
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 June 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.687625
Biofilms are a frequent cause of food contamination of potentially pathogenic bacteria, such as Staphylococcus aureus. Given its vast role in human disease, the possible impact of biofilm-producing S. aureus isolates in a food processing environment is evident. Sixty-nine S. aureus isolates collected from one firm following multiple staphylococcal food poisoning outbreak investigations were utilized for this analysis. Strain evaluations were performed to establish virulence determinants and the evolutionary relationships using data generated by shotgun whole-genome sequencing (WGS), along with end point polymerase chain reaction (PCR) and in vitro phenotypic assessments. S. aureus isolates were grouped into six well-supported clades in the phylogenetic tree, with the relationships within the clades indicating a strong degree of clonal structure. Our analysis identified four major sequence types 47.8% ST1, 31.9% ST45, 7.2% ST5, and 7.2% ST30 and two major spa types 47.8% t127 and 29.0% t3783. Extrapolated staphylococcal enterotoxin (SE) analysis found that all isolates were positive for at least 1 of the 23 SEs and/or SE-like toxin genes. Enterotoxigenic assessments found that 93% of the isolates expressed a classical SE(A–E). SE gene concurrence was observed at 96.2%, based on PCR and WGS results. In total, 46 gene targets were distinguished. This included genes that encode for adhesion and biofilm synthesis such as clfA, clfB, bbp, ebpS, ica, bap and agr. Our evaluation found agr group III to be the most prevalent at 55%, followed by 35% for agr group I. All isolates harbored the complete intercellular adhesion operon that is recognized to contain genes responsible for the adhesion step of biofilm formation by encoding proteins involved in the syntheses of the biofilm matrix. Phenotypic characterization of biofilm formation was evaluated three times, with each test completed in triplicate and accomplished utilizing the microtiter plate method and Congo red agar (CRA). The microtiter plate results indicated moderate to high biofilm formation for 96% of the isolates, with 4% exhibiting weak to no biofilm development. CRA results yielded all positive to intermediate results. The potential to inadvertently transfer pathogenic bacteria from the environment into food products creates challenges to any firm and may result in adulterated food.
Staphylococcus aureus is the known cause of many diseases and is considered one of the most significant pathogens for humans. It is a frequent source of infections, such as pneumonia, postoperative wound infections, endocarditis, and bacteremia. The secreted toxins and cell wall-associated virulence factors contribute to the bacterium’s ability to adhere to, proliferate in, and potentially destroy host cells and tissue matrix (Seo and Bohach, 2007). Enterotoxigenic Staphylococcus strains are also a common cause of staphylococcal food poisoning (SFP) that affects an estimated 241,148 individuals the United States annually (Scallan et al., 2011).
Staphylococcus species are ubiquitous in the environment and is often found in the air, dust, water, and food, and exist on humans and animals (Hennekinne et al., 2010). S. aureus and S. epidermidis are commensal inhabitants of the human skin and consequently have increased probability to be present in foods handled directly by humans and on food contact surfaces. Inadequate hygiene practices in food processing plants may result in contamination of food products with an enterotoxin-producing strain of S. aureus (Hait et al., 2014). Biofilms are a frequent source of food contamination of potentially pathogenic bacteria, such as S. aureus. Biofilms are the most common bacterial lifestyle in nature, and as the cells multiply, they secrete a matrix of extracellular polymeric substances (EPS) that wraps the cells (Gutiérrez et al., 2012). The most commonly found bacteria on food contact surfaces in dairy environments are Enterobacter, Lactobacillus, Listeria, Micrococcus, Streptococcus, Staphylococcus, Bacillus, and Pseudomonas (Gutiérrez et al., 2012). The presence of biofilm-forming strains of S. aureus in food processing environments and in food are equally important as biofilms are involved in cross-contamination events.
The two-step process in biofilm development first involves the bacteria adherence to a surface followed by the bacteria multiplying to form a multilayered biofilm associated with the production of polysaccharide intercellular adhesion (PIA) (Nasr et al., 2012). PIA is produced and secreted by the proteins encoded in the intercellular adhesion (icaABCD) operon (Miao et al., 2019). S. aureus adherence is facilitated by protein adhesions of the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which is most frequently anchored to the cell wall peptidoglycan (Foster and Höök, 1998). S. aureus genes that encode MSCRAMMs include bone sialoprotein binding protein (bbp), collagen binding protein (cna), elastin binding protein (ebpS), encoding laminin binding protein (eno), encoding fibrinogen binding protein (fib), fibronectin binding proteins A (fnbA), fibronectin binding proteins A (fnbB), clumping factors A (clfA), and clumping factors B (clfB) genes (Tristan et al., 2003). The icaABCD operon is responsible for the slime production in S. aureus and S. epidermidis (Cucarella et al., 2001). This operon contains ica genes, responsible in the icaABCD step of biofilm formation by encoding proteins involved in the syntheses of the biofilm matrix polysaccharide–poly-N-succinyl β-1-6 glucosamine (PIA–PNSG) (Cucarella et al., 2001). The icaA and icaD genes are known to play a significant role in biofilm formation. The icaA gene encodes N-acetylglucosaminyltransferase, an enzyme involved in PIA synthesis, and the icaD gene has been described as critical in the maximal expression of N-acetylglucosaminyltransferase resulting in complete phenotypic expression of the capsular polysaccharide (Nasr et al., 2012). The DtlA and Bap proteins have been recognized to influence biofilm formation for S. aureus. The staphylococcal accessory regulator (SarA) is an essential element that controls production of S. aureus virulence factors and was found to be vital in a study for biofilm development (Valle et al., 2003). The accessory gene regulator (agr) quorum-sensing system has been described as central to the role in S. aureus pathogenesis (Yarwood and Schlievert, 2003). Biofilm synthesis is mainly encoded by ica, bap, and agr genes, with clfA, clfB, bbp, and ebpS genes also involved (Notcovich et al., 2018).
