
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 October 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.729369
Bacterial membrane vesicles (MVs) are produced by both Gram-positive and Gram-negative bacteria during growth in vitro and in vivo. MVs are nanoscale vesicular structures with diameters ranging from 20 to 400 nm. MVs incorporate bacterial lipids, proteins, and often nucleic acids, and can effectively stimulate host immune response against bacterial infections. As vaccine candidates and drug delivery systems, MVs possess high biosafety owing to the lack of self-replication ability. However, wild-type bacterial strains have poor MV yield, and MVs from the wild-type strains may be harmful due to the carriage of toxic components, such as lipopolysaccharides, hemolysins, enzymes, etc. In this review, we summarize the genetic modification of vesicle-producing bacteria to reduce MV toxicity, enhance vesicle immunogenicity, and increase vesicle production. The engineered MVs exhibit broad applications in vaccine designs, vaccine delivery vesicles, and drug delivery systems.
Both eukaryotic and prokaryotic cells can produce extracellular membrane vesicles (MVs), which are nanoscale structures secreted by cells during growth and proliferation (Brown et al., 2015; Gill et al., 2019). Gram-negative (G–) bacteria can secret MVs directly from their outer membrane, thus called outer membrane vesicles (OMVs) (DeVoe and Gilchrist, 1973). By contrast, a Gram-positive (G+) bacterium has only one cellular membrane covered by a thick layer of peptidoglycan, and its ability to produce MVs was not discovered until Lee et al. (2009). Bacteria can release MVs into the extracellular space in all environments, however they are most easily observed in bacterial culture media (Kaparakis-Liaskos and Ferrero, 2015). The typical MVs are nanoscale bilayer lipid membrane structures with diameters of 20–400 nm (Toyofuku et al., 2019). Bacterial MVs can contain proteins (membrane proteins, lipoproteins, and bacterial toxins), lipopolysaccharides (LPS), and nucleic acids (plasmids, chromosome fragments, and RNA) (Lee et al., 2009; Koeppen et al., 2016; Yuan et al., 2018; Li M. et al., 2020). MVs exhibit important functions, including the transfer of DNA and RNA (Fulsundar et al., 2014; Koeppen et al., 2016), transport of virulence factors (Olaya-Abril et al., 2014; Yuan et al., 2018), interception of bacteriophages (Manning and Kuehn, 2011; Tzipilevich et al., 2017), communication among bacterial populations (Li et al., 2016; Toyofuku et al., 2017), and interaction with host cells (Elmi et al., 2012; Mondal et al., 2016). The inherent characteristics of bacterial MVs make them good candidates for a broad range of applications (Pathirana and Kaparakis-Liaskos, 2016). Firstly, as products secreted by bacterial strains, MVs do not have the ability to grow and reproduce. Thus, the usage of MVs will not cause infections (Kaparakis-Liaskos and Ferrero, 2015). Secondly, plenty of bacterial antigens can be displayed on the surface or sealed inside MVs to stimulate innate and adaptive immune responses. Thus, MVs can be used as vaccines (Parikh et al., 2016; Haque et al., 2021; Li et al., 2021). Thirdly, MVs can incorporate exogenous substances and can easily be used as a vaccine or drug carriers (Tian et al., 2021; Yang et al., 2021). However, some challenges in the application of MVs exist, such as potential biotoxicity (Yuan et al., 2018), insufficient immunogenicity (Wang et al., 2020b), and low natural yield (Cao and Liu, 2020). With the development of bioengineering technology and the deepening knowledge on MVs, further improvement in the safety issue, immunogenicity, or production of MVs is possible through the manipulation of the host bacteria. This review focuses on the engineering of MV-producing bacteria to attenuate MV toxicity, improve MV immunogenicity, and increase MV production. The tremendously improved potential of engineered MVs in vaccine development and drug delivery is discussed.
Membrane vesicles produced by the wild-type bacteria contain toxic components, may carry limited immunogens, and have low yield, resulting in safety problems, inefficiency, and high costs during MV production (Liu et al., 2016a; Cao and Liu, 2020; Yang et al., 2020). Genetic modifications have been widely applied to bacteria for the purposes of MV toxicity reduction, immunogenicity enhancement, and production improvement.
Both G+ and G– pathogenic bacteria produce toxic molecules that play important roles in bacterial infections (Rivera et al., 2010). As the secreted products of bacteria, MVs may incorporate the toxic molecules during MV formation (Table 1). Detoxification of the toxic components in the MVs is the basic requirement for MV application.
G– bacteria-produced MVs can have lipopolysaccharides (LPS), adhesins, and other virulence factors (Olofsson et al., 2010; Liu et al., 2016a). As the main component of G– bacterial outer membrane, LPS can stimulate a strong inflammatory response in humans through the toll-like receptor 4 (TLR4)–MD2–CD14 pathway (Kong et al., 2012). The direct incorporation of LPS increases the virulence of MVs and limits their application. MV toxicity could be greatly reduced by altering and modifying the structure of LPS, including acylation and phosphorylation of lipid A, synthesis and transport of core oligosaccharides, and polymerization of O-antigen polysaccharides (Yang et al., 2020).
Lipopolysaccharides consists of lipid A, core oligosaccharide, and O-antigen (Raetz and Whitfield, 2002). Lipid A, which is the toxic group of LPS, consists of a hexacylated diglucosamine, six acyl chains, and two phosphate groups (Raetz et al., 2007). LpxM, LpxL, and PagL are vital acyl transferases involved in lipid-A modification in bacteria, such as Escherichia coli and Salmonella (Raetz et al., 2007; Bertani and Ruiz, 2018). Ranallo et al. (2010) reported that the deletion of msbB (lpxM) in Shigella flexneri results in the formation of penta-acylated lipid A, which could serve as a TLR4-antagonist. The mortality rate was reduced to 37–50% in mice challenged with penta-acylated lipid A for 72 h compared with mice challenged with wild-type lipid A (100%) (Ranallo et al., 2010). Lee et al. (2017) demonstrated that the mice infected with the phosphatase gene lpxF-mutated E. coli exhibited less weight loss and slighter lung inflammation than the mice infected with the wild-type strain. To eliminate the effect of LPS in the MVs, the lpxL1 gene for LPS biosynthesis in Neisseria meningitidis was genetically deleted, and the toxicity of MVs was attenuated (van de Waterbeemd et al., 2010). However, the growth rate of the lpxL1 mutant was remarkably affected compared with that of the wild-type strain. Therefore, this mutant may not be suitable for application due to the growth defect.
In most G– bacteria, the genes for core oligosaccharide and O-antigen synthesis are integrated into two operons, namely, waa and wba (rfb) (Whitfield et al., 2003; Frirdich and Whitfield, 2005). The lack of full-length O-antigens and/or incomplete core polysaccharides leads to the truncation of LPS (Liu et al., 2016b). Liu et al. (2016a) found that the mice infected with MVs produced by waaC-, rfaH-, or rfbP-deleted S. Typhimurium mutants presented a higher survival rate of 16.7–33.3% compared with the mice inoculated with MVs produced by the wild-type strain. The incomplete structure of LPS caused by the engineered remolding of the key genes for LPS biosynthesis is an important way to attenuate MV toxicity, but such approaches have usually let to decrease in MV yield. Therefore, optimizing the strategy to achieve knock-out mutants with normal growth is a quite significant issue for application of the engineered MVs.
In addition to LPS, MVs from G– bacteria can package numerous other virulence factors, such as bacterial adhesins, proteases, and cytotoxins (Table 1). Li et al. (2021) consecutively deleted 14 genes encoding variant virulence factors in Pseudomonas aeruginosa PA103 to generate a PA-m14 mutant (ΔexoU/ΔexoA/ΔexoT/ΔlasA/ΔlasB/ΔwbjA/ΔpchA/Δ phzM/Δalg/ΔRhlAB/ΔpvdA/ΔplcH/ΔphoA/ΔlpxL). The sizes of MVs produced by PA-m14 were greatly smaller than those from the wild-type PA103. Intramuscular injection with 50 μg MVs from PA-m14 mutant did not cause any death in BALB/c mice, in contrast 100% of mice challenged with wild-type MVs died after 3 days. Such consecutive deletion of genes encoding different virulence factors in bacteria is an effective strategy to attenuate bacterial MVs, whereas this method takes time and effort. To prepare MVs with reduced toxicity, new fast and effective strategies for bacterial engineering are urgently needed.
In G+ bacteria, the genetic manipulation of genes for virulence factors may result in the detoxification of MVs. In Staphylococcus aureus, the expression of virulence factors is controlled by a complex regulatory network that responds to host and environmental changes. The well-studied regulatory elements in S. aureus strains are the accessory gene regulatory system (Agr) and the SaeR/S two-component system (SaeR/S TCS). Agr encodes a quorum sensing system to control the expression of major virulence factors, including exotoxin up-regulation and surface protein down-regulation (Jenul and Horswill, 2019). The SaeR/S TCS consists of four genes (saeP, saeQ, saeR, and saeS) controlled by two promoters (P1 and P3), which play a major role in regulating the production of more than 20 virulence factors in S. aureus (Liu et al., 2016c). In a study conducted to determine the effect of Agr and SaeR/S TCS on the virulence of S. aureus-secreted MVs, Yuan et al. (2014) found that the mortality of mice challenged with engineered MVs derived from S. aureus strain RN4220-Δagr was remarkably reduced compared with that of mice stimulated with wild-type MVs. In addition, Wang et al. (2018) found that the single mutant of global regulator agr in S. aureus strain JE2 (JE2Δagr) remarkably reduced the mRNA expressions of genes that encode all nine subunits of staphylococcal leukocidins and the gene hla that encodes alpha toxin. Immunization of female Swiss Webster mice with 5μg MVs produced by the double mutant of agr and spa (encoding protein A) in S. aureus JE2 (JE2-ΔagrΔspa) provided significant protection against fatal sepsis caused by a heterologous USA300 isolate, FPR3757 (Wang et al., 2018). MVs produced by S. aureus JE2-ΔagrΔsae remarkably reduced the cytotoxicity to THP-1 macrophages (Wang et al., 2020a). Thus, engineering bacteria by deletion of regulatory systems controlling virulence gene expression is promising for MV detoxification.
