- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 3School of Science, Mae Fah Luang University, Chiang Rai, Thailand
- 4Department of Health Sciences, Faculty of Medicine and Health Sciences, University of Mauritius, Réduit, Mauritius
- 5Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Sciences, Chiang Mai University, Chiang Mai, Thailand
- 6CIFOR-ICRAF China Program, World Agroforestry (ICRAF), Kunming, China
- 7Honghe Center for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Sciences, Honghe, China
- 8Centre for Mountain Futures (CMF), Kunming Institute of Botany, Kunming, China
- 9Engineering Research Center of Chinese Ministry of Education for Edible and Medicinal Fungi, Jilin Agricultural University, Changchun, China
- 10Innovative Institute of Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 11Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 12Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Immotthia is a poorly known genus, and currently, no DNA sequence data are available to ascertain its proper phylogenetic placement and evolutionary relationships with other bitunicate fungi. To date, there are only two species accepted in the genus. During our ongoing research study of bambusicolous fungi in southwest China and Thailand, a fungus associated with stromata of Hypoxylon sp. was found on dead bamboo culms in Loei Province, Thailand. Preliminary morphological identification revealed that the fungal collection belongs to Immotthia. A novel species, Immotthia bambusae, is introduced herein based on a comparison of morphological characteristics with the type specimen of I. hypoxylon (≡ Amphisphaeria hypoxylon Ellis and Everh.), a synonym of I. atrograna (Cooke and Ellis) M. E. Barr. Phylogenetic analyses of a concatenated ITS, LSU, SSU, and TEF1-α DNA sequence matrix showed that Immotthia belongs to Dictyosporiaceae, Pleosporales. Despite I. bambusae strains constituting a supported subclade, they are nested with the genus Pseudocoleophoma. Pseudocoleophoma clematidis is morphologically different from all other Pseudocoleophoma species, while its conidial characteristics are similar to Cyclothyriella. Multigene phylogenetic analyses showed that P. clematidis formed a clade basal to Immotthia, separated from Pseudocoleophoma with strong statistical support. Therefore, we introduce a monotypic genus, Pseudocyclothyriella Phukhams. and Phookamsak, gen. nov. to accommodate the single species, Pseudocyclothyriella clematidis (Phukhams. and K. D. Hyde) Phukhams. and Phookamsak, comb. nov. Detailed descriptions, color micrographs, and phylogenetic trees to show the placement of the new taxa are provided. In addition, an updated taxonomic treatment of the genera Immotthia and Pseudocyclothyriella is also provided based on the study of the type materials and phylogeny generated from DNA sequence data.
Introduction
Immotthia was introduced by Barr (1987a) with I. hypoxylon (Ellis and Everh.) M. E. Barr (≡ Amphisphaeria hypoxylon Ellis and Everh.) as the type species. Through examinations of the type material of I. hypoxylon and Australian collections of I. atrograna (Cooke and Ellis) M. E. Barr (≡ Sphaeria atrograna Cooke and Ellis), Jaklitsch et al. (2002) concluded that these two taxa are conspecific. To date, two species are accepted in this genus, viz. I. atrograna and I. atroseptata (Piroz.) M. E. Barr (Species Fungorum, 2020) based on morphology, but no DNA sequence data are available to confirm their phylogenetic placement (Hyde et al., 2017; Doilom et al., 2018).
Immotthia is characterized by small- to medium-sized, globose to subglobose ascomata, forming on blackened hypostroma, bitunicate, fissitunicate, cylindrical asci, and brown to reddish brown, ellipsoidal to fusiform, 1-septate, smooth or slightly verrucose ascospores (Hyde et al., 2017; Hongsanan et al., 2020). The asexual morph of Immotthia has been reported as coelomycetous, identified as Coniothyrium parasitans (Berk. and Ravenel) Tassi which formed enteroblastic, phialidic, doliiform to ampulliform, or cylindrical, smooth, hyaline conidiogenous cells bearing brown, ellipsoidal, smooth, and aseptate conidia (Hyde et al., 2017; Hongsanan et al., 2020). However, the link between Immotthia and C. parasitans has not yet been proven based on DNA sequence analyses. Immotthia has been reported as hyperparasites on stromata of Annulohypoxylon, Hypoxylon, and Pestalopezia, or forms compressed ascostromata on decorticated wood (Cooke and Ellis, 1879; Ellis and Everhart, 1886; Pirozynski, 1973; Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Hongsanan et al., 2020).
Immotthia was assigned to Dacampiaceae (Pleosporales, Dothideomycetes) by Barr (1987a, b) and this taxonomic treatment was followed by Akulov and Hayova (2016). Barr (2002) transferred non-lichenicolous genera from the Dacampiaceae to Teichosporaceae (Pleosporales, Dothideomycetes), where Immotthia was also included along and transferred to Teichosporaceae. Recently, Immotthia was tentatively placed in Roussoellaceae (Pleosporales, Dothideomycetes) based on similar morphological features of the asci, ascospores, and coelomycetous asexual morph which largely resemble taxa in Roussoellaceae (Hyde et al., 2017; Doilom et al., 2018; Hongsanan et al., 2020).
A well-studied genus Pseudocoleophoma Kaz. Tanaka and K. Hiray. was introduced by Tanaka et al. (2015) to accommodate two novel species having asexual morphology similar to Coleophoma from Japan. The genus is characterized by scattered to clustered, immersed to erumpent, globose to subglobose ascomata, with ostiolar neck, thin-walled peridium, composed of brown to dark brown, polygonal to rectangular cells, cylindrical to clavate, fissitunicate asci, with short-pedicellate, and hyaline, fusiform, 1-septate ascospores, with a conspicuous sheath (Tanaka et al., 2015; Jayasiri et al., 2019). Pseudocoleophoma has coelomycetous, coleophoma-like asexual morphs forming pycnidial, subglobose conidiomata, phialidic, doliiform to lageniform conidiogenous cells, and cylindrical or oblong, hyaline, aseptate, smooth-walled conidia (Tanaka et al., 2015). The genus belongs to Dictyosporiaceae based on phylogenetic evidence (Tanaka et al., 2015).
Two holomorphic species, Pseudocoleophoma calamagrostidis Kaz. Tanaka and K. Hiray. and P. polygonicola Kaz. Tanaka and K. Hiray., were initially accommodated in this genus (Tanaka et al., 2015). Later, six other species were accommodated based on morphological and phylogenetic support, viz. P. bauhiniae Jayasiri, E. B. G. Jones and K. D. Hyde (on Bauhinia sp., Thailand); P. clematidis Phukhams. and K. D. Hyde (on Clematis vitalba, Italy); P. flavescens (Gruyter, Noordel. and Boerema) W. J. Li and K. D. Hyde (from soil, rhizosphere of Solanum tuberosum, Netherlands); P. rusci W. J. Li, Camporesi and K. D. Hyde (on Ruscus aculeatus, Italy); P. typhicola Kamolhan, Banmai, Boonmee, E. B. G. Jones and K. D. Hyde (on decaying submerged Typha latifolia, Great Britain); and P. zingiberacearum Tennakoon, D. J. Bhat, C. H. Kuo and K. D. Hyde (on Hedychium coronarium, Taiwan) (Tanaka et al., 2015; Hyde et al., 2016; Jayasiri et al., 2019; Tennakoon et al., 2019; Li et al., 2020; Phukhamsakda et al., 2020).
Most of the Pseudocoleophoma species have been represented by their asexual morphs (Hyde et al., 2016; Tennakoon et al., 2019; Li et al., 2020; Phukhamsakda et al., 2020). Only three species have been reported for both sexual and asexual morphs, viz. P. bauhiniae, P. calamagrostidis, and P. polygonicola (Tanaka et al., 2015; Jayasiri et al., 2019). Currently, species of Pseudocoleophoma are only known from Europe (Great Britain, Italy, and Netherlands) and Asia (Taiwan and Thailand), and they were found as saprobes on various hosts and substrates from both terrestrial and freshwater habitats (Tanaka et al., 2015; Hyde et al., 2016; Jayasiri et al., 2019; Tennakoon et al., 2019; Li et al., 2020; Phukhamsakda et al., 2020).
In the present study, a fresh collection of Immotthia is examined and compared with other Immotthia species based on morphological characteristics. The new collection is described as a novel species in Immotthia and illustrated. Through DNA sequencing of the fresh material, we also resolved the phylogenetic placement of Immotthia for the first time, based on maximum likelihood and Bayesian inference analyses. In addition, the novel genus Pseudocyclothyriella is also introduced to accommodate Pseudocyclothyriella clematidis comb. nov. (≡ Pseudocoleophoma clematidis) based on morphological distinctiveness and multigene phylogenetic analyses.
Materials and Methods
Sample Collection, Specimen Examination, and Preservation
Dead bamboo culms were collected from Loei Province, Thailand, in 2020. The specimens were kept in a paper bag and returned to the laboratory for observation and examination. Fungal fruiting bodies on the host substrate were observed with a Motic SMZ 140 series dissecting stereoscope, and a centrum was mounted in sterilized distilled water for morphological examination and captured using a Nikon ECLIPSE Ni compound microscope connected with a Canon EOS 600D digital camera. Tarosoft (R) Image Frame Work version 0.9.7 was used to measure the size of ascomata, peridium, pseudoparaphyses, asci, and ascospores. In addition, holotypic specimens of Immotthia atroseptata [United States, North Carolina, behind N. C. Department of Agriculture, Nursery Inspection Station, 1 mile west of Linville, Avery Co., on apothecia of Pestalopezia rhododendri on the leaves of Rhododendron maximum L. (Ericaceae), March 21, 1972, Neli Lapp DAOM 139001], I. hypoxylon (United States, Louisiana, Plaquemines Parish, on decaying wood, December 30, 1885, A. B. Langlois 138, NY0083004), and Pseudocoleophoma clematidis (Italy, Arezzo Province, Badia Tega—Ortignano Raggiolo, on dead aerial branch of Clematis vitalba, March 9, 2013, E. Camporesi, IT 1110, MFLU 16-0280) were also re-examined and illustrated. Adobe Photoshop CS6 software (Adobe Systems Inc., United States) was used to edit and provide the photographic plates based on captured pictures of the fungal structures. Good practices for morphological examinations as outlined by Senanayake et al. (2020b) were followed for the morphological study, while phylogenetic methods as outlined by Dissanayake et al. (2020) were followed for phylogenetic analyses. The holotype is deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), Yunnan, China. The isotype was stored in Herbarium Mycologicum Academiae Sinicae (HMAS), Beijing, China. The Facesoffungi and Index Fungorum numbers are registered for the newly described taxa (Jayasiri et al., 2015; Index Fungorum, 2020). New species are established based on the guidelines provided by Jeewon and Hyde (2016).
