
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 October 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.692707
This article is part of the Research TopicFlagellar Motors and Force Sensing in BacteriaView all 12 articles
Assembly of the bacterial flagellar rod, hook, and filament requires penetration through the peptidoglycan (PG) sacculus and outer membrane. In most β- and γ-proteobacteria, the protein FlgJ has two functional domains that enable PG hydrolyzing activity to create pores, facilitating proper assembly of the flagellar rod. However, two distinct proteins performing the same functions as the dual-domain FlgJ are proposed in δ- and ε-proteobacteria as well as spirochetes. The Lyme disease spirochete Borrelia burgdorferi genome possesses a FlgJ and a PG lytic SLT enzyme protein homolog (BB0259). FlgJ in B. burgdorferi is crucial for flagellar hook and filament assembly but not for the proper rod assembly reported in other bacteria. However, BB0259 has never been characterized. Here, we use cryo-electron tomography to visualize periplasmic flagella in different bb0259 mutant strains and provide evidence that the E580 residue of BB0259 is essential for PG-hydrolyzing activity. Without the enzyme activity, the flagellar hook fails to penetrate through the pores in the cell wall to complete assembly of an intact periplasmic flagellum. Given that FlgJ and BB0259 interact with each other, they likely coordinate the penetration through the PG sacculus and assembly of a functional flagellum in B. burgdorferi and other spirochetes. Because of its role, we renamed BB0259 as flagellar-specific lytic transglycosylase or LTaseBb.
In most bacterial species, the flagellum is a rotary nanomachine framed in a supramolecular structure that initiates assembly from the cell envelope and extends into the extracellular space. However, in motile spirochetes, this migratory organelle resides in the periplasmic space and is therefore called the periplasmic flagellum (Wolgemuth et al., 2006; Charon et al., 2009, 2012). Despite differences in location, the external flagella found in the model organisms Escherichia coli and Salmonella enterica serovar Typhimurium share a remarkably high degree of similarity with respect to overall structure, as the key components are conserved among the external flagella and the periplasmic flagella of spirochetes (Liu et al., 2009; Chen et al., 2011; Zhao et al., 2013; Terashima et al., 2017; Carroll and Liu, 2020). The supramolecular structure is composed of more than 25 distinct protein subunits and self-assembles to final lengths of up to 20μm in the model organisms. The individual flagellum consists of three major substructures: a basal body, hook, and filament (reviewed in Nakamura and Minamino, 2019; Carroll and Liu, 2020). Synthesis of the flagellum begins with the assembly of a basal body composed of an MS ring embedded in the cell membrane and a rod. The MS ring is comprised of multiple copies of FliF and acts as a platform for assembly of the rod, C-ring, torque-generating stator, and flagellum-specific type III secretion system (fT3SS). The rod is composed of five proteins, FliE, FlgB, FlgC, FlhO/FlgF, and FlgG. These rod proteins are transported from the cytoplasm to the periplasmic space via the fT3SS (Saijo-Hamano et al., 2019) and then assemble onto the MS ring—in an ordered fashion from the proximal rod to the distal rod (Zhao et al., 2013; Terashima et al., 2017; Carroll and Liu, 2020). The distal rod serves as the template for subsequent assembly of the P-ring and hook (B. burgdorferi lacks the L-ring; Zhao et al., 2013). The filaments are subsequently polymerized onto the hook to complete assembly of a flagellum. Hook and filament assembly is mediated by the hook cap FlgD and filament cap FliD protein, respectively (Homma and Iino, 1985; Ohnishi et al., 1994; Yonekura et al., 2000).
In order for the assembly of the rod with 30nm×4nm to penetrate the outer membrane in E. coli or S. enterica, a hole must be created in the peptidoglycan sacculus, as its mesh diameter is narrower than 2nm (Demchick and Koch, 1996; Herlihey et al., 2014). This PG hydrolytic activity is accomplished by a dual-domain protein FlgJ of S. enterica and E. coli. FlgJ is composed of an N-terminal scaffolding domain required for polymerization of the distal rod and a C-terminal domain required to form a hole in the PG sacculus. The C-terminal domain of FlgJ proteins belongs to the glycoside hydrolase family in which these β-N-acetyl-glucosaminidases cleave the β-1,4 glycosidic linkage between the N-acetylglucosamine and N-acetylmuramic acid residues of bacterial cell wall peptidoglycan, with Glu184 and Glu223 serving as the catalytic residues (Nambu et al., 1999; Herlihey et al., 2014; Zaloba et al., 2016).
Once the enzyme has made a hole in the cell wall, FlgG proteins are secreted and assemble in two stacks to make an 11nm-long rod just underneath the FlgJ rod cap, thereby spanning the remaining distance between the cell wall and outer membrane (Jones and Macnab, 1990; Cohen and Hughes, 2014). The ejection of the FlgJ rod cap allows the FlgD hook cap to assemble at the tip of the rod and results in the transition from rod to hook polymerization (Nambu et al., 1999; Cohen and Hughes, 2014). While dual-domain FlgJ proteins are found in most β- and γ-proteobacteria, two distinct proteins performing similar functions as the dual-domain FlgJ are reported in diverse bacterial genomes, including δ- and ε-proteobacteria as well as spirochete species B. burgdorferi, Leptospira interrogans, and Treponema (Nambu et al., 1999, 2006; Gonzalez-Pedrajo et al., 2002; Zaloba et al., 2016; Garcia-Ramos et al., 2018). Moreover, the single-domain flagellar scaffolding protein FlgJ is reported to interact with PG-lytic enzymes, such as soluble lytic transglycosylases (SLT) proteins (Garcia-Ramos et al., 2018). These specialized cell wall-degrading enzymes allow the efficient assembly and anchoring of supramolecular structures in the cell envelope. The SLTs represent one class of autolysins that act like a lysozyme (muramidase) to cleave the β-(1→4) glycosidic linkage between N-acetylmuramyl and N-acetylglucosaminyl residues of cell wall peptidoglycan. Unlike lysozymes, however, SLTs are not hydrolases but cleave the β-glycosidic linkages, with simultaneous production of 1,6-anhydromuramyl residues (Holtje et al., 1975; van Asselt et al., 1999; Blackburn and Clarke, 2001; Koraimann, 2003).
As B. burgdorferi FlgJ is reported to be a single-domain protein that lacks a PG-hydrolyzing enzymatic domain, we sought to investigate the genome to identify a protein that harbors this domain (Zhang et al., 2012). BB0531 and BB0259 identified in this communication are both predicted to possess PG-hydrolyzing enzyme activities. BB0531 possesses a glucosaminidase domain; however, subsequent analysis indicates that this protein is not involved in flagellar pore-forming activities. BB0259 possesses an SLT homolog at its C-terminal domain, and further analyses indicate that FlgJ specifically interacts with BB0259. Because of its apparent flagellar specific peptidoglycan lytic enzyme function in the spirochete, BB0259 is renamed as LTaseBb. FlgJ and LTaseBb appear to function distinctively compared to their counterparts from other bacteria. Our model describes how B. burgdorferi FlgJ and LTaseBb work synchronously to form a functional periplasmic flagellum.
Borrelia burgdorferi B31-A is a high-passage noninfectious clone used as the wild-type (WT) strain (Bono et al., 2000; Tilly et al., 2001). The isogenic bb0259 mutant and complemented strains were constructed as described below. All B. burgdorferi strains were grown in Barbour–Stoenner–Kelly II (BSK-II) liquid medium or on soft agarose plates at 35°C in the presence of 2.5% CO2 (Motaleb et al., 2007). Antibiotics, when needed, were supplied in BSK-II medium at proper concentrations: 200μg/ml kanamycin and/or 40μg/ml gentamycin. All Escherichia coli strains were cultured in Luria-Bertani (LB) liquid medium or on LB agar plates (Bertani, 1951, 2004). When required, 100μg/ml ampicillin or 4μg/ml gentamycin was supplemented into the LB medium.
Basic local alignment search tool (BLAST) was used to determine protein homologs from the sequence database (Altschul et al., 1990, 1997). For the BLAST analysis, a significant homolog is determined based on their E-value [the lower an E-value (the closer it is to zero), the more significant the score is]. Signal peptide was predicted using SignalP 5.0 (Almagro Armenteros et al., 2019) and Phobius (Kall et al., 2004, 2005, 2007). Analysis of bacterial lipoprotein was performed using DOLOP (Madan Babu and Sankaran, 2002; Babu et al., 2006). Clustal Omega was utilized to align multiple amino acid sequences (Sievers et al., 2011). FFAS server was used as described elsewhere.
