
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 August 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.720381
Butterflies represent one of the largest animal groups on Earth, yet antimicrobial peptides (AMPs) of this group are less studied in comparison with their moth counterparts. This study employed an integrated bioinformatics approach to survey natural AMPs from publicly available genomic datasets. Numerous AMPs, including cecropins, defensins, and moricins, were identified and subsequently used as templates for the design of a series of synthetic AMPs that mimicked the naturally occurring sequences. Despite differing biological effects among the various sequences, the synthetic AMPs exhibited potent antibacterial and antifungal activities in vitro and in vivo, without inducing hemolysis, which implied their therapeutic potential in infectious diseases. Electron and confocal fluorescence microscopies revealed that the AMPs induced distinct morphological and biophysical changes on microbial cell membranes and nuclei, suggesting that the antimicrobial effects were related to a mechanism of membrane penetration and nucleic acid binding by the peptides. In conclusion, this study not only offers insights into butterfly AMPs but also provides a practical strategy for high-throughput natural AMP discoveries that will have implications for future research in this area.
Infectious diseases were a major problem in human history until the development of various efficacious antibiotics that target the causative microorganisms. However, to adapt an aphorism of Friedrich Nietzsche, that which does not kill them makes them stronger, consequently, antimicrobial resistance emerged thereafter and has become a significant threat to public health worldwide. Declining numbers of new antibiotic candidates has further worsened concern about the shortage of antibiotics in the near future, and development of new classes of antimicrobial agents is therefore urgently required (Roope et al., 2019).
Antimicrobial peptides (AMPs) are a heterogeneous group of small amino acid molecules that are evolutionarily conserved in the genome and ubiquitously produced throughout the kingdoms of life to combat microbial infections (Unckless and Lazzaro, 2016). As effector molecules of the innate host defense, AMPs are vital to all living organisms, especially simpler organisms that only depend on innate and humoral immunity to prevent the onset of infection (Jasper and Bohmann, 2002). In recent decades, AMPs have received considerable attention as a promising group of molecules due to their unique mechanism of rapid physical disruption of microbial membranes, although this dogma is now being challenged, as alternative targets have also been reported (Manniello et al., 2021). This mechanism is preferable for suppression of the deteriorating resistance problem because microbes are eliminated regardless of antibiotic sensitivity or resistance, and also AMPs are not prone to inducing resistant mutants (Zharkova et al., 2019). Furthermore, other desirable properties such as broad-spectrum activity and low host toxicity make AMPs appropriate alternatives to antibiotics (Hazam et al., 2019).
To date, numerous AMPs have been researched as drug candidates, and many are currently in clinical trials, the majority of which are naturally derived (Greber and Dawgul, 2017). As one of the most diverse and abundant orders of animals on Earth, Lepidoptera, which comprises butterflies and moths, is a likely major source of natural AMPs (Badapanda and Chikara, 2016). Among the order Lepidoptera, Papilionoidea (butterflies), which comprises typical butterflies (Lycaenidae, Nymphalidae, Papilionidae, Pieridae, and Riodinidae), skippers (Hesperiidae), and moth-butterflies (Hedylidae) (Kawahara and Breinholt, 2014), is one of the most speciose superfamilies, but there are limited studies on AMPs of this superfamily. However, most available genomic datasets of lepidopteran species belong to the butterflies, which are characterized by their relatively small genome size and relatively low repeat content when compared with other eukaryote genomes (Triant et al., 2018). Therefore, identification of the transcripts and gene-encoded peptidyl sequences from butterfly genomes is a feasible approach to investigate whether high-throughput sequencing data are a potential repository for natural butterfly AMP sequences.
In this study, an integrative bioinformatics analysis was conducted to discover naturally occurring AMPs from butterfly genomes. Based on the sequence and structural characteristics of these butterfly AMPs, a series of synthetic analogs were designed and synthesized. The antimicrobial activities and mechanisms of these synthetic molecules were studied and subsequently discussed (Supplementary Figure 1).
Complete genome sequences of Papilionoidea species available in the Gene Expression Omnibus (GEO) database were downloaded (Supplementary Table 1). Ab initio gene predictions were performed using Augustus 3.3.11 (Stanke et al., 2008) trained on the Heliconius melpomene genome model (parameters: augustus --softmasking = true --species = heliconius_melpomene1 genome.fasta), and complete predicted open reading frames (ORFs) of up to 400 amino acids were retained. The predicted encoded amino acid sequences were aligned against AMP sequences obtained from the RefSeq protein database2 using BLASTp (O’Leary et al., 2016), with the highest scoring homologies considered as final annotation results. Amino acid sequences of the predicted genes were also matched with the corresponding hidden Markov models (HMMs) of AMPs retrieved from the Pfam database3 with an e-value threshold of 10–04 (Mistry et al., 2020). The results of all these analyses were merged as a putative AMP candidates set. In addition, other available sequences of other AMPs and AMP-encoding transcripts derived from Papilionoidea species in public databases, as well as AMPs identified from our next-generation transcriptomic data (GSE142679) of Papilio clytia and Atrophaneura mencius following lipopolysaccharide (LPS) challenges and verified by Sanger sequencing, were also included for further analyses. The sequences of mature peptides were deduced by alignment with the APD3 database (Wang et al., 2016) and identification of potential cleavage sites in the putative prepropeptides.
The deduced butterfly AMPs and homologous sequences available from public databases were subjected to phylogenetic analyses (Supplementary Table 2). Phylogenetic trees were reconstructed using MEGA X (version 10.1.7) software (Kumar et al., 2018), based on peptidyl sequence similarities and differences aligned by ClustalW2 (version 2.1) program4 (Larkin et al., 2007, 0) using multiple alignment parameters with default settings.
To reveal the evolutionarily conserved positions of the AMPs, entropy calculations were performed, and the residue preference of putative butterfly-derived natural AMPs was investigated and visualized using the WebLogo program5 (Crooks et al., 2004). A series of peptide analogs were designed based on the sequence conservation and amino acid frequency at particular positions. Hydropathic characteristics of the amino acids at evolutionarily variable positions were also considered, which retained a consistent hydrophilicity or hydrophobicity to respective positions at the amphipathic α-helices. Estimated physicochemical characteristics of the newly designed AMPs were calculated using PepCalc tool6 (Lear and Cobb, 2016). Secondary structures were constructed by homology modeling using the SWISS-MODEL server7 (Waterhouse et al., 2018).
The designed AMPs were synthesized by the standard Fmoc-based solid-phase peptide synthesis (SPPS) methodology (GL Biochem Ltd., Shanghai, China) on respective Wang resins and Rink Amide MBHA resin for C-terminal amidated peptides. Intramolecular disulfide bond formation was achieved by air-mediated oxidation. Fluorescence-labeled AMPs were synthesized using the same protocol with conjugation of fluorescein isothiocyanate (FITC) and an ε-aminocaproic (Acp) linker at the N-terminus. Synthesized peptides were purified using a high-performance liquid chromatography (HPLC) system (LC3000, Beijing Chuangxin Tongheng, Beijing, China), and the identity of the peptides was confirmed by mass spectrometry (6125B, Agilent Technologies, Santa Clara, CA, United States).
In vitro antimicrobial activities of the synthetic peptides against different microorganisms, including the Gram-negative bacteria Escherichia coli (ATCC 25922), the Gram-positive bacteria Staphylococcus aureus (ATCC 6538P), and the fungus Candida albicans (ATCC 10231), were determined by microtiter broth dilution assays (Wiegand et al., 2008). Briefly, the peptides were dissolved and diluted with respective culture mediums to a gradient concentration range of 0.01–32 μmol⋅L–1 and were then applied to mid-logarithmic phase microbes with cell densities of approximately 1.0 × 108 CFU⋅ml–1 and cultured for 24 h. The absorbance of the cultures was evaluated at 600 nm using a microplate reader (Pulang, China). The minimum inhibitory concentration (MIC) was defined as the lowest concentration of each peptide that exhibited apparent growth suppression on the microbes, while the minimum bactericidal concentration (MBC) was defined as the lowest peptide concentration that resulted in no obvious colony growth of the microorganisms.
Biofilms of the aforementioned microbes were allowed to form in microtiter plates under respective culture conditions in the absence (eradication) or presence (prevention) of AMPs. After incubation, the culture supernatant was discarded, and each well was gently rinsed to remove planktonic bacteria. Biofilms were fixed with methanol and stained with 0.1% crystal violet for 20 min; then the dye bound to the cells was resolubilized with ethanol, and the absorbance of the solution was evaluated at 570 nm using a microplate reader (Abouelhassan et al., 2017). The minimal biofilm inhibitory concentration (MBIC) was defined as the lowest concentration of each peptide that exhibited apparent biofilm inhibition, while the minimal biofilm eradication concentration (MBEC) was defined as the lowest peptide concentration that resulted in biofilm eradication.
Hemolysis activity of the synthesized peptides was tested using freshly drawn mouse erythrocytes, which were rinsed three times with phosphate-buffered saline (PBS) and then cultured with peptides at concentrations of 4, 8, 16, 32, and 64 μmol⋅L–1 for 2 h (Lin et al., 2013). After centrifugation, 100 μl of the supernatant from each sample was transferred to a microtiter plate, and the cell-free hemoglobin, representing hemolysis, was detected at 550 nm with a microplate reader (Pulang, China).
To evaluate the potential cytotoxicity on the normal cells, human embryonic kidney (HEK293) cells were used, and the cell viability was assessed by cell counting kit (CCK)-8 assays following the manufacturer’s protocol (Dojindo Inc., Kumamoto, Japan) (Wang et al., 2018). Briefly, the cells were seeded in microtiter plates at a density of 1 × 104 cells per well for 24 h and cultured with increasing concentrations of AMPs ranging from 4 to 64 μmol⋅L–1 and cultured for 24 h. The CCK-8 solution was then added to each well, and the cells were incubated for an additional 3 h. The optical density (OD) was measured at 450 nm.
