
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 16 September 2021
Sec. Virology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.725074
This article is part of the Research Topic Emerging and Important Infectious Diseases: the Cutting-Edged Studies on Animal Models and Immunology View all 21 articles
Animal models and cell culture in vitro are primarily used in virus and antiviral immune research. Whereas the limitation of these models to recapitulate the viral pathogenesis in humans has been made well aware, it is imperative to introduce more efficient systems to validate emerging viruses in both domestic and wild animals. Organoids ascribe to representative miniatures of organs (i.e., mini-organs), which are derived from three-dimensional culture of stem cells under respective differential conditions mimicking endogenous organogenetic niches. Organoids have broadened virological studies in the human context, particularly in recent uses for COVID19 research. This review examines the status and potential for cross-species applied organotypic culture in validating emerging animal, particularly zoonotic, viruses in domestic and wild animals.
Organoids, also known as “mini-organs,” are three-dimensional (3D) constructs; these constructs differentiate from stem cells to recapitulate the cellular architecture and functionality of native organs (Kaushik et al., 2018; Lehmann et al., 2019; Kim et al., 2020). Using both embryonic pluripotent stem cells (PSCs) and somatic adult stem cells (ASCs), organoids have been generated by culturing stem cells under conditions mimicking the in vivo developmental and differential cues of their respective tissues (Kaushik et al., 2018; Lehmann et al., 2019; Corrò et al., 2020; Kim et al., 2020). The cornerstone of organoid systems are stem cells, and relevant research signifies the origin of organoid systems. Further elucidation of biochemical (e.g., growth factors and other bioactive molecules) and physical cues (e.g., mechanic stimuli and topography applied from the extracellular matrix) serves as a “cow pony,” shaping organoid culture to regenerate multi-cellular tissue proxies of original tissues and organs (Yin et al., 2016; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Hofer and Lutolf, 2021).
As per the virus-host interactions, animal models and two-dimensional (2D) cell cultures represent major biological experimental systems in biomedical studies. Animal models directly replicate animal infections in nature, and most of these models holistically recapitulate human pathophysiology. However, they are limited by individual variations, inter-species biological differences, interference with other microorganisms (pathogenic or common flora), animal welfare and ethical concerns; obstacles in throughput and convenience for microimaging and analytic readouts are also present (Shanks et al., 2009; Mead and Karp, 2019; Corrò et al., 2020; Kim et al., 2020; Hofer and Lutolf, 2021). In contrast, 2D monocultures of cell lines offer a consistent and reproducible in vitro model supporting viral infections, and they are widely used in virus-cell interaction studies. However, cancerous cell lines are genetically instable and over-simplistic to recapitulate the complex inter-cell and cell-matrix interactions that are required to model viral pathogenesis and particularly antiviral reactions in tissues (Shanks et al., 2009; Mead and Karp, 2019; Corrò et al., 2020; Kim et al., 2020; Hofer and Lutolf, 2021). Different alternatives of cell culture systems including co-culture of cell lines or primary cells and cell aggregate cultures of targeted tissues have both shown improved features compared to monotype cell culture. However, they lose the relevance to tissue organization in vivo—and even cell integrity—due to 2D culture adaptation. In addition, tissue explant cultures—which require rushed handling after surgeries to transiently retain physiologic relevance of cell organization/interaction— undergo cell phenotypic loss and senescence in order to align with an adequate time-window for performing studies in virology and antiviral immunology (Shanks et al., 2009; Mead and Karp, 2019; Corrò et al., 2020; Schutgens and Clevers, 2020; Hofer and Lutolf, 2021). Hence, these model systems each exhibit a meaningful gap to recapitulate the cellular organization and physiology of original organs; this is especially true for studying virus-host interactions, which generally involve more than one cell type in the targeted tissues (Barrila et al., 2018; Ramani et al., 2018; Bar-Ephraim et al., 2020; Sridhar et al., 2020; Yuki et al., 2020). Upon thorough examination of recent advent and peer reviews in both humans and animals organoid research, it is clear that an urgent need has emerged to harness better biological systems; a balance between practicability and faithfulness is needed to assess the cross-species potential of viral pathogens in both humans and animals (Shanks et al., 2009; Barrila et al., 2018; Ramani et al., 2018; Montes-Olivas et al., 2019; Bar-Ephraim et al., 2020; Corrò et al., 2020; Sridhar et al., 2020; Yuki et al., 2020). The status and perspective of organoid systems has been particularly reviewed, demonstrating potency and comparative advantages in order to fulfill most needs for in vitro modeling of viral infections and pathogenesis in humans (Barrila et al., 2018; Ramani et al., 2018; Lehmann et al., 2019; Montes-Olivas et al., 2019; Bar-Ephraim et al., 2020; Kim et al., 2020; Sridhar et al., 2020; Yuki et al., 2020). Table 1 summarizes the pros and cons of 3D organoid cultures; they are compared with classical 2D cell cultures and animal models regarding their complexity, cost, controllability, and downstream biomedical applications (Shanks et al., 2009; Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Mead and Karp, 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021). As each type of model generally possesses inherent peculiarities, increasing the scale entails higher system complexity and challenges for the success of controlled cultures; hence, a reduced accessibility emerges when analyzing approaches at molecular and cellular levels (Lehmann et al., 2019; Kim et al., 2020). Organoid models render a platform to coordinate moderate system complexity and reproducibility. It also simultaneously enables the probing of the structure and function of a given biological system. Even when compared with 3D cultures of tissue explants, organoid systems model better genetic stability and cell-cell/cell-matrix interactions; they also simultaneously permit for long-term cultures based on regeneration from stem cells and maintenance of biological cues (Yin et al., 2016; Ramani et al., 2018; Mead and Karp, 2019; Fuchs and Blau, 2020). Organoids bridge the gap between 2D cell culture and animal models by providing a culturable system; this allows manipulation of cell cultures while representing better mimicking of in vivo cellularity and physiology (Lehmann et al., 2019; Kim et al., 2020). As implicated in several pilot studies and eminent reviews, we emphasize that cross-species investigation of organoid cultures in domestic and wild animals, will significantly promote virology studies, particularly for efficiently validating the infection loop of epizootic and zoonotic viruses in different animal species, including humans (Barrila et al., 2018; Ramani et al., 2018; Lehmann et al., 2019; Montes-Olivas et al., 2019; Bar-Ephraim et al., 2020; Duque-Correa et al., 2020; Kim et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Yuki et al., 2020; Hofer and Lutolf, 2021; Kar et al., 2021; Pain, 2021).
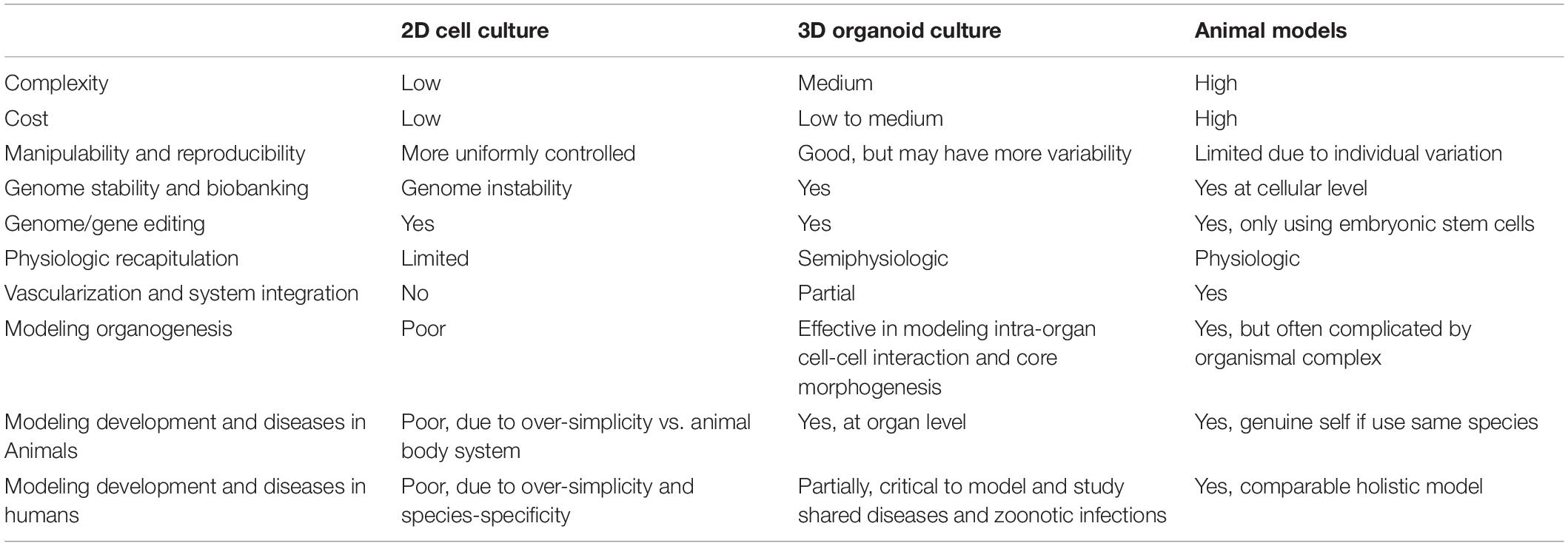
Table 1. Comparison of 3D organoid culture with conventional cell cultures and animal models (Shanks et al., 2009; Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Mead and Karp, 2019; Moghaddam et al., 2019; Corrò et al., 2020; Duque-Correa et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021; Kar et al., 2021; Pain, 2021).
Multiple types of organoids have been cultured primarily from human and mouse stem cells, and this has revolutionized the research and clinical practice in human medicine; this is true in terms of applications in biobanking, drug screening, genotyping (i.e., cell typing) for precision medicine, and disease modeling involving pathogenic infections (Figure 1; Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021). As demonstrated from a variety of pioneering studies, organoid technology can be adopted in most vertebrate species to retain its strength in modeling animal diseases; this is particularly true in the case of the microbe-host interaction in infectious cases (Barrila et al., 2018; Ramani et al., 2018; Montes-Olivas et al., 2019; Bar-Ephraim et al., 2020; Duque-Correa et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Yuki et al., 2020). In this regard, tissue/organ resources are readily available for establishing organoid culture, particularly in livestock animals; this also addresses the concerns of animal welfare, reducing the number of animals that may succumb to experimentally pathogenic infections and disease modeling (Kar et al., 2021; Pain, 2021). Organoid-relevant studies have been boosted among animals in the last 10 years (Derricott et al., 2019; Corrò et al., 2020; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021), with multiple studies using organoids to promote COVID-19 investigation (Clevers, 2020; Monteil et al., 2020; Yang et al., 2020; Han et al., 2021). Despite some types of organoid culture having been validated in multiple animals—particularly domestic species—there is an overall lack of robust procedures and dependent resources to generate animal organoids (Derricott et al., 2019; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021). These forms of organoid research in animals are advantageous, and they provide dual roles and benefits in promoting the wellbeing of animals and humans. Advantages include more reliable offal resources than medical biopsy samples, easier handling regarding organ size as compared to mice, and more physiological similarity in modeling human organs (Derricott et al., 2019; Lehmann et al., 2019; Kim et al., 2020; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021). It is also worth noting that the genetic stability retained by stem cells make organoid systems a promising tool for genetic rescue of endangered animal species.
