- 1Department of Biochemistry and Molecular and Cellular Biology, Georgetown University Medical Center, Washington, DC, United States
- 2Lombardi Comprehensive Cancer Center, Georgetown University Medical Center, Washington DC, United States
In Escherichia coli, repression of phosphatidylglycerol synthase A gene (pgsA) lowers the levels of membrane acidic phospholipids, particularly phosphatidylglycerol (PG), causing growth-arrested phenotype. The interrupted synthesis of PG is known to be associated with concomitant reduction of chromosomal content and cell mass, in addition to accumulation of unprocessed outer membrane lipoprotein intermediate, pro-Lpp, at the inner membrane. However, whether a linkage exists between the two altered-membrane outcomes remains unknown. Previously, it has been shown that pgsA+ cells overexpressing mutant Lpp(C21G) protein have growth defects similar to those caused by the unprocessed pro-Lpp intermediate in cells lacking PG. Here, we found that the ectopic expression of DnaA(L366K) or deletion of fis (encoding Factor for Inversion Stimulation) permits growth of cells that otherwise would be arrested for growth due to accumulated Lpp(C21G). The DnaA(L366K)-mediated restoration of growth occurs by reduced expression of Lpp(C21G) via a σE-dependent small-regulatory RNA (sRNA), MicL-S. In contrast, restoration of growth via fis deletion is only partially dependent on the MicL-S pathway; deletion of fis also rescues Lpp(C21G) growth arrest in cells lacking physiological levels of PG and cardiolipin (CL), independently of MicL-S. Our results suggest a close link between the physiological state of the bacterial cell membrane and DnaA- and Fis-dependent growth.
Introduction
In Escherichia coli, DnaA protein initiates chromosomal DNA replication once, and only once, per cell cycle (Bramhill and Kornberg, 1988; Marszalek and Kaguni, 1994; Baker and Bell, 1998). DnaA shows a high affinity for both ATP (KD = 0.03 μM) and ADP (KD = 0.1 μM) (Sekimizu et al., 1987). Early biochemical studies established the role of acidic phospholipids in the in vitro rejuvenation of inactive ADP-DnaA to active ATP-DnaA (Sekimizu and Kornberg, 1988; Yung and Kornberg, 1988; Castuma et al., 1993). Limited proteolysis of DnaA in the presence of acidic phospholipids to generate functional fragments revealed a specific region of DnaA involved in membrane interaction (Garner and Crooke, 1996). Independently, the same region was found to insert into fluid acidic phospholipid bilayers when probed with a photoactivatable lipid analog (Garner et al., 1998). Cytolocalization studies involving an allelic replacement of the chromosomal copy of dnaA with a gene encoding GFP-DnaA demonstrated a discrete, longitudinal helical arrangement of GFP-DnaA along the cell periphery (Boeneman et al., 2009), supporting that DnaA is located along the inner leaflet of the cytoplasmic membrane.
Genetic studies on acidic phospholipid synthesis revealed that the absence of a functional gene for phosphatidylglycerol phosphate synthase, pgsA (Heacock and Dowhan, 1987; Xia and Dowhan, 1995), results in arrested growth due to reduced levels of phosphatidylglycerol (PG) and cardiolipin (CL). Site-directed mutagenesis of the membrane-binding region of DnaA offered a novel perspective on membrane–DnaA initiator protein interactions: a single-point mutation in DnaA at the hydrophobic face of the amphipathic helix, DnaA(L366K), allows growth in cells with depleted acidic phospholipid content (Zheng et al., 2001). DnaA(L366K) cannot serve as the only source of DnaA to initiate DNA synthesis both in vivo (Zheng et al., 2001) and in vitro (Saxena et al., 2011) and requires limiting levels of wild-type DnaA to function. Biochemical characterization of DnaA(L366K) demonstrated that it behaves like DnaA with respect to nucleotide-binding affinities, ATP hydrolysis, and specificity for PG and CL in promoting nucleotide exchange (Li et al., 2005). In vitro DMS-footprinting demonstrated that DnaA(L366K) fails to saturate low-affinity recognition sites I2 and I3 irrespective of its adenine nucleotide-bound state. However, in the presence of a limiting amount of wild-type DnaA, DnaA(L366K) promotes the formation of replication-competent DnaA–oriC complexes (Saxena et al., 2011). Moreover, it has also been shown that membrane-mediated DnaA rejuvenation is strongly cooperative with respect to DnaA membrane occupancy. In vitro, when DnaA remains present on dioleolylPG liposomes, it could transit between two occupancy states: high-density occupancy (state I) and low-density occupancy (state II) states (Aranovich et al., 2006, 2015). Only low-density occupancy state II is efficient in facilitating the nucleotide exchange that is presumably promoted by macromolecular crowding (Aranovich et al., 2006, 2015). DnaA(L366K) and is shown to require relatively lower amounts of membrane acidic phospholipids to facilitate nucleotide exchange (Aranovich et al., 2007).
In addition to DnaA–acidic membrane association, PG is also required for the biogenesis of outer membrane lipoprotein, Lpp (also known as Braun’s lipoprotein), which is the most abundant protein in the E. coli cell envelope (Inouye, 1979). The maturation of pro-lipoprotein (pro-Lpp) involves a series of enzyme-catalyzed reactions starting with lipoprotein diacylglyceryl transferase (Lgt), which catalyzes the transfer of diacylglycerol (DAG) from PG to cysteine 21 of pro-Lpp (Braun, 1975; Sankaran and Wu, 1994; Mao et al., 2016). Signal peptidase II (Lsp) recognizes the diacylglycerated-cysteine and cleaves the signal sequence from pro-Lpp to form apolipoprotein (Paetzel et al., 2002). Previous work has indicated the requirement for equimolar levels of PG to Lpp in the maturation process of pro-Lpp (Suzuki et al., 2002).
In sum, existing findings suggest that inhibited growth of E. coli cells lacking membrane acidic phospholipids arises from either defective DnaA-mediated initiation of replication in the vicinity of the cytoplasmic membrane, faulty maturation of major outer membrane lipoprotein, or the intersection of both processes (Inouye et al., 1983; Cronan, 2003; Saxena et al., 2013). However, with respect to the latter, the manner by which immature pro-Lpp might affect normal, DnaA-dependent normal growth is still unclear.
The recent understanding of outer membrane homeostasis highlights RNA polymerase sigma factor RpoE (σ24/σE)-dependent, MicL-S small RNA-mediated regulation of endogenous Lpp levels for cell survival at the early stationary phase (Guo et al., 2014; Nicoloff et al., 2017). The extracytoplasmic function (ECF) sigma factor σE responds to outer membrane disorders in a complex signal cascade involving degradation of the inner membrane anti-sigma factor, RseA, and subsequent release of σE into the cytoplasm (Raivio and Silhavy, 2001; Chaba et al., 2007; Lima et al., 2013; Klein and Raina, 2019). Active σE transcribes multiple small RNAs, like MicA, RybB, and MicL to translationally inhibit outer membrane porins and lipoproteins (Johansen et al., 2006; Udekwu and Wagner, 2007; Guo et al., 2014). MicL-S is an 80-bp small RNA derived from MicL (308 bp), present at the 3′UTR adjacent to the gene-coding region for copper homeostasis protein (cutC), and targets specifically lpp mRNA (Guo et al., 2014). Furthermore, the capacity of a suppressor mutation in lpp (lpp-2) or deletion of lpp to restore growth to pgsA-null cells with a lower acidic phospholipid content supports the requisite for PG to allow maturation of pro-Lpp (Miyazaki et al., 1985; Kikuchi et al., 2000). Related, inhibited growth caused by expression of Lpp(C21G) substantiates the need for the thioester diacylglycerol modification required for proper Lpp maturation (Inouye et al., 1983).
The bacterial nucleoid resides in the vicinity of the cytoplasmic side of the inner membrane, which also serves as an important site for Lpp biogenesis. The nucleoid is dynamic and flexible in nature and undergoes various changes to regulate complex processes, including DNA replication, DNA recombination, and gene expression (Dorman and Deighan, 2003; Luijsterburg et al., 2006; Dillon and Dorman, 2010). The organization and structure of the bacterial nucleoid are affected by small nucleoid-associated proteins (NAPs), which bind to chromosomal DNA (R.T Dame, 2005; Dillon and Dorman, 2010). NAPs include proteins such as histone-like protein HupA, HupB (functions in DNA topology management) (Varshavsky et al., 1977), H-NS protein (reinforces negative supercoiling to facilitate DNA–protein–DNA bridges) (Dorman et al., 1999; S. Rimsky, 2004), and DNA bending–binding protein such as integration host factor IhfA, IhfB (S Khrapunov et al., 2006), and Fis (Factor for Inversion Stimulation) (Gille et al., 1991). Particularly, Fis protein, which is abundantly present during exponential growth, binds to numerous sites on the chromosome and acts as a global transcription factor (Cho et al., 2008). In vitro studies indicated that Fis also binds to specific chromosomal sites present in oriC (Wold et al., 1996) and DARSs (DnaA-reactivating sequences) (Kasho et al., 2014); the latter appears to play a role in rejuvenation of ADP-DnaA to ATP-DnaA (Fujimitsu et al., 2009). Fis and IHF proteins complex with these regions and act as a pleiotropic regulator of initiation of replication to inhibit untimely initiations (Wold et al., 1996; Kasho et al., 2014), probably via altering of the DNA supercoiling (Margulies and Kaguni, 1998). In vivo, E. coli lacking fis are able to grow when cultured in minimal media; however, when cultured in rich media, the rapidly growing cells had increased mass but possessed fewer origins per cell, having initiated replication in an asynchronous manner (Flatten and Skarstad, 2013).
Although reports indicate that NAPs are needed to regulate several important housekeeping genes, whether they serve in a manner to link two membrane-associated outcomes in acidic phospholipid-deficient cells, perturbed chromosomal replication and defective biogenesis of outer membrane lipoproteins, has not been addressed. Here, we investigate whether accumulated Lpp(C21G) intermediates poison DnaA-dependent oriC-mediated replication initiation in E. coli with normal levels of acidic phospholipids, and if any auxiliary initiation factors play a role in linking the two membrane-associated processes. We also examine the possible role of the σE-MicL/Lpp protective loop in the ability of DnaA(L366K) to facilitate growth of cells carrying mutant Lpp(C21G) or lacking the ability to synthesize the acidic phospholipids.
Materials and Methods
Restriction enzymes were purchased from New England Biolabs (NEB). All polymerase chain reactions, unless mentioned otherwise, were performed using Q5 High fidelity DNA polymerase (NEB). PCR primers were designed using GeneRunner program Version 6.5.521 Primers used in the study were custom synthesized from Integrated DNA Technologies. Various ingredients to reconstitute minimal media (M9) or Luria broth (LB) were purchased from Sigma or VWR.
Media, Bacterial Strain, and Plasmids
Minimal media supplemented with 0.4% glucose and 0.5% Casamino acids were used to grow cells both in liquid culture and in agar plates. Supplementary Table 1 showed E. coli strains and the plasmids used in the study. E. coli strain BW25113 (lpp+) or JW1667-5 (lpp–) was obtained from E. coli genetic stock center2 and their different derivatives (used in this study) were constructed in laboratory using plasmid-inducible lambda-red recombinase-mediated genome modification system.
Plasmid Construction
Plasmid pSC were generated by replacing pBR322 origin of replication and the ampicillin-resistance gene from plasmid pBAD24c (Guzman et al., 1995) with p15A origin of replication and tetracycline-resistant gene from the pACYC184 vector (Chang and Cohen, 1978; Rose, 1988). The dnaA or dnaAL366K allele was removed from previously described plasmids, pZL606 and pZL607 (Zheng et al., 2001), by digesting with NdeI and StyI, and inserting into the same restriction enzyme site in pSC plasmid. An open reading frame for fis gene was amplified by PCR using chromosomal DNA as a template and a pair of primers (Supplementary Table 2) carrying NdeI and StyI. The amplified product was inserted into the corresponding sites of plasmid pSC to place fis allele under arabinose-inducible promoter. Similarly, plasmid pC2 (Nakamura and Inouye, 1982, we received as a gift from Dr. Thomas Silhavy) carries lpp(C21G) gene cloned between XbaI and EcoRI, and placed under control of an IPTG-inducible promoter. Wild-type lpp or lpp(ΔK58) was amplified by PCR using chromosomal DNA as a template and a pair of primers containing XbaI and EcoRI restriction sites. The amplified fragments were inserted into the corresponding sites of the pC2 plasmids. The transformants were screened to select for the integration of the desired products by restriction digestion analysis, which were later confirmed by the automated Sanger DNA sequencing method (Genewiz).
Lambda-Red [λ-Red] Recombinase-Mediated Genetic Recombineering
For insertional-deletion of target genes, the relevant bacterial strains were electroporated with one of the two pKD46 (Datsenko and Wanner, 2000; Baba et al., 2006) derivatives carrying λ-red recombinase genes (exo, beta, and gam): pKD-sg-ack (Reisch and Prather, 2015) or pSIJ8 (Jensen et al., 2015) for the insertion of antibiotic resistance gene cassette. For the homology-directed recombination (HDR), donor DNA was designed to carry (1) 50–60 base-pair-long 5′-upstream and 3′-downstream DNA sequence of the gene to be deleted, and (2) reverse DNA sequence of the antibiotic resistance gene cassette to maintain expression of the antibiotic resistance gene through its promoter. All donor DNA for HDR with desired antibiotic cassette were amplified using plasmid pCas9cr4 (Reisch and Prather, 2015), for which primers were designed based on its gene sequence.
