
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 May 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.660871
This article is part of the Research Topic Mycotoxins: From Prediction to Management and Control View all 6 articles
 Silvia Valente1,2
Silvia Valente1,2 Edoardo Piombo1,2
Edoardo Piombo1,2 Volker Schroeckh3
Volker Schroeckh3 Giovanna Roberta Meloni1,2
Giovanna Roberta Meloni1,2 Thorsten Heinekamp3
Thorsten Heinekamp3 Axel A. Brakhage3,4*
Axel A. Brakhage3,4* Davide Spadaro1,2*
Davide Spadaro1,2*Penicillium polonicum, commonly found on food matrices, is a mycotoxigenic species able to produce a neurotoxin called verrucosidin. This methylated α-pyrone polyketide inhibits oxidative phosphorylation in mitochondria and thereby causes neurological diseases. Despite the importance of verrucosidin as a toxin, its biosynthetic genes have not been characterized yet. By similarity analysis with the polyketide synthase (PKS) genes for the α-pyrones aurovertin (AurA) and citreoviridin (CtvA), 16 PKS genes for putative α-pyrones were identified in the P. polonicum genome. A single PKS gene, verA, was found to be transcribed under verrucosidin-producing growth conditions. The annotated functions of the genes neighboring verA correspond to those required for verrucosidin biosynthesis. To prove the involvement of verA in verrucosidin biosynthesis, the clustered regularly interspaced short palindrome repeats (CRISPR) technology was applied to P. polonicum. In vitro reconstituted CRISPR-Cas9 was used to induce targeted gene deletions in P. polonicum. This approach allowed identifying and characterizing the verrucosidin biosynthetic gene cluster. VerA deletion mutants were no longer able to produce verrucosidin, whereas they were displaying morphological characteristics comparable with the wild-type strain. The available CRISPR-Cas9 technology allows characterizing the biosynthetic potential of P. polonicum as a valuable source of novel compounds.
Penicillium polonicum is a ubiquitous fungus, found as a contaminant of food matrices, such as meat (Sunesen and Stahnke, 2003; Wigmann et al., 2018), nuts (Prencipe et al., 2018), and fresh fruit, like grape berries and strawberries (Jensen et al., 2013; Santini et al., 2014). It is also reported as a postharvest pathogen on apple (Ouhibi et al., 2018), pear (Scholtz and Korsten, 2016), onion (Duduk et al., 2014), chestnut (Prencipe et al., 2018), and cactus pear (Faedda et al., 2015). The fungus can grow as saprophyte in diverse environments (Sonjak et al., 2006). P. polonicum is able to produce a variety of secondary metabolites (SMs), including mycotoxins, which exhibit toxic effects on animals and humans. Characteristic compounds produced by P. polonicum are penicillic acid, aspterric acid, verrucofortine, cyclopenins, nephrotoxic glycopeptides, and verrucosidin (Frisvad et al., 2004), a neurotoxic compound initially isolated from Penicillium verrucosum var. cyclopium, and later from other Penicillium species, such as Penicillium expansum (Burka et al., 1983; Frisvad et al., 2004; Kim et al., 2016).
Penicillium polonicum strain X6 was previously identified, by adopting a polyphasic approach based on molecular, morphological, and chemical characterization (Prencipe et al., 2018). A multilocus phylogenetic analysis was performed using internal transcribed spacer (ITS) region, calmodulin, and β-tubulin partial genes. Morphological analysis was conducted on three different media: malt extract agar (MEA), czapek yeast agar (CYA), and yeast extract sucrose (YES). Finally, the SMs produced in vivo were analyzed, confirming the ability of this strain to produce verrucosidin.
From the chemical point of view, verrucosidin is a highly reduced polyketide, composed of a methylated α-pyrone, a polyene linker, and an epoxidated tetrahydrofuran ring. Many polyketides share the α-pyrone structure, such as citreoviridin and aurovertins, and have been extensively studied because of their ability to inhibit mitochondrial oxidative phosphorylation, resulting in cytotoxicity and potential antitumor activity (Li et al., 2018). Verrucosidin acts on the central nervous system, causing neurological diseases, first diagnosed in cattle and experimentally confirmed in mice (Hodge et al., 1988; Fink-Gremmels et al., 1991). Verrucosidin is the most cytotoxic and genotoxic compound among tremorgenic mycotoxins (Núñez et al., 2000; Sabater-Vilar et al., 2003). Based on its cytotoxicity, verrucosidin has been evaluated for its effect against cancer cells (Park et al., 2007; Thomas et al., 2013), making this compound interesting for medical purposes. Despite the importance of this metabolite for human health and its potential benefit in medicine, only few studies have been conducted to elucidate its biosynthesis. Núñez et al. (2000) determined favorable growth conditions of P. polonicum for verrucosidin production, whereas Aranda et al. (2002) screened verrucosidin-producer strains to develop a molecular probe to identify mycotoxigenic fungi in foodstuff, which was further used by Rodríguez et al. (2012) to design a TaqMan probe. Nowadays, the elucidated biosyntheses of the structurally related α-pyrone polyenes citreoviridin, aurovertins, and aspernidgulenes (Lin et al., 2016, 2019; Li et al., 2018), available fungal genome sequences (Weber and Kim, 2016; Nielsen and Nielsen, 2017), and bioinformatic tools open up the possibility to identify the biosynthetic genes responsible for verrucosidin biosynthesis.
Biosynthetic gene clusters (BGCs) (Brakhage, 2013; Keller, 2019) consist of central biosynthetic genes, often PKS or/and non-ribosomal peptide synthetase (NRPSs) genes, and are associated with additional genes encoding so-called tailoring enzymes, which are required for the modification of the carbon structure or encode transporters, transcription factors, and resistance proteins. Thousands of gene clusters have been described in fungal genomes, a number that is higher compared to the chemically identified compounds (Yu et al., 2015). Most of these uncharacterized BGCs are often “silent,” meaning their genes are not transcribed under laboratory conditions and a number of strategies exist for their activation (Bergmann et al., 2007, 2010; Fischer et al., 2016). On the other hand, gene deletion on active BGCs can be used to characterize the function of the tailoring genes and to assign a BGC to a compound (Valente et al., 2020).
The clustered regularly interspaced short palindrome repeats (CRISPR) technology is by now the method of choice in genome studies of filamentous fungi (Nødvig et al., 2015; Fang and Tyler, 2016; Pohl et al., 2016; Deng et al., 2017; Krappmann, 2017; Nagy et al., 2017; Nielsen et al., 2017; Weber et al., 2017; Song et al., 2019; Tong et al., 2019; Wang and Coleman, 2019). It allows marker-free genome editing by generating mutations at specific sites in the genome. CRISPR is based on the endonuclease Cas9, that, when associated with small sequences of crRNA (CRISPR-RNA) and tracrRNA (Trans-activating crRNA), usually referred to as guide RNA (gRNA), recognizes genomic protospacer sequences and site-specifically cuts the double strand of DNA (Tong et al., 2019).
In the present manuscript, we described the bioinformatic identification of a number of α-pyrone polyketide synthases (PKSs) in P. polonicum. One BGC was selected based on the putative function of the biosynthetic genes of the cluster, and the verrucosidin-encoding PKS was confirmed to be expressed in verrucosidin-producing Penicillium strains. Besides, we established the CRISPR-Cas9 technology in P. polonicum X6, which helped us characterize the verrucosidin BGC in P. polonicum.
Penicillium polonicum strain X6 and P. crustosum strain CAL64 were isolated from chestnut production chain (Prencipe et al., 2018). Penicillium aurantiogriseum CBS 112021 was obtained from the Westerdijk Fungal Biodiversity Institute and used as a positive control of verrucosidin production. The strains were grown on Potato Dextrose Agar plates (PDA, Merck KGaA, Darmstadt, Germany) with 50 μg/ml streptomycin (Merck KGaA) in the dark at 25°C for 7–10 days. P. polonicum mutants obtained were grown on PDA supplemented with 100 μg/ml of hygromycin B (Thermo Fischer Scientific, Waltham, MA, United States) under the same conditions. All the strains were maintained in glycerol stock at –80°C.
Conidial suspensions were obtained by adding 5 ml of sterile water with 0.01% (v/v) Tween-20 and gently scraping the surface of fungal cultures grown on Petri dishes, as in Spadaro et al. (2013). The final conidia concentration was measured using a hemocytometer and adjusted by dilution to different concentrations depending on each assay.
