- 1Department of Microbiology and Infectious Diseases, Nara Medical University, Nara, Japan
- 2Livestock Food Agriculture Course, Soo High School Kagoshima, Kagoshima, Japan
- 3Department of Microbiology and Immunology, Teikyo University School of Medicine, Tokyo, Japan
Colistin is used to treat infectious diseases in humans and livestock; it has also been used as a feed additive for livestock for approximately 50 years. Since the mcr-1 plasmid-mediated colistin resistance gene was discovered in China in 2015, it has been detected worldwide, mainly in livestock. In this study, we investigated the prevalence and characteristics of mcr-mediated colistin-resistant Escherichia coli in livestock and farmers in Japan. We collected fecal samples from 295 healthy livestock (202 cattle and 93 swine) and 62 healthy farmers from 72 livestock farms (58 cattle farms and 14 swine farms) between 2013 and 2015. Twenty-eight mcr-1-harboring E. coli strains were isolated from 25 livestock (six cattle and 19 swine) and three farmers (two cattle farmers and one swine farmer). The prevalence rates of mcr-1-harboring E. coli in livestock and farmers were 8.47 and 4.84%, respectively. Of the 28 strains, the resistance genes of three were transferable via the mcr-1-coding plasmids to E. coli J53 at low frequencies (10−7–10−8). Six strains coharbored mcr-1 with CTX-M β-lactamases (CTX-M-14, CTX-M-27, or CTX-M-156). Of the isolates obtained from livestock and farmers in four farms (farms C, I, N, and P), nine strains had the same genotypical characteristics (sequence types and pulsed-field gel electrophoresis band patterns), plasmid characteristics (incompatibility group and plasmid transferability), and minimum inhibitory concentrations. Thus, the findings suggested that clonal strains could spread among livestock and farmers within farms. To our knowledge, this is the first study to detect clonal relatedness of mcr-1-mediated colistin-resistant E. coli in livestock and farmers. It is suggested that farmers are at a higher risk of acquiring mcr-1-harboring strains, calling for our attention based on the One Health concept.
Introduction
Colistin is a cationic antimicrobial peptide that damages bacterial cell membranes by targeting the lipid A moiety of lipopolysaccharides (LPSs), which are present in the outer cell membrane of Gram-negative bacteria. Although the clinical usage of colistin induces strong adverse effects (i.e., renal dysfunction and neurotoxicity), it is still an extremely important antibiotic against infections caused by multidrug-resistant Gram-negative bacteria resistant to carbapenems and fluoroquinolones (Falagas and Kasiakou, 2005). In animals, colistin sulfate has been used as a therapeutic drug and feed additive worldwide. There is a concern that drug-resistant bacteria that have increased due to the use of antibiotics in livestock will not only make it difficult to treat infectious diseases in livestock but also make it difficult to treat infectious diseases in humans acquired through the consumption of livestock products. Therefore, the Food Safety Commission of Japan conducted a risk assessment of colistin sulfate for livestock in 2017 and estimated the risk posed by the drug as “Medium” (Food Safety Commission of Japan, 2017). In response to this estimation, the Japanese government shifted colistin sulfate for livestock as a therapeutic drug to a second-choice drug and withdrew colistin sulfate as a feed additive (Makita et al., 2020).
Colistin resistance is mediated by chromosomes or plasmids. The most common mechanism of chromosomally mediated colistin resistance is mainly through modification of the bacterial outer membrane through alteration of LPSs, by the addition of 4-amino-4-deoxy-l-arabinose and/or phosphoethanolamine, via amino acid changes in the PmrA/PmrB and PhoP/PhoQ two-component regulatory systems (Olaitan et al., 2014; Poirel et al., 2017; Aghapour et al., 2019). The other mechanism is the overexpression of efflux-pump systems or overproduction of capsule polysaccharide (Bengoechea and Skurnik, 2000; Campos et al., 2004). Plasmid-mediated colistin resistance is attributable to the transmissible gene mcr-1, which is a phosphoethanolamine transferase, leading to a more cationic LPS structure and consequently resistance to polymyxins (Liu et al., 2016). It was first discovered in swine in China in 2015; subsequently, mcr-1-harboring Escherichia coli has been reported worldwide, mainly in livestock and meat and even in humans (Nang et al., 2019). In addition, there are reports of the chromosomal localization of mcr-1 in E. coli (Zurfluh et al., 2016; Li et al., 2018; Peng et al., 2019).
