
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 October 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.722369
A correction has been applied to this article in:
Corrigendum: Alkalihalobacterium elongatum gen. nov. sp. nov.: An Antibiotic-Producing Bacterium Isolated From Lonar Lake and Reclassification of the Genus Alkalihalobacillus Into Seven Novel Genera
A Gram-stain positive, long, rod-shaped, motile, and spore-forming bacterium (MEB199T) was isolated from a sediment sample collected from Lonar Lake, India. The strain was oxidase and catalase positive. The strain grew optimally at pH 10, NaCl concentration of 3.5% at 37°C. The major fatty acids were iso-C15:0, iso-C16:0, anteiso-C15:0, and iso-C17:0. The peptidoglycan contained meso-diaminopimelic acid (meso-DAP). Phosphatidylethanolamine, diphosphatidylglycerol, and phosphatidylglycerol were the major polar lipids of MEB199T. Phylogenetic analysis based on 16S rRNA gene sequence showed that strain MEB199T belonged to the family Bacillaceae and exhibited a distinctive position among the members of the genus Alkalihalobacillus (Ahb.). Strain MEB199T shared the highest 16S rRNA gene sequence similarity with Alkalihalobacillus alkalinitrilicus ANL-iso4T (98.36%), whereas with type species Ahb. alcalophilus DSM 485T, it is 94.91%, indicating that strain MEB199T is distinctly related to the genus Alkalihalobacillus. The G + C content of genomic DNA was 36.47 mol%. The digital DNA–DNA hybridization (dDDH) (23.6%) and average nucleotide identity (ANI) (81%) values between strain MEB199T and Ahb. alkalinitrilicus ANL-iso4T confirmed the novelty of this new species. The pairwise identity based on the 16S rRNA gene sequence between the species of genus Alkalihalobacillus ranges from 87.4 to 99.81% indicating the heterogeneity in the genus. The different phylogenetic analysis based on the genome showed that the members of the genus Alkalihalobacillus separated into eight distinct clades. The intra-clade average amino acid identity (AAI) and percentage of conserved proteins (POCP) range from 52 to 68% and 37 to 59%, respectively, which are interspersed on the intra-genera cutoff values; therefore, we reassess the taxonomy of genus Alkalihalobacillus. The phenotypic analysis also corroborated the differentiation between these clades. Based on the phylogenetic analysis, genomic indices, and phenotypic traits, we propose the reclassification of the genus Alkalihalobacillus into seven new genera for which the names Alkalihalobacterium gen. nov., Halalkalibacterium gen. nov., Halalkalibacter gen. nov., Shouchella gen. nov., Pseudalkalibacillus gen. nov., Alkalicoccobacillus gen. nov., and Alkalihalophilus gen. nov. are proposed and provide an emended description of Alkalihalobacillus sensu stricto. Also, we propose the Ahb. okuhidensis as a heterotypic synonym of Alkalihalobacillus halodurans. Based on the polyphasic taxonomic analysis, strain MEB199T represents a novel species of newly proposed genus for which the name Alkalihalobacterium elongatum gen. nov. sp. nov. is proposed. The type strain is MEB199T (= MCC 2982T, = JCM 33704T, = NBRC 114256T, = CGMCC 1.17254T).
The genus Bacillus is an extremely diverse group of bacteria within the phylum Firmicutes whose members currently exhibit great phylogenetic and phenotypic diversity. Numerous species that are part of this genus are unrelated to the type species as they do not share a common evolutionary history (La Duc et al., 2004). Recently, using phylogenomic approaches resolved the issue of the phylogenetic heterogeneity of the genus Bacillus by reclassifying existing species into novel genera, such as Alkalihalobacillus (Ahb.), Cytobacillus, Mesobacillus, Metabacillus, Neobacillus, and Peribacillus (Patel and Gupta, 2020). Among all these genera, the genus Alkalihalobacillus consists of rod-shaped, endospore-forming, and Gram-stain-variable bacteria included in the family Bacillaceae with the type species Alkalihalobacillus alcalophilus. Based on phylogenomic studies, it was proposed that most of the members of the genus Alkalihalobacillus exclusively shared 10 CSIs found in the different proteins (Patel and Gupta, 2020). The genus Alkalihalobacillus contains 39 species.1 Most species of this genus are aerobic, but some members are facultative anaerobic and anaerobic. Species are found to be motile by peritrichous flagella, while a few members are non-motile. Members of genus Alkalihalobacillus were isolated from diverse environments including Soda lake soil/sediment, saltpan, hypersaline lake, mushroom compost, seawater, sea urchin, guts of larvae, feces, rhizosphere soil, non-saline forest soil, mud goldmine, mangrove sediment, mural paintings, etc., (Vedder, 1934; Nielsen et al., 1995; Li et al., 2002; Heyrman et al., 2003; Yumoto et al., 2003, 2005; Ivanova et al., 2004; Santini et al., 2004; Yoon et al., 2004; Nogi et al., 2005; Olivera et al., 2005; Vargas et al., 2005; Nowlan et al., 2006; Borchert et al., 2007; Ghosh et al., 2007; Sorokin et al., 2008; Aizawa et al., 2010; Denizci et al., 2010; Borsodi et al., 2011, 2017; Chen et al., 2011a, b; Madhaiyan et al., 2011; Zhang et al., 2011; Nedashkovskaya et al., 2012; Lei et al., 2014; Zhu et al., 2014; Reddy et al., 2015; Dou et al., 2016; Song et al., 2016; Singh et al., 2018; Liu et al., 2019; Gupta et al., 2020; Mo et al., 2020; Patel and Gupta, 2020; Shin et al., 2020). The majority of species from this genus are alkaliphilic and can grow in the pH range of 6–11 with optimum growth at pH 9–10. Some of the members are found to be obligately alkaliphilic in nature. The members are halotolerant or halophilic in nature as they grow in the presence of 1–5% w/v NaCl concentration. Members of this genus are mesophilic and grow at a temperature from 4 to 45°C with optimum growth at 25–37°C. Several species from this genus are of considerable industrial interest due to the production of enzymes such as cellulases, xylanases, proteases, and cyclodextrin glucanotransferase. Ahb. rhizosphaerae are diazotrophic, while Ahb. clausii exhibit probiotic activity (Nielsen et al., 1995; Madhaiyan et al., 2011).
While exploring the bacterial diversity of alkaline Lonar Lake, an antimicrobial compound producing alkaliphilic, moderately halophilic bacterial strain designated as MEB199T was isolated from the sediment sample. Its taxonomic position was determined by employing a polyphasic taxonomic approach including whole genome-based analysis. During the assessment of the taxonomic status of the strain MEB199T, it was observed that the genome-based phylogenetic analysis and overall genome relatedness index (OGRI) indicated that the genus Alkalihalobacillus is composed of heterogeneous members, and its reclassification is required. Apart from having phylogenetic differences, members of the genus Alkalihalobacillus also differ in phenotypic characters such as morphology, growth requirement, polar lipids, and fatty acid composition. Based on phenotypic characteristics, phylogenetic analysis, and OGRI, we propose the reclassification of genus Alkalihalobacillus into seven new genera and provide an emended description of the genus Alkalihalobacillus sensu stricto. Similarly, the combined phenotypic and genotypic analysis indicate that the strain MEB199T represents a novel species of the newly proposed genus Alkalihalobacterium gen. nov., for which the name Alkalihalobacterium elongatum gen. nov. sp. nov. is proposed. Based on digital DNA–DNA hybridization (dDDH) and ANI value, it was noticed that Ahb. halodurans DSM 497T and Ahb. okuhidensis DSM 13666T belong to the same species. Therefore, we propose Ahb. okuhidensis as a heterotypic synonym of Ahb. halodurans.
Strain MEB199T was isolated from a sediment sample collected from Lonar, an Indian soda lake situated at Buldhana District, Maharashtra, India, at a depth of 0.46 m (1.5 ft) on October 27, 2010. At the time of sampling, the pH of the sample was found to be 9.8 and temperature was 28°C. The sediment sample was serially diluted, spread on nutrient agar (pH 9.8; HiMedia, catalog no. M001), and incubated aerobically at 30°C. Bacterial colonies were observed after 2 days, which were purified after three successive transfers to a fresh medium. Ahb. alkalinitrilicus DSM 22532T was procured from DSMZ German Collection of Microorganisms and Cell Cultures GmbH. Desertibacillus haloalkaliphilus KJ1-10-99T was shared with us by Dr. Hitarth B. Bhatt, Saurashtra University, Rajkot, Gujarat, India, as a gratis. All strains were grown on nutrient agar (pH 9.8) and preserved as glycerol (20% v/v) stock, which was stored at −80°C and in liquid nitrogen.
The phenotypic characterization of MEB199T and Ahb. alkalinitrilicus DSM 22532T was carried out under the same laboratory conditions. Morphological characteristics were studied following the growth on nutrient agar (pH 9.8) media (HiMedia, catalog no. M001) plates incubated at 37°C for 48–72 h. Gram staining and spore staining were performed following standard procedures. Catalase and oxidase tests was carried as described earlier (Smibert and Krieg, 1994). Motility was checked by the hanging drop method. Hydrolysis of casein, starch, gelatin, nitrate, and nitrite were tested separately as reported previously (Smibert and Krieg, 1994). API 20E, API 20NE, API 50CH, API ZYM strips (bioMérieux, France), and BIOLOG GEN III plate were used to study the activities of constitutive enzymes, fermentation/oxidation profile, acid production, and substrate utilization as sole carbon and energy sources at 37°C for 48 h according to the instructions of the manufacturers. The temperature range for growth was determined on nutrient agar (pH 10) plates by incubating cultures at 4–45°C (4, 10, 20, 28, 37, and 45°C) for 72–96 h. Tolerance to various NaCl concentrations and pH were investigated using salt basal medium (SBM) as described earlier (Dimitriu et al., 2005) by measuring the optical densities (wavelength 600 nm) at 37°C up to 96 h. Tolerance to NaCl was tested using SBM with various NaCl concentrations (0–10%, w/v, at intervals of 0.5%). Growth was assessed in SBM adjusted to pH 7–11 at intervals of 0.5 pH unit by KH2PO4/K2HPO4 or Na2CO3/NaHCO3 buffer system. All parameters (temperature, NaCl concentration, and pH of the medium) were tested in triplicate.
For cellular fatty acid analysis, strain MEB199T and Ahb. alkalinitrilicus DSM 22532T were grown on nutrient agar (pH 10) plates at 37°C for 16 h and collected at the same physiological age (at a logarithmic phase of growth). Cellular fatty acid methyl esters (FAMEs) were obtained from cells by saponification, methylation, and extraction following the protocol of MIDI. Cellular FAMEs were separated by gas chromatography (7890N, Agilent Technologies) and analyzed using the Sherlock Microbial Identification System (MIDI with database RTSBA6) according to the protocol described by the Sherlock Microbial Identification System. Cell wall samples were prepared from approximately 3 g of wet cells. Whole-cell hydrolyzates were prepared (6 M HCl, 100°C, 18 h) and examined by thin layer chromatography (TLC) on cellulose plates using n-butanol:water:acetic acid (50:25:25, v/v) as the solvent system.
Polar lipids were extracted from both the strains and analyzed. The cultures were harvested at a logarithmic phase, and the pellet was used for polar lipid extraction with methanol/chloroform/0.3% sodium chloride (2:1:0.8, by vol.) as described by Bligh and Dyer (1959) considering the modifications of Card (1973). Lipids were separated using silica gel TLC (Kieselgel 60 F254; Merck) by two-dimensional chromatography using chloroform:methanol:water (65:25:4 by vol.) in the first dimension and chloroform:acetic acid:methanol:water (40:7.5:6:2, by vol.) in the second dimension (Minnikin et al., 1984). The dried plates were subjected to spraying with 5% ethanolic phosphomolybdic acid for total lipids and further characterized by spraying with ninhydrin (specific for amino groups), molybdenum blue (specific for phosphates), Dragendorff (quaternary nitrogen), or α-naphthol (specific for sugars).
For DNA extraction, strain MEB199T and Desertibacillus haloalkaliphilus KJ1-10-99T were grown on nutrient agar (pH 10) medium and incubated at 37°C for 48–96 h. Genomic DNA was extracted as described earlier, and 16S rRNA genes were amplified using the universal primer set 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1488R (5′-CGGTTACCTTGTTACGACTTCACC-3′) (Brosius et al., 1978). Purified PCR products were sequenced on both strands on an ABI 3730 xl DNA analyzer using the Big Dye terminator kit (Applied Biosystems). The sequence was compared with the 16S rRNA gene sequence available at the EzBioCloud database (Yoon et al., 2017).
The genomes of MEB199T and Desertibacillus haloalkaliphilus KJ1-10-99T were sequenced on the Illumina MiSeq (250 × 2 chemistry) platform. The reads were assembled using SPAdes (version 3.1.3.0), and the quality of the assembly was checked using QUAST (version 5.0.2). The obtained genome sequence was subsequently deposited in NCBI. The Gene prediction was performed using GeneMarkS and validated with the prokaryotic annotation pipeline of NCBI, PGAP. The Rapid Annotations using Subsystem Technology (RAST) server was used for the genome annotations.2 The GenBank accession numbers for the 16S rRNA gene sequence and draft genome sequence of strain MEB199T are KX171019 and WMKZ00000000, respectively.
