- 1Fujian Provincial Key Laboratory of Applied Genetics, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, China
Mitogen-activated protein kinase (MAPK) pathways play a vital role in multiple plant processes, including growth, development, and stress signaling, but their involvement in response to Ralstonia solanacearum is poorly understood, particularly in pepper plants. Herein, CaMAPK7 was identified from the pepper genome and functionally analyzed. The accumulations of CaMAPK7 transcripts and promoter activities were both significantly induced in response to R. solanacearum strain FJC100301 infection, and exogenously applied phytohormones, including methyl jasmonate (MeJA), brassinolide (BR), salicylic acid (SA), and ethephon (ETN), were decreased by abscisic acid (ABA) treatment. Virus-induced gene silencing (VIGS) of CaMAPK7 significantly enhanced the susceptibility of pepper plants to infection by R. solanacearum and downregulated the defense-related marker genes, including CaDEF1, CaPO2, CaSAR82A, and CaWRKY40. In contrast, the ectopic overexpression of CaMAPK7 in transgenic tobacco enhanced resistance to R. solanacearum and upregulated the defense-associated marker genes, including NtHSR201, NtHSR203, NtPR4, PR1a/c, NtPR1b, NtCAT1, and NtACC. Furthermore, transient overexpression of CaMAPK7 in pepper leaves triggered intensive hypersensitive response (HR)-like cell death, H2O2 accumulation, and enriched CaWRKY40 at the promoters of its target genes and drove their transcript accumulations, including CaDEF1, CaPO2, and CaSAR82A. Taken together, these data indicate that R. solanacearum infection induced the expression of CaMAPK7, which indirectly modifies the binding of CaWRKY40 to its downstream targets, including CaDEF1, CaPO2, and CaSAR82A, ultimately leading to the activation of pepper immunity against R. solanacearum. The protein that responds to CaMAPK7 in pepper plants should be isolated in the future to build a signaling bridge between CaMAPK7 and CaWRKY40.
Introduction
As sessile organisms, plants are frequently subjected to unfavorable environmental challenges, including biotic and abiotic stresses, and have to adapt their metabolism, growth, and development accordingly (Berriri et al., 2012). Among the environmental challenges, attack by pathogens is a major plant stress that disturbs cellular homeostasis, leads to serious retardation of crop growth and development, and ultimately causes the dramatic loss of crop yield and quality (Ali et al., 2020a; Bargues-Ribera and Gokhale, 2020). To protect themselves against invasion by pathogens, plants have evolved a sophisticated and economical defense system. A well-established model of plant immunity is composed of two interconnected pathway layers, including pathogen-associated microbial patterns (PAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). The recognition of PAMPs by membrane-targeted pattern recognition receptors (PRRs) in plants triggers an array of signaling pathways, such as calcium flux, the production of reactive oxygen species (ROS), and the activation of mitogen-activated protein kinases (MAPKs). These early events in turn activate the expression of downstream target proteins, such as transcription factors, and eventually lead to the activation of defense-related genes, cell wall strengthening, the biosynthesis of camalexin and other phytoalexins, hypersensitive response (HR)-like cell death mediated by ROS, and induced resistance against pathogens (Zhang and Zhou, 2010; Jiang et al., 2017; Kawasaki et al., 2017; Luo et al., 2017). A successful pathogen is capable of secreting and delivering effectors to the plant apoplast and cytoplasm and suppressing PTI, leading to the invasion of host. To overcome PTI impairment, plants further evolved intracellular receptors with nucleotide (NB)-Leu-rich repeat (LRR) domains that are capable of recognizing effectors and triggering ETI (Jones and Dangl, 2006). ETI is often accompanied by the accumulation of phytohormones, such as salicylic acid (SA) and a local HR-like cell death.
The plant immune system possesses several integrated signaling networks that predominantly involve protein kinases and phosphatases that perceive and respond to the different stimuli (Berriri et al., 2012; Jalmi and Sinha, 2016). One of the most important protein-kinase-based amplification cascades is the MAPK cascade. The MAPK cascade is an evolutionarily conserved signal transduction module that is involved in transducing extracellular signals to the nucleus for appropriate cellular adjustment (Li et al., 2017). The MAPK is composed of three protein kinases that sequentially activate each other by phosphorylation: a MAP kinase kinase kinase (MAPKKK) activates a MAP kinase kinase (MAPKK), which, in turn, activates a MAPK (Takahashi et al., 2007; Berriri et al., 2012). An activated MAPK phosphorylates specific substrates, such as transcription factors and enzymes and subsequently triggers cellular responses. MAPKs are activated when both tyrosine and threonine residues in the TEY/TDY motif are phosphorylated by dual-specificity kinases (MAPKKs), which, in turn, are activated by MAPKKKs via phosphorylation of the conserved Thr/Ser motif (Song and Goodman, 2002; Yanagawa et al., 2016). A genome-wide analysis reveals that 20 putative MAPKs, 10 MAPKKs, and 80 MAPKKKs were contained in the genome of Arabidopsis, with a similar repertoire of genes found in other plant species (Kong et al., 2013; Lu et al., 2015; Wang et al., 2016; Jing et al., 2017). Our previous study indicated that a total of 19 MAPKs and 5 MAPKKs were identified in the pepper genome (Liu Z. et al., 2015). Quantitative RT-PCR results revealed that CaMAPK7 was transcriptionally upregulated by inoculation with Ralstonia solanacearum primarily at later time points (Liu Z. et al., 2015).
Mitogen-activated protein kinase cascades have been found to participate in the regulation of defense against biotic stresses or against the tolerance of abiotic stress, and their functions are conserved across plant species (Teardo et al., 2019; He et al., 2021). For example, Arabidopsis MPK3, MPK4, and MPK6 are reported to be induced by pathogen elicitors and play a vital role in innate immune responses (Takahashi et al., 2007; Berriri et al., 2012; Eschen-Lippold et al., 2016). GmMPK4 negatively regulates the accumulation of SA and defense responses of soybean against downy mildew (Peronospora manshurica) and soybean mosaic virus (Liu et al., 2011). The clade-A MAPKs MPK3 and MPK6 were dramatically induced by wounding and function in wounding stress (Sozen et al., 2020). A given MAPK can be involved in multiple processes. For example, Arabidopsis MPK4 not only participates in innate immunity against pathogens (Qiu et al., 2008; Berriri et al., 2012; Eschen-Lippold et al., 2016) but is also involved in cytokinesis and cytoskeleton organization (Beck et al., 2010, 2011; Zeng et al., 2011). However, the functions of majority of MAPK members and their possible roles in defense remain largely unknown, particularly in non-model plants.
Pepper (Capsicum annuum) is one of the most important vegetables worldwide. As a typical solanaceous plant, pepper frequently suffers from various soil-borne pathogens, including R. solanacearum, particularly when its growing environment exhibits high temperature and high humidity (Dang et al., 2013; Ali et al., 2020b). Bacterial wilt caused by the phytopathogenic bacterium R. solanacearum is one of the most important plant diseases worldwide (Hayward, 1991). This disease causes dramatic losses in the yield and quality of peppers, which lead to serious economic losses for pepper farmers.
Only a small number of MAPK proteins have been characterized in pepper. In pepper plants, ABA functions through CaMPK17-1-mediated MAPK signaling (Liu et al., 2016). Pepper CaMK1 and CaMK2 encode stress-inducible protein kinases that can contribute to the response to wounding, UV-C, and cold (Shin et al., 2001). Furthermore, CaMK1 and CaMK2 were found to interact with CaWRKYa and phosphorylate the SP clusters but not the MAPK docking (D) domain of CaWRKYa (Huh et al., 2015). However, the majority of MAPK kinases have not been characterized. In this study, a MAPK from the pepper genome was identified as CaMAPK7 and functionally analyzed. The expression of CaMAPK7 is upregulated by R. solanacearum infection and acts as a positive regulator in the pepper defense response against R. solanacearum by modifying the binding of CaWRKY40 to downstream defense-associated marker genes.
Materials and Methods
Plant Materials and Pathogen Inoculation
The seeds of pepper (C. annuum cv GZ03) and tobacco plants (Nicotiana benthamiana and Nicotiana tobacum K326) were provided by the Pepper Breeding Group of Fujian Agriculture and Forestry University, Fuzhou, China).1 The seeds were first germinated in sterilized ddH2O, and the seedlings were transferred into plastic pots and placed in the greenhouse at 25°C, 70–80 μmol photons m–2 s–1, with 70% relative humidity and a 16-h light/8-h dark photoperiod. R. solanacearum strain FJC100301 was isolated from wilted samples of pepper from the Fujian province (China) and cultured as described by Dang et al. (2013). For pathogen inoculation, the roots of CaMAPK7-silenced pepper or 6-week-old transgenic tobacco plants that ectopically expressed CaMAPK7 were manually damaged by inserting a knife into the soil three times, and 10 ml of 108 colony-forming units (CFU)/ml R. solanacearum FJC100301 suspended in 10 mM MgCl2 was infiltrated into the damaged spot using a syringe with a needle. The R. solanacearum-infected plants were maintained at 28°C in the greenhouse and were harvested at the indicated time points for further experiments.
Treatment of Pepper Plants With Exogenously Applied Phytohormones and R. solanacearum
The leaves of 6-week-old pepper plants were sprayed with 100 μM ethylene (ET), 1 mM SA, 100 μM methyl jasmonate (MeJA), 100 μM abscisic acid (ABA), and 10 μM brassinolide (BR), respectively (Dang et al., 2013; Li et al., 2016). Pepper leaves treated with the corresponding solvent were used as a control. To study the expression pattern of CaMAPK7 in response to infection with R. solanacearum, 6-week-old pepper plants were inoculated with a 10-μl suspension of FJC100301 (108 CFU/ml). The phytohormone- or FJC100301-treated pepper leaves were harvested at corresponding time points for total RNA extraction and further reverse transcription assay as previously described (Liu Z. Q. et al., 2015).
Subcellular Localization of CaMAPK7 in N. benthamiana Plants
The full length of CaMAPK7 was amplified from the pepper cDNA library and cloned into the Gateway vector pMDC83 to generate 35S:CaMAPK7-GFP for the subcellular localization assay. GV3101 cells harboring 35S:CaMAPK7-GFP or 35S:GFP were cultured in LB media overnight, and the pellets were harvested by centrifugation. The pellets were suspended in infiltration media (10 mM MgCl2, 10 mM MES, pH 5.7, and 200 μm acetosyringone) and adjusted to an OD595 of 0.8. The suspensions were infiltrated into leaves of N. benthamiana plants with a needleless syringe and maintained in the greenhouse. At 48 h post-infiltration, the infiltrated leaves were harvested and subjected to fluorescent signal detection using a confocal laser scanning microscope (SP8, Leica, Wetzlar, Germany). The emission and excitation wavelengths were 488 and 510–520 nm, respectively.
Histochemical Staining
Trypan blue staining and 3,3′-diaminobenzidine (DAB) staining were performed as described by Choi and Hwang (2011) with slight modifications (Choi and Hwang, 2011). For trypan blue staining, the pepper leaves were harvested and boiled in lactophenol/ethanol/trypan blue solution (10 ml of lactic acid, 10 ml of glycerol, 10 g of phenol, 10 ml of absolute ethanol, and 10 mg of trypan blue dissolved in 10 ml of distilled water) for 5 min and incubated at room temperature overnight. The samples were then destained in 2.5 g/ml chloral hydrate in ddH2O. For DAB staining, the detached pepper leaves were immersed in 1 mg/ml DAB staining buffer, incubated at 25°C overnight, and cleared with 70% ethanol. Images of pepper leaves with trypan blue and DAB staining were photographed using a dissecting microscope (Leica).
