
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 August 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.715802
This article is part of the Research TopicWhat Is known and What Remains To Be Discovered About Bacterial Outer Membrane Vesicles, Volume IIView all 10 articles
 Irina V. Kudryakova1
Irina V. Kudryakova1 Alexey S. Afoshin1
Alexey S. Afoshin1 Tanya V. Ivashina2
Tanya V. Ivashina2 Natalia E. Suzina3
Natalia E. Suzina3 Elena A. Leontyevskaya1
Elena A. Leontyevskaya1 Natalia V. Leontyevskaya (Vasilyeva)1*
Natalia V. Leontyevskaya (Vasilyeva)1*Outer membrane vesicles (OMVs) produced by Gram-negative bacteria constitute important factors in defining interactions with the extracellular milieu. Lysobacter sp. XL1 produces OMVs capable of lysing microbial cells due to the presence in their cargo of bacteriolytic protease L5 (AlpB). Although protein L5 has been functionally and biochemically characterized (including aspects of its packing into OMVs), its role in vesicle biogenesis through genetic deletion of alpB had not been studied previously. Here, we have successfully deleted alpB by allelic replacement and show that the alpB deletion mutant produces a significantly lower amount of OMVs that lack bacteriolytic activity and display altered ultrastructural characteristics in relation to the OMVs produced by the wild-type strain. These results confirm that, as previously proposed, protein L5 participates in OMV production through a mechanism that is not yet fully understood.
Outer membrane vesicles (OMVs) are structures 50–300 nm in diameter that all Gram-negative bacteria form from their outer membrane (OM). OMVs perform a number of vital activities in relation to nutrient uptake, microbe–microbe interactions (including horizontal gene transfer), pathogen–host interactions, symbiotic interactions, and cell protection like phage and toxin release (Mashburn-Warren and Whiteley, 2006; Avila-Calderón et al., 2015; Schwechheimer and Kuehn, 2015; Guerrero-Mandujano et al., 2017; Nagakubo et al., 2020). In light of this, the topicality of vesicle studies is evident.
At present, one of the most intriguing issues in vesicle subject matter is OMV biogenesis. At first, vesicles were considered to form due to spontaneous evagination of the OM, followed by the release of these fragments (Knox et al., 1966). Later on, three models of vesicle biogenesis were formulated. According to the first model, vesicles form in the region of a decreased concentration of lipoproteins (Hoekstra et al., 1976; Deatherage et al., 2009; Schwechheimer et al., 2014). The second model explains the presence of cell debris [misfolded proteins, peptidoglycan and lipopolysaccharide (LPS) fragments] in vesicles (Zhou et al., 1998; Hayashi et al., 2002; Schwechheimer et al., 2014). According to the third model, the formation of vesicles occurs under the action of curvature-inducing molecules (B-type LPS, signalling molecule 2-heptyl-3-hydroxy-4-quinolone (PQS) (Kadurugamuwa and Beveridge, 1995; Mashburn-Warren and Whiteley, 2006; Schertzer and Whiteley, 2012). Thus, all these models indicate that vesicles form from OM destabilization loci, and their contents are periplasmic components randomly captured in the process of vesicle formation.
With time, the view of vesicle biogenesis began to change. Data supporting a model for the specific sorting of vesicle components emerged (Horstman and Kuehn, 2000; Kato et al., 2002; Haurat et al., 2011, 2015; Schwechheimer and Kuehn, 2015). This contributed to the emergence of the theory of the formation of various vesicle groups (subpopulations) produced by one cell type (Olofsson et al., 2010; Rompikuntal et al., 2012; Kunsmann et al., 2015; Kudryakova et al., 2015; Afoshin et al., 2020). It should be noted that this theory enables understanding the ability of vesicles to perform various vital activities of the bacterial cell. It is evident that a relation between vesicle biogenesis processes and component sorting is bound to exist. It is hypothesized that these components themselves are factors contributing to vesicle biogenesis. We addressed this hypothesis by studying of vesicle biogenesis in Lysobacter sp. XL1.
The Gram-negative bacterium Lysobacter sp. XL1 has been studied at our laboratory for about 45 years. This bacterium is a potent producer of a complex of extracellular bacteriolytic enzymes. To date, five bacteriolytic enzymes (L1–L5) have been isolated and partially characterized. By the specificity of their action on bacterial peptidoglycans, the lytic enzymes of Lysobacter sp. XL1 are endopeptidases (L1, L4, L5), amidase (L2), and a muramidase (L3). The bacteriolytic complex is highly efficient in breaking down competitive cells of bacteria, yeasts, mycelial fungi and some protozoa (Kulaev et al., 2006). This property makes Lysobacter sp. XL1 an active participant of microbial biocenoses in nature and a biotechnologically promising strain producing novel antimicrobial agents. In 2008, we found Lysobacter sp. XL1 to form vesicles that lysed cells of Gram-positive bacteria, yeasts, mycelial fungi and possessed a curative action with respect to staphylococcal and anthrax infections. The lytic action of vesicles is determined by bacteriolytic enzyme L5, which is a component of their cargo (Vasilyeva et al., 2008, 2014). Research into the OMV biogenesis of Lysobacter sp. XL1 found their formation to occur in OM segments enriched with cardiolipin (Kudryakova et al., 2017). Fractionation of vesicles revealed a group/subpopulation of vesicles containing protein L5. Electron immunocytochemistry of Lysobacter sp. XL1 cell sections established protein L5 to concentrate in certain loci of the periplasm at the inner leaflet of the OM; it is from these loci that vesicles form (Kudryakova et al., 2015). We hypothesized that protein L5 could participate in vesicle biogenesis. To study this, the gene of protein L5 (alpB) was expressed in cells of a phylogenetically close genus Pseudomonas fluorescens Q2-87/B. It was found that in the recombinant strain, protein L5 was contained in vesicles. Cells of the recombinant strain were shown to form a larger number of vesicles as compared with the parent strain (Kudryakova et al., 2017). All these data indirectly confirmed the involvement of bacteriolytic protein L5 in the biogenesis of that group of vesicles by means of which it was released into the extracellular milieu. To substantiate this model, the goal of the present study to investigate OMV production by an alpB mutant strain compared with that of the wild-type strain. We have deleted alpB in Lysobacter sp. XL1, which in itself is an important experimental accomplishment mainly because the ill-developed molecular tools available for conducting genetic studies on Lysobacter had not previously permitted gene knockouts. Here, we report that the quantity of vesicles produced by the mutant strain was significantly lower than that of the wild-type strain and the lytic properties of mutant strain vesicles were lost practically completely, confirming that protein L5 influences OMV formation.
All bacterial strains and plasmids used are listed in Table 1.
Escherichia coli strain XL1-Blue used for molecular cloning was grown in a Luria–Bertani (LB) medium containing (g/l): tryptone, 10; yeast extract, 5; NaCl, 10; pH 7.0 at 37°C. Wild-type Lysobacter sp. XL1, Lysobacter sp. XL1ΔalpB::tet, and Lysobacter sp. XL1ΔalpB::alpB strains were grown at 29°C in modified LB (LB-M) broth (in g/l: peptone, 5; yeast extract, 5; NaCl, 5; pH 7.5). Lysobacter cultures were grown for 20 h, which corresponds to the end of the exponential growth phase. Antibiotics were added as necessary at the following final concentrations (μg/ml): for E. coli: tetracycline (Tc), 10; gentamicin (Gm), 10; ampicillin (Ap), 100; for Lysobacter sp. XL1: Tc, 10; Gm, 20. For screening clones that had undergone double crossover events, sucrose was added to a final concentration of 10%. When plates of solid media were required, agar at a concentration of 15 g/L was added to the corresponding broths.