Cell Viability Assay
E. coli cultures were prepared by growing cells overnight (12–14 h) in M9+CAA+Glu media supplemented with appropriate antibiotics (ampicillin: 100 μg/ml, tetracycline: 12.5 μg/ml, and kanamycin: 50 μg/ml) at 37°C. Overnight grown cultures were diluted to inoculate an equal number of cells (∼10,000 cfu/ml) in fresh M9+CAA+Glu media supplemented with appropriate antibiotics in the presence of inducers. To perform growth curve analysis, optical densities (OD at 600 nm) were recorded after every 60 min (±10) for at least 12–18 h. Additionally, samples (1 ml) were drawn at different time point intervals from separate conditions to analyze cell viability and level of protein expression. For viability assays, OD (600 nm) values for cell suspensions were normalized to 0.6 × 109 cells/ml, which were then serially diluted by 10-fold dilutions up to 10–5. A fixed volume (25 μl) of each serial dilution was plated on M9+CAA+Glu agar plates to obtain 200–500 colonies/plates. Viability was also assessed using a spotting assay in which serially diluted cell suspensions (2.5 μl) grown in the absence of inducer were spotted on M9+CAA+Glu-agar plates carrying various concentrations of isopropyl-β-D-galactopyranoside (IPTG), or L-Arabinose (Sigma), or both. One-way analysis of variance (ANOVA) with multiple comparison tests was performed to compare viability among groups using Prism 8 (GraphPad).
Immunoblotting
Based on the OD (600 nm), equal numbers of cells were pelleted by centrifugation at 16,000 × g for 10 min at 4°C. Cell pellets were resuspended in 30 μl of PBS and 12 μl (4×) of SDS sample buffer and subsequently boiled for 5 min before resolving proteins with 19% SDS-polyacrylamide-urea gels. Proteins were transferred to PVDF membranes and visualized by Ponceau S staining to evaluate the total protein content. Expressions of Lpp and Lpp(C21G) proteins were detected using anti-Lpp antiserum, and the expression of DnaA(L366K) was detected using the anti-DnaA antiserum. Subsequently, immunoblots were treated with secondary stabilized peroxidase-conjugated goat anti-rabbit antibody (Thermo Fisher Scientific) and processed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) using AI600 Imager (GE Healthcare Biosciences). Experiments were performed with at least three biological replicates. ImageJ (Version 1.51) software was used to quantitate the band intensity of proteins detected by immunoblots and Ponceau S staining.
Lipidomic Analysis
For lipidomic analysis, MDL12 and MDL12Δfis cells were grown to obtain 10–50 million cells/ml (cell number was determined by dilution plating). Bacterial cells were collected (13,500 rpm at 4°C for 10 min), and the cell pellets were flash-frozen and stored at −80°C until further analyzed. Frozen cell pellets were thawed on ice and resuspended in 40 μl of chilled water/methanol/IPA [35:25:40]. The mixture was subjected to freezing (plunging the tube containing mixture into dry ice for 30 s) and rapid heating (plunging tubes into a 37°C water bath for 90 s) twice before sonication at 30 kHz for 30 s mixed with 100 μl of ice-chilled isopropyl alcohol containing internal standards. Subsequently, the samples were placed on ice for 30 min and transferred to −20°C for 30 min following centrifugation at 13,500 rpm at 4°C for 20 min. Supernatants were collected to perform mass spectrometric (MS) analysis using the targeted LCMS-MS approach where they were resolved on an Xbridge Amide 3.5 μm, 4.6 × 100 mm column (Phenomenex) online with a triple quadrupole mass spectrometer (5500 QTRAP, SCIEX, United States) operating in the multiple reaction monitoring (MRM) mode. The declustering potential, collision energies, cell exit potential, and entrance potential optimized for each metabolite to obtain maximum ion intensity for parent and daughter ions via manual tuning in Analyst 1.6.3 software (SCIEX, United States). Signal intensities from all MRM Q1/Q3 ion pairs for the analyte were ranked to ensure the selection of the most intense precursor and fragment ion pair for MRM-based quantitation. This approach resulted in the selection of declustering potential, collision energies, cell exit potential, and entrance potential that maximized the generation of each fragment ion species. The metabolite ratios were calculated by normalizing the peak area of endogenous metabolites within samples normalized to the internal standard for every class of lipid molecule. The sample queue was randomized, and solvent blanks were injected to assess sample carryover. Pooled quality control (pooled QC) samples were injected after every eight samples to check for instrumental variation. Like pooled QC samples, Sciex standard QC plasma samples were also injected for lipidomic data analysis. For CL data acquisition, bovine heart extract was used as quality control. Data normalized to QC variance. QC normalized data and imputed MRM data were processed using MultiQuant 3.0.3 (Sciex). The relative quantification values of analytes were determined by calculating the ratio of peak areas of transitions of samples normalized to the peak area of the internal standard specific for every class. All statistical analyses were performed using Prism 8 (GraphPad). Two or more groups were compared with one-way ANOVA, and data for selective 124 lipid metabolites were represented in the heatmap. Multiple t-tests were performed using log-transformed values of metabolite peak ratio and compared using volcano plot. For volcano plots, the p-value cutoff was set at p = 0.05, and log2 fold change (fold difference) that corresponds to the statistical significance was set at ± 2.
Results
Induction of Lpp(C21G) Expression Inhibits Growth That Is Restored by Ectopic Expression of DnaA(L366K)
E. coli cells lacking acidic membranes, particularly PG, caused growth-arrested phenotype. Insufficient levels of PG interrupt two important cellular processes, (i) DnaA-mediated process of replication initiation at the chromosomal origins (Fingland et al., 2012), and (ii) the maturation and proper sorting of outer membrane lipoprotein, Lpp (Matsuyama et al., 1995). Overexpression of a mutant DnaA, DnaA(L366K) protein, suppresses the growth defect in cells unable to synthesize PG. A previous study (Inouye et al., 1983) had demonstrated that bacterial cells carrying cellular levels of PG but overproducing mutant outer membrane lipoprotein, Lpp(C21G), which is unable to be processed by PG, also resulted in growth-arrested phenotype. Wondering whether there is a link of some fashion between the two growth-arrest phenotypes led us to assess the capacity of DnaA(L366K) to restore growth in bacterial cells exogenously overproducing Lpp(C21G). For this, we developed a two-plasmid system for the simultaneous expression of the Lpp(C21G) and DnaA(L366K) (see “Materials and Methods”). The genes for Lpp and Lpp(C21G) were cloned under dual Lpp and IPTG-inducible LacUV5 promoters (Inouye et al., 1983; Supplementary Table 1). Lpp or Lpp(C21G) was expressed either via the constitutive lpp promoter in the absence of IPTG or via the combined lpp and lac promoters in the presence of IPTG in a lacI+ (Nakamura and Inouye, 1982; Supplementary Table 1) genetic background. The genes for DnaA and DnaA(L366K) were cloned under control of the L-arabinose-inducible pBAD promoter.
As reported before, we confirmed that E. coli lpp-null cells carrying plasmid-borne mutant lpp(C21G) grown in the presence of inducer (as low as 50 μM IPTG) resulted in a 27.4-fold reduction (p < 0.001) in cell viability (Supplementary Figures 1C,D). The bacterial cells overproducing Lpp(C21G) when transformed with plasmid vector pSC in the absence or presence of inducer (0.2% arabinose) did not show any changes in the restricted growth (Figures 1A,B). The ectopic expression of DnaA(L366K), however, restored growth, with cells retaining viability (p < 0.04, Figures 1C,D). In contrast, overexpression of DnaA(WT) either alone, as reported elsewhere (Atlung et al., 1987; Pierucci et al., 1989; Flatten et al., 2015), or in conjunction with Lpp(C21G) led to poor growth (Supplementary Figures 2A,B). As a control, similar overexpression of DnaA(L366K) in cells expressing Lpp(WT) had no effect (Figures 1E,F).
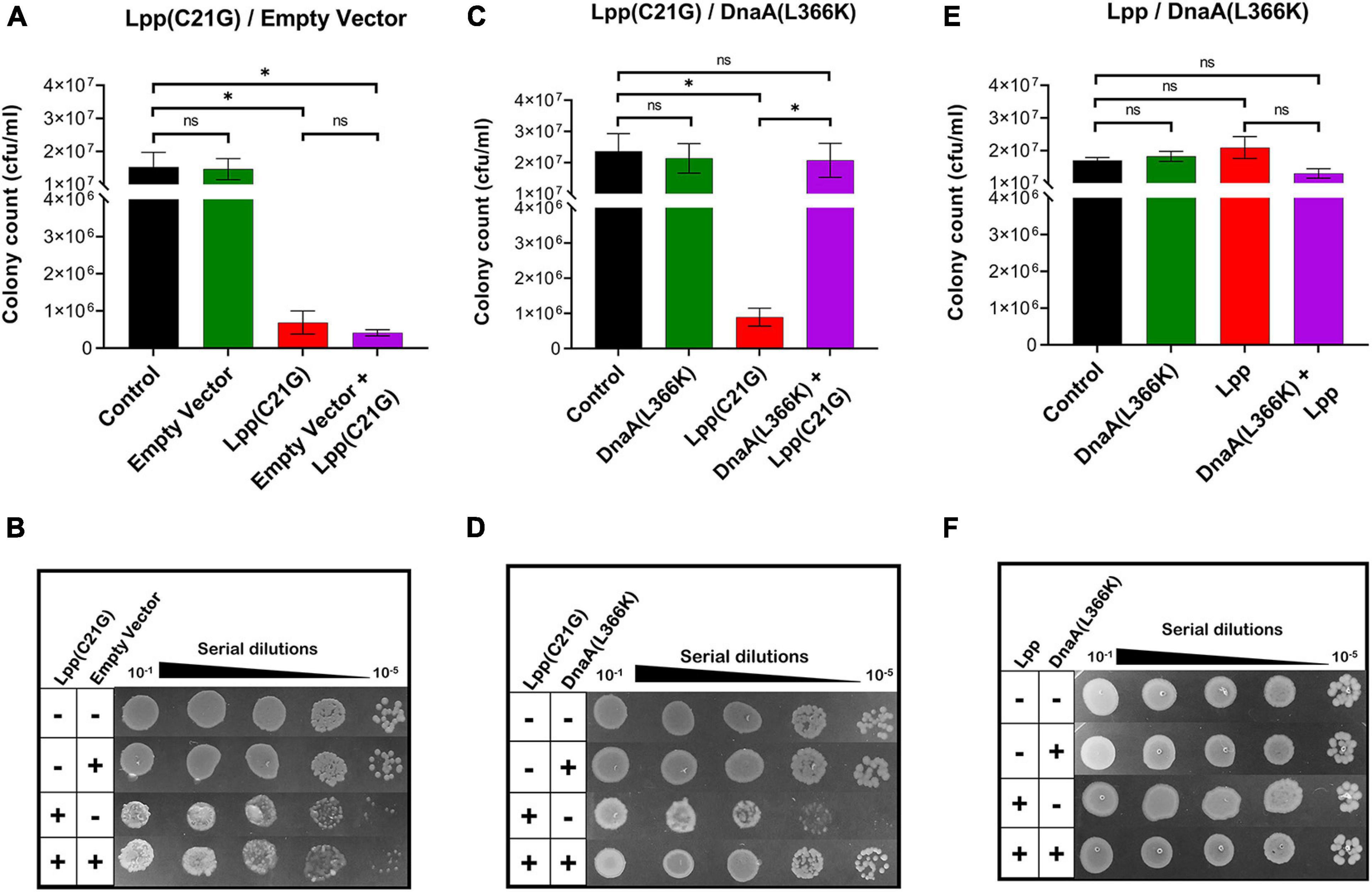
Figure 1. DnaA(L366K) overexpression rescues growth in cells expressing Lpp(C21G). (A) Viability of lpp-null cells transformed with empty vector, and plasmids expressing Lpp(C21G). No inducer control (black), in the presence of 0.2% Arabinose (green) for empty vector, 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and empty vector expression in M9+CAA+Glu media. (B) Serial dilutions of lpp-null cells, capable of expressing plasmid-derived Lpp(C21G) and empty vector, on agar plates. (C) Viability of lpp-null cells expressing DnaA(L366K) and Lpp(C21G). No inducer control (black), in the presence of 0.2% Arabinose (green) for DnaA(L366K), 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and DnaA(L366K) expression in M9+CAA+Glu media. (D) Serial dilutions of lpp-null cells, capable of expressing plasmid-derived Lpp(C21G) and DnaA(L366K), on agar plates. (E) Viability of lpp-null cells expressing DnaA(L366K) and Lpp. No inducer control (black), in the presence of 0.2% Arabinose (green) for DnaA(L366K), 50 μM IPTG (red) for Lpp, and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp and DnaA(L366K) expression in M9+CAA+Glu media. (F) Serial dilutions of lpp-null cells, capable of expressing plasmid-derived Lpp and DnaA(L366K), on agar plates. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗p < 0.05, ns p > 0.05 in one-way ANOVA with Dunnett’s multiple comparison test.
