
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 25 May 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.669396
This article is part of the Research TopicRoles of Regulatory RNAs in Bacterial PathogensView all 13 articles
Bacteria adapt to the different environments encountered by rapid and tightly controlled regulations involving complex networks. A first line of control is transcriptional with regulators such as two-component systems (TCSs) that respond to physical and chemical perturbations. It is followed by posttranscriptional regulations in which small regulatory RNAs (sRNAs) may affect RNA translation. Streptococci are opportunistic pathogens for humans and farm animals. The TCS CiaRH is highly conserved among this genus and crucial in bacterial survival under stressful conditions. In several streptococcal species, some sRNAs belong to the CiaRH regulon and are called csRNAs for cia-dependent sRNAs. In this review, we start by focusing on the Streptococcus species harboring a CiaRH TCS. Then the role of CiaRH in streptococcal pathogenesis is discussed in the context of recent studies. Finally, we give an overview of csRNAs and their functions in Streptococci with a focus on their importance in bacterial adaptation and virulence.
Due to their importance in the regulation of gene expression, small non-coding regulatory RNAs (sRNAs) are present in all kingdoms of life. The sRNAs were discovered in prokaryotes long before the first short interfering RNAs (siRNAs) and microRNAs (miRNAs) in eukaryotes. Adaptation to the environment involves a complex regulatory network in which sRNAs play an essential role. A decade ago, the high number of sRNAs discovered in various bacterial species was surprising (Brantl, 2009). Interestingly, these sRNAs differ in length, structure, and mode of action (Gottesman and Storz, 2011). However, sRNAs, 50–500 nucleotides long molecules, are often involved in the regulation of several cellular pathways and allow bacteria to adapt and survive under stressful conditions. All sRNAs are classified in several groups according to their location in the genome and their modes of action (Storz et al., 2011). In 1984, the first chromosomally encoded sRNA was discovered in Escherichia coli: MicF. This sRNA inhibits the translation of OmpF messenger RNA (mRNA) encoding the major membrane porin, OmpF (Mizuno et al., 1984). To respond to environmental changes, bacteria must first sense these changes, and two-component regulatory systems (TCS) are known to perform this function (Stock et al., 2000).
Streptococcal species infect humans and farm animals. Although some of them are commensal, other are responsible for severe infections in humans (Krzyściak et al., 2013). In Streptococci, many TCSs have been found. The TCS CiaRH was identified to be involved in natural competence and general virulence (Patenge et al., 2013). It is widespread among Streptococci but not found in another bacterial genus. Interestingly, it controls the expression of sRNAs called cia-dependent sRNAs (csRNAs) (Halfmann et al., 2007). This review concerns csRNAs identified in streptococci. It starts by highlighting the most important streptococcal species harboring a CiaRH and then analyzes the CiaRH TCS roles. Finally, this review focuses on all csRNAs identified until now and their functions.
The Streptococcus genus is composed of chain-forming gram-positive bacteria including a large number of species (>100). Although this genus includes beneficial species such as Streptococcus thermophilus, used in the food industry for the production of yogurt (Blomqvist et al., 2006), streptococci are opportunistic pathogens, often involved in severe diseases in humans and farm animals. The major species in human infections are Streptococcus pneumoniae, S. pyogenes, and S. agalactiae (Krzyściak et al., 2013). S. pneumoniae is the main cause of community-acquired pneumonia, meningitis, and acute otitis media. S. pyogenes (Group A Streptococcus), an exclusively human pathogen, is involved in mild (pharyngitis, skin infections) to severe fatal invasive infections, such as necrotizing fasciitis and streptococcal toxic shock syndrome. Groups C and G Streptococci, such as Streptococcus dysgalactiae subsp. equisimilis and Streptococcus equi, are microbiologically similar to S. pyogenes (Baracco, 2019). S. agalactiae (Group B Streptococcus), a commensal bacterium of the gastrointestinal and genitourinary tracts, is the leading cause of neonatal infections, causing pneumonia, bacteremia, and meningitis via maternal transmission. As S. pneumoniae, Streptococcus mutans, Streptococcus sanguinis, Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis belong to the physiological flora in the human oral cavity. S. mutans is an opportunistic commensal species responsible for biofilm formation causing dental caries but also infective endocarditis. Conversely, S. gordonii and S. sanguinis are non-cariogenic colonizers. Streptococcus gallolyticus (and less frequently Streptococcus lutetiensis), an opportunistic bacterium inhabiting the gastrointestinal tract, is one of the main causes of infective endocarditis and is strongly associated with colorectal cancer (Pasquereau-Kotula et al., 2018). Among Streptococci, including S. agalactiae and S. equi mentioned above, some species can also infect animals, as Streptococcus suis, responsible for severe invasive, and often lethal diseases in swine and humans and Streptococcus uberis, main agent of mastitis in dairy cows (Keane, 2019). These bacteria must colonize, invade, and persist in the host. But above all, they must adapt to environmental changes and the various types of stress they encounter. One of the mechanisms that bacteria use to adapt and survive is the regulation of gene expression through the sRNA-mediated two-component regulatory systems.
