
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 September 2021
Sec. Systems Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.724980
This article is part of the Research Topic Multi-omics Study on Gut Microbiota Related to Faecal Microbiota Transplantation View all 13 articles
 Shuntian Cai1,2†
Shuntian Cai1,2† Yanyun Fan1,2†
Yanyun Fan1,2† Bangzhou Zhang1,2†
Bangzhou Zhang1,2† Jinzhou Lin1
Jinzhou Lin1 Xiaoning Yang1
Xiaoning Yang1 Yunpeng Liu1
Yunpeng Liu1 Jingjing Liu1,2
Jingjing Liu1,2 Jianlin Ren1,2*
Jianlin Ren1,2* Hongzhi Xu1,2*
Hongzhi Xu1,2*Recent research has revealed the importance of the appendix in regulating the intestinal microbiota and mucosal immunity. However, the changes that occur in human gut microbial communities after appendectomy have never been analyzed. We assessed the alterations in gut bacterial and fungal populations associated with a history of appendectomy. In this cross-sectional study, we investigated the association between appendectomy and the gut microbiome using 16S and ITS2 sequencing on fecal samples from 30 healthy individuals with prior appendectomy (HwA) and 30 healthy individuals without appendectomy (HwoA). Analysis showed that the gut bacterial composition of samples from HwA was less diverse than that of samples from HwoA and had a lower abundance of Roseburia, Barnesiella, Butyricicoccus, Odoribacter, and Butyricimonas species, most of which were short-chain fatty acids-producing microbes. The HwA subgroup analysis indicated a trend toward restoration of the HwoA bacterial microbiome over time after appendectomy. HwA had higher gut fungi composition and diversity than HwoA, even 5 years after appendectomy. Compared with those in samples from HwoA, the abundance correlation networks in samples from HwA displayed more complex fungal–fungal and fungal–bacterial community interactions. This study revealed a marked impact of appendectomy on gut bacteria and fungi, which was particularly durable for fungi.
The human appendix was traditionally considered an evolutionary remnant with limited biological function. It is typically removed upon the development of appendicitis or even removed preventatively (D’Souza and Nugent, 2016). However, increasing evidence has revealed that the human appendix plays important biological roles in regulating the intestinal immune system and microbiome (Randal Bollinger et al., 2007; Laurin et al., 2011; Masahata et al., 2014; Kooij et al., 2016; Girard-Madoux et al., 2018; Vitetta et al., 2019). Moreover, studies suggest that prior appendectomy may be associated with increased risk of many diseases, such as sarcoidosis, antibiotic-resistant bacteria-mediated bacteremia caused by biliary tract infection, gallstones, pyogenic liver abscesses, gastrointestinal cancers, Parkinson’s disease (PD), and rheumatoid arthritis (Tzeng et al., 2015; Chung et al., 2016; Liao et al., 2016; Song et al., 2016; Kawanishi et al., 2017; Sawahata et al., 2017; Rubin, 2019). However, other studies have indicated no overall increase in cancer incidence several years after appendectomy (Mellemkjaer et al., 1998; Cope et al., 2003). The role of the appendix must continue to be reevaluated and further investigated to reveal its roles in human health and disease.
Similar to the colon, the healthy appendix is inhabited by diverse microorganisms, predominantly composed of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria species (Guinane et al., 2013; Vitetta et al., 2019). Appendicitis is associated with altered microbiota in the appendix. Interestingly, some microbial taxa that are infrequently found in the distal gut, including the oral pathogens Gemella, Parvimonas, and Fusobacterium, have been identified in surgically removed appendices from patients with acute appendicitis (Guinane et al., 2013). Studies have demonstrated differences in the intestinal microbiota of patients with appendicitis and healthy controls, such as a greater abundance of Fusobacteria species in the setting of appendicitis (Swidsinski et al., 2011; Zhong et al., 2014; Rogers et al., 2016). However, data about differences in the microbiome based on disease severity are inconsistent (Peeters et al., 2019; The et al., 2019). Using culture methods, the diversity of anaerobes in the appendix differs between individuals with and without appendicitis (Hattori et al., 2019).
Recent studies have demonstrated crucial roles of the appendix in regulating intestinal microecology, possibly acting as a reserve or sanctuary for the gut microbiota that promotes the recovery of gut microecological homeostasis after intestinal perturbation (Randal Bollinger et al., 2007; Vitetta et al., 2019). Appendectomized mice show delayed accumulation of IgA+ cells in the large intestine with altered fecal microbiota composition compared with sham-operated mice (Masahata et al., 2014). However, few studies have focused on intestinal bacterial changes after appendectomy, with inconsistent results (Goedert et al., 2014; Masahata et al., 2014). To our knowledge, there is no research on the relationship between appendectomy and intestinal fungi (Underhill and Iliev, 2014). Although the gut microbiota is populated mainly by bacteria, it also contains less than 1% of fungi. Intestinal fungal dysbiosis occurs in or contributes to many diseases, including colitis, alcoholic liver disease, primary sclerosing cholangitis, pancreatic cancer, and colon cancer (Iliev et al., 2012; Sokol et al., 2017; Yang et al., 2017; Aykut et al., 2019; Coker et al., 2019; Lemoinne et al., 2020).
To explore the alterations of gut bacterial and fungal communities associated with appendectomy, we firstly recruited and collected fecal samples from 30 healthy individuals with prior appendectomy and 30 healthy individuals without appendectomy. We examined the diversity and community structure of gut bacteria and fungi and evaluated their inter-kingdom interactions using 16S and ITS2 amplicon metagenomics in this study.
Healthy individuals with appendectomy (HwA; n = 30, 15 men and 15 women) were recruited from April 2016 to June 2017 from the local population of Xiamen, China. All participants were healthy, not on medication, had no clinically significant disease at the inception of the study, and had undergone appendectomy > 6 months ago. We also enrolled 30 healthy individuals without appendectomy (HwoA, 17 men and 13 women) as controls between February 2018 and May 2018 from the physical examination center in the outpatient department of Zhongshan Hospital Affiliated to Xiamen University (Xiamen, China). Written informed consent was obtained from all participants before stool donation. The study was approved by the local Ethical Review Board of Zhongshan Hospital Affiliated to Xiamen University(IRB2015014).
The exclusion criteria for both groups were as follows: < 18 years of age, antibiotic or proton pump inhibitor (PPI) treatment within 30 days, gastrointestinal surgery (except appendectomy for HwA), or diseases known to affect the gut microbiota. The following clinical data were recorded: age, body mass index (BMI), sex, and years since appendectomy.
Fecal samples from each participant were collected and immediately frozen at −80°C until DNA extraction. The samples were thawed and homogenized, and total DNA was extracted from each sample (0.25 g) using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s protocol. The resulting DNA yield and quality were assessed with a MultiskanTM GO spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Bacterial and fungal communities were determined by amplicon metagenomics targeting the 16S rRNA gene and ITS2 fragments, respectively. Briefly, the forward primer targeting the 16S rRNA gene V3 and V4 regions was 5′-CCTACGGGNBGCASCAG-3′, and the reverse primer was 5′-GGACTACNVGGGTWTCTAAT-3′. The forward primer targeting ITS2 was 5′-GCATCGATGAAGAACGCAGC-3′, and the reverse primer was 5′-TCCTCCGCTTATTGATATGC-3′. The polymerase chain reaction (PCR) products were purified and assessed with Qubit 3.0 (Thermo Fisher Scientific, Waltham, MA, United States). Sequencing was performed by the Xiamen Treatgut Biotechnology Co., using a 250-bp paired-end sequencing protocol on a HiSeq 2500 platform (Illumina, San Diego, CA, United States). Raw sequences were deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA655569.
