
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 September 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.709942
This article is part of the Research TopicWorldwide Emergence of Drug Resistant Fungi: from Basic to ClinicView all 24 articles
Fusarium species exhibit significant intrinsic resistance to most antifungal agents and fungicides, resulting in high mortality rates among immunocompromised patients. Consequently, a thorough characterization of the antifungal resistance mechanism is required for effective treatments and for preventing fungal infections and reducing antifungal resistance. In this study, an isolate of Fusarium oxysporum (wild-type) with broadly resistant to commonly antifungal agents was used to generate 1,450 T-DNA random insertion mutants via Agrobacterium tumefaciens-mediated transformation. Antifungal susceptibility test results revealed one mutant with increased sensitivity to azoles. Compared with the resistant wild-type, the mutant exhibited low MICs to KTZ, ITC, VRC, POS, and PCZ (0.125, 1, 0.06, 0.5, and 0.125μg/ml, respectively). The T-DNA insertion site of this mutant was characterized as involving two adjacent genes, one encoding a hypothetical protein with unknown function and the other encoding the NADPH-cytochrome P450 reductase, referred as CPR1. To confirm the involvement of these genes in the altered azole susceptibility, the independent deletion mutants were generated and the Cpr1 deletion mutant displayed the same phenotypes as the T-DNA random mutant. The deletion of Cpr1 significantly decreased ergosterol levels. Additionally, the expression of the downstream Cyp51 gene was affected, which likely contributed to the observed increased susceptibility to azoles. These findings verified the association between Cpr1 and azole susceptibility in F. oxysporum. Furthermore, this gene may be targeted to improve antifungal treatments.
Fusarium species, which are well-known filamentous ascomycetous fungi, include many agriculturally important plant pathogens and opportunistic pathogens of humans and other animals (Ma et al., 2013; Al-Hatmi et al., 2016; Tupaki-Sreepurna and Kindo, 2018; Zhao et al., 2021). Fusarium species usually cause local infections, including fungal keratitis, which often leads to blindness. However, over the last few decades, the number of dangerously invasive infections has increased in immunocompromised individuals, especially cancer patients with prolonged neutropenia and patients with hematological disorders. These infections can spread to the lungs, heart, liver, kidneys, and central nervous system (Tupaki-Sreepurna and Kindo, 2018; Lockhart and Guarner, 2019; Batista et al., 2020; Hof, 2020). As emerging fungal pathogens, some Fusarium species, such as Fusarium oxysporum and Fusarium solani, are now among the most common pathogenic molds associated with significant morbidity and mortality, behind only Aspergillus and Mucorales molds (Miceli and Lee, 2011; Guarro, 2013; Tortorano et al., 2014; Al-Hatmi et al., 2016; Lockhart and Guarner, 2019; Hof, 2020).
Antifungal therapy is necessary for successful disease management. However, because of intrinsic resistance and selection pressure, infections caused by Fusarium species are relatively difficult to treat. Most species of this genus are typically resistant to a broad range of antifungal agents developed for clinical use, including azoles, polyenes, and echinocandin. They are also minimally susceptible to agricultural fungicides (Azor et al., 2007; Miceli and Lee, 2011; Ma et al., 2013; Ribas et al., 2016; Sharma and Chowdhary, 2017; Batista et al., 2020; Hof, 2020). In vitro studies have indicated amphotericin B and echinocandin are relatively ineffective for controlling Fusarium species, whereas triazoles, such as voriconazole and posaconazole, are effective against almost 50% of isolates (Azor et al., 2007; Miceli and Lee, 2011; Tortorano et al., 2014). Therefore, the mechanisms underlying the antifungal resistance of Fusarium species must be characterized.
Most of the studies on the antifungal resistance of pathogenic fungi conducted to date have focused on the genera Candida and Aspergillus. There has been relatively little related research regarding Fusarium species, with most studies examining the susceptibility of the species to antifungal agents. The few studies analyzing resistance mechanisms have mostly involved plant pathogens and investigations of the changes in the amino acid sequence encoded by the Fks1 gene or the effects of overexpressing the Cyp51 gene or the genes encoding ABC efflux pumps (Katiyar and Edlind, 2009; Abou Ammar et al., 2013; Zhang et al., 2021; Zhao et al., 2021).
To identify genes related to the antifungal resistance of Fusarium species, Agrobacterium tumefaciens-mediated transformation (ATMT) was used to construct T-DNA random insertion mutants. The 1,450 generated mutants from a broadly resistant isolate of F. oxysporum included FOM1123, which exhibited altered susceptibility to azoles. We functionally characterized the genes interrupted by the T-DNA insertion and clarified their regulatory roles related to antifungal resistance.
Wild-type F. oxysporum JLCC31768, which was originally isolated from a patient with fungal keratitis in Jilin province, China, was used to construct T-DNA random insertion mutants. The antifungal susceptibility test (AFST) results revealed it is broadly resistant to different azoles, amphotericin B, and caspofungin commonly used in clinical settings (Table 1).