Given that DnaA(L366K) restored growth when the inducer for Lpp(C21G) was present, we next questioned whether DnaA(L366K) affects the cellular levels of Lpp(C21G). Immunoblots revealed that pSC vector alone did not affect the expression of Lpp(C21G) (lanes 4–5, Figures 2A,B); however, overproduction of DnaA(L366K) caused an approximate sevenfold reduction in levels of Lpp(C21G) (lane 5, Figures 2C,D), as compared to cells expressing Lpp(C21G) alone (lane 4, Figure 2C). When compared to the reduction of Lpp(C21G) levels, we observed significantly less reduction in Lpp arising from overexpression of DnaA(L366K) (lanes 4–5, Figures 2E,F), suggesting a lpp mutation-specific response to DnaA(L366K).
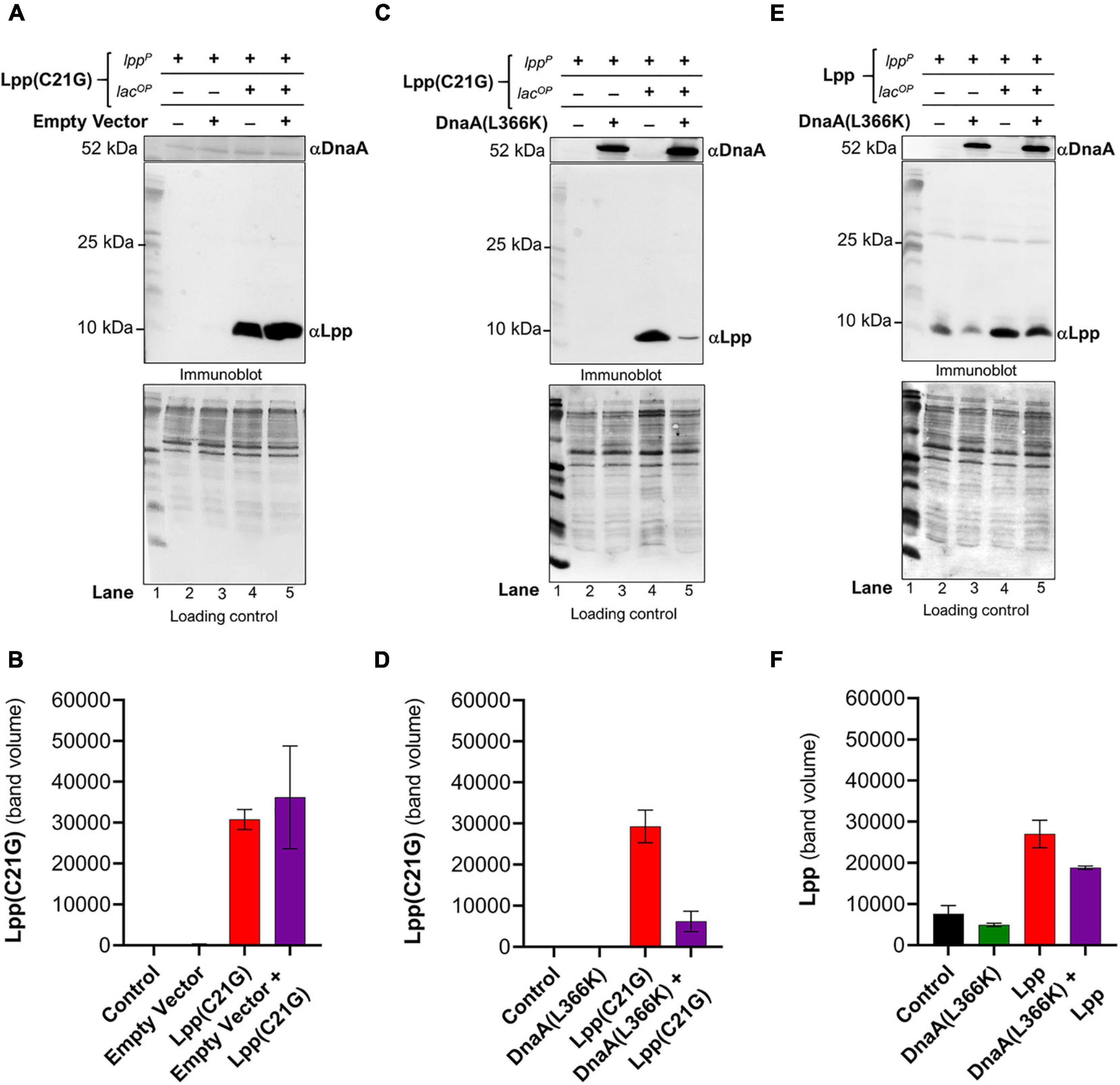
Figure 2. Ectopic expression of DnaA(L366K) reduces levels of Lpp(C21G) even when the inducer is present. (A,B) Immunoblotting for the detection of DnaA and Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (C,D) Immunoblotting for the detection of DnaA(L366K) and Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (E,F) Immunoblotting for the detection of DnaA(L366K) and Lpp and quantitative analysis of Lpp levels. Lpp(C21G) and Lpp are expressed via Lpp (lppP) and Lac (lacUV5OP) promoters. Loading controls for immunoblotting are ponceau-S staining for total protein normalization presented in grayscale. Data are means ± SEM of at least three independent experiments.
Separately, we confirmed the differences in exogenous expression of Lpp and Lpp(C21G) in lpp-null cells (Supplementary Figures 1G,H). We observed approximately threefold higher Lpp(C21G) levels (lane 3, Supplementary Figure 1H) when compared with Lpp (lane 3, Supplementary Figure 1G) in the presence of 50 μM IPTG inducer in a manner similar to the expression of Lpp(C21G) (lane 4, Figure 2C; and lane 4, Figure 2E). At the same time, the Lpp(C21G) levels (lane 3, Supplementary Figure 1H) were still observed to be approximately 50-fold less than the endogenous levels of Lpp in lpp+ cells (lane 6, Supplementary Figure 1G). These results further confirm that inhibited growth in lpp-null cells resulted from the accumulation of unprocessed Lpp(C21G), but not Lpp, as shown elsewhere (Inouye et al., 1983).
Absence of the Promoter Region for MicL-S Small RNA Prevents the Ability of DnaA(L366K) to Decrease Lpp(C21G) Expression and Only Partially Restore Growth
The E. coli transcription factor σE has been recognized as an envelope stress-responsive sigma factor that senses an abnormality of the outer membrane integrity. It has been previously reported that abundance of Lpp results in the σE activity and that MicL and Lpp comprise a new regulatory loop that opposes membrane stress (Guo et al., 2014). Interestingly, when cells were grown without IPTG, we only observed expression via the lpp promoter of Lpp (lanes 2–3, Figures 2E,F) but not Lpp(C21G) (lanes 2–3, Figures 2A,D). This observation indicated the possible role of σE-dependent MicL-S-mediated envelope protective mechanisms in preventing expression of Lpp(C21G) from the lpp promoter in cells harboring the empty vector (lanes 2–3, Figure 2A) or expressing DnaA(L366K) (lanes 2–3, Figure 2C). However, when cells were treated with IPTG to induce Lpp(C21G), the ectopic expression of DnaA(L366K) was required to decrease Lpp(C21G) levels (lane 5, Figure 2C) and allow growth (Figure 1C), as opposed to the empty pSC vector (lane 5, Figures 1A, 2A). The ability of DnaA(L366K) to reduce levels of Lpp(C21G) led us to question whether the ectopic expression of DnaA(L366K) translationally inhibits Lpp(C21G) via early activation of the σE-dependent MicL-S-mediated envelope protective loop. To test this, we chose an E. coli strain lacking the chromosome region for cutC (cutC-null, Supplementary Table 1); cutC encodes for a copper homeostasis protein. The region for cutC comprises an σE binding site for the transcription of MicL small RNA (Guo et al., 2014; Nicoloff et al., 2017). Therefore, the lack of the cutC disrupts the σE-dependent envelope protective loop by preventing transcription of the MicL-S sRNA. Studies suggest that lack of cutC leads to increased levels of endogenous Lpp and affects membrane integrity (Guo et al., 2014). So, for this study, we deleted the gene-encoding region for lpp in cells already lacking cutC (ΔcutC) to test the effect of Lpp(C21G) on cell growth.
In cutC-null cells, we observed an approximate 250-fold reduction (p < 0.02) in cell viability (Figures 3A,B) by the limited (50 μM IPTG) expression of Lpp(C21G) as compared to that for cells not induced for Lpp(C21G) expression. In line with this observation, we co-expressed in cutC-null cells Lpp(C21G) along with the DnaA(L366K). DnaA(L366K) overproduction resulted in an approximate 16-fold increase in cell viability, even when Lpp(C21G) expression was induced (Figure 3A). Although, in comparison with cutC+ cells (Figures 1C,D), cutC-null cells co-expressing Lpp(C21G) and DnaA(L366K) showed limited capacity to restore cell viability (Figures 3A,B).
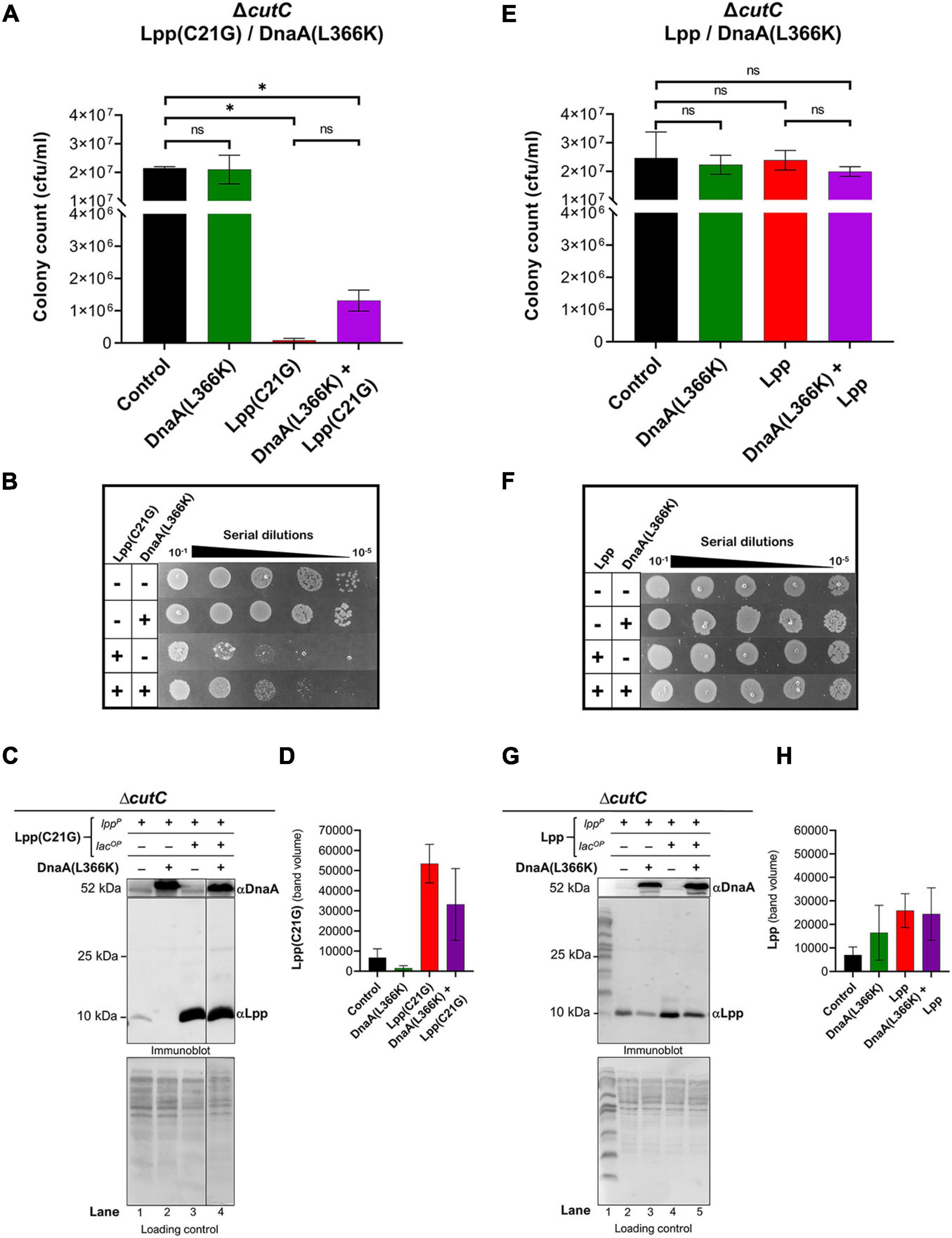
Figure 3. Overexpression DnaA(L366K) restores limited growth but fails to reduce levels of Lpp(C21G) when the promoter for MicL-S small RNA is absent. (A) Viability of cutC-null cells expressing DnaA(L366K) and Lpp(C21G). No inducer control (black), in the presence of 0.2% Arabinose (green) for DnaA(L366K), 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and DnaA(L366K) expression in M9+CAA+Glu media. (B) Serial dilutions of cutC-null cells, capable of expressing Lpp(C21G) and DnaA(L366K), on agar plates. (C,D) Immunoblotting for the detection of DnaA(L366K) and Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (E) Viability of cutC-null cells expressing DnaA(L366K) and Lpp. No inducer control (black), in the presence of 0.2% Arabinose (green) for DnaA(L366K), 50 μM IPTG (red) for Lpp, and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp and DnaA(L366K) expression in M9+CAA+Glu media. (F) Serial dilutions of cutC-null cells, capable of expressing plasmid-derived Lpp and DnaA(L366K), on agar plates. (G,H) Immunoblotting for the detection of DnaA(L366K) and Lpp and quantitative analysis of Lpp levels. Lpp(C21G) and Lpp are expressed via Lpp (lppP) and Lac (lacUV5OP) promoters. All loading controls are ponceau-S staining for total protein normalization presented in grayscale. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗p < 0.05, ns p > 0.05 in one-way ANOVA with Dunnett’s multiple comparison test.
