
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 November 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.684565
 Rafael Vicentini Popin1
Rafael Vicentini Popin1 Danillo Oliveira Alvarenga1,2
Danillo Oliveira Alvarenga1,2 Raquel Castelo-Branco1,3
Raquel Castelo-Branco1,3 David Peter Fewer1
David Peter Fewer1 Kaarina Sivonen1*
Kaarina Sivonen1*Microbial natural products are compounds with unique chemical structures and diverse biological activities. Cyanobacteria commonly possess a wide range of biosynthetic gene clusters (BGCs) to produce natural products. Although natural product BGCs have been found in almost all cyanobacterial genomes, little attention has been given in cyanobacterial research to the partitioning of these biosynthetic pathways in chromosomes and plasmids. Cyanobacterial plasmids are believed to disperse several natural product BGCs, such as toxins, by plasmids through horizontal gene transfer. Therefore, plasmids may confer the ability to produce toxins and may play a role in the evolution of diverse natural product BGCs from cyanobacteria. Here, we performed an analysis of the distribution of natural product BGCs in 185 genomes and mapped the presence of genes involved in the conjugation in plasmids. The 185 analyzed genomes revealed 1817 natural products BGCs. Individual genomes contained 1–42 biosynthetic pathways (mean 8), 95% of which were present in chromosomes and the remaining 5% in plasmids. Of the 424 analyzed cyanobacterial plasmids, 12% contained homologs of genes involved in conjugation and natural product biosynthetic pathways. Among the biosynthetic pathways in plasmids, manual curation identified those to produce aeruginosin, anabaenopeptin, ambiguine, cryptophycin, hassallidin, geosmin, and microcystin. These compounds are known toxins, protease inhibitors, odorous compounds, antimicrobials, and antitumorals. The present study provides in silico evidence using genome mining that plasmids may be involved in the distribution of natural product BGCs in cyanobacteria. Consequently, cyanobacterial plasmids have importance in the context of biotechnology, water management, and public health risk assessment. Future research should explore in vivo conjugation and the end products of natural product BGCs in plasmids via chemical analyses.
Microbial natural products originate from secondary metabolism and exhibit a wide range of chemical structures and biological activities (Woodruff, 1980). These metabolites can act as antibiotics, anticancer agents, antivirals, and toxins and can be used as enzyme inhibitors, polymers, or surfactants (Demain, 2014). The enzymes involved in the biosynthesis of natural products are commonly encoded in biosynthetic gene clusters (BGCs) located in contiguous stretches of DNA (Stone and Williams, 1992; Osbourn, 2010; Jensen, 2016). Natural product BGCs usually include core biosynthesis, regulatory and resistance, and tailoring genes (Daum et al., 2009; Nett et al., 2009). Among accessory enzymes, 4-phosphopantetheinyl transferases (PPTs) play a major role in the biosynthesis of several types of natural products (Lambalot et al., 1996; Beld et al., 2014; Yang et al., 2017).
Understanding of the genetic diversity and distribution of natural product BGCs has greatly increased in the last decade due to the enormous expansion in the number of sequenced bacterial genomes in the last decade (Land et al., 2015; Jensen, 2016). Genome mining, which uses bioinformatics techniques to identify genes encoding enzymes possibly involved in natural products biosynthesis, has led to the discovery of novel compounds (Corre and Challis, 2009; Zerikly and Challis, 2009; Bachmann et al., 2014). Cyanobacteria are among several phyla of bacteria that are commonly explored using these techniques (Micallef et al., 2015).
Since the genome of Synechocystis sp. PCC 6803 was sequenced in 1996 (Kaneko et al., 1996), the number of complete cyanobacterial genomes deposited in the NCBI GenBank has slowly increased in comparison to other bacteria (Alvarenga et al., 2017). Despite their underrepresentation in public databases, cyanobacterial genomes were successfully investigated from evolutionary, ecological, and taxonomic perspectives (Sciuto and Moro, 2015). Cyanobacteria are recognized as a source of diverse natural products with applications in pharmacology, biotechnology, and bioenergy production (Burja et al., 2001; Angermayr et al., 2009; Kehr et al., 2011). A considerable portion of these molecules is produced by non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) (Tan, 2007; Dittmann et al., 2015). Other classes of broadly distributed cyanobacterial natural products include ribosomally synthesized and post-translationally modified peptides (RiPPs), alkaloids, and terpenoids, among others (Hillwig et al., 2014; Martins and Vasconcelos, 2015; Pattanaik and Lindberg, 2015; Jain et al., 2017). Genome mining of cyanobacterial genomes has helped unravel the diversity of BGCs involved in the production of various natural products (Singh et al., 2010; Wang et al., 2011; Micallef et al., 2015).
Cyanobacterial natural product BGCs are mostly concentrated in the genomes of late-branching cyanobacteria, mainly in the orders Oscillatoriales and Nostocales, although they are found in almost all cyanobacterial genomes (Shih et al., 2013; Calteau et al., 2014; Dittmann et al., 2015). Several studies have mapped the distribution of BGCs in these organisms (Dittmann et al., 2015). However, in cyanobacterial studies, little attention has been given to BGCs located in plasmids (Ehrenreich et al., 2005; Wang et al., 2011; Shih et al., 2013; Calteau et al., 2014; Dittmann et al., 2015). Nevertheless, horizontal gene transfer events are linked to the dissemination and evolution of many cyanobacterial natural product BGCs, including toxins such as cylindrospermopsin, microcystin, anatoxin-a, and saxitoxin (Lawrence and Roth, 1996; Ginolhac et al., 2005; Dittmann et al., 2013). This hypothesis places importance on cyanobacterial plasmids, as they would be directly involved in differentiating toxic and non-toxic strains, and in production of several natural products with economic, environmental, and public health importance (Tooming-Klunderud et al., 2008; Dittmann et al., 2013).
Plasmids play a key role in horizontal gene transfer, and conjugation is one of the processes that can transfer genetic material (Boto, 2010; Harrison and Brockhurst, 2012). The most frequent mechanism of DNA conjugation in gram-negative bacteria involves a relaxome, which includes a relaxase and a type IV coupling protein (T4CP) encoded by mobility genes (MOB), and a transferosome assembled by a type IV secretion system (T4SS) that is encoded by mating pair formation genes (MPF) (De La Cruz et al., 2010; Smillie et al., 2010). During conjugation, the relaxase cleaves and covalently binds itself to the transferring DNA on a site called oriT (Garcillán-Barcia et al., 2009). The T4SS is believed to then act as a secretor protein by transferring DNA and the relaxase to the recipient cell (Christie, 2004). For this purpose, the T4CP recognizes, energizes, and delivers the nucleoprotein to the T4SS (Zechner et al., 2012). Plasmids encoding these three components are called self−transmissible or conjugative, while mobilizable plasmids usually encode just the MOB and a T4SS and are transmitted only in the presence of a helper conjugative plasmid (Garcillán-Barcia et al., 2009). Most cyanobacterial plasmids were predicted to lack the necessary genes to be conjugative (Smillie et al., 2010). However, no concomitant analysis of the presence of BGCs in plasmids and the mobility of these replicons is currently available for cyanobacteria.
Thus, the present study screened 184 complete genomes publicly available in the GenBank database (Clark et al., 2016) from the phylum Cyanobacteria and one from Candidatus Melainabacteria, a phylum that is closely related to cyanobacteria (Di Rienzi et al., 2013). We used genome mining to provide evidence that several known natural product BGCs are in plasmids, some of which might be conjugative.
“Cyanobacteria/Melainabacteria group” genomes deposited until January 14, 2020 in the NCBI GenBank (Clark et al., 2016) at “Complete” and “Chromosome” assembly levels were analyzed. Altogether, they included 184 genomes from the phylum Cyanobacteria and 1 genome from the closely related Candidatus Melainabacteria (Supplementary Table 1; Di Rienzi et al., 2013). Statistics of the genome assemblies were obtained from the NCBI GenBank website (Clark et al., 2016). Means, p-value, averages, standard deviations, and boxplot and scatter graphs were generated using standard analyses in Microsoft Excel v16.0.6742.2048 (Microsoft, Redmond, WA). Pearson and Spearman’s rank correlation coefficients were calculated in R Project for Statistical Computing version 4.0.5.1
BGCs involved in natural product biosynthetic pathways were automatically annotated using the “Bacterial” sequence analysis in antiSMASH v5.1.1 (Blin et al., 2019). For this analysis, the standard “relaxed” strictness was applied, and no extra future was activated. Manual annotation and curation of BGCs were performed in the program Artemis v18.1.0 (Carver et al., 2012). This manual step involved comparisons of automatically annotated BGCs to diverse previously described reference gene clusters from cyanobacterial strains using the protein searches with BLASTp (default parameters were used; see Supplementary Table 1 for alignment scores; Altschul et al., 1990). Searches for PPTs were performed using BLASTp searches of candidate proteins in the analyzed genomes (default parameters; Altschul et al., 1990) using the 4’-phosphopantetheinyl transferase from Nostoc sp. PCC 7120 (NCBI accession number P37695.2) as reference. The best identified possible homologs (proteins with the highest E-value) were compared using BLASTp (default parameters; Altschul et al., 1990) to the previously described 4’-phosphopantetheinyl transferase type Sfp from Bacillus cereus (WP_002185911.1) and holo-acyl carrier protein synthase (type A, accession number NP_388343.1) from Bacillus subtilis subsp. subtilis str. 168 (see Supplementary Table 2 for search scores). Identification of relaxase (accession number in NCBI: MBD2275468.1), VirB4 (BAB78290.1), and VirD4 (MBD2275482.1) were performed with BLASTp protein searches (default parameters; Altschul et al., 1990) using genes of the known conjugative plasmid pCC7120α from Nostoc sp. PCC 7120 (see Supplementary Table 3 for scores of manually curated best candidate genes in plasmids with known BGCs and Supplementary Table 4 for automatic searches for candidate genes in the other plasmids using cutoff of coverage 80%, identity 30%, and E-value 1.00 E-20).
