- 1Department of Veterinary Medicine, Institute of Preventive Veterinary Sciences, Zhejiang University College of Animal Sciences, Hangzhou, China
- 2National Institutes for Food and Drug Control, Beijing, China
- 3Hygiene and Zoonoses Department, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 4Bacterial Epidemiology and Antimicrobial Resistance Research Unit, United States National Poultry Research Center, United States Department of Agriculture, Agricultural Research Service (USDA-ARS), Athens, GA, United States
- 5College of Veterinary Medicine, Henan University of Animal Husbandry and Economy, Zhengzhou, China
- 6Hainan Institute of Zhejiang University, Sanya, China
- 7Zhejiang Provincial Key Laboratory of Preventive Veterinary Medicine, Hangzhou, China
- 8State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Salmonella spp. is recognized as an important zoonotic pathogen. The emergence of antimicrobial resistance in Salmonella enterica poses a great public health concern worldwide. While the knowledge on the incidence and the characterization of different S. enterica serovars causing chick embryo death remains obscure in China. In this study, we obtained 45 S. enterica isolates from 2,139 dead chick embryo samples collected from 28 breeding chicken hatcheries in Henan province. The antimicrobial susceptibility assay was performed by the broth microdilution method and the results showed that 31/45 (68.8%) isolates were multidrug-resistant (≥3 antimicrobial classes). Besides the highest resistance rate was observed in the aminoglycoside class, all the isolates were susceptible to chloramphenicol, azithromycin, and imipenem. Furthermore, genomic characterization revealed that S. Enteritidis (33.33%; 15/45) was a frequent serovar that harbored a higher number of virulence factors compared to other serovars. Importantly, genes encoding β-lactamases were identified in three serovars (Thompson, Enteritidis, and Kottbus), whereas plasmid-mediated quinolone resistance genes (qnrB4) were detected in certain isolates of S. Thompson and the two S. Kottbus isolates. All the examined isolates harbored the typical virulence factors from Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2). Additionally, a correlation analysis between the antimicrobial resistance genes, phenotype, and plasmids was conducted among Salmonella isolates. It showed strong positive correlations (r < 0.6) between the different antimicrobial-resistant genes belonging to certain antimicrobial classes. Besides, IncF plasmid showed a strong negative correlation (r > −0.6) with IncHI2 and IncHI2A plasmids. Together, our study demonstrated antimicrobial-resistant S. enterica circulating in breeding chicken hatcheries in Henan province, highlighting the advanced approach, by using genomic characterization and statistical analysis, in conducting the routine monitoring of the emerging antimicrobial-resistant pathogens. Our findings also proposed that the day-old breeder chicks trading could be one of the potential pathways for the dissemination of multidrug-resistant S. enterica serovars.
Introduction
Salmonella is a Gram-negative bacterium and a member of the Enterobacteriaceae family (Boyle et al., 2007). S. enterica is an imperative foodborne pathogens and is capable of causing enteric gastroenteritis (CDC, 2013; Liu et al., 2020). S. enterica subsp. enterica includes more than 2,600 serovars and can infect both humans and animals (Biswas et al., 2019; Paudyal et al., 2019). S. enterica is a widespread pathogen in poultry farms that can be disseminated horizontally and vertically, causing massive economic losses to the poultry industries (Bengtsson and Greko, 2014; Jajere, 2019; Xu et al., 2020b).
S. enterica serovars have been previously reported in many countries as a direct reason for a high number of poultry outbreaks (European Food Safety Authority and European Centre for Disease Prevention and Control, 2017; Hazards EPanelo et al., 2019; Wibisono et al., 2020). Poultry and poultry products are one of the potential pools of bacterial pathogens carrying multidrug resistance. The frequent utilization of antimicrobials in agriculture for growth promotion and bacterial infection treatment can lead to the evolution of the susceptible to antimicrobial-resistant pathogens (Crump et al., 2015; Jajere, 2019). In several reports, poultry products have been proposed as a common reservoir to multidrug-resistant (MDR) S. enterica serovars (Vo et al., 2006; Boyle et al., 2007; Jiang et al., 2019).
Although China is one of the key producers and consumers of poultry products across the world (USDA, 2020), still there is a knowledge gap about the frequency of S. enterica serovars causing chick embryo death in China (Yue et al., 2020). In this study, we investigated the prevalence of different S. enterica serovars isolated from dead chick embryos in Henan province, China. Additionally, we performed antimicrobial susceptibility testing to investigate the antimicrobial resistance phenotypes of the examined S. enterica serovars. Furthermore, genomic characterization, phylogenomic tree, and correlation analyses were conducted in order to scrutinize the potential relationship between the bacterial resistance phenotype, antimicrobial-resistant (AR) determinants, and accompanying plasmids. Here, as a part of the surveillance program, we demonstrated that routine genomic sequencing combined with an advanced analytic approach could accelerate the recognition of novel threats with significant public health concerns.
Materials and Methods
Sample Collection
The methods of sampling and isolation were described in our previous published studies (Wang et al., 2019; Xu et al., 2020b; Jiang et al., 2021). Between August 2014 and April 2015, a total of 2,139 dead chick embryo samples were collected from 28 randomly selected breeding chicken hatcheries from nine cities in Henan province: Zhengzhou, Xuchang, Pingdingshan, Hebi, Anyang, Zhoukou, Shangqiu, Xinyang, and Luohe (Figure 1A). The surface of the embryo was placed on the clean bench's sterile tray after disinfection with ethanol. Sterile forceps and scissors were used to find and extract the yolk sac of the chicken embryo samples. The yolk sac solution was enriched in 100 mL of buffered peptone water (BPW) and incubated overnight at 37°C. The solution was placed on Salmonella-Shigella agar and streaked with disposable sterile inoculating loops, and the plates were then incubated at 37°C for 24 h (Xu et al., 2020b). Salmonella was considered presumptive of the translucent colorless or black center colonies (Xu et al., 2020b; Jiang et al., 2021).
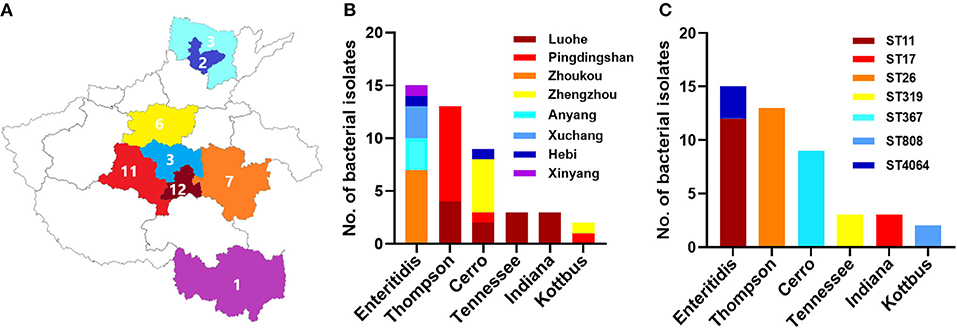
Figure 1. The distribution of different serovars among eight cities in Henan province, China. (A) The geographical distribution of the Salmonella isolates in Henan province with eight cities which were examined in current investigations. N.B., The number indicates the numbers of isolates collected from individual city. (B) The distribution of six serovars of Salmonella isolates. The dominant serovars are S. Enteritidis, S. Thompson, and S. Cerro. (C) The prevalence of individual serovar with their sequence type (ST) detected in this study.
Salmonella Isolation and Identification
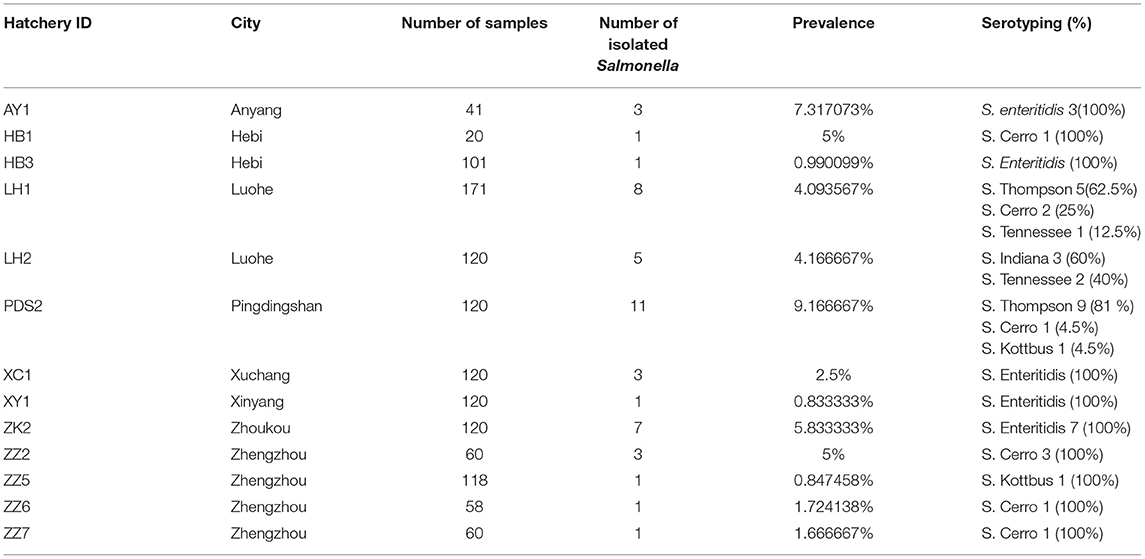
In total, 45 Salmonella isolates were obtained among the samples. These isolates were conducted for further antimicrobial susceptibility testing and genomic characterization analysis. The distribution of the 45 selected isolates in different cities was as follows: 12 isolates from Luohe, 11 isolates from Pingdingshan, 7 isolates from Zhoukou, 6 isolates from Zhengzhou, 3 isolates from Anyang, three isolates from Xuchang, 2 isolates from Hebi, and 1 isolate from Xinyang.
