- Institute of Microbiology, University of Innsbruck, Innsbruck, Austria
Mortierellaceae species are among the most frequent and globally distributed soil fungi. However, the factors shaping their diversity and distribution remain obscure. Several species have been reported to be associated to bacteria, but the kind and frequency of such associations were not addressed up to now. We hypothesized that such associations could be important for Mortierellaceae ecology. Therefore, our aim was to understand the driving factors responsible for the Mortierellaceae diversity, community composition and bacterial associations in alpine and subalpine habitats. For answering our question, we collected both snow-free and snow-covered soil at sampling sites from different habitats: bare alpine soil in a glacier forefield, alpine dwarf-willow habitats, and high-altitude Pinus cembra forests. The isolations were carried out by direct cultivation without any antibiotics to the isolation media. Altogether, we obtained 389 Mortierellaceae isolates representing 29 operational taxonomic units (OTUs). Many OTUs could be placed to the genera Mortierella sensu stricto, Dissophora, Entomortierella, Gamsiella, Linnemannia, and Podila, but others could not unambiguously be assigned to a genus. Our results demonstrate that both, the distribution as well as the diversity of the Mortierellaceae species, were significantly influenced by habitat, soil pH, and snow-cover. We noticed that >30% of our isolates were associated to a non-contaminant bacterium. The bacteria associated to our Mortierellaceae isolates belonged to seven different genera. Pseudomonas was the most frequently detected genus associated to the isolated Mortierellaceae species and it was found to be species-specific. Mortierellaceae–bacteria pairs, including those with Pseudomonas, were influenced by location, habitat, and snow-cover. The majority of the fungus–bacterium associations were potentially epihyphal, but we also detected potential endohyphal bacterial species belonging to Mycoavidus, Burkholderiaceae, and Paraburkholderia. Taken together, the non-random associations we detected suggest that fungus–bacterium associations are ecologically meaningful – an interesting path that needs to be investigated further.
Introduction
Mortierellaceae is a widespread and frequent fungal genus belonging to Mortierellomycotina. It has a global distribution and is a member of the core fungal soil community (Tedersoo et al., 2014). It is a well-studied genus, with a solid database allowing for reliable species identification. Most of the type strains have been sequenced and reference sequences are available in UNITE or other sequence databases. However, the most comprehensive molecular phylogenetic analysis for this group (Wagner et al., 2013) revealed that only a part of the Mortierellaceae diversity is currently known.
A recent study showed that, based on a phylogenomic approach, the genus Mortierella is polyphyletic, justifying its splitting and the description of several new genera (Vandepol et al., 2020). We have used this revised taxonomy throughout the manuscript where possible. However, the correct generic placement of many Mortierella species is still obscure. In this study, this is the case for M. angusta, M. bainieri, M. gemmifera, M. pseudozygospora, M. solitaria and M. zonata. For the sake of clarity and consistency, we refer to these species as Mortierella sensu lato (s.l.) (Mortierella in the classical sense). Mortierella, as defined in a modern taxonomic concept (Vandepol et al., 2020) is addressed as Mortierella sensu stricto (s.s.).
Mortierellaceae are very often detected in mycobiome sequencing data obtained from different environments. However, their diversity can often not be resolved, as mycobiome studies are usually based on a short region of the ITS region, in which members of the Mortierellaceae are highly similar. As cultivation data are also scarce (Hibbett and Glotzer, 2011), the ecology of this interesting group of fungi is not well understood. To overcome the problem of lacking type collections, Hibbett and Glotzer (2011) proposed sequence-based classification. Sequence-based classification would be fast and high-throughput, compared to classification based on a physical specimen. However, this methodology has the disadvantage of lacking information on the morphology and function. Given that Mortierellaceae are usually easily cultivable in the lab, studies based on isolates can contribute tremendously to understanding their ecology, because the thread of cultivation bias is lower compared to other less-easily cultivable organisms, although it might not be negligible. In addition, physical specimens, especially living fungal isolates, allow thorough studies on the physiology, metabolite production, or interactions of organisms representing a species concept. This can open interesting and new perspectives concerning the ecological function and interactions of these fungi.
Mortierellaceae, as an example, are classically regarded as saprobial soil fungi. As saprotrophs in the soil, Mortierellaceae drive soil carbon cycling, P-dissolution and immobilization, lipid metabolism, and chitin degradation (Uehling et al., 2017; Zhang et al., 2020). However, Mortierellaceae have also been reported to occur in the rhizosphere of plants (Kwaśna, 2001; Summerbell, 2005a), and in close association to ectomycorrhizae (Ohara, 1967; Summerbell, 2005b). Similar to arbuscular mycorrhizal fungi, Mortierellaceae spp. support phosphate uptake into plants, which may contribute to increased biomass production (Zhang et al., 2011; Johnson et al., 2019). The exact mechanisms of how these beneficial effects are triggered are still unclear, but a dual lifestyle of selected Mortierellaceae spp. as plant-endophytes and saprotrophs could constitute an important trait (Zhang et al., 2020).
On the other hand, Mortierellaceae spp. are not only interacting with other fungi and plants, but they are also tightly interacting with different species of bacteria (Bonfante and Desiro, 2017). Very little information in this regard is currently available, but a recent study (surveying 238 isolates) proved that 20 species of Mortierellaceae isolated from Japanese soil ecosystems harbor Burkholderiaceae-related endobacteria (Takashima et al., 2018). A comparative genomic analysis suggested an ancient origin of the association between Linnemannia elongata and its endosymbiont Mycoavidus cysteinexigens likely underpinning their close interaction (Uehling et al., 2017).
Synergistic or mutualistic associations with bacteria appear to be comparatively common in Mucoromycotina and Glomeromycotina (Bonfante and Desiro, 2017). Bifiguratus, a recently discovered monotypic fungal genus of Mucoromycotina, is also characterized by a dual, endophytic, and soil-dwelling lifestyle. This fungus is associated with a wide variety of endo- and epihyphal bacteria (Torres-Cruz et al., 2017). This publication adds further evidence to the hypothesis, that fungus–bacteria associations are common and ecologically important. However, reports are usually limited to individual fungal host species or selected groups of associated bacteria, and a more general overview about the distribution and frequency across habitats of such fungus–bacteria associations is still lacking.
Altogether, the currently available data indicate that associations could generally be one important key-factor for the frequent occurrence and global distribution of Mortierellaceae. The ability to grow at temperatures around 0 °C could constitute another key-factor for the global success of this fungal lineage. Mortierellaceae belongs to the most active fungal taxa in snow-covered soil (Kuhnert et al., 2012; Dresch et al., 2019). During winter, they are not only part of the soil microbiota but also have been reported to grow on the soil surface, forming dense under-snow mats, as reported from subalpine forests in Colorado (United States). In vitro, these snow-mat fungi (eight unidentified Mortierellaceae spp.) had very high growth rates at −2 °C (Schmidt et al., 2008).
Thus, we aimed to address the following questions with this extensive study on Mortierellaceae: Are environmental conditions and habitat properties influencing the Mortierellaceae distribution? How does snow-cover influence the Mortierellaceae diversity and distribution? And, most interestingly, we wanted to address the question of whether or not Mortierellaceae spp. are associated to bacteria, and if such associations might be species-specific. For addressing these questions, we isolated Mortierellaceae spp. without using antibiotics from a wide range of sites in the Alps, and from both, snow-free and snow-covered soil. We surveyed high-altitude stone pine (Pinus cembra) forests, alpine dwarf-willow (Salix retusa, S. reticulata, Dryas octopetala) habitats, and alpine bare terrain.
Materials and Methods
Sampling and Isolation of Mortierellaceae Isolates
Mortierellaceae pure cultures were isolated from ten different sampling sites in subalpine and alpine habitats of the Austrian Alps at an altitudinal range from 1541 to 2450 m. These included the following habitat types: bare terrain in the glacier forefield, alpine chalk dumps (Kalkschutthalden), alpine dwarf-willow communities (Salix herbacea, S. reticulata, Dryas octopetala), high-altitude Pinus cembra forests, and subalpine mixed P. cembra/Picea abies forests (Table 1). To study seasonal dynamics, at each location sampling was carried out from both snow-covered and snow-free soil. All locations are usually snow-covered in winter (November – May) and snow-free during summer (June – October).
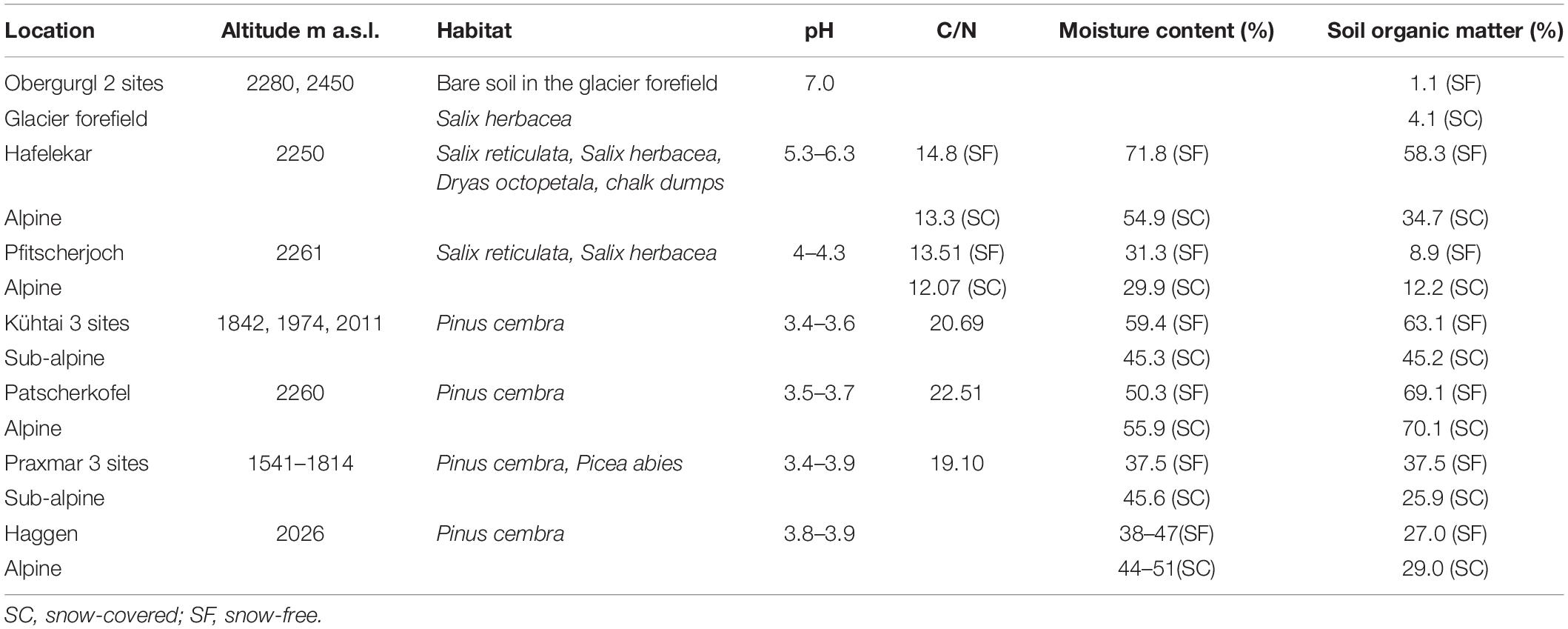
Table 1. Sampling sites in Austria with information on the origin of isolates the dominating plant communities and sampling time.
