- 1State Key Laboratory of Crop Stress Biology for Arid Areas, Shaanxi Key Laboratory of Agricultural and Environmental Microbiology, College of Life Sciences, Northwest A&F University, Yangling, China
- 2Department of Entomology, College of Plant Protection, Northwest A&F University, Yangling, China
Aerobactin is a citrate-hydroxamate siderophore that is critical for the virulence of pathogenic enteric bacteria. However, although the aerobactin-producing iucABCD-iutA operon is distributed widely in the genomes of Yersinia species, none of the pathogenic Yersinia spp. was found to produce aerobactin. Here, we showed that the iucABCD-iutA operon in the food-borne enteric pathogen Yersinia pseudotuberculosis YPIII is a functional siderophore system involved in iron acquisition. The expression of the operon was found to be directly repressed by the ferric uptake regulator (Fur) in an iron concentration-dependent manner. In addition, we demonstrated that the aerobactin-mediated iron acquisition contributes to bacterial growth under iron-limited conditions. Moreover, we provided evidence that aerobactin plays important roles in biofilm formation, resistance to oxidative stress, ROS removal, and virulence of Y. pseudotuberculosis. Overall, our study not only uncovered a novel strategy of iron acquisition in Y. pseudotuberculosis but also highlighted the importance of aerobactin in the pathogenesis of Y. pseudotuberculosis.
Introduction
Iron is an irreplaceable metal for most of living organisms, as it is necessary for the activity of functional proteins or regulators that are involved in many cellular processes such as the tricarboxylic acid (TCA) cycle, DNA precursor synthesis, and oxygen metabolism (Neilands, 1981; Galaris and Pantopoulos, 2008). However, although iron is one of the most abundant elements on earth, its bioavailability is extremely restricted because it forms insoluble ferric hydroxide complexes under aerobic conditions and neutral pH. In higher organisms, iron is further restricted by the formation of high-affinity complexes with proteins such as ferritin, transferrin, and lactoferrin, an effective immune mechanism termed nutritional immunity (Cassat and Skaar, 2013; Nairz and Weiss, 2020). To overcome iron restriction, bacteria have evolved many effective strategies to scavenge iron from their surroundings (Neilands, 1981; Weinberg, 1989; Lin et al., 2017), and the most commonly used iron scavenging strategy is the production and secretion of siderophores.
Siderophores are low-molecular-weight (500–1,500 Da) high-affinity iron-chelating compounds for solubilization and transport of ferric iron into bacterial cells (Neilands, 1995). In the extracellular milieu, secreted siderophores form soluble iron–siderophore complexes with ferric iron. The soluble iron–siderophore complexes are actively transported into bacterial cells via specific outer membrane receptors, then ferric iron is released and reduced to ferrous iron, which can be used for cellular needs (Chakraborty et al., 2007). Siderophores are not only essential for the growth of most pathogenic bacteria but also play important roles in non-iron metal transport, protection against oxidative stress, antibiotic activity, interspecies interactions, and virulence (Kramer et al., 2020). The production of siderophores is strictly regulated in an iron concentration-dependent manner to maintain iron homeostasis. Ferric uptake regulator (Fur) is the key regulator, which acts as a transcriptional repressor of siderophore synthesis genes by utilizing ferrous iron as a corepressor (Ratledge and Dover, 2000; Miethke and Marahiel, 2007). In iron-rich environments, the ferrous iron-Fur dimer binds to the promoter regions of siderophore synthesis genes to block transcription. In iron-deplete environments, Fur no longer contains ferrous iron; therefore, it is detached from the siderophore gene promoter to relieve repression and the eventual synthesis of siderophores (Troxell and Hassan, 2013).
Siderophores show a high variety in structure and function, which can be divided into four types depending on their chemical nature. These are catecholates, hydroxamates, carboxylates, and phenolates, and mixtures of at least two classes are also common (Hider and Kong, 2010). Aerobactin, a citrate-hydroxamate siderophore, is produced by many pathogenic bacteria. The aerobactin operon encodes four biosynthetic enzymes (IucABCD) and a transmembrane transporter (IutA) involved in aerobactin siderophore biosynthesis and transport. First, monooxygenase IucD catalyzes N6 hydroxylation of L-lysine to generate N6-hydroxy-Llysine (hLys). Second, acetyltransferase IucB transfers an acetyl from acetyl CoA to hLys, which subsequently yields N6-acetyl-N6-hydroxy-L-lysine (ahLys). Finally, aerobactin synthetase IucA adds one ahLys to a primary carboxylate of citrate to form citryl-ahLys, and a second ahlys is added to citryl-ahLys by the other aerobactin synthetase IucC to produce aerobactin siderophore (Di Lorenzo and Stork, 2014). Then the outer membrane receptor IutA transports ferric-aerobactin into the periplasm in a TonB-dependent manner (Di Lorenzo and Stork, 2014). The aerobactin pathway has been detected in a number of pathogenic enteric bacteria including Escherichia, Vibrio, Salmonella, and Shigella, which enhances the virulence of many of these bacteria (Sheldon et al., 2016).
Yersinia pseudotuberculosis is a food-borne enterobacterial pathogen, which is one of the three human-pathogenic Yersinia species (Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis) (Achtman et al., 1999). Like many other pathogens, pathogenic Yersinia contains numerous iron acquisition systems to ensure optimal iron uptake (Forman et al., 2010), including three ferrous transporters (YfeABC transporter, Yfe; Feo transporter, Feo; and Fet transporter, Fet), three ferric transporters (YfuABC transporter, Yfu; YiuABC transporter, Yiu; and heme transporter, Hmu), and three siderophore-dependent systems (yersiniabactin, Ybt; pseudochelin, Pch; and yersiniachelin, Ych) (Rakin et al., 2012; Perry et al., 2015). So far, five of these (Ybt, Yfe, Yfu, Yiu, and Hmu) have been proved functional in Y. pestis. However, Ybt, Yfe, and Hmu systems are the only functional iron transport systems that have been tested in Y. pseudotuberculosis (Forman et al., 2010; Schwiesow et al., 2018), and these systems have not been extensively studied. Meanwhile, bioinformatics studies highlight putative iron transport systems in Y. pseudotuberculosis, of which the roles in iron uptake functionality have not been identified.
In this study, we identified a functional aerobactin-producing iucABCD-iutA operon in Y. pseudotuberculosis YPIII, which is directly regulated by the Fur regulator. Our results demonstrated that the aerobactin-mediated iron transport system plays crucial roles in iron uptake, biofilm formation, oxidative stress resistance, and virulence of Y. pseudotuberculosis. Our study not only uncovered a novel strategy of iron acquisition in Y. pseudotuberculosis but also highlighted the importance of aerobactin in the pathogenesis of Y. pseudotuberculosis.
Results
Aerobactin Production From the iucABCD-iutA Operon in Yersinia pseudotuberculosis
Genome analysis of Y. pseudotuberculosis YPIII identified a putative aerobactin-producing iucABCD-iutA operon, which is similar to the functional characterized iucABCD-iutA operon in Escherichia coli, Shigella flexneri, Vibrio mimicus, and Klebsiella pneumoniae in the operon structure (Figure 1A; Payne, 1980; Nassif and Sansonetti, 1986; Okujo and Yamamoto, 1994; Peigne et al., 2009). In this Y. pseudotuberculosis iucABCD-iutA operon, the first gene iucA (ypk_0786) encodes a IucA ortholog with 63% identity to an E. coli IucA that is implicated in couple ahLys onto the primary carboxylates of citrate to synthesize aerobactin siderophore (Figure 1A; Neilands, 1992; Marchler-Bauer et al., 2013). Downstream of iucA, aerobactin biosynthesis-related genes iucB (ypk_0785), iucC (ypk_0784), and iucD (ypk_0783) were present, which encode acetyltransferase, aerobactin synthase, and monooxygenase, respectively. In addition, ypk_0782 encodes a putative outer membrane receptor protein for the ferric–aerobactin complex, which shared 67% amino acid sequence identity with the TonB-dependent membrane receptor IutA in E. coli (Figure 1A; Marchler-Bauer et al., 2013).
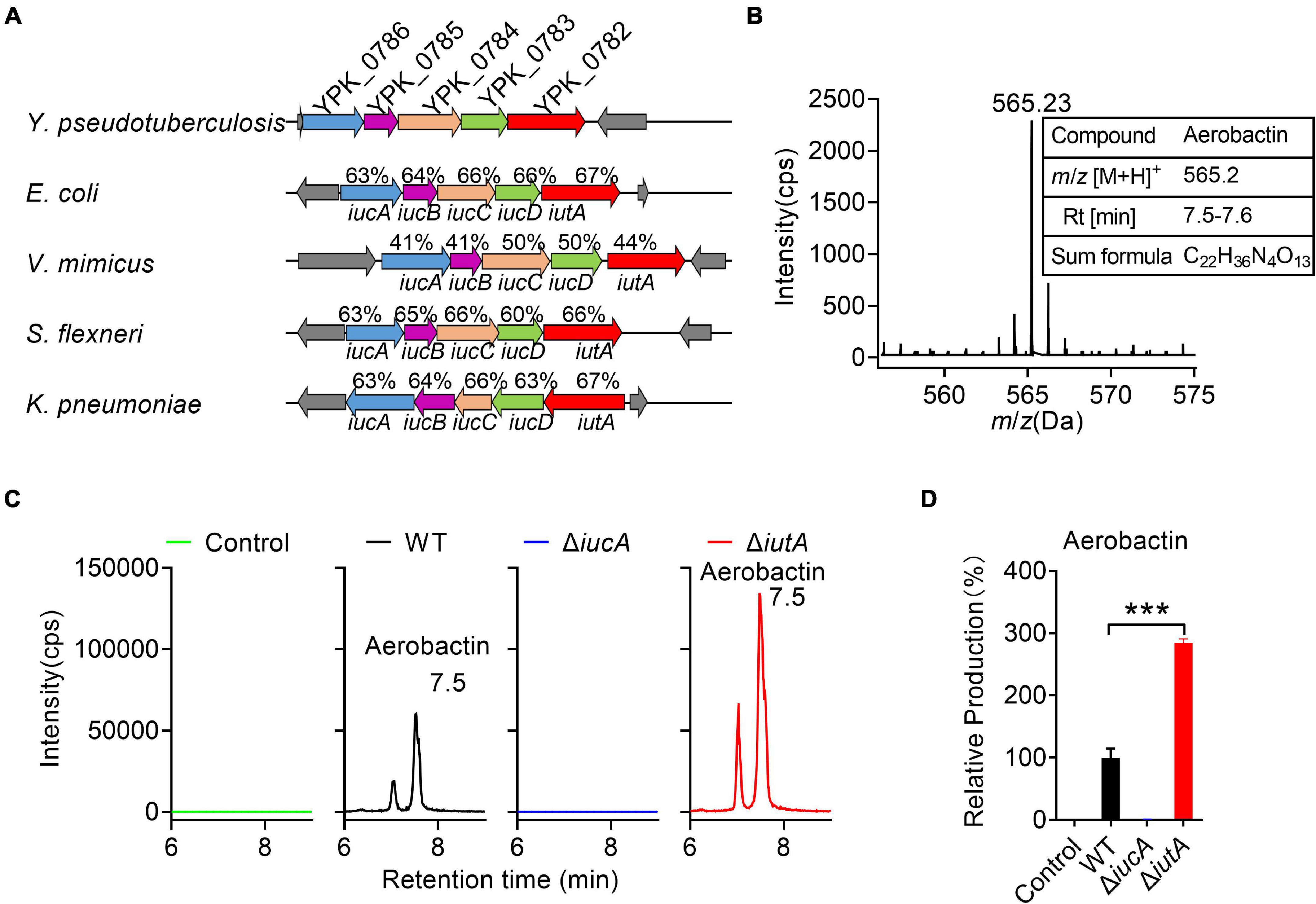
Figure 1. Aerobactin production from the iucABCD-iutA operon in Yersinia pseudotuberculosis. (A) Comparison of IucABCD-IutA amino acid sequences from different species. The accession numbers of SwissProt accession numbers of proteins from different species: Y. pseudotuberculosis YPIII (gi:169749569, 169749568, 169749567, 169749566, 169749565); Escherichia coli O45:K1:H7 S88 (gi:7119436, 7119437, 7119438, 7119439, 7119366); Vibrio mimicus (gi:991835833, 991835832, 991835831, 991835830, 991835829); Shigella flexneri 2457T (gi:30043313, 30043312, 30043311, 30043310, 30043309); Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 (gi:238549846, 238549845, 238549844, 238549843, 238549842). (B) High-performance liquid chromatography combined with tandem mass spectrometry (HPLC-MS/MS) aerobactin siderophore profiles of Y. pseudotuberculosis wild-type (WT) in iron-limited medium. Y. pseudotuberculosis WT was grown in M9G minimal media with and grown for 18 h at 26°C with shaking. Then aerobactin from culture media supernatant was extracted and identified with the following specific transition 565 > 205 m/z. Information of aerobactin siderophore. RT, retention time. (C) HPLC-MS/MS analysis of aerobactin production of Y. pseudotuberculosis WT, ΔiucA, and ΔiutA mutant strains in iron-limited medium. Control represents fresh culture media supernatant. (D) Relative production of aerobactin measured by HPLC-MS/MS. Results are expressed in relation to the production of the WT strain which represent 100%. Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Data represent the mean ± SEM of three biological replicates, each of which was performed with three technical replicates. ***p < 0.001.