Salmonella Serotyping
Salmonella isolates serotyping was conducted by use of a classic slide agglutination assay with anti-O and anti-H serum (Tianrun Bio-Pharmaceutical, Ningbo, China). For those isolates with inconsistent results when comparing with the in silico prediction, we further evaluated these isolates by using serum purchased from Denmark (SSI Diagnostica, Denmark). The results were analyzed and interpreted according to the Kauffmann-White scheme, as conducted by previous publications (Pan et al., 2019).
Antimicrobial Susceptibility Testing
The isolates were subjected to an antimicrobial susceptibility testing assay using the broth microdilution method, as described previously (Elbediwi et al., 2020b; Xu et al., 2020b; Yu et al., 2020). In total, 14 antimicrobials belonging to 10 classes were used for this assay. The cut-off values recommended by CLSI 2019 guidelines (Clinical Laboratory Standards Institute, 2019) were used for the categorization of results, and the intermediate strain, if available, was merged with the resistant strains for ease of analysis. The concentration range (μg/ml) of antimicrobials used in this assay were as follows: ampicillin (AMP): 0.5–64; amoxicillin/clavulanate potassium (AMC): 0.5/0.25–64/32; gentamicin (GEN): 0.25–32, kanamycin (KAN): 0.5–64, streptomycin (STR): 0.5–64; tetracycline (TET): 0.5–64; ciprofloxacin (CIP): 0.015–8; nalidixic (NAL): 0.5–64; chloramphenicol (CHL): 0.5–64; ceftiofur (CF): 0.015–8; cefoxitin (CX): 0.5–64; trimethoprim/sulfamethoxazole (TST): 0.06/1.19–32/608; azithromycin (AZI): 0.5–64; and imipenem (IMP): 0.015–8. We defined the isolates which could resist three antimicrobial classes or more as multidrug-resistant (MDR). The Two quality control strains, including E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853, were used to validate the antimicrobial susceptibility testing.
DNA Extraction, Genome Sequencing, and Bioinformatics Analysis
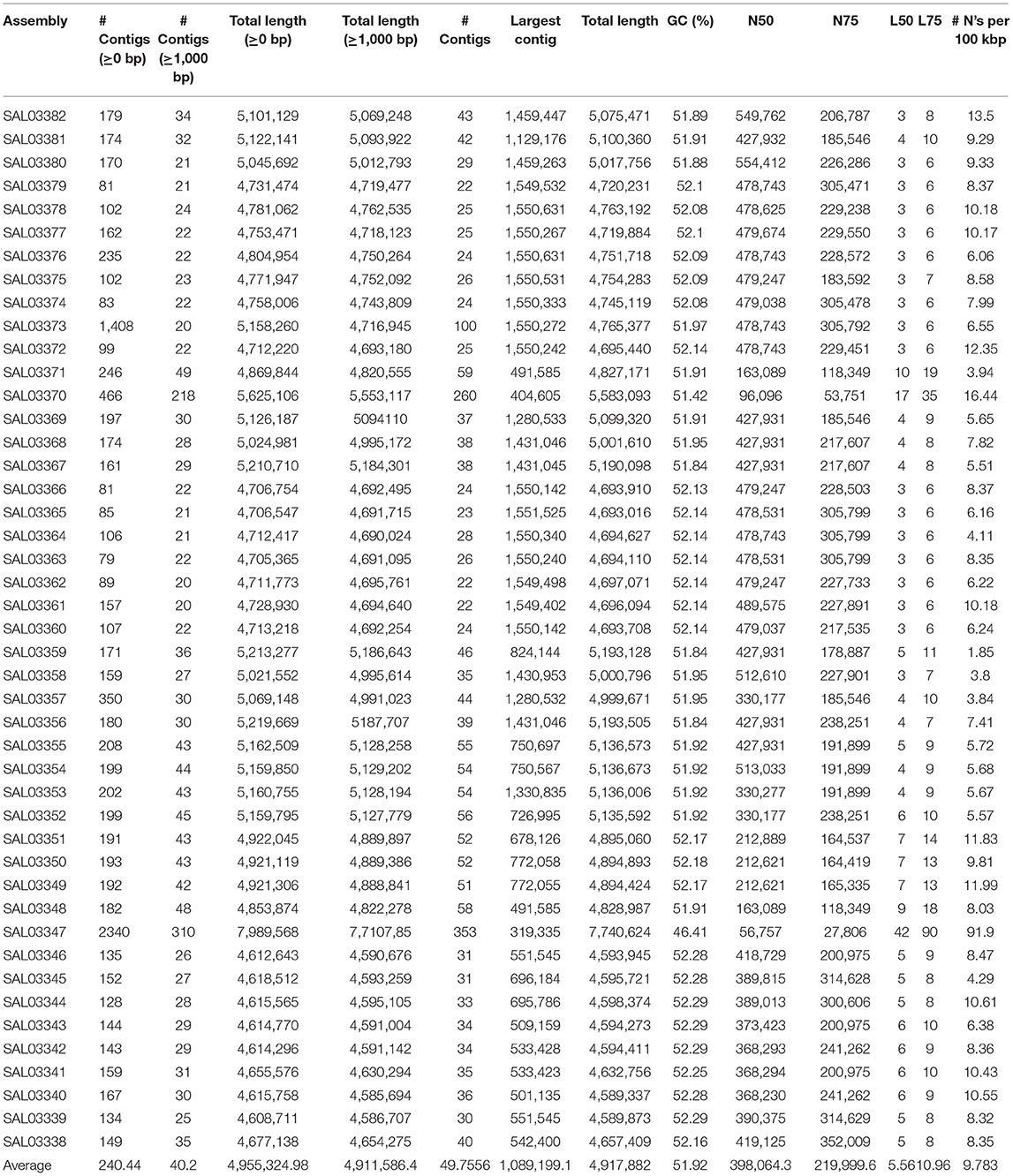
The genomic DNA of the isolates was extracted and purified using QIAamp DNA mini-Kit (German Qiagen Company, Art. No. 51304), according to the manufacturer's recommended protocols. The Genomic DNA library was constructed using Nextera XT DNA library construction kit (American Illumina Company, model: FC-131-1024); then the genomic sequencing was conducted using Miseq Reagent Kit v2 300cycle kit (American Illumina Company, model: MS-102-2002). High-throughput genome sequencing was accomplished by an Illumina Miseq sequencing platform. After the sequencing results were returned, all subsequent biological information analysis was performed on the in-house Galaxy platform as described previously (Liu et al., 2021). The quality of sequencing and trimming was checked with a fast QC toolkit, while low-quality sequences and joint sequences were removed with trimmomatic (Bolger et al., 2014). The raw sequence data were under quality check and assembled by using SPAdes 4.0.1 (Bankevich et al., 2012) using the “careful correction” option to reduce the number of mismatches in the final assembly with automatically chosen k-mer values by SPAdes. The QUAST (Gurevich et al., 2013) tool was used to evaluate the assembled genomes through basic statistics generation, including the total number of contigs, contig length, and N50. After the completion of data assembly, serovar prediction was performed with SISTR v1.0.2 (Yoshida et al., 2016) and Seqsero v1.0.0 (www.denglab.info/SeqSero) by using default parameters, and MLST (https://github.com/tseemann/mlst) tools in the local Galaxy platform were used to analyze the bacterial genotype.
Furthermore, the virulence genes, antimicrobial resistance genes, and plasmid types were detected using the abricate tool (https://github.com/tseemann/abricate). This phylogenomic tree was conducted through whole-genome multilocus sequence typing (wgMLST) using the cano-wgMLST_BacCompare software that was reported recently (Liu et al., 2019) using default parameters.
Statistical Analyses
The Pearson's correlation (r) between antimicrobial resistance genes, phenotypes, and plasmids among Salmonella isolates was measured. Resistance phenotype against the individual antimicrobial drug, antimicrobial resistance gene, and plasmid presence received scores of 1, whereas susceptibility to antimicrobials and the absence of (AR) genes or the plasmids received scores of 0. Binary data (0/1) were imported into R software (version 3.6.1; https://www.r-project.org), and correlation was determined using the “cor” function and visualized using the “corrplot” function. The significance of correlation (P < 0.05) was also determined using “cor.mtest” function. The correlation was considered strong if the r ≥ 0·6, moderate if the r value was between 0.4 and 0.6, and weak if r < 0.4 (Kirch, 2008).