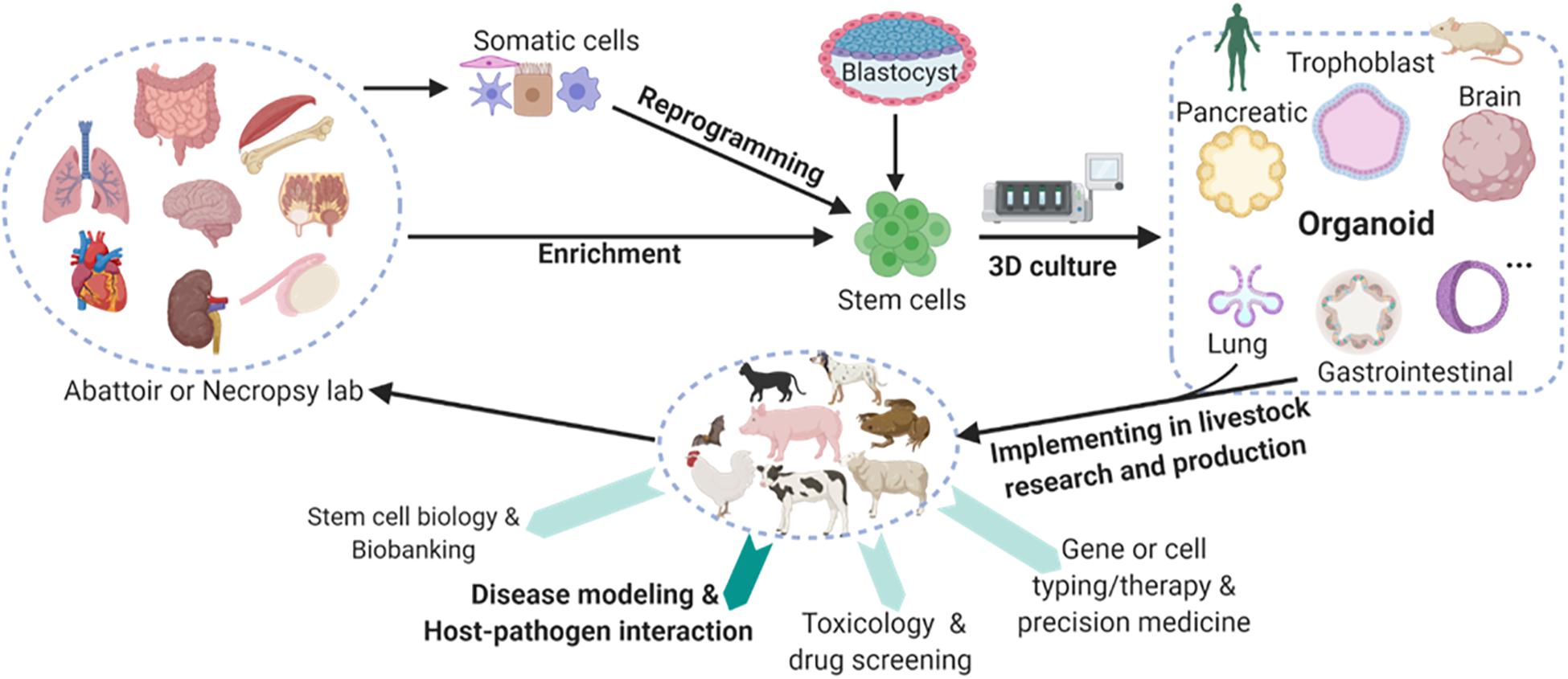
Figure 1. Schematic of mainstream organoid culture in humans and mice and their multifaceted potential in the promotion of domestic and wild animals studies. Organoids, also known as “mini-organs,” are three-dimensional (3D) constructs that differentiate from stem cells to recapitulate cellular architecture and functionality of native organs. Multiple types of organoids have been cultured primarily in humans and mice, and they have profoundly revolutionized the research and practice in human medicine. Some types of organoid culture have also been validated in livestock species. However, these cultures must be extended for promoting research and production in agricultural animals. Collectively, organoid research in domestic animals will provide dual roles and dual benefits in promoting the wellbeing of both the animals and humans with several advantages. These advantages include reliable offal resources, easier handling regarding organ size when compared to that of a mouse, and more physiological similarity in modeling human organs. Animal organoid studies will reveal unknown species-specific prospects in terms of organ development, stem cell biology, mechanistic regulation at gene and cellular levels, and particularly disease modeling and host-pathogen interaction for epizootic and zoonotic viral infections. Created with BioRender.com.
Nevertheless, animal organoid studies are also essential for modeling epizootic and zoonotic diseases that generally involve multiple cell types. Relevant studies can reveal unknown species-specific prospects regarding organ development, stem cell biology, host-microbe interaction, and mechanistic regulation at the gene and cellular levels (Figure 1; Montes-Olivas et al., 2019; Bar-Ephraim et al., 2020; Yuki et al., 2020). Table 2 summarizes the status of organoid research in animals with highlighted applications in studying viral diseases (Derricott et al., 2019; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021). Intestinal organoids have been replicated in major domestic animal species and horseshoe bats (Derricott et al., 2019; Seeger, 2020; Zhou et al., 2020; Beaumont et al., 2021). Some studies further utilized intestinal organoids to successfully model enteric viral infections in cats and pigs (Li et al., 2019a,b, 2020; Luo et al., 2020; Tekes et al., 2020; Yin et al., 2020). Organoids of multiple other organs have also been established. These include canine organoids from the skin (Wiener et al., 2018), prostate gland (Usui et al., 2017), urinary bladder (Elbadawy et al., 2019), kidney (Chen et al., 2019) and liver (Nantasanti et al., 2015), as well as porcine organoids from the esophagus submucosal gland (von Furstenberg et al., 2017), rectum (Adegbola et al., 2017), and testicle (Sakib et al., 2019; Vermeulen et al., 2019). It also includes those from the mammary gland, pancreatic duct, and endometrium in cattle (Martignani et al., 2018), sheep (Liu et al., 2020) and horse (Thompson et al., 2020), respectively. Notably, no respiratory or lung organoids have been reported in animals. However, they have been reported in humans. There are only a few studies of organoids in wild animals, which include two recent reports in the bat intestine and snake venom glands (Post et al., 2020; Zhou et al., 2020). There is an imperative need of respiratory/lung organoids to model respiratory viral infections in animals; for this reason, we expect an increase in studies pertaining to animal respiratory organoids from active research in COVID-19 and other zoonotic respiratory viruses (Miller et al., 2019; Sachs et al., 2019; Han et al., 2021).
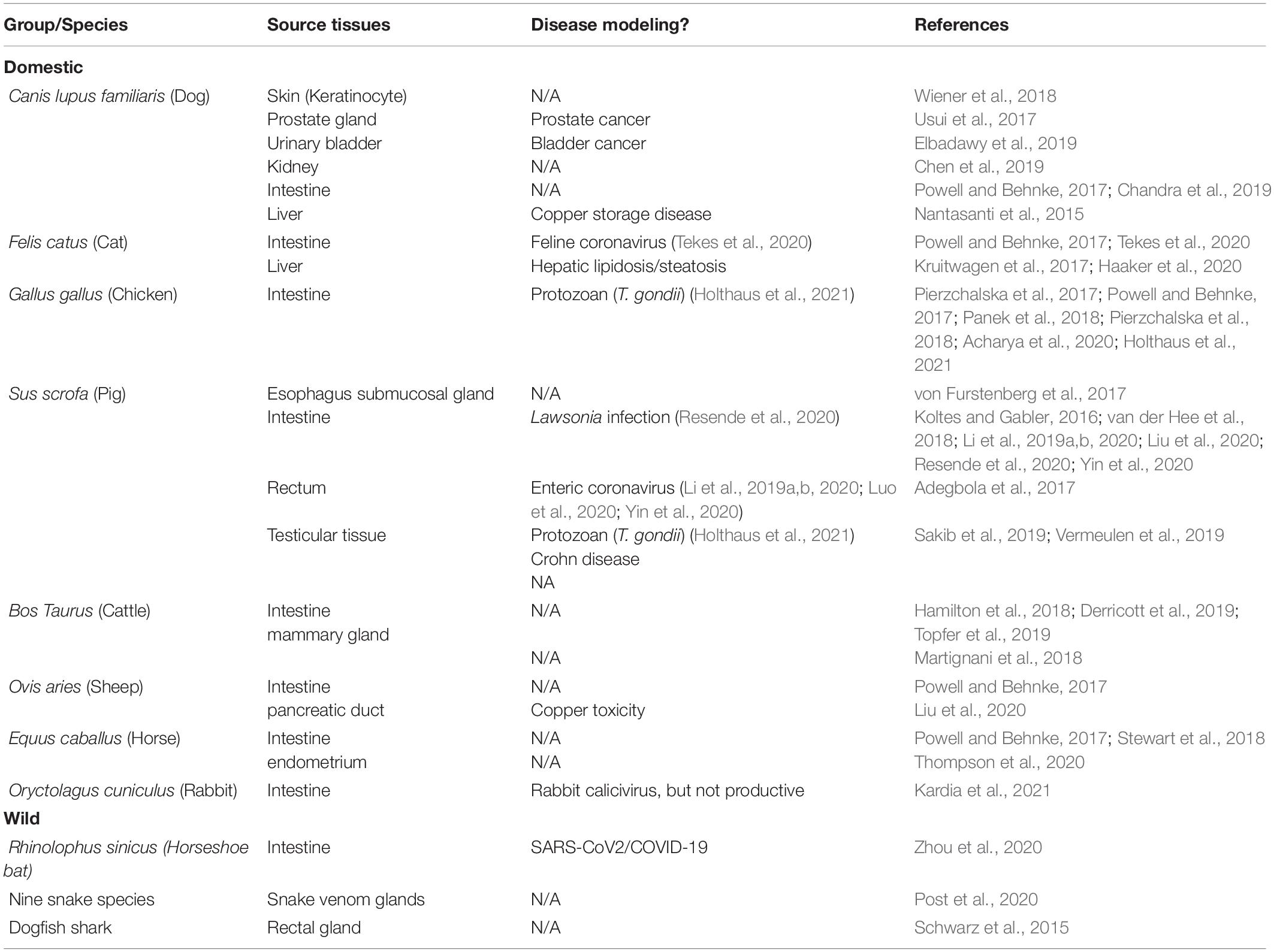
Table 2. Organoid cultures validated in domestic and wild animal species (Bourdon et al., 2021; Kar et al., 2021; Pain, 2021).
Figure 2 illustrates major historical stages in the development of organoid cultures. The status and perspective to apply these to modeling viral diseases in humans, domestic animals, and wild animals is also shown. Foundational work regarding the observation and elucidation of stem cells’ role in tissue, organ, and organism regenerations dates back to the early last century; Dr. Henry Wilson scientifically demonstrated that dissociated sponge cells could self-organize to regenerate an entire organism in vitro (Wilson, 1907; Corrò et al., 2020). Later, similar dissociation-reaggregation experiments were successfully performed using dissociated amphibian pronephros (Holtfreter, 1944) and chick embryos (Weiss and Taylor, 1960) to generate different types of organs; this implies a differential adhesion hypothesis for stem cell-derived self-regeneration (Steinberg, 1964). Pioneering studies centered on stem cell handling began with two groups in 1981, when PSCs were first isolated and established from mouse embryos (Evans, 1981; Martin, 1981). Human PSCs were first isolated and cultured from human blastocysts in 1998 (Thomson et al., 1998; Corrò et al., 2020). Both mouse- and human-induced PSCs (iPSCs) were subsequently obtained by reprogramming fibroblasts or somatic cells in the mid-2000s; this burgeoned the research in both stem cell biology and application for organoid systems in vitro (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007).
