- LEAF—Linking Landscape, Environment, Agriculture and Food—Research Center, Instituto Superior de Agronomia, Universidade de Lisboa, Lisbon, Portugal
Introduction
Over the last 20 years, numerous definitions have been proposed for the term microbiome (Berg et al., 2020). One of the most cited, albeit not the earliest (Whipps et al., 1988), was that introduced by Joshua Lederberg, who referred to the microbiome as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space […]” (Lederberg and Mccray, 2001). This definition became especially popular as, over the following years, its meaning shifted from the organisms as taxonomical units (i.e., microbiota) to their collective genetic material. Four years after Lederberg's publication, the word microbiome began to be employed in the scientific literature (Nicholson et al., 2005), and to date, it has been used in tens of thousands of scientific publications. However, as its popularity increased, numerous definitions for the word microbiome appeared in scientific literature, sparking a hot debate over the birth and evolution of the meaning of this word (Prescott, 2017; Morar and Bohannan, 2019; Berg et al., 2020).
Over the last decade, advancements in next-generation sequencing allowed for a new understanding of the microbial complexity tightly associated with living beings and environments. Recent research demonstrated that microbiomes play a key role in the health of the organisms whom they are associated with, influencing, among others, their physiology, biochemistry, and reproductive success (Dinan and Cryan, 2017; Bai et al., 2020; Compant et al., 2020). In fact, the disruption of microbial homeostasis leads to dysbiosis, a condition which plays a major role in disease in humans and other animals (Liu et al., 2020), and decline in plants (Bettenfeld et al., 2020). Undoubtedly, much research has yet to be carried out to further understand the composition, functions, and resilience to stressors of microbiomes (e.g., anthropic activities and climate change). However, in recent years, microbiome research has branched out into a new direction. The acquired awareness over the role that microbial communities play in the health of biological systems led to the search for strategies to exploit microbiomes and microbial blends to achieve specific goals, such as restoring compromised systems (e.g., reversing a condition of dysbiosis) or enhancing existing ones.
Microbiomes and Microbial Blends
The employment of microbiomes to produce positive effects on individuals or environments has been recently proven possible. One of the most popular examples is that of fecal transplants in humans. This technique is currently used to treat recurrent Clostridium difficile infections, and it showed promising results in treating inflammatory bowel diseases (Vindigni and Surawicz, 2017). Microbiome transplants have been successfully tested also in plant protection. For example, leaf endophytes of healthy Phyllostegia hirsuta were transferred on Phyllostegia kaalaensis leaves, significantly reducing disease severity caused by Neoerysiphe galeopsidis (Zahn and Amend, 2017). In another study, a soil microbiome transplantation significantly decreased the disease incidence in the Solanum lycopersicum–Ralstonia solanacearum pathosystem (Wei et al., 2019). Despite these promising results, there are numerous limitations still preventing the full exploitation of this technology. Microbiomes are often composed of hundreds, if not thousands, of species of microorganisms, many of which have not yet been identified (e.g., uncultivable species) and many more are found in very low abundances (rare taxa). Moreover, the composition of microbiomes is known to vary in time and space, complicating the access to reliable and stable sources of specific microbiomes (Lawson et al., 2019; Berg et al., 2020).
Unlike microbiomes, microbial blends are intelligently designed to study their component behavior (e.g., ecological studies) or to carry out well-defined tasks (e.g., biotechnology and biological control). Most frequently, microbial blends are composed of a mixture of bacterial species or strains (Voges et al., 2019), although those based on fungal species (Del Frari et al., 2019) or a combination of fungi and bacteria (Shahab et al., 2018) are becoming increasingly popular. Contrary to microbiomes, microbial blends are exact and reproducible, originating from microorganisms cultivable in vitro. For this reason, when employing a microbial blend, the outcome of its predicted function has a much greater reproducibility when compared to that of microbiomes. However, presently, microbial blends are relatively simple mixtures often composed of a few species and cannot match the results such as those achieved by microbiome transplants. It is worth remembering that, despite the fact that microbial blends are engineered with the intent of containing exclusively specific microorganisms, under some circumstances, endohyphal bacteria (Arora and Riyaz-ul-hassan, 2018), bacteriophages, and/or mycoviruses may be unwillingly added to the final mixture.
