- 1Department of Environment and Resource Sciences, University of Miyazaki, Miyazaki, Japan
- 2Thai Nguyen University of Agriculture and Forestry, Thai Nguyen, Vietnam
- 3Department of Infectious Disease, Faculty of Medicine, University of Miyazaki, Miyazaki, Japan
- 4Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan
Tellurite is highly toxic to most bacteria owing to its strong oxidative ability. However, some bacteria demonstrate tellurite resistance. In particular, some Escherichia coli strains, including Shiga toxin-producing E. coli O157:H7, are known to be resistant to tellurite. This resistance is involved in ter operon, which is usually located on a prophage-like element of the chromosome. The characteristics of the ter operon have been investigated mainly by genome analysis of pathogenic E. coli; however, the distribution and structural characteristics of the ter operon in other E. coli are almost unknown. To clarify these points, we examined 106 E. coli strains carrying the ter operon from various animals. The draft genomes of 34 representative strains revealed that ter operons were clearly classified into four subtypes, ter-type 1–4, at the nucleotide sequence level. Complete genomic sequences revealed that operons belonging to three ter-types (1, 3, and 4) were located on the prophage-like elements on the chromosome, whereas the ter-type 2 operon was located on the IncHI2 plasmid. The positions of the tRNASer, tRNAMet, and tRNAPhe indicated the insertion sites of elements carrying the ter operons. Using the PCR method developed in this study, 106 strains were classified as type 1 (n = 66), 2 (n = 13), 3 (n = 8), and 4 (n = 17), and two strains carried both types 1 and 2. Furthermore, significant differences in the minimum inhibitory concentration (MIC) of tellurite were observed between strains carrying ter-type 4 and the others (p < 0.05). The ter-type was also closely related to the isolation source, with types 2 and 4 associated with chickens and deer, respectively. This study provided new insights related not only to genetic characteristics of the ter operons, but also to phenotypic and ecological characteristics that may be related to the diversity of the operon.
Introduction
Tellurium (Te) is a rare element on Earth (Ibers, 2009) and is found in metal ores such as gold and copper (Terziev, 1966; Adams, 2016). Tellurite (TeO32–), the oxyanion of tellurium, is highly toxic to most bacteria owing to its strong oxidative ability. Tellurite toxicity is related to the generation of reactive oxygen species (ROS) (Turner et al., 2001; Pérez et al., 2007; Tremaroli et al., 2007; Calderón et al., 2009; Díaz-Vásquez et al., 2014; Sandoval et al., 2015), which then induce damage to key cell components (Cabiscol et al., 2000; Imlay, 2003; Ling and Söll, 2010; Molina-Quiroz et al., 2013), and consequently affect bacterial growth. However, some bacteria demonstrate tellurite resistance. In particular, most Shiga toxin-producing Escherichia coli (STEC) belonging to O157:H7 and other serotypes such as O26:H11, O103:H2, O111:H8, O121:H19, O145:H28, and O45:H2, which are responsible for numerous foodborne disease outbreaks, are known to possess tellurite resistance. Therefore, when STEC is efficiently isolated from patient and food samples, selective agar plates containing potassium tellurite (K2TeO3), such as cefixime-tellurite sorbitol MacConkey (CT-SMAC) agar and CHROMagar STEC are routinely used in laboratories.
Several genes involved in arsenic efflux (arsABC) (Turner et al., 1992), cysteine metabolism (cysK, cysM, csdB, IscS) (Vásquez et al., 2001; Lithgow et al., 2004; Rojas and Vásquez, 2005), nitrate reduction (narGHIJ) (Avazéri et al., 1997; Sabaty et al., 2001), superoxide dismutation (sodA, sodB, soxS) (Tantaleán et al., 2003; Pérez et al., 2007), hydrogen peroxide detoxification (katG) (Calderón et al., 2006), tellurite resistance proteins (tehAB, kilA, telAlB) (Walter et al., 1991; Turner et al., 1995), and glucose-phosphate dehydrogenase (zwf) (Sandoval et al., 2011; Sandoval et al., 2015) have been found to be related with tellurite resistance. Additionally, the tellurite resistance gene (ter) operon (Whelan et al., 1995) is involved in tellurite resistance at relatively high concentrations; the ter operon was first identified from a conjugative plasmid of an Alcaligenes strain (Jobling and Ritchie, 1987, 1988). Although the detailed functions of the ter operon underlying tellurite resistance are not fully understood, experiments such as cloning have revealed that six genes (terZABCDE) are directly involved in tellurite resistance (Whelan et al., 1995; Kormutakova et al., 2000; Valková et al., 2007). Genome sequences deposited in databases have revealed that the ter operon is widely distributed in gram-negative bacteria including Escherichia, Salmonella, Yersinia, Pseudomonas, and Proteus species (Supplementary Tables 1, 2).
STECs belonging to the seven major serotypes described above are known to carry the ter operon. For example, the STEC O157:H7 Sakai strain carried the ter operon on a prophage-like 86-kb element, named SpLE1 on the chromosome (Hayashi et al., 2001; Ohnishi et al., 2002). SpLE1 also contains genes encoding integrase, urease (ure), and iron-regulated gene A (IrgA)-homolog adhesin (iha) (Hayashi et al., 2001). Comparative genome analyses indicated that the ter operons and their carrier elements are highly conserved among STEC strains belonging to the major serotypes (Ogura et al., 2009; Lorenz et al., 2017). Some STEC isolates, such as STEC O104:H4 and O178:H19, and non-STEC E. coli isolates also carry ter operons and/or show significant tellurite resistance (Orth et al., 2007; Gouali et al., 2013; Miko et al., 2014; Ferdous et al., 2015; Verhaegen et al., 2015; Kalule et al., 2018; Nguyen et al., 2021). A uropathogenic E. coli isolate was found to carry the ter operon on a large conjugative plasmid (Soltys et al., 2018). These reports suggest that the ter operon is widely distributed in E. coli including intestinal and extraintestinal pathogenic E. coli, as well as non-pathogenic E. coli. However, except for some STECs, the genetic characteristics of ter operons and their carrier elements have not been investigated in detail. In this study, in addition to STECs belonging to seven major serotypes, detailed analysis of ter operons and their carrier elements were performed in E. coli isolates belonging to more than 70 serotypes isolated from humans, cattle, pigs, chickens, and deer.
Materials and Methods
E. coli Strains
In total, 106 terA-positive E. coli strains isolated in Japan were used in this study (Supplementary Table 3). Human-derived STECs were isolated from patients and asymptomatic carriers through routine laboratory inspection. Twenty-four cattle-derived STEC strains, isolated in our previous study, were also used (Nguyen et al., 2021). Other E. coli strains from cattle, pigs, and deer were isolated from the rectal feces of slaughtered healthy animals. Feces were pre-cultured in modified E. coli broth (mEC) (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) at 37°C for 18–20 h and then inoculated on deoxycholate hydrogen sulfide lactose (DHL) (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) or CHROMagar STEC (CHROMagarTM Paris, France) agar plates. E. coli strains from chickens were isolated from colibacillosis infected sites (such as under the skin and organs) of chickens on farms and slaughterhouses. The swabs were directly inoculated onto DHL agar plates. After incubation at 37°C for 18–20 h, E. coli-like single colonies were picked from each plate. DNA for PCR was purified using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, United States). E. coli species were confirmed by PCR using E. coli-specific gyrB sequences (Iguchi et al., 2015). The presence of the ter operon was screened by PCR using a universal primer pair (Supplementary Table 4), designed based on the terA sequences collected from the DNA database (GenBank/ENA/DDBJ) (Supplementary Table 1). The thermal conditions for PCR included 25 cycles of 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s. PCR was conducted using Kapa Taq DNA polymerase (Kapa Biosystems, Woburn, MA, United States) or Dream Taq DNA polymerase (Thermo Fisher Scientific, Inc.) following the manufacturer’s instructions. The PCR products were electrophoresed on 2% agarose gels in 0.5 × Tris-borate-EDTA (TBE) (25 mM Tris-borate and 0.5 mM EDTA) followed by staining with ethidium bromide (1 mg/mL), and imaging under UV light.
DNA-Based O and H Serotyping
DNA-based E. coli O and H serotyping (Og- and Hg-typing) were conducted as described previously (Iguchi et al., 2015; Banjo et al., 2018; Iguchi et al., 2020). Briefly, 25 multiplex PCR kits (MP-1 to MP-25) containing 195 primer pairs targeting O-antigen encoding genes and 10 multiplex PCR kits (MP-A to MP-J) containing 52 primer pairs targeting flagellin (H-antigen) encoding genes were used for broad Og- and Hg-typing, respectively. If no PCR product was obtained using any of the multiplex PCR kits, the sample was determined as OgUT (Og untypeable) or HgUT (Hg untypeable).
Prevalence of Virulence Related Genes
The virulence profiles of stx1, stx2, and eae were screened using PCR. The primers and PCR conditions for stx1, stx2, and eae were previously described by Cebula et al. (1995) and Nguyen et al. (2021), respectively.