DNA Extraction, PCR Amplification, and Sequencing
Fungal genomic DNA was extracted from fruiting bodies using Forensic DNA Kit (Omega®, United States) following the manufacturer’s instructions and protocols outlined by Doilom et al. (2017) and Zeng et al. (2018). DNA was extracted from five duplicates of different fruiting bodies on the holotypic specimen of Immotthia bambusae (KUN-HKAS 112012) to allow verification of correct DNA sequence data. DNA amplification was performed by polymerase chain reaction (PCR). The primer pairs ITS5/ITS4 (White et al., 1990), LR0R/LR5 (Vilgalys and Hester, 1990), NS1/NS4 (White et al., 1990), and EF1-983F/EF1-2218R (Rehner, 2001) were used to amplify the PCR fragments of the internal transcribed spacers (ITS1-5.8S-ITS2), the 28S large subunit rDNA (LSU), the 18S small subunit rDNA (SSU), and the translation elongation factor 1-alpha (TEF1-α), respectively. PCR reactions were conducted in 25 μl total volume containing 2 μl of fungal genomic DNA template, 1 μl of each forward and reverse primer, 12.5 μl of 2 × Power Taq PCR Master Mix (mixture of EasyTaqTM DNA Polymerase, dNTPs, and optimized buffer; Beijing BioTeke Corporation, China), and 8.5 μl of sterilized double-distilled water (ddH2O). The PCR thermal cycle profiles for ITS, LSU, SSU, and TEF1-α gene were set up following Jiang H. B. et al. (2020). PCR products were sent to TsingKe Biological Technology (Beijing) Co., Ltd., China, for PCR purification and sequencing.
Alignment and Phylogenetic Analyses
The newly generated ITS, LSU, SSU and TEF1-α sequences of the new taxon were subjected to the BLASTn search tool1 for initial verification and search of reference taxa for further analyses. Similarity indices based on BLASTn search showed that five new strains are closely related to Pseudocoleophoma Kaz. Tanaka and K. Hiray (Dictyosporiaceae, Pleosporales). In order to investigate the phylogenetic status of the new taxa, a combined dataset of taxa including members of the Dictyosporiaceae was analyzed based on DNA sequence data available in recent publications (Iturrieta-González et al., 2018; Yang et al., 2018; Hyde et al., 2020a, b; Phukhamsakda et al., 2020). DNA sequences of representative taxa used are shown in Table 1. Individual DNA sequence alignments were initially performed via the online platform MAFFT v. 7.4742 (Katoh et al., 2019) and were improved manually using BioEdit v. 5.0.6 (Hall, 2001). Preliminary phylogenetic analyses of a concatenated LSU–SSU–TEF1-α–RPB2–ITS sequence matrix represented the relationships of Immotthia in Dictyosporiaceae with other families in Pleosporales (Supplementary Figure 3), and a concatenated ITS–LSU–TEF1-α sequence dataset (Supplementary Figure 1) was also analyzed by maximum-likelihood (ML) analysis via the web portal CIPRES Science Gateway v. 3.3 (Miller et al., 2010), with the help of the tool RAxML-HPC v.8 on XSEDE (8.2.12).
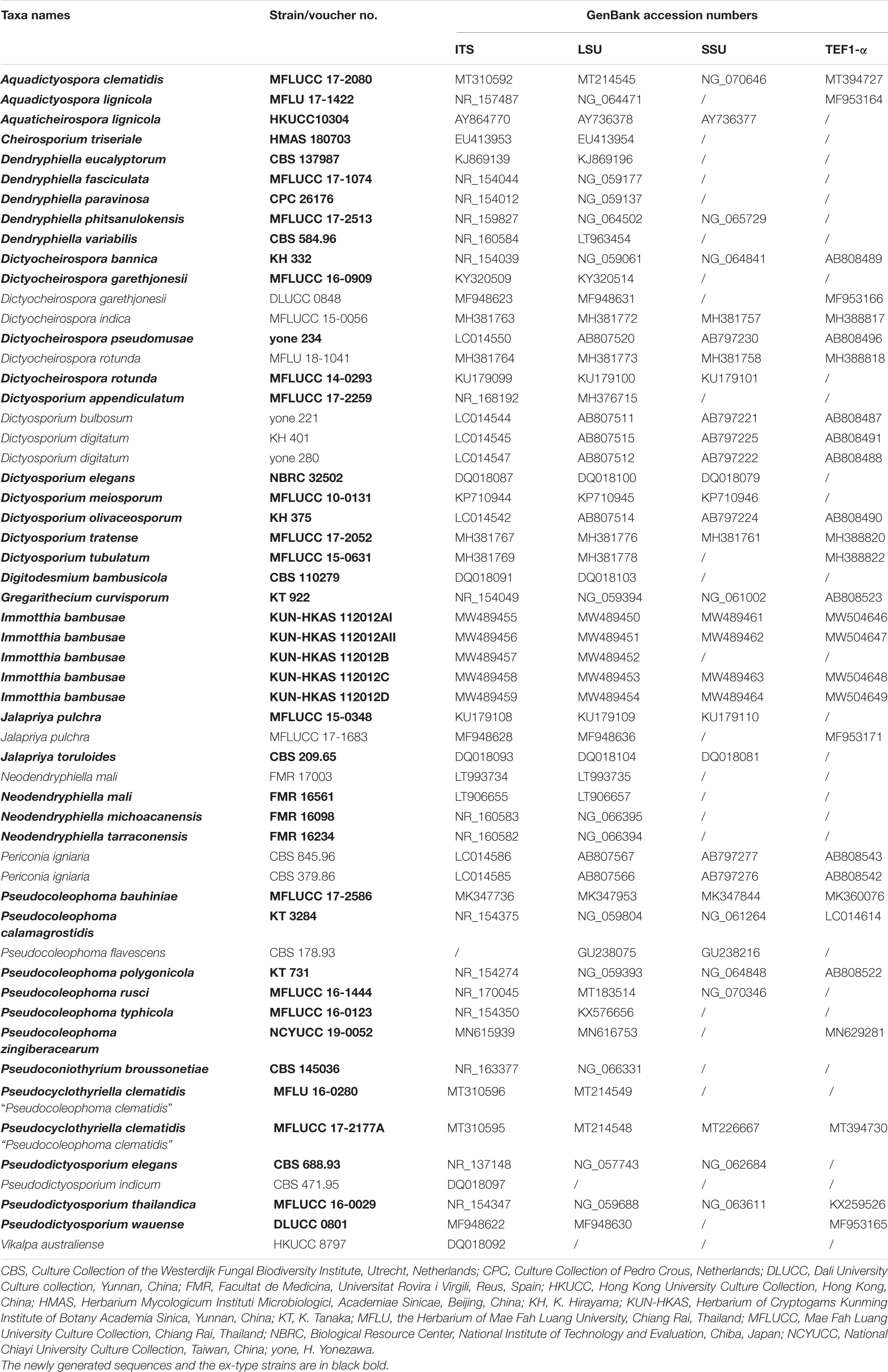
Table 1. Taxa names, strain numbers, and GenBank accession numbers of taxa used for the present phylogenetic analyses.
ML analysis using the GAMMA model of nucleotide substitution was performed via the web portal CIPRES Science Gateway v.3.3 (Miller et al., 2010), with the help of the tool RAxML-HPC v.8 on XSEDE (8.2.12). The evolutionary model of nucleotide substitution for Bayesian inference (BI) analysis was selected independently for each locus using MrModeltest 2.3 (Nylander, 2008). GTR + I + G is the best fit for ITS and LSU loci under the Akaike information criterion (AIC), while the HKY + I substitution model is the best fit for SSU and TEF1-α loci. BI analysis was carried out by MrBayes v. 3.2.6 (Ronquist and Huelsenbeck, 2003). Markov chain Monte Carlo sampling (MCMC) was used to decide posterior probabilities (PP) (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002). Six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled every 100th generation. The 0.15 “temperature” value was set in the MCMC heated chain. All sampled topologies beneath the asymptote (20%) were discarded as part of a burn-in procedure, and the remaining 8,000 trees were used for calculating posterior probabilities (PP) in the 50% majority rule consensus tree (when the split frequency was lower than 0.01). Tree topology of BI analysis is represented in Supplementary Figure 2.
Tree topologies generated in this study were visualized on FigTree v. 1.4.03, and Microsoft Office PowerPoint 2016 (Microsoft Inc., United States) was used to edit and redraw the phylogram. New sequences generated from the present study are deposited in GenBank (Table 1). The final alignment and phylogram were submitted in TreeBASE4.
Results
Taxonomy
Immotthia M. E. Barr, Mycotaxon 29: 504 (1987a), Figure 1.

Figure 1. Morphological characteristics of Immotthia. (c,d,g,h,j,k,q–s) Immotthia atroseptata (DAOM 13900; holotype of Didymosphaeria atroseptata). (a,b,e,f,i–k,n–p) Immotthia hypoxylon (NY00830041, holotype of Amphisphaeria hypoxylon). (a,c) Herbarium label and specimens. (b,d) Appearance of ascomata on host substrates. (e–g) Section through ascomata. (h) Exterior of ascoma. (i) Pseudoparaphyses. (j–m) Asci. (n–s) Ascospores. Scale bars: (e,f) = 100 μm, (g,h) = 30 μm, (i–m) = 20 μm, and (n–s) = 5 μm.