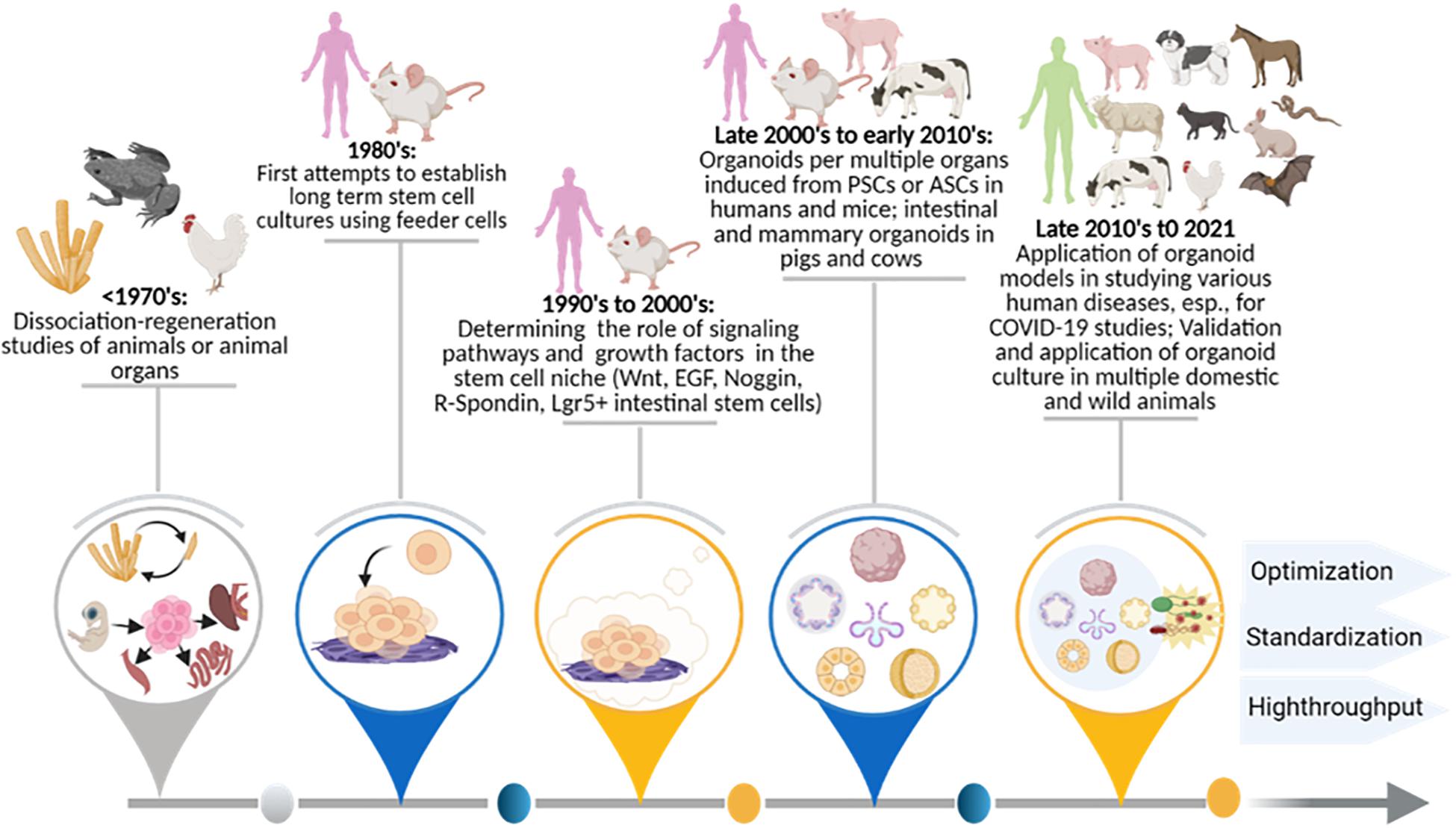
Figure 2. Historical stages for the development of organoid cultures, as well as the status and perspective for application in viral diseases in humans and animals. Summaries are shown of organoid research and technological advent benchmark breakthroughs in order to facilitate cross-species modeling of epizootic and zoonotic viral infections post-prospective development. The involvement of human and animal species are diagrammed at the top along with the major temporal events summarized below. ASC, adult stem cells; COVID-19, novel coronavirus disease 2019; EGF, epidermal growth factor; Lgr5+, Leucine Rich Repeat Containing G Protein-Coupled Receptor 5; Noggin, NOG gene product; PSC, pluripotent stem cell; R-Spondin, Rspo1 gene product; Wnt, Wingless and Int- signaling pathway.
Following the discovery of stem cells in the 1980s, studies began to boom in harnessing stem cell culture and differentiation; this was done by simulating relevant microenvironment cues in vivo (Corrò et al., 2020). Using extracellular matrix (ECM) reconstructed from an Engelbreth-Holm-Swarm tumor (EHS), Li et al. (1987) were able to maintain mammary epithelia growth and induce the formation of 3D milk-secreting duct-lumen structures; these structures would have otherwise rapidly lost their ability to secrete milk protein in the monocellular culture. Similarly, the presence of ECM of EHS also helped alveolar type II epithelial cells to retain their cell morphology; it helped them maintain function as compared to cells cultured on plastic dishes (Shannon et al., 1987). This highlights the vital role of both cell-cell and cell-matrix interactions in maintaining cell/tissue function in vitro. Eiraku et al. (2008) recapitulated early corticogenesis in order to generate cerebral cortex tissue from both human and mouse ESCs; this was done using a 3D aggregation culture method. Around the same time, Sato et al. (2009) identified adult intestinal stem cells as leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5)-expressing cells at the bottoms of small-intestinal crypts; they further established intestinal organoids through cultured single crypts or sorted Lgr5(+) stem cells in Matrigel (Sato et al., 2009). This study established 3D organoid culture from ASC-containing tissues or a single ASC; and in the late 2000s and early 2010s, this bookmarked the flourish of organoid studies in humans and various animal species (Kaushik et al., 2018; Lehmann et al., 2019; Corrò et al., 2020; Kim et al., 2020). Generated using either tissues that containing ASCs/PSCs or stem cells themselves, organoid systems have been successfully established in different organ systems, including the mesendoderm-type (e.g., stomach, liver, pancreas, lung, and kidney) and the neuroectoderm-type (brain and retina) in mice and humans (Figure 2; Kaushik et al., 2018; Lehmann et al., 2019; Corrò et al., 2020; Kim et al., 2020). The most recent success includes establishing organoid culture simulating snake venom glands for venom peptide production (Post et al., 2020) and application of various organoid systems for disease modeling; this is emphasized to efficiently model infections by viruses, a group of cell-obligated infectious agents challenging the classical 2D cell cultures (Barrila et al., 2018; Ramani et al., 2018; Sachs et al., 2019; Bar-Ephraim et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Yuki et al., 2020).
Animal models and cell cultures (including both primary cells and immortalized cell lines) are broadly used in most aspects of virological studies; this includes assays involving virus entry, titration, cytopathy, and rescue (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). However, these classical models present obvious limitations (Barrila et al., 2018; Ramani et al., 2018; Bar-Ephraim et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Yuki et al., 2020). These limitations are particularly related to physiological similarity and inter-cell interaction of natural hosts. Of the noted advantages, organoids have been used to extend virological studies in several aspects (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). First, organoids recapitulate viral infections more efficiently from naturally infected tissue samples in the laboratory. When compared to the general need for extensive screening and serial adaptation using cell cultures (especially cancerous cell lines from other species), organoids might support the replication of the wild-type virus when they originate from relevant organs where clinical isolates are surgically obtained (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). It has been shown that viruses from most infected human samples (e.g., nasopharyngeal swabs, blood, or feces) were more readily grown in organoids or airway epithelial cultures (Palmer et al., 2012; Tapparel et al., 2013; Bochkov et al., 2016; Sridhar et al., 2020). Animal viruses are often empirically cultured and studied using cell lines of primate or other species origins; and this is due to the unavailability or lack of support regarding cell cultures from the species of a viral natural host. For examples, swine arterivirus (Cafruny et al., 2006; Calvert et al., 2007) and coronaviruses were replicated and studied using cell lines from the monkey kidney (Hofmann and Wyler, 1988; Govorkova et al., 1996; Li et al., 2019ab, 2020; Luo et al., 2020; Yin et al., 2020) and MDCK, a canine kidney cell line used widely for studies of influenza virus from human and various animal resources (Takada et al., 2019). Given the species-tropism of viruses, extensive targeting is needed for adaption and mutation using animal models to replicate productive infections of human viruses; this kind of cross-species adaptation increases biosafety concerns (Simmonds et al., 2019; Takada et al., 2019; Dai et al., 2020; Kuchipudi et al., 2021). Nonetheless, viral tropism, receptor usage, and virus-cell interaction in these species-distinct cells or animal models may divert from genuine virus-host interaction in the natural host (Hofmann and Wyler, 1988; Govorkova et al., 1996; Simmonds et al., 2019; Takada et al., 2019; Dai et al., 2020; Kuchipudi et al., 2021).
Second, organoids enable the culture of viruses that are unculturable in cells. As with a number of virological study cases, it is futile to find suitable cell lines to replicate and rescue a wild-type virus that causes active infection in vivo (Leland and Ginocchio, 2007; Rabenau et al., 2007; Hodinka, 2013). Organoid systems constitute a convenient alternative for culturing these viruses under laboratory conditions (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). Despite extensive tests using available cell lines, viruses including human norovirus, human coronavirus HKU1, bocavirus, and rhinovirus C remain futile for productive replication; this is possibly due to certain viruses’ need for inter-cell or cell-matrix interaction for effective infection. Studies showed that human intestinal organoids supported norovirus and rotavirus infection; human airway epithelial cultures allowed culture and characterization of human coronaviruses HKU1 and NL63, human bocavirus, and human rhinovirus C, which implicated the potential of relevant respiratory organoids (Dijkman et al., 2009; Pyrc et al., 2010; Hao et al., 2012; Tapparel et al., 2013; Farsani et al., 2015). Therefore, multiple cell types differentiated in organoid systems provide updated culturable biological platforms, and these can be utilized for further virological characterization and rescuing of viruses from clinical isolates (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020).
Third, organoid systems allow the determination of authentic virus-host interaction and recapitulation findings in vivo; this is still in its infancy in terms of virological application. Epithelia that line the skin, blood vessels (specified as endothelia), and internal tracts/glands of digestive and respiratory systems provide primary portals for virus infection initiation. In the body, epithelial cells are polarized as apical (oriented toward the lumen) and basolateral (oriented away from the lumen) sides with differential expression of various surface molecules; this can be exploited as entering receptors for viral infections (Bomsel and Alfsen, 2003; Compans and Herrler, 2005). Contrary to the loss or difference of surface molecules expressed in 2D-cultured cells and animal models of other species, organoids of relevant tissues reconstruct the cellular organization/polarization; this allows dissecting the polarized entry and an in vitro spread of viruses in the epithelial barrier (Bomsel and Alfsen, 2003; Compans and Herrler, 2005; Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). The characterization of measles virus (MV) cellular receptors provides an eloquent example. This calls for increased use of “vivo” models—such as organoids—for characterizing the pathogenesis of wild-type viruses. Previous studies using vaccine- and laboratory-adapted MV strains and cell cultures resulted in the discovery of the MV cellular receptor as CD46. This was a cell-surface molecule widely expressed by nucleated cells. The discovery was based on in vitro findings induced by the thought that MV infection entered the apical side of respiratory epithelia (Dorig et al., 1993). Recent studies incorporating ex vivo and in vivo approaches determined nectin-4 as a genuine receptor of wild-type MV strains. Nectin-4 is restrictively expressed at the basolateral side of epithelial cells. This indicated that respiratory epithelial cells are infected later from the basolateral side post-epithelial damage. The virus is then released into the respiratory tract. This contrasted the previous proposal of MV initiating epithelial infection through its apical side (Ludlow et al., 2010; Laksono et al., 2020).
Organoid systems also potentially demonstrate clinical signs, as observed in viral infections of natural hosts. For instance, 5–14% of children born with Zika virus infections suffered severe neurological complications, including inducing microcephaly or abnormally small heads (Qian et al., 2017). Recently, this clinical observation was recapitulated using human-brain organoids derived from induced PSCs (iPSCs). It was shown that Zika virus infection reduced organoid size in mimicking the virus-caused microcephaly in vivo. Studies using human brain organoids also potentiated insight of neurotropism and pathogenesis of other neurotropic viruses; this including neonatal herpes simplex virus (HSV) and congenital-cytomegalovirus, causing broad neurological defects such as microcephaly (Wen et al., 2017; Brown et al., 2019; D’Aiuto et al., 2019; Depla et al., 2020; Sridhar et al., 2020). Studies using porcine enteroids—generated from different segments of the intestine—successfully recapitulated the segment- and cell-dependent tropism. Using porcine enteroids, the antiviral interferon (IFN) response as observed in vivo was demonstrated during infections with two porcine enteric coronaviruses, porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV) (Li et al., 2019a,b, 2020; Luo et al., 2020; Yin et al., 2020). In addition, the virus-organoid models can be readily applied to drug screening procedures, and this leads to effective therapeutic interventions for mitigating viral infection (Ramani et al., 2018; Depla et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Figure 1).
Fourth, organoids extend the capacity of a culture system to support replication of multiple viruses (including pathogens of other phyla) that have different cell tropisms. This allows for studying ecological interaction among co-infected agents and with the host systems. While these kinds of co-infection models better reflect the complexity of natural infections, they are increasingly necessary. An updated characterization is also need in regards to the microbiome’s contribution in shaping viral infections and antiviral immunity. For example, human enteroids showed susceptibility to infection with several enteroviruses, including echovirus 11 (E11), coxsackievirus B (CVB), and enterovirus 71 (EV71). However, they depended on different cell lineages—contained by the enteroids—to induce virus-specific antiviral and inflammatory responses. Enteroids therefore provide an ecological hub for characterization of virus-specific pathogenesis; this also includes antiviral responses with the presence of interacting microbes (Sridhar et al., 2020). The use of organoids to model viral co-infections promotes better understanding and therapeutic interventions for syndromes or disease complexes; these are centered by one to several etiological viruses commonly observed in the digestive, respiratory, and reproductive tracts (Kim et al., 2020; Min et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021).
