- Department of Biological Sciences, University of Delaware, Newark, DE, United States
Factor for inversion stimulation (Fis) is a global regulator that is highly expressed during exponential phase growth and undetectable in stationary phase growth. Quorum sensing (QS) is a global regulatory mechanism that controls gene expression in response to changes in cell density and growth phase. In Vibrio parahaemolyticus, a marine species and a significant human pathogen, the QS regulatory sRNAs, Qrr1 to Qrr5, are expressed during exponential growth and negatively regulate the high cell density QS master regulator OpaR. OpaR is a positive regulator of capsule polysaccharide (CPS) formation, which is required for biofilm formation, and is a repressor of lateral flagella required for swarming motility. In V. parahaemolyticus, we show that Fis is a positive regulator of the qrr sRNAs expression. In an in-frame fis deletion mutant, qrr expression was repressed and opaR expression was induced. The Δfis mutant produced CPS and biofilm, but swarming motility was abolished. Also, the fis deletion mutant was more sensitive to polymyxin B. Swarming motility requires expression of both the surface sensing scrABC operon and lateral flagella laf operon. Our data showed that in the Δfis mutant both laf and scrABC genes were repressed. Fis controlled swarming motility indirectly through the QS pathway and directly through the surface sensing pathway. To determine the effects of Fis on cellular metabolism, we performed in vitro growth competition assays, and found that Δfis was outcompeted by wild type in minimal media supplemented with intestinal mucus as a sole nutrient source. The data showed that Fis positively modulated mucus components L-arabinose, D-gluconate and N-acetyl-D-glucosamine catabolism gene expression. In an in vivo colonization competition assay, Δfis was outcompeted by wild type, indicating Fis is required for fitness. Overall, these data demonstrate a global regulatory role for Fis in V. parahaemolyticus that includes QS, motility, and metabolism.
Introduction
The factor for inversion stimulation (Fis) is a nucleoid associated protein (NAP) that has two major functions in bacteria, chromosome organization and gene regulation (Azam et al., 1999; Ishihama, 2010). Fis, along with other NAPs, is an important regulator of ribosome, tRNA and rRNA expression (Bokal et al., 1997; Auner et al., 2003; Schneider et al., 2003; Dennis et al., 2004). As a transcriptional regulator, it can act as both an activator and repressor of a large number of genes (Keane and Dorman, 2003; Kelly et al., 2004; Browning et al., 2005; Grainger et al., 2006). As an activator, Fis can directly bind to RNA polymerase to affect transcription, or indirectly control transcription via DNA supercoiling at promoters (Bokal et al., 1997; McLeod et al., 2002; Auner et al., 2003; Cróinín et al., 2006). Fis controls DNA topology by regulating DNA gyrases (gyrA and gyrB) and DNA topoisomerase I (topA), required for DNA negative supercoiling in Escherichia coli and Salmonella enterica (Schneider et al., 1999; Keane and Dorman, 2003; Weinstein-Fischer and Altuvia, 2007). In enteric species, Fis was shown to be a global regulator that responded to growth phases and abiotic stresses (González-Gil et al., 1996; Goldberg et al., 2001; Kelly et al., 2004; Browning et al., 2005; Bradley et al., 2007; Lautier and Nasser, 2007; Steen et al., 2010; Zhang et al., 2012; Wang et al., 2013; Lv et al., 2018). In E. coli, Fis is one of the most abundant proteins, highly expressed in early exponential phase cells and absent in stationary phase cells, under aerobic growth conditions (Osuna et al., 1995; Mallik et al., 2004). Fis binds to specific sites, however with limited sequence conservation (Finkel and Johnson, 1992). Fis is known as a nucleoid structuring protein that causes DNA bending, allowing for changes in gene expression (Finkel and Johnson, 1992; Bétermier et al., 1994; Skoko et al., 2006). Genome wide studies have shown over a 1,000 binding peaks for Fis in E. coli controlling a fifth of chromosomal genes (Grainger et al., 2006; Cho et al., 2008; Kahramanoglou et al., 2011). It has also been shown that many regulatory regions contain multiple and sometimes overlapping Fis binding sites (BS; Hengen et al., 2003; Shao et al., 2008). Genetic, biochemical and structural analysis in E. coli of Fis high affinity binding sites has demonstrated the presence of a 15-bp core sequence flanked by G/C (position −7) and C/G (position +7) base-pairs with a central A/T rich region (position 0) (Cho et al., 2008; Shao et al., 2008; Kahramanoglou et al., 2011; Hancock et al., 2016).
In E. coli and S. enterica, studies have shown that Fis can control virulence, motility, and metabolism (González-Gil et al., 1996; Kelly et al., 2004; Grainger et al., 2006; Bradley et al., 2007). These studies identified 100s of genes whose expression in vivo is either enhanced or repressed by Fis. In S. enterica, the polar flagellum genes, and genes within several pathogenicity islands, were differentially expressed between a Δfis mutant and wild type (Osuna et al., 1995; Cróinín et al., 2006; Wang et al., 2013). In the plant pathogen, Dickeya zeae, a Δfis mutant strain showed a total of 490 genes significantly regulated by Fis (Ouafa et al., 2012; Prigent-Combaret et al., 2012; Lv et al., 2018). In Vibrio cholerae, it has been shown that Fis modulates expression of the quorum sensing non-coding regulatory sRNAs (Qrr), qrr1 to qrr4 (Lenz and Bassler, 2007). Quorum sensing (QS) is a term used to describe bacterial communication mediate by chemical signals that allows bacteria to control global gene expression in response to cell density changes (Nealson et al., 1970; Fuqua et al., 1994; Gray et al., 1994; Bassler et al., 1997; Bassler, 1999; Miller and Bassler, 2001; Swift et al., 2001; Miller et al., 2002). In V. cholerae, it was proposed that Fis acts along with the QS response regulator LuxO, a sigma factor-54 activator, to transcribe the four qrr1 to qrr4 sRNAs. The Qrr sRNAs repressed the master QS high cell density (HCD) regulator HapR and activated the QS low cell density (LCD) regulator AphA. In a Δfis mutant in this species, the qrr sRNAs were repressed and hapR was expressed at wild type levels (Lenz and Bassler, 2007). This is the only other study to examine the role of Fis among Vibrio species.
Vibrio parahaemolyticus is a marine halophile and the leading cause of bacterial seafood-borne gastroenteritis worldwide with increased incidences of infection due to climate change (Nair et al., 2007; Su and Liu, 2007; Froelich and Noble, 2016; Froelich and Daines, 2020). A V. parahaemolyticus infection causes inflammatory diarrhea and its main virulence factors are two type 3 secretions systems and their effector proteins (Makino et al., 2003; O’Boyle and Boyd, 2014; Kodama et al., 2015; De Souza Santos and Orth, 2019; Miller et al., 2019). Unlike V. cholerae that only produces a single polar flagellum, V. parahaemolyticus produces both a polar flagellum and lateral flagella expressed from the Flh (Fli) and Laf loci, respectively (Belas et al., 1986; McCarter et al., 1988; Stewart and McCarter, 2003). The polar flagellum, required for swimming motility, is produced in cells grown in liquid media and is under the control of sigma factor RpoN (σ54) and its activator FlaK, and sigma factor FliAP (σ28). The lateral flagella, required for swarming motility on solid surfaces, are under the control of RpoN and its activator LafK, and a second σ28 factor FliAL (McCarter and Wright, 1993; Stewart and McCarter, 2003; Jaques and McCarter, 2006; Gode-Potratz et al., 2011; Kernell Burke et al., 2015). Disruption of rpoN abolishes all motility, whereas deletion of either of the two σ28 sigma factors, FliAP (FliA) or FliAL, abolishes swimming and swarming, respectively (Stewart and McCarter, 2003; Whitaker et al., 2014). Overall, control of motility in the dual flagellar system of V. parahaemolyticus differs significantly from monoflagellar systems of enteric species (Klose and Mekalanos, 1998; McCarter, 1999; Prouty et al., 2001). Previously, it was demonstrated that bacterial motility and metabolism require a functional QS pathway in V. parahaemolyticus (Kalburge et al., 2017). In this species, it was shown that deletion of the QS response regulator luxO resulted in repression of the five qrr genes and constitutive expression of opaR (the hapR homolog). The ΔluxO mutant had reduced swimming motility, but swarming motility was abolished, while a ΔopaR mutant was hyper-motile and swarming proficient (Kalburge et al., 2017). The QS regulator OpaR is a direct repressor of the laf operon, required for lateral flagellum synthesis and swarming motility (Güvener and McCarter, 2003; Jaques and McCarter, 2006; Gode-Potratz and McCarter, 2011; Kernell Burke et al., 2015). OpaR is an activator of the cps operon, required for capsular polysaccharide (CPS) formation, an important component of biofilm (McCarter, 1998; Boles and McCarter, 2002; Kim and McCarter, 2007; Kalburge et al., 2017). Additionally, studies have shown that the V. parahaemolyticus surface sensing operon scrABC activates swarming motility and represses CPS formation by reducing the intracellular levels of c-di-GMP (Boles and McCarter, 2002; Ferreira et al., 2008; Trimble and McCarter, 2011). A deletion of the scrABC operon induces high c-di-GMP levels that repress the laf operon and induce cps gene expression (Boles and McCarter, 2002; Ferreira et al., 2008; Trimble and McCarter, 2011). The surface colonization regulatory (Scr) program in V. parahaemolyticus contains a 100 genes, 70 genes of which are involved in swarming motility and 30 genes involved in biofilm formation that are controlled by intracellular c-di-GMP levels (Boles and McCarter, 2002; Kim and McCarter, 2007; Gode-Potratz and McCarter, 2011; Ferreira et al., 2012). In addition, QS can also modulate c-di-GMP levels to control swarming behavior (Gode-Potratz et al., 2011).
Here, we characterized the role of Fis in V. parahaemolyticus, a marine halophile and gastrointestinal pathogen. This work shows that Fis connects the QS and surface sensing signaling pathways in this species to control swarming motility. We determined the expression pattern of fis across the growth curve and constructed an in-frame Δfis deletion mutant to examine its role in V. parahaemolyticus physiology. For example, the fis mutant produced more capsule and was more sensitive to the antimicrobial peptide polymyxin B. We demonstrated a role of Fis in the QS pathway, specifically its modulation of the five non-coding regulatory sRNAs, qrr1 to qrr5, as well as the QS HCD master regulator, OpaR. The effects of a fis deletion on swimming and swarming motility were determined and an essential role for Fis in swarming motility was uncovered. DNA binding assays and green fluorescent protein (GFP) reporter assays demonstrated Fis regulation of the qrr sRNA genes, the laf lateral flagellum operon and the surface sensing scrABC operon. We performed in vitro growth competition assays and an in vivo colonization assay between the Δfis mutant and a lacZ knock-in WT strain, WBWlacZ, to demonstrate a fitness effect when fis is deleted. Further, our data show that Fis positively modulates the expression of L-arabinose, D-gluconate and N-acetyl-D-glucosamine (NAG) metabolism genes. This study demonstrates that Fis integrates the QS and surface sensing pathways to control swarming motility and is important for control of metabolism and in vivo fitness.
Materials and Methods
Bacterial Strains, Media, and Culture Conditions
All strains and plasmids used in this study are listed in Supplementary Table S1. A streptomycin-resistant clinical isolate V. parahaemolyticus RIMD2210633 was used in this study. Unless stated otherwise, all V. parahaemolyticus strains were grown in lysogeny broth (LB) medium (Fischer Scientific, Pittsburgh, PA) containing 3% NaCl (LBS) at 37°C with aeration or M9 minimal media (Sigma Aldrich, St. Louis, MO) supplemented with 3% NaCl (M9S). Antibiotics were added to growth media at the following concentrations: ampicillin (Amp), 100μg/ml, streptomycin (Sm), 200μg/ml, tetracycline (Tet), 1μg/ml, and chloramphenicol (Cm), 12.5μg/ml when required.
Construction of Δfis Mutant in V. parahaemolyticus RIMD2210633
Splicing by overlap extension (SOE) PCR and an allelic exchange method (Ho et al., 1989) were used to construct an in-frame, non-polar deletion mutant of fis (VP2885) in V. parahaemolyticus RIMD2210633. Briefly, primers were designed using V. parahaemolyticus RIMD2210633 genomic DNA as a template. All primers used in this study are listed in Supplementary Table S2. SOE PCR was conducted to obtain an 18bp-truncated version of VP2885 (297-bp). The Δfis PCR fragments were cloned into the suicide vector pDS132 (Philippe et al., 2004) and named pDSΔfis. pDSΔfis was then transformed into E. coli strain β2155 λpir (Dehio and Meyer, 1997), and conjugated into V. parahaemolyticus RIMD2210633. Conjugation was conducted by cross streaking both strains onto LB plates containing 0.3mm diaminopimelic acid. The colonies were verified for single crossover via PCR. The colonies that had undergone a single crossover were grown overnight in LBS with no antibiotic added and plated onto LBS containing 10% sucrose to select for double crossover deletion mutants. The gene deletion was confirmed by PCR and sequencing.
Phenotype Assays
To observe CPS, heart Infusion media containing 1.5% agar, 2.5mm CaCl2, and 0.25% Congo red dye was used and plates were incubated at 30°C, as previously described (Güvener and McCarter, 2003). Biofilm assays were conducted using crystal violet staining. Cultures were grown overnight in LBS and then used to inoculate (1:40 dilution) a 96 well plate, grown static at 37°C. After 24h, the wells are washed with PBS, stained with crystal violet for 30min and then accessed for biofilm formation. The biofilms are then dissolved in DMSO and OD595 was measured. Swimming assays were conducted in LB 2% NaCl with 0.6% agar and swarming assays were conducted in heart infusion (HI) media with 2% NaCl and 1.5% agar (Whitaker et al., 2014). To study swimming behavior, a single colony of the bacterium was stabbed into the center of the plate, and plates were incubated at 37°C for 24h. For the swarming assay, plates were spot inoculated on the surface of the media and grown at 30°C for 48h (Whitaker et al., 2014). For polymyxin B sensitivity assays, overnight bacterial cultures were diluted (1:50) into LBS and grown for 2h and then spun down and suspended in 5ml LBS. Polymyxin B sulphate (Sigma-Aldrich) was added to the cultures (final concentration of 40μg/ml) and incubated at 37°C for 1h. For the zero minute time point, an aliquot was obtained before adding the polymyxin B, and then aliquots were taken at 30min and 60min time points. The aliquots were serially diluted and plated to determine the colony forming units (CFUs) at each specific time point. Percent survival was calculated by dividing the CFUs at 30min and 60min with that of 0min and multiplied by 100. For disk assay, similar growth condition as above were used, with 2h growth cultures spread plated onto LBS plates. Polymyxin B disks (100μg of polymyxin in each disk) were placed on each plate and incubated for 24h at 37°C before the zone of inhibitions were measured. Three technical replicates and two biological replicates were performed for each strain.
