- 1Microbiology Laboratory, National Center for Children’s Health, Beijing Pediatric Research Institute, Beijing Children’s Hospital, Capital Medical University, Beijing, China
- 2Shenzhen Children’s Hospital, Shenzhen, China
- 3Beijing Chaoyang Hospital Affiliated to the Capital Medical University, Beijing, China
Streptococcus pyogenes is a bacterial pathogen that causes a wide spectrum of clinical diseases exclusively in humans. The distribution of emm type, antibiotic resistance and virulence gene expression for S. pyogenes varies temporally and geographically, resulting in distinct disease spectra. In this study, we analyzed antibiotic resistance and resistance gene expression patterns among S. pyogenes isolates from pediatric patients in China and investigated the relationship between virulence gene expression, emm type, and disease categories. Forty-two representative emm1.0 and emm12.0 strains (n = 20 and n = 22, respectively) isolated from patients with scarlet fever or obstructive sleep apnea-hypopnea syndrome were subjected to whole-genome sequencing and phylogenetic analysis. These strains were further analyzed for susceptibility to vancomycin. We found a high rate and degree of resistance to macrolides and tetracycline in these strains, which mainly expressed ermB and tetM. The disease category correlated with emm type but not superantigens. The distribution of vanuG and virulence genes were associated with emm type. Previously reported important prophages, such as φHKU16.vir, φHKU488.vir, Φ5005.1, Φ5005.2, and Φ5005.3 encoding streptococcal toxin, and integrative conjugative elements (ICEs) such as ICE-emm12 and ICE-HKU397 encoding macrolide and tetracycline resistance were found present amongst emm1 or emm12 clones from Shenzhen, China.
Introduction
Streptococcus pyogenes (group A Streptococcus, GAS) is an important gram-positive bacteria that ranks among the 10 main causes of death from infectious diseases worldwide, with more than 517,000 deaths annually (Carapetis et al., 2005). GAS causes a wide spectrum of clinical diseases ranging from mild pharyngitis to life-threatening invasive infections (Carapetis et al., 2005; Walker et al., 2014). Although antibiotics are effective and widely used for treating GAS infections, antibiotic resistance, especially to macrolides, is increasing in several countries (Lu et al., 2017; Bhardwaj et al., 2018). The rise of antibiotic resistance leads to an increase in mortality, which has become a public health issue of global concern (Wajima et al., 2014; Silva-Costa et al., 2015). This issue has also received close attention in China (You et al., 2018, 2020). Genotyping is an effective method for monitoring bacterial strains in microbiology research (Steer et al., 2009), and sequence analysis of the emm gene is currently used for GAS genotyping (Facklam et al., 1999; Gherardi et al., 2018). In recent years, emm cluster analysis has also been widely used in GAS molecular epidemiology analysis. The type-specific M protein, encoded by the emm gene, and superantigens (SAgs), encoded by the sAg genes, are important virulence factors in S. pyogenes (Golińska et al., 2016). Currently, stains of S. pyogenes are often tested for the expression of speA, speC, speG, speH, speI, speJ, speK, speL, speM, ssa, smez, and the enzyme-encoding speB and speF genes as genes encoding SAgs, even though speB and speF have been confirmed to encode cysteine protease and streptococcal DNase proteins (Strus et al., 2017). However, the correlations between sAg distribution, emm type, and disease spectrum for GAS have not yet been established (Rantala et al., 2012; Imöhl et al., 2017; González-Abad and Alonso Sanz, 2020). In this study, the antimicrobial sensitivity of GAS strains as well as the relationships among sAg distribution, emm types, and disease categories were analyzed. Moreover, 42 representative strains of the two main epidemic emm types were analyzed for population structure, genetic diversity, phylogeny, and susceptibility to vancomycin.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
A total of 342 GAS strains were isolated from children aged < 18 years who were admitted to Shenzhen Children’s Hospital from 2016 to 2018 for treatment of one of 15 diseases. Of these strains, 87 were isolated in 2016 (4 invasive and 83 non-invasive strains), 138 were isolated in 2017 (13 invasive and 125 non-invasive strains), and 117 were isolated in 2018 (15 invasive and 102 non-invasive strains). The strains were isolated from 262 throat swabs, 47 sputum samples, 21 pus samples (abscess, surgical wound infections, and skin burn infections), 5 wound secretions, 3 vulvar secretions, 3 blood samples, and 1 urine sample. The streptococcus grouping kit (Oxoid Limited) was used to identify these strains, as previously reported (Liang et al., 2012).
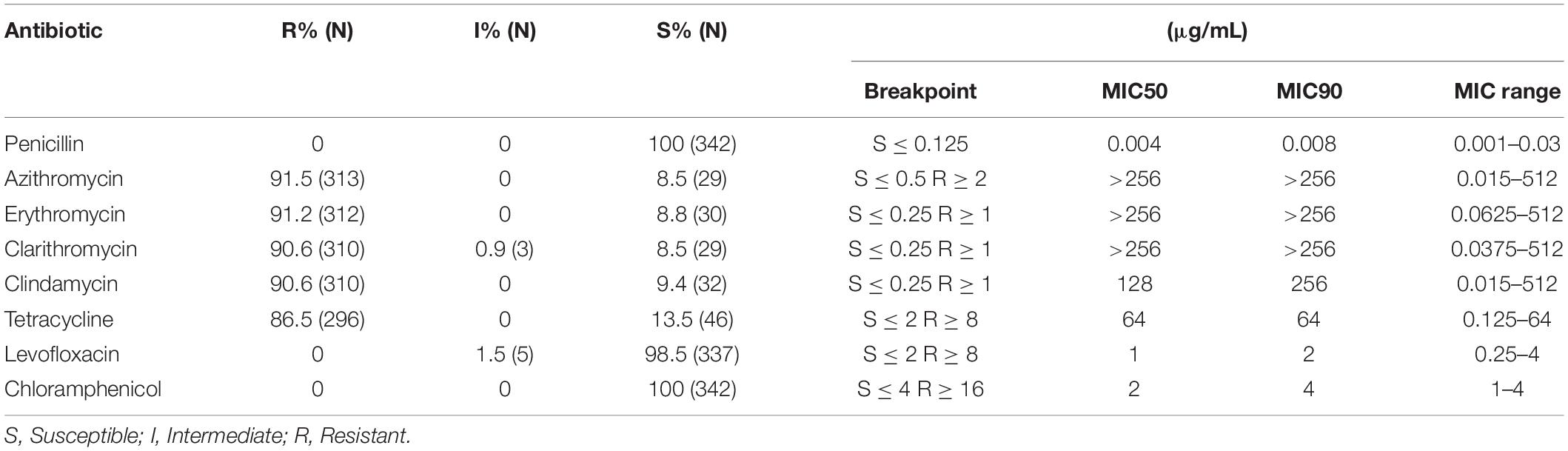
We tested the susceptibility of the 342 strains to antimicrobial agents including penicillin, azithromycin, erythromycin, clarithromycin, clindamycin, tetracycline, levofloxacin, and chloramphenicol (Oxoid Limited). Susceptibility to vancomycin was analyzed in 42 representative strains. Minimum inhibitory concentration (MIC) values were determined according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (2019) by using the broth dilution method. Quality control was performed using Streptococcus pneumoniae ATCC 49619, which was provided by the Clinical Test Center of the Ministry of Health and maintained by the Microbiology Laboratory of Shenzhen Children’s Hospital.
DNA Extraction and Detection of Superantigens and Macrolide and Tetracycline Resistance Genes
Genomic DNA was obtained from freshly grown GAS using a Chelex-based DNA extracting kit for genetic analysis. Thirteen SAg-coding genes (speA, speB, speC, SpeF, speG, speH, speI, speJ, speK, speL, speM, ssa, and smeZ), three genes encoding resistance to macrolides (ermB, ermA, and mefA), and tetracycline (tetM) were detected by polymerase chain reaction (PCR) using a previously reported protocol (Chatellier et al., 2000; Igwe et al., 2003; Rivera et al., 2006; Lintges et al., 2007; Pérez-Trallero et al., 2007). The DNA extraction kit, PCR reagents, and primers were all obtained from Shanghai SBS Genetech Co., Ltd. (China).