TCS CiaRH was first identified in S. pneumoniae while selecting for cefotaxime resistance in spontaneous laboratory mutants. CiaH is a histidine protein kinase anchored in the cytoplasmic membrane that receives information from the environment. It transmits the information to CiaR, a cytoplasmic response regulator that translates the signal into a cellular response by regulating the expression of targeted genes (Figure 1; Guenzi et al., 1994). The amino-acid sequence identity of CiaH and CiaR from different species ranges between 48–71 and 77–85%, respectively (Riani et al., 2007). In several species, CiaRH is involved in biofilm formation. In fact, the presence of SpeA (streptococcal pyogenic exotoxin A) in S. pyogenes leads to down-regulation of CiaRH expression genes and attenuates the biofilm-forming capacity, suggesting a link between TCS expression and biofilm formation (Babbar et al., 2019). In S. sanguinis, the deletion of the ciaR gene up-regulates the expression of arginine biosynthesis genes resulting in the formation of a fragile biofilm (Zhu et al., 2017). In S. gordonii, the inactivation of SdbA, a thiol-disulfide oxidoreductase, up-regulates CiaRH, which in turn leads to enhanced biofilm formation (Davey et al., 2016a). In S. mutans, the inactivation of CiaH gene affects the dental biofilm formation, abolishes bacteriocin production and competence development, suggesting the involvement of CiaRH in these phenotypes (Qi et al., 2004). Actually, the up-regulation of CiaRH in sdbA mutant S. gordonii strain leads to bacteriocin expression shutdown whereas inactivation of CiaRH restores bacteriocin production. Involvement of the TCS in bacteriocin expression indicates its importance in bacterial competition in order to colonize the host (Davey et al., 2016b). CiaRH is also known to influence streptococcal stress tolerance. TCS is involved in tolerance to acid and thermal stress in S. mutans (Liu and Burne, 2009b). In S. gordonii, mutation of the TCS leads a greater susceptibility of the mutant to low pH and oxidative stress than the wild type (Liu and Burne, 2009a). Moreover, CiaRH is involved in resistance to the immune system, intracellular survival, and virulence. Actually, in CiaR-deficient S. agalactiae strains, resistance to the immune system and intracellular survival are affected (Quach et al., 2009; Mu et al., 2016). The deletion of CiaRH in S. suis enhances the bactericidal activity of macrophages and attenuates bacterial virulence in animal models (Li et al., 2011; Zaccaria et al., 2016). Furthermore, the transcription level of the TCS is significantly higher in virulent than in strains of low virulence (Dong et al., 2015). As for S. pneumoniae, the CiaRH system prevents autolysis triggered by different conditions and allows the maintenance of a stationary growth phase (Mascher et al., 2006). In S. pyogenes, a ciaH mutant strain binds more efficiently to HEp-2 epithelial cells than the wild type. The internalization rate of the mutant is increased within the same range (Riani et al., 2007). Conversely, the deletion of CiaRH in S. suis exhibits a decrease in adherence to HEp-2 epithelial cells (Li et al., 2011). These conflicting results could be explained in two different ways. First, CiaRH-mediated regulation can be different between streptococcal species and under different conditions. Second, inactivation of only one gene of the TCS (ciaH) does not allow for obtaining the same phenotype as when both are inactive. In fact, when ciaH is inactive, ciaR may respond to other regulators whereas when only ciaR is inactive, ciaH may regulate other sensors. To summarize, CiaRH is involved in many cellular processes, including β-lactam resistance, lytic processes, biofilm formation, bacteriocin production, natural competence, virulence, and resistance to the immune system (Dagkessamanskaia et al., 2004; Sebert et al., 2005; Quach et al., 2009; Li et al., 2011). In S. pneumoniae, a direct repeat sequence, TTTAAG-N5-TTTAAG, has been identified by in vitro and in vivo transcriptional mapping as essential for the binding of CiaR. Fifteen promoters are controlled by CiaR, five of them (the strongest) drive the expression of sRNAs, which are designated csRNAs for cia-dependent sRNAs (Halfmann et al., 2007).