Results
Prevalence and Geographical Distribution of Salmonella Serovars
A total of 45 (2.1%) Salmonella isolates were obtained from 2,139 chick embryo yolk samples (Supplementary Material). These 45 Salmonella isolates were identified in 13 out of 28 hatcheries (Supplementary Table 1). We noticed that the prevalence of Salmonella isolates in the hatcheries ranged between (0.8 and 9.1%), and the most contaminated hatchery was PDS2 in Pingdingshan city. The serotyping was conducted by using SISTR v1.0.2, Seqsero v1.0.0 tools, and slide agglutination assay with anti-O and anti-H serum. According to the two different in silico analyses, delivering identical results, we found that these 45 isolates belonged to six serovars. Additionally, the serum agglutination assay has confirmed this result. The isolates were widely distributed among Henan province cities, and the city with the highest frequent distribution was Luohe with 12 isolates followed by Pingdingshan with 11 isolates (Figure 1A). Moreover, the highest frequent serovar was S. Enteritidis (33.33%; 15/45) that was predominately identified in 5 out of 13 hatcheries, followed by S. Thompson (28.89%; 13/45), S. Cerro (20.00%; 9/45), S. Tennessee (6.67%; 3/45), S. Indiana (6.67%; 3/45), and S. Kottbus (4.44%; 2/45) (Figure 1B, Table 1).
Antimicrobial Phenotype and Related Resistant Determents
Antimicrobial resistance profiles of the examined Salmonella isolates were determined by the broth microdilution method; accordingly, the antimicrobial resistance genes were predicted by using whole-genome sequencing data. The results obtained were classified as susceptible, intermediate resistance, and resistant. Additionally, in order to facilitate results interpretation, intermediate isolates have been considered resistant. These results showed that the higher resistance was observed in the aminoglycoside class, including STR (n = 36, 80%), KAN (n = 35, 77.7%); GEN (n = 30, 66.6%), while all isolates showed susceptibility for IMP, CHL and AZI (Figure 2). The antimicrobial susceptibility assay analysis showed that 68.9% (31/45) of the studied isolates were MDR. The results also revealed that S. Thompson obtained from the hatchery LH1 (Luohe city) and S. Kottbus obtained from the hatchery PDS2 (Pingdingshan city) had the highest resistance rate among all serovars, showing a resistance profile toward 11 antimicrobial drugs (STR, KAN, GEN, AMC, AMP, TST, CF, CX, TET, CIP, and NAL) (Figure 3). S. Cerro isolates were observed to be resistant against the aminoglycoside class (Figure 3).
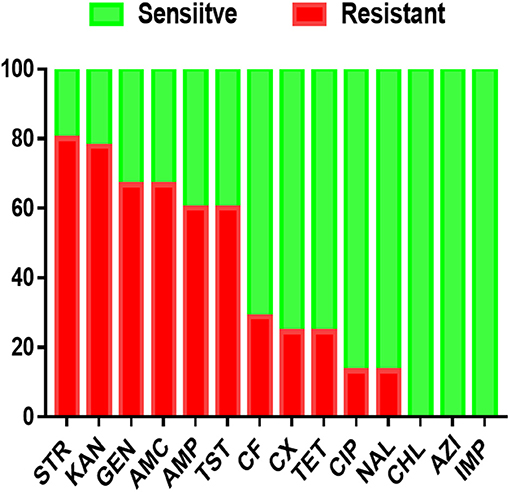
Figure 2. The antimicrobial resistance profile of S. enterica isolates was identified in this study. N.B., X-axis indicates antimicrobials and Y-axis indicates Salmonella isolates number.
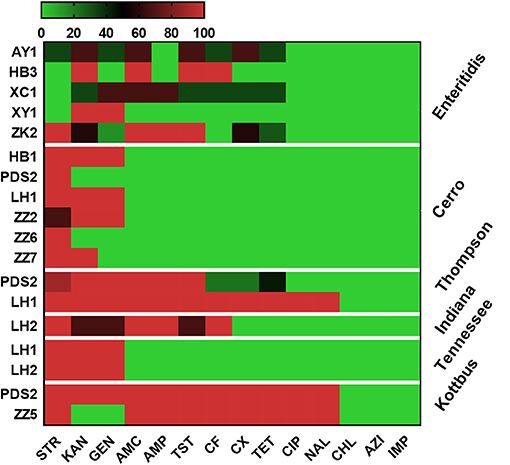
Figure 3. Heat map showing the antimicrobial resistance profile of different serovars isolated from different hatcheries. N.B., X-axis indicates antimicrobials and Y-axis indicates Salmonella serovars and Hatcheries. The antimicrobials used in this assay were as follows: ampicillin (AMP), amoxicillin and clavulanate potassium (AMC), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), tetracycline (TET), ciprofloxacin (CIP), nalidixic (NAL), chloramphenicol (CHL), ceftiofur (CF), cefoxitin (CX), trimethoprim and sulfamethoxazole (TST), azithromycin (AZI), and imipenem (IMP). The abbreviations on the left side of the Y axis refer to the hatcheries located in Henan city. AY1 in Anyang city, HB1 and HB3 in Hebi city, LH1 and LH2 in Luohe city, PDS2 in Pingdingshan city, XC1 in Xuchang city, XY1 in Xinyang city, ZK2 in Zhoukou, ZZ2, ZZ5, ZZ6, and ZZ7 in Zhengzhou city. The scale showed the percentage of resistance for the isolates (from 0 to 100%). The strength of the colors corresponds to the numerical value of the prevalence of resistant isolates. Green revealed to susceptible and red revealed to resistant isolates.
Furthermore, the screening for the resistance determinants in the whole genomic sequence (WGS) of these isolates was in concordance with the phenotypic resistance. WGS analysis showed that all the examined isolates harbored the aac(6′)-Iaa gene, conferring resistance to aminoglycoside (Figure 4). Importantly, plasmid-mediated quinolone resistance genes (qnrB4) were detected in certain S. Thompson and all S. Kottbus isolates. Genes encoding β-lactamases were identified in three serovars (S. Thompson, S. Enteritidis, S. Kottbus). The gene sul1, which encodes resistance to sulfonamide, was only found in S. Thompson and S. Kottbus, while Sul2 was only detected in S. Enteritidis serovar. Regarding serovars distribution, our results founded that the prevalence of antimicrobial resistance genes in S. Thompson was the highest compared to the other serovars.
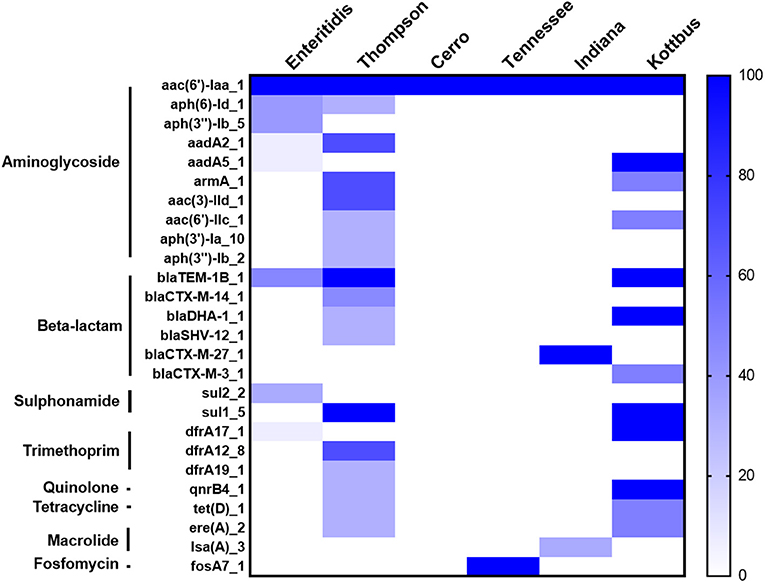
Figure 4. The heatmap of antimicrobial resistance (AR) genes in the recovered S. enterica isolates. All the studied isolates harbored aac(6′)-Iaa gene encoding resistance to aminoglycoside, and S. Cerro isolates only carried it. The scale showed the prevalence of AR genes among the isolates (from 0 to 100%), The strength of the colors corresponds to the numerical value of AR genes prevalence. Dark blue color revealed to high prevalence and white color revealed gene absence.