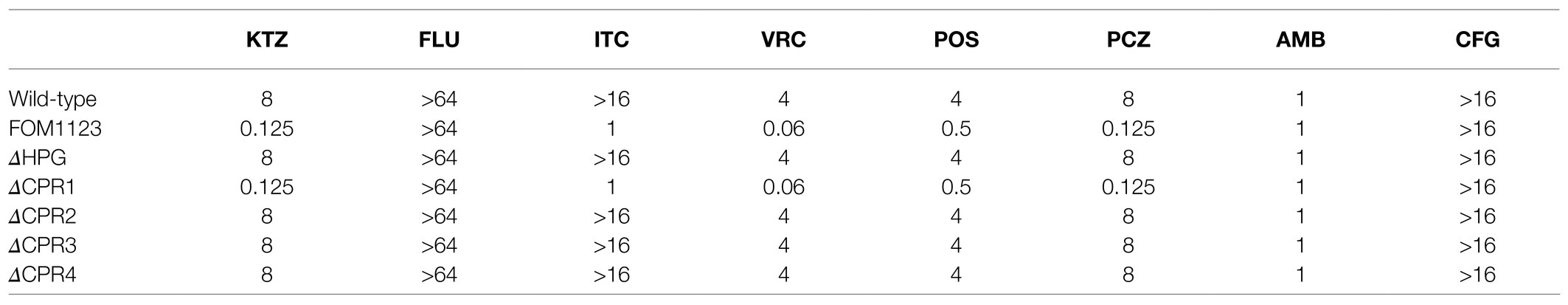
Table 1. Antifungal susceptibility test results for the wild-type Fusarium oxysporum and the mutants (MIC, μg/ml).
Plasmids pXEH and pXEN (containing neomycin and kanamycin resistance tags) as well as A. tumefaciens Agr0 and AgrN (containing pXEN) were used to generate F. oxysporum mutants. All strains and plasmids were preserved at the Jilin University Mycology Research Center (Jilin, China).
Antifungal resistance tests indicated that wild-type F. oxysporum is sensitive to geneticin (G418). Accordingly, geneticin was selected as a resistance tag. The geneticin phosphotransferase II gene (Neo) mediating G418 resistance was ligated to pXEH to construct the pXEN recombinant plasmid. A. tumefaciens Agr0 cells were transformed with pXEN to obtain the AgrN strain, which was used for ATMT.
The F. oxysporum T-DNA insertion mutants were generated as previously described (Fan et al., 2016). Briefly, fungal spores (1×104CFU/ml) were mixed with an equal volume (1ml) of AgrN cells (OD600nm=0.8). A Millipore filter was placed on the surface of solid induction medium containing 200μm acetosyringone. A 200μl aliquot of the spore–AgrN mixture was spread evenly on the filter. After incubating for 48h at 25°C in darkness, the filter was transferred to selection medium (PDA containing 200μm cefotaxime sodium and 100μg/ml G418) and incubated at 25°C. The mutants were used to inoculate PDA slants in tubes.
Genomic DNA was extracted from randomly selected mutants using the TIANgel Rapid Mini Plasmid Kit (Tiangen Biotech, Beijing, China) for a PCR amplification using the neoF and neoR primers specific for the Neo gene (Table 2). The amplified products were sequenced by Comate Bioscience Co., Ltd (Jilin, China), after which the sequences were analyzed to determine whether the T-DNA was inserted into the F. oxysporum genome. After multiple transformations, many T-DNA insertion mutants were preserved for further research.
The AFST was performed using the CLSI broth microdilution method as described in M38-Ed3 (Clinical and Laboratory Standards Institute, 2017). The following antifungal agents, including azole fungicides, were tested: fluconazole (FLU; NICPBP, Beijing, China), itraconazole (ITC; Sigma, St. Louis, MO, United States), voriconazole (VRC; Sigma), posaconazole (POS; Sigma), amphotericin B (AMB; Sigma), caspofungin (CFG; Meilunbio, Dalian, China), ketoconazole (KTZ; NICPBP), and propiconazole (PCZ; NICPBP). The antifungal agents were diluted 10 times (2-fold dilutions) for the following concentration ranges: FLU, 0.125–64μg/ml; ITC, VRC, POS, AMB, CFG, KTZ, and PCZ, 0.03–16μg/ml. As recommended by CLSI, Candida krusei ATCC6258 and Candida parapsilosis ATCC22019 were used as quality control strains. The MIC endpoint for AMB was defined as the lowest concentration with 100% growth inhibition relative to the antifungal-free control. For the other antifungal agents, the MICs were defined as the lowest concentration with a prominent decrease in growth (almost 50%) relative to the control. After a comparison with the wild-type F. oxysporum, the mutants with altered antifungal susceptibility were selected.
The sequences flanking the inserted T-DNA were amplified by touchdown-TAIL-PCR using previously described primers (Table 2; Gao et al., 2016). To determine the insertion sites, the flanking sequences were aligned with the F. oxysporum f. sp. lycopersici genome (taxid: 426428) using the Basic Local Alignment Search Tool (BLAST).1 Relevant information regarding the interrupted genes was obtained from the NCBI, KEGG, and UniProtKB databases.
By exploiting homologous genetic recombination, the genes interrupted by the T-DNA insertion, including Hpg as well as Cpr1 and its homologs (Cpr2, Cpr3, and Cpr4), were targeted using specific primers (Table 2) to produce F. oxysporum deletion mutants. The Hpg, Cpr1, Cpr2, Cpr3, and Cpr4 genes were replaced by Neo in pXEN, which was then inserted into Agr0 cells. The ΔHPG, ΔCPR1, ΔCPR2, ΔCPR3, and ΔCPR4 deletion mutants were generated by ATMT.
To compare colony morphologies, PDA medium was inoculated with the wild-type F. oxysporum and the deletion mutants and then incubated at 25°C for 5days. Slide cultures were prepared for these strains and then examined using a microscope after lactophenol cotton blue staining. Additionally, the susceptibility of the deletion mutants to antifungal agents was tested as described in M38-Ed3 (Clinical and Laboratory Standards Institute, 2017).