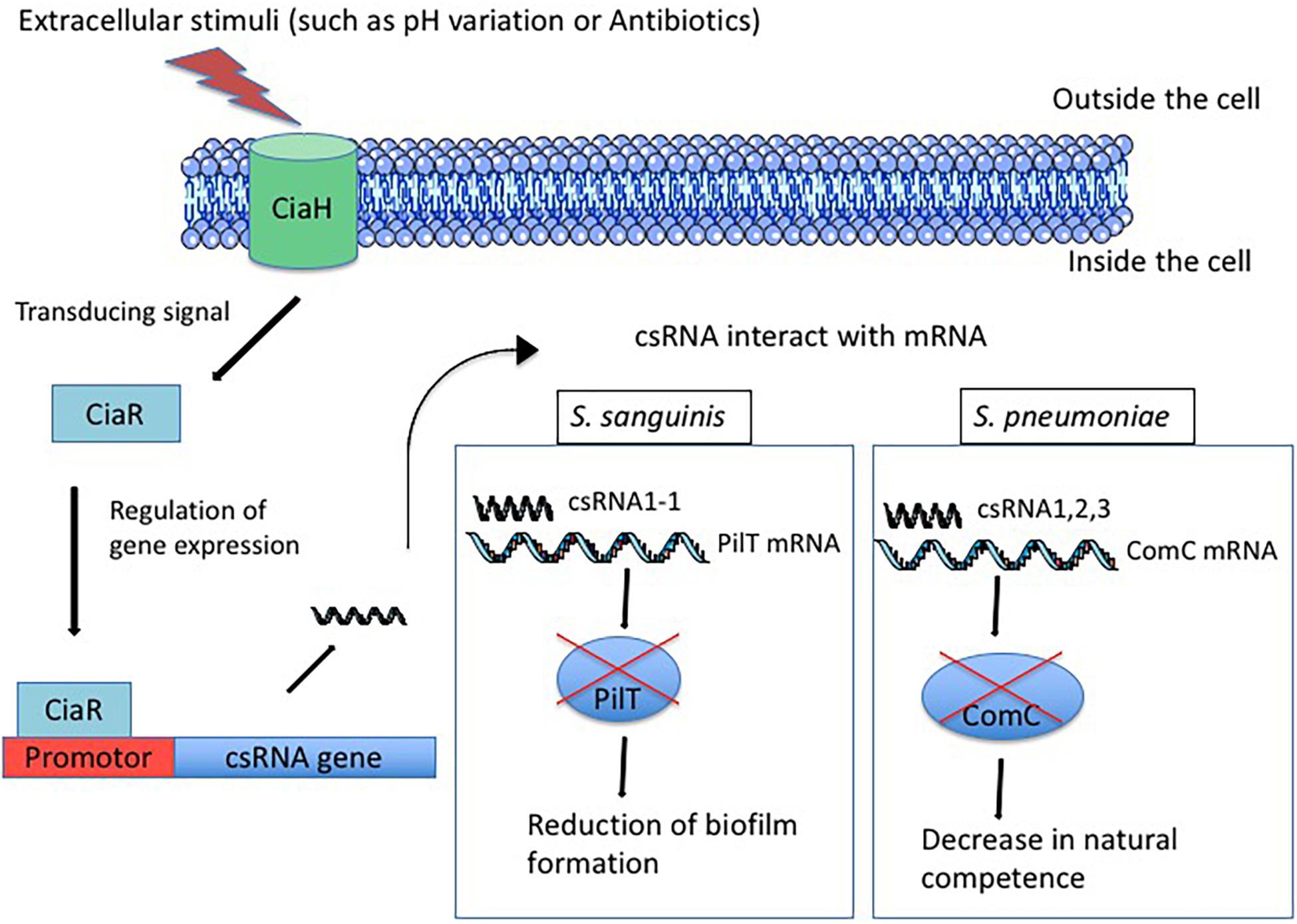
Figure 1. Regulation of csRNAs expression by TCS CiaRH after exposure of the cell to extracellular stimuli. First, the sensor CiaH detects extracellular stimuli such as pH variation and antibiotics exposure. CiaH transduces the signal to the regulator CiaR that interacts with promotor and allows csRNA expression. Then, csRNA interacts with target mRNA and allows or represses the translation. For example, in S. sanguinis, the csRNA1-1 interacts with the PilT mRNA and represses the translation of PilT protein inducing the reduction of biofilm formation. In S. pneumoniae, csRNA1 combined with csRNA2 and csRNA3 allows the repression of ComC translation, which reduces the natural competence of bacteria.