The raw paired-end reads were assembled and filtered using FLASH with default parameters except parameters of –M = 200 and –x = 0.15 (Magoc and Salzberg, 2011). The resulting high quality reads were checked for chimeras and clustered to generate operational taxonomic units (OTUs) based on 97% similarity cutoff with USEARCH (Edgar, 2013). The representative OTU sequences were classified against the SILVA database for 16S data and against the UNITE database for ITS2 data using RDP Classifier with a confidence threshold of 50% (Wang et al., 2007; Henderson et al., 2019; Nilsson et al., 2019). Bacterial and fungal data were re-sampled to 31,509 and 30,924 reads/sample, respectively, for downstream analyses.
Alpha diversity indices, including richness (observed), Shannon diversity (Shannon), and Pielou’s evenness (evenness) were computed based on the OTU table using the vegan package. The significances of differences in the diversity indices and individual taxa were tested using a non-parametric Wilcoxon rank-sum test for two groups or Kruskal–Wallis rank-sum test with Benjamini–Hochberg corrections for multiple groups using the agricolae package. Beta diversity was measured using Bray–Curtis distance, and significance was determined with PERMANOVA with 9,999 permutations using adonis in the R package vegan. Correlations between bacterial and fungal genera were computed and tested using the Hmisc package. Finally, the results were visualized using the custom R script based on ggplot2 or VennDiagram, and the network figures were generated with Gephi v0.9.2. The analyses were performed in R v3.3.2.
The study population included two groups: HwA (n = 30) and HwoA (n = 30). There were no significant differences in the age and sex between the HwA and HwoA groups. The BMI of HwA was less than that of HwoA (20.8 ± 3.0 vs. 22.3 ± 2.5, p = 0.017), but both were within normal limits (18.5 ≤ BMI < 25). The HwA group was further divided into subgroups based on number of years post-appendectomy: cutoff at 2 years (2Y; < 2Y vs. ≥ 2Y), at 2 and 5 years [< 2Y, < 5 years (5Y) to ≥ 2Y, and ≥ 5Y], and at 5 years (< 5Y vs. ≥ 5Y) (Table 1).
Gut bacterial communities were analyzed by 16S V3–V4 sequencing. Alpha diversity, assessed with the observed, Shannon, and evenness indices, did not significantly differ between the HwA and HwoA groups (Supplementary Figure 1A). However, individuals with post-appendectomy periods shorter than 2 years (HwA_ < 2Y) had significantly lower gut bacterial evenness values than HwoA and HwA with a post-appendectomy period longer of at least 2 years (HwA_ ≥ 2Y) (Figure 1A; p < 0.05). HwA_ < 2Y also showed marginally lower Shannon diversity (Figure 1A; p = 0.08). There were no significant differences in the diversity indices between the other subgroups (Supplementary Figures 1B,C). The Venn diagram displays the 314 “universal” OTUs (of 486 total OTUs) shared among the three groups; a larger proportion (15.4%) of OTUs in HwA_ ≥ 2Y than HwA_ < 2Y (5.7%) was shared with the HwoA group (Figure 1B). Beta diversity analysis revealed that the gut bacterial communities in the samples from HwoA significantly differed from those in samples from HwA (PERMANOVA, F = 3.1526, p < 0.001) and from the subgroups with a cutoff of 2 years (PERMANOVA, F = 2.1526, p < 0.001). Interestingly, the HwA subgroups (HwA_ < 2Y and HwA_ ≥ 2Y) tended to have greater microbial ecological similarity with HwoA over time (Figure 1C), even with no significant differences detected between these two subgroups (PERMANOVA, F = 1.1646, p = 0.184). These results suggest appendectomy disrupted the gut bacteria composition, which was restored over time.
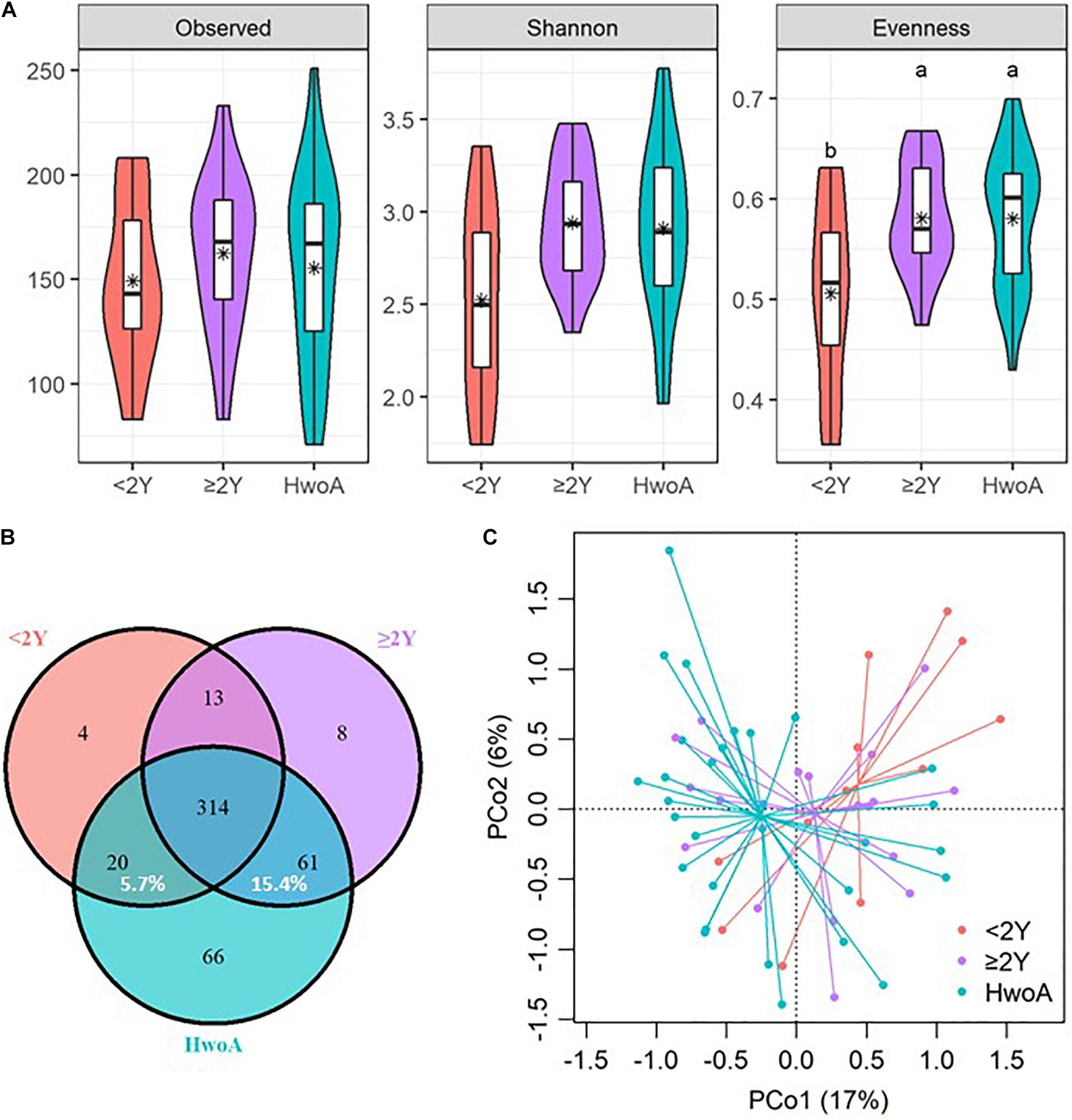
Figure 1. Alterations of gut bacterial diversity and communities, based on 16S V3–V4 sequencing data from HwoA, HwA_ < 2Y, and HwA_ ≥ 2Y. (A) Alpha diversity estimated by richness (observed OTUs), Shannon diversity, and Pielou’s evenness. Letters indicate the grouping (p < 0.05) by Kruskal–Wallis rank-sum test with Benjamini–Hochberg corrections. (B) Venn diagram of OTUs shared by and exclusive to the three groups. Corresponding percentages are noted for relevant overlaps. (C) Differences in gut bacterial community structures among the groups, assessed by principal coordinate (PCo) analysis of Bray–Curtis distance (p < 0.001). OTUs, operational taxonomic units. *Average.