Tobacco Transformation
Tobacco was transformed as described by Muller et al. (1984). In brief, Agrobacterium GV3101 cells harboring the CaMAPK7 overexpression vector were used to infect tobacco leaf disks for transformation. Murashige and Skoog (MS) media supplemented with 500 mg/L carbenicillin and 75 mg/L hygromycin (Roche Diagnostics Corporation, Indianapolis, IN, United States2) was used to screen the potential tobacco transformants. The regenerated plants were further confirmed by PCR using specific primers of CaMAPK7. Lines #3 and #4 with a high level of expression were selected for further functional analysis.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described with slight modifications (Cai et al., 2015). The full length of CaMAPK7 was cloned into pEarleyGate 201 to generate 35S:CaMAPK7-HA using standard Gateway technology. Agrobacterium GV3101 cells harboring 35S:CaMAPK7-HA or 35S:HA (negative control) were infiltrated into 8-week-old pepper leaves. At 48 h post-infiltration, the infiltrated pepper leaves were sampled and cross-linked with 1% formaldehyde for 10 min, and 3 M glycine was added to stop the reaction and incubated for 5 min. Nuclear extracts were isolated and used for immunoprecipitation assays. Micrococcal nuclease was used to shear the isolated chromatin to a length of 200–500 bp. Anti-HA antibody was used to immunoprecipitate (IPed) the sheared chromatin, washed three times with Tris-buffered saline Tween 20 (TBST) buffer, and reversed cross-linked, and finally, the IPed DNA was eluted with sterile ddH2O. Both immunoprecipitated and input DNA were analyzed by PCR or real-time PCR with specific primers. The ChIP efficiency was calculated as percentage of input as previously described (Haring et al., 2007).
VIGS Assay
The VIGS assay was performed as described by Liu et al. (2002) with slight modifications (Liu et al., 2002). A specific fragment of CaMAPK7 whose specificity had been confirmed by BLASTn in the pepper genome database3 was amplified from the pepper cDNA library using gene-specific primers (Supplementary Table 1) and cloned into the VIGS vector TRV2 to generate TRV2:CaMAPK7. Transformed Agrobacterium cells harboring TRV1 or TRV2:CaMAPK7 were grown, harvested, and resuspended in the infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.7, and 200 μM acetosyringone). The resuspended Agrobacterium cells harboring TRV1 and TRV2 were mixed in a 1:1 ratio. At 3–4 h post-incubation at 25°C, the mixture was infiltrated into the cotyledons of pepper plants at the four-leaf stage with a needleless syringe. The Agrobacterium-infiltrated pepper plants were maintained in a growth chamber at 16°C for the first 56 h, and then the temperature was increased to 25°C for further growth. The CaMAPK7-silenced and unsilenced pepper plants were used for further experiments at 3 weeks post-agroinfiltration.
Real-Time PCR Assay
To determine the relative transcript accumulation of target genes, real-time PCR was performed as previously described (Liu Z. et al., 2015) with specific primers (Supplementary Table 1). Briefly, the total RNA of pepper or tobacco plants was extracted using the TRIzol reagent, and DNase I was used to digest the genomic DNA in isolated RNA samples. The total RNA was reverse-transcribed using a cDNA First Strand Synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. A Bio-Rad real-time PCR system (Bio-Rad, Hercules, CA, United States) and the SYBR premix Ex Taq II system were used for real-time PCR analysis. The pepper CaActin, Ca18SrRNA, or tobacco NtEF1α were used as internal controls. Three technical replicates were performed for at least three independent biological replicates for the real-time PCR.
Results
The Sequence Analysis of CaMAPK7 and Its Promoter
The genome-wide identification of pepper MAPK members was performed in our previous study, and a total of 19 MAPKs were identified in the genome of pepper variety CM334 using the pepper genome database (Liu Z. et al., 2015). Similar to other pepper MAPKs, CaMAPK7, studied herein, contains 11 domains (I–XI) that are conserved in the serine/threonine protein kinases along with a TEY motif (Figure 1). The deduced amino acid sequences of CaMAPK7 were 370 amino acid residues in length. The size and theoretical pI of the predicted protein were 42.74 kDa and 8.29, respectively. The protein sequence of CaMAPK7 and its corresponding orthologs from other species were compared and analyzed, and the results showed that CaMAPK7 shares 79.73, 80.54, 96.76, 78, and 77% of amino acid identities with AtMAPK7 (Arabidopsis thaliana), NtMAPK15 (Nicotiana tabacum), SlMAPK8 (Solanum lycopersicum), OsMAPK4 (Oryza sativa), and PsMAPK2 (Pisum sativum), respectively (Figure 1). In addition, the motifs contained in promoter of CaMAPK7 was scanned using the PLANTCARE database, and various hormone- and defense-associated motifs were included, including ERE, the TCA-motif, W-box, HSE, and LTR (Supplementary Figure 1). The diverse motifs present in the CaMAPK7 promoter indicated that CaMAPK7 may participate in the responses of pepper against biotic and abiotic stresses.
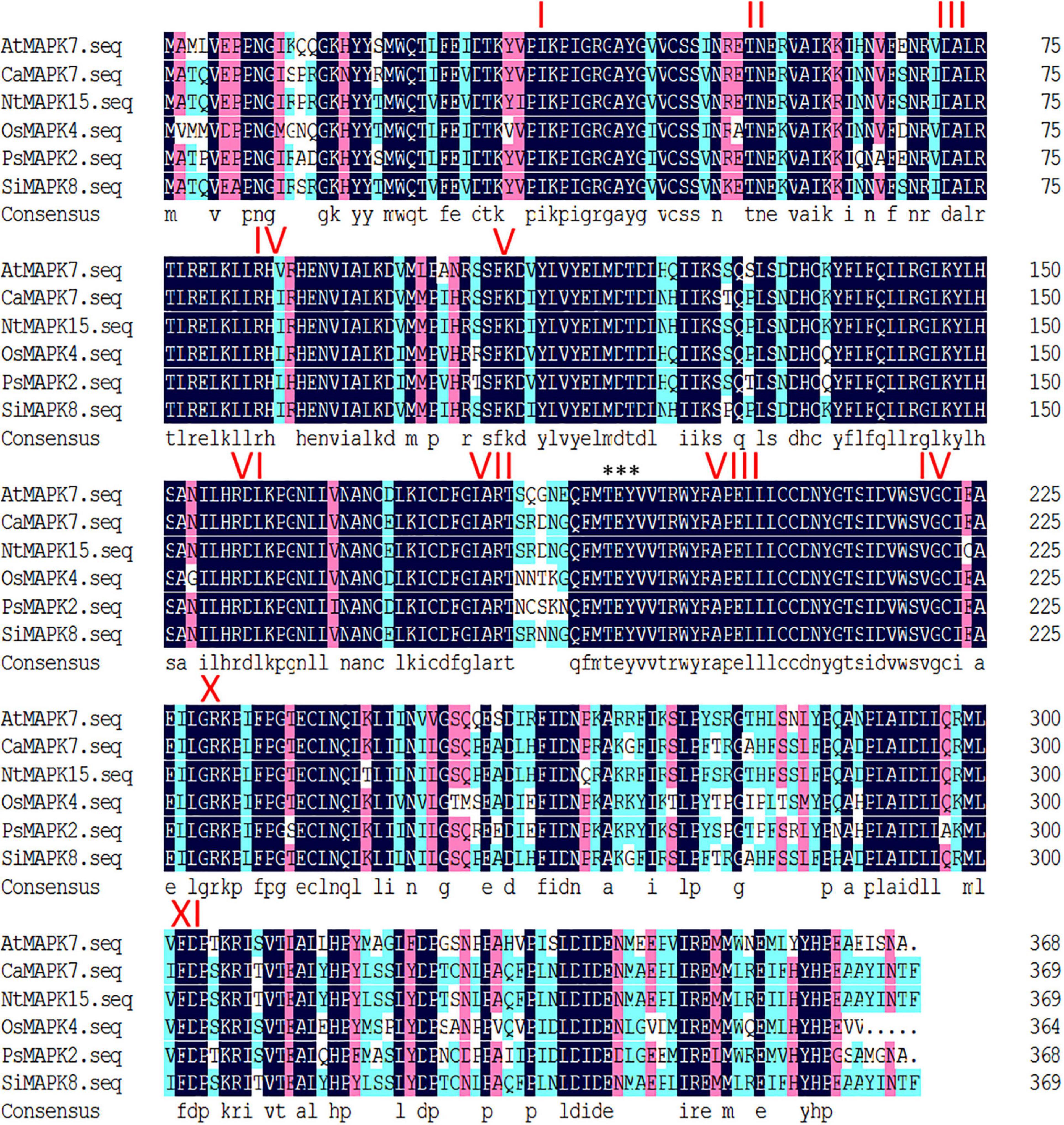
Figure 1. Amino acid sequence arrangements and phylogenetic analysis with the CaMAPK7 protein and its orthologs. Amino acid sequence alignments of CaMAPK7 and its orthologs from Arabidopsis thaliana (AtMAPK7), Oryza sativa (OsMAPK4), Pisum sativum (PsMAPK2), Solanum lycopersicum (SlMAPK8), and Nicotiana tabacum (NtMAPK15). The 11 conserved domains (I–XI) present in the serine/threonine protein kinases are denoted by Roman numerals. The conserved threonine and tyrosine residues are indicated by asterisks (***).
The Expression Profile and Promoter Analysis of CaMAPK7 Against R. solanacearum Infection and Exogenous Applied Phytohormones
Signaling pathways mediated by phytohormones, such as ABA, JA, BR, SA, and ET, are involved in plant defense responses against biotic and abiotic stresses. To test if CaMAPK7 was involved in signaling pathways, the profile of expression of CaMAPK7 in response to applied ABA, JA, BR, SA, and ET was studied. The results revealed that the CaMAPK7 transcript exhibited a decrease after treatment with 100 μM ABA from 1 to 24 hpt (hours post treatment) (Figure 2A). In contrast, the relative abundance of CaMAPK7 was enhanced after treatment with 100 μM MeJA and 1 μM BR from 12 to 48 hpt and exhibited its highest level at 48 and 12 hpt, respectively (Figures 2B,C). The exogenous SA resulted in significant increases in the accumulation of CaMAPK7 from 12 to 24 hpt (Figure 2D) and exhibited its highest level at 24 hpt. For ethephon (ETN) treatment, the relative abundance of CaMAPK7 began to increase at 3 hpt and lasted until 12 hpt (Figure 2E). The presence of a myriad of putative immunity-related motifs in the promoter of CaMAPK7 implied its potential inducible expression upon pathogen attack. To confirm our hypothesis, qRT-PCR was performed to investigate the pattern of expression of CaMAPK7 against R. solanacearum inoculation (RSI). The CaMAPK7 transcript was significantly enhanced against R. solanacearum infection, compared with the mock-treated pepper leaves (Figure 2F).