Fungal spores (1×106CFU) were used to inoculate 100ml PDB medium, which was then incubated at 25°C for 48h with shaking (180rpm). In the antifungal treatment group, VRC (amount corresponding to the 0.5 MIC) was added after 24h. Next, 100mg mycelia were collected and resuspended in 5ml deionized water before being disrupted for 20min using the Scientz-IID ultrasonic cell disrupter (SCIENTZ, Ningbo, China). A 5ml aliquot of the solution was mixed with 20ml ether. The absorbance of the resulting extract was measured at 281.5nm. The ergosterol content was calculated using a standard curve.
Mycelia were disrupted by grinding in liquid nitrogen. Total RNA was extracted from the ground material using the RNAiso Plus kit (Takara, Shiga, Japan). The RNA concentration was determined using the NanoDrop One spectrophotometer (Thermo Fisher, San Jose, CA, United States). The RNA served as the template for synthesizing cDNA using the HiScript II Q RT SuperMix for qPCR (Vazyme, Nanjing, China). Quantitative real-time PCR was performed using the AceQ qPCR SYBR Green Master Mix (Vazyme), gene-specific primers (Table 3), and the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The expression levels of genes related to ergosterol synthesis (e.g., Cpr, Cytb5, and Cyp51) were normalized against the expression of the 18S rRNA housekeeping gene. Relative gene expression levels were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Data are presented as the mean±standard deviation of at least three replicated measurements. Differences between the VRC-treated and untreated samples were evaluated by a one-way ANOVA followed by the T test using SPSS 2.0 (p<0.05 was set as the threshold for significance).
Using an established ATMT system, wild-type F. oxysporum was transformed, with an efficiency of 250 mutants per 104CFU. The PCR amplification of the Neo gene resulted in a single specific amplicon (approximately 700~750bp) for all T-DNA random mutants generated in this study (Figure 1). The sequenced fragment was 99% similar to the Neo gene, indicating the T-DNA containing the G418 resistance tag was inserted into the F. oxysporum genome. After multiple transformations, 1,450 mutants were obtained.
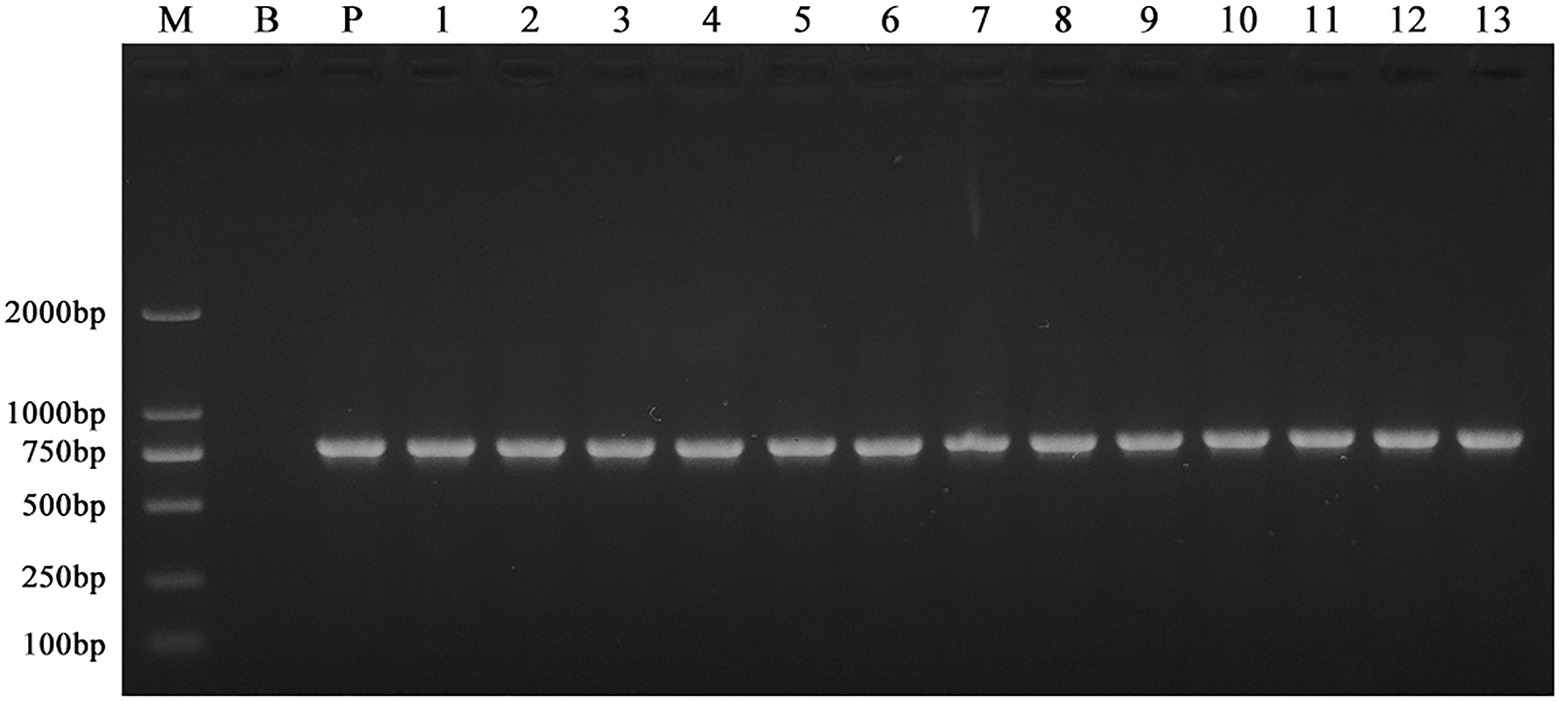
Figure 1. PCR amplification of the Neo gene in the wild-type F. oxysporum and the T-DNA insertion mutants. Genomic DNA of the mutants grown on the selection medium containing G418 was amplified using the neoF and neoR primers. All the mutants generated in this study produced a specific amplicon (approximately 700~750bp). Here, only showed the results of 13 different mutants selected randomly. These indicated the T-DNA containing the G418 resistance tag was inserted into the F. oxysporum genome. M: Trans 2 K marker; B: wild-type; P: pXEN; and lanes 1–13: 13 mutants with different T-DNA insertion.