Gut bacteria were dominated by Bacteroidetes, Firmicutes, Proteobacteria, and Fusobacteria at the phylum level (Figure 2A and Supplementary Figure 2A) and by Bacteroidaceae, Ruminococcaceae, Prevotellaceae, Acidaminococcaceae, Lachnospiraceae, Enterobacteriaceae, and Veillonellaceae at the family level (Figure 2B and Supplementary Figure 2B). Further analyses at the genus level revealed significantly higher abundances of Escherichia-Shigella, Veillonella, Klebsiella, Megasphaera, Flavonifractor, the Ruminococcus gnavus group, and Streptococcus in HwA subgroups than in HwoA (Supplementary Figure 3), with a trend toward restoration of the HwoA level with time after appendectomy (Figure 2C). On the other hand, Roseburia, Barnesiella, Butyricicoccus, Odoribacter, and Butyricimonas were significantly more abundant in the HwoA group than in the HwA subgroup (Supplementary Figure 3). Roseburia, Butyricicoccus, Odoribacter, and Butyricimonas became more abundant over time after appendectomy (Figure 2C).
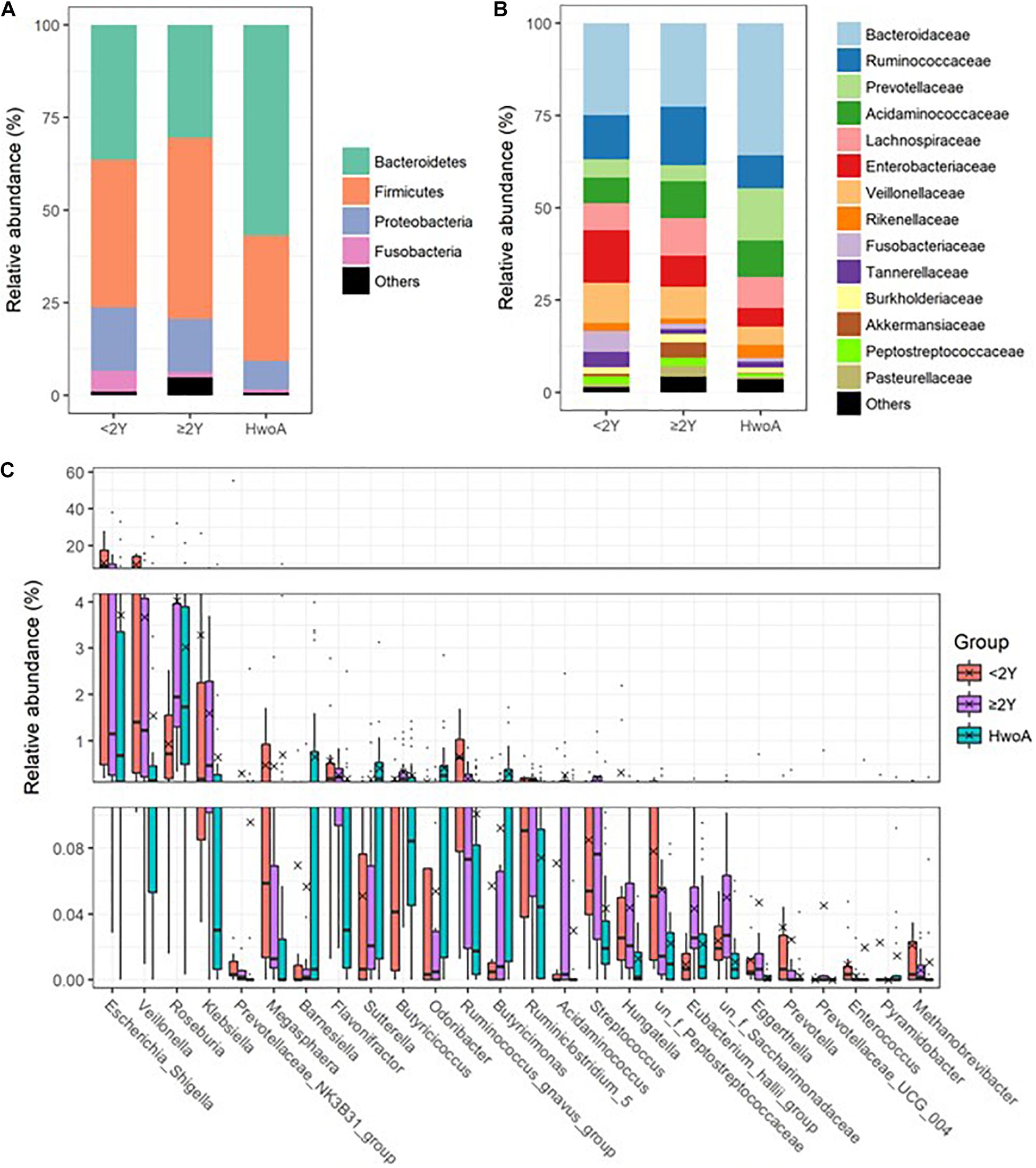
Figure 2. Gut bacteria compositions and differences in the HwoA and HwA subgroups. The overall bacterial structures of the three groups at (A) phylum and (B) family levels, expressed as the relative abundance of OTUs in each group. (C) The relative abundances of major (> 0.01%) bacterial genera significantly differed among HwA subgroups (< 2Y and ≥ 2Y) and HwoA (p < 0.05). The “un_f” in bacterial nomenclature means unclassified family at genus level. OTUs, operational taxonomic units.
Gut fungal communities were analyzed by ITS2 sequencing. The alpha diversity indices of the gut fungal communities, including the observed, Shannon, and evenness indices, were significantly higher in samples from HwA than those from HwoA (Supplementary Figure 4A). This difference was also observed for all HwA subgroups (Supplementary Figures 4B,C), even for individuals who had undergone appendectomy at least 5 years prior to the study (HwA_ ≥ 5Y) (Figure 3A). The Venn diagram shows a larger proportion of OTUs in HwA_ ≥ 5Y (25.7%) than in HwA_ < 5Y (17.6%) were shared with HwoA. Conversely, HwA_ < 5Y contained a higher proportion (63.7%) of exclusive OTUs than HwA_ ≥ 5Y (31.5%) (Figure 3B). Beta diversity analysis showed that the samples from HwoA were clearly separated from those from HwA (PERMANOVA, F = 4.030, p < 0.001) and from the subgroups divided at 5 years (PERMANOVA, F = 2.532, p < 0.001). Moreover, the HwA subgroups did not display increasing similarity to HwoA over time (Figure 3C). These results suggest that the effects of appendectomy on the gut fungal community persisted for at least 5 years, without obvious restoration over time. The fungal communities were dominated by Ascomycota and Basidiomycota at the phylum level (Figure 4A) and by Saccharomycetaceae, Aspergillaceae, and unclassified Ascomycota and Basidiomycetes at the family level (Figure 4B and Supplementary Figure 5). Further analyses at the genus level revealed that the abundances of many genera were significantly different in HwA and HwoA fecal samples (Supplementary Figure 6). Interestingly, the abundances of Hanseniaspora, Alternaria, Chaetomium, Fusarium, Paraphoma, Mycosphaerella, and Penicillium decreased with time post-appendectomy (Figure 4C), whereas the abundances of Aspergillus and unclassified Microascaceae increased over time after appendectomy (Figure 4C).
