
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 September 2021
Sec. Terrestrial Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.729047
This article is part of the Research Topic Microbial Communities and Functions Contribute to Plant Performance under Various Stresses View all 25 articles
 Ming Yuan1
Ming Yuan1 Taobing Yu2,3
Taobing Yu2,3 Qihan Shi2,3
Qihan Shi2,3 Dongwei Han1
Dongwei Han1 Kanchao Yu1
Kanchao Yu1 Lianxia Wang1
Lianxia Wang1 Shurong Wang1
Shurong Wang1 Hao Xiang4
Hao Xiang4 Ronghui Wen5
Ronghui Wen5 Hai Nian2,3*
Hai Nian2,3* Tengxiang Lian2,3*
Tengxiang Lian2,3*The continuous planting of soybeans leads to soil acidification, aggravation of soil-borne diseases, reduction in soil enzyme activity, and accumulation of toxins in the soil. Microorganisms in the rhizosphere play a very important role in maintaining the sustainability of the soil ecosystem and plant health. In this study, two soybean genotypes, one bred for continuous cropping and the other not, were grown in a Mollisol in northeast China under continuous cropping for 7 and 36years in comparison with soybean–maize rotation, and microbial communities in the rhizosphere composition were assessed using high-throughput sequencing technology. The results showed that short- or long-term continuous cropping had no significant effect on the rhizosphere soil bacterial alpha diversity. Short-term continuous planting increased the number of soybean cyst nematode (Heterodera glycines), while long-term continuous planting reduced these numbers. There were less soybean cyst nematodes in the rhizosphere of the tolerant genotypes than sensitive genotypes. In addition, continuous cropping significantly increased the potential beneficial bacterial populations, such as Pseudoxanthomonas, Nitrospira, and Streptomyces compared to rotation and short-term continuous cropping, suggesting that long-term continuous cropping of soybean shifts the microbial community toward a healthy crop rotation system. Soybean genotypes that are tolerant to soybean might recruit some microorganisms that enhance the resistance of soybeans to long-term continuous cropping. Moreover, the network of the two genotypes responded differently to continuous cropping. The tolerant genotype responded positively to continuous cropping, while for the sensitive genotype, topology analyses on the instability of microbial community in the rhizosphere suggested that short periods of continuous planting can have a detrimental effect on microbial community stability, although this effect could be alleviated with increasing periods of continuous planting.
As one of the most important soil resources in China, Mollisols in the northeast play a crucial role in maintaining domestic food demand. Soybean [Glycine max (L.) Merrill] is one of the most important food crops in the world, providing a large amount of oil and protein for humans and animals (Chigen et al., 2018; Liu et al., 2020). There is a large demand for soybeans, especially in China (Liu et al., 2020). Due to the limited arable land, climatic conditions, and large proportion of arable land with other crops, soybeans are often continuously planted in this region (Liu et al., 2012). On some farms, soybeans have been planted continuously for 40years or even longer (Liu et al., 2020). The continuous planting of soybeans leads to soil acidification, aggravation of soil-borne diseases, reduction in soil enzyme activity, and accumulation of toxins in the soil (Zhan et al., 2004; Yan et al., 2012; Pérez-Brandán et al., 2014; Bai et al., 2015). Several studies have clarified that these changes are significantly related to biological factors in the soil (Ji et al., 1996; Dias et al., 2015; Liu et al., 2020).
Rhizosphere microorganisms play a very important role in maintaining the sustainability of the soil ecosystem and plant health (Waldrop et al., 2000; Avidano et al., 2005). Different cropping systems can significantly change the microbial community structure (Meriles et al., 2009; Zhou et al., 2018; Liu et al., 2019). However, these changes depend on the type of cropping system, soil type, and crop species. For instance, both Tang et al. (2009) and Zhu et al. (2014) found that, compared with crop rotation systems, the abundance of Actinobacteria significantly decreased under the continuous cropping of soybean. Xu et al. (1995) found that continuous cropping decreased the abundance of Penicillium sp., while it increased the abundance of Fusarium sp. However, another study showed that, compared with crop rotation, continuous soybean cropping did not change the microbial community structure (Hu and Wang, 1996). The inconsistencies between these studies are mainly due to the differences in soil types, research methods, crop rotation systems, and years of continuous cropping. At the same time, the mechanisms associated with continuous cropping obstacles are complex and need to be explored in greater depth under different conditions.
Soybean is one of the most sensitive crops to continuous cropping. Studies have shown that continuous cropping of soybean for 3years could reduce yield by approximately 30% (Liu et al., 2020). In addition, compared to crop rotation, soybean root rot diseases and cyst nematodes in soybean fields increased significantly in a short-term continuously cropped system (Cai et al., 2015). However, root rot and cystic nematode disease in soybean fields might be weakened after long-term continuous cropping (Song et al., 2017). This might be attributed to the fact that long-term continuous cropping could increase the population of beneficial microorganisms, which could help inhibit the colonization and development of pathogens in the soil (Wei et al., 2015; Liu et al., 2020). For example, Liu et al. (2020) found that 13years of long-term continuously cropped soybean increased beneficial bacteria, such as Bradyrhizobium sp., Gemmatimonas sp., Mortierella sp., and Paecilomyces sp. and decreased the pathogenic fungi of Fusarium sp. However, some researchers also found that long-term continuous of soybeans may reduce the populations of beneficial microorganisms such as Trichoderma, Colloids, and Pseudomonas fluorescens (Pérez-Brandán et al., 2014). Therefore, the change in the microbial community caused by continuous or rotational cropping is crucial to reveal the mechanisms underlying obstacles to continuous soybean cropping. In addition, the inconsistency of these studies indicates that more research on the soil microbial community needs to be performed.
The rhizosphere microbial community of crops is also affected by genotypes (Kaisermann et al., 2017; Lian et al., 2019). Resistant genotypes can recruit some beneficial microorganisms to help the host resist various stresses (Lebeis et al., 2015; Kwak et al., 2018). For example, tomato genotypes that are resistant to Fusarium wilt can recruit a large number of Flavobacterium to help alleviate the symptoms of Fusarium wilt (Kwak et al., 2018). Our previous studies have also shown that aluminium-resistant soybeans can recruit Tumebacillus and Burkholderia and alleviate aluminium toxicity (Lian et al., 2019). Regarding the continuous cropping of soybean, although microorganisms are known to play a very important role in alleviating or aggravating continuous cropping obstacles, the comparison of the microbial community in the rhizosphere among different genotypes has not been reported. Understanding the characteristics of the rhizosphere microbial community of continuous-resistant genotype soybeans can help us understanding how those resistant genotypes could better adapt to continuous cropping.
Here, we aimed to characterize the rhizosphere microorganisms of different soybean genotypes, one bred for continuous cropping and one not, grown in a Mollisol in northeast China under continuous cropping for 7 and 36years with soybean–maize rotation. Based on the higher adaptability of continuous cropping-tolerant (CC-T) soybean genotypes to continuous cropping, we hypothesized that (1) the bacterial diversity in the rhizosphere of CC-T genotypes would be higher than that of continuous cropping-sensitive (CC-S) genotypes and that (2) the response of bacterial community structure to continuous cropping between CC-T and CC-S soybean genotypes would be different. CC-T may recruit beneficial microbial species to alleviate the occurrence of soil-borne diseases.
To assess the responses of continuous cropping-tolerant and cropping-sensitive soybean genotypes soil used in this study was a Mollisol, according to USDA soil taxonomy. Soybeans used in the experiment included Qinong1 and Qinong5, which were continuous cropping-tolerant (CCT), and Heihe43 and Henong76, which were continuous cropping-sensitive (CCS).