SYTOX Green uptake assays were performed according to the instructions of the manufacturer (Thermo Fisher Scientific, Waltham, MA, United States) to estimate the membrane permeabilization effect of the AMPs (Jin et al., 2016). Briefly, 1.0 × 108 CFU⋅ml–1 microorganisms in logarithmic phase were incubated with AMPs at concentrations of 4× the corresponding MIC value for 1 h. Microbial cells were then collected, washed in Hanks’ balanced salt solution (Solarbio, Beijing, China), and incubated with SYTOX Green Nucleic Acid Stain. The fluorescent intensity, which reflects uptake of SYTOX Green, was measured using a fluorescence microplate reader (Thermo Fisher Scientific), at wavelengths of 485 and 520 nm for excitation and emission, respectively.
Samples for electron microscopy were prepared according to a previously described method (Hartmann et al., 2010). In general, 1.0 × 108 CFU⋅ml–1 exponential growth phase cultures of E. coli, S. aureus, and C. albicans were incubated with AMPs at 4× the corresponding MIC value for 1 h. Microbial cells were then pelleted by centrifugation, fixed with glutaraldehyde solution (Phygene, Fuzhou, China), and stained with osmium tetroxide (OsO4). Morphological and ultrastructural changes in the microorganisms were observed and photographed using scanning electron microscopy (SEM; SU8020, Hitachi, Tokyo, Japan) and transmission electron microscopy (TEM; JEM-1200EX, JEOL, Tokyo, Japan).
Based on the electron microscopic findings and to further investigate the mechanism of the bacteriostatic activity of AMPs on C. albicans, fluorescence-labeled peptides were employed to localize potential targets of the synthetic AMPs. Briefly, C. albicans cultures were treated with FITC-labeled AMPs at 4× the corresponding MIC value for 1 h and were then sequentially stained with propidium iodide (PI) and 4′, 6-diamidino-2-phenylindole (DAPI). Images were acquired with a confocal microscope (Ti2-E A1R+, Nikon, Tokyo, Japan).
Genomic DNA was isolated from C. albicans using a fungal genomic DNA extraction kit (Solarbio) following the instructions of the manufacturer. The quantity of the resulting DNA was assessed by NanoDrop 2,000 μ-volume spectrophotometer (Thermo Fisher Scientific). An aliquot of the prepared DNA was then mixed with an equivalent volume of synthetic AMPs at 4× the concentration of respective MIC value. DNA-binding activity of the peptides was detected by measuring the circular dichroism (CD) spectra in quartz cuvettes of 1-mm path length and wavelength ranging from 240 to 330 nm using a Chirascan spectrometer (Applied Photophysics, Leatherhead, United Kingdom) (Garbett et al., 2007).
In vivo antimicrobial activities of the designed AMPs were examined in neutropenic mice following subcutaneous inoculation of 100 μl of 1.0 × 108 CFU⋅ml–1 E. coli, S. aureus, or C. albicans (Lee et al., 2005). At 12 h post-infection, 100 μl of AMPs at the respective MIC value was applied by subcutaneous injection each day. Mice were sacrificed on day 4, and the infected area was carefully excised and homogenized. The CFU count was determined after overnight incubation of the homogenate aliquots. All animal care and experiments were performed in compliance with institutional and government guidelines and were approved by the Peking Union Medical College Hospital Animal Care and Use Committee.
All data represent the means ± standard deviations (SDs) of triplicate determinations in three independent experiments. Statistical analyses were performed by t-test or one-way analysis of variance (ANOVA) with the least significant difference (LSD) post hoc test for multiple comparisons using SPSS Statistics 19.0 software (IBM Corp., Armonk, NY, United States); p < 0.05 was considered statistically.
Available genomic sequencing datasets from 32 Papilionoidea species of six families (one species from Hesperiidae, one from Lycaenidae, 20 from Nymphalidae, four from Papilionidae, four from Pieridae, and two from Riodinidae) deposited in the GEO database up to January 2019 were downloaded for analyses. From these sequences, 434,811 genes encoding small peptides (<400 residues) were retrieved by Augustus prediction, despite differences in the integrity of the genomic data. Alignment of these sequences with AMPs from RefSeq and Pfam databases resulted in 183 computational translated peptides being annotated as AMPs. Thirty-six of these deduced peptides, including 28 predicted cecropins, five defensins, and three moricins, with high sequence similarities had their presumed propeptide cleavage sites confirmed manually. In addition, two cecropins and one defensin identified from P. clytia and A. mencius transcriptomic data were also included. This set of peptide sequences was merged with available sequences obtained by database query and literature research (four moricins from Danaus plexippus, Papilio polytes, Pieris rapae, and Bicyclus anynana). However, no glycine-rich peptides (such as attacins or gloverins) or proline-rich peptides (such as lebocins) were found in the butterfly genome datasets. Amino acid sequences of the putative AMPs and theoretical physicochemical properties of the deduced mature peptides are displayed in Table 1.
As the first discovered and one of the most intensely studied classes of AMPs, cecropins are characterized by their cationic-residue-rich and alpha-helical structures, and their potent and wide-spectrum antimicrobial activities against different microbes (Bulet et al., 2004). Cecropins consist of a signal peptide and propeptide at the N-terminus of the mature peptide, and most cecropins also contain tryptophan (Trp) at the first or second position and a carboxamide modification at the N-terminus of the mature peptide (Yi et al., 2014). Based on the bioinformatics analysis and data from previous studies, 30 cecropins were identified from different Papilionoidea species. These cecropins had an average length of 39.23 ± 2.67 amino acids and an average molecular weight (MW) of 4,290.86 ± 289.78 g⋅mol–1. All deduced sequences were presumed to possess a Trp residue at the N1 or N2 position, while one-third of the sequences were predicted to have C-terminal amidation by the presence of a glycine (Gly) residue at the C1 position. A tetrapeptide motif AGPA that formed the hinge region between the two α-helices featured consistently in almost all cecropins. According to the alignment results, the identified cecropins could be subdivided into two categories that contained prominent sequence differences around the Trp residue (Figures 1A,B). Phylogenetic classification further suggested that these two subclasses of cecropins had diphyletic origin, although the result also illustrated that the two clusters of peptides and other lepidopteran cecropins could be derived from a single ancestral sequence (Figure 2A).
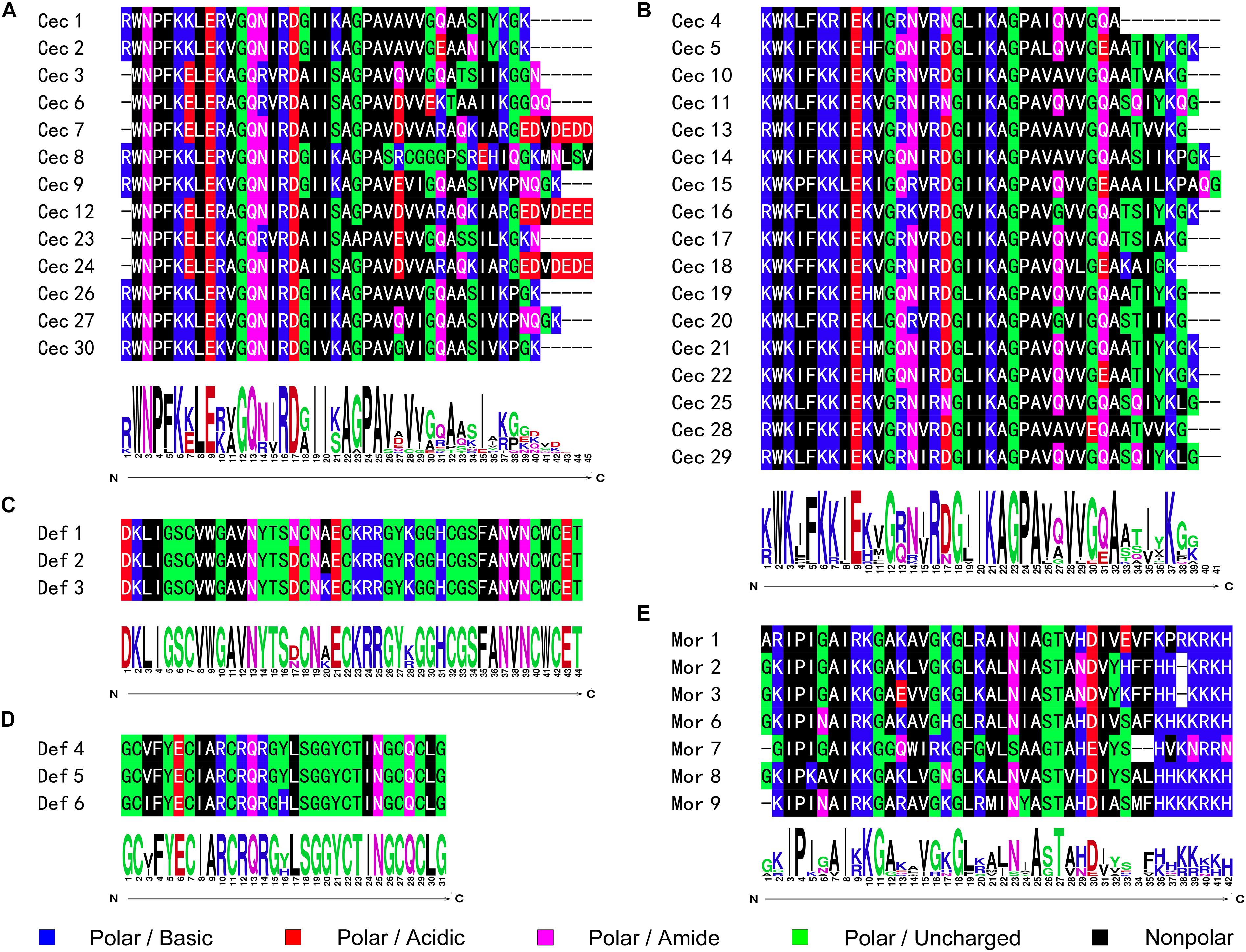
Figure 1. Sequence alignment and logo analysis of naturally occurring AMPs from butterflies. The sequence differences therein suggested that both cecropins and defensins could be further subclassed into two categories, respectively. The peptidyl sequence similarities and site-specific preference of (A) cecropins subclass A; (B) cecropins subclass B; (C) defensins subclass A; (D) defensins subclass B; and (E) moricins were analyzed and displayed as graphics.