Previous studies reported that Y. pestis and Y. pseudotuberculosis are incapable of producing aerobactin even though they possess the iucABCD-iutA operon (Forman et al., 2007). We wondered whether the putative aerobactin synthetase gene cluster (the iucABCD-iutA operon) in Y. pseudotuberculosis YPIII is functional. Therefore, we immediately determined the ability to produce aerobactin of Y. pseudotuberculosis YPIII wild-type (WT) strain. After cultivation in nutrient-limited M9G minimal medium, the culture was prepared and analyzed via HPLC-MS/MS (Figure 1B). The mass with m/z 565.23 [M+H]+, which is equal to the protonated mass of aerobactin was detected in the culture (Figure 1B). Comparison with previously published MS2 spectra of aerobactin (Kupper et al., 2006) confirmed that this compound produced by Y. pseudotuberculosis YPIII is very likely to be aerobactin, since both compounds showed a characteristic fragment of m/z 205.1 [M+H]+ (Supplementary Figures 1A,B). From the HPLC-MS/MS measurements, a sum formula of C22H36N4O13 was determined for the compound (m/z 565.23 [M+H]+), further confirming the sum formula and the number of double bond equivalents of aerobactin (Figure 1B). These results confirmed that Y. pseudotuberculosis YPIII can produce aerobactin under iron-limited conditions.
Neilands and co-workersde Lorenzo et al. (1986) deciphered the aerobactin biosynthetic pathway, in which IucA represents the enzymes to ligate ahLys with citrate, and IutA represents the outer membrane receptor for ferric–aerobactin. To further study the function of the iucABCD-iutA operon in aerobactin biosynthesis and transport, we created iucA and iutA deletion mutants. We then compared aerobactin concentration in culture supernatants of Y. pseudotuberculosis WT, ΔiucA, and ΔiutA mutant strains in M9G minimal medium by HPLC-MS/MS (Figure 1C), and the relative quantification results are shown in Figure 1D. Although the WT strain obviously showed aerobactin production, no aerobactin was detected in the ΔiucA mutant (Figure 1D), which indicated that the iucA gene is essential for aerobactin production and secretion. Interestingly, the ΔiutA mutant produced even more aerobactin in the supernatant compared with that in the WT (284.5%). We reasoned that this further increase in the ΔiutA mutant may result from continually producing and secreting aerobactin because it fails to transport the ferric–aerobactin complex back into the cell.
Meanwhile, we detected aerobactin levels in culture supernatants of Y. pseudotuberculosis WT, ΔiucA, and ΔiutA mutant strains in nutrient-rich Yersinia–Luria–Bertani (YLB) medium by HPLC-MS/MS, but no aerobactin was detected neither in the WT nor in the mutant strains (Supplementary Figures 2A,B), indicating that Y. pseudotuberculosis YPIII is not able to produce aerobactin under iron-rich conditions. Collectively, these results provided evidence that the iucABCD-iutA operon is responsible for aerobactin production and transportation only under iron-limited conditions in Y. pseudotuberculosis YPIII.
Aerobactin Produced by the iucABCD-iutA Operon Is Crucial for Iron Acquisition in Yersinia pseudotuberculosis
Iron is one of the most important metals for life. In order to determine whether the iucABCD-iutA operon is required for the growth of Y. pseudotuberculosis especially under iron-limited conditions, growth curves were determined for Y. pseudotuberculosis WT and aerobactin siderophore biosynthetic and transport mutants ΔiucA and ΔiutA under different conditions. Whereas the ΔiucA and ΔiutA mutants had no difference in growth compared with the WT in iron-rich YLB medium, their growth significantly decreased when 20 or 80 μM iron chelator EDDHA [ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid)] was added to the YLB medium (Figure 2A). Moreover, the growth defects of ΔiucA and ΔiutA mutants could be restored by complementation with a plasmid expressing iucA or iutA, respectively, or by supplying 200 μM Fe3+ into the YLB medium containing 80 μM EDDHA (Figure 2A). These results suggested that aerobactin produced by the iucABCD-iutA operon plays a critical role in promoting the growth of Y. pseudotuberculosis under iron-limited conditions.
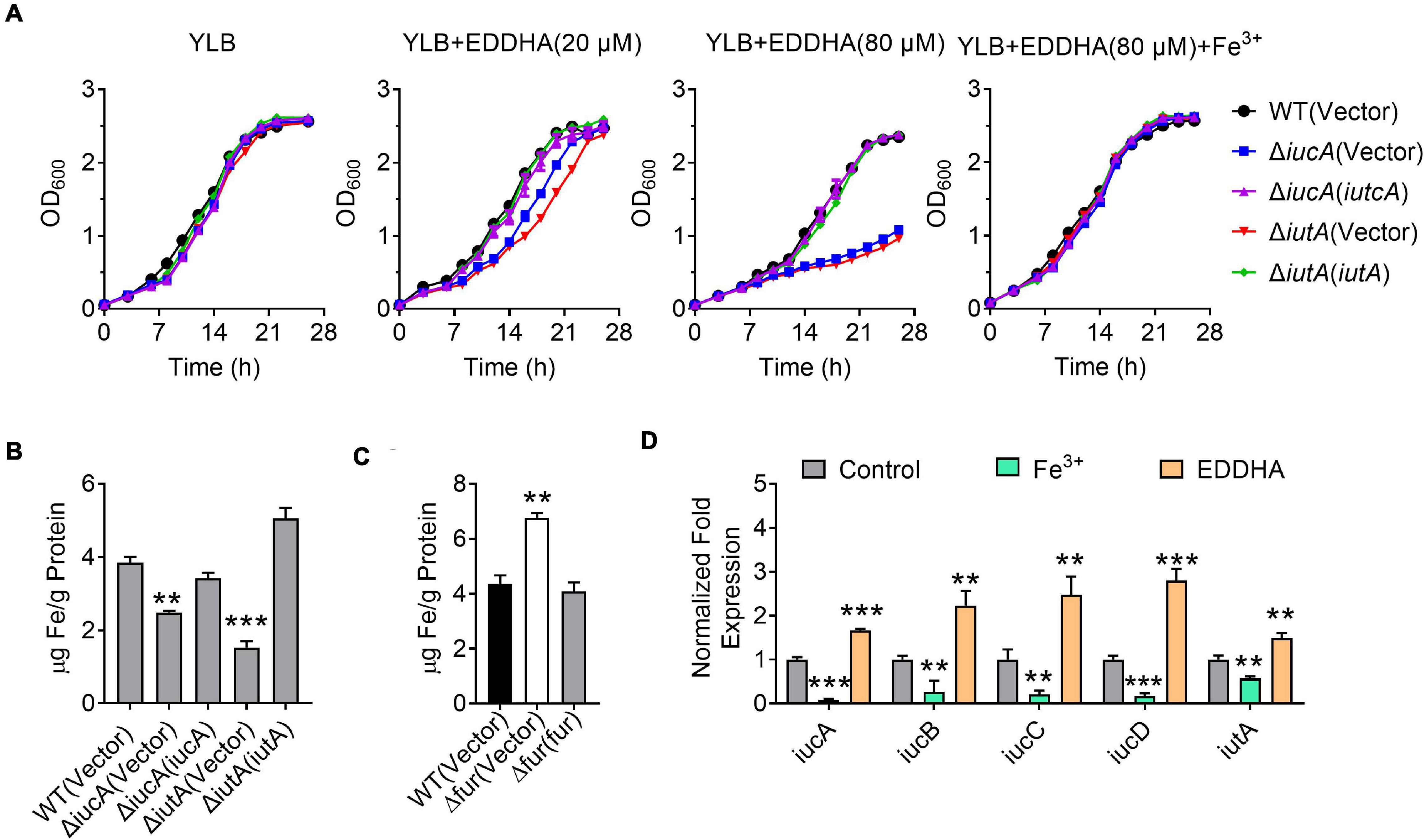
Figure 2. Aerobactin produced by the iucABCD-iutA operon is crucial for iron acquisition in Y. pseudotuberculosis. (A) Saturated bacterial cultures were diluted to an OD600 of 0.1 in Yersinia–Luria–Bertani (YLB) medium, YLB medium with 20 μM EDDHA, YLB medium with 80 μM EDDHA, or YLB medium with 80 μM EDDHA and 200 μM Fe3+. The growth of the cultures was monitored by measuring OD600 at indicated time points. (B) Iron uptake requires aerobactin. Relevant Y. pseudotuberculosis strains were grown overnight in M9G minimal medium to the end of logarithmic phase, the intracellular iron associated with bacterial cells was measured by inductively coupled plasmon resonance atomic absorption spectrometry (ICP-MS). (C) The Δfur mutant accumulates intracellular iron. Intracellular iron was measured by ICP-MS in the Y. pseudotuberculosis WT, Δfur mutant, and complemented Δfur(fur) mutant strains grown to the end of logarithmic phase in YLB medium. (D) Expression of iucABCD-iutA responds to different iron concentration. Y. pseudotuberculosis WT grown in YLB medium with 50 μM EDDHA and 200 μM Fe3+, and qRT-PCR analysis of the expression of iucA, iucB, iucC, iucD, and iutA. Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Data represent the mean ± SEM of three biological replicates, each of which was performed with three technical replicates. **p < 0.01; ***p < 0.001.