Over a dozen types of organoids have been successfully established per human organs, including the brain, intestine, kidney, liver, lung, and pancreas (Kaushik et al., 2018; Lehmann et al., 2019; Corrò et al., 2020; Kim et al., 2020). Most of these human organoids were also applied in disease modeling; several of these were piloted to study diseases caused by viruses, including human rotavirus, coronaviruses HKU1 and VL63, bocavirus, rhinovirus C, measles virus, enteroviruses, and the Zika virus (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). The outbreak and ongoing pandemic of the new coronavirus 2019 disease (COVID-19) drove the wide application of organoids; this was done to recapitulate the disease and validate its virological insights of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Clevers, 2020; Sridhar et al., 2020). The lung organoids—or organoid-like alveolar epithelia (HAE)—containing specified airway ciliated cells are productive in supporting SARS-CoV-2 replication, as clinically observed in vivo (Sridhar et al., 2020; Yang et al., 2020; Han et al., 2021). The respiratory organoids and the HAE model also demonstrated virus-caused epithelial damage and death processes underlying the plaque-like cytopathies observed in the lung of COVID-19 pneumonia (Milewska et al., 2020; Yang et al., 2020; Han et al., 2021). For therapeutic evaluation, Pizzorno et al. (2020) showed that remdesivir and remdesivir–diltiazem were effective against SARS-CoV-2 infection in both nasal and bronchial epithelia cultures (Pizzorno et al., 2020; Sridhar et al., 2020).
SARS-CoV-2 transmission among humans occurs principally through exposure to virus-containing respiratory fluids (Clevers, 2020; Han et al., 2021). It must be noted that effective transmission may occur through mucous membranes in the mouth, nose, or eyes. This can happen as a result of either passive deposition or active inhalation. This indicates that the epithelial lining of these body organs are generally susceptible to the virus. After initial infection in the airway epithelia, SARS-CoV-2 may spread systemically and cause dysfunction in multiple organs such as the gut, liver, kidney, testicle, and brain. Vasculitis or endothelitis ascribes inflammation of blood vessels or vascular cells (endothelia), which consist of a major systemic symptom underlying most organ-specific manifestations in COVID-19 patients. Therefore, COVID-19 was proposed to be a vascular disease, and this suggests that SARS-CoV-2 causes direct endothelial infection/injury while mediating multi-system dysfunction (Siddiqi et al., 2021). Penninger et al. established both capillary organoids and kidney organoids from human iPSCs, and they demonstrated that SARS-CoV-2 could directly infect endothelial cells therein (Monteil et al., 2020). These findings explain the systemic spread of the virus; it also explains renal dysfunction in patients with severe COVID-19 (Monteil et al., 2020; Siddiqi et al., 2021). The potential of SARS-CoV-2 to cause secondary infections in multiple organs was also demonstrated using organoid models. Several studies showed productive infection of SARS-CoV-2 in human enteroids or intestinal organoids; and demonstrated the sentinel role of Type III interferons (IFNs)—a group of epithelia-specific IFNs—in controlling the viral infection at an early phase (Lamers et al., 2020; Stanifer et al., 2020). Findings implicated direct gut infection and potential of fecal transmission of SARS-CoV-2, and rationalized the prolonging detection of SARS-CoV-2 in fecal samples during clinical screening of convalescent patients (Clevers, 2020; Sridhar et al., 2020; Wu et al., 2020).
Neurological manifestations occur in about 36.4% of COVID-19 patients. Relevant complications include loss of smell, headaches, ischemic stroke, muscle weakness, and encephalitis. Neuropathic pain has even been found as a post-COVID-19 symptom. These observations have promoted the use of brain organoids for assessing potential SARS-CoV-2 neuro-invasion. Two studies demonstrated that SARS-CoV-2 targeted neuron cells to cause direct infection of human brain organoids (Pellegrini et al., 2020; Ramani et al., 2020; Zhang et al., 2020). Using choroid plexus organoids, Pellegrini et al. (2020) demonstrated that SARS-CoV-2 was capable of causing brain infection through targeting the network of brain blood vessels; this explained the presence of the virus in the cerebrospinal fluid (CSF) and other parts of the brain. Therefore, studies using tissue-specific organoids of the human brain provide eloquent evidence regarding neurological complications of SARS-CoV-2 infections, as clinically observed in a portion of COVID-19 patients (Clevers, 2020; Pellegrini et al., 2020; Ramani et al., 2020; Sridhar et al., 2020; Zhang et al., 2020). Further studies increasing throughput using patient-originated organoids may critically address the broad diversity of COVID-19 manifestations as well as the association with various preexisting comorbidities in the patients (Clevers, 2020; Sridhar et al., 2020).
SARS-CoV-2 seems to have a zoonotic origin evolutionarily from relevant coronaviruses in horseshoe bats; however, there is no scientific consensus about certain intermediate host(s) to bridge SARS-CoV-2 transmission to human beings (Conceicao et al., 2020; Sang et al., 2020, 2021). Remarkably, several animal species—including dogs, cats, minks, ferrets, hamsters, lions, and tigers—of both domestic and wild groups have been shown to be infected by SARS-CoV-2 through a zooanthroponotic (or reverse-zoonotic) way (Conceicao et al., 2020; Sang et al., 2020, 2021). It is impractical to experimentally validate viral susceptibility in most relevant animals, especially in wild species; however, organoid cultures and epidemiological prediction can be integrated to handle this task, especially after essential optimization from the established human systems (Figure 3; Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). In this regard, Zhou et al. (2020) established intestinal organoids from a China horseshoe bat species using the culture condition for the human counterpart. As expected, the bat intestinal organoids were readily infected by SARS-CoV-2, in contrast to abortive trials using cell cultures (Zhou et al., 2020).
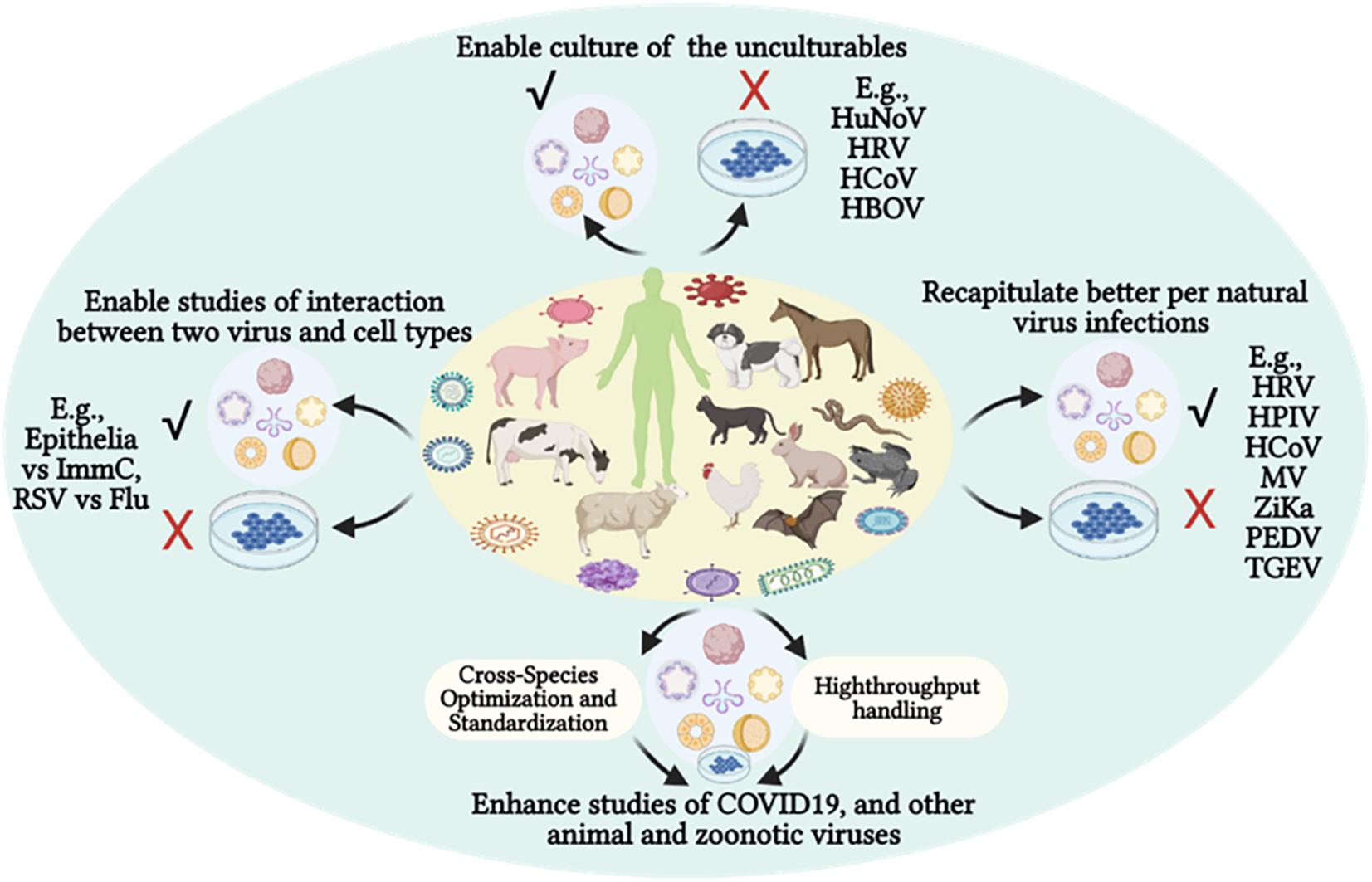
Figure 3. Organoid culture potentiates virological studies, providing a better system to enable recapitulating of the natural viral infections regarding productive culture (√) of previous unculturable viruses using cell lines (X) and mimicking in vivo clinical signs and cell interactions. The advantages of organoid cultures in virological studies have recently been demonstrated using multiple human viruses—particularly in COVID-19 studies—and have shaped prospective cross-species modeling of epizootic or zoonotic viral infections. COVID-19, new coronavirus disease 2019; Flu, influenza virus; HBOV, human bocavirus; HCoV, endemic human respiratory coronaviruses; HPIV, human parainfluenza virus; HuNoV, human norovirus; HRV, human rhinovirus; ImmC, immune cells; MV, measles virus; PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis coronavirus; RSV, respiratory syncytial virus; Zika, Zika virus.
Along with emerging virome studies, viruses of known and unknown families were profiled from animal fecal samples as well as other animal or environmental samples at the genome level (Cadwell, 2015; Ramani et al., 2018; Wang, 2020). Nonetheless, most of these viruses are incapable of replication using cell culture or even laboratory animal models (Cadwell, 2015; Wang, 2020). This bat and other animal organoid studies are promising indications that culture conditions for human organoids should generally be applicable for most mammalian—and even vertebrate—species (Table 2; Derricott et al., 2019; Seeger, 2020; Zhou et al., 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021). Cross-species development of comparable organoid cultures will provide an effective biological system for validating the zoonotic (and reverse zoonotic) potential of newly emerging viruses. This potentiates cross-species assessment of virus-susceptibility in both domestic and wild animals in order to prevent zooanthroponotic and re-emerging zoonotic cycles (Conceicao et al., 2020; Sang et al., 2020, 2021). Moreover, organoids generated from animal organs—aligned with their respective virome studies—will allow culturing of novel viruses that are difficult to culture in established cell lines (Cadwell, 2015; Ramani et al., 2018; Moghaddam et al., 2019; Wang, 2020).