The numerous advantages of using microbial blends over microbiomes or single strains have been attracting a growing interest in several fields of science (Compant et al., 2019; Santos et al., 2019). Among the numerous applications of microbial blends, we find probiotics (El Hage et al., 2019), biotechnology (Christiaens et al., 2019), bioremediation (Brune and Bayer, 2012), biocontrol (Del Frari et al., 2019), ecology (Voges et al., 2019), human disease (Pereira et al., 2020), crop enhancement (Allaga et al., 2020), and more. In addition, recent advances in microbial blend engineering (Lawson et al., 2019) suggest that this technology will strongly contribute to re-shape microbiological research in the near future.
Are Microbial Blend-Related Terms Synonyms?
Over the last 10 years, a variety of terms have been employed to refer to microbial blends, some of which are often used interchangeably (Mabwi et al., 2021), namely:
(a) microbial consortium, also referred to as “bacterial consortium” or “fungal consortium,”
(b) microbial inoculant, also referred to as “bioinoculant,”
(c) synthetic microbial community, also referred to as synthetic community or synthetic microbial consortium,
(d) microbial cocktail,
(e) synthetic microbiome.
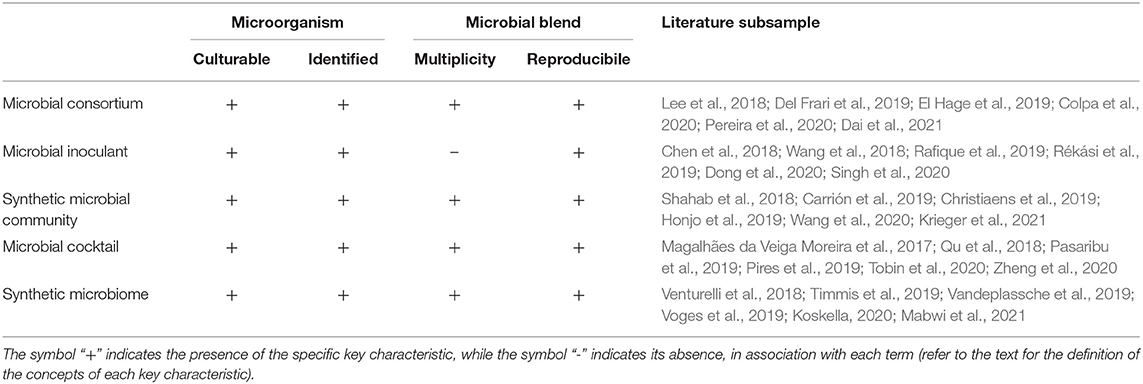
To assess the similarities and differences among these terms, we examined 90 recently published (2017–2021) peer-reviewed scientific articles (both original research and review articles), and we focused on four key characteristics:
1. Cultivability of microorganisms: All microorganisms used to produce the final blend originate from mother cultures cultivated in vitro.
2. Taxonomical identification of microorganisms: All microorganisms are identified at least down to species level. Alternatively, microorganisms identified at the genus level are given an isolated identification code (e.g., strain ID).
3. Microbial multiplicity: The microbial composition involves multiple microorganisms (two or more).
4. Blend reproducibility: The microbial composition, qualitative and quantitative, is exact and reproducible (e.g., based on colony-forming unit and optical density).
In addition, we summarily looked at the fields of science in which these terms were most frequently used.
The results of our literature survey are shown in Table 1, where, for ease of visualization, we report only a subsample of literature references.
The literature survey revealed that four out of the five terms under examination, which are commonly employed to refer to microbial blends, share the four key characteristics of cultivability of microorganisms, taxonomical identification, microbial multiplicity and blend reproducibility. The only exception refers to the term microbial inoculant, which does not share, in all cases, the microbial multiplicity characteristic. Even though, in the last decade, microbial inoculants containing multiple microorganism species have been increasing in number, this term also refers to inoculants that contain a single microorganism species (Santos et al., 2019). In addition, the majority of studies that make use of this term, although not the entirety, are context specific, as it is used to indicate the addition of microorganisms to seeds or soil. All other terms have been employed in several fields of science. From our examination, some terms seem to be preferentially used in specific contexts—for example, the term microbial cocktail is often used in biotechnological research, while the term synthetic microbiome is frequently used in medical research. However, giving an accurate estimate of context specificity for each term is virtually unfeasible and certainly goes beyond the scope of this opinion article.
