- 1Division of Bacterial and Parasitic Disease, National Institute of Animal Health, National Agriculture and Food Research Organization, Tsukuba, Japan
- 2Advanced Analysis Center, National Agriculture and Food Research Organization, Tsukuba, Japan
- 3Institute of Crop Science, National Agriculture and Food Research Organization, Tsukuba, Japan
- 4The United Graduate School of Veterinary Sciences, Gifu University, Gifu, Japan
American foulbrood (AFB) is the most serious bacterial disease of honey bee brood. Spores of the causative agent Paenibacillus larvae are ingested by bee larvae via brood foods and germinated cells proliferate in the larval midgut. In Japan, a macrolide antibiotic, tylosin, is used as the approved prophylactic for AFB. Although tylosin-resistant P. larvae has yet to be found in Japan, it may emerge in the future through the acquisition of macrolide resistance genes from other bacteria, and bacteria latent in brood foods, such as honey, may serve as a source of resistance genes. In this study, to investigate macrolide resistance genes in honey, we attempted to isolate tylosin-resistant bacteria from 53 Japanese honey samples and obtained 209 isolates from 48 samples in the presence of 1 μg/ml of tylosin. All isolates were Gram-positive spore-forming bacteria mainly belonging to genera Bacillus and Paenibacillus, and 94.3% exhibited lower susceptibility to tylosin than Japanese P. larvae isolates. Genome analysis of 50 representative isolates revealed the presence of putative macrolide resistance genes in the isolates, and some of them were located on mobile genetic elements (MGEs). Among the genes on MGEs, ermC on the putative mobilizable plasmid pJ18TS1mac of Oceanobacillus strain J18TS1 conferred tylosin and lincomycin resistance to P. larvae after introducing the cloned gene using the expression vector. Moreover, pJ18TS1mac was retained in the P. larvae population for a long period even under non-selective conditions. This suggests that bacteria in honey is a source of genes for conferring tylosin resistance to P. larvae; therefore, monitoring of bacteria in honey may be helpful to predict the emergence of tylosin-resistant P. larvae and prevent the selection of resistant strains.
Introduction
American foulbrood (AFB) is a highly contagious bacterial disease of honey bee brood and causes the collapse of honey bee colonies (Genersch, 2010). AFB is found in every area in which apiculture is practiced (Ellis and Munn, 2005). In Japan, AFB has also become widespread and the disease occurs every year throughout the country. As honey bees, in particular Apis mellifera, are the most important commercial pollinator contributing to the production of a variety of agricultural and horticultural crops, AFB is recognized as one of the most economically important diseases in apiculture worldwide.
The etiological agent of AFB is the Gram-positive, rod-shaped bacterium Paenibacillus larvae. Strains of this bacterium have been classified into five ERIC types (ERIC I–V) by repetitive-element PCR (Genersch et al., 2006; Beims et al., 2020) and 30 sequence types (STs) by multilocus sequence typing (MLST) (Morrissey et al., 2015)1. Strains with different genotypes differ in phenotypes, including virulence, at both the larval and colony levels (Genersch et al., 2005, 2006; Rauch et al., 2009; Beims et al., 2020). Among the five ERIC genotypes, ERIC I and II are the major types among the strains isolated from AFB cases. In Japan, ERIC I and II are the only genotypes found in isolates from AFB cases, and isolation of ERIC II strains has recently increased (Ueno et al., 2018). Similar to other Paenibacillus bacteria, P. larvae forms spores, which are the only infectious form of this organism (Genersch, 2010).
Honey bee larvae cannot get food by themselves and are fed by young worker bees called nurse bees. In a hive, worker larvae are fed worker jelly, a mixture of the hypopharyngeal and mandibular gland secretions of nurse bees, and then additionally fed some pollen and honey on or after the third day of larval life (Winston, 1987). Larvae are the most susceptible to P. larvae infection during the early larval stages, i.e., 12–36 h after egg hatching, and become infected through the ingestion of brood foods contaminated with P. larvae spores (Genersch, 2008). Ingested spores germinate and massively proliferate in the larval midgut. P. larvae vegetative cells then penetrate the peritrophic membrane, breach the midgut epithelium via the paracellular route, enter the haemocoel and kill the larvae (Genersch, 2008, 2010; Yue et al., 2008). Dead brood becomes ropy, dries out and then forms hard dark scales. The scales contain millions of P. larvae spores and drive disease transmission within and between colonies (Sturtevant, 1932; Bailey and Ball, 1991; Genersch, 2008). P. larvae spores may also exist in clinically healthy colonies. Indeed, analysis of honey samples harvested some years before the outbreak of AFB revealed that colonies were contaminated with spores several years before the detection of clinical symptoms (von der Ohe, 2003).
P. larvae spores are highly resistant to heat and chemical agents and can survive for many years in dried larval scales, hive products and equipment (OIE, 2018); therefore, burning of diseased colonies and contaminated hive materials is considered to be the most effective control measure for AFB. In several countries, antimicrobials have also been used as an alternative control measure. Oxytetracycline (OTC) is the most commonly used antimicrobial in the control of AFB (Reybroeck et al., 2012). However, OTC- and tetracycline-resistant P. larvae strains have been detected in several countries, including the United States, Canada and Argentina (Alippi, 2000; Miyagi et al., 2000; Evans, 2003; Krongdang et al., 2017). In previous studies, approximately 40% of Argentinian and 16% North American P. larvae isolates tested were OTC resistant (Alippi, 2000; Krongdang et al., 2017). The United States Food and Drug Administration additionally approved a macrolide antibiotic (tylosin tartrate [TS]) in 2005 and a lincosamide antibiotic (lincomycin hydrochloride [LCM]) in 2012 for the control of this disease; however, in the United States, TS- and/or LCM-resistant P. larvae were isolated from AFB cases in 2007, 2010 and 2013 (Krongdang et al., 2017).
OTC-resistant phenotypes observed in P. larvae develop, at least in part, due to plasmids carrying the tetracycline resistance gene [tet(L)], and the plasmids may be transferable across bacterial species (Murray and Aronstein, 2006; Alippi et al., 2007, 2014; Murray et al., 2007). In honey, many bacteria, including Bacillus and Paenibacillus species, are known to exist (Snowdon and Cliver, 1996; Alippi et al., 2004; Sinacori et al., 2014; Floyd et al., 2020). In a previous study, López et al. (2008) isolated Bacillus cereus strains carrying a variety of tetracycline and OTC resistance genes, including tet(L) and tet(K), from honey, and suggested that bacteria in honey are a reservoir for tetracycline resistance genes. In addition, the genome sequence analysis by Abrahamovich et al. (2018) revealed that Bacillus thuringiensis strain m401, a tetracycline-resistant isolate recovered from honey in Argentina, possessed a tetracycline resistance gene with high homology to tet45. As honey is a brood food for worker larvae, such antimicrobial-resistant bacteria in honey may contact P. larvae in the larval gut, and P. larvae may have acquired the tetracycline resistance genes from the honey-derived bacteria in the larval gut and become resistant to OTC.
In Japan, diseased colonies and contaminated hive materials have to be burned when AFB occurs; therefore, antimicrobials cannot be used for the treatment of AFB. However, to prevent AFB, 16-membered ring macrolides (mirosamicin [MRM] and TS) have been used in honey bee colonies for more than 20 years (Ueno et al., 2018). MRM was approved in 1999 by the Ministry of Agriculture, Forestry and Fisheries of Japan, and TS was additionally approved in 2017. As the sale of the MRM-containing prophylactic drug (Apiten for Honey bees; FEED ONE, Yokohama, Japan) was discontinued, TS is the only approved prophylactic available for controlling AFB at present. TS is currently used by dusting methods in Japan, and 150 mg of TS is used for 10,000 adult bees; i.e., the mixture consisting of 5 g of confectioner’s sugar and 50 mg of TS is sprinkled over the hive’s top bars three times at one-week intervals, and the dose can be scaled up depending on the number of adult bees. Although macrolide prophylactics have been used for a long time, macrolide-resistant P. larvae has yet to be found in Japan (Ueno et al., 2018). However, as mentioned above, TS-resistant P. larvae strains have been detected in North America (Krongdang et al., 2017). Although TS-resistant mechanisms of the North American P. larvae strains are unknown, we cannot exclude the possibility that TS-resistant P. larvae will emerge in Japan in the future through the acquisition of macrolide resistance genes, and the only approved prophylactic for AFB may therefore lose its effectiveness. As implied in the cases of OTC-resistant P. larvae, bacteria latent in honey may serve as a reservoir of antimicrobial resistance genes and P. larvae may acquire macrolide resistance genes from them in the larval gut. As best as we know, there is no information regarding the contamination of macrolide-resistant bacteria in honey. Therefore, we investigated the presence of TS-resistant bacteria in commercially available Japanese honey in this study. We also searched for macrolide resistance genes on mobile genetic elements (MGEs) by whole genome sequencing of representative TS-resistant honey-derived isolates and evaluated their potential for conferring TS resistance to P. larvae.
Materials and Methods
Japanese Honey Used in This Study
The 53 Japanese honey samples used in this study are listed in Supplementary Table 1 in Supplementary Materials. All of them were commercially available honey from different origins (i.e., different apiaries, different producers and/or different floral origins) and purchased between 2017 and 2018. Four honey samples (J12, J15, J24 and J42) were produced by Japanese honey bees, Apis cerana japonica, and the others were produced by European honey bees, A. mellifera. All samples were stored at room temperature until use.
Isolation and Identification of Bacteria From Japanese Honey
Isolation of Bacteria From Honey Samples
For isolation of bacteria from honey, 20 ml of each honey sample was diluted to 50% (v/v) with sterile water and centrifuged at 11,000 rpm for 45 min. The supernatant was removed and the sediment was suspended with 1 ml of sterile water. One hundred microliters of each suspension was plated on Columbia blood agar (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) containing 5% sheep blood (CSA), KSBHI agar (Arai et al., 2012), Lactobacilli MRS agar (Becton, Dickinson and Company) and modified GAM agar (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) containing 5% sheep blood (GAMSA). For selective isolation of TS-resistant bacteria, all agar media used for the isolation were supplemented with 1 μg/ml of TS. CSA was incubated at 35°C under air plus 5% CO2 conditions for two days, and the other agar media were incubated at 35°C under anaerobic conditions for four days. Colonies grown on the agar media were subcultured for purification. Pure-cultured bacterial isolates were suspended in LB broth (Becton, Dickinson and Company) containing 30% glycerol and stored at −80°C until use.
Identification of Honey-Derived Bacteria
To identify bacterial species of the isolates from Japanese honey, genomic DNA was extracted using InstaGene Matrix (Bio-Rad Laboratories, Inc., Hercules, CA, United States) and 16S rRNA gene sequences of the isolates were determined as described previously (Arai et al., 2012). Briefly, an approximately 1.5-kbp region of the 16S rRNA gene was amplified from the genomic DNA by TaKaRa ExTaq DNA polymerase (Takara Bio, Kusatsu, Japan) using primers F1 (5′-GAGTTTGATCCTGGCTCAG-3′) and R13 (5′-AGAAAGGAGGTGATCCAGCC-3′) (Dorsch and Stackebrandt, 1992). The amplified fragments were purified using QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced by a BigDye terminator v3.1 cycle sequencing kit using a 3130xl Genetic Analyzer (Applied Biosystems, Tokyo, Japan). Sequencher ver. 5.4.6 (Gene Codes Corp., Ann Arbor, MI, United States) was used to assemble the sequences. The species or genus of the isolates was identified by analyzing the 16S rRNA gene sequences using the EzBioCloud server2 (Yoon et al., 2017). As 98.65% was proposed as the threshold of 16S rRNA gene sequence similarity for differentiating two species (Kim et al., 2014), when type strains with 98.65% or higher sequence similarity were not found by EzBioCloud, identification of the isolates was carried out only on a genus level. In addition, when more than one closest type strains were found by EzBioCloud, the isolates were also not identified on a species level but instead on a genus level. The 16S rRNA gene sequences determined in this study were deposited in the DNA Data Bank of Japan (DDBJ)/GenBank/European Molecular Biology Laboratory (EMBL) database under the accession numbers LC588427– LC588635. The presence of the cereulide synthetase gene was investigated in some Bacillus isolates using the Bacillus cereus (CRS gene) PCR Detection Kit (Takara Bio) according to the manufacturer’s instructions.