The spread of antimicrobial resistance genes should be controllable using the One Health concept, which involves the collaborative effort of health science professionals from multiple spheres toward attaining optimal health conditions for people, domestic animals, wildlife, plants, and the environment. The drivers of antimicrobial resistance include antimicrobial use and abuse in humans, animals, and the environment and the spread of resistant bacteria and resistance determinants within and between these spheres and around the world (McEwen and Collignon, 2018). Even in Japan, mcr-1-harboring E. coli strains have been isolated from livestock, imported and domestic meat, human clinical specimens, and the environment (Kusumoto et al., 2016; Nishino et al., 2017; Ohsaki et al., 2017; Tada et al., 2017; Hayashi et al., 2019). The possibility of crossing of the strains among the spheres is considered. Therefore, the relatedness of mcr-1-harboring strains from these different spheres needs to be determined based on the One Health concept.
The clonal relatedness of mcr-1-harboring E. coli from livestock on farms has been previously reported (Zheng et al., 2019). However, the clonal spread and relatedness of isolates of mcr-harboring E. coli from other spheres (e.g., among humans, livestock, and environment) remain unclear. To evaluate the relatedness of mcr-1-harboring E. coli isolated from livestock and farmers, we identified and compared the characteristics [sequence types (STs), pulsed-field gel electrophoresis (PFGE) band patterns, plasmid incompatibility (Inc) groups, plasmid transferability, and minimum inhibitory concentrations (MICs)] of the isolates. In this study, we investigated the prevalence, characteristics, and relatedness of mcr-mediated colistin-resistant E. coli in livestock (cattle and swine) and farmers in Japan.
Materials and Methods
Bacterial Isolates
Between 2013 and 2015, fecal samples from 295 healthy livestock (202 cattle and 93 swine) and 62 healthy livestock farmers (53 cattle farmers and nine swine farmers) were collected from 72 livestock farms (58 cattle farms and 14 swine farms) in the southern part of the Kyushu island, a major production area of cattle farming and swine farming in Japan. The livestock included 80 calves, 101 parent cattle, 21 fattening cattle (1–10 cattle per cattle farm), 26 piglets, 29 parent swine, and 38 fattening swine (3–10 swine per swine farm). Only one E. coli isolate was obtained per sample upon selection using Deoxycholate-hydrogen sulfide-lactose agar. The isolates were identified using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS; Vitek MS system; bioMérieux, Co., Ltd.). The studies involving human participants were reviewed and approved by the Ethical Review Committee at the Teikyo University School of Medicine (no.13–118). The participants provided written informed consent to participate in this study.
Antimicrobial Susceptibility Testing
The antimicrobial susceptibilities to β-lactamase and levofloxacin (except for colistin) were determined using the agar dilution method (Clinical and Laboratory Standards Institute, 2020), and quality control was performed using E. coli ATCC 25922. The colistin concentration was determined using the broth dilution method. MICs were interpreted according to the breakpoints defined by the Clinical and Laboratory Standards Institute (CLSI) guidelines.
Detection of Antimicrobial Resistance Genes
The presence of mcr genes, extended-spectrum beta-lactamase (ESBL)-encoding genes, and carbapenemase-encoding genes was determined using PCR and sequencing as described in previous studies (Dallenne et al., 2010; Poirel et al., 2011; Ye et al., 2016). DNA sequencing was conducted using the BigDye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, United States) and an Applied Biosystems ABI3730xl analyzer (Thermo Fisher Scientific K.K.). BLAST version 1.121 was used to process the sequencing data and identify the genes. Sequences were deposited in the GenBank database under accession numbers LC618530 through LC618535 for bla CTX-M genes and LC618536 through LC618563 for mcr-1 gene.
Plasmid Incompatibility Groups and Conjugation Experiments
Plasmid Inc. groups were identified using PCR-based replicon typing (Carattoli et al., 2005; Johnson et al., 2012). Conjugation experiments were performed using a broth-mating method with mcr-1-harboring E. coli as the donor and sodium azide-resistant E. coli J53 as the recipient, as described previously (Nakano et al., 2004). Transconjugants were selected on Luria-Bertani agar plates containing colistin (4 mg/L) and sodium azide (100 mg/L).
