- 1Fujian Provincial Key Laboratory of Agroecological Processing and Safety Monitoring, School of Life Sciences, Fujian Agriculture and Forestry University, Fujian, China
- 2Key Laboratory of Crop Ecology and Molecular Physiology (Fujian Agriculture and Forestry University), Fujian Province University, Fujian, China
- 3Department of Biochemistry and Molecular Biology, School of Life Sciences, Pondicherry University, Puducherry, India
- 4Department of Microbiology, M.R. Government Arts College, Tamil Nadu, India
- 5PG Research and Department of Microbiology, St. Joseph’s College of Arts and Science (Autonomous), Tamil Nadu, India
- 6Key Laboratory of Marine Biotechnology of Fujian Province, Institute of Oceanology, Fujian Agriculture and Forestry University, Fujian, China
Biofilm formation is a major concern in various sectors and cause severe problems to public health, medicine, and industry. Bacterial biofilm formation is a major persistent threat, as it increases morbidity and mortality, thereby imposing heavy economic pressure on the healthcare sector. Bacterial biofilms also strengthen biofouling, affecting shipping functions, and the offshore industries in their natural environment. Besides, they accomplish harsh roles in the corrosion of pipelines in industries. At biofilm state, bacterial pathogens are significantly resistant to external attack like antibiotics, chemicals, disinfectants, etc. Within a cell, they are insensitive to drugs and host immune responses. The development of intact biofilms is very critical for the spreading and persistence of bacterial infections in the host. Further, bacteria form biofilms on every probable substratum, and their infections have been found in plants, livestock, and humans. The advent of novel strategies for treating and preventing biofilm formation has gained a great deal of attention. To prevent the development of resistant mutants, a feasible technique that may target adhesive properties without affecting the bacterial vitality is needed. This stimulated research is a rapidly growing field for applicable control measures to prevent biofilm formation. Therefore, this review discusses the current understanding of antibiotic resistance mechanisms in bacterial biofilm and intensely emphasized the novel therapeutic strategies for combating biofilm mediated infections. The forthcoming experimental studies will focus on these recent therapeutic strategies that may lead to the development of effective biofilm inhibitors than conventional treatments.
Introduction
Biofilms are severe health concerns due to their multidrug resistance abilities, host defense, and other stresses (De La Fuente-Nunez et al., 2013). Therefore, it leads to chronic bacterial infections worldwide (Subhadra et al., 2018; Sharma et al., 2019). Bacterial biofilm is a multifaceted structure of communities with diverse bacterial colonies of cells in a group (Kostakioti et al., 2013). Biofilm referred to the intricate three-dimensional (3-D) aggregation of bacteria attached to a surface and buried inflexibly in an Extracellular Polymeric Substance (EPS) matrix. Bacteria form biofilms in every substratum, and their associated infections in plants, animals, and humans (Lebeaux et al., 2014; Padmavathi et al., 2017; Kannappan et al., 2020). Besides, biofilms also play destructive roles in industrial pipelines corrosion (Lenhart et al., 2014). Bacterial biofilms can attach to various materials such as metals, glass surfaces, plastic wares, tissues, and clinical devices. Bacterial communities also produce biofilm, especially on all medical implants, including vascular grafts, heart valves, intrauterine devices, pacemakers, prosthetic joints, catheters, sutures, and contact lenses to acute infections (Kannappan et al., 2017b).
Bacteria inside the biofilm can also withstand harsh conditions and hold secreted polymers such as polysaccharides, extracellular DNA (e-DNA), proteins, and amyloidogenic proteins (Sharma et al., 2019). The pathogenesis and persistence of bacterial pathogens are dependent on the formation and maintenance of intact biofilms (Xu et al., 2000; Stewart and Costerton, 2001; Pang et al., 2013; Wilkins et al., 2014). Bacterial cells swathed in the biofilm are up to 1000 fold resistant to antibiotic agents. In this biofilm form, bacteria are more resistant to various antibacterial and chemical treatments. Biofilms offer the guard to the bacteria from pH, nutrients deficiency, and mechanical forces (Bryers, 1993; Sutherland, 2001; Singh et al., 2017). Therefore, the biofilm matrix gives additional resistance to bacteria, leading to bad bug’s infections like drug resistant bacteria.
Novel biofilm inhibitors have been investigated from a various sources in order to prevent biofilm formation and eliminate persistent biofilms. Although the research focused on identifying compounds able to target and inhibit this biofilm mode of bacterial growth is explicitly still inadequate (Peng et al., 2015; van Tilburg Bernardes et al., 2015; Kannappan et al., 2017a; Srinivasan et al., 2018). As a result, new therapeutic options are needed for controlling the biofilm associated infections. At present, the study of biofilm and its strategies to eliminate without any resistant development is one of the utmost significant fields of research. Several reviews on biofilm inhibitors have already been reported, but our review mainly focuses on significant novel strategies to control biofilm mediated bacterial infections.
Biofilm Formation
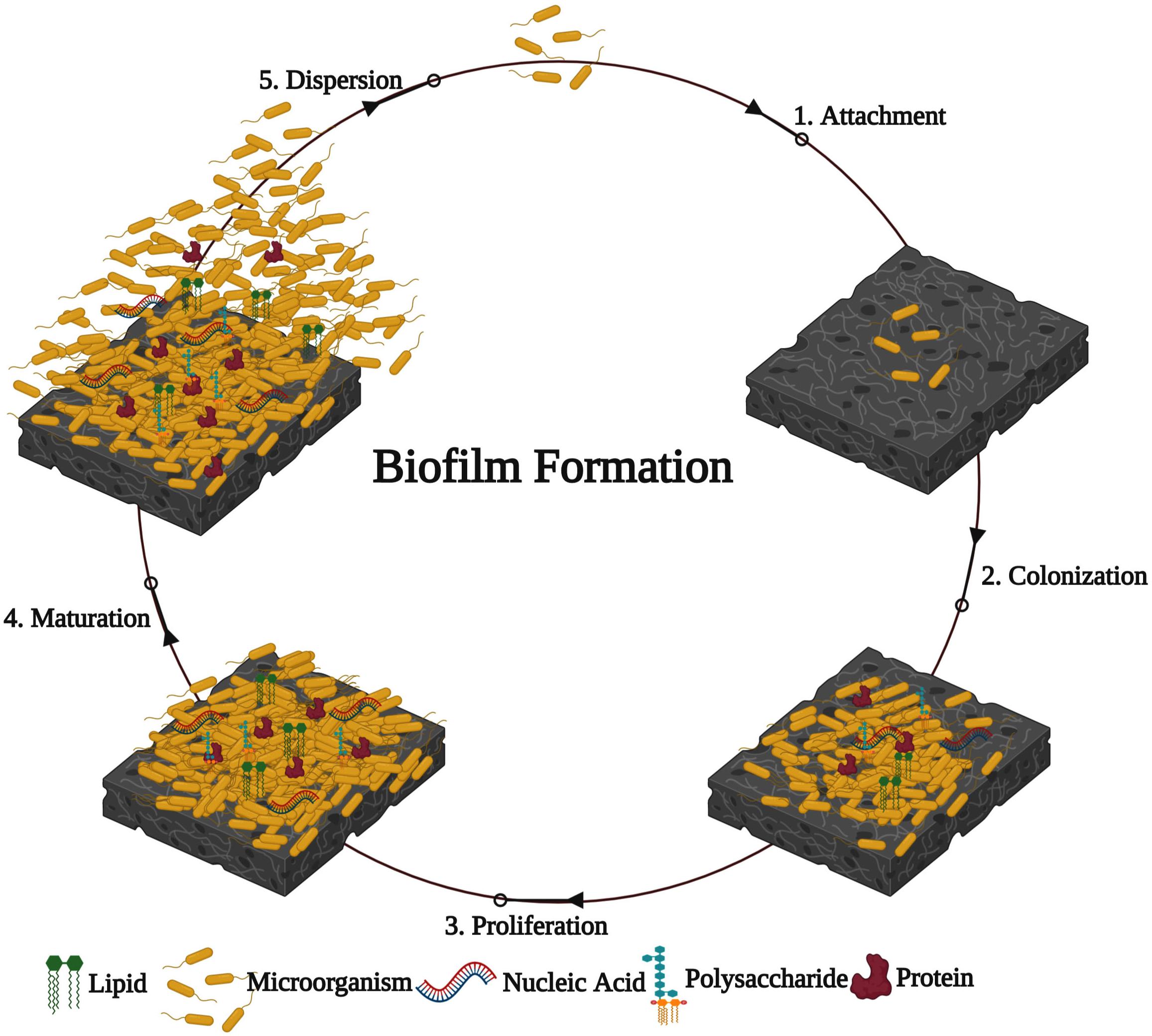
Generally, biofilm formation by bacterial pathogens on any substratum/layer involves five major stages (Kostakioti et al., 2013; Yin et al., 2019). (1) Attachment: at an initial stage, free-swimming planktonic cells reversibly attach to the biotic or abiotic surfaces through weak interactions such as acid-base, hydrophobic, Van der Waals, and electrostatic forces. (2) Colonization: bacterial pathogens irreversibly attach to the surface through stronger interactions such as collagen-binding adhesive proteins, lipopolysaccharides, flagella, and pili. (3) Proliferation: the multilayered bacterial cells are profoundly accumulated, and the enormous amounts of EPS are produced. (4) Maturation: the attached multilayered bacterial cells grown into the matured biofilm with the typical 3D biofilm structure. (5) Dispersion: after the complete development of biofilm, it is disassembled or dispersed using mechanical and active processes (Figure 1).
Characteristics of Biofilm Formation
The formation of biofilm is a progressive process. Primarily, bacterial cells move onto a surface and adhere reversibly to the surface. In the second step, irreversible adherence occurs with the microcolonies expansion that produces an EPS matrix. Subsequently, the progress of the mature 3-D biofilm architecture emerges. Matured biofilms are more resistant to the host immune defenses and the action of antibacterial agents. During the dispersal of biofilm, the cells endure lysis and discharge from the biofilm community. Inside the host, bacteria produce biofilm on a biotic or an abiotic layer. The abiotic surface is typically coated with proteins or other biological molecules, forming a habituation film that changes cells adhesion. In biofilm formation, host cells can develop a fundamental part, and their components can be assimilated into the biofilm matrix (Lynch and Robertson, 2008; Romling and Balsalobre, 2012).
Components in the Biofilm Matrix
Biofilms are a group of microorganisms in which microbes produced EPS such as proteins (<1–2%), polysaccharides (1–2%), DNA (<1%), and RNA (<1%). In addition to these components, water (up to 97%) is the key portion of biofilm, distributed in a non-homogenous pattern and mainly accountable for the movement of nutrients inside the biofilm matrix (Batoni et al., 2016; Nazir et al., 2019). The capability to build and conserve an organized biofilm community mainly depends on EPS matrix components (Sutherland, 2001; Branda et al., 2005; Limoli et al., 2015). The EPS in the biofilm matrix commands a charter for the biofilms. The biofilm inhabitants are always shielded from the atmosphere (competitive microbes, temperature, host cells, antimicrobials, and desiccation) while also having access to nutrients and the capacity to react environmental changes. Bacteria generate multiple types of EPS to handle these needs in different ways. EPS can help the bacteria to adhere on many different surfaces and hosts; provide protection from the environment and reservoirs for nutrient acquisition (Ramanathan et al., 2018; Kannappan et al., 2019a).
Role of EPS in Biofilm Formation
EPS is a superglue that accounts for the biofilm communities slimy nature and is a complex blend of biopolymers, including polysaccharides, proteins, e-DNA, and phospholipids (Nazir et al., 2019). In general, EPS composition changes with the type of pathogens, biofilm age, and environmental conditions (desiccation, pH, oxygen, nitrogen, temperature, and nutrients availability) (Mayer et al., 1999; Kostakioti et al., 2013). The bacteria existing in biofilm suggest that they can respond to their surroundings by modifying their EPS composition and adhesion. EPS provides a physical framework for the attachment among cells and surfaces. It also acts as a blockade between biofilm cells and surroundings (Mitchell et al., 2016). It protects microbes from antimicrobial compounds, chemicals, desiccation, radiation, and unfavorable environmental conditions. They are also cherishing bacterial cells inside the biofilm with a constant supply of nutrients and keeping their capability to respond environmental variations. As related to a protein in EPS composition, polysaccharides are extremely sticky and fundamental for biofilm maintenance and its environment. Similarly, proteins from EPS matrix modify the cell wall assets, adherence, virulence, and morphogenesis; protect cells from harmful conditions and phagocytes (Chaffin et al., 1998; Bridier et al., 2011). Another vital component of EPS is e-DNA, which increases biofilm structural integrity, exchange of genetic information, nutrients provision, biofilm stability, and drug resistance (Donlan, 2002; Martins et al., 2010).
Biofilm Mediated Bacterial Infections
The level of clinical care has advanced dramatically over the few years, but bacterial biofilm infections continue to pose a significant threat to public health. Hoiby, Lam and his colleagues were the first to identify a direct correlation between the formation of biofilms and recurrent infections, especially with Pseudomonas aeruginosa in cystic fibrosis patients (Hoiby, 1977; Lam et al., 1980). The decades that followed embraced the idea that biofilms are an important source of tissue related infections (Lebeaux et al., 2013). There are various sites in the human body where biofilm infections may occur due to either a pre-existing condition or a hospital acquired infection. Further, tissue related bacterial biofilm infections have been noted to occur more often in immunocompromised patients, and patients with underlying chronic illness such as cardiovascular disease, diabetes, skin barrier breakage, cancer, or especially if the infection is severe or starts early in the course of the illness (Sivaranjani et al., 2018). In addition, it was understood that the usage of different forms of embedded medical devices would favor adhesion and the colonization of bacteria, resulting in infections (Marrie et al., 1982; Donlan and Costerton, 2002). Further, several types of embedded medical devices are associated with the development of bacterial biofilms (Figure 2). More, ventilator pneumonia, central line bloodstream infections, urinary, pacemaker, and peripheral vascular catheter infections are the utmost common device related bacterial biofilm infections (Kamaruzzaman et al., 2018; Kannappan et al., 2019b).
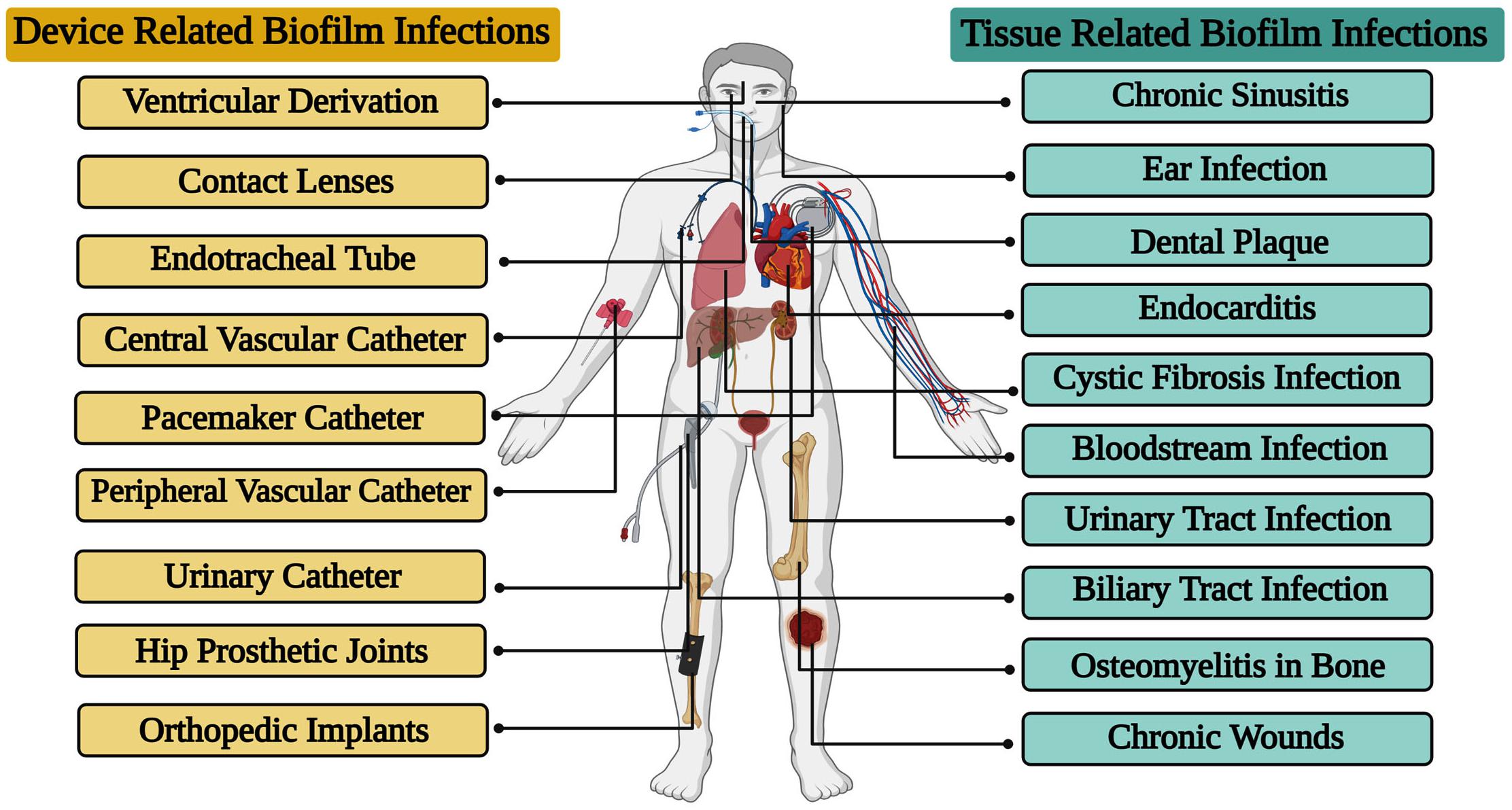
Figure 2. Various types of tissue and device related biofilm infections caused by bacterial pathogens.
