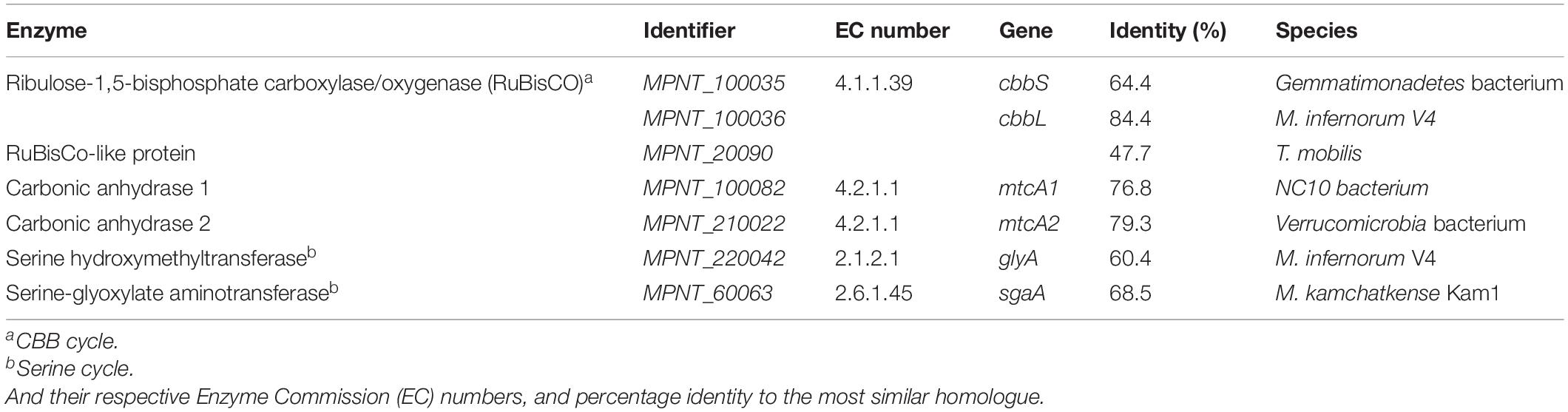
- 1Department of Microbiology, Institute for Water and Wetland Research (IWWR), Radboud University, Nijmegen, Netherlands
- 2Istituto Nazionale di Geofisica e Vulcanologia, Sezione di Palermo, Palermo, Italy
- 3Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, Palermo, Italy
Verrucomicrobial methanotrophs are a group of aerobic bacteria isolated from volcanic environments. They are acidophiles, characterized by the presence of a particulate methane monooxygenase (pMMO) and a XoxF-type methanol dehydrogenase (MDH). Metagenomic analysis of DNA extracted from the soil of Favara Grande, a geothermal area on Pantelleria Island, Italy, revealed the presence of two verrucomicrobial Metagenome Assembled Genomes (MAGs). One of these MAGs did not phylogenetically classify within any existing genus. After extensive analysis of the MAG, we propose the name of “Candidatus Methylacidithermus pantelleriae” PQ17 gen. nov. sp. nov. The MAG consisted of 2,466,655 bp, 71 contigs and 3,127 predicted coding sequences. Completeness was found at 98.6% and contamination at 1.3%. Genes encoding the pMMO and XoxF-MDH were identified. Inorganic carbon fixation might use the Calvin-Benson-Bassham cycle since all genes were identified. The serine and ribulose monophosphate pathways were incomplete. The detoxification of formaldehyde could follow the tetrahydrofolate pathway. Furthermore, “Ca. Methylacidithermus pantelleriae” might be capable of nitric oxide reduction but genes for dissimilatory nitrate reduction and nitrogen fixation were not identified. Unlike other verrucomicrobial methanotrophs, genes encoding for enzymes involved in hydrogen oxidation could not be found. In conclusion, the discovery of this new MAG expands the diversity and metabolism of verrucomicrobial methanotrophs.
Introduction
Verrucomicrobial methanotrophs are a group of aerobic bacteria usually found in the acidic soil of geothermal active regions (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008; Sharp et al., 2014; van Teeseling et al., 2014). Their genomes all encode one or up to three particulate methane monooxygenase enzymes (pMMO) for the conversion of methane to methanol and a XoxF-type methanol dehydrogenase (MDH) to transform methanol to formate. The peculiarity of their XoxF-MDH is the strict dependence on rare earth elements (REEs), which are present in the active site together with the pyrroloquinoline quinone (PQQ) cofactors (Pol et al., 2014). Formate is ultimately converted to CO2 by a formate dehydrogenase (Picone and Op den Camp, 2019).
Inorganic carbon is fixed autotrophically using the Calvin-Benson-Bassham (CBB) cycle rather than the serine- or ribulose monophosphate (RuMP) pathways used by proteobacterial methanotrophs (Khadem et al., 2011; van Teeseling et al., 2014). Two verrucomicrobial methanotrophs were shown to be able to grow in the absence of methane when supplied with a mixture of carbon dioxide and hydrogen (Mohammadi et al., 2017a; Schmitz et al., 2020). Moreover, they are capable of nitrogen fixation and partial denitrification (Khadem et al., 2010; Mohammadi et al., 2017b). The current classification divides verrucomicrobial methanotrophs into two genera: Methylacidimicrobium, generally mesophilic and extremely acidophilic and the thermophilic but less acidophilic Methylacidiphilum (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008; Sharp et al., 2014; van Teeseling et al., 2014; Picone et al., 2021).
Verrucomicrobial methanotrophs were detected in different geothermal ecosystems, including the Favara Grande, a volcanic area on Pantelleria Island, Italy. In particular, pmo-containing bacteria closely related to Methylacidiphilum fumariolicum SolV, were detected in site FAV2 (Gagliano et al., 2014). This site was characterized by pH values of 4–4.5 and temperature ranging from 60°C in the top layer of the soil, to 92°C at 50 cm depth. Ammonia was scarce, whereas high emissions of carbon dioxide (CO2), hydrogen (H2), and methane (CH4) were recorded (Gagliano et al., 2016). A recent metagenomic analysis of site FAV2 revealed a methanotrophic community composed of Proteobacteria and Verrucomicrobia (Picone et al., 2020), supporting the findings of Gagliano et al. (2014). Two Metagenome Assembled Genomes (MAGs) that belonged to the phylum Verrucomicrobia were retrieved. One of these MAGs (MAG5) was a novel Methylacidimicrobium species (Picone et al., 2021). MAG9, instead, did not classify as Methylacidiphilum or Methylacidimicrobium, indicating that it may represent a novel genus.
In this study we determine the phylogeny of this new verrucomicrobial methanotroph species and analyze its genome to predict the metabolic potential.
Materials and Methods
Sampling
Samples were collected in June 2017 in the area of Favara Grande on Pantelleria Island, Italy (23°21′77′′N; 40°73′160′′E). Soil samples were taken using a core sampler (diameter 1.5 cm) and deposited in sterile 50 mL tubes. Tubes were stored at 4°C till further analysis. For a more extensive description of the sampling site (see Picone et al., 2020).