Plasmid representations were generated using the standard parameters of the “BLAST analysis” in the server Gview (Petkau et al., 2010) and default analysis in BRIG v0.95 (Alikhan et al., 2011). The program Inkscape v0.92 was used for drawing BGCs.2
The phylogenetic analyses of the concatenated amino acid sequences from the BGCs and nucleotide sequences of 16S rRNA were created with 5,000,000 generations in MrBayes 3.2.7a (Ronquist et al., 2012). The best substitution model for each protein in the BGCs was predicted using the default BIC calculation in ProtTest 3.4.2 (Darriba et al., 2011). Default BIC calculation was also used in jModelTest v2.1.10 (Darriba et al., 2012) for the 16S rRNA phylogenetic analyses; and the model HKY + I + G was predicted as the best. Tree visualization was performed with FigTree v1.4.43 and editing with Inkscape 1.0 (see text footnote 2). Cyanobacterial order was assigned according to a polyphasic taxonomic system (Komarek et al., 2014).
A maximum likelihood tree was constructed in RAxML v8.2.12 (Stamatakis, 2014) using 1,000 bootstrap samples. The model LG + G + I was identified as the best-fitting model by default BIC calculation in ProtTest 3.4.2 (Darriba et al., 2011). The phylogenomic placement was based on concatenated amino acid sequences of 120 bacterial single-copy conserved marker genes identified by GTDBTk 1.0.2 (default parameters; Parks et al., 2018). The tree was visualized in FigTree v1.4.4 (see text footnote 3) and figure editing was performed in Inkscape 1.0 (see text footnote 2).
The taxonomic distribution of analyzed genomes was initially explored. Following the latest proposed system of cyanobacteria, 39% of the analyzed genomes belong to the order Synechococcales (mainly Synechococcus, a polyphyletic genus with representatives also allocated in Oscillatoriales and Prochlorococcus), followed by Nostocales (31% of genomes; largely represented by Nostoc and Calothrix) (Figure 1). The remaining 30% of the genomes were distributed in the orders Oscillatoriales, Chroococcales, Pleurocapsales, Gloeobacterales, Gloeomargaritales, and Chroococcidiopsidales (Figure 1). No representative genome of the order Spirulinales was analyzed here due to unavailability. The Miscellaneous category includes three unclassified cyanobacterial genomes and the Candidatus Melainabacteria strain MEL.A1.
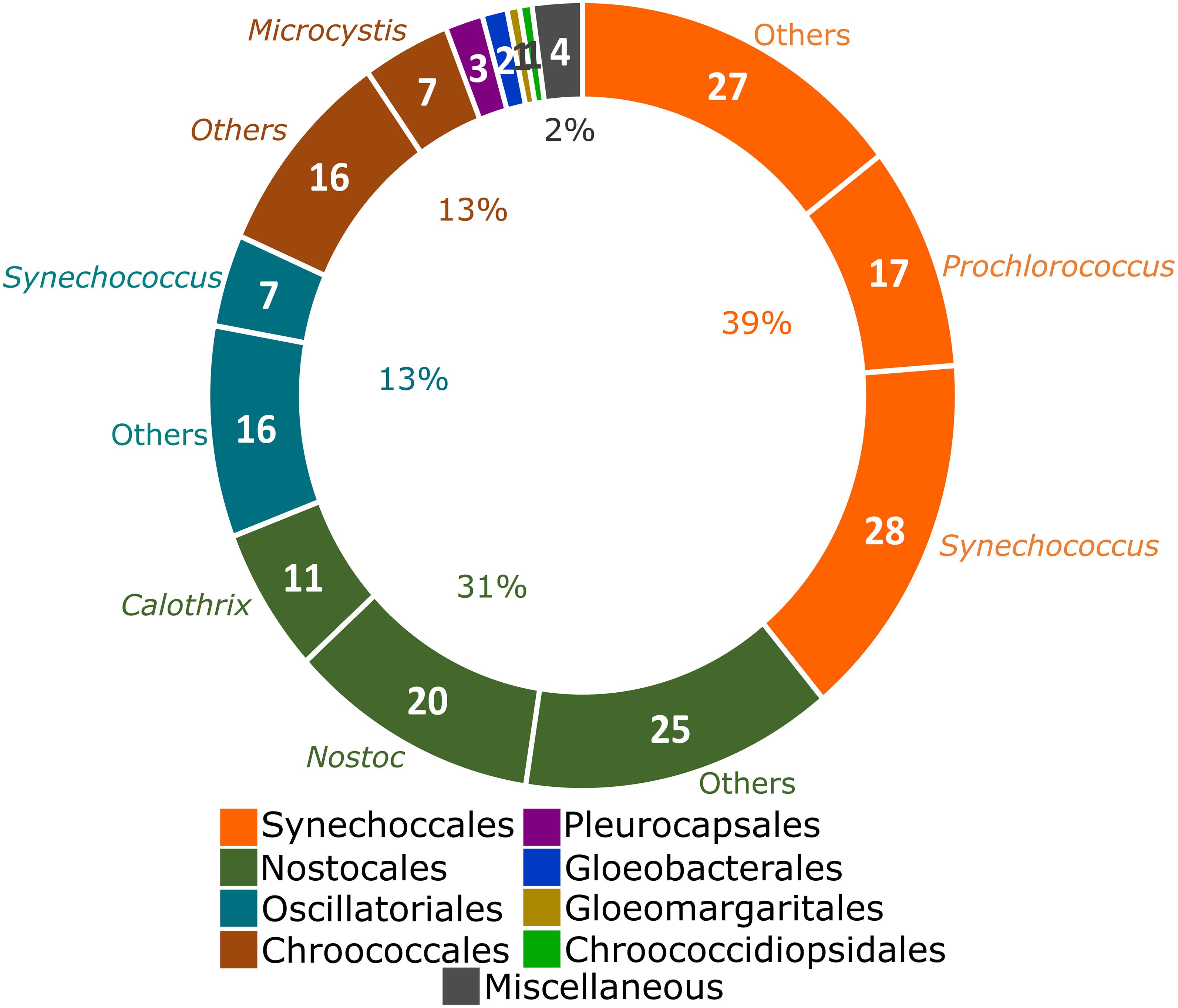
Figure 1. Taxonomic distribution of the 184 complete genomes deposited in the NCBI Genbank (Clark et al., 2016) from Cyanobacteria (Komarek et al., 2014) and one from the closely related phylum Candidatus Melainabacteria (Di Rienzi et al., 2013). Strains that remain unclassified in any cyanobacterial order and the Candidatus Melainabacteria strain MEL.A1 were allocated as Miscellaneous. Synechococcus is a polyphyletic genus (Komarek et al., 2020). Genera over-represented in each order are indicated. The number of genomes from over-represented genera is presented and the percentages were calculated in relationship to the total of 185 genomes.
From the 52 genera represented in the retrieved dataset, 27 included more than one genome. For these genera, basic statistical analyses (averages and standard deviation) were used and revealed insights into the genomic differences of cyanobacterial orders (Table 1). According to the NCBI GenBank database, these cyanobacterial genomes consisted of up to two chromosomes (calculated median of 1 per genome) and 14 plasmids (median of 1; Supplementary Table 5). Chromosomes represented from 97 to 100% of the strain’s genomic DNA, while plasmids represented up to 3% (Supplementary Table 5). Genome sizes ranged from 1.65 to 12.05 Mb (median 5.02 Mb), GC content from 30.8 to 68.7% (median of 42.2%), and the number of genes varied from 1816 to 11674 (median 4553) (Supplementary Table 5). The number of BGCs automatically annotated by antiSMASH v5.1.1 in chromosomes ranged from 1 to 42 (mean 8); up to five were found located in plasmids (median 0).
