
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 September 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.703014
Seagrasses are marine angiosperms that can live completely or partially submerged in water and perform a variety of significant ecosystem services. Like terrestrial angiosperms, seagrasses can reproduce sexually and, the pollinated female flower develop into fruits and seeds, which represent a critical stage in the life of plants. Seed microbiomes include endophytic microorganisms that in terrestrial plants can affect seed germination and seedling health through phytohormone production, enhanced nutrient availability and defence against pathogens. However, the characteristics and origins of the seagrass seed microbiomes is unknown. Here, we examined the endophytic bacterial community of six microenvironments (flowers, fruits, and seeds, together with leaves, roots, and rhizospheric sediment) of the seagrass Halophila ovalis collected from the Swan Estuary, in southwestern Australia. An amplicon sequencing approach (16S rRNA) was used to characterize the diversity and composition of H. ovalis bacterial microbiomes and identify core microbiome bacteria that were conserved across microenvironments. Distinct communities of bacteria were observed within specific seagrass microenvironments, including the reproductive tissues (flowers, fruits, and seeds). In particular, bacteria previously associated with plant growth promoting characteristics were mainly found within reproductive tissues. Seagrass seed-borne bacteria that exhibit growth promoting traits, the ability to fix nitrogen and anti-pathogenic potential activity, may play a pivotal role in seed survival, as is common for terrestrial plants. We present the endophytic community of the seagrass seeds as foundation for the identification of potential beneficial bacteria and their selection in order to improve seagrass restoration.
It is well established that vascular plants form complex interactions and mutualistic relationships with bacteria that play a critical role in supporting plant growth and fitness (Turner et al., 2013; Fitzpatrick et al., 2018). This work, conducted largely on terrestrial vascular plants, has demonstrated that such diverse functions as boosted growth and protection from pathogens, can be attributed to interactions between plants and their unique microbial communities (microbiomes; Compant et al., 2005; Gopalakrishnan et al., 2015; Enebe and Babalola, 2019). In addition to improving our mechanistic understandings of plant physiology, plant-microbe mutualisms and their manipulation have become the focus of research into plant husbandry with applications ranging from crop improvement to vegetation restoration (Chaparro et al., 2012; O’Callaghan, 2016; Castro et al., 2018; Compant et al., 2019).
Seagrasses are marine plants characterised by several eco-physiological traits that allow them to live submerged in water (Les et al., 1997; Hemminga and Duarte, 2000; Kuo and den Hartog, 2006). They are globally distributed throughout the coastlines of all continents (except Antarctica), and are key players in sequestration of carbon dioxide, coastal protection and support of human communities by food provision and tourism (Costanza et al., 1997; Mtwana Nordlund et al., 2016). By comparison to terrestrial plants, understanding of the nature of seagrass-bacteria interactions, and the mechanisms through which bacteria are acquired is limited. This is astonishing, given the attention that plant microbiomes have received from the past two decades, and the significant value of seagrasses ($AUD 3.9–5.4 billion for carbon sequestration; Lavery et al., 2013).
As seen in terrestrial vascular plants, distinct seagrass-associated bacterial communities inhabit discrete microenvironments within and around the plant, including the internal tissue of leaves (phyllosphere), roots (endosphere) and the layer of sediment directly influenced by the plant activity (rhizosphere) (Crump and Koch, 2008; Garcias-Bonet et al., 2012; Sun et al., 2015; Hurtado-McCormick et al., 2019). The different physico-chemical conditions within these microenvironments, such as oxygen and nutrient concentrations, are likely to favour the divergent bacterial communities (McRoy and Goering, 1974; Kilminster and Garland, 2009; Chaparro et al., 2014; Martin et al., 2019). Seagrass microbiomes are involved in many processes that benefit the plants (Ugarelli et al., 2017; Tarquinio et al., 2019), including nutrient supply (e.g., nitrogen fixation associated with seagrass leaves and roots) and detoxification from harmful compounds (e.g., hydrogen sulphides around seagrass roots; Welsh, 2000; Jensen et al., 2007; Hamisi et al., 2009). Indeed, nitrogen fixing bacteria are estimated to provide up to 50% of the nitrogen required by seagrasses (O’Donohue et al., 1991), while sulphate oxidising bacteria alleviate seagrass below ground tissues from the toxic effects of hydrogen sulphide (Brodersen et al., 2018; Martin et al., 2019).
Recently, studies on terrestrial plants have focused on the microbiomes associated with seeds, and particularly their manipulation in order to improve plant health. Since the bacterial community established within seeds will be carried by the seedling and possibly the adult plant (Hardoim et al., 2015; Truyens et al., 2015), the quality of the seed microbiome is likely to impact the future plant fitness (Berg et al., 2017). Seed-borne bacteria are localised within the integument, endosperm and embryo of seeds (Shade et al., 2017) and their manipulation has been particularly significant to reduce, for example, the incidence of plant disease (Adhikari et al., 2001; Bloemberg and Lugtenberg, 2001), increase agricultural production (O’Callaghan, 2016), improve seedling development (Compant et al., 2010; Johnston-Monje and Raizada, 2011; White et al., 2019), promote germination and induce plant defence mechanisms (Weyens et al., 2009; Sánchez-López et al., 2018).
The characteristics and origins of the seagrass seed microbiome is largely unexamined. Seagrasses can be monoecious, with flowers containing male and female organs, or dioecious, with separate male and female plants. During the reproductive season, male flowers produce a hydrophilic pollen that fertilises ovules in female flowers, which can produce up to 60 seeds, depending on the species (Orth et al., 2007). Seeds represent a crucial phase in the life cycle of flowering plants, including seagrasses, because they maintain genetic diversity within a population (Kendrick et al., 2017) and, in some species, can persist for years in a dormant state developing into a new plant only when the appropriate conditions are met (Kuo and Kirkman, 1992; Orth et al., 2000). Halophila ovalis is a dioecious marine plant present in tropical Indo-Pacific waters as well as Australian temperate waters (Short et al., 2007, 2010) and is a foundation species in the Swan River (Southwestern Australia) where it plays an important role in terms of sediment stability and ecology (Hillman et al., 1995). H. ovalis exhibits seasonal colonisation, and, as with other opportunistic species, relies mainly on sexual reproduction and seed recruitment for the annual regeneration of meadows (Statton et al., 2017). The interest in exploring H. ovalis seed microbiome extends beyond that of exploring a potential biodiversity niche, as seed borne bacteria are likely to impact seedling germination and development, therefore affecting meadows enduring.
When endophytic bacteria form spatially and temporally stable associations with their host we refer to them as core microbiome (Astudillo-García et al., 2017). Core microbiomes can be defined by taxonomy or functionality (i.e., metagenomic or metatranscriptomic analyses used to predict shared functions). Moreover, we can define core microbiomes at a population level (i.e., the bacteria shared between plants of the studied population) or at a species level (i.e., the bacteria shared between plants from different populations; Vandenkoornhuyse et al., 2015). In each case, the plant core microbiome is considered to contain key microbial taxa that are critical for plant health and fitness (Lemanceau et al., 2017). For seagrasses, few studies have explored the existence of a core microbiome (Hurtado-McCormick et al., 2019; Martin et al., 2019).
In this study, we characterised the diversity and role of endophytic bacteria associated with a population of the seagrass Halophila ovalis collected from the Swan River. In particular, we characterised the composition and role of the bacterial community for six H. ovalis microenvironments; leaves, roots, flowers, fruits, seeds and the rhizosphere. We also investigated the presence of a H. ovalis core microbiome at a population level, within the six H. ovalis microenvironments and followed the changes of core bacteria within the reproductive tissues.
Seagrass samples were collected from a female H. ovalis meadow located at Pelican Point, Swan-Canning Estuary, in southwestern Australia (−32.026S; 115.7580E). H. ovalis is dioecious, with separate female and male flowers that develop during the austral summer (between December and February). Fruits start to develop from February till May and each fruit can contain up to 60 seeds (each about 2-mm diameter) which are released into the surrounding sediment once they reach maturity. Plant material was collected during two different occasions: February 2019 when the plants had immature flowers (Time 1) and April 2019, when the plants had developed mature fruits (Time 2). Flowers and mature fruits were collected while still attached to the adult plant. Leaves, roots and sediment were collected during each occasion (five replicates on each occasion) while flowers, fruits and seeds were collected once (each represented by five replicates), reflecting the reproductive phenology of H. ovalis.
Replicates were collected using a sterile PVC core of 10-cm diameter (1 mt depth), at a distance of 0.5 m apart; cores were stored in sterile zip-sealed plastic bags, in an ice box filled with ice until delivery to the laboratory, where they were frozen. The rhizospheric sediment was kept attached to the roots within the zip-sealed plastic bags, detached from the roots in the laboratory and kept in 2.5-ml sterile tubes at −20°C.
All the seagrass samples were processed in order to remove plant epiphytes and obtain only microbial endophytic DNA. All samples were rinsed in 0.2 μm filtered water collected from the river and, sediment-free tissues were then immerged in 70% ethanol for 30 s followed by a rinse in sterile Milli-Q water (MoBio Laboratories, Carlsbad, CA, United States). This procedure was repeated three times, with a final rinse in Milli-Q water performed three times to remove any excess of ethanol (Saldierna Guzmán et al., 2020). Fruits were surface sterilised as described above prior to opening with a sterilised razor blade in order to remove the seeds. Seeds were then placed in sterile 2.5 ml tubes and surface sterilised as described above.
Between 0.132 and 0.333 wet weight grams of samples were weighed for each replicate and tissue type and DNA was extracted using a PowerSoil Kit (MoBio) following the manufacturer’s instructions (on average, sediments = 0.27 g, roots = 0.25 g, leaves = 0.24 g, flowers = 0.24 g, fruits = 0.26 g, and seeds = 0.23 g). For each replicate of H. ovalis tissue, 15 flowers, between 8 and 13 fruits, between 120 and 180 seeds, 5 to 6 leaves and about 15 roots were pooled in order to obtain enough material for the DNA extraction. Final extracted DNA concentration was checked by using the QIAxpert machine from QIAGEN (which calculates DNA concentration of samples from the measured UV/VIS absorption spectrum) at CSIRO facility. Extracted DNA was then eluted and kept in 50 μL nuclease-free water.
PCRs were performed on extracted samples to screen for bacterial presence. GoTaq Green Master Mix (Promega) was used for the PCR reactions and primer concentration was 0.7 μM. Primer pairs used for the present study were 515F 5′ – GTGCCAGCMGCCGCGGTAA -3′ and 806R 5′ – GGACT ACHVGGGTWTCTAAT – 3′ primers which target the V4 hypervariable region of the 16S rRNA gene (Caporaso et al., 2012). PCR cycling conditions involved an initial step at 95°C during 120 s followed by 32 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 60 s; followed by a final extension step at 72°C for 10 min in accordance to the guideline of the Taq DNA furnisher’s (Promega).
Microbial communities were sequenced by the Australian Genome Research Facility (AGRF) on the Illumina MiSeq platform, using Nextera XT v2 indices and 300 bp paired end sequencing chemistry. Bioinformatic analysis of sequence reads were performed in MOTHUR (v1.39.3; Department of Microbiology and Immunology, The University of Michigan)1 using the Standard Operating Procedure (SOP, page accessed on September 2019) (Schloss et al., 2009; Kozich et al., 2013). Paired end reads were assembled by aligning the forward and reverse amplicon sequence reads, which joined paired reads using the make.contigs() command with the option trimoverlap = T. Sequences with ambiguous bases and and/or containing a homopolymer stretch longer than eight bases were removed. Unique contigs were aligned to the SILVA database (version 132) (Cole et al., 2014) using the align.seqs() command with default settings. Following alignment, sequences were pre-clustered (diffs = 2) for further error reduction and chimera.uchime() command (Edgar et al., 2011) was used for de novo removal of chimeric reads. Reads were clustered using the default Opti clustering method in MOTHUR at 97% similarity to produce Operational Taxonomic Units (OTUs). A “blank” sample of reaction buffers was also sequenced and OTUs present in the control (predominantly Clostridium_XlVa) were removed from all other samples. Also, OTUs represented by a single sequence and/or not present in at least three samples were removed. The remaining OTUs were classified using the SILVA taxonomy database method = wang (cutoff probability value = 80; Wang et al., 2007; Quast et al., 2013) and OTUs identified as chloroplast, mitochondria or algae (12.4% of total sequences) were removed. The most abundant unclassified OTUs were using megablast against the nucleotide database (nr/nt) using the NBCI (National Center for Biotechnology Information, U.S. National Library of Medicine) web portal2 to assign them a taxonomy using the best blast hit.