Genome Sequencing and Assembly
DNA for genome sequencing was extracted and purified using the Qiagen Genomic DNA Preparation Kit (Qiagen) or Maxwell® DNA Purification Kit (Promega). Illumina short-read libraries were prepared using 100 ng of extracted DNA using the Nextera DNA Library Prep Kit, and paired-end reads were generated using the MiSeq Reagent Kit (v3-600) on MiSeq (Illumina). MinION long-read libraries were prepared from 300 ng of genomic DNA using the Rapid Barcoding kit (SQK-RBK004) and were sequenced using R9 flow cells (FLO-MIN106) on MinION (Oxford Nanopore Technologies) to produce > 100 Mb of data per strain. The Illumina data were preprocessed using Trimmomatic (v0.36) to remove the adapter and low-quality sequences (Bolger et al., 2014). The quality of the Nanopore sequencing data was assessed using NanoPack (De Coster et al., 2018) a hybrid assembly with the Illumina and Nanopore reads was performed using Unicycler (v0.4.6) with the default parameters (Wick et al., 2017). The sequence data have been deposited in NCBI under the accession number, BioProject PRJDB10561.
Genome Analysis
Genome sequences were annotated using the online tool DFAST (Tanizawa et al., 2017) and were then visualized using an in silico Molecular Cloning software (in silico biology, Inc.). Plasmids were identified using PlasmidFinder 2.1 (Carattoli et al., 2014). The family of transposase genes was identified using the online tool ISfinder (Siguier et al., 2006). DNA sequences were aligned using the ClustalW program provided by DNA Data Bank of Japan1. A phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates in MEGAX (Stecher et al., 2020). Genetic comparison and dot plot analysis of the elements was conducted using GenomeMatcher 3.14 (Ohtsubo et al., 2008).
Subtyping PCR for the ter Operon
The PCR primer sequences used for ter operon subtyping are shown in Supplementary Table 4. The thermal conditions for PCR were the same as those of PCR using the universal primers targeting terA.
Multilocus Sequence Typing
Multilocus sequence typing (MLST) was performed using the internal sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) (Wirth et al., 2006). The primers used were described in a previous study (Nguyen et al., 2021). Sequence types (STs) were determined according to the Enterobase MLST website2. Concatenated sequences (3,423 bp) of the seven genes were used to construct a phylogenetic tree.
Minimum Inhibitory Concentration (MIC) of Tellurite
Tellurite resistance was evaluated on Mueller-Hinton agar plates containing twofold serial dilutions of potassium tellurite (K2TeO3) ranging from 0.125 to 512 μg/mL, according to the general steps of the agar dilution method (Clinical and Laboratory Standards Institutes [CLSI], 2012). Three independent experiments and two replicates were performed for each experiment. For resistant strains, the median log2 transformed MIC values of six replicates were used for statistical analysis. The difference in log2 transformed MIC was analyzed by Tukey’s t-test and statistical significance was set at p ≤ 0.05.
Results
terA-Positive E. coli Strains
In total, 106 terA-positive E. coli strains from humans (26 strains), cattle (34 strains), pigs (6 strains), chickens (8 strains), and deer (32 strains) were collected (see Supplementary Table 3). There was no duplication of the Og:Hg-type in the strains isolated from the same sample. The strains were classified into 78 Og:Hg types, including an OgUT:Hg2 strain. terA-positive E. coli strains contained eae-positive STEC (positive for either or both stx1 and stx2) (42 strains), eae-negative STEC (28 strains), enteropathogenic E. coli (EPEC) (negative for both stx1 and stx2 and positive for eae) (4 strains), and others (all negative for stx1, stx2, and eae) (32 strains).
Subtypes of ter Operon
First, we determined the draft genomes of 34 representative strains using the MiSeq system, which were randomly selected after sorting out so that the Og:Hg types did not overlap (Supplementary Table 3) and extracted all ter operons (4,546 − 4,551 bp) containing six genes terZ, terA, terB, terC, terD, and terE (Figure 1A). When a phylogenetic tree was created using the ter operon sequences, it became clear that four groups were formed, named ter-type 1–4 (Figure 1B). In particular, the nucleotide sequences of types 1, 3, and 4 were highly conserved within their respective groups. The phylogenetic trees using the sequences of each gene of 34 strains, terZ (582 bp), terA (1,158 bp), terB (456 bp), terC (1,041 bp), terD (579 bp), and terE (576 bp) showed that type-3 and type-4 formed independent groups for all genes, whereas type-1 and type-2 roughly formed their respective groups for terZ, terA, terD, and terE (Supplementary Figure 1). When a phylogenetic tree was created by adding 13 types of ter operons possessed by bacteria other than E. coli (Supplementary Table 2), three operons from Yersinia pseudotuberculosis, Salmonella enterica, and Klebsiella pneumoniae belonged to type-2, operons from seven Enterobacteriaceae genera formed an additional clade inside the group of E. coli-derived operons but no E. coli strain belonged to this clade, and the remaining three from Proteus mirabilis, Yersinia pestis, and Pseudomonas aeruginosa were distant from the E. coli-derived group (Supplementary Figure 2).
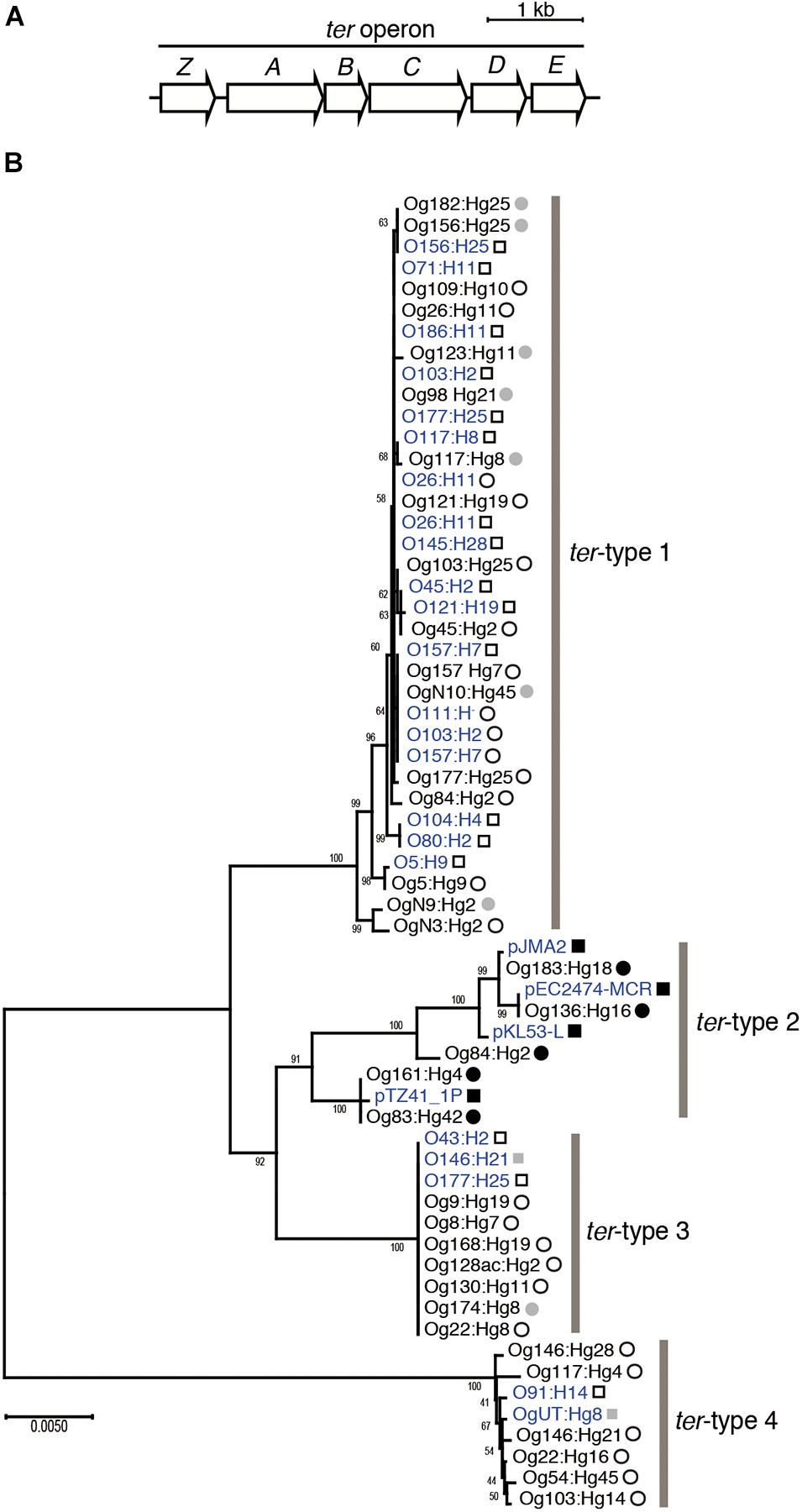
Figure 1. Structure of the ter operon (A). Phylogenetic tree of the ter operon (B). The tree was created based on the ter operon sequences (4,546–4,551 bp) from 34 Escherichia coli strains used in this study, and 27 E. coli genomes registered in DNA database. Four of them were isolated in Japan and the remaining 23 were isolated outside Japan. Strains isolated in Japan and outside Japan are indicated by circles and squares, respectively. Open and fulfilled circles or squares indicate that the ter operon is located on the chromosome and on the plasmid, respectively. Those whose location is unknown are shown in gray. DNA sequences obtained from the DNA database (GenBank/ENA/DDBJ) are shown in blue color. Bootstrap values are displayed at the branching points.
Comparison of Elements Carrying ter Operons
To analyze the structure of the ter operon as well as the surrounding region in detail, complete or almost complete genome sequences were obtained from the short- and long-read sequences of the 26 strains (Supplementary Table 3). When the attributes of ter operons were confirmed from the surrounding sequence information, the operons classified as ter-type 1, 3, and 4 were located on the chromosomes, whereas all operons classified as ter-type 2 were located on plasmids (Figure 1 and Supplementary Figure 2).