The sRNAs are classified into four major groups: CRISPRs (clustered regulatory interspaced, short palindromic repeats), riboswitches, protein-binding RNAs, and trans-acting RNAs. And the csRNAs belong to the trans-acting class and more precisely to the trans-encoded subclass. Trans-encoded RNAs are often expressed by genes located at a different locus from their target genes and thus share only a short and imperfect complementarity sequence with their target mRNAs. This imperfect complementarity allows trans-encoded RNAs to control more than one target mRNA and, therefore, to be part of complex regulatory networks. Trans-encoded RNAs, by forming a base association, affect the translation or the stability of the mRNAs. The interaction between an sRNA and its target mRNA leads to the inhibition of protein translation by blocking the ribosome binding site (RBS) or by inducing the degradation of the sRNA-mRNA duplex by RNases (Hui et al., 2014). Trans-encoded RNAs can also prevent mRNA degradation or activate mRNA translation by preventing the formation of an inhibitory structure that hides the RBS (Prévost et al., 2007). The FasX sRNA of S. pyogenes allows bacterial dissemination through two different mechanisms: first, by increasing ska mRNA stability, allowing the overexpression of streptokinase, and second, by hiding the RBS and decreasing pilus mRNA translation (Ramirez-Peña et al., 2010; Danger et al., 2015).
The five csRNAs, discovered in S. pneumoniae and validated by Northern blot analyses, are between 87 and 151 nucleotides long and have a high degree of similarity to each other. The presence of sequences complementary to RBS in these csRNAs indicates that they may control the initiation of translation. csRNA1 was shown to be a negative regulator of competence development (Tsui et al., 2010). The deletion of csRNA4 and csRNA5 revealed their role in autolysis control (Halfmann et al., 2007), and a mutant of csRNA5 was defective in a lung infection (Mann et al., 2012). The targets of these csRNAs were investigated by computational predictions with targetRNA and IntaRNA (Tjaden et al., 2006; Busch et al., 2008). Thirty-three predicted genes were tested by translational fusion, and six of them are possibly regulated by S. pneumoniae csRNAs (Schnorpfeil et al., 2013). The spr0081, spr0371, spr0551, and spr1097 genes encode membrane-spanning proteins that belong to different transporter families. The spr0159 gene encodes a protein harboring a DNA-binding domain and therefore is most likely to be a transcriptional regulator. The last one, spr2043 (ComC), encodes the competence-stimulating peptide precursor (CSP), suggesting a link between CiaRH and competence control, mediated by csRNAs (Figure 1; Håvarstein et al., 1995; Schnorpfeil et al., 2013). It has been shown that each of the csRNAs down-regulates the comC gene, but they are not as effective alone as they are all together (Schnorpfeil et al., 2013). However, the combination of three csRNAs, csRNA1, 2, 3, or csRNA1, 2, 4, is sufficient to decrease the competence of S. pneumoniae (Laux et al., 2015). Interestingly, duplicated csRNA was observed in Hungarian S. pneumoniae serotype 19A isolate. Indeed, an internal sequence duplication is the cause of the carriage and expression of longer version of csRNA5 (Brantl and Brückner, 2014).
In the study of Marx et al. (2010), the presence of the CiaR binding site located in the intergenic regions and followed by transcriptional terminator was investigated in 14 streptococcal genomes. Thus, 61 candidate genes potentially express csRNAs. Among them, four were predicted in all S. agalactiae strains: csRNA10, csRNA11, csRNA12, and csRNA13. Their expression was confirmed in NEM316 by RNA sequencing and the first three were also validated by Northern blot. The genes were renamed srn015, srn024, srn070, and srn085, respectively. The corresponding csRNAs were overexpressed at low pH (5.2), suggesting they could contribute to acid stress resistance (Rosinski-Chupin et al., 2015). However, no function and no targets have been assigned to them yet.
In S. sanguinis SK36, six csRNAs were predicted (csRNA1-1, csRNA1-2, csRNA1-3, csRNA2, csRNA7, and csRNA8) and confirmed by Northern blot (Marx et al., 2010). Target prediction and a luciferase reporter assay allowed the identification of the pilT gene, a constituent of the type IV pilus gene cluster, to be the target of S. sanguinis csRNA1-1. The interaction between csRNA1-1 and the pilT mRNA was proved by RNA-RNA electrophoretic mobility shift assay (EMSA). Furthermore, csRNA1-1 and csRNA1-2 arranged in tandem in S. sanguinis genome are probably duplicated genes that negatively regulate biofilm formation (Ota et al., 2017; Figure 1). This observation suggests the implication of csRNAs in the host colonization by this species and provides further evidence concerning the involvement of csRNAs in bacterial adaptation.