According to this analysis and in light of the fact that they are often used interchangeably, we conclude that the terms microbial consortium, synthetic microbial community, microbial cocktail, and synthetic microbiome can be considered synonyms. Nevertheless, we examined a relatively small number of recent papers (90); therefore, we recognize that, among the immense volume of literature where these terms are employed, exceptions may be occasionally encountered.
Discussion
Scientific vocabulary is at the core of science, as it is our means to share abstract novel concepts and ideas with our peers. Therefore, its diversity must be considered a strength. However, in recent years, in the fields of applied microbiology, biotechnology, and microbial ecology, numerous terms have been employed to identify “a blend of multiple microorganisms engineered to carry out a specific function.” Microbial consortium, synthetic microbiome, microbial cocktail, and synthetic microbial community are some of the terms that normally refer to such a broad definition. Similarly to the word microbiome, which gained considerable popularity over the last 10 years, the immense potential represented by microbial blends suggests that the term chosen to refer to them may become as much popular in the near future. However, the presence of multiple terms, some of which we demonstrated to be synonyms, may complicate communication both within the scientific community and with the greater public.
Therefore, why should we consider introducing a new term?
(1) Simplification: The process of simplification in scientific terminology, which invariably leads to the coinage of neologisms, aims to reduce to a single word abstract ideas that can only be defined with multiple words (e.g., see Raad, 1989; Gibson et al., 2017). To date, all terms that refer to microbial blends are composed of two or three words. Therefore, creating a single-worded neologism will significantly simplify communication in the medium and long terms.
(2) A clear definition: A single-worded term is easier to use, provided that it is clearly defined. Within this definition, all four key characteristics associated with the current terms that refer to microbial blends must be addressed, limiting the room for individual interpretation.
(3) Set a standard: Offering a single-worded, well-defined term may discourage more synonyms from being coined and introduced in the scientific literature.
Within this context, we would like to introduce and propose, as a potential candidate, the word skopobiota, a neologism built from the Greek word skopós, which translates in “purpose.” The term skopobiota is defined as:
“A purposefully designed, exact, and reproducible blend of multiple species of taxonomically identified microorganisms, whose components may act additively or synergistically to accomplish a predefined function in or on a specific environment.”
This single-worded term puts emphasis on the purpose for which different microbial blends are engineered as well as its quantitative and qualitative composition, addressing all four key characteristics associated with microbial blends.
Our undertaking goes beyond facilitating communication within and beyond academia. As a famous quote by philosopher Ludwig Wittgenstein (1922) goes, “the limits of my language mean the limits of my world”, the limits of science depend on scientific terminology as well. From this perspective, the word skopobiota may have a significant impact on future microbiome and microbial blend research. It may shape scientific approaches and future terminology and prevent controversies (see the word microbiome, Berg et al., 2020). However, we recognize that our effort has its limits as we agree with Raad (1989), who stated that “perhaps more and more scientists are beginning to realize that inaccuracy is rooted in the nature of knowledge, as we uncover more of it, rather than in the words used to transmit that knowledge.”
To conclude, in our opinion, the introduction of novel terms in science tends to follow five main sequential steps. First, there must be a need to describe a novel abstract idea; second, such idea is shared with peers using current scientific vocabulary; third, simplification leads to the coinage of a single-worded term (neologism); fourth, such neologism is publicly proposed in scientific literature; and fifth, the proposed neologism may or may not gain popularity. With respect to microbial blends, we have just reached the fourth step, and only time and the best judgment of scientists will decide which direction scientific terminology shall take.
Author Contributions
GDF conceived the idea and wrote the manuscript. RF revised the manuscript.