Infection tests in animals provide direct evidence for cross-species and species-specific infectivity of emerging animal and zoonotic viruses. Limitations of animal models owe to individual diversity and concerns on animal welfare. However, organoid systems offer a great substitute to document cross-species and species-different infectivity of targeted viruses (Sridhar et al., 2020; Hofer and Lutolf, 2021). In this regard, studies using human organoids have proven to be efficient prospects for investigating the species-specific susceptibility of influenza viruses. Influenza viruses—particularly the type A subfamily (IAV)—infect a broad range of vertebrate species; they also present a paradigm for studying emerging viruses that possess both epizootic and zoonotic threats (Long et al., 2019; Hofer and Lutolf, 2021). Human airway organoids from iPSCs contained the major airway epithelial cells types including ciliated cells, goblet cells, club cells, and basal cells. When applied to the modeling of IAV infections, these airway organoids provided discernment of different infectivity of emerging flu viruses, particularly those of avian or human origin. As shown in one comparative study, human airway organoids behaved similarly to the ex vivo cultured bronchus explants; this regarded tissue/cell tropism, virus production, and cytokine response to either human or avian IAV strains (Long et al., 2019; Sridhar et al., 2020; Hofer and Lutolf, 2021). With the procedural development for successful cryopreservation, subculture, and imaging resolution, human organoids have the potential to examine zoonotic threats of influenza and other emerging viruses (Wiley et al., 2016; Tsai et al., 2018; Dekkers et al., 2019; Tysoe et al., 2019). An estimation of over 60% of known infectious diseases in humans are zoonotic in origin; prominent diseases have the organ tropism for the respiratory or digestive systems, and can even span multiple systems (Jones et al., 2008). Most organoid cultures validated in animals are of the intestinal type (Table 2). Urgent and increasing public concerns for studying emerging animal and zoonotic viruses should be considered; it is imperative to adapt human organoid technology and develop animal organoid systems for respiratory and other organ types (Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Miller et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020). However, further challenges for the application of organoids in virological studies await substantial species-specific optimization and cross-species standardization to harness organoid cultures in a high-throughput platform (Figure 3).
Until recently, studies of stem cell biology and organoid systems have been performed primarily in humans and mice (Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020). Despite practices for establishing intestinal and other organoids in most domestic and few wild animal species (Table 2), species-specific optimization of culture conditions is required for maintaining and differentiating organoids (Derricott et al., 2019; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Kar et al., 2021; Pain, 2021). Even for the generation of human or mouse intestinal organoids, empirically conditioned media are used to provide essential growth factors including Wnt3a, Noggin, and R-spondin; these factors foster the growth and differentiation of stem cells into organotypic structures (Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021). This introduces uncertainties for reproducing some organoid cultures in different laboratories within the same species (Sridhar et al., 2020; Hofer and Lutolf, 2021). Most growth factors for organoid cultures can be specified using recombinant proteins from reliable commercial suppliers. However, a recent study using side-by-side comparison indicated that Wnt3a-conditioned media was more supportive for the long-term survival of human or mouse colon organoids (Wilson et al., 2021). In addition, the availability of relevant recombinant proteins is questionable for most animal species (Dawson et al., 2020). Hence, in organoid cultures of another species, the use of growth factors (and conditioned media) with a human- or mouse-origin requires a substantial optimization process; it may even work better at different dosages or regimens (Shanks et al., 2009; Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Mead and Karp, 2019; Moghaddam et al., 2019; Corrò et al., 2020; Dawson et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021).
After establishing an organoid culture, it is subjected to characterization— particularly for cell heterogeneity and lineage differentiation (gene expression)—in mimicking respective organ proxies in addition to dynamically monitoring the morphology and growth property of the organoid. This requires immunoreagents to determine various cell markers; these have been historically developed in humans and mice and are very limited for most animal species (Faldyna et al., 2007; Sopp et al., 2007; Dawson et al., 2020). If no cross-reactive immunoreagents can be identified from the available resources, it is necessary to target the development of these species-specific immunoreagents and growth factors; this provides the basis for the authentic characterization of animal organoids (Faldyna et al., 2007; Sopp et al., 2007; Dawson et al., 2020). Because of the ultimate goal—namely, usage in virological studies—intensive molecular and cellular characterization may be performed in parallel or secondary to virus infection tests; this can determine functional potential in virological research (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020).
Variation inherited from starting materials and conditioned media generated by different laboratories introduce challenges, particularly for cross-species standardizing of a given type of organoid culture, or even solely in humans (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). Previous studies have shown that human intestinal organoids—derived in different medium formulations or using different starting tissues—had different levels of marker gene expression and cellular composition (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). More applicable criteria and guidelines may be needed for cross-species organoid cultures to reduce variability; however, the supportive fact for a given virus infection in the organoids should be a major consideration, provided the major tissue proxy is recapitulated (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). In this context, a harmonized system for cross-species culture of organoids is proposed for modeling animal infectious diseases. Holthaus et al. (2021) generated intestinal organoids for four species—human, mouse, pig, and chicken—which are common hosts of Apicomplexa and other protozoa of zoonotic concern. This study unified the culture of intestinal organoids from different species into a comparable system for studying the protozoan infections. In addition, they provided straightforward guidelines and a simple medium formulation for cross-species generation of intestinal organoids from respective intestinal crypts. Using the organoid-derived monolayer culture in a Transwell system, they demonstrated the suitability of this system to support the protozoan infections as well as the potential to study parasite-host interactions for relevant intestinal protozoan co-infections (Holthaus et al., 2021). Currently, no virological studies have reported a form of comparison using cross-species organoids; however, it warrants relevant studies to determine cross-species susceptibility of emerging animal and zoonotic viruses (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020; Holthaus et al., 2021).
Adequate scale-up defines the maturity of organoid systems that target cross-species validation of viral infections and antiviral responses. Aside from essential species-specific optimization and inter-species comparison, increasing throughput of handling comprises a further challenge for efficient use of organoid technology, particularly in screening of virus susceptibility and therapeutic antivirals (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). This must be arranged to master resource suppliers like American Tissue and Cell Culture (ATCC); it must also unify standards for quality control in regards to the complex nature of the organoid systems. In addition to the existing commercial settings and multiplex formats developed primarily for cell cultures, the invention and utilization of multiplex culture formats/scaffolds are needed for culturing and analyzing organoids. This kind of high-throughput adaptation will make organoid application compatible with extensive omics technology, facilitating studies on system virology that target the goal of one-health initiative (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021).
Most organoids mainly recapitulate a single-organ system representing the epithelial capsule of specified functions (Yin et al., 2016; Kaushik et al., 2018; Lehmann et al., 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Hofer and Lutolf, 2021). For instance, gut organoids comprise the villus-bearing enterocytes, goblet cells, and enteroendocrine cells. Lung organoids contain ciliated cells, club cells, goblet cells, and basal cells. In general, organoids (with the exception of organoids for lymph nodes or tonsils) lack the mesenchymal or immune-cell niches that are integrated with their respective organs (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). This prevents studying inter-system reactions that are commonly observed in antiviral responses involving interacting parts in vivo (Bar-Ephraim et al., 2020; Yuki et al., 2020). Tissue stromal cells and especially immune cells—such as macrophages for porcine arterivirus and bovine viral diarrhea virus—commonly serve as susceptible cells to define a virus tropism (Sang et al., 2015). Lack of these supportive cells in organoids may result in non-productive infection and virological modeling failure (Sang et al., 2015; Bar-Ephraim et al., 2020; Yuki et al., 2020). Both tissue stromal cells and immune cells function in secreting cytokines to shape the local microenvironment niche and potentiate antiviral immunity; this, in turn, affects the susceptible status of epithelial cells in the organoids (Sang et al., 2015; Bar-Ephraim et al., 2020; Yuki et al., 2020). This may provide a rationale for the inability to infect rabbit enteroids using a rabbit poxvirus (Kardia et al., 2021). Several approaches have been proposed to include stromal and immune cells in organoids. First, epithelial organoids may be co-cultured with tissue stromal cells, immune cells (including macrophages, dendritic cells, and T cells), or even the cell population from organoids of blood vessels or lymph nodes. Using this co-culture system, several studies have successfully demonstrated the interaction of epithelial cells and immune cells in antiviral reactions (Miranda et al., 2018; Holloway et al., 2019; Duzagac et al., 2021; Hofer and Lutolf, 2021). Unlike their ASC-derived counterparts, PSC-derived epithelial organoids may contain a mesenchymal layer, as shown in some human and mouse studies. Most animal organoids were ASC-derived due to the lack of specific PSCs and the comparative convenience of culture handling over the PSC-dependent protocol (Derricott et al., 2019; Seeger, 2020; Beaumont et al., 2021; Bourdon et al., 2021; Hofer and Lutolf, 2021; Kar et al., 2021; Pain, 2021). Second, the organ-on-a-chip system can be used to incorporate different organotypic functions. The organ-on-a-chip system is based on a reductionist engineering approach to culture major tissue cells; this uses a 3D microfluidic cell culture chip to capture key functions of an organ (Miranda et al., 2018; Holloway et al., 2019; Duzagac et al., 2021). Different types of organoids provide functional organ components in order to model the complexity of viral pathogenesis and antiviral immunity at a multi-system level. This multi-system organization of organoids may be suitable for investigating immunometabolic and immunoneurologic interaction underlying antiviral regulation (Bar-Ephraim et al., 2020; Yuki et al., 2020; Hofer and Lutolf, 2021). Third, multi-organ organoids—or organoids that include different physiological niches—can be developed through advanced organoid engineering technology (Min et al., 2020; Hofer and Lutolf, 2021).
The microbiota represents another functional niche coating both internal organ tracts and external skin; this plays a critical role in propelling pathogenic infections and training antiviral immunity (Rooks and Garrett, 2016; Li et al., 2019c). Due to the sterile condition of the stem cell and organoid cultures and the complexity of animal microbiota, it is not practical to include even most commensal microbes (including viruses that are numerously prominent there) during organotypic cultures. Recent studies have shown that immune education by microbiota can be rebuilt; this can be done by introducing only one to few representative microbes in the gut of germ-free mice. It was proposed that Escherichia coli, Helicobacter pylori, or rotavirus might be included to recapitulate the immune education role of microbiota in intestinal organoids (Min et al., 2020).
Despite its success and potential as demonstrated, both organoid technology and its application in disease modeling are still in infancy (Shanks et al., 2009; Yin et al., 2016; Kaushik et al., 2018; Ramani et al., 2018; Lehmann et al., 2019; Mead and Karp, 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). Table 3 lists major technological limitations/challenges and relevant resolutions of current organoid systems in virological applications. In summary, one inherent limitation stems from the short lifespan of most organoids (especially those generated from various organ ASCs) and the consequential immaturity compared to the adult organs in vivo. Given the need for modeling virus infections at the adult and even senescent status, some physical or biochemical stimuli may be required to induce maturation of most organoids (Kim et al., 2020; Sridhar et al., 2020; Hofer and Lutolf, 2021). The newly developed protocol for cryopreservation then sub-passage of an established organoid may physically elongate the duration, particularly for using the “colonized” organoid and increasing reproducibility of relevant tests (Wiley et al., 2016; Tsai et al., 2018; Dekkers et al., 2019; Tysoe et al., 2019). Otherwise, virus adaption may be necessary for a provided organoid system in a relatively juvenile state. Secondly, most organoids may form encapsulated epithelial structures within the enclosed lumen and lumen-facing apical side. This causes a limited exchange of nutrients and waste removal from the lumen side. It requires special handling (such as injection or physical poration) to introduce viruses inside the lumen, as most epithelia-tropic viruses infect from the apical side (Sridhar et al., 2020; Hofer and Lutolf, 2021). An alternative is to culture organoid cells to form 2D cell layers in a Transwell system; this will be analogical to the classical 2D culture for efficient cell exposure to virus infection and facilitate downstream optical analyses—among others—based on cells (Wiley et al., 2016; Tsai et al., 2018; Dekkers et al., 2019; Tysoe et al., 2019). In addition, a recent study developed an apical-out intestinal organoid in pigs; this was used to successfully model the infection of porcine TGEV and study local interferon responses (Li et al., 2020). Finally, the recent application of engineering principles to organoid technology enables inter-system incorporation (such as vascular systems, immune cells, and microbiota niches); it also increases experimental robustness, which is applicable to promoting organoid uses for virus research (Miranda et al., 2018; Holloway et al., 2019; Sridhar et al., 2020; Duzagac et al., 2021; Hofer and Lutolf, 2021).
