- 1State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University, Beijing, China
- 3Children’s Hospital of Fudan University, Shanghai, China
- 4School of Light Industry, Beijing Technology and Business University, Beijing, China
- 5Jiangxi Provincial Key Laboratory of Preventive Medicine, School of Public Health, Nanchang University, Nanchang, China
- 6Department of Microbiology, Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China
Salmonella is the primary cause of community-acquired foodborne infections, so its resistance to antimicrobials, such as aminoglycosides, is a public health issue. Of concern, aminoglycoside resistance in Salmonella is increasing rapidly. Here, we performed a retrospective study evaluating the prevalence of Salmonella harboring armA-mediated aminoglycoside resistance in community-acquired infections and in food or environmental sources. The prevalence rates of armA-harboring Salmonella strains were 1.1/1,000 (13/12,095) and 8.7/1,000 (32/3,687) in outpatient and food/environmental isolates, respectively. All the armA-harboring Salmonella strains were resistant to multiple drugs, including fluoroquinolone and/or extended-spectrum cephalosporins, and most (34/45) belonged to serovar Indiana. The armA gene of these strains were all carried on plasmids, which spanned five replicon types with IncHI2 being the dominant plasmid type. All the armA-carrying plasmids were transferable into Escherichia coli and Acinetobacter baumannii recipients. The conjugation experiment results revealed that the armA-harboring S. Indiana strains had a relatively higher ability to acquire armA-carrying plasmids. The low similarity of their pulsed field gel electrophoresis patterns indicates that the armA-harboring Salmonella strains were unlikely to have originated from a single epidemic clone, suggesting broad armA spread. Furthermore, the genetic backgrounds of armA-harboring Salmonella strains isolated from outpatients exhibited higher similarity to those isolated from poultry than to those isolated from swine, suggesting that poultry consumption maybe an infection source. These findings highlight an urgent need to monitor the prevalence and transmission of armA-harboring Salmonella, especially S. Indiana, to better understand the potential public health threat and prevent the further spread of these strains.
Introduction
Aminoglycoside antibiotics are widely used in hospitals for the treatment of infections, especially those caused by Gram-negative bacteria (Doi et al., 2004), and can be used synergistically with many other antibiotics (Xia et al., 2016). Aminoglycosides were also used extensively in food-producing animals to prevent bacterial infections (Chen et al., 2007); several clinical first-line aminoglycoside agents were misused in conventional broiler chicken and swine production facilities for the prevention of infections, as well as for growth promotion (Qin et al., 2012; Xia et al., 2016).
Bacterial resistance to aminoglycosides was initially found to be due to the production of aminoglycoside-modifying enzymes, including acetyltransferases, phosphotransferases, and nucleotidyltransferases, which may modify aminoglycosides into non-functional forms (Ramirez and Tolmasky, 2010). 16S rRNA methylation enzymes were thought to be confined to environmental strains until the discovery of armA gene-mediated aminoglycoside resistance in Klebsiella pneumoniae in 2003 (Galimand et al., 2003). To date, ten different 16S rRNA methylase genes, armA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, and npmA, have been reported in various Enterobacteriaceae strains, and the 16S rRNA methylases encoded by these genes confer high-level resistance to various aminoglycosides (Galimand et al., 2012; Wachino and Arakawa, 2012; Bueno et al., 2013; O’Hara et al., 2013). Because the 16S rRNA methylase genes are located on plasmids, they can be easily transferred to other bacteria (Wachino et al., 2007). Therefore, these genes should be monitored by surveillance as a public health precaution.
Salmonella spp., which are members of the family Enterobacteriaceae, cause a broad range of diseases in humans, mainly diarrhea and systemic infections, and are one of the greatest food safety threats (LaRock et al., 2015). These bacteria usually live in the intestinal tracts of various animals and humans and can be excreted into the environment via feces (Michael and Schwarz, 2016). Salmonella spp. often exhibit resistance to multiple antibiotics and are recognized as very important bacterial vectors of drug-resistance genes in the spread of antibiotic resistance (Crump et al., 2015; Kariuki et al., 2015; Wen et al., 2017). In several aminoglycoside-resistant Enterobacteriaceae (Ma et al., 2009), including Salmonella (Naas et al., 2011; Du et al., 2012), the most common 16S rRNA methylase gene is armA. Since the report of its discovery in K. pneumoniae strains isolated from patients in 2003 (Galimand et al., 2003), plasmids containing armA have been detected in diverse bacterial species (Du et al., 2012; Wang et al., 2017b; Uechi et al., 2018). The plasmids carrying rmtB were found in S. Typhimurium isolated from food-animal products, and found could co-spread with qepA and blaCTX-M-27 in S. Indiana strains from water husbandry (Fang et al., 2019). In addition, other 16S rRNA methylase genes such as rmtC and rmtD were observed in Salmonella enterica, S. Indiana and S. California strains from chicken samples or people, but are less common (Folster et al., 2009; Hopkins et al., 2010; Wang et al., 2017c). The other 16S rRNA methylase genes such as rmtE, rmtF, rmtG, and rmtH have never been reported in Salmonella strains.
However, the epidemic and transmission trends of this gene in Salmonella are less defined. Plasmids carrying armA may allow easy transmission of this gene among various bacterial species, including pathogenic strains.
The prevalence of Salmonella harboring plasmids containing armA needs to be monitored in people, food, and environmental sources. Here, we conducted a retrospective study to characterize the epidemic status and transmission of armA in Salmonella strains isolated from outpatients as well as from food and environmental sources.
Materials and Methods
Bacterial Strains
Beginning in 2005, Salmonella infections in diarrheal outpatients were surveyed yearly in Shanghai. Following the expansion of the participating surveillance laboratories in June 2010, the annual number of isolated strains increased from 2011. Of the 15,782 Salmonella strains used in this study: 12,095 strains were collected from outpatient stools in Shanghai between 2005 and 2016; 3,206 strains were collected from various food or environmental samples, including chicken, pork, water, and seafood, in Shanghai between 2009 and 2015; and 481 strains were collected from various food or environmental samples, including chicken, pork, water, and seafood, in Guangdong between 2013 and 2015. All strains were recovered from the strain pool and isolated in CHROMagar Salmonella agar (CHROMagar Company, Paris, France). Suspected isolates were identified with the Vitek-II system (BioMerieux, Lyon, France).