Continuous cropping of two soybean genotypes for 7 and 36years, as well as treatment of soybean-corn rotations for 10years, were selected for this study. The total area of the experimental field (47°15′N, 123°40′E) was 2ha, including 1.3ha of 36-year continuous cropping and 0.3ha each of 7-year continuous and rotation cropping (Supplementary Figure S1). A total of 72 rhizosphere soil samples (six treatments×12 replicates) were collected at the flowering stage of soybean on July 15, 2020. All rhizosphere soil samples were collected by gently shaking the plant root to remove loosely attached soil, and then the soil adhering to the root system was transported to an aseptic bag. Two grams of rhizosphere soil from each sample was placed in a sterilized microcentrifuge tube and stored at −80°C for DNA extraction. The shoots and roots were separated for biomass measurements. All the soybean cyst nematodes (Heterodera glycines) and nodules from the roots were counted after removal from the root system. In brief, after the soybean cyst nematode growth broke through the root surface of soybean (35days after seedling), 10 soybean plants were randomly selected for the soybean cyst nematodes counting. The yield per square meter was measured after soybean maturity.
In total, 72 rhizosphere soil samples were used for sequencing. Soil total DNA was extracted using the Fast DNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA). Primer sequences of 341F/805R were used to amplify the V4 (V3–V4) hypervariable regions of the 16S rRNA. The PCR products were purified and then sequenced using the MiSeq Illumina platform (Illumina, United States) at Icongene (Wuhan) Gene Technology Co., Ltd. (Wuhan, China). All sequences were deposited into the GenBank short-read archive PRJNA732989.
After sequencing, the raw FASTQ files were processed using QIIME Pipeline Version 1.19.1. Briefly, all sequence reads were assigned to each sample based on the barcodes. Sequences with low quality (length<200bp and average base quality score<20) were removed before further analysis. The chimera of trimmed sequences was detected and removed using the UCHIME algorithm (Edgar et al., 2011). The sequences were phylogenetically assigned according to their best matches to sequences in the RDP database using the RDP classifier (Cole et al., 2009). Operational taxonomic units (OTUs) were classified at 97% sequence similarity using CD-HIT (Li and Godzik, 2015). Chao1 richness and Shannon’s diversity index were calculated in QIIME. Constrained principal coordinate analysis (CPCoA) and significance tests (Adonis test and mantel test) were performed in program R version 3.5.1 for Windows with the “vegan” package. Ternary plots were constructed to show the abundance comparison of OTUs (>5%) for rotation system, and continuous cropping for 7 and 36years of the two genotypes soybeans, using the “vcd” package (Friendly and Meyer, 2015). One-way analysis of variance (ANOVA) was used in GenStat 13 (VSN International, Hemel Hempstead, United Kingdom) to assess the relative abundances of different taxonomic levels of bacteria. Additionally, a Pearson bivariate correlation analysis was performed to access the correlations between pH, soybean cyst nematode, yield, and the genera with higher relative abundance.
Core bacteria, which contain a list of OTUs observed in 60% of all rhizosphere samples, were obtained by MicrobiomeAnalyst (Chong et al., 2020). Bacterial co-occurrence networks were analyzed for each cropping system. To study the network structure of OTUs with high abundance, we selected OTUs with more than 0.2% relative abundance to calculate Spearman’s rank correlation coefficients. The correlations between OTUs were selected at p<0.05 and Spearman’s correlation coefficient of more than 0.8 (Mendes et al., 2018). The nodes and edges represent bacterial OTUs and the correlations between bacterial OTUs, respectively. Statistical analyses were calculated using the “psych” package in R and then visualized in Gephi (Jiang et al., 2017). Keystone species were defined according to high node degree, high betweenness centrality, and high closeness centrality (Berry and Widder, 2014; Agler et al., 2016). The NetShift analysis was performed using the NetShift Software tool1 to compare the networks of different treatments and find the “driver microbes” between the treatment of soybean-corn rotations and 7, 36years continuous cropping for the sensitive and tolerant genotypes, respectively (Kuntal et al., 2019).
Both cropping systems and soybean genotypes significantly affected the soybean cyst nematode number and yield (Table 1). There was a higher cyst nematode number in the sensitive genotypes than in the tolerant genotypes. In particular, for the sensitive genotypes, the soybean cyst nematode number significantly increased in the 7-year continuous cropping field but markedly decreased in the 36-year continuous cropping field. However, continuous cropping had no significant effect on the tolerant genotypes. In addition, the yield increased in the 7-year continuous cropping field but decreased in the 36-year continuous cropping field, with a higher yield for the tolerant genotypes than the sensitive genotypes (Table 1).
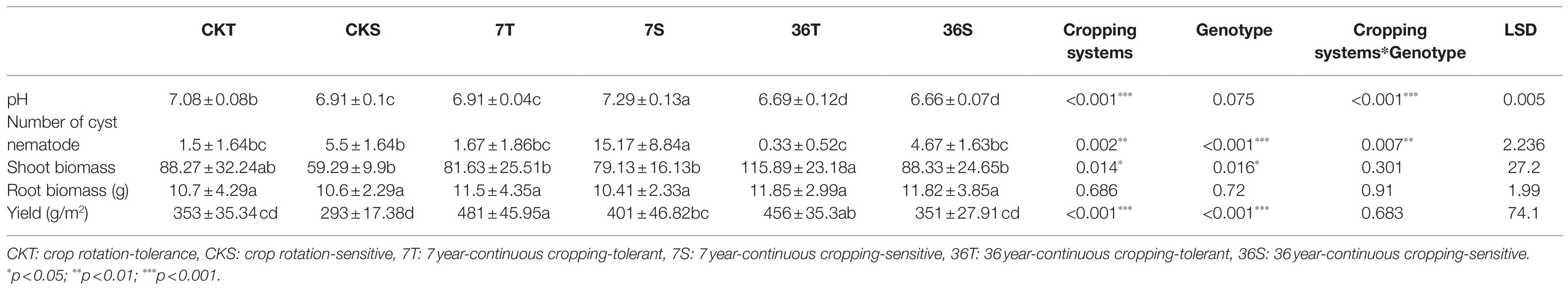
Table 1. Soil pH, number of cyst nematode, shoot and root biomass, and yield of different treatments.
Both cropping systems and soybean genotypes had no significant effect on the alpha diversity of rhizobacterial communities (one-way ANOVA, p>0.05; Figures 1A,B). Regarding beta diversity, CPCoA and two-way PERMANOVA revealed that cropping systems and soybean genotypes interactively affected the rhizobacterial communities (PERMANOVA, p<0.05; Figure 2; Supplementary Figure S2; Table 2). In more detail, rhizobacterial community structures differed between tolerant and sensitive soybean genotypes irrespective of cropping systems (Supplementary Figure S3; Table 2).