Antimicrobial Susceptibility Tests for Bacterial Isolates From Japanese Honey
Two hundred and nine bacterial isolates derived from Japanese honey were cultured under the appropriate culture conditions listed in Supplementary Table 2 in Supplementary Materials and used for antimicrobial susceptibility tests. TS (Combi-Blocks Inc., San Diego, CA, United States), OTC (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) and LCM (Sigma-Aldrich, St. Louis, MO, United States) were used in this study. Except for the Clostridium beijerinckii isolate, minimum inhibitory concentrations (MICs) of these antimicrobials were determined by broth microdilution methods according to standard methods of Clinical and Laboratory Standards Institute (CLSI, M07-A10). Cation adjusted Mueller Hinton (MH) II broth (Becton, Dickinson and Company) was used for the tests and MICs were measured after a 24-h incubation at 35°C under aerobic conditions. For the quality control of each test, Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 cultured on MH agar (Becton, Dickinson and Company) for 20 h at 37°C under aerobic conditions were also employed. MICs for C. beijerinckii J33TS5 were measured by agar dilution methods according to CLSI, M07-A10, except that MICs were measured on GAMSA after a 24-h incubation at 35°C under anaerobic conditions. The antimicrobial concentration range employed for the tests was 0.25–256 μg/ml.
Search for Antimicrobial Resistance Genes in Honey-Derived Bacteria by Genome Sequence Analysis
Genomic DNA Extraction
Bacterial cells cultured on agar media under appropriate culture conditions (Supplementary Table 2) were harvested and suspended in 550 μl of TE (10 mM Tris–HCl [pH 8.0] and 1 mM EDTA [pH 8.0]). The bacterial suspensions were then mixed with 60–80 μl of a mixed solution of two enzymes (100 mg/ml of lysozyme [Sigma-Aldrich] and 500 U/ml of mutanolysin [Sigma-Aldrich]) and incubated for 1 h at 37°C. After incubation, bacterial cells were lysed by adding 60–70 μl of 10% sodium dodecyl sulfate, and the lysates were extracted with an equal volume of phenol, phenol-chloroform-isoamyl alcohol (25:24:1) (PCI) and chloroform at least once, three times and once, respectively. Nucleic acids were then precipitated by ethanol, rinsed with 70% ethanol and dissolved in sterile H2O. DNA was further treated with 100 μg/ml of RNase (NIPPON GENE Co., Ltd., Tokyo, Japan) at 37°C for 2 h, extracted with PCI and chloroform twice and once, respectively, and precipitated by ethanol. Extracted DNA was rinsed with 70% ethanol, dissolved in 10 mM Tris–HCl (pH 8.5) and stored at −20°C until use.
Genome Sequencing
Among the 209 isolates from Japanese honey, 50 were selected for whole genome shotgun sequencing in this study (Table 1). Genome sequencing data were obtained using the Miseq and/or Novaseq 6000 platforms (Illumina, Inc., San Diego, CA, United States). Sequencing by the Miseq platform (300-bp paired-end reads) was performed at the Institute of Crop Science, National Agriculture and Food Research Organization as recommended by the manufacturer. Genomic libraries for the Miseq sequencing were prepared by the TruSeq Nano DNA low-throughput library preparation kit (Illumina). Sequencing by the Novaseq 6000 platform (151-bp paired-end reads) was performed by Macrogen Japan Corp (Tokyo, Japan). Genomic libraries for the Novaseq 6000 sequencing were prepared by the TruSeq DNA PCR Free kit (Illumina). Adapter sequences and low-quality bases in Illumina paired end reads were removed using Trimmomatic v.0.36 (Bolger et al., 2014) with options of ‘ILLUMINACLIP:TruSeq3-PE.fa:3:40:15 LEADING:20 TRAILING:20 MINLEN:20’. Whole genome de novo assembly was conducted using SPAdes v.3.13.2 with options of –careful and –cov-cutoff auto (Nurk et al., 2013). Genomes were annotated using Prokka v.1.14.5 (Seemann, 2014). The genome sequences were deposited in the DDBJ/GenBank/EMBL database under the DRA accession number DRA011480, BioProject accession number PRJDB11087 and BioSample accession numbers SAMD00277588–SAMD00277637.
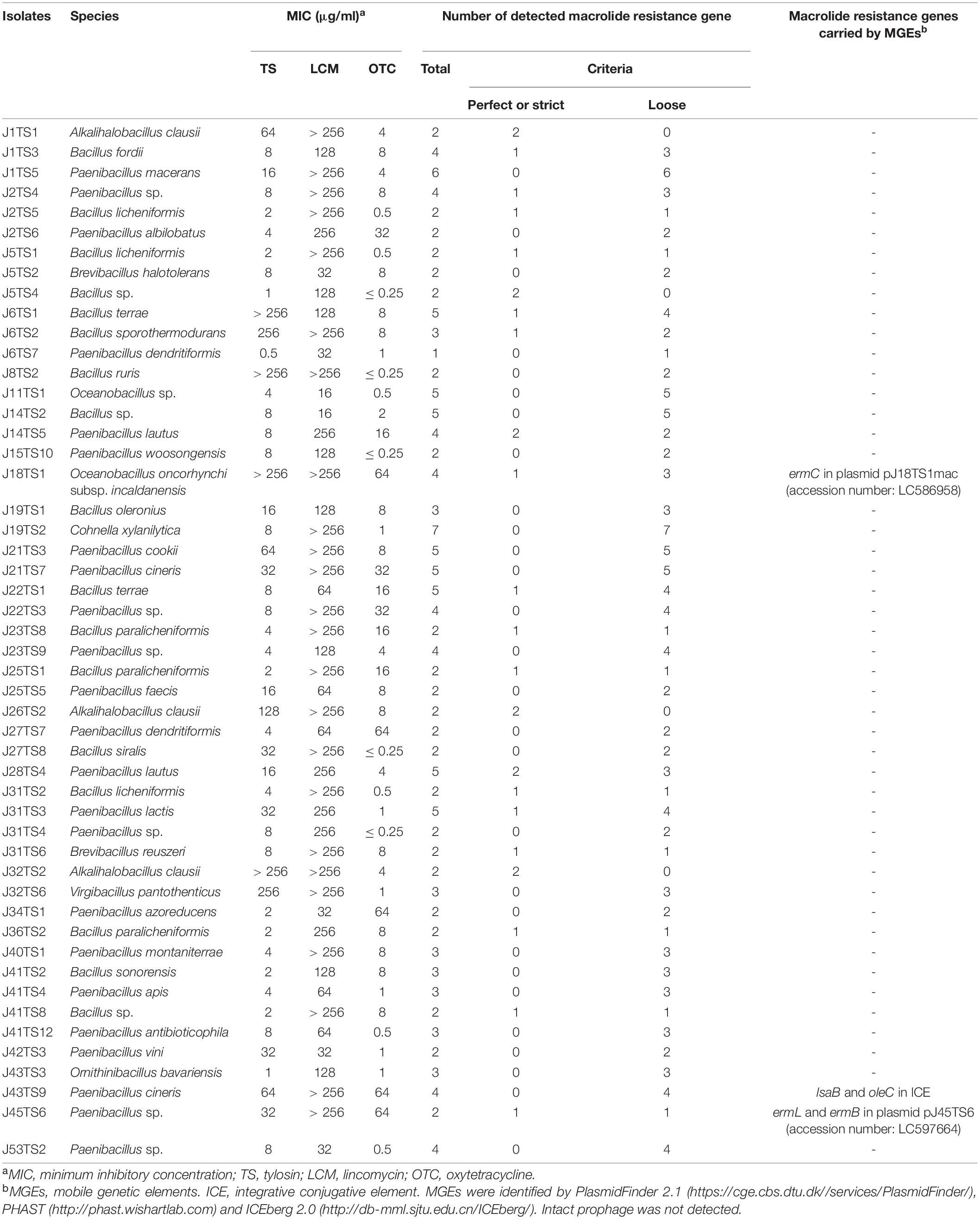
Table 1. Bacterial isolates used for genome sequencing and macrolide resistance genes detected by Resistance Gene Identifier (https://card.mcmaster.ca/analyze/rgi).
Screening of Antimicrobial Resistance Genes and MGEs From Genome Sequence Data
Putative antimicrobial resistance genes in the genome sequences were searched using Resistance Gene Identifier (RGI) in the Comprehensive Antibiotic Resistance Database (CARD)3 (McArthur et al., 2013). In this study, genes that exhibited > 50% best identity (percent identity of match to top hit in CARD) and percentage length of 60–140% with the reference sequence (for further details, see4) were selected as antimicrobial resistance genes. Integrative conjugative elements (ICEs) and prophages were searched using ICEberg 2.05 (Liu et al., 2019) and PHAST6 (Zhou et al., 2011), respectively. Scaffolds including plasmid replicons were selected by PlasmidFinder 2.17 (Carattoli et al., 2014), and complete sequences of the plasmids were determined by PCR and primer walking using the primers and conditions listed in Supplementary Table 3 in Supplementary Materials. The plasmid sequences were deposited in the DDBJ/GenBank/EMBL database under the accession numbers LC586958 (pJ18TS1mac), LC596398 (pJ18TS1tet) and LC597664 (pJ45TS6).
Construction of Putative Macrolide Resistance Gene Expression Vectors and Introduction of the Vectors Into P. larvae
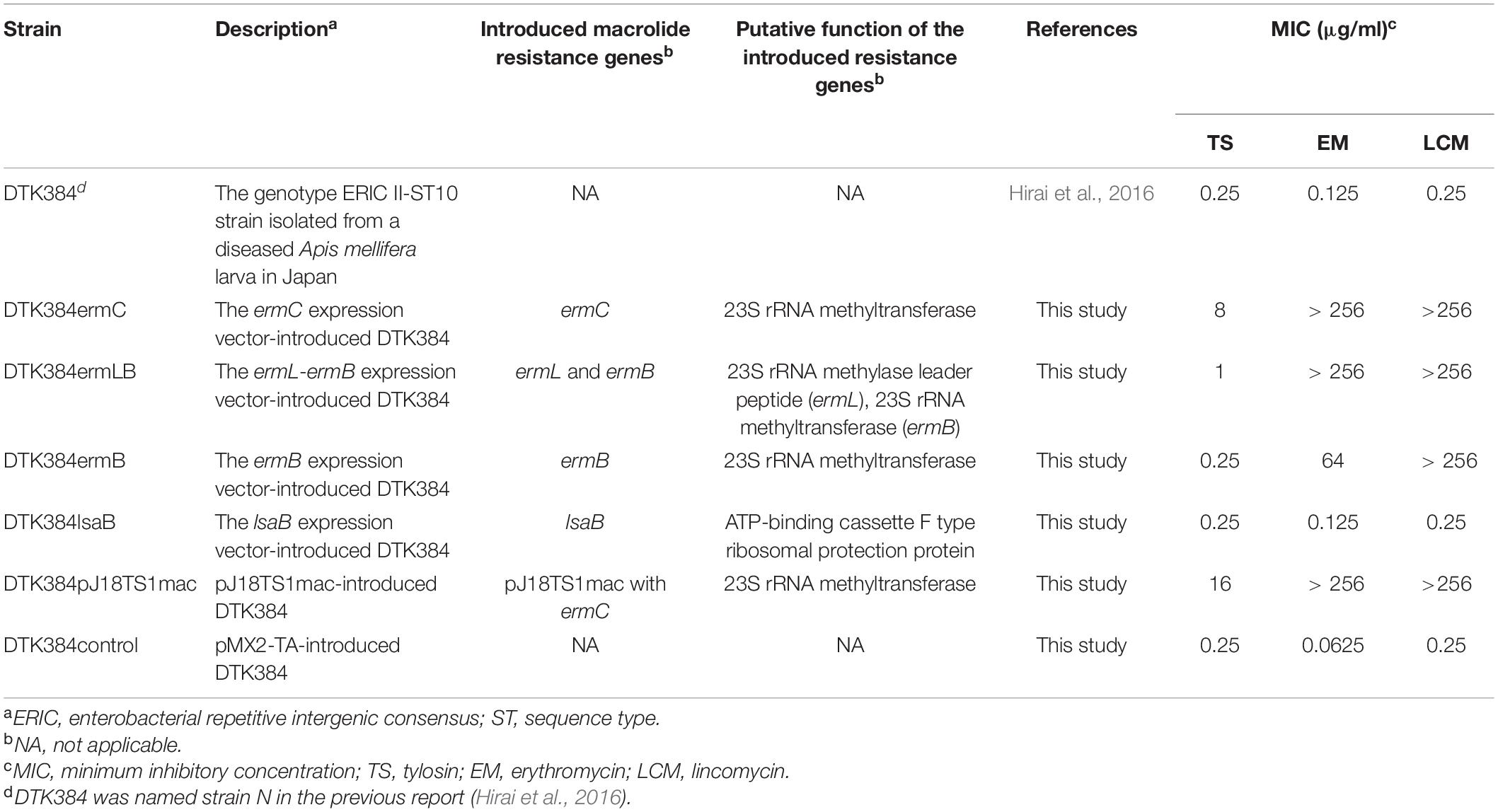
To construct putative macrolide resistance gene expression vectors, the ermC region in plasmid pJ18TS1mac from isolate J18TS1, the ermL-ermB and ermB regions in plasmid pJ45TS6 from isolate J45TS6, and the lsaB and oleC regions carried by a putative ICE in the isolate J43TS9 chromosome were amplified from the genomic DNA of the respective isolates by PCR using the primers and enzymes listed in Supplementary Table 3. The primers were designed to produce amplicons that contained the antimicrobial resistance gene and its Shine-Dalgarno sequence and/or putative promoter region. In this study, pMX2-TA, which was originally developed as a stable gene expression vector for the European foulbrood pathogen, Melissococcus plutonius, and contains the Gram-positive bacterial promoter sequence upstream of the multiple cloning sites (Takamatsu et al., 2020), was used. The PCR products were cloned into this plasmid vector via EcoRI and PstI or BamHI and PstI sites in E. coli MC1061 (Casadaban and Cohen, 1980). E. coli was cultured in LB (Becton, Dickinson and Company) agar at 37°C under aerobic conditions, and chloramphenicol (CP) was added to the media at 16 μg/ml for maintenance of the expression vectors. Accuracy of the cloned sequences was confirmed by PCR and sequencing analyses. The resultant plasmids were then introduced into an ERIC II P. larvae strain, DTK384 (strain N in Hirai et al., 2016), by electroporation according to the procedures described by de Graaf et al. (2013) except that electrotransformed P. larvae cells were incubated in MYPGP broth at 37°C for 2 h 30 min–7 h 30 min without shaking, and transformants were selected on MYPGP agar supplemented with 16 μg/ml of CP. As a control, DTK384 possessing only pMX2-TA (DKT384control) was also constructed by introducing pMX2-TA into the strain. Resultant P. larvae strains are listed in Table 2.