To evaluate verrucosidin production in vitro, 1 ml of conidial suspension (108 conidia/ml) was inoculated in 30 ml of malt extract broth (MEB, w/v: 2% malt extract, 2% glucose, 0.1% peptone) and in 30 ml of Czapek yeast broth (CYB, w/v: 0.5% yeast extract, 3.5% Czapek broth). Flasks were kept at 26°C for 10 days, 55% RH with 12 h of light and 12 h of dark. After 10 days of incubation, the fungal mycelium was separated from the liquid media through a double layer of sterile gauze and half of the mycelium was immediately frozen in liquid nitrogen, stored at –80°C, and used for RNA extraction. The remaining fungal tissue was used for verrucosidin extraction.
To identify BGCs in the genomes of P. polonicum strain IBT4502 (GCA_002072265.1) and P. polonicum strain hy4 (GCA_003344595.1), antiSMASH (Weber et al., 2015) was used and only clusters containing a putative PKS similar to both CtvA protein (Q0C9L7.1) and AurA (A0A0M4L8I7.1) were further considered (query coverage ≥50% and e-value > e–5). All genes found to be highly similar to the putative PKS genes were considered as part of the gene cluster whereas proteins with unknown function encoded by genes found far away from the core gene were omitted. The genes showing 70% query coverage and identity with differentially expressed genes (DEGs) of Kim et al. (2016) were also reported. The proteins in these clusters were additionally blasted against P. expansum (ALJY00000000.1) to verify their presence. BLAST using Non-Redundant database (Madden, 2013) and Interproscan (Quevillon et al., 2005) was used to find functional domains and predict a putative function.
Fungal DNA was extracted from mycelium using E.Z.N.A. ®Fungal DNA Mini Kit (Omega Bio-tek, Norcross, GA, United States) with brief modifications. The mycelium was lysed by adding the required amount of lysis buffer and two tungsten beads in a TissueLyser II (Qiagen, Hilden, Germany) at 20.00 Hz speed for 20 min. DNA concentration and purity were checked by a spectrophotometer (Nanodrop 2000, Thermo Scientific, Wilmington, DE, United States).
To amplify the genomic DNA (gDNA) of Penicillium spp., Taq DNA Polymerase (Qiagen) was used with PCR mixture containing 1 × PCR buffer, 0.2 mM dNTPs, 0.4 μM of each primer, 0.5 U of polymerase, and 10 ng of gDNA. PCR parameters to amplify fragments between 100 and 400 bp were as follows: 3 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 58°C and 30 s at 72°C; 5 min at 72°C. PCR parameters to amplify fragments between 2 and 3 kbp were as follows: 5 min at 95°C; 35 cycles of 30 s at 95°C, 45 s at 65°C and 3 min at 72°C; 5 min at 72°C.
To amplify DNA repair template from plasmid pUChph1 (Liebmann et al., 2004), the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fischer Scientific) was used according to the manufacturer’s instructions. PCR parameters were as follows: 10 s at 98°C; 30 cycles of 10 s at 98°C, 30 s at 60°C and 20 s at 72°C; 1 min at 72°C. The amplified PCR product was loaded onto an agarose gel, cut out, and extracted using Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, United States), according to the manufacturer’s instruction. All primer sequences used in the PCR are listed in Supplementary Table 1.
RNA was extracted from 100 mg of fungal mycelium using Spectrum Plant Total RNA (Sigma-Aldrich, St. Louis, MO, United States), following the manufacturer’s instructions. Mycelium was placed in a 2-ml tube with two tungsten beads and tubes were immersed in liquid nitrogen for 1 min. Then, samples were immediately lysed using TissueLyser II at 20.00 Hz for 1 min.
DNase treatment and first-strand cDNA synthesis were performed according to Valente et al. (2020) using a TURBO DNA-freeTM Kit (Thermo Fischer Scientific) and a High Capacity cDNA Reverse Transcription Kit (Thermo Fischer Scientific).
RT-qPCR was performed with StepOneTM and StepOnePlusTM Real-Time PCR System with Power SYBRTM Green PCR Master Mix (Thermo Fischer Scientific); cycling conditions were 5 min at 95°C, followed by 45 cycles of 10 s at 95°C, 30 s at 58°C, and 30 s at 72°C. In order to determine relative gene expression (RGE), the 2ΔΔcq method (Pfaffl, 2001) was used with cDNA of samples, by comparing the amplification of β-tubulin gene with the amplification of the target gene. P. aurantiogriseum was used as reference strain for verrucosidin production, as it is reported to be a verrucosidin producer (Fink-Gremmels et al., 1991). All primer sequences used in RT-qPCR reactions are listed in Supplementary Table 1.
Verrucosidin produced in vitro was extracted from approximately 0.5 g of fungal mycelium. The mycelium was placed in 2-ml tubes, and 1.5 ml of MeOH:chloroform (1:2, v/v) was added to the samples, which were then subjected to ultrasound for 30 min. After centrifugation at 4452 × g for 5 min, the liquid phase was transferred into a new tube. The mycelium pellet was subjected again to two further extractions: the first with ethyl acetate and the second with isopropanol. The extracts were combined and concentrated at 45°C (Eppendorf concentrator 5301, Hamburg, Germany). The dry extracts were then resuspended in 500 μl of H2O:acetonitrile (1:1, v/v) and transferred into a HPLC vial for HPLC-MS/MS analysis.
Verrucosidin produced on apples was extracted from 5 g of decayed tissue around the inoculation site, samples were homogenized with 5 ml of water, and the protocol described in Valente et al. (2020) was adopted to extract verrucosidin from clear juice.
The HPLC-MS/MS system consisted of a binary pump and a vacuum degasser (1260 Agilent Technologies, Santa Clara, CA, United States) connected to a Varian auto-sampler Model 410 Prostar (Varian, Palo Alto, CA, United States), equipped with a 20-μl loop and coupled with a Varian 310-MS TQ Mass Spectrometer.
To characterize the metabolic profile of wild-type and knockout strains, 10 μl of each extract was analyzed using a C18 analytical column (Luna 3 μm, 150 × 2 mm, 100 Å, Phenomenex, Torrance, CA, United States) and the chromatographic separation was achieved by gradient conditions for 45 min at a flow rate of 300 μl/min. Solvent A was water and solvent B was acetonitrile both containing 0.1% formic acid. The gradient was programmed as follows: 0–3 min isocratic 5% B, followed by a linear gradient to 100% B, ending at 40 min and from 40 to 45 min isocratic 100% B. Full-scan mass spectra were acquired in the positive-ion mode over the m/z range from 100 to 700 using the TQ mass analyzer.
To perform the verrucosidin qualitative analysis, the same HPLC-MS/MS system was used. HPLC was equipped with a Pursuit XRs ULTRA 2.8 μm C18 (100 × 2 mm, Varian) column and a binary mixture as a mobile phase: solvents A and B were composed of 40 of 0.05% formic acid and 60% of acetonitrile, respectively. The isocratic mode was used at a flow rate of 0.2 L/min for 5 min. A mass spectrometer was equipped with an electrospray ionization (ESI) source operating in positive ion mode, whereas Product Ion Scan (PS) mode was used for triple-quadrupole: m/z 417→100–427. The collision gas (Ar) pressure was set at 2 mbar.
Protocols used to obtain protoplasts from Aspergillus fumigatus (Weidner et al., 1998), Ophiostoma piceae (Wang et al., 1999), Penicillium nalgiovense (Fierro et al., 2004), Penicillium paxilli (Young et al., 1998; McMillan et al., 2003), Penicillium chrysogenum (Pohl et al., 2016), Penicillium crustosum, and Penicillium janthinellium (Nicholson et al., 2015) were used as the basis to design a protocol for protoplast preparation of P. polonicum. Fungal mycelium was obtained by inoculating 2 ml of conidial suspension (1.25 × 108 conidia/ml) in 50 ml of YGG (8 g/L KCl, 16 g/L glucose, 6.6 g/L yeast nitrogen base, 1.5 g/L citric acid, 6 g/L KH2PO4, and 2 g/L yeast extract). Flasks were shaken on a rotary shaker (180 rpm) at 26°C for 20 h. Fungal mycelium was filtered through Miracloth (Merck KGaA) and washed with MgSO4 0.6 M. A pre-treatment was conducted by mixing 5 g of mycelium in 50-ml tubes with 5 mM Na2EDTA and 25 mM 2-mercaptoethanol; tubes were kept in horizontal position in a rotary shaker (80 rpm) at 30°C for 20 min. Fungal mycelium was washed with 0.6 M MgSO4 and then digested with 2 g of Vinotaste®Pro (Lamothe-Abiet, Canéjan, France) and 0.1 g of Lysing Enzymes from Trichoderma harzianum (Sigma-Aldrich) suspended in 15 ml of Osmo Solution (1.2 M MgSO4, 10 mM sodium phosphate buffer). The lytic solution was kept at 30°C, 80 rpm. Every 30 min, the mycelium-lysis solution was mixed using a serological pipette and visually checked under a microscope; about 3–4 h were necessary to digest the cell wall and obtain protoplasts. The suspension was filtered through Miracloth in order to separate and remove undigested mycelia. Protoplasts were then recovered using Trapping Buffer (0.6 M sorbitol, 0.1 M Tris–HCl, pH 7) and by centrifuging for 25 min. The intermediate layer was separated and washed twice with STC Buffer (2.4 M sorbitol, 10 mM CaCl2, and 10 mM Tris–HCl, pH 7.5). All centrifugation steps were performed at 3,000 × g at 4°C. Finally, protoplasts were resuspended in 1 ml of STC Buffer and kept on ice until transformation with PEG.