Staphylococcus aureus are capable of harboring a plethora of virulence factors. This includes the staphylococcal enterotoxin (SE) superfamily of secreted virulence factors that share both functional and structural similarities (Fisher et al., 2018). There are SEs and SE-like toxins (SEI); SEA, SEB, SEC1,2,3, SED, SEE, SEG, SEH, SEI, SElJ, SEK, SEL, SEM, SEN, SEO, SEP, SEQ, SER, SES, SET, SElU, SElW SElV, SElX, and SElY (Fisher et al., 2018). The SE group is recognized to exhibit emetic activity, while the SEl toxins have not been established to cause emesis or gastroenteritis in primates (Fisher et al., 2018). For example, previous analysis determined SElJ to be non-emetic, while SElU, SElV, SElW, SElX, and SElY have not been tested in non-human primates (Fisher et al., 2018). Using a comparison of the nucleotide and amino acid sequences, SEs and SEls are grouped into four evolutionary groups: SEA group 1 (SEA, SED, SEE, SElJ, SEH, SEN, SEO, SEP, and SES), SEB group 2 (SEB, SECs, SEG, SER, SElU, and SElW), SEI group 3 (SEI, SEK, SEL, SEQ, SEM, and SElV), and SElX group 4 (TSST-1, SET, SElX, and SElY) (Fisher et al., 2018). Overall, these enterotoxin and enterotoxin-like proteins are described as single-chain proteins with molecular weights ranging from 22 to 29 kDa and are known to exhibit superantigenic activity resulting in non-specific T-cell proliferation (Hennekinne et al., 2010). These toxins can be encoded in plasmids, prophages, and on chromosomal pathogenicity islands (Rajkovic et al., 2019). SE genes on mobile genetic elements create an additional risk factor for transmission due to the potential of horizonal gene transfer (Rajkovic et al., 2019).
Ingestion of SEs produced by some S. aureus strains are the etiological agent of common illness called SFP. The extracellular SEs are resistant to various environmental conditions and are resistant to proteolytic enzymes such as trypsin and pepsin meaning the SEs persist in the digestive tract after ingestion (Hait et al., 2014). SFP has a rapid onset of 1–7 h following ingestion. The SE intoxication results in nausea, emesis, abdominal cramping, and diarrhea that typically last less than 24–48 h (Hennekinne et al., 2010).
In this study, an evaluation between two tests for phenotypic biofilm formation and the correlation of the prevalence of genes involved in biofilm production/formation were investigated for 69 S. aureus isolates from outbreak investigations. The genomic sequencing tools were used to identify virulence gene targets in these outbreak-linked S. aureus isolates. A total of 601 environmental swabs and ingredient samples were collected and analyzed, in which 14% were confirmed as S. aureus. Whole-genome sequencing (WGS) and biofilm analysis were conducted on 69 of these S. aureus isolates. The isolates were collected on three dates, the initial investigation date December 27, 2010, the post-clean up investigation date January 10, 2011, and the final investigation date November 18, 2011. The collection locations or sources in the bakery include assembly production room, baking production room, final product storage room, ingredients storage room, and products.
This study aimed to investigate the phylogenetic relationships and virulence determinants of 69 outbreak-linked environmental and food acquired S. aureus isolates. S. aureus phenotypes such as adhesion and biofilm formation are important since they promote the colonization of food environments (Gutiérrez et al., 2012). All 69 isolates were screened for the genes associated with biofilm formation bap, icaA, and icaD, and for genes that encode MSCRAMMs bbp, cna, ebpS, eno, fib, fnbA, fnbB, clfA, and clfB genes. Sequence-based analysis was used to identify 23 SE genes as well as additional virulence factors such as pvl (Panton–Valentine leukocidin). PVL is a cytotoxin composed of two components, LukS-PV and LukF-PV. These secreted components cause neutrophil lysis after assembling into a pore-forming heptamer on neutrophil membranes (Shallcross et al., 2013). In total, 46 genes were investigated. Phenotype testing was accomplished to demonstrate biofilm formation capabilities and the expression of SEs SEA–SEE.
The outbreak isolates of S. aureus (n = 69) used in this study are maintained in the strain collection of FDA’s Center for Food Safety and Applied Nutrition. These isolates were confirmed phenotypically using Baird Parker + Rabbit Plasma Fibrinogen (RPF) agar as the selective plating (Item number: 43531, bioMérieux, Marcy-l’Etoile, France) and biochemical testing using the VITEK® 2 Identification of Gram-positive card (bioMérieux, Marcy-l’Etoile,France). Sixty-nine of the S. aureus isolates collected were assessed for genotype and phenotype virulence factors. These evaluations used a combination of end point polymerase chain reaction (PCR), WGS analysis, biofilm assays, and enterotoxin testing. The outbreak summaries previously published describe the PCR primers and method utilized for SE and SE-like gene identification and enterotoxigenic potential outcomes of these isolates (Hait et al., 2014). In this earlier research, each isolate was tested using the VIDAS Staph Enterotoxin II (bioMérieux, Marcy-l’Etoile, France) to determine the expression of SEA; SEB; SEC 1, 2, 3; SED; and SEE. The enterotoxigenic assessments established that 93% (64/69) of the isolates expressed a classical SE(A–E). The PCR results previously performed will be used to evaluate congruency with WGS results.
