
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 September 2021
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.713347
This article is part of the Research Topic Recent Advances in the Controversial Human Pathogens Pneumocystis, Microsporidia and Blastocystis, Volume II View all 9 articles
Blastocystis is the most frequently isolated protozoan from human stool. Its role in human health is still debated, and a high prevalence was reported in irritable bowel syndrome (IBS) subjects, suggesting a potential link with microbiota. In the present study, we aimed to investigate prokaryotic and eukaryotic microbiota in both IBS-C (constipated) and healthy individuals. We recruited 35 IBS-C patients and 23 healthy subjects, from which 12 and 11 carried Blastocystis, respectively. We performed 16S and 18S rRNA high-throughput sequencing on feces. Whereas we did not observe differences between infected and non-infected controls, several phyla were significantly modified in IBS-C patients according to the presence of Blastocystis. Tenericutes phylum and Ruminococcaceae family were especially increased in Blastocystis carriers. Furthermore, colonization with Blastocystis was associated with discrete changes in the microbial eukaryome, particularly among the Fungi taxa. Depending on the group of patients considered, the mycobiota changes do not go in the same direction and seem more deleterious in the IBS-C group. These results encourage further in vivo and in vitro investigations concerning the role of Blastocystis in the gut environment.
Blastocystis is the most prevalent intestinal parasite found in human worldwide, even in industrialized countries; for example, the prevalence is approximately 17% in France (El Safadi et al., 2016). Twenty-two subtypes (ST) of the parasite have been described using a 600-bp barcode sequence of the 18S rRNA encoding gene (Stensvold and Clark, 2020). Among them, ST1 to ST9 and ST12 were identified in human stools, with ST1 to ST4 being the most frequent (Alfellani et al., 2013; Ramírez et al., 2016). Blastocystis pathogenicity remains debated as most of the colonized people are asymptomatic. A potential link between Blastocystis and irritable bowel syndrome (IBS), a functional chronic disorder, was suspected in several studies on the basis of prevalence data and potential virulence factors produced by the parasite (Poirier et al., 2012; Nourrisson et al., 2014, 2016). Four symptom-based subgroups of IBS can be distinguished according to the predominant bowel habit: IBS-C for patients with constipation, IBS-D for patients with diarrhea, IBS-M for patients with alternating diarrhea and constipation, and IBS-U for patients with unsubtyped IBS.
Investigations in asymptomatic subjects carrying Blastocystis are required, given (i) the large number of colonized people, and (ii) questions about the infectious risks link with fecal microbiota transplantation (FMT) and its consequences on the health of recipients. Indeed, Blastocystis-colonized people should be excluded from donation, even if they are otherwise in good health, since the safety of transplanting the parasite has not been demonstrated (Cammarota et al., 2017).
These concerns have led to the publication of a growing number of studies on the impact of Blastocystis on the intestinal microbiota, focusing mainly on its prokaryotic component (reviewed in Deng et al., 2021). Many studies support that Blastocystis is associated with higher bacterial diversity and richness, and that the parasite could be considered as a commensal of a healthy gut environment. However, the debate remains wide open since other works identify a decrease of the abundance of beneficial bacteria such as Bifidobacterium sp. (Nourrisson et al., 2014). A decrease in Bifidobacterium and Lactobacillus was also observed in mice infected with ST7 (Yason et al., 2019). Moreover, Defaye et al. (2020) recently demonstrated that Blastocystis infection in rat is responsible for microbiota changes associated with visceral hypersensitivity. These changes included an increase in the relative abundance of Oscillospira and a decrease in Clostridium.
Works studying the eukaryotic microbiota are rarer. One study reported that Blastocystis colonization is associated with an increase in fungal diversity of intestinal tract (Nieves-Ramírez et al., 2018).
Facing these contradictory results and the need of additional data for microbiota modification associated with Blastocystis colonization, and especially interactions with eukaryotic microbiota, we conducted in the present work a prospective study in both healthy and IBS-C subjects.
From January 2014 to July 2017, patients suffering from IBS-C and healthy subjects were recruited in a prospective study. Both IBS-C (fulfilling the Rome III criteria, Longstreth et al., 2006) and healthy subjects beneficiated of a medical consultation with a gastroenterologist in the Gastroenterology unit of the University Hospital of Clermont-Ferrand (France). All included subjects were men or women over the age of 18. Subjects who used antibiotics or probiotics less than 2 months before stool collection were not included in the study, as are control subjects with functional digestive disorders or other known intestinal disease. IBS symptom severity score (IBS-SSS, also known as Francis score), BMI, sex, and age were collected. This clinical study was approved by the research ethics committees of the Clermont-Ferrand Hospital (“Comité de Protection des Personnes Sud-Est VI,” France) with the reference number 2013-A00031-44.
Stool specimens from included subjects were processed in less than 4 h after emission. Approximately 200 mg of stools was mechanically ground with 0.5-mm-diameter glass beads on Tissue Lyser (Qiagen) during 3 min at 30 Hz/s. Total DNA extraction was then performed with the QIAamp® DNA Stool Mini Kit (Qiagen) and eluted in a final volume of 200 μl according to the manufacturer’s recommendations. DNA extracts were stored at –80°C until the next steps.
Specific quantitative PCR (qPCR) to detect and subtype Blastocystis was carried out using BL18SPPF1/BL18SR2PP primers (Supplementary Table 1) that target a conserved region of the SSU rRNA gene and allow discrimination between subtypes as previously described (Poirier et al., 2011). Blastocystis subtypes were assigned with a query coverage > 98% with exact match or identity > 98%.
Illumina high-throughput sequencing was performed by MRDNA lab (1 Shallowater, TX, United States) on a MiSeq (Macrogen, Inc.) following the manufacturer’s guidelines. Briefly, the variable regions V3–V5 of the bacterial 16S rRNA gene were amplified using the broad-range forward primer 515F and the reverse primer 909R (Li et al., 2016; Supplementary Table 1). Similarly, a portion of the sequence of the 18S rRNA gene was amplified using the primers 515F_Euk and 1119R previously described (Parfrey et al., 2014; Supplementary Table 1). For each target, a 28-cycle PCR (5 cycles used on PCR products) was performed with barcoded forward primers and using the HotStarTaq Plus Master Mix Kit (Qiagen, Germantown, MD, United States) under the following conditions: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, after which a final elongation step at 72°C for 5 min was performed. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Multiple samples were pooled together (e.g., 100 samples) in equal proportions based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads. Then, Illumina sequencing was performed and DNA libraries were built according to the Illumina TruSeq DNA library preparation protocol.
Sequence data were processed using QIIME microbiome analysis package (Quantitative Insights into Microbial Ecology QIIME, version 1.8.02) (Caporaso et al., 2010).
In summary, sequences were demultiplexed to remove barcodes and primer sequences. Sequences < 250 bp or > 600 bp or with ambiguous base calls were removed. Operational taxonomic units (OTUs) based on 97% specific 16S rRNA gene sequence identities were generated with the SILVA database (release 1383) using uclust, and chimeras were removed using ChimeraSlayer. Final OTUs were taxonomically classified using BLASTn against SILVA database. Data sets were filtered to exclude singletons and mitochondrion and chloroplast sequences. Following filtering, a cutoff of 650 reads per sample was applied. All 16S rRNA gene samples passed the cutoff.
In summary, sequences were demultiplexed to remove barcodes and primer sequences. Sequences < 200 bp or > 1,200 bp or with ambiguous base calls were removed. Sequences were clustered in OTUs using an open reference strategy with the SILVA database (release 138), and chimeras were removed using ChimeraSlayer. Final OTUs were taxonomically classified using BLASTn against SILVA database. Data sets were filtered to exclude singletons and mammalian and plant sequences. Following filtering, a cutoff of 650 reads per sample was applied.
R package Tax4Fun4 was used to predict the functional capabilities of the microbial communities based on 16S rRNA data (Aßhauer et al., 2015). The OTU table generated with QIIME was imported to Tax4Fun (R version 4.0.3) and the SILVA123 database was used to predict functional capabilities. Briefly, the linear relationship between the SILVA classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) database prokaryotic classification realized the prediction of the microbial community function. A heat map was built based on gene abundances of the enzymes of interest extracted from the output tables of Tax4Fun.