Bioinformatics Analysis to Identify Putative Fis Binding Sites
The regulatory region of each gene cluster of interest from V. parahaemolyticus RIMD210633 was obtained using NCBI nucleotide database. Virtual footprint was used to identify putative Fis binding sites using the E. coli Fis consensus binding sequence (Münch et al., 2003). The 229-bp, 416-bp, 371-bp, 385-bp, 153-bp, 385-bp, and 545bp DNA regions upstream of flhA (VP2235-VP2231), lafB (VPA1550-VPA1557), araB (VPA1674), nagB (VPA0038), gntK (VP0063), and scrABC (VPA1513) respectively, were used as inputs for Fis binding. The regulatory regions of qrr1 (193-bp), qrr2 (338-bp), qrr3 (162-bp), qrr4 (287-bp), and qrr5 (177-bp) were also used as inputs. Default settings were used to obtain putative Fis binding sites. A 130-bp sequence of VPA1424 regulatory region was used as a negative control that contains no Fis binding sites. A 229-bp sequence of gyrA regulatory region was used as a positive control for Fis binding, which was previously shown to contain Fis binding sites and to be directly regulated by Fis (Schneider et al., 1999; Keane and Dorman, 2003).
Fis Protein Purification
Fis was purified using a method previously described with modifications as necessary (Carpenter et al., 2015; Kalburge et al., 2017). Briefly, Fis was cloned into the pMAL-c5x expression vector in which a 6X His-tag maltose binding protein (MBP) was fused to fis separated by a tobacco etch virus (TEV) protease cleavage site (Liu et al., 2013). Primer pair FisFWDpMAL and FisREVpMAL (Supplementary Table S2) and V. parahaemolyticus RIMD2210633 genomic DNA were used to amplify fis (VP2885). The fis PCR product along with purified pMAL-c5x, were digested with NcoI and BamHI, ligated with T4 ligase, and transformed into DH5α. The vector pMAL-c5xfis was purified, sequenced, and then transformed into E. coli BL21 (DE3). A 10ml portion of E. coli BL21 pMAL-c5xfis overnight cultures were used to inoculate 1L of fresh LB supplemented with 100μg/ml ampicillin and 0.2% glucose and grown at 37°C until the OD reached 0.4, at which point, the culture was induced by adding 0.5mm IPTG. The cells were grown overnight at 18°C. Cells were pelleted at 2,800×g and resuspended in 15ml of column buffer (50mm sodium phosphate, 200mm NaCl, pH 7.5) supplemented with 0.5mm benzamidine, and 1mm phenylmethylsulphonyl fluoride. Bacterial cells were lysed using a microfluidizer, spun down at 25,000×g for 60min, and the supernatant was collected. The supernatant was passed through a 20ml amylose resin (New England BioLabs) and washed with 10 column volumes (CVs) of column buffer. Fis fused with 6X His-MBP was then eluted with three CVs of column buffer supplemented with 20mm maltose. Using 6X His-TEV protease (1:10, TEV:protein in 50mm sodium phosphate, 200mm NaCl, 10mm imidazole, 5mm BME, pH 7.5) the fused protein was cleaved at the TEV cleavage site. The cleaved protein was adjusted to 20mm imidazole and run through an immobilized metal affinity chromatography column using HisPur Ni-NTA resin to remove the cleaved 6X His-MBP and the 6X His-TEV protease. Mass spectrometry was performed to confirm Fis protein molecular weight and SDS-PAGE was conducted to determine its purity.
Electrophoretic Mobility Shift Assays
The regulatory regions of genes of interest were used as probes in electrophoretic mobility shift assays (EMSA). The regulatory regions of all five qrr sRNAs were analyzed for binding of Fis. A 193-bp fragment of Pqrr1, a 338-bp fragment of Pqrr2, a 162-bp fragment of Pqrr3, a 287-bp fragment of Pqrr4, and a 177-bp fragment of Pqrr5 regulatory regions were used as probes. A 130-bp probe of VPA1424 regulatory region was used as a negative control and a 229-bp probe of PgyrA regulatory region was used as a positive control. Analysis of Fis binding to flagellum gene clusters included a 161-bp probe of PflhA (VP2235-VP2231) and a 244-bp probe of PlafB (VPA1550-VPA1557). The 545-bp regulatory region of scrABC (VPA1513-VPA1515) operon was divided into three probes, probe 1147-bp, probe 2136-bp, and probe 3139-bp. For analysis of metabolism, 138-bp and 152-bp probes of ParaB (VPA1674, L-ribulokinase, L-arabinose catabolism), 154-bp and 120-bp probes of PnagB (VPA0038, glucosamine-6-phosphate isomerase, D-glucosamine catabolism), and a138-bp probe of PgntK (VP0063, gluconokinase, D-gluconate catabolism) were used in EMSAs. The EMSA probes were PCR amplified using Phusion Hifidelity Polymerase in 50μl reaction mixture using respective primers sets listed in Supplementary Table S2 and V. parahaemolyticus RIMD2210633 genomic DNA as template. Various molar ratios of purified Fis were incubated with 30ng of target DNA in binding buffer (10mm Tris, 150mm KCL, 0.1mm dithiothreitol, 0.1mm EDTA, 5% PEG, pH 7.4) for 20min at room temperature. A native acrylamide 6% gel was prepared and pre-run for 2h (200V at 4°C) with 1x Tris-acetate-EDTA (TAE) buffer, and then 10μl of the target DNA-protein mixture was loaded into consecutive lanes. The gel was run at 200V for 2h in 1X TAE buffer at 4°C, which was then stained in an ethidium bromide bath (0.5μg/ml) for 20min and imaged. To examine the specificity of Fis binding sites, first, we used an approach using SOE PCR to create mutations in a Fis BS within the PnagB probe 2. Using this approach, we mutated 5 -sites which resulted in less Fis binding, but did not abolish binding since, in this region, there were multiple potential Fis BS. Therefore, for the other regions of interest we used a different approach, designing shorter DNA probes for each regulatory region that contained a single Fis BS. A second mutated probe was also designed to contain mutations at key nucleotide positions (Supplementary Table S3). EMSA were performed on wild type and mutated probes using the same ratios and run on the same gel.
Transcription Reporter Assays
Green fluorescent protein reporter assays were conducted in V. parahaemolyticus RIMD2210633 and Δfis strains. Reporter plasmids were constructed with the regulatory regions of motility genes, flhA and lafB, and metabolic genes, araB, nagB and gntK, upstream of a promoterless gfp gene, as previously described (Gregory et al., 2019). Briefly, primers were designed to amplify the regulatory region upstream of each gene or gene cluster with primer pairs listed in Supplementary Table S2. Each amplified regulatory region was then ligated with the promoterless parent vector pRU1064 (Karunakaran et al., 2005), which had been linearized prior with SpeI, using NEBuilder High Fidelity (HiFi) DNA Assembly Master Mix (New England Biolabs, Ipswich, MA) via Gibson Assembly Protocol (Gibson, 2011). Overlapping regions for Gibson Assembly are indicated in lower case letters in the primer sequence in Supplementary Table S2. Reporter plasmid PflhA-gfp encompasses 269-bp of the regulatory region upstream of flhA. Reporter plasmid PlafB-gfp encompasses 456-bp of the regulatory region upstream of lafB. Reporter plasmid ParaB-gfp encompasses 411-bp of the regulatory region upstream of araB. Reporter plasmid PnagB-gfp encompasses 434-bp of the regulatory region upstream of nagB. Reporter plasmid PgntK-gfp encompasses 193-bp of the regulatory region upstream of gntK. Reporter plasmid PscrABC-gfp encompasses 545-bp of the regulatory region upstream of the scrABC operon. Additionally, the regulatory region of qrr1 to qrr5 were amplified and cloned into the pRU1064 reporter plasmid. The plasmids were transformed into E. coli Dh5α, purified and sequenced. Plasmids were then conjugated into wild type and the Δfis mutant for further analysis.
Strains were grown overnight with aeration at 37°C in LBS with Tet (1μg/ml). Cells were then pelleted, washed two times with 1X PBS, and diluted 1:100 in LBS. Strains containing the Pqrr and PopaR reporters were grown to LCD (0.4–0.45 OD), washed two times with 1X PBS, and resuspended to a final OD of 1.0 before measuring relative fluorescence. The metabolism gene reporters were grown for 20h with antibiotic selection. ParaB-gfp was grown in LBS supplemented with 10mm D-arabinose, PnagB-gfp was grown in LBS supplemented with 10mm D-glucosamine, PgntK-gfp was grown in LBS supplemented with 10mm D-gluconate. Cells were pelleted and resuspended in 1X PBS. The pRUPlafB reporter assay was performed using cells grown on heart infusion (HI) plates for 16h. Colonies were scraped from the plate and resuspended in 1xPBS to a final OD595 0.5. GFP fluorescence was measured with excitation at 385 and emission at 509nm in black, clear-bottom 96-well plates on a Spark microplate reader with Magellan software (Tecan Systems Inc.). Specific fluorescence was calculated for each sample by normalizing relative fluorescence to OD595. At least two biological replicates, in triplicate, were performed for each assay. Statistics were calculated using an unpaired Student’s t-test.
In vitro Growth Competition Assays
In vitro growth competition assays were performed by diluting an inoculum 1:50 into LBS broth, and separately in M9S supplemented with mouse intestinal mucus or 10mm of individual carbon sources, D-glucose, L-arabinose, L-ribose, D-gluconate, D-glucosamine, and NAG. A β-galactosidase knock in V. parahaemolyticus RIMD2210633 strain, WBWlacZ, which was previously shown to grow similarly to wild type in vitro and in vivo, was used for all the competition assays (Whitaker et al., 2012, 2014). The culture was incubated at 37°C for 24h, serially diluted and plated on LBS plus streptomycin and 5-bromo-4-chloro-3-indolyl-B-D-galactoside (X-gal). The competitive index (CI) was determined using the following equation: CI=ratio out (Δfis/WBWlacZ)/ratio in (Δfis/WBWlacZ). A CI of <1 indicates WBWlacZ outcompetes the Δfis mutant, a CI of >1 indicates that the Δfis mutant outcompetes WBWlacZ. The ratio of Δfis to WBWlacZ in the inoculum mixture is termed as “Ratio in” and the ratio of Δfis to WBWlacZ colonies recovered from the mouse intestine is referred as “Ratio out.”
In vivo Colonization Competition Assays
All mice experiments were approved by the University of Delaware Institutional Animal Care and Use Committee (Whitaker et al., 2012, 2014). Inoculum for competition assays was prepared using overnight cultures of WBWlacZ and Δfis diluted into fresh LBS media and grown for 4h. Exponential phase cultures were then pelleted by centrifugation at 4,000×g, washed and resuspended in PBS. One ml of WBWlacZ and one ml of Δfis were prepared, corresponding to 1×1010CFU of each strain, based on the previously determined OD and CFU ratio. A 500μl aliquot of Δfis was combined with 500μl of the WBWlacZ, yielding a total bacterial concentration of 1×1010CFU/ml. The inoculum was serially diluted and plated on LBS agar plate supplemented with 200μg/ml streptomycin and 8μg/ml of X-gal to determine the exact ratio of the inoculum. Male C57BL/6 mice aged 6 to 10weeks were housed under specific-pathogen-free conditions in standard cages in groups (5 per group) and provided standard mouse feed and water ad libitum. Pretreatment of mice with streptomycin was performed as previously described (Whitaker et al., 2012, 2014; Haines-Menges et al., 2014; Kalburge et al., 2017). Mice were inoculated with 100μl of the bacterial suspension and 24h post-infection, mice were sacrificed, and the entire gastrointestinal tract was harvested. Samples were placed in 8ml of sterile 1x PBS, mechanically homogenized and serially diluted in 1xPBS. Diluted samples were plated for CFUs on LBS, supplemented with streptomycin and X-gal for a blue (WBWlacZ) versus white (Δfis) screen of colonies after incubation at 37°C overnight. The competitive index (CI) was determined as described above.
Results
fis Expression Is Controlled in a Growth Dependent Manner
In V. parahaemolyticus RIMD2210633, locus tag VP2885 is annotated as a Fis protein homolog, a 98 amino acid protein that shows 100% protein identity to Fis from V. cholerae and 82% amino acid identity to Fis from E. coli. Fis is an abundant protein in E. coli, highly expressed in exponential phase cells and undetected in stationary phase cells grown under aerobic conditions (Azam et al., 1999; Mallik et al., 2004). In V. parahaemolyticus RIMD2210633, we determined the expression pattern of fis across the growth curve, via RNA isolated from wild type cells grown in LBS at 37°C aerobically at various optical densities (ODs). Using quantitative real time PCR analysis, fis showed highest expression levels in exponential cells at Ods 0.15, 0.25, and 0.5 and then rapidly declined at Ods 0.8 and 1.0, as cells entered stationary phase (Supplementary Figure S1). These data show that fis in V. parahaemolyticus has a similar expression pattern to fis in E. coli and also what has been demonstrated in V. cholerae (Azam et al., 1999; Mallik et al., 2004; Lenz and Bassler, 2007).