Type strains of 41 species of the genus Alkalihalobacillus (28 type strains), Desertibacillus, and Anaerobacillus and type species of the different genera of the family Bacilla, whose genomes were available in the public database, were considered in this study. Fifty genomes of non-type strains of genus Alkalihalobacillus were also included in this study. Streptococcus gordonii ATCC 10558T and Streptococcus agalactiae ATCC 13813T genomes were used to root (outgroup) the phylogenetic tree. The genome of strain MEB199T and Desertibacillus haloalkaliphilus KJ1-10-99T were sequenced under this study, and all the other genomes were downloaded from the PATRIC database (Supplementary Table 1). The functional annotations of each genome was carried out using EggNOG-mapper v2 (Cantalapiedra et al., 2021). The pathway was mapped using the KEGG Orthology (KO) Database3 for all the selected genomes. Heatmap to visualize the distribution of pathways across all the members of the genus Alkalihalobacillus was constructed using heatmapper.4
The antiSMASH bacterial version 6.0 was used to understand the secondary metabolite biosynthetic gene clusters in strain MEB199T (Blin et al., 2021). To check the antibacterial activity of the compound produced by MEB199T, the aqueous extract of strain MEB199T was tested for its activity against four multidrug-resistant (MDR, resistant to three or more antimicrobial classes) pathogens: Acinetobacter baumannii BAC01, Escherichia coli BAC03, Staphylococcus aureus MCC 2043T, and Klebsiella pneumoniae BAC02 using the agar overlay method. All the above pathogens used in the present study are clinical isolates resistant to more than six antibiotics. The strain MEB199T was grown aerobically in nutrient broth (pH 10) medium under shaking conditions (150 rev min–1) for 96 h at 37°C. After incubation, the culture broth was centrifuged at 16,770 × g for 30 min. The supernatant was filtered through a filter of 0.22-μm pore size. The filtrate was concentrated 10-fold by lyophilization. The antimicrobial activity of the extract was carried out using a well diffusion method where 50 μl of the concentrated filtrate was added to wells (6-mm diameter) in Mueller–Hinton agar plates containing pathogenic indicator strains and incubated at 37°C for 96 h. The inhibition of growth was expressed as the diameter of the zone of inhibition around the well. All tests were carried out in triplicate.
The 16S rRNA gene sequences of all the Alkalihalobacillus spp. and related members were retrieved from the NCBI database. Using the Up-to-date bacterial core gene (UBCG) tool, 92 core genes were extracted from all the genomes including two outgroups i.e., Streptococcus gordonii ATCC 10558T and Streptococcus agalactiae ATCC 13813T (Na et al., 2018). A concatenated sequence was used to construct the phylogenetic tree using MEGA7. The distance was calculated with Kimura two-parameter as a model of nucleotide substitution, in pairwise deletion procedure, Poisson model. The phylogenetic tree was constructed using the neighbor-joining (NJ), maximum-parsimony (MP), and maximum-likelihood (ML) methods with bootstrap analysis of 1,000 resamplings in the MEGA7 software package (Kumar et al., 2016). To assess the taxonomic position of the strain MEB199T, a codon tree based on 500 single-copy genes was reconstructed using amino acid and nucleotide sequences as described in Suresh et al. (2019). The Genome Taxonomy Database toolkit (GTDB-Tk) was used to construct the phylogenetic tree from the genome sequences (Chaumeil et al., 2019).
The pan-genome of the species of the genus Alkalihalobacillus was analyzed by the Bacterial Pan Genome Analysis (BPGA) software (Chaudhari et al., 2016). To understand the interspecies variation and core genome, BPGA was used at its default parameters. Similarly, the pan-genome of the proposed genera was analyzed independently. Conserved signature indels (CSIs) were identified using protein sequences from the core proteins present in the members of the genus Alkalihalobacillus as described by Gupta (2014). Multiple sequence alignments were performed using Clustal Omega.5 The alignments were visually inspected for sequence gaps (insertion or deletion) of fixed lengths. Average nucleotide identity (ANI) analysis between the species of the genus Alkalihalobacillus was performed by using the ANI calculator.6 The dDDH was calculated using the genome-to-genome distance calculator using the HSP length, and formula 2 values were considered in this analysis (Goris et al., 2007). The percentage of conserved proteins (POCP) and average amino acid identity (AAI) for the genus level delineation were calculated. The POCP was calculated as described by Qin et al. (2014), and the AAI was computed using an online ANI/AAI-Matrix calculator.7
During the exploration of bacterial diversity of the alkaline saline Lonar Lake, a strain MEB199T was isolated in nutrient agar (pH 10) from a sediment sample. Cells of strain MEB199T showed 2- to 5-mm, cream-colored, flat, and dry colonies with irregular margins on nutrient agar (pH 10) medium after 48 h at 37°C. The cells of the strain MEB199T was Gram stain positive, motile, long thick rods (6.4–16.5 × 0.6–2.4 μm), and spore forming. The strain was oxidase and catalase positive. The strain MEB199T is alkaliphilic and halophilic, grew optimally at pH 10, at an NaCl concentration of 3.5%.
The strain MEB199T and Ahb. alkalinitrilicus DSM 22532T were tested for utilization and assimilation of various carbon sources and enzyme activity against different substrates by API (bioMérieux, France) and BIOLOG GEN III plate. The differential morphological, physiological, and biochemical characteristics of strain MEB199T and Ahb. alkalinitrilicus DSM 22532T are given in Table 1. In the BIOLOG GEN III plate, MEB199T was positive for various substrates like gentiobiose, D-melibiose, α-D-glucose, D-fructose, D-galactose, 3-methyl glucose, D-fucose, L-fucose, L-rhamnose, D-fructose-6-PO4, D-galacturonic acid, D-glucuronic acid, glucuronamid, and sodium butyrate. The strain is negative for acetoacetic acid and acetic acid, while its closest phylogenetic neighbor, Ahb. alkalinitrilicus DSM 22532T, was positive for those substrates. The strain MEB199T showed a weak positive reaction for propionic acid, while Ahb. alkalinitrilicus DSM 22532T showed negative activity for that substrate. In the API ZYM system, both the strains under study tested positive for the production of enzymes like leucine arylamidase, valine arylamidase, α-chymotrypsin, napthol AS-BI-phosphohydrolase, and α-glucosidase. Ahb. alkalinitrilicus DSM 22532T could be able to produce enzymes like esterase (C4) and esterase lipase (C8), while MEB199T could not. Acid phosphatases and ß-glucosidase were produced by MEB199T and not found in Ahb. alkalinitrilicus DSM 22532T; this differentiated the novel strain from the type strain DSM 22532T. In the API 20E system, MEB199T showed negative results for Voges Proskauer and sucrose fermentation, while Ahb. alkalinitrilicus showed positive results for both tests. Strain MEB199T reduced nitrate to nitrite, hydrolyze esculin, and could assimilate mannitol and malate, while Ahb. alkalinitrilicus showed negative results for all those substrates but could assimilate maltose in the API 20 NE system. The physiological characteristics using BIOLOG GEN III and API analyses provided further support to investigate strain MEB199T for its unique taxonomic position.
Chemotaxonomic characteristics of strain MEB199T were consistent with those of members of the family Bacillaceae. The cellular fatty acid composition of strain MEB199T showed a spectrum of 12 fatty acids with a pronounced dominance of iso-C15:0 (27.4%), iso-C16:0 (13.4%), anteiso-C15:0 (11.8%), and iso-C17:0 (10.5%). The fatty acids were dominated by branched and monounsaturated fatty acids. Ahb. alkalinitrilicus DSM 22532T showed anteiso-C15:0, iso-C15:0, -iso-C16:0, and iso-C14:0 as primary fatty acids with anteiso-C15:0 as the dominant one, while MEB199T has iso-C15:0 the dominant fatty acid. Apart from the major fatty acids in strain MEB199T, other qualitative and quantitative differences were observed in the reference strains with respect to other minor fatty acids (Table 2). The polar lipid profile of strain MEB199T was found to contain diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and an unidentified phospholipid lipid (Supplementary Figure 1). Polar lipids of strain MEB199T differed from strain DSM 22532T by the absence of unidentified aminolipid (APL) and unidentified phospholipid (PL2, PL3, PL4, and PL5). The peptidoglycan of strain MEB199T contained meso-diaminopimelic acid (meso-DAP) as the cell wall diamino acid. The peptidoglycan diamino acid of the cell wall of strains MEB199T and Ahb. alkalinitrilicus DSM 22532T were similar to those found in members of the genus Alkalihalobacillus.
Paired-end sequencing resulted in about 1,983,695 quality-filtered reads with an average read length of 224.33 bp. Assembly of reads resulted in 61 contigs with a total sequence length of 4.81 Mbp. The sequencing coverage was approximately 232X. The DNA G + C content was determined from the genome sequence, which is 36.7%. Annotation of the genome consisted of 4,926 coding sequences. The protein-coding genes of MEB199T have an average length of 776 bases, ranging from 56 to 7,274 bases. Strain MEB199T harbor only one copy of the 16S rRNA gene (1,551 bp). Out of 4,926 open reading frames (ORFs) identified, 3,156 (64.06%) were functionally annotated, with 1,770 (35.93%) being hypothetical genes. The 16S rRNA gene sequence extracted from whole-genome was compared with that determined by PCR and Sanger sequencing (KX171019). Both sequences were found identical.
The complete (1,551 bp) 16S rRNA gene sequence of strain MEB199T was used for sequence and phylogenetic analysis. Based on EzTaxon-e search analysis, the closest phylogenetic neighbor of strain MEB199T is Ahb. alkalinitrilicus DSM 22532T, with which it shared 98.36% sequence similarity, followed by Desertibacillus haloalkaliphilus KJ1-10-99T (97.10%), Anaerobacillus alkaliphilus B16-10T (96.72%), and Anaerobacillus isosaccharinicus NB2006T (96.59%). It showed a similarity of <96% with other species. However, the pairwise sequence identity level between strain MEB199T and Ahb. alcalophilus DSM485T, the type species of the genus Alkalihalobacillus, was 94.91%, which indicates that strain MEB199T might not be a member of the genus Alkalihalobacillus.
The phylogenetic tree based on the 16S rRNA gene placed strain MEB199T and a closely related Ahb. alkalinitrilicus DSM 22532T in a separate clade (Figure 1). The phylogenetic trees constructed using MP and ML methods revealed a similar tree topology with the common node with Ahb. alkalinitrilicus DSM 22532T, which confirmed a close similarity between these two strains. The genus Alkalihalobacillus was separated into eight different clades, which were referred to as Clade I, Clade II, Clade III, Clade IV, Clade V, Clade VI, Clade VII, and Clade VIII in the subsequent discussion (Figure 1). It is interesting to point out that the 16S rRNA gene sequence similarities between species of the genus Alkalihalobacillus ranged from 87.40 to 99.81% (Supplementary Table 2). The pairwise distance of 16S rRNA gene sequence identity value of <95 indicates affiliation with different genera (Rosselló-Móra and Amann, 2015). This wide range (87.40–99.81%) of 16S rRNA gene sequence similarity indicates heterogeneity in the genus Alkalihalobacillus and signposts the need to reassess the taxonomy of the genus. The pairwise sequence identity among the Alkalihalobacillus species showed that 16S rRNA gene sequences have limited power, and to better resolve the taxonomic affiliation in the members of the genus Alkalihalobacillus, other approaches available in the genomic era have to be investigated.