The genomic DNA was extracted using the DNAeasy Blood and Tissue Kit (Qiagen, Inc., Valencia, CA, United States) following an overnight grown culture incubated at 37°C in Trypticase Soy Broth (TSB) (Becton, Dickinson, Franklin Lakes, NJ, United States). The DNA concentration was measured using a Qubit 3.0 fluorometer (Life Technologies, Frederick, MD, United States). The libraries were prepared following the Illumina Nextera XT or Nextera Flex Library Prep protocols (Illumina, San Diego, CA, United States). The prepared libraries were sequenced on the Illumina MiSeq (Illumina, San Diego, CA, United States) using MiSeq Reagents Kits v2 (500 cycles) or MiSeq Reagents Kits v3 (600 cycles). Raw reads quality was assessed by the following parameters: cluster density (1200–1400 k/mm2) and percentage of clusters passing filters (>80%). Raw reads were trimmed using Trimmomatic (Bolger et al., 2014) and assembled de novo using SKESA v2.2 with minimal contig length as 500 bp (Souvorov et al., 2018). Annotations of assemblies were processed using the Prokka v1.14.5 (Seemann, 2014). Virulence genes were identified here using WGS data of these isolates (Hait et al., 2021). BLAST (Altschul et al., 1990)-based approach was used to identify presence–absence of enterotoxin genes SEA–SElX, virulence factors, and biofilm-associated genes. kSNP 3.0 (Gardner et al., 2015) was used to generate the single-nucleotide polymorphism (SNP) matrix with the optimum k-mer size of 19. RAxML v8.2.9 (Stamatakis, 2014) was used to construct the maximum-likelihood phylogenetic tree (GTRGAMMA with 1000 rapid bootstrapping searches), and ggtree was used to visualize the phylogenetic tree (de Vienne et al., 2012; Wood and Salzberg, 2014; Yu et al., 2017). MLST 2.0 was used to identify the sequence types (Larsen et al., 2012) and spaTyper 1.0 was used to identify spa types (Bartels et al., 2014). GenBank accession numbers for the 69 Staphylococcus aureus isolates is provided in the Supplementary Table 1. In addition, we selected six S. aureus isolates from NCBI’s Pathogen Detection,1 which were isolated from the United States in recent years (SA1428_cattle_IA_2017, 1522_gallus_AR_2019, LS1_environment_WI_2019, CFSAN092696_rat feces_IL_2018, WAPHLSTAP00047_water_HI_2020, and WAPHLSTAP00089_ water_HI_2020, Figure 2) as reference in the phylogenetic analysis.
Biofilm analysis was conducted to determine the ability of each isolate to form a biofilm. This was accomplished using two previously described methods, the microtiter plate method and observations on Congo red agar (CRA) (Mathur et al., 2006; de Castro Melo et al., 2013). CRA was composed of 37 g/L of brain–heart infusion broth (Becton Dickinson, Franklin Lakes, NJ, United States), 50 g/L of sucrose (Sigma, St. Louis, MO, United States), 10 g/L of agar (Becton Dickinson, Franklin Lakes, NJ, United States), and 0.8 g/L of Congo red (Fisher Scientific, Waltham, MA, United States). Phenotypic characterization of biofilm formation of each isolate along with controls was evaluated three times, with each done in triplicate. American Type Culture Collection (ATCC, Manassas, VA, United States)-acquired control strains were utilized for biofilm determinations in the microtiter plate assay and to distinguish colonies on CRA. S. epidermidis ATCC 35984 is characterized as a high-level biofilm formation strain with black crystalline colonies on CRA, and S. epidermidis ATCC 12228 is a non-biofilm producer with pink colonies on CRA. Phenotypic biofilm examination using CRA identified biofilm producers as black colonies with dry crystalline consistency and non-biofilm-producing colonies developed smooth colonies that ranged from pink to red in color. Weak and moderate biofilm formers produced darkened brownish or black smooth colonies.
A microtiter plate assay was performed to test biofilm formation in vitro following a described method (de Castro Melo et al., 2013). Biofilm detection using the microtiter plate method used a sterile flat-bottom 96-well plate. These plates were inoculated with TSB (Becton Dickinson, Franklin Lakes, NJ, United States) containing 25% glucose and overnight cultures diluted at 1:100. These plates were incubated aerobically for 18–24 h at 37°C and then the wells were emptied followed by three consecutive washes using phosphate buffered saline solution, pH 7.2. The washed microtiter plates were dried for 1 h at 60°C and then stained with 1% crystal violet (Fisher Scientific, Waltham, MA, United States) for 1 min. The stained wells were washed three times with distilled water and allowed to dry at room temperature. Using a microplate reader, the optical density (OD) values measured at 490 nm were utilized as an index of surface bacteria adherence and biofilm formation. The uninoculated wells containing TSB with 25% glucose were the blanks used to correct the mean OD absorbance values for each S. aureus isolate. Based on the resulting absorbance values, the isolates were classified into three categories: positive result (high-level biofilm former) OD > 0.240, moderate result (intermediate-level biofilm former) OD = 0.240–0.120, and negative result (non-biofilm former) OD < 0.120.
The presence/absence of virulence factor encoding genes was determined for the outbreak-linked isolates using WGS analysis to identify 23 SE genes as well as additional virulence genes such as pvl, tsst, and biofilm-associated genes. These individual WGS results are illustrated in the heatmap exhibited in Figure 1. The overall prevalence of these virulence factor- and biofilm-associated genes is summarized in Table 1. The assessment of the agr group established that the agr group III was the most prevalent at 55%, followed by 35% for agr group I, and 10% for agr group II, while agr group IV was not present in the isolates tested. All of the isolates harbored the icaABCD genes icaA, icaB, icaC, and icaD. The clumping factor B gene, clfB, was detected in all of the isolates, while clfA was found in 99% of the outbreak isolates tested. None of the isolates harbored the biofilm-associated gene bap, while the genes encoding the ebpS and the bbp were present in all but one isolate, resulting in 99% prevalence. The incidence of additional genes that encode MSCRAMM was determined. The genes encoding fibronectin binding proteins A and B (fnbA and fnbB) were detected in all of the isolates, fib encoding fibrinogen binding protein were found in 96%, the eno gene was present in all but one isolate and cna encoding the collagen binding protein was found in 88% of the isolates.