The csRNA genes in S. pyogenes MGAS315 (csRNA14, csRNA15, and csRNA25) were also predicted, and the expression of csRNA15 and csRNA25 was confirmed by RNA sequencing (Marx et al., 2010; Le Rhun et al., 2016).
Five csRNAs in S. mitis B6 (csRNA1, csRNA2, csRNA3, csRNA4, and csRNA5) and five other csRNAs in S. oralis Uo5 (csRNA1, csRNA2, csRNA3, csRNA4, and csRNA6) were predicted and confirmed by Northern blot (Marx et al., 2010).
Other csRNAs have been predicted in S. mutans UA159, S. suis 05ZYH33, S. gordonii, S. gallolyticus, S. dysgalactiae, S. equi, S. uberis, and S. thermophilus but not confirmed so far (Marx et al., 2010; Table 1 and Supplementary Table 1). The identification of csRNAs of new viridans streptococci obtained from primates indicates that all csRNAs predicted previously in S. mitis, S. gallolyticus, S. gordonii, and S. oralis are present in the new strains studied. Two species with unknown csRNAs contain csRNAs from other species. Indeed, S. infantis harbors four of the five S. oralis Uo5 csRNAs, and S. lutetiensis harbors the S. gallolyticus UCN34 csRNAs except for csRNA40 (Denapaite et al., 2016).
Except for S. pneumoniae and S. sanguinis, few studies regarding the role and targets of these csRNAs in other streptococci were conducted, although the importance of RNAs is highlighted.
The aim of this review is to carry out an inventory of the sRNAs regulated by the two-component regulatory system CiaRH present in streptococci. CiaRH TCS is conserved in all streptococci and controls many cellular processes including natural competence, virulence, and resistance to the immune system (Dagkessamanskaia et al., 2004; Sebert et al., 2005; Quach et al., 2009; Li et al., 2011). The csRNAs increase the regulatory networks of CiaR, which already directly controls more than 20 other genes (Halfmann et al., 2007). Promoters that drive the expression of the five csRNAs of S. pneumoniae are strongest in the CiaR regulon (Halfmann et al., 2007). The high proportion of sRNAs compared with other genes controlled by CiaRH indicates the importance of these csRNAs in bacterial regulation. Although csRNAs are predicted in various Streptococcus species and their importance highlighted, for most of them, no role or target has been identified until now. So far, only the csRNAs of S. pneumoniae and S. sanguinis have been investigated (Schnorpfeil et al., 2013; Laux et al., 2015; Ota et al., 2017). The study of csRNAs in those species has allowed the identification of different metabolic pathways in which csRNAs may be involved. Indeed, S. pneumoniae harbors five csRNAs, all implicated in competence development and thus, probably in horizontal transfer (Halfmann et al., 2007; Tsui et al., 2010). Moreover, two S. pneumoniae csRNAs (csRNA4 and csRNA5) seem to control bacterial autolysis (Halfmann et al., 2007). The involvement of csRNA5 in lung infection as well shows that each csRNA may be involved in different regulatory pathways (Mann et al., 2012). In this case, csRNA5 is on the one hand involved in competence development and on the other hand in virulence.
The investigation of S. pneumoniae csRNAs targets also allowed identifying different regulation pathways. According to the competence regulation previously mentioned, one target (ComC), encoding the competence-stimulating peptide precursor (CSP) was identified. This identification adds a proof concerning the involvement of S. pneumoniae csRNAs in horizontal transmission pathways. The Spr0159 target is most likely a transcriptional regulator: this suggests the involvement of the csRNAs in complex regulatory networks. Other identified targets (spr0081, spr0371, spr0551, and spr1097), encoding membrane spanning, belonging to different transporter families, indicate the possible involvement of csRNAs in stress resistance (Schnorpfeil et al., 2013). The four csRNAs identified in S. agalactiae NEM316 strain (srn015, srn024, srn070, and srn085) are overexpressed at low pH (5.2), suggesting their role in acid stress resistance (Rosinski-Chupin et al., 2015). Thus, the possible implication of csRNAs in stress tolerance in S. pneumoniae and S. agalactiae reveals a new regulation pathway in which csRNAs may play a role. In S. sanguinis, csRNAs are involved in host colonization by biofilm formation (Ota et al., 2017). This regulation of colonization by csRNAs has not yet been observed in other streptococcal species. Analysis of S. pneumoniae, S. agalactiae, and S. sanguinis csRNAs demonstrates that they are involved in a wide range of regulatory pathways. Indeed, the colonization, the virulence, the horizontal transfer, and maybe the resistance to environmental stress is affected by csRNAs. The various regulatory pathways in which csRNAs are involved can be explained by the diversity of csRNAs in each species and between streptococcus species. Moreover, as observed in S. pneumoniae, one csRNA can be involved in different regulatory pathways, thus increasing the complexity of regulatory networks.