The AFST results for the 1,450 confirmed mutants revealed one mutant (FOM1123) with altered antifungal susceptibility. More specifically, this mutant exhibited significantly increased susceptibility to azoles (except for FLU) with low MICs to KTZ, ITC, VRC, POS, and PCZ (0.125, 1, 0.06, 0.5, and 0.125μg/ml, respectively), compared with the resistant wild-type with high MICs (8,>16, 4, 4, and 8μg/ml, respectively). In contrast, its susceptibility to the polyene AMB and the echinocandin CFG was unchanged (Table 1). These observations suggested that the gene interrupted by T-DNA insertion in this mutant might be related to azole resistance in the wild-type F. oxysporum.
The sequences flanking the inserted T-DNA in the mutant FOM1123 were amplified by touchdown-TAIL-PCR and sequenced. The subsequent BLAST analysis of the F. oxysporum genome indicated the T-DNA fragment replaced a 5,312bp sequence from 2,932,119bp to 2,937,431bp on chromosome 2 between the initiation regions of genes FOXG_08273 and FOXG_08274 (Figure 2). The FOXG_08273 gene encodes a hypothetical protein (HPG) comprising 2,548 amino acids. Its function is unknown, and no homologs were identified. The FOXG_08274 gene encodes an NADPH-cytochrome P450 reductase (CPR1) consisting of 692 amino acids. This gene, which is related to ergosterol biosynthesis, contributes to the delivery of electrons to P450 enzymes, similar to Cytb5 (cytochrome b5). It has three homologs, namely, FOXG_07461 (Cpr2 on chromosome 4), FOXG_03206 (Cpr3 on chromosome 8), and FOXG_04834 (Cpr4 on chromosome 7).
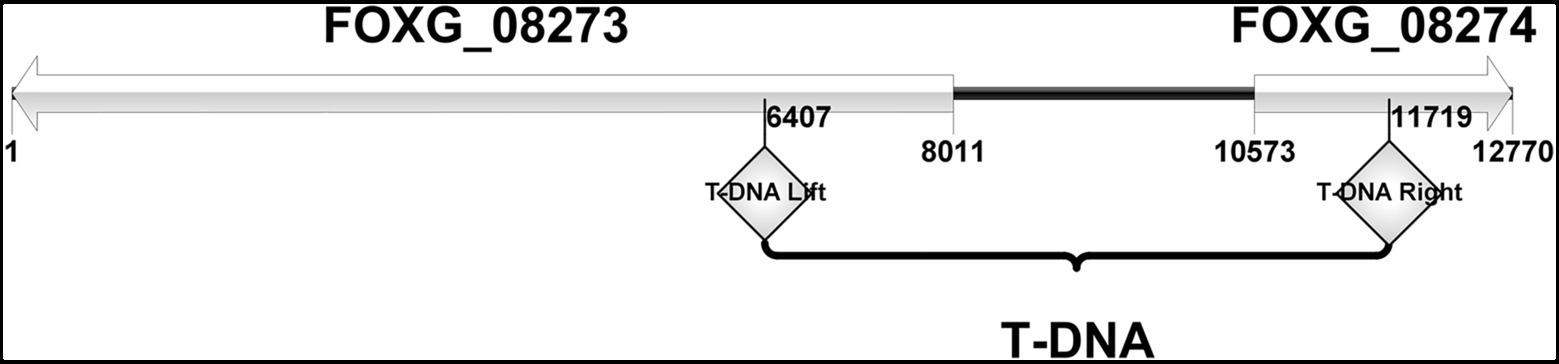
Figure 2. The site of T-DNA insertion of mutant FOM1123. It was characterized as involving 2 adjacent genes, FOXG_08273 and FOXG_08274. The inserted T-DNA replaced a 5,312bp sequence between the initiation regions of these two genes, from 2,932,119bp to 2,937,431bp on F. oxysporum chromosome 2.
Compared with the wild-type F. oxysporum, the T-DNA insertion mutant FOM1123 and the deletion mutants ΔHPG, ΔCPR1, ΔCPR2, ΔCPR3, and ΔCPR4 had no obvious differences regarding colony and microscopic morphological characteristics, including mycelial growth, pigment production, spore germination, and spore structure (Figure 3).