DNA Extraction and Sequencing
DNA was extracted from soil samples using two different methods: Fast DNA Spin kit for soil (MP Biomedicals, Santa Ana, California), according to manufacturer’s instructions, and CTAB DNA extraction (Allen et al., 2006). Cell lysis within the CTAB method was performed by incubating 250 mg of soil with 675 μl of CTAB buffer (100 mM Tris, 100 mM EDTA, 100 mM Na2HPO4, 1.5 M NaCl and 1% CTAB), 50 μl of lysozyme (10 mg/ml, 66,200 U/mg), and 30 μl of Rnase A (10 mg/ml) for 30 min at 37°C. Fifty microliter of Proteinase K (10 mg/ml, 20 U/mg) was added to the sample and incubated for 30 min at 37°C. Next, 150 μl of 10% SDS was added and the mixture was incubated for 2 h at 65°C. DNA was extracted by adding 1 volume of phenol/chloroform/isoamylalcohol (25:24:1) and incubating the sample for 20 min at 65°C. Supernatant was treated with 1 volume of chloroform/isoamylalcohol (24:1) and centrifuged for 10 min at 20,000 × g. Next, 0.6 volume of isopropanol was added to the aqueous phase and DNA was precipitated by centrifuging the sample for 15 min at 20,000 × g. The DNA pellet was washed using ice cold 70% ethanol and centrifuged for 10 min at 20,000 × g. The pellet was air-dried and resuspended in 30 μl of MilliQ water. DNA was quantified with the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA) and sequenced on the Illumina sequencing platform. For library preparation the Nextera XT kit (Illumina, San Diego, California) was used according to the manufacturer’s instructions. Enzymatic tagmentation was performed starting with 1 ng of DNA, followed by incorporation of the indexed adapters and amplification of the library. After purification of the amplified library using AMPure XP beads (Beckman Coulter, Indianapolis), the two libraries were checked for quality and size distribution using the Agilent 2100 Bioanalyzer and the High sensitivity DNA kit. Quantitation of the libraries was performed by Qubit using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts). The two libraries were pooled, denatured and sequenced with the Illumina Miseq sequence machine (San Diego, California). Paired end sequencing of 2 × 300 base pairs was performed using the MiSeq Reagent Kit v3 (Illumina, San Diego, California) according the manufacturers protocol. Sequencing resulted in a total of 55,685,154 and 56,928,602 reads for CTAB and PowerSoil extracted DNA, respectively.
Genome Assembly, Binning, and Annotation
Reads were trimmed using BBDuk (BBMap), assembled by MEGAHIT v1.0.3 (Li et al., 2015) and binned using a combination of different algorithms, namely BinSanity (Graham et al., 2017), COCACOLA (Lu et al., 2017), CONCOCT (Alneberg et al., 2014), MaxBin 2.0 (Wu et al., 2016), and MetaBAT 2 (Kang et al., 2019). DAS Tool 1.0 was used for consensus binning (Sieber et al., 2018), and CheckM was used to assess the MAG quality (Parks et al., 2015). The average nucleotide identity using BLAST (ANIb) was calculated using JSpeciesWS software with standard settings (Richter et al., 2016). The genome of Bin 9 was integrated in the Microscope platform (Vallenet et al., 2006, 2009) and annotated as described elsewhere (Lücker et al., 2010).
Phylogenetic Analysis
16S rRNA gene sequences were identified in the NCBI database by BLAST, aligned with the sequences obtained in this study by MUSCLE32 and used to build phylogenetic trees (with the Maximum Likelihood method and 500 bootstraps in Mega 7 (Kumar et al., 2016). Average Amino acid Identity (AAI) values were calculated using the AAI calculation tool developed by the Kostas lab (Rodriguez-R and Konstantinidis, 2014).
Results and Discussion
Proteobacterial and Verrucomicrobial Methanotrophs in the Soil of Favara Grande
The Favara Grande is an area on Pantelleria Island characterized by hydrothermal activity with gas emissions of CO2, H2, and CH4 (Picone et al., 2020). Within the bacterial community, methanotrophs belonging to the Gammaproteobacteria and Verrucomicrobia phyla could be identified through pmoA sequencing (Gagliano et al., 2014). 16S rRNA gene amplicon sequencing analysis, instead, did not detect Verrucomicrobia in the soil of Favara Grande, but potential methanotrophy was mainly attributed to Gammaproteobacteria (Gagliano et al., 2016). Recent Illumina metagenomic sequencing at a much higher resolution could show the presence of five MAGs related to methanotrophs (Picone et al., 2020; Supplementary Figure 1). MAG2 resembled a novel gammaproteobacterial Methylobacter species (Hogendoorn et al., 2021) and MAG8 and MAG16 were related to Methylococcus sp. The two remaining MAGs clustered within the phylum Verrucomicrobia. A detailed description of Methylacidimicrobium thermophilum AP8, an isolated representative of MAG5, was recently published (Picone et al., 2021). 16S rRNA analysis of MAG9 revealed a species phylogenetically distant to other known verrucomicrobial methanotrophs. The closest cultured relatives were Methylacidiphilum sp. RTK17 and Methylacidiphilum infernorum V4, that shared only 89.9% 16S rRNA identity to MAG9. This MAG was analyzed in detail.
Phylogenetic analysis showed that the 16S rRNA gene of MAG9 clustered in between Methylacidimicrobium and Methylacidiphilum species (Figure 1; Dunfield et al., 2007; Pol et al., 2007; Op den Camp et al., 2009; van Teeseling et al., 2014; Schmitz et al., 2021). The 16S rRNA and AAI values (Table 1 and Supplementary Table 1) fell below the threshold for species delimitation (95% for AAI and 98.7–99% for 16S rRNA) (Stackebrandt and Ebers, 2006; Thompson et al., 2013). Considering the AAI thresholds proposed by Konstantinidis et al. (2017) for uncultivated microorganisms (45–65% for the same family, 65–95% for the same genus and 95–100% for the same species), these results classified MAG9 as representing a new species of a new genus, for which we propose the name “Candidatus Methylacidithermus pantelleriae” sp. PQ17. This “Ca. Methylacidithermus” genus is the third genus of methanotrophic Verrucomicrobia within the family Methylacidiphilaceae, that adds to the previously described Methylacidiphilum and Methylacidimicrobium genera (Op den Camp et al., 2009; van Teeseling et al., 2014; Schmitz et al., 2021).