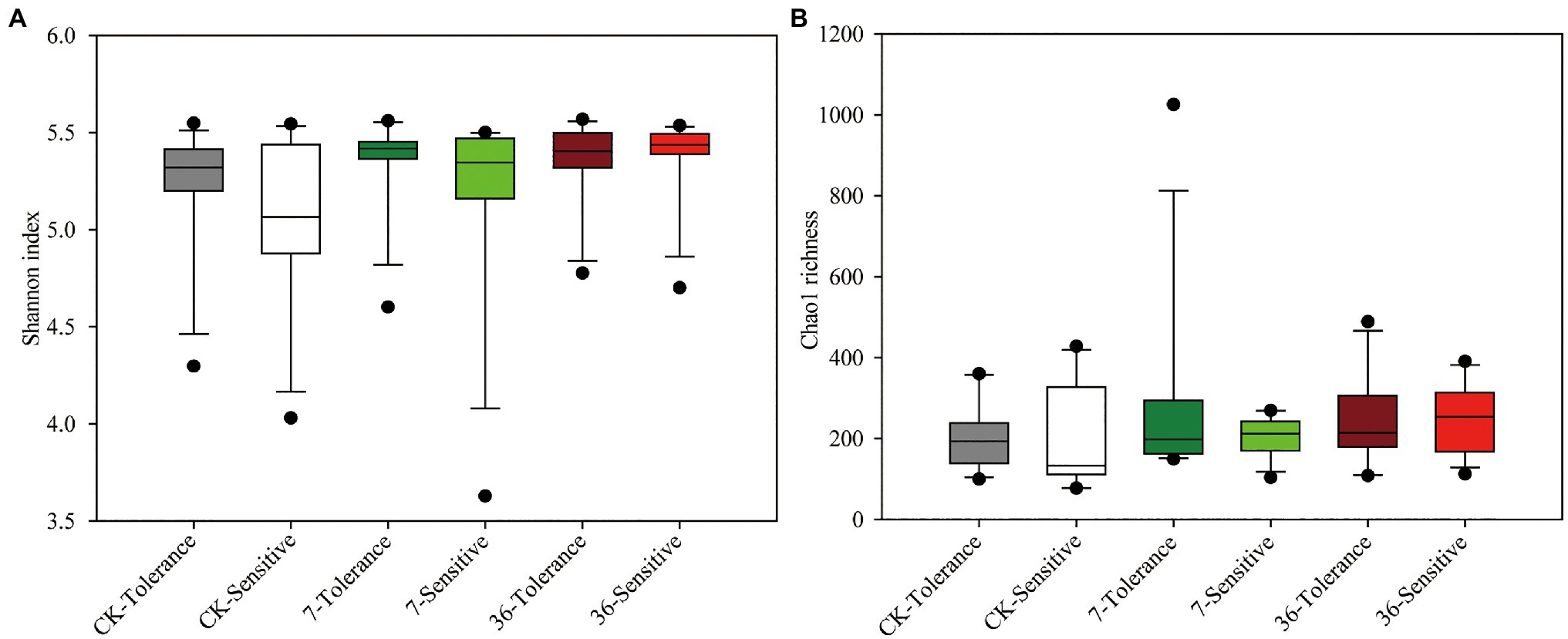
Figure 1. Rhizobacterial community Shannon index (A) and Chao1 richness (B) of different treatments. One-way ANOVA, n=12, p<0.05.
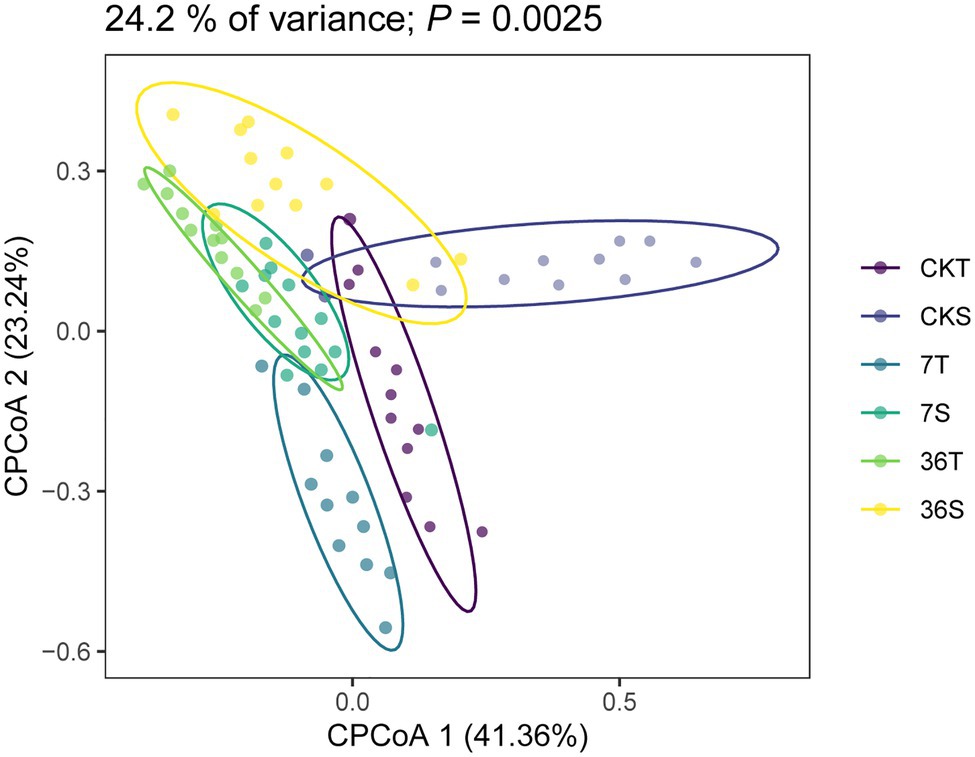
Figure 2. Constrained Principal coordinate analysis (CPCoA) based on Bray-Curtis dissimilarities of 16S rRNA diversity in the rhizosphere.
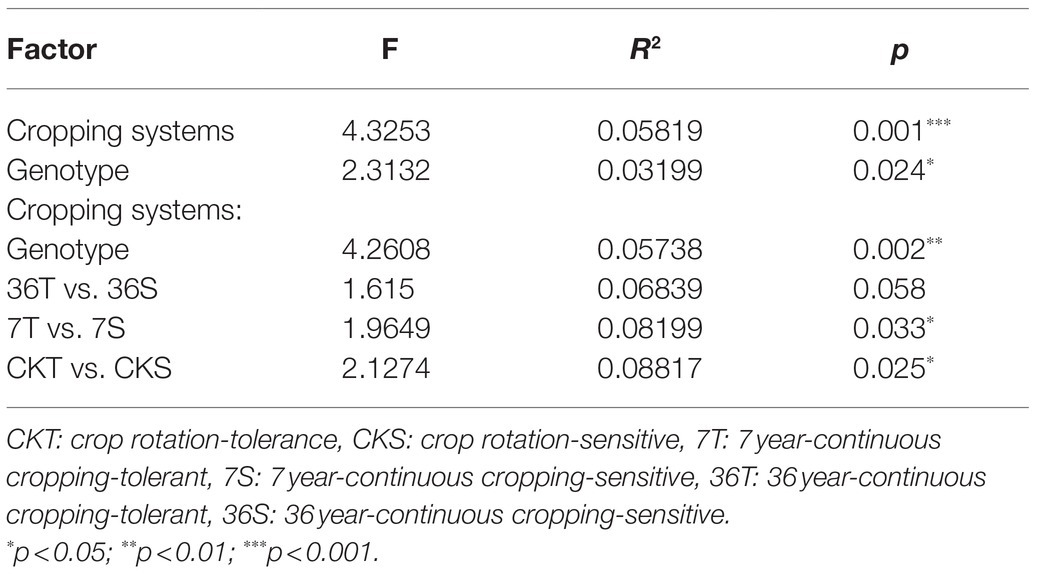
Table 2. Effects of cropping systems and soybean genotypes on the differentiation of bacterial communities based on PERMANOVA.