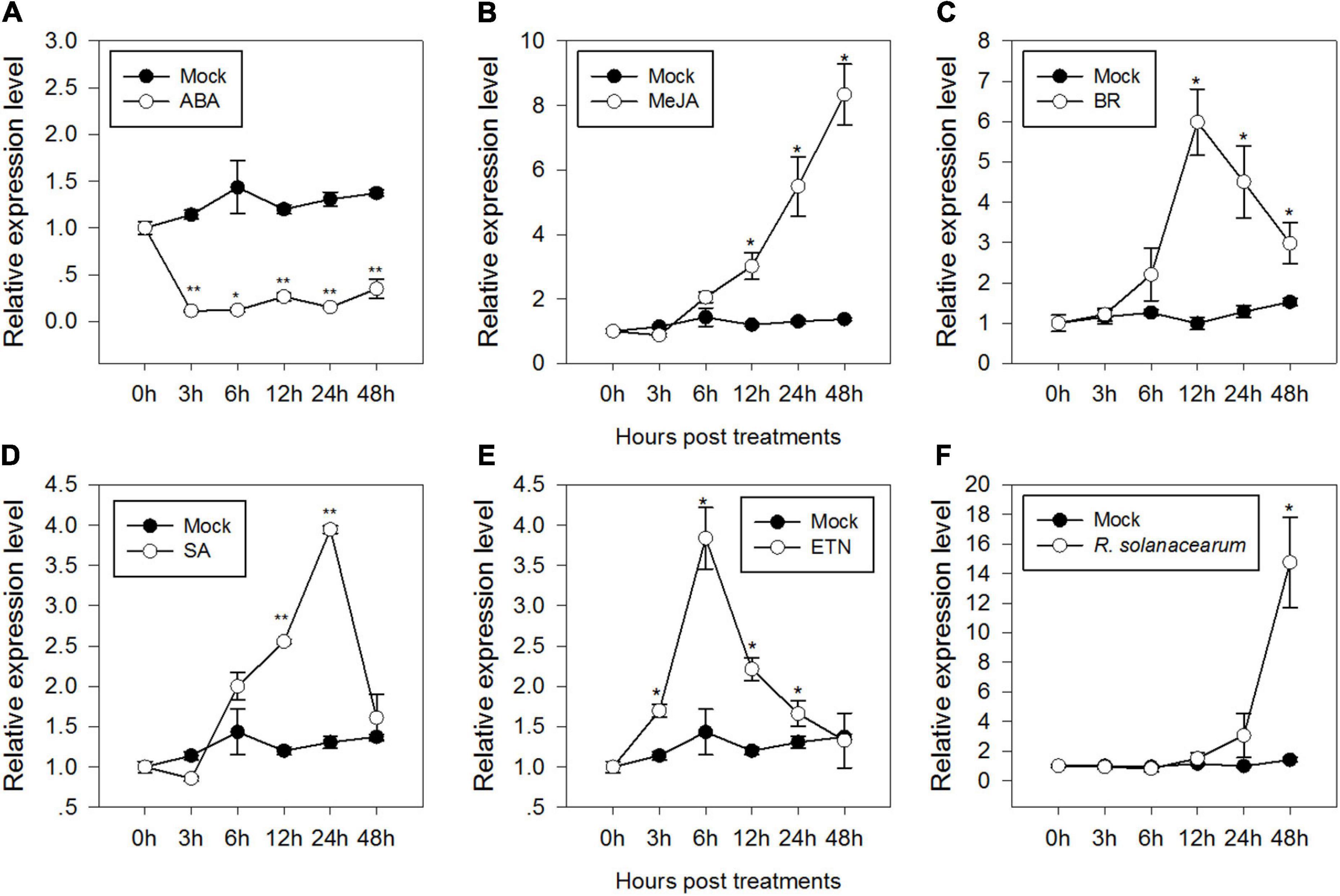
Figure 2. Relative CaMAPK7 transcript levels in pepper plants in response to Ralstonia solanacearum infection and exogenously applied phytohormone treatments. (A–F) Relative CaMAPK7 transcript levels in pepper leaves at various time periods after treatment with 100 μM ABA (A), 100 μM MeJA (B), 10 μM brassinolide (C), 1 mM salicylic acid SA (D), 100 μM ethephon (E), and R. solanacearum (F). (A–F) CaMAPK7 transcript levels in pepper plants after treatment with R. solanacearum or hormones were compared with those in mock-treated control plants (normalized to a relative expression level of “1”). Asterisks indicate significant differences from three independent experiments based on the LSD test (*p < 0.01; **p < 0.05).
To study the promoter activity of CaMAPK7 in response to the applied exogenous phytohormones described above and inoculation with R. solanacearum, a pCaMAPK7-driven GUS reporter vector was generated and expressed in pepper leaves by agroinfiltration. The Agro-infiltrated pepper leaves were treated with phytohormones and RSI and followed by harvest for the quantification of GUS activity. GUS expression driven by pCaMAPK7 was upregulated by JA, BR, SA, ET, and RSI, although it was downregulated by treatment with ABA (Supplementary Figure 2).
Subcellular Localization of CaMAPK7 in N. benthamiana Plants
The subcellular localization of a protein is closely associated with its potential function. To determine the subcellular localization of CaMAPK7, a CaMAPK7-GFP fusion construct driven by the constitutive CaMV35S promoter was generated and expressed in the leaves of N. benthamiana plant by agroinfiltration. At 48 hpt, the infiltrated leaves were harvested for the detection of fluorescence using a laser scanning confocal microscope (SP8, Leica). CaMAPK7-GFP, similar to the control GFP, was located throughout the whole plant cell, including the cytoplasm and nucleus (Figure 3A). A Western blotting assay was performed to determine the size of the CaMAPK-GFP protein (Figure 3B) expressed in the N. benthamiana leaves.
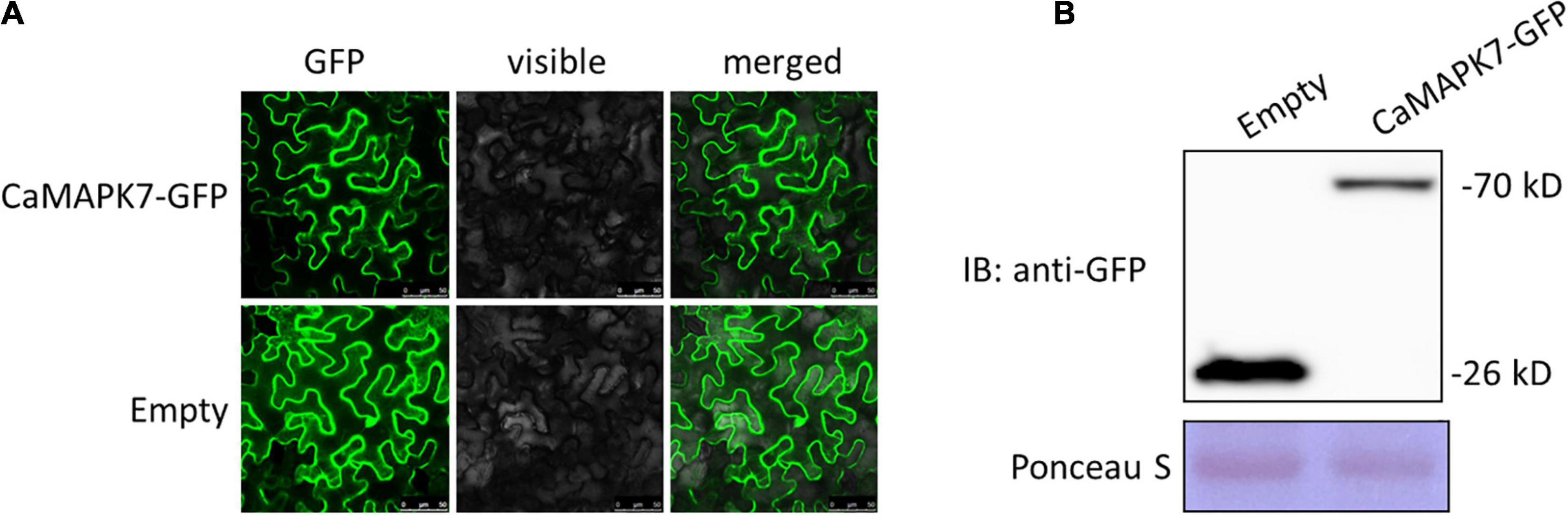
Figure 3. Subcellular localization of CaMAPK7 in Nicotiana benthamiana leaves. (A) CaMAPK7-GFP, similar to the empty vector, was localized throughout the whole cells. Images were taken using Leica confocal microscopy at 48 h post-agroinfiltration. Bars = 50 μm. (B) Immunoblot analysis of the expression of CaMAPK7 protein in N. benthamiana leaves. Immunoblotting used an anti-GFP antibody. Ponceau S was used to equal the loading. Empty, empty vector; GFP, green fluorescent protein; IB, immunoblotting.
Silencing of CaMAPK7 in Pepper Plants Enhanced Its Susceptibility in Response to R. solanacearum Infection
To investigate the function of CaMAPK7 in pepper immunity against R. solanacearum infection, a loss of function of CaMAPK7 was performed using a VIGS assay (Liu et al., 2002; Lim and Lee, 2016). The specific coding sequence (CDS) fragment of CaMAPK7 whose specificity had been evaluated by BLASTn in NCBI was cloned into the virus vector (pTRV2) to yield pTRV2:CaMAPK7. Agrobacterium tumefaciens GV3101 harboring the pTRV1 vector and the pTRV2 vector with or without CaMAPK7 were mixed at a ratio of 1:1 and infiltrated into the cotyledons of pepper plants at a stage of three to four leaves. The phytoene desaturase gene results in a phytobleaching phenotype, and pepper plants transfected with TRV:pds and TRV:gfp were used as the positive and negative control, respectively. Each of the 80 plants of TRV:gfp and TRV:CaMAPK7 was obtained and used for further experiments. Six CaMAPK7-silenced and unsilenced pepper plants were randomly selected for silencing efficiency detection. The transcript levels of CaMAPK7 in CaMAPK7-silenced pepper plants were reduced to 25% of those in the unsilenced plants (Figure 4A), suggesting the success of silencing. R. solanacearum FJC100301 (Genin and Boucher, 2004) was used to infect the CaMAPK7-silenced and unsilenced pepper plants. Phenotypic observation showed that CaMAPK7-silencing enhanced the susceptibility of pepper plants in response to R. solanacearum infection as manifested by more obvious wilting symptoms on the TRV:CaMAPK7 pepper plants (Figure 4B). To accurately quantify the extent of disease in R. solanacearum-infected plants, we determined the relative disease indices at 8–20 dpi (Figure 4C) in the TRV:CaMAPK7 and unsilenced plants. qRT-PCR was performed to detect the transcriptional levels of defense-associated marker genes, and the accumulation of transcripts of the defense-related marker genes, including CaDEF1, CaPO2, CaSAR82A, and CaWRKY40, was lessened in FJC100301-infected CaMAPK7-silenced pepper plants at 24 hpi, compared with the unsilenced pepper plants (Figure 4D).