Resistance Gene Amplification
All Salmonella isolates were tested for the presence of the armA gene by using real-time polymerase chain reaction (PCR) (Supplementary Table 1). All armA-harboring Salmonella strains were then subjected to additional PCR assays to assess the presence of aminoglycoside-resistant genes (ant(2”)-Ia, aph(3′)-Ia, aac(3)-Ia, aac(3)-IIa, aac(6′)-Ib, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, and npmA), extended spectrum β-lactamase (ESBL)/AmpC genes (blaDHA, blaTEM, blaCMY, blaOXA, blaCTX–M, and blaSHV), plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrC, qnrD, qnrS, oqxA, oqxB, and qepA), the colistin-resistant gene mcr-1, and the carbapenem-resistant gene blaNDM. Primers (Wu et al., 2009; Gopalakrishnan et al., 2018; Taylor et al., 2018) used in this study were showed in Supplementary Table 1. The variants of blaCTX–M were confirmed by Sanger sequencing. The PCR amplification products were also sequenced, and the resulting DNA sequences were analyzed by using the BLAST program1.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed on all the armA-positive isolates using the reference broth microdilution method with custom plates (PRCDCN2, Thermo) (Clinical and Laboratory Standards Institute [CLSI], 2015) for the following 20 antibiotics: ampicillin, tetracycline, erythromycin, chloramphenicol, cefazolin, ciprofloxacin, trimethoprim/sulfamethoxazole, ceftazidime, imipenem, nalidixic acid, cefoxitin, cefotaxime, gentamicin, azithromycin, ceftazidime, amikacin, tobramycin, cefepime, colistin, and tigecycline. The results were assessed with the CLSI (2015) breakpoints (Clinical and Laboratory Standards Institute [CLSI], 2015). The control strain is ATCC 25922.
Pulsed Field Gel Electrophoresis (PFGE) Analysis
All armA-harboring Salmonella strains were analyzed by pulsed field gel electrophoresis (PFGE) to determine their genetic homology. Bacterial DNA was digested with the restriction enzyme XbaI and then analyzed by electrophoresis for 19 h at 6 V/cm, with a pulse angle of 120°, a temperature of 14°C, and pulse times ranging from 2.16 to 63.8 s. Comparison of the resulting PFGE patterns was performed with Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) based on the Tenover’s criteria (Tenover et al., 1995). Isolates were allocated into genetic similarity clusters using a cut-off value of 80%.
Plasmid and Southern Blot Analyses
To detect the sizes of the armA-carrying plasmids, agarose gel plugs containing total cellular DNA were prepared and digested with S1 nuclease (TaKaRa, Dalian, China) as described previously (Barton et al., 1995). Digested plugs were subjected to PFGE using a CHEF-Mapper system (pulse times, 2.16–63.8 s; running time, 19 h; 6 V/cm). Gels were blotted onto nylon membranes (Millipore, United States). The membranes were hybridized with a digoxigenin-labeled probe directed against armA.
Based on the Southern hybridization results, the gel slices resulting from PFGE following digestion with S1 nuclease found to contain the armA fragment were excised. The DNA fragments from the gels were purified and used as templates for identifying the plasmid replicon type by PCR. A PBRT kit (Diatheva, Italy) was applied for plasmid molecular typing (Garcia-Fernandez et al., 2009).
Plasmid Elimination Test
For the SDS treatment method, 50 μl of armA-positive Salmonella suspension (OD = 0.5–0.7) was inoculated into 5 ml of LB medium with 10% SDS and then incubated in a thermostatic shaker at 37°C for 18–24 h; an SDS concentration one-half of that in which growth was first observed was chosen as the working concentration for the elimination test (Wangkheimayum et al., 2017). The products were diluted ten times with saline and then selected on agar plates containing 1000 μg/ml amikacin and on plates without antibiotics. The plasmid-eliminated strains were identified by real-time PCR.
Plasmid Conjugation
The conjugation test was performed with the broth mating method using all 45 aminoglycoside-resistant Salmonella strains as the donor strains and sodium azide-resistant Escherichia coli J53 or streptomycin-resistant Acinetobacter baumannii as the recipient strains. The transconjugants were selected on agar plates containing 30 μg/ml amikacin and 400 μg/ml sodium azide when using E. coli J53 as the recipient and on LB agar plates containing 30 μg/ml amikacin and 5,000 μg/ml streptomycin when using A. baumannii as the recipient. The resulting transconjugants were identified by real-time PCR targeting the armA gene. The transfer frequency is expressed as the number of transconjugants per total recipients.
To test the ability of the Salmonella strains from different serotypes to obtain an armA-carrying plasmid, we performed conjugation experiments using three S. Derby strains, five S. Enteritidis strains, four S. Typhimurium strains, four armA-negative S. Indiana strains, and three S. Indiana strains in which the armA-carrying plasmid had been eliminated as recipients and using three E. coli J53 strains with different armA-carrying plasmid lengths as donors. In total, 57 independent conjugative experiments were performed. The resulting transformants were selected on LB agar with amikacin (30 μg/ml) and streptomycin (4,000 μg/ml), and successful transconjugants were confirmed by real-time PCR. The transfer frequency is expressed as the number of transconjugants per total recipients. The transfer frequency data were analyzed with a multiway analysis of variance using SPSS 19 statistic software.
Results
armA-Harboring Salmonella Emerged in the Diarrheal Outpatients in Shanghai District Around 2011
A total of 45 armA-harboring strains were detected in the 15,782 tested Salmonella strains. Of these, 13 (positive rate: 0.11%) were from the 12,095 outpatient strains collected in Shanghai (human source), and 32 (1.00%) were from the 3,687 food/environmental isolates.
The first armA-harboring strain was found in 2011 (Supplementary Table 3). Except in 2014, two or three armA-harboring Salmonella strains were identified each year. After the initial appearance of armA-harboring Salmonella in 2011, the prevalence rate of these strains in community-acquired diarrheal cases in Shanghai was 0.83/1,000 (13/10,234). Of the armA-harboring food/environmental-source strains, 22 (positive rate: 0.69%) were from the 3,206 strains collected in Shanghai and 10 (2.07%) were from the 481 strains collected in Guangdong.
To detect other concomitant aminoglycoside-resistant genes in these 45 armA-harboring Salmonella strains, PCR and qPCR were performed. Of the known aminoglycoside-modifying enzymes genes, ant(2”)-Ia, aph(3’)-Ia, aac(3)-Ia, aac(3)-IIa, and aac(6’)-Ib were detected. The positive rates of these genes were 31.1% (14/45), 11.1% (5/45), 68.9% (31/45), 8.9% (4/45), and 86.7% (39/45), respectively. The positive rates of rmtB, rmtC, and rmtD were 15.6% (7/45), 2.2% (1/45), and 44.4% (20/45), respectively; in contrast, the rmtA, rmtE, rmtF, rmtG, rmtH, and npmA genes were not detected in any of the isolates.