Escherichia coli Genotyping
For multilocus sequence typing (MLST), the protocol reported by Achtman et al. was applied to E. coli (Wirth et al., 2006). Seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were sequenced. DNA sequence variations were analyzed using the MLST database for E. coli 2 to determine STs. The similarity of the isolates was compared by PFGE of XbaI-digested genomic DNA (XbaI-PFGE) with a CHEF-MAPPER System (BioRad Laboratories, Japan), as previously described (Yano et al., 2013). PFGE was performed at 14°C for 20 h at 6 V/cm with a pulse time of 5.3–49.9 s, and the angle was set at 120°C. PFGE patterns were interpreted according to the criteria described by Tenover et al. (1995): “indistinguishable,” isolate with only one band different from that of the reference strain; “closely related,” isolate with 2–3 bands different from those of the reference strain; and “possibly related,” isolate with up to six bands different from those of the reference strain.
Results
Prevalence of mcr-1-Harboring E. coli Among the Livestock and Farmers
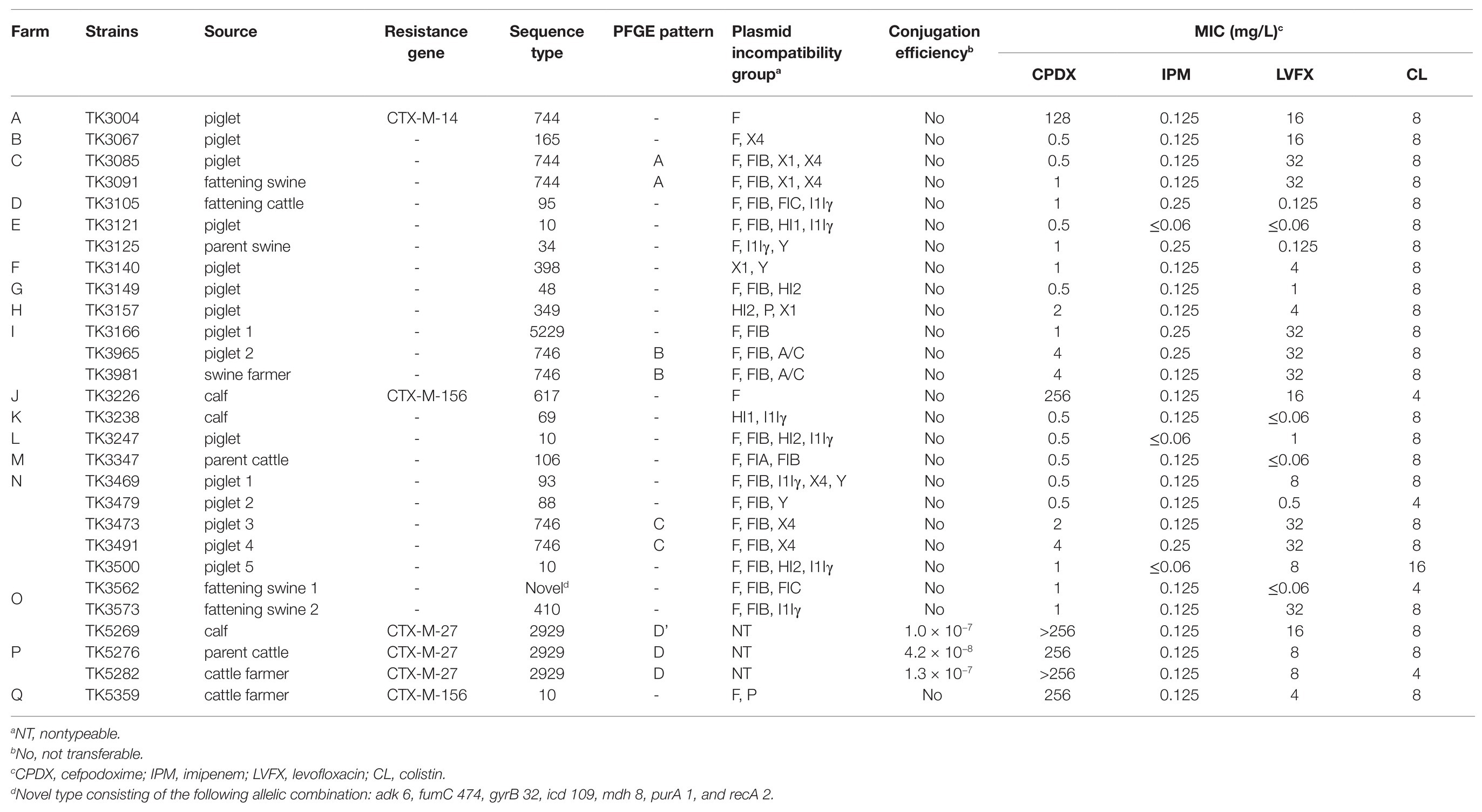
Colistin-resistant E. coli strains were isolated from 25 (8.47%) of the 295 livestock. The 25 strains were isolated from six cattle (three calves, two parent cattle, and one fattening cattle) from five farms and 19 swine (15 piglets, one parent swine, and three fattening swine) from 11 farms. All strains harbored mcr-1, as confirmed by PCR and sequencing. The prevalence of mcr-1-harboring E. coli strains was 2.97% (6/202 strains) for cattle and 20.43% (19/93 strains) for swine. mcr-1-harboring E. coli strains were also isolated from three healthy farmers (two cattle farmers and one swine farmer; 4.84%) among 62 farmers. The prevalence of mcr-1-harboring E. coli strains was 3.77% (2/53 strains) for cattle farmers and 11.11% (1/9 strains) for swine farmers.
Antimicrobial Susceptibility and Resistance Genes
The antimicrobial susceptibilities of 28 mcr-1-harboring E. coli strains are shown in Table 1. All the strains were resistant to colistin (4–16 mg/L), and 16 strains were co-resistant to levofloxacin (≥8 mg/L). Carbapenem-resistant strains were not found, and six strains were resistant to cefpodoxime (≥4 mg/L) and coharbored the CTX-M β-lactamase gene. The bla CTX-M genotype belongs to the CTX-M-9 group (three CTX-M-27 strains and one CTX-M-14 strain) and CTX-M-1 group (two CTX-M-156 strains).