RNA Extraction From P. larvae DTK384 and Its Derivatives, and Confirmation of Introduced-Antimicrobial Resistance Gene Expression by Reverse Transcription PCR (RT-PCR)
Total RNA of P. larvae was extracted from bacterial cells cultured on MYPGP agar (de Graaf et al., 2013) containing 16 μg/ml of CP at 35°C for two days under air plus 5% CO2 conditions using the RNeasy Mini kit supplemented with RNAprotect Bacteria Reagent and RNase-Free DNase Set, as recommended by the manufacturer (QIAGEN) with the following modifications: harvested and RNAprotect Bacteria Reagent-treated P. larvae cells were incubated in TE containing 10 mg/ml of lysozyme (Sigma-Aldrich) and 50 U/ml of mutanolysin (Sigma-Aldrich) at 37°C for 20 min and then lysed by β-mercaptoethanol-supplemented Buffer RLT in the RNeasy Mini kit. Extracted RNA was transcribed to single-strand cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio). Expression of the introduced-antimicrobial resistance gene was confirmed by PCR using cDNA made from 10 ng of total RNA. To confirm the removal of DNA, the reverse transcriptase-untreated RNA was used as a template. All primers, polymerase and PCR conditions are listed in Supplementary Table 3.
Filter Mating Test
In this study, isolates J18TS1 and J45TS6, which possess macrolide resistance plasmids, were used as donors, and MICs of mitomycin C (FUJIFILM Wako Pure Chemical Corp.) for the isolates were measured by broth microdilution methods using MYPGP broth as described above. As the recipient, rifampicin (RIF)-resistant P. larvae was used. To prepare the recipient strain, P. larvae DTK384 was cultured on MYPGP agar containing 2 μg/ml of RIF (FUJIFILM Wako Pure Chemical Corp.), and a spontaneously generated RIF-resistant strain was isolated and named strain M20. The MIC of RIF for this strain measured by agar dilution methods was 128 μg/ml. For mating tests, J18TS1 and J45TS6 cultured on MYPGP agar with 1 μg/ml of TS at 35°C for approximately 24 h under aerobic conditions and M20 cultured on MYPGP agar with 4 μg/ml RIF at 35°C for 48 h under air plus 5% CO2 conditions were separately inoculated into MYPGP broth and cultured at 35°C with shaking (200 rpm). For donors, TS was added to the broth at a final concentration of 1 μg/ml. When the optical density of the cultures at 600 nm reached 0.4–0.6, donor cells from 5 ml of the culture were further incubated in fresh MYPGP broth (no mitomycin C), and that containing 1/2 and 1/4 MIC of mitomycin C. After incubation at 35°C for 1 h, the donor cells were collected by centrifugation (11,000 rpm 10 min), washed three times with sterile saline to remove mitomycin C and suspended in 5 ml of fresh MYPGP broth. The donor and recipient cultures were then mixed at a ratio of 1:1, and the mixture was filtered through a 0.45-μm nitrocellulose filter (25 mm diameter). The filters were placed on MYPGP agar and incubated at 35°C for 24 h and 48 h under air plus 5% CO2 conditions. To select recipient cells that acquired macrolide resistant plasmids, the donor and recipient cells on the cultured filters were suspended in 1 ml of sterile saline. One hundred microliters of the suspension was placed on MYPGP agar containing both RIF (4 μg/ml) and TS (1 μg/ml), and incubated at 35°C for 48 h under air plus 5% CO2 conditions.
Plasmid Extraction From J18TS1 and Introduction of pJ18TS1mac Into P. larvae
The plasmid pJ18TS1mac harbored by isolate J18TS1 was extracted using the illustra plasmidPrep Mini Spin Kit (GE Healthcare, Buckinghamshire, United Kingdom) according to the manufacturer’s instructions with the following modifications. Bacteria suspended in lysis buffer type 7 were treated with 0.2–1 mg/ml of lysozyme (Sigma-Aldrich) for 3–4 min at room temperature and then lysed by adding lysis buffer type 8. Extracted plasmids were introduced into P. larvae DTK384 by electroporation as described above. Transformants carrying pJ18TS1mac were selected on MYPGP agar containing 1 μg/ml of TS and a representative transformant (DTK384pJ18TS1mac) was used for further analyses (Table 2).
Antimicrobial Susceptibility Tests for Putative Macrolide Resistance Gene Expression Vector/pJ18TS1mac- Introduced P. larvae
Putative macrolide resistance gene expression vector-introduced P. larvae DTK384 and pMX2-TA-introduced DTK384 (DTK384control) were cultured on MYPGP agar supplemented with 16 μg/ml of CP for two days under air plus 5% CO2 conditions. The pJ18TS1mac-introduced DTK384 (DTK384pJ18TS1mac) was cultured on MYPGP agar supplemented with 2 μg/ml of TS under the same conditions. Antimicrobial susceptibility tests of the strains for TS, erythromycin (EM; FUJIFILM Wako Pure Chemical Corp.) and LCM were performed by agar dilution methods according to CLSI, M07-A10, except that MICs were measured on MYPGP agar plates after a 48-h incubation at 35°C under air plus 5% CO2 conditions. The antimicrobial concentration range tested was 0.0031–256 μg/ml. S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 cultured on MH agar for 20 h at 37°C under aerobic and air plus 5% CO2 conditions, respectively, were employed as the quality control strains, and antimicrobials used were confirmed to have acceptable potency after a 20-h incubation of E. faecalis- and S. aureus-inoculated MH agar plates at 35°C under aerobic conditions.
pJ18TS1mac Stability Tests in P. larvae
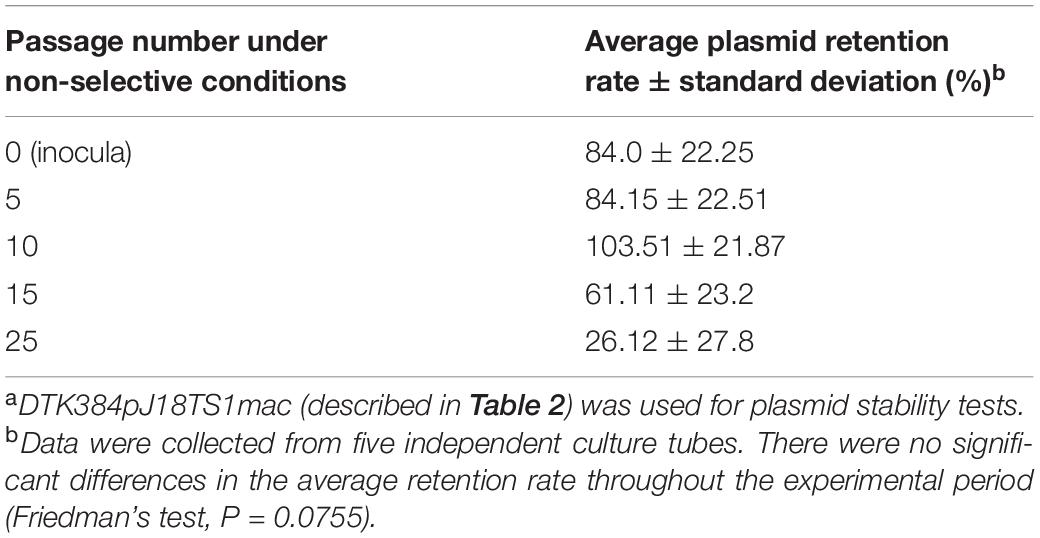
To investigate pJ18TS1mac stability in P. larvae under non-selective (no antimicrobial) conditions, DTK384pJ18TS1mac cultured for approximately two days on MYPGP agar containing 2 μg/ml of TS at 35°C under air plus 5% CO2 conditions was suspended in MYPGP broth to an optical density at 600 nm of approximately 0.2, and 10 μl of the suspension (inocula) was inoculated into 2 ml of non-selective MYPGP broth. After an approximately 24-h culture at 35°C with shaking (200 rpm), 1–2 μl of the culture was inoculated into 2 ml of the fresh non-selective MYPGP broth and further cultured for 24 h under the same conditions. After 5, 10, 15 and 25 cycles of the non-selective broth culture steps, serial dilutions of the culture were inoculated onto selective (2 μg/ml of TS) or non-selective MYPGP agar plates. After 2–3-day culture of the plates at 35°C under air plus 5% CO2 conditions, plasmid retention rates were calculated by the following formula: the plasmid retention rate = the number of colonies grown on selective MYPGP agar plate/the number of colonies grown on non-selective MYPGP agar plate × 100%. Plasmid retention rates of inocula were also measured similarly. Data were collected from five independent culture tubes. Differences in the average retention rate of pJ18TS1mac among inocula and bacteria at 5, 10, 15, and 25 passages were analyzed by Friedman’s test. All tests were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan) (Kanda, 2013), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria). A value of P < 0.05 was considered as the threshold for significance.