Cultivation of Mortierellaceae was carried out either directly from the soil, or using in-growth mesh bags (MBs) containing sterile quartz sand. Mesh bags (4 × 3 cm) were sewed from nylon mesh (72 μm mesh size) and filled with acid-washed, sterilized quartz sand. They were buried for 2 months during the vegetation period, and for the whole snow-covered period (6–8 months) during winter. MBs were stored at −80 °C. For cultivation by direct plating, few particles of soil grains or quartz sand from MBs were plated on agar plates containing potato dextrose agar (PDA) or LCA media (Takashima et al., 2019) without antibiotics, and incubated at 10 °C for at least a week. The plates were monitored daily and pure cultures were separated to new plates based on their growth. Several consecutive isolation steps were carried out to obtain fungal pure culture isolates.
Sequencing of rDNA ITS Regions of Mortierellaceae Isolates
Molecular identification of the obtained fungal isolates was carried out by sequencing the rDNA-ITS region. Colony PCR was performed using aerial mycelium from 7-day-old cultures based on Walch et al. (2016), with the following modifications: The template for the identification of the fungi was prepared by harvesting a little amount of mycelium with sterile water, homogenized and incubated at 85 °C for 15 min. The rDNA-ITS region was amplified by standard primers ITS1 and ITS4 (White et al., 1990). Microsynth AG (Balgach, Switzerland) carried out the sequencing of the PCR products. GenBank accession numbers for all ITS sequences are provided in Supplementary Table 1. Sequences were binned to operational taxonomic units (OTUs) based on 99% sequence similarity using Sequencher 5.2.3. OTUs were used for further analysis. If OTUs with identical species annotation and location inside the phylogenetic tree did not differ in terms of their location distribution and seasonal occurrences, they were summarized to reduce the high number of different Mortierellaceae observed here, which simplifies the accessibility and presentation of the data (especially in the figures).
Phylogenetic Analysis of Mortierellaceae Isolates
A total of 389 ITS sequences were newly generated in the current study. The obtained sequences were first quality checked, then preliminarily identified based on blastn searches and finally used for phylogenetic analysis. The longer ITS sequences obtained from few isolates were trimmed to avoid misalignment of the species level phylogenetic clade. Along with our ITS sequences, 36 sequences from type material and environmental sequences retrieved from GenBank were included in the alignment. Sequences were then aligned using MAFFT v.7 (Katoh et al., 2017). The alignment was checked using Mega X (Kumar et al., 2018). The best Maximum Likelihood (ML) model was calculated in MEGA X before carrying out an ML analysis. To evaluate branch robustness of trees, Maximum Parsimony based bootstrap analyses were applied. Bootstrap analyses (1000 replicates) were conducted by Subtree-Pruning-Regrafting (SPR) algorithm level 3 in which the initial trees were obtained. For the ML analysis and the bootstrap search, all positions with less than 95% site coverage were eliminated.
Additionally, branch robustness was tested with Bayesian Inference in MrBayes 3.2.6 (Ronquist et al., 2012). For the Markov Chain Monte Carlo (MCMC) analyses, four chains were run for 10 million generations, with trees being sampled every 5000 generations. The analysis was stopped as the convergence diagnostic (average standard deviation of split frequencies) was below 0.05 after 10 million generations. From the 20000 sampled trees (for each of the 2 runs) 25% were discarded as burn-in before summary statistics were calculated (using sump and sumt commands). Diagnostic plots, as well as the convergence diagnostics EES (Estimated Sample Size; min ESS around 10 K) and PSRF (Potential Scale Reduction Factor; 1.000 for all parameters), indicated stationarity. Phylogenetic trees were drawn with Figtree 1.4.4 (Rambaut, 2009).
Detection and Identification of Mortierellaceae Associated Bacteria
Mortierellaceae pure culture isolates did not show any sign of bacterial contaminations when growing under standard conditions. The presence/absence of associated bacteria was targeted by sequencing of the bacterial 16S rRNA region. The same template as prepared for the fungal colony PCR was used to identify the associated bacteria as well. The PCR amplification of the 16S region was as follows: 25 μl of a PCR mixture containing 2.5 μl of ten-fold buffer S (15 mM MgCl2, 100 mM Tris, 500 mM KCl), 2.5 μl of 10% BSA, 1 μl of primers 8f and 1492r, each (10 mM), 0.5 μl of dNTPs (10 mM each), 1 μl of DNA template and 0.15 μl of LabQ Taq-pol (5 units/μl), and 16.35 μl of sterile PCR water. The bacterial PCR cycling conditions were according to Takashima et al. (2018). PCR products were sequenced using the primer 27F by Microsynth AG (Balgach, Switzerland). All sequences obtained in the study are available in GenBank (Supplementary Table 1). Closely related sequences were downloaded from GenBank. To identify the bacteria on a species level, the threshold was set to 99% sequence similarity. Sequences were aligned, and phylogenetic analyses were carried out in Mega X. A Maximum likelihood analysis was carried out based on the best model (Kimura 2-parameter model) as suggested by Modeltest. For the branch robustness Maximum parsimony bootstrapping analyses was done with 1000 replicates with 95% site coverage was eliminated. Bacterial sequences were binned to OTUs based on 99% sequence similarity using Sequencher 5.2.3.
Soil Properties
Moisture content was determined based on the weight loss of the sieved soil sample when incubated at 105 °C overnight. The soil organic matter (SOM) was determined based on loss of ignition at 550 °C for 7 h. The pH of the samples was measured with 0.01 M Calcium chloride (CaCl2) potentiometrically. The soil properties were measured as suggested by Kuhnert et al. (2012). The carbon and nitrogen element analysis was performed by an organic elemental analysis flash combustion instrument (Thermo Fischer Scientific, Austria).
Statistical Analysis
All statistical analyses of the data were performed in R 4.0.2 (R Core Team, 2020). For alpha diversity analysis, package vegan v2.5-6 (Oksanen et al., 2019) was used. In order to test for influences of environmental variables (pH, C/N, moisture, soil organic matter) and environmental factors [season (snow-cover vs. snow-free) and habitat (alpine bare terrain, dwarf-willow communities, P. cembra forest)] permutational multivariate analysis of variances was applied on a Bray–Curtis distance matrix using adonis function (999 permutations). For clustering of fungal occurrences, bacterial associations, and Mortierellaceae–bacterial pairs as well as the clustering of Mortierellaceae species based on their association to Pseudomonas OTUs, several clustering algorithms were tested and compared. For fungal clustering, the distance matrix was based on Bray–Curtis dissimilarity. All other variables were inherently dependent on the distribution of the Mortierellaceae species and on the total number of observations in this sample group. Therefore, we calculated the distance between two sample groups as the number of shared observations normalized by the maximum number of possible shared observations. For example, if there were two different fungus–bacterium pairs observed in location A and five different fungus–bacterium pairs in location B and only one of those associations would be found in both locations, the distance between location A and B would be 0.5. For each clustering, different clustering algorithms were applied. For all clusterings, the ward algorithm best represented all other algorithms and, consequently, it was chosen for display. Networks were drawn from an edgelist containing all observed Mortierellaceae–bacteria associations using R package igraph (Csardi and Nepusz, 2006). The dataset is available as Supplementary Table 2.
Our dataset contained several observations that were only made once. While for alpha diversity calculations, these have not been disregarded, for visualization, these one-time occurrences have been omitted from display.
Results
Mortierellaceae Composition Is Diverse, Widespread, and Affected by Habitat, Location, and Snow-Cover
In this study, a total number of 389 Mortierellaceae pure cultures were isolated and analyzed (Figure 1). Phylogenetically, these clustered into 24 terminal clades based on their rDNA-ITS sequences, representing OTUs with ≥99% sequence similarity (Figure 2). The majority of our isolates could unambiguously be identified based on available type sequences (19 clades: 348 isolates). Podila humilis and P. verticillata could not be differentiated based on their ITS sequences and were summarized to one species complex (Figure 2). Isolates, that could not be identified fell into five distinct clades. Those likely represent new species that remain to be described, and we addressed them as Mortierellaceae sp. 1–5 in our study.
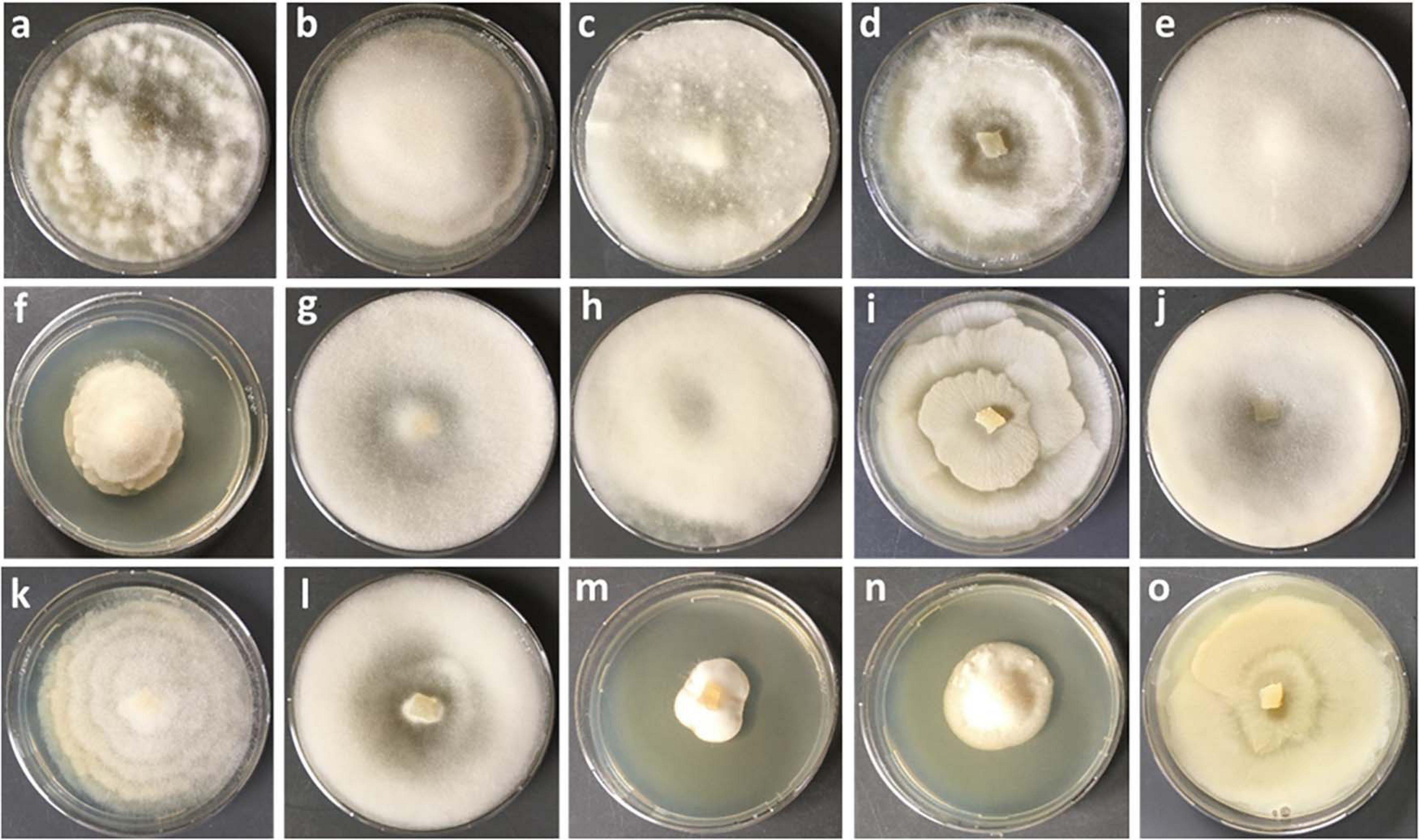
Figure 1. Culture characteristics of different species of Mortierellaceae isolated on PDA. (a) Podila verticillata, (b) Linnemannia elongata, (c) M. pseudozygospora (s.l.), (d) M. angusta (s.l.), (e) P. minutissima, (f) M. solitaria, (g) M. zonata (s.l.), (h) L. hyalina, (i) L. gamsii, (j) M. gemmifera (s.l.), (k) Entomortierella parvispora, (l) M. bainieri (s.l.), (m) M. globalpina (s.s.), (n) M. alpina (s.s.), (o) L. exigua.