CRISPR RNAs were designed using Alt-R Custom Cas9 crRNA (CRISPR RNA) Design Tool1 and are listed in Supplementary Table 1. The off-target analysis was performed using Blast. Alt-R® CRISPR-Cas9 crRNA, Alt-R® CRISPR-Cas9 tracrRNA (trans-activating crRNA), and Alt-R® S.p. Cas9 nuclease V3 were purchased from IDT (Integrated DNA Technologies, Inc., Coralville, IA, United States) and were combined to obtain a ribonucleoprotein (RNP) complex according to the manufacturer’s instructions. Briefly, crRNA (2 nM) and tracrRNA (2 nM) were heated at 95°C for 5 min and cooled at room temperature to obtain gRNA. Three microliters of each gRNA was mixed with 4 μl of Cas9 at room temperature for 20 min. The entire mixture of RNP together with 10 μl of repair template (3 μg) and 40 μl of PEG20 solution (20% PEG6000) were gently mixed with 80 μl of protoplasts (107 protoplasts/ml). As a control, instead of DNA, 10 μl of water was used. The mixtures were kept on ice for 30 min, and then 900 μl of PEG60 solution (60% of PEG6000) was added. After 30 min of incubation on ice, protoplasts were gently spread on Petri dishes with regeneration media (RM; w/v: Malt extract 2%, peptone 1%, glucose 2%; sucrose 0.8 M) supplemented with or without hygromycin.
Mutants were selected on PDA supplemented with hygromycin B. Conidia were obtained in order to assure uniformity in the genetic material. The deletion event was confirmed by PCR, sequencing PCR products, and Southern blot. Southern blot analysis was performed according to Stroe et al. (2020), and the probes used are listed in Supplementary Table 1.
Deletion mutants were compared with wild-type P. polonicum X6 by inoculating 5 μl of spore suspension (1 × 106 conidia/ml) on PDA and incubating the plates at 25°C in the dark. Colony diameter (cm) and number of asexual spores (conidia/plate) were measured up to 7 days post inoculation (dpi). Additionally, deletion mutants were inoculated in liquid media (CYB and MEB) as previously described in order to examine verrucosidin production.
Apples cv. Ambrosia and cv. Opal were surface disinfected and wounded as described in Zhang et al. (2012). Ten microliters of conidial suspension (1 × 106 conidia/ml) of each strain was pipetted into each wound, whereas controls were inoculated with deionized Ringer solution. Inoculated apples were placed in plastic trays, covered with a transparent polyethylene film, and stored for 14 days at room temperature. To verify the pathogenicity of P. polonicum X6, apples cv. Gala were used.
All statistical analyses were performed with one-way ANOVA followed by Tukey’s b multiple comparison test using IBM SPSS statistics software version 24 (SPSS Inc., Chicago, IL, United States); p < 0.05 was considered significant.
SnapGene software (from Insightful Science; available at snapgene.com) was used to draw gene clusters and visualize annotated DNA sequences.
Using the PKS genes for aurovertin (AurA) and citreoviridin (CtvA) biosyntheses as matrix, 16 putative BGCs (Supplementary Figure 1) were found in the genome of P. polonicum. Of them, 10 clusters were present in both available P. polonicum genomes, whereas clusters 4 (later identified as verrucosidin BGC), 12, and 13 were only present in P. polonicum IBT 4502 and clusters 14, 15, and 16 in P. polonicum strain hy4 genome (Supplementary Table 2). Cluster 4 was further investigated due to the presence of genes encoding putative tailoring enzymes involved in verrucosidin biosynthesis (Figure 1): a putative methyltransferase (named cl4B or verB), two Flavin adenine dinucleotide (FAD)-dependent monooxygenases (cl4C1 and cl4C2 or verC1 and verC2), an acyltransferase (cl4G or verG), and a cytochrome P450 (cl4H or verH) gene. To monitor verrucosidin production, P. polonicum X6, P. aurantiogriseum CBS 112021, and P. crustosum CAL64 were grown in induction media. As shown in Supplementary Figure 2, P. polonicum and P. aurantiogriseum were able to produce verrucosidin, whereas P. crustosum failed to produce this compound. Consistently, we could verify the corresponding PKS gene sequence in the genomes of P. polonicum X6 and P. aurantiogriseum CBS 112021 by PCR, but not in the P. crustosum genome (Figure 2A). Moreover, the expression of the verA gene was confirmed both in P. polonicum X6 and in P. aurantiogriseum CBS 112021 by qPCR (Figure 2B).
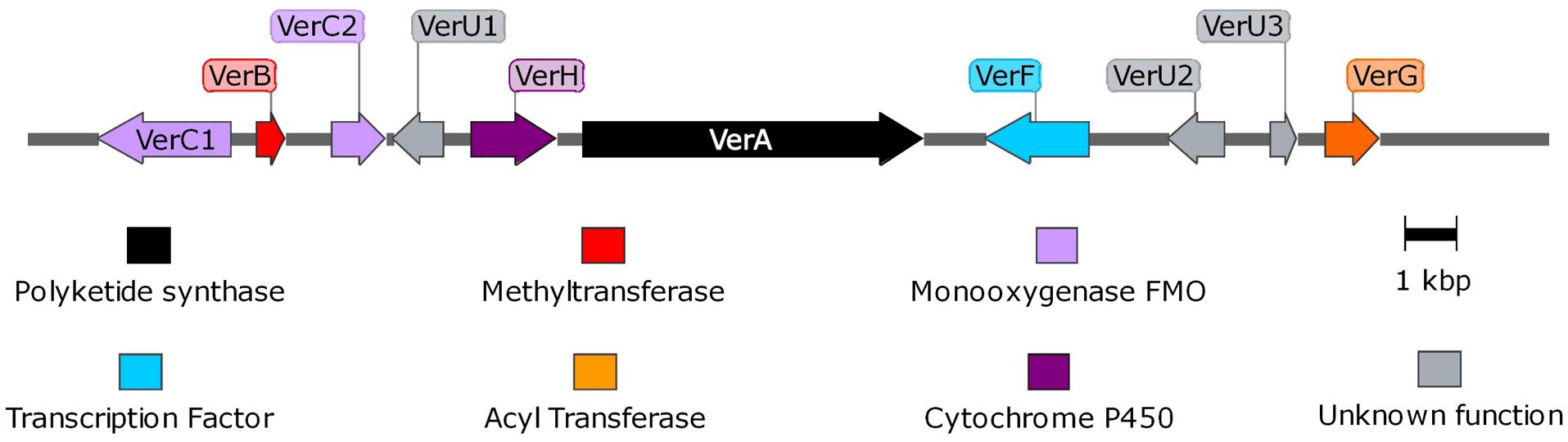
Figure 1. Representation of verrucosidin gene cluster in P. polonicum. Maps were obtained with SnapGene software.