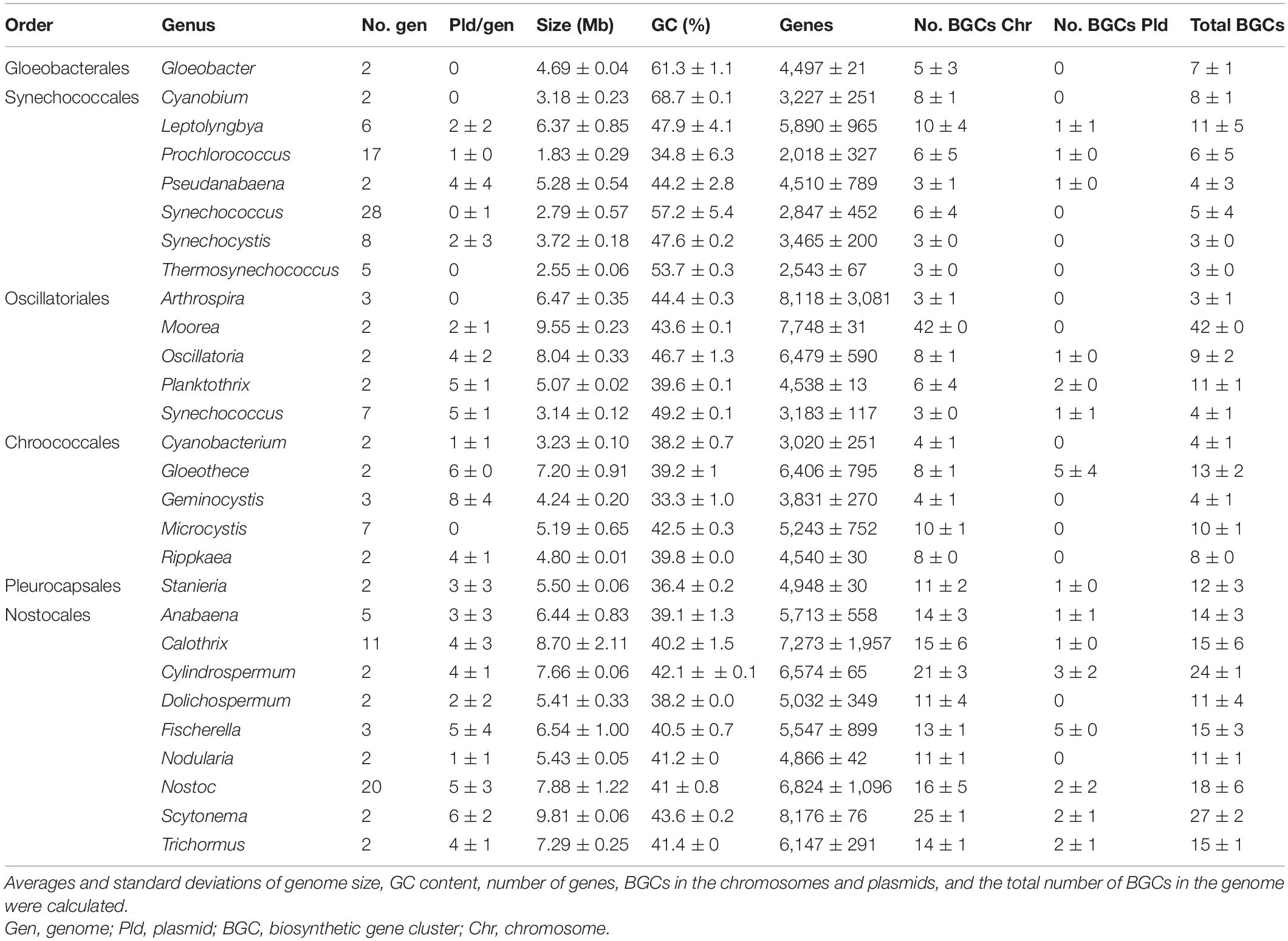
Table 1. Genome statistics of cyanobacterial genera with more than one complete sequence in the NCBI GenBank (Clark et al., 2016).
The analyzed cyanobacterial genomes were automatically annotated and a total of 1817 BGCs were identified (Supplementary Table 5). Synechococcus sp. JA-2-3B’a(2–13), Candidatus Melainabacteria MEL.A1, and Synechococcus sp. JA-3-3Ab had only one natural product BGC and thus were the genomes with the lowest number. In contrast, Moorea producens JHB and Moorea producens PAL-8-15-08-1 had 42 BGCs each (Supplementary Figure 1). Nostocales genomes were among those with the highest average number of natural product BGCs (Supplementary Figure 1). A positive correlation between the number of natural product BGCs and genome size was found (Pearson r = 0.75, p < 0.001; Spearman’s rho = 0.71, p < 0.001; Figure 2).
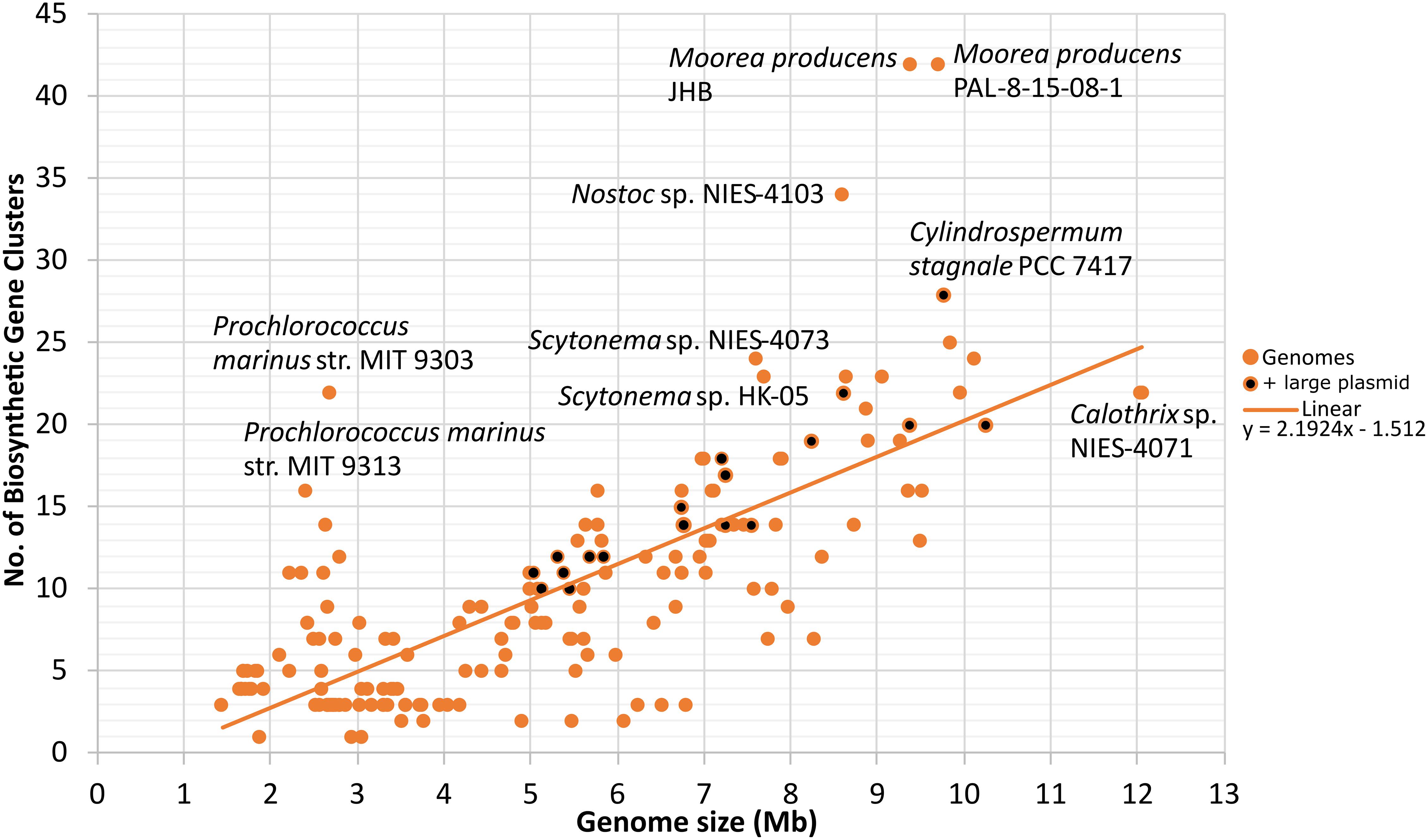
Figure 2. Number of automatically predicted natural product biosynthetic gene clusters plotted against genome size (Mb). Genomes with large plasmids (>500 kb) are also indicated (diCenzo and Finan, 2017). Linear trend and its corresponding linear correlation coefficient are presented. N = 185.