The Detected armA-Harboring Salmonella Strains Show a Highly Skewed Serotype Distribution
Among the 45 armA-harboring Salmonella strains, nine different serotypes were identified. The serotype distribution was heavily skewed toward serovar Indiana, with 34 strains (75.6%) identified as S. Indiana. Other identified serotypes included Thompson (three strains), Kottbus (two strains), and six different serovars with one strain each (Agona, Singapore, Schwerin, Corvallis, Goldcoast, and Infantis).
The Detected armA-Harboring Salmonella Strains Were All Multi-Antibiotic Resistant, and Most Carried Extended Spectrum β-Lactamase (ESBL)/Plasmid-Mediated Quinolone Resistance (PMQR) Genes
The results reveal that all these strains were multidrug resistant (MDR), i.e., they exhibited high levels of resistance to more than three classes of antibiotics (Supplementary Table 3). Resistance to amikacin, erythromycin, tobramycin, and gentamicin was observed in all 45 armA-harboring Salmonella isolates. Additionally, 40 of the strains (including 13 from outpatients and 27 from food/environmental samples) were found to be fluoroquinolone-resistant, and 35 of these fluoroquinolone-resistant strains (including 10 from outpatients and 25 from food/environmental samples) were identified as ESBL producers. In contrast, all 45 armA-harboring Salmonella strains were susceptible to both imipenem and tigecycline. The ampicillin-resistant rates of the armA-harboring Salmonella strains isolated from poultry or outpatients were much higher than that of armA-harboring Salmonella strains isolated from swine (36/37 vs. 2/4).
Among the 45 armA-harboring Salmonella isolates, the prevalence of ESBL/AmpC genes blaTEM–1B, blaCMY, blaOXA, and blaSHV–12 were 46.7% (21/45), 2.2% (1/45), 68.9% (31/45), and 2.2% (1/45), respectively. The prevalence of blaCTX–M was 68.9% (31/45). Notably, the extent of resistance to third generation cephalosporin mediated by different blaCTX–M subtypes is distinct; we found 21 strains with blaCTX–M–65, 6 strains with blaCTX–M–55, 3 strains with blaCTX–M–3, 2 strains with blaCTX–M–27, and 1 strain with blaCTX–M–14. The blaDHA gene was not detected in these strains (Supplementary Table 3).
The prevalence of PMQR genes qnrA, qnrD, qnrS, oqxA, oqxB, and qepA was 2.2% (1/45), 11.1% (5/45), 15.6% (7/45), 48.9% (22/45), 48.9% (22/45), and 2.2% (1/45), respectively. The qnrB and qnrC genes were not detected. Additionally, neither the colistin-resistant mcr-1 gene nor the carbapenem-resistant blaNDM gene was found to be present in any of the 45 armA-harboring Salmonella strains.
The Detected armA-Harboring Salmonella Strains Exhibited Diverse Genomic Subtyping Patterns
All 45 armA-harboring Salmonella strains had different PFGE profiles, which generally had low similarity with one another, suggesting that these isolates are unlikely to have originated from a single clone of Salmonella. Only two PFGE clusters were found; one pattern group included strains SH13SF082, SH13SF466, and SH13SF540, which were isolated from food in 2013, and the other included strains SH15SF180 and SH15SF181, which were isolated from food in 2015. The strains in each cluster had indistinguishable PFGE patterns (Figure 1). The PFGE patterns of S. Indiana strains isolated from outpatients were more similar to those of the strains isolated from poultry (e.g., SH12G1005 and SH15SF559, SH13G1614 and SH15SF180/SH15SF181, SH12SF039 and SH16G1356) than to those of strains isolated from swine.
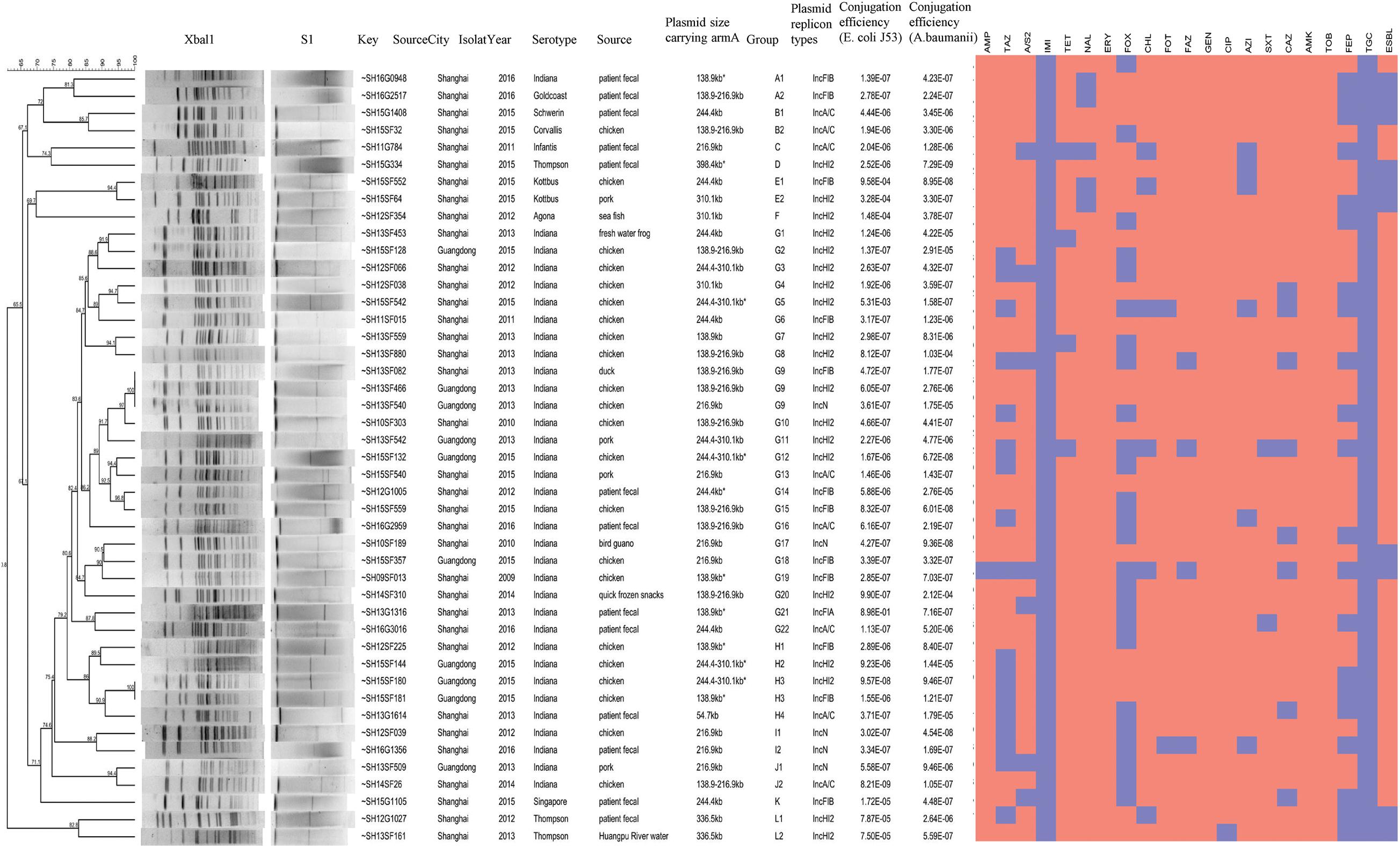
Figure 1. Generated dendrograms showing the cluster analysis of XbaI-digested PFGE patterns of the armA-harboring Salmonella strains isolated in this study.
