherichia coli Isolates
A total of 132 blaCTX–M–1–group-positive ESBLs-producing E. coli (35.29%) isolates were obtained from all 374 ESBLs-producing E. coli clinical isolates. Overall, 6 blaCTX–M–3-positive isolates (1.60%), 37 blaCTX–M–15-positive isolates (9.89%), and 89 blaCTX–M–55-positive isolates (23.80%) were identified. In this study, 89 blaCTX–M–55-positive isolates were further analyzed, and the distribution of their specimen source and collection year are shown in Figure 1. The blaCTX–M–55-positive isolates were derived mainly from patient urine (56%), while other sources included blood (14%), purulent secretion (10%), sputum (9%), wound secretion (9%), and abdominal drainage fluid (2%).
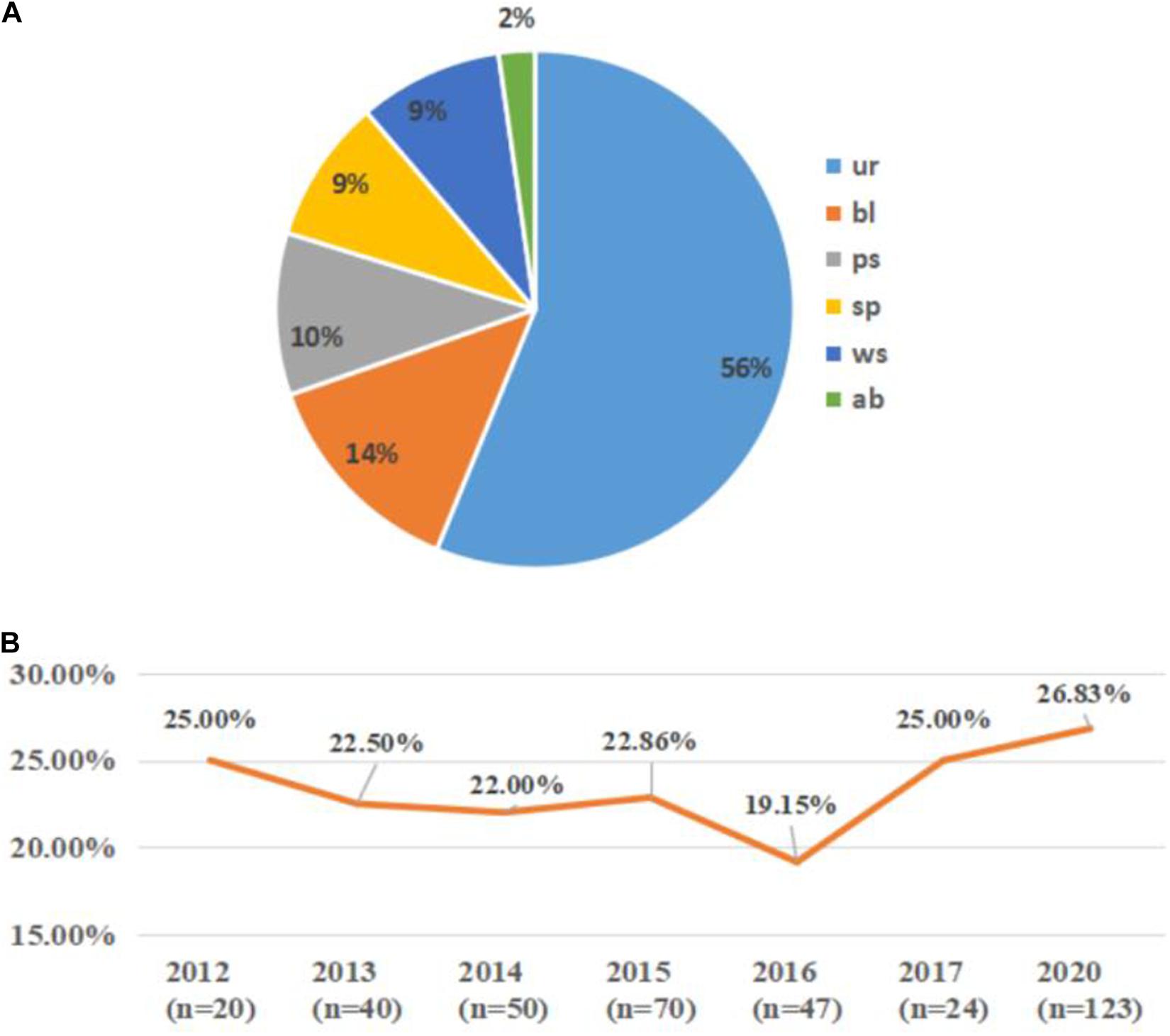
Figure 1. The distribution of specimen sources and collection years of the blaCTX–M–55-positive isolates. (A) The specimen source distribution of all the blaCTX–M–55-positive isolates. ur, urine; bl, blood; ps, purulent secretion; sp, sputum; ws, wound secretion; ab, abdominal drainage fluid. (B) The collection year’s distribution of the blaCTX–M–55-positive isolates among all the isolates. n indicates the number of isolates collected in that year; orange line indicates the proportion of blaCTX–M–55-positive isolates among the ESBL-producing E. coli isolates collected in that year (missing data from 2018 to 2019).
MLST Profile
Thirty-two MLST profiles were determined from the 89 blaCTX–M–55-positive isolates (Supplementary Table 2), including three novel STs (ST12284, ST12285, and ST12303), of which sequences have been submitted to the PubMLST database.2 In addition to the relatively high detection rate of ST1193 (19.10%, 17/89), other ST types were scattered. It is worth noting that ST1193 isolates mainly emerged in 2020 (10/17) and were distributed in different years (Supplementary Table 2). By cgMLST, among 17 ST1193, except for 3 pairs of strains that showed the same alleles, the rest ST1193 strains isolated from different years still had different alleles (Figure 2).