Emm Sequence Typing
The emm sequence types were determined using a previously reported protocol1. Amplicons were sequenced by Guangzhou BGI Genomics Co. Ltd. Emm type was determined based on the sequence identity (> 95%) of the first 180 bp of the emm gene between the tested sequence and the reference emm gene.
Whole-Genome Sequencing and Phylogenetic Analysis
We selected 42 strains that represented the major emm types for WGS according to the following criteria: (i) the major emm types (emm1.0, emm12.0), (ii) the two most common diseases [scarlet fever and obstructive sleep apnea syndrome (OSAS)], and (iii) 3 strains each year from 2016 to 2018. For each subtype of emm1 and emm12, we choose one strain from each year, and then chose strains from different seasons in each year. Forty-two strains isolated from children with OSAS or scarlet fever (20 emm1.0 strains and 22 emm12.0 strains) were used for the WGS analysis. Genomic DNA was sequenced using the DNBSEQ platform (BGI-Shenzhen, China). DNA quality was assessed by electrophoresis, and then the DNA was fragmented and processed by end repair, A-tailing, adapter ligation, DNA size selection, circulation, and DNA nanoball formation according to an in-house library. DNA libraries with an insert size of 300 bp were sequenced using paired-end 100-bp reads (PE100). Low-quality sequences were trimmed using SOAPnuke (Chen et al., 2018). The remaining short reads were assembled into contigs using SPAdes version 3.11.1 (Lagesen et al., 2007).
Genes were predicted using Glimmer 3.02 (Delcher et al., 2007). Virulence genes were identified by searching against the Virulence Factor Database (Liu et al., 2019), Antibiotic Resistance Genes Database (Liu and Pop, 2009), and Comprehensive Antibiotic Resistance Database (Alcock et al., 2020). The core genes of the sequenced and reference genomes were identified by clustering proteins with a sequence identity > 50% and a coverage > 70% using CD-HIT (Fu et al., 2012). Single nucleotide polymorphisms (SNPs) among the core genes were aligned pairwise and subjected to phylogenetic tree inference using TreeBeST and the NJ method (TreeSoft, 2021).
Statement of Ethics
This study was approved by the research ethics committee of the Shenzhen Children’s Hospital. Informed consent was obtained from patients or their guardians before sample collection.
Statistical Analysis
Data were analyzed using SPSS version 22.0. Differences in the distributions of diseases and emm types and comparisons between diseases and a specific emm type were analyzed using the independent-samples Kruskal-Wallis test. The exact Mann-Whitney U-test was applied to identify differences in distributions between streptococcal sAg expression, emm types, and disease categories. Two-sided P-values < 0.05 indicated a statistically significant difference between groups. Values of Padj < 0.05/m indicated that a within group difference was statistically significant, where m represents the total number of Bonferroni corrections within the group.
Results
Antimicrobial Susceptibility Patterns and Resistance Genes
All strains were highly susceptible to penicillin and chloramphenicol, whereas 98.5% of the tested strains were sensitive to levofloxacin. The results regarding the rates and degree of resistance to azithromycin, erythromycin, clarithromycin, clindamycin, and tetracycline are shown in Table 1. In addition, 90.9 and 6.1% of strains expressed ermB and ermA, respectively. Additionally, 16.4% were mefA-positive, and 85.4% were tetM-positive. All 42 representative strains were susceptible to vancomycin.
Distribution of emm Genotypes and Their Correlation With Disease Categories
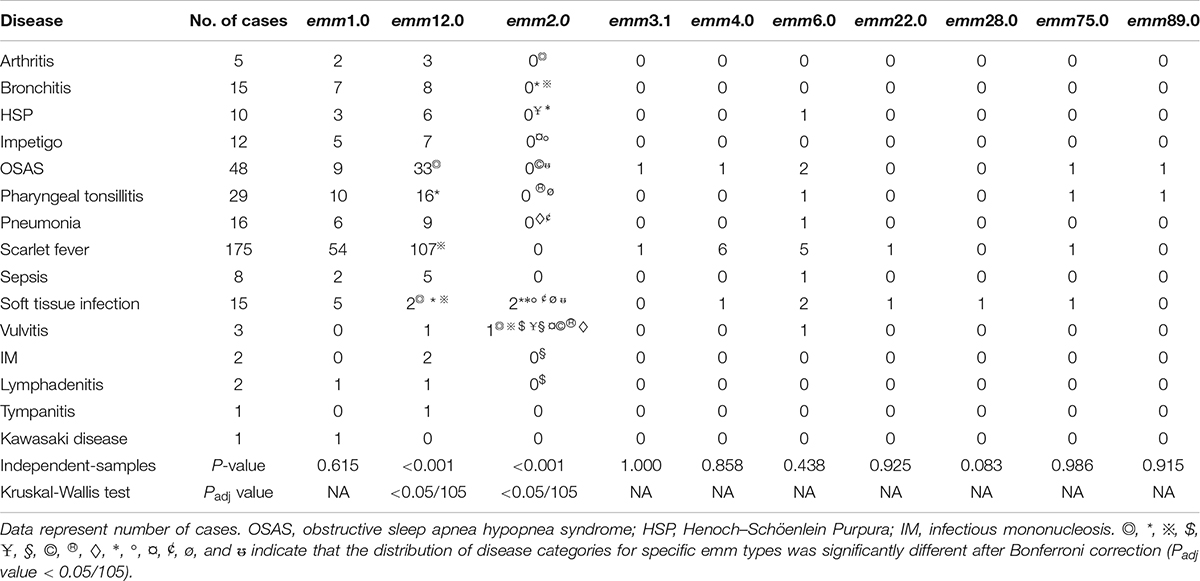
In this survey, 10 emm types, including 7 subtypes, were identified among the 342 GAS strains from 15 different diseases. Figures 1, 2 show the details of the 10 emm types, as well as the 7 subtypes, for the different disease categories in strains collected between 2016 and 2018. The distribution of disease categories differs significantly between emm12.0 and emm2.0 stains (Table 2).
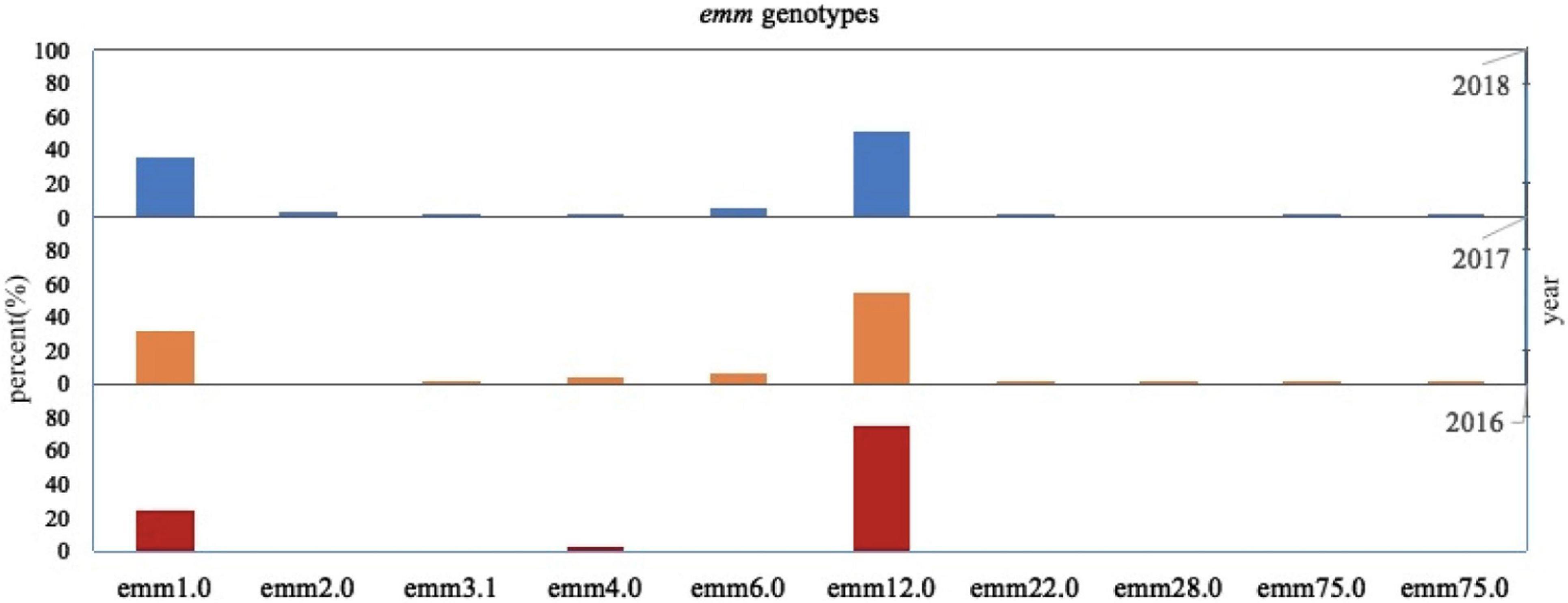
Figure 1. Distribution of Streptococcus pyogenes emm genotypes in isolates collected from pediatric patients in China from 2016 to 2018.