The diversity of csRNAs between streptococcus species is remarkable (Table 1 and Supplementary Table 1). However, some species contain csRNAs from other species (S. infantis harbors csRNAs of S. oralis Uo5 and S. lutetiensis harbors the S. gallolyticus UCN34 csRNAs) (Denapaite et al., 2016). Moreover, S. oralis strains contain duplicated csRNAs genes. A genetic island of four genes is present between them but absent in strains without csRNAs gene duplication. Furthermore, this genetic island is integrated in S. infantis DD18 between two csRNAs (Denapaite et al., 2016). These data suggest that csRNAs are not only involved in gene regulation but may also contribute to horizontal gene transfers improving bacterial adaptation.
In some species, csRNAs display a high degree of similarity to each other (Figure 2). This similarity is observed more particularly in regions complementary to RBSs and AUG start codons, suggesting that csRNAs bind to mRNA target and inhibit translational initiation. This would be fully consistent with the regulatory mechanism most commonly associated with sRNAs (Gottesman and Storz, 2011; Storz et al., 2011).

Figure 2. Alignments of csRNAs sequences by species by MultAlin. Nucleotides in red correspond to highly conserved sequences. Nucleotides in blue correspond to conserved sequences. Nucleotides in black correspond to non-conserved sequences.
Other bacterial regulators also control multiple sRNA genes. For example, LuxO of Vibrio harveyi controls the expression of five sRNA genes (Tu and Bassler, 2007). The presence of regulators involved in sRNA regulation in various species suggests the importance of these sRNAs in bacterial adaptation and, beyond that, in bacterial survival.
Virulence and resistance to antibiotics and to the immune system mediated by CiaRH are possibly carried out through csRNAs. The discovery of Cbf1 protein that stabilizes all csRNAs in S. pneumoniae provides additional proof of the importance of csRNAs (Hör et al., 2020). In conclusion, understanding the csRNA-dependent regulatory network may contribute to the development of strategies against bacterial infections by targeting these sRNAs (Warner et al., 2018).
NJ and M-FL wrote the manuscript. Both authors contributed to the article and approved the submitted version.
NJ was supported by a doctoral fellowship of the Région Centre (France).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Philippe Bouloc for his critical reading of the manuscript and helpful discussion. We thank the reviewers for their critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.669396/full#supplementary-material
Supplementary Table 1 | Streptococcus csRNAs sequences.
Babbar, A., Barrantes, I., Pieper, D. H., and Itzek, A. (2019). Superantigen SpeA attenuates the biofilm forming capacity of Streptococcus pyogenes. J. Microbiol. 57, 626–636. doi: 10.1007/s12275-019-8648-z
Baracco, G. J. (2019). Infections caused by group C and G Streptococcus (Streptococcus dysgalactiae subsp. equisimilis and others): epidemiological and clinical aspects. Microbiol. Spectr. 7, 1–2. doi: 10.1128/microbiolspec.GPP3-0016-2018
Blomqvist, T., Steinmoen, H., and Håvarstein, L. S. (2006). Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 72, 6751–6756. doi: 10.1128/AEM.01156-06
Brantl, S. (2009). Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 4, 85-103. doi: 10.2217/17460913.4.1.85
Brantl, S., and Brückner, R. (2014). Small regulatory RNAs from low-GC gram-positive bacteria. RNA Biol. 11, 443-456. doi: 10.4161/rna.28036
Busch, A., Richter, A. S., and Backofen, R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinforma 24, 2849–2856. doi: 10.1093/bioinformatics/btn544
Dagkessamanskaia, A., Moscoso, M., Hénard, V., Guiral, S., Overweg, K., Reuter, M., et al. (2004). Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51, 1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x
Danger, J. L., Makthal, N., Kumaraswami, M., and Paul Sumby, P. (2015). The FasX small regulatory RNA negatively regulates the expression of two fibronectin-binding proteins in group A Streptococcus. J. Bacteriol. 197, 3720–3730. doi: 10.1128/JB.00530-15