Alpha diversity, i.e., the richness of single microbial taxa within a sample, was measured using observed OTU, Chao 1, and Shannon’s and Simpson’s indexes. Observed OTU measurements were determined with QIIME using an OTU table rarefied at various depths. Boxplots showing alpha diversity were created in QIIME. Monte Carlo permutations were used to calculate the p-values.
Beta diversity, i.e., the variation in microbiota composition between individual samples, was assessed based on both weighted and unweighted UniFrac distances metrics between samples computed with the rarefied OTUs count table. Principal coordinates analysis (PCoA), generated in QIIME, was used to further assess and visualize beta diversity. PCoA was created in R software (4.0.3).
LefSe (LDA Effect Size) was used to investigate bacterial members that drive differences between groups (Segata et al., 2011).
For non-Gaussian data, comparisons were performed by the non-parametric Mann–Whitney U test (unpaired data). A p-value ≤ 0.05 was considered statistically significant. All statistical analyses were conducted in R software (4.0.3).
Thirty-five patients suffering from IBS-C (8 males and 27 females, sex ratio 0.30) and 23 healthy subjects (10 males and 13 females, sex ratio 0.77) were recruited (Supplementary Table 2). Blastocystis was detected among 12 patients of the IBS-C group (34.3%) and among 11 subjects of the control group (47.8%). In the IBS-C group, the most frequent subtype (ST) was ST4 (n = 4), followed by ST3 (n = 3), ST2 (n = 3 including one co-infection), ST1 (n = 1), ST5 (n = 1 co-infection), and ST7 (n = 1). In the control group, the ST distribution was as follows: 4 ST1, 3 ST3, 3 ST4, and 1 ST7. Body mass index (BMI) was not significantly different between the two groups (23.4 in the IBS group and 26.2 in the control group, Mann–Whitney test, p = 0.096) or according to Blastocystis carriage (IBS group: 24.8 in Blastocystis carriers and 23.1 in non-carriers, p = 1.000; control group: 24.0 in Blastocystis carriers and 28.7 in non-carriers, p = 0.141). In the IBS group, Francis score was not significantly different between carriers and non-carriers of Blastocystis (277 and 324, respectively, Mann–Whitney test, p = 0.270).
We first analyzed the 16S rRNA sequence data set. Our analyses revealed a significant decrease in bacterial richness (Chao 1 and observed number of OTUs) in IBS-C compared with controls (Figures 1A,B). Diversity (Shannon function and Simpson’s index) was not significantly different between IBS-C and control groups, but there was a trend to Shannon diversity decrease, which depends more on less abundant species than Simpson’s index (Figures 1C,D). There was no significant difference of alpha diversity metrics within the control and IBS groups depending on whether Blastocystis was present or not (Figures 1E–H). However, bacterial richness was significantly decreased in IBS-C compared to both carriers and non-carriers in control groups. Interestingly, both bacterial richness and diversity tend to be increased in IBS-C Blastocystis-positive subjects compared to -negative subjects, but the difference was not significant.
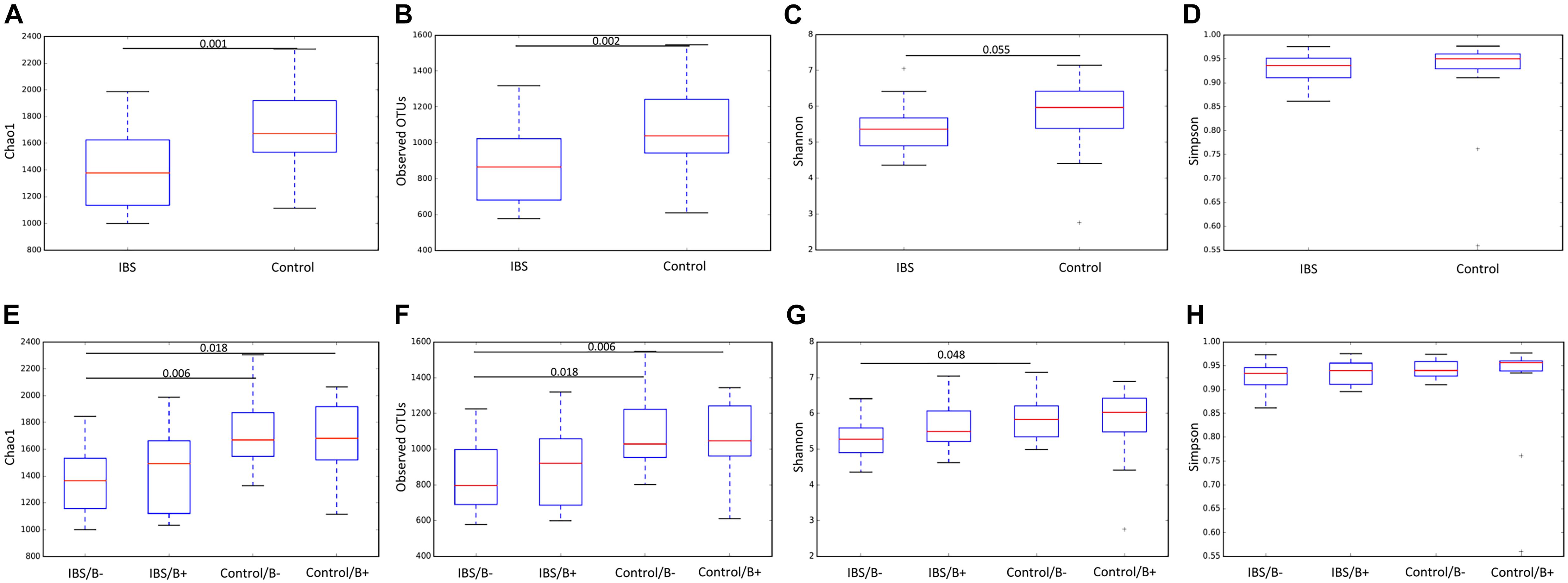
Figure 1. Comparison of alpha diversity indices of the fecal bacterial microbiota between controls (E–H) and IBS-C subjects (A–D) according to Blastocystis carriage. (A,E) Chao 1, (B,F) observed number of OTUs, (C,G) Shannon diversity, and (D,H) Simpson’s index. Significant (or close to significance) p-values were reported on boxplots. IBS/B-: Blastocystis-negative IBS patients; IBS/B + : Blastocystis-positive IBS patients; Control/B-: Blastocystis-negative control subjects; Control/B + : Blastocystis-positive control subjects.
At the phylum level, the Firmicutes/Bacteroidetes ratio was decreased in the IBS-C group (Figure 2A). Firmicutes were significantly decreased in IBS-C subjects (Mann–Whitney test, p = 0.012), whereas Proteobacteria and Verrucomicrobia were significantly increased (Mann–Whitney test, p = 0.046 and 0.035, respectively). Moreover, β-diversity analyses confirmed differences between bacterial communities of IBS-C and controls at the OTU level (based on Bray–Curtis dissimilarities, Figure 2B). The Firmicutes/Bacteroidetes ratio tended to be increased in Blastocystis carriers from both IBS-C and control groups when compared to non-carrier groups (Mann–Whitney test, p = 0.085 and p = 0.449, respectively, Figure 3A). Interestingly, whereas no phylum was significantly different between Blastocystis-negative and -positive controls (Table 1), significant differences were observed within IBS subjects. Tenericutes phylum was significantly expanded among Blastocystis carriers of the IBS-C group (Figure 3B). In the Blastocystis-positive control group, a trend to an increase of Tenericutes was also observed (Figure 3B). Enrichment and depletion of several bacterial taxa were further confirmed by LEfSe (linear discriminant analysis effect size) analysis (p < 0.05, LDA > | 2|; Figures 3C,D and Supplementary Figure 1). Interestingly, Ruminococcaceae were enriched in the control group when subjects were positive for Blastocystis (Figure 3C). In IBS-C patients, Lactobacillus were decreased when subjects were colonized with Blastocystis (Figure 3D). The PCoA based on Bray–Curtis dissimilarities and unweighted or weighted Unifrac distances showed a modest clustering of the samples from IBS-C according to Blastocystis carriage, but not for controls (Figures 3E,F).