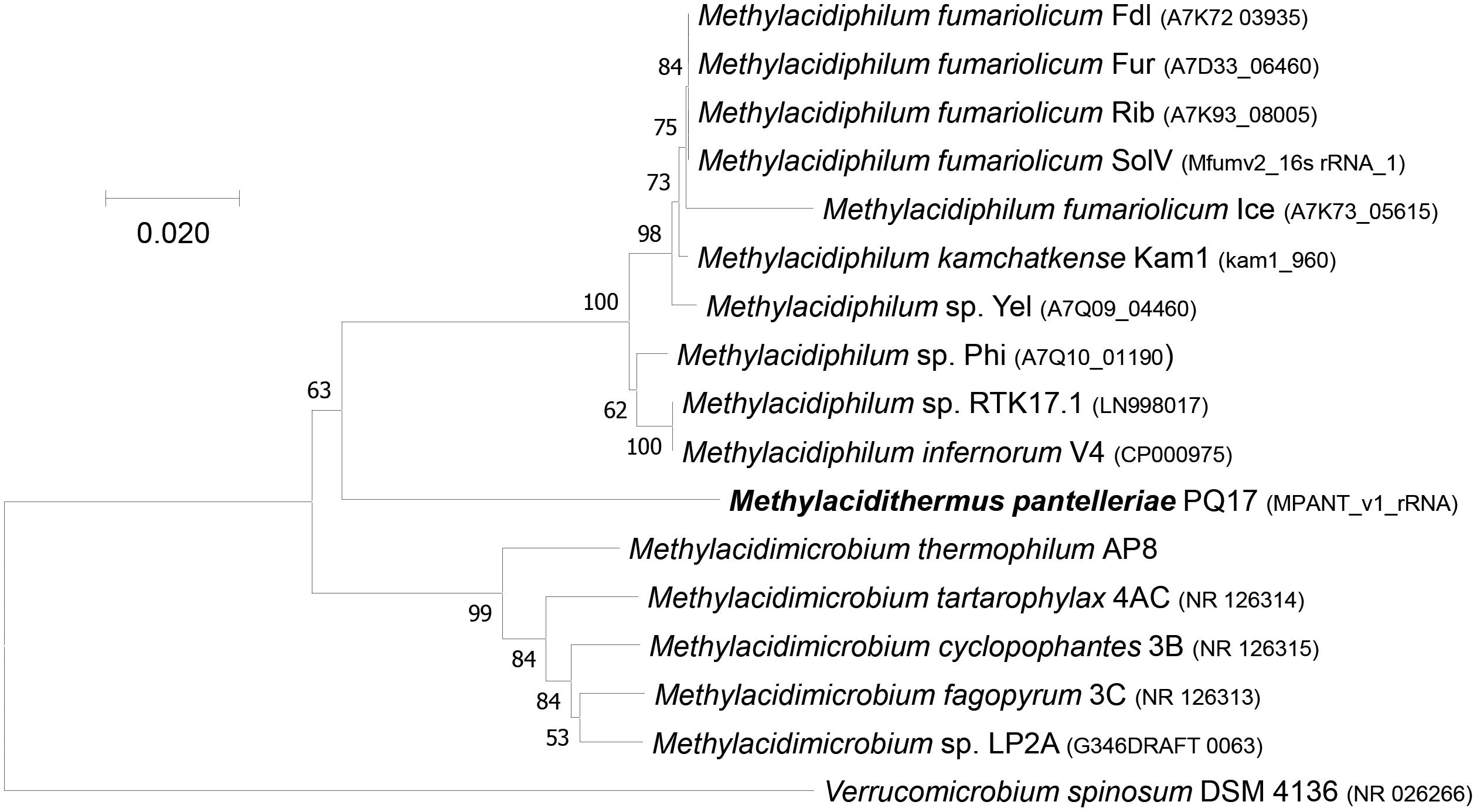
Figure 1. 16S rRNA gene-based phylogenetic tree of methanotrophic Verrucomicrobia. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 0.41438253 is shown. The percentage of replicate trees (>50%) in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 17 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1,575 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
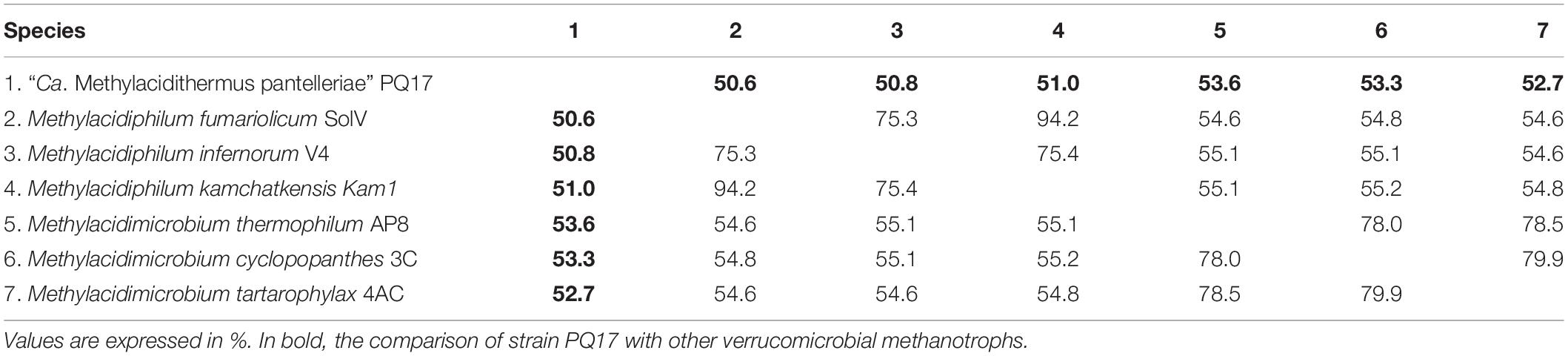
Table 1. Average amino acid identity (AAI) value comparison between different verrucomicrobial methanotroph species.
Genomic Characterization of “Ca. Methylacidithermus Pantelleriae”
The draft genome of strain PQ17 was analyzed in details to gain a better understanding about its metabolic potential and its role in the geothermal soil of Pantelleria. MAG9 consisted of 71 contigs ranging from 401,379 to 2,075 bp, for a total of 2,466,655 bp, containing 3,127 predicted CDSs and an overall 55.2% GC-content. CheckM analysis revealed a completeness of 98.6, 1.3% contamination and no strain heterogeneity (Supplementary Figure 2). A total of 3,204 genes could be identified, 3,127 of which were protein coding genes and 77 were RNA genes. One 16S and two 23S and 5S rRNA genes were retrieved, indicating that one 16S rRNA copy was probably missing from the draft genome. Functions could be assigned to 2,164 protein coding genes (Table 2). 47.4% of the predicted genes were allocated into Clusters of Orthologous Groups (Supplementary Table 2).
Genes involved in carbon, nitrogen and sulfur metabolism were analyzed in detail. Their pathways will be described in the upcoming sections and a schematic representation of the metabolism of strain PQ17 can be found in Figure 2.
