
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 July 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.698478
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
The rapidly increasing prevalence of Klebsiella pneumoniae carbapenemase 2 (KPC-2)-producing bacteria has become a serious challenge to public health. Currently, the blaKPC–2 gene is mainly disseminated through plasmids of different sizes and replicon types. However, the plasmids carrying the blaKPC–2 gene have not been fully characterized. In this study, we report the complete genome sequences of two novel blaKPC–2-harboring incompatibility group U (IncU) plasmids, pEC2341-KPC and pEC2547-KPC, from international high-risk clones of Escherichia coli isolated from Zhejiang, China. Two KPC-2-producing E. coli isolates (EC2341 and EC2547) were collected from clinical samples. Whole-genome sequencing (WGS) analysis indicated that EC2341 and EC2547 belonged to the ST410 and ST131 clones, respectively. S1-nuclease pulsed-field gel electrophoresis (S1-PFGE), Southern blot and conjugation experiments confirmed the presence of the blaKPC–2 gene on the pEC2341-KPC plasmid and that this was a conjugative plasmid, while the blaKPC–2 gene on the pEC2547-KPC plasmid was a non-conjugative plasmid. In addition, plasmid analysis further revealed that the two blaKPC–2-harboring plasmids have a close evolutionary relationship. To the best of our knowledge, this is the first report of E. coli strains carrying the blaKPC–2 gene on IncU plasmids. The emergence of the IncU-type blaKPC–2-positive plasmid highlights further dissemination of blaKPC–2 in Enterobacteriaceae. Therefore, effective measures should be taken immediately to prevent the spread of these blaKPC–2–positive plasmids.
The rapidly increasing prevalence of KPC-producing bacteria has become a serious challenge to public health (Suay-García and Pérez-Gracia, 2019). At the time of writing (April 2021), 82 variants of KPC enzymes (KPC-1 to KPC-82) have been identified among gram-negative bacteria worldwide1. Among these carbapenemases, KPC-2 was first identified from a Klebsiella pneumoniae strain in the United States in 2003 (Smith Moland et al., 2003) and attracted extensive attention because of its rapid worldwide dissemination. Currently, the blaKPC–2 gene is prevalent in K. pneumoniae strains, and the sequence type 258 (ST258) clone has successfully spread worldwide (Munoz-Price et al., 2013).
Although not as common as in K. pneumoniae, the blaKPC–2 gene has also been identified in Escherichia coli strains. Some reports, including two from our group, have recently found that the blaKPC–2 gene was present in the ST131-type E. coli strains, which are international multidrug-resistant high-risk clones (Du et al., 2020; Wang et al., 2020). KPC-2-producing E. coli strains were isolated not only from humans but also from animals, such as cattle (Vikram and Schmidt, 2018), swine (Liu et al., 2018) and cats (Sellera et al., 2018). Unfortunately, blaKPC–2 has also been identified in environmental samples [urban rivers (Xu et al., 2015), drinking water (Mahmoud et al., 2020), and vegetables (Wang et al., 2018)], indicating its presence in the environment. In addition, blaKPC–2 was further disseminated through plasmids of different sizes and replicon types (Mathers et al., 2017), such as the pKpQIL-like plasmid (Chen et al., 2014b), the IncFIA plasmid (Chen et al., 2014a), the IncI2 plasmid (Chen et al., 2013), the IncX3 plasmid (Fuga et al., 2020), the IncP-6 plasmid (Hu et al., 2019) and the IncN plasmid (Schweizer et al., 2019). The movement of blaKPC plasmids into E. coli strains that are known pathogens of urinary tract and intra-abdominal infections raises clinical concerns (Bratu et al., 2007). Plasmid transfer will further lead to continued spread of resistance and limit clinical treatment options (Chen et al., 2014). However, plasmids carrying the blaKPC–2 gene have not been fully characterized.
In the present study, we reported the complete sequences of two novel blaKPC–2-harboring IncU plasmids from international high-risk clones of E. coli ST131 and ST410 isolates from China. In addition, the whole genome sequence revealed that the two blaKPC–2-positive plasmids have a close evolutionary relationship.