The immunoblotting analysis further showed no significant decrease in levels of Lpp(C21G) in cutC-null cells in the absence or presence of DnaA(L366K) (lanes 3–4, Figures 3C,D), as opposed to cells with cutC+ cells (lanes 4–5, Figures 2C,D). Of note, we also observed a limited expression of Lpp(C21G) (lane 1, Figure 3C) from the lpp promoter in cells with DnaA(L366K). Interestingly, with the wild-type Lpp control, we witnessed no change in viability (Figures 3E,F). Moreover, there were no significant differences in Lpp expression in the presence or absence of DnaA(L366K) overexpression (lanes 4–5, Figures 3G,H), indicating that DnaA(L366K) selectively decreases the expression of Lpp(C21G), but not Lpp, in a σE-dependent MicL-S-mediated manner.
Δfis Cells Show the Ability to Grow Even When the Lpp(C21G) Expression Is Induced
The capacity of DnaA(L366K) to still rescue growth to some extent in the absence of MicL-S sRNA led us to then question whether the lack of acidic phospholipid-mediated accumulation of pro-lipoprotein and the ectopic expression of Lpp(C21G) poisons the DnaA-dependent, oriC-mediated DNA replication initiation process. Considering that DNA replication occurs in the vicinity of inner cellular membranes, where the outer membrane lipoprotein accumulates due to faulty maturation, we wondered whether the accumulated, unprocessed pro-Lpp intermediate creates stress on the chromatin structure, causing growth arrest linked to DNA replication. To characterize the adverse effect of Lpp(C21G) on the initiation, we employed a loss-of-function study targeting NAPs that participate in DnaA-dependent initiation of DNA replication at oriC (Riber et al., 2016). E. coli strains lacking gene-encoding regions for HU-α, HU-β, Ihf-A, Ihf-B, Fis, and SeqA (Supplementary Table 1) were used to see which of those proteins might be adversely affected by the accumulation of the Lpp intermediate by testing which NAP deletions allowed growth upon the overproduction of Lpp(C21G). Cells lacking HU-α, HU-β, Ihf-A, Ihf-B, and SeqA (Supplementary Figures 3A–J) were still adversely affected by the overproduction of Lpp(C21G), thus suggesting that the Lpp(C21G) does not directly affect these NAPs. In contrast, the cells lacking Fis were able to retain approximately 84 and 64% viability even when Lpp(C21G) expression was induced with IPTG at 50 and 1,000 μM, respectively. It was noted, however, that colonies were large, opaque, and irregularly lobate (Supplementary Figures 3K,L).
A two-plasmid system introduced Fis into fis-null cells expressing Lpp(C21G). As was expected, no adverse effects of Lpp(C21G) on cell viability (Figures 4A–D) were observed in the fis-null cells. However, when Fis was exogenously expressed in cells also expressing Lpp(C21G), a 57.4-fold (p < 0.006) loss of cell viability (Figures 4C,D) was observed. We also noticed that the ectopic expression of Fis alone led to a 15.7-fold (p < 0.01) reduction in cell viability (Figure 4C). However, the combined expression of Lpp(C21G) and Fis [pFis+Lpp(C21G), Figure 4C] had a more severe effect on viability than the expression of Fis alone (pFis, Figure 4C: approximately fourfold reduction). As a control, we tested the effect of Fis in cells expressing wild-type Lpp. Cell viability (Figures 4E,F) remained unaffected following the overexpression of Lpp alone. Like cells expressing Lpp(C21G), the overexpression of Fis alone affected viability in fis-null cells carrying a plasmid for Lpp. The co-expression of Lpp and Fis (pFis+Lpp, Figure 4E) resulted in increased viability when compared with Fis expression alone (pFis, Figure 4E), as opposed to the co-expression of Lpp(C21G) and Fis [pFis+Lpp(C21G), Figure 4C].
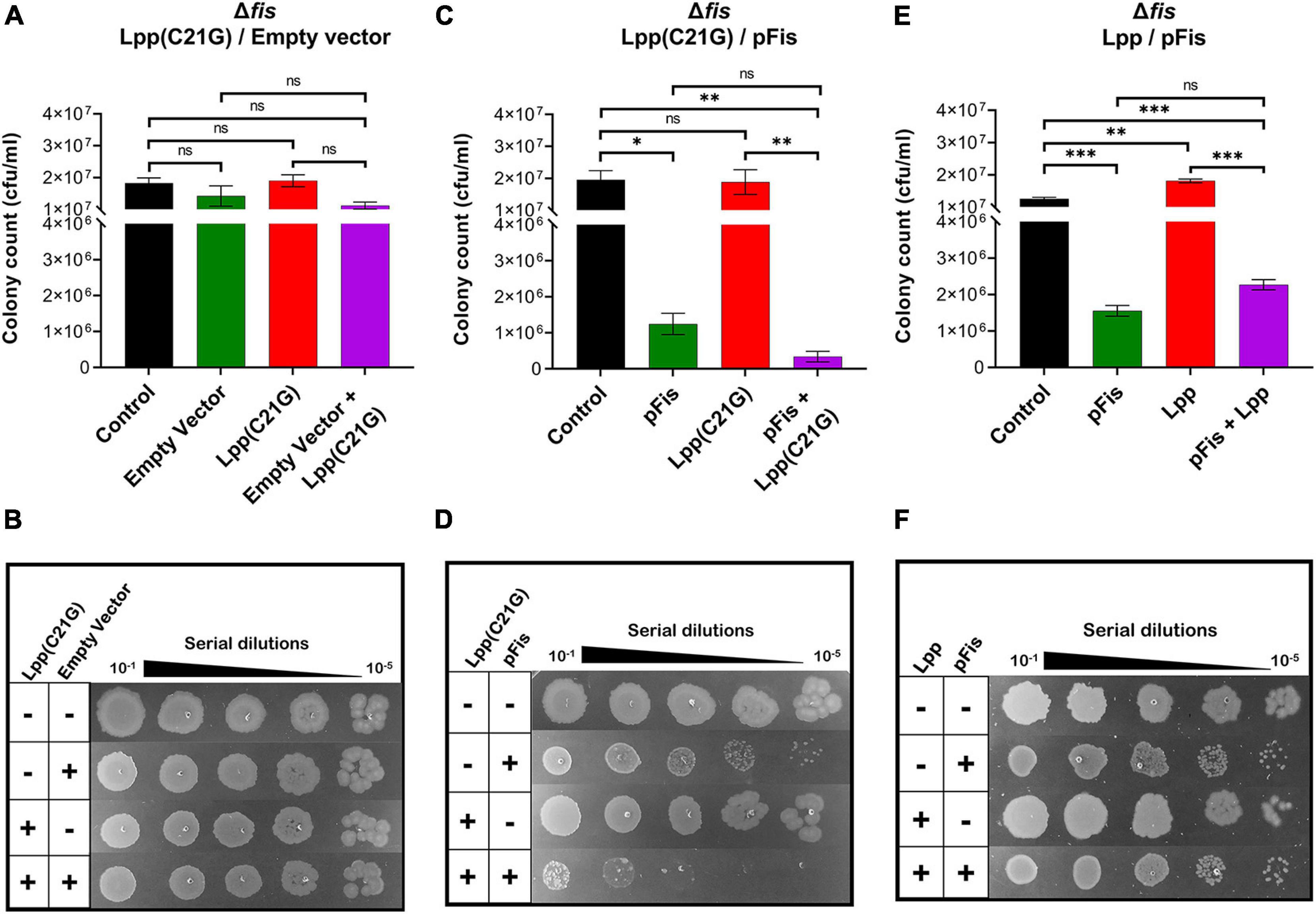
Figure 4. Cells lacking fis can grow even when the inducer for Lpp(C21G) is present. (A) Viability of fis-null cells transformed with empty vector and plasmids expressing Lpp(C21G). No inducer control (black), in the presence of 0.2% Arabinose (green) for empty vector, 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and empty vector expression in M9+CAA+Glu media. (B) Serial dilutions of fis-null cells, capable of expressing plasmid-derived Lpp(C21G) and empty vector, on agar plates. (C) Viability of fis-null cells expressing Fis (pFis) and Lpp(C21G). No inducer control (black); in the presence of 0.2% Arabinose (green) for Fis, 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and Fis expression in M9+CAA+Glu media. (D) Serial dilutions of fis-null cells, capable of expressing plasmid-derived Lpp(C21G) and Fis on agar plates. (E) Viability of fis-null cells expressing Fis (pFis) and Lpp. No inducer control (black), in the presence of 0.2% Arabinose (green) for Fis, 50 μM IPTG (red) for Lpp, and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp and Fis expression in M9+CAA+Glu media. (F) Serial dilutions of fis-null cells, capable of expressing plasmid-derived Lpp and Fis, on agar plates. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns p > 0.05 in one-way ANOVA with Dunnett’s multiple comparison test.
Cells Lacking Fis Have Reduced Levels of Lpp(C21G) but Not Lpp
Seeing that the DnaA(L366K) reduces levels of Lpp(C21G) but not the wild-type Lpp, as mentioned above, we wanted to test whether fis-null cells restore growth to Lpp(C21G)-expressing cells, in a manner similar to the ectopic expression of DnaA(L366K), by reducing Lpp(C21G) expression but not Lpp. Surprisingly, in the cells carrying vector control, we did not detect any Lpp(C21G) when expressed from either the lpp promoter (lanes 2–3, Figures 5A,B) and only limited expression of Lpp(C21G) was seen from the combined lpp and lacUV5 promoters (lanes 4–5, Figures 5A,B). The expression of Lpp(C21G) was evident only in the presence of Fis (lanes 2 and 4, Figures 5C,D). In contrast, for cells that carry plasmids for Fis and wild-type Lpp, we detected Lpp from both lpp (lanes 2–3, Figures 5E,F) and combined lpp and lacUV5 promoters (lanes 4–5, Figures 5E,F), as opposed to Lpp(C21G) (lane 3, Figures 5C,D). We also noted that in cells lacking fis, ectopic expression of Lpp through the lpp promoter (lanes 2–3, Figure 5E) was similar to that caused by DnaA(L366K) (lanes 2–3, Figure 2E).