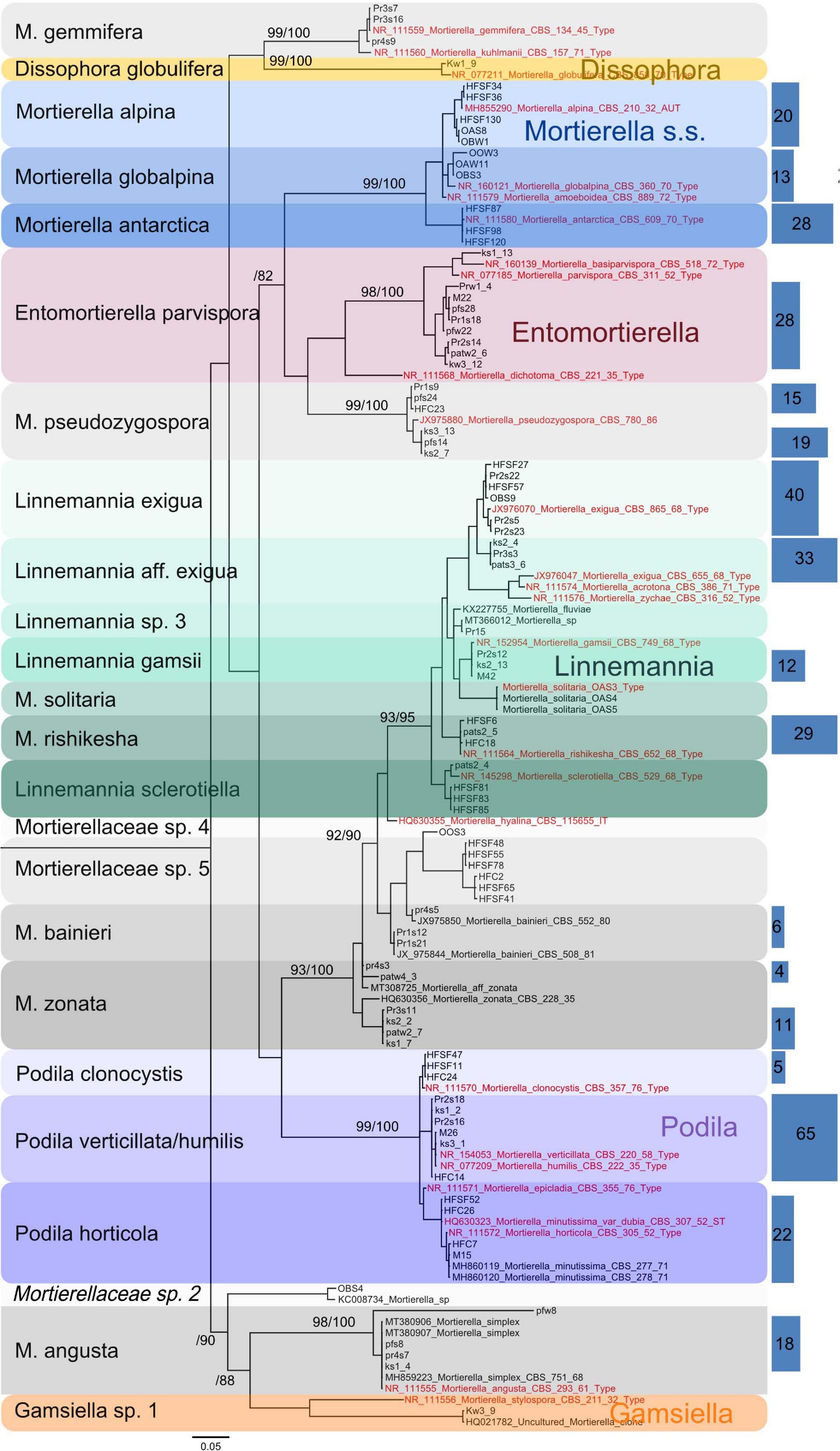
Figure 2. Phylogenetic tree of the Mortierellaceae species isolated in this study (n = 389 isolates) (loglikelihood –3810.59) based on rDNA-ITS sequences. The isolates fall into 24 well-supported clades. For branches close to the origin, branch ≥70 is shown above the branches as Bayesian posterior probabilities/Parsimony-based bootstrap values. Sequences originating from type specimens are highlighted in red. In order to reduce the size of the tree, all isolates’ sequences were binned to 99% OTUs and a representative number of sequences for each OTU were shown. Blue rectangles on the right illustrate the total number of isolates represented by the sequences they cover (same height). The abundance of single sequences that are directly represented in the tree (n = 1) is not illustrated by rectangles. Rectangle areas are proportional to each other and to isolate abundance, which is also given in the rectangles.
Mortierellaceae spp. were isolated from all habitats and occurred in both snow-covered and snow-free soil. The numbers of both isolates and species isolated per location varied strongly (Table 2). The species composition of Mortierellaceae differed between sampling sites (Figure 3), especially between dwarf-willow habitats and P. cembra forests (33% variance, pAdonis = 0.031, Supplementary Figure 1). Moreover, Mortierellaceae composition was also correlated to soil pH (41% variance, pAdonis = 0.001), and to a minor extent to C/N ratio (20% variance, pAdonis = 0.063). Furthermore, snow-cover (seasonality) seemed to influence the species composition (Figure 3).
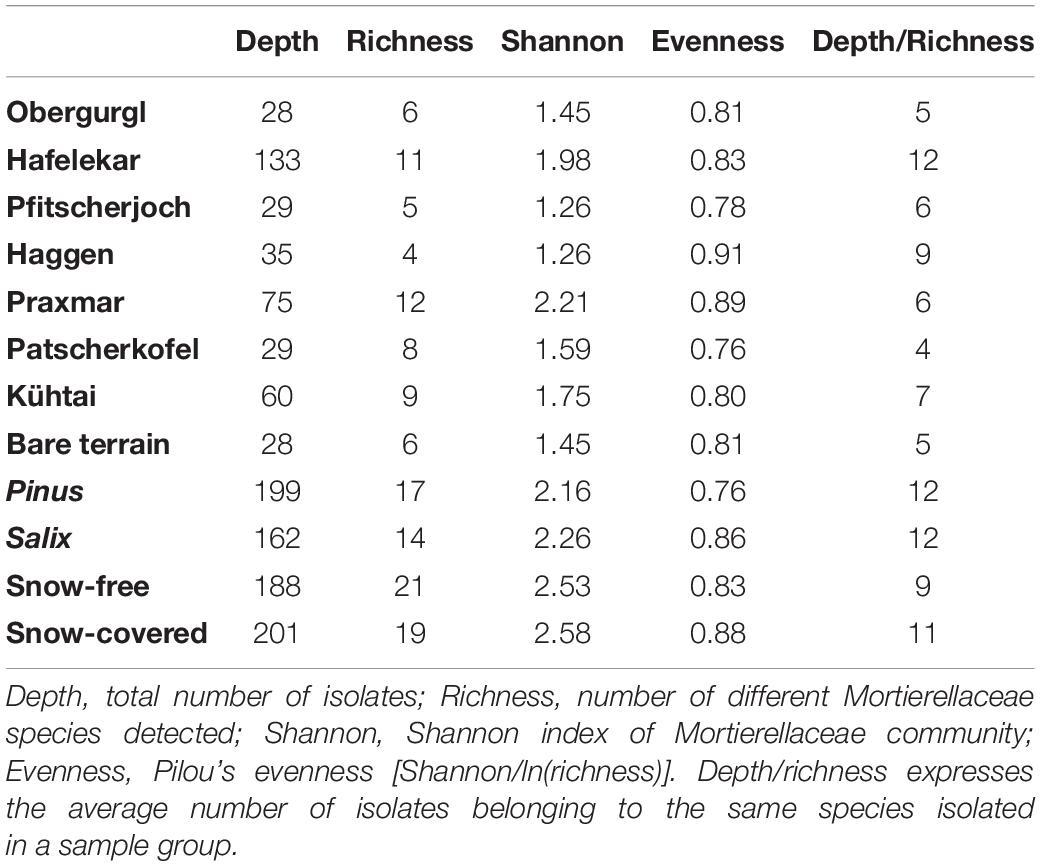
Table 2. Overview of Mortierellaceae composition at different locations, in different habitats, and in snow-covered and snow-free soil.
