
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 July 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.703993
This article is part of the Research Topic Campylobacter-associated Food Safety View all 32 articles
Campylobacter, a major foodborne pathogen, is susceptible to oxygen. Recently, aerotolerant Campylobacter with enhanced tolerance to aerobic stress has become a major concern in food safety. However, the aerotolerance of Campylobacter coli from pigs has not been studied extensively. Here, we sought to investigate the prevalence of C. coli across multiple swine groups in farms, including weaning, growing, and fattening pigs in production stages and pregnant sows. Additionally, we analyzed C. coli aerotolerance, quinolone resistance, virulence potential, and multilocus sequence typing (MLST) genotypes. Finally, we compared the characteristics of C. coli according to the aerotolerance levels. In total, we obtained 124 (66.3%) C. coli isolates from 187 swine fecal samples across six swine farms. The pathogen was prevalent in weaning (45.5%), growing (68.3%), and fattening (75.4%) pigs, and pregnant sows (66.7%). Hyper-aerotolerant HAT C. coli (13.7% of 124 isolates) was present in all swine groups, with the highest proportion in the pregnant sows (27.3%). All HAT isolates possessed diverse virulence-related genes such as flaA, cadF, pldA, ceuE, and cdtA. All C. coli isolates were resistant to quinolones, and 12 (10%) presented high-level ciprofloxacin resistance (MIC ≥ 32 μg/mL). The proportion of C. coli isolates with a high-level ciprofloxacin resistance was the highest in HAT C. coli (18.8%). Furthermore, six MLST sequence types (STs) (ST827, ST830, ST854, ST1016, ST1068, and ST1096) of swine-derived C. coli were in common with human-derived C. coli (PubMLST). The proportion of C. coli belonging to such shared STs at each aerotolerance level was the highest in HAT C. coli (HAT vs. oxygen-sensitive; OR = 3.13). In conclusion, quinolone resistance of C. coli may be distributed throughout in all swine groups in farms. HAT C. coli is likely to remain in pig farms and re-infect other pigs in the farms. Furthermore, swine-derived HAT C. coli could be transmitted to humans easily through the food chain owing to its aerotolerance, and it could pose a threat to public health owing to its high-level ciprofloxacin resistance and virulence. This study highlights the need to develop management practices that prevent the transmission of swine-derived HAT C. coli to humans.
Campylobacter, a major foodborne pathogen, causes gastrointestinal infections in humans, with symptoms such as diarrhea, fever, abdominal pain, vomiting, and prostration (Kaakoush et al., 2015; O’Kane and Connerton, 2017; Riedel et al., 2020). This bacterium harbors diverse virulence-related genes responsible for its motility, cell adhesion, colonization, invasion, iron uptake, cytotoxin production, secretion, and Guillain–Barré syndrome expression (Bacon et al., 2000; Ziprin et al., 2001; Datta et al., 2003; Bolton, 2015; Corcionivoschi et al., 2015; Koolman et al., 2015). While campylobacteriosis with gastrointestinal infections is usually self-limiting, it may also lead to severe illnesses including bacteremia, meningitis, irritable bowel syndrome, Guillain–Barré syndrome, or arthritis (Azrad et al., 2018; Centers for Disease Control and Prevention, 2019). Antibiotics are prescribed for clinical treatment of severe Campylobacter infections in humans (Tang et al., 2017; Centers for Disease Control and Prevention, 2019), and fluoroquinolones are the most frequently used (Sato et al., 2004; Sifré et al., 2015; Tang et al., 2020; Hull et al., 2021). However, an increasing trend of Campylobacter strains with fluoroquinolone resistance has become a major concern (Engberg et al., 2001; Moore et al., 2006; Sahin et al., 2015; Bolinger and Kathariou, 2017; Sproston et al., 2018).
Campylobacter is an obligate microaerophile and capnophile, optimally growing at low oxygen (5–10%) and high carbon dioxide (5–10%) concentrations (Bronowski et al., 2014). This bacterium is generally highly susceptible to atmospheric oxygen (Kaakoush et al., 2007). However, recently, the high prevalence of aerotolerant (AT) and hyper-aerotolerant (HAT) Campylobacter jejuni strains with enhanced tolerance to aerobic stress has been identified in retail chickens. Moreover, these strains cluster mostly into a few major multilocus sequence typing (MLST) clonal complexes that are often involved in human Campylobacter infections (Oh et al., 2015). In addition, most human clinical C. jejuni strains were reported to be HAT, with tolerance to multiple stress, including disinfectant, freeze-thaw, and heat treatments (Oh et al., 2018). In a previous study, we found HAT Campylobacter coli to be prevalent in duck carcasses and meat (Guk et al., 2019). Based on MLST sequence types (STs), most of the HAT C. coli strains from ducks are genetically related to C. coli isolates from humans (Guk et al., 2019). These findings suggest that the aerotolerance of Campylobacter is likely related with infections in humans. For these reasons, the aerotolerance of Campylobacter has become a major concern.
Pigs only become sub-clinically infected with Campylobacter spp., especially C. coli, making them a potential source of Campylobacter infections in humans through the consumption of contaminated pork meat (Mataragas et al., 2008; Kempf et al., 2017; Di Donato et al., 2020; Riedel et al., 2020). In addition, contamination of carcasses or meat with swine feces during processing or slaughtering can pose risks to food safety (Abley et al., 2012; Nisar et al., 2018; Di Donato et al., 2020). Considering these findings and the possibility that aerotolerance may be involved in human Campylobacter infections, studies on the aerotolerance of swine-derived C. coli are needed.
The aims of this study were: (1) to investigate the prevalence of C. coli in pigs in different swine groups, including pigs in the production stages (weaning, growing, and fattening stage) and pregnant sows, at pig farms; (2) to analyze the characteristics, such as aerotolerance, quinolone resistance, virulence, and genetic relatedness, of C. coli in different swine groups; and (3) to compare its quinolone resistance, virulence, and genetic relatedness according to the aerotolerance levels. This study is the first to elucidate the different characteristics of C. coli across swine groups in pig farms and according to the aerotolerance levels.
A total of 187 fresh swine fecal samples were collected from six swine farms from March 2018 to January 2019. The number of samples at each farm ranged from 26 to 34, and the number of samples from each swine group was as follows: 5–6 in the weaning pigs (4–12 weeks old), 8–11 in the growing pigs (12–20 weeks old), 8–11 in the fattening pigs (21–30 weeks old), and 5–6 in the pregnant sows. The number of swine fecal samples according to the swine groups in each farm is shown in Supplementary Table 1. Swine fecal samples were collected from healthy, randomly selected pigs, and the number of fecal samples collected for each swine group was determined considering the number of pigs per swine group in the farms. Each fecal sample was collected using a sterilized cotton swab, stored in a sterilized tube, and then transported from the swine farms to our laboratory on the same day. In addition, enrichment of the samples was performed to isolate C. coli immediately after the samples were transported. Except for the pregnant sows, which resided on the farms for breeding, most pigs were raised in these swine farms until the fattening stage and then shipped to slaughterhouses. The samples were classified into four different swine groups, including weaning pigs (n = 33), growing pigs (n = 60), and fattening pigs (n = 61) in the production stages and pregnant sows (n = 33). The samples were enriched in Preston broth (Oxoid Ltd., Basingstoke, United Kingdom) at 42°C for 24 h in microaerobic conditions (6% O2, 7.1% CO2, 3.6% H2, and 83.3% N2). Then, aliquots (100 μL) of the enriched broth were spread and streaked on modified charcoal-cefoperazone-deoxycholate agar plates and incubated at 42°C for 24 h in microaerobic conditions. Following incubation, 3–5 presumptive Campylobacter colonies per plate were selected, and DNA was extracted. C. coli was identified using polymerase chain reaction (PCR) with primers targeting Campylobacter 16S rDNA and ask genes (Supplementary Table 2).
Aerotolerance levels of the C. coli isolates were investigated following previous methods (Oh et al., 2015; Guk et al., 2019). The isolates were cultured on Mueller-Hinton (MH) agar at 42°C for 24 h in microaerobic conditions and resuspended in fresh MH broth to a McFarland scale of 1.0 (3 × 108 CFU/mL). Then, the bacterial suspension was cultured aerobically at 42°C with shaking at 200 rpm. Serial dilutions of the suspension were prepared, and inoculation on MH agar was performed at 0, 12, and 24 h after exposure to aerobic shaking. The inoculation concentrations of the suspension ranged from 100 to 107 dilutions, and 5 μL of each dilution was inoculated on the plates. C. coli isolates that lost their viability within 12 h of aerobic shaking were determined to be oxygen-sensitive (OS) strains, whereas the isolates that maintained their viability for 12–24 h were determined as AT strains. Those that remained viable for more than 24 h were considered HAT strains. Experiments were conducted in triplicate.