The elements carrying ter-type 1 from Og157:Hg7, Og121:Hg19, Og26:Hg11, Og84:Hg2, and Og177:Hg25 (76,233–96,657 bp) were highly conserved with SpLE1 in the O157:H7 Sakai strain (Figure 2A and Supplementary Figure 3). In addition to the ter operon, these elements also carried the conserved integrase gene, the ure operon, iha, and some transposase genes including the IS66, IS3, and IS110 families; however, the integrase gene was not found in Og177:Hg25. The elements carrying ter-type 1 in Og103:Hg25, Og45:Hg2, Og5:Hg9, Og109:Hg10, and OgN3:Hg2 were different from those in the SpLE1 group. Although these elements were conserved in the ter operon and in some regions, there were many indels, inversions, and recombination (Figure 2B and Supplementary Figure 3).
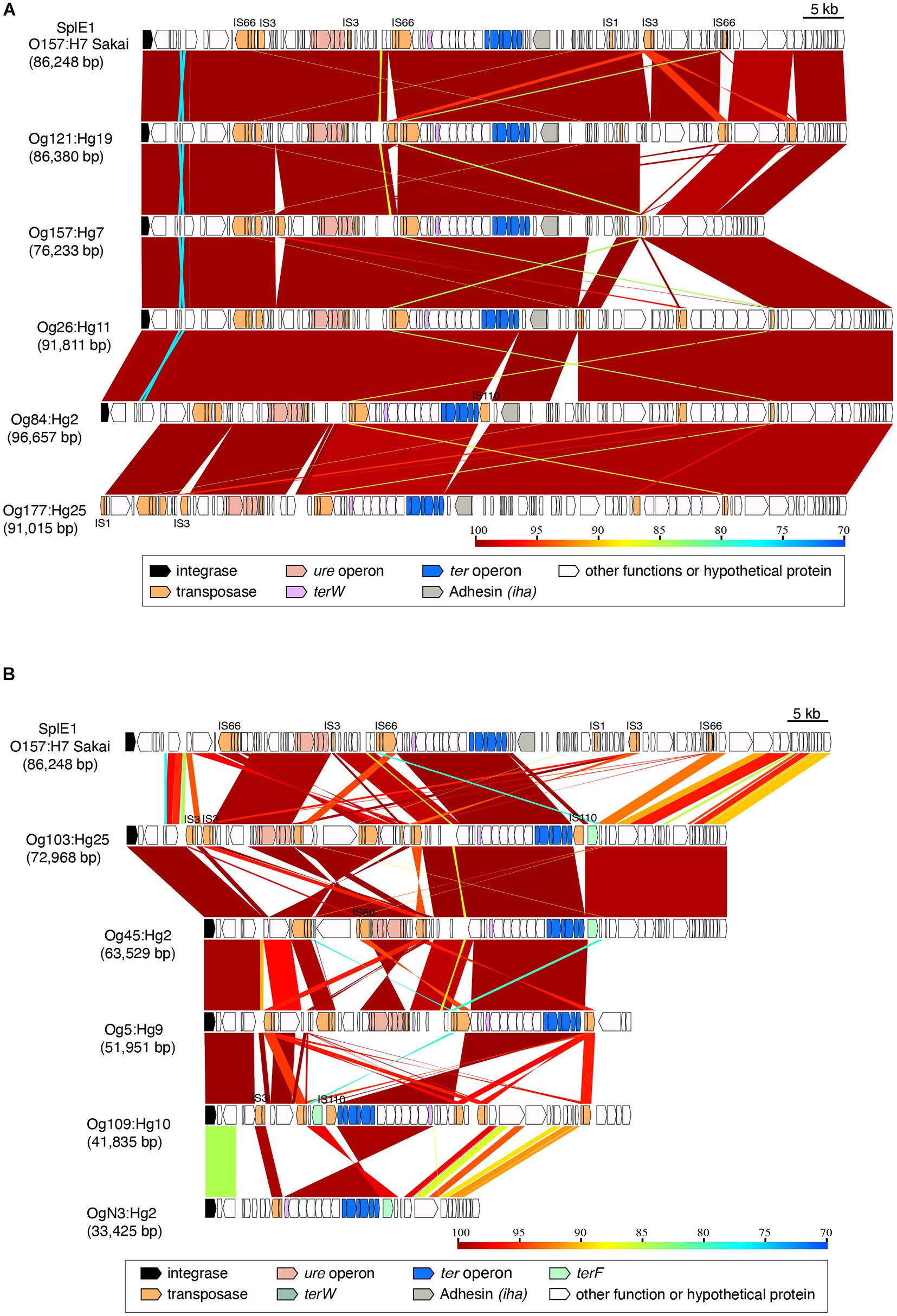
Figure 2. Genomic comparison of the integrating elements carrying ter-type 1. (A) Comparison between SplE1 and SplE1-like elements; (B) comparison among SplE1 with SplE1-like elements and unknown elements. The approximate sizes of the elements are indicated in parentheses. Genes are colored based on their functional characteristics as described in the legend. The nucleotide sequence identities between the elements (cutoff ≥ 70% identity) are indicated by color shading according to the scale shown at the bottom of the figure.
The elements carrying ter-type 3 from Og22:Hg8, Og8:Hg7, Og128ac:Hg2, Og130:Hg11, Og168:Hg19, and Og9:Hg19 (52,460–68,691 bp) were almost identical, except for the upstream region in Og168:Hg19 and Og9:Hg19 (Figure 3 and Supplementary Figure 3). It included an adhesin gene and some transposase genes, including the IS66 and IS4 families.
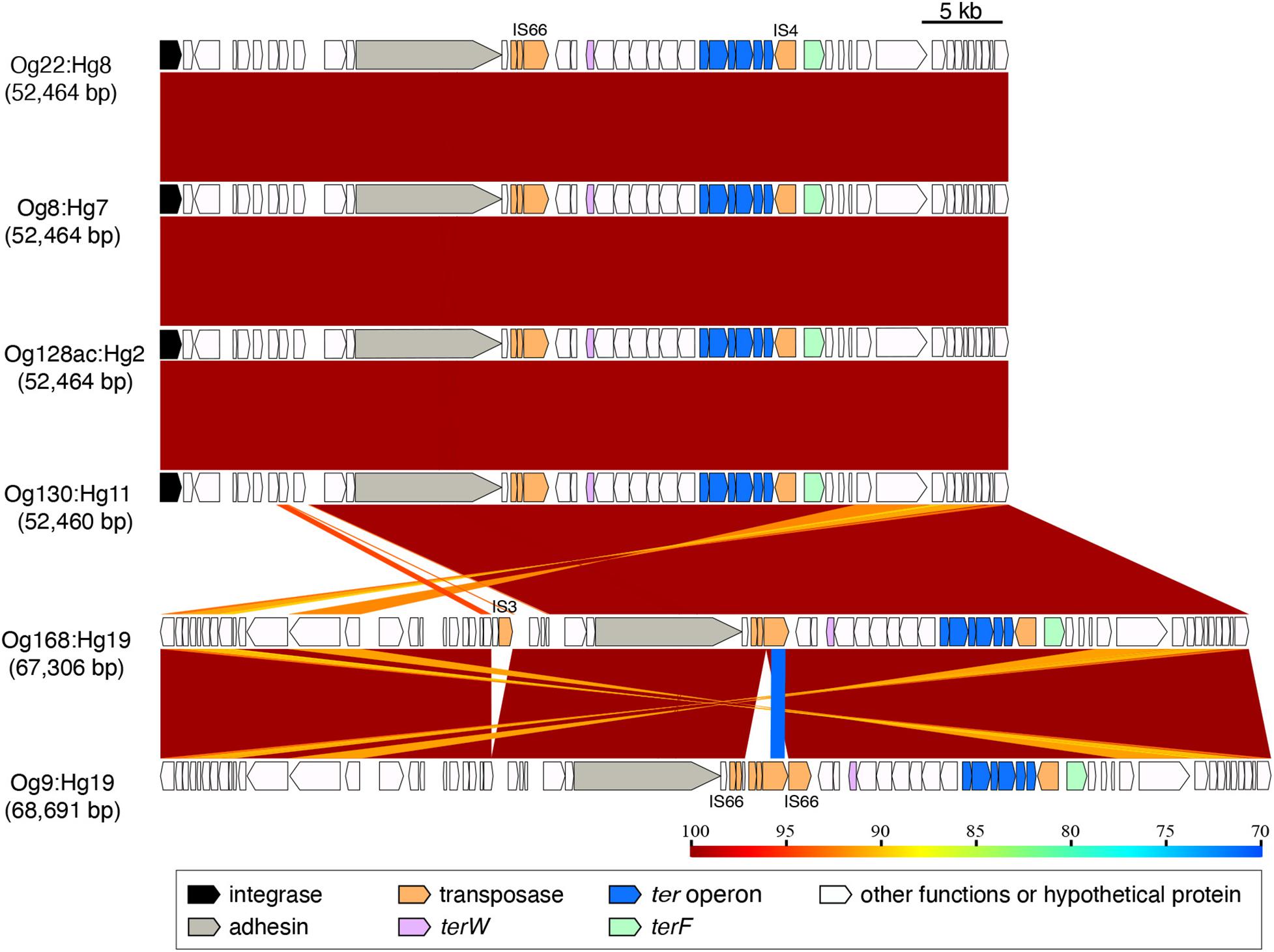
Figure 3. Genomic comparison of the integrating elements carrying ter-type 3. The approximate sizes of the elements are indicated in parentheses. The genes are colored based on the functional characteristics as described in the figure legend. The nucleotide sequence identities between the elements (cutoff ≥ 70% identity) are indicated by color shading according to the scale shown at the bottom of the figure.