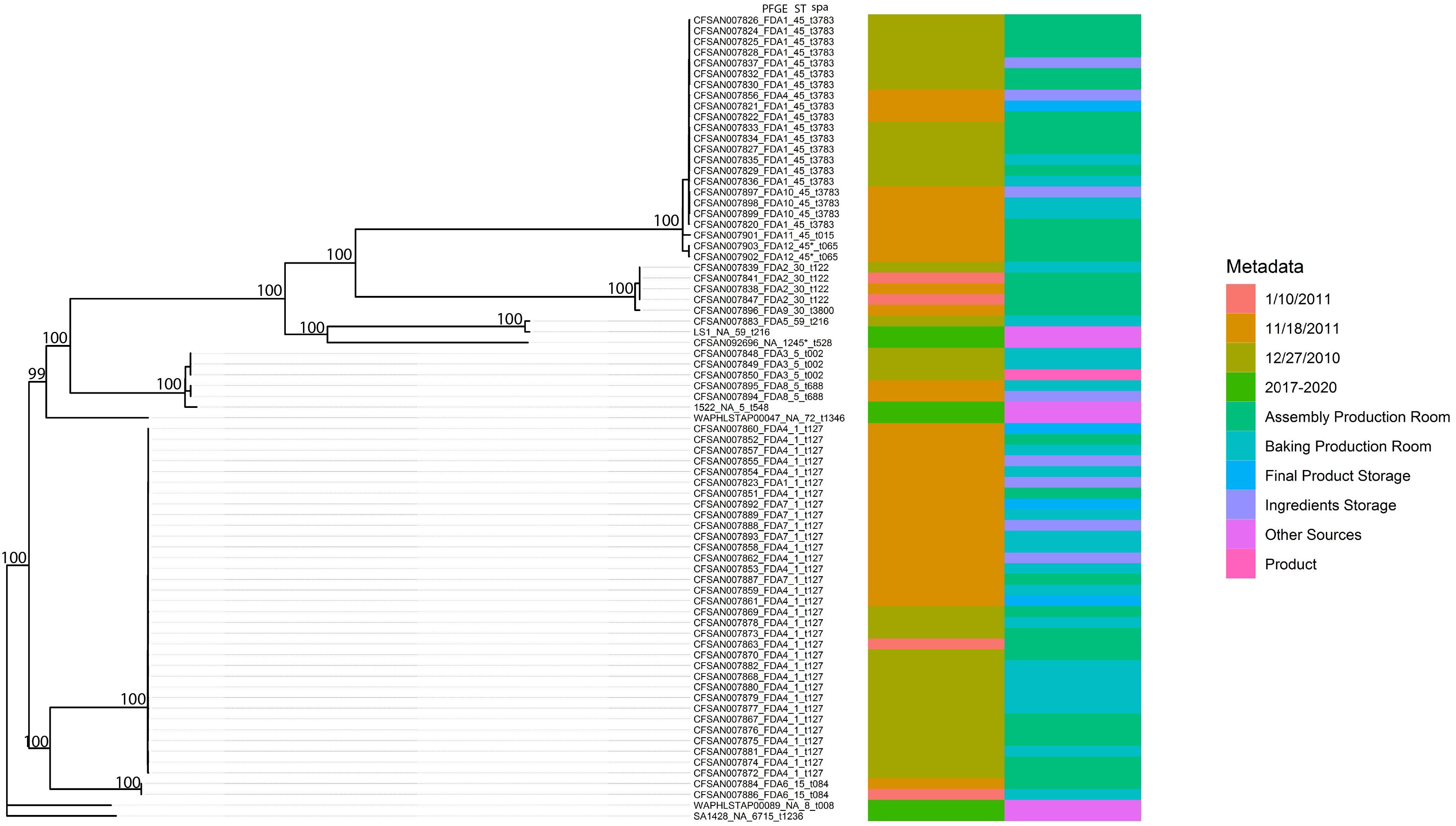
Figure 2. Phylogenetic tree inferred from a SNP matrix generated with kSNP3 and RAxML. Branch lengths are inferred number of substitutions per site and numbers at nodes represent percent bootstrap support.
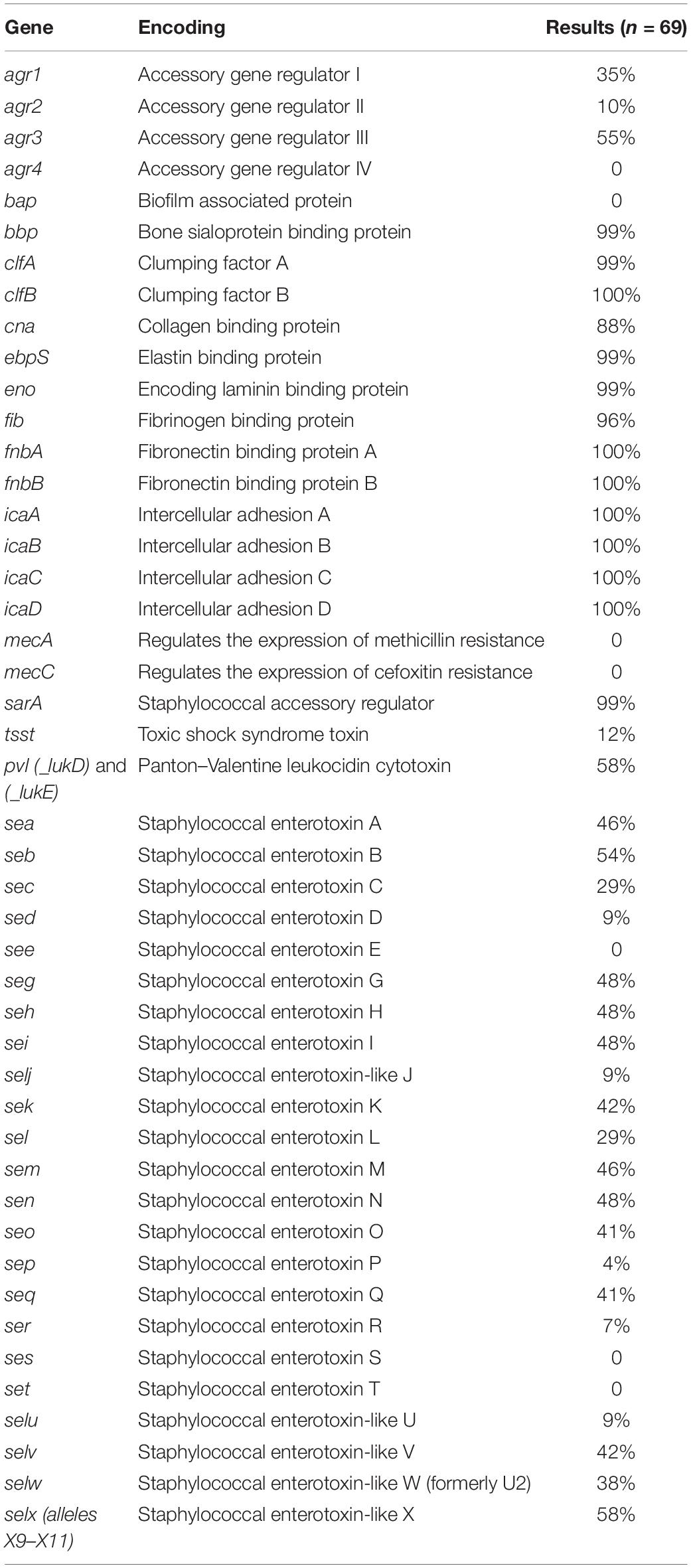
Table 1. Incidence of virulence factors, including enterotoxin genes, for 69 outbreak-linked isolates using WGS analysis.