The quinolone resistance of C. coli isolates was tested against nalidixic acid (NAL) and ciprofloxacin (CIP). Minimum inhibitory concentration (MIC) values of the two antibiotics were determined using the broth dilution method with a Sensititre CAMPY2 plate (TREK Diagnostic Systems, Cleveland, OH, United States) and cation-adjusted Müller Hinton broth with TES (CAMHBT, TREK Diagnostic Systems). The NAL and CIP in the plate were diluted twofold with a range of 4–64 and 0.015–64 μg/mL, respectively. C. coli isolates were cultured on MH agar at 42°C for 24 h in microaerobic conditions, and then suspended in the cation-adjusted MH broth with TES to a McFarland standard of 0.5 (1.5 × 108 CFU/mL). The suspension (100 μL) was added to a mixture of cation-adjusted MH broth with TES and 5% laked horse blood and mixed well. Then, 100 μL of the new suspension was inoculated into each well of the CAMPY2 plate containing different concentrations of the quinolone antibiotics. After the inoculation, the plates were cultured at 42°C for 24 h in microaerobic conditions. Resistance to the antibiotics was determined according to the National Antimicrobial Resistance Monitoring System (NARMS, 2019). Furthermore, high-level resistance to NAL (MIC ≥ 64 μg/mL) and CIP (MIC ≥ 32 μg/mL) was investigated by previously published methods (Segreti et al., 1992; Engberg et al., 2001). In this experiment, C. jejuni ATCC 33560 was used for quality control.
The prevalence of nine virulence-related genes in C. coli isolates was investigated using PCR. The primers and PCR conditions used in this study are presented in Supplementary Table 2. The virulence-related genes selected in this study were as follows: flaA, involved in motility; cadF, in cell adhesion; pldA, in colonization; iamA, in invasion; ceuE, in iron uptake system; cdtA, in cytotoxin production; wlaN, in expression of Guillain–Barré syndrome; hcp, in the type VI secretion system (T6SS); and virB11, in the type IV secretion system (T4SS) (Bacon et al., 2000; Ziprin et al., 2001; Datta et al., 2003; Bolton, 2015; Corcionivoschi et al., 2015; Koolman et al., 2015).
The MLST genotypes of the C. coli isolates were determined according to the PubMLST protocol1. In brief, the sequences of the housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) were submitted to PubMLST, and an allele was assigned for each housekeeping gene. MLST STs were defined by using the allelic profiles of the seven housekeeping genes.
Cluster analysis of C. coli isolates by MLST STs was performed using the minimum spanning tree (MST) method based on the allelic profiles of the housekeeping genes with BioNumerics version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium). A dendrogram was generated using the unweighted-pair group method with arithmetic mean (UPGMA) using the BioNumerics software, based on the allelic profiles of the MLST housekeeping genes for characteristics, including aerotolerance and virulence potential, of the C. coli isolates belonging to each MLST ST.
MLST data of human C. coli isolates were obtained from the PubMLST database and used to analyze the genetic relatedness between C. coli from swine and humans.
MLST data of 3,545 human C. coli isolates belonging to 809 MLST STs were obtained from PubMLST (accessed on 27 January 2021). Of these, 43 MLST STs with more than 10 C. coli isolates per ST were selected as representative MLST genotypes of human-derived C. coli isolates (PubMLST). The proportion of swine-derived C. coli isolates that shared MLST genotypes with human-derived C. coli was calculated and compared according to the four groups of swine and to the isolates’ aerotolerance levels.
Based on the allelic profiles of the MLST housekeeping genes, cluster analysis of C. coli isolates from swine and humans was conducted in BioNumerics version 6.6, following the MST method to illustrate the genetic relatedness of C. coli from swine and humans.
The prevalence of C. coli and the distribution of aerotolerance levels in C. coli were calculated with 95% confidence interval (95% CI) considering swine farms as a hierarchical level. In addition, a Generalized linear mixed-effects regression model (GLMM) with the swine farms as a random effect was applied to determine whether the prevalence of C. coli differed according to the swine groups using the lme4 package in the RStudio software version 1.4.1103 (Bates et al., 2014; RStudio Team, 2021). Using the same method, genetic relatedness between swine-derived C. coli and human C. coli isolates (PubMLST) was analyzed according to the swine groups and the aerotolerance levels based on MLST genotypes. Additionally, the same statistical method was used to verify whether there was a difference in characteristics, including the proportion of high-level CIP resistance and virulence-related genes, between the swine groups or between the aerotolerance levels. In the GLMM with the swine farms as a random effect, the p-value of 0.1 was used as the significance level.
A total of 124 (66.3%, 95% CI 52.9–80.8%) C. coli isolates were obtained from 187 swine fecal samples, and the prevalence of C. coli in each swine group was: 45.5% (15/33, 95% CI 22.4–67.6%) in the weaning pigs, 68.3% (41/60, 95% CI 51.2–88.3%) in the growing pigs, 75.4% (46/61, 95% CI 60.5–90.8%) in the fattening pigs, and 66.7% (22/33, 95% CI 49.9–85.6%) in the pregnant sows, showing that C. coli was more predominant in the fattening pigs than in the other groups. However, C. coli prevalence was not statistically different between the swine groups. The prevalence of C. coli according to the swine groups at each swine farm is in Supplementary Table 1.
We categorized the isolates according to the aerotolerance levels and determined that approximately 58.1% (n = 72), 28.2% (n = 35), and 13.7% (n = 17) of the 124 C. coli isolates were OS, AT, and HAT, respectively. Information on the aerotolerance levels of C. coli isolated at each swine group is presented in Table 1. As shown, the proportion of HAT C. coli was the highest in the pregnant sows (27.3%, 6/22) and the lowest in the weaning pigs (6.7%, 1/15). Information on the distribution of aerotolerance levels in C. coli isolates from swine feces at farm levels is provided in Supplementary Table 3.
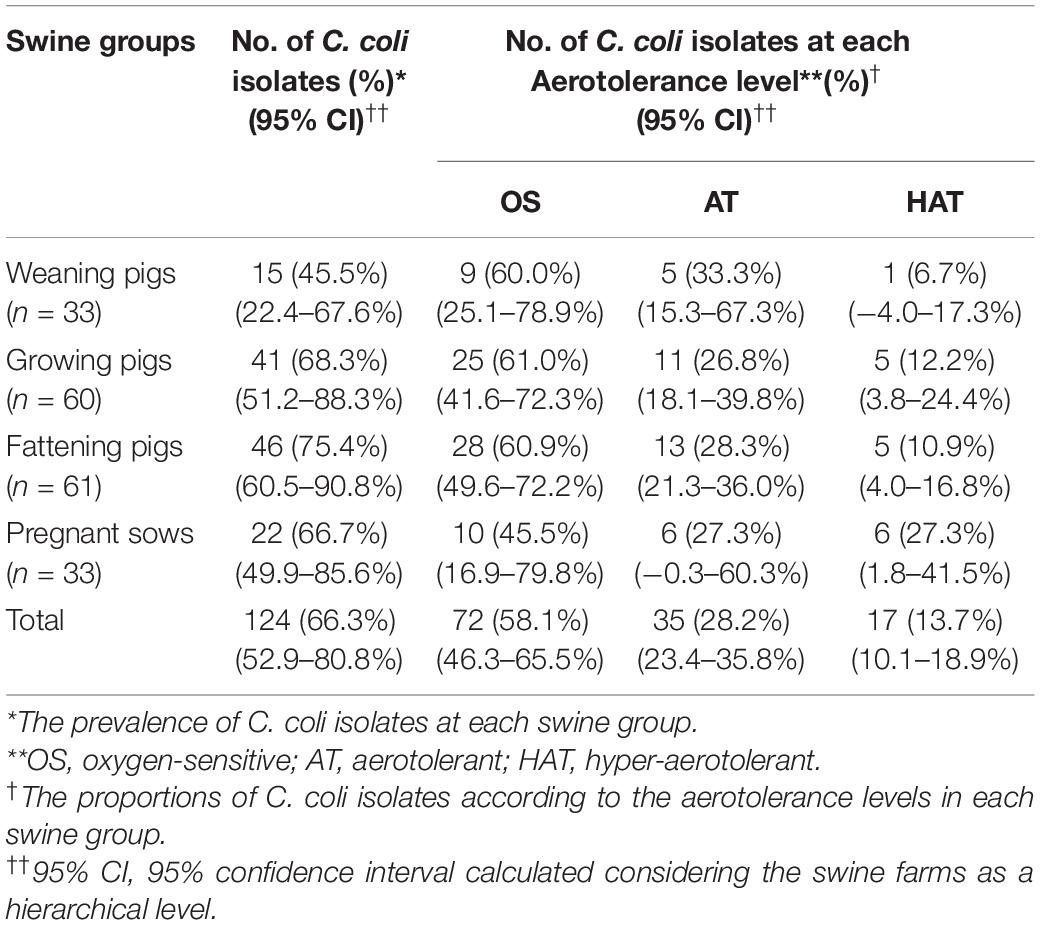
Table 1. Prevalence of Campylobacter coli isolates according to the aerotolerance levels in each swine group.