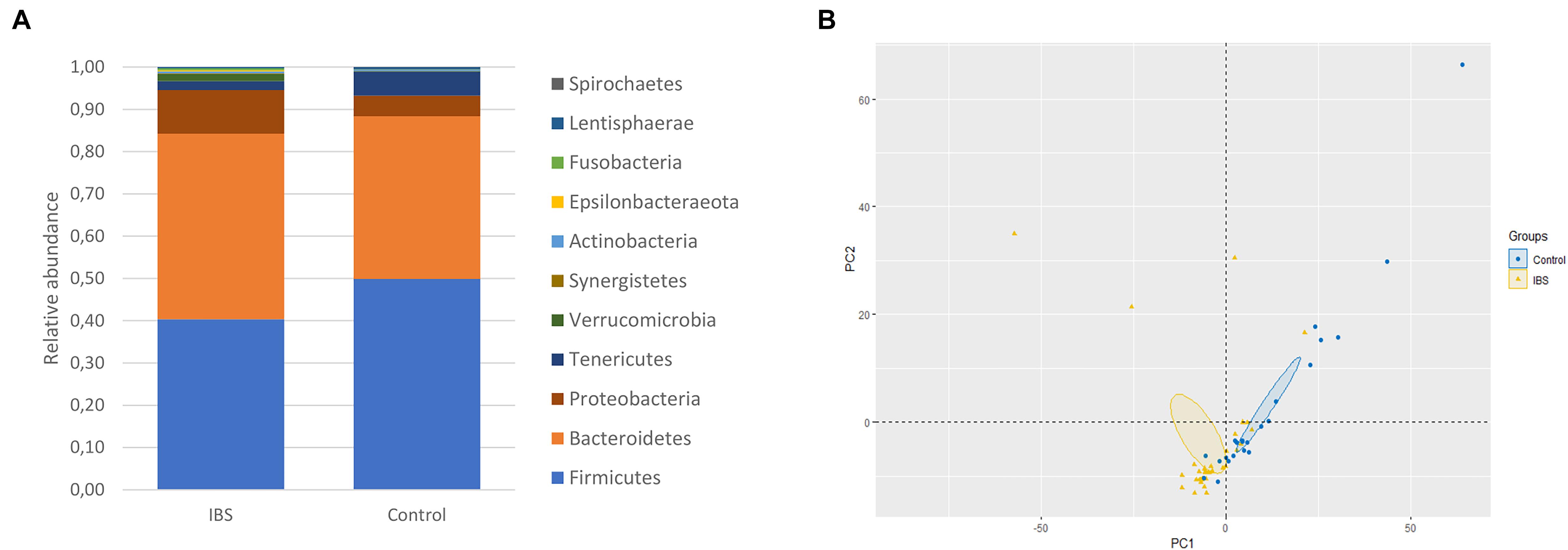
Figure 2. Bacterial alterations in IBS. (A) Comparison of the fecal bacterial microbiota composition between controls and IBS-C subjects at the phylum level. (B) PCoA of the unweighted UniFrac distance of control (blue plots) and IBS (yellow plots) subjects.
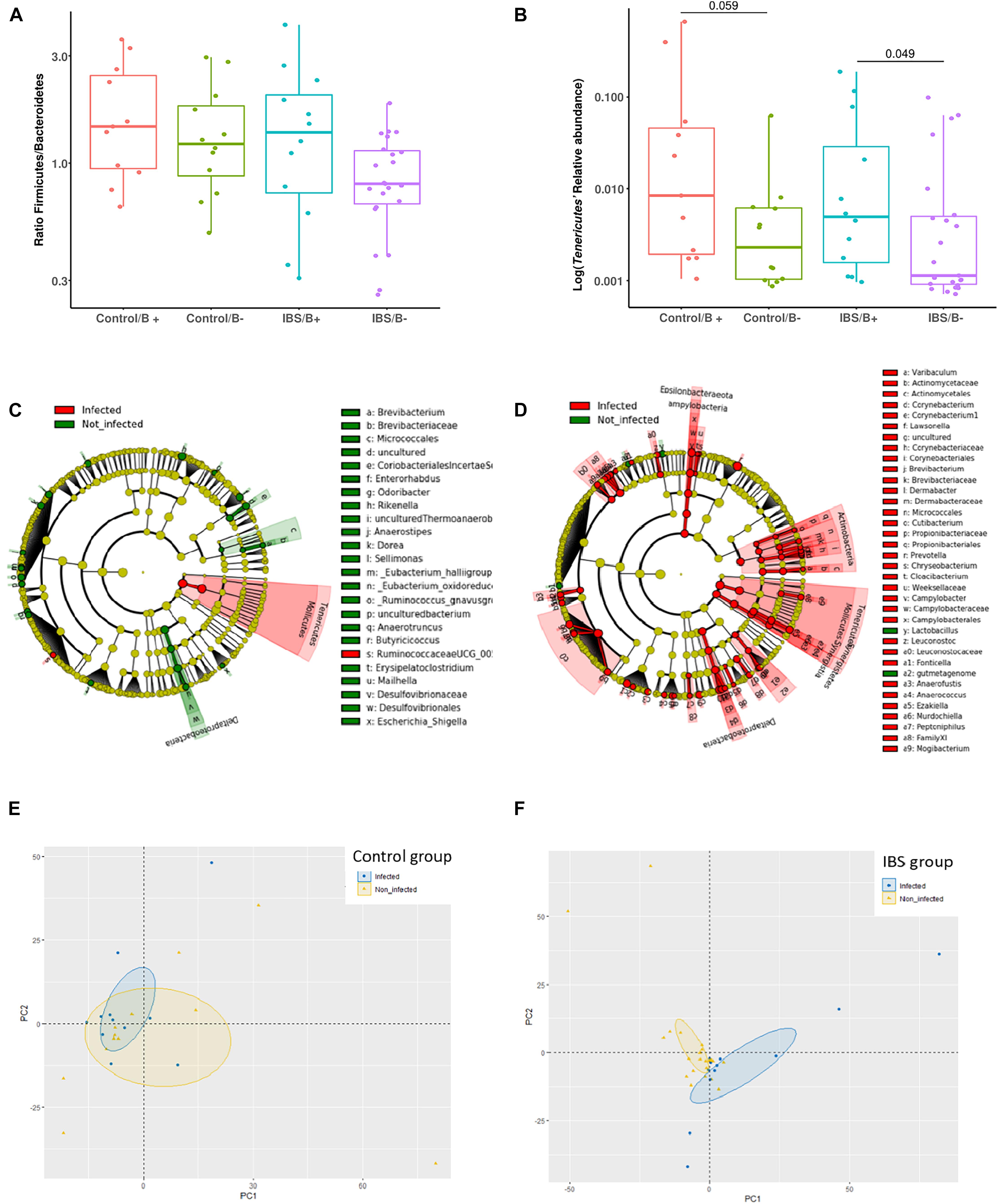
Figure 3. Investigations about bacterial phyla/members that drive differences in each group of patients according to Blastocystis carriage. (A,B) Analysis at the phyla level: (A) Firmicutes/Bacteroidetes ratio in controls and IBS according to Blastocystis carriage. (B) Relative abundance of Tenericutes. Statistical significance was determined by Mann–Whitney test. Significant (or close to significance) p-values were reported on boxplots. IBS/B-: Blastocystis-negative IBS patients; IBS/B + : Blastocystis-positive IBS patients; Control/B-: Blastocystis-negative control subjects; Control/B + : Blastocystis-positive control subjects. (C,D) LefSe (LDA Effect Size) analysis in the control group (C) and the IBS group (D). Red, taxa higher in Blastocystis-infected subjects; green, taxa higher in non-Blastocystis-infected subjects. Statistical significance was determined by LefSe analysis with FDR correction (only those species with q values < 0.05 and LDA effect size > 2 are shown). (E,F) Beta diversity analysis: PCoA of the unweighted UniFrac distance of non-carriers (yellow plots) and carriers (blue plots) of Blastocystis in the control group (E) and the IBS group (B).