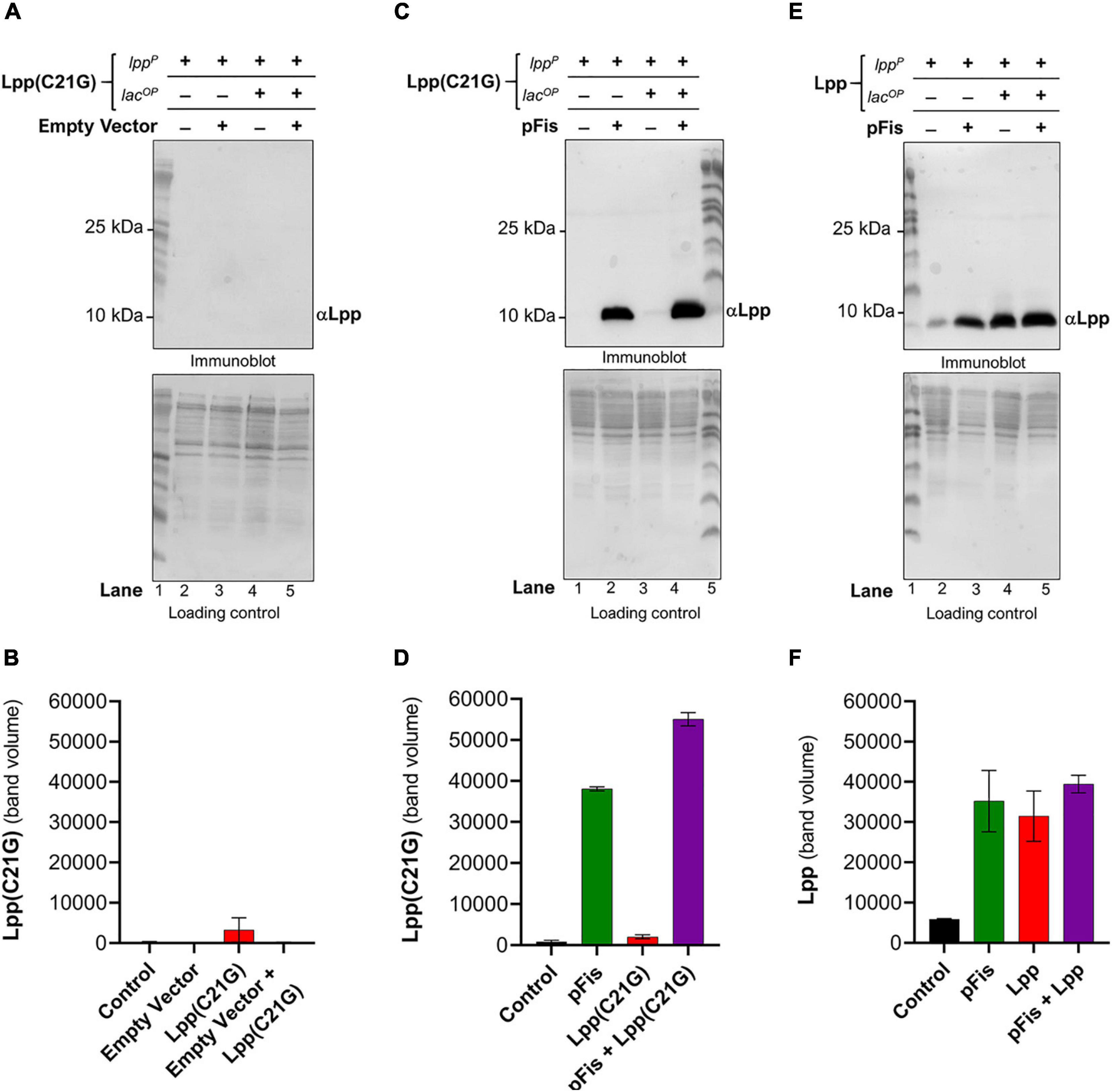
Figure 5. Cells lacking fis reduces levels of Lpp(C21G) even when the inducer is present. (A,B) Immunoblotting of fis-null cells carrying an empty vector and a plasmid expressing Lpp(C21G) to detect Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (C,D) Immunoblotting of fis-null cells carrying plasmid expressing Fis (pFis) and Lpp(C21G) to detect Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (E,F) Immunoblotting of fis-null cells carrying plasmid expressing Fis (pFis) and Lpp to detect Lpp and quantitative analysis of Lpp levels. Lpp(C21G) and Lpp are expressed via Lpp (lppP) and Lac (lacUV5OP) promoters. All loading controls are ponceau-S staining for total protein normalization presented in grayscale. Data are means ± SEM of at least three independent experiments.
fis-null Cells Lacking the Promoter Region for MicL-S Small RNA Only Partially Express Lpp(C21G) but Still Are Able to Grow
We examined the role of MicL-S small RNA in the reduction of Lpp(C21G) levels in fis-null cells. In cells lacking Fis and the promoter region for MicL-S small RNA (ΔcutCΔfis, Supplementary Table 1), induction of Lpp(C21G) (Figures 6A,B) and Lpp (Figures 6E,F) alone did not adversely affect cell viability, in a manner similar to cutC+ fis-null cells (Figures 4C–F). Interestingly, immunoblotting analysis detected an approximate 1.8-fold higher expression of Lpp(C21G) (lane 3, Figures 6C,D) when expressed alone, as opposed to that in cutC+ fis-null cells (lane 3, Figures 5C,D). However, the levels of Lpp(C21G) in cutC-null, fis-null cells (lane 3, Figures 6C,D) were still 16-fold less in comparison with the co-expression of Lpp(C21G) and Fis (lane 4, Figures 6C,D).
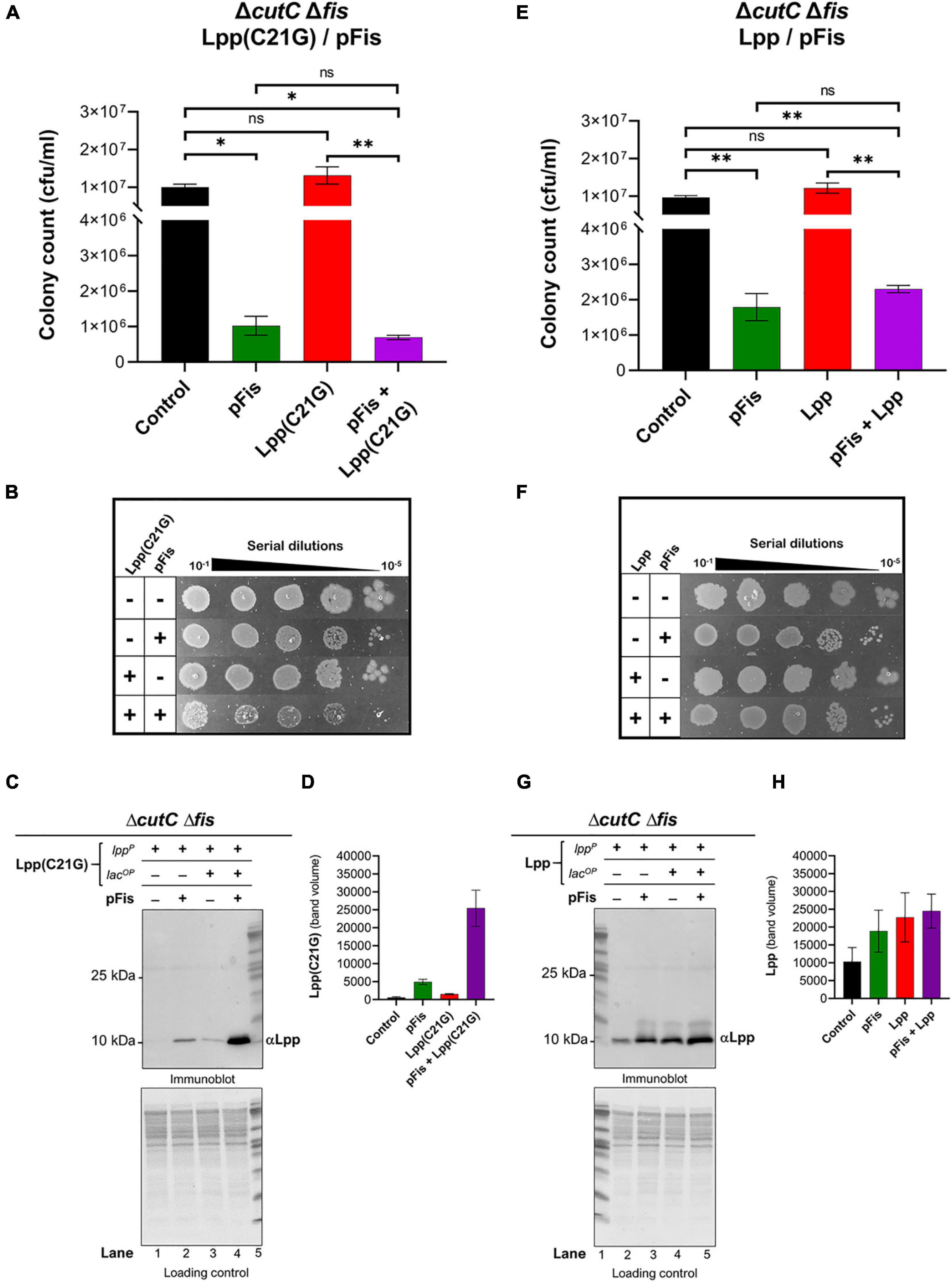
Figure 6. fis-null cells lacking promoter for MicL-S small RNA can grow by expressing only low levels Lpp(C21G). (A) Viability of cutC-null fis-null cells expressing Fis (pFis) and Lpp(C21G). No inducer control (black); in the presence of 0.2% Arabinose (green) for Fis, 50 μM IPTG (red) for Lpp(C21G), and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp(C21G) and Fis expression in M9+CAA+Glu media. (B) Serial dilutions of cutC-null fis-null cells, capable of expressing plasmid-derived Lpp(C21G) and Fis, on agar plates. (C,D) Immunoblotting for the detection of Lpp(C21G) and quantitative analysis of Lpp(C21G) levels. (E) Viability of cutC-null fis-null cells expressing Fis (pFis) and Lpp. No inducer control (black), in the presence of 0.2% Arabinose (green) for Fis, 50 μM IPTG (red) for Lpp, and 50 μM IPTG + 0.2% Arabinose (purple) for both Lpp and Fis expression in M9+CAA+Glu media. (F) Serial dilutions of cutC-null fis-null cells, capable of expressing plasmid-derived Lpp and Fis, on agar plates. (G,H) Immunoblotting for the detection of Lpp and quantitative analysis of Lpp levels. Lpp(C21G) and Lpp are expressed via Lpp (lppP) and Lac (lacUV5OP) promoters. All loading controls are ponceau-S staining for total protein normalization presented in grayscale. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ns p > 0.05 in one-way ANOVA with Dunnett’s multiple comparison test.
The ectopic expression of Fis alone still caused growth inhibition (Figures 6A,E). At the same time, co-expression of Fis and Lpp(C21G) caused a 14.5-fold reduction (p = 0.02, Figure 6A) in the cell viability, as opposed to a 4.2-fold decrease (p = 0.009, Figure 6E) when Fis and wild-type Lpp were co-expressed. Intriguingly, expression of Lpp was unaffected by the lack of fis (lanes 3–5, Figures 5E,F) or fis and cutC (lanes 3–5, Figures 6G,H), as was seen for cells expressing DnaA(L366K) (lanes 3–5, Figures 2E,F), showing a mutation-specific decrease in the gene expression of the Lpp(C21G) but not Lpp.
fis-null Cells Unable to Synthesize PG and CL Are Viable
Previously, it was seen that the ectopic expression of DnaA(L366K) allows cell growth even when acidic phospholipids are absent. We now questioned whether deletion of fis permitted growth in lpp+ cells lacking normal levels of acidic phospholipids. E. coli MDL12 cells, which have chromosomal pgsA under control of the lac promoter, undergo growth arrest in the absence of inducer (IPTG) due to diminished levels of anionic phospholipids (Fingland et al., 2012; Figures 7A,B). MDL12 fis-null cells (Supplementary Table 1) were able to grow even when the inducer IPTG was absent (Figures 7E,F). To confirm that this outcome was due to the loss of Fis, we exogenously expressed Fis in MDL12 fis-null cells. We noted that the ectopic expression of Fis indeed resulted in severely diminished viability (Figures 7G,H) when acidic phospholipid biosynthesis was not induced.
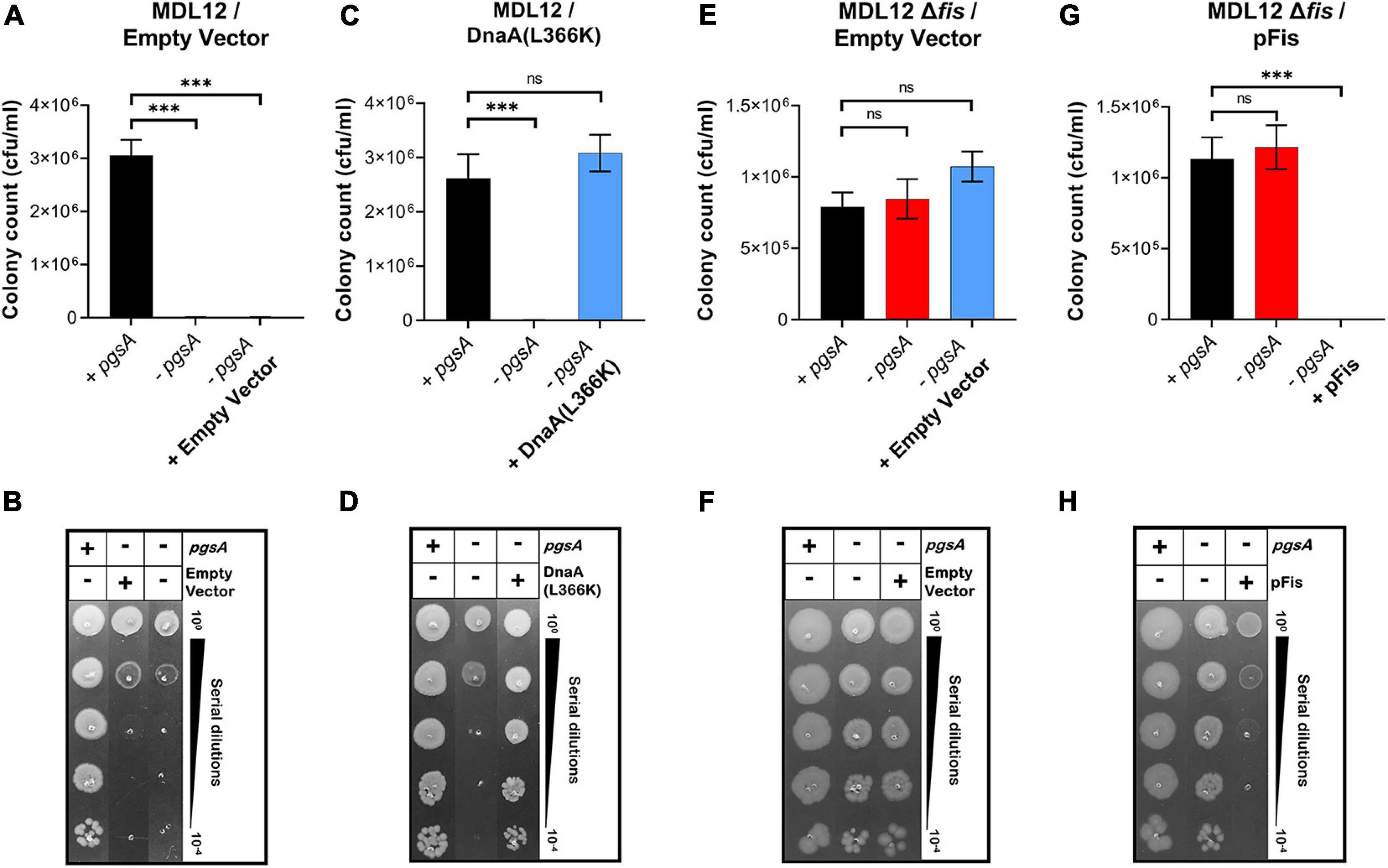
Figure 7. Lack of fis in cells deficient in phosphatidylglycerol (PG) synthesis permits growth. (A,B) Viability of MDL12 cells carrying empty vector with 1 mM IPTG for the expression of pgsA (black), no inducer control (red), and 0.2% Arabinose control (blue) in M9+CAA+Glu media. (C,D) Viability of MDL12 cells carrying plasmid for DnaA(L366K) with 1 mM IPTG for the expression of pgsA (black), no inducer control (red), 0.2% Arabinose for DnaA(L366K) (blue) in M9+CAA+Glu media. (E,F) Viability of MDL12 fis-null cells carrying empty vector with 1 mM IPTG for the expression of pgsA (black), no inducer control (red), and 0.2% Arabinose control (blue) in M9+CAA+Glu media. (G,H) Viability of MDL12 fis-null cells carrying plasmid for Fis (pFis) with 1 mM IPTG for the expression of pgsA (black), no inducer control (red), and 0.2% Arabinose for Fis (blue) in M9+CAA+Glu media. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗∗∗p < 0.001, ns p > 0.05 in one-way ANOVA with Dunnett’s multiple comparison test.