Multiple attempts to express soluble B. burgdorferi BB0259 with variable lengths using short-affinity tags (His6 or Strep) failed, as all these recombinant BB0259 proteins with short affinity tags formed inclusion bodies in the E. coli host. In order to produce soluble recombinant BB0259 protein, a DNA fragment harboring the BB0259 open reading frame (ORF) without signal peptide (1-49 aa) was polymerase chain reaction (PCR)-amplified from chromosomal DNA of B. burgdorferi B31-A using the primers PF rBB0259-BamHI (GGATCC TGGTTATGGAATTTTGATTATAC) and PR rBB0259-PstI (CTGCAGTTAATTTTTGGGGAATTCGCCC; restriction sites are underlined) and cloned into the pMAL c5x (NEB Inc.) through BamHI and PstI restriction sites to produce maltose binding protein (MBP)-tagged BB0259. Similarly, the 1xFLAG (DYKDDDDK) tagged FlgJ with gene locus number (BB0771a, also named BB0858) was constructed for far-western or affinity blotting. A DNA fragment of full-length flgJ fused with 1xFLAG tag coding sequence (GACTACAAAGACGATGACGACAAG) at C-terminus was amplified by PCR with primers PF MBPFlgJBamHI (CGTCGACGGATCCGAAACCAAAATTAAT TCACAAAATC) and PR MBPFlgJPstI (TAATTACCTGCAG TTACTTGTCGTCATCGTCTTTGTAGTCTTTACTTTTTTGTA ATTGATTGTA) and cloned into the pMAL c5x using BamHI and PstI restriction sites. MBP tagged FlaB (BB0147), FlgE (BB0283), FlgD (BB0284), FlgG (BB0774), and FlgL (BB0182) were also prepared to determine protein–protein interactions. The coding regions of FlaB, FlgE, FlgG, and FlgL were amplified by PCR with primers PF MBPflaB_NotI (GTCCATGGGCGG CCGCATGATTATCAATCATAATACATC) and PR MBPflaB_BamHI (GGAATTCGGATCCTTATCTAAGCAATGACAAAA CATATTG), PF MBPflgE_NotI (GTCCATGGGCGGCCGC ATGAT GAGGTCTTTATATTCTGG) and PR MBPflgE_BamHI (GGAATTCGGATCCTTAATTTTTCAATCTTACAAGTTCTTG), PF MBPflgG_NotI (GTCCA TGGGCG GCCGCATGATGAGA GCATTATGGACAGC) and PR MBPflgG_BamHI (GGAATTC GGATCC TTATTGCCTTTTTAAGTTATTTGC), PF MBPflgL_NotI (GTCCATGGGCGGCCGCATGATAAATAGAGTAAGTCA TCC) and PR MBPflgL_BamHI (GGAATTCGGATCCTTATTT TATAAAATCTAATAAAGTCG), respectively, and cloned into the pMAL c5x (NEB Inc.) using NotI and BamHI restriction sites. Recombinant FlgD construct was similarly prepared using NdeI-NcoI restriction enzymes with primers PF MBPflgD_NdeI (GCTGGACGACATATGAATAAAATAAACGGTGTTGAAAATG) and PR MBPflgD_NcoI (TGCTGGGCACCATGG TTATTCC TCCAAACCTACCGATAAT). All E. coli DH5α strains carrying the pMAL c5x constructs for expressing BB0259, FlaB, FlgE, FlgG, FlgL, FlgD, 1xFLAG tagged FlgJ were induced with 0.25mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at room temperature, and purifications of recombinant proteins were performed with amylose resin (NEB Inc.) according to the manufacturer’s protocol.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as mentioned previously (Motaleb et al., 2000). Exponentially growing B. burgdorferi cells were harvested and washed with phosphate-buffered saline (PBS) and resuspended in the same buffer to process the preparation of cell lysate for SDS-PAGE. Immunoblotting was performed with B. burgdorferi FlaB, FlgE, MotB, FliL, and DnaK-specific antibodies (Motaleb et al., 2000, 2011; Sal et al., 2008; Sultan et al., 2015) and Pierce™ ECL or ECL 2 substrate (Thermo Fisher Scientific Inc.). Concentration of the proteins was determined using a Bio-Rad protein assay kit with bovine serum albumin (BSA) as the standard. Unless specified, approximately 10μg of cell lysates was subjected to SDS-PAGE.
Far-western or affinity blotting assay with recombinant proteins was performed as described previously (Toker and Macnab, 1997; Kariu et al., 2015; Moon et al., 2016, 2018; Xu et al., 2019). Briefly, 1μg purified recombinant proteins was subjected to SDS-PAGE and Coomassie blue staining or transferred to polyvinylidene difluoride (PVDF) membrane. The PVDF membranes were blocked in the blocking solution (5% BSA, 10mM Tris, 150mM NaCl and 0.3% Tween 20, pH 7.4) with gentle shaking for 4 to 6h at room temperature, and then the membranes were incubated with or without the purified FLAG-FlgJ protein at the concentration 2μg/ml in blocking solution overnight. The membranes were washed 3 times with the washing buffer (10mM Tris, 150mM NaCl and 0.3% Tween 20, pH 7.4) and then immunoblotted with monoclonal anti-FLAG® M2 antibody (Sigma-Aldrich Co. LLC) followed by ECL 2 detection, as mentioned above. The X-ray films were exposed to the membranes for less than a minute.
Construction of the bb0259 and bb0531 inactivation plasmids, electroporation, and plating of B. burgdorferi were as described earlier (Motaleb et al., 2000; Sultan et al., 2010, 2013; Moon et al., 2016; Xu et al., 2017). bb0259 was inactivated by PflgB-kanamycin resistance cassette (PflgB-aph1), which possessed the flgB promoter driving kanamycin resistance gene (Ge et al., 1997; Bono et al., 2000; Li et al., 2010; Zhang et al., 2012; Moon et al., 2016; Xu et al., 2017, 2019). In short, DNA fragment including upstream 394-bp region of bb0259, bb0259 containing five HindIII sites, and downstream 324-bp region of bb0259 was amplified by PCR from chromosomal DNA of wild-type B. burgdorferi strain B31-A through Expand™ High Fidelity PCR System (Sigma-Aldrich Inc.) using primers BB0259-KO-F (GTTGAGTATATTGACGAGAAG) and BB0259-KO-R (TCCCAACAA CTCCGGTAACA), which was further TA cloned into pGEM-T Easy (Promega Inc.), yielding pGEM-T Easy::bb0259. PflgB-Kan was introduced into pGEM-T Easy::bb0259 through the first and last HindIII sites located within bb0259 gene, creating pGEM-T Easy::bb0259KO-PflgB-Kan, of which the direction of transcription of PflgB-Kan was confirmed to be the same as that of bb0259. Plasmid pGEM-T Easy::bb0259KO-PflgB-Kan was linearized by NotI restriction digestion and electroporated into competent B. burgdorferi B31-A cells to create ∆bb0259 mutant cells. bb0531 mutant was similarly prepared using a promoter-less kanamycin cassette (Pl-Kan), as described (Sultan et al., 2010). Kanamycin-resistant transformants were screened by PCR to confirm the genotypes of the mutants.
The bb0259 mutant was complemented in trans using the B. burgdorferi-E. coli shuttle vector pBSV2G (Elias et al., 2003). In short, bb0259 was PCR amplified using primers BB0259 Comp F (GATCCATATGTTTAATAGAAGTTCTTG; underlined sequence is NdeI site) and BB0259 Comp R (CTGCAGTTAATTT TTGGGGAATTCGCCC) and ligated into pGEM-T Easy, yielding plasmid bb0259ORF-Easy. DNA fragment containing bb0259 ORF was then released using NdeI restriction digestion from bb0259ORF-Easy, and further ligated into NdeI-treated PflgB-motB-Easy vector (Sultan et al., 2015), creating plasmid PflgB-bb0259-Easy. Finally, this plasmid was digested with NotI and cloned into the same restriction site of the shuttle vector pBSV2G, generating pBSV2G::PflgB-bb0259. The newly constructed shuttle vector pBSV2G::PflgB-bb0259 was electroporated into the ∆bb0259 mutant cells, followed by selection with both kanamycin and gentamycin. The resistant clones, namely bb0259com, were analyzed by PCR for confirmation of the presence of plasmid pBSV2G::PflgB-bb0259.
Borrelia burgdorferi bb0259-E580Q (E glutamate to Q glutamine) and bb0259-D606N (D aspartate to N asparagine) point-mutated strains were constructed through allelic exchange mutagenesis (Yang et al., 2003). Simply, using B31-A chromosomal DNA as the template, the left arm (882bp, partial bb0259 gene), promoter-less kanamycin cassette (Pl-Kan; 846bp), and right arm (564bp, including 88bp leftover from bb0259 gene for potential ribosome binding and 476bp of partial bb0258 gene) were PCR amplified, respectively, using primer pairs P1F BB0259PointMuKOplKan (CCTAACGTAAGCGGAGAATACA AGAGTCTTTTGCATTCTG) and P1R BB0259Point MuKO plKan (TAAAATTGCTTTTAACTATTAATTTTTGGGGAATTC GCCC), P2F BB0259PointMuKOplKan (GGGCGAATTCCCC AAAAATTAATAGTTAAAAGCAATTTTA) and P2R BB0259 PointMuKOplKan (CTTTTCATACAAAGCATCATTTAGAAA AACTCATCGAGC), P3F BB0259PointMuKOplKan (GCTCGA TGAGTTTTTCTAAATGATGCTTTGTATGAAAAG) and P3R BB0259PointMuKOplKan (CCCAAGCCTTGCATCAGCCCCA TAAAAATTCCTGCTAAC). Through overlapping PCR, Pl-Kan was inserted between the left arm and right arm, and further cloned into pGEM-T Easy, creating vector pGEM-T Easy:: bb0259PointMuKO-Pl-Kan. Using pGEM-T Easy::bb0259 PointMuKO-Pl-Kan as the template, point mutations of BB0 259E580Q were introduced by QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies Inc.) using the primers PF BB0259E580Q (CTTTAAT AAAAGCACAA AGTAG CTTTG AAAAAAATG) and PR BB0259E580Q (CATTTTTTTCAA AGCTACTTTGT GCTTTTATTAAAG; bold and underlined sequences indicate the point mutation), to create pGEM-T Easy::bb0259E580QKO-Pl-Kan. Similarly, through site-directed mutagenesis pGEM-T Easy::bb0259D606NKO-Pl-Kan was generated with primers PF BB0259D606N (GCCATCAACAGC AAATAATATTTCTAAA GAACTTAAG) and PR BB0259D606N (CTTAAGTTCTTTA GAAATATTATTTGC TGTTGATGGC). These two vectors were linearized by NotI digestion and electroporated into the competent B. burgdorferi B31 A cells to generate bb0259-E590Q and bb0259-D606N mutant strains, of which the point mutations of bb0259 were further verified by sequencing.
Exponentially growing B. burgdorferi cells were observed using a Zeiss Axio Imager M1 dark-field microscope to determine bacterial morphology and motility. To evaluate the motility of B. burgdorferi cells, swarm plate assays were performed following our well-established protocol (Motaleb et al., 2000, 2007, 2011).
Frozen-hydrated B. burgdorferi specimens were prepared as described previously (Zhao et al., 2013). In short, the bacterial culture was centrifuged at 5,000×g for 5min, and the resulting pellets were suspended in PBS to achieve a cell concentration ~1×108/ml. After adding 10-nm gold marker solution, 5μl of the cell suspension was placed on freshly glow-discharged holey carbon grid (Quantifoil Cu R2/2, 200 mesh) for 25s. The grids were blotted with filter paper for 3 to 5s and rapidly frozen in liquid ethane, using a homemade plunger apparatus as described previously (Zhao et al., 2013).