To further explore the impact of microbiota differences, a functional prediction of the microbial capabilities within each group was conducted in silico. An important difference was observed between IBS patients and healthy subjects, where two major categories of functional groups emerged. Cluster I fitted with a functional enrichment increase in IBS group, whereas in cluster II, there was a decrease in enrichment (Figure 4). Genes concerning metabolism of cofactors and vitamins, or amino acid metabolism were found in both clusters. There were a number of genes annotated to functions involved in the metabolism of complex carbohydrates, most of these are in Cluster I, but genes implicated in glycolysis/gluconeogenesis were identified in cluster II. Genes linked with lipid metabolism such as glycerolipid metabolism and fatty acid biosynthesis were only present in Cluster I. The only significant difference in functional capabilities according to the presence of Blastocystis in a same group (IBS vs. healthy) concerns glycerolipid metabolism among healthy subjects (1.027 for Blastocystis carriers vs. 0.964 for non-carriers, Student’s t-test p = 0.032). No difference was observed for this pathway in the IBS group according to carriage or not of Blastocystis (p = 0.919).
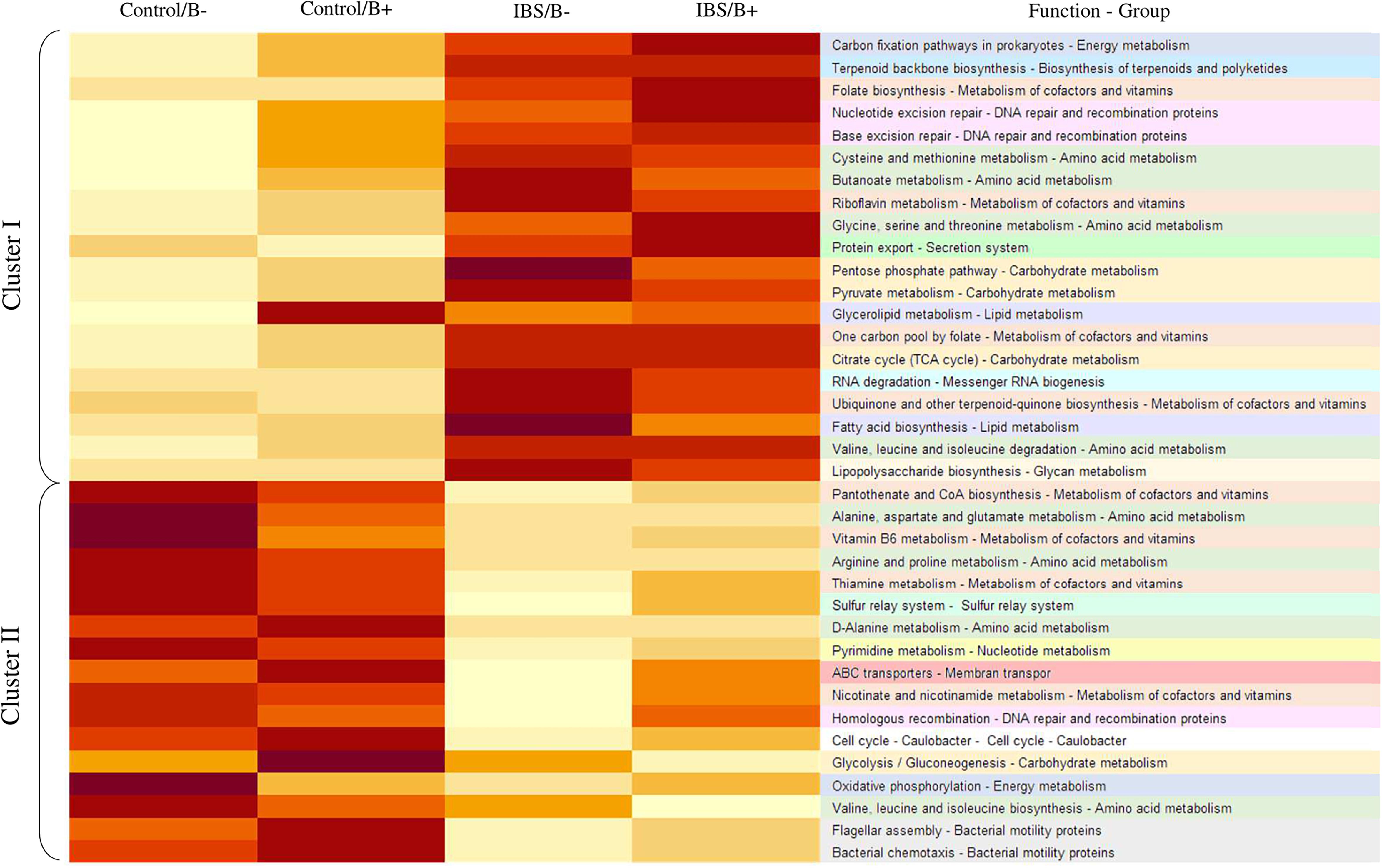
Figure 4. Heat map illustrating changes in functional capabilities in the four groups of patients. The color scale ranges from light yellow to dark red: light yellow indicates a low functional enrichment, and dark red indicates a high functional enrichment of the averaged individual KEGG function.
In the 18S rRNA sequence data set, 13 samples had fewer than 650 sequences per sample after applying filtering steps, which prompted their removal from the data set. Thus, 45 samples (25 IBS and 20 controls) were retained for ecological analysis of eukaryotes.
As expected, the Fungi class accounted for the largest fraction of the eukaryotic microbiota in IBS-C and control groups (75 and 64%, respectively, Figure 5). Blastocystis was obviously the most abundant protist as we selected carriers for the study. Other protists identified in our study were Dientamoeba fragilis (Parabasalia class) and Entamoeba sp. (Entamoeba class). We further conducted an analysis of the Fungi class after filtering all the others eukaryota. Then, 14 more samples had fewer than 650 sequences per sample, which prompted their removal from the data set. Thus, 31 samples (17 IBS and 14 controls) were retained. Alpha diversity metrics were not significantly different between IBS and controls (Figures 6A–D). Blastocystis carriage had no impact on alpha diversity (Figures 6E–H). At the family level, Dipodascaceae, Aspergillaceae, and Saccharomycetaceae were the most abundant (64, 17, and 16% in the IBS group, and 31, 41, and 18% in controls, respectively; Figure 7A). Within the two groups of patients, Dipodascaceae were increased among Blastocystis carriers, while Aspergillaceae were decreased (Figure 7A; Table 2). Saccharomycetaceae were more than twofold higher in controls carrying Blastocystis compared to controls not colonized (39 and 15%, respectively). Considering the 10 most prevalent families, Aspergillaceae tended to be increased among controls not carrying Blastocystis compared to positive controls (Table 2). Metschnikowiaceae were increased among IBS subjects carrying Blastocystis compared to non-carriers (Table 2). Geotrichum, Aspergillus, Saccharomyces, and Yarrowia were the most abundant genera (51, 16, 9, and 13% in the IBS group, and 23, 38, 17, and 8% in controls, respectively; Figure 7B). Clavispora relative abundance was significantly decreased among IBS patients compared to healthy subjects (1.47 × 10–5 and 2.52 × 10–3, respectively, Mann–Whitney test, p = 0.031). In the same way, there was also a decrease close to significance of Penicillium relative abundance (6.04 × 10–3 and 3.09 × 10–2, respectively, Mann–Whitney test, p = 0.063). The relative abundance and p-values of the most abundant genera in our study and of the genera of the core mycobiota are reported in Table 2. Considering the relative abundance, Penicillium were significantly reduced in Blastocystis carriers of the control group. Clavispora and Trichosporon were significantly increased in Blastocystis carriers of the IBS group (Table 2).