Biofilm in Antibiotic Tolerance and Persister Formation
Usually, chronic tissue and device related bacterial infections are difficult to treat because the patient is exposed to the risk of recurrence (Lewis, 2007). Bacterial biofilms can spread to other parts of the body or around the infection source if planktonic bacteria originate from the biofilm. Planktonic bacteria may be eradicated by combined action of host immune responses and antibiotics. However, a subset of biofilm bacteria those are not destroyed by the antibiotic treatment and can able to trigger the recurrence of infection (Lebeaux et al., 2014).
Inside the biofilm, bacterial cells reveal morphological, and physiological changes assisted by differential gene expression due to the gradient in toxic components, diffusible gasses, or nutritional pressure (Stewart and Costerton, 2001). Depleted oxygen and nutrients within biofilm stimulate asynchronous growth, which exhibits variations in the level of gene expression and may lead to drug tolerance. Phenotypic variety of bacteria within biofilm augments greater coordination, empowers genes for reprogramming, and involves the efflux of toxins, lipid biosynthesis, iron sequestration, DNA repair, and host immune modulation, etc. (Li et al., 2016). It provides persistence and selective dissemination of resilient cells enduring stress. Nowadays, resistance to antibacterial agents is the most crucial cause of non-effective therapy of biofilm-associated bacterial infections. The reason behind increased antibiotic resistance of bacteria is (1) Difficulty for the diffusion of antibiotics into the biofilm and electrostatic charge of the EPS, which attract oppositely charged antibiotics; (2) A slower growth rate; (3) Variations in phenotype acquired by bacteria forming biofilms and (4) Inactivation of antibiotics by enzymes secreted by bacteria (Figure 3; Lewis, 2001; Lewis, 2008, 2010; Sharma et al., 2019).
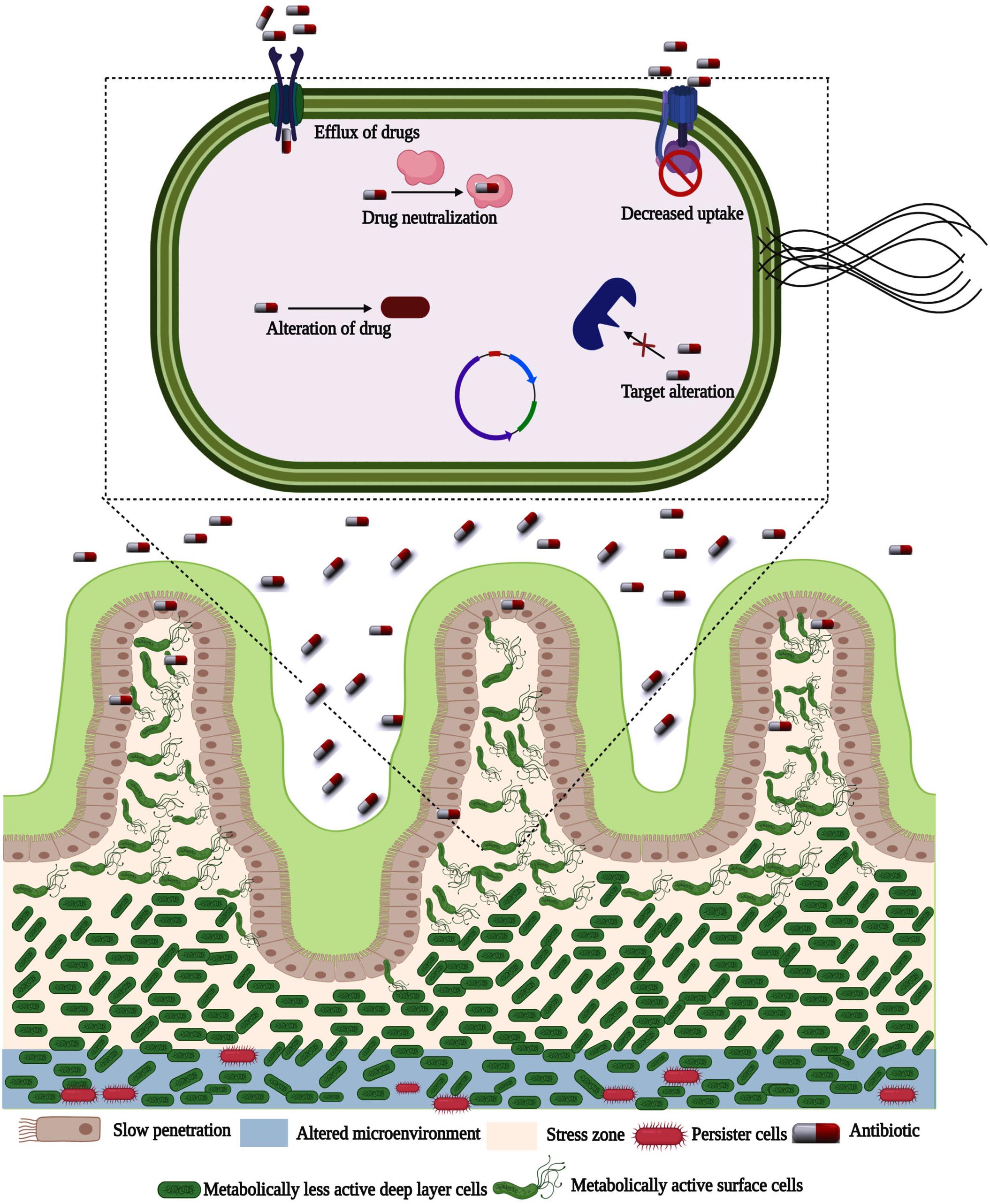
Figure 3. Antibiotic resistance and persister formation in bacterial biofilm. In the presence of EPS, antibiotic penetration is slowed. In reaction to antibiotic stress, certain bacteria in the biofilm alter their behavior. To resist biofilm eradication, the microenvironment in deeper parts is changed. Biofilms have a more concentration of persister cells in the altered microenvironment.
Several promising mechanisms are underlying the phenotypic resistance, which may be influenced by the type of antibiotic treatment and the host, growth rate of biofilm, transformed metabolism, and the presence of an oxygen gradient that prevents the action of some antibiotics (Li et al., 2016; Cheng et al., 2019). Besides, biofilms contain a great population of persister cells, which endure against antibiotics treatment. A limited dispersal of antibiotics into biofilms has been proposed, but in most occurrences, no direct evidence has been provided (O’Toole et al., 2000; Lewis, 2010).
Signaling in Biofilm Formation: How Bacteria “Talk to Each Other”
Bacterial cells in biofilm communicate with each other and coordinate their behavior through the signal molecules. This cell-cell communication system is called Quorum Sensing (QS), which goes beyond bacterial cell density (Miller and Bassler, 2001). Mostly, roles of QS are classified into four kinds such as (1) Cell maintenance and division (exoenzymes and siderophores production), (2) Horizontal gene transfer (conjugation), (3) Host-pathogen interactions (antibiotic and bioluminescence production), and most importantly (4) Behavior (movement and biofilm formation). The QS mediated biofilm formation has well documented in several Gram-negative and Gram-positive bacterial pathogens (Carniol and Gilmore, 2004; Labbate et al., 2004; Kong et al., 2006; Longo et al., 2014; Brindhadevi et al., 2020).
Generally, QS facilitates the physiological status of the microbial population and controls the biofilm formation through signal molecules called N-acyl-homoserine lactones (AHL) or auto-inducing peptides (AIP) in Gram-negative and Gram-positive bacterial pathogens, respectively (Whitehead et al., 2001; Papenfort and Bassler, 2016; Rama Devi et al., 2016; Bhatt, 2018). According to the bacterial species, varieties of AHL exist in Gram-negative bacterial pathogens, and some may vary as the strain varies. Usually, the AHL are synthesized by AHL synthase gene luxI. The luxI gene is transcriptionally expressed to the basal level at low population density. Hence, the AHL molecules are scattered in the field. At the high cell density, the LuxR family of receptor proteins senses the AHL molecules. Then, the signal molecule attaches to the receptor protein until AHL hits a particular threshold concentration. At that time, the activated LuxR-AHL complex forms multimers with other activated LuxR-AHL complexes. Finally, these multimers control the transcription of QS regulated biofilm formation in several bacterial pathogens (Rasmussen and Givskov, 2006).
Recent Therapeutic Strategies for Biofilm Inhibition
Besides the conventional antibiotics, certain promising underlying strategies extended by the prevailing biofilm inhibitors hinder the biofilm formation and reduce microorganisms’ virulence. Most biofilm forming microorganisms are responsible for 80% of human infections (Shunmugaperumal, 2010) and their ill-health. Due to the EPS matrix of the biofilm, they resist the immune system of humans. Some antimicrobial peptides like defensins or existence biofilm inhibitors have an extensive part in acting upon the matrix (Lewis, 2001). However, many novel and interesting tactics or lines of attack combating against biofilms were identified, and their progression at the current scenario described in detail.
Quorum Sensing (QS) Blockage Strategy
Considering the QS system as the noteworthy comportment of biofilm synthesis by microorganisms, many researches contributed to recognizing the QS system’s blockage as a vital strategy to prevent biofilm. More, bacterial pathogens in the host can activate the QS signals for biofilm formation and virulence factors production. Therefore, inhibiting this bacterial communication through QS inhibitors makes the bacterial pathogens more susceptible to the host immune system and antibiotic responses (Ravindran et al., 2018; Jiang et al., 2019; Li et al., 2020). Consequently, it facilitates the targeting of QS as a therapeutic target for controlling biofilm mediated bacterial infections. The phenomenon of down-regulating or silencing the QS system is referred to as quorum quenching. Generally, blocking the QS system of Gram-negative bacterial pathogens can be done through three essential strategies: 1. Blocking the AHL molecule biosynthesis, 2. AHL inactivation or degradation, and 3. Interference with the signal receptor (Figure 4).
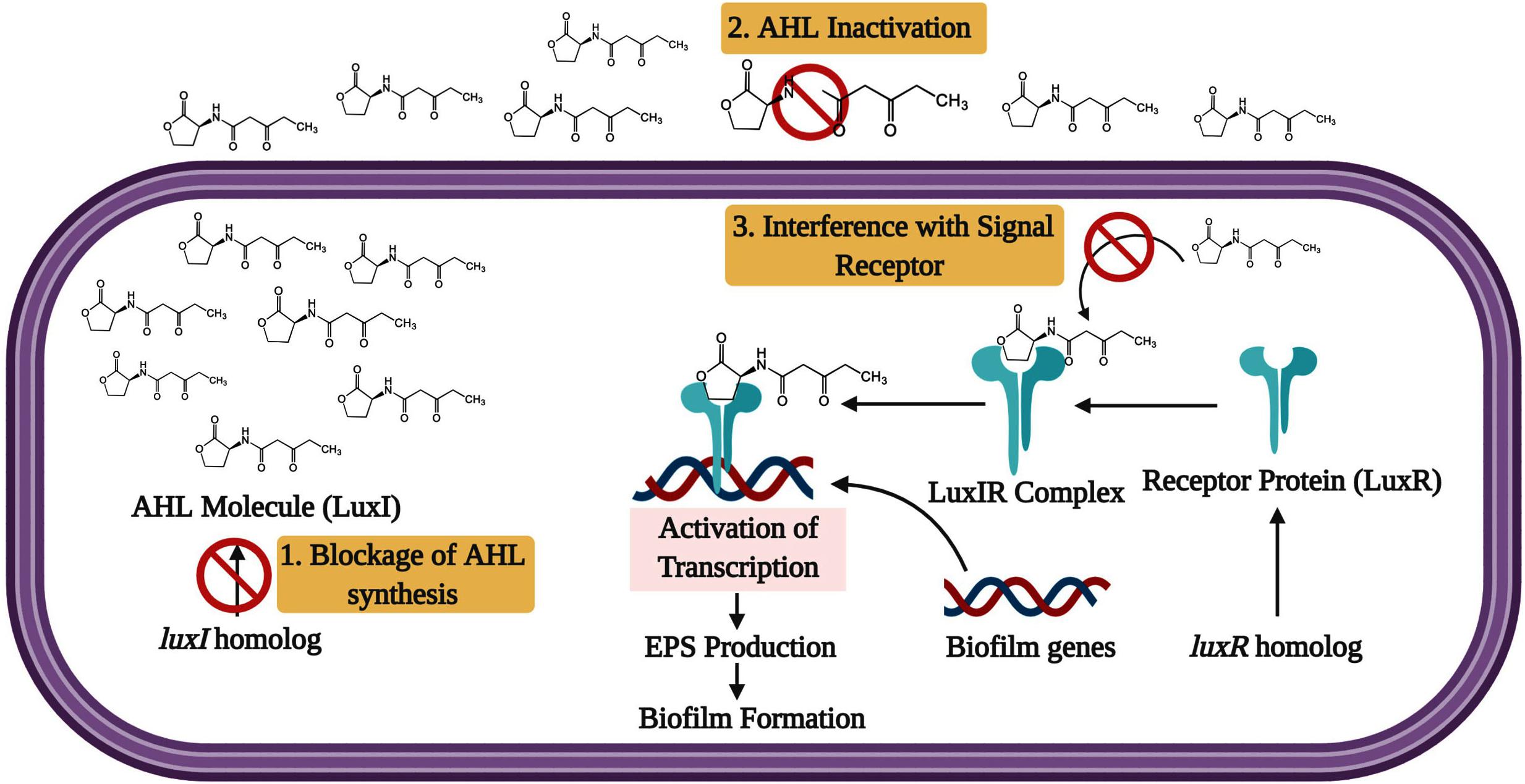
Figure 4. The inhibition approaches for LuxI/LuxR QS system. The signaling molecule AHL is produced by the luxI synthase gene and freely diffuses from each cell. When critical concentration is reached, the synthesized signal molecules diffuse back inside the bacterial cell and binds with LuxR. Then the QS transcription is activated by the LuxR-AHL complex. The target stages for inhibition are (1) Blockage of AHL molecule synthesis, (2) Degradation of the AHL molecule, and 3. Interference with the signal receptor.