Examination for virulence determinant genes, such as sarA, tsst, pvl, mecA, and mecC, and SEs were also investigated using WGS during this study (Table 1). This investigation revealed that all except one of the isolates carried sarA. The gene encoding toxic shock syndrome toxin was present in 12% of the outbreak-associated isolates; 58% of the isolates carried the pvl gene, and mecA and mecC were not found in the 69 isolates. Genomic SE screening established that all isolates were positive for at least 1 of the 23 SEs and/or SEI genes. As illustrated in Table 1, the most abundant genes were seb and selx, found in greater than half of the isolates. The incidence of SE genes, seg, seh, sei, and sen, was observed at 48% and that of sea and sem was at 46%. The remaining SE genes were distinguished at a lesser frequency, with see, ses, and set genes absent in the isolates tested here. In Table 2, the congruency of each enterotoxin gene detected by both the PCR and WGS analysis was determined. Here, a higher number suggests a greater level of disagreement between the two assays.
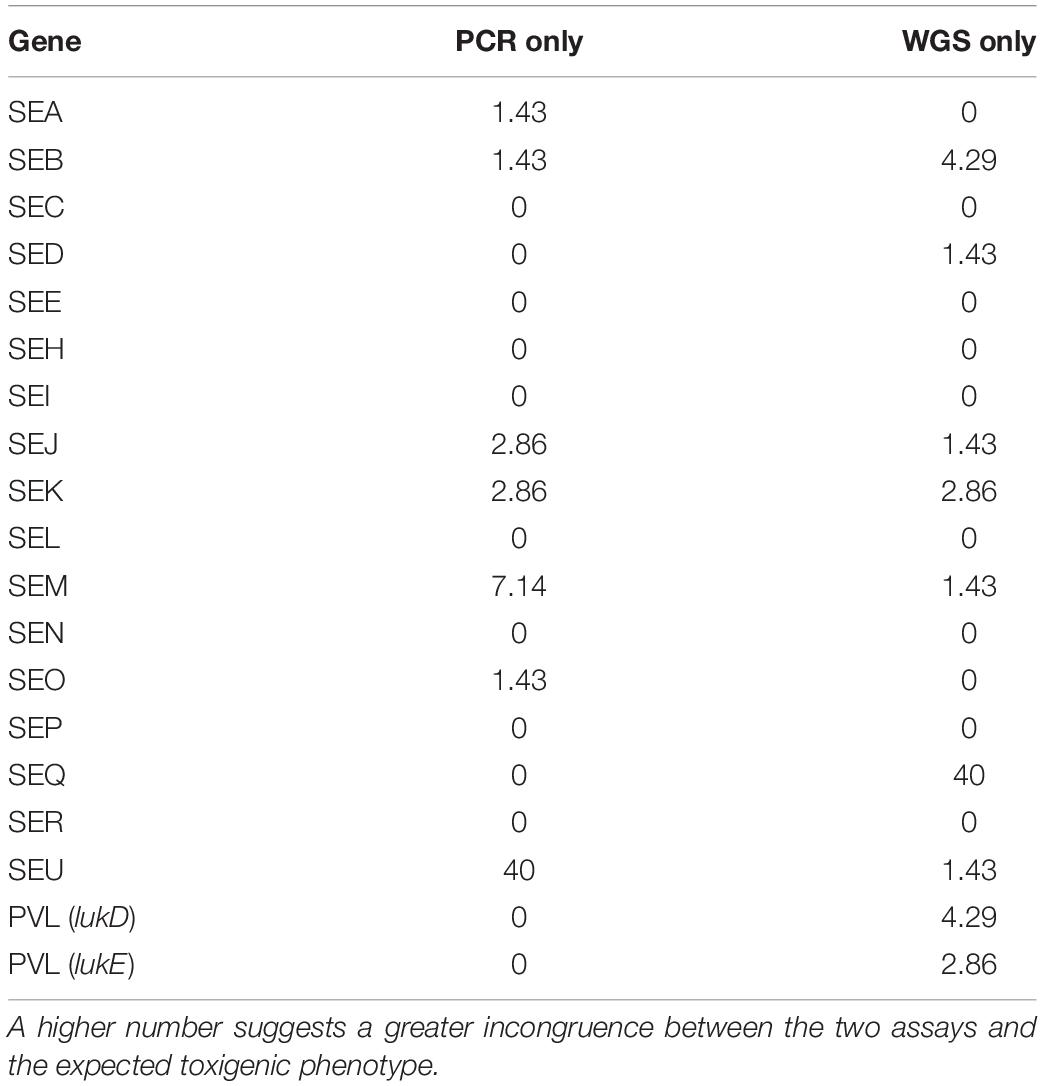
Table 2. The congruence of enterotoxin genes detected by both the PCR assay and WGS analysis are expressed here as a percentage.