The immunogenicity of MV-contained antigens is crucial to the successful development of an MV vaccine (Yuan et al., 2018). The key to inducing an effective immune response is the ingestion of antigen-containing particles by antigen-presenting cells (APCs). Therefore, engineering bacteria to load more target antigens into MVs and manipulating MV nanoparticles are effective ways to enhance MV immunogenicity. Here, we discuss four useful strategies applied to enhance the immunogenicity of MVs, which improved the application prospect of MVs (Figure 1).
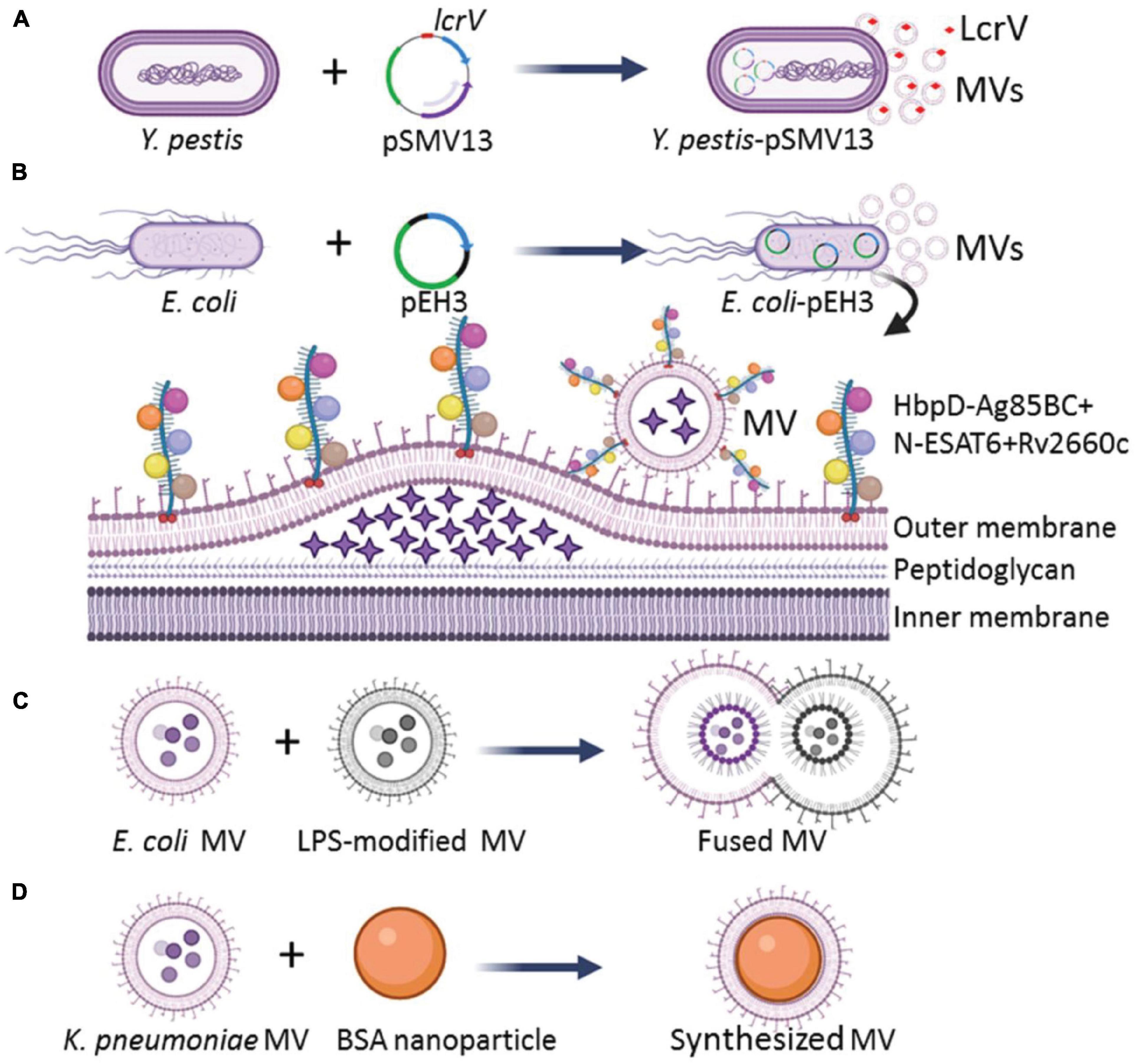
Figure 1. Strategies for engineering of bacterial MVs with enhanced immunogenicity. (A) Engineering Y. pestis by transformation of an Asd+ plasmid pSMV13 to express antigen LcrV, and MVs produced by the engineered Y. pestis loaded more LcrV than those derived from the wild-type strain. (B) E. coli transformed with a recombinant plasmid pEH3 to express HbpD+ M. tuberculosis Ag85BC+N-ESAT6 + Rv2660c chimeric antigens, and the expressed HbpD-Ag85BC+N-ESAT6-Rv2660c chimeric antigen could be effectively integrated into the MVs. The asterisks in the lumen of MVs represent bacterial proteins. (C) The MVs with enhanced immunogenicity could be prepared in vitro by the aggregation and fusion of MVs derived from different bacterial strains or species. The circle dots colored with diverse blue in the lumen of MVs represent bacterial proteins, DNA, and RNA. (D) Larger MVs could be constructed by depositing of MVs onto BSA nanoparticles.
First, overexpression of the protective antigens in a harmless bacterium was used to generate MVs with reduced toxicity. The immune response induced by Yersinia pestis MVs depends on the immune dominant LcrV. To strengthen the immunogenicity of MVs, Wang et al. (2020b) constructed a host-vector balanced lethal system based on an essential bacterial gene encoding aspartate β-semialdehyde dehydrogenase (Asd) to overexpress the LcrV antigen of Y. pestis and to reduce bacterial toxicity. The wild-type Y. pestis isolates carry a virulent plasmid pCD1 with genes to encode virulence effectors (YopE, YopJ, YopH, YopM, and YopT) and protective antigen LcrV. The pCD1-deficient Y. pestis was engineered to overexpress the LcrV antigen with Asd+ plasmid pSMV13 carrying a chimeric gene to encode the N-terminal-lactamase signal peptide fused with LcrV (Figure 1A). Increased amounts of LcrV antigen enclosed in the MVs were observed. The engineered MV-immunized mice produced high titers of IgG against LcrV and higher levels of Th1 cytokines (IFN-γ, IL-2, IL-17, and TNF-α) than those from recombinant LcrV/alhydrogel-immunized mice (Wang et al., 2020b). Therefore, the overexpression of LcrV antigen with a recombinant plasmid in pCD1-free Y. pestis can significantly enhance the immunogenicity of MVs.
Antigens of pathogens are fused with the MV-loaded proteins of otherwise harmless bacteria to mitigate toxicity but still induce a specific immune response. Several bacterial proteins, such as E. coli ClyA (a hemolytic protein that forms small pores) (Chen et al., 2010), adhesin involved in diffuse adherence (AIDA-I) auto-transporter domain (Rizos et al., 2003), hemoglobin protease (Hbp) (Daleke-Schermerhorn et al., 2014), and N. meningitidis factor H binding protein (FHbp) (Shirley and Taha, 2018; Findlow et al., 2020) were studied for their potential to carry and display heterologous antigens on the MV surface. Hbp consists of an N-terminal cleavable signal sequence, a secreted passenger domain, and a C-terminal β-domain. Mature Hbp often folds into ∼100-Å β-helical stem structure, which acts as a stable scaffold for the five salient lateral domains (D1 to D5) (Otto et al., 2005). Daleke-Schermerhorn et al. (2014) replaced the side domains (D1, D2, D4, and D5) with Mycobacterium tuberculosis antigens Ag85BC, Ag85BN, ESAT6, and Rv2660c, respectively, and the fusion protein HbpD-Ag85BC+N-ESAT6-Rv2660c was successfully expressed in the recombinant E. coli. All M. tuberculosis antigens were loaded to the surface of E. coli MVs by the Hbp auto-transporter platform (Figure 1B). The engineered MVs could induce a CD4+ T cell response against the M. tuberculosis infections in mice (Prados-Rosales et al., 2014a). van den Berg van Saparoea et al. (2018) modified the Hbp display platform with a SpyTag/SpyCatcher protein ligation system. The fusion of SpyTag to the Hbp did not impair the display on bacterial MVs, and the addition of purified proteins fused to the SpyCatcher domain could efficiently couple to Hbp-SpyTag. Thus, multiple antigen modules (SpyCatcher domain-fused) could easily be ligated to Hbp on MVs. Such engineered MVs could effectively stimulate the immune response (van den Berg van Saparoea et al., 2018, 2020). Therefore, the fusion of multiple antigens of a pathogen to the MV-contained bacterial transporters and proteins is another effective strategy to increase immunogenicity of the MVs.
The diversity of heterogeneous antigens loaded by bacterial MVs can be enhanced by the fusion of MV populations. Aggregation and fusion can be performed with MVs from different bacterial strains or species. Gnopo et al. (2020) prepared the MVs of native E. coli Nissle strain 1917 (EcN MV) and its LPS-modified strain ClearColi (CC MV). Then, the aggregation and fusion of MVs were performed by adding equal volumes of EcN MV and CC MV and inducing at low pH value of 3.6, as well as modulating ion composition and concentration to form a multifunctional vesicle (Figure 1C). The fusion efficiency approached ∼25%, and the MV-fusion strategy facilitates the design of multi-antigen vaccines that can elicit effective immune responses (Gnopo et al., 2020). A high fusion efficiency of bacterial MVs may be achieved with decreased pH and increased salt concentration (Gnopo et al., 2020), however, the optimizing conditions for an ideal fusion efficiency needed to make large-scale applications, as well as the detailed composition and architecture of the fused vesicles require further investigation.