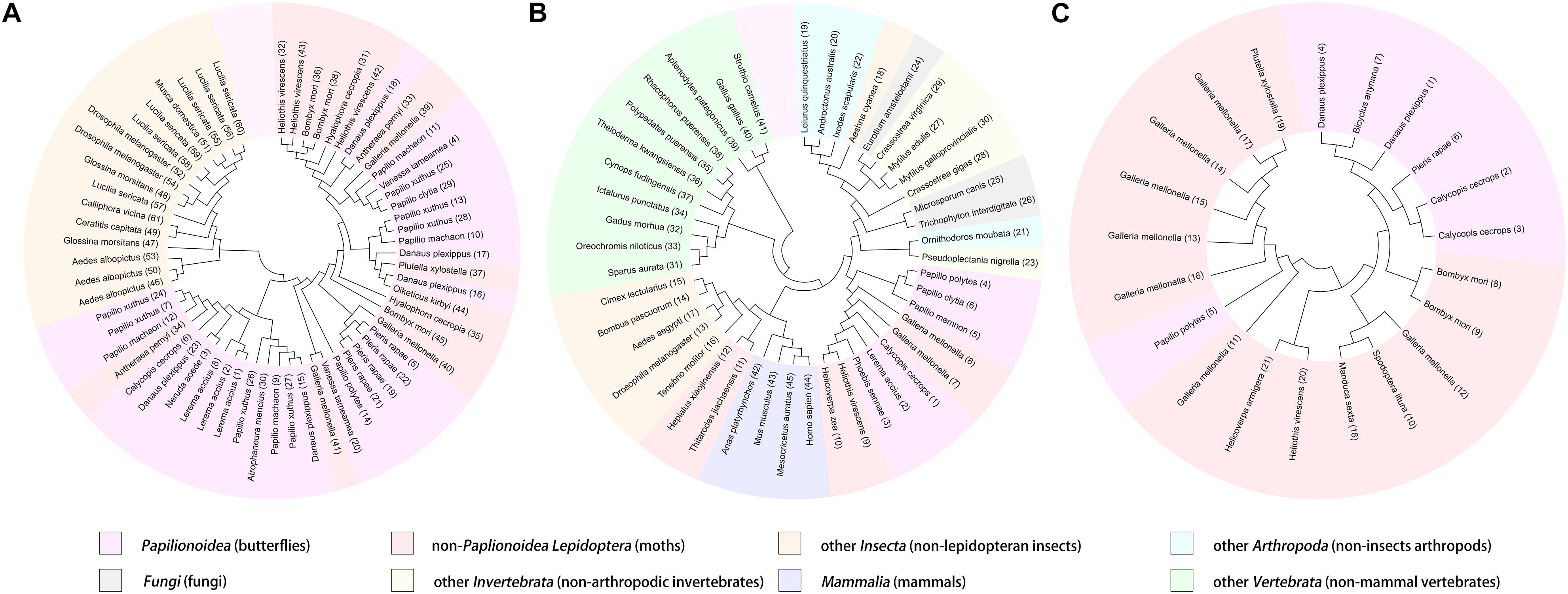
Figure 2. Phylogenetic classification and visualization of butterfly AMPs. The analysis was conducted based on the amino acid sequences of cecropins (A), defensins (B), and moricins (C), among butterfly AMPs and their homologous sequences.
Defensins represent a class of evolutionarily ancient AMPs that are present in nearly all multicellular organisms and play important roles in humoral defense reactions (Machado and Ottolini, 2015). The predominant characteristic of defensins is the highly conserved motif of cysteine (Cys) residues, through which intracellular disulfide bridges are formed to maintain the secondary structure despite differences in the localization of the Cys residues and the pairing patterns thereof [14]. In contrast to cecropins, the overall sequence similarity of the defensins was not apparent, and the phylogenetic analysis revealed complex relationships among diverse organisms (Figure 2B). However, the amino acid sequences of the deduced defensins in the Papilionoidea superfamily were highly conserved. For example, three transcripts from Calycopis cecrops were predicted to differ by nucleotide sequences and corresponding precursor peptides but shared the same mature peptide sequences, which were also identical to those of a defensin from Archaeoprepona demophon (Landon et al., 2004). This suggested that certain sequences might be preferable to the host defense system of butterflies. After removal of duplicated sequences, six unique defensins were identified from the genomic and transcriptomic data (Table 1) and could be categorized into two subgroups according to the alignment results and theoretical physicochemical parameters (Figures 1C,D). Sequences of the first cluster of butterfly defensins contained 44 residues and had an average MW of 4,826.31 ± 29.04 g⋅mol–1, while sequences of the other subgroup were composed of 30 amino acids and had a mean MW of 3,326.83 ± 6.93 g⋅mol–1. The Cys motifs in the two subgroups were C-X10-C-X3-C-X9-C-X7-C-X1-C and C-X4-C-X3-C-X10-C-X4-C-X1-C, which were within the range of reported Cys patterns of invertebrate defensins (Seufi et al., 2011). The distinct Cys motifs and primary sequences of the two subgroups of butterfly defensins also reflected the far molecular distance in the phylogenetic analysis, whereas the high sequence similarity within each respective cluster indicated that a tight phylogenetic relationship exists between defensins in the same subgroup (Figure 2B).
Moricins are a class of highly positively charged AMPs that are broadly effective against diverse microorganisms and feature a long α-helix secondary structure and a cluster of basic amino acid residues in the C-terminal region (Hemmi et al., 2002). Moricin peptides, initially isolated from the hemolymph of Bombyx mori larva under bacterial challenge and later discovered from several other moth species, are considered to only be expressed in members of the order Lepidoptera (Xu et al., 2019). Although limited moricin sequences of butterfly origin have been reported, the current study identified a total of seven moricins by genomic analysis and database retrieval. These moricin sequences had an average amino acid length of 41.14 ± 1.07, an MW of 4,460.66 ± 140.66 g⋅mol–1, and a calculated mean isoelectric point of 11.51 ± 0.67, which was greater than that of the cecropins (10.32 ± 2.12) and defensins (7.95 ± 0.16). Despite the limited number of published moricin sequences, the multiple sequence alignment and phylogenetic analyses revealed significant sequence similarity and a monophyletic origin among all naturally occurring moricins (Figure 2C). The amphipathic segment of the α-helix at the N-terminus of the moricins is critical for the antibacterial activities of these peptides, but the function of the positively charged segment at the C-terminus remains unknown (Hemmi et al., 2002; Oizumi et al., 2005). However, amino acid frequency analysis at the relative region in the current study suggested that net positive charge rather than site-specific preference could control the activities of moricins (Figure 1E).
Analysis of the sequence conservation and amino acid frequency at particular positions facilitated the design of a series of peptides with antimicrobial potential, including two cecropins [cecropin A (CecA) and cecropin B (CecB)], two defensins [defensin A (DefA) and defensin B (DefB)], and one moricin [moricin A (MorA)], corresponding to aforementioned butterfly AMP classifications. The amino acid length, theoretical MW, and isoelectric point of all designed peptides were within the range of corresponding parameters analyzed (Table 2 and Supplementary Table 3). The homology modeling results displayed classical secondary structural characteristics of cecropins, defensins, and moricins, which were in accordance with the corresponding conservative structural features described above (Figure 3). Following this in silico analysis, the peptides were chemically synthesized for further evaluation (Supplementary Figure 2).