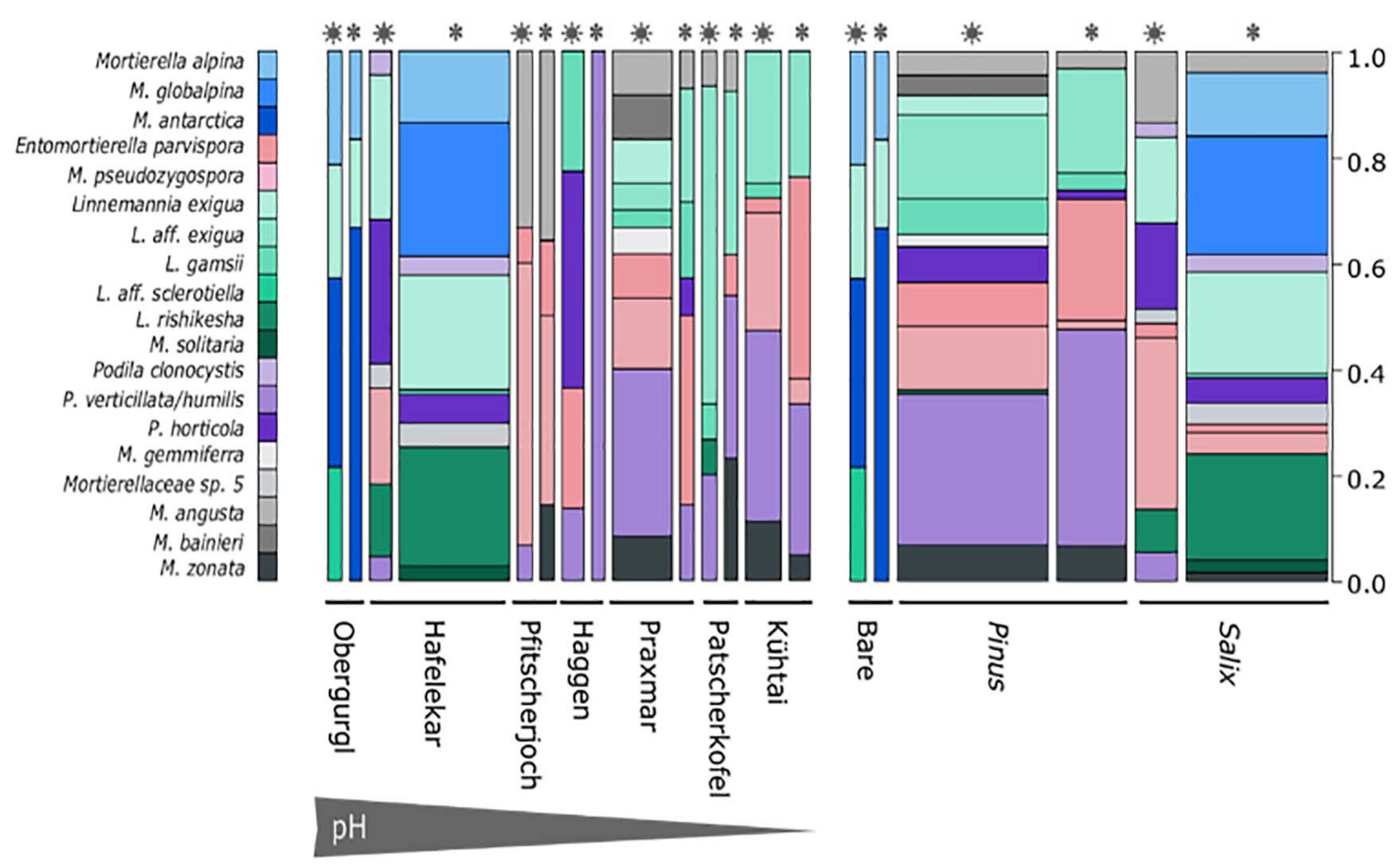
Figure 3. Mortierellaceae species distribution across sampling sites (left) and habitats (right). Snowflakes and stars indicate whether the sample, from which the strain was isolated, was covered with snow or not. Species represented by one single species were excluded from the illustration. Bar width is proportionate to the sample size of the sample group in the overall data set. M. alpina, M. globalpina and M. antarctica represent Mortierella s.s., all other Mortierella s.l.; E., Entomortierella; L., Linnemannia; M., Mortierella; P., Podila.
Independent of the species’ abundances, that is the number of isolates at a site or in a habitat, the majority of species were isolated from a single location or showed habitat specificity. Mortierella alpina (s.s.) and P. clonocystis were isolated from different locations but exclusively found in dwarf-willow habitats. M. antarctica (s.s.), Linnemannia rishikesha, and an unknown Mortierellaceae sp. 5 were obtained only from the dwarf-willow habitats at one calcareous site (Hafelekar). Mortierella globalpina (s.s.) and M. solitaria (s.l.) were isolated solely from alpine bare terrain in the glacier forefield (Obergurgl). Other species are typical for P. cembra forests: With few exceptions, L. gamsii, Entomortierella parvispora, M. bainieri (s.l.), M. gemmifera (s.l.) and M. zonata (s.l.) were typically isolated from this habitat. The latter two species were exclusively obtained from Praxmar. However, we detected also generalist species: For M. angusta (s.l.), P. horticola, and M. pseudozygospora (s.l.) no location or habitat specificity could be detected.
The genus Linnemannia appears to be especially abundant in subalpine and alpine soil habitats. Linnemannia exigua was the most abundant species observed in our dataset (73 isolates). According to the ITS phylogeny, almost half of these isolates (33 isolates) formed a subclade (Figure 2). Interestingly, all isolates belonging to the subclade L. affinis (aff.) exigua were isolated solely from P. cembra forests (Figure 3). The isolates belonging to clade L. exigua were isolated across different locations and habitats but showed a higher abundance in dwarf-willow habitats.
Linnemannia sclerotiella originated from a snow-free P. cembra forest soil, while three sister group isolates were obtained from a snow-covered dwarf-willow habitat (Hafelekar). According to the ITS-based phylogeny, this sister-group might represent a new species. A higher sampling size would be necessary for confirming the environmental preferences of this species (Figure 3).
The second largest group of isolates is represented by the genus Podila (P. humilis/verticillata clade; 65 isolates). They, too, were found across different locations, but were most frequently isolated in P. cembra forests (Figure 3).
Several species were isolated once or a few times, only, e.g., E. basiparvispora, Dissophora globulifera, Gamsiella sp. 1, and M. solitaria (s.l.). Consequently, their distribution could not be determined.
While there was little difference in Mortierellaceae compositions between seasons in the alpine sites Obergurgl and Pfitscherjoch, the Mortierellaceae species compositions of the forest sites exhibited a clear seasonal variation (snow-covered soil vs. snow-free soil) (Figure 3). Especially the lowest site in terms of altitude, Praxmar, appeared to have a specific summer and a specific winter Mortierellaceae community. Some Mortierella s.l. species, M. solitaria, M. bainieri, and M. gemmifera, were only isolated from snow-free soil types. M. antarctica (s.s.) occurred only in snow-covered soil (Figure 3).
The Mortierellaceae richness of a sampling site ranges between 4 and 12 species per site. The richness increases with decreasing altitude and is highest in P. cembra forests. The richness did not differ significantly between dwarf-willow habitats and P. cembra forests. There were no significant differences in richness between snow-covered and snow-free soil (Table 2). The total number of isolates differed between sampling sites (Table 2), although the sampling effort was comparable. The total number of isolates obtained from one sampling site influenced Mortierellaceae richness, that is number of Mortierellaceae OTUs, and Mortierellaceae diversity, here expressed by Shannon index (r = 0.76, p = 0.049). The calcareous alpine site with dwarf-willow vegetation, Hafelekar, exhibited unique features compared to all other sampling sites. Given a high number of 133 isolates obtained from this site, only 11 different species were detected. In contrast, at the P. cembra site Patscherkofel and in bare glacier foreland terrain (Obergurgl), where not even 30 isolates were obtained, eight and six different species, respectively, were detected (Table 2). In terms of both richness and diversity, there was no pattern detectable among locations, despite the compositional pattern following soil pH and C/N ratio. For the majority of the sampling sites, the Mortierellaceae composition was uneven, with the exception of the pure P. cembra plantation at Haggen (Figure 3).
Mortierellaceae spp. Are Frequently Associated With Bacteria
Out of 389 Mortierellaceae isolates studied here, 126 were found to be associated with bacteria (32%), all of them Proteobacteria, and all Mortierellaceae species independently isolated at least three times were associated with bacteria (n = 18). In order to illustrate their relationship and diversity, a phylogenetic tree of the bacteria associated with the isolated strains was calculated based on their full-length 16S sequence. According to the terminal clades of the resulting tree (Bootstrap values and Bayesian Posterior probabilities ≥70%), the associated bacteria belonged to seven different genera (Figure 4). Two bacteria could not be identified on a genus level: one is a Proteobacterium, and the other is a member of Enterobacteriaceae. The discrimination among closely related bacteria was difficult due to their high sequence similarity, especially for Pseudomonas species. Hence, bacterial sequences were binned into OTUs (99% sequence similarity) and a representative number of sequences for each OTU were included in the phylogenetic tree (Figure 4). Although the phylogeny was generally well supported, for some OTUs (5/20 OTUs, Supplementary Table 2), their representative sequences were not clustered in direct neighborhood. Therefore and as this study did not target phylogenetic questions, we used well supported lineages for further analysis.
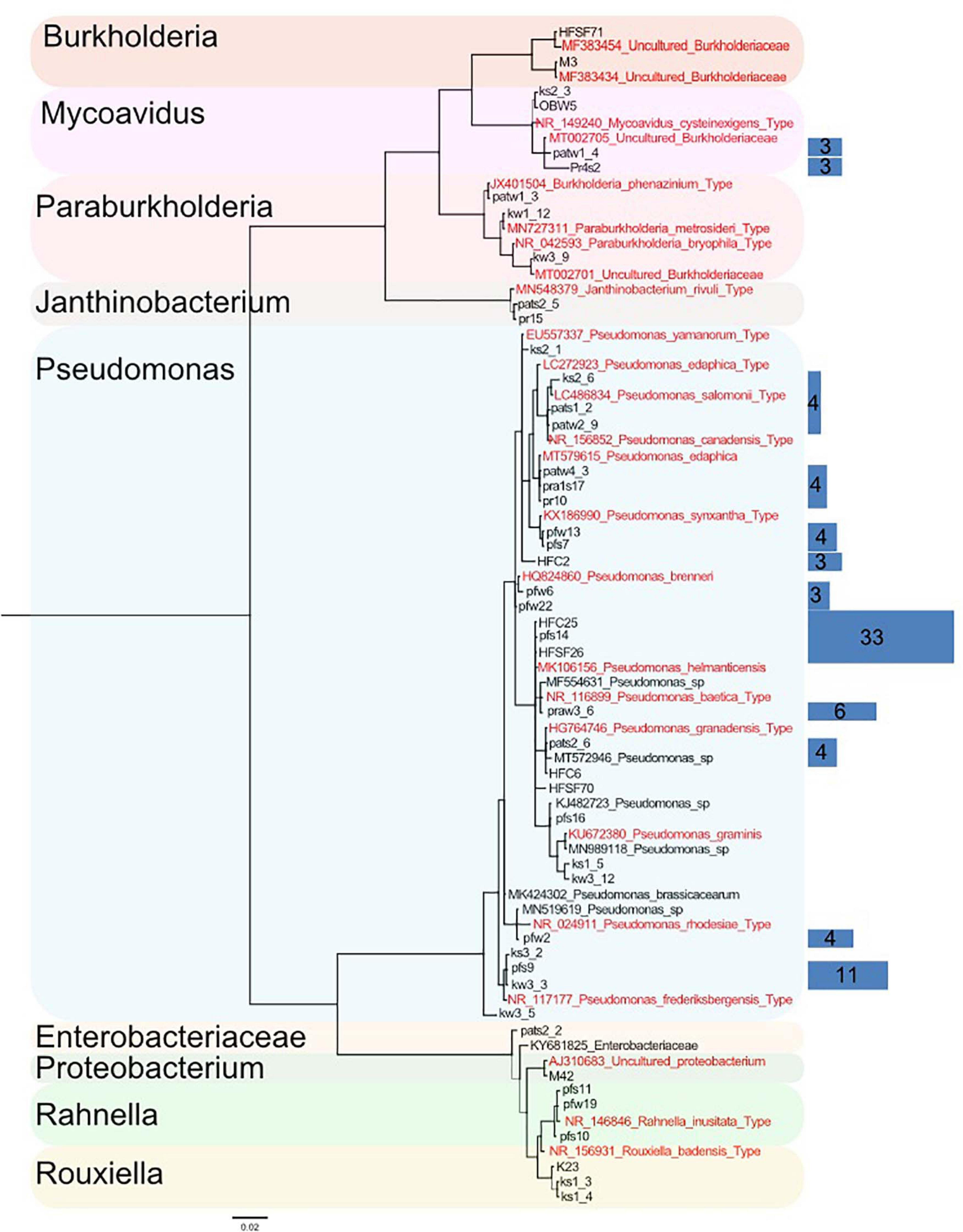
Figure 4. Phylogenetic tree of a representative subset of Mortierellaceae-associated bacteria based on their 16S region (log-likelihood –3459.28). A total of 126 bacteria were associated to Mortierellaceae isolates. They were binned into operational taxonomic units (OTUs) based on their 16S sequence (99% sequence similarity). From each OTU, a representative number of sequences were chosen, from which the phylogeny was constructed. The detected bacteria fall into nine lineages. Relationships are generally well-supported. Support values [Bayesian posterior probabilities (BS)/Parsimony-based bootstrap values (BPP)] are provided for the nine major lineages besides the respective nodes. Thin lines represent relationships with BS and/or BPP < 80%. Sequences originating from type specimens are highlighted in red. Blue rectangles on the right illustrate the total number of isolates represented by the sequences they cover (same height). The abundance of single sequences that are directly represented in the tree is not illustrated by rectangles (n = 1). Rectangle areas are proportional to each other and isolate abundance, which is also provided as numbers in the rectangles.