In a retrospective study, 109 carbapenem-resistant Enterobacteriaceae strains were isolated from June 2018 to September 2019. Common carbapenemase genes (blaKPC, blaNDM, blaVIM, and blaIMP) were amplified, and the positive products were sequenced. Two KPC-2-producing E. coli strains were included in this study and further identified by the VITEK MS system (bioMérieux, Marcy-l’Etoile, France).
Antimicrobial susceptibility testing was carried out using the broth microdilution method according to the protocol of CLSI guidelines (CLSI, 2020). Minimum inhibitory concentrations (MICs) were interpreted according to the guideline document established by Clinical and Laboratory Standards Institute (CLSI, 2020). For tigecycline and polymyxin E, the MIC results were categorized in accordance with the breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing criteria2. E. coli ATCC 25922 was used as a quality control strain.
The plasmid location of the blaKPC–2 gene was determined by Southern blot experiments according to the previous study (Wang et al., 2020). Briefly, whole chromosomal DNA was digested with S1-nuclease (TaKaRa, Japan). The digested fragments were electrophoresed on a CHEF-mapper XA pulsed-field gel electrophoresis (PFGE) system (Bio-Rad, United States) for 18 h at 14°C. The DNA fragments were transferred to a positively charged nylon membrane (Millipore, United States) and then hybridized with a digoxigenin-labeled blaKPC–2-specific probe. The fragments was detected by an NBT/BCIP color detection kit (Roche, Germany). The Salmonella enterica serotype Braenderup H9812 was used as the size marker.
A filter-mating experiment was performed with E. coli J53 as the recipient strain and blaKPC–2–positive isolates as the donor strains. Transconjugants were selected on Mueller-Hinton agar plates supplemented with 300 mg/L sodium azide and 100 mg/L ampicillin. The transconjugants were confirmed by PCR sequencing and antimicrobial susceptibility testing.
Total genomic DNA extraction and analysis were carried out according to previously described methods (Wang et al., 2020). Briefly, the QIAamp DNA MiniKit (Qiagen, Valencia, CA, United States) was used to extract the genomic DNA of two strains for genome sequencing. A NextEra XT DNA library preparation kit (Illumina, Inc., Cambridge, United Kingdom) was used to prepare the DNA library. Genomic DNA was sequenced on an Illumina HiSeqTM 4000 instrument with a 150-bp paired-end approach at a depth of approximately 200×. The CLC Genomics Workbench 10.0 was used to assemble the raw reads of the strains into draft genomes using. In addition, a Pacific Biosciences RSII DNA sequencing system (PacBio, Menlo Park, CA, United States) was used to obtain the complete genomes of strains EC2341 and EC2547. The resulting sequences were de novo assembled using the Hierarchical Genome Assembly Process (HGAP_Assembly.2) with the default settings of the SMRT Analysis v2.3.0 software package.
The Rapid Annotation using Subsystems Technology (RAST) annotation website server3 was used to annotate the genomes. A schematic map of the linear comparison of the two blaKPC–2-positive plasmids and their related plasmids was generated with EasyFig 2.2.2 (Sullivan et al., 2011). Multi-locus sequence typing (MLST) of the strain and incompatibility typing of the blaKPC–2–positive plasmid were performed with the assistance of the PlasmidFinder-1.3 server and the MLST 2.0 server, which are available at the Center for Genomic Epidemiology4.
In addition, plasmid stability was determined according to a previous study (Li et al., 2018).
The complete sequences of the plasmids pEC2341-KPC (accession number CP072979) and pEC2547-KPC (accession number CP072981) were deposited in DDBJ/EMBL/GenBank.
In the present study, two KPC-2-producing isolates were collected from a teaching hospital in Zhejiang, China. E. coli strains EC2341 and EC2547 were isolated from urine and sputum, respectively. The antimicrobial susceptibility testing results showed that the blaKPC–2-positive isolates were resistant to carbapenems, cephalosporins, amoxicillin/clavulanate, ciprofloxacin, and amikacin but were susceptible to colistin, tigecycline and ceftazidime-avibactam (Table 1).