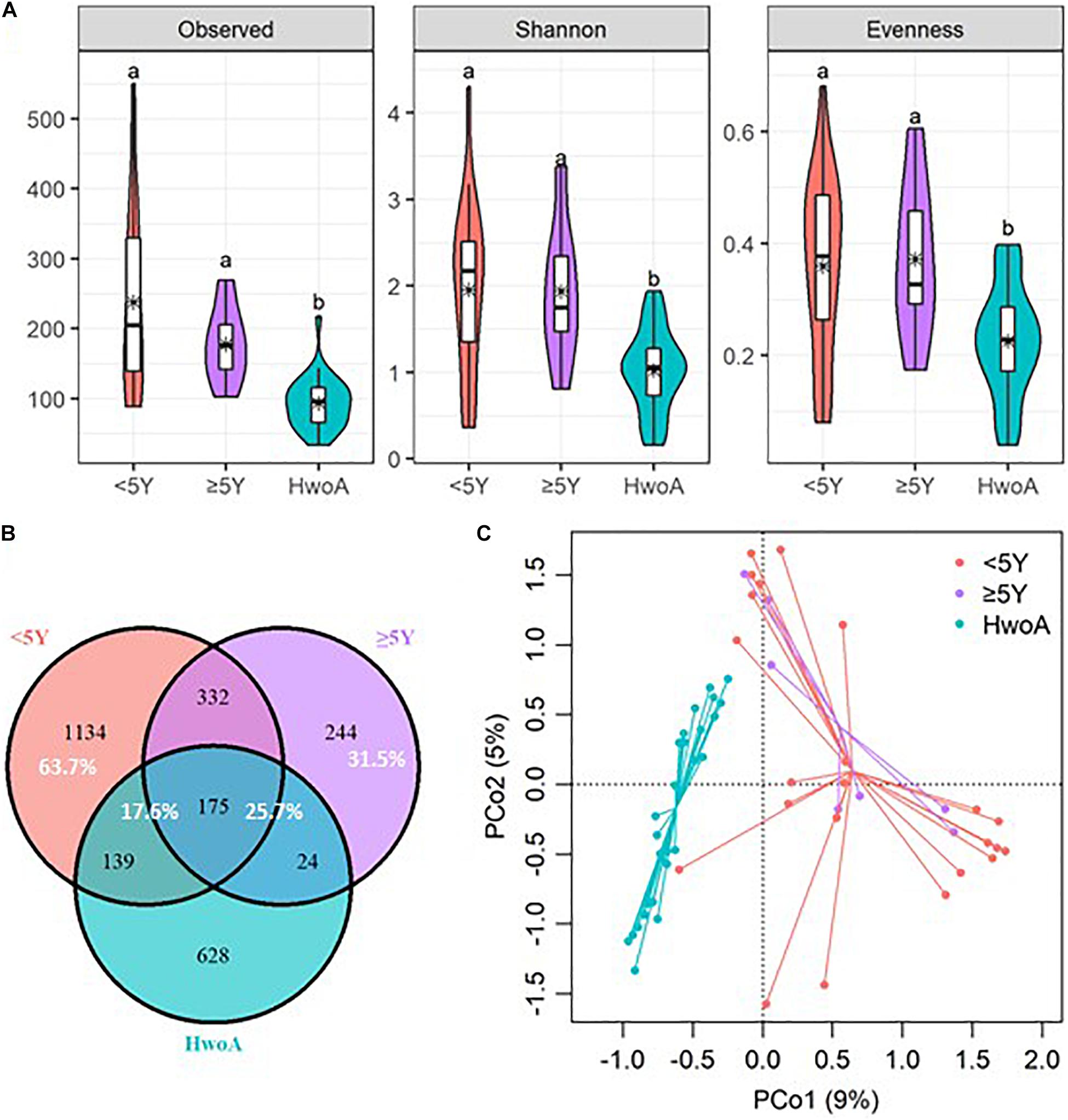
Figure 3. Alterations of gut fungal diversity and communities, based on ITS2 sequencing data of HwoA, HwA_ < 5Y, and HwA_ ≥ 5Y. (A) Alpha diversity estimated by the observed, Shannon, and evenness indices. Letters indicate the grouping by Kruskal–Wallis rank-sum test with Benjamini–Hochberg corrections (p < 0.05). (B) Venn diagram of the OTUs shared by and exclusive to the three groups. Corresponding percentages are noted for relevant overlaps. (C) Differences in gut fungal community structures among the groups, assessed by principal coordinate (PCo) analysis of Bray–Curtis distance (p < 0.001). OTUs, operational taxonomic units.
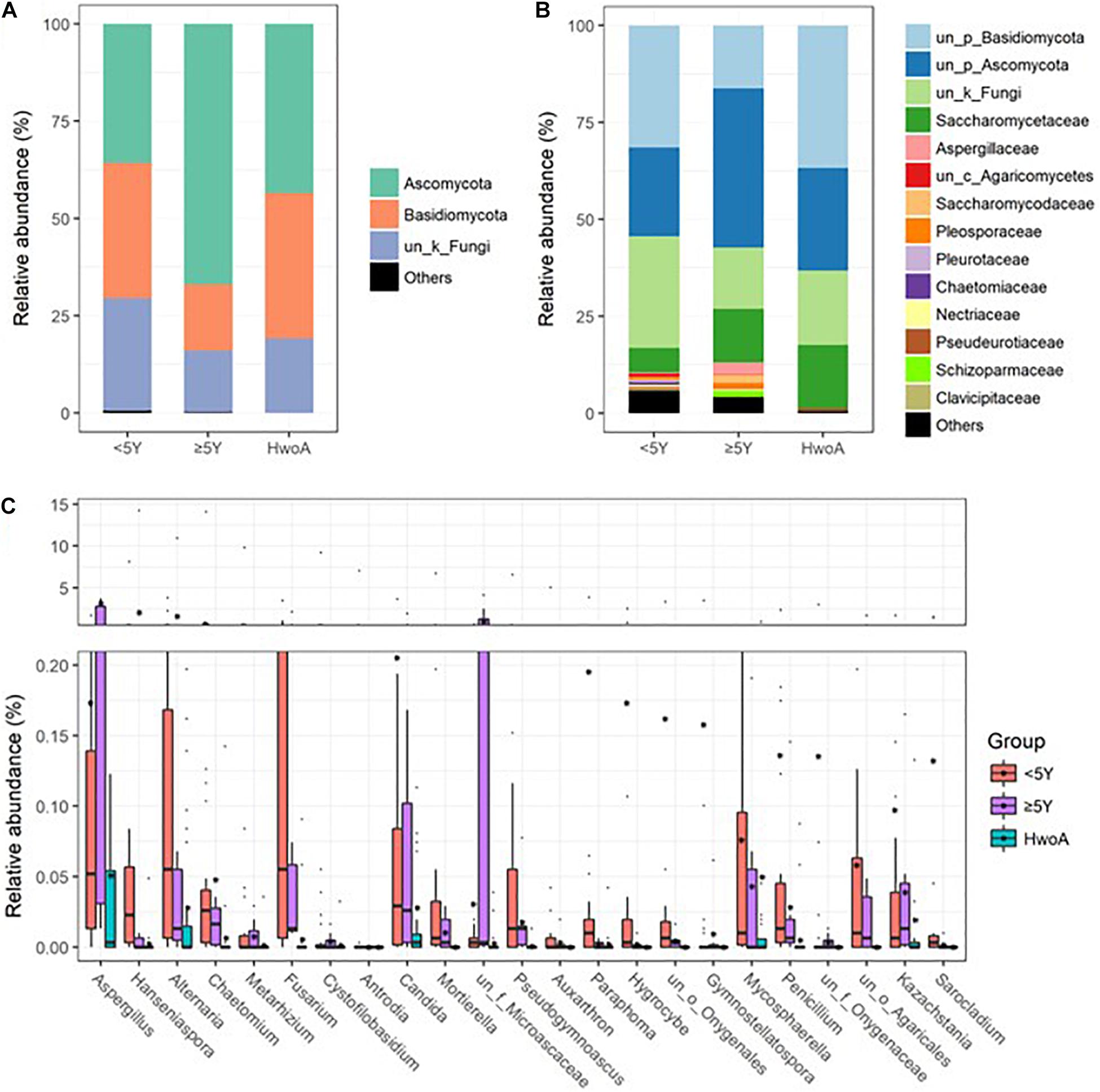
Figure 4. Gut fungal community compositions and differences in the HwoA and HwA subgroups. The overall fungal structures of the three groups at (A) phylum and (B) family levels, expressed as the relative abundance of OTUs in each group. (C) Relative abundances of major (> 0.05%) fungal genera significantly differed among HwA subgroups (< 2Y and ≥ 2Y) and HwoA (p < 0.05). OTUs, operational taxonomic units.