Hindering the AHL Signal Molecule Biosynthesis
Previously, the in vitro analysis has been performed on the catalysis of AHL molecule biosynthesis in sequentially ordered reaction. The S-adenosyl methionine (SAM) is used as the amino donor to produce homoserine lactone ring moiety. An adequately charged acyl carrier protein (ACP) is used as the precursor for producing the acyl side chain of the AHL signal (Parsek et al., 1999; Hentzer and Givskov, 2003; Li et al., 2016). Other studies made by Zano et al., 2013 and Masevicius et al., 2016 have shown that several Gram-negative bacterial pathogens can synthesize the S-adenosyl-L-methionine (AdoMet) as the primary methyl donor for several methylation processes. Zano et al. (2013) have revealed that this AdoMet may also act as a precursor for the production of two different QS signal molecules; therefore, targeting the hindrance of AdoMet can lead to inhibiting the biofilm formation in various Gram-negative bacterial pathogens. So, QS inhibitors that target AHL molecule biosynthesis can be developed using knowledge of signal generation. Various analogs of SAM have been continuously revealed to be an effective inhibitor of AHL molecule biosynthesis. Some antagonists of SAM, such as S-adenosylcysteine, and S-adenosylhomocysteine have shown to ensure the capacity to effectively inhibit the AHL synthesis, which is facilitated by the P. aeruginosa RhlI protein (Parsek et al., 1999). Further, Christensen et al. (2013) have screened the QS inhibitors to target the AHL molecule biosynthesis against Proteobacteria such as Burkholderia mallei and Yersinia pestis through high-throughput screening. Some earlier experiments have shown that macrolide antibiotics such as azithromycin and erythromycin administered at sub inhibitory concentrations have the capacity to suppress the P. aeruginosa AHL molecule biosynthesis and thereby inhibited their virulence factors and biofilm formation (Sofer et al., 1999; Pechere, 2001; Tateda et al., 2001).
AHL Signal Molecule Biodegradation or Alteration
Searching for enzymes capable of breaking down the AHL signal molecules is a promising strategy to eradicate the biofilm mediated bacterial infections altogether. AHL molecules are enzymatically destroyed by various forms of enzymes, eliminating AHL accumulation in the system. Generally, the enzymatic degradation or alteration of AHL signal molecules can be catalyzed by six major classes of enzymes rendering to their catalytic sites (Figure 5).
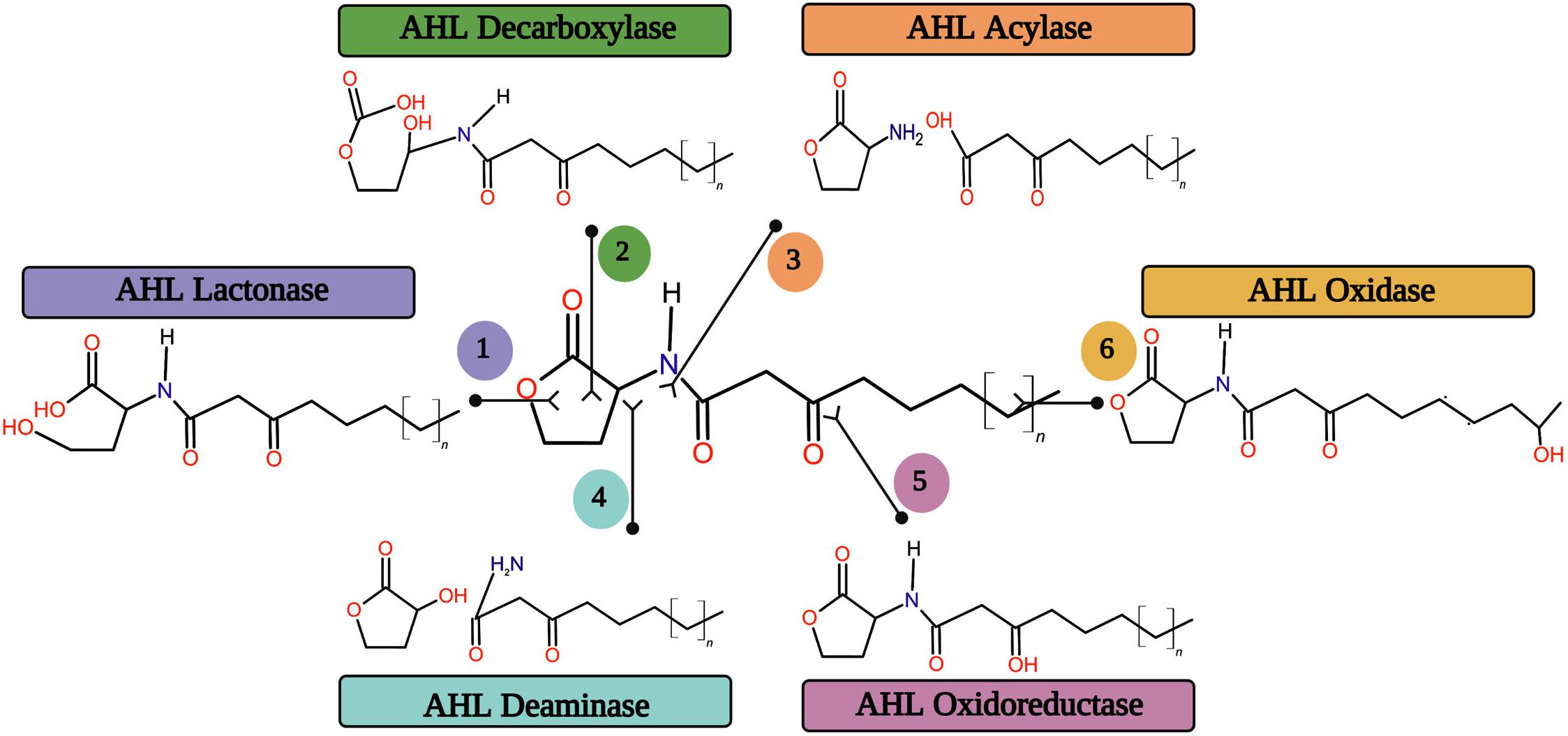
Figure 5. The six major quorum quenching enzymes on the degradation or alteration of AHL signal molecule.
The AHL lactonases are capable of opening the homoserine lactone ring by way of breaking the bond on the leftward of the double bonded oxygen. Further, the enzyme decarboxylases are also capable of doing the same by way of breaking the bond on the rightward of the double bonded oxygen without disturbing the rest of the AHL molecule structure. AiiA 24B1, the product of the aiiA gene from Bacillus spp. 24B1, hydrolyzes the lactone ring in the homoserine moiety of AHL and which is one of the first identified and well characterized AHL lactonase (Dong et al., 2000). A recent study made by Shastry et al. (2019) has revealed the biofilm inhibitory efficacy of AHL lactonase enzyme on the Aeromonas hydrophila biofilm formation. The AHL acylase is a family of enzyme that corresponds to the Ntn hydrolase superfamily. It hydrolyzes the AHL signal molecules, as their names imply (Utari et al., 2017). The AHL acylase enzyme was first exposed in the Variovorax paradoxus strain VAI-C. It hydrolyzes the amide bond between the homoserine lactone and acyl side chain in AHL molecules, releasing homoserine lactone (HSL) and free fatty acid (Leadbetter and Greenberg, 2000). Several AHL acylase enzymes were identified from different bacterial sources for their biofilm inhibitory potential (Paul et al., 2009; Christiaen et al., 2014). The acyl side chain may also be cleaved from the HSL ring by deaminase but at a different location. Deamination encodes final products as an acyl side with NH2 and homoserine lactone with OH (Kose-Mutlu et al., 2019). The AHL oxidase catalyzes the carbon atoms oxidation in acyl chains of AHL signal molecules (Gao et al., 2013). The AHL oxidoreductases oxidize or reduce the carboxyl group of the third carbon to attack the side chain of AHL molecules. This type of enzymes does not break down the signal, but rather it alters the AHL molecule, thereby modifying the binding efficacy of receptor proteins with signal molecules (Vogel and Quax, 2019). The AHL oxidoreductase enzyme has recently been reported for its inhibitory potential on the autoinducer-2 mediated biofilm formation in Gram-negative bacterial pathogens by modifying the AHL signal molecule (Weiland-Brauer et al., 2016).
Interference With Receptor Proteins by Analog Compounds
The membrane receptors can be interrupted by binding the antagonistic molecules so that the receptors are unavailable to bind with AHL signaling molecules. If there is no signal recognition, then there is a variation in the bacterial population’s physiological behavior, especially in reducing biofilm activity, less virulence, and low antibacterial tolerance. Furanone has been first identified analog compound as the potent QS inhibitor, which effectively inhibits the biofilm formation of Staphylococcus epidermidis (Hume et al., 2004). Further, the study of Lonn-Stensrud et al. (2009) has highlighted the antagonistic activity of furanone with a drastic interlude in the QS signaling, isolated from the marine algae, Delisea pulchra. The QS system of Vibrio harveyi was silenced by a marine strain Halobacillus salinus, which synthesize an antagonistic molecule that suppress the activation of lux gene and therefore hindering the signaling molecule biosynthesis (Teasdale et al., 2009). Similarly, in yet another study by Choi et al. (2012), honaucins was synthesized by Leptolyngbya crosbyana, which affects cell-to-cell communication by inhibiting the QS. Furthermore, bacterial communication of QS can also be blocked by the fungal metabolite such as patulin and penicillic acid from different Penicillium strains, respectively. For instance, the reports of Wagner et al. (2004); Rasmussen et al. (2005); Abraham (2005) have revealed that the QS in P. aeruginosa could be inhibited by the penicillinic acid and patulin. The antagonistic potential of phytol on biofilm mediated infections in Serratia marcescens was confirmed through in vitro studies by Alexpandi et al. (2019) and Srinivasan et al. (2016). Further, the pre-clinical trial has been performed in a mouse model and confirmed these antagonistic molecules activity as efficient QS inhibitors. Similarly, studies made by Srinivasan et al. (2017); Arunachalam et al. (2018) have revealed the biofilm inhibitory potential of antagonistic molecules such as phytol and geraniol on S. marcescens associated acute pyelonephritis infection and S. epidermidis associated endocarditis infection in animal models, respectively.
Apart from these three main strategies, some antimicrobial peptides could inhibit the QS system either by affecting the signal molecules transport within or outside the cell, thereby affecting the signal transduction cascade and biofilm formation. More, a newer tool in recent research is employed to block the expression of the luxS gene of Escherichia coli during QS signaling to reduce the biofilm formation by CRISPRi technology (Zuberi et al., 2017b; Sharma et al., 2019).
Biofilm Degradation by Electrochemical Method
The electrochemical method is one of the striking and promising strategies employed to put forth a great hindrance in bacterial biofilm formation. The electrochemical approach is the combinatorial effect of applying the lower dose of antibiotics in a weak electric field to disintegrate the biofilm formation or mature biofilm (Figure 6), which is also denoted as the ‘Bioelectric effect’. Several reports acknowledged that the electric potential lowers the antibiotics dosage to inactivate the biofilm and exerts a lethal effect on the biofilm organisms.
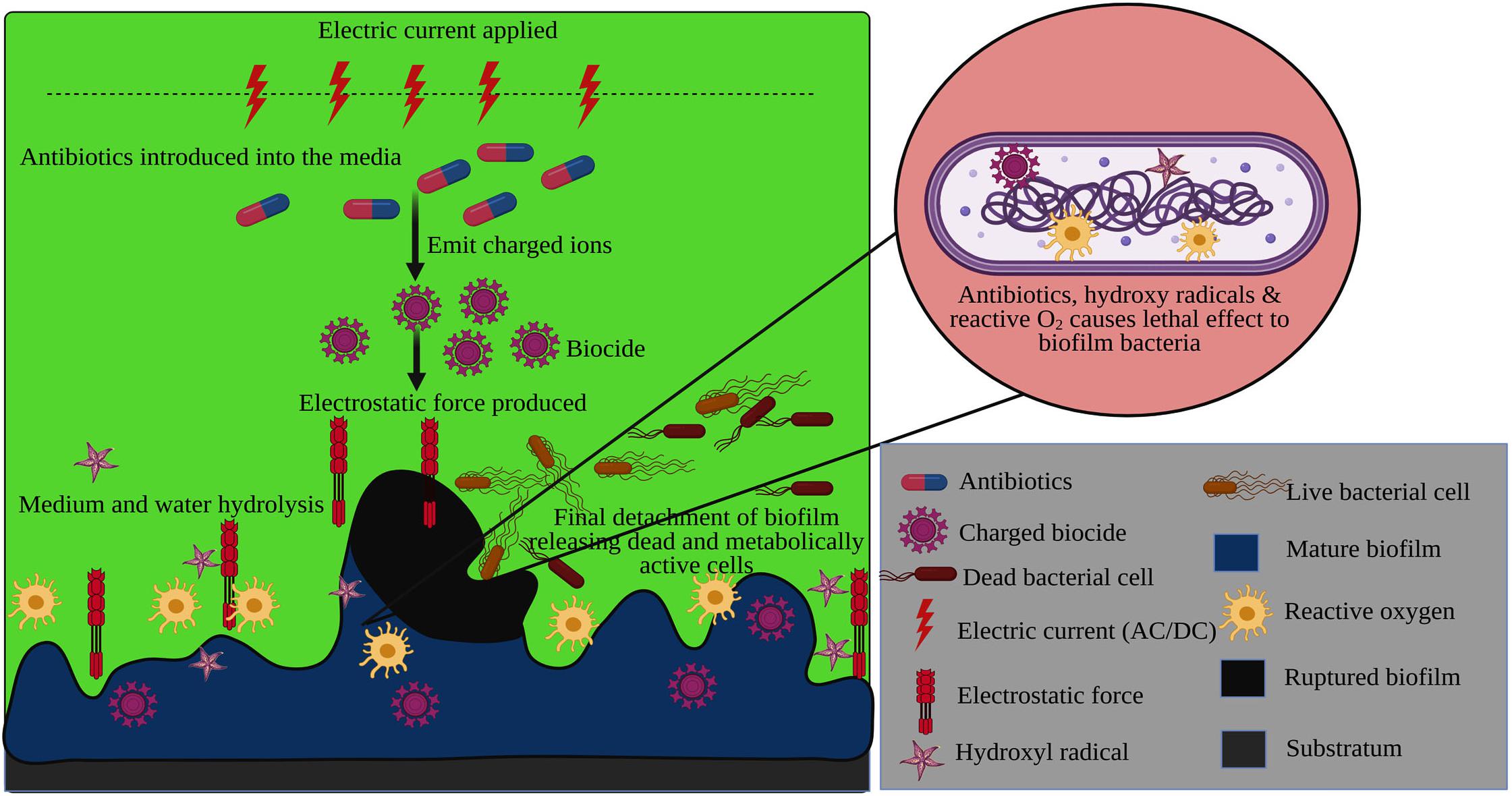
Figure 6. Electrochemical degradation of bacterial mature biofilm using the electrostatic force generated by an electric current with antibiotics.
A contrary report has been stated by Shirtliff et al. (2005) when eliminating biofilm formation by mixed-species underwater. According to the application field, diverging hypothesis has been suggested for employing the electrochemical method to disperse the biofilm formation. The underlying principle behind electrochemical approach is that the antimicrobial binding and transport towards the biofilm matrix are enhanced due to electrostatic force under direct current and thereby it augments the efficacy of biofilm detachment (Blenkinsopp et al., 1992; Van Der Borden et al., 2004). Owing to the electric field, the media’s hydrolysis occurs, resulting in the release of charged ions and hyperoxygenation with thermal stimuli (Del Pozo et al., 2008).
Usually, it is very tedious for the antibiotics to penetrate the biofilm matrix. Under the influence of the electrical field, the antimicrobial agents lead to the discharge of the biocide ions attributed to the alteration in biofilm permeability. The influx of those biocide ions into the biofilm matrix consequentially inactivates the biofilm. It destroys the bacterial cells via electrophoresis and electro-osmosis even at a low concentration (Chang et al., 1995; Stewart et al., 1999). It has been clear evidence from the study of Blenkinsopp et al. (1992) that the electric current does not have any such effect over the biofilm unless it exhibits a synergistic action with the antimicrobial agents. The hydrolysis of water and the pH change in the electrical impulses expresses the high production of oxygen molecules contributing to the improved level of minimal inhibitory concentration, thereby increases antibiotic susceptibility among biofilm and drug resistant bacteria (Borriello et al., 2004; Del Pozo et al., 2009b; Wolfmeier et al., 2018). Furthermore, the hydrated ions create an electrostatic repulsive force that aids in detachment of the biofilm from the substratum surface (Poortinga et al., 2000).