Genetic Characteristics of mcr-1-Harboring E. coli
All mcr-1-harboring E. coli strains were genotyped using MLST analysis (Table 1). Six mcr-1-harboring E. coli strains from cattle belonged to five different STs (two strains belonged to ST2929 and one strain each to ST69, ST95, ST106, and ST617). Nineteen mcr-1-harboring E. coli strains from swine belonged to 13 different STs (three to ST10, three to ST744, and three to ST746, followed by 10 STs). The STs of E. coli isolated from cattle completely differed from those of E. coli isolated from swine. mcr-1-harboring E. coli strains from three farmers belonged to three different STs (ST10, ST746, and ST2929). All mcr-1-harboring E. coli strains possessed diverse types of plasmid Inc. Of the 28 strains, 22 possessed F replicon regions, 17 possessed IncFIB, eight possessed IncI1Iγ, six possessed IncX4, four possessed IncHI2, four possessed IncX1, four possessed IncY, three possessed nontypeable, two possessed IncA/C, two possessed IncFIC, two possessed IncHI1, and two possessed IncP. By performing conjugation experiments, transconjugants containing the mcr-1-encoding plasmid were obtained at low frequencies (1.0 × 10−7 to 4.2 × 10−8) from three strains. These three strains coharbored the CTX-M-27 β-lactamase gene with a nontypeable Inc. plasmids and were isolated from farm P.
Clonal Relatedness of mcr-1-Harboring E. coli Among Livestock and Farmers
Nine strains from four farms were found to have similar CTX-M types, STs, Inc. group, plasmid transferability, and MICs. The strains were as follows: TK3085 and TK3091 from farm C; TK3965 and TK3981 from farm I; TK3473 and TK3491 from farm N; and TK5269, TK5276, and TK5282 from farm P. XbaI-PFGE was used to confirm the clonal relatedness of the strains among the farms (Table 1). Five PFGE band patterns were obtained for the nine strains. Each PFGE pattern represented only one farm: Pattern A was noted in the farm C strains; pattern B in the farm I strains; pattern C in the farm N strains; and patterns D and D’ in the farm P strains. The PFGE D’ band patterns differed by three bands from the D band patterns; the strain with the D’ band patterns may have been derived from that with the D band patterns. The strains with similar PFGE band patterns were considered to represent clonal strains.