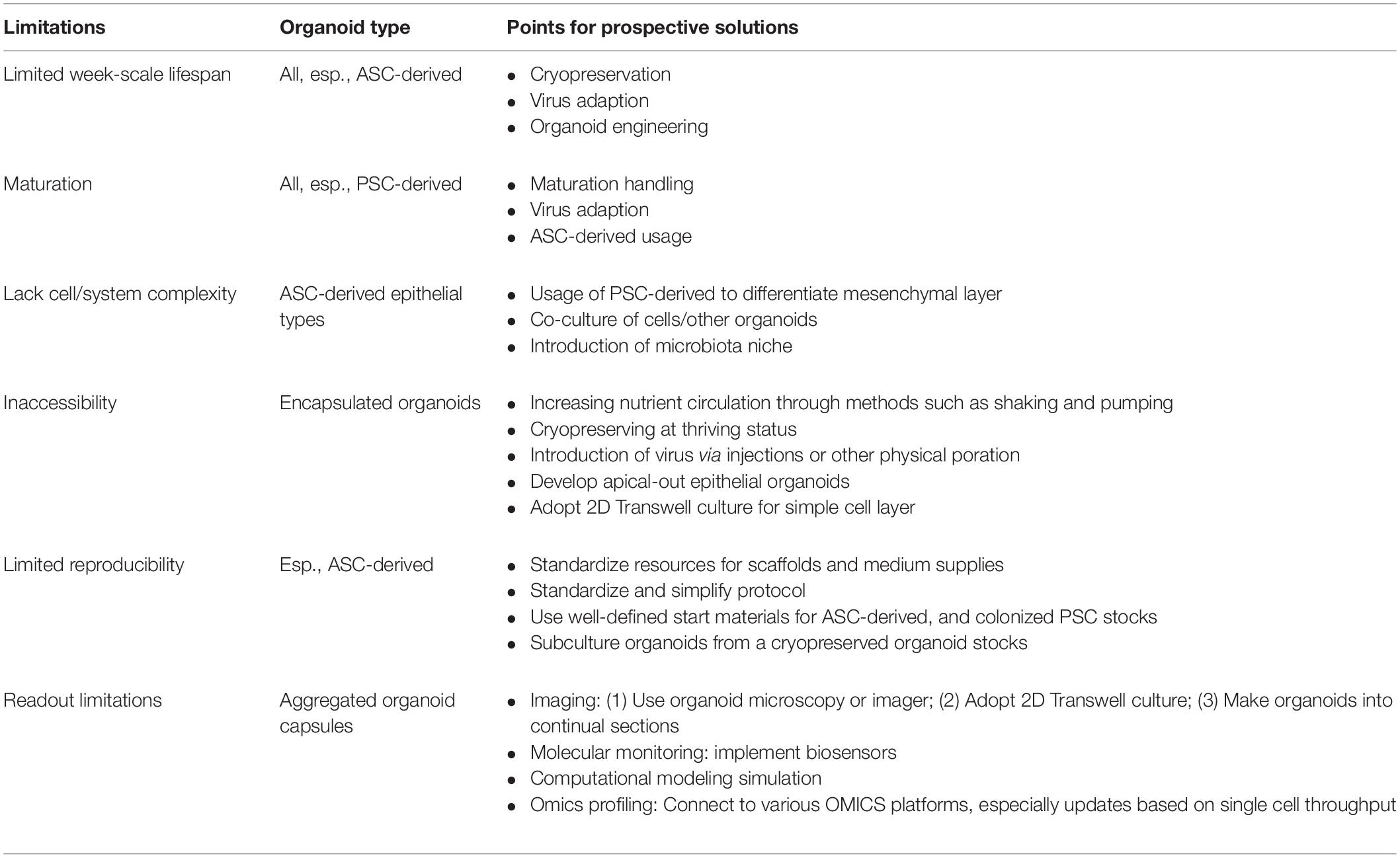
Table 3. Limitations of current organoid systems and potential solutions for virus research (Shanks et al., 2009; Lehmann et al., 2019; Kim et al., 2020; Hofer and Lutolf, 2021).
Organoid systems utilize the self-organizing properties of stem cells to model multi-cellular proxies of organ tissues. Possessing properties intermediate between conventional cell culture and animal models, organoids have gained enormous interest for modeling disease, personalized medicine, drug screening, and organ therapy in humans (Shanks et al., 2009; Yin et al., 2016; Kaushik et al., 2018; Ramani et al., 2018; Lehmann et al., 2019; Mead and Karp, 2019; Moghaddam et al., 2019; Corrò et al., 2020; Fuchs and Blau, 2020; Kim et al., 2020; Schutgens and Clevers, 2020; Hofer and Lutolf, 2021). There are limitations presented in cell culture and animal models to recapitulate viral pathogenesis and host antiviral responses; for this reason, organoid systems are attracting attention in emerging, authentically-validating animal and zoonotic viruses across animals and humans (Ramani et al., 2018; Schutgens and Clevers, 2020; Sridhar et al., 2020). Organoids have already broadened virological studies in the human context, particularly with recent use in COVID-19 research (Clevers, 2020; Sridhar et al., 2020). Despite considerable success in modeling viral infections, limitations and challenges remain in virological studies involving emerging animal and zoonotic viruses (Ramani et al., 2018; Clevers, 2020; Schutgens and Clevers, 2020; Sridhar et al., 2020). Further studies are needed to standardize culture protocols, apply engineering principles, and increase both the reproducibility and utility in virological research and therapies (Kim et al., 2020; Hofer and Lutolf, 2021; Holthaus et al., 2021).
YS conducted overall conceptualization, reference collection and process, digestion, draft writing and finalization, and funding acquisition. LM, RN, and LG-L contributed to the conception, discussion, and proofreading. All authors contributed to the article and approved the submitted version.
This work was supported by USDA NIFA Evans-Allen-1013186 and NIFA 2018-67016-28313 to YS, and in part through reagent sharing of NIFA AFRI 2020-67016-31347 and NSF-IOS-1831988 to YS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acharya, M., Arsi, K., Donoghue, A. M., Liyanage, R., and Rath, N. C. (2020). Production and characterization of avian crypt-villus enteroids and the effect of chemicals. BMC Vet. Res. 16:179. doi: 10.1186/s12917-020-02397-1
Adegbola, S., Moore, J., Sahnan, K., Tozer, P., Phillips, R., Warusavitarne, J., et al. (2017). Establishing a porcine model to translate anorectal stem cell organoid models to elucidate the aetiology of perianal Crohn’s fistulae. J. Crohns. Colitis 11, S81–S82.
Bar-Ephraim, Y. E., Kretzschmar, K., and Clevers, H. (2020). Organoids in immunological research. Nat. Rev. Immunol. 20, 279–293. doi: 10.1038/s41577-019-0248-y
Barrila, J., Crabbé, A., Yang, J., Franco, K., Nydam, S. D., Forsyth, R. J., et al. (2018). Modeling host-pathogen interactions in the context of the microenvironment: three-dimensional cell culture comes of age. Infect Immun. 86:e00282-18. doi: 10.1128/IAI.00282-18
Beaumont, M., Blanc, F., Cherbuy, C., Egidy, G., Giuffra, E., Lacroix-Lamandé, S., et al. (2021). Intestinal organoids in farm animals. Vet. Res. 52:33.
Bochkov, Y. A., Watters, K., Basnet, S., Sijapati, S., Hill, M., Palmenberg, A. C., et al. (2016). Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa–E8 cells. Virology 499, 350–360. doi: 10.1016/j.virol.2016.09.025
Bomsel, M., and Alfsen, A. (2003). Entry of viruses through the epithelial barrier: pathogenic trickery. Nat. Rev. Mol. Cell Biol. 4, 57–68. doi: 10.1038/nrm1005
Bourdon, G., Cadoret, V., Charpigny, G., Couturier-Tarrade, A., Dalbies-Tran, R., Flores, M.-J., et al. (2021). Progress and challenges in developing organoids in farm animal species for the study of reproduction and their applications to reproductive biotechnologies. Vet. Res. 52:42.
Brown, R. M., Rana, P. S. J. B., Jaeger, H. K., O’Dowd, J. M., Balemba, O. B., and Fortunato, E. A. (2019). Human cytomegalovirus compromises development of cerebral organoids. J. Virol. 93:e00957-19. doi: 10.1128/JVI.00957-19
Cadwell, K. (2015). The virome in host health and disease. Immunity 42, 805–813. doi: 10.1016/j.immuni.2015.05.003
Cafruny, W. A., Duman, R. G., Wong, G. H., Said, S., Ward-Demo, P., Rowland, R. R., et al. (2006). Porcine reproductive and respiratory syndrome virus (PRRSV) infection spreads by cell-to-cell transfer in cultured MARC-145 cells, is dependent on an intact cytoskeleton, and is suppressed by drug-targeting of cell permissiveness to virus infection. Virol. J. 3:90.
Calvert, J. G., Slade, D. E., Shields, S. L., Jolie, R., Mannan, R. M., Ankenbauer, R. G., et al. (2007). CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81, 7371–7379. doi: 10.1128/JVI.00513-07
Chandra, L., Borcherding, D. C., Kingsbury, D., Atherly, T., Ambrosini, Y. M., Bourgois-Mochel, A., et al. (2019). Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biol. 17:33. doi: 10.1186/s12915-019-0652-6
Chen, T. C., Neupane, M., Chien, S. J., Chuang, F. R., Crawford, R. B., Kaminski, N. E., et al. (2019). Characterization of adult canine kidney epithelial stem cells that give rise to dome-forming tubular cells. Stem Cells Dev. 28, 1424–1433. doi: 10.1089/scd.2019.0049
Clevers, H. (2020). COVID-19: organoids go viral. Nat. Rev. Mol. Cell Biol. 21, 355–356. doi: 10.1038/s41580-020-0258-4
Compans, R. W., and Herrler, G. (2005). Virus infection of epithelial cells. Mucosal Immunol. 9, 769–782. doi: 10.1016/B978-012491543-5/50045-0
Conceicao, C., Thakur, N., Human, S., Kelly, J. T., Logan, L., Bialy, D., et al. (2020). The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 18:e3001016. doi: 10.1371/journal.pbio.3001016
Corrò, C., Novellasdemunt, L., and Li, V. S. W. (2020). A brief history of organoids. Am. J. Physiol. Cell Physiol. 319, C151–C165. doi: 10.1152/ajpcell.00120.2020
Dai, X., Zhang, X., Ostrikov, K., and Abrahamyan, L. (2020). Host receptors: the key to establishing cells with broad viral tropism for vaccine production. Crit. Rev. Microbiol. 46, 147–168. doi: 10.1080/1040841X.2020.1735992
D’Aiuto, L., Bloom, D. C., Naciri, J. N., Smith, A., Edwards, T. G., McClain, L., et al. (2019). Modeling herpes simplex virus 1 infections in human central nervous system neuronal cells using two- and three-dimensional cultures derived from induced pluripotent stem cells. J. Virol. 93:e00111-19. doi: 10.1128/JVI.00111-19
Dawson, H. D., Sang, Y., and Lunney, J. K. (2020). Porcine cytokines, chemokines and growth factors: 2019 update. Res. Vet. Sci. 131, 266–300. doi: 10.1016/j.rvsc.2020.04.022
Dekkers, J. F., Alieva, M., Wellens, L. M., Ariese, H. C. R., Jamieson, P. R., Vonk, A. M., et al. (2019). High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771. doi: 10.1038/s41596-019-0160-8
Depla, J. A., Sogorb-Gonzalez, M., Mulder, L. A., Heine, V. M., Konstantinova, P., van Deventer, S. J., et al. (2020). Cerebral organoids: a human model for AAV capsid selection and therapeutic transgene efficacy in the brain. Mol. Ther. Meth. Clin. D 18, 167–175. doi: 10.1016/j.omtm.2020.05.028
Derricott, H., Luu, L., Fong, W. Y., Hartley, C. S., Johnston, L. J., Armstrong, S. D., et al. (2019). Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res. 375, 409–424. doi: 10.1007/s00441-018-2924-9
Dijkman, R., Koekkoek, S. M., Molenkamp, R., Schildgen, O., and van der Hoek, L. (2009). Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83, 7739–7748. doi: 10.1128/jvi.00614-09
Dorig, R. E., Marcil, A., Chopra, A., and Richardson, C. D. (1993). The human Cd46 molecule is a receptor for measles-virus (edmonston strain). Cell 75, 295–305. doi: 10.1016/0092-8674(93)80071-l
Duque-Correa, M. A., Maizels, R. M., Grencis, R. K., and Berriman, M. (2020). Organoids - new models for host-helminth interactions. Trends Parasitol. 36, 170–181. doi: 10.1016/j.pt.2019.10.013
Duzagac, F., Saorin, G., Memeo, L., Canzonieri, V., and Rizzolio, F. (2021). Microfluidic organoids-on-a-chip: quantum leap in cancer research. Cancers 13:737. doi: 10.3390/cancers13040737
Eiraku, M., Watanabe, K., Matsuo-Takasaki, M., Kawada, M., Yonemura, S., Matsumura, M., et al. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. doi: 10.1016/j.stem.2008.09.002
Elbadawy, M., Usui, T., Mori, T., Tsunedomi, R., Hazama, S., Nabeta, R., et al. (2019). Establishment of a novel experimental model for muscle-invasive bladder cancer using a dog bladder cancer organoid culture. Cancer Sci. 110, 2806–2821. doi: 10.1111/cas.14118
Evans, M. (1981). Origin of mouse embryonal carcinoma cells and the possibility of their direct isolation into tissue culture. J. Reprod. Fertil. 62, 625–631.