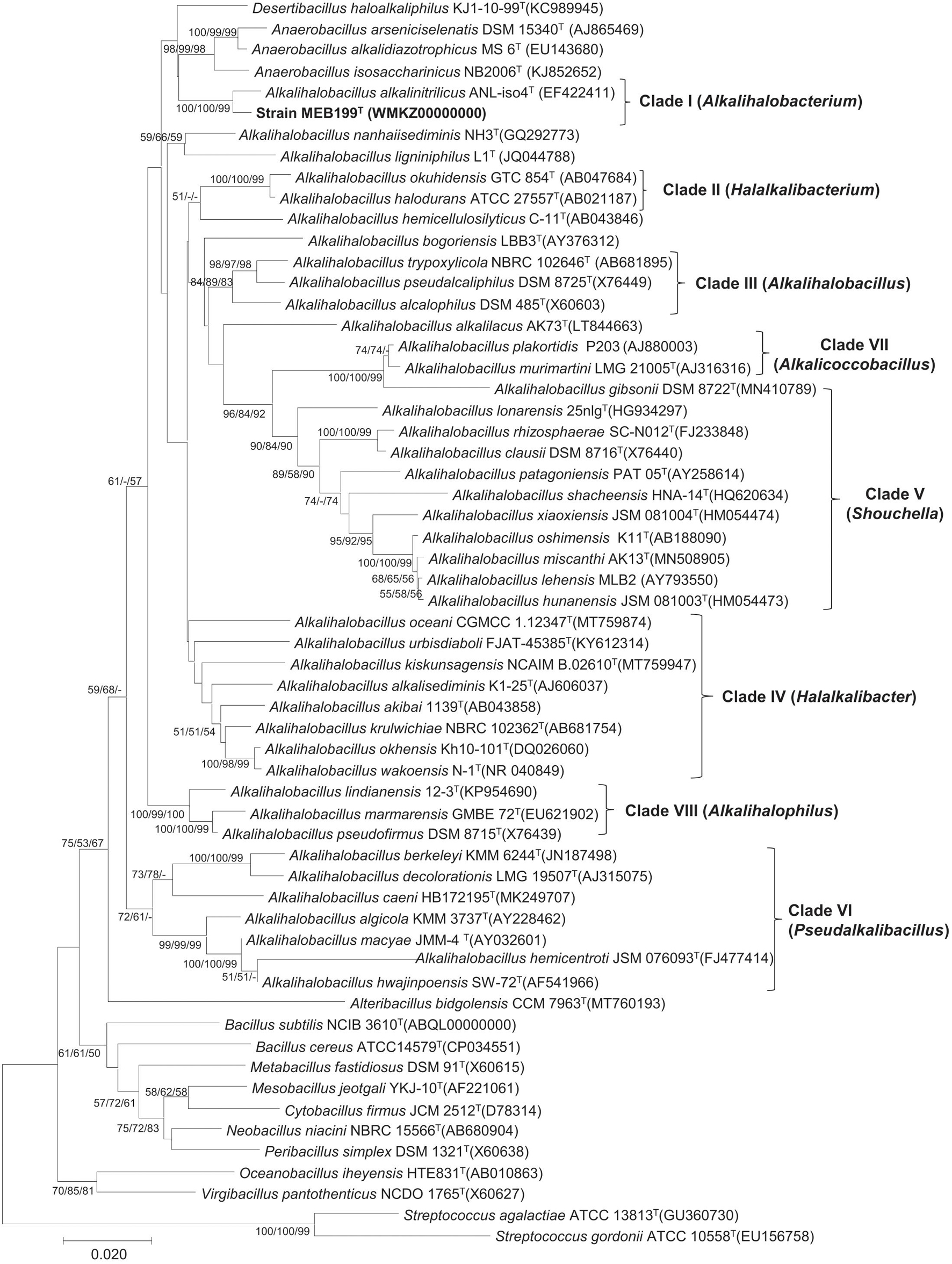
Figure 1. Phylogenetic tree based on 16S rRNA gene sequence showing the phylogenetic relationship between the members of the genus Alkalihalobacillus. The tree was constructed using MEGA7 and Streptococcus gordonii ATCC 10558T, and Streptococcus agalactiae ATCC 13813T was used as an outgroup. The 16S rRNA gene bank accession number is shown in parentheses. The bootstrap percentage refers to minimum-evolution (ME)/neighbor-joining (NJ)/maximum-likelihood (ML) analysis. The bootstrap values only above 50 for each node are indicated. Scale bar indicates the number of substitutions per site.
Phylogenomic analysis based on the core genome made up of 92 genes and codon tree of the recently described genera by Patel and Gupta (2020) as well as the remaining species in the genus Alkalihalobacillus formed well-supported eight clades (Figure 2 and Supplementary Figure 2). The phylogenomic tree based on 120 ubiquitous single-copy proteins was also constructed using GTDB-Tk, which also separates the genus Alkalihalobacillus into eight clades (Supplementary Figure 3). Strain MEB199T clustered together with Ahb. alkalinitrilicus DSM 22532T and Ahb. bogoriensis ATCC BAA-922T by forming a separate clade distinguishable from genus Alkalihalobacillus. Based on the phylogenomic analysis, Clade I, Clade II, Clade III, Clade IV, Clade V, Clade VI, Clade VII, and Clade VIII were observed similar to the 16S rRNA gene sequence-based analysis. Ahb. murimartini LMG 21005T was present in Clade VII in the 16S rRNA gene-based tree within the members of genus Alkalihalobacillus, but in phylogenomic analysis, it was completely outgrouped from the genus Alkalihalobacillus (Figure 2).
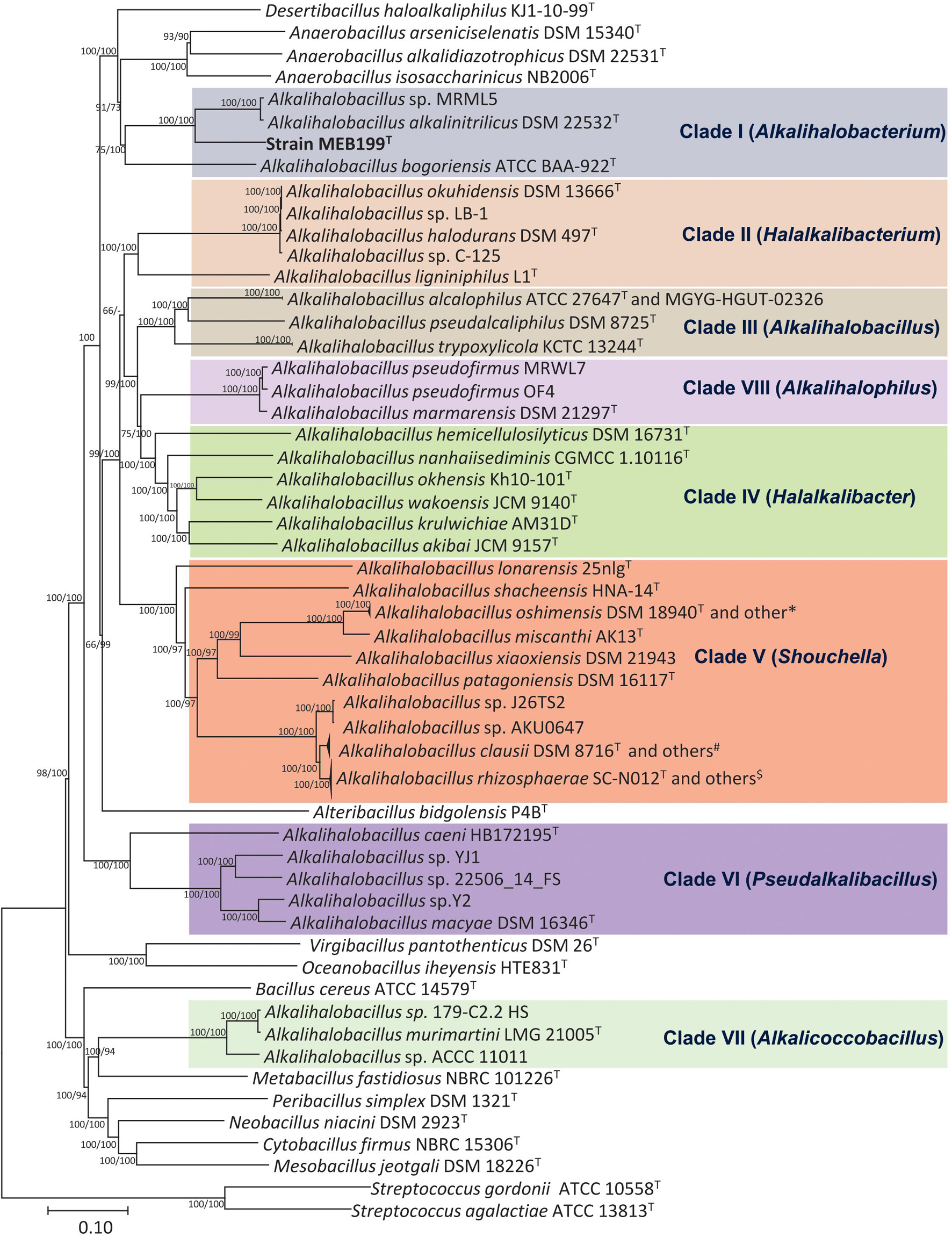
Figure 2. Phylogenetic tree constructed using the 92 bacterial core gene sequences showing the relationships of the members of genus Alkalihalobacillus and nearest genera. The 92 gene sequences were extracted using Up-to-date bacterial core gene (UBCG) tool, which is a widely used resource for delineating the phylogeny of bacteria, and the phylogenetic tree was constructed using MEGA7 with NJ and ME algorithms. Bar, 0.1 nucleotide substitution per position. *Strains DSM 19099, G25-134, G1, DSM 19153; #Strains 7520-2, 7540-2, 7547-G, 179-F 5B1 HS, 7535-K, 7541, 7538, 7523-2, 088AE, BC112, 7522; $Strains 7894-1, UBBC-07, J32TS2, 7529, KSM-K16, 7540-1, 7539, 7537-T, J1TS1, UBBC-08/C, UBBC-08/T, UBBC-08/R, GMN, B637/NM, B619/R, B603/Nb, CSI08, ENTPro, UBBC-08/S, B106, 7543.
Bacterial pan-genome analysis was carried out between the type strains of the genus Alkalihalobacillus. In the 28 analyzed species of the genus Alkalihalobacillus, 598 genes were identified as core genes indicating that members of the genus Alkalihalobacillus share a very small number of core genes. The core genes encode for the shikimate pathway, isoprenoid biosynthesis (non-mevalonate pathway), thiamine biosynthesis, tetrahydrofolate biosynthesis, pantothenate biosynthesis, lysine biosynthesis, riboflavin biosynthesis, inosine monophosphate biosynthesis, uridine monophosphate biosynthesis, F-type ATPase, heme biosynthesis, pyrimidine deoxyribonucleotide biosynthesis, citrate cycle (TCA cycle, Krebs cycle), lysine biosynthesis, coenzyme A biosynthesis, glycolysis (Embden–Meyerhof pathway), pantothenate biosynthesis, gluconeogenesis, glycolysis, pyruvate oxidation, adenine ribonucleotide biosynthesis, NAD biosynthesis, guanine ribonucleotide biosynthesis, UDP-N-acetyl-D-glucosamine biosynthesis, and dicarboxylate–hydroxybutyrate cycle. The core genome analysis was also carried out for the individual clade formed in the phylogenomic tree. Clade I shares 1,991 as core genes with 3,024 genes as accessory genomes. Clade II shares 1,873 core genes with 3,128 accessory genes (Figure 3 and Table 3). Clade III shares 2,070 core genes and 1,942 as accessory genes. Clade IV shares 1,482 as core genes, and Clade V shares 1,195 core genes. Clade VI consists of only two members of the genus Alkalihalobacillus, which shares 2,233 core genes between them (Figure 3 and Table 3). In the clade-wise pan-genome analysis, there was an increase in the number of core genes, which showed the divergence in inter-clade genomes.