To further confirm the role of the iucABCD-iutA operon in iron acquisition, we detected the total metal contents in bacterial cells under iron-limited conditions (M9G) using inductively coupled plasma mass spectrometry (ICP-MS). As shown in Figure 2B, the ΔiucA and ΔiutA mutants showed significantly lower intracellular iron contents compared with the WT, and these defects were almost fully recovered by complementation of iucA or iutA genes, respectively. In addition, we found that the Δfur (Δypk_2991) mutant accumulated more intracellular iron than the WT and Δfur(fur) complemented strain (Figure 2C). In contrast, the accumulation of Na+ and Ca2+ were not affected in these mutants (Supplementary Figures 3A,B). These results indicated that the aerobactin-producing system helps bacteria to obtain iron under iron-limited conditions.
The observation that the aerobactin-producing system is involved in iron uptake points to the notion that the expression of the iucABCD-iutA operon should respond to iron concentration. We therefore performed qRT-PCR to measure the expression level of the iucABCD-iutA operon in Y. pseudotuberculosis WT strain under different iron concentrations. Indeed, the addition of exogenous iron repressed the expression of the iucABCD-iutA operon, while chelating iron from the medium with EDDHA led to robust expression (Figure 2D). This result indicated that iucABCD-iutA expression is responsive to the levels of iron in the environment, which is consistent with its role in acquiring iron from the extracellular milieu. Altogether, these results demonstrated that the aerobactin-producing system is crucial for Y. pseudotuberculosis to acquire iron under iron-limited conditions, and the expression of the iucABCD-iutA operon is regulated by iron concentration.
Fur Directly Represses the Expression of the iucABCD-iutA Operon in Yersinia pseudotuberculosis
Fur is a well- known regulator that represses siderophore synthesis in bacteria to maintain intracellular iron homeostasis (Escolar et al., 1998; Troxell and Hassan, 2013; Di Lorenzo and Stork, 2014; Li et al., 2019; Banerjee et al., 2020). Interestingly, analysis of the promoter region of the iucABCD-iutA operon identified a putative Fur-binding site (Figure 3A). The 20-bp Fur-binding site (TGATAATGATAACCACTATT) is highly similar to the Fur box identified in E. coli (Figure 3B; de Lorenzo et al., 1987; Stojiljkovic et al., 1994). To explore whether Fur regulates the iucABCD-iutA operon directly, we examined the interaction between Fur and the promoter of the iucABCD-iutA operon by using electrophoretic mobility shift assay (EMSA). As shown in Figure 3C, incubation of a probe harboring the iucABCD-iutA promoter (PiucA) sequence [-1 to −139 relative to the ATG start codon of the first ORF (Open Reading Frame) of the iucABCD-iutA operon] with His6-Fur led to the formation of DNA–protein complexes, while an unrelated DNA fragment did not. Consistently, replacing this 20-bp binding site in the iucABCD-iutA promoter probe with arbitrary mutation abolished the formation of DNA–protein complexes in the EMSA assay (Supplementary Figure 4). Therefore, these results confirmed that Fur specifically recognizes the promoter of the iucABCD-iutA operon.
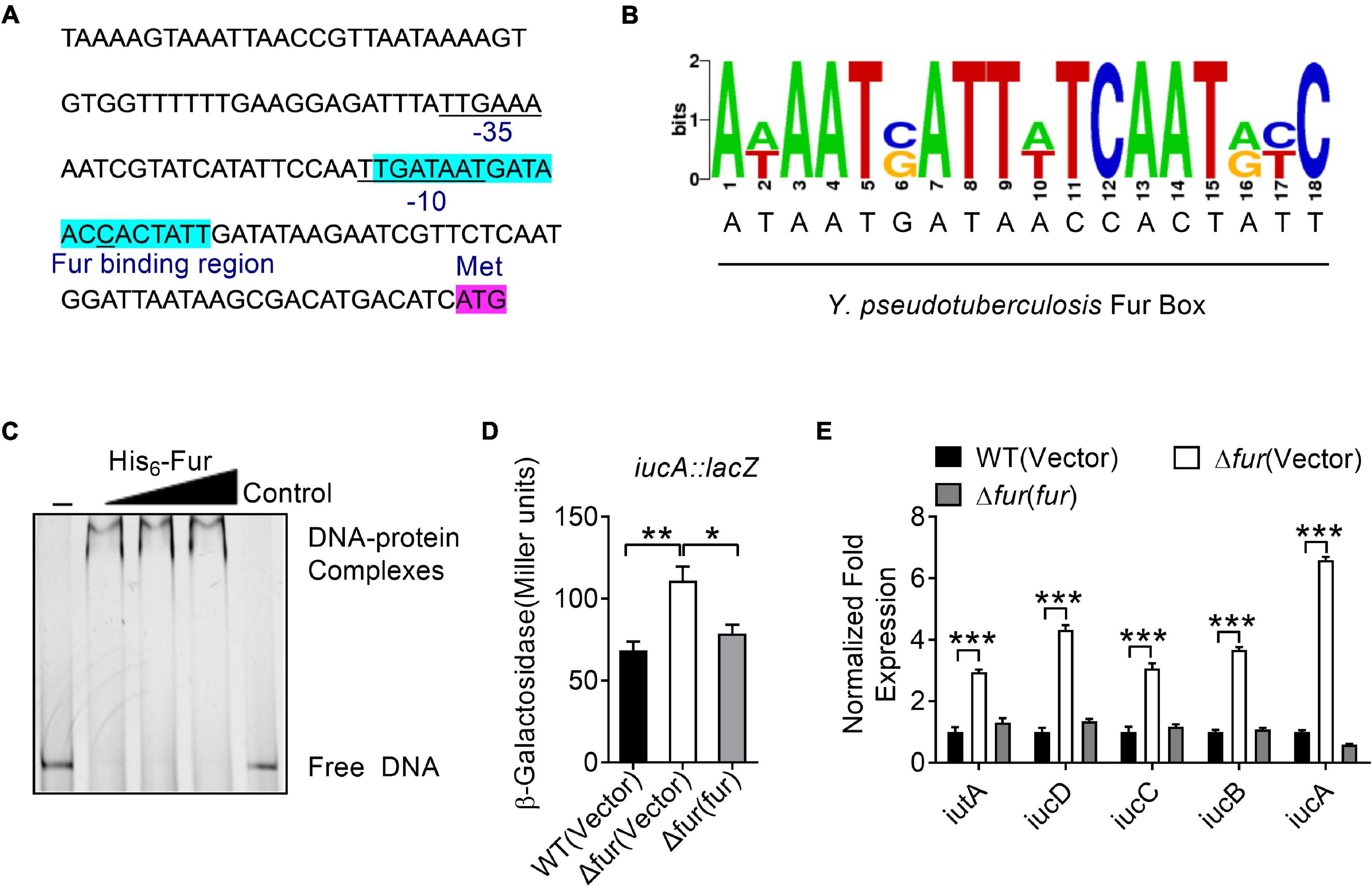
Figure 3. Ferric uptake regulator (Fur) negatively regulates the iucABCD-iutA operon expression in Y. pseudotuberculosis. (A) Identification of a Fur-binding site in the promoter region of iucABCD-iutA. The putative Fur-binding site identified by the online software Virtual Footprint is indicated by blue highlighting. Putative −35 and −10 elements of the iucABCD-iutA promoter are underlined. The ATG start codon of the first ORF of the iucABCD-iutA operon is shown. (B) Fur box sequence upstream of iucA. Virtual footprint analysis of the Y. pseudotuberculosis Fur-binding sequence. Letters represent position weight matrix based on E. coli K-12 consensus sequence for Fur binding. The Y-axis represents relative nucleotide probability and the X-axis represents nucleotide position. Y. pseudotuberculosis Fur box sequence is located at −64 bp of iucA and has a probability score of 28.50/0 (max score = 28.50). (C) Electrophoretic mobility shift assay (EMSA) was performed to analyze the interactions between His6-Fur and the promoter. Increasing amounts of Fur (0, 0.24, 0.48, and 0.72 μM) and 5 ng DNA fragment were used. As a negative control, a 5 ng of 151-bp unrelated DNA fragment was used in control. (D) Fur represses the expression of iucABCD-iutA. β-galactosidase analyses of iucABCD-iutA promoter activities were performed using the transcriptional PiucA::lacZ chromosomal fusion reporter expressed in the Y. pseudotuberculosis WT, Δfur mutant, and complemented Δfur(fur) strains grown to stationary phase in YLB medium. (E) qRT-PCR analysis of mRNA levels of iucABCD-iutA. Cells of relevant Y. pseudotuberculosis strains were grown to mid-exponential phase in YLB medium, and the expression of iucA, iucB, iucC, iucD, and iutA were measured by qRT-PCR. Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Data represent the mean ± SEM of three biological replicates, each of which was performed with three technical replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
To further verify the role of Fur in the regulation of the iucABCD-iutA operon, a single-copy PiucA::lacZ fusion reporter was introduced into the chromosomes of the WT, Δfur mutant, and complemented Δfur(fur) strains, respectively. By quantitatively measuring the LacZ activity of the resulting strains, we found that deletion of fur significantly increased the activity of the iucABCD-iutA promoter, which was fully restored to the WT level by complementation with a plasmid expressing fur (pKT100-fur), confirming that Fur negatively regulates iucABCD-iutA expression (Figure 3D). Negative regulation of the iucABCD-iutA operon by Fur was further confirmed by qRT-PCR, which indicated that the expression levels of iucA, iucB, iucC, iucD, and iutA were significantly increased in the Δfur mutant, and that such increases could be completely reversed by fur complementation (Figure 3E). Taken together, we demonstrated that Fur directly represses iucABCD-iutA expression through binding to its promoter.