Resistance of the C. coli isolates to quinolone antibiotics, including NAL and CIP, was investigated using the broth dilution method. Among the 124 C. coli isolates, four did not grow in the MIC tests, whereas the remaining 120 isolates exhibited resistance to both antibiotics, irrespective of the swine groups and aerotolerance levels (Supplementary Figure 1). In addition, 12 (10%) and 118 (98.3%) C. coli isolates showed high-level resistance to CIP (MIC ≥ 32 μg/mL) and NAL (MIC ≥ 64 μg/mL), respectively (Table 2). The proportion of C. coli isolates showing high-level CIP resistance at each group was: 21.4% (3/14) in the weaning pigs, 12.8% (5/39) in the growing pigs, 8.9% (4/45) in the fattening pigs, and 0% (0/22) in the pregnant sows. In terms of aerotolerance levels, the proportion of C. coli isolates with high-level CIP resistance was the highest in HAT C. coli (18.8%, 3/16), followed by AT (8.8%, 3/34) and OS (8.6%, 6/70) C. coli. Statistical differences were not found in the proportion of C. coli isolates showing high-level CIP resistance according to the swine groups and according to the aerotolerance levels.
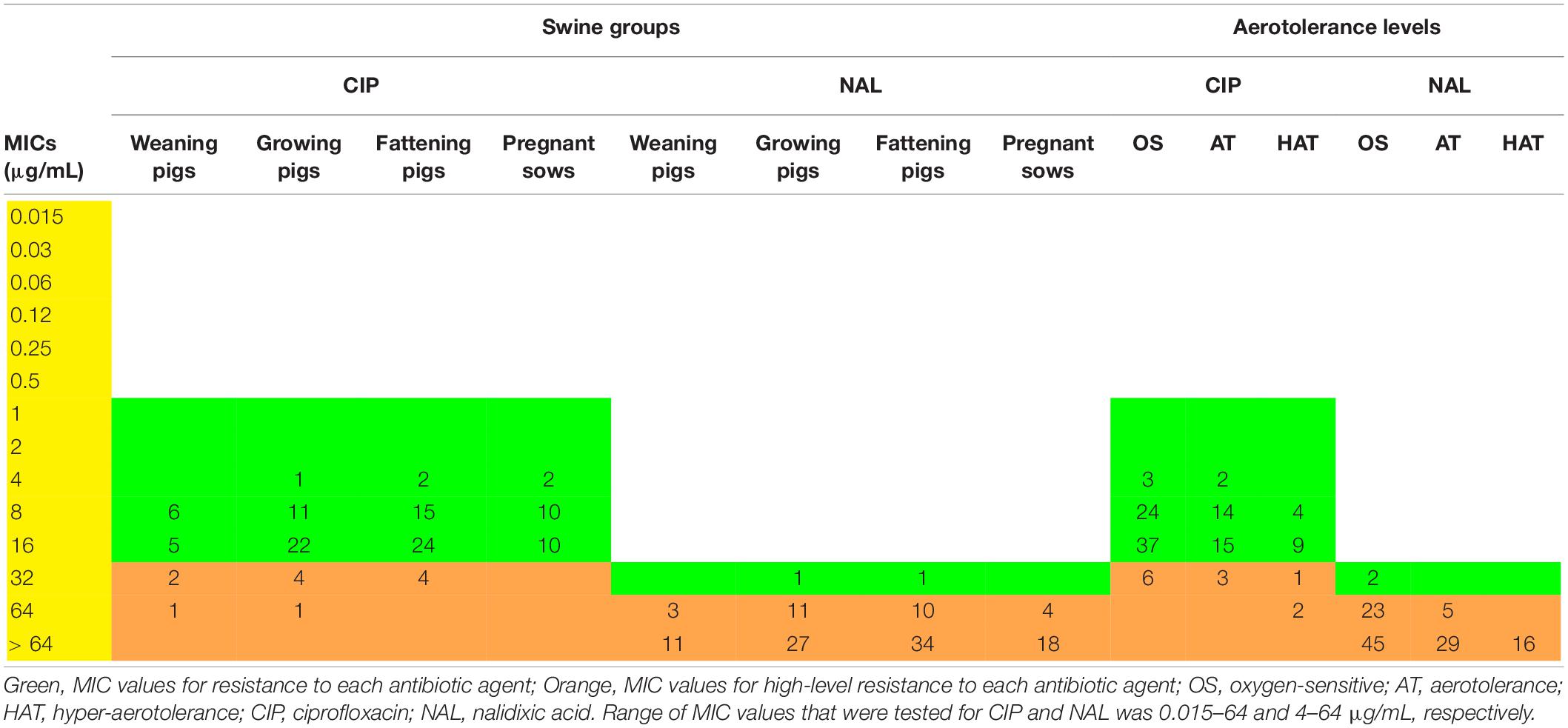
Table 2. Distribution of minimum inhibitory concentration (MIC) values (μg/mL) of Campylobacter coli isolates according to the swine groups and aerotolerance levels.
The virulence potential of C. coli isolates was investigated using nine major virulence-related genes. Of these, flaA (93.5%, 116/124), cadF (99.2%, 123/124), pldA (97.6%, 121/124), ceuE (93.5%, 116/124), and cdtA (100%, 124/124) genes were predominant (Table 3). The prevalence of virulence-related genes according to the swine groups was generally similar, except for some genes. However, for the iamA gene, significant differences in its prevalence according to the swine groups were identified (GLMM, p = 0.035, variance of the random effect = 0.432). The prevalence of the iamA gene was significantly higher in the fattening pigs than in the weaning pigs (fattening pigs vs. weaning pigs; GLMM, OR = 3.96; 95% CI 0.92–17.03, p = 0.064). In addition, the prevalence of the ceuE gene was different according to the swine groups (GLMM, p = 0.056, variance of the random effect = 0.855). The ceuE gene was more prevalent in the growing pigs than in the weaning pigs (growing pigs vs. weaning pigs; GLMM, OR = 9.49; 95% CI 0.84–107.33, p = 0.069). The hcp gene showed a difference in its distribution according to the swine groups (GLMM, p = 0.012, variance of the random effect = 0.915), and its prevalence was significantly higher in the pregnant sows than in the weaning pigs (pregnant sows vs. weaning pigs; GLMM, OR = 7.72; 95% CI 1.24–47.94, p = 0.028). In addition, the prevalence of virulence-related genes was generally similar according to the aerotolerance levels of C. coli. However, the prevalence of virulence-related genes, except for the hcp gene, was the highest in HAT C. coli. All HAT C. coli isolates possessed diverse virulence-related genes including flaA, cadF, pldA, ceuE, and cdtA genes.