Furthermore, we examined whether the growth-arrested phenotype of E. coli MDL12 cells lacking cutC gene could be suppressed by either overexpression of DnaA(L366K) or deletion of chromosomal fis. Our result suggested that the growth of cells lacking the chromosomal cutC allele is dependent on cellular levels of acidic phospholipids (Figures 8A,B). Whereas cells carrying the intact pgsA allowed growth when cutC is not present, the absence of pgsA allele adversely affected growth of cells lacking cutC (Figures 8A,B). Moreover, overexpression of DnaA(L366K) cannot suppress the growth-arrested phenotype when cutC-null cells are unable to synthesize acidic phospholipids (Figures 8C,D). In contrast, removal of the chromosomal fis allele allowed growth in a pgsA independent manner (Figures 8E,F). These results agree with our findings that either the cells overexpressing mutant Lpp protein or the lack of cellular levels of acidic phospholipids resulted in the accumulation of cytotoxic form of Lpp(C21G), causing a growth-arrested phenotype.
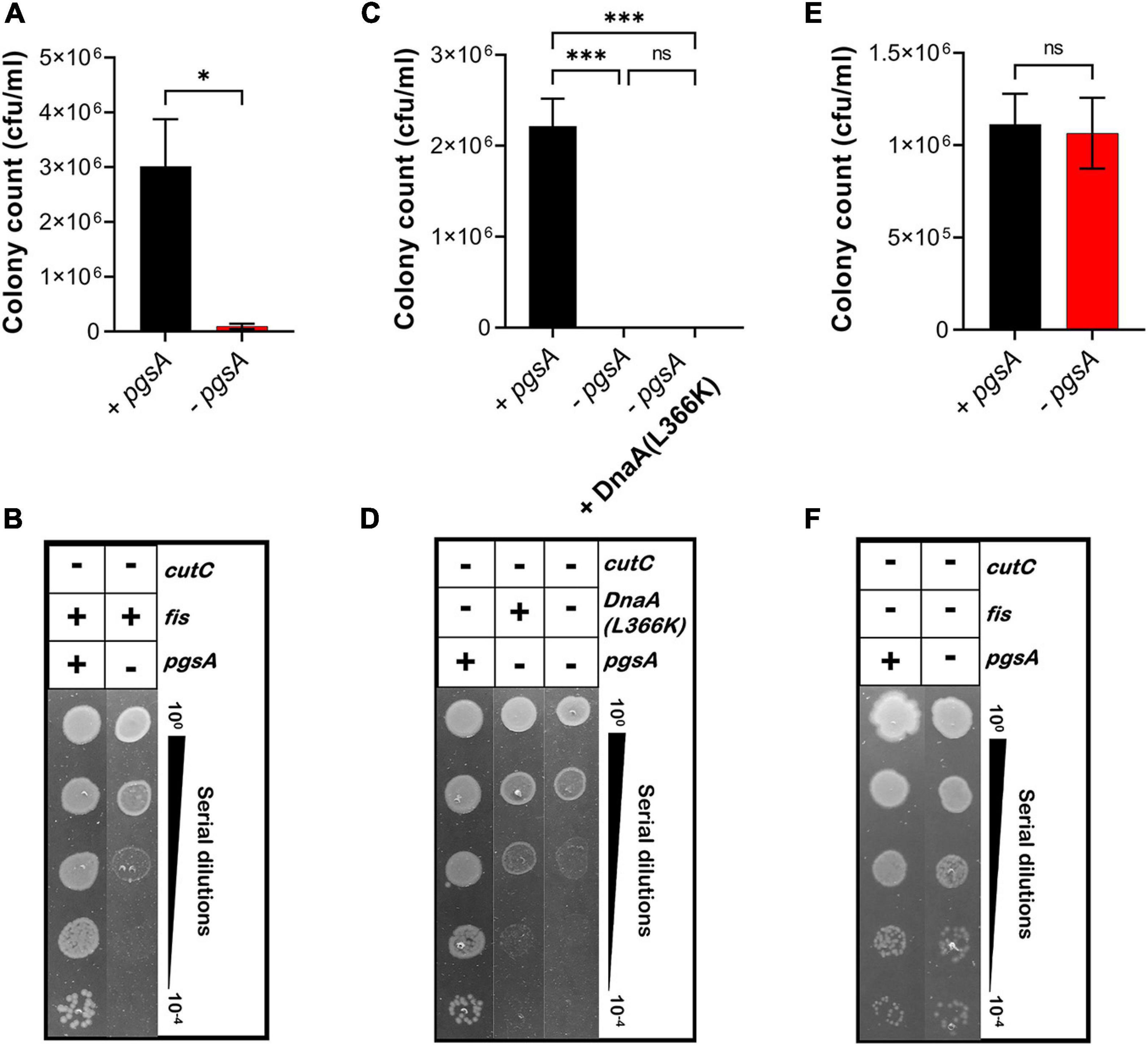
Figure 8. Growth of fis-null cells lacking promoter for MicL-S small RNA does not depend on cellular levels of acidic phospholipids. (A,B) Viability of MDL12 cutC-null cells treated with 1 mM IPTG for the expression of pgsA (black), no inducer control (red) in M9+CAA+Glu medium. (C,D) Viability of MDL12 cutC-null fis-null cells treated with 1 mM IPTG for the expression of pgsA (black), no inducer control (red) in M9+CAA+Glu medium. (E,F) Viability of MDL12 cutC-null cells carrying plasmid for DnaA(L366K) with 1 mM IPTG for the expression of pgsA (black), no inducer control (red), and 0.2% Arabinose control (blue) in M9+CAA+Glu medium. Viability is expressed in cfu/ml and presented on a linear scale. Data are means ± SEM of at least three independent experiments. ∗p < 0.05, ∗∗∗p < 0.001, ns p > 0.05 in Student’s t-test for analysis in plots A and C, and one-way ANOVA with Dunnett’s multiple comparison test.
fis-null Cells Unable to Synthesize PG Have an Altered Lipidomic Profile
We assessed the lipid profile of MDL12 cells with and without Fis to confirm that the absence of Fis did not somehow bestow a normal lipid composition. A targeted LC-MS approach to estimate total lipid content detected no evident change in phospholipid content in pgsA-expressing cells with or without endogenous levels of Fis (Figure 9A). However, in cells lacking fis and pgsA, we observed that out of 191 lipid metabolites, 60 phospholipid species were differentially produced as compared to fis+ and pgsA+ cells with levels of PG significantly reduced in the absence of active pgsA and fis (Figures 9B,C). The depletion in the levels of PG was accompanied by significantly higher levels of phosphatidic acid (PA), phosphatidylethanolamine (PE), diacylglycerol (DAG), and lysophosphatidylethanolamine (LPE) in comparison with the fis+ and pgsA+ cells.
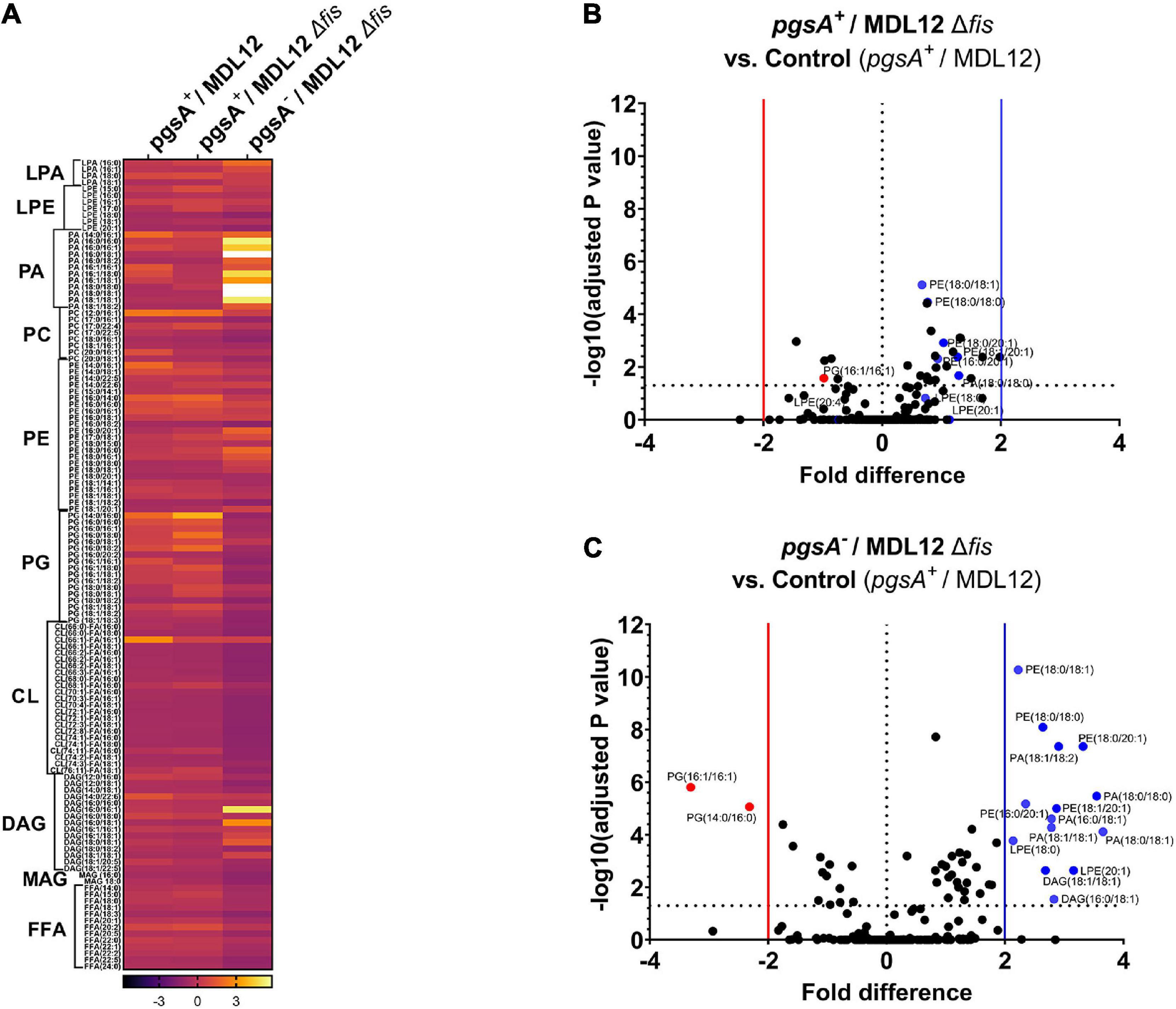
Figure 9. Lipidomic analysis of MDL12 cells in the presence or absence of Fis. (A) Lipidomic analysis represented in heatmap comparing MDL12 cells with 1 mM IPTG for the expression of pgsA (pgsA+/MDL12), MDL12 fis-null cells with 1 mM IPTG for the expression of pgsA (pgsA+/MDL12Δfis), and MDL12 fis-null cells without expression of pgsA (pgsA– /MDL12Δfis). Each row indicates the ratio values of individual lipid species for at least three biological replicates. ANOVA was performed to compare statistical differences between each group. Volcano plots comparing distribution of 191 lipids species among. (B) MDL12 fis-null cells expressing pgsA (pgsA+/MDL12Δfis) and control [MDL12 cells expressing of pgsA (pgsA+/MDL12)]. (C) MDL12 fis-null cells without expression of pgsA (pgsA– /MDL12Δfis) and control [MDL12 cells expressing of pgsA (pgsA+/MDL12)]. Horizontal dotted line indicates statistical significance with p = 0.05 for at least three biological replicates. A vertical red line separates red dots indicating data points with statistically significant fold decrease. A vertical blue line separates blue dots, indicating data points with statistically significant fold increase.