The size, shape, and rigidity of MV nanoparticles can affect the APC uptake, antigen presentation, and activation (Benne et al., 2016). Adjustment of the properties of MV nanoparticles is also a valuable strategy to enhance the immune response. Shima et al. (2013) found that when poly γ-glutamic acid-graft-L-phenylalanine (γ-PGA-Phe) nanoparticles with sizes of 40, 100, and 200 nm were subcutaneously injected into mice, respectively, the 40 nm nanoparticles distributed more rapidly to lymph nodes of the challenged mice and were taken up by a greater number of dendritic cells (DCs) compared with the 100 and 200 nm γ-PGA-Phe nanoparticles. This finding indicates that smaller-sized nanoparticles are taken more effectively by APCs than larger-sized ones. Therefore, the immune effect of MV nanoparticles can be maximized by properly controlling the nanoparticle sizes. Wu et al. (2020) deposited the hollow-structured MVs produced by carbapenem-resistant Klebsiella pneumoniae onto 70 nm bovine serum albumin (BSA) nanoparticles (BN) to synthesize 100 nm BN-MV by a mechanical extrusion process (Figure 1D), and the BN-MV increased the expression of CD11c, CD40, CD80, CD86, and MHC-II by cell line of DC 2.4 compared with those stimulated with the wild-type bacterial MVs. Taken together, the structure optimization of MV nanoparticles can effectively improve the immune efficacy of bacterial MVs for vaccine development.
When considering MVs for medical applications, MV yield of bacteria under natural conditions is generally low, which is one of the most important factors limiting MV application (Wang et al., 2018; Cao and Liu, 2020). MV production can be increased by regulating bacterial growth, increasing the accumulation of components in the bacterial outer membrane, changing the fluidity of the cell membrane, and reducing the degree of cross-linking of peptidoglycan (Toyofuku et al., 2019).
There is increasing evidence that MV production is strongly affected by the growth conditions of bacteria. Many environmental factors influence the rate of bacterial MV formation, including media composition, growth phase, culture temperature, iron concentration, oxygen availability, and antibiotics exposure (Kulp and Kuehn, 2010; Orench-Rivera and Kuehn, 2016). For example, a study conducted by Choi et al. (2014) revealed that MV production of Pseudomonas putida KT2440 in Luria Bertani (LB) broth was increased more than three-fold than that in the minimal medium with 10 mM succinate or minimal medium with 5 mM benzoate. Anoxic cultures of P. aeruginosa PAO1 with LB media produced up to six-fold more MVs in comparison to the aerobic conditions (Toyofuku et al., 2014). In the cases of Helicobacter pylori and M. tuberculosis, MV productions were enhanced in iron limiting conditions (Keenan and Allardyce, 2000; Prados-Rosales et al., 2014b). Maredia et al. (2012) demonstrated that P. aeruginosa treated with ciprofloxacin increased MV production by 100-fold in comparison to the untreated bacteria. When treated with β-lactam antibiotics (flucloxacillin and ceftaroline), S. aureus increased the MV production in both a lysogenic and a virus-free strain. Ciprofloxacin triggered MV production in the lysogenic S. aureus isolates but not in their phage-free counterparts (Andreoni et al., 2019). Optimizing the conditions to increase bacterial MV production may be strain- or species-dependent, however, it is worth to be investigated for MV yield improvement.
In addition to environmental factors, lots of bacterial molecules were found to be associated with MV production. Genetic manipulation of certain molecules in target bacteria has been performed to greatly improve MV production (Table 2). A study by Obana et al. (2017) revealed that the spore formation pathway of Clostridium perfringens is related to MV production. The phosphorylation of a conserved aspartic acid residue (Asp58) in the Spo0A protein encoded by the spore formation regulatory gene spo0A is essential for MV production. Meanwhile, sporulation-related sensor kinases promote the MV production. Sensor kinases, such as CPE1316 and ReeS, can regulate the production of MVs through the phosphorylation of the C. perfringens Spo0A protein. MV production of spo0A knock-out strain is reduced by about five times compared with the wild-type strain, while overexpression of the spo0A gene in C. perfringens increases MV production by four times (Obana et al., 2017). In Group A Streptococcus (GAS), the CovRS two-component system negatively regulates the production of MVs. Deletion of the covRS gene in GAS increased MV production (Resch et al., 2016). In M. tuberculosis, MV production is regulated through a Pst/SenX3-RegX3 signal transduction pathway (White et al., 2018). Knock-out of the pstA1 gene, which encodes the membrane-spanning component of the phosphate-specific transport (PST) system, weakened the inhibitory effect of the PST system, and resulted in the activation of SenX3-RegX3 two-component system and an approximately 15-fold increase in MV production (White et al., 2018). Wen et al. (2021) used functional genomics to identify genes associated with MV production in Streptococcus mutans and found that sfp, bacA, bacA2, dac, and pdeA genes affected bacterial MV production. In Listeria monocytogenes, the MV yield of the sigB-mutant strain is approximately nine times lower than that of the wild-type strain (Lee et al., 2013).
The first step in the release of MVs is the budding of the cell membrane, which can be promoted by altering cell membrane fluidity and lipoproteins, which play important roles in maintaining fluidity. Schlatterer et al. (2018) found that S. aureus MVs contain many cytoplasmic proteins, and phenol-soluble modulins (PSMs) can mobilize lipoproteins from the cytoplasmic membrane to increase membrane fluidity, resulting in the formation of MVs. In the agr-deficient S. aureus strain SA113 that does not express PSMs or psmα1-4 gene-deleted strain USA300, the MV yield is substantially decreased. Overexpression of psmα1-4 genes in S. aureus SA113 with a vector pTX16-psmα1-4, the MV release of recombinant strain increased 4.2-fold compared with that of the SA113 carrying an empty pTX16 (Schlatterer et al., 2018). Meanwhile, the lack of lipoproteins can increase cytoplasmic membrane fluidity. Lipoprotein diacylglyceryl transferases (Lgt) catalyze the acylation of lipoproteins and play an important role in lipoprotein lipidation and maturation (Figure 2A; Kovacs-Simon et al., 2011). After the knock-out of lgt, the production of S. aureus MVs increases (Wang et al., 2020a). Deletion of the tolR gene in E. coli IHE3034 results in substantial increase of MV production without loss of membrane integrity (Berlanda Scorza et al., 2008).
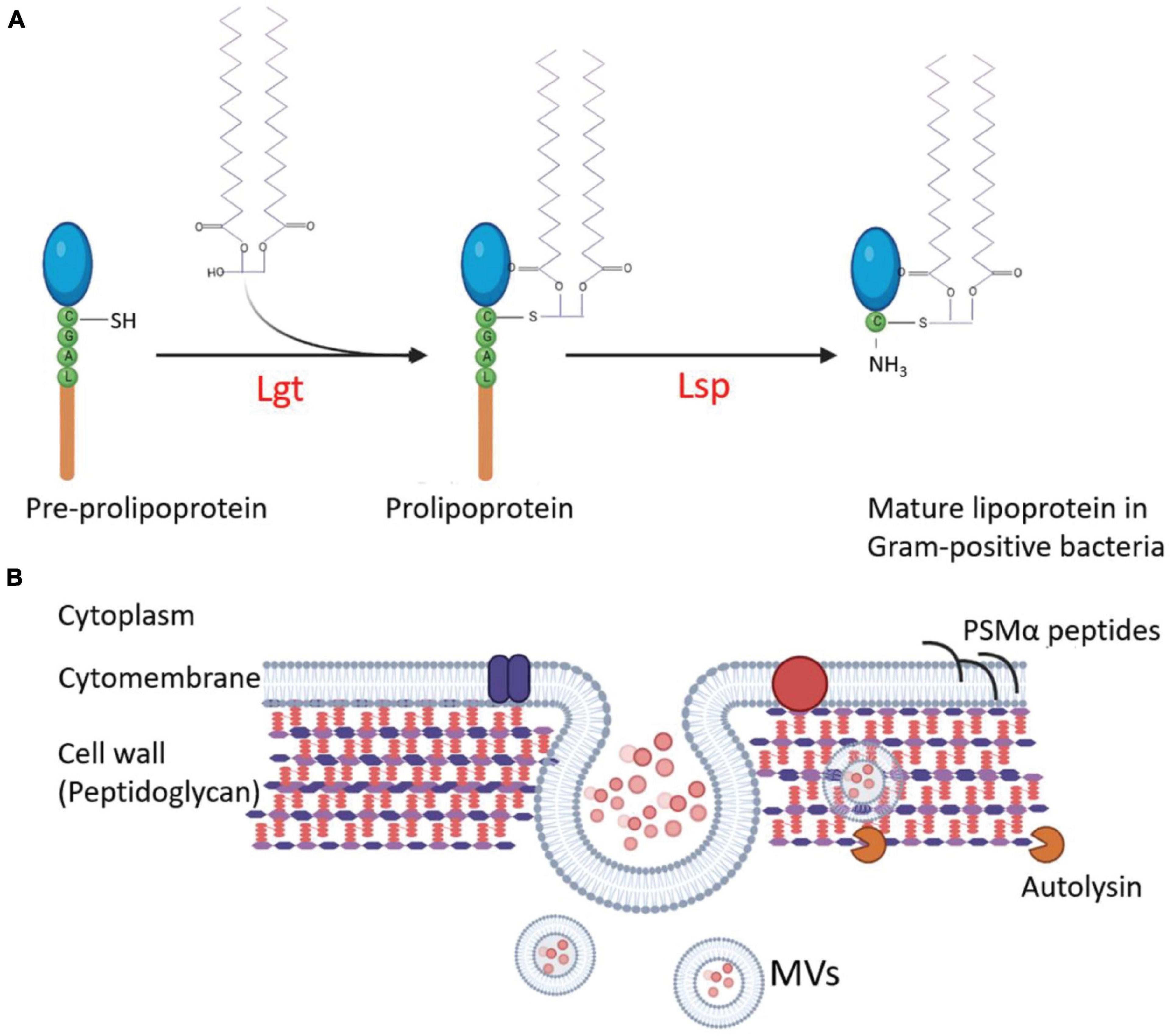
Figure 2. Regulation of the MV production in G+ bacteria. (A) Interference of lipoprotein maturation increases MV production. During lipoprotein maturation of G+ bacteria, the sulfhydryl group of prolipoprotein cysteine is modified and lipidated by lipoprotein diacylglyceryl transferase (Lgt) and then the lipoprotein signal peptidase (Lsp) cleaves the signal peptide to make cysteine a new amino terminal residue to form mature lipoproteins. Knocking out of lgt increases MV production of the S. aureus mutant. (B) The strategies involved in the regulation of MV production in S. aureus. The MVs are produced from the plasma membrane of S. aureus, and staphylococcal lipoproteins can be mobilized from the cell membrane to increase the fluidity of the membrane and promote MV formation. At the same time, MVs must traverse the highly cross-linked peptidoglycan layer to release. The degradation of the peptidoglycan layer or the reduction of cross-linking promotes MV yield. Autolysins, such as Sle1, promote MV release by hydrolyzing peptidoglycan of S. aureus cells. The circle dots colored with diverse pink in the lumen of MVs represent bacterial proteins, DNA, and RNA.