The armA Genes in armA-Positive Salmonella Strains Were Located on Plasmids
Pulsed field gel electrophoresis using XbaI and S1 nuclease digestion followed by Southern-hybridization analysis revealed that all 45 isolates, regardless of their conjugative status, carried only a single armA-carrying plasmid. Based on the length calculations for the hybridized bands, all the armA-carrying plasmids were approximately 54.7–398.4 kb in size.
We performed plasmid elimination on these 45 armA-harboring Salmonella strains via treatment with SDS and found that all the armA-carrying plasmids could be successfully eliminated, i.e., the armA gene was not detected in the plasmid-eliminated strains, which supports the conclusion that the armA genes in these strains were located only on plasmids. To determine the replicon type(s) of the armA-carrying plasmids, replicon-typing PCR, which detects the replicon sequences of plasmid types, was performed. The results show that the armA-carrying plasmids included IncHI2 (19 strains, 42.2%), IncFIB (12 strains, 26.7%), IncA/C (8 strains, 17.8%), IncN (5 strains, 11.1%), and IncFIA (1 strain, 2.2%). IncHI2 was the dominant type of armA-carrying plasmid in these Salmonella strains.
The armA-Carrying Plasmids Were Transferable, and S. Indiana Strains Acquired the armA-Carrying Plasmids More Easily
Conjugation experiments revealed that all 45 armA-harboring Salmonella isolates were able to transfer their armA-carrying plasmids to both E. coli J53 and A. baumannii, respectively. The transfer frequencies were low, ∼10–6 CFU/donor for both E. coli J53 and A. baumannii (Supplementary Table 2 and Figure 2).
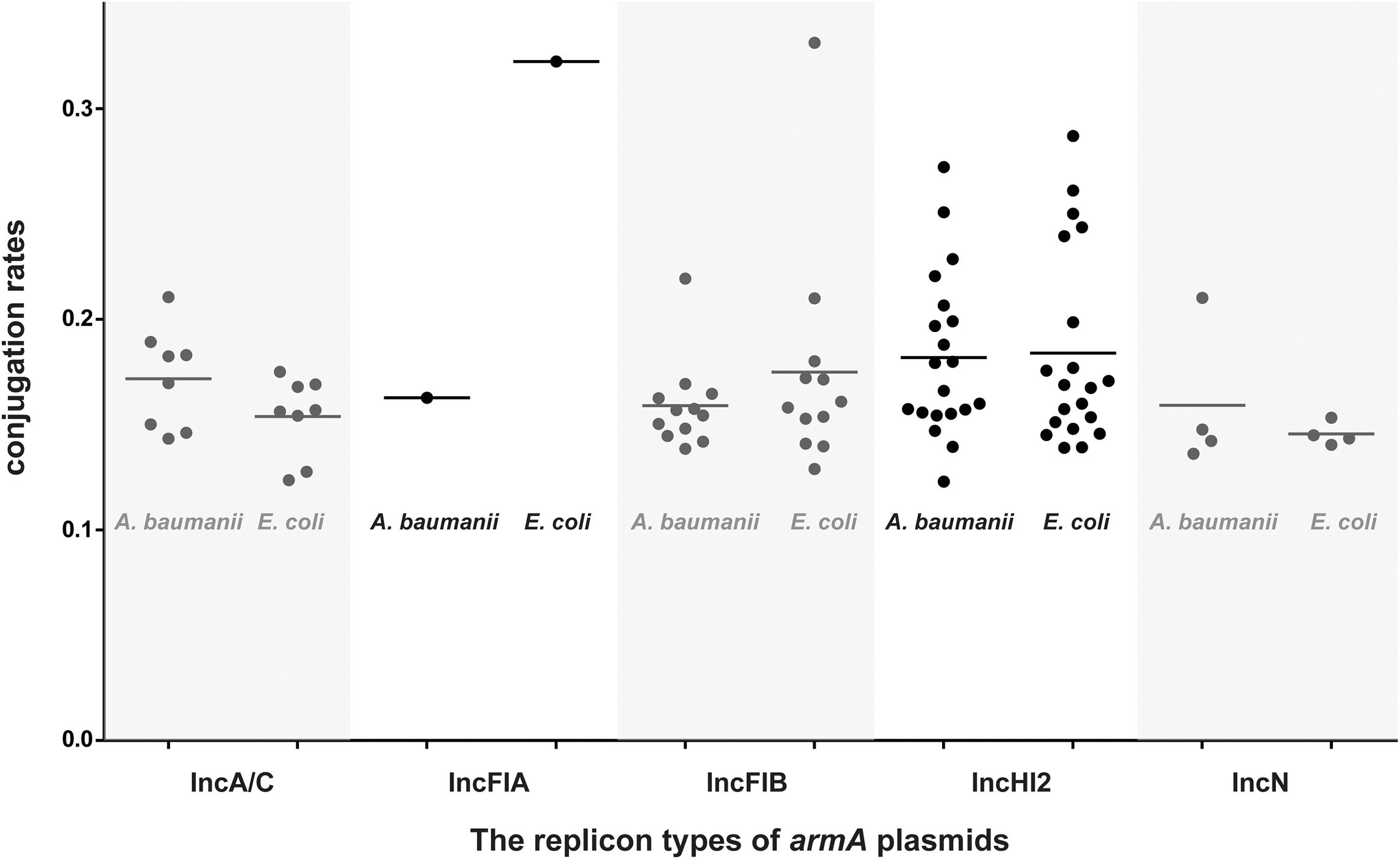
Figure 2. Conjugation rates of the armA-harboring plasmids isolated in this study. All 45 detected armA-harboring Salmonella strains were used as the donors, and either E. coli J53 or A. baumannii was used as the recipient. Black circles show the 1/abs (log10 transformation of the transfer frequency) data.