The MLST results showed that E. coli strains EC2547 and EC2341 belonged to ST131 and ST410, respectively. The ST131 clone-type E. coli strain emerged in the mid-2000s and has spread worldwide (Can et al., 2015). Similar to clone lineage of ST131, the E. coli ST410 strain has been confirmed as another successful clone in E. coli (Schaufler et al., 2016). Furthermore, these two clone-type E. coli strains have gained a further selective advantage due to acquisition of carbapenem resistance (Du et al., 2020; Lee and Choi, 2020). In addition, other resistance genes, such as blaCTX–M–3, blaCTX–M–27, fosA3, and qnrS1, were also detected in the E. coli strains by analysis of the genome sequences. Multiple resistance genes were identified in the ST410 and ST131 strains, indicating that these two clone-type strains might be more capable of acquiring resistance genes.
Notably, these two international high-risk clones have caused a wide variety of clinical infections (Roer et al., 2018; Wang et al., 2020) and are associated with treatment failure because of their high virulence potential (Can et al., 2015). In the present study, multiple potential virulence factors were identified by VirulenceFinder analysis of E. coli EC2341 and EC2547 strains, such as ompA (outer membrane protein A), fdeC (adhesin), and fepC (iron-enterobactin transporter). blaKPC–2 was present in the ST131 and ST410 strains, further supporting the results that these two clone types may become a successful lineage of KPC-2-producing E. coli strains.
To ascertain the plasmid location of the blaKPC–2 gene, S1-PFGE was performed followed by Southern blot experiments. The blaKPC–2 gene was located on two plasmids of different sizes, ca. 80 Kb and ca. 100 Kb (data not shown). The transferability of the two blaKPC–2-positive plasmids was further determined by filter mating experiments. The EC2341 isolate tested could successfully transfer its carbapenem-resistance to E. coli strain J53 (Table 1), while the EC2547 isolate could not transfer its carbapenem resistance. Additionally, the blaKPC–2-positive plasmids were both stable in the two isolates by plasmid stability experiments. In the absence of antibiotics, the randomly selected strains all carried the blaKPC–2-positive plasmid that was identical to the parental isolate after 12 rounds of subculture on MH agar.
Incompatibility plasmid classification showed that the two blaKPC–2-positive plasmids were both grouped into IncU replicon types. The IncU plasmid incompatibility group was assigned in 1981 (Sirgel et al., 1981) and is a unique group of mobile elements with highly conserved backbone functions and variable antibiotic resistance gene cassettes (Tschäpe et al., 1981; Rhodes et al., 2000). The IncU incompatibility group has been isolated from a number of Aeromonas spp. and E. coli strains from natural and clinical environments (Tschäpe et al., 1981; Sandaa and Enger, 1994; Adams et al., 1998; Rhodes et al., 2000). Various resistance genes have also been described for IncU plasmids, such as qnrS2, aac(6′)-Ib-cr, aadA1 and aadA2, sulI and sulII, dfrA16 dfrIIc (dfrB3) and catAII (Sørum et al., 2003). However, carbapenem-resistant IncU plasmids have not been found previously. In this study, the blaKPC–2 gene was confirmed to be carried on the IncU plasmids. To the best of our knowledge, this is the first report of E. coli strains carrying the blaKPC–2 gene on IncU plasmids. Our study further demonstrated that plasmids harboring the blaKPC–2 gene were diverse.
Two entire sequences were obtained to further characterize the IncU plasmids carrying blaKPC–2. Sequence analysis showed that plasmid pEC2341_KPC was 76,952 bp in size, had 51.9% G + C content, and harbored 133 predicted ORFs (Figure 1A). The core region of pEC2341_KPC includes a replication module (repE), one transfer (tra) system, and a stability operon (stbAB and umuCD). Four antimicrobial resistance genes, qnrS1, blaCTX–M–13, blaTEM–1, and drfA14, were detected in this plasmid except for the blaKPC–2 gene. In addition, a class 1 integron-like element was also detected in this plasmid. The element is a dfrA14 gene with its 3′-conserved sequence truncated by the insertion of an IS6100 element. Sequence alignments revealed that the plasmid sequences were almost identical to those previously reported plasmids pECN-580 (KF914891) of E. coli ECN580 (97% coverage, 99.97% identity) in China (Chen et al., 2014c) and pCRKP-1-KPC (KX928750) of K. pneumoniae CRKP-1-KPC (96% coverage, 99.90% identity) in China (unpublished data) (Figure 2).