The Δbb0531, Δbb0259, and complemented bb0259com strains were imaged at −170°C using a Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a 4,096-by-4,096 charge-coupled device (16-megapixel) camera (TVIPS Gmbh, Germany). The microscope was operated at 300kV with a magnification of ×31,000, resulting in an effective pixel size of 5.7Å. Using the FEI batch tomography program, low-dose single-axis tilt series were collected from each bacterium at ~8μm defocus with a cumulative dose of ~100 e−/Å2 distributed over 65 images. In total, 80 tomographic reconstructions of Δbb0259 cell tips and 10 tomographic reconstructions of complemented bb0259 cells were generated.
The bb0259 point mutants (bb0259-E580Q and bb0259-D606N) were imaged with Krios electron microscope (Thermo Fisher) with a field emission gun, a volta phase plate, and a direct electron detector (Gatan K3 Summit). SerialEM was used to collect tilt series at focus. A total dose of 55 e−/Å2 is distributed among 35 tilt images covering angles from −51° to +51° at tilt steps of 3°. For every single tilt series collection, dose-fractionated mode was used to generate 11 frames per projection image. Collected dose-fractionated data were first subjected to the MotionCor2 to generate drift-corrected files (Zheng et al., 2017). IMOD software was used to align the tilt series and to generate tomograms (Kremer et al., 1996). In total, 240 tomographic reconstructions of bb0259-E580Q cells and 10 tomographic reconstructions of bb0259-D606N cells were generated.
A total of 646 flagellar motor subtomograms were picked from Δbb0259 tomograms. The initial orientation of each particle was manually estimated by the C-ring and hook. The subtomograms were used to generate the averaged structure as described previously (Zhao et al., 2013).
The software package EMAN2 was used for 3D segmentation (Chen et al., 2017). Segmentation of tomograms was performed using supervised machine learning to segment features of interest. The outer membrane, inner membrane, peptidoglycan layer, and flagella were segmented in this manner and manually cleaned using ChimeraX (Goddard et al., 2018). An existing motor model derived from subtomogram averaging was manually inserted into the segmented model. ChimeraX was used for visualization.
The single-domain FlgJ of B. burgdorferi plays roles in hook and filament assembly but not in rod assembly in S. enterica (Nambu et al., 1999; Cohen and Hughes, 2014; Zaloba et al., 2016), as flgJ mutant cells possess an intact rod and P-ring but form partial hook and filament structures (Zhang et al., 2012). Given that an enzymatic activity is required for pore formation in the PG layer for assembly of the flagellar hook and filament, and that B. burgdorferi FlgJ lacks a peptidoglycan-hydrolyzing enzyme domain, we sought to determine an ORF containing an enzymatic domain in the Lyme disease spirochetal genome. Using the S. enterica FlgJ with GenBank accession number NP_460153 as a query in BLAST search, we found no significant homologs in B. burgdorferi (not shown; Nambu et al., 1999; Hirano et al., 2001; Cohen and Hughes, 2014). However, using the FFAS server,1 BB0531 was identified as the only significant homolog of FlgJ (homology score −41.70; a lower FFAS score indicates higher confidence of the prediction; Rychlewski et al., 2000; Jaroszewski et al., 2005, 2011). BB0531 is annotated as a hypothetical protein with unknown function in the B. burgdorferi genome; however, it is predicted to possess a mannosyl-glycoprotein endo-β-N-acetylglucosaminidase domain responsible for peptidoglycan remodeling or hydrolyzing activity (Pfam: PF01832). BB0531 shares only 15% amino acid sequence identity with S. enterica FlgJ and alignment of amino acid sequences of S. enterica FlgJ and BB0531 from FFAS shows that the catalytic residues of FlgJ are not well conserved in BB0531 leading us to speculate that BB0531 is not involved in peptidoglycan hydrolyzing activity for flagellar penetration (not shown).
Because B. burgdorferi lacks a significant homolog of S. enterica FlgJ, we used the model organism’s SLT domain as a query in the BLAST search and identified BB0259 as a homolog in B. burgdorferi. BB0259 shares 35–41% amino acid sequence identity with significant E-value at the SLT domain (Figure 1; Pfam: PF01464; Thunnissen et al., 1994; van Asselt et al., 1999; Koraimann, 2003). Importantly, the motifs and amino acid residues required for peptidoglycan lytic enzymatic activity are highly conserved in BB0259 (Figure 1). BB0259 possesses a signal sequence (1-24 aa) with lipobox consensus residues LVSC, an unknown domain with no homologs (25-558 aa), and an SLT domain at the C-terminal region (559-717 aa; Figure 1). The presence of these features led us to hypothesize that BB0259 is secreted into the periplasmic space using its signal sequence to create pores in the PG layer, thereby completing assembly of the flagellar hook–filament.
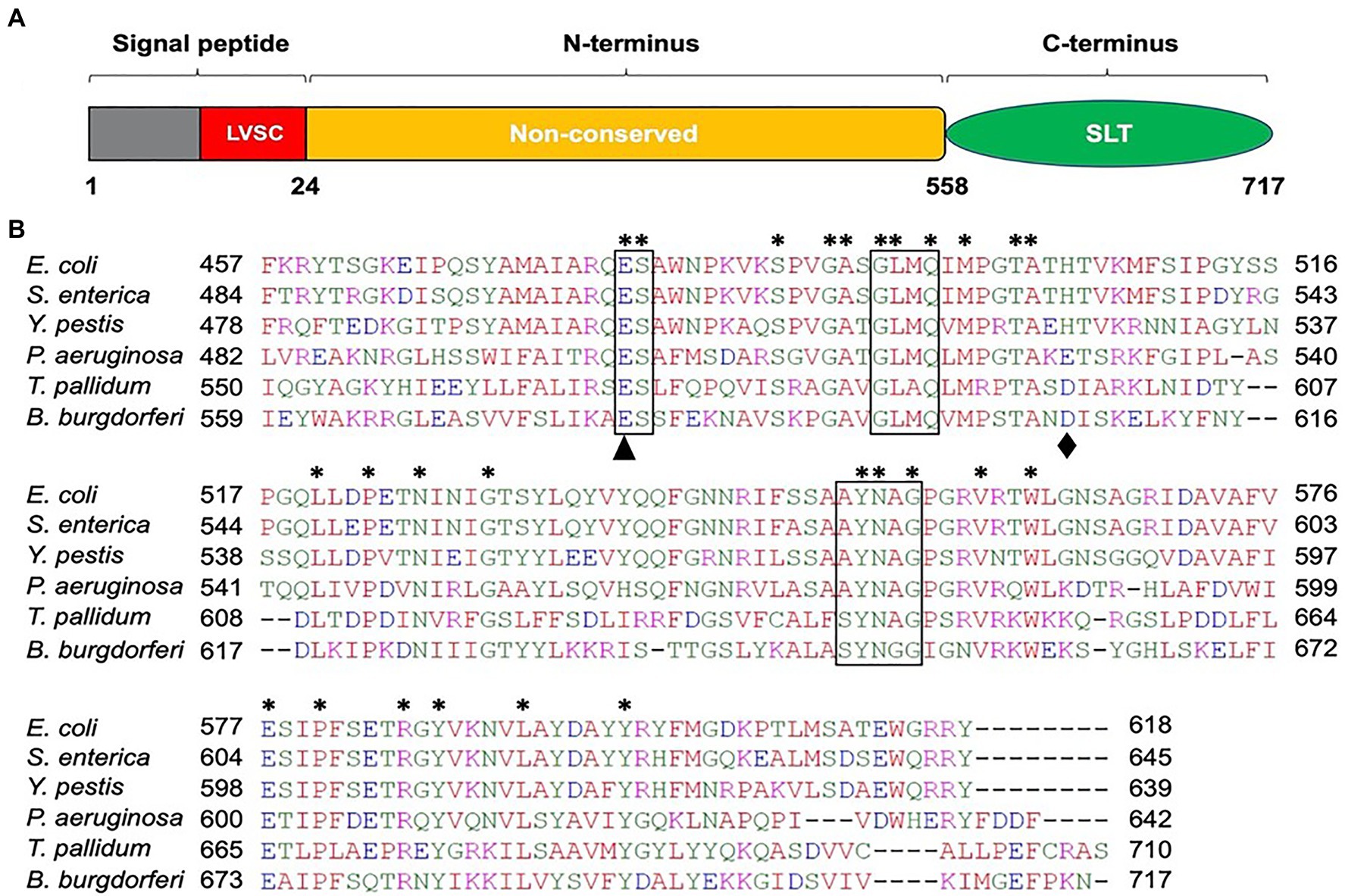
Figure 1. Amino acid sequence analysis of B. burgdorferi BB0259 (GenBank accession no. NP_212393). (A) Domain architecture of BB0259. BB0259 consists of 717 amino acids and is predicted to contain a lipobox consensus sequence (LVSC) in the signal peptide region (1-24 aa) determined by DOLOP program: https://www.mrc-lmb.cam.ac.uk/genomes/dolop/analysis.shtml. BB0259 also possesses a non-conserved N-terminal domain (25-558 aa) and a conserved C-terminal soluble lytic transglycosylase (SLT) domain (559-717 aa). Diagram is not in scale. (B) Multiple sequence alignment of SLT domains with GenBank accession no. 1QSA_A from E. coli, APW71272 from S. enterica, ANW15826 from Y. pestis, AID74248 from P. aeruginosa, and WP_010881492 from Treponema pallidum using Clustal Omega program: https://www.ebi.ac.uk/Tools/msa/clustalo/. Asterisks indicate identical residues. Boxed regions denote the conserved motifs ES-GLMQ-AYNAG among the SLT-domain proteins. S. enterica SLT domain shares 35% identity with the SLT domain of BB0259 with E-value 6e−25. Black triangle and diamond demonstrate the catalytic (E580) and non-conserved (D606) residues, respectively.