The transfer frequency data were analyzed by a multiway analysis of variance, and the factors used in this analysis were: five different recipients (S. Derby, S. Enteritidis, S. Typhimurium, armA-negative S. Indiana, and S. Indiana strains in which the armA-carrying plasmid was eliminated), and armA-carrying plasmids of three different lengths (∼140, ∼220, and ∼300 kb) as donors. There was no significant difference among the different serotypes in their ability to acquire armA-carrying plasmids. In contrast, the S. Indiana strains in which the armA-carrying plasmids were eliminated were able to re-acquire armA-carrying plasmids with higher frequencies compared with the other recipients (p < 0.01, multi-way ANOVA) (Figure 3).
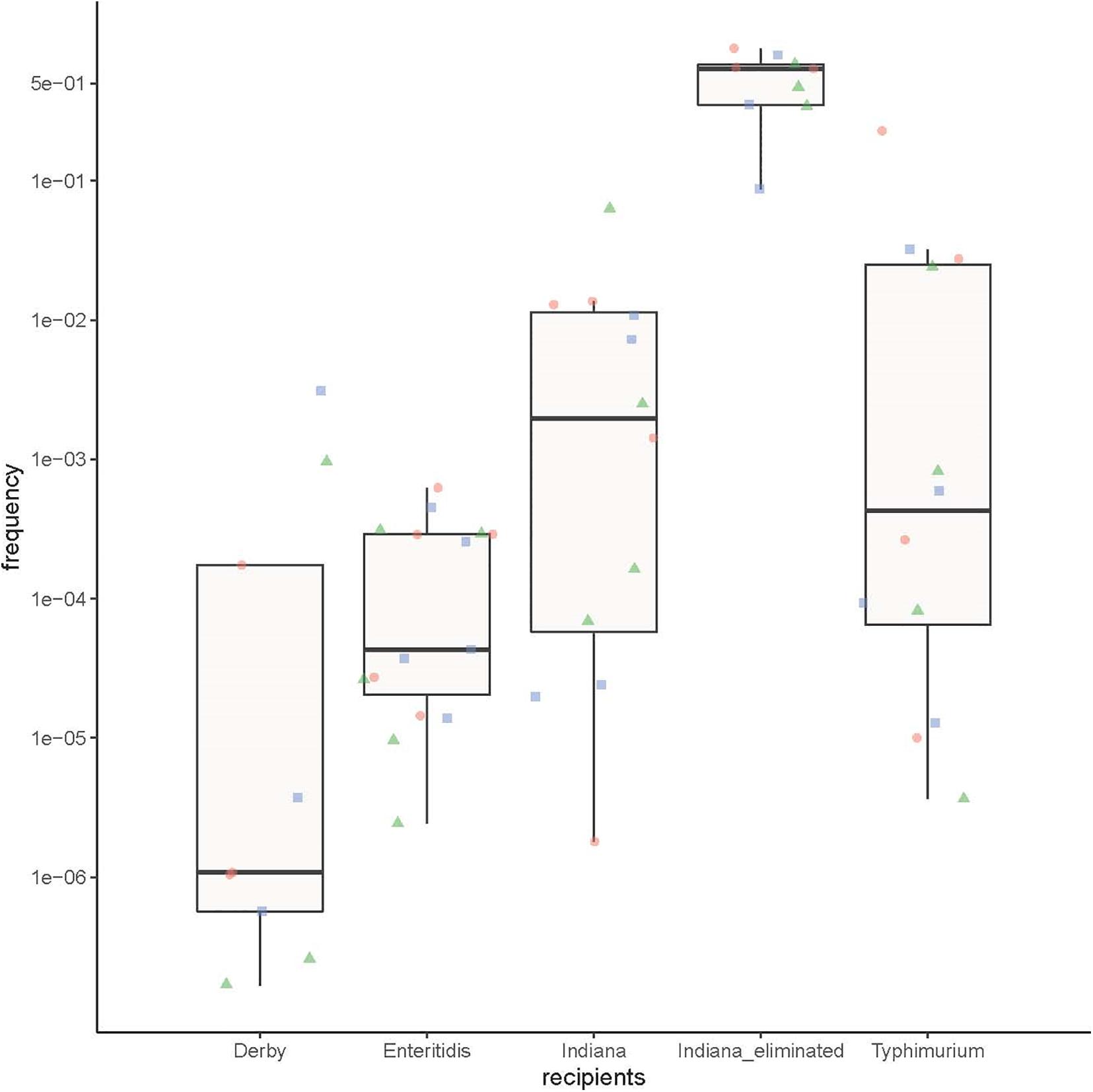
Figure 3. The ability of the different Salmonella serotype strains to obtain an armA-carrying plasmid. Three S. Derby strains, five S. Enteritidis strains, four S. Typhimurium strains, four naturally armA-negative S. Indiana strains, and three S. Indiana strains in which the armA-carrying plasmid was artificially eliminated were used as recipients, and three E. coli J53 strains with different lengths of armA-carrying plasmid (∼140 kb: green triangles, ∼220 kb: red circles, and ∼300 kb: blue squares) were used as donors.
Discussion
Aminoglycosides are the antibiotics used most commonly in clinical treatment and veterinary applications. Unfortunately, aminoglycoside use in economically important animals has impelled the emergence and spread of aminoglycoside-resistant bacteria, especially members of the Enterobacteriaceae family. Plasmids carrying 16S rRNA methylase genes, which are responsible for aminoglycoside resistance, have been widely recognized in Enterobacteriaceae family members; however, there are no reports of an epidemiology-based analysis of the armA gene prevalence in Salmonella spp. isolated from outpatients. Here, we examined samples from a systemic laboratory-based salmonellosis surveillance that was performed yearly from 2005 in diarrheal outpatients in Shanghai; the application of this study design should allow for a reasonable epidemiologic estimate of the prevalence of armA-carrying Salmonella strains.
The first isolation of an armA-harboring Salmonella strain occurred in 2011; fortuitously, no increasing trend for armA presence was found in the community-acquired infections in Shanghai. As with other clinical Enterobacteriaceae isolates (Samadi et al., 2015), the armA prevalence rate in clinical Salmonella isolates from the present work was found to remain low over the study period. However, the presence of other aminoglycoside-resistance genes was not assessed, and antimicrobial susceptibility testing was not performed on all the isolated strains from this study, only the strains with armA, so strains with other aminoglycoside resistance mechanisms may have remained undetected.