Aerobactin-Mediated Iron Acquisition Influences Biofilm Formation
Like many pathogens, Y. pseudotuberculosis is capable of forming biofilm (Darby et al., 2002), which contributes to environmental survival, transmission of microorganism, host interaction, and virulence (Hinnebusch and Erickson, 2008; Zhou and Yang, 2011; Martinez-Chavarria and Vadyvaloo, 2015; Calder et al., 2020). Previous studies have revealed that iron is essential for biofilm formation (Chung, 2016; Oliveira et al., 2017), and less biofilm forms under low-iron conditions (Liu et al., 2016). Also, siderophore-mediated iron acquisition systems have been shown to enhance biofilm formation in different bacteria, including yersiniabactin and enterobactin in E. coli (Hancock et al., 2008; Keogh et al., 2016), pyoverdine and pyochelin in P. aeruginosa (Banin et al., 2005; Yang et al., 2009), exochelin in Mycobacterium smegmatis (Ojha and Hatfull, 2007), and cupriabactin in Cupriavidus necator (Li et al., 2019). To further explore the role of the aerobactin-producing system in biofilm formation in Y. pseudotuberculosis, we examined the biofilm-forming capacity of WT and aerobactin siderophore biosynthetic and transport mutants by using the crystal violet assay under different conditions. Notably, no significant differences on biofilm formation were observed between WT and ΔiucA or ΔiutA mutant in nutrient-rich YLB medium (Supplementary Figure 5). However, in nutrient-limited M9G medium, the aerobactin mutants ΔiucA and ΔiutA showed obvious defects in biofilm formation (Figure 4A), and the biofilm-formation capacity was restored to WT levels by complementation with iucA or iutA, respectively (Figure 4A). Meanwhile, we also compared the biofilm formation capacity of the WT, Δfur mutant, and complemented Δfur(fur) strains. Increased biofilm formation was observed in the Δfur mutant (Figure 4B), which produces more aerobactin and contains more intracellular iron. Thus, these results indicated that aerobactin-mediated iron acquisition system plays pivotal roles in biofilm formation under iron-limited conditions in Y. pseudotuberculosis.
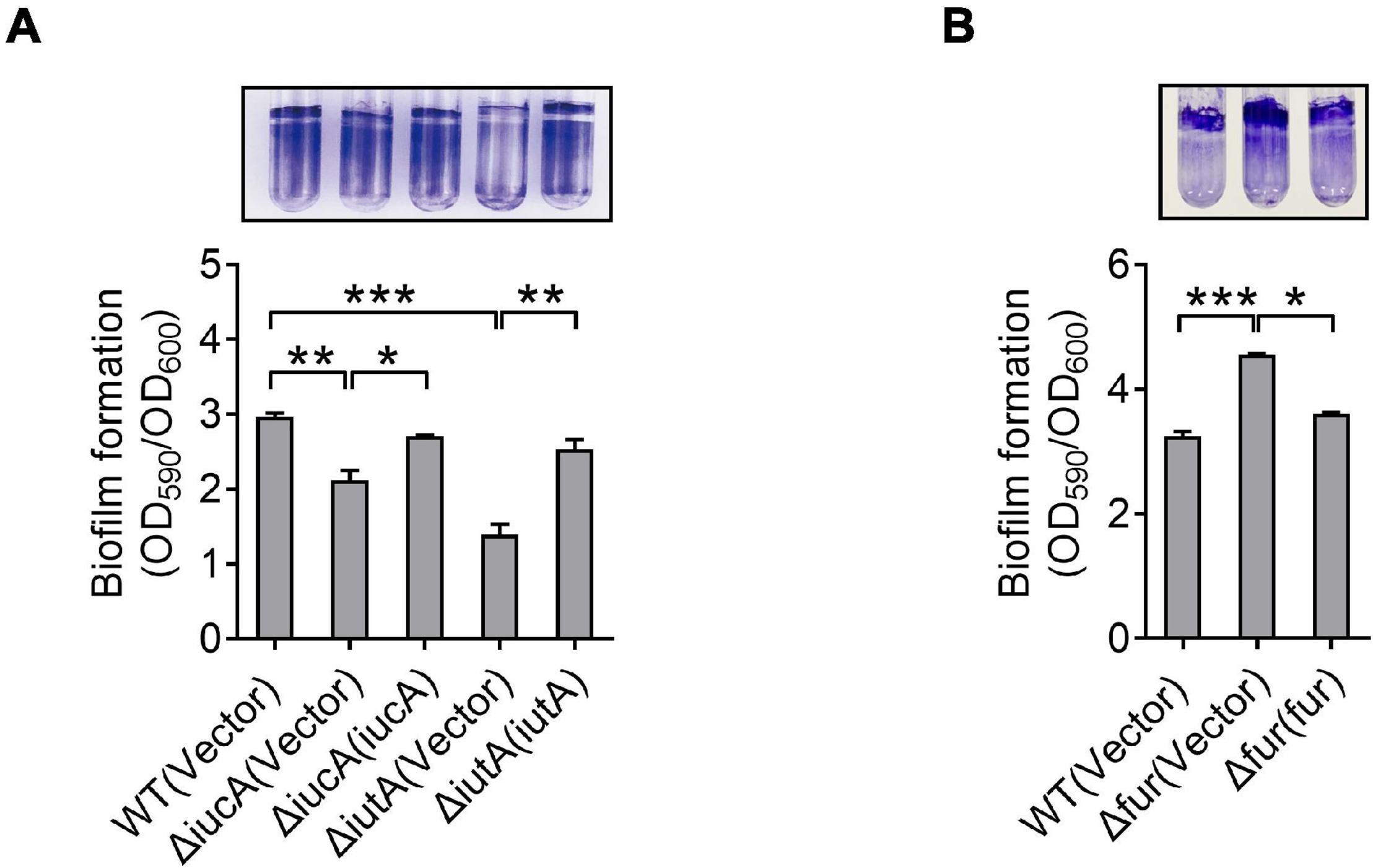
Figure 4. Aerobactin-mediated iron acquisition influences biofilm formation. (A,B) Effect of aerobactin system (A) and Fur (B) on biofilm formation in iron-limiting media. Saturated bacterial cultures were diluted 100-fold in fresh M9G minimal medium. After vertical incubation for 3 days with shaking at 150 rpm in 26°C, biofilm formation of the strains was determined by crystal violet staining and quantified using optical density measurement. Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Data represent the mean ± SEM of three biological replicates, each of which was performed with three technical replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
Aerobactin-Mediated Iron Transport System Is Required for Resistance to Oxidative Stress in Yersinia pseudotuberculosis
Siderophores were reported to offer protection against oxidative stress by reducing the reactive oxygen species (ROS) levels in some bacteria, for example, enterobactin in E. coli (Adler et al., 2014), pyoverdine and pyocyanin in P. aeruginosa (Rada and Leto, 2013; Jin et al., 2018), and catecholate in S. Typhimurium (Achard et al., 2013). To examine whether aerobactin plays a role in protection against oxidative stress in Y. pseudotuberculosis, we first determined the expression of the iucABCD-iutA operon in the Y. pseudotuberculosis WT by qRT-PCR after H2O2 challenge, and the results showed that the expression level of iucA, iucB, iucC, iucD, and iutA genes was enhanced by 2.5—5.3-fold by addition of 5 mM H2O2 (Figure 5A). We also determined the effects of the aerobactin-producing system on bacterial resistance to oxidative stress by measuring the viability of the iucABCD-iutA operon mutants after H2O2 challenge. The results showed that the survival rates of the ΔiucA and ΔiutA mutants were significantly more sensitive to H2O2 than the WT (Figure 5B). Meanwhile, the survival rates of all complemented strains were almost completely restored to the WT level (Figure 5B), further supporting the role of the aerobactin-producing system in combating oxidative stress. To further examine the effect of the aerobactin-mediated iron transport system on ROS reduction upon oxidative stress, we assessed the intracellular ROS levels in Y. pseudotuberculosis WT, ΔiucA, and ΔiutA mutant strains challenged with H2O2 by using fluorescent reporter dye 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA). As shown in Figure 5C, ΔiucA and ΔiutA mutants had significantly higher ROS levels than the WT after exposure to H2O2, indicating that the aerobactin-mediated iron transport system is critical in reducing ROS accumulation in Y. pseudotuberculosis under oxidative stress conditions. Note that the ROS-induced fluorescence signals were specific because no signal was detected in the control samples treated with dyes but not treated with H2O2 (Supplementary Figure 6). Altogether, these data indicated that the aerobactin-mediated iron transport system is induced and important for survival under oxidative stress in Y. pseudotuberculosis.
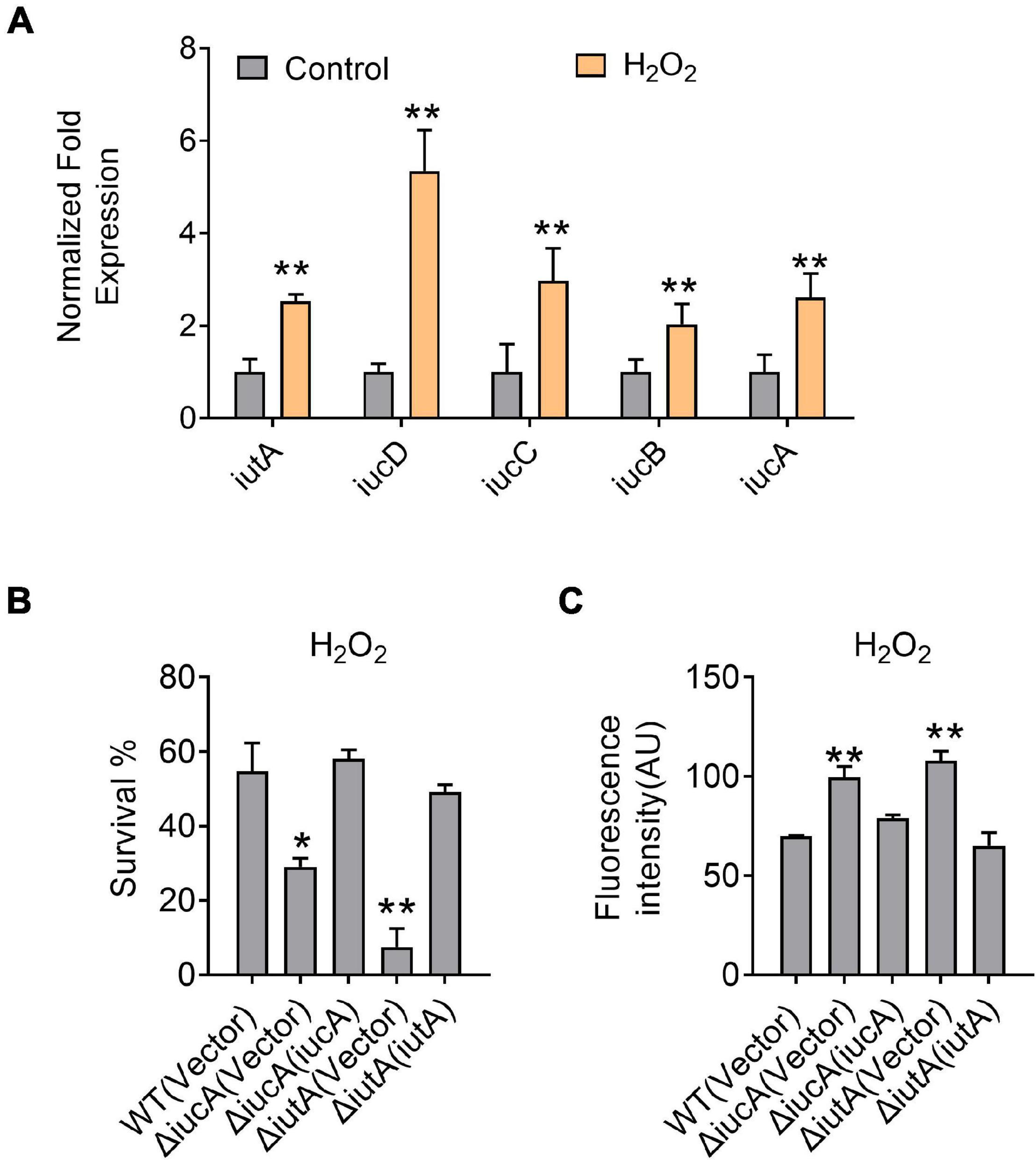
Figure 5. Aerobactin-mediated iron acquisition is involved in oxidative stress resistance. (A) Aerobactin operon expression responds to oxidative stress. Y. pseudotuberculosis WT was grown to mid-exponential phase in YLB medium, then saturated bacterial cultures were diluted 100-fold in fresh M9G minimal medium with or without 5 mM H2O2, qRT-PCR analysis of the expression level of iucA, iucB, iucC, iucD and iutA. (B) Alleviation of the sensitivity of Y. pseudotuberculosis strains to H2O2. The viability of mid-exponential phase Y. pseudotuberculosis strains was determined after exposure to 1 mM H2O2 for 40 min in M9G medium. (C) Deletion of iucA or iutA leads to accumulation of intracellular reactive oxygen species (ROS) under oxidative conditions. The intracellular levels of ROS were determined with the 2′, 7′-dichlorodihydro-fluorescein diacetate (H2DCFDA) probe after exposure of stationary phase Y. pseudotuberculosis strains to 1 mM H2O2 in M9G medium. Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Data represent the mean ± SEM of three biological replicates, each of which was performed with three technical replicates. *p < 0.05; **p < 0.01.