Completion of the appropriately assembled flagellar rod, hook, and filament is critical for the spirochete motility and morphology (Motaleb et al., 2000, 2015; Sal et al., 2008; Sultan et al., 2013; Zhao et al., 2013). If the SLT domain of BB0259 or the glucosaminidase domain of BB0531 displays PG-hydrolyzing activities, which are specific for flagellar assembly, then a deletion mutant in bb0259 or bb0531 is expected to be unable to create pores in the PG layer, resulting in defective rod/hook/filament assembly and a motility-deficient phenotype. To test this proposition, we created deletion mutants Δbb0259 and Δbb0531 (Supplementary Figures S1, S2, and S5). While the Δbb0531 cells lack any observable phenotype with respect to motility and morphology, the Δbb0259 mutant cells observed under a dark-field microscope are completely non-motile (Figure 2A and not shown). Swarm plate motility assays also indicated that Δbb0259 mutant cells are deficient in swarming out from the initial site of inoculation in soft agarose plates (Figure 2B). Furthermore, while wild-type cells show characteristic flat-wave morphology, Δbb0259 mutant cells are rod-shaped (Figure 2A). To demonstrate that the phenotypes described here are devoid of a polar effect on downstream gene expression or a secondary mutation elsewhere in the genome, we complemented the mutant in trans using a shuttle vector (Supplementary Figures S1, S2). Complemented bb0259 (bb0259com) cells observed under a microscope as well as in swarm plate motility assays indicated morphology and motility phenotypes restored to wild-type level (Figures 2A,B). Furthermore, SLT proteins important for hydrolyzing the PG sacculus for flagellar insertion and subsequent assembly of an intact flagellum are reported to be secreted in the periplasm using their Sec-signal peptide (Garcia-Ramos et al., 2018). Consistent with this observation, a recent study reported that BB0259 is associated with the inner membrane (Toledo et al., 2018). To determine whether the Sec-signal sequence is required for BB0259 proteins to be secreted in the periplasm, where they create pores in the PG sacculus for flagellar assembly and thereby provide motility to the cell, we complemented the non-motile Δbb0259 mutant without the signal sequence (bb0259Δsec). As described above, while the full-length bb0259 was able to complement the Δbb0259 mutant and therefore restored the morphology and motility phenotypes (Figures 2A,B), the Sec-signal sequence-deleted variant bb0259Δsec was unable to restore any of the phenotypes (data not shown). These results suggest that the signal sequence is essential for BB0259 proteins to be secreted in the periplasm.
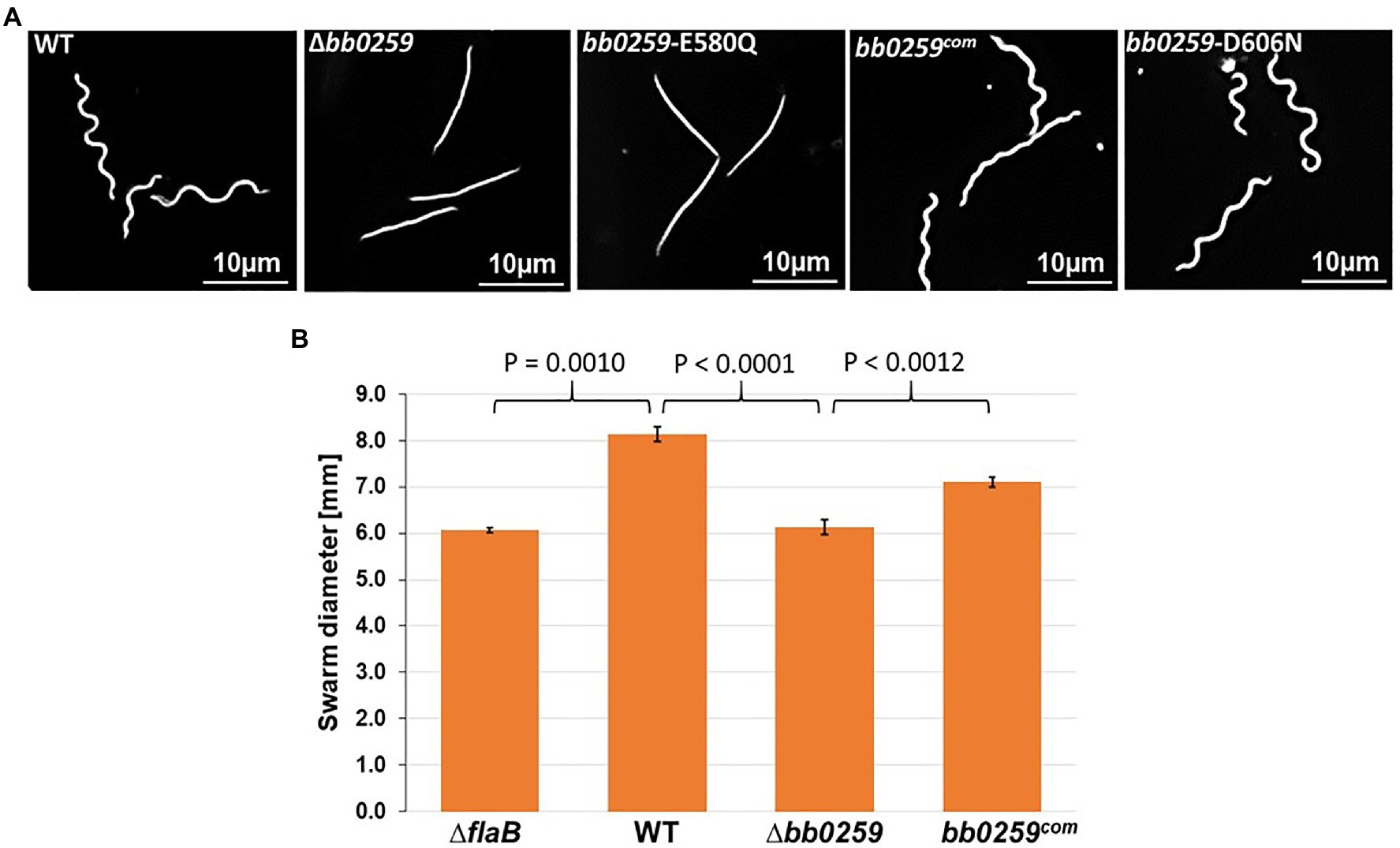
Figure 2. Morphology and motility phenotypes of the bb0259 mutant cells. (A) Dark-field microscopic images showing the characteristic flat-wave morphology of the WT, complemented bb0259com and bb0259-D606N cells and rod-shaped morphology of the Δbb0259 and bb0259-E580Q mutants. (B) Average swarm diameters from three swarm plates are shown in millimeter scale. bb0259-E580Q and bb0259-D606N mutants show motility phenotypes similar to the Δbb0259 and WT cells, respectively, and are not shown. A non-motile flagellar filament mutant ΔflaB was used as a control. Bars represent mean±standard deviation of the mean from three plates. Values of p between samples are shown at the top. A value of p<0.05 was considered as significantly different.
The altered motility and morphology phenotypes exhibited by the deletion mutant suggest that the rod/hook/filament structure was not properly assembled due to a lack of the SLT enzymatic activity of BB0259 that would hydrolyze the PG layer for flagellar structures to assemble through the PG sacculus and provide motility to the bacterial cell. B. burgdorferi cells lacking the gene-encoding filament FlaB or hook FlgE were reported to be non-motile and rod-shaped (Motaleb et al., 2000; Sal et al., 2008; Sultan et al., 2013). We therefore tested whether these gene products were synthesized in the Δbb0259 cells. Western blotting results (Supplementary Figure S3) indicate that the mutant cells synthesized very few FlaB proteins, with no effect on synthesis of the hook protein FlgE. Lack of the filaments in the Δbb0259 cells may thus have resulted in the motility-deficient and rod-shaped morphology (Figures 2A,B).
The SLT enzyme domain proteins possess a conserved ES-GLMQ-AYNAG motif in which the glutamate [E] residue is reported to be the catalytic residue (Figure 1; Thunnissen et al., 1994; van Asselt et al., 1999; Koraimann, 2003). The SLT domain identified in the C-terminal region of BB0259 possesses an E residue at position 580. To determine whether the E residue is the catalytic site, we created a point mutant in E580 of BB0259, resulting in bb0259-E580Q B. burgdorferi (Supplementary Figures S1, S2). Furthermore, the E223 and D248 residues of the S. enterica FlgJ enzymatic domain were reported to be important for PG-hydrolyzing activities (Nambu et al., 1999), and we noticed that the BB0259 SLT domain also encompasses similarly distanced E580 and D606 residues (Figure 1). Even though BB0259 shares limited homology with S. enterica FlgJ, and E580 and D606 do not align with E223 and D248 residues of FlgJ (not shown), we created a bb0259-D606N B. burgdorferi point mutant to determine whether this non-conserved D606 residue codes for any enzyme function (Supplementary Figures S1, S2). The bb0259-E580Q point mutant B. burgdorferi cells exhibited phenotypes identical to those of the deletion mutant Δbb0259 with respect to morphology and motility, whereas the bb0259-D606N cells were devoid of any detectable phenotype (Figures 2A,B). These results indicate that E580 of BB0259 is the catalytic site residue required for BB0259 function, while the D606 residue is not important for PG-hydrolyzing activities.