Bacterial cultures used in lysis tests (Staphylococcus aureus 209P, Micrococcus luteus B-1813, Micrococcus roseus B-1236, and Bacillus cereus B-454) were grown at 29°C as confluent lawns on plates of IBPM RAS medium (in g/l: yeast extract, 1; soybean extract, 30; tryptone, 5; aminopeptide, 60; agar, 15; pH 7.2).
DNA recombinant techniques were performed according to standard procedures (Sambrook and Russell, 2001). All restriction endonucleases, T4 polynucleotide kinase, shrimp alkaline phosphatase, and T4 DNA ligase were obtained from Thermo Fisher Scientific (United States) and used according to the manufacturer’s recommendations. TaqSE DNA polymerase was from SybEnzyme (Russia), and Q5 high-fidelity DNA polymerase was purchased from New England Biolabs (United States). Plasmid DNA was isolated and purified using a Quantum Prep Plasmid Miniprep Kit (Bio-Rad, United States). The PCR reactions (total volume is 50 μl) were conducted in the following conditions: 200 μM dNTPs, 0.5 μM forward and reverse primers, 0.05 ng/μl Lysobacter sp. XL1 genomic DNA (for amplification of a 924-bp DNA fragment containing the 3′ end of alpB and downstream region, an 825-bp DNA fragment containing the 5′ end of alpB and upstream region and the full-length alpB gene with downstream and upstream genome sequences) or 7 ng/μl pBR322 plasmid (for amplification of 1.43 kb TcR cassette), 0.02 U/μl Q5 high-fidelity DNA polymerase (New England Biolabs) in 1 × reaction buffer containing 2 mM MgCl2. The thermo cycles were programmed according to the manufacturer’s protocol: initial denaturation at 98°C for 30 s, followed by 30 cycles of 98°C for 10 s, annealing temperature 60°C for 20 s, 72°C for time that was determined by amplicon length (extension times are 30 s per kb), and a final extension at 72°C for 2 min. Amplicons were separated in gels of 0.8% agarose (Merck, Germany) in Tris-acetate-ethylenediaminetetraacetic acid (TAE) buffer containing 0.5 μg/ml ethidium bromide, visualized in a Bio-Print ST4 gel documentation system (Vilber lourmat, France), at 354 nm, and purified by QIAquick spin-column (Qiagen, United States). The concentration of DNA was measured using NanoPhotometer P360 (IMLEN, Germany) and by electrophoretic assay in 0.8% agarose gel.
Deletion of alpB was done by allelic replacement using a suicide plasmid construct (Figure 1). Briefly, a 924-bp DNA fragment containing the 3′ end of alpB and its downstream region was amplified from the genomic DNA of Lysobacter sp. XL1 with a pair of primers L5_SacI(for) and L5_SmaI(rev). All primers are listed in Supplementary Table 1. The resulting amplicon was ligated into SacI/SmaI-digested suicide vector pJQ200SK to form pJQ200SKΔ5′alpB. Subsequently, an 825-bp DNA fragment containing the 5′ end of alpB and its upstream region was amplified with primers L5_SmaI(for) and L5_XhoI(rev) and ligated into SmaI/XhoI-digested pJQ200SKΔ5′alpB to give pJQ200SKΔalpB. In pJQ200SKΔalpB, the alpB gene carries an 880-bp deletion in its central region. Finally, the deletion allele was marked by the insertion of a 1.43-kb TcR cassette amplified from pBR322 with primers Tc(for) and Tc(rev), which was phosphorylated with T4 polynucleotide kinase and ligated into SmaI-digested pJQ200SKΔalpB pretreated with shrimp alkaline phosphatase to produce the suicide vector plasmid pJQ200SKΔalpB::tet (Figure 1 and Supplementary Figure 1A).
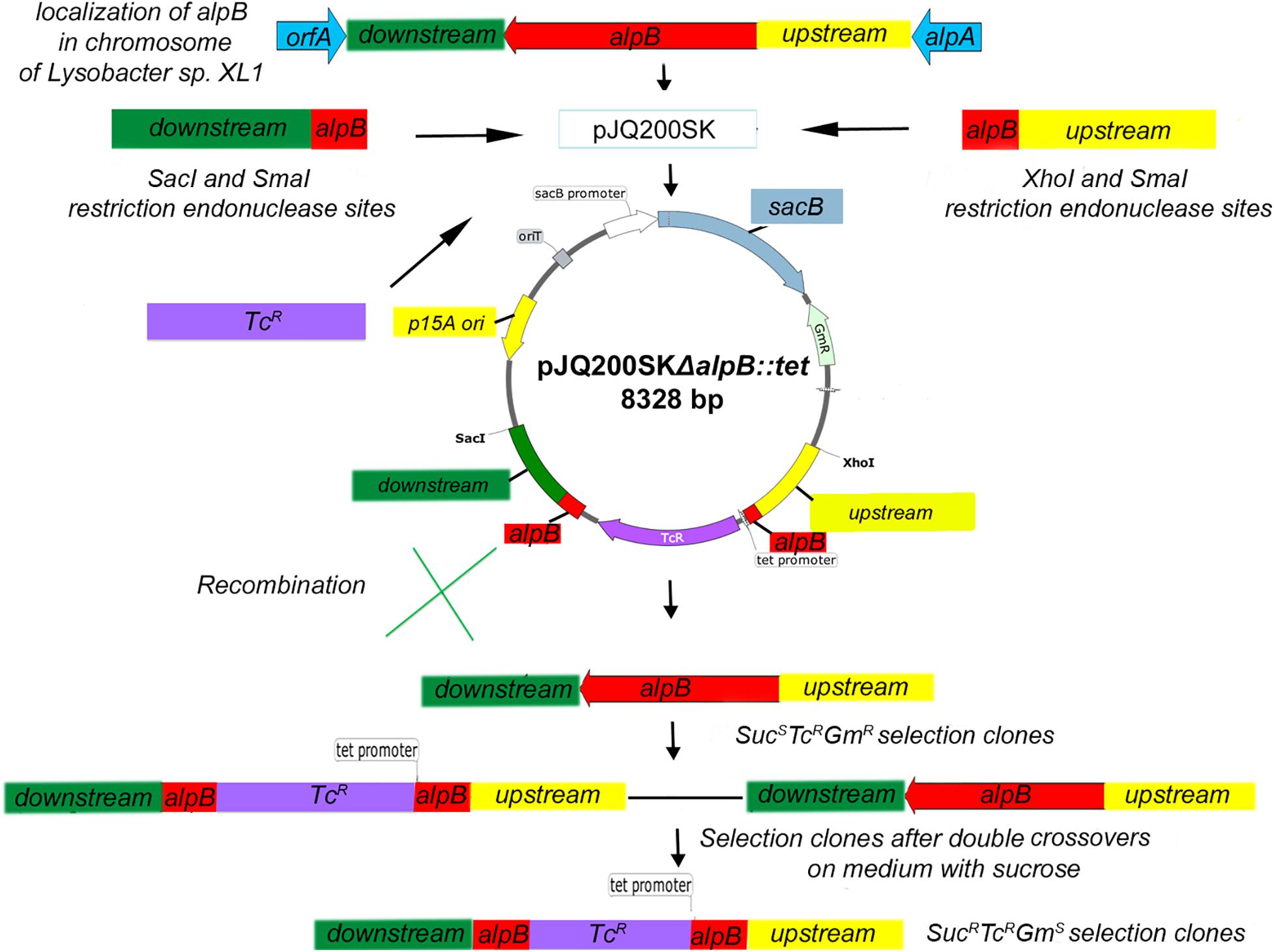
Figure 1. Scheme of inserting a mutation into the alpB gene of Lysobacter sp. XL1. The pJQ200SKΔalpB::tet recombinant plasmid was introduced into cells of Lysobacter sp. XL1 via electroporation. Integration of the plasmid into the chromosome was testified by the resistance of transformants to gentamicin (plasmid marker) and tetracycline (cassette marker inserted into the deletion locus) and by the sensitivity to sucrose. As a result of merodiploid resolution, SucRTcRGmS clones with double crossovers between the mutant and wild-type alleles of the alpB gene were selected.