Funding
The Open Access is funded by national funds through FCT –Fundação para a Ciência e a Tecnologia, I.P., in the scope of the project Linking Landscape, Environment, Agriculture and Food Research Centre (Ref. UIDB/04129/2020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allaga, H., Bóka, B., Poór, P., Nagy, V. D., Sz, A., Stankovics, I., et al. (2020). A composite bioinoculant based on the combined application of beneficial bacteria and fungi. Agronomy 10:220. doi: 10.3390/agronomy10020220
Arora, P., and Riyaz-ul-hassan, S. (2018). Endohyphal bacteria; the prokaryotic modulators of host fungal biology. Fungal Biol. Rev. 33, 72–81. doi: 10.1016/j.fbr.2018.08.003
Bai, S., Yao, Z., Raza, M. F., Cai, Z., and Zhang, H. (2020). Regulatory mechanisms of microbial homeostasis in insect gut. Insect Sci. 28, 286–301. doi: 10.1111/1744-7917.12868
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Bettenfeld, P., Fontaine, F., Trouvelot, S., Fernandez, O., and Courty, P. (2020). Woody plant declines. What's wrong with the microbiome? Trends Plant Sci. 25, 381–394. doi: 10.1016/j.tplants.2019.12.024
Brune, K. D., and Bayer, T. S. (2012). Engineering microbial consortia to enhance biomining and bioremediation. Front. Microbiol. 3:203. doi: 10.3389/fmicb.2012.00203
Carrión, V. J., Perez-jaramillo, J., Cordovez, V., Tracanna, V., Gomez-exposito, R., Elsayed, S. S., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Chen, D., Zhao, X., Miao, X., Chen, J., Ye, J., Cheng, Z., et al. (2018). A solid composite microbial inoculant for the simultaneous removal of volatile organic sulfide compounds: preparation, characterization, and its bioaugmentation of a biotrickling filter. J. Hazard Mater. 342, 589–596. doi: 10.1016/j.jhazmat.2017.08.079
Christiaens, M. E. R., De Paepe, J., Ilgrande, C., De, J., Barys, J., Teirlinck, P., et al. (2019). Urine nitrification with a synthetic microbial community. Syst. Appl. Microbiol. 42:126021. doi: 10.1016/j.syapm.2019.126021
Colpa, D. I., Zhou, W., Wempe, J. P., Tamis, J., Stuart, M. C. A., Krooneman, J., et al. (2020). Thauera aminoaromatica MZ1T identified as a polyhydroxyalkanoate-producing bacterium within a mixed microbial consortium. Bioengineering 7:19. doi: 10.3390/bioengineering7010019
Compant, S., Cambon, M. C., Vacher, C., Mitter, B., Samad, A., and Sessitsch, A. (2020). The plant endosphere world – bacterial life within plants. Environ. Microbiol. 23, 1812–1829. doi: 10.1111/1462-2920.15240
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Dai, X., Lv, J., Guo, S., and Wei, W. (2021). Heavy oil biodegradation by mixed bacterial consortium of biosurfactant-producing and heavy oil-degrading bacteria. Pol. J. Environ. Stud. 30, 71–80. doi: 10.15244/pjoes/120769
Del Frari, G., Gobbi, A., Aggerbeck, M. R., Oliveira, H., Hansen, L. H., and Ferreira, R. B. (2019). Fungicides and the grapevine wood mycobiome: a case study on tracheomycotic ascomycete Phaeomoniella chlamydospora reveals potential for two novel control strategies. Front. Plant Sci. 10:1405. doi: 10.3389/fpls.2019.01405
Dinan, T. G., and Cryan, J. F. (2017). Brain–gut–microbiota axis — mood, metabolism and behaviour. Nat. Rev. Gastroenterol. Hepatol. 14, 69–70. doi: 10.1038/nrgastro.2016.200
Dong, M., Li, Q., Xu, F., Shuyang, W., Chen, J., and Li, W. (2020). Effects of microbial inoculants on the fermentation characteristics and microbial communities of sweet sorghum bagasse silage. Sci. Rep. 10:837. doi: 10.1038/s41598-020-57628-0
El Hage, R., Hernandez-sanabria, E., Arroyo, M. C., Props, R., and Van De Wiele, T. (2019). Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front. Microbiol. 10:1206. doi: 10.3389/fmicb.2019.01206
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Honjo, H., Iwasaki, K., Soma, Y., Tsuruno, K., Hamada, H., and Hanai, T. (2019). Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab. Eng. 55, 268–275. doi: 10.1016/j.ymben.2019.08.007
Krieger, A., Zhang, J., and Lin, X. N. (2021). Temperature regulation as a tool to program synthetic microbial community composition. Biotechnol. Bioeng. 118, 1381–1392. doi: 10.1002/bit.27662
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Löffler, F. E., O'Malley, M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi: 10.1038/s41579-019-0255-9
Lederberg, J., and Mccray, A. (2001). ‘Ome Sweet ‘Omics–A genealogical treasury of words. Scientist 15:8.