The morphology and motility phenotypes of the mutant cells led us to speculate that the periplasmic flagellar rod/hook/filament is not able to assemble properly due to the lack of PG-hydrolyzing activity of the Δbb0259 and bb0259-E580Q cells. Despite using the best possible assays to determine whether BB0259 is able to hydrolyze PG, as this protein possesses an SLT domain, our numerous attempts to demonstrate such activity resulted in little success. Attempts to determine the PG-hydrolyzing enzyme assays using various published assay conditions were unsuccessful even when utilizing the purified B. burgdorferi PG as a substrate (Jutras et al., 2016). As an alternative to enzyme assays, we used cryo-electron tomography (cryo-ET) to directly visualize the mutant’s flagella in situ to determine whether the rod/hook/filament assembles or whether the flagella are able to penetrate the PG-layer in the mutant cells. As shown in cryo-ET images (Supplementary Figure S4), the Δbb0259 mutant cells appear able to synthesize or assemble the rod, P-ring, and hook structures but lack the filament structure (Supplementary Figures S4A,B). The hook length of the Δbb0259 mutant cells was measured to be the same as that of wild-type cells (~51nm; n=20), and the hooks of the mutant cells look to be underneath the peptidoglycan layer (Figures 3E–H, 4D). However, similar cryo-ET data from the wild-type and complemented bb0259com cell tips show the intact rod, P-ring, and hook as well as filament (Supplementary Figures S4D,E,G). The averaged motor structure from the mutant cells appears to be identical to the wild-type motor structure, without any visible defect in the rod, P-ring, stator, C-ring, export apparatus, or collar structure (Supplementary Figures S4C,F). Importantly, the hooks from the bb0259-E580Q or Δbb0259 cells were found to be underneath the peptidoglycan layer (Figures 3E–H, 4D), whereas an intact rod-hook–filament structure was clearly assembled in the wild-type, bb0259-D606N point mutant, and complemented bb0259com cells (Figures 3A–D, 4C). The Δbb0259 or bb0259-E580Q mutant completed assembly of all the components of the flagella except the filament, and its hooks were not able to penetrate through the PG (Figures 3E–H, 4D). These results suggest that BB0259 possesses PG-lytic activity; that residue E580, but not D606, is essential for the catalytic activity of the enzyme; and that hydrolyzing the PG sacculus to allow the hook to penetrate through the cell wall is essential for completion of the assembly of an intact flagellum. We therefore renamed the BB0259 protein flagellar-specific LTaseBb. On the other hand, cryo-ET images of the Δbb0531 mutant cells show that the periplasmic flagella are completely assembled like the wild-type cells without any detectable defect in penetrating the PG layer or assembly of the flagella indicating that BB0531 is not involved in PG hydrolytic activity for flagellar penetration (Supplementary Figure S6).
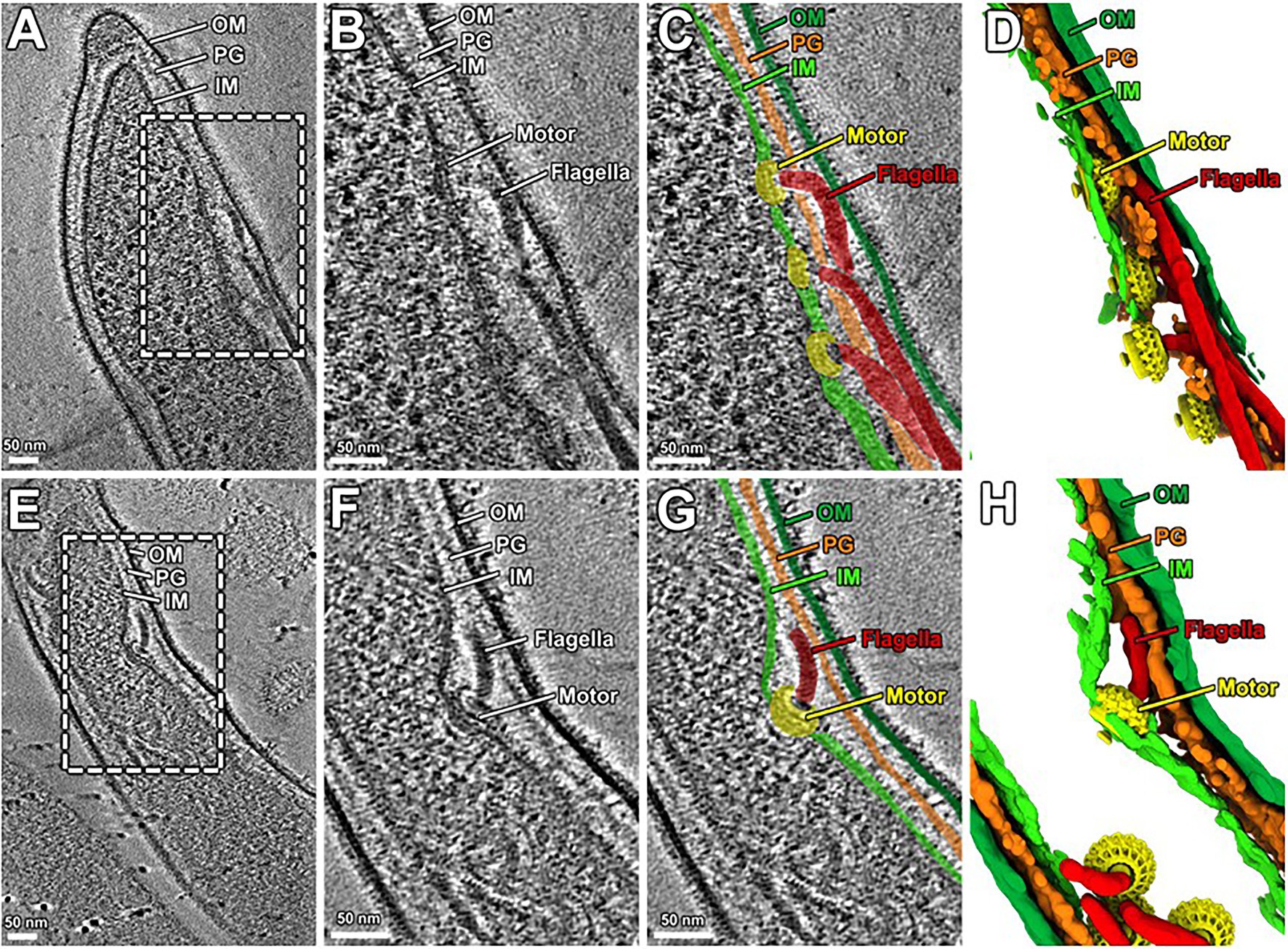
Figure 3. Cryo-ET analysis of bb0259-D606N and bb0259-E580Q. (A) A representative tomographic slice from a bb0259-D606N cell tip shows multiple intact flagella, including the motor, hook, and the filament. (B) A zoom-in view. (C) The motors are colored in yellow, and the hooks and filaments are in red. The inner membrane (light green), outer membrane (dark green), and PG (orange) are colored differently. (D) A 3D view shows the motors (yellow) and the hooks and filaments (red). (E) A representative tomographic slice from a bb0259-E580Q cell tip shows multiple flagella without filament. (F) A zoom-in view of one motor and its hook. (G) The motor is colored in yellow, and the hook is in red. (H) A 3D view shows the motors (yellow) and the hooks (red).
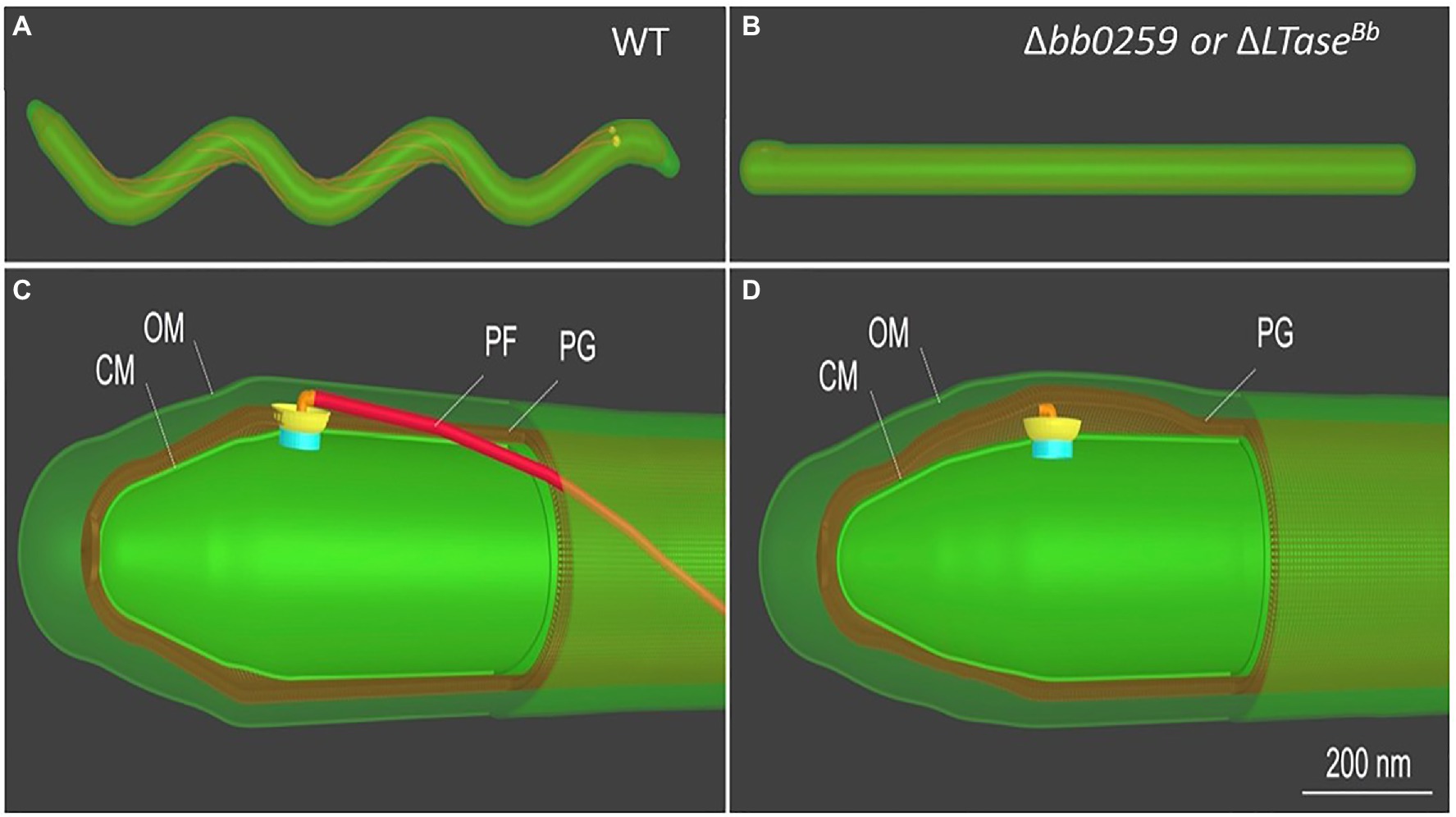
Figure 4. Schematic of B. burgdorferi WT and Δbb0259 mutant cells. B. burgdorferi cells summarizing the results from Figure 3. Δbb0259 is alternatively called ΔLTaseBb. (A) A WT B. burgdorferi cell has a flat-wave shape with multiple periplasmic flagella, which reside and rotate in the periplasmic space. (B) A ΔLTaseBb mutant cell is rod-shaped because of the lack of flagellar filaments. (C) A zoom-in view of the WT cell tip shows one motor and its long flagellar filament in the periplasmic space as its hook was able to penetrate the PG sacculus to complete assembly of an intact flagellum. (D) A zoom-in view of the ΔLTaseBb mutant cell shows a hook underneath the PG layer lacking the filament as its hook failed to penetrate the cell wall.