Results
Bacteria Isolated From Japanese Honey and Their Antimicrobial Susceptibility
In this study, we obtained 209 bacterial isolates from 48 of the 53 (90.6%) Japanese honey samples under 1 μg/ml-TS conditions (Supplementary Table 2). As of September 2020, the 209 isolates were classified into 10 genera by 16S rRNA gene sequencing, and at least 52 species existed in the isolates (Figure 1A and Supplementary Table 2). Bacillus (89 isolates, 42.6%) and Paenibacillus (84 isolates, 40.2%) were the two most common genera found in the isolates, followed by Alkalihalobacillus (21 isolates, 10.0%), Cytobacillus (5 isolates, 2.39%), Oceanobacillus (3 isolates, 1.44%), Brevibacillus (2 isolates, 0.96%) and Cohnella (2 isolates, 0.96%) (Figure 1A). Clostridium, Ornithinibacillus and Virgibacillus were also found, but only in one isolate each (Figure 1A). All isolates were Gram-positive spore-forming bacteria. Except for the anaerobic bacterium Clostridium beijerinckii J33TS5, all isolates were aerobic or facultatively anaerobic. At the species level, Bacillus licheniformis, Paenibacillus cineris, Alkalihalobacillus clausii and Paenibacillus dendritiformis were frequently isolated, and found in 45.3, 35.8, 30.2, and 26.4% of honey samples, respectively. Under the 1 μg/ml-TS conditions, neither P. larvae nor M. plutonius was isolated from the Japanese honey. Clostridium botulinum, cereulide synthetase gene-positive Bacillus cereus and Bacillus anthracis, which may cause public health problems, were also not isolated in this study.
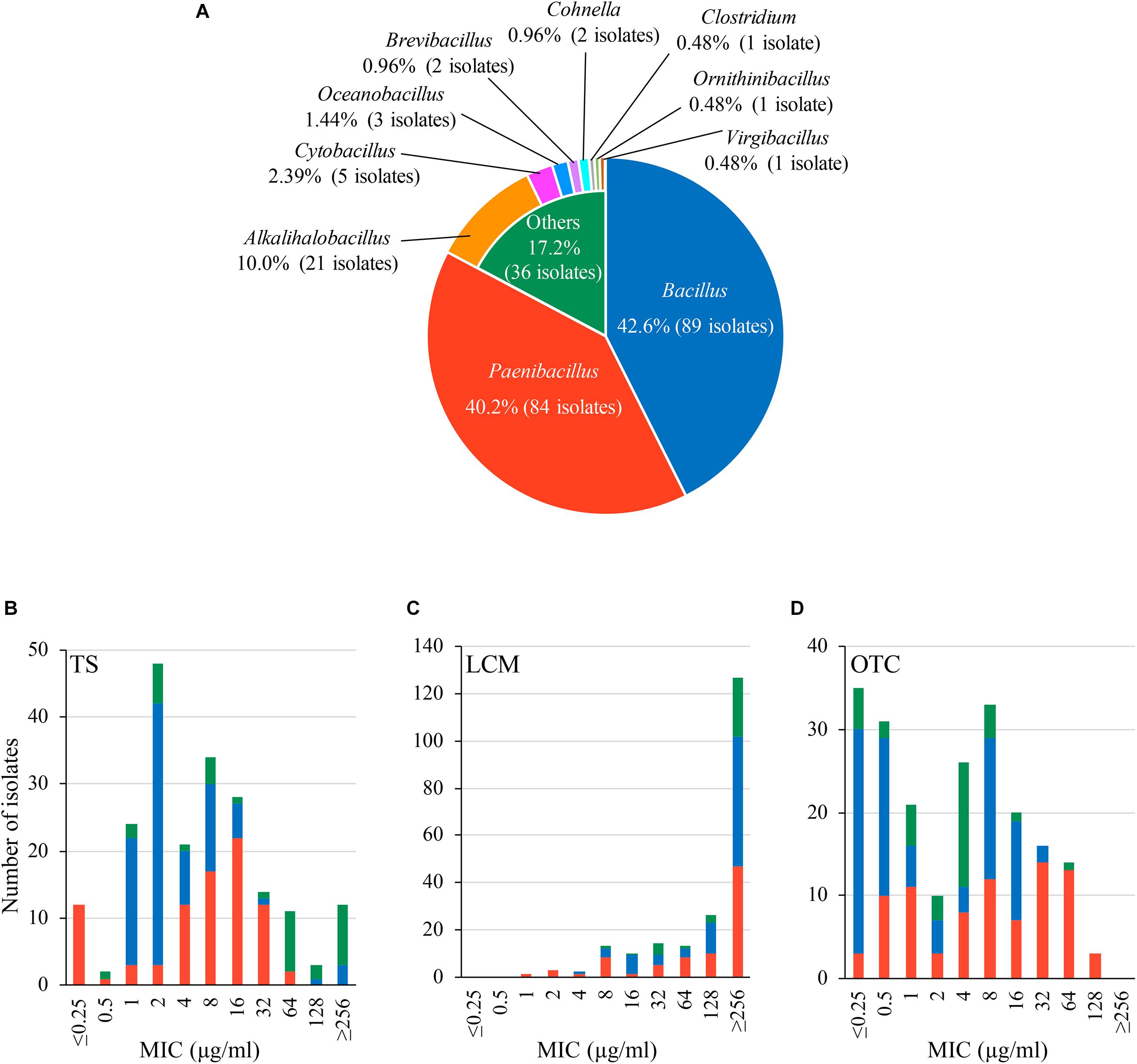
Figure 1. Characteristics of bacteria isolated from Japanese honey on 1 μg/ml-tylosin (TS)-containing agar media. (A) Proportion of genera found in the bacterial isolates. Genera/species of the isolates was identified by 16S rRNA gene sequencing. (B–D) Susceptibility of the isolates to TS, lincomycin (LCM) and oxytetracycline (OTC), respectively. Minimum inhibitory concentrations (MICs) of TS, LCM and OTC were measured by broth microdilution/agar dilution methods. The proportion of isolates with different genera in each MIC is indicated by different colors according to (A).
Results of the antimicrobial susceptibility tests are shown in Figures 1B–D and Supplementary Table 2. In addition to TS, LCM and OTC are used for controlling AFB in other countries (Reybroeck et al., 2012; Krongdang et al., 2017). Although LCM and OTC are not approved drugs in Japan, the use of these drugs may be considered in the future if TS-resistant P. larvae will spread in Japan. Therefore, we also employed LCM and OTC for the tests. MICs of the antimicrobials for the honey-derived isolates were widely distributed, and the MIC range of TS, LCM and OTC was ≤ 0.25– ≥ 256, 1– ≥ 256 and ≤ 0.25–128 μg/ml, respectively. Although bacteria in honey were screened under 1 μg/ml-TS conditions, MICs of TS were 1 μg/ml or lower in some isolates. However, most (94.3%) of the isolates exhibited lower susceptibility to TS than Japanese P. larvae strains (MIC of TS, 0.125–0.25 μg/ml) (Ueno et al., 2018), and 12 isolates had a MIC of 256 μg/ml or higher (Figure 1B and Supplementary Table 2). Most of the isolates also demonstrated low susceptibility (MIC50, ≥ 256 μg/ml) to LCM, which is structurally different but functionally similar to macrolides (Figure 1C and Supplementary Table 2). Moreover, in 122 (58.4%) isolates, MICs of OTC were higher than those for Japanese P. larvae strains (MIC, ≤ 1 μg/ml) (Ueno et al., 2018) (Figure 1D and Supplementary Table 2). This suggests a wide distribution of antimicrobial resistance genes in honey-derived bacteria.
Antimicrobial Resistance Genes in Honey-Derived Bacteria Detected by Genome Sequence Analysis
In order to identify genes conferring antimicrobial resistant phenotypes of the honey-derived isolates, we selected 50 isolates for genome sequencing (Table 1). The isolates were selected from various species. When there were multiple isolates in one species, 1–3 isolate(s), which demonstrated low susceptibility to TS and/or LCM, were selected as representatives. The summary of the genome sequencing results is shown in Supplementary Table 4 in Supplementary Materials. The sizes of the assembled genome of the isolates ranged from 3.48 to 7.44 Mbp, and there were 3,463–6,685 predicted coding sequences (Supplementary Table 4). RGI detected 379 macrolide, lincosamide and/or tetracycline resistance genes from the 50 isolates, and 59 met the perfect or strict criteria of the analysis3 (Supplementary Table 5 in Supplementary Materials). Among the resistance genes, 156, 166 and 207 genes, including 28, 46 and 11 perfect or strict hit genes, were predicted to confer macrolide, lincosamide and tetracycline resistance, respectively (Table 1 and Supplementary Table 5).
PlasmidFinder and additional sequence analysis identified three plasmids carrying antimicrobial resistance genes (Table 1). Two of the three plasmids (pJ18TS1mac and pJ18TS1tet), which carry the macrolide resistance gene ermC and tetracycline resistance gene tetL, respectively, were found in Oceanobacillus oncorhynchi subsp. incaldanensis isolate J18TS1. Both plasmids were relatively small in size (less than 4.7 kbp) and possessed the mob gene (Figure 2, accession number LC586958 [pJ18TS1mac] and LC596398 [pJ18TS1tet]), suggesting that they are mobilizable plasmids. Another plasmid pJ45TS6 found in Paenibacillus sp. isolate J45TS6 was 51.4 kbp in size, and carried the macrolide resistance gene ermB and tetracycline resistance gene tetL. Although not detected by RGI, ermL, which was predicted to control the expression of ermB, was identified in the upstream region of the ermB gene by Blast analysis8 (Figure 3). In addition, two putative macrolide resistance genes, lsaB and oleC, were identified in an ICE region of Paenibacillus cineris isolate J43TS9 (Table 1). In contrast, no intact prophages with antimicrobial resistance genes were identified in this study.
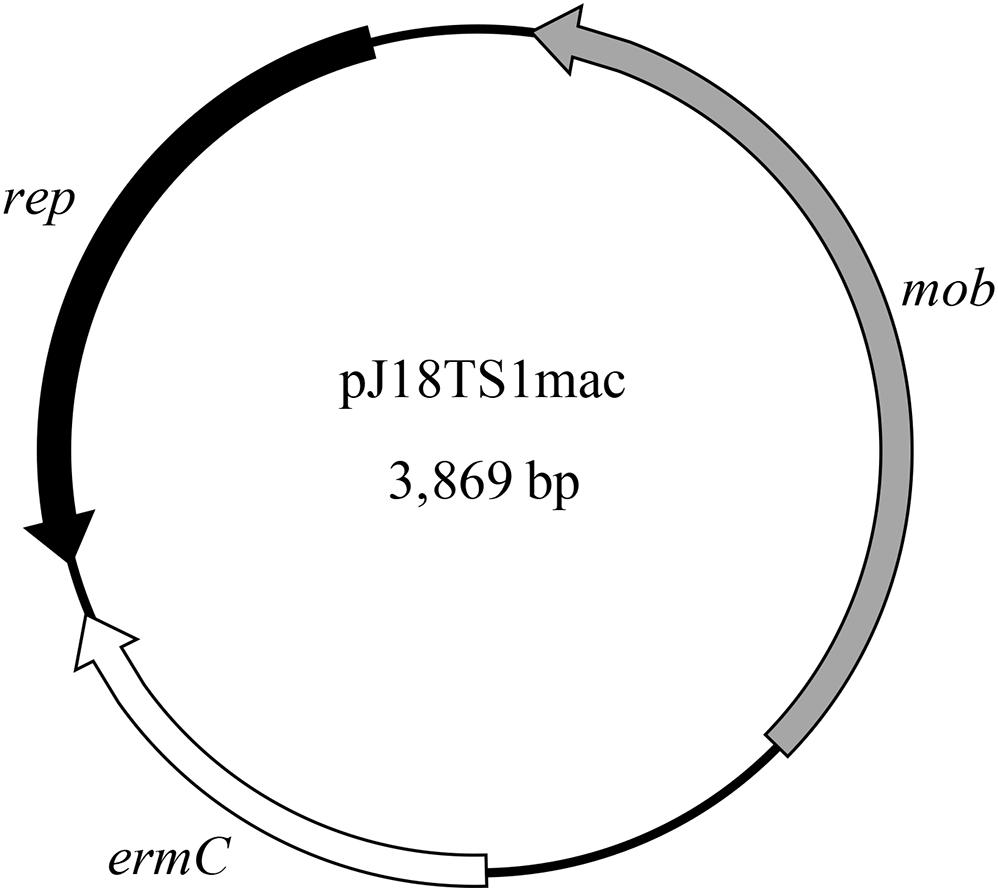
Figure 2. Map of pJ18TS1mac. rep, replication protein gene; mob, mobilization protein gene; ermC, 23S rRNA methyltransferase gene (macrolide resistance gene).