The highly cross-linked peptidoglycan (PGN) layers are the main barrier for MV release, mostly in G+ bacteria. The methods for PGN degradation or reduction of the cell wall cross-linking may promote the production of MVs. After treating S. aureus with a sublethal concentration of penicillin G (PenG), the PGN cross-linking decreased, and MV yield increased by about 10 times compared with the untreated strain (Wang et al., 2018; Andreoni et al., 2019). The deletion of genes associated with bacterial cell wall synthesis, such as pbp4 and tagO, which encodes an N-acetyl glucosamine-phosphate transferase enzyme to catalyze the biosynthesis of wall teichoic acid (a PGN-anchored glycopolymer and a major component of the S. aureus cell wall), can also lead to a decrease in the cross-linking of S. aureus PGN and a 3-fold to 4-fold increase in MV production (Wang et al., 2018). The sle1 gene product is a PGN hydrolase of S. aureus strains (Figure 2B). Deletion of sle1 in S. aureus reduced MV production. When sle1 was overexpressed, MV production could remarkably be increased (Wang et al., 2018). In addition, many endolysins produced by bacteriophages have PGN hydrolase activities. During bacteriophage biosynthesis in a bacterial cell, the endolysins can destroy the cell wall from the inside to facilitate MV release and to promote MV production (Toyofuku et al., 2017).
During MV production, the stability of MVs is a crucial hurdle for their application. The antigens in bacterial MVs could be released through the destruction of lipid membrane of MVs by surfactants or functional enzymes. Reducing MV damage may promote their accumulation and result in a high yield. Sfp is a 4′-phosphopantetheinyl transferase; it is crucial in lipopeptide surfactant biosynthesis in Bacillus subtilis. Brown et al. (2014) found that the MV yield of B. subtilis strain harboring a functional sfp gene was less than that of a strain with non-functional sfp gene. PSMs can promote MV production in S. aureus (Wang et al., 2018). However, PSMs have surfactant-like activities, they can destroy MVs at the concentrations more than 12.5 μg/ml (Schlatterer et al., 2018). This finding correlates with the fact that S. aureus MVs are mostly isolated from culture supernatants collected between 6 and 8 h of cultivation when the PSM concentration in cultures was below 12.5 μg/ml (Schlatterer et al., 2018). Therefore, avoidance of disruption is another important issue in the large-scale preparation of MVs for application.
Although the detailed mechanisms underlying the formation of bacterial MVs are still not fully elucidated (Chen Q. et al., 2020), the application of either naturally produced or engineered bacterial MVs is promising mostly in vaccine development and delivery system construction.
Vaccines are suspensions of inactivated, weakened, or fragmented toxic antigens or disease-causing agents, such as bacteria, viruses, and parasites or of antibodies or lymphocytes that are vaccinated for disease prevention. In the field of bacterial MV vaccines, N. meningitidis MV vaccine is the most widely studied. Two meningococcal serogroup B (MenB) MV vaccines, namely, MenB-4C and MenB-FHbp, have been currently approved in Europe to prevent invasive meningococcal disease (IMD) (Basta et al., 2016; Parikh et al., 2016; De Wals et al., 2017; Grogan and Roos, 2017; Rappuoli et al., 2018; Shirley and Taha, 2018). The response induced by the monovalent MV vaccine is ascribed to the immune dominant PorA, which has a high degree of sequence diversity and has limitations in covering various MenB strains. FHbp is a surface-exposed protein that is widely distributed in the meningococcal isolates and has high immunogenicity. The MenB-4C and MenB-FHbp MV vaccines could protect against infections caused by the 14 pathogenic meningococcal strains tested (Findlow et al., 2020). Li et al. (2021) prepared P. aeruginosa MV vaccine (OMV-PH) enclosed the recombinant PcrV-HitAT (PH) bivalent antigen. Vaccination with this engineered OMV-PH vaccine in BALB/c mice exhibited 70% protection from the intranasal infection with 6.5 × 106 colony forming unit of P. aeruginosa PA103, while immunization of mice with MVs in absence of PH antigen failed to afford effective protection against the same dose of PA103 challenge (Li et al., 2021).
The MVs from Streptococcus pneumoniae and M. tuberculosis are rich in bacterial lipoproteins, which can induce humoral immunity to produce antibodies against infections caused by S. pneumoniae and M. tuberculosis, respectively (Olaya-Abril et al., 2014; Prados-Rosales et al., 2014a). Vaccination with S. aureus MVs can activate Th1 and Th17 cells to induce cellular response in mice and can also stimulate B cells to produce antibody response against S. aureus infection (Choi et al., 2015). In addition, staphylococcal MVs can up-regulate the expression of co-stimulatory molecules, such as IL-12 and IL-6. Wang et al. (2018) prepared highly immunogenic attenuated MVs by knocking out the genes agr and spa and expressing non-toxic HlaH35L and LukE antigens in the engineered S. aureus strain. Such engineered MVs elicited effective protection against lethal sepsis caused by S. aureus strain USA300 LAC (Wang et al., 2018). Both naturally occurring bacterial MVs and engineered MVs can be developed as new vaccines.
The powerful delivery capabilities of MVs for exogenous antigens make bacterial MVs promising vaccine delivery vehicles. The MVs produced by G– bacteria for loading native antigens, heterologously expressed proteins, or fused molecules on the surface and in the lumen have been extensively investigated. Kesty and Kuehn (2004) tested whether a heterologously expressed protein would be delivered into E. coli MVs. They expressed an outer membrane adhesin Ail from Yersinia enterocolitica in E. coli strains DH5α, HB101, and MC4100 with a recombinant plasmid encoded Ail. The Ail was successfully delivered into the MVs of all three strains tested. The authors proposed that the expressed exogenous proteins were firstly secreted into the periplasmic space of bacteria. MVs might take in the heterologous proteins from the periplasmic space and integrate them into the vesicles during MV release and maturation. To evaluate the delivery efficiency of exogenous antigens, several MV-enriched endogenous molecules were screened as carriers to deliver vaccine candidates by a protein fusion strategy. E. coli ClyA, Hbp, AIDA, N. meningitidis FHbp, and S. aureus Mntc, Eno, and PdhB are experimentally verified bacterial molecules capable of delivery of foreign antigens (Benz and Schmidt, 1989; Chen et al., 2010; Daleke-Schermerhorn et al., 2014; Yuan et al., 2018). Huang et al. (2016) constructed a ClyA-Omp22 fusion protein in E. coli strain DH5α, and the MVs produced by the engineered bacteria contained the Omp22 antigen of Acinetobacter baumannii. The mice immunized with the engineered MVs produced a strong Omp22-specific humoral immune response that protect mice from lethal A. baumannii attacks (Huang et al., 2016). Yang et al. (2021) fused the receptor binding domain (RBD) of SARS-Cov-2 to ClyA and expressed the Cly RBD protein in E. coli BL21, then a bacterial biomimetic vesicle (BBV) was generated with MVs of the engineered bacteria extra loaded with polymerized RBD (RBD-BBV) by a high-pressure (1,200 bar) homogenization technology. Subcutaneously injection of RBD-BBVs could stimulate SARS-CoV-2-specific immune responses in murine models. Irene et al. (2019) used lipoprotein transport pathways to prepare MVs with heterologously expressed proteins. The coding genes of five S. aureus antigens, namely, HlaH35L, SpAKKAA, LukE, Csa1A, and FhuD2, were fused with the lipoprotein leader sequence, and these recombinant proteins were expressed in E. coli BL21-ΔompAΔmsbBΔpagP. Immunization with MVs derived from the engineered bacteria could protect mice from infection caused by S. aureus strain Newman (Irene et al., 2019).
The proteins carried by the MVs from G+ bacteria, such as S. pneumoniae, M. tuberculosis, and S. aureus, are highly immunogenic, and they can induce effective immune responses in animal models (Olaya-Abril et al., 2014; Prados-Rosales et al., 2014a; Choi et al., 2015; Bitto and Kaparakis-Liaskos, 2017). However, studies on the loading of heterologous antigens in G+ MVs are few, probably due to the thickened cell wall that may hamper the MV’s release from G+ bacteria. We have used a 3 × FLAG protein as an exogenous antigen molecule to test the delivery potential of S. aureus proteins by fusing several protein genes with the coding sequence of 3 × FLAG (Yuan et al., 2018). In the S. aureus strain RN4220, at least four candidates, namely, PdhB, Eno, Mntc, and PdhA, can be fused with heterologous 3 × FLAG. The fusion proteins can be displayed on MVs observed with immunoelectron microscopy. Furthermore, when NS1 and two degenerated protective antigens EDIIIconA and EDIIIconB of dengue virus were individually fused to Mntc, Eno, and PdhB encoding genes in S. aureus strain RN4220-Δagr, the resultant MVs could induce protective antibodies against all four serotypes of the dengue virus (Benz and Schmidt, 1989; Yuan et al., 2018). Chen G. et al. (2020) constructed multiple antigen vaccines by coating S. aureus MVs on the indocyanine green (ICG)-loaded magnetic mesoporous silica nanoparticles (MSN) to achieve EV/ICG/MSN, which could improve CD8+ T cell responses by activating MHC-I expression and promote CD4+ T cell response by up-regulating the expressions of costimulatory molecules, MHC-II molecules, and cytokines. Such engineered vaccines delivered by bacterial MVs could prevent skin/soft tissue infections caused by S. aureus and reduce bacterial invasion (Chen G. et al., 2020). The MV-enriched components are potential carrier molecules to load heterologous antigens to the MVs, however, the loading efficiency may be varied and must be experimentally determined during the development of a vaccine delivery vehicle.