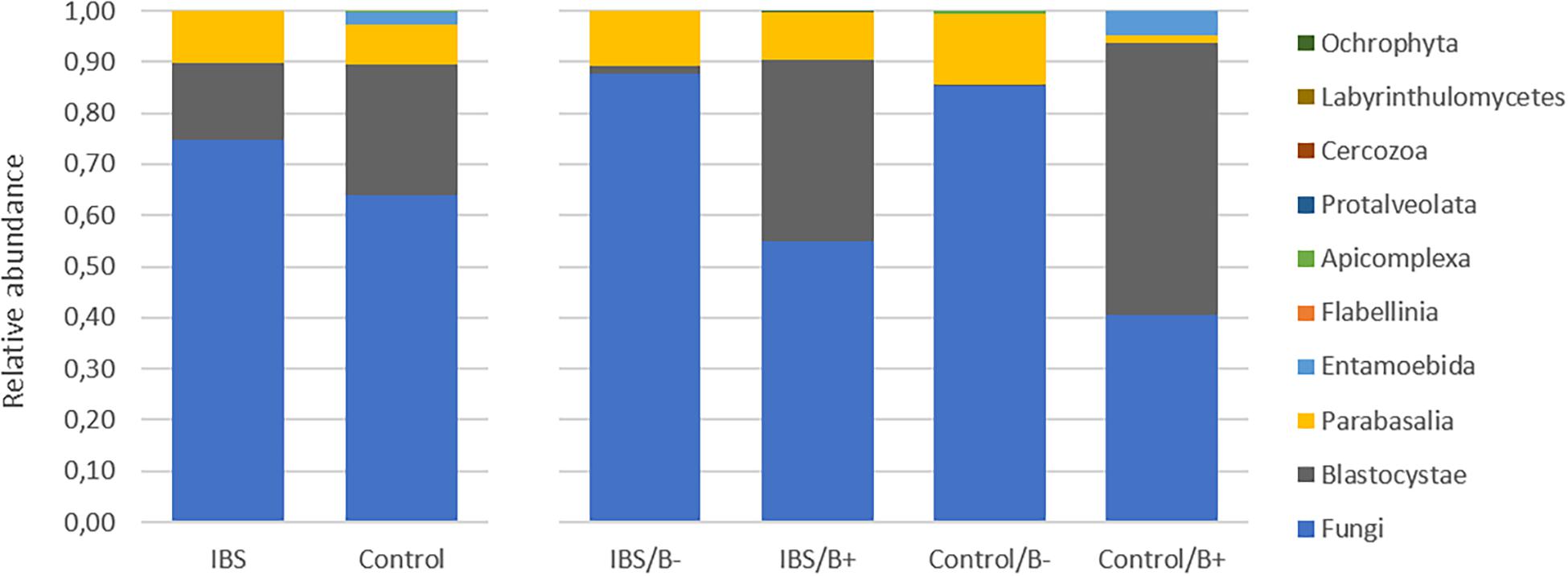
Figure 5. Comparison of the fecal eukaryotic microbiota composition between controls and IBS subjects at the class level. IBS/B-: Blastocystis-negative IBS patients; IBS/B + : Blastocystis-positive IBS patients; Control/B-: Blastocystis-negative control subjects; Control/B + : Blastocystis-positive control subjects.
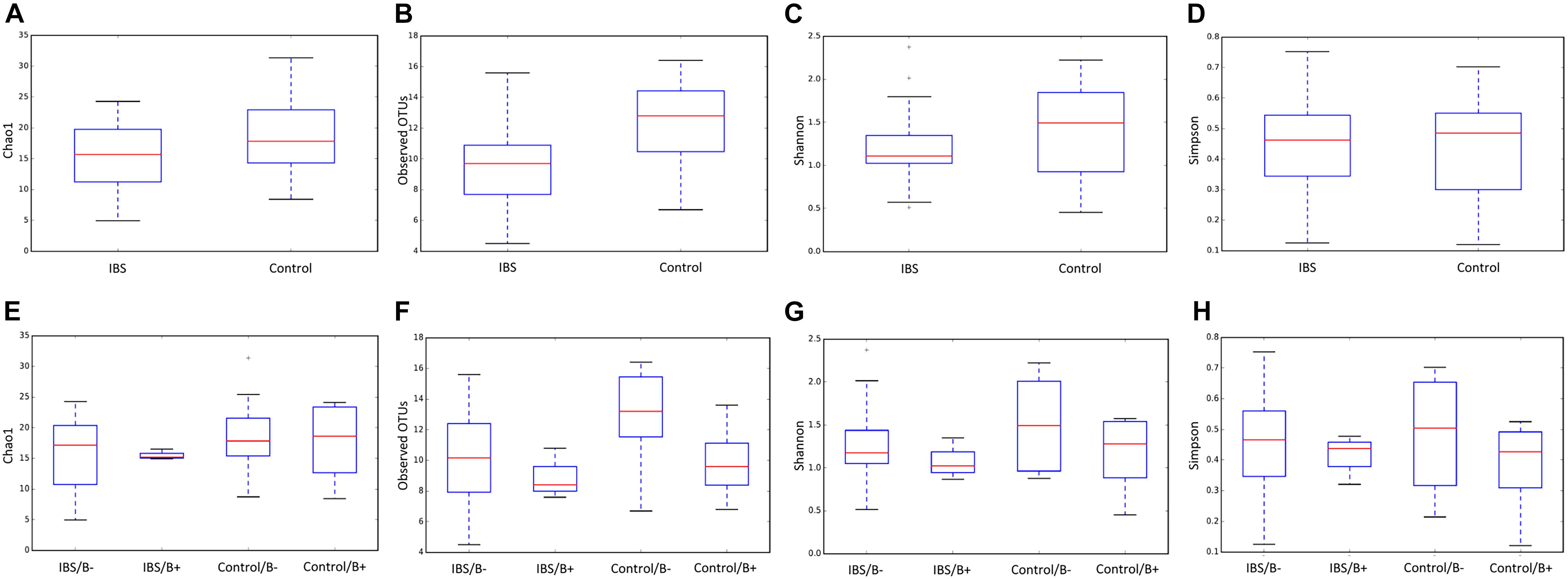
Figure 6. Comparison of alpha diversity indices of the fecal fungal microbiota between controls (E–H) and IBS-C subjects (A–D) according to Blastocystis carriage. (A,E) Chao 1, (B,F) observed number of OTUs, (C,G) Shannon diversity, and (D,H) Simpson’s index. Significant (or close to significance) p-values were reported on boxplots. IBS/B-: Blastocystis-negative IBS patients; IBS/B + : Blastocystis-positive IBS patients; Control/B-: Blastocystis-negative control subjects; Control/B + : Blastocystis-positive control subjects.
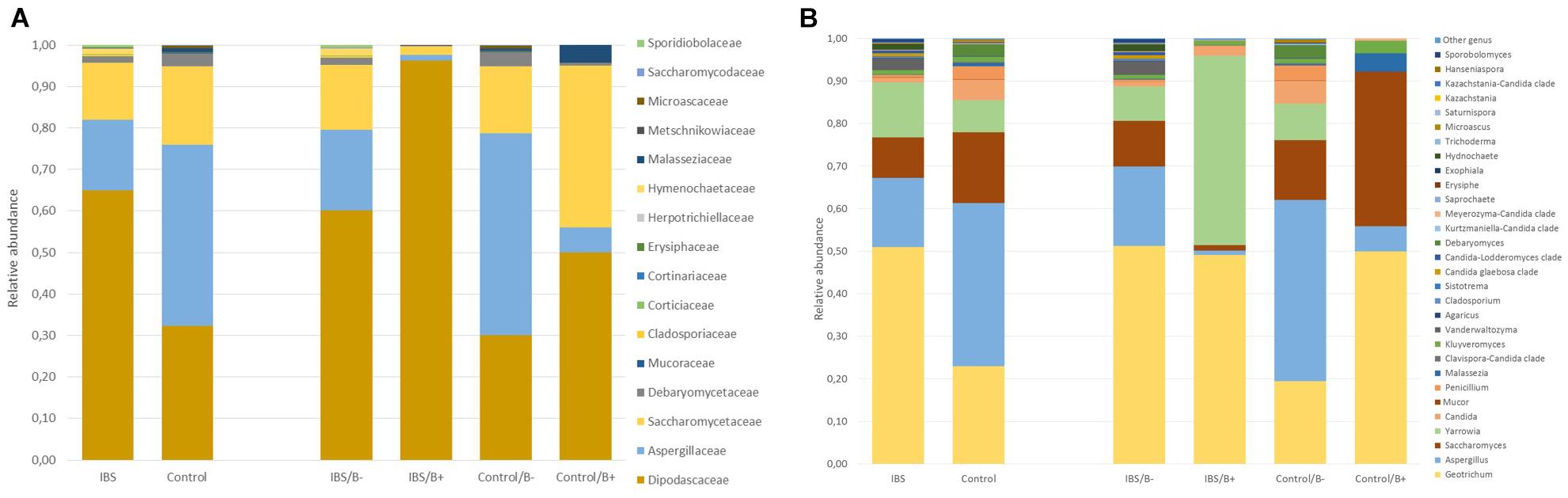
Figure 7. Comparison of the fecal mycobiota composition between controls and IBS subjects at family (A) and genus (B) levels. IBS/B-: Blastocystis-negative IBS patients; IBS/B+: Blastocystis-positive IBS patients; Control/B-: Blastocystis-negative control subjects; Control/B+: Blastocystis-positive control subjects.