When the distribution of the automatically annotated BGCs in specific replicons was considered, most BGCs were identified in chromosomes (1718) and represented 95% of the total (1817). RiPPs were the most widespread class of natural product BGC products in these replicons (526 representatives, 31% of the chromosomal BGCs). Terpenes were the second most widespread products (470 representatives, 27% of the natural product BGCs in chromosomes) and were absent only in Arthrospira platensis C1, Candidatus Melainabacteria bacterium MEL.A1, and Nostoc sphaeroides CCNUC1. PKS genes were most frequently found associated with other classes, whereas only 49 single class PKS BGCs representatives were found.
A total of 424 plasmids were identified from the 185 genomes, of which 73 (17% of the total number of plasmids) had at least one natural product BGC (Supplementary Table 5). Ninety-nine BGCs were found in cyanobacterial plasmids. While most plasmids included only one natural product BGC, Gloeothece verrucosa PCC 7822 Cy782201 (0.88 Mb) had five (Supplementary Figure 2). Hybrid NRPS/PKS corresponded to more than half of the natural product BGCs located on plasmids (26 BGCs). NRPS consisted of 20 representatives, followed by bacteriocin (15 representatives). In contrast to chromosomes, terpenes were one of the least frequently observed products with biosynthetic pathways found on plasmids. The natural product BGCs identified as known biosynthetic pathways were further explored by manual curation.
Following the automatic annotation, BGCs assigned to known biosynthetic pathways were manually curated to identify false positives. Interestingly, several of the known natural product BGCs were in large replicons that were nevertheless named in the database as plasmids (here > 500 kb, reaching a maximum of about 1.78 Mb in Nostoc linckia NIES-25 plasmid1, Supplementary Table 5). These large replicons contained 1–5 natural product BGCs. The pathways for the known natural products ambiguine (in Fischerella sp. NIES-4106 plasmid 2, 550 kb), anabaenopeptin (Scytonema sp. HK-05 plasmid 1, 831 kb), geosmin (Nostoc linckia NIES-25 plasmid1, 1.78 Mb), and microcystin (Fischerella sp. NIES-4106 plasmid1, 550 kb) were encoded in large plasmids (Supplementary Figure 3 and Supplementary Table 5).
Other known natural product BGCs were located in smaller plasmids: i.e., aeruginosin in Cylindrospermum sp. NIES-4074 plasmid1 (340 kb); anabaenopeptin in Gloeothece citriformis PCC 7424 plasmid pP742401 (328 kb) and Gloeothece verrucose PCC 7822 plasmid Cy782201 (879 kb), cryptophycin in Nostoc sp. ATCC 53789 plasmid pNSP_c (219 kb), geosmin in Nostoc sp. NIES-2111 plasmid2 (320 kb), and hassallidin in Aulosira laxa NIES-50 plasmid1 (292 kb) and Tolypothrix tenuis PCC 7101 plasmid1 (292 kb) (Supplementary Table 5). Although the core enzymes for biosynthesis were found in these natural product BGCs in small plasmids, some of them are missing accessory genes that are present in the references and may produce compounds with distinct structures (Supplementary Table 1). To provide further evidence that the known natural product BGCs in plasmids are likely functional, the presence of key enzyme PPTs in the genomes was investigated.
A total of 165 possible homologs of PPTs were found (Supplementary Table 3). From the 185 genomes analyzed here, 157 had at least one copy of PPT homologs (84% of the total number of genomes). Most of these genomes encoded only 1 enzyme (149 genomes), while 6 genomic sequences encoded 2 enzymes and 1 genome (Halomicronema hongdechloris C2206) had 3 copies of PPTs. The size of these enzymes ranged from 137 (one of the two copies located in Chroococcidiopsis thermalis PCC 7203) to 339 aa (one of the three copies in Halomicronema hongdechloris C2206). However, 90% of the enzymes were between 200 and 280 aa (147 PPTs).
Interestingly, Acaryochloris marina MBIC11017 had a chromosomal PPT that was more like an Sfp-type PPT, and another in plasmid pREB1 that was the only one predicted as an AcpS-type PPT. The remaining 163 PPTs in the analyzed cyanobacterial genomes were likely Sfp-type.
To investigate the hypothesis of plasmids carrying natural product BGCs through conjugation, the 424 plasmids from the analyzed genomes were searched for the presence of homologs of relaxases, VirB4 (alr7206, a coupling protein, T4CP), and VirD4 (alr7213, a secretion system protein, T4SS). The reference plasmid pCC7120alpha from Nostoc sp. PCC 7120 harbored a similar gene cluster as that found in plasmid2 in Nostoc sp. NIES-2111 (Figure 3). This latter replicon was the sole plasmid encoding homolog proteins of a known natural product BGC (geosmin) to be predicted as conjugative. The results from the manual curation of known gene clusters in 10 other plasmids can be found in Supplementary Table 3.
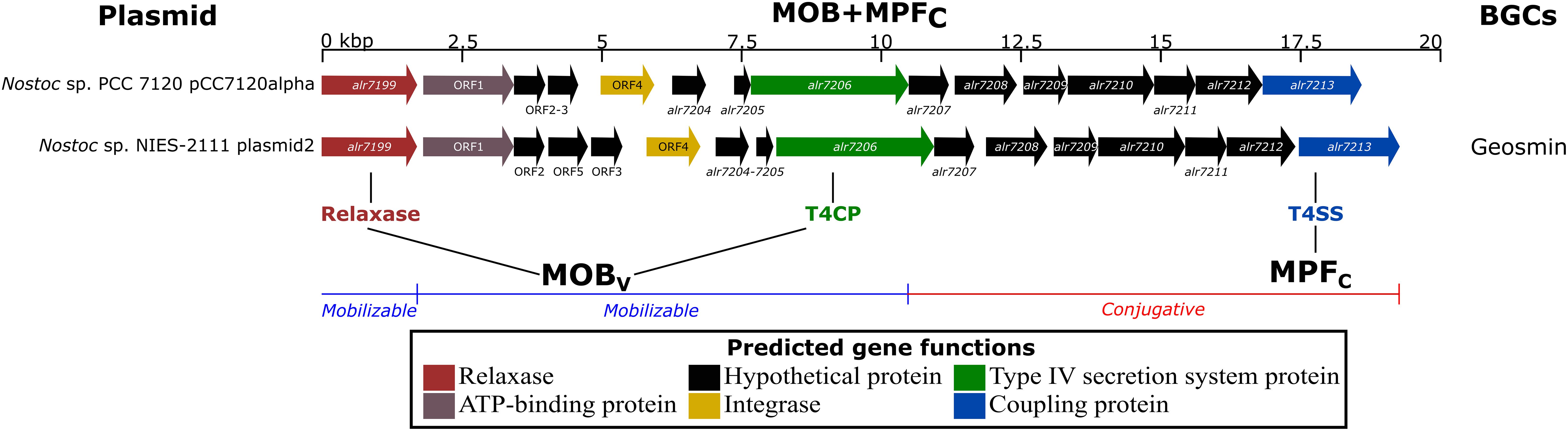
Figure 3. Genomic region of Nostoc sp. PCC 7120 plasmid pCC7120alpha and Nostoc sp. NIES-2111 plasmid2 encoding homologs of the main protein involved in the plasmid conjugative apparatus [relaxase- alr7199, virB4- alr7206 (T4CP) and virD4- alr7213 (T4SS)]. Previous studies characterized the mobility of plasmids (Smillie et al., 2010), MOBv (Garcillán-Barcia et al., 2009), and MPFc (Guglielmini et al., 2011). See Supplementary Table 4 for annotation of the relaxase, T4CP, and T4SS proteins in the 185 genomes involved in the present work.
Automatic annotation was used to predict the mobility of the remaining 414 plasmids (Supplementary Table 4). In total, 23 plasmids from Nostocales (genera Calothrix, Cylindrospermum, Fischerella, Nodularia, Nostoc, Trichormus) and 1 from Oscillatoriales (Microcoleus) contained relaxases, VirB4, and VirD4; and were predicted as conjugative. This represents 6% of the 424 analyzed cyanobacterial plasmids. Another plasmid from Chroococcales (Gloeocapsa), 24 from Nostocales (Anabaena, Aulosira, Calothrix, Nostoc, Trichormus, Tolypothrix), and 1 from Oscillatoriales (Crinalium) were predicated to be mobilizable as they encoded the relaxase but not Virb4 and VirD4. They also harbored 6% of the 424 evaluated cyanobacterial plasmids. The remaining 374 (88%) plasmids had only Virb4 or VirD4 and are likely immobile according to the current model.
Therefore, our results indicated that 15 possibly conjugative plasmids harbored a total of 18 natural product BGCs (including the known biosynthetic pathways for geosmin) and 4 other mobilizable plasmids with 1 natural product BGC each.