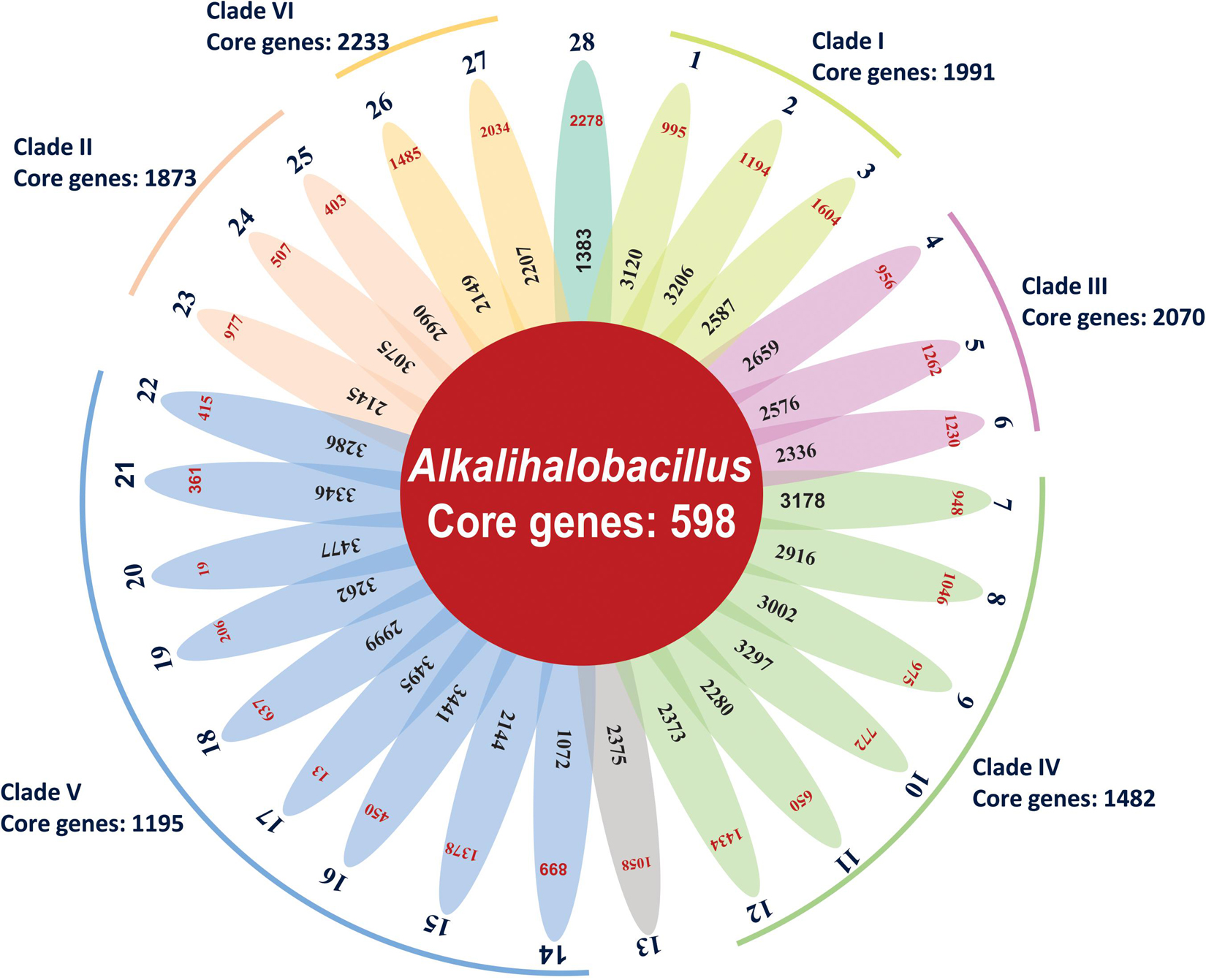
Figure 3. Flower-pot diagram representing core, accessory, and unique genomes of type strains of genus Alkalihalobacillus (1, Strain MEB199T; 2, Ahb. alkalinitrilicus DSM 22532T; 3, Ahb. bogoriensis ATCC BAA-922T; 4, Ahb. alcalophilus ATCC 27647T; 5, Ahb. pseudalcaliphilus DSM 8725T; 6, Ahb. trypoxylicola KCTC 13244T; 7, Ahb. akibai JCM 9157T; 8, Ahb. krulwichiae AM31DT; 9, Ahb. wakoensis JCM 9140T; 10, Ahb. okhensis Kh10-101T; 11, Ahb. nanhaiisediminis CGMCC 1.10116T; 12, Ahb. hemicellulosilyticus DSM 16731T; 13, Ahb. marmarensis DSM 21297T; 14, Ahb. lonarensis 25nlgT; 15, Ahb. shacheensis HNA-14T; 16, Ahb. clausii DSM 8716T; 17, Ahb. oshimensis DSM 18940T; 18, Ahb. patagoniensis DSM 16117T; 19, Ahb. lehensis DSM 19099; 20, Ahb. plakortidis DSM 19153; 21, Ahb. rhizosphaerae SC-N012T; 22, Ahb. miscanthi AK13T; 23, Ahb. ligniniphilus L1T; 24, Ahb. okuhidensis DSM 13666T; 25, Ahb. halodurans DSM 497T; 26, Ahb. macyae DSM 16346T; 27, Ahb. caeni HB172195T and 28, Ahb. murimartini LMG 21005T).
The members of the genus Alkalihalobacillus are an industrially important group of bacteria, which have been isolated from diverse ecological niches (Jones et al., 1998; Grant, 2003). They are likely to play an important but yet unexplored role in the functional stability and maintenance of the ecosystem. Several species from this genus are of considerable industrial interest due to their production of enzymes such as cellulases, proteases for inclusion in laundry detergents, xylanases for use in the pulp paper industry, and cyclodextrin glucanotransferase for manufacture of cyclodextrin from starch (Horikoshi, 2006). The genus Alkalihalobacillus is attracting interest because its members have a great biotechnological potential for producing compatible solutes or hydrolytic enzymes (Horikoshi, 1999; Margesin and Schinner, 2001; Arahal and Ventosa, 2002; Krulwich et al., 2007). Some of these bacterial species are believed to have industrial potential as a source of alkali-stable enzymes (Gessesse and Gashe, 1997). Ahb. patagoniensis, Ahb. lehensis, and Ahb. marmarensis are producers of alkaline proteases, while Ahb. lonarensis and Ahb. oshimensis could produce various protease, lipase, and xylanase enzymes. Obligately alkaliphilic species, Ahb. krulwichiae, can degrade aromatic compounds in alkaline conditions, while Ahb. ligniniphilus, a halotolerant alkaliphilic bacterium, is used to degrade lignin (Zhu et al., 2017). Ahb. rhizosphaerae, which is diazotrophic and can fix atmospheric nitrogen to ammonia and other species such as Alkalihalobacillus clausii, exhibits probiotic activity due to the production of antimicrobial compounds (Nielsen et al., 1995; Madhaiyan et al., 2011). The strain MEB199T also showed an antimicrobial compound-producing activity.
The heatmap clustering based on the distributions of metabolic pathways in the genomes is shown in Figure 4. The clusters formed in the heatmap corroborate the clades in phylogenomic analysis except Ahb. ligniniphilus L1T, Ahb. hemicellulosilyticus DSM 16731T, and Ahb. lonarensis 25nlgT. The genome mining showed that the 11 subunits of NADH:quinone oxidoreductase (nuoA, nuoB, nuoC, nuoD, nuoH, nuoI, nuoJ, nuoK, nuoL, nuoM, and nuoN) was exclusively present in the members of Clade I, Clade VI, and Alkalihalobacillus ligniniphilus L1T and absent in all the other members of genus Alkalihalobacillus. The genetic capability to synthesize the isoprenoid compounds and terpenoid was screened in the genomes. All the members of the genus Alkalihalobacillus harbors methylerythritol phosphate (MEP) i.e., non-mevalonate pathway for the synthesis of isoprenoid precursors isopentenyl pyrophosphate (IPP). IPP is the precursor for the various isoprenoid molecules playing diverse roles in different processes in the bacterial cell. Menaquinones, a lipid-soluble quinone, are isoprenoid compounds that participate in the electron transport chain. All the members of the genus Alkalihalobacillus has the menaquinone (MK) biosynthesis pathway, but an alternative futalosine pathway to convert chorismate to 1,4-dihydroxy-6-naphthoate requires four enzymes encoded by mqnABCD, which are absent in Clade VI (Ahb. macyae DSM 16346T and Ahb. caeni HB172195T) and Ahb. murimartini LMG 21005T. The classical MK pathway is exclusively present in all aerobic and facultatively anaerobic bacteria in contrast to the futalosine pathway, which is present in aerobic and anaerobic bacteria. All the Clade I, Clade VI, and VII members have sporulenol synthase gene, which gives the genetic capability to cyclize tetraprenyl beta-curcumene into sporulenol (C35 terpenes), a pentacyclic sesquarterpene (Sato et al., 2011). Sporulenol, produced during the sporulation, is present in the spores and increases the resistance to reactive oxygen species (Kontnik et al., 2008).
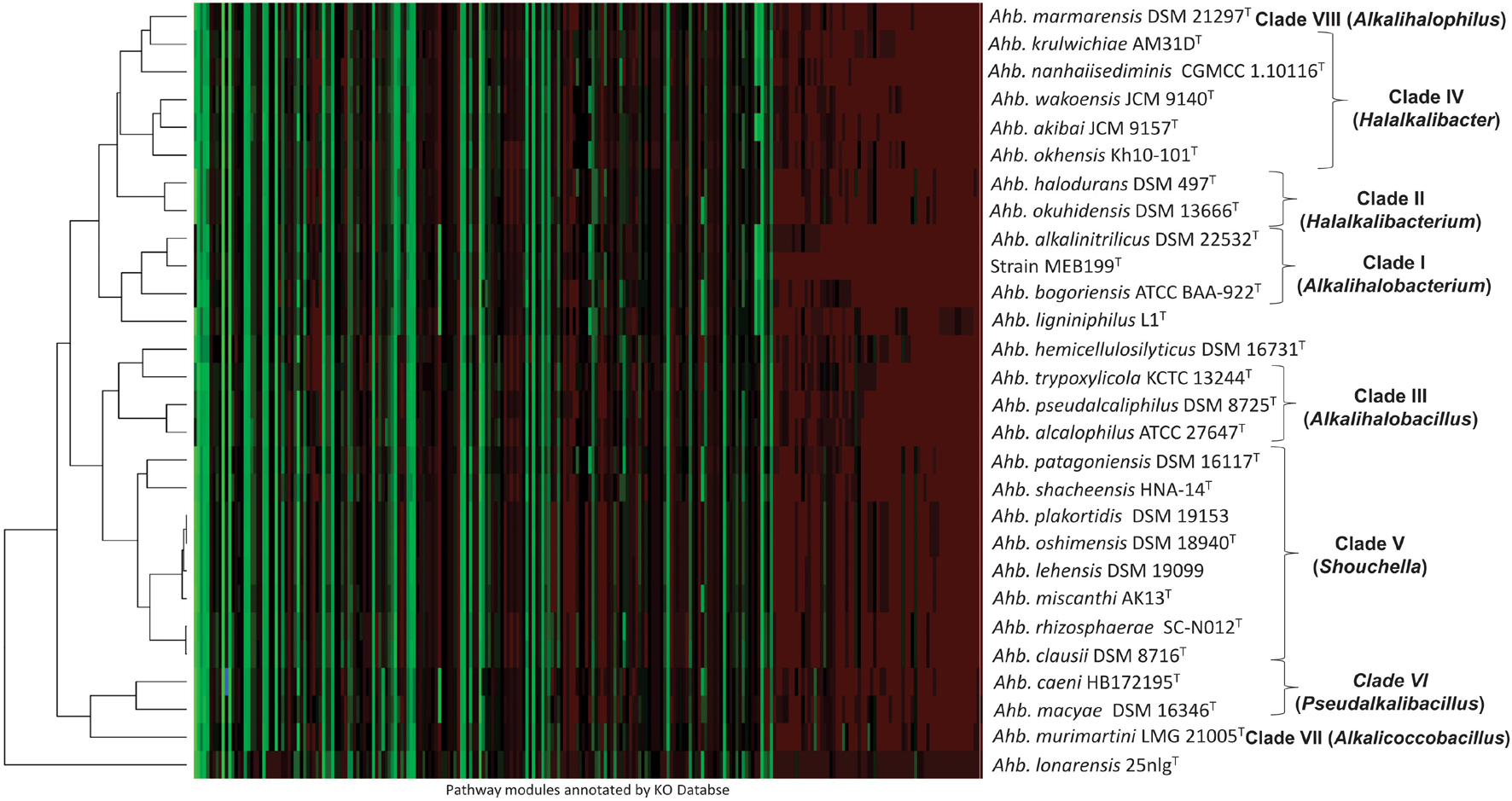
Figure 4. Heatmap showing functional potential of all the members of the genus Alkalihalobacillus. Functional annotations were performed using EggNOG, and pathways were reconstructed using the KEGG Orthology (KO) Database server, and heatmap was generated by the Heatmapper using average linkage clustering method and Spearman rank correlation distance measurement method. The color variations depict the relative abundance of genes in the pathways wherein red denotes the maximally abundant pathways, and green represents the least abundant pathways.
The bacterial strain MEB199T produced antibacterial metabolites against MDR pathogens (Acinetobacter baumannii BAC01, Escherichia coli BAC03, Staphylococcus aureus MCC 2043T, and Klebsiella pneumoniae BAC02) and was shown by the zone of inhibition (Supplementary Figure 4). The antiSMASH analysis showed that the strain MEB199T has 12 secondary metabolite biosynthetic gene clusters for the synthesis of siderophore, terpene (carotenoids and thailanstatin A), lasso peptide (paeninodin), lanthipeptide-class-I (streptin), beta lactone (fengycin), type III polyketide synthases (T3PKS) (7-deoxypactamycin), ribosomally synthesized and post-translationally modified peptides [linear azole(in)e-containing peptides LAP], and ectoine, whereas Ahb. alkalinitrilicus DSM 22532T has 8 biosynthetic gene clusters for the synthesis of siderophore, terpene (carotenoid), LAP (RiPP-like), lasso peptide (paeninodin), beta-lactone (fengycin), T3PKS (7-deoxypactamycin), and ectoine but the aqueous extract of strain DSM22532T did not show antimicrobial activity against MDR strains, which separate it from the strain MEB199T. All the genomes of genus Alkalihalobacillus were screened for the presence of secondary metabolite biosynthetic gene clusters. All members of the genus Alkalihalobacillus have multiple biosynthetic gene clusters ranging from 3 to 12 (Supplementary Table 3). All the studied members of genus Alkalihalobacillus have T3PKS biosynthetic gene cluster coding for 7-deoxypactamycin, a new member of the pactamycin group except Ahb. bogoriensis ATCC BAA-922T. Ahb. murimartini LMG 21005T has 12 biosynthetic gene clusters out of which 8 are unique and not present in other members of genus Alkalihalobacillus. There is heterogeneity in the distribution of biosynthetic gene clusters in the genus Alkalihalobacillus. There is no clade-wise pattern observed in the antiSMASH analysis as the secondary metabolite production is strain-specific character.