Data visualisation and statistical analyses were performed in R (version 4.0.2) (R Core Team, 2020) using the phyloseq (McMurdie and Holmes, 2013), ggplot2 (Wickham, 2016) and vegan (v 2.5–6; Oksanen et al., 2019) packages. Considering the number of reads (or number of sequences) per sample for a specific taxon as a proxy of its abundance (Calle, 2019), OTUs with a relative abundance across the full dataset of <1% were removed from the analyses to eliminate the influence of possible sequencing artefacts. A final number of 710 taxa (OTUs) from the six seagrass microenvironments (i.e., five types of tissue – leaf, root, flower, fruit, seed – and sediment) were kept for downstream analyses.
Bacterial diversity within samples (alpha diversity) was estimated using richness (number of observed OTUs) and Shannon’s diversity (Shannon and Weaver, 1964). Analysis of Variance (ANOVA) was performed to test hypotheses about differences in diversity between seagrass microenvironments and sampling occasions. A Tukey’s Honest Significant Difference (HSD) test was used (Tukey, 1953; Kramer, 1956, 1957) to identify which pairwise comparisons were significant.
Principal coordinate analysis (PCoA) was used to visualise patterns in the microbial assemblage (i.e., beta diversity) among the seagrass microenvironments, based on Bray-Curtis dissimilarities (Bray and Curtis, 1957) calculated from log10-transformed OTU abundances. To test hypotheses about differences in the bacterial assemblage among microenvironments, a permutational multivariate analysis of variance (PERMANOVA) was performed using the adonis function from the vegan package in R with 999 permutations (Anderson, 2017).
To explore which taxa most contributed to patterns, we used similarity percentage analysis (SIMPER, 999 permutations, log10 +1 transformed abundance; Clarke, 1993) to. The first two OTUs with the highest contribution to the dissimilarity between pairwise tests were extracted and fitted onto the PCoA with the function envfit from the package vegan (Oksanen et al., 2019).
To explore the possible role of bacteria within the seagrass holobiont, we were able to assign a function to 247 taxa based on their family or species taxonomic identity using the database FAPROTAX (Louca et al., 2016) and literature search (Supplementary Table 1). The two major limitations in applying FAPROTAX or other trait databases, are (1) databases are limited/incomplete in terms of their ability to capture all organisms. They are biased toward more heavily studied and cultured organisms, and (2) the assumption that if cultured bacterial species are able to perform a particular function, then all members of the same taxon also share and exhibit this phenotype. Despite these limitations, we were able to identify in this analysis bacteria involved in nitrogen cycling (nitrogen fixing, nitrifying, and denitrifying bacteria), sulphur cycling (sulphide-oxidising and sulphur-reducing bacteria) and/or with known plant growth promoting properties (PGPB: bacteria able to produce phytohormones).
Stable associations between H. ovalis and specific taxa were investigated through an exploratory analysis of the core microbiome. OTUs present (relative abundance >0%) in all samples of each microenvironment were considered part of their core microbiome. Moreover, we tested for the presence of a seagrass core microbiome present across samples of vegetative tissues (i.e., leaves and roots) and among reproductive parts (flowers, fruits and seeds), the latter to show which OTUs were conserved from the flower to the seed stage.
Seven hundred and ten OTUs were identified from high quality 16S rRNA gene sequences of the 45 samples. The majority of these OTUs (426 out 710, Figure 1A) belonged to the phylum Proteobacteria which was the dominant lineage within all six microenvironments, representing 87%, 84%, 83%, 83%, 78%, and 63% of the OTUs within seed, fruit, leaf, root, flower, and sediment, respectively (Supplementary Table 2). Gamma-, Delta-, and Alpha-proteobacteria were the dominant proteobacterial classes (97, 131, and 173 OTUs, respectively), compared to Oligoflexia and Beta- and Epsilon-proteobacteria (3, 9, and 13 OTUs, respectively).
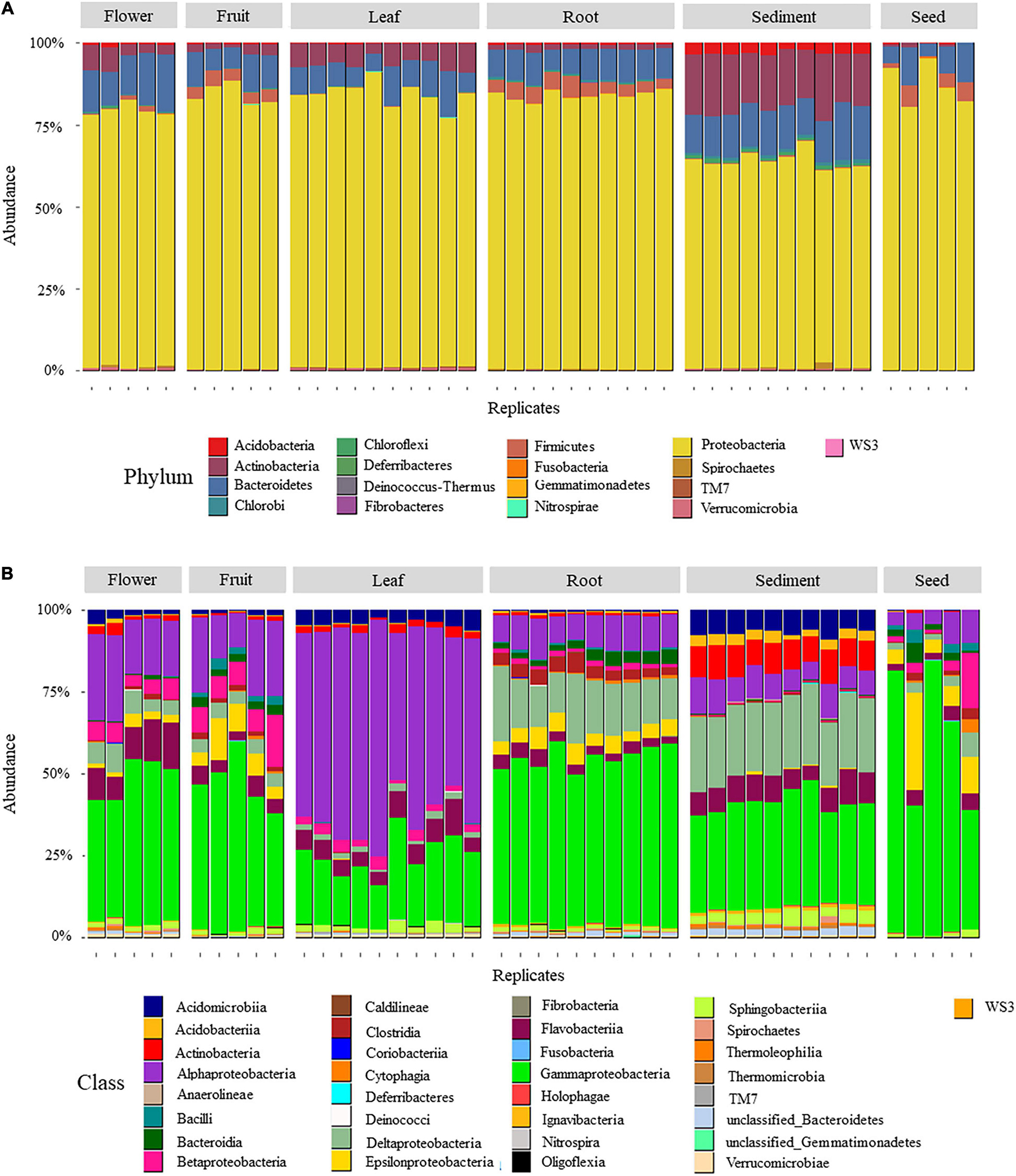
Figure 1. Relative abundance of OTUs aggregated by phylum (A) and class (B) in six H. ovalis microenvironments (rhizospheric sediment, roots, leaves, flowers, fruits, and seed). Numbers are presented in percentage.
With 140 unique OTUs, the Bacteroides phylum contained the second highest number of OTUs associated with H. ovalis microenvironments (Figure 1A and Supplementary Table 2). Bacteroides represented from 8 to 14% of the bacterial community in different microenvironments. Flavobacteriia (phylum Bacteroides) was the most relatively abundant Bacteroides class, being relatively prevalent in flowers (11%), sediment (8%) and leaves (6%, Figure 1B), while Sphingobacteriia, the second most abundant Bacteroides class, was most relatively abundant in sediment (3.5%). The remaining phyla were less relatively abundant, but they showed a clear association with specific seagrass microenvironments. For example, Acidobacteria (31 unique OTUs) and Actinobacteria (46 unique OTUs) were mainly present in the rhizosphere and almost absent in seagrass tissues (Figure 1B).
Alpha diversity (based on a total number of 1565233 sequences, Supplementary Figure 1) measured as Shannon’s diversity index and observed OTUs, varied significantly between seagrass microenvironments (ANOVA, pobserved < 0.0001, MSobserved = 3.7, Fobserved = 27.7; pshannon < 0.0001, MSshannon = 71418, Fshannon = 34.6). However, alpha diversity did not vary substantially among the two sampling occasions (ANOVA, pobserved = 0.8375, pshannon = 0.8752) and so this was excluded from further statistical analyses. In general, diversity and richness within seeds was lower than all other microenvironments. Sediment was characterised by the highest diversity and richness among microenvironments and was significantly different from H. ovalis tissues, except for fruits and flowers (Supplementary Figure 1 and Supplementary Table 3). Bacterial community diversity and richness of roots, flowers, leaves and fruits were not significantly different although these parameters tended to be smaller for the leaf bacterial community.
The first two axes of a PCoA comparison of bacterial diversity explaining 55% of bacterial variability among microenvironments. Clear partitioning of the communities belonging to the leaves, roots and sediment and a less accentuated separation for flowers, fruits and seeds was evident (Figure 2A). These patterns of differences in bacterial community structure according to the plant microenvironment was also supported by PERMANOVA, (p < 0.0001, Supplementary Table 4), where all bacterial communities were significantly different from each other. The different physical and chemical conditions within discrete seagrass microenvironments are likely to favour microscale heterogeneity in bacterial diversity and community structure (Borum et al., 2007; Ettinger et al., 2017; Brodersen et al., 2018; Hurtado-McCormick et al., 2019).