As demonstrated in Figure 2, the phylogenetic relationships among the 69 outbreak-related isolates and six reference genomes indicated a strong degree of population substructure where well-supported clades of nearly identical isolates were identified. In addition, in previous work (Hait et al., 2014), we identified 12 PFGE patterns among these isolates. The original 12 PFGE designations are exhibited in Figure 2, along with strain identifiers, the sample collection dates and locations, sequence type, and spa type. Most of the outbreak-related isolates belong to ST1 (33/69, 47.8%), ST45 (22/69, 31.9%), ST5 (5/69, 7.2%), and ST30 (5/69, 7.2%). Isolate CFSAN007883 belong to ST59 and CFSAN007902 and CFSAN007903 were new ST similar to ST45. In addition, we identified two major spa types: t127 (33/69, 47.8%) and t3783 (20/69, 29.0%). In this study, these isolates were grouped into six well-supported clades in the phylogenetic tree. Though some reference isolates from recent years (isolates 1522 and LS1) showed a close relationship with certain outbreak-related isolates, the majority of outbreak-related isolates in the current work presented clonal structure, such as the patterns FDA1 and FDA4 clades. Some PFGE patterns exclusively represent the isolates in some well-supported clades. For example, isolates CFSAN007848, CFSAN007849, and CFSAN007850 were collected on the same date, which all belong to Pattern FDA3 and shared the common ancestor by forming a cluster in the phylogenetic tree. This also applied to Patterns FDA2, FDA6, FDA8, FDA10, and FDA12. As expected, WGS data showed discrepancies compared to the PFGE results. Though most Pattern FDA1 isolates were grouped together with FDA10, FDA11, and FDA12 in the same well-supported clade, the FDA1 isolate CFSAN007823 was located in the clade mainly consisting of Patterns FDA4 and FDA7 isolates. Similarly, the FDA4 isolate CFSAN007856 was found in the clade mainly consisting of FDA1 isolates. In addition, we observed that closely related isolates were collected from different dates. The pattern FDA4 clade had isolates collected in December 2010, January 2011, and November 2011. This also applied to the pattern FDA2 cluster, which consisted of four isolates (CFSAN007838, CFSAN007839, CFSAN007841, and CFSAN007847) collected from the Assembly Production Room and Baking Production Room but different dates.
The microtiter plate assay used the mean blank-corrected absorbance values for each S. aureus isolate to classify them as positive (OD > 0.240), moderate (OD = 0.240–0.120), and negative (OD < 0.120) with regard to their ability to form a functional biofilm. This analysis indicated that 4.3% of the isolates examined resulted in weak to no biofilm formation, 36.2% were characterized as moderate, and 59.4% were high biofilm producers (Table 3). CRA examinations yielded 52.2% positive results identified by black colonies with dry crystalline consistency; 47.8% intermediate results differentiated by black/brownish smooth colonies and red smooth colonies characteristic of weak to non-biofilm producers were not identified in the isolates tested (Table 3).
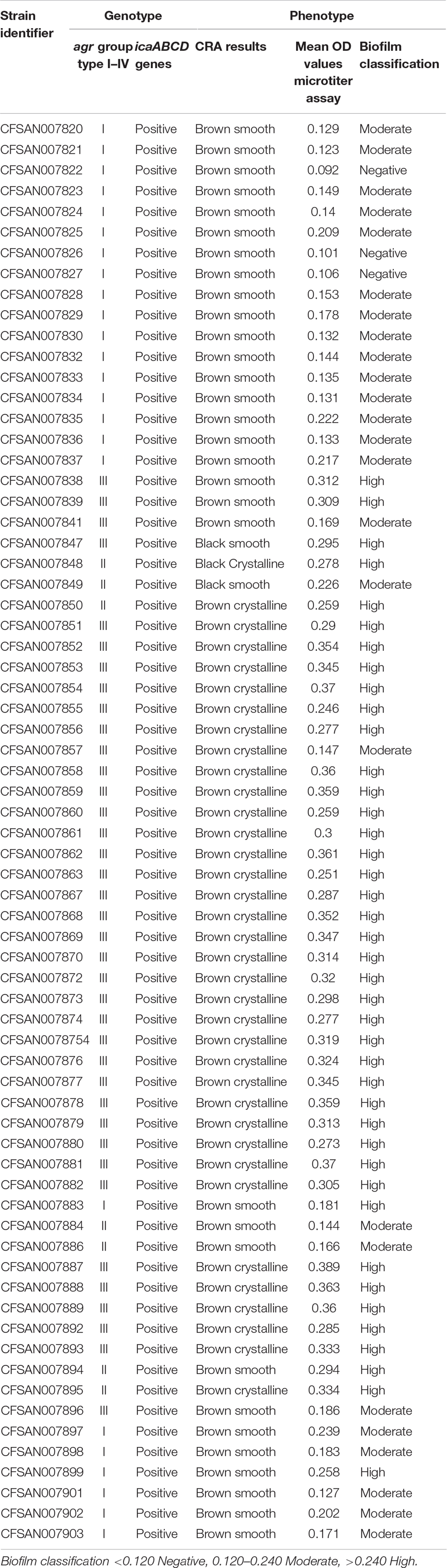
Table 3. Genotypic identification of agr groups and icaABCD genes along with phenotypic classification of biofilm formation using CRA and microtiter plate assays for the S. aureus outbreak recovered isolates.
Staphylococcus aureus has an immense role in human disease. This investigation studied virulence determinants and the evolutionary relationships for 69 outbreak-linked S. aureus isolates using WGS data and in vitro phenotypic assessments. Central to the role in S. aureus pathogenesis, agr is characterized in four allelic groups numbered I–IV. The agr locus influences the expression of many S. aureus virulence factors (Robinson et al., 2005). This includes roles in regulating genes for biofilm formation and antimicrobial resistance (Tan et al., 2018). The association between biofilm formation and the agr groups has been extensively examined. For example, strains of agr groups II and III are reported as the main biofilm producers of the four agr groups I–IV (Tan et al., 2018). The association among agr groups and antibiotic resistance was reported in a study by Nichol et al. (2011) which found a significant association of agr group I and community-acquired methicillin-resistant Staphylococcus aureus (MRSA), and correlated agr group II with hospital acquired MRSA. The agr group III was the most prevalent in our analysis, followed by agr group I, agr group II, and agr group IV, which was not found in the isolates tested. Additionally, antibiotic resistance indicator genes mecA and mecC (Paterson et al., 2014) were not distinguished in our analysis. In a study of mainly clinical isolates of S. aureus published by Jarraud et al. (2002), the four agr groups were found in relatively even distribution. Jarraud et al. (2002) documented that the agr group distribution robustly correlated with the genetic background of the strains tested, and consequently also indirectly with certain disease profiles.
