
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 April 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.654783
This article is part of the Research Topic Antimicrobial Resistance in Aquatic Environments View all 15 articles
A correction has been applied to this article in:
Corrigendum: Reactive Oxygen Species-Related Ceftazidime Resistance Is Caused by the Pyruvate Cycle Perturbation and Reverted by Fe3+ in Edwardsiella tarda
Reactive oxygen species (ROS) are related to antibiotic resistance and have been reported in bacteria. However, whether ROS contribute to ceftazidime resistance and plays a role in ceftazidime-mediated killing is unknown. The present study showed lower ROS production in ceftazidime-resistant Edwardsiella tarda (LTB4-RCAZ) than that in LTB4-sensitive E. tarda (LTB4-S), two isogenic E. tarda LTB4 strains, which was related to bacterial viability in the presence of ceftazidime. Consistently, ROS promoter Fe3+ and inhibitor thiourea elevated and reduced the ceftazidime-mediated killing, respectively. Further investigation indicated that the reduction of ROS is related to inactivation of the pyruvate cycle, which provides sources for ROS biosynthesis, but not superoxide dismutase (SOD) and catalase (CAT), which degrade ROS. Interestingly, Fe3+ promoted the P cycle, increased ROS biosynthesis, and thereby promoted ceftazidime-mediated killing. The Fe3+-induced potentiation is generalizable to cephalosporins and clinically isolated multidrug-resistant pathogens. These results show that ROS play a role in bacterial resistance and sensitivity to ceftazidime. More importantly, the present study reveals a previously unknown mechanism that Fe3+ elevates ROS production via promoting the P cycle.
Edwardsiella tarda is known for causing diseases in both humans and fish, in both of which these diseases can potentially be fatal if untreated. In aquaculture, the bacterium targets at a wide range of fish species and thereby leads to extensive economic losses in the industry (Wang et al., 2011; Abayneh et al., 2013). Various antibiotics are used to prevent and control the infections caused by the bacterium. Unfortunately, the overuse of antibiotics has inadvertently promoted the emergence and rapid spread of antibiotic-resistant bacteria (Cabello et al., 2016). The emergence of antibiotic-resistant bacteria poses a major challenge for health practitioners and a huge threat to human health and aquaculture since antibiotic-resistant bacteria are insensitive to antibiotics. As the process of developing new pharmaceutical agents to control antibiotic-resistant pathogens is slow and not a viable approach to manage the growing infectious diseases, further understanding of antibiotic resistance mechanisms for control of these antibiotic-resistant pathogens is an important scientific issue and becomes a major research focus (Defoirdt et al., 2011; Blair et al., 2015).
A line of evidences has indicated that microbial metabolic environment confounds antibiotic sensitivity (Peng et al., 2015; Yao et al., 2016; Cheng et al., 2019; Stokes et al., 2019; Jiang et al., 2020a; Li et al., 2020), where reactive oxygen species (ROS) are related to bacterial resistance to antibiotics and antibiotic-mediated killing efficacy (Dwyer et al., 2009; Van Acker and Coenye, 2017; Zhao and Drlica, 2014). The tricarboxylic acid cycle (TCA) cycle plays a crucial role in ROS formation (Van Acker and Coenye, 2017). Therefore, antibiotic-resistant bacteria exhibit the occurrence of reduced or fluctuated TCA cycle and decreased ROS (Ye et al., 2018; Zhang et al., 2020), indicating that low ROS concentrations induce resistance (Van Acker and Coenye, 2017). In the antibiotic-mediated killing mechanisms, antibiotics belonging to different classes activate the TCA cycle, supporting the formation of ROS (superoxide and hydrogen peroxide) via hyperactivation of the electron transport chain (Dwyer et al., 2009; Van Acker and Coenye, 2017). Fe3+ causes the activation of the Fenton reaction to generate abundant ROS against methicillin−resistant Staphylococcus aureus (MRSA) infection (Song et al., 2020). Our recent publications have showed that the ROS induced by exogenous metabolites elevate aminoglycoside-mediated killing efficacy to EIB202 and Vibrio alginolyticus (Ye et al., 2018; Zhang et al., 2020). Reports also indicate that the beta-lactam stress increased the intracellular ROS level (Rosato et al., 2014; Huang et al., 2019), but whether ROS promote beta-lactam-mediated killing is unknown. Therefore, further understanding of ROS role is required for control of beta-lactam-resistant pathogens.
Ceftazidime is a semisynthetic, broad-spectrum, beta-lactam antibiotic, playing a bactericidal action by inhibiting enzymes responsible for cell wall synthesis, primarily penicillin-binding protein 3 (PBP3). The drug is a commonly used antibiotic in clinics. However, with a wide use of cephalosporins in clinics, resistance to cephalosporins including ceftazidime is predominant. Inactivation of β-lactams by β-lactamases, failure in binding to penicillin-binding proteins, and alteration of binding affinity to penicillin-binding proteins are identified as the three common mechanisms of resistance to β-lactams (Peng et al., 2019). However, whether ROS play a role in ceftazidime sensitivity and resistance is largely unknown. Furthermore, information regarding mechanisms for Fe3+-mediated ROS is not available.
In this study, we showed that the intracellular ROS production was lower in LTB4-RCAZ than that in LTB4-S due to a decrease of ROS generation. The decrease of ROS generation was attributed to inactivation of the pyruvate cycle (the P cycle) (Ye et al., 2018). Fe3+ promoted the P cycle for elevation of ROS production, thereby elevating ceftazidime-mediated killing. These results are described below.
Edwardsiella tarda LTB4 used in this study was obtained from Professor Xiaohua Zhang, Ocean University of China University. LTB4 was grown at 30°C for 24 h in 50 ml Luria-Bertani (LB) broth in 250-ml flasks.
Measurement of minimum inhibitory concentration (MIC) was performed as previously described (National Committee for Clinical Laboratory Standards, 1999). In brief, LTB4 was cultured in LB medium with twofold serially diluted ceftazidime (CAZ, Guangzhou QiYun Biological Technology) ranging from 0.01 to 160 μg/ml. Overnight bacterial cultures were diluted 1:100 in fresh LB medium and cultured at 30°C to an OD600 of 0.5. The tray contained a series of twofold dilutions of antibiotics. Ninety microliters of LB containing CAZ and 10 μl of logarithmic phase cells with 107 CFU/ml of LTB4-RCAZ or LTB4-RCAZ were incubated in a microwell plate for 24 h at 30°C. The lowest concentration showing no visible growth was recorded as the MIC. At least three biologic replicates were performed.
Real-time quantitative PCR (qRT-PCR) was carried out as previously described (Li et al., 2016). Bacterial cells were harvested at OD600 = 1.0. The total RNA of each sample was isolated with TRIzol (Invitrogen, United States). Reverse transcription-PCR was carried out on a PrimeScriptTM RT Reagent Kit with gDNA eraser (Takara, Japan) with 1 μg of total RNA according to manufacturer’s instructions. qRT-PCR was performed in 384-well plates, and each well contained a total volume of 10 ml liquid including 5 ml 2X SYBR Premix Ex TaqTM, 2.6 μl PCR-grade water, 2 μl cDNA template, and 0.2 μl each pair of primers (10 mM). The primers are listed in Supplementary Table 1. All the assays were performed on the LightCycler 480 system (Roche, Germany) according to the manufacturer’s instructions, and four independent samples were assayed for both the control group and the test group. The cycling parameters were listed as follows: 95°C for 30 s to activate the polymerase; 40 cycles of 95°C for 10 s; and 60°C for 30 s. Fluorescence measurements were performed at 70°C for 1 s during each cycle. Cycling was terminated at 95°C with a calefactive velocity of 5°C per second, and a melting curve was obtained. To analyze the relative expression level of the target gene, we converted the data to percentages relative to the value of no-treatment group. At least triplicate biological repeats were carried out.