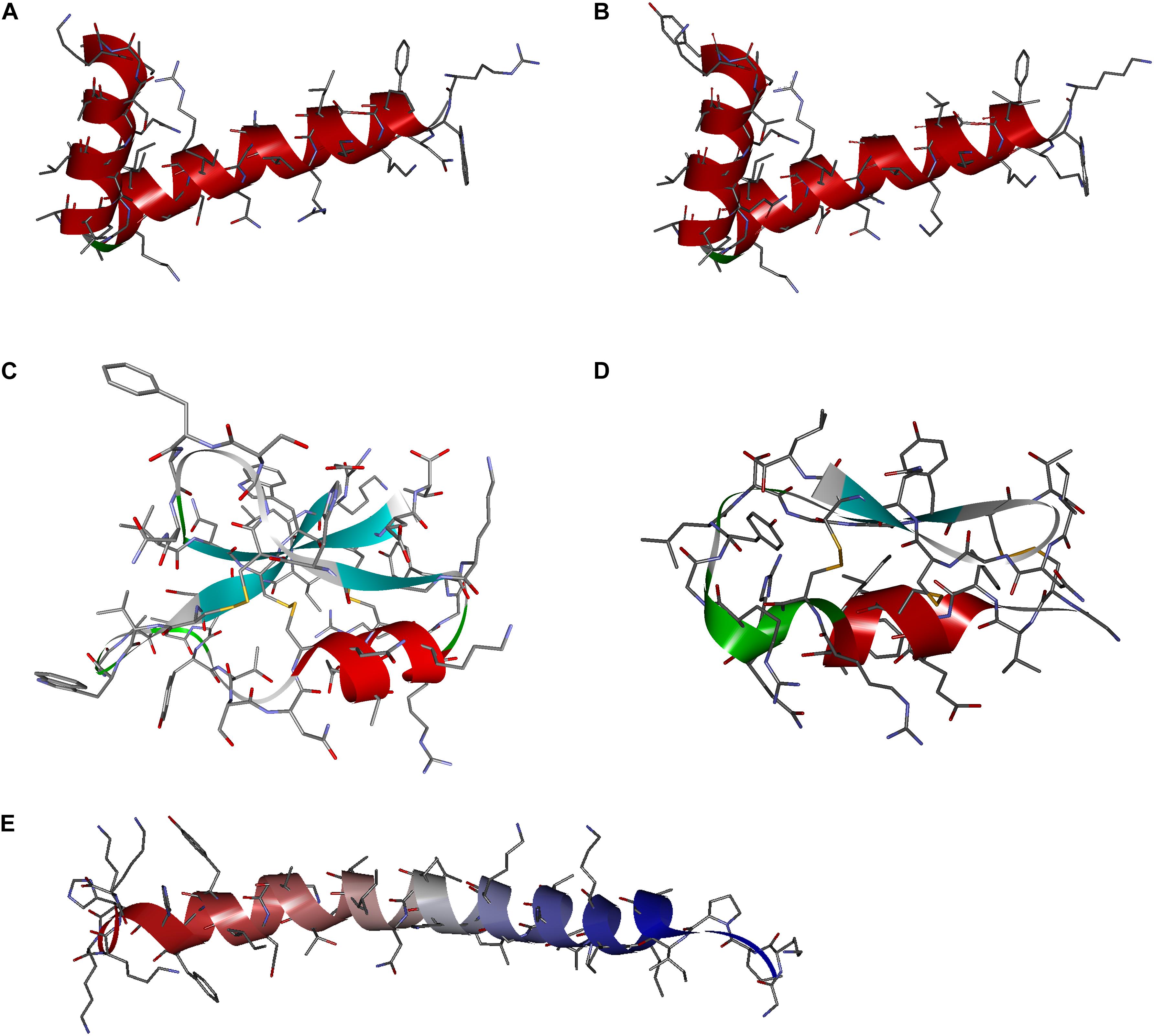
Figure 3. Secondary structures of the designed peptide. The three-dimensional shapes of the synthetic molecules, e.g., (A) cecropin A, (B) cecropin B, (C) denfensin A, (D) denfensin B, and (E) moricin A, were constructed by homology modeling, which demonstrated typical structural features of respective classes of antimicrobial peptides (AMPs).
In vitro antimicrobial activities of the peptides were examined by determination of the MIC and MBC values against representative microorganisms (E. coli, S. aureus, and C. albicans) of public concern using microtiter broth dilution assays. Despite different biological effects among the AMPs, the synthesized cecropins, defensins, and moricins all exhibited potent bactericidal actives against both Gram-positive and Gram-negative bacteria, with MIC and MBC values in micromolar and sub-micromolar concentration ranges; cecropins were the most effective agents. In addition, while cecropins exhibited modest antifungal effects, the synthetic moricin possessed relatively strong fungicidal activities, with low micromolar MIC and MBC values (Table 3). No apparent hemolysis was detected at concentrations exceeding the respective MIC and MBC values (Supplementary Figure 3).
The biofilm inhibitive and degradative effects of the synthetic peptides were quantitated by measuring the MBIC and MBEC values against E. coli, S. aureus, and C. albicans, which represent Gram-negative bacteria, Gram-positive bacteria, and fungi, respectively. All the synthetic peptides prevented the formation of biofilms of both Gram-negative and Gram-positive bacteria and eliminated pre-formed biofilms of both bacterial species. Congruent with the results of the antifungal assays, neither of the defensins displayed visible antibiofilm activity against C. albicans. In contrast, MorA displayed the greatest antibiofilm activities among all the designed AMPs (Table 4).
The membrane disruption effects of the synthesized peptides on the representative microorganisms were analyzed using a SYTOX Green uptake assay as well as SEM and TEM. SYTOX Green, a membrane integrity stain, is unable to enter intact cell membranes; hence, the fluorescence intensity from this stain is positively related to membrane permeability. SYTOX Green staining indicated significant alteration of cell membrane permeability in the assayed microorganisms exposed to the synthetic AMPs (Figure 4). Observations of both Gram-negative and Gram-positive bacteria by electron microscopy revealed distinct morphological and biophysical changes induced by all of the synthetic AMPs (Figure 5). These observations were indicative of a typical mechanism of AMPs binding to and disrupting the cell membranes, resulting in leakage of cell contents. However, images of C. albicans showed only a slightly changed cell morphology and a relatively dense internal structure, which indicated that an alternative mechanism might be involved in the antifungal activities of the synthetic AMPs (Figure 6).
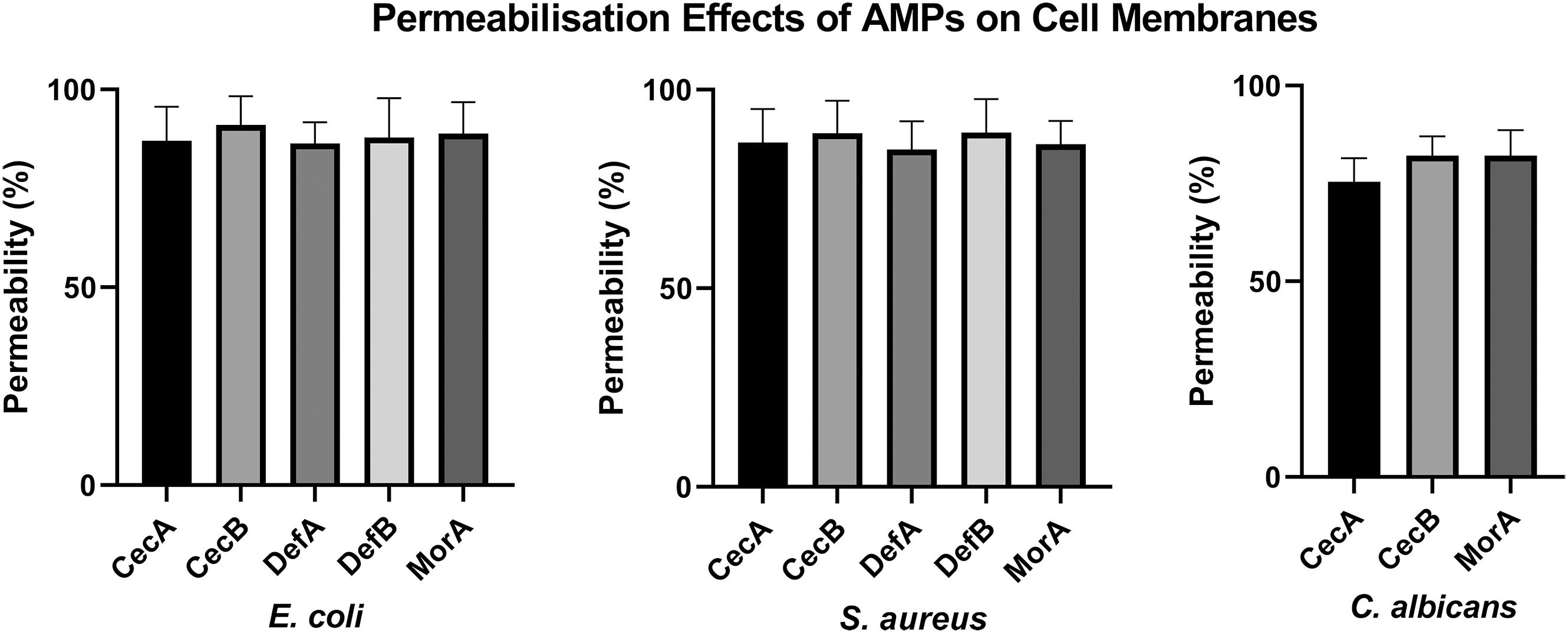
Figure 4. Evaluation of membrane destruction effects of antimicrobial peptides (AMPs) by SYTOX Green uptake assay. Results demonstrated the synthetic peptides induced significant membrane permeability changes against different microorganisms.