To replace the alpB::tet allele in the deletion mutant with the wild-type alpB gene, the full-length alpB gene (which is 1,200 bp long) plus its immediate downstream and upstream sequences was amplified using primers L5_SacI(for) and L5_XhoI(rev) followed by ligation of the amplicon into the SacI/XhoI-digested pJQ200SK to yield pJQ200SKΔalpB::alpB (Supplementary Figure 1B). All cloned DNA fragments were sequenced at the Evrogen JSC (Russia) to confirm the correctness of inserts and the absence of random mutations.
E. coli cells were transformed by the RbCl method according to Hanahan (1983). Lysobacter sp. XL1 electro-competent cells were prepared according to Lin and McBride (1996). Briefly, 20-μl aliquots of Lysobacter sp. competent cells in 10% ice-cold glycerol were mixed with 250 ng of pJQ200SKΔalpB::tet or pJQ200SKΔalpB::alpB plasmid DNA in 2-mm gapped cuvettes (Bio-Rad, United States). Electroporation was performed using a Gene Pulser apparatus (Bio-Rad, United States) under conditions at 12.5 kV/cm. Immediately after the electric pulse, 1 ml of an LB-M medium was added to the pulsed bacterial cell suspension, which was then incubated for 3 h at 29°C without agitation, and subsequently plated on solid medium containing the appropriate antibiotics.
After electroporation of pJQ200SKΔalpB::tet into Lysobacter sp. XL1 (or of pJQ200SKΔalpB::alpB into the Lysobacter sp. XL1 alpB::tet mutant), single crossover (SCO) clones with the SucSTcRGmR phenotype were screened (Figure 1). Double crossover clones (DCOs) with the SucRTcRGmS phenotype were counter-selected after growing one SCO clone in LB-M broth containing sucrose for 3 h at 29°C, followed by plating and cultivation for 72 h at 29°C on LB-M agar containing Tc. DCOs were confirmed by PCR for the presence of the wild-type or deletion allele using primers in Supplementary Table 1, which amplify a 2.62-kb product from the wild-type allele, or a 3.17-kb product from the mutant allele. The sites of recombination in DCOs were confirmed by sequencing.
OMVs were isolated by ultracentrifugation from equal volumes of the culture liquids of Lysobacter sp. XL1, Lysobacter sp. XL1ΔalpB::tet, and Lysobacter sp. XL1ΔalpB::alpB strains grown in LB-M liquid medium. Briefly, cells from a 0.3-L culture were pelleted by centrifugation at 7,500 × g for 20 min at 4°C, and the supernatant was recovered. Vesicles were then pelleted from 300-ml supernatants by centrifugation in an L5-50 ultracentrifuge (Beckman, United States) at 113,000 × g for 2 h at 4°C. The OMV pellet was washed once with 50 mM Tris-HCl, pH 8.0, by resuspension and centrifugation at the same speed for 1 h. The washed vesicle pellet was resuspended in 600 μl of 50 mM Tris-HCl, pH 8.0, and stored at –20°C.
For the electrophoretic assay, preparations of wild-type Lysobacter sp. XL1 OMVs and Lysobacter sp. XL1ΔalpB::tet OMVs were sedimented with trichloroacetic acid at a final concentration of 10%. Preparations of OMVs were equal and contained 0.04 mg of total proteins. Protein residues were analyzed by electrophoresis with 0.1% sodium dodecyl sulfate (SDS) in 12.5% polyacrylamide gel (PAG). Electrophoresis in stacking gel was run at 60 V; in separating gel, at 180 V. Protein bands in gels were revealed by staining with a solution of Coomassie Brilliant Blue R-250 (Serva, Germany). As molecular weight markers, we used SM0431 (Thermo Fisher Scientific, United States): β-galactosidase (116 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), lactate dehydrogenase (35 kDa), REase Bsp981 (25 kDa), β-lactoglobulin (18.4 kDa), and lysozyme (14.4 kDa).
Preparations of wild-type Lysobacter sp. XL1 OMVs and Lysobacter sp. XL1ΔalpB::tet OMVs were aligned by mass of protein (20 μg) for phospholipid extraction. Phospholipids were extracted from both preparations with chloroform–methanol mixture (1:2 v/v) according to the method of Ames (1968). Individual phospholipids were separated by two-dimensional chromatography on silica gel plates 60 F254 (HPTLC; Merck, Germany) using CHCl3–CH3OH–H2O (65:25:4 v/v) mixture as the first-dimension separation system and CHCl3–CH3OH–CH3COOH–H2O (40:7.5:6:1.8 v/v) as the second-dimension separation system. Phospholipids were visualized using molybdenum blue solution.
The total protein concentration in OMV preparations was measured by the Lowry method (Lowry et al., 1951) against a standard curve done with bovine serum albumin (Sigma, United States) in the 2–20 μg/ml range.
The concentration of 2-Keto-3-deoxyoctanate (Kdo, a core LPS component) was determined by the reaction with thiobarbituric acid, exactly as described by Karkhanis et al. (1978). The standard curve was done with an aqueous solution of Kdo (Sigma, United States) in the 1.85–29.60 μg/ml range.
The total bacteriolytic activities of culture supernatants were determined by turbidimetry as previously reported (Vasilyeva et al., 2014). Briefly, liquid cultures of Lysobacter strains in LB-M were centrifuged at 7,500 × g for 20 min at 4°C to pellet bacterial cells. Then, 25 μl of the clear supernatant were added to 975 μl of heat-killed cells of S. aureus 209P suspended to an OD540 = 0.5 in 10 mM Tris-HCl, pH 8.0, and incubated at 37°C for 5 min. The reaction was arrested by placing test tubes on ice, and the OD540 of the suspension was measured in a Beckman DU 730 (United States) spectrophotometer. A decrease in 0.01 optical units/min (at 37°C) was taken as a lytic unit (LU). Therefore, LUs were calculated by the following formula: {[0.5 (initial OD540 of suspension) – final OD540] × 1,000 μl (total reaction volume)}/[5 min (time of reaction) × 25 μl (volume of sample) × 0.01 (correction coefficient for OD reduction per minute)].