Proteobacteria, Bacteroidetes, Firmicutes, Acidobacteria, Actinobacteria, and Verrucomicrobia were the most abundant bacterial phyla living in the rhizosphere across all treatments, accounting for 80.57–82.98% of the whole community (Figure 3; Supplementary Table S1). Analysis of the phylum abundance with two-way ANOVA (Supplementary Table S1) showed that 10, 7, and 10 phyla were significantly (p<0.05) affected by cropping systems. In addition, they were significantly (p<0.05) affected by cropping systems, soybean genotypes, and their interaction (Supplementary Table S1). Moreover, the response to continues cropping between the two genotypes was different. For instance, the relative abundances of Acidobacteria were significantly increased in the 7-year continuous cropping field but decreased relative abundances of both soybean genotypes in the 36-year continuous cropping field, and with a higher relative abundance for the sensitive genotypes than the tolerant genotypes. The relative abundances of Proteobacteria significantly decreased and then increased in these cropping systems for the tolerant genotypes, while it continuously decreased for the sensitive genotypes. At the genus level, 16, 20, and 22 genera were significantly (p<0.05) affected by cropping systems, soybean genotypes, and their interaction, respectively (Table 3). Among them, some genera in different genotypes respond differently to continuous cropping. For example, Pseudomonas, which belong to Proteobacteria, in tolerant genotype rhizosphere increased and then decreased with continuous cropping time, while an opposite trend was observed in the sensitive genotypes. However, there are also some genera that respond to continuous cropping in a consistent trend between the two genotypes. For instance, Streptomyces and Bacillus, which belong to Actinobacteria and Firmicutes, respectively, decreased in the 7-year continuous cropping field but increased in the 36-year continuous cropping field, with higher relative abundances in the sensitive genotypes. A linear model analysis was used to identify bacterial OTUs significantly enriched in tolerant and sensitive genotypes of rhizosphere soil in the crop rotation system, 7-year continuous cropping system and 36-year continuous cropping system. For the tolerant genotypes, OTUs belonging to Chitinophaga, Nitrospira, and Saprospirales dominated in the rhizospheres of the crop rotation system, 7-year continuous cropping system and 36-year continuous cropping system, respectively (Figure 4A). For the sensitive genotypes, OTUs belonging to Phyllobacteriaceae, Comamonadaceae, and Cytophagaceae dominated in the rhizospheres of the crop rotation system, 7-year continuous cropping system and 36-year continuous cropping system, respectively (Figure 4B). More detailed information is available in Supplemental files (Supplementary Table S2).
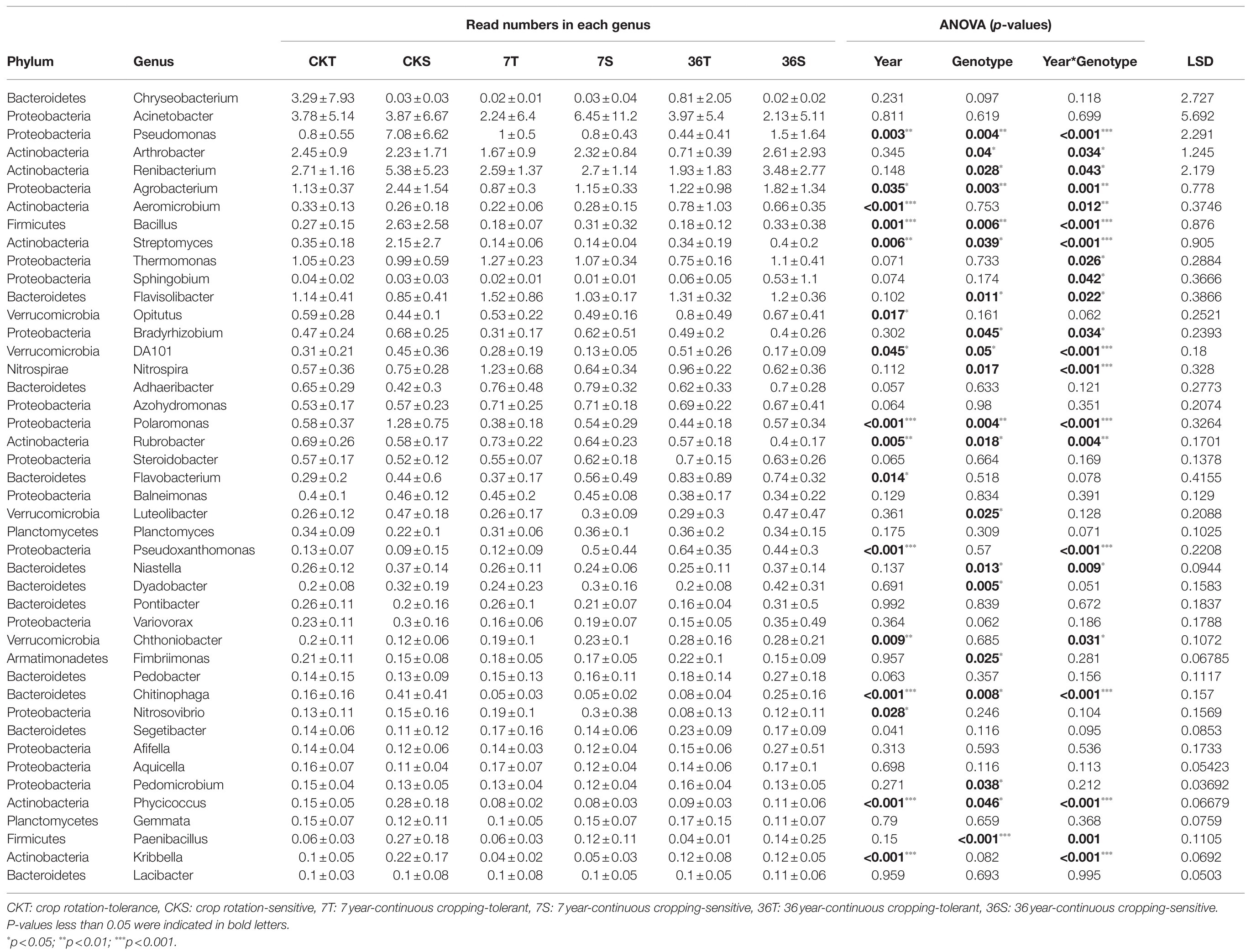
Table 3. Cropping systems, soybean genotypes, and their interactive effects on the relative abundance of bacteria at genus level.