Index Fungorum Number: IF 25106
Facesoffungi Number: FoF 08362
Hyperparasitic on Annulohypoxylon, Hypoxylon, and Pestalopezia, or saprobic on decorticated wood. Sexual morph: Hypostroma dark brown to black, crust under ascomata, patch-like, composed of thick-walled cells of textura angularis, covering the surface of host stromata, difficult to distinguished from the peridium of ascomata, sometimes forming dark brown hyphae at the lowest layer penetrated with the host of Hypoxylon which was interpreted to belong to the Hypoxylon host. Ascomata dark brown to black, subglobose to obpyriform, or inequilateral, gregarious to densely aggregated, superficial, uniloculate, or carbonaceous when dry, glabrous, surface rough with protruding cells, arising from a large hypostroma, usually with a pore-like, inconspicuous ostiole or minute papilla. Peridium thin- to thick-walled, of unequal thickness, composed of several layers, inner layers comprising hyaline to dark brown, pseudoparenchymatous cells, of textura angularis, outer layers composed of thick, dark brown to black cells, arranged in a textura angularis. Hamathecium composed of dense, septate, branched, anastomosing, cellular pseudoparaphyses embedded in a gelatinous matrix. Asci (4–) (6–)–8-spored, bitunicate, fissitunicate, cylindrical or cylindric-clavate, subsessile to short-pedicellate with obtuse knob-like or furcate pedicel, apically rounded with a well-developed ocular chamber. Ascospores overlapping 1–2-seriate, pale yellowish to brown, or reddish brown, ellipsoidal to fusiform, with rounded ends, 1-septate, asymmetrical, with the upper cell slightly larger than the lower cell, slightly constricted at the septum, smooth- to rough-walled, verrucose. Asexual morph: Coelomycetous, chaetophoma-, coniothyrium-, microsphaeropsis-, or pyrenochaeta-like, associated with the sexual morph on natural substrate. Conidiomata pycindial, similar to ascomata but differ in having a smaller size, black, carbonaceous, immersed to erumpent, becoming superficial, globose to obpyriform, uni- to multiloculate, glabrous, with indistinct ostiolate. Pycnidial wall thin- to thick-walled, composed of several layers of brown to dark brown pseudoparenchymatous cells, of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, phialidic, discrete, determinate, ampulliform, or cylindric, smooth, hyaline, with minute collarette and conspicuous periclinal thickening. Conidia ellipsoidal, rounded at both ends, one-celled, at first hyaline, becoming brown at maturity, smooth-walled or finely verrucose (adapted from Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Doilom et al., 2018).
Type Species: Immotthia hypoxylon (Ellis and Everh.) M. E. Barr, Mycotaxon 29: 504 (1987a).
≡ Amphisphaeria hypoxylon Ellis and Everh., J. Mycol. 2(4): 41 (1886).
Current Name: Immotthia atrograna (Cooke and Ellis) M. E. Barr (1993), Mycotaxon 46: 71.
≡ Sphaeria atrograna Cooke and Ellis, Grevillea 8(no. 45): 15 (1879).
Life Mode and Known Distribution: Hyperparasitic on Annulohypoxylon, Hypoxylon, and Pestalopezia on various host substrates and saprobic on decaying wood. Immotthia is presently known from Austria, Belgium, China, France, Lithuania, Norway, Poland, Puerto Rico, Russia, Sweden, Switzerland, Ukraine, United States, and Venezuela (Pirozynski, 1973; Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Doilom et al., 2018; Farr and Rossman, 2020).
Notes: The phylogenetic affinities of Immotthia have never been investigated in previous studies, and the genus was accommodated in different families by different authors based only on morphological characteristics (Barr, 1987a, 2002; Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Doilom et al., 2018). In the present study, Immotthia is phylogenetically close to Pseudocoleophoma in Dictyosporiaceae. The sexual morph of Immotthia differs from Pseudocoleophoma in morphology and habitat. Immotthia forms dense, superficial ascomata on hypostoma, with cylindrical to cylindric-clavate asci, ellipsoidal to fusiform, light brown to reddish brown, asymmetrical ascospores, lacking mucilaginous sheath and usually found as hyperparasites on hypoxylon-like stroma. On the other hand, Pseudocoleophoma forms scattered or in groups, immersed to erumpent ascomata, with cylindrical to clavate asci, fusiform, hyaline ascospores, surrounded by mucilaginous sheath, and mostly found as saprobes in a terrestrial environment (Tanaka et al., 2015; Jayasiri et al., 2019; Tennakoon et al., 2019; Li et al., 2020). The asexual morph of Immotthia differs from Pseudocoleophoma in having multiloculate, carbonaceous conidiomata and ellipsoidal, brown, aseptate conidia (Hyde et al., 2017), whereas Pseudocoleophoma has uniloculate conidiomata and cylindrical to subcylindrical or fusiform, hyaline, 0–1-septate conidia.
Immotthia bambusae H. B. Jiang and Phookamsak, sp. nov., Figure 2.
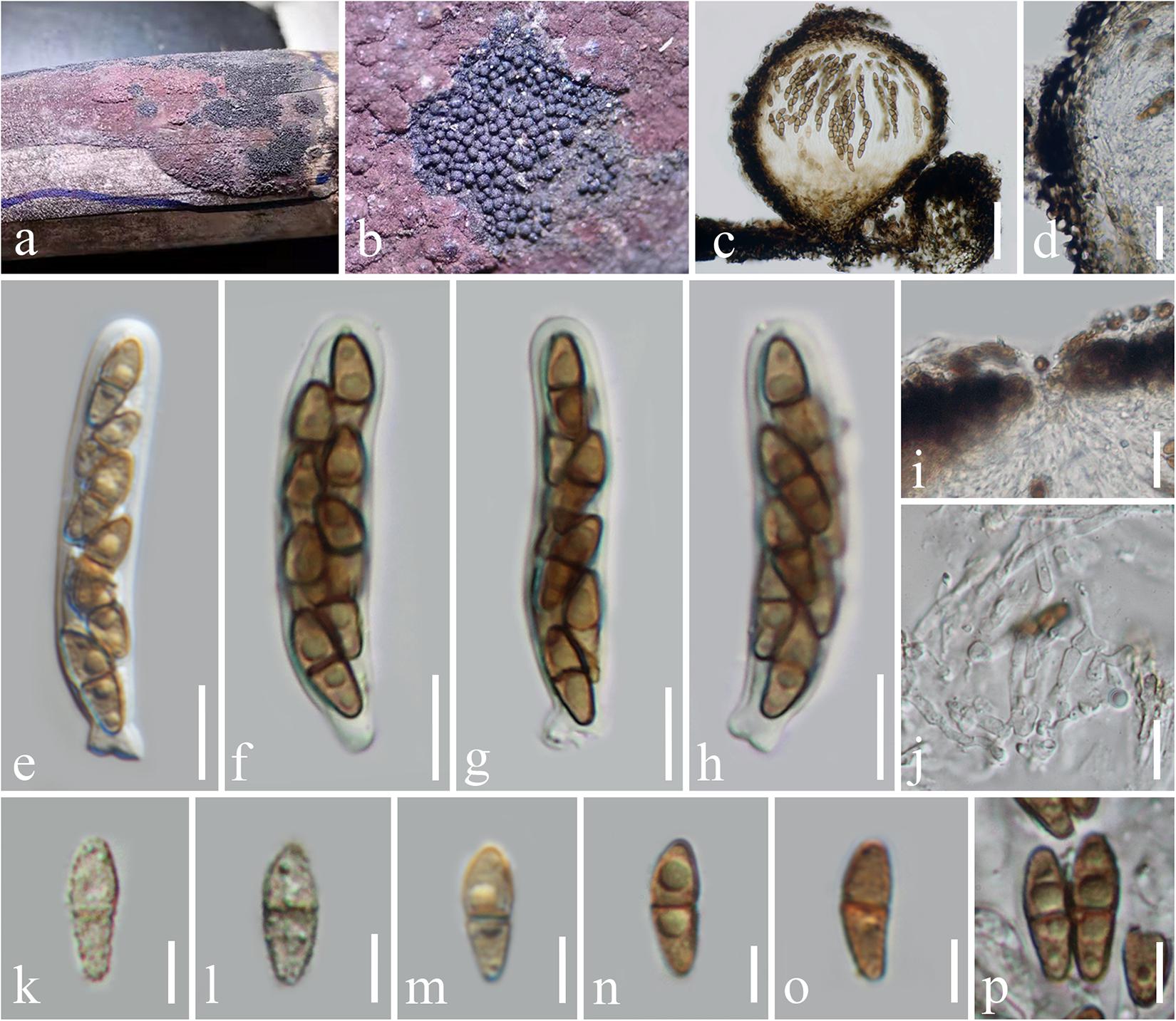
Figure 2. Immotthia bambusae (KUN-HKAS 112012, holotype). (a,b) Appearance of hypostromata on Hypoxylon sp. associated with dead bamboo culms. (c) Vertical section of ascoma. (d) Peridium. (e–h) Asci. (i) Pore-like ostiole. (j) Pseudoparaphyses. (k–p) Ascospores. Scale bars: (c) = 50 μm, (d) = 20 μm, (i) = 15 μm, (e–h,j) = 10 μm, and (k–p) = 5 μm.
Index Fungorum Number: IF 557242
Facesoffungi Number: FoF 09538
Etymology: The specific epithet “bambusae” refers to the host, bamboo, of which the new species was collected.
Holotype: KUN-HKAS 112012
Hyperparasitic on Hypoxylon sp. on dead bamboo culms. Sexual morph: Hypostroma effuse, black, with numerous, superficial ascomata, composed of thin layered, of blackened, pseudoparenchymatous cells of textura angularis. Ascomata 130–210 μm high, 150–220 μm diam., dark brown to black, scattered, gregarious, globose to subglobose, or obpyriform, arising from the hypostroma, carbonaceous, brittle when dry, easily dispersed, uniloculate, glabrous, rough-walled, with a pore-like, inconspicuous ostiole. Peridium 15–30 μm wide, of unequal thickness, slightly thick at the base, composed of several layers of pseudoparenchymatous cells, outer layers composed of thick-walled, blackened cells, arranged in textura angularis to textura globulosa, inner layers composed of flattened, brown cells, of textura angularis to textura prismatica, paler toward the inner layers. Hamathecium composed of dense, 1.5–2 μm broad, cellular pseudoparaphyses, septate, branched, anastomosing among the asci, embedded in gelatinous matrix. Asci 45–60(–62) × 7.5–10 μm (x̄ = 53 × 9 μm, n = 15), (6–)8-spored, bitunicate, fissitunicate, cylindrical, subsessile, with knob-like or furcate pedicel, apically rounded with well-developed ocular chamber. Ascospores 9–11 × 3–4.5 μm (x̄ = 10 × 3.7 μm, n = 20), overlapping 1–2-seriate, light brown to brown, ellipsoidal to fusiform, with rounded ends, lower cell slightly longer and narrower than the upper, 1-septate, rough-walled, finely verrucose, initially presented small to large guttules, disappeared when mature, without sheath. Asexual morph: Undetermined.