Faldyna, M., Samankova, P., Leva, L., Cerny, J., Oujezdska, J., Rehakova, Z., et al. (2007). Cross-reactive anti-human monoclonal antibodies as a tool for B-cell identification in dogs and pigs. Vet. Immunol. Immunopathol. 119, 56–62. doi: 10.1016/j.vetimm.2007.06.022
Farsani, S. M. J., Deijs, M., Dijkman, R., Molenkamp, R., Jeeninga, R. E., Ieven, M., et al. (2015). Culturing of respiratory viruses in well-differentiated pseudostratified human airway epithelium as a tool to detect unknown viruses. Influenza Other Resp. 9, 51–57. doi: 10.1111/irv.12297
Fuchs, E., and Blau, H. M. (2020). Tissue stem cells: architects of their niches. Cell Stem Cell. 27, 532–556. doi: 10.1016/j.stem.2020.09.011
Govorkova, E. A., Murti, G., Meignier, B., de Taisne, C., and Webster, R. G. (1996). African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J. Virol. 70, 5519–5524. doi: 10.1128/JVI.70.8.5519-5524.1996
Haaker, M. W., Kruitwagen, H. S., Vaandrager, A. B., Houweling, M., Penning, L. C., Molenaar, M. R., et al. (2020). Identification of potential drugs for treatment of hepatic lipidosis in cats using an in vitro feline liver organoid system. J. Vet. Intern. Med. 34, 132–138. doi: 10.1111/jvim.15670
Hamilton, C. A., Young, R., Jayaraman, S., Sehgal, A., Paxton, E., Thomson, S., et al. (2018). Development of in vitro enteroids derived from bovine small intestinal crypts. Vet. Res. 49:54.
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275.
Hao, W. D., Bernard, K., Patel, N., Ulbrandt, N., Feng, H., Svabek, C., et al. (2012). Infection and propagation of human rhinovirus c in human airway epithelial cells. J. Virol. 86, 13524–13532.
Hodinka, R. L. (2013). Point: is the era of viral culture over in the clinical microbiology laboratory? J. Clin. Microbiol. 51, 2–4. doi: 10.1128/JCM.02593-12
Hofer, M., and Lutolf, M. P. (2021). Engineering organoids [published online ahead of print, 2021 Feb 19]. Nat. Rev. Mater. [Epub ahead of print]. doi: 10.1038/s41578-021-00279-y
Hofmann, M., and Wyler, R. (1988). Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 26, 2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988
Holloway, E. M., Capeling, M. M., and Spence, J. R. (2019). Biologically inspired approaches to enhance human organoid complexity. Development 146:dev166173. doi: 10.1242/dev.166173
Holtfreter, J. (1944). Experimental studies on the development of the pronephros. Rev. Can. Biol. 3, 220–250.
Holthaus, D., Delgado-Betancourt, E., Aebischer, T., Seeber, F., and Klotz, C. (2021). Harmonization of protocols for multi-species organoid platforms to study the intestinal biology of toxoplasma gondii and other protozoan infections. Front. Cell Infect. Microbiol. 10:610368. doi: 10.3389/fcimb.2020.610368
Jones, K., Patel, N., Levy, M., Storeygard, A., Balk, D., Gittleman, J. L., et al. (2008). Global trends in emerging infectious diseases. Nature 451, 990–993. doi: 10.1038/nature06536
Kar, S. K., Wells, J. M., Ellen, E. D., Te Pas, M. F. W., Madsen, O., Groenen, M. A. M., et al. (2021). Organoids: a promising new in vitro platform in livestock and veterinary research. Vet. Res. 52:43.
Kardia, E., Frese, M., Smertina, E., Strive, T., Zeng, X.-L., Estes, M., et al. (2021). Culture and differentiation of rabbit intestinal organoids and organoid-derived cell monolayers. Sci. Rep. 11:5401.
Kaushik, G., Ponnusamy, M. P., and Batra, S. K. (2018). Concise review: current status of three-dimensional organoids as preclinical models. Stem Cells 36, 1329–1340. doi: 10.1002/stem.2852
Kim, J., Koo, B. K., and Knoblich, J. A. (2020). Human organoids: model systems for human biology and medicine [published online ahead of print, 2020 Jul 7]. Nat. Rev. Mol. Cell Biol. 21, 571–584. doi: 10.1038/s41580-020-0259-3
Koltes, D. A., and Gabler, N. K. (2016). Characterization of porcine intestinal enteroid cultures under a lipopolysaccharide challenge. J. Anim. Sci. 94, 335–339. doi: 10.2527/jas.2015-9793
Kruitwagen, H. S., Oosterhoff, L. A., Vernooij, I., Schrall, I. M., van Wolferen, M. E., Bannink, F., et al. (2017). Long-term adult feline liver organoid cultures for disease modeling of hepatic steatosis. Stem Cell Rep. 8, 822–830. doi: 10.1016/j.stemcr.2017.02.015
Kuchipudi, S. V., Nelli, R. K., Gontu, A., Satyakumar, R., Surendran Nair, M., and Subbiah, M. (2021). Sialic acid receptors: the key to solving the enigma of zoonotic virus spillover. Viruses 13:262. doi: 10.3390/v13020262
Laksono, B. M., de Vries, R. D., Duprex, W. P., and de Swart, R. L. (2020). Measles pathogenesis, immune suppression and animal models. Curr. Opin. Virol. 41, 31–37. doi: 10.1016/j.coviro.2020.03.002
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. doi: 10.1126/science.abc1669
Lehmann, R., Lee, C. M., Shugart, E. C., Benedetti, M., Charo, R. A., Gartner, Z., et al. (2019). Human organoids: a new dimension in cell biology. Mol. Biol. Cell 30, 1129–1137. doi: 10.1091/mbc.E19-03-0135
Leland, D. S., and Ginocchio, C. C. (2007). Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 20, 49–78. doi: 10.1128/cmr.00002-06
Li, L., Fu, F., Guo, S., Wang, H., He, X., Xue, M., et al. (2019a). Porcine intestinal enteroids: a new model for studying enteric coronavirus porcine epidemic diarrhea virus infection and the host innate response. J. Virol. 93:e01682.
Li, L., Xue, M., Fu, F., Yin, L., Feng, L., and Liu, P. (2019b). IFN-Lambda 3 mediates antiviral protection against porcine epidemic diarrhea virus by inducing a distinct antiviral transcript profile in porcine intestinal epithelia. Front. Immunol. 10:2394. doi: 10.3389/fimmu.2019.02394
Li, M. L., Aggeler, J., Farson, D. A., Hatier, C., Hassell, J., and Bissell, M. J. (1987). Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 84, 136–140. doi: 10.1073/pnas.84.1.136
Li, N., Ma, W. T., Pang, M., Fan, Q. L., and Hua, J. L. (2019c). The commensal microbiota and viral infection: a comprehensive review. Front. Immunol. 10:1551. doi: 10.3389/fimmu.2019.01551
Li, Y., Yang, N., Chen, J., Huang, X., Zhang, N., Yang, S., et al. (2020). Next-Generation porcine intestinal organoids: an apical-out organoid model for swine enteric virus infection and immune response investigations. J. Virol. 94, e01006–e01020.
Liu, M., Yu, W., Jin, J., Ma, M., An, T., Nie, Y., et al. (2020). Copper promotes sheep pancreatic duct organoid growth by activation of an antioxidant protein 1-dependent MEK-ERK pathway. Am. J. Physiol. Cell Physiol. 318, C806–C816.
Long, J. S., Mistry, B., Haslam, S. M., and Barclay, W. S. (2019). Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 17, 67–81. doi: 10.1038/s41579-018-0115-z
Ludlow, M., Rennick, L. J., Sarlang, S., Skibinski, G., McQuaid, S., Moore, T., et al. (2010). Wild-type measles virus infection of primary epithelial cells occurs via the basolateral surface without syncytium formation or release of infectious virus. J. Gen. Virol. 91, 971–979. doi: 10.1099/vir.0.016428-0
Luo, H., Zheng, J., Chen, Y., Wang, T., Zhang, Z., Shan, Y., et al. (2020). Utility evaluation of porcine enteroids as PDCoV infection model in vitro. Front. Microbiol. 11:821. doi: 10.3389/fmicb.2020.00821
Martignani, E., Accornero, P., Miretti, S., and Baratta, M. (2018). Bovine mammary organoids: a model to study epithelial mammary cells. Methods Mol. Biol. 1817, 137–144. doi: 10.1007/978-1-4939-8600-2_14
Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638. doi: 10.1073/pnas.78.12.7634
Mead, B. E., and Karp, J. M. (2019). All models are wrong, but some organoids may be useful. Genome Biol. 20:66. doi: 10.1186/s13059-019-1677-4
Milewska, A., Kula-Pacurar, A., Wadas, J., Suder, A., Szczepanski, A., Dabrowska, A., et al. (2020). Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. J. Virol. 94:e00957-20.
Miller, A. J., Dye, B. R., Ferrer-Torres, D., Hill, D. R., Overeem, A. W., Shea, L. D., et al. (2019). Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540.
Min, S., Kim, S., and Cho, S. W. (2020). Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp. Mol. Med. 52, 227–237. doi: 10.1038/s12276-020-0386-0
Miranda, C. C., Fernandes, T. G., Diogo, M. M., and Cabral, J. M. S. (2018). Towards multi-organoid systems for drug screening applications. Bioengineering 5:49. doi: 10.3390/bioengineering5030049
Moghaddam, M. M., Bonakdar, S., Shariatpanahi, M. R., Shokrgozar, M. A., and Faghihi, S. (2019). The effect of physical cues on the stem cell differentiation. Curr. Stem Cell Res. Ther. 14, 268–277. doi: 10.2174/1574888x14666181227120706
Monteil, V., Kwon, H., Prado, P., Hagelkrüys, A., Wimmer, R. A., Stahl, M., et al. (2020). Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905.e7–913.e7.
Montes-Olivas, S., Marucci, L., and Homer, M. (2019). Mathematical models of organoid cultures. Front. Genet. 10:873. doi: 10.3389/fgene.2019.00873
Nantasanti, S., Spee, B., Kruitwagen, H. S., Chen, C., Geijsen, N., Oosterhoff, L. A., et al. (2015). Disease modeling and gene therapy of copper storage disease in canine hepatic organoids. Stem Cell Rep. 5, 895–907. doi: 10.1016/j.stemcr.2015.09.002
Pain, B. (2021). Organoids in domestic animals: with which stem cells? Vet. Res. 52:38. doi: 10.1186/s13567-021-00911-3
Palmer, S. G., Porotto, M., Palermo, L. M., Cunha, L. F., Greengard, O., and Moscona, A. (2012). Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. mBio 3:e00137-12. doi: 10.1128/mBio.00137-12
Panek, M., Grabacka, M., and Pierzchalska, M. (2018). The formation of intestinal organoids in a hanging drop culture. Cytotechnology 70, 1085–1095. doi: 10.1007/s10616-018-0194-8
Pellegrini, L., Bonfio, C., Chadwick, J., Begum, F., Skehel, M., and Lancaster, M. A. (2020). Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 369:eaaz5626. doi: 10.1126/science.aaz5626
Pierzchalska, M., Panek, M., Czyrnek, M., Gielicz, A., Mickowska, B., and Grabacka, M. (2017). Probiotic Lactobacillus acidophilus bacteria or synthetic TLR2 agonist boost the growth of chicken embryo intestinal organoids in cultures comprising epithelial cells and myofibroblasts. Comp. Immunol. Microbiol. Infect. Dis. 53, 7–18. doi: 10.1016/j.cimid.2017.06.002
Pierzchalska, M., Panek, M., and Grabacka, M. (2018). The migration and fusion events related to ROCK activity strongly influence the morphology of chicken embryo intestinal organoids. Protoplasma 256, 575–581. doi: 10.1007/s00709-018-1312-3
Pizzorno, A., Padey, B., Julien, T., Trouillet-Assant, S., Traversier, A., Errazuriz-Cerda, E., et al. (2020). Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep. Med. 1:100059. doi: 10.1016/j.xcrm.2020.100059
Post, Y., Puschhof, J., Beumer, J., Kerkkamp, H. M., de Bakker, M. A. G., Slagboom, J., et al. (2020). Snake venom gland organoids. Cell 180, 233.e21–247.e21.