Fungal abundance correlation networks were constructed to evaluate the ecosystem structure. A richer, more complex network of correlations between fungal communities was observed in HwA than in HwoA samples (Figure 5A). As expected, the density of the fungal correlation network decreased over time in the HwA subgroups, as attested by decreased relative connectedness and fewer neighbors. However, there was a significantly higher density in HwA_ ≥ 5Y samples than in HwoA samples for these two parameters, as well as more nodes (OTUs) and edges (connections) in the networks (Figures 5A,B).
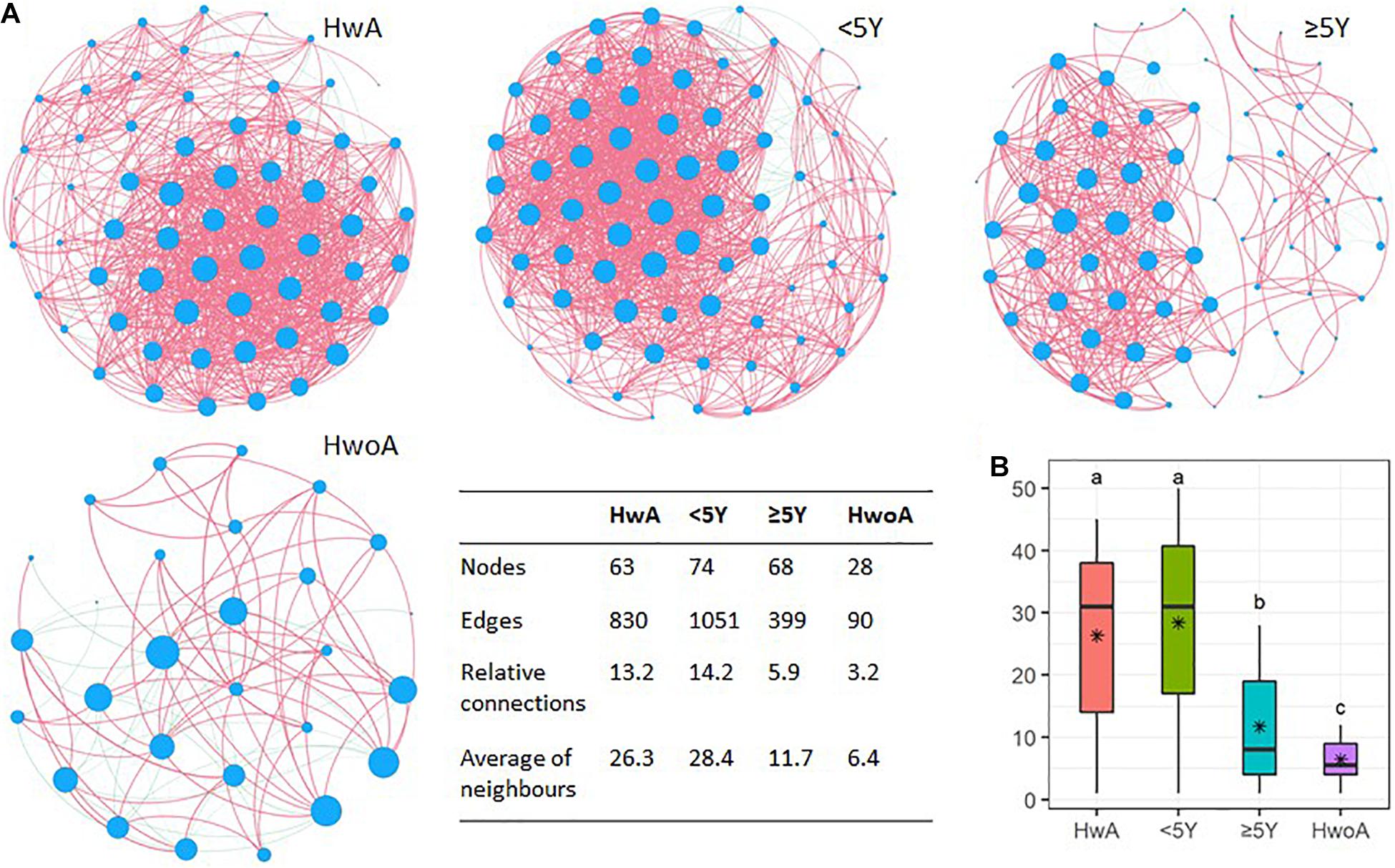
Figure 5. Gut fungal microbiota correlation networks. (A) Abundance correlation networks of HwA, HwA subgroups (< 5Y and ≥ 5Y), and HwoA analyzed by Spearman’s test with Benjamini–Hochberg corrections. Each node represents an OTU, and its size is scaled to the number of indirect edges within each network. Edges indicate correlations (positive in red and negative in green). Only OTUs present in > 50% of samples in the group were considered, and only significant correlations (p < 0.05) are shown. The table in the inset shows the network parameters. The relative connectedness is the ratio between the number of edges and the number of nodes in the network. (B) Neighbors of each node within the network. Black stars indicate mean values. Letters indicate the grouping by Kruskal–Wallis rank-sum test with Benjamini–Hochberg corrections (p < 0.05). OTUs, operational taxonomic units. *Average.
To gain an overview of the gut microbial shifts after appendectomy, we first addressed the equilibrium between fungal and bacterial diversity by determining the fungi-to-bacteria diversity ratio. The ratios of the observed, Shannon, and evenness indices were all significantly increased in the HwA group (Figure 6A; p < 0.05), suggesting a more prominent influence of appendectomy on the fungal community than the bacterial community. Furthermore, abundance correlation networks of bacterial and fungal interactions at the genus level were constructed to explore the interkingdom interactions. Compared with the HwoA group, the HwA group had a denser, obviously disrupted fungi–bacteria network, as illustrated by the increased relative connectedness and more neighbors (Figure 6B). Indeed, significantly more neighbors were observed for each node in the HwA samples than for those of the HwoA samples (Figure 6C). These results indicate that appendectomy is associated with alterations of bacterial–fungal interactions in terms of diversity and taxa relative abundances.