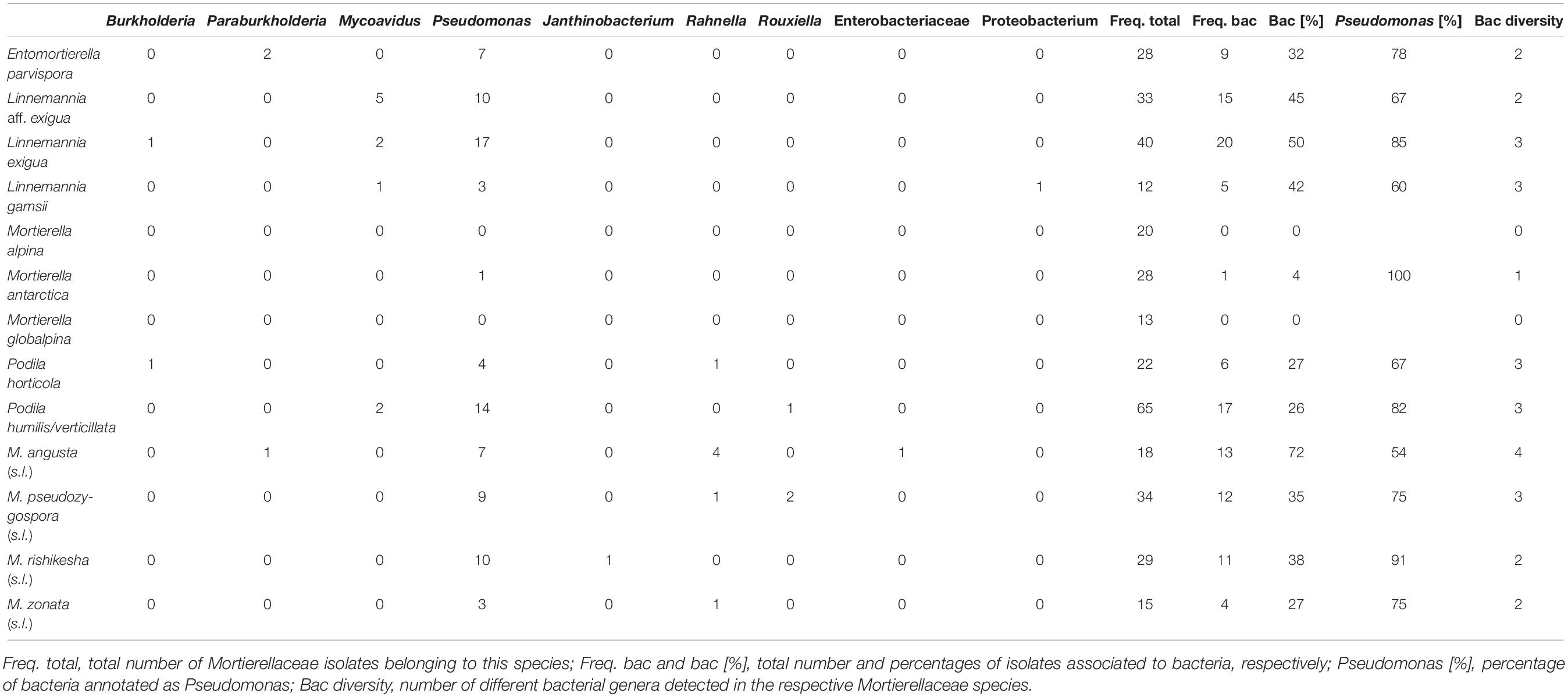
The vast majority of bacteria associated with Mortierellaceae isolates represented species of the genus Pseudomonas (96/126 = 76%, Table 3). Moreover, we detected species of the genera Mycoavidus (10/126), Rahnella (7/126), Burkholderia (3/126), Paraburkholderia (3/126), Rouxiella (3/126), and Janthinobacterium (2/126).
The frequency of associations depended on the fungal species (Supplementary Figure 2). For some Mortierellaceae species, around half of their isolates were associated with bacteria, indicating there was no selection in terms of whether or not such fungus was associated with a bacterium (Table 3). For other species, associations were rare (<25%). For M. alpina and M. globalpina (both s.s.), no bacterial associations were detected. In contrast, M. angusta (s.l.) was more frequently associated with bacteria than the other Mortierellaceae (>75%). Generally, Mortierellaceae species with a representative number of both isolates and bacteria-associated isolates were associated with more than one bacterial genus (Table 3).
The dependency of Mortierellaceae species distribution on the environmental factors, location, habitat and season, was inherently reflected in the distribution of associations to bacteria (Figure 5 and Supplementary Figures 1, 3, 4). However, Mortierellaceae–bacteria pairs were not randomly distributed across the occurrence spectrum of Mortierellaceae species. They, too, were influenced by those environmental factors. Pseudomonas (Pseudomonadales) was detected in association with Mortierellaceae species across all locations (Supplementary Figures 3, 4).
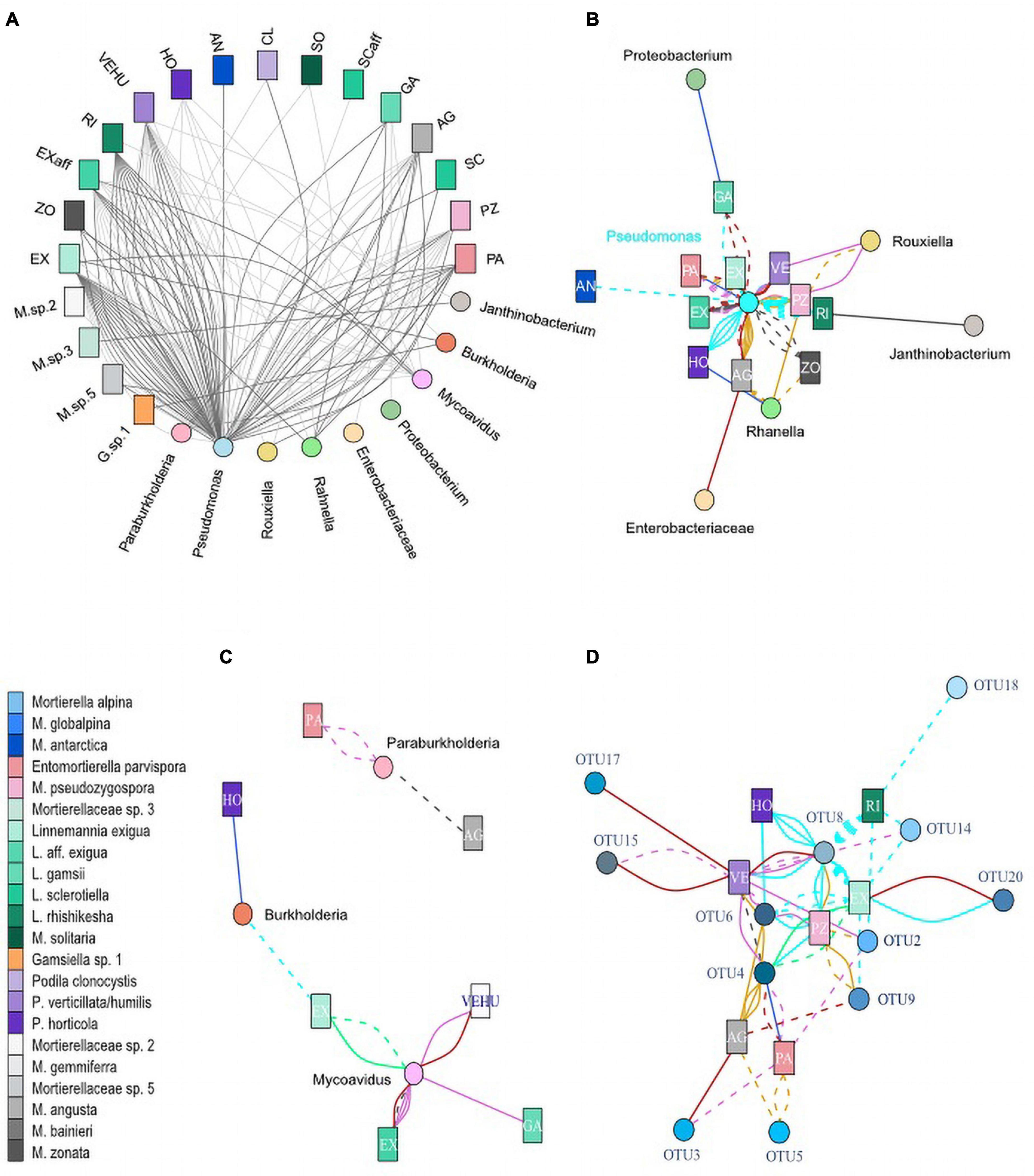
Figure 5. Co-occurrences of Mortierellaceae isolates and bacteria. Each line represents an isolated Mortierellaceae strain with an associated bacterium. (A) Seasonal dependence of co-occurrences on snow-cover. Dark gray lines, isolate was obtained from snow-free soil; light gray lines, isolate was obtained from snow-covered soil. (B) Co-occurrences of isolated Mortierellaceae strains and Burkholderiales. (C) Co-occurrences of isolated Mortierellaceae strains and Pseudomonadales and Enterobacteriales. (D) Co-occurrences of isolated Mortierellaceae strains and Pseudomonas OTUs. For panels (B–D), line colors indicate the location of strain isolation: cyan, Hafelekar; royalblue, Haggen; orchid, Kühtai; springgreen, Obergurgl; purple, Patscherkofel; goldenrod, Pfitscherjoch; firebrick, Praxmar. Solid lines, isolate was obtained from snow-covered soil; dashed lines, isolate was obtained from snow-free soil. AG, M. angusta (s.l.); AL, M. alpina (s.s.); AN, M. antarctica (s.s.); BA, M. bainieri (s.l.); CL, Podila clonocystis; Ex, Linnemannia exigua; Exaff, L. aff. exigua; GA, L. gamsii; GL, M. globalpina (s.s.); HO, P. horticola; M. sp. 5, Mortierellaceae species complex 5; PA, Enteromortierella parvispora; PZ, M. pseudozygospora (s.l.); RI, L. rishikesha; SCaff, L. aff. sclerotiella; SC, L. sclerotiella; SO, M. solitaria (s.l.); VEHU, P. verticillata/humilis; ZO, M. zonata (s.l.).