Discussion
Cellular membranes have long been hypothesized to be involved in chromosomal replication, including accumulating evidence indicating that membranes have an influence on DnaA protein function. E. coli pgsA-null cells unable to synthesize PG and CL contain decreased chromosomal DNA content due to reduced frequency of initiation from chromosomal origin and thus undergo growth-arrested phenotype (Fingland et al., 2012). In addition, cells deficient in PG and CL are unable to process outer membrane lipoprotein intermediates, with the intermediates accumulating at the inner membrane. Alternatively, defective lipoprotein sorting in bacterial cells containing an intact pgsA allele can be achieved by overproducing mutant Lpp protein in which the Cys-21 residue is substituted with Gly (Figures 10A,B). Although two not obviously related processes, initiation of replication and the biogenesis of lipoproteins are associated with the cellular composition of bacterial membranes, and any linkage among them is unknown, yet. We hypothesized the possibility that unprocessed pro-Lpp intermediate might poison the DnaA–oriC-mediated process of replication of initiation.
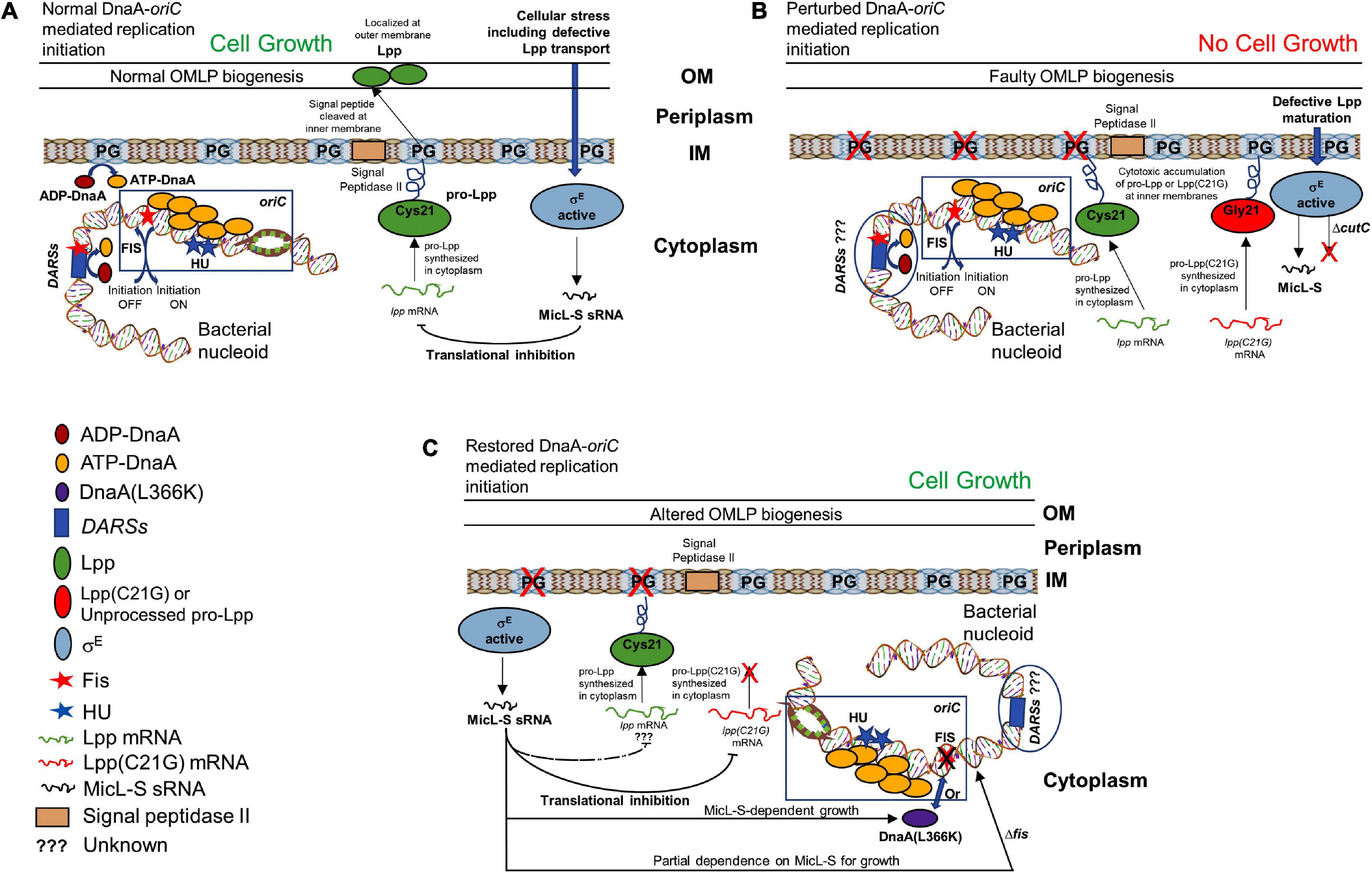
Figure 10. Schematic representation of how membrane stress caused by unprocessed outer membrane lipoprotein intermediate pro-Lpp affects DnaA and Fis-dependent growth. (A) Cells carrying normal cellular levels of inner membrane acidic phospholipids, PG, and CL support biogenesis of outer membrane lipoprotein (Lpp) and initiate DNA replication at the chromosomal origins. ATP-DnaA binds at specific oriC sites, and in the presence of HU protein, unwinds the duplex DNA, leading to the formation of a functional replisome. Acidic membranes and the specific chromosomal site DARSs play a role in the rejuvenation of replication-defective ADP-DnaA to replication-proficient ATP-DnaA. Binding of Fis protein at oriC and DARSs helps maintain timely inhibition of replication initiation. Stress to the cellular envelope stimulates the activation of an alternative sigma factor (such as sigma factor E, σE), which regulates the production of Lpp protein through MicL-S sRNA in response to the defective Lpp localization. (B) Cells unable to synthesize PG or that accumulate cytotoxic mutant Lpp(C21G) on the inner membrane are negatively affected for growth. (C) Cells expressing Lpp(C21G) or lacking PG can grow in a σE-dependent MicL-S sRNA manner, when overexpressing DnaA(L366K). In addition, restoration of growth can occur in cells lacking fis, independently of MicL-S sRNA.
A specific point mutant of chromosomal replication initiator protein, DnaA(L366K), when working in concert with wild-type DnaA, can initiate DNA synthesis both in vivo (Zheng et al., 2001) and in vitro (Saxena et al., 2011) and suppress the arrested growth of pgsA null cells. However, any mechanistic insights into how that occurs remain unknown. Secondly, the inability of cells to synthesize acidic membranes results in accumulation of unprocessed pro-lpp protein. That leads to the question of whether accumulating, unprocessed pro-Lpp poisons DnaA-dependent replication. Therefore, a valid question to ask is whether DnaA(L366K) can allow growth in cells carrying mutant Lpp(C21G). We found that the bacterial cells expressing both DnaA(L366K) and Lpp(C21G) (Figures 1C,D) can survive, but only with a marked decrease in the expression of the latter (lane 5, Figures 2C,D). There is evidence that the cellular abundance of Lpp is regulated by RNA polymerase sigma factor σE-dependent, small RNA, MicL-S (Guo et al., 2014). This led us to further test if cells overexpressing mutant Lpp(C21G) but lacking the σE-dependent MicL-S-mediated regulatory loops are capable of growth. If so, it would seem that DnaA(L366K) contains the potential to override cellular toxicity independent of MicL-S. Our results indicate that overexpression of DnaA(L366K) can bestow partial growth when Lpp(C21G) is present in growth-arresting amounts even when the σE-dependent envelope-protective mechanism is inactive (Figure 3A). Earlier work indicates that DnaA overproduction increases polA (encodes DNA polymerase I) expression in stationary-phase cultures (Quinones et al., 1997). The stimulation effect was independent of rpoS, which encodes the sigma factor for stationary-phase-inducible genes, which also include σE involved in the transcription of MicL.
From the subsequent loss-of-function study to determine whether Lpp(C21G) adversely affects the orisome, we found that cells lacking Fis were able to grow even when the inducer for Lpp(C21G) expression is present. However, the combined expression of Fis and Lpp(C21G) (Figures 4C,D) was highly inhibitory for growth in contrast to co-expression of Fis and Lpp (Figures 4E,F). These observations confirmed that Fis, at least when overproduced, is adversely affected by the inner membrane perturbed by accumulating Lpp intermediates. Strikingly, the near-complete depletion of Lpp(C21G) (lane 4, Figure 5A, and lane 3, Figure 5C) but not Lpp (lane 4, Figure 5E) in fis-null cells, similar to the ectopic expression of DnaA(L366K) (lane 4, Figure 2C, and lane 4, Figure 2E), underscores a concurrence in the restoration of growth with a decrease in expression of Lpp(C21G). DnaA(L366K), however, shows a strong dependence on σE-mediated MicL-S-dependent envelope protective loop to decrease Lpp(C21G) expression (lane 4, Figure 3C), while fis-null cells exhibit only a partial dependence on MicL-S-mediated translational inhibition of Lpp(C21G) (lane 3, Figure 6C) and thus indicates the pleiotropic role of Fis.
Cells lacking Fis grow even when normal levels of PG are absent (Figures 7E–H), confirming that the perturbation of inner membranes affects growth in a Fis-dependent manner. Our targeted LC-MS lipidomic approach to determine changes in total phospholipid content in the presence or absence of Fis indicates a significant fold increase in PA and other phospholipid species such as PE, DAG, and LPE when both pgsA and fis are absent (Figures 9A–C). This observation agrees with a previous study involving the ectopic expression of DnaA(L366K) to rescue cell growth in PG-deficient cells, accumulating higher levels of PA (Zheng et al., 2001). However, the elevated levels of PA do not fully substitute for the normal combined levels of PG and CL (Heacock and Dowhan, 1987; Figures 7A,B). Intervention, such as (1) high levels of DnaA(L366K) (Zheng et al., 2001; Figures 7C,D) or (2) lack of Fis (Figures 7E–H), may serve to reorganize the orisome. In conclusion, we now suggest that the poisoning of orisome by unprocessed immature pro-lipoprotein present at the inner membranes causes growth inhibition when cells are lacking normal levels of acidic phospholipids (Figure 10). This report suggests an intricate network between the physiological state and the composition of bacterial membranes and the orisome for the proper initiation of the chromosomal DNA replication (Figure 10C).
The role of auxiliary replication initiation factor Fis is well-established as a global transcription factor undertaking several functions (Cho et al., 2008). In addition to acting as a global transcription factor, Fis, like other nucleoid-associated proteins, remains associated with bacterial nucleoid and serves as a DNA bending–binding protein that helps to maintain the proper confirmation of oriC DNA (Gille et al., 1991). The binding of Fis protein to oriC DNA prevents untimely initiation of DNA replication in the cells, therefore serving as a negative regulator of DnaA-mediated replication at oriC (Wold et al., 1996). At the time of initiation, Fis needs to be removed and replaced with DnaA protein to form replication-proficient DnaA–oriC complexes (Margulies and Kaguni, 1998). Certain synthetic oriC sequences carrying mutations in DnaA recognition sites as well as Fis or IHF binding sites cause asynchronous initiations (Weigel et al., 2001). In addition to binding oriC, Fis binding to DARSs may regulate timely initiation of replication (Kasho et al., 2014). It remains to be determined whether the lack of fis might alter any ongoing primary or secondary mechanisms required to maintain cellular levels of replication-proficient ATP-DnaA, and help bacterial cells to escape membrane-mediated cytotoxicity. Considering the important roles that Fis plays in both nucleoid structure and gene regulation, further studies are also needed to see how these roles are impacted in cells either stressed from insufficient acidic phospholipid levels or accumulating Lpp intermediates. These studies include how the cellular level of Fis protein controls the expression of the σE sigma factor, the processing of the small RNA such as MicL-S, and the levels and function of other important proteins such as different cellular proteases, which are known to upregulate when cells are stressed.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
RS and DP conceived the idea along with EC. DP, RS, and DX performed experiments. MS provided the assistance. DP, RS, and EC analyzed the results and wrote the manuscript. DP, SB, and AC performed and analyzed lipidomic experiments. All authors have reviewed the results and approved the final version of the manuscript.