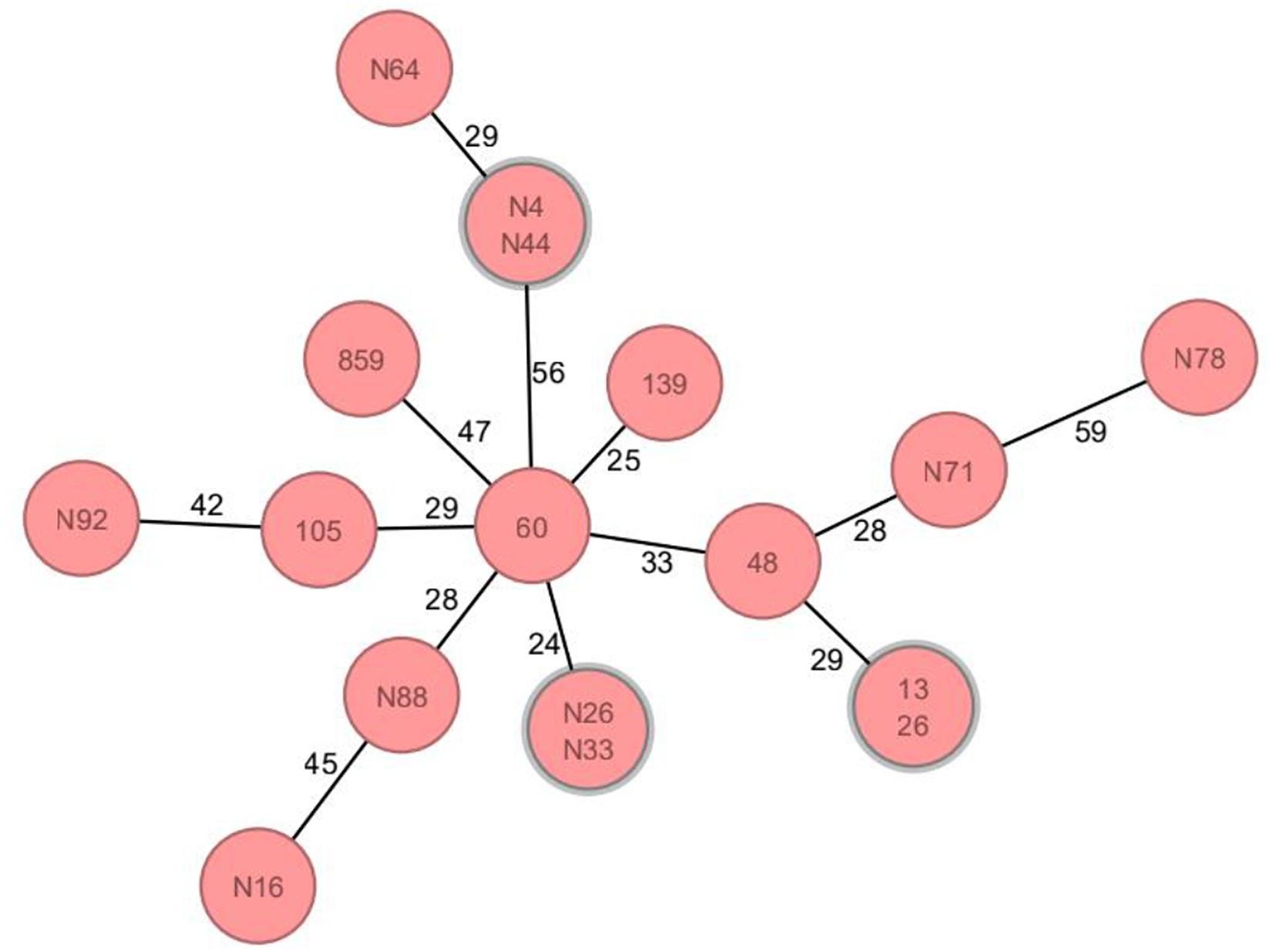
Figure 2. Core genome multilocus sequence typing analysis of ST1193 isolates carrying blaCTX–M–55 gene. Circles represent ST1193 isolates, and the number on each circle connecting line is the allele difference between the two isolates. Clonally related genetic complexes (=15 alleles difference) containing more than one patient are encircled in gray. There were no differential alleles between three pairs of isolates (N4&N44, N26&N33, and 13&26); furthermore, the number of differential alleles in other isolates was more than 15.
Antimicrobial Susceptibility Profiles
All of the blaCTX–M–55-positive isolates were resistant to CRO, and 88.76 and 76.40% were resistant to CAZ and FEP, respectively (Supplementary Table 3). The resistance rates to LVX and SXT were 66.29 and 59.55%, respectively. However, the blaCTX–M–55-positive isolates presented high susceptibility rates to AMC, TZP, and AMK, with sensitivity rates of 95.95, 95.40, and 94.38%, respectively. All blaCTX–M–55-positive isolates were sensitive to IPM and ETP. Meanwhile, we found that blaTEM (coding β-lactamase genes), sul, and dfrA (mediating sulfonamide resistance), tet (mediating tetracycline resistance), aph(3″)-Ib and aph(6′)-Ib (mediating aminoglycoside resistance) were abundant in blaCTX–M–55-positive isolates (Figure 3 and Supplementary Table 3). The 16S rRNA methylase gene rmtB (mediating aminoglycoside antibiotic resistance) was detected in five isolates, all of which were resistant to AMK (Supplementary Table 3). There were sixteen isolates containing genes mediating fluoroquinolone resistance, of which three were aac(6′)-Ib-cr and thirteen were qnrS (Figure 3 and Supplementary Table 3). dfrA or sul genes were detected in almost all SXT resistant isolates (52/53), and 46 isolates were detected in both of them.
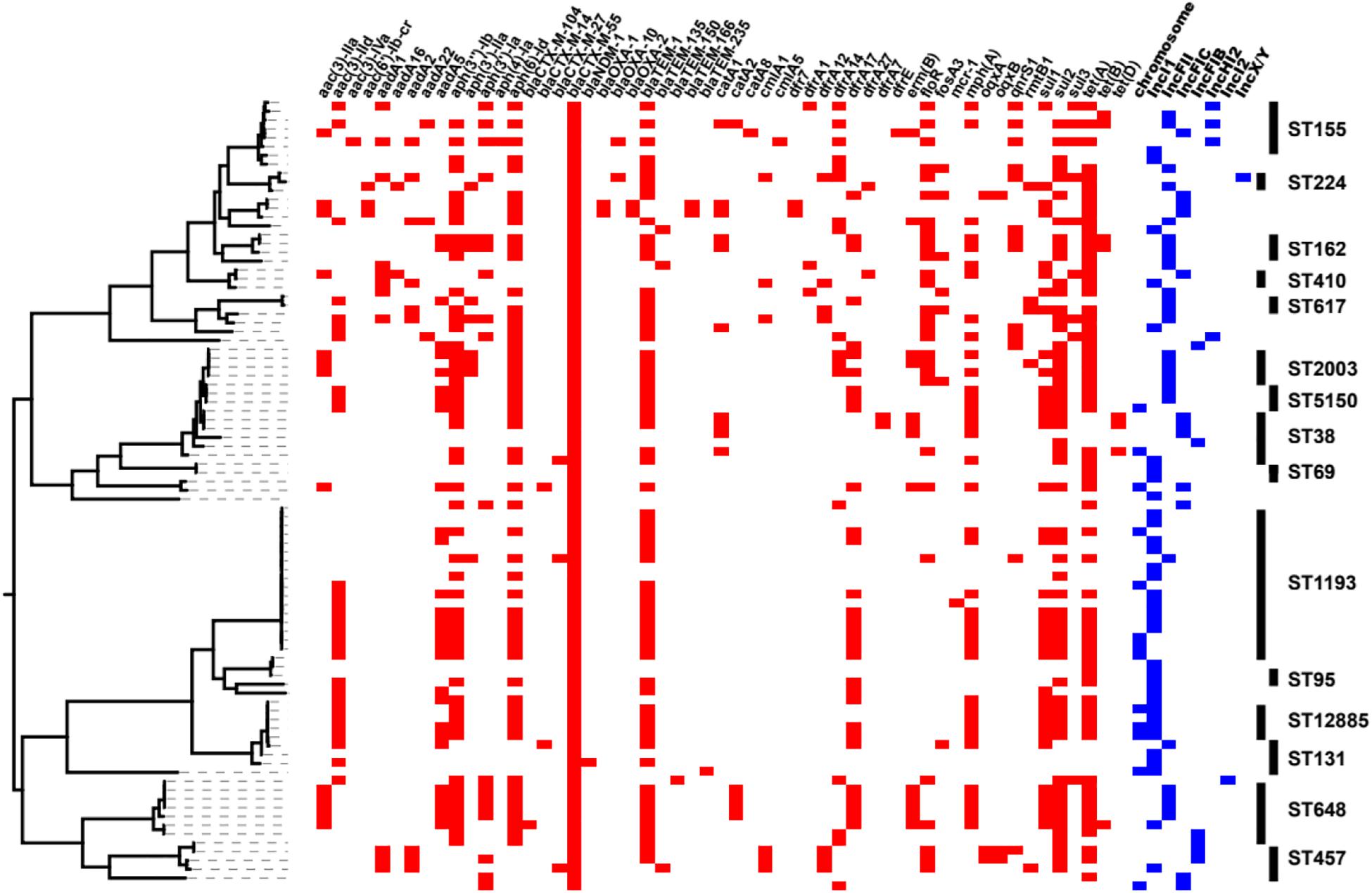
Figure 3. Heatmaps of partial resistance genes, ST types and blaCTX–M–55 gene locations combined with the phylogenetic tree of the blaCTX–M–55 isolates. The phylogenetic tree was drawn using PARSNP software. The red box indicates that the isolate contains relevant resistance gene, and the blue box indicates the location of the blaCTX–M–55 gene in the isolate. The rightmost end of the figure shows the ST type (n ≥ 2) of the isolates.