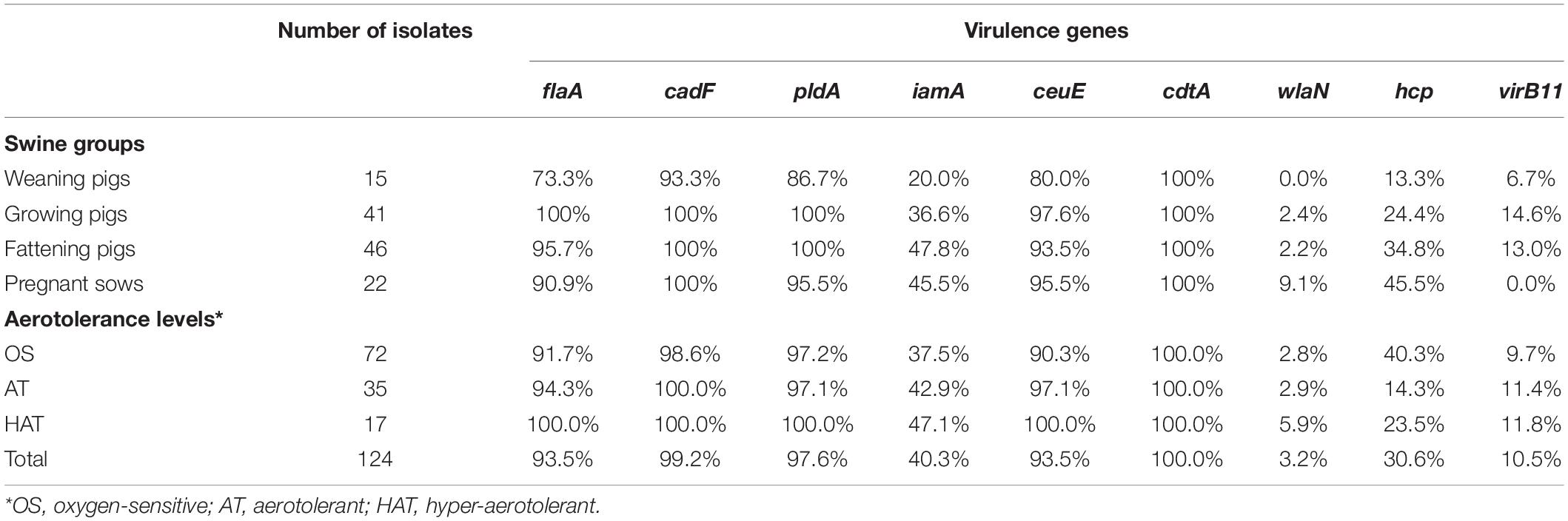
Table 3. Prevalence of virulence genes of Campylobacter coli isolates from swine sources according to the swine groups and aerotolerance levels.
From the 124 swine-derived C. coli isolates, 28 MLST STs were identified. Generally, all isolates were closely clustered, regardless of the swine groups and aerotolerance levels (Figure 1). Most (87.9%, 109/124) C. coli strains belonged to the same MLST clonal complex (CC-828), except for C. coli, whose MLST CCs had not been determined yet. In addition, three main MLST genotypes (ST827, ST854, ST887) that covered all swine groups were identified in 32 C. coli isolates (25.8%) (Figure 1A). The distribution of MLST genotypes in C. coli isolates from swine feces at farm levels is provided in Supplementary Table 4. The UPGMA dendrogram showed that most of the isolates possessed various virulence-related genes, regardless of the MLST genotype (Figure 2). Information on the MLST genotype of each C. coli isolate is listed in Supplementary Table 5.
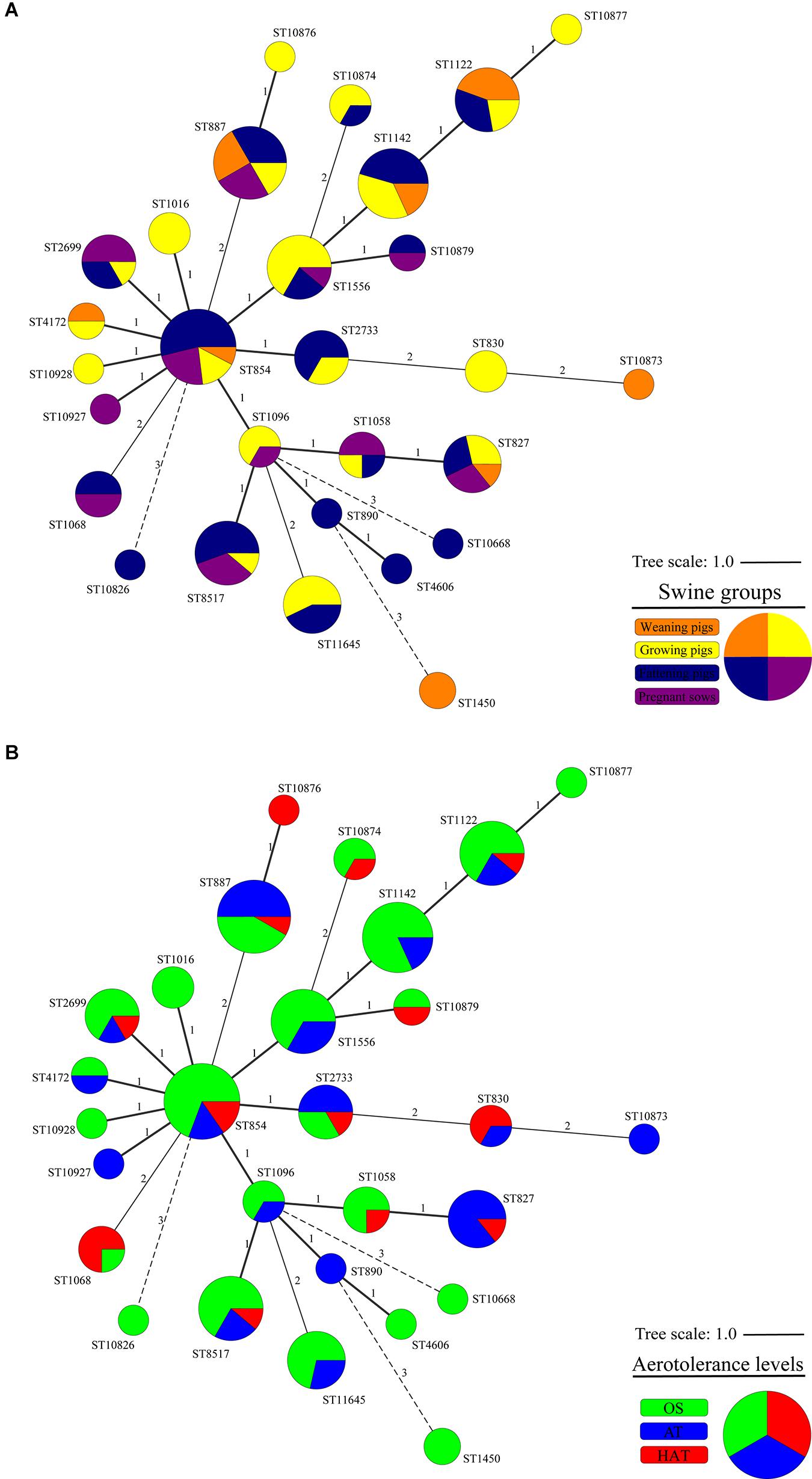
Figure 1. Cluster analysis of 124 Campylobacter coli isolates by multilocus sequence typing (MLST) sequence types (STs) according to (A) the swine groups and (B) the aerotolerance levels of C. coli isolates. A minimum spanning tree (MST) was constructed based on the allelic profiles of MLST housekeeping genes for 124 C. coli isolates. Each node represents the MLST ST, and the size of each node indicates the number of isolates belonging to each MLST ST. Colors indicate (A) the swine groups from which C. coli isolates were isolated (orange, weaning pigs; yellow, growing pigs; navy, fattening pigs; purple, pregnant sows) and (B) aerotolerance levels of C. coli (green, oxygen-sensitive, OS; blue, aerotolerant, AT; and red, hyper-aerotolerant, HAT). The numbers on the branches present the degree of difference in allelic profiles for housekeeping genes.