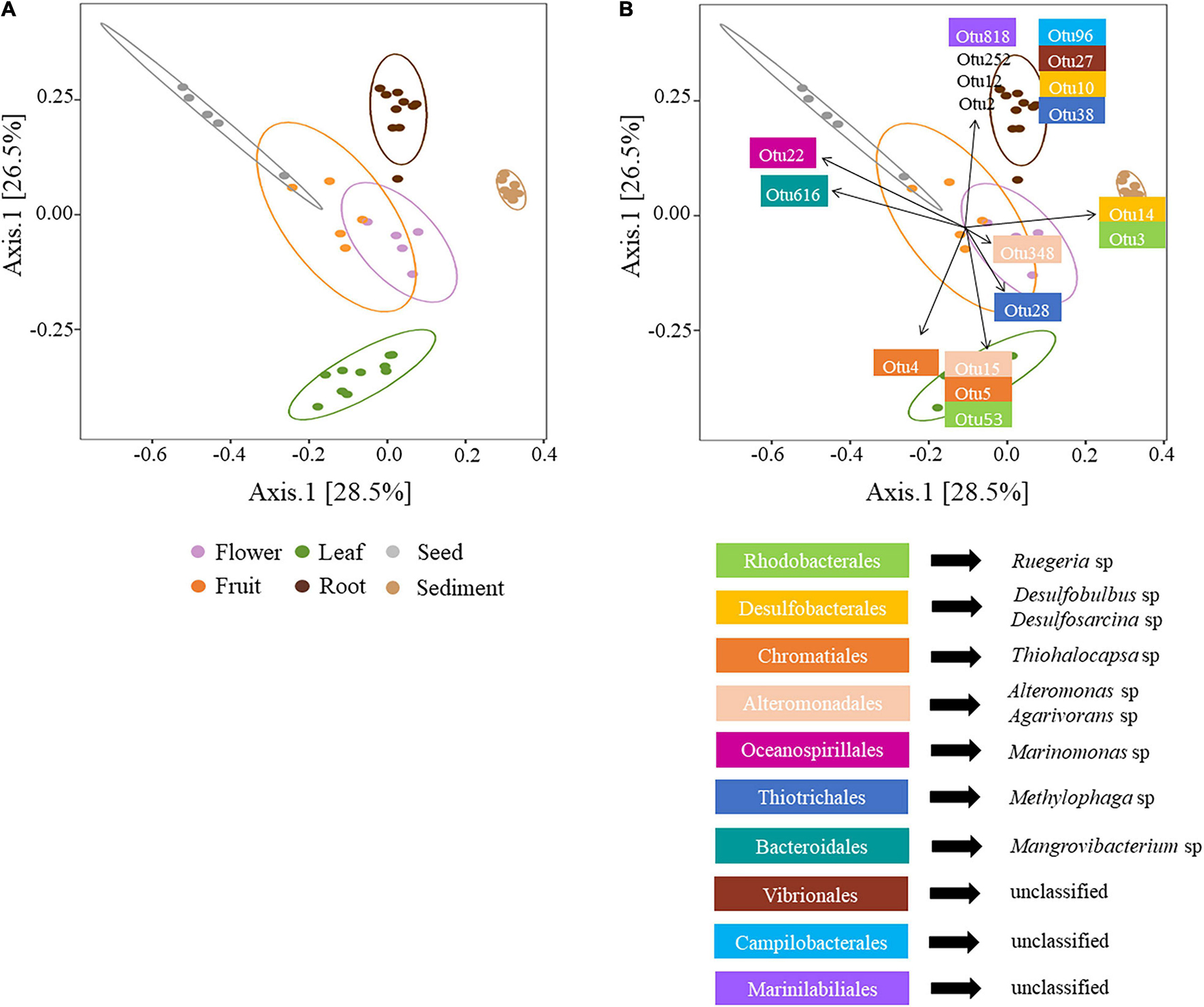
Figure 2. (A) Principal coordinates analysis (PCoA) of microbial communities associated with H. ovalis rhizospheric sediment, roots, leaves, flowers, fruits, and seeds. The percentages in parentheses refer to the proportions of variation explained by each ordination axis. The confidence ellipses (with a 95% confidence level) corresponding to each microenvironment are shown (B) PCoA with envfit object displaying the first two OTUs with the highest contribution to the dissimilarity between pairwise tests from SIMPER. OTUs identity is reported below the PCoA. OTU 2, 12, and 252 are unclassified Gammaproteobacteria.
The bacterial assemblage inhabiting H. ovalis rhizosphere represented a clearly distinguished cluster from the other microenvironments (Figure 2A). The bacterial community found in this microenvironment was, on average, the richer (400.6 ± 6.56 total number of OTUs) and more diverse than those associated with H. ovalis tissues. This is not surprising since in terrestrial systems, the rhizosphere is characterised by an increased diversity compared to host-associated communities (Edwards et al., 2015) and our study is consistent with many studies of seagrass microbial diversity (Jensen et al., 2007; Mejia et al., 2016; Fahimipour et al., 2017; Martin et al., 2018, 2020).
The main driver of the clustering of the rhizospheric bacterial community was the high abundance of Desulfobacterales (especially Desulfosarcina sp.) and Chromatiales (Chromatiaceae) which accounted for 17%, and 17.5% of rhizospheric sequences, respectively (Figure 2B and Supplementary Table 5). Members of these two taxa have been shown to dominate the rhizosphere of seagrasses (Sun et al., 2015; Ettinger et al., 2017; Cúcio et al., 2018) and salt marsh plants (Thomas et al., 2014), where they influence sulphur and nitrogen cycling (Cúcio et al., 2016). Actinobacteria was the third most relatively abundant phylum in sediment (17%), represented by the classes Actinobacteria (9%) and Acidimicrobiia (7%), while its average abundance was less than 6.5% of sequences within H. ovalis tissues. In general, within the oxygenic sediment layer and/or at the root-sediment interface, members of the Actinobacteria, Flavobacteriales, and Myxococcales, can play a prominent role in degrading organic substrates (Lu et al., 2006; Bowman et al., 2012; Větrovský et al., 2014; Lasa et al., 2019). For example, bacteria belonging to the genera Eudoraea and Maribacter (Flavobacteriaceae) found in the present study, can grow aerobically utilising several organic substrates (e.g., organic acids, amino acids, and carbohydrates; Alain et al., 2008; Handley and Lloyd, 2013). On the contrary, within the anoxic layers of seagrass sediment, decomposing processes can be based on the activity of obligate fermentative Clostridium species, which provide substrates to other organisms, such as sulphate-reducing bacteria (SRB; Küsel et al., 1999).
Sulphate reduction is one of the main processes in marine sediments related to the anoxic mineralisation of organic matter (Jørgensen, 1982) and it is reported as high in seagrass meadows, where it is fuelled by organic exudates from the plant roots (Isaksen and Finster, 1996; Holmer and Nielsen, 1997; Hansen et al., 2000; Cúcio et al., 2016). This process it also likely to be significant in eutrophic environments such as the Swan-Canning Estuary, where sediments have been characterised by nutrient and organic matter enrichment (Kelsey and Western Australia, Department of Water, 2010). SRB gain energy for cell synthesis and growth by coupling the oxidation of organic compounds to the reduction of different sulphate/sulphur molecules to hydrogen sulphide (H2S, HS–; Dworkin et al., 2006; Camacho, 2009). H. ovalis rhizospheric sediment contained a diverse assemblage of OTUs belonging to Desulfobacterales, Desulfovibrionales and Desulfuromonadales known to reduce elemental sulphur to H2S (Muyzer and Stams, 2008; Varon-Lopez et al., 2014). Among them Desulfosarcina sp. was associated only with the sediment community in our study (Figure 2B). However, sulphides that are produced from the reduction of sulphur, in particular H2S, are toxic to plants (Koch and Erskine, 2001; Borum et al., 2005; Lamers et al., 2013). Several studies have described that the association of seagrass below-ground parts with sulphide-oxidising bacteria (SOB) can release the marine plants from sulphide toxic stress (van der Heide et al., 2012; Fahimipour et al., 2017; Martin et al., 2019; Van Der Geest et al., 2020). In fact, SOB by using H2S for their metabolism demand could help to reduce sulphide concentration, as suggested for Zostera species (Hansen et al., 2000; Nielsen et al., 2001; van der Heide et al., 2012; Brodersen et al., 2018). In our study, four rhizospheric OTUs (relative abundance of 0.15) were putatively involved in the oxidisation of sulphides such as Rhodovulum sp., (Rhodobacterales) and Thiohalocapsa sp. (Chromatiales) which is able to oxidize both sulphide and elemental sulphur (Straub et al., 1999; Hamilton et al., 2014).
Microbially mediated denitrification is a key ecosystem service provided by seagrass associated bacteria (Reynolds et al., 2016), which acts to remove the excess of nitrogen from eutrophic systems (Stohr and Ullrich, 2002). This may be especially important in estuarine systems which usually present high concentrations of nutrients. For example, Caldithrix sp. (Deferribacterales) is an anaerobic nitrate reducer, and Saccharospirillum sp. (Oceanospirillales) is able to catalyse the three steps of the denitrification process (Miroshnichenko et al., 2003; Zhang et al., 2019). OTUs affiliated with Ignavibacterium sp. were mostly present in sediment samples; Ignavibacterium species are able to reduce ammonium to dinitrogen gas (N2) that could be used by nitrogen fixing bacteria present within the seagrass ecosystem. While organic nitrogen recycling in the rhizospheric sediments can provide a large proportion of the plant’s nitrogen demand, it may be insufficient to meet the annual N requirement of the plant, due to the limited nutrient diffusion and root uptake rates (Lepoint et al., 2002). Nitrogen fixation might represent another important source of nitrogen used by seagrasses and high rates of nitrogen fixation have been recorded in sediments colonised by diverse seagrass species (Welsh, 2000; Bagwell et al., 2002). Some members of Desulfobulbaceae and Desulfobulbacteraceae could be involved in anaerobic nitrogen fixation (Herbert, 1999; Sun et al., 2015) as well as Cohaesibacter sp. which has already been found associated with H. ovalis roots in the Swan River Estuary (Martin et al., 2020). Finally, Nitrosococcus sp. (Chromatiales), oxidises nitrites (Bock and Wagner, 2013), which are toxic to plant tissues, to nitrates which can be up taken by seagrass roots and translocated to leaves for storage in vacuoles (Touchette and Burkholder, 2000).
Bacterial assemblages characterised from roots were distinct from those of other microenvironments (Figure 2A). Nonetheless, bacterial diversity was comparable to that of reproductive tissues and leaves, consistent with results found for Zostera marina (Ettinger et al., 2017). The root bacterial community mainly constituted Proteobacteria, in which Gamma- and Deltaproteobacteria were highly abundant (52% and 15.5% of the sequences, respectively). Thiotrichales, Vibrionales (Gammaproteobacteria), Campylobacterales (Epsilonproteobacteria), and Clostridiales (Firmicutes), were highly abundant in roots and reproductive tissues, but absent or almost absent in leaves and sediment (Figure 2B). Desulfobacterales were abundant in roots and sediment, but not in other tissues while Fibrobacterales were only present within the roots.
A number of studies have shown that the order Clostridiales is often part of the microbiome of seagrass species (e.g., Z. marina, Halodule wrightii; Küsel et al., 1999; Cúcio et al., 2016; Garcias-Bonet et al., 2020). This association between Clostridiales and seagrasses could be a potential aid for seagrass health, since some members of this order can be involved in nitrogen fixation (Minamisawa et al., 2004), fermentation (Cato et al., 1986), as well as sulphate reduction (Widdel, 2006). As for the sediment community, an ability to reduce sulphate and sulphur was a common feature of a high proportion of root-associated bacteria, in particular Desulfobacterales (13.5%; Supplementary Table 5). Interestingly, some genera within the family Desulfobacteraceae are also able to oxidize alcohols. During the night cycle, the lack of oxygen around the roots leads seagrass root tissues to switch to fermentation, which causes the release of ethanol to the rhizosphere (Smith et al., 1988). Cúcio et al. (2016) suggested that this association might represent a fair trade between host and microbes, where bacteria use ethanol as an electron donor at night and remove the alcohol from the surrounding of the roots (Kuever et al., 2005).
However, reduction of sulphur and sulphate may be toxic to plants and, in the case of seagrass roots, Campylobacterales may serve as detoxicant. In the present study, the families Campylobacteraceae and Helicobacteraceae were part of the root community (nine OTUs with a relative abundance of 5%; Figure 2B). Helicobacter sp. as well as Arcobacter sp. and Sulfurimonas sp. are important SOB commonly found within below-ground parts of several seagrasses (i.e., Z. marina and H. ovalis, Küsel et al., 2006; Jensen et al., 2007; Lu et al., 2020; Martin et al., 2020); the high relative abundance of Helicobacter sp. OTUs contributed to the distinct clustering of the root community. Moreover, some SOB may be involved in the recycling of nitrogen in seagrass beds. For example, one OTU from our study matched at 99% with Arcobacter nitrofigilis, a nitrogen fixing bacterium isolated from the roots of the salt marsh Spartina alterniflora (McClung et al., 1983). Likewise, some Sulfurimonas species are able to use nitrate and/or nitrite as electron acceptors (Han and Perner, 2014). Other SOB associated with H. ovalis roots belonged to Thiotrichales and were relatively abundant (5.5%). Thiotrichales are filamentous sulphur oxidising bacteria (Garrity et al., 2005) which have been found abundant in the rhizosphere of seagrasses from Portugal (Cúcio et al., 2016) and Z. marina leaves (Ettinger et al., 2017).