The blaCTX–M–55 Gene Location
The predicted plasmid components carrying the blaCTX–M–55 gene were of variable replicon types including IncI1, IncFIB, IncFII, IncFIC, IncHI2, IncI2, and IncX/Y (Figure 4A). The blaCTX–M–55 gene of 33 isolates was located on the IncI1 plasmid, among which 11 were carried by ST1193. The other blaCTX–M–55-positive plasmid was identified, 24 on the IncFII plasmid, 12 on the IncFIC plasmid, 6 on the IncFIB plasmid, 3 on the IncHI2 plasmid, 1 on the IncI2 or IncX/Y plasmid, and 1 simultaneously on the IncHI2 and IncFII plasmids (Supplementary Table 2). These plasmids were randomly present in different ST isolates. We also found that in thirteen of the 89 isolates containing the blaCTX–M–55 gene, BLSTA in NCBI suggested a chromosomal location (Figure 3 and Supplementary Table 2). Among them, the blaCTX–M–55 gene of three isolates was simultaneously located on both the chromosome and the IncI1 plasmid, and two were simultaneously located on the chromosome and the IncFIC plasmid. The isolates with the chromosome-encoding blaCTX–M–55 gene seemed to increase annually (Figure 4B). Our data (Figure 4B) showed that the total diversity of the plasmid types carrying the blaCTX–M–55 gene gradually increased between 2012 and 2020 (missing data from 2018 to 2019). Especially, the IncI1 and IncFII plasmids were continuously identified with higher detection rates. The IncFIB and IncHI2 emerged in the last 5 years, and the detection rate for these plasmids was still relatively low.
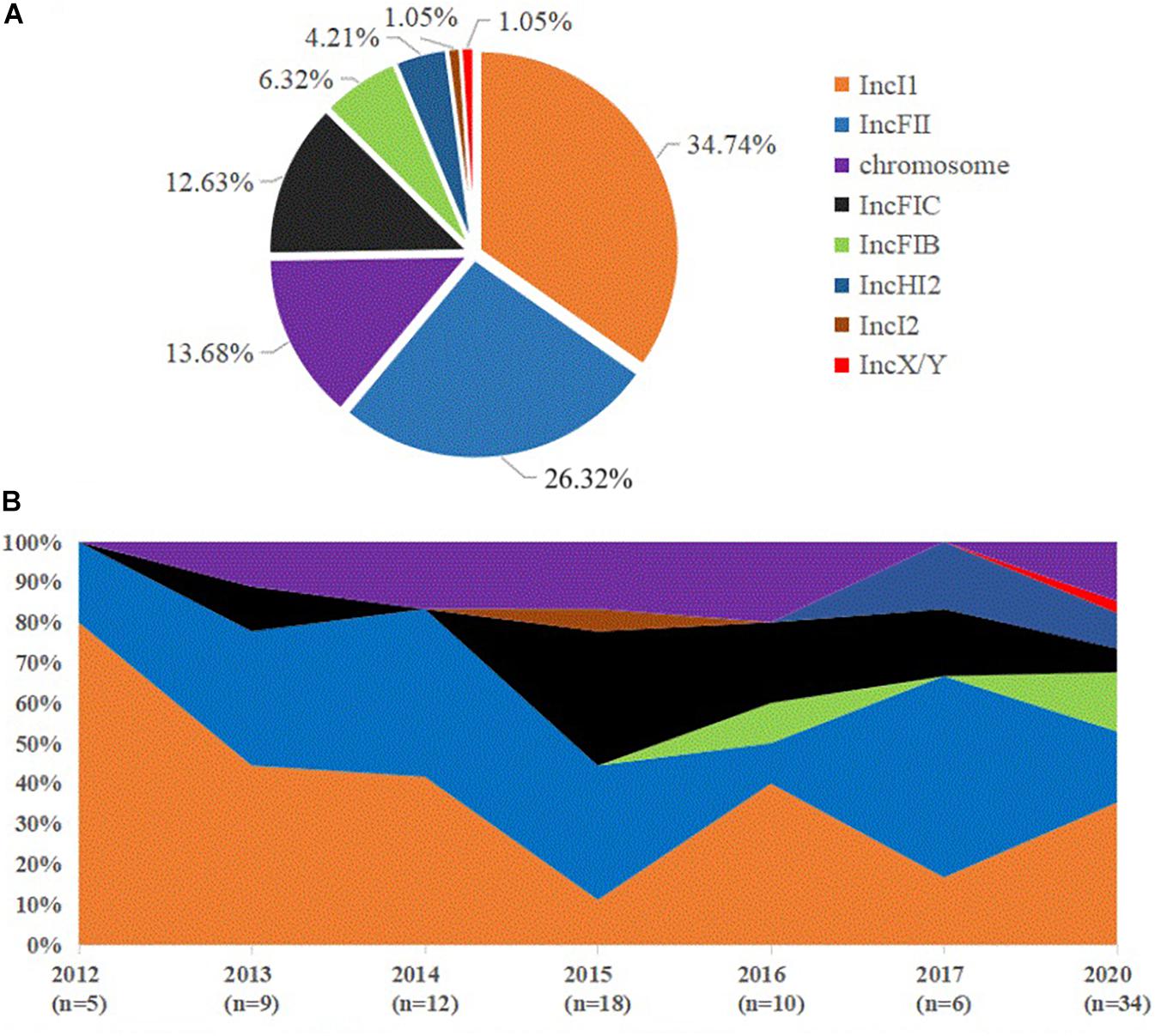
Figure 4. Distribution of blaCTX–M–55 gene location and dynamic changes in the incompatible group of plasmids in all the blaCTX–M–55-positive E. coli isolates. (A) The percentage of blaCTX–M–55 gene location found in all blaCTX–M–55-positive E. coli isolates. (B) Percentage dynamics in major incompatible plasmids and chromosomes per year (missing data from 2018 to 2019).