Whole Genome Sequencing and Bioinformatics Analysis
After conducting the whole genome sequencing and genomic assembly of the Salmonella isolates, the number of contigs was calculated to be between 22 and 100 contigs. Genome sequencing and assembly results parameters are summarized in Table 2. The average genome size of draft assemblies was 4,917,882 bp. Furthermore, the average N50 was 398,064 bp. With further WGS analysis, we found that MLST patterns were assigned based on the allelic profile of each isolate, and the results showed that the Salmonella isolates belonged to seven Sequence Types (STs) (Figure 1C). More than half of the isolates (55.56%) were assigned to ST26 (28.9%) and ST11 (26.7%). Our results also showed that 80% of S. Enteritidis isolates belonged to ST11 and 20% belonged to ST4064. All S. Thompson isolates belonged to ST26, and S. Tennessee, S. Indiana, and S. Kottbus belonged to ST319, ST17, ST808, respectively.
Moreover, WGS also showed that the detection of 16 different plasmid replicons in the recovered Salmonella strains. Among the whole data, there were few plasmids carried by all strains, and the characteristics of plasmids carried by strains with different serovars were obvious. The largest number of plasmid replicons with different types have been detected in S. Cerro (five different plasmids), followed by S. Thompson and S. Enteritidis (four different plasmids). However, the major plasmids found in S. Enteritidis were IncFIB(S)_1 and IncFII(S)_1, whereas, in S. Thompson and S. Kottbus, they were IncHI2 and IncHI2A; S. Cerro, S. Tennessee, and S. Indiana carried the only detected plasmids Col440I_1, IncFIB(pHCM2)1_pHCM2, and p0111_1, respectively (Figure 5).
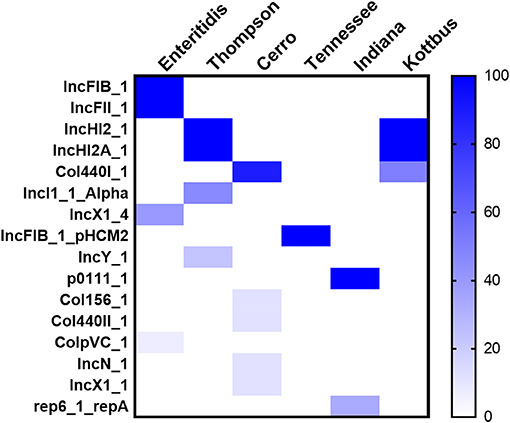
Figure 5. The heatmap of plasmids distribution in different S. enterica isolates. The scale showed the prevalence of the plasmids among the isolates (from 0 to 100%). The strength of the colors corresponds to the numerical value of the prevalence of the plasmids. Dark blue color indicates high prevalence and white color for gene absence.
Prediction of virulence genes was performed on the basis of the virulence factors database (VFDB) by using the abricate web tool, and the results are presented in Supplementary Table 2. In total, 111 virulence genes implicated in different mechanisms of virulence and pathogenicity were detected (Figure 6). Our results showed that S. Enteritidis harbored a higher number of virulence genes compared to other serovars. Fimbrial adherence factor Pef, serum resistance gene rck, stress adaptation gene sodCl, and the plasmid-borne spv locus in addition to the ssel gene, which plays a role in the type three secretion system, were detected only in S. Enteritidis isolates (Figure 6, Supplementary Table 2). We also noticed sspH1 gene has been only detected in S. Cerro isolates. Importantly, we also found that one S. Thompson isolate (SAL03370) and two S. Indiana (SAL03347, SAL03348) isolates harbored the gene cdtB encoding typhoid toxin. Interestingly, pltA and pltB genes, which are essential for producing cytolethal distending toxin (CDT), were not found. All the isolates harbored Salmonella pathogenicity island 1 and 2 (SPI-1 and SPI-2) virulence factors (Figure 6).
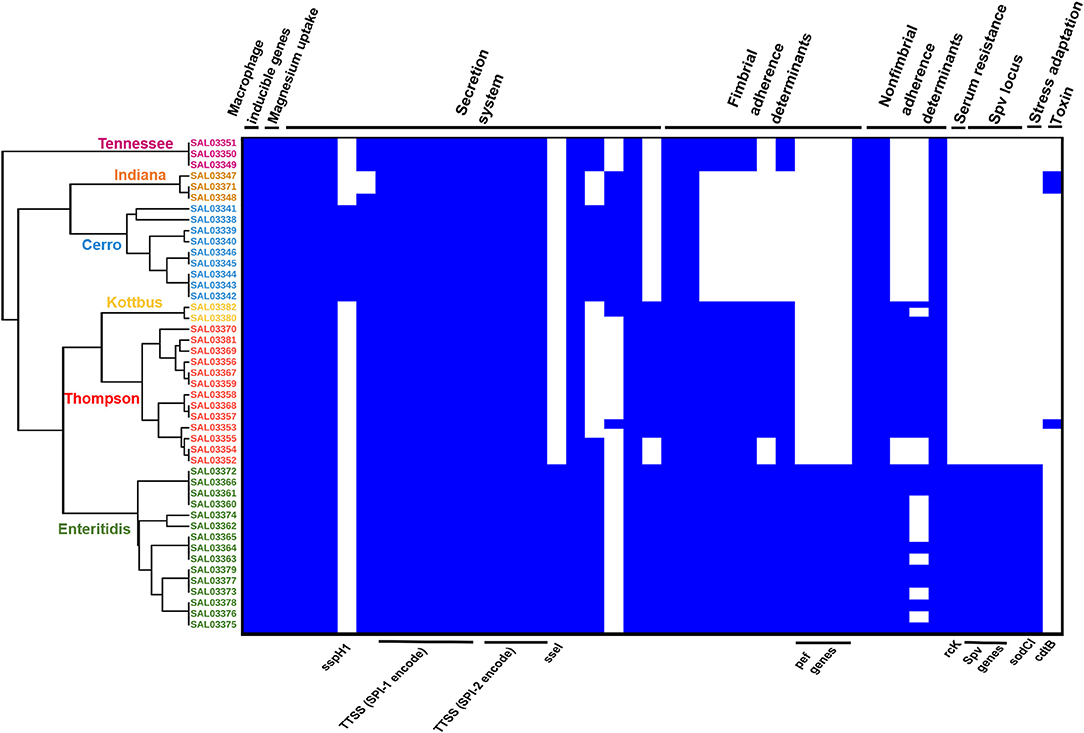
Figure 6. The combinatorial graph of the wgMLST phylogenomic evolutionary tree and virulence genes in the S. enterica isolates. The figure showed that S. enteritidis harbored a higher number of virulence genes compared with other serovars. In the heat map of the virulence genes, blue color refers to “presence,” white color refers to “absence.” The figure showed that pef, rck, sodCl, and spv loci in addition to ssel genes were detected only in the S. enteritidis isolates. Besides, one S. thompson isolate and two S. Indiana isolates harbored the gene cdtB encoding typhoid toxins. All the isolates harbored Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2) virulence factors. N.B., According to the phylogenomic tree, green color refers to S. Enteritidis isolates, orange color refers to S. Cerro isolates, brown color refers to S. Kottbus isolates, gray color refers to S. Indiana isolates, and red color refers to S. Thompson isolates.
To further investigate the genomic relationship among the isolates, we performed a phylogenomic analysis. Importantly, wgMLST analysis (Figure 6) revealed that the isolates belonging to the same serotypes are very close to each other, clustering in the same sub-clade. The phylogenomic tree suggested a strong association between S. Indiana and S. Cerro isolates and between S. Thompson and S. Kottbus isolates, which were grouped in the same sub-clade with a very similar pattern for virulence factor cassettes (Figure 6).
Association of Phenotypic Resistance, Resistance Associated Gene, and Plasmid Among the Salmonella Isolates
Correlation analysis of antimicrobial resistance phenotype, antimicrobial resistance gene, and plasmid among the examined Salmonella isolates was performed. All observed correlation results were statistically significant (p < 0.05) as indicated in Figure 7. The results showed strong positive correlations (r < 0.6) between antimicrobial-resistant genes which belonging to different antimicrobial classes such as those remarked for aminoglycosides resistant genes (apha.3.lb, apha.3.la, aac.6.lic), bla genes (blaSHV−12 and blaDHA−1), tetD, and qnrB (Figure 7A). We also noticed that these genes showed a week correlation with IncHI2/A plasmid types and sul1. Additionally, IncHI2 and IncHI2A plasmids showed a strong positive correlation with sul1, dfrA12, armA, and blaTEM.1B genes. apha.6.ld also showed strong positive correlations with apha.3.lb, sul1, and IncX1 (Figure 7A). Interestingly, the IncF plasmid was suggested to display a strong negative correlation (r > −0.6) with IncHI2 and IncHI2A plasmids (Figure 7A). Figure 7B showed the correlation analysis between the antimicrobial resistance phenotypes and antimicrobial-resistant genes. Interestingly, we found that CIP and NAL showed a strong positive correlation with each other. Besides, both of them showed a strong positive correlation with aac.6.lic, ereA, blaDHA, and qnrB4 genes. AMP was found to be a strong positive correlation with AMC and KAN showed a strong positive correlation with GEN (Figure 7B).