Synthetic nanomaterials, such as polymers, liposomes, and metal nanoparticles, have been extensively studied as drug carriers (Lin et al., 2018). However, the interaction between such delivery materials and the mammal cells is ambiguous. Bacterial MVs are made up of a bilayer lipid membrane and can effectively interact with living cells by passively accumulating at the site of infection or actively targeting host immune cells, such as macrophages (Schlatterer et al., 2018; Wang et al., 2020b). MVs can be loaded with therapeutic drugs and serve as engineered treatment agents during active infection. Gao et al. (2019) found that a nanoparticle coated with bacterial MVs (NP@EV) is an active targeting carrier that can successfully be delivered to the infectious sites in vitro and in vivo. The NP@EV carriers prepared with S. aureus MVs are internalized more efficiently by the S. aureus-infected macrophage than the un-infected counterpart. NP@EV particles constructed with E. coli MVs are more effectively internalized by the E. coli-challenged macrophage than the un-infected counterpart, but not the NP@EV agents prepared with S. aureus MVs (Gao et al., 2019). In mice with S. aureus infections, the intravenously injected rifampicin-loaded NP@EV particles constructed with S. aureus MVs conferred striking therapeutic efficiency (Gao et al., 2019). The active targeting abilities of bacterial MVs to their homologous pathogen-infected cells make them a promising drug delivery platform for engineering drug nanoparticles to control bacterial infections, especially infections caused by drug-resistant superbugs. However, owing to the intrinsic complexity, size heterogeneity, and component inhomogeneity, the inherent risks of bacterial MV as a drug-loaded platform are higher than those of well-established liposomes (Herrmann et al., 2021). Drug-loading methods for MVs should be also optimized and initiated in the industrial production.
The role of bacterial MVs in anti-tumor drug delivery for treatment has attracted attention in recent years (Cao and Liu, 2020). Compared with most traditional drug delivery vehicles, MVs have several unique advantages as anti-tumor drug carriers for cancer treatment. Firstly, bacterial MVs have a large anti-tumor drug loading space like synthetic nanoparticles. The protein drugs such as fibroblast growth factors were presented on the surface of MVs (Huang et al., 2020), while siRNA drugs were loaded into MV lumen by electroporation (Gujrati et al., 2014). Secondly, nano-sized MVs are more rigid and they present less leakage during host circulation than traditional liposomes. Thirdly, bacterial MVs have natural cell targeting capabilities. MVs derived from E. coli and S. Typhimurium contain adhesin molecules which could make MVs to be recognized and endocytosed by cells in the gastrointestinal tract (Benz and Schmidt, 1989; Liu et al., 2016a). Lastly, bacterial MVs carry various immune-stimulating molecules such as LPS that can initiate anti-tumor immune response (Cao and Liu, 2020). Chen Q. et al. (2020) coated MVs produced by Salmonella on drug-loaded polymeric micelles to activate the host’s immune response for cancer immunotherapy. The engineered MVs provided effective immune protection against melanoma and significantly inhibited the growth of tumors, thereby prolonging the survival of melanoma mice (Chen Q. et al., 2020).
Bacterial MVs have been widely used to deliver different kinds of anti-tumor drugs, including chemo-therapeutic agents, thermo-therapeutic molecules, and immuno-stimulatory elements (Liu et al., 2013; Chen Q. et al., 2020; Huang et al., 2020). A clinic trial has revealed that paclitaxel-loaded bacterial MVs were safe in patients carrying solid tumors and exhibited a modest clinical treatment efficacy (Solomon et al., 2015). The doxorubicin-carried MVs could deliver drugs to the neuroblastoma in vivo (Sagnella et al., 2018). Gujrati et al. (2019) genetically modified E. coli K12 to generate MVs loaded with biopolymer-melanin, and the resulting MVs were successfully used for optoacoustic imaging and thermal therapy of mice carrying subcutaneous 4T1 mammary gland tumors. Genetic modification technology was also applied to E. coli DH5α to assemble MVs surface with murine fibroblast growth factor (FGF). The persistent autoantibodies against FGF could be stimulated in mice after three subcutaneous injections of the engineered MVs, and the growth and metastasis of TC-1 and B16F10 xenograft tumors were effectively inhibited in mice vaccinated with MVs (Huang et al., 2020). Overall, these studies above demonstrate that bacterial MVs can provide targeted loading and delivery of a range of anti-tumor drugs in a highly effective way.
The nano-sized and lipid membrane structure of bacterial MVs make them become a promising platform for broad application prospects. Genetic modifications of target bacteria have been verified to be one of the most effective strategies to optimize bacterial MVs for applications. Detoxification of bacterial MVs by consecutively deleting virulence factor genes one by one is inefficient, manipulation of pathogenicity island or global regulators that control the expression of virulence factors provides new options. Studies have shown the non-homogenous distribution of antigens and lipids in bacterial MVs. Further investigations to uncover the mechanisms of vesiculation would facilitate the generation of engineered MVs enriched in ideal components for application. Furthermore, the quantification of bacterial MVs is complicated and varies in different studies, including Braford protein assay, Lowry protein assay, phospholipid assay, KDO assay, FM1-43 assay, 14C-labeled radioactive assay, etc. (Table 2). A universal methodology to quantify bacterial MVs would be required for the fields of MV research and application. In addition, the biological safety, loading capacity, relative purity, structural homogeneity, cell-targeting ability, and tissue distribution of MV-coated particles need further investigation for creating more effective MV agents and improving human health.
RZ and XR contributed to the conception and design of the review. LQ wrote the first draft of the manuscript. YR edited the manuscript and the figures. XR, RZ, and KZ critically read and corrected the manuscript. All authors contributed to manuscript revision, editing, and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (Grant Numbers 81971565 and 82071857).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abe, K., Toyofuku, M., Nomura, N., and Obana, N. (2021). Autolysis-mediated membrane vesicle formation in Bacillus subtilis. Environ. Microbiol. 23, 2632–2647. doi: 10.1111/1462-2920.15502
Andreoni, F., Toyofuku, M., Menzi, C., Kalawong, R., Mairpady Shambat, S., et al. (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob. Agents Chemother. 63, e01439–18. doi: 10.1128/aac.01439-18
Antenucci, F., Magnowska, Z., Nimtz, M., Roesch, C., Jänsch, L., and Bojesen, A. M. (2019). Immunoproteomic characterization of outer membrane vesicles from hyper-vesiculating Actinobacillus pleuropneumoniae. Vet. Microbiol. 235, 188–194. doi: 10.1016/j.vetmic.2019.07.001
Augustyniak, D., Seredyński, R., McClean, S., Roszkowiak, J., Roszniowski, B., Smith, D. L., et al. (2018). Virulence factors of Moraxella catarrhalis outer membrane vesicles are major targets for cross-reactive antibodies and have adapted during evolution. Sci. Rep. 8:4955. doi: 10.1038/s41598-018-23029-7
Basta, N. E., Mahmoud, A. A. F., and Borrow, R. (2016). Meningococcal B vaccine during a university outbreak. N. Engl. J. Med. 375:1595. doi: 10.1056/NEJMc1610666
Benne, N., van Duijn, J., Kuiper, J., Jiskoot, W., and Slütter, B. (2016). Orchestrating immune responses: how size, shape and rigidity affect the immunogenicity of particulate vaccines. J. Control. Release. 234, 124–134. doi: 10.1016/j.jconrel.2016.05.033
Benz, I., and Schmidt, M. A. (1989). Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57, 1506–1511. doi: 10.1128/IAI.57.5.1506-1511.1989
Berlanda Scorza, F., Colucci, A. M., Maggiore, L., Sanzone, S., Rossi, O., Ferlenghi, I. P., et al. (2012). High yield production process for Shigella outer membrane particles. PLoS One 7:e35616. doi: 10.1371/journal.pone.0035616
Berlanda Scorza, F., Doro, F., Rodríguez-Ortega, M. J., Stella, M., Liberatori, S., Taddei, A. R., et al. (2008). Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DtolR IHE3034 mutant. Mol. Cell Proteomics 7, 473–485. doi: 10.1074/mcp.M700295-MCP200
Bertani, B., and Ruiz, N. (2018). Function and biogenesis of lipopolysaccharides. EcoSal. Plus 8, 1–33. doi: 10.1128/ecosalplus.ESP-0001-2018
Bielaszewska, M., Rüter, C., Bauwens, A., Greune, L., Jarosch, K. A., Steil, D., et al. (2017). Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog. 13:e1006159. doi: 10.1371/journal.ppat.1006159
Bitto, N. J., and Kaparakis-Liaskos, M. (2017). The Therapeutic benefit of bacterial membrane vesicles. Int. J. Mol. Sci. 18:1287. doi: 10.3390/ijms18061287
Brown, L., Kessler, A., Cabezas-Sanchez, P., Luque-Garcia, J. L., and Casadevall, A. (2014). Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol. Microbiol. 93, 183–198. doi: 10.1111/mmi.12650
Brown, L., Wolf, J. M., Prados-Rosales, R., and Casadevall, A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630. doi: 10.1038/nrmicro3480
Cao, Z., and Liu, J. (2020). Bacteria and bacterial derivatives as drug carriers for cancer therapy. J. Control. Release. 326, 396–407. doi: 10.1016/j.jconrel.2020.07.009
Chen, D. J., Osterrieder, N., Metzger, S. M., Buckles, E., Doody, A. M., DeLisa, M. P., et al. (2010). Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. U. S. A. 107, 3099–3104. doi: 10.1073/pnas.0805532107
Chen, G., Bai, Y., Li, Z., Wang, F., Fan, X., and Zhou, X. (2020). Bacterial extracellular vesicle-coated multi-antigenic nanovaccines protect against drug-resistant Staphylococcus aureus infection by modulating antigen processing and presentation pathways. Theranostics 10, 7131–7149. doi: 10.7150/thno.44564
Chen, Q., Bai, H., Wu, W., Huang, G., Li, Y., Wu, M., et al. (2020). Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 20, 11–21. doi: 10.1021/acs.nanolett.9b02182
Choi, C. W., Park, E. C., Yun, S. H., Lee, S. Y., Lee, Y. G., Hong, Y., et al. (2014). Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. J. Proteome Res. 13, 4298–4309. doi: 10.1021/pr500411d