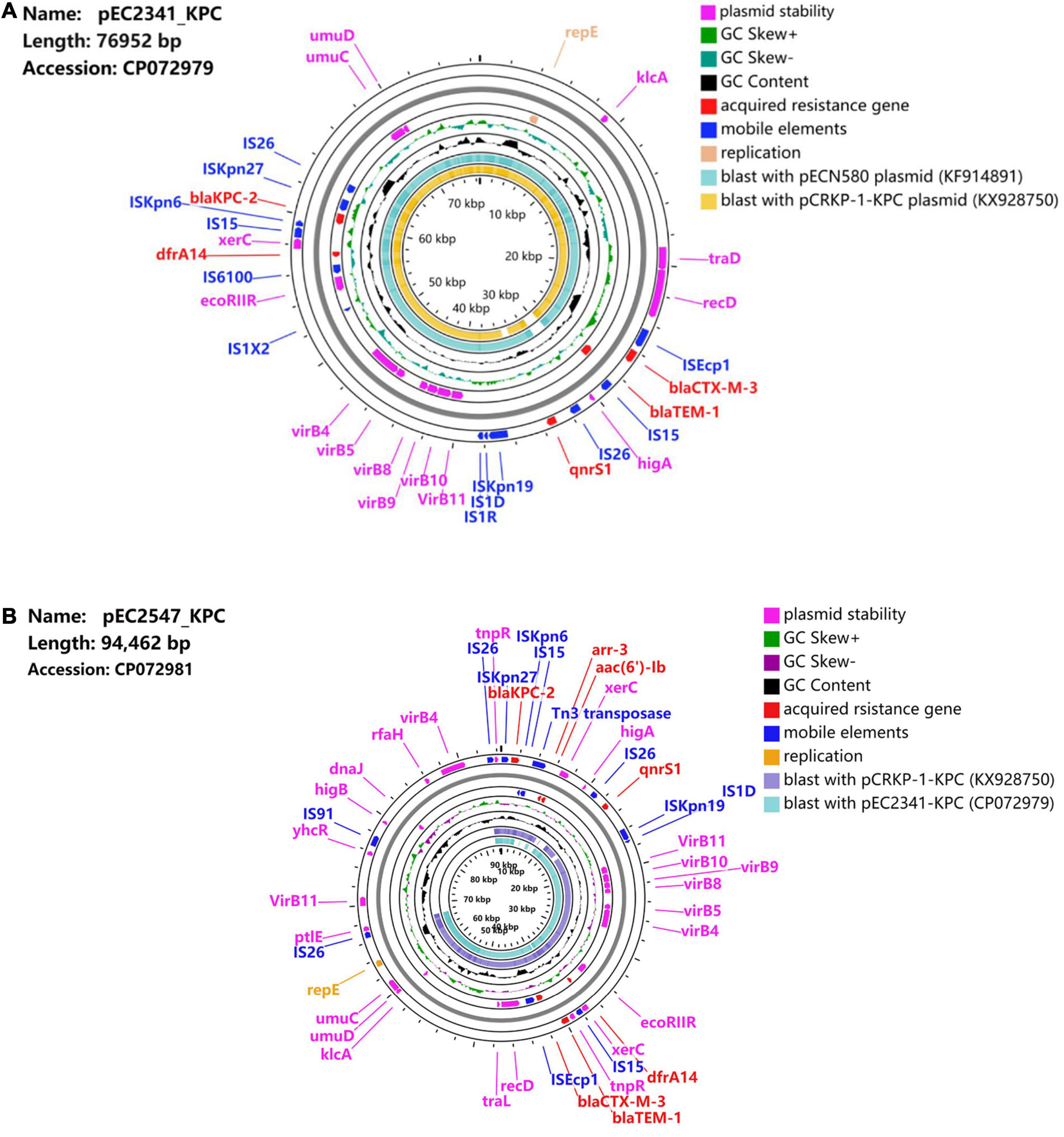
Figure 1. (A) Schematic map of plasmid pEC2341_KPC. Sequence alignment between pECN580 (accession number KF914891) and pCRKP-1-KPC (accession number KX928750) is the outer circle in orange and teal. (B) Schematic map of plasmid pEC2547_KPC. Sequence alignment between pEC2341-KPC (accession number CP072979) and pCRKP-1-KPC (accession number KX928750) is the outer circle in teal and blue.