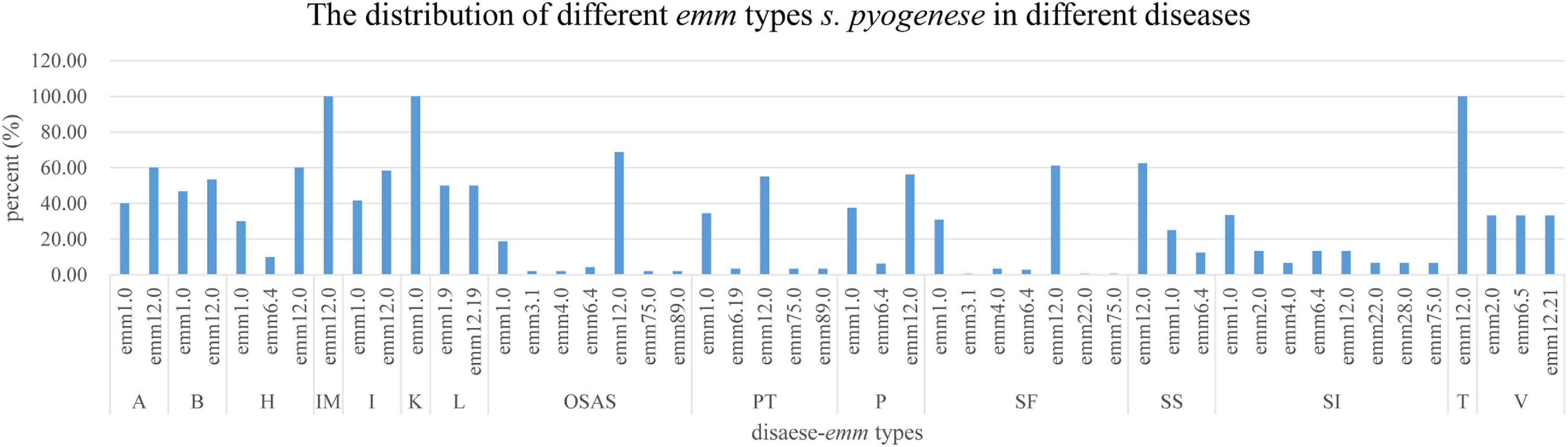
Figure 2. Distribution of Streptococcus pyogenes emm types in isolates from children with different diseases.
Correlations Between Superantigen Expression Profiles, Disease Categories, and emm Types
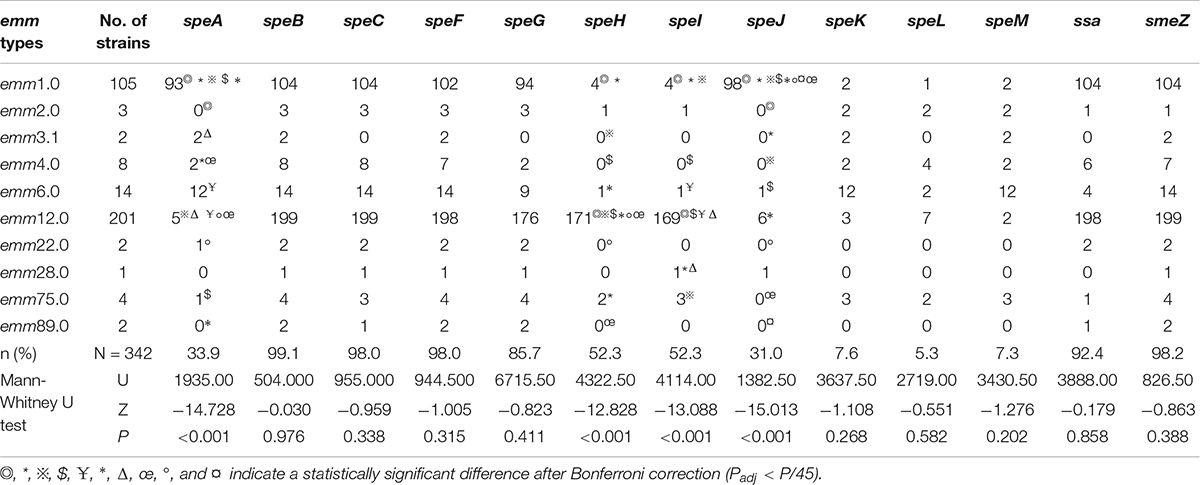
The positivity rates for speA, speB, speC, speF, speG, speH, speI, speJ, speK, speL, speM, ssa, and smeZ differed among the strains, and the distributions of speA, speH, speI, and speJ expression were related to the emm types (Table 3). The distributions of sAgs in different disease categories did not differ significantly (Table 4).
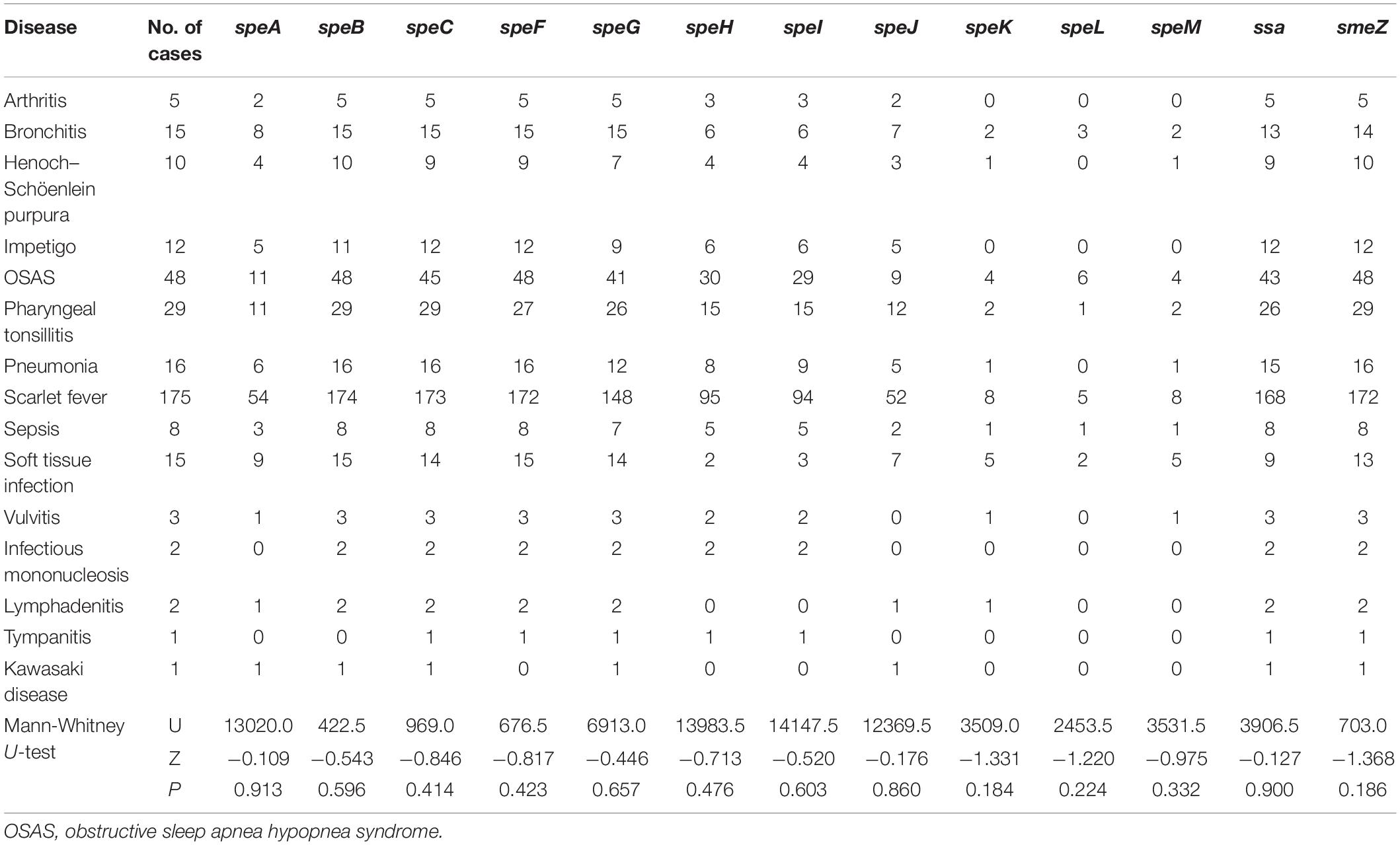
Table 4. Distribution of sAgs in S. pyogenes strains isolated from children with different diseases.