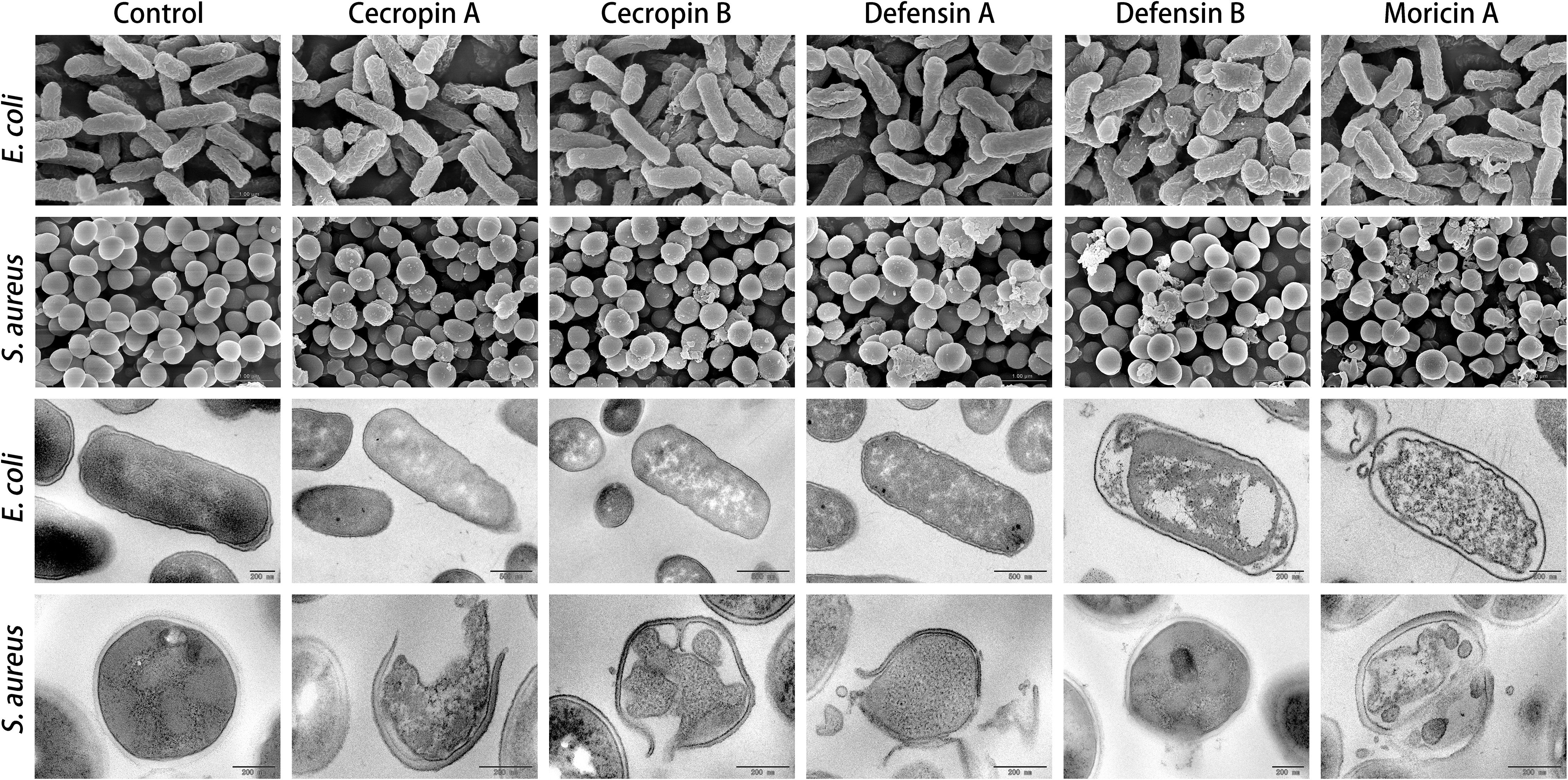
Figure 5. Electron microscopic analysis of bacteria treated with synthetic antimicrobial peptides (AMPs). Representative images of both scanning electron microscopy (SEM; upper panels) and transmission electron microscopy (TEM; lower panels) showed significant changes on cell morphology induced by AMPs at 4× of corresponding minimum inhibitory concentration (MIC) value, compared with the intact smooth surfaces of the untreated control group (left panel). The TEM assay further displayed marked decreases in cell densities that resulted from peptide intervention. These findings suggest membrane permeabilization mechanism being involved in bactericidal activities of the synthesized butterfly AMPs.
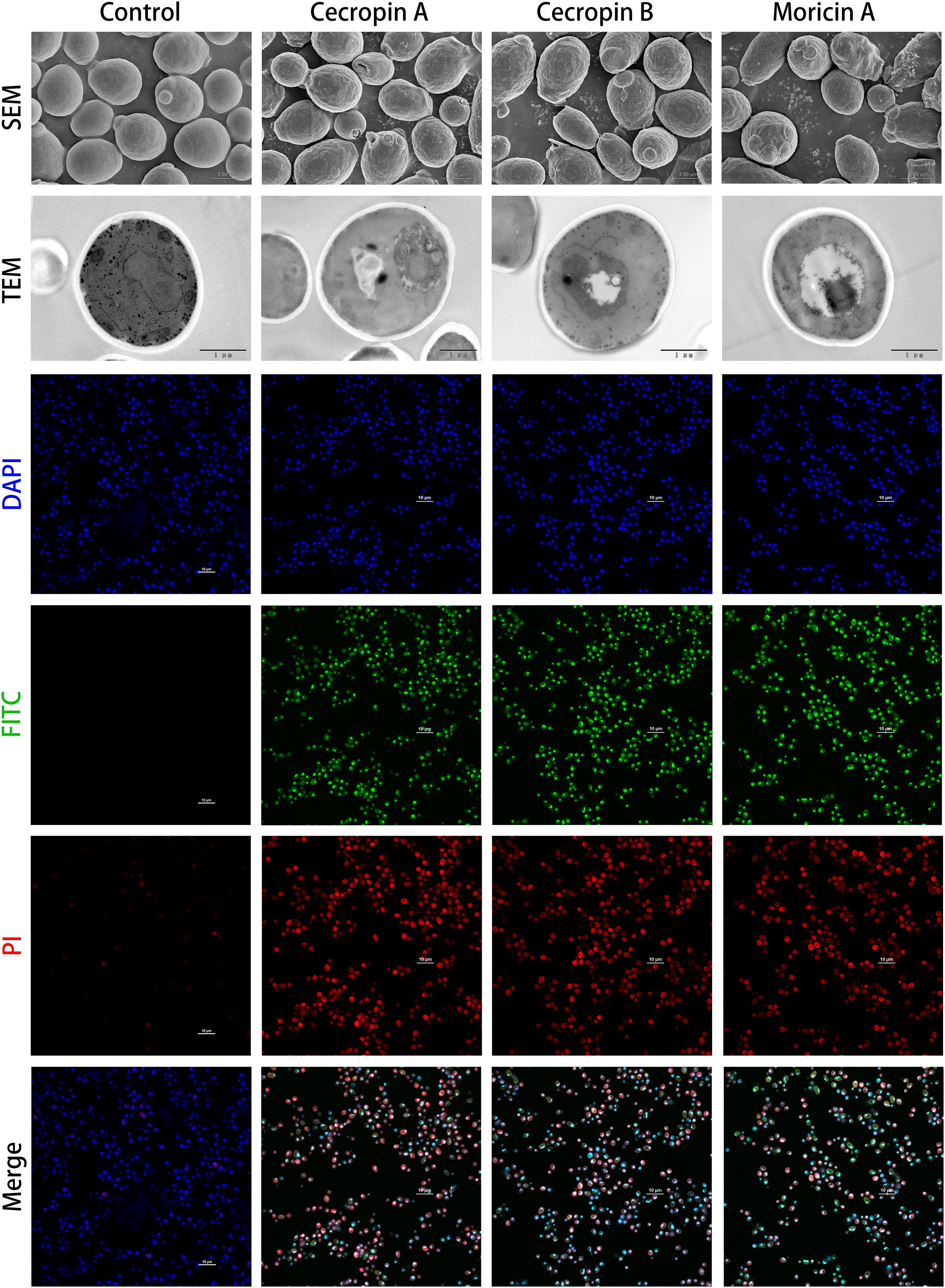
Figure 6. Microscopic investigation of antifungal capabilities of designed antimicrobial peptides (AMPs). The antifungal effects of cecropins and moricins, which had been confirmed by microtiter broth dilution assays (Table 3), were analyzed by combination of scanning electron microscopy (SEM; first upper panel), transmission electron microscopy (TEM; second upper panel), and confocal fluorescence microscopies (lower panels). Although there were no apparent morphological changes observed by electron microscopies, the confocal microscopy demonstrated the fluorescein isothiocyanate (FITC) (green) labeled peptides colocalized with DAPI (blue)-stained fungus nucleus, and the FITC fluorophore showed high spatial overlaps with propidium iodide (PI) (red) fluorescence, which indicates a probable cell death induction mechanism caused by DNA binding.
To further elucidate the underlying antifungal mechanism of the designed AMPs, FITC-labeled peptides were synthesized and used for localization of the potential binding targets of the synthetic AMPs by confocal fluorescence microscopy. The synthetic cecropins and moricin targeted the DAPI-stained nucleus of C. albicans (Figure 6). In addition, the FITC fluorescence showed strong colocalization with the PI signal. DNA-binding activities of the synthetic AMPs were verified by CD spectroscopy, which indicated detectable DNA structural changes (Supplementary Figure 4). Therefore, it was hypothesized that the cationic AMPs exert their antifungal activities by binding to the anionic nucleic acids, thus interfering with bioprocesses and resulting in cell death.
Finally, the in vivo antimicrobial activities of the most promising AMPs against bacteria (CecB) and fungi (MorA) were evaluated in a skin infection mouse model. CFU counts for the bacteria and fungus in the presence of the tested AMPs were significantly less than those of the respective controls. This demonstrated that the synthetic cecropin could attenuate growth of both E. coli and S. aureus in the animal model, while the synthetic moricin could suppress proliferation of C. albicans in vivo (Figure 7). These findings suggested that CecB and MorA may have therapeutic potential under different infectious circumstances.