The list of other significant factors related to the bioelectric effects also depends on the voltage and electric current during the electrical stimulation as it affects the cell membrane, cellular process, behavior, and electrophysiology (Sabelnikov et al., 1991; Hancock and Rozek, 2002). Certain research suggested implementing either Alternating Current (AC) or Direct Current (DC) (Stewart et al., 1999; Del Pozo et al., 2009a) or even both (Kim et al., 2015) to produce electrical impulse for bioelectric effect with the low dosage of antibiotics. In which, the AC contributes to the direct electrostatic force, and the DC attributes to the increased permeability as a result of charged molecules vibrations. In an in vivo experiment of Sultana et al. (2015), a drastic reduction of the biofilm has been observed. The biofilm of Acinetobacter baumannii has been developed on the porcine explant, which is superimposed with the electrochemical-scaffold. When an electric field is introduced using Ag/AgCl electrodes at the constant potential of about 600mVAg/AgCl, the biofilm formation is much reduced due to the production of H2O2, consequentially resulting in the synthesis of hydroxyl radicals leading to the lethal effect of the cell. In an another report using P. aeruginosa with a similar experimental setup employing glass-bottomed petri dishes as a biofilm model, a 105-fold reduction in the biofilm formation has been demonstrated (Sultana et al., 2016). The action of plasma under low current influences a decline in the EPS intensity surrounding the bacterial cells resulting in low cellular adhesion (Kovalova et al., 2016). Another innovative technique for eradicating the biofilm employing the electrochemical method is electrospray. The dispersion of liquid from the high energy potential is carried out to obtain a sterile polymer surface devoid of biofilm (Kovalova et al., 2014).
Surface Modulation of Bacterial Adhesion
There are numerous series of issues are found in marine, medical, food, and industrial fields due to biofilm formation. Especially in the marine environment, antifouling has attracted the utmost importance in recent years. Because biofouling created a severe threat in the marine environment and consequentially marine industries faced tremendous challenges. Biodeposition leads to the alteration of the nutritive source in the surrounding ecosystem, resulting in the disarray of the ecological niche in the benthic zone (Weise et al., 2009). Numerous studies have been carried out in search of innovative and novel technologies regarding the antifouling components. Antifouling compounds prevent or counteract the buildup of barnacles and other deposits, including microbial biofilms on the surface undersea. Dusane et al. (2011) demonstrated an antifouling compound from the marine strain S. marcescens, producing glycolipid surfactant that inhibits certain biofouling marine bacterial species such as Bacillus pumilus, Candida albicans, and P. aeruginosa.
Perhaps, the discovery of meticulous interaction outlay between the biofouling microbes and its substratum gives an alternative strategy against the biofouling by marine micro and macro-organisms. Modulation of the substratum or the surface to which the microbes attach is one interesting approach that has an impact over a few years in this field. Rogers et al. (1994) has sorted out various surface materials in the biofilm formation. The biocides recovered from certain microbes are implemented to coat on the surfaces to avoid the formation of biofilms, especially in the field of medicine. Due to the toxicity and other limitations, many regulations inclusive of European Union, Biocidal Products Regulation has been laid and restricted many biocides applications (Norcy et al., 2017). Tributyltin formulations are widely used in paints in earlier days, but their usage has been prohibited owing to their toxic nature towards the marine ecosystem. Instead, natural potent biofilm inhibitors have been focused. Multispecies biofilm formed by E. coli, S. aureus, and P. aeruginosa have been successfully eliminated using this technology by coating silver oxynitrate effectively (Lemire et al., 2017). Further, the numerous in vitro studies using silver coating materials have been extensively studied by various researchers (Lansdown, 2006; Stobie et al., 2008; Lemire et al., 2015; Kalan et al., 2017).
In the case of food industries, coating of the surface using non-toxic, nonstick components like silicones and fluoropolymer derivatives are preferred, because of its ability to form non-porous surface due to the association of hydrophobicity along with low surface free energy as well as the microroughness (Sadekuzzaman et al., 2015). Other attention grabbing criteria are the absorption of nanoparticles to the surface prevents the formation of biofilm. Such implementation has a vast impact in recent years in medicine and domestic purposes like pipelines, etc. The nanoparticles attach to the microbial surface, providing them with a larger surface area and thereby react with the protein and cellular DNA, eventually resulting in the inhibition of DNA replication and gene expression (Yamanaka et al., 2005; Sadekuzzaman et al., 2015). Hetrick et al. (2009) has demonstrated the biofilm inhibitory potential of nanoparticles embedded materials towards the pathogens such as E. coli, P. aeruginosa, S. aureus, and S. epidermidis. More, coating certain components like antibiotics, quaternary ammonium salts, polyethylene glycol, and silver ions towards the surface keeps away from bacterial colonization by weakening the cell membrane and the cellular activities (Park et al., 1998).
The application of bacteriocin is a promising approach for the control of biofilms due to their biofilm inhibitory property. Further, the biofilm formation of Listeria monocytogenes on the stainless steel has been destroyed using the bacteriocin derived from Lactobacillus sakei (Winkelstroter et al., 2011). Certain varieties of bacteriocin affect bacterial adhesion and biofilm formation even at the sub inhibitory concentration. The bovicin HC5 and nisin are the bacteriocin, which targeted the microbial cellular attachment by varying the microbial cell’s hydrophobicity and the substratum at the sub-inhibitory concentration (de Jesus Pimentel-Filho et al., 2014). This study also identified that bacteriocin inhibits the expression of icaD, fnbA, clfB, and rnaIII genes related to biofilm formation in S. aureus. The indwelling medical device surfaces coated with bactericidal compound like aryl rhodanines has prevented the biofilm development (Chung and Toh, 2014). Hence, the surface modulation allows the prevention of bacterial adhesion towards the substratum and thereby sets an obstacle for biofilm formation.
Disruption of the Cell Membrane by Antimicrobial Peptides (AMP)
Recent research works continuously highlight that among the various biofilm inhibition strategies, the possible use of AMP, also known as host defense peptides, may denote a promising approach (Batoni et al., 2011; Jorge et al., 2012). The biofilm is disturbed through the different AMP by the transmembrane pore mechanism, which will lead to the final condition of cell death. The study of Pulido et al. (2016) reveals that the total permeabilization effect was visualized by Confocal Laser Scanning Microscopy (CLSM) and Sytox Green permeabilization assay, where the analysis of RN3 (5-17P22-36), an AMP, at higher concentration established the permeabilizing effect in biofilm cellular population. Indeed, the AMP action over the membrane depolarization and permeabilization facilitating the antimicrobial and biofilm inhibitory activities.
Interestingly, certain AMP binds to the bacterial cells of the biofilm structure, encouraging its cellular agglutination and membrane interaction. The peptides reorganization of the membrane determinants for the lipopolysaccharides lie at the N-terminus portion on coding sequence and thus expresses a higher specificity for the affinity towards the lipopolysaccharides on the cell membrane of Gram-negative bacteria (Bhunia et al., 2009; Torrent et al., 2009). The mechanism of AMP has been described in the models named ‘carpet’, ‘barrel-stave’ or ‘toroidal-pore’, which highlights the factors such as cationic charge, amphipathicity, amino acid composition, and size perusing the peptide attachment, translocation, and altering membrane permeability through an alternation in cytoplasmic membrane configuration.
In yet another report, the cationic peptide interaction towards the Gram-negative bacteria occurs to the anionic surface with the lipopolysaccharides layer on the outer membrane. It disturbs the structural configuration of the cell membrane, promoting the cellular leakage causing cell death (Moore et al., 1986). Further, Zhang et al. (2000) has accepted this view, insisting that the AMP interacts to the specific divalent cationic binding site at the lipopolysaccharides of the outer cell membrane bringing up the transposition through self-promoted uptake. Peptides are rendering interaction with the cell membrane based on its charge moiety and its hydrophobic interactions. Membrane targeting peptides like RT2, KT2, and magainin II enables their hydrophobic portion to interact with the anionic moiety in the lipid head of E. coli cell membrane and thereby launches them in the hydrophobic core (Anunthawan et al., 2015). Therefore, the structure, shape, and design of AMP are crucial for the electrostatic interaction to interrupt cell membrane and biofilm.
Antimicrobial Lipids (AML) as Biofilm Inhibitors
Antimicrobial lipids (AML) are known as single-chain lipid amphiphiles, including fatty acids and monoglycerides (Verderosa et al., 2019). Later the 1,800s, after Koch and his colleagues first reported the growth inhibitory effects of soap, the antibacterial properties of fatty acids in soap had been identified and subsequently shown to inhibit B. anthracis growth, the causative agent of anthrax (Thormar, 2011). The antimicrobial potential of monoglycerides and fatty acids has been continuously exposed against various pathogens (Desbois and Smith, 2010; Desbois, 2012; Li et al., 2013; Karthikeyan et al., 2014; Wang et al., 2020). It is well understood that AML function through several pathways, such as enhanced membrane permeability, form temporary or permanent membrane pores, target the bacterial surface signal transduction system, electron transport chain destruction, cell lysis, and bacterial enzyme inhibition (Hyldgaard et al., 2012; Schlievert and Peterson, 2012; Verderosa et al., 2019).
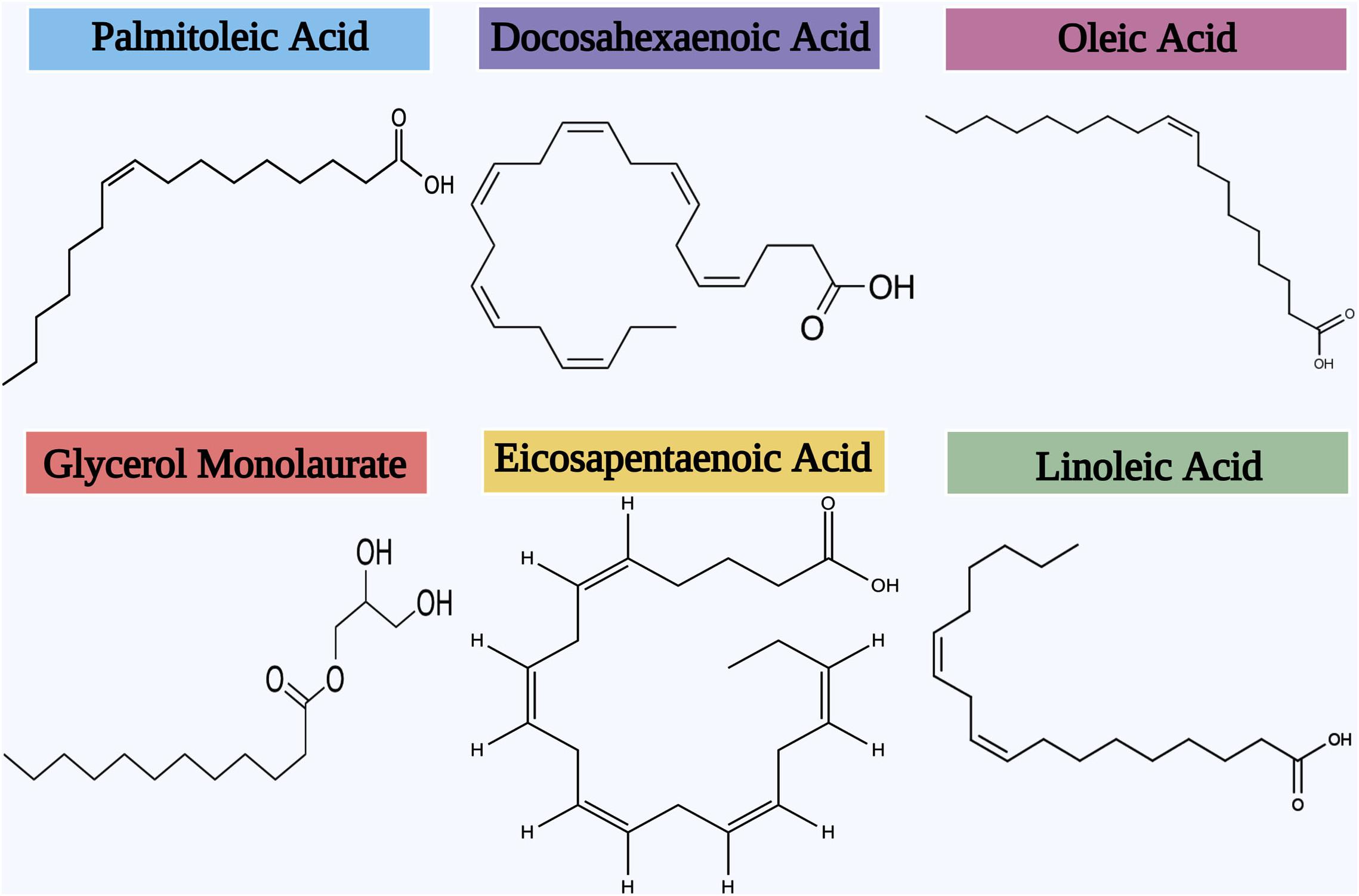
Several studies have continuously stated the biofilm inhibitory efficacy of AML at low doses against various bacterial biofilm formations (Figure 7). Oh and Marshall, 1995 has first explored the biofilm inhibitory efficacy of AML (Glycerol monolaurate) against L. monocytogenes biofilm formation. Glycerol monolaurate (GML) has antimicrobial or immunomodulatory effects, a fatty acid composed of glycerol and lauric acid (Witcher et al., 1996; Vetter and Schlievert, 2005). GML is currently used as a dietary and cosmetic ingredient, which is generally recognized as safe (GRAS) by the U.S. Food and Drug Administration. Schlievert and Peterson (2012) also validated the biofilm inhibitory efficacy of GML against Haemophilus influenzae and S. aureus biofilm formation. They have endorsed the bactericidal effects of GML on the mature biofilm formation of both bacterial pathogens. Recently, Lopes et al. (2019) substantiates the biofilm inhibitory potential of GML nanocapsules against P. aeruginosa biofilm by Atomic Force Microscopy.
A study made by Harvey et al. (2019) has revealed the biofilm inhibitory efficacy of antibacterial lipids on the cariogenic organism Streptococcus mutans by Fluorescent Microscopy using Alexa Fluor® 647 and SYTO® 9-labeled dextran conjugate. Further, the biofilm development of Gram-positive bacterial pathogens such as S. epidermidis, S. aureus, and S. mutans has been effectively inhibited by some unsaturated fatty acids such as oleic acid, linoleic acid, and palmitoleic acid (Yuyama et al., 2020). The inhibitory efficacy of the two fatty acids includes docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against Fusobacterium nucleatum and Porphyromonas gingivalis biofilms were recently studied by Sun et al. (2016). Obtained results of this study displayed that both DHA and EPA significantly eradicated the mature biofilm of P. gingivalis to the level of 61 and 47%, respectively. Further, both these two fatty acids were tested for action against S. mutans biofilm formation in a follow-up publication by the same research group (Sun et al., 2017). It was observed that both EPA and DHA significantly weakened the outer membrane of residing biofilm cells and thereby decreased the thickness of biofilm in S. mutans.
Degradation of the EPS Matrix of Biofilm
The major components of the EPS matrix of biofilm are the polysaccharides, proteins, and nucleic acids (e-DNA), and thus one of the biofilm inhibition strategies includes attack over the biofilm matrix. The biofilm degradation is possible by disrupting the EPS matrix components. The biofilm EPS matrix components vary according to the microbial strain, age, and other environmental factors like pH, oxygen tension, and nutrient abundance (Flemming and Wingender, 2010). Lipopolysaccharide, alginate, Psl (Polysaccharide synthesis locus), and Pel (Pellicle) are the major EPS of the P. aeruginosa biofilm (Rabin et al., 2015). Other significant mechanisms regarding the action over the dissolution of the EPS matrix of biofilm include alginate lyase, DNase, and hydrolase-based approach. However, Dispersin B (DspB) protein acts upon the biofilm to disperse the EPS matrix in Actinobacillus pleuropneumoniae (Kaplan et al., 2004b). Similarly, DspB protein derived from the A. actinomycetemcomitans dissolute the biofilm matrix of S. epidermidis (Kaplan et al., 2004a,b). Further, DspB protein also affects the linkage of the glycosidic bond in the EPS matrix’s polysaccharide, and, hence the biofilm architectures are detached (Izano et al., 2007). Besides, certain phages and phage-derived enzymes like polysaccharide depolymerase can invade the EPS matrix and demolish the biofilm architecture (Sadekuzzaman et al., 2015).