Circle diagrams were drawn for plasmid backbone comparison based on the known full-sequence plasmids in this study as reference. The IncI1 (Figure 5A), IncFIB (Figure 5B), IncI2 (Figure 5C), IncX/Y (Figure 5D), and IncHI2 (Figure 5E) plasmids had only one backbone type, while the IncFIC plasmid had two backbone types (Figures 5G,H). Most plasmids carrying blaCTX–M–55 with the same replicon were relatively conserved in this study, such as IncI1, IncFIB and IncHI2. In particular, almost all of the IncI1 plasmids were highly similar to p2474-3 (CP021208) in clinical E. coli isolated from a hospital in Anhui Province. Incidentally, we found that the sequence similarity between the IncI1 plasmid p628-CTXM (KP987217) isolated from K. pneumoniae, pST53-2 (CP050747.1) isolated from Salmonella enteritidis and p2474-3 was higher than 95%, and there was only one resistance gene blaCTX–M–55 between them. In contrast, the backbones of the four IncFII plasmids (Figure 5F) were slightly different, with a distinct region of approximately 24 kbp containing 24 coding sequences (CDSs). This region included some mobile genes (IS26, ISEc36, ISKpn19, IS6100, ISSBbo1, IS50R, and Tn2), the neo gene encoding aminoglycoside 3′-phosphotransferase, the hin gene encoding a specific recombinase, the hacb gene encoding a dehydrogenase, the tet(M) gene mediating tetracycline resistance, the tap gene encoding a multidrug efflux pump, and eight hypothetical protein. Meanwhile, we found that the IncFII plasmid carrying the blaCTX–M–55 gene also contains other drug resistance genes, such as blaTEM–1, fos(A), qnrS1, tet, dfrA, or catA genes by analyzing the long read sequence (Supplementary Tables 2, 3).
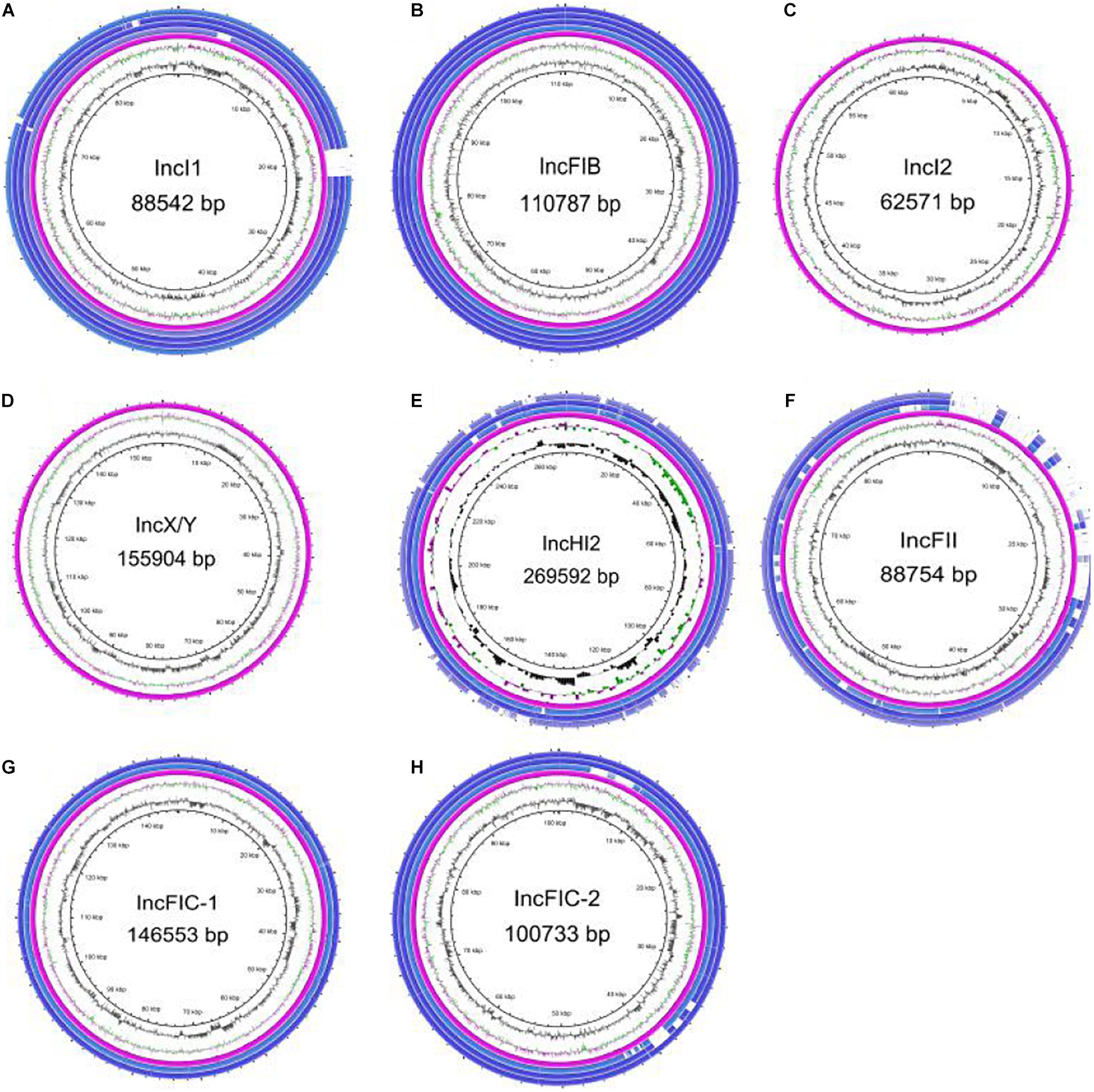
Figure 5. Plasmid backbone comparison between different blaCTX–M–55-carrying plasmids. All types of plasmid circles were based on the known full-sequence plasmids as reference sequences from this study, and several isolates with the same type of plasmid were selected for drawing through BRIG. Among them, the IncI2 and IncX/Y plasmids contained only reference plasmids, and the IncFIC plasmid had two backbone types. (A) IncI1 plasmid. (B) IncFIB plasmid. (C) IncI2 plasmid. (D) IncX/Y plasmid. (E) IncHI2 plasmid. (F) IncFII plasmid. (G) IncFIC plasmid, the first backbone. (H) IncFIC plasmid, the second backbone.
Transconjugants of the blaCTX–M–55-Positive Isolates
In total, 45 transconjugants were successfully obtained from 89 blaCTX–M–55-positive isolates through conjugation experiments, and the transfer success rate was 50.56% (45/89). Combined with the blaCTX–M–55 gene location and the plasmid replicon typing results of transconjugants (Supplementary Table 2), it was found that most of the IncI1 (24/33, 72.72%) and IncFII plasmids (16/25, 64%) could be transferred into E. coli C600, while the transfer success rate of the IncFIC and IncFIB plasmids was lower than 33.33%. In this study, the IncHI2 (0/4), IncI2 (0/1), and IncX/Y (0/1) plasmids carrying the blaCTX–M–55 gene failed to transfer through conjugation experiments. Meanwhile, there is not only one plasmid in some transconjugants. All transconjugants were resistant to CRO. The transconjugants exhibited 95.56 and 82.22% resistance rates to CAZ and FEP, respectively. Only 4.44% of the transconjugants were resistant to AMC. In addition, the resistance rates of transconjugants to SXT, LEV, and AMK were 17.78, 6.67, and 2.22%, respectively. All transconjugants were sensitive to TZP, IPM, and ETP (Supplementary Table 3).