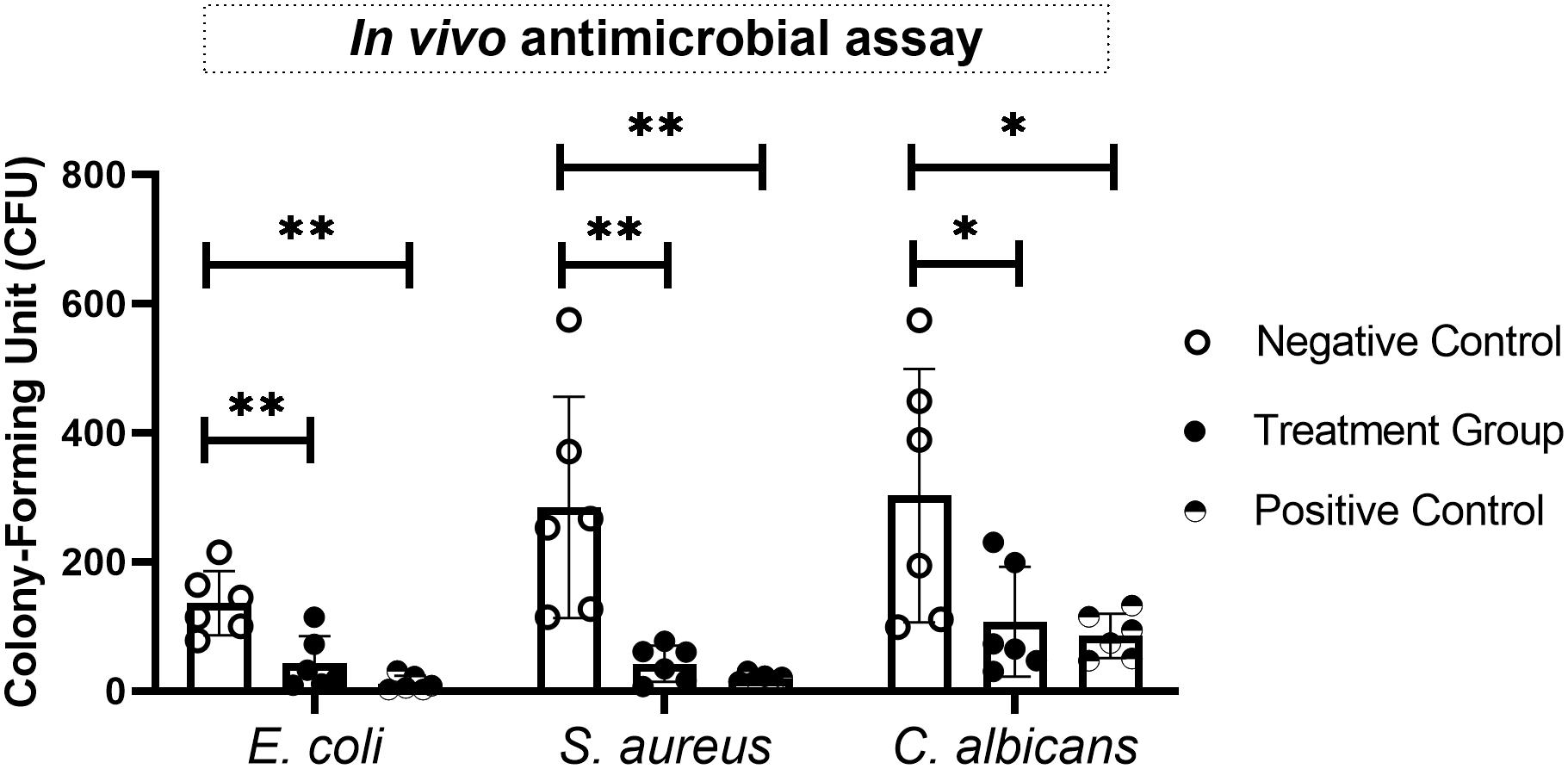
Figure 7. Evaluation of in vivo antimicrobial effects of synthetic antimicrobial peptides (AMPs) on various bacteria and fungi in a skin infection mice model. The most effective antibacterial (CecB against Escherichia coli and Staphylococcus aureus) and antifungal (MorA against Candida albicans) agents in the in vitro assays were examined in this study, and the antimicrobial activities were expressed as CFU per skin sample. Statistical analyses were performed by t-test, and asterisks indicate significant difference (∗p ≤ 0.05, ∗∗p ≤ 0.01).
Numerous AMPs have been identified from natural resources, and many more have been synthetically created by modification and ab initio design based on sequence and structural features of the naturally occurring molecules (Hale and Hancock, 2007). Many AMPs are currently in preclinical or clinical development for the treatment of various infectious diseases, and most of these AMPs were of natural origin (Greber and Dawgul, 2017). Conventionally, naturally occurring AMPs are predominantly identified by cDNA cloning and sequencing and/or de novo peptide sequencing. Moreover, by taking advantage of precursor sequence conservation of known AMPs within Pisces and Amphibia (fishes and amphibians), a more productive shotgun cloning approach and strategies for database retrieval have been developed to facilitate the identification of homologous sequences (Wang et al., 2012; Liu et al., 2015). However, precursor sequence-based methodologies are not applicable for screening butterfly AMPs, since such sequence conservation has not been revealed among butterfly species. Nevertheless, dozens of naturally occurring AMPs have been recognized from various lepidopteran insects (moths and butterflies) insects, although only a few are derived from Papilionoidea species (butterflies). Butterfly species could therefore be a hidden treasure for natural AMP discoveries and may provide potential templates for peptide design. The availability of an increasing number of high-throughput sequencing datasets from butterflies facilitated the design of an integrated bioinformatics analysis method for the efficient discovery of naturally occurring AMPs.
To process large-scale genome data, many computational tools have been developed that can perform sequence assembly and gene prediction and identification. In the context of the current study and identifying AMPs, protein-encoding portions of transcripts are deducible by ORF analysis, which can be accomplished by using software such as ORFfinder. Next, the signal peptide of propeptides can be predicted by calculation of the potential cleavage sites, which is achieved by using a program such as SignalP. Furthermore, despite the lack of software available to analyze other post-translational modifications, the mature peptide can still be identified by homologous sequence alignment and prediction of cleavage sites (Wang and Hu, 2017). In general, the N-terminus of a pro-AMP undergoes a two-step proteolytic cleavage by signal peptidase and dipeptidyl aminopeptidase (Boman et al., 1989), while peptides with a Gly residue present at the C-terminus are subjected to an amide modification by a two-step sequential catalytic reaction with peptidylglycine α-amidating monooxygenase (PAM) (Kim and Seong, 2001). Based on this bioinformatics analysis method, a total of 36 new, naturally occurring AMPs of butterfly origin were identified from the published genomic data used in the current study. This was substantially greater than the number of previously reported, naturally occurring butterfly AMPs. In addition, the designed peptides, which acted as representatives of the discovered natural AMPs, exhibited potent and wide-spectrum antimicrobial effects with typical membrane-disruption activities. Therefore, the genome-wide in silico screening is suggested to be an effective and practical method for identification of naturally occurring AMPs.
The findings from this study add to previous discoveries, and it is our understanding that three classes of AMPs have been found from Papilionoidea species; these classes are cecropins, defensins, and moricins. Of the three classes, cecropins are predominantly expressed by lepidopterans and dipterans and are primarily responsible for the host defense of lepidopterans. Although α-helical AMPs nominated as cecropins have been identified from other species, the sequences of these molecules exhibit a substantially lower degree of similarity to insect cecropins. Defensins have rarely been discovered from moths or butterflies but are ubiquitously expressed throughout multicellular organisms. In contrast, moricins have been identified only in Lepidoptera species. Another class of AMPs unique to the order Lepidoptera is the lebocins; however, no butterfly lebocin sequences were found in the current study. Phylogenetic analyses revealed a monophyletic origin within butterfly cecropins and moricins, which is yet another demonstration of the monophyletic origin of the superfamily Papilionoidea. The molecular distances among defensins of butterfly origin were relatively far, which was expected since it has been proposed that higher variabilities of insect defensins exist among closely related species instead of among distantly related insects (Bulet et al., 2004).
The molecular design of the synthetic peptides in this study was generally based on the mimicry of natural AMPs. The pronounced sequence and structural features, e.g., α-helical structures, C-terminal amidation, disulfide bridges, and the positively charged C-terminal segment, were included in the synthetic peptide design (Table 1 and Figure 3). However, due to the possibility of producing the designed AMPs by microorganism expression systems in the future, more complex chemical modification was not considered in this research. It is recognized that the positively charged amphipathic α-helix of cecropins plays a dominant role in their antimicrobial activities, allowing them to partition into the phospholipid bilayers and disrupt microbial membranes (Hancock and Rozek, 2002). The C-terminal hydrophobic helix of cecropins is believed to have a synergistic effect and provide selectivity against Gram-negative bacteria (Lee et al., 2015). Moricins, which possess an N-terminal amphipathic segment in the one large α-helix structure, were presumed to share a similar mechanism as that of the cecropins. The amphipathicity of the α-helices of the designed peptides was visualized via helical wheel projection (Supplementary Figure 5). Furthermore, C-terminal amidation is suggested to be a common modification of the cecropins and is presumed to contribute to the broad-spectrum antimicrobial activity of these peptides (Yi et al., 2014). Among the previously discovered butterfly cecropins, papiliocin from Papilio xuthus (Kim et al., 2010) and DAN1 and DAN2 from D. plexippus (Duwadi et al., 2018) were proposed to have amidated C-termini, although a non-Gly amino acid as the ultimate residue in DAN2 indicated a natural, non-amidated terminus. On the contrary, hinnavin I and hinnavin II (Yoe et al., 2006) from P. rapae were reported to have carboxyl C-termini, even if the presence of a Gly residue at the C-terminus of hinnavin II suggested a potential amidated modification. There were no apparent C-terminal amidation tendencies within the naturally occurring butterfly cecropins examined in the current study (Table 1). Moreover, antimicrobial activities of the amidated analog of CecB were not augmented when compared with the natural prototype, which suggested that C-terminal amidation may not be required for cecropins derived from butterflies. A C-terminal amidation modification has also been observed in a group of natural butterfly defensins, and this modification was relatively rare among other species (Bruhn et al., 2009). C-terminal amidation could be postulated to neutralize the negative charge and increase resistance to exopeptidase hydrolyzation. Further studies are required to elucidate the underlying functions of C-terminal amidation of AMPs. A more conserved character of the defensins is the Cys-stabilized αβ (CSαβ) motif signature, which is considered as the main functional structural basis, despite certain mammalian defensins lacking α-helices (de Oliveira Dias and Franco, 2015). Moreover, the positively charged segment at the C-termini of moricins is the most distinct feature distinguishing them from other α-helical AMPs. Although tetra- and penta-peptide motifs at the positively charged part of moricin have been proposed in other species (Hemmi et al., 2002; Oizumi et al., 2005), the butterfly moricin sequences showed diverse residual propensities in this region. Combined with the confocal microscopy and CD spectroscopy results, which demonstrated that moricins could penetrate the cell membrane and bind to the cell nucleus (Figure 6 and Supplementary Figure 3), it could be arguably hypothesized that the basic amino acid cluster significantly increased the net charge of moricins and hence facilitated the interaction with the negatively charged nucleic acids.