Material Examined: Thailand, Loei Province, a wild bamboo forest (17°15′44.68″N, 101°8′39.32″E, altitude 364.09 m), on Hypoxylon sp. associated with dead culms of bamboo, February 25, 2020, H. B. Jiang and R. Phookamsak, BBL07-2-1 (KUN-HKAS 112012, holotype; HMAS 249866, isotype).
Life Mode and Known Distribution: Hyperparasitic on Hypoxylon sp. associated with bamboo culms (Thailand).
Notes: The type specimens of all Immotthia species were compared; I. bambusae shows the most similarity to I. hypoxylon, a synonym of I. atrograna. However, the novel taxon differs from I. atrograna (= I. hypoxylon) in having a smaller size of ascomata, asci, and ascospores. The lower cell of ascospores of I. bambusae is longer and narrower than the upper cell, while in I. atrograna (= I. hypoxylon), the upper cell of ascospores is larger than the lower cell. Immotthia bambusae was collected on the stromata of Hypoxylon associated with bamboo in Thailand (tropical region), whereas the type specimen of I. hypoxylon was collected from Hypoxylon truncatum colonizing dead canes of Rosa in Louisiana, United States (subtropical region). The other different features between I. bambusae and the other Immotthia species are provided in Table 2. Therefore, I. bambusae is introduced as a new species in this study based on both morphology and multigene phylogeny.
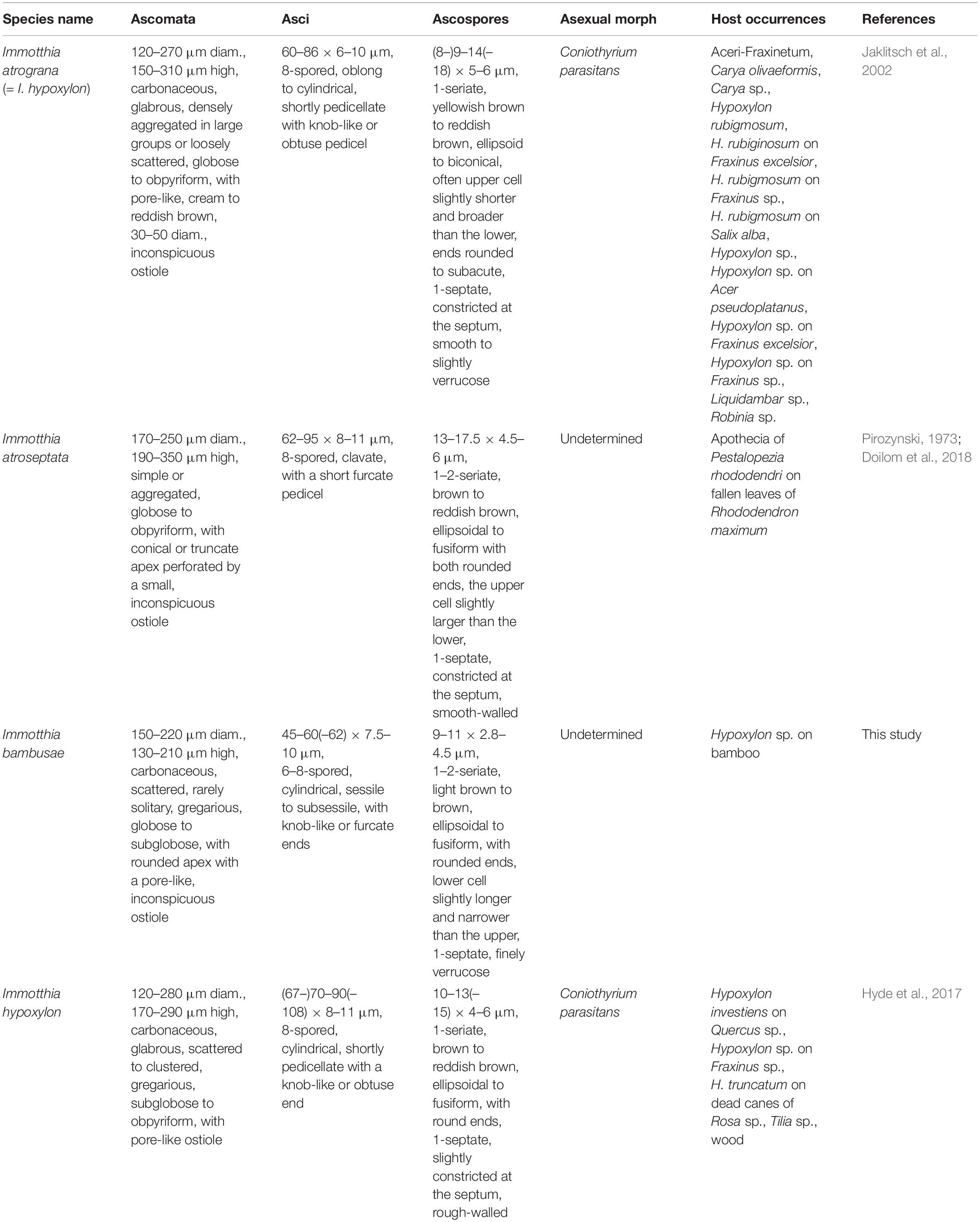
Table 2. Synopsis of Immotthia species based on type studies (I. hypoxylon is listed separately from I. atrograna in this study for a better understanding).
In this study, we sequenced five different fruiting bodies and DNA sequence similarity (ITS regions) revealed that they have identical nucleotides (100% similarity). Even our phylogeny depicts a close relationship (100% support) among these five strains. This ensures the correctness of the new generated sequences from the direct DNA extraction of fruiting bodies.
Pseudocyclothyriella Phukhams. and Phookamsak, gen. nov.
Index Fungorum Number: IF 557441
Facesoffungi Number: FoF 09539
Etymology: The generic epithet “Pseudocyclothyriella” refers to the resemblance of conidial morphology of the new genus to the genus Cyclothyriella.
Saprobic on Clematis vitalba (Ranunculaceae). Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial, solitary to gregarious, uniloculate, immersed to erumpent, laterally becoming superficial, visible as black, shiny on host, subglobose to subconical, coriaceous, subcoriaceous at the outer layers, glabrous, dark brown to black, ostiolate, papillate. Ostioles central, ovoid, with minute papilla, filled with hyaline periphyses. Pycnidial wall thick-walled of equal thickness, composed of multilayered scleroplectenchymatous cells, outer layer composed of several layers of thick-walled, dark brown to black cells of textura angularis to textura globulosa, inner layer composed of hyaline to pale brown cells, bearing conidiogenous cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, phialidic, determinate, discrete, cylindrical to subcylindrical, or ampulliform, hyaline, aseptate, smooth-walled, arising from the inner layers of conidioma. Conidia oval to oblong, hyaline to yellowish brown, slightly curved toward the ends, aseptate, smooth-walled.
Type Species: Pseudocyclothyriella clematidis (Phukhams. and K. D. Hyde) Phukhams. and Phookamsak.
Life Mode and Known Distribution: Pseudocyclothyriella is reported as a saprobe on Clematis vitalba (Ranunculaceae). The genus is presently known from Italy (Phukhamsakda et al., 2020).
Notes: Based on morphological distinctiveness and multigene phylogenetic analyses, a monotypic genus Pseudocyclothyriella is introduced herein to accommodate a single coelomycetous species, P. clematidis which was previously described as Pseudocoleophoma clematidis by Phukhamsakda et al. (2020). Pseudocyclothyriella formed an independent clade basal to Immotthia and Pseudocoleophoma with strong statistical supports (92% ML, 0.98 PP). Pseudocyclothyriella is similar to Cyclothyriella in having cylindrical, oblong to ellipsoid, aseptate, hyaline to pigmented conidia (Jaklitsch and Voglmayr, 2016). However, Pseudocyclothyriella can be distinguished from Cyclothyriella based on the conidiomatal characteristics and phylogenetic evidence. Pseudocyclothyriella is characterized by solitary to gregarious, immersed to erumpent, black, shiny, subglobose to subconical conidiomata, with oval, papilla, ostiolar canal, and pycnidial wall composed of thick-walled, scleroplectenchymatous cells. On the other hand, Cyclothyriella has black, more or less globose pycnidia, clustered in valsoid configuration, with brightly colored, disc-like ostiole, and pycnidial wall composed of pseudoparenchymatous cells (Jaklitsch and Voglmayr, 2016). Cyclothyriella belongs to its own family Cyclothyriellaceae, whereas Pseudocyclothyriella belongs to Dictyosporiaceae.
Pseudocoleophoma differs from Pseudocyclothyriella in pycnidial wall composed of thin-walled, brown to dark brown pseudoparenchymatous cells and oblong, cylindrical to subcylindrical, or rod-shaped, 0–1-septate, smooth-walled conidia (Tanaka et al., 2015; Jayasiri et al., 2019; Tennakoon et al., 2019; Li et al., 2020). Pseudocyclothyriella is morphologically similar to a presumably coniothyrium-like asexual morph of Immotthia in having black, carbonaceous pycnidia, and pigmented, aseptate, smooth-walled conidia (Hyde et al., 2017). However, Immotthia forms pycnidia in groups, or multiloculate conidiostromata associated with the ascomata of Immotthia on natural host substrates and has ellipsoidal conidia as well as having hyperparasitic life mode (Hyde et al., 2017), while Pseudocyclothyriella forms solitary to gregarious, uniloculate pycidia and has oval to oblong conidia as well as having saprobic life mode (Phukhamsakda et al., 2020). The sexual morph of Pseudocyclothyriella is undetermined; thus, the sexual morphologies of Pseudocyclothyriella and Immotthia could not be compared.