Associations of Mortierellaceae species to members of the Enterobacterales were limited to both a low number of observations and locations (Supplementary Figure 2). Rahnella was found associated with P. horticola, M. pseudozygospora (s.l.), M. zonata (s.l.), and M. angusta (s.l.) in Pfitscherjoch (dwarf-willow) and Haggen (P. cembra) (Supplementary Figure 4). Rouxiella was found associated in the sampling sites Pfitscherjoch (dwarf-willow) and Kühtai (P. cembra). Associations were limited to P. humilis/verticillata and M. pseudozygospora (s.l.).
In addition to Pseudomonadales and Enterobacterales, we found Burkholderiales members associated to Mortierellaceae species. Mycoavidus, which was the second frequent bacterial genus associated with Mortierellaceae, was detected in Obergurgl and all P. cembra forests, except Haggen (Supplementary Figures 2, 4), which might be due to sampling size. Mycoavidus was associated with L. exigua, L. aff. exigua, and P. humilis/verticillata (Figure 5C). Burkholderia was found in Hafelekar and also in Haggen. Paraburkholderia were detected only in P. cembra forests, Kühtai, and Patscherkofel. They were mainly associated with E. parvispora (Figure 5C). Against the odds, L. aff. exigua, L. exigua, P. humilis/verticillata, and E. parvispora were frequently associated to bacteria from different Proteobacteria orders (Figures 5A,C). Mortierella angusta (s.l.), M. zonata (s.l.), M. pseudozygospora (s.l.), and L. rishikesha (the latter appeared associated only with Pseudomonas) were not found associated to Burkholderiales (Figure 5B and Supplementary Figures 2, 3).
Altitude not only affected the occurrences of associations but generally impacted on the association patterns between Mortierellaceae and bacterial lineages (Figures 5B,C). Entomortierella parvispora, M. pseudozygospora (s.l.), and M. zonata (s.l.), who were all detected across several locations (Figure 3), were associated to bacteria only in those locations at higher altitude and associations were usually absent in Praxmar, which was the location with the lowest altitude in this dataset (Table 1 and Figure 5). The exception of this rule was P. humilis/verticillata, for which it was the other way around. Linnemannia exigua, who was detected across all habitats (Figure 3), was associated with Burkholderia only at high altitude locations [e.g., Obergurgl (bare ground)] (Figure 5).
Altitude appeared to have a stronger impact on Mortierellaceae–bacteria associations compared to habitat (dwarf-willow community vs. P. cembra forest), as E. parvispora and M. zonata (s.l.) were both isolated more frequently from P. cembra forests than dwarf-willow habitats (Figure 3), while associations to bacteria were found equally frequently in both habitats (M. zonata (s.l.): P. cembra = 2/13, dwarf-willow = 2/3; M. zonata (s.l.): P. cembra = 7/25, dwarf-willow = 2/3) (Figure 5).
In addition to altitude/location and habitat, also the season influenced the associations between Mortierellaceae and bacteria (Figure 5A). While there was no seasonal effect on the distribution of M. angusta (s.l.), M. zonata (s.l.), P. horticola, and E. parvispora (Figure 3), the snow-cover affected whether or not these species were found associated with bacteria or to which bacterial genera they were associated. Under snow-cover, M. angusta (s.l.) was more frequently associated with Rahnella (3/4) than with Pseudomonas (2/7), while in snow-free sites, the pattern was reversed. M. zonata (s.l.) (4/4) and E parvispora (8/9) were more frequently associated with bacteria under snow-cover compared to snow-free conditions. For P. horticola, associations were detected only in snow-free conditions (6/6) (Figure 5A).
As Pseudomonas was associated with all Mortierellaceae species investigated here, and these associations were detected across all locations, we asked for a potential specificity on 99% bacterial OTU level (Supplementary Table 2). Strikingly, almost all Mortierellaceae species with more than one association to Pseudomonas were located in one big branch, including Podila and Linnemannia, carrying the highest diversity and most closely related species. With regards to Pseudomonas OTUs, no Mortierellaceae species specificity could be detected (Figure 5D). However, clustered by their association to Pseudomonas OTUs, especially the phylogenetic relationship among Linnemannia species, but also the placement of P. clonocystis and P. humilis/verticillata (Figure 2) was loosely conserved (Supplementary Figure 5).
Discussion
Taxonomy and Nomenclature of Mortierellaceae
Mortierellaceae species have been extensively studied (Linnemann, 1941; Milko, 1974; Gams, 1977; Wagner et al., 2013) but the evolutionary relationship remained unresolved. A recent study now could resolve the phylogeny of the Mortierellaceae based on multigene phylogenetics and phylogenomics (Vandepol et al., 2020). However, several of our isolated Mortierellaceae have not yet been recombined into new genera, and their exact taxonomic position still needs to be resolved, which is far beyond the scope of this paper. In order to clearly differentiate between Mortierella species that have not been placed into the new phylogeny and those that have, we will indicate the former by sensu lato (s.l.) and the latter by sensu stricto (s.s.). Species identification of Mortierellaceae, especially for the genera Mortierella (s.s.), Podila and Linnemannia, is not always possible by ITS sequence due to their high sequence similarity (Vandepol et al., 2020). We used OTUs with 99% sequence identity for our fungal isolates, which can then clearly be differentiated, and were in agreement with most recent taxonomy. Here, we used phylogenetic analysis and information solely in order to show the diversity of our isolates and to illustrate and support their membership to existing and likely undescribed genera, respectively. For these purposes and for constructing species representing-clades, the sequence of the ITS region usually suffices.
Factors Shaping Mortierellaceae Communities
The main aim of our study was to understand the factors shaping the distribution and diversity of Mortierellaceae and their associations with bacteria in the alpine region. The prevalence and geographical distribution of species belonging to this family suggests that these fungi are playing a key role in the soil microbiota of alpine and subalpine habitats. Our results show that Mortierellaceae species are widely distributed and quite abundant in alpine and subalpine forests. Species distribution across all the sampling sites showed habitat specificity, suggesting that habitat properties determined Mortierellaceae communities. Some of the obtained species were exclusively found in the bare terrain of the glacier forefield sites, others exclusively in P. cembra forests, clearly indicating that these Mortierellaceae species are habitat-specific. For example, M. solitaria (s.l.) and M. globalpina (s.s.) were isolated from bare terrain, whereas Entomortierella parvispora, Linnemannia gamsii, M. gemmifera (s.l.), M. bainieri (s.l.), and M. zonata (s.l.) were isolated from P. cembra forests. In the phylogenetic tree calculated from ITS sequences, these latter three species were located in the Linnemannia branch. They were not included in the study of Vandepol et al. (2020). According to Wagner et al. (2013) their taxonomic placement needs revision (M. gemmifera, M. zonata) and both M. gemmifera and M. zonata were placed into group 7, from which several species have been recombined into Linnemannia by Vandepol et al. (2020). Taking together this information and the close phylogenetic relation between Linnemannia and Entomortierella (Vandepol et al., 2020), our data indicate a preference of P. cembra forests of some members of these genera. Further research on these potential relations is necessary, which are beyond the scope and results of this study.
In addition, altitude impacted on both, the richness and the species composition of Mortierellaceae communities. This is in line with data reported for the soil and rhizosphere microbial diversity of Ranunculus glacialis habitats along a high-altitudinal gradient. Mortierellaceae communities were altitude dependent, changed with altitude, and had the highest abundances in the nival zone (Praeg et al., 2019). However, the factors habitat and altitude also imply drastic changes in environmental factors such as soil pH, soil organic matter (SOM), duration of snow-cover, and C/N, which are characteristic for habitat type and sampling sites (Siles and Margesin, 2016).
In our dataset soil pH showed a pronounced effect on the Mortierellaceae communities in alpine habitats. Certain species of Mortierellaceae colonize acidic soils, whilst other species prefer calcareous soil with neutral pH (Yamanaka, 2003; Ning et al., 2020). The investigated dwarf-willow habitats had a higher pH variation and plant diversity compared to P. cembra forests, thus explaining the higher diversity and variation in Mortierellaceae species composition. Our dataset also suggested that C/N ratio and seasonality influence the distribution of Mortierellaceae species. This further explains the abundance and species richness at the studied sites and habitats (Table 2).
Seasonal dynamics have also been reported to impact on the microbial communities, including those in alpine habitats (Bardgett et al., 2005; Žifčáková et al., 2016; Dresch et al., 2019) but this has never been investigated for distinct fungal lineages, like Mortierellaceae. The presence of snow-cover is an important factor for soil physiological processes and soil organic matter (SOM) build-up. Snow-cover favors undisturbed saprobial activity under constant conditions, with reduced competition or predation. In contrast, rhizosphere processes related to the photosynthetic activity of plants are highly important during the vegetation processes. They are the first step for manifold synergistic interactions in the soil, and they are also known to be crucial for SOM formation in boreal forests (Clemmensen et al., 2013), which is also reflected by the high values of SOM in the P. cembra forests investigated here. It is evident with our dataset that the seasonal changes did influence the distribution of Mortierellaceae species in their given habitats. For instance, the Mortierella s.l. species M. solitaria, M. bainieri, and M. gemmifera, were only isolated from snow-free soil, whereas M. antarctica (s.s.) was isolated only from snow-covered soil. As argued above, these former three species need taxonomic reclassification and might be members of the genus Linnemannia. Their isolation from solely snow-free and snow-covered soil, respectively, might provide another ecological distinction criteria between the genera Mortierella (s.s.) and Linnemannia.
Several functions have been attributed to Mortierellaceae: they are saprobial soil fungi able to grow under nutrient-poor (oligotrophic) conditions (Bergero et al., 1999) and at high elevations (Schmidt et al., 2012), but are also known as plant endophytes with a dual lifestyle (Zhang et al., 2020). The species-specific physiological features of individual Mortierella species are probably contributing to habitat preferences and seasonal dynamics. The patterns by location, habitat and season detected here among Mortierellaceae genera across species based on ITS sequence similarity support their placement based on phylogenetics and phylogenomics (Vandepol et al., 2020) and indicates a potential phylogenetic conservation of some ecological features among species within different genera.
Mortierellaceae Species Are Regularly Associated With Bacteria
Our expectation was that Mortierellaceae species are associated with different taxa of bacteria, and that such associations are widespread in nature. We detected associated bacteria in almost all our frequently isolated Mortierellaceae species. On average, >30% of the isolates within a species (n = 19) were associated to a bacterium, indicating that these associations were facultative. In general, fungi and bacteria co-exist in the environment. Some bacteria can colonize hyphae and move by gliding on the hyphal surface (Simon et al., 2017), or they can invade fungi and dwell inside the fungus (Deveau et al., 2018). Due to this close co-habitation of bacteria and fungi in the soil, the associations between Mortierellaceae and bacteria observed here might seem like artifacts. The possibility of random co-isolations of Mortierellaceae and bacteria during the fungal isolation by direct plating cannot be excluded, but seems unlikely to us for several reasons: (i) Random associations would be taxonomically more diverse and not limited to Proteobacteria as observed here. This is underpinned by the fact that the vast majority of soil bacteria are usually not part of this taxonomic group. (ii) Random associations would not follow environmental trajectories. (iii) Random associations would not show a species-specific pattern (Table 3). Although fungal hyphae are known as bacterial highways (Simon et al., 2017; Jiang et al., 2021), we did not detect any bacterial species known as frequent passengers of Fusarium and Chaetomium (e.g., Variovorax soli, Olivibacter soli, Acinetobacter calcoaceticus, and Stenotrophomonas, Achromobacter and Ochrobactrum) (Simon et al., 2015) in our Mortierellaceae strains, supporting the specificity of fungal–bacterial associations.