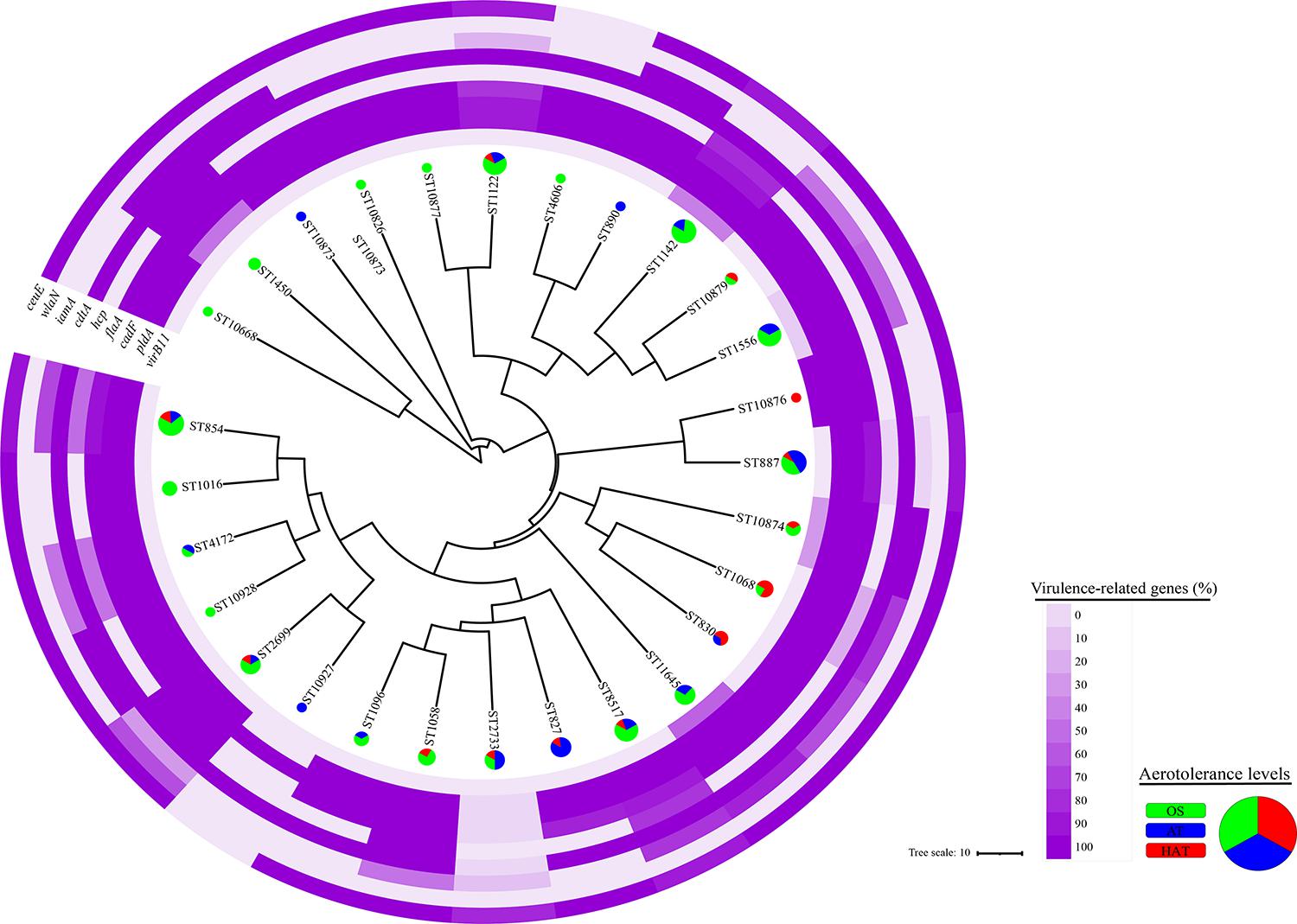
Figure 2. Clonal distribution of 124 Campylobacter coli isolates based on the allelic profiles for the multilocus sequence typing (MLST) housekeeping genes. An unweighted-pair group method with arithmetic mean (UPGMA) dendrogram was constructed based on the allelic profiles of the MLST housekeeping genes for 124 C. coli isolates. The UPGMA dendrogram shows the distribution of the aerotolerance levels and proportions of nine virulence genes of C. coli isolates in each MLST sequence type. Each color in the pie chart indicates an aerotolerance level (green, oxygen-sensitive, OS; blue, aerotolerant, AT; and red, hyper-aerotolerant, HAT) of C. coli isolates.
Forty-three MLST STs, among the 809 MLST genotypes obtained from the PubMLST database, containing 2,314 C. coli isolates, were selected as representative genotypes of human C. coli isolates and used in the analysis of genetic relatedness with swine-derived C. coli isolates. Six of the 28 MLST STs from swine (ST827, ST830, ST854, ST1016, ST1068, ST1096) were shared with humans (Figure 3), including approximately 26.6% (33/124) and 35.6% (823/2314) of C. coli isolates from swine and humans, respectively.
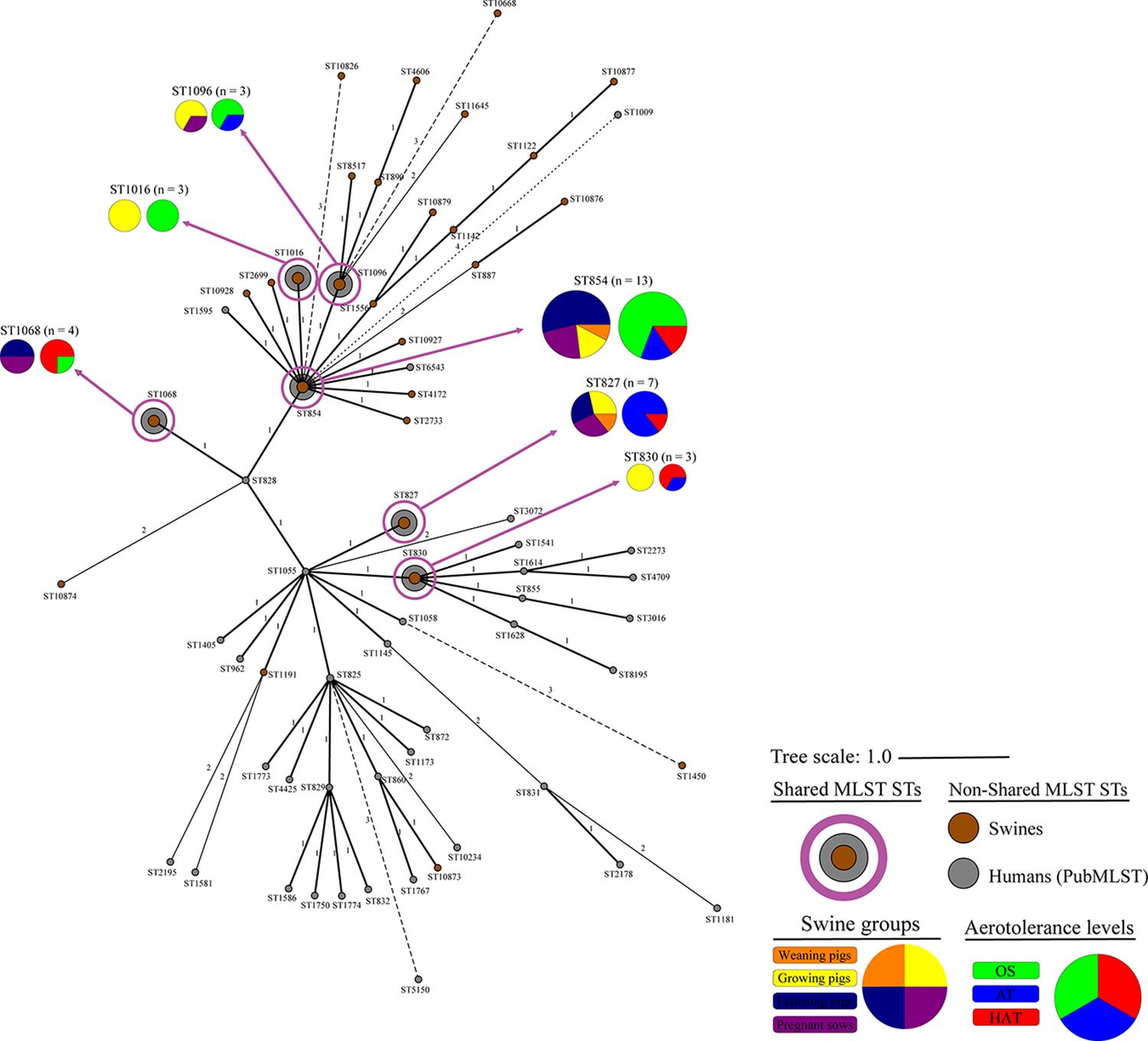
Figure 3. Genetic relatedness of swine-derived Campylobacter coli isolates in this study with human C. coli isolates registered in the PubMLST. Cluster analysis of C. coli isolates from swine and humans was conducted using the MST method, based on the allelic profiles of the MLST housekeeping genes. Colors indicate the sources (brown, swine from this study; and gray, humans from PubMLST), the swine groups (orange, weaning pigs; yellow, growing pigs; navy, fattening pigs; and purple, pregnant sows), and aerotolerance levels of C. coli isolates (green, oxygen-sensitive, OS; blue, aerotolerant, AT; and red, hyper-aerotolerant, HAT). The numbers on the branches present the difference degree in allelic profiles for housekeeping genes.