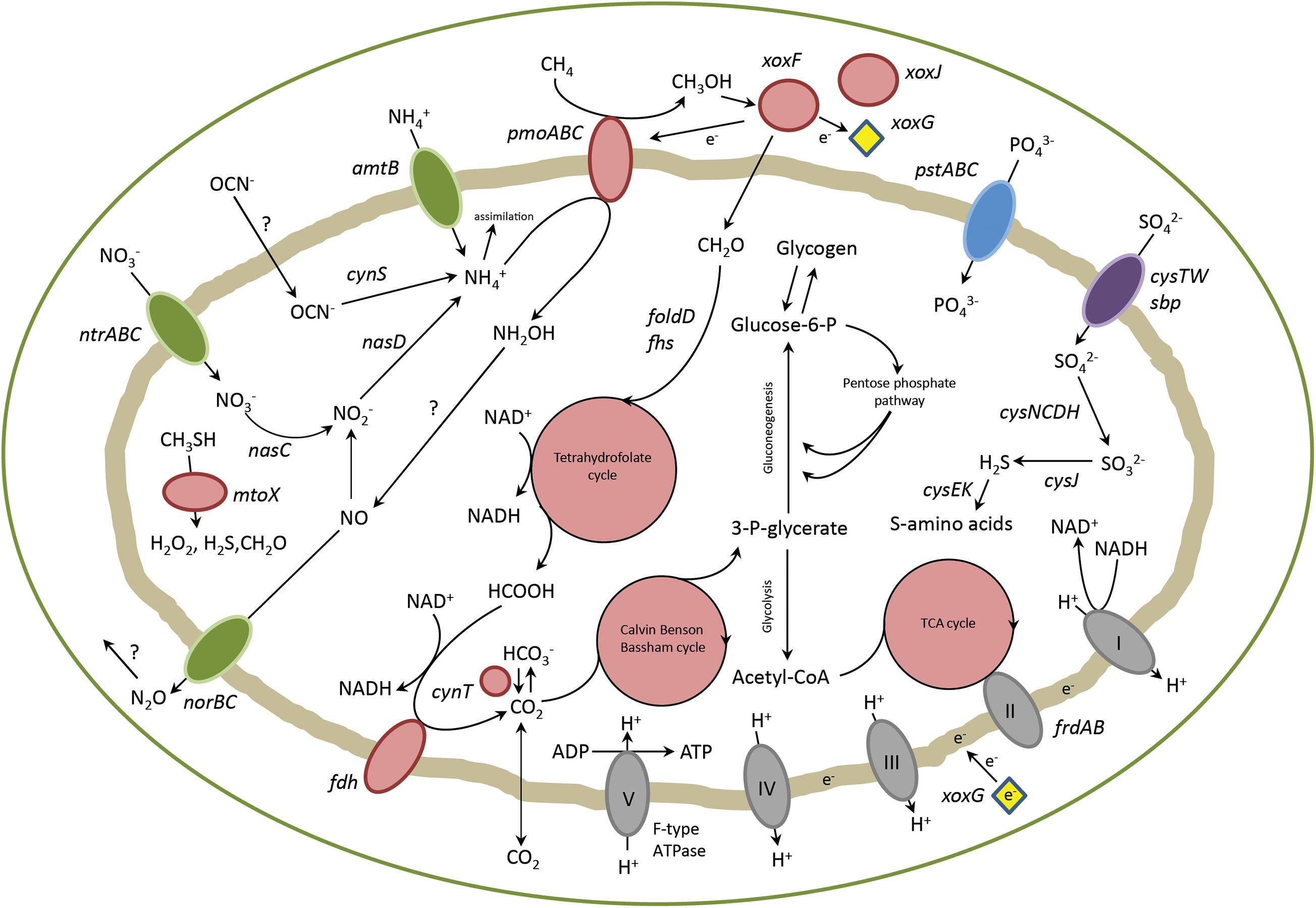
Figure 2. Overview of metabolic pathways in “Ca. M. pantelleriae.” Colors of enzymes and transporters indicate nitrogen metabolism (green), carbon metabolism (red), sulfate metabolism (purple), phosphate metabolism (blue) and complexes of the respiratory chain (gray). Genes: pmoABC, methane monoxygenase; xoxF, methanol dehydrogenase, xoxG, cytochrome cL; xoxJ, periplasmic binding protein; folD, bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase; fhs, formate-tetrahydrofolate ligase; fdh, formate dehydrogenase; cynT, carbonic anhydrase; frdAB, fumarate reductase; pstABC, phosphate transporter; cysTW/sbp, sulfate transporter; amtB, ammonia transporter; ntrABC, nitrate transporter; nasC, assimilatory nitrate reductase; nasD, assimilatory nitrite reductase; norBC, nitric oxide reductase; cynS, cyanase; cysCDH, adenylyl-sulfate kinase, sulfate adenylyltransferase, phosphoadenosine phosphosulfate reductase; cysJ, sulfite reductase; cysK, cysteine synthase; cysE, serine acetyltransferase; mtoX, methanethiol oxidase.
Methanotrophy
As a first step in methane oxidation, methane monooxygenases use molecular oxygen to break the energetically strong C-H bond of methane to form methanol (Sirajuddin and Rosenzweig, 2015). So far, two types of this enzyme have been described: a membrane-bound pMMO and a soluble methane monooxygenase (sMMO). One single pmoCAB operon and a pmoD subunit, encoding a copper-binding protein (Fisher et al., 2018), were found in MAG9, whereas no other mmo genes were identified (Table 3). This is in line with previously described verrucomicrobial methanotrophs, although the number of pmo operons seems to be variable (Op den Camp et al., 2009; van Teeseling et al., 2014; Erikstad et al., 2019; Picone et al., 2020; Schmitz et al., 2021). The PmoA phylogenetic tree including strain PQ17 (Supplementary Figure 3) supports the phylogeny derived from 16S rRNA gene analysis.
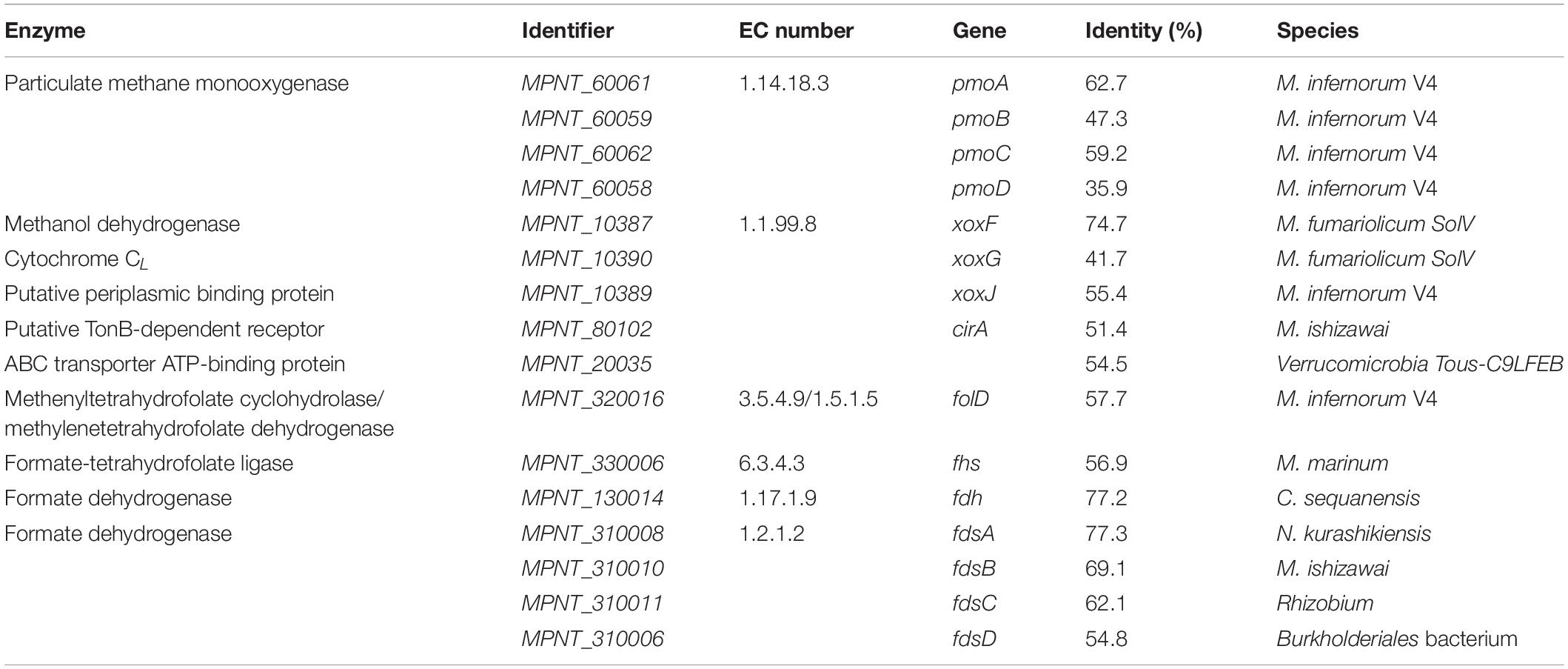
Table 3. Genes encoding for enzymes involved in methane oxidation pathway, along with their Enzyme Commission (EC) numbers and percentage identity to the most similar homologue.