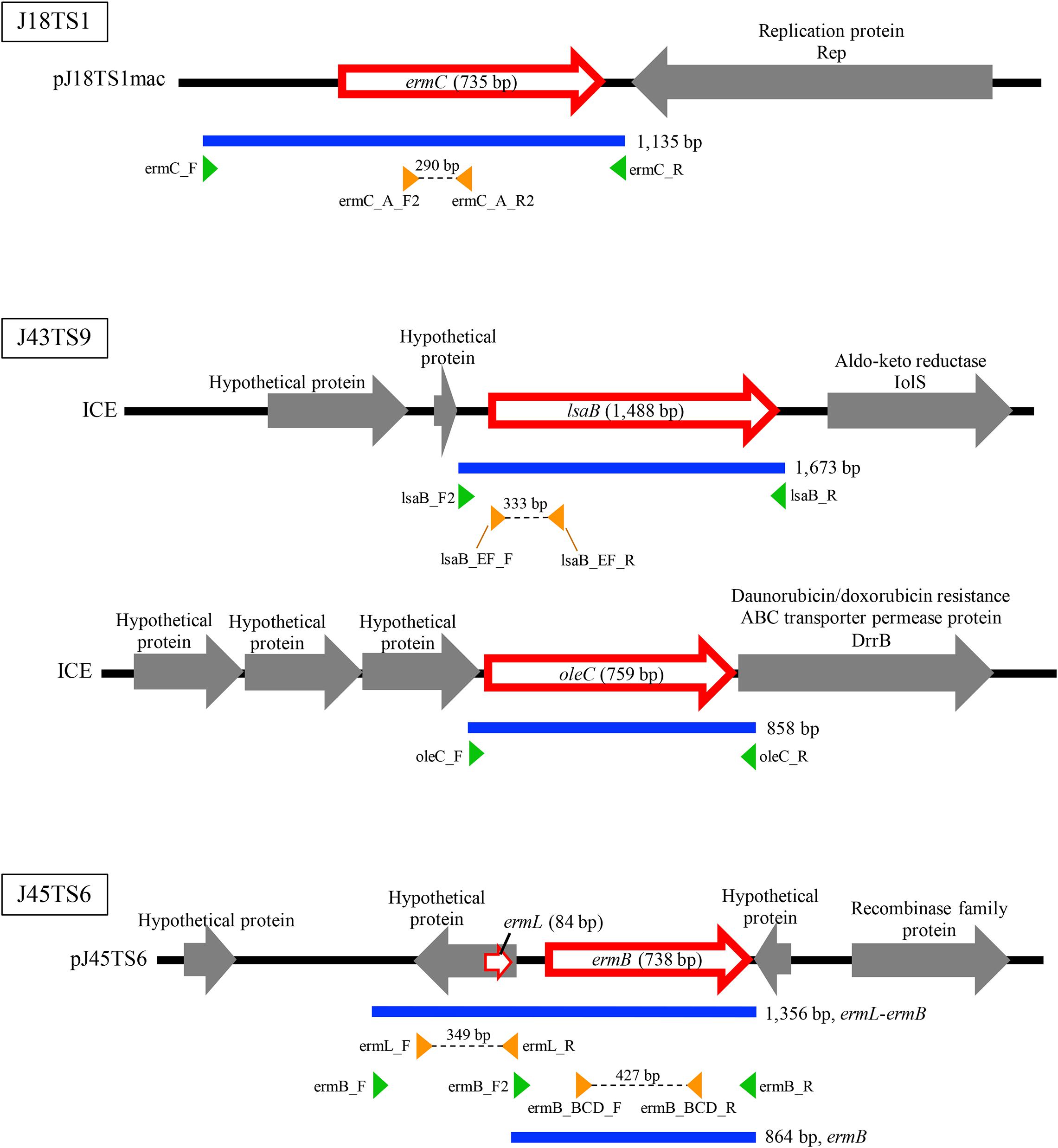
Figure 3. Schematic representation of macrolide resistance gene regions found in putative mobile genetic elements of honey-derived isolates. Blue lines indicate regions amplified by PCR for construction of antimicrobial resistance gene expression vectors, and green arrowheads represent primers used for PCR. Orange arrowheads represent primers used to confirm expression of the introduced macrolide resistance genes in P. larvae.
The ermC and ermL-ermB Regions Possessed by Plasmids pJ18TS1mac and pJ45TS6, Respectively, Confer Macrolide and Lincosamide Resistance to P. larvae
To investigate whether the putative macrolide resistance genes found in MGEs (Figure 3 and Table 1) have the potential to confer macrolide resistance to P. larvae, we introduced them into P. larvae using pMX2-TA and assessed antimicrobial susceptibility of the transformants. As isolation of ERIC II strains from AFB cases is recently increasing in Japan (Ueno et al., 2018), an ERIC II strain, DTK384, was used as a representative host for the analysis. Among the five target resistance genes/region (blue lines in Figure 3), the oleC gene of isolate J43TS9, which encodes ATP-binding cassette (ABC) antibiotic efflux pump, was unable to be cloned into pMX2-TA in E. coli, possibly due to toxicity of the product to E. coli; therefore, this gene was not used for further analysis. As shown in Figure 4, the ermC, ermL-ermB, ermB and lsaB genes were introduced into and expressed in P. larvae DTK384, and the resultant P. larvae strains were designated DTK384ermC, DTK384ermLB, DTK384ermB and DTK384lsaB, respectively (Table 2). As a control, DTK384 with the empty vector pMX2-TA (DTK384control) was also prepared. In addition to the 16-membered ring macrolide TS, EM was used as a representative 14-membered ring macrolide for antimicrobial susceptibility tests. As macrolide resistance genes may confer cross-resistance to lincosamides, LCM was also used for the tests.
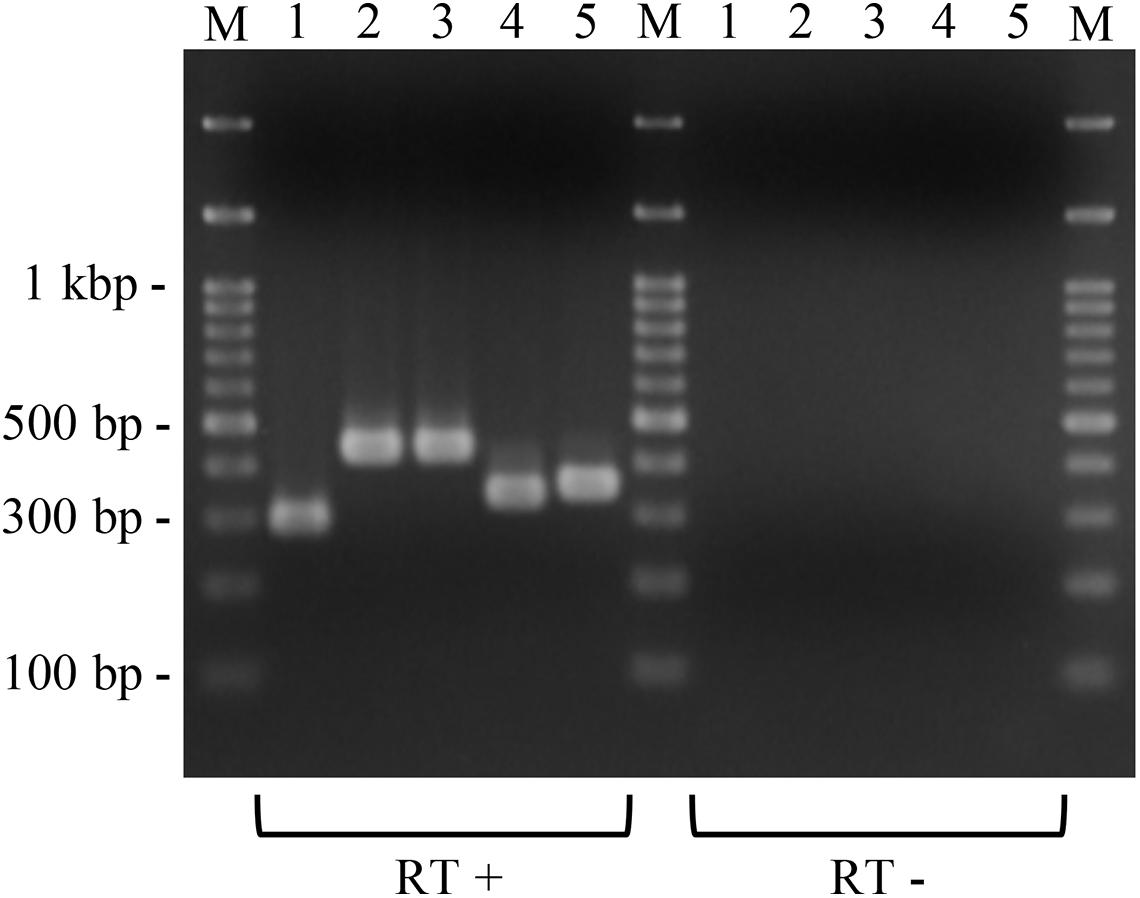
Figure 4. Expression of the introduced putative macrolide resistance genes in P. larvae. Total RNA was extracted from P. larvae strains cultured on MYPGP agar containing 16 μg/ml of chloramphenicol, and the expression of macrolide resistance genes was assessed by reverse transcription (RT)-PCR. To exclude DNA contamination in the RNA samples, total RNA, which was not treated with reverse transcriptase (RT -), was also used as the template for PCR. Lane 1: ermC expression in DTK384ermC; lane 2: ermB expression in DTL384ermLB; lane 3: ermB expression in DTK384ermB; lane 4: lsaB expression in DTK384lsaB; lane 5: ermL expression in DTL384ermLB. All introduced putative macrolide resistance genes were expressed in P. larvae.
As expected, the antimicrobial susceptibility of DTK384control (MICs of TS, EM and LCM; 0.25, 0.0625, and 0.25 μg/ml, respectively) was almost the same as that of the parent strain DTK384 (MICs of TS, EM and LCM; 0.25, 0.125, and 0.25 μg/ml, respectively) (Table 2). In contrast, introduction of ermC into P. larvae DTK384 significantly reduced the susceptibility of the strain (DTK384ermC) to all the three antimicrobials tested (MICs of TS, EM and LCM; 8, > 256 and > 256 μg/ml, respectively) (Table 2). Introduction of ermB into DTK384 also significantly reduced the susceptibility of the strain (DTK384ermB) to EM and LCM (MICs; 64 and > 256 μg/ml, respectively). In addition, co-introduction of ermL and ermB into the strain made P. larvae (DTK384ermLB) more resistant to EM (MIC, > 256 μg/ml) than DTK384ermB. However, DTK384ermLB was still relatively highly susceptible to TS (MIC, 1 μg/ml) even in the presence of both ermL and ermB (Table 2). On the other hand, lsaB did not confer macrolide resistance to P. larvae (MICs of TS, EM and LCM for DTK384lsaB; 0.25, 0.125, and 0.25 μg/ml, respectively) (Table 2). Thus, among the genes tested, the ermC genes in pJ18TS1mac, and ermL and ermB genes in pJ45TS6 have the potential to confer macrolide and lincosamide resistance to P. larvae.
The Macrolide-Resistant Plasmid pJ18TS1mac Can Be Stably Inherited in the P. larvae Population
As J18TS1 possesses putative ICEs, which encode type IV secretion system components, in the chromosome (Supplementary Table 6 in Supplementary Materials), pJ18TS1mac carrying the mob gene (Figure 2) may be horizontally transferable to other bacterial strains/species using the type IV secretion systems. Isolate J45TS6 also possesses putative ICEs in the chromosome (Supplementary Table 6). Although pJ45TS6 was not considered to be a transferable plasmid due to the absence of the mob gene, we performed filter mating tests using both J18TS1 and J45TS6 as donors and P. larvae M20 as the recipient in order to investigate whether the isolates can directly transfer their macrolide-resistant plasmids to P. larvae. However, neither J18TS1 nor J45TS6 transferred the plasmids to P. larvae under the experimental conditions used in this study.