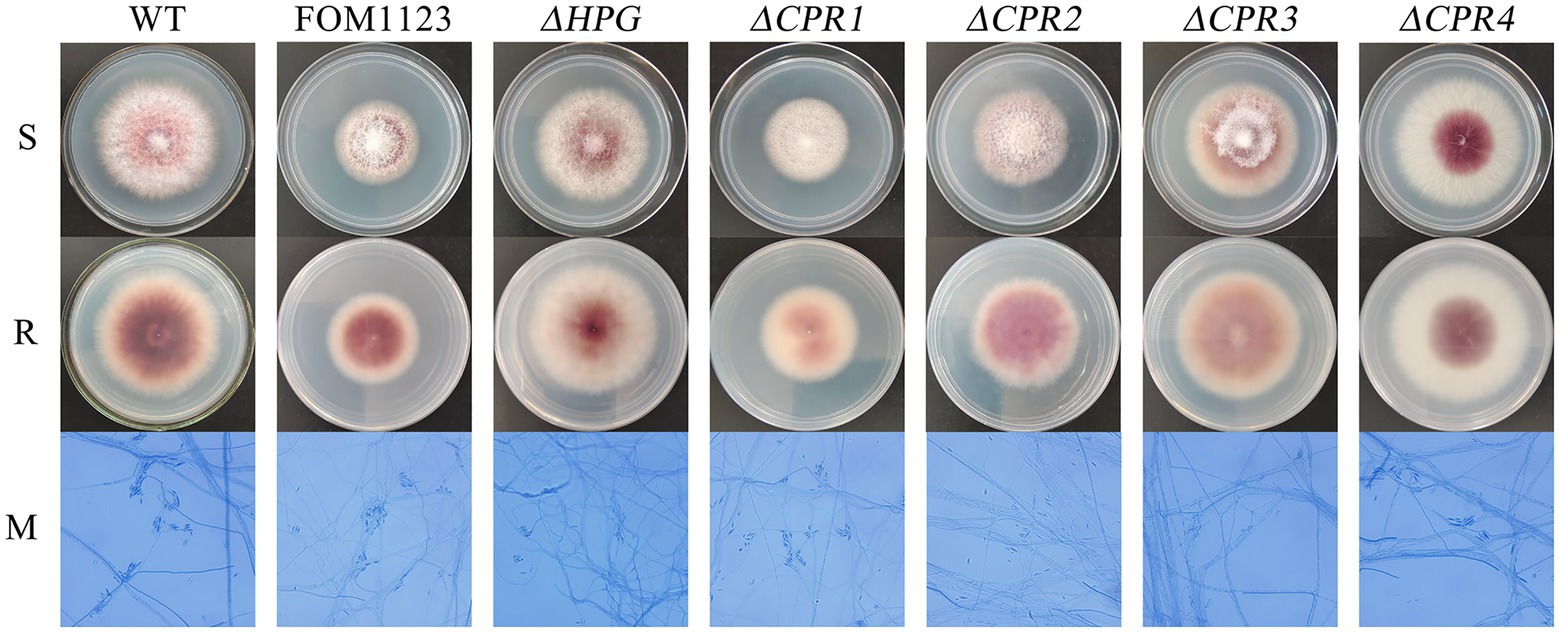
Figure 3. Colony and microscopic morphology of the wild-type F. oxysporum and the mutants. Strains cultured on PDA medium were incubated at 25°C for 5days. Slide cultures were examined after lactophenol cotton blue staining (PDA, 25°C, 3days, 400×magnification). S: surface of colony, R: reverse of colony, and M: micromorphology.
On the basis of the AFST results, ΔCPR1 had the same phenotypes as that of T-DNA mutant FOM1123 displaying low MICs to azoles (except for FLU). In contrast, the other deletion mutants (ΔHPG, ΔCPR2, ΔCPR3, and ΔCPR4) had the same phenotypes as that of the wild-type F. oxysporum, implying the corresponding genes were unrelated to antifungal resistance (Table 1). Accordingly, of the examined genes, only CPR1 appears to be associated with azole resistance.
To clarify the regulatory effects of CPR1 on ergosterol synthesis in cell membranes, we measured the ergosterol content. Without any treatment, the ergosterol content was lower in ΔCPR1 than in the wild-type control. In response to the VRC treatment, the ergosterol contents of the examined strains decreased, and the ergosterol content in ΔCPR1 remained low (Table 4).
To analyze the expression-level changes to the genes involved in the ergosterol biosynthesis pathway, we analyzed the relative expression of Cpr, Cytb5, and Cyp51. Following the VRC treatment, the Cpr1 and Cpr2 expression levels increased by about 7-fold, whereas Cpr3 was almost unexpressed and Cpr4 was unaffected in the wild-type F. oxysporum. In the deletion mutant ΔCPR1, the Cpr2 expression level was about 2-fold higher than the corresponding level in the wild-type control, whereas Cpr3 and Cpr4 were almost unexpressed. When ΔCPR1 was treated with VRC, the expression of Cpr2 increased by about 8.5-fold, whereas Cpr3 and Cpr4 expression levels were unaffected (Figure 4A).