Aerobactin-Mediated Iron Acquisition System Contributes to the Pathogenicity of Yersinia pseudotuberculosis
Iron contributes an important branch to bacterial infection because many pathogens need iron for virulence (Drakesmith and Prentice, 2012; Cassat and Skaar, 2013; Nakashige et al., 2015). Siderophore-mediated systems are one of the most important tools to uptake iron for bacteria, which play a vital role in virulence, such as salmochelin (Lemaitre et al., 2012) and yersiniabactin (Chaturvedi et al., 2012). Therefore, we examined whether the aerobactin-producing system is involved in the virulence of Y. pseudotuberculosis. C57BL/6 mice were orogastrically infected with the WT, ΔiucA, or ΔiutA mutants, respectively, and the survival rate of each group was analyzed. The results showed that infection with the WT led to more than 90% death within 3 weeks of infection (Figure 6A), and the lethality rates slightly but substantially decreased in the ΔiucA mutant- and ΔiutA mutant-infected group (Figure 6A). Next, the bacterial loads recovered from the feces, cecum, intestine, colon, spleen, and liver were counted at 96 h post-infection with Y. pseudotuberculosis strains. Consistently, mice infected with aerobactin siderophore biosynthetic mutants ΔiucA and ΔiutA had significantly fewer loads compared with WT-infected mice (Figures 6B–G). These results indicated that the aerobactin-producing system contributes to the pathogenicity of Y. pseudotuberculosis by enhancing the ability of colonization in mice.
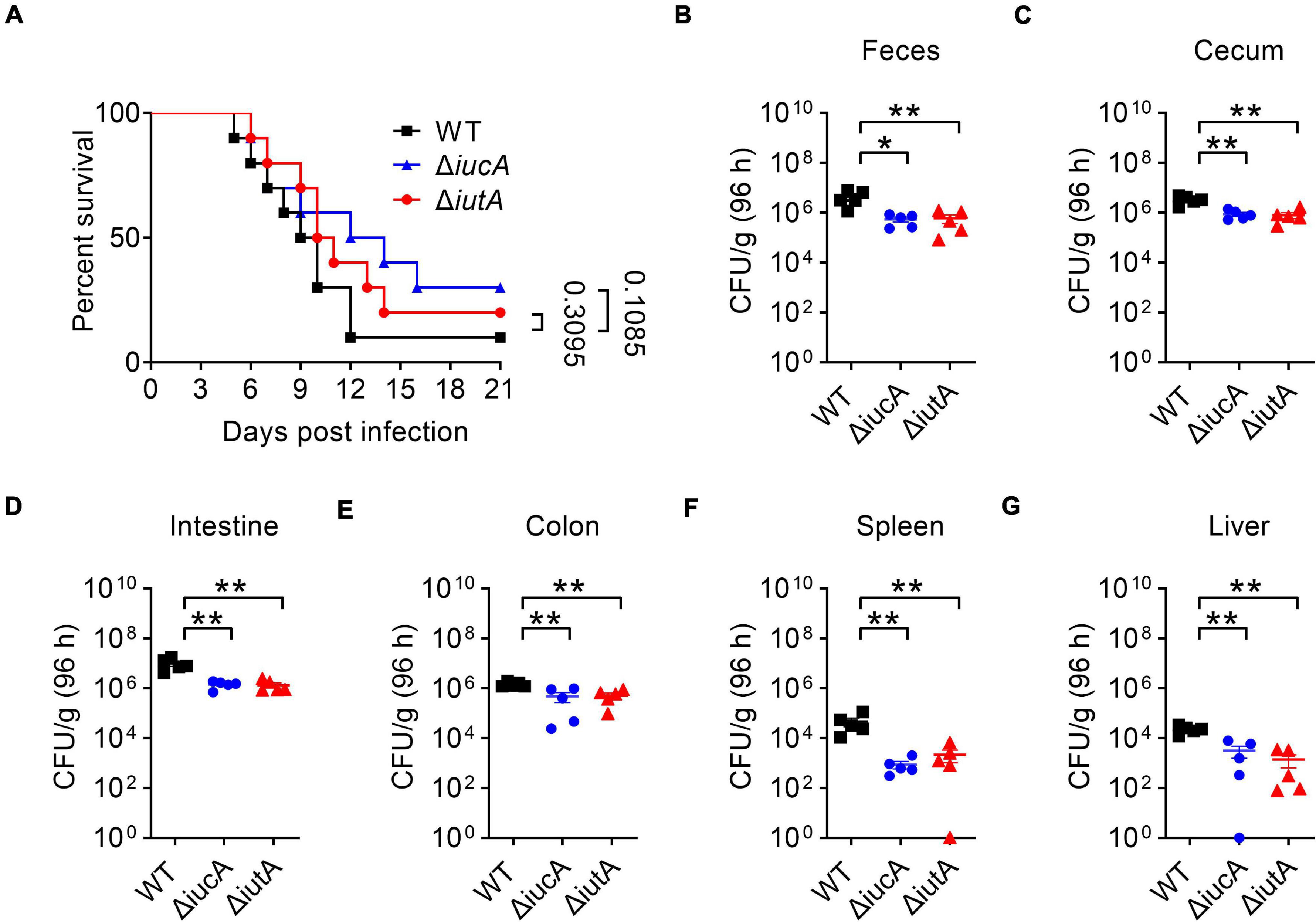
Figure 6. Aerobactin-mediated active iron acquisition contributes to the pathogenicity of Y. pseudotuberculosis. Relevant Y. pseudotuberculosis strains grown in YLB were washed twice in sterilized PBS and used for orogastric infection of 6- to 7-week-old female C57BL/6 mice using a ball-tipped feeding needle. For survival assays, 1 × 109 bacteria of each strain were applied to different groups of mice (n = 10/strain), and the survival rate of the mice was determined by monitoring the survival daily for 3 weeks (A). Enumeration of bacterial burdens in the feces (B), cecum (C), intestine (D), colon (E), spleen (F), and liver (G) of infected C57BL/6 mice at 3 days post-infection by CFU assays (n = 5). Similar results were obtained in three independent experiments, and data shown are from one representative experiment done in triplicate (A). Statistical analyses were performed by Log-Rank test (A). The statistical significances were determined by the Mann–Whitney test (B–G). *p < 0.05; **p < 0.01.
Discussion
Aerobactin, a hydroxamate type of siderophore, was first discovered in the supernatant of Aerobacter aerogenes 62-I and was later shown to be encoded by plasmid (Gibson and Magrath, 1969; McDougall and Neilands, 1984). Subsequently, the aerobactin cluster was identified in many pathogenic bacteria, including E. coli, Salmonella, Klebsiella, and Shigella, which was found to be encoded in both plasmids and chromosomes (Di Lorenzo and Stork, 2014). However, whether pathogenic Yersinia can produce aerobactin remains enigmatic. Although the aerobactin biosynthetic (iucA-D) and outer membrane receptor (iutA) locus are widely distributed in the genomes of Yersinia species, only some non-pathogenic strains, such as Yersinia frederiksenii, Yersinia kristensenii, and Yersinia intermedia were found to produce aerobactin (Stuart et al., 1986). Surprisingly, none of the three pathogenic Yersinia species was found to produce aerobactin. Even though the Y. pestis genome contains the homologous aerobactin cluster (iucABCD-iutA), it was reported to have lost the ability to synthesize aerobactin due to frameshift mutation in iucA (Supplementary Figure 7; Forman et al., 2007). Stuart et al. (1986) reported that all 50 examined Y. enterocolitica strains failed to produce aerobactin, suggesting the absence of aerobactin in any of the Y. enterocolitica strains. Interestingly, some examined Y. pseudotuberculosis strains, which encode intact aerobactin biosynthetic genes, were also unable to synthesize aerobactin (Stuart et al., 1986; Forman et al., 2007). However, because the number of Y. pseudotuberculosis strains tested was too small, it seems difficult to conclude that Y. pseudotuberculosis does not synthesize aerobactin.
In this study, we examined whether the aerobactin operon iucABCD-iutA is functional in Y. pseudotuberculosis YPIII. Strikingly, we observed that Y. pseudotuberculosis YPIII can secrete and transport aerobactin siderophore by biosynthetic enzymes (IucABCD) and outer membrane receptor (IutA) (Figure 1), and aerobactin was further proved to enhance the bacterial growth under iron-limited conditions (Figure 2A). The production of aerobactin was confirmed with HPLC-MS/MS analysis and mutation analysis (Figures 1C,D). To the best of our knowledge, this is the first time to report that pathogenic Yersinia can produce aerobactin for iron acquisition. We compared the homolog of aerobactin-producing functional iucABCD-iutA operon in Y. pseudotuberculosis YPIII with that in all other Y. pseudotuberculosis strains and Y. pestis KIM10+ (Supplementary Figure 7). Although the aerobactin biosynthetic gene cluster iucABCD has no obvious defects and shows high similarities in all Y. pseudotuberculosis strains, inexplicably, previous studies found that none of Y. pseudotuberculosis PB1/0 or any of other five Y. pseudotuberculosis isolates obtained from diseased poultry and sheep in Australia produced aerobactin (Stuart et al., 1986; Forman et al., 2007). Regrettably, we still do not know how, generally, Y. pseudotuberculosis strains make aerobactin, or whether Y. pseudotuberculosis YPIII is an anomaly.