Funding
The authors would like to acknowledge the Metabolomics Shared Resource at Georgetown University, which is partially supported by the NIH/NCI/CCSG grant P30-CA051008. The financial support to conduct this research was provided by the Office of the Dean of Research, Georgetown University Medical Center.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Thomas Silhavy for sharing plasmid pC2 and anti-Lpp antiserum, Tsutomu Katayama for sharing Escherichia coli KP7364 strain, Kyle Divito for careful reading of the manuscript, and Hiroshi Nakai for early discussion in bacterial growth and quorum sensing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.677812/full#supplementary-material
Footnotes
References
Aranovich, A., Braier-Marcovitz, S., Ansbacher, E., Granek, R., Parola, A. H., and Fishov, I. (2015). N-terminal-mediated oligomerization of DnaA drives the occupancy-dependent rejuvenation of the protein on the membrane. Biosci. Rep. 35:e00250.
Aranovich, A., Gdalevsky, G. Y., Cohen-Luria, R., Fishov, I., and Parola, A. H. (2006). Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J. Biol. Chem. 281, 12526–12534. doi: 10.1074/jbc.m510266200
Aranovich, A., Parola, A. H., and Fishov, I. (2007). The reactivation of DnaA(L366K) requires less acidic phospholipids supporting their role in the initiation of chromosome replication in Escherichia coli. FEBS Lett. 581, 4439–4442. doi: 10.1016/j.febslet.2007.08.019
Atlung, T., Løbner-Olesen, A., and Hansen, F. G. (1987). Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol. Gen. Genet. MGG 206, 51–59. doi: 10.1007/bf00326535
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006–2008.
Baker, T. A., and Bell, S. P. (1998). Polymerases and the replisome: machines within machines. Cell 92, 295–305. doi: 10.1016/s0092-8674(00)80923-x
Boeneman, K., Fossum, S., Yang, Y., Fingland, N., Skarstad, K., and Crooke, E. (2009). Escherichia coli DnaA forms helical structures along the longitudinal cell axis distinct from MreB filaments. Mol. Microbiol. 72, 645–657. doi: 10.1111/j.1365-2958.2009.06674.x
Bramhill, D., and Kornberg, A. (1988). A model for initiation at origins of DNA replication. Cell 54, 915–918. doi: 10.1016/0092-8674(88)90102-x
Braun, V. (1975). Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 415, 335–377. doi: 10.1016/0304-4157(75)90013-1
Castuma, C. E., Crooke, E., and Kornberg, A. (1993). Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J. Biol. Chem. 268, 24665–24668. doi: 10.1016/s0021-9258(19)74517-7
Chaba, R., Grigorova, I. L., Flynn, J. M., Baker, T. A., and Gross, C. A. (2007). Design principles of the proteolytic cascade governing the σE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 21, 124–136. doi: 10.1101/gad.1496707
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978
Cho, B. K., Knight, E. M., Barrett, C. L., and Palsson, B. O. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910. doi: 10.1101/gr.070276.107
Cronan, J. E. (2003). Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57, 203–224. doi: 10.1146/annurev.micro.57.030502.090851
Dame, R. T. (2005). The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56, 858–870. doi: 10.1111/j.1365-2958.2005.04598.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dorman, C. J., and Deighan, P. (2003). Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13, 179–184. doi: 10.1016/s0959-437x(03)00025-x
Dorman, C. J., Hinton, J. C., and Free, A. (1999). Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7, 124–128. doi: 10.1016/s0966-842x(99)01455-9
Fingland, N., Flåtten, I., Downey, C. D., Fossum-Raunehaug, S., Skarstad, K., and Crooke, E. (2012). Depletion of acidic phospholipids influences chromosomal replication in Escherichia coli. Microbiol. Open 1, 450–466. doi: 10.1002/mbo3.46
Flatten, I., Fossum-Raunehaug, S., Taipale, R., Martinsen, S., and Skarstad, K. (2015). The DnaA protein is not the limiting factor for initiation of replication in Escherichia coli. PLoS Genet. 11:e1005276. doi: 10.1371/journal.pgen.1005276
Flatten, I., and Skarstad, K. (2013). The Fis protein has a stimulating role in initiation of replication in Escherichia coli in vivo. PLoS One 8:e83562. doi: 10.1371/journal.pone.0083562
Fujimitsu, K., Senriuchi, T., and Katayama, T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. doi: 10.1101/gad.1775809
Garner, J., and Crooke, E. (1996). Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 15, 2313–2321. doi: 10.1002/j.1460-2075.1996.tb00585.x
Garner, J., Durrer, P., Kitchen, J., Brunner, J., and Crooke, E. (1998). Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J. Biol. Chem. 273, 5167–5173. doi: 10.1074/jbc.273.9.5167
Gille, H., Egan, J. B., Roth, A., and Messer, W. (1991). The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 19, 4167–4172. doi: 10.1093/nar/19.15.4167
Guo, M. S., Updegrove, T. B., Gogol, E. B., Shabalina, S. A., Gross, C. A., and Storz, G. (2014). MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634. doi: 10.1101/gad.243485.114
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. O. N. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Heacock, P. N., and Dowhan, W. (1987). Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J. Biol. Chem. 262, 13044–13049. doi: 10.1016/s0021-9258(18)45164-2
Inouye, M. (1979). “Lipoprotein of the outer membrane of Escherichia coli,” in Biomembranes, ed. L. A. Manson (Boston, MA: Springer), 141–208. doi: 10.1007/978-1-4615-6564-2_4
Inouye, S., Franceschini, T., Sato, M., Itakura, K., and Inouye, M. (1983). Prolipoprotein signal peptidase of Escherichia coli requires a cysteine residue at the cleavage site. EMBO J. 2, 87–91. doi: 10.1002/j.1460-2075.1983.tb01386.x
Jensen, S. I., Lennen, R. M., Herrgård, M. J., and Nielsen, A. T. (2015). Seven gene deletions in seven days: fast generation of Escherichia coli strains tolerant to acetate and osmotic stress. Sci. Rep. 5:17874.
Johansen, J., Rasmussen, A. A., Overgaard, M., and Valentin-Hansen, P. (2006). Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364, 1–8. doi: 10.1016/j.jmb.2006.09.004
Kasho, K., Fujimitsu, K., Matoba, T., Oshima, T., and Katayama, T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. doi: 10.1093/nar/gku1051
Khrapunov, S., Brenowitz, M., Rice, P. A., and Catalano, C. E. (2006). Binding then bending: a mechanism for wrapping DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 19217–19218. doi: 10.1073/pnas.0609223103
Kikuchi, S., Shibuya, I., and Matsumoto, K. (2000). Viability of an Escherichia coli pgsANull mutant lacking detectable phosphatidylglycerol and cardiolipin. J. Bacteriol. 182, 371–376. doi: 10.1128/jb.182.2.371-376.2000
Klein, G., and Raina, S. (2019). Regulated assembly of LPS, its structural alterations and cellular response to LPS defects. Int. J. Mol. Sci. 20:356. doi: 10.3390/ijms20020356
Li, Z., Kitchen, J. L., Boeneman, K., Anand, P., and Crooke, E. (2005). Restoration of growth to acidic phospholipid-deficient cells by DnaA (L366K) is independent of its capacity for nucleotide binding and exchange and requires DnaA. J. Biol. Chem. 280, 9796–9801. doi: 10.1074/jbc.m413923200
Lima, S., Guo, M. S., Chaba, R., Gross, C. A., and Sauer, R. T. (2013). Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340, 837–841. doi: 10.1126/science.1235358
Luijsterburg, M. S., Noom, M. C., Wuite, G. J., and Dame, R. T. (2006). The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156, 262–272. doi: 10.1016/j.jsb.2006.05.006
Mao, G., Zhao, Y., Kang, X., Li, Z., Zhang, Y., Wang, X., et al. (2016). Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat. Commun. 7:10198.
Margulies, C., and Kaguni, J. M. (1998). The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 26, 5170–5175. doi: 10.1093/nar/26.22.5170
Marszalek, J., and Kaguni, J. M. (1994). DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269, 4883–4890. doi: 10.1016/s0021-9258(17)37627-5
Matsuyama, S., Tajima, T., and Tokuda, H. (1995). A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14, 3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x
Miyazaki, C., Kuroda, M., Ohta, A., and Shibuya, I. (1985). Genetic manipulation of membrane phospholipid composition in Escherichia coli: pgsA mutants defective in phosphatidylglycerol synthesis. Proc. Natl. Acad. Sci. U.S.A. 82, 7530–7534. doi: 10.1073/pnas.82.22.7530
Nakamura, K., and Inouye, M. (1982). Construction of versatile expression cloning vehicles using the lipoprotein gene of Escherichia coli. EMBO J. 1, 771–775. doi: 10.1002/j.1460-2075.1982.tb01244.x
Nicoloff, H., Gopalkrishnan, S., and Ades, S. E. (2017). Appropriate regulation of the σE-dependent envelope stress response is necessary to maintain cell envelope integrity and stationary-phase survival in Escherichia coli. J. Bacteriol. 199, 1–17.
Paetzel, M., Karla, A., Strynadka, N. C., and Dalbey, R. E. (2002). Signal peptidases. Chem. Rev. 102, 4549–4580.
Pierucci, O. L. G. A., Rickert, M., and Helmstetter, C. E. (1989). DnaA protein overproduction abolishes cell cycle specificity of DNA replication from oriC in Escherichia coli. J. Bacteriol. 171, 3760–3766. doi: 10.1128/jb.171.7.3760-3766.1989
Quinones, A., Wandt, G., Kleinstäuber, S., and Messer, W. (1997). DnaA protein stimulates polA gene expression in Escherichia coli. Mol. Microbiol. 23, 1193–1202. doi: 10.1046/j.1365-2958.1997.2961658.x
Raivio, T. L., and Silhavy, T. J. (2001). Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55, 591–624. doi: 10.1146/annurev.micro.55.1.591
Reisch, C. R., and Prather, K. L. (2015). The no-SCAR (scarless Cas9 assisted recombineering) system for genome editing in Escherichia coli. Sci. Rep. 5:15096.
Riber, L., Frimodt-Møller, J., Charbon, G., and Løbner-Olesen, A. (2016). Multiple DNA binding proteins contribute to timing of chromosome replication in E. coli. Front. Mol. Biosci. 3:29. doi: 10.3389/fmolb.2016.00029
Rimsky, S. (2004). Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7, 109–114. doi: 10.1016/j.mib.2004.02.001
Rose, R. E. (1988). The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. doi: 10.1093/nar/16.1.355
Sankaran, K., and Wu, H. C. (1994). Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706. doi: 10.1016/s0021-9258(17)32077-x
Saxena, R., Fingland, N., Patil, D., Sharma, A. K., and Crooke, E. (2013). Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 14, 8517–8537. doi: 10.3390/ijms14048517
Saxena, R., Rozgaja, T., Grimwade, J., and Crooke, E. (2011). Remodeling of nucleoprotein complexes is independent of the nucleotide state of a mutant AAA+ protein. J. Biol. Chem. 286, 33770–33777. doi: 10.1074/jbc.m111.223495
Sekimizu, K., Bramhill, D., and Kornberg, A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. doi: 10.1016/0092-8674(87)90221-2
Sekimizu, K., and Kornberg, A. (1988). Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 263, 7131–7135. doi: 10.1016/s0021-9258(18)68615-6
Suzuki, M., Hara, H., and Matsumoto, K. (2002). Envelope disorder of Escherichia coli cells lacking phosphatidylglycerol. J. Bacteriol. 184, 5418–5425. doi: 10.1128/jb.184.19.5418-5425.2002
Udekwu, K. I., and Wagner, E. G. H. (2007). Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35, 1279–1288. doi: 10.1093/nar/gkl1154
Varshavsky, A. J., Nedospasov, S. A., Bakayev, V. V., Bakayeva, T. G., and Georgiev, G. P. (1977). Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 4, 2725–2745. doi: 10.1093/nar/4.8.2725
Weigel, C., Messer, W., Preiss, S., Welzeck, M., and Morigen, B. E. (2001). The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40, 498–507. doi: 10.1046/j.1365-2958.2001.02409.x
Wold, S., Crooke, E., and Skarstad, K. (1996). The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 24, 3527–3532. doi: 10.1093/nar/24.18.3527
Xia, W., and Dowhan, W. (1995). In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 783–787. doi: 10.1073/pnas.92.3.783
Yung, B. Y., and Kornberg, A. (1988). Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 85, 7202–7205. doi: 10.1073/pnas.85.19.7202
Keywords: Escherichia coli, DNA replication, acidic phospholipids, lipoprotein biogenesis, DnaA, Fis, MicL-S
Citation: Patil D, Xun D, Schueritz M, Bansal S, Cheema A, Crooke E and Saxena R (2021) Membrane Stress Caused by Unprocessed Outer Membrane Lipoprotein Intermediate Pro-Lpp Affects DnaA and Fis-Dependent Growth. Front. Microbiol. 12:677812. doi: 10.3389/fmicb.2021.677812
Received: 08 March 2021; Accepted: 29 April 2021;
Published: 07 June 2021.
Edited by:
Morigen Morigen, Inner Mongolia University, ChinaReviewed by:
Julia Grimwade, Florida Institute of Technology, United StatesTsutomu Katayama, Kyushu University, Japan
Copyright © 2021 Patil, Xun, Schueritz, Bansal, Cheema, Crooke and Saxena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rahul Saxena, cnM0MjZAZ2VvcmdldG93bi5lZHU=
 Digvijay Patil
Digvijay Patil Dan Xun1
Dan Xun1 Amrita Cheema
Amrita Cheema Elliott Crooke
Elliott Crooke Rahul Saxena
Rahul Saxena