Lee, Y., Jeong, S. E., Hur, M., Ko, S., Jeon, C. O., and Edwards, E. A. (2018). Construction and evaluation of a Korean native microbial consortium for the bioremediation of diesel fuel-contaminated soil in Korea. Front. Microbiol. 9:2594. doi: 10.3389/fmicb.2018.02594
Liu, N., Ma, Q., Ge, Y., Yi, C., Wei, L., Tan, J., et al. (2020). Microbiome dysbiosis in lung cancer: from composition to therapy. Npj Precis. Oncol. 4:33. doi: 10.1038/s41698-020-00138-z
Mabwi, H. A., Kim, E., Song, D., Yoon, S. H., Pan, C., Komba, E. V. G., et al. (2021). Synthetic gut microbiome: advances and challenges. Comput. Struct. Biotechnol. J. 19, 363–371. doi: 10.1016/j.csbj.2020.12.029
Magalhães da Veiga Moreira, I., de Figueiredo Vilela, L., da Cruz Pedroso Miguel, M. G., Santos, C., Lima, N., and Freitas Schwan, R. (2017). Impact of a microbial cocktail used as a starter culture on cocoa fermentation and chocolate flavor. Molecules 22:766. doi: 10.3390/molecules22050766
Morar, N., and Bohannan, B. J. M. (2019). The conceptual ecology of the human microbiome. Q. Rev. Biol. 94, 149–175. doi: 10.1086/703582
Nicholson, J. K., Holmes, E., and Wilson, I. D. (2005). Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3, 431–438. doi: 10.1038/nrmicro1152
Pasaribu, T., Laconi, E. B., and Kompiang, I. P. (2019). Evaluation of the nutrient contents of palm kernel cake fermented by microbial cocktails as a potential feedstuff for poultry. J. Indones. Trop. Anim. Agric. 44, 295–302. doi: 10.14710/jitaa.44.3.295-302
Pereira, F. C., Wasmund, K., Cobankovic, I., Jehmlich, N., Herbold, C. W., Lee, K. S., et al. (2020). Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat. Commun. 11:5104. doi: 10.1038/s41467-020-18928-1
Pires, J. F., Schwan, R. F., and Silva, C. F. (2019). Assessing the efficiency in assisted depuration of coffee processing wastewater from mixed wild microbial selected inoculum. Environ. Monit. Assess. 191:284. doi: 10.1007/s10661-019-7398-z
Prescott, S. L. (2017). History of medicine: origin of the term microbiome and why it matters. Hum. Microbiome J. 4, 24–25. doi: 10.1016/j.humic.2017.05.004
Qu, H., Cao, J., Chen, Y., Li, R., Wang, P., Chen, M., et al. (2018). Enhancement of biogas production from bundled rice straw solid-state fermentation by adding microbial agents. Bioresources 13, 8723–8737. doi: 10.15376/biores.13.4.8723-8737
Raad, B. L. (1989). Modern trends in scientific terminology: morphology and metaphor. Am. Speech 64, 128–136. doi: 10.2307/455039
Rafique, M., Ortas, I., Ahmed, I. A. M., Rizwan, M., Siddique, M., Sultan, T., et al. (2019). Potential impact of biochar types and microbial inoculants on growth of onion plant in differently textured and phosphorus limited soils. J. Environ. Manage 247, 672–680. doi: 10.1016/j.jenvman.2019.06.123
Rékási, M., Szili-kovács, T., Takács, T., Bernhardt, B., Kovács, R., Kutasi, J., et al. (2019). Improving the fertility of sandy soils in the temperate region by combined biochar and microbial inoculant treatments. Arch. Agron. Soil Sci. 65, 44–57. doi: 10.1080/03650340.2018.1482536
Santos, M. S., Nogueira, M. A., and Hungria, M. (2019). Microbial inoculants: reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 9:205. doi: 10.1186/s13568-019-0932-0
Shahab, R. L., Luterbacher, J. S., Brethauer, S., Hans,-, M., and Studer, P. (2018). Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal-bacterial consortium. Biotechnol. Bioeng. 115, 1207–1215. doi: 10.1002/bit.26541