Phylogenetic trees were built with the manually curated known natural product BGCs found both in plasmids and chromosomes to investigate divergent evolutionary history (Supplementary Figures 4B–E). Phylogenetic trees based on hassallidin, geosmin, and anabaenopeptin biosynthetic enzymes showed that BGCs from plasmids appear to be more closely related than the chromosomal natural product BGCs. Thus, natural product BGCs from plasmids possibly face different evolutionary pressures than those present in chromosomes. A single microcystin BGC was found in plasmids included in the present study and formed a clade with a BGC from a chromosome (Supplementary Figure 4D).
A phylogenomic tree was used to map the taxonomical distribution of natural product BGCs, PPTs, and mobile plasmids (conjugative and mobilizable) in the analyzed cyanobacterial genomes (Figure 4). Natural product BGCs and PPTs were detected in all the cyanobacterial orders included in the present study. However, later-branching cyanobacteria from Nostocales had most of the natural product BGCs and a clear majority of the mobile plasmids. In contrast, early branching cyanobacteria, especially those from Gloeobacterales, Synechococcales, and Gloeomargaritales, had fewer natural product BGCs (mainly terpenes and RiPPs) and no mobile plasmids. A supplementary 16S rRNA phylogenetic tree of the analyzed genomes is shown in Supplementary Figure 4A.
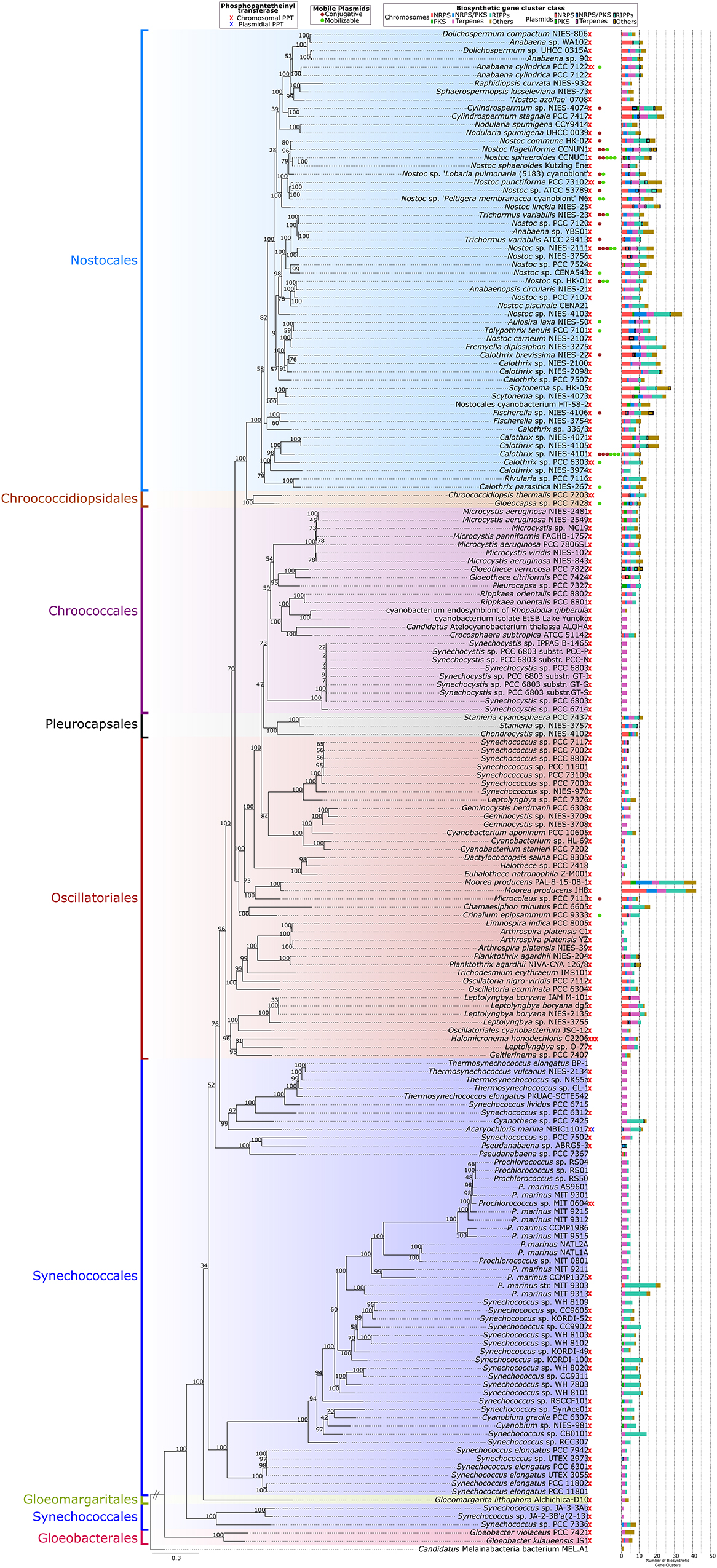
Figure 4. Phylogenomic tree using 120 conserved proteins from the 185 complete genomes in this study using Candidatus Melainabacteria bacterium MEL.A1 as outgroup. The number of phosphopantetheinyl transferases, mobilizable and conjugative plasmids, and biosynthetic gene clusters in the genomes are presented for each strain (x-axis).
Based on the model Gram-negative bacteria Escherichia coli, cyanobacteria are commonly assumed to be monoploid (Griese et al., 2011). However, these organisms can become oligoploid during rapid growth (Skarstad et al., 1983; Blank and Sánchez-Baracaldo, 2010). Some cyanobacteria may contain multiple chromosome copies throughout their life cycles (Griese et al., 2011; Sargent et al., 2016; Watanabe, 2020). Previous studies reported polyploidy in several genera, such as Anabaena (Nostoc) (Hu et al., 2007), Synechococcus (Chen et al., 2012), and Synechocystis (Schneider et al., 2007). Here, Nostoc sphaeroides CCNUC1, Crocosphaera subtropica ATCC 51142 (Welsh et al., 2008), and Anabaena (Dolichospermum) sp. 90 (Wang et al., 2012) were found to possess more than one chromosome. In contrast, large plasmids (considered here as > 500 kb) were found in 11 genomes from 4 of the 6 analyzed cyanobacterial orders and could correspond to previously described “chromids” (diCenzo et al., 2019).
Chromids are large, plasmid-like replicons that were previously found in approximately 10% of bacterial genomes (Harrison et al., 2010). Chromids possess replication systems that are similar to plasmids and can carry essential genes for cell viability (diCenzo and Finan, 2017). One of the proposed functions of these large replicons is to increase genome plasticity through the rapid acquisition or loss of genes by horizontal gene transfer (diCenzo et al., 2019). Chromids occurred in approximately 15% of the analyzed cyanobacterial genomes and contained several natural product BGCs automatically annotated.
Tools for automatic annotation of natural product BGCs were initially proposed to be a solution to the labor-intensive and time-consuming task of manual curation and, currently, antiSMASH is widely used in cyanobacterial research (Medema et al., 2011; Calteau et al., 2014; Micallef et al., 2015; Leao et al., 2017). The algorithm behind the identification of the biosynthetic pathways has been constantly updated to improve detection and prevent false positives (Weber et al., 2015; Blin et al., 2017, 2019). For instance, more pathways for the biosynthesis of RiPPs and PKS BGCs were detected using the latest versions of the tool compared with initial versions (Weber et al., 2015; Blin et al., 2017). Therefore, the fact that the analyzed genomes contained on average eight natural product BGCs in comparison to the previous estimation of five, should be approached with caution (Dittmann et al., 2015). Here, most of the automatically annotated natural products were predicted to have RiPPs and terpenes as end products.
Natural product BGCs encoding enzymes for RiPPs were found in almost all analyzed genomes. These molecules are products of post-translational modification of ribosomally synthesized precursor peptides (Arnison et al., 2013). Thus far, over 20 families of compounds that possess unique chemical features have been proposed (Arnison et al., 2013). Cyanobacteria encode the machinery to produce several RiPPs, including cyanobactins (Sivonen et al., 2010), lanthipeptides (Cubillos-Ruiz et al., 2017), lasso peptides (Tietz et al., 2017), and microviridins (Ziemert et al., 2010). Considering that automated tools are being improved to better predict genes involved in the biosynthesis of these compounds, future studies may expand the known repertoire of RiPPs produced by cyanobacteria (Blin et al., 2017, 2019).
Like RiPPs, genes for the biosynthesis of terpenes are widespread in cyanobacterial genomes (Shih et al., 2013). Although terpenes are commonly isolated from plants and fungi, terpene BGCs are widely found in bacterial genomes (Yamada et al., 2015). These compounds are essential in primary metabolism, such as for photosynthesis and respiration, but also have roles as secondary metabolites (Breitmaier, 2006). These roles likely explain why genes encoding enzymes involved in the biosynthesis of terpenes are present in cyanobacterial genomes (Leao et al., 2017). The repertoire of terpenes produced by cyanobacteria is possibly larger than currently known, as various cryptic terpene synthases are commonly found in their genomes (Shih et al., 2013; Yamada et al., 2015). Thus far, geosmin and 2-methylisoborneol are widely studied terpenes as they are odorous metabolites that impact drinking water quality (Jüttner, 1995; Suurnäkki et al., 2015; Oh et al., 2017). A pathway for geosmin production was among the known natural product BGCs located in plasmids.