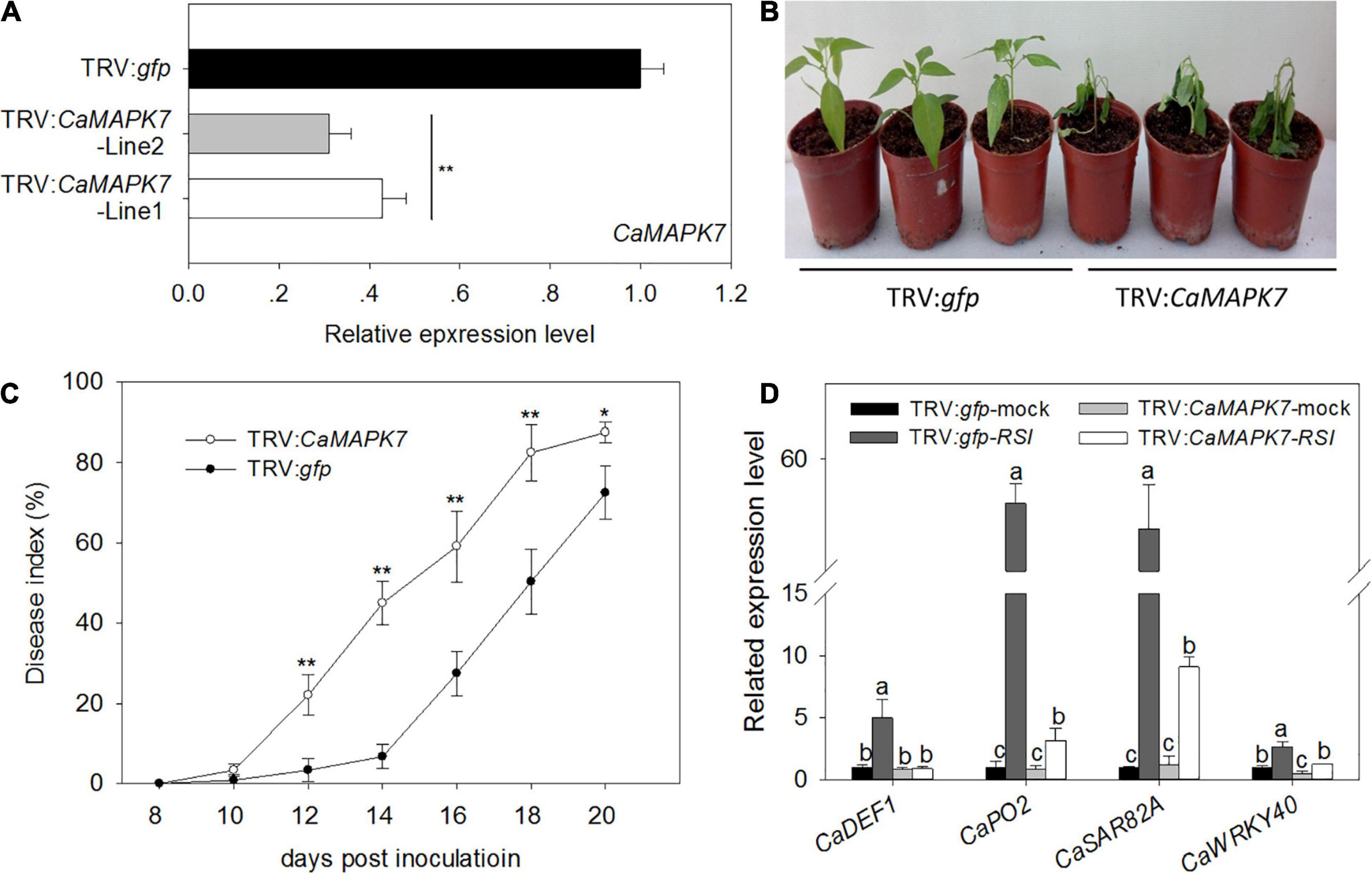
Figure 4. Increased susceptibility of CaMAPK7-silenced pepper plants to Ralstonia solanacearum infection. (A) Relative CaMAPK7 transcript levels in empty vector control (TRV:gfp) and silenced (TRV:CaMAPK7) plants investigated by qRT-PCR. (B) The phenotype of CaMAPK7-silenced and unsilenced pepper plants 14 days post-R. solanacearum inoculation. (C) R. solanacearum-inoculated leaves scored every 2 days using a disease index ranging from 0 to 4: 0 (no wilting), 1 (1–25% wilted), 2 (26–50% wilted), 3 (51–75% wilted), and 4 (76–100% wilted or dead). The averages presented are based on three biological replicates each comprising 10 plants. (A–C) Error bars indicate the standard error. Asterisks indicate a significant difference as determined by the two-tailed t-test (*p < 0.05; **p < 0.01). (D) Real-time RT-PCR analysis of defense marker gene expression in CaMAPK7-silenced and unsilenced pepper plants 48 h post-inoculation with R. solanacearum. The relative expression of defense marker genes in unsilenced pepper leaves without pathogen inoculation was set to “1.” Data represent the means ± SD from three biological replicates. Different letters indicate significant differences in gene expression, as determined by Fisher’s protected LSD test (p < 0.05). dai, days after inoculation; SD, standard deviation.
Transient Overexpression of CaMAPK7 Elicits Intensive HR-Like Cell Death and the Accumulation of H2O2 in Pepper Leaves
The CaMAPK7-silencing experiment in pepper plants indicates that CaMAPK7 acts as a positive regulator in pepper immunity in response to R. solanacearum infection. To further confirm our hypothesis, a transient overexpression assay was conducted to study the effect of CaMAPK7 transient overexpression on the induction of HR-like cell death in pepper leaves. GV3101 cells that harbored CaMAPK7-HA were infiltrated into the leaves of pepper plants for transient overexpression. The qRT-PCR results showed that CaMAPK7-HA was successfully overexpressed in pepper leaves (Figure 5A). An intensive HR-like cell death was observed in pepper leaves that transiently overexpressed CaMAPK7-HA, manifested by darker trypan blue staining and higher electrolyte leakage release. In addition, CaMAPK7 transient overexpression triggered obvious H2O2 accumulation, confirmed by DAB staining (Figures 5B,C). qRT-PCR was further performed to examine the transcript accumulation of defense-associated marker genes, and the overexpression of CaMAPK7 could elicit the upregulation of many marker genes, including CaDEF1, CaPO2, and CaSAR82A (Figure 5D). Interestingly, we found that the overexpression of CaMAPK7 can also induce the transcriptional level of CaWRKY40 (Figure 5D), a transcription factor reported in our previous study that plays a vital role in the immunity of pepper against R. solanacearum (Dang et al., 2013).
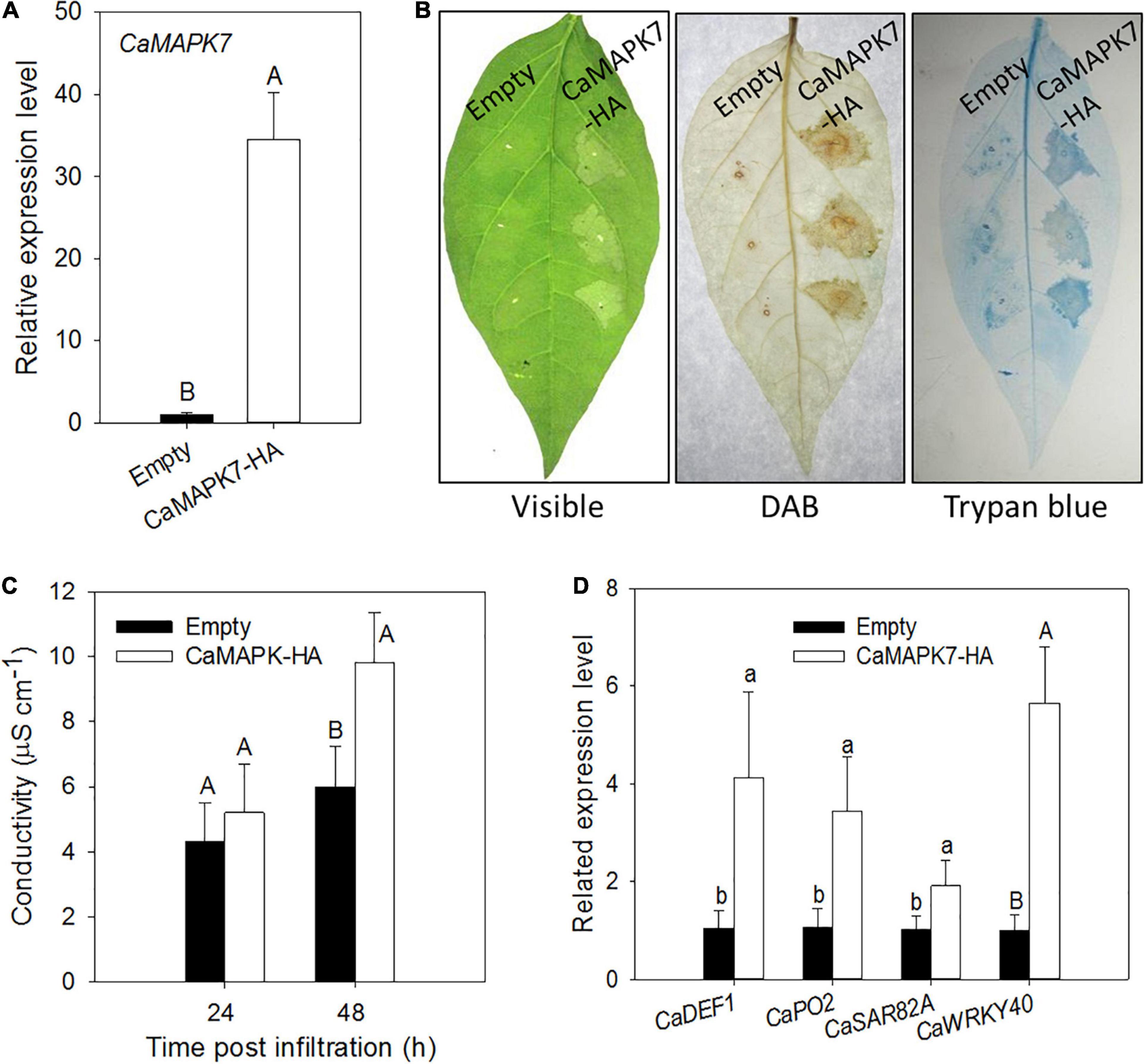
Figure 5. Effects of the transient expression of CaMAPK7 on the cell death response of pepper leaves. (A) qRT-PCR analysis of transiently expressed CaMAPK7 in pepper leaves at 24 h post-agroinfiltration. (B) Induction of cell death and H2O2 accumulation by transient CaMAPK7 expression in pepper leaves. The images of phenotype (left), trypan blue staining (middle), and DAB staining (right) were photographed 2 days post-agroinfiltration. (C) Electrolyte leakage from pepper leaves transiently expressing CaMAPK7 at 24 and 48 h after inoculation. (D) Real-time analysis of defense-associated marker genes in CaMAPK7-transient expression pepper leaves. The relative expression level of defense marker genes in pepper plants transiently expressed empty vector were set to “1.” (C,D) Error bars indicate the standard error. Different letters above the bar show a significant difference between the means of the three biological replicates based on two-tailed t-test: uppercase letters, p < 0.01; lowercase letters, p < 0.05. DAB, diaminobenzidine.
Ectopic Overexpression of CaMAPK7 in Tobacco Plants Enhanced Disease Resistance Against R. solanacearum Inoculation
To further confirm the results of VIGS and transient overexpression, transgenic tobacco lines that ectopically overexpressed CaMAPK7 driven by two copies of the CaMV35S promoter were generated. More than 10 T3 lines were acquired, and two lines of 10 (lines 3 and 4) were selected for further functional analysis. Semi-quantitative RT-PCR was performed to detect the CaMAPK7 transcript in CaMAPK7-OX-3, CaMAPK7-OX-4, and wild-type (WT) tobacco plants (Figure 6A). Transgenic tobacco plants and the WT were subjected to R. solanacearum infection. At 8 dpi, an obvious wilting symptom was observed in the WT tobacco plants, whereas the CaMAPK7-OX lines only wilted slightly (Figure 6B). The CFU of R. solanacearum in the CaMAPK7-OX line and WT were calculated at 3 dpi, and the ectopic expression of CaMAPK7 in tobacco plants significantly suppressed the growth of R. solanacearum, compared with that in the WT tobacco plants (Figure 6C). Trypan blue and DAB staining were performed to evaluate the local defense response in CaMAPK7-OX and WT tobacco plants. R. solanacearum infection in the CaMAPK7-OX lines elicited more severe HR-like cell death and H2O2 accumulation than that in WT tobacco plants (Figure 6D).