The lytic action of OMVs against live Gram-positive bacteria was determined by the spot assay with S. aureus 209P, M. roseus B-1236, M. luteus B-1813, and B. cereus B-454 (Vasilyeva et al., 2014). Three independent experiments were carried out. Vesicle preparations (as 12-μl aliquots) were applied on a pre-grown lawn of the target bacterium on an agar plate after the concentration of OMVs had been adjusted to have identical amounts (0.09 μg) of Kdo. Applied OMV samples were allowed to absorb into the lawn for 30 min, and plates were then incubated at 29°C overnight (16 h). The lytic action of the preparations was then assessed by simple observation where the presence of a clear lysis zone indicated strong activity, a hazy lysis zone indicated weak activity, and the absence of a lysis zone indicated no activity.
Samples of OMVs (obtained from two independent isolation runs) of Lysobacter sp. XL1, Lysobacter sp. XL1ΔalpB::tet, and Lysobacter sp. XL1ΔalpB::alpB were diluted fivefold in 50 mM Tris-HCl, pH 8.0, and 10 μl were placed on top of a formvar-coated copper grid. The applied sample was allowed to adsorb for 2 min, and sample excess was then removed using filter paper. After air-drying, the samples were stained with 10 μl of 0.3% aqueous solution of uranyl acetate (pH 4.0), placed on the grids, and immediately removed using filter paper. Negatively stained preparations were examined with a JEM-1400 transmission electron microscope (JEOL, Japan) at an accelerating voltage of 80 kV, and random images of representative fields of observation were captured with an 11-Megapixel TEM Camera MORADA G2 (EMSIS GmbH, Germany). All micrographs were done with 25,000 magnification. The six micrographs were obtained from each experiment. All vesicles of wild-type Lysobacter sp. XL1, Lysobacter sp. XL1ΔalpB::tet, and Lysobacter sp. XL1ΔalpB::alpB in all obtained micrographs were studied. Ultrastructural observations, including various degrees of density (full and empty vesicles), and measurements were done on the acquired images. The ratio of full/empty vesicles was estimated by visual analysis of micrographs: full OMVs have compact packaging or local crystal packing, and in the case of the empty OMVs, it was possible to distinguish the backing film of the grid.
All experiments were in triplicates. Statistical analysis was done using GraphPad Prism (version 9.1.0; GraphPad Software, San Diego, CA). The analytical results were compared by Student’s t-test. Data are presented as the mean ± standard deviation. Statistical significance was considered at p < 0.05.
The deletion of alpB in Lysobacter sp. XL1ΔalpB::tet was genetically confirmed by the production of a 3,177-bp PCR amplicon (Figure 2) that was subsequently sequenced to show that the TcR cassette had indeed replaced, as expected, the region between 117 and 997 bp of the alpB gene. Lysobacter sp. XL1ΔalpB::tet grew well in LB-M (Table 2), indicating that alpB is not an essential gene and does not affect the gross overall physiology of the bacterium. The mutation was confirmed biochemically and functionally by the reduction in bacteriolytic activity of the mutant strain (Table 2 and Supplementary Figure 4). Since the total lytic activity of Lysobacter sp. XL1 is attributed to several secreted bacteriolytic enzymes (reviewed by Vasilyeva et al., 2013), it seems reasonable to surmise that the observed difference in lytic activities between Lysobacter sp. XL1 and Lysobacter sp. XL1ΔalpB::tet corresponds to the activity contributed by protein L5 (AlpB).
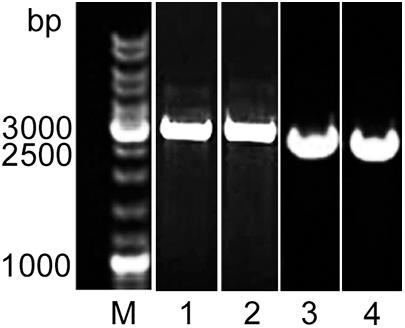
Figure 2. Molecular evidence of the deletion of alpB in Lysobacter XL1. Agarose gel stained with ethidium bromide after the electrophoresis of PCR amplicons produced with primers L5_SacI(for) and L5_XhoI(rev). The PCR amplicon for the mutant allele is 3,177 bp long (including alpB flanking sequences + the TcR cassette), whereas the wild-type allele produces a 2,623-bp product (including the full-length alpB gene + adjacent regions). M, SM0331 GeneRuler DNA Ladder Mix (Thermo Fisher Scientific, United States) (lane taken from Supplementary Figure 3); (1) Lysobacter sp. XL1ΔalpB::tet clone (from Supplementary Figure 2); (2) pJQ200SKΔalpB::tet plasmid (from Supplementary Figure 2); (3) Lysobacter sp. XL1ΔalpB::alpB clone (from Supplementary Figure 3); and (4) Lysobacter sp. XL1 clone (from Supplementary Figure 3).
Vesicle preparations obtained from equal volumes of the culture liquids of the wild-type, complemented strain, and mutant strains were analyzed by TEM and characterized biochemically.
In the preparation of mutant strain vesicles, the predominant species are larger vesicles 100–150 nm (47%) and 150–190 nm (30%) in diameter (Figure 3B and Supplementary Figure 5A). Vesicles 50–100 nm in diameter are few (14%). Vesicles in the preparation produced from the wild-type culture liquid are predominantly 50–100 nm (73%) and 100–150 nm (24%) in diameter (Figure 3A and Supplementary Figure 5B). Large vesicles 150–190 nm in diameter are 3% of the total number of vesicles occurring in all fields of vision. Vesicles in the preparation produced from the complemented strain are two groups as wild-type: 100–150 nm (46%) and 50–100 nm (33%) (Figure 3C and Supplementary Figure 5C). Vesicles 150–190 nm and 190–250 nm in diameter are few (17 and 4%, respectively).