Bacterial sample preparation was carried out as previously described (Zhang et al., 2019). In brief, 10 ml OD600 = 1.0 cells were quenched with cold methanol and sonicated for 5 min at 200 W. Samples were centrifuged at 12,000 rpm for 10 min. Supernatant, containing 1 μg/ml ribitol (Sigma–Aldrich) as internal analytical standard, was transferred into a new tube and dried by vacuum centrifugation device (LABCONCO). The dried extracts were then incubated with 80 μl methoxyamine hydrochloride (20 mg/ml, Sigma–Aldrich) in pyridine (Sigma–Aldrich) for 90 min at 37°C and derivatization was done with an identical volume of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma–Aldrich) for another 30 min. Samples were centrifuged at 12,000 rpm for 10 min, and the supernatant was transferred into new tubes. Gas chromatography-mass spectrometry (GC-MS) analysis was performed with an Agilent GC-MS instrument. Spectral deconvolution and calibration were performed using AMDIS and internal standards as previously described. A retention time (RT) correction was performed for all the samples, and then the RT was used as a reference against which the remaining spectra were queried, and a file containing the abundance information for each metabolite in all the samples was assembled. Metabolites from the GC-MS spectra were identified by searching in the National Institute of Standards and Technology (NIST11.L) Mass Spectral Library. Among the detected peaks of all chromatograms, compound peaks were considered as endogenous metabolites and the same metabolite names were merged. The resulting data matrix was normalized by the concentrations of added internal standards and the total intensity. This file was then used for subsequent statistical analyses. The abundance of a metabolite was scaled by total abundance of all metabolites in a sample as its relative abundance for further analysis. Hierarchical clustering was completed in the R platform with the package gplots1 using the distance matrix. Multivariate statistical analysis included principal component analysis (SIMCA-P 12.0.1), which was used to discriminate sample patterns. GraphPad Prism 7 was used to draw figures.
Measurement of enzyme activity was performed as previously described with a few modifications (Zhang et al., 2019; Jiang et al., 2020c). In brief, the harvested cells were collected at OD600 = 1.0 and then re-suspended in sterile saline to OD600 = 1.0 after washing. Samples of 30 ml were collected by centrifugation at 8,000 rpm for 5 min. Pellets were re-suspended in phosphate-buffered saline (PBS) and broke down by sonication for 2 min at a 200-W power setting on ice, and then centrifuged at 12,000 rpm for 10 min to remove insoluble materials. Supernatants containing 200 μg of total proteins were transferred to a pyruvate dehydrogenase (PDH) reaction mix (0.15 mM MTT, 1 mM MgCl2, 0.5 mM PMS, 0.2 mM TPP, 2 mM sodium pyruvate, and 50 mM PBS), an α-ketoglutarate dehydrogenase (KGDH) reaction mix (0.15 mM MTT, 1 mM MgCl2, 0.5 mM PMS, 0.2 mM TPP, 50 mM α-ketoglutaric acid potassium salt, and 50 mM PBS), a succinate dehydrogenase (SDH) reaction mix (0.15 mM MTT, 1 mM PMS, 5 mM sodium succinate, and 50 mM PBS), and a malate dehydrogenase (MDH) reaction mix (0.15 mM MTT, 1 mM PMS, 50 mM PBS, and 50 mM malate) to a final volume of 200 μl in a 96-well plate. Subsequently, the plate was incubated at 37°C for 5 min for PDH/KGDH/SDH/MDH assay and then measured at 566 nm for colorimetric readings. The plate was protected from light during the incubation. The dehydrogenase activity was assayed in quadruplicate.
The activity of superoxide dismutase (SOD) and catalase (CAT) was determined by commercial kits (Nanjing Jiancheng, China). The process is as follows: 10 ml of bacteria with OD600 = 1.0 was collected, washed twice with PBS, and suspended in PBS. The suspension was broke down by sonication for 2 min at a 200-W power setting on ice and then centrifuged at 12,000 rpm for 10 min to remove insoluble materials. The supernatant was taken, and the protein concentration was determined. The activity of SOD was measured with 150 μg total protein. The follow-up test was carried out according to the manufacturer’s instructions. Then, SOD activity was measured at 450 nm, and CAT activity was measured at 405 nm. The plate was protected from light during the incubation.
Detection of ATP was determined by a BacTiter-GloTM Microbial Cell Viability Assay (Cat. G8231, Promega, Madison, WI, United States) as previously described (Cheng et al., 2019). In brief, bacterial cells were harvested in the OD600 of 1.0 by centrifugation for 10 min at 12,000 g and washed twice by centrifugation with sterile saline. Cells were resuspended with saline solution and adjusted the OD600 to 1.0. Then, 50-μl samples were added to a 96-well plate and mixed with an equal volume of the kit solution. Then, the absorbance was measured using VICTOR X5 (PerkinElmer, Turku, Finland) according to the manufacturer’s instructions. The concentration of ATP was calculated according to the standard curve of ATP.
Antibacterial assay was carried out as described previously (Peng et al., 2015). Overnight bacterial cultures were diluted 1:100 in fresh LB medium and cultured at 30°C to an OD600 of 1.0. Bacterial cells were collected by centrifugation at 8,000 rpm for 5 min. The samples were then washed with sterile saline three times; suspended in M9 minimal media containing 10 mM acetate, 1 mM MgSO4, and 100 μM CaCl2; and diluted to an OD600 of 0.2. Fe3+ or/and ceftazidime were added and incubated at 30°C and 200 rpm for 6 h. To determine bacterial count, 100 μl of cultures was obtained and then serially diluted. An aliquot of 10μl of each dilution was plated in TSB agar plates and incubated at 30°C for 22 h. The plates only with 20–200 colonies were counted, and the colony-forming unit per milliliter was calculated.
Bacterial membrane potential was measured by BacLight Bacterial Membrane Potential Kit (Invitrogen). In brief, bacteria were diluted to 106 CFU/ml and stained with 10 μl of 3 mM DiOC2, followed by incubation for 30 min. Samples were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, United States). The green/red fluorescence was detected with 488–530/610 nm. The membrane potential was determined and normalized as the intensity ratio of the red fluorescence and the green fluorescence. Relative proton motive force (PMF) was determined by test samples compared with control samples.
LTB4 was cultured in LB medium with or without twofold serially diluted ceftazidime, leading to LTB4-RCAZ and LTB4-S, respectively. The MIC of LTB4-S was 0.039 μg/ml CAZ, while that of LTB4-RCAZ was 0.625 μg/ml CAZ. There was a 16-fold difference between the two strains (Figure 1A). Consistently, a higher viability was detected in LTB4-RCAZ than LTB4-S (Figure 1B). To explore whether LTB4-RCAZ had a reduced ROS production, ROS production was detected in LTB4-RCAZ and LTB4-S. A lower ROS production was determined in LTB4-RCAZ than LTB4-S (Figure 1C). These results indicate that ROS production is reduced in LTB4-RCAZ compared with LTB4-S.
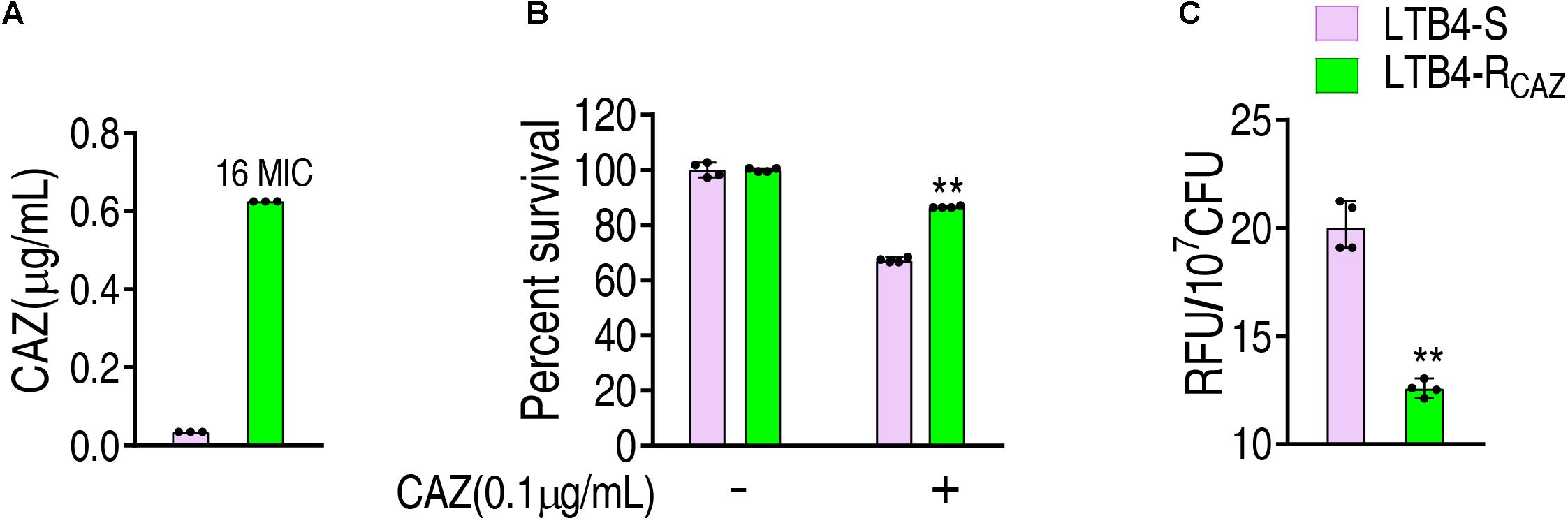
Figure 1. Ceftazidime resistance and reactive oxygen species (ROS) level in LTB4-RCAZ. (A) Minimum inhibitory concentration (MIC) of LTB4-S and LTB4-RCAZ. (B) Killing efficiency of ceftazidime to LTB4-S and LTB4-RCAZ. (C) ROS of LTB4-S and LTB4-RCAZ. Results (B,C) are displayed as mean ± SEM, as determined by two-tailed Student’s t-test. Four biological repeats are carried out. **P < 0.01.