Fis Positively Regulates the QS Non-coding qrr sRNAs
One other study has examined the role of Fis in Vibrio and this work showed that in V. cholerae Fis was a positive regulator of the QS non-coding regulatory sRNA genes qrr1 to qrr4 (Lenz and Bassler, 2007). The qrr1 to qrr4 genes in V. parahaemolyticus were homologous to those present in V. cholerae and showed identical genome locations in both species for each qrr gene. To determine the role of Fis in the regulation of qrr1 to qrr5 in V. parahaemolyticus, we identified multiple putative Fis binding sites within the regulatory regions of all five sRNAs (Figure 1). To demonstrate Fis binding, we purified the Fis protein and constructed DNA probes of the regulatory region of each qrr sRNA and performed EMSA with increasing concentrations of purified Fis. Fis binding was shown in a concentration dependent manner in all five Pqrr-Fis EMSAs that resulted in a complete shift of the probe (Figures 1A–E). Smearing in the wells at higher Fis concentrations was noted but was not present in a protein only control lane. Slight smearing in the lane at the highest ratios of DNA:protein, is potentially due to multimerization of Fis protein that can occur, but we cannot rule out minute DNA contamination. The regulatory region of gyrA was used as a Fis binding positive control and showed specific concentration dependent binding with a complete shift in the probe in all five qrr genes (Figure 1F). A DNA fragment without a putative Fis binding site was used as a non-binding control and showed non-specific, weak binding, and no complete shift of the probe at any concentration (Figure 1G). To examine the specificity of binding further, we aligned the putative Fis binding sites in the regulatory regions of the qrr genes, most of which had multiple and sometimes overlapping binding sites. However, we identified a single Fis binding motif in the coding region of qrr3 and mutated this conserved sites to examine Fis binding. In these EMSAs, no banding shift was observed in the mutated probe (Figure 1H).
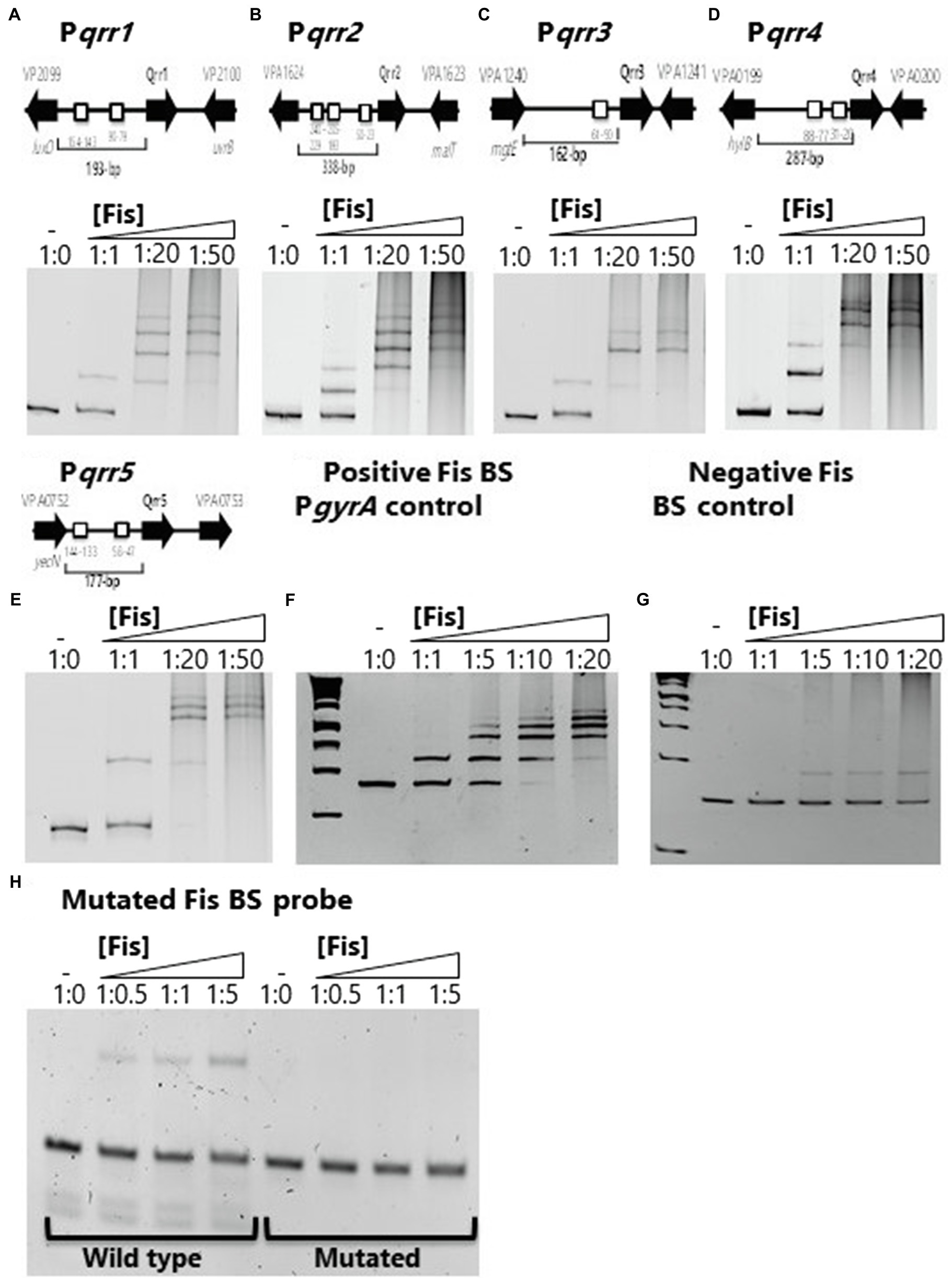
Figure 1. Factor for inversion stimulation binds to the regulatory regions of V. parahaemolyticus regulatory sRNAs (A) qrr1, (B) qrr2, (C) qrr3, (D) qrr4, and (E) qrr5. A putative Fis binding site (BS) is shown as a box in the regulatory region of qrr1 to qrr5. Electrophoretic mobility shift assays (EMSA) using purified Fis and the regulatory regions of qrr1 to qrr5. DNA:protein ratios are as follows: 1:0, 1:1, 1:20, 1:50. (F) Positive Fis BS control in gyrA regulatory region showing complete shift at all concentrations. (G) Fis non-binding negative control using DNA with no putative Fis BS showing weak non-specific binding and no shift at lowest concentration. (H) qrr3 probe containing a single Fis BS (wild type) and a mutated BS demonstrating specificity of Fis binding.
To further examine the role of Fis in V. parahaemolyticus qrr gene expression, an in-frame deletion of fis was constructed by deleting 279-bp of VP2885. We examined growth of the Δfis mutant in LBS broth and found that it grew identical to wild type V. parahaemolyticus (Supplementary Figure S2A). However, on LBS agar plates, the Δfis mutant formed a small colony morphology compared to wild type. This phenotype was rescued by complementation with a functional copy of fis under the control of an IPTG inducible promoter, restoring wild type colony morphology (Supplementary Figures S2B,C). Next, we performed transcriptional GFP reporter assays using the regulatory region of each qrr to determine whether deletion of fis affects expression. Cells were grown to 0.4–0.45 OD and GFP levels measured using relative fluorescence normalized to OD (specific fluorescence). In Δfis, the overall expression of Pqrr2-gfp to Pqrr4-gfp was significantly downregulated compared to wild type (p<0.01 and 0.001, respectively), indicating that Fis is a positive regulator of qrr2, qrr3, and qrr4 in V. parahaemolyticus (Figures 2B–D). We observed a reduction in Pqrr1-gfp and Pqrr5-gfp expression in the Δfis mutant relative to wild type, but this reduction was less significant, which may not be physiologically relevant (Figures 2A,E). Next, we examined whether opaR expression was changed in the Δfis mutant using GFP reporter expression assays under the control of the opaR regulatory region. In these assays, PopaR-gfp activity was significantly upregulated (p<0.01) in the Δfis mutant compared to wild type (Figure 2F). Overall, the data suggest that Fis plays a role in the QS pathway, but is not an essential component, instead it likely modulates expression of the qrr sRNAs and opaR in early exponential phase cells. The data also suggest that other factors are required to control qrr expression. For example, studies have shown that, AphA, LuxO, OpaR, and LuxT also significantly control qrr expression in Vibrio species (Rutherford et al., 2011; Zhang et al., 2012; Kalburge et al., 2017; Eickhoff et al., 2021; Simpson et al., 2021).
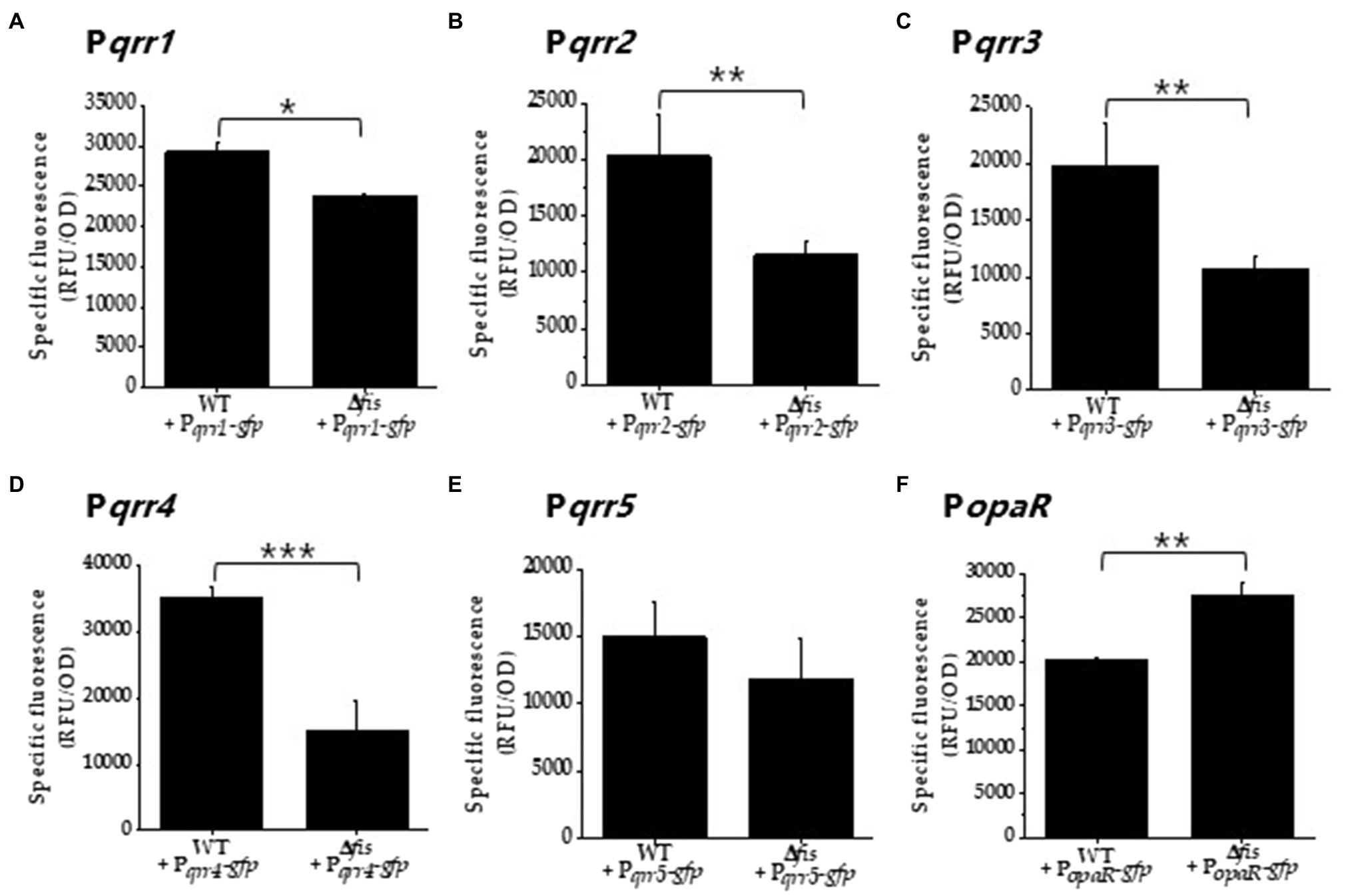
Figure 2. Fis is a positive regulator of the qrr genes. (A) Pqrr1, (B) Pqrr2, (C) Pqrr3, (D) Pqrr4, (E) Pqrr5, and (F) PopaR green fluorescent protein (GFP) transcriptional reporter assays in wild type and the Δfis mutant in cultures grown to OD 0.4–0.45 and measured for specific fluorescence (RFU/OD). Means and standard deviations of three biological replicates are plotted. Statistics calculated using a Student’s t-test (*p<0.05, **p<0.01, ***p<0.001).
Fis Controls Multiple Phenotypes in V. parahaemolyticus
OpaR, the QS master regulator, is a positive regulator of CPS required for biofilm formation. We examined the effects of a fis deletion on this phenotype given the changes in expression of opaR in the Δfis mutant. CPS production in V. parahaemolyticus manifests as an opaque rough wrinkly colony morphology also known as rugose morphology that is also an indirect measure of c-di-GMP levels that oppositely control CPS and swarming motility (McCarter, 1998; Boles and McCarter, 2002; Trimble and McCarter, 2011; Ferreira et al., 2012; Jones and Wozniak, 2017). In the wild-type strain on Congo red plates, cells formed large opaque wrinkly raised colonies, whereas a ΔopaR mutant formed a large smooth colony morphology, indicating CPS is lacking (Figure 3A). The Δfis mutant produced a much smaller highly wrinkly colony morphology (Figure 3A). This colony morphology is indicative of overproduction of CPS in bacteria (Jones and Wozniak, 2017).