The study of Powell et al. (2018) revealed the disruption of biofilm matrix through the implementation of alginate oligosaccharide (OligoG). The effect has been visualized using Lectin and ConA staining with CLSM imaging. The effects visualized by Stokniene et al. (2020) in CLSM images were similar to what was revealed in previous experiments: CLSM imaging visualized the OligoG and colistin conjugates disrupting the P. aeruginosa biofilm matrix. On the other hand, the interaction of divalent ions like Ca and Mg ions brings about the variation in EPS matrix through OligoG as these ions play a vital role in regulating the association of EPS and e-DNA in the mucoid matrix (Flemming and Wingender, 2010). These ions overcome the electrostatic repulsion of the negatively charged biofilm components and maintain biofilm physiology (Govan and Deretic, 1996; Whitchurch et al., 2002). It has been proved that Ca ions possess the efficacy in the stability and maintenance of the biofilm of marine species like V. cholerae (Kierek and Watnick, 2003). Furthermore, OligoG also influences the thickness of the biofilm (Strathmann et al., 2002).
Inhibition of Alarmone Scheme
Under stress conditions like nutrient depletion, bacteria exhibit the stringent response, which is considered to be a crucial part for the synthesis of certain molecules in the alarmone scheme such as guanidine 3′diphosphate 5′triphosphate (pppGpp) and guanidine 3′5′bis-diphosphate (ppGpp), which are jointly expressed as (p)ppGpp through the cassette of RelA and SpoT. In Eubacteria, the stringent response is regulated by the signal molecule (p)ppGpp, which is produced by RelA and hydrolyzed by SpoT (Metzger et al., 1989), especially in Gram-negative bacteria and in Gram-positive bacteria with the aid of bifunctional enzyme, Rel/Spo for both the hydrolysis and synthesis (Wendrich and Marahiel, 1997). These molecules attribute the stringent response mediation that regulates the biofilm formation in E. coli and S. mutans (Lemos et al., 2004; Aberg et al., 2006).
The AMP 1,018 degrades alarmone ((p)ppGpp) signal by acting as the biofilm inhibitor. The biofilm inhibitory efficiency of the peptides 1,018 has been reported in earlier study (De La Fuente-Nunez et al., 2014). In the same study, the genes conferring the synthesis of alarmone (p)ppGpp signal in P. aeruginosa have been identified as relA and spoT. The gene expression and repression suggesting their part in the biofilm formation, maintenance, and outcome of the downregulation of those genes have also been documented through the in vitro studies of mutant deficient with the alarmone (p) ppGpp signal. The peptide 1,018 has more potential than the conventional antibiotics as it only suppresses the synthesis of alarmone (p) ppGpp signal rather than degradation. In addition, certain antibiotics have also been reported to affect the alarmone scheme, which eventually acts upon the alarmone (p) ppGpp signal synthesis, leading to adaptive resistance (Ikehara et al., 1985; Gilbert et al., 1990).
The exclusive investigation of He et al. (2012) provides insight into the regulation of the biofilm forming genes during stringent response in V. cholerae. It has been confirmed that due to the insufficiency in the synthesis of alarmone (p) ppGpp synthases, the shortage in the biofilm formation occurs. The stringent response is liable for certain regulatory factors responsible for the expression of relA, spoT, and relV in V. cholerae. It has been proved that the regulation of alarmone (p)ppGpp signal system of vpsR and vpsT in V. cholerae is accomplished through the transcriptional factor rpoS. Indeed, the sole source of alarmone (p) ppGpp synthase essential for the activation of the biofilm formation gene vpsT as it depends on RelA. The investigation of a novel compound named relacin has been designed to hold back the synthesis of alarmone (p)ppGpp signal molecules by inhibiting the RelA as it prompts the stringent response (Wexselblatt et al., 2012). During environmental stress like nutrient starvation, RelA induces the stringent response further affected by relacin and thereby down-regulated the biofilm formation in bacteria. Due to the reduction in alarmone (p) ppGpp synthesis, there is a low level of inosine monophosphate dehydrogenase synthesis, leading to low GTP consequences. The pyrophosphate’s ribosomal transfer describes the alarmone (p) ppGpp signal synthesis from ATP to GTP/GDP. Thus the relacin plays a major role in the biofilm formation pathway and other developmental pathways in B. subtilis.
Other peptides like LL-37, derived from natural peptides like human cathelicidin and 1,037, a synthetic cationic peptide, have also been reported for their biofilm inhibitory activity in earlier studies (Overhage et al., 2008; De La Fuente-Nunez et al., 2012). The other peptide includes the protease-resistant D-enantiomeric peptides DJK-5 and DJK-6 are noteworthy in describing the antagonistic activity against the biofilm formation in P. aeruginosa through inflowing into the biofilm cells and by degrading the intracellular nucleotides (p) ppGpp even at the low concentration range of 0.5–0.8 μg/mL below the MIC (De La Fuente-Nunez et al., 2015). Further, their variation and similarity in structure, function, and association are attributed to immune-modulatory, antimicrobial, antibiofilm, and anticancer properties. The AMP, which exerts a distinctive mode of action other than the conventional antimicrobial agents, are suggested for the biofilm inhibitory activity by hindering the alarmone system.
Enzyme Mediated Biofilm Control
Certain enzymes are mediating the disruption of the biofilm of various bacterial species. Nevertheless, the bacteria themselves would synthesis certain endogenous matrix-degrading enzymes like glycosidases, proteases and DNase as it may induce the dispersion of the biofilm. DNase has been known for its biofilm inhibitory activity against Gram-positive and Gram-negative bacteria (Tetz et al., 2009). Further, DNase involve in breaking down of phosphodiester linkage in the backbone of e-DNA molecules in the formed biofilm, as the e-DNA is essential for the initial attachment and aggregation of the EPS onto the surface to make it an intact biofilm for a more extended period. Moreover, the e-DNA is one of the necessary factors for the biofilm formation, stability, and regulation (Das et al., 2010; Periasamy et al., 2012; Chagnot et al., 2013).
The synergistic action of DNase with metronidazole antibiotic established the disintegration of the biofilm formed by Gardnerella vaginalis at 100 μg/mL concentration. The report of Eckhart et al. (2007) also provided evidence for the suppression of biofilm through DNase in P. aeruginosa and S. aureus. For the first time, Monnappa et al. (2014) have been reported the extracellular protease and DNase from a host-independent Bdellovibrio bacteriovorus restrain the biofilm of Gram-positive bacteria especially, S. aureus. Furthermore, DNase (NucB) used as the feasible tool for dispersing e-DNA within the biofilm’s EPS matrix has been depicted by Nijland et al. (2010). Similarly, the study of Bugli et al. (2016) suggested the concentration of 100 mg/L DNase can able to eliminate the biofilm formed by Helicobacter pylori, while Torelli et al. (2017) gives insight on the DNase and alginate lyase as effective in disintegrating the biofilm of Enterococcus faecalis and E. faecium by acting over the EPS matrix of biofilm.
A significant biofilm inhibitory effect of proteases has been extensively described by Mukherji et al. (2015). The study of Hangler et al. (2009) discloses that Esperase HPF, a protease efficacy in preventing the biofilm of the species such as Dokdonia donghaensis, Shewanella japonica, Microbacterium phyllosphaerae, and A. lwoffi. Further, the commercially available proteases such as proteinase K, trypsin, and chymotrypsin (serine proteases category), and serratiopeptidase, carboxypeptidase A (metalloproteases category) have been reported for their biofilm inhibitory potential against Staphylococcus spp. (Artini et al., 2013). Craigen et al. (2011) has revealed the inactivation and removal of biofilm formation by S. aureus with the aid of alpha-amylase. Alginate lyase is a potent enzyme involved in the dissolution of biofilm of certain bacteria (Germoni et al., 2016). Many studies such as Rahman et al., 2010; Singh et al., 2011; Wang et al., 2013; Daboor et al., 2019 have indicated that the marine bacterium serves as a renowned source for the production of enzyme alginate lyase, as a powerful agent in disassembling the biofilm exerted by pathogens. It may be related to the fact that marine bacteria are exposed to substantial quantities of the alginate present in their surroundings (Zhu et al., 2016). The enzyme alginate lyase is recovered from the marine bacterium Pseudoalteromonas active in exhibiting biofilm inhibitory activity against P. aeruginosa, E. coli, and S. enterica (Dheilly et al., 2010; Dufourcq et al., 2014).
Mechanical Eradication of Biofilm Formation Through a Photodynamic Approach
The photodynamic approach is the recent innovative method adapted for the disintegration of biofilms. The underlying principle behind this approach is the implementation of photosensitizing molecules that absorb the light intensity of a specific wavelength and, by binding to the target cellular components like lipid, protein, and nucleic acid. It produces reactive oxygen radicals, which in turn will give rise to hydrogen peroxide, hydroxyl radicals and superoxide anion resulting in the lethal/toxic effect to the target. There are the latest attempts for evidencing biofilm inhibition in this regard (Darmani et al., 2018; Gholibegloo et al., 2018). The photosensitizer and the light source implemented are crucial for this kind of mechanical eradication of biofilms through photodynamic therapy. The frequently employed photosensitizer includes methylene blue, toluidine blue, or toluidine blue O. Other photosensitizer comprises 23H-porphine, tetrakis (1-methyl-pyridino)-21H, tetrakis (phenylthio)-29H, 31H-phthalocyanine, and tetra-p-tosylate salt (Collins et al., 2010; Junqueira et al., 2012; Sadekuzzaman et al., 2015).
The absorption spectrum of methylene blue is at 664 nm, while toluidine blue is at 638 nm and lies within the UV-visible range of 600–1000 nm wavelength. Though methylene blue photosensitizer is effective against various bacterial pathogens, it has more potential to eliminate Gram-positive bacteria. It is due to the reason that it constitutes additional effectiveness in transferring the negative charge towards the target cell with a negative charge cell wall owing to the presence of teichoic acid (Mirouze et al., 2018; de Oliveira et al., 2019). Although light emission is accomplished through the diode laser while employing toluidine blue as photosensitizer (Zanin et al., 2006), the study of Soukos et al. (2000) has reported that the light emission from helium/neon laser light employing toluidine blue as a photosensitizer in eradicating the 95% of oral biofilm formation. Furthermore, Andrade et al. (2013) has suggested that pre-radiating time over the target influences the elimination of the microorganism. Considering future viewpoints, more attempts have to be made to establish the complete knowledge and detailed mechanism of action for successful photodynamic therapy in eradicating bacterial biofilm formation.
Interruption/Down-Regulation of Molecular System of Biofilm Formation
Transcriptional regulation of a gene is far most important for gene expression to occur. The regulatory proteins and regulatory binding sites are also promising factors for regulatory machinery. The detailed study regarding gene expression profiling will pay for the appropriate up-regulation or down-regulation mechanism. Considering such approach, the biofilm formation can be interrupted or blocked through the downregulation of biofilm synthesis genes via certain components. Nal-P-113 is an AMP that acted against the biofilm formation of P. gingivalis W83 by deregulating the genes that confer biofilm formation, thereby controls the infection (Wang et al., 2017). These authors evaluated and described that when the gene PG0282 and PG1663 gets down-regulated through the mediation of Nal-P-113 peptide, it directly influences the ABC transporter, which plays a critical role in the biofilm formation machinery.
Upregulation of the ABC transporter is necessary for the initial biofilm formation when evaluating the potB gene coded for ABC transporter in P. putida (Sauer and Camper, 2001). This view has also been accepted by Hinsa et al. (2003). Later they have evaluated the association between the biofilm formation and the ABC transporter system. Though lapEBC cassette or lap gene encoded for the ABC system, the mechanism and the genes coding vary between species and strain. Furthermore, down-regulation of certain genes reduces the biofilm formation, as narrated in a few studies (Whiteley et al., 2001; Overhage et al., 2007). The study of De La Fuente-Nunez et al., 2012 highlighted the genes conferring swimming and swarming motility to be down-regulated to influence biofilm elimination using the AMP 1,037.
Several earlier studies also illustrated that the down-regulation of certain genes including flgB, nirS, norC, and nosZ (which are responsible for anaerobic biofilm formation), fimX (encoding for Type IV pilus assembly), rhlB (regulating QS and involved in the rhamnolipid synthesis), lecB (involved in the fucose binding of lectin) hinders the biofilm formation in different bacterial pathogens (Schuster et al., 2003; Johansson et al., 2008; De La Fuente-Nunez et al., 2012). The novel gene rsaL has been earlier demonstrated by De Kievit et al. (1999), which involved in the synthesis of the harmful regulatory protein that impacts the suppression of lasI gene expression. The gene product of rsaL has a negative transcription regulation of lasI operon in P. aeruginosa, thereby obstructing the QS cascade, leading to biofilm inhibition. The other approaches include altering the QS signaling cascade to prevent the biofilm formation is CRISPRi technology for deregulating the luxS gene expression (Seed et al., 1995; Lonn-Stensrud et al., 2009; Zuberi et al., 2017b), which is involved in QS signaling and fimbriae associated gene (fimH) expression, so that, it has an influencing effect to suppress the biofilm formation (Zuberi et al., 2017a).
Conclusion and Future Prospective
In nature, several bacteria live in the form of biofilms. For the medical community, biofilms constitute a serious problem since not only they are linked with numerous infections in humans, but they are often particularly challenging to handle, because of their tolerance to antibiotics and immune responses. Biofilm mediated infections are difficult to control due to their complexity and increasing antibiotic resistance. It is necessary to prevent their surface colonization to restrict biofilm development, as this is the first step in the formation of biofilms. In this review, we have discussed several emerging strategies and potential perceptions for developing enhanced therapeutics to control biofilm mediated bacterial infections. The alternative approaches for preventing biofilm formation in the medical devices or marine environment are also explored in depth, with special emphasis on surface modulation of bacterial adhesion. Overall, all these recent therapeutic biofilm inhibition strategies can open up new prospects for controlling biofilm development in diverse sectors. Future prospective of improved biofilm eradication strategies may aim to commercial intake of certain biofilm inhibitors like enzymes, AMP, AML, and QS inhibitors would make it possible as a tremendous tool to achieve the target. Nevertheless, in-depth research is necessary to clarify the effect of these biofilm inhibitors during biofilm infection in the host and prove their applicability to humans. Meanwhile, biofilm inhibitors may not cause antibiotic resistance; they have a lot of promise in the future for treating biofilm based infections in healthcare settings.
Author Contributions
RS, SS, and LX conceived the concept of the review. RS, SS, PP, and MG drafted the manuscript. RS prepared the figures. RS and LX coordinated the work and acquired funding. RS, SS, MD, and LX reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors RS and LX thankfully acknowledges China Post-doctoral Science Foundation (Grant No. 2019M662214), Key projects of Natural Science Foundation of Fujian Province (Grant No. 2020J02023), Program for Innovative Research Team in Fujian Agricultural and Forestry University (Grant No. 712018009), Key Laboratory of Marine Biotechnology of Fujian Province (Grant No. 2020MB04), and the Fujian-Taiwan Joint Innovative Center for Germplasm Resources and Cultivation of Crop (FJ 2011 Program, Grant No. 2015-75) for the financial support to carry out the Post-doctoral research work. Author SS sincerely thanks the Science and Engineering Research Board, Department of Science and Technology, Government of India for financial support in the National Post-Doctoral Fellowship (NPDF Order No. PDF/2018/001518). The authors like to acknowledge that all the figures in this review were created using BioRender (https://biorender.com/) platform except Figures 5, 7, which were created using ChemSketch (https://www.acdlabs.com/resources/freeware/chemsketch/index.php) software.
References
Aberg, A., Shingler, V., and Balsalobre, C. (2006). (p) ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 60, 1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x
Abraham, W. R. (2005). Controlling pathogenic gram-negative bacteria by interfering with their biofilm formation. Drug Des. Rev. Online (Discontinued) 2, 13–33. doi: 10.2174/1567269053390257
Alexpandi, R., Prasanth, M. I., Ravi, A. V., Balamurugan, K., Durgadevi, R., Srinivasan, R., et al. (2019). Protective effect of neglected plant Diplocyclos palmatus on quorum sensing mediated infection of Serratia marcescens and UV-A induced photoaging in model Caenorhabditis elegans. J. Photochem. Photobiology B Biol. 201, 111637. doi: 10.1016/j.jphotobiol.2019.111637
Andrade, M. C., Ribeiro, A. P. D., Dovigo, L. N., Brunetti, I. L., Giampaolo, E. T., Bagnato, V. S., et al. (2013). Effect of different pre-irradiation times on curcumin-mediated photodynamic therapy against planktonic cultures and biofilms of Candida spp. Arch. Oral Biol. 58, 200–210. doi: 10.1016/j.archoralbio.2012.10.011
Anunthawan, T., De La Fuente-Nunez, C., Hancock, R. E., and Klaynongsruang, S. (2015). Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochimica. Biophys. Acta (BBA) Biomembranes 1848, 1352–1358. doi: 10.1016/j.bbamem.2015.02.021
Artini, M., Papa, R., Scoarughi, G., Galano, E., Barbato, G., Pucci, P., et al. (2013). Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 114, 266–277. doi: 10.1111/jam.12038
Arunachalam, K., Ramar, M., Ramanathan, S., Govindaraju, A., Shunmugiah, K. P., Kandasamy, R., et al. (2018). In vivo protective effect of geraniol on colonization of Staphylococcus epidermidis in rat jugular vein catheter model. Pathog. Dis. 76:fty055.