The requirement of PG-hydrolyzing activity for proper assembly of periplasmic flagella and consequently for motility suggests that LTaseBb is essential for creating space within the peptidoglycan sacculus for insertion of cell-envelope spanning structures such as flagella. Given that the single-domain FlgJ protein lacks a PG-hydrolyzing enzyme domain, LTaseBb is postulated to interact with FlgJ as the enzyme creates pores in the PG sacculus for the hook to assemble through the layer, subsequently allowing the completion of hook–filament structures for cells to be mobile. To test this proposition or to determine which proteins FlgJ interacts with, we performed far-western or affinity blotting. Recombinant FlgJ protein was used as a probe to incubate a PVDF membrane transferred with the distal rod protein FlgG, hook protein FlgE, hook cap protein FlgD, hook–filament junction protein FlgL, filament protein FlaB, and LTaseBb. As shown in Figure 5, FlgJ specifically interacts with LTaseBb and the hook cap protein FlgD.
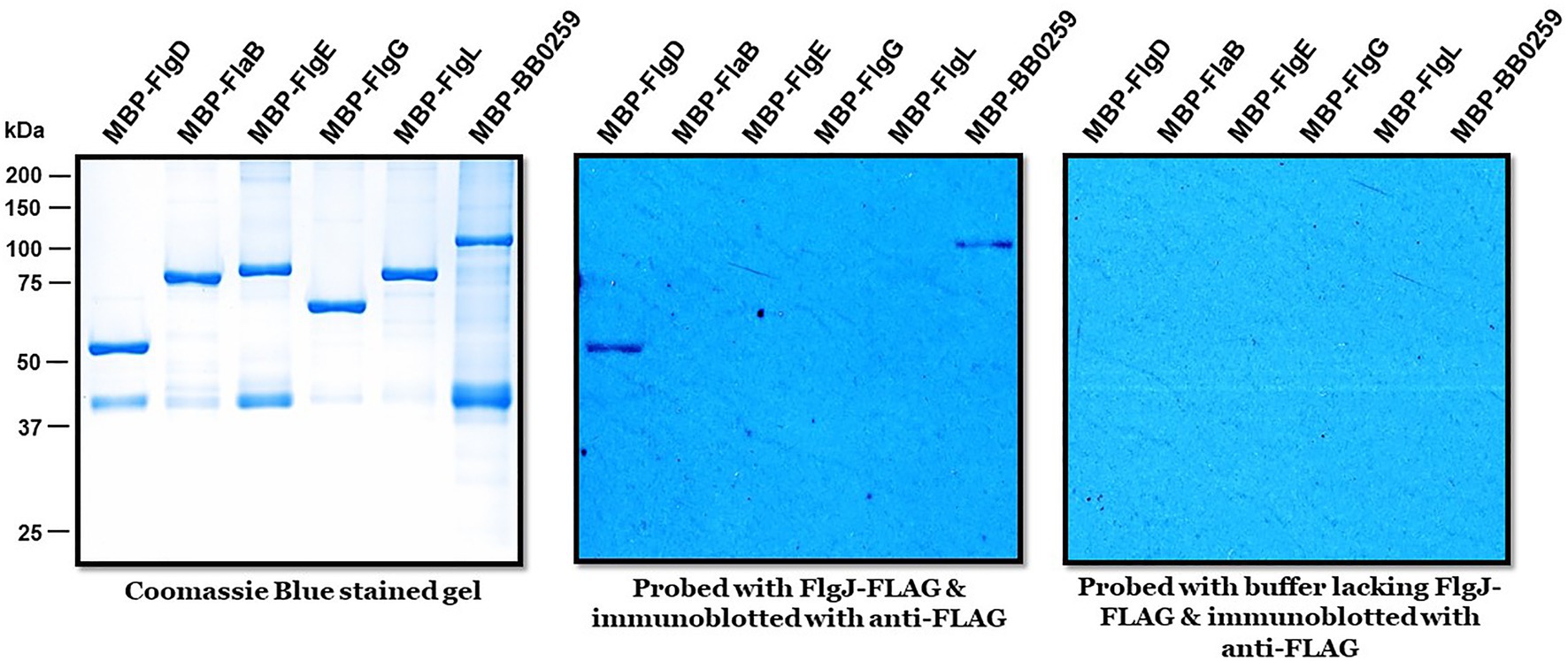
Figure 5. Biophysical interactions of FlgJ with BB0259/LTaseBb and FlgD. Approximately 1μg of maltose binding protein (MBP)-tagged proteins shown on top of each panel were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and stained with Coomassie blue (left) or transferred to a polyvinylidene difluoride membrane (middle & right panels). The membranes were incubated with or without FLAG-FlgJ and then immunoblotted with anti-FLAG monoclonal antibodies.
Soluble lytic transglycosylases cleave the glycosidic linkage between N-actetylmuramoyl and N-acetylglucosaminyl residues with the simultaneous production of a 1,6-anydromuramoyl product (Holtje et al., 1975; Herlihey et al., 2014). These enzymes, transglycosylases, are abundant in bacteria, found in different forms, and crucial for generating pores by hydrolyzing the PG sacculus for biosynthesis and recycling of PG, bacterial cell division, and the insertion of cell membrane-spanning structures such as flagella (Herlihey et al., 2014). B. burgdorferi LTaseBb appears similar to the S. enterica and E. coli SLT proteins as it possesses the conserved ES-GLMQ-AYNAG motif, with E580 being the catalytic residue (Figure 1). Furthermore, the C-terminal domain with the enzymatic motif encompassing the amino acid residues 559-717 is also similar in size to the PG-hydrolytic domains of other bacteria (de la Mora et al., 2012). While a confirmatory enzymatic assay is required to state whether BB0259 in fact possesses any enzymatic activity, our numerous attempts at such an assay were unsuccessful. It is noteworthy that measuring the peptidoglycan-degrading or lytic activity of FlgJ/SLT proteins poses significant technical drawbacks. The lack of a defined substrate, the heterogeneity of both the substrate and resulting hydrolyzing products in terms of peptide chain composition and extent of PG sacculus cross-linking, and any modification to the glycan chains contribute to the complexity of analysis of the peptidoglycan. However, genetic studies concomitant with direct visualization of the flagella in situ by cryo-ET enabled us to propose that BB0259 possesses peptidoglycan pore-forming activities and that E580 is the catalytic residue for the peptidoglycan lytic activities, as the bb0259-E580Q mutant’s hooks were unable to penetrate the peptidoglycan sacculus (Figure 3).
LTaseBb possesses an N-terminal peptide with unknown function (25-558 aa). As the C-terminal domain possesses the enzymatic motif, we speculate that the N-terminal domain is involved in binding to FlgJ, as LTaseBb secretes into the periplasm using its Sec-signaling sequence. Since LTaseBb appears to possess peptidoglycan lytic activities and FlgG and FlgJ are required for proper rod/hook/filament assembly (Zhang et al., 2012; Zhao et al., 2013), we propose that these proteins physically interact to facilitate the insertion of the flagella through the PG layer as the enzyme creates pores in the sacculus (Figure 6). Our direct visualization of the mutant’s flagella in situ by cryo-ET shows that the hooks are unable to pass through the PG sacculus, and therefore, these structures were detected just underneath the PG layer (Figures 3E–H, 4D). Consequently, the flagella did not assemble properly, leading to the non-motile and rod-shaped morphology (Figures 2A,B).
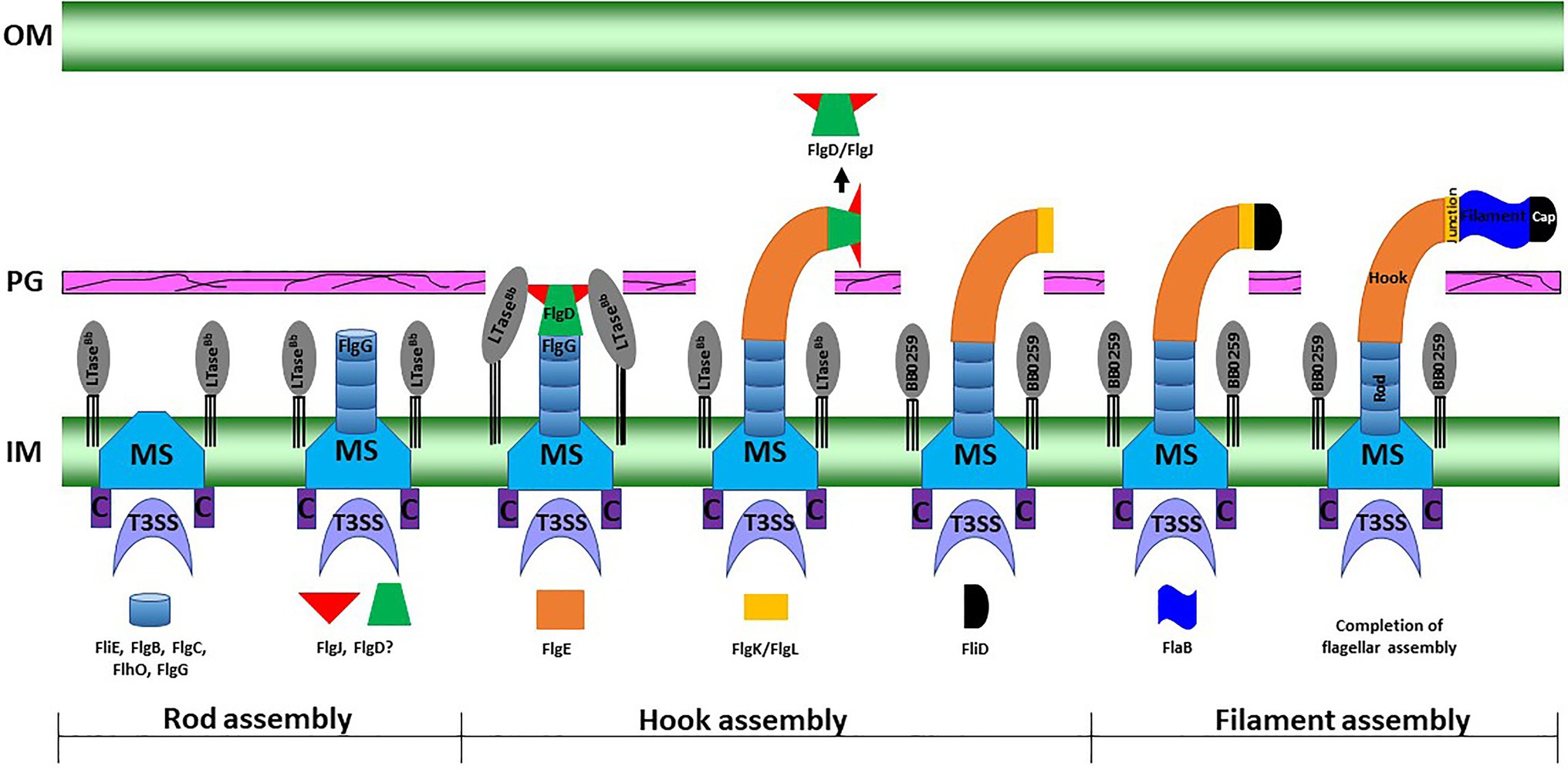
Figure 6. A working model of periplasmic flagellar hook–filament assembly in the spirochete B. burgdorferi. At the onset of rod assembly, many flagellar components assemble, such as the MS ring, C ring, export apparatus, stators, and collar. Subsequently, the rod components (FliE, FlgB, FlgC, FlhO/FlgF, and FlgG) are sequentially secreted into the periplasmic space through flagellar T3SS and assemble (Zhao et al., 2013). The hook cap protein FlgD and its chaperone or interacting protein FlgJ form at the distal end of the rod and initiate hook assembly. Binding of FlgJ with the pre-localized SLT enzyme BB0259 or LTaseBb in the periplasmic leaflet of the cell wall promotes hydrolysis of the sacculus as the hook completes assembly through the cell wall. Assembly of the filament (FlaA and FlaB) is promoted by the filament cap.