Discussion
mcr-1-harboring E. coli has increased worldwide. The prevalence of mcr-1-harboring E. coli in healthy livestock has been reported to be as follows: 0.6% in Great Britain in 2013–2015 (from swine); 4.45% in Thailand (from swine); 71.43, 68.86, and 87.58% in China in 2015–2016 (from cattle, swine, and chickens, respectively; Duggett et al., 2017; Zhang et al., 2019; Khine et al., 2020); and 0.42% in Japan in 2000–2014 (cattle, swine, and chickens; Kawanishi et al., 2017). Our study revealed a high prevalence of mcr-1-harboring E. coli in healthy livestock (8.47%); the prevalence in swine (20.43%) was higher than that in cattle (2.97%). Furthermore, we found that the prevalence was 4.84% in farmers in Japan. A previous study showed that the prevalence rates of mcr-1-harboring E. coli strains were 1.43 and 0.65% in inpatients, wherein it was associated with infection, and healthy volunteers, respectively, in China in 2007–2015 (Wang et al., 2017); our study showed comparatively higher distribution among livestock farmers.
The high prevalence is likely to be associated with colistin use (Tong et al., 2018). Continuous selection pressure might have promoted the dissemination of mcr-1, ultimately resulting in its exceedingly high prevalence in livestock. Colistin is no longer used as a feed additive in some countries, such as the United States of America and the EU (European Commission, 2001; European Medicines Agency, 2016). Similarly, the use of colistin as a feed additive in livestock farms was banned from July 2018 in Japan; 52.5% of test farms showed a decrease in the prevalence of E. coli with plasmid-mediated colistin resistance 12 months after colistin use was banned in Japan (Makita et al., 2020).
We determined the genetic characteristics of 28 mcr-1-harboring E. coli strains isolated from livestock and farmers. The genotypes of the 28 strains consisted of 18 different STs; dominant STs were not observed. Major genotypes of ESBL-producing E. coli in humans, such as ST131 (Nicolas-Chanoine et al., 2014), were not detected in this study. Seven strains (four strains, ST10; one strain, ST34; one strain, ST48; and one strain, ST617) were found to belong to clonal complex 10 (CC10), including two strains of CTX-M-156 coharboring E. coli CC10 (ST10 and ST617), which is considered to have a broad range of hosts, including humans, animals, vegetable, wastewater, and urban streams (Manges et al., 2015; Varela et al., 2015; Reid et al., 2019; Massella et al., 2020). Furthermore, the strains harboring antimicrobial resistance genes were frequently coharboring virulence-associated genes. There is growing concern about the spread of E. coli CC10 across the spheres.
Three mcr-1-harboring E. coli strains (TK5269, TK5276, and TK5282) isolated from a calf, parent cattle, and cattle farmer in farm P had transferable resistance genes to E. coli. J53, which is considered as a plasmid-mediated colistin-resistant E. coli. We also isolated mcr-1-harboring Klebsiella pneumonia from the same individual (calf of farm P; data not shown). Herein, we selected only one E. coli from one sample, even though there was a possibility that mcr-1-harboring E. coli strains with different characteristic (STs, Inc. groups, or PFGE band patterns) were present in these samples. There is also a possibility of mcr-1-coding gene transfer across different species and strains by horizontal gene transfer, such as conjugation and transformation. A recent study reported that E. coli strains harboring chromosomally encoded mcr-1 were isolated from 3.5% of mcr-1-positive E. coli in China from 2016 to 2018 (Shen et al., 2020). Therefore, the whole genome of the other 25 strains of mcr-1-positive E. coli should be analyzed to characterize the genetic construction and determine whether the mcr-1 gene is encoded on a plasmid or chromosome in the future.
Since resistance genes (e.g., mcr-1, ESBL-encoding genes, and carbapenemase-encoding genes) are being detected in humans, animals, and the environment worldwide, it is necessary to take comprehensive measures to prevent the spread based on the One Health concept (Baquero et al., 2019). It is considered that one of the epidemic routes of resistant bacteria is from livestock to humans, but ESBL-harboring strains from livestock and humans show low relatedness (Dorado-García et al., 2018; Mughini-Gras et al., 2019). On the contrary, ESBL-harboring strains from companion animals and humans show high relatedness (Hong et al., 2019). Our study revealed the clonal relatedness of mcr-1-harboring E. coli isolated from livestock and farmers. It is suggested that farmers are at a higher risk than individuals living outside farms because farmers come into close contact with livestock every day, such as companion animals.