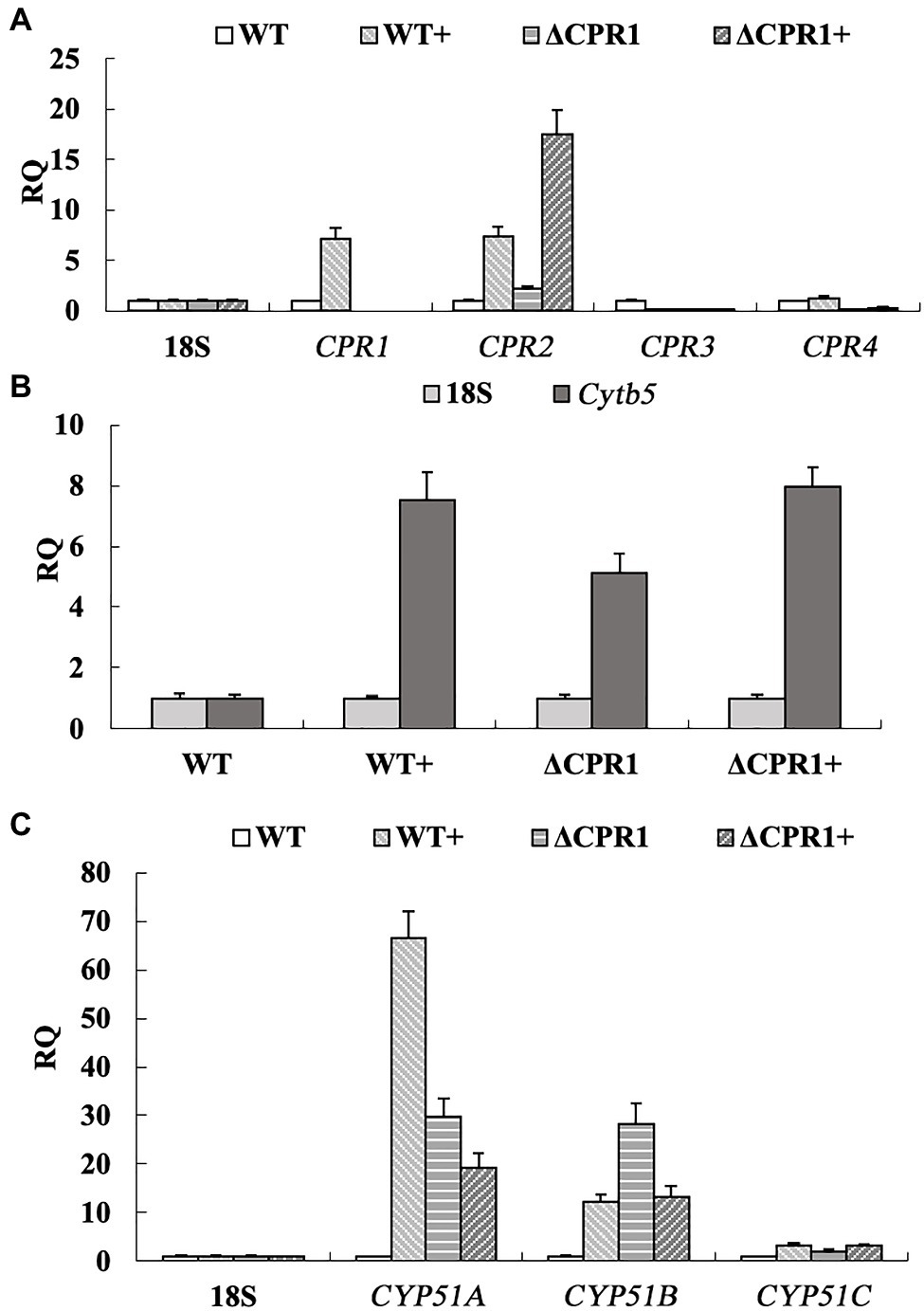
Figure 4. Relative expression levels of genes involved in ergosterol biosynthesis were calculated, using the 18S rRNA gene as the internal reference. (A) Expression of Cpr1 and its homologs. (B) Expression of Cytb5. (C) Expression of Cyp51. “+” represents strains were treated with VRC.
The Cytb5 expression level was about 5-fold higher in ΔCPR1 than in the wild-type F. oxysporum. The VRC treatment upregulated Cytb5 expression by about 7.5-fold and 8-fold in the wild-type control and ΔCPR1, respectively (Figure 4B). Three homologous genes (Cyp51A, Cyp51B, and Cyp51C) encode proteins targeted by azole antifungal agents. These proteins may receive electrons from CPR and Cytb5. The Cyp51A and Cyp51B expression levels were about 30-fold higher in ΔCPR1 than in the wild-type control. The exposure to VRC increased the Cyp51A expression level by about 67-fold in the wild-type F. oxysporum. In contrast, Cyp51A and Cyp51B expression levels were downregulated by about 33 and 57%, respectively, in ΔCPR1. There were no significant changes to Cyp51C expression in any sample (Figure 4C).
Fusarium species are resistant to multiple common antifungal agents (Azor et al., 2007; Al-Hatmi et al., 2016; Tupaki-Sreepurna and Kindo, 2018; Zhao et al., 2021). Analyses of their resistance mechanisms are critical for improving clinical treatments and for preventing or mitigating antifungal resistance. Previous research confirmed F. solani exhibits intrinsic resistance to echinocandin, likely because of a mutation to the gene (Fks1) encoding the catalytic subunit of β-1,3-glucan synthase (Katiyar and Edlind, 2009; Al-Hatmi et al., 2016).
The histidine kinase III gene (Fhk1) in F. oxysporum helps regulate the Hog1-MAPK signaling pathway during stress responses. Deleting this gene leads to increased resistance to phenylpyrrole and dicarboximide fungicides (Rispail and Di Pietro, 2010). Mutations in the genes encoding β1-tubulin and β2-tubulin, which are targeted by benzimidazole fungicides, can lead to increased resistance to carbendazim in various Fusarium species, including Fusarium graminearum, Fusarium fujikuroi, and Fusarium asiaticum (Suga et al., 2011; Chen et al., 2014; Zhou et al., 2016). Hence, the antifungal resistance of Fusarium species is complex and regulated by multiple genes. The precise mechanisms mediating this antifungal resistance remain to be investigated.