Among the 342 strains, 79.5% expressed six or more sAgs. The five major gene profiles (A–E) associated with emm types were identified according to the sAg combinations (Figure 3).
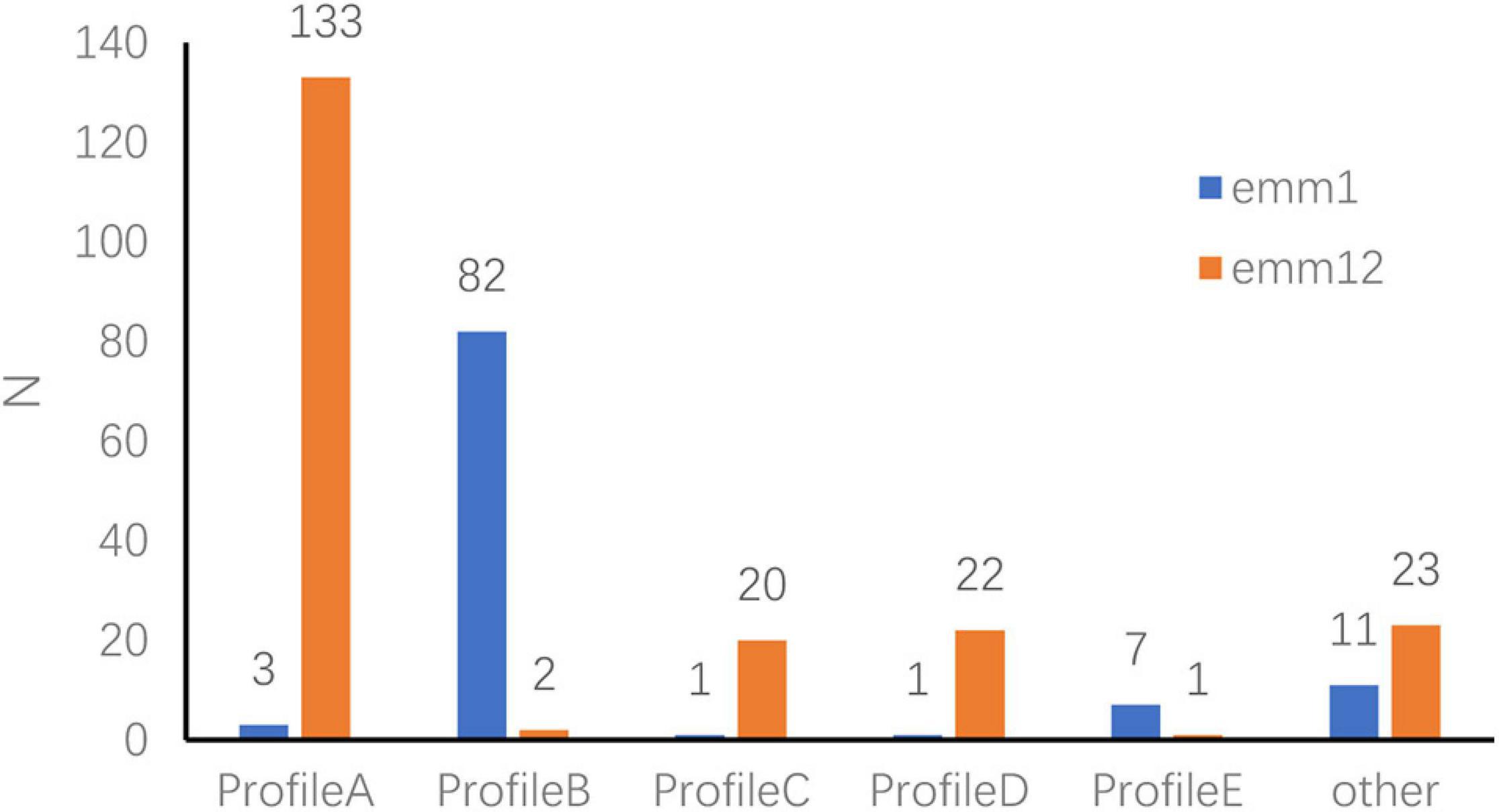
Figure 3. Superantigen-encoding gene expression profiles in emm1.0 and emm12.0 types. Profile A: speA–, speB+, speC+, speF+, speG+, speH+, speI+, speJ–, speL–, speM–, ssa+, smeZ+, speK–. Profile B: speA+, speB+, speC+, speF+, speG+, speH–, speI–, speJ+, speL–, speM–, ssa+, smeZ+, speK–. Profile C: speA–, speB+, speC+, speF+, speG+, speH–, speI–, speJ–, speL–, speM–, ssa+, smeZ+, speK–. Profile D: speA–, speB+, speC+, speF+, speG–, speH+, speI+, speJ–, speL–, speM–, ssa+, smeZ+, speK–. Profile E: speA+, speB+, speC+, speF+, speG–, speH–, speI–, speJ+, speL–, speM–, ssa+, smeZ+, speK–.
Genetic Diversity in Representative Emm1.0 and Emm12.0 Clones
The analysis of antimicrobial resistance gene expression in the genome sequences identified 10 genes (ermB, tetM, pbp2x, bcrA, bacA, pmrA, lmrP, vanB, vanrG, and vanuG), which included two copies of bcrA and a single copy of each of the other nine genes. The frequency of vanuG expression was significantly higher in emm12.0 strains (18/22) than in emm1.0 strains (11/20) (Figures 4B,D).