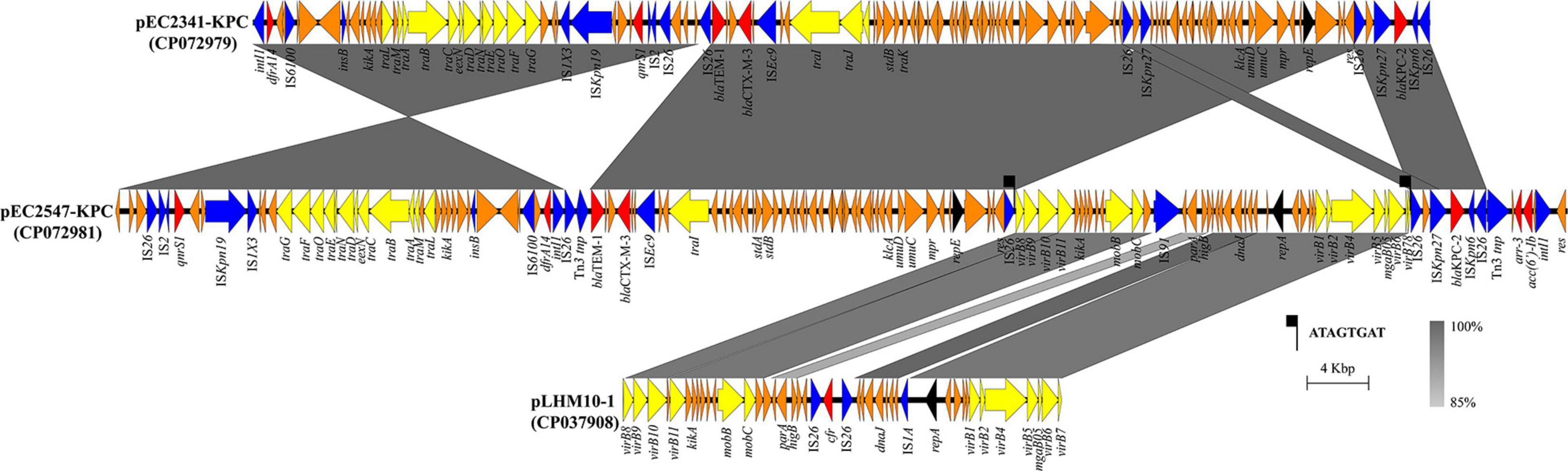
Figure 2. Linear characterization of the plasmids pEC2341-KPC and pEC2547-KPC with closely related plasmid. The gray regions between plasmids indicate nucleotide identity (85–100%) by BLASTn. Arrows indicate predicted ORFs. Primary structural characteristics of plasmids pEC2341-KPC (CP072979), pEC2547-KPC (CP072981), and pLHM10-1 (CP037908). Colored arrows represent open reading frames, with red, yellow, blue, and orange representing antibiotic resistance gene, plasmid transfer related genes, mobile elements and common function genes, respectively.
Plasmid pEC2547 contained blaKPC–2 and was 94,462 bp in size, with an average G + C content of 49.3% (Figure 1B). Compared with plasmid pEC2341_KPC, two other antimicrobial resistance genes, aar-3 and acc(6′)Ib, were identified in this plasmid. Two class 1 integron-like elements were identified in pEC2547_KPC. The first element is same as that in pEC2341_KPC. The second element is an IntI1-aac(6′)-Ib-cr-aar-3-Tn3 gene cassette located downstream of the blaKPC–2 gene.