In conclusion, we determined the prevalence of mcr-1-harboring E. coli in healthy livestock (8.47%) and farmers (4.84%). Some strains with the same genotype characteristics (ST and PFGE band patterns), plasmid characteristics (Inc groups and plasmid transferability), and MICs were isolated from livestock and farmers at the farms. Thus, the findings suggest the spread of mcr-1-mediated colistin-resistant E. coli among livestock and farmers within the farms studied. To our knowledge, this is the first study to identify the clonal relatedness of mcr-1-mediated colistin-resistant E. coli in livestock and farmers. Based on the concept of One Health, it is required to identify the association of mcr-1-harboring E. coli among healthy individuals, hospital patients, animals, wastewater of the livestock, and river water by characterizing the isolates.
Data Availability Statement
The data presented in the study are deposited in the GeneBank database repository, accession numbers (LC618530 through LC618563).
Ethics Statement
The studies involving human participants were reviewed and approved by Ethical Review Committee at the Teikyo University School of Medicine (no.13–118). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AN and RNa: conceptualization, methodology, investigation, and writing – original draft. RNi: conceptualization and resources. YS, SH, TK-U, and TU: validation and data curation. YO and HY: supervision and project administration. All authors contributed to the article and approved the submitted version.
Funding
The authors declare that this study received funding from JSPS KAKENHI (grant numbers; 17K16228 and 20K10433). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Takako Nakano, Keimi Nakano, Kaori Nishisouzu, Kazuyuki Makiguchi, and Shigehiro Shimokariya and all the farmers for collecting the fecal samples.
Footnotes
References
Aghapour, Z., Gholizadeh, P., Ganbarov, K., Bialvaei, A. Z., Mahmood, S. S., Tanomand, A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 12, 965–975. doi: 10.2147/IDR.S199844
Baquero, F., Coque, T. M., Martínez, J. L., Aracil-Gisbert, S., and Lanza, V. F. (2019). Gene transmission in the one health microbiosphere and the channels of antimicrobial resistance. Front. Microbiol. 10:2892. doi: 10.3389/fmicb.2019.02892
Bengoechea, J. A., and Skurnik, M. (2000). Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 37, 67–80. doi: 10.1046/j.1365-2958.2000.01956.x
Campos, M. A., Vargas, M. A., Regueiro, V., Llompart, C. M., Albertí, S., and Bengoechea, J. A. (2004). Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72, 7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Clinical and Laboratory Standards Institute (2020). Performance standards for antimicrobial susceptibility testing. 30th Edn. Wayne, PA: CLSI.
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dorado-García, A., Smid, J. H., van Pelt, W., Bonten, M. J., Fluit, A. C., van den Bunt, G., et al. (2018). Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother. 73, 339–347. doi: 10.1093/jac/dkx397
Duggett, N. A., Sayers, E., AbuOun, M., Ellis, R. J., Nunez-Garcia, J., Randall, L., et al. (2017). Occurrence and characterization of mcr-1-harbouring Escherichia coli isolated from pigs in Great Britain from 2013 to 2015. J. Antimicrob. Chemother. 72, 691–695. doi: 10.1093/jac/dkw477
European Commission (2001). Scientific Steering Committee. 2nd Opinion on Antimicrobial Resistance. Adopted on May 10–11.
European Medicines Agency (2016). Updated Advice on the Use of Colistin Products in Animals Within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. EMA/CVMP//CHMP/231573/2016.