The second step in methane oxidation is the conversion of methanol to formaldehyde or formate. Two types of pyrroloquinoline quinone (PQQ)-containing MDHs are generally found in methanotrophs and methylotrophs: The MxaFI and the XoxF type MDH. Whereas MxaFI depends on calcium for its catalysis, XoxF was found to contain lanthanides in the active site (Pol et al., 2014). Analysis of the MDH sequence of strain PQ17 showed that this enzyme presented a conserved Asp residue required for the coordination of lanthanides in the active site (Keltjens et al., 2014; Pol et al., 2014; Good et al., 2020). Furthermore, it exhibited 74% amino acid identity to XoxF from Methylacidiphilum fumariolicum SolV, confirming that this protein was a XoxF type and it belonged to group XoxF1 (Keltjens et al., 2014). xoxG and xoxJ genes were also found in the genome of strain PQ17 (Table 3). xoxG encodes a cytochrome CL that functions as electron acceptor for XoxF, whereas xoxJ encodes a periplasmic binding protein that is proposed to be involved in the activation of XoxF and, more specifically, in the insertion of the PQQ cofactor in apo-XoxF (Zheng et al., 2018; Featherston et al., 2019; Versantvoort et al., 2019). In the Methylacidiphilum species SolV and Kam1 these proteins are exceptionally present as the fusion protein XoxG/J (Islam et al., 2008; Versantvoort et al., 2019).
Several genes have been proposed as candidates for lanthanide incorporation in bacterial cells (Ochsner et al., 2019). The gene cirA, encoding a TonB-dependent receptor, and a component of the ABC transport system have been identified in other Verrucomicrobia (Picone et al., 2021) and were also shown to be present in the genome of strain PQ17 (Table 3). The lanthanide binding protein lanmodulin described in M. extorquens, instead, could not be found (Cotruvo et al., 2018).
XoxF from strain SolV is known to convert methanol to formate in vitro in a four electron process (Pol et al., 2014). In Methylobacterium extorquens AM instead, XoxF generated formaldehyde (Good et al., 2018), which is converted to formate by formaldehyde dehydrogenase. Similarly to strain SolV, no formaldehyde dehydrogenase was detected in “Ca. M. pantelleriae.” If formaldehyde is produced, different detoxification pathways are known. The tetrahydrofolate pathway was the only pathway identified in strain PQ17. The first step in this cycle is a spontaneous reaction which couples formaldehyde to tetrahydrofolate forming 5,10-methylenetetrahydrofolate. The bifunctional enzyme FolD catalyzes the second and third steps of this cycle, converting 5,10-methylenetetrahydrofolate via 5,10-methenyltetrahydrofolate to 10-formyltetrahydrofolate. The last reaction step is catalyzed by formate tetrahydrofolate ligase (Fhs), in which formate is produced and tetrahydrofolate is regenerated (Table 3 and Figure 2; Vorholt, 2002). Formaldehyde could also be produced by methanethiol oxidase (MtoX, MPNT_180031), an enzyme apparently conserved in verrucomicrobial methanotrophs (Eyice et al., 2018; Picone et al., 2021).
In the last step of methane oxidation, formate is converted to CO2 by formate dehydrogenase (FDH). In the bacterial kingdom, a large variety of FDH exist, which are all highly diverse regarding cofactor usage and mechanism (Hartmann et al., 2015). For “Ca. M. pantelleriae,” two different FDHs were found in the genome, one was cytoplasmic and the other was predicted to be a membrane-bound enzyme complex composed of four subunits (Table 3).
Central Carbon Metabolism
Methanotrophs assimilate carbon into their metabolism using different pathways. Verrucomicrobia are generally able to fix CO2 via the Calvin-Benson-Bassham (CBB) cycle (Khadem et al., 2011). This cycle is initiated by the reaction of carbon dioxide with ribulose bisphosphate, which is catalyzed by ribulose bisphosphate carboxylase (RuBisCO) (Tabita, 2007). The small and a large subunit of this enzyme could be identified in the genome, together with two carbonic anhydrases (Table 4). The products of RuBisCO are two molecules of 3-phosphoglycerate (3-PG), which are converted back to ribulose bisphosphate through a series of gluconeogenic and pentose phosphate pathway reactions. Every three CO2 molecules yield net one molecule of 3-PG, which can be incorporated in central carbon metabolism.
Beside the CBB cycle, the Serine and the RuMP pathways are other strategies for carbon incorporation in microorganisms (Chistoserdova, 2011). Some genes involved in the Serine pathway could be found in the genome of strain PQ17 (Table 4), but four essential genes were lacking (hprA EC 1.1.1.29, gckA 2.7.1.165, mtkB EC 6.1.2.9, mcl EC 4.1.3.24). Likewise, two genes required for the RuMP pathway were also absent (hxlA EC 4.1.2.43, hxlB EC 5.3.1.27). Therefore, it is highly unlikely for “Ca. M. pantelleriae” to fix carbon using these pathways.
All glycolytic, gluconeogenic and pentose phosphate pathway genes could be retrieved from MAG9 (Supplementary Table 3), except for phosphofructokinase (EC 2.7.1.11). The other two genera of verrucomicrobial methanotrophs, Methylacidiphilum and Methylacidimicrobium have genes encoding this protein. All genes for the citric acid cycle were found. Moreover, genes for synthesis and degradation of glycogen were identified.
Energy Conservation and Respiration
“Ca. M. pantelleriae” uses O2 as electron acceptor. Complex I of the respiratory chain (nuoABCDEFGHIKLMN) was found in the genome. For complex II, subunits a, b and c of succinate dehydrogenase were found, but subunit d was lacking. Subunits for a canonical complex III could not be retrieved. The verrucomicrobial methanotrophs, including strain PQ17, possess genes encoding an Alternative Complex III, a structurally different protein complex with similar function (MPNT_10279-10285) (Refojo et al., 2012; Schmitz et al., 2021). Finally, the electrons are transferred to complex IV and the F0F1 ATP synthase (complex V) generating ATP using the proton motive force (Supplementary Table 3). “Ca. M. pantelleriae” possesses genes encoding two distinct Complexes IV: aa3-type and a ba3-type. The genomes of the other verrucomicrobial methanotrophs encode for an additional cbb3-type Complex IV (Schmitz et al., 2021).
Amino Acid Biosynthesis
Pathways for the biosynthesis of 12 amino acids (alanine, isoleucine, leucine, proline, valine, phenylalanine, tyrosine, tryptophan, arginine, lysine, threonine, and cysteine) were completely present in the genome. For histidine, only one gene encoding the biosynthesis protein HisE was absent.