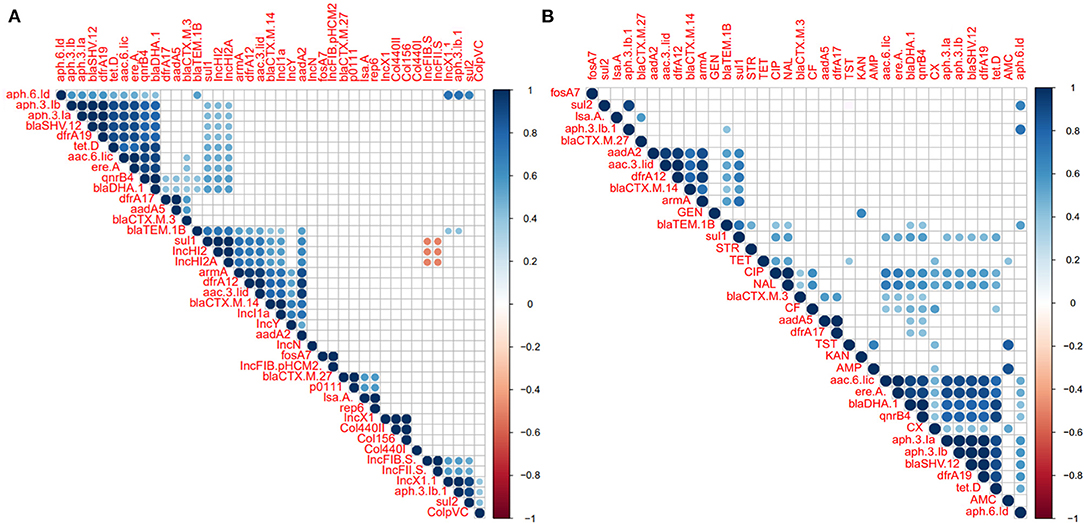
Figure 7. Correlation analysis between antimicrobial resistance (AR) phenotypes, AR determinants, and plasmids among the S. enterica isolates. (A) Correlation analysis between plasmids and AR determinants. (B) Correlation analysis between AR phenotypes and AR determinants. The blue and orange colors of boxes indicate positive and negative correlation, respectively. The strength of the colors corresponds to the numerical value of the correlation coefficient (r). All correlation results were statistically significant (p < 0.05). The results showed positive correlations (r < 0.6) between the different AR genes belonging to different antimicrobial classes such as those apha.3.lb, apha.3.la, aac.6.lic, blaSHV−12, and blaDHA−1, tetD, and qnrB. We also noticed that these genes showed a week correlation with IncHI2/A plasmid types and sul1. Additionally, IncHI2/A plasmid types showed a strong positive correlation with sul1, dfrA12, armA, and blaTEM.1B genes. Interestingly, IncF plasmid showed a strong negative correlation (r > −0.6) with IncHI2/A plasmid.
Discussion
Recently, A meta-analysis study displayed that Salmonella is one of the most widespread pathogenic foodborne bacteria in Chinese food products (Paudyal et al., 2018). Avian salmonellosis is listed as one of the 16 priority animal diseases according to the national medium- and long-term plan for prevention and control of animal diseases in China (2012–2020). Several studies reported that avian salmonellosis can be disseminated through fertilized eggs, leading to the chicken embryo death (EFSA, 2007; Gantois et al., 2009; Zhao et al., 2021). However, several previous studies have reported a high prevalence of Salmonella Gallinarum from poultry and poultry products in China. In this study, we focus on Non-Gallinarum serovars (Xu et al., 2020b). Accordingly, to better describe the actual disease burden in breeders caused by S. enterica, we aimed to pick up the dead embryos instead of the normal embryo samples.
Vertical transmission or transovarian transmission is one of the key routes of Salmonella transmission to the egg. It occurs when the egg content is contaminated with Salmonella during the egg formation (Messens et al., 2005). Vertical transmission is common in host restricted Salmonella serovars, such as S. Gallinarum and S. Pullorum, but has also been reported in typical foodborne pathogen including S. Enteritidis (Poppe et al., 1998). Transmission via this route is directly related to the affinity of certain serovars in the reproductive tract of the hens (EFSA, 2010). Several studies reported that S. Enteritidis is the predominant serovar to colonize the reproductive organs of mature laying hens and cause severe outbreaks for humans due to egg contamination across the world (Okamura et al., 2001; Gantois et al., 2009). In this study, 45 (2.1%) S. enterica isolates were identified from yolk samples. These isolates were subjected to antimicrobial susceptibility assay and further genomic characterization. The positive rate found in this study was lower than that observed in eggs collected from poultry farms in Yangling, Shaanxi province (6.6%) (Li et al., 2020a), and Shandong province (Zhao et al., 2021) and higher than that obtained by Li et al. (0.5%) across China in 2016 (Li et al., 2020b).
The serovars identified in this study suggested a wide range of Salmonella diversified serovars within chick embryos. S. Enteritidis is a common serovar of non-typhoid Salmonella, and is also an invasive pathogenic foodborne pathogen. Our results showed that S. Enteritidis is the dominant serovar, which is consistent with those found in breeder chick farms (Fei et al., 2018). Additionally, Li et al. reported that S. Enteritidis was the dominant serovar isolated from diseased or dead chicken samples in Jiangsu, Henan, Heilongjiang, and Shandong provinces in 2012 (Li et al., 2018). S. Enteritidis was also found to be the cause of numerous foodborne outbreaks in Henan province, China (Xia et al., 2009). Globally, several studies also reported that S. Enteritidis was the dominant serovar in diseased or dead chicken from Uruguay (Betancor et al., 2010), the United States (Schutze et al., 1996), India (Suresh et al., 2006), Japan (Sasaki et al., 2011), Algeria (Ayachi et al., 2015), and Canada (Poppe, 1994). Our results were somewhat contradicted with those obtained from Shandong province, reporting the dominance of S. Thompson in dead-in-shell chicken embryos (Zhao et al., 2021). While S. Indiana was found to be dominant in Shandong (Lu et al., 2011), and S. Weltevreden was the dominant serovar in poultry farms in Vietnam (Lettini et al., 2016). Of the remaining isolates, S. Thompson, S. Indiana, S. Kottbus, S. Tennessee, and S. Cerro were also commonly recovered from poultry-based products and humans in China (Bai et al., 2015; Chen et al., 2020; Yang et al., 2020). These reports provide strong evidence about the increasing prevalence of S. Enteritidis in the poultry farms as well as the poultry supply chain in China. Some literature proposed that the decreasing prevalence of S. Gallinarum and S. Pullorum in the global poultry industry may have allowed S. Enteritidis to disseminate in poultry flocks (Rabsch et al., 2001). Collectively, the epidemic severity of S. Enteritidis in China's chicken industry chain is of great concern.
Moreover, all isolates showed high drug resistance, of these, 68% (31/45) were multidrug-resistant, and the frequency of the aminoglycoside resistance was the highest. All strains harbored the aac(6′)-Iaa gene, which conferring aminoglycoside resistance. This inspection can be clarified by the misuse and/or overuse of antibiotics in the agriculture and veterinary sectors, where antibiotics are applied for therapeutic practices, prophylactic, and growth improvement purposes. This study also showed that the examined Salmonella serovars had significant behavior differences against a range of antimicrobials. Most of our examined Salmonella isolates exhibited resistance to three or more antimicrobials. The surge prevalence of antimicrobial-resistant Salmonella isolates is recognized as a crucial public health issue. The antimicrobial resistance of S. Thompson and S. Kottbus was alarming and showed high antimicrobial resistance rates among all isolates, with a widespread antimicrobial resistance spectrum. This type of bacterial resistance has been previously detected in S. Thompson obtained from eggs in Shaanxi province, China (Li et al., 2020a).
Although they are not the dominant serovars, multiple antimicrobial selection pressure can play an important role in converting the serovar prevalence to dominant serovars over time by increasing their virulence (Boyle et al., 2007). In contrast, the overall resistance rate of S. Cerro is low, presenting only resistance to the aminoglycoside drugs. These results were in agreement with several worldwide reports on the dissemination of MDR among the Salmonella isolates (Li et al., 2013; Abdeen et al., 2018; Chuah et al., 2018; Elbediwi et al., 2021).