All SE designations have been proven to induce emesis, and numerous outbreak incidences have been reported involving these non-classical SEs. This signifies the need for routine testing to include SEs beyond the classical SEA–SEE. Our analysis identified all SE and SE-like gene targets. Here, we identified six isolates — CFSAN007884, CFSAN007886, CFSAN007896, CFSAN007901, CFSAN007902, and CFSAN007903 — that lacked classical enterotoxins but harbored one or more of the non-classical or SEI gene seg–selx. This accounts for 9% of the total number of S. aureus isolates evaluated in this study. Other examples include a mass SFP outbreak in Osaka, Japan, where greater than 10,000 cases were linked to the consumption of contaminated reconstituted milk due to the presence of small quantities of nearly equal amounts of both SEA and SEH toxins (Ikeda et al., 2005). A different outbreak involving contaminated milk in Romania sickened 30 children. In this outbreak, the S. aureus isolates recovered from milk, patients, and food handlers lacked classical enterotoxins A–E and exhibited the same enterotoxin gene profile of seg, seh, sei, and sem (Coldea et al., 2015).
Fisher et al. (2018) reported that of the S. aureus isolates found in clinical, SFP outbreaks and commensal environments, approximately 80% carry an average of five to six SE genes. Using sequencing analysis, we characterized as many as 11 SEs and SEI genes in several of the S. aureus isolates. Both seb and selx were identified in greater than 50% of the isolates at 54 and 57%, respectively. The enterotoxin genes pattern sea, seb, seh, sek, seq, and selx was the most prevalent among the isolates in this study, while see, ses, and set genes were not detected at all. The toxic shock syndrome toxin gene was identified in 12% and pvl was distinguished in 58% for the isolates. These outcomes were compared to previously studied PCR and VIDAS® SET2 results. Results for the functional assays, an end point PCR assay, and WGS were largely in agreement with the exception of new variants of SElU2, SElV, and SElX that were not detected with original primers. The incongruence of SElU is presumably due to gene similarity with SElU2. SE gene concurrence was observed at 96.2%, based on PCR and WGS results.
The determined genes associated with biofilm formation composed of bap, icaA, and icaD, and the genes that encode MSCRAMMs bbp, cna, ebpS, eno, fib, fnbA, fnbB, clfA, and clfB genes. WGS assessment identified the presence of icaA, icaB, icaC, and icaD genes in all outbreak isolates. Cucarella et al. (2001) found that all the staphylococcal isolates harboring bap were pathogenic and highly adherent biofilm producers. Bap is a surface protein involved in biofilm formation. Our sequencing analyses determined that all isolates were negative for bap S. aureus gene V329. However, biofilm-associated genes: clfA, clfB, el1S, bb1, fnbA, fnbB, and eno were all present in 99% to all of the isolates examined. Here, bap presence was not found to be correlated with expected virulence or biofilm formation ability.
The two methods most commonly described for biofilm detection are the microtiter plate and CRA. The microtiter plate method was developed to replace the first macroscopic method estimation utilizing the tube method. The microtiter plate assay produces qualitative results using a spectrophotometer to measure OD of the 96-well plate containing the stained biofilms. The CRA analysis uses direct colony observation for the characterization of non-slime-producing strains yielding red-colored colonies versus slime-producing strains resulting in black-colored colonies. Here, CRA spot inoculation yielded 52.2% positive results and 47.8% intermediate outcomes, and weak to non-biofilm producers were not identified. The microtiter plate assay distinguished 4.3% of the isolates as weak to no biofilm formation, 36.2% moderate, and 59.4% high biofilm formation among the isolates examined. FDA4-designated isolates were collected during all three inspection dates, with 96% of these isolates being characterized as high biofilm producers. These FDA4 grouped isolates accounted for approximately 40% of the S. aureus contamination detected during this investigation. These collated outcomes establish that a significant level of high biofilm-producing strains of S. aureus likely assisted in the contamination difficulties found in this bakery environment, particularly given the increased resistance to antimicrobial agents and disinfectants that are characteristic of microorganisms proliferating within a biofilm. Biofilms aid the ability of S. aureus to survive in extreme environments.
In addition, the WGS was performed on all isolates from which phylogenetic analysis and genomic examination for virulence factors and biofilm-associated genes were conducted. Importantly, with the detailed metadata information and addition of reference genomes, our sequencing data identified six well-supported distinct clades consisting of the majority of outbreak-related isolates and demonstrated the presence of resident pathogens and multiple introductions in the same facility.
The evidence indicates that a considerable amount of diversity was observed across the contamination events, which is explained by a combination of resident pathogens with clonal structure and multiple independent introductions. This investigation emphasizes the complexities of eradicating potentially lethal pathogens from food production facilities.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JH and ST contributed to the study design and concept. JH, ST, GC, GK, and JP contributed to the collection of data. JH, GC, JP, and LY contributed to analysis and interpretation of data. JH contributed to drafting of the manuscript. ST, GC, and JP contributed to critical revision. All authors contributed to the manuscript and approved the submitted version.
This study was funded by the U.S. Food and Drug Administration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.687625/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bartels, M., Petersen, A., Worning, P., Nielsen, J., Larner-Svensson, H., Johansen, H., et al. (2014). Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Microbiol. 52, 4305–4308. doi: 10.1128/JCM.01979-14
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Coldea, I., Zota, L., Dragomirescu, C., Lixandru, B., Dragulescu, E., Sorokin, M., et al. (2015). Staphylococcus aureus harboring egc cluster coding for non-classic enterotoxins involved in a food-poisoning outbreak, Romania 2012 DE GRUYTER. Rev. Rom. Med. Lab. 23, 285–294. doi: 10.1515/rrlm-2015-0026
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penades, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/jb.183.9.2888-2896.2001
de Castro Melo, P., Ferreira, L. M., Filho, A. N., Zafalon, L. F., Vicente, H. I., and de Souza, V. (2013). Comparison of methods for detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Braz. J. Microbiol. 44, 119–124. doi: 10.1590/S1517-83822013005000031
de Vienne, D. M., Ollier, S., and Aguileta, G. (2012). Phylo-MCOA : a fast and efficient method to detect outlier genes and species in phylogenomics using multiple co-inertia analysis. Mol. Biol. Evol. 29, 1587–1598. doi: 10.1093/molbev/msr317
Fisher, E. L., Otto, M., and Cheung, G. Y. C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 9:436. doi: 10.3389/fmicb.2018.00436
Foster, T. J., and Höök, M. (1998). Surface proteins adhesins of Staphylococcus aureus. Trends Microbiol. 6, 484–488.