Borrelia burgdorferi FlgJ and the SLT domain-containing LTaseBb are distinct from their counterparts in S. enterica. For example, FlgJ from S. enterica is involved in flagellar rod formation or required for the transition from rod to hook formation (Cohen and Hughes, 2014). However, cryo-ET data from the B. burgdorferi ΔflgJ mutant show that the mutant completed assembly of the rod and P-ring but assembled a partial hook and filament structures (Zhang et al., 2012). It is noteworthy that, unlike the model organisms, the rod and P-ring in this spirochete are not able to contact the PG layer, as the rod is significantly smaller than in the model organisms (17nm vs. 30nm in E. coli; Zhao et al., 2013; Saijo-Hamano et al., 2019). It is the spirochete-specific collar that contacts the PG sacculus, while the hook penetrates the peptidoglycan layer to complete assembly and formation of the filament in B. burgdorferi (Zhao et al., 2013). FlgJ in the spirochete is therefore likely important for assembly of the hook/filament during the hook-to-filament transition (Zhang et al., 2012). The mutants of bb0259 (or LTaseBb) also assemble the rod, P-ring, and hook but are unable to synthesize/assemble the filament (Figures 3, 4, Supplementary Figure S3). The hooks are not able to penetrate the PG layer due to the lack of PG lytic activity in Δbb0259 or bb0259-E580Q mutants (ΔLTaseBb or LTaseBb-E580Q, respectively). This result also enforces the notion that FlgJ and LTaseBb are crucial for hook/ filament assembly and/or for hook-to-filament transition (Figure 6).
Δbb0259/ΔLTaseBb or bb0259-E580Q/LTaseBb-E580Q mutant phenotypes appear to be very similar to that of the ΔflaB mutant (Zhao et al., 2013). In both cases, the mutants complete assembly and formation of the motor, rod, P-ring, and hook but lack the filament (compare Figures 3F,G with Supplementary Figure S4H or Figure 5A from ref. Zhao et al., 2013). However, in the flaB mutant, the hook is able to penetrate the PG layer (Supplementary Figure S4H) as the PG enzyme activity of BB0259 is believed to be retained in ΔflaB cells, again reinforcing that BB0259 or LTaseBb is a PG hydrolyzing enzyme. Moreover, the hook length of the ΔLTaseBb, ΔflaB, and wild-type cells is the same (~51nm; n=20) and consistent with the previously reported hook length (Zhao et al., 2013), suggesting that LTaseBb is not involved in hook assembly. All these data suggest that FlgJ and LTaseBb are distinct from their counterparts in other flagellated bacteria reported to date. Importantly, in S. enterica, FlgJ interacts with the rod proteins including the distal rod FlgG as FlgJ is considered a rod-capping or scaffolding protein (Hirano et al., 2001; Cohen and Hughes, 2014). However, that is not the case in B. burgdorferi. FlgJ interaction with FlgG is undetectable (Figure 5), and the ΔflgJ mutant’s rod structures are intact in B. burgdorferi (Zhang et al., 2012).
Based on reports of FlgJ and this communication, we propose a model for how LTaseBb and FlgJ work synergistically with the hook cap protein FlgD to complete assembly of the hook–filament in B. burgdorferi (Figure 6). In this model, LTaseBb uses Sec-dependent pathway to be secreted into the periplasm. Using the flagellar type III secretion system, FlgJ and hook-capping protein FlgD compile onto the rod as the rod completes assembly onto the MS-ring. Interactions of FlgJ with BB0259 or LTaseBb and FlgD (Figure 5) enable the hook to initiate and complete its assembly through the PG holes created by LTaseBb wherein FlgJ chaperones or interacts with FlgD for the hook to complete its assembly in the periplasm (Figure 6). Without the FlgJ, the hook FlgE synthesis or assembly is therefore incomplete (Zhang et al., 2012) due to a lack of interaction between FlgD and FlgJ or lack of chaperone functions of FlgJ for FlgD. We speculate that the hook-to-filament assembly occurs as in other bacteria, as we have reported (Zhao et al., 2013). Additional in-depth analysis is warranted to better understand the spatiotemporal synthesis of the FlgJ, FlgD, and LTaseBb proteins and hook–filament assemblies in B. burgdorferi.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
HX, BH, and DAF conducted the experiments. JL and MM designed the experiments. All authors contributed to the article and approved the submitted version.
JL was supported by grants GM107629 and R01AI087946. MM was supported by R01AI132818 from National Institutes of Health (NIH). Part of cryo-ET data were collected at Yale CryoEM resource that is funded in part by the NIH grant 1S10OD023603-01A1.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Jennifer Aronson for critical reading of the manuscript. We thank David Burton for modeling and illustration and Michelle LaHair for technical helps.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.692707/full#supplementary-material
Almagro Armenteros, J. J., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Babu, M. M., Priya, M. L., Selvan, A. T., Madera, M., Gough, J., Aravind, L., et al. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188, 2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006
Bertani, G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300. doi: 10.1128/jb.62.3.293-300.1951
Bertani, G. (2004). Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186, 595–600. doi: 10.1128/JB.186.3.595-600.2004
Blackburn, N. T., and Clarke, A. J. (2001). Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 52, 78–84. doi: 10.1007/s002390010136
Bono, J. L., Elias, A. F., Kupko, J. J. 3rd, Stevenson, B., Tilly, K., and Rosa, P. (2000). Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182, 2445–2452. doi: 10.1128/JB.182.9.2445-2452.2000
Carroll, B. L., and Liu, J. (2020). Structural conservation and adaptation of the bacterial flagella motor. Biomol. Ther. 10:1492. doi: 10.3390/biom10111492
Charon, N. W., Cockburn, A., Li, C., Liu, J., Miller, K. A., Miller, M. R., et al. (2012). The unique paradigm of spirochete motility and chemotaxis. Annu. Rev. Microbiol. 66, 349–370. doi: 10.1146/annurev-micro-092611-150145
Charon, N. W., Goldstein, S. F., Marko, M., Hsieh, C., Gebhardt, L. L., Motaleb, M. A., et al. (2009). The flat ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J. Bacteriol. 191, 600–607. doi: 10.1128/JB.01288-08
Chen, S., Beeby, M., Murphy, G. E., Leadbetter, J. R., Hendrixson, D. R., Briegel, A., et al. (2011). Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981. doi: 10.1038/emboj.2011.186
Chen, M., Dai, W., Sun, S. Y., Jonasch, D., He, C. Y., Schmid, M. F., et al. (2017). Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14, 983–985. doi: 10.1038/nmeth.4405
Cohen, E. J., and Hughes, K. T. (2014). Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J. Bacteriol. 196, 2387–2395. doi: 10.1128/JB.01580-14
de la Mora, J., Osorio-Valeriano, M., Gonzalez-Pedrajo, B., Ballado, T., Camarena, L., and Dreyfus, G. (2012). The C terminus of the flagellar muramidase SltF modulates the interaction with FlgJ in Rhodobacter sphaeroides. J. Bacteriol. 194, 4513–4520. doi: 10.1128/JB.00460-12
Demchick, P., and Koch, A. L. (1996). The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768–773. doi: 10.1128/jb.178.3.768-773.1996
Elias, A. F., Bono, J. L., Kupko, J. J. 3rd, Stewart, P. E., Krum, J. G., and Rosa, P. A. (2003). New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6, 29–40. doi: 10.1159/000073406
Garcia-Ramos, M., de la Mora, J., Ballado, T., Camarena, L., and Dreyfus, G. (2018). Biochemical and phylogenetic study of SltF, a flagellar lytic transglycosylase from Rhodobacter sphaeroides. J. Bacteriol. 200:e00397-18. doi: 10.1128/JB.00397-18
Ge, Y., Old, I. G., Saint Girons, I., and Charon, N. W. (1997). Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J. Bacteriol. 179, 2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997
Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H., et al. (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. doi: 10.1002/pro.3235
Gonzalez-Pedrajo, B., de la Mora, J., Ballado, T., Camarena, L., and Dreyfus, G. (2002). Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta 1579, 55–63. doi: 10.1016/s0167-4781(02)00504-3
Herlihey, F. A., Moynihan, P. J., and Clarke, A. J. (2014). The essential protein for bacterial flagella formation FlgJ functions as a beta-N-acetylglucosaminidase. J. Biol. Chem. 289, 31029–31042. doi: 10.1074/jbc.M114.603944
Hirano, T., Minamino, T., and Macnab, R. M. (2001). The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312, 359–369. doi: 10.1006/jmbi.2001.4963
Holtje, J. V., Mirelman, D., Sharon, N., and Schwarz, U. (1975). Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 124, 1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975
Homma, M., and Iino, T. (1985). Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J. Bacteriol. 162, 183–189. doi: 10.1128/jb.162.1.183-189.1985