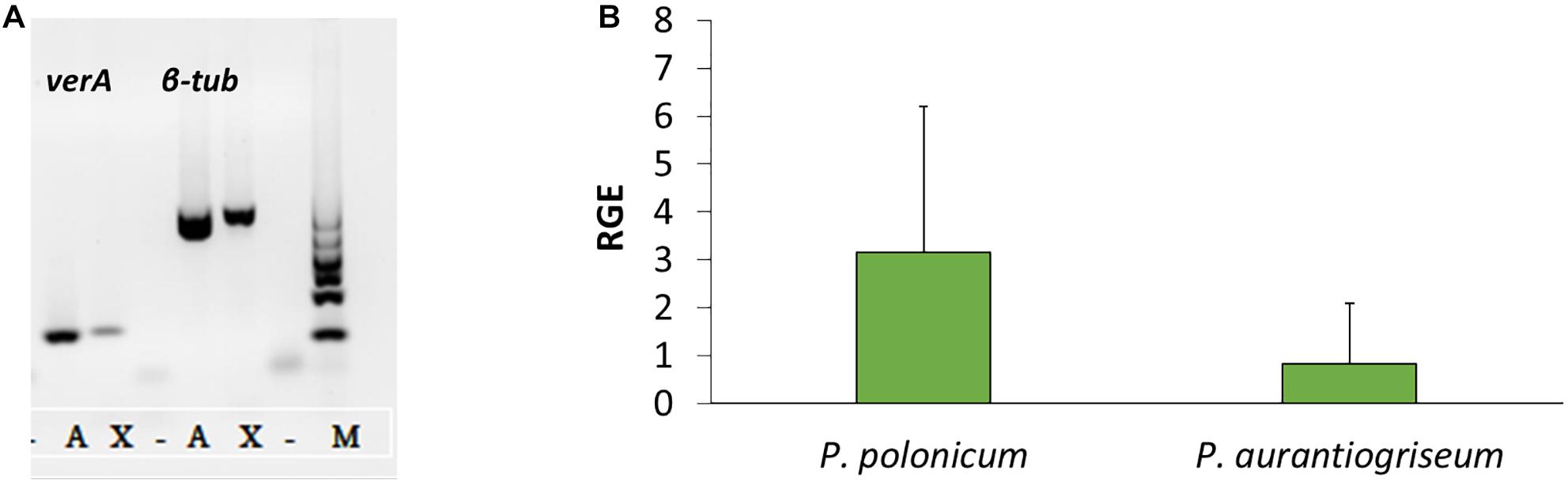
Figure 2. Presence and expression of verA gene in Penicillium spp. Amplification of verA and β-tubulin genes from gDNA (A): X, P. polonicum X6; A, P. aurantiogriseum CBS121001; M, GelPilot 100 bp Ladder; –, negative control (PCR mix without DNA). Relative gene expression (RGE) of verA (cl4A) gene in P. polonicum and P. aurantiogriseum (B) was evaluated 10 days post inoculation on CYB. The expression is relative to the expression of the β-tubulin gene, and P. aurantiogriseum was used as reference strain.
To investigate the role of verA in verrucosidin biosynthesis, the gene was deleted using CRISPR-Cas9. For this purpose, a method to obtain protoplasts for P. polonicum strain X6 was developed. The method allowed obtaining 1 × 107 protoplasts/ml starting from 5 g of mycelium.
In order to delete target genes with the endonuclease Cas9, the protospacers were designed based on the sequenced promoter and terminator of target genes in P. polonicum X6. To obtain the deletion mutants, two sets of gRNA were used, allowing to cut both the promoter and the terminator and to excise the target gene. The endonuclease Cas9 was mixed in vitro with gRNA and a repair DNA template encoding the hygromycin resistance cassette. The repair DNA template was amplified using primers with an additional tail of 50 bp providing at both flanking regions DNA micro-homology close to the PAM site. This way, the correct integration of the repair DNA through homology-mediated end joining (MMEJ) was achieved. Obtained knockout mutants were assessed through PCR, amplifying both the hygromycin resistance cassette and the target gene (Figures 3A–C). Sixteen mutants, in that the hygromycin resistance cassette was successfully amplified whereas amplification of the verA PKS gene failed, were randomly chosen and the correct integration of the repair template was confirmed using PCR with primer pairs designed inside the hph gene and in the promoter and terminator region of the deleted PKS gene (Figures 3A,D,E). In most cases, correct ΔverA mutants were found (Figures 3D,E). Some of the positive transformants were further confirmed by sequencing the amplified PCR products (Supplementary Figure 3) and by Southern blot analysis (Supplementary Figure 4). The results showed that one of the strains displayed a repetition of 50 bp of micro-homology in the promoter region (Supplementary Figure 3), whereas Δcl4A_C10, Δcl4A_C11, and Δcl4A_C12 mutants displayed a correct integration of the hph gene and lack of ectopic integrations (Supplementary Figures 3, 4) and were therefore further studied.
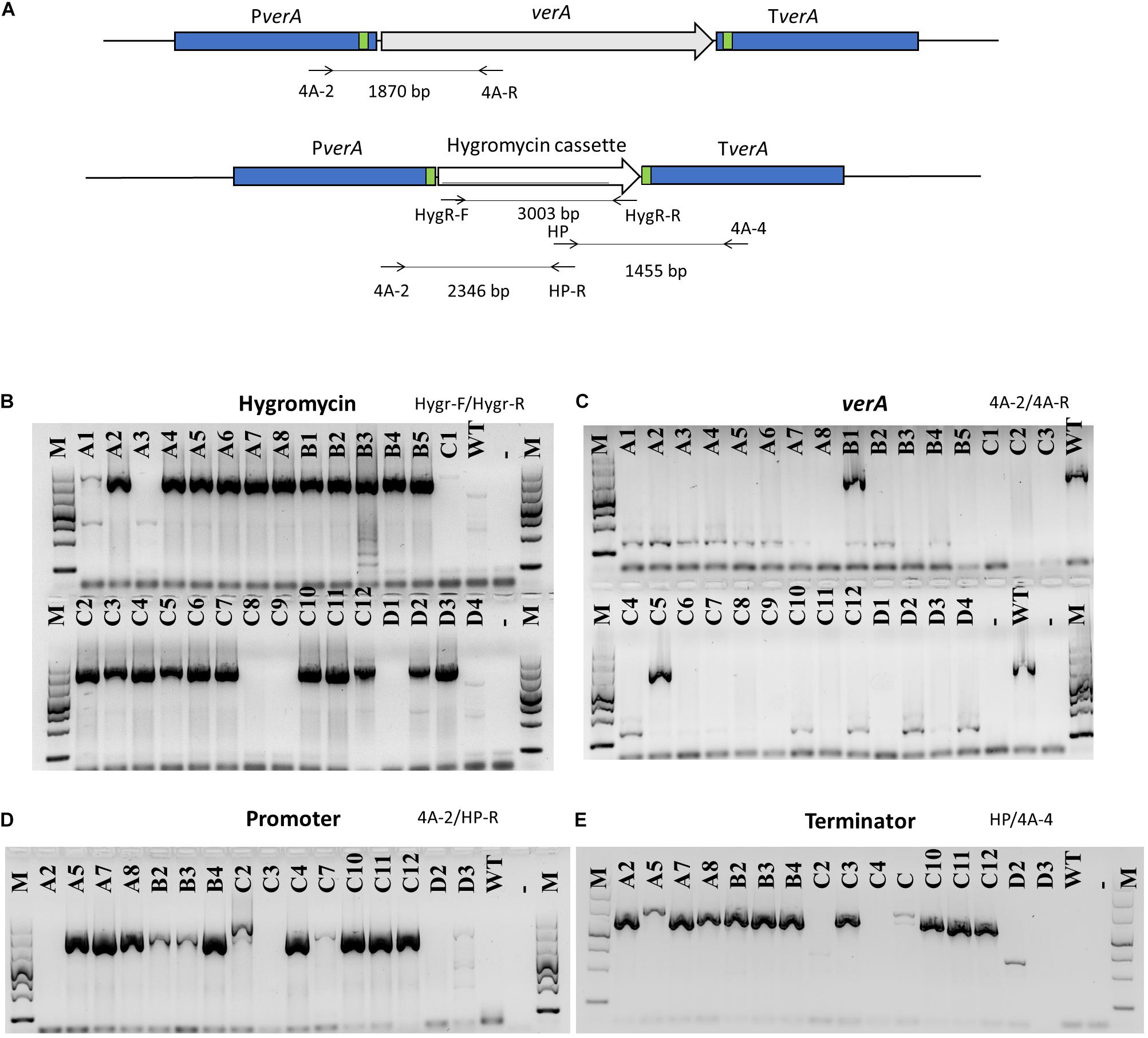
Figure 3. PCR analysis of verA mutants. Schematic presentation of the verA (cl4A) locus in the wild-type and deletion mutants; homology sequences (50 bp) are indicated in green, and primers used are marked by arrows (A). Amplification of hygromycin resistance cassette (B) and verA gene (C); confirmation of orientation of inserted repair DNA (D,E); M, GelPilot Wide Range Ladder; WT, wild-type P. polonicum X6; A–D, deletion mutants for verA; –, negative control (PCR mix without DNA).