A total of 10 transconjugants were resistant to at least one antibiotic in LEV or SXT. Interestingly, the blaCTX–M–55 gene of seven transconjugants was located on IncFII, which means that in addition to the blaCTX–M–55 gene, IncFII in these transconjugants also carries genes that mediate LVX or SXT resistance (Supplementary Tables 2, 3). Analysis of drug resistance genes in transconjugants showed that the dfrA17/dfrA14 gene are the main reason for the resistance of transconjugants to SXT. There were three transconjugants resistant to LEV, and qnrS1 gene was detected in all of them. Only one transconjugant carried the rmtB gene and was resistant to amikacin.
Genetic Environment Surrounding the blaCTX–M–55 Gene
The genetic environment surrounding the blaCTX–M–55 gene is presented in Figure 6. Five structures were obtained by analyzing mobile elements around the blaCTX–M–55 gene and named type I to V. The mobile elements located upstream of blaCTX–M–55 mainly included ISEcp1 (complete or incomplete) and IS26. Downstream of the blaCTX–M–55 genes ORF477 was consistently found. Among them, type II “ISEcp1-blaCTX–M–55-ORF477” was the predominant (63.16%, 60/95) genetic environment of the blaCTX–M–55 gene and plasmids containing this structure included IncI1, IncFIB, IncFIC, IncFII, IncHI2, and IncI2 (Figure 6). Likewise, the genetic environment of the blaCTX–M–55 gene on the chromosome (12/13) was almost type II, the other is type I. Compared with type II, only a large deletion (489 to 1140 bp) of ISEcp1 was found in type I. Moreover, the blaCTX–M–55 genes of isolate 75, 128, and 173 were found on both the chromosome and the IncI1 plasmid, and both of the genetic environments between them belong to type II. The blaCTX–M–55 gene of isolate N18 was found on both the chromosome and the IncFIC plasmid, among which the genetic environment on the chromosome was type II, and that on the IncFIC plasmid was type III “IS26-ΔISEcp1-blaCTX–M–55-ORF477.” The occurrence of the type III structure was similar to that of the type II structure, but ISEcp1 of the type III structure was disrupted by IS26. Interestingly, IS26 mainly emerged upstream of the blaCTX–M–55 gene in the IncFIC and IncFII plasmids. Type IV “IS26-blaCTX–M–55-ORF477” mainly exists in IncFII plasmids (15/17).
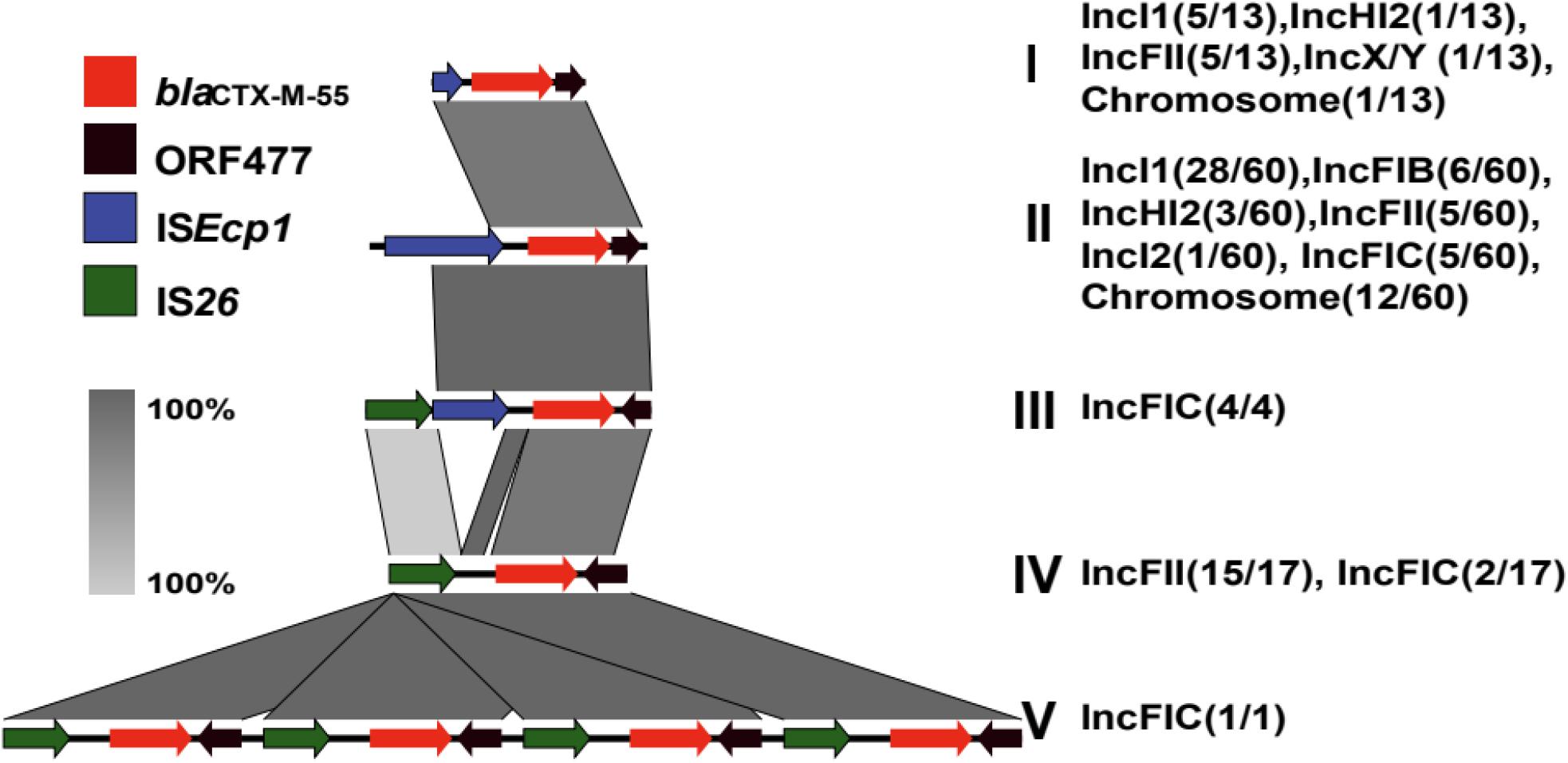
Figure 6. Genetic environment surrounding the blaCTX–M–55 gene. The colored arrow represents the ORFs and the arrow direction indicates the transcription direction. A total of 6 blaCTX–M–55 genetic structures were obtained, and different genetic environments surrounding the blaCTX–M–55 gene exist in different types of plasmids. The architecture of type I (13/95) is “ΔISEcp1-blaCTX–M–55-ORF477”; type II (60/95) is “ISEcp1-blaCTX–M–55-ORF477”; type III (4/95) is “IS26-ΔISEcp1-blaCTX–M–55-ORF477”; type IV (17/95) is “IS26-blaCTX–M–55-ORF477”; type V (1/95) is “IS26-blaCTX–M–55-ORF477-IS26-blaCTX–M–55-ORF477- IS26-blaCTX–M–55-ORF477- IS26-blaCTX–M–55-ORF477.”