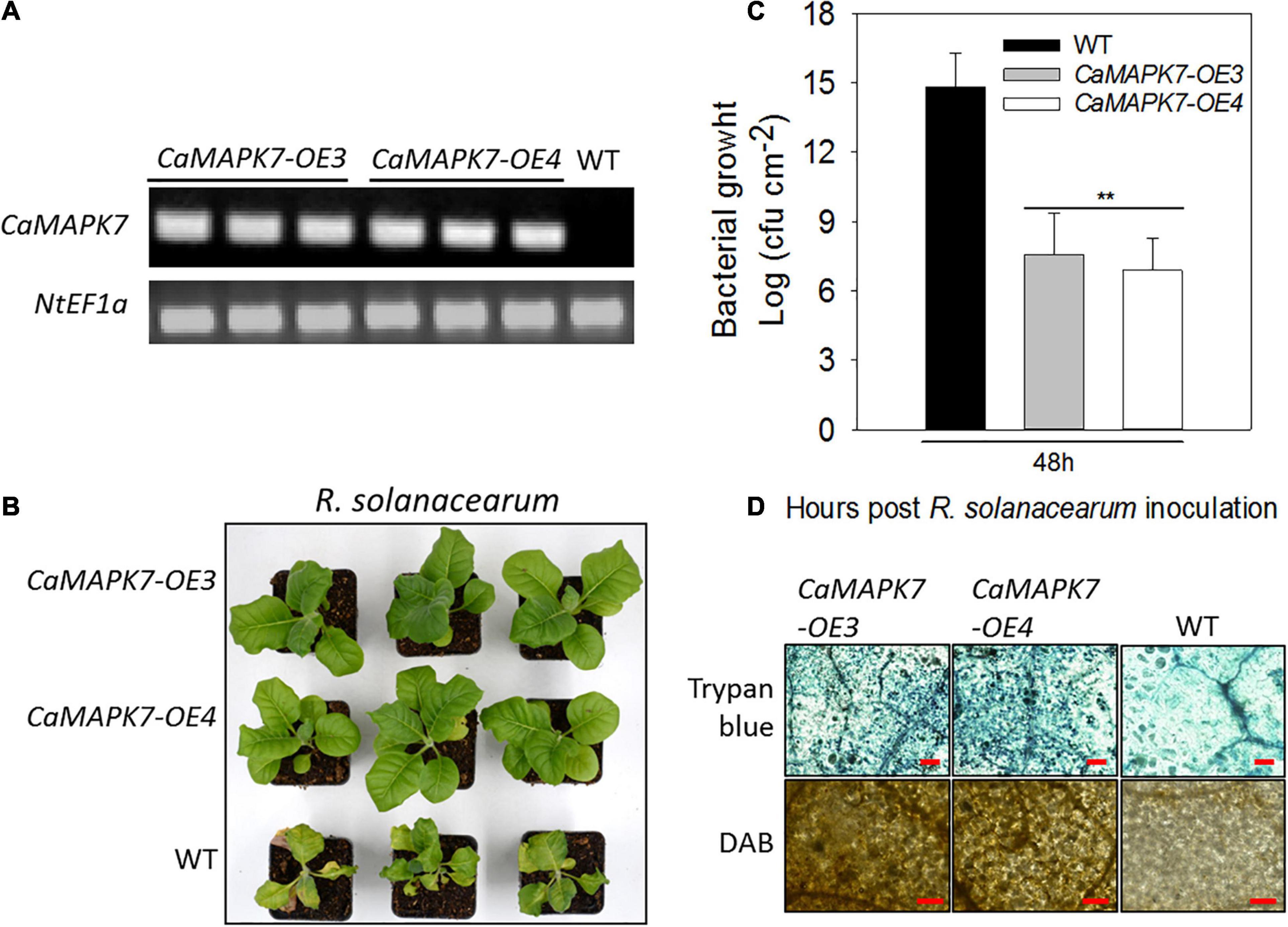
Figure 6. Transgenic T3 tobacco plants ectopically overexpressing CaMAPK7 exhibit enhanced resistance to Ralstonia solanacearum. (A) Transcript accumulation of CaMAPK7 in CaMAPK7-OX lines (#3 and #4) and WT detected by semi-quantitative PCR. (B) The disease phenotype of CaMAPK7-OX lines and WT 10 dpi with R. solanacearum strain FJC100301 in the plant roots. (C) The growth of R. solanacearum in leaves of CaMAPK7-OX lines and WT at 48 h post-R. solanacearum inoculation. Values are average CFU based on three biological replicates. Asterisk indicates significant differences determined by the Fisher’s protected LSD test (**p < 0.01). (D) HR-like cell death and H2O2 accumulation detected in CaMAPK7-OX lines and WT induced by R. solanacearum infection. Bar = 500 μm. dpi, days post-inoculation; HR, hypersensitive response; WT, wild-type.
To further confirm the role of CaMAPK7 in disease resistance and investigate its possible molecular mode of action, the transcriptional response of well-known defense-associated marker genes in the CaMAPK7-OX lines and WT tobacco plants were detected using qRT-PCR. We examined the accumulation of transcripts of the HR-associated genes NtHSR201 and NtHSR203, JA-responsive genes NtPR1b and NtPR4, SA-responsive genes NtPR1a/c, ROS-associated NtCAT1, and the ethylene-related gene NtACC. All the marker genes tested in the CaMAPK7-OE lines above were shown to be significantly higher than those in WT tobacco plants (Figure 7).
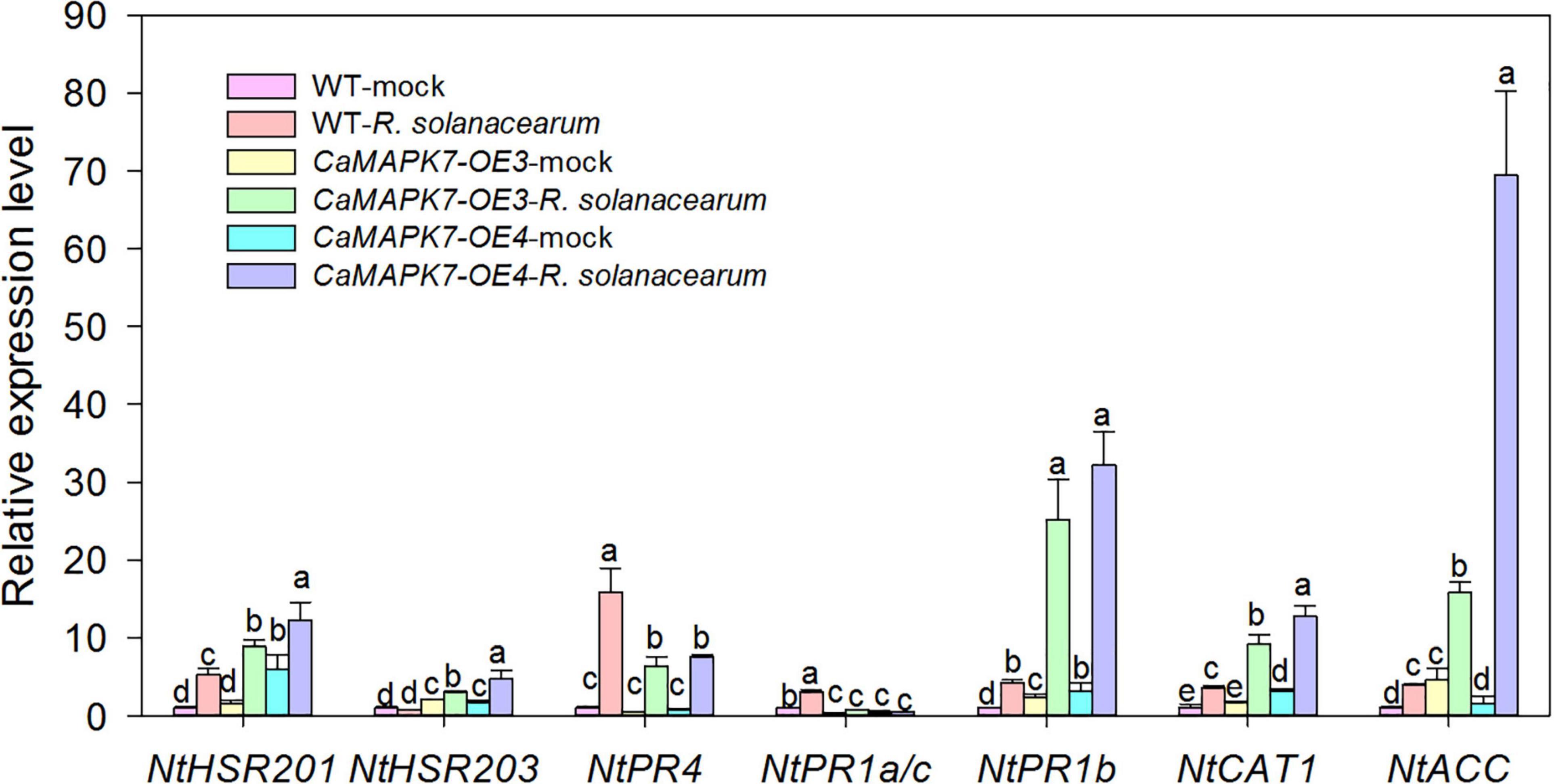
Figure 7. qRT-PCR analyses of relative expression levels of defense marker genes in CaMAPK7-OX lines compared with WT tobacco plants. Defense-related transcript accumulation of WT tobacco plants were used as reference, which was set to “1.” Different letters above the bar show a significant difference between the means of the three biological replicates based on Fisher’s protected LSD test: lowercase letters, p < 0.05. WT, wild-type.
The Effect of CaMAPK7 Transient Overexpression on the Binding of CaWRKY40 to Its Target Genes
The transient overexpression assay revealed that CaMAPK7 overexpression triggered the upregulation of CaWRKY40, a WRKY transcription factor involved in pepper immunity against R. solanacearum; moreover, one or two W-box elements were found to be contained in the promoters of the defense-related marker genes CaSAR82A, CaPO2, and CaDEF1 regulated by CaMAPK7 (Figures 8A,B). We hypothesize that CaMAPK7 regulates the expression of marker genes above via the modification of the binding of CaWRKY40 to the promoter of marker genes described above. ChIP and PCR assays were performed to verify our hypothesis. To conduct this experiment, GV3101 cells harboring CaMAPK7-HA or an empty vector were infiltrated into the leaves of 6-week-old pepper plants and maintained in the greenhouse. At 48 hpi, the infiltrated pepper leaves were harvested and cross-linked with 1% formaldehyde, and the chromatin was isolated as described in our previous study (Shen et al., 2016a) with slight modifications. Primers flanking the W-box of the marker genes were used for ChIP-PCR. For promoters with more than one W-box, the primer pairs were screened for product amplification and used in the real-time PCR measurements of specific CaWRKY40 binding to the promoter. The binding of CaWRKY40 to the promoters of CaSAR82A, CaPO2, and CaDEF1 was significantly enhanced by the overexpression of CaMAPK7 (Figure 8C). CaWRKY40-silenced pepper plants were acquired to study the effect of CaWRKY40-silencing on the regulation of CaMAPK7 on downstream marker genes described above. VIGS and qRT-PCR assays showed that in the unsilenced pepper plants, CaMAPK7 overexpression significantly increased the expression of defense-marker genes, including CaPO2, CaDEF1, and CaSAR82A, whereas the increase was partially abolished in CaWRKY40-silenced pepper plants (Figure 8D).