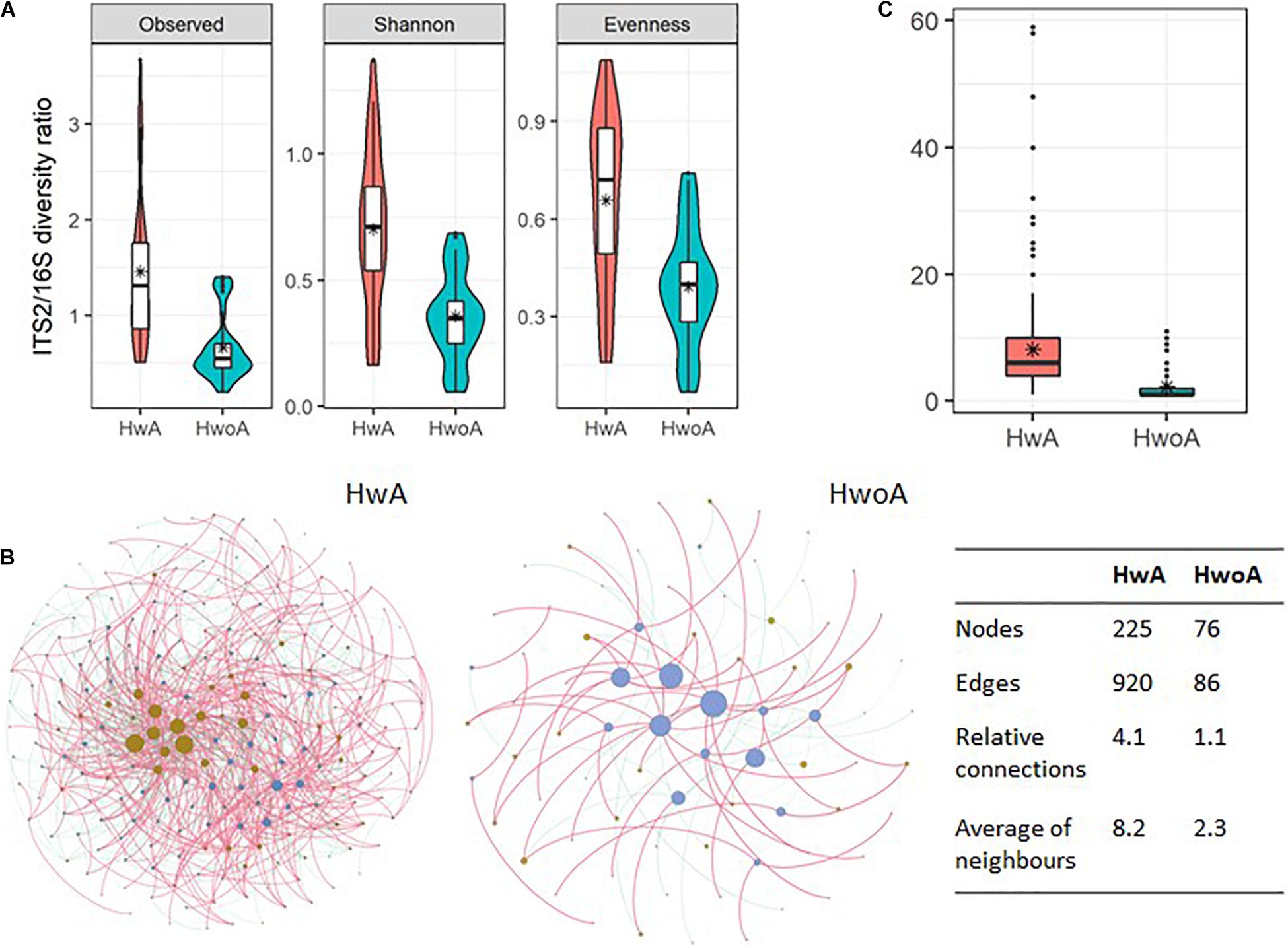
Figure 6. Interactions between gut fungal and bacterial communities. (A) Fungi-to-bacteria diversity ratios of the observed, Shannon, and evenness indices. (B) Abundance correlation networks of gut fungal and bacterial communities analyzed by Spearman’s test. Each node represents a genus, with bacteria in brown and fungi in blue/violet. Node size is scaled to the number of indirect edges within each network. Edges indicate correlations (positive in red and negative in green). Only genera present in ≥ 20% of samples in the group were considered, and only significant correlations (p < 0.05) are shown. The table in the inset shows the network parameters. The relative connectedness is the ratio between the number of edges and the number of nodes in the network. (C) Neighbors of each node within the network. Black stars are mean values. *Average.
In this study, fecal 16S and ITS2 sequences were used to investigate the gut microbiota of individuals with and without a history of appendectomy. We demonstrated that both gut bacterial and fungal communities in healthy subjects with a history of appendectomy are apparently distinct from those in healthy controls. Studies indicated that, 4 weeks after conventionalization, appendectomized germ-free mice have a distinct, less diverse bacterial composition than sham-operated germ-free mice (Masahata et al., 2014). On the contrary, a previous study reported that a history of appendectomy was not associated with beta diversity and that 22 taxa that were more abundant after appendectomy were not statistically different after adjustment (Goedert et al., 2014). In our study, alpha diversity indices did not significantly differ between HwA and HwoA, but beta diversity revealed that the gut bacterial composition of HwA was significantly separated from that of HwoA. Since the literature on this topic is limited, a more complete understanding remains to be gained through additional studies. Notably, in accordance with a previous report, our results suggested that the HwA subgroups tended to gain bacterial ecological similarity to HwoA over time after appendectomy. Similarly, at 8 weeks after conventionalization, the alteration of fecal microbiota composition in appendectomized mice was no longer apparent, and the numbers of colonic IgA-secreting cells normalized (Masahata et al., 2014). Interestingly, our HwA subgroup analysis revealed that gut fungal composition did not shift toward that observed in HwoA over time. Thus, the effect of appendectomy on gut fungi may be more persistent than that on bacteria. Our research indicated that appendectomy had different impact on fecal fungi and bacteria over time.
In our study, gut bacteria were dominated by Bacteroidetes, Firmicutes, Proteobacteria, and Fusobacteria at the phylum level in both HwA and HwoA. Further analyses at the genus level revealed that Roseburia, Barnesiella, Butyricicoccus, Odoribacter, and Butyricimonas were significantly more abundant in HwoA samples than HwA subgroup samples. Importantly, these more abundant bacteria were identified as short-chain fatty acid (SCFA)-producing microbes (Louis and Flint, 2017). In the gut, SCFAs such as butyric acid, propionic acid, and acetic acid are speculated to play key roles in immune regulation, intestinal mucosal protection, protection against inflammation, and epithelial cell energy provision (Rios-Covian et al., 2016). Some epidemiological studies have shown that removal of the appendix may increase the risk of type 2 diabetes (T2D) and PD (Killinger et al., 2018; Rubin, 2019). Though these associations are controversial and the mechanisms are unclear, alterations in microorganism communities may contribute to post-appendectomy disease occurrence (Killinger et al., 2018; Killinger and Labrie, 2019; Rubin, 2019). Disorders of propionate, an SCFA, in the gut are associated with an increased risk of T2D (Sanna et al., 2019). Studies of fecal microbiota in patients with PD have revealed lower levels of fiber-degrading bacterial strains and less SCFA production than observed in matched healthy controls (Unger et al., 2016; Li et al., 2017). Moreover, the long-term side effects of antibiotics can decrease the concentration of SCFAs (Holota et al., 2019). However, the roles of SCFA production are contradictory, as they can both benefit the host and lead to metabolic diseases (Schwiertz et al., 2010; Zhao et al., 2018; Pingitore et al., 2019). These results suggest that increasing SCFA-producing microbes due to appendectomy could contribute to the development of specific diseases.