Batoni, G., Maisetta, G., and Esin, S. (2016). Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochimica. Biophys. Acta (BBA) Biomembranes 1858, 1044–1060. doi: 10.1016/j.bbamem.2015.10.013
Batoni, G., Maisetta, G., Lisa Brancatisano, F., Esin, S., and Campa, M. (2011). Use of antimicrobial peptides against microbial biofilms: advantages and limits. Curr. Med. Chem. 18, 256–279. doi: 10.2174/092986711794088399
Bhatt, V. S. (2018). “Quorum sensing mechanisms in gram positive bacteria,” in Implication of Quorum Sensing System in Biofilm Formation and Virulence, eds G. V. Bramhachari, (Cham: Springer), 297–311. doi: 10.1007/978-981-13-2429-1_20
Bhunia, A., Mohanram, H., Domadia, P. N., Torres, J., and Bhattacharjya, S. (2009). Designed β-boomerang antiendotoxic and antimicrobial peptides: structures and activities in lipopolysaccharide. J. Biol. Chem. 284, 21991–22004. doi: 10.1074/jbc.m109.013573
Blenkinsopp, S. A., Khoury, A. E., and Costerton, J. W. (1992). Electrical enhancement of biocide efficacy against Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 58, 3770–3773. doi: 10.1128/aem.58.11.3770-3773.1992
Borriello, G., Werner, E., Roe, F., Kim, A. M., Ehrlich, G. D., and Stewart, P. S. (2004). Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial Agents Chemother. 48, 2659–2664. doi: 10.1128/aac.48.7.2659-2664.2004
Branda, S. S., Vik, A., Friedman, L., and Kolter, R. (2005). Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26. doi: 10.1016/j.tim.2004.11.006
Bridier, A., Briandet, R., Thomas, V., and Dubois-Brissonnet, F. (2011). Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27, 1017–1032. doi: 10.1080/08927014.2011.626899
Brindhadevi, K., Lewisoscar, F., Mylonakis, E., Shanmugam, S., Verma, T. N., and Pugazhendhi, A. (2020). Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Proc. Biochem. 96, 49–57 doi: 10.1016/j.procbio.2020.06.001
Bugli, F., Palmieri, V., Torelli, R., Papi, M., De Spirito, M., Cacaci, M., et al. (2016). In vitro effect of clarithromycin and alginate lyase against Helicobacter pylori biofilm. Biotechnol. Progr. 32, 1584–1591.
Carniol, K., and Gilmore, M. S. (2004). Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J. Bacteriol. 186, 8161–8163. doi: 10.1128/jb.186.24.8161-8163.2004
Chaffin, W. L., Lopez-Ribot, J. L., Casanova, M., Gozalbo, D., and Martinez, J. P. (1998). Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62, 130–180. doi: 10.1128/mmbr.62.1.130-180.1998
Chagnot, C., Zorgani, M. A., Astruc, T., and Desvaux, M. (2013). Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 4:303.
Chang, Y. H. D., Grodzinsky, A. J., and Wang, D. I. (1995). Augmentation of mass transfer through electrical means for hydrogel-entrapped Escherichia coli cultivation. Biotechnol. Bioeng. 48, 149–157. doi: 10.1002/bit.260480209
Cheng, Z. X., Guo, C., Chen, Z. G., Yang, T. C., Zhang, J. Y., Wang, J., et al. (2019). Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. commun. 10, 1–17.
Choi, H., Mascuch, S. J., Villa, F. A., Byrum, T., Teasdale, M. E., Smith, J. E., et al. (2012). Honaucins A– C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships. Chemi. Biol. 19, 589–598. doi: 10.1016/j.chembiol.2012.03.014
Christensen, Q. H., Grove, T. L., Booker, S. J., and Greenberg, E. P. (2013). A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc. Natl. Acad. Sci.U.S.A. 110, 13815–13820. doi: 10.1073/pnas.1313098110
Christiaen, S. E., Matthijs, N., Zhang, X.-H., Nelis, H. J., Bossier, P., and Coenye, T. (2014). Bacteria that inhibit quorum sensing decrease biofilm formation and virulence in Pseudomonas aeruginosa PAO1. Pathog. Dis. 70, 271–279. doi: 10.1111/2049-632x.12124
Chung, P. Y., and Toh, Y. S. (2014). Anti-biofilm agents: recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog. Dis. 70, 231–239. doi: 10.1111/2049-632x.12141
Collins, T. L., Markus, E. A., Hassett, D. J., and Robinson, J. B. (2010). The effect of a cationic porphyrin on Pseudomonas aeruginosa biofilms. Curr. Microbiol. 61, 411–416. doi: 10.1007/s00284-010-9629-y
Craigen, B., Dashiff, A., and Kadouri, D. E. (2011). The use of commercially available alpha-amylase compounds to inhibit and remove Staphylococcus aureus biofilms. Open Microbiol. J. 5, 21. doi: 10.2174/1874285801105010021
Daboor, S. M., Raudonis, R., Cohen, A., Rohde, J. R., and Cheng, Z. (2019). Marine bacteria, a source for alginolytic enzyme to disrupt Pseudomonas aeruginosa biofilms. Marine drugs 17, 307. doi: 10.3390/md17050307
Darmani, H., Tawalbeh, K. H., Al-Hiyasat, A. S., and Al-Akhras, M.-A. (2018). Comparison of the photosensitivity of biofilms of different genera of cariogenic bacteria in tooth slices. Polish J. Microbiol. 67, 455. doi: 10.21307/pjm-2018-053
Das, T., Sharma, P. K., Busscher, H. J., Van Der Mei, H. C., and Krom, B. P. (2010). Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76, 3405–3408. doi: 10.1128/aem.03119-09
de Jesus Pimentel-Filho, N., De Freitas Martins, M. C., Nogueira, G. B., Mantovani, H. C., and Vanetti, M. C. D. (2014). Bovicin HC5 and nisin reduce Staphylococcus aureus adhesion to polystyrene and change the hydrophobicity profile and Gibbs free energy of adhesion. Int. J. Food Microbiol. 190, 1–8. doi: 10.1016/j.ijfoodmicro.2014.08.004
De Kievit, T., Seed, P. C., Nezezon, J., Passador, L., and Iglewski, B. H. (1999). RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181, 2175–2184. doi: 10.1128/jb.181.7.2175-2184.1999
De La Fuente-Nunez, C., Korolik, V., Bains, M., Nguyen, U., Breidenstein, E. B., Horsman, S., et al. (2012). Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrobial Agents Chemother. 56, 2696–2704. doi: 10.1128/aac.00064-12
De La Fuente-Nunez, C., Reffuveille, F., Fernández, L., and Hancock, R. E. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
De La Fuente-Nunez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152
De La Fuente-Nunez, C., Reffuveille, F., Mansour, S. C., Reckseidler-Zenteno, S. L., Hernández, D., Brackman, G., et al. (2015). D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205. doi: 10.1016/j.chembiol.2015.01.002
de Oliveira, A. B., Ferrisse, T. M., Marques, R. S., De Annunzio, S. R., Brighenti, F. L., and Fontana, C. R. (2019). Effect of photodynamic therapy on microorganisms responsible for dental caries: a systematic review and meta-analysis. In. J. Mol. Sci. 20:3585. doi: 10.3390/ijms20143585
Del Pozo, J., Rouse, M., and Patel, R. (2008). Bioelectric effect and bacterial biofilms. A Syst. Rev. 31, 786–795. doi: 10.1177/039139880803100906
Del Pozo, J. L., Rouse, M. S., Mandrekar, J. N., Sampedro, M. F., Steckelberg, J. M., and Patel, R. (2009a). Effect of electrical current on the activities of antimicrobial agents against Pseudomonas aeruginosa, Staphylococcus aureus, and Staphylococcus epidermidis biofilms. Antimicrobial Agents Chemother. 53, 35–40. doi: 10.1128/aac.00237-08
Del Pozo, J. L., Rouse, M. S., Mandrekar, J. N., Steckelberg, J. M., and Patel, R. (2009b). The electricidal effect: reduction of Staphylococcus and Pseudomonas biofilms by prolonged exposure to low-intensity electrical current. Antimicrobial Agents Chemother. 53, 41–45. doi: 10.1128/aac.00680-08
Desbois, A. P. (2012). Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Patents Anti-Infective Drug Discov. 7, 111–122. doi: 10.2174/157489112801619728
Desbois, A. P., and Smith, V. J. (2010). Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 85, 1629–1642. doi: 10.1007/s00253-009-2355-3
Dheilly, A., Soum-Soutera, E., Klein, G. L., Bazire, A., Compere, C., Haras, D., et al. (2010). Antibiofilm activity of the marine bacterium Pseudoalteromonas sp. strain 3J6. Appl. Environ. Microbiol. 76, 3452–3461. doi: 10.1128/aem.02632-09
Dong, Y. H., Xu, J. L., Li, X. Z., and Zhang, L. H. (2000). AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proceedings of the National Academy of Sciences 97, 3526–3531. doi: 10.1073/pnas.97.7.3526
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emer. Infect. Dis. 8:881. doi: 10.3201/eid0809.020063
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/cmr.15.2.167-193.2002
Dufourcq, R., Chalkiadakis, E., Fauchon, M., Deslandes, E., Kerjean, V., Chanteau, S., et al. (2014). Isolation and partial characterization of bacteria (Pseudoalteromonas sp.) with potential antibacterial activity from a marine costal environment from New Caledonia. Lett. Appl. Microbiol. 58, 102–108. doi: 10.1111/lam.12162
Dusane, D. H., Pawar, V. S., Nancharaiah, Y., Venugopalan, V. P., Kumar, A. R., and Zinjarde, S. S. (2011). Anti-biofilm potential of a glycolipid surfactant produced by a tropical marine strain of Serratia marcescens. Biofouling 27, 645–654. doi: 10.1080/08927014.2011.594883
Eckhart, L., Fischer, H., Barken, K. B., Tolker-Nielsen, T., and Tschachler, E. (2007). DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 156, 1342–1345. doi: 10.1111/j.1365-2133.2007.07886.x
Gao, A., Mei, G. Y., Liu, S., Wang, P., Tang, Q., Liu, Y. P., et al. (2013). High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase. Acta Crystallographica Section D 69, 82–91. doi: 10.1107/s0907444912042369
Germoni, L., Bremer, P., and Lamont, I. (2016). The effect of alginate lyase on the gentamicin resistance of Pseudomonas aeruginosa in mucoid biofilms. J. Appl. Microbiol. 121, 126–135. doi: 10.1111/jam.13153
Gholibegloo, E., Karbasi, A., Pourhajibagher, M., Chiniforush, N., Ramazani, A., Akbari, T., et al. (2018). Carnosine-graphene oxide conjugates decorated with hydroxyapatite as promising nanocarrier for ICG loading with enhanced antibacterial effects in photodynamic therapy against Streptococcus mutans. J. Photochem. Photobiol. B Biol. 181, 14–22. doi: 10.1016/j.jphotobiol.2018.02.004
Gilbert, P., Collier, P. J., and Brown, M. (1990). Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrobial Agents Chemother. 34:1865. doi: 10.1128/aac.34.10.1865
Govan, J. R., and Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574. doi: 10.1128/mr.60.3.539-574.1996
Hancock, R. E., and Rozek, A. (2002). Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206, 143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x
Hangler, M., Burmolle, M., Schneider, I., Allermann, K., and Jensen, B. (2009). The serine protease Esperase HPF inhibits the formation of multispecies biofilm. Biofouling 25, 667–674. doi: 10.1080/08927010903096008
Harvey, V., Brady, D., and Kealey, C. (2019). Effect of antibacterial lipids on biofilm formation by Streptococcus mutans. Access. Microbiol. 1:871.
He, H., Cooper, J. N., Mishra, A., and Raskin, D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. doi: 10.1128/jb.00014-12
Hentzer, M., and Givskov, M. (2003). Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest. 112, 1300–1307. doi: 10.1172/jci20074
Hetrick, E. M., Shin, J. H., Paul, H. S., and Schoenfisch, M. H. (2009). Anti-biofilm efficacy of nitric oxide-releasing silica nanoparticles. Biomaterials 30, 2782–2789. doi: 10.1016/j.biomaterials.2009.01.052
Hinsa, S. M., Espinosa-Urgel, M., Ramos, J. L., and O’toole, G. A. (2003). Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49, 905–918. doi: 10.1046/j.1365-2958.2003.03615.x
Hoiby, N. (1977). Acta pathologica et microbiologica scandinavica. supplement: Pseudomonas aeruginosa Infection in cystic fibrosis: diagnostic and prognostic significance of Pseudomonas Aeruginosa precipitins determined by means of crossed immunoelectrophoresis. Scand. J. Respir Dis. 58, 65–79.
Hume, E., Baveja, J., Muir, B., Schubert, T., Kumar, N., Kjelleberg, S., et al. (2004). The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones. Biomaterials 25, 5023–5030. doi: 10.1016/j.biomaterials.2004.01.048
Hyldgaard, M., Sutherland, D. S., Sundh, M., Mygind, T., and Meyer, R. L. (2012). Antimicrobial mechanism of monocaprylate. Appl. Environ. Microbiol. 78, 2957–2965. doi: 10.1128/aem.07224-11
Ikehara, K., Kamitani, E., Koarata, C., and Ogura, A. (1985). Induction of stringent response by streptomycin in Bacillus subtilis cells. J. Biochem. 97, 697–700. doi: 10.1093/oxfordjournals.jbchem.a135107
Izano, E., Wang, H., Ragunath, C., Ramasubbu, N., and Kaplan, J. (2007). Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J. Dent. Res. 86, 618–622. doi: 10.1177/154405910708600707
Jiang, Q., Chen, J., Yang, C., Yin, Y., and Yao, K. (2019). Quorum sensing: a prospective therapeutic target for bacterial diseases. BioMed. Res. Int. 2019:2015978.