Operational Taxonomic Units from Vibrionales that were present in the roots (6%) were almost completely absent in the sediment and phyllosphere (0.5 and 0.6%, respectively) and contributed to the clustering of the root community (Figure 2B). Members of the Vibrionales have been found within seagrass sediments (Enhalus acoroide and Thalassia hemprichii; Liu et al., 2018) or associated with seagrass roots (Martin et al., 2020). Although the role of Vibrionales within the seagrass microbiome is not clear and our OTUs may be simply related to potential pathogenic species (e.g., Vibrio sp.; Blazer et al., 1988; Liu et al., 2018), some members might be involved in nitrous oxide (N2O) reduction (Hanke et al., 2016). Bacteria from the Fibrobacterales (Fibrobacter sp.) which have been found associated with the roots of H. wrightii, but not with the surrounding sediment (Stuij, 2018), were also only found in root samples in the present study. They are cellulolytic bacteria that can degrade lignocellulose and this capability has been hypothesised to be an essential feature related to the ability of those bacteria to colonise plant roots (Jose et al., 2014; Kandel et al., 2017; Liu et al., 2017).
The bacterial community in H. ovalis leaves was significantly different from the microbiomes associated with the rhizospheric sediment and seagrass reproductive tissues (Figure 2A). Alphaproteobacteria were the most abundant bacterial phylum in leaf samples. Some of these, such as Caulobacterales and Sphingomonadales, were almost absent from the other microenvironments, while others were present more generally in the plant (e.g., Rhodobacterales; Figure 2B and Supplementary Table 5).
The family Rhodobacteraceae (Rhodobacterales) has been associated with the phyllosphere of several seagrass species (Z. muelleri and Thalassia hemprichii; Jiang et al., 2015; Hurtado-McCormick et al., 2019; Rotini et al., 2020) and comprises photosynthetic purple non-sulphur bacteria which are primary surface colonizers in marine habitats (Palacios and Newton, 2005; Dang et al., 2008). Interestingly, the ability of colonising and growing on surfaces may be linked to their capacity to produce antibacterial compounds which could prevent other bacteria/pathogens from colonising the surface and/or form a biofilm, therefore protecting their host (Dang et al., 2008). Some members of Rhodobacterales are also able to denitrify. For example, Labrenzia sp. found in the present study, which has been previously isolated among epiphytes of P. oceanica leaves and endophytes of H. ovalis roots, is involved in conversion of N2O to N2 (Blanchet et al., 2017; Martin et al., 2020). Species of Labrenzia are also known to be plant growth promoters in terrestrial environments because they can produce auxin which might be of a particular importance for supporting plant growth (Liu et al., 2014; Fidalgo et al., 2016).
Other bacteria involved in nitrogen cycling were present within the leaf endophyte community. For example, Filomicrobium sp. (Rhizobiales) is able to utilize organic molecules (e.g., ammonia) as the N source for growth (Wu et al., 2009). Flavobacteriales (phylum Bacteroidetes) such as Muricauda sp. Aquimarina sp. and Maribacter sp. may detoxify seagrasses from N2O through its reduction to N2 (Xu et al., 2015; Nakagawa et al., 2019). Finally, five OTUs belonged to Sphingomonadales and Caulobacterales. Among them, one OTU matching with Altererythrobacter sp. is able to resist metal contamination (Wu et al., 2014), a trait that could be useful in the phyllosphere of H. ovalis since high metal concentrations in Swan-Canning River have been detected (Kelsey and Western Australia, Department of Water, 2010).
Alteromonadales and Chromatiales (Gammaproteobacteria) were also relatively abundant within leaves compared to the other seagrass tissues and seemed to be of particular importance for the clustering of leaf community in our study (Figure 2B). Granulosicoccus sp. and Ruegeria sp., harbour many genes involved in sulphur metabolism comprising a gene for dimethylsulfoniopropionate (DMSP) demethylase (Kang et al., 2018; Wirth et al., 2020). DMSP can be produced by macroalgae and higher plants as osmoregulator, herbivore deterrent and antioxidant against reactive oxygen species (Jonkers et al., 2000; Husband et al., 2012) and represent an important source of reduced carbon and sulphur for marine bacteria (Wirth et al., 2020). Genetic evidence indicates that several bacteria commonly found within seagrass leaves share the capacity to utilise DMSP that may represent a trait characteristic of seagrass leaf associated bacteria (Crump and Koch, 2008; Lucas-Elío et al., 2012).
Flower and fruit communities were mainly composed by bacteria from the phyla Proteobacteria and Bacteroidetes. Although fundamental knowledge of flower-associated microbiota remains largely unknown, when Shade et al. (2013) studied the microbiome of apple flowers they found diverse taxa affiliated to Proteobacteria, Actinobacteria, and Bacteroidetes. In the present study, the Proteobacterial families Piscirickettsiaceae (Methylophaga sp.), Campylobacteraceae, Helicobacteraceae, and Methylophilaceae were highly represented in these tissues. Methylophaga species have been isolated from diverse marine environments (Janvier et al., 1985; Doronina et al., 2007) where they are likely involved in denitrification processes (Auclair et al., 2010). Also, nitrogen fixation is likely to occur in those communities, since Campylobacterales were identified as nitrogen fixers when isolated from Spartina sp. roots (McClung and Patriquin, 1980). Marinomonas sp. (Oceanospirillaceae), a plant growth promoting bacteria found associated with flower and fruit microbiomes is known to play a key role in P. oceanica seedling growth (Celdrán et al., 2012). Alteromonas sp., associated with the flower community, but not the fruits, can have algicidal proprieties (Sakami et al., 2017) and associations between seagrass and algicidal bacteria have already been reported (Inaba et al., 2017). Finally, Diskin et al. (2017) reported the family Chitinophagaceae (Bacteroidetes) to be the most abundant one within the mango fruit microbiome. Some species from this family degrade chitin, while others hydrolyse cellulose (Rosenberg, 2014). In our study one OTU belonged to Chitinophagaceae and it was found only in fruits and seeds but not in flowers.
Bacteria belonging to Deinococcus-Thermus and TM7 phyla were also members of the apple flower microbiome, although they were not as abundant as in the present study. This discrepancy may be link to a different abundance of those phyla related to the age of flowers. For example, TM7 taxa are contenders for colonisation of closed flowers, where they survive but do not grow until flowers open and then grow rapidly and competitively on open flowers (Shade et al., 2013). Interestingly, Truepera sp. (Deinococcus-Thermus) found in this study, is also associated with apple flowers (Shade et al., 2013), which may suggest a possible important role within the flower microbiome. Truepera sp. seems to possess important adaptations to different environmental stress, such as resistance to desiccation, ultraviolet radiation, high salinity, and high temperature (Blasius et al., 2008), which can be experience in the Swan River during summer time when H. ovalis meadows became exposed to air and water temperatures can raise up to 30°C in the hottest months (Kelsey and Western Australia, Department of Water, 2010).
Seed endophytes have intensively been study in terrestrial plants, in particular crops (Johnston-Monje and Raizada, 2011; Adam et al., 2018; Chen et al., 2020), since they can assist seedling germination and cope with environmental stresses, such as drought or salinity (Truyens et al., 2015; Nelson, 2018). H. ovalis seeds were characterised by the lowest number of OTUs compared to the other microenvironments, and were mainly composed of Gammaproteobacteria. A similar pattern in terms of OTU abundance and diversity has been found for the seeds of the pumpkin Cucurbita pepo (Adam et al., 2018) and the spinach Spinacia oleracea (Lopez-Velasco et al., 2013). Three OTU belonged to the genus Marinomonas (Figure 2B and Supplementary Table 5); M. posidonica may enhance the growth of P. oceanica seedlings, induce changes in the epiphytic bacterial community and have a regulatory effect on macro-epiphyte structure (Celdrán et al., 2012). Two OTUs were related to M. communis which exhibits grow-promoting traits when found in plants (Mesa et al., 2015). Another OTU belonged to the genus Mangrovibacterium (Bacteroidales), a nitrogen-fixing bacterium isolated previously from mangrove sediment (Huang et al., 2014). Moreover, members of the family Pseudoalteromonodaceae were more abundant in the seed bacterial community compared to other tissues. Species of the genus Pseudoalteromonas are common within the marine environment and have been found associated with corals, sponges, seagrass surface and within molluscs (Romanenko et al., 2008). P. luteoviolacea (found in this study) is a globally distributed marine bacterium that can induce the metamorphosis of tubeworm and coral larvae (Alker et al., 2020) Finally, one OTU matching with Methylophaga thiooxydans was found in the present study within the seed microenvironments. Methanol-consuming bacteria are common in marine environments and include members of the genera Methylophaga and Methylobacter. Angiosperms, and so seagrasses, produce methanol as a by-product of cell-wall synthesis (Nemecek-Marshall et al., 1995), yet it has been reported that methanol could inhibit germination and/or negatively affect the growth of angiosperm seedlings (Abanda-Nkpwatt et al., 2006). In strawberry plants, this negative effect may be mitigated by methanol-consuming bacteria (e.g., Methylobacterium extorquens; Abanda-Nkpwatt et al., 2006; Kurilenko et al., 2010). Moreover, Methylophaga species isolated from rhizospheric sediments are also capable of producing auxins and therefore may be able to boost the seedling growth. However, more studies related to seagrass seed-borne bacteria are needed to understand how seagrass seed acquire bacteria, the diversity of seed-borne bacteria and potential role in seedling germination and growth.
The number of taxa in core microbiomes varied substantially between microenvironments, ranging from 10 OTUs for seeds, up to 238 OTUs for rhizospheric sediment (Figure 3). Although our criterion was quite strict (i.e., OTUs present in 100% of samples from a microenvironment), the number of OTUs was high relative to those found in Zostera muelleri (i.e., 102 OTUs in sediment, 61 OTUs in roots, two OTUs in leaves; Hurtado-McCormick et al., 2019). However, it needs to be taken in consideration that previously works have investigated seagrass core microbiomes at a species level rather than population, thus environmental variability across sampling sites could reflect the high variability of bacterial microbiomes. For example, Martin et al. (2020) found out that the root microbiomes of H. ovalis plants sampled in different locations within the Swan River presented a greater divergence of microbial communities than populations sampled around the Leschenault Peninsula. For the present study, the core microbiome of the six microenvironments included 63 families from 44 bacterial orders, across 10 phyla (Supplementary Table 6). Only one OTU, belonging to the family Methylophilaceae, was observed in all 45 samples, indicating that the seagrass microenvironments represent markedly different microbial niches. Our results provide evidence of a clear differentiation of core bacterial communities across the different microenvironments within the seagrass, instead of a unified seagrass core microbiome. The existence of discrete core microbiomes across the different H. ovalis microenvironments is consistent with patterns found in terrestrial plants, for which the rhizosphere, the phyllosphere and the root endospheres host communities that are both distinct from each other and the surrounding soils (Coleman-Derr et al., 2016; Fonseca-García et al., 2016; Maggini et al., 2019). The patterns observed here are also consistent with other benthic marine organisms including corals, where distinct microbial communities colonize different microenvironments within the coral colony, coral polyps and coral tissue (Sweet et al., 2011).