The ter-type 4 operons from Og54:Hg45, Og22:Hg16, Og146:Hg21, Og117:Hg4, and Og103:Hg14 were located on elements of different sizes ranging from 116,493 to 256,442 bp (Figure 4). However, the downstream regions of about 100 kb including the ter operon, were conserved, except for that of Og103:Hg14 (Figure 4 and Supplementary Figure 3). Conserved regions contained a plasmid-conjugative system (tra genes), a fatty acid biosynthesis gene cluster, site-specific tyrosine recombinase genes, and some transposase genes, including the IS256 family.
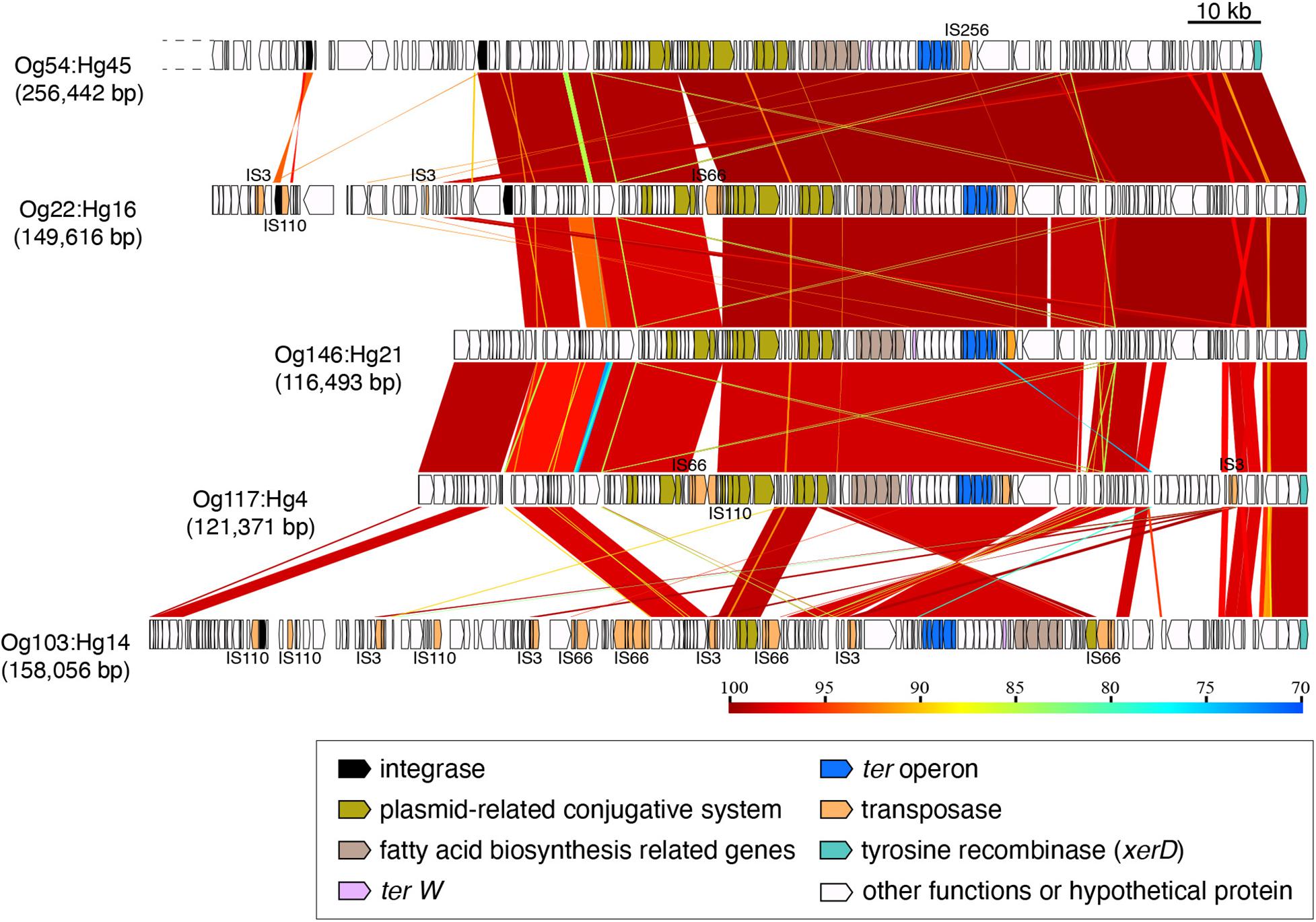
Figure 4. Genomic comparison of the integrating elements carrying ter-type 4. Dashed lines indicate cutoff sequences. The approximate sizes of the elements are indicated in parentheses. Genes are colored based on their functional characteristics as described in the figure legend. The nucleotide sequence identities between the elements (cutoff ≥ 70% identity) are indicated by color shading according to the scale shown at the bottom of the figure.
The operon of ter-type 2 from Og136:Hg16, Og183:Hg18, Og161:Hg4, Og183:Hg42, and Og84:Hg2 was located on IncHI2 plasmids, including the conjugative system (tra genes) and some antibiotic resistant genes [aadA, aph (3’), bla, ble, cml, eptA, and tetA], with sizes ranging from 165,509 to 249,877 bp (Figure 5 and Supplementary Figure 3). Except for the central regions downstream of the ter operons, where multiple recombination and inversions were confirmed, these plasmids roughly retained homologous gene structures. Based on the coverage of genomic sequences, the copy number of plasmids carrying the ter-operons was approximately the same as that of the chromosomes (1 vs. 0.85–1.74).
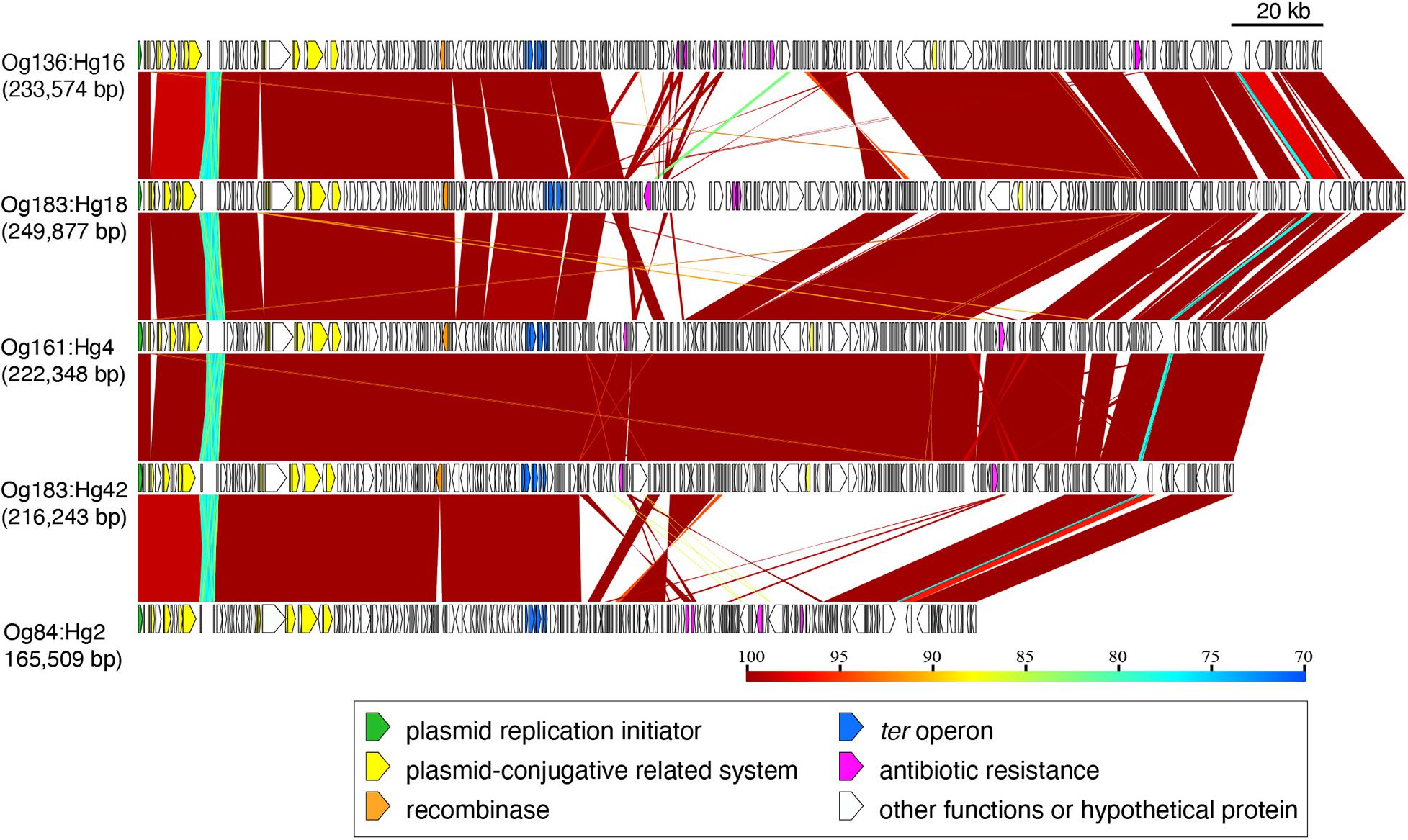
Figure 5. Genomic comparison of the plasmids carrying ter-type 2. The plasmid sizes are indicated in parentheses. Genes are colored based on their functional characteristics as described in the figure legend. The nucleotide sequence identities between the elements (cutoff ≥ 70% identity) are indicated by color shading according to the scale shown at the bottom of the figure.