AMPs are active against a broad range of microorganisms, but selectivity is also evident toward respective microbes. The activity of cecropins is thought to be particularly directed against Gram-negative bacteria, while that of defensins and moricins is mainly targeted toward Gram-positive bacteria (Hara and Yamakawa, 1995). However, the in vitro antimicrobial assay in the current study revealed that all designed peptides, particularly cecropins, exerted potent but generally undifferentiated activities against Gram-negative and Gram-positive bacteria, with cecropins and moricin exhibiting slightly higher antimicrobial preference against Gram-negative bacteria and defensins showing a stronger effect on Gram-positive bacteria (Table 3). It was not surprising that the designed moricin exhibited predominant antimicrobial activity against Gram-negative bacteria, since the high amphipathicity and net charge of this class of AMP are thought to be positively associated with the activities toward Gram-negative bacteria (Wang et al., 2017). In accordance with this, both SEM and TEM illustrated that the designed peptides in the current study could exert their bactericidal activities through a mechanism of membrane binding and disruption. Furthermore, the synthetic cecropins and moricin, especially the latter, also demonstrated antifungal potential through a possible DNA-binding mechanism. The in vivo experiment provided further evidence that the designed peptide CecB had important therapeutic functions in bacterial elimination, while also showing that the synthetic MorA could inhibit fungal growth to some extent (Figure 7). These preliminary findings suggest that the synthetic AMPs, primarily CecB and MorA, are promising antimicrobial agents that warrant further investigation. All the synthetic sequences generated in the current study demonstrated potent antimicrobial activities, which further supports the rationality and necessity of selecting sequences and structures that have been phylogenetically conserved through evolution. The characteristics of natural butterfly AMPs could be explored further through more extensive sequence and structural optimization as well as considering the possibility of peptide expression. Furthermore, the lower antimicrobial activity of defensins does not mean that these AMPs are less important; there is accumulating evidence for the existence of immunoregulatory properties of defensins on host immunity and resultant therapeutic potential, as well as the occurrence of functional synergies between a defensin and other AMPs or antimicrobial agents (Pazgier et al., 2007; Lazzaro et al., 2020). Examination of these theories was beyond the scope of the current study.
The limited numbers of representative microorganisms applied in the current study mean the antimicrobial spectrum and bioactivities of the AMPs require further evaluation to obtain a more detailed characterization of these molecules. For example, AMPs have been found that can target and lyse phosphatidylserine (PS), thereby exposing negatively charged tumor cells, and these AMPs are even active against drug-resistant tumor cell variants and may not affect their healthy counterparts (Riedl et al., 2011). In addition, some AMPs were reported to inhibit and degrade biofilm formation by coating the bacterium or the biomaterial surface (Segev-Zarko et al., 2015); however, the mechanism of antibiofilm activity was not investigated in this study. AMPs have also demonstrated antiviral effects on different viruses, ranging from influenza to human immunodeficiency virus (HIV), through multiple mechanisms (Vilas Boas et al., 2019). Antiviral effects of the synthetic AMPs were not explored in the current study. Despite these limitations, the current study provides valuable information on butterfly AMPs and an efficient and practical methodology for AMP screening and design.
This study presented an overview of the landscape of butterfly AMPs and insights into their diverse sequence and structural features. Based on these sequences, a series of potent and wide-spectrum peptides with respective antimicrobial mechanisms were successfully designed. This study not only provided a practical strategy for high-throughput natural AMP discoveries but also produced a rational methodology for AMP design, and this will be of importance for future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Peking Union Medical College Hospital Animal Care and Use Committee, Peking Union Medical College Hospital.
MW and XH conceived the study. WZ, SL, and XH performed the in silico experiment. MW and ZZ carried out the wet lab experiment. MW prepared the draft manuscript. XH reviewed the draft manuscript. All authors read and approved the final manuscript.
This work was funded by the National Natural Science Foundation of China (Nos. 81801635 and 81803848) and China Postdoctoral Science Foundation (No. 2017M610812). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.720381/full#supplementary-material
Supplementary Figure 1 | Schematic illustration of the experimental design.
Supplementary Figure 2 | HPLC profiles and spectrometric data of the synthetic AMPs.
Supplementary Figure 3 | Evaluation of hemolytic and cytotoxic activities of synthetic AMPs. The hemolysis effects (left) and cytotoxicity on normal cells (right) of the synthetic peptides at different concentration were tested on mice erythrocytes and HEK293 cells, respectively. PBS and Triton-X100 were used as the negative and positive control. No apparent hemolysis or cytotoxicity was detected among groups.
Supplementary Figure 4 | CD spectrum of the designed AMPs interacted with fungal genomic DNA. The top curve corresponded to free DNA, while the lower curves were that mixed with respective AMPs. The decreased intensities of genomic DNA molar ellipticity exhibited occurrence conformational alteration of DNA that induced by AMPs, which suggested a DNA-binding mechanism of the designed sequences.
Supplementary Figure 5 | Helical wheel projection analysis of the designed peptide. (A) N-terminal amphiphilic helix of cecropin A; (B) C-terminal hydrophobic helix of cecropin A; (C) N-terminal amphiphilic helix of cecropin B; (D) C-terminal hydrophobic helix of cecropin B; (E) helical segment of defensin A; (F) helical segment of defensin B; (G) helical segment of moricin A.
Supplementary Table 1 | Papilionoidea species and their high-throughput sequencing data used in this study.
Supplementary Table 2 | The AMP sequences for phylogenetic analysis in this study.
Supplementary Table 3 | Sequence alignment of the synthetic and naturally occurring AMPs.
Abouelhassan, Y., Yang, Q., Yousaf, H., Nguyen, M. T., Rolfe, M., Schultz, G. S., et al. (2017). Nitroxoline: a broad-spectrum biofilm-eradicating agent against pathogenic bacteria. Int. J. Antimicrob. Agents 49, 247–251. doi: 10.1016/j.ijantimicag.2016.10.017
Badapanda, C., and Chikara, S. K. (2016). “Lepidopteran antimicrobial peptides (AMPs): overview, regulation, modes of action, and therapeutic potentials of insect-derived AMPs,” in Short Views on Insect Genomics and Proteomics: Insect Proteomics, Vol.2 Entomology in Focus, eds C. Raman, M. R. Goldsmith, and T. A. Agunbiade (Cham: Springer International Publishing), 141–163. doi: 10.1007/978-3-319-24244-6_6
Boman, H. G., Boman, I. A., Andreu, D., Li, Z. Q., Merrifield, R. B., Schlenstedt, G., et al. (1989). Chemical synthesis and enzymic processing of precursor forms of cecropins A and B. J. Biol. Chem. 264, 5852–5860. doi: 10.1016/S0021-9258(18)83628-6
Bruhn, O., Paul, S., Tetens, J., and Thaller, G. (2009). The repertoire of equine intestinal alpha-defensins. BMC Genomics 10:631. doi: 10.1186/1471-2164-10-631
Bulet, P., Stöcklin, R., and Menin, L. (2004). Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198, 169–184. doi: 10.1111/j.0105-2896.2004.0124.x
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
de Oliveira Dias, R., and Franco, O. L. (2015). Cysteine-stabilized αβ defensins: from a common fold to antibacterial activity. Peptides 72, 64–72. doi: 10.1016/j.peptides.2015.04.017
Duwadi, D., Shrestha, A., Yilma, B., Kozlovski, I., Sa-Eed, M., Dahal, N., et al. (2018). Identification and screening of potent antimicrobial peptides in arthropod genomes. Peptides 103, 26–30. doi: 10.1016/j.peptides.2018.01.017
Garbett, N. C., Ragazzon, P. A., and Chaires, J. B. (2007). Circular dichroism to determine binding mode and affinity of ligand–DNA interactions. Nat. Protoc. 2, 3166–3172. doi: 10.1038/nprot.2007.475
Greber, K. E., and Dawgul, M. (2017). Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 17, 620–628. doi: 10.2174/1568026616666160713143331
Hale, J. D. F., and Hancock, R. E. W. (2007). Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert. Rev. Anti. Infect. Ther. 5, 951–959. doi: 10.1586/14787210.5.6.951
Hancock, R. E. W., and Rozek, A. (2002). Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206, 143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x