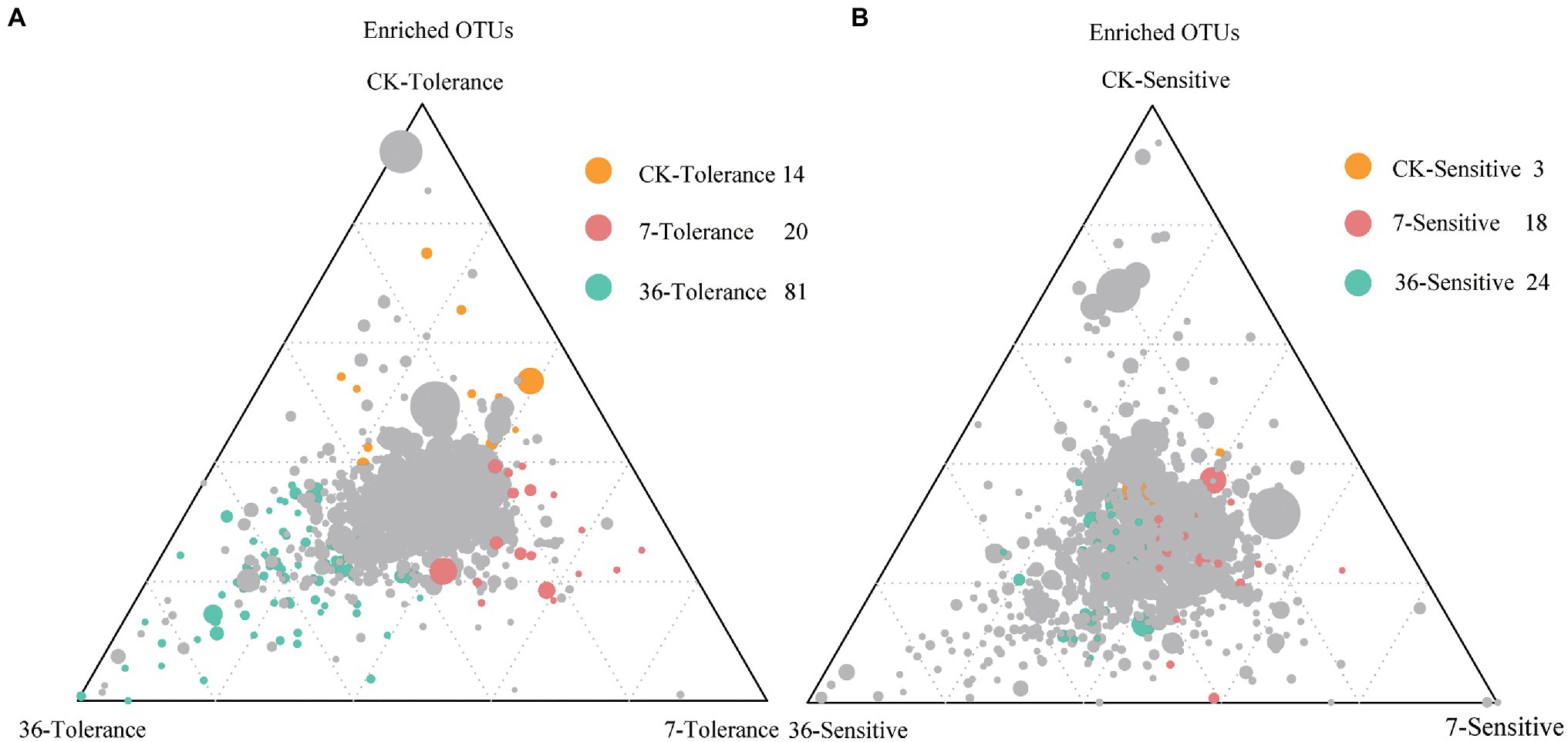
Figure 4. Ternary plots depicting compartments showing the distributions of community differentiation among different cropping systems of tolerant (A) and sensitive (B) genotypes. The position of each circle depends on its contribution to the total relative abundance. Colored circles represent OTUs that enriched in different cropping systems, which the green for the rotation cropping system; red and green for 7- and 36-year continuous cropping systems, respectively. The gray circles represent the OTUs that are not significantly enriched within a given range.
Correlations between rhizosphere soil microbiota with higher relative abundance and pH, soybean cyst nematode, and yield were obtained via Pearson’s correlation analysis (Supplementary Figure S4). Soil pH was positively correlated with Acinetobacter and Bradyrhizobium, while it was negatively correlated with other genera. Cyst nematode was negatively correlated with Thermomonas, Flavisolibacter, and Opitutus, while it was positively correlated with Pseudomonas, Renibacterium, Agrobacterium, Bacillus, and Streptomyces. Moreover, the yield was positively correlated with Sphingobium, Flavisolibacter, and Opitutus.
Among the 12,436OTUs, we found that tolerant and sensitive genotypes shared almost the same core microorganisms, such as OTU3795 (Thermomonas), OTU6911 (norank_Sphingomonadaceae), OTU8585 (norank_Sphingomonadaceae), OTU9044 (norank_Erythrobacteraceae), OTU9116 (norank_Methylophilaceae), OTU9219 (Agrobacterium), OTU4334 (Nitrospira), OTU6274 (norank_Chitinophagaceae), OTU6533 (Flavisolibacter), OTU10447 (Renibacterium), OTU4280 (norank_ii1-15), OTU4827 (norank_Ellin6075), OTU8697 (norank_RB41), and OTU10068 (norank_ii1-15; Figure 5). Moreover, OTU7122 (Pseudomonas) was the core species specific to the sensitive genotypes (Figure 5B). The relative abundances of the total core OTUs of tolerant and sensitive Proteobacteria were 7.77% and 11.48, respectively (Supplementary Table S3).
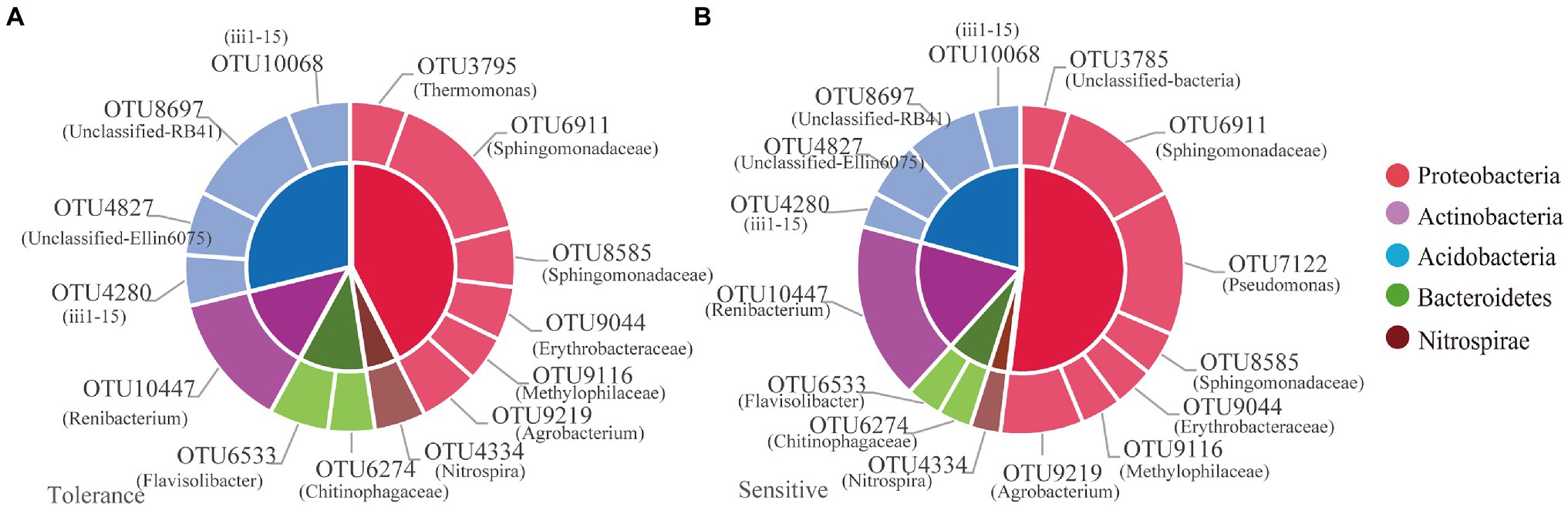
Figure 5. The relative abundance of the core microbiome for tolerant (A) and sensitive (B) genotypes.
