
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 August 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.706934
This article is part of the Research TopicRoles of Regulatory RNAs in Bacterial PathogensView all 13 articles
Virulence gene expression of Yersinia pseudotuberculosis changes during the different stages of infection and this is tightly controlled by environmental cues. In this study, we show that the small protein YmoA, a member of the Hha family, is part of this process. It controls temperature- and nutrient-dependent early and later stage virulence genes in an opposing manner and co-regulates bacterial stress responses and metabolic functions. Our analysis further revealed that YmoA exerts this function by modulating the global post-transcriptional regulatory Csr system. YmoA pre-dominantly enhances the stability of the regulatory RNA CsrC. This involves a stabilizing stem-loop structure within the 5′-region of CsrC. YmoA-mediated CsrC stabilization depends on H-NS, but not on the RNA chaperone Hfq. YmoA-promoted reprogramming of the Csr system has severe consequences for the cell: we found that a mutant deficient of ymoA is strongly reduced in its ability to enter host cells and to disseminate to the Peyer’s patches, mesenteric lymph nodes, liver and spleen in mice. We propose a model in which YmoA controls transition from the initial colonization phase in the intestine toward the host defense phase important for the long-term establishment of the infection in underlying tissues.
Enteropathogenic yersiniae, Yersinia enterocolitica and Yersinia pseudotuberculosis are fecal-oral pathogens that can cause food-borne infections in animals and humans with symptoms ranging from self-limiting enteritis, mesenterial lymphadenitis to autoimmune responses (Bottone, 1997; Koornhof et al., 1999; Galindo et al., 2011). Both Yersinia species initiate infection after oral uptake in the gastrointestinal tract by tight adhesion to the mucosal surface of the intestine, which is followed by rapid internalization and translocation through M-cells of the intestinal epithelium (Pepe and Miller, 1993; Marra and Isberg, 1997; Koornhof et al., 1999). Migration through M-cells leads to the accumulation of the bacteria in underlying lymphatic tissues (Peyer’s patches) and their dissemination to mesenteric lymph nodes, liver and spleen (Barnes et al., 2006; Trülzsch et al., 2007).
Upon ingestion, Yersinia encounters changing growth conditions including elevated temperature as well as variations of nutrients and ions within the intestinal tract. Yersinia senses changes of environmental parameters to determine its localization and adapt expression of pathogenicity factors and survival strategies accordingly (Marceau, 2005; Heroven and Dersch, 2014; Chen et al., 2016). Some of the most important virulence properties are encoded on the virulence plasmid pYV (pIB1). They are predominantly expressed at 37°C upon contact with host immune cells after the bacteria entered the gut-associated lymphatic tissues (Straley and Perry, 1995; Kusmierek et al., 2019; Volk et al., 2019; Schneiders et al., 2021). Among them are the adhesion factor YadA, the structural components of a functional type three secretion system (T3SS) and the antiphagocytic Yersinia outer proteins (Yops) - the effectors which are secreted and translocated by the T3S machinery (Cornelis, 2006; Bliska et al., 2013). Expression of these virulence genes is activated by the AraC-type transcriptional regulator LcrF (VirF) (Lambert de Rouvroit et al., 1992; Hoe and Goguen, 1993; Böhme et al., 2012; Schwiesow et al., 2015) (Figure 1).
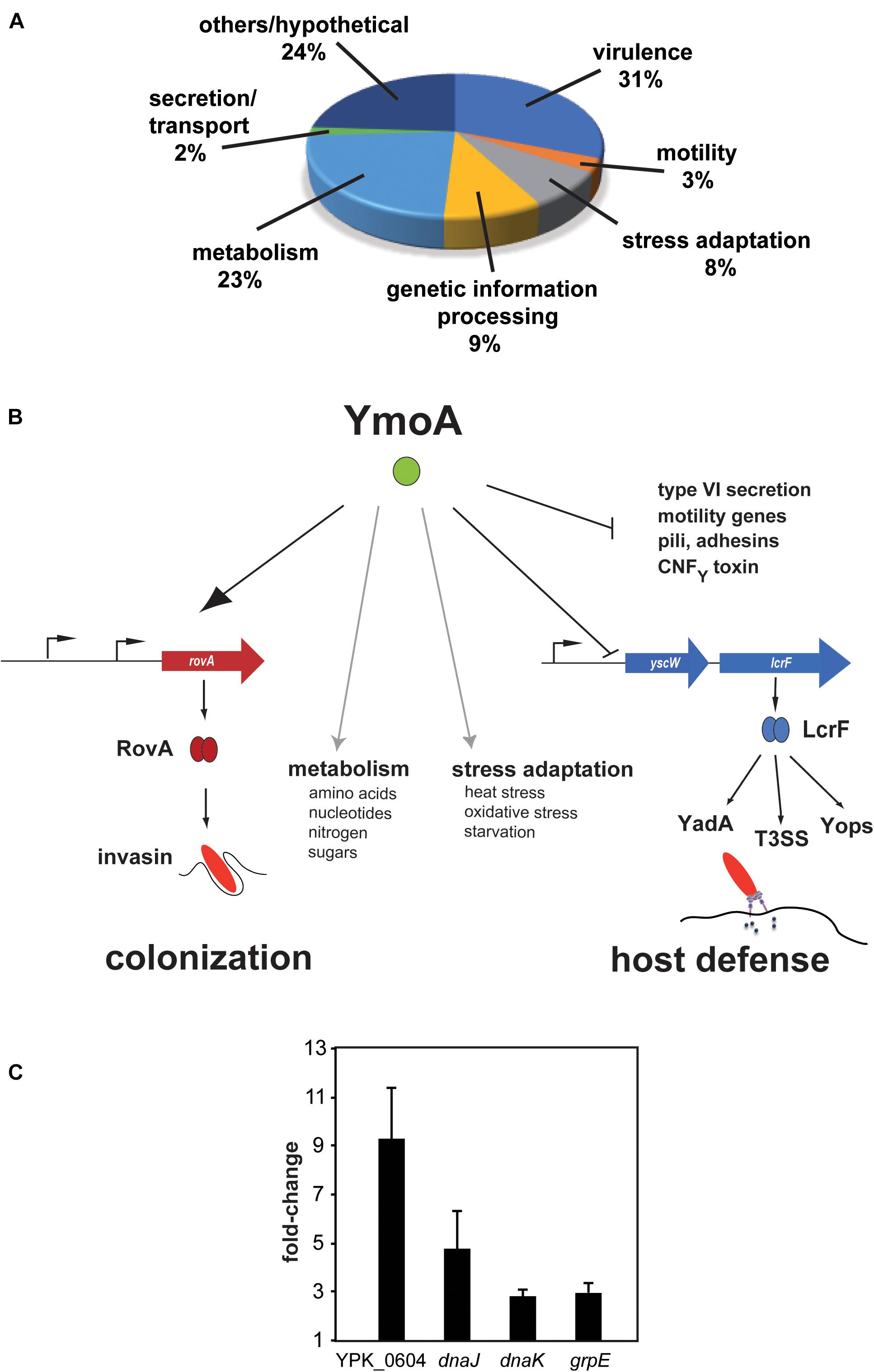
Figure 1. Influence of YmoA on the global gene expression in Y. pseudotuberculosis. (A) Proportion of gene classes up- or downregulated by YmoA. Genes showing overall fold-changes ≥ 2.0 are included in the displayed diagram of gene classes. (B) Schematic scheme of the global influence of YmoA on virulence-relevant genes. YmoA activates the synthesis of the early-phase virulence RovA, leading to expression of colonization factors such as invasin. In contrast, later-stage virulence factors, e.g., YadA, Yops, or T3SS, which are essential to combat triggered immune responses, are silenced by YmoA through repression of the transcriptional activator LcrF. Moreover, YmoA is implicated in many other virulence-relevant metabolic and stress adaptation processes. This control network allows switching of virulence gene expression by variations of YmoA levels. The dark arrow displays activation of the gene expression or protein synthesis; T represents repression or inactivation. Gray arrows indicate control of metabolic and stress adaptation processes by YmoA. (C) RNA of early stationary cultures of strains YPIII and YP50 (ΔymoA) in LB medium at 25°C was prepared and six independent samples were pooled. qRT-PCR was carried out in triplicates. Gene expression levels were normalized to levels of the sopB transcript and are given as relative values ΔymoA/wild-type. Mean ± SD of three independent experiments are displayed.
Other virulence factors, including smooth lipopolysaccharides, flagellar motility, iron uptake and storage, enterotoxin (Yst) and the cell internalization factor invasin are predominantly expressed at moderate temperatures during stationary phase, conditions, which are more typical for the environmental life-style of the bacteria (Straley and Perry, 1995; Marceau, 2005; Heroven and Dersch, 2014; Chen et al., 2016). This class of virulence factors appears to be required for the initial stages of infection and seems to assure rapid and efficient colonization and penetration of the intestinal epithelial layer shortly after infection. The molecular mechanisms, regulating the expression of the virulence genes during the different stages of the infection are still far from being understood. However, over the last years it became evident that a complex regulatory network including the carbon storage regulator (Csr) system plays a crucial role in the control of virulence-associated traits initiating intestinal colonization and dissemination into deeper tissues (Heroven et al., 2008, 2012a; Nuss et al., 2014, 2017a, b; Kusmierek and Dersch, 2017; Kusmierek et al., 2019).
The Csr system is a global regulatory system that consists of (i) the dimeric RNA-binding protein CsrA, that binds to and thus regulates translation and stability of target mRNAs, and (ii) the untranslated regulatory Csr-RNAs CsrB and CsrC that antagonize CsrA function (Heroven et al., 2012a; Kusmierek and Dersch, 2017; Romeo and Babitzke, 2018; Pourciau et al., 2020). Both CsrB and CsrC bind multiple CsrA dimers. This sequestration of CsrA reduces the pool of free CsrA which is able to interact with other target mRNAs without changing the overall level of CsrA. The Csr system was first identified to control the expression of the virulence regulator RovA of Y. pseudotuberculosis (Heroven et al., 2008). RovA, a member of the SlyA/Hor family of DNA-binding proteins, acts as an activator of the primary cell internalization factor invasin, the PsaA adhesin (pH6 antigen) and many other factors contributing to Yersinia virulence in the early stages of the infection (Nagel et al., 2001; Cathelyn et al., 2006, 2007; Yang et al., 2010). CsrA-mediated influence on rovA expression occurs through indirect control of the LysR-type regulator RovM (Heroven and Dersch, 2006; Heroven et al., 2008). Further analysis revealed a complex control system in which the regulatory RNAs CsrC and CsrB are regulated by the global cAMP-binding repressor protein (Crp) and the two-component system BarA/UvrY or PhoQ/PhoP in response to nutrients/metabolites and ions (Heroven et al., 2008, 2012b; Bücker et al., 2014; Nuss et al., 2014).
Virulence-promoting processes are activated by the CsrABC-RovM-RovA cascade in the very early colonization stages of the infection. Once bacteria have reached the lymphatic tissues these virulence traits are no longer needed and hence are downregulated during ongoing infection stages. In this phase, virulence plasmid-encoded LcrF-induced defensive pathogenicity factors are upregulated to protect the pathogen against the attack of the host immune system. How reprogramming of virulence gene expression is controlled in the course of the infection is still unknown, however, it is emerging that the small protein YmoA (Yersinia modulator A; 9 kDa, 67 aa) participates in this process. YmoA shares extensive homology with the Escherichia coli Hha protein, and belongs to a group of low-molecular-weight proteins involved in the regulation of bacterial pathogenicity and virulence-associated physiological traits (Cornelis et al., 1991; de la Cruz et al., 1992; Mikulskis and Cornelis, 1994; Madrid et al., 2002, 2007; Lawal et al., 2013). YmoA was first identified as a thermosensitive repressor of lcrF, encoding the main transcriptional activator of the pYV-encoded later-stage virulence genes, and the Y. enterocolitica enterotoxin gene yst (Cornelis et al., 1991; de la Cruz et al., 1992; Cornelis, 1993; Mikulskis et al., 1994; Jackson et al., 2004; Böhme et al., 2012). Thermal control is mediated by rapid degradation of YmoA by Lon and Clp proteases at a host temperature of 37°C (Jackson et al., 2004; Böhme et al., 2012). Later on, several studies demonstrated that YmoA interacts with a small portion of the nucleoid-structuring and global regulatory protein H-NS, and this generates YmoA/H-NS heteromeric complexes with somewhat different target specificity (Madrid et al., 2002; Nieto et al., 2002; Ellison and Miller, 2006). This suggests that YmoA has a more global role and evolved to fine-tune the activity of H-NS to adapt expression of virulence-associated traits in response to temperature. More recently, is has been reported that YmoA/Hha together with YmoB/TomB(YbaJ) bears the characteristics of a bonafide type II toxin-antitoxin (TA) system, which is involved in biofilm and persister cell formation in E. coli and in persistence and programmed cell death in Salmonella enterica serovar Typhimurium (García-Contreras et al., 2008; Marimon et al., 2016).
In this study, we demonstrate that YmoA plays a key role in the control Yersinia pathogenesis. YmoA is part of a complex regulatory network and controls more than 289 genes, many of them implicated in virulence. In addition to its inhibitory effect on antiphagocytic T3SS-associated virulence factors, YmoA was found to promote the induction of the CsrABC-RovM-RovA regulatory cascade, which is important for the expression of early-stage virulence genes through control of CsrB and CsrC RNA levels. We postulate that YmoA in Y. pseudotuberculosis plays a crucial role switching expression from RovA-activated early-stage virulence genes toward LcrF-induced virulence genes important for host defense.
Overnight cultures of E. coli were routinely grown at 37°C, Yersinia strains were grown at 25°C or 37°C in LB (Luria Bertani) broth if not indicated otherwise. The antibiotics used for bacterial selection were as follows: ampicillin 100 μg ml–1, chloramphenicol 30 μg ml–1, tetracycline 5 μg ml–1, kanamycin 50 μg ml–1, and gentamicin 50 μg ml–1. HEp-2 cells were cultured in RPMI 1640 media (Sigma) supplemented with 5% new-born calf serum (Invitrogen) and 2 mM glutamine at 37°C in the presence of 5% CO2.
All DNA manipulations, restriction digestions, ligations and transformations were performed using standard genetic and molecular techniques (Miller, 1992; Sambrook, 2001). Plasmid DNA was purified using a Qiagen kit. Restriction and DNA-modifying enzymes were obtained from Roche, Promega or New England Biolabs. The oligonucleotides used for amplification by PCR, sequencing and primer extension were purchased from Metabion (Martinsried, Germany). PCR reactions were performed routinely in a 100 μl mix for 25 cycles using Taq polymerase or Phusion High-Fidelity DNA polymerase (New England Biolabs) according to the manufacturer’s instructions. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Germany) before and after digestion of the amplification product. Sequencing reactions were performed by Agowa (Berlin, Germany) or GATC (Konstanz, Germany).
Plasmids used in this study are listed in Supplementary Table 1 and primers for plasmid generation are listed in Supplementary Table 2. Plasmids pAKH106, pAKH107, pKB20, pAKH76 and pAKH104 carry PCR generated fragments harboring csrC promoter regions from nucleotide −355 (primer 3) to nucleotide +39 (primer 5), +61 (primer 6), +71 (primer 7), +81 (primer 8) and +254 (primer 9), respectively. The fragments were digested with EcoRI and SalI and cloned into the corresponding sites of pHT124. The hfq+ plasmids pAKH115 and pAKH119 were constructed by insertion of a PCR fragment amplified with primer 10 and primer 11 into the vector pACYC184 and pHSG575 digested with SalI and BamHI.