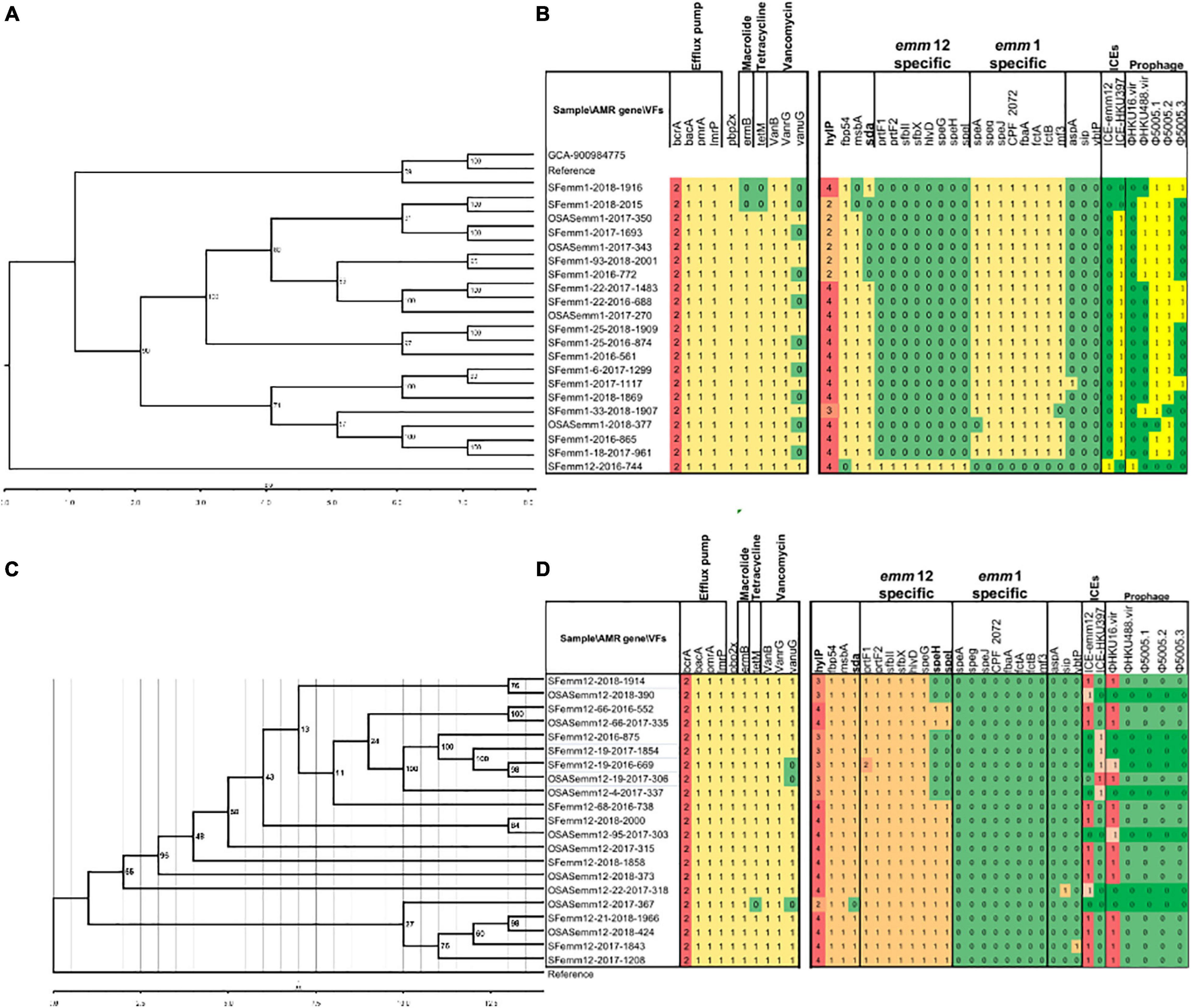
Figure 4. Phylogenetic relationship of selective emm1 and emm12 strains using whole genome core-gene SNPs (A,C) and their antimicrobial drug resistance gene and virulence factor variation profiles (b,d). References indicate S. pyogenes strain MGAS5005 (Genbank accession CP000017.2) and S. pyogenes strain TJ11-001 (CP028148.1), respectively (B,C). The S. pyogenes strain Bra048, assembly accession GCA_900984775, is also included in the phylogeny of emm1 for reference. Strains containing 2 copies (emm1) and 3 copies (emm12), rather than the majority 4 copies, formed clusters and are indicated in bold font. The numbers in cells (B,D) indicate gene copy numbers.
The average number of streptococcal virulence genes expressed in all sequenced emm1.0 and emm12.0 strains was 125. The distribution of virulence genes varied among the strains, but was conserved and unique according to emm type (Figures 4B,D). Clades without speH and speI expression were found in emm12.0 strains (Figures 4C,D, highlighted in yellow). Genomic similarity analysis showed that sAgs were more conserved in emm1.0 strains than in the others. The frequency of sda expression in emm12.0 strains (20/22) was significantly higher than that in emm1.0 strains (15/20). Two copies of the bacteriophage encoding hyaluronate lyases hylP were present in emm1.0 strains, whereas three copies of hylP were observed in the emm12.0 strains (Figures 4B,D). Streptococcal toxin-encoding prophages and macrolide and tetracycline resistance integrative conjugative elements (ICEs) were found in both emm1 or emm12 isolates from Shenzhen with the following positive rates: φHKU16.vir (38.1%), φHKU488.vir (16.7%), Φ5005.1 (45.2%), Φ5005.2 (45.2%), and Φ5005.3 (11.9%), ICE-emm12 (35.7%), and ICE-HKU397 (54.8%).
Phylogenetic analysis using 33,067 core-gene SNPs revealed that emm1.0 and emm12.0 strains were clustered independently and diverged without recombination (Supplementary Figure 1). Additionally, for emm1.0 and emm12.0 strains, no clear independent cluster correlating to patients with OSAS or scarlet fever was found. The genetic diversity of emm12.0 strains (2902 SNPs) was higher than that of emm1.0 strains (384 SNPs) (Figures 4A,C). This difference in genetic diversity was also confirmed by the finding that emm12.0 strains presented more allelic variants (i.e., 12 allelic variants) than emm1.0 strains (i.e., 8 allelic variants).
Discussion
GAS is a gram-positive bacterial pathogen that causes a wide range of clinical diseases, and antibiotics are effective agents for treating GAS infections. Although no GAS strains resistant to β-lactam antibiotics have been found, strains that are highly resistant to macrolides, lincosamides and streptomycin B have been identified globally since 1990 (Gherardi et al., 2015). All strains analyzed in this study were susceptible to penicillin and chloramphenicol, and most were susceptible to levofloxacin. Therefore, penicillin can remain the first-line drug for treating GAS infections in pediatric patients in China. The use of chloramphenicol and levofloxacin in children is limited due to the side effects of these antibiotics on the hematopoietic system and bone and joint development, respectively. Due to the high rate and degree of resistance to macrolides observed for GAS, these antibiotics cannot be used to effectively treat children who are allergic to lactam antibiotics. The use of tetracycline is also limited by a high rate and level of resistance among GAS as well as the side effects of tetracycline on bone and teeth. In the present study, 98.5% of the isolated GAS strains were sensitive to levofloxacin, and this finding is consistent with the rate observed among GAS strains isolated from children in Shanghai, China, but significantly different from the rate observed among GAS strains isolated from Chinese adults (Shen et al., 2018). The reason for this difference may be related to the medication habits of Chinese people. Levofloxacin is not widely prescribed to children due to its side effects on bone and joint development.
The PCR and WGS results in the present study indicate that macrolide-resistant and tetracycline-resistant strains mainly expressed ermB and tetM, respectively, which is consistent with the findings of previous studies (Feng et al., 2010; Liang et al., 2012; Tsai et al., 2020). Unexpectedly, expression of lmrP, a broad-spectrum drug efflux gene, and expression of pmrA, which potentially confers resistance to fluoroquinolone through drug efflux, were identified in all 42 strains by WGS. No penicillin-resistant strains or mutations in pbp2x were found. Nonetheless, a relationship exists between pbp2x gene variations and MIC values (Hayes et al., 2020; Musser et al., 2020; Vannice et al., 2020). Our WGS and MIC results showed that the vanB+ vanrG+ vanuG– and vanB+ vanrG+ vanuG+ strains were susceptible to vancomycin. These results underscore the need to better understand the relationship between antibiotic resistance phenotypes and resistance genes. ICEs have been proposed to play major roles in the selection and expansion of emm12 scarlet-fever outbreak lineages in Hong Kong and mainland China where antimicrobial usage patterns are elevated, highlighting their importance for S. pyogenes population structure (Davies et al., 2015; Shen et al., 2018; You et al., 2018; Jespersen et al., 2020). However, the role of ICEs in driving the global population structure of S. pyogenes has not been fully explored (Jespersen et al., 2020). The majority of characterized S. pyogenes exotoxins are carried by prophages, i.e., bacteriophages integrated in the bacterial chromosome. These SAg toxins, termed streptococcal pyrogenic exotoxins (spe), streptococcal mitogenic exotoxin Z (smeZ) and streptococcal superantigen (ssa), are amongst the most potent known activators of T cells (Blake et al., 2019).