The ANI value between MEB199T and Ahb. alkalinitrilicus strain DSM 22532T was 81%. The in silico dDDH was carried out using the genome-to-genome distance calculator8 between the strain MEB199T and Ahb. alkalinitrilicus DSM 22532T. The dDDH analysis using HSP length showed 24% relatedness between MEB199T and Ahb. alkalinitrilicus DSM 22532T. From ANI and dDDH analysis, it can be inferred that MEB199T is a novel species. On the other hand, the ANI level between strain MEB199T and the type species of the genus Alkalihalobacillus was determined as 76% indicating its distant affiliation to the genus Alkalihalobacillus. The results of the calculations of the ANI and dDDH among the studied genomes are given in Supplementary Table 4. The results of ANI and dDDH calculations showed that the genomes grouped into the same clusters observed by the analyses of core genes and phylogenomics. Intra-clade ANI and dDDH values ranges from 75 to 95% and 18 to 63%, respectively (Table 3). All the ANI values between the species of genus Alkalihalobacillus was >96 except ANI between Ahb. okuhidensis DSM 13666T and Ahb. halodurans DSM 497T, which is 99%. This explicitly indicates that defined species of the genus are delineating from each other except Ahb. okuhidensis DSM 13666T and Ahb. halodurans DSM 497T, which failed to delineate from each other at the species level and belong to the same species. Each of the eight clusters showed that inter-clade ANI values ranged between 73.26 and 89.0% (Figure 5 and Table 3). These values are relatively similar to those reported by Qin et al. that found 68–82% interspecies ANI values among the different genera (Qin et al., 2014). The dDDH values for the species of the genus ranged from 17.8 to 63.2%, which are <70% except between Ahb. okuhidensis DSM 13666T and Ahb. halodurans DSM 497T. The dDDH between Ahb. okuhidensis DSM 13666T and Ahb. halodurans DSM 497T is 94%, which indicates that these two belong to the same species of the genus Alkalihalobacillus.
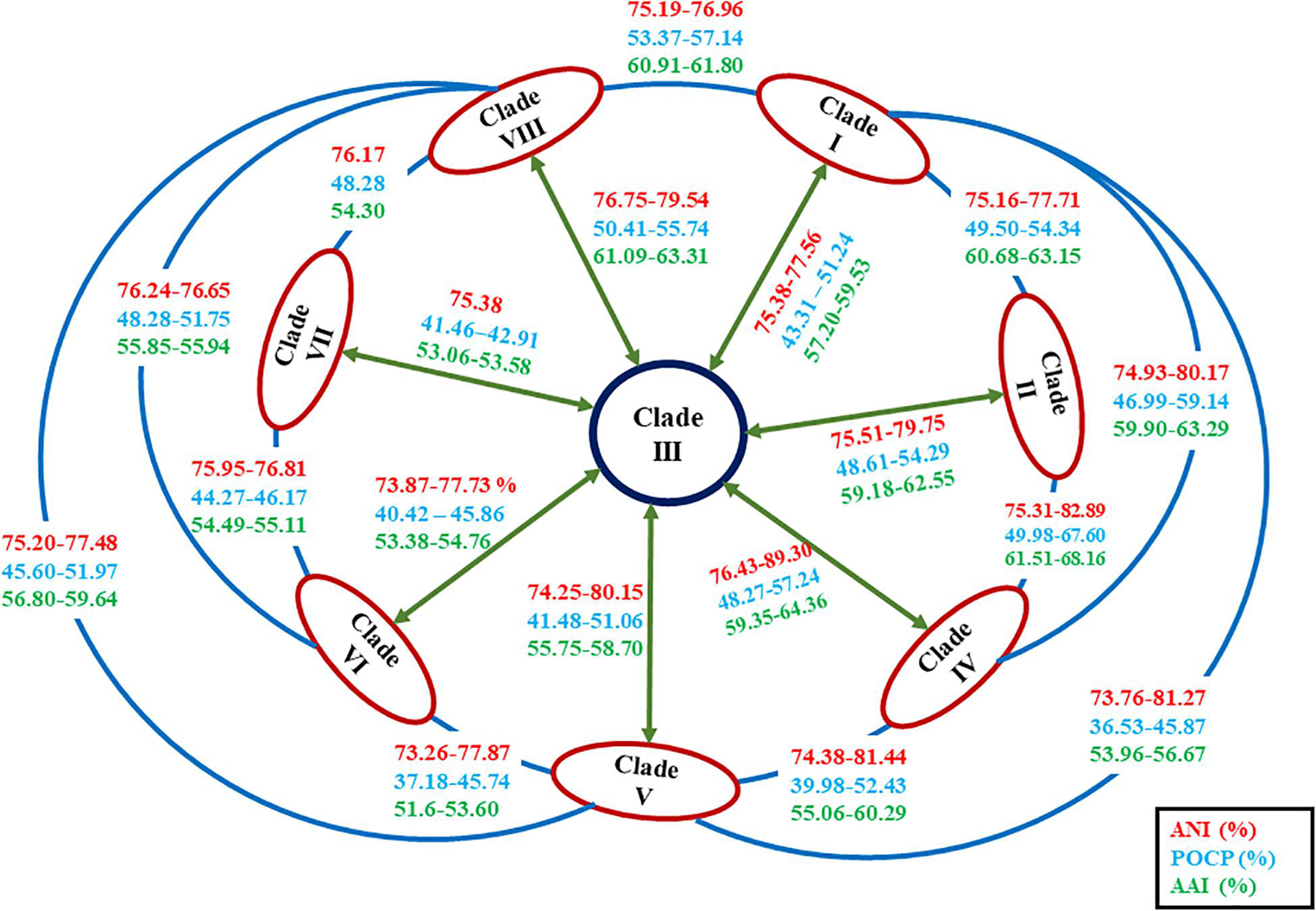
Figure 5. Diagrammatic representation of the average nucleotide identity (ANI), average amino acid identity (AAI), and percentage of conserved proteins (POCP) values in between the members of each clade of the genus Alkalihalobacillus.
In order to reassess the taxonomic position of the species belonging to the genus Alkalihalobacillus and newly isolated strain MEB199T, genome-based comparisons for conserved protein-coding genes were performed by calculating AAI levels between strain MEB199T and its close neighbors using the AAI matrix calculator. AAI values between strain MEB199T and its closest neighbors Ahb. alkalinitrilicus DSM 22532Tand Ahb. bogoriensis ATCC BAA-922T were calculated as 81 and 64%, respectively, while AAI values between MEB199T and other type strains of the genus Alkalihalobacillus were lower than 60% (Supplementary Table 5). POCP values between strain MEB199T and its closest neighbors Ahb. alkalinitrilicus DSM 22532T and Ahb. bogoriensis ATCC BAA-922T were calculated as 72 and 57%, respectively, while POCP values were <50% for the type species of the genus Alkalihalobacillus as well as between the newly described genera in the family Bacillaceae (Figure 5 and Supplementary Table 5). Consequently, strain MEB199T, together with its close phylogenetic neighbors, i.e., Ahb. alkalinitrilicus DSM 22532T and Ahb. bogoriensis ATCC BAA-922T, are considered to represent a novel genus within the family Bacillaceae.
To confirm whether the clades observed in the phylogenomic tree might represent different genera, the genomic indices POCP and AAI were also calculated with the species of genus Alkalihalobacillus (Supplementary Table 5). Considering recent work on genus delineation based on mean protein sequence similarity of all protein-coding genes, members of the family Bacillaceae can be distinguished by 65–70% AAI value at the genus level (Aliyu et al., 2016). The inter-clade AAI values ranged from 52 to 68% indicating that the genus Alkalihalobacillus is divergent and polyphyletic (Figure 5 and Supplementary Table 5). Although Qin et al. (2014) proposed <50% POCP to delineate the genera, POCP values between different newly proposed Bacillus genera ranged from 34.8 to 69.8% (Aliyu et al., 2016; Patel and Gupta, 2020). The inter-clade POCP values ranged from 37 to 68% (Figure 5 and Supplementary Table 5). The AAI and POCP values also indicate that the members of the genus Alkalihalobacillus are divergent, and there is a need for reclassification of the genus Alkalihalobacillus. Each clade in phylogenomic analysis represents a novel genus. A detailed survey of the phenotypic characters was carried out to determine if the description of new taxa at the genus level is possible, or such clades were only clusters or genomovars within the genus Alkalihalobacillus. Because the genomic analysis of 52 Arcobacter cryaerophilus strains indicate four different genomospecies, but the phenotypic study failed to delineate the species, therefore, they were considered genomovars of the same species (Pérez-Cataluña et al., 2018).
Phylogenetic analysis (16S rRNA gene-based and genome-based) indicated the existence of polyphyletic clades of genus Alkalihalobacillus. The conserved signature insertions and deletions (CSIs) in the proteins are the rare genetic changes that are exclusively shared by evolutionary linked organisms. CSIs are useful molecular signatures for evolutionarily and taxonomic studies. Therefore, the CSIs specific for the novel clades of members of the Alkalihalobacillus was studied. Patel and Gupta have reported that six CSIs are the signature of the genus Alkalihalobacillus (Patel and Gupta, 2020), whereas four CSI signatures in protein transcription–repair coupling factor, tRNA uridine-5-carboxymethylaminomethyl (34) synthesis enzyme (mnmG), 50S ribosomal protein L11 methyltransferase, and homoserine kinase were exclusively shared by all the members of the genus Alkalihalobacillus except Ahb. alkalinitrilicus and Ahb. bogoriensis. These results separate Clade I from the rest of the members of the genus Alkalihalobacillus. In the present study, CSIs that are distinctive characteristics of other clades were identified. The species of Clade II harbor the CSI of a two-amino acid insertion in the protein translocase subunit (secD) protein, which is absent in other members of the genus Alkalihalobacillus (Supplementary Figure 5). Members of Clade III consist of the CSI of a two-amino acid insertion in the UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2, 6-diaminopimelate ligase (murE) protein, which is absent in other members of the genus Alkalihalobacillus (Supplementary Figure 6). Two CSIs exclusively present in the members of Clade IV and absent in other species of the genus Alkalihalobacillus were identified. Insertion of two amino acids in DNA mismatch repair (mutL) protein and one amino acid deletion in ATP-dependent protease ATPase subunit (hslU) protein were observed in the species of Clade IV (Supplementary Figures 7, 8). Similarly, two CSIs were exclusively present in the species of Clade V, which are five-amino acid insertion in the protein translocase subunit (secY) protein and one-amino acid insertion in ATP-dependent protease ATPase subunit (hslU) protein (Supplementary Figures 9, 10). These CSIs in Clades I, II, III, IV, and V also showed the genetic distinctness of these clades formed in phylogenetic analysis and support the reclassification. We were not able to identify any CSIs for Clades VI, VII, and VIII.
Phylogenomic analysis as well as genomic indices, indicated that Alkalihalobacillus species are too divergent to be placed into the same genus. To support this further, phenotypic and chemotaxonomic markers were also surveyed to further understand its taxonomic position. The comparison of phenotypic characters between the clades and nearest genera is given in Table 4. The phenotypic characters are also able to distinguish these clades, as the members of Clade I are isolated from a soda lake, alkaliphilic, long thick rods, Gram-stain positive, G + C content range from 35.1 to 37.5%, and strictly aerobic in nature, whereas members of Clade II are facultative anaerobes, Gram-stain variable, G + C content range from 40.76 to 43.9 mol%, and motile (Table 4; Nielsen et al., 1995; Li et al., 2002; Vargas et al., 2005; Sorokin et al., 2008; Zhu et al., 2014). Members of Clade IV are alkaliphilic, quinone (MK-5, MK-6, or MK-7), the optimum temperature is 37°C, oxidase negative, and contain glycolipid, which makes it unique from the rest of the members of the genus Alkalihalobacillus (Yumoto et al., 2003; Nogi et al., 2005; Nowlan et al., 2006; Borsodi et al., 2011, 2017; Zhang et al., 2011; Song et al., 2016; Liu et al., 2019; Gupta et al., 2020). Clade V is composed of mesophilic and neutrophilic organisms that differentiate them from the rest of the Alkalihalobacillus species (Nielsen et al., 1995; Olivera et al., 2005; Yumoto et al., 2005; Ghosh et al., 2007; Chen et al., 2011a; Madhaiyan et al., 2011; Lei et al., 2014; Reddy et al., 2015; Singh et al., 2018; Gupta et al., 2020; Shin et al., 2020). Members of Clade VI can grow both aerobically and anaerobically and alkalitolerant with optimum growth at pH 7, whereas other members are alkaliphilic in nature (Heyrman et al., 2003; Ivanova et al., 2004; Santini et al., 2004; Yoon et al., 2004; Chen et al., 2011b; Nedashkovskaya et al., 2012; Mo et al., 2020). Ahb. murimartini LMG 21005T is a neutrophilic, coccoidal-shaped bacterium that separates it from the rest of the members of the genus Alkalihalobacillus (Borchert et al., 2007). These phenotypic differences also indicate that there is a need for reclassification of the genus Alkalihalobacillus.