All ter operons used in this study, which were extracted from the genomes of strains other than E. coli, were located on plasmids, except for a Salmonella-derived ter-type 2 operon, and Proteus- and Yersinia-derived operons, which are on the chromosome (Supplementary Figure 2).
The Insertion Sites of Integrating Elements Carrying the ter Operon on the Chromosome
The insertion sites of elements carrying the ter operon are summarized in Figure 6. tRNASerX and tRNAPheV were the only insertion sites of elements carrying ter-type 1 and 4 operons, respectively. Two positions of tRNAMet (ypjC–ygaQ and mug–yqhH) were the insertion sites for the elements of the ter-type 1 and 3 operons. Although the types of elements carrying the ter-type 1 and 3 operons were different, a homology (98% amino acid sequence) was confirmed in the integrase genes located on the elements inserted in tRNAMet (Supplementary Figure 4). Similarly, the nucleotide sequences of the integrase in the element carrying ter-type 4 (Og54:Hg45 and Og22:Hg16 strain) were highly homologous (upper 97% amino acid sequence) with the integrases in the other integrating elements or the locus of enterocyte effacement that was inserted in the tRNAPhe positions in seven major STEC genomes (Supplementary Table 1) (data not presented). In addition, the element carrying the ter-type 1 operon of Og177:Hg25 was inserted into tRNASerW, the element carrying the ter-type 3 operon of Og9:Hg19 was inserted into tRNAPheU, and the element carrying the ter-type 4 operon of Og54:Hg45 was in tRNAPheU. Although tRNA was absent in the insertion site of the ter operon-carrying elements in the Og5:Hg9 and Og109:Hg10 strains, a corresponding region surrounding the indicated tRNA was present in each strain (Supplementary Figure 5). There was an element other than the ter operon carrier inserted in the tRNAPheU position of the Og9:Hg19 strain (Supplementary Figure 5).
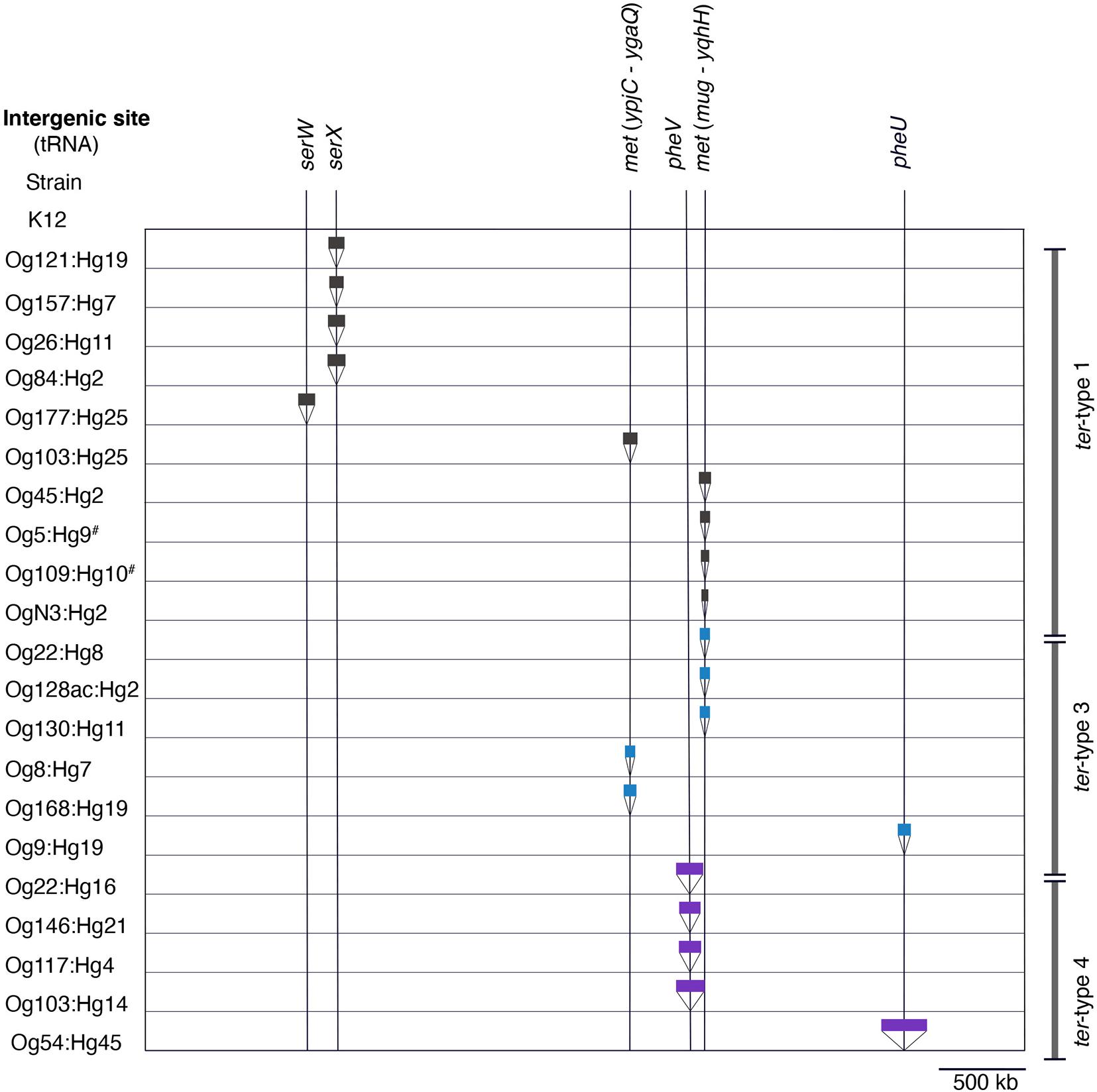
Figure 6. Insertion sites of elements carrying the ter operon in the chromosome. The sites are based on the positions of tRNA in the genome of the K-12 MG1655 strain. Black, blue, and purple bars indicate elements carrying ter-type 1, 3, and 4, respectively. The width of the bars is proportionate with the element size. Hash key indicates the absence of tRNA at the site.
PCR for ter Operon Subtyping
Based on the sequences of the ter operons belonging to four subtypes, we designed type-specific PCR primers, and confirmed that these primers could specifically determine each type (Supplementary Table 4 and Supplementary Figure 6). Using PCR, all 106 ter-positive strains tested were successfully classified into 4 ter-types: type 1 (n = 66), type 2 (n = 13), type 3 (n = 8), and type 4 (n = 17), except for two strains belonging to Og84:Hg2, which were positive for both ter-type 1 and 2 (Supplementary Table 3). All eae-positive strains, including seven major STEC serotype strains, were positive for ter-type 1 (n = 46), whereas eae-negative E. coli strains, including STEC and non-STEC, were positive for ter-type 2 (n = 13), type 3 (n = 8), and type 4 (n = 17), in addition to ter-type 1 (n = 21) (Supplementary Table 3).
Distribution of ter-Types in Escherichia coli Linages
The phylogenetic tree of 80 terA-positive E. coli strains (one representative strain of the same O:H genotype and the same ter-type was selected from 106 strains) is shown in Figure 7. Overall, four ter-types were distributed in various lineages of E. coli including A, B1, B2, E, D, A × B1, and ABD (Supplementary Table 3), although some strains with the same ter-type formed several small clades.