Genomic analysis of Salmonella isolates showed the detection of different antimicrobial resistance genes, which could correlate with the high level of antimicrobial resistance. All strains harbored the aac(6′)-Iaa gene, which was believed to be responsible for aminoglycoside resistance. Additionally, other aminoglycoside resistance genes were also detected in the present study (Figure 3). Among different mechanisms of aminoglycoside resistance, enzymatic modification is the most prevalent in pathogenic bacteria, including Salmonella spp. (Ramirez and Tolmasky, 2010; Biswas et al., 2019). Resistance to β-lactam antimicrobials is controlled by bla genes, and these genes are generally hydrolyzing the β-lactam ring, leading to antibiotic inactivation. bla resistance genes were found in ampicillin-resistant S. Enteritidis, S. Thompson, and S. Kottbus isolates and one S. Indiana isolate in this study (Figure 5). The dominant bla gene conferring ampicillin resistance in most of the Salmonella serovars was found to carry a few different types of blaTEM (de Toro et al., 2011; Eguale et al., 2017; García et al., 2018). TEM-2, SHV, and OXA-1 (β -lactamases) are also widespread in Enterobacteriaceae, although they are much rarer than TEM-1 (Livermore, 1995). We also found sul1 and sul2 antibiotic resistance genes in addition to other important gene dfrA12-17-19 in trimethoprim-sulfamethoxazole-resistant isolates. It has been reported in several studies that resistance to sulphonamide antibiotics is mainly mediated by the sul1, sul2, and sul3 genes (Antunes et al., 2007; Wang et al., 2014). Besides, the main mechanism of trimethoprim resistance, which is mainly mediated by dfrA genes, is the existence of integron-borne dihydrofolate reductases (Miko et al., 2005; Krauland et al., 2010; El-Sharkawy et al., 2017; Wang et al., 2017). Resistance to tetracyclines and quinolones antibiotics, which was conferred due to the presence of the tetD gene and the plasmid-mediated quinolones-resistant gene qnrB4, was also identified in S. Thompson and S. Kottbus serovars, posing a great public health concern. While both of them are considered critical antimicrobials, quinolones are currently preferred as a front-line drug of choice for the treatment of salmonellosis.
The genomic analysis also demonstrated that the high resistant serovars (Thompson and Kottbus) harbored IncHI2 and IncHI2A plasmids, as well as the hypervirulent serovar Enteritidis harbored IncF plasmid. Interestingly, these two plasmids were reported to be the predominant types in the MDR and hypervirulent isolates (Chen et al., 2016; Elbediwi et al., 2019, 2020a). IncHI2, IncHI2A, and IncF plasmids are often linked with persistence, as well as harboring virulence and antimicrobial resistance genes, which play a vital role in contributing to bacterial fitness (Ibarra and Steele-Mortimer, 2009; Villa et al., 2010; Coelho et al., 2012; Chen et al., 2016; Biswas et al., 2020; Xu et al., 2020a).
Considering the virulence factors that were investigated, S. Enteritidis isolates displayed a broader range of pathogenicity determinants as compared to other serovars. S. Enteritidis isolates were found to be harboring pef fimbrial genes, rck gene, spv locus, and sodCl gene. Serum resistance gene rck is encoding an outer membrane protein, which enhances the bacterial adhesion and invasion, also confers high resistance to the bactericidal activity of complement, by hindering the polymerization of complement component in the outer bacterial membrane (Guiney et al., 1995; Rosselin et al., 2010). Fimbrial adherence Pef and spv locus were previously reported as plasmid-borne determinants and have an important role in the Salmonella adhesion to the host gut epithelium (Guiney et al., 1995; Ledeboer et al., 2006; Yue et al., 2012, 2015). Stress adaptation gene sodCl is known to be involved in the Salmonella protection from phagocytic superoxide during infection (Sly et al., 2002; Uzzau et al., 2002). Regarding the sspH1 gene, which has been only identified in S. Cerro isolates, this gene was found to play an essential role in bacterial invasion of the epithelial cells and stimulation of the intestinal inflammatory response (Haraga and Miller, 2003). Otherwise, all the examined isolates harbored the typical virulence factors from Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2). SPI-1/-2 encodes a type three secretion system (T3SS), which is important for the injection of effector proteins into the host cells for modulation of the course of Salmonella infection (Que et al., 2013). SPI-1 genes are responsible for the invasion of the host cells, and the host immune response regulation. Besides, it encodes transcription factors that regulate the expression of some virulence factors (Raffatellu et al., 2005; Lou et al., 2019). Moreover, the development of the systemic disease is dependent on a type III secretion system (T3SS) encoded by SPI-2. SPI-2 harboring genes that are essential for bacterial intracellular survival (Hensel, 2000; Figueira et al., 2013).
The correlation analysis displayed strong positive associations between the different antimicrobial-resistant genes, exhibiting phenotypic resistance to different antimicrobial classes, such as those remarked for aminoglycosides resistant genes, bla genes, tetD, and qnrB, suggesting selective pressure exerted by the misusage of these antimicrobials in poultry production (Xu et al., 2020b). Additionally, the co-occurrence of these antimicrobial-resistant genes together either on the chromosome (same gene cassette) or plasmids, could be also another potential reason for this strong relation (Wales and Davies, 2015). The analysis also showed a strong positive correlation between different antimicrobial-resistant genes and IncHI2, IncHI2A, and IncF plasmids, confirming the vital role of these plasmids in antimicrobial-resistant genes dissemination, as mentioned earlier (Elbediwi et al., 2019).
Collectively, we found a considerable diversity of Salmonella serovars circulation in dead chick embryos in Henan, China, with multi-drug resistance potentials. Therefore, it is essential to continue monitoring the Salmonella serovars and implement the relevant strategic plans for prevention and control. Systematically and continuously monitoring the antimicrobial resistance of Salmonella, and application of an antimicrobial management plan for rational uses of essential antimicrobials in chicken farms are essential to improve food safety and prevent the MDR bacteria emergence. Direct whole-genome sequencing surveillance could provide a more comprehensive understanding of Salmonella population diversity, which might project public health awareness in the course of the next epidemics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA727934, PRJNA727934.
Author Contributions
ME and YT analyzed the data and drafted the manuscript. YT did the experiments and finalized the figures. HR did the statistical analysis work and review the paper. DS, SX, YX, and YL provided essential comments and helped with the edition of the manuscript. MY conceived the idea and assisted with data analysis and writing. All authors read, revised, and approved the final manuscript.
Funding
This study was supported by the National Program on Key Research Project of China (2019YFE0103900 and 2017YFC1600103) as well as the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement No 861917—SAFFI, Zhejiang Provincial Natural Science Foundation of China (LR19C180001) and Zhejiang Provincial Key R&D Program of China (2020C02032 and 2021C02008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.684400/full#supplementary-material
Supplementary Table 1. Salmonella isolates serotyping by sera agglutination assay and in silico analysis by using Seqsero v1.0.1 and SISTR v1.0.0.
Supplementary Table 2. The virulence factor profiling for all the examined Salmonella isolates in this study.
References
Abdeen, E., Elmonir, W., Suelam, I. I. A., and Mousa, W. S. (2018). Antibiogram and genetic diversity of Salmonella enterica with zoonotic potential isolated from morbid native chickens and pigeons in Egypt. J. Appl. Microbiol. 124, 1265–1273. doi: 10.1111/jam.13697
Antunes, P., Machado, J., and Peixe, L. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3' conserved sequence region among Salmonella isolates. Antimicrobiol Agents Chemother. 51, 1545–1548. doi: 10.1128/AAC.01275-06
Ayachi, A., Bennoune, O., Heleili, N., and Alloui, N. (2015). Minor Salmonella: potential pathogens in eggs in Algeria. J. Infect. Dev. Ctries. 9, 1156–1160. doi: 10.3855/jidc.6526
Bai, L., Lan, R., Zhang, X., Cui, S., Xu, J., Guo, Y., et al. (2015). Prevalence of Salmonella isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica Serovar Indiana in Henan, China. PLoS ONE. 10:e0144532. doi: 10.1371/journal.pone.0144532
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bengtsson, B., and Greko, C. (2014). Antibiotic resistance–consequences for animal health, welfare, and food production. Upsala J. Med. Sci. 119, 96–102. doi: 10.3109/03009734.2014.901445
Betancor, L., Pereira, M., Martinez, A., Giossa, G., Fookes, M., Flores, K., et al. (2010). Prevalence of Salmonella enterica in poultry and eggs in uruguay during an epidemic due to Salmonella enterica Serovar Enteritidis. J. Clin. Microbiol. 48, 2413–2423. doi: 10.1128/JCM.02137-09
Biswas, S., Elbediwi, M., Gu, G., and Yue, M. (2020). Genomic characterization of new variant of hydrogen sulfide (H2S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in China dagger. Antibiotics 9:80. doi: 10.3390/antibiotics9020080
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella typhimurium monophasic Clone ST34. Microorganisms 7:298. doi: 10.3390/microorganisms7090298
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyle, E. C., Bishop, J. L., Grassl, G. A., and Finlay, B. B. (2007). Salmonella: from pathogenesis to therapeutics. J. Bacteriol. 189, 1489–1495. doi: 10.1128/JB.01730-06
CDC (2013). Incidence and trends of infection with pathogens transmitted commonly through food — foodborne diseases active surveillance network,10 U.S. Sites, 1996–2012. MMWR Morb. Mortal. Wkly. Rep. 62, 283–287.