These above results motivated us to speculate that ROS plays an important role in ceftazidime resistance. To demonstrate this, the viability of LTB4-RCAZ was detected in the presence or absence of ROS promoter or/and inhibitor with ceftazidime. There was a stronger resistance to H2O2 in LTB4-S than Escherichia coli K12 and thereby 5 mM H2O2 was used (Supplementary Figure 1). The promoters Fe3+ and H2O2 (positive control) potentiated ceftazidime-mediated killing. However, the inhibitor thiourea not only eliminated the potentiation caused by the promoters but also inhibited the ceftazidime-mediated killing (Figure 2A). The effect of Fe3+ was increased in a dose-dependent manner and was related to the concentration of ceftazidime used (Figure 2B). When the concentration of Fe3+ was fixed, the killing efficacy was ceftazidime dose dependent (Figure 2C). To validate the role of the ROS promoter or/and inhibitor in the ceftazidime-mediated killing, we measured the ROS production of LTB4-RCAZ in the presence or absence of the ROS promoter or/and inhibitor. ROS production was elevated and reduced in the presence of the inhibitor thiourea and promoters Fe3+, H2O2, or ceftazidime alone, respectively. Comparatively, the elevated ROS ranked as H2O2 > ceftazidime > Fe3+. When the synergistic use of ceftazidime with one of the promoters or/and the inhibitor was performed, they promoted and inhibited ROS level, respectively (Figure 2D). These results support the conclusion that ROS is related to ceftazidime resistance and promotes ceftazidime-mediated killing.
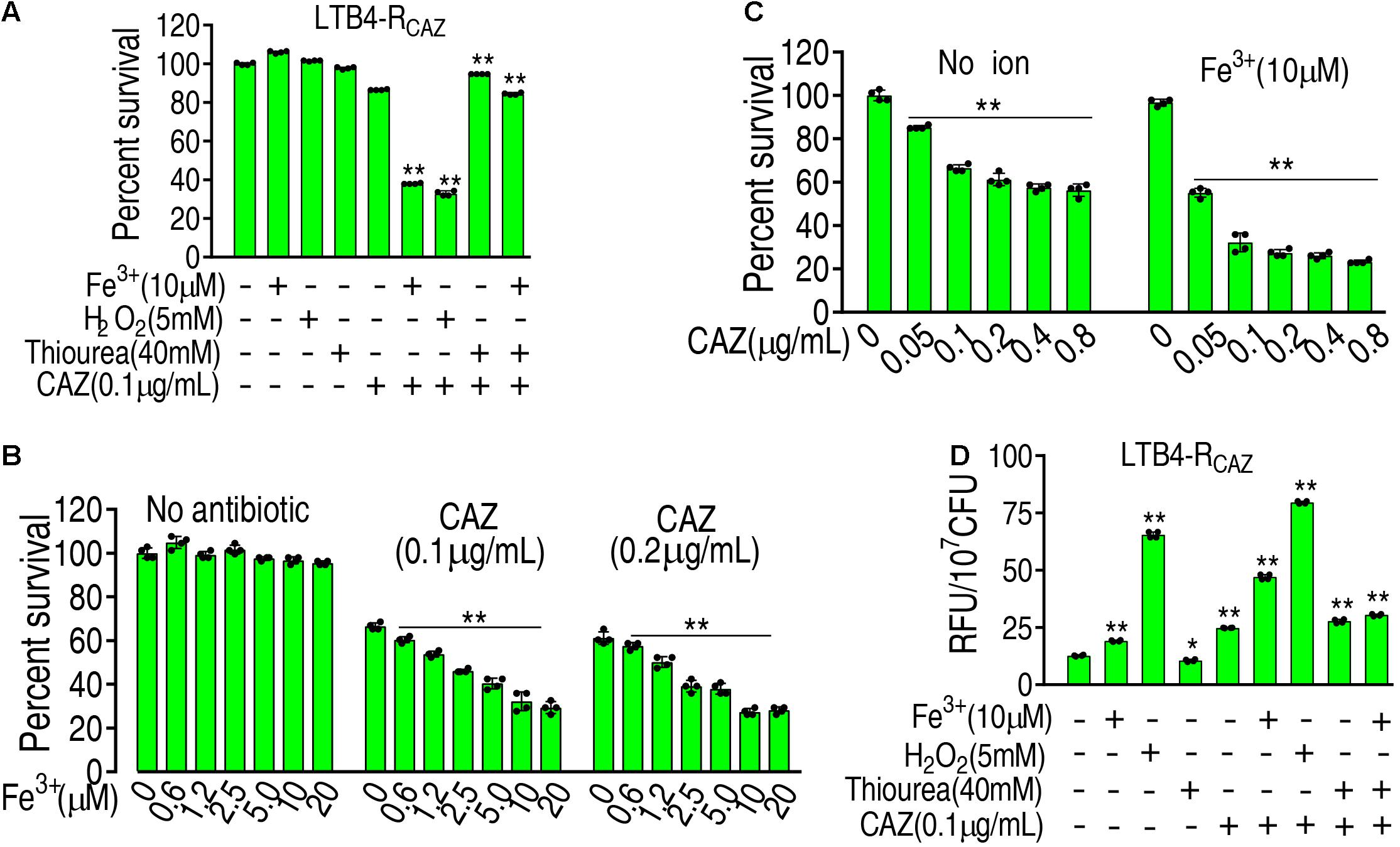
Figure 2. The role of ROS in ceftazidime-mediated killing. (A) ROS was quantified in LTB4-RCAZ in the absence or presence of ceftazidime plus Fe3+, H2O2, or/and thiourea as indicated by fluorescence. (B) Percent survival of LTB4-RCAZ in the presence of ceftazidime and the indicated concentrations of Fe3+. (C) Percent survival of LTB4-RCAZ in the presence or absence of Fe3+ plus the indicated concentrations of ceftazidime. (D) ROS level in the presence or absence of ROS promoter and inhibitor plus ceftazidime. Results are displayed as mean ± SEM, as determined by two-tailed Student’s t-test. Four biological repeats are carried out. * < 0.05 and ** < 0.01.
To understand why ROS are reduced in LTB4-RCAZ, the P cycle and SOD degradation were investigated. The P cycle is a recently illustrated cycle, which provides respiratory energy in E. tarda (Su et al., 2018), and is related to ROS biosynthesis (Figure 3A). For the investigation of the P cycle, qRT-PCR was used to measure the expression of genes in the P cycle. Among the 18 genes detected, 11, 2, and 5 were reduced, elevated, and unchanged, respectively. Specifically, the 11 reduced genes encode all enzymes detected except for citrate synthase (CS) and isocitrate dehydrogenase (ICDH). The two elevated genes encode PDH and MDH. The five unchanged genes encode CS, ICDH, SDH, fumarate reductase (FRD), and pyruvate kinase (PK) (Figures 3B,C). Consistently, a lower activity of PDH, KGDH, and SDH was detected in LTB4-RCAZ than that in LTB4-S (Figure 3D). NADH, membrane potential, and ATP were reduced in LTB4-RCAZ compared with LTB4-S (Figures 3E–G). These results indicate that the inactivation of the P cycle forms a characteristic feature in LTB4-RCAZ, which is related to the reduced ROS production. For the investigation of ROS degradation, the activity of ROS degradation enzymes was measured. Among these enzymes working for the ROS degradation, SOD and CAT are widely employed to indicate the antioxidant response. Therefore, the activity of SOD and CAT was detected in LTB4-RCAZ and LTB4-S. A similar activity of SOD and CAT was measured between LTB4-RCAZ and LTB4-S (Figures 3H,I). These results indicate that antioxidant response is not changed in LTB4-RCAZ. Taken together, the lower ROS in LTB4-RCAZ is attributed to a reduction of ROS generation instead of an increase of ROS degradation.