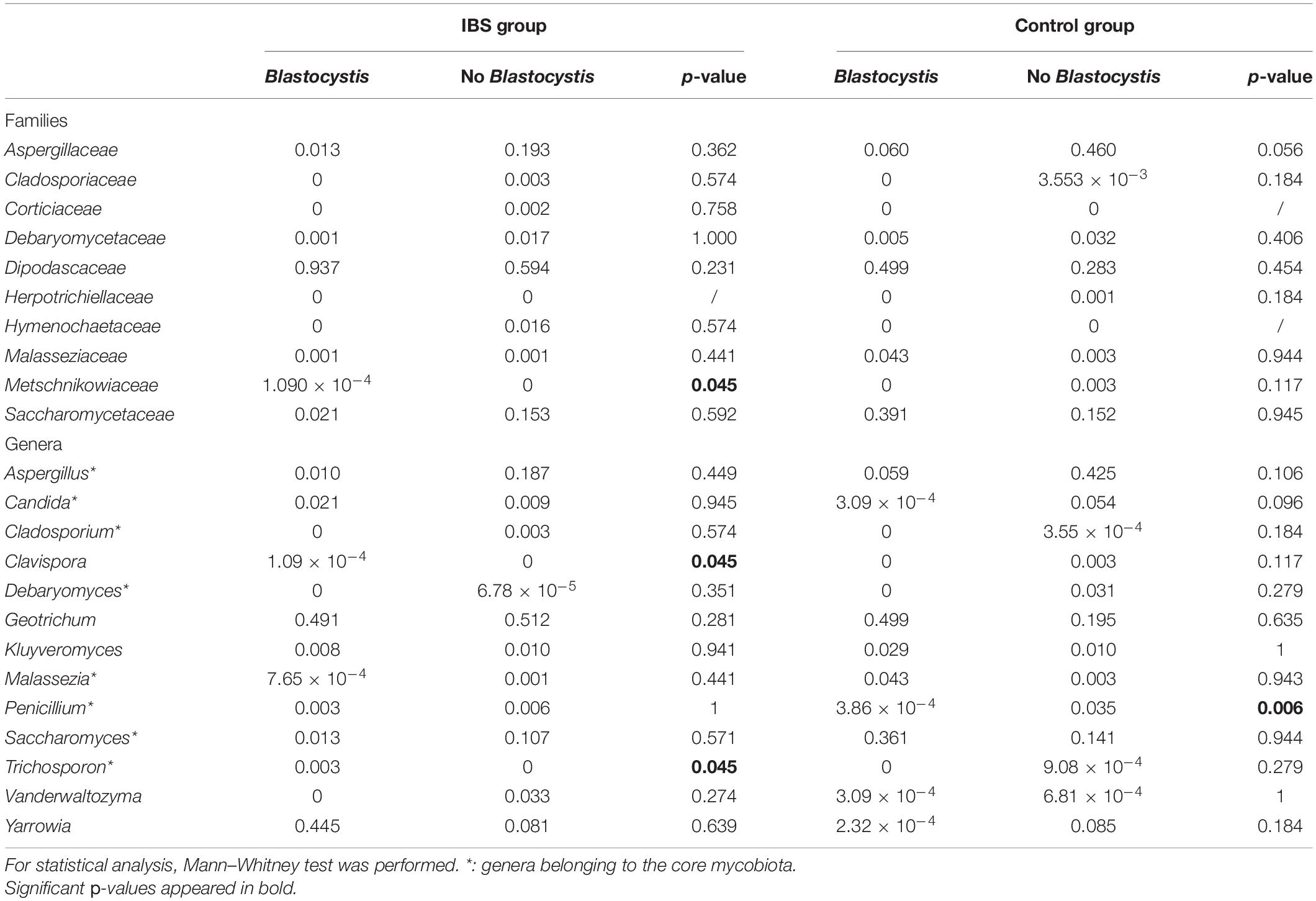
Table 2. Relative abundances of the 10 most prevalent fungal families and the most commonly detected genera in gut mycobiome studies (core mycobiota) and in this study.
Our study was motivated by the literature of the last decade reporting high prevalence of Blastocystis in IBS patients, the concerns about the risk associated with Blastocystis in fecal microbiota transplantation, and recent studies in animal models (Nourrisson et al., 2014; Terveer et al., 2019; Defaye et al., 2020). Recent works have studied the impact of Blastocystis in IBS-D patients, but none in IBS-C (Nagel et al., 2016). As each type of IBS is considered to be profoundly different from each other, we decided to focus on IBS-C patients. So, our data are the first to describe both prokaryotic and eukaryotic microbiota in IBS-C patients colonized with Blastocystis.
Classical IBS-C symptoms were described by patients, mostly female as expected with IBS, whether or not they were carriers of Blastocystis. Particularly, severity, based on Francis score, was not different according to Blastocystis carriage. All patients consume conventional treatments (antispasmodic, laxatives, transit accelerators, anti-bloating, etc.), periodically or over the long term, so it was difficult to compare them with each other. Taking probiotics was a non-inclusion criterion in order not to induce bias during the study of the microbiota. Thus, patients enrolled in this study are permitted to constitute comparable groups.
As previously reported, we observed a decrease in the prokaryotic microbiota richness of IBS-C subjects. Decrease in alpha diversity was reported from numerous studies interested in chronic intestinal or extra-intestinal diseases and is suspected to be associated with an altered function of gut microbiota (Le Chatelier et al., 2013). Conversely, a higher bacterial diversity is commonly associated with good health and lower incidence of inflammatory diseases (Loh and Blaut, 2012). Blastocystis was reported to be strongly associated with broad shifts in the gut-resident bacterial community and an increase in bacterial alpha diversity (Deng et al., 2021). In our study, we did not find significant increase of alpha diversity in Blastocystis carriers from the control group, but a trend was observed in IBS-C. Facing this increase in alpha diversity associated with Blastocystis carriage, some authors suggested that Blastocystis may be a component of a healthy microbiota (Deng et al., 2021). This point remains to be clarified as Blastocystis is reported to be more frequent in IBS patients, which present a dysbiosis. Moreover, pathogenic protozoan, such as Giardia duodenalis, Entamoeba histolytica, or Cryptosporidium sp., were associated with an increase in gut bacterial diversity or with altered microbiome profiles compared to uninfected people (Burgess and Petri, 2016; Fekete et al., 2020).