Salmonella strains isolated from food and food animal samples were also investigated in our study. These samples were obtained in Shanghai and Guangzhou, and the resulting isolates showed higher armA-positive rates compared with the outpatient isolates, suggesting that food and food animals may be a potential threat for the spread of armA-mediated aminoglycoside resistance to humans. Although the overall prevalence of strains harboring armA was low in this study, all 45 of the armA-harboring Salmonella isolates were MDR, and 35/45 strains were resistant to both cephalosporins and fluoroquinolone; notably, this indicates a high level of resistance by these strains to the antibiotics that are commonly used in the treatment of salmonellosis patients. The coexistence of the armA gene together with ESBL/PMQR genes in the Salmonella strains of animal origin may also present a potential risk to human health if the plasmids containing these genes are disseminated via contaminated food. Other studies have similarly found evidence for the coexistence of ESBL/PMQR genes with the armA gene in Salmonella strains from both food and human sources (Bouzidi et al., 2011; Fang et al., 2019). This situation may pose threats to both food safety and health management owing to two factors: (1) the active horizontal transfer of MDR genes among Salmonella and into other Enterobacteriaceae species, and (2) the preservation and possible spread of MDR genes through co-selection with other antibiotics that are commonly used for diarrhea treatment or for improved growth in food-producing animals. The complete sequences could provide more information about the amrA-bearing plasmids. Therefore, to provide more characteristic about the amrA-positive plasmid, we are looking forward to obtain the complete plasmid sequence base on long-reads sequencing in the future, with the cost of long-reads sequencing decline.
In the present work, we obtained Salmonella strains isolated from three different sources (humans, poultry, and swine). The S. Indiana strains isolated from outpatients had similar PFGE patterns compared with the strains isolated from poultry, suggesting that Salmonella with armA-mediated aminoglycoside resistance may be carried by poultry and transmitted from poultry products to humans. Antibiotics are used frequently and extensively in the poultry industry, which has led to the emergence of MDR Salmonella (Witte, 1998), and strains of MDR Salmonella originating from poultry have the potential to be spread to humans through the food chain (Afema et al., 2016). Poultry and poultry products are recognized as the major vehicles for Salmonella transmission to humans (Mezal et al., 2013). In this study, the ampicillin resistance rates of Salmonella strains isolated from poultry or humans were much higher than that of strains isolated from swine, which may be related to the excessive use of ampicillin antibiotics in poultry breeding and human medicine (Roth et al., 2019); this finding further supports the idea that the armA gene may be transmitted to humans via poultry products.
Pulsed field gel electrophoresis assessment of the 45 armA-harboring Salmonella strains obtained in this study revealed diverse genetic clones, suggesting that these strains are genetically unrelated and that clonal spread has not been the major mechanism of armA gene transmission in Salmonella. Thus, compared with the clonal spread of armA-harboring Salmonella, the transmission of armA-carrying plasmids may be more common. Additionally, our results indicate that the armA genes in all 45 Salmonella isolates were located on plasmids, and plasmids from these strains could be successfully transferred into E. coli and A. baumannii strains. The armA-harboring plasmids comprised five types; the major type, IncHI2, is an MDR plasmid that has been associated with a range of antibiotic resistance genes in Salmonella spp. and E. coli isolated from humans and food-producing animals (Zhong et al., 2017). The detection of diverse genetic clones and plasmids provides insight into the complicated and likely frequent transmission of armA-carrying plasmids among Salmonella strains and other Enterobacteriaceae species.
In the present study, 34 of the 45 armA-harboring Salmonella strains belong to serovar Indiana, raising the question of whether S. Indiana is more capable of obtaining these plasmids compared with Salmonella strains belonging to other serovars. We compared the plasmid acquisition abilities among S. Indiana and some other common Salmonella serotypes and found that the group of armA plasmid-carrying S. Indiana strains from the present study had a greater ability to obtain these plasmids. Interestingly, this level was even higher compared with the tested group of S. Indiana strains that lacked armA-carrying plasmids, suggesting that a specific genetic characteristic in these armA-harboring S. Indiana strains is required for the acquisition and maintenance of armA-carrying plasmids. In recent years, S. Indiana has been frequently isolated from chicken and broilers, as well as from poultry-farming workers, and it has gradually become one of the most common serotypes that cause animal and human salmonellosis (Xia et al., 2009; Lai et al., 2013; Gong et al., 2016). S. Indiana isolates that have concurrent resistance to cefotaxime, amikacin, and ciprofloxacin are widespread among chickens in China (Wang et al., 2017b). The MDR S. Indiana isolates in China belong to a wide variety of genotypes, suggesting that they probably evolved from many widely dispersed areas rather than from a few more local sources (Luque et al., 2009; Punia et al., 2010; Yang et al., 2010; Lai et al., 2014; Yan et al., 2014; Zhang et al., 2016). Other studies have demonstrated that S. Indiana isolates usually carry one or more drug resistance plasmids (Lai et al., 2013; Wang et al., 2016, 2017a), which further supports the important role of S. Indiana in public health.
Conclusion
We conducted a retrospective study using samples collected between 2005 and 2016 from diarrheal outpatients who participated in a continuous Salmonella infection surveillance in Shanghai Municipality, China. Our data reveal that, since 2011, the prevalence of Salmonella with armA-containing plasmids in community-acquired diarrhea cases was low and steady but that all the armA-harboring Salmonella strains were MDR and were notably resistant to fluoroquinolone and/or extended-spectrum cephalosporins.
The use of aminoglycoside antibiotics is contributing to the rise of MDR Salmonella, which poses an increasing public health threat and presents a considerable challenge for the treatment of clinical infections. Future surveillance is necessary to monitor the prevalence and transmission of armA-harboring Salmonella, especially S. Indiana, both in humans and animals and to better understand the potential threat to public health posed by these strains.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XL, MZ, and NZ wrote the manuscript. MW, BG, JL, HJ, and WX provided the technical assistance. ZL, HZa, HZo, and ZPL performed the data analysis and prepared the resources. JX, XX, and BK edited the manuscript. All authors read and approved the article.
Funding
This work was supported by the National Science and Technology Major Project (2018ZX10714002) and the Foundation for Young Scholars from China CDC (2018A103).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Katie Oakley, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.663210/full#supplementary-material
Supplementary Table 1 | Primers used in this study for polymerase chain reaction.
Supplementary Table 2 | The conjugation rates of the armA-harboring plasmids.
Supplementary Table 3 | Basic information about the 45 armA-harboring Salmonella strains identified in this study and their armA-carrying plasmids.