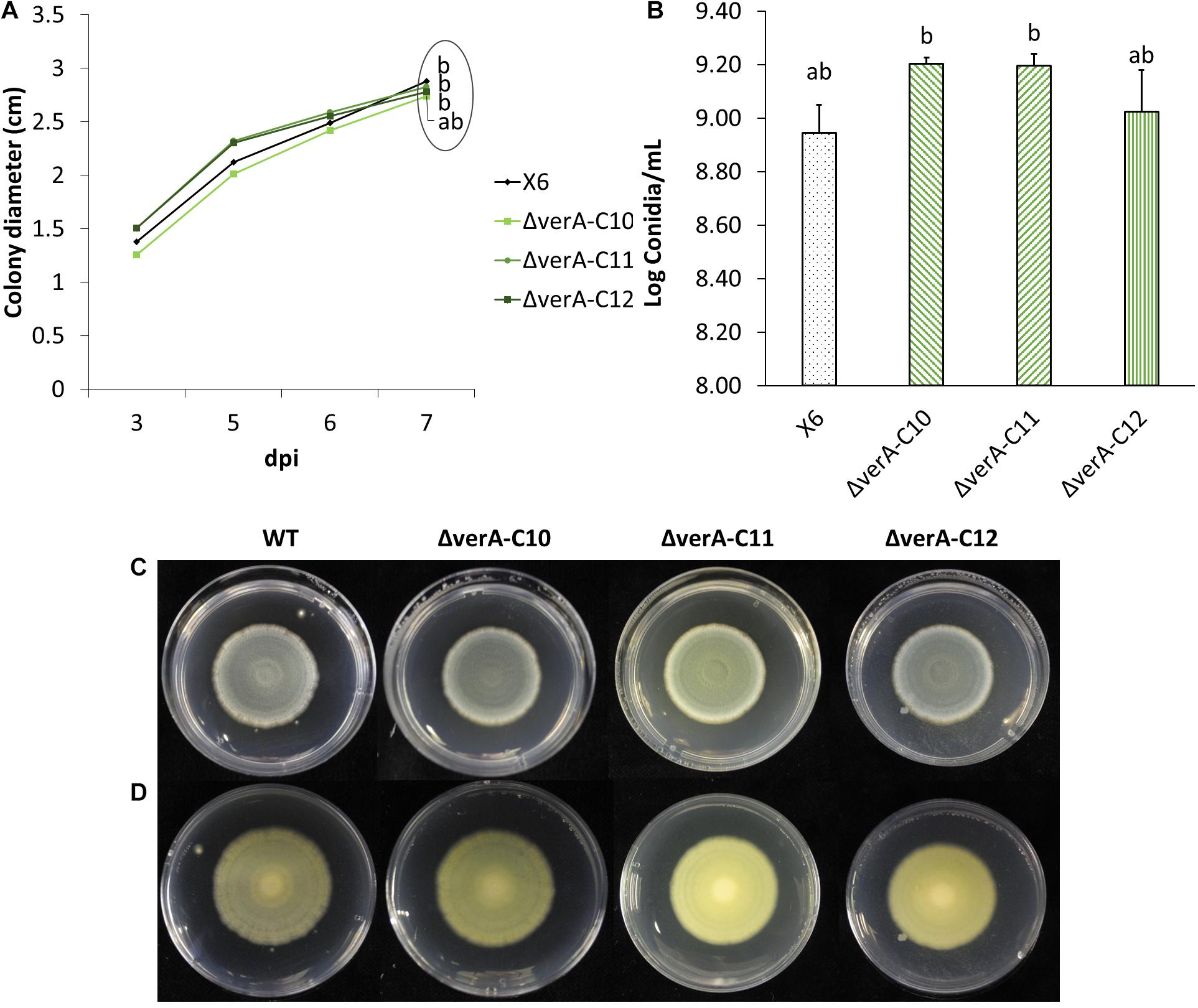
Figure 4. Effect of verA deletion on P. polonicum growth in vitro. Colony diameter after 3 to 7 days post inoculation (A) conidia production (B) and plate view (front C and reverse D) at 7 days post inoculation (dpi) on PDA. WT, wild-type P. polonicum X6; ΔverA, deletion mutants for verA (cl4A). Values followed by the same letter are not statistically different by Tukey’s b multiple comparison test (p < 0.05).
Three P. polonicum X6 verA mutants were phenotypically characterized in vitro and on apples. Compared to wild-type strain, the mutant strains displayed the same ability to grow (Figures 4A,C,D) and no significant differences in conidiation (Figure 4B). Analyzing the metabolic profile of wild-type strain and knockout mutants, the verrucosidin chromatographic peak was absent in deletion mutants (Supplementary Figure 5), independently of the used induction medium (Table 1 and Supplementary Figure 6). By contrast, verrucosidin formation in P. polonicum X6 was not influenced, when the PKS gene of another α-pyrone BGC, cl3A, was deleted (data not shown).
The pathogenicity of the wild-type strain X6 of P. polonicum was previously verified on apple (Supplementary Table 3). The effect of gene deletion on the virulence of P. polonicum was assessed on apple, as it is a host species for this pathogen and the virulence of the fungal strain can be quantified by measuring the lesion diameter, whereas on chestnut, the symptoms are mild and not easily measured. The virulence was tested on two apple cultivars. On apples cv. Ambrosia, verA deletion mutants displayed a slightly reduced virulence 7 days after the inoculation (Supplementary Table 4), whereas this behavior was not observed after 14 days of storage (Figure 5A) or on apples cv. Opal (Figure 5B).
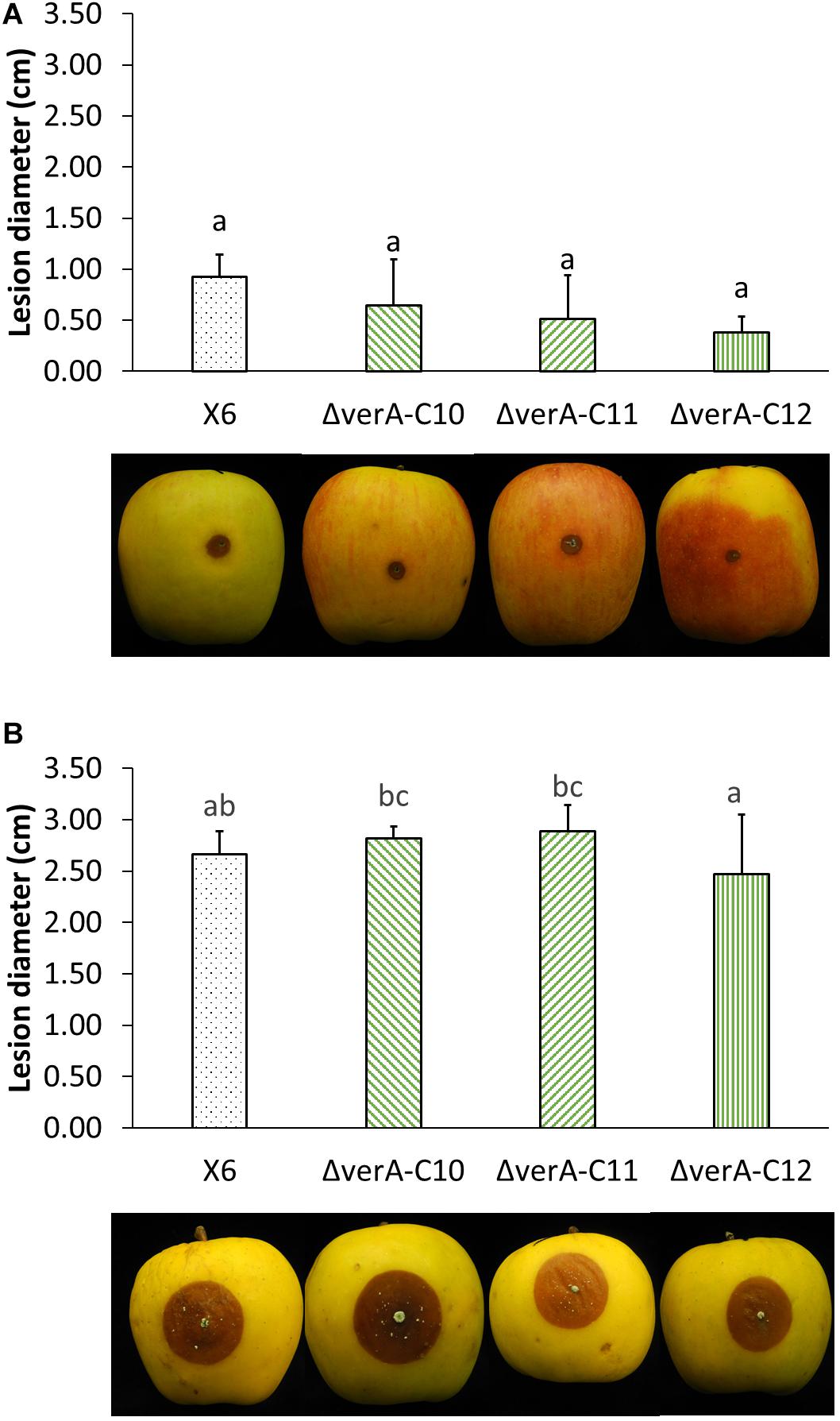
Figure 5. Analysis of verA deletion on the virulence of P. polonicum in vivo. Lesion diameter (cm) and pictures of apples cv. Ambrosia (A) and Opal (B) were recorded 14 days after inoculation. WT, wild-type P. polonicum X6; ΔverA, deletion mutants for verA (cl4A). Values followed by the same letter are not statistically different by Tukey’s b multiple comparison test (p < 0.05).