The complete pathways for asparagine/aspartate and glutamine/glutamate biosynthesis could not be fully resolved but the enzyme for the conversion of oxaloacetate to aspartate was identified (aspC MPNT_250010). Genes encoding enzymes for the formation of asparagine, instead, could not be retrieved. Glutamate could be formed from 2-oxoglutarate via glutamate synthase (GltB, MPNT_40080) or from gamma-aminobutyric acid through glutamate decarboxylase (MPNT_510001). Glutamate dehydrogenase (GDH) was not identified, whereas glutamine synthetase and glutamate synthase (GS-GOGAT) were both present (Supplementary Table 3). GDH and GS-GOGAT pathways are also used for ammonia incorporation into biomass. Ammonia incorporation via GS-GOGAT usually happens under low ammonia concentrations (Tyler, 1978; Bellion and Bolbot, 1983). The presence of an alanine dehydrogenase (MPNT_50137) in the genome indicates that ammonia could also be incorporated though alanine, starting from pyruvate and NH4+.
The pathways for glycine and serine biosynthesis are less straightforward. If serine is synthetized from 3-PG, only D-3-phosphoglycerate dehydrogenase/2-oxoglutarate reductase was present (serA, MPNT_20138), whereas phosphoserine aminotransferase (serC, EC 2.6.1.52) and phosphoserine phosphatase (serB, EC 3.1.3.3) were absent. We cannot exclude that these reactions are still performed in vivo, but catalyzed by unknown enzymes. The fragmentation of the genome could also prevent us from retrieving these genes. Assuming that these pathways are actually missing in strain PQ17, serine can still be synthesized in a one-step reaction catalyzed by serine hydroxymethyltransferase (glyA, MPNT_220042) using 5,10-methylenetetrahydrofolate and glycine. For this to be a feasible strategy, “Ca. M. pantelleriae” should be able to synthesize its glycine from a different source than serine. As all four subunits of the glycine cleavage system are present in its genome (MPNT_20097, 20098, 20099, 420008), we propose that “Ca. M. pantelleriae” could use this machinery in reverse to synthesize glycine from ammonia, carbon dioxide and 5,10-methylenetetrahydrofolate (Kikuchi et al., 2008), which has also been described previously for Clostridium acidiurici (Gariboldi and Drake, 1984). Furthermore, glycine can be synthetized from glyoxylate (agxt2, MPNT_100077) and from sarcosine (dauA, MPNT_10078).
Nitrogen Metabolism
Ammonium from the environment can be imported directly into the cell using either of two AmtB transporters (MPNT_100073, 250005). Alternatively, nitrogen can be obtained by uptake of nitrate via a NrtABC transporter (Supplementary Table 3), followed by reduction to ammonia by NasC and NasD (Table 5 and Supplementary Table 3). Interesting is the presence of the gene cynS, which encodes cyanase, an enzyme converting cyanate and bicarbonate to carbon dioxide and ammonium. Cyanate can act as energy and nitrogen source for nitrifiers (Palatinszky et al., 2015) and its presence has been detected in other verrucomicrobial methanotrophs (Picone et al., 2021). Unlike other verrucomicrobial methanotrophs, genes encoding a nitrogenase enzyme could not be found (Op den Camp et al., 2009; Khadem et al., 2010; Schmitz et al., 2021). Fixed ammonium is mostly used for biosynthetic purposes, but some ammonium is also converted into hydroxylamine by pmoA, which is a structural homolog of ammonium monooxygenase amoA (Sirajuddin and Rosenzweig, 2015). As hydroxylamine is toxic to the cell, it must be further metabolized into less harmful compounds. However, hydroxylamine oxidoreductase (hao), which is present in ammonia oxidizers but also in other methanotrophs such as M. fumariolicum SolV (Pol et al., 2007), could not be retrieved from the MAG. The nitric oxide reductase encoded by norBC (Table 5) was identified, whereas other denitrification genes, like narB (EC 1.7.5.1) and nosZ (EC 1.7.2.5), were not detected.
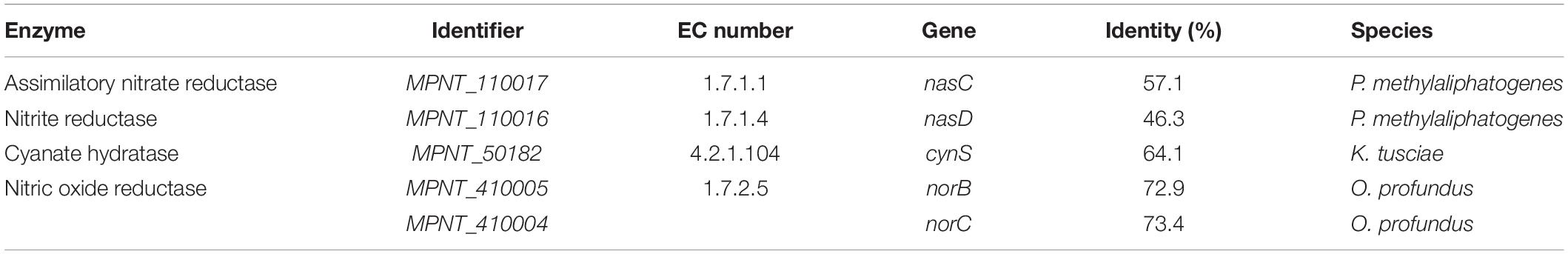
Table 5. Genes encoding for enzymes involved in nitrogen metabolism, along with their Enzyme Commission (EC) numbers, and percentage identity to the most similar homologue.
Sulfur and Phosphate Metabolism
The primary way to fix sulfur for “Ca. M. pantelleriae” is to reduce sulfate to biologically available sulfide. For this, sulfate needs to be transported into the cell using the sulfate ABC-transporter sbp/cysTW (MPNT_580001-580004). Subsequently, sulfate can be reduced to sulfite via adenylyl sulfate and 3′-phosphoadenylyl sulfate intermediates by the genes, catalyzed by cysD (MPNT_10354), cysC (MPNT_10355) and cysH (MPNT_20190), respectively. Finally, sulfite can be further reduced to H2S by cysJ (MPNT_10061, MPNT_40049) or sir1 (MPNT_20189) and used for cysteine biosynthesis (cysK (MPNT_110064). Phosphate can be transported directly over the membrane using the ABC-transporter encoded by pstABCS (Supplementary Table 3) and does not require further conversions. The presence of polyphosphate particles has been observed in verrucomicrobial methanotrophs (van Teeseling et al., 2014). Polyphosphate storage is likely in strain PQ17 as genes encoding polyphosphate kinase (MPNT_190035) and exopolyphosphatase (MPNT_40183) were identified.