However, we cannot exclude the possibility that the macrolide-resistant plasmids, in particular pJ18TS1mac, will be horizontally transferred to P. larvae under natural environmental conditions. If transferred plasmids are stably maintained in P. larvae, TS-resistant P. larvae may spread in Japan like OTC-resistant strains in other countries. Therefore, in order to investigate whether pJ18TS1mac can replicate in and be stably inherited by P. larvae, we introduced the plasmid into P. larvae DTK384 by electroporation and investigated plasmid retention rates in the strain under non-selective culture conditions.
pJ18TS1mac was introduced into P. larvae DTK384, and the strain (DTK384 pJ18TS1mac) became resistant to macrolide and lincosamide antibiotics as expected (MICs of TS, EM and LCM for DTK384pJ18TS1mac: 16, > 256 and > 256 μg/ml, respectively) (Table 2). Even after 10 passages under non-selective (no antibiotic) culture conditions, almost all P. larvae cells stably retained the plasmid. After 15 passages, the average plasmid retention rate decreased to 61.11 ± 23.2%, but the rate remained approximately 70% in four of the five test culture tubes. Although the average retention rate decreased to 26.12 ± 27.80% after 25 passages, the rate varied depending on the culture tubes (0.22–77.78%) and there were no significant differences in the average retention rate throughout the experimental period (Friedman’s test, P = 0.0755) (Table 3 and Figure 5). This suggested that the macrolide resistance plasmid can be inherited in the P. larvae population for a long period even under non-selective conditions after acquiring the plasmid.
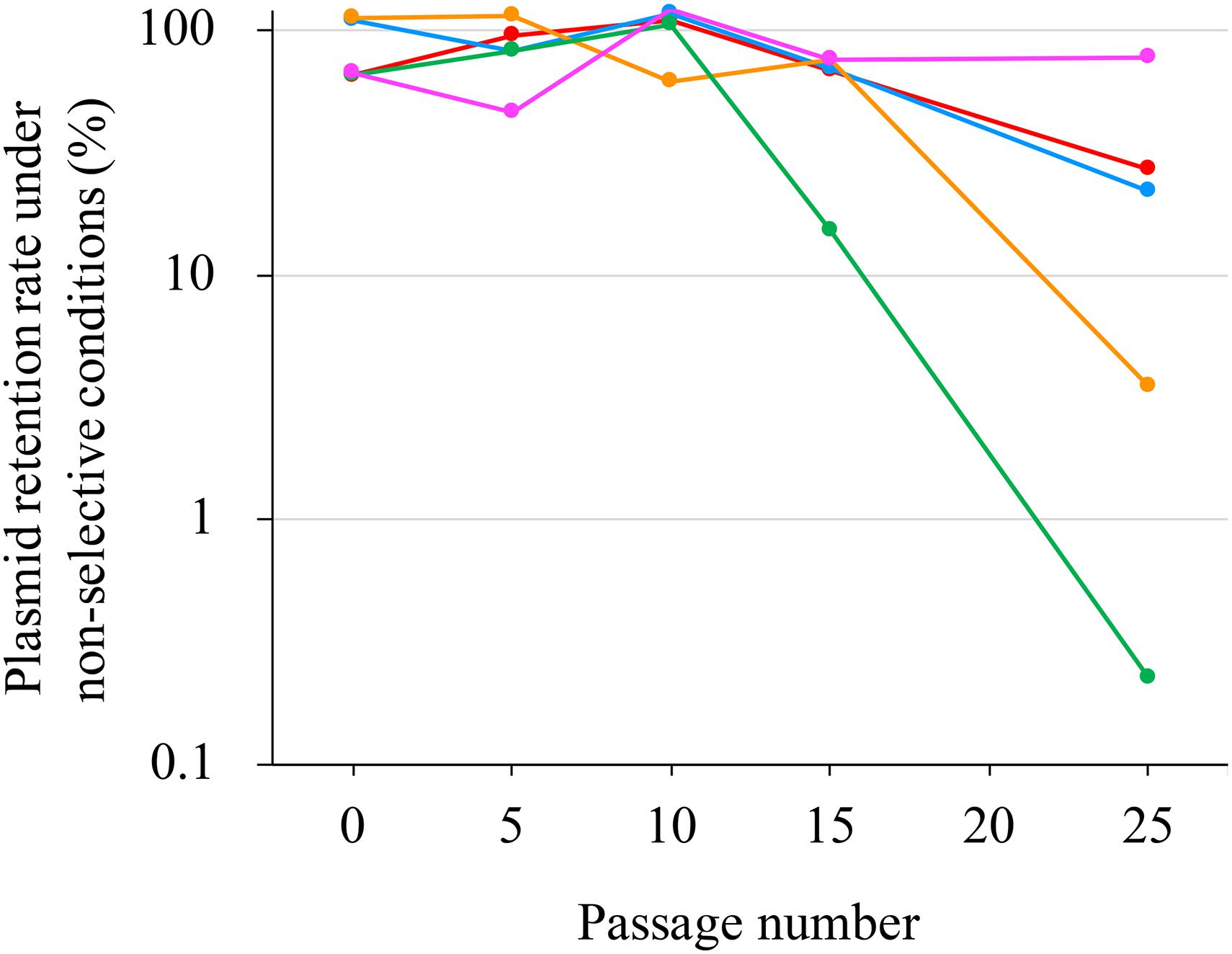
Figure 5. Stability of pJ18TS1mac in P. larvae. pJ18TS1mac-introduced DTK384 (DTK384 pJ18TS1mac) was cultured at 35°C in MYPGP broth under non-selective conditions, and bacteria were subcultured every 24 h by inoculating 1–2 μl of the culture into a fresh non-selective MYPGP broth. After 5, 10, 15 and 25 cycles of the culturing steps, serial dilutions of the culture were inoculated onto selective (2 μg/ml of TS) and non-selective MYPGP agar plates, and plasmid retention rates were calculated by the following formula: the plasmid retention rate = the number of colonies grown on selective MYPGP plate/the number of colonies grown on non-selective MYPGP plate × 100%. Plasmid retention rates of inocula for the first culture step were also measured. Data was collected from five independent culture tubes and results of each culture tube are shown in different colors.
Discussion
OTC and tetracycline-resistant P. larvae strains have been isolated in several countries. The resistant strains were suggested to develop, at least in part, by horizontal gene transfer of tetracycline resistance genes via MGEs (Evans, 2003; Murray and Aronstein, 2006; Alippi et al., 2007, 2014; Murray et al., 2007; Krongdang et al., 2017), and bacteria in honey may be a source of the antimicrobial resistance genes (López et al., 2008). Therefore, elucidation of the resistome in honey is important to predict the future evolution of antimicrobial resistance in foulbrood pathogens. In this study, we isolated bacteria from Japanese honey for the resistome analysis and found macrolide resistance genes on MGEs, which may confer TS-resistance if transmitted to P. larvae.
In the recent 16S rRNA gene amplicon sequencing for investigating microbiota in honey, Lactobacillus kunkeei (Apilactobacillus kunkeei), Parasacharribacter apium (Bombella apis), Fructobacillus fructosus, Gilliamella apicola, Xanthomonadales and Actinomycetales were detected as major bacterial species/groups at the DNA level (Floyd et al., 2020). Other previous studies reported that aerobic spore-forming bacteria of the genera Paenibacillus and Bacillus are commonly found in honey (Snowdon and Cliver, 1996; Alippi et al., 2004; López and Alippi, 2007; López et al., 2008). In this study, Paenibacillus and Bacillus were also the two major genera found in the isolates from Japanese honey, and all isolates from the Japanese honey were spore-forming bacteria. Although it is unknown why major bacterial species/groups detected by 16S rRNA gene amplicon sequencing (Floyd et al., 2020) were not isolated in this study, the use of 1 μg/ml-TS-containing media at the first bacterial isolation step and the antimicrobial characteristics of honey itself may have affected the repertoire of live bacteria recovered by the culture methods. Alternatively, some honey samples used in this study may have been heated after harvesting and the heating step may have affected non-spore forming bacteria in the honey such as A. kunkeei, B. apis and F. fructosus.
In this study, P. larvae and M. plutonius were not isolated from Japanese honey under the 1 μg/ml-TS conditions. This suggests that Japan is still free from TS-resistant foulbrood pathogens. However, we cannot rule out the possibility that we have missed TS-resistant foulbrood pathogens in honey samples. For isolation of foulbrood pathogens from samples with other bacteria, it is necessary to use selective media supplemented with appropriate antimicrobials such as nalidixic acid and pipemidic acid (de Graaf et al., 2013; Forsgren et al., 2013). In this study, we did not use such selective media, and this may have caused overgrowth of other honey-derived bacteria, making the recovery of the pathogens difficult. In addition, culture methods are known to not be effective for detecting foulbrood pathogens from honey (Nordström et al., 2002; Gillard et al., 2008; Forsgren et al., 2013; Bassi et al., 2018). Therefore, further investigation using selective media and/or molecular techniques is needed to clarify the contamination status of P. larvae and M. plutonius in Japanese honey.
Although TS-containing media was used, some honey-derived isolates were highly susceptible to TS. In particular, the TS susceptibility of 12 isolates was at a similar level to that of Japanese P. larvae strains (i.e., MIC of TS, ≤ 0.25 μg/ml). As suggested by previous studies (Snowdon and Cliver, 1996; Alippi et al., 2004; López and Alippi, 2007; López et al., 2008), the presence of spore-forming bacteria in honey was expected. Therefore, in order to germinate as many spores as possible, we cultured honey sediment-inoculated agar media for 2–4 days at the first isolation step. During the relatively long cultivation step, the potency of TS may have decreased gradually, and TS-susceptible bacteria may have grown on the media. Alternatively, because of efficient TS inactivation by macrolide-resistant bacteria coexisting in the same honey, the antibiotic may not be able to inhibit the growth of these TS-susceptible bacteria. Indeed, our genome analysis suggested the presence of bacteria in honey, which produce enzymes involved in macrolide inactivation (macrolide phosphotransferases) (Supplementary Table 5). However, such highly TS-susceptible bacteria were the minority among the honey-derived isolates retrieved in this study. MICs of TS for the honey-derived isolates varied (≤ 0.25– ≥ 256 μg/ml), and the majority of the isolates exhibited lower TS susceptibility than Japanese P. larvae strains (Ueno et al., 2018). Although breakpoints for antimicrobials tested have not yet been established for the bacterial species identified in this study, some isolates were highly resistant to TS (Figure 1 and Supplementary Table 2). These suggested the presence of multiple macrolide resistance mechanisms in honey-derived bacteria. Indeed, a variety of putative macrolide resistance genes involved in macrolide efflux, inactivation, target alteration and target protection were found in the representative isolates analyzed by genome sequencing (Supplementary Table 5).
In addition to macrolide resistance genes, a variety of lincosamide and tetracycline resistance genes were also detected in the genomes of the representative isolates by RGI (Supplementary Table 5). Consistent with this, many isolates from honey exhibited low susceptibility to LCM and OTC. In particular, 60.8% of the isolates were highly resistant to LCM (MICs, ≥ 256 μg/ml). Although the details were not reported in this study, RGI also detected many putative resistance genes for other drug classes, including aminoglycoside, phenicol, fluoroquinolone, glycopeptide and β-lactam antibiotics, in the genome of the honey-derived isolates (an example of RGI analysis results is shown in Supplementary Table 7 in the Supplementary Materials). This suggests the presence of many (and probably multidrug) resistant bacteria in honey and support the previous hypothesis that bacteria in honey are a reservoir of resistance genes (López et al., 2008).
Macrolides and lincosamides are antibiotics that inhibit protein synthesis, particularly the elongation step, by targeting the bacterial ribosome. In the ribosome, the peptidyl-transferase center (PTC) and adjacent nascent peptide exit tunnel (NPET) are the key players in protein elongation and function in catalyzing the peptide bond formation and the emergence of the nascent chain, respectively. Macrolides bind to a site within the NPET, whereas lincosamides are PTC-targeting antibiotics (Schlünzen et al., 2001; Wilson, 2009; Dunkle et al., 2010; Kannan and Mankin, 2011). Among the four genes tested, lsaB located in an ICE of P. cineris J43TS9 is considered to encode an ATP-binding cassette F (ABC-F) type ribosomal protection protein. ABC-F proteins are known to confer resistance to a number of clinically relevant antibiotics targeting the ribosome PTC/NPET region by interacting with the ribosome and displacing the bound drug (i.e., ribosomal protection mechanism) (Kerr et al., 2005; Reynolds and Cove, 2005; Sharkey et al., 2016). However, expression of lsaB in P. larvae affected neither macrolide nor lincosamide susceptibility (Table 2). ABC-F proteins have been classified into three groups based on antibiotic resistance (Msr homologs [macrolides and streptogramin B], Vga/Lsa/Sal homologs [lincosamides, pleuromutilins and streptogramin A] and OptrA homologs [phenicols and oxazolidinones]) (Sharkey et al., 2016; Su et al., 2018). Although the gene of J43TS9 was detected as lsaB by RGI, its product was only loosely similar to the reference sequence of LsaB in CARD (Table 2 and Supplementary Table 5). Based on this relatively low similarity to LsaB and our results, neither macrolides nor lincosamides are the targets of the lsaB product of J43TS9.