We report the identification of the BGC for the α-pyrone polyketide verrucosidin in the genome of P. polonicum, a known verrucosidin producer (Frisvad et al., 2004). Initial similarity analyses with the PKS genes for aurovertin (AurA) and citreoviridin (CtvA) had ended up with 16 BGCs putatively encoding the biosynthesis of α-pyrone polyketides. Besides P. polonicum, we confirmed P. aurantiogriseum to be able to produce verrucosidin (Fink-Gremmels et al., 1991), whereas P. crustosum did not produce this compound, as expected (Frisvad et al., 2004). Under the used laboratory conditions, meaning cultivation in different induction media, the verA gene was expressed and verrucosidin formation was verified in both producer species P. polonicum X6 and P. aurantiogriseum CBS 112021.
To delete verA, encoding a putative highly reducing polyketide synthase (HR-PKS), for the first time an in vitro method for CRISPR-Cas9 gene deletion was established in P. polonicum X6. Up to the present, CRISPR-Cas9 technology was adopted only in the Penicillium species P. chrysogenum. Protoplasts of a strain harboring the Cas9 gene on an AMA1 plasmid were transformed with a synthesized gRNA (Pohl et al., 2016). In this work, the method of Al Abdallah et al. (2017) was followed, who successfully transformed protoplasts of A. fumigatus with RNP and a repair template harboring antibiotic resistance to select mutants. This strategy does not require the generation of plasmids or the expression of the endonuclease Cas9, but is based on transforming protoplasts with the endonuclease protein Cas9 together with crRNA and tracrRNA. Therefore, there is an immediate cleavage activity when the RNP is targeted into the cells and the RNP is degraded by the host cell within a short period, reducing the possibility of off-target activity (Wang and Coleman, 2019). This strategy requires both accurate design of the crRNA and a protocol to transform fungi. A method to obtain protoplasts in P. polonicum X6 was optimized based on available protocols used for other filamentous fungi (Weidner et al., 1998; Young et al., 1998; Wang et al., 1999; McMillan et al., 2003; Fierro et al., 2004; Nicholson et al., 2015; Pohl et al., 2016) and allows obtaining a high amount of protoplasts. Mutants for putative PKS gene were obtained, confirming that the CRISPR-Cas9 method is effective and could be used for gene editing in P. polonicum. The finding of a small number of mutants displaying incorrect insertion of the foreign DNA necessitate screening and confirmation of mutants by molecular techniques such as PCR and Southern blot analysis. As an example, when sequencing the terminator region of the deleted verA gene in the mutant B4, we observed an unexpected insertion, that suggested correct cutting of the double-strand DNA by the endonuclease, but missing MMEJ. To our knowledge, there were no previous reports about MMEJ occurring only at one excision site. Therefore, the integration of repair DNA could have occurred by non-homologous end-joining (Krappmann, 2017). The efficiency of homologous recombination could be improved by adjusting the length of the homology sequence or by changing the amount of donor DNA, as previously shown in A. fumigatus (Al Abdallah et al., 2017).
Deletion mutants for the HR-PKS encoded by verA were no longer able to produce verrucosidin in vitro confirming that verA is responsible for the biosynthesis of this compound. The here identified gene cluster was bioinformatically hypothesized as verrucosidin cluster in P. polonicum by Li et al. (2018), but this prediction was not experimentally proven. Surprisingly, the verrucosidin gene cluster was absent in the genome of P. polonicum strain hy4, confirming high variability in secondary metabolism between strains of the same species (Ballester et al., 2015). The biosynthetic genes were also absent in P. expansum strain NRRL 62431. This fungal genome was initially deposited as P. aurantiogriseum and later assigned to another species (Ballester et al., 2015). Due to the lack of verrucosidin gene cluster, we can conclude that both P. expansum strains NRRL 62431 and P. polonicum strain hy4 are not able to produce this SM.
Secondary metabolites are known to play a central role in many biological processes such as growth development (Calvo et al., 2002) and pathogenesis (Scharf et al., 2014; Macheleidt et al., 2016), which were extensively reviewed (Fox and Howlett, 2008; Keller, 2019). In this work, the deletion of the verA gene did not affect P. polonicum in terms of growth rate and conidiation on PDA. Concerning the effect of deletion on the virulence of the strains, apples were used as preferential host, as P. polonicum is reported as a postharvest pathogen on pome fruit (Scholtz and Korsten, 2016; Ouhibi et al., 2018). The pathogenicity of the wild-type strain X6 of P. polonicum was previously verified on apple, where the virulence is easily quantified by measuring the lesion diameter. Slight differences in virulence of ΔverA mutants were observed on apple cv. Ambrosia, whereas this behavior was not observed on apples cv. Opal. These results are not surprising, as the apple cultivar is known to be a key factor in the virulence, such as in the case of patulin biosynthesis, which may be considered a cultivar-dependent aggressiveness factor (Banani et al., 2016; Snini et al., 2016). However, a reduced virulence of ΔverA mutants was observed only during the first steps of blue mold development on apple fruit; therefore, further studies are needed to confirm these preliminary observations.