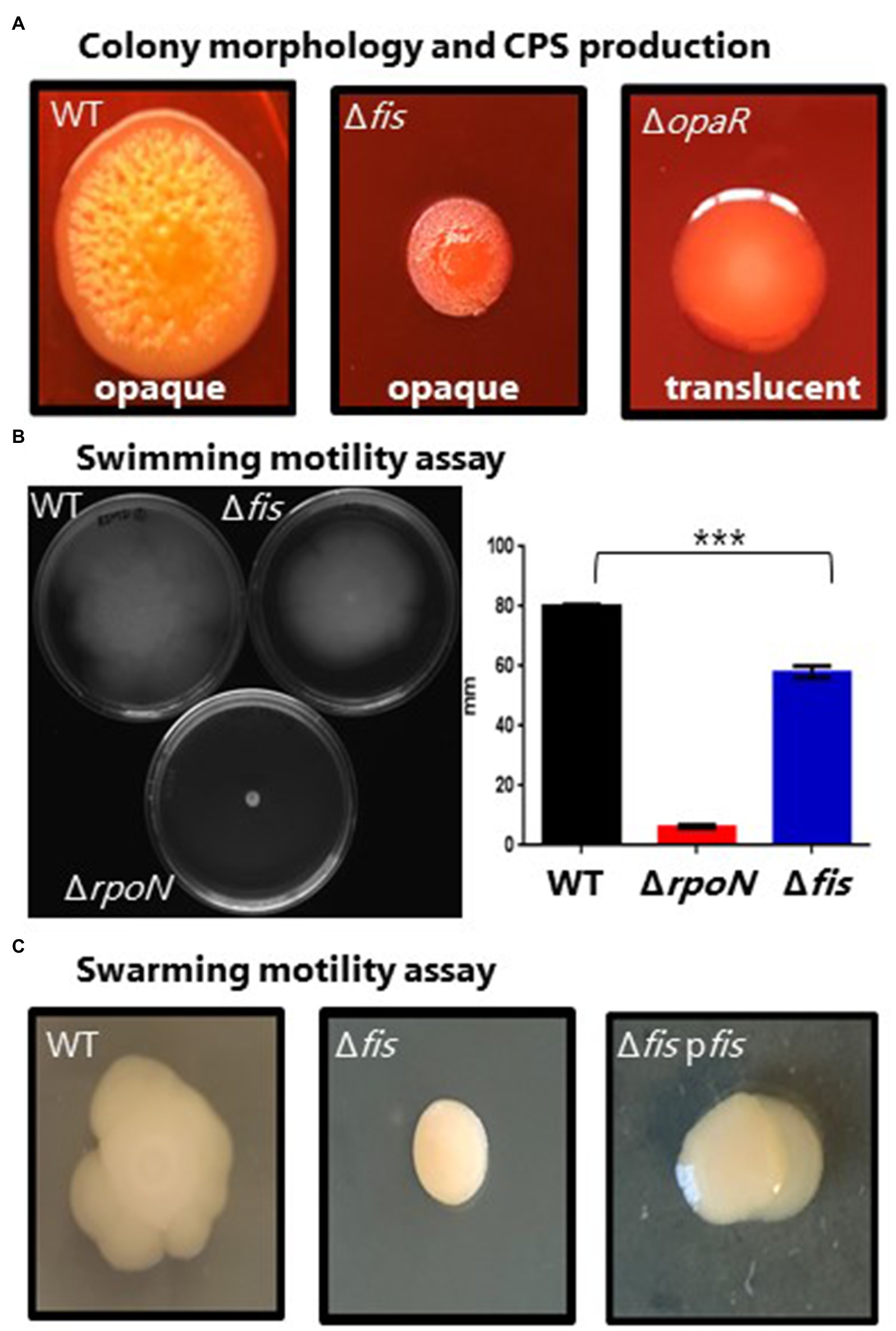
Figure 3. Phenotypic analysis of the Δfis mutant. (A) Capsule polysaccharide (CPS) production in wild type (WT), Δfis and ΔopaR mutants was observed after incubation for 48h at 30°C. Images are an example of three biological replicates performed in triplicate. (B) Swimming motility assays and quantification. Two biological replicates were measured, performed in triplicate. Statistics calculated using a Student’s t-test (***p<0.001). (C) Swarming motility assay of V. parahaemolyticus wild type and the Δfis mutant and Δfis complementation strain. Complementation assays were grown in the presence of Cm and IPTG. All images are examples from three biological replicates.
To investigate whether Fis has any role in antimicrobial peptide resistance, the Δfis strain was examined for polymyxin B sensitivity using disk diffusion and survival assays. Previous work in Vibrio has shown that changes in the bacterial cell wall can result in increased sensitivity to polymyxin B (Haines-Menges et al., 2014; Lubin et al., 2015; McDonald et al., 2018). We used ΔrpoE as a control, which lacks the sigma factor RpoE that is required for the cell envelope stress response and was previously shown to be polymyxin B sensitive (Haines-Menges et al., 2014). For the disk diffusion assay, the Δfis and ΔrpoE mutants had significantly larger zones of inhibition compared to wild type indicating these mutants are more sensitive to polymyxin B (Figure 4A). To verify this further, we conducted survival assay using 200μg of total polymyxin B in 5ml of LBS media. CFUs were counted at 0, 30 and 60min post exposure. Significantly lower CFUs for the Δfis were recovered at 30 and 60min compared to wild type (Figure 4B). The data demonstrated that Fis is required for polymyxin B resistance in V. parahaemolyticus, although the mechanism remains unknown.
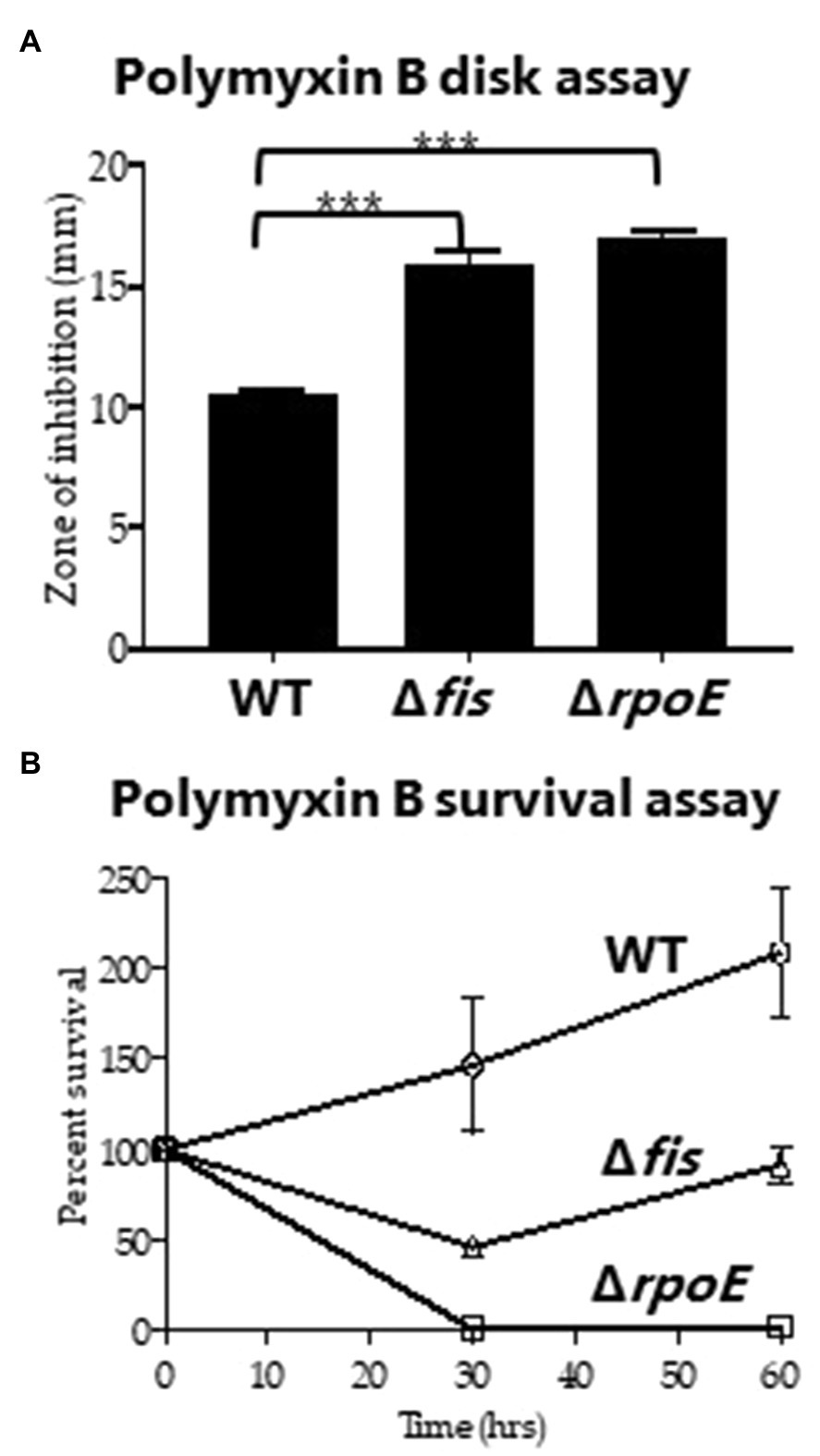
Figure 4. Polymyxin B sensitivity assay. (A) Disk containing 100μg of total polymyxin B was used to identify the zone of inhibition for WT, Δfis and ΔrpoE and was quantified by measuring the diameter of the zone of inhibition. (B) Survival assays were conducted using WT, Δfis, and ΔrpoE after treating the bacterial cultures with polymyxin B (final concentration of 40μg/ml) and calculating percent survival at 30min and 60min. The disk assay was performed in duplicate using two biological replicates. The survival assay was performed in triplicate using two biological replicates. Unpaired t-test was conducted to determine the value of p. ***p<0.001.
Next, we examined whether deletion of fis affected motility in V. parahaemolyticus, a species that produces both polar and lateral flagella, and where OpaR is a negative regulator of the laf operon (Jaques and McCarter, 2006). Swimming assays demonstrated that the Δfis mutant had a defect in motility compared to wild type with reduced spreading across the agar plate (Figure 3B). An ΔrpoN mutant was also examined as a negative control, and this mutant, as expected, showed no motility (Figure 3B). In swarming assays, the wild-type strain produced a typical swarming cauliflower colony morphology, whereas in the Δfis deletion mutant, swarming motility was abolished suggesting lateral flagella are absent (Figure 3C). Complementation of the Δfis mutant with a functional copy of the fis gene under the control of an IPTG-inducible promoter rescued swarming and swimming motility (Supplementary Figure S3).
Fis Is Necessary for Motility in V. parahaemolyticus
To further investigate Fis control of swimming and swarming motility, using bioinformatics, we identified multiple putative Fis binding sites within the regulatory regions of both the polar and lateral flagella biosynthesis operons (Figures 5A,E). We performed EMSA analysis using purified Fis and DNA probes of the regulatory regions of the polar flagellum operon (flh loci VP2235-PV2231) and the lateral flagellum operon (laf loci VPA1550-VPA1557) and demonstrated binding (Figures 5B,F). To determine the specificity of Fis binding further, we created two DNA probes with mutations in a single Fis binding motif and examined binding. In these EMSAs, the wild type sequence showed weak binding for both wild type probes and no change in the mutated probe (Figures 5C,G). These data suggest that Fis binds at these sites in a non-specific manner. To examine Fis regulation of motility further, GFP reporter expression assays were performed. In these assays, the Δfis mutant relative to wild type did not show differential expression of PflhA-gfp (Figure 5D). In contrast, in reporter assays of swarming cells, PlafB-gfp was significantly repressed (p<0.001) in the Δfis mutant relative to wild type (Figure 5H). Thus, the data suggest that Fis controls lateral flagella gene expression and is an important positive regulator of swarming motility.