Using the combined 16S rRNA data of the two soybean genotypes grown in different cropping systems, the co-occurrence network in the rhizosphere showed marked differences between the treatments (Figure 6; Supplementary Table S4). For the tolerant genotype, the number of negative correlations and modularity (M) increased with continuous cropping years, suggesting that there were more coupling, cooperation, and exchange events among the dominant bacterial genera. Moreover, there was no significant change in the average clustering coefficient (avgCC) or average degree (avgK). For the sensitive genotype, the modularity (M) increased with continuous cropping years. However, the number of negative correlations, average clustering coefficient (avgCC), and average degree (avgK) increased in the 7-year continuous cropping systems field but decreased in the 36-year continuous cropping systems field. The keystone species of the bacterial network in the rhizosphere were identified by calculating node degree, closeness centrality, and betweenness centrality for all nodes in the network (Supplementary Table S5). In general, OTU11321 (Luteolibacter), OTU6533 (Flavisolibacter), OTU1003 (Chitinophagaceae), OTU4362 (norank_RB41), OTU881 (Balneimonas), and OTU3461 (norank_Ellin6075) were identified as keystone species for the two genotypes grown in the different cropping systems. Using NetShift analysis, the common sub-network plot showed the “driver microbes” were OTU2820 (Unclassified Chitinophagaceae), OTU881 (Unclassified oc28), and OTU3463 (Unclassified Acidobacteria-6) for the network between rotations and 7years continuous cropping system, and OTU881 (Unclassified oc28), OTU3487 (Unclassified Gemm-1), OTU9219 (Agrobacterium), and OTU3461 (Unclassified Ellin6075) for the network between rotations and 7years continuous cropping system. For the sensitive soybean genotype, the “drivers microbes” were OTU2166 (Unclassified Piscirickettsiaceae) and OTU9044 (Unclassified Erythrobacteraceae) for the network between rotations and 7years continuous cropping system, and OTU4813 (Unclassified Saprospiraceae), OTU3714 (Unclassified Oxalobacteraceae), OTU4607 (Unclassified Cytophagaceae), OTU3463 (Unclassified Acidobacteria-6), and OTU3416 (Unclassified OPB35) for the network between rotations and 7years continuous cropping system (Figure 7).

Figure 6. Network analysis of the rhizosphere bacterial community from different treatments. Nodes represent OTUs coded by phyla and are scaled by the number of connections (node degree). Connections are plotted at r>0.8 (positive correlation, red) or r<−0.8 (negative correlation, blue) and p<0.05.
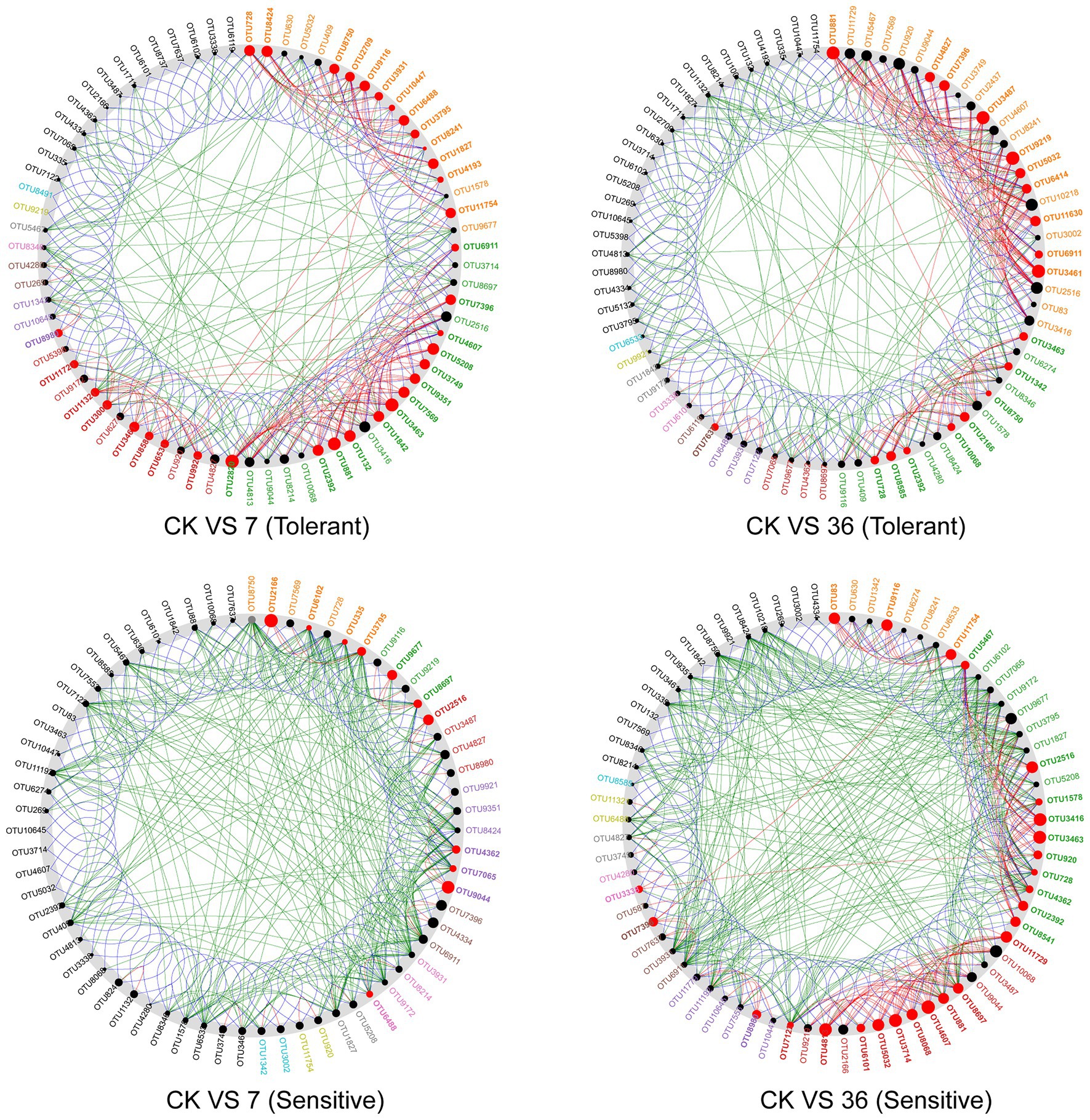
Figure 7. A common sub-network view with the “driver” node highlighted. All nodes belonging to the same community have been randomly assigned a similar color. Gray OTU nodes represent nodes that are present in both cases, but interact directly with the common subnetwork in either the case or control group. The size of the nodes was proportional to their NESH scores, and a node was colored red if its spacing between the control and study groups increased. Large and red nodes are particularly important “drivers.” CK VS 7 (36; tolerant or sensitive), network comparison between soybean-corn rotations and 7 (36) years continuous cropping for tolerant or sensitive genotype.
Our initial hypotheses were that soybean with a tolerant genotype would have higher rhizosphere bacterial diversity than soybean with a sensitive genotype, and tolerant soybean would recruit microorganisms that benefited continuous cropping tolerance. Our results showed that there was no significant difference in microbial diversity between the two genotypes soybean in all cropping systems. In addition, our second hypothesis was tested: tolerant genotypes recruited specific microorganisms that may help soybean mitigate soil-borne diseases. Short period of continuous cropping is unfavorable for soybean, but long periods of continuous cropping will mitigate this unfavorability. Moreover, different genotypes of soybean respond differently to continuous cropping systems, and microorganisms recruited by tolerant soybeans might play an important role in mitigating the adverse effects of continuous cropping.
In the present study, we found that there was no significant difference in bacterial community diversity among the treatments. Previous studies have shown that an increase in crop type could lead to an increase in soil microbial diversity. Liu et al. (2020) found that rhizosphere bacterial diversity was reduced with continuous cropping of soybean compared to soybean-corn rotation cropping systems. In addition, Zhu et al. (2017) and Liu et al. (2020) found that soil bacterial diversity increased with increasing years of continuous cropping. However, Li et al. (2010) found no significant difference in soil microbial diversity between the soil of continuous soybean and soybean-corn rotations. This might be attributed to the different soil pH in these studies, as Liu et al. conducted their research on acidic soil, while our study was conducted on neutral soil. Changes in soil pH and other physical and chemical properties were significantly associated with microbial diversity, and this variation was related to changes in root secretions, such as organic acids, phenols, and flavonoids, which were significantly affected by the cropping systems (Venter et al., 2016; Liu et al., 2020). For beta diversity, we found that cropping systems and soybean genotypes were the main factors that changed the bacterial communities (Adonis tests, p<0.05). This result is consistent with the results reported by Zhu et al. (2017) and Lian et al. (2019), who indicated that there were significant changes in soil bacterial communities in long-term and short-term continuous soybean cropping systems and in different soybean genotypes.