To generate pKB4, the ymoA gene was amplified from chromosomal DNA of YPIII using primer 1 and 2 and inserted into the SalI/BamHI sites of pHSG575. To generate pKB17, a csrC fragment harboring an internal deletion from nucleotide +24 to +57 was amplified using primers 3 and 12 and cloned into the EcoRI/SalI sites of pHT124. To express the csrC gene under the control of the tet promoter, csrC was amplified with primers 13 (including the tet promoter sequence) and 14. The resulting fragment was cloned into the SalI/BamHI sites of vector pHSG575, generating pKB47. The csrC gene, including the 5′-regulatory region, was amplified with primers 15 and 16 and inserted into the BamHI/SalI site of pHSG576, resulting in pKB59. Subsequently, the region from position +24 to +57 within the csrC gene was deleted by three-step-PCR. First, the upstream region was amplified by primers 15 and 17 (encoding the deleted region), and the downstream region was amplified with primer 18 (encoding the deleted region) and primer 16. Next, up- and downstream fragments were mixed with primers 15 and 16 to generate the csrC deletion fragment, which was inserted into the SalI/BamHI sites resulting in pKB49. The Ptet:lacZ containing vector pTT1 was constructed by the insertion of a DNA fragment amplified with primers 23 and 24 into the PstI/EcoRV sites of pTS03. The sequence and the correct orientation of all cloned fragments were proven by DNA sequencing.
The Y. pseudotuberculosis mutant strains were constructed with the RED recombinase system as described previously (Datsenko and Wanner, 2000; Derbise et al., 2003) and are all derived from wild-type strain YPIII. First, a kanamycin cassette was amplified by PCR with primers homologous to the resistance gene encoded on pKD4 followed by homologous sequences of adjacent regions of the target gene (for primer see Supplementary Table 3). The PCR fragment was transformed into Y. pseudotuberculosis YPIII pKD46. Chromosomal integration of the fragment was selected by plating on LB supplemented with kanamycin. Subsequently, mutant derivatives were cured of the temperature-sensitive plasmid pKD46 by cultivation at 37°C. To remove the resistance gene at its FLP recognition sites the mutants were transformed with the helper plasmid pCP20 encoding the FLP recombinase. For thermal induction of FLP synthesis and subsequent removal of the temperature-sensitive plasmid pCP20, mutants were incubated at 37°C.
To generate YP73 and YP75, a ΔymoA fragment was inserted into YP69 (YPIII ΔcsrB) and YP72 (YPIII ΔrovM), respectively. First, the kanamycin resistance gene was amplified using primer pairs 25/26 for YP73 and YP75 (see Supplementary Table 3). Next, the Yersinia genomic DNA was used as a template to amplify 500-bp regions flanking the target genes ymoA (see Supplementary Table 3). The upstream fragment was amplified with a primer pair of which the reverse primer contained additional 20 nt at the 5′-end which were homologous to the start of the kanamycin resistance gene, the downstream fragment was amplified with a primer pair of which the forward primer contained additional 20 nt at the 5′-end which were homologous to the end of the kanamycin resistance gene (for primer see Supplementary Table 4). In the next step, a PCR reaction was performed with the forward primer and the reverse primer using the upstream and downstream PCR products of the target gene and the kan gene fragment as templates. The PCR fragment was transformed into Y. pseudotuberculosis YPIII pKD46 and chromosomal integration of the fragments was selected by plating on LB supplemented with kanamycin. The selection of the mutants and removal of the kanamycin resistance gene was performed as described (Datsenko and Wanner, 2000).
Overnight cultures were grown to stationary phase (OD600 of 3). 2.5 ml culture were withdrawn, mixed with 0.2 volume of stop solution (5% water-saturated phenol, 95% ethanol) and snap-frozen in liquid nitrogen. After thawing on ice, bacteria were pelleted by centrifugation (2 min, 14.000 rpm, 4°C), and RNA was isolated using the SV total RNA purification kit (Promega) as described by the manufacturer. RNA concentration and quality were determined by measurement of A260 and A280. Total cellular RNA (10 μg) was mixed with loading buffer (0.03% bromophenol blue, 4 mM EDTA, 0.1 mg/ml EtBr, 2.7% formaldehyde, 31% formamide, 20% glycerol in 4 × MOPS buffer) and was separated on agarose gels (1.2%), transferred overnight onto positively charged membranes (Roche) in 20 × SSC and UV cross-linked. Prehybridization, hybridization to DIG-labeled DNA probes and membrane washing were conducted using the DIG luminescent Detection kit (Roche) according to the manufacturer’s instructions. The csrC and csrB transcripts were detected with a DIG-labeled PCR fragment (DIG-PCR nucleotide mix, Roche) with primer pair 23/24 and 25/26 (Supplementary Table 2), respectively.
RNA stability assay was used to compare degradation of the CsrC RNA between wild-type and different mutant strains. A YPIII overnight culture was mixed with rifampicin at a final concentration of 500 μg/ml to inhibit transcription. At certain time points after blockage of transcription, samples were withdrawn and total RNA was prepared for Northern blot analysis as described above. The half-life of CsrC was calculated by least squares analysis of semi-logarithmic plots of RNA concentration versus time.
Primer extension analysis was performed to determine the steady-state level and the stability of the CsrB and CsrC RNA from strains YPIII and YP80 (YPIII Δhfq). At an OD600 of 2.0 (early stationary phase), rifampicin was added to a final concentration of 500 μg ml–, after 0, 10, 20, 30, and 60 min, 2 ml aliquots were withdrawn and total RNA was extracted of the samples using the SV total RNA purification kit (Promega) as described by the manufacturer. Annealing was performed with 5 μg extracted RNA and the 5′-Dig-labeled oligonucleotides (primer 5′-CTGAAGACACATCTTCC-3′ for CsrB, primer 5′- CCTGAGTAACTGTGCTCC-3 for CsrC, and primer 5′-CCCACACTACCATCGGCGC-3′ for 5S RNA) in 20 μl of 1x First Strand Buffer (Invitrogen) by slow cooling of the sample (0.01°C/sec) including 8 mM dNTPs with 200 U Superscript II reverse transcriptase (Invitrogen) was added and incubated for 1 h at 42°C. The size of the Dig-labeled reaction products was determined on a denaturing 4% DNA sequencing gel by a detection procedure as described (Heroven et al., 2008).
KB4 (Δhns, ΔstpA, and Δhha) transformed with pAKH77 or pAKH11 was grown at 37°C in LB broth to an A600 of 0.6. Anhydrotetracycline was added (0.2 μg/ml) to induce the expression of YmoA-Strep-Tag and/or 2 mM IPTG was used to induce H-NS-His6 expression. For purification of the YmoA-H-NS heterodimer KB4 transformed with pAKH77 and pAKH11 was used for overexpression of the YmoA and H-NS protein. The cells were grown for an additional 3 h before being harvested. The purification procedure for the Strep-tagged YmoA protein was performed according to the manufacturer’s instructions (IBA GmbH, Germany). H-NS purification was performed as described (Heroven et al., 2004). The purity of the YmoA and the H-NS protein was estimated to be >95%.
For RNA-binding studies the purified YmoA and H-NS proteins were dialyzed against the RNA-binding buffer (10 mM Tris-HCl pH 7.5; 3 mM M DTT; 7,5% glycerol; 100 mM KCl; 100 mM MgCl2). The CsrC RNA for RNA band shift analysis was obtained by in vitro transcription from a PCR fragment as described (Heroven et al., 2008; Böhme et al., 2012). The csrC transcript (extending 150 nt downstream of the transcriptional start site of csrC) and the control transcript (extending 117 nt downstream of the transcriptional start site of the 5S rRNA gene) were synthesized in vitro using the Fermentas TranscriptAid T7 High Yield Transcription Kit from PCR products of the target regions. Primers 29/30 and 31/32 were used for amplification of the csrC and 5S RNA control fragment from chromosomal DNA of YPIII. The RNA transcripts were extracted with phenol:chloroform, precipitated with ethanol and stored in DEPC-treated water. The RNA-binding reactions included the CsrC (1.1 μM) and 5S rRNA transcripts (1 μM), 1× RNA-binding buffer and increasing concentrations of the H-NS and YmoA proteins. In the following, they were incubated for 30 min at room temperature and immediately loaded on 8% polyacrylamide gels.
For DNA-binding studies, the purified YmoA and H-NS proteins were dialyzed against the DNA-binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5 mM dithiothreitol, 5% glycerol, 10 mM NaCl, 1 mM MgCl2, 100 μg/ml BSA). Defined PCR fragments, carrying portions of the csrC regulatory region and the csiD region of E. coli K-12 CC118λpir (negative control), were mixed in an equimolar ratio and incubated with increasing amounts of purified YmoA and/or H-NS for 20 min at room temperature and used for DNA band shift assays as described by (Böhme et al., 2012).
Sequences used for the design of the microarrays (Agilent, 8 × 15K format), containing three different 60 nt oligonucleotides for all 4,172 chromosomal genes (ORFs > 30 codons) of the Y. pseudotuberculosis YPIII genome and six probes for the 92 genes of the virulence plasmid pYV of Y. pseudotuberculosis strain IP32953, were obtained from the NCBI Genome Genbank (NC_010465 and NC_006153). The ORF-specific oligonucleotides were designed using the web design application eArray from Agilent1. 16 independent cultures of Y. pseudotuberculosis YPIII and the ymoA mutant strain were grown in LB at 25°C to OD600 0.8, total RNA was isolated from samples using the SV total RNA purification kit (Promega), and RNA concentration and quality was determined with an Agilent 2100 Bioanalyzer using the RNA Nano6000 kit as described by the manufacturers. Total RNA of four independent samples was pooled. 1 μg of the pooled samples was used for RNA-labeling with Cy5 (for wild-type RNA) and Cy3 (for mutant RNA) using the ULSTM Fluorescent Labeling Kit for Agilent Arrays (Kreatech). Non-incorporated Cy5/Cy3 was removed by KREApure purification columns as suggested by the manufacturers. The degree of labeling was determined by a Nanodrop (Peqlab). Subsequently, 300 ng Cy5-labeled RNA and 300 ng Cy3-labeled RNA were mixed, fragmented and hybridized to custom-made Agilent microarray slides (8 × 15K) using the Agilent gene expression hybridization kit as described by the manufacturer. Direct use of labeled RNA for array hybridization was chosen to avoid bias obtained by cDNA library formation. In general, four biological replicates were used for each experiment. After washing and drying of the microarray slide, data were scanned using Axon GenePix Personal 4100A scanner and array images were captured using the software package GenePix Pro 6.015.
The processing of the resulting microarray data was performed using the software package R2 in combination with the ‘‘Bioconductor’’ software framework3 (Gentleman et al., 2004). Preprocessing based on the marray package employing the read.GenePix function. Control code for probe selection was adapted to the custom-made Agilent microarray system and a quality control was performed to check for hybridization artifacts and large-scale differences between the microarrays of one experiment. A two-color intensity-dependent normalization (“Lowess” normalization) was applied and if necessary supplemented by scale normalization between different microarrays as described (Yang et al., 2002). Differentially expressed genes were obtained using the limma package and the lmFit function for linear modeling. eBayes was used for significance calculations (Smyth, 2004). The overall fold-changes of a gene represented by at least three probes are given as median values for all probes. A cut-off of fold-change ≥ 2 (P-value of >0.001) was chosen to determine YmoA-dependent genes/operons and the set of resulting differential expressed genes was analyzed employing the topGO package for Gene Ontology (GO) term enrichment (Alexa et al., 2006). MIAME compliant array data were deposited in Gene Expression Omnibus (GEO) database and are available via the following accession number: GSE35043.
One step real-time RT-PCR was performed in triplicate with RNA preparations of six independent cultures using a Rotor-Gene Q thermo cycler (Qiagen). For qRT-PCR analysis RNA was prepared from bacterial cells grown to early stationary phase at 25°C using Qiazol and the chloroform/phenol extraction method. Quantitative RT-PCR was carried out with the SensiFast SYBR No-ROX One-Step Kit (Bioline, Germany) applying the 3-step cycling protocol according to the manufacturer. Gene specific-primers used for qRT-PCR amplification are listed in Supplementary Table 3 and were designed to produce a 200–250 bp amplicon with Y. pseudotuberculosis YPIII cDNA as template. The amount of PCR product was quantified by measuring fluorescence of SYBR Green dye. Reported gene expression levels were normalized to levels of the sopB transcript. This gene was used as it exhibited identical expression levels in the wild-type and the ymoA mutant under used experimental conditions. Standard curves were detected during every run for each gene tested and established by comparing transcript levels in serial dilutions of total RNA from a control sample. The relative expression of each gene was calculated as described (Pfaffl, 2001).
The activity of the inv-phoA fusion encoded on plasmid pPD297 and the β-galactosidase activity of the lacZ fusion constructs were measured in permeabilized cells as described previously (Manoil, 1990; Miller, 1992). The activities were calculated as follows: β-galactosidase activity OD420 ⋅ 6.75 ⋅ OD600–1 ⋅ Δt (min)–1 ⋅ Vol (ml)–1; alkaline phosphatase activity OD420 ⋅ 6.46 ⋅ OD578–1 ⋅ Δt (min)–1 ⋅ Vol (ml)–1.
For immunological detection of the YmoA, RovA, RovM, and invasin proteins, Y. pseudotuberculosis cultures were grown under specific environmental conditions as described. Cell extracts of equal amounts of the bacteria were prepared and separated on a 15% (RovA, Hfq), 12% (RovM) or 10% (invasin) SDS-PAGE (Sambrook, 2001). The low molecular weight proteins (YmoA and CsrA) were separated by 20% TRICINE-PAGE as described recently (Schagger, 2006).
Subsequently, the samples were transferred onto an Immobilon-P membrane (Millipore) and probed with a polyclonal antibodies directed against RovA, RovM, Hfq, CsrA, and YmoA, or a monoclonal antibody 3A2 directed against invasin as described recently (Heroven and Dersch, 2006). The cell extracts used for Western blotting were also separated by SDS-PAGE and stained with Coomassie blue to ensure that the protein concentrations in the different cell extracts are comparable; about 10 μg protein was applied of each sample.
In preparation of the cell adhesion and uptake assay, 5 x 104 HEp-2 cells were seeded and grown overnight in individual wells of 24-well cell culture plates (Nunc). Cell monolayers were washed three times with phosphate-buffered saline (PBS) and incubated in binding buffer (RPMI 1640 medium supplemented with 20 mM HEPES pH7.0 and 0.4% bovine serum albumin) before infection with bacteria. Approximately 106 bacteria were added to the monolayer and incubated at 37°C. Bacterial uptake was assessed 30 min after infection as the percentage of bacteria, which survived a gentamicin treatment versus the input cell number as described previously (Dersch and Isberg, 1999). The experiments were routinely performed in triplicate.
Yersinia pseudotuberculosis YPIII (wild-type) and YP50 (ymoA mutant) were grown overnight at 25°C, washed in sterile PBS and used for intragastrical inoculation of 6–8 weeks old female BALB/c mice (Janvier, France) using a ball-tipped feeding needle. To assess the impact of an ymoA deletion in Y. pseudotuberculosis on tissue colonization, different groups of BALB/c mice were infected with 5 × 108 bacteria of each strain. In co-infection experiments mice were orally inoculated with an equal mixture of strains YPIII and YP50, each at a dosage of 5 × 108 CFU. Three days post infection, mice were sacrificed using CO2. Mesenterial lymph nodes, liver and spleen were recovered. Isolated Peyer’s patches were washed with sterile PBS, incubated in 100 μg/ml gentamycin for 30 min and washed intensively with sterile PBS three-times. Organs were weighed and homogenized for 30 s in sterile PBS using a Polytron PT 2100 homogenizer (Kinematica, Switzerland). Subsequently, they were plated on Yersinia selective agar (Oxoid, Germany) in three serial dilutions with or without kanamycin. Colony forming units were determined and are given in cfu per gram organ/tissue. The competitive index in comparison to the wild-type strain YPIII was calculated as described (Monk et al., 2008).
Animal work was performed in strict accordance with the German regulations of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). The animal study was reviewed and approved by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit: animal licensing committee permission no. 33.9.42502-04-055/09.