Notably, sequencing analysis further indicated that pEC2547 might evolve from plasmid pEC2341_KPC of E. coli EC2341 (67% coverage, 100% identity) (Figure 2). Compared with plasmid pEC2341_KPC, an approximately 24-kb sequence flanked by two IS26 elements was carried on plasmid pEC2547, which resulted in disruption of the transfer systems of this plasmid. Consistent with our conjugate transfer results, the EC2547 isolate could not transfer its carbapenem resistance to E. coli strain J53. The 24-kb sequence was further aligned to an unnamed plasmid of E. coli strain LHM10-1 (GenBank accession number CP037908) with 89% coverage and 96.52% identity. This 24-kb composite transposon-like element flanking with two IS26 elements undergoes replicative transposition by the 8-bp target site duplication (TSD) (ATAGTGAT). IS26 elements have been demonstrated to undergo frequent intramolecular transposition and facilitate recombination between the plasmid or the chromosome (He et al., 2015). These findings suggest that the plasmid pEC2547 was composed of the pEC2341_KPC plasmid and an unnamed plasmid of E. coli strain LHM10-1, which was a composite transposon formed by IS26 (Figure 2).
In addition, the blaKPC–2 gene carried on the two plasmids was preceded by IS26, ISKpn27, and ISKpn6, and followed by IS26. In China, blaKPC–2 genetic environments can be classified into three main types: Tn4401 with the ISKpn7-blaKPC–2-ISKpn6 core structure, Tn1722-based unit transposons with the ISKpn27-blaKPC–2-ISKpn6 core structure and IS26-based composite transposons with the ISKpn27-blaKPC–2-ISKpn6 core structure (Wang et al., 2015). In this study, blaKPC–2 genes were both located in an approximately 5-kb composite transposon-like element with the ISKpn27 insertion sequence upstream and the ISKpn6 insertion sequence downstream of the element and flanked by two IS26 elements bracketed by IS26, which belonged to the IS26-based composite transposon. IS26-based composite transposons are mainly carried by IncN-type plasmids. Our plasmids belonged to the IncU type, which led to speculation that the IS26 elements may promote recombination between the plasmids and explain the movement of the new IncU regions.
Overall, we describe here the complete sequences of two novel blaKPC–2-positive IncU plasmids from E. coli isolates. The two blaKPC–2-harboring plasmids have a close evolutionary relationship, which highlighted the diversity of these highly promiscuous plasmids. The spread of blaKPC–2 harboring multidrug-resistant plasmids, e.g., pEC2341-KPC and pEC2547-KPC, into the international high-risk clones E. coli ST131 and ST410, presents tremendous challenges for clinicians. It is important for the IncU-type plasmid to further disseminate blaKPC–2 in Enterobacteriaceae in order for it to be maintained. Therefore, effective measures should be taken immediately to prevent the spread of these blaKPC–2–positive plasmids.
The complete sequences of the plasmids pEC2341-KPC (accession number CP072979) and pEC2547-KPC (accession number CP072981) were deposited in DDBJ/EMBL/GenBank).
The Ethics Committee of the Zhejiang Provincial People’s Hospital exempted this study from review because the present study focused on bacteria.
XL and HP conceived and designed the experiments. WW, LL, and WF performed the experiments. CC and DJ analyzed the data. WW and XL wrote the manuscript. All authors read and approved the final manuscript.