In this study, an isolate of F. oxysporum with broadly resistant to commonly antifungal agents and an ATMT-based random insertional mutagenesis method was used to construct T-DNA insertion mutants. And a total of 1,450 T-DNA insertion mutants were obtained. According to the AFST results, compared with the resistant wild-type, one mutant (FOM1123) exhibited significantly increased susceptibility to azoles other than FLU (Table 1). It indicated that the gene interrupted by T-DNA insertion in this mutant might be related to the resistance of wild-type.
Bioinformatics analyses indicated that the T-DNA insertion might have affected two genes (Hpg and Cpr1; Figure 2). To confirm the involvement of these genes to the changes with azole susceptibility, the independent mutants (ΔHPG and ΔCPR1) were generated. And the AFST results revealed that only ΔCPR1 mutant had the same phenotypes with MICs as the T-DNA mutants (Table 1). It suggested that the Cpr1 gene might be related to the resistance of F. oxysporum.
The Cpr1 gene encodes NADPH-cytochrome P450 reductase, which is important for electron transport in various organisms. In fungi, it also participates in ergosterol biosynthesis. In an earlier study by Sutter and Loper (1989), the deletion of the CPR-encoding gene in Saccharomyces cerevisiae resulted in increased susceptibility to KTZ. The F. oxysporum genome includes four Cpr homologs. The independent mutants (ΔCPR2, ΔCPR3, and ΔCPR4) were generated in this study, and the AFST results implied these three genes do not influence antifungal resistance (Table 1). Accordingly, only Cpr1 is associated with azole resistance in F. oxysporum.
Because of its function related to electron transport, CPR1 can affect the function of CYP51, which is targeted by azole antifungal agents, in the ergosterol biosynthesis pathway. Previous studies proved that deleting CYP51A can lead to increased susceptibility to azoles in Magnaporthe oryzae, Aspergillus fumigatus, and F. graminearum (Mellado et al., 2007; Yan et al., 2011). Unlike other fungi, the genomes of Fusarium species contain three Cyp51 genes (Cyp51A, Cyp51B, and Cyp51C). Fan et al. (2013) heterologously expressed three F. graminearum Cyp51 genes in S. cerevisiae. They revealed that Cyp51A is associated with azole susceptibility, whereas Cyp51B and Cyp51C are not. Additionally, Cyp51A expression is reportedly induced by ergosterol depletion. Moreover, it is responsible for the intrinsic variation in azole susceptibility. These findings imply CYP51A might be the main target regulated by CPR1.
Because both CPRs and Cytb5 can deliver electrons to CYP51s, we analyzed the expression of the corresponding genes. In this study, when the wild-type F. oxysporum was treated with VRC, the Cpr1, Cpr2, and Cytb5 expression level increased (Figures 4A,B). Subsequently, the expression of Cyp51A and Cyp51B was upgraduated (Figure 4C). In response to the VRC treatment, Cytb5 expression in ΔCPR1 was not significantly different from that in the wild-type control (Figure 4B), indicating Cyp51 was unaffected by Cytb5. At the same time, though the expression of Cpr2 increased (Figure 4A), the electron supply to CYP51 was insufficient owing to Cpr1 deletion, lead to the expression of Cyp51A and Cyp51B was downgraduated than that in the wild-type (Figure 4C). Consequently, ergosterol biosynthesis was restricted, the ergosterol levels decreased significantly in Cpr1 deletion mutant than the wild-type (Table 4), which contributed to the increase in azole susceptibility.
In conclusion, our results indicate that CPR1 plays an important role in the ergosterol biosynthesis pathway. Furthermore, it is the only NADPH-cytochrome P450 reductase related to azole resistance in F. oxysporum. The increased expression in the CPR1 content may ensure sufficient electrons are supplied to CYP51s for the biosynthesis of ergosterol. This may help to explain why the wild-type fungus was resistant to all tested azoles. Thus, it represents a novel therapeutic target for fungal infections.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
DH and LW: conceptualization and design. ZF, SG, and SH: methodology and experiments. YW: data analysis. DH: original manuscript. LW: review and editing. All authors have read and approved the manuscript for publication.
This study was supported by grants from the National Natural Science Foundation of China (81772162 and U1704283) and the Foundation of Jilin Education Committee (JJKH20211150KJ).