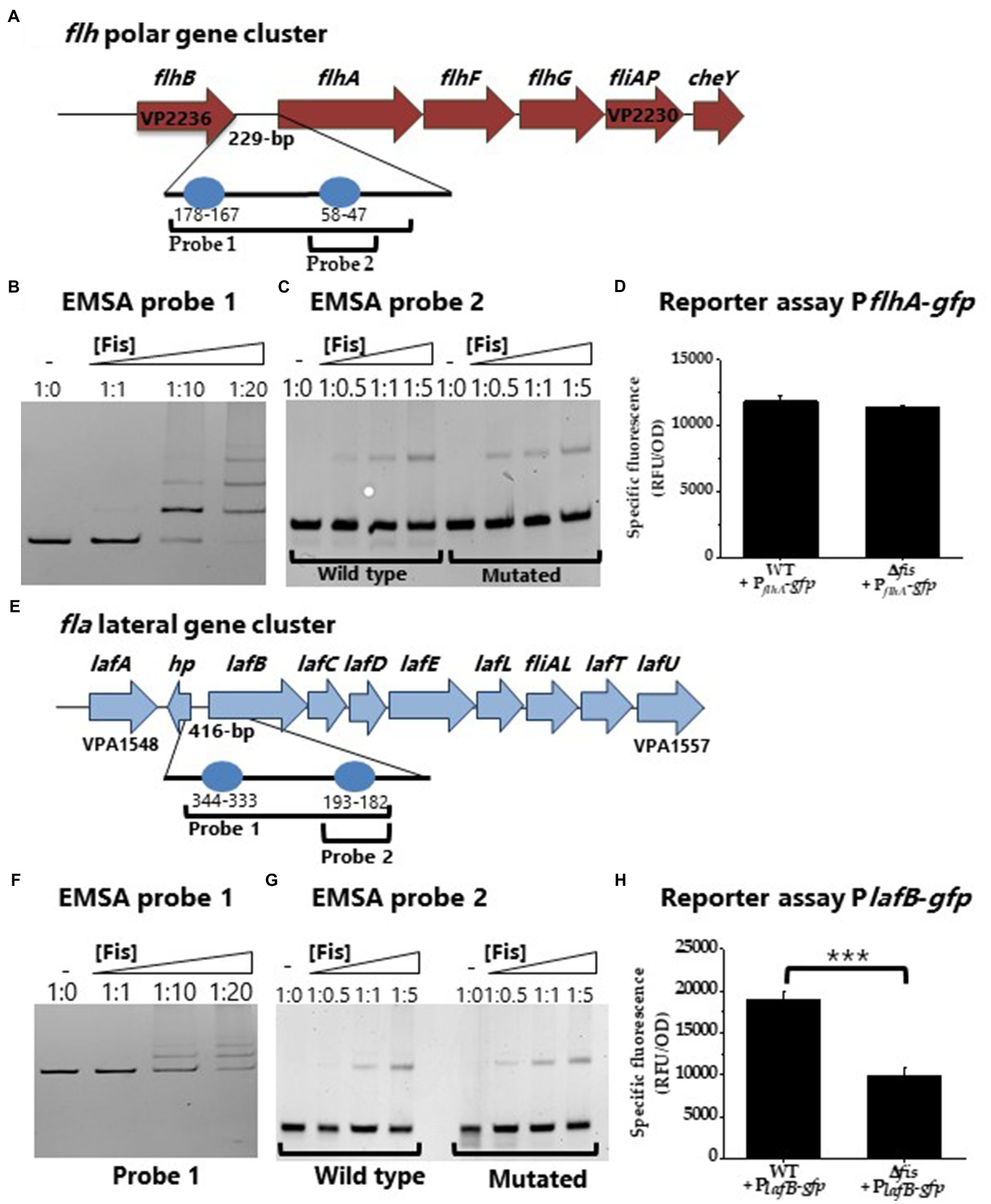
Figure 5. Regulation of lateral flagella biosynthesis by Fis. (A) Regulatory region of polar flagellum flh genes with putative Fis binding sites (BS) depicted as blue circles. (B) EMSA of Fis bound to PflhA in a concentration dependent manner. DNA: protein ratios are as follows: 10, 1:1, 1:10, 1:20. (C) Probe 2 containing a single Fis BS (WT) was mutated. DNA: protein ratios are as follows: 0, 1:0.5, 1:1, 1:5. (D) Transcriptional GFP reporter assay of PflhA-gfp in the Δfis mutant relative to wild type. (E) Regulatory region of lateral flagellum laf genes with putative Fis binding sites. (F) EMSA of Fis bound to Plaf DNA probe. (G) Probe 2 containing a single Fis BS and a mutated probe 2 Fis BS. DNA: protein ratios are as follows: 0, 1:0.5, 1:1, 1:5. (H) Transcriptional GFP reporter assay of PlafB-gfp between wild type and Δfis (***p<0.001).
Fis Is a Positive Regulator of the Surface Sensing Operon scrABC
The scrABC operon has been shown to positively control swarming motility and negatively control CPS by controlling c-di-GMP levels (Boles and McCarter, 2002; Kim and McCarter, 2007; Gode-Potratz et al., 2011; Ferreira et al., 2012). We reasoned that Fis might also regulate this operon to co-ordinate with the QS pathway in the control of this phenotype. First, using bioinformatics, we identified multiple and sometimes overlapping putative Fis binding sites in the regulatory region of the scrABC operon (Figure 6A). We performed EMSAs with three probes that contained Fis BS and demonstrated Fis binding to all three probes in a concentration dependent manner (Figure 6B). To show Fis BS specificity, we created a DNA probe from probe 3 that contains one Fis BS, mutated this site, and performed an EMSA. In this EMSA, Fis still bound to the mutated probe, but at a reduced level (Figure 6C). In GFP reporter assays, P scrABC-gfp activity was significantly downregulated (p<0.001) in the Δfis mutant compared to wild type, indicating that Fis is a direct positive regulator of this operon (Figure 6D). Overall, the data suggest that loss of swarming motility is likely due to Fis direct regulation of the scrABC operon and the lateral flagella biosynthesis laf operon and indirect regulation of opaR through modulation of qrr expression in V. parahaemolyticus.
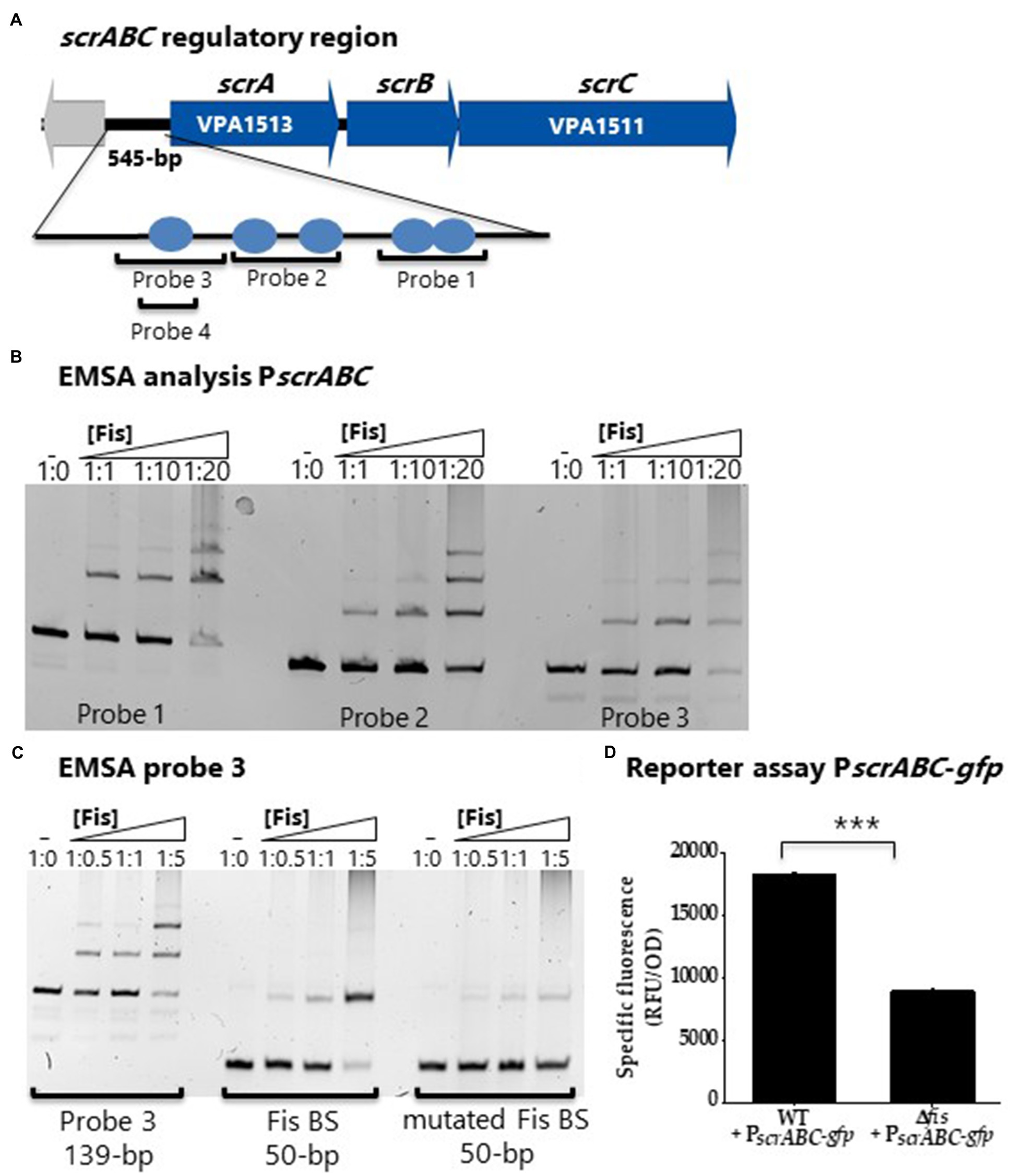
Figure 6. Fis is a positive regulator of the surface sensing operon, scrABC. (A) Putative Fis binding sites identified in the regulatory region of the scrABC surface sensing operon. (B) EMSA using purified Fis protein and probes 1, 2 and 3 encompassing the regulatory region of scrABC. DNA: protein ratios are as follows: 10, 1:1, 1:10, 1:20, 1:50. (C) Probe 3 containing a single Fis BS and a mutated probe 3 Fis BS. DNA: protein ratios are as follows: 0, 1:0.5, 1:1, 1:5. (D) GFP reporter assay of PscrABC-gfp in wild type and the Δfis mutant. Specific fluorescence was calculated (RFU/OD) for three biological replicates and plotted as mean and standard deviation. Statistics were calculated using a Student’s t-test (***p<0.001).
Fis Modulates Expression of Carbon Catabolism Genes
To determine the effects of Fis on overall metabolism, we conducted growth competition assays between wild type and Δfis in various carbon sources. For the in vitro competition assays, we used a β-galactosidase knock-in strain of RIMD2210633, strain WBWlacZ, which was previously demonstrated to behave identically to wild type in in vitro and in vivo studies (Whitaker et al., 2012, 2014; Haines-Menges et al., 2014; Kalburge et al., 2017). Comparisons of WBWlacZ and the Δfis mutant allows for a blue/white colony screen in the presence of IPTG. First, in vitro growth competition assays were performed in LB 3% NaCl (LBS) resulting in a competitive Index (CI) of 0.99 demonstrating under these conditions that neither strain had a competitive advantage. Similarly, in vitro competition assays in M9 3%NaCl (M9S) supplemented with glucose showed neither strain had a competitive advantage. However, in M9S supplemented with mouse intestinal mucus or individual mucus components as sole carbon sources, Δfis was significantly outcompeted by WBWlacZ. In mouse intestinal mucus, the Δfis mutant had a CI of 0.61, and in mucus components L-arabinose a CI of 0.4, NAG a CI of 0.68 and D-gluconate a CI of 0.78 (Figure 7).
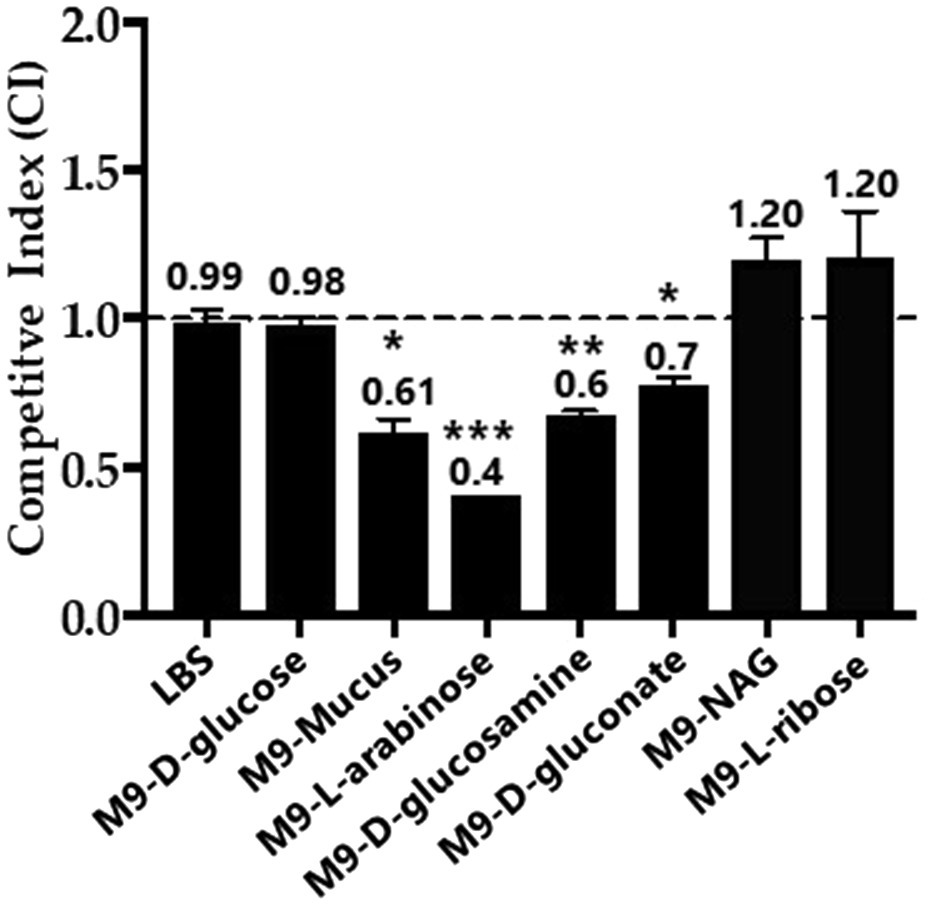
Figure 7. In vitro growth competition assay between wild type and the Δfis mutant. WBWlacZ and Δfis were grown in co-culture (1:1 ratio) for 24h in LBS, M9 supplemented with 100μg/ml intestinal mucus, 10mm of D-glucose, L-arabinose, D-glucosamine, D-gluconate, N-acetyl-D-glucosamine (NAG), or D-ribose. WBWlacZ outcompetes Δfis, <1.00, and Δfis outcompetes WBWlacZ when >1.00. The assay was conducted in two biological replicates in triplicates. Error bar indicates SEM. Unpaired Student’s t-test was conducted. The significant difference is denoted by asterisks (*p<0.05, **p<0.01, ***p<0.001).
Putative Fis binding sites were identified in the regulatory region of the araBDAC (L-arabinose catabolism) operon (Figure 8A). EMSAs were performed using 138-bp and 152-bp DNA probes containing two and one putative Fis binding sites, respectively (Figure 8A). In these assays, Fis bound to the regulatory region of araBDAC and the binding was concentration dependent (Figure 8B). To examine the specificity of Fis binding further, we created probe 3 that contained a single Fis BS, mutated this site, and performed an EMSA. The wild type probe 3 showed Fis binding, whereas the mutated probe showed no binding, demonstrating Fis binding specificity (Figure 8C). In GFP transcriptional reporter assays, ParaB-gfp showed lower activity in the Δfis mutant compared to wild type, although the fold change difference is small suggesting it may not be physiologically relevant at this time-point (Figure 8D).