Footnotes
References
Afema, J. A., Byarugaba, D. K., Shah, D. H., Atukwase, E., Nambi, M., and Sischo, W. M. (2016). Potential sources and transmission of Salmonella and antimicrobial resistance in Kampala, Uganda. PLoS One 11:e0152130. doi: 10.1371/journal.pone.0152130
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Bouzidi, N., Aoun, L., Dekhil, M., Granier, S. A., Poirel, L., Brisabois, A., et al. (2011). Co-occurrence of aminoglycoside resistance gene armA in non-Typhi Salmonella isolates producing CTX-M-15 in Algeria. J. Antimicrob. Chemother. 66, 2180–2181. doi: 10.1093/jac/dkr237
Bueno, M. F., Francisco, G. R., O’Hara, J. A., de Oliveira Garcia, D., and Doi, Y. (2013). Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum beta-lactamases in Klebsiella pneumoniae. J. Antimicrob. Chemother. 57, 2397–2400. doi: 10.1128/aac.02108-12
Chen, L., Chen, Z. L., Liu, J. H., Zeng, Z. L., Ma, J. Y., and Jiang, H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. doi: 10.1093/jac/dkm065
Clinical and Laboratory Standards Institute [CLSI] (2015). M07-A10. Methods for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically, 10th Edn, Wayne, PA: CLSI.
Crump, J. A., Sjolund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/cmr.00002-15
Doi, Y., Yokoyama, K., Yamane, K., Wachino, J., Shibata, N., Yagi, T., et al. (2004). Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48, 491–496. doi: 10.1128/aac.48.2.491-496.2004
Du, X. D., Li, D. X., Hu, G. Z., Wang, Y., Shang, Y. H., Wu, C. M., et al. (2012). Tn1548-associated armA is co-located with qnrB2, aac(6’)-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. J Antimicrob Chemother 67, 246–248. doi: 10.1093/jac/dkr407
Fang, L. X., Deng, G. H., Jiang, Q., Cen, D. J., Yang, R. S., Feng, Y. Y., et al. (2019). Clonal expansion and horizontal transmission of epidemic F2:A1:B1 plasmids involved in co-spread of rmtB with qepA and blaCTX-M-27 in extensively drug-resistant Salmonella enterica serovar Indiana isolates. J. Antimicrob. Chemother. 74, 334–341. doi: 10.1093/jac/dky441
Folster, J. P., Rickert, R., Barzilay, E. J., and Whichard, J. M. (2009). Identification of the aminoglycoside resistance determinants armA and rmtC among non-Typhi Salmonella isolates from humans in the United States. Antimicrob. Agents Chemother. 53, 4563–4564. doi: 10.1128/aac.00656-09
Galimand, M., Courvalin, P., and Lambert, T. (2003). Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47, 2565–2571. doi: 10.1128/aac.47.8.2565-2571.2003
Galimand, M., Courvalin, P., and Lambert, T. (2012). RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob. Agents Chemother. 56, 3960–3962. doi: 10.1128/aac.00660-12
Garcia-Fernandez, A., Fortini, D., Veldman, K., Mevius, D., and Carattoli, A. (2009). Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63, 274–281. doi: 10.1093/jac/dkn470
Gong, J., Kelly, P., and Wang, C. (2016). Prevalence and antimicrobial resistance of Salmonella enterica Serovar Indiana in China (1984-2016). Zoonoses Public Health 64, 239–251.
Gopalakrishnan, S., Kamalanathan, A., Rajan, S., Bhagat, V. M., and Ali, M. K. S. (2018). Emergence of armA and rmtB genes among VIM, NDM, and IMP metallo-β-lactamase-producing multidrug-resistant Gram-negative pathogens. Acta Microbiol. Immunol. Hung. 65, 107–118. doi: 10.1556/030.64.2017.027
Hopkins, K. L., Escudero, J. A., Hidalgo, L., and Gonzalez-Zorn, B. (2010). 16S rRNA methyltransferase RmtC in Salmonella enterica serovar Virchow. Emerg. Infect. Dis. 16, 712–715. doi: 10.3201/eid1604.090736
Kariuki, S., Gordon, M. A., Feasey, N., and Parry, C. M. (2015). Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 33(Suppl. 3), C21–C29.
Lai, J., Wang, Y., Shen, J., Li, R., Han, J., Foley, S. L., et al. (2013). Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from chicken in China. Foodborne Pathog. Dis. 10, 581–588. doi: 10.1089/fpd.2012.1455
Lai, J., Wu, C., Wu, C., Qi, J., Wang, Y., Wang, H., et al. (2014). Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Intern. J. Food Microbiol. 180, 30–38. doi: 10.1016/j.ijfoodmicro.2014.03.030
LaRock, D. L., Chaudhary, A., and Miller, S. I. (2015). Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13, 191–205. doi: 10.1038/nrmicro3420
Luque, I., Echeita, A., León, J., Herrera-León, S., Tarradas, C., González-Sanz, R., et al. (2009). Salmonella Indiana as a cause of abortion in ewes: genetic diversity and resistance patterns. Vet. Microbiol. 134, 396–399. doi: 10.1016/j.vetmic.2008.08.015
Ma, L., Lin, C. J., Chen, J. H., Fung, C. P., Chang, F. Y., Lai, Y. K., et al. (2009). Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 53, 104–111. doi: 10.1128/aac.00852-08
Mezal, E. H., Rossina, S., and Khan, A. A. (2013). Isolation and molecular characterization of Salmonella enterica serovar Javiana from food, environmental and clinical samples. Intern. J. Food Microbiol. 164, 113–118. doi: 10.1016/j.ijfoodmicro.2013.03.021
Michael, G. B., and Schwarz, S. (2016). Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin. Microbiol. Infect. 22, 968–974. doi: 10.1016/j.cmi.2016.07.033
Naas, T., Bentchouala, C., Cuzon, G., Yaou, S., Lezzar, A., Smati, F., et al. (2011). Outbreak of Salmonella enterica serotype infantis producing ArmA 16S RNA methylase and CTX-M-15 extended-spectrum beta-lactamase in a neonatology ward in Constantine, Algeria. Int. J. Antimicrob. Agents 38, 135–139. doi: 10.1016/j.ijantimicag.2011.04.012
O’Hara, J. A., McGann, P., Snesrud, E. C., Clifford, R. J., Waterman, P. E., Lesho, E. P., et al. (2013). Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob. Agents Chemother. 57, 2413–2416. doi: 10.1128/aac.00266-13
Punia, P., Hampton, M. D., Ridley, A. M., Ward, L. R., Rowe, B., and Threlfall, E. J. (2010). Pulsed-field electrophoretic fingerprinting of Salmonella indiana and its epidemiological applicability. J. Appl. Microbiol. 84, 103–107. doi: 10.1046/j.1365-2672.1997.00325.x
Qin, S., Wang, Y., Zhang, Q., Chen, X., Shen, Z., Deng, F., et al. (2012). Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 56, 5332–5339. doi: 10.1128/aac.00809-12
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171.