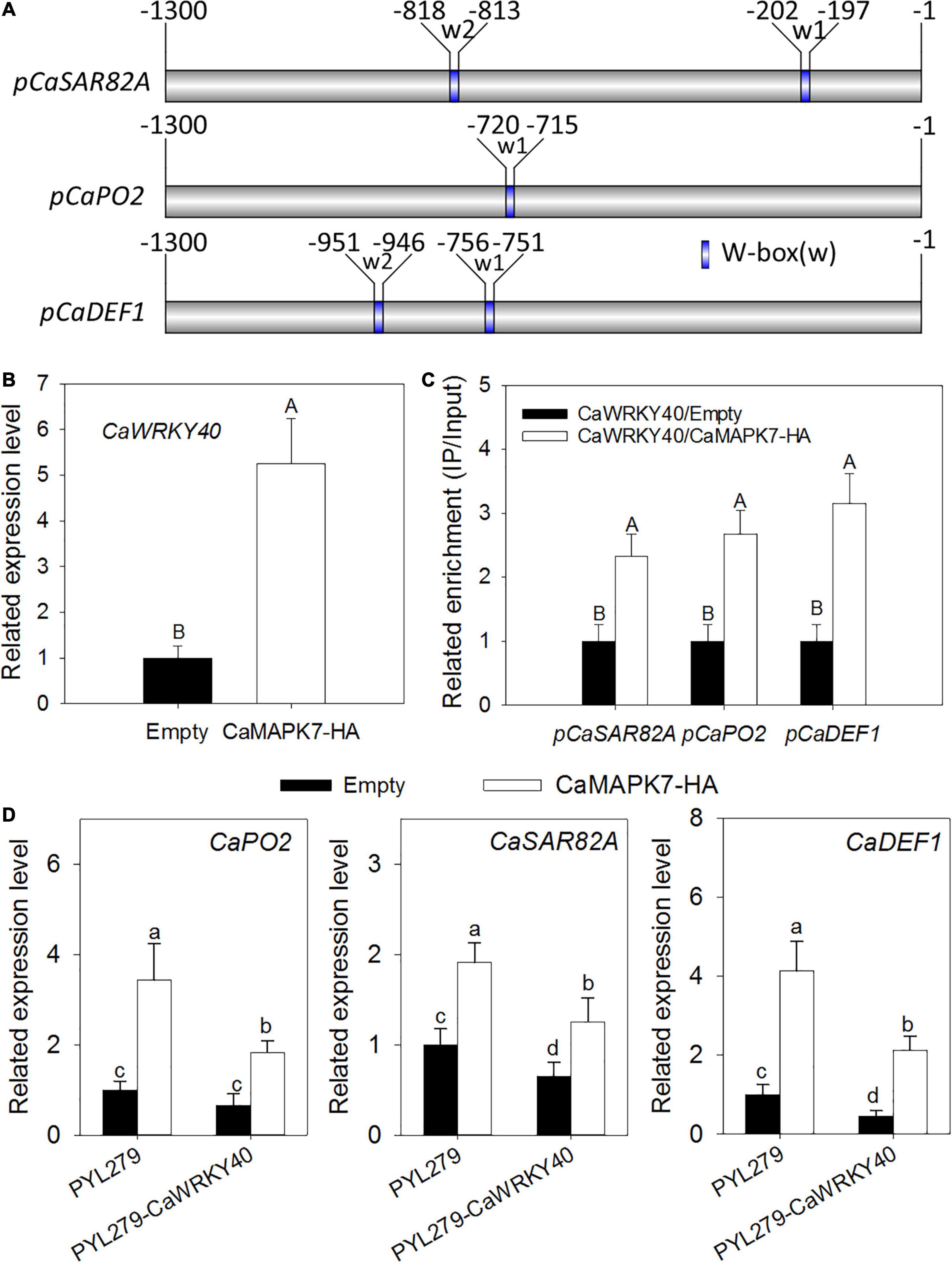
Figure 8. The transcriptional expression of CaWRKY40 triggered by CaMAPK7 overexpression and the bindings of CaWRKY40 to the promoters of its target genes were enhanced by CaMAPK7 expression. (A) Schematic representation of a typical W-box in the promoters of CaWRKY40 downstream defense marker genes. (B) CaWRKY40 transcript level triggered by CaMAPK7 overexpression in pepper leaves. The CaWRKY40 transcript level in pepper leaves transient expressed empty vector was set to “1.” (C) The binding of CaWRKY40 to the promoters of its target genes was enhanced by CaMAPK7 transient expression in pepper plants. Relative enrichment levels of samples of the CaWRKY40 transient overexpression were set to “1” after normalization by input. (D) Real-time PCR analysis of the defense-associated marker genes in CaWRKY40-silenced pepper leaves transiently overexpressing CaMAPK7. The expression of defense-related marker genes in unsilenced pepper leaves transiently expressing vector was set to “1.” (B–D) Data are the means ± SD from at least three independent experiments. Different letters above the bar show a significant difference between the means of the three biological replicates based on Fisher’s protected LSD test: uppercase letters, p < 0.01; lowercase letters, p < 0.05. SD, standard deviation.
Discussion
CaMAPK7 Is a Positive Regulator of Defense Responses and Plant Cell Death in Pepper Plants
At the extreme downstream of MAPKs cascade, MAPKs were reported to play vital roles in plant growth, development, and responses to several environmental challenges via the phosphorylation of a target substrate. However, most of the studies of MAPKs have focused on model plants, such as Arabidopsis and rice. Therefore, the roles of MAPK in non-model plants such as pepper remain to be elucidated. The most extensively studied plant MAPKs are Arabidopsis AtMPK6, AtMPK3, and AtMPK4, all of which are activated by several stimuli, including biotic and abiotic stresses (Ichimura et al., 2000; Droillard et al., 2002; Pitzschke et al., 2009; Schikora et al., 2011; Dutilleul et al., 2012). Nineteen MAPKs were identified in the pepper genome in our previous study, although only 2 MAPKs out of 19, MK1 and MK2 (designated CaMPK3 and CaMPK6-1 in our previous study), were partially functionally analyzed (Shin et al., 2001). MK1 and MK2 encode stress-inducible protein kinases that can contribute to the response to wounding, UV-C and cold. The identification and functional analysis of other MAPK members in pepper plants will benefit from the dissection of MAPK cascades in pepper. In this study, a novel pepper MAPK CaMAPK7 was identified, and its function in response to R. solanacearum was analyzed. Pathogen infection induces the expression of many MAPK genes (Lumbreras et al., 2010). Of the 19 pepper MAPK genes, 12 were found to be differentially regulated following treatment with R. solanacearum (Liu Z. et al., 2015). We found CaMAPK7 transcripts to be induced by inoculation with R. solanacearum. Gene expression is strictly regulated by the upstream promoter, and the motifs contained in the promoter were associated with the potential functions of target gene. The results analyzed on PLANTCARE showed that diverse motifs, including stress-related motifs, were contained in the CaMAPK7 promoter, including the transcription factor WRKY binding motif W-box (4), ethylene-responsive ERE box (1), transcription factor MYB binding motif MBS (1), low-temperature-responsive element LTR (1), and heat shock element HSE (1). These cis-elements have been reported to be involved with the plant signaling pathway and immunity responses. We further investigated the activities of the CaMAPK7 promoter (pCaMAPK7), and the data showed that pCaMAPK7 significantly induces GUS activities in response to infection with R. solanacearum (Supplementary Figure 1). The overexpression of CaMAPK7 enhanced resistance of transgenic tobacco to inoculation with R. solanacearum, suggesting a positive contribution of CaMAPK7 to resistance against bacterial pathogens. Consistent with this, the resistance of pepper plants to inoculation with R. solanacearum was attenuated in CaMAPK7-silenced pepper plants. In Arabidopsis, MPK7 was activated by upstream MKK3 in response to ABA, but poorly activated by flagellin-derived flg22 peptide treatment, suggesting that MPK7 participates in ABA signaling pathway and plays a negative role in PTI.
A typical event of a defense response is the accumulation of transcripts of HR-associated and pathogen-responsive (PR) genes and usually serves as a marker for the activation of plant immunity. In tobacco, the expression of NtHSR201 and NtHSR203 is frequently upregulated during defense responses against bacterial pathogens (Czernic et al., 1996; Pontier et al., 1998; Lee et al., 2004). The overexpression of CaMAPK7 in transgenic tobacco constitutively upregulates the expression of NtHSR201 and NtHSR203. The immunity mediated by CaMAPK7 against R. solanacearum is likely based on the prompting effect of CaMAPK7 on transcription of the HR-associated and PR genes. Furthermore, the transient overexpression of CaMAPK7 in pepper leaves triggers insensitive HR-like cell death accompanied with a significant burst of ROS. It is worth noting that NtHSR203, a negative regulator of plant hypersensitive cell death (Pontier et al., 1998; Tronchet et al., 2001), was upregulated in tobacco plants by R. solanacearum infection and also by CaMAPK7 overexpression in tobacco plants uninoculated or inoculated with R. solanacearum. This finding that NtHSR203 was upregulated in transgenic tobacco plants that ectopically overexpressed CaWRKY40 during the defense response of pepper against R. solanacearum was also observed in our previous study (Dang et al., 2013). We hypothesize that the upregulation of NtHSR203 triggered by R. solanacearum and CaMAPK7 overexpression serves to avoid the inappropriate activation of defense responses. Collectively, these results suggest that CaMAPK7 functions as a positive regulator of defense response and in HR-like cell death in pepper leaves.
CaMAPK7 May Play a Vital Role in Defense Signaling Mediated by SA, JA, ET, and BR
The production of SA, JA, ET, and BR is frequently induced in response to various pathogens. The balance of these hormones is indispensable in fine-tuning appropriate defense responses (Dong, 1998; Dangl and Jones, 2001; Divi et al., 2010). SA, JA, ET, and BR have been reported to activate numerous plant PR genes and act synergistically or antagonistically during defense signaling dependent on their concentrations (Bari and Jones, 2009; Yang et al., 2015, 2019; Pan et al., 2018). We found CaMAPK7 transcripts to be induced in response to the exogenous application of SA, JA, ET, and BR. The overexpression of CaMAPK7 in pepper leaves led to the upregulation of JA-responsive CaDEF1, SA-associated CaSAR82A, and ROS-related CaPO2. In addition, the overexpression of CaMAPK7 in transgenic tobacco enhanced the levels of transcripts of SA-responsive NtPR1a/c, JA-responsive NtPR4 and NtPR-b, the ROS detoxification-associated gene NtCAT1, and the ethylene production-associated NtACC in response to inoculation with R. solanacearum. It is worth noting that ABA treatment decreased the level of transcript of CaMAPK7, which supports an antagonistic relationship between ABA and SA in host–pathogen interactions (Nahar et al., 2012). Therefore, we hypothesize that SA, JA, ET, and BR regulate the expression of CaMAPK7, leading to the expression of downstream defense-associated marker genes.