Chen, W., Fang, T., Zhou, X., Zhang, D., Shi, X., and Shi, C. (2016). IncHI2 plasmids are predominant in antibiotic-resistant Salmonella isolates. Front. Microbiol. 7:1566. doi: 10.3389/fmicb.2016.01566
Chen, Z., Bai, J., Wang, S., Zhang, X., Zhan, Z., Shen, H., et al. (2020). Prevalence, antimicrobial resistance, virulence genes and genetic diversity of Salmonella isolated from retail duck meat in Southern China. Microorganisms 8:444. doi: 10.3390/microorganisms8030444
Chuah, L. O., Shamila Syuhada, A. K., Mohamad Suhaimi, I., Farah Hanim, T., and Rusul, G. (2018). Genetic relatedness, antimicrobial resistance and biofilm formation of Salmonella isolated from naturally contaminated poultry and their processing environment in northern Malaysia. Food Res. Int. 105, 743–751. doi: 10.1016/j.foodres.2017.11.066
Clinical and Laboratory Standards Institute (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho, A., Piedra-Carrasco, N., Bartolom,é, R, Quintero-Zarate, J. N., Larrosa, N., Cornejo-Sánchez, T., et al. (2012). Role of IncHI2 plasmids harbouring blaVIM-1, blaCTX-M-9, aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d'Hebron, Barcelona, Spain. Int. J. Antimicrobiol. Agents 39, 514–517. doi: 10.1016/j.ijantimicag.2012.01.006
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
de Toro, M., Sáenz, Y., Cercenado, E., Rojo-Bezares, B., García-Campello, M., Undabeitia, E., et al. (2011). Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int. Microbiol. 14, 173–181. doi: 10.2436/20.1501.01.146
EFSA (2007). The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J. 130, 34–117. doi: 10.2903/j.efsa.2007.130r
EFSA (2010). Scientific opinion on a quantitative estimation of the public health impact of setting a new target for the reduction of Salmonella in laying hens. EFSA J. 8:1546. doi: 10.2903/j.efsa.2010.1546
Eguale, T., Birungi, J., Asrat, D., Njahira, M. N., Njuguna, J., Gebreyes, W. A., et al. (2017). Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrobiol. Resist. Infect. Control 6, 13–16. doi: 10.1186/s13756-017-0171-6
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980–2018). Microorganisms 7:461. doi: 10.3390/microorganisms7100461
Elbediwi, M., Pan, H., Biswas, S., Li, Y., and Yue, M. (2020a). Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerge. Microbes Infect. 9, 535–538. doi: 10.1080/22221751.2020.1733439
Elbediwi, M., Pan, H., Jiang, Z., Biswas, S., Li, Y., and Yue, M. (2020b). Genomic Characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China. Front. Bioeng. Biotechnol. 8:663. doi: 10.3389/fbioe.2020.00842
Elbediwi, M., Pan, H., Zhou, X., Rankin, S. C., Schifferli, D. M., and Yue, M. (2021). Detection of mcr-9-harbouring ESBL-producing Salmonella newport isolated from an outbreak in a large-animal teaching hospital in the USA. J. Antimicrobiol Chemother. 76, 1107–1109. doi: 10.1093/jac/dkaa544
El-Sharkawy, H., Tahoun, A., El-Gohary, A. E.G. A., El-Abasy, M., El-Khayat, F., Gillespie, T., et al. (2017). Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 9:8. doi: 10.1186/s13099-017-0157-1
European Food Safety Authority and European Centre for Disease Prevention and Control (2017). The European Union summary report on trends and sources of zoonoses. EFSA J. 15:e05077. doi: 10.2903/j.efsa.2017.5077
Fei, X., Yin, K., Yin, C., Hu, Y., Li, J., Zhou, Z., et al. (2018). Analyses of prevalence and molecular typing reveal the spread of antimicrobial-resistant Salmonella infection across two breeder chicken farms. Poult. Sci. 97, 4374–4383. doi: 10.3382/ps/pey305
Figueira, R., Watson, K. G., Holden, D. W., and Helaine, S. (2013). Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of Salmonella enterica serovar typhimurium: implications for rational vaccine design. mBio. 4, e00065–e00013. doi: 10.1128/mBio.00065-13
Gantois, I., Ducatelle, R., Pasmans, F., Haesebrouck, F., Gast, R., Humphrey, T. J., et al. (2009). Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol. Rev. 33, 718–738. doi: 10.1111/j.1574-6976.2008.00161.x
García, V., Vázquez, X., Bances, M., Herrera-León, L., Herrera-León, S., and Rodicio, M. R. (2018). Molecular characterization of Salmonella enterica serovar enteritidis, genetic basis of antimicrobial drug resistance and plasmid diversity in ampicillin-resistant isolates. Microbiol. Drug Resist. 25, 219–226. doi: 10.1089/mdr.2018.0139
Guiney, D. G., Fang, F. C., Krause, M., Libby, S., Buchmeier, N. A., Fierer, J., et al. (1995). Biology and clinical significance of virulence plasmids in Salmonella serovars. Clin. Infect. Dis. 21 (Supplement_2):S146–S51. doi: 10.1093/clinids/21.Supplement_2.S146
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Haraga, A., and Miller, S. I. (2003). A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 71, 4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003
Hazards EPanelo, B., Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., et al. (2019). Salmonella control in poultry flocks and its public health impact. EFSA J. 17:e05596. doi: 10.2903/j.efsa.2019.5596
Hensel, M. (2000). Salmonella pathogenicity island 2. Mol. Microbiol. 36, 1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x
Ibarra, J. A., and Steele-Mortimer, O. (2009). Salmonella–the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11, 1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x
Jajere, S. M. (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 12, 504–521. doi: 10.14202/vetworld.2019.504-521
Jiang, Z., Anwar, T. M., Peng, X., Biswas, S., Elbediwi, M., Li, Y., et al. (2021). Prevalence and antimicrobial resistance of Salmonella recovered from pig-borne food products in Henan, China. Food Control 121:107535. doi: 10.1016/j.foodcont.2020.107535
Jiang, Z., Paudyal, N., Xu, Y., Deng, T., Li, F., Pan, H., et al. (2019). Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front. Microbiol. 10:1513. doi: 10.3389/fmicb.2019.01513
Kirch, W (ed). (2008). “Pearson's correlation coefficient,” in Encyclopedia of Public Health (Dordrecht: Springer), 1090–1091. doi: 10.1007/978-1-4020-5614-7_2569
Krauland, M., Harrison, L., Paterson, D., and Marsh, J. (2010). Novel integron gene cassette arrays identified in a global collection of multi-drug resistant non-typhoidal Salmonella enterica. Curr. Microbiol. 60, 217–223. doi: 10.1007/s00284-009-9527-3
Ledeboer, N. A., Frye, J. G., McClelland, M., and Jones, B. D. (2006). Salmonella enterica serovar typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immunity 74, 3156–3169. doi: 10.1128/IAI.01428-05
Lettini, A. A., Vo Than, T., Marafin, E., Longo, A., Antonello, K., Zavagnin, P., et al. (2016). Distribution of Salmonella serovars and antimicrobial susceptibility from poultry and swine farms in central Vietnam. Zoonoses Public Health 63, 569–576. doi: 10.1111/zph.12265
Li, R., Lai, J., Wang, Y., Liu, S., Li, Y., Liu, K., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Li, W., Li, H., Zheng, S., Wang, Z., Sheng, H., Shi, C., et al. (2020a). Prevalence, serotype, antibiotic susceptibility, and genotype of Salmonella in eggs from poultry farms and marketplaces in Yangling, Shaanxi Province, China. Front. Microbiol. 11:1482. doi: 10.3389/fmicb.2020.01482
Li, W. W., Bai, L., Zhang, X. L., Xu, X. J., Tang, Z., Bi, Z. W., et al. (2018). [Prevalence and antimicrobial susceptibility of Salmonella isolated from broiler whole production process in four provinces of China]. Zhonghua Yu Fang Yi Xue Za Zhi 52, 352–357. doi: 10.3760/cma.j.issn.0253-9624.2018.04.005
Li, Y., Yang, X., Zhang, H., Jia, H., Liu, X., Yu, B., et al. (2020b). Prevalence and antimicrobial susceptibility of Salmonella in the commercial eggs in China. Int. J. Food Microbiol. 325:108623. doi: 10.1016/j.ijfoodmicro.2020.108623
Liu, Q., Chen, W., Elbediwi, M., Pan, H., Wang, L., Zhou, C., et al. (2020). Characterization of Salmonella resistome and plasmidome in pork production system in Jiangsu, China. Front. Vet. Sci. 7:617. doi: 10.3389/fvets.2020.00617
Liu, Y., Jiang, J., Ed-Dra, A., Li, X., Peng, X., Xia, L., et al. (2021). Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res. Int. 142:110198. doi: 10.1016/j.foodres.2021.110198
Liu, Y.-Y., Lin, J.-W., and Chen, C.-C. (2019). cano-wgMLST_BacCompare: a bacterial genome analysis platform for epidemiological investigation and comparative genomic analysis. Front. Microbiol. 10:1687. doi: 10.3389/fmicb.2019.01687
Livermore, D. M. (1995). Beta-Lactamases in laboratory and clinical resistance. Clin. Microbial. Rev. 8, 557–584. doi: 10.1128/CMR.8.4.557
Lou, L., Zhang, P., Piao, R., and Wang, Y. (2019). Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell Infect. Microbiol. 9:270. doi: 10.3389/fcimb.2019.00270
Lu, Y., Wu, C. M., Wu, G. J., Zhao, H. Y., He, T., Cao, X. Y., et al. (2011). Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 8, 45–53. doi: 10.1089/fpd.2010.0605
Messens, W., Grijspeerdt, K., and Herman, L. (2005). Eggshell penetration by Salmonella: a review. Worlds Poultry Sci. J. 61, 71–86. doi: 10.1079/WPS200443
Miko, A., Pries, K., Schroeter, A., and Helmuth, R. (2005). Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrobiol. Chemother. 56, 1025–1033. doi: 10.1093/jac/dki365
Okamura, M., Kamijima, Y., Miyamoto, T., Tani, H., Sasai, K., and Baba, E. (2001). Differences among six Salmonella serovars in abilities to colonize reproductive organs and to contaminate eggs in laying hens. Avian Dis. 45, 61–69. doi: 10.2307/1593012
Pan, H., Zhou, X., Chai, W., Paudyal, N., Li, S., Zhou, X., et al. (2019). Diversified sources for human infections by Salmonella enterica serovar newport. Transboundary Emerg. Dis. 66, 1044–1048. doi: 10.1111/tbed.13099
Paudyal, N., Pan, H., Elbediwi, M., Zhou, X., Peng, X., Li, X., et al. (2019). Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 19:226. doi: 10.1186/s12866-019-1598-0
Paudyal, N., Pan, H., Liao, X., Zhang, X., Li, X., Fang, W., et al. (2018). A meta-analysis of major foodborne pathogens in chinese food commodities between 2006 and 2016. Foodborne Pathog. Dis. 15, 187–197. doi: 10.1089/fpd.2017.2417
Poppe, C. (1994). Salmonella enteritidis in Canada. Int. J. Food Microbiol. 21, 1–5. doi: 10.1016/0168-1605(94)90193-7
Poppe, C., Duncan, C. L., and Mazzocco, A. (1998). Salmonella contamination of hatching and table eggs: a comparison. Can. J. Vet. Res. 62, 191–198.