This study was supported by the Public Technology Research Projects of Zhejiang Province, China (LGD21H190001), Natural Science Foundation of Zhejiang Province, China (Q18H010007), and Medical and Health Research Project (2020KY420). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LL was employed by company Adicon Clinical Laboratories.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adams, C. A., Austin, B., Meaden, P. G., and Mcintosh, D. (1998). Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64, 4194–4201. doi: 10.1128/aem.64.11.4194-4201.1998
Bratu, S., Brooks, S., Burney, S., Kochar, S., Gupta, J., Landman, D., et al. (2007). Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44, 972–975. doi: 10.1086/512370
Can, F., Azap, O. K., Seref, C., Ispir, P., Arslan, H., and Ergonul, O. (2015). Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin. Infect. Dis. 60, 523–527. doi: 10.1093/cid/ciu864
Chen, L., Chavda, K. D., Al Laham, N., Melano, R. G., Jacobs, M. R., Bonomo, R. A., et al. (2013). Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 57, 5019–5025. doi: 10.1128/aac.01397-13
Chen, L., Chavda, K. D., Melano, R. G., Hong, T., Rojtman, A. D., Jacobs, M. R., et al. (2014a). Molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 58, 2289–2294. doi: 10.1128/aac.02749-13
Chen, L., Chavda, K. D., Melano, R. G., Jacobs, M. R., Koll, B., Hong, T., et al. (2014b). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob. Agents Chemother. 58, 2871–2877. doi: 10.1128/aac.00120-14
Chen, L., Hu, H., Chavda, K. D., Zhao, S., Liu, R., Liang, H., et al. (2014c). Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/aac.02587-13
Chen, L., Mathema, B., Chavda, K. D., Deleo, F. R., Bonomo, R. A., and Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Du, X., Huang, J., Wang, D., Zhu, Y., Lv, H., and Li, X. (2020). Whole genome sequence of an Escherichia coli ST131 strain isolated from a patient with bloodstream infection in China co-harbouring bla(KPC-2), bla(CTX-M-3), bla(CTX-M-14), qnrS1, aac(3)-IIa and aac(6′)-Ib-cr genes. J. Glob. Antimicrob. Resist. 22, 700–702. doi: 10.1016/j.jgar.2020.06.027
Fuga, B., Ferreira, M. L., Cerdeira, L. T., De Campos, P. A., Dias, V. L., Rossi, I., et al. (2020). Novel small IncX3 plasmid carrying the bla(KPC-2) gene in high-risk Klebsiella pneumoniae ST11/CG258. Diagn. Microbiol. Infect. Dis. 96:114900. doi: 10.1016/j.diagmicrobio.2019.114900
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762.
Hu, X., Yu, X., Shang, Y., Xu, H., Guo, L., Liang, Y., et al. (2019). Emergence and characterization of a novel IncP-6 plasmid harboring bla (KPC-2) and qnrS2 genes in Aeromonas taiwanensis isolates. Front. Microbiol. 10:2132. doi: 10.3389/fmicb.2019.02132
Lee, M., and Choi, T. J. (2020). Species transferability of Klebsiella pneumoniae Carbapenemase-2 isolated from a high-risk clone of Escherichia coli ST410. J. Microbiol. Biotechnol. 30, 974–981. doi: 10.4014/jmb.1912.12049
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al. (2018). Dissemination of bla(NDM-5) gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59.
Liu, X., Liu, H., Wang, L., Peng, Q., Li, Y., Zhou, H., et al. (2018). Molecular characterization of extended-spectrum β-lactamase-producing multidrug resistant Escherichia coli from Swine in Northwest China. Front. Microbiol. 9:1756. doi: 10.3389/fmicb.2018.01756
Mahmoud, N. E., Altayb, H. N., and Gurashi, R. M. (2020). Detection of carbapenem-resistant genes in Escherichia coli isolated from drinking water in khartoum, Sudan. J. Environ. Public Health 2020:2571293.
Mathers, A. J., Stoesser, N., Chai, W., Carroll, J., Barry, K., Cherunvanky, A., et al. (2017). Chromosomal integration of the Klebsiella pneumoniae carbapenemase gene, bla(KPC), in klebsiella species is elusive but not rare. Antimicrob. Agents Chemother. 61:e01823-16.
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/s1473-3099(13)70190-7
Rhodes, G., Huys, G., Swings, J., Mcgann, P., Hiney, M., Smith, P., et al. (2000). Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet A. Appl. Environ. Microbiol. 66, 3883–3890. doi: 10.1128/aem.66.9.3883-3890.2000
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18.