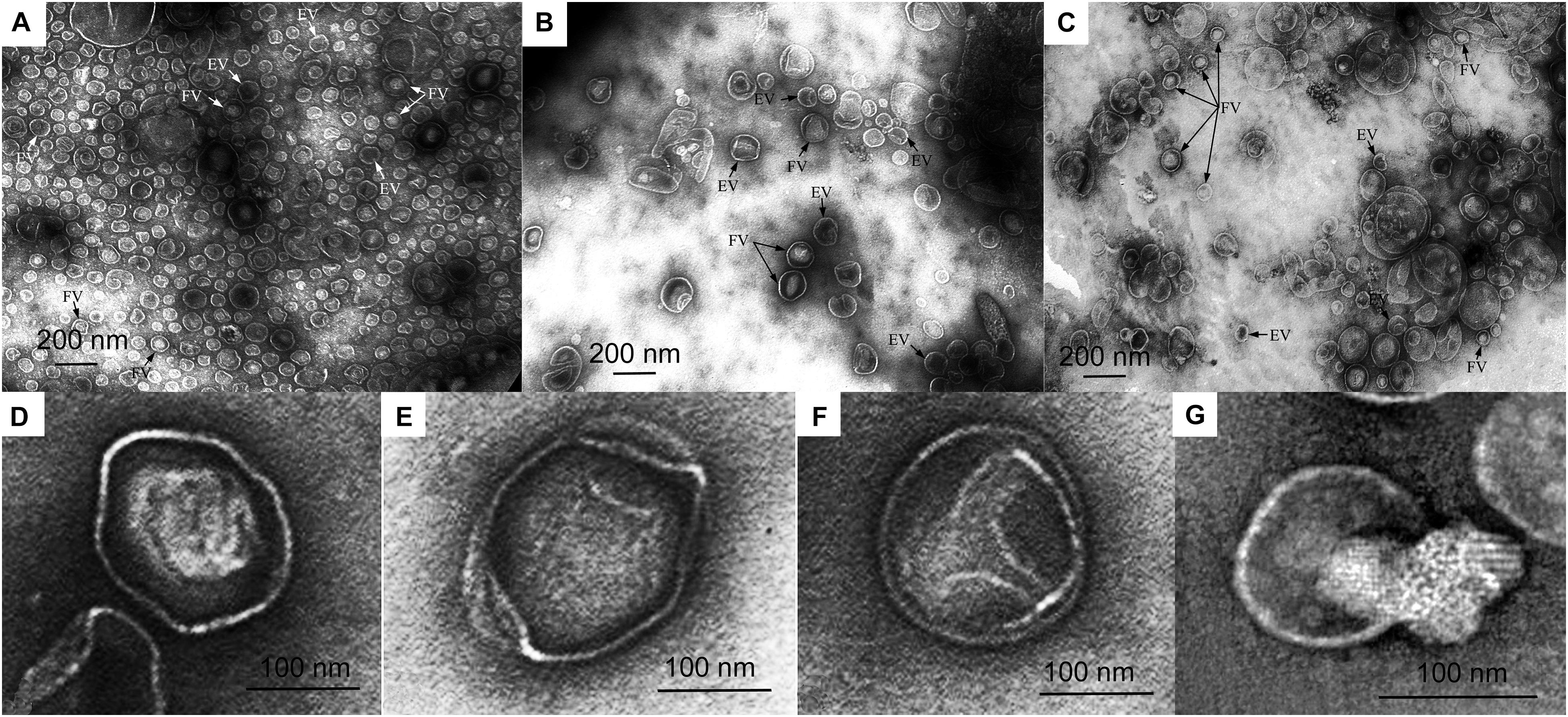
Figure 3. Electron microscopy of vesicle preparations. (A) Wild-type Lysobacter sp. XL1 vesicles. (B) Lysobacter sp. XL1ΔalpB::tet mutant strain vesicles. (C) Lysobacter sp. XL1ΔalpB::alpB complemented strain vesicles. (D) Representative outer membrane vesicle (OMV) with a clear-cut dense polygonal core. (E) Representative OMV with diffusely distributed low-density contents. (F) Representative OMV with small contents in the form of a small loose granule at the periphery inside the vesicle. (G) Representative empty (or broken) vesicle that seems to have spilled its contents. Vesicles from (D–G) are from the mutant strain. EV, empty vesicle; FV, full vesicle.
The main distinction of the preparations is the various degrees of density (full and empty vesicles). The micrographs in Figures 3D–G show OMVs with representative ultrastructures observed in all preparations. Vesicles with clear-cut dense cores of polygonal shapes occur (Figure 3D). Such structures bear resemblance to crystals and may form as a result of a dense packing of their contents. Besides these, vesicles with diffusely distributed contents of low densities (Figure 3E) and with small contents in the form of a small loose granule at the periphery inside the vesicle (Figure 3F) occur, as well as broken vesicles with their contents released outside (Figure 3G). Although all these types of vesicles were observed in all preparations, they were present in very different proportions. TEM obtained that the preparations of wild-type OMVs and complemented strain OMVs contain more full vesicles than that of mutant strain vesicles. Thus, the ratio of full/empty vesicles in the wild-type strain and complemented strain is 2, whereas in the mutant strain, it is 0.7. This analysis considered only intact vesicles.
The main components of vesicles are proteins, lipids, and LPS. By the content of protein and Kdo (the essential constituent of LPS) in the preparations, we can assess the quantity of formed vesicles. The general analysis of protein revealed that its content in the vesicle preparation of the mutant strain is 2.4 times and 1.6 times lower than that in the vesicle preparation of the wild-type strain and complemented strain, respectively (Table 3 and Supplementary Figure 6). The content of Kdo in the vesicle preparation of the mutant strain is 2.1 times and 1.6 times lower than in the vesicle preparation of the wild-type strain and complemented strain, respectively (Table 3 and Supplementary Figure 7).
The general electrophoretic pattern for the distribution of major proteins in the vesicle preparation of the mutant strain did not differ from that in the wild-type strain. The phospholipid composition of mutant strain vesicles did not change either (Supplementary Figure 8). The main phospholipid of vesicles of the wild-type and mutant strains is cardiolipin.
In summary, the knockout of the alpB gene led to a decrease of the quantity of formed vesicles and to a change in the degree to which they are filled.
The obtained results confirm the influence of protein L5 on vesicle biogenesis of Lysobacter sp. XL1.
To study the effect of alpB gene knockout on the lytic activity of vesicles, as test cultures, we used living cells of several bacteria (Table 4 and Figure 4).
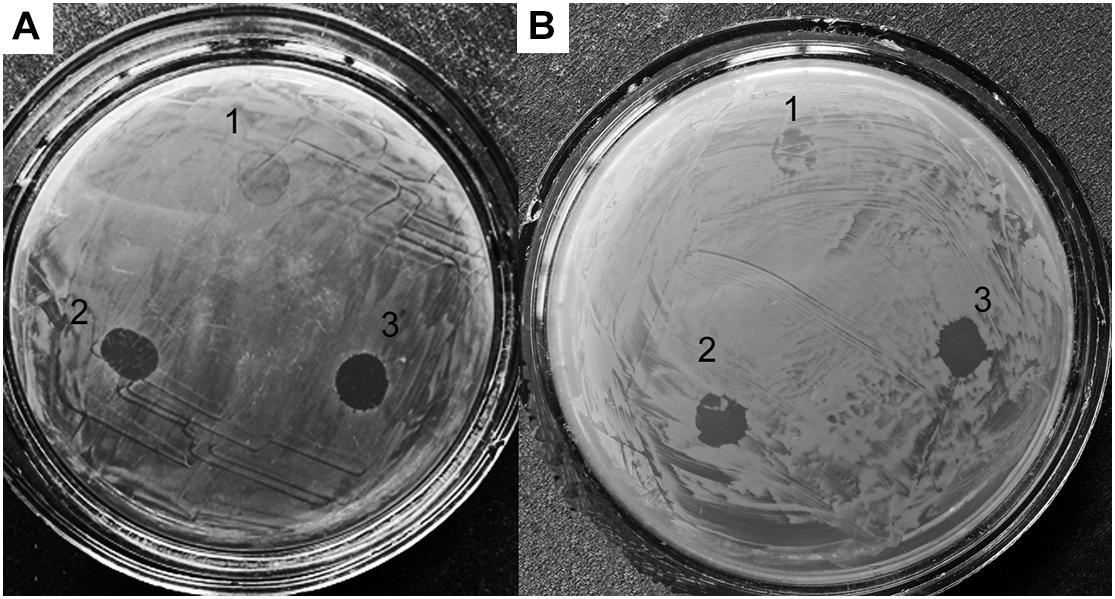
Figure 4. Lytic activities of Lysobacter sp. XL1ΔalpB::tet (1), Lysobacter sp. XL1ΔalpB::alpB (2), and wild-type Lysobacter sp. XL1 (3) vesicle preparations with respect to live test cultures. (A) S. aureus 209P. (B) M. luteus B-1813.