CaMAPK7 Transient Overexpression Modifies the Binding of CaWRKY40 to Its Target Genes
Although MAPKs and WRKYs are both reported to participate in plant immune response to pathogens, the molecular linkage between these two proteins has not been fully established. Our previous study indicated that the CaWRKY40 transcript was upregulated by inoculation with R. solanacearum and acts as a positive regulator of pepper immunity against R. solanacearum. The fact that CaMAPK7 and CaWRKY40 exhibited similar patterns of expression in response to R. solanacearum and applied exogenous hormones, including SA, MeJA, and ET, implies a potential relationship between these two proteins. This hypothesis was supported by the result that the CaWRKY40 transcript was significantly induced by the overexpression of CaMAPK7 and downregulated in CaMAPK7-silenced pepper plants, compared with the unsilenced pepper plants. CaMAPK7, located in the plasma membrane and cytoplasm, is believed to be involved in the early signaling pathway and participates in pepper defense response against R. solanacearum infection. CaWRKY40, activated by upstream CaWRKY6 and CaWRKY40b in our previous study (Cai et al., 2015; Ifnan Khan et al., 2018), is expected to function in R. solanacearum-induced pepper immunity via regulation of the downstream target genes, such as CaC3H14, CabZIP63, and CaCDPK15 (Shen et al., 2016a, b; Qiu et al., 2018). Of note is that CabZIP63 and CaCDPK15 directly or indirectly, respectively, regulate the expression of CaWRKY40 at both the transcriptional and post-transcriptional level, forming a positive feedback loop with CaWRKY40 during the response of pepper to RSI or high temperature and high humidity (Shen et al., 2016a, b). Similar molecular linkages between MAPKs and WRKYs have been reported previously. For example, a study by Adachi et al. (2015) showed that four WRKY transcription factors were phosphorylated by MEK2 to regulate a plant immune NADPH oxidase in N. benthamiana (Adachi et al., 2015). Ishihama et al. (2011) provided evidence that the N. benthamiana WRKY8 transcription factor is physiologically phosphorylated by SIPK, NTF4, and WIPK, and the phosphorylation of WRKY8 increased its DNA binding activity to the cognate W-box sequence (Ishihama et al., 2011) and triggered the expression of downstream genes. Furthermore, the VIGS and qRT-PCR assays in this study showed that in unsilenced pepper plants, the overexpression of CaMAPK7 significantly increased the expression of defense-marker genes, including CaPO2, CaDEF1, and CaSAR82A, whereas the increase was partially abolished in CaWRKY40-silenced pepper plants. Based on the results described above and our previous study, we hypothesize that CaMAPK7 may regulate the expression of defense-related marker genes, CaDEF1, CaPO2, and CaSAR82A, in a CaWRKY40-dependent manner. In addition, the inducible expression of CaWRKY40 in response to R. solanacearum was significantly attenuated in CaMAPK7-silenced pepper plants, suggesting that CaWRKY40 functions in pepper defense against R. solanacearum probably in a CaMAPK7-dependent manner. We hypothesize that CaMAPK7 may interact with CaWRKY40 and directly modify the regulation of CaWRKY40 on downstream marker genes. Unexpectedly, our co-IP assay showed that no binding was observed between CaMAPK7 and CaWRKY40 (data not shown). One explanation is that CaMAPK7 might phosphorylate and activate other component that directly targets CaWRKY40, and the phosphorylated component acts as a bridge between CaMAPK7 and CaWRKY40. Further identification of possible CaMAPK7 interactors that subsequently target CaWRKY40 could provide new insights into the mechanism of pepper immunity mediated by CaMAPK7 and CaWRKY40. Taken together, we propose a working model that infection with R. solanacearum induces the accumulation of SA, JA, BR, and ET and activates the expression of CaMAPK7, which subsequently modifies the transcriptional activity of CaWRKY40, ultimately leading to cell death and disease resistance in pepper plants (Figure 9).
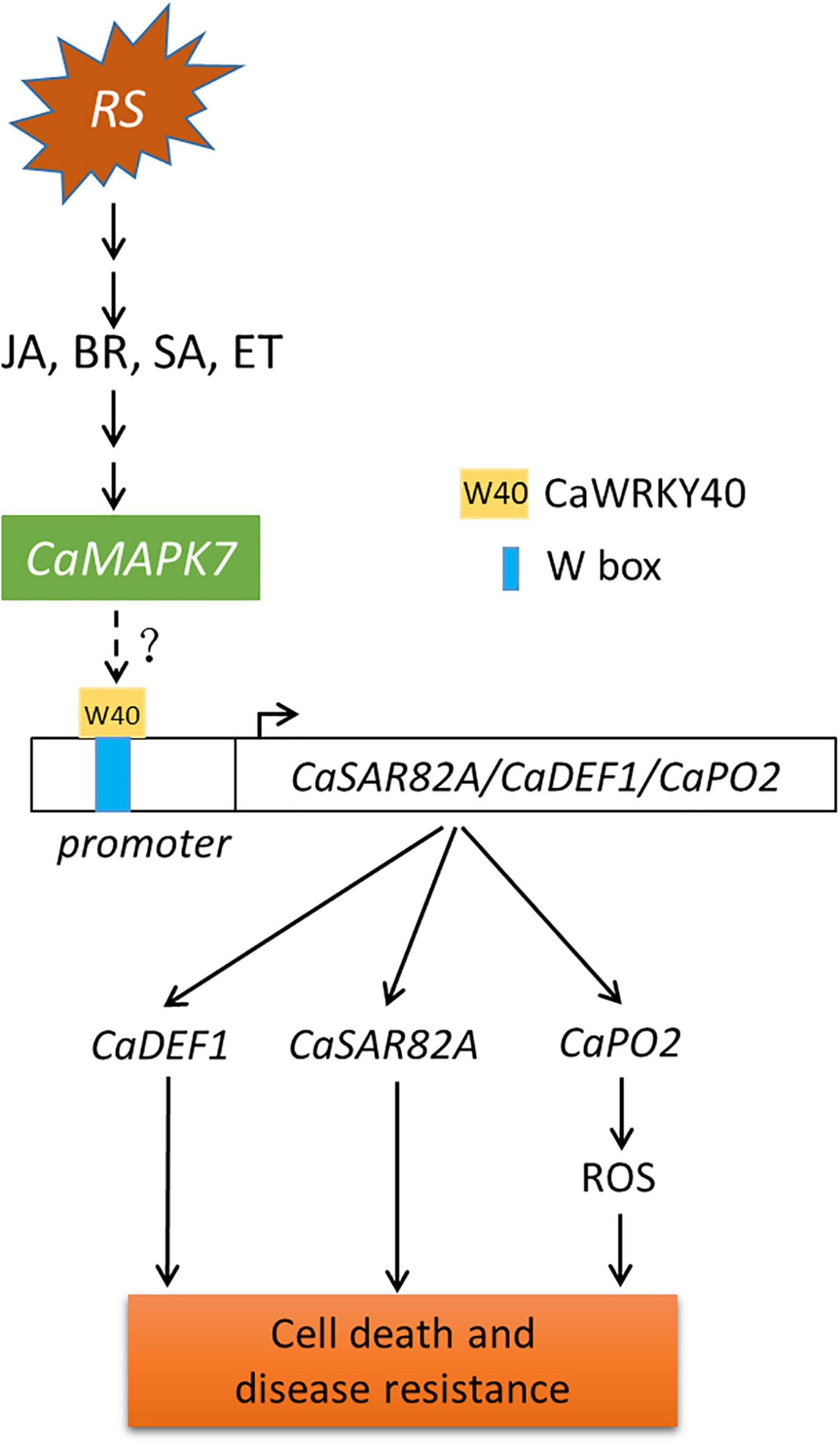
Figure 9. Proposed model of CaMAPK7 in response to inoculation with Ralstonia solanacearum in pepper plants. The pepper CaMAPK7 has a positive effect on the trans-activation of CaWRKY40, which regulates the expression of defense-associated marker genes, ultimately leading to cell death and disease resistance. Arrows indicate positive regulation, and dotted ends denote unknown regulation. RS, Ralstonia solanacearum.
In summary, our data demonstrate that CaMAPK7 responds to infection with R. solanacearum and plays a positive role in resistance to R. solanacearum by modifying the trans-activation of CaWRKY40. These results suggest a possible molecular mechanism involving CaMAPK7 in R. solanacearum responses and provides insight into the MAPK cascade in pepper plants.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LS, SH, and ZL designed the experiments. LS, KZ, LX, MY, and BX performed the experiments and the data analyses. LS, SH, and ZL prepared the figures and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Fujian Province, China (2020J01532), the Excellent Youth Foundation of Fujian Agriculture and Forestry University (xjq201913), and the National Natural Science Foundation of China (31501767, 30971718, and 31372061). The funders had no role in the study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.664926/full#supplementary-material
Supplementary Figure 1 | The distribution of potential motifs in CaMAPK7 promoter CAATBOX, cis-acting regulatory element involved in light responsiveness; W-box, WRKY transcription factor binding site; ERE, ethylene-responsive element; TCA, cis-acting regulatory element involved in salicylic acid responsiveness; ARE, cis-acting regulatory elements essential for the anaerobic induction; MBS, MYB binding site; HSE, cis-acting regulatory element involved in heat stress responsiveness; LTR, cis-acting element involved in low-temperature responsiveness; TATA-Box, core promoter of the transcription start site.
Supplementary Figure 2 | Relative GUS activities driven by the CaMAPK7 promoter in transgenic tobacco plants in response to Ralstonia solanacearum inoculation and the application of exogenous phytohormones. (A–F) Relative GUS activities in tobacco plants harboring pCaMAPK7:GUS at various time periods after treatment with 100 μM ABA (A), 100 μM MeJA (B), 10 μM Brassinolide (C), mM SA (D), 100 μM ET (E), 1 and R. solanacearum (F). The GUS activity of transgenic tobacco plants carrying CaMAPK7:GUS without treatments were set to “1.” Error bars indicate the standard error. Different letters above the bar show a significant difference between the means of three biological replicates based on Fisher’s protected LSD test: uppercase letters, p < 0.01; lowercase letters, p < 0.05.
Supplementary Table 1 | Oligonucleotides for the plasmid constructs and PCR used in this study.
Footnotes
References
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663. doi: 10.1105/tpc.15.00213
Ali, M., Muhammad, I., Ul Haq, S., Alam, M., Khattak, A. M., Akhtar, K., et al. (2020a). The CaChiVI2 gene of Capsicum annuum L. Confers resistance against heat stress and infection of Phytophthora capsici. Front. Plant Sci. 11:219. doi: 10.3389/fpls.2020.00219
Ali, M., Tumbeh Lamin-Samu, A., Muhammad, I., Farghal, M., Khattak, A. M., Jan, I., et al. (2020b). Melatonin mitigates the infection of Colletotrichum gloeosporioides via modulation of the chitinase gene and antioxidant activity in Capsicum annuum L. Antioxidants 10:7. doi: 10.3390/antiox10010007
Bargues-Ribera, M., and Gokhale, C. S. (2020). Eco-evolutionary agriculture: host-pathogen dynamics in crop rotations. PLoS Comput. Biol. 16:e1007546. doi: 10.1371/journal.pcbi.1007546
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Beck, M., Komis, G., Muller, J., Menzel, D., and Samaj, J. (2010). Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22, 755–771. doi: 10.1105/tpc.109.071746
Beck, M., Komis, G., Ziemann, A., Menzel, D., and Samaj, J. (2011). Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189, 1069–1083. doi: 10.1111/j.1469-8137.2010.03565.x
Berriri, S., Garcia, A. V., Freidit Frey, N., Rozhon, W., Pateyron, S., Leonhardt, N., et al. (2012). Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24, 4281–4293. doi: 10.1105/tpc.112.101253
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Choi, D. S., and Hwang, B. K. (2011). Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–842. doi: 10.1105/tpc.110.082081
Czernic, P., Huang, H. C., and Marco, Y. (1996). Characterization of hsr201 and hsr515, two tobacco genes preferentially expressed during the hypersensitive reaction provoked by phytopathogenic bacteria. Plant Mol. Biol. 31, 255–265. doi: 10.1007/bf00021788
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Divi, U. K., Rahman, T., and Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10:151. doi: 10.1186/1471-2229-10-151
Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. doi: 10.1016/1369-5266(88)80053-0
Droillard, M., Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2002). Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 527, 43–50. doi: 10.1016/s0014-5793(02)03162-9
Dutilleul, C., Benhassaine-Kesri, G., Demandre, C., Reze, N., Launay, A., Pelletier, S., et al. (2012). Phytosphingosine-phosphate is a signal for AtMPK6 activation and Arabidopsis response to chilling. New Phytol. 194, 181–191. doi: 10.1111/j.1469-8137.2011.04017.x
Eschen-Lippold, L., Jiang, X., Elmore, J. M., Mackey, D., Shan, L., Coaker, G., et al. (2016). Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 171, 2223–2238. doi: 10.1104/pp.16.00336
Genin, S., and Boucher, C. (2004). Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42, 107–134. doi: 10.1146/annurev.phyto.42.011204.104301
Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., and Stam, M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3:11. doi: 10.1186/1746-4811-3-11
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
He, Z. D., Tao, M. L., Leung, D. W. M., Yan, X. Y., Chen, L., Peng, X. X., et al. (2021). The rice germin-like protein OsGLP1 participates in acclimation to UV-B radiation. Plant Physiol. 186, 1254–1268. doi: 10.1093/plphys/kiab125
Huh, S. U., Lee, G. J., Jung, J. H., Kim, Y., Kim, Y. J., and Paek, K. H. (2015). Capsicum annuum transcription factor WRKYa positively regulates defense response upon TMV infection and is a substrate of CaMK1 and CaMK2. Sci. Rep. 5:7981.