Cyanobacterial plasmids have previously been shown to contain genes encoding RiPPs and are associated with the production of toxic and odorous compounds (Bolch et al., 1997; Wang et al., 2011). These replicons are also believed to play a major role in the distribution and evolution of toxin biosynthetic pathways in cyanobacteria (Dittmann et al., 2015). Plasmid-encoded natural product BGCs were manually curated in the present study and contained all the core genes for the biosynthetic pathways of the hepatotoxin microcystin, the antifungal hassallidin, and the odorous terpenoid geosmin (Kaebernick et al., 2000; Vestola et al., 2014; Suurnäkki et al., 2015). A biosynthetic pathway of the antiproliferative cytotoxin cryptophycin is located in a smaller plasmid of Nostoc sp. strain ATCC 53789 and is known to produce the natural product (Panda et al., 1998; Tippelt et al., 2020). Therefore, chemical analyses of the strains included in this study could determine that the biosynthetic pathways located in plasmids could be producing the compounds. Supporting this hypothesis, PPTs were also located in almost all analyzed cyanobacterial genomes.
PPTs have an important role in the biosynthesis of most analyzed classes of natural products in the present study (Hopwood, 1997; Marahiel et al., 1997). Previous mapping of PPTs in cyanobacteria indicated that only one copy occurred per genome (Copp and Neilan, 2006). Here, up to three different PPTs were found. Corroborating our results, other bacteria also contain multiple copies of these enzymes (Beld et al., 2014; Kim et al., 2018). Moreover, most of the predicted enzymes in the analyzed cyanobacterial genomes corresponded to the known lengths of PPTs (115–230 amino acids; Lambalot et al., 1996; Quadri et al., 1998). Two main families of PPTs are known, namely AcpS-type PPTs, which are involved in activating carrier proteins involved in primary metabolism, and Sfp-like PPTs as part of secondary metabolism pathways (Lambalot et al., 1996; Quadri et al., 1998). Sfp-like PPTs were the sole type found in cyanobacterial genomes (Copp and Neilan, 2006). However, Acaryochloris marina MBIC11017 possibly possesses a representative of an AcpS-like PPT in a plasmid. Consistent with our results, plasmids from other bacterial phyla have also been found to encode PPTs (Liu et al., 2005; De Lay and Cronan, 2008). This enzyme is in a cyanobacterial plasmid and may have originated from a horizontal gene transfer.
The replicon pCC7120α from Nostoc sp. PCC7120 is the only cyanobacterial plasmid that has been reported to be transmissible in vivo (Muro-Pastor et al., 1994). A previous study using automatic annotation found no homologs of T4SS in cyanobacteria and hypothesized that an unknown mechanism of conjugation could be present in these organisms (Smillie et al., 2010). Later studies investigated some of these possible mechanisms, such as integrative conjugative elements (ICE) and origin-of-transfer (oriT) sequences. ICEs are conjugative elements integrated into chromosomes (Guglielmini et al., 2011), whereas oriT are present in non-conjugative plasmids that can be mobilized by the relaxase in conjugative plasmids (O’Brien et al., 2015). Considering that ICEs and oriT are relatively less explored than the genes involved in conjugation, the mobility of the analyzed cyanobacterial plasmids was predicted exclusively on the genes for conjugation (Smillie et al., 2010).
We have provided evidence using manual curation that some plasmids in cyanobacteria may be mobile and contain natural product BGCs. These should be tested in vivo to expand the number of known conjugative plasmids from those organisms and to determine if the predicted cyanobacterial natural product BGCs are being transferred through conjugation. Toxic natural products produced by other bacteria, such as botulinum toxin from Clostridium botulinum and cereulide from Bacillus cereus were also found on plasmids (Ehling-Schulz et al., 2006; Carter et al., 2014). In the case of the botulinum toxin, horizontal gene transfer of BGC by large conjugative plasmids is also believed to be possible (Skarin and Segerman, 2011).
The availability of complete genomes allowed mapping of many natural product BGCs in cyanobacterial plasmids. Manual curation identified the biosynthetic pathways of known toxins (microcystin), odorous metabolites (geosmin), protease inhibitors (anabaenopeptin, aeruginosin), and antimicrobial (ambiguine and hassalidin), and antitumor (cryptophycin) compounds. These biosynthetic pathways in plasmids included core genes necessary for biosynthesis. PPTs, a key enzyme for biosynthesis of many of these natural products, was found in most of the genomes. Therefore, we predict that these plasmid-based biosynthetic pathways are likely able to produce natural products. Moreover, the plasmid containing the geosmin BGC and others with cryptic natural product BGCs were predicted to be mobile. This is novel in silico evidence that plasmids are involved in the dissemination and evolution of diverse natural product biosynthetic pathways in cyanobacteria. If confirmed, the transmission of natural product BGCs among cyanobacteria by conjugation would present new biotechnological opportunities but also several risks. Cyanobacterial taxa which are not known to be problematic could acquire genes for toxin biosynthesis and cause economic losses, threats to public health, and damage to natural environments. Future research should investigate in vivo the conjugation of cyanobacterial plasmids and the potential transmission of BGCs among cyanobacteria. Likewise, chemical analyses of the strains included in the present study may lead to the discovery of novel natural products and chemical variants of known compounds.
The datasets presented in this study can be found in the Supplementary Material.
RP, DA, and DF conceptualized the study. RP, DA, and RC-B performed the analyses. RP wrote the manuscript. DF and KS were responsible for supervision. KS managed funding acquisition. All authors participated in reviewing and editing the manuscript and agreed to the published version of the manuscript.
RP was funded by the Doctoral Programme in Microbiology and Biotechnology of the University of Helsinki. DA was supported by a São Paulo Research Foundation post-doctoral fellowship (FAPESP #2018/01563-2). RC-B acknowledges financial support from Fundação para a Ciência e a Tecnologia (FCT) fellowship SFRH/BD/136367/2018. Grants from Nordforsk (NordAqua) and the Jane and Aatos Erkko Foundation to KS supported this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.684565/full#supplementary-material
Supplementary Figure 1 | Boxplot of the number of automatically annotated biosynthetic gene clusters identified in the cyanobacterial genera with more than one representative deposited in the NCBI GenBank (Clark et al., 2016). The genera are presented in increasing order of the average number of gene clusters (represented by “x”).
Supplementary Figure 2 | Visualization of Gloeothece verrucosa PCC 7822 plasmid Cy782201. The position of the five automatically annotated biosynthetic gene clusters are indicated. Similarities with known BGCs calculated by antiSMASH v5.1.1 (Blin et al., 2019) are also indicated.
Supplementary Figure 3 | Known natural products biosynthetic gene clusters (BGCs) and proteins involved in conjugation (MOB + MPFc) identified in cyanobacterial plasmids. The location and organization of BGCs in their respective plasmids are shown. See Supplementary Table 1 for information on gene annotation. The positions of manually annotated BGCs in the genomic maps are highlighted in blue or red.
Supplementary Figure 4 | 16S rRNA phylogeny of the 185 analyzed genomes (A) and representation of known BGCs found in plasmids (shown in bold) and chromosomes (B–E). The posterior probability of clades is shown in the trees. NCBI GenBank (Clark et al., 2016) accession numbers are presented in parentheses. 16S rRNA genes of Synechococcus elongatus PCC 11801 and Synechococcus elongatus PCC 11802 were not available in publicly available genomes.
Supplementary Table 1 | Manual curation of known biosynthetic gene clusters found in cyanobacterial plasmids. See “Materials and Methods” section for further information.
Supplementary Table 2 | Annotation of 4-phosphopantetheinyl transferases in the analyzed cyanobacterial genomes. See the “Materials and Methods” section for further information.
Supplementary Table 3 | Manual curation of genomic regions with relaxase, virB4, and virD4 genes. Previous studies characterized the mobility of plasmids (Smillie et al., 2010), MOBv (Garcillán-Barcia et al., 2009), and MPFc (Guglielmini et al., 2011).
Supplementary Table 4 | Predicted mobility of cyanobacterial plasmids. See the “Materials and Methods” section for further information.
Supplementary Table 5 | List of public genomes deposited in the NCBI GenBank (Clark et al., 2016) used in the present work and the results of automatic annotation performed with antiSMASH v5.1 (Blin et al., 2019). See the “Materials and Methods” section and Supplementary Table 1 for information on the manually curated genes.