The proportion of C. coli belonging to the six shared STs at each swine group was the highest in the pregnant sows (36.4%, 8/22) (Table 4). In addition, the proportion at each aerotolerance level was the highest in HAT C. coli (47.1%, 8/17), followed by AT (28.6%, 10/35) and OS C. coli (20.8%, 15/72) (Table 4). The genetic relatedness between swine-derived C. coli and human C. coli isolates according to the swine groups was statistically different between the swine groups (GLMM, p = 0.014, variance of the random effect = 0.605). The proportion of C. coli in the shared STs was significantly higher in the pregnant sows than in the weaning pigs (pregnant sows vs. weaning pigs; GLMM, OR = 5.55; 95% CI 0.85–36.39, p = 0.074). Furthermore, the genetic relatedness between swine-derived C. coli and human C. coli isolates according to the aerotolerance levels was significantly different (GLMM, p = 0.001, variance of the random effect = 0.343). The proportion of C. coli in the shared STs in HAT C. coli was significantly higher than in OS C. coli (HAT vs. OS; GLMM, OR = 3.13; 95% CI 0.99–9.96, p = 0.053) (Table 4).
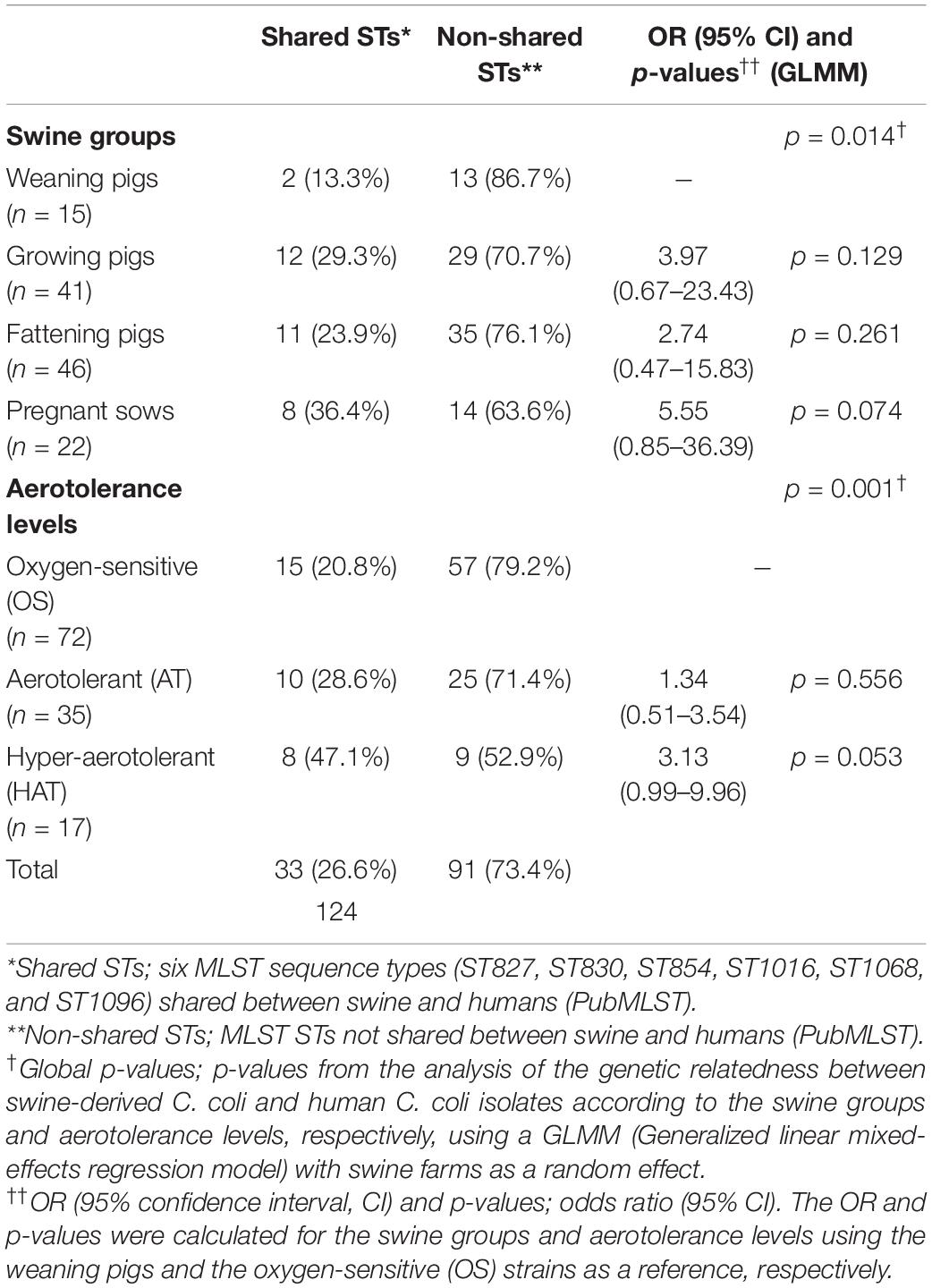
Table 4. Campylobacter coli in the shared multilocus sequence typing (MLST) sequence types (STs) with human C. coli isolates (PubMLST) according to the swine groups and aerotolerance levels of C. coli.
The aerotolerance of Campylobacter has been particularly concerning recently because aerotolerant strains are prevalent in poultry, and these strains are closely related to human Campylobacter infections (Oh et al., 2015; Karki et al., 2018; Guk et al., 2019; Song et al., 2020). However, the aerotolerance of C. coli isolated from pigs, another important host of C. coli, has not been studied yet (Kempf et al., 2017; Riedel et al., 2020). In this study, we investigated the prevalence of C. coli in six pig farms according to four swine groups including pigs in three production stages (weaning, growing, fattening) and pregnant sows. Campylobacter coli characteristics, including aerotolerance, quinolone resistance, virulence, and MLST genotype, were compared for each swine group. In addition, we evaluated the potential impacts of swine-derived HAT C. coli on food safety by analyzing quinolone resistance, virulence potential, and genetic relatedness of C. coli according to aerotolerance levels. This is the first study to compare the prevalence and characteristics of swine-derived C. coli according to the swine groups and aerotolerance levels.
We identified that the majority of swine fecal samples were positive for C. coli (66.3%, 124/187, 95% CI 52.9–80.8%). The prevalence of C. coli in pigs in this study was higher than that in other livestock, including cattle (7.4–15.0%) and poultry, such as chickens (26.4–40.2%) and ducks (46.6–57.9%) (Bae et al., 2005; Kang et al., 2006; Little et al., 2008; Torralbo et al., 2015; Tang et al., 2017; Guk et al., 2019; Karama et al., 2020). This was similar to the results of a previous study reporting a high prevalence (56–77%) of C. coli in swine colon samples (Kempf et al., 2017), indicating that pigs are a major reservoir of C. coli. The prevalence of C. coli according to the swine groups was the highest in the fattening pigs (75.4%). Pigs are grown in swine farms until the fattening stage and then shipped to slaughterhouses. Therefore, this high prevalence of C. coli in the fattening pigs suggests that C. coli might be spread out of the farms when fattening pigs are sent to slaughterhouses. In addition, it suggests the possibility of C. coli transmissions to humans during the farm-to-table process.
HAT C. coli was less prevalent (13.7%) in pigs compared with OS and AT C. coli. This finding was contrary to previous studies reporting high proportions (49.1–50%) of HAT C. coli in poultry, including chicken livers and duck sources (Karki et al., 2018; Guk et al., 2019). However, we identified that the proportion of HAT C. coli was the highest in pregnant sows. This suggests that HAT C. coli is likely to remain in pig farms and re-infect other pigs in the same farms through fecal-to-oral transmission, considering that pregnant sows reside in the pig farms. Further studies on transmission dynamics of HAT C. coli in pig farms are required to confirm if re-infection in these farms is plausible.