As seen in the figure and table, vesicles of the mutant strain practically lost their ability to lyse living target cells, whereas vesicles of the wild-type and complemented strains possess a strong lytic action. Thus, knockout of the alpB gene was accompanied by a significant decrease of vesicles’ lytic properties, in addition to the reduction of the antimicrobial potential of Lysobacter sp. XL as a whole.
The results presented in this work continue the investigation of the influence of bacteriolytic enzyme L5 on the vesicle biogenesis in the Gram-negative bacterium Lysobacter sp. XL1.
Based on our earlier work, we proposed that protein L5 could be involved in the biogenesis of the subpopulation of vesicles by means of which it was released into the extracellular milieu and proposed a model of the process. Immediately, a question arose how enzyme L5 could initiate the formation of vesicles. One of the existing models of vesicle biogenesis in Gram-negative bacteria assumes a pressure of cell debris (misfolded proteins, peptidoglycan, and LPS fragments) on the inner leaflet of the OM, which provokes vesicle formation (Zhou et al., 1998; Hayashi et al., 2002; Schwechheimer et al., 2014). At some point, we assumed that vesicle biogenesis in Lysobacter sp. XL1 could occur according to that model. However, on the premise that the OM is a dynamic and well-regulated cell structure of complex organization, it is impossible to imagine that a pressure caused by accumulation of protein at the inner leaflet of the OM could evoke vesicle formation. In our view, this single condition is insufficient.
Further investigation of the biogenesis revealed that the main phospholipid of Lysobacter sp. XL1 vesicles was cardiolipin (Kudryakova et al., 2017). That meant that vesicles formed from OM loci enriched with this phospholipid. The hydrophilic head of cardiolipin carries two negative charges, and, due to their intermolecular repulsion, the rigidity of the OM can be disturbed in loci enriched in this phospholipid, which leads to the emergence of a destabilization locus. According to two more known models, vesicles form from certain loci of OM destabilization, which can emerge due to a decreased lipoprotein content (Hoekstra et al., 1976; Deatherage et al., 2009; Schwechheimer et al., 2014) or an increased content of curvature-inducing molecules (B-type LPS, signaling molecule PQS) (Kadurugamuwa and Beveridge, 1995; Mashburn-Warren and Whiteley, 2006; Schertzer and Whiteley, 2012). We note here once again that the cell envelope has a complex organization, and the occurrence of destabilization loci leading to spontaneous evagination and release of OM fragments is not probable. Thus, it is also hard to consider that destabilization loci in the OM are the sole condition for vesicle formation.
Instead, we showed that Lysobacter sp. XL1 vesicles formed from cardiolipin-enriched OM loci, and a particular subpopulation contained bacteriolytic protein L5 (Figure 5). Based on these results, we suggested that vesicle formation was due to a set of several factors. An obligatory factor is the occurrence of an OM destabilization locus, and a factor initiating vesicle formation is a component of the inner contents; in the case of Lysobacter sp. XL1, it is bacteriolytic enzyme L5. We should single out here one more vesicle biogenesis model based on specific sorting of vesicles’ inner components, proposed by American scientists Amanda Horstman and Meta Kuehn (Horstman and Kuehn, 2000). Their idea is intensively developed now (Haurat et al., 2011, 2015; Evans et al., 2012; Elhenawy et al., 2014; Schwechheimer and Kuehn, 2015). In our opinion, at present, this model can be considered to be the most significant for understanding vesicle biogenesis. It is proposed to be supplemented by the idea that vesicles form in the OM destabilization locus, and additionally, a vesicle-formation initiator is a sorted vesicle component. Other results can be taken to be in favor of this concept. Haurat et al. (2011) present possible models of selective protein sorting due to the occurrence of domains, which can interact with B-type LPS (from which destabilization loci form). The work considers the selective sorting of gingipain into Porphyromonas gingivalis vesicles. Besides, the same authors assume the occurrence of a sorting factor, which binds protein directed into vesicles to B-type LPS (Haurat et al., 2015). In this model, they draw an analogy with the role of galectin in protein sorting in the formation of eukaryotic exosomes. Those authors were the first to show a relation of the prospective vesicle component (the sorting component) with B-type LPS, which leads to OM destabilization. Our model assumes that alkaline protein L5 possesses some affinity to the acid cardiolipin. Notably, studies of the structural and functional features of this enzyme have revealed the formation of amyloid-like structures with increased concentration. It has been found that, in vesicles, this enzyme is namely in its amyloid-like form (Kudryakova et al., 2018). This property can be assumed to be important for initiating the formation of vesicle in the destabilization locus. However, proof of this requires integrated molecular, genetic, and structural studies, which will enable understanding the accurate mechanism of the process.