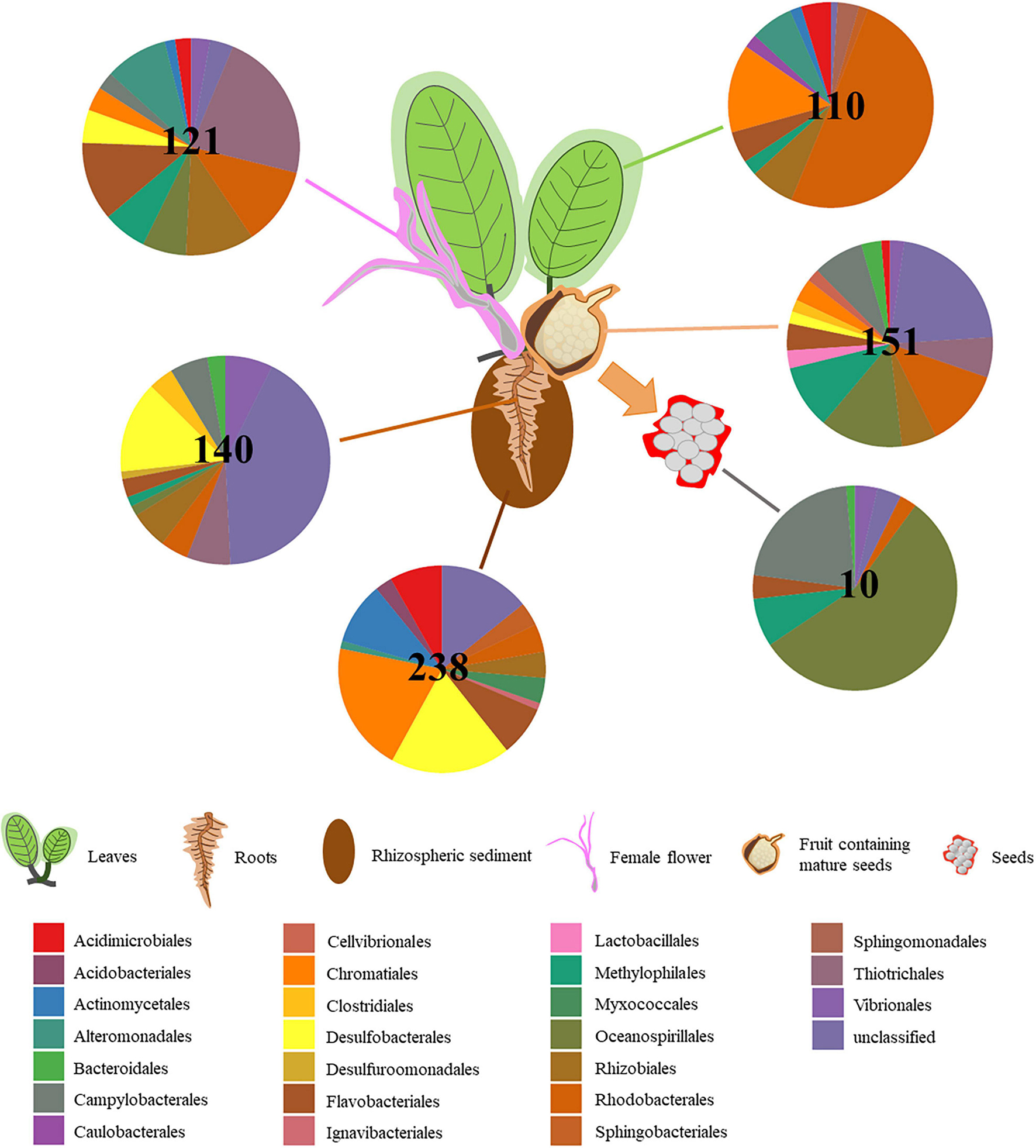
Figure 3. Core microbiome composition varies depending on H. ovalis microenvironment. Pie charts show the relative abundances of the major bacterial orders that are detected within the core microbiome of the rhizospheric sediment, rhizosphere, root, leaf, flower, fruit, and seed. Only orders that represented >0.1% of the total population are included. In the centre of the pie charts is reported the number of OTUs present in the core microbiome of each microenvironment.
The rhizospheric sediment core microbiome (238 OTUs; Figure 3) displayed the greatest abundance of OTUs. The most abundant order was represented by Chromatiales (19%), although a relatively high number of OTUs was unclassified (12%). Desulfobacterales was the second most abundant (17.5%) order present in the sediment core microbiome and it was primarily composed of Desulfobacteraceae (54%) and Desulfobulbaceae (46%).
One hundred and forty OTUs were present across all ten root replicates and were mainly related to Desulfobacterales (13.5%, comprising Desulfobacteraceae and Desulfobulbaceae), and Chromatiales. Likewise, Gamma- and Deltaproteobacteria have previously been found to be among the most abundant members of the core microbiome of below-ground structures in the seagrass H. stipulacea (Mejia et al., 2016) and several others seagrasses (Cúcio et al., 2016). 96 OTUs were shared between all the root and sediment replicates and belonged mainly to Desulfobacterales, followed by Acidimicrobiales, Flavobacteriales, and Rhodobacterales.
Those core microbiomes are likely to be involved in sulphur processes and nitrogen cycle in the core seagrass rhizobiome that influence the decomposition of organic material and ultimately the health of the host (Varon-Lopez et al., 2014; Lehnen et al., 2016).
The leaf core microbiome mainly consisted of Alpha- and Gamma- Proteobacteria (Figure 3), which is also consistent with previous studies (Hurtado-McCormick et al., 2019). Almost half of the 110 OTUs constituting the leaf core microbiome belonged to the Rhodobacteraceae (Rhodobacterales, 49%). The Chromatiales was second most represented in the H. ovalis leaf core microbiome (13.5%) and the family Chromatiaceae (11.5%) was the most abundant within Chromatiales. Members of the Rhodobacteraceae, and Chromatiaceae may favour inhabiting the H. ovalis phyllosphere, where they exploit the oxic conditions and high levels of dissolved organic carbon on the leaf surface (McRoy and Goering, 1974; Rotini et al., 2017; Trevathan-Tackett et al., 2020).
121 OTUs comprised the flower core microbiome and belonged mainly to the Piscirickettsiaceae (Thiotrichales, 21.5%). Rhodobacteraceae (Rhodobacterales, 11%) and Flavobacteriaceae (Flavobacteriales, 11%) were the two other families abundant in the flower core microbiome (Supplementary Table 6). As discussed above, they could be involved in nitrogen cycle (Jones et al., 2008; Nakagawa et al., 2019) and Rhodobacteraceae has been described as colonizers in marine habitats (Palacios and Newton, 2005; Dang et al., 2008). Although, fruits and flowers were likely to have similarities in bacterial community composition, the fruit core microbiome showed a high abundance of unclassified Gammaproteobacteria as well as Oceanospirillaceae, which contain plant growth promoting bacteria and Rhodobacteraceae.
The fruit core microbiome (151 OTUs) comprised bacteria belonging to Oceanospirillaceae (Oceanospirillales, 12.5%), Rhodobacteraceae (Rhodobacterales, 12%) and Methylophilaceae (Methylophilales, 9%). However, the most abundant bacteria were unclassified Gammaproteobacteria (20.5%) to which we were unable to assign taxonomy even after blasting and literature searching.
Finally, the 10 OTUs found in the seed core microbiome belonged mainly to Oceanospirillaceae (Oceanospirillales, 55.5%) and Helicobacteraceae (Campylobacterales, 17.5%; Figure 3). Interestingly, the seed core microbiome was composed of OTUs belonging to the genera Marinomonas and Mangrovibacterium. Moreover, one OTU belonging to Labrenzia sp. was found across the five replicates from seed microenvironment and is known to have plant growth promoting activities. The persistence of the association between endophytic bacteria that display plant growth promoting traits and H. ovalis seeds highlight the potential critical function performed by those bacteria in seedling development (Puente et al., 2009; Verma et al., 2017).
Samples of all three reproductive microenvironments combined (i.e., flowers, fruits, and seeds) contained 7 OTUs. This suggests the possibility that these taxa might be passed from the flowers through fruits and into seeds. Three of these OTUs were unclassified and four belonged to Labrenzia sp., Marinomonas sp., Amphritea sp. and Helicobacter sp. Excluding seeds (which contained the fewest taxa), flower and fruit core microbiomes shared 70 OTUs. The existence of complex mechanisms of bacterial transmission from flower to fruit and then to seeds appears likely and warrants further investigation. Recent studies from the terrestrial environment have shown that endophytic bacteria associated with flowers may colonize developing ovules and ultimately end up in fruits and seeds. Compant et al. (2011) demonstrated that several Pseudomonas and Bacillus species that were present in flowers and inside the xylem vessels of ovaries were also present within seeds. Furthermore, the inoculation of flowers with known bacteria resulted in significant levels of those strains within seeds (Dutta et al., 2014; Mitter et al., 2017).
Microbial assemblages in six discrete microenvironments associated with Halophila ovalis plants were distinct and the taxa present within the microenvironments were related to those that are likely involved in ecologically important processes that benefit plants. Although the specific mechanisms of interaction between bacterial endophytes and the host remain unknown in seagrasses, bacteria previously associated with plant growth promoting characteristics (e.g., ability to produce auxin and cytoxin), were mainly found within reproductive tissues. Seagrass seed-borne bacteria that exhibit growth promoting traits, the ability to fix nitrogen and anti-pathogenic potential activity, may play a pivotal role in seed survival, as is common for terrestrial plants. Therefore, our findings are consistent with research performed on terrestrial plants and highlight the potential beneficial role of seagrass associated bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.25919/9dga-ra29.
FT conducted the field work together with DNA extraction, sequencing, downstream analyses, and led the manuscript writing. OA conducted statistical analyses and contributed to the literature search on bacterial putative function. MV, OB, and AB provided critical input for data interpretation. All the authors contributed to the manuscript production.
This work was supported by the CSIRO Environomics Future Science Platform.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.703014/full#supplementary-material
Abanda-Nkpwatt, D., Krimm, U., Coiner, H. A., Schreiber, L., and Schwab, W. (2006). Plant volatiles can minimize the growth suppression of epiphytic bacteria by the phytopathogenic fungus Botrytis cinerea in co-culture experiments. Environ. Exp. Bot. 56, 108–119. doi: 10.1016/j.envexpbot.2005.01.010
Adam, E., Bernhart, M., Müller, H., Winkler, J., and Berg, G. (2018). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422, 35–49. doi: 10.1007/s11104-016-3113-9
Adhikari, T. B., Joseph, C. M., Yang, G., Phillips, D. A., and Nelson, L. M. (2001). Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice. Can. J. Microbiol. 47, 916–924. doi: 10.1139/w01-097
Alain, K., Intertaglia, L., Catala, P., and Lebaron, P. (2008). Eudoraea adriatica gen. nov., sp. nov., a novel marine bacterium of the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 58, 2275–2281. doi: 10.1099/ijs.0.65446-0
Alker, A. T., Delherbe, N., Purdy, T. N., Moore, B. S., and Shikuma, N. J. (2020). Genetic examination of the marine bacterium Pseudoalteromonas luteoviolacea and effects of its metamorphosis-inducing factors. Environ. Microbiol. 22, 4689–4701. doi: 10.1111/1462-2920.15211
Anderson, M. J. (2017). “Permutational multivariate analysis of variance (PERMANOVA),” in Wiley StatsRef: Statistics Reference Online, eds N. Balakrishnan, T. Colton, B. Everitt, W. Piegorsch, F. Ruggeri, and J. L. Teugels (Chichester: John Wiley & Sons, Ltd), 1–15. doi: 10.1002/9781118445112.stat07841
Astudillo-García, C., Bell, J. J., Webster, N. S., Glasl, B., Jompa, J., Montoya, J. M., et al. (2017). Evaluating the core microbiota in complex communities: a systematic investigation: core microbiota in complex communities. Environ. Microbiol. 19, 1450–1462. doi: 10.1111/1462-2920.13647
Auclair, J., Lépine, F., Parent, S., and Villemur, R. (2010). Dissimilatory reduction of nitrate in seawater by a Methylophaga strain containing two highly divergent narG sequences. ISME J. 4, 1302–1313. doi: 10.1038/ismej.2010.47
Bagwell, C. E., Rocque, J. R., Smith, G. W., Polson, S. W., Friez, M. J., Longshore, J. W., et al. (2002). Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39, 113–119. doi: 10.1111/j.1574-6941.2002.tb00912.x
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050.
Blanchet, E., Prado, S., Stien, D., Oliveira da Silva, J., Ferandin, Y., Batailler, N., et al. (2017). Quorum sensing and quorum quenching in the Mediterranean seagrass Posidonia oceanica microbiota. Front. Mar. Sci. 4:218. doi: 10.3389/fmars.2017.00218
Blasius, M., Hübscher, U., and Sommer, S. (2008). Deinococcus radiodurans: what belongs to the survival kit? Crit. Rev. Biochem. Mol. 43, 221–238. doi: 10.1080/10409230802122274
Blazer, V. S., Austin, B., and Austin, D. A. (1988). Bacterial fish pathogens. Environ. Biol. Fish. 21, 77–79.