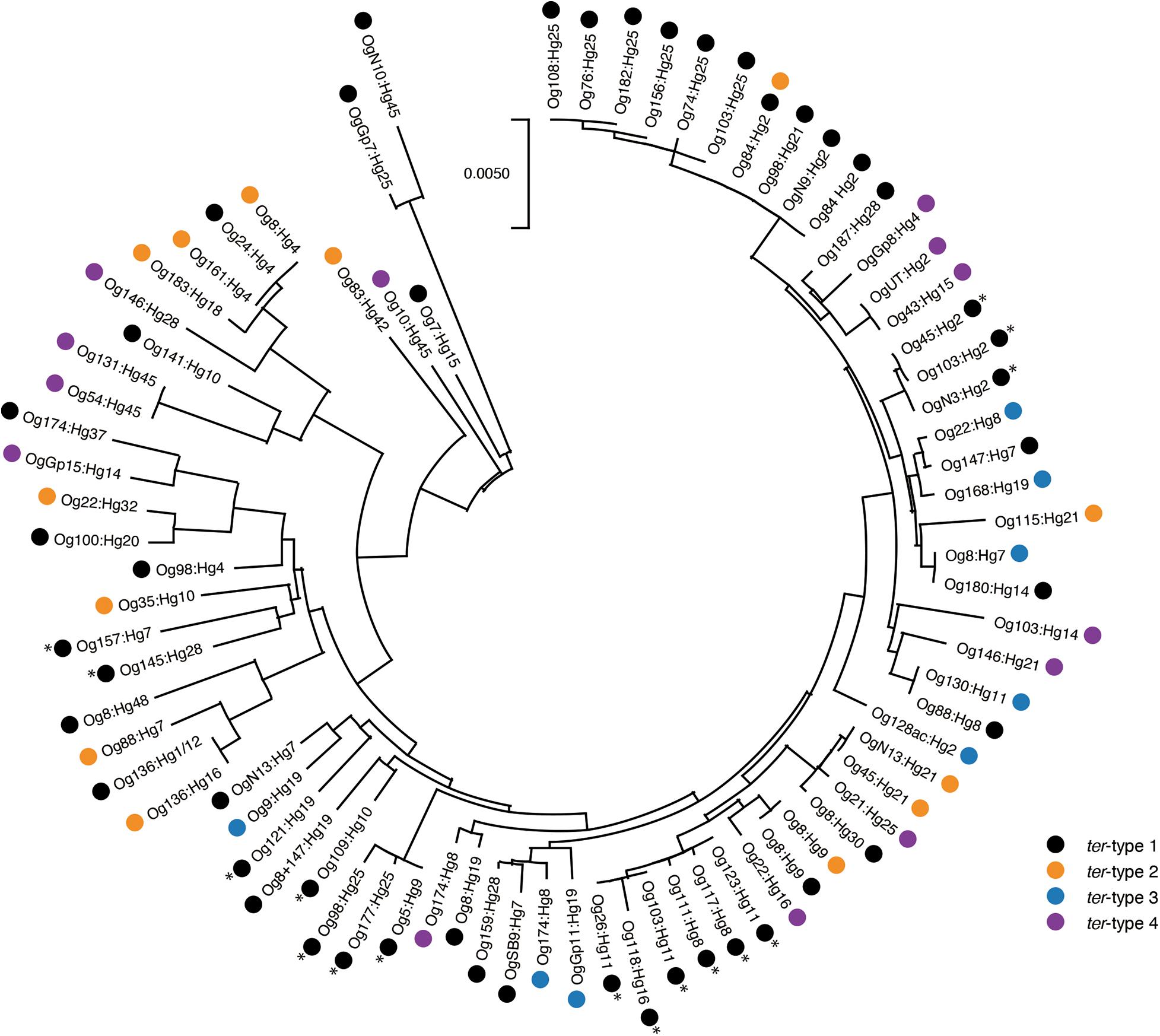
Figure 7. Phylogenetic tree of 80 terA-positive Escherichia coli strains. The tree was built based on the internal concatenated sequences of seven housekeeping genes (3,423 bp). The asterisk indicates eae-positive strains.
Tellurite Resistance
The 106 ter-positive E. coli strains showed MICs ranging from 16 to 256 μg/mL of potassium tellurite (Supplementary Table 5), except for a ter-type 1 strain (KAP39, Og98:Hg4), which was tellurite-sensitive. The genome sequence of this strain had an IS5 inserted on the terZ due to which the gene was disrupted (Supplementary Figure 7). Based on the ter subtype, seventy-five percent of the strains were classified as ter-type 1, 2, and 3 were resistant to potassium tellurite concentrations of 64–128 μg/mL or higher, whereas only one strain (6%) in ter-type 4 was resistant to 64–128 μg/mL (Supplementary Table 5). There was no significant difference in the resistance among ter-type 1, 2, and 3 groups, whereas the resistance activity of the ter-type 4 group was significantly lower than that of the other three types (p < 0.05) (Figure 8).
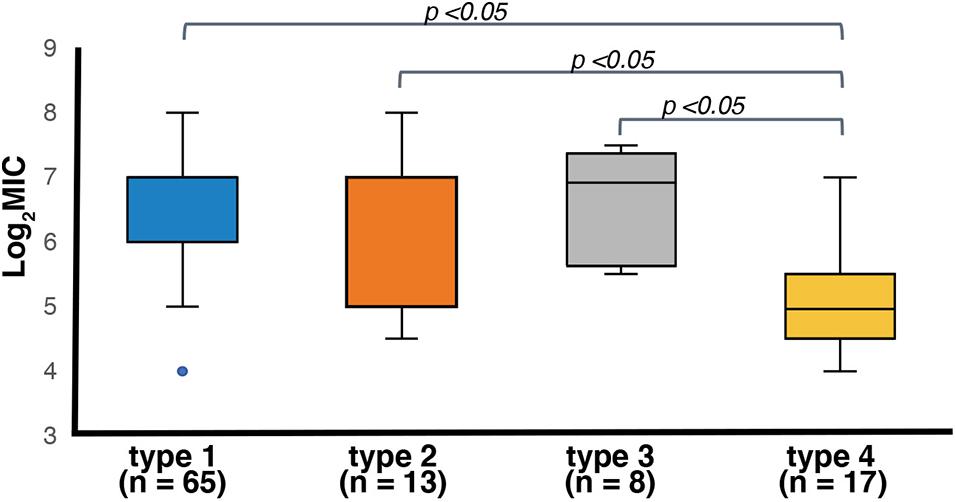
Figure 8. Box plots of log2 transformed MIC distributions of potassium tellurite (K2TeO3) based on the ter operon type. Boxes indicate the 25th, 50th, and 75th percentiles. Whiskers indicate the lowest and highest data point. Outliers are shown as blue circles. P-values of statistically significant differences are shown (p < 0.05).
Discussion
The gene composition and sequence homology of the ter operon carrier elements on the chromosomes were roughly conserved for each of the ter-types, 1, 3, and 4 (Supplementary Figure 3), and strains belonging to different phylogenetic lineages carried the same types of elements (Figure 7), suggesting that at least three different types of chromosomal elements carrying the ter operons have spread horizontally within E. coli species. Six tRNA genes on the chromosome were confirmed as insertion sites for these elements, and were classified into three tRNA types: tRNASer, tRNAMet, and tRNAPhe. The integrase genes on the elements carrying the ter-type 1 operon were divided into two types according to the sequence homology: elements with one type of integrase gene were inserted into tRNASer (as seen in strains belonging to O157:H7, and so on) and elements with the other type of integrase gene were inserted into tRNAMet (as seen in strains belonging to O103:H25, and so on) (Figures 2, 6). The latter integrase gene showed high homology with that of ter-type 3 operon-carrying elements (Supplementary Figure 4), and all elements with the ter-type 3 operon were also inserted into tRNAMet, except for one strain (A150083 of Og9:Hg19) (Figure 6). Although the integrase gene was found only in two of the five ter-type 4 operon carrier elements used in this study, these two were conserved and highly homologous with the integrase in other elements of other strains that were all inserted into tRNAPhe. These results suggest that the insertion site of these elements on the chromosome depends strongly on the integrase genes and not on the basic structure of the elements. Transposases such as IS66 and IS256 were located upstream and/or downstream of the ter operons, and especially in some ter-type 1 operons, they coincided with the recombination and inversion junction points, suggesting that IS transposase was involved in the transfer of small fragments, including ter operons, to other elements.
The ter-type 2 operons were located on the IncHI2 plasmids of E. coli strains, and were also found on the IncHI2 plasmids carried by Y. pseudotuberculosis and K. pneumoniae (Supplementary Figure 2), suggesting that this type of operon was horizontally transferred in E. coli as well as within the Enterobacteriaceae through plasmids. The conjugative IncHI2 plasmid is widely found in Enterobacteriaceae and is known to mediate various drug resistance genes (Herrera-León et al., 2011; Roberts et al., 2020). In fact, the plasmids found to harbor the ter-type 2 operons in this study carried three or more drug resistance genes (Figure 5). In this study, most of the ter-type 2 operons were found in isolates from chickens (47%, 7/15). In recent years, large amounts of antibacterial drugs have been used in broiler production, and the resulting emergence and spread of drug-resistant bacteria has become a global public health concern (Nhung et al., 2017; Mehdi et al., 2018; Yang et al., 2019). The IncHI2 plasmid carrying drug resistance genes may have thus been selected in the gastrointestinal tract of chickens after many years of antibacterial drug use, and the ter operon coexisting on the plasmid was co-selected simultaneously.
All E. coli strains used in this study were isolated in Japan. However, the sequences of the ter operons obtained from the DNA database used in Figure 1 originated from E. coli isolated outside Japan (14 strains belonging to type 1, 4 strains to type 2, 3 strains to type 3, and 2 strains to type 4), except for 4 strains belonging to type 1 (O157:H7, O103:H2, 0111: H-, and O26:H11) (see Supplementary Table 1). These results suggested that the characteristics of the ter operon revealed in this study were not only those found in E. coli isolated in Japan, but also common to E. coli isolated outside Japan.
Strains carrying the ter-type 4 operon showed significantly lower MICs of potassium tellurite compared to the other three types. The function of Ter proteins has been suggested to involve their mutual association, as they interact at the interface of the inner plasma membrane and the cytosol (Valkovicova et al., 2013; Turkovicova et al., 2016). The nucleotide and amino acid sequences of genes on the ter-type 4 operon were significantly different from those of the other three types (Supplementary Figure 1), suggesting that these differences directly affected the functionality of Ter proteins. Most of the ter-type 4 operons were found in isolates from wild deer (94%, 16/17). Of the isolation sources used in this study, deer were the only animals from the wild environment, and their feces were collected from a wide area ranging from Hokkaido in the north, to Kyushu in the south of Japan. In addition to tellurite resistance, the ter operon is also involved in resistance to infection by various bacteriophages and to membrane-pore forming colicins (Whelan et al., 1995, 1997; Alonso et al., 2000), tolerance to oxidative stresses (Valková et al., 2007), resistance to phagocytosis by macrophages (Ponnusamy and Clinkenbeard, 2015), the ability of cells to adhere to epithelial cells in vitro (Yin et al., 2009), and filamentous bacterial cellular morphology (Whelan et al., 1997). In addition, a ter homolog from Clostridium acetobutylicum showed resistance to methyl methane sulfonate and mitomycin C when expressed in an E. coli recA mutant (Azeddoug and Reysset, 1994). Bacterial acquisition of the ter operon may thus be beneficial for colonization of the host intestinal tract in the presence of various stresses and attacks. Although the reason for the strong relationship between the ter type-4 operon and deer is currently unknown, food and water in the wild environment, and the wild deer gut microbiome formed by their effects, may select strains carrying this type of ter-operon.