In a previous study we showed that YmoA directly represses transcription of the regulator of lcrF, which is encoded on the Yersinia virulence plasmid (pYV) and controls expression of the antiphagocytic T3SS machinery and effectors (Böhme et al., 2012). However, despite its influence on lcrF expression, the overall influence of YmoA on Y. pseudotuberculosis virulence was still unknown. In the context of a previous study addressing the influence of different early-stage virulence regulators (CsrA, Crp, and RovA) on gene expression of Y. pseudotuberculosis YPIII (Bücker et al., 2014), we also investigated the effect of YmoA on the Yersinia transcriptome. The analysis was performed with Y. pseudotuberculosis wildtype strain YPIII and the isogenic ymoA mutant (YP50) grown at 25°C to late exponential phase, conditions under which YmoA production was found to be high (Supplementary Figure 1). In total 289 genes showed twofold or greater difference in transcript abundance between the wild-type and the ΔymoA strain. Among them, 227 genes (79%) were up-regulated and 62 (21%) were down-regulated in the ymoA mutant (Supplementary Table 4), indicating that YmoA is an important global regulator implicated in the control of fitness- and virulence-relevant processes. Classification according to genome annotation of Y. pseudotuberculosis YPIII showed that altered genes belong to several functional categories (Figure 1A and Supplementary Table 4). About 31% of all YmoA-dependent transcripts (90 genes) are related to virulence. As expected from previous studies (de la Cruz et al., 1992; Cornelis, 1993), 42 genes of the virulence plasmid encoding the structural components of the type III secretion system (yscA-L, yscO-S, and yscU), the intracellularly delivered Yop effector proteins and their chaperones (yopE, sycE, yopH, yopJ, yopO, sycO, yopK, and yopM), regulators of the secretion system (yscW-lcrF, lcrQ, yopN-sycN-tyeA-yscXY, and lcrDRGVH-yopBD), and the adhesin YadA were upregulated in the ymoA mutant. Notably, multiple virulence plasmid-encoded putative resolvases and transposase genes were also upregulated in the ymoA mutant (Supplementary Table 4), suggesting that YmoA did not only control expression of the T3SS-Yop injectisome via its influence on LcrF (Böhme et al., 2012), but seems to have a more global effect.
In addition, many chromosomally encoded virulence genes were differentially regulated in the absence of YmoA. Among them were genes for potential adhesins (ail, ailB, psaA, yadC, and yadF) and fimbriae/pili (ybgP, fimA1-3, fimC, fimD, smfA1-2, and csgG), the cnfY gene encoding a cytotoxic necrotizing factor and the insecticidal toxin genes tcaAB, as well as genes for iron/heme acquisition (hasA and bfr), LPS/O-antigen synthesis (wzz, manB, gne, wbyL, manC, fcl, gmd, and wbyK) and autoinducer synthesis (lsrRBFG, ypsI/ytbI). In addition, genes encoding Hcp1 family effectors and different operons for type VI secretion systems (hcp1 and imp genes), flagella assembly and chemotaxis genes (flhDC, fliETFZ, and flgDCB) were upregulated in the ymoA deficient strain (Figures 1A,B and Supplementary Table 4). These data were validated with a subset of identified YmoA-dependent genes using qRT-PCR with sopB as control as sopB was not affected by YmoA according to the microarray analysis [Figure 1C, YPK_0604 (l-psp; 9.3-fold), grpE (3-fold), dnaJ (4.8-fold), and dnaK (2.8-fold)].
Among the identified YmoA-dependent virulence genes were early-stage pathogenicity factors of Yersinia, which are predominantly expressed at lower temperature in vitro. Several of them, including the virulence regulator gene rovA (Supplementary Table 4), have been shown to be controlled by the Csr system (Heroven et al., 2008, 2012a; Bücker et al., 2014). This suggested that YmoA might be implicated in the control of the CsrABC-RovM-RovA regulatory cascade (Figure 2A). To prove this assumption, we first compared intracellular YmoA, RovM and RovA levels between wild-type (YPIII), the ymoA mutant (YP50) and the complemented strain YP50 (ΔymoA) pAKH71 (ymoA+). As shown in Figures 2B–D, expression of the negative regulator RovM is significantly increased in the absence of YmoA, which in turn leads to a strong reduction of the amount of RovA. Influence on RovM and RovA levels could be fully complemented by the ymoA+ plasmid. Interestingly, the intracellular amount of YmoA is very low in Y. pseudotuberculosis YPIII under growth conditions simulating the early-stage of infection (Figure 2B and Supplementary Figure 1), but this concentration seems sufficient to promote RovA synthesis to levels similar to pymoA+ containing strains (Figure 2D), with significantly higher amounts of YmoA (Figure 2B).
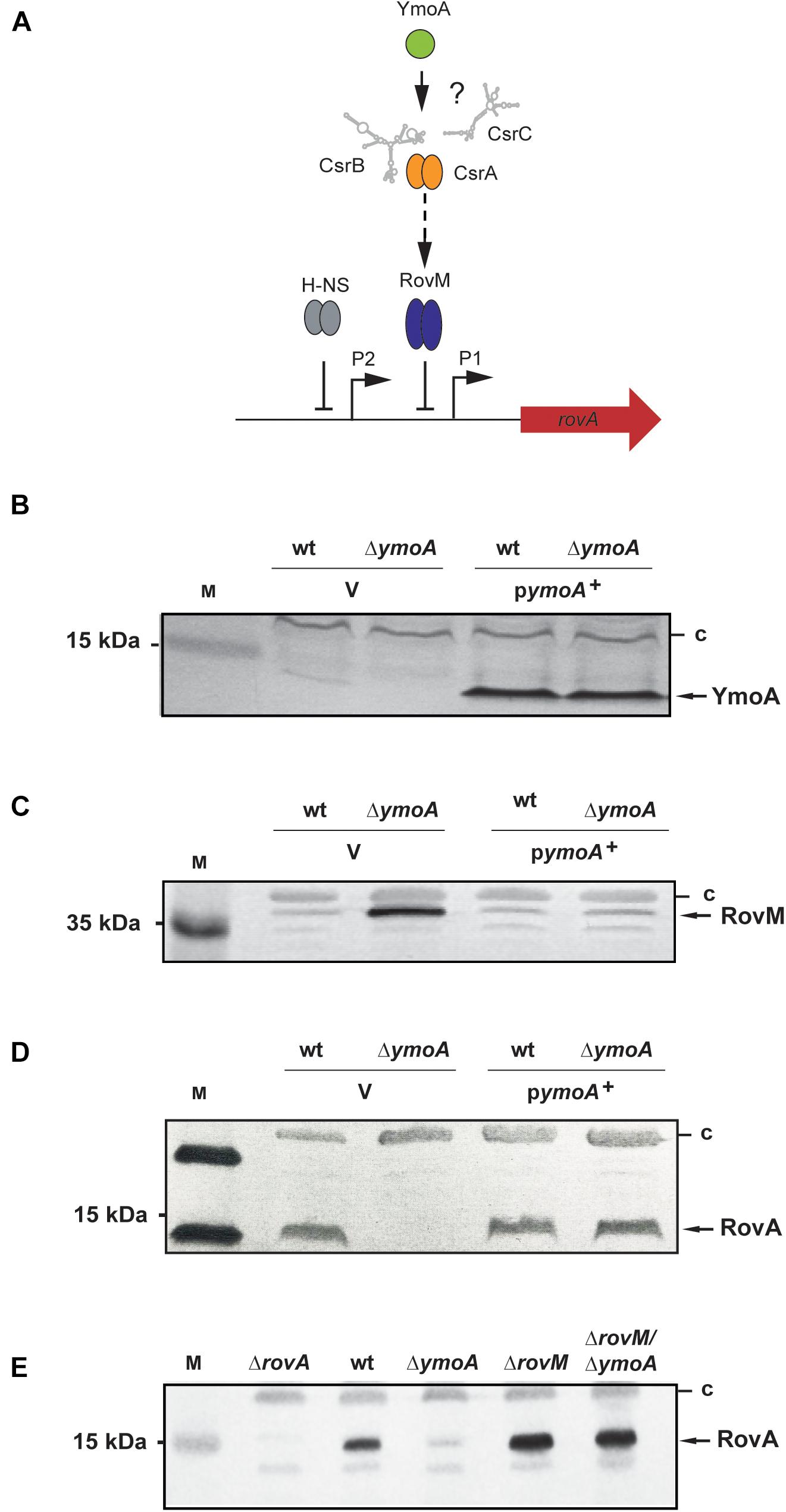
Figure 2. Analysis of RovA and RovM levels in Y. pseudotuberculosis in the presence or absence of YmoA. (A) Regulatory components controlling rovA expression in Y. pseudotuberculosis are illustrated and assumed influence of YmoA on the regulatory cascade is indicated by ‘?’. (B–D) Whole-cell extracts from overnight cultures of Y. pseudotuberculosis wild-type and mutant strain YP50 (ΔymoA) transformed with pACYC184 (V) or pAKH71 (ymoA+) grown at 25°C were prepared and analyzed by Western blotting with a polyclonal antibody directed against YmoA (B), RovM (C), and RovA (D). A molecular weight marker is loaded on the left. (E) Whole-cell extracts from overnight cultures of the Y. pseudotuberculosis wild-type (YPIII), and the rovA (YP3), ymoA (YP50), rovM (YP41), and rovM/ymoA (YP73) mutant strains were prepared. RovA in the extracts were detected by Western blotting using polyclonal antibodies directed against these proteins. A prestained molecular weight marker is loaded on the left. Unspecifically detected proteins (c) were used as loading control.
While a deletion of ymoA alone abrogated RovA synthesis, it had no influence on rovA expression in the rovM/ymoA double mutant; in the contrary higher RovA level were detected similar to the rovM mutant (Figure 2E). This demonstrated that YmoA exerts is effect mainly via RovM. This was somewhat surprising as YmoA was found to interact with the nucleoid-structuring protein H-NS (Nieto et al., 2002) which is known to control rovA expression directly (Heroven et al., 2004) (Figure 2A). In fact, overexpression of a dominant-negative N-terminal H-NS fragment (H-NS∗) – a knock-out is deleterious to Y. pseudotuberculosis – reduced H-NS-mediated rovA silencing as shown previously (Heroven et al., 2004). This led to an increase of RovA levels in the Y. pseudotuberculosis wild-type, but not in the ymoA mutant (Supplementary Figure 2).
Expression of the rovM gene was shown to be under control of the Yersinia Csr system, comprising the two regulatory ncRNAs, CsrB, and CsrC, and the RNA-binding protein CsrA (Heroven et al., 2008) (Figure 3A). The CsrB and CsrC RNAs both sequester the CsrA RNA-binding protein resulting in reduced RovM synthesis. To determine whether YmoA acts on rovM via the Yersinia Csr system, we compared the amount of CsrA and expression of the csr RNA genes between wild-type and the ymoA mutant. While no changes of CsrA levels were detectable (Figure 3B), a mild increase in CsrB and a strong reduction of CsrC could be observed in a ΔymoA strain compared to the wild-type (Figures 3C,D).
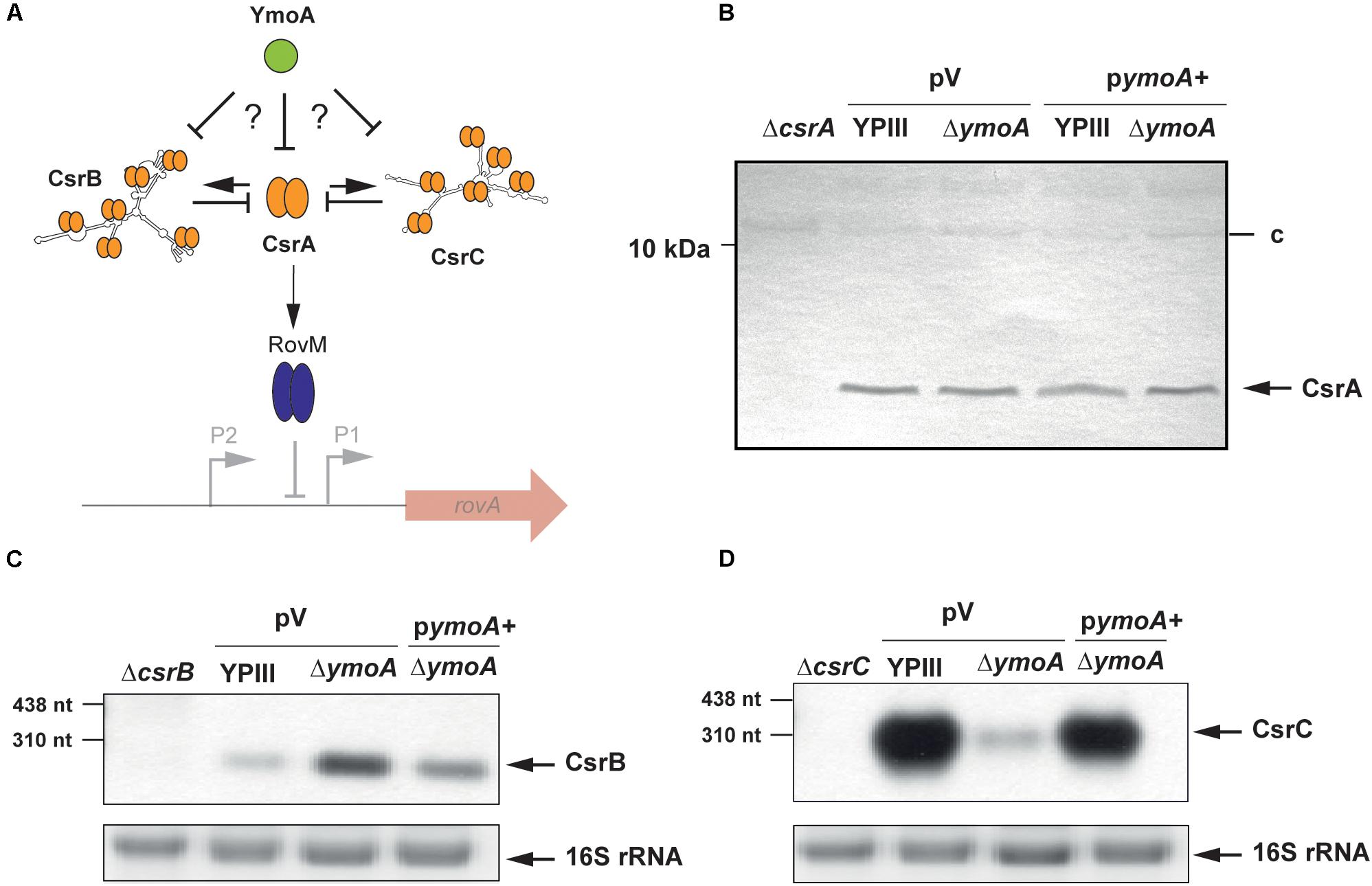
Figure 3. Influence of YmoA on the synthesis of CsrA and the Csr-type RNAs of Y. pseudotuberculosis. (A) Illustration of potential YmoA influence on RovM synthesis via the CsrABC system. (B) Whole-cell extracts from overnight cultures of Y. pseudotuberculosis wild-type and mutant strain YP50 (ΔymoA) transformed with pACYC184 (V) or pAKH71 (ymoA+) grown at 25°C were prepared and analyzed by Western blotting with as a polyclonal antibody directed against CsrA YP50 (ΔymoA). YP53 (ΔcsrA) was used a negative control, and an unspecifically detected protein (c) served as loading control. (C,D) Total RNA of Y. pseudotuberculosis YPIII and YP50 (ΔymoA) carrying the empty vector pAKH85 (V) and YP50 pAKH71 (pymoA+), YP51 (ΔcsrB) and YP48 (ΔcsrC) was prepared, separated on agarose gels and a CsrB- or CsrC-specific probe was used to detect CsrB (C) or CsrC (D) by Northern blotting. The 16S rRNAs are shown as RNA loading control. Used RNA marker is indicated on the left.
As both Csr-RNAs have a negative influence on each other (Heroven et al., 2008), we first addressed whether YmoA affects only the expression of CsrC RNA, which in turn inhibits the synthesis of CsrB. In fact, identical changes were observed on CsrC and RovM levels in a ymoA and a csrB/ymoA mutant strain, implying that YmoA exerts its major effect on the Csr regulon through regulation of CsrC (Supplementary Figure 3).