Bloemberg, G. V., and Lugtenberg, B. J. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350. doi: 10.1016/s1369-5266(00)00183-7
Bock, E., and Wagner, M. (2013). “Oxidation of inorganic nitrogen compounds as an energy source,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer). doi: 10.1007/978-3-642-30141-4_64
Borum, J., Pedersen, O., Greve, T. M., Frankovich, T. A., Zieman, J. C., Fourqurean, J. W., et al. (2005). The potential role of plant oxygen and sulphide dynamics in die-off events of the tropical seagrass, Thalassia testudinum. J. Ecol. 93, 148–158. doi: 10.1111/j.1365-2745.2004.00943.x
Borum, J., Sand-Jensen, K., Binzer, T., Pedersen, O., and Greve, T. M. (2007). “Oxygen movement in seagrasses,” in Seagrasses: Biology, Ecology and Conservation, eds A. W. D. Larkum, J. R. Orth, and C. M. Duarte (Dordrecht: Springer), 255–270. doi: 10.1007/978-1-4020-2983-7_10
Bowman, J. S., Rasmussen, S., Blom, N., Deming, J. W., Rysgaard, S., and Sicheritz-Ponten, T. (2012). Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 6, 11–20. doi: 10.1038/ismej.2011.76
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Brodersen, K. E., Siboni, N., Nielsen, D. A., Pernice, M., Ralph, P. J., Seymour, J., et al. (2018). Seagrass rhizosphere microenvironment alters plant-associated microbial community composition: seagrass rhizosphere microbes. Environ. Microbiol. 20, 2854–2864. doi: 10.1111/1462-2920.14245
Calle, M. L. (2019). Statistical analysis of metagenomics data. Genomics Inform. 17:e6. doi: 10.5808/GI.2019.17.1.e6
Camacho, A. (2009). “Sulfur bacteria,” in Encyclopedia of Inland Waters, ed. G. E. Likens (New York, NY: Academic Press), 261–278. doi: 10.1016/B978-012370626-3.00128-9
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Castro, R. A., Dourado, M. N., Almeida, J. R. D., Lacava, P. T., Nave, A., Melo, I. S. D., et al. (2018). Mangrove endophyte promotes reforestation tree (Acacia polyphylla) growth. Braz. J. Microbiol. 49, 59–66. doi: 10.1016/j.bjm.2017.04.002
Cato, E. P., George, W. L., and Finegold, S. M. (1986). “Genus Clostridium,” in Bergey’s Manual of Systematic Bacteriology, Vol. 2, eds P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (Baltimore, MD: The Williams & Wilkins Co.), 1141–1200.
Celdrán, D., Espinosa, E., Sánchez-Amat, A., and Marín, A. (2012). Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica. Mar. Ecol. Prog. Ser. 456, 21–27. doi: 10.3354/meps09672
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chaparro, J. M., Sheflin, A. M., Manter, D. K., and Vivanco, J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chen, X., Krug, L., Yang, H., Li, H., Yang, M., Berg, G., et al. (2020). Nicotiana tabacum seed endophytic communities share a common core structure and genotype-specific signatures in diverging cultivars. Comput. Struct. Biotechnol. J. 18, 287–295. doi: 10.1016/j.csbj.2020.01.004
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acid Res. 42, D633–D642.
Coleman-Derr, D., Desgarennes, D., Fonseca-Garcia, C., Gross, S., Clingenpeel, S., Woyke, T., et al. (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 209, 798–811. doi: 10.1111/nph.13697
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. J. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Compant, S., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197. doi: 10.1007/s00248-011-9883-y
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Costanza, R., d’Arge, R., de Groot, R., Farber, S., Grasso, M., Hannon, B., et al. (1997). The value of the world’s ecosystem services and natural capital. Nature 387, 253–260. doi: 10.1038/387253a0
Crump, B. C., and Koch, E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 74, 5948–5957. doi: 10.1128/AEM.00952-08
Cúcio, C., Engelen, A. H., Costa, R., and Muyzer, G. (2016). Rhizosphere microbiomes of European seagrasses are selected by the plant but are not species specific. Front. Microbiol. 7:440. doi: 10.3389/fmicb.2016.00440
Cúcio, C., Overmars, L., Engelen, A. H., and Muyzer, G. (2018). Metagenomic analysis shows the presence of bacteria related to free-living forms of sulfur-oxidizing chemolithoautotrophic symbionts in the rhizosphere of the seagrass Zostera marina. Front. Mar. Sci. 5:171. doi: 10.3389/fmars.2018.00171
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Diskin, S., Feygenberg, O., Maurer, D., Droby, S., Prusky, D., and Alkan, N. (2017). Microbiome alterations are correlated with occurrence of postharvest stem-end rot in mango fruit. Phytobiomes 1, 117–127. doi: 10.1094/pbiomes-05-17-0022-r
Doronina, N. V., Trotsenko, Y. A., and Kim, S. W. (2007). Methylophaga aminisulfidivorans sp. nov., a restricted facultatively methylotrophic marine bacterium. Int. J. Syst. Evol. Microbiol. 57, 2096–2101. doi: 10.1099/ijs.0.65139-0
Dutta, B., Gitaitis, R., Smith, S., and Langston, D. Jr. (2014). Interactions of seedborne bacterial pathogens with host and non-host plants in relation to seed infestation and seedling transmission. PLoS One 9:e99215. doi: 10.1371/journal.pone.0099215
Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., and Stackebrandt, E. (2006). The Prokaryotes. New York, NY: Springer New York. doi: 10.1007/0-387-30742-7
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Enebe, M. C., and Babalola, O. O. (2019). The impact of microbes in the orchestration of plants’ resistance to biotic stress: a disease management approach. Appl. Microbiol. Biotechnol. 103, 9–25. doi: 10.1007/s00253-018-9433-3
Ettinger, C. L., Voerman, S. E., Lang, J. M., Stachowicz, J. J., and Eisen, J. A. (2017). Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5:e3246. doi: 10.7717/peerj.3246
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., and Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83:e03391-16. doi: 10.1128/AEM.03391-16
Fidalgo, C., Henriques, I., Rocha, J., Tacão, M., and Alves, A. (2016). Culturable endophytic bacteria from the salt marsh plant Halimione portulacoides: phylogenetic diversity, functional characterization, and influence of metal(loid) contamination. Environ. Sci. Pollut. Res. 23, 10200–10214. doi: 10.1007/s11356-016-6208-1
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165.
Fonseca-García, C., Coleman-Derr, D., Garrido, E., Visel, A., Tringe, S. G., and Partida-Martínez, L. P. (2016). The cacti microbiome: interplay between habitat-filtering and host-specificity. Front. Microbiol. 7:150. doi: 10.3389/fmicb.2016.00150
Garcias-Bonet, N., Arrieta, J. M., de Santana, C. N., Duarte, C. M., and Marbà, N. (2012). Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica). Front. Microbiol. 3:342. doi: 10.3389/fmicb.2012.00342
Garcias-Bonet, N., Eguíluz, V. M., Díaz-Rúa, R., and Duarte, C. M. (2020). Host-association as major driver of microbiome structure and composition in Red Sea seagrass ecosystems. Environ. Microbiol. 23, 2021–2034. doi: 10.1111/1462-2920.15334
Garrity, G. M., Bell, J. A., and Lilburn, T. (2005). “Order V. Thiotrichales ord. nov,” in Bergey’s Manual of Systematic Bacteriology: (The Proteobacteria), part B (The Gammaproteobacteria), 2nd Edn. Vol. 2, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (New York, NY: Springer), 131. doi: 10.1007/0-387-28022-7_5
Gopalakrishnan, S., Sathya, A., Vijayabharathi, R., Varshney, R. K., Gowda, C. L., and Krishnamurthy, L. (2015). Plant growth promoting rhizobia: challenges and opportunities. Biotech 5, 355–377. doi: 10.1007/s13205-014-0241-x
Hamilton, T. L., Bovee, R. J., Thiel, V., Sattin, S. R., Mohr, W., Schaperdoth, I., et al. (2014). Coupled reductive and oxidative sulfur cycling in the phototrophic plate of a meromictic lake. Geobiology 12, 451–468. doi: 10.1111/gbi.12092
Hamisi, M., Lyimo, T., Muruke, M., and Bergman, B. (2009). Nitrogen fixation by epiphytic and epibenthic diazotrophs associated with seagrass meadows along the Tanzanian coast, Western Indian Ocean. Aquat. Microb. Ecol. 57, 33–42. doi: 10.3354/ame01323
Han, Y., and Perner, M. (2014). The role of hydrogen for Sulfurimonas denitrificans’ metabolism. PLoS One 9:e106218. doi: 10.1371/journal.pone.0106218
Handley, K. M., and Lloyd, J. R. (2013). Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 4:136. doi: 10.3389/fmicb.2013.00136
Hanke, A., Berg, J., Hargesheimer, T., Tegetmeyer, H. E., Sharp, C. E., and Strous, M. (2016). Selective pressure of temperature on competition and cross-feeding within denitrifying and fermentative microbial communities. Front. Microbiol. 6:1461. doi: 10.3389/fmicb.2015.01461
Hansen, J., Udy, J., Perry, C., Dennison, W., and Lomstein, B. (2000). Effect of the seagrass Zostera capricorni on sediment microbial processes. Mar. Ecol. Prog. Ser. 199, 83–96. doi: 10.3354/meps199083
Hardoim, P. R., Van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/mmbr.00050-14
Herbert, R. A. (1999). Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23, 563–590. doi: 10.1111/j.1574-6976.1999.tb00414.x
Hillman, K., McComb, A. J., and Walker, D. I. (1995). The distribution, biomass and primary production of the seagrass Halophila ovalis in the Swan/Canning Estuary, Western Australia. Aquat. Bot. 51, 1–54. doi: 10.1016/0304-3770(95)00466-D
Holmer, M., and Nielsen, S. (1997). Sediment sulfur dynamics related to biomass- density patterns in Zostera marina (eelgrass) beds. Mar. Ecol. Prog. Ser. 146, 163–171. doi: 10.3354/meps146163
Huang, X. F., Liu, Y. J., Dong, J. D., Qu, L. Y., Zhang, Y. Y., Wang, F. Z., et al. (2014). Mangrovibacterium diazotrophicum gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from a mangrove sediment, and proposal of Prolixibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 64, 875–881. doi: 10.1099/ijs.0.052779-0
Hurtado-McCormick, V., Kahlke, T., Petrou, K., Jeffries, T., Ralph, P. J., and Seymour, J. R. (2019). Regional and microenvironmental scale characterization of the Zostera muelleri seagrass microbiome. Front. Microbiol. 10:1011. doi: 10.3389/fmicb.2019.01011
Husband, J. D., Kiene, R. P., and Sherman, T. D. (2012). Oxidation of dimethylsulfoniopropionate (DMSP) in response to oxidative stress in Spartina alterniflora and protection of a non-DMSP producing grass by exogenous DMSP+ acrylate. Environ. Exp. Bot. 79, 44–48. doi: 10.1016/j.envexpbot.2012.01.006
Inaba, N., Trainer, V. L., Onishi, Y., Ishii, K.-I., Wyllie-Echeverria, S., and Imai, I. (2017). Algicidal and growth-inhibiting bacteria associated with seagrass and macroalgae beds in Puget Sound, WA, USA. Harmful Algae 62, 136–147. doi: 10.1016/j.hal.2016.04.004
Isaksen, M., and Finster, K. (1996). Sulphate reduction in the root zone of the seagrass Zostera noltii on the intertidal flats of a coastal lagoon (Arcachon, France). Mar. Ecol. Prog. Ser. 137, 187–194. doi: 10.3354/meps137187
Janvier, M., Frehel, C., Grimont, F., and Gasser, F. (1985). Methylophaga marina gen. nov., sp. nov. and Methylophaga thalassica sp. nov., marine methylotrophs. Int. J. Syst. Bacteriol. 35, 131–139. doi: 10.1099/00207713-35-2-131
Jensen, S. I., Kühl, M., and Priemé, A. (2007). Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina: root-associated bacteria. FEMS Microbiol. Ecol. 62, 108–117. doi: 10.1111/j.1574-6941.2007.00373.x
Jiang, Y. F., Ling, J., Dong, J. D., Chen, B., Zhang, Y. Y., Zhang, Y. Z., et al. (2015). Illumina-based analysis the microbial diversity associated with Thalassia hemprichii in Xincun Bay, South China Sea. Ecotoxicology 24, 1548–1556. doi: 10.1007/s10646-015-1511-z
Johnston-Monje, D., and Raizada, M. N. (2011). “Plant and endophyte relationships,” in Comprehensive Biotechnology, ed. M. Moo-Young (Amsterdam: Elsevier), 713–727. doi: 10.1016/B978-0-08-088504-9.00264-6
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966. doi: 10.1093/molbev/msn146
Jonkers, H. M., Bergeijk, S. A., and Gemerden, H. (2000). Microbial production and consumption of dimethyl sulfide (DMS) in a seagrass (Zostera noltii) dominated marine intertidal sediment ecosystem (Bassin d’Arcachon, France). FEMS Microbiol. Ecol. 31, 163–172. doi: 10.1111/j.1574-6941.2000.tb00681.x
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296, 643–645. doi: 10.1038/296643a0
Jose, P. A., Sivakala, K. K., and Jebakumar, S. R. D. (2014). Molecular Phylogeny and Plant Growth Promoting Traits of Endophytic Bacteria Isolated from Roots of Seagrass Cymodocea serrulata. Niscair Publications. Available online at: http://nopr.niscair.res.in/handle/123456789/28647 doi: 10.1038/296643a0
Kandel, S. L., Joubert, P. M., and Doty, S. L. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. doi: 10.3390/microorganisms5040077
Kang, I., Lim, Y., and Cho, J. C. (2018). Complete genome sequence of Granulosicoccus antarcticus type strain IMCC3135T, a marine gammaproteobacterium with a putative dimethylsulfoniopropionate demethylase gene. Mar. Genomics 37, 176–181. doi: 10.1016/j.margen.2017.11.005
Kelsey, P. J., and Western Australia, Department of Water (2010). Hydrological and Nutrient Modelling of the Swan Canning Coastal Catchments: Coastal Catchment Initiative Project. Perth, WA: Dept. of Water.