Pseudocyclothyriella clematidis (Phukhams. and K. D. Hyde) Phukhams. and Phookamsak, comb. nov., Figure 3
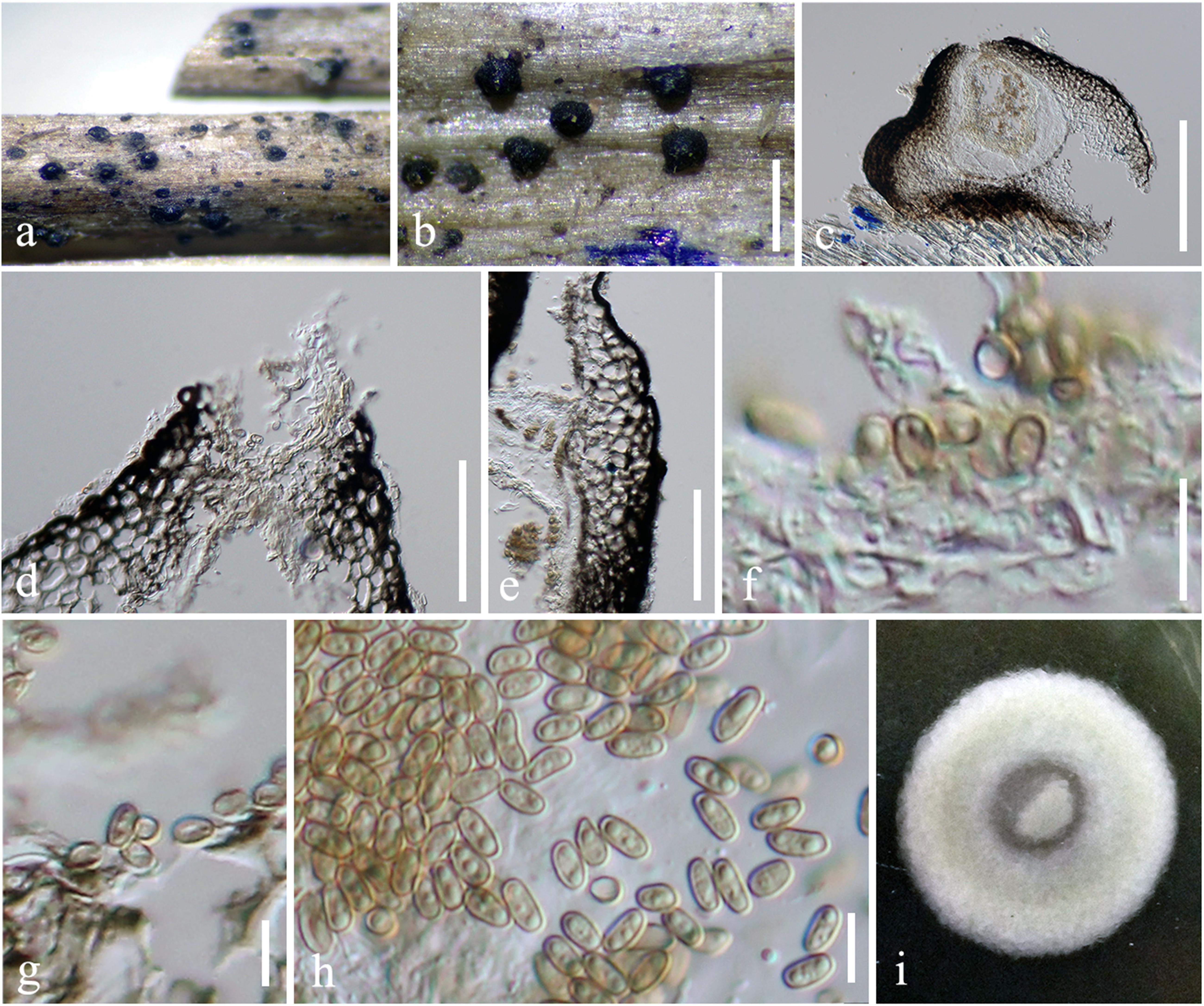
Figure 3. Pseudocyclothyriella clematidis (MFLU 16-0280, holotype). (a,b) Appearance of pycnidial conidiomata on host substrates. (c) Vertical section of conidioma. (d) Ostiole. (e) Pycnidial wall. (f,g) Conidiogenous cells with attached conidia. (h) Conidia. (i) Culture colony on MEA (upper view). Scale bars: (b) = 500 μm, (c) = 200 μm, (d,e) = 20 μm, and (f–h) = 5 μm.
Index Fungorum Number: IF 557442
Facesoffungi Number: FoF 09540
≡ Pseudocoleophoma clematidis Phukhams. and K. D. Hyde, in Phukhamsakda et al., Fungal Divers (102): 21 (2020).
Holotype Details: Italy, Arezzo Province, Badia Tega—Ortignano Raggiolo, on dead aerial branch of Clematis vitalba, March 9, 2013, E. Camporesi, IT 1110 (MFLU 16-0280), ex-type living culture, MFLUCC 17-2177.
Description and Illustration: See Phukhamsakda et al. (2020).
Ecology and Known Distribution: As a saprobe, Pseudocyclothyriella clematidis has contributions in the cycle of the material. To date, the species is just reported in Italy (Phukhamsakda et al., 2020).
Notes: Pseudocyclothyriella clematidis was previously treated in Pseudocoleophoma based on phylogenetic evidence (Phukhamsakda et al., 2020). Pseudocyclothyriella clematidis differs from the other Pseudocoleophoma species in having yellowish brown, oval to oblong, aseptate conidia (Phukhamsakda et al., 2020). A morphological comparison of P. clematidis with other Pseudocoleophoma species is provided in Table 3. The two strains of Pseudocyclothyriella clematidis (MFLU 16-0280 and MFLUCC 17-2177A) formed a strongly supported clade in this study. Phukhamsakda et al. (2020) introduced Pseudocoleophoma clematidis based on phylogenetic evidence and reported the species was poorly supported in a subclade in between the main clade of Pseudocoleophoma and P. typhicola (MFLUCC 16-0123). However, in our multigene phylogeny, we recovered the same taxon as basal to Pseudoconiothyrium broussonetiae.
Crous et al. (2019) mentioned that the closest hits of P. broussonetiae using the ITS sequence had highest similarity to Pseudocoleophoma typhicola (MFLUCC 16-0123). However, P. typhicola is morphologically different from P. broussonetiae, but is typical of Pseudocoleophoma. We rechecked the BLASTn search result based on ITS and LSU sequences of P. typhicola (MFLUCC 16-0123) available in GenBank and noted that the DNA sequences from the ITS regions of P. typhicola are similar to the endophytic Pleosporales sp. isolate MBD_4078 (MK595603) and the uncultured fungus clone ITS_S7_clon2 (HQ873356) with 90.50% similarities and matches with P. broussonetiae strain CBS 145036 (NR_163377) with 90.23% similarity which is far away from Pseudocoleophoma. On the other hand, the BLASTn search result based on LSU sequence showed that the species is closely related with Pseudocoleophoma, and hence, these may be erroneous. In this study, we therefore excluded the ITS sequence of P. typhicola from our aligned sequence dataset and the phylogenetic results showed that P. typhicola clusters within Pseudocoleophoma (Figure 4). However, P. typhicola needs to be resequenced as well as obtaining more reliable genes (only ITS and LSU are available in GenBank) for a better phylogenetic resolution is needed.
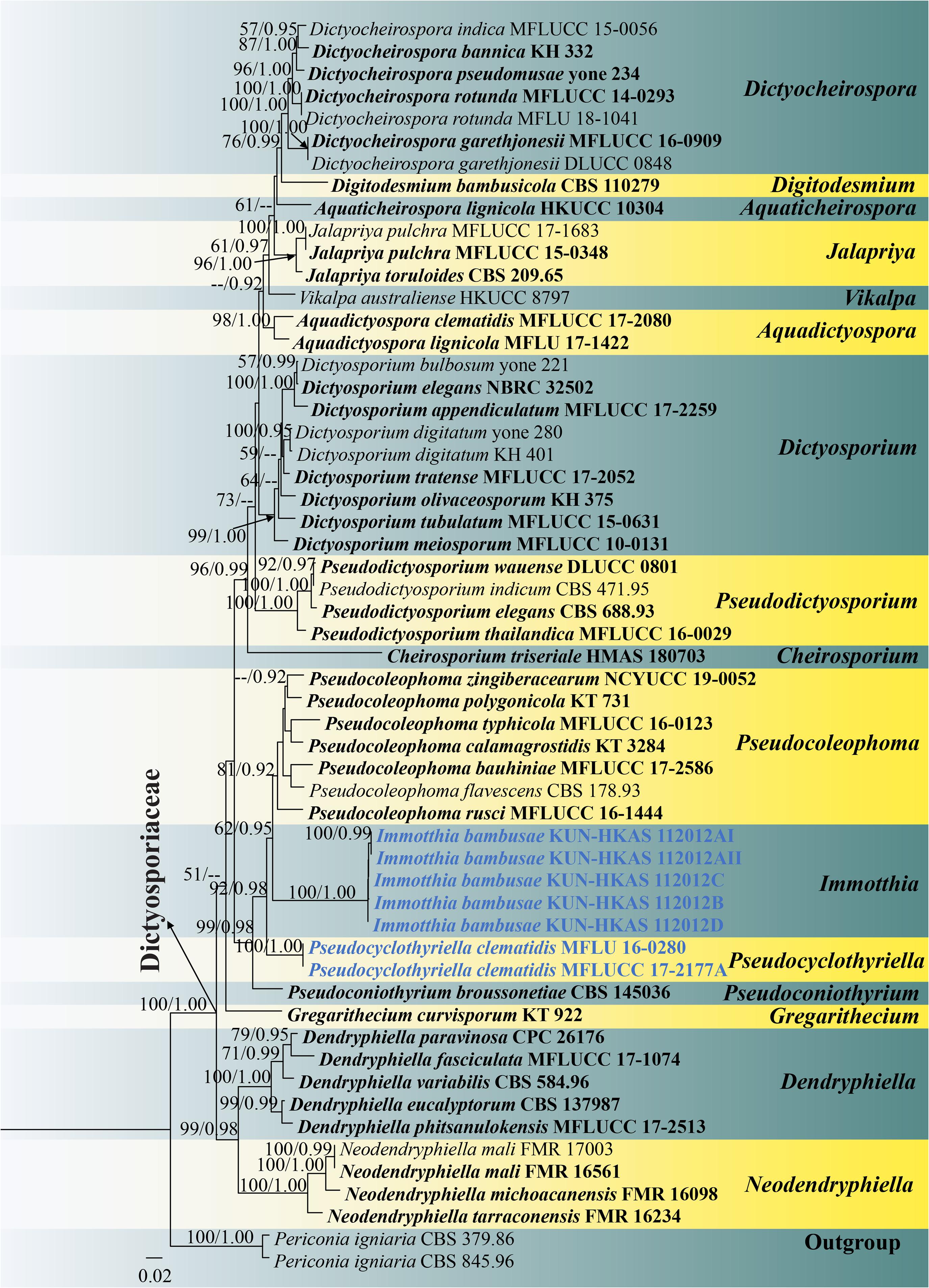
Figure 4. RAxML tree based on a combined ITS, LSU, SSU, and TEF1-α sequence matrix represented the phylogenetic relationships of taxa in Dictyosporiaceae. The tree is rooted to Periconia igniaria (CBS 845.96 and CBS 379.86). Bootstrap support values for ML equal to or greater than 50% and the Bayesian posterior probabilities equal to or higher than 0.90 PP are indicated above the nodes as ML/PP. Ex-type strains are in black bold and the new species and new combinations are indicated in blue bold.