Singh, S., Singh, U. B., Malviya, D., Paul, S., Sahu, P. K., Trivedi, M., et al. (2020). Seed biopriming with microbial inoculant triggers local and systemic defense responses against Rhizoctonia solani causing banded leaf and sheath blight in maize (Zea mays L.). Int. J. Environ. Res. Public Health 17:1396. doi: 10.3390/ijerph17041396
Timmis, K., Timmis, J. K., Brussow, H., and Fernandez, L. A. (2019). Crystal ball synthetic consortia of nanobody-coupled and formatted bacteria for prophylaxis and therapy interventions targeting microbiome dysbiosis-associated diseases and co-morbidities. Microb. Biotechnol. 12, 58–65. doi: 10.1111/1751-7915.13355
Tobin, H. M., Lele, S. R., Cutter, C. N., Anantheswaran, R. C., and Laborde, L. F. (2020). Hot water sanitization of a commercial mushroom disk slicer to inactivate Listeria monocytogenes. Food Control 109:106900. doi: 10.1016/j.foodcont.2019.106900
Vandeplassche, E., Sass, A., Lemarcq, A., Dandekar, A. A., Coenye, T., and Crabbé, A. (2019). In vitro evolution of Pseudomonas aeruginosa AA2 biofilms in the presence of cystic fibrosis lung microbiome members. Sci. Rep. 9:12859. doi: 10.1038/s41598-019-49371-y
Venturelli, O. S., Carr, A. V., Fisher, G., Hsu, R. H., Lau, R., Bowen, B. P., et al. (2018). Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14:e8157. doi: 10.15252/msb.20178157
Vindigni, S. M., and Surawicz, C. M. (2017). Fecal microbiota transplantation. Gastroenterol. Clin. North Am. 46, 171–185. doi: 10.1016/j.gtc.2016.09.012
Voges, M. J. E. E. E., Bai, Y., Schulze-lefert, P., and Sattely, E. S. (2019). Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U.S.A. 116, 12558–12565. doi: 10.1073/pnas.1820691116
Wang, H., Jiang, K., Zhu, Z., Jiang, W., Yang, Z., Zhu, S., et al. (2018). Optimization of fed-batch fermentation and direct spray drying in the preparation of microbial inoculant of acetochlor-degrading strain Sphingomonas sp. DC-6. 3 Biotech 8:294. doi: 10.1007/s13205-018-1324-x
Wang, S., Zhang, L., Qi, L., Liang, H., Lin, X., Li, S., et al. (2020). Effect of synthetic microbial community on nutraceutical and sensory qualities of kombucha. Int. J. Food Sci. Technol. 55, 3327–3333. doi: 10.1111/ijfs.14596
Wei, Z., Gu, Y., Friman, V., Kowalchuk, G. A., Xu, Y., Shen, Q., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw0759
Whipps, J., Lewis, K., and Cooke, R. (1988). “Mycoparasitism and plant disease control,” in Fungi Biol Control Syst., ed M. Burge (Manchester University Press, 161–187.
Zahn, G., and Amend, A. S. (2017). Foliar microbiome transplants confer disease resistance in a critically- endangered plant. PeerJ 5:e4020. doi: 10.7717/peerj.4020
Keywords: microbiome, microbial consortium, microbial inoculant, synthetic microbiome, synthetic community, microbial cocktail
Citation: Del Frari G and Ferreira RB (2021) Microbial Blends: Terminology Overview and Introduction of the Neologism “Skopobiota”. Front. Microbiol. 12:659592. doi: 10.3389/fmicb.2021.659592
Received: 27 January 2021; Accepted: 07 June 2021;
Published: 02 July 2021.
Edited by:
Franz Baumdicker, University of Tübingen, GermanyReviewed by:
Davina Höll, Eberhard Karls University, GermanyJohannes Sikorski, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), Germany
Copyright © 2021 Del Frari and Ferreira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Del Frari, Z2RlbGZyYXJpQGlzYS51bGlzYm9hLnB0
 Giovanni Del Frari
Giovanni Del Frari Ricardo Boavida Ferreira
Ricardo Boavida Ferreira