Ichimura, K., Mizoguchi, T., Yoshida, R., Yuasa, T., and Shinozaki, K. (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24, 655–665. doi: 10.1046/j.1365-313x.2000.00913.x
Ifnan Khan, M., Zhang, Y., Liu, Z., Hu, J., Liu, C., Yang, S., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19:1403. doi: 10.3390/ijms19051403
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., and Yoshioka, H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23, 1153–1170. doi: 10.1105/tpc.110.081794
Jalmi, S. K., and Sinha, A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 6:37974.
Jiang, L., Wan, Y., Anderson, J. C., Hou, J., Islam, S. M., Cheng, J., et al. (2017). Genetic dissection of Arabidopsis MAP kinase phosphatase 1-dependent PAMP-induced transcriptional responses. J. Exp. Bot. 68, 5207–5220. doi: 10.1093/jxb/erx335
Jing, T., Wang, L., Liu, H., Wuyun, T. N., and Du, H. (2017). Genome-wide identification of mitogen-activated protein kinase cascade genes and transcriptional profiling analysis during organ development in Eucommia ulmoides. Sci. Rep. 7:17732.
Kawasaki, T., Yamada, K., Yoshimura, S., and Yamaguchi, K. (2017). Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant Signal. Behav. 12:e1361076. doi: 10.1080/15592324.2017.1361076
Kong, X., Lv, W., Zhang, D., Jiang, S., Zhang, S., and Li, D. (2013). Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS One 8:e57714. doi: 10.1371/journal.pone.0057714
Lee, D. E., Lee, I. J., Han, O., Baik, M. G., Han, S. S., and Back, K. (2004). Pathogen resistance of transgenic rice plants expressing mitogen-activated protein kinase 1, MK1, from Capsicum annuum. Mol. Cells 17, 81–85.
Li, J., Yang, P., Kang, J., Gan, Y., Yu, J., Calderon-Urrea, A., et al. (2016). Transcriptome analysis of pepper (Capsicum annuum) revealed a role of 24-epibrassinolide in response to chilling. Front. Plant Sci. 7:1281. doi: 10.3389/fpls.2016.01281
Li, K., Yang, F., Zhang, G., Song, S., Li, Y., Ren, D., et al. (2017). AIK1, a mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 173, 1391–1408. doi: 10.1104/pp.16.01386
Lim, C. W., and Lee, S. C. (2016). Pepper protein phosphatase type 2C, CaADIP1 and its interacting partner CaRLP1 antagonistically regulate ABA signalling and drought response. Plant Cell Environ. 39, 1559–1575. doi: 10.1111/pce.12721
Liu, J. Z., Horstman, H. D., Braun, E., Graham, M. A., Zhang, C., Navarre, D., et al. (2011). Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157, 1363–1378. doi: 10.1104/pp.111.185686
Liu, P., Li, B., Lin, M., Chen, G., Ding, X., Weng, Q., et al. (2016). Phosphite-induced reactive oxygen species production and ethylene and ABA biosynthesis, mediate the control of Phytophthora capsici in pepper (Capsicum annuum). Funct. Plant Biol. 43, 563–574. doi: 10.1071/fp16006
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313x.2002.01394.x
Liu, Z., Shi, L., Liu, Y., Tang, Q., Shen, L., Yang, S., et al. (2015). Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front. Plant Sci. 6:780. doi: 10.3389/fpls.2015.00780
Liu, Z. Q., Qiu, A. L., Shi, L. P., Cai, J. S., Huang, X. Y., Yang, S., et al. (2015). SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698. doi: 10.1093/jxb/erv161
Lu, K., Guo, W., Lu, J., Yu, H., Qu, C., Tang, Z., et al. (2015). Genome-wide survey and expression profile analysis of the mitogen-activated protein kinase (MAPK) gene family in Brassica rapa. PLoS One 10:e0132051. doi: 10.1371/journal.pone.0132051
Lumbreras, V., Vilela, B., Irar, S., Sole, M., Capellades, M., Valls, M., et al. (2010). MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 63, 1017–1030. doi: 10.1111/j.1365-313x.2010.04297.x
Luo, X., Xu, N., Huang, J., Gao, F., Zou, H., Boudsocq, M., et al. (2017). A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol. 174, 2501–2514. doi: 10.1104/pp.17.00404
Muller, A., Manzara, T., and Lurquin, P. F. (1984). Crown gall transformation of tobacco callus cells by cocultivation with Agrobacterium tumefaciens. Biochem. Biophys. Res. Commun. 123, 458–462. doi: 10.1016/0006-291x(84)90252-3
Nahar, K., Kyndt, T., Nzogela, Y. B., and Gheysen, G. (2012). Abscisic acid interacts antagonistically with classical defense pathways in rice-migratory nematode interaction. New Phytol. 196, 901–913. doi: 10.1111/j.1469-8137.2012.04310.x
Pan, G., Liu, Y., Ji, L., Zhang, X., He, J., Huang, J., et al. (2018). Brassinosteroids mediate susceptibility to brown planthopper by integrating with the salicylic acid and jasmonic acid pathways in rice. J. Exp. Bot. 69, 4433–4442. doi: 10.1093/jxb/ery223
Pitzschke, A., Schikora, A., and Hirt, H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12, 421–426. doi: 10.1016/j.pbi.2009.06.008
Pontier, D., Tronchet, M., Rogowsky, P., Lam, E., and Roby, D. (1998). Activation of hsr203, a plant gene expressed during incompatible plant-pathogen interactions, is correlated with programmed cell death. Mol. Plant Microbe Interact. 11, 544–554. doi: 10.1094/mpmi.1998.11.6.544
Qiu, A., Lei, Y., Yang, S., Wu, J., Li, J., Bao, B., et al. (2018). CaC3H14 encoding a tandem CCCH zinc finger protein is directly targeted by CaWRKY40 and positively regulates the response of pepper to inoculation by Ralstonia solanacearum. Mol. Plant Pathol. 19, 2221–2235. doi: 10.1111/mpp.12694
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Schikora, A., Schenk, S. T., Stein, E., Molitor, A., Zuccaro, A., and Kogel, K. H. (2011). N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 157, 1407–1418. doi: 10.1104/pp.111.180604
Shen, L., Liu, Z., Yang, S., Yang, T., Liang, J., Wen, J., et al. (2016a). Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 67, 2439–2451. doi: 10.1093/jxb/erw069
Shen, L., Yang, S., Yang, T., Liang, J., Cheng, W., Wen, J., et al. (2016b). CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci. Rep. 6:22439.
Shin, H. J., Lee, D. E., Shin, D. H., Kim, K. U., Kim, H. Y., Ohashi, Y., et al. (2001). Molecular cloning and cultivar specific expression of MAP kinases from Capsicum annuum. Mol. Cells 11, 48–54.
Song, F., and Goodman, R. M. (2002). OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215, 997–1005. doi: 10.1007/s00425-002-0794-5
Sozen, C., Schenk, S. T., Boudsocq, M., Chardin, C., Almeida-Trapp, M., Krapp, A., et al. (2020). Wounding and insect feeding trigger two independent MAPK pathways with distinct regulation and kinetics. Plant Cell 32, 1988–2003. doi: 10.1105/tpc.19.00917
Takahashi, F., Yoshida, R., Ichimura, K., Mizoguchi, T., Seo, S., Yonezawa, M., et al. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19, 805–818. doi: 10.1105/tpc.106.046581
Teardo, E., Carraretto, L., Moscatiello, R., Cortese, E., Vicario, M., Festa, M., et al. (2019). A chloroplast-localized mitochondrial calcium uniporter transduces osmotic stress in Arabidopsis. Nat. Plants 5, 581–588. doi: 10.1038/s41477-019-0434-8
Tronchet, M., Ranty, B., Marco, Y., and Roby, D. (2001). HSR203 antisense suppression in tobacco accelerates development of hypersensitive cell death. Plant J. 27, 115–127. doi: 10.1046/j.1365-313x.2001.01072.x
Wang, M., Yue, H., Feng, K., Deng, P., Song, W., and Nie, X. (2016). Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genomics 17:668. doi: 10.1186/s12864-016-2993-7
Yanagawa, Y., Yoda, H., Osaki, K., Amano, Y., Aono, M., Seo, S., et al. (2016). Mitogen-activated protein kinase 4-like carrying an MEY motif instead of a TXY motif is involved in ozone tolerance and regulation of stomatal closure in tobacco. J. Exp. Bot. 67, 3471–3479. doi: 10.1093/jxb/erw173
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
Yang, Y. X., Ahammed, G. J., Wu, C., Fan, S. Y., and Zhou, Y. H. (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 16, 450–461. doi: 10.2174/1389203716666150330141638
Zeng, Q., Chen, J. G., and Ellis, B. E. (2011). AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 67, 895–906. doi: 10.1111/j.1365-313x.2011.04642.x
Keywords: Capsicum annuum, mitogen-activated protein kinase, Ralstonia solanacearum, WRKY transcription factor, pathogenicity
Citation: Shi L, Zhang K, Xie L, Yang M, Xie B, He S and Liu Z (2021) The Pepper Mitogen-Activated Protein Kinase CaMAPK7 Acts as a Positive Regulator in Response to Ralstonia solanacearum Infection. Front. Microbiol. 12:664926. doi: 10.3389/fmicb.2021.664926
Received: 06 February 2021; Accepted: 07 June 2021;
Published: 06 July 2021.
Edited by:
Marco Scortichini, Council for Agricultural and Economics Research (CREA), ItalyCopyright © 2021 Shi, Zhang, Xie, Yang, Xie, He and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuilin He, c2hsaGVmYWZ1QDE2My5jb20=; Zhiqin Liu, bHpxZnVqaWFuQDEyNi5jb20=
†These authors have contributed equally to this work
 Lanping Shi1,2†
Lanping Shi1,2† Kan Zhang
Kan Zhang Zhiqin Liu
Zhiqin Liu