The relative abundance of Proteobacteria was significantly decreased in the rhizospheric soils of the continuous cropping system field compared to those of the rotation system, indicating that those bacteria were decreased with low available nutrients (Fierer et al., 2007; Li et al., 2014). The relative abundances of Pseudomonas, Streptomyces, and Bacillus decreased in the 7-year continuous cropping field but increased in the 36-year continuous cropping field, with a higher relative abundance in the sensitive genotypes. Pseudomonas can improve the solubilization of fixed soil phosphorus and applied phosphates, resulting in higher crop yields (Nautiyal, 1999). Moreover, the metabolites of Pseudomonas could increase amino acid secretion in alfalfa, maize, and wheat roots (Phillips et al., 2004). Streptomyces can help plants resist diseases such as wilt and root rot (Lian et al., 2019; Shi et al., 2020). In addition, some bacteria, such as Thermomonas, Flavisolibacter, and Opitutus were negatively associated with nematodes, which suggested that these bacteria might inhibit the soybean cyst nematode. Thus, based on the functions of the species mentioned above, we speculated that the changed relative abundances of these bacteria might be associated with the antagonistic activity of these taxa against plant pathogens and the improvement of soil nutrients.
We used differential OTU abundance analysis in the treatments and observed a high relative abundance of the OTUs enriched in continuous cropping systems of the tolerant genotypes that belonged to Chitinophagaceae, Pseudoxanthomonas, Nitrospira, and Streptomyces (Figure 4; Table 3). This finding is consistent with our hypothesis that tolerant genotypes recruited some microorganisms that play an important role in the resistance of soybeans to long-term continuous cropping. OTU4434 (Nitrospira), OTU751 (Pseudoxanthomonas), and OTU10218 (Streptomyces) showed higher relative abundances in the continuous cropping system than in the rotation cropping system in the present study (Figure 4; Table 3). Many studies have reported that Nitrospira can promote the nitrogen cycle and increase the absorption and utilization of nitrogen by plants (Zeng et al., 2014). Therefore, the higher abundances of these bacteria may improve soybean tolerance in the rhizosphere. However, the response of these genera to long-term continuous cropping warrants further investigation of their functional significance in response to different long-term continuous cropping systems.
An association network analysis was performed to gain a more integrated understanding of the bacterial community composition and compare the complexities of the networks operating in the rhizosphere soils of continuous cropping-tolerant and continuous cropping-sensitive soybean genotypes (Jiang et al., 2017). The networks of the two genotypes responded differently to continuous cropping. The modularity and negative correlations in the network of the tolerant genotype responded positively to continuous cropping, suggesting that there were more coupling, cooperation, and exchange events among the dominant bacterial genera (Coyte et al., 2015). For the sensitive genotype, the negative correlations and average degree (avgK) increased in the 7-year continuous cropping system field but decreased in the 36-year continuous cropping system field, suggesting that the instability of the sensitive genotype and short periods of continuous planting can have a detrimental effect on microbial community stability, but this effect could be alleviated with increasing periods of continuous planting. This result was consistent with a previous study, which found that a long-term continuous cropping system has similar microbial interactions to those of a healthy crop rotation system (Liu et al., 2020). Additionally, the important microbial taxa in the networks which serve as “drivers” from the rotations to short or long continue cropping were different between the two soybean genotypes, which indicated that the response of rhizosphere microbial community under different soybean genotypes to short- and long-term continuous cropping was different.
In summary, short- or long-term continuous cropping had no significant effect on the rhizosphere soil bacterial alpha diversity. Short-term continuous planting increased the number of soybean cyst nematodes, while long-term continuous planting reduced these numbers, and tolerant genotypes had lower soybean cyst nematodes than sensitive genotypes. In addition, continuous cropping significantly increased potentially beneficial bacterial populations compared to rotation and short-term continuous cropping, suggesting that long-term continuous cropping of soybean shifts the microbial community into that of a healthy crop rotation system. Soybean genotypes that are tolerant might recruit some microorganisms that play an important role in the resistance of soybeans to long-term continuous cropping. However, further experiment with species isolation and verification is needed to confirm these findings. Moreover, the networks of the two genotypes responded differently to continuous cropping. The tolerant genotype responded positively to continuous cropping, while for the sensitive genotype, topology suggested that the instability of the sensitive genotype and short periods of continuous planting can have a detrimental effect, but this effect could be alleviated with increasing periods of continuous planting.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
TL, HN, and MY designed the research. MY, KY, LW, DH, and SW performed the research. TY, QS, HX, and RW analyzed the data. MY and TL analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by China Agriculture Research System of MOF and MARA (CARS-04), and the Laboratory independent research projects (SKLCUSA-a201913).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Chai Xiaocui and Qiao Zhixian at the Analysis and Testing Center of Institute of Hydrobiology, Chinese Academy of Sciences for their assistance with PCR amplification, sequencing, and data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.729047/full#supplementary-material
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S. T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Avidano, L., Gamalero, E., Cossa, G. P., and Carraro, E. (2005). Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30, 21–33. doi: 10.1016/j.apsoil.2005.01.003
Bai, L., Cui, J. Q., Jie, W. G., and Cai, B. Y. (2015). Analysis of the community compositions of rhizosphere fungi in soybeans continuous cropping fields. Microbiol. Res. 180, 49–56. doi: 10.1016/j.micres.2015.07.007
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Cai, B. Y., Wang, L. Y., Hu, W., Jie, W. G., and Ling, H. Z. (2015). Analysis of community structure of root rot pathogenic fungi in seedling stage of soybean continuous cropping. Chin. Agr. Sci. Bull. 31, 92–98.
Chigen, T., Muhammad, A. N., Ayaka, K., Bao, L., Jeong, D. L., Eunho, S., et al. (2018). Isoflavone profile diversity in Korean wild soybeans (Glycine soja Sieb. & Zucc.). Turk. J. Agric. For. 42, 248–261. doi: 10.3906/tar-1709-119
Chong, J., Liu, P., Zhou, G., and Xia, J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 5, 799–821. doi: 10.1038/s41596-019-0264-1
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, 141–145. doi: 10.1093/nar/gkn879
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Dias, T., Dukes, A., and Antunes, P. M. (2015). Accounting for soil biotic effects on soil health and crop productivity in the design of crop rotations. J. Sci. Food Agric. 95, 447–454. doi: 10.1002/jsfa.6565
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Friendly, M., and Meyer, D. (2015). Discrete Data Analysis With R: Visualization and Modeling Techniques for Categorical and Count Data. Boca Raton: CRC Press, 63–108.
Hu, J. C., and Wang, S. J. (1996). Study on soil sickness by soybean continuous cropping I. Effect of mycotoxin produced by Penicillium purpurogenum. Chin. J. Appl. Ecol. 7, 396–400.
Ji, S. N., Xiao, Y. Z., Tian, H. M., and Zhao, F. H. (1996). Effects of sterilized soil from soybean and corn stubble field on the growth development of soybean. J. Northeast. Agr. 27, 326–329.