Bacterial biofilms are communities of microorganisms, which attach to the surface of environments and host (Flemming et al., 2016). Rather than existing as individual planktonic cells, most pathogenic bacteria prefer to form biofilm to enhance its survival and defense in the host, such as P. aeruginosa, M. tuberculosis, Y. pestis, and Y. pseudotuberculosis (Darby et al., 2002; Kumar et al., 2017). Previous studies suggested that iron metabolism is important in biofilm formation in several pathogens. For example, in Bacillus velezensis, iron acquisition mediated by the FeuABC transporter promotes biofilm development (Xu et al., 2019). Iron has also been shown to play an important role for biofilm formation in S. maltophilia (Kalidasan et al., 2018). In this study, our data indicated that aerobactin produced by the iucABCD-iutA operon is crucial for iron acquisition in Y. pseudotuberculosis (Figure 2B). Therefore, we examined whether aerobactin affects the biofilm formation (Figure 4). As expected, the result suggested that aerobactin-mediated iron acquisition contributes to biofilm formation under iron-limited conditions in Y. pseudotuberculosis.
Bacteria have been reported to uptake iron to protect against oxidative damage because iron is a catalyst for ROS. For instance, siderophore-mediated iron acquisition plays important roles in ROS detoxification and cellular resistance to oxidative stress in Alternaria alternata (Chen et al., 2013). Catecholate and enterobactin siderophores respond to iron limitation, which were reported to play a role in the protection of S. typhimurium from oxidative stress and ROS (Bogomolnaya et al., 2020). Our previous study also demonstrated that the cupriabactin-mediated iron acquisition contributes to the resistance to oxidative stress in C. necator JMP134 (Li et al., 2019). Similarly, this study found that aerobactin plays vital roles in oxidative stress resistance and ROS removal in Y. pseudotuberculosis (Figures 5B,C). We reason that this ROS protection was likely due to the iron acquisition ability mediated by the aerobactin, and the process of ROS scavenging was subsequently completed by the functional catalase and other iron-dependent antioxidant enzymes. These findings allowed us to conclude that aerobactin-mediated iron acquisition plays important roles in biofilm formation, oxidative stress resistance, and ROS removal in Y. pseudotuberculosis.
Apart from maintaining microbial life, iron acquisition ability is essential for a part of siderophores to mediate the full virulence of pathogens, and this process is controlled by Fur. During infection, the host-related environment is maintained in an iron-limited state because of the anemia of inflammation and nutritional immunity exerted by the host (Troxell and Hassan, 2013; Begg, 2019). Fur derepresses the expression of multiple iron acquisition systems as soon as it senses that iron is depleted, and the pathogenic bacteria counterattack through robbing host iron sources via producing siderophores, such as pyoverdine, salmochelin, and staphyloferrin, which further result in virulence increase during infection by enhancing its proliferation (Saha et al., 2013), regulating the production of virulence factors (Llamas et al., 2014), and evading host innate immune response (Palmer and Skaar, 2016). In pathogenic Y. pseudotuberculosis, yersiniabactin system is the only siderophore-mediated iron transport system that has been tested and found to affect the virulence of this organism until now (Forman et al., 2010). However, only Y. pseudotuberculosis serotype O1 strains possess the yersiniabactin system (Buchrieser et al., 1999), such as PB1/+(serotype 1B), 1, IP32593 (serotype 1), IP32593 (serotype I), and MD67. In this study, we first found that Y. pseudotuberculosis YPIII, which belongs to serotype O3 strains of Y. pseudotuberculosis lacking full yersiniabactin synthesis genes, could produce aerobactin siderophore. Consistent with previous reports that aerobactin plays important roles in virulence of K. pneumoniae, Pantoea stewartia, and S. flexneri (Sheldon et al., 2016), we showed that Y. pseudotuberculosis YPIII aerobactin biosynthetic and transport mutants ΔiucA and ΔiutA were attenuated in virulence in the mice infection model (Figure 6), confirming its important roles in virulence in Y. pseudotuberculosis YPIII.
In conclusion, we provided evidence that pathogenic Y. pseudotuberculosis has the ability to produce the aerobactin siderophore, which was demonstrated to play important roles not only in growth under iron-limited conditions but also in oxidative stress resistance, biofilm formation, and virulence. This study has improved our knowledge on iron acquisition strategies of pathogenic Y. pseudotuberculosis, providing an opportunity to deepen our understanding of the multiple functions of siderophore-mediated iron transport system.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. E. coli strains were grown in Luria–Bertani (LB) with appropriate antibiotics at 37°C. Y. pseudotuberculosis strains were cultured in Yersinia–Luria–Bertani (YLB) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or M9G minimal medium (Na2HPO4, 6 g L–1; KH2PO4, 3 g L–1; NaCl, 0.5 g L–1; NH4Cl, 1 g L–1; MgSO4, 1 mM; CaCl2, 0.1 mM; glucose 0.4%, pH 7.0) at 26°C with appropriate antibiotics when necessary. Y. pseudotuberculosis YPIII was the parent of all derivatives used in this study. In-frame deletions were generated as described previously (Xu et al., 2014). Cellular growth was monitored based on the optical density (OD) at 600 nm. All chemicals were of analytical reagent grade purity or higher. Antibiotics were added at the following concentrations: nalidixic acid, 20 μg ml–1; kanamycin, 50 μg ml–1; and chloramphenicol, 20 μg ml–1.
Plasmid Construction
Primers used in this study are listed in Supplementary Table 2, respectively. The plasmid pDM4-ΔiucA (ypk_0786) was used to construct the ΔiucA in-frame deletion mutant of Y. pseudotuberculosis. A 795-bp upstream fragment and an 820-bp downstream fragment of iucA were amplified using the primer pairs iucA-1F-BglII/iucA-1R and iucA-2F/iucA-2R-SalI, respectively. The upstream and downstream PCR fragments were ligated by overlapping PCR. The resulting PCR products were digested with BglII and SalI and inserted into the BglII/SalI sites of pDM4 to produce pDM4-ΔiucA. The knock-out plasmids pDM4-ΔiutA (ypk_0782) and pDM4-Δfur (ypk_2991) were constructed in a similar method by using primers list in Supplementary Table 2. To complement the ΔiucA mutant, primers iucA-F-BamHI/iucA-R-SalI were used to amplify the iucA gene from the Y. pseudotuberculosis genome DNA. The PCR product of iucA was digested with BamHI/SalI and inserted into the BamHI/SalI sites of pKT100 to produce pKT100-iucA. The complementation plasmids pKT100-iutA and pKT100-fur were similarly constructed by using primers list in Supplementary Table 2. To express His6-tagged Fur, plasmid pET28a-fur was constructed. Briefly, primers fur-F-BamHI and fur-R-SalI were used to amplify the fur gene fragment from the Y. pseudotuberculosis genome. The PCR product of fur was digested with BamHI/SalI and inserted into the BamHI/SalI sites of pET28a to generate pET28a-fur. For complementation, complementary plasmids pKT100-iucA, pKT100-iutA, and pKT100-fur were introduced into respective mutants by electroporation. The integrity of the insert in all constructs was confirmed by DNA sequencing.
Determination of Intracellular Ion Contents
Intracellular ion content was determined as described previously (Wang et al., 2015; Si et al., 2017). Briefly, cells were grown in M9G minimal medium until mid-exponential phase. After 20-ml culture solutions were collected and washed with M9 two times, the cell pellet weight was measured, and bacteria were chemically lysed using Bugbuster (Novagen, Madison, WI, United States) according to the manufacturer’s instructions. Bacteria were resuspended in Bugbuster solution by pipetting and incubation on a rotating mixer at a slow setting for 12 h. Total protein for each sample was measured by using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) according to the manufacturer’s instructions. Each sample was diluted 10-fold in 2% molecular grade nitric acid to a total volume of 5 ml at a slow setting for 12 h. Samples were analyzed by inductively coupled plasma mass spectrometry (ICP-MS, Varian 802-MS), and the result was corrected using the appropriate buffers for reference and dilution factors. Triplicate cultures of each strain were analyzed during a single experiment, and the experiment was repeated at least three times.
Biofilm Formation Assay
Biofilm formation was determined as described previously (O’Toole and Kolter, 1998; Zhang et al., 2020). Briefly, overnight bacterial cultures were diluted 100-fold in 5 ml of fresh M9G or YLB medium with appropriate antibiotics when necessary. After vertical incubation for 3 days with the shake of 150 rpm at 26°C, the bacterial cultures were removed after measuring the OD600, and the test tubes were washed twice with fresh M9. Cells that adhered to the test tubes were stained with 0.1% crystal violet for 30 min and then washed twice with M9. The cell-bound dye was eluted in 6 ml of 95% ethanol, and the absorbance of the eluted solution was measured at 590 nm using a microplate reader.
Overexpression and Purification of Recombinant Protein
To express and purify soluble His6-tagged recombinant proteins, the plasmid pET28a-fur was transformed into BL21 (DE3). Bacteria were cultured at 37°C in LB medium to an OD600 of 0.5, shifted to 24°C and induced with 0.2 mM IPTG, and then cultivated for an additional 12 h at 24°C. Harvested cells were disrupted by sonication, and proteins were purified with the His•Bind Ni-NTA resin (Novagen, Madison, WI, United States) according to the instructions of the manufacturer. Eluted recombinant proteins were dialyzed against buffer (50 mM Tris, 137 mM NaCl, 10% glycerol, pH 7.5) at 4°C. The resulting proteins were stored at −80°C until use. Protein concentrations were determined using the Bradford assay according to the instructions of the manufacturer (Bio-Rad, Hercules, CA, United States) with bovine serum albumin as standard.
Construction of Chromosomal Fusion Reporter Strains and β-Galactosidase Assays
The lacZ fusion reporter vector pDM4-PiucA::lacZ was transformed into E. coli S17-1 λpir and mated with Y. pseudotuberculosis strains as described previously (Zhang et al., 2013). The lacZ fusion reporter strains were grown to stationary phase in YLB at 26°C, and β-galactosidase activity was assayed using ONPG (o-nitrophenyl β-D-galactopyranoside) as the substrate. These assays were performed in triplicate at least three times, and error bars represent standard deviations.
Bacterial Survival Assays
Mid-exponential phase Y. pseudotuberculosis strains grown in YLB medium were collected, washed, and diluted 50-fold into M9G medium, and treated with or without H2O2 (1.0 mM) for 35 min at 26°C. After treatment, the cultures were serially diluted and plated onto YLB agar plates, and colonies were counted after 36 h growth at 26°C. Percentage survival was calculated by dividing the number of CFU of stressed cells by the number of CFU of cells without stress. All these assays were performed in triplicate at least three times.
Fluorescence Dye-Based Intracellular Reactive Oxygen Species Detection
To detect intracellular ROS, the fluorescent reporter dye 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Invitrogen) was used as previously described (Dong et al., 2015). Briefly, 1-ml samples were collected, washed with PBS, and then resuspended in 1 ml of M9G containing 10 μM H2DCFDA. Samples were incubated in the dark for 20 min at 26°C. The cells were then pelleted, the supernatant removed, and were resuspended in 1 ml M9 medium containing 0.4% glucose with or without H2O2 (1 mM). After 30-min treatment at 26°C, the cells were pelleted, washed with PBS, resuspended in 1 ml of M9, and then 200 μl of the resultant cell suspension was transferred to a dark 96-well plate. Fluorescence signals were measured using a SpectraMax M2 Plate Reader (Molecular Devices) with excitation/emission wavelengths of 495/520 nm.