Phylogenetic Analyses
The closest query cover identification of five new strains (KUN-HKAS 112012AI, KUN-HKAS 112012AII, KUN-HKAS 112012B, KUN-HKAS 112012C, and KUN-HKAS 112012D) on BLASTn search tool indicated that our new Immotthia taxon is similar to Pseudocoleophoma rusci (MFLUCC 16-1444), P. calamagrostidis (KT 3284), P. polygonicola (KT 731), and Dictyocheirospora pseudomusae (yone 234) when DNA sequences (ITS, LSU, SSU, and TEF1-α) were compared.
Phylogenetic analyses of a combined ITS, LSU, SSU, and TEF1-α sequence matrix were performed based on 56 strains of taxa in Dictyosporiaceae and two strains of Periconia igniaria (CBS 845.96 and CBS 379.86) as outgroup. This dataset consists of 3,397 total characters including gaps (ITS: 1–585 bp, LSU: 586–1,445 bp, SSU: 1,446–2,473 bp, TEF1-α: 2,474–3,397 bp). The best scoring ML tree was selected to represent the phylogenetic relationships of the new taxon with other representative taxa in Dictyosporiaceae (Figure 4), with the final ML optimization likelihood value of −17,968.180137 (ln). All free model parameters were estimated by GTRGAMMA model, with 1,026 distinct alignment patterns and 33.65% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240488, C = 0.242841, G = 0.269410, and T = 0.247261, with substitution rates AC = 1.638750, AG = 3.305690, AT = 2.424291, CG = 0.898018, CT = 8.545431, and GT = 1.000000. The gamma distribution shape parameter alpha = 0.169318 and the Tree-Length = 1.822628. The final average standard deviation of split frequencies at the end of total MCMC generations was calculated as 0.009979 in BI analysis.
Preliminary phylogenetic analysis based on a concatenated LSU–SSU–TEF1-α–RPB2–ITS sequence matrix of representative families in Pleosporales depicts that Immotthia belongs to the family Dictyosporiaceae (Supplementary Figure 3). The phylograms from ML and BI analyses (Figure 4 and Supplementary Figure 2) of a concatenated ITS–LSU–SSU–TEF1-α sequence matrix were similar in overall topologies and were also similar to the tree topology of a concatenated ITS–LSU–TEF1-α sequence dataset (Supplementary Figure 1). Our five new strains of I. bambusae constitute a strongly supported independent subclade basal to Pseudocoleophoma (62% ML, 0.95 PP). Two strains of Pseudocyclothyriella clematidis (≡ Pseudocoleophoma clematidis, MFLU 16-0280 and MFLUCC 17-2177A) formed an independent subclade basal to Immotthia with strong statistical supports (92% ML, 0.98 PP). Based on current phylogenetic status and morphological distinctiveness compared with the other Pseudocoleophoma species, Pseudocoleophoma clematidis is transferred to the novel genus Pseudocyclothyriella as P. clematidis.
Discussion
An updated taxonomic treatment of bambusicolous fungi has been conducted since the last decade by Japanese mycologists (Tanaka et al., 2009, 2015) and followed by many Asian mycologists (Liu et al., 2011, 2012, 2014, 2015; Dai et al., 2012, 2014a, 2014b, 2014c, 2017, 2018, 2019; Phookamsak et al., 2014, 2015, 2018; Adamčík et al., 2015; Ariyawansa et al., 2015; Senanayake et al., 2015, 2020a; Jiang et al., 2018, 2019a,2019b, 2019c; Sommai et al., 2019; Yang et al., 2019a,b; Jiang H. B. et al., 2020; Jiang N. et al., 2020; Hyde et al., 2020a, b; Monkai et al., 2020; Sun et al., 2020; Tang et al., 2020; Xie et al., 2020; Zhang et al., 2020). To date, more than 1,300 bambusicolous fungi have been reported consisting of 150 basidiomycetes and 1,150 ascomycetes. However, the taxonomic placements of many described species have yet to be verified based on DNA sequence phylogeny (Dai et al., 2018).
In this study, we collected a fungus associated with Hypoxylon stromata on bamboo from northeastern Thailand. Based on morphological examination comparable with the type specimens, the species is identified as a typical Immotthia but largely different to warrant the establishment of a new species. Thus, we introduce a novel species I. bambusae and this is also the first report of Immotthia associated with Hypoxylon stromata on bamboo in Thailand. Furthermore, it is the first time that DNA sequence data of Immotthia are obtained and its phylogenetic affinity within the Dictyosporiaceae was investigated. In addition, a novel genus Pseudocyclothyriella is introduced as a monotypic genus to accommodate P. clematis during the phylogenetic investigation of Immotthia.
Multigene phylogenetic analyses showed that Immotthia formed a well-resolved clade within Dictyosporiaceae in all analyses (Figure 4 and Supplementary Figures 1–3). The genus clustered with Pseudocoleophoma in all analyses with significant support in BI analysis (0.95 PP; Figure 4), but low support in ML analysis (62% ML; Figure 4). However, Immotthia is also morphologically different from Pseudocoleophoma (see notes under generic description).
Immotthia is widely distributed from tropical to temperate regions including Austria, Belgium, China, France, Lithuania, Norway, Poland, Puerto Rico, Russia, Sweden, Switzerland, Ukraine, United States, and Venezuela (Pirozynski, 1973; Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Doilom et al., 2018; Farr and Rossman, 2020). Immotthia does not seem to exhibit a hyperparasitic lifestyle on Hypoxylon, but species of this genus were also reported as saprobes on various decayed hardwoods (Jaklitsch et al., 2002). Immotthia bambusae did not germinate on potato dextrose agar (PDA) medium, suggesting that the species has possibly an obligate parasitic life mode, which is in agreement with Jaklitsch et al. (2002).
Jaklitsch et al. (2002) treated the type species of Immotthia, I. hypoxylon, as a synonym of I. atrograna after they examined the type materials of these two species. They found that the basionym of both I. hypoxylon and I. atrograna shared similar size range of ascomata, asci, and ascospores and it does not show any convincing difference on the ascomata, although these two species occurred on different hosts and habitats (Jaklitsch et al., 2002). Hyde et al. (2017) re-examined the type material of I. hypoxylon and compared it with other collections from North America and reported similar morphology, but the latter have larger ascomata and shorter asci than the type (Hyde et al., 2017). A comparison of the type examination between I. hypoxylon (Hyde et al., 2017) and I. atrograna (Jaklitsch et al., 2002) shows that I. atrograna has a larger ascomata, smaller asci, and overlapped size range of ascospores. However, these two species still lack DNA sequence data, and hence, their taxonomic status with regards to whether they are conspecific warrants further investigation. Herein, we follow the treatment of Jaklitsch et al. (2002) until the epitypes of these two species are designated and their taxonomy is revisited.
Dictyosporiaceae comprises 17 genera, including Immotthia and Pseudocyclothyriella. Asexual morph of Immotthia is recognized as chaetophoma-, coniothyrium-, microsphaeropsis-, or pyrenochaeta-like (Jaklitsch et al., 2002; Akulov and Hayova, 2016; Hyde et al., 2017; Doilom et al., 2018), which is also similar to coelomycetous asexual morph of Roussoella (Hyde et al., 2017). Hyde et al. (2017) tentatively placed Immotthia in Roussoellaceae based on the morphological similarity to Roussoella. However, Immotthia is phylogenetically distinct from Coniothyrium (Coniothyriaceae) and Roussoella (Roussoellaceae). The genus is transferred from Roussoellaceae to Dictyosporiaceae in this study. The coelomycetous asexual morph of Immotthia was found on natural substrates associated with the sexual morph. However, the link between sexual and asexual morph of Immotthia has not yet been proven, although Jaklitsch et al. (2002) attempted to elucidate the connection based on cultural experiments. The ascospores of Immotthia do not germinate on artificial media due to its obligate parasitic life mode. The connection of the sexual–asexual morph needs to be confirmed based on DNA sequence data obtained from direct DNA extraction of fruiting bodies as well as on culture-based studies.
Asexual morphs of most genera in Dictyosporiaceae are hyphomycetes, except for Immotthia, Pseudocoleophoma, Pseudocyclothyriella, and Pseudoconiothyrium which are coelomycetous asexual morphs (Tanaka et al., 2015; Crous et al., 2019; Hongsanan et al., 2020). Based on morphological characteristics, Immotthia is most similar to Pseudoconiothyrium in having hyaline, smooth, doliiform to ampulliform, phialidic conidiogenous cells and ellipsoidal, brown conidia (Crous et al., 2019). However, the genus can be distinguished from Pseudoconiothyrium by multiloculate, smaller conidiomata, smooth-walled conidia, whereas Pseudoconiothyrium has uniloculate, larger conidiomata, verrucose conidia (Crous et al., 2019). Two strains of Pseudocoleophoma clematidis formed a stable clade basal to Immotthia and separated from the main clade of Pseudocoleophoma in all analyses (Figure 4 and Supplementary Figures 1–3). We, therefore, re-examined the holotype specimen of P. clematidis and found that the species was morphologically different from the other Pseudocoleophoma (see Table 3). Thus, Pseudocyclothyriella is introduced herein based on the evidence from both morphology and phylogeny. Pseudocyclothyriella is also similar to Pseudoconiothyrium but differs in having thick-walled, scleroplectenchymatous cells of pycnidial wall. Based on our current phylogenetic results, Immotthia and Pseudocyclothyriella are clearly distinct from Pseudoconiothyrium.