Johansson, E. M., Crusz, S. A., Kolomiets, E., Buts, L., Kadam, R. U., Cacciarini, M., et al. (2008). Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB. Chemistry & biology 15, 1249–1257. doi: 10.1016/j.chembiol.2008.10.009
Jorge, P., Lourenco, A., and Pereira, M. O. (2012). New trends in peptide-based anti-biofilm strategies: a review of recent achievements and bioinformatic approaches. Biofouling 28, 1033–1061. doi: 10.1080/08927014.2012.728210
Junqueira, J. C., Jorge, A., Barbosa, J., Rossoni, R., Vilela, S., Costa, A., et al. (2012). Photodynamic inactivation of biofilms formed by Candida spp., Trichosporon mucoides, and Kodamaea ohmeri by cationic nanoemulsion of zinc 2, 9, 16, 23-tetrakis (phenylthio)-29H, 31H-phthalocyanine (ZnPc). Lasers Med. Sci. 27, 1205–1212. doi: 10.1007/s10103-012-1050-2
Kalan, L. R., Pepin, D. M., Ul-Haq, I., Miller, S. B., Hay, M. E., and Precht, R. J. (2017). Targeting biofilms of multidrug-resistant bacteria with silver oxynitrate. Int. J. Antimicrobial Agents 49, 719–726. doi: 10.1016/j.ijantimicag.2017.01.019
Kamaruzzaman, N. F., Tan, L. P., Mat Yazid, K. A., Saeed, S. I., Hamdan, R. H., Choong, S. S., et al. (2018). Targeting the bacterial protective armour; challenges and novel strategies in the treatment of microbial biofilm. Materials 11:1705. doi: 10.3390/ma11091705
Kannappan, A., Balasubramaniam, B., Ranjitha, R., Srinivasan, R., Packiavathy, I. A. S. V., Balamurugan, K., et al. (2019a). In vitro and in vivo biofilm inhibitory efficacy of geraniol-cefotaxime combination against Staphylococcus spp. Food Chem. Toxicol. 125, 322–332. doi: 10.1016/j.fct.2019.01.008
Kannappan, A., Durgadevi, R., Srinivasan, R., Lagoa, R. J. L., Packiavathy, I. A. S. V., and Pandian, S. K. (2020). 2-Hydroxy-4-methoxybenzaldehyde from Hemidesmus indicus is antagonistic to Staphylococcus epidermidis biofilm formation. Biofouling 36, 549–563. doi: 10.1080/08927014.2020.1777989
Kannappan, A., Gowrishankar, S., Srinivasan, R., Pandian, S. K., and Ravi, A. V. (2017a). Antibiofilm activity of Vetiveria zizanioides root extract against methicillin-resistant Staphylococcus aureus. Microbial. Pathog. 110, 313–324. doi: 10.1016/j.micpath.2017.07.016
Kannappan, A., Sivaranjani, M., Srinivasan, R., Rathna, J., Pandian, S. K., and Ravi, A. V. (2017b). Inhibitory efficacy of geraniol on biofilm formation and development of adaptive resistance in Staphylococcus epidermidis RP62A. J. Med. Microbiol. 66, 1506–1515. doi: 10.1099/jmm.0.000570
Kannappan, A., Srinivasan, R., Nivetha, A., Annapoorani, A., Pandian, S. K., and Ravi, A. V. (2019b). Anti-virulence potential of 2-hydroxy-4-methoxybenzaldehyde against methicillin-resistant Staphylococcus aureus and its clinical isolates. Appl. Microbiol. Biotechnol. 103, 6747–6758. doi: 10.1007/s00253-019-09941-6
Kaplan, J. B., Ragunath, C., Velliyagounder, K., Fine, D. H., and Ramasubbu, N. (2004a). Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrobial Agents Chemother. 48, 2633–2636. doi: 10.1128/aac.48.7.2633-2636.2004
Kaplan, J. B., Velliyagounder, K., Ragunath, C., Rohde, H., Mack, D., Knobloch, J. K.-M., et al. (2004b). Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186, 8213–8220. doi: 10.1128/jb.186.24.8213-8220.2004
Karthikeyan, S. C., Velmurugan, S., Donio, M. B. S., Michaelbabu, M., and Citarasu, T. (2014). Studies on the antimicrobial potential and structural characterization of fatty acids extracted from Sydney rock oyster Saccostrea glomerata. Ann. Clin. Microbiol. Antimicrobials 13, 1–11.
Kierek, K., and Watnick, P. I. (2003). The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. 100, 14357–14362. doi: 10.1073/pnas.2334614100
Kim, Y. W., Subramanian, S., Gerasopoulos, K., Ben-Yoav, H., Wu, H.-C., Quan, D., et al. (2015). Effect of electrical energy on the efficacy of biofilm treatment using the bioelectric effect. Npj Biofilms Microbiomes 1, 1–8.
Kong, K.-F., Vuong, C., and Otto, M. (2006). Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296, 133–139. doi: 10.1016/j.ijmm.2006.01.042
Kose-Mutlu, B., Ergon-Can, T., Koyuncu, I., and Lee, C.-H. (2019). Quorum quenching for effective control of biofouling in membrane bioreactor: a comprehensive review of approaches, applications, and challenges. Environ. Eng. Res. 24, 543–558. doi: 10.4491/eer.2018.380
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harbor Perspect. Med. 3:a010306. doi: 10.1101/cshperspect.a010306
Kovalova, Z., Leroy, M., Kirkpatrick, M. J., Odic, E., and Machala, Z. (2016). Corona discharges with water electrospray for Escherichia coli biofilm eradication on a surface. Bioelectrochemistry 112, 91–99. doi: 10.1016/j.bioelechem.2016.05.002
Kovalova, Z., Zahoran, M., Zahoranová, A., and Machala, Z. (2014). Streptococci biofilm decontamination on teeth by low-temperature air plasma of dc corona discharges. J. Phys. D Appl. Phys. 47:224014. doi: 10.1088/0022-3727/47/22/224014
Labbate, M., Queck, S. Y., Koh, K. S., Rice, S. A., Givskov, M., and Kjelleberg, S. (2004). Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186, 692–698. doi: 10.1128/jb.186.3.692-698.2004
Lam, J., Chan, R., Lam, K., and Costerton, J. (1980). Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28, 546–556.
Lansdown, A. B. (2006). Silver in health care: antimicrobial effects and safety in use. Biofunctional Textiles Skin 33, 17–34. doi: 10.1159/000093928
Leadbetter, J. R., and Greenberg, E. (2000). Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182, 6921–6926. doi: 10.1128/jb.182.24.6921-6926.2000
Lebeaux, D., Chauhan, A., Rendueles, O., and Beloin, C. (2013). From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356. doi: 10.3390/pathogens2020288
Lebeaux, D., Ghigo, J. M., and Beloin, C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543. doi: 10.1128/mmbr.00013-14
Lemire, J. A., Kalan, L., Bradu, A., and Turner, R. J. (2015). Silver oxynitrate, an unexplored silver compound with antimicrobial and antibiofilm activity. Antimicrobial Agents Chemother. 59, 4031–4039. doi: 10.1128/aac.05177-14
Lemire, J. A., Kalan, L., Gugala, N., Bradu, A., and Turner, R. J. (2017). Silver oxynitrate–an efficacious compound for the prevention and eradication of dual-species biofilms. Biofouling 33, 460–469. doi: 10.1080/08927014.2017.1322586
Lemos, J. A., Brown, T. A., and Burne, R. A. (2004). Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72, 1431–1440. doi: 10.1128/iai.72.3.1431-1440.2004
Lenhart, T. R., Duncan, K. E., Beech, I. B., Sunner, J. A., Smith, W., Bonifay, V., et al. (2014). Identification and characterization of microbial biofilm communities associated with corroded oil pipeline surfaces. Biofouling 30, 823–835. doi: 10.1080/08927014.2014.931379
Lewis, K. (2001). Riddle of biofilm resistance. Antimicrobial Agents Chemother. 45, 999–1007. doi: 10.1128/aac.45.4.999-1007.2001
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Lewis, K. (2008). Multidrug tolerance of biofilms and persister cells. Bacterial Biofilms 322, 107–131. doi: 10.1007/978-3-540-75418-3_6
Li, D., Ramanathan, S., Wang, G., Wu, Y., Tang, Q., and Li, G. (2020). Acetylation of lysine 7 of AhyI affects the biological function in Aeromonas hydrophila. Microbial Pathog. 140:103952. doi: 10.1016/j.micpath.2019.103952
Li, W., Yao, Z., Sun, L., Hu, W., Cao, J., Lin, W., et al. (2016). Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J. Proteome Res. 15, 1810–1820. doi: 10.1021/acs.jproteome.5b01127
Li, Z., Yuan, P., Xing, M., He, Z., Dong, C., Cao, Y., et al. (2013). Fatty acid conjugation enhances the activities of antimicrobial peptides. Recent Patents food Nutr. Agric. 5, 52–56. doi: 10.2174/2212798411305010008
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbial Biofilms 3, 223–247. doi: 10.1128/9781555817466.ch11
Longo, F., Vuotto, C., and Donelli, G. (2014). Biofilm formation in Acinetobacter baumannii. New Microbiol 37, 119–127.
Lonn-Stensrud, J., Landin, M. A., Benneche, T., Petersen, F. C., and Scheie, A. A. (2009). Furanones, potential agents for preventing Staphylococcus epidermidis biofilm infections? J. Antimicrobial Chemother. 63, 309–316. doi: 10.1093/jac/dkn501
Lopes, L. Q. S., De Almeida Vaucher, R., Giongo, J. L., Gündel, A., and Santos, R. C. V. (2019). Characterisation and anti-biofilm activity of glycerol monolaurate nanocapsules against Pseudomonas aeruginosa. Microbial Pathog. 130, 178–185. doi: 10.1016/j.micpath.2019.03.007
Lynch, A. S., and Robertson, G. T. (2008). Bacterial and fungal biofilm infections. Annu. Rev. Med. 59, 415–428. doi: 10.1146/annurev.med.59.110106.132000
Marrie, T. J., Nelligan, J., and Costerton, J. W. (1982). A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation 66, 1339–1341. doi: 10.1161/01.cir.66.6.1339
Martins, M., Uppuluri, P., Thomas, D. P., Cleary, I. A., Henriques, M., Lopez-Ribot, J. L., et al. (2010). Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169, 323–331. doi: 10.1007/s11046-009-9264-y
Masevicius, V., Nainyte, M., and Klimasauskas, S. (2016). Synthesis of s-adenosyl-l-methionine analogs with extended transferable groups for methyltransferase-directed labeling of DNA and RNA. Curr. Prot. Nucl. Acid Chem. 64, 1.36.1–31.36.13.
Mayer, C., Moritz, R., Kirschner, C., Borchard, W., Maibaum, R., Wingender, J., et al. (1999). The role of intermolecular interactions: studies on model systems for bacterial biofilms. Int. J. Biol. Macromol. 26, 3–16. doi: 10.1016/s0141-8130(99)00057-4
Metzger, S., Sarubbi, E., Glaser, G., and Cashel, M. (1989). Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J. Biol. Chem. 264, 9122–9125. doi: 10.1016/s0021-9258(18)60500-9
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Ann. Rev. Microbiol. 55, 165–199.
Mirouze, N., Ferret, C., Cornilleau, C., and Carballido-Lopez, R. (2018). Antibiotic sensitivity reveals that wall teichoic acids mediate DNA binding during competence in Bacillus subtilis. Nat. Commun. 9, 1–11.
Mitchell, K. F., Zarnowski, R., and Andes, D. R. (2016). Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 12:e1005828. doi: 10.1371/journal.ppat.1005828
Monnappa, A. K., Dwidar, M., Seo, J. K., Hur, J.-H., and Mitchell, R. J. (2014). Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci. Rep. 4, 1–8. doi: 10.1007/978-3-319-23534-9_1
Moore, R. A., Bates, N. C., and Hancock, R. (1986). Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrobial Agents Chemother. 29, 496–500. doi: 10.1128/aac.29.3.496
Mukherji, R., Patil, A., and Prabhune, A. (2015). Role of extracellular proteases in biofilm disruption of gram positive bacteria with special emphasis on Staphylococcus aureus biofilms. Enz. Eng. 4:126.
Nazir, R., Zaffar, M. R., and Amin, I. (2019). Bacterial biofilms: the remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes. Freshwater Microbiol. 2019, 307–340.
Nijland, R., Hall, M. J., and Burgess, J. G. (2010). Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS One 5:e15668. doi: 10.1371/journal.pone.0015668
Norcy, T. L., Niemann, H., Proksch, P., Linossier, I., Vallee-Rehel, K., Hellio, C., et al. (2017). Anti-biofilm effect of biodegradable coatings based on hemibastadin derivative in marine environment. Int. J. Mol. sci. 18:1520. doi: 10.3390/ijms18071520
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Ann. Rev. Microbiol. 54, 49–79.
Oh, D. H., and Marshall, D. L. (1995). Destruction of Listeria monocytogenes biofilms on stainless steel using monolaurin and heat. J. Food Prot. 58, 251–255. doi: 10.4315/0362-028x-58.3.251
Overhage, J., Campisano, A., Bains, M., Torfs, E. C., Rehm, B. H., and Hancock, R. E. (2008). Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182. doi: 10.1128/iai.00318-08
Overhage, J., Lewenza, S., Marr, A. K., and Hancock, R. E. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189, 2164–2169. doi: 10.1128/jb.01623-06
Padmavathi, A. R., Bakkiyaraj, D., and Pandian, S. K. (2017). “Biofilm inhibition by natural products of marine origin and their environmental applications,” in Biofilms in Plant and Soil Health, eds I. Ahmad and F. M. Husain (Hoboken NJ: Wiley), 465. doi: 10.1002/9781119246329.ch23
Pang, H. Y., Zhang, X. Z., Wu, Z. H., Jian, J. C., Cai, S. H., and Liang, J. (2013). Identification of novel immunogenic proteins of Vibrio alginolyticus by immunoproteomic methodologies. Aquac. Res. 44, 472–484. doi: 10.1111/j.1365-2109.2012.03150.x
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14:576. doi: 10.1038/nrmicro.2016.89
Park, K. D., Kim, Y. S., Han, D. K., Kim, Y. H., Lee, E. H. B., Suh, H., et al. (1998). Bacterial adhesion on PEG modified polyurethane surfaces. Biomaterials 19, 851–859. doi: 10.1016/s0142-9612(97)00245-7
Parsek, M. R., Val, D. L., Hanzelka, B. L., Cronan, J. E., and Greenberg, E. (1999). Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci.U.S.A. 96, 4360–4365.
Paul, D., Kim, Y. S., Ponnusamy, K., and Kweon, J. H. (2009). Application of quorum quenching to inhibit biofilm formation. Environ. Eng. Sci. 26, 1319–1324. doi: 10.1089/ees.2008.0392
Pechere, J. C. (2001). Azithromycin reduces the production of virulence factors in Pseudomonas aeruginosa by inhibiting quorum sensing. Jpn. J. Antibiot. 54, 87–89.
Peng, B., Su, Y.-B., Li, H., Han, Y., Guo, C., Tian, Y.-M., et al. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Periasamy, S., Joo, H. S., Duong, A. C., Bach, T. H. L., Tan, V. Y., Chatterjee, S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci.U.S.A. 109, 1281–1286. doi: 10.1073/pnas.1115006109
Poortinga, A. T., Bos, R., and Busscher, H. J. (2000). Controlled electrophoretic deposition of bacteria to surfaces for the design of biofilms. Biotechnol. Bioeng. 67, 117–120. doi: 10.1002/(sici)1097-0290(20000105)67:1<117::aid-bit14>3.0.co;2-6
Powell, L. C., Pritchard, M. F., Ferguson, E. L., Powell, K. A., Patel, S. U., Rye, P. D., et al. (2018). Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 4, 1–10.
Pulido, D., Prats-Ejarque, G., Villalba, C., Albacar, M., González-López, J. J., Torrent, M., et al. (2016). A novel RNase 3/ECP peptide for Pseudomonas aeruginosa biofilm eradication that combines antimicrobial, lipopolysaccharide binding, and cell-agglutinating activities. Antimicrobial Agents Chemother. 60, 6313–6325. doi: 10.1128/aac.00830-16
Rabin, N., Zheng, Y., Opoku-Temeng, C., Du, Y., Bonsu, E., and Sintim, H. O. (2015). Biofilm formation mechanisms and targets for developing antibiofilm agents. Fut. Med. Chem. 7, 493–512. doi: 10.4155/fmc.15.6
Rahman, M. M., Inoue, A., Tanaka, H., and Ojima, T. (2010). Isolation and characterization of two alginate lyase isozymes, AkAly28 and AkAly33, from the common sea hare Aplysia kurodai. Comparative Biochem Physiol. Part B 157, 317–325. doi: 10.1016/j.cbpb.2010.07.006
Rama Devi, K., Srinivasan, R., Kannappan, A., Santhakumari, S., Bhuvaneswari, M., Rajasekar, P., et al. (2016). In vitro and in vivo efficacy of rosmarinic acid on quorum sensing mediated biofilm formation and virulence factor production in Aeromonas hydrophila. Biofouling 32, 1171–1183. doi: 10.1080/08927014.2016.1237220
Ramanathan, S., Arunachalam, K., Chandran, S., Selvaraj, R., Shunmugiah, K., and Arumugam, V. (2018). Biofilm inhibitory efficiency of phytol in combination with cefotaxime against nosocomial pathogen Acinetobacter baumannii. J. Appl. Microbiol. 125, 56–71. doi: 10.1111/jam.13741
Rasmussen, T. B., and Givskov, M. (2006). Quorum sensing inhibitors: a bargain of effects. Microbiology 152, 895–904. doi: 10.1099/mic.0.28601-0
Rasmussen, T. B., Skindersoe, M. E., Bjarnsholt, T., Phipps, R. K., Christensen, K. B., Jensen, P. O., et al. (2005). Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151, 1325–1340. doi: 10.1099/mic.0.27715-0
Ravindran, D., Ramanathan, S., Arunachalam, K., Jeyaraj, G., Shunmugiah, K., and Arumugam, V. (2018). Phytosynthesized silver nanoparticles as antiquorum sensing and antibiofilm agent against the nosocomial pathogen Serratia marcescens: an in vitro study. J. Appl. microbiol. 124, 1425–1440. doi: 10.1111/jam.13728
Rogers, J., Dowsett, A., Dennis, P., Lee, J., and Keevil, C. (1994). Influence of plumbing materials on biofilm formation and growth of Legionella pneumophila in potable water systems. J. Appl. Environ. Microbiol. 60, 1842–1851. doi: 10.1128/aem.60.6.1842-1851.1994
Romling, U., and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J.Int. Med. 272, 541–561. doi: 10.1111/joim.12004
Sabelnikov, A., Cymbalyuk, E., Gongadze, G., and Borovyagin, V. (1991). Escherichia coli membranes during electrotransformation: an electron microscopy study. Biochimica Biophysica Acta (BBA) Biomembranes 1066, 21–28. doi: 10.1016/0005-2736(91)90245-4
Sadekuzzaman, M., Yang, S., Mizan, M., and Ha, S. (2015). Current and recent advanced strategies for combating biofilms. Comprehensive Revi. Food Sci. Food Saf. 14, 491–509. doi: 10.1111/1541-4337.12144
Sauer, K., and Camper, A. K. (2001). Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183, 6579–6589. doi: 10.1128/jb.183.22.6579-6589.2001
Schlievert, P. M., and Peterson, M. L. (2012). Glycerol monolaurate antibacterial activity in broth and biofilm cultures. PLoS One 7:e40350. doi: 10.1371/journal.pone.0040350
Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185, 2066–2079. doi: 10.1128/jb.185.7.2066-2079.2003
Seed, P. C., Passador, L., and Iglewski, B. H. (1995). Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177, 654–659. doi: 10.1128/jb.177.3.654-659.1995
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrobial Resistance Infect. Control 8, 1–10.