Roth, N., Kasbohrer, A., Mayrhofer, S., Zitz, U., Hofacre, C., and Domig, K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 98, 1791–1804. doi: 10.3382/ps/pey539
Samadi, N., Pakzad, I., Monadi Sefidan, A., Hosainzadegan, H., and Tanomand, A. (2015). Study of aminoglycoside resistance genes in Enterococcus and Salmonella strains isolated from ilam and milad hospitals, iran. Jundishapur J. Microbiol. 8:e18102.
Taylor, E., Sriskandan, S., Woodford, N., and Hopkins, K. L. (2018). High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int. J. Antimicrob. Agents 52, 278–282. doi: 10.1016/j.ijantimicag.2018.03.016
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Uechi, K., Tada, T., Shimada, K., Nakasone, I., Sonozaki, T., Kirikae, T., et al. (2018). Emergence of ArmA, a 16S rRNA methylase in highly aminoglycoside-resistant clinical isolates of Klebsiella pneumoniae and Klebsiella oxytoca in Okinawa, Japan. J. Infect. Chemother. 24, 68–70. doi: 10.1016/j.jiac.2017.09.006
Wachino, J., and Arakawa, Y. (2012). Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist. Updat. 15, 133–148. doi: 10.1016/j.drup.2012.05.001
Wachino, J., Shibayama, K., Kurokawa, H., Kimura, K., Yamane, K., Suzuki, S., et al. (2007). Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob. Agents Chemother. 51, 4401–4409. doi: 10.1128/aac.00926-07
Wang, J., Li, X., Li, J., Hurley, D., Bai, X., Yu, Z., et al. (2017a). Complete genetic analysis of a Salmonella enterica serovar Indiana isolate accompanying four plasmids carrying mcr-1, ESBL and other resistance genes in China. Vet. Microbiol. 210, 142–146. doi: 10.1016/j.vetmic.2017.08.024
Wang, J., Ma, Z. B., Zeng, Z. L., Yang, X. W., Huang, Y., and Liu, J. H. (2017b). The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 38, 55–80.
Wang, Y., Zhang, A., Yang, Y., Lei, C., Jiang, W., Liu, B., et al. (2017c). Emergence of Salmonella enterica serovar Indiana and California isolates with concurrent resistance to cefotaxime, amikacin and ciprofloxacin from chickens in China. Int. J. Food Microbiol. 262, 23–30. doi: 10.1016/j.ijfoodmicro.2017.09.012
Wang, W., Liu, F., Peng, Z., Li, F., and Ma, A. (2016). Complete genome sequence of Salmonella enterica subsp. enterica Serovar Indiana C629, a carbapenem-resistant bacterium isolated from chicken carcass in China. Genome Announc. 4:e0662-16.
Wangkheimayum, J., Paul, D., Dhar, D., Nepram, R., Chetri, S., Bhowmik, D., et al. (2017). Occurrence of acquired 16S rRNA Methyltransferase-mediated aminoglycoside resistance in clinical isolates of Enterobacteriaceae within a tertiary referral hospital of Northeast India. Antimicrob. Agents Chemother. 61:e01037-16.
Wen, S. C., Best, E., and Nourse, C. (2017). Non-typhoidal Salmonella infections in children: review of literature and recommendations for management. J. Paediatr. Child Health 53, 936–941. doi: 10.1111/jpc.13585
Wu, Q., Zhang, Y., Han, L., Sun, J., and Ni, Y. (2009). Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob. Agents Chemother. 53, 271–272.
Xia, J., Sun, J., Cheng, K., Li, L., Fang, L. X., Zou, M. T., et al. (2016). Persistent spread of the rmtB 16S rRNA methyltransferase gene among Escherichia coli isolates from diseased food-producing animals in China. Vet. Microbiol. 188, 41–46.
Xia, S., Hendriksen, R. S., Xie, Z., Huang, L., Zhang, J., Guo, W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47, 401–409.
Yan, L., Hongyu, Z., Jian, S., Yuqi, L., Xuping, Z., Beier, R. C., et al. (2014). Characterization of multidrug-resistant Salmonella enterica serovars Indiana and enteritidis from chickens in Eastern China. PLoS One 9:e96050. doi: 10.1371/journal.pone.0096050
Yang, B., Qu, D., Zhang, X., Shen, J., Cui, S., Shi, Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Intern. J. Food Microbiol. 141, 63–72.
Zhang, W. H., Lin, X. Y., Xu, L., Gu, X. X., Yang, L., Li, W., et al. (2016). CTX-M-27 Producing Salmonella enterica Serotypes typhimurium and Indiana are prevalent among food-producing animals in China. Front. Microbiol. 7:436. doi: 10.3389/fmicb.2016.00436
Zhong, L. L., Phan, H. T. T., Shen, C., Doris-Vihta, K., Sheppard, A. E., Huang, X., et al. (2017). High rates of human fecal carriage of mcr-1-positive multi-drug resistant Enterobacteriaceae isolates emerge in China in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Keywords: Salmonella, armA, multi-drug resistant, poultry, swine
Citation: Lu X, Zeng M, Zhang N, Wang M, Gu B, Li J, Jin H, Xiao W, Li Z, Zhao H, Zhou H, Li Z, Xu J, Xu X and Kan B (2021) Prevalence of 16S rRNA Methylation Enzyme Gene armA in Salmonella From Outpatients and Food. Front. Microbiol. 12:663210. doi: 10.3389/fmicb.2021.663210
Received: 02 February 2021; Accepted: 06 April 2021;
Published: 25 May 2021.
Edited by:
Aloysius Wong, Kean University-Wenzhou, ChinaReviewed by:
Jing Han, National Center for Toxicological Research (FDA), United StatesRuichao Li, Yangzhou University, China
Copyright © 2021 Lu, Zeng, Zhang, Wang, Gu, Li, Jin, Xiao, Li, Zhao, Zhou, Li, Xu, Xu and Kan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biao Kan, a2FuYmlhb0BpY2RjLmNu; Xuebin Xu, eHhiNzJAc2luYS5jb20=; Jialiang Xu, eHVqaWFsaWFuZ0BidGJ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xin Lu
Xin Lu Mei Zeng
Mei Zeng Ning Zhang1,2,4†
Ning Zhang1,2,4† Mengyu Wang
Mengyu Wang Zhenpeng Li
Zhenpeng Li Jialiang Xu
Jialiang Xu Xuebin Xu
Xuebin Xu Biao Kan
Biao Kan