Only four genera in Dictyosporiaceae have been reported for their sexual morphs, viz. Dictyosporium Corda, Gregarithecium Kaz. Tanaka and K. Hiray., Immotthia, and Pseudocoleophoma. Immotthia is morphologically different from these three genera in having ellipsoidal to fusiform, light brown to reddish brown, asymmetrical ascospores, lacks mucilaginous sheath, and exhibits a hyperparasitic life mode (Jaklitsch et al., 2002; Hyde et al., 2017). On the other hand, the other three genera have hyaline, fusiform to narrowly fusiform ascospores, with or without mucilaginous sheath and have been reported as saprobes in terrestrial and freshwater habitats (Tanaka et al., 2015; Boonmee et al., 2016). The familial descriptions for Dictyosporiaceae have previously been restricted to hyphomycetous asexual morphs (Boonmee et al., 2016; Hongsanan et al., 2020). We recommend that the descriptions and illustrations of Dictyosporiaceae should include both coelomycetous and hyphomycetous asexual morphs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
H-BJ and RP: conceptualization, data curation, and formal analysis. SL, SCK, and RP: funding acquisition. H-BJ, RP, CP, and MD: investigation, methodology, and writing—original draft. RP and NS: project administration. SL, SCK, RJ, and PK: supervision. NS, PK, RJ, SL, and SCK: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Key Research Project, Agroforestry Systems for Restoration and Bio-industry Technology Development (Grant No. 2017YFC0505101), Ministry of Sciences and Technology of China (Grant No. 2017YFC0505100), National Science Foundation of China (NSFC) project code 31850410489 (Grant No. Y81I982211), and Chiang Mai University. RP thanks CAS President’s International Fellowship Initiative (PIFI) for young staff (Grant No. Y9215811Q1), the National Science Foundation of China (NSFC) project code 31850410489 (Grant No. Y81I982211), and Chiang Mai University for financial support. SCK thanks CAS President’s International Fellowship Initiative (PIFI) young staff under the grant number: 2020FYC0002 and the National Science Foundation of China (NSFC) for funding this work under the project code 31851110759.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Biology Experimental Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the facilities of molecular laboratory. Shaun Pennycook at Manaaki Whenua – Landcare Research is thanked for his help in naming the new fungal species. Austin Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing. Sinang Hongsanan at Guangdong Provincial Key Laboratory for Plant Epigenetics, Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Science and Oceanography, Shenzhen University, is thanked for advising on phylogenetic analyses. H-BJ would like to thank Mae Fah Luang University for the Ph.D. scholarship. RJ would like to thank the University of Mauritius for research support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.656235/full#supplementary-material
Supplementary Figure 1 | RAxML tree based on ITS, LSU and TEF1-α sequence matrix represented the phylogenetic relationships of taxa in Dictyosporiaceae. The tree is rooted to Periconia igniaria (CBS 845.96 and CBS 379.86). Bootstrap support values for ML are indicated above the nodes. Ex-type strains are in black bold and the new species and new combinations are indicated in blue bold.
Supplementary Figure 2 | Phylogram generated by Bayesian inference (BI) analysis based on a combined ITS, LSU, SSU and TEF1-α sequence matrix represented the phylogenetic relationships of taxa in Dictyosporiaceae. The tree is rooted to Periconia igniaria (CBS 845.96 and CBS 379.86). Bayesian posterior probabilities are indicated above the nodes. Ex-type strains are in black bold and the new species and new combinations are indicated in blue bold.
Supplementary Figure 3 | RAxML tree based on LSU, SSU, TEF1-α, RPB2 and ITS sequence matrix represented the phylogenetic relationships of Immotthia in Dictyosporiaceae with other families in Pleosporales. The tree is rooted to Capondium coffeae (CBS 147.52). Bootstrap support values for ML are indicated above the nodes. The placement of Immotthia in Dictyosporiaceae is indicated by yellow background.
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://mafft.cbrc.jp/alignment/server/
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ https://www.treebase.org/, submission ID: 27606.
References
Adamčík, S., Cai, L., Chakraborty, D., Chen, X. H., Cotter, H. V. T., Dai, D. Q., et al. (2015). Fungal biodiversity profiles 1–10. Cryptogam. Mycol. 36, 121–166. doi: 10.7872/crym/v36.iss2.2015.121
Akulov, O. Y., and Hayova, V. P. (2016). Immotthia atrograna (Dacampiaceae, Ascomycota), a new for Ukraine fungicolous fungus from Carpathians. Ukr. Bot. J. 73, 84–89. doi: 10.15407/ukrbotj73.01.084
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. W. T., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s13225-015-0346-5
Boonmee, S., D’souza, M. J., Luo, Z., Pinruan, U., Tanaka, K., Su, H., et al. (2016). Dictyosporiaceae fam. nov. Fungal Divers. 80, 457–482. doi: 10.1007/s13225-016-0363-z
Crous, P. W., Schumacher, R. K., Akulov, A., Thangavel, R., Hernández-Restrepo, M., Carnegie, A. J., et al. (2019). New and interesting fungi. 2. Fungal Syst. Evol. 3, 57–134. doi: 10.3114/fuse.2019.03.06
Dai, D. Q., Bahkali, A. H., Li, Q. R., Bhat, D. J., Wijayawardene, N. N., Li, W. J., et al. (2014a). Vamsapriya (Xylariaceae) re-described, with two new species and molecular sequence data. Cryptogam. Mycol. 35, 339–357. doi: 10.7872/crym.v35.iss4.2014.339
Dai, D. Q., Bhat, D. J., Liu, J. K., Chukeatirote, E., Zhao, R. L., and Hyde, K. D. (2012). Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogam. Mycol. 33, 363–379. doi: 10.7872/crym.v33.iss3.2012.363
Dai, D. Q., Phookamsak, R., Wijayawardene, N. N., Li, W. J., Bhat, D. J., Xu, J. C., et al. (2017). Bambusicolous fungi. Fungal Divers. 82, 1–105. doi: 10.1007/s13225-016-0367-8
Dai, D. Q., Tang, L. Z., and Wang, H. B. (2018). “A review of bambusicolous ascomycetes,” in Bamboo: Current and Future Prospects, ed. H. P. S. Abdul Khalil (London: IntechOpen). doi: 10.5772/intechopen.76463
Dai, D. Q., Wijayawardene, N. N., Bhat, D. J., Chukeatirote, E., Bahkali, A. H., Zhao, R. L., et al. (2014b). Pustulomyces gen. nov. accommodated in Diaporthaceae, Diaporthales, as revealed by morphology and molecular analyses. Cryptogam. Mycol. 35, 63–72. doi: 10.7872/crym.v35.iss1.2014.63
Dai, D. Q., Wijayawardene, N. N., Bhat, D. J., Chukeatirote, E., Zhao, R. L., Wang, Y., et al. (2014c). The phylogenetic placement of Eriosporella bambusicola sp. nov. in Capnodiales. Cryptogam. Mycol. 35, 41–49. doi: 10.7872/crym.v35.iss1.2014.41
Dai, D. Q., Wijayawardene, N. N., Tang, L. Z., Liu, C., Han, L. H., Chu, H. L., et al. (2019). Rubroshiraia gen. nov., a second hypocrellin-producing genus in Shiraiaceae (Pleosporales). MycoKeys 58, 1–26. doi: 10.3897/mycokeys.58.36723
De Gruyter, J., Noordeloos, M. E., and Boerema, G. H. (1993). Contributions towards a monograph of Phoma (Coelomycetes)—I. 2. Section Phoma: additional taxa with very small conidia and taxa with conidia up to 7 μm long. Persoonia 15, 369–400.
Dissanayake, A. J., Bhunjun, C. S., Maharachchikumbura, S. S. N., and Liu, J. K. (2020). Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11, 2652–2676. doi: 10.5943/mycosphere/11/1/18
Doilom, M., Dissanayake, A. J., Wanasinghe, D. N., Boonmee, S., Liu, J. K., Bhat, D. J., et al. (2017). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 82, 107–182. doi: 10.1007/s13225-016-0368-7
Doilom, M., Hyde, K. D., Phookamsak, R., Dai, D. Q., Tang, L. Z., Hongsanan, S., et al. (2018). Mycosphere notes 225–274: types and other specimens of some genera of Ascomycota. Mycosphere 9, 647–754. doi: 10.5943/mycosphere/9/4/3
Ellis, J. B., and Everhart, B. M. (1886). New species of fungi from various localities. J. Mycol. 2, 37–42.
Farr, D. F., and Rossman, A. Y. (2020). Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Available online at: https://nt.ars-grin.gov/fungaldatabases/ (accessed November 1, 2020).
Hongsanan, S., Hyde, K. D., Phookamsak, R., Wanasinghe, D. N., McKenzie, E. H. C., Sarma, V. V., et al. (2020). Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11, 1553–2107. doi: 10.5943/mycosphere/11/1/13
Hyde, K. D., de Silva, N. I., Jeewon, R., Bhat, D. J., Phookamsak, R., Doilom, M., et al. (2020a). AJOM new records and collections of fungi: 1–100. Asian J. Mycol. 3, 22–294. doi: 10.5943/ajom/3/1/3
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. B. G., et al. (2020b). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., Norphanphoun, C., Abreu, V. P., Bazzicalupo, A., Chethana, K. W. T., Clericuzio, M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3
Index Fungorum (2020). Index Fungorum. Available online at: http://www.indexfungorum.org/Names/Names.asp (accessed Oct 27, 2020)
Iturrieta-González, I., Gené, J., Guarro, J., Castañeda-Ruiz, R. F., and García, D. (2018). Neodendryphiella, a novel genus of the Dictyosporiaceae (Pleosporales). MycoKeys 37, 19–38. doi: 10.3897/mycokeys.37.27275
Jaklitsch, W. M., and Voglmayr, H. (2016). Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 85, 35–64. doi: 10.1016/j.simyco.2016.09.002
Jaklitsch, W. M., Scheuer, C., and Voglmayr, H. (2002). Notes on the genus Immotthia (Pleosporales, Ascomycetes), including some type studies. Osterr. Z. Pilzkd. 11, 93–106.