Quinolone resistance in Campylobacter and the incidence of Campylobacter infections in humans have been dramatically increasing worldwide (Engberg et al., 2001; Moore et al., 2006; Alfredson and Korolik, 2007; Luangtongkum et al., 2009). Indeed, the occurrence of resistance to CIP in Campylobacter from humans was in a range from high to extremely high according to a previous report by the European Food Safety Authority and European Centre for Disease Prevention and Control (2021). Moreover, Campylobacter was listed as one of the high-priority antimicrobial-resistant pathogens by World Health Organization owing to the increase of fluoroquinolone resistance, and fluoroquinolone-resistant Campylobacter was identified as a serious public health threat by the Centers for Disease Control and Prevention (World Health Organization, 2017; Centers for Disease Control and Prevention, 2019). In this study, all strains, except for four C. coli strains that did not grow in the MIC tests, showed resistance to both CIP and NAL, suggesting that resistance to quinolones may be distributed in C. coli in swine throughout the swine groups, irrespective of the aerotolerance levels. Furthermore, the proportion of high-level CIP-resistant C. coli was the highest in HAT C. coli (18.8%, 3/16). These findings indicate that quinolone-resistant C. coli could be transmitted to humans from swine, potentially making it difficult to treat human Campylobacter infections because quinolones are used in campylobacteriosis therapy (Sifré et al., 2015; Tang et al., 2017; Guk et al., 2019). Moreover, it could be even more difficult to treat patients infected with HAT C. coli strains because these strains tend to be highly resistant to fluoroquinolones such as CIP.
Most virulence-related genes in this study were distributed similarly in swine-derived C. coli strains, irrespective of the swine groups and the aerotolerance levels. Additionally, all HAT C. coli strains possessed various major virulence-related genes which are associated with motility, cell adhesion, colonization, iron uptake, and toxin production (Ziprin et al., 2001; Bolton, 2015). This suggests that the HAT C. coli from pigs might be more pathogenic than other C. coli strains owing to their high-level CIP resistance and virulence. This finding is similar to that of our previous study that reported a high prevalence (75–96.4%) of diverse virulence-related genes in HAT C. coli strains from ducks (Guk et al., 2019). Similarly, there was no significant difference in the proportions of virulence-related genes between HAT strains and other strains, as seen in C. coli isolated from ducks (Guk et al., 2019). However, a previous study on C. jejuni from chickens reported that the prevalence of several virulence-related genes was significantly higher in HAT C. jejuni than in OS C. jejuni (Oh et al., 2017). This could be attributed to the species differences between C. coli and C. jejuni or to differences in adaptation to hosts. However, further studies on interspecies differences in Campylobacter species or their adaptations in host environments are needed.
All C. coli strains were closely clustered regardless of the swine groups and the aerotolerance levels. Of the 32 strains belonging to the three major MLST STs (ST827, ST854, and ST887) that covered all swine groups, most (62.5%) C. coli belonged to ST827 and ST854, which were identified as major STs in the human isolates (PubMLST) and accounted for 32.5% of human C. coli isolates. These findings indicate that C. coli belonging to certain MLST genotypes may circulate throughout all swine groups in pig farms by horizontal or vertical transmission. Moreover, these results imply the possibility that C. coli—which may be genetically related to human C. coli isolates—is likely to be present in pig farms, indicating the possible transmission of C. coli from swine to humans. Furthermore, HAT C. coli strains from swine constituted a considerable proportion of the shared STs (ST827, ST830, ST854, ST1016, ST1068, and ST1096) with human C. coli isolates compared with OS C. coli strains, indicating a potential high genetic relatedness between swine-derived HAT C. coli and human C. coli isolates. This was consistent with the results of our previous study on aerotolerance of C. coli in ducks; we found that HAT C. coli from ducks accounted for a higher proportion of the shared MLST genotypes with the human C. coli isolates (PubMLST) compared with OS C. coli (Guk et al., 2019). Taken together, it can be speculated that the aerotolerance of C. coli might affect Campylobacter infections in humans, and that HAT C. coli might be transmitted to humans through the food chain. Our speculation is supported by a previous study, which revealed that most C. jejuni strains from human clinical cases were HAT (Oh et al., 2018).
In conclusion, resistance to quinolones may be distributed in our C. coli isolates in swine throughout all swine groups. In addition, HAT C. coli was predominant in the pregnant sows, suggesting that HAT C. coli is likely to remain in pig farms and might re-infect other pigs in the same farms. All HAT C. coli strains, including some of them showing high-level CIP resistance, showed quinolone resistance and harbored various virulence-related genes associated with motility, cell adhesion, colonization, iron uptake, and toxin production. These findings imply that swine-derived HAT C. coli could be a potential threat to public health owing to their high-level quinolone resistance and virulence. Furthermore, HAT C. coli from swine constituted a highest proportion of the shared MLST genotypes with human isolates (PubMLST), indicating that HAT C. coli might be transmitted to humans through the food chain given its aerotolerance. Further studies are needed to elucidate the mechanisms underlying the transmission of swine-derived HAT C. coli to humans through the farm-to-table process. This study highlights the need to improve management practices to diminish the transmission of swine-derived HAT C. coli to humans.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
SC and W-HK conceived and designed the study. HS, SY, J-UA, SL, and J-HG performed the sampling and experiments. J-HG analyzed the data and made a great contribution to the experiments, data analysis, and preparing the manuscript. SC, W-HK, and J-HG prepared and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Research Foundation of Korea (NRF-2021R1A2C2005907).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.703993/full#supplementary-material
Abley, M. J., Wittum, T. E., Moeller, S. J., Zerby, H. N., and Funk, J. A. (2012). Quantification of campylobacter in swine before, during, and after the slaughter process. J. Food Prot. 75, 139–143. doi: 10.4315/0362-028x.Jfp-11-334
Alfredson, D. A., and Korolik, V. (2007). Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277, 123–132. doi: 10.1111/j.1574-6968.2007.00935.x
Azrad, M., Tkhawkho, L., Isakovich, N., Nitzan, O., and Peretz, A. (2018). Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli: comparison between Etest and a broth dilution method. Ann. Clin. Microbiol. Antimicrob. 17:23. doi: 10.1186/s12941-018-0275-8
Bacon, D. J., Alm, R. A., Burr, D. H., Hu, L., Kopecko, D. J., Ewing, C. P., et al. (2000). Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68, 4384–4390.
Bae, W., Kaya, K. N., Hancock, D. D., Call, D. R., Park, Y. H., and Besser, T. E. (2005). Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington state. Appl. Environ. Microbiol. 71, 169–174. doi: 10.1128/aem.71.1.169-174.2005
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2014). Fitting linear mixed-effects models using lme4. arXiv [preprint] arXiv:1406.5823,
Bolinger, H., and Kathariou, S. (2017). The current state of macrolide resistance in Campylobacter spp.: trends and impacts of resistance mechanisms. Appl. Environ. Microbiol. 83:e00416-17. doi: 10.1128/aem.00416-17
Bolton, D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. doi: 10.1016/j.fm.2014.11.017
Bronowski, C., James, C. E., and Winstanley, C. (2014). Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 356, 8–19. doi: 10.1111/1574-6968.12488
Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC.
Corcionivoschi, N., Gundogdu, O., Moran, L., Kelly, C., Scates, P., Stef, L., et al. (2015). Virulence characteristics of hcp+ Campylobacter jejuni and Campylobacter coli isolates from retail chicken. Gut Pathog. 7:20.
Datta, S., Niwa, H., and Itoh, K. (2003). Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 52(Pt 4), 345–348. doi: 10.1099/jmm.0.05056-0
Di Donato, G., Marotta, F., Nuvoloni, R., Zilli, K., Neri, D., Di Sabatino, D., et al. (2020). Prevalence, population diversity and antimicrobial resistance of Campylobacter coli isolated in Italian Swine at Slaughterhouse. Microorganisms 8:222. doi: 10.3390/microorganisms8020222
Engberg, J., Aarestrup, F. M., Taylor, D. E., Gerner-Smidt, P., and Nachamkin, I. (2001). Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7, 24–34. doi: 10.3201/eid0701.010104
European Food Safety Authority and European Centre for Disease Prevention and Control (2021). The European Union Summary Report on Antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 19:e06490. doi: 10.2903/j.efsa.2021.6490
Guk, J. H., Kim, J., Song, H., Kim, J., An, J. U., Kim, J., et al. (2019). Hyper-aerotolerant Campylobacter coli from duck sources and its potential threat to public health: virulence, antimicrobial resistance, and genetic relatedness. Microorganisms 7:579. doi: 10.3390/microorganisms7110579
Hull, D. M., Harrell, E., van Vliet, A. H. M., Correa, M., and Thakur, S. (2021). Antimicrobial resistance and interspecies gene transfer in Campylobacter coli and Campylobacter jejuni isolated from food animals, poultry processing, and retail meat in North Carolina, 2018-2019. PLoS One 16:e0246571. doi: 10.1371/journal.pone.0246571
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720.