To determine whether this positive effect of YmoA on CsrC occurs on the transcriptional or post-transcriptional level (Figure 4A), we analyzed expression of csrC-lacZ transcriptional fusions harboring csrC fragments with varying 3′ endpoints (+4, +39, +61, +71, +81, or +254). In general, elongation of the csrC fragment led to a continuous decrease of csrC-lacZ expression (Figure 4B), suggesting that csrC sequences destabilize or reduce translation of the lacZ RNA and/or lead to a premature termination of transcription. Furthermore, we found that absence of YmoA had no significantly different effect on csrC-lacZ reporter fusions which included the first ≤61 nucleotides of csrC, whereas expression of fusions, containing the first 71 nucleotides or more, was more than twofold reduced. Computational secondary structure analyses of the csrC transcript predict that nucleotides from position +24 to +57 form a hairpin structure (Figure 4C). Expression of mutant variants of csrC(+81)-lacZ fusion and csrC in which nucleotides +24 to +57 were deleted became independent of YmoA (Figures 4B,D). This suggests the presence of (i) a stabilizing element or (ii) a transcriptional terminator within the 5′-sequences of the CsrC RNA which is affected by YmoA. The fact that (i) the 5′-hairpin region has a positive influence on the overall expression of csrC and (ii) truncated CsrC species could not be detected in the ymoA mutant suggested that YmoA enhances CsrC RNA stability through the stem loop segment. To confirm YmoA-mediated post-transcriptional control of CsrC, csrC was expressed under the control of a plasmid-encoded tetracycline promoter (Ptet). To ensure expression was only derived from the plasmid, culturing conditions were chosen under which the chromosomal csrC gene is fully repressed (Heroven et al., 2008). While expression of the control gene lacZ under Ptet control was identical in the wildtype and the ymoA mutant (Supplementary Figure 4A), considerably higher levels of CsrC were detectable in the wild-type when expressed from the identical Ptet expression plasmid (Supplementary Figure 4B). This also suggested that YmoA acts on CsrC on the post-transcriptional level.
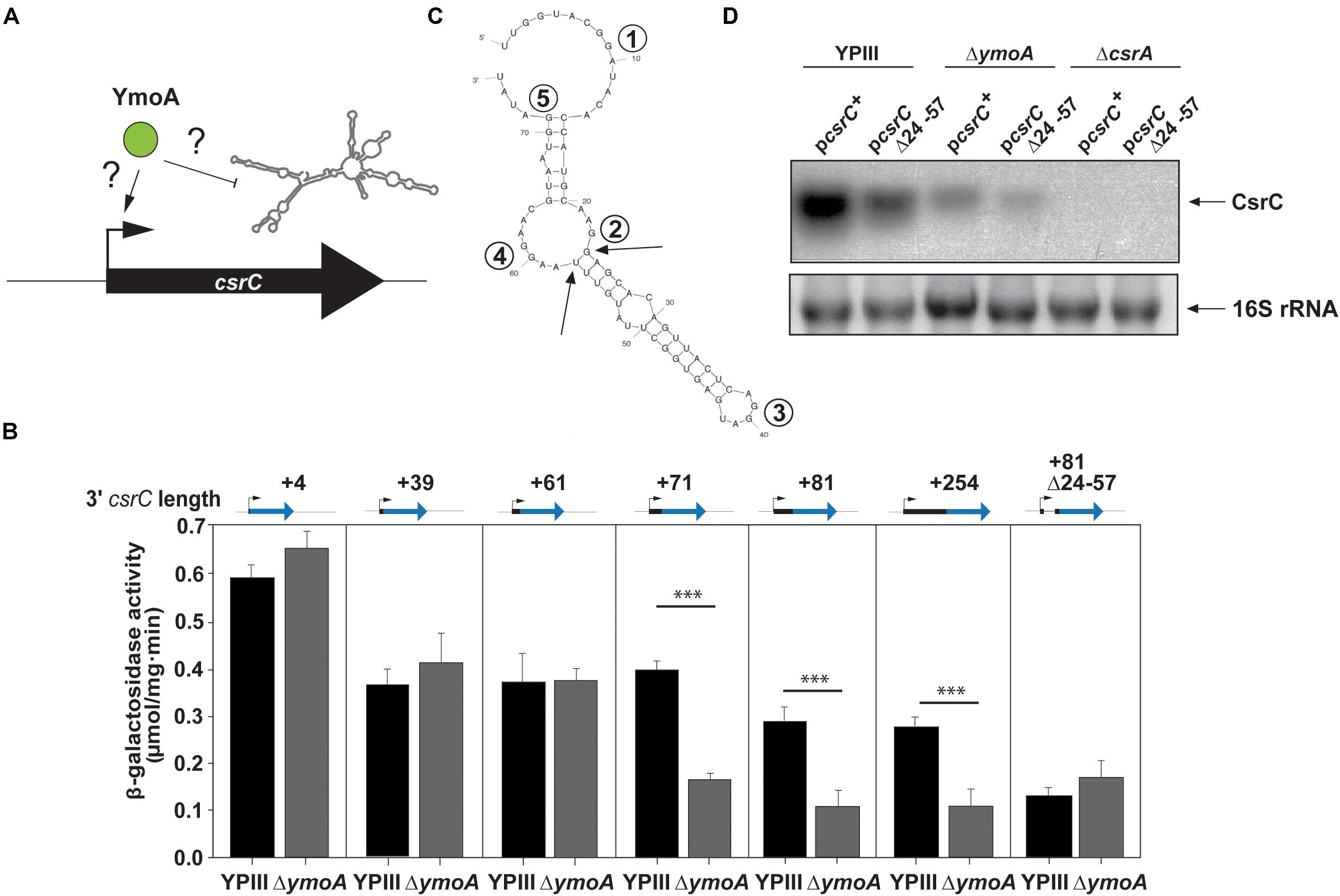
Figure 4. YmoA-mediated control of CsrC occurs on the post-transcriptional level. (A) Model for YmoA influence on CsrC on the transcriptional or post-transcriptional level. Increased CsrC levels can result from increased transcription or decreased degradation of CsrC. (B) Plasmids harboring transcriptional csrC-lacZ fusions with varying 3′-ends of the csrC gene and a csrC(+81)-lacZ fusion with a Δ(+24–+57) deletion (pKB17) were transformed into Y. pseudotuberculosis YPIII and YP50 (ΔymoA). Strains were grown overnight at 25°C and transcriptional activity of the different fusions was determined by β-galactosidase assays. β-galactosidase activity is given in μmol min–1 mg–1 for comparison. The data represent the average ± SD from at least three different experiments each done in duplicate. The statistical significances between the wild-type and the ymoA mutant were determined by the ANOVA test. P-values: ∗∗∗: <0.001. (C) Secondary structure prediction of the CsrC RNA from nucleotide +1 to +76 generated by Mfold. The single-stranded regions including the most highly conserved GGA element of CsrA binding sites are indicated by numbers. The arrows indicate the 5′ and 3′ end of the Δ(+24–+57) deletion. (D) Y. pseudotuberculosis strains YPIII (wild-type), YP50 (ΔymoA), and YP53 (ΔcsrA) harboring pKB59 (csrC+) or pKB49 (csrCΔ+24–+57) were grown in LB at 25°C overnight. Total RNA of the strains was extracted, separated on agarose gels and a CsrC-specific probe was used to detect the CsrC RNA by Northern blotting. The 16S rRNAs are shown as RNA loading control.
To further analyze the influence of YmoA on CsrC (Figure 5A), CsrC levels were determined by Northern blotting after transcription of Ptet:csrC was blocked by rifampicin. As shown in Figure 5B, the CsrC RNA was slowly degraded in the Y. pseudotuberculosis wild-type strain with a half-life of about 100 min, whereas CsrC was less stable in the ymoA mutant and decayed with a half-life of about 45–50 min. Presence of five potential CsrA-binding sites (GGA motif) within the 5′-hairpin region of the CsrC transcript (Figure 4C), suggested that the interaction and sequestration of CsrA could also increase the stability of the CsrC RNA. Northern blot analysis further revealed that CsrA is important for CsrC abundance as CsrC is not detectable in the ΔcsrA mutant (Figure 4D). To further characterize CsrA influence on CsrC stability, we induced csrC expression under the control of Ptet by anhydrotetracycline (AHT). After transcription was blocked by rifampicin, the CsrC transcript was much more rapidly degraded in the csrA mutant compared to wildtype (Figure 5C). In comparison, the influence of CsrA on CsrC stability was much more pronounced than that of YmoA (Figures 5B,C). Hence, it seemed possible that presence of YmoA influences the function of CsrA. To test this assumption, we tested whether CsrA overexpression is able to complement a ymoA mutant strain or whether YmoA overproduction can complement csrA deficiency by comparing CsrC levels in the different recombinant strains. As shown in Figure 5D, overexpression of YmoA was not able to complement csrA deficiency, whereas CsrC RNA synthesis was partially complemented when CsrA was overproduced in the ymoA mutant. This suggested that loss of YmoA can be overcome by an increase of CsrA-binding to the CsrC RNA. Based on these results, it is possible that YmoA has an influence on the activity of CsrA, e.g., by controlling an RNA chaperone assisting in CsrA-binding or folding of the CsrC RNA.
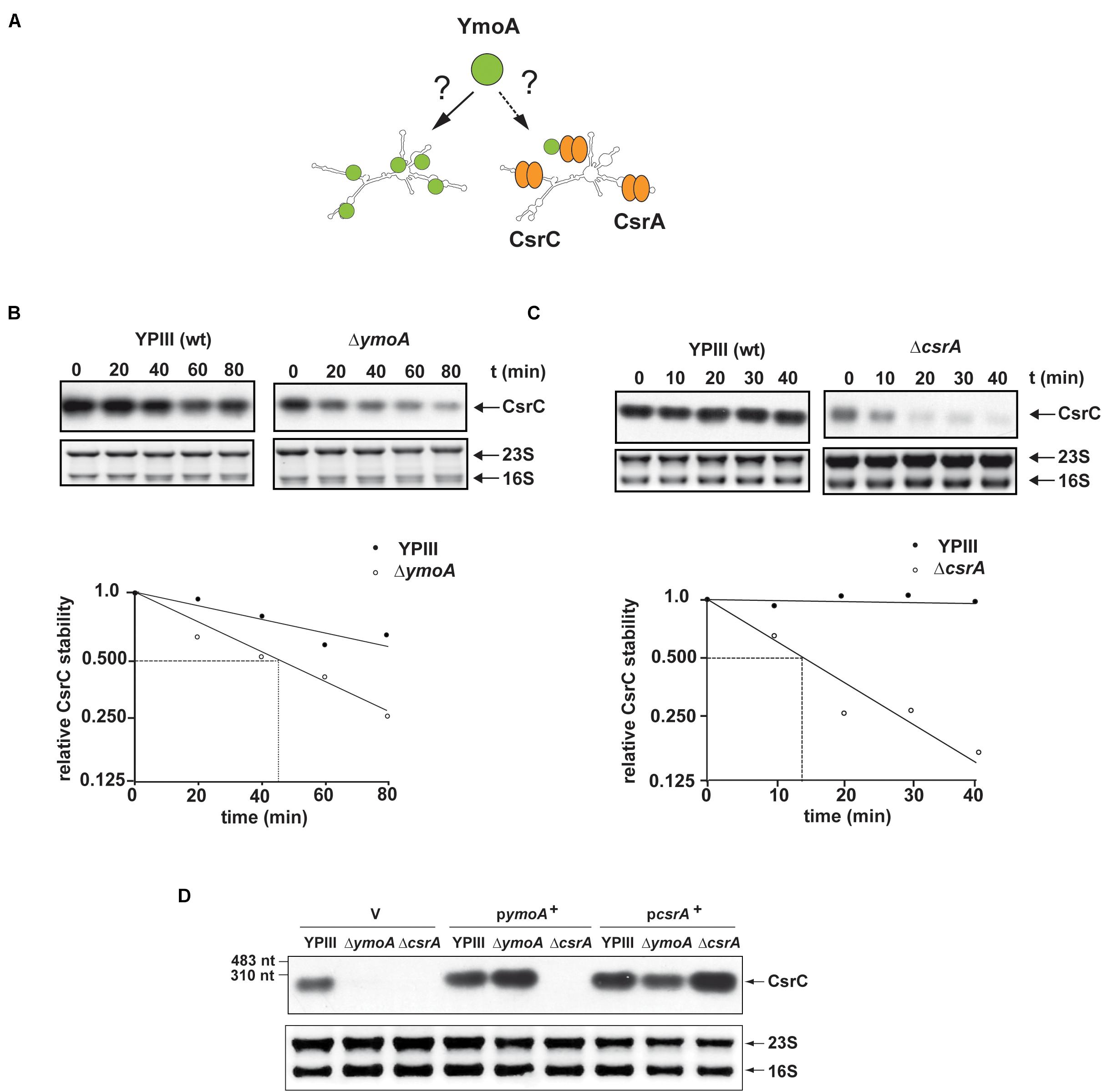
Figure 5. YmoA and CsrA increase the stability of the CsrC RNA. (A) Model for YmoA-dependent CsrC stabilization. (B,C) The stability of the CsrC RNA expressed under the control of the tet promoter was compared between Y. pseudotuberculosis strain YPIII (wild-type) and YP50 (ΔymoA) (B) or YP53 (ΔcsrA) (C) and CsrC RNA stability was analyzed over time by Northern blotting with a CsrC- specific probe. A representative Northern blot of three independent experiments is shown in the upper panel. 16S and 23S rRNA were used as loading controls. The band corresponding to the CsrC transcript was quantified using the BioRAD Image LabTM Software and was plotted versus time of extraction (in minutes) to calculate the half-life of the sRNA indicated by dotted lines (lower panel). (D) Y. pseudotuberculosis strains YPIII, YP50 (ΔymoA), and YP53 (ΔcsrA) harboring vector pHSG576 (V), a ymoA+ (pKB4) or csrA+ derivative (pKB60) were cultivated overnight in LB medium at 25°C. Total RNA of the strains was extracted, separated on agarose gels and a CsrC-specific probe was used to detect the CsrC ncRNA by Northern blotting. Used RNA marker is marked on the left. The 16S and 23S rRNAs are shown as RNA loading control.
One molecule that has been shown to assist in folding of Csr/Rsm-type RNAs, CsrA/RsmA binding and increases the stability and abundance of Csr/Rsm-type RNAs in Pseudomonas aeruginosa is the ubiquitous hexameric RNA-chaperone Hfq (Sonnleitner et al., 2006; Sorger-Domenigg et al., 2007). Hence, it seemed possible that YmoA exerts its stabilizing influence on CsrC via upregulation of Hfq (Figure 6A). As shown in Figure 6B, no or only very low amounts of the CsrC RNA were detected in an Δhfq mutant strain. This resulted in a dramatic increase of RovM and decrease of RovA levels and all these effects could be complemented by an hfq+ plasmid (Supplementary Figure 5). To elucidate how Hfq affects the Csr-RNAs, we performed a primer extension analysis to test whether absence of Hfq affects the abundance of CsrB and CsrC when transcription was blocked. Although slightly more CsrB transcript was found in the hfq mutant, the CsrB RNA seemed to be more rapidly degraded in the absence of Hfq (Figure 6C). In contrast, overall CsrC levels were significantly reduced in the Δhfq strain, but the half-life of the transcript was not influenced by Hfq (Figure 6C). This strongly indicated that Hfq has a positive influence on csrC transcription, but not on CsrC RNA stability. In agreement with this assumption we found that expression of the csrC(+4)-lacZ fusion harboring only the first four nucleotides of the csrC gene was downregulated in the hfq mutant (Figure 6D). Complementation assays further demonstrated that neither YmoA nor Hfq could compensate the effect of the hfq or the ymoA knock-out mutation on the abundance of the CsrC RNA (Figure 6E). Based on this observation, we concluded that Hfq and YmoA act differently and independently on CsrC.