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, D. J., Buyck, B., Cai, L., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jayasiri, S. C., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Jeewon, R., Phillips, A. J. L., et al. (2019). Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10, 11–86. doi: 10.5943/mycosphere/10/1/1
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Jiang, H. B., Hyde, K. D., Doilom, M., Karunarathna, S. C., Xu, J. C., and Phookamsak, R. (2019a). Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. Asian J. Mycol. 2, 254–268. doi: 10.5943/ajom/2/1/16
Jiang, H. B., Hyde, K. D., Jayawardena, R. S., Doilom, M., Xu, J., and Phookamsak, R. (2019b). Taxonomic and phylogenetic characterizations reveal two new species and two new records of Roussoella (Roussoellaceae, Pleosporales) from Yunnan, China. Mycol. Prog. 18, 577–591. doi: 10.1007/s11557-019-01471-9
Jiang, H. B., Phookamsak, R., Bhat, D. J., Khan, S., Bahkali, A., Elgorban, A. B., et al. (2018). Vamsapriya yunnana, a new species of Vamsapriya (Xylariaceae, Xylariales) associated with bamboo from Yunnan, China. Phytotaxa 356, 61–70. doi: 10.11646/phytotaxa.356.1.5
Jiang, H. B., Phookamsak, R., Doilom, M., Mortimer, P. E., Xu, J. C., Lumyong, S., et al. (2019c). Taxonomic and phylogenetic characterizations of Keissleriella bambusicola sp. nov. (Lentitheciaceae, Pleosporales) from Yunnan, China. Phytotaxa 423, 129–144. doi: 10.11646/phytotaxa.423.3.2
Jiang, H. B., Phookamsak, R., Xu, J. C., Karunarathna, S. C., Mortimer, P. E., and Hyde, K. D. (2020). Taxonomic and phylogenetic characterizations reveal three new species of Mendogia (Myriangiaceae, Myriangiales). Mycol. Prog. 19, 41–51. doi: 10.1007/s11557-019-01540-z
Jiang, N., Liang, Y. M., and Tian, C. M. (2020). A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia 72, 77–83. doi: 10.12905/0380.sydowia72-2020-0077
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). Mafft online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Li, W. J., McKenzie, E. H. C., Liu, J. K., Bhat, D. J., Dai, D. Q., Camporesi, E., et al. (2020). Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. 100, 279–801. doi: 10.1007/s13225-020-00440-y
Liu, J. K., Hyde, K. D., Gareth, E. B. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1– 110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Liu, J. K., Phookamsak, R., Dai, D. Q., Tanaka, K., Jones, E. B. G., Xu, J. C., et al. (2014). Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa 181, 1–33. doi: 10.11646/phytotaxa.181.1.1
Liu, J. K., Phookamsak, R., Jones, E. B. G., Zhang, Y., Ko-Ko, T. W., Hu, H. L., et al. (2011). Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 51, 135–154. doi: 10.1007/s13225-011-0142-9
Liu, Y., Liu, Z., and Wongkaew, S. (2012). Developing characteristics and relationships of Shiraia bambusicola with Bamboo. Songklanakarin. J. Sci. Technol. 34, 17–22.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), (New Orleans, LA), 1–8.
Monkai, J., Boonmee, S., Ren, G. C., Wei, D. P., Phookamsak, R., and Mortimer, P. E. (2020). Distoseptispora hydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 459, 93–107. doi: 10.11646/phytotaxa.459.2.1
Nylander, J. (2008). MrModeltest2 v. 2.3 (Program for Selecting DNA Substitution Models using PAUP∗). Uppsala: Evolutionary Biology Centre.
Phookamsak, R., Liu, J. K., Manamgoda, D. S., Wanasinghe, D. N., Ariyawansa, H., Mortimer, P. E., et al. (2014). Epitypification of two bambusicolous fungi from Thailand. Cryptogam. Mycol. 35, 239–256. doi: 10.7872/crym.v35.iss3.2014.239
Phookamsak, R., Lu, Y. Z., Hyde, K. D., Jeewon, R., Li, J., Doilom, M., et al. (2018). Phylogenetic characterization of two novel Kamalomyces species in Tubeufiaceae (Tubeufiales). Mycol. Prog. 17, 647–660. doi: 10.1007/s11557-017-1365-2
Phookamsak, R., Norphanphoun, C., Tanaka, K., Dai, D. Q., Luo, Z. L., Liu, J. K., et al. (2015). Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov and Astrosphaeriellopsis, gen. nov. Fungal Divers. 74, 143–197. doi: 10.1007/s13225-015-0352-7
Phukhamsakda, C., McKenzie, E. H. C., Phillips, A. J. L., Jones, E. B. G., Bhat, D. J., Stadler, M., et al. (2020). Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 102, 1–203. doi: 10.1007/s13225-020-00448-4
Pirozynski, K. A. (1973). Three hyperparasites of ascomycetes. Mycologia 65, 761–767. doi: 10.2307/3758515
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Rehner, S. (2001). Primers for Elongation Factor 1-Alpha (EF1-Alpha). Available online at: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf (accessed June 20, 2020)
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Senanayake, I. C., Bhat, D. J., Cheewangkoon, R., and Xie, N. (2020a). Bambusicolous Arthrinium species in Guangdong Province, China. Front. Microbiol. 11:602773. doi: 10.3389/fmicb.2020.602773
Senanayake, I. C., Maharachchikumbura, S. S. N., Hyde, K. D., Bhat, J. D., Jones, E. B. G., McKenzie, E. H. C., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 73, 73–144. doi: 10.1007/s13225-015-0340-y
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020b). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Sommai, S., Nuankaew, S., Khamsuntorn, P., Suetrong, S., and Pinruan, U. (2019). Tamhinispora saraburiensis sp. nov. (Tubeufiaceae, Dothideomycetes) on bamboo in Thailand based on morphology & phylogenetic analysis. Phytotaxa 402, 1–2. doi: 10.11646/phytotaxa.402.1.1
Species Fungorum (2020). Species Fungorum. Available online at: http://www.speciesfungorum.org/Names/Names.asp (assessed Oct 27, 2020)
Sun, Y., Goonasekara, I. D., Thambugala, K. M., Jayawardena, R. S., Wang, Y., and Hyde, K. D. (2020). Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 8:e53678. doi: 10.3897/BDJ.8.e53678
Tanaka, K., Hirayama, K., Yonezawa, H., Hatakeyama, S., Harada, Y., Sano, T., et al. (2009). Molecular taxonomy of bambusicolous fungi: tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 64, 175–209. doi: 10.3114/sim.2009.64.10
Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., et al. (2015). Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 82, 75–136. doi: 10.1016/j.simyco.2015.10.002
Tang, X., Goonasekara, I. D., Jayawardena, R. S., Jiang, H. B., Li, J. F., Hyde, K. D., et al. (2020). Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers. Data J. 8:e58755. doi: 10.3897/BDJ.8.e58755
Tennakoon, D. S., Bhat, D. J., Kuo, C. H., and Hyde, K. D. (2019). Leaf litter saprobic Dictyosporiaceae (Pleosporales, Dothideomycetes): Pseudocoleophoma zingiberacearum sp. nov. from Hedychium coronarium. Kavaka 53, 1–7. doi: 10.36460/Kavaka/53/2019/1-7
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic), 315–322.
Xie, X., Liu, L. L., Shen, X. C., Kang, Y. Q., Hyde, K. D., Kang, J. C., et al. (2020). Contributions to species of Xylariales in China-3. Collodiscula tubulosa (Xylariaceae). Phytotaxa 428, 122–130. doi: 10.11646/phytotaxa.428.2.6
Yang, C. L., Baral, H. O., Xu, X., and Liu, Y. (2019a). Parakarstenia phyllostachydis, a new genus and species of non-lichenized Odontotremataceae (Ostropales, Ascomycota). Mycol. Prog. 18, 833–845. doi: 10.1007/s11557-019-01492-4
Yang, C. L., Xu, X. L., Wanasinghe, D. N., Jeewon, R., Phookamsak, R., Liu, Y. G., et al. (2019b). Neostagonosporella sichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachys heteroclada (Poaceae) from Sichuan Province, China. MycoKeys 46, 119–150. doi: 10.3897/mycokeys.46.32458
Yang, J., Liu, J. K., Hyde, K. D., Jones, E. B. G., and Liu, Z. Y. (2018). New species in Dictyosporium, new combinations in Dictyocheirospora and an updated backbone tree for Dictyosporiaceae. MycoKeys 36, 83–105. doi: 10.3897/mycokeys.36.27051
Zeng, X. Y., Jeewon, R., Wen, T. C., Hongsanan, S., Boonmee, S., and Hyde, K. D. (2018). Simplified and efficient DNA extraction protocol for Meliolaceae specimens. Mycol. Prog. 17, 403–415. doi: 10.1007/s11557-017-1361-6
Zhang, J. Y., Phookamsak, R., Boonmee, S., Hyde, K. D., Dai, D. Q., and Lu, Y. Z. (2020). Roussoella guttulata (Roussoellaceae, Pleosporales), a novel bambusicolous ascomycete from Thailand. Phytotaxa 471, 221–233. doi: 10.11646/phytotaxa.471.3.4
Keywords: bambusicolous fungi, Dothideomycetes, Pseudocoleophoma, taxonomy, two new taxa
Citation: Jiang H-B, Jeewon R, Karunarathna SC, Phukhamsakda C, Doilom M, Kakumyan P, Suwannarach N, Phookamsak R and Lumyong S (2021) Reappraisal of Immotthia in Dictyosporiaceae, Pleosporales: Introducing Immotthia bambusae sp. nov. and Pseudocyclothyriella clematidis comb. et gen. nov. Based on Morphology and Phylogeny. Front. Microbiol. 12:656235. doi: 10.3389/fmicb.2021.656235
Received: 20 January 2021; Accepted: 22 March 2021;
Published: 07 May 2021.
Edited by:
Jadson Diogo Pereira Bezerra, Universidade Federal de Goiás, BrazilReviewed by:
Sinang Hongsanan, Shenzhen University, ChinaRoger Melo, Federal University of Pernambuco, Brazil
Copyright © 2021 Jiang, Jeewon, Karunarathna, Phukhamsakda, Doilom, Kakumyan, Suwannarach, Phookamsak and Lumyong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rungtiwa Phookamsak, am9tamFtLnJwMkBnbWFpbC5jb20=; Saisamorn Lumyong, c2Nib2kwMDlAZ21haWwuY29t
 Hong-Bo Jiang
Hong-Bo Jiang Rajesh Jeewon
Rajesh Jeewon Samantha C. Karunarathna
Samantha C. Karunarathna Chayanard Phukhamsakda
Chayanard Phukhamsakda Mingkwan Doilom
Mingkwan Doilom Pattana Kakumyan
Pattana Kakumyan Nakarin Suwannarach
Nakarin Suwannarach Rungtiwa Phookamsak
Rungtiwa Phookamsak Saisamorn Lumyong
Saisamorn Lumyong