Kaakoush, N. O., Miller, W. G., De Reuse, H., and Mendz, G. L. (2007). Oxygen requirement and tolerance of Campylobacter jejuni. Res. Microbiol. 158, 644–650.
Kang, Y. S., Cho, Y. S., Yoon, S. K., Yu, M. A., Kim, C. M., Lee, J. O., et al. (2006). Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from raw chicken meat and human stools in Korea. J. Food Prot. 69, 2915–2923. doi: 10.4315/0362-028x-69.12.2915
Karama, M., Kambuyi, K., Cenci-Goga, B. T., Malahlela, M., Jonker, A., He, C., et al. (2020). Occurrence and antimicrobial resistance profiles of Campylobacter jejuni, Campylobacter coli, and Campylobacter upsaliensis in beef cattle on cow-calf operations in South Africa. Foodborne Pathog. Dis. 17, 440–446. doi: 10.1089/fpd.2019.2703
Karki, A. B., Marasini, D., Oakey, C. K., Mar, K., and Fakhr, M. K. (2018). Campylobacter coli from retail liver and meat products is more aerotolerant than Campylobacter jejuni. Front. Microbiol. 9:2951. doi: 10.3389/fmicb.2018.02951
Kempf, I., Kerouanton, A., Bougeard, S., Nagard, B., Rose, V., Mourand, G., et al. (2017). Campylobacter coli in organic and conventional pig production in France and Sweden: prevalence and antimicrobial resistance. Front. Microbiol. 8:955. doi: 10.3389/fmicb.2017.00955
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12, 424–432.
Little, C. L., Richardson, J. F., Owen, R. J., de Pinna, E., and Threlfall, E. J. (2008). Prevalence, characterisation and antimicrobial resistance of Campylobacter and Salmonella in raw poultrymeat in the UK, 2003-2005. Int. J. Environ. Health Res. 18, 403–414. doi: 10.1080/09603120802100220
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Mataragas, M., Skandamis, P. N., and Drosinos, E. H. (2008). Risk profiles of pork and poultry meat and risk ratings of various pathogen/product combinations. Int. J. Food Microbiol. 126, 1–12. doi: 10.1016/j.ijfoodmicro.2008.05.014
Moore, J. E., Barton, M. D., Blair, I. S., Corcoran, D., Dooley, J. S., Fanning, S., et al. (2006). The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 8, 1955–1966. doi: 10.1016/j.micinf.2005.12.030
NARMS (2019). Antibiotics Tested by NARMS. Available: https://www.cdc.gov/narms/antibiotics-tested.html (accessed 2020, July 15)
Nisar, M., Ahmad, M. U. D., Mushtaq, M. H., Shehzad, W., Hussain, A., Nasar, M., et al. (2018). Occurrence of Campylobacter in retail meat in Lahore, Pakistan. Acta Trop. 185, 42–45. doi: 10.1016/j.actatropica.2018.04.030
Oh, E., Chui, L., Bae, J., Li, V., Ma, A., Mutschall, S. K., et al. (2018). Frequent implication of multistress-tolerant Campylobacter jejuni in human infections. Emerg. Infect. Dis. 24:1037.
Oh, E., McMullen, L., and Jeon, B. (2015). High prevalence of hyper-aerotolerant Campylobacter jejuni in retail poultry with potential implication in human infection. Front. Microbiol. 6:1263. doi: 10.3389/fmicb.2015.01263
Oh, E., McMullen, L. M., Chui, L., and Jeon, B. (2017). Differential survival of hyper-aerotolerant Campylobacter jejuni under different gas conditions. Front. Microbiol. 8:954. doi: 10.3389/fmicb.2017.00954
O’Kane, P. M., and Connerton, I. F. (2017). Characterisation of aerotolerant forms of a robust chicken colonizing Campylobacter coli. Front. Microbiol. 8:513. doi: 10.3389/fmicb.2017.00513
Riedel, C., Förstner, K. U., Püning, C., Alter, T., Sharma, C. M., and Gölz, G. (2020). Differences in the transcriptomic response of Campylobacter coli and Campylobacter lari to heat stress. Front. Microbiol. 11:523. doi: 10.3389/fmicb.2020.00523
Sahin, O., Kassem, I. I., Shen, Z., Lin, J., Rajashekara, G., and Zhang, Q. (2015). Campylobacter in poultry: ecology and potential interventions. Avian Dis. 59, 185–200. doi: 10.1637/11072-032315-Review
Sato, K., Bartlett, P. C., Kaneene, J. B., and Downes, F. P. (2004). Comparison of prevalence and antimicrobial susceptibilities of Campylobacter spp. isolates from organic and conventional dairy herds in Wisconsin. Appl. Environ. Microbiol. 70, 1442–1447. doi: 10.1128/aem.70.3.1442-1447.2004
Segreti, J., Gootz, T. D., Goodman, L. J., Parkhurst, G. W., Quinn, J. P., Martin, B. A., et al. (1992). High-level quinolone resistance in clinical isolates of Campylobacter jejuni. J. Infect. Dis. 165, 667–670. doi: 10.1093/infdis/165.4.667
Sifré, E., Salha, B. A., Ducournau, A., Floch, P., Chardon, H., Mégraud, F., et al. (2015). EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J. Microbiol. Methods 119, 206–213. doi: 10.1016/j.mimet.2015.10.018
Song, H., Kim, J., Guk, J. H., An, J. U., Lee, S., and Cho, S. (2020). Complete genome sequence and comparative genomic analysis of hyper-aerotolerant Campylobacter lari strain SCHS02 isolated from duck for its potential pathogenicity. Microb. Pathog. 142:104110. doi: 10.1016/j.micpath.2020.104110
Sproston, E. L., Wimalarathna, H. M. L., and Sheppard, S. K. (2018). Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 4:e000198. doi: 10.1099/mgen.0.000198
Tang, M., Zhou, Q., Zhang, X., Zhou, S., Zhang, J., Tang, X., et al. (2020). Antibiotic resistance profiles and molecular mechanisms of Campylobacter from chicken and pig in China. Front. Microbiol. 11:592496. doi: 10.3389/fmicb.2020.592496
Tang, Y., Sahin, O., Pavlovic, N., LeJeune, J., Carlson, J., Wu, Z., et al. (2017). Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci. Rep. 7:494. doi: 10.1038/s41598-017-00584-z
Torralbo, A., Borge, C., García-Bocanegra, I., Méric, G., Perea, A., and Carbonero, A. (2015). Higher resistance of Campylobacter coli compared to Campylobacter jejuni at chicken slaughterhouse. Comp. Immunol. Microbiol. Infect. Dis. 39, 47–52. doi: 10.1016/j.cimid.2015.02.003
World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: WHO Press.
Keywords: aerotolerance, Campylobacter coli, genetic relatedness, quinolone resistance, swine, virulence potential, production stages
Citation: Guk J-H, Song H, Yi S, An J-U, Lee S, Kim W-H and Cho S (2021) Hyper-Aerotolerant Campylobacter coli From Swine May Pose a Potential Threat to Public Health Based on Its Quinolone Resistance, Virulence Potential, and Genetic Relatedness. Front. Microbiol. 12:703993. doi: 10.3389/fmicb.2021.703993
Received: 01 May 2021; Accepted: 24 June 2021;
Published: 16 July 2021.
Edited by:
Xiaonan Lu, McGill University, CanadaReviewed by:
Zhiming Pan, Yangzhou University, ChinaCopyright © 2021 Guk, Song, Yi, An, Lee, Kim and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo-Hyun Kim, aWNlbWFuMmJAc251LmFjLmty; Seongbeom Cho, Y2hvc2VAc251LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.