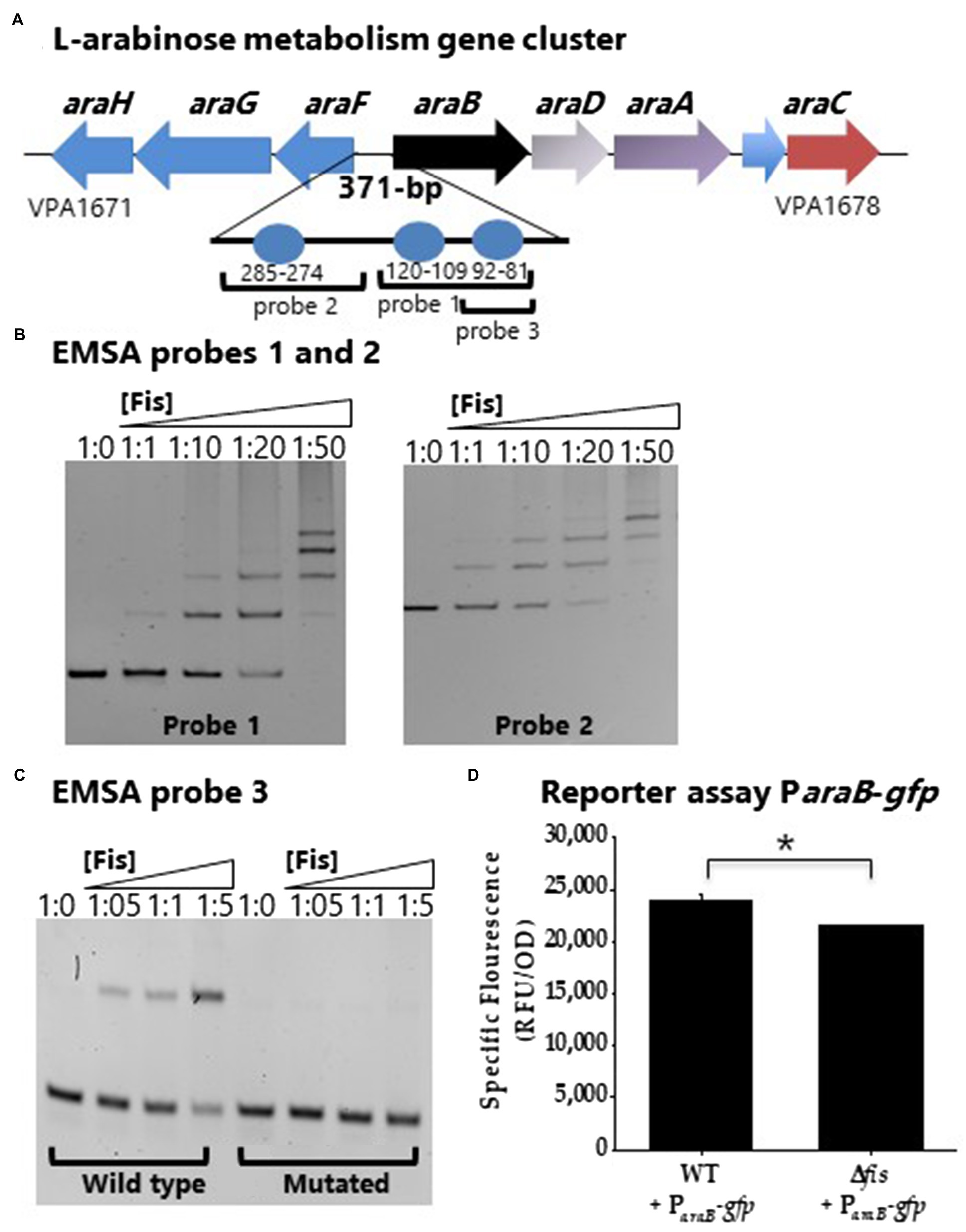
Figure 8. Fis is a positive regulator of the L-arabinose operon araBCDA. (A) Fis binding sites identified in the regulatory region of the araBDAC operon depicted as blue circles. (B) ParaB was divided into two probes, probe 1 and probe 2 for EMSA analysis using purified Fis protein. (C) EMSA analysis probe 3 wild type with a single Fis BS and a mutated probe 3. DNA: protein ratios were as follows: 10, 1:1, 1:10, 1:20, 1:50. (D) GFP transcriptional reporter assay of the araBDAC regulatory region in wild type and the Δfis mutant. *p<0.05.
Factor for inversion stimulation binding sites were also identified in the regulatory region of gntK (D-gluconate catabolism; Figure 9A). A DNA probe encompassing PgntK showed Fis binding (Figure 9B). A second probe with only a single putative Fis BS was constructed and the putative Fis BS was mutated and EMSAs performed. In this assay a slight reduction in binding is noted, but non-specific binding cannot be ruled out (Figure 9C). To examine Fis control of D-gluconate metabolism further, a GFP reporter assay of PgntK was performed. In these assays, PgntK showed significantly lower expression levels in the Δfis mutant compared to wild type indicating that Fis is a positive regulator of expression (Figure 9D).
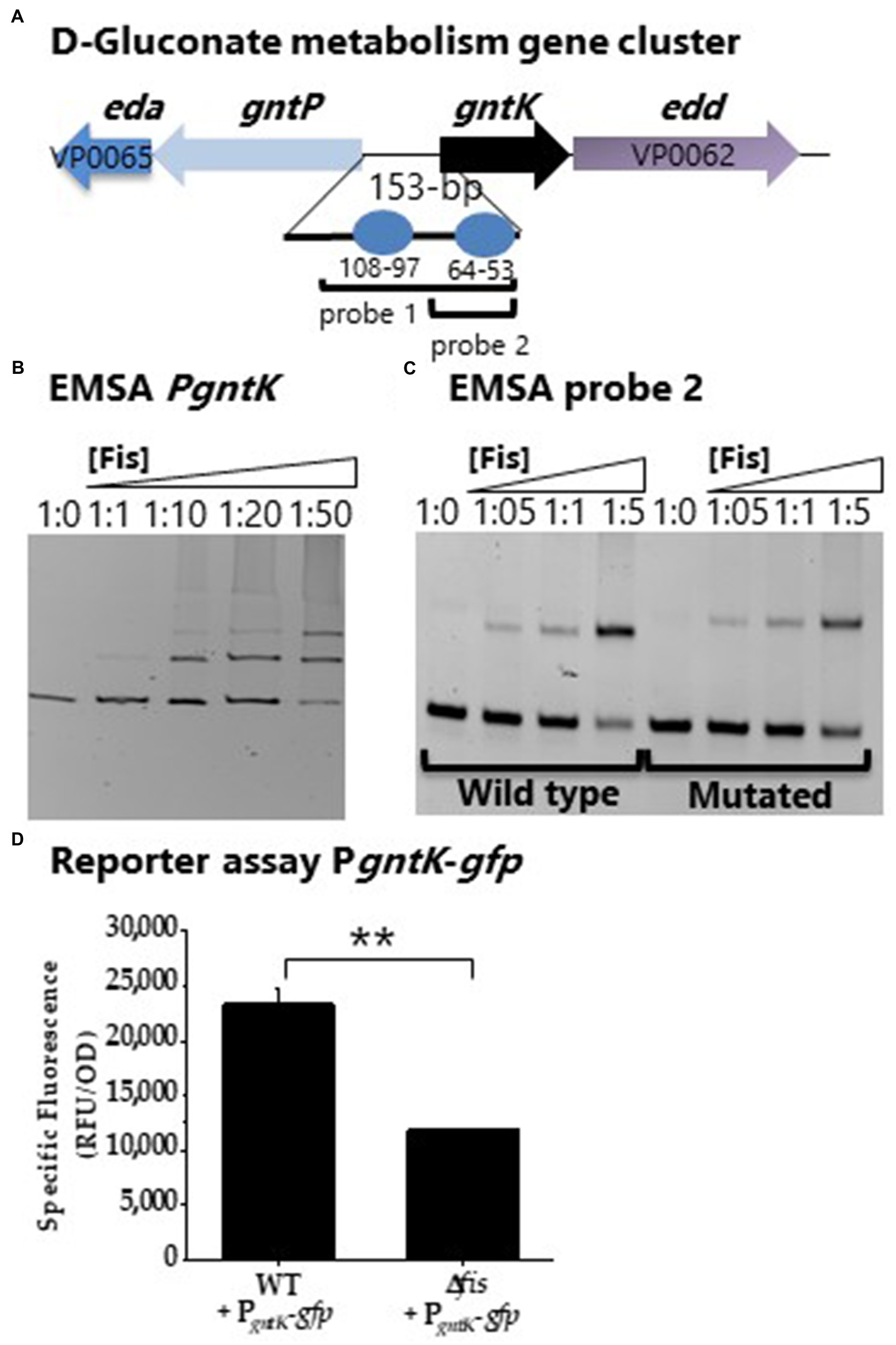
Figure 9. Fis is a positive regulator of gntK. (A) Putative Fis binding sites in the regulatory region of gntK. (B) EMSA analysis with purified Fis protein and PgntK probe 1. DNA: protein ratios were as follows: 10, 1:1, 1:10, 1:20, 1:50. (C) EMSA analysis of probe 2 containing a single Fis BS and a mutated Fis BS probe 2. (D) GFP transcriptional reporter assay of gntK regulatory region in wild type and Δfis. **p<0.01.
Multiple overlapping Fis binding sites were identified in the regulatory region of nagB (NAG catabolism; Figure 10A). EMSA analysis with two probes encompassing the Fis sites showed specific binding in a concentration dependent manner (Figures 10B,C). To examine specificity of binding further we created a probe 2 with mutations in 5 sites of the Fis BS motif. In this analysis, Fis bound to the mutated probe, but the binding was reduced, suggesting Fis is a direct regulator of nagB (Figure 10C). In addition, PnagB showed significantly lower activity in the GFP reporter assay in the Δfis mutant compared to wild type (Figure 10D). Overall, our results demonstrated that Fis is a positive regulator of catabolism genes, araB, gntK, and nagB in V. parahaemolyticus.
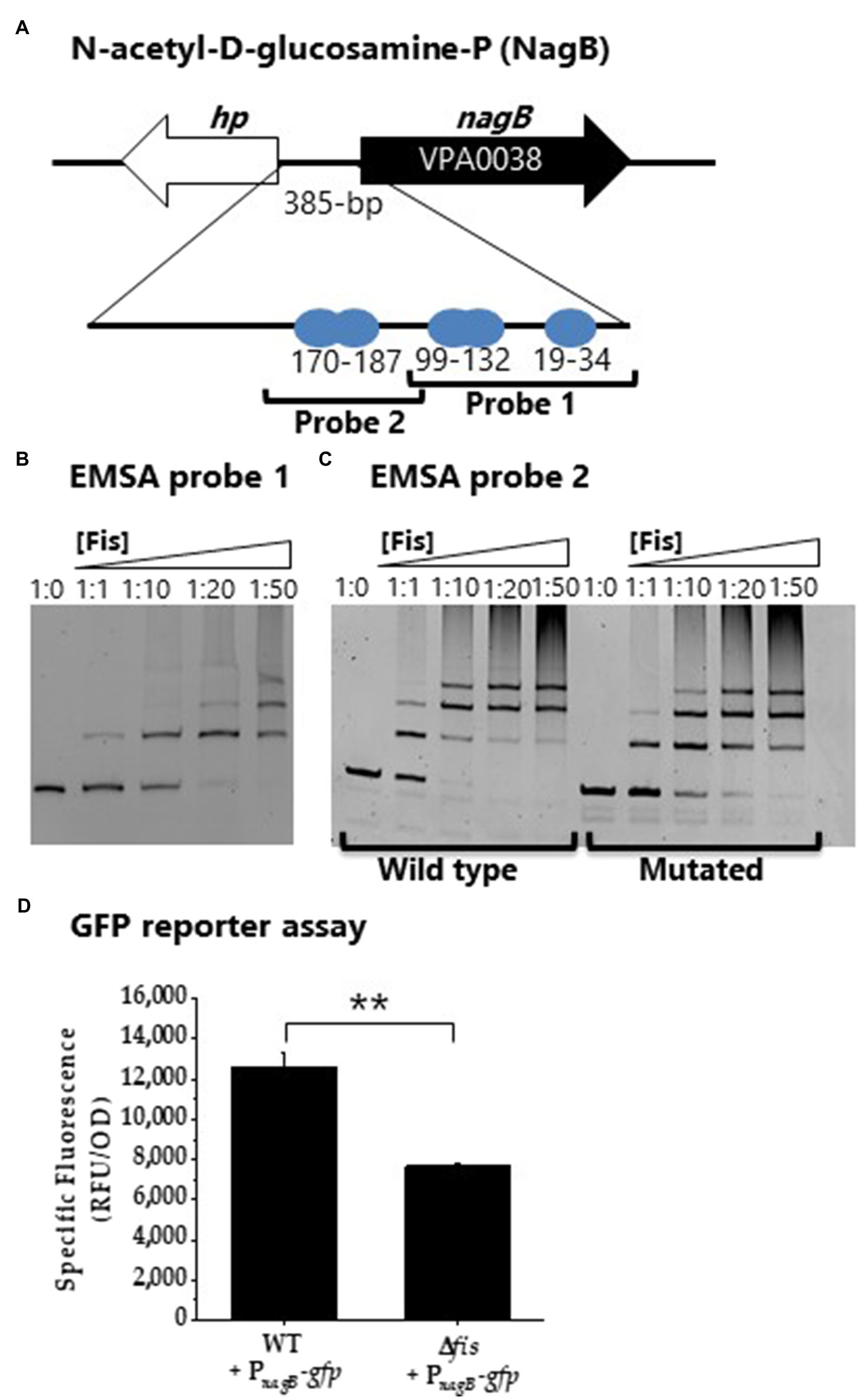
Figure 10. Fis is a positive regulator of NAG gene nagB. (A) Fis binding sites identified in the regulatory region of nagB. (B) EMSA analysis of Fis binding to probe 1. (C) EMSA analysis of probe 2 and a mutated Fis BS probe 2. DNA: protein molar ratios were as follows: 10, 1:1, 1:10, 1:20, 1:50. (D) GFP transcriptional reporter assay with PnagB-gfp. Means and standard deviations of two biological replicates are shown. Statistics calculated using a Student’s t-test (**p<0.01).