The verrucosidin biosynthesis BGC was identified by bioinformatic and genetic approaches. Sixteen HR-PKS gene-containing BGCs were discovered in the available genomes of P. polonicum. These BGCs are probably involved in the biosynthesis of α-pyrone type polyketides, which could have many biological activities, such as cytotoxic, antitumor, antimicrobial, and anti-germination activities and are therefore a potential source of novel compounds (Schäberle, 2016; Li et al., 2018; Stroe et al., 2020). Based on the putative function of the genes present in the clusters, one of these BGCs was further characterized through deletion, in order to investigate its involvement in verrucosidin biosynthesis. To delete the selected PKS gene, CRISPR-Cas9 was adopted in P. polonicum. This allowed obtaining mutants by replacing the target genes with repair template sequences with 50 bp homology to chromosomal sequences. By applying this technology, the verrucosidin BGC was confirmed by creating deletion mutants for verA gene coding for HR-PKS known to be the key enzyme of the biosynthesis. The availability of the CRISPR-Cas technology will allow characterizing the biosynthetic potential of this interesting fungal species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SV, EP, VS, TH, AB, and DS conceived and designed the study. SV performed the experiments. EP, VS, and GM performed bioinformatic, Southern blot, and chemical analyses, respectively. SV and EP retrieved the literature and drafted the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by the German Academic Exchange Service (DAAD) with a scholarship in the program Research Grants – Short-Term Grants, 2019.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Christina Täumer from the Leibniz Institute for Natural Product Research and Infection Biology for technical assistance, and the students at the University of Torino who helped characterize the mutants.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.660871/full#supplementary-material
Al Abdallah, Q., Ge, W., and Fortwendel, J. R. (2017). A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro -Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. mSphere 2, 446–417e. doi: 10.1128/msphere.00446-17
Aranda, E., Rodríguez, M., Benito, M. J., Asensio, M. A., and Córdoba, J. J. (2002). Molecular cloning of verrucosidin-producing Penicillium polonicum genes by differential screening to obtain a DNA probe. Int. J. Food Microbiol. 76, 55–61. doi: 10.1016/S0168-1605(02)00008-9
Ballester, A.-R., Marcet-Houben, M., Levin, E., Sela, N., Selma-Lázaro, C., Carmona, L., et al. (2015). Genome, Transcriptome, and Functional Analyses of Penicillium expansum Provide New Insights Into Secondary Metabolism and Pathogenicity. Mol. Plant Microbe Interact. 28, 232–248. doi: 10.1094/MPMI-09-14-0261-FI
Banani, H., Marcet-Houben, M., Ballester, A. R., Abbruscato, P., González-Candelas, L., Gabaldón, T., et al. (2016). Genome sequencing and secondary metabolism of the postharvest pathogen Penicillium griseofulvum. BMC Genomics 17:19. doi: 10.1186/s12864-015-2347-x
Bergmann, S., Funk, A. N., Scherlach, K., Schroeckh, V., Shelest, E., Horn, U., et al. (2010). Activation of a Silent Fungal Polyketide Biosynthesis Pathway through Regulatory Cross Talk with a Cryptic Nonribosomal Peptide Synthetase Gene Cluster. Appl. Environ. Microbiol. 76, 8143–8149. doi: 10.1128/AEM.00683-10
Bergmann, S., Schümann, J., Scherlach, K., Lange, C., Brakhage, A. A., and Hertweck, C. (2007). Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3, 213–217. doi: 10.1038/nchembio869
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Burka, L. T., Ganguli, M., and Wilson, B. J. (1983). Verrucosidin, a Tremorgen from Penicillium verrucosum var. cyclopium. J. Chem. Soc. Chem. Commun. 1670, 544–545. doi: 10.1039/C39830000544
Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002). Relationship between Secondary Metabolism and Fungal Development. Microbiol. Mol. Biol. Rev. 66, 447–459. doi: 10.1128/mmbr.66.3.447-459.2002
Deng, H., Gao, R., Liao, X., and Cai, Y. (2017). CRISPR system in filamentous fungi: Current achievements and future directions. Gene 627, 212–221. doi: 10.1016/j.gene.2017.06.019
Duduk, N., Vasiæ, M., and Vico, I. (2014). First report of Penicillium polonicum causing blue mold on stored onion (Allium cepa) in Serbia. Plant Dis. 98:1440. doi: 10.1094/PDIS-05-14-0550-PDN
Faedda, R., Pane, A., Cacciola, S. O., Granata, G., Salafia, L., and Sinatra, F. (2015). Penicillium polonicum causing a postharvest soft rot of cactus pear fruits. Acta Hortic. 1067, 193–197. doi: 10.17660/ActaHortic.2015.1067.26
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Fierro, F., Laich, F., García-Rico, R. O., and Martín, J. F. (2004). High efficiency transformation of Penicillium nalgiovense with integrative and autonomously replicating plasmids. Int. J. Food Microbiol. 90, 237–248. doi: 10.1016/S0168-1605(03)00306-4
Fink-Gremmels, J., Henning, J., and Leistner, A. (1991). Quantitative determination of verrucosidin producing Penicillium aurantiogriseum. Microbiologie, Aliments. Nutr. Microbiol. Foods Feed. Nutr. 5, 155–160.
Fischer, J., Schroeckh, V., and Brakhage, A. A. (2016). “Awakening of Fungal Secondary Metabolite Gene Clusters,” in Gene Expression Systems in Fungi: Advancements and Applications, eds M. Schmoll and C. Dattenböck (Cham: Springer), 253–273. doi: 10.1007/978-3-319-27951-0_11
Fox, E. M., and Howlett, B. J. (2008). Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11, 481–487. doi: 10.1016/j.mib.2008.10.007
Frisvad, J. C., Smedsgaard, J., Larsen, T. O., and Samson, R. A. (2004). Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 49, 201–241.
Hodge, R. P., Harris, C. M., and Harris, T. M. (1988). Verrucofortine, a major metabolite of Penicillium verrucosum var. cyclopium, the fungus that produces the mycotoxin verrucosidin. J. Nat. Prod. 51, 66–73. doi: 10.1021/np50055a008
Jensen, B., Knudsen, I. M. B., Andersen, B., Nielsen, K. F., Thrane, U., Jensen, D. F., et al. (2013). Characterization of microbial communities and fungal metabolites on field grown strawberries from organic and conventional production. Int. J. Food Microbiol. 160, 313–322. doi: 10.1016/j.ijfoodmicro.2012.11.005
Keller, N. P. (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180. doi: 10.1038/s41579-018-0121-1
Kim, H. Y., Heo, D. Y., Park, H. M., Singh, D., and Lee, C. H. (2016). Metabolomic and transcriptomic comparison of solid-state and submerged fermentation of Penicillium expansum KACC 40815. PLoS One 11:e0149012. doi: 10.1371/journal.pone.0149012
Krappmann, S. (2017). CRISPR-Cas9, the new kid on the block of fungal molecular biology. Med. Mycol. 55, 16–23. doi: 10.1093/mmy/myw097
Li, W., Ma, Z., Chen, L., and Yin, W. B. (2018). Synthesis and production of the antitumor polyketide aurovertins and structurally related compounds. Appl. Microbiol. Biotechnol. 102, 6373–6381. doi: 10.1007/s00253-018-9123-1
Liebmann, B., Müller, M., Braun, A., and Brakhage, A. A. (2004). The cyclic AMP-dependent protein kinase A network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72, 5193–5203. doi: 10.1128/IAI.72.9.5193-5203.2004
Lin, T. S., Chen, B., Chiang, Y. M., and Wang, C. C. C. (2019). Discovery and Elucidation of the Biosynthesis of Aspernidgulenes: Novel Polyenes from Aspergillus Nidulans by Using Serial Promoter Replacement. ChemBioChem 20, 329–334. doi: 10.1002/cbic.201800486
Lin, T. S., Chiang, Y. M., and Wang, C. C. C. (2016). Biosynthetic Pathway of the Reduced Polyketide Product Citreoviridin in Aspergillus terreus var. aureus Revealed by Heterologous Expression in Aspergillus nidulans. Org. Lett. 18, 1366–1369. doi: 10.1021/acs.orglett.6b00299
Macheleidt, J., Mattern, D. J., Fischer, J., Netzker, T., Weber, J., Schroeckh, V., et al. (2016). Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 50, 371–392. doi: 10.1146/annurev-genet-120215-035203
Madden, T. (2013). “The BLAST sequence analysis tool,” in The NCBI Handbook, 2nd Edn, (Maryland: National Center for Biotechnology Information).