Our study also found that, compared with HwoA, HwA had increased fungal biodiversity and relative changes in the abundance of many fungal groups, which lasted for at least 5 years. Ascomycota and Basidiomycota predominated among the intestinal fungi in both the HwA and control groups. The Basidiomycota-to-Ascomycota ratio in HwA was lower than that in HwoA, and the ratio dropped with time after appendectomy. The gut microbiota plays an important role in the pathogenesis of ulcerative colitis (UC) and colorectal cancer (CRC) (Gao et al., 2017; Ni et al., 2017; Raskov et al., 2017; Khan et al., 2019). It has also been reported that undergoing appendectomy in early life, before the onset of UC, may reduce the risk of colectomy and UC-related hospital admissions (Myrelid et al., 2017). However, it is still unclear if patients with UC can benefit from appendectomy (Park et al., 2014; Parian et al., 2017; Sahami et al., 2019). The relationship between appendectomy and CRC is inconclusive (Grobost et al., 1991; Mellemkjaer et al., 1998; Cope et al., 2003; Wu et al., 2015). The roles of immunoregulation and the microbiota in these associations require clarification. The fecal fungal microbiota of UC and CRC patients are also dominated by Ascomycota and Basidiomycota. In contrast to our findings, the Basidiomycota-to-Ascomycota ratio is higher in individuals with active UC and CRC than in healthy individuals, indicating intestinal fungal imbalance. We demonstrated that the correlation networks of fungal–fungal and fungal–bacterial interactions were denser and obviously disrupted in HwA. Alterations of intrafungal and interkingdom bacteria–fungi interactions are also observed in the settings of CRC and UC (Sokol et al., 2017; Coker et al., 2019). Taken together, these clinical observations and our results provide microbial insights in future research on the mechanism of appendectomy and related diseases.
However, there are some limitations to our study. This was a single-center observational study with a relatively small sample size. In addition, changes in gut immunity and microbial metabolism were not investigated. However, we enrolled individuals who were at various stages post-appendectomy, which allowed us to preliminarily analyze the duration of microecological changes after surgery. We also provided a first analysis of the fecal fungal profiles of individuals with a history of appendectomy.
We conclude that bacterial and fungal gut microbiota are altered after appendectomy. Moreover, our study elucidates that removal of the appendix alters intrafungal and bacteria–fungi interactions. It appears that the effects of appendectomy on the fecal fungal community are more marked and durable than on bacteria. However, the underlying mechanisms through which appendectomy alters the gut microbiota and the biological consequences of these changes remain to be explored.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA655569.
The studies involving human participants were reviewed and approved by the Ethical Review Board of Zhongshan Hospital Affiliated to Xiamen University. The patients/participants provided their written informed consent to participate in this study.
SC, YF, HX, JR, JLin, and BZ designed the study, analyzed the data, and wrote the manuscript. JLin, XY, YL, and JLiu undertook data collection and performed the literature search. All authors read and approved the final manuscript.
This work was supported by the Natural Science Foundation of Fujian Province, China (No. 2020J05286), National Natural Science Foundation of China (No. 81800517), and the China Postdoctoral Science Foundation funded project (No. 2018M632588). The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank all study participants and medical staff for their involvement and support in the study. We thank Editage (www.editage.cn) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.724980/full#supplementary-material
Aykut, B., Pushalkar, S., Chen, R., Li, Q., Abengozar, R., Kim, J. I., et al. (2019). The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. doi: 10.1038/s41586-019-1608-2
Chung, S. D., Huang, C. C., Lin, H. C., Tsai, M. C., and Chen, C. H. (2016). Increased Risk of Clinically Significant Gallstones following an Appendectomy: A Five-Year Follow-Up Study. PLoS One 11:e0165829. doi: 10.1371/journal.pone.0165829
Coker, O. O., Nakatsu, G., Dai, R. Z., Wu, W. K. K., Wong, S. H., Ng, S. C., et al. (2019). Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68, 654–662. doi: 10.1136/gutjnl-2018-317178
Cope, J. U., Askling, J., Gridley, G., Mohr, A., Ekbom, A., Nyren, O., et al. (2003). Appendectomy during childhood and adolescence and the subsequent risk of cancer in Sweden. Pediatrics 111(6 Pt 1), 1343–1350. doi: 10.1542/peds.111.6.1343
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Gao, R., Gao, Z., Huang, L., and Qin, H. (2017). Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 36, 757–769. doi: 10.1007/s10096-016-2881-8
Girard-Madoux, M. J. H., Gomez, de Aguero, M., Ganal-Vonarburg, S. C., Mooser, C., Belz, G. T., et al. (2018). The immunological functions of the Appendix: An example of redundancy? Semin. Immunol. 36, 31–44. doi: 10.1016/j.smim.2018.02.005
Goedert, J. J., Hua, X., Yu, G., and Shi, J. (2014). Diversity and composition of the adult fecal microbiome associated with history of cesarean birth or appendectomy: Analysis of the American Gut Project. EBioMed. 1, 167–172. doi: 10.1016/j.ebiom.2014.11.004
Grobost, O., Boutron, M. C., Arveux, P., Bedenne, L., Chatrenet, P., and Faivre, J. (1991). Appendectomy, cholecystectomy, cholelithiasis and colorectal cancer. A retrospective case control study at the Cote-d’Or. Gastroenterol. Clin. Biol. 15, 594–599.
Guinane, C. M., Tadrous, A., Fouhy, F., Ryan, C. A., Dempsey, E. M., Murphy, B., et al. (2013). Microbial composition of human appendices from patients following appendectomy. mBio 4:12. doi: 10.1128/mBio.00366-12
Hattori, T., Yuasa, N., Ikegami, S., Nishiyama, H., Takeuchi, E., Miyake, H., et al. (2019). Culture-based bacterial evaluation of the appendix lumen in patients with and without acute appendicitis. J. Infect. Chemother. 25, 708–713. doi: 10.1016/j.jiac.2019.03.021
Henderson, G., Yilmaz, P., Kumar, S., Forster, R. J., Kelly, W. J., Leahy, S. C., et al. (2019). Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ. 7:e6496. doi: 10.7717/peerj.6496
Holota, Y., Dovbynchuk, T., Kaji, I., Vareniuk, I., Dzyubenko, N., Chervinska, T., et al. (2019). The long-term consequences of antibiotic therapy: Role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PLoS One 14:e0220642. doi: 10.1371/journal.pone.0220642
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. doi: 10.1126/science.1221789
Kawanishi, K., Kinoshita, J., Abe, H., Kakimoto, T., Yasuda, Y., Hara, T., et al. (2017). Appendectomy as a Risk Factor for Bacteremic Biliary Tract Infection Caused by Antibiotic-Resistant Pathogens. Biomed. Res. Int. 2017:3276120. doi: 10.1155/2017/3276120
Khan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., et al. (2019). Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 8:3. doi: 10.3390/pathogens8030126
Killinger, B., and Labrie, V. (2019). The Appendix in Parkinson’s Disease: From Vestigial Remnant to Vital Organ? J. Parkinsons Dis. 9, S345–S358. doi: 10.3233/JPD-191703
Killinger, B. A., Madaj, Z., Sikora, J. W., Rey, N., Haas, A. J., Vepa, Y., et al. (2018). The vermiform appendix impacts the risk of developing Parkinson’s disease. Sci. Transl. Med. 10:465. doi: 10.1126/scitranslmed.aar5280
Kooij, I. A., Sahami, S., Meijer, S. L., Buskens, C. J., and Te Velde, A. A. (2016). The immunology of the vermiform appendix: a review of the literature. Clin. Exp. Immunol. 186, 1–9. doi: 10.1111/cei.12821