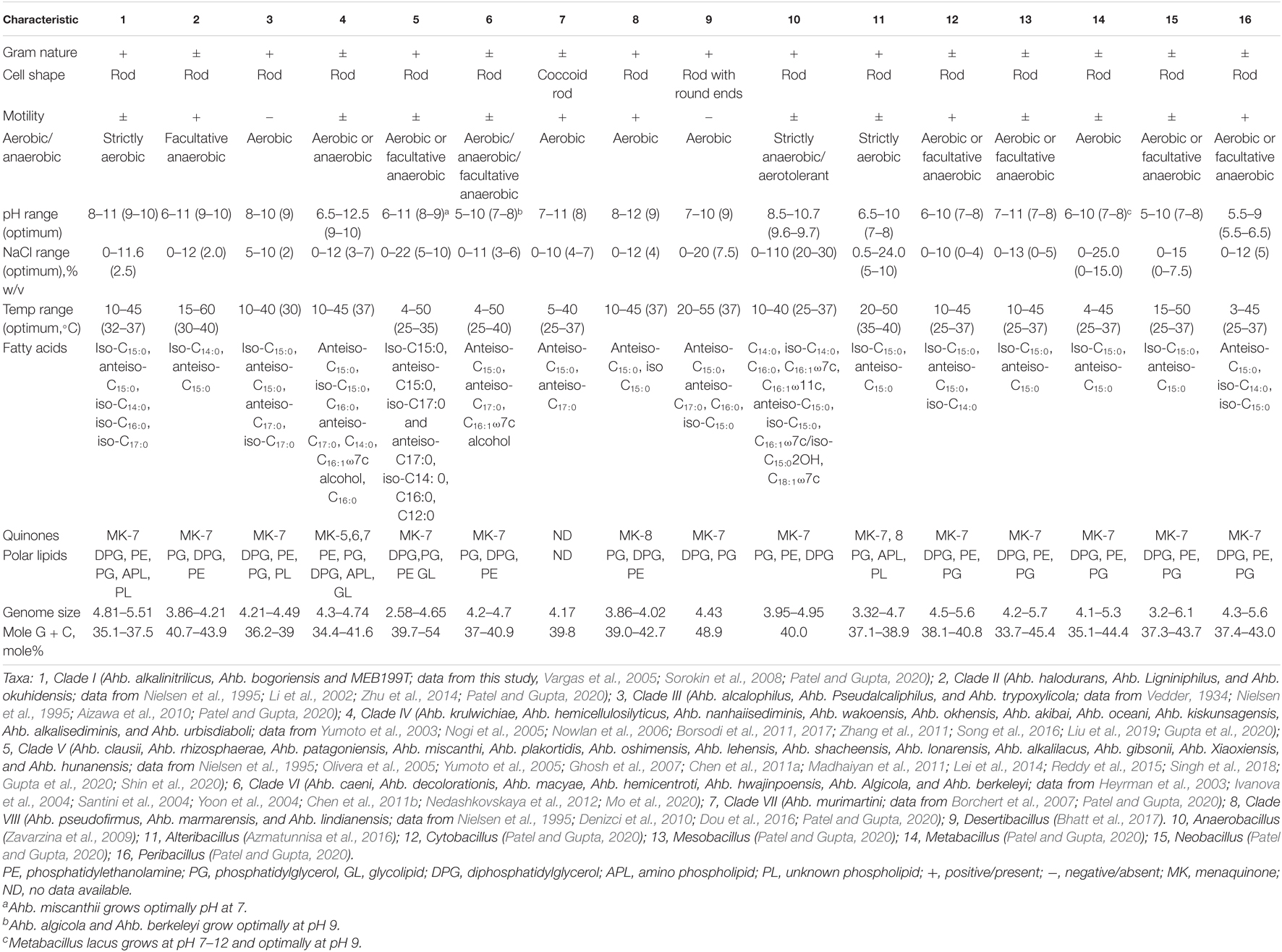
Table 4. Comparison of seven newly proposed genera with genus Alkalihalobacillus sensu stricto and members of closely related genera of the family Bacillaceae.
Based on phenotypic, genomic, phylogenetic, and chemotaxonomic characteristics, we propose the reclassification of genus Alkalihalobacillus into seven new genera with an emended description of the genus Alkalihalobacillus sensu stricto. We propose members of Clade I to be classified into a new genus for which we propose the name Alkalihalobacterium gen. nov. Though genomic indices showed that Ahb. bogoriensis is distinctly related to members of Clade I, we prefer to transfer Ahb. bogoriensis into Alkalihalobacterium gen. nov. for practical reasons to separate new genera until more strains related to this taxon become available. Therefore, Alkalihalobacterium alkalinitrilicus comb. nov. is proposed as type species of the newly proposed genus Alkalihalobacterium gen. nov. Clade II showed that the AAI, POCP, and phenotypic traits are showing clear delineation from other members. Hence, we propose that the members of Clade II be classified into the genus Halalkalibacterium gen. nov. with Halalkalibacterium halodurans as type species of the genus. Moreover, as Ahb. okuhidensis DSM 13666T and Ahb. halodurans DSM 497T have high genomic indices (ANI, dDDH), we concluded that they are not different species. Based on the phenotypic differences, we propose Ahb. okuhidensis as a heterotypic synonym of Ahb. halodurans. Clade III harbors Ahb. alcalophilus ATCC 27647T, which is the type species of the genus Alkalihalobacillus; therefore, the members of Clade III are included in the genus Alkalihalobacillus sensu stricto. Clade IV encompasses Ahb. hemicellulosilyticus, Ahb. nanhaiisediminis, Ahb. wakoensis, Ahb. okhensis, Ahb. krulwichiae, Ahb. akibai, Ahb. oceani, Ahb. kiskunsagensis, Ahb. alkalisediminis, and Ahb. urbisdiaboli for we propose Halalkalibacter gen. nov. For Clade V, we propose Shouchella gen. nov. to accommodate Ahb. rhizosphaerae, Ahb. clausii, Ahb. patagoniensis, Ahb. miscanthi, Ahb. plakortidis, Ahb. oshimensis, Ahb. lehensis, Ahb. shacheensis, Ahb. lonarensis, Ahb. alkalilacus, Ahb. gibsonii, Ahb. xiaoxiensis, and Ahb. hunanensis species. Genomic analysis also corroborated a previous finding that Ahb. plakortidis DSM 19153 and Ahb. lehensis DSM 19099 are heterotypic synonyms of Ahb. oshimensis DSM 18940T. For Clade VI, we propose Pseudalkalibacillus gen. nov. to accommodate Ahb. caeni, Ahb. macyae, Ahb. hemicentroti, Ahb. hwajinpoensis, Ahb. algicola, Ahb. berkeleyi, and Ahb. decolorationis species. To accommodate Ahb. murimartini LMG 21005T, we propose the new genus Alkalicocobacillus gen. nov. The type strains, Ahb. lindianensis 12-3T, Ahb. marmarensis GMBE 72T, and Ahb. pseudofirmus DSM 8715T, formed a distinct clade in 16S rRNA gene sequence-based phylogeny. In genome-based phylogeny, Ahb. marmarensis DSM 21297T gets outgrouped to the Clade IV. AAI between Ahb. marmarensis DSM 21297T and other members of the Clade IV ranges from 63 to 68%, which indicates that it belongs to a distinct genus. Therefore, we propose members of Clade VIII as a novel genus for which name Alkalihalophilus gen. nov. is proposed. The description of the newly proposed genera is given below, and a description of all names and new combinations is given in Table 5.
Members can be isolated from the guts of larvae, soil, and feces. Cells are rod shaped, Gram-stain positive, endospore forming, aerobic, and motile, and tolerate NaCl concentration up to 5–10% with optimum growth at 2% w/v. Growth occurs in the range 10–40°C, with optimum growth at 30°C; alkaliphilic with growth in the range of pH 8–10 with optimum growth at pH 9. The major isoprenoid quinone is MK-7. The major fatty acids are iso-C15:0, anteiso-C15:0, anteiso-C17:0, and iso-C17:0. The polar lipid profile contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and unidentified phospholipids. meso-DAP is cell wall diamino acid. The DNA G + C content is 36.2–39.0 mol%.
The type species is Alkalihalobacillus alcalophilus.
Based on ANI and dDDH analysis, Alkalihalobacillus okuhidensis is not a distinct species as the differences between Ahb. halodurans and Ahb. okuhidensis represent intra-species divergence. It should be noted that Ahb. halodurans and Ahb. okuhidensis differ with regard to fatty acid content, G + C content, growth at pH 6 and 11, utilization of carbon sources, hydrolysis of hippurate, tween 20, tween 40, and tween 60 (Li et al., 2002). Though there are minor differences in Ahb. halodurans and Ahb. okuhidensis but not enough for delineating two species. Therefore, based on the physiological, chemotaxonomic, and genotypic analysis, we propose Ahb. okuhidensis as a heterotypic synonym of Alkalihalobacillus halodurans.
Alkalihalobacterium [Al.ka.li.ha.lo.bac.te’ri.um. N.L. n. alkali, alkali (from Arabic article al, the; Arabic n. qaliy, ashes of saltwort); N.L. neut. n. bacterium, a small rod; N.L. neut. n. Alkalihalobacterium, bacterium living under alkaline-saline conditions].
Long rod-shaped cells, Gram-stain positive, aerobic, and endospore forming, have been isolated from Soda lake soil/sediment, motile or non-motile, tolerate NaCl concentration up to 11.6% with optimum growth at 2.5–3.5% (w/v). Growth occurs in the range 10–45°C, with optimum growth temperature in the range 28–37°C. All members of this genus are alkaliphilic with growth at pH 8–11 (optimum 9). The major isoprenoid quinone is MK-7. The polar lipid profile contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, aminophospholipid, and unidentified phospholipids. The major fatty acids are iso-C15:0, anteiso-C15:0, iso-C14:0, iso-C16:0, and iso-C17:0. meso-DAP is cell wall diamino acid. The DNA G + C content 35.1–37.5%.
The type species is Alkalihalobacterium alkalinitrilicum.
Alkalihalobacterium elongatum (e.lon.ga’tum. L. neut. part. adj. elongatum elongated).
Cells are motile, long, and thick rod shaped. Size is 6.4–16.5 μm × 0.6–2.4 μm (l × w). Gram-stain positive, aerobic, and forming subterminal oval endospore. The strain produced faint cream colored, dry, flat colonies with rhizoidal margins on nutrient agar (pH 10) medium, obligately alkaliphilic, and growth occurs between pH 8–11 with an optimum at pH 10. The optimum temperature for growth is 37°C and can grow at 10–45°C. The range of NaCl concentration is 0–10.5% (w/v) with optimum of 3.5% (w/v). Oxidase and catalase positive. Able to hydrolyze casein and esculin. Nitrate is reduced to nitrite. Not able to hydrolyze starch, gelatin, urea, and tween 40. Citrate is not utilized; methyl red, H2S, acetoin, or indole are not produced. Leucine arylamidase, valine arylamidase, α chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α glucosidase, and ß glucosidase activities are present. Alkaline phosphatase, esterase, esterase lipase, cysteine arylamidase, b-galactosidase, and N-acetyl-b-glucosaminidase activities are present. Lipase (C14), trypsin, α galactosidase ß glucuronidase, α-mannosidase and a-fucosidase activities are absent in API ZYM. In BIOLOG GEN III plate, the strain is negative for dextrin, D-maltose, D-trehalose, D-cellobiose, sucrose, turanose, stachyose, D-raffinose, α-D-lactose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine N-acetyl-D-galactosamine, N-acetyl neuraminic acid, D-mannose, inosine, D-sorbitol, D-arabitol, D-mannitol, myo-inositol, glycerol D-glucose-6-PO4, D-aspartic acid, D-serine, gelatin, Glycyl-L-proline, L-alanine, L-arginine, L-aspartic acid, L-glutamic acid, L-histidine, L-pyroglutamic acid L-serine, pectin, L-galactonic acid lactone, D-gluconic acid, mucic acid, qunic acid, D-saccharic acid, p-hydroxy-phenylacetic acid, methyl pyruvate, D-lactic acid methyl ester, L-lactic acid, citric acid, α-keto-glutaric acid, D-malic acid, L-malic acid, tween 40 bromo-succinic acid, Υ-amino-butryric acid, α-hydroxy-butyric acid, β-hydroxy-D, L-butyric acid, α-keto-butyric acid, acetoacetic acid, acetic acid, and formic acid. The major cellular fatty acids are iso-C15:0, iso-C16:0, anteiso-C15:0, and iso-C17:0. Phosphatidylethanolamine, phosphatidylglycerol, and diphosphatidylglycerol as major polar lipids.
The type strain, MEB199T (= MCC 2982T = CGMCC 1.17254T = JCM 33704), was isolated from a sediment sample collected from alkaline Lonar Lake, India. The DNA G + C content of the type strain is 36.47 mol%. The GenBank accession numbers for the 16S rRNA gene and draft genome sequence of strain MEB199T are KX171019 and WMKZ00000000, respectively.
Halalkalibacterium (Hal.al.ka.li.bac.te’ri.um. Gr. masc. n. hals, salt; Arabic n. al-qalyi, soda ash; N.L. neut. n. bacterium, a small rod; N.L. neut. n. Halalkalibacterium, bacterium living under alkaline saline conditions).