Davey, L., Halperin, S. A., and Lee, S. F. (2016a). Mutation of the Streptococcus gordonii thiol-disulfide oxidoreductase SdbA leads to enhanced biofilm formation mediated by the CiaRH two-component signaling system. PLoS One 11:e0166656. doi: 10.1371/journal.pone.0166656
Davey, L., Halperin, S. A., and Lee, S. F. (2016b). Mutation of the Thiol-Disulfide Oxidoreductase SdbA Activates the CiaRH two-component system, leading to bacteriocin expression shutdown in Streptococcus gordonii. J. Bacteriol. 198, 321–331. doi: 10.1128/JB.00800-15
Denapaite, D., Rieger, M., Köndgen, S., Brückner, R., Ochigava, I., Kappeler, P., et al. (2016). Highly variable Streptococcus oralis strains are common among viridans Streptococci isolated from primates. mSphere 1:e00041-15. doi: 10.1128/mSphere.00041-15
Dong, W., Ma, J., Zhu, Y., Zhu, J., Yuan, L., Wang, Y., et al. (2015). Virulence genotyping and population analysis of Streptococcus suis serotype 2 isolates from China. Infect. Genet. Evol. 36, 483–489. doi: 10.1016/j.meegid.2015.08.021
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Guenzi, E., Gasc, A. M., Sicard, M. A., and Hakenbeck, R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12, 505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x
Halfmann, A., Kovács, M., Hakenbeck, R., and Brückner, R. (2007). Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66, 110–126. doi: 10.1111/j.1365-2958.2007.05900.x
Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Hör, J., Garriss, G., Di Giorgio, S., Hack, L. M., Vanselow, J. T., Förstner, K. U., et al. (2020). Grad-seq in a Gram-positive bacterium reveals exonucleolytic sRNA activation in competence control. EMBO J. 39:e103852. doi: 10.15252/embj.2019103852
Hui, M. P., Foley, P. L., and Belasco, J. G. (2014). Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 48, 537–559. doi: 10.1146/annurev-genet-120213-092340
Keane, O. M. (2019). Symposium review: Intramammary infections-Major pathogens and strain-associated complexity. J. Dairy Sci. 102, 4713–4726. doi: 10.3168/jds.2018-15326
Krzyściak, W., Pluskwa, K. K., Jurczak, A., and Kościelniak, D. (2013). The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1361–1376. doi: 10.1007/s10096-013-1914-9
Laux, A., Sexauer, A., Sivaselvarajah, D., Kaysen, A., and Brückner, R. (2015). Control of competence by related non-coding csRNAs in Streptococcus pneumoniae R6. Front. Genet. 6:246. doi: 10.3389/fgene.2015.00246
Le Rhun, A., Beer, Y. Y., Reimegård, J., Chylinski, K., and Charpentier, E. (2016). RNA sequencing uncovers antisense RNAs and novel small RNAs in Streptococcus pyogenes. RNA Biol. 13, 177–195. doi: 10.1080/15476286.2015.1110674
Li, J., Tan, C., Zhou, Y., Fu, S., Hu, L., Hu, J., et al. (2011). The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet. Microbiol. 148, 99–104. doi: 10.1016/j.vetmic.2010.08.005
Liu, Y., and Burne, R. A. (2009a). Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J. Bacteriol. 191, 7353–7362. doi: 10.1128/JB.01053-09
Liu, Y., and Burne, R. A. (2009b). Multiple two-component systems of Streptococcus mutans regulate agmatine deiminase gene expression and stress tolerance. J. Bacteriol. 191, 7363–7366. doi: 10.1128/JB.01054-09
Mann, B., van Opijnen, T., Wang, J., Obert, C., Wang, Y.-D., Carter, R., et al. (2012). Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 8:e1002788. doi: 10.1371/journal.ppat.1002788
Marx, P., Nuhn, M., Kovács, M., Hakenbeck, R., and Brückner, R. (2010). Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 11:661. doi: 10.1186/1471-2164-11-661
Mascher, T., Heintz, M., Zähner, D., Merai, M., and Hakenbeck, R. (2006). The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188, 1959–1968. doi: 10.1128/JB.188.5.1959-1968.2006
Mizuno, T., Chou, M. Y., and Inouye, M. (1984). A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U.S.A. 81, 1966–1970. doi: 10.1073/pnas.81.7.1966
Mu, R., Cutting, A. S., Del Rosario, Y., Villarino, N., Stewart, L., Weston, T. A., et al. (2016). Identification of CiaR regulated genes that promote group B Streptococcal virulence and interaction with brain endothelial cells. PLoS One 11:e153891. doi: 10.1371/journal.pone.0153891