Notably, an IncFIC plasmid contained four copies of the blaCTX–M–55 gene (Type V) harbored in isolate 110, and the structure of the four-duplicated segment was “IS26-blaCTX–M–55-ORF477.” The plasmid backbone was similar to that of IncFIC-2 (Figure 5H) and 151563 bp in size. This plasmid cannot be transferred through conjugation. The MIC of isolate 110 for CRO was higher than 256 μg/mL, and the MICs for CAZ and FEP were 64 μg/mL. Compared to the common blaCTX–M–55-positive isolates with CRO ≥ 64 μg/mL, and the MIC50 for CAZ was 32 μg/mL, and the MIC50 for FEP was only 16 μg/mL.
Discussion
Previous studies (Lartigue et al., 2004; Bevan et al., 2017) have shown that the isolation rate of Enterobacteriaceae producing CTX-M-55 in China has increased significantly in recent years, in aquaculture animals and in clinical patients, which demonstrates the rapid dissemination of blaCTX–M–55, especially in South China (Zhang et al., 2014). In that study (Zhang et al., 2014), the CTX-M-55 (21/38, 55.26%) positive rate in clinical ESBLs-producing isolates from Guangdong Province (in South China) was much higher than that from other provinces in China (range from 2.56 to 31.25%). Previous study (Zhang et al., 2014) have shown that the percentage of blaCTX–M–55-positive E. coli reported in China was 18.40% (47/256), exceeding the percentage of blaCTX–M–15-positive E. coli (31/256, 12.1%), which has been reported as the most widespread CTX-M gene in E. coli in China. In this study, we obtained similar results with a more significant trend: the detection rate of blaCTX–M–55-positive isolates reached 23.80% (89/374), which was higher than that of blaCTX–M–15-positive isolates (37/374, 9.89%).
Most blaCTX–M–55-positive isolates have been reported co-harboring blaTEM (Hu et al., 2018), and similar results were shown in this study. Except for blaTEM, aph (3′)-Ib, aph(6′)-Ib, dfrA, sul, and tet mediating different antibiotic resistances emerged frequently in the blaCTX–M–55-positive isolates. All of the blaCTX–M–55-positive isolates were resistant to CRO, and 88.76 and 76.40% were resistant to CAZ and FEP, respectively. The production of ESBLs is the main mechanism of resistance against cephalosporin in E. coli, and CTX-M-55 producing E. coli are known to be resistant to the second and third generations of cephalosporin (Wang et al., 2013). Cefepime is the first fourth-generation cephalosporin approved for use in China, showing low toxicity and high activity against third-generation cephalosporin-resistant Enterobacteriaceae (Chapman and Perry, 2003). However, the resistance rates of blaCTX–M–55-positive isolates to FEP reached 76.40% in this study, which showed that the resistance phenomenon of blaCTX–M–55-positive isolates to cephalosporins became tougher. In addition, blaCTX–M–55-positive isolates often carry sul and dfrA genes, which mediate their resistance to SXT up to 66.09%. Even though there are few blaCTX–M–55-positive isolates carrying qnrS or aac(6′)-Ib-cr gene, the resistance to fluoroquinolone reached 59.55%. Because fluoroquinolones are mainly mediated by plasmid mediated drug resistance genes (PMQRs) and mutations in quinolone resistance determining regions (QRDRs), and PMQR only confers low-level fluoroquinolone resistance (Robicsek et al., 2006; Strahilevitz et al., 2009; Zhang et al., 2019). Fortunately, the sensitivity of blaCTX–M–55-positive isolates to AMC, TZP, and AMK was higher than 90%, and all blaCTX–M–55-positive isolates were sensitive to IPM and ETP. Therefore, β-lactamase inhibitors, amikacin, and carbapenems can be used as a treatment strategy for blaCTX–M–55-positive isolates. The results of resistance analysis indicate that the resistance genes carried by blaCTX–M–55-positive clinical isolates are diverse and complex, which will bring great challenges to clinical treatment.
Previous studies (Russo and Johnson, 2003) showed that the population structure of CTX-M-producing E. coli is dominated by the high-risk clone ST131, and other important epidemic clones included ST405, ST38, ST648, ST410, and ST1193 (Coque et al., 2008). However, the detection rate of ST131 in this study was only 4.94% (4/89). ST1193 (19.10%) was detected much higher than that of other strains. ST1193 was reported as a fluoroquinolone-resistant E. coli clone (Platell et al., 2012; Kim Y. et al., 2017; Tchesnokova et al., 2019) and it is commonly coresistant to sulfonamides, β-lactams, and tetracyclines (Tchesnokova et al., 2019). The blaCTX–M–55-positive E. coli isolates carried by patients with urinary tract infections was reported in China (Xia et al., 2017), ST1193 (18%) was also the most common ST. ST1193 is often associated with clinical isolates of urinary tract infections (Ding et al., 2021). The blaCTX–M–55-positive isolates were obtained from multiple clinical specimens in this study. Of the 17 ST1193 isolates, 10 were derived from urine specimens. The results of cgMLST analysis showed that, except for the three pairs of strains, the other blaCTX–M–55-positive ST1193 clones isolated from different time and departments exist allelic difference, which indicate that although the genetic relationship of these ST1193 strains were closely related, they still had different degrees of mutation over time. We also found that a total of 10 isolates of ST1193 were detected in 2020 and 7 isolates in 2012–2017 (Supplementary Table 2). Therefore, we speculate that ST1193 may become a potential epidemic clone among clinical isolates carrying blaCTX–M–55 in the future, but it still needs further observation. Furthermore, this study showed that the blaCTX–M–55 gene is located on the chromosome with an increasing trend, and a study of blaCTX–M–55 from Salmonella (Zhang et al., 2019) also showed that blaCTX–M–55 mainly existed in the chromosome (10/11), which suggests that we should pay close attention to the vertical transmission of blaCTX–M–55-positive strains.
Conjugative plasmids play a key role in the horizontal transfer of drug resistance genes among E. coli (Carattoli, 2013), which is an important reason for blaCTX–M transmission. In recent years, blaCTX–M–55 in China has mainly been located on the IncI1 plasmid and sporadically emerged on the IncF and IncP plasmids (Bevan et al., 2017; Xia et al., 2017). The plasmid types carrying the blaCTX–M–55 gene found in this study included IncI1, IncFII, IncFIC, IncFIB, IncHI2, IncI2, and IncX/Y. Among them, the detection rates of IncI1 and IncFII plasmids were the highest. Meanwhile, most IncI1 and IncFII plasmids can be transferred to E. coli C600, but the transfer success rate of other plasmids was low. We also found plasmids similar to IncI1 in K. pneumoniae and S. enteritidis (Fu et al., 2020), and the sequence identity was higher than 95%, indicating that the IncI1 plasmid carrying blaCTX–M–55 can be spread in different strains. The IncI1 plasmid carrying only one drug resistance gene of blaCTX–M–55 in Enterobacteriaceae is still relatively conserved after several years of transmission. In this study, 64.71% (11/17) of ST1193 contained IncI1 plasmids, while 88% (14/16) of the ST1193 isolates studied by Xia et al. (2017) also contained IncI1 plasmids. In addition, many IncI1 plasmids (22/33, 66.67%) were distributed in other ST types. Our results suggest that the dominant plasmid IncI1 may be one of the main reasons for the widespread dissemination of the blaCTX–M–55 gene in E. coli. Additionally, the IncFII plasmid can be spread in many ST isolates. Meanwhile, it should be noted that the IncFII plasmid carrying the blaCTX–M–55 gene also contains other drug resistance genes, such as blaTEM–1, fos(A), qnrS1, tet, dfrA, or catA genes by analyzing the long read sequence (Supplementary Tables 2, 3). Likewise, the analysis of the results of transconjugants showed that the coexistence of the blaCTX–M–55 gene and other resistance genes (dfrA17, dfrA14, qnrS1, tet, and rmtB) seemed to be the most common on the IncFII plasmid. Therefore, isolates containing the IncFII plasmid carrying blaCTX–M–55 were more resistant to clinical antibiotics, and it should also be considered in the clinic.