Rod shaped, Gram-stain-variable. Members are aerobic or facultative anaerobes; endospore forming; have been isolated from sediments of the South China Sea/hot spa area, Motile, Tolerate NaCl concentration up to 12% with optimum growth at 2% (w/v). Growth occurs in the range 15–60°C, with optimum growth temperature in the range 30–40°C. All members of this genus are alkaliphilic with growth in the range of pH 6–11 with optimum growth at pH (9–10). The predominant polar lipids are diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine. The major isoprenoid quinone is MK-7. The major fatty acids are iso-C14:0 and anteiso-C15:0. meso-DAP is cell wall diamino acid. The DNA G + C content is 40.76–43.9 mol%.
The type species is Halalkalibacterium halodurans.
Halalkalibacter (Hal.al.ka.li.bac’ter. Gr. masc. n. hals, salt; Arabic n. al-qalyi, soda ash; N.L. masc. n. bacter, rod; N.L. masc. n. Halalkalibacter briny and alkaline media-loving rod-shaped cells).
Cells are rod shaped, Gram-stain-variable, endospore forming, motile or non-motile, Aerobic or facultative anaerobic in nature, have been isolated from mushroom compost, sediment sample from the sea, seawater, soda pond, saltpan/soil, tolerate NaCl concentration up to 12% with optimum growth at 3–7% (w/v). Growth occurs in the range 10–50°C, with optimum growth at 30–37°C. Alkaliphilic with growth in the range of pH 6.5–12.5 with optimum growth at pH (9–10). The major isoprenoid quinone are MK-5/MK-6/MK-7. The major fatty acids are anteiso-C15:0 and iso-C15:0, C16:0 and anteiso-C17:0, C14:0, C16:1ω7c-alcohol, C16:0. Polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine with a minor amount of aminophospholipid, and one glycolipid. meso-DAP is cell wall diamino acid. The DNA G + C content is 34.4–46.3 mol%.
The type species is Halalkalibacter krulwichiae.
Shouchella (Shou.chel’la. N.L. fem. dim. n. Shouchella, named after Dr. Yogesh Shouche, an eminent Indian microbiologist and taxonomist who has made a significant contribution in the field of microbial systematics and genomics of extremophilic bacteria from various extreme environments).
Cells are rod shaped, Gram-stain positive, endospore-forming, motile, or non-motile. All members are aerobic with few facultative anaerobes, have been isolated from rhizosphere soil of sugarcane/perennial shrub Atriplex lampa or Miscanthus sacchariflorus or soil or sediment sample from saline-alkaline habitat and non-saline forest soil. All the members are moderately halophilic and can tolerate NaCl concentrations up to 22% with optimum growth at 5–10% (w/v). Growth occurs in the range of 4–50°C, with optimum growth at 25–35°C. All members of this genus are alkalitolerant with growth in the range of pH 6.5–11 with optimum growth at pH 8–9. The major isoprenoid quinone is MK-7. The major fatty acids are iso-C15:0, anteiso-C15:0, iso-C17:0, and anteiso-C17:0, iso-C14:0, C16:0, and C12:0. The polar lipid profile contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and glycolipid. meso-DAP is cell wall diamino acid. The DNA G + C content is 39.7–54 mol%.
The type species is Shouchella clausii.
Pseudalkalibacillus (Pseud.al.ka.li.ba.cil’lus. Gr. masc. adj. pseudês, false; N.L. masc. n. Alkalibacillus a bacterial genus; N.L. masc. adj. Pseudalkalibacillus a false Alkalibacillus because it only tolerates alkaline pH but does not grow optimally at higher pH).
Cells are rod shaped, Gram-stain positive, or Gram variable, endospore forming, motile, or non-motile, aerobic, facultative anaerobes, or anaerobic, have been isolated from the mud a goldmine/mangrove sediment, sea urchin, seawater, and mural paintings. All the members can tolerate NaCl concentrations up to 11% with optimum growth at 3–6% (w/v). Growth occurs in the range 4–50°C with optimum growth at 25–40°C. Most of the members of this genus can tolerate pH in the range of 5–10 with optimum growth at pH 7–8 and, thus, are alkalitolerant in nature. The major isoprenoid quinone is MK-7. The major fatty acids are anteiso-C15:0, anteiso-C17:0, and C16:1ω7c alcohol. Diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine are the major polar lipids. meso-DAP is cell wall diamino acid. The DNA G + C content is 37–40.9 mol%.
The type species is Pseudalkalibacillus decolorationis.
Alkalicoccobacillus (Al.ka.li.coc.co.ba.cil’lus. N.L. n. alkali, alkali; from Arabic article al, the; from Arabic n. qaly, ashes of saltwort; Gr. masc. n. kokkos a berry; L. masc. n. bacillus a small rod; N.L. masc. n. Alkalicoccobacillus, a cocobacillary rod living in basic surroundings).
Cells are coccoid rod shaped, Gram-stain variable, endospore forming, and motile, isolated from mural paintings, discolored by microbial growths. It can tolerate NaCl concentrations up to 10% with optimum growth at 4–7% (w/v). Growth occurs in the range 5–40°C, with optimum growth at 25–37°C. The member is alkalitolerant, which grows in the pH range of 7–11 with optimum growth at pH 8. The major fatty acids are anteiso-C15:0 and anteiso-C17:0. The DNA G + C content is 39⋅8 mol%.
The type species is Alkalicoccobacillus murimartini.
Alkalihalophilus (Al.ka.li.ha.lo’phi.lus. N.L. n. alkali, alkali (from Arabic article al the; Arabic n. qaliy ashes of saltwort); Gr. masc. n. hals (gen. halos), salt; Gr. masc. adj. philos loving; N.L. masc. n. Alkalihalophilus, bacterium liking alkaline and saline environment).
Cells are rod shaped, Gram-stain positive, aerobic, endospore-forming, and motile, have been isolated from saline and alkaline soils, mushroom compost, and animal manure. It can tolerate NaCl concentrations up to 12% with optimum growth at 4% (w/v). Growth occurs in the range 10–45°C, with optimum growth at 37°C. All the members are obligate alkaliphilic in nature and can grow in the pH range of 8–12, and no growth is found at pH 7 with optimum growth at pH 9. The major fatty acids are anteiso-C15:0, iso-C15:0 and anteiso-C17:0. Diphosphatidylglycerol, phosphatidylethanolamine, and phosphatidylglycerol are the major polar lipids. meso-DAP is cell wall diamino acid. The DNA G + C content is 39.0–42.7 mol%.
The type species is Alkalihalophilus pseudofirmus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, WMKZ00000000.
AJ and TL designed the work. ST and AJ carry out the morphological, biochemical, physiological and molecular characterisation of novel strain and maintained the bacterial cultures. TL carried out the genomic data retrieval from databases, phylogenomic, phylogenetic data analysis, and calculated the genomic indices. NJ carried out the FAME analysis and PK did the polar lipids profiling. All authors contributed to writing the manuscript and accepted it for publication.
This work was funded by the Department of Biotechnology, DBT, Government of India, under grant no. DBT, grant BT/Coord.II/01/03/2016. TL acknowledges the funding as a Start-up Research grant by the Department of Science and Technology, Government of India, under grant number SRG/2019/001818.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We greatly acknowledge the support of Aharon Oren and Bernhard Schink for their expert advice concerning the genus and species epithet and Latin etymology. We thank Hitharth Bhatt for sharing Desertibacillus haloalkaliphilus culture as a gratis. We also thank Nikeeta Chavan for the assembly of the MEB199T genome.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.722369/full#supplementary-material
Aizawa, T., Urai, M., Iwabuchi, N., Nakajima, M., and Sunairi, M. (2010). Bacillus trypoxylicola sp. nov., xylanase-producing alkaliphilic bacteria isolated from the guts of Japanese horned beetle larvae (Trypoxylus dichotomus septentrionalis). Int. J. Syst. Evol. Microbiol. 60, 61–66. doi: 10.1099/ijs.0.005843-0
Aliyu, H., Lebre, P., Blom, J., Cowan, D., and Maayer, P. D. (2016). Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 39, 527–533. doi: 10.1016/j.syapm.2016.09.004
Arahal, D. R., and Ventosa, A. (2002). “Moderately halophilic and halotolerant species of Bacillus genera,” in Applications and Systematics of Bacillus and Relatives, eds R. C. W. Berkeley, M. Heyndrickx, N. Logan, and P. de Vos (Oxford: Blackwell Publishing), 83–99. doi: 10.1002/9780470696743.ch7
Azmatunnisa, B. M., Varshini, V., Rahul, K., Chandana, A., Sasikala, C., and Ramana, C. V. (2016). Description of Alteribacillus alkaliphilus sp. nov., reassignment of Bacillus iranensis (Bagheri et al. 2012) as Alteribacillus iranensis comb. nov. and emended description of the genus Alteribacillus. Int. J. Syst. Evol. Microbiol. 66, 4772–4778. doi: 10.1099/ijsem.0.001428
Bhatt, H. B., Azmatunnisa, B. M., Chintalapati, S., Chintalapati, V. R., and Singh, S. P. (2017). Desertibacillus haloalkaliphilus gen. nov., sp. nov., isolated from a saline desert. Int. J. Syst. Evol. Microbiol. 67, 4435–4442. doi: 10.1099/ijsem.0.002310
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Weezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkab335
Borchert, M. S., Nielsen, P., Graeber, I., Kaesler, I., Szewzyk, U., Pape, T., et al. (2007). Bacillus plakortidis sp. nov. and Bacillus murimartini sp. nov., novel alkalitolerant members of rRNA group 6. Int. J. Syst. Evol. Microbiol. 57, 2888–2893. doi: 10.1099/ijs.0.65177-0
Borsodi, A. K., Toth, E., Aszalos, J. M., Barany, A., Schumann, P., Sproer, C., et al. (2017). Bacillus kiskunsagensis sp. nov., a novel alkaliphilic and moderately halophilic bacterium isolated from soda soil. Int. J. Syst. Evol. Microbiol. 67, 3490–3495. doi: 10.1099/ijsem.0.002149
Borsodi, A. K., Pollak, B., Keki, Z., Rusznyak, A., Kovacs, A. L., Spröer, C., et al. (2011). Bacillus alkalisediminis sp. nov., an alkaliphilic and moderately halophilic bacterium isolated from sediment of extremely shallow soda ponds. Int. J. Syst. Evol. Microbiol. 61, 1880–1886. doi: 10.1099/ijs.0.019489-0
Boyer, E. W., Ingle, M., Mercer, G. D. (1973). Bacillus alcalophilus subsp. halodurans subsp. nov.: an alkaline amylase producing alkalophilic organism. Int. J. Syst. Bacteriol. 23, 238–242. doi: 10.1099/00207713-23-3-238
Brosius, J., Palmer, M. L., Kennedy, P. J., and Noller, H. F. (1978). Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 75, 4801–4805. doi: 10.1073/pnas.75.10.4801
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. bioRxiv [Preprint]. doi: 10.1101/2021.06.03.446934
Card, G. L. (1973). Metabolism of phosphatidylglycerol, phosphatidylethanolamine, and cardiolipin of Bacillus stearothermophilus. J. Bacteriol. 114, 1125–1137. doi: 10.1128/jb.114.3.1125-1137.1973
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, Y. G., Zhang, Y. Q., He, J. W., Klenk, H. P., Xiao, J. Q., Zhu, H. Y., et al. (2011a). Bacillus hemicentroti sp. nov., a moderate halophile isolated from a sea urchin. Int. J. Syst. Evol. Microbiol. 61, 2950–2955. doi: 10.1099/ijs.0.026732-0
Chen, Y. G., Zhang, Y. Q., Chen, Q. H., Klenk, H., He, J. W., Tang, S. K., et al. (2011b). Bacillus xiaoxiensis sp. nov., a slightly halophilic bacterium isolated from non-saline forest soil. Int. J. Syst. Evol. Microbiol. 61, 2095–2100. doi: 10.1099/ijs.0.026286-0
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Denizci, A. A., Kazan, D., and Erarslan, A. (2010). Bacillus marmarensis sp. nov.,an alkaliphilic, protease-producing bacterium isolated from mushroom compost. Int. J. Syst. Evol. Microbiol. 60, 1590–1594. doi: 10.1099/ijs.0.012369-0
Dimitriu, P. A., Shukla, S. K., Conradt, J. M., Márquez, M. C., Ventosa, A., Maglia, A., et al. (2005). Nitrincola lacisaponensis gen. nov., sp. nov., a novel alkaliphilic bacterium isolated from an alkaline, saline lake. Int. J. Syst. Evol. Microbiol. 55, 2273–2278. doi: 10.1099/ijs.0.63647-0
Dou, G., Liu, H., He, W., and Ma, Y. (2016). Bacillus lindianensis sp. nov., a novel alkaliphilic and moderately halotolerant bacterium isolated from saline and alkaline soils. Antonie van Leeuwenhoek 109, 149–158. doi: 10.1007/s10482-015-0616-y
Gessesse, A., and Gashe, B. A. (1997). Production of alkaline protease by an alkaliphilic bacteria isolated from an alkaline soda lake. Biotechnol. Lett. 19:479.