Kendrick, G. A., Orth, R. J., Statton, J., Hovey, R., Ruiz Montoya, L., Lowe, R. J., et al. (2017). Demographic and genetic connectivity: the role and consequences of reproduction, dispersal and recruitment in seagrasses. Biol. Rev. 92, 921–938. doi: 10.1111/brv.12261
Kilminster, K., and Garland, J. (2009). Aerobic heterotrophic microbial activity associated with seagrass roots: effects of plant type and nutrient amendment. Aquat. Microb. Ecol. 57, 57–68. doi: 10.3354/ame01332
Koch, M. S., and Erskine, J. M. (2001). Sulfide as a phytotoxin to the tropical seagrass Thalassia testudinum: interactions with light, salinity and temperature. J. Exp. Mar. Biol. Ecol. 266, 81–95. doi: 10.1016/S0022-0981(01)00339-2
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kramer, C. Y. (1956). Extension of multiple range tests to group means with unequal numbers of replications. Biometrics 12, 307–310. doi: 10.2307/3001469
Kramer, C. Y. (1957). Extension of multiple range tests to group correlated adjusted means. Biometrics 13, 13–18. doi: 10.2307/3001898
Kuever, J., Rainey, F. A., and Widdel, F. (2005). “Genus II. Desulfocapsa,” in Bergey’s Manual of Systematic Bacteriology: (The Proteobacteria), Part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), 2nd Edn. Vol. 2, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (New York, NY: Springer), 992–994.
Kuo, J., and den Hartog, C. (2006). “Seagrass morphology, anatomy, and ultrastructure,” in Seagrasses: Biology, Ecology and Conservation, eds A. W. D. Larkum, R. J. Orth, and C. M. Duarte (Dordrecht: Springer Netherlands), 51–87. doi: 10.1007/1-4020-2983-7_3
Kuo, J., and Kirkman, H. (1992). Fruits, seeds and germination in the seagrass Halophila ovalis (Hydrocharitaceae). Bot. Mar. 35, 197–204. doi: 10.1515/botm.1992.35.3.197
Kurilenko, V. V., Christen, R., Zhukova, N. V., Kalinovskaya, N. I., Mikhailov, V. V., Crawford, R. J., et al. (2010). Granulosicoccus coccoides sp. nov., isolated from leaves of seagrass (Zostera marina). Int. J. Syst. Evol. Microbiol. 60, 972–976. doi: 10.1099/ijs.0.013516-0
Küsel, K., Pinkart, H. C., Drake, H. L., and Devereux, R. (1999). Acetogenic and sulfate-reducing bacteria inhabiting the rhizoplane and deep cortex cells of the sea grass Halodule wrightii. Appl. Environ. Microbiol. 65, 5117–5123. doi: 10.1128/aem.65.11.5117-5123.1999
Küsel, K., Trinkwalter, T., Drake, H. L., and Devereux, R. (2006). Comparative evaluation of anaerobic bacterial communities associated with roots of submerged macrophytes growing in marine or brackish water sediments. J. Exp. Mar. Biol. Ecol. 337, 49–58. doi: 10.1016/j.jembe.2006.06.004
Lamers, L. P. M., Govers, L. L., Janssen, I. C. J. M., Geurts, J. J. M., Van der Welle, M. E. W., Van Katwijk, M. M., et al. (2013). Sulfide as a soil phytotoxin—a review. Front. Plant Sci. 4:268. doi: 10.3389/fpls.2013.00268
Lasa, A. V., Mašínová, T., Baldrian, P., and Fernández-López, M. (2019). Bacteria from the endosphere and rhizosphere of Quercus spp. use mainly cell wall-associated enzymes to decompose organic matter. PLoS One 14:e0214422. doi: 10.1371/journal.pone.0214422
Lavery, P. S., Mateo, M. Á, Serrano, O., and Rozaimi, M. (2013). Variability in the carbon storage of seagrass habitats and its implications for global estimates of blue carbon ecosystem service. PLoS One 8:e73748. doi: 10.1371/journal.pone.0073748
Lehnen, N., Marchant, H. K., Schwedt, A., Milucka, J., Lott, C., Weber, M., et al. (2016). High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. Syst. Appl. Microbiol. 39, 476–483. doi: 10.1016/j.syapm.2016.08.004
Lemanceau, P., Blouin, M., Muller, D., and Moënne-Loccoz, Y. (2017). Let the core microbiota be functional. Trends Plant Sci. 22, 583–595. doi: 10.1016/j.tplants.2017.04.008
Lepoint, G., Millet, S., Dauby, P., Gobert, S., and Bouquegneau, J. (2002). Annual nitrogen budget of the seagrass Posidonia oceanica as determined by in situ uptake experiments. Mar. Ecol. Prog. Ser. 237, 87–96. doi: 10.3354/meps237087
Les, D. H., Cleland, M. A., and Waycott, M. (1997). Phylogenetic studies in Alismatidae, II: evolution of marine angiosperms (Seagrasses) and Hydrophily. Syst. Bot. 22, 443–463. doi: 10.2307/2419820
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M., et al. (2017). Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552. doi: 10.3389/fmicb.2017.02552
Liu, S., Jiang, Z., Deng, Y., Wu, Y., Zhang, J., Zhao, C., et al. (2018). Effects of nutrient loading on sediment bacterial and pathogen communities within seagrass meadows. Microbiologyopen 7:e00600. doi: 10.1002/mbo3.600
Liu, X. L., Liu, S. L., Liu, M., Kong, B. H., Liu, L., and Li, Y. H. (2014). A primary assessment of the endophytic bacterial community in a xerophilous moss (Grimmia montana) using molecular method and cultivated isolates. Braz. J. Microbiol. 45, 165–173. doi: 10.1590/S1517-83822014000100022
Lopez-Velasco, G., Carder, P. A., Welbaum, G. E., and Ponder, M. A. (2013). Diversity of the spinach (Spinacia oleracea) spermosphere and phyllosphere bacterial communities. FEMS Microbiol. Lett. 346, 146–154. doi: 10.1111/1574-6968.12216
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lu, W., Tomas, F., and Mueller, R. S. (2020). Nutrient enrichment increases size of Zostera marina shoots and enriches for sulfur and nitrogen cycling bacteria in root-associated microbiomes. FEMS Microbiol. Ecol. 96:fiaa129. doi: 10.1093/femsec/fiaa129
Lu, Y., Rosencrantz, D., Liesack, W., and Conrad, R. (2006). Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ. Microbiol. 8, 1351–1360. doi: 10.1111/j.1462-2920.2006.01028.x
Lucas-Elío, P., Goodwin, L., Woyke, T., Pitluck, S., Nolan, M., Kyrpides, N. C., et al. (2012). Complete genome sequence of the melanogenic marine bacterium Marinomonas mediterranea type strain (MMB-1T). Stand. Genomic Sci. 6, 63–73. doi: 10.4056/sigs.2545743
Maggini, V., Mengoni, A., Gallo, E. R., Biffi, S., Fani, R., Firenzuoli, F., et al. (2019). Tissue specificity and differential effects on in vitro plant growth of single bacterial endophytes isolated from the roots, leaves and rhizospheric soil of Echinacea purpurea. BMC Plant Biol. 19:284. doi: 10.1186/s12870-019-1890-z
Martin, B. C., Alarcon, M. S., Gleeson, D., Middleton, J. A., Fraser, M. W., Ryan, M. H., et al. (2020). Root microbiomes as indicators of seagrass health. FEMS Microbiol. Ecol. 96:fiz201. doi: 10.1093/femsec/fiz201
Martin, B. C., Bougoure, J., Ryan, M. H., Bennett, W. W., Colmer, T. D., Joyce, N. K., et al. (2019). Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 13, 707–719. doi: 10.1038/s41396-018-0308-5
Martin, B. C., Gleeson, D., Statton, J., Siebers, A. R., Grierson, P., Ryan, M. H., et al. (2018). Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front. Microbiol. 8:2667. doi: 10.3389/fmicb.2017.02667
McClung, C. R., and Patriquin, D. G. (1980). Isolation of a nitrogen-fixing Campylobacter species from the roots of Spartina alterniflora Loisel. Can. J. Microbiol. 26, 881–886. doi: 10.1139/m80-153
McClung, C. R., Patriquin, D. G., and Davis, R. E. (1983). Campylobacter nitrofigilis sp. nov., a nitrogen-fixing bacterium associated with roots of Spartina alterniflora Loisel. Int. J. Syst. Bacteriol. 33, 605–612. doi: 10.1099/00207713-33-3-605
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
McRoy, C. P., and Goering, J. J. (1974). Nutrient transfer between the seagrass Zostera marina and its epiphytes. Nature 248, 173–174. doi: 10.1038/248173a0
Mejia, A. Y., Rotini, A., Lacasella, F., Bookman, R., Thaller, M. C., Shem-Tov, R., et al. (2016). Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecol. Indic. 60, 1150–1163. doi: 10.1016/j.ecolind.2015.09.014
Mesa, J., Mateos-Naranjo, E., Caviedes, M. A., Redondo-Gómez, S., Pajuelo, E., and Rodríguez-Llorente, I. D. (2015). Endophytic cultivable bacteria of the metal bioaccumulator Spartina maritima improve plant growth but not metal uptake in polluted marshes soils. Front. Microbiol. 6:1450. doi: 10.3389/fmicb.2015.01450
Minamisawa, K., Nishioka, K., Miyaki, T., Ye, B., Miyamoto, T., You, M., et al. (2004). Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70, 3096–3102. doi: 10.1128/aem.70.5.3096-3102.2004
Miroshnichenko, M. L., Kostrikina, N. A., Chernyh, N. A., Pimenov, N. V., Tourova, T. P., Antipov, A. N., et al. (2003). Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int. J. Syst. Evol. Microbiol. 53, 323–329. doi: 10.1099/ijs.0.02390-0
Mitter, B., Pfaffenbichler, N., Flavell, R., Compant, S., Antonielli, L., Petric, A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8:11. doi: 10.3389/fmicb.2017.00011
Mtwana Nordlund, L., Koch, E. W., Barbier, E. B., and Creed, J. C. (2016). Seagrass ecosystem services and their variability across genera and geographical regions. PLoS One 11:e0163091. doi: 10.1371/journal.pone.0163091
Muyzer, G., and Stams, A. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Nakagawa, T., Tsuchiya, Y., Ueda, S., Fukui, M., and Takahashi, R. (2019). Eelgrass sediment microbiome as a nitrous oxide sink in brackish Lake Akkeshi, Japan. Microbes Environ. 34, 13–22. doi: 10.1264/jsme2.ME18103
Nelson, E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Nemecek-Marshall, M., MacDonald, R. C., Franzen, J. J., Wojciechowski, C. L., and Fall, R. (1995). Methanol emission from leaves (enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development). Plant Physiol. 108, 1359–1368. doi: 10.1104/pp.108.4.1359
Nielsen, L. B., Finster, K., Welsh, D. T., Donelly, A., Herbert, R. A., De Wit, R., et al. (2001). Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 3, 63–71. doi: 10.1046/j.1462-2920.2001.00160.x
O’Callaghan, M. (2016). Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl. Microbiol. Biotechnol. 100, 5729–5746. doi: 10.1007/s00253-016-7590-9
O’Donohue, M. J., Moriarty, D. J. W., and Mac Rae, I. C. (1991). Nitrogen fixation in sediments and the rhizosphere of the seagrass Zostera capricorni. Microb. Ecol. 22, 53–64. doi: 10.1007/bf02540212
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R Package Version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan
Orth, R. J., Harwell, M. C., Bailey, E. M., Bartholomew, A., Jawad, J. T., Lombana, A. V., et al. (2000). A review of issues in seagrass seed dormancy and germination: implications for conservation and restoration. Mar. Ecol. Prog. Ser. 200, 277–288. doi: 10.3354/meps200277
Orth, R. J., Harwell, M. C., and Inglis, G. J. (2007). “Ecology of seagrass seeds and seagrass dispersal processes,” in Seagrasses: Biology, Ecology and Conservation, eds A. W. D. Larkum, R. J. Orth, and C. M. Duarte (Dordrecht: Springer), 111–133. doi: 10.1007/1-4020-2983-7_5
Palacios, R., and Newton, W. E. (2005). Genomes and Genomics of Nitrogen-Fixing Organisms, Nitrogen Fixation. Dordrecht: Springer.