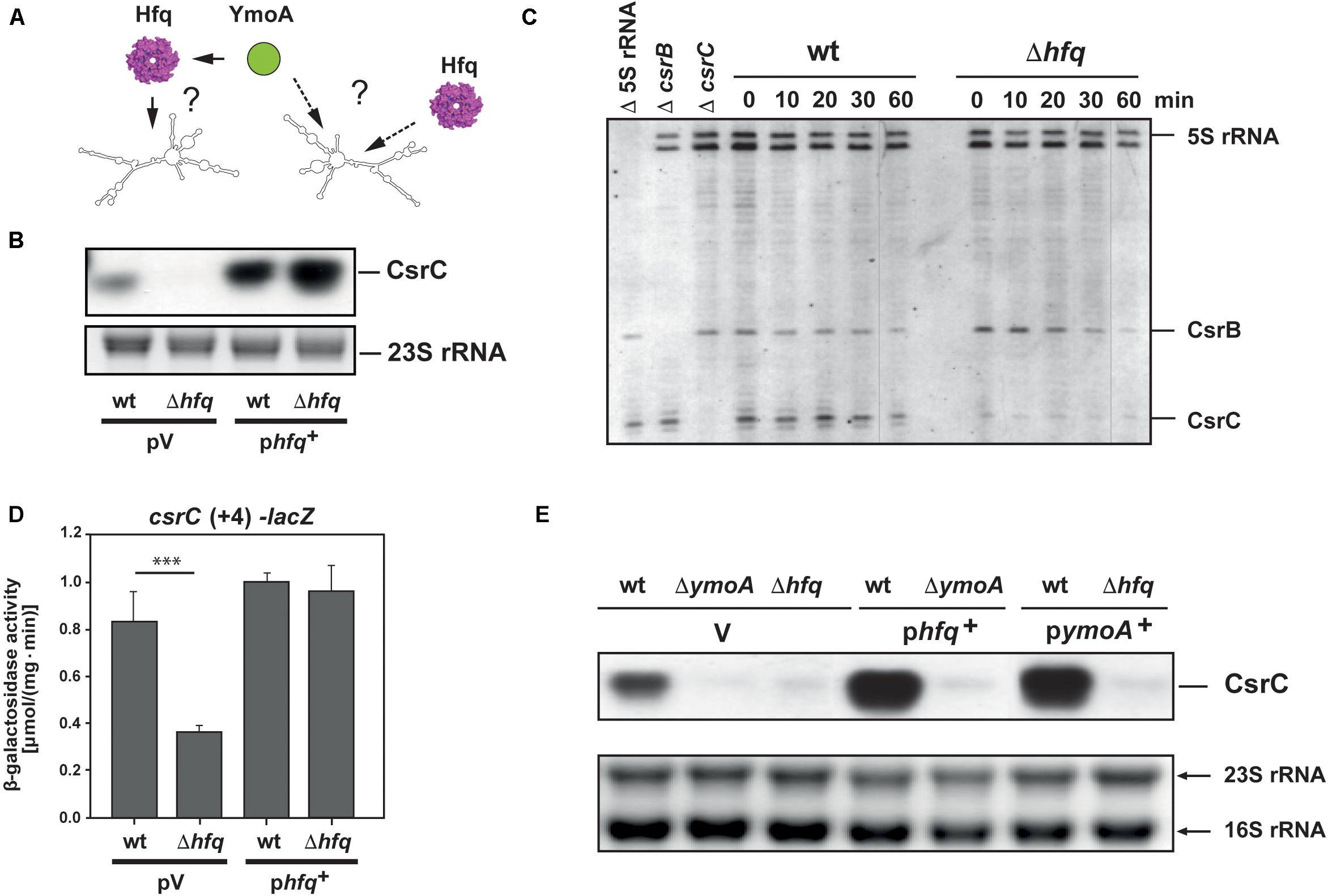
Figure 6. Hfq influence on CsrB and CsrC stability and cooperation with YmoA. (A) The model illustrates possible influence of YmoA on CsrC abundance via or independent of Hfq. (B) Y. pseudotuberculosis strain YPIII (wt) and YP80 (Δhfq) carrying the vector pAKH85 (pV) or its hfq+ derivative (pAKH115) were grown overnight in LB medium. Total RNA of the samples was isolated, separated on agarose gels and the CsrC RNA was detected by Northern blotting using a CsrC-specific probe. The 23S rRNAs is shown as RNA loading control. (C) Y. pseudotuberculosis strain YPIII (wt) and YP80 (Δhfq) were grown in LB at 25°C to early stationary phase, rifampicin was added in a final concentration of 500 μg ml–1 and aliquots were withdrawn at indicated time points. Total RNA was extracted and used for primer extension analysis. Samples without addition of labeled primers for 5S rRNA (Δ 5S rRNA), csrB (ΔcsrB), and CsrC (ΔcsrC) were used as controls. The reaction products were separated on 4% DNA sequencing gels. The positions of the transcripts are indicated. (D) Y. pseudotuberculosis strain YPIII (wt) and YP80 (Δhfq) carrying the vector pAKH85 (pV) or its hfq+ derivative (pAKH115) and the csrC-lacZ fusion (pAKH103) were grown in LB at 25°C overnight. β-Galactosidase activity of the cultures was determined (upper panels) and is given in μmol min–1 mg–1 for comparison. The data represent the average ± SD from at least three different experiments each done in duplicate. The statistical significances between the wild-type and the ymoA mutant were determined by the ANOVA test. P-values: ∗∗∗: <0.001. (E) Y. pseudotuberculosis strains YPIII, YP50 (ΔymoA) and YP80 (Δhfq) harboring vector pHSG575 (V), a ymoA+ (pKB4) or hfq+ derivative (pAKH119) were cultivated overnight in LB medium at 25°C. Total RNA of the strains was prepared, separated on agarose gels and a specific probe was used to detect the CsrC RNAs by Northern blotting. The 16S and 23S rRNAs are shown as RNA loading control.
As members of the YmoA/Hha protein family are able to form heteromeric complexes with H-NS (Nieto et al., 2002), we wanted to know whether YmoA acts through H-NS on CsrC. If so, changes in the active amounts of H-NS should influence CsrC RNA levels in Y. pseudotuberculosis. As shown in Figure 7A, the intracellular CsrC level was reduced in the presence of the dominant negative hns∗ allele and increased upon hns overexpression. Moreover, low abundance of CsrC in the ymoA mutant was further reduced by hns∗, whereas overexpression of H-NS is able to compensate for the loss of YmoA and led to CsrC levels similar to the wild-type strain (Figure 7A). Furthermore, a significant reduction of csrC(+81)-lacZ expression, was observed in the absence of ymoA which could be compensated by overexpression of hns (Figure 7B). However, the identical changes of YmoA and H-NS levels had no influence on csrC(+4)-lacZ expression, indicating that both regulatory factors act through the csrC fragment including positions +5 to +81.
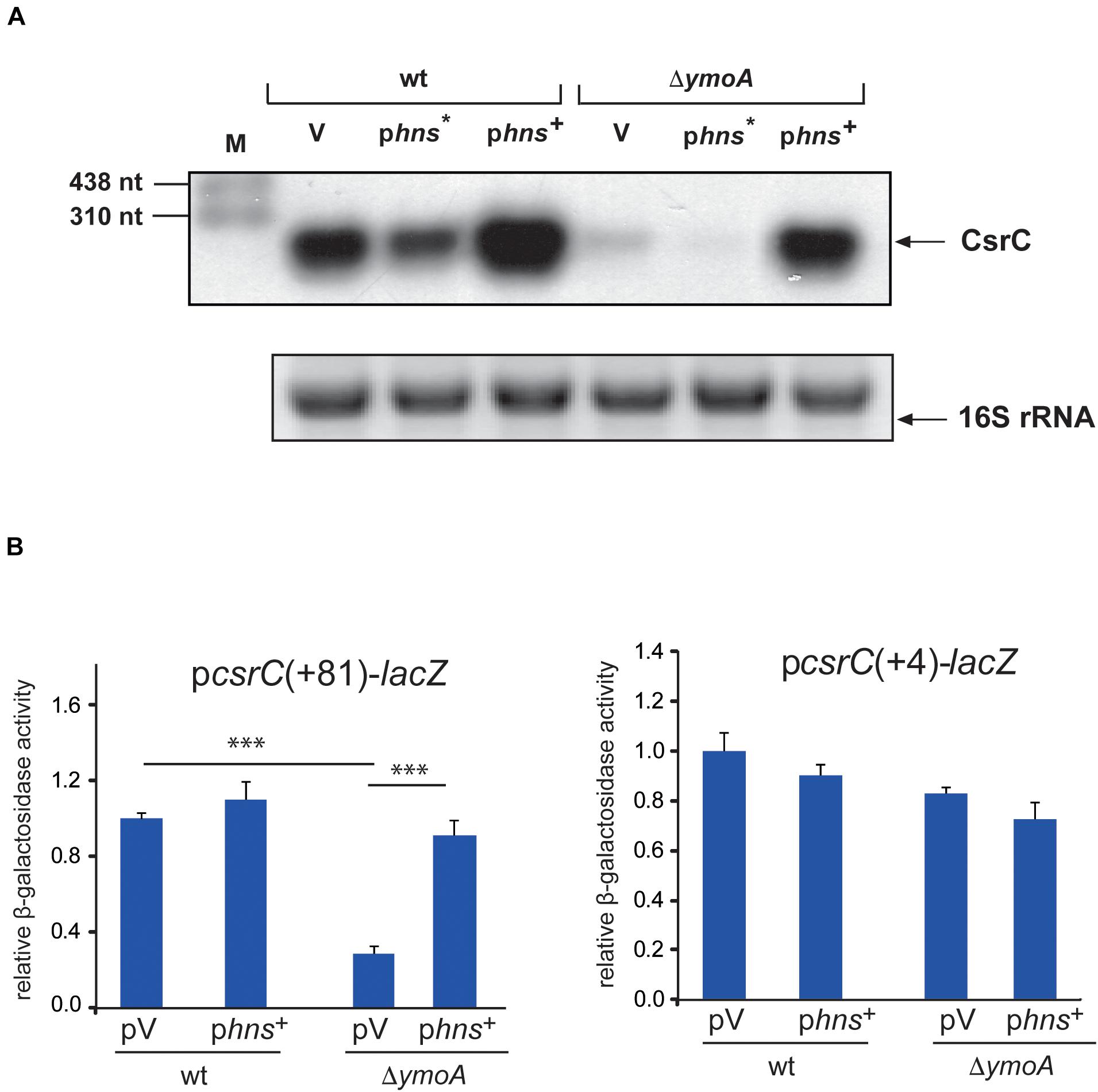
Figure 7. H-NS has a positive influence on csrC. (A) Y. pseudotuberculosis strain YPIII and YP50 (ΔymoA) harboring the empty vector (V), a hns* (pAKH31) or hns+ (pAKH74) overexpression construct were cultivated overnight in LB medium at 25°C. Total RNA of the strains was prepared, separated on agarose gels and a specific probe was used to detect the CsrC RNA by Northern blotting. The 16S rRNA is shown as RNA loading control. (B) Expression of csrC(+4)-lacZ and csrC(+81)-lacZ was analyzed in overnight cultures of Y. pseudotuberculosis strains YPIII (wild-type) and YP50 (ΔymoA) harboring vector pACYC184, pAKH85 or its hns+ derivative pAKH74 grown in LB at 25°C. β-galactosidase activity from the overnight cultures was determined and is given as relative expression for comparison. The data represent the average ± SD from at least three different experiments each done in duplicate. Data were analyzed by the ANOVA test, ∗∗∗P < 0.001.
As loss of YmoA did not influence H-NS levels (Supplementary Figure 6), we first assumed that YmoA enhances H-NS activity through YmoA-H-NS complex formation, and this could have a positive influence on CsrC level. Although YmoA and H-NS were shown to act predominantly on the transcriptional level, it has also been demonstrated that H-NS and paralogous proteins such as StpA can bind specifically to RNA and affect their folding and/or susceptibility to nuclease cleavage by RNases (Zhang et al., 1996; Mayer et al., 2002; Brescia et al., 2004). We tested whether YmoA and/or H-NS of Y. pseudotuberculosis could bind to the CsrC RNA in vitro. For this purpose, purified CsrC RNA transcript was incubated with increasing concentrations of the Yersinia H-NS and/or YmoA proteins. Both proteins were overexpressed and purified from E. coli strain KB4 deficient of all E. coli H-NS/Hha family proteins. However, neither H-NS alone nor in complex with YmoA was found to bind specifically to the CsrC RNA (Supplementary Figure 7A), whereas specific H-NS-dependent high molecular weight complexes were identified with a yscW promoter control fragment as reported previously (Böhme et al., 2012), demonstrating that the H-NS protein is active (Supplementary Figure 7B). This strongly suggests that YmoA and H-NS influence on CsrC is indirect, e.g., occurs through a factor controlling CsrC stability.
Next, we analyzed how YmoA and CsrA cooperate in the control of CsrC in response to different environmental parameters. To do so, we monitored the amount of CsrA and YmoA in the wild-type under different growth conditions. Synthesis of CsrA was slightly dependent on growth phase control resulting in moderately higher protein levels during stationary phase, but temperature and media composition had no considerable effect (Supplementary Figure 1). In contrast, YmoA protein levels were strongly dependent on temperature and growth phase, and at 37°C also on nutrient composition of the medium. Maximal amount of YmoA were observed at moderate temperature, in complex medium during exponential growth whereas no YmoA was detectable at 37°C in minimal medium (Supplementary Figure 1). To analyze the influence of the two regulatory factors in more detail, we determined CsrA, YmoA, and CsrC levels along the bacterial growth curve at 25 and 37°C in complex medium. At 25°C, highest CsrC levels were detected during stationary phase (8–12 h), whereas at 37°C CsrC levels were maximal during late exponential phase (6 h), but decreased upon entry into stationary phase (Figure 8). As, the intracellular concentration of CsrA remained at the same or even higher level, the decrease of CsrC amounts seems to result mainly from the strong reduction of YmoA levels during stationary phase, which is more pronounced at 37°C than at 25°C. The latter is the result of temperature-regulated proteolysis of YmoA mainly by the Lon protease but to a minor extent also by the ClpP protease (Supplementary Figure 8), similar to what has been described for the YmoA protein of Yersinia pestis (Jackson et al., 2004).