We determined a Fis consensus sequence from the regulatory regions of genes under Fis control. To accomplish this, we examined a 50-bp region encompassing each putative Fis binding site identified in this study using multiple EM for motif elicitation (MEME) analysis. This analysis identified a 15-bp core motif highly similar to what has been identified in E. coli (Figure 11A). This motif is aligned in each sequence and maps to the Fis binding sites identified via virtual footprint (Figure 11B). Similar to the sequence motif used by virtual footprint, along with previous reports of Fis consensus sequences, the motif we present has a stretch of T/A bps, characteristic of Fis binding (Cho et al., 2008).

Figure 11. Analysis of Fis consensus sequence. (A) A Fis DNA binding motif was created using MEME analysis. Sequences containing a putative Fis binding site were used. (B) An alignment of the motifs found in each sequence, along with the corresponding values of p.
Fis Is Required for in vivo Fitness in V. parahaemolyticus
To determine whether Fis contributes to in vivo fitness of V. parahaemolyticus, in vivo colonization competition assays were performed using a streptomycin pretreated adult mouse model of colonization (Whitaker et al., 2012, 2014; Haines-Menges et al., 2014; Yang et al., 2019). The in vivo competition assay determined the ability of WBWlacZ and the Δfis mutant to co-colonize the intestinal tract of streptomycin pretreated adult mice. Competition assays were performed in adult C57BL/6 mice pretreated with an orogastric dose of streptomycin (20mg/animal) 24h prior to orogastric co-inoculation with a 1:1 mixture of V. parahaemolyticus WBWlacZ and Δfis (n=9). In an in vitro assay in LBS using the same inoculum, the WBWlacZ vs. Δfis had a CI of 1.17 whereas in vivo the WBWlacZ vs. Δfis CI was 0.57 indicating that the mutant was significantly (p<0.01) outcompeted by WBWlacZ (Supplementary Figure S4). This indicates that Δfis has decreased fitness in vivo compared to the wild type strain.
Discussion
Nucleoid associated proteins such as Fis, bind and bend DNA to aid in DNA compaction, and are also important global regulators of gene expression. In V. parahaemolyticus we show, similar to enteric species, that fis is expressed in early to mid-exponential phase cells and declines in late exponential and stationary phase cells. The only other study examining the role of Fis in a Vibrio species, showed a direct role of Fis in the QS pathway in V. cholerae. They demonstrated Fis activation of qrr1 to qrr4 sRNAs and hapR (opaR homolog) constitutive expression in a fis deletion mutant (Lenz and Bassler, 2007). In V. parahaemolyticus, Fis bound to all five qrr sRNAs regulatory regions in a concentration dependent manner. Our work showed that Fis is a positive regulator of the QS regulatory sRNAs. We show that in the fis deletion mutant qrr1 to qrr5 were repressed and opaR was induce. It is of interest to note that in our study, the expression of each of the qrr sRNA differed in the Δfis mutant compared to wild type with significant repression of qrr2, qrr3 and qrr4, whereas both qrr1 and qrr5 showed reduced expression. The importance of this remains to be determine since we do not know whether in V. parahaemolyticus the Qrr sRNAs are redundant as is the case in V. cholerae or additive as is the case in V. harveyi. Our data suggest that other regulators are also involved in modulating qrr expression and that Fis is one of several factors controlling expression. This is not too surprising as studies have shown that, although Fis may bind to a large number of regulatory regions, only a portion of these sites are significantly regulated by Fis (Kahramanoglou et al., 2011; Monteiro et al., 2020). For example, in E. coli, ChIP-seq analysis uncovered 1,464 Fis binding sites, but only 462 genes were differential regulated by Fis under the conditions examined (Kahramanoglou et al., 2011). This suggests that Fis has a regulatory role, however other factors are likely involved and that the Fis regulon varies with changes in growth conditions, growth phase, binding affinity, amongst other factors.
In V. parahaemolyticus, we showed that Fis is a positive regulator of swarming motility through modulated expression of the QS regulator, OpaR, and control of the lateral flagellum laf operon and the surface sensing scrABC operon. The scrABC operon in V. parahaemolyticus controls the transition between swarming motility and adhesion to a surface by altering gene expression of the laf and cps operons (Boles and McCarter, 2002; Kim and McCarter, 2007). Together, ScrA, ScrB, and ScrC modulate the level of c-di-GMP in the cell, a secondary messenger that controls numerous downstream processes. More specifically, ScrC contains both GGDEF and EAL enzymatic activity, making it a unique bifunctional enzyme (Kim and McCarter, 2007; Ferreira et al., 2012). ScrA produce the extracellular S-signal, which represses CPS gene expression. In the presence of ScrA interaction with ScrB, ScrC acts as a phosphodiesterase to degrade c-di-GMP. High levels of c-di-GMP promote CPS production, while low levels of c-di-GMP promote swarming motility (Trimble and McCarter, 2011; Ferreira et al., 2012). In the Δfis mutant, we observed repression of both the laf and scrABC operons in the GFP reporter assays, indicating that Fis is a positive regulator of these operons. We suggest that the Δfis mutant swarming defect is through repression of both the laf and scrABC operons and through derepression of opaR, which is a repressor of swarming motility (Figure 12). As shown in Supplementary Figure S1, fis expression is induced at early exponential phase cells and decreases as the cells transition to HCD. We propose that at LCD, Fis activates the qrr sRNAs, which repress opaR, and activates the surface sensing operon. The scrABC operon activates the laf operon, stimulating motility, while repressing the cps operon. As the cells transition to HCD, fis expression decreases while opaR expression increases, as it has been shown to be maximally expressed in stationary phase cells. OpaR represses the laf operon and activates expression of the cps operon, inducing CPS formation. Therefore, Fis integrates both the QS and surface sensing signals by positively regulating qrr sRNAs and scrABC to induce swarming motility and repress CPS formation until the cells enter HCD (Figure 12).
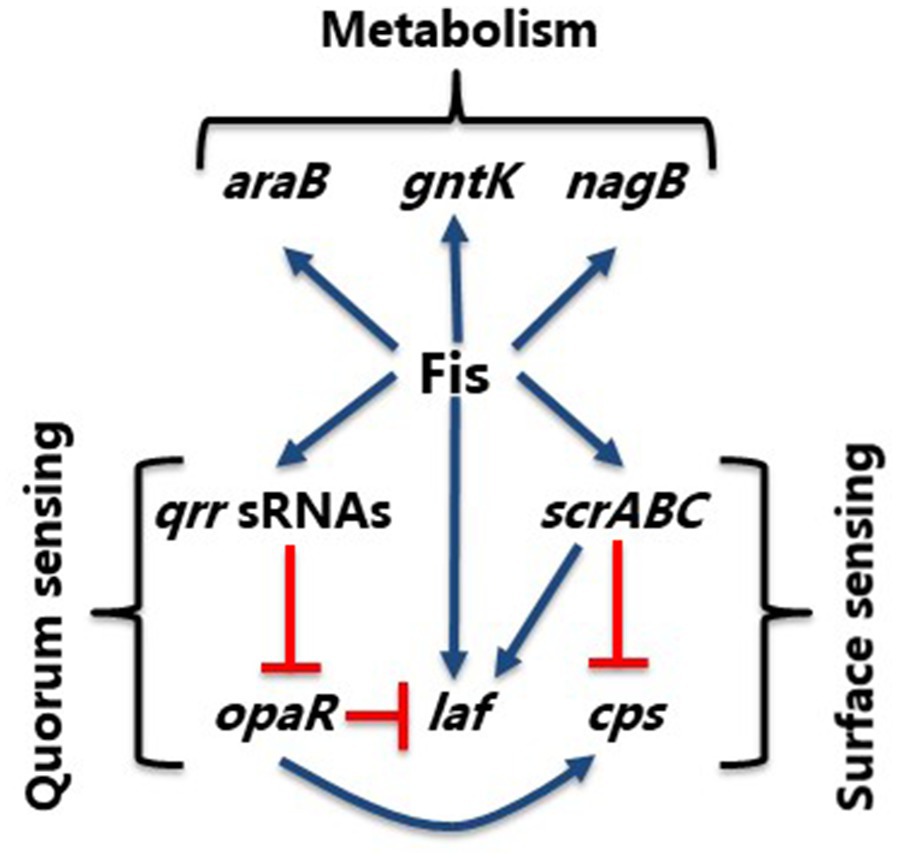
Figure 12. Model of Fis integration of QS and surface sensing pathways. Model shows control of swarming (laf) and CPS (cps) production in V. parahaemolyticus. Blue arrows show positive regulation and orange hammers show negative regulation.
In E. coli, using global approaches, Fis was shown to regulate genes involved in, but not limited to, two-component systems, biofilm formation, and pilus organization. It was determined that Fis regulates genes required for sugar metabolism as well, including NAG as shown in our study (Gawade et al., 2020). In S. enterica, Fis was shown to negatively regulate genes contributing to virulence and metabolism in the mammalian gut (Kelly et al., 2004). In this species, it was demonstrated that Fis was a negative regulator of acetate metabolism, biotin synthesis, fatty acids metabolism and propanediol utilization, amongst others. The authors speculated that this could be important for intestinal colonization and/or systemic infection (Kelly et al., 2004). In V. parahaemolyticus, the in vivo competition assays between the Δfis and WBWLacZ strain showed that the Δfis strain was outcompeted demonstrating that Fis is required for in vivo fitness. In addition, in vitro growth competition assays in intestinal mucus and mucus components demonstrated that Δfis was again outcompeted by wild type. We speculate that the Δfis mutant is outcompeted by wild type in vivo at least in part due to its inability to efficiently utilize nutrient sources. Carbon metabolism was previously implicated as important for colonization of V. parahaemolyticus in a streptomycin pretreated adult mouse model (Whitaker et al., 2014; Kalburge et al., 2017). Additionally, we showed that Fis is important for resistance to antimicrobial peptides prevalent in animal intestines. The fis deletion mutant was highly sensitive to polymyxin B, which could also account for its reduced fitness in vivo. In summary, Fis modulates expression of genes involved in QS, motility, and metabolism in V. parahaemolyticus and it will be of interest to determine the different mechanisms used to modulate expression by this NAP.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by University of Delaware Institutional Animal Care and Use Committee.
Author Contributions
EB, JT, and AR designed the study and analyzed the results. JT, GG, and AR performed the experiments. All authors contributed to writing and review the manuscript and approved the submitted version.