Choi, D. S., Kim, D. K., Choi, S. J., Lee, J., Choi, J. P., Rho, S., et al. (2011). Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11, 3424–3429. doi: 10.1002/pmic.201000212
Choi, S. J., Kim, M. H., Jeon, J., Kim, O. Y., Choi, Y., Seo, J., et al. (2015). Active immunization with extracellular vesicles derived from Staphylococcus aureus effectively protects against Staphylococcal lung infections, mainly via Th1 cell-mediated immunity. PLoS One 10:e0136021. doi: 10.1371/journal.pone.0136021
Coelho, C., Brown, L., Maryam, M., Vij, R., Smith, D. F. Q., Burnet, M. C., et al. (2019). Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J. Biol. Chem. 294, 1202–1217. doi: 10.1074/jbc.RA118.006472
Daleke-Schermerhorn, M. H., Felix, T., Soprova, Z., Ten Hagen-Jongman, C. M., Vikström, D., Majlessi, L., et al. (2014). Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl. Environ. Microbiol. 80, 5854–5865. doi: 10.1128/AEM.01941-14
Davies, C., Taylor, A. J., Elmi, A., Winter, J., Liaw, J., Grabowska, A. D., et al. (2019). Sodium taurocholate stimulates Campylobacter jejuni outer membrane vesicle production via down-regulation of the maintenance of lipid asymmetry pathway. Front. Cell Infect. Microbiol. 9:177. doi: 10.3389/fcimb.2019.00177
DeVoe, I. W., and Gilchrist, J. E. (1973). Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J. Exp. Med. 138, 1156–1167. doi: 10.1084/jem.138.5.1156
De Wals, P., Deceuninck, G., Lefebvre, B., Tsang, R., Law, D., De Serres, G., et al. (2017). Impact of an immunization campaign to control an increased incidence of serogroup B meningococcal disease in one region of Quebec, Canada. Clin. Infect. Dis. 64, 1263–1267. doi: 10.1093/cid/cix154
Donato, G. M., Goldsmith, C. S., Paddock, C. D., Eby, J. C., Gray, M. C., and Hewlett, E. L. (2012). Delivery of Bordetella pertussis adenylate cyclase toxin to target cells via outer membrane vesicles. FEBS Lett. 586, 459–465. doi: 10.1016/j.febslet.2012.01.032
Eberlein, C., Starke, S., Doncel, ÁE., Scarabotti, F., and Heipieper, H. J. (2019). Quantification of outer membrane vesicles: a potential tool to compare response in Pseudomonas putida KT2440 to stress caused by alkanols. Appl. Microbiol. Biotechnol. 103, 4193–4201. doi: 10.1007/s00253-019-09812-0
Echeverría-Bugueño, M., Espinosa-Lemunao, R., Irgang, R., and Avendaño-Herrera, R. (2020). Identification and characterization of outer membrane vesicles from the fish pathogen Vibrio ordalii. J. Fish Dis. 43, 621–629. doi: 10.1111/jfd.13159
Eddy, J. L., Gielda, L. M., Caulfield, A. J., Rangel, S. M., and Lathem, W. W. (2014). Production of outer membrane vesicles by the plague pathogen Yersinia pestis. PLoS One 9:e107002. doi: 10.1371/journal.pone.0107002
Elmi, A., Watson, E., Sandu, P., Gundogdu, O., Mills, D. C., Inglis, N. F., et al. (2012). Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 80, 4089–4098. doi: 10.1128/IAI.00161-12
Findlow, J., Bayliss, C. D., Beernink, P. T., Borrow, R., Liberator, P., and Balmer, P. (2020). Broad vaccine protection against Neisseria meningitidis using factor H binding protein. Vaccine 38, 7716–7727. doi: 10.1016/j.vaccine.2020.08.031
Friedrich, V., Gruber, C., Nimeth, I., Pabinger, S., Sekot, G., Posch, G., et al. (2015). Outer membrane vesicles of Tannerella forsythia: biogenesis, composition, and virulence. Mol. Oral. Microbiol. 30, 451–473. doi: 10.1111/omi.12104
Frirdich, E., and Whitfield, C. (2005). Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11, 133–144. doi: 10.1179/096805105x46592
Fulsundar, S., Harms, K., Flaten, G. E., Johnsen, P. J., Chopade, B. A., and Nielsen, K. M. (2014). Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 80, 3469–3483. doi: 10.1128/AEM.04248-13
Gao, F., Xu, L., Yang, B., Fan, F., and Yang, L. (2019). Kill the real with the fake: eliminate intracellular Staphylococcus aureus using nanoparticle coated with its extracellular vesicle membrane as active-targeting drug carrier. ACS Infect. Dis. 5, 218–227. doi: 10.1021/acsinfecdis.8b00212
Gerritzen, M. J. H., Stangowez, L., van de Waterbeemd, B., Martens, D. E., Wijffels, R. H., and Stork, M. (2019). Continuous production of Neisseria meningitidis outer membrane vesicles. Appl. Microbiol. Biotechnol. 103, 9401–9410. doi: 10.1007/s00253-019-10163-z
Gill, S., Catchpole, R., and Forterre, P. (2019). Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 43, 273–303. doi: 10.1093/femsre/fuy042
Gnopo, Y. M. D., Misra, A., Hsu, H. L., DeLisa, M. P., Daniel, S., and Putnam, D. (2020). Induced fusion and aggregation of bacterial outer membrane vesicles: experimental and theoretical analysis. J. Colloid Interface Sci. 578, 522–532. doi: 10.1016/j.jcis.2020.04.068
Grogan, J., and Roos, K. (2017). Serogroup B meningococcus outbreaks, prevalence, and the case for standard vaccination. Curr. Infect. Dis. Rep. 19:30. doi: 10.1007/s11908-017-0587-4
Gujrati, V., Kim, S., Kim, S. H., Min, J. J., Choy, H. E., Kim, S. C., et al. (2014). Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 8, 1525–1537. doi: 10.1021/nn405724x
Gujrati, V., Prakash, J., Malekzadeh-Najafabadi, J., Stiel, A., Klemm, U., Mettenleiter, G., et al. (2019). Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 10:1114. doi: 10.1038/s41467-019-09034-y
Haque, S., Swami, P., and Khan, A. (2021). S. Typhi derived vaccines and a proposal for outer membrane vesicles (OMVs) as potential vaccine for typhoid fever. Microb. Pathog. 158:105082. doi: 10.1016/j.micpath.2021.105082
Herrmann, I. K., Wood, M. J. A., and Fuhrmann, G. (2021). Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 16, 748–759. doi: 10.1038/s41565-021-00931-2
Huang, W., Shu, C., Hua, L., Zhao, Y., Xie, H., Qi, J., et al. (2020). Modified bacterial outer membrane vesicles induce autoantibodies for tumor therapy. Acta Biomater. 108, 300–312. doi: 10.1016/j.actbio.2020.03.030
Huang, W., Wang, S., Yao, Y., Xia, Y., Yang, X., Li, K., et al. (2016). Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci. Rep. 6:37242. doi: 10.1038/srep37242
Irene, C., Fantappiè, L., Caproni, E., Zerbini, F., Anesi, A., Tomasi, M., et al. (2019). Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc. Natl. Acad. Sci. U. S. A. 116, 21780–21788. doi: 10.1073/pnas.1905112116
Jang, K. S., Sweredoski, M. J., Graham, R. L., Hess, S., and Clemons, W. M. Jr. (2014). Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J. Proteomics 98, 90–98. doi: 10.1016/j.jprot.2013.12.014
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 6, 1–21. doi: 10.1128/microbiolspec.GPP3-0031-2018
Jeon, H., Oh, M. H., Jun, S. H., Kim, S. I., Choi, C. W., Kwon, H. I., et al. (2016). Variation among Staphylococcus aureus membrane vesicle proteomes affects cytotoxicity of host cells. Microb. Pathog. 93, 185–193. doi: 10.1016/j.micpath.2016.02.014
Kaparakis-Liaskos, M., and Ferrero, R. L. (2015). Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 15, 375–387. doi: 10.1038/nri3837
Karthikeyan, R., Gayathri, P., Gunasekaran, P., Jagannadham, M. V., and Rajendhran, J. (2019). Comprehensive proteomic analysis and pathogenic role of membrane vesicles of Listeria monocytogenes serotype 4b reveals proteins associated with virulence and their possible interaction with host. Int. J. Med. Microbiol. 309, 199–212. doi: 10.1016/j.ijmm.2019.03.008
Keenan, J. I., and Allardyce, R. A. (2000). Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur. J. Gastroenterol. Hepatol. 12, 1267–1273. doi: 10.1097/00042737-200012120-00002
Kengmo Tchoupa, A., and Peschel, A. (2020). Staphylococcus aureus Releases Proinflammatory Membrane Vesicles To Resist Antimicrobial Fatty Acids. mSphere 5, e804–e820. doi: 10.1128/mSphere.00804-20
Kesty, N. C., and Kuehn, M. J. (2004). Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279, 2069–2076. doi: 10.1074/jbc.M307628200
Kim, S. Y., Kim, M. H., Kim, S. I., Son, J. H., Kim, S., Lee, Y. C., et al. (2019). The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii. BMC Microbiol. 19:301. doi: 10.1186/s12866-019-1679-0
Koeppen, K., Barnaby, R., Jackson, A. A., Gerber, S. A., Hogan, D. A., and Stanton, B. A. (2019). Tobramycin reduces key virulence determinants in the proteome of Pseudomonas aeruginosa outer membrane vesicles. PLoS One 14:e0211290. doi: 10.1371/journal.pone.0211290
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 12:e1005672. doi: 10.1371/journal.ppat.1005672
Kong, Q., Six, D. A., Liu, Q., Gu, L., Wang, S., Alamuri, P., et al. (2012). Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infect. Immun. 80, 3215–3224. doi: 10.1128/IAI00123-12
Kovacs-Simon, A., Titball, R. W., and Michell, S. L. (2011). Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561. doi: 10.1128/IAI.00682-10
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Kunsmann, L., Rüter, C., Bauwens, A., Greune, L., Glüder, M., Kemper, B., et al. (2015). Virulence from vesicles: novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain. Sci. Rep. 5:13252. doi: 10.1038/srep13252
Kwon, S. O., Gho, Y. S., Lee, J. C., and Kim, S. I. (2009). Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 297, 150–156. doi: 10.1111/j.1574-6968.2009.01669.x
Lee, E. Y., Choi, D. Y., Kim, D. K., Kim, J. W., Park, J. O., Kim, S., et al. (2009). Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9, 5425–5436. doi: 10.1002/pmic.200900338
Lee, J., Kim, S. H., Choi, D. S., Lee, J. S., Kim, D. K., Go, G. P., et al. (2015). Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics 15, 3331–3337. doi: 10.1002/pmic.201500037
Lee, J. H., Choi, C. W., Lee, T., Kim, S. I., Lee, J. C., and Shin, J. H. (2013). Transcription factor σB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS One 8:e73196. doi: 10.1371/journal.pone.0073196
Lee, T. Y., Kim, C. U., Bae, E. H., Seo, S. H., Jeong, D. G., Yoon, S. W., et al. (2017). Outer membrane vesicles harboring modified lipid A moiety augment the efficacy of an influenza vaccine exhibiting reduced endotoxicity in a mouse model. Vaccine 35, 586–595. doi: 10.1016/j.vaccine.2016.12.025
Li, J., Azam, F., and Zhang, S. (2016). Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ. Microbiol. 18, 3850–3866. doi: 10.1111/1462-2920.13344
Li, M., Zhou, H., Yang, C., Wu, Y., Zhou, X., Liu, H., et al. (2020). Bacterial outer membrane vesicles as a platform for biomedical applications: an update. J. Control. Release. 323, 253–268. doi: 10.1016/j.jconrel.2020.04.031
Li, P., Wang, X., Sun, X., Cimino, J., Guan, Z., and Sun, W. (2021). Recombinant Pseudomonas bio-nanoparticles induce protection against pneumonic Pseudomonas aeruginosa infection. Infect. Immun. IAI0039621. doi: 10.1128/IAI.00396-21 [Epub ahead of print].