Laurin, M., Everett, M. L., and Parker, W. (2011). The cecal appendix: one more immune component with a function disturbed by post-industrial culture. Anat. Rec. 294, 567–579. doi: 10.1002/ar.21357
Lemoinne, S., Kemgang, A., Ben Belkacem, K., Straube, M., Jegou, S., Corpechot, C., et al. (2020). Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 69, 92–102. doi: 10.1136/gutjnl-2018-317791
Li, W., Wu, X., Hu, X., Wang, T., Liang, S., Duan, Y., et al. (2017). Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 60, 1223–1233. doi: 10.1007/s11427-016-9001-4
Liao, K. F., Lai, S. W., Lin, C. L., and Chien, S. H. (2016). Appendectomy correlates with increased risk of pyogenic liver abscess: A population-based cohort study in Taiwan. Medicine 95:e4015. doi: 10.1097/MD.0000000000004015
Louis, P., and Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41. doi: 10.1111/1462-2920.13589
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Masahata, K., Umemoto, E., Kayama, H., Kotani, M., Nakamura, S., Kurakawa, T., et al. (2014). Generation of colonic IgA-secreting cells in the caecal patch. Nat. Commun. 5:3704. doi: 10.1038/ncomms4704
Mellemkjaer, L., Johansen, C., Linet, M. S., Gridley, G., and Olsen, J. H. (1998). Cancer risk following appendectomy for acute appendicitis (Denmark). Cancer Causes Control 9, 183–187. doi: 10.1023/a:1008834311514
Myrelid, P., Landerholm, K., Nordenvall, C., Pinkney, T. D., and Andersson, R. E. (2017). Appendectomy and the Risk of Colectomy in Ulcerative Colitis: A National Cohort Study. Am. J. Gastroenterol. 112, 1311–1319. doi: 10.1038/ajg.2017.183
Ni, J., Wu, G. D., Albenberg, L., and Tomov, V. T. (2017). Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584. doi: 10.1038/nrgastro.2017.88
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Parian, A., Limketkai, B., Koh, J., Brant, S. R., Bitton, A., Cho, J. H., et al. (2017). Appendectomy does not decrease the risk of future colectomy in UC: results from a large cohort and meta-analysis. Gut 66, 1390–1397. doi: 10.1136/gutjnl-2016-311550
Park, S. H., Loftus, E. V. Jr., and Yang, S. K. (2014). Appendiceal skip inflammation and ulcerative colitis. Dig. Dis. Sci. 59, 2050–2057. doi: 10.1007/s10620-014-3129-z
Peeters, T., Penders, J., Smeekens, S. P., Galazzo, G., Houben, B., Netea, M. G., et al. (2019). The fecal and mucosal microbiome in acute appendicitis patients: an observational study. Future Microbiol. 14, 111–127. doi: 10.2217/fmb-2018-0203
Pingitore, A., Gonzalez-Abuin, N., Ruz-Maldonado, I., Huang, G. C., Frost, G., and Persaud, S. J. (2019). Short chain fatty acids stimulate insulin secretion and reduce apoptosis in mouse and human islets in vitro: Role of free fatty acid receptor 2. Diabetes Obes Metab. 21, 330–339. doi: 10.1111/dom.13529
Randal Bollinger, R., Barbas, A. S., Bush, E. L., Lin, S. S., and Parker, W. (2007). Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J. Theor. Biol. 249, 826–831. doi: 10.1016/j.jtbi.2007.08.032
Raskov, H., Burcharth, J., and Pommergaard, H. C. (2017). Linking Gut Microbiota to Colorectal Cancer. J. Cancer 8, 3378–3395. doi: 10.7150/jca.20497
Rios-Covian, D., Ruas-Madiedo, P., Margolles, A., Gueimonde, M., de Los Reyes-Gavilan, C. G., and Salazar, N. (2016). Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 7:185. doi: 10.3389/fmicb.2016.00185
Rogers, M. B., Brower-Sinning, R., Firek, B., Zhong, D., and Morowitz, M. J. (2016). Acute Appendicitis in Children Is Associated With a Local Expansion of Fusobacteria. Clin. Infect. Dis. 63, 71–78. doi: 10.1093/cid/ciw208
Rubin, R. (2019). Uncovering a Link Between the Appendix and Parkinson Disease Risk. JAMA 322, 293–294. doi: 10.1001/jama.2019.9041
Sahami, S., Wildenberg, M. E., Koens, L., Doherty, G., Martin, S., D’Haens, G., et al. (2019). Appendectomy for Therapy-Refractory Ulcerative Colitis Results in Pathological Improvement of Colonic Inflammation: Short-Term Results of the PASSION Study. J. Crohns Colitis 13, 165–171. doi: 10.1093/ecco-jcc/jjy127
Sanna, S., van Zuydam, N. R., Mahajan, A., Kurilshikov, A., Vich Vila, A., Vosa, U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605. doi: 10.1038/s41588-019-0350-x
Sawahata, M., Nakamura, Y., and Sugiyama, Y. (2017). Appendectomy, tonsillectomy, and risk for sarcoidosis - A hospital-based case-control study in Japan. Respir. Investig. 55, 196–202. doi: 10.1016/j.resinv.2016.12.004
Schwiertz, A., Taras, D., Schafer, K., Beijer, S., Bos, N. A., Donus, C., et al. (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195. doi: 10.1038/oby.2009.167
Sokol, H., Leducq, V., Aschard, H., Pham, H. P., Jegou, S., Landman, C., et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Song, H., Abnet, C. C., Andren-Sandberg, A., Chaturvedi, A. K., and Ye, W. (2016). Risk of Gastrointestinal Cancers among Patients with Appendectomy: A Large-Scale Swedish Register-Based Cohort Study during 1970-2009. PLoS One 11:e0151262. doi: 10.1371/journal.pone.0151262
Swidsinski, A., Dorffel, Y., Loening-Baucke, V., Theissig, F., Ruckert, J. C., Ismail, M., et al. (2011). Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60, 34–40. doi: 10.1136/gut.2009.191320
The, S. M. L., Bakx, R., Budding, A. E., de Meij, T. G. J., van der Lee, J. H., Bunders, M. J., et al. (2019). Microbiota of Children With Complex Appendicitis: Different Composition and Diversity of The Microbiota in Children With Complex Compared With Simple Appendicitis. Pediatr. Infect. Dis. J. 38, 1054–1060. doi: 10.1097/INF.0000000000002434
Tzeng, Y. M., Kao, L. T., Kao, S., Lin, H. C., Tsai, M. C., and Lee, C. Z. (2015). An appendectomy increases the risk of rheumatoid arthritis: a five-year follow-up study. PLoS One 10:e0126816. doi: 10.1371/journal.pone.0126816
Underhill, D. M., and Iliev, I. D. (2014). The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416. doi: 10.1038/nri3684
Unger, M. M., Spiegel, J., Dillmann, K. U., Grundmann, D., Philippeit, H., Burmann, J., et al. (2016). Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism. Relat. Disord. 32, 66–72. doi: 10.1016/j.parkreldis.2016.08.019
Vitetta, L., Chen, J., and Clarke, S. (2019). The vermiform appendix: an immunological organ sustaining a microbiome inoculum. Clin. Sci. 133, 1–8. doi: 10.1042/CS20180956
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wu, S. C., Chen, W. T., Muo, C. H., Ke, T. W., Fang, C. W., and Sung, F. C. (2015). Association between appendectomy and subsequent colorectal cancer development: an Asian population study. PLoS One 10:e0118411. doi: 10.1371/journal.pone.0118411
Yang, A. M., Inamine, T., Hochrath, K., Chen, P., Wang, L., Llorente, C., et al. (2017). Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841. doi: 10.1172/JCI90562
Zhao, L., Zhang, F., Ding, X., Wu, G., Lam, Y. Y., Wang, X., et al. (2018). Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156. doi: 10.1126/science.aao5774
Keywords: gut bacteria, gut fungi, appendectomy, short-chain fatty acids, community interactions
Citation: Cai S, Fan Y, Zhang B, Lin J, Yang X, Liu Y, Liu J, Ren J and Xu H (2021) Appendectomy Is Associated With Alteration of Human Gut Bacterial and Fungal Communities. Front. Microbiol. 12:724980. doi: 10.3389/fmicb.2021.724980
Received: 14 June 2021; Accepted: 19 August 2021;
Published: 16 September 2021.
Edited by:
Yanling Wei, Army Medical University, ChinaReviewed by:
Hailong Cao, Tianjin Medical University General Hospital, ChinaCopyright © 2021 Cai, Fan, Zhang, Lin, Yang, Liu, Liu, Ren and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianlin Ren, amlhbmxpbi5yZW5AMTI2LmNvbQ==; Hongzhi Xu, eHVob25nemhpQHhtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.