A previous study evaluated time-dependent changes in the emm type prevalence of S. pyogenes (Meisal et al., 2010). In the present study, 10 different emm types, including 7 subtypes, were identified. The most prevalent types and subtypes were emm12.0 and emm12, followed by the emm1.0 and emm1 (Figure 1). The prevalence of these two emm types is known to change over time (Ma et al., 2009; Liang et al., 2012; Li et al., 2020; You et al., 2020; Yu et al., 2020).
Previous studies confirmed the relationship between diseases caused by GAS infection and emm types (González-Abad and Alonso Sanz, 2020), although some other studies did not support this relationship. In the present study, the distribution of 15 diseases among the emm12.0 and emm2.0 types differed significantly. The emm2.0 strains were isolated from patients with 15 different diseases, whereas the emm12.0 strains were isolated mainly from patients with three diseases. These results indicate that the emm types of GAS strains infecting children in Shenzhen from 2016 to 2018 were associated with specific diseases.
S. pyogenes can cause infection by crossing human mucosal membranes and skin barriers. Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing S. pyogenes (Brouwer and Barnett, 2020). The M protein and SAgs play a crucial role in the pathogenesis of S. pyogenes infections, and a close relationship exists between emm type and sAg distribution (Imöhl et al., 2017). The results of this study revealed differences in the frequency of sAg expression among emm types. Indeed, five sAg profiles were identified among GAS strains carrying six or more sAgs. Several sAg profiles were observed for each emm type, but only one or two genes were predominant for each type. Profiles B and E were most common among emm1.0 and its subtypes, whereas profiles A, C, and D were most common among emm12.0 and its subtypes. Thus, sAgs and their combinations are closely related to emm types. In this study, only a few strains were isolated for some diseases. We believe this could be related to the pathogenicity of S. pyogenes or to a lack of attention by researchers. In the past 30 years, most investigators have focused only on the relationship between S. pyogenes and postpartum infections, while the relationship with neonatal infections and vaginitis in girls has not received sufficient attention. Recent research on S. pyogenes shows that the pathogenicity of S. pyogenes in the vulva and vaginal mucosa of girls has become a concern (Donders et al., 2021; Hu et al., 2021). Although the amount of relevant information in this study is limited, the results provide insight into the relationship between emm type and vulvitis and vaginitis. More comprehensive research is needed.
The pathogenicity of GAS is related to the various virulence factors it produces. Pathogenic GAS has evolved to generate a large number of virulence factors, which promote its adhesion to host cells and invasion of deep tissues, ultimately leading to disease. SpeB can promote the spread of bacteria and their products in host tissues by degrading the tissue structure and can also degrade proteins and antimicrobial peptide LL-37 in order to resist immunity (Walker et al., 2014). SpeF is the main cause of pulmonary vascular permeability, which is sufficient to cause acute respiratory distress syndrome under the conditions of toxic shock-like syndrome caused by S. pyogenes (Matsumoto et al., 1999). Among all virulence factors, SAg is of particular concern. SAgs are proteins synthesized by ribosomes that have a relatively low molecular mass (∼22–28 kDa) and contain classic signaling peptides that are cleaved after secretion to release the mature toxin. SAgs function by activating T cells and are among the most powerful T-cell activators identified to date. At present, at least 14 genetically distinct SAgs have been characterized, and many of them are encoded within lysogenic bacteriophage or putative bacteriophage elements (Blake et al., 2019). Therefore, different strains encode different repertoires typically consisting of 3–6 distinct SAgs (Blake et al., 2019). Some studies reported a strong correlation between SAgs and disease categories (Smoot et al., 2002; Türk Dağı et al., 2018), and additional research showed that the distribution of sAgs correlates with disease categories and differs considerably among emm types (Murakami et al., 2002; Gergova et al., 2019). However, in the present study, we found that the sAg distributions were closely correlated to emm types but not to disease categories. The emm12.0 and especially emm2.0 strains were significantly associated with disease categories. Therefore, we speculate that variation in the distribution of sAgs is mainly due to the emm type, rather than the disease category.
In the present study, we generated the genome sequences of 42 S. pyogenes emm1 and emm12 strains. Streptococcal toxin-encoding prophage φHKU16.vir, φHKU488.vir, Φ5005.1, Φ5005.2, and Φ5005.3 in addition to the macrolide- and tetracycline-resistant ICE-emm12 and ICE-HKU397 elements were found amongst the Shenzhen strains. These results confirm our previous findings (You et al., 2018) of φHKU.vir, ICE-emm12 and ICE-HKU397 elements amongst multi-clonal emm12 strains of mainland China. Sequencing of more strains from China in the future will be important to determine the evolutionary pathway and population structure of the predominant emm1 and emm12 S. pyogenes strains.
The virulence factor streptococcal DNase sda1 was previously shown to interfere with the entrapment of bacteria through neutrophil extracellular traps and Toll-like receptor 9 (TLR9) signaling. This factor impairs plasmacytoid dendritic cell recruitment by reducing interferon (IFN)-1 levels at the site of infection and destabilizes DNA via the host protein HMGB1 (high mobility group box 1), which may decrease IFN-1 levels at the site of infection (1 levels at the site of infection (Uchiyama et al., 2012; Keller et al., 2019). Our WGS data showed that sda was specifically expressed by emm12.0 strains. In both emm12.0 and emm1.0 strains, the expression of sda showed no correlation with disease. hylP increases the virulence of GAS via the digestion of hyaluronic acid capsules (Singh et al., 2014). Our WGS data showed that the hylP expression in both emm12.0 and emm1.0 strains was specifically correlated with emm genotype but not with disease. Two copies of hylP were mainly distributed in the emm1.0 strains, and three copies of hylP were mainly distributed in the emm12.0 strains (Figure 4). Our study showed that variations in hylP might play roles in epidemic cloning expansion, but this needs to be further investigated.
In the present study, emm12.0 and its subtypes were present in 58.8% (201/342) of strains, and emm1.0 and its subtypes were present in 30.7% (105/342) of strains, which suggests that emm1.0 and emm12.0 were widespread and causing diseases in Chinese children from 2016 to 2018. The population structure and genetic diversity of these emm types were characterized by sequence typing and WGS (Supplementary Figure 1). The results showed that emm1.0 and emm12.0 each contained a stable clone, suggesting they had genetically diverged without recombination. However, the emm12.0 strains showed higher genetic diversity than the emm1.0 strains, suggesting that longer circulation of the former led to several robust clades with > 700 bootstrap replicates (Supplementary Figure 1). These observations are consistent with their epidemiological history. Horizontal transmission of virulence genes can occur between emm types. This should be considered in future GAS surveillance studies.
Conclusion
In conclusion, antimicrobial agents commonly used to treat GAS infections are highly active against clinical strains. However, increasing macrolide resistance warrants analysis of the epidemiological characteristics of these strains. The molecular epidemiology of GAS in China has shown many differences from earlier reports, based on the present study along with the above-mentioned reports evaluating the molecular profiles of GAS strains collected in different time periods and geographical regions. In the future, multicenter studies including various diseases are necessary to assess whether our findings are affected by temporal and geographical changes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA743366.
Ethics Statement
The studies involving human participants were reviewed and approved by this study was approved by the Research Ethics Committee of the Shenzhen Children’s Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/nex of kin.
Author Contributions
YZ and YY contributed to conception, design, and administrative support. QM, RZ, and YC provided study materials and patients. DY, YL, QL, WW, LH, and YB contributed to the collection and assembly of data, data analysis, and interpretation. DY and YL contributed to the manuscript writing. All authors contributed the final approval of manuscript.