Jaroszewski, L., Li, Z., Cai, X. H., Weber, C., and Godzik, A. (2011). FFAS server: novel features and applications. Nucleic Acids Res. 39, W38–W44. doi: 10.1093/nar/gkr441
Jaroszewski, L., Rychlewski, L., Li, Z., Li, W., and Godzik, A. (2005). FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 33, W284–W288. doi: 10.1093/nar/gki418
Jones, C. J., and Macnab, R. M. (1990). Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J. Bacteriol. 172, 1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990
Jutras, B. L., Scott, M., Parry, B., Biboy, J., Gray, J., Vollmer, W., et al. (2016). Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc. Natl. Acad. Sci. U. S. A. 113, 9162–9170. doi: 10.1073/pnas.1610805113
Kall, L., Krogh, A., and Sonnhammer, E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. doi: 10.1016/j.jmb.2004.03.016
Kall, L., Krogh, A., and Sonnhammer, E. L. (2005). An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl. 1), i251–i257. doi: 10.1093/bioinformatics/bti1014
Kall, L., Krogh, A., and Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–W432. doi: 10.1093/nar/gkm256
Kariu, T., Sharma, K., Singh, P., Smith, A. A., Backstedt, B., Buyuktanir, O., et al. (2015). BB0323 and novel virulence determinant BB0238: Borrelia burgdorferi proteins that interact with and stabilize each other and are critical for infectivity. J. Infect. Dis. 211, 462–471. doi: 10.1093/infdis/jiu460
Koraimann, G. (2003). Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60, 2371–2388. doi: 10.1007/s00018-003-3056-1
Kremer, J. R., Mastronarde, D. N., and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. doi: 10.1006/jsbi.1996.0013
Li, C., Xu, H., Zhang, K., and Liang, F. T. (2010). Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75, 1563–1576. doi: 10.1111/j.1365-2958.2010.07078.x
Liu, J., Lin, T., Botkin, D. J., McCrum, E., Winkler, H., and Norris, S. J. (2009). Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 191, 5026–5036. doi: 10.1128/JB.00340-09
Madan Babu, M., and Sankaran, K. (2002). DOLOP–database of bacterial lipoproteins. Bioinformatics 18, 641–643. doi: 10.1093/bioinformatics/18.4.641
Moon, K. H., Hobbs, G., and Motaleb, M. A. (2016). Borrelia burgdorferi CheD promotes various functions in chemotaxis and the pathogenic life cycle of the spirochete. Infect. Immun. 84, 1743–1752. doi: 10.1128/IAI.01347-15
Moon, K. H., Zhao, X., Xu, H., Liu, J., and Motaleb, M. A. (2018). A tetratricopeptide repeat domain protein has profound effects on assembly of periplasmic flagella, morphology and motility of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 110, 634–647. doi: 10.1111/mmi.14121
Motaleb, M. A., Corum, L., Bono, J. L., Elias, A. F., Rosa, P., Samuels, D. S., et al. (2000). Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. U. S. A. 97, 10899–10904. doi: 10.1073/pnas.200221797
Motaleb, M. A., Liu, J., and Wooten, R. M. (2015). Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Curr. Opin. Microbiol. 28, 106–113. doi: 10.1016/j.mib.2015.09.006
Motaleb, M. A., Miller, M. R., Bakker, R. G., Li, C., and Charon, N. W. (2007). Isolation and characterization of chemotaxis mutants of the Lyme disease Spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol. 422, 421–437. doi: 10.1016/S0076-6879(06)22021-4
Motaleb, M. A., Pitzer, J. E., Sultan, S. Z., and Liu, J. (2011). A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J. Bacteriol. 193, 3324–3331. doi: 10.1128/JB.00202-11
Nakamura, S., and Minamino, T. (2019). Flagella-driven motility of bacteria. Biomol. Ther. 9:279. doi: 10.3390/biom9070279
Nambu, T., Inagaki, Y., and Kutsukake, K. (2006). Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet. Syst. 81, 381–389. doi: 10.1266/ggs.81.381
Nambu, T., Minamino, T., Macnab, R. M., and Kutsukake, K. (1999). Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181, 1555–1561. doi: 10.1128/JB.181.5.1555-1561.1999
Ohnishi, K., Ohto, Y., Aizawa, S. I., Macnab, R. M., and Iino, T. (1994). FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176, 2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994
Rychlewski, L., Jaroszewski, L., Li, W., and Godzik, A. (2000). Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 9, 232–241. doi: 10.1110/ps.9.2.232
Saijo-Hamano, Y., Matsunami, H., Namba, K., and Imada, K. (2019). Architecture of the bacterial flagellar distal rod and hook of Salmonella. Biomol. Ther. 9:260. doi: 10.3390/biom9070260
Sal, M. S., Li, C., Motalab, M. A., Shibata, S., Aizawa, S., and Charon, N. W. (2008). Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190, 1912–1921. doi: 10.1128/JB.01421-07
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sultan, S. Z., Manne, A., Stewart, P. E., Bestor, A., Rosa, P. A., Charon, N. W., et al. (2013). Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect. Immun. 81, 2012–2021. doi: 10.1128/IAI.01228-12
Sultan, S. Z., Pitzer, J. E., Miller, M. R., and Motaleb, M. A. (2010). Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol. Microbiol. 77, 128–142. doi: 10.1111/j.1365-2958.2010.07191.x
Sultan, S. Z., Sekar, P., Zhao, X., Manne, A., Liu, J., Wooten, R. M., et al. (2015). Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect. Immun. 83, 1765–1777. doi: 10.1128/IAI.03097-14
Terashima, H., Kawamoto, A., Morimoto, Y. V., Imada, K., and Minamino, T. (2017). Structural differences in the bacterial flagellar motor among bacterial species. Biophys. Physicobiol. 14, 191–198. doi: 10.2142/biophysico.14.0_191
Thunnissen, A. M., Dijkstra, A. J., Kalk, K. H., Rozeboom, H. J., Engel, H., Keck, W., et al. (1994). Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature 367, 750–753. doi: 10.1038/367750a0
Tilly, K., Elias, A. F., Errett, J., Fischer, E., Iyer, R., Schwartz, I., et al. (2001). Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183, 5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001
Toker, A. S., and Macnab, R. M. (1997). Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273, 623–634. doi: 10.1006/jmbi.1997.1335
Toledo, A., Huang, Z., Coleman, J. L., London, E., and Benach, J. L. (2018). Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins. Mol. Microbiol. 108, 63–76. doi: 10.1111/mmi.13914
van Asselt, E. J., Thunnissen, A. M., and Dijkstra, B. W. (1999). High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J. Mol. Biol. 291, 877–898. doi: 10.1006/jmbi.1999.3013
Wolgemuth, C. W., Charon, N. W., Goldstein, S. F., and Goldstein, R. E. (2006). The flagellar cytoskeleton of the spirochetes. J. Mol. Microbiol. Biotechnol. 11, 221–227. doi: 10.1159/000094056
Xu, H., He, J., Liu, J., and Motaleb, M. A. (2019). BB0326 is responsible for the formation of periplasmic flagellar collar and assembly of the stator complex in Borrelia burgdorferi. Mol. Microbiol. 113, 418–429. doi: 10.1111/mmi.14428
Xu, H., Sultan, S., Yerke, A., Moon, K. H., Wooten, R. M., and Motaleb, M. A. (2017). Borrelia burgdorferi CheY2 is dispensable for chemotaxis or motility but crucial for the infectious life cycle of the spirochete. Infect. Immun. 85:e00264-16. doi: 10.1128/IAI.00264-16
Yang, X. F., Alani, S. M., and Norgard, M. V. (2003). The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 100, 11001–11006. doi: 10.1073/pnas.1834315100
Yonekura, K., Maki, S., Morgan, D. G., DeRosier, D. J., Vonderviszt, F., Imada, K., et al. (2000). The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290, 2148–2152. doi: 10.1126/science.290.5499.2148
Zaloba, P., Bailey-Elkin, B. A., Derksen, M., and Mark, B. L. (2016). Structural and biochemical insights into the peptidoglycan hydrolase domain of FlgJ from Salmonella typhimurium. PLoS One 11:e0149204. doi: 10.1371/journal.pone.0149204
Zhang, K., Tong, B. A., Liu, J., and Li, C. (2012). A single-domain FlgJ contributes to flagellar hook and filament formation in the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 194, 866–874. doi: 10.1128/JB.06341-11
Zhao, X., Zhang, K., Boquoi, T., Hu, B., Motaleb, M. A., Miller, K. A., et al. (2013). Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 110, 14390–14395. doi: 10.1073/pnas.1308306110
Keywords: Lyme disease, flagella, peptidoglycan hydrolases, cryo-elecron tomography, spirochete, Borrelia
Citation: Xu H, Hu B, Flesher DA, Liu J and Motaleb MA (2021) BB0259 Encompasses a Peptidoglycan Lytic Enzyme Function for Proper Assembly of Periplasmic Flagella in Borrelia burgdorferi. Front. Microbiol. 12:692707. doi: 10.3389/fmicb.2021.692707
Received: 09 April 2021; Accepted: 19 August 2021;
Published: 01 October 2021.
Edited by:
Matthew Arthur Baker, University of New South Wales, AustraliaReviewed by:
Eli Cohen, Imperial College London, United KingdomCopyright © 2021 Xu, Hu, Flesher, Liu and Motaleb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Md A. Motaleb, bW90YWxlYm1AZWN1LmVkdQ==; Jun Liu, amxpdUB5YWxlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.