We were intrigued by the fact that the proportion of bacteria-associated isolates varied depending on the particular Mortierellaceae species. Some species almost never had associated bacteria, despite a high number of isolates (Table 3). For others, such as M. angusta (s.l.), almost 80% of all isolates were associated with bacteria. In addition, to an apparent preference of the fungal species in terms of bacterial associations, the environmental conditions, including season and habitat, affected Mortierellaceae–bacterial pairs. For most Mortierellaceae species, higher habitat altitude resulted in a higher number of bacterial associations, indicating that associations might provide an advantage under these environmental conditions.
In our dataset, M. alpina and M. globalpina (both s.s.) were never found in association with bacteria. This is in contrast to a study from Kai et al. (2012) reporting an association of M. alpina and endobacteria. In our study, those species were isolated only from Obergurgl (alpine bare terrain in the glacier forefield) and in the case of M. alpina, from Hafelekar (dwarf-willow habitat). It is possible that these species did not form any associations with bacteria due to the environmental factors given in the specific habitat/location. Further studies involving M. alpina and M. globalpina isolates from a range of different locations are necessary to address this question.
From an ecological perspective, the associations between Mortierellaceae species and bacteria and their potential functions are very interesting. It is also interesting to ask for benefits and burdens for both the fungus and the bacterium for establishing an association and situations, in which these might provide a selective advantage, such as the exchange of metabolites in nutrient limited conditions. Until now, research has been focused on specific, endosymbiotic bacterial lineages. For a more mechanistic understanding of these associations, further studies analyzing numerous isolates from different environmental conditions and screening for a broad range of bacteria, including co-cultivation studies are necessary.
Mortierellaceae Is Associated to Different Bacteria
Pseudomonas was the most widespread and most frequent bacterial taxon associated to Mortierellaceae in this study. Moreover, Mortierellaceae species appeared promiscuous in terms of Pseudomonas. However, we observed a phylogenetically conserved clustering of species belonging to Podila and Linnemannia by Pseudomonas OTUs (Supplementary Figure 5). Only M. angusta (s.l.) and E. parvispora, which were the two species with the highest number of exceptions from the rule in our study, did not follow the congruency in Pseudomonas clustering and phylogenetic relationships. Although being a weak indication, we consider a potential co-evolution as an interesting option, which should be mentioned as it might be worth further investigations. The phylogenetically conserved clustering could indicate that these fungal–bacterial associations have provided an evolutionary advantage. Independent of co-evolution, the association to Pseudomonas might be advantageous in alpine areas. Complete genome sequence of P. frederiksbergensis, one of our frequently detected Pseudomonas OTUs (Figure 1), revealed that this bacterium owns the genetic basis for survival at high altitudes (Kumar et al., 2019), and efficient lipid metabolism. Cold adaptation and UV protection are important traits for survival at high altitudes and could possibly be provided by the bacterium in exchange for lipids, which are usually abundantly produced by Mortierellaceae species (Sakuradani et al., 2009). A focused, experimental approach is needed for addressing these topics, which we regard as fundamental for high-altitude ecosystem establishment.
In addition to Pseudomonas, we found Mortierellaceae associated to bacteria: Rahnella, Rouxiella, and Janthinobacterium. While some studies showed a positive influence of Pseudomonas, Rahnella, and Janthinobacterium on plant growth (Kang et al., 2007; Vyas et al., 2010; Chatterjee et al., 2017), such information remain scarce for fungal–bacterial associations. Particularly, Pseudomonas and Rahnella were reported to use fungal hyphae as highways (Palmieri et al., 2020; Steffan et al., 2020), while they were not reported to occur within fungal hyphae. The question of mutualism between fungi, more specifically Mortierella, and epihyphal bacteria has not been fully addressed yet.
For several fungal phyla, including Glomeromycotina (arbuscular mycorrhizal fungi), Mucoromycotina, and Mortierellomycotina, bacterial endohyphal symbionts have been extensively studied so far. For Mortierellomycotina, both Burkholderia- and Mycoplasma-related endohyphal symbionts have been reported (Uehling et al., 2017; Desirò et al., 2018; Takashima et al., 2018). Here, the genera Mycoavidus, Burkholderia, and Paraburkholderia were detected. The epi- or endohyphal location of the bacteria has not been within the aim of this study, which solely focused on distribution patterns. Interestingly, despite the high number of strains included in this study, Mycoplasma-related bacteria were not detected. This seems to be in agreement with Takashima et al. (2018) who also only reported Burkholderia-related bacteria in their study screening >200 Mortierellaceae strains isolated from cold, temperate and subtropical soils. Possibly, geography and environmental conditions are important factors shaping fungus–bacterial associations. Moreover, also isolation conditions might have played a role as both studies used LCA and our study in addition PDA medium for isolation. According to Takashima et al. (2018), however, the percentage of Mortierellaceae harboring Burkholderia-related endobacteria was 22%. Here, the fraction of such associations was 13%. Different habitats offer a possible explanation for the different percentages. The different species compositions among habitats, might be a further explanation, as the overlap of species considered in these studies is small. Here, higher altitude was positively associated with the frequency of associations. Taking together this information and owing to the hypothesis of species-specificity, different Mortierellaceae genera and species might associate with Burkholderia-related bacteria, depending on environmental conditions, which are also likely to play a role in shaping these associations.
Independent comparative phylogenomic analyses of fungal and bacterial genomes are consistent with an ancient origin for L. elongata – Mycoavidus cysteinexigens symbiosis, most likely originating over 350 million years ago and concomitant with the colonization of terrestrial habitats on Earth, and the diversification of plants and fungi (Uehling et al., 2017). If endohyphal associations have an ancient origin, the ability to form associations with bacteria could be a common synapomorphic trait in the Mortierellaceae lineage. We detected at least four bacterial species, which, based on their taxonomic placement, are possibly endohyphal (Ohshima et al., 2016; Uehling et al., 2019; Okrasiñska et al., 2021). Mycoavidus cysteinexigens was the most frequently detected one. This is the first time that M. cysteinexigens associations have been shown for alpine habitats (alpine bare terrain and P. cembra forests). Based on our results, M. cysteinexigens can be associated with a wide range of Mortierellaceae species, such as P. horticola/verticillata, L. gamsii, and L. exigua, all falling into the Linnemannia/Podila lineage (Figure 2). Endobacteria depend on the carbon and nitrogen supply by the Mortierellaceae host (Uehling et al., 2017; Desirò et al., 2018). Harboring endobacteria would come with fitness costs based on metabolomics and proteomics data (Li et al., 2017). As a fair trade, the endosymbiont would provide certain metabolites (e.g., amino acids) and chemical weapons to the host, repaying the fungi for sustaining and transporting them (Lackner et al., 2011; Steffan et al., 2020). However, it is still unclear how recognition and partner acquisition work, and whether or not bacterial endosymbionts can be horizontally transmitted. A possible reason for these endobacteria to associate with Mortierellaceae could be the ability of these fungi to grow under oligotrophic, or cold environmental conditions. Moreover, they are very frequent and are one of the dominant soil fungal groups on a global scale irrespective of the habitats.
In summary, our study provides evidence in favor of a selective facultative association hypothesis, which is established based on the presence of a suitable partner, and environmental conditions. Fungi belonging to the phylum Mortierellomycotina do not only occasionally associate with bacteria, but can also establish a wide range of associations with different bacteria, especially with selected Pseudomonas OTUs.
Conclusion
Mortierellaceae species are diverse in alpine and high-altitude habitats. Their diversity depended on environmental factors, namely habitat and snow-cover, as well as environmental conditions, such as soil pH. We frequently observed associations between Mortierellaceae isolates and gram-negative bacteria belonging to seven different genera. The associations we observed were partially specific for Mortierellaceae species and genera and also driven by environmental factors, including habitat, altitude, and season. Especially the effect of altitude might indicate that these interactions are ecologically meaningful and need to be investigated further.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AT, MP, and UP conceived the study design. AT isolated and characterized all Mortierella strains and was responsible for the wet lab experiments. MP contributed to the data analysis and data interpretation. AT, MP, and UP are responsible for manuscript writing and editing. UP contribute to the funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed by the FWF and by the Land Tirol (project Microbial Interactions in Snow-covered soil MICINSNOW- P31038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Gry Björg Larsen for the C/N analysis and Bettina Schneidhofer for her help with analyzing soil properties.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669784/full#supplementary-material
References
Bardgett, R. D., Bowman, W. D., Kaufmann, R., and Schmidt, S. K. (2005). A temporal approach to link aboveground and belowground ecology. Trends Ecol. Evol. 20, 634–641. doi: 10.1016/j.tree.2005.08.005
Bergero, R., Girlanda, M., Varese, G. C., Intili, D., and Luppi, A. M. (1999). Psychrooligotrophic fungi from Arctic soils of Franz Joseph Land. Polar Biol. 21, 361–368. doi: 10.1007/s003000050374
Bonfante, P., and Desiro, A. (2017). Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. Isme J. 11, 1727–1735. doi: 10.1038/ismej.2017.21
Chatterjee, P., Samaddar, S., Anandham, R., Kang, Y., Kim, K., Selvakumar, G., et al. (2017). Beneficial soil bacterium Pseudomonas frederiksbergensis Os261 augments salt tolerance and promotes red pepper plant growth. Front. Plant Sci. 8:705. doi: 10.3389/fpls.2017.00705
Clemmensen, K. E., Bahr, A., Ovaskainen, O., Dahlberg, A., Ekblad, A., Wallander, H., et al. (2013). Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339, 1615–1618. doi: 10.1126/science.1231923
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9.