Funding
This research was funded by the Sanming Project of Medicine in Shenzhen (No. SZSM201512030), the Shenzhen Key Medical Discipline Construction Fund (No. SZXK032), the Guangdong Medical Research Fund (No. A2021437), the Hospital Level Project of Shenzhen Children’s Hospital (No. ynkt2020-zz19), the Shenzhen Fund for Guangdong Provincial High-level Clinical Key Specialties (No. SZGSP012), and the Project of the Expert Committee on Clinical Application and Drug Resistance Evaluation of Antimicrobial Drugs of the National Health Commission (No. KJYWZWH-OT-02-2021-06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the physicians who participated in this study and the professionals involved in sample collection and culture maintenance, especially the healthcare workers, for their contribution to disease control. We wish to thank the timely help given by Shengzhe Bian of the School of Public Health of Sun Yat-sen University (Guangzhou, China) for obtaining the accession number. We thank Liqiang Li and Yuanhai You for their helpful works on genomic data analysis and manuscript writing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.722225/full#supplementary-material
Footnotes
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Bhardwaj, N., Mathur, P., Behera, B., Mathur, K., Kapil, A., and Misra, M. C. (2018). Antimicrobial resistance in beta-haemolytic streptococci in India: a four-year study. Indian J. Med. Res. 147, 81–87. doi: 10.4103/ijmr.IJMR_1517_16
Blake, A. S., John, K. M., and Schlievert, P. M. (2019). Toxins and superantigens of group A streptococci. Microbiol. Spect. 7, 1–15.
Brouwer, S., and Barnett, T. C. (2020). Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat. Commun. 11:5018. doi: 10.1038/s41467-020-18700-5
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/s1473-3099(05)70267-x
Chatellier, S., Ihendyane, N., Kansal, R. G., Khambaty, F., Basma, H., Norrby-Teglund, A., et al. (2000). Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68, 3523–3534. doi: 10.1128/iai.68.6.3523-3534.2000
Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, 1–6. doi: 10.1093/gigascience/gix120
Clinical and Laboratory Standards Institute (CLSI) (2019). Performance Standards for Antimicrobial Susceptibility Testing M100, 30th Edn. Available online at: https://webstore.ansi.org/preview-pages/CLSI/preview_CLSI+M100-Ed30.pdf
Davies, M. R., Holden, M. T., Coupland, P., Chen, J. H. K., Venturini, C., Barnett, T. C., et al. (2015). Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat. Genet. 47, 84–87. doi: 10.1038/ng.3147
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Donders, G., Greenhouse, P., Donders, F., Engel, U., Paavonen, J., and Mendling, W. (2021). Genital tract GAS infection ISIDOG guidelines. J. Clin. Med. 10:2043. doi: 10.3390/jcm10092043
Facklam, R., Beall, B., Efstratiou, A., Fischetti, V., Johnson, D., Kaplan, E., et al. (1999). emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis. 5, 247–253. doi: 10.3201/eid0502.990209
Feng, L., Lin, H., Ma, Y., Yang, Y., Zheng, Y., Fu, Z., et al. (2010). Macrolide-resistant Streptococcus pyogenes from Chinese pediatric patients in association with Tn916 transposons family over a 16-year period. Diagn. Microbiol. Infect. Dis. 67, 369–375. doi: 10.1016/j.diagmicrobio.2010.03.014
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gergova, R., Muhtarova, A., Mitov, I., Setchanova, L., Mihova, K., Kaneva, R., et al. (2019). Relation between emm types and virulence gene profiles among Bulgarian Streptococcus pyogenes clinical isolates. Infect. Dis. (Lond) 51, 668–675. doi: 10.1080/23744235.2019.1638964
Gherardi, G., Petrelli, D., Di Luca, M. C., Pimentel de Araujo, F., Bernaschi, P., Repetto, A., et al. (2015). Decline in macrolide resistance rates among Streptococcus pyogenes causing pharyngitis in children isolated in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1797–1802. doi: 10.1007/s10096-015-2414-x
Gherardi, G., Vitali, L. A., and Creti, R. (2018). Prevalent emm Types among Invasive GAS in Europe and North America since Year 2000. Front. Public Health 6:59. doi: 10.3389/fpubh.2018.00059
Golińska, E., van der Linden, M., Więcek, G., Mikołajczyk, D., Machul, A., Samet, A., et al. (2016). Virulence factors of Streptococcus pyogenes strains from women in peri-labor with invasive infections. Eur. J. Clin. Microbiol. Infect. Dis. 35, 747–754. doi: 10.1007/s10096-016-2593-0
González-Abad, M. J., and Alonso Sanz, M. (2020). [Invasive Streptococcus pyogenes infections (2011-2018): emm-type and clinical presentation]. An. Pediatr. (Engl Ed) 92, 351–358. doi: 10.1016/j.anpedi.2019.10.014
Hayes, A., Lacey, J. A., Morris, J. M., and Davies, M. R. (2020). Restricted sequence variation in streptococcus pyogenes penicillin binding proteins. mSphere 5, e90–e20. doi: 10.1128/mSphere.00090-20
Hu, B., Hua, C., Sun, L., Chao-Fang, M., and Zhou, M. (2021). Microbiological findings of symptomatic vulvovaginitis in Chinese prepubertal girls. J. Pediatr. Adolesc. Gynecol. doi: 10.1016/j.jpag.2021.05.012 [Epub ahead of print].
Igwe, E. I., Shewmaker, P. L., Facklam, R. R., Farley, M. M., van Beneden, C., and Beall, B. (2003). Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol. Lett. 229, 259–264. doi: 10.1016/s0378-1097(03)00842-5
Imöhl, M., Fitzner, C., Perniciaro, S., and van der Linden, M. (2017). Epidemiology and distribution of 10 superantigens among invasive Streptococcus pyogenes disease in Germany from 2009 to 2014. PLoS One 12:e0180757. doi: 10.1371/journal.pone.0180757
Jespersen, M. G., Lacey, J. A., Tong, S. Y. C., and Davies, M. R. (2020). Global genomic epidemiology of Streptococcus pyogenes. Infect. Genet. Evol. 86:104609. doi: 10.1016/j.meegid.2020.104609
Keller, N., Woytschak, J., Heeb, L. E. M., Marques Maggio, E., Mairpady Shambat, S., Snäll, J., et al. (2019). Group a streptococcal DNase Sda1 impairs plasmacytoid dendritic Cells’ Type 1 interferon response. J. Invest. Dermatol. 139, 1284–1293. doi: 10.1016/j.jid.2018.11.027
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Li, H., Zhou, L., Zhao, Y., Ma, L., Xu, J., Liu, Y., et al. (2020). Epidemiological analysis of Group A Streptococcus infections in a hospital in Beijing, China. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2361–2371. doi: 10.1007/s10096-020-03987-5
Liang, Y., Liu, X., Chang, H., Ji, L., Huang, G., Fu, Z., et al. (2012). Epidemiological and molecular characteristics of clinical isolates of Streptococcus pyogenes collected between 2005 and 2008 from Chinese children. J. Med. Microbiol. 61(Pt 7), 975–983. doi: 10.1099/jmm.0.042309-0
Lintges, M., Arlt, S., Uciechowski, P., Plümäkers, B., Reinert, R. R., Al-Lahham, A., et al. (2007). A new closed-tube multiplex real-time PCR to detect eleven superantigens of Streptococcus pyogenes identifies a strain without superantigen activity. Int. J. Med. Microbiol. 297, 471–478. doi: 10.1016/j.ijmm.2007.03.015