Sandaa, R. A., and Enger, O. (1994). Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl. Environ. Microbiol. 60, 4234–4238. doi: 10.1128/aem.60.12.4234-4238.1994
Schaufler, K., Semmler, T., Wieler, L. H., Wöhrmann, M., Baddam, R., Ahmed, N., et al. (2016). Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410–another successful pandemic clone? FEMS Microbiol. Ecol. 92:fiv155. doi: 10.1093/femsec/fiv155
Schweizer, C., Bischoff, P., Bender, J., Kola, A., Gastmeier, P., Hummel, M., et al. (2019). Plasmid-mediated transmission of KPC-2 carbapenemase in Enterobacteriaceae in critically Ill patients. Front. Microbiol. 10:276. doi: 10.3389/fmicb.2019.00276
Sellera, F. P., Fernandes, M. R., Ruiz, R., Falleiros, A. C. M., Rodrigues, F. P., Cerdeira, L., et al. (2018). Identification of KPC-2-producing Escherichia coli in a companion animal: a new challenge for veterinary clinicians. J. Antimicrob. Chemother. 73, 2259–2261.
Sirgel, F. A., Coetzee, J. N., Hedges, R. W., and Lecatsas, G. (1981). Phage C-1: an IncC group; plasmid-specific phage. J. Gen. Microbiol. 122, 155–160. doi: 10.1159/000149385
Smith Moland, E., Hanson, N. D., Herrera, V. L., Black, J. A., Lockhart, T. J., Hossain, A., et al. (2003). Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51, 711–714. doi: 10.1093/jac/dkg124
Sørum, H., L’abée-Lund, T. M., Solberg, A., and Wold, A. (2003). Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47, 1285–1290. doi: 10.1128/aac.47.4.1285-1290.2003
Suay-García, B., and Pérez-Gracia, M. T. (2019). Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics 8:122. doi: 10.3390/antibiotics8030122
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tschäpe, H., Tietze, E., and Koch, C. (1981). Characterization of conjugative R plasmids belonging to the new incompatibility group IncU. J. Gen. Microbiol. 127, 155–160. doi: 10.1099/00221287-127-1-155
Vikram, A., and Schmidt, J. W. (2018). Functional bla(KPC-2) sequences are present in U.S. beef cattle feces regardless of antibiotic use. Foodborne Pathog. Dis. 15, 444–448. doi: 10.1089/fpd.2017.2406
Wang, D., Mu, X., Chen, Y., Zhao, D., Fu, Y., Jiang, Y., et al. (2020). Emergence of a clinical Escherichia coli sequence type 131 strain carrying a chromosomal bla (KPC-2) gene. Front. Microbiol. 11:586764. doi: 10.3389/fmicb.2020.586764
Wang, J., Yao, X., Luo, J., Lv, L., Zeng, Z., and Liu, J. H. (2018). Emergence of Escherichia coli co-producing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J. Antimicrob. Chemother. 73, 252–254. doi: 10.1093/jac/dkx335
Wang, L., Fang, H., Feng, J., Yin, Z., Xie, X., Zhu, X., et al. (2015). Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 6:838. doi: 10.3389/fmicb.2015.00838
Keywords: E. coli, KPC-2, IncU plasmid, high-risk clones, whole genome sequencing
Citation: Wu W, Lu L, Fan W, Chen C, Jin D, Pan H and Li X (2021) Complete Genome Sequences of Two Novel KPC-2-Producing IncU Multidrug-Resistant Plasmids From International High-Risk Clones of Escherichia coli in China. Front. Microbiol. 12:698478. doi: 10.3389/fmicb.2021.698478
Received: 21 April 2021; Accepted: 23 June 2021;
Published: 21 July 2021.
Edited by:
Yi-Wei Tang, Cepheid, United StatesReviewed by:
Jeong Hwan Shin, Inje University Busan Paik Hospital, South KoreaCopyright © 2021 Wu, Lu, Fan, Chen, Jin, Pan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Li, bGl4aV8wNjExQDE2My5jb20=; Hongying Pan, aHlwYW56anNybXl5QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.