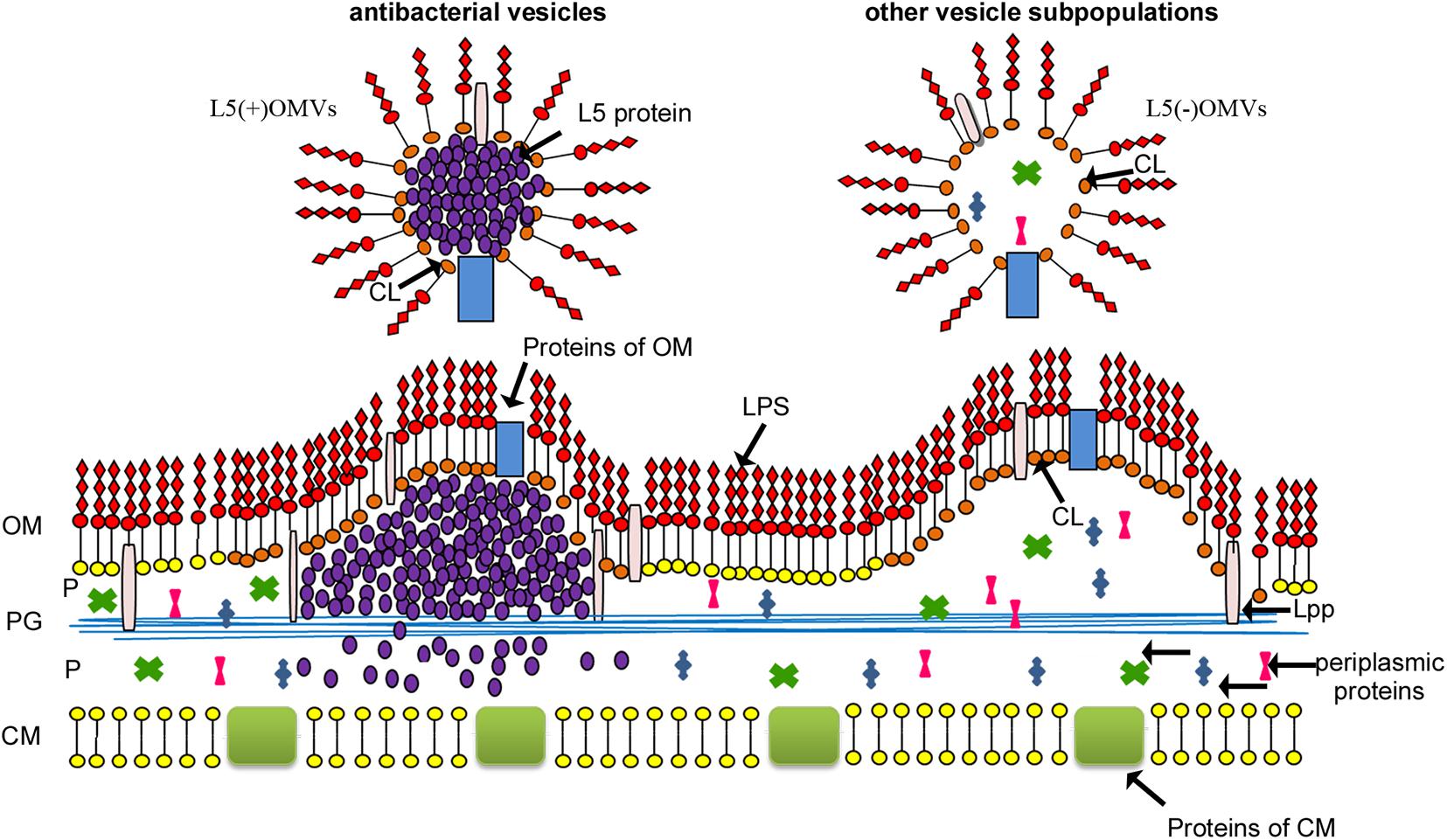
Figure 5. Model of Lysobacter sp. XL1 vesicle biogenesis (Kudryakova et al., 2015) with modification. Lysobacter sp. XL1 forms several subpopulations of vesicles from OM loci enriched with CL. The hydrophilic head of cardiolipin carries two negative charges, and, due to their intermolecular repulsion, the rigidity of the OM can be disturbed in loci enriched with this phospholipid, which leads to the initiation of vesicle formation (Kudryakova et al., 2017). One of the vesicle subpopulations contains bacteriolytic enzyme L5. In the process of its topogenesis, this protein accumulates in the periplasm at the inner leaflet of the OM (Kudryakova et al., 2015) in the form of amyloid-like structures (Kudryakova et al., 2018). This may lead to a pressure on the cardiolipin-enriched inner leaflet of the OM (the destabilization locus) and contribute to the formation of a subpopulation of antibacterial vesicles containing protein L5. This vesicle subpopulation is a “bacterial bomb” for competitive bacteria and is involved in invasion expansion of ecological niches for Lysobacter sp. XL1. L5(+)OMVs, antibacterial subpopulation of vesicles containing protein L5; L5(–)OMVs, other subpopulations of vesicles; LPS, lipopolysaccharide; OM, outer membrane; P, periplasm; CM, cytoplasmic membrane; Lpp, lipoprotein; PG, peptidoglycan; CL, cardiolipin.
Additional confirmation that protein L5 possesses an ability to initiate vesicle formation is the result of the expression of enzyme L5 in the recombinant strain P. fluorescens Q2-87/B (Kudryakova et al., 2017). As the result of expression of this protein gene, the recombinant strain formed a larger number of vesicles, which produced enzyme L5 and acquired lytic properties.
To confirm the role of protein L5 in Lysobacter sp. XL1 vesicle biogenesis, we lacked genetic studies, in particular, of the knockout of a corresponding gene. The generation of gene knockouts in Lysobacter spp. constitutes a very recent development, as the availability of molecular genetic tools for this bacterium has evolved slowly. Only a few investigations to date are known to involve the deletion of Lysobacter genes, like the chitinase gene and the genes responsible for the biosynthesis of antibiotics and for regulation of this process in L. enzymogenes OH11 (Qian et al., 2012; Xu et al., 2015; Wang et al., 2016, 2017). Therefore, the successful deletion of alpB represents an important contribution to this field of genetic investigation.
In this work, we present the results of research into the effect of bacteriolytic enzyme L5 gene knockout on vesicle formation in Lysobacter sp. XL1. As a result of this gene knockout, the total bacteriolytic activity of the culture liquid in the mutant strain decreased. The following results became crucial for our hypothesis: the mutant strain formed fewer vesicles, the degree of their filling decreased, the lytic properties of vesicles were lost practically completely. The latter result indicates that, in the mutant strain, there is no subpopulation of vesicles containing enzyme L5. However, other subpopulations of vesicles continued to form.
Thus, we conclude that bacteriolytic enzyme L5 influences the formation of vesicle subpopulations that contain it. The molecular mechanisms of this effect are yet to be established by future research.
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation.
IK, AA, and NL contributed to planning the experiments. IK, AA, and TI contributed to the experimental work. EL contributed to the purification of vesicles. NS contributed to the electron microscopy. NL contributed to the project administration. IK contributed to the funds acquisition. IK and NL contributed to the writing the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Russian Science Foundation (Project No. 19-74-00086).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Victor Selivanov for professional English translation. Electron microscopy was carried out at the UNIQEM Collection (Research Center of Biotechnology, Russian Academy of Sciences, Moscow, Russia).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.715802/full#supplementary-material
Afoshin, A. S., Kudryakova, I. V., Borovikova, A. O., Suzina, N. E., Toropygin, I. Y., Shishkova, N. A., et al. (2020). Lytic potential of Lysobacter capsici VKM B-2533T: bacteriolytic enzymes and outer membrane vesicles. Sci. Rep. 10:9944. doi: 10.1038/s41598-020-67122-2
Ames, G. F. (1968). Lipids of Salmonella typhimurium and Escherichia coli: structure and metabolism. J. Bacteriol. 95:833843. doi: 10.1128/jb.95.3.833-843.1968
Avila-Calderón, E. D., Araiza-Villanueva, M. G., Cancino-Diaz, J. C., López-Villegas, E. O., Sriranganathan, N., Boyle, S. M., et al. (2015). Roles of bacterial membrane vesicles. Arch. Microbiol. 197, 1–10. doi: 10.1007/s00203-014-1042-7
Balbás, P., Soberón, X., Merino, E., Zurita, M., Lomeli, H., Valle, F., et al. (1986). Plasmid vector pBR322 and its special-purpose derivatives – a review. Gene 50, 3–40. doi: 10.1016/0378-1119(86)90307-0
Bullock, W. O., Fernandez, J. M., and Short, J. M. (1987). XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5, 376–378.