At the taxonomic level, our results were congruent with the literature, as we observed in IBS-C patients an increase in the Bacteroidetes phylum leading to an inversion of the Firmicutes/Bacteroidetes ratio, and an increase in the Proteobacteria phylum (Pittayanon et al., 2019). The Firmicutes/Bacteroidetes ratio was increased in Blastocystis carriers of both IBS-C and control groups, which is in favor of a less inflammatory gut environment. In our study, Blastocystis carriage was associated with a high relative abundance of the Tenericutes phylum, which is composed of commensals and pathogens (Wang et al., 2020). Even though this phylum is frequently modified in microbiota studies, little is known about its involvement in human health. In control group, we also observed in Blastocystis-positive individuals an increase in Ruminococcaceae, which are key symbionts of the gut ecosystem. This result was previously observed in subjects infected with E. histolytica or E. dispar (Morton et al., 2015). Interestingly, we recently described in rat an increase of Tenericutes phylum after experimental infections with Blastocystis, suggesting that this phylum may be specifically modified by the parasite, independently of the underlying intestinal disease (Defaye et al., 2020).
Functional predictions of the microbial capabilities were strongly impacted by the IBS pathology, irrespective of Blastocystis status. Due to this significant impact of IBS, it did not seem relevant to us to look for the differences in functional capacities by taking into account only the presence of Blastocystis. KEGG orthology pathway of glycerolipid metabolism was found to be significantly enriched in healthy subjects carrying Blastocystis compared to non-carrier healthy subjects. The role of the parasite on enrichment of genes involved in this metabolism has to be confirmed with further studies.
In the gut ecosystem, fungi and bacteria directly interact with each other (Lynch and Pedersen, 2016). Despite being a minor component of the gut microbial community, evidences indicated that fungi can play a role in gut diseases, as demonstrated in inflammatory bowel disease (Sokol et al., 2017). Whereas extensive literature is available about prokaryotic dysbiosis associated with IBS, few studies were interested in eukaryoma. Fungal dysbiosis has been reported in IBS with an enrichment of Saccharomycetes and a decrease of alpha diversity (Botschuijver et al., 2017; Sciavilla et al., 2021). Considering the fecal eukaryoma, and more precisely the Fungi class, the relative abundance of Dipodascaceae was twofold higher in IBS-C subjects compared to controls. Clavispora and Penicillium genera were decreased in IBS-C compared to healthy subjects.
With the work by Nieves- Ramirez, our study is the second to address intestinal eukaryotic microflora associated with Blastocystis carriage. In our study, Blastocystis colonization was associated with more discrete differences in the eukaryotic microbiota compared to prokaryotic shifts, without modification of alpha or beta diversity. Nieves-Ramirez and colleagues reported recently a modification of fungal microbiota of a rural Mexican population colonized with Blastocystis (Nieves-Ramírez et al., 2018). They reported an increase in yeast and fungal species (Debaryomyces hansenii, Mucor mucedo, Aspergillus flavus, Mucor racemosus, and Issatchenkia terricola) and a decrease of Hymenolepis nana in patients colonized with Blastocystis. The transposition of these results is not possible to a population of industrialized countries, where infections due to H. nana are absent. We did not observe significant modifications of Debaryomyces and Mucor genera according to Blastocystis carriage. These differences could be explained by a different region of the 18S rRNA gene targeted in the two studies. Diet could also have impacted proportions of some fungi (Richard and Sokol, 2019). Interestingly, in our results, Dipodascaceae family was increased in the presence of the parasite in the two groups of patients, whereas Aspergillaceae, Aspergillus, and Penicillium were decreased. At the opposite end, the modifications of some groups were different depending on whether one considered the IBS or the control groups. Clavispora were significantly increased in IBS patients carrying the parasite, whereas in the control group, this genus was only encountered in negative subjects. Clavispora has been reported to be increased in patients suffering from Crohn’s disease (Lewis et al., 2015). Interestingly, when Blastocystis was present in the IBS group, the modifications were the same as previously described in Dectin-1 knockout mice; i.e., the proportion of opportunistic pathogenic fungi including Candida and Trichosporon increased, whereas non-pathogenic Saccharomyces decreased (Iliev et al., 2012). These mice experienced more severe DSS-induced colitis than wild-type mice, suggesting that this mycobiota pattern could be linked to inflammatory environment in predisposed animals. These modifications were not observed in healthy subjects in whom Saccharomyces genus was increased when Blastocystis was present. Saccharomyces are known to limit inflammatory response and increase immune health; these yeasts have also been used as probiotics (Kourelis et al., 2010). No differences were observed among the other genera that may constitute the core mycobiota (i.e., gut residents) previously described (Richard and Sokol, 2019).
We recognize that our study presents some limitations. First, a limited number of patients were included. This impacts on the analysis of secondary criteria. Also, we did not address the link between Blastocystis STs and microbiota modifications because we were limited by the number of subjects included. The role of Blastocystis ST was mentioned several times, but this hypothesis needs further investigations (Yason et al., 2019; Deng et al., 2021). Francis score subgroup analysis could not be performed either. However, it would be interesting to investigate a correlation between changes in the microbiota of patients and the severity of their symptoms. Moreover, the presence of other parasites may also have slightly affected our results. Indeed, two patients of the control group were positive for Entamoeba sp. and a low rate of Blastocystis/D. fragilis co-carriages was detected in both IBS-C and control groups. Nevertheless, it underlies the difficulty to interpret to what degree Blastocystis contributes to microbiota changes in human studies. So, our results suggest that when studying the link between Blastocystis (or other parasites) and prokaryotic microbiota in human, we should also perform analyses of associated eukaryoma.
To conclude, Blastocystis is associated with modifications of the fecal microbiota, but which do not go in the same direction depending on whether we consider healthy people or IBS-C patients, particularly for the mycobiota. Even though animal models have highlighted some modifications specifically attributable to this parasite, we still do not know for most of the microbiota changes if they are due to the presence of Blastocystis. So, further experimentations with animal models using various Blastocystis ST, and/or humanized microbiota models, could help to understand the role of Blastocystis in the gut environment.
The datasets generated for this study can be found in NCBI PRJNA730687.
The studies involving human participants were reviewed and approved by this clinical study was approved by the Research Ethics Committees of the Hospital of Clermont-Ferrand (“Comité de Protection des Personnes Sud-Est VI,” France) with the reference number 2013-A00031-44. The patients/participants provided their written informed consent to participate in this study.
CN, MD, and PP: conceptualization. CN, JS, and PP: methodology. CN and PP: software, formal analysis, and visualization. CN, FD, MD, and PP: validation. CN, JS, JB, and MD: investigation. FD and PP: resources. CN: data curation and writing – original draft preparation. JS, JB, FD, MD, and PP: writing, review, and editing. FD, MD, and PP: supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
This study was funded by “GIRCI Auvergne Rhône Alpes 2013.”
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to the Mésocentre Clermont Auvergne University for providing help and storage resources. Computations have been performed on the supercomputer facilities of the Mésocentre Clermont Auvergne University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.713347/full#supplementary-material
Supplementary Figure 1 | LDA effect size, log10 transformed q value (FDR-adjusted p value) and species annotation are shown. (A) Control group, (B) IBS group. Green bars indicate species enriched in non-infected subjects, while red bars indicate species enriched in Blastocystis-infected subjects. Statistical significance was determined by LefSe analysis with FDR correction (only those species with q values < 0.05 and LDA effect size > 2 are shown).
Supplementary Table 1 | Sequences of the primers used in the study.
Supplementary Table 2 | Demographic characteristics and clinical data of patients included. F, female; M, male; nd, not determined; na, not applicable.