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarenga, D. O., Fiore, M. F., and Varani, A. M. (2017). A metagenomic approach to cyanobacterial genomics. Front. Microbiol. 8:809. doi: 10.3389/fmicb.2017.00809
Angermayr, S. A., Hellingwerf, K. J., Lindblad, P., and Teixeira de Mattos, M. J. (2009). Energy biotechnology with cyanobacteria. Curr. Opin. Biotechnol. 20, 257–263. doi: 10.1016/j.copbio.2009.05.011
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/C2NP20085F
Bachmann, B. O., Van Lanen, S. G., and Baltz, R. H. (2014). Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 41, 175–184. doi: 10.1007/s10295-013-1389-9
Beld, J., Sonnenschein, E. C., Vickery, C. R., Noel, J. P., and Burkart, M. D. (2014). The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 31, 61–108. doi: 10.1039/C3NP70054B
Blank, C. E., and Sánchez-Baracaldo, P. (2010). Timing of morphological and ecological innovations in the cyanobacteria – a key to understanding the rise in atmospheric oxygen: timing of evolutionary innovations in the cyanobacteria. Geobiology 8, 1–23. doi: 10.1111/j.1472-4669.2009.00220.x
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). Antismash 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., et al. (2017). Antismash 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41. doi: 10.1093/nar/gkx319
Bolch, C. J. S., Blackburn, S. I., Jones, G. J., Orr, P. T., and Grewe, P. M. (1997). Plasmid content and distribution in the toxic cyanobacterial genus microcystis kützing ex lemmermann (Cyanobacteria: Chroococcales). Phycologia 36, 6–11. doi: 10.2216/i0031-8884-36-1-6.1
Boto, L. (2010). Horizontal gene transfer in evolution: facts and challenges. Proc. R. Soc. B Biol. Sci. 277, 819–827. doi: 10.1098/rspb.2009.1679
Burja, A. M., Banaigs, B., Abou-Mansour, E., Grant Burgess, J., and Wright, P. C. (2001). Marine cyanobacteria—a prolific source of natural products. Tetrahedron 57, 9347–9377. doi: 10.1016/S0040-4020(01)00931-0
Calteau, A., Fewer, D. P., Latifi, A., Coursin, T., Laurent, T., Jokela, J., et al. (2014). Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genomics 15:977. doi: 10.1186/1471-2164-15-977
Carter, A. T., Austin, J. W., Weedmark, K. A., Corbett, C., and Peck, M. W. (2014). Three classes of plasmid (47–63 kb) carry the type B neurotoxin gene cluster of group II Clostridium botulinum. Genome Biol. Evol. 6, 2076–2087. doi: 10.1093/gbe/evu164
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Chen, A. H., Afonso, B., Silver, P. A., and Savage, D. F. (2012). Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One 7:e47837. doi: 10.1371/journal.pone.0047837
Christie, P. J. (2004). Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694, 219–234. doi: 10.1016/j.bbamcr.2004.02.013
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2016). GenBank. Nucleic Acids Res. 44, D67–D72. doi: 10.1093/nar/gkv1276
Copp, J. N., and Neilan, B. A. (2006). The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl. Environ. Microbiol. 72, 2298–2305. doi: 10.1128/AEM.72.4.2298-2305.2006
Corre, C., and Challis, G. L. (2009). New natural product biosynthetic chemistry discovered by genome mining. Nat. Prod. Rep. 26:977. doi: 10.1039/b713024b
Cubillos-Ruiz, A., Berta-Thompson, J. W., Becker, J. W., van der Donk, W. A., and Chisholm, S. W. (2017). Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, E5424–E5433. doi: 10.1073/pnas.1700990114
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Daum, M., Peintner, I., Linnenbrink, A., Frerich, A., Weber, M., Paululat, T., et al. (2009). Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. Chembiochem 10, 1073–1083. doi: 10.1002/cbic.200800823
De La Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
De Lay, N. R., and Cronan, J. E. (2008). Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178, 1327–1337. doi: 10.1534/genetics.107.081836
Demain, A. L. (2014). Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 41, 185–201. doi: 10.1007/s10295-013-1325-z
Di Rienzi, S. C., Sharon, I., Wrighton, K. C., Koren, O., Hug, L. A., Thomas, B. C., et al. (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2:e01102. doi: 10.7554/eLife.01102
diCenzo, G. C., and Finan, T. M. (2017). The divided bacterial genome: structure, function, and evolution. Microbiol. Mol. Biol. Rev. 81:e00019. doi: 10.1128/MMBR.00019-17
diCenzo, G. C., Mengoni, A., and Perrin, E. (2019). Chromids aid genome expansion and functional diversification in the family Burkholderiaceae. Mol. Biol. Evol. 36, 562–574. doi: 10.1093/molbev/msy248
Dittmann, E., Fewer, D. P., and Neilan, B. A. (2013). Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 37, 23–43. doi: 10.1111/j.1574-6976.2012.12000.x
Dittmann, E., Gugger, M., Sivonen, K., and Fewer, D. P. (2015). Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 23, 642–652. doi: 10.1016/j.tim.2015.07.008
Ehling-Schulz, M., Fricker, M., Grallert, H., Rieck, P., Wagner, M., and Scherer, S. (2006). Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20. doi: 10.1186/1471-2180-6-20
Ehrenreich, I. M., Waterbury, J. B., and Webb, E. A. (2005). Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 71, 7401–7413. doi: 10.1128/AEM.71.11.7401-7413.2005
Garcillán-Barcia, M. P., Francia, M. V., and de La Cruz, F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. doi: 10.1111/j.1574-6976.2009.00168.x
Ginolhac, A., Jarrin, C., Robe, P., Perrière, G., Vogel, T. M., Simonet, P., et al. (2005). Type I polyketide synthases may have evolved through horizontal gene transfer. J. Mol. Evol. 60, 716–725. doi: 10.1007/s00239-004-0161-1
Griese, M., Lange, C., and Soppa, J. (2011). Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323, 124–131. doi: 10.1111/j.1574-6968.2011.02368.x
Guglielmini, J., Quintais, L., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. C. (2011). The repertoire of ice in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. doi: 10.1371/journal.pgen.1002222
Harrison, E., and Brockhurst, M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. doi: 10.1016/j.tim.2012.04.003
Harrison, P. W., Lower, R. P. J., Kim, N. K. D., and Young, J. P. W. (2010). Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 18, 141–148. doi: 10.1016/j.tim.2009.12.010
Hillwig, M. L., Zhu, Q., and Liu, X. (2014). Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem. Biol. 9, 372–377. doi: 10.1021/cb400681n
Hopwood, D. A. (1997). Genetic contributions to understanding polyketide synthases. Chem. Rev. 97, 2465–2498. doi: 10.1021/cr960034i
Hu, B., Yang, G., Zhao, W., Zhang, Y., and Zhao, J. (2007). MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120: chromosome partitioning in a cyanobacterium. Mol. Microbiol. 63, 1640–1652. doi: 10.1111/j.1365-2958.2007.05618.x
Jain, S., Prajapat, G., Abrar, M., Ledwani, L., Singh, A., and Agrawal, A. (2017). Cyanobacteria as efficient producers of mycosporine-like amino acids. J. Basic Microbiol. 57, 715–727. doi: 10.1002/jobm.201700044
Jensen, P. R. (2016). Natural products and the gene cluster revolution. Trends Microbiol. 24, 968–977. doi: 10.1016/j.tim.2016.07.006
Jüttner, J. (1995). Physiology and biochemistry of odorous compounds from freshwater cyanobacteria and algae. Water Sci. Technol. 31, 69–78. doi: 10.1016/0273-1223(95)00458-Y
Kaebernick, M., Neilan, B. A., Börner, T., and Dittmann, E. (2000). Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 66, 3387–3392. doi: 10.1128/AEM.66.8.3387-3392.2000
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136.
Kehr, J.-C., Gatte Picchi, D., and Dittmann, E. (2011). Natural product biosyntheses in cyanobacteria: a treasure trove of unique enzymes. Beilstein J. Org. Chem. 7, 1622–1635. doi: 10.3762/bjoc.7.191
Kim, J. H., Komatsu, M., Shin-ya, K., Omura, S., and Ikeda, H. (2018). Distribution and functional analysis of the phosphopantetheinyl transferase superfamily in Actinomycetales microorganisms. Proc. Natl. Acad. Sci. U.S.A. 115, 6828–6833. doi: 10.1073/pnas.1800715115
Komarek, J., Johansen, J. R., Smarda, J., and Strunecky, O. (2020). Phylogeny and taxonomy of Synechococcus-like cyanobacteria. Fottea 20, 171–191. doi: 10.5507/fot.2020.006
Komarek, J., Kastovsky, J., Mares, J., and Johansen, J. (2014). Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86, 295–335.
Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., et al. (1996). A new enzyme superfamily — the phosphopantetheinyl transferases. Chem. Biol. 3, 923–936. doi: 10.1016/S1074-5521(96)90181-7
Land, M., Hauser, L., Jun, S.-R., Nookaew, I., Leuze, M. R., Ahn, T.-H., et al. (2015). Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. doi: 10.1007/s10142-015-0433-4
Lawrence, J. G., and Roth, J. R. (1996). Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143, 1843–1860.