Que, F., Wu, S., and Huang, R. (2013). Salmonella pathogenicity island 1(SPI-1) at Work. Curr. Microbiol. 66, 582–587. doi: 10.1007/s00284-013-0307-8
Rabsch, W., Tschäpe, H., and Bäumler, A. J. (2001). Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3, 237–247. doi: 10.1016/S1286-4579(01)01375-2
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect. Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Rosselin, M., Virlogeux-Payant, I., Roy, C., Bottreau, E., Sizaret, P.-Y., Mijouin, L., et al. (2010). Rck of Salmonella enterica, subspecies enterica serovar enteritidis, mediates Zipper-like internalization. Cell Res. 20, 647–664. doi: 10.1038/cr.2010.45
Sasaki, Y., Tsujiyama, Y., Asai, T., Noda, Y., Katayama, S., and Yamada, Y. (2011). Salmonella prevalence in commercial raw shell eggs in Japan: a survey. Epidemiology. Infect. 139, 1060–1064. doi: 10.1017/S0950268810002153
Schutze, G. E., Fawcett, H. A., Lewno, M. J., Flick, E. L., and Kirby, R. S. (1996). Prevalence of Salmonella enteritidis in poultry shell eggs in Arkansas. South. Med. J. 89, 889–891. doi: 10.1097/00007611-199609000-00008
Sly, L. M., Guiney, D. G., and Reiner, N. E. (2002). Salmonella enterica serovar typhimurium periplasmic superoxide dismutases SodCI and SodCII are required for protection against the phagocyte oxidative burst. Infect. Immun. 70:5312. doi: 10.1128/IAI.70.9.5312-5315.2002
Suresh, T., Hatha, A. A., Sreenivasan, D., Sangeetha, N., and Lashmanaperumalsamy, P. (2006). Prevalence and antimicrobial resistance of Salmonella enteritidis and other salmonellas in the eggs and egg-storing trays from retail markets of Coimbatore, South India. Food Microbiol. 23, 294–299. doi: 10.1016/j.fm.2005.04.001
Uzzau, S., Bossi, L., and Figueroa-Bossi, N. (2002). Differential accumulation of Salmonella[Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol. Microbiol. 46, 147–156. doi: 10.1046/j.1365-2958.2002.03145.x
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrobiol. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Vo, A. T. T., van Duijkeren, E., Fluit, A. C., Heck, M. E. O. C., Verbruggen, A., Maas, H. M. E., et al. (2006). Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella typhimurium phage type 90. Vet. Microbiol. 113, 153–158. doi: 10.1016/j.vetmic.2005.10.034
Wales, A., and Davies, R. (2015). Co-Selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. doi: 10.3390/antibiotics4040567
Wang, N., Yang, X., Jiao, S., Zhang, J., Ye, B., and Gao, S. (2014). Sulfonamide-Resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS ONE 9:e112626. doi: 10.1371/journal.pone.0112626
Wang, W., Peng, Z., Baloch, Z., Hu, Y., Xu, J., Zhang, W., et al. (2017). Genomic characterization of an extensively-drug resistance Salmonella enterica serotype Indiana strain harboring bla(NDM-1) gene isolated from a chicken carcass in China. Microbiol Res. 204:48–54. doi: 10.1016/j.micres.2017.07.006
Wang, X., Biswas, S., Paudyal, N., Pan, H., Li, X., Fang, W., et al. (2019). Antibiotic resistance in Salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 10:985. doi: 10.3389/fmicb.2019.00985
Wibisono, F., Wibisono, F., Effendi, M., Plumeriastuti, H., Hidayatullah, A. R., Hartadi, E., et al. (2020). A review of salmonellosis on poultry farms: public health importance. Syst. Rev. Pharm. 11, 481–486. doi: 10.31838/srp.2020.9.69
Xia, S., Hendriksen, R. S., Xie, Z., Huang, L., Zhang, J., Guo, W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47, 401–409. doi: 10.1128/JCM.01099-08
Xu, X., Biswas, S., Gu, G., Elbediwi, M., Li, Y., and Yue, M. (2020a). Characterization of Multidrug resistance patterns of emerging Salmonella enterica serovar rissen along the food chain in China. Antibiotics 9:660. doi: 10.3390/antibiotics9100660
Xu, Y., Zhou, X., Jiang, Z., Qi, Y., Ed-dra, A., and Yue, M. (2020b). Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell Infect. Microbiol. 10:497. doi: 10.3389/fcimb.2020.00497
Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C., et al. (2020). Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P. J., Nash, J. H. E., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE. 11:e0147101. doi: 10.1371/journal.pone.0147101
Yu, H., Elbediwi, M., Zhou, X., Shuai, H., Lou, X., Wang, H., et al. (2020). Epidemiological and Genomic Characterization of Campylobacter jejuni Isolates from a Foodborne Outbreak at Hangzhou, China. Int. J. Mol. Sci. 21:3001. doi: 10.3390/ijms21083001
Yue, M., Han, X., Masi, L. D., Zhu, C., Ma, X., Zhang, J., et al. (2015). Allelic variation contributes to bacterial host specificity. Nat. Commun. 6:8754. doi: 10.1038/ncomms9754
Yue, M., Rankin, S. C., Blanchet, R. T., Nulton, J. D., Edwards, R. A., and Schifferli, D. M. (2012). Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS ONE 7:e38596. doi: 10.1371/journal.pone.0038596
Yue, M., Song, H., and Bai, L. (2020). Call for special issue papers: food safety in china: current practices and future needs. Foodborne Pathog. Dis. 17:295. doi: 10.1089/fpd.2020.29013.cfp5
Keywords: Salmonella, chicken embryo, antimicrobial resistance, correlation, genomic characterization, virulence factor
Citation: Elbediwi M, Tang Y, Shi D, Ramadan H, Xu Y, Xu S, Li Y and Yue M (2021) Genomic Investigation of Antimicrobial-Resistant Salmonella enterica Isolates From Dead Chick Embryos in China. Front. Microbiol. 12:684400. doi: 10.3389/fmicb.2021.684400
Received: 23 March 2021; Accepted: 14 June 2021;
Published: 23 August 2021.
Edited by:
Bing Gu, Guangdong Provincial People's Hospital, ChinaReviewed by:
Assaf Rokney, Ministry of Health, IsraelJing Han, National Center for Toxicological Research (FDA), United States
Copyright © 2021 Elbediwi, Tang, Shi, Ramadan, Xu, Xu, Li and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Yue, bXl1ZUB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Mohammed Elbediwi
Mohammed Elbediwi Yanting Tang1†
Yanting Tang1† Dawei Shi
Dawei Shi Hazem Ramadan
Hazem Ramadan Sihong Xu
Sihong Xu Yan Li
Yan Li Min Yue
Min Yue