High-Performance Liquid Chromatography Combined With Tandem Mass Spectrometry Analysis of Siderophores From Culture Supernatants
Strains were suspended to an OD600 of 0.05 in 5 ml of M9G medium and grown for 18 h at 26°C with shaking. The cells were pelleted by centrifugation at 4,500 rpm for 10 min, and the supernatant was filtered through a 0.22-μm filter (Millipore, MA, United States). One milliliter of supernatant was evaporated to dryness under a gentle stream of nitrogen at 45°C for 6 h, and dry residues were frozen at −20°C for subsequent analysis.
The level of siderophores from culture supernatants were analyzed through high-performance liquid chromatography combined with tandem mass spectrometry (HPLC-MS/MS). LC-MS/MS analyses were performed on a tandem quadrupole time-of-flight mass spectrometer (TripleTOF5600, SCIEX) equipped with an electrospray ionization (ESI) interface and HPLC system comprising a binary LC-30AD pump, a SIL-20AHT autosampler, and a column oven (Shimadzu, Tokyo, Japan). Dry residues were dissolved in 50 μl of 5% acetonitrile in water, and 5 μl was directly injected for analysis by HPLC-MS/MS (Bailey et al., 2018). Separation was carried out on a Shim-pack XR-ODS C18 column (100 mm × 2.0 mm, 2.2 μm) using gradient elution with mobile phases at 40°C with a flow rate of 0.3 ml/min. The mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The HPLC program was used as follows: 5% B, 2 min; from 5 B to 20% B, 9 min; 20% B, 2 min; from 20 B to 98% B, 2 min; 98% B, 2 min; from 98 B to 5% B, 2 min; 5% B, 2 min. For LC–MS/MS analyses, the ESI interface was used in positive ion mode with the following settings: temperature (TEM) 550°C, the nebulizer gas (GS1) was air at 50 psi, the heater gas (GS2) was air at 50 psi, the ion spray voltage was 5,500 V, scan range (MS): 100–1,000 m/z, scan range (MS/MS): 50–1,000 m/z.
Electrophoretic Mobility Shift Assay
EMSA was performed as described by Zhang and colleagues (Zhang et al., 2013). The iuc promoter probe (151 bp) was amplified from the Y. pseudotuberculosis YPIII genome with primers PiucA-F and PiucA-R. Increasing concentrations of purified His6-Fur (0.24, 0.48, and 0.72 μM) were incubated with 5 ng DNA probes in EMSA buffer (20 mM Tris-HCl, pH 7.4, 4 mM MgCl2, 100 mM NaCl, 100 μM MnCl2, 1 mM dithiothreitol, 10% glycerol). After incubation for 20 min at room temperature, the binding reaction mixture was subjected to electrophoresis on a 6% native polyacrylamide gel containing 5% glycerol in 0.5 × TBE (Tris-borate-EDTA) electrophoresis buffer, and the DNA probe was detected using SYBR Green. As negative controls, a 151-bp fragment amplified from clpV4 (ypk_3559) coding region using primers control-F/control-R (Supplementary Table 2) was included in the binding assay.
Quantitative Real-Time-PCR
Bacteria were harvested during the mid-exponential phase, and RNA was extracted using the RNAprep Pure Cell/Bacteria Kit and treated with RNase-free DNase (TIANGEN, Beijing, China). The purity and concentration of the RNA were determined by gel electrophoresis and spectrophotometer (NanoDrop, Thermo Fisher Scientific). The first-strand cDNA was reverse transcribed from 1 μg of total RNA with the TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Quantitative real-time PCR (qRT-PCR) was performed in CFX96 Real-Time PCR Detection System (Bio-Rad, United States) with TransStart Green qPCR SuperMix (TransGen Biotech, Beijing, China). For all primer sets (Supplementary Table 2), the following cycling parameters were used: 95°C for 30 s followed by 40 cycles of 94°C for 15 s, 50°C for 30 s. For standardization of results, the relative abundance of 16S rRNA was used as the internal standard. All samples were analyzed in triplicate, and the expression of target genes was calculated as relative fold values using the 2–ΔΔCT method. These assays were performed in triplicate at least three times, and error bars represent standard error of the mean.
Mouse Infections
All mice were maintained and handled in accordance with the animal welfare assurance policy issued by Northwest A&F University. The mouse assay was performed as previously described (Schweer et al., 2013; Song et al., 2021). Mid-exponential phase Y. pseudotuberculosis strains grown in YLB medium at 26°C were washed twice in sterilized PBS and used for orogastric infection of 6—7-week-old female C57BL/6 mice using a ball-tipped feeding needle. For survival assays, 1 × 109 bacteria of each strain were applied to different groups of mice, and the survival rate of the mice was determined by monitoring the everyday survival for 21 days. For the analysis of the bacterial load in the feces, the feces were sampled from individual living mice at specific time points, weighed, and homogenized in PBS. For the analysis of the bacterial load in the cecum, colon, small intestine, spleen, and liver, the mice were sacrificed by carbon dioxide asphyxiation followed by cervical dislocation at specific time points after infection, the tissues were weighed and homogenized in PBS, and serial dilutions of the homogenates were plated on YLB plates with 20 μg ml–1 of nalidixic acid. The colony-forming units (CFU) were counted and are given as CFU per g organ/tissue. C57BL/6 mice were purchased from the Animal Center of Xi’An JiaoTong University (SCXK: Shan 2012-003, Xi’an, China). All mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China.
Statistical Analysis
Experimental data analyzed for significance were performed by using GraphPad Prism 8 (GraphPad Software, San Diego, CA, United States). The p-values for mice survival were calculated using log-rank (Mantel–Cox) test. The p-values for bacterial CFU in mouse tissues were calculated using Mann–Whitney test (I). Statistical analyses for the rest of the assays were performed using unpaired two-tailed Student’s t-test. Error bars represent ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Northwest A&F University.
Author Contributions
CL, LZ, and XS designed the research and drafted the manuscript. CL, DP, LZ, ML, LS, DY, YZ, KW, YL, and ZW performed the experimental work. CL, DP, and LZ analyzed the data. YW and ZL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grant of the National Key R&D Program of China (2018YFA0901200 to XS), the National Natural Science Foundation of China (31725003 and 31670053 to XS and 31970114 and 31671292 to YW), and the China Postdoctoral Science Foundation (2020M673501 to LZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Life Science Research Core Services (LSRCS), NWAFU (LL), for the technical support to the HPLC-MS/MS assay.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.699913/full#supplementary-material
References
Achard, M. E., Chen, K. W., Sweet, M. J., Watts, R. E., Schroder, K., Schembri, M. A., et al. (2013). An antioxidant role for catecholate siderophores in Salmonella. Biochem. J. 454, 543–549. doi: 10.1042/BJ20121771
Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A., and Carniel, E. (1999). Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 96, 14043–14048. doi: 10.1073/pnas.96.24.14043
Adler, C., Corbalan, N. S., Peralta, D. R., Pomares, M. F., de Cristobal, R. E., and Vincent, P. A. (2014). The alternative role of enterobactin as an oxidative stress protector allows Escherichia coli colony development. PLoS One 9:e84734. doi: 10.1371/journal.pone.0084734
Bailey, D. C., Alexander, E., Rice, M. R., Drake, E. J., Mydy, L. S., Aldrich, C. C., et al. (2018). Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J. Biol. Chem. 293, 7841–7852. doi: 10.1074/jbc.RA118.002798
Banerjee, R., Weisenhorn, E., Schwartz, K. J., Myers, K. S., Glasner, J. D., Perna, N. T., et al. (2020). Tailoring a global iron regulon to a uropathogen. mBio 11:e00351-20. doi: 10.1128/mBio.00351-20
Banin, E., Vasil, M. L., and Greenberg, E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081. doi: 10.1073/pnas.0504266102
Begg, S. L. (2019). The role of metal ions in the virulence and viability of bacterial pathogens. Biochem. Soc. Trans. 47, 77–87. doi: 10.1042/BST20180275
Bogomolnaya, L. M., Tilvawala, R., Elfenbein, J. R., Cirillo, J. D., and Andrews-Polymenis, H. L. (2020). Linearized siderophore products secreted via MacAB efflux pump protect Salmonella enterica Serovar Typhimurium from oxidative stress. mBio 11:e00528-20. doi: 10.1128/mBio.00528-20
Buchrieser, C., Rusniok, C., Frangeul, L., Couve, E., Billault, A., Kunst, F., et al. (1999). The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67, 4851–4861. doi: 10.1128/IAI.67.9.4851-4861.1999
Calder, J. T., Christman, N. D., Hawkins, J. M., and Erickson, D. L. (2020). A trimeric autotransporter enhances biofilm cohesiveness in Yersinia pseudotuberculosis but not in Yersinia pestis. J. Bacteriol. 202:e00176-20. doi: 10.1128/JB.00176-20
Cassat, J. E., and Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. doi: 10.1016/j.chom.2013.04.010
Chakraborty, R., Storey, E., and van der Helm, D. (2007). Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli. Biometals 20, 263–274. doi: 10.1007/s10534-006-9060-9
Chaturvedi, K. S., Hung, C. S., Crowley, J. R., Stapleton, A. E., and Henderson, J. P. (2012). The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736. doi: 10.1038/nchembio.1020
Chen, L. H., Lin, C. H., and Chung, K. R. (2013). A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol. Plant Pathol. 14, 497–505. doi: 10.1111/mpp.12021
Chung, P. Y. (2016). The emerging problems of Klebsiella pneumoniae infections: carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 363:fnw219. doi: 10.1093/femsle/fnw219
Darby, C., Hsu, J. W., Ghori, N., and Falkow, S. (2002). Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244. doi: 10.1038/417243a
de Lorenzo, V., Bindereif, A., Paw, B. H., and Neilands, J. B. (1986). Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165, 570–578. doi: 10.1128/jb.165.2.570-578.1986
de Lorenzo, V., Wee, S., Herrero, M., and Neilands, J. B. (1987). Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169, 2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987
Di Lorenzo, M., and Stork, M. (2014). Plasmid-encoded iron uptake systems. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.PLAS-0030-2014
Dong, T. G., Dong, S., Catalano, C., Moore, R., Liang, X., and Mekalanos, J. J. (2015). Generation of reactive oxygen species by lethal attacks from competing microbes. Proc. Natl. Acad. Sci. U.S.A. 112, 2181–2186. doi: 10.1073/pnas.1425007112