The tellurite-resistant phenotypes were variable within each type. For example, five strains of Og177: Hg25 with the ter-type 1 operon showed MICs of potassium tellurite ranging from 16 to 128 μg/mL. Furthermore, the amino acid sequences of the ter-type 1 operon genes of major STEC O26:H11 (11,368 strain) was identical to those of Og121:Hg19 (TS23_1 strain) (data not show). However, the resistance level of the latter strain (16–32 μg/mL of potassium tellurite) was roughly lower than that of the former (64–128 μg/mL). A previous study has also shown that even O157:H7 serotype strains differ in their MICs of potassium tellurite (64–1,024 μg/mL) (Taylor et al., 2002). The above results indicate that the ter operon as well as other genetic factors influence tellurite resistance.
Conclusion
This study compared the details of ter operons responsible for tellurite resistance among various serotypes and sources of E. coli. Comparative analysis clearly distinguished the ter operon sequences into four subtypes, namely, ter-type 1–4 operons. The amino acid sequences of the ter-type 4 operon genes were distant from those of the other types, further, strains with the ter-type 4 operons showed significantly lower MICs compared to the other types. Furthermore, genomic analysis using long reads indicated the structural details of the integrating elements carrying the ter operons and insertion sites on the chromosome. Genomic comparison and phylogenetic analysis showed that at least three highly conserved elements were spread horizontally within E. coli species. Type 2 was found to be located on plasmids distributed in E. coli as well as in some Enterobacteriaceae species. The ter-type was also closely related to the source of isolation, the cause of which was unknown; however, types 2 and 4 were associated with chickens and deer, respectively. This study provided new insights related not only to genetic characteristics of the ter operons in E. coli, but also to phenotypic and ecological differences that may be related to them. In the future, by clarifying the unknown function of the Ter proteins, it is expected that the relationship between the diversity of the ter operon and the phenotypic and ecological characteristics revealed in this study will be clarified.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TN, AI, and SI conceived and designed the experiments. TN performed the experiments. TN, TK, and TT analyzed the data. TN and AI contributed to reagents, materials, analysis tools, critical revision of the manuscript for important intellectual content, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was partially supported by AMED under grant no. JP20fk0108065.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yuiko Kato for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.681175/full#supplementary-material
Footnotes
References
Alonso, G., Gomes, C., González, C., and Rodríguez Lemoine, V. (2000). On the mechanism of resistance to channel-forming colicins (PacB) and tellurite, encoded by plasmid Mip233 (IncHI3). FEMS Microbiol. Lett. 192, 257–261. doi: 10.1111/j.1574-6968.2000.tb09391.x
Avazéri, C., Turner, R. J., Pommier, J., Weiner, J. H., Giordano, G., and Verméglio, A. (1997). Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology (Reading) 143, 1181–1189. doi: 10.1099/00221287-143-4-1181
Azeddoug, H., and Reysset, G. (1994). Cloning and sequencing of a chromosomal fragment from Clostridium acetobutylicum strain ABKn8 conferring chemical-damaging agents and UV resistance to E. coli recA strains. Curr. Microbiol. 29, 229–235. doi: 10.1007/bf01570159
Banjo, M., Iguchi, A., Seto, K., Kikuchi, T., Harada, T., Scheutz, F., et al. (2018). Escherichia coli H-genotyping PCR: a complete and practical platform for molecular H typing. J. Clin. Microbiol. 56, e190–e218. doi: 10.1128/jcm.00190-18
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cabiscol, E., Tamarit, J., and Ros, J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8.
Calderón, I. L., Arenas, F. A., Pérez, J. M., Fuentes, D. E., Araya, M. A., Saavedra, C. P., et al. (2006). Catalases are NAD(P)H-dependent tellurite reductases. PLoS One 1:e70. doi: 10.1371/journal.pone.0000070
Calderón, I. L., Elías, A. O., Fuentes, E. L., Pradenas, G. A., Castro, M. E., Arenas, F. A., et al. (2009). Tellurite-mediated disabling of [4Fe–4S] clusters of Escherichia coli dehydratases. Microbiology (Reading) 155, 1840–1846. doi: 10.1099/mic.0.026260-0
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents. Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Cebula, T. A., Payne, W. L., and Feng, P. (1995). Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33, 248–250. doi: 10.1128/jcm.33.1.248-250.1995
Clinical and Laboratory Standards Institutes [CLSI] (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard, CLSI document M07-A9, 9th Edn. Wayne, PA: CLSI.
De Coster, W., D’hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Díaz-Vásquez, W. A., Abarca-Lagunas, M. J., Arenas, F. A., Pinto, C. A., Cornejo, F. A., Wansapura, P. T., et al. (2014). Tellurite reduction by Escherichia coli NDH-II dehydrogenase results in superoxide production in membranes of toxicant-exposed cells. Biometals 27, 237–246. doi: 10.1007/s10534-013-9701-8
Ferdous, M., Zhou, K., De Boer, R. F., Friedrich, A. W., Kooistra-Smid, A. M. D., and Rossen, J. W. A. (2015). Comprehensive characterization of Escherichia coli O104:H4 isolated from patients in the Netherlands. Front. Microbiol. 6:1348. doi: 10.3389/fmicb.2015.01348
Gouali, M., Ruckly, C., Carle, I., Lejay-Collin, M., and Weill, F.-X. (2013). Evaluation of CHROMagar STEC and STEC O104 chromogenic agar media for detection of Shiga toxin-producing Escherichia coli in stool specimens. J. Clin. Microbiol. 51, 894–900. doi: 10.1128/JCM.03121-12
Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., et al. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22. doi: 10.1093/dnares/8.1.11
Herrera-León, S., González-Sanz, R., Herrera-León, L., and Echeita, M. A. (2011). Characterization of multidrug-resistant Enterobacteriaceae carrying plasmid-mediated quinolone resistance mechanisms in Spain. J. Antimicrob. Chemother 66, 287–290. doi: 10.1093/jac/dkq423
Iguchi, A., Iyoda, S., Seto, K., Morita-Ishihara, T., Scheutz, F., and Ohnishi, M. (2015). Escherichia coli O-Genotyping PCR: a comprehensive and practical platform for molecular O serogrouping. J. Clin. Microbiol. 53, 2427–2432. doi: 10.1128/jcm.00321-15
Iguchi, A., Nishii, H., Seto, K., Mitobe, J., Lee, K., Konishi, N., et al. (2020). Additional Og-typing PCR techniques targeting E. coli-novel and Shigella-unique O-antigen biosynthesis gene clusters. J. Clin. Microbiol. 58, e1493–e1520. doi: 10.1128/JCM.01493-20
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
Jobling, M. G., and Ritchie, D. A. (1987). Genetic and physical analysis of plasmid genes expressing inducible resistance of tellurite in Escherichia coli. Mol. Gen. Genet. 208, 288–293. doi: 10.1007/bf00330455
Jobling, M. G., and Ritchie, D. A. (1988). The nucleotide sequence of a plasmid determinant for resistance to tellurium anions. Gene 66, 245–258. doi: 10.1016/0378-1119(88)90361-7
Kalule, J. B., Keddy, K. H., and Nicol, M. P. (2018). Characterisation of STEC and other diarrheic E. coli isolated on CHROMagarTM STEC at a tertiary referral hospital, Cape Town. BMC Microbiol. 18:55. doi: 10.1186/s12866-018-1195-7
Kormutakova, R., Klucar, L., and Turna, J. (2000). DNA sequence analysis of the tellurite-resistance determinant from clinical strain of Escherichia coli and identification of essential genes. Biometals 13, 135–139. doi: 10.1023/a:1009272122989
Ling, J., and Söll, D. (2010). Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 107, 4028–4033. doi: 10.1073/pnas.1000315107
Lithgow, J. K., Hayhurst, E. J., Cohen, G., Aharonowitz, Y., and Foster, S. J. (2004). Role of a cysteine synthase in Staphylococcus aureus. J. Bacteriol. 186, 1579–1590. doi: 10.1128/jb.186.6.1579-1590.2004
Lorenz, S. C., Gonzalez-Escalona, N., Kotewicz, M. L., Fischer, M., and Kase, J. A. (2017). Genome sequencing and comparative genomics of enterohemorrhagic Escherichia coli O145:H25 and O145:H28 reveal distinct evolutionary paths and marked variations in traits associated with virulence & colonization. BMC Microbiol. 17:183. doi: 10.1186/s12866-017-1094-3
Mehdi, Y., Létourneau-Montminy, M.-P., Gaucher, M.-L., Chorfi, Y., Suresh, G., Rouissi, T., et al. (2018). Use of antibiotics in broiler production: global impacts and alternatives. Anim. Nut. 4, 170–178. doi: 10.1016/j.aninu.2018.03.002