Hara, S., and Yamakawa, M. (1995). Moricin, a novel type of antibacterial peptide isolated from the silkworm Bombyx Mori. J. Biol. Chem. 270, 29923–29927. doi: 10.1074/jbc.270.50.29923
Hartmann, M., Berditsch, M., Hawecker, J., Ardakani, M. F., Gerthsen, D., and Ulrich, A. S. (2010). Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 54, 3132–3142. doi: 10.1128/AAC.00124-10
Hazam, P. K., Goyal, R., and Ramakrishnan, V. (2019). Peptide based antimicrobials: design strategies and therapeutic potential. Prog. Biophys. Mol. Biol. 142, 10–22. doi: 10.1016/j.pbiomolbio.2018.08.006
Hemmi, H., Ishibashi, J., Hara, S., and Yamakawa, M. (2002). Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori. FEBS Lett. 518, 33–38. doi: 10.1016/S0014-5793(02)02637-6
Jasper, H., and Bohmann, D. (2002). Drosophila innate immunity: a genomic view of pathogen defense. Mol. Cell 10, 967–969. doi: 10.1016/S1097-2765(02)00752-9
Jin, L., Bai, X., Luan, N., Yao, H., Zhang, Z., Liu, W., et al. (2016). A designed tryptophan- and lysine/arginine-rich antimicrobial peptide with therapeutic potential for clinical antibiotic-resistant Candida albicans vaginitis. J. Med. Chem. 59, 1791–1799. doi: 10.1021/acs.jmedchem.5b01264
Kawahara, A. Y., and Breinholt, J. W. (2014). Phylogenomics provides strong evidence for relationships of butterflies and moths. Proc. R. Soc. B. 281:20140970. doi: 10.1098/rspb.2014.0970
Kim, K.-H., and Seong, B. L. (2001). Peptide amidation: production of peptide hormonesin vivo andin vitro. Biotechnol. Bioprocess Eng. 6, 244–251. doi: 10.1007/BF02931985
Kim, S. R., Hong, M. Y., Park, S. W., Choi, K. H., Yun, E. Y., Goo, T. W., et al. (2010). Characterization and cDNA cloning of a cecropin-like antimicrobial peptide, papiliocin, from the swallowtail butterfly Papilio xuthus. Mol. Cells 29, 419–423. doi: 10.1007/s10059-010-0050-y
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Landon, C., Barbault, F., Legrain, M., Menin, L., Guenneugues, M., Schott, V., et al. (2004). Lead optimization of antifungal peptides with 3D NMR structures analysis. Protein Sci. 13, 703–713. doi: 10.1110/ps.03404404
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lazzaro, B. P., Zasloff, M., and Rolff, J. (2020). Antimicrobial peptides: application informed by evolution. Science 368:eaau5480. doi: 10.1126/science.aau5480
Lear, S., and Cobb, S. L. (2016). Pep-Calc.com: a set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J. Comput. Aided Mol. Des. 30, 271–277. doi: 10.1007/s10822-016-9902-7
Lee, E., Kim, J. K., Jeon, D., Jeong, K. W., Shin, A., and Kim, Y. (2015). Functional roles of aromatic residues and helices of papiliocin in its antimicrobial and anti-inflammatory activities. Sci. Rep. 5:12048. doi: 10.1038/srep12048
Lee, P. H. A., Ohtake, T., Zaiou, M., Murakami, M., Rudisill, J. A., Lin, K. H., et al. (2005). Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc Natl Acad Sci U S A 102, 3750–3755. doi: 10.1073/pnas.0500268102
Lin, M.-C., Hui, C.-F., Chen, J.-Y., and Wu, J.-L. (2013). Truncated antimicrobial peptides from marine organisms retain anticancer activity and antibacterial activity against multidrug-resistant Staphylococcus aureus. Peptides 44, 139–148. doi: 10.1016/j.peptides.2013.04.004
Liu, H., Lei, M., Du, X., Cui, P., and Zhang, S. (2015). Identification of a novel antimicrobial peptide from amphioxus Branchiostoma japonicum by in silico and functional analyses. Sci. Rep. 5:18355. doi: 10.1038/srep18355
Machado, L. R., and Ottolini, B. (2015). An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Front. Immunol. 6:115. doi: 10.3389/fimmu.2015.00115
Manniello, M. D., Moretta, A., Salvia, R., Scieuzo, C., Lucchetti, D., Vogel, H., et al. (2021). Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell Mol. Life Sci. 78, 4259–4282. doi: 10.1007/s00018-021-03784-z
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2020). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Oizumi, Y., Hemmi, H., Minami, M., Asaoka, A., and Yamakawa, M. (2005). Isolation, gene expression and solution structure of a novel moricin analogue, antibacterial peptide from a lepidopteran insect, Spodoptera litura. Biochim. Biophys. Acta 1752, 83–92. doi: 10.1016/j.bbapap.2005.07.013
Pazgier, M., Li, X., Lu, W., and Lubkowski, J. (2007). Human defensins: synthesis and structural properties. Curr. Pharm. Des. 13, 3096–3118. doi: 10.2174/138161207782110381
Riedl, S., Rinner, B., Asslaber, M., Schaider, H., Walzer, S., Novak, A., et al. (2011). In search of a novel target — Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta (BBA) Biomembr. 1808, 2638–2645. doi: 10.1016/j.bbamem.2011.07.026
Roope, L. S. J., Smith, R. D., Pouwels, K. B., Buchanan, J., Abel, L., Eibich, P., et al. (2019). The challenge of antimicrobial resistance: what economics can contribute. Science 364:eaau4679. doi: 10.1126/science.aau4679
Segev-Zarko, L., Saar-Dover, R., Brumfeld, V., Mangoni, M. L., and Shai, Y. (2015). Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem. J. 468, 259–270. doi: 10.1042/BJ20141251
Seufi, A. M., Hafez, E. E., and Galal, F. H. (2011). Identification, phylogenetic analysis and expression profile of an anionic insect defensin gene, with antibacterial activity, from bacterial-challenged cotton leafworm, Spodoptera littoralis. BMC Mol. Biol. 12:47. doi: 10.1186/1471-2199-12-47
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013
Triant, D. A., Cinel, S. D., and Kawahara, A. Y. (2018). Lepidoptera genomes: current knowledge, gaps and future directions. Curr. Opin. Insect. Sci. 25, 99–105. doi: 10.1016/j.cois.2017.12.004
Unckless, R. L., and Lazzaro, B. P. (2016). The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150291. doi: 10.1098/rstb.2015.0291
Vilas Boas, L. C. P., Campos, M. L., Berlanda, R. L. A., de Carvalho Neves, N., and Franco, O. L. (2019). Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 76, 3525–3542. doi: 10.1007/s00018-019-03138-w
Wang, C.-K., Shih, L.-Y., and Chang, K. Y. (2017). Large-scale analysis of antimicrobial activities in relation to amphipathicity and charge reveals novel characterization of antimicrobial peptides. Molecules 22:E2037. doi: 10.3390/molecules22112037
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wang, M., and Hu, X. (2017). Antimicrobial peptide repertoire of Thitarodes armoricanus, a host species of Ophiocordyceps sinensis, predicted based on de novo transcriptome sequencing and analysis. Infect. Genet. Evol. 54, 238–244. doi: 10.1016/j.meegid.2017.07.011
Wang, M., Li, Y., and Hu, X. (2018). Chebulinic acid derived from triphala is a promising antitumour agent in human colorectal carcinoma cell lines. BMC Comp. Altern. Med. 18:342. doi: 10.1186/s12906-018-2412-5
Wang, M., Wang, L., Chen, T., Walker, B., Zhou, M., Sui, D., et al. (2012). Identification and molecular cloning of a novel amphibian Bowman Birk-type trypsin inhibitor from the skin of the Hejiang Odorous Frog Odorrana hejiangensis. Peptides 33, 245–250. doi: 10.1016/j.peptides.2012.01.003
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wiegand, I., Hilpert, K., and Hancock, R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Xu, X., Zhong, A., Wang, Y., Lin, B., Li, P., Ju, W., et al. (2019). Molecular identification of a moricin family antimicrobial peptide (px-mor) from plutella xylostella with activities against the opportunistic human pathogen Aureobasidium pullulans. Front. Microbiol. 10:2211. doi: 10.3389/fmicb.2019.02211
Yi, H.-Y., Chowdhury, M., Huang, Y.-D., and Yu, X.-Q. (2014). Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 98, 5807–5822. doi: 10.1007/s00253-014-5792-6
Yoe, S. M., Kang, C. S., Han, S. S., and Bang, I. S. (2006). Characterization and cDNA cloning of hinnavin II, a cecropin family antibacterial peptide from the cabbage butterfly, Artogeia rapae. Comp. Biochem. Physiol. Part B 144, 199–205. doi: 10.1016/j.cbpb.2006.02.010
Zharkova, M. S., Orlov, D. S., Golubeva, O. Yu, Chakchir, O. B., Eliseev, I. E., Grinchuk, T. M., et al. (2019). Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? Front. Cell Infect. Microbiol. 9:128. doi: 10.3389/fcimb.2019.00128
Keywords: antimicrobial peptide, butterfly, bioinformatics, identification, characterization
Citation: Wang M, Zhou Z, Li S, Zhu W and Hu X (2021) Identification and Characterization of Antimicrobial Peptides From Butterflies: An Integrated Bioinformatics and Experimental Study. Front. Microbiol. 12:720381. doi: 10.3389/fmicb.2021.720381
Received: 04 June 2021; Accepted: 21 July 2021;
Published: 26 August 2021.
Edited by:
Marc Maresca, Aix-Marseille Université, FranceReviewed by:
Séverine Zirah, Muséum National d’Histoire Naturelle, FranceCopyright © 2021 Wang, Zhou, Li, Zhu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Wang, dml2aWFuLTA4MTUyMDAzQGhvdG1haWwuY29t; Xianda Hu, aGVsbG9jZWFuQGhvdG1haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.