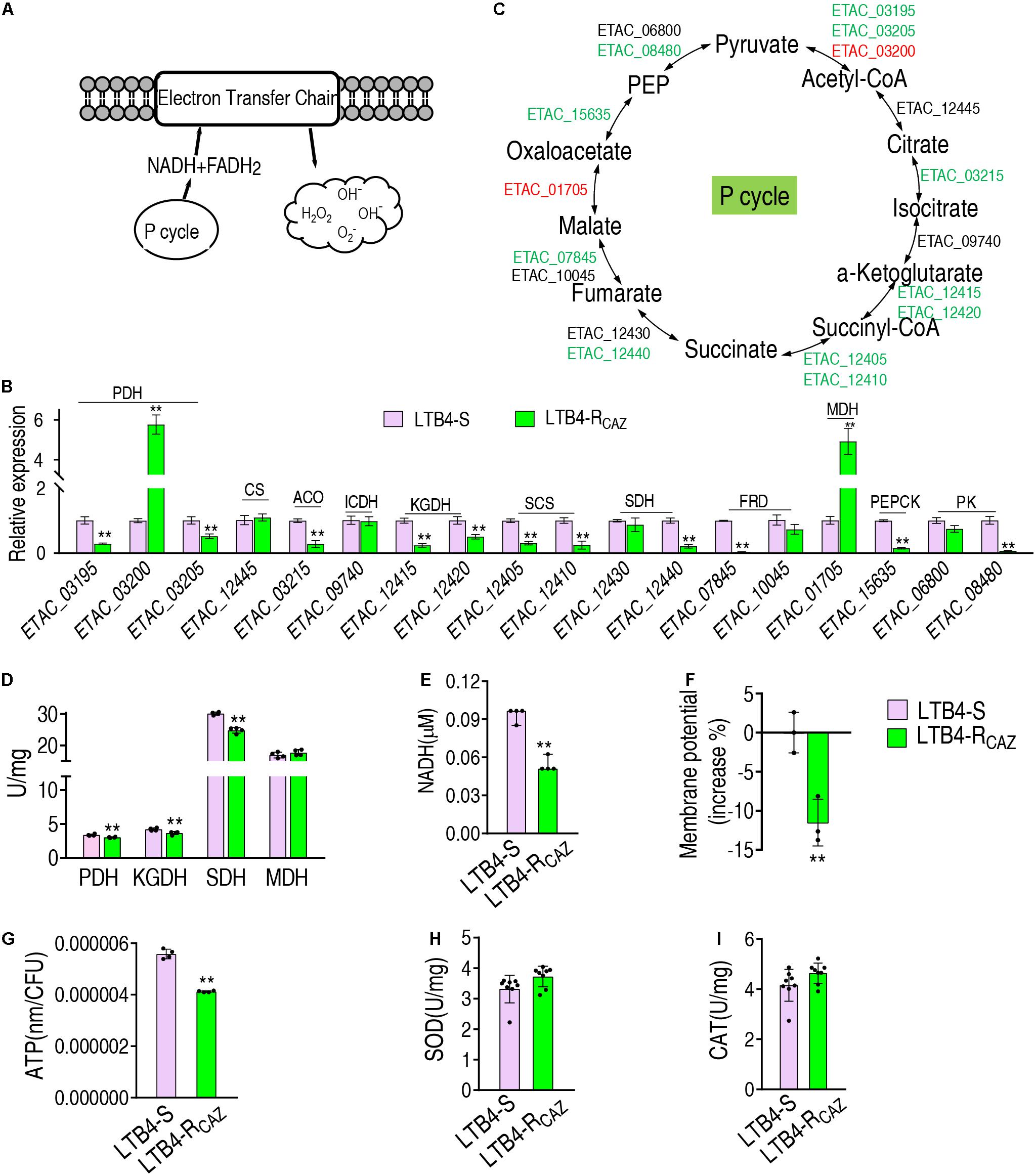
Figure 3. The P cycle, NADH, and ATP in LTB4-S and LTB4-RCAZ. (A) Sketch diagram describing the effect of the P cycle on ROS. (B) Real-time quantitative PCR (qRT-PCR) for the expression of the P cycle-related genes in LTB4-RCAZ compared to LTB4-S. (C) A global view for transcription level of the P cycle genes. Red and blue indicate upregulated and downregulated expression, respectively. (D) The activity of PDH, KGDH, SDH, and MDH in the P cycle. NADH (E), membrane potential (F), and ATP (G) concentrations in LTB4-S and LTB4-RCAZ. The activity of superoxide dismutase (SOD) (H) and catalase (CAT) (I) in LTB4-S and LTB4-RCAZ. Results (B,D–I) are displayed as mean ± SEM, as determined by two-tailed Student’s t-test. Four biological repeats are carried out. **P < 0.01.
The above results motivated us to explore whether Fe3+ promotes global metabolism including the P cycle to elevate ROS level and potentiate ceftazidime-mediated killing. To do this, GC-MS-based metabolomics was used to compare metabolic profiles between media with or without Fe3+. Four biological and two technical replicates were performed in each group, yielding eight data sets with 65 metabolites in a sample. Among the 65 metabolites, 33 (50.8%) showed differential abundance (p < 0.05), with 27 at higher abundance and 6 at lower abundance in the presence of Fe3+ (Figure 4A). Fourteen pathways were enriched, of which alanine, aspartate, and glutamate metabolism; the TCA cycle; and aminoacyl-tRNA biosynthesis were the top three pathways by impact (Figure 4B). Differential metabolites at abundance are listed in Figure 4C, where all detected metabolites were elevated in the TCA cycle (Figure 4C). Principal component analysis identified two principal components, where component t[1] distinguished LTB4-RCAZ from LTB4-RCAZ + Fe3+ (Figure 4D). Discriminating variables were identified on an S-plot (Figure 4E). Cutoff values were ≥ 0.05 for absolute value of the covariance p and ≥ 0.5 for correlation p (corr). Nine putative biomarkers were identified, including decreased abundance of 9-hexadecenoic acid, acetic acid, and myo-inositol and elevated abundance of citric acid, oxalic acid, putrescine, butanoic acid, threonine, aspartic acid, and palmitic acid. Among the elevated metabolites, citric acid plays a role in the P cycle. These results indicate that Fe3+ impacts the metabolic profile, where the P cycle is elevated.