Deatherage, B. L., Lara, J. C., Bergsbaken, T., Rassoulian Barrett, S. L., Lara, S., and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x
Elhenawy, W., Debelyy, M. O., and Feldman, M. F. (2014). Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5, e909–e914. doi: 10.1128/mBio.00909-14
Evans, A. G. L., Davey, H. M., Cookson, A., Currinn, H., Cooke-Fox, G., Stanczyk, P. J., et al. (2012). Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158, 2742–2752. doi: 10.1099/mic.0.060343-0
Guerrero-Mandujano, A., Hernández-Cortez, C., Ibarra, J. A., and Castro-Escarpulli, G. (2017). The outer membrane vesicles: secretion system type zero. Traffic 18, 425–432. doi: 10.1111/tra.12488
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/s0022-2836(83)80284-8
Haurat, M. F., Aduse-Opoku, J., Rangarajan, M., Dorobantu, L., Gray, M. R., Curtis, M. A., et al. (2011). Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286, 1269–1276. doi: 10.1074/jbc.M110.185744
Haurat, M. F., Elhenawy, W., and Feldman, M. F. (2015). Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol. Chem. 396, 95–109. doi: 10.1515/hsz-2014-0183
Hayashi, J., Hamada, N., and Kuramitsu, H. K. (2002). The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol. Lett. 216, 217–222. doi: 10.1111/j.1574-6968.2002.tb11438.x
Hoekstra, D., van der Laan, J. W., de Leij, L., and Witholt, B. (1976). Release of outer membrane fragments from normally growing Escherichia coli. Biochim. Biophys. Acta 455, 889–899. doi: 10.1016/0005-2736(76)90058-4
Horstman, A. L., and Kuehn, M. J. (2000). Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275, 12489–12496. doi: 10.1074/jbc.275.17.12489
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Karkhanis, Y. D., Zeltner, J. Y., Jackson, J. J., and Carlo, D. J. (1978). A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of Gram-negative bacteria. Anal. Biochem. 85, 595–601. doi: 10.1016/0003-2697(78)90260-9
Kato, S., Kowashi, Y., and Demuth, D. R. (2002). Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 32, 1–13. doi: 10.1006/mpat.2001.0474
Knox, K. W., Vesk, M., and Work, E. (1966). Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92, 1206–1217. doi: 10.1128/JB.92.4.1206-1217.1966
Kudryakova, I. V., Gabdulkhakov, A. G., Tishchenko, S. V., Lysanskaya, V. Y., Suzina, N. E., Tsfasman, I. M., et al. (2018). Structural and functional properties of antimicrobial protein L5 of Lysîbacter sp. XL1. Appl. Microbiol. Biotechnol. 102, 10043–10053. doi: 10.1007/s00253-018-9364-z
Kudryakova, I. V., Suzina, N. E., and Vasilyeva, N. V. (2015). Biogenesis of Lysobacter sp. XL1 vesicles. FEMS Microbiol. Lett. 362:fnv137. doi: 10.1093/femsle/fnv137
Kudryakova, I. V., Suzina, N. E., Vinokurova, N. G., Shishkova, N. A., and Vasilyeva, N. V. (2017). Studying factors involved in biogenesis of Lysobacter sp. XL1 outer membrane vesicles. Biochemistry 82, 501–509. doi: 10.1134/S0006297917040125
Kulaev, I. S., Stepnaya, O. A., Tsfasman, I. M., Tchermenskaja, T. S., Ledova, L. A., Zubrizkaja, L. G., et al. (2006). Bacteriolytic complex, method for producing said complex and strain for carrying out said method. PatentNo: US 7,150,985 B2.
Kunsmann, L., Rüter, C., Bauwens, A., Greune, L., Glüder, M., Kemper, B., et al. (2015). Virulence from vesicles: novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain. Sci. Rep. 5:13252. doi: 10.1038/srep13252
Lin, D., and McBride, M. J. (1996). Development of techniques for the genetic manipulation of the gliding bacteria Lysobacter enzymogenes and Lysobacter brunescens. Can. J. Microbiol. 42, 896–902. doi: 10.1139/m96-115
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275.
Mashburn-Warren, L. M., and Whiteley, M. (2006). Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61, 839–846. doi: 10.1111/j.1365-2958.2006.05272.x
Nagakubo, T., Nomura, N., and Toyofuku, M. (2020). Cracking open bacterial membrane vesicles. Front. Microbiol. 10:3026. doi: 10.3389/fmicb.2019.03026
Olofsson, A., Vallström, A., Petzold, K., Tegtmeyer, N., Schleucher, J., Carlsson, S., et al. (2010). Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 77, 1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x
Qian, G., Wang, Y., Qian, D., Fan, J., Hu, B., and Liu, F. (2012). Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J. Microbiol. Biotechnol. 28, 549–557. doi: 10.1007/s11274-011-0846-8
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Rompikuntal, P. K., Thay, B., Khan, M. K., Alanko, J., Penttinen, A. M., Asikainen, S., et al. (2012). Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect. Immun. 80, 31–42. doi: 10.1128/IAI.06069-11
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
Schertzer, J. W., and Whiteley, M. (2012). A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3, e297–e211. doi: 10.1128/mBio.00297-11
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Schwechheimer, C., Kulp, A., and Kuehn, M. J. (2014). Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol. 14:324. doi: 10.1186/s12866-014-0324-1
Vasilyeva, N. V., Shishkova, N. A., Marinin, L. I., Ledova, L. A., Tsfasman, I. M., Muranova, T. A., et al. (2014). Lytic peptidase L5 of Lysobacter sp. XL1 with broad antimicrobial spectrum. J. Mol. Microbiol. Biotechnol. 24, 59–66. doi: 10.1159/000356838
Vasilyeva, N. V., Tsfasman, I. M., Suzina, N. E., Stepnaya, O. A., and Kulaev, I. S. (2008). Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J. 275, 3827–3835. doi: 10.1111/j.1742-4658.2008.06530.x
Vasilyeva, N. V., Tsfasman, I. M., Kudryakova, I. V., Suzina, N. E., Shishkova, N. A., Kulaev, I. S., et al. (2013). The role of membrane vesicles in secretion of Lysobacter sp. bacteriolytic enzymes. J. Mol. Microbiol. Biotechnol. 23, 142–151. doi: 10.1159/000346550
Wang, R., Xu, H., Du, L., Chou, S. H., Liu, H., Liu, Y., et al. (2016). TonB-dependent receptor regulates antifungal HSAF biosynthesis in Lysobacter. Sci. Rep. 6:26881. doi: 10.1038/srep26881
Wang, R., Xu, H., Zhao, Y., Zhang, J., Yuen, G. Y., Qian, G., et al. (2017). Lsp family proteins regulate antibiotic biosynthesis in Lysobacter enzymogenes OH11. AMB Express 7:123. doi: 10.1186/s13568-017-0421-2
Xu, G., Zhao, Y., Du, L., Qian, G., and Liu, F. (2015). Hfq regulates antibacterial antibiotic biosynthesis and extracellular lytic-enzyme production in Lysobacter enzymogenes OH11. Microb. Biotechnol. 8, 499–509. doi: 10.1111/1751-7915.12246
Keywords: biogenesis of vesicles, Lysobacter sp. XL1, antimicrobial potency of vesicles, deletion in gene alpB, bacteriolytic protease L5
Citation: Kudryakova IV, Afoshin AS, Ivashina TV, Suzina NE, Leontyevskaya EA and Leontyevskaya (2021) NV (2021) Deletion of alpB Gene Influences Outer Membrane Vesicles Biogenesis of Lysobacter sp. XL1. Front. Microbiol. 12:715802. doi: 10.3389/fmicb.2021.715802
Received: 27 May 2021; Accepted: 16 July 2021;
Published: 16 August 2021.
Edited by:
Alejandro J. Yañez, Austral University of Chile, ChileReviewed by:
Yoshihiro Ojima, Osaka City University, JapanCopyright © 2021 Kudryakova, Afoshin, Ivashina, Suzina, Leontyevskaya and Leontyevskaya (Vasilyeva). This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalia V. Leontyevskaya (Vasilyeva), dmFzaWx5ZXZhbnZAcmFtYmxlci5ydQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.