Leao, T., Castelão, G., Korobeynikov, A., Monroe, E. A., Podell, S., Glukhov, E., et al. (2017). Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proc. Natl. Acad. Sci. U.S.A. 114, 3198–3203. doi: 10.1073/pnas.1618556114
Liu, Q., Ma, Y., Zhou, L., and Zhang, Y. (2005). Gene cloning, expression and functional characterization of a phosphopantetheinyl transferase from Vibrio anguillarum serotype O1. Arch. Microbiol. 183, 37–44. doi: 10.1007/s00203-004-0745-6
Marahiel, M. A., Stachelhaus, T., and Mootz, H. D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674.
Martins, J., and Vasconcelos, V. (2015). Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge. Mar. Drugs 13, 6910–6946. doi: 10.3390/md13116910
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). Antismash: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. doi: 10.1093/nar/gkr466
Micallef, M. L., D’Agostino, P. M., Sharma, D., Viswanathan, R., and Moffitt, M. C. (2015). Genome mining for natural product biosynthetic gene clusters in the Subsection V cyanobacteria. BMC Genomics 16:669. doi: 10.1186/s12864-015-1855-z
Muro-Pastor, A. M., Kuritz, T., Flores, E., Herrero, A., and Wolk, C. P. (1994). Transfer of a genetic marker from a megaplasmid of Anabaena sp. strain PCC 7120 to a megaplasmid of a different Anabaena strain. J. Bacteriol. 176, 1093–1098. doi: 10.1128/JB.176.4.1093-1098.1994
Nett, M., Ikeda, H., and Moore, B. S. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26:1362. doi: 10.1039/b817069j
O’Brien, F. G., Yui Eto, K., Murphy, R. J. T., Fairhurst, H. M., Coombs, G. W., Grubb, W. B., et al. (2015). Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res. 43, 7971–7983. doi: 10.1093/nar/gkv755
Oh, H.-S., Lee, C. S., Srivastava, A., Oh, H.-M., and Ahn, C.-Y. (2017). Effects of environmental factors on cyanobacterial production of odorous compounds: geosmin and 2-methylisoborneol. J. Microbiol. Biotechnol. 27, 1316–1323. doi: 10.4014/jmb.1702.02069
Osbourn, A. (2010). Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 26, 449–457. doi: 10.1016/j.tig.2010.07.001
Panda, D., DeLuca, K., Williams, D., Jordan, M. A., and Wilson, L. (1998). Antiproliferative mechanism of action of cryptophycin-52: kinetic stabilization of microtubule dynamics by high-affinity binding to microtubule ends. Proc. Natl. Acad. Sci. U.S.A. 95, 9313–9318. doi: 10.1073/pnas.95.16.9313
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Pattanaik, B., and Lindberg, P. (2015). Terpenoids and their biosynthesis in cyanobacteria. Life 5, 269–293. doi: 10.3390/life5010269
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Quadri, L. E. N., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595. doi: 10.1021/bi9719861
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sargent, E. C., Hitchcock, A., Johansson, S. A., Langlois, R., Moore, C. M., LaRoche, J., et al. (2016). Evidence for polyploidy in the globally important diazotroph Trichodesmium. FEMS Microbiol. Lett. 363:fnw244. doi: 10.1093/femsle/fnw244
Schneider, D., Fuhrmann, E., Scholz, I., Hess, W. R., and Graumann, P. L. (2007). Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8:39. doi: 10.1186/1471-2121-8-39
Sciuto, K., and Moro, I. (2015). Cyanobacteria: the bright and dark sides of a charming group. Biodivers. Conserv. 24, 711–738. doi: 10.1007/s10531-015-0898-4
Shih, P. M., Wu, D., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058. doi: 10.1073/pnas.1217107110
Singh, S. P., Klisch, M., Sinha, R. P., and Häder, D.-P. (2010). Genome mining of mycosporine-like amino acid (MAA) synthesizing and non-synthesizing cyanobacteria: a bioinformatics study. Genomics 95, 120–128. doi: 10.1016/j.ygeno.2009.10.002
Sivonen, K., Leikoski, N., Fewer, D. P., and Jokela, J. (2010). Cyanobactins—ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 86, 1213–1225. doi: 10.1007/s00253-010-2482-x
Skarin, H., and Segerman, B. (2011). Horizontal gene transfer of toxin genes in Clostridium botulinum: involvement of mobile elements and plasmids. Mob. Genet. Elements 1, 213–215. doi: 10.4161/mge.1.3.17617
Skarstad, K., Steen, H. B., and Boye, E. (1983). Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J. Bacteriol. 154, 656–662.
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P. C., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stone, M. J., and Williams, D. H. (1992). On the evolution of functional secondary metabolites (natural products). Mol. Microbiol. 6, 29–34. doi: 10.1111/j.1365-2958.1992.tb00834.x
Suurnäkki, S., Gomez-Saez, G. V., Rantala-Ylinen, A., Jokela, J., Fewer, D. P., and Sivonen, K. (2015). Identification of geosmin and 2-methylisoborneol in cyanobacteria and molecular detection methods for the producers of these compounds. Water Res. 68, 56–66. doi: 10.1016/j.watres.2014.09.037
Tan, L. T. (2007). Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 68, 954–979. doi: 10.1016/j.phytochem.2007.01.012
Tietz, J. I., Schwalen, C. J., Patel, P. S., Maxson, T., Blair, P. M., Tai, H.-C., et al. (2017). A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478. doi: 10.1038/nchembio.2319
Tippelt, A., Busche, T., Rückert, C., and Nett, M. (2020). Complete genome sequence of the cryptophycin-producing cyanobacterium Nostoc sp. Strain ATCC 53789. Microbiol. Resour. Announc. 9:e00040. doi: 10.1128/MRA.00040-20
Tooming-Klunderud, A., Fewer, D. P., Rohrlack, T., Jokela, J., Rouhiainen, L., Sivonen, K., et al. (2008). Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera. BMC Evol. Biol. 8:256. doi: 10.1186/1471-2148-8-256
Vestola, J., Shishido, T. K., Jokela, J., Fewer, D. P., Aitio, O., Permi, P., et al. (2014). Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E1909–E1917. doi: 10.1073/pnas.1320913111
Wang, H., Fewer, D. P., and Sivonen, K. (2011). Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PLoS One 6:e22384. doi: 10.1371/journal.pone.0022384
Wang, H., Sivonen, K., Rouhiainen, L., Fewer, D. P., Lyra, C., Rantala-Ylinen, A., et al. (2012). Genome-derived insights into the biology of the hepatotoxic bloom-forming cyanobacterium Anabaena sp. strain 90. BMC Genomics 13:613. doi: 10.1186/1471-2164-13-613
Watanabe, S. (2020). Cyanobacterial multi-copy chromosomes and their replication. Biosci. Biotechnol. Biochem. 84, 1309–1321. doi: 10.1080/09168451.2020.1736983
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Welsh, E. A., Liberton, M., Stockel, J., Loh, T., Elvitigala, T., Wang, C., et al. (2008). The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. U.S.A. 105, 15094–15099. doi: 10.1073/pnas.0805418105
Woodruff, H. (1980). Natural products from microorganisms. Science 208, 1225–1229. doi: 10.1126/science.7375932
Yamada, Y., Kuzuyama, T., Komatsu, M., Shin-ya, K., Omura, S., Cane, D. E., et al. (2015). Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 857–862. doi: 10.1073/pnas.1422108112
Yang, G., Zhang, Y., Lee, N. K., Cozad, M. A., Kearney, S. E., Luesch, H., et al. (2017). Cyanobacterial Sfp-type phosphopantetheinyl transferases functionalize carrier proteins of diverse biosynthetic pathways. Sci. Rep. 7:11888. doi: 10.1038/s41598-017-12244-3
Zechner, E. L., Lang, S., and Schildbach, J. F. (2012). Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 367, 1073–1087. doi: 10.1098/rstb.2011.0207
Zerikly, M., and Challis, G. L. (2009). Strategies for the discovery of new natural products by genome mining. Chembiochem 10, 625–633.
Keywords: cyanobacteria, conjugation, genome mining, toxins, geosmin
Citation: Popin RV, Alvarenga DO, Castelo-Branco R, Fewer DP and Sivonen K (2021) Mining of Cyanobacterial Genomes Indicates Natural Product Biosynthetic Gene Clusters Located in Conjugative Plasmids. Front. Microbiol. 12:684565. doi: 10.3389/fmicb.2021.684565
Received: 23 March 2021; Accepted: 11 October 2021;
Published: 04 November 2021.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Jan Zrimec, National Institute of Biology (NIB), SloveniaCopyright © 2021 Popin, Alvarenga, Castelo-Branco, Fewer and Sivonen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaarina Sivonen, a2FhcmluYS5zaXZvbmVuQGhlbHNpbmtpLmZp
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.