Powell, R. H., and Behnke, M. S. (2017). WRN conditioned media is sufficient for in vitro propagation of intestinal organoids from large farm and small companion animals. Biol. Open 6, 698–705.
Pyrc, K., Sims, A. C., Dijkman, R., Jebbink, M., Long, C., Deming, D., et al. (2010). Culturing the Unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 84, 11255–11263. doi: 10.1128/jvi.00947-10
Qian, X. Y., Nguyen, H. N., Jacob, F., Song, H. J., and Ming, G. L. (2017). Using brain organoids to understand Zika virus-induced microcephaly. Development 144, 952–957. doi: 10.1242/dev.140707
Rabenau, H. F., Kessler, H. H., Kortenbusch, M., Steinhorst, A., Raggam, R. B., and Berger, A. (2007). Verification and validation of diagnostic laboratory tests in clinical virology. J. Clin. Virol. 40, 93–98. doi: 10.1016/j.jcv.2007.07.009
Ramani, A., Muller, L., Ostermann, P. N., Gabriel, E., Abida-Islam, P., Muller-Schiffmann, A., et al. (2020). SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 39:e106230.
Ramani, S., Crawford, S. E., Blutt, S. E., and Estes, M. K. (2018). Human organoid cultures: transformative new tools for human virus studies. Curr. Opin. Virol. 29, 79–86. doi: 10.1016/j.coviro.2018.04.001
Resende, T. P., Medida, R. L., Vannucci, F. A., Saqui-Salces, M., and Gebhart, C. (2020). Evaluation of swine enteroids as in vitro models for Lawsonia intracellularis infection. J. Anim. Sci. 98:skaa011.
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Sachs, N., Papaspyropoulos, A., Zomer-van Ommen, D. D., Heo, I., Böttinger, L., Klay, D., et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO J. 38:e100300. doi: 10.15252/embj.2018100300
Sakib, S., Yu, Y., Voigt, A., Ungrin, M., and Dobrinski, I. (2019). Generation of porcine testicular organoids with testis specific architecture using microwell culture. J. Vis. Exp. 152:e60387. doi: 10.3791/60387
Sang, E. R., Tian, Y., Gong, Y., Miller, L. C., and Sang, Y. (2020). Integrate structural analysis, isoform diversity, and interferon-inductive propensity of ACE2 to predict SARS-CoV2 susceptibility in vertebrates. Heliyon 6:e04818. doi: 10.1016/j.heliyon.2020.e04818
Sang, E. R., Tian, Y., Miller, L. C., and Sang, Y. (2021). Epigenetic evolution of ACE2 and IL-6 genes: non-canonical interferon-stimulated genes correlate to COVID-19 susceptibility in vertebrates. Genes 12:154. doi: 10.3390/genes12020154
Sang, Y., Miller, L. C., and Blecha, F. (2015). Macrophage polarization in virus-host interactions. J. Clin. Cell Immunol. 6:311.
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi: 10.1038/nature07935
Schutgens, F., and Clevers, H. (2020). Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. 15, 211–234. doi: 10.1146/annurev-pathmechdis-012419-032611
Schwarz, J. S., de Jonge, H. R., and Forrest, J. N. Jr. (2015). Value of organoids from comparative epithelia models. Yale J. Biol. Med. 88, 367–374.
Seeger, B. (2020). Farm animal-derived models of the intestinal epithelium: recent advances and future applications of intestinal organoids. Altern. Lab. Anim. 48, 215–233. doi: 10.1177/0261192920974026
Shanks, N., Greek, R., and Greek, J. (2009). Are animal models predictive for humans? Philos. Ethics Humanit. Med. 4, 1–20.
Shannon, J. M., Mason, R. J., and Jennings, S. D. (1987). Functional differentiation of alveolar type II epithelial cells in vitro: effects of cell shape, cell-matrix interactions and cell-cell interactions. Biochim. Biophys. Acta 931, 143–156. doi: 10.1016/0167-4889(87)90200-x
Siddiqi, H. K., Libby, P., and Ridker, P. M. (2021). COVID-19 - A vascular disease. Trends Cardiovasc. Med. 31, 1–5. doi: 10.1016/j.tcm.2020.10.005
Simmonds, P., Aiewsakun, P., and Katzourakis, A. (2019). Prisoners of war — host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 17, 321–328. doi: 10.1038/s41579-018-0120-2
Sopp, P., Werling, D., and Baldwin, C. (2007). Cross-reactivity of mAbs to human CD antigens with cells from cattle. Vet. Immunol. Immunopathol. 119, 106–114. doi: 10.1016/j.vetimm.2007.06.014
Sridhar, A., Simmini, S., Ribeiro, C. M. S., Tapparel, C., Evers, M. M., Pajkrt, D., et al. (2020). A perspective on organoids for virology research. Viruses 12:1341. doi: 10.3390/v12111341
Stanifer, M. L., Kee, C., Cortese, M., Zumaran, C. M., Triana, S., Mukenhirn, M., et al. (2020). Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 32:107863. doi: 10.1016/j.celrep.2020.107863
Steinberg, M. S. (1964). “The problem of adhesive selectivity in cellular interactions,” in Cellular Membranes in Development, ed. M. Locke (London: Academic Press).
Stewart, A. S., Freund, J. M., and Gonzalez, L. M. (2018). Advanced three-dimensional culture of equine intestinal epithelial stem cells. Equine Vet. J. 50, 241–248. doi: 10.1111/evj.12734
Takada, K., Kawakami, C., Fan, S., Chiba, S., Zhong, G., Gu, C., et al. (2019). A humanized MDCK cell line for the efficient isolation and propagation of human influenza viruses. Nat. Microbiol. 4, 1268–1273. doi: 10.1038/s41564-019-0433-6
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. doi: 10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Tapparel, C., Sobo, K., Constant, S., Huang, S., Van Belle, S., and Kaiser, L. (2013). Growth and characterization of different human rhinovirus C types in three-dimensional human airway epithelia reconstituted in vitro. Virology 446, 1–8. doi: 10.1016/j.virol.2013.06.031
Tekes, G., Ehmann, R., Boulant, S. A.-O., and Stanifer, M. A.-O. (2020). Development of feline ileum- and colon-derived organoids and their potential use to support feline coronavirus infection. Cells 9:2085. doi: 10.3390/cells9092085
Thompson, R. E., Johnson, A. K., Dini, P., Turco, M. Y., Prado, T. M., Premanandan, C., et al. (2020). Hormone-responsive organoids from domestic mare and endangered Przewalski’s horse endometrium. Reproduction 160, 819–831. doi: 10.1530/rep-20-0266
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. doi: 10.1126/science.282.5391.1145
Topfer, E., Pasotti, A., Telopoulou, A., Italiani, P., Boraschi, D., Ewart, M. A., et al. (2019). Bovine colon organoids: from 3D bioprinting to cryopreserved multi-well screening platforms. Toxicol. In Vitro 61:104606. doi: 10.1016/j.tiv.2019.104606
Tsai, Y. H., Czerwinski, M., Wu, A., Dame, M. K., Attili, D., Hill, E., et al. (2018). A method for cryogenic preservation of human biopsy specimens and subsequent organoid culture. Cell Mol. Gastroenterol. Hepatol. 6, 218.e7–222.e7. doi: 10.1016/j.jcmgh.2018.04.008
Tysoe, O. C., Justin, A. W., Brevini, T., Chen, S. E., Mahbubani, K. T., Frank, A. K., et al. (2019). Isolation and propagation of primary human cholangiocyte organoids for the generation of bioengineered biliary tissue. Nat. Protoc. 14, 1884–1925. doi: 10.1038/s41596-019-0168-0
Usui, T., Sakurai, M., Nishikawa, S., Umata, K., Nemoto, Y., Haraguchi, T., et al. (2017). Establishment of a dog primary prostate cancer organoid using the urine cancer stem cells. Cancer Sci. 108, 2383–2392. doi: 10.1111/cas.13418
van der Hee, B., Loonen, L. M. P., Taverne, N., Taverne-Thiele, J. J., Smidt, H., and Wells, J. M. (2018). Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res. 28, 165–171. doi: 10.1016/j.scr.2018.02.013
Vermeulen, M., Del Vento, F., Kanbar, M., Pyr Dit Ruys, S., Vertommen, D., Poels, J., et al. (2019). Generation of organized porcine testicular organoids in solubilized hydrogels from decellularized extracellular matrix. Int. J. Mol. Sci. 20:5476. doi: 10.3390/ijms20215476
von Furstenberg, R. J., Li, J., Stolarchuk, C., Feder, R., Campbell, A., Kruger, L., et al. (2017). Porcine esophageal submucosal gland culture model shows capacity for proliferation and differentiation. Cell Mol. Gastroenterol. Hepatol. 4, 385–404. doi: 10.1016/j.jcmgh.2017.07.005
Wang, D. (2020). 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog. 16:e1008318. doi: 10.1371/journal.ppat.1008318
Weiss, P., and Taylor, A. C. (1960). Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc. Natl. Acad. Sci. U.S.A. 46, 1177–1185. doi: 10.1073/pnas.46.9.1177
Wen, Z. X., Song, H. J., and Ming, G. L. (2017). How does Zika virus cause microcephaly? Gene Dev. 31, 849–861. doi: 10.1101/gad.298216.117
Wiener, D. J., Basak, O., Asra, P., Boonekamp, K. E., Kretzschmar, K., Papaspyropoulos, A., et al. (2018). Establishment and characterization of a canine keratinocyte organoid culture system. Vet. Dermatol. 29:375. doi: 10.1111/vde.12541
Wiley, L. A., Beebe, D. C., Mullins, R. F., Stone, E. M., and Tucker, B. A. (2016). A Method for Sectioning and Immunohistochemical Analysis of Stem Cell-Derived 3-D Organoids. Curr. Protoc. Stem Cell Biol. 37:1. doi: 10.1002/cpsc.3
Wilson, H. V. (1907). A new method by which sponges may be artificially reared. Science 25, 912–915. doi: 10.1126/science.25.649.912
Wilson, S. S., Mayo, M., Melim, T., Knight, H., Patnaude, L., Wu, X., et al. (2021). Optimized culture conditions for improved growth and functional differentiation of mouse and human colon organoids. Front. Immunol. 11:547102. doi: 10.3389/fimmu.2020.547102
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., et al. (2020). Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435. doi: 10.1016/S2468-1253(20)30083-2
Yang, L., Han, Y., Nilsson-Payant, B. E., Gupta, V., Wang, P., Duan, X., et al. (2020). A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27, 125.e7–136.e7. doi: 10.1016/j.stem.2020.06.015
Yin, L., Chen, J., Li, L., Guo, S., Xue, M., Zhang, J., et al. (2020). Aminopeptidase N expression, not interferon responses, determines the intestinal segmental tropism of porcine deltacoronavirus. J. Virol. 94:e00480.
Yin, X., Mead, B. E., Safaee, H., Langer, R., Karp, J. M., and Levy, O. (2016). Engineering stem cell organoids. Cell Stem Cell 18, 25–38. doi: 10.1016/j.stem.2015.12.005
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920.
Yuki, K., Cheng, N., Nakano, M., and Kuo, C. J. (2020). Organoid models of tumor immunology. Trends Immunol. 41, 652–664. doi: 10.1016/j.it.2020.06.010
Zhang, B. Z., Chu, H., Han, S., Shuai, H., Deng, J., Hu, Y. F., et al. (2020). SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 30, 928–931. doi: 10.1038/s41422-020-0390-x
Keywords: organoid, viruses, disease modeling, domestic and wild animals, zoonosis
Citation: Sang Y, Miller LC, Nelli RK and Giménez-Lirola LG (2021) Harness Organoid Models for Virological Studies in Animals: A Cross-Species Perspective. Front. Microbiol. 12:725074. doi: 10.3389/fmicb.2021.725074
Received: 14 June 2021; Accepted: 27 August 2021;
Published: 16 September 2021.
Edited by:
Haitao Guo, University of Pittsburgh, United StatesReviewed by:
Eve-Isabelle Pecheur, INSERM U1052 Center de Recherche en Cancerologie de Lyon, FranceCopyright © 2021 Sang, Miller, Nelli and Giménez-Lirola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongming Sang, eXNhbmdAdG5zdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.