Puente, M. E., Li, C. Y., and Bashan, Y. (2009). Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 66, 402–408. doi: 10.1016/j.envexpbot.2009.04.007
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R. Foundation for Statistical Computing.
Reynolds, L. K., Waycott, M., McGlathery, K. J., and Orth, R. J. (2016). Ecosystem services returned through seagrass restoration: restoration of ecosystem services. Restor. Ecol. 24, 583–588. doi: 10.1111/rec.12360
Romanenko, L. A., Uchino, M., Kalinovskaya, N. I., and Mikhailov, V. V. (2008). Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol. Res. 163, 633–644. doi: 10.1016/j.micres.2006.10.001
Rosenberg, E. (2014). “The family Chitinophagaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer Berlin Heidelberg), 493–495. doi: 10.1007/978-3-642-38954-2_137
Rotini, A., Conte, C., Seveso, D., Montano, S., Galli, P., Vai, M., et al. (2020). Daily variation of the associated microbial community and the Hsp60 expression in the Maldivian seagrass Thalassia hemprichii. J. Sea Res. 156:101835. doi: 10.1016/j.seares.2019.101835
Rotini, A., Mejia, A. Y., Costa, R., Migliore, L., and Winters, G. (2017). Ecophysiological plasticity and bacteriome shift in the seagrass Halophila stipulacea along a depth gradient in the Northern Red Sea. Front. Plant Sci. 7:2015. doi: 10.3389/fpls.2016.02015
Sakami, T., Sakamoto, S., Takagi, S., Inaba, N., and Imai, I. (2017). Distribution of three algicidal Alteromonas sp. strains in seagrass beds and surrounding areas in the Seto Inland Sea, Japan. Fish. Sci. 83, 113–121. doi: 10.1007/s12562-016-1048-y
Saldierna Guzmán, J. P., Nguyen, K., and Hart, S. C. (2020). Simple methods to remove microbes from leaf surfaces. J. Basic Microbiol. 60, 730–734. doi: 10.1002/jobm.202000035
Sánchez-López, A. S., Pintelon, I., Stevens, V., Imperato, V., Timmermans, J. P., González-Chávez, C., et al. (2018). Seed endophyte microbiome of Crotalaria pumila unpeeled: identification of plant-beneficial methylobacteria. Int. J. Mol. Sci. 19:291. doi: 10.3390/ijms19010291
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Shade, A., Jacques, M. A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shade, A., McManus, P. S., and Handelsman, J. (2013). Unexpected diversity during community succession in the apple flower microbiome. mBio 4:e00602-12. doi: 10.1128/mBio.00602-12
Shannon, C. E., and Weaver, W. (1964). The Mathematical Theory of Communication. Urbana, IL: University of Illinois Press, 131.
Short, F., Carruthers, T., Dennison, W., and Waycott, M. (2007). Global seagrass distribution and diversity: a bioregional model. J. Exp. Mar. Biol. Ecol. 350, 3–20. doi: 10.1016/j.jembe.2007.06.012
Short, F. T., Moore, G. E., and Peyton, K. A. (2010). Halophila ovalis in the tropical Atlantic Ocean. Aquat. Bot. 93, 141–146. doi: 10.1016/j.aquabot.2010.05.001
Smith, R. D., Pregnall, A. M., and Alberte, R. S. (1988). Effects of anaerobiosis on root metabolism of Zostera marina (eelgrass): implications for survival in reducing sediments. Mar. Biol. 98, 131–141. doi: 10.1007/BF00392668
Statton, J., Sellers, R., Dixon, K. W., Kilminster, K., Merritt, D. J., and Kendrick, G. A. (2017). Seed dormancy and germination of Halophila ovalis mediated by simulated seasonal temperature changes. Estuar. Coast. Shelf Sci. 198, 156–162. doi: 10.1016/j.ecss.2017.08.045
Stohr, C., and Ullrich, R. W. (2002). Generation and possible roles of NO in plant roots and their apoplastic space. J. Exp. Bot. 53, 2293–2303. doi: 10.1093/jxb/erf110
Straub, K. L., Rainey, F. A., and Widdel, F. (1999). Rhodovulum iodosum sp. nov. and Rhodovulum robiginosum sp. nov., two new marine phototrophic ferrous-iron-oxidizing purple bacteria. Int. J. Syst. Bacteriol. 49, 729–735. doi: 10.1099/00207713-49-2-729
Stuij, T. (2018). Distinct Microbiomes in Three Tropical Seagrasses Around the Island of Curaçao: Halophila stipulacea, Halodule wrightii and Thalassia testudinum. Master’s thesis. Faro: Universidade do Algarve, 22.
Sun, F., Zhang, X., Zhang, Q., Liu, F., Zhang, J., and Gong, J. (2015). Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Appl. Environ. Microbiol. 81, 6901–6914. doi: 10.1128/AEM.01382-15
Sweet, M. J., Croquer, A., and Bythell, J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52. doi: 10.1007/s00338-010-0695-1
Tarquinio, F., Hyndes, G. A., Laverock, B., Koenders, A., and Säwström, C. (2019). The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 366:fnz057. doi: 10.1093/femsle/fnz057
Thomas, F., Giblin, A. E., Cardon, Z. G., and Sievert, S. M. (2014). Rhizosphere heterogeneity shapes abundance and activity of sulfur-oxidizing bacteria in vegetated salt marsh sediments. Front. Microbiol. 5:309. doi: 10.3389/fmicb.2014.00309
Touchette, B. W., and Burkholder, J. M. (2000). Review of nitrogen and phosphorus metabolism in seagrasses. J. Exp. Mar. Biol. Ecol. 250, 133–167. doi: 10.1016/S0022-0981(00)00195-7
Trevathan-Tackett, S. M., Allnutt, T. R., Sherman, C. D., Richardson, M. F., Crowley, T. M., and Macreadie, P. I. (2020). Spatial variation of bacterial and fungal communities of estuarine seagrass leaf microbiomes. Aquat. Microb. Ecol. 84, 59–74. doi: 10.3354/ame01926
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants: bacterial seed endophytes. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Tukey, J. W. (1953). “The problem of multiple comparisons,” in The Collected Works of John W. Tukey, Vol. 8, ed. H. I. Braun (New York, NY: Chapman & Hall).
Turner, T. R., James, E. K., and Poole, P. S. (2013). The plant microbiome. Genome Biol. 14:209. doi: 10.1186/gb-2013-14-6-209
Ugarelli, K., Chakrabarti, S., Laas, P., and Stingl, U. (2017). The seagrass holobiont and its microbiome. Microorganisms 5:81. doi: 10.3390/microorganisms5040081
Van Der Geest, M., Van Der Heide, T., Holmer, M., and De Wit, R. (2020). First field-based evidence that the seagrass-lucinid mutualism can mitigate sulfide stress in seagrasses. Front. Mar. Sci. 7:11. doi: 10.3389/fmars.2020.00011
van der Heide, T., Govers, L. L., de Fouw, J., Olff, H., van der Geest, M., van Katwijk, M. M., et al. (2012). A three-stage symbiosis forms the foundation of seagrass ecosystems. Science 336, 1432–1434. doi: 10.1126/science.1219973
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Varon-Lopez, M., Dias, A. C. F., Fasanella, C. C., Durrer, A., Melo, I. S., Kuramae, E. E., et al. (2014). Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments: sulphur cycling in mangroves. Environ. Microbiol. 16, 845–855. doi: 10.1111/1462-2920.12237
Verma, S. K., Kingsley, K., Irizarry, I., Bergen, M., Kharwar, R. N., and White, J. F. (2017). Seed-vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 122, 1680–1691. doi: 10.1111/jam.13463
Větrovský, T., Steffen, K. T., and Baldrian, P. (2014). Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS One 9:e89108. doi: 10.1371/journal.pone.0089108
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Welsh, D. T. (2000). Nitrogen fixation in seagrass meadows: regulation, plant-bacteria interactions and significance to primary productivity. Ecol. Lett. 3, 58–71. doi: 10.1046/j.1461-0248.2000.00111.x
Weyens, N., van der Lelie, D., Taghavi, S., Newman, L., and Vangronsveld, J. (2009). Exploiting plant–microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 27, 591–598. doi: 10.1016/j.tibtech.2009.07.006
White, J. F., Kingsley, K. L., Butterworth, S., Brindisi, L., Gatei, J. W., Elmore, M. T., et al. (2019). “Seed-vectored microbes: their roles in improving seedling fitness and competitor plant suppression,” in Seed Endophytes, eds S. Verma and J. White Jr. (Cham: Springer), 3–20. doi: 10.1007/978-3-030-10504-4_1
Widdel, F. (2006). “The genus Desulfotomaculum,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. L. Schleifer, and E. Stackebrandt (Berlin: Springer Berlin Heidelberg), 787–794. doi: 10.1007/0-387-30744-3_25
Wirth, J. S., Wang, T., Huang, Q., White, R. H., and Whitman, W. B. (2020). Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria species. mBio 11:e00329-20. doi: 10.1128/mBio.00329-20
Wu, X. L., Yu, S. L., Gu, J., Zhao, G. F., and Chi, C. Q. (2009). Filomicrobium insigne sp. nov., isolated from an oil-polluted saline soil. Int. J. Syst. Evol. Microbiol. 59, 300–305. doi: 10.1099/ijs.0.65758-0
Wu, Y. H., Xu, L., Meng, F. X., Zhang, D. S., Wang, C. S., Oren, A., et al. (2014). Altererythrobacter atlanticus sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 64, 116–121. doi: 10.1099/ijs.0.052951-0
Xu, T., Yu, M., Lin, H., Zhang, Z., Liu, J., and Zhang, X. H. (2015). Genomic insight into Aquimarina longa SW024 T: its ultra-oligotrophic adapting mechanisms and biogeochemical functions. BMC Genomics 16:772. doi: 10.1186/s12864-015-2005-3
Keywords: Halophila ovalis, seagrass restoration, seed microbiome, core, bacteria, amplicon sequencing
Citation: Tarquinio F, Attlan O, Vanderklift MA, Berry O and Bissett A (2021) Distinct Endophytic Bacterial Communities Inhabiting Seagrass Seeds. Front. Microbiol. 12:703014. doi: 10.3389/fmicb.2021.703014
Received: 30 April 2021; Accepted: 12 July 2021;
Published: 21 September 2021.
Edited by:
Akiko Sugio, Institut de Génétique, Environnement et Protection des Plantes, Institut National de la Recherche Agronomique, FranceReviewed by:
Ram Naresh Bharagava, Babasaheb Bhimrao Ambedkar University, IndiaCopyright © 2021 Tarquinio, Attlan, Vanderklift, Berry and Bissett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flavia Tarquinio, ZmxhdmlhLnRhcnF1aW5pb0Bjc2lyby5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.