Jiang, Y., Li, S., Li, R., Zhang, J., Liu, Y., Lv, L., et al. (2017). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109, 145–155. doi: 10.1016/j.soilbio.2017.02.010
Kaisermann, A., Vries, F. T., Grifths, R. I., and Bardget, R. D. (2017). Legacy effects of drought on plant-soil feedbacks and plant-plant interactions. New Phytol. 215, 1413–1424. doi: 10.1111/nph.14661
Kuntal, B. K., Chandrakar, P., Sadhu, S., and Mande, S. S. (2019). ‘NetShift:’ a methodology for understanding ‘driver microbes’ from healthy and disease microbiome datasets. ISME J. 13, 442–454. doi: 10.1038/s41396-018-0291-x
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Li, W., and Godzik, A. (2015). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1007/978-1-4899-7478-5_221
Li, C. G., Li, X. M., Kong, W. D., Wu, Y., and Wang, J. G. (2010). Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 330, 423–433. doi: 10.1007/s11104-009-0216-6
Li, X. Z., Rui, J. P., Mao, Y. J., Yannarell, A., and Mackie, R. (2014). Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401. doi: 10.1016/j.soilbio.2013.10.017
Lian, T. X., Ma, Q. B., Shi, Q. H., Cai, Z. D., Zhang, Y. F., Cheng, Y. B., et al. (2019). High aluminum stress drives different rhizosphere soil enzyme activities and bacterial community structure between aluminum-tolerant and aluminum-sensitive soybean genotypes. Plant Soil 440, 409–425. doi: 10.1007/s11104-019-04089-8
Liu, X. B., Burras, C. L., Kravchenko, Y. S., Duran, A., Huffman, T., Morras, H., et al. (2012). Overview of Mollisols in the world:distribution, land use and management. Can. J. Soil Sci. 92, 383–402. doi: 10.4141/cjss2010-058
Liu, Z. X., Liu, J. J., Yua, Z. H., Yao, Q., Li, Y. S., Liang, A. Z., et al. (2020). Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 197:104503. doi: 10.1016/j.still.2019.104503
Liu, H., Pan, F. J., Han, X. Z., Song, F. B., Zhang, Z. M., Yan, J., et al. (2019). Response of soil fungal community structure to long-term continuous soybean cropping. Front. Microbiol. 9:3316. doi: 10.3389/fmicb.2018.03316
Mendes, L. W., Mendes, R., Raaijmakers, J. M., and Tsai, S. M. (2018). Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 12, 3038–3042. doi: 10.1038/s41396-018-0234-6
Meriles, J. M., Gil, S. V., Conforto, C., Figoni, G., Lovera, E., March, G. J., et al. (2009). Soil microbial communities under different soybean cropping systems: characterization of microbial population dynamics, soil microbial activity, microbial biomass, and fatty acid profiles. Soil Tillage Res. 103, 271–281. doi: 10.1016/j.still.2008.10.008
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Pérez-Brandán, C., Huidobro, J., Grümberg, B., Scandiani, M. M., Luque, A. G., Meriles, J. M., et al. (2014). Soybean fungal soil-borne diseases: a parameter for measuring the effect of agricultural intensification on soil health. Can. J. Microbiol. 60, 73–84. doi: 10.1139/cjm-2013-0792
Phillips, D., Fox, T. C., King, M., Bhuvaneswari, T. V., and Teuber, L. R. (2004). Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 136, 2887–2894. doi: 10.1104/pp.104.044222
Shi, Q. H., Liu, Y. T., Shi, A. Q., Cai, Z. D., Nian, H., Hartmann, M., et al. (2020). Rhizosphere soil fungal communities of aluminum-tolerant and -sensitive soybean genotypesrespond differently to aluminum stress in an acid soil. Front. Microbiol. 11:1177. doi: 10.3389/fmicb.2020.01177
Song, J., Li, S. X., Wei, W., Xu, Y. L., and Yao, Q. (2017). Assessment of parasitic fungi for reducing soybean cyst nematode with suppressive soil in soybean fields of Northeast China. Acta Agr. Scand. B Soil Plant 67, 730–736. doi: 10.1080/09064710.2017.1343377
Tang, H., Xiao, C. H., Ma, J. Z., Yu, M., Li, Y. M., Wang, G. L., et al. (2009). Prokaryotic diversity in continuous cropping and rotational cropping soybean soil. FEMS Microbiol. Lett. 298, 267–273. doi: 10.1111/j.1574-6968.2009.01730.x
Venter, Z. S., Jacobs, K., and Hawkins, H. J. (2016). The impact of crop rotation on soil microbial diversity: a meta-analysis. Pedobiologia 59, 215–223. doi: 10.1016/j.pedobi.2016.04.001
Waldrop, M. P., Balser, T. C., and Firestone, M. K. (2000). Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 32, 1837–1846. doi: 10.1016/S0038-0717(00)00157-7
Wei, W., Xu, Y. L., Li, S. X., Zhu, L., and Song, J. (2015). Developing suppressive soil for root diseases of soybean with continuous long-term cropping of soybean in black soil of Northeast China. Acta Agr. Scand. B Soil Plant 65, 279–285. doi: 10.1080/09064710.2014.992941
Xu, Y. L., Wang, G. H., and Han, X. Z. (1995). Relationship between soil microbial ecological distribution characteristics and soybean root disease in soybean monocropping and rotation. Syst. Sciemces Compr. Stud. Agr. 11, 311–314.
Yan, M. C., Xu, T. T., Song, P. H., and Dai, J. J. (2012). Effects of different cropping patterns of soybean and maize seedlings on soil enzyme activities and MBC and MBN. J. Northeast. Agr. Univ. 19, 42–47. doi: 10.1016/S1006-8104(13)60049-5
Zeng, W., Bai, X., Zhang, L., Wang, A., and Peng, Y. (2014). Population dynamics of nitrifying bacteria for nitritation achieved in Johannesburg (JHB) process treating municipal wastewater. Bioresour. Technol. 162, 30–37. doi: 10.1016/j.biortech.2014.03.102
Zhan, X. M., Han, X. R., Yang, J. F., and Gao, Z. Q. (2004). The effect of succession cropping and soybean stubble on soybean root exudates. Chin. J. Soil Sci. 35, 631–635. doi: 10.1300/J064v24n01_09
Zhou, X. G., Wang, Z. L., Jia, H. T., Li, L., and Wu, F. Z. (2018). Continuously monocropped Jerusalem artichoke changed soil bacterial community composition and ammoniaoxidizing and denitrifying bacteria abundances. Front. Microbiol. 9:705. doi: 10.3389/fmicb.2018.00705
Zhu, Y. B., Shi, F. Y., Zhang, R. J., and Wu, Y. P. (2014). Comparison of bacterial diversity in rotational and continuous soybean cropping soils in Heilongjiang. Acta Phytophy. Sin. 41, 403–409.
Keywords: continuous cropping, soybean, Mollisol, rhizosphere microorganisms, network
Citation: Yuan M, Yu T, Shi Q, Han D, Yu K, Wang L, Wang S, Xiang H, Wen R, Nian H and Lian T (2021) Rhizosphere Soil Bacterial Communities of Continuous Cropping-Tolerant and Sensitive Soybean Genotypes Respond Differently to Long-Term Continuous Cropping in Mollisols. Front. Microbiol. 12:729047. doi: 10.3389/fmicb.2021.729047
Received: 22 June 2021; Accepted: 16 August 2021;
Published: 13 September 2021.
Edited by:
Hai-Ming Zhao, Jinan University, ChinaReviewed by:
Shuai Zhao, Chinese Academy of Sciences, ChinaCopyright © 2021 Yuan, Yu, Shi, Han, Yu, Wang, Wang, Xiang, Wen, Nian and Lian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tengxiang Lian, bGlhbnR4QHNjYXUuZWR1LmNu; Hai Nian, aG5pYW5Ac2NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.