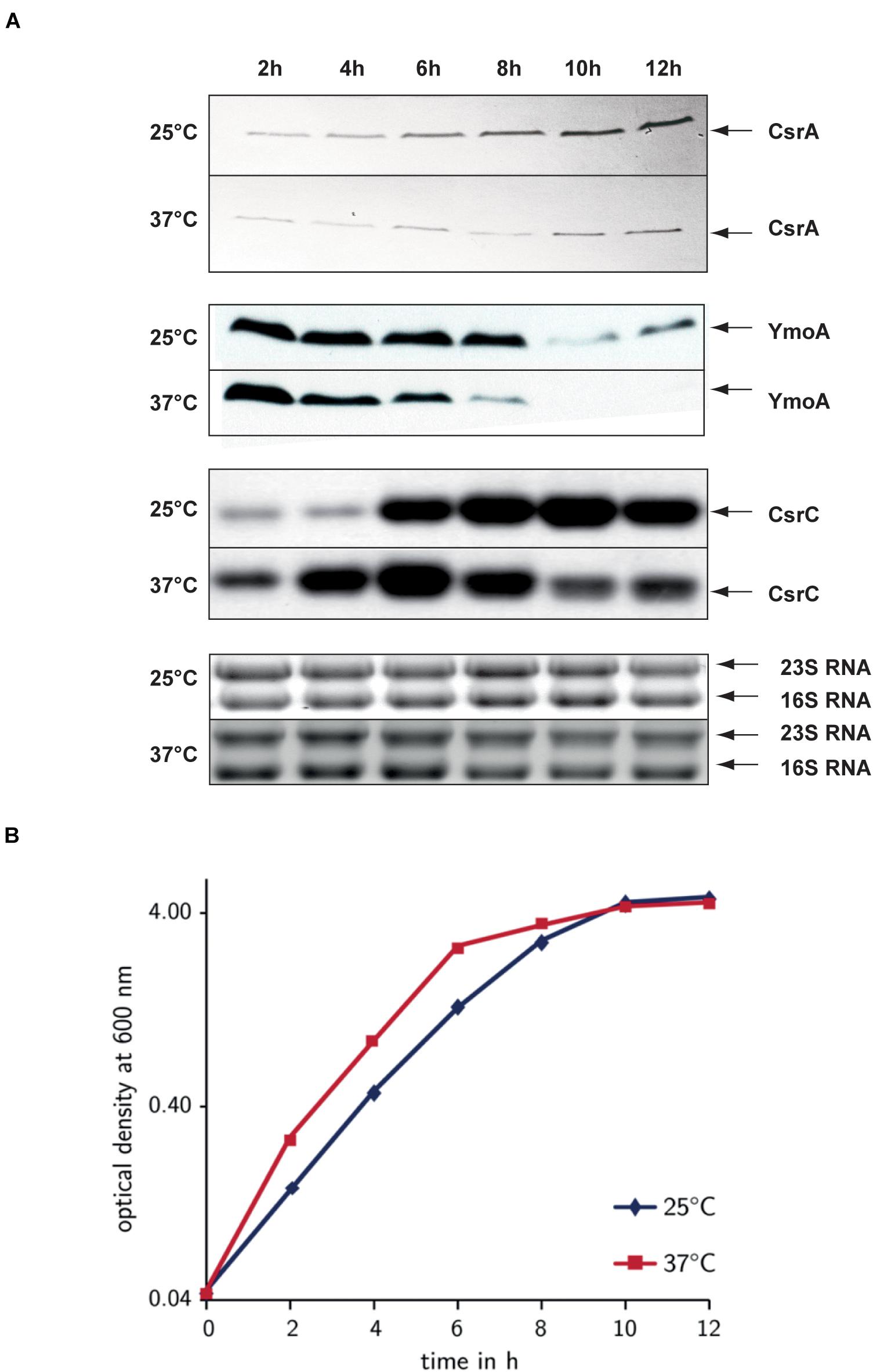
Figure 8. Comparison of the environmental control of CsrA, YmoA, and CsrC synthesis. Y. pseudotuberculosis strain YPIII was diluted 1:100 from an overnight culture and was grown in LB at 25 and 37°C. Every 2 h, samples were withdrawn for whole cell extract and total RNA preparations (A) and the OD600 of the cultures was determined to monitor growth (B). (A) Total RNA of the samples was separated on agarose gels and the CsrC RNA was detected by Northern blotting using a CsrC-specific probe. Whole cell extracts were separated on 18% TRICINE polyacrylamide gels and analyzed by Western blotting with a polyclonal antibody directed against YmoA and CsrA. The CsrC RNA band and the YmoA and CsrA protein bands are indicated by arrows. (B) OD600 of the cultures used for the upper analysis was plotted over time to illustrate and compare the different growth phases.
Transcriptome analysis and subsequent investigation revealed that YmoA not only inhibits the expression of the pYV-encoded antiphagocytic T3SS-associated virulence factors important to resist innate immune cell attacks through, but also modulates the expression of multiple colonization factors crucial for the initial infection phase (Supplementary Table 4). In fact, absence of YmoA strongly reduced synthesis of the RovA-dependent internalization factor invasin (InvA) (Figures 9A,B) which decreased the ability of Y. pseudotuberculosis to invade human cells by 50% (Figure 9C). This suggested that YmoA acts as a key virulence modulator which supports reprogramming from the initial colonization into the host defense mode.
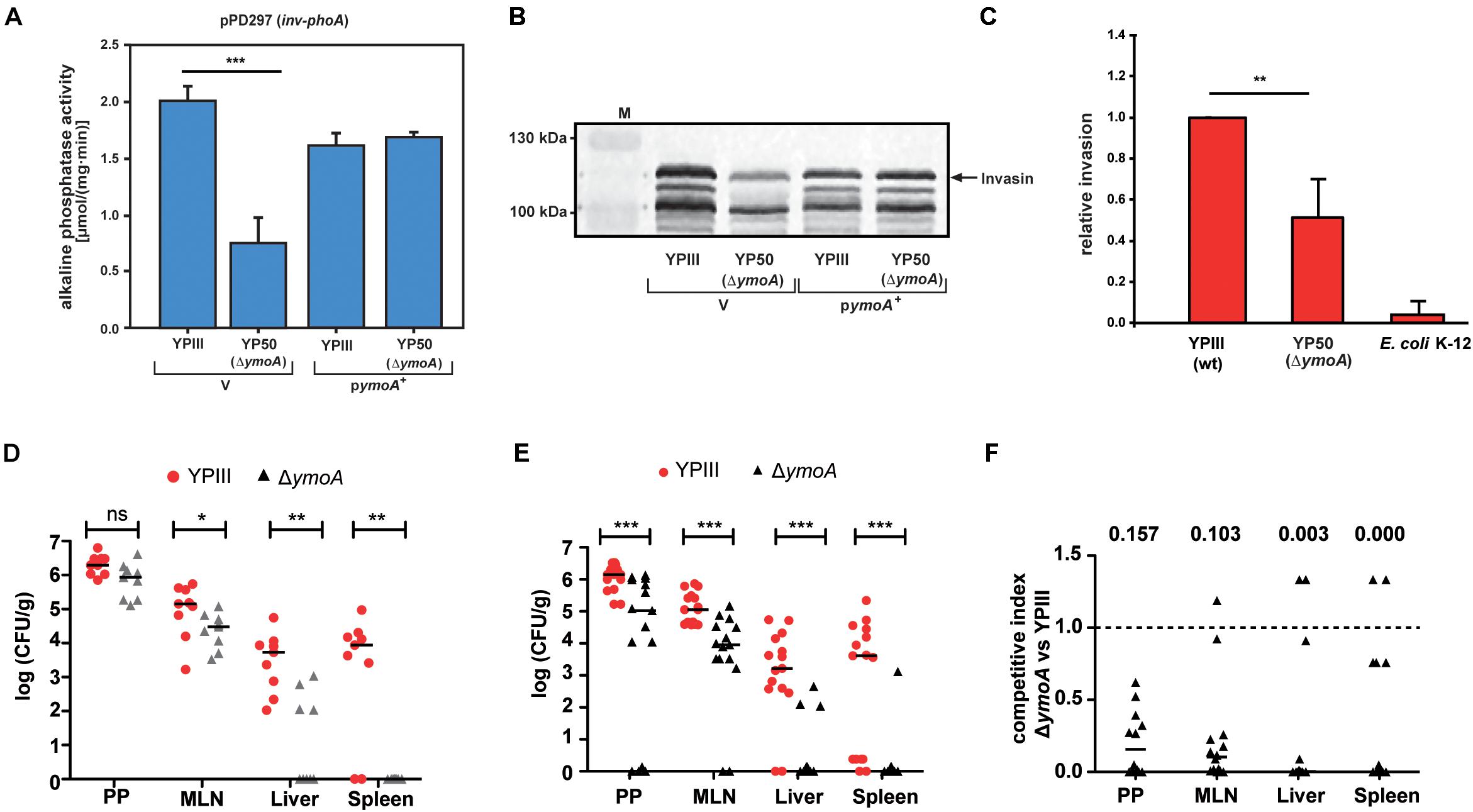
Figure 9. Influence of ymoA on the virulence of Y. pseudotuberculosis. Y. pseudotuberculosis wild-type strain YPIII and the mutant strain YP50 (ΔymoA) harboring the inv-phoA fusion plasmid pPD297 and the empty vector control pACYC184 (V) or the ymoA+ plasmid pAKH71 were grown overnight in LB medium at 25°C. (A) Alkaline phosphatase activity from overnight cultures was determined and is given in μmol min–1 mg–1. The data represent the average SD from at least three different experiments each done in duplicate. The statistical significances between the wild-type and the ymoA mutant were determined by the ANOVA test. P-values: ***: <0.001. (B) In parallel, whole cell extracts of the strains were prepared and analyzed by Western blotting with a monoclonal antibody directed against invasin. A molecular weight marker is loaded on the left. (C) HEp-2 cells were infected with wild-type Y. pseudotuberculosis, ΔymoA Y. pseudotuberculosis and E. coli K-12 and relative invasion was calculated. Each bar represents the average of three independent experiments relative to the invasion rate promoted by the Y. pseudotuberculosis wild-type strain, defined as 1.0. The statistical significances between the wild-type and the ymoA mutant were determined by the Students-test. P-values: **: <0.01. (D) BALB/c mice were infected intragastrically with an inoculum of 5⋅108 CFU of Y. pseudotuberculosis wild-type YPIII and the ymoA mutant YP50. Colonization of Peyer’s patches (PP), mesenteric lymph nodes (MLN), liver and spleen after 3 days of infection is displayed. (E) For co-infection experiments, an inoculum with 1⋅109 CFU of an equal mixture comprising Y. pseudotuberculosis wild-type YPIII and the ymoA mutant strain YP50 was infected into BALB/c mice via the orogastric route. After 3 days of infection, mice were sacrificed and the number of bacteria in homogenized host tissues and organs was determined by plating. Data are represented in scatter plots of numbers of CFU per gram as determined by counts of viable bacteria on plates. The statistical significances between the wild-type and the ymoA mutant were determined by the Mann–Whitney-test. P-values: *: <0.05; **: <0.01; ***: <0.001. (F) Data are graphed as competitive index values for the tissue samples from one mouse. Solid lines indicate median values. A competitive index score of 1 denotes no difference in the virulence compared to YPIII. Underlined values indicated the competitive index scores of the ymoA mutant relative to the wild-type.
To further define the overall influence of ymoA on bacterial pathogenesis, we compared the ability to colonize lymphatic tissues and organs between the Y. pseudotuberculosis wild-type strain YPIII and an isogenic ymoA-deficient strain (YP50) in a mouse infection model. First, we examined success of the infection in BALB/c mice 3 days after oral uptake of 5 × 108 bacteria by quantifying the number of bacteria that reached and survived in the Peyer’s patches (PP), the mesenterial lymph nodes (MLN), liver and spleen (Figure 9D). Significantly reduced numbers of the ymoA mutant bacteria were recovered from the MLNs and the organs. Attenuation of the ymoA mutant strain was even more evident when competition experiments were performed with 1 × 109 bacteria in an inoculum of an equal mixture of the wild-type and the ymoA-deficient strain (Figure 9E). Ten times less bacteria of the mutant were identified in the PPs and the MLNs, and only few numbers of bacteria were able to reach liver and spleen. Calculations of the competitive index of the mutant relative to wild-type clearly indicated that presence of YmoA is advantageous for the colonization of Y. pseudotuberculosis in all tested organs (Figure 9F).
Rapid adaptation of the gene expression profile by bacterial pathogens to changing environments within their hosts is a prerequisite to persist and establish a successful infection. Complex regulatory networks including global transcriptional and post-transcriptional regulatory factors orchestrate global cellular changes and control expression of virulence factors in close association with stress adaptation and metabolic functions. The carbon storage regulator (Csr) system composed of the RNA-binding protein CsrA and at least two regulatory RNAs (CsrB and CsrC) play a central role in the control of many host-pathogen interactions (Lucchetti-Miganeh et al., 2008; Kusmierek and Dersch, 2017; Romeo and Babitzke, 2018; Pourciau et al., 2020). In this report, we show that the Yersinia modulating protein YmoA is crucial for the control of Csr-type regulatory RNAs and reprograms virulence-relevant functions important for different stages of the infection (Figure 1B).
The YmoA protein of Yersinia belongs to a class of small proteins which modulate virulence gene expression in Enterobacteriaceae in response to environmental changes, e.g., temperature (Madrid et al., 2007). YmoA has a high amino acid sequence identity to and a similar overall fold consisting of four helices to Hha of E. coli which was identified as repressor of α-hemolysin production and other virulence genes (Mourino et al., 1996; Sharma and Zuerner, 2004; McFeeters et al., 2007). Also, the Yersinia YmoA protein is a key regulator of pathogenicity factors. It was identified as a thermosensitive transcriptional silencer for lcrF expression. LcrF is a key regulator for the pYV-encoded temperature-induced T3SS and yop effector genes important to prevent the immune defense after entry of the gut-associated lymphatic tissues (Mikulskis et al., 1994; Jackson et al., 2004; Böhme et al., 2012). Hence, by controlling lcrF expression, YmoA regulates one of the most crucial pathogenicity factors for infection of the human host. In this study, we show that YmoA also activates expression of virulence genes important for host tissue colonization during early stages of the infection. YmoA induces expression of the virulence regulator gene rovA and the RovA-dependent invA gene by downregulation of the LysR repressor RovM, and this is mediated through YmoA-dependent changes of the global post-transcriptional Csr system (Figure 10). YmoA was found to enhance the stability of the CsrC RNA and this was shown to involve sequences within the 5′-region of the CsrC RNA predicted to form an extended stem-loop.
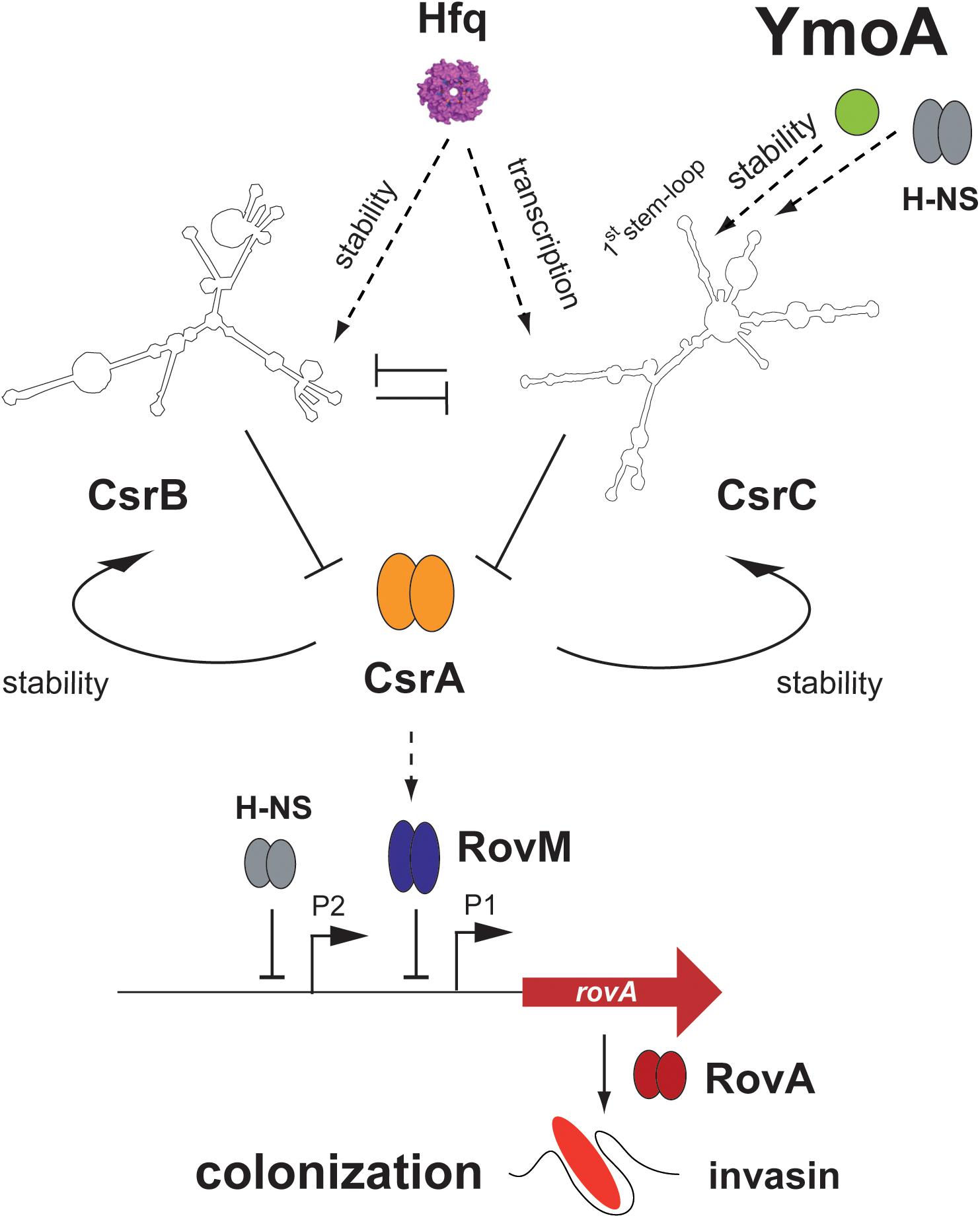
Figure 10. Influence of YmoA on the CsrABC-RovM-RovA-InvA cascade in Y. pseudotuberculosis. YmoA is a global virulence regulator of Y. pseudotuberculosis which is expressed under environmental conditions reflecting the initial stages of infection. It activates the synthesis of early-phase cell internalization factor invasin (InvA) which is important for colonization. This seems to occur together with H-NS, which is known to form heteromeric complexes with YmoA, through regulation of the degradation of the regulatory RNA CsrC at the first stem-loop structure of the CsrC RNA. Influence of YmoA on CsrC occurs independently of Hfq, which was also involved in Csr system control. YmoA and H-NS influence on CsrC is indirect and most likely occurs through RNA-processing factors. As a consequence of YmoA-mediated CsrC upregulation the CsrA-RovM-RovA regulator cascade is activated, leading to expression of colonization factors such as invasin. Dark arrows display direct activation, T represents repression, and dashed arrows indicate (indirect) activation by an unknown molecular mechanism.