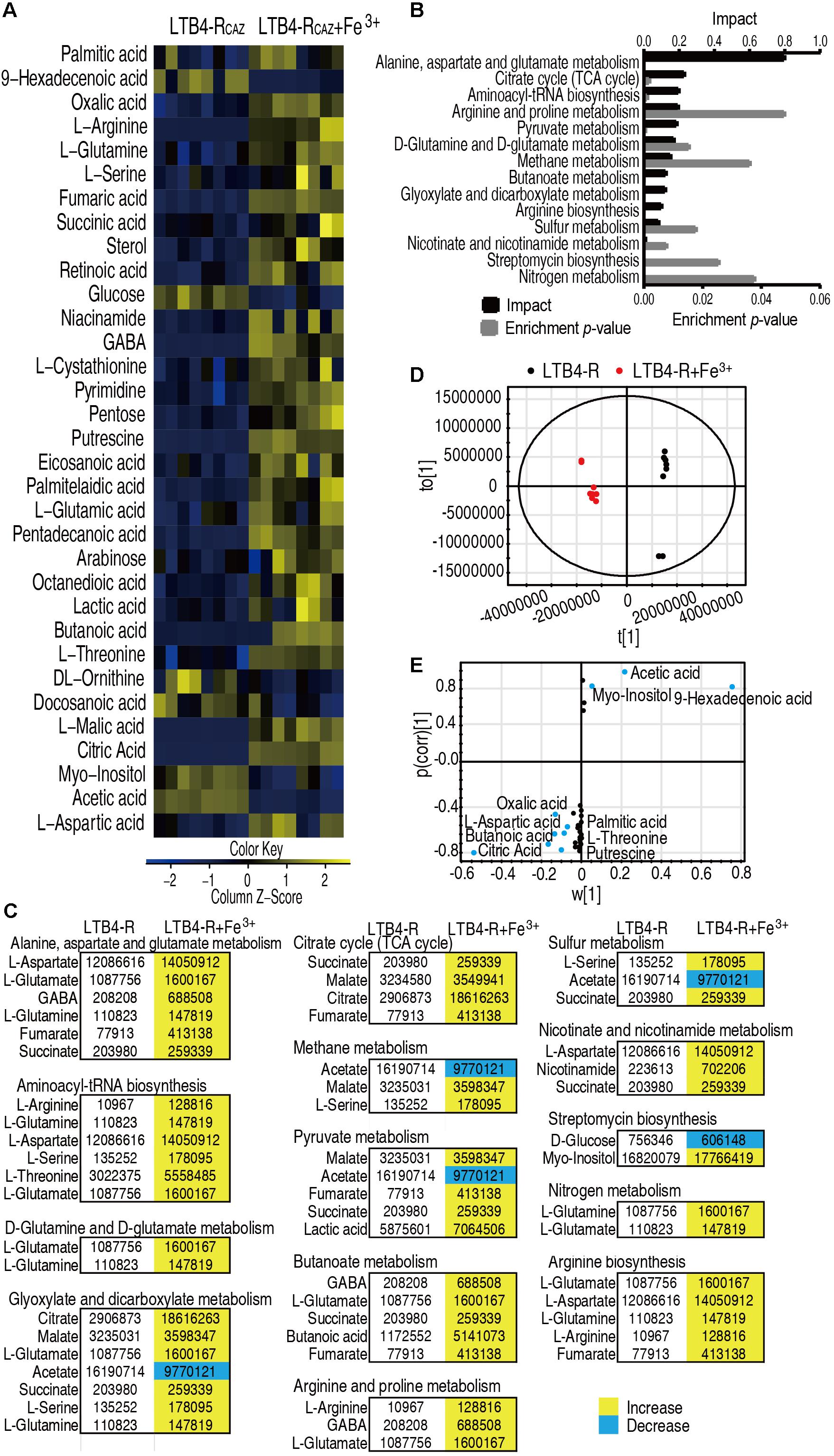
Figure 4. Differential metabolomics of LTB4-RCAZ in response to Fe3+. (A) Heat map showing differential abundance of metabolites. Yellow and blue indicate increase and decrease of metabolites relative to the median metabolite level of the control, respectively (see color scale). (B) Pathway enrichment of varied metabolites in LTB4-RCAZ. (C) Integrative analysis of metabolites in significantly enriched pathways. Yellow and blue indicate increased and decreased metabolites, respectively. (D) PCA of LTB4-S and LTB4-RCAZ. Each dot represents the technical replicate analysis of samples in the plot. (E) S-plot generated from OPLS-DA. Predictive component p [1] and correlation p (corr) [1] differentiate LTB4-RCAZ from LTB4-S. Dot represents metabolites, and candidate biomarkers are highlighted in blue.
To further demonstrate the Fe3+-mediated activation, gene expression and enzyme activity were measured in the P cycle. qRT-PCR analysis showed that out of 18 genes tested, 5, 3, and 10 were elevated, reduced, and unchanged, respectively. In detail, the five elevated genes encode PDH, ACO, ICDH, SCS, and FRD, and the reduced genes encode KGDH, SDH, and PK (these enzymes are encoded by two genes) (Figures 5A,B). The activity of PDH, KGDH, SDH, and MDH was elevated (Figure 5C), which was further supported by increased NADH, membrane potential, and ATP (Figures 5D–F). These results indicate that Fe3+ promotes the P cycle but does not affect the activity of SOD and CAT (Figure 5G).
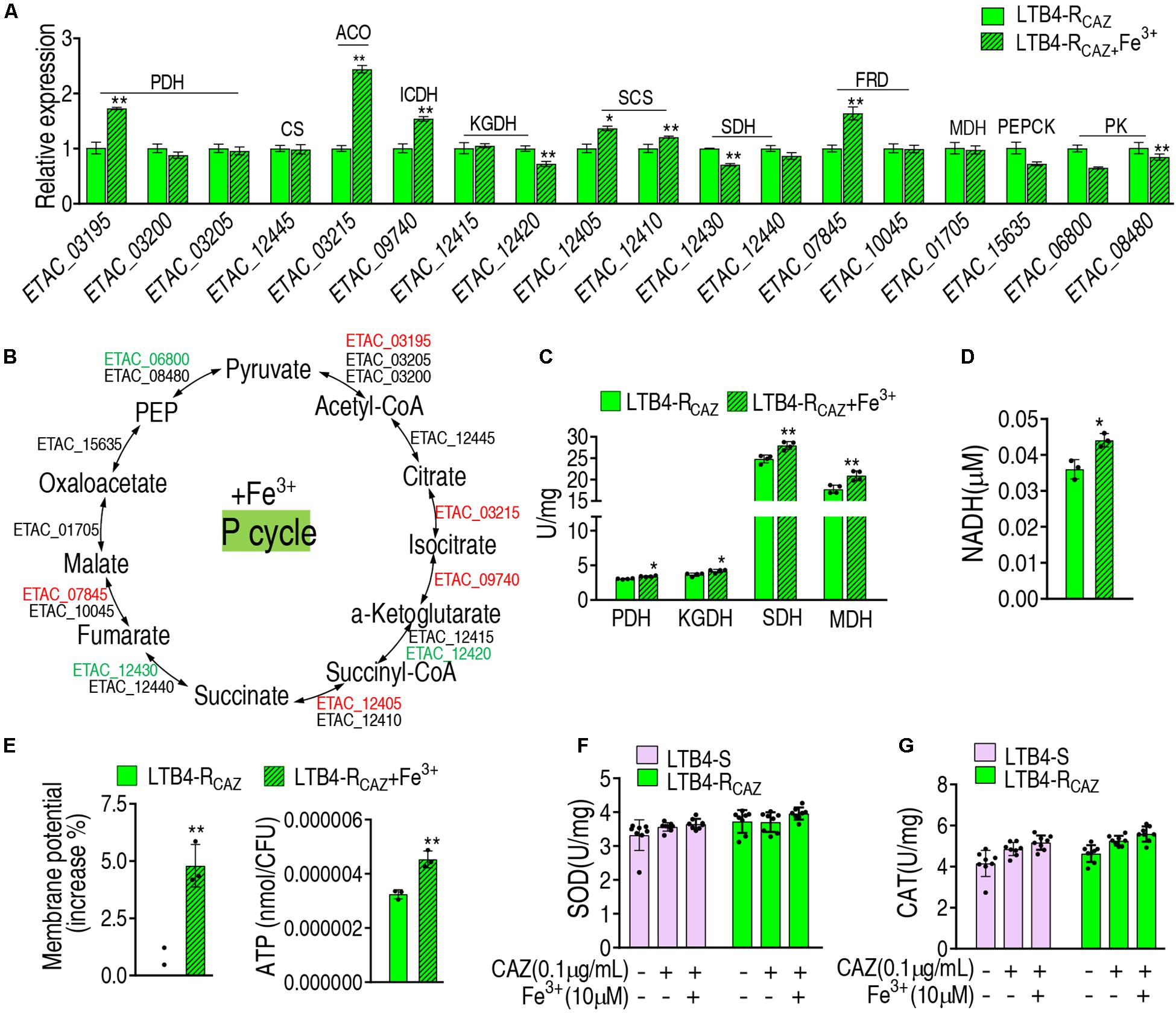
Figure 5. The effect of Fe3+ on the P cycle. (A) qRT-PCR for expression of the P cycle genes in the presence of Fe3+. (B) A global view for transcription level of the P cycle genes. Red and blue indicate upregulated and downregulated expression, respectively. The activity of enzymes of the P cycle (C), NADH level (D), membrane potential (E), and ATP (F) of LTB4-RCAZ in the presence or absence of Fe3+. (G) The activity of SOD and CAT in the presence or absence of ceftazidime or/and Fe3+. Results (A,C–G) are displayed as mean ± SEM, as determined by two-tailed Student’s t-test. Four biological repeats are carried out. *p < 0.05 and **p < 0.01.