Mobile sequences such as insertion sequences (ISs) and transposons (Tns) are important elements that mediate the horizontal transmission of the blaCTX–M gene (Cullik et al., 2010; Ma et al., 2011). ISEcp1 is often located upstream of the CTX-M gene and is responsible for the movement of all blaCTX–M genotypes (Zhao and Hu, 2013). At present, there are few studies exploring the genetic structure of the blaCTX–M–55 gene in E. coli. Five types of genetic structure were obtained by analyzing the environment surrounding blaCTX–M–55 in the isolates. The upstream elements were mainly ISEcp1 and IS26. Similar to previous studies (Cao et al., 2011; Fu et al., 2020), the most common genetic environment for blaCTX–M–55 is type II “ISEcp1-blaCTX–M–55-ORF477.” Studies (Wang et al., 2013) have shown that ISEcp1 seems to be a strong activator of blaCTX–M–55 expression. Additionally, IS26, IS903, and ORF477 are also detected frequently around them (Lartigue et al., 2006; Poirel et al., 2008; Hu et al., 2018). In this study, we found that the blaCTX–M–55 gene existed on both chromosomes and plasmids of the same isolate. Interestingly, the genetic environments of isolates 75, 128, and 173 on the chromosome and on the IncI1 plasmid were the same, both of which were type II. When comparing the genetic environments of the chromosome and IncFIC plasmid of isolate N18, only one copy of ISEcp1 on the plasmid was disrupted by IS26. It has also been reported (Ensor et al., 2006; Zhang et al., 2019) that the disruption of the ISEcp1 element by IS26 is related to the transmission of the blaCTX–M gene. ISEcp1 can mobilize the CTX-M gene to different types of plasmids with IS26 (Canton et al., 2012). This result showed that under certain conditions, the mobile element may be able to mediate the transfer of the blaCTX–M–55 gene between the plasmid and the chromosome. Although IS903 was not detected, we found that ISEcp1 and IS26 were upstream of the blaCTX–M–55 gene. More interestingly, there were four copies of “IS26-blaCTX–M–55-ORF477” in isolate 110. Generally, the presence of multi-copy blaCTX–M–55 genes may be associated with increased cephalosporins resistance. The transposition mechanism of IS26 is generally regarded to involve replicative transposition and co integrate formation (Jiang et al., 2020). We can speculate that IS26 mediated transposon unequal crossover, produced four copies of IS26 composite transposons in a row, which has also been reported in previous studies (Lee et al., 2012; Jiang et al., 2020). Therefore, we should strengthen the detection and analysis of mobile genetic element IS26 to monitor the transmission trend of plasmids carrying the blaCTX–M–55 gene with related elements in the clinical environment.
Conclusion
Most of the isolates carrying the blaCTX–M–55 gene are highly resistant to cephalosporins, but are still highly sensitive to amikacin, β-lactamase inhibitors and carbapenems, which can be used as the choice of clinical medication. In this study, prevalent ST clones were not detected. However, it remains to be seen whether ST1193 carrying the blaCTX–M–55 gene can become a popular clone in this region. Meanwhile, we should pay close attention to the vertical transmission of blaCTX–M–55-positive isolates. The epidemic plasmid IncI1 and IncFII, which have the highest detection rate and transfer efficiency, may play an important role in the spread of the blaCTX–M–55 gene. In particular, the IncFII plasmid usually carries more drug resistance genes. Mobile elements such as ISEcp1 and IS26 may be the main factors leading to the spread and prevalence of CTX-M-55 genotypes on chromosomes and plasmids.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, JAHSRN000000000-JAHSSE000000000.
Author Contributions
XL conceived and designed the study. CZ and AW provided the samples. XL and CZ funded the project. SZ and LH carried out the experiment. SZ, XC, and JL steered the literature search, data collection, and analysis. SZ drafted the manuscript. AW, XL, and CZ reviewed and approved the submission of the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This project was funded by the Youth Foundation of the National Natural Science Foundation of China (81902104), the Medical Science and Technology Research Project of Foshan Science and Technology Innovation Program (1920001000717), and the International Cooperation and Exchange Program of the National Natural Science Foundation of China (81861138056).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Kai Zhou (The First Affiliated Hospital of Southern University of Science and Technology, China) and Yan Jiang (The Sir Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China) for their suggestions and revisions to this study, and Jingjie Song for his technical assistance in the genome sequencing data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.730012/full#supplementary-material
Footnotes
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bauernfeind, A., Grimm, H., and Schweighart, S. (1990). A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18, 294–298. doi: 10.1007/BF01647010
Bernard, H., Tancrede, C., Livrelli, V., Morand, A., Barthelemy, M., and Labia, R. (1992). A novel plasmid-mediated extended-spectrum beta-lactamase not derived from TEM- or SHV-type enzymes. J. Antimicrob. Chemother. 29, 590–592. doi: 10.1093/jac/29.5.590
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Cao, X., Cavaco, L. M., Lv, Y., Li, Y., Zheng, B., Wang, P., et al. (2011). Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J. Clin. Microbiol. 49, 2496–2501. doi: 10.1128/JCM.02503-10
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Chapman, T. M., and Perry, C. M. (2003). Cefepime: a review of its use in the management of hospitalized patients with pneumonia. Am. J. Respir. Med. 2, 75–107. doi: 10.1007/BF03256641
Clinical and Laboratory Standards Institute [CLSI] (2021). Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement. CLSI/NCCLS M100-S30, 30th Edn. Wayne PA: Clinical and Laboratory Standards Institute.