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Food Safety Commission of Japan (2017). Antimicrobial-resistant bacteria arising from the use of colistin sulfate in the livestock (antimicrobial-resistant bacteria). Food Saf. 5, 24–28. doi: 10.14252/foodsafetyfscj.2016033s
Hayashi, W., Tanaka, H., Taniguchi, Y., Iimura, M., Soga, E., Kubo, R., et al. (2019). Acquisition of mcr-1 and cocarriage of virulence genes in avian pathogenic Escherichia coli isolates from municipal wastewater influents in Japan. Appl. Environ. Microbiol. 85:e01661–19. doi: 10.1128/AEM.01661-19
Hong, J. S., Song, W., Park, H. M., Oh, J. Y., Chae, J. C., Shin, S., et al. (2019). Clonal spread of extended-spectrum cephalosporin-resistant Enterobacteriaceae between companion animals and humans in South Korea. Front. Microbiol. 18:1371. doi: 10.3389/fmicb.2019.01371
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kawanishi, M., Abo, H., Ozawa, M., Uchiyama, M., Shirakawa, T., Suzuki, S., et al. (2017). Prevalence of colistin resistance gene mcr-1 and absence of mcr-2 in Escherichia coli isolated from healthy food-producing animals in Japan. Antimicrob. Agents Chemother. 61:e02057–16. doi: 10.1128/AAC.02057-16
Khine, N. O., Lugsomya, K., Kaewgun, B., Honhanrob, L., Pairojrit, P., Jermprasert, S., et al. (2020). Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated colistin resistance: mcr-1 and mcr-3 genes in contracted pig farms in Thailand. Front. Vet. Sci. 7:582899. doi: 10.3389/fvets.2020.582899
Kusumoto, M., Ogura, Y., Gotoh, Y., Iwata, T., Hayashi, T., and Akiba, M. (2016). Colistin-resistant mcr-1-positive pathogenic Escherichia coli in swine, Japan, 2007–2014. Emerg. Infect. Dis. 22, 1315–1317. doi: 10.3201/eid2207.160234
Li, R., Yu, H., Xie, M., Chen, K., Dong, N., Lin, D., et al. (2018). Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 51, 578–585. doi: 10.1016/j.ijantimicag.2017.11.015
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Makita, K., Fujimoto, Y., Sugahara, N., Miyama, T., Usui, M., Asai, T., et al. (2020). Quantitative release assessment of mcr-mediated colistin-resistant Escherichia coli from Japanese pigs. Food Saf. 8, 13–33. doi: 10.14252/foodsafetyfscj.D-20-00004
Manges, A. R., Harel, J., Masson, L., Edens, T. J., Portt, A., Reid-Smith, R. J., et al. (2015). Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog. Dis. 12, 302–310. doi: 10.1089/fpd.2014.1860
Massella, E., Reid, C. J., Cummins, M. L., Anantanawat, K., Zingali, T., Serraino, A., et al. (2020). Snapshot study of whole genome sequences of Escherichia coli from healthy companion animals, livestock, wildlife, humans and food in Italy. Antibiotics 9:782. doi: 10.3390/antibiotics9110782
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 6, 521–547. doi: 10.1128/microbiolspec.ARBA-0009-2017
Mughini-Gras, L., Dorado-García, A., van Duijkeren, E., van den Bunt, G., Dierikx, C. M., Bonten, M. J., et al. (2019). Attributable sources of community-acquired carriage of Escherichia coli containing beta-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet. Health 3, e357–e369. doi: 10.1016/S2542-5196(19)30130-5
Nakano, R., Okamoto, R., Nakano, Y., Kaneko, K., Okitsu, N., Hosaka, Y., et al. (2004). CFE-1, a novel plasmid-encoded AmpC beta-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob. Agents Chemother. 48, 1151–1158. doi: 10.1128/AAC.48.4.1151-1158.2004
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Nishino, Y., Shimojima, Y., Suzuki, Y., Ida, M., Fukui, R., Kuroda, S., et al. (2017). Detection of the mcr-1 gene in colistin-resistant Escherichia coli from retail meat in Japan. Microbiol. Immunol. 61, 554–557. doi: 10.1111/1348-0421.12549
Ohsaki, Y., Hayashi, W., Saito, S., Osaka, S., Taniguchi, Y., Koide, S., et al. (2017). First detection of an Escherichia coli strain harboring the mcr-1 gene in retail domestic chicken meat in Japan. Jpn. J. Infect. Dis. 70, 590–592. doi: 10.7883/yoken.JJID.2016.572