Ghosh, A., Bhardwaj, M., Satyanarayana, T., Khurana, M., Mayilraj, S., and Jain, R. K. (2007). Bacillus lehensis sp. nov., an alkalitolerant bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 57, 238–242. doi: 10.1099/ijs.0.64617-0
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gupta, R. S. (2014). “Identification of conserved indels that are useful for classification and evolutionary studies,” in Methods in Microbiology, eds M. Goodfellow, I. Sutcliffe, and J. Chun (Oxford: Academic Press), 153–182. doi: 10.1016/bs.mim.2014.05.003
Gupta, R. S., Patel, S., Saini, N., and Chen, S. (2020). Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the subtilis and cereus clades of species. Int. J. Syst. Evol. Microbiol. 70, 5753–5798. doi: 10.1099/ijsem.0.004475
Grant, W. D. (2003). “Alkaline environments and biodiversity,” in Extremophiles (Life Under Extreme External Conditions), eds C. Gerday and N. Glansdorff (Oxford: Eolss Publishers).
Heyrman, J., Balcaen, A., Rodriguez-Diaz, M., Logan, N. A., Swings, J., and De Vos, P. (2003). Bacillus decolorationis sp. nov., isolated from biodeteriorated parts of the mural paintings at the servilia tomb (Roman necropolis of Carmona, Spain) and the saint-catherine chapel (Castle Herberstein, Austria). Int. J. Syst. Evol. Microbiol. 53, 459–463. doi: 10.1099/ijs.0.02452-0
Horikoshi, K. (1999). Alkaliphiles: some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 63, 735–750. doi: 10.1128/MMBR.63.4.735-750.1999
Horikoshi, K. (2006). Alkaliphiles – from an industrial point of view. FEMS Microbiol. Rev. 18, 259–270. doi: 10.1016/0168-6445(96)00017-4
Ivanova, E. P., Alexeeva, Y. A., Zhukova, N. V., Gorshkova, N. M., Buljan, V., Nicolau, D. V., et al. (2004). Bacillus algicola sp. nov., a novel filamentous organism isolated from brown alga Fucus evanescens. Syst. Appl. Microbiol. 27, 301–307. doi: 10.1078/0723-2020-00269
Jones, B. E., Grant, W. D., Duckworth, A. W., and Owenson, G. G. (1998). Microbial diversity of soda lakes. Extremophiles 2, 191–200. doi: 10.1007/s007920050060
Kontnik, R., Bosak, B., Butcher, R. A., Brocks, J. J., Losick, R., Clardy, J., et al. (2008). Sporulenes, heptaprenyl metabolites from Bacillus subtilis spores. Org. Lett. 10, 3551–3554. doi: 10.1021/ol801314k
Krulwich, T. A., Hick, D. B., Swartz, T., and Ito, M. (2007). “Bioenergetics adaptation that support alkaliphyliy,” in Physiology and Biochemistry of Extremophiles, eds C. Gerday and N. Glansodorff (Washington, DC: ASM Press), 311–329. doi: 10.1128/9781555815813.ch24
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
La Duc, M. T., Satomi, M., Agata, N., and Venkateswaran, K. (2004). gyrB as a phylogenetic discriminator for members of the Bacillus anthracis-cereus-thuringiensis group. J. Microbiol. Methods. 56, 383–394. doi: 10.1016/j.mimet.2003.11.004
Lei, Z., Qiu, P., Ye, R., Tian, J., Liu, Y., Wang, L., et al. (2014). Bacillus Shacheensis sp. nov.,a moderately halophilic bacterium isolated from a saline-alkalisoil. J. Gen. Appl. Microbiol. 60, 101–105. doi: 10.2323/jgam.60.101
Li, Z., Kawamura, Y., Shida, O., Yamagata, S., Deguchi, T., and Ezaki, T. (2002). Bacillus okuhidensis sp. nov., isolated from the Okuhida spa area of Japan. Int. J. Syst. Evol. Microbiol. 52, 1205–1209. doi: 10.1099/00207713-52-4-1205
Liu, B., Liu, G. H., Wang, X. Y., Wang, J. P., Chen, Z., Chen, M. C., et al. (2019). Bacillus urbisdiaboli sp. nov., isolated from soil sampled in Xinjiang. Int. J. Syst. Evol. Microbiol. 69, 1591–1596. doi: 10.1099/ijsem.0.003363
Madhaiyan, M., Poonguzhali, S., Lee, J. S., Lee, K. C., and Hari, K. (2011). Bacillus rhizosphaerae sp. nov., an novel diazotrophic bacterium isolated from sugarcane rhizosphere soil. Antonie van Leeuwenhoek 100, 437–444. doi: 10.1007/s10482-011-9600-3
Margesin, R., and Schinner, F. (2001). Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5, 73–83. doi: 10.1007/s007920100184
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Mo, K., Huang, H., Bao, S., and Hu, Y. (2020). Bacillus caeni sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 70, 1503–1507. doi: 10.1099/ijsem.0.003853
Na, S. I., Kim, Y. O., Yoon, S., Ha, S., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Nedashkovskaya, O. I., Van Trappen, S., Frolova, G. M., and De Vos, P. (2012). Bacillus berkeleyi sp. nov., isolated from the sea urchin Strongylocentrotus intermedius. Arch. Microbiol. 194, 215–221. doi: 10.1007/s00203-011-0771-0
Nielsen, P., Fritze, D., and Priest, F. G. (1995). Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 141, 1745–1761. doi: 10.1099/13500872-141-7-1745
Nogi, Y., Takami, H., and Horikoshi, K. (2005). Characterization of alkaliphilic Bacillus strains used in industry: proposal of five novel species. Int. J. Syst. Evol. Microbiol. 55, 2309–2315. doi: 10.1099/ijs.0.63649-0
Nowlan, B., Dodia, M. S., Singh, S. P., and Patel, B. K. C. (2006). Bacillus okhensis sp.nov., a halotolerant and alkalitolerant bacterium from an Indian saltpan. Int. J. Syst. Evol. Microbiol. 56, 1073–1077. doi: 10.1099/ijs.0.63861-0
Olivera, N., Siñeriz, F., and Breccia, J. D. (2005). Bacillus patagoniensis sp. nov.,a novel alkalitolerant bacterium from the rhizosphere of Atriplex lampa in Patagonia, Argentina. Int. J. Syst. Evol. Microbiol. 55, 443–447. doi: 10.1099/ijs.0.63348-0
Patel, S., and Gupta, R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Pérez-Cataluña, A., Collado, L., Salgado, O., Lefiñanco, V., and Figueras, M. J. (2018). A polyphasic and taxogenomic evaluation uncovers Arcobacter cryaerophilus as a species complex that embraces four genomovars. Front. Microbiol. 9:805. doi: 10.3389/fmicb.2018.00805
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Reddy, S. V., Thirumala, M., Farooq, M., Sasikala, C., and Ramana, C. V. (2015). Bacillus lonarensis sp. nov., an alkalitolerant bacterium isolated from a soda lake. Arch. Microbiol. 197, 27–34. doi: 10.1007/s00203-014-1040-9
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Santini, J. M., Streimann, I. C. A., and vanden Hoven, R. N. (2004). Bacillus macyae sp.nov., an arsenate-respiring bacterium isolated from an Australian gold mine. Int. J. Syst. Evol. Microbiol. 54, 2241–2244. doi: 10.1099/ijs.0.63059-0
Sato, T., Yoshida, S., Hoshino, H., Tanno, M., Nakajima, M., and Hoshino, T. (2011). Sesquarterpenes (C35 terpenes) biosynthesized via the cyclization of a linear C35 isoprenoid by a tetraprenyl-β-curcumene synthase and a tetraprenyl-β-curcumene cyclase: identification of a new terpene cyclase. J. Am. Chem. Soc. 133, 9734–9737. doi: 10.1021/ja203779h
Shin, B., Park, C., Lee, B. H., Lee, K. E., and Park, W. (2020). Bacillus miscanthi sp. nov., a alkaliphilic bacterium from the rhizosphere of Miscanthus sacchariflorus. Int. J. Syst. Evol. Microbiol. 70, 1843–1849. doi: 10.1099/ijsem.0.003982
Singh, H., Kaur, M., Sharma, S., Kaur, L., Mishra, S., Tanuku, N. R. S., et al. (2018). Bacillus alkalilacus sp. nov., isolated from a sediment sample from a lake in India. Int. J. Syst. Evol. Microbiol. 68, 1665–1671. doi: 10.1099/ijsem.0.002726
Smibert, R. M., and Krieg, N. R. (1994). “Phenotypic characterization,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (Washington, DC: American Society for Microbiology), 611–651.
Song, L., Liu, H., Wang, J., Huang, Y., Dai, X., Han, X., et al. (2016). Bacillus oceani sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 66, 796–800. doi: 10.1099/ijsem.0.000793
Sorokin, D. Y., van Pelt, S., and Tourova, T. P. (2008). Utilization of aliphatic nitriles under haloalkaline conditions by Bacillus alkalinitrilicus sp. nov. isolated from soda solonchak soil. FEMS Microbiol. Lett. 288, 235–240. doi: 10.1111/j.1574-6968.2008.01353.x
Suresh, G., Lodha, T. D., Indu, B., Sasikala, Ch, and Ramana, ChV (2019). Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front. Microbiol. 10:2480. doi: 10.3389/fmicb.2020.01111
Vargas, V. A., Delgado, O. D., Hatti-Kaul, R., and Mattiasson, B. (2005). Bacillus bogoriensis sp. nov., a novel alkaliphilic, halotolerant bacterium isolated from a Kenyan soda lake. Int. J. Syst. Evol. Microbiol. 55, 899–902. doi: 10.1099/ijs.0.63318-0
Vedder, A. (1934). Bacillus alcalophilus n. sp.; benevens enkele ervaringen met sterk alcalische voedingsbodems. Antonie van Leeuwenhoek J. Microbiol. Serol. 1, 141–147. doi: 10.1007/BF02543931
Yoon, J. H., Kim, I. G., Kang, K. H., Oh, T. K., and Park, Y. H. (2004). Bacillus hwajinpoensis sp. nov. and an unnamed Bacillus genomospecies, novel members of Bacillus rRNA group 6 isolated from sea water of the East Sea and the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 54, 803–808. doi: 10.1099/ijs.0.02678-0
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., and Seo, H. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yumoto, I., Hirota, K., Goto, T., Nodasaka, Y., and Nakajima, K. (2005). Bacillus oshimensis sp. nov., a moderately halophilic, non-motile alkaliphile. Int. J. Syst. Evol. Microbiol. 55, 907–911. doi: 10.1099/ijs.0.63488-0
Yumoto, I., Yamaga, S., Sogabe, Y., Nodasaka, Y., Matsuyama, H., and Nakajima, K. (2003). Bacillus krulwichiae sp. nov., a halotolerant obligate alkaliphile that utilizes benzoate and m-hydroxybenzoate. Int. J. Syst. Evol. Microbiol. 53, 1531–1536. doi: 10.1099/ijs.0.02596-0
Zavarzina, D. G., Tourova, T. P., Kolganova, T. V., Boulygina, E. S., and Zhilina, T. N. (2009). Description of Anaerobacillus alkalilacustre gen. nov., sp. nov. strictly anaerobic diazotrophic bacillus isolated from Soda Lake and transfer of Bacillus arseniciselenatis, Bacillus macyae, and Bacillus alkalidiazotrophicus to Anaerobacillus as the new combinations A. arseniciselenatis comb. nov., A. macyae comb. nov., and A. alkalidiazotrophicus comb. nov. Microbiology 78, 723–731. doi: 10.1134/S0026261709060095
Zhang, J., Wang, J., Song, F., Fang, C., Xin, Y., and Zhang, Y. (2011). Bacillus nanhaiisediminis sp. nov., an alkalitolerant member of Bacillus rRNA group 6. Int. J. Syst. Evol. Microbiol. 61, 1078–1083. doi: 10.1099/ijs.0.023671-0
Zhu, D., Tanabe, S. H., Xie, C., Honda, D., Sun, J., and Ai, L. (2014). Bacillus ligniniphilus sp. nov., an alkaliphilic and halotolerant bacterium isolated from sediments of the South China Sea. Int. J. Syst. Evol. Microbiol. 64, 1712–1717. doi: 10.1099/ijs.0.058610-0
Keywords: alkaliphilic bacteria, Alkalihalobacterium, Alkalihalobacillus, taxogenomic, Lonar lake, Shouchella, Halalkalibacterium, Halalkalibacter
Citation: Joshi A, Thite S, Karodi P, Joseph N and Lodha T (2021) Alkalihalobacterium elongatum gen. nov. sp. nov.: An Antibiotic-Producing Bacterium Isolated From Lonar Lake and Reclassification of the Genus Alkalihalobacillus Into Seven Novel Genera. Front. Microbiol. 12:722369. doi: 10.3389/fmicb.2021.722369
Received: 08 June 2021; Accepted: 06 September 2021;
Published: 11 October 2021.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Christopher Dunlap, Agricultural Research Service, United States Department of Agriculture, United StatesCopyright © 2021 Joshi, Thite, Karodi, Joseph and Lodha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tushar Lodha, dHVzaGFyLmxvZGhhQG5jY3MucmVzLmlu; Amaraja Joshi, YW1hcmFqYUBuY2NzLnJlcy5pbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.