Shastry, R. P., Rekha, P., and Rai, V. R. (2019). Biofilm inhibitory activity of metallo-protein AHL-lactonase from cell-free lysate of endophytic Enterobacter species isolated from Coscinium fenestratum Gaertn. Biocatalysis Agric. Biotechnol. 18:101009. doi: 10.1016/j.bcab.2019.01.047
Shirtliff, M. E., Bargmeyer, A., and Camper, A. K. (2005). Assessment of the ability of the bioelectric effect to eliminate mixed-species biofilms. J. Appl. Environ. Microbiol. 71, 6379–6382. doi: 10.1128/aem.71.10.6379-6382.2005
Shunmugaperumal, T. (2010). Biofilm Eradication and Prevention: A Pharmaceutical Approach to Medical Device Infections. Hoboken NJ: John Wiley & Sons.
Singh, R. P., Gupta, V., Kumari, P., Kumar, M., Reddy, C., Prasad, K., et al. (2011). Purification and partial characterization of an extracellular alginate lyase from Aspergillus oryzae isolated from brown seaweed. Journal of Applied Phycology 23, 755–762. doi: 10.1007/s10811-010-9576-9
Singh, S., Singh, S. K., Chowdhury, I., and Singh, R. (2017). Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 11:53. doi: 10.2174/1874285801711010053
Sivaranjani, M., Srinivasan, R., Aravindraja, C., Karutha Pandian, S., and Veera Ravi, A. (2018). Inhibitory effect of α-mangostin on Acinetobacter baumannii biofilms–an in vitro study. Biofouling 34, 579–593. doi: 10.1080/08927014.2018.1473387
Sofer, D., Gilboa-Garber, N., Belz, A., and Garber, N. C. (1999). ‘Subinhibitory’erythromycin represses production of Pseudomonas aeruginosa lectins, autoinducer and virulence factors. Chemotherapy 45, 335–341. doi: 10.1159/000007224
Soukos, N. S., Socransky, S. S., Mulholland, S. E., Lee, S., and Doukas, A. G. (2000). Photomechanical drug delivery into bacterial biofilms. Pharm. Res. 17, 405–409.
Srinivasan, R., Devi, K. R., Kannappan, A., Pandian, S. K., and Ravi, A. V. (2016). Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J. Ethnopharmacol. 193, 592–603. doi: 10.1016/j.jep.2016.10.017
Srinivasan, R., Mohankumar, R., Kannappan, A., Karthick Raja, V., Archunan, G., Karutha Pandian, S., et al. (2017). Exploring the anti-quorum sensing and antibiofilm efficacy of phytol against Serratia marcescens associated acute pyelonephritis infection in Wistar rats. Front. Cell. Infect. Microbiol. 7:498.
Srinivasan, R., Vigneshwari, L., Rajavel, T., Durgadevi, R., Kannappan, A., Balamurugan, K., et al. (2018). Biogenic synthesis of silver nanoparticles using Piper betle aqueous extract and evaluation of its anti-quorum sensing and antibiofilm potential against uropathogens with cytotoxic effects: an in vitro and in vivo approach. Environ. Sci. pollut. Res. 25, 10538–10554. doi: 10.1007/s11356-017-1049-0
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/s0140-6736(01)05321-1
Stewart, P. S., Wattanakaroon, W., Goodrum, L., Fortun, S. M., and Mcleod, B. R. (1999). Electrolytic generation of oxygen partially explains electrical enhancement of tobramycin efficacy against Pseudomonas aeruginosa biofilm. Antimicrobial Agents Chemother. 43, 292–296. doi: 10.1128/aac.43.2.292
Stobie, N., Duffy, B., Mccormack, D. E., Colreavy, J., Hidalgo, M., Mchale, P., et al. (2008). Prevention of Staphylococcus epidermidis biofilm formation using a low-temperature processed silver-doped phenyltriethoxysilane sol–gel coating. Biomaterials 29, 963–969. doi: 10.1016/j.biomaterials.2007.10.057
Stokniene, J., Powell, L. C., Aarstad, O. A., Aachmann, F. L., Rye, P. D., Hill, K. E., et al. (2020). Bi-Functional Alginate Oligosaccharide–Polymyxin Conjugates for Improved Treatment of Multidrug-Resistant Gram-Negative Bacterial Infections. Pharmaceutics 12:1080. doi: 10.3390/pharmaceutics12111080
Strathmann, M., Wingender, J., and Flemming, H. C. (2002). Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 50, 237–248. doi: 10.1016/s0167-7012(02)00032-5
Subhadra, B., Kim, D. H., Woo, K., Surendran, S., and Choi, C. H. (2018). Control of biofilm formation in healthcare: Recent advances exploiting quorum-sensing interference strategies and multidrug efflux pump inhibitors. Materials 11:1676. doi: 10.3390/ma11091676
Sultana, S. T., Atci, E., Babauta, J. T., Falghoush, A. M., Snekvik, K. R., Call, D. R., et al. (2015). Electrochemical scaffold generates localized, low concentration of hydrogen peroxide that inhibits bacterial pathogens and biofilms. Sci. Rep. 5, 1–10.
Sultana, S. T., Call, D. R., and Beyenal, H. (2016). Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility. NPJ biofilms and microbiomes 2, 1–8.
Sun, M., Dong, J., Xia, Y., and Shu, R. (2017). Antibacterial activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against planktonic and biofilm growing Streptococcus mutans. Microbial pathogenesis 107, 212–218. doi: 10.1016/j.micpath.2017.03.040
Sun, M., Zhou, Z., Dong, J., Zhang, J., Xia, Y., and Shu, R. (2016). Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microbial Pathog. 99, 196–203. doi: 10.1016/j.micpath.2016.08.025
Sutherland, I. W. (2001). The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 9, 222–227. doi: 10.1016/s0966-842x(01)02012-1
Tateda, K., Comte, R., Pechere, J.-C., Kohler, T., Yamaguchi, K., and Van Delden, C. (2001). Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrobial Agents Chemother. 45, 1930–1933. doi: 10.1128/aac.45.6.1930-1933.2001
Teasdale, M. E., Liu, J., Wallace, J., Akhlaghi, F., and Rowley, D. C. (2009). Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl. Environ. Microbiol. 75, 567–572. doi: 10.1128/aem.00632-08
Tetz, G. V., Artemenko, N. K., and Tetz, V. V. (2009). Effect of DNase and antibiotics on biofilm characteristics. Antimicrobial Agents Chemother. 53, 1204–1209. doi: 10.1128/aac.00471-08
Thormar, H. (2011). Antibacterial Effects of Lipids: Historical Review (1881 to 1960). Lipids and Essential oils as Antimicrobial Agents. Hoboken, NJ: John Wiley & Sons, Ltd, 25–45.
Torelli, R., Cacaci, M., Papi, M., Sterbini, F. P., Martini, C., Posteraro, B., et al. (2017). Different effects of matrix degrading enzymes towards biofilms formed by E. faecalis and E. faecium clinical isolates. Colloids Surf. B Biointerfaces 158, 349–355. doi: 10.1016/j.colsurfb.2017.07.010
Torrent, M., De La Torre, B. G., Nogués, V. M., Andreu, D., and Boix, E. (2009). Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochem. J. 421, 425–434. doi: 10.1042/bj20082330
Utari, P. D., Vogel, J., and Quax, W. J. (2017). Deciphering physiological functions of AHL quorum quenching acylases. Front. Microbiol. 8:1123.
Van Der Borden, A. J., Van Der Werf, H., Van Der Mei, H. C., and Busscher, H. J. (2004). Electric current-induced detachment of Staphylococcus epidermidis biofilms from surgical stainless steel. Appl. Environ. Microbiol. Microbiol. 70, 6871–6874. doi: 10.1128/aem.70.11.6871-6874.2004
van Tilburg Bernardes, E., Lewenza, S., and Reckseidler-Zenteno, S. (2015). Current research approaches to target biofilm infections. Postdoc J. A J. Postdoctoral Res. Postdoctoral Affairs 3:36.
Verderosa, A. D., Totsika, M., and Fairfull-Smith, K. E. (2019). Bacterial biofilm eradication agents: a current review. Front. Chem. 7:824.
Vetter, S. M., and Schlievert, P. M. (2005). Glycerol monolaurate inhibits virulence factor production in Bacillus anthracis. Antimicrobial Agents Chemother. 49, 1302–1305. doi: 10.1128/aac.49.4.1302-1305.2005
Vogel, J., and Quax, W. J. (2019). “Enzymatic quorum quenching in biofilms,” in Quorum Sensing, ed. G. Tommonaro (Amsterdam: Elsevier), 173–193. doi: 10.1016/b978-0-12-814905-8.00007-1
Wagner, V. E., Gillis, R. J., and Iglewski, B. H. (2004). Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeruginosa. Vaccine 22, S15–S20.
Wang, H. Y., Lin, L., Tan, L. S., Yu, H. Y., Cheng, J. W., and Pan, Y. P. (2017). Molecular pathways underlying inhibitory effect of antimicrobial peptide Nal-P-113 on bacteria biofilms formation of Porphyromonas gingivalis W83 by DNA microarray. BMC Microbiol. 17:1–7.
Wang, W., Wang, R., Zhang, G., Chen, F., and Xu, B. (2020). In vitro Antibacterial Activities and Mechanisms of Action of Fatty Acid Monoglycerides against Four Foodborne Bacteria. Journal of food protection 83, 331–337. doi: 10.4315/0362-028x.jfp-19-259
Wang, Y., Guo, E. W., Yu, W. G., and Han, F. (2013). Purification and characterization of a new alginate lyase from a marine bacterium Vibrio sp. Biotechnol. Lett. 35, 703–708. doi: 10.1007/s10529-012-1134-x
Weiland-Brauer, N., Kisch, M. J., Pinnow, N., Liese, A., and Schmitz, R. A. (2016). Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 7:1098.
Weise, A. M., Cromey, C. J., Callier, M. D., Archambault, P., Chamberlain, J., and Mckindsey, C. W. (2009). Shellfish-DEPOMOD: modelling the biodeposition from suspended shellfish aquaculture and assessing benthic effects. Aquaculture 288, 239–253. doi: 10.1016/j.aquaculture.2008.12.001
Wendrich, T. M., and Marahiel, M. A. (1997). Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26, 65–79.
Wexselblatt, E., Oppenheimer-Shaanan, Y., Kaspy, I., London, N., Schueler-Furman, O., Yavin, E., et al. (2012). Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog 8:e1002925.
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295, 1487–1487.
Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J., and Salmond, G. P. (2001). Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404.
Whiteley, M., Bangera, M. G., Bumgarner, R. E., Parsek, M. R., Teitzel, G. M., Lory, S., et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864.
Wilkins, M., Hall-Stoodley, L., Allan, R. N., and Faust, S. N. (2014). New approaches to the treatment of biofilm-related infections. J. Infect. 69, S47–S52.
Winkelstroter, L. K., Gomes, B. C., Thomaz, M. R., Souza, V. M., and De Martinis, E. C. (2011). Lactobacillus sakei 1 and its bacteriocin influence adhesion of Listeria monocytogenes on stainless steel surface. Food Control 22, 1404–1407.
Witcher, K. J., Novick, R. P., and Schlievert, P. M. (1996). Modulation of immune cell proliferation by glycerol monolaurate. Clin. Diagnostic Lab. Immunol. 3, 10–13.
Wolfmeier, H., Pletzer, D., Mansour, S. C., and Hancock, R. E. (2018). New perspectives in biofilm eradication. ACS Infect. Dis. 4, 93–106.
Xu, K. D., Mcfeters, G. A., and Stewart, P. S. (2000). Biofilm resistance to antimicrobial agents. Microbiology 146, 547–549.
Yamanaka, M., Hara, K., and Kudo, J. (2005). Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl. Environ. Microbiol. 71, 7589–7593.
Yin, W., Wang, Y., Liu, L., and He, J. (2019). Biofilms: the microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 20:3423.
Yuyama, K. T., Rohde, M., Molinari, G., Stadler, M., and Abraham, W.-R. (2020). Unsaturated fatty Acids control biofilm formation of staphylococcus aureus and other gram-positive bacteria. Antibiotics 9:788.
Zanin, I. C., Lobo, M. M., Rodrigues, L. K., Pimenta, L. A., Höfling, J. F., and Gonçalves, R. B. (2006). Photosensitization of in vitro biofilms by toluidine blue O combined with a light-emitting diode. Eur J Oral Sci. 114, 64–69.
Zano, S. P., Bhansali, P., Luniwal, A., and Viola, R. E. (2013). Alternative substrates selective for S-adenosylmethionine synthetases from pathogenic bacteria. Arch. Biochem. Biophys. 536, 64–71.
Zhang, L., Scott, M. G., Yan, H., Mayer, L. D., and Hancock, R. E. (2000). Interaction of polyphemusin I and structural analogs with bacterial membranes, lipopolysaccharide, and lipid monolayers. Biochemistry 39, 14504–14514.
Zhu, Y., Wu, L., Chen, Y., Ni, H., Xiao, A., and Cai, H. (2016). Characterization of an extracellular biofunctional alginate lyase from marine Microbulbifer sp. ALW1 Antioxidant Activity Enz. Hydrolysates. Microbiol. Res. 182, 49–58.
Zuberi, A., Ahmad, N., and Khan, A. U. (2017a). CRISPRi induced suppression of fimbriae gene (fimH) of a uropathogenic Escherichia coli: an approach to inhibit microbial biofilms. Front. Immunol. 8:1552.
Keywords: antibiotics, bacterial biofilm, biofilm inhibitors, biofilm mediated infections, multidrug resistance, persistence, therapeutic strategies
Citation: Srinivasan R, Santhakumari S, Poonguzhali P, Geetha M, Dyavaiah M and Xiangmin L (2021) Bacterial Biofilm Inhibition: A Focused Review on Recent Therapeutic Strategies for Combating the Biofilm Mediated Infections. Front. Microbiol. 12:676458. doi: 10.3389/fmicb.2021.676458
Received: 05 March 2021; Accepted: 14 April 2021;
Published: 12 May 2021.
Edited by:
Palanivel Velmurugan, Alagappa University, IndiaReviewed by:
Arulkumar Abimannan, Alagappa University, IndiaZhe Zhao, Hohai University, China
Periasamy Anbu, Inha University, South Korea
Copyright © 2021 Srinivasan, Santhakumari, Poonguzhali, Geetha, Dyavaiah and Xiangmin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramanathan Srinivasan, dmFzYW5iaW90ZWNoQGdtYWlsLmNvbQ==; Lin Xiangmin, eGlhbmdtaW5AZmFmdS5lZHUuY24=
 Ramanathan Srinivasan
Ramanathan Srinivasan Sivasubramanian Santhakumari
Sivasubramanian Santhakumari Pandurangan Poonguzhali4
Pandurangan Poonguzhali4 Lin Xiangmin
Lin Xiangmin