Conclusion and Ecological Role
“Ca. Methylacidithermus pantelleriae” PQ17 presents most of the typical characteristics of verrucomicrobial methanotrophs. This microorganism was detected in a thermoacidophilic volcanic environment and its genome predicts it to be an aerobic bacterium able to fix carbon via the CBB cycle. Methane oxidation to methanol may use the methane monooxygenase encoded by the pmoCAB operon and the conversion of methanol could be carried out by the XoxF-type MDH. Contrary to other verrucomicrobial methanotrophs, the genome of strain PQ17 does not encode genes for nitrogen fixation, nor for the oxidation of hydrogen, a common energy substrate for verrucomicrobial methanotrophs. These features, together with phylogenetic analysis, suggest that “Ca. M. pantelleriae” has evolved differently from other verrucomicrobial methanotrophs. This bacterium probably utilizes exclusively methane or methanol for energy production and provides nitrogen for biomass mainly via nitrate and ammonia and not by fixing N2 gas.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJEB38823 (genome accession number CAJNOB000000000), https://mage.genoscope.cns.fr/microscope/mage/viewer.php?O_id=8619, 8619.
Author Contributions
NP, AP, MJ, and HO designed the projects and experiments. NP, CH, AP, AG, WD’A, PQ, and HO sampled the geothermal soils. NP and CH performed the DNA isolation. TA, JF, NP, and PB performed sequencing, assembly and annotation. NP, PB, AP, and HO carried out the data analysis. NP, PB, and HO wrote the manuscript. All authors contributed to revision of the manuscript, and read and approved the submitted version.
Funding
NP, CH, and HO were supported by the European Research Council (ERC Advanced Grant Project VOLCANO 669371). MJ was supported by the European Research Council (ERC Advanced Grant Project Eco_MoM 339880) and the Soehngen Institute of Anaerobic Microbiology (SIAM 024002002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.666929/full#supplementary-material
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Alneberg, J., Bjarnason, B. S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Bellion, E., and Bolbot, J. A. (1983). Nitrogen assimilation in facultative methylotrophic bacteria. Curr. Microbiol. 9, 37–44. doi: 10.1016/B978-0-12-809633-8.20680-8
Chistoserdova, L. (2011). Modularity of methylotrophy, revisited. Environ. Microbiol. 13, 2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x
Cotruvo, J. A., Featherston, E. R., Mattocks, J. A., Ho, J. V., and Laremore, T. N. (2018). Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J. Am. Chem. Soc. 140, 15056–15061. doi: 10.1021/jacs.8b09842
Dunfield, P. F., Yuryev, A., Senin, P., Smirnova, A. V., Stott, M. B., Hou, S., et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–882. doi: 10.1038/nature06411
Erikstad, H.-A., Ceballos, R. M., Smestad, N. B., and Birkeland, N.-K. (2019). Global biogeographic distribution patterns of thermoacidophilic Verrucomicrobia methanotrophs suggest allopatric evolution. Front. Microbiol. 10:1129. doi: 10.3389/fmicb.2019.01129
Eyice, Ö, Myronova, N., Pol, A., Carrión, O., Todd, J. D., Smith, T. J., et al. (2018). Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 12, 145–160. doi: 10.1038/ismej.2017.148
Featherston, E. R., Rose, H. R., McBride, M. J., Taylor, E. M., Boal, A. K., and Cotruvo, J. A. Jr. (2019). Biochemical and structural characterization of XoxG and XoxJ and their roles in lanthanide-dependent methanol dehydrogenase activity. Chem. Bio. Chem. 20, 2360–2372. doi: 10.1002/cbic.201900184
Fisher, O. S., Kenney, G. E., Ross, M. O., Ro, S. Y., Lemma, B. E., Batelu, S., et al. (2018). Characterization of a long overlooked copper protein from methane- and ammonia-oxidizing bacteria. Nat. Commun. 9:4276. doi: 10.1038/s41467-018-06681-5
Gagliano, A. L., D’Alessandro, W., Tagliavia, M., Parello, F., and Quatrini, P. (2014). Methanotrophic activity and diversity of methanotrophs in volcanic geothermal soils at Pantelleria (Italy). Biogeosciences 11, 5865–5875. doi: 10.5194/bg-11-5865-2014
Gagliano, A. L., Tagliavia, M., D’Alessandro, W., Franzetti, A., Parello, F., and Quatrini, P. (2016). So close, so different: geothermal flux shapes divergent soil microbial communities at neighbouring sites. Geobiology 14, 150–162. doi: 10.1111/gbi.12167
Gariboldi, R. T., and Drake, H. L. (1984). Glycine synthase of the purinolytic bacterium, Clostridium acidiurici. Purification of the glycine-CO2 exchange system. J. Biol. Chem. 259, 6085–6089.
Good, N. M., Fellner, M., Demirer, K., Hu, J., Hausinger, R. P., and Martinez-Gomez, N. C. (2020). Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function. J. Biol. Chem. 295, 8272–8284. doi: 10.1074/jbc.RA120.013227
Good, N. M., Walser, O. N., Moore, R. S., Suriano, C. J., Huff, A. F., and Martinez-Gomez, N. C. (2018). Investigation of lanthanide-dependent methylotrophy uncovers complementary roles for alcohol dehydrogenase enzymes. bioRxiv [Preprint] doi: 10.1101/329011 bioRxiv 329011,
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2017). Bin Sanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 5, e3035. doi: 10.7717/peerj.3035
Hartmann, T., Schwanhold, N., and Leimkühler, S. (2015). Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. Biochim. Biophys. Acta Proteins Proteomics 1854, 1090–1100. doi: 10.1016/j.bbapap.2014.12.006
Hogendoorn, C., Picone, N., van Hout, F., Vijverberg, S., Poghosyan, L., van Alen, T. A., et al. (2021). Draft genome of a novel methanotrophic Methylobacter sp. from the volcanic soils of Pantelleria Island. Antonie van Leeuwenhoek 114, 313–324. doi: 10.1007/s10482-021-01525-7
Islam, T., Jensen, S., Reigstad, L. J., Larsen, Ø, and Birkeland, N.-K. (2008). Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105, 300–304. doi: 10.1073/pnas.0704162105
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J. 7:e7359. doi: 10.7717/peerj.7359
Keltjens, J. T., Pol, A., Reimann, J., and Op den Camp, H. J. M. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98, 6163–6183. doi: 10.1007/s00253-014-5766-8
Khadem, A. F., Pol, A., Jetten, M. S. M., and Op den Camp, H. J. M. (2010). Nitrogen fixation by the verrucomicrobial methanotroph “Methylacidiphilum fumariolicum” SolV. Microbiology 156, 1052–1059. doi: 10.1099/mic.0.0360610
Khadem, A. F., Pol, A., Wieczorek, A., Mohammadi, S. S., Francoijs, K.-J., Stunnenberg, H. G., et al. (2011). Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the calvin-benson-bassham cycle for carbon dioxide fixation. J. Bacteriol. 193, 4438–4446. doi: 10.1128/JB.00407-11
Kikuchi, G., Motokawa, Y., Yoshida, T., and Hiraga, K. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Natl. Acad. Sci. U.S.A. 84, 246–263. doi: 10.2183/pjab/84.246
Konstantinidis, K. T., Rosselló-Móra, R., and Amann, R. (2017). Uncultivated microbes in need of their own taxonomy. ISME J. 11, 2399–2406. doi: 10.1038/ismej.2017.113