Ota, C., Morisaki, H., Nakata, M., Arimoto, T., Fukamachi, H., Kataoka, H., et al. (2017). Streptococcus sanguinis noncoding cia-dependent small RNAs negatively regulate expression of Type IV Pilus retraction ATPase PilT and biofilm formation. Infect. Immun. 86:e00894-17. doi: 10.1128/IAI.00894-17
Pasquereau-Kotula, E., Martins, M., Aymeric, L., and Dramsi, S. (2018). Significance of Streptococcus gallolyticus subsp. gallolyticus association with colorectal cancer. Front. Microbiol. 9:614. doi: 10.3389/fmicb.2018.00614
Patenge, N., Fiedler, T., and Kreikemeyer, B. (2013). Common regulators of virulence in streptococci. Curr. Top. Microbiol. Immunol. 368, 111–153. doi: 10.1007/82_2012_295
Pichon, C., du Merle, L., Caliot, M. E., Trieu-Cuot, P., and Le Bouguénec, C. (2012). An in silico model for identification of small RNAs in whole bacterial genomes: characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Res. 40, 2846–2861. doi: 10.1093/nar/gkr1141
Prévost, K., Salvail, H., Desnoyers, G., Jacques, J.-F., Phaneuf, E., and Massé, E. (2007). The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 64, 1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x
Qi, F., Merritt, J., Lux, R., and Shi, W. (2004). Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72, 4895–4899. doi: 10.1128/IAI.72.8.4895-4899.2004
Quach, D., van Sorge, N. M., Kristian, S. A., Bryan, J. D., Shelver, D. W., and Doran, K. S. (2009). The CiaR response regulator in group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J. Bacteriol. 191, 2023–2032. doi: 10.1128/JB.01216-08
Ramirez-Peña, E., Treviño, J., Liu, Z., Perez, N., and Sumby, P. (2010). The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska MRNA transcript. Mol. Microbiol. 78, 1332-1347. doi: 10.1111/j.1365-2958.2010.07427.x
Riani, C., Standar, K., Srimuang, S., Lembke, C., Kreikemeyer, B., and Podbielski, A. (2007). Transcriptome analyses extend understanding of Streptococcus pyogenes regulatory mechanisms and behavior toward immunomodulatory substances. Int. J. Med. Microbiol. 297, 513–523. doi: 10.1016/j.ijmm.2007.04.005
Rosinski-Chupin, I., Sauvage, E., Sismeiro, O., Villain, A., Da Cunha, V., Caliot, M.-E., et al. (2015). Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics 16:419. doi: 10.1186/s12864-015-1583-4
Schnorpfeil, A., Kranz, M., Kovács, M., Kirsch, C., Gartmann, J., Brunner, I., et al. (2013). Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol. Microbiol. 89, 334–349. doi: 10.1111/mmi.12277
Sebert, M. E., Patel, K. P., Plotnick, M., and Weiser, J. N. (2005). Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187, 3969–3979. doi: 10.1128/JB.187.12.3969-3979.2005
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Tjaden, B., Goodwin, S. S., Opdyke, J. A., Guillier, M., Fu, D. X., Gottesman, S., et al. (2006). Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 34, 2791–2802. doi: 10.1093/nar/gkl356
Tsui, H.-C. T., Mukherjee, D., Ray, V. A., Sham, L.-T., Feig, A. L., and Winkler, M. E. (2010). Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192, 264–279. doi: 10.1128/JB.01204-09
Tu, K. C., and Bassler, B. L. (2007). Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 21, 221–233. doi: 10.1101/gad.1502407
Warner, K. D., Hajdin, C. E., and Weeks, K. M. (2018). Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 17, 547–558. doi: 10.1038/nrd.2018.93
Zaccaria, E., Cao, R., Wells, J. M., and van Baarlen, P. (2016). A Zebrafish larval model to assess virulence of porcine Streptococcus suis strains. PLoS One 11:e0151623. doi: 10.1371/journal.pone.0151623
Keywords: Streptococci, regulation, CiaRH, regulatory RNAs, csRNAs
Citation: Jabbour N and Lartigue M-F (2021) An Inventory of CiaR-Dependent Small Regulatory RNAs in Streptococci. Front. Microbiol. 12:669396. doi: 10.3389/fmicb.2021.669396
Received: 18 February 2021; Accepted: 30 March 2021;
Published: 25 May 2021.
Edited by:
Olga Soutourina, UMR 9198 Institut de Biologie Intégrative de la Cellule (I2BC), FranceReviewed by:
Nadja Patenge, University of Rostock, GermanyCopyright © 2021 Jabbour and Lartigue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie-Frédérique Lartigue, bGFydGlndWVAdW5pdi10b3Vycy5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.