Drakesmith, H., and Prentice, A. M. (2012). Hepcidin and the iron-infection axis. Science 338, 768–772. doi: 10.1126/science.1224577
Escolar, L., Perez-Martin, J., and de Lorenzo, V. (1998). Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283, 537–547. doi: 10.1006/jmbi.1998.2119
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Forman, S., Nagiec, M. J., Abney, J., Perry, R. D., and Fetherston, J. D. (2007). Analysis of the aerobactin and ferric hydroxamate uptake systems of Yersinia pestis. Microbiology 153, 2332–2341. doi: 10.1099/mic.0.2006/004275-0
Forman, S., Paulley, J. T., Fetherston, J. D., Cheng, Y. Q., and Perry, R. D. (2010). Yersinia ironomics: comparison of iron transporters among Yersinia pestis biotypes and its nearest neighbor, Yersinia pseudotuberculosis. Biometals 23, 275–294. doi: 10.1007/s10534-009-9286-4
Galaris, D., and Pantopoulos, K. (2008). Oxidative stress and iron homeostasis: mechanistic and health aspects. Crit. Rev. Clin. Lab. Sci. 45, 1–23. doi: 10.1080/10408360701713104
Gibson, F., and Magrath, D. I. (1969). The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta 192, 175–184. doi: 10.1016/0304-4165(69)90353-5
Hancock, V., Ferrieres, L., and Klemm, P. (2008). The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology (Reading) 154, 167–175. doi: 10.1099/mic.0.2007/011981-0
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Hinnebusch, B. J., and Erickson, D. L. (2008). Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Top. Microbiol. Immunol. 322, 229–248. doi: 10.1007/978-3-540-75418-3_11
Jin, Z., Li, J., Ni, L., Zhang, R., Xia, A., and Jin, F. (2018). Conditional privatization of a public siderophore enables Pseudomonas aeruginosa to resist cheater invasion. Nat. Commun. 9:1383. doi: 10.1038/s41467-018-03791-y
Kalidasan, V., Azman, A., Joseph, N., Kumar, S., Awang Hamat, R., and Neela, V. K. (2018). Putative iron acquisition systems in Stenotrophomonas maltophilia. Molecules 23:2048. doi: 10.3390/molecules23082048
Keogh, D., Tay, W. H., Ho, Y. Y., Dale, J. L., Chen, S., Umashankar, S., et al. (2016). Enterococcal metabolite cues facilitate interspecies niche modulation and polymicrobial infection. Cell Host Microbe 20, 493–503. doi: 10.1016/j.chom.2016.09.004
Kramer, J., Ozkaya, O., and Kummerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Kumar, A., Alam, A., Rani, M., Ehtesham, N. Z., and Hasnain, S. E. (2017). Biofilms: survival and defense strategy for pathogens. Int. J. Med. Microbiol. 307, 481–489. doi: 10.1016/j.ijmm.2017.09.016
Kupper, F. C., Carrano, C. J., Kuhn, J. U., and Butler, A. (2006). Photoreactivity of iron(III)-aerobactin: photoproduct structure and iron(III) coordination. Inorg. Chem. 45, 6028–6033. doi: 10.1021/ic0604967
Lemaitre, C., Bidet, P., Bingen, E., and Bonacorsi, S. (2012). Transcriptional analysis of the Escherichia coli ColV-Ia plasmid pS88 during growth in human serum and urine. BMC Microbiol. 12:115. doi: 10.1186/1471-2180-12-115
Li, C., Zhu, L., Pan, D., Li, S., Xiao, H., Zhang, Z., et al. (2019). Siderophore-mediated iron acquisition enhances resistance to oxidative and aromatic compound stress in Cupriavidus necator JMP134. Appl. Environ. Microbiol. 85:e01938-18. doi: 10.1128/AEM.01938-18
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Liu, S., Gunawan, C., Barraud, N., Rice, S. A., Harry, E. J., and Amal, R. (2016). Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ. Sci. Technol. 50, 8954–8976. doi: 10.1021/acs.est.6b00835
Llamas, M. A., Imperi, F., Visca, P., and Lamont, I. L. (2014). Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597. doi: 10.1111/1574-6976.12078
Marchler-Bauer, A., Zheng, C., Chitsaz, F., Derbyshire, M. K., Geer, L. Y., Geer, R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352. doi: 10.1093/nar/gks1243
Martinez-Chavarria, L. C., and Vadyvaloo, V. (2015). Yersinia pestis and Yersinia pseudotuberculosis infection: a regulatory RNA perspective. Front. Microbiol. 6:956. doi: 10.3389/fmicb.2015.00956
McDougall, S., and Neilands, J. B. (1984). Plasmid- and chromosome-coded aerobactin synthesis in enteric bacteria: insertion sequences flank operon in plasmid-mediated systems. J. Bacteriol. 159, 300–305. doi: 10.1128/JB.159.1.300-305.1984
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/MMBR.00012-07
Nairz, M., and Weiss, G. (2020). Iron in infection and immunity. Mol. Aspects Med. 75:100864. doi: 10.1016/j.mam.2020.100864
Nakashige, T. G., Zhang, B., Krebs, C., and Nolan, E. M. (2015). Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol. 11, 765–771. doi: 10.1038/nchembio.1891
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608. doi: 10.1128/IAI.54.3.603-608.1986
Neilands, J. B. (1981). Iron absorption and transport in microorganisms. Annu. Rev. Nutr. 1, 27–46. doi: 10.1146/annurev.nu.01.070181.000331
Neilands, J. B. (1992). Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30). Can. J. Microbiol. 38, 728–733. doi: 10.1139/m92-119
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Ojha, A., and Hatfull, G. F. (2007). The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 66, 468–483. doi: 10.1111/j.1365-2958.2007.05935.x
Okujo, N., and Yamamoto, S. (1994). Identification of the siderophores from Vibrio hollisae and Vibrio mimicus as aerobactin. FEMS Microbiol. Lett. 118, 187–192. doi: 10.1111/j.1574-6968.1994.tb06824.x
Oliveira, F., Franca, A., and Cerca, N. (2017). Staphylococcus epidermidis is largely dependent on iron availability to form biofilms. Int. J. Med. Microbiol. 307, 552–563. doi: 10.1016/j.ijmm.2017.08.009
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Palmer, L. D., and Skaar, E. P. (2016). Transition metals and virulence in bacteria. Annu. Rev. Genet. 50, 67–91. doi: 10.1146/annurev-genet-120215-035146
Payne, S. M. (1980). Synthesis and utilization of siderophores by Shigella flexneri. J. Bacteriol. 143, 1420–1424. doi: 10.1128/JB.143.3.1420-1424.1980
Peigne, C., Bidet, P., Mahjoub-Messai, F., Plainvert, C., Barbe, V., Medigue, C., et al. (2009). The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 77, 2272–2284. doi: 10.1128/IAI.01333-08
Perry, R. D., Bobrov, A. G., and Fetherston, J. D. (2015). The role of transition metal transporters for iron, zinc, manganese, and copper in the pathogenesis of Yersinia pestis. Metallomics 7, 965–978. doi: 10.1039/c4mt00332b
Rada, B., and Leto, T. L. (2013). Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 21, 73–81. doi: 10.1016/j.tim.2012.10.004
Rakin, A., Schneider, L., and Podladchikova, O. (2012). Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol. 2:151. doi: 10.3389/fcimb.2012.00151
Ratledge, C., and Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Saha, R., Saha, N., Donofrio, R. S., and Bestervelt, L. L. (2013). Microbial siderophores: a mini review. J. Basic Microbiol. 53, 303–317. doi: 10.1002/jobm.201100552
Schweer, J., Kulkarni, D., Kochut, A., Pezoldt, J., Pisano, F., Pils, M. C., et al. (2013). The cytotoxic necrotizing factor of Yersinia pseudotuberculosis (CNFY) enhances inflammation and Yop delivery during infection by activation of Rho GTPases. PLoS Pathog. 9:e1003746. doi: 10.1371/journal.ppat.1003746
Schwiesow, L., Mettert, E., Wei, Y., Miller, H. K., Herrera, N. G., Balderas, D., et al. (2018). Control of hmu heme uptake genes in Yersinia pseudotuberculosis in response to iron sources. Front. Cell. Infect. Microbiol. 8:47. doi: 10.3389/fcimb.2018.00047
Sheldon, J. R., Laakso, H. A., and Heinrichs, D. E. (2016). Iron acquisition strategies of bacterial pathogens. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.VMBF-0010-2015
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Song, L., Pan, J., Yang, Y., Zhang, Z., Cui, R., Jia, S., et al. (2021). Contact-independent killing mediated by a T6SS effector with intrinsic cell-entry properties. Nat. Commun. 12:423. doi: 10.1038/s41467-020-20726-8
Stojiljkovic, I., Baumler, A. J., and Hantke, K. (1994). Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236, 531–545. doi: 10.1006/jmbi.1994.1163
Stuart, S. J., Prpic, J. K., and Robins-Browne, R. M. (1986). Production of aerobactin by some species of the genus Yersinia. J. Bacteriol. 166, 1131–1133. doi: 10.1128/jb.166.3.1131-1133.1986
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Weinberg, E. D. (1989). Cellular regulation of iron assimilation. Q. Rev. Biol. 64, 261–290. doi: 10.1086/416359
Xu, S., Peng, Z., Cui, B., Wang, T., Song, Y., Zhang, L., et al. (2014). FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 16, 1090–1104. doi: 10.1111/1462-2920.12222
Xu, Z., Mandic-Mulec, I., Zhang, H., Liu, Y., Sun, X., Feng, H., et al. (2019). Antibiotic bacillomycin D affects iron acquisition and biofilm formation in Bacillus velezensis through a Btr-mediated FeuABC-dependent pathway. Cell Rep. 29, 1192.e–1202.e. doi: 10.1016/j.celrep.2019.09.061
Yang, L., Nilsson, M., Gjermansen, M., Givskov, M., and Tolker-Nielsen, T. (2009). Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74, 1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11:5371. doi: 10.1038/s41467-020-19243-5
Zhang, W., Wang, Y., Song, Y., Wang, T., Xu, S., Peng, Z., et al. (2013). A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 15, 557–569. doi: 10.1111/1462-2920.12005
Keywords: Yersinia pseudotuberculosis, Fur, aerobactin, siderophore, iron acquisition, oxidative stress, biofilm formation, virulence
Citation: Li C, Pan D, Li M, Wang Y, Song L, Yu D, Zuo Y, Wang K, Liu Y, Wei Z, Lu Z, Zhu L and Shen X (2021) Aerobactin-Mediated Iron Acquisition Enhances Biofilm Formation, Oxidative Stress Resistance, and Virulence of Yersinia pseudotuberculosis. Front. Microbiol. 12:699913. doi: 10.3389/fmicb.2021.699913
Received: 24 April 2021; Accepted: 09 June 2021;
Published: 15 July 2021.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyRunhua Han, University of Texas at Austin, United States
David Erickson, Brigham Young University, United States
Copyright © 2021 Li, Pan, Li, Wang, Song, Yu, Zuo, Wang, Liu, Wei, Lu, Zhu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingfang Zhu, bGluZ2Zhbmd6aHVAbndhZnUuZWR1LmNu; Xihui Shen, eGlodWlzaGVuQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Changfu Li1,2†
Changfu Li1,2† Yao Wang
Yao Wang Luting Song
Luting Song Zhiqiang Lu
Zhiqiang Lu Xihui Shen
Xihui Shen