McMillan, L. K., Carr, R. L., Young, C. A., Astin, J. W., Lowe, R. G. T., Parker, E. J., et al. (2003). Molecular analysis of two cytochrome P450 monooxygenase genes required for paxilline biosynthesis in Penicillium paxilli, and effects of paxilline intermediates on mammalian maxi-K ion channels. Mol. Genet. Genomics 270, 9–23. doi: 10.1007/s00438-003-0887-2
Nagy, G., Szebenyi, C., Csernetics, Á, Vaz, A. G., Tóth, E. J., Vágvölgyi, C., et al. (2017). Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-17118-2
Nicholson, M. J., Eaton, C. J., Stärkel, C., Tapper, B. A., Cox, M. P., and Scott, B. (2015). Molecular cloning and functional analysis of gene clusters for the biosynthesis of indole-diterpenes in Penicillium crustosum and P. janthinellum. Toxins 7, 2701–2722. doi: 10.3390/toxins7082701
Nielsen, J. C., and Nielsen, J. (2017). Development of fungal cell factories for the production of secondary metabolites: Linking genomics and metabolism. Synth. Syst. Biotechnol. 2, 5–12. doi: 10.1016/j.synbio.2017.02.002
Nielsen, M. L., Isbrandt, T., Rasmussen, K. B., Thrane, U., Hoof, J. B., Larsen, T. O., et al. (2017). Genes Linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PLoS One 12:1–9. doi: 10.1371/journal.pone.0169712
Nødvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:1–18. doi: 10.1371/journal.pone.0133085
Núñez, F., Díaz, M. C., Rodríguez, M., Aranda, E., Martín, A., and Asensio, M. A. (2000). Effects of substrate, water activity, and temperature on growth and verrucosidin production by Penicillium polonicum isolated from dry-cured ham. J. Food Prot. 63, 231–236. doi: 10.4315/0362-028X-63.2.231
Ouhibi, S., Santos, C., Ghali, R., Soares, C., Hedhili, A., Paterson, R., et al. (2018). Penicillium tunisiense sp. nov., a novel species of Penicillium section Ramosa discovered from Tunisian orchard apples. Int. J. Syst. Evol. Microbiol. 68, 3217–3225. doi: 10.1099/ijsem.0.002962
Park, H. R., Ryoo, I. J., Choo, S. J., Hwang, J. H., Kim, J. Y., Cha, M. R., et al. (2007). Glucose-deprived HT-29 human colon carcinoma cells are sensitive to verrucosidin as a GRP78 down-regulator. Toxicology 229, 253–261. doi: 10.1016/j.tox.2006.11.049
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pohl, C., Kiel, J. A. K. W., Driessen, A. J. M., Bovenberg, R. A. L., and Nygård, Y. (2016). CRISPR/Cas9 Based Genome Editing of Penicillium chrysogenum. ACS Synth. Biol. 5, 754–764. doi: 10.1021/acssynbio.6b00082
Prencipe, S., Siciliano, I., Gatti, C., Garibaldi, A., Gullino, M. L., Botta, R., et al. (2018). Several species of Penicillium isolated from chestnut flour processing are pathogenic on fresh chestnuts and produce mycotoxins. Food Microbiol. 76, 396–404. doi: 10.1016/j.fm.2018.07.003
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: Protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Rodríguez, A., Córdoba, J. J., Werning, M. L., Andrade, M. J., and Rodríguez, M. (2012). Duplex real-time PCR method with internal amplification control for quantification of verrucosidin producing molds in dry-ripened foods. Int. J. Food Microbiol. 153, 85–91. doi: 10.1016/j.ijfoodmicro.2011.10.020
Sabater-Vilar, M., Nijmeijer, S., and Fink-Gremmels, J. (2003). Genotoxicity assessment of five tremorgenic mycotoxins (fumitremorgen B, paxilline, penitrem A, verruculogen, and verrucosidin) produced by molds isolated from fermented meats. J. Food Prot. 66, 2123–2129. doi: 10.4315/0362-028X-66.11.2123
Santini, A., Mikušová, P., Sulyok, M., Krska, R., Labuda, R., and Šrobárová, A. (2014). Penicillium strains isolated from Slovak grape berries taxonomy assessment by secondary metabolite profile. Mycotoxin Res. 30, 213–220. doi: 10.1007/s12550-014-0205-3
Schäberle, T. F. (2016). Biosynthesis of α-pyrones. Beilstein J. Org. Chem. 12, 571–588. doi: 10.3762/bjoc.12.56
Scharf, D. H., Heinekamp, T., and Brakhage, A. A. (2014). Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PLoS Pathog. 10:e1003859. doi: 10.1371/JOURNAL.PPAT.1003859
Scholtz, I., and Korsten, L. (2016). Profile of Penicillium species in the pear supply chain. Plant Pathol. 65, 1126–1132. doi: 10.1111/ppa.12494
Snini, S. P., Tannous, J., Heuillard, P., Bailly, S., Lippi, Y., Zehraoui, E., et al. (2016). Patulin is a cultivar-dependent aggressiveness factor favouring the colonization of apples by Penicillium expansum. Mol. Plant Pathol. 17, 920–930. doi: 10.1111/mpp.12338
Song, R., Zhai, Q., Sun, L., Huang, E., Zhang, Y., Zhu, Y., et al. (2019). CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl. Microbiol. Biotechnol. 103, 6919–6932. doi: 10.1007/s00253-019-10007-w
Sonjak, S., Frisvad, J. C., and Gunde-Cimerman, N. (2006). Penicillium mycobiota in Arctic subglacial ice. Microb. Ecol. 52, 207–216. doi: 10.1007/s00248-006-9086-0
Spadaro, D., Lorè, A., Garibaldi, A., and Gullino, M. L. (2013). A new strain of Metschnikowia fructicola for postharvest control of Penicillium expansum and patulin accumulation on four cultivars of apple. Postharv. Biol. Technol. 75, 1–8. doi: 10.1016/j.postharvbio.2012.08.001
Stroe, M. C., Netzker, T., Scherlach, K., Krüger, T., Hertweck, C., Valiante, V., et al. (2020). Targeted induction of a silent fungal gene cluster encoding the bacteria-specific germination inhibitor fumigermin. Elife 9:e52541. doi: 10.7554/eLife.52541
Sunesen, L. O., and Stahnke, L. H. (2003). Mould starter cultures for dry sausages - Selection, application and effects. Meat Sci. 65, 935–948. doi: 10.1016/S0309-1740(02)00281-4
Thomas, S., Sharma, N., Gonzalez, R., Pao, P. W., Hofman, F. M., Chen, T. C., et al. (2013). Repositioning of Verrucosidin, a Purported Inhibitor of Chaperone Protein GRP78, as an Inhibitor of Mitochondrial Electron Transport Chain Complex I. PLoS One 8:1–14. doi: 10.1371/journal.pone.0065695
Tong, Y., Weber, T., and Lee, S. Y. (2019). CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 36, 1225–1372. doi: 10.1039/c8np00089a
Valente, S., Cometto, A., Piombo, E., Meloni, G. R., Ballester, A. R., González-Candelas, L., et al. (2020). Elaborated regulation of griseofulvin biosynthesis in Penicillium griseofulvum and its role on conidiation and virulence. Int. J. Food Microbiol. 328:108687. doi: 10.1016/j.ijfoodmicro.2020.108687
Wang, H. L., Kim, S. H., Siu, H., and Breuil, C. (1999). Transformation of sapstaining fungi with hygromycin B resistance plasmids pAN7-1 and pCB1004. Mycol. Res. 103, 77–80. doi: 10.1017/S0953756298006698
Wang, Q., and Coleman, J. J. (2019). Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi. Comput. Struct. Biotechnol. J. 17, 761–769. doi: 10.1016/j.csbj.2019.06.007
Weber, J., Valiante, V., Nødvig, C. S., Mattern, D. J., Slotkowski, R. A., Mortensen, U. H., et al. (2017). Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth. Biol. 6, 62–68. doi: 10.1021/acssynbio.6b00203
Weber, T., and Kim, H. U. (2016). The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 1, 69–79. doi: 10.1016/j.synbio.2015.12.002
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Weidner, G., D’Enfert, C., Koch, A., Mol, P. C., and Brakhage, A. A. (1998). Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5’-monophosphate decarboxylase. Curr. Genet. 33, 378–385. doi: 10.1007/s002940050350
Wigmann, ÉF., Jahn, R. C., Scherer, C. D., Saccomori, F., Alcano-González, M., de, J., et al. (2018). Detection and identification of Penicillium spp. in a frozen chicken nuggets production facility. Food Microbiol. 70, 42–48. doi: 10.1016/j.fm.2017.09.002
Young, C., Itoh, Y., Johnson, R., Garthwaite, I., Miles, C. O., Munday-Finch, S. C., et al. (1998). Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 33, 368–377. doi: 10.1007/s002940050349
Yu, J., Jurick, M. W., and Bennett, J. W. (2015). Current status of genomics research on mycotoxigenic fungi. Int. J. plant Biol. Res. 3:1035.
Zhang, D., Spadaro, D., Valente, S., Garibaldi, A., and Gullino, M. L. (2012). Cloning, characterization, expression and antifungal activity of an alkaline serine protease of Aureobasidium pullulans PL5 involved in the biological control of postharvest pathogens. Int. J. Food Microbiol. 153, 453–464. doi: 10.1016/j.ijfoodmicro.2011.12.016
Keywords: secondary metabolites, Penicillium, mycotoxins, CRISPR-Cas, alpha-pyrone polyketides
Citation: Valente S, Piombo E, Schroeckh V, Meloni GR, Heinekamp T, Brakhage AA and Spadaro D (2021) CRISPR-Cas9-Based Discovery of the Verrucosidin Biosynthesis Gene Cluster in Penicillium polonicum. Front. Microbiol. 12:660871. doi: 10.3389/fmicb.2021.660871
Received: 29 January 2021; Accepted: 15 April 2021;
Published: 21 May 2021.
Edited by:
Esther Garcia-Cela, University of Hertfordshire, United KingdomReviewed by:
Jéssica Gil-Serna, Complutense University of Madrid, SpainCopyright © 2021 Valente, Piombo, Schroeckh, Meloni, Heinekamp, Brakhage and Spadaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Axel A. Brakhage, QXhlbC5CcmFraGFnZUBoa2ktamVuYS5kZQ==; Davide Spadaro, ZGF2aWRlLnNwYWRhcm9AdW5pdG8uaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.