In contrast, introduction of the ermC and ermB genes located in plasmids of J18TS1 and J45TS6, respectively, into P. larvae DTK384 markedly reduced the susceptibility of the strain to EM and LCM (Table 2). This was not surprising because the putative products of the two genes were similar to the methyltransferases that modify a specific adenine residue (A2058, E. coli numbering) of 23S rRNA and confer macrolide-lincosamide-streptogramin B (MLSB) resistance (Franceschi et al., 2004; Weisblum, 1995). In the upstream region of ermB, an open reading frame (ermL) encoding the leader peptide was present, suggesting that expression of ermB is inducible. Leader peptide and inducible erm genes are known to constitute an operon, and expression of the erm gene is normally repressed due to sequestration of the erm ribosome binding site in the mRNA secondary structure. In the presence of macrolides, ribosomes stall during the translation of the leader peptide. The stall triggers a conformational rearrangement in mRNA, resulting in the opening of the erm ribosome binding site and activation of erm translation (Gaynor and Mankin, 2003). As expected from this, introduction of ermL further reduced the EM susceptibility of P. larvae (MIC of EM increased from 64 μg/ml to > 256 μg/ml); however, the 16-membered macrolide TS was still relatively effective against P. larvae (MIC, 1 μg/ml) (Table 2). Most erm genes are known to be induced by the 14- or 15-membered macrolides and not by 16-membered macrolides (Fyfe et al., 2016); therefore, in P. larvae, expression of the ermB gene may not have been efficiently induced by TS. Of note, the hypothetical protein gene located in the ermL region was also introduced into P. larvae in this study (Figure 3); however, antimicrobial susceptibility of DTK384ermLB suggests that the gene does not have the ability to greatly reduce the TS susceptibility of P. larvae.
In contrast to ermB in pJ45TS6, no leader peptide gene was found in the upstream region of the ermC gene in pJ18TS1mac, suggesting constitutive ermC expression in bacteria. Indeed, the MIC of TS for P. larvae DTK384 increased 32-fold only by introducing the ermC gene (DTK384ermC in Table 2). However, TS was still more effective in DTK384ermC (MIC, 8 μg/ml) than EM (> 256 μg/ml). Macrolides consist of a macrocyclic lactone ring carrying one or more sugar moieties (Gaynor and Mankin, 2003). EM carries two monosaccharides, cladinose and desosamine, at the C3 and C5 positions of the ring, respectively. When it binds to the 50S subunit of ribosomes, the macrocyclic lactone ring associates the NPET wall formed by positions 2057–2059 of 23S rRNA, whereas the monosaccharide moiety at the C5 position points toward the PTC. However, it cannot span the distance to the catalytic center (Hansen et al., 2002; Franceschi et al., 2004). In contrast, TS has a disaccharide at the C5 position of the ring and reaches further toward to the PTC (Hansen et al., 2002; Franceschi et al., 2004). Such structural features of TS may have increased the affinity of the drug with ribosomes even in the presence of methylated A2058 in 23S rRNA and exhibited stronger antimicrobial effects against P. larvae than EM.
However, proliferation of ermC-positive P. larvae in larvae will hardly be arrested even by TS. In the previous study by Reynaldi et al. (2017), beehives with 25,000 adult bees were treated with TS by dusting methods; i.e., the mixture consisting of 114 g of confectioner’s sugar, 5 g of cherry jelly and 1 g of TS was divided into four, 250 mg of TS to 30-g portions, and each portion was sprinkled over the ends of the hives’ top bars for four weeks at one-week intervals. According to the article, the mean concentration of TS in 2-day-old larvae measured by microbiological assay with Geobacillus stearothermophilus ATCC 12980 was always less than 4 μg/ml (Reynaldi et al., 2017). In another previous study, honey bee colonies containing around 30,000 worker bees were fed 500 g of synthetic honey with 200 mg or 400 mg of TS for 48 h, and the maximum concentration of TS in 48-h-old larvae was less than 1 μg/g even when treated with 400 mg of TS (Bernal et al., 2011). Similar to the study by Reynaldi et al. (2017), TS is used by dusting methods in Japan, and only 150 mg of TS is used for 10,000 adult bees. The dose of TS in Japan is lower than and almost equivalent to those in the studies by Reynaldi et al. (2017) and Bernal et al. (2011), respectively; therefore, the TS concentration in young larvae is expected to be less than 1–4 μg/ml. In contrast, MICs of TS for DTK384ermC and DTK384pJ18TS1mac were 8–16 μg/ml, suggesting that ermC-positive P. larvae is clinically resistant to TS. Therefore, acquisition of ermC by P. larvae is one of the greatest threats to apiculture in Japan.
As the ermC-positive plasmid pJ18TS1mac possesses the mob gene (Figure 2), it is considered a mobilizable plasmid and may transfer to other bacterial cells using conjugation machineries encoded by other MGEs. Although pJ18TS1mac in O. oncorhynchi subsp. incaldanensis J18TS1 did not transfer to P. larvae DTK384 under the mating conditions tested in this study, we cannot exclude the possibility that the plasmid transfers to P. larvae under specific conditions in the natural environment. In Bergey’s manual (Heyrman and Vos, 2015), O. oncorhynchi subsp. incaldanensis is described to be susceptible to EM and LCM; however, isolate J18TS1 was resistant to TS and LCM, and ermC in pJ18TS1mac was able to confer EM resistance. Therefore, J18TS1 is also considered to have acquired pJ18TS1mac from other bacteria horizontally, and thus, it is also possible that P. larvae acquires pJ18TS1mac or similar plasmids from other bacteria in the natural environment. As demonstrated in this study, pJ18TS1mac can be maintained in the P. larvae population for a long period. Stable plasmid inheritance can be achieved by several means such as (i) integration into the host chromosome, (ii) incorporating genes that participate in plasmid stable inheritance, like the toxin-antitoxin systems, into the plasmid, (iii) incorporating partitioning systems such as the ParABS systems and (iv) reducing plasmid size and increasing plasmid copy number in bacterial cells (Pluta and Espinosa, 2018). As pJ18TS1mac encodes a rolling circle replication initiator protein (Rep) (Figure 2; accession number LC586958), it is considered a rolling circle replicating-plasmid. Rolling circle replicating-plasmids usually do not carry specialized partitioning systems but ensure their stable inheritance because of their elevated plasmid copy numbers so that the probability of plasmid loss is reduced (Pluta and Espinosa, 2018). Indeed, pJ18TS1mac possesses neither the toxin-antitoxin systems nor the ParABS systems and its size is small (3,869 bp) (Figure 2). Therefore, pJ18TS1mac may have been maintained stably in P. larvae because of its high copy number in the cells. Due to the stable inheritance, once P. larvae acquires this plasmid in the field, TS-resistant P. larvae may be selected by the use of the prophylactic for AFB and spread throughout Japan. A putative mobilizable plasmid carrying the tetL gene (pJ18TS1tet) was also found in J18TS1. Although the use of OTC in apiculture is not allowed in Japan, OTC-resistant P. larvae may also be selected and spread in Japan if the drug is used illegally. Furthermore, the use of these drugs as prophylactics may affect the antimicrobial susceptibility of another foulbrood pathogen, M. plutonius, as seen by the use of MRM in Japan (Takamatsu et al., 2018). Although microorganisms in honey primarily originate from pollen, the digestive tracts of honey bees, dust, air, earth and nectar, honey can also be contaminated with microorganisms during and after harvesting process by humans, equipment and buildings (Snowdon and Cliver, 1996). Therefore, the isolates and their resistance genes found in commercially available honey in this study may not have necessarily existed in comb honey. However, our present study suggests the possibility of the future emergence of antimicrobial-resistant P. larvae by acquiring resistance genes from bacteria in honey. Therefore, it is necessary to use the prophylactic more carefully to prevent or delay the emergence. Development of novel molecular methods to detect resistance genes on MGEs together with foulbrood pathogens in honey will be helpful to use the prophylactic more carefully. Elucidation of the TS and LCM resistance mechanisms of North American P. larvae (Krongdang et al., 2017) is also required to predict and prevent the further emergence of antimicrobial resistant P. larvae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DT and MO designed the study and mainly performed the experiments. HK obtained genome sequence data, and MK analyzed the genome data. All authors contributed to the preparation of the manuscript and approved the final version.
Funding
This study was conducted under the regulatory research projects for food safety, animal health and plant protection (JPJ008617.17935699) funded by the Ministry of Agriculture, Forestry and Fisheries of Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A part of the genome sequence analysis was supported by the Science Research Supporting Programs of the Advanced Analysis Center and Institute of Crop Science, National Agriculture and Food Research Organization (NARO). We thank Ken Katsuda for helpful suggestions regarding the article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.667096/full#supplementary-material
Footnotes
- ^ https://pubmlst.org/plarvae/
- ^ https://www.ezbiocloud.net
- ^ https://card.mcmaster.ca/analyze/rgi
- ^ https://github.com/arpcard/rgi
- ^ http://db-mml.sjtu.edu.cn/ICEberg/
- ^ http://phast.wishartlab.com
- ^ https://cge.cbs.dtu.dk//services/PlasmidFinder/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Abrahamovich, E., Franco, M. E. E., López, A. C., Alippi, A. M., and Balatti, P. A. (2018). Draft genome sequence of Bacillus thuringiensis strain m401, isolated from honey in Argentina. Microbiol. Resour. Announc. 7, e1158-18. doi: 10.1128/MRA.01158-18
Alippi, A. M. (2000). Is Terramycin R losing its effectiveness against AFB? The Argentinian experience. Bee. Biz. 11, 27–29.
Alippi, A. M., León, I. E., and López, A. C. (2014). Tetracycline-resistance encoding plasmids from Paenibacillus larvae, the causal agent of American foulbrood disease, isolated from commercial honeys. Int. Microbiol. 17, 49–61. doi: 10.2436/20.1501.01.207
Alippi, A. M., López, A. C., Reynaldi, F. J., Grasso, D. H., and Aguilar, O. M. (2007). Evidence for plasmid-mediated tetracycline resistance in Paenibacillus larvae, the causal agent of American Foulbrood (AFB) disease in honeybees. Vet. Microbiol. 125, 290–303. doi: 10.1016/j.vetmic.2007.05.018
Alippi, A. M., Reynaldi, F. J., López, A. C., De Giusti, M. R., and Aguilar, O. M. (2004). Molecular epidemiology of Paenibacillus larvae larvae and incidence of American foulbrood in Argentinean honeys from Buenos Aires province. J. Apic. Res. 43, 135–143. doi: 10.1080/00218839.2004.11101124
Arai, R., Tominaga, K., Wu, M., Okura, M., Ito, K., Okamura, N., et al. (2012). Diversity of Melissococcus plutonius from honeybee larvae in Japan and experimental reproduction of European foulbrood with cultured atypical isolates. PLoS One 7:e33708. doi: 10.1371/journal.pone.0033708
Bassi, S., Carpana, E., Bergomi, P., and Galletti, G. (2018). Detection and quantification of Paenibacillus larvae spores in samples of bees, honey and hive debris as a tool for American foulbrood risk assessment. Bull. Insectol. 71, 235–241.