Turnover of regulatory RNAs is a highly regulated process. Potential pathways for modulation include recruitment or inhibition of RNA decay enzymes (RNases), formation of stabilizing RNA secondary structures and/or alteration of the expression, activity or localization of RNA-binding factors. In E. coli and Salmonella enterica serovar Typhimurium, CsrB and CsrC decay involves the essential enzyme RNase E, a single-strand-specific endoribonuclease, which associates with the 3′–5′ processive exonuclease polynucleotide phosphorylase (PNPase), the glycolytic enzyme enolase, and an RNA helicase (RhlB/CsdA) to form an RNA-degrading complex (degradosome) (Suzuki et al., 2006; Viegas et al., 2007). Processing of the Salmonella CsrC RNA at one of the longer 3′ stems is also RNase III-dependent (Viegas et al., 2007). Here, we show that a 5′-terminal segment between nt +24 and +57 of the Yersinia CsrC RNA which is able to form a stem-loop structure is important for its stabilization. It is likely that the accessibility of potential ribonuclease cleavage sites is inhibited by this structured RNA element. However, none of the genes for RNase III (rnc), PNPase (pnp), and RNase E (rne) were differently expressed in the ΔymoA mutant (Supplementary Table 4). As this part of the RNA varies significantly from E. coli and Salmonella CsrC and no smaller CsrC transcripts were detectable, it seems that CsrC decay mechanism in Yersinia differs from the other CsrC RNAs.
As absence of the hairpin sequence in the csrC gene eliminates YmoA influence on CsrC stabilization, it is also possible that YmoA controls RNA-binding proteins, protecting CsrC from decay. Although the abundance of CsrB and CsrC of Y. pseudotuberculosis is positively affected by the RNA chaperone Hfq, we show that YmoA acts independently of Hfq. As Hfq is still able to induce csrC(+4)-lacZ fusion, it is more likely that Hfq activates csrC expression indirectly through control of (an)other sensory or regulatory RNA(s). We further found that also the RNA-binding protein CsrA is crucial for CsrC abundance in Yersinia, and absence of YmoA could be partly overcome by CsrA overexpression. This implies that stabilization of CsrC by CsrA might either impede the activity of a YmoA-dependent destabilizing factor or CsrA might act as a stabilization factor itself. The most highly conserved element of CsrA binding sites is a GGA sequence. Five GGA motifs are located within the stabilizing 5′ region of the CsrC RNA, one in the loop, two at the 5′- and 3′-end of the hairpin structure and one at the start and end of the sequence (Figure 4C), which could assist in the formation of the stabilizing RNA element and/or protection of RNase cleavage sites. In this case, sequestration of CsrA to other target sequences would destabilze CsrC. Although overall CsrA levels remain unchanged in the absence of YmoA (Figure 3B), it is possible that global transcriptional changes of 289 genes detected in the ΔymoA mutant – the great majority (79%) of which were upregulated – are associated with a significant increase of alternative CsrA target transcripts, which would reduce the availability of free CsrA to bind and stabilize the CsrC transcript. Similarly, CsrB and CsrC synthesis in E. coli is positively regulated by CsrA, but their stability was essentially identical between wild-type and the csrA mutant. This indicated that CsrA does not directly affect CsrB and CsrC RNA degradation in this organism (Gudapaty et al., 2001; Weilbacher et al., 2003).
Several years ago, Saxena and Gowrishankar (2011) showed that overexpression of the YmoA/Hha paralog YdgT suppressed the phenotypes of rho and nusG mutants. They suggest a model whereby YdgT modulates Rho-dependent transcription termination, by compensating for Rho loss (Saxena and Gowrishankar, 2011). Following the E. coli model, YmoA would increase transcriptional termination, what is not observed with csrC. Nonetheless, the hairpin structure of CsrC could be involved in premature transcriptional termination which could be inhibited by YmoA. However, no short, truncated CsrC transcripts have been identified with CsrC-specific probes.
Several studies further indicate that the global YmoA/Hha modulators exert their effect through heterocomplex formation with H-NS (Nieto et al., 2002; Madrid et al., 2007). As YmoA was also found to be structurally similar to H-NS, it was further suggested that YmoA may intercalate into higher-order structures by substituting for an H-NS dimer (McFeeters et al., 2007). H-NS is a highly abundant DNA-binding protein, which binds and polymerizes along AT-rich, curved DNA sites and acts as global regulator of gene expression in response to environmental signals (Bouffartigues et al., 2007; Fang and Rimsky, 2008). Heterocomplex formation with YmoA modifies the biological activity of H-NS and reduces or increases its ability to repress transcription (Nieto et al., 2002; Banos et al., 2008). Alike YmoA, H-NS controls the amount of CsrC and complements the decrease of CsrC levels in an ymoA mutant, indicating that YmoA and H-NS most likely acts in a cooperative manner to modulate transcription of factors controlling CsrC decay.
More recently, YmoA/Hha, together with YmoB/TomB(YbaJ) encoded in the same operon, was found to constitute a type II toxin-antitoxin (TA), which is implicated in the formation of persister cells and biofilms by repressing type I fimbriae expression in related Enterobacteriaceae (García-Contreras et al., 2008; Marimon et al., 2016). In E. coli, TomB(YbaJ) transiently interacts with the YmoA homolog Hha and initiates spontaneous oxidation of a conserved Cys residue leading to a destabilization of Hha. Its function can be replaced by the TomB homolog YmoB of Yersinia (Marimon et al., 2016). Interestingly, the abundance of the ymoB transcript is strongly (81-fold) increased in the absence of ymoA (Supplementary Table 4), indicating a negative autoregulatory loop. This reveals also the possibility that enhanced degradation of CsrC and changes of RovM, RovA and InvA levels in the ΔymoA mutant could be promoted through upregulation of YmoB. The overall contribution of the TA system is not yet fully understood, but induction of the TA loci by environmental stress conditions led to the idea that these systems enhance bacterial fitness under adverse conditions and play roles in pathogenesis (Norton and Mulvey, 2012). Our work showing that YmoA manipulates the global post-transcriptional Csr system, influences multiple fitness- and virulence-relevant components and has a strong effect on virulence, supports this assumption.
Our transcriptome analysis revealed a global impact of YmoA on early and later stage virulence genes. This includes virulence traits important for host tissue colonization such as cell adhesion and invasion factors (invasin, Ail, PsaA adhesin, and fimbriae), toxins and type VI secretion system components as well as immune cell defense strategies (T3SS-Yops). Moreover, a variety of genes for stress responses experienced in the initial stage of the infection (oxidative stress, starvation, and pH) are activated by YmoA, whereas factors promoting translation and proper folding of proteins (e.g., heat stress/shock genes) are repressed. In addition, metabolic activities related to the amino acid/nitrogen metabolism and assimilation are activated, whereas the metabolism of many carbon sources is repressed by YmoA (Figures 1B,C and Supplementary Table 4). This reprogramming implicates control of the Csr system which (i) plays a central role coordinating virulence gene expression with adaptative responses functions for different nutritional demands and stress resistance during infections (Lucchetti-Miganeh et al., 2008; Timmermans and Van Melderen, 2010; Bücker et al., 2014; Kusmierek and Dersch, 2017) and (ii) promotes transition between different physiological stages of pathogens, e.g., the switch from an acute to a chronic infection or from a transmission into an intracellular replication state (Molofsky and Swanson, 2003; Goodman et al., 2004; Laskowski and Kazmierczak, 2006; Mulcahy et al., 2008; Brencic and Lory, 2009; Sahr et al., 2009).
One important environmental parameter, which induce global alterations in the gene expression and reprogramming between the different virulence-related stages of Yersinia, is temperature (Nuss et al., 2015; Schmühl et al., 2019). As YmoA turnover by the ClpP and Lon proteases is modulated by temperature (Jackson et al., 2004) (Supplementary Figure 8), it is tempting to speculate that YmoA plays a crucial role adjusting the Csr regulon in response to temperature which is pivotal for the establishment of a successful infection.
Based on our analysis we assume that thermosensitive YmoA promotes expression of very early-stage virulence-relevant traits to promote initiation of the infection. Subsequent degradation of YmoA at 37°C within the host will then trigger an upregulation of pathogenicity factors, i.e., toxin CNFY and the antiphagocytic T3SS/Yop machinery, and improve use of carbon sources, which are crucial for the establishment of the infection in the intestine and underlying lymphoid tissues. Co-induction of mechanisms that promote protein translation and folding may guarantee a rapid and highly efficient production of these virulence traits. This global impact of YmoA on virulence was visible in non-competitive mouse infection studies and was even more noticeable in competitive assays in which the ymoA mutant was clearly outcompeted by the wild-type. Many questions concerning the molecular interactions between YmoA and the Csr system, and significance of YmoA-controlled genes for virulence remain to be solved. Yet, it is foreseeable that the YmoA-Csr control network is a valuable target for antibacterial therapeutics to block the bacteria at specific stages during infection.
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit: animal licensing committee permission no. 33.9.42502-04-055/09.
AH, KB, and PD designed the research. AH, KB, and SL performed the experiments. AH, KB, SL, PD, ASS, and YG analyzed the data. AH, KB, and PD interpreted the data. AH and PD wrote the manuscript. AH and ASS edited the manuscript. PD acquired funding. All authors contributed to the article and approved the submitted version.
This work was supported by Deutsche Forschungsgemeinschaft Grant DE616/3 and DE616/7. YG has a stipend of the China Scholarship Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Martin Fenner for helpful discussions. Further we thank Tatjana Stolz and Tanja Krause for technical support, Rebekka Steinmann and Janika Viereck for construction of plasmids, and Jana Melzer, Johannes Klein, and Richard Münch for support in the analysis of the microarray data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.706934/full#supplementary-material
Supplementary Figure 1 | Influence of the growth medium, growth phase and temperature on YmoA synthesis. Y. pseudotuberculosis strain YPIII was grown in LB or minimal medium A (MMA) to exponential (OD600 = 0.8) or stationary phase at 25 and 37°C. Whole cell extracts of the cultures were prepared, proteins were separated on 18% TRICINE polyacrylamide gels and analyzed by Western blotting with a polyclonal antibody directed against YmoA and CsrA. Cell extracts of an ymoA and a csrA mutant strain were used as controls. A used prestained molecular weight marker is indicated on the left. Unspecifically detected proteins (c) were used as loading control.
Supplementary Figure 2 | YmoA has no influence on rovA expression through H-NS. Y. pseudotuberculosis strain YPIII and YP50 (ΔymoA) harboring the empty vector (V), or a dominant negative hns∗ allele (pAKH31) were cultivated overnight in LB medium at 25°C. Whole cell extracts were prepared, separated by SDS-PAGE and analyzed by Western blotting with a polyclonal antibody directed against RovA. Cell extracts of a rovA mutant was used as control. Unspecifically detected proteins (c) were used as loading control.
Supplementary Figure 3 | Influence of YmoA on the Csr system occurs through CsrC. Y. pseudotuberculosis strains YPIII (wild-type), YP50 (ΔymoA), YP69 (ΔcsrB), and YP75 (ΔcsrB and ΔymoA) were grown in LB at 25°C overnight. (A) Total RNA of the strains was extracted, and CsrC transcripts were detected by Northern blotting with a CsrC-specific probe. The 16S rRNAs are shown as RNA loading control. (B) In parallel, whole-cell extracts were prepared from the cultures and analyzed by Western blotting with a polyclonal antibody directed against RovM. A molecular weight marker is loaded on the left. Unspecifically detected proteins (c) were used as loading control.
Supplementary Figure 4 | YmoA has a positive effect on CsrC stability. (A) Y. pseudotuberculosis strains YPIII (wild-type) and YP50 (ΔymoA) harboring a Ptet:lacZ expression construct (pTT1) were grown to exponential phase in minimal medium at 25°C in the presence of 0.5 nM anhydrotetracycline. β-galactosidase activity from the overnight cultures was determined and is given in μmol min–1 mg–1 for comparison. The data represent the average ± SD from at least three different experiments each done in duplicate. (B) Y. pseudotuberculosis strains YPIII (wild-type) and YP50 (ΔymoA) harboring the empty vector (pHSG576) or a Ptet:csrC expression construct (pKB47) were grown to exponential phase in minimal medium at 25°C in the presence of 0.5 nM anhydrotetracycline. Total RNA of the strains was extracted and CsrC was detected by Northern blotting with a CsrC-specific probe. The 16S and 23S rRNAs are shown as RNA loading control.
Supplementary Figure 5 | Hfq influence on RovA and RovM levels. Y. pseudotuberculosis strain YPIII and YP80 (Δhfq) carrying the vector pAKH85 (pV) or its hfq+ (pAKH115) were grown in LB at 25°C overnight. Whole-cell extracts from the overnight cultures were prepared and analyzed by Western blotting with polyclonal antibodies directed against RovA or RovM. Unspecifically detected proteins (c) were used as loading control.
Supplementary Figure 6 | YmoA influence on Hfq production. Y. pseudotuberculosis strain YPIII and YP50 (ΔymoA) carrying the vector pAKH85 (pV) or its ymoA+ derivative (pAKH71) were grown in LB at 25°C overnight. Whole-cell extracts from the overnight cultures were prepared and analyzed by Western blotting with a polyclonal antibody directed against Hfq. An unspecifically detected protein (c) was used as loading control.
Supplementary Figure 7 | Analysis of YmoA and H-NS binding to CsrC and yscW. (A) CsrC RNA (+1 to +150) and a 5S RNA fragment serving as a negative control as well as (B) the yscW-lcrF promoter fragment and a csiD fragment of E. coli serving as negative control were incubated without or with increasing amounts of purified Y. pseudotuberculosis YmoA in the presence or absence of H-NS or H-NS alone. The samples were separated on 8% polyacrylamide gels. The positions of the gene fragments are indicated, and the formation of higher molecular complexes is shown by an arrow.
Supplementary Figure 8 | Temperature-dependent YmoA degradation. Whole cell extracts from overnight cultures of Y. pseudotuberculosis strains YPIII (wild-type), YP63 (ΔclpP), YP64 (Δlon) and YP67 (ΔclpP Δlon) grown in LB at 25 and 37°C were prepared, separated by 20% TRICINE-PAGE and analyzed by Western blotting using a polyclonal anti-YmoA antibody. Whole cell extract of an overnight culture of YP50 (ΔymoA) was used as negative control. A molecular weight marker was loaded on the left, the YmoA protein is indicated by an arrow. Unspecifically detected proteins (c) were used as loading control.