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, D., Liu, C.-M., Luo, R., Sadakane, K., and Lam, T.-W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Lu, Y. Y., Chen, T., Fuhrman, J. A., and Sun, F. (2017). COCACOLA: binning metagenomic contigs using sequence COm position, read cover age, CO-alignment and paired-end read link age. Bioinformatics 33, 791–798. doi: 10.1093/bioinformatics/btw290
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
Mohammadi, S., Pol, A., Van Alen, T. A., Jetten, M. S. M., and Op den Camp, H. J. M. (2017a). Methylacidiphilum fumariolicum SolV, a thermoacidophilic “Knallgas” methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11, 945–958. doi: 10.1038/ismej.2016.171
Mohammadi, S. S., Pol, A., van Alen, T., Jetten, M. S. M., and Op den Camp, H. J. M. (2017b). Ammonia oxidation and nitrite reduction in the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. Front. Microbiol. 8:1901. doi: 10.3389/fmicb.2017.01901
Ochsner, A. M., Hemmerle, L., Vonderach, T., Nüssli, R., Bortfeld-Miller, M., Hattendorf, B., et al. (2019). Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol. Microbiol. 111, 1152–1166. doi: 10.1111/mmi.14208
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Palatinszky, M., Herbold, C., Jehmlich, N., Pogoda, M., Han, P., von Bergen, M., et al. (2015). Cyanate as an energy source for nitrifiers. Nature 524, 105–108. doi: 10.1038/nature14856
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Picone, N., Blom, P., Wallenius, A. J., Hogendoorn, C., Mesman, R., Cremers, G., et al. (2021). Methylacidimicrobium thermophilum AP8, a novel methane- and hydrogen-oxidizing bacterium isolated from volcanic soil on Pantelleria Island. Italy. Front. Microbiol. 12:637762. doi: 10.3389/fmicb.2021.637762
Picone, N., Hogendoorn, C., Cremers, G., Poghosyan, L., Pol, A., van Alen, T. A., et al. (2020). Geothermal gases shape the microbial community of the volcanic soil of Pantelleria. Italy. mSystems 5, e517–e520. doi: 10.1128/mSystems.00517-20
Picone, N., and Op den Camp, H. J. M. (2019). Role of rare earth elements in methanol oxidation. Curr. Opin. Chem. Biol. 49, 39–44. doi: 10.1016/j.cbpa.2018.09.019
Pol, A., Barends, T. R. M., Dietl, A., Khadem, A. F., Eygensteyn, J., Jetten, M. S. M., et al. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 16, 255–264. doi: 10.1111/1462-2920.12249
Pol, A., Heijmans, K., Harhangi, H. R., Tedesco, D., Jetten, M. S. M., and Op den Camp, H. J. M. (2007). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–878. doi: 10.1038/nature06222
Refojo, P. N., Teixeira, M., and Pereira, M. M. (2012). The alternative complex III: properties and possible mechanisms for electron transfer and energy conservation. Biochim. Biophys. Acta Bioenergetics 1817, 1852–1859. doi: 10.1016/j.bbabio.2012.05.003
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe 9, 111–118. doi: 10.1128/microbe.9.111.1
Schmitz, R. A., Peeters, S. H., Versantvoort, W., Picone, N., Pol, A., Jetten, M. S. M., et al. (2021). Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles. FEMS Microbiol. Rev. fuab007. doi: 10.1093/femsre/fuab007 [Epub ahead of print].
Schmitz, R. A., Pol, A., Mohammadi, S. S., Hogendoorn, C., van Gelder, A. H., Jetten, M. S. M., et al. (2020). The thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV oxidizes subatmospheric H2 with a high-affinity, membrane-associated [NiFe] hydrogenase. ISME J. 14, 1223–1232. doi: 10.1038/s41396-020-0609-3
Sharp, C. E., Smirnova, A. V., Graham, J. M., Stott, M. B., Khadka, R., Moore, T. R., et al. (2014). Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 16, 1867–1878. doi: 10.1111/1462-2920.12454
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Sirajuddin, S., and Rosenzweig, A. C. (2015). Enzymatic oxidation of methane. Biochemistry 54, 2283–2294. doi: 10.1021/acs.biochem.5b00198
Stackebrandt, E., and Ebers, J. (2006). Taxonomic parameter revised: tarnishes gols standards. Microbiol. Today 33, 153–155.
Tabita, F. R. (2007). Rubisco: the enzyme that keeps on giving. Cell 129, 1039–1040. doi: 10.1016/j.cell.2007.06.002
Thompson, C. C., Chimetto, L., Edwards, R. A., Swings, J., Stackebrandt, E., and Thompson, F. L. (2013). Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913
Tyler, B. (1978). Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 47, 1127–1162. doi: 10.1146/annurev.bi.47.070178.005403
Vallenet, D., Engelen, S., Mornico, D., Cruveiller, S., Fleury, L., Lajus, A., et al. (2009). Micro scope: a platform for microbial genome annotation and comparative genomics. Database 2009:ba021. doi: 10.1093/database/bap021
Vallenet, D., Labarre, L., Rouy, Z., Barbe, V., Bocs, S., Cruveiller, S., et al. (2006). MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34, 53–65. doi: 10.1093/nar/gkj406
van Teeseling, M. C. F., Pol, A., Harhangi, H. R., van der Zwart, S., Jetten, M. S. M., Op den Camp, H. J. M., et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 80, 6782–6791. doi: 10.1128/AEM.01838-14
Versantvoort, W., Pol, A., Daumann, L. J., Larrabee, J. A., Strayer, A. H., Jetten, M. S. M., et al. (2019). Characterization of a novel cytochrome cGJ as the electron acceptor of XoxF-MDH in the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV. Biochim. Biophys. Acta - Proteins Proteomics 1867, 595–603. doi: 10.1016/j.bbapap.2019.04.001
Vorholt, J. A. (2002). Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178, 239–249. doi: 10.1007/s00203-002-0450-2
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Keywords: Verrucomicrobia, acidophilic, methanotroph, Ca. Methylacidithermus pantelleriae, volcanic soil
Citation: Picone N, Blom P, Hogendoorn C, Frank J, van Alen T, Pol A, Gagliano AL, Jetten MSM, D’Alessandro W, Quatrini P and Op den Camp HJM (2021) Metagenome Assembled Genome of a Novel Verrucomicrobial Methanotroph From Pantelleria Island. Front. Microbiol. 12:666929. doi: 10.3389/fmicb.2021.666929
Received: 11 February 2021; Accepted: 20 April 2021;
Published: 19 May 2021.
Edited by:
Sari Peura, Swedish University of Agricultural Sciences, SwedenReviewed by:
Nils-Kaare Birkeland, University of Bergen, NorwayNikolai Ravin, Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, Russia
Copyright © 2021 Picone, Blom, Hogendoorn, Frank, van Alen, Pol, Gagliano, Jetten, D’Alessandro, Quatrini and Op den Camp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huub J. M. Op den Camp, aC5vcGRlbmNhbXBAc2NpZW5jZS5ydS5ubA==
 Nunzia Picone
Nunzia Picone Pieter Blom1
Pieter Blom1 Carmen Hogendoorn
Carmen Hogendoorn Jeroen Frank
Jeroen Frank Theo van Alen
Theo van Alen Arjan Pol
Arjan Pol Antonina L. Gagliano
Antonina L. Gagliano Mike S. M. Jetten
Mike S. M. Jetten Walter D’Alessandro
Walter D’Alessandro Paola Quatrini
Paola Quatrini Huub J. M. Op den Camp
Huub J. M. Op den Camp