Miko, A., Rivas, M., Bentancor, A., Delannoy, S., Fach, P., and Beutin, L. (2014). Emerging types of Shiga toxin-producing E. coli (STEC) O178 present in cattle, deer, and humans from Argentina and Germany. Front. Cell. Infect. Microbiol. 4:78. doi: 10.3389/fcimb.2014.00078
Molina-Quiroz, R. C., Loyola, D. E., Muñoz-Villagrán, C. M., Quatrini, R., Vásquez, C. C., and Pérez-Donoso, J. M. (2013). DNA, cell wall and general oxidative damage underlie the tellurite/cefotaxime synergistic effect in Escherichia coli. PloS One 8:e79499. doi: 10.1371/journal.pone.0079499
Nguyen, N., Iguchi, A., Ohata, R., Kawai, H., Ooka, T., Nakajima, H., et al. (2021). Distribution of novel Og-types in Shiga toxin-producing Escherichia coli isolated from healthy cattle. J. Clin. Microbiol. 59, e02624–e02630. doi: 10.1128/JCM.02624-20
Nhung, N. T., Chansiripornchai, N., and Carrique-Mas, J. J. (2017). Antimicrobial resistance in bacterial poultry pathogens: a review. Front. Vet. Sci. 4:126. doi: 10.3389/fvets.2017.00126
Ogura, Y., Ooka, T., Iguchi, A., Toh, H., Asadulghani, M., Oshima, K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Nat. Acad. Sci. U.S.A. 106, 17939–17944. doi: 10.1073/pnas.0903585106
Ohnishi, M., Terajima, J., Kurokawa, K., Nakayama, K., Murata, T., Tamura, K., et al. (2002). Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. U.S.A. 99, 17043–17048. doi: 10.1073/pnas.262441699
Ohtsubo, Y., Ikeda-Ohtsubo, W., Nagata, Y., and Tsuda, M. (2008). GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinform. 9:376. doi: 10.1186/1471-2105-9-376
Orth, D., Grif, K., Dierich, M. P., and Wurzner, R. (2007). Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res. Microbiol. 158, 105–111. doi: 10.1016/j.resmic.2006.10.007
Pérez, J. M., Calderón, I. L., Arenas, F. A., Fuentes, D. E., Pradenas, G. A., Fuentes, E. L., et al. (2007). Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. doi: 10.1371/journal.pone.0000211
Ponnusamy, D., and Clinkenbeard, K. D. (2015). Role of tellurite resistance operon in filamentous growth of Yersinia pestis in macrophages. PLoS One 10:e0141984. doi: 10.1371/journal.pone.0141984
Roberts, L. W., Catchpoole, E., Jennison, A. V., Bergh, H., Hume, A., Heney, C., et al. (2020). Genomic analysis of carbapenemase-producing Enterobacteriaceae in Queensland reveals widespread transmission of blaIMP–4 on an IncHI2 plasmid. Microb. Genom. 6:e000321. doi: 10.1099/mgen.0.000321
Rojas, D. M., and Vásquez, C. C. (2005). Sensitivity to potassium tellurite of Escherichia coli cells deficient in CSD, CsdB and IscS cysteine desulfurases. Res. Microbiol. 156, 465–471. doi: 10.1016/j.resmic.2004.12.010
Sabaty, M., Avazeri, C., Pignol, D., and Vermeglio, A. (2001). Characterization of the reduction of selenate and tellurite by nitrate reductases. Appl. Environ. Microbiol. 67, 5122–5126. doi: 10.1128/aem.67.11.5122-5126.2001
Sandoval, J. M., Arenas, F. A., García, J. A., Díaz-Vásquez, W. A., Valdivia-González, M., Sabotier, M., et al. (2015). Escherichia coli 6-phosphogluconate dehydrogenase aids in tellurite resistance by reducing the toxicant in a NADPH-dependent manner. Microbiol. Res. 177, 22–27. doi: 10.1016/j.micres.2015.05.002
Sandoval, J. M., Arenas, F. A., and Vásquez, C. C. (2011). Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS One 6:e25573. doi: 10.1371/journal.pone.0025573
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Soltys, K., Vavrova, S., Budis, J., Palkova, L., Minarik, G., and Grones, J. (2018). Draft genome sequence of Escherichia coli KL53. Genome Announc. 6, e220–e318. doi: 10.1128/genomeA.00220-18
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239. doi: 10.1093/molbev/msz312
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2017). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Tantaleán, J. C., Araya, M. A., Saavedra, C. P., Fuentes, D. E., Pérez, J. M., Calderón, I. L., et al. (2003). The Geobacillus stearothermophilus V iscS gene, encoding cysteine desulfurase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 185, 5831–5837. doi: 10.1128/jb.185.19.5831-5837.2003
Taylor, D. E., Rooker, M., Keelan, M., Ng, L.-K., Martin, I., Perna, N. T., et al. (2002). Genomic variability of O islands encoding tellurite resistance in Enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184, 4690–4698. doi: 10.1128/JB.184.17.4690-4698.2002
Tremaroli, V., Fedi, S., and Zannoni, D. (2007). Evidence for a tellurite-dependent generation of reactive oxygen species and absence of a tellurite-mediated adaptive response to oxidative stress in cells of Pseudomonas pseudoalcaligenes KF707. Arch. Microbiol. 187, 127–135. doi: 10.1007/s00203-006-0179-4
Turkovicova, L., Smidak, R., Jung, G., Turna, J., Lubec, G., and Aradska, J. (2016). Proteomic analysis of the TerC interactome: novel links to tellurite resistance and pathogenicity. J. Proteom. 136, 167–173. doi: 10.1016/j.jprot.2016.01.003
Turner, R. J., Aharonowitz, Y., Weiner, J. H., and Taylor, D. E. (2001). Glutathione is a target in tellurite toxicity and is protected by tellurite resistance determinants in Escherichia coli. Can. J. Microbiol. 47, 33–40.
Turner, R. J., Hou, Y., Weiner, J. H., and Taylor, D. E. (1992). The arsenical ATPase efflux pump mediates tellurite resistance. J. Bacteriol. 174, 3092–3094. doi: 10.1128/jb.174.9.3092-3094.1992
Turner, R. J., Weiner, J. H., and Taylor, D. E. (1995). The tellurite-resistance determinants tehA tehB and klaAklaBtelB have different biochemical requirements. Microbiology 141, 3133–3140. doi: 10.1099/13500872-141-12-3133
Valková, D., Valkovičová, L., Vávrová, S., Kováčová, E., Mravec, J., and Turňa, J. (2007). The contribution of tellurite resistance genes to the fitness of Escherichia coli uropathogenic strains. Cen. Eur. J. Biol. 2, 182–191. doi: 10.2478/s11535-007-0019-9
Valkovicova, L., Vavrova, S. M., Mravec, J., Grones, J., and Turna, J. (2013). Protein-protein association and cellular localization of four essential gene products encoded by tellurite resistance-conferring cluster “ter” from pathogenic Escherichia coli. Antonie Van Leeuwenhoek 104, 899–911. doi: 10.1007/s10482-013-0009-z
Vásquez, C. C., Saavedra, C. P., Loyola, C. A., Araya, M. A., and Pichuantes, S. (2001). The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43, 418–423. doi: 10.1007/s002840010331
Verhaegen, B., De Reu, K., Heyndrickx, M., and De Zutter, L. (2015). Comparison of six chromogenic agar media for the isolation of a broad variety of non-O157 Shiga toxin-producing Escherichia coli (STEC) serogroups. Int. J. Environ. Res. Public Health 12, 6965–6978. doi: 10.3390/ijerph120606965
Walter, E. G., Thomas, C. M., Ibbotson, J. P., and Taylor, D. E. (1991). Transcriptional analysis, translational analysis, and sequence of the kilA-tellurite resistance region of plasmid RK2Ter. J. Bacteriol. 173, 1111–1119. doi: 10.1128/jb.173.3.1111-1119.1991
Whelan, K. F., Colleran, E., and Taylor, D. E. (1995). Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 177, 5016–5027. doi: 10.1128/jb.177.17.5016-5027.1995
Whelan, K. F., Sherburne, R. K., and Taylor, D. E. (1997). Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J. Bacteriol. 179:63. doi: 10.1128/jb.179.1.63-71.1997
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Yang, Y., Ashworth, A. J., Willett, C., Cook, K., Upadhyay, A., Owens, P. R., et al. (2019). Review of antibiotic resistance, ecology, dissemination, and mitigation in U.S. broiler poultry systems. Front. Microbiol. 10:2639. doi: 10.3389/fmicb.2019.02639
Yin, X., Wheatcroft, R., Chambers, J. R., Liu, B., Zhu, J., and Gyles, C. L. (2009). Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75, 5779–5786. doi: 10.1128/aem.00507-09
Keywords: ter operon, tellurite resistance, Shiga toxin-producing Escherichia coli, integrating genetic element, horizontal genetic transfer
Citation: Nguyen TTH, Kikuchi T, Tokunaga T, Iyoda S and Iguchi A (2021) Diversity of the Tellurite Resistance Gene Operon in Escherichia coli. Front. Microbiol. 12:681175. doi: 10.3389/fmicb.2021.681175
Received: 16 March 2021; Accepted: 30 April 2021;
Published: 28 May 2021.
Edited by:
Ruiting Lan, University of New South Wales, AustraliaReviewed by:
Guojie Cao, Center for Food Safety and Applied Nutrition, United StatesMarcello Trevisani, University of Bologna, Italy
David Needleman, Delaware Valley University, United States
Copyright © 2021 Nguyen, Kikuchi, Tokunaga, Iyoda and Iguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atsushi Iguchi, aWd1Y2hpQG1lZC5taXlhemFraS11LmFjLmpw
 Thi Thu Huong Nguyen
Thi Thu Huong Nguyen Taisei Kikuchi
Taisei Kikuchi Tadaaki Tokunaga
Tadaaki Tokunaga Sunao Iyoda4
Sunao Iyoda4 Atsushi Iguchi
Atsushi Iguchi