Desirò, A., Hao, Z., Liber, J. A., Benucci, G. M. N., Lowry, D., Roberson, R., et al. (2018). Mycoplasma-related endobacteria within Mortierellomycotina fungi: diversity, distribution and functional insights into their lifestyle. ISME J. 12, 1743–1757. doi: 10.1038/s41396-018-0053-9
Deveau, A., Bonito, G., Uehling, J., Paoletti, M., Becker, M., Bindschedler, S., et al. (2018). Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 42, 335–352. doi: 10.1093/femsre/fuy008
Dresch, P., Falbesoner, J., Ennemoser, C., Hittorf, M., Kuhnert, R., and Peintner, U. (2019). Emerging from the ice-fungal communities are diverse and dynamic in earliest soil developmental stages of a receding glacier. Environ. Microbiol. 21, 1864–1880. doi: 10.1111/1462-2920.14598
Hibbett, D., and Glotzer, D. (2011). Where are all the undocumented fungal species? A study of Mortierella demonstrates the need for sequence-based classification. New Phytol. 191, 592–596. doi: 10.1111/j.1469-8137.2011.03819.x
Jiang, F., Zhang, L., Zhou, J., George, T. S., and Feng, G. (2021). Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytol. 230, 304–315. doi: 10.1111/nph.17081
Johnson, J. M., Ludwig, A., Furch, A. C. U., Mithöfer, A., Scholz, S., Reichelt, M., et al. (2019). The beneficial root-colonizing fungus mortierella hyalina promotes the aerial growth of arabidopsis and activates calcium-dependent responses that restrict alternaria brassicae-induced disease development in roots. Mol. Plant Microbe Interact. 32, 351–363. doi: 10.1094/MPMI-05-18-0115-R
Kai, K., Furuyabu, K., Tani, A., and Hayashi, H. (2012). Production of the quorum-sensing molecules N- Acylhomoserine lactones by endobacteria associated with Mortierella alpina A-178. ChemBioChem 13, 1776–1784. doi: 10.1002/cbic.201200263
Kang, S. H., Cho, H. S., Cheong, H., Ryu, C. M., Kim, J. F., and Park, S. H. (2007). Two bacterial entophytes eliciting both plant growth promotion and plant defence on pepper (Capsicum annuum L.). J. Microbiol. Biotechnol. 17, 96–103.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kuhnert, R., Oberkofler, I., and Peintner, U. (2012). Fungal growth and biomass development is boosted by plants in snow-covered soil. Microb. Ecol. 64, 79–90. doi: 10.1007/s00248-011-0001-y
Kumar, R., Acharya, V., Mukhia, S., Singh, D., and Kumar, S. (2019). Complete genome sequence of Pseudomonas frederiksbergensis ERDD5:01 revealed genetic bases for survivability at high altitude ecosystem and bioprospection potential. Genomics 111, 492–499. doi: 10.1016/j.ygeno.2018.03.008
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwaśna, H. (2001). Fungi in the rhizosphere of common oak and its stumps and their possible effect on infection by Armillaria. Appl. Soil Ecol. 17, 215–227. doi: 10.1016/S0929-1393(01)00137-8
Lackner, G., Moebius, N., Partida-Martinez, L. P., Boland, S., and Hertweck, C. (2011). Evolution of an endofungal Lifestyle: deductions from the Burkholderia rhizoxinica Genome. BMC Genomics 12:210. doi: 10.1186/1471-2164-12-210
Li, Z., Yao, Q., Dearth, S. P., Entler, M. R., Castro Gonzalez, H. F., Uehling, J. K., et al. (2017). Integrated proteomics and metabolomics suggest symbiotic metabolism and multimodal regulation in a fungal-endobacterial system. Environ. Microbiol. 19, 1041–1053. doi: 10.1111/1462-2920.13605
Linnemann, G. (1941). in Pflanzenforschung: Die Mucorineen-Gattung Mortierella COEMANS. Heft 23, ed. R. Kolkwitz (Jena: Gustav Fischer), doi: 10.1002/ardp.19422800509
Ning, Q., Chen, L., Jia, Z., Zhang, C., Ma, D., Li, F., et al. (2020). Multiple long-term observations reveal a strategy for soil pH-dependent fertilization and fungal communities in support of agricultural production. Agric. Ecosyst. Environ. 293:106837. doi: 10.1016/j.agee.2020.106837
Ohara, H. (1967). Disappearance of bacteria from the zone of active mycorrhizas in Tricholoma matsutake (s. Ito et Imai) Singer. Nature 213, 528–529. doi: 10.1038/213528a0
Ohshima, S., Sato, Y., Fujimura, R., Takashima, Y., Hamada, M., Nishizawa, T., et al. (2016). Mycoavidus cysteinexigens gen. Nov., sp. Nov., an endohyphal bacterium isolated from a soil isolate of the fungus Mortierella elongata. Int. J. Syst. Evol. Microbiol. 66, 2052–2057. doi: 10.1099/ijsem.0.000990
Okrasiñska, A., Bokus, A., Duk, K., Gęsiorska, A., Sokołowska, B., Miłobędzka, A., et al. (2021). New endosymbiotic relationships between Mucoromycota and Burkholderiaceae representatives. Appl. Environ. Microbiol. 87:e02707-20. doi: 10.1128/aem.02707-20
Oksanen, J., Guillaume Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R Package Version 2.5-6. Avaliable at: https://CRAN.R-project.org/package=vegan
Palmieri, D., Vitale, S., Lima, G., Di Pietro, A., and Turrà, D. (2020). A bacterial endophyte exploits chemotropism of a fungal pathogen for plant colonization. Nat. Commun. 11, 1–11. doi: 10.1038/s41467-020-18994-5
Praeg, N., Pauli, H., and Illmer, P. (2019). Microbial diversity in bulk and rhizosphere soil of Ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 10:1429. doi: 10.3389/fmicb.2019.01429
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sakuradani, E., Ando, A., Ogawa, J., and Shimizu, S. (2009). Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl. Microbiol. Biotechnol. 84, 1–10. doi: 10.1007/s00253-009-2076-7
Schmidt, S. K., Naff, C. S., and Lynch, R. C. (2012). Fungal communities at the edge: ecological lessons from high alpine fungi. Fungal Ecol. 5, 443–452. doi: 10.1016/j.funeco.2011.10.005
Schmidt, S. K., Wilson, K. L., Meyer, A. F., Gebauer, M. M., and King, A. J. (2008). Phylogeny and ecophysiology of opportunistic “snow molds” from a subalpine forest ecosystem. Microb. Ecol. 56, 681–687. doi: 10.1007/s00248-008-9387-6
Siles, J. A., and Margesin, R. (2016). Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: what are the driving factors? Microb. Ecol. 72, 207–220. doi: 10.1007/s00248-016-0748-2
Simon, A., Bindschedler, S., Job, D., Wick, L. Y., Filippidou, S., Kooli, W. M., et al. (2015). Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol. Ecol. 91, 1–13. doi: 10.1093/femsec/fiv116
Simon, A., Hervé, V., Al-Dourobi, A., Verrecchia, E., and Junier, P. (2017). An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol. 93, 1–9. doi: 10.1093/femsec/fiw217
Steffan, B. N., Venkatesh, N., and Keller, N. P. (2020). Let’s get physical: bacterial-fungal interactions and their consequences in agriculture and health. J. Fungi 6, 1–18. doi: 10.3390/jof6040243
Summerbell, R. C. (2005a). From Lamarckian fertilizers to fungal castles: recapturing the pre-1985 literature on endophytic and saprotrophic fungi associated with ectomycorrhizal root systems. Stud. Mycol. 53, 191–256. doi: 10.3114/sim.53.1.191
Summerbell, R. C. (2005b). Root endophyte and mycorrhizosphere fungi of black spruce, Picea mariana, in a boreal forest habitat: influence of site factors on fungal distributions. Stud. Mycol. 53, 121–145. doi: 10.3114/sim.53.1.121
Takashima, Y., Degawa, Y., and Narisawa, K. (2019). Mortierella oedorhiza, a new species forming a dichotomously branched rhizoid at the sporangiophore base. Mycoscience 60, 361–365. doi: 10.1016/j.myc.2019.06.007
Takashima, Y., Seto, K., Degawa, Y., Guo, Y., Nishizawa, T., Ohta, H., et al. (2018). Prevalence and intra-family phylogenetic divergence of Burkholderiaceae-related endobacteria associated with species of Mortierella. Microbes Environ. 33, 417–427. doi: 10.1264/jsme2.ME18081
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688
Torres-Cruz, T. J., Tobias, T. L. B., Almatruk, M., Hesse, C. N., Kuske, C. R., Desirò, A., et al. (2017). Bifiguratus adelaidae, gen. et sp. nov., a new member of Mucoromycotina in endophytic and soil-dwelling habitats. Mycologia 109, 363–378. doi: 10.1080/00275514.2017.1364958
Uehling, J., Gryganskyi, A., Hameed, K., Tschaplinski, T., Misztal, P. K., Wu, S., et al. (2017). Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ. Microbiol. 19, 2964–2983. doi: 10.1111/1462-2920.13669
Uehling, J. K., Entler, M. R., Meredith, H. R., Millet, L. J., Timm, C. M., Aufrecht, J. A., et al. (2019). Microfluidics and metabolomics reveal symbiotic bacterial–fungal interactions between Mortierella elongata and Burkholderia include metabolite exchange. Front. Microbiol. 10:2163. doi: 10.3389/fmicb.2019.02163
Vandepol, N., Liber, J., Desirò, A., Na, H., Kennedy, M., Barry, K., et al. (2020). Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 104, 267–289. doi: 10.1007/s13225-020-00455-5
Vyas, P., Joshi, R., Sharma, K. C., Rahi, P., Gulati, A., and Gulati, A. (2010). Cold-adapted and rhizosphere-competent strain of Rahnella sp. with broad-spectrum plant growth-promotion potential. J. Microbiol. Biotechnol. 20, 1724–1734.
Wagner, L., Stielow, B., Hoffmann, K., Petkovits, T., Papp, T., Vágvölgyi, C., et al. (2013). A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia Mol. Phylogeny Evol. Fungi 30, 77–93. doi: 10.3767/003158513X666268
Walch, G., Knapp, M., Rainer, G., and Peintner, U. (2016). Colony-PCR is a rapid method for DNA amplification of hyphomycetes. J. Fungi 2:12. doi: 10.3390/jof2020012
White, T. J., Bruns, T., Lee, S., Taylor, J., Innis, M. A., Gelfand, D. H., et al. (1990). PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press.
Yamanaka, T. (2003). The effect of pH on the growth of saprotrophic and ectomycorrhizal ammonia fungi in vitro. Mycologia 95, 584–589. doi: 10.1080/15572536.2004.11833062
Zhang, G.-Y., Zhang, L.-P., Wei, M.-F., Liu, Z., Fan, Q.-L., Shen, Q.-R., et al. (2011). Effect of arbuscular mycorrhizal fungi, organic fertilizer and soil sterilization on maize growth. Acta Ecol. Sin. 31, 192–196. doi: 10.1016/j.chnaes.2011.04.005
Zhang, K., Bonito, G., Hsu, C. M., Hameed, K., Vilgalys, R., and Liao, H. L. (2020). Mortierella elongata increases plant biomass among non-leguminous crop species. Agronomy 10, 1–8. doi: 10.3390/agronomy10050754
Keywords: Mortierellomycotina, bacterial interactions, winter, pH, altitude, alpine, sub-alpine, psychrotolerant
Citation: Telagathoti A, Probst M and Peintner U (2021) Habitat, Snow-Cover and Soil pH, Affect the Distribution and Diversity of Mortierellaceae Species and Their Associations to Bacteria. Front. Microbiol. 12:669784. doi: 10.3389/fmicb.2021.669784
Received: 26 February 2021; Accepted: 10 June 2021;
Published: 01 July 2021.
Edited by:
Saskia Bindschedler, Université de Neuchâtel, SwitzerlandReviewed by:
Gregory M. Bonito, Michigan State University, United StatesJessie Uehling, University of California, Berkeley, United States
Copyright © 2021 Telagathoti, Probst and Peintner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ursula Peintner, VXJzdWxhLlBlaW50bmVyQHVpYmsuYWMuYXQ=
 Anusha Telagathoti
Anusha Telagathoti Maraike Probst
Maraike Probst Ursula Peintner
Ursula Peintner