Li, S., Chen, D. Q., Ji, L., Sun, S., Jin, Z., Jin, Z. L., et al. (2020). Development of different methods for preparing Acinetobacter baumannii outer membrane vesicles vaccine: impact of preparation method on protective efficacy. Front. Immunol. 11:1069. doi: 10.3389/fimmu.2020.01069
Li, Z. T., Zhang, R. L., Bi, X. G., Xu, L., Fan, M., Xie, D., et al. (2015). Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microb. Pathog. 81, 46–52. doi: 10.1016/j.micpath.2015.03.009
Lin, L. C. W., Chattopadhyay, S., Lin, J. C., and Hu, C. M. J. (2018). Advances and opportunities in nanoparticle- and nanomaterial-based vaccines against bacterial infections. Adv. Healthc. Mater. 7:e1701395. doi: 10.1002/adhm.201701395
Liu, J., Hsieh, C. L., Gelincik, O., Devolder, B., Sei, S., Zhang, S., et al. (2019). Proteomic characterization of outer membrane vesicles from gut mucosa-derived Fusobacterium nucleatum. J. Proteomics 195, 125–137. doi: 10.1016/j.jprot.2018.12.029
Liu, Q., Liu, Q., Zhao, X., Liu, T., Yi, J., Liang, K., et al. (2016b). Immunogenicity and cross-protective efficacy induced by outer membrane proteins from Salmonella Typhimurium mutants with truncated LPS in mice. Int. J. Mol. Sci. 17:416. doi: 10.3390/ijms17030416
Liu, Q., Liu, Q., Yi, J., Liang, K., Liu, T., Roland, K. L., et al. (2016a). Outer membrane vesicles derived from Salmonella Typhimurium mutants with truncated LPS induce cross-protective immune responses against infection of Salmonella enterica serovars in the mouse model. Int. J. Med. Microbiol. 306, 697–706. doi: 10.1016/j.ijmm.2016.08.004
Liu, Q., Yeo, W. S., and Bae, T. (2016c). The SaeRS two-component system of Staphylococcus aureus. Genes 7:81. doi: 10.3390/genes7100081
Liu, Q., Yi, J., Liang, K., Zhang, X., and Liu, Q. (2017). Salmonella Choleraesuis outer membrane vesicles: proteomics and immunogenicity. J. Basic Microbiol. 57, 852–861. doi: 10.1002/jobm.201700153
Liu, Y., Ai, K., Liu, J., Deng, M., He, Y., and Lu, L. (2013). Dopamine-melanin colloidal nanospheres: an efficient near-infrared photothermal therapeutic agent for in vivo cancer therapy. Adv. Mater. 25, 1353–1359. doi: 10.1002/adma.201204683
Manning, A. J., and Kuehn, M. J. (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. doi: 10.1186/1471-2180-11-258
Mantri, C. K., Chen, C. H., Dong, X., Goodwin, J. S., Pratap, S., Paromov, V., et al. (2015). Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. Microbiologyopen 4, 53–65. doi: 10.1002/mbo3.221
Maredia, R., Devineni, N., Lentz, P., Dallo, S. F., Yu, J., Guentzel, N., et al. (2012). Vesiculation from Pseudomonas aeruginosa under SOS. ScientificWorldJournal. 2012:402919. doi: 10.1100/2012/402919
McBroom, A. J., and Kuehn, M. J. (2007). Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558. doi: 10.1111/j.1365-2958.2006.05522.x
McCaig, W. D., Koller, A., and Thanassi, D. G. (2013). Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 195, 1120–1132. doi: 10.1128/JB.02007-12
McCaig, W. D., Loving, C. L., Hughes, H. R., and Brockmeier, S. L. (2016). Characterization and vaccine potential of outer membrane vesicles produced by Haemophilus parasuis. PLoS One 11:e0149132. doi: 10.1371/journal.pone.0149132
McMahon, K. J., Castelli, M. E., Vescovi, E. G., and Feldman, M. F. (2012). Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J. Bacteriol. 194, 3241–3249. doi: 10.1128/JB.00016-12
Mondal, A., Tapader, R., Chatterjee, N. S., Ghosh, A., Sinha, R., Koley, H., et al. (2016). Cytotoxic and inflammatory responses induced by outer membrane vesicle-associated biologically active proteases from Vibrio cholerae. Infect. Immun. 84, 1478–1490. doi: 10.1128/IAI.01365-15
Monnappa, A. K., Bari, W., Seo, J. K., and Mitchell, R. J. (2018). The cytotoxic necrotizing factor of Yersinia pseudotuberculosis (CNFy) is carried on extracellular membrane vesicles to host cells. Sci. Rep. 8:14186. doi: 10.1038/s41598-018-32530-y
Moon, D. C., Choi, C. H., Lee, J. H., Choi, C. W., Kim, H. Y., Park, J. S., et al. (2012). Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J. Microbiol. 50, 155–160. doi: 10.1007/s12275-012-1589-4
Mullaney, E., Brown, P. A., Smith, S. M., Botting, C. H., Yamaoka, Y. Y., Terres, A. M., et al. (2009). Proteomic and functional characterization of the outer membrane vesicles from the gastric pathogen Helicobacter pylori. Proteomics Clin. Appl. 3, 785–796. doi: 10.1002/prca.200800192
Obana, N., Nakao, R., Nagayama, K., Nakamura, K., Senpuku, H., and Nomura, N. (2017). Immunoactive clostridial membrane vesicle production is regulated by a sporulation factor. Infect. Immun. 85, e00096–17. doi: 10.1128/IAI.00096-17
Olaya-Abril, A., Prados-Rosales, R., McConnell, M. J., Martín-Peña, R., González-Reyes, J. A., Jiménez-Munguía, I., et al. (2014). Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteomics 106, 46–60. doi: 10.1016/j.jprot.2014.04.023
Olofsson, A., Vallström, A., Petzold, K., Tegtmeyer, N., Schleucher, J., Carlsson, S., et al. (2010). Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 77, 1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x
Orench-Rivera, N., and Kuehn, M. J. (2016). Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol. 18, 1525–1536. doi: 10.1111/cmi.12676
Otto, B. R., Sijbrandi, R., Luirink, J., Oudega, B., Heddle, J. G., Mizutani, K., et al. (2005). Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280, 17339–17345. doi: 10.1074/jbc.M412885200
Parikh, S. R., Andrews, N. J., Beebeejaun, K., Campbell, H., Ribeiro, S., Ward, C., et al. (2016). Effectiveness and impact of a reduced infant schedule of 4CMenB vaccine against group B meningococcal disease in England: a national observational cohort study. Lancet 388, 2775–2782. doi: 10.1016/s0140-6736 (16)31921-3
Pasqua, M., Zennaro, A., Trirocco, R., Fanelli, G., Micheli, G., Grossi, M., et al. (2021). Modulation of OMV Production by the lysis module of the DLP12 defective prophage of Escherichia coli K12. Microorganisms 9:369. doi: 10.3390/microorganisms9020369
Pathirana, R. D., and Kaparakis-Liaskos, M. (2016). Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol. 18, 1518–1524. doi: 10.1111/cmi.12658
Pérez-Cruz, C., Cañas, M. A., Giménez, R., Badia, J., Mercade, E., Baldomà, L., et al. (2016). Membrane vesicles released by a hypervesiculating Escherichia coli Nissle 1917 tolR mutant are highly heterogeneous and show reduced capacity for epithelial cell interaction and entry. PLoS One 11:e0169186. doi: 10.1371/journal.pone.0169186
Prados-Rosales, R., Carreño, L. J., Batista-Gonzalez, A., Baena, A., Venkataswamy, M. M., Xu, J., et al. (2014a). Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5, e01921–14. doi: 10.1128/mBio.01921-14
Prados-Rosales, R., Weinrick, B. C., Piqué, D. G., Jacobs, W. R., Casadevall, A., and Rodriguez, G. M. (2014b). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196, 1250–1256. doi: 10.1128/JB.01090-13
Raetz, C. R. H., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Ranallo, R. T., Kaminski, R. W., George, T., Kordis, A. A., Chen, Q., Szabo, K., et al. (2010). Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infect. Immun. 78, 400–412. doi: 10.1128/IAI.00533-09
Rappuoli, R., Pizza, M., Masignani, V., and Vadivelu, K. (2018). Meningococcal B vaccine (4CMenB): the journey from research to real world experience. Expert Rev. Vaccines 17, 1111–1121. doi: 10.1080/14760584.2018.1547637
Rath, P., Huang, C., Wang, T., Wang, T., Li, H., Prados-Rosales, R., et al. (2013). Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 110, E4790–E4797. doi: 10.1073/pnas.1320118110
Reimer, S. L., Beniac, D. R., Hiebert, S. L., Booth, T. F., Chong, P. M., Westmacott, G. R., et al. (2021). Comparative analysis of outer membrane vesicle isolation methods with an Escherichia coli tolA mutant reveals a hypervesiculating phenotype with outer-inner membrane vesicle content. Front. Microbiol. 12:628801. doi: 10.3389/fmicb.2021.628801
Resch, U., Tsatsaronis, J. A., Le Rhun, A., Stübiger, G., Rohde, M., Kasvandik, S., et al. (2016). A two-component regulatory system impacts extracellular membrane-derived vesicle production in Group A Streptococcus. mBio 7, e00207–16. doi: 10.1128/mBio.00207-16
Rivera, J., Cordero, R. J., Nakouzi, A. S., Frases, S., Nicola, A., and Casadevall, A. (2010). Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107, 19002–19007. doi: 10.1073/pnas.1008843107