Alexa, A., Rahnenfuhrer, J., and Lengauer, T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. doi: 10.1093/bioinformatics/btl140
Banos, R. C., Pons, J. I., Madrid, C., and Juarez, A. (2008). A global modulatory role for the Yersinia enterocolitica H-NS protein. Microbiology 154, 1281–1289. doi: 10.1099/mic.0.2007/015610-0
Barnes, P. D., Bergman, M. A., Mecsas, J., and Isberg, R. R. (2006). Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J. Exp. Med. 203, 1591–1601. doi: 10.1084/jem.20060905
Bliska, J. B., Wang, X., Viboud, G. I., and Brodsky, I. E. (2013). Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell. Microbiol. 15, 1622–1631. doi: 10.1111/cmi.12164
Böhme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., Berger, E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Bottone, E. J. (1997). Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10, 257–276. doi: 10.1128/cmr.10.2.257-276.1997
Bouffartigues, E., Buckle, M., Badaut, C., Travers, A., and Rimsky, S. (2007). H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14, 441–448. doi: 10.1038/nsmb1233
Brencic, A., and Lory, S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x
Brescia, C. C., Kaw, M. K., and Sledjeski, D. D. (2004). The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 339, 505–514. doi: 10.1016/j.jmb.2004.03.067
Bücker, R., Heroven, A. K., Becker, J., Dersch, P., and Wittmann, C. (2014). The pyruvate-tricarboxylic acid cycle node: a focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J. Biol. Chem. 289, 30114–30132. doi: 10.1074/jbc.M114.581348
Cathelyn, J. S., Crosby, S. D., Lathem, W. W., Goldman, W. E., and Miller, V. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 13514–13519. doi: 10.1073/pnas.0603456103
Cathelyn, J. S., Ellison, D. W., Hinchliffe, S. J., Wren, B. W., and Miller, V. L. (2007). The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol. Microbiol. 66, 189–205. doi: 10.1111/j.1365-2958.2007.05907.x
Chen, S., Thompson, K. M., and Francis, M. S. (2016). Environmental regulation of Yersinia pathophysiology. Front. Cell. Infect. Microbiol. 6:25. doi: 10.3389/fcimb.2016.00025
Cornelis, G. R. (1993). Role of the transcription activator virF and the histone-like protein YmoA in the thermoregulation of virulence functions in yersiniae. Zentralbl. Bakteriol. 278, 149–164. doi: 10.1016/s0934-8840(11)80833-9
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. doi: 10.1038/nrmicro1526
Cornelis, G. R., Sluiters, C., Delor, I., Geib, D., Kaniga, K., Lambert de Rouvroit, C., et al. (1991). ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5, 1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
de la Cruz, F., Carmona, M., and Juarez, A. (1992). The Hha protein from Escherichia coli is highly homologous to the YmoA protein from Yersinia enterocolitica. Mol. Microbiol. 6, 3451–3452. doi: 10.1111/j.1365-2958.1992.tb02214.x
Derbise, A., Lesic, B., Dacheux, D., Ghigo, J. M., and Carniel, E. (2003). A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38, 113–116. doi: 10.1016/s0928-8244(03)00181-0
Dersch, P., and Isberg, R. R. (1999). A region of the Yersinia pseudotuberculosis invasin protein enhances integrin-mediated uptake into mammalian cells and promotes self-association. EMBO J. 18, 1199–1213. doi: 10.1093/emboj/18.5.1199
Ellison, D. W., and Miller, V. L. (2006). H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188, 5101–5112. doi: 10.1128/jb.00862-05
Fang, F. C., and Rimsky, S. (2008). New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11, 113–120. doi: 10.1016/j.mib.2008.02.011
Galindo, C. L., Rosenzweig, J. A., Kirtley, M. L., and Chopra, A. K. (2011). Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in human yersiniosis. J. Pathog. 2011:182051. doi: 10.4061/2011/182051
García-Contreras, R., Zhang, X.-S., Kim, Y., and Wood, T. K. (2008). Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage kller genes. PLoS One 3:e2394. doi: 10.1371/journal.pone.0002394
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80.
Goodman, A. L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R. S., and Lory, S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754. doi: 10.1016/j.devcel.2004.08.020
Gudapaty, S., Suzuki, K., Wang, X., Babitzke, P., and Romeo, T. (2001). Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183, 6017–6027. doi: 10.1128/jb.183.20.6017-6027.2001
Heroven, A. K., and Dersch, P. (2006). RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 62, 1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x
Heroven, A. K., and Dersch, P. (2014). Coregulation of host-adapted metabolism and virulence by pathogenic Yersiniae. Front. Cell Infect. Microbiol. 4:146. doi: 10.3389/fcimb.2014.00146
Heroven, A. K., Bohme, K., and Dersch, P. (2012a). The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol 9, 379–391. doi: 10.4161/rna.19333
Heroven, A. K., Bohme, K., Rohde, M., and Dersch, P. (2008). A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68, 1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x
Heroven, A. K., Nagel, G., Tran, H. J., Parr, S., and Dersch, P. (2004). RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53, 871–888. doi: 10.1111/j.1365-2958.2004.04162.x
Heroven, A. K., Sest, M., Pisano, F., Scheb-Wetzel, M., Steinmann, R., Bohme, K., et al. (2012b). Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front. Cell Infect. Microbiol. 2:158. doi: 10.3389/fcimb.2012.00158
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
Jackson, M. W., Silva-Herzog, E., and Plano, G. V. (2004). The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54, 1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x
Koornhof, H. J., Smego, R. A., and Nicol, M. (1999). Yersiniosis. II: The pathogenesis of Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18, 87–112. doi: 10.1007/s100960050237
Kusmierek, M., and Dersch, P. (2017). Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr. Opin. Microbiol. 41, 58–67. doi: 10.1016/j.mib.2017.11.022
Kusmierek, M., Hoßmann, J., Witte, R., Opitz, W., Vollmer, I., Volk, M., et al. (2019). A bacterial secreted translocator hijacks riboregulators to control type III secretion in response to host cell contact. PLoS Pathog. 15:e1007813. doi: 10.1371/journal.ppat.1007813
Lambert de Rouvroit, C., Sluiters, C., and Cornelis, G. R. (1992). Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6, 395–409. doi: 10.1111/j.1365-2958.1992.tb01483.x
Laskowski, M. A., and Kazmierczak, B. I. (2006). Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 74, 4462–4473. doi: 10.1128/iai.00575-06
Lawal, A., Kirtley, M. L., van Lier, C. J., Erova, T. E., Kozlova, E. V., Sha, J., et al. (2013). The effects of modeled microgravity on growth kinetics, antibiotic susceptibility, cold growth, and the virulence potential of a Yersinia pestis ymoA-deficient mutant and Its isogenic parental strain. Astrobiology 13, 821–832. doi: 10.1089/ast.2013.0968
Lucchetti-Miganeh, C., Burrowes, E., Baysse, C., and Ermel, G. (2008). The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154, 16–29. doi: 10.1099/mic.0.2007/012286-0
Madrid, C., Balsalobre, C., Garcia, J., and Juarez, A. (2007). The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 63, 7–14. doi: 10.1111/j.1365-2958.2006.05497.x
Madrid, C., Nieto, J. M., and Juarez, A. (2002). Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med Microbiol. 291, 425–432. doi: 10.1078/1438-4221-00149
Manoil, C. (1990). Analysis of protein localization by use of gene fusions with complementary properties. J. Bacteriol. 172, 1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990
Marceau, M. (2005). Transcriptional regulation in Yersinia: an update. Curr. Issues Mol. Biol. 7, 151–177.
Marimon, O., Teixeira, J. M. C., Cordeiro, T. N., Soo, V. W. C., Wood, T. L., Mayzel, M., et al. (2016). An oxygen-sensitive toxin-antitoxin system. Nat. Commun. 7:13634.
Marra, A., and Isberg, R. R. (1997). Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer’s patch intestinal epithelium. Infect. Immun. 65, 3412–3421. doi: 10.1128/iai.65.8.3412-3421.1997
Mayer, O., Waldsich, C., Grossberger, R., and Schroeder, R. (2002). Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 30, 1175–1180. doi: 10.1042/bst0301175
McFeeters, R. L., Altieri, A. S., Cherry, S., Tropea, J. E., Waugh, D. S., and Byrd, R. A. (2007). The high-precision solution structure of Yersinia modulating protein YmoA provides insight into interaction with H-NS. Biochemistry 46, 13975–13982. doi: 10.1021/bi701210j
Mikulskis, A. V., and Cornelis, G. R. (1994). A new class of proteins regulating gene expression in enterobacteria. Mol. Microbiol. 11, 77–86. doi: 10.1111/j.1365-2958.1994.tb00291.x
Mikulskis, A. V., Delor, I., Thi, V. H., and Cornelis, G. R. (1994). Regulation of the Yersinia enterocolitica enterotoxin Yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol Microbiol 14, 905–915. doi: 10.1111/j.1365-2958.1994.tb01326.x
Miller, J. H. (1992). A Short Course In Bacterial Genetic: A Laboratory Manual And Handbook For Escherichia Coli And Related Bacteria. Cold Spring Habor, New York: Cold Spring Harbor Laboratory Press.
Molofsky, A. B., and Swanson, M. S. (2003). Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50, 445–461. doi: 10.1046/j.1365-2958.2003.03706.x
Monk, I. R., Casey, P. G., Cronin, M., Gahan, C. G., and Hill, C. (2008). Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC; a new site-specific integrative vector. BMC Microbiol. 8:96. doi: 10.1186/1471-2180-8-96
Mourino, M., Madrid, C., Balsalobre, C., Prenafeta, A., Munoa, F., Blanco, J., et al. (1996). The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect. Immun. 64, 2881–2884. doi: 10.1128/iai.64.7.2881-2884.1996
Mulcahy, H., O’Callaghan, J., O’Grady, E. P., Macia, M. D., Borrell, N., Gomez, C., et al. (2008). Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 76, 632–638. doi: 10.1128/iai.01132-07
Nagel, G., Lahrz, A., and Dersch, P. (2001). Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41, 1249–1269. doi: 10.1046/j.1365-2958.2001.02522.x
Nieto, J. M., Madrid, C., Miquelay, E., Parra, J. L., Rodriguez, S., and Juarez, A. (2002). Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184, 629–635. doi: 10.1128/jb.184.3.629-635.2002
Norton, J. P., and Mulvey, M. A. (2012). Toxin-Antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 8:e1002954. doi: 10.1371/journal.ppat.1002954
Nuss, A. M., Beckstette, M., Pimenova, M., Schmühl, C., Opitz, W., Pisano, F., et al. (2017a). Tissue dual RNA-seq: a fast discovery path for infection-specific functions and riboregulators shaping host-pathogen transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 114, E791–E800.
Nuss, A. M., Heroven, A. K., and Dersch, P. (2017b). RNA regulators: formidable modulators of YersinIia virulence. Trends Microbiol. 25, 19–34. doi: 10.1016/j.tim.2016.08.006
Nuss, A. M., Heroven, A. K., Waldmann, B., Reinkensmeier, J., Jarek, M., Beckstette, M., et al. (2015). Transcriptomic profiling of Yersinia pseudotuber-culosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet. 11:e1005087. doi: 10.1371/journal.pgen.1005087
Nuss, A. M., Schuster, F., Kathrin Heroven, A., Heine, W., Pisano, F., and Dersch, P. (2014). A direct link between the global regulator PhoP and the Csr regulon in Y. pseudotuberculosis through the small regulatory RNA CsrC. RNA Biol. 11, 580–593. doi: 10.4161/rna.28676
Pepe, J. C., and Miller, V. L. (1993). Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. U.S.A. 90, 6473–6477. doi: 10.1073/pnas.90.14.6473
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Pourciau, C., Lai, Y.-J., Gorelik, M., Babitzke, P., and Romeo, T. (2020). Diverse mechanisms and circuitry for global regulation by the RNA-Binding protein CsrA. Front. Microbiol. 11:601352. doi: 10.3389/fmicb.2020.601352
Romeo, T., and Babitzke, P. (2018). Global regulation by CsrA and is RNA antagonists. Microbiol. Spectr. 6:2. doi: 10.1128/microbiolspec.RWR-0009-2017
Sahr, T., Bruggemann, H., Jules, M., Lomma, M., Albert-Weissenberger, C., Cazalet, C., et al. (2009). Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72, 741–762. doi: 10.1111/j.1365-2958.2009.06677.x
Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories.
Saxena, S., and Gowrishankar, J. (2011). Compromised factor-dependent transcription termination in a nusA mutant of Escherichia coli: spectrum of termination efficiencies generated by perturbations of Rho, NusG, NusA, and H-NS family proteins. J. Bacteriol. 193, 3842–3850. doi: 10.1128/JB.00221-11
Schmühl, C., Beckstette, M., Heroven, A. K., Bunk, B., Spröer, C., McNally, A., et al. (2019). Comparative transcriptomic profiling of Yersinia enterocolitica O:3 and O:8 reveals major expression differences of fitness- and virulence-relevant genes indicating ecological separation. mSystems 4:e00239–18. doi: 10.1128/mSystems.00239-18
Schneiders, S., Hechard, T., Edgren, T., Avican, K., Fällman, M., Fahlgren, A., et al. (2021). Spatiotemporal variations in growth rate and virulence plasmid copy number during Yersinia pseudotuberculosis infection. Infect. Immun. 89, e710–e720. doi: 10.1128/IAI.00710-20
Schwiesow, L., Lam, H., Dersch, P., and Auerbuch, V. (2015). Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198, 604–614. doi: 10.1128/JB.00686-15
Sharma, V. K., and Zuerner, R. L. (2004). Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186, 7290–7301. doi: 10.1128/jb.186.21.7290-7301.2004
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. doi: 10.2202/1544-6115.1027
Sonnleitner, E., Schuster, M., Sorger-Domenigg, T., Greenberg, E. P., and Blasi, U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x
Sorger-Domenigg, T., Sonnleitner, E., Kaberdin, V. R., and Blasi, U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773. doi: 10.1016/j.bbrc.2006.11.084
Straley, S. C., and Perry, R. D. (1995). Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3, 310–317. doi: 10.1016/s0966-842x(00)88960-x
Suzuki, K., Babitzke, P., Kushner, S. R., and Romeo, T. (2006). Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20, 2605–2617. doi: 10.1101/gad.1461606
Timmermans, J., and Van Melderen, L. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell Mol. Life Sci. 67, 2897–2908. doi: 10.1007/s00018-010-0381-z
Trülzsch, K., Oellerich, M. F., and Heesemann, J. (2007). Invasion and dissemination of Yersinia enterocolitica in the mouse infection model. Adv. Exp. Med. Biol. 603, 279–285. doi: 10.1007/978-0-387-72124-8_25
Viegas, S. C., Pfeiffer, V., Sittka, A., Silva, I. J., Vogel, J., and Arraiano, C. M. (2007). Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35, 7651–7664. doi: 10.1093/nar/gkm916
Volk, M., Vollmer, I., Heroven, A. K., and Dersch, P. (2019). Transcriptional and post-transcriptional regulatory mechanisms controlling type III secretion. Curr. Top. Microbiol. Immunol. 427, 11–33. doi: 10.1007/82_2019_168
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, S., Morozov, I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Yang, F., Ke, Y., Tan, Y., Bi, Y., Shi, Q., Yang, H., et al. (2010). Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS One 5:e12840. doi: 10.1371/journal.pone.0012840
Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J., et al. (2002). Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15.
Keywords: Yersinia, regulatory RNA, gene regulation, virulence, CsrA, YmoA, Hha
Citation: Böhme K, Heroven AK, Lobedann S, Guo Y, Stolle AS and Dersch P (2021) The Small Protein YmoA Controls the Csr System and Adjusts Expression of Virulence-Relevant Traits of Yersinia pseudotuberculosis. Front. Microbiol. 12:706934. doi: 10.3389/fmicb.2021.706934
Received: 08 May 2021; Accepted: 12 July 2021;
Published: 03 August 2021.
Edited by:
Olga Soutourina, UMR 9198 Institut de Biologie Intégrative de la Cellule (I2BC), FranceReviewed by:
Kai Papenfort, Friedrich Schiller University Jena, GermanyCopyright © 2021 Böhme, Heroven, Lobedann, Guo, Stolle and Dersch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Dersch, cGV0cmEuZGVyc2NoQHVuaS1tdWVuc3Rlci5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.