To further validate that Fe3+ promotes ROS via the P cycle, inhibitors of the P cycle were used to investigate whether the inhibition affects ROS and viability of LTB4-RCAZ in the presence of both Fe3+ and ceftazidime. Furfural is a non-competitive inhibitor for PDH, and malonic acid is competitive against SDH. Lower ROS were detected in the presence than the absence of furfural or malonic acid, while higher ROS were determined in the medium with than without Fe3+ in the presence or absence of ceftazidime and furfural or malonic acid (Figure 6A), suggesting that Fe3+ partly reverts the inhibition mediated by the two inhibitors. Consistently, a lower viability was detected in the medium with than without Fe3+ in the presence of furfural or malonic acid (Figure 6B). To understand the mechanisms by which Fe3+ promotes the P cycle, we supposed that Fe3+ activates the activity of enzymes in the P cycle. To explore this, the activity of PDH, KGDH, SDH, and MDH was measured in a medium with the indicated concentrations of Fe3+. The activity of the four enzymes was increased in a Fe3+ dose-dependent manner in vitro (Figure 6C). These results indicate that Fe3+ promotes the activity of PDH, KGDH, SDH, and MDH in the P cycle directly, which is related to the elevation of ROS level in the presence of Fe3+.
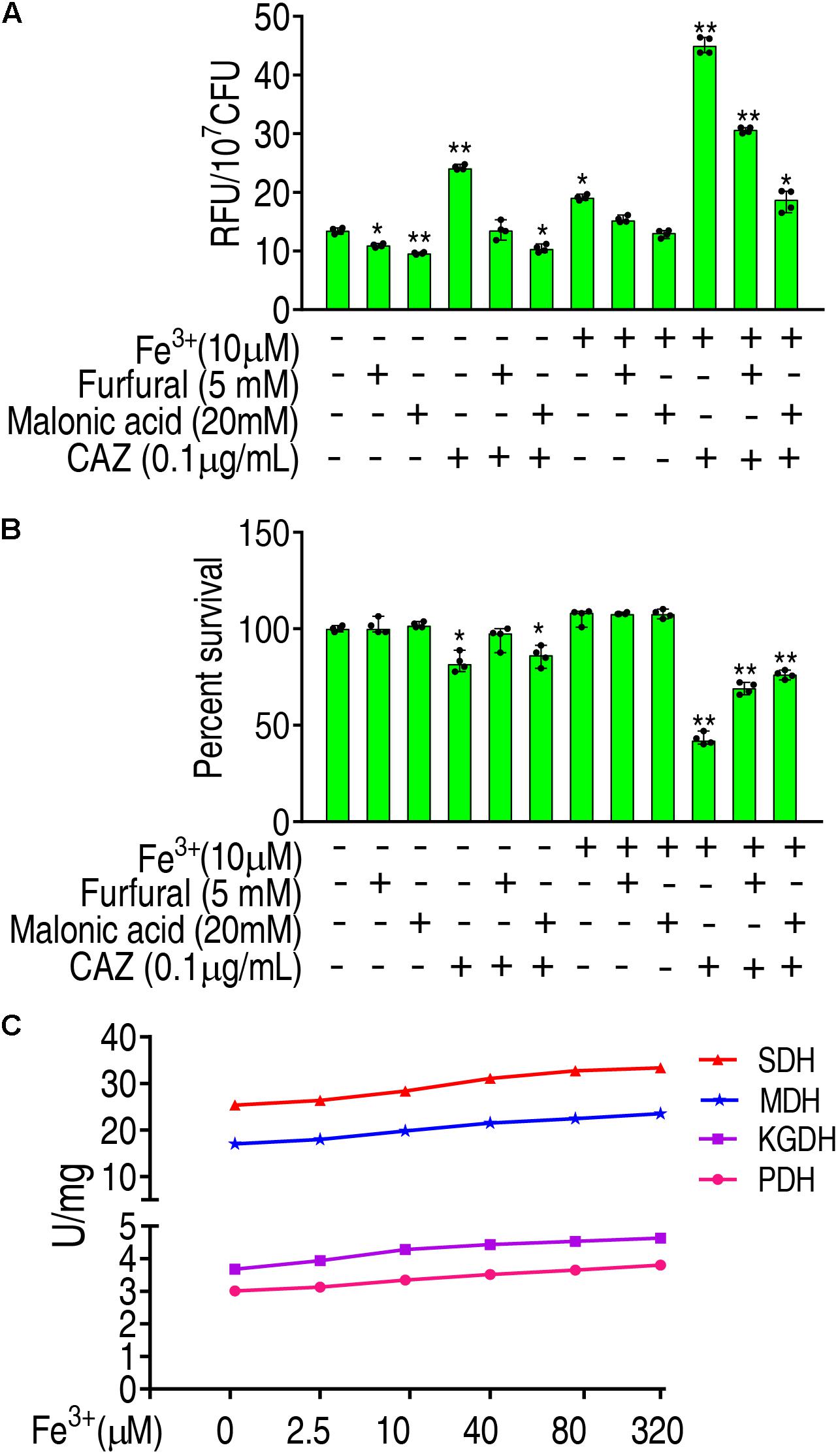
Figure 6. Promotion of Fe3+ to the P cycle. (A) ROS of LTB4-RCAZ in the presence of Fe3+ and CAZ plus inhibitors of the P cycle. (B) Percent survival of LTB4-RCAZ in the presence of Fe3+ and CAZ plus inhibitors of the P cycle. (C) The activity of PDH, KGDH, SDH, and MDH in the presence of Fe3+. Results are displayed as mean ± SEM, as determined by two-tailed Student’s t-test. Four biological repeats are carried out. *p < 0.05 and **p < 0.01.
To ensure the Fe3+-induced potentiation is generalizable to cephalosporins and clinically isolated pathogens, three types of cephalosporins (ceftazidime, cefoperazone, and cefazolin) and 12 strains of bacterial pathogens (E. tarda, E. coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa) were used. The 12 strains of pathogens are resistant to at least three classes of antibiotics and thereby belong to multidrug-resistant bacteria. However, Fe3+ effectively promotes the three drugs to kill all pathogens (Figure 7). These results indicate that the Fe3+-induced potentiation is generalizable to the drugs and bacterial pathogens.
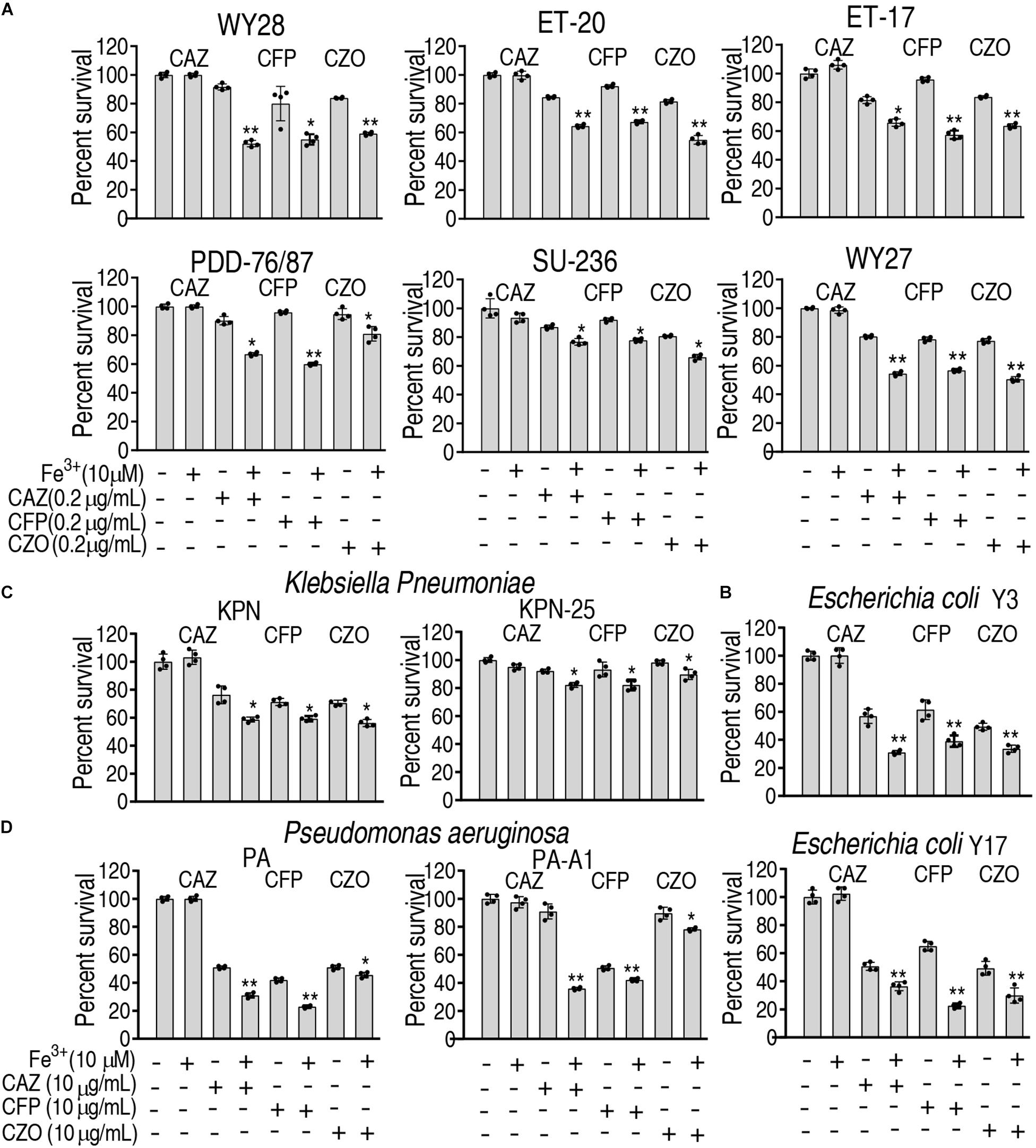
Figure 7. Percent survival of clinically isolated pathogens in the presence of Fe3+ and ceftazidime, cefoperazone, or cefazolin. (A) E. tarda. (B) E. coli. (C) K. pneumoniae. (D) P. aeruginosa.