Alfellani, M. A., Taner-Mulla, D., Jacob, A. S., Imeede, C. A., Yoshikawa, H., Stensvold, C. R., et al. (2013). Genetic diversity of blastocystis in livestock and zoo animals. Protist 164, 497–509. doi: 10.1016/j.protis.2013.05.003
Aßhauer, K. P., Wemheuer, B., Daniel, R., and Meinicke, P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884. doi: 10.1093/bioinformatics/btv287
Botschuijver, S., Roeselers, G., Levin, E., Jonkers, D. M., Welting, O., Heinsbroek, S. E. M., et al. (2017). Intestinal fungal dysbiosis associates with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology 153, 1026–1039. doi: 10.1053/j.gastro.2017.06.004
Burgess, S. L., and Petri, W. A. (2016). The intestinal bacterial microbiome and e. histolytica infection. Curr. Trop. Med. Rep. 3, 71–74. doi: 10.1007/s40475-016-0083-1
Cammarota, G., Ianiro, G., Tilg, H., Rajilić-Stojanović, M., Kump, P., Satokari, R., et al. (2017). European consensus conference on faecal microbiota transplantation in clinical practice. Gut 66, 569–580. doi: 10.1136/gutjnl-2016-313017
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Defaye, M., Nourrisson, C., Baudu, E., Lashermes, A., Meynier, M., Meleine, M., et al. (2020). Fecal dysbiosis associated with colonic hypersensitivity and behavioral alterations in chronically blastocystis-infected rats. Sci. Rep. 10:9146. doi: 10.1038/s41598-020-66156-w
Deng, L., Wojciech, L., Gascoigne, N. R. J., Peng, G., and Tan, K. S. W. (2021). New insights into the interactions between blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 17:e1009253. doi: 10.1371/journal.ppat.1009253
El Safadi, D., Cian, A., Nourrisson, C., Pereira, B., Morelle, C., Bastien, P., et al. (2016). Prevalence, risk factors for infection and subtype distribution of the intestinal parasite blastocystis sp. from a large-scale multi-center study in France. BMC Infect. Dis. 16:451. doi: 10.1186/s12879-016-1776-8
Fekete, E., Allain, T., Siddiq, A., Sosnowski, O., and Buret, A. G. (2020). Giardia spp. and the gut microbiota: dangerous liaisons. Front. Microbiol. 11:618106. doi: 10.3389/fmicb.2020.618106
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-Type lectin receptor dectin-1 influence colitis. Science 336, 1314–1317. doi: 10.1126/science.1221789
Kourelis, A., Kotzamanidis, C., Litopoulou-Tzanetaki, E., Papaconstantinou, J., Tzanetakis, N., and Yiangou, M. (2010). Immunostimulatory activity of potential probiotic yeast strains in the dorsal air pouch system and the gut mucosa. J. Appl. Microbiol. 109, 260–271. doi: 10.1111/j.1365-2672.2009.04651.x
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
Lewis, J. D., Chen, E. Z., Baldassano, R. N., Otley, A. R., Griffiths, A. M., Lee, D., et al. (2015). Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500. doi: 10.1016/j.chom.2015.09.008
Li, H., Qu, J., Li, T., Li, J., Lin, Q., and Li, X. (2016). Pika population density is associated with the composition and diversity of gut microbiota. Front. Microbiol. 7:758. doi: 10.3389/fmicb.2016.00758
Loh, G., and Blaut, M. (2012). Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 3, 544–555. doi: 10.4161/gmic.22156
Longstreth, G. F., Thompson, W. G., Chey, W. D., Houghton, L. A., Mearin, F., and Spiller, R. C. (2006). Functional bowel disorders. Gastroenterology 130, 1480–1491. doi: 10.1053/j.gastro.2005.11.061
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Morton, E. R., Lynch, J., Froment, A., Lafosse, S., Heyer, E., Przeworski, M., et al. (2015). Variation in rural african gut microbiota is strongly correlated with colonization by entamoeba and subsistence. PLoS Genet. 11:e1005658. doi: 10.1371/journal.pgen.1005658
Nagel, R., Traub, R. J., Allcock, R. J. N., Kwan, M. M. S., and Bielefeldt-Ohmann, H. (2016). Comparison of faecal microbiota in blastocystis-positive and blastocystis-negative irritable bowel syndrome patients. Microbiome 4:47. doi: 10.1186/s40168-016-0191-0
Nieves-Ramírez, M. E., Partida-Rodríguez, O., Laforest-Lapointe, I., Reynolds, L. A., Brown, E. M., Valdez-Salazar, A., et al. (2018). Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. Msystems 3:e00007. doi: 10.1128/mSystems.00007-18
Nourrisson, C., Scanzi, J., Pereira, B., NkoudMongo, C., Wawrzyniak, I., Cian, A., et al. (2014). Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PloS One 9:e111868. doi: 10.1371/journal.pone.0111868
Nourrisson, C., Wawrzyniak, I., Cian, A., Livrelli, V., Viscogliosi, E., Delbac, F., et al. (2016). On blastocystis secreted cysteine proteases: a legumain-activated cathepsin b increases paracellular permeability of intestinal caco-2 cell monolayers. Parasitology 143, 1713–1722. doi: 10.1017/S0031182016001396
Parfrey, L., Wegener, W. A., Walters, C. L., Lauber, J. C., Clemente, D., Berg-Lyons, C., et al. (2014). Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 5:298. doi: 10.3389/fmicb.2014.00298
Pittayanon, R., Lau, J. T., Yuan, Y., Leontiadis, G. I., Tse, F., Surette, M., et al. (2019). Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology 157, 97–108. doi: 10.1053/j.gastro.2019.03.049
Poirier, P., Wawrzyniak, I., Albert, A., El Alaoui, H., Delbac, F., and Livrelli, V. (2011). Development and evaluation of a real-time PCR assay for detection and quantification of blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J. Clin. Microbiol. 49, 975–983. doi: 10.1128/JCM.01392-10
Poirier, P., Wawrzyniak, I., Vivarès, C. P., Delbac, F., and El Alaoui, H. (2012). New insights into Blastocystis spp.: a potential link with irritable bowel syndrome. PLoS Pathog. 8:e1002545. doi: 10.1371/journal.ppat.1002545
Ramírez, J. D., Sánchez, A., Hernández, C., Flórez, C., Consuelo Bernal, M., Giraldo, J. C., et al. (2016). Geographic distribution of human blastocystis subtypes in South America. Infect. Genet. Evol. 41, 32–35. doi: 10.1016/j.meegid.2016.03.017
Richard, M. L., and Sokol, H. (2019). The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 16, 331–345. doi: 10.1038/s41575-019-0121-2
Sciavilla, P., Strati, F., Di Paola, M., Modesto, M., Vitali, F., Cavalieri, D., et al. (2021). Gut microbiota profiles and characterization of cultivable fungal isolates in IBS patients. Appl. Microbiol. Biotechnol. 105, 3277–3288. doi: 10.1007/s00253-021-11264-4
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sokol, H., Leducq, V., Aschard, H., Pham, H.-P., Jegou, S., Landman, C., et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Stensvold, C. R., and Clark, C. G. (2020). Pre-empting pandora’s box: blastocystis subtypes revisited. Trends Parasitol. 36, 229–232. doi: 10.1016/j.pt.2019.12.009
Terveer, E. M., van Gool, T., Ooijevaar, R. E. I, Sanders, M. J. G., Boeije-Koppenol, E., Keller, J. J., et al. (2019). Human transmission of blastocystis by fecal microbiota transplantation without development of gastrointestinal symptoms in recipients. Clin. Infect. Dis. 71, 2630–2636. doi: 10.1093/cid/ciz1122
Wang, Y., Huang, J.-M., Zhou, Y.-L., Almeida, A., Finn, R. D., Danchin, A., et al. (2020). Phylogenomics of expanding uncultured environmental tenericutes provides insights into their pathogenicity and evolutionary relationship with bacilli. BMC Genomics 21:408. doi: 10.1186/s12864-020-06807-4
Keywords: Blastocystis, gut microbiota, irritable bowel syndrome, IBS-C, eukaryome, 16S/18S ribosomal RNA gene analysis
Citation: Nourrisson C, Scanzi J, Brunet J, Delbac F, Dapoigny M and Poirier P (2021) Prokaryotic and Eukaryotic Fecal Microbiota in Irritable Bowel Syndrome Patients and Healthy Individuals Colonized With Blastocystis. Front. Microbiol. 12:713347. doi: 10.3389/fmicb.2021.713347
Received: 22 May 2021; Accepted: 26 August 2021;
Published: 17 September 2021.
Edited by:
Olga Matos, New University of Lisbon, PortugalReviewed by:
Xiaojun Zhuang, The First Affiliated Hospital of Sun Yat-sen University, ChinaCopyright © 2021 Nourrisson, Scanzi, Brunet, Delbac, Dapoigny and Poirier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Poirier, cHBvaXJpZXJAY2h1LWNsZXJtb250ZmVycmFuZC5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.