Rizos, K., Lattemann, C. T., Bumann, D., Meyer, T. F., and Aebischer, T. (2003). Autodisplay: efficacious surface exposure of antigenic UreA fragments from Helicobacter pylori in Salmonella vaccine strains. Infect. Immun. 71, 6320–6328. doi: 10.1128/IAI.71.11.6320-6328.2003
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7:10515. doi: 10.1038/ncomms10515
Roy, K., Hamilton, D. J., Munson, G. P., and Fleckenstein, J. M. (2011). Outer membrane vesicles induce immune responses to virulence proteins and protect against colonization by enterotoxigenic Escherichia coli. Clin. Vaccine Immunol. 18, 1803–1808. doi: 10.1128/CVI.05217-11
Sagnella, S. M., Trieu, J., Brahmbhatt, H., MacDiarmid, J. A., MacMillan, A., Whan, R. M., et al. (2018). Targeted doxorubicin-loaded bacterially derived nano-cells for the treatment of neuroblastoma. Mol. Cancer Ther. 17, 1012–1023. doi: 10.1158/1535-7163.MCT-17-0738
Schlatterer, K., Beck, C., Hanzelmann, D., Lebtig, M., Fehrenbacher, B., Schaller, M., et al. (2018). The mechanism behind bacterial lipoprotein release: phenol-soluble modulins mediate toll-like receptor 2 activation via extracellular vesicle release from Staphylococcus aureus. mBio 9, e01851–18. doi: 10.1128/mBio.01851-18
Shima, F., Uto, T., Akagi, T., Baba, M., and Akashi, M. (2013). Size effect of amphiphilic poly (γ-glutamic acid) nanoparticles on cellular uptake and maturation of dendritic cells in vivo. Acta Biomater. 9, 8894–8901. doi: 10.1016/j.actbio.2013.06.010
Shirley, M., and Taha, M. K. (2018). MenB-FHbp meningococcal group B vaccine (Trumenba®): a review in active immunization in individuals aged = 10 Years. Drugs 78, 257–268. doi: 10.1007/s40265-018-0869-7
Siljamäki, P., Varmanen, P., Kankainen, M., Sukura, A., Savijoki, K., and Nyman, T. A. (2014). Comparative exoprotein profiling of different Staphylococcus epidermidis strains reveals potential link between nonclassical protein export and virulence. J. Proteome Res. 13, 3249–3261. doi: 10.1021/pr500075j
Solomon, B. J., Desai, J., Rosenthal, M., McArthur, G. A., Pattison, S. T., Pattison, S. L., et al. (2015). A first-time-in-human phase I clinical trial of bispecific antibody-targeted, paclitaxel-packaged bacterial minicells. PLoS One 10:e0144559. doi: 10.1371/journal.pone.0144559
Thay, B., Wai, S. N., and Oscarsson, J. (2013). Staphylococcus aureus α-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One 8:e54661. doi: 10.1371/journal.pone.0054661
Tian, H., Li, B., Xu, T., Yu, H., Chen, J., Yu, H., et al. (2021). Outer membrane vesicles derived from Salmonella Typhimurium can deliver Shigella flexneri 2a O-polysaccharide antigen to prevent Shigella flexneri 2a infection in mice. Appl. Environ. Microbiol. AEM0096821. doi: 10.1128/AEM.00968-21 [Online ahead of print]
Toyofuku, M., Morinaga, K., Hashimoto, Y., Uhl, J., Shimamura, H., Inaba, H., et al. (2017). Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509. doi: 10.1038/ismej.2017.13
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Toyofuku, M., Zhou, S., Sawada, I., Takaya, N., Uchiyama, H., and Nomura, N. (2014). Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 16, 2927–2938. doi: 10.1111/1462-2920.12260
Tzipilevich, E., Habusha, M., and Ben-Yehuda, S. (2017). Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168, 186–199. doi: 10.1016/j.cell.2016.12.003
van de Waterbeemd, B., Streefland, M., van der Ley, P., Zomer, B., van Dijken, H., Martens, D., et al. (2010). Improved OMV vaccine against Neisseria meningitidis using genetically engineered strains and a detergent-free purification process. Vaccine 28, 4810–4816. doi: 10.1016/j.vaccine.2010.04.082
van den Berg van Saparoea, H. B., Houben, D., de Jonge, M. I., Jong, W. S. P., and Luirink, J. (2018). Display of recombinant proteins on bacterial outer membrane vesicles by using protein ligation. Appl. Environ. Microbiol. 84, e02567–17. doi: 10.1128/AEM.02567-17
van den Berg van Saparoea, H. B., Houben, D., Kuijl, C., Luirink, J., and Jong, W. S. P. (2020). Combining protein ligation systems to expand the functionality of semi-synthetic outer membrane vesicle nanoparticles. Front. Microbiol. 11:890. doi: 10.3389/fmicb.2020.00890
Veith, P. D., Chen, Y. Y., Gorasia, D. G., Chen, D., Glew, M. D., O’Brien-Simpson, N. M., et al. (2014). Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J. Proteome Res. 13, 2420–2432. doi: 10.1021/pr401227e
Vipond, C., Suker, J., Jones, C., Tang, C., Feavers, I. M., and Wheeler, J. X. (2006). Proteomic analysis of a meningococcal outer membrane vesicle vaccine prepared from the group B strain NZ98/254. Proteomics 6, 3400–3413. doi: 10.1002/pmic.200500821
Wagner, T., Joshi, B., Janice, J., Askarian, F., Škalko-Basnet, N., Hagestad, O. C., et al. (2018). Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteomics 187, 28–38. doi: 10.1016/j.jprot.2018.05.017
Wai, S. N., Lindmark, B., Söderblom, T., Takade, A., Westermark, M., Oscarsson, J., et al. (2003). Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115, 25–35. doi: 10.1016/s0092-8674(03)00754-2
Wang, X., Eagen, W. J., and Lee, J. C. (2020a). Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proc. Natl. Acad. Sci. U. S. A. 117, 3174–3184. doi: 10.1073/pnas.1915829117
Wang, X., Singh, A. K., Zhang, X., and Sun, W. (2020b). Induction of protective antiplague immune responses by self-adjuvanting bionanoparticles derived from engineered Yersinia pestis. Infect. Immun. 88, e00081–20. doi: 10.1128/IAI.00081-20
Wang, X., Thompson, C. D., Weidenmaier, C., and Lee, J. C. (2018). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 9:1379. doi: 10.1038/s41467-018-03847-z
Wen, Z. T., Jorgensen, A. N., Huang, X., Ellepola, K., Chapman, L., Wu, H., et al. (2021). Multiple factors are involved in regulation of extracellular membrane vesicle biogenesis in Streptococcus mutans. Mol. Oral Microbiol. 36, 12–24. doi: 10.1111/omi.12318
Wessel, A. K., Liew, J., Kwon, T., Marcotte, E. M., and Whiteley, M. (2013). Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J. Bacteriol. 195, 213–219. doi: 10.1128/JB.01253-12
White, D. W., Elliott, S. R., Odean, E., Bemis, L. T., and Tischler, A. D. (2018). Mycobacterium tuberculosis Pst/SenX3-RegX3 regulates membrane vesicle production independently of ESX-5 activity. mBio 9, e00778–18. doi: 10.1128/mBio.00778-18
Whitfield, C., Kaniuk, N., and Frirdich, E. (2003). Molecular insights into the assembly and diversity of the outer core oligosaccharide in lipopolysaccharides from Escherichia coli and Salmonella. J. Endotoxin Res. 9, 244–249. doi: 10.1179/096805103225001440
Wu, G., Ji, H., Guo, X., Li, Y., Ren, T., Dong, H., et al. (2020). Nanoparticle reinforced bacterial outer-membrane vesicles effectively prevent fatal infection of carbapenem-resistant Klebsiella pneumoniae. Nanomedicine 24:102148. doi: 10.1016/j.nano.2019.102148
Yang, A., Hua, L., Yang, M., Liu, S. Q., Shen, J., Li, W., et al. (2021). RBD-Modified Bacterial Vesicles Elicited Potential Protective Immunity against SARS-CoV-2. Nano Lett. 21, 5920–5930. doi: 10.1021/acs.nanolett.1c00680
Yang, J., Li, B., Cai, R., Song, S., Gou, H., Chu, P., et al. (2020). Advance in effects of LPS modification on virulence of G– bacteria and biological characteristics of OMVs. Prog. Vet. Med. 41, 98–103. doi: 10.16437/j.cnki.1007-5038.2020.02.019
Yoon, H., Ansong, C., Adkins, J. N., and Heffron, F. (2011). Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79, 2182–2192. doi: 10.1128/IAI.01277-10
Yuan, J., Yang, J., Hu, Z., Yang, Y., Shang, W., Hu, Q., et al. (2018). Safe staphylococcal platform for the development of multivalent nanoscale vesicles against viral infections. Nano Lett. 18, 725–733. doi: 10.1021/acs.nanolett.7b03893
Keywords: extracellular vesicles, vesicle production, vesicle immunogenicity, vaccine, delivery system, genetic modification
Citation: Qiao L, Rao Y, Zhu K, Rao X and Zhou R (2021) Engineered Remolding and Application of Bacterial Membrane Vesicles. Front. Microbiol. 12:729369. doi: 10.3389/fmicb.2021.729369
Received: 23 June 2021; Accepted: 31 August 2021;
Published: 08 October 2021.
Edited by:
Elisa Michelini, University of Bologna, ItalyReviewed by:
Mariola J. Edelmann, University of Florida, United StatesCopyright © 2021 Qiao, Rao, Zhu, Rao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiancai Rao, cmFveGlhbmNhaUAxMjYuY29t; Renjie Zhou, emhvdV9yakBhbGl5dW4uY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.