The present study first investigated whether ceftazidime resistance is related to ROS level and then explored whether Fe3+-potentiated ROS elevation in ceftazidime-mediated killing is related to the P cycle, which contributes to ROS biosynthesis. For this purpose, ROS level was compared between LTB4-S and LTB4-RCAZ. Lower ROS was detected in LTB4-RCAZ than in LTB4-S. The reduction is accompanied with the inactivation of the P cycle. Since the P cycle is related to ROS generation, the inactivated P cycle should be one reason by which ROS is reduced in LTB4-RCAZ. Moreover, the role of ROS in ceftazidime-mediated killing was demonstrated by the ROS promoter Fe3+ and inhibitor thiourea. They increased and decreased the ceftazidime-mediated killing, respectively, suggesting that the ROS promoter and inhibitor affect the killing via regulation of ROS biosynthesis. These results indicate that the ceftazidime-mediated resistance and killing are regulated by ROS level. We further show that the Fe3+-induced potentiation is generalizable to ceftazidime and other cephalosporins to kill clinically multidrug-resistant pathogens E. tarda, E. coli, K. pneumoniae,and P. aeruginosa.
It has been reported that ROS level is a characteristic feature as a consequence of antibiotic resistance and sensitivity (Ma et al., 2016; Ye et al., 2018). Rosato et al. indicated that TCA cycle-mediated generation of ROS is a key mediator for MRSA survival under beta-lactam antibiotic exposure (Rosato et al., 2014). Hayakawa et al. (2019) demonstrated that anti-ROS agent prevents the acquisition of multi-drug resistance in clinical isolates of P. aeruginosa. Thomas et al. (2013) showed that a dysfunctional TCA cycle enables Staphylococcus epidermidis to resist oxidative stress and alter its cell surface properties, making it less susceptible to beta-lactam antibiotics. In addition, Battán et al. (2004) found that resistance to oxidative stress caused by ceftazidime and piperacillin in a biofilm of Pseudomonas is related to bacterial strains. On the other hand, ROS act as an antibiotic sensitizer for the treatment of antibiotic-resistant bacteria involved in infectious diseases (Guo et al., 2018). Recent observations have linked ROS production with bactericidal action of antibiotics, pointing to antibiotic-induced TCA cycle- and respiratory chain-dependent ROS production as playing a role in cell death (Dwyer et al., 2009; Van Acker and Coenye, 2017; Ye et al., 2018). The present study identified that both actions play a role. Specifically, decreased ROS production is a characteristic feature in LTB4-RCAZ, and ceftazidime-induced ROS production is required for ceftazidime-mediated killing in LTB4-RCAZ. These results indicate that the decreased ROS production contributes to the insensitivity to ceftazidime, and thereby reduction of ROS production is a mechanism by which E. tarda resist to ceftazidime.
The present study further explored why ROS production is reduced in LTB4-RCAZ based on the P cycle-mediated generation and ROS degradation. It is documented that the reduction of ROS production is attributed to the decreased biosynthesis caused by inactivation of the P cycle instead of the degradation. The understanding of the biosynthesis of ROS is especially important for promoting antibiotic-mediated killing since ROS production contributes to antibiotic resistance. Wang et al. (2014) showed that loss of sigma(s) rendered stationary-phase E. coli more sensitive to the bactericidal antibiotic gentamicin due to a weakened antioxidant defense. Our recent publication indicates that alanine enhances aminoglycoside-induced ROS production through promoting ROS biosynthesis pathways and repressing transcription of antioxidant-encoding genes (Ye et al., 2018). Thus, a synergistic use of antibiotics with ROS generation promoter or/and ROS degradation inhibitor will elevate the antibiotic-mediated killing. In addition, metabolites promoting hosts to eliminate bacterial pathogens have been reported (Jiang et al., 2019a, b, 2020b; Gong et al., 2020a, b; Yang et al., 2020), where ROS are involved (Sarr et al., 2018; Gong et al., 2020b). Thus, it is possible to select the promoters or/and inhibitors that simultaneously elevate the ROS level of both hosts and bacteria. This approach will make both the host’s anti-infective ability and antibiotic-mediated killing play a role, having the most effect on the elimination of bacterial pathogens via ROS-mediated pathways.
A line of evidence has indicated that Fe3+ reduces ROS via the Fenton system (Song et al., 2020). However, information regarding whether Fe3+ regulates ROS by other ways is absent in bacteria. The present study showed that Fe3+ reverts the inactivity of the P cycle, which elevates ROS biosynthesis. Further evidences include the elevated activity of PDH, KGDH, SDH, and MDH in the presence of Fe3+ in vitro, suggesting that Fe3+ is an activator of these enzymes. Therefore, Fe3+ as a ROS promoter plays a role in the P cycle and Fenton system, which is not reported before in bacteria.