Beims, H., Bunk, B., Erler, S., Mohr, K. I., Spröer, C., Pradella, S., et al. (2020). Discovery of Paenibacillus larvae ERIC V: phenotypic and genomic comparison to genotypes ERIC I-IV reveal different inventories of virulence factors which correlate with epidemiological prevalences of American Foulbrood. Int. J. Med. Microbiol. 310:151394. doi: 10.1016/j.ijmm.2020.151394
Bernal, J., Martín, M. T., Toribio, L., Martín-Hernández, R., Higes, M., Bernal, J. L., et al. (2011). Determination of tylosins A, B, C and D in bee larvae by liquid chromatography coupled to ion trap-tandem mass spectrometry. J. Chromatogr. B 879, 1596–1604. doi: 10.1016/j.jchromb.2011.03.052
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Carattoli, A., Zankari, E., García-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Casadaban, M. J., and Cohen, S. N. (1980). Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207. doi: 10.1016/0022-2836(80)90283-1
de Graaf, D. C., Alippi, A. M., Antúnez, K., Aronstein, K. A., Budge, G., De Koker, D., et al. (2013). Standard methods for American foulbrood research. J. Apic. Res. 52, 1–28. doi: 10.3896/IBRA.1.52.1.11
Dorsch, M., and Stackebrandt, E. (1992). Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J. Microbiol. Methods 16, 271–279. doi: 10.1016/0167-7012(92)90017-X
Dunkle, J. A., Xiong, L., Mankin, A. S., and Cate, J. H. D. (2010). Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 107, 17152–17157. doi: 10.1073/pnas.1007988107
Ellis, J. D., and Munn, P. A. (2005). The worldwide health status of honey bees. Bee World 86, 88–101.
Evans, J. D. (2003). Diverse origins of tetracycline resistance in the honey bee bacterial pathogen Paenibacillus larvae. J. Invertebr. Pathol. 83, 46–50. doi: 10.1016/S0022-2011(03)00039-9
Floyd, A. S., Mott, B. M., Maes, P., Copeland, D. C., McFrederick, Q. S., and Anderson, K. E. (2020). Microbial ecology of European foul brood disease in the honey bee (Apis mellifera): towards a microbiome understanding of disease susceptibility. Insects 11:555. doi: 10.3390/insects11090555
Forsgren, E., Budge, G. E., Charrière, J.-D., and Hornitzky, M. A. Z. (2013). Standard methods for European foulbrood research. J. Apic. Res. 52, 1–14. doi: 10.3896/IBRA.1.52.1.12
Franceschi, F., Kanyo, Z., Sherer, E. C., and Sutcliffe, J. (2004). Macrolide resistance from the ribosome perspective. Curr. Drug Targets Infect. Disord. 4, 177–191. doi: 10.2174/1568005043340740
Fyfe, C., Grossman, T. H., Kerstein, K., and Sutcliffe, J. (2016). Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 6:a025395. doi: 10.1101/cshperspect.a025395
Gaynor, M., and Mankin, A. S. (2003). Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 3, 949–960. doi: 10.2174/1568026033452159
Genersch, E. (2008). Paenibacillus larvae and American foulbrood-long since known and still surprising. J. Verbr. Lebensm. 3, 429–434. doi: 10.1007/s00003-008-0379-8
Genersch, E. (2010). American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 103, S10–S19. doi: 10.1016/j.jip.2009.06.015
Genersch, E., Ashiralieva, A., and Fries, I. (2005). Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, a bacterial pathogen causing American foulbrood disease in honeybees. Appl. Environ. Microbiol. 71, 7551–7555. doi: 10.1128/AEM.71.11.7551-7555.2005
Genersch, E., Forsgren, E., Pentikäinen, J., Ashiralieva, A., Rauch, S., Kilwinski, J., et al. (2006). Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Microbiol. 56, 501–511. doi: 10.1099/ijs.0.63928-0
Gillard, M., Charriere, J. D., and Belloy, L. (2008). Distribution of Paenibacillus larvae spores inside honey bee colonies and its relevance for diagnosis. J. Invertebr. Pathol. 99, 92–95. doi: 10.1016/j.jip.2008.05.010
Hansen, J. L., Ippolito, J. A., Ban, N., Nissen, P., Moore, P. B., and Steitz, T. A. (2002). The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10, 117–128. doi: 10.1016/S1097-2765(02)00570-1
Heyrman, J., and Vos, P. D. (2015). Oceanobacillus, in Bergey’s Manual of Syst. Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons Ltd.
Hirai, Y., Suzuki, T., Inaba, N., Minoguchi, N., and Takamatsu, D. (2016). Existence of Paenibacillus larvae genotypes ERIC I-ST2, ERIC I-ST15 and ERIC II-ST10 in the western region of Aichi prefecture, Japan. J. Vet. Med. Sci. 78, 1195–1199. doi: 10.1292/jvms.16-0041
Kanda, Y. (2013). Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 48, 452–458. doi: 10.1038/bmt.2012.244
Kannan, K., and Mankin, A. S. (2011). Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann. N.Y. Acad. Sci. 1241, 33–47. doi: 10.1111/j.1749-6632.2011.06315.x
Kerr, I. D., Reynolds, E. D., and Cove, J. H. (2005). ABC proteins and antibiotic drug resistance: is it all about transport? Biochem. Soc. Trans. 33, 1000–1002. doi: 10.1042/BST0331000
Kim, M., Oh, H.-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Krongdang, S., Evans, J. D., Pettis, J. S., and Chantawannakul, P. (2017). Multilocus sequence typing, biochemical and antibiotic resistance characterizations reveal diversity of North American strains of the honey bee pathogen Paenibacillus larvae. PLoS One 12:e0176831. doi: 10.1371/journal.pone.0176831
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
López, A. C., and Alippi, A. M. (2007). Phenotypic and genotypic diversity of Bacillus cereus isolates recovered from honey. Int. J. Food Microbiol. 117, 175–184. doi: 10.1016/j.ijfoodmicro.2007.03.007
López, A. C., de Ortúzar, R. V. M., and Alippi, A. M. (2008). Tetracycline and oxytetracycline resistance determinants detected in Bacillus cereus strains isolated from honey samples. Rev. Argent. Microbiol. 40, 231–236.
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Miyagi, T., Peng, C. Y. S., Chuang, R. Y., Mussen, E. C., Spivak, M. S., and Doi, R. H. (2000). Verification of oxytetracycline-resistant American foulbrood pathogen Paenibacillus larvae in the United States. J. Invertebr. Pathol. 75, 95–96.
Morrissey, B. J., Helgason, T., Poppinga, L., Fünfhaus, A., Genersch, E., and Budge, G. E. (2015). Biogeography of Paenibacillus larvae, the causative agent of American foulbrood, using a new multilocus sequence typing scheme. Environ. Microbiol. 17, 1414–1424. doi: 10.1111/1462-2920.12625
Murray, K. D., and Aronstein, K. A. (2006). Oxytetracycline-resistance in the honey bee pathogen Paenibacillus larvae is encoded on novel plasmid pMA67. J. Apic. Res. 45, 207–214. doi: 10.1080/00218839.2006.11101349
Murray, K. D., Aronstein, K. A., and de León, J. H. (2007). Analysis of pMA67, a predicted rolling-circle replicating, movilizable, tetracycline-resistance plasmid from the honey bee pathogen, Paenibacillus larvae. Plasmid 58, 89–100. doi: 10.1016/j.plasmid.2007.02.001
Nordström, S., Forsgren, E., and Fries, I. (2002). Comparative diagnosis of American foulbrood using samples of adult honey bees and honey. J. Apic. Sci. 46, 5–12.
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
OIE (2018). Terrestrial Manual. Available online at: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.02.02_AMERICAN_FOULBROOD.pdf (accessed April 16, 2021).
Pluta, R., and Espinosa, M. (2018). Antisense and yet sensitive: copy number control of rolling circle-replicating plasmids by small RNAs. WIREs RNA 9:e1500. doi: 10.1002/wrna.1500
Rauch, S., Ashiralieva, A., Hedtke, K., and Genersch, E. (2009). Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the etiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 75, 3344–3347. doi: 10.1128/AEM.02839-08
Reybroeck, W., Daeseleire, E., De Brabander, H. F., and Herman, L. (2012). Antimicrobials in beekeeping. Vet. Microbiol. 158, 1–11. doi: 10.1016/j.vetmic.2012.01.012
Reynaldi, F., Albo, G., Avellaneda, E., and Rule, R. (2017). Evaluation of kinetic behaviour of two preparations of tylosin administered in beehives for American foulbrood control. Bulg. J. Vet. Med. 20, 264–270. doi: 10.15547/bjvm.1024
Reynolds, E., and Cove, J. H. (2005). Enhanced resistance to erythromycin is conferred by the enterococcal msrC determinant in Staphylococcus aureus. J. Antimicrob. Chemother. 55, 260–264. doi: 10.1093/jac/dkh541
Schlünzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, A., et al. (2001). Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821. doi: 10.1038/35101544
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharkey, L. K. R., Edwards, T. A., and O’Neill, A. J. (2016). ABC-F proteins mediate antibiotic resistance through ribosomal protection. Mbio 7:e01975-15. doi: 10.1128/mBio.01975-15
Sinacori, M., Francesca, N., Alfonzo, A., Cruciata, M., Sannino, C., Settanni, L., et al. (2014). Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 38, 284–294. doi: 10.1016/j.fm.2013.07.013
Snowdon, J. A., and Cliver, D. O. (1996). Microorganisms in honey. Int. Food Microbiol. 31, 1–26. doi: 10.1016/0168-1605(96)00970-1
Sturtevant, A. P. (1932). Relation of commercial honey to the spread of American foulbrood. J. Agr. Res. 45, 257–285.
Su, W., Kumar, V., Ding, Y., Ero, R., Serra, A., Lee, B. S. T., et al. (2018). Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. U.S.A. 115, 5157–5162. doi: 10.1073/pnas.1803313115
Takamatsu, D., Okumura, K., Tabata, A., Okamoto, M., and Okura, M. (2020). Transcriptional regulator SpxA1a controls the resistance of the honey bee pathogen Melissococcus plutonius to the antimicrobial activity of royal jelly. Environ. Microbiol. 22, 2736–2755. doi: 10.1111/1462-2920.15125
Takamatsu, D., Yoshida, E., Watando, E., Ueno, Y., Kusumoto, M., Okura, M., et al. (2018). A frameshift mutation in the rRNA large subunit methyltransferase gene rlmAII determines the susceptibility of a honey bee pathogen Melissococcus plutonius to mirosamicin. Environ. Microbiol. 20, 4431–4443. doi: 10.1111/1462-2920.14365
Ueno, Y., Yoshida, E., Misumi, W., Watando, E., Suzuki, K., Hirai, Y., et al. (2018). Population structure and antimicrobial susceptibility of Paenibacillus larvae isolates from American foulbrood cases in Apis mellifera in Japan. Environ. Microbiol. Rep. 10, 210–216. doi: 10.1111/1758-2229.12623
von der Ohe, W. (2003). Control of American foulbrood by using alternatively eradication method and artificial swarms. APIACTA 38, 137–139.
Weisblum, B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585. doi: 10.1128/AAC.39.3.577
Wilson, D. N. (2009). The A–Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44, 393–433. doi: 10.3109/10409230903307311
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yue, D., Nordhoff, M., Wieler, L. H., and Genersch, E. (2008). Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ. Microbiol. 10, 1612–1620. doi: 10.1111/j.1462-2920.2008.01579.x
Keywords: American foulbrood, antimicrobial susceptibility, bacteria in honey, macrolide and lincosamide resistance, mobile genetic elements, tylosin, Paenibacillus larvae
Citation: Okamoto M, Kumagai M, Kanamori H and Takamatsu D (2021) Antimicrobial Resistance Genes in Bacteria Isolated From Japanese Honey, and Their Potential for Conferring Macrolide and Lincosamide Resistance in the American Foulbrood Pathogen Paenibacillus larvae. Front. Microbiol. 12:667096. doi: 10.3389/fmicb.2021.667096
Received: 12 February 2021; Accepted: 06 April 2021;
Published: 29 April 2021.
Edited by:
Satoru Suzuki, Ehime University, JapanCopyright © 2021 Okamoto, Kumagai, Kanamori and Takamatsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Takamatsu, cDEwMTNkdEBhZmZyYy5nby5qcA==
 Mariko Okamoto
Mariko Okamoto Masahiko Kumagai
Masahiko Kumagai Hiroyuki Kanamori
Hiroyuki Kanamori Daisuke Takamatsu
Daisuke Takamatsu