Liu, B., and Pop, M. (2009). ARDB–antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Lu, B., Fang, Y., Fan, Y., Chen, X., Wang, J., Zeng, J., et al. (2017). High prevalence of macrolide-resistance and molecular characterization of Streptococcus pyogenes isolates circulating in China from 2009 to 2016. Front. Microbiol. 8:1052. doi: 10.3389/fmicb.2017.01052
Ma, Y., Yang, Y., Huang, M., Wang, Y., Chen, Y., Deng, L., et al. (2009). Characterization of emm types and superantigens of Streptococcus pyogenes isolates from children during two sampling periods. Epidemiol. Infect. 137, 1414–1419. doi: 10.1017/s0950268809002118
Matsumoto, M., Ishikawa, N., Saito, M., Shibayama, K., Horii, T., Sato, K., et al. (1999). Streptococcal pyrogenic exotoxin F (SpeF) causes permeabilization of lung blood vessels. Infect. Immun. 67, 4307–4311. doi: 10.1128/iai.67.9.4307-4311.1999
Meisal, R., Andreasson, I. K., Høiby, E. A., Aaberge, I. S., Michaelsen, T. E., and Caugant, D. A. (2010). Streptococcus pyogenes isolates causing severe infections in Norway in 2006 to 2007: emm types, multilocus sequence types, and superantigen profiles. J. Clin. Microbiol. 48, 842–851. doi: 10.1128/jcm.01312-09
Murakami, J., Kawabata, S., Terao, Y., Kikuchi, K., Totsuka, K., Tamaru, A., et al. (2002). Distribution of emm genotypes and superantigen genes of Streptococcus pyogenes isolated in Japan, 1994-9. Epidemiol. Infect. 128, 397–404. doi: 10.1017/s0950268802006854
Musser, J. M., Beres, S. B., Zhu, L., Olsen, R. J., Vuopio, J., Hyyryläinen, H. L., et al. (2020). Reduced in vitro susceptibility of streptococcus pyogenes to β-Lactam antibiotics associated with mutations in the pbp2x gene is geographically widespread. J. Clin. Microbiol. 58, e1993–e1919. doi: 10.1128/jcm.01993-19
Pérez-Trallero, E., Montes, M., Orden, B., Tamayo, E., García-Arenzana, J. M., and Marimón, J. M. (2007). Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob. Agents Chemother. 51, 1228–1233. doi: 10.1128/aac.01054-06
Rantala, S., Vähäkuopus, S., Siljander, T., Vuopio, J., Huhtala, H., Vuento, R., et al. (2012). Streptococcus pyogenes bacteraemia, emm types and superantigen profiles. Eur. J. Clin. Microbiol. Infect. Dis. 31, 859–865. doi: 10.1007/s10096-011-1385-9
Rivera, A., Rebollo, M., Miró, E., Mateo, M., Navarro, F., Gurguí, M., et al. (2006). Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J. Med. Microbiol. 55(Pt 8), 1115–1123. doi: 10.1099/jmm.0.46481-0
Shen, Y., Cai, J., Davies, M. R., Zhang, C., Gao, K., Qiao, D., et al. (2018). Identification and characterization of fluoroquinolone non-susceptible Streptococcus pyogenes clones harboring tetracycline and macrolide resistance in Shanghai, China. Front. Microbiol. 9:542. doi: 10.3389/fmicb.2018.00542
Silva-Costa, C., Friães, A., Ramirez, M., and Melo-Cristino, J. (2015). Macrolide-resistant Streptococcus pyogenes: prevalence and treatment strategies. Expert Rev. Anti Infect. Ther. 13, 615–628. doi: 10.1586/14787210.2015.1023292
Singh, S. K., Bharati, A. P., Singh, N., Pandey, P., Joshi, P., Singh, K., et al. (2014). The prophage-encoded hyaluronate lyase has broad substrate specificity and is regulated by the N-terminal domain. J. Biol. Chem. 289, 35225–35236. doi: 10.1074/jbc.M113.507673
Smoot, L. M., McCormick, J. K., Smoot, J. C., Hoe, N. P., Strickland, I., Cole, R. L., et al. (2002). Characterization of two novel pyrogenic toxin superantigens made by an acute rheumatic fever clone of Streptococcus pyogenes associated with multiple disease outbreaks. Infect. Immun. 70, 7095–7104. doi: 10.1128/iai.70.12.7095-7104.2002
Steer, A. C., Law, I., Matatolu, L., Beall, B. W., and Carapetis, J. R. (2009). Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9, 611–616. doi: 10.1016/s1473-3099(09)70178-1
Strus, M., Heczko, P. B., Golińska, E., Tomusiak, A., Chmielarczyk, A., Dorycka, M., et al. (2017). The virulence factors of group A streptococcus strains isolated from invasive and non-invasive infections in Polish and German centres, 2009–2011. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1643–1649. doi: 10.1007/s10096-017-2978-8
TreeSoft (2021). Available online at: http://treesoft.sourceforge.net/treebest.shtml. (accessed February 18, 2021).
Tsai, W., Shen, C., Lin, Y., Shen, F., Tsai, P., Wang, S., et al. (2020). Emergence of macrolide-resistant Streptococcus pyogenes emm12 in southern Taiwan from 2000 to 2019. J. Microbiol. Immunol. Infect. [Online ahead of print] S1684–S1182. doi: 10.1016/j.jmii.2020.08.019
Türk Dağı, H., Yüksekkaya, S., Seyhan, T., Fındık, D., Tuncer, I., and Arslan, U. (2018). [Investigation of Streptococcus pyogenes virulence factors and typing by multiple locus variable number tandem repeat fingerprinting (MLVF) method]. Mikrobiyol. Bul. 52, 233–246. doi: 10.5578/mb.66841
Uchiyama, S., Andreoni, F., Schuepbach, R. A., Nizet, V., and Zinkernagel, A. S. (2012). DNase Sda1 allows invasive M1T1 Group A streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 8:e1002736. doi: 10.1371/journal.ppat.1002736
Vannice, K. S., Ricaldi, J., Nanduri, S., Fang, F. C., Lynch, J. B., Bryson-Cahn, C., et al. (2020). Streptococcus pyogenes pbp2x mutation confers reduced susceptibility to β-Lactam antibiotics. Clin. Infect. Dis. 71, 201–204. doi: 10.1093/cid/ciz1000
Wajima, T., Chiba, N., Morozumi, M., Shouji, M., Sunaoshi, K., Sugita, K., et al. (2014). Prevalence of macrolide resistance among group A streptococci isolated from pharyngotonsillitis. Microb. Drug. Resist. 20, 431–435. doi: 10.1089/mdr.2013.0213
Walker, M. J., Barnett, T. C., McArthur, J. D., Cole, J. N., Gillen, C. M., Henningham, A., et al. (2014). Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin. Microbiol. Rev. 27, 264–301. doi: 10.1128/cmr.00101-13
You, Y., Davies, M. R., Protani, M., McIntyre, L., Walker, M. J., and Zhang, J. (2018). Scarlet fever epidemic in China caused by streptococcus pyogenes serotype M12: epidemiologic and molecular analysis. EBioMedicine 28, 128–135. doi: 10.1016/j.ebiom.2018.01.010
You, Y., Peng, X., Yang, P., Wang, Q., and Zhang, J. (2020). 8-year M type surveillance of Streptococcus pyogenes in China. Lancet Infect. Dis. 20, 24–25. doi: 10.1016/s1473-3099(19)30694-2
Keywords: Streptococcus pyogenes, disease, China, child, superantigen, antibiotic resistance, emm type
Citation: Yu D, Liang Y, Lu Q, Meng Q, Wang W, Huang L, Bao Y, Zhao R, Chen Y, Zheng Y and Yang Y (2021) Molecular Characteristics of Streptococcus pyogenes Isolated From Chinese Children With Different Diseases. Front. Microbiol. 12:722225. doi: 10.3389/fmicb.2021.722225
Received: 09 December 2021; Accepted: 11 November 2021;
Published: 09 December 2021.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Jianzhong Zhang, Chinese Center for Disease Control and Prevention, ChinaLuiza Guilherme, Universidade de São Paulo, Brazil
Kenji Gotoh, Kurume University, Japan
Copyright © 2021 Yu, Liang, Lu, Meng, Wang, Huang, Bao, Zhao, Chen, Zheng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunsheng Chen, Y2hlbnl1bnNoZW5nbHdAMTYzLmNvbQ==; Yuejie Zheng, eXVlamllekBzaW5hLmNvbQ==; Yonghong Yang, eXloNjI4NjI4QHNpbmEuY29t
†These authors have contributed equally to this work
 Dingle Yu1,2†
Dingle Yu1,2† Yunmei Liang
Yunmei Liang Yonghong Yang
Yonghong Yang