Gardner, G., Slezak, T., and Hall, B. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gutiérrez, D., Delgado, S., Vázquez-Sánchez, D., Martínez, B., López Cabo, M., Rodríguez, A., et al. (2012). Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl. Environ. Microbiol. 78, 8547–8554. doi: 10.1128/AEM.02045-12
Hait, J., Cao, G., Balkey, M., Kastanis, G., Curry, P., Yin, L., et al. (2021). Draft genome sequences of 62 Staphylococcus aureus isolates associated with four foodborne outbreaks in the United States. Microbiol Resour. Announc. 10, e118–e121. doi: 10.1128/MRA.00118-21
Hait, J., Tallent, S., Melka, D., Keys, C., and Bennett, R. (2014). Prevalence of enterotoxins and toxin gene profiles of Staphylococcus aureus isolates recovered from a bakery involved in a second staphylococcal food poisoning occurrence. J. Appl. Microbiol. 117, 866–875. doi: 10.1111/jam.12571
Hennekinne, J.-A., Ostyn, A., Guillier, F., Herbin, S., Prufer, A.-L., and Dragacci, S. (2010). How should Staphylococcal food poisoning outbreaks be characterized? Toxins 2, 2106–2116. doi: 10.3390/toxins2082106
Ikeda, T., Tamate, N., Yamaguchi, K., and Makino, S. I. (2005). Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl. Environ. Microbiol. 71, 2793–2795. doi: 10.1128/aem.71.5.2793-2795.2005
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., and Meugnier, H. (2002). Relationships between genetic relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70, 631–641. doi: 10.1128/iai.70.2.631-641.2002
Larsen, M., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R., et al. (2012). Multilocus sequence typing of total genome sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Mathur, T., Singhal, S., Khan, S., Upadhyay, D. J., Fatma, T., and Rattan, A. (2006). Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J. Med. Microbiol. 24, 25–29. doi: 10.4103/0255-0857.19890
Miao, J., Lin, S., Soteyome, T., Peters, B. M., Li, Y., Chen, H., et al. (2019). Biofilm formation of Staphylococcus aureus under food heat processing conditions: first report on CML production within biofilm. Sci. Rep. 9:1312. doi: 10.1038/s41598-018-35558-2
Nasr, R. A., AbuShady, H. M., and Hussein, H. S. (2012). Biofilm formation and presence of icaAD gene in clinical isolates of staphylococci. Egypt. J. Med. Hum. Genet. 13, 269–274. doi: 10.1016/j.ejmhg.2012.04.007
Nichol, K. A., Adam, H. J., Hussain, Z., Mulvey, M. R., McCracken, M., Mataseje, L. F., et al. (2011). Comparison of community-associated and health care-associated methicillin-resistant Staphylococcus aureus in Canada: results of the CANWARD 2007–2009 study. Diagn. Microbiol. Infect. Dis. 69, 320–325. doi: 10.1016/j.diagmicrobio.2010.10.028
Notcovich, S., DeNicolo, G., Flint, S. H., Williamson, N. B., Gedye, K., Grinberg, A., et al. (2018). Biofilm-forming potential of Staphylococcus aureus isolated from clinical mastitis cases in New Zealand. Vet. Sci. 5, 8. doi: 10.3390/vetsci5010008
Paterson, G. K., Harrison, E. M., and Holmes, M. A. (2014). The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 22, 42–47. doi: 10.1016/j.tim.2013.11.003
Rajkovic, A., Jovanovic, J., Monteiro, S., Decleer, M., Andjelkovic, M., Foubert, A., et al. (2019). Detection of toxins involved in foodborne diseases caused by Gram-positive bacteria. Compr. Rev. Food Sci. Food Saf. 19, 1605–1657. doi: 10.1111/1541-4337.12571
Robinson, D. A., Monk, A. B., Cooper, J. E., Feil, E. J., and Enright, M. C. (2005). Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J. Bacteriol. 187, 8312–8321. doi: 10.1128/JB.187.24.8312-8321.2005
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seo, K. S., and Bohach, G. A. (2007). “Staphylococcus aureus,” in Food Microbiology, 3rd Edn, eds M. P. Doyle and L. R. Beuchat (Washington, DC: ASM Press), 493–518.
Shallcross, L. J., Fragaszy, E., Johnson, A. M., and Hayward, A. C. (2013). The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. 13, 43–54. doi: 10.1016/S1473-3099(12)70238-4
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tan, L., Li, S. R., Jiang, B., Hu, X. M., and Li, S. (2018). Therapeutic targeting of Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 9:55. doi: 10.3389/fmicb.2018.00055
Tristan, A., Ying, L., Bes, M., Etienne, J., Vandenesch, F., and Lina, G. (2003). Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 41, 4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J. M., Amorena, B., Penades, J. R., et al. (2003). SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
Yarwood, J. M., and Schlievert, P. M. (2003). Quorum sensing in Staphylococcus infections. J. Clin. Invest. 112, 1620–1625. doi: 10.1172/JCI200320442
Keywords: staphylococcal food poisoning, staphylococcal enterotoxins, WGS, biofilm, Staphylococcus aureus
Citation: Hait JM, Cao G, Kastanis G, Yin L, Pettengill JB and Tallent SM (2021) Evaluation of Virulence Determinants Using Whole-Genome Sequencing and Phenotypic Biofilm Analysis of Outbreak-Linked Staphylococcus aureus Isolates. Front. Microbiol. 12:687625. doi: 10.3389/fmicb.2021.687625
Received: 29 March 2021; Accepted: 24 May 2021;
Published: 29 June 2021.
Edited by:
Evandro L. de Souza, Federal University of Paraíba, BrazilReviewed by:
Daniel F. M. Monte, São Paulo State University, BrazilCopyright © 2021 Hait, Cao, Kastanis, Yin, Pettengill and Tallent. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer M. Hait, amVubmlmZXIuaGFpdEBmZGEuaGhzLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.