Authors SG and SH were employed by Beijing ZhongKai TianCheng Bio-technology Co. Ltd., (Beijing, China).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all members of our research center for helpful discussions. We also thank Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Abou Ammar, G., Tryono, R., Doll, K., Karlovsky, P., Deising, H. B., and Wirsel, S. G. (2013). Identification of ABC transporter genes of Fusarium graminearum with roles in azole tolerance and/or virulence. PLoS One 8:e79042. doi: 10.1371/journal.pone.0079042
Al-Hatmi, A. M., Meis, J. F., and de Hoog, G. S. (2016). Fusarium: molecular diversity and intrinsic drug resistance. PLoS Pathog. 12:e1005464. doi: 10.1371/journal.ppat.1005464
Azor, M., Gene, J., Cano, J., and Guarro, J. (2007). Universal in vitro antifungal resistance of genetic clades of the Fusarium solani species complex. Antimicrob. Agents Chemother. 51, 1500–1503. doi: 10.1128/AAC.01618-06
Batista, B. G., Chaves, M. A., Reginatto, P., Saraiva, O. J., and Fuentefria, A. M. (2020). Human fusariosis: An emerging infection that is difficult to treat. Rev. Soc. Bras. Med. Trop. 53:e20200013. doi: 10.1590/0037-8682-0013-2020
Chen, Z., Gao, T., Liang, S., Liu, K., Zhou, M., and Chen, C. (2014). Molecular mechanism of resistance of Fusarium fujikuroi to benzimidazole fungicides. FEMS Microbiol. Lett. 357, 77–84. doi: 10.1111/1574-6968.12504
Clinical and Laboratory Standards Institute (2017). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi—Approved Standard CLSI Document M38-Ed3. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
Fan, J., Urban, M., Parker, J. E., Brewer, H. C., Kelly, S. L., Hammond-Kosack, K. E., et al. (2013). Characterization of the sterol 14alpha-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 198, 821–835. doi: 10.1111/nph.12193
Fan, Z., Yu, H., Guo, Q., He, D., Xue, B., Xie, X., et al. (2016). Identification and characterization of an anti-oxidative stress-associated mutant of Aspergillus fumigatus transformed by Agrobacterium tumefaciens. Mol. Med. Rep. 13, 2367–2376. doi: 10.3892/mmr.2016.4839
Gao, S., He, D., Li, G., Zhang, Y., Lv, H., and Wang, L. (2016). A method for amplification of unknown flanking sequences based on touchdown PCR and suppression-PCR. Anal. Biochem. 509, 79–81. doi: 10.1016/j.ab.2016.07.001
Guarro, J. (2013). Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1491–1500. doi: 10.1007/s10096-013-1924-7
Katiyar, S. K., and Edlind, T. D. (2009). Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 53, 1772–1778. doi: 10.1128/AAC.00020-09
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lockhart, S. R., and Guarner, J. (2019). Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 36, 177–181. doi: 10.1053/j.semdp.2019.04.010
Ma, L. J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O'Donnell, K., Trail, F., et al. (2013). Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. doi: 10.1146/annurev-micro-092412-155650
Mellado, E., Garcia-Effron, G., Alcazar-Fuoli, L., Melchers, W. J., Verweij, P. E., Cuenca-Estrella, M., et al. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-06
Miceli, M. H., and Lee, S. A. (2011). Emerging moulds: epidemiological trends and antifungal resistance. Mycoses 54, e666–e678. doi: 10.1111/j.1439-0507.2011.02032.x
Ribas, E. R. A. D., Spolti, P., Del Ponte, E. M., Donato, K. Z., Schrekker, H., and Fuentefria, A. M. (2016). Is the emergence of fungal resistance to medical triazoles related to their use in the agroecosystems? A mini review. Braz. J. Microbiol. 47, 793–799. doi: 10.1016/j.bjm.2016.06.006
Rispail, N., and Di Pietro, A. (2010). The two-component histidine kinase Fhk1 controls stress adaptation and virulence of Fusarium oxysporum. Mol. Plant Pathol. 11, 395–407. doi: 10.1111/j.1364-3703.2010.00612.x
Sharma, C., and Chowdhary, A. (2017). Molecular bases of antifungal resistance in filamentous fungi. Int. J. Antimicrob. Agents 50, 607–616. doi: 10.1016/j.ijantimicag.2017.06.018
Suga, H., Nakajima, T., Kageyama, K., and Hyakumachi, M. (2011). The genetic profile and molecular diagnosis of thiophanate-methyl resistant strains of Fusarium asiaticum in Japan. Fungal Biol. 115, 1244–1250. doi: 10.1016/j.funbio.2011.08.009
Sutter, T. R., and Loper, J. C. (1989). Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole. Biochem. Biophys. Res. Commun. 160, 1257–1266. doi: 10.1016/S0006-291X(89)80139-1
Tortorano, A. M., Prigitano, A., Esposto, M. C., Arsic Arsenijevic, V., Kolarovic, J., Ivanovic, D., et al. (2014). European Confederation of Medical Mycology (ECMM) epidemiological survey on invasive infections due to Fusarium species in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1623–1630. doi: 10.1007/s10096-014-2111-1
Tupaki-Sreepurna, A., and Kindo, A. J. (2018). Fusarium: The versatile pathogen. Indian J. Med. Microbiol. 36, 8–17. doi: 10.4103/ijmm.IJMM_16_24
Yan, X., Ma, W. B., Li, Y., Wang, H., Que, Y. W., Ma, Z. H., et al. (2011). A sterol 14alpha-demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 48, 144–153. doi: 10.1016/j.fgb.2010.09.005
Zhang, Y., Mao, C. X., Zhai, X. Y., Jamieson, P. A., and Zhang, C. Q. (2021). Mutation in cyp51b and overexpression of cyp51a and cyp51b confer multiple resistant to DMIs fungicide prochloraz in Fusarium fujikuroi. Pest Manag. Sci. 77, 824–833. doi: 10.1002/ps.6085
Zhao, B., He, D., and Wang, L. (2021). Advances in Fusarium drug resistance research. J. Glob. Antimicrob. Resist. 24, 215–219. doi: 10.1016/j.jgar.2020.12.016
Keywords: Fusarium oxysporum, NADPH-cytochrome P450 reductase, azole resistance, Agrobacterium tumefaciens-mediated transformation, ergosterol biosynthesis, antifungal susceptibility
Citation: He D, Feng Z, Gao S, Wei Y, Han S and Wang L (2021) Contribution of NADPH-cytochrome P450 Reductase to Azole Resistance in Fusarium oxysporum. Front. Microbiol. 12:709942. doi: 10.3389/fmicb.2021.709942
Received: 14 May 2021; Accepted: 18 August 2021;
Published: 14 September 2021.
Edited by:
Weihua Pan, Shanghai Changzheng Hospital, ChinaCopyright © 2021 He, Feng, Gao, Wei, Han and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Wang, d2xpOTlAamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.