Coque, T. M., Baquero, F., and Canton, R. (2008). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13:19044. doi: 10.2807/ese.13.47.19044-en
Cullik, A., Pfeifer, Y., Prager, R., von Baum, H., and Witte, W. (2010). A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 59, 580–587. doi: 10.1099/jmm.0.016188-0
Ding, Y., Zhang, J., Yao, K., Gao, W., and Wang, Y. (2021). Molecular characteristics of the new emerging global clone ST1193 among clinical isolates of Escherichia coli from neonatal invasive infections in China. Eur. J. Clin. Microbiol. Infect. Dis. 40, 833–840. doi: 10.1007/s10096-020-04079-0
Ensor, V. M., Shahid, M., Evans, J. T., and Hawkey, P. M. (2006). Occurrence, prevalence and genetic environment of CTX-M beta-lactamases in Enterobacteriaceae from Indian hospitals. J. Antimicrob. Chemother. 58, 1260–1263. doi: 10.1093/jac/dkl422
Fu, Y., Xu, X., Zhang, L., Xiong, Z., Ma, Y., Wei, Y., et al. (2020). Fourth generation cephalosporin resistance among Salmonella enterica Serovar Enteritidis Isolates in Shanghai, China Conferred by blaCTX–M–55 Harboring Plasmids. Front. Microbiol. 11:910. doi: 10.3389/fmicb.2020.00910
Hounmanou, Y. M. G., Bortolaia, V., Dang, S. T. T., Truong, D., Olsen, J. E., and Dalsgaard, A. (2021). ESBL and AmpC beta-Lactamase Encoding Genes in E. coli from pig and pig farm workers in vietnam and their association with mobile genetic elements. Front. Microbiol. 12:629139. doi: 10.3389/fmicb.2021.629139
Hu, X., Gou, J., Guo, X., Cao, Z., Li, Y., Jiao, H., et al. (2018). Genetic contexts related to the diffusion of plasmid-mediated CTX-M-55 extended-spectrum beta-lactamase isolated from Enterobacteriaceae in China. Ann. Clin. Microbiol. Antimicrob. 17:12. doi: 10.1186/s12941-018-0265-x
Jiang, Y., Wang, Y., Hua, X., Qu, Y., Peleg, A. Y., and Yu, Y. (2020). Pooled plasmid sequencing reveals the relationship between mobile genetic elements and antimicrobial resistance genes in clinically isolated Klebsiella pneumoniae. Genomics Proteomics Bioinformatics 18, 539–548. doi: 10.1016/j.gpb.2020.12.002
Junemann, S., Sedlazeck, F. J., Prior, K., Albersmeier, A., John, U., Kalinowski, J., et al. (2013). Updating benchtop sequencing performance comparison. Nat. Biotechnol. 31, 294–296. doi: 10.1038/nbt.2522
Kim, J. S., Kim, S., Park, J., Shin, E., Yun, Y. S., Lee, D. Y., et al. (2017). Plasmid-mediated transfer of CTX-M-55 extended-spectrum beta-lactamase among different strains of Salmonella and Shigella spp. in the Republic of Korea. Diagn. Microbiol. Infect. Dis. 89, 86–88. doi: 10.1016/j.diagmicrobio.2017.03.014
Kim, Y., Oh, T., Nam, Y. S., Cho, S. Y., and Lee, H. J. (2017). Prevalence of ST131 and ST1193 Among Bloodstream Isolates of Escherichia coli not Susceptible to Ciprofloxacin in a Tertiary Care University Hospital in Korea, 2013 - 2014. Clin. Lab. 63, 1541–1543. doi: 10.7754/Clin.Lab.2017.170319
Kiratisin, P., Apisarnthanarak, A., Saifon, P., Laesripa, C., Kitphati, R., and Mundy, L. M. (2007). The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58, 349–355. doi: 10.1016/j.diagmicrobio.2007.02.005
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lartigue, M. F., Poirel, L., Aubert, D., and Nordmann, P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50, 1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006
Lartigue, M. F., Poirel, L., and Nordmann, P. (2004). Diversity of genetic environment of bla(CTX-M) genes. FEMS Microbiol. Lett. 234, 201–207. doi: 10.1111/j.1574-6968.2004.tb09534.x
Lee, S. Y., Park, Y. J., Yu, J. K., Jung, S., Kim, Y., Jeong, S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67, 2843–2847. doi: 10.1093/jac/dks319
Ma, L., Siu, L. K., and Lu, P. L. (2011). Effect of spacer sequences between bla(CTX-M) and ISEcp1 on bla(CTX-M) expression. J. Med. Microbiol. 60, 1787–1792. doi: 10.1099/jmm.0.033910-0
Platell, J. L., Trott, D. J., Johnson, J. R., Heisig, P., Heisig, A., Clabots, C. R., et al. (2012). Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob. Agents Chemother. 56, 3898–3904. doi: 10.1128/AAC.06120-11
Poirel, L., Naas, T., and Nordmann, P. (2008). Genetic support of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14, Suppl 1, 75–81. doi: 10.1111/j.1469-0691.2007.01865.x
Robicsek, A., Jacoby, G. A., and Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640. doi: 10.1016/S1473-3099(06)70599-0
Rossolini, G. M., D’Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1), 33–41. doi: 10.1111/j.1469-0691.2007.01867.x
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/S1286-4579(03)00049-2
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Spanu, T., Sanguinetti, M., Tumbarello, M., D’Inzeo, T., Fiori, B., Posteraro, B., et al. (2006). Evaluation of the new VITEK 2 extended-spectrum beta-lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J. Clin. Microbiol. 44, 3257–3262. doi: 10.1128/JCM.00433-06
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tchesnokova, V. L., Rechkina, E., Larson, L., Ferrier, K., Weaver, J. L., Schroeder, D. W., et al. (2019). Rapid and extensive expansion in the united states of a new multidrug-resistant Escherichia coli Clonal Group, Sequence Type 1193. Clin. Infect. Dis. 68, 334–337. doi: 10.1093/cid/ciy525
Thomsen, M. C., Ahrenfeldt, J., Cisneros, J. L., Jurtz, V., Larsen, M. V., Hasman, H., et al. (2016). A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718
Wang, Y., Song, C., Duan, G., Zhu, J., Yang, H., Xi, Y., et al. (2013). Transposition of ISEcp1 modulates blaCTX-M-55-mediated Shigella flexneri resistance to cefalothin. Int. J. Antimicrob. Agents 42, 507–512. doi: 10.1016/j.ijantimicag.2013.08.009
Wingett, S. W., and Andrews, S. (2018). FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 7:1338. doi: 10.12688/f1000research.15931.1
Xia, L., Liu, Y., Xia, S., Kudinha, T., Xiao, S. N., Zhong, N. S., et al. (2017). Prevalence of ST1193 clone and IncI1/ST16 plasmid in E-coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci. Rep. 7:44866. doi: 10.1038/srep44866
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located blaCTX–M–55 in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Keywords: CTX-M-55, IncI1 plasmid, ISEcp1, ST1193, E. coli, IncFII plasmid
Citation: Zeng S, Luo J, Chen X, Huang L, Wu A, Zhuo C and Li X (2021) Molecular Epidemiology and Characteristics of CTX-M-55 Extended-Spectrum β-Lactamase-Producing Escherichia coli From Guangzhou, China. Front. Microbiol. 12:730012. doi: 10.3389/fmicb.2021.730012
Received: 24 June 2021; Accepted: 20 September 2021;
Published: 11 October 2021.
Edited by:
Catherine Neuwirth, Centre Hospitalier Regional Universitaire De Dijon, FranceReviewed by:
Yaovi Mahuton Gildas Hounmanou, University of Copenhagen, DenmarkHua Zhou, Zhejiang University, China
Copyright © 2021 Zeng, Luo, Chen, Huang, Wu, Zhuo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aiwu Wu, aiwwu66@163.com; Chao Zhuo, Chao_sheep@263.net; Xiaoyan Li, xiaoyanli@gzhmu.edu.cn
†These authors have contributed equally to this work
 Shihan Zeng
Shihan Zeng Jiajun Luo
Jiajun Luo Xiankai Chen
Xiankai Chen LiShao Huang
LiShao Huang Aiwu Wu
Aiwu Wu Chao Zhuo
Chao Zhuo