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Peng, Z., Hu, Z., Li, Z., Li, X., Jia, C., Zhang, X., et al. (2019). Characteristics of a colistin-resistant Escherichia coli ST695 harboring the chromosomally-encoded mcr-1 gene. Microorganisms 7:558. doi: 10.3390/microorganisms7110558
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Reid, C. J., Demaere, M. Z., and Djordjevic, S. P. (2019). Australian porcine clonal complex 10 (CC10) Escherichia coli belong to multiple sublineages of a highly diverse global CC10 phylogeny. Microb. Genom. 5:e000225. doi: 10.1099/mgen.0.000225
Shen, C., Zhong, L. L., Ma, F., El-Sayed, M. A. E. G., Doi, Y., Zhang, G., et al. (2020). Genomic patterns and characterizations of chromosomally-encoded mcr-1 in Escherichia coli populations. Gut Pathog. 12:55. doi: 10.1186/s13099-020-00393-2
Tada, T., Uechi, K., Nakasone, I., Shimada, K., Nakamatsu, M., Kirikae, T., et al. (2017). Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int. J. Infect. Dis. 63, 21–22. doi: 10.1016/j.ijid.2017.07.023
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995
Tong, H., Liu, J., Yao, X., Jia, H., Wei, J., Shao, D., et al. (2018). High carriage rate of mcr-1 and antimicrobial resistance profiles of mcr-1-positive Escherichia coli isolates in swine faecal samples collected from eighteen provinces in China. Vet. Microbiol. 225, 53–57. doi: 10.1016/j.vetmic.2018.09.018
Varela, A. R., Manageiro, V., Ferreira, E., Guimarães, M. A., da Costa, P. M., Caniça, M., et al. (2015). Molecular evidence of the close relatedness of clinical, gull and wastewater isolates of quinolone-resistant Escherichia coli. J. Glob. Antimicrob. Resist. 3, 286–289. doi: 10.1016/j.jgar.2015.07.008
Wang, Y., Tian, G. B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/S1473-3099(16)30527-8
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Yano, H., Uemura, M., Endo, S., Kanamori, H., Inomata, S., Kakuta, R., et al. (2013). Molecular characteristics of extended-spectrum beta-lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS One 8:e64359. doi: 10.1371/journal.pone.0064359
Ye, H., Li, Y., Li, Z., Gao, R., Zhang, H., Wen, R., et al. (2016). Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio 7:e00177. doi: 10.1128/mBio.00177-16
Zhang, X., Zhang, B., Guo, Y., Wang, J., Zhao, P., Liu, J., et al. (2019). Colistin resistance prevalence in Escherichia coli from domestic animals in intensive breeding farms of Jiangsu Province. Int. J. Food Microbiol. 291, 87–90. doi: 10.1016/j.ijfoodmicro.2018.11.013
Zheng, B., Xu, H., Huang, C., Yu, X., Guo, L., Han, H., et al. (2019). Occurrence and genomic characterization of two MCR-1-producing Escherichia coli isolates from the same mink farmer. mSphere 4:e00602–19. doi: 10.1128/mSphere.00602-19
Keywords: colistin resistance, mcr-1, Escherichia coli , livestock, farmer, genotype, one health
Citation: Nakano A, Nakano R, Nishisouzu R, Suzuki Y, Horiuchi S, Kikuchi-Ueda T, Ubagai T, Ono Y and Yano H (2021) Prevalence and Relatedness of mcr-1-Mediated Colistin-Resistant Escherichia coli Isolated From Livestock and Farmers in Japan. Front. Microbiol. 12:664931. doi: 10.3389/fmicb.2021.664931
Edited by:
Azucena Mora Gutiérrez, University of Santiago de Compostela, SpainReviewed by:
Mikhail Edelstein, Smolensk State Medical University, RussiaRuichao Li, Yangzhou University, China
Copyright © 2021 Nakano, Nakano, Nishisouzu, Suzuki, Horiuchi, Kikuchi-Ueda, Ubagai, Ono and Yano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryuichi Nakano, cm5ha2Fub0BuYXJhbWVkLXUuYWMuanA=
†These authors have contributed equally to this work
 Akiyo Nakano
Akiyo Nakano Ryuichi Nakano
Ryuichi Nakano Ryuji Nishisouzu2
Ryuji Nishisouzu2 Yuki Suzuki
Yuki Suzuki Takane Kikuchi-Ueda
Takane Kikuchi-Ueda Tsuneyuki Ubagai
Tsuneyuki Ubagai Yasuo Ono
Yasuo Ono