Funding
This research was supported in part by a National Science Foundation grant (award IOS-1656688) to EB. JT and GG were funded by University of Delaware graduate fellowship awards.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank members of the Boyd Group for constructive feedback on the manuscript and the reviewers for their suggestions and comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669447/full#supplementary-material
References
Auner, H., Buckle, M., Deufel, A., Kutateladze, T., Lazarus, L., Mavathur, R., et al. (2003). Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J. Mol. Biol. 331, 331–344. doi: 10.1016/S0022-2836(03)00727-7
Azam, T. A., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370. doi: 10.1128/JB.181.20.6361-6370.1999
Bassler, B. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. doi: 10.1016/S1369-5274(99)00025-9
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Belas, R., Simon, M., and Silverman, M. (1986). Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167, 210–218. doi: 10.1128/jb.167.1.210-218.1986
Bétermier, M., Galas, D., and Chandler, M. (1994). Interaction of Fis protein with DNA: bending and specificity of binding. Biochimie 76, 958–967. doi: 10.1016/0300-9084(94)90021-3
Bokal, A. J., Ross, W., Gaal, T., Johnson, R. C., and Gourse, R. L. (1997). Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J. 16, 154–162. doi: 10.1093/emboj/16.1.154
Boles, B. R., and McCarter, L. L. (2002). Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184, 5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002
Bradley, M. D., Beach, M. B., de Koning, A. P., Pratt, T. S., and Osuna, R. (2007). Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153, 2922–2940. doi: 10.1099/mic.0.2007/008565-0
Browning, D. F., Grainger, D. C., Beatty, C. M., Wolfe, A. J., Cole, J. A., and Busby, S. J. (2005). Integration of three signals at the Escherichia coli nrf promoter: a role for Fis protein in catabolite repression. Mol. Microbiol. 57, 496–510. doi: 10.1111/j.1365-2958.2005.04701.x
Carpenter, M., Rozovsky, S., and Boyd, E. (2015). Pathogenicity island cross talk mediated by recombination directionality factors facilitates excision from the chromosome. J. Bacteriol. 198, 766–776. doi: 10.1128/JB.00704-15
Cho, B., Knight, E., Barrett, C., and Palsson, B. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910. doi: 10.1101/gr.070276.107
Cróinín, T. O., Carroll, R. K., Kelly, A., and Dorman, C. J. (2006). Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 62, 869–882. doi: 10.1111/j.1365-2958.2006.05416.x
De Souza Santos, M., and Orth, K. (2019). The role of the type III secretion system in the intracellular lifestyle of enteric pathogens. Microbiol. Spectr. 7: doi: 10.1128/microbiolspec.BAI-0008-2019
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540. doi: 10.1128/jb.179.2.538-540.1997
Dennis, P. P., Ehrenberg, M., and Bremer, H. (2004). Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68, 639–668. doi: 10.1128/MMBR.68.4.639-668.2004
Eickhoff, M., Fei, C., Huang, X., and Bassler, B. (2021). LuxT controls specific quorum-sensing-regulated behaviors in Vibrionaceae spp. via repression of qrr1, encoding a small regulatory RNA. PLoS Genet. 17:e1009336. doi: 10.1371/journal.pgen.1009336
Ferreira, R. B., Antunes, L. C., Greenberg, E. P., and McCarter, L. L. (2008). Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190, 851–860. doi: 10.1128/JB.01462-07
Ferreira, R. B., Chodur, D. M., Antunes, L. C., Trimble, M. J., and McCarter, L. L. (2012). Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J. Bacteriol. 194, 914–924. doi: 10.1128/JB.05807-11
Finkel, S., and Johnson, R. (1992). The Fis protein: it’s not just for DNA inversion anymore. Mol. Microbiol. 6, 3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x
Froelich, B., and Daines, D. (2020). In hot water: effects of climate change on vibrio-human interactions. Environ. Microbiol. 22, 4101–4111. doi: 10.1111/1462-2920.14967
Froelich, B., and Noble, R. (2016). Vibrio bacteria in raw oysters: managing risks to human health. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 371:20150209. doi: 10.1098/rstb.2015.0209
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gawade, P., Gunjal, G., Sharma, A., and Ghosh, P. (2020). Reconstruction of transcriptional regulatory networks of Fis and H-NS in Escherichia coli from genome-wide data analysis. Genomics 112, 1264–1272. doi: 10.1016/j.ygeno.2019.07.013
Gibson, D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361. doi: 10.1016/b978-0-12-385120-8.00015-2
Gode-Potratz, C. J., Kustusch, R. J., Breheny, P. J., Weiss, D. S., and McCarter, L. L. (2011). Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79, 240–263. doi: 10.1111/j.1365-2958.2010.07445.x
Gode-Potratz, C. J., and McCarter, L. L. (2011). Quorum sensing and silencing in Vibrio parahaemolyticus. J. Bacteriol. 193, 4224–4237. doi: 10.1128/JB.00432-11
Goldberg, M. D., Johnson, M., Hinton, J. C., and Williams, P. H. (2001). Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41, 549–559. doi: 10.1046/j.1365-2958.2001.02526.x
González-Gil, G., Bringmann, P., and Kahmann, R. (1996). FIS is a regulator of metabolism in Escherichia coli. Mol. Microbiol. 22, 21–29. doi: 10.1111/j.1365-2958.1996.tb02652.x
Grainger, D. C., Hurd, D., Goldberg, M. D., and Busby, S. J. (2006). Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34, 4642–4652. doi: 10.1093/nar/gkl542
Gray, K. M., Passador, L., Iglewski, B. H., and Greenberg, E. P. (1994). Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 176, 3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994
Gregory, G. J., Morreale, D. P., Carpenter, M. R., Kalburge, S. S., and Boyd, E. F. (2019). Quorum sensing regulators AphA and OpaR control expression of the operon responsible for biosynthesis of the compatible solute ectoine. Appl. Environ. Microbiol. 85, 1543–1519. doi: 10.1128/aem.01543-19
Güvener, Z., and McCarter, L. (2003). Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185, 5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003
Haines-Menges, B., Whitaker, W. B., and Boyd, E. F. (2014). Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect. Immun. 82, 3667–3677. doi: 10.1128/IAI.01854-14
Hancock, S., Stella, S., Cascio, D., and Johnson, R. (2016). DNA sequence determinants controlling affinity, stability and shape of DNA complexes bound by the nucleoid protein Fis. PLoS One 11:e0150189. doi: 10.1371/journal.pone.0150189
Hengen, P., Lyakhov, I., Stewart, L., and Schneider, T. (2003). Molecular flip-flops formed by overlapping Fis sites. Nucleic Acids Res. 31, 6663–6673. doi: 10.1093/nar/gkg877
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Ishihama, A. (2010). Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 34, 628–645. doi: 10.1111/j.1574-6976.2010.00227.x
Jaques, S., and McCarter, L. (2006). Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 188, 2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006
Jones, C., and Wozniak, D. (2017). Congo red stain identifies matrix overproduction and is an indirect measurement for c-di-GMP in many species of bacteria. Methods Mol. Biol. 1657, 147–156. doi: 10.1007/978-1-4939-7240-1_12
Kahramanoglou, C., Seshasayee, A. S. N., Prieto, A. I., Ibberson, D., Schmidt, S., Zimmermann, J., et al. (2011). Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39, 2073–2091. doi: 10.1093/nar/gkq934
Kalburge, S. S., Carpenter, M. R., Rozovsky, S., and Boyd, E. F. (2017). Quorum sensing regulators are required for metabolic fitness in Vibrio parahaemolyticus. Infect. Immun. 85, 930–916. doi: 10.1128/iai.00930-16
Karunakaran, R., Mauchline, T. H., Hosie, A. H., and Poole, P. S. (2005). A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology 151, 3249–3256. doi: 10.1099/mic.0.28311-0
Keane, O. M., and Dorman, C. J. (2003). The gyr genes of Salmonella enterica serovar Typhimurium are repressed by the factor for inversion stimulation, Fis. Mol. Gen. Genomics. 270, 56–65. doi: 10.1007/s00438-003-0896-1
Kelly, A., Goldberg, M. D., Carroll, R. K., Danino, V., Hinton, J. C. D., and Dorman, C. J. (2004). A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150, 2037–2053. doi: 10.1099/mic.0.27209-0
Kernell Burke, A., Guthrie, L. T. C., Modise, T., Cormier, G., Jensen, R. V., McCarter, L. L., et al. (2015). OpaR controls a network of downstream transcription factors in Vibrio parahaemolyticus BB22OP. PLoS One 10:e0121863. doi: 10.1371/journal.pone.0121863
Kim, Y. K., and McCarter, L. L. (2007). ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189, 4094–4107. doi: 10.1128/JB.01510-06
Klose, K. E., and Mekalanos, J. J. (1998). Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–520. doi: 10.1046/j.1365-2958.1998.00809.x
Kodama, T., Hiyoshi, H., Okada, R., Matsuda, S., Gotoh, K., and Iida, T. (2015). Regulation of Vibrio parahaemolyticus T3SS2 gene expression and function of T3SS2 effectors that modulate actin cytoskeleton. Cell. Microbiol. 17, 183–190. doi: 10.1111/cmi.12408
Lautier, T., and Nasser, W. (2007). The DNA nucleoid-associated protein Fis co-ordinates the expression of the main virulence genes in the phytopathogenic bacterium Erwinia chrysanthemi. Mol. Microbiol. 66, 1474–1490. doi: 10.1111/j.1365-2958.2007.06012.x
Lenz, D. H., and Bassler, B. L. (2007). The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 63, 859–871. doi: 10.1111/j.1365-2958.2006.05545.x
Liu, J., Li, F., and Rozovsky, S. (2013). The intrinsically disordered membrane protein selenoprotein S is a reductase in vitro. Biochemist 52, 3051–3061. doi: 10.1021/bi4001358
Lubin, J., Lewis, W., Gilbert, N., Weimer, C., Almagro-Moreno, S., Boyd, E., et al. (2015). Host-like carbohydrates promote bloodstream survival of Vibrio vulnificus in vivo. Infect. Immun. 83, 3126–3136. doi: 10.1128/IAI.00345-15
Lv, M., Chen, Y., Liao, L., Liang, Z., Shi, Z., Tang, Y., et al. (2018). Fis is a global regulator critical for modulation of virulence factor production and pathogenicity of Dickeya zeae. Sci. Rep. 8:341. doi: 10.1038/s41598-017-18578-2
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Mallik, P., Pratt, T. S., Beach, M. B., Bradley, M. D., Undamatla, J., and Osuna, R. (2004). Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J. Bacteriol. 186, 122–135. doi: 10.1128/JB.186.1.122-135.2004
McCarter, L. L. (1998). OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180, 3166–3173. doi: 10.1128/JB.180.12.3166-3173.1998
McCarter, L. (1999). The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1, 51–57.PMID:
McCarter, L., Hilmen, M., and Silverman, M. (1988). Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54, 345–351. doi: 10.1016/0092-8674(88)90197-3
McCarter, L. L., and Wright, M. E. (1993). Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J. Bacteriol. 175, 3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993
McDonald, N., DeMeester, K., Lewis, A., Grimes, C., and Boyd, E. (2018). Structural and functional characterization of a modified legionaminic acid involved in glycosylation of a bacterial lipopolysaccharide. J. Biol. Chem. 293, 19113–19126. doi: 10.1074/jbc.RA118.004966
McLeod, S. M., Aiyar, S. E., Gourse, R. L., and Johnson, R. C. (2002). The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 316, 517–529. doi: 10.1006/jmbi.2001.5391
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K., and Bassler, B. L. (2002). Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. doi: 10.1016/S0092-8674(02)00829-2
Miller, K. A., Tomberlin, K. F., and Dziejman, M. (2019). Vibrio variations on a type three theme. Curr. Opin. Microbiol. 47, 66–73. doi: 10.1016/j.mib.2018.12.001
Monteiro, L. M. O., Sanches-Medeiros, A., Westmann, C. A., and Silva-Rocha, R. (2020). Unraveling the complex interplay of Fis and IHF through synthetic promoter engineering. Front. Bioeng. Biotechnol. 8:510. doi: 10.3389/fbioe.2020.00510
Münch, R., Hiller, K., Barg, H., Heldt, D., Linz, S., Wingender, E., et al. (2003). PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31, 266–269. doi: 10.1093/nar/gkg037
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Nealson, K. H., Platt, T., and Hastings, J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322. doi: 10.1128/jb.104.1.313-322.1970
O’Boyle, N., and Boyd, A. (2014). Manipulation of intestinal epithelial cell function by the cell contact-dependent type III secretion systems of Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 3:114. doi: 10.3389/fcimb.2013.00114
Osuna, R., Lienau, D., Hughes, K. T., and Johnson, R. C. (1995). Sequence, regulation, and functions of fis in Salmonella typhimurium. J. Bacteriol. 177, 2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995
Ouafa, Z. A., Reverchon, S., Lautier, T., Muskhelishvili, G., and Nasser, W. (2012). The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 40, 4306–4319. doi: 10.1093/nar/gks014
Philippe, N., Alcaraz, J. P., Coursange, E., Geiselmann, J., and Schneider, D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. doi: 10.1016/j.plasmid.2004.02.003
Prigent-Combaret, C., Zghidi-Abouzid, O., Effantin, G., Lejeune, P., Reverchon, S., and Nasser, W. (2012). The nucleoid-associated protein Fis directly modulates the synthesis of cellulose, an essential component of pellicle-biofilms in the phytopathogenic bacterium Dickeya dadantii. Mol. Microbiol. 86, 172–186. doi: 10.1111/j.1365-2958.2012.08182.x
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Rutherford, S. T., van Kessel, J. C., Shao, Y., and Bassler, B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011
Schneider, D. A., Ross, W., and Gourse, R. L. (2003). Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6, 151–156. doi: 10.1016/S1369-5274(03)00038-9
Schneider, R., Travers, A., Kutateladze, T., and Muskhelishvili, G. (1999). A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34, 953–964. doi: 10.1046/j.1365-2958.1999.01656.x
Shao, Y., Feldman-Cohen, L., and Osuna, R. (2008). Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 376, 771–785. doi: 10.1016/j.jmb.2007.11.101
Simpson, C., Petersen, B., Haas, N., Geyman, L., Lee, A., Podicheti, R., et al. (2021). The quorum-sensing systems of Vibrio campbellii DS40M4 and BB120 are genetically and functionally distinct. Environ. Microbiol. 23, 5412–5432. doi: 10.1111/1462-2920.15602
Skoko, D., Yoo, D., Bai, H., Schnurr, B., Yan, J., McLeod, S., et al. (2006). Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 364, 777–798. doi: 10.1016/j.jmb.2006.09.043
Steen, J. A., Harrison, P., Seemann, T., Wilkie, I., Harper, M., Adler, B., et al. (2010). Fis is essential for capsule production in Pasteurella multocida and regulates expression of other important virulence factors. PLoS Pathog. 6:e1000750. doi: 10.1371/journal.ppat.1000750
Stewart, B. J., and McCarter, L. L. (2003). Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185, 4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Swift, S., Downie, J., Whitehead, N., Barnard, A., Salmond, G., and Williams, P. (2001). Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45, 199–270. doi: 10.1016/s0065-2911(01)45005-3
Trimble, M. J., and McCarter, L. L. (2011). Bis-(3'-5')-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U. S. A. 108, 18079–18084. doi: 10.1073/pnas.1113790108
Wang, H., Liu, B., Wang, Q., and Wang, L. (2013). Genome-wide analysis of the salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PLoS One 8:e64688. doi: 10.1371/journal.pone.0064688
Weinstein-Fischer, D., and Altuvia, S. (2007). Differential regulation of Escherichia coli topoisomerase I by Fis. Mol. Microbiol. 63, 1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x
Whitaker, W. B., Parent, M. A., Boyd, A., Richards, G. P., and Boyd, E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80, 1834–1845. doi: 10.1128/IAI.06284-11
Whitaker, W. B., Richards, G. P., and Boyd, E. F. (2014). Loss of sigma factor RpoN increases intestinal colonization of Vibrio parahaemolyticus in an adult mouse model. Infect. Immun. 82, 544–556. doi: 10.1128/IAI.01210-13
Yang, H., de Souza Santos, M., Lee, J., Law, H. T., Chimalapati, S., Verdu, E. F., et al. (2019). A novel mouse model of enteric Vibrio parahaemolyticus infection reveals that the type III secretion system 2 effector VopC plays a key role in tissue invasion and gastroenteritis. mBio 10, 2608–2619. doi: 10.1128/mBio.02608-19
Keywords: Fis, quorum sensing, motility, metabolism, swarming
Citation: Tague JG, Regmi A, Gregory GJ and Boyd EF (2021) Fis Connects Two Sensory Pathways, Quorum Sensing and Surface Sensing, to Control Motility in Vibrio parahaemolyticus . Front. Microbiol. 12:669447. doi: 10.3389/fmicb.2021.669447
Edited by:
Felipe Cava, Umeå University, SwedenReviewed by:
Tom Defoirdt, Ghent University, BelgiumSteven Rutherford, Genentech, Inc., United States
Copyright © 2021 Tague, Regmi, Gregory and Boyd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: E. Fidelma Boyd, ZmJveWRAdWRlbC5lZHU=
 Jessica G. Tague
Jessica G. Tague Abish Regmi
Abish Regmi Gwendolyn J. Gregory
Gwendolyn J. Gregory E. Fidelma Boyd
E. Fidelma Boyd