In summary, ROS production is decreased as a crucial characteristic of LTB4-RCAZ, which is related to inactivation of the P cycle. Fe3+ promotes the activation of the P cycle and thereby elevates ROS level. The elevated ROS potentiates ceftazidime-mediated killing. When ROS inhibitor is used, the killing is reduced (Figure 8). These results expand our understanding for the role of Fe3+-induced ROS in antibiotic resistance. They may also provide tools and/or knowledge for future new strategies to stop infections by multidrug-resistant human pathogens.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
HL conceived the study. JY and YS conducted the experiments. JY, YS, and HL analyzed the data. HL and XP wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the NSFC project (31902414), the China Postdoctoral Science Foundation-funded project (2019M663216), and the Guangzhou Science and Technology Key Project (201904020042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.654783/full#supplementary-material
Abayneh, T., Colquhoun, D. J., and Sørum, H. (2013). Edwardsiella piscicida sp. nov. a novel species pathogenic to fish. J. Appl. Microbiol. 114, 644–654. doi: 10.1111/jam.12080
Battán, P. C., Barnes, A. I., and Albesa, I. (2004). Resistance to oxidative stress caused by ceftazidime and piperacillin in a biofilm of Pseudomonas. Luminescence 19, 265–270. doi: 10.1002/bio.779
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Cabello, F. C., Godfrey, H. P., Buschmann, A. H., and Dölz, H. J. (2016). Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 16, e127–e133. doi: 10.1016/S1473-3099(16)00100-6
Cheng, Z. X., Guo, C., Chen, Z. G., Yang, T. C., Zhang, J. Y., et al. (2019). Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 10:3325. doi: 10.1038/s41467-019-11129-5
Defoirdt, T., Sorgeloos, P., and Bossier, P. (2011). Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 14, 251–258. doi: 10.1016/j.mib.2011.03.004
Dwyer, D. J., Kohanski, M. A., and Collins, J. J. (2009). Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12, 482–489. doi: 10.1016/j.mib.2009.06.018
Gong, Q. Y., Yang, D. X., Jiang, M., Zheng, J., and Peng, B. (2020a). L-aspartic acid promotes fish survival against Vibrio alginolyticus infection through nitric oxide-induced phagocytosis. Fish Shellfish Immunol. 97, 359–366. doi: 10.1016/j.fsi.2019.12.061
Gong, Q. Y., Yang, M. J., Yang, L. F., Chen, Z. G., Jiang, M., et al. (2020b). Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb. Biotechnol. 13, 796–812. doi: 10.1111/1751-7915.13553
Guo, L., Xu, R., Zhao, Y., Liu, D., Liu, Z., et al. (2018). Gas plasma pre-treatment increases antibiotic sensitivity and persister eradication in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 9:537. doi: 10.3389/fmicb.2018.00537
Hayakawa, S., Kawamura, M., Sato, T., Hirano, T., Kikuchi, T., et al. (2019). An α-Lipoic acid derivative, and anti-ROS agent, prevents the acquisition of multi-drug resistance in clinical isolates of Pseudomonas aeruginosa. J. Infect. Chemother. 25, 28–33. doi: 10.1016/j.jiac.2018.10.003
Huang, Y. W., Huang, H. H., Huang, K. H., Chen, W. C., Lin, Y. T., et al. (2019). AmpI functions as an iron exporter to alleviate β-Lactam-mediated reactive oxygen species stress in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 63:e02467-18. doi: 10.1128/AAC.02467-18
Jiang, M., Chen, Z. G., Zheng, J., and Peng, B. (2019a). Metabolites-enabled survival of crucian carps infected by Edwardsiella tarda in high water temperature. Front. Immunol. 10:1991. doi: 10.3389/fimmu.2019.01991
Jiang, M., Gong, Q. Y., Lai, S. S., Cheng, Z. X., Chen, Z. G., et al. (2019b). Phenylalanine enhances innate immune response to clear ceftazidime-resistant Vibrio alginolyticus in Danio rerio. Fish Shellfish Immunol. 84, 912–919. doi: 10.1016/j.fsi.2018.10.071
Jiang, M., Kuang, S. F., Lai, S. S., Zhang, S., Yang, J., et al. (2020a). Na(+)NQR confers aminoglycoside resistance via the regulation of L-alanine netabolism. mBio 11:e02086-20. doi: 10.1128/mBio.02086-20
Jiang, M., Yang, L., Chen, Z. G., Lai, S. S., Zheng, J., et al. (2020b). Exogenous maltose enhances Zebrafish immunity to levofloxacin-resistant Vibrio alginolyticus. Microb. Biotechnol. 13, 1213–1227. doi: 10.1111/1751-7915.13582
Jiang, M., Yang, L., Zheng, J., Chen, Z. G., and Peng, B. (2020c). Maltose promotes crucian carp survival against Aeromonas sobrial infection at high temperature. Virulence 11, 877–888. doi: 10.1080/21505594.2020.1787604
Li, L., Su, Y. B., Peng, B., Peng, X. X., and Li, H. (2020). Metabolic mechanism of colistin resistance and its reverting in Vibrio alginolyticus. Environ. Microbiol. 22, 4295–4313. doi: 10.1111/1462-2920.15021
Li, W., Yao, Z., Sun, L., Hu, W., Cao, J., et al. (2016). Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J. Proteome Res. 15, 1810–1820. doi: 10.1021/acs.jproteome.5b01127
Ma, L. N., Mi, H. F., Xue, Y. X., Wang, D., and Zhao, X. L. (2016). The mechanism of ROS regulation of antibiotic resistance and antimicrobial lethality. Yi Chuan 38, 902–909. doi: 10.16288/j.yczz.16-157
National Committee for Clinical Laboratory Standards (1999). Methods for Determining Bactericidal Activity of Antimicrobial Agents. Approved Guideline. Document M26-A. Villanova, Pa: National Committee for Clinical Laboratory Standards.
Peng, B., Li, H., and Peng, X. X. (2019). Proteomics approach to understand bacterial antibiotic resistance strategies. Expert Rev. Proteomics 16, 829–839. doi: 10.1080/14789450.2019.1681978
Peng, B., Su, Y. B., Li, H., Han, Y., Guo, C., et al. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Rosato, R. R., Fernandez, R., Paz, L. I., Singh, C. R., and Rosato, A. E. (2014). TCA Cycle-mediated generation of ROS is a key mediator for HeR-MRSA survival under β-Lactam antibiotic exposure. PLoS One 9:e99605. doi: 10.1371/journal.pone.0099605
Sarr, D., Toth, E., Gingerich, A., and Rada, B. (2018). Antimicrobial actions of dual oxidases and lactoperoxidase. J. Microbiol. 56, 373–386. doi: 10.1007/s12275-018-7545-1
Song, M., Cheng, Y., Tian, Y., Chu, C., Zhang, C., et al. (2020). Sonoactivated chemodynamic therapy: a robust ROS generation nanotheranostic eradicates multidrug-resistant bacterial infection. Adv. Funct. Mater. 30:2003587. doi: 10.1002/adfm.202003587
Stokes, J. M., Gutierrez, A., Lopatkin, A. J., Andrews, I. W., French, S., et al. (2019). A multiplexable assay for screening antibiotic lethality against drug-tolerant bacteria. Nat. Methods 16, 303–306. doi: 10.1038/s41592-019-0333-y
Su, Y. B., Peng, B., Li, H., Cheng, Z. X., Zhang, T. T., et al. (2018). Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U S A. 115, E1578–E1587. doi: 10.1073/pnas.1714645115
Thomas, V. C., Kinkead, L. C., Janssen, A., Schaeffer, C. R., Woods, K. M., et al. (2013). A dysfunctional tricarboxylic acid cycle enhances fitness of staphylococcus epidermidis during β-Lactam stress. mBio 4:e00437-13. doi: 10.1128/mBio.00437-13
Van Acker, H., and Coenye, T. (2017). The role of reactive oxygen species in antibiotic-Mediated killing of bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008
Wang, J. H., Singh, R., Benoit, M., Keyhan, M., Sylvester, M., et al. (2014). Sigma S-dependent antioxidant defense protects stationary-phase Escherichia coli against the bactericidal antibiotic gentamicin. Antimicrob. Agents Chemother. 58, 5964–5975. doi: 10.1128/AAC.03683-14
Wang, Y. M., Wang, Q. Y., Xiao, J. F., Liu, Q., Wu, H. Z., et al. (2011). Genetic relationships of Edwardsiella strains isolated in China aquaculture revealed by rep-PCR genomic fingerprinting and investigation of Edwardsiella virulence genes. J. Appl. Microbiol. 111, 1337–1348. doi: 10.1111/j.1365-2672.2011.05166.x
Yang, M. J., Xu, D., Yang, D. X., Li, L., Peng, X. X., et al. (2020). Malate enhances survival of zebrafish to Vibrio alginolyticus infection in the same manner of taurine. Virulence 11, 349–364. doi: 10.1080/21505594.2020.1750123
Yao, Z., Li, W., Lin, Y., Wu, Q., Yu, F., et al. (2016). Proteomic analysis reveals that metabolic flows affect the susceptibility of Aeromonas hydrophila to antibiotics. Sci. Rep. 6:39413. doi: 10.1038/srep39413
Ye, J. Z., Su, Y. B., Lin, X. M., Lai, S. S., Li, W., et al. (2018). Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 9:29. doi: 10.3389/fmicb.2018.00029
Zhang, S., Wang, J., Jiang, M., Xu, D., Peng, B., et al. (2019). Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ. Microbiol. 21, 4724–4739. doi: 10.1111/1462-2920.14811
Zhang, S., Yang, M. J., Peng, B., Peng, X. X., and Li, H. (2020). Reduced ROS mediated antibiotic resistance and its reverting by glucose in Vibrio alginolyticus. Environ. Microbiol. 22, 4367–4380. doi: 10.1111/1462-2920.15085
Keywords: antibiotic resistance, reactive oxygen species, Edwardsiella tarda, the pyruvate cycle, ceftazidime
Citation: Ye J, Su Y, Peng X and Li H (2021) Reactive Oxygen Species-Related Ceftazidime Resistance Is Caused by the Pyruvate Cycle Perturbation and Reverted by Fe3 + in Edwardsiella tarda. Front. Microbiol. 12:654783. doi: 10.3389/fmicb.2021.654783
Received: 17 January 2021; Accepted: 23 March 2021;
Published: 28 April 2021.
Edited by:
Lorena Rodriguez-Rubio, University of Barcelona, SpainReviewed by:
Yong-